Abstract
The trithorax group of genes (trxG) was identified in mutational screens that examined developmental phenotypes and suppression of Polycomb mutant phenotypes. The protein products of these genes are primarily involved in gene activation, although some can also have repressive effects. There is no central function for these proteins. Some move nucleosomes about on the genome in an ATP-dependent manner, some covalently modify histones such as methylating lysine 4 of histone H3, and some directly interact with the transcription machinery or are a part of that machinery. It is interesting to consider why these specific members of large families of functionally related proteins have strong developmental phenotypes.
Trithorax-group proteins are primarily involved in gene activation but have diverse activities. Some modify histones or remodel chromatin; others interact with the transcription machinery or are a part of that machinery.
Overview
All cells in an organism must be able to “remember” what type of cell they are meant to be. This process, referred to as “cellular memory” or “transcriptional memory,” requires two basic classes of mechanisms. The first class, discussed in Grossniklaus and Paro (2014), functions to maintain an “off” state for genes that, if turned on, would specify an inappropriate cell type. The Polycomb-group (PcG) proteins have as their primary function a repressive role in cellular memory. The second class of mechanisms is composed of those that are required to maintain key genes in an “on” state. Any cell type requires the expression of master regulatory proteins that direct the specific functions required for that cell type. The genes that encode these master regulatory proteins must be maintained in an “on” state throughout the lifetime of an organism to maintain the proper cell types within that organism.
The proteins that are involved in maintaining the “on” state are called trithorax-group (trxG) proteins in honor of the trithorax gene, the founding member of this group of regulatory proteins. A large group of proteins with diverse functions make up the trxG. The roles these proteins play in the epigenetic mechanisms that maintain the “on” state appear more complex at this juncture than the roles for PcG proteins in repression. The first complexity is that a very large number of proteins and mechanisms are needed to actively transcribe RNA from any gene. Thus, in contrast to repression, which might be accomplished by comparatively simple mechanisms that block access of all proteins, activation of a gene requires numerous steps, any of which might play a role in maintaining an “on” state. Thus, there are numerous possible stages in which a trxG protein might work.
A second complexity in thinking about trxG proteins is that proteins that function in activation can also, in different contexts, function in repression. This might appear counterintuitive, but depending on the precise architecture of a gene, the same protein performing its function might in one case help a gene become activated, and in another case help a different gene become repressed. At this time it does not appear that trxG proteins are dedicated solely to the maintenance of gene expression, but that these proteins can also play multiple roles in the cell. These complexities make for several interesting unanswered questions. Why are only some of the proteins needed to activate transcription also critical for maintenance of transcription? Do these proteins have functions that are uniquely suited to maintaining the active state? Or are some of these proteins needed for maintenance solely because an evolutionary accident that made them key regulators of gene(s) particularly important to development?
As we will see when we discuss mechanisms of action, some of the trxG proteins are involved in regulating chromatin structure in opposition to the mechanisms used by the PcG proteins. trxG proteins can place covalent posttranslational modifications (PTMs) on chromatin or can alter chromatin by changing the structure and position of the nucleosomes that are the building blocks of chromatin. Other trxG proteins function as part of the transcription machinery. Thus, these proteins are found in a wider variety of complexes than the PcG proteins and are likely to play more complicated roles in epigenetic mechanisms.
1. INTRODUCTION
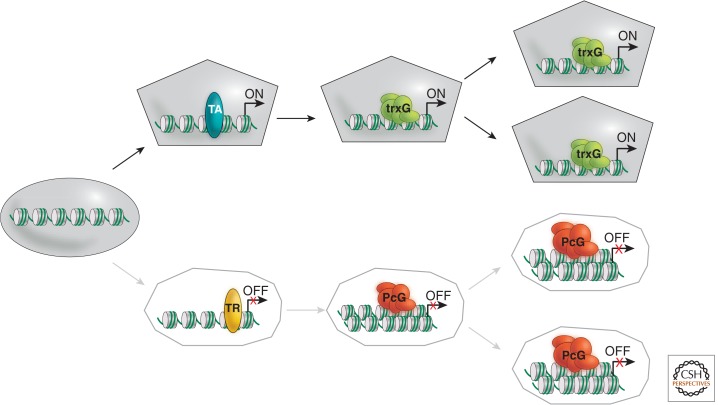
Numerous developmental decisions—including the determination of cell fates—are made in response to transient positional information in the early embryo. These decisions are dependent on changes in gene expression. This allows cells with identical genetic blueprints to acquire unique identities and follow distinct pathways of differentiation. The changes in gene expression underlying the determination of cell fates are heritable; a cell’s fate rarely changes once it is determined, even after numerous cell divisions and lengthy periods of developmental time (Fig. 1). Understanding the molecular mechanisms underlying the maintenance of the determined state has long been a goal of developmental and molecular biologists.
Figure 1.
The concept of cellular memory. Schematic illustration highlighting the role of trxG complexes in maintaining heritable states of active gene expression in contrast to heritable silencing by PcG complexes, as defined originally for the Drosophila homeotic (Hox) gene cluster.
Many of the regulatory proteins involved in the maintenance of heritable states of gene expression were identified in studies of Drosophila homeotic (Hox) genes. Hox genes encode homeodomain transcription factors that regulate the transcription of batteries of downstream target genes, which in turn specify the identities of body segments (Gellon and McGinnis 1998). In Drosophila, Hox genes are found in two gene complexes: the Antennapedia complex (ANT-C), which contains the Hox genes labial (lab), Deformed, Sex combs reduced (Scr), and Antennapedia (Antp); and the bithorax complex, which contains the Hox genes Ultrabithorax (Ubx), abdominalA (abdA), and AbdominalB (AbdB; Duncan 1987; Kaufman et al. 1990). Each Hox gene specifies the identity of a particular segment, or group of segments, along the anterior–posterior axis of the developing fly. For example, Antp specifies the identity of the second thoracic segment, including the second pair of legs, whereas Ubx specifies the identity of the third thoracic segment, including the balancer organs located behind the wings. Thus, the transcription factors encoded by Hox genes function as master regulatory switches that direct the choice between alternative pathways of development.
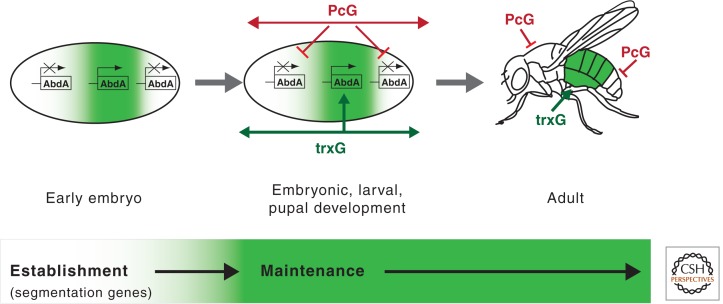
The transcription of Hox genes must be regulated precisely because dramatic alterations in cell fates can result from their inappropriate expression (Simon 1995; Simon and Tamkun 2002). For example, the derepression of Antp in head segments transforms antennae into legs, whereas the inactivation of Ubx in thoracic segments transforms balancer organs into wings. In Drosophila, the initial patterns of Hox transcription are established early in embryogenesis by transcription factors encoded by segmentation genes. The proteins encoded by segmentation genes—including the gap, pair-rule, and segment polarity genes—subdivide the early embryo into 14 identical segments. These proteins also establish the initial patterns of Hox transcription, the first step toward the development of segments with distinct identities and morphology. The majority of segmentation genes, however, are transiently expressed during early development. Once established, the segmentally restricted patterns of Hox transcription must be maintained throughout subsequent embryonic, larval, and pupal stages to maintain the identities of the individual body segments. This function is performed by two other groups of regulatory proteins: the Polycomb group of repressors (PcG) and the trithorax group of transcriptional regulators (trxG). The regulation of Hox transcription therefore consists of at least two distinct phases: establishment (by segmentation genes) and maintenance (by PcG and trxG genes; Fig. 2).
Figure 2.
Regulation of Hox transcription. The boundaries of abd-A transcription and other Hox genes are established by segmentation proteins. These include the products of gap and pair-rule genes, which subdivide the embryo into 14 identical segments. During subsequent development, the “off” or “on” states of Hox transcription are maintained by the ubiquitously expressed members of the trxG of activators and the PcG of repressors via mechanisms that remain poorly understood.
1.1. Identification of Genes Involved in the Maintenance of the Determined State
Because of their roles in the maintenance of cell fates, Drosophila PcG and trxG genes have been the subject of intense study for decades. As discussed in Grossniklaus and Paro (2014), the majority of PcG genes were identified by mutations that cause homeotic transformations due to the failure to maintain repressed states of Hox transcription. A classic example of a phenotype associated with PcG mutations is the transformation of second and third legs to first legs. This homeotic transformation results from the derepression of the ANT-C gene, Scr, and is manifested by the appearance of first leg bristles known as sex comb teeth on the second and third legs of the adult. This “Polycomb” or “extra sex combs” phenotype—together with other homeotic transformations resulting from the failure to maintain repression of Hox genes—led to the identification of more than a dozen PcG genes in Drosophila. The majority of PcG genes encode subunits of two complexes involved in transcriptional repression: Polycomb repressive complex (PRC) 1 and PRC2 (Levine et al. 2004). PRC1 and PRC2 are targeted to the vicinity of Hox (and other) promoters via cis-regulatory elements known as Polycomb-response elements (PREs) in the Drosophila genome (see Grossniklaus and Paro 2014). A large body of evidence suggests that PcG complexes repress transcription by modulating chromatin structure (Francis and Kingston 2001; Ringrose and Paro 2004).
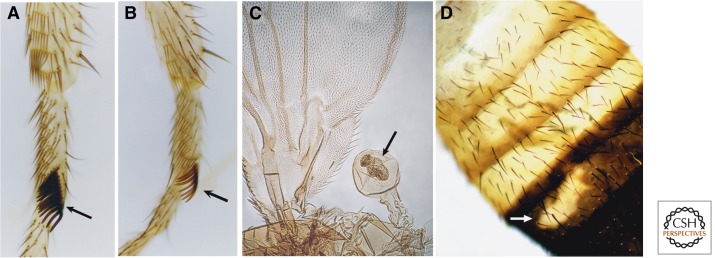
Members of the trxG were initially identified by mutations that mimic loss-of-function Hox mutations in Drosophila (Fig. 3) (Kennison 1995). Genes identified by this phenotype include trithorax (trx); absent, small, or homeotic 1 (ash1), absent, small, or homeotic 2 (ash2), and female-sterile homeotic (fsh). Mutations in trx—the founding member of the trxG—for example, cause the partial transformation of halteres to wings (because of decreased Ubx transcription), first legs to second legs (because of decreased Scr transcription), and posterior abdominal segments to more anterior identities (because of decreased abdA and AbdB transcription). Numerous other trxG members were identified in screens for extragenic suppressors of Pc (Su(Pc)) mutations (Kennison and Tamkun 1988). The rationale behind these genetic screens was that a reduction in the level of a protein that maintains an active state should compensate for a reduction in the level of a PcG repressor (Fig. 4). brahma (brm) and numerous other Su(Pc) loci were identified using this approach, bringing the total number of trxG members to more than 16 (Table 1). Many other proteins have been classified as trxG members based on other, less stringent criteria, including sequence homology with known trxG proteins, physical association with trxG proteins, biochemical activity, or effects on Hox transcription in vitro or in vivo.
Figure 3.
Examples of developmental cell fate transformations associated with mutations in Drosophila trxG genes. (A) Wild-type first leg. The sex comb, unique to the first leg, is marked by an arrow. (B) A patch of kis mutant tissue (marked by an arrow) is partially transformed from the first leg to the second leg because of decreased Scr transcription, albeit incomplete, as evidenced by a reduction in the number of sex comb teeth. (C) A patch of mor mutant tissue (marked by an arrow) displays the partial transformation from balancer organ to wing because of decreased Ubx expression. (D) A patch of kis mutant tissue (marked by an arrow) in the fifth abdominal segment is partially transformed to a more anterior identity because of decreased Abd-B expression, as evidenced by the loss of the dark pigmentation characteristic of this segment. (A,B,D, Reprinted from Daubresse et al. 1999.)
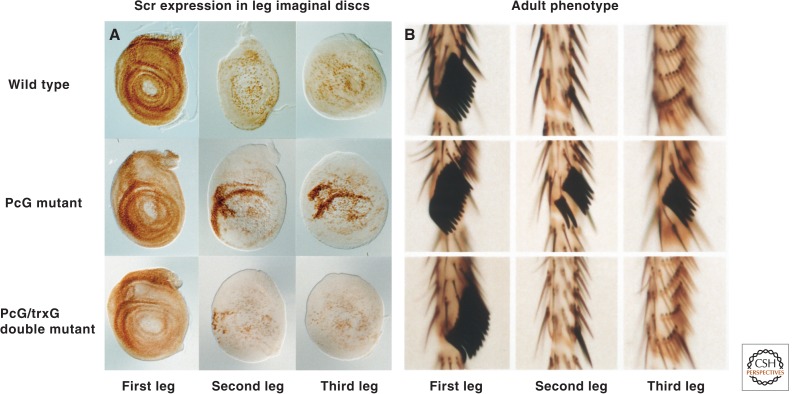
Figure 4.
trxG mutations block the derepression of Hox genes in PcG mutants. (A) Leg imaginal discs stained with antibodies against the protein encoded by the Hox gene, Scr, which specifies the identity of the labial and first thoracic segments, including the first leg. (B) Basitarsal segments of the legs of wild-type and mutant adults. Note the presence of sex comb teeth on the first leg, but not the second and third legs of wild-type adults. The Scr gene is partially derepressed in the second and third leg discs, in which it is normally silent, in individuals heterozygous for mutations in PcG genes leading to the appearance of ectopic sex comb teeth on the second and third legs. These phenotypes are suppressed by mutations in brm and many other trxG genes. (A, Reprinted, with permission, from Tamkun et al. 1992, © Elsevier; B, portion modified, with permission, from Kennison 2003, © Elsevier.)
Table 1.
Biochemical functions of trxG proteins
| Known function | Organism | Complexed with non-trxG proteins? | ||
|---|---|---|---|---|
| Drosophila | Human | Yeast | ||
| ATP-dependent chromatin remodeling | BRM | BRG1/HBRM | Swi2/Snf2, Sth1 | Yes (5–10)a |
| OSA | BAF250 | Swi1/Adr6 | Yes (5–10) | |
| MOR | BAF155, BAF170 | Swi3, Rsc8 | Yes (5–10) | |
| SNR1 | hSNF5/INI1 | Snf5, Sfh1 | Yes (5–10) | |
| Kismet (KIS) | CHD7 | – | NK | |
| Histone methyltransferases | Trithorax (TRX) | MLL1, MLL2, MLL3 | Set1 | Yes (5–20) |
| Absent, small or homeotic 1(ASH1) | MlLL4, hSET1 hASH1 | – | NK | |
| Mediator subunits | Kohtalo (KTO) | TRAP230 | Srb8 | Yes (13–24) |
| Skuld (SKD) | TRAP240 | Srb9 | Yes (13–24) | |
| Cohesin subunit | Verthandi (VTD) | Rad21 | Scc1/Rad21 | Yes (>3) |
| Transcription factor | Trithorax-like (TRL) | BTBD14B | – | No |
| Growth factor receptor | Breathless (BTL) | FGFR3 | – | NK |
| Other | Sallimus (SLS) | Titin | NK | |
| ASH2 | hASH2Lb | Bre2 | Yes (5–20) | |
aBRM, OSA, MOR, and SNR1 can all be found in stable association with each other in a single complex.
bRelatively low sequence similarity to ASH2.
NK, not known.
The functional relationship between members of the trxG is complicated, as is the mechanistic connection between trxG function and the maintenance of cell fate. There are numerous mechanisms via which a protein might maintain an appropriately high level of expression of a homeotic gene (the genetic definition of a trxG protein) without being a devoted transcriptional activator, or a protein devoted to epigenetic control. Formal possibilities for trxG function (in addition to the ability to directly activate transcription) include the ability to increase function of direct activators, to block function of PcG repressors, and to create a “permissive” chromatin state that facilitates the function of numerous other regulatory complexes. Furthermore, as discussed in Section 2, some trxG proteins play complicated mechanistic roles that on some genes contribute to activation and that on other genes can contribute to repression. The evolutionary conservation of this family and the conserved functions of this family offer hints concerning what types of mechanisms are needed to maintain the appropriate level of activation of master regulatory genes that determine cell fate.
1.2. trxG Members in Other Organisms
Functional counterparts of virtually all Drosophila trxG proteins are present in mammals, including humans (Table 1). Genetic and biochemical studies have shown that the fly and mammalian proteins play highly conserved roles in both gene expression and development. A good example of the functional conservation of trxG proteins is provided by MLL1, one of the four mammalian orthologs of Drosophila trx. Mutations in MLL1 cause homeotic transformations of the axial skeleton of mice because of the failure to maintain active transcription of Hox genes (Yu et al. 1995; Yu et al. 1998). Both mixed lineage leukemia (MLL) proteins and trx function as histone lysine (K) methyltransferases (KMTs), and direct evidence of functional homology between the two proteins was provided by the use of human MLL to partially rescue developmental defects in flies resulting from the loss of trx function (Muyrers-Chen et al. 2004). Thus, the mechanisms underlying the maintenance of the determined state have been highly conserved during evolution.
1.3. trxG Proteins Play Diverse Roles in Eukaryotic Transcription
The trxG of activators is a large and functionally diverse group of regulatory proteins. This may reflect the complexity of eukaryotic transcription, which involves highly regulated interactions between gene-specific transcriptional activators, the numerous components of the general transcription machinery, and the DNA template that is transcribed. Transcriptional activation involves the binding of sequence-specific activating proteins, the recruitment of the general transcription machinery by those proteins, the formation of a preinitiation complex in which RNA polymerase II is bound to the promoter, the opening of the DNA helix near the promoter, the efficient escape of RNA polymerase from the promoter, and efficient elongation of RNA polymerase through the gene.
The ability to maintain an active transcriptional state might involve any of the numerous steps required for activation, as on any given gene, different steps might play a rate-determining role for transcriptional activity. The packaging of eukaryotic DNA into chromatin also provides another level at which trxG proteins can regulate transcription. Nucleosomes and other components of chromatin tend to inhibit the binding of general and gene-specific transcription factors to DNA, as well as inhibiting the elongation of RNA polymerase. Alterations in chromatin structure, including changes in the structure or positioning of nucleosomes, can influence virtually every step in the process of transcription.
Any protein that is required for transcription is required for the maintenance of the active state. Indeed, some trxG proteins play relatively general roles in transcription and are not dedicated solely to the maintenance of the determined state. Other trxG proteins, however, may play specialized roles in this process either by directly counteracting PcG repression or maintaining heritable states of gene activity through DNA replication and mitosis. The latter class of trxG proteins is of particular interest to developmental biologists.
2. CONNECTIONS BETWEEN trxG PROTEINS AND CHROMATIN
Genetic studies indicating that trxG genes play key roles in transcription and development has stimulated significant work to understand the biochemical function of their products. Many of these experiments have used, as their conceptual basis, the hypothesis that chromatin will be the biologically relevant substrate of trxG proteins. All genes are packaged into chromatin and that packaging can create a compacted and inaccessible state, or can be in an open and permissive state. Both the permissive and inaccessible states may conceivably be heritable. These considerations led to the simple hypothesis that trxG proteins might modulate chromatin structure to affect regulation. Furthermore, as trxG genes were cloned and sequenced it became apparent that some of their products are related to proteins involved in ATP-dependent chromatin remodeling (covered in detail in Becker and Workman 2013) or the covalent modification of nucleosomal histones in other organisms, including the yeast Saccharomyces cerevisiae. Thus, although yeast lack either Hox genes or PcG repressors, this organism has provided valuable clues about potential roles for trxG proteins in eukaryotic transcription.
One of the first connections between trxG proteins and chromatin was provided by the discovery that the Drosophila trxG gene brahma (brm) is highly related to yeast SWI2/SNF2 (Tamkun et al. 1992). SWI2/SNF2 was identified in screens for genes involved in mating-type switching (switch [swi] genes) and sucrose-fermentation (sucrose nonfermenting [snf] genes). It was subsequently shown to be required for the activation of numerous inducible yeast genes (Holstege et al. 1998; Sudarsanam et al. 2000). The transcription defects observed in swi2/snf2 mutants are suppressed by mutations in nucleosomal histones, an early observation that first suggested that SWI/2/SNF2 activates transcription by counteracting chromatin repression (Kruger et al. 1995). Biochemical studies conducted in the early 1990s confirmed this hypothesis; SWI2/SNF2 and many other proteins identified in the swi/snf screens function as subunits of a large protein complex called SWI/SNF that uses the energy of ATP hydrolysis to increase the ability of proteins to bind to nucleosomal DNA (Cote et al. 1994; Imbalzano et al. 1994; Kwon et al. 1994). SWI2/SNF2 has ATPase activity and thus functions as the “engine” of this chromatin-remodeling machine; other subunits of the SWI/SNF complex mediate interactions with regulatory proteins or its chromatin substrate (Phelan et al. 1999).
Another connection between trxG and chromatin was suggested by the presence of SET domains in the trxG proteins, Trithorax (TRX) and Absent, small or homeotic (ASH1). The SET domain was originally defined by a stretch of amino acids that shows homology between Su(var)3-9, Enhancer of Zeste and TRX, the latter two proteins being, respectively, PcG and trxG members. In the late 1990s the SET family of proteins was shown to have lysine methyltransferase (KMT) activity. Su(var)3-9 methylates histone H3K9 (H3K9me), whereas Enhancer of Zeste methylates H3K27 (H3K27me; Rea et al. 2000; Levine et al. 2004; Ringrose and Paro 2004). As discussed elsewhere, H3K9 methylation promotes heterochromatin assembly, whereas H3K27 methylation appears to be required for PcG repression (see Elgin and Reuter 2013 and Grossniklaus and Paro 2014, respectively, for more detailed discussion). The presence of SET domains in trxG proteins suggested that the methylation of histone tails might also be important for the maintenance of active transcriptional states.
These findings, together with the growing realization that chromatin-remodeling and -modifying enzymes play key roles in transcriptional activation, motivated biochemists to identify protein complexes that contain trxG proteins and examine the effect of these complexes on chromatin structure in vitro. Other experiments tested the hypothesis that trxG proteins might interact directly with the transcriptional machinery, another well established method of affecting regulation. As described below, these studies revealed that some trxG proteins impact regulation by modifying chromatin structure whereas others function via direct interactions with components of the transcription machinery.
2.1. trxG Proteins Involved in ATP-Dependent Chromatin Remodeling
Chromatin-remodeling complexes have been implicated in a wide variety of biological processes including transcriptional repression and activation, chromatin assembly, the regulation of higher-order chromatin structure, and cellular differentiation (see Becker and Workman 2013). Two trxG members, BRM and KIS, have ATPase domains and members of evolutionarily conserved families of proteins with chromatin-remodeling activity (Table 1). BRM is part of the SWI2/SNF2 subfamily of remodelers, whereas KIS is a member of the CHD7 subfamily. These two subfamilies are both able to move nucleosomes (see Becker and Workman 2013), but differ in mode of action; CHD7 requires naked DNA adjacent to the nucleosome, and thus loosely spaced nucleosomes, to function, whereas the SWI/SNF complexes do not require adjacent naked DNA and are able to function on tightly packed nucleosomes (Bouazoune and Kingston 2012).
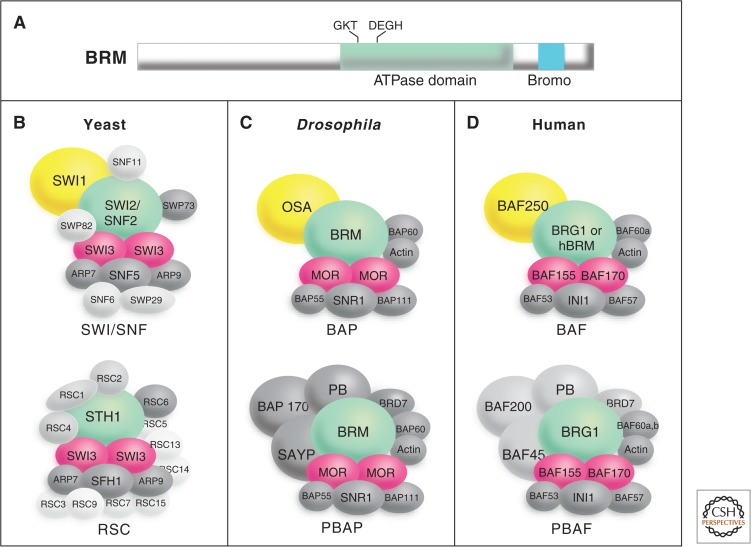
The most extensively studied trxG proteins involved in chromatin remodeling are BRM and its human counterparts, BRG1 and HBRM. As predicted, these proteins function as the ATPase subunits of complexes that are highly related to yeast SWI/SNF (Kwon et al. 1994; Wang et al. 1996). SWI/SNF complexes contain between eight and 15 subunits and have been highly conserved during evolution (Fig. 5). A second Drosophila trxG gene identified in the screens for suppressors of Pc mutations, moira (mor), encodes another key member of this ATP-dependent remodeling complex. Homologs of BRM and MOR interact directly to form a functional core of SWI/SNF in humans (Phelan et al. 1999). SWI/SNF complexes are abundant in higher eukaryotes. For example, each mammalian nucleus contains about 25,000 copies of SWI/SNF-subfamily complexes. They are potent remodelers in that they are able to efficiently create access to sites at the center of a mononucleosome, which is energetically difficult because sites at the center of the nucleosome have ∼70 bp of constrained nucleosomal DNA on both sides.
Figure 5.
The SWI/SNF family of remodeling complexes. Each complex contains a member of SNF2/SWI2 family of ATPases and at least eight other subunits. (A) Schematic diagram of the BRM protein showing the location of the ATPase domain and carboxy-terminal bromodomain (which shows affinity to acetylated lysine residues in histone tails), which are conserved in all SNF2/SWI2 family members. SWI/SNF complexes in yeast (B), Drosophila (C), and humans (D) are shown. Drosophila trxG proteins (BRM, MOR, and OSA) and their counterparts in other organisms are shown in color. Further information about these complexes and their subunits may be found in Mohrmann and Verrijzer (2005).
Each species studied has at least two distinct SWI/SNF complexes, all of which contain BRM or a highly related chromatin-remodeling ATPase. Another trxG protein, OSA, provides distinction between the complexes, in that one class of complexes (Brahma associate proteins complex [BAP]) contains OSA and another evolutionarily conserved complex (polybromo-containing BAP complex [PBAP]) contains a Polybromo-domain protein (Fig. 5) (Mohrmann and Verrijzer 2005). Further evidence distinguishing these complexes comes from fractionation studies that show that several other proteins, including SAYP and BAP170, are PBAP-specific subunits (Chalkley et al. 2008). SAYP has trxG properties, implying that the PBAP complex is directly involved in trxG function. There are many related SWI/SNF family subcomplexes in flies and mammals, and an interesting question, not yet fully resolved, is how each of these contributes to differentiation and trxG functions.
The identification of three different members of the SWI/SNF subfamily of complexes in the trxG screens highlights the importance of this diverse family of large complexes during developmental processes. It is believed that this primarily reflects the role for this complex in activating transcription, although this family of complexes can also be involved in repressive mechanisms (Becker and Workman 2013), SWI/SNF complexes have been implicated in transcriptional activation in every species examined. They can be targeted to genes by interactions with transcriptional activators, remodel nucleosomes to assist in the initial binding of general transcription factors and RNA polymerase II (RNA Pol II), and become targeted later in the activation process to assist with transcriptional elongation. Thus, SWI/SNF complexes appear to function at every step in the process of transcriptional activation, although there appears to be an emphasis on function at the early steps that lead to loading of RNA Pol II.
The trxG gene kis, like brm, mor, and osa, was identified in a screen for extragenic suppressors of Pc. This suggested that it acts antagonistically to PcG proteins to maintain active states of Hox transcription (Kennison and Tamkun 1988). Genetic studies revealed that kis is required for both segmentation and maintenance of Hox transcription during Drosophila development (Daubresse et al. 1999). The molecular analysis of kis revealed that it encodes several large proteins, including a ∼575 kDa isoform (KIS-L), which contains an ATPase domain characteristic of chromatin-remodeling factors (Daubresse et al. 1999; Therrien et al. 2000). Other conserved domains outside the ATPase domain (e.g., bromodomains and chromodomains) contribute to the functional specificity of chromatin-remodeling factors by mediating interactions with nucleosomes or other proteins. BRM and all ATPase subunits of the SWI/SNF subfamily contain a single bromodomain—a protein motif associated with the binding of certain acetylated histones (Fig. 5A). KIS-L contains two chromodomains (protein motifs that bind certain methylated histones are covered in Cheng 2014 and Patel 2014) and is therefore more similar to Mi2 and other members of the CHD family of chromatin-remodeling factors. The large size of KIS-L (∼575 kDa) has made it difficult to analyze this protein biochemically. Its sequence implies that it is an ATP-dependent nucleosome-remodeling protein, and, as discussed below, its apparent human homolog has been shown to have remodeling activity.
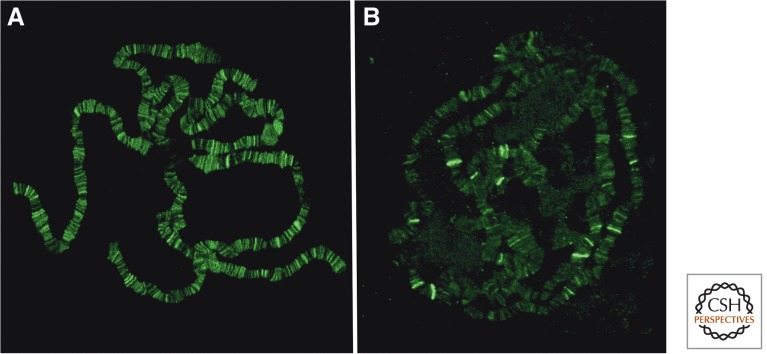
KIS-L is not physically associated with BRM and may function as a monomer or in a distinct protein complex (Srinivasan et al. 2005). The two proteins overlap extensively with each other and RNA Pol II on polytene chromosomes, however, suggesting that both play relatively global roles in transcription (Fig. 6) (Armstrong et al. 2002; Srinivasan et al. 2005). Loss of BRM function in the larval salivary gland blocks a relatively early step in transcription (Armstrong et al. 2002), whereas the loss of KIS-L function leads to a decrease in the level of elongating, but not initiating forms of RNA Pol II (Srinivasan et al. 2005, 2008). These findings suggest that BRM and KIS-L facilitate distinct steps in transcription by RNA Pol II by catalyzing ATP-dependent alterations in nucleosome structure or spacing.
Figure 6.
Chromosomal distribution of trxG proteins. The genome-wide distribution of trxG proteins was examined by staining Drosophila salivary gland polytene chromosomes with antibodies against BRM (A) or TRX (B). Consistent with a relatively global role in transcriptional activation, BRM is associated with hundreds of sites in a pattern that overlaps extensively with RNA Pol II. In contrast, strong TRX signals are detected at a much smaller number of sites on polytene chromosomes.
Recent studies on the CHD7 gene in mammals, which is homologous to kis, highlight the importance of this protein in development and show that its product functions in a distinct manner to the BRM family of remodelers. Mutation of human CHD7 leads to CHARGE syndrome and other related developmental syndromes that impact approximately one in 10,000 live births (Vissers et al. 2004). Infants with CHARGE syndrome show serious defects in multiple tissues and organs, including the central nervous system, eye, ear, nose, heart, and genitalia (Jongmans et al. 2006). These phenotypes are due to altered gene expression in neural crest cells, where CHD7 and other human trxG proteins synergistically regulate gene-expression programs critical for development (Bajpai et al. 2010). Mutations in CHD7 also lead to a variety of other human developmental defects and more than 500 distinct mutations in CHD7 have been identified in human patients, emphasizing the importance of this trxG protein to human development and human pathologies (Janssen et al. 2012).
These dramatic developmental impacts are likely to be related to defects in the ability of CHD7 to remodel nucleosomes. The CHD7 protein is a potent ATP-dependent remodeler that does not require other proteins for function in vitro. As expected by its difference in domain structure from the BRM-related proteins, and its distinct impacts on gene expression, this protein shows several functional differences in remodeling behavior from the SWI/SNF subfamily of remodelers (Bouazoune and Kingston 2012). CHD7 requires excess DNA to remodel a nucleosome, whereas BRM family proteins do not. CHD7 is also less adept at opening sites internal to the nucleosome and “slides” nucleosomes in a different manner than BRM family proteins. Many of the mutations found in human patients with CHARGE syndrome truncate the CHD7 protein in a manner that is expected to impair or eliminate remodeling activity. Missense CHD7 mutations in patients were shown to affect remodeling activity, suggesting it is needed for appropriate human development. The comparison between CHD7 and SWI/SNF function shows that trxG phenotypes can be caused by mutations that affect two distinct classes of remodeling machinery operating through different mechanisms, implying that both of these remodeling functions are required for appropriate developmental regulation.
An important goal for future research is to unravel the role that ATP-dependent remodeling plays in the maintenance of the activated state. It is intriguing that four trxG members (BRM, MOR, OSA, and KIS) are known members of two distinct ATP-dependent chromatin-remodeling complexes, but that none of the other numerous ATP-dependent remodeling complexes have been identified in genetic screens for Drosophila trxG proteins. Two predominant hypotheses that could explain this, which are not mutually exclusive, are (1) the BRM and KIS chromatin-remodeling complexes are targeted to genes important for developmental progression, and (2) they have special remodeling characteristics that are uniquely required for maintenance. Although the BRM family of remodeling complexes shows distinct mechanism from other families, consistent with the second possibility, CHD7 and, presumably, KIS function similarly to remodeling proteins in the ISWI family, yet generate distinct phenotypes when mutated from those generated by ISWI mutations, which do not impact maintenance. Thus, it is possible that generic ATP-dependent remodeling is required for all active states, and maintenance of the active state of developmentally important genes happens to require these trxG members because they are targeted to these genes.
It is also intriguing to think about the mechanisms that remodelers might use to contribute to epigenetic regulation of the active state. At least three classes of mechanisms can be envisioned that might apply. First, remodeling functions might be required in a somewhat indirect manner to facilitate the binding (or rebinding following replication) of gene-specific activating proteins that are needed to maintain active transcription. In this case, the remodelers would not be the “brains” of the epigenetic mechanism, but instead would act as a necessary tool to allow the proteins required to function efficiently. Second, remodelers could work alone or with histone chaperones to evict nucleosomes from a region, and this lack of occupancy by nucleosomes would hypothetically cause the region to remain nonnucleosomal following replication. As mentioned above, the ability of the replication/nucleosome deposition machinery to accurately recapitulate nucleosome modification or location is an important unanswered issue in epigenetics (covered in more detail in Almouzni and Cedar 2014). Finally, remodeling machineries could reposition nucleosomes to create a structure that is amenable to activation. This latter mechanism has experimental support from studies of the albumin gene (Chaya et al. 2001; Cirillo et al. 2002). Several DNA-binding factors are required to maintain activity of this key gene in the liver. One of these factors, FoxA, binds directly to a nucleosome and the specific nucleosome-FoxA architecture is key to maintaining the active state of the albumin gene. Although it is not clear whether there is a required role for ATP-dependent remodeling to position this specific nucleosome in the liver, this example shows the potential for specific nucleosome positioning to play a key epigenetic role.
2.2. trxG Proteins That Covalently Modify Nucleosomal Histones
A second common method of regulating gene expression involves covalent PTM of the amino-terminal tails of the core histones that comprise the protein component of the nucleosome. These tails, which protrude from the surface of the nucleosome, can mediate interactions with other nucleosomes as well as with a wide variety of structural and regulatory proteins (illustrated in Fig. 12 of Allis et al. 2014). The covalent modification of histone tails by acetylation, methylation, or phosphorylation can help target regulatory complexes to chromatin and also directly change the ability of nucleosomes to compact into repressive structures by changing the charge on the tails. Covalent modification might also provide a mark to help maintain a specific regulated state, as covalently modified histones have the potential to divide to the two daughter strands and thereby propagate the information contained in the covalent mark to both mother and daughter cells following replication. The involvement of covalent modification in maintenance remains controversial, as the time frame for turnover of histones in vivo is under investigation and might be too fast for this type of mechanism to function, and the mechanisms that regulate transfer of histones to the daughter strand are also not understood (see also Almouzni and Cedar 2014).
Several trxG proteins are able to covalently modify histone tails, and these proteins are frequently found in complexes that are able to perform more than one type of modification reaction. For example, Drosophila TRX and its counterparts in other organisms methylate histone H3 at lysine 4 (H3K4); this covalent mark is tightly associated with active genes in a wide variety of organisms including yeast, flies, and humans (Fig. 7A). The yeast homolog of Drosophila TRX, Set1, is a subunit of large complex (COMPASS or Set1C) that methylates H3K4. COMPASS contains six other subunits, including the yeast counterpart of a second trxG protein, ASH2 (Miller et al. 2001; Roguev et al. 2001). Six KMTs related to Drosophila TRX are present in humans, including hSET1A, hSET1B, and the MLL1-4 proteins. hSET1A and hSET1B are the histone KMT subunits of human COMPASS, whereas MLL1-4 are subunits of related complexes (Shilatifard 2012). By promoting H3K4 trimethylation (H3K4me3) in the vicinity of promoters, COMPASS is thought to promote the transition of RNA Pol II to active elongation (Ardehali et al. 2011). H3K4 methylation has also been implicated in the maintenance of active gene expression in yeast, based on the timing of its appearance and removal at active genes (Santos-Rosa et al. 2002; Pokholok et al. 2005).
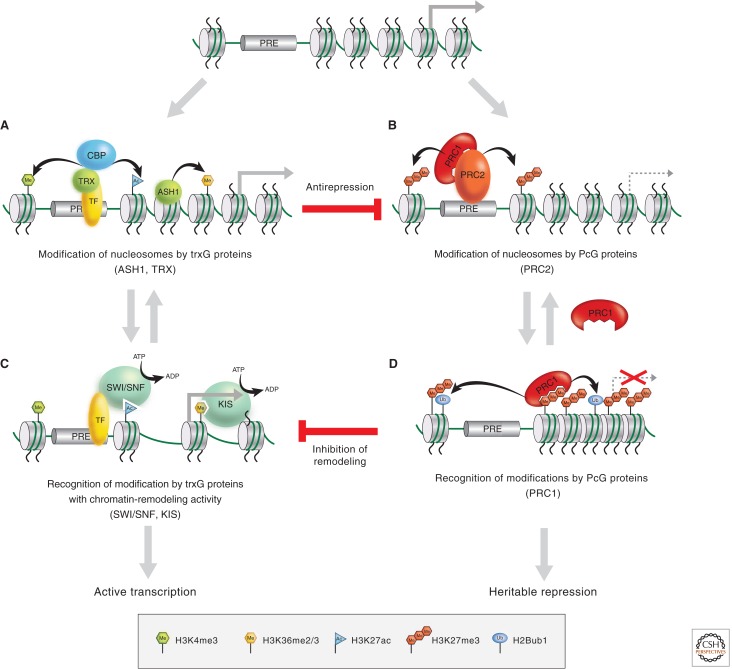
Figure 7.
trxG and PcG functions and interactions. Both trxG and PcG families include proteins that covalently modify histones and those that noncovalently modify chromatin. Covalent modifications on histones can promote or block the binding or activity of trxG complexes (e.g., SWI/SNF and KIS), PcG complexes (e.g., PRC1 and PRC2), or other factors involved in the maintenance of active or repressed states. Binding by these latter complexes has the potential to lead to further covalent modification, thus leading to iterative cycles of covalent modification and recognition of the covalent marks.
It has widely been assumed that TRX maintains active states of transcription by methylating H3K4 in the vicinity of promoters, based on the above observations. The dSet1 KMT (a subunit of Drosophila COMPASS) is responsible for the bulk of H3K4me3 in flies, however, and the loss of TRX function does not significantly alter H3K4me3 (Ardehali et al. 2011; Hallson et al. 2012). These findings suggest that the histone KMT activity of TRX is not as important for the maintenance of the active state as initially suspected.
The proteolytic cleavage of TRX leads to the production of two proteins: TRX-C and TRX-N. TRX-C, which retains the SET domain, is associated with PREs and promoters (Schuettengruber et al. 2009; Schwartz et al. 2010). TRX-N, which lacks histone KMT activity, is associated with broad regions of active genes (Schuettengruber et al. 2009; Schwartz et al. 2010), suggesting that it activates transcription via mechanisms other than histone methylation. There is some evidence that TRX/MLL complexes are also involved in the acetylation of histones. In flies, TRX is associated with dCBP, a histone lysine (K) acetyltransferase (frequently abbreviated to HAT or KAT) with broad specificity that is involved in activation (Petruk et al. 2001; Tie et al. 2009). Acetylation is also known to prevent the methylation of residues such as H3K9 and H3K27 that direct repression of the template, providing yet another mechanism for maintaining active states (Fig. 7A,B).
The trxG protein ASH1 also has histone KMT activity. ASH1 methylates H3K4, H3K9, H4K20, and H3K36 in vitro (Beisel et al. 2002; Byrd and Shearn 2003; Tanaka et al. 2007), but its H3K36 methyltransferase activity appears to be particularly important for the maintenance of active states (Yuan et al. 2011; Dorighi and Tamkun 2013). ASH1 colocalizes with TRX-N over broad domains of derepressed PcG targets (Schwartz et al. 2010), but it is not understood how the activities of ASH1 and TRX are coordinated. ASH1 has been seen to colocalize and associate with the CBP family of acetyltransferases (Bantignies et al. 2000), once again suggesting that methylation and acetylation go hand in hand.
A distinct type of modification, most normally found on the carboxy-terminal tail of histones H2A and H2B, is monoubiquitination. As opposed to the covalent modifications discussed above, ubiquitin is a large peptide that not only impacts charge but has the potential for significant steric effects. The OSA subunit of the PBAP family of complexes has been implicated in promoting ubiquitination of histone H2B on lysine 120 (Li et al. 2010), a modification associated with activated transcription. The mechanism via which H2B ubiquitination functions during transcriptional activation is not understood, although it appears to be an ancient function as this role is conserved from S. cerevisiae through humans.
There are numerous fascinating questions concerning how covalent modification of histones might contribute to trxG function. What functional role do the marks play? Covalent modification can contribute to epigenetic regulation via a wide spectrum of mechanisms. Methylation and acetylation marks might serve to directly alter chromatin compaction (sometimes termed cis-effects, as in Fig. 14 of Allis et al. 2014). The ability of chromatin to enter a compacted state, which is generally assumed to be repressive for transcription, is influenced by the charge distribution on the histone tails. Modifications that occur on lysine (e.g., acetylation) can eliminate the positive charge normally found with this residue, and therefore might directly decrease the ability of nucleosomes to form compacted structures, thus increasing the ability of the template to be transcribed (discussed in Marmorstein and Zhou 2014).
Histone PTMs have been proposed to create strong binding sites for complexes that direct transcriptional activation. These covalent modifications are able to create specific “knobs” on the surface of the nucleosome that fit into pockets on the complexes that promote activation, thus increasing binding energy and function of these complexes. For example, acetylation of histone tails increases binding by homologs of the BRM protein, thus promoting ATP-dependent remodeling of acetylated templates (Fig. 7C) (Hassan et al. 2001). This type of mechanism, frequently referred to as the “histone code” or trans-effects of covalent histone modifications, has the potential to be a central epigenetic function. Further studies are needed to determine which marks created by trxG proteins enhance binding of which complexes, and to determine the extent to which the energy of binding to a single modified residue can influence function and targeting. More research is also needed to determine the temporal order of addition of the marks and whether they are maintained across mitosis.
The flip side of this mechanism is that the marks could inhibit binding by repressive complexes. A covalent mark on a key residue required for optimal binding by a repressive complex could strongly inhibit binding by the repressive complex. For example, it is known that binding by repressive complexes is increased by methylation of histone H3 at K9 and K27 (Khorasanizadeh 2004). Acetylation of these residues would both block methylation and create an ill-shaped knob on the histone that impairs binding by the repressive complex. As discussed in Section 6, the ability of modifications to influence function of other complexes can cut in both directions, increasing the potency of this potential mode of epigenetic regulation.
These mechanisms by which PTMs achieve an effect are not mutually exclusive but, in fact, are likely to work together to help maintain an active state. Marks that chemically increase the ability to form a compacted state (a cis-effect) might also increase the ability of complexes to bind (a trans-effect), further promoting a compacted state. Conversely, marks that chemically decrease compaction might increase binding of complexes that also decompact nucleosomes. This mechanistically parsimonious use of covalent marks to alter several characteristics of chromatin structure and the ability of regulatory complexes to bind could create a powerful means of maintaining an active state.
3. CONNECTIONS BETWEEN trxG PROTEINS AND THE GENERAL TRANSCRIPTION MACHINERY
The theme that trxG proteins frequently are found in the same complex is continued with the skuld (skd) and kohtalo (kto) proteins. These two Drosophila proteins are homologs of the mammalian proteins identified biochemically as TRAP240 (Skuld) and TRAP230 (Kohtalo), which are both members of the Mediator complex (Janody et al. 2003). The Mediator complex is a large complex that functions at the interface between gene-specific activator proteins and formation of the preinitiation complex that contains RNA Pol II (Lewis and Reinberg 2003). Thus, these proteins are involved in general activation processes, much in the same way as SWI/SNF-family remodelers are involved in general activation. SKD and KTO might have some special function involved in maintenance, as other components of the Mediator complex were not identified in screens for trxG genes. The observation that SKD and KTO interact with each other, and skd kto double mutants have the same phenotype as either single mutant, has led to the hypothesis that the two proteins together form a functional module that somehow alters Mediator action (Janody et al. 2003).
4. CONNECTIONS BETWEEN trxG PROTEINS AND COHESIN
trxG proteins can influence levels of chromatin organization above the level of the nucleosome, as evidenced by the discovery that the Drosophila trxG gene, verthandi (vtd), encodes the Rad21 subunit of cohesin (Hallson et al. 2008). The four core subunits of cohesin, Rad21 (Scc1/Mcd1), Smc1, Smc3, and stromalin (SA/Scc3/Stag2), form a ring that encircles DNA. Cohesin is essential for sister chromatid cohesion during mitosis and meiosis (Haering et al. 2008; Nasmyth and Haering 2009). Cohesin also plays important roles in transcriptional activation and repression in interphase cells by promoting interactions between insulators, enhancers, and promoters via the formation of DNA loops (illustrated in Fig. 1 of Kim et al. 2014; also Wood et al. 2010; Dorsett 2011; Fay et al. 2011; Seitan and Merkenschlager 2012). These and other findings raise the intriguing possibility that the stabilization of long-range chromosomal interactions by cohesin contributes to the maintenance of the active state (Cunningham et al. 2012; Schaaf et al. 2013).
5. BIOCHEMICAL FUNCTIONS OF OTHER trxG PROTEINS
The biochemical activities of the majority of other trxG proteins remain relatively mysterious. Brd2/Ring3, a human counterpart of the Drosophila trxG gene fsh, encodes a nuclear protein kinase with two bromodomains, implicated in cell-cycle progression and leukemogenesis, but the substrates of the kinase are currently unknown (Denis and Green 1996). The trxG gene Tonalli (Tna) encodes a protein related to SP-RING finger proteins involved in sumoylation, suggesting that it may regulate transcription via the covalent modification of proteins other than histones (Gutierrez et al. 2003; Monribot-Villanueva et al. 2013). The trxG gene sallimus (sls) was identified in a screen for extragenic suppressors of Pc and subsequently found to encode Drosophila Titin (Machado and Andrew 2000). Like its vertebrate counterpart, Drosophila Titin helps maintain the integrity and elasticity of the muscle sarcomere. In addition, Titin is a chromosomal protein that is required for chromosome condensation and segregation (Machado and Andrew 2000). These findings provide additional evidence of potential roles for trxG proteins in the regulation of higher-order chromatin structure.
6. FUNCTIONAL INTERACTIONS BETWEEN trxG AND PcG PROTEINS
As the basic biochemical activities of many trxG and PcG members have been identified, attention has shifted toward looking at how their activities are coordinated in regulating transcription and maintaining the active state. In spite of the lack of in vitro systems for studying the maintenance of the determined state, good progress has been made toward addressing this issue. One popular hypothesis is that the trxG and PcG members facilitate a sequence of dependent events required for the maintenance of the active or repressed state.
Support for this idea has come from studies of the PcG complexes PRC1 and PRC2; by methylating H3K27, the E(z) histone KMT subunit of PRC2 creates a covalent mark that is directly recognized by the chromodomain of the Pc subunit of PRC1 (Jacobs and Khorasanizadeh 2002; Min et al. 2003). Thus, one PcG complex appears to directly promote the binding of another PcG complex to chromatin. This simple model has been complicated by recent studies showing that not all complexes in the PRC1 family contain Pc and that methylation of H3K27, although involved in targeting PRC1, is neither necessary nor sufficient for targeting (Margueron and Reinberg 2011; Tavares et al. 2012). The basic concept that one complex can help target the function of another complex nonetheless remains an important paradigm.
By analogy with the PcG complex paradigm described in the previous paragraph, it is possible that trxG-mediated covalent modification of nucleosomes by members with histone KMT or KAT activities (e.g., TRX or ASH1) could directly regulate the activities of trxG members involved in ATP-dependent chromatin remodeling (e.g., the BRM or KIS proteins) (Fig. 7). Consistent with this possibility, BRM and other subunits of SWI/SNF complexes contain bromodomains that can directly interact with acetylated histone tails. KIS and its human homolog CHD7 contain chromodomains that may directly interact with methylated histone tails. Indeed global ChIP analyses have detected colocalization of CHD7 with methylated H3K4, leading to the possibility that there could be a direct interaction between CHD7 and this mark (Schnetz et al. 2009, 2010). This model, which is supported by studies of chromatin-remodeling factors in both yeast and mammals (Agalioti et al. 2000; Hassan et al. 2001), is particularly attractive because it provides a mechanism by which a heritable histone modification could perpetuate a constitutively “open” chromatin configuration that is permissive for active transcription.
Another important issue concerns the functional relationship between PcG repressors and trxG activators. Do these regulatory proteins have independent roles in activation and repression or do they act in direct opposition to maintain the heritable state? One interesting property of PcG proteins is that they are capable of repressing transcription when tethered near virtually any gene transcribed by RNA Pol II. trxG members that play global roles in transcription, including BRM, KIS, and other trxG members involved in chromatin remodeling, are thus excellent candidates for direct targets of PcG repressors (Fig. 7). PRC1 does apparently block access of the SWI/SNF-family complex to the chromatin template (Francis et al. 2001). This is consistent with the notion that a mechanism to achieve PcG repression is to prevent ATP-dependent remodeling by trxG members. The Brahma complex and PRC1 are further connected by the fact that both directly interact with the Zeste protein, which plays a complicated role in the regulation of gene expression in Drosophila by somehow helping to direct cross-talk between the two complexes.
A second protein that connects PcG proteins and trxG proteins is the GAGA factor, which is encoded by the Trithorax-like (Trl) gene and is thus a trxG member (Farkas et al. 1994). This protein can function as a sequence-specific activator protein at some promoters, but also is a prominent member of the proteins that bind to the PRE. PRE sequences direct PcG function, and at least one PRE can act as a memory module when affixed to a reporter construct, emphasizing the importance of these sequences. PRE sequences bind the GAGA factor and play an important role in PRE function; they tether the GAGA protein to DNA, which has been proposed to enhance binding and function of the PRC1 complex (Mahmoudi and Verrijzer 2001). Thus, the GAGA factor may play key roles in the maintenance of activation (via its transcriptional activating properties) and repression (via interactions with the PcG proteins). An important issue for future research is to understand the rationale for why proteins such as GAGA and Zeste appear to interact with both the activating and repressing machineries of maintenance.
Genetic studies have shown that removal of PcG complexes will reactivate genes even in the absence of TRX and ASH1 (Klymenko and Muller 2004), suggesting that some trxG proteins might use their activity to function as PcG antirepressors, as opposed to direct activators (Fig. 7). How do TRX and ASH1 then counteract PcG repression? trxG proteins may directly antagonize PcG repression by interfering with the binding of PRC2 to nucleosomes via methylated H3K27, inhibiting its histone KMT activity, directly blocking H3K27 methylation, or removing this repressive mark. Recent studies have shown that trxG proteins use each of these mechanisms to antagonize PcG repression. For example, a complex containing TRX and the histone acetyltransferase CBP acetylates H3K27, which prevents H3K27 methylation by PRC2 (Fig. 7) (Tie et al. 2009). H3K4me3, another histone modification catalyzed by trxG proteins, blocks the interaction of PRC2 with its nucleosome substrate (Schmitges et al. 2011). Finally, the methylation of H3K36, a modification catalyzed by the trxG protein ASH1, inhibits the H3K27 methyltransferase activity of PRC2 (Tanaka et al. 2007; Schmitges et al. 2011; Yuan et al. 2011). These findings explain why repressive H3K27 methylation does not spread over actively transcribed genes and provides a satisfying molecular explanation for the functional antagonism between PcG and trxG proteins that has fascinated geneticists for decades.
7. NONCODING RNAs AND THE trxG
Recent studies suggest that long noncoding RNAs (lncRNAs) might be involved in modulating trxG targeting and function. This proposed mechanism would function in addition to targeting mechanisms that use covalently modified histones and those described in the voluminous literature on sequence-specific DNA-binding factors targeting different members of the trxG family, such as Mediator and SWI/SNF. The use of lncRNAs in targeting factors involved in maintenance is attractive because these RNAs would be expected to segregate to daughter cells and thus help faithfully maintain a regulated state.
Examples of lncRNAs that have been proposed to modulate trxG function include HOTTIP and DBE-T. HOTTIP is expressed from the HOXA cluster and has been proposed to regulate methylation of H3K4 via the interaction with a component (WDR5) of several MLL complexes (Wang et al. 2011). DBE-T has been proposed to directly target Ash1L to the facioscapulohumeral dystrophy (FSHD) locus to guide methylation of H3K36 and thereby assist in derepressing that locus in muscular dystrophy (Cabianca et al. 2012). Interesting areas for future work on these and other candidate lncRNAs include the identification and characterization of the domains that they bind to in trxG complexes, how they function to modulate the activity of trxG complexes, and whether they are able to target complexes in trans via direct interaction with targeting sequences in the DNA.
8. trxG PROTEINS AND HUMAN DISEASE
Cancer and other human diseases can result from the failure to maintain heritable states of gene expression. Not surprisingly, many human PcG and trxG genes function as proto-oncogenes or tumor-suppressor genes. For example, the human trxG gene MLL, was originally identified by 11q23 chromosome translocations associated with acute lymphoblastic or myeloid leukemia. Mutations in other mammalian trxG genes are also associated with a variety of cancers (Roberts and Orkin 2004; see Beaudet and Zoghbi 2014 for more detail). For example, BRG1, the human counterpart of Drosophila brm, physically interacts with the retinoblastoma tumor-suppressor protein; disruption of this interaction leads to increased cell division and malignant transformation in certain human tumor cell lines (Dunaief et al. 1994; Strober et al. 1996). Consistent with a role of BRG1 in tumor suppression, mice heterozygous for mutations in this gene are prone to develop a variety of tumors (Bultman et al. 2000). Mutations in INI1, the human counterpart of the Drosophila trxG gene SNF5-related gene 1 (SNR1), also predispose individuals to cancers and have been identified in a large percentage of malignant rhabdoid tumors, an aggressive cancer of children (Versteege et al. 1998; Wilson et al. 2010). Drugs that alter the expression or function of trxG proteins might therefore prove to be useful therapeutic agents in treating human cancer.
9. CONCLUSION AND OUTLOOK
Two of the major questions surrounding the function of trxG proteins remain a matter for conjecture. First, what is the rationale for why a relatively small subset of proteins required for transcriptional activation score genetically as being important for maintenance of the active state? Is this because these proteins play global roles in transcription but are expressed in limiting quantities, or happen, by evolutionary serendipity, to be especially important for developmentally important genes? Second, how can the active state be maintained across replication and mitosis? Replication will create two daughter strands that must both be regulated identically, and mitosis requires condensation and thereby inhibition of transcription of most genes in a cell. What mechanisms create the epigenetic mark(s) that ensure reactivation of a gene on both daughter strands following mitosis?
The majority of trxG proteins are part of complexes that are broadly used in gene expression and most of these complexes also contain many other proteins not in the trxG (see Table 1). This raises the important question as to whether there are special functions that are used for maintenance of active gene expression. It is possible that SWI/SNF remodelers are able to perform a special remodeling function, H3K4 methylation targets special complexes and/or chromatin conformations, and Skuld/Kohtalo alter function of Mediator in a specific manner important for maintenance. Alternatively, it is possible that each of these proteins performs a reaction that is normally used in activation of all types of genes, and these complexes are among those that have emerged as being important for maintenance for a relatively uninteresting reason (e.g., because even relatively subtle changes in the expression of Drosophila Hox genes cause homeotic transformations). To resolve these issues, considerably more information is needed about the precise mechanisms that each of these proteins use in activation. For example, do the SWI/SNF complexes harness the energy of ATP hydrolysis in the same manner as other ATP-dependent remodeling complexes, or do they differ in an important way in how this energy is used to alter nucleosome structure? Structural techniques including crystallography, biophysical techniques such as single-molecule analysis and FRET (fluorescence resonance energy transfer), and detailed imaging in vivo might help to shed light on whether there are mechanisms specially designed for epigenetic maintenance of activation. The initial functional studies that have been performed with trxG complexes on simple model templates are just the beginning of the process for answering these important questions.
The epigenetic mechanisms that might maintain an active state are even less well understood. Are covalent marks distributed to help create an active mark? Are nucleosome positions maintained following replication to create open stretches of chromatin, or specially positioned nucleosomes, that increase binding of activators? Does trxG function cause active genes to compartmentalize within the nucleus to regions that favor active transcription? These are all viable hypotheses; more hypotheses exist, and others have not yet even been envisioned. The incredible complexity of the machinery that transcribes DNA offers numerous possibilities for regulation and the development of mechanisms that allow an epigenetic maintenance of active transcription. This intersection of two fields rich in intellectual history, transcriptional activation and epigenetic mechanism, will provide fertile ground for experimentalists for many years.
Footnotes
Editors: C. David Allis, Marie-Laure Caparros, Thomas Jenuwein, and Danny Reinberg
Additional Perspectives on Epigenetics available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell 103: 667–678 [DOI] [PubMed] [Google Scholar]
- *.Allis CD, Jenuwein T, Reinberg D 2014. Overview and concepts. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018739 [DOI] [Google Scholar]
- *.Almouzni G, Cedar H 2014. Maintenance of epigenetic information. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardehali MB, Mei A, Zobeck KL, Caron M, Lis JT, Kusch T 2011. Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription. EMBO J 30: 2817–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong JA, Papoulas O, Daubresse G, Sperling AS, Lis JT, Scott MP, Tamkun JW 2002. The Drosophila BRM complex facilitates global transcription by RNA polymerase II. EMBO J 21: 5245–5254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajpai R, Chen DA, Rada-Iglesias A, Zhang J, Xiong Y, Helms J, Chang CP, Zhao Y, Swigut T, Wysocka J 2010. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature 463: 958–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantignies F, Goodman RH, Smolik SM 2000. Functional interaction between the coactivator Drosophila CREB-binding protein and ASH1, a member of the trithorax group of chromatin modifiers. Mol Cell Biol 20: 9317–9330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Beaudet A, Zoghbi H 2014. Epigenetics and human disease. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Becker PB, Workman JL 2013. Nucleosome remodeling and epigenetics. Cold Spring Harb Perspect Biol 5: a017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel C, Imhof A, Greene J, Kremmer E, Sauer F 2002. Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature 419: 857–862 [DOI] [PubMed] [Google Scholar]
- Bouazoune K, Kingston RE 2012. Chromatin remodeling by the CHD7 protein is impaired by mutations that cause human developmental disorders. Proc Natl Acad Sci 109: 19238–19243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, et al. 2000. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell 6: 1287–1295 [DOI] [PubMed] [Google Scholar]
- Byrd KN, Shearn A 2003. ASH1, a Drosophila trithorax group protein, is required for methylation of lysine 4 residues on histone H3. Proc Natl Acad Sci 100: 11535–11540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabianca DS, Casa V, Bodega B, Xynos A, Ginelli E, Tanaka Y, Gabellini D 2012. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell 149: 819–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalkley GE, Moshkin YM, Langenberg K, Bezstarosti K, Blastyak A, Gyurkovics H, Demmers JA, Verrijzer CP 2008. The transcriptional coactivator SAYP is a trithorax group signature subunit of the PBAP chromatin remodeling complex. Mol Cell Biol 28: 2920–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaya D, Hayamizu T, Bustin M, Zaret KS 2001. Transcription factor FoxA (HNF3) on a nucleosome at an enhancer complex in liver chromatin. J Biol Chem 276: 44385–44389 [DOI] [PubMed] [Google Scholar]
- *.Cheng X 2014. Structural and functional coordination of DNA and histone methylation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS 2002. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell 9: 279–289 [DOI] [PubMed] [Google Scholar]
- Cote J, Quinn J, Workman JL, Peterson CL 1994. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 265: 53–60 [DOI] [PubMed] [Google Scholar]
- Cunningham MD, Gause M, Cheng Y, Noyes A, Dorsett D, Kennison JA, Kassis JA 2012. Wapl antagonizes cohesin binding and promotes Polycomb-group silencing in Drosophila. Development 139: 4172–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubresse G, Deuring R, Moore L, Papoulas O, Zakrajsek I, Waldrip WR, Scott MP, Kennison JA, Tamkun JW 1999. The Drosophila kismet gene is related to chromatin-remodeling factors and is required for both segmentation and segment identity. Development 126: 1175–1187 [DOI] [PubMed] [Google Scholar]
- Denis GV, Green MR 1996. A novel, mitogen-activated nuclear kinase is related to a Drosophila developmental regulator. Genes Dev 10: 261–271 [DOI] [PubMed] [Google Scholar]
- Dorighi KM, Tamkun JW 2013. The trithorax group proteins Kismet and ASH1 promote H3K36 dimethylation to counteract Polycomb group repression in Drosophila. Development 140: 4182–4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D 2011. Cohesin: Genomic insights into controlling gene transcription and development. Curr Opin Genet Dev 21: 199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunaief JL, Strober BE, Guha S, Khavari PA, Alin K, Luban J, Begemann M, Crabtree GR, Goff SP 1994. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 79: 119–130 [DOI] [PubMed] [Google Scholar]
- Duncan I 1987. The bithorax complex. Annu Rev Genet 21: 285–319 [DOI] [PubMed] [Google Scholar]
- *.Elgin SCR, Reuter G 2013. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb Perspect Biol 5: a017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas G, Gausz J, Galloni M, Reuter G, Gyurkovics H, Karch F 1994. The Trithorax-like gene encodes the Drosophila GAGA factor. Nature 371: 806–808 [DOI] [PubMed] [Google Scholar]
- Fay A, Misulovin Z, Li J, Schaaf CA, Gause M, Gilmour DS, Dorsett D 2011. Cohesin selectively binds and regulates genes with paused RNA polymerase. Curr Biol 21: 1624–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE 2001. Mechanisms of transcriptional memory. Nat Rev Mol Cell Biol 2: 409–421 [DOI] [PubMed] [Google Scholar]
- Francis NJ, Saurin AJ, Shao Z, Kingston RE 2001. Reconstitution of a functional core polycomb repressive complex. Mol Cell 8: 545–556 [DOI] [PubMed] [Google Scholar]
- Gellon G, McGinnis W 1998. Shaping animal body plans in development and evolution by modulation of Hox expression patterns. Bioessays 20: 116–125 [DOI] [PubMed] [Google Scholar]
- *.Grossniklaus U, Paro R 2014. Transcriptional silencing by Polycomb-group proteins. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L, Zurita M, Kennison JA, Vazquez M 2003. The Drosophila trithorax group gene tonalli (tna) interacts genetically with the Brahma remodeling complex and encodes an SP-RING finger protein. Development 130: 343–354 [DOI] [PubMed] [Google Scholar]
- Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K 2008. The cohesin ring concatenates sister DNA molecules. Nature 454: 297–301 [DOI] [PubMed] [Google Scholar]
- Hallson G, Syrzycka M, Beck SA, Kennison JA, Dorsett D, Page SL, Hunter SM, Keall R, Warren WD, Brock HW, et al. 2008. The Drosophila cohesin subunit Rad21 is a trithorax group (trxG) protein. Proc Natl Acad Sci 105: 12405–12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallson G, Hollebakken RE, Li T, Syrzycka M, Kim I, Cotsworth S, Fitzpatrick KA, Sinclair DA, Honda BM 2012. dSet1 is the main H3K4 di- and tri-methyltransferase throughout Drosophila development. Genetics 190: 91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan AH, Neely KE, Workman JL 2001. Histone acetyltransferase complexes stabilize SWI/SNF binding to promoter nucleosomes. Cell 104: 817–827 [DOI] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728 [DOI] [PubMed] [Google Scholar]
- Imbalzano AN, Kwon H, Green MR, Kingston RE 1994. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature 370: 481–485 [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Khorasanizadeh S 2002. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295: 2080–2083 [DOI] [PubMed] [Google Scholar]
- Janody F, Martirosyan Z, Benlali A, Treisman JE 2003. Two subunits of the Drosophila mediator complex act together to control cell affinity. Development 130: 3691–3701 [DOI] [PubMed] [Google Scholar]
- Janssen N, Bergman JE, Swertz MA, Tranebjaerg L, Lodahl M, Schoots J, Hofstra RM, van Ravenswaaij-Arts CM, Hoefsloot LH 2012. Mutation update on the CHD7 gene involved in CHARGE syndrome. Hum Mutat 33: 1149–1160 [DOI] [PubMed] [Google Scholar]
- Jongmans MC, Admiraal RJ, van der Donk KP, Vissers LE, Baas AF, Kapusta L, van Hagen JM, Donnai D, de Ravel TJ, Veltman JA, et al. 2006. CHARGE syndrome: The phenotypic spectrum of mutations in the CHD7 gene. J Med Genet 43: 306–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman TC, Seeger MA, Olsen G 1990. Molecular and genetic organization of the antennapedia gene complex of Drosophila melanogaster. Adv Genet 27: 309–362 [DOI] [PubMed] [Google Scholar]
- Kennison JA 1995. The Polycomb and trithorax group proteins of Drosophila: Trans-regulators of homeotic gene function. Annu Rev Genet 29: 289–303 [DOI] [PubMed] [Google Scholar]
- Kennison JA 2003. Introduction to Trx-G and Pc-G genes. Meth Enzymol 377: 61–70 [DOI] [PubMed] [Google Scholar]
- Kennison JA, Tamkun JW 1988. Dosage-dependent modifiers of Polycomb and Antennapedia mutations in Drosophila. Proc Natl Acad Sci 85: 8136–8140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khorasanizadeh S 2004. The nucleosome: From genomic organization to genomic regulation. Cell 116: 259–272 [DOI] [PubMed] [Google Scholar]
- *.Kim T-K, Hemberg M, Gray JM 2014. Enhancer RNAs: A class of long noncoding RNAs synthesized at enhancers. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymenko T, Muller J 2004. The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep 5: 373–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger W, Peterson CL, Sil A, Coburn C, Arents G, Moudrianakis EN, Herskowitz I 1995. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev 9: 2770–2779 [DOI] [PubMed] [Google Scholar]
- Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR 1994. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature 370: 477–481 [DOI] [PubMed] [Google Scholar]
- Levine SS, King IF, Kingston RE 2004. Division of labor in Polycomb group repression. Trends Biochem Sci 29: 478–485 [DOI] [PubMed] [Google Scholar]
- Lewis BA, Reinberg D 2003. The mediator coactivator complex: Functional and physical roles in transcriptional regulation. J Cell Sci 116: 3667–3675 [DOI] [PubMed] [Google Scholar]
- Li XS, Trojer P, Matsumura T, Treisman JE, Tanese N 2010. Mammalian SWI/SNF—A subunit BAF250/ARID1 is an E3 ubiquitin ligase that targets histone H2B. Mol Cell Biol 30: 1673–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado C, Andrew DJ 2000. D-Titin: A giant protein with dual roles in chromosomes and muscles. J Cell Biol 151: 639–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi T, Verrijzer CP 2001. Chromatin silencing and activation by Polycomb and trithorax group proteins. Oncogene 20: 3055–3066 [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D 2011. The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Marmorstein R, Zhou M-M 2014. Writers and readers of histone acetylation: Structure, mechanism and inhibition. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A 2001. COMPASS: A complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci 98: 12902–12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Zhang Y, Xu RM 2003. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev 17: 1823–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrmann L, Verrijzer CP 2005. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim Biophys Acta 1681: 59–73 [DOI] [PubMed] [Google Scholar]
- Monribot-Villanueva J, Juarez-Uribe RA, Palomera-Sanchez Z, Gutierrez-Aguiar L, Zurita M, Kennison JA, Vazquez M 2013. TnaA, an SP-RING protein, interacts with Osa, a subunit of the chromatin remodeling complex BRAHMA and with the SUMOylation pathway in Drosophila melanogaster. PloS One 8: e62251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyrers-Chen I, Rozovskaia T, Lee N, Kersey JH, Nakamura T, Canaani E, Paro R 2004. Expression of leukemic MLL fusion proteins in Drosophila affects cell cycle control and chromosome morphology. Oncogene 23: 8639–8648 [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH 2009. Cohesin: Its roles and mechanisms. Annu Rev Genet 43: 525–558 [DOI] [PubMed] [Google Scholar]
- *.Patel DJ 2014. A structural perspective on readout of epigenetic histone DNA methylation marks. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Smith S, Tillib S, Kraevski V, Nakamura T, Canaani E, Croce CM, Mazo A 2001. Trithorax and dCBP acting in a complex to maintain expression of a homeotic gene. Science 294: 1331–1334 [DOI] [PubMed] [Google Scholar]
- Phelan ML, Sif S, Narlikar GJ, Kingston RE 1999. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell 3: 247–253 [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122: 517–527 [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406: 593–599 [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R 2004. Epigenetic regulation of cellular memory by the Polycomb and trithorax group proteins. Annu Rev Genet 38: 413–443 [DOI] [PubMed] [Google Scholar]
- Roberts CW, Orkin SH 2004. The SWI/SNF complex—Chromatin and cancer. Nat Rev Cancer 4: 133–142 [DOI] [PubMed] [Google Scholar]
- Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF 2001. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J 20: 7137–7148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419: 407–411 [DOI] [PubMed] [Google Scholar]
- Schaaf CA, Misulovin Z, Gause M, Koenig A, Gohara DW, Watson A, Dorsett D 2013. Cohesin and polycomb proteins functionally interact to control transcription at silenced and active genes. PLoS Genet 9: e1003560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, et al. 2011. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 42: 330–341 [DOI] [PubMed] [Google Scholar]
- Schnetz MP, Bartels CF, Shastri K, Balasubramanian D, Zentner GE, Balaji R, Zhang X, Song L, Wang Z, Laframboise T, et al. 2009. Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res 19: 590–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnetz MP, Handoko L, Akhtar-Zaidi B, Bartels CF, Pereira CF, Fisher AG, Adams DJ, Flicek P, Crawford GE, Laframboise T, et al. 2010. CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet 6: e1001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Ganapathi M, Leblanc B, Portoso M, Jaschek R, Tolhuis B, van Lohuizen M, Tanay A, Cavalli G 2009. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol 7: e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Stenberg P, Ohno K, Bourgon R, Pirrotta V 2010. Alternative epigenetic chromatin states of Polycomb target genes. PLoS Genet 6: e1000805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitan VC, Merkenschlager M 2012. Cohesin and chromatin organisation. Curr Opin Genet Dev 22: 93–100 [DOI] [PubMed] [Google Scholar]
- Shilatifard A 2012. The COMPASS family of histone H3K4 methylases: Mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem 81: 65–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J 1995. Locking in stable states of gene expression: Transcriptional control during Drosophila development. Curr Opin Cell Biol 7: 376–385 [DOI] [PubMed] [Google Scholar]
- Simon JA, Tamkun JW 2002. Programming off and on states in chromatin: Mechanisms of Polycomb and trithorax group complexes. Curr Opin Genet Dev 12: 210–218 [DOI] [PubMed] [Google Scholar]
- Srinivasan S, Armstrong JA, Deuring R, Dahlsveen IK, McNeill H, Tamkun JW 2005. The Drosophila trithorax group protein Kismet facilitates an early step in transcriptional elongation by RNA Polymerase II. Development 132: 1623–1635 [DOI] [PubMed] [Google Scholar]
- Srinivasan S, Dorighi KM, Tamkun JW 2008. Drosophila Kismet regulates histone H3 lysine 27 methylation and early elongation by RNA polymerase II. PLoS Genet 4: e1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strober BE, Dunaief JL, Guha, Goff SP 1996. Functional interactions between the hBRM/hBRG1 transcriptional activators and the pRB family of proteins. Mol Cell Biol 16: 1576–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsanam P, Iyer VR, Brown PO, Winston F 2000. Whole-genome expression analysis of SNF/SWI mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci 97: 3364–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkun JW, Deuring R, Scott MP, Kissinger M, Pattatucci AM, Kaufman TC, Kennison JA 1992. brahma: A regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell 68: 561–572 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Katagiri Z, Kawahashi K, Kioussis D, Kitajima S 2007. Trithorax-group protein ASH1 methylates histone H3 lysine 36. Gene 397: 161–168 [DOI] [PubMed] [Google Scholar]
- Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, et al. 2012. RYBP-PRC1 complexes mediate H2A ubiquitylation at Polycomb target sites independently of PRC2 and H3K27me3. Cell 148: 664–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therrien M, Morrison DK, Wong AM, Rubin GM 2000. A genetic screen for modifiers of a kinase suppressor of Ras-dependent rough eye phenotype in Drosophila. Genetics 156: 1231–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ 2009. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 136: 3131–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, Aurias A, Delattre O 1998. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394: 203–206 [DOI] [PubMed] [Google Scholar]
- Vissers LELM, van Ravenswaaij CMA, Admiraal R, Hurst JA, de Vries BBA, Janssen IM, van der Vliet WA, Huys EHLPG, de Jong PJ, Hamel BC, et al. 2004. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet 36: 955–957 [DOI] [PubMed] [Google Scholar]
- Wang W, Cote J, Xue Y, Zhou S, Khavari PA, Biggar SR, Muchardt C, Kalpana GV, Goff SP, Yaniv M, et al. 1996. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J 15: 5370–5382 [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. 2011. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472: 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BG, Wang X, Shen X, McKenna ES, Lemieux ME, Cho YJ, Koellhoffer EC, Pomeroy SL, Orkin SH, Roberts CW 2010. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell 18: 316–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, Severson AF, Meyer BJ 2010. Condensin and cohesin complexity: The expanding repertoire of functions. Nat Rev Genet 11: 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ 1995. Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378: 505–508 [DOI] [PubMed] [Google Scholar]
- Yu BD, Hanson RD, Hess JL, Horning SE, Korsmeyer SJ 1998. MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc Natl Acad Sci 95: 10632–10636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B 2011. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem 286: 7983–7989 [DOI] [PMC free article] [PubMed] [Google Scholar]