Abstract
Coalescent theory provides powerful models for population genetic inference and is now increasingly important in estimates of divergence times and speciation research. We use molecular data and methods based on coalescent theory to investigate whether genetic evidence supports the hypothesis of A. pretrei and A. tucumana as separate species and whether genetic data allow us to assess which allopatric model seems to better explain the diversification process in these taxa. We sampled 13 A. tucumana from two provinces in northern Argentina and 28 A. pretrei from nine localities of Rio Grande do Sul, Brazil. A 491 bp segment of the mitochondrial gene cytochrome c oxidase I was evaluated using the haplotype network and phylogenetic methods. The divergence time and other demographic quantities were estimated using the isolation and migration model based on coalescent theory. The network and phylogenetic reconstructions showed similar results, supporting reciprocal monophyly for these two taxa. The divergence time of lineage separation was estimated to be approximately 1.3 million years ago, which corresponds to the lower Pleistocene. Our results enforce the current taxonomic status for these two Amazon species. They also support that A. pretrei and A. tucumana diverged with little or no gene flow approximately 1.3 million years ago, most likely after the establishment of a small population in the Southern Yungas forest by dispersion of a few founders from the A. pretrei ancestral population. This process may have been favored by habitat corridors formed in hot and humid periods of the Quaternary. Considering that these two species are considered threatened, the results were evaluated for their implications for the conservation of these two species.
Introduction
Speciation remains an important question in science, even after nearly two centuries of research and intense debate owing to the evolutionary theories of Darwin and Wallace [1]. Three models have been proposed to explain the origin of new species of animals with sexual reproduction: allopatric, parapatric and sympatric (for review, see [2]). Allopatric speciation can occur basically in two models: vicariant speciation [3] and dispersion/founder effect [4]. Under the first model, a widely distributed species becomes subdivided into two or more relatively large populations due to extrinsic barriers, and genetic differences begin to accumulate between isolates as each population responds to its own array of selective forces and tracks its ever-changing environment. Under the second model, species evolve through the establishment of small demes often peripherally isolated by dispersing a few founders from an ancestral population.
There are numerous studies of phylogenetic analyses based on molecular data that support vicariance (e.g., [5]), dispersal (e.g., [6], [7]), or both (e.g., [8]) as causes of distribution patterns in Neotropical birds. Similarly, empirical studies have increasingly found dispersion/colonization as a force behind the most common allopatric speciation events [9]–[13]. Estimates of divergence times based on the molecular clock can be used to correlate the time of cladogenesis of species with paleogeographical vicariant events. Depending on the relative ages of species divergence and vicariant events, assessments can be made of whether a dispersal hypothesis (barrier older than species divergence) or vicariant hypothesis (barrier age and species divergence similar) better explains the observed distributions [6]. In many cases, it has been shown that the study group is too young to have been affected by the alleged vicariant barrier, suggesting a recent history of dispersal instead of vicariance of the taxa [6], [14]–[16]. In recent years, methods based on coalescent theory have been applied to estimate divergence times among populations and/or species, enabling pure isolation with subsequent independent evolution to be distinguished from isolation with subsequent gene flow [17].
The Spectacled Amazon (Amazona pretrei) and the Tucuman Amazon (A. tucumana) are Neotropical parrots which were shown to be sister species in phylogenetic reconstructions [18]. However, the Tucuman Amazon was previously regarded as a race of A. pretrei [19], but was separated as a full species based on a (most likely erroneous [20]) record of sympatry in the Misiones province of Argentina. These two forms seem to differ in their adult and juvenile plumage, with A. pretrei being sexually dimorphic, while the sexes are alike in A. tucumana [21], [22]. Different feeding requirements also tend to support the view that these two amazons should be treated as two species, although other evidence is needed to confirm their true relationship [23].
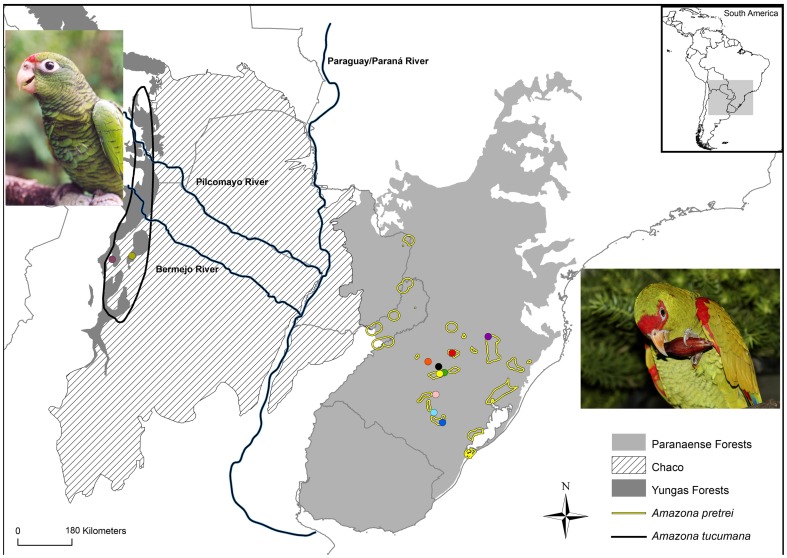
Currently, the A. pretrei distribution area comprises the state of Rio Grande do Sul and southern Santa Catarina in Brazil, with some records in the Misiones province in Argentina (Fig. 1). This species is observed mainly at altitudes between 500 and 1,000 meters in environments marked by the presence of Araucaria (Araucaria angustifolia) forests and savannas with Araucaria [24]. A. tucumana is restricted to the Southern Yungas forests, a montane forest of the Andes of northwestern Argentina and eastern Bolivia, particularly in areas 1,500–2,200 meters in altitude [25]. These two forest formations are currently separated by approximately 700 km of xerophytic Chaco woodland. The latter is an alluvial plain formed by sediments from the Andes [26] and is part of diagonal open biomes in South America, covering an area of approximately 840,000 km2 in northern Argentina, western Paraguay, southeastern Bolivia and the extreme west of Mato Grosso do Sul, Brazil [27]. The flora of this region is considered a relict of the Pliocene or lower Pleistocene, stabilized on salinic soils left after two events of marine transgressions [28], which occurred between 15 and 5 million years ago [29]. Currently, it has a semi-arid climate with intensely hot summers and severe frosts in winter [26]. However, during the interglacial periods of the Quaternary, the Chaco area may have been more humid than today [30]–[32].
Figure 1. Geographic distribution of Amazona pretrei (black circle) and A. tucumana (yellow circles).
The geographical distribution maps of species were kindly donated by IUCN [71]). Collection localities of samples of Amazona pretrei and A. tucumana analyzed in this study were indicated by colored dots. For more details of the collection sites, see Table 1.
According to Nores [33], forests advanced from the southern Yungas and the Paranense region along the Bermejo and Pilcomayo rivers deep enough to form a continuous forest bridge between both regions. Furthermore, Nores [33] noted that some bird species presumably expanded to form a continuous distribution along this continuous forest bridge and became isolated again in the subsequent drier period. This connection-interruption process most likely occurred several times during the Pleistocene and Holocene periods [30], [34]–[36].
Having considered the biogeographical scenario described above and several pairs of species and subspecies with disjunct populations in the Southern Yungas forests and Paraná state (Brazil), Nores [33] described four different levels of bird speciation currently observable in both forests. These different levels of speciation were most likely related to the time elapsed since the species crossed the forest bridge between these two regions. The author classified A. pretrei and A. tucumana as belonging to an allospecies group, in which differentiation most likely began very early in the wet-dry cycles, although it is possible that isolation and speciation began before the Quaternary period. However, the author noted that future studies applying new methodologies might assist in the review of this classification.
In the present study, we use molecular data and methods based on coalescent theory to investigate whether genetic evidence supports the hypothesis of A. pretrei and A. tucumana as separate species, and whether genetic data allow us to assess which allopatric model seems to better explain the diversification process of A. pretrei and A. tucumana. Based on allopatry by vicariance, we should expect that separation of these species occurred before the Quaternary climatic fluctuations, possibly due to the formation of vegetation in the Chaco province estimated to have started during the Pliocene. In allopatry via the dispersion/founder effect or peripheral isolation [4], we should expect that separation occurred subsequently to the formation of the Chaco by a dispersal event of a few individuals which could have been facilitated by the forest corridor, estimated to have occured during the Pleistocene. Finally, considering that the two species in this study are considered to be threatened at a global level [37] or endangered at a national levels [38], [39], the results have been evaluated for their implications for the conservation of these species. For endangered species, knowledge of these historical processes is critical for correct interpretation of current patterns of genetic variation. Furthermore, this information should guide the assignment of conservation priorities and the implementation and elaboration of subsequent conservation strategies.
Materials and Methods
Taxon sampling
We sampled 13 A. tucumana from two provinces located in northern Argentina and 28 A. pretrei from nine localities of Rio Grande do Sul, Brazil (Table 1, Fig. 1). All samples were obtained from nestlings found in natural nests with the permission of the respective environmental agencies (DPMAyRN license n° 082/2006 and APN license n° 04/2008 in Argentina, and SISBIO license n° 25694-3 in Brazil). Only one nestling from each nest was considered for genetic analysis. Three to four drops of blood were taken from each nestling via puncture of the brachial wing vein using needles and glass capillaries, subsequently stored in microtubes containing absolute ethanol. All the samples were stored frozen at the Laboratory of Molecular Genetics and Bird Evolution at the Institute of Biosciences of the University of São Paulo.
Table 1. Taxa used in this study: species, sample number, sampled locality, haplotype name and GenBank accession number for COI sequences.
| Species | Sample | Country | State/Province | Locality | Coordinates | Haplotype | Genbank |
| A. pretrei | 3388 | Brazil | Rio Grande do Sul | Fortaleza dos Valos | 28°48′30.32″S/53°15′34.28″W | pre 7 | KF697712 |
| 3390 | |||||||
| 1249 | Santana da Boa vista | 30°52′58.74″S/53°07′11.97″W | pre 1 | KF697709 | |||
| 1248 | pre 6 | KF697708 | |||||
| 1234 | Jacuizinho | 29°01′58.74″S/53°03′04.75″W | pre 3 | KF697710 | |||
| 2257 | pre 5 | KF697707 | |||||
| 2261 | |||||||
| 1255 | Restinga seca | 29°50′37.26″S/53°22′15.26″W | pre 5 | KF697707 | |||
| 2257 | Carazinho | 28°18′13.63″S/52°45′30.35″W | pre 1 | KF697709 | |||
| 2259 | |||||||
| 2260 | |||||||
| 3380 | |||||||
| 3382 | |||||||
| 4699 | |||||||
| 4700 | |||||||
| 4846 | |||||||
| 4701 | pre 2 | KF697713 | |||||
| 3383 | pre 4 | KF697711 | |||||
| 3384 | pre 5 | KF697707 | |||||
| 2267 | |||||||
| 5683 | |||||||
| 5676 | Cruz alta | 28°37′39.83″S/53°39′00.38″W | pre 5 | KF697707 | |||
| 5693 | Caçapava do sul | 30°30′40.26″S/53°26′25.26″W | pre 5 | KF697707 | |||
| 2252 | Salto do Jacuí | 29°04′18.18″S/53°13′21.64″W | pre 1 | KF697709 | |||
| 2253 | |||||||
| 2255 | |||||||
| 4702 | |||||||
| 5675 | Barracão | 27°41′57.32″S/51°25′32.59″W | pre 1 | KF697709 | |||
| A. tucumana | UFG442 | Argentina | Salta | Parque Nacional El Rey | 24°42′27.56″S/64°37′09.37″W | tuc 1 | KF697704 |
| UFG446 | |||||||
| UFG440 | tuc 2 | KF697706 | |||||
| UFG450 | tuc 3 | KF697705 | |||||
| UFG413 | Jujuy | Santa Barbara | 24°50′06.55″S/65°21′10.19″W | tuc 1 | KF697704 | ||
| UFG416 | |||||||
| UFG419 | |||||||
| UFG420 | |||||||
| UFG424 | |||||||
| UFG435 | |||||||
| UFG430 | |||||||
| AMCC110751 | AY301459 | ||||||
| UFG428 | tuc 2 | KF697706 |
Sample size
For evaluation of whether sequence samples were representative of the genetic constitution of the species, we estimated P = [(k−1)/(k+1)], which represents the probability that a sample of size k and the whole population share the most recent ancestor [40]. P can be interpreted as the probability of the sample being representative of the genetic diversity of the population.
DNA extraction and sequencing
Total genomic DNA was extracted using a standard proteinase K/SDS and phenol–chloroform protocol [41]. Using a Lambda DNA marker (Biosciences) as a reference, the DNA samples were quantified in a 1.5% agarose gel stained with ethidium bromide.
A segment of the mitochondrial gene cytochrome c oxidase I (COI) was amplified using COIA and COIF primers [42] by polymerase chain reaction (PCR) as follows: final volume of 10 µL containing 0.5 mM of each primer, 0.5 U of Taq DNA polymerase (Phoneutria, CA), 1 mM dNTP, 1× buffer (10 mM Tris-HCl pH 8.3, 50 mM KCl, 1.5 mM MgCl2) and 20 to 50 ng DNA. The reactions were subjected to a cycle with a temperature of 95°C for the initial denaturation, followed by 35 cycles at 95°C for denaturation, 53°C for primer annealing and extension at 72°C, ending with one cycle at 72°C for 10 minutes for the final extension. The samples were purified by enzymatic reaction with the enzymes exonuclease I (EXO, USB) and Shrimp Alkaline Phosphatase (SAP, USB). The purified PCR products were sequenced using the DYEnamic ET Terminator kit (Amersham Bioscience) in an automated sequencer ABI 3100 (Applied Biosystems) according to manufacturer's recommendations. The sequences obtained were evaluated for quality and checked for ambiguities using SeqScape v.2.1 (Applied Biosystems). The alignment of the sequences was performed with ClustalW algorithm [43] using the MEGA 5.0 software [44]. The COI haplotype sequences generated and used in this work were deposited in the GenBank (accession numbers KF697704–KF697713).
Sequence statistics, genetic diversity, and phylogenetic analysis
The nucleotide and haplotype diversity for each species were estimated using the DNAsp 5.10.01 program [45]. The relationship between distinct genetic sequences or haplotypes was evaluated by the haplotype network via median joining method [46] using Network 4.1 (Fluxus Technology Ltd). To verify whether the sequences conformed to neutral expectations of nucleotide substitution, we computed Tajima's D- [47], Fu and Li D* and F* values [48] for each gene using DNAsp 5.10.01.
Phylogenetic relationships among sequences were reconstructed based on the maximum parsimony using MEGA 5.0 [44] and on the Bayesian analyses by Markov Chain Monte Carlo (MCMC) using MrBayes 3.2 [49]. Maximum parsimony analysis was performed considering all the character states as unordered and equally weighted. A heuristic search was implemented with ten random addition sequence replicates, tree bisection-reconnection branch swapping, and a maximum of 100 trees saved per round. To evaluate relative robustness of the clades found in the most parsimonious trees, the bootstrap analysis [50] employed 1,000 replicates using the same heuristic search settings and a maximum of 100 trees were saved per round. Bayesian inference analysis was performed using HKY models. The best-fit model of nucleotide evolution was selected with the Akaike information Criterion (AIC) in JModeltest 2.1.4 [51]. Four Markov chains were run for 2,000,000 generations (sampling every 100 generations) to allow adequate time for convergence. Posterior probabilities of the nodes were computed for all Bayesian analyses across the sampled trees after burn-in. The number of generations required to reach stationary of the posterior distribution was determined by examining marginal probabilities plotted as a time series in Tracer v1.5 [52]. The burn-in period was set to 10%. All MCMCs were run twice to confirm consistent approximation of the posterior parameter distributions. In both analyses, a homologous sequence of Amazona vinacea (Genbank AY301462) was used as outgroup. This species was chosen as outgroup based on phylogeny of Russello and Amato [18].
Divergence time and other demographic quantities of the two species were estimated using the isolation and migration model described by Nielsen and Wakeley [17] employing IMa2 [53]. This method uses a Markov Chain Monte Carlo to estimate jointly the posterior distribution of the model parameters and the demographic quantities: θ (Neμ, where Ne is the effective population size of females and μ is the mutation rate per sequence per generation), M (Nem, where m is the proportion of effective number of migrants per generation) and t (time since population splitting) and TMRCA (time to the most recent common ancestor). For mitochondrial COI, we used the substitution rate of 0.772%/lineage/myr, based on the mean for the Neoaves clade derived in a mitogenomic timescale for birds [54].This rate was converted to 3.8×10−6 substitutions/locus/year by multiplying by the number of base pairs of the locus (491), and transforming from millions of years to years. Generation time was set as five years [55]. Several preliminary runs were performed in IMa2 with different priors and heating schemes to find the conditions that allowed proper mixing among chains to avoid local optimum parameter values. When good conditions were achieved, three runs were performed to verify whether the estimated parameters were converging to similar results. Final IMa2 analyses were run for 10,000,000 generations after a burn-in of 10,000 steps using geometric heating, with high heating parameters (g1 = 0.9 and g2 = 0.95), and 20 chains.
Finally, we also evaluated the historical demography by calculating the R2 statistic [56] in DNAsp version 4.0. Significant and low values of R2 suggest demographic expansion if neutrality was not rejected.
Results
Genetic variation, haplotype network and phylogenetic analysis
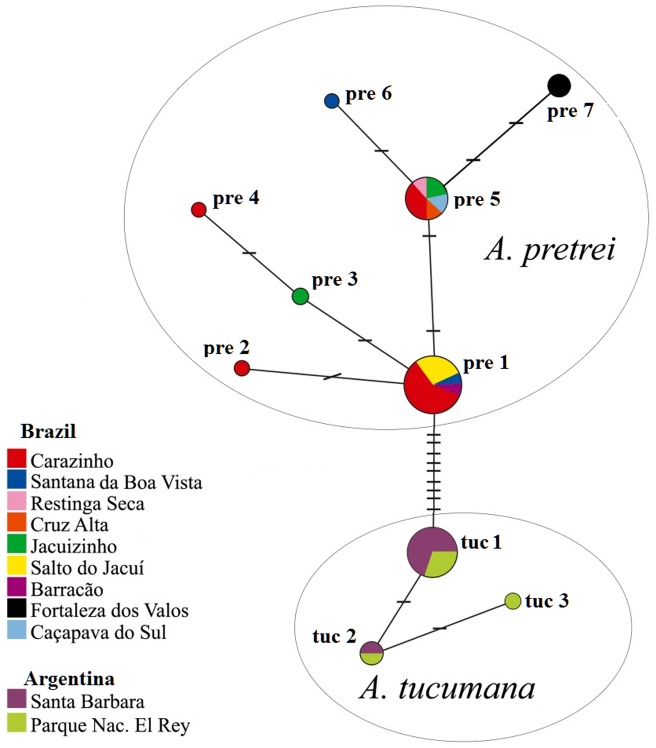
We analyzed a segment of 491 bp of the COI for all individuals. Translation of the protein-coding gene COI did not reveal stop codons or frame-shift mutations, and third codon positions were more variable than first and second codon positions, as expected in functional genes. In the haplotype network, two haplotype groups were clearly identified, which correspond to the haplotypes of the two taxonomically recognized species (Fig. 2).Ten unique haplotypes defined by seven variable sites (five are parsimony informative) were identified in A. pretrei, while only three haplotypes defined by two variable sites (only one parsimony informative) were identified in A. tucumana. These two species did not share haplotypes (Fig. 2). Singleton COI haplotypes differed from one another by one to four mutational steps in A. pretrei, and by one or two mutational steps in A. tucumana. The number of mutational steps separating these species was nine. Considering the genetic variability, A. pretrei presented haplotype diversity (DH = 0.68) greater than in A. tucumana (DH = 0.41), as well as an average genetic distance (DM) three times greater compared to A. tucumana (DM = 0.003 in A. pretrei and DM = 0.001 in A. tucumana). The average genetic distance interspecific was DM = 0.022. When compared to the outgroup (A. vinacea), the average genetic distance was DN = 0.057. The COI sequences have an 85% and 93% probability of having correctly sampled the size of the gene genealogy of the A. tucumana and A. pretrei, respectively. Therefore, our gene samples are considered suitable for the subsequent studies.
Figure 2. Relationships among the seven Amazona pretrei and three A. tucumana haplotypes based on the median-joining network of 491 bp of mitochondrial DNA COI sequences.
Sizes of circles are proportional to the haplotype frequency. The number of variable sites between haplotypes is indicated by dashes between them.
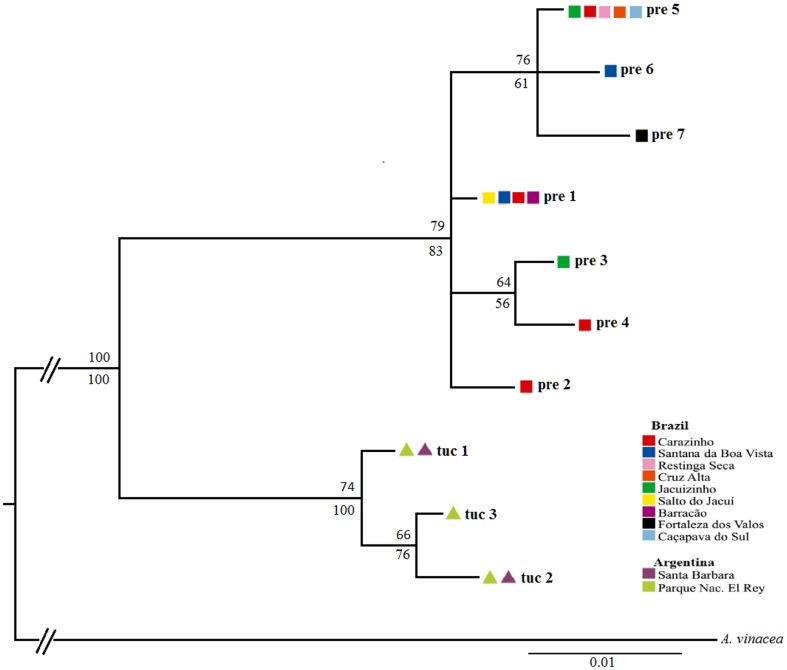
The phylogenetic reconstructions based on maximum parsimony (data not shown) and Bayesian analysis (Fig. 3) showed similar results to those obtained by the haplotype network, showing reciprocal monophyly for these two species, with 100% of bootstrap support value and posterior probability of 1.
Figure 3. Bayesian tree of 491 bp alignment of mitochondrial DNA COI haplotypes of Amazona pretrei (colored squares) and Amazona tucumana (colored triangle).
Tree is a 50% majority-rule consensus of 18,000 trees. The numbers indicate Bayesian posterior probabilities (above the branches) and maximum parsimony bootstrap (below the branches) values.
Coalescent analyses
Sequence data sets for both species had nonsignificant values (p>0.10) of Tajima's D values and Fu and Li's D* and F* statistics, suggesting that the COI gene is selectively neutral. Therefore, it is appropriate to use it to estimate gene flow and other parameters with neutral coalescent methods.
The effective population size of females for A. pretrei and A. tucumana was approximately 357,000 and 94,000 individuals, respectively (Table 2). The effective size of the ancestral population could not be estimated properly with the analyzed COI fragment. The divergence between lineages occurred with an absence or low level of gene flow because the effective number of female migrants among populations was estimated to be less than one individual per generation (Table 2). The divergence time of lineage separation was estimated to be approximately 1.3 million years ago (Table 2), which corresponds to the lower Pleistocene. In relation to the time of the most recent common ancestor, it was estimated to be 1.6 million years ago (ranging between 562,016 and 1,996,124 years ago). Assuming a rate of 0.772%/lineage/myr, and based on 5.7% genetic divergence between A. vinacea and A. pretrei/A. tucumana lineages, we estimate that the separation between these lineages occurred at least 3.96 million years ago.
Table 2. Demographic and population parameters for A. pretrei and A. tucumana based on analysis of 491 bp fragment of the mitochondrial gene cytochrome oxidase I (COI) using IMa2 program.
| Species | NeF (CI) | m1>m2 (CI) | m2>m1 (CI) | t (CI) |
| A. pretrei | 94,913 | 0.00 | 1,339,474 | |
| (m1) | (36,192 to 220,785) | (0.00–4.38) | (265,504 to 1,735,876) | |
| A. tucumana | 27,471 | 0.00 | ||
| (m2) | (2,762 to 135,901) | (0.00–1.86) |
NeF - effective number of females; CI - confidence interval of 95%; m1>m2 - effective number of females migrants/generation from m1 to m2; m2>m1 - effective number of females migrants/generation from m2 to m1; t - divergence time between A. pretrei and A. tucumana.
According to the analysis of demographic expansion (R2), the values were not statistically significant (R2 = 0.861, p>0.05), suggesting population stability for both species.
Discussion
Species delimitation
Our results indicate that the two amazon species studied here are clearly genetically differentiated. The average genetic distance between these two amazon species was seven fold of those observed within them, including nine fixed nucleotide substitutions. Sequences from the COI fragment indicated a clear subdivision with haplotypes confined to each clade in both network and phylogenetic analysis, indicating reciprocal monophyly for these two species. Coalescent analysis suggested that divergence with little or no gene flow occurred between the two groups in the Lower Pleistocene.
The unified species concept [57] suggests treating independently evolving population lineages as different species if independent lines of evidence are available to support their reciprocal isolation [20]. Thus, the current taxonomic status for these two amazon species is reinforced, since genetic evidence and differences in morphology and ecology (see Introduction) indicate A. tucumana lineage is evolving independently from A. pretrei lineage.
Speciation and biogeography
It is not always easy to identify whether two sister species have resulted from vicariance or dispersal events. The vicariance model is supported if the sister species were separated by an intermittent barrier or a barrier that is younger than the estimated age of the ancestral population. In contrast, the dispersion/founder effect model is supported if the sister species were differentiated by means of a permanent barrier, which is considered to be older than the pair of species [58].
According to our analysis, A. pretrei and A. tucumana diverged with little or no gene flow when the current barrier (xerophytic Chaco vegetation) that separates these two species had already formed (see Introduction). Most likely, this divergence occurred after the establishment of a small population in the Southern Yungas forest through dispersion of a few founders from the A. pretrei ancestral population. Thus, this scenario suggests that the allopatry by the dispersion/founder effect model (peripheral isolation) seems to be more likely than by the vicariance model to explain the diversification of these two Amazon species, as discussed below.
According to the phylogeny proposed by Russello and Amato [18], Amazona vinacea is the sister species of A. pretrei/A. tucumana lineage. Considering that A. vinacea such as A. pretrei occupy the Araucaria forests of southern Brazil [59], it is acceptable to assume that the A. pretrei/A. tucumana lineage arose in this region. The divergence between A. vinacea and A. pretrei/A. tucumana lineages should have occurred at least 3.96 million years ago when the vegetation of the Chaco was most likely still in training on salty soil, due to the last marine transgression in South America [29], [60]. This non-forest vegetation may have limited the spread of the A. pretrei/A. tucumana lineage in this period.
Based on our results, the divergence between A. tucumana and A. pretrei occurred approximately 1.3 million years ago, possibly when the xerophytic vegetation of the Chaco was already similar to current vegetation, limiting the occupation of this area by a typical forest species. It is possible that the formation of more suitable habitat corridors, such as those proposed along the banks of the Bermejo and Pilcomayo rivers across the Chaco during interglacial periods of the Quaternary [33], allowed the passage of amazons through the Chaco arid zone, reaching the wetlands of the tropical forests of the Yungas forest. The isolation between those lineages most likely occurred after the disappearance of these corridors, establishing a condition of isolation between the A. pretrei ancestral population and the A. tucumana peripheral population. Willis [61] previously also raised the subject of the use of corridors, commenting on the possible use of a south-Andean route (Andean-south route) by A. pretrei and A. tucumana.
The haplotype diversity observed in A. tucumana is lower than in A. pretrei, and the relationship pattern among the A. tucumana haplotypes in the network, composed by a frequent and internal haplotype and other derivative thereof, can be interpreted as indicative of a founder effect, supporting the peripheral isolation model. However, there is a difference between the sample sizes: the smallest sample has at least 85% genetic variability, moreover, when comparing equivalent numbers of individuals of the two species, variability is still higher in A. pretrei. As an example, considering only the 13 spectacled Amazons (same number of A. tucumana) from Carazinho, RS, a wider range of haplotypes (DH = 0.64, four haplotypes) was still found in this locality.
Ecological observation also supports the origin of A. tucumana in the Pleistocene. A. pretrei seems to be dependent on Podocarpus lambertii seeds during the breeding season [62] mainly in southern Rio Grande do Sul (savannah with Araucaria), while in the Southern Yungas forests of northwestern Argentina, A. tucumana feed their nestlings mainly with Podocarpus parlatorei seeds, with 95% of the crop content consisting of seeds (L. O. Rivera, personal observation). Moreover, an increase in nest density was found in years with a high production of P. parlatorei fruits. All the populations of A. tucumana recorded along its distribution range are found in forests of P. parlatorei; this tree is a very good predictor of the presence of A. tucumana (L. O. Rivera, personal communication). Ledru et al. [63] found high genetic homogeneity in P. lambertii, which occurs mainly in the southernmost latitudes of Brazil, and also noted that such high genetic homogeneity most likely reflects the recent expansion of this genus in the area. This observation, and the affinity between southeastern Podocarpus populations and P. parlatorei from Argentina and Bolivia, indicates an ancient connection with other South American regions. They suggested that this finding supported the hypothesis of a wider pre-Quaternary distribution of P. lambertii on the South American continent, most likely before the formation of the Andes. Thus, it could be possible that a western portion of the continuous extension of P. lambertii forest became isolated before the Quaternary and that this forest was then colonized in the Quaternary by the dispersion of some A. pretrei individuals, giving origin to A. tucumana.
In conclusion, our genetic data indicate that the taxa studied here should be considered both distinct and worthy of conservation efforts. However, the scenario of speciation described in this paper was constructed based on a single marker (mitochondrial DNA) with maternal inheritance. Several deviations related to the estimated divergence time and levels of gene flow between populations during this process may be different if other independent markers are observed (see review in Nichols [64]). Thus, it is important to assess this scenario based on other markers to corroborate our biogeographical hypothesis of speciation for these two taxa.
Conservation implications
The conservation of genetic diversity within populations is always desirable to ensure that they are genetically sustainable, particularly in response to future environmental changes. Furthermore, genetic diversity is broadly correlated with population size, hence conservation efforts should seek to maintain or create large populations. The main factors affecting the two amazon species studied here are natural habitat destruction and fragmentation, as well as the illegal trafficking of nestlings and adults [24], [25]. Although these two species are suffering similar human pressures, A. pretrei seems to have a better conservation status than its sister species.
Systematic monitoring, which began in 1991, indicates that the A. pretrei population is stable at approximately 19,000 amazons [65]. Additionally, the genetic variability observed in this species (DH = 0.68) is similar to the one observed (DH = 0.75) in the non-endangered amazon Amazona aestiva [55]. This can also be interpreted by comparative analysis made from the analysis of microsatellite loci [65], where A. pretrei also shows similar genetic variability to other non-endangered parrots. Thus, the high genetic variability observed in these species can be seen as indicative that anthropogenic interferences have not yet drastically affected its genetic variability. Based on this, it is possible to suggest that the conservation measures to be adopted for this species should focus on the conservation of the remaining forest and restoration of degraded areas, thus ensuring conditions to maintain the current population size and hence genetic variability. In this direction, several actions were proposed in the National Plan of Action for the Conservation of Atlantic Parrots [66] to ensure the conservation of this species, among which we highlight the need for the conservation of remaining forests for nesting and feeding by amazons.
In contrast, A. tucumana has a smaller population size in relation to its sister species, with population estimates have identified approximately 8,000 individuals, 80% of which are in Argentina in the northern and central distribution [25], [67]. The low genetic variability observed in this species (DH = 0.41) when compared with its sister species may be mainly the result of its evolutionary history, due to the founding event in the early formation of this species. Furthermore, perhaps in recent centuries, it could have also been influenced by the different negative impacts of human actions, making the species more genetically vulnerable than its sister species does. Thus, conservation efforts for the Tucuman Amazon should prioritize the northern and central regions of Southern Yungas in Argentina, where 80% of the remaining Tucuman Amazon population occurs, with high priority given to the creation of public and private reserves [25]. Furthermore, this species may require actions aimed at increasing the population size, such as eliminating nest poaching and retaining nest and food trees in forestry operations [68], [69]. Finally, the low genetic variability and genetic distinctiveness in A. tucumana revealed in our study can be seen as very valuable information given the recent change of the categorization of this species on the IUCN Red List from Near Threatened to Vulnerable [70].
Acknowledgments
We thank the collaboration of Juliana Gondim de Albuquerque Lima and Vivian Ribeiro in performing some laboratory analyses, Cassia Alves Lima for her help in designing the figures, and Natan Medeiros, Lilian Giugliano and two anonymous referees for providing important suggestions for the manuscript. We are grateful to the National Park Administration of Argentina and the Direction of Natural Resources of the Jujuy Province that granted the research permits.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.
Funding Statement
This work is supported by graduate scholarships for A. V. Rocha by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Capes. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Darwin CR, Wallace AR (1858) Proceedings of the meeting of the Linnean Society held on July 1st, 1858. J Proc Linn Soc Zool 3: 54–56 (J Proc Linn Soc Zool van Wyhe John, editor). 2002. The complete work of Charles Darwin online. Available: http://darwin-online.org.uk/ [Google Scholar]
- 2. Bush GL (1975) Modes of animal speciation. Annu Rev Ecol Evol Syst 6: 339–364. [Google Scholar]
- 3.Dobzhansky T (1970) Genetic of the evolutionary process. New York: Columbia university press. 505 p. [Google Scholar]
- 4.Mayr E (1942) Systematics and the origin of species. New York: Columbia University Press. 334 p. [Google Scholar]
- 5. Quintero E, Ribas CC, Cracraft J (2013) The Andean Hapalopsittaca parrots (Psittacidae, Aves): an example of montane-tropical lowland vicariance. Zool Scr 42 1: 28–43. [Google Scholar]
- 6. Voelker G (1999) Dispersal, vicariance and clocks: historical biogeography and speciation in a cosmopolitan passerine genus (Anthus: Motacillidae). Evolution 53: 1536–1552. [DOI] [PubMed] [Google Scholar]
- 7. Voelker G (2002) Systematics and historical biogeography of wagtails (Aves: Motacilla): dispersal versus vicariance revisited. Condor 104: 725–739. [Google Scholar]
- 8. Weir JT, Price M (2011) Andean uplift promotes lowland speciation through vicariance and dispersal in Dendrocincla woodcreepers. Mol Ecol 20 21: 4550–4563. [DOI] [PubMed] [Google Scholar]
- 9.Cunnningham CW, Collins TM (1998) Beyond area relationships: extinction and recolonization in molecular marine biogeography. In: Schierwater B, Streit B, Wagner GP, Desalle R, editors. Molecular Ecology and Evolution: Approaches and Applications: Birkhauser, Basel, Switzerland. pp. 297–322. [Google Scholar]
- 10. Waters JM, Dijkstra LH, Wallis GP (2000a) Biogeography of a southern hemisphere freshwater fish: how important is a marine dispersal? Mol Ecol 9: 1815–1821. [DOI] [PubMed] [Google Scholar]
- 11. De Queiroz K (2005) The resurrection of oceanic dispersal in historical biogeography. Trends Ecol Evol 20: 68–73. [DOI] [PubMed] [Google Scholar]
- 12. Noonam BP, Chippindale PT (2006) Vicariant origin of Malagasy reptiles supports late cretaceous Antarctic landbrige. Am Nat 168: 730–774. [DOI] [PubMed] [Google Scholar]
- 13. Yoder AD, Nowak MD (2006) Has vicariance or dispersal been the predominant biogeographic force in Madagascar? Only time will tell. Annu Rev Ecol Evol Syst 37: 405–431. [Google Scholar]
- 14. Waters JM, Lopez A, Wallis GP (2000b) Molecular phylogenetics and biogeography of galaxiid fishes (Osteichthyes: Galaxiidae): dispersal, vicariance and the position of Lepidogalaxias salamandroides. Syst Biol 49: 777–795. [DOI] [PubMed] [Google Scholar]
- 15. Sanmartín I, Enghoff H, Ronquist F (2001) Patterns of animal dispersal, vicariance and diversification in the Holarctic. Biol J Linn Soc 73: 345–390. [Google Scholar]
- 16. Johnson NK, Cicero C (2002) The role of ecological diversification in sibling speciation of Empidonax flycatchers (Tyrannidae): multigene evidence from mtDNA. Mol Ecol 11: 2065–2081. [DOI] [PubMed] [Google Scholar]
- 17. Nielsen R, Wakeley J (2001) Distinguishing migration from isolation: a Markov Chain Monte Carlo approach. Genetics 158: 885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Russello MA, Amato G (2004) A molecular phylogeny of Amazona: implications for neotropical parrot biogeography, taxonomy, and conservation. Mol Phylogenet Evol 30: 421–437. [DOI] [PubMed] [Google Scholar]
- 19.Peters JL (1937) Checklist of birds of the world. Cambridge: Harvard University Press. 318 p. [Google Scholar]
- 20.Ridgely RS (1981) A guide to the birds of Panama. Princeton: Princeton University Press. 412 p. [Google Scholar]
- 21.Forshaw JM (1977) Parrots of the World. 3rd ed, Willoughby: Lansdowne Press. 584 p. [Google Scholar]
- 22. Diefenbach KH, Goldhammer SP (1986) Biologie und okologie der rotzchwanzamazone Amazona brasilienis . Trochilus 7: 72–78. [Google Scholar]
- 23.Collar NJ, Gonzaga LP, Krabbe N, Madroño Nieto A, Naranjo LG, et al.. (1992) Threatened birds of the Americas: the ICBP/IUCN Red Data Book. International Council for Bird Preservation, Cambridge, U.K. [Google Scholar]
- 24.Martinez J, Prestes NP (2008) Biologia da conservação: estudo de caso com papagaio-charão e outros papagaios brasileiros.Passo Fundo:Editora UPF. 287 p. [Google Scholar]
- 25. Rivera L, Politi N, Bucher EH (2007) Decline of the Tucuma' n parrot Amazona tucumana in Argentina: present status and conservation needs. Oryx 41: 101–105. [Google Scholar]
- 26. Prado D (1993) What is the Gran Chaco vegetation in South America?. II. A redefinition. Contribution to the study of flora and vegetation of the Chaco. Candollea 48: 615–629. [Google Scholar]
- 27. Pennington RT, Prado DE, Pendry CA (2000) Neotropical seasonally dry forestsand Quaternary vegetation changes. J Biogeogr 27: 261–273. [Google Scholar]
- 28. Iriondo M (1993) Geomorphology and late Quaternary of the Chaco (South America). Geomorphology 7: 289–303. [Google Scholar]
- 29. Hernández RM, Jordan TE, Farjat AD, Echavarría L, Idleman BD, et al. (2005) Age, distribution, tectonics, and eustatic controls of the Paranense and Caribbean marine transgressions in southern Bolivia and Argentina. J S Am Earth Sci 19: 495–512. [Google Scholar]
- 30. Vuilleumier F (1965) Relationships and evolution within the Cracidae. Bull of the Mus Comp Zool 134: 1–27. [Google Scholar]
- 31. Short LL (1975) A zoogeographical analysis of the South American Chaco avifauna. Bull Amer Mus Nat Hist 154: 163–352. [Google Scholar]
- 32.Simpson BB (1979) Quaternary biogeography of the righ montane regions of South America. In: Duelman WE editor. The South American Herpetofauna: its origin, evolution, and dispersal. Monograph of the Museum of Natural History. Laurence Kansas: University of Kansas. pp. 157–188. [Google Scholar]
- 33. Nores M (1992) Bird speciation in subtropical South America in relation to forest expansion and retraction. Auk 109: 346–357. [Google Scholar]
- 34. Van der Hammen T (1974) The Pleistocene changes of vegetation and climate in tropical South America. J Biogeogr 1: 3–26. [Google Scholar]
- 35.Van der hammen T (1983) The palaeoecology and palaeogeography of savannas. In: Bourliere F, editor. Tropical savannas. Amsterdam: Elsevier. pp. 19–35. [Google Scholar]
- 36.Fitzpatrick JW (1980) Some aspects of speciation bin South American flycatchers. In: Short LL, editor. Symposium on speciation in South American birds. Actis 17 Congressus Internationalis Ornithologici. Berlin: Deutsche Ornithologen-gesellschaft. pp. 1273–1279. [Google Scholar]
- 37.IUCN (2013) The IUCN Red List of Threatened Species. v. 2013.1. Available: http://www.iucnredlist.org. Accessed 2 July 2013.
- 38.Machado AB, Drummond GM, Paglia AP (org.) (2008) Livro vermelho da fauna brasileira ameaçada de extinção. Ministério do Meio Ambiente, Brasília, DF, v.2: . 1420 p. [Google Scholar]
- 39.López-Lanús B, Grilli P, Coconier E, Di Giacom A, Banchs R (2008) Categorización de las aves de la Argentina según su estado de conservación.Informe de Aves Argentinas/AOP y Secretaría de Ambiente y Desarrollo Sustentable. Buenos Aires, Argentina. [Google Scholar]
- 40.Hein J, Schierup MH, Wiuf C (2005) Gene genealogies, variation and evolution: a primer in coalescent theory. Oxford: Oxford University Press. [Google Scholar]
- 41.Brudford MW, Hannote O, Brookfield JFY, Burke T (1992) Single locus and multilocus DNA fingerprinting. In: Hoezel CAR, editor. Molecular genetic analyses of populations: a pratical approach. New York: Oxford University Press. pp. 225–269. [Google Scholar]
- 42.Palumbi SR (1996) Nucleic acids II: The polymerase chain reaction. In: Hillis DM, Moritz DM, Mable BK, editors. Molecular systematics. Sunderland: Sinauer Associates pp. 205–247. [Google Scholar]
- 43. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The clustalx windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tamura N, Peterson D, Peterson N, Skecher S, Nei M, et al. (2011) MEGA 5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximun Parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rozas J, Sánchez-De I, Barrio JC, Messeguer X, Rozas R (2003) DNAsp, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- 46. Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16: 37–48. [DOI] [PubMed] [Google Scholar]
- 47. Tajima F (1989) Statistical method for testing the neutral mutation hypothesis. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fu Y, Li W (1993) Statistical tests of neutrality of mutations. Genetics 133: 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of Phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- 50. Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- 51. Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9 8: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rambaut A, Drummond AJ (2007). TRACER v1.5. Available: http://beast.bio.ed.ac.uk/Tracer.
- 53. Hey J, Nielsen R (2004) Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis . Genetics 167: 747–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pereira SL, Baker AJ (2006) A mitogenomic timescale for birds detects variable phylogenetic rates of molecular evolution and refutes the standard molecular clock. Mol Biol Evol 23: 1731–1740. [DOI] [PubMed] [Google Scholar]
- 55. Caparroz R, Seixas GHF, Berkunsky I, Collevatti RG (2009) The role of demography and climatic events in shaping the phylogeography of Amazona aestiva (Psittaciformes, Aves) and definition of management units for conservation. Diversity Distrib 15: 459–468. [Google Scholar]
- 56. Ramos-Onsins SF, Rozas J (2002) Statistical properties of new neutrality test against population growth. Mol Biol Evol 19: 2092–2100. [DOI] [PubMed] [Google Scholar]
- 57. De Queiroz K (2007) Species concepts and species delimitation. Syst Biol 56: 879–886. [DOI] [PubMed] [Google Scholar]
- 58.Newton I (2003) The speciation and biogeography of birds. London: Academic Press. 668 p. [Google Scholar]
- 59.Mata JRR, Erize F, Rumboll M (2006) Birds of South America non-passerines: Rheas to Woodpeckers. Oxford: Princeton University Press. 376 p. [Google Scholar]
- 60. Hulka C, Gräfe KU, Sames CE, Uba C, Heubeck C (2006) Depositional setting of the middle to Late Miocene Yecua formation of the Chaco Foreland Basin, southern Bolivia. J S Am Earth Sci 21: 135–150. [Google Scholar]
- 61. Willis EO (1992) Zoogeographical origins of eastern Brazilian birds. Ornitol Neotrop 3: 1–15. [Google Scholar]
- 62.Martinez J, Prestes NP (2002) Ecologia e conservação do papagaio-charão Amazona pretrei. In: Galetti M, Pizo MA, editors. Ecologia e conservação de psitacídeos no Brasil. Belo Horizonte: Melopsittacus Publicações Científicas. pp. 173–192. [Google Scholar]
- 63. Ledru M-P, Salatino MLF, Ceccantini G, Salatino A, Pinheiro F, et al. (2007) Regional assessment of the impacto f climatic change on the distribution of a tropical conifer in the lowlands of South America. Diversity Distrib 13: 761–771. [Google Scholar]
- 64. Nichols R (2011) Gene trees and species trees are not the same. Trends Ecol Evol 16 7: 358–364. [DOI] [PubMed] [Google Scholar]
- 65.Prestes NP, Martinez J (2011) Papagaio- charão. In: Schunck F, Samenzari M, Lugarini R, Soares ES, editors. Plano de ação nacional para a conservação dos papagaios da Mata Atlântica. ICMBio/MMA Série Especial (20). pp. 34–40. [Google Scholar]
- 66.Schunck F, Samenzari M, Lugarini R, Soares ES (org) (2011) Plano de ação nacional para a conservação dos papagaios da Mata Atlântica. ICMBio/MMA Série Especial (20). 128 p. [Google Scholar]
- 67. Rivera L, Llanos RR, Politi N, Hennessey B, Bucher EH (2010) The Near Threatened Tucumán Parrot Amazona tucumana in Bolivia: insights for a global assessment. Oryx 44: 110–113. [Google Scholar]
- 68. Rivera L, Politi N, Bucher EH (2012) Nesting habitat of the Tucuman Parrot Amazona tucumana in an old-growth cloud-forest of Argentina. Bird Conserv Internat 22: 398–410. [Google Scholar]
- 69. Rivera L, Politi N, Bucher EH, Pidgeon A (2013) Nesting success and productivity of Tucuman Parrots (Amazona tucumana) in high-altitude forests of Argentina: do they differ from lowland Amazona parrots? Emu 114 1: 41–49. [Google Scholar]
- 70.IUCN (2012) IUCN Red List of Threatened Species. Version 2012.1. Available: http://www.iucnredlist.org. Downloaded on 15 November 2013.
- 71.Birdlife International (2013) Species factsheet: Tucuman Amazon Amazona tucumana Available: http://www.birdlife.org/datazone/speciesfactsheet.Php?Id=1667. Accessed 25 October 2013.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.