Abstract
MicroRNAs (miRNAs) are short non-coding RNAs that regulate gene output at the post-transcriptional level by targeting degenerate elements primarily in 3′untranslated regions (3′UTRs) of mRNAs. Individual miRNAs can regulate networks of hundreds of genes, yet for the majority of miRNAs few, if any, targets are known. Misexpression of miRNAs is also a major contributor to cancer progression, thus there is a critical need to validate miRNA targets in high-throughput to understand miRNAs' contribution to tumorigenesis. Here we introduce a novel high-throughput assay to detect miRNA targets in 3′UTRs, called Luminescent Identification of Functional Elements in 3′UTRs (3′LIFE). We demonstrate the feasibility of 3′LIFE using a data set of 275 human 3′UTRs and two cancer-relevant miRNAs, let-7c and miR-10b, and compare our results to alternative methods to detect miRNA targets throughout the genome. We identify a large number of novel gene targets for these miRNAs, with only 32% of hits being bioinformatically predicted and 27% directed by non-canonical interactions. Functional analysis of target genes reveals consistent roles for each miRNA as either a tumor suppressor (let-7c) or oncogenic miRNA (miR-10b), and preferentially target multiple genes within regulatory networks, suggesting 3′LIFE is a rapid and sensitive method to detect miRNA targets in high-throughput.
INTRODUCTION
MicroRNAs (miRNAs) are short non-coding RNAs that bind to sequence elements primarily in the 3′untranslated regions (3′UTRs) of messenger RNAs (mRNAs), resulting in mRNA degradation or translational repression (1). MiRNAs are expressed in nearly every eukaryotic cell type investigated to date, where they regulate fundamental biological processes such as development and morphogenesis (2). Furthermore, miRNA misregulation has also been observed in a wide range of cancers (3), where they can function as tumor suppressors or oncogenic miRNAs (oncomiRs) (4), based solely on the function of target genes. The misexpression of miRNAs can have substantial consequences in development and disease, highlighting the powerful role miRNAs play in influencing cell behavior. In human, each miRNA is predicted to target hundreds to thousands of genes based on complementarity with mRNAs (5), thus identifying and experimentally validating gene targets is central to understanding an miRNA function. To analyze the large number of putative miRNA/mRNA interactions, bioinformatic and next-gen sequencing approaches are commonly used to predict and identify miRNA target sites throughout the genome. Each approach has inherent strengths and shortfalls.
Many algorithms have been developed to predict miRNA target sites throughout the transcriptome by searching for regions of complementarity with mRNAs by Watson–Crick and G:U wobble base pairing (1). These interactions may utilize as little as six consecutive nucleotides near the 5′end, or seed region, of the mature miRNA (5–7). Perfect complementarity with the seed region is considered the canonical indicator of an miRNA target site, but can be strengthened by additional pairing with the 3′end of the miRNA. Due to the small size and degenerate nature of target sequence elements, most algorithms predict hundreds to thousands of putative target genes for each miRNA. These algorithms typically apply parameters such as stringent seed pairing, cross-species conservation and thermodynamic stability to identify high-confidence targets (5,8). However, the analysis of the predictive performance of several of the most prominent algorithms reports extremely high false-negative (9–11) (Supplementary Figure S1) and false-positive rates (∼66%) (12,13). These error rates reflect that these algorithms typically do not predict poorly conserved species-specific interactions or non-canonical targets not reliant on perfect seed complementarity, which recent evidence suggests may be widespread (11–12,14–20). These algorithms are useful for candidate gene approaches to identify putative miRNA targets, but the high error rates make systematic target detection challenging.
Several recently developed techniques allow the experimental detection of miRNA targets on a transcriptome wide level, bypassing target prediction software. One of the most used methods combines the cross-link and immunoprecipitation of argonaute (AGO) family members with next-generation sequencing (AGO-HITS-CLIP). AGO proteins directly bind mature miRNAs and guide the large multi-protein complex called the RNA-induced silencing complex (RISC) to target mRNAs. By sequencing the mRNA population bound to AGO, it is possible to map the sequence footprints occupied by the RISC on specific genes (21). However, the physical immunoprecipitation of AGO from cell lysates is often challenging to execute and may add unwanted noise, impacting reproducibility and complicating downstream analysis. Importantly, while this approach produces hundreds to thousands of targets, it does not provide information about which miRNA led the RISC to that location, which has to be extracted using bioinformatic approaches (22). Furthermore, AGO-HITS-CLIP fails to measure the functional impact of miRNA targeting on the level of protein synthesis. The result of targeting by the RISC can range from translational repression to mRNA degradation (13), and it is not currently clear if these alternative mechanisms bias the results of AGO-HITS-CLIP toward target sites that are held in translational repression. While AGO-HITS-CLIP is an excellent method to detect miRNA target sites in high-throughput in a specific biological context, the limitations with assigning an miRNA and the lack of a functional measure of targeting limit this approach.
To address these gaps, we have developed a high-throughput assay to probe for interactions between miRNAs and their targets in 3′UTRs. This assay, named Luminescent Identification of Functional Elements in 3′UTRs (3′LIFE), is capable of measuring direct interactions between a test 3′UTR and a query miRNA using a dual luciferase approach. Luciferase assays rely on the fusion of the 3′UTR of a gene of interest to a luciferase reporter gene. The reporter is cotransfected with a query miRNA in cell culture. Targeting is measured as a relative change between the test 3′UTR reporter and a second, non-targeted luciferase reporter. Luciferase assays offer several advantages over other methods to identify miRNA targets such as mRNA sequencing and protein quantification approaches. Quantification of the luciferase assay occurs at the protein level, but reflects interactions at the RNA level between the miRNA and 3'UTR, bypassing differences in mRNA degradation/translational repression, and changes in protein abundance independent of 3'UTR based regulation. Luciferase assays are sensitive and scalable, yet use in high-throughput screens is greatly limited by (i) high costs associated with consumable reagents, (ii) lack of publically available 3′UTR reporter libraries and (iii) the absence of standardized dual luciferase protocols to perform large-scale screens, leading to difficulties in comparing functional repression across multiple data sets.
The 3′LIFE assay enhances the dual luciferase assay by placing emphasis on the rapid, sensitive, standardized and high-throughput screening for miRNA targets. These advances include the development of a test 3′UTR library, Gateway™ compatible miRNA expression and 3′UTR reporter plasmids, non-consumable transfection reagents and non-commercial dual-luciferase assay reagents (23). Importantly, the scalability of the 3′LIFE assay allows screens of individual miRNAs against a large 3′UTR library without biasing the screen toward candidate genes identified bioinformatically (24), allowing the identification of genes targeted via non-canonical and poorly conserved interactions. Importantly, the sensitivity of the luciferase reporters combined with the inherently large number of negative miRNA/mRNA interactions in 3′LIFE allows for detection of the subtle effects of miRNA targeting, a benefit not possible with low throughput, candidate gene or other selection based reporter assays (25).
To test the ability of the 3′LIFE assay to detect biologically relevant target genes, we studied the targets of two cancer relevant miRNAs, let-7c and miR-10b. The let-7 family members are broadly characterized as tumor suppressors; reduction in the expression of multiple family members is observed in many cancers and correlates with poor survival outcomes (26–29). let-7 miRNAs target genes that promote cell proliferation [RAS (30), HMGA2 (26), E2F5 (31,32)] and invasion and metastasis [MMP11, PBX3 (33)]. Each let-7 miRNA is bioinformatically predicted to target over 1000 genes (5), with overlapping target sets. miR-10b was initially described in the context of late-stage breast cancer, where overexpression initiated malignancy in vivo (34). Subsequently, miR-10b overexpression has been observed in over a dozen late-stage tumor types (35–38). There are several validated targets of miR-10b including tumor suppressors HOXD10, KLF4 and NCOR2 (34,39–41). Despite the significant body of research (283 publications in PubMed) on these miRNAs, it is striking that the number of targets with direct experimental validation equates to less than 3% of bioinformatically predicted targets (5,40).
We tested the feasibility of the 3′LIFE assay by querying let-7c and miR-10b against 275 test human 3′UTRs. Our results confirmed 80% of previously validated targets and identified many novel targets for these two miRNAs. In comparing the 3′LIFE assay to TargetScan, a widely utilized target prediction software, we identify that 63% of predicted targets have some degree of repression in the 3′LIFE screen, yet 69% of top targets were not predicted, and ∼27% did not contain a canonical seed target. We also observed that many miRNA target genes contain unpredicted canonical seed elements and demonstrate this is likely due to poor conservation within 3′UTRs of unpredicted genes, and not at miRNA target sites per se. Taken together, our results suggest that the 3′LIFE assay is a powerful tool to rapidly and systematically identify miRNA targets in high-throughput, validate bioinformatic predictions, identify novel non-canonical targets and highlights the complexity of miRNA targeting mechanisms.
MATERIALS AND METHODS
MiRNA target analysis of target prediction software
We have extracted the predicted targets and their prediction scores for the miRNAs let-7c, miR-10b, miR-125a, miR-138 and miR-22 from the TargetScan (5), PicTar (8) and Diana-microT (42) websites. For TargetScan, the default conservation filter was used, although we include both ‘Conserved sites’ and ‘Poorly conserved sites’ in our analysis. The target score for each gene was normalized using the mean target score per miRNA. A list of the validated targets was compiled using Tarbase (43), miRTarBase (40) and a manual literature search using the gene name and all aliases listed in NCBI gene database. A gene was assigned as a validated target if it had at least two distinct experimental methods demonstrating direct miRNA regulation.
pLIFE-3′UTR vector construction
The original luciferase vector T7 DLP was kindly provided by Dr John Chaput. The SV40 3′UTR was amplified from the pcDNA3.1/V5-his vector and introduced downstream of the Renilla luciferase open reading frame by introducing the restriction sites for NsiI and XmaJI (Supplementary Figure S2A). The internal ribosome entry site (IRES) was replaced with the phosphoglycerate kinase 1 (PGK) promoter by introducing a PstI restriction site and ligated using BamHI and PstI restriction sites upstream of firefly luciferase. The P2R-P3 Gateway cassette was amplified from the pDONR P2R-P3 (Invitrogen) and cloned downstream of firefly luciferase gene by introducing BglII and NheI restriction sites using QuikChange site-directed mutagenesis (Agilent). MS2 repeats were amplified from pSL-MS2–6x (AddGene Clone ID: 27118). Four MS2 repeats were cloned downstream of firefly luciferase using BglII and ApaI restriction sites for use in downstream RNA isolation protocols. This vector is available through DNASU (http://dnasu.org Clone ID EvNO00601503) (44).
pLIFE-miRNA vector construction
The miRNA expression vector (pLIFE-miRNA) was adapted from the pCAG-RFP-miRint plasmid (45). We replaced the cytomegalovirus early enhancer/chicken beta actin (CAG) promoter with the cytomegalovirus (CMV) promoter by introducing SpeI and SacI restriction sites upstream of the DSRed2 open reading frame using QuikChange site-directed mutagenesis (Agilent). let-7c and miR-10b were amplified from human genomic DNA and cloned into the pLIFE-miR vector using the AsiSI and NotI restriction sites. We included ∼200 nucleotides upstream and downstream from the surrounding genomic locus to replicate endogenous miRNA processing. We also created a Gateway compatible miRNA expression plasmid, which contains the L2R3 Gateway cassette cloned into AsiSI and MluI restriction sites (Supplementary Figure S2B). This vector is available through DNASU (http://dnasu.org, Clone ID EvNO00601504) (44). To prepare the pLIFE-PGK miRNA expression vector, which produces low expression level for the test miRNAs, we amplified the PGK promoter from the pLIFE-3′UTR plasmid and ligated it into the pLIFE-miRNA plasmid using SpeI and XhoI restriction sites.
Test 3′UTR library preparation
We designed primers to amplify 384 3′UTRs from various protein-coding genes based on the RefSeq annotation (HG19) and custom Perl scripts (46). Forward primers were anchored to the 3′end of the terminal exon. Reverse primers anchor ∼150 nucleotides beyond the end of the longest annotated 3′UTR to include putative elements important for mRNA processing (47). The attB2 and attB3 Gateway cloning elements were added to the ends for the forward and reverse primers, respectively. Primers were arrayed based on annealing temperature and expected amplicon size (Supplementary Table S1). Genomic DNA was isolated from HEK293T cells using DNAzol (Invitrogen), per manufacturer's protocol. Touchdown-polymerase chain reaction (PCR) was conducted using Taq polymerase (Invitrogen) in 96-well plates with 100-ng genomic DNA per PCR reaction. Cycling conditions were based on manufacturers protocol, with the addition of ∼15 touchdown cycles, where the annealing temperature started at 14°C above the lowest primer Tm on each plate, decreasing 1°C per cycle for 15 cycles. Remaining cycles were carried out at 55°. Amplicons were then recombined into the pLIFE-3′UTR vector using BP Clonase reactions (Invitrogen) per manufacturer's protocol. Cloning reactions were incubated overnight at room temperature, and 1 μl of the cloning reaction was transformed into DH5-α bacteria cells and plated on 48-well LB-amp agar plates. We initially picked one colony per well, and individual colonies were screened for correct size using universal primers that flank the P2R-P3 cassette in colony PCR experiments. Twenty percent were randomly picked and sequence verified. Clones that passed these tests were further used in the 3′LIFE assay. In our positive and negative control experiments, we recombined the SV40 3′UTR into the pLIFE-3′UTR vector using BP Clonase (Invitrogen), and introducing control miRNA targets for let-7c (AACCATACAACCTACTACCTCA) and miR-10b (ACAAATTCGGTTCTACAGGGTA) using QuikChange site-directed mutagenesis (Agilent). Putative miRNA seeds in RhoB and HOXD11 were deleted from the 3′UTR luciferase reporter plasmids using QuikChange site-directed mutagenesis.
Plasmid DNA preparation
pLIFE-miRNA plasmids were prepared using Maxi Preps (Promega) and the recovered DNA was resuspended to a concentration of 500 ng/μl. Bacterial stocks of pLIFE-3′UTR plasmids were arrayed in 96-well 2-ml blocks and grown for ∼16 h in 1.3-ml of Terrific Broth. Plasmids were purified using the Nucleospin 96-well mini-prep kit (Macherey-Nagel) and the Biomek FX (Beckman Coulter) liquid handling robot according to manufacturers' protocol. DNA was eluted in 50 μl of nuclease-free water. DNA concentrations were then normalized to ∼100 ng/μl using the Biomek FX liquid handling robot. To facilitate the assay in high-throughput, we have also optimized this step using in-house HighRes Biosolutions fully automated DNA miniprep systems that include three Liconic automated 800RPM/37C incubators, one automated Liconic automated −20°C freezer, three Denso fully articulated robotic arms, two Heraeus robotic centrifuges, two Thermo Wellmate bulk dispensers, one Thermo combi bulk dispenser, two KBio Wasp automated plate sealers, one Nexus automated plate pealer, one Molecular Devices DTX880 Plate Reader, one Beckman Biomek FX dual arm 96/SPAN 8 liquid Handler, one EL405 plate washer and two ambient automated plate storage hotels.
Cell culture
HEK293T cells were kindly provided by Dr Josh LaBaer. Cells were cultured in Dulbecco's modified Eagle's medium (Sigma), supplemented with 10% fetal bovine serum (Fisher) and 1% penicillin/streptomycin (Fisher).
Nucleofection buffer test
A significant cost associated with high-throughput cell-based assays is consumable reagents associated with transfection, thus we tested a panel of buffers for efficient transfection and cell survival using the 96-well Shuttle Device by Lonza. An initial panel of 11 buffers and the SF cell line solution (Lonza) were used to transfect 1 × 105 HEK293T cells with 100-ng pmaxGFP plasmid (Lonza). Each buffer was tested using several different nucleofection pulse codes. As the conditions of these pulse codes are proprietary, exact pulse conditions are unknown. The pulse codes tested were ER100, CU123, CM102, EO115, DT130, FF120, CN114, CM130, CB150, DS150, CA123 and DS138. We tested the following buffer solutions: phosphate buffered saline (PBS) (pH 7.4), PBS + 10% HEPES (pH 7.0), PBS + 10% HEPES (pH 7.4), PBS + 5% HEPES (pH 7.0), PBS + 1.5% HEPES (pH 7.0), HEPES buffered saline (HBS), 50% PBS/50% HBS, BD Perm Wash, electroporation buffer and phosphate buffered sucrose. HBS solution contains 140-mM NaCl + 1.5-mM Na2HPO4•2H2O + 50-mM HEPES. Electroporation buffer contains 15-mM potassium phosphate + 1-mM MgCl2 + 250-mM sucrose + 10-mM HEPES adjusted to pH 7.3 using concentrated HCl. Phosphate buffered sucrose solution contains 272-mM sucrose + 7-mM K2HPO4 and is pH adjusted to 7.4 with phosphoric acid. HBS and PBS HEPES consistently produced high transfection efficiency and/or cell survival, so we performed secondary experiments using permutations of pH and buffer composition: 75% PBS/25% HBS, 50% PBS/50% HBS, 25% PBS/75% HBS, PBS 1% HEPES (pH 7.0), PBS 1.5% HEPES (pH 7.0), PBS 1.5% HEPES (pH 6.4), PBS 1.5% HEPES (pH 7.4) and PBS 2% HEPES (pH 7.0). Because PBS 1.5% HEPES (pH 7.0) in combination with the FF120 pulse code yielded relatively few nucleofection errors, had high transfection efficiency, and relatively high cell survival with HEK293T cells, this protocol was used for all subsequent experiments.
Nucleofection protocol
A detailed 3′LIFE protocol is included in the Supplementary Materials and Methods and updates are available at www.mangonelab.com. In brief, cells were grown to ∼90% confluency and passaged between 24 and 48 h prior to transfection to be in logarithmic growth phase at the time of transfection. Cells were removed from a 145-mm parent culture plate with 0.25% trypsin (Fisher) for 3 min, washed with an equal volume of media and spun for 5 min at 300 g. Media/trypsin was eluted and cell pellet was resuspended to a density of ∼3.0 × 107 cells/ml. For each well, 100 000 cells were pelleted and resuspended in 20 μl of nucleofection buffer/plasmid solution, in the presence of no more than 2 μl of plasmid DNA (10% total volume). Each well contained a plasmid solution composed of 500 ng of pLIFE-miRNA and 100 ng of pLIFE-3′UTR. Cell/buffer solution was loaded into a 96-well shuttle plate and transfected via nucleofection using the 96-well Shuttle System according to manufacturer's conditions (Lonza). Immediately following nucleofection, 80 μl of pre-warmed complete media was added into each well. Transfected cells were then transferred to a 96-well culture plate to a total volume of 200-μl media. Each plate containing human 3′UTRs cloned into pLIFE-3′UTR vector was transfected in quadruplicate, with each replicate co-transfected with either the empty miRNA vector control, pLIFE-miR let-7c or miR-10b. Transfected cells were incubated for 48–72 h post transfection to minimize non-specific effects caused by cascades of changing gene expression due to exogenous miRNAs. Cell are then imaged using fluorescence microscopy and dsRed2 protein to identify wells with failed transfections, and subjected to the luciferase assay.
Luciferase assay
Prior to luciferase assay, the media was removed and cells were lysed for 30 min in the presence of 26 μl of passive lysis buffer (Promega) and gentle rocking. 25 μl of total lysate was removed from the culture plate and transferred to opaque white 96-well plates and used in the luminescence assay. The non-commercial dual-luciferase buffer was prepared as previously described (23). Coelenterazine (Promega) was resuspended in 5-mM acidified methanol and stored at −80°C. Beetle luciferin (Promega) was resuspended in nuclease-free water and stored at −80°C. Individual buffer components were prepared and stored at 10X concentration. Buffers were prepared fresh with stored reagents prior to each luciferase assay. Luminescence was measured using the Glomax 96 Microplate Luminometer with dual injectors (Promega). 100-μl of firefly luciferase and Renilla buffers were injected sequentially, followed by a 5-s delay and a 10-s measurement time.
Statistical analysis of luciferase assay
Raw data from firefly and Renilla luciferase luminescence assay were recorded for each well. We calculated the ratio between firefly and Renilla using the relative light units obtained for each well, to control variation in DNA concentrations, cell number and transfection efficiency. The ratio of the firefly and Renilla luciferase values between the negative miRNA control and its correspondent miRNA experiment was then used to calculate the absolute repression (A) for each well using the following formula (1):
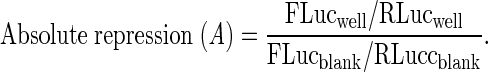 |
(1) |
The normalized repression value (N) was obtained by averaging the repression of every well across a plate and used to normalize each well (2):
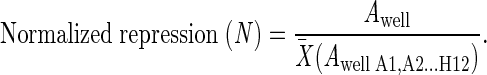 |
(2) |
This transformation measures repression as a function of all the 3′UTRs on each plate and allows the comparison of replicates of the experiment, removing fluctuations in raw FLuc/Ren ratios caused by subtle variation in the luciferase buffers and luminometers. Each plate is performed in quadruplicate on different days, and normalized repression ratios for each well are averaged to obtain miRNA repression index (RI) (3):
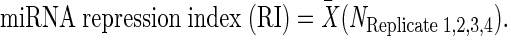 |
(3) |
We then rank the results based on the RI value, and putative hits are defined as 3′UTRs with an RI <0.80, and obtain a statistically significant repression (P < 0.05) using student's t-test.
pLIFE-PGK secondary screen
To quantify PGK expression levels, pmiRNA-LIFE or pmiRNA-LIFE-PGK was cotransfected with the pmaxGFP plasmid (Lonza). Fluorescent images for RFP and GFP were obtained, and expression levels were calculated using ImageJ (NIH) with standard parameters. To test the effect of miRNA overexpression on positive hits, one plate containing 87 3′UTRs was screened for targeting by let-7c and miR-10b driven by the PGK promoter. Experiments were performed with four replicates. Results were analyzed as above.
MiRNA target literature review
The genes with an RI score <0.8 and P-value <0.05 were analyzed in a literature search for potential contributions to tumorigenesis. In the case of a gene having roles in tumors of different origins, emphasis was placed on tumors originating in breast tissues due to the relevance of let-7c and miR-10b to breast cancer progression. Criteria for placing each gene into a category as either a positive or negative driver of tumorigenesis included evidence of overexpression or downregulation, if its role was classified in at least two publications, and was associated with a defined mechanism of action. Cases where a gene has opposing roles in distinct tissues of origin are noted with dual color annotation. References used in this analysis are provided in Supplementary Table S3.
MiRNA target alignment
The 3′UTRs of top putative hits were analyzed for potential miRNA target sites using ClustalW (48). To identify both canonical and likely non-canonical targets, individual alignments were attempted for nucleotides 2–7, 1–9, 10–22, and the entirety of the miRNA, against each 3′UTR. The gap open and gap extension parameters were adjusted between 10–25 and 0.5–5, respectively, to obtain the best alignment in terms of seed binding and 3′ compensatory complementarity. To create the miRNA binding footprint we used the WebLogo software (49). Gaps in the binding footprint corresponding to loops in the mRNA were removed leaving only the nucleotides that interact with the miRNA. G:U wobbles were accounted for by first running the software with all normal sequences. Second, we temporarily replaced all U's in mRNA sequence with C if the interacting miRNA nucleotide corresponded with a G and replaced all G's with A if the corresponding miRNA nucleotide was a U. Third, we reran the software, measuring the height of the combined wobble peaks, and fourth, corrected the original WebLogo with the heights corresponding to the number of bits expected if G:U wobbles are accounted for.
Comparison with AGO-HITS-CLIP CLASH data set from Helwak et al. (2013)
The complete Supplementary data set from (50) was downloaded from www.cell.com/cell/fulltext/S0092-8674(13)00439-X. Chimeric reads from all let-7 and miR-10 family members were searched for genes that were also present in the 3′LIFE library. Genes whose reads mapped to regions other than the 3′UTR were binned separately. Genes with chimeric reads in the 3′UTR were compared against the scores from the 3′LIFE assay.
Analysis of 3′UTR conservation using PhyloP
For each hit in the 3′LIFE assay that contained a canonical seed, defined as perfect complementarity with no G:U wobbles in nucleotides 2–7 of the miRNA, conservation scores were identified using the 100 vertebrates Basewise Conservation by PhyloP track (51) in the UCSC genome browser. For each gene conservation scores for three loci were obtained: the six nucleotide seed, 200 nucleotides flanking the seed (score includes seed site), and up to 200 nucleotides of the terminal exon. The conservation score for each nucleotide in these loci were averaged, and this mean conservation score for each locus was plotted as a schematic against a hypothetical gene model.
RESULTS
MiRNA target predictions have high false-negative rates
Several algorithms have been developed to predict miRNA/mRNA interactions in 3′UTRs corresponding to each gene in the genome (7). Predictions for miRNA targets are generally based on the characteristics of previously validated miRNA target elements and utilize additional parameters, such as site conservation, to refine putative target lists. These algorithms generate hundreds to thousands of predicted interactions for each miRNA, and thus can be used to analyze miRNA targets on a systems level. However, several groups have reported high false-positive (12,13) and false-negative rates (9,10). We measured the efficacy of three widely utilized algorithms, TargetScan (5), Diana-microT (42) and PicTar (8) to identify the most predictive algorithm to compare against results from the 3′LIFE assay. We selected five miRNAs based on their significant presence in the literature and relevance to cancer and compared the predicted targets with experimentally validated targets from the literature (Supplementary Figure S1A–C). Despite the wide body of literature on these miRNAs (thousands in PubMed), there are only 64 validated targets, representing ∼2% of all bioinformatic predictions. TargetScan correctly predicted 66% of experimentally validated targets with the conservation filter enabled, while Diana-microT and PicTar predicted 53% and 47% of validated targets, respectively (Supplementary Figure S1D). However, TargetScan predicted 3236 targets, compared to 2001 by PicTar, which may account for differences in false-negative rates. We compared all hits from the 3′LIFE assay against predictions from TargetScan because it contained the lowest false-negative rate.
3′LIFE is a scalable method to rapidly detect miRNA targets in high-throughput
Luciferase-based assays are widely used to directly detect miRNA targeting due to their high sensitivity. Commercial high-throughput luciferase reagents are available, but are cost prohibitive for large-scale approaches. To overcome these limitations, we have developed a cost-effective high-throughput assay that systematically screens miRNA/mRNA interactions in an unbiased and standardized fashion. 3′LIFE is an adaptation of the dual luciferase reporter assay performed in high-throughput (Figure 1). The dual luciferase assay relies on the fusion of a test 3′UTR to a luminescent reporter gene. Targeting and translational repression of the test 3′UTR by a probe miRNA is identified by a decrease in the luciferase::3′UTR signal (firefly luciferase) relative to a second normalization signal (Renilla luciferase) (Figure 1A, red spots). To account for endogenous miRNAs that may target the luciferase reporter, we include a no miRNA negative control, which provides a baseline from which to detect targeting by the exogenous test miRNAs. 3′LIFE is designed to rapidly identify functional targets of a given miRNA in a panel of hundreds of test 3′UTRs in co-transfection experiments.
Figure 1.
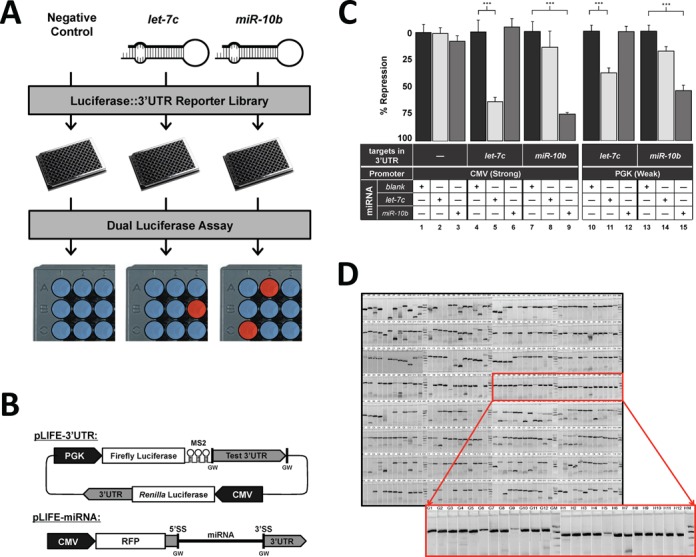
The 3′LIFE Assay. (A) Schematic of the 3′LIFE assay. A reporter library of luciferase::3′UTR fusion genes is cotransfected in HEK293T cells with miRNAs of interest and a negative control in 96-well format. Following transfection cell lysate is used in a dual luciferase assay, and target genes are identified by a significant repression of the luciferase gene compared to the negative control (B) 3′LIFE Vector Design. (Top) Gateway compatible dual luciferase expression vector. The 3′UTR of interest is cloned downstream of firefly luciferase using Gateway cloning. Both luciferase genes are located on the same plasmid to prevent fluctuations in the luciferase ratio across experiments. (Bottom) Gateway compatible miRNA expression plasmid. The miRNA of interest is cloned within an intron in the 3′UTR of RFP using Gateway cloning. This approach functions dually as a visual marker for transfection efficiency and miRNA expression. (C) Positive controls for let-7c and miR-10b. The SV40 3′UTR containing a scrambled miR-10b target site (blank) or let-7c and miR-10b targets were cloned into the pLIFE-3′UTR and tested in quadruplicate using a dual luciferase assay. Results are normalized to the Blank condition of each 3′UTR. Y-axis measures the percentage of repression compared to the Blank condition. Error bars are standard error unless otherwise noted. * denotes statistical significance (Student's t-test, *P < 0.05, **P < 0.01 and ***P < 0.001). (D) Gel electrophoresis of 384 3′UTRs amplified from human genomic DNA using touchdown PCR for use in the 3′LIFE assay.
3′LIFE uses two specially designed vectors to express the test 3′UTRs (pLIFE-3′UTR) and the miRNA (pLIFE-miRNA) (Figure 1B). In the pLIFE-3′UTR vector, the two luciferase genes were cloned into the same plasmid to prevent fluctuations in their molar ratios across separate transfections. We have also included a Gateway-compatible recombination cassette (GW) cloned downstream of the firefly luciferase gene to efficiently shuttle test 3′UTRs into the pLIFE-3′UTR vector (Figure 1B (top) and Supplementary Figure S2A). In the pLIFE-miRNA vector, the miRNA is cloned within an intron in the 3′UTR of the red fluorescent protein (45) (Figure 1B (bottom) and Supplementary Figures S2B and S3), dually functioning as a marker for transfection efficiency and miRNA expression (Supplementary Figure S3B).
To test the processing and functional activity of the miRNA expression vectors, we introduced perfect target sites for each miRNA into the SV40 3′UTR and co-transfected the pLIFE-SV40 vectors with a negative miRNA control, let-7c or miR-10b. Each miRNA strongly repressed only the 3′UTR containing its target site, confirming the processing of the miRNA and demonstrating the specificity of the targeting (Figure 1C, panels 4–9).
Overexpression of exogenous miRNAs may cause non-specific or dosage-dependent effects. To test this hypothesis, we replaced the CMV promoter present in the pLIFE-miRNA vector with the weaker PGK promoter (pLIFE-PGK). In HEK293T cells, the CMV promoter was previously reported to be approximately five times stronger than PGK (52), a result echoed by our own analysis of fluorescence expression (Supplementary Figure S4). Importantly, the PGK promoter is also used to drive expression of the firefly luciferase::3′UTR reporter, thus expression levels of these two genes should be equivalent in the pmiRNA-PGK experiment. Repeating the SV40 targeting experiments with the pLIFE-PGK miRNA expression vector yielded decreased translational repression (Figure 1C, panels 10–15), although not to the extent expected by decreasing expression levels 80% (Figure 1C, compare panel 5 with panel 11, and panel 9 with panel 15). While it is possible that the translation of the RFP may be effected by unknown 5'UTR elements provided by the CMV and PGK promoters, the strong functional differences between the same miRNA driven by these two promoters suggests that there are transcriptional differences (Figure 1C, Supplementary Figure 3).
The feasibility of high-throughput dual luciferase assays is limited by high costs associated with transfection and luciferase assay reagents, and the lack of a publicly available human 3′UTR library. Furthermore, genome-wide screens for miRNA targets are challenged by the need for appropriate high-throughput technologies and pipelines. 3′LIFE assay utilizes low-cost non-proprietary transfection and dual-luciferase reagents (23) (Supplementary Figure S5). We have designed each step of the 3′UTR cloning pipeline and the 3′LIFE assay to be highly automated (Supplementary Figure S6). PCR, cloning, plasmid DNA preparation, cell culture, transfection and luciferase assays are performed in 96-well format using multi-channel micropipettes, dedicated liquid handling robots and other high-throughput instrumentation.
To test the 3′LIFE assay, we chose a pilot library of 384 human 3′UTRs containing experimentally validated targets (n = 10) and bioinformatically predicted targets (n = 47) of let-7c and miR-10b, as well as genes that play regulatory roles in tumorigenesis (Figure 1D and Supplementary Table S2). This human 3′UTR clone collection is publically available through the DNASU plasmid repository (http://dnasu.org) (44). More clones are periodically added to the library, with the goal of covering the entire human 3′UTRome.
Here, we screened 275 of these 3′UTRs for potential targeting by each miRNA, performing four replicates of each experiment using the pLIFE-miRNA vector (totaling ∼3500 transfection reactions and luciferase assays). To detect miRNA targeting, each 3′UTR was co-transfected with the miRNA, and the luciferase ratio was compared to a negative control. Putative hits are defined as those that have an RI below the mean standard error of the assay (0.15 for let-7c and 0.16 for miR-10b). These putative hits were enriched with bioinformatically predicted targets and previously validated targets for both let-7c (E2F5, RTCA, PBX3, TRIM71) and miR-10b (SDC1, NCOR2, HOXD10) (Figure 2A, arrows). Surprisingly, we detected repression in only 62% of genes predicted by TargetScan.
Figure 2.
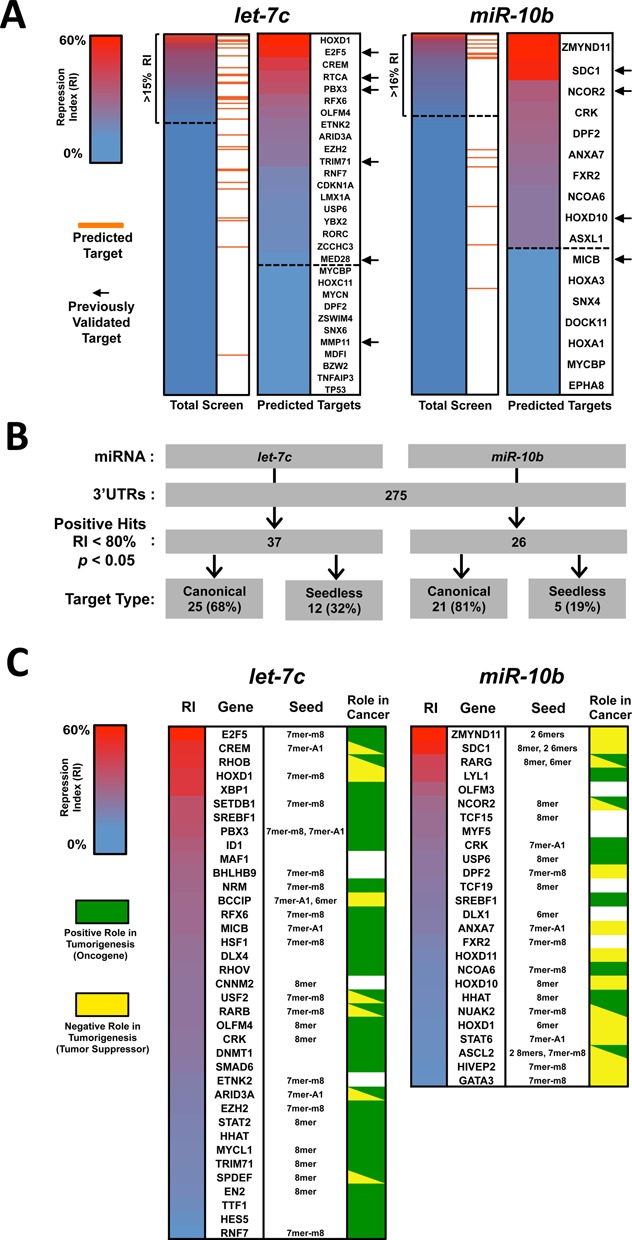
Results of the 3′LIFE pilot study. (A) Two hundred and seventy five 3′UTRs were probed for targeting by let-7c (left) or miR-10b (right). RI values (see the Materials and Methods section) represent the average of four replicates. 3′UTRs were considered putative hits if the RI falls below the standard error for each miRNA (dashed line). Bioinformatically predicted genes are extracted and ranked (right of each panel), demonstrating an enrichment for predicted targets among putative hits. Previously validated targets are marked with black arrows. (B) Top hits obtained are defined as those with RI < 0.80 and P < 0.05. let-7c had 37 and miR-10b had 26 top hits, the majority of which contain canonical target sites. (C) List of top hits in 3′LIFE assay. Putative seed target site is listed as defined in (5). Each gene was identified as having a positive role (green), negative role, conflicting roles (red/green) or no identifiable or established role in tumorigenesis based on a manual literature review (see the Materials and Methods section and Supplementary Table S3).
To further refine our putative hits to only high-confidence targets, we set a cutoff for genes with an RI <0.80 and P-value <0.05. Using this criterion we obtained 37 high-confidence hits for let-7c and 26 for miR-10b (Figure 2B). To identify putative miRNA target sites within these top hits, we scanned each 3′UTR for elements highly complementary to the miRNA (Supplementary Table S1). While 63% of these 3′UTRs contained perfect seed elements, only 32% were predicted by TargetScan, presumably because they contain either poorly conserved canonical seeds or non-canonical target sites.
Strong miRNA and weak miRNA overexpression yield comparable results
We were then interested in studying the extent of non-specific or dosage-dependent targeting caused by miRNA overexpression. We rescreened one plate of 3′UTRs (n = 87) using the pLIFE-PGK miRNA expression vector, repeating the assay in quadruplicate (a total of 1152 transfection reactions and luciferase assays) (Supplementary Figure S7A). The RI of each gene tested against varying extents of miRNA overexpression yielded largely comparable results, with the majority of genes having some degree of repression in both, or neither (Supplementary Figure S7A). Among the top hits obtained with pLIFE-miRNA vectors, 77% of targets were also repressed with the pLIFE-PGK vectors, albeit generally to a lesser degree (Supplementary Figure S7B and C). We obtained three genes not repressed with the pLIFE-miRNA screen that show significant repression (RI < 0.80, P < 0.05) by miR-10b in the pLIFE-PGK experiment (AATK, HOXA1, KRT1). HOXA1 and KRT1 contain perfect seed matches for miR-10b, while AATK contains a highly complementary non-canonical target (Supplementary Figure S7D). In conclusion, these data suggest that while there may be a dosage effect for a small portion of miRNA targets, the majority of targets show consistent repression regardless of the degree of miRNA overexpression.
Putative miRNA target genes have functions consistent with known role of each miRNA in cancer
We next sought to identify potential correlations between the functional role of each miRNA in tumorigenesis and the novel targets identified by our pilot 3′LIFE assay. We performed a literature review for the top gene targets to determine if our ‘bona fide’ targets were previously recognized to have positive or negative contributions to tumorigenesis (Figure 2C and Supplementary Table S3). let-7c targets identified by the 3′LIFE assay are generally overexpressed in tumors and have positive roles in tumorigenesis, which is consistent with the tumor suppressor function of let-7c. Conversely, miR-10b target genes are typically downregulated in late-stage tumors and inhibit cancer progression and metastasis, which is also consistent with miR-10b as an indicator of late-stage, aggressive tumors (Figure 2C).
Each miRNA exhibits a unique binding footprint
Analysis for enriched nucleotides in these putative miRNA target sites revealed that the seed was the least variant region within the target element of both miRNAs (Figure 3A). The 3′end of the miRNA also showed consistent interaction with the mRNA compared to the central nucleotides. let-7c also possesses a more even distribution of interacting nucleotides compared to miR-10b, which has two distinct regions at the terminal ends. The mean bit score gives a measure of the extent to which each nucleotide interacts with the target mRNA, and in let-7c it is slightly higher than in miR-10b (1.03 compared to 0.86), suggesting that let-7c interacts with more nucleotides in each target mRNA than miR-10b. As G:U base pairing expands the flexibility of these nucleotides to interact with more than one base, we compared the bit score of G/U and A/C nucleotides. G/U's outside the seed region were twice as likely to interact with corresponding mRNA nucleotides than A/C (0.85 compared to 0.43), which is expected given that the number of nucleotides that G/U can pair with is double that of A/C. The five 3′-most nucleotides of both miRNAs are G/U's, which may account for the targeting peaks in this region. Of note is the high percentage of G/U nucleotides in let-7c (82%) compared to miR-10b (52%), which may contribute to the overall higher bit score, the increased number of bioinformatically predicted targets (Supplementary Figure S1A) and the larger number miRNA targets detected by the 3′LIFE screen (Figure 2B) for let-7c compared to miR-10b.
Figure 3.
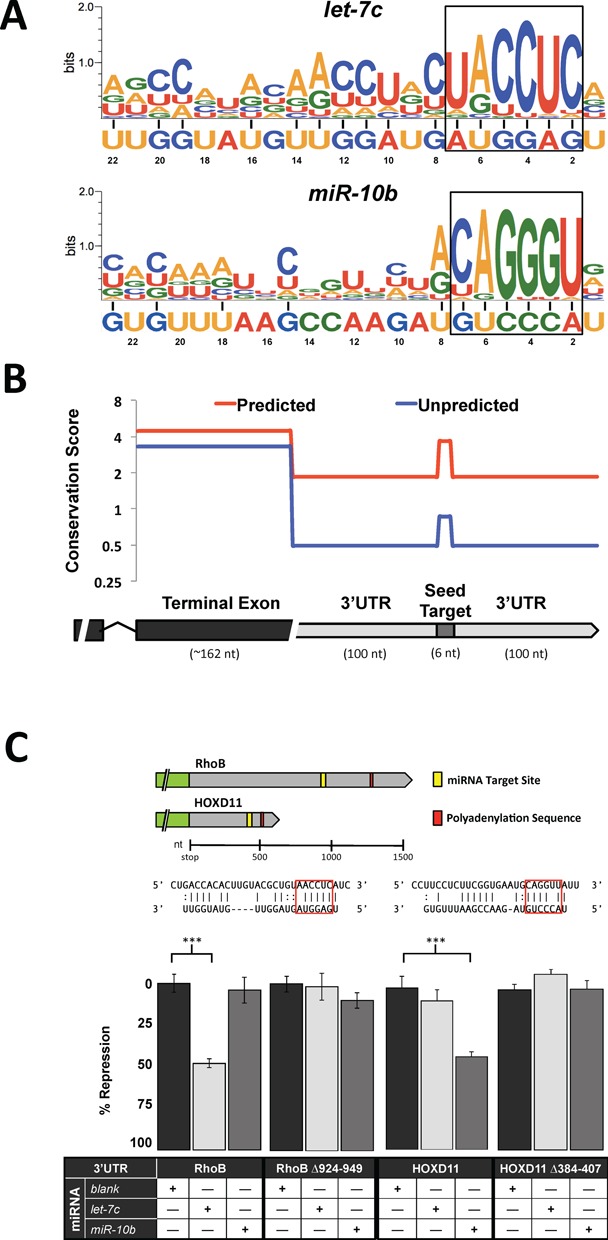
Characterization of target sites within top 3′LIFE hits. (A) Target signature motif of putative miRNA target sites within top hits identified using ClustalW alignments. The black box represents the miRNA seed region. Black horizontal lines indicate nucleotides within the mRNA with a bit-score >0.8. (B) Mean conservation scores of the nucleotides in terminal exon, miRNA target site, and flanking 3’UTR of genes possessing perfect seed matches. Scores are plotted log(2) scale along a hypothetical gene model. 3’UTRs are separated based on the TargetScan prediction status of each gene. Conservation score is obtained using the PhyloP vertebrate conservation track on UCSC genome browser. (C) Deletion analysis of two top non-canonical miRNA target sites identified in 3′LIFE assay. Map showing the relation of the miRNA target site within the 3′UTR relative to polyadenylation sequences (AAUAAA). Red boxes indicate canonical seed nucleotides. Each 3′UTR containing either the wild-type or deleted miRNA target site is cotransfected with Blank miRNA control, let-7c or miR-10b.
Seed elements are conserved compared to surrounding 3′UTR sequence
Thirty percent of 3′UTRs in the target library contain canonical seed elements. Within this group, more than half were among 3′LIFE top hits, suggesting that 3′LIFE hits are enriched with genes containing canonical seeds. Among the 3′LIFE top hits, 73% contain a canonical seed (46 out of 63), yet only 32% were predicted bioinformatically (20 out of 63). We reasoned that most of these canonical targets escaped prediction because the seed element is poorly conserved. To address this discrepancy we measured the sequence conservation among vertebrates for both predicted and unpredicted canonical seed elements within the 3′LIFE top hits. We then compared the results to the conservation levels in the open reading frames and in the 3′UTRs of the same genes (Figure 3B). Not surprisingly, the seed region in predicted targets was four times more conserved than in non-predicted targets. However, in both the terminal exon and the 3′UTR of non-predicted targets we observed far less conservation compared to the same regions in predicted targets. This suggests that when comparing conservation of genes as a whole, the degree of conservation of unpredicted genes may automatically exclude any canonical targets from prediction by algorithms using conservation criteria. Furthermore, the seed region in unpredicted targets is twice as conserved as the surrounding 3′UTR, a jump similar in magnitude to that of predicted seed targets. This suggests that despite the relatively poor conservation of unpredicted targets, these elements are possibly functional as there is indeed some selective pressure to maintain them. Taken together, these results suggest that the lack of conservation in the seed region is not solely responsible for the failure of TargetScan to predict the miRNA targets that are detected by the 3′LIFE assay, but that the lack of conservation in the 3′UTR itself plays a role in increasing false-negative rates of prediction softwares.
To further validate non-canonical targets sites among the top hits of 3′LIFE, we selected two genes, RhoB and HOXD11, and deleted the putative target sites for let-7c and miR-10b, respectively (Figure 3C). Deleting these elements completely rescued each 3′UTR from repression by their respective miRNAs, demonstrating that the 3′LIFE assay is capable of identifying non-canonical miRNA/mRNA interactions (Figure 3C).
DISCUSSION
3′LIFE is a highly sensitive method to detect miRNA targets in high-throughput
In this study we present a novel, scalable and sensitive method to identify miRNA targets in high-throughput, which addresses a critical need in miRNA biology. Currently, bioinformatic and RISC-IP approaches are the only tools to investigate miRNA targets in a high-throughput manner.
Bioinformatic approaches utilize a suite of parameters to predict highly likely targets, such as target composition (i.e. miRNA seed regions), site conservation and thermodynamic stability, and benefit from the ability to identify sites throughout the entire genome. While these algorithms are indispensable tools to identify candidate genes for experimental validation, their high false-positive and false-negative rates suggest that the parameters upon which these algorithms are built do not yet account for all the mechanisms miRNAs use to recognize target sites. Genome wide bioinformatic predictions are only useful in the absence of experimental tools that deliver results at a comparable scale. AGO-HITS-CLIP provides a high-throughput experimental approach to identify miRNA targets in a specific context, i.e. cell line or tissue. When we compare our results with data obtained from similar approaches, we note that IP-based assays limit the findings to the set of abundantly expressed transcripts and that occupancy of an miRNA at a specific target site may not necessarily result in translational repression (Supplementary Figure S8). Dual luciferase reporter assays are rapid, sensitive and quantitative and measure the functional output of miRNA interactions at the protein level. Despite the widespread use of dual luciferase assays to validate miRNA targets, the notable lack of use in initial high-throughput screens speaks to the substantial technical and methodological barriers to such applications. These barriers include the lack of a high-quality publicly available 3′UTR reporter library, the high costs associated with transfection and luciferase assay reagents and the absence of standardized high throughput protocols and data analysis pipelines.
The 3′LIFE assay overcomes the above challenges by (i) measuring the effect of miRNA targeting at the protein level, (ii) utilizing a reporter assay that is quantitative and highly sensitive to subtle fluctuations in protein concentration, (iii) does not rely on prior assumptions about miRNA targets to generate candidate gene libraries and (iv) by using a high-throughput screening approach it becomes possible to detect subtle repression in a large number of genes due to the large number of negative interactions built into the assay. This last point is of critical importance, in that evidence suggests that miRNAs exert significant influence on the transcriptome not by strongly repressing individual genes, but by subtly repressing many targets (see below).
In this ‘proof-of-principle’ screen we tested 275 3′UTRs for targeting by let-7c and miR-10b and observed repression in a large number of novel putative targets, eight of 10 previously validated targets, and 62% of the bioinformatically predicted genes included in this 3′UTR library (Figure 2A). Among the top hits in the 3′LIFE assay were genes targeted by non-canonical target sites (27%) (Figure 2C), poorly conserved canonical seeds (Figure 3B) and genes frequently utilizing G:U wobble pairing (Figure 3A and Supplementary Table S1). These degenerate targeting principles expand the repertoire of potential miRNA target sites, contribute to the observation that only 32% of the top 3′LIFE hits were bioinformatically predicted and support the use of unbiased high-throughput experimental approaches to identify miRNA targets.
Surprisingly, 20% of previously validated targets and 38% of bioinformatically predicted targets showed no detectable degree of repression. Previous groups have reported similar false-positive rates for prediction algorithms (12,13), suggesting that algorithm error rates may account for the lack of targeting in this screen. However, false negatives could also be explained by several biological factors. First, sequence elements flanking putative canonical target sites that possess high degrees of secondary structure may have a critical impact on target recognition by restricting the access of the miRNA silencing machinery to the target site (53,54). Second, cooperative repression by multiple miRNA target sites (55) or interactions with trans-acting factors such as RNA-binding proteins (56) can contribute to miRNA target recognition that requires multiple trans-acting factors, resulting in targeting only in specific contexts. Third, recent studies indicate that a large portion of eukaryotic mRNAs are reversibly methylated, preferentially within non-coding regulatory regions, and potentially contributing to the evasion of miRNA targeting (57). Lastly, alternative polyadenylation may increase the false-negative rate in the 3′LIFE assay. Fifty percent of human genes contain multiple polyadenylation signals within 3′UTRs that signal transcription termination, cleavage and polyadenylation (58,59). Rapidly proliferating and tumor cells were shown to possess, on average, shorter 3′UTR isoforms, a mechanism resulting in the evasion of miRNA regulation (60–62). Alternative polyadenylation may also result in the exclusion of miRNA target sites in a context-dependent manner, by producing short 3′UTR isoforms only in certain cell lines or tissues. Thus, context-specific targeting mechanisms, including interactions with trans-acting factors and alternative polyadenylation, may contribute to false negatives in the 3′LIFE assay.
Conversely, overexpression of regulatory factors, such as miRNAs, may yield false-positive results due to non-specific repression of the reporter gene or supraphysiological expression levels driving specific, yet biologically irrelevant repression. The first scenario results from cascades of regulatory interactions that may alter the expression levels of other regulatory genes, which in turn target the reporter. Such false positives are a major caveat to overexpression experiments and, in general, are difficult to detect. Non-specific effects are particularly pronounced in experiments utilizing RNA-Seq or proteomics approaches to identify miRNA targets among endogenous genes, in that the repression can occur both pre- and post-transcriptionally, and be driven by sequence elements at any region of the mRNA. Because 3′LIFE utilizes only the 3′UTR in the reporter genes, non-specific effects are minimized to downstream factors that regulate elements within the 3′UTR, such as miRNAs and RNA-binding proteins. 3′LIFE minimizes these effects by performing the luciferase assay shortly (48–72 h) following transfection, reducing the time in which any changes may occur. Any non-specific targeting effects would have to occur following a chain of events, including transcription and processing of the exogenous miRNA, miRNA targeting and any subsequent downstream events (i.e. transcription and translation/miRNA processing). Finally, these changes would need to result in the downregulation of the reporter 3′UTR to an extent comparable with the strongly expressed exogenous miRNA. Several pieces of evidence suggest that the top hits detected by 3′LIFE suffer little from such false positives. First, 73% of the top hits detected by 3′LIFE contain canonical miRNA target sites, a significant enrichment compared to the remainder of the library (∼17%) (Figure 2B). Second, among the non-canonical putative target genes, we have experimentally validated direct targeting of two genes by deletion analysis (Figure 3C), suggesting that 3′LIFE can identify direct repression of genes targeted via non-canonical sites. Lastly, we observe relative conservation of the canonical seed elements, both predicted and unpredicted, within our top hits, suggesting that these are evolutionarily maintained functional target sites (Figure 3B). While false positives due to indirect regulatory interactions are certainly plausible events, the above evidence suggests that the top hits from 3′LIFE are ‘bona fide’ direct miRNA targets.
The second scenario yielding false positives, where miRNA overexpression results in specific and direct regulatory interactions that only occur at supraphysiological levels, is a primary concern in generalizing results from experiments that rely on miRNA overexpression. The relative abundance between miRNAs and their target mRNA can influence the degree of repression, as well as the phenotypic consequences of miRNA targeting, thus experimental approaches that mimic physiological miRNA/mRNA levels may yield less such false positives. To address this concern we screened a portion of the 3′UTR library with miRNAs driven by a relatively weak promoter that drives both the luciferase reporter and miRNA genes (Supplementary Figure S7). The weak promoter repressed 10 of the 13 top hits identified by the strong promoter, albeit to a lesser extent. Interestingly, there were also three genes that were significantly repressed by the weak promoter and not the strong promoter (Supplementary Figure S7D). Of these three genes, two contain perfect seed matches, and the third a perfect seed shifted one nucleotide outside of the canonical position.
These data suggest that while the majority of 3′UTRs are consistently repressed regardless of the degree of overexpression in the 3′LIFE assay, miRNA overexpression may yield both false-positive and false-negative results. In conclusion, the context, i.e. cell lines, used to identify miRNA targets is typically dependent on the biological questions being asked, therefore caution must be taken when generalizing results across cellular contexts.
MiRNAs target multiple genes within biological pathways
The effect of miRNA targeting on protein production is generally understood to result in modest translational repression (7,13), and can be influenced by several factors. These factors include the number of target sites in the mRNA, target site characteristics, with interactions guided by canonical seeds and high degrees of complementarity in the 3′end of the miRNA resulting in stronger repression, and position in the 3′UTR relative to the polyadenylation site, with more distal target sites generally showing stronger repression (7). These observations led to the hypothesis that a primary effect of gene regulation via miRNAs is not to serve as a switch, turning protein production on or off, but instead functions as a mechanism to fine-tune protein output, protect against aberrant levels of gene expression and provide robustness to cell-specific programs (63,64). Thus, the discrepancy between the modest impact an miRNA has on any single message and the powerful role specific miRNAs have in diverse biological processes suggests that miRNAs must target multiple genes at various nodes in these networks in order to obtain more vigorous regulation. For the emergence of such a multi-faceted mechanism, miRNAs and/or their targets would require a high degree of evolvability. In support of this notion is the observation that in metazoan genomes miRNAs have undergone three major expansions in recent evolutionary history, with the number of miRNAs correlating with organismal complexity (65). These expansions are driven primarily by the duplication and divergence of miRNAs, resulting in miRNA families, members of which are distinguished by only a few nucleotides in the mature miRNA. Furthermore, the small size and degenerate nature of miRNA target elements, combined with the relatively rapid evolutionary flexibility of 3′UTRs (66), suggests that miRNAs are capable of readily evolving novel targets (67). Consistent with these hypotheses, the 3′LIFE assay identified an enrichment of specific pathways among the top hits targeted by each miRNA, including central members, upstream regulators, and downstream effectors of RAS signaling for let-7c, and the retinoic acid (RA) signaling pathway for miR-10b.
let-7c targets multiple genes within the RAS signaling pathway
Loss of let-7 family members correlates with poor survival rates in many cancers (26–29), in part by inhibiting the RAS signaling pathway by targeting several elements within the 3′UTRs of KRAS and NRAS (30,33,68). Consistent with the hypothesis that miRNAs function by targeting multiple members of the same pathway, the top hits for let-7c identified by 3′LIFE were enriched for multiple genes within the RAS signaling pathway. Among these genes were two small GTPases within the RAS superfamily (RhoB, RhoV). RhoB has conflicting reports of its role in tumorigenesis dependent on tumor type (69–71). let-7c targets RhoB at a single non-canonical target site (Figure 3C). Recent evidence also suggests that RhoV overexpression may contribute to tumorigenesis (72). In addition to these central signaling components, 3′LIFE also identified several downstream effectors of the RAS signal (ID1, HSF1, CRK, DNMT1, ARID3A, EZH2). Inhibitor of differentiation 1 (ID1) regulates the activity of several transcription factors, is positively correlated with tumor progression, is transcriptionally activated in a KRAS-dependent manner in culture (73), is a downstream effector or RAS signaling in fibrosarcomas (74) and cooperates with RAS in metastatic breast tumors in vivo (75). Heat shock factor 1 (HSF1) is a transcription factor and downstream effector of RAS signaling, and is required for malignant transformation induced by oncogenic RAS (76–80). V-CRK avian sarcoma virus oncogene (CRK) is an adaptor protein that binds to several tyrosine-phosphorylated proteins, and by interacting with multiple signaling pathways suppresses cell adhesion and contributes to transformation in the presence of oncogenic RAS (81,82). AT-rich interacting domain 3A (ARID3A) is a transcription factor that has been shown to rescue RAS-induced senescence, promoting cell survival and immortalization (83,84). DNA methyltransferase 1 (DNMT1) is overexpressed in a variety of tumors and is required for RAS-dependent epigenetic silencing of several tumor suppressors (85–87). Enhancer of zeste homolog 2 (EZH2) is a member of the polycomb family of transcriptional repressor proteins, its expression is an initiating event of a variety of tumors induced by oncogenic RAS and results in a wide range of downstream effects including transformation and metastasis (88–90). Among these eight let-7c target genes, ARID3A and EZH2 are the only bioinformatically predicted targets (Figure 2A), while CRK, ARID3A, EZH2 and HSF1 contain canonical seed elements within their 3′UTRs (Figure 2C).
miR-10b targets are enriched for multiple genes within the RA signaling pathway
Recent studies link the miR-10 family to the RA signaling pathway, yet report conflicting roles of these miRNAs in tumor progression. In neuroblastoma miR-10a and miR-10b positively regulate the activity of RA signaling and promote differentiation (36), while the ability of miR-10a to promote metastasis in pancreatic ductal adenocarcinoma is inhibited by RA treatment (91). While context is critical in relation to the response of a cell to RA signaling, the overexpression of miR-10 in various late-stage tumors and its induction of invasion and migration is well documented (92,93). In line with these observations, 3′LIFE confirmed three previously validated targets (SDC1, NCOR2 and HOXD10) and identified seven novel targets of miR-10b with various roles in the RA signaling pathway. These include upstream regulatory effectors of the RA response (RARG, NCOA6, NCOR2, ASCL2), as well as downstream transcriptional targets (HOXD1, HOXD10, HOXD11, SDC1, STAT6, MYF5).
The effectors of the RA response, the RA receptors (RARs), are ligand-dependent transcription factors that are central regulators of a wide range of biological processes. RA is a potent inhibitor of tumorigenesis by activating networks of genes via RARs (94) and generally promotes differentiation in various contexts (95). The RAR gamma (RARG) was a top hit in 3′LIFE and has been linked to conflicting aspects of tumorigenesis. RARG promotes differentiation and cell cycle arrest in keratinocytes (96) and neuroblastoma (97), and differentiation, apoptosis (98) and inhibition of invasiveness in melanomas (99). Conversely, RARG has been shown to have oncogenic properties in carcinomas by activating the Akt/NF-kβ and Wnt/β-catenin pathways (100,101). NCOR2/SMRT and NCOA6/ASC2 are both nuclear receptor cofactors that physically interact with the RARs. NCOR2/SMRT has conflicting reports of its contribution to tumorigenesis dependent on tumor type, but is responsible for integrating estrogen and RAR signals by recruiting chromatin remodeling complexes that repress target gene transcription in the absence of the ligand hormone (102). In contrast, NCOA6/ASC2 is a nuclear receptor coactivator that outcompetes nuclear corepressors in binding nuclear receptors in the presence of RA, resulting in transcriptional activation of RAR target genes. NCOA6/ASC2 is also frequently upregulated in various tumors (103,104). ASCL2 is a transcription factor that is critical in the maintenance of adult intestinal stem cells and interacts with RARs via direct interaction with NCOA6 (105).
Interestingly, we also identified several downstream transcriptional targets of RA signaling, including HOXD1, HOXD10 and HOXD11, whose genomic locus also contains the miR-10b gene itself. The entire HOXD gene cluster responds to RA treatment by sequentially activating the transcription of the HOXD genes, generally promoting the differentiation of a variety of tissues (106,107). HOXD10 is the only bioinformatically predicted member of the HOXD cluster and was the first experimentally validated target of miR-10b, yet we identify three members of this cluster as targets of miR-10b and demonstrate that miR-10b targets HOXD11 by a single non-canonical target site (Figure 3C). SDC1/CD138 is a cell surface proteoglycan that plays a role in adhesion to the extracellular matrix (108), is frequently downregulated in multiple myelomas (109) and other cancers and is upregulated in response to RA treatment (110,111). STAT6 is a transcription factor that is a key effector of IL4-mediated signaling and promotes growth inhibition, apoptosis and differentiation in breast tumors (112,113). STAT6 is also transcriptionally activated in response to RA treatment in T-helper cells (113) and requires interaction with a nuclear receptor coactivator (NCOA1) to enact the IL4 response in hepatocytes (114). Lastly, MYF5, a transcription factor central to skeletal muscle differentiation, is also activated in response to RA in the developing limb bud (115). Among these 10 targets of miR-10b, NCOR2, NCOA6, HOXD10 and SDC1 are the only bioinformatically predicted targets (Figure 2A), and RARG, NCOR2, NCOA6, HOXD1, HOXD10, SDC1 and STAT6 contain canonical seed elements within their 3′UTRs (Figure 2C).
The identification of target genes associated with RAS signaling (let-7c) and RA signaling (miR-10b) by 3′LIFE is consistent with the known roles of these miRNAs as either negative or positive regulators of tumorigenesis, respectively, and potentially broadens the scope by which they target these pathways. Furthermore, the novel, non-canonical and unpredicted targets within these pathways suggest that unbiased, high-throughput approaches to identify miRNA targets is a productive strategy to identify regulatory networks targeted by miRNAs.
In conclusion, our study suggests that 3′LIFE is a powerful method to identify novel and non-canonical miRNA targets, and to identify mechanisms by which miRNAs contribute to biological processes. 3′LIFE conveys several advantages over current methods. First, 3′LIFE is a comprehensive screen because it utilizes all functional elements present in each 3′UTR that are targeted by the query miRNA. Second, 3′LIFE is unbiased, since it probes one interaction at a time and does not rely on prior assumptions about target genes. Third, the detection of an miRNA target requires direct translational inhibition of the 3′UTR in the presence of the miRNA, providing a functional measure of targeting. Fourth, the sensitivity of the assay is greatly improved by its high-throughput nature, revealing subtle targeting of multiple genes within regulatory networks. Lastly, 3′LIFE is flexible in that it can be adapted to detect other functional elements in 3′UTRs targeted by non-coding RNAs and RNA-binding proteins. Although 3′LIFE is a powerful experimental tool to detect miRNA targets in high-throughput, the complementary wet-lab and bioinformatic approaches described above are informative to refine, validate and expand the results obtained by this assay. While the biological relevance of miRNA targets identified by 3′LIFE cannot be assigned, this assay provides rapid detection and initial validation of direct miRNA/3′UTR target interaction at a scale not possible with current methods.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [1–116].
Acknowledgments
We thank John Chaput for providing initial vectors needed for the experiments; Stephen Blazie, Cody Babb, Carine Otto, Karen Anderson, Josh LaBaer, John Chaput and Yung Chang for advice and discussion; Karen Anderson, John Chaput and Josh LaBaer for sharing reagents and instrumentation; and Michael Gaskin and Andrea Throop for technical advice and protocols. Justin Wolter is a Maher scholar and thanks the Maher family for their generous support.
FUNDING
College of Liberal Arts and Science; Biodesign Institute at Arizona State University. Funding for open access charge: College of Liberal Arts and Science, School of Life Sciences at Arizona State University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 3.Garzon R., Marcucci G., Croce C.M. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat. Rev. Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esquela-Kerscher A., Slack F.J. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 5.Friedman R.C., Farh K.K., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 7.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krek A., Grun D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 9.Easow G., Teleman A.A., Cohen S.M. Isolation of microRNA targets by miRNP immunopurification. RNA. 2007;13:1198–1204. doi: 10.1261/rna.563707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selbach M., Schwanhausser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 11.Zhou P., Xu W., Peng X., Luo Z., Xing Q., Chen X., Hou C., Liang W., Zhou J., Wu X., et al. Large-scale screens of miRNA-mRNA interactions unveiled that the 3’UTR of a gene is targeted by multiple miRNAs. PLoS ONE. 2013;8:e68204. doi: 10.1371/journal.pone.0068204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chi S.W., Hannon G.J., Darnell R.B. An alternative mode of microRNA target recognition. Nat. Struct. Mol. Biol. 2012;19:321–327. doi: 10.1038/nsmb.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baek D., Villen J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Azzouzi I., Moest H., Winkler J., Fauchere J.C., Gerber A.P., Wollscheid B., Stoffel M., Schmugge M., Speer O. MicroRNA-96 directly inhibits gamma-globin expression in human erythropoiesis. PLoS ONE. 2011;6:e22838. doi: 10.1371/journal.pone.0022838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cevec M., Thibaudeau C., Plavec J. NMR structure of the let-7 miRNA interacting with the site LCS1 of lin-41 mRNA from Caenorhabditis elegans. Nucleic Acids Res. 2010;38:7814–7821. doi: 10.1093/nar/gkq640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J., Zhang X., Lentz C., Abi-Daoud M., Pare G.C., Yang X., Feilotter H.E., Tron V.A. miR-193b Regulates Mcl-1 in Melanoma. Am. J. Pathol. 2011;179:2162–2168. doi: 10.1016/j.ajpath.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiao L.R., Frampton A.E., Jacob J., Pellegrino L., Krell J., Giamas G., Tsim N., Vlavianos P., Cohen P., Ahmad R., et al. MicroRNAs targeting oncogenes are down-regulated in pancreatic malignant transformation from benign tumors. PLoS ONE. 2012;7:e32068. doi: 10.1371/journal.pone.0032068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lal A., Navarro F., Maher C.A., Maliszewski L.E., Yan N., O'Day E., Chowdhury D., Dykxhoorn D.M., Tsai P., Hofmann O., et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3’UTR microRNA recognition elements. Mol. Cell. 2009;35:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu L., Belasco J.G. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol. Cell. Biol. 2005;25:9198–9208. doi: 10.1128/MCB.25.21.9198-9208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shin C., Nam J.W., Farh K.K., Chiang H.R., Shkumatava A., Bartel D.P. Expanding the microRNA targeting code: functional sites with centered pairing. Mol. Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M., Jr, Jungkamp A.C., Munschauer M., et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Majoros W.H., Lekprasert P., Mukherjee N., Skalsky R.L., Corcoran D.L., Cullen B.R., Ohler U. MicroRNA target site identification by integrating sequence and binding information. Nat. Methods. 2013;10:630–633. doi: 10.1038/nmeth.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dyer B.W., Ferrer F.A., Klinedinst D.K., Rodriguez R. A noncommercial dual luciferase enzyme assay system for reporter gene analysis. Anal. Biochem. 2000;282:158–161. doi: 10.1006/abio.2000.4605. [DOI] [PubMed] [Google Scholar]
- 24.Boutz D.R., Collins P.J., Suresh U., Lu M., Ramirez C.M., Fernandez-Hernando C., Huang Y., Abreu Rde S., Le S.Y., Shapiro B.A., et al. Two-tiered approach identifies a network of cancer and liver disease-related genes regulated by miR-122. J. Biol. Chem. 2011;286:18066–18078. doi: 10.1074/jbc.M110.196451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaken J., Mohamedali A.M., Jiang J., Malik F., Stangl D., Smith A.E., Chronis C., Kulasekararaj A.G., Thomas N.S., Farzaneh F., et al. A functional assay for microRNA target identification and validation. Nucleic Acids Res. 2012;40:e75. doi: 10.1093/nar/gks145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shell S., Park S.M., Radjabi A.R., Schickel R., Kistner E.O., Jewell D.A., Feig C., Lengyel E., Peter M.E. Let-7 expression defines two differentiation stages of cancer. Proc. Natl Acad. Sci. U.S.A. 2007;104:11400–11405. doi: 10.1073/pnas.0704372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Childs G., Fazzari M., Kung G., Kawachi N., Brandwein-Gensler M., McLemore M., Chen Q., Burk R.D., Smith R.V., Prystowsky M.B., et al. Low-level expression of microRNAs let-7d and miR-205 are prognostic markers of head and neck squamous cell carcinoma. Am. J. Pathol. 2009;174:736–745. doi: 10.2353/ajpath.2009.080731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takamizawa J., Konishi H., Yanagisawa K., Tomida S., Osada H., Endoh H., Harano T., Yatabe Y., Nagino M., Nimura Y., et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 29.Sakurai M., Miki Y., Masuda M., Hata S., Shibahara Y., Hirakawa H., Suzuki T., Sasano H. LIN28: a regulator of tumor-suppressing activity of let-7 microRNA in human breast cancer. J. Steroid Biochem. Mol. Biol. 2012;131:101–106. doi: 10.1016/j.jsbmb.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 30.Johnson S.M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K.L., Brown D., Slack F.J. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 31.Papaioannou G., Inloes J.B., Nakamura Y., Paltrinieri E., Kobayashi T. let-7 and miR-140 microRNAs coordinately regulate skeletal development. Proc. Natl Acad. Sci. U.S.A. 2013;110:E3291–E3300. doi: 10.1073/pnas.1302797110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Umemura S., Shirane M., Takekoshi S., Kusakabe T., Itoh J., Egashira N., Tokuda Y., Mori K., Osamura Y.R. Overexpression of E2F-5 correlates with a pathological basal phenotype and a worse clinical outcome. Br. J. Cancer. 2009;100:764–771. doi: 10.1038/sj.bjc.6604900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han H.B., Gu J., Zuo H.J., Chen Z.G., Zhao W., Li M., Ji D.B., Lu Y.Y., Zhang Z.Q. Let-7c functions as a metastasis suppressor by targeting MMP11 and PBX3 in colorectal cancer. J. Pathol. 2012;226:544–555. doi: 10.1002/path.3014. [DOI] [PubMed] [Google Scholar]
- 34.Ma L., Teruya-Feldstein J., Weinberg R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 35.Bourguignon L.Y., Wong G., Earle C., Krueger K., Spevak C.C. Hyaluronan-CD44 interaction promotes c-Src-mediated twist signaling, microRNA-10b expression, and RhoA/RhoC up-regulation, leading to Rho-kinase-associated cytoskeleton activation and breast tumor cell invasion. J. Biol. Chem. 2010;285:36721–36735. doi: 10.1074/jbc.M110.162305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foley N.H., Bray I., Watters K.M., Das S., Bryan K., Bernas T., Prehn J.H., Stallings R.L. MicroRNAs 10a and 10b are potent inducers of neuroblastoma cell differentiation through targeting of nuclear receptor corepressor 2. Cell Death Differ. 2011;18:1089–1098. doi: 10.1038/cdd.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gabriely G., Teplyuk N.M., Krichevsky A.M. Context effect: microRNA-10b in cancer cell proliferation, spread and death. Autophagy. 2011;7:1384–1386. doi: 10.4161/auto.7.11.17371. [DOI] [PubMed] [Google Scholar]
- 38.Nakata K., Ohuchida K., Mizumoto K., Kayashima T., Ikenaga N., Sakai H., Lin C., Fujita H., Otsuka T., Aishima S., et al. MicroRNA-10b is overexpressed in pancreatic cancer, promotes its invasiveness, and correlates with a poor prognosis. Surgery. 2011;150:916–922. doi: 10.1016/j.surg.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 39.Sun L., Yan W., Wang Y., Sun G., Luo H., Zhang J., Wang X., You Y., Yang Z., Liu N. MicroRNA-10b induces glioma cell invasion by modulating MMP-14 and uPAR expression via HOXD10. Brain Res. 2011;1389:9–18. doi: 10.1016/j.brainres.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 40.Hsu S.D., Lin F.M., Wu W.Y., Liang C., Huang W.C., Chan W.L., Tsai W.T., Chen G.Z., Lee C.J., Chiu C.M., et al. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011;39:D163–D169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian Y., Luo A., Cai Y., Su Q., Ding F., Chen H., Liu Z. MicroRNA-10b promotes migration and invasion through KLF4 in human esophageal cancer cell lines. J. Biol. Chem. 2010;285:7986–7994. doi: 10.1074/jbc.M109.062877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paraskevopoulou M.D., Georgakilas G., Kostoulas N., Vlachos I.S., Vergoulis T., Reczko M., Filippidis C., Dalamagas T., Hatzigeorgiou A.G. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41:W169–W173. doi: 10.1093/nar/gkt393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vergoulis T., Vlachos I.S., Alexiou P., Georgakilas G., Maragkakis M., Reczko M., Gerangelos S., Koziris N., Dalamagas T., Hatzigeorgiou A.G. TarBase 6.0: capturing the exponential growth of miRNA targets with experimental support. Nucleic Acids Res. 2012;40:D222–D229. doi: 10.1093/nar/gkr1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seiler C.Y., Park J.G., Sharma A., Hunter P., Surapaneni P., Sedillo C., Field J., Algar R., Price A., Steel J. DNASU plasmid and PSI:Biology-Materials repositories: resources to accelerate biological research. Nucleic Acids Res. 2014;42:D1253–D1260. doi: 10.1093/nar/gkt1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qiu L., Wang H., Xia X., Zhou H., Xu Z. A construct with fluorescent indicators for conditional expression of miRNA. BMC Biotechnol. 2008;8:77. doi: 10.1186/1472-6750-8-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mangone M., Manoharan A.P., Thierry-Mieg D., Thierry-Mieg J., Han T., Mackowiak S.D., Mis E., Zegar C., Gutwein M.R., Khivansara V., et al. The landscape of C. elegans 3’UTRs. Science. 2010;329:432–435. doi: 10.1126/science.1191244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takagaki Y., Manley J.L. RNA recognition by the human polyadenylation factor CstF. Mol. Cell. Biol. 1997;17:3907–3914. doi: 10.1128/mcb.17.7.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 49.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Helwak A., Kudla G., Dudnakova T., Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qin J.Y., Zhang L., Clift K.L., Hulur I., Xiang A.P., Ren B.Z., Lahn B.T. Systematic comparison of constitutive promoters and the doxycycline-inducible promoter. PLoS ONE. 2010;5:e10611. doi: 10.1371/journal.pone.0010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Farh K.K., Grimson A., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 55.Nielsen C.B., Shomron N., Sandberg R., Hornstein E., Kitzman J., Burge C.B. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA. 2007;13:1894–1910. doi: 10.1261/rna.768207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhattacharyya S.N., Habermacher R., Martine U., Closs E.I., Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 57.Fu Y., Dominissini D., Rechavi G., He C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat. Rev. Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 58.Shepard P.J., Choi E.A., Lu J., Flanagan L.A., Hertel K.J., Shi Y. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA. 2011;17:761–772. doi: 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yan J., Marr T.G. Computational analysis of 3’-ends of ESTs shows four classes of alternative polyadenylation in human, mouse, and rat. Genome Res. 2005;15:369–375. doi: 10.1101/gr.3109605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mayr C., Bartel D.P. Widespread shortening of 3’UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sandberg R., Neilson J.R., Sarma A., Sharp P.A., Burge C.B. Proliferating cells express mRNAs with shortened 3’ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin Y., Li Z., Ozsolak F., Kim S.W., Arango-Argoty G., Liu T.T., Tenenbaum S.A., Bailey T., Monaghan A.P., Milos P.M. An in-depth map of polyadenylation sites in cancer. Nucleic Acids Res. 2012;40:8460–8471. doi: 10.1093/nar/gks637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ebert M.S., Sharp P.A. Roles for MicroRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pelaez N., Carthew R.W. Biological robustness and the role of microRNAs: a network perspective. Curr. Top. Dev. Biol. 2012;99:237–255. doi: 10.1016/B978-0-12-387038-4.00009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Berezikov E. Evolution of microRNA diversity and regulation in animals. Nat. Rev. Genet. 2011;12:846–860. doi: 10.1038/nrg3079. [DOI] [PubMed] [Google Scholar]
- 66.Xu J., Zhang R., Shen Y., Liu G., Lu X., Wu C.I. The evolution of evolvability in microRNA target sites in vertebrates. Genome Res. 2013;23:1810–1816. doi: 10.1101/gr.148916.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen K., Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat. Rev. Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 68.Chin L.J., Ratner E., Leng S., Zhai R., Nallur S., Babar I., Muller R.U., Straka E., Su L., Burki E.A., et al. A SNP in a let-7 microRNA complementary site in the KRAS 3’ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Medale-Giamarchi C., Lajoie-Mazenc I., Malissein E., Meunier E., Couderc B., Berge Y., Filleron T., Keller L., Marty C., Lacroix-Triki M., et al. RhoB modifies estrogen responses in breast cancer cells by influencing expression of the estrogen receptor. Breast Cancer Res. 2013;15:R6. doi: 10.1186/bcr3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kazerounian S., Gerald D., Huang M., Chin Y.R., Udayakumar D., Zheng N., O'Donnell R.K., Perruzzi C., Mangiante L., Pourat J., et al. RhoB differentially controls Akt function in tumor cells and stromal endothelial cells during breast tumorigenesis. Cancer Res. 2013;73:50–61. doi: 10.1158/0008-5472.CAN-11-3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ridley A.J. RhoA, RhoB and RhoC have different roles in cancer cell migration. J. Microsc. 2013;251:242–249. doi: 10.1111/jmi.12025. [DOI] [PubMed] [Google Scholar]
- 72.Shepelev M.V., Korobko I.V. The RHOV gene is overexpressed in human non-small cell lung cancer. Cancer Genet. 2013;206:393–397. doi: 10.1016/j.cancergen.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 73.Pillai S., Rizwani W., Li X., Rawal B., Nair S., Schell M.J., Bepler G., Haura E., Coppola D., Chellappan S. ID1 facilitates the growth and metastasis of non-small cell lung cancer in response to nicotinic acetylcholine receptor and epidermal growth factor receptor signaling. Mol. Cell. Biol. 2011;31:3052–3067. doi: 10.1128/MCB.01311-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kalas W., Yu J.L., Milsom C., Rosenfeld J., Benezra R., Bornstein P., Rak J. Oncogenes and Angiogenesis: down-regulation of thrombospondin-1 in normal fibroblasts exposed to factors from cancer cells harboring mutant ras. Cancer Res. 2005;65:8878–8886. doi: 10.1158/0008-5472.CAN-05-1479. [DOI] [PubMed] [Google Scholar]
- 75.Swarbrick A., Roy E., Allen T., Bishop J.M. Id1 cooperates with oncogenic Ras to induce metastatic mammary carcinoma by subversion of the cellular senescence response. Proc. Natl Acad. Sci. U.S.A. 2008;105:5402–5407. doi: 10.1073/pnas.0801505105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dai C., Santagata S., Tang Z., Shi J., Cao J., Kwon H., Bronson R.T., Whitesell L., Lindquist S. Loss of tumor suppressor NF1 activates HSF1 to promote carcinogenesis. J. Clin. Invest. 2012;122:3742–3754. doi: 10.1172/JCI62727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dai C., Whitesell L., Rogers A.B., Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005–1018. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xi C., Hu Y., Buckhaults P., Moskophidis D., Mivechi N.F. Heat shock factor Hsf1 cooperates with ErbB2 (Her2/Neu) protein to promote mammary tumorigenesis and metastasis. J. Biol. Chem. 2012;287:35646–35657. doi: 10.1074/jbc.M112.377481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stanhill A., Levin V., Hendel A., Shachar I., Kazanov D., Arber N., Kaminski N., Engelberg D. Ha-ras(val12) induces HSP70b transcription via the HSE/HSF1 system, but HSP70b expression is suppressed in Ha-ras(val12)-transformed cells. Oncogene. 2006;25:1485–1495. doi: 10.1038/sj.onc.1209193. [DOI] [PubMed] [Google Scholar]
- 80.Mivechi N.F., Giaccia A.J. Mitogen-activated protein kinase acts as a negative regulator of the heat shock response in NIH3T3 cells. Cancer Res. 1995;55:5512–5519. [PubMed] [Google Scholar]
- 81.Liu Y., Hiraiwa Y., Liu E., Kurata H., Thant A.A., Matsuda S., Hamaguchi M. Suppression of cell spreading by v-Crk requires Ras-MEK-MAP kinase signaling. Oncogene. 2001;20:5908–5912. doi: 10.1038/sj.onc.1204738. [DOI] [PubMed] [Google Scholar]
- 82.Mochizuki N., Ohba Y., Kobayashi S., Otsuka N., Graybiel A.M., Tanaka S., Matsuda M. Crk activation of JNK via C3G and R-Ras. J. Biol. Chem. 2000;275:12667–12671. doi: 10.1074/jbc.275.17.12667. [DOI] [PubMed] [Google Scholar]
- 83.Peeper D.S., Shvarts A., Brummelkamp T., Douma S., Koh E.Y., Daley G.Q., Bernards R. A functional screen identifies hDRIL1 as an oncogene that rescues RAS-induced senescence. Nat. Cell Biol. 2002;4:148–153. doi: 10.1038/ncb742. [DOI] [PubMed] [Google Scholar]
- 84.Fukuyo Y., Takahashi A., Hara E., Horikoshi N., Pandita T.K., Nakajima T. E2FBP1 antagonizes the p16(INK4A)-Rb tumor suppressor machinery for growth suppression and cellular senescence by regulating promyelocytic leukemia protein stability. Int. J. Oral Sci. 2011;3:200–208. doi: 10.4248/IJOS11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wajapeyee N., Malonia S.K., Palakurthy R.K., Green M.R. Oncogenic RAS directs silencing of tumor suppressor genes through ordered recruitment of transcriptional repressors. Genes Dev. 2013;27:2221–2226. doi: 10.1101/gad.227413.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bechtel W., McGoohan S., Zeisberg E.M., Muller G.A., Kalbacher H., Salant D.J., Muller C.A., Kalluri R., Zeisberg M. Methylation determines fibroblast activation and fibrogenesis in the kidney. Nat. Med. 2010;16:544–550. doi: 10.1038/nm.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gazin C., Wajapeyee N., Gobeil S., Virbasius C.M., Green M.R. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature. 2007;449:1073–1077. doi: 10.1038/nature06251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fujii S., Fukamachi K., Tsuda H., Ito K., Ito Y., Ochiai A. RAS oncogenic signal upregulates EZH2 in pancreatic cancer. Biochem. Biophys. Res. Commun. 2012;417:1074–1079. doi: 10.1016/j.bbrc.2011.12.099. [DOI] [PubMed] [Google Scholar]
- 89.Cai H., Memarzadeh S., Stoyanova T., Beharry Z., Kraft A.S., Witte O.N. Collaboration of Kras and androgen receptor signaling stimulates EZH2 expression and tumor-propagating cells in prostate cancer. Cancer Res. 2012;72:4672–4681. doi: 10.1158/0008-5472.CAN-12-0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Min J., Zaslavsky A., Fedele G., McLaughlin S.K., Reczek E.E., De Raedt T., Guney I., Strochlic D.E., Macconaill L.E., Beroukhim R., et al. An oncogene-tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-kappaB. Nat. Med. 2010;16:286–294. doi: 10.1038/nm.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Weiss F.U., Marques I.J., Woltering J.M., Vlecken D.H., Aghdassi A., Partecke L.I., Heidecke C.D., Lerch M.M., Bagowski C.P. Retinoic acid receptor antagonists inhibit miR-10a expression and block metastatic behavior of pancreatic cancer. Gastroenterology. 2009;137:2136–2145;. doi: 10.1053/j.gastro.2009.08.065. e2131–2137. [DOI] [PubMed] [Google Scholar]
- 92.Tehler D., Hoyland-Kroghsbo N.M., Lund A.H. The miR-10 microRNA precursor family. RNA Biol. 2011;8:728–734. doi: 10.4161/rna.8.5.16324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ma L. Role of miR-10b in breast cancer metastasis. Breast Cancer Res. 2010;12:210. doi: 10.1186/bcr2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hua S., Kittler R., White K.P. Genomic antagonism between retinoic acid and estrogen signaling in breast cancer. Cell. 2009;137:1259–1271. doi: 10.1016/j.cell.2009.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Niederreither K., Dolle P. Retinoic acid in development: towards an integrated view. Nat. Rev. Genet. 2008;9:541–553. doi: 10.1038/nrg2340. [DOI] [PubMed] [Google Scholar]
- 96.Chen C.F., Goyette P., Lohnes D. RARgamma acts as a tumor suppressor in mouse keratinocytes. Oncogene. 2004;23:5350–5359. doi: 10.1038/sj.onc.1207682. [DOI] [PubMed] [Google Scholar]
- 97.Goranov B.B., Campbell Hewson Q.D., Pearson A.D., Redfern C.P. Overexpression of RARgamma increases death of SH-SY5Y neuroblastoma cells in response to retinoic acid but not fenretinide. Cell Death Differ. 2006;13:676–679. doi: 10.1038/sj.cdd.4401824. [DOI] [PubMed] [Google Scholar]
- 98.Spanjaard R.A., Ikeda M., Lee P.J., Charpentier B., Chin W.W., Eberlein T.J. Specific activation of retinoic acid receptors (RARs) and retinoid X receptors reveals a unique role for RARgamma in induction of differentiation and apoptosis of S91 melanoma cells. J. Biol. Chem. 1997;272:18990–18999. doi: 10.1074/jbc.272.30.18990. [DOI] [PubMed] [Google Scholar]
- 99.Zhao X., Graves C., Ames S.J., Fisher D.E., Spanjaard R.A. Mechanism of regulation and suppression of melanoma invasiveness by novel retinoic acid receptor-gamma target gene carbohydrate sulfotransferase 10. Cancer Res. 2009;69:5218–5225. doi: 10.1158/0008-5472.CAN-09-0705. [DOI] [PubMed] [Google Scholar]
- 100.Huang G.L., Luo Q., Rui G., Zhang W., Zhang Q.Y., Chen Q.X., Shen D.Y. Oncogenic activity of retinoic acid receptor gamma is exhibited through activation of the Akt/NF-kappaB and Wnt/beta-catenin pathways in cholangiocarcinoma. Mol. Cell. Biol. 2013;33:3416–3425. doi: 10.1128/MCB.00384-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yan T.D., Wu H., Zhang H.P., Lu N., Ye P., Yu F.H., Zhou H., Li W.G., Cao X., Lin Y.Y., et al. Oncogenic potential of retinoic acid receptor-gamma in hepatocellular carcinoma. Cancer Res. 2010;70:2285–2295. doi: 10.1158/0008-5472.CAN-09-2968. [DOI] [PubMed] [Google Scholar]
- 102.Battaglia S., Maguire O., Campbell M.J. Transcription factor co-repressors in cancer biology: roles and targeting. Int. J. Cancer. 2010;126:2511–2519. doi: 10.1002/ijc.25181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee S., Lee D.K., Dou Y., Lee J., Lee B., Kwak E., Kong Y.Y., Lee S.K., Roeder R.G., Lee J.W. Coactivator as a target gene specificity determinant for histone H3 lysine 4 methyltransferases. Proc. Natl Acad. Sci. U.S.A. 2006;103:15392–15397. doi: 10.1073/pnas.0607313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kong H.J., Yu H.J., Hong S., Park M.J., Choi Y.H., An W.G., Lee J.W., Cheong J. Interaction and functional cooperation of the cancer-amplified transcriptional coactivator activating signal cointegrator-2 and E2F-1 in cell proliferation. Mol. Cancer Res. 2003;1:948–958. [PubMed] [Google Scholar]
- 105.Goo Y.H., Sohn Y.C., Kim D.H., Kim S.W., Kang M.J., Jung D.J., Kwak E., Barlev N.A., Berger S.L., Chow V.T., et al. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol. Cell. Biol. 2003;23:140–149. doi: 10.1128/MCB.23.1.140-149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zha Y., Ding E., Yang L., Mao L., Wang X., McCarthy B.A., Huang S., Ding H.F. Functional dissection of HOXD cluster genes in regulation of neuroblastoma cell proliferation and differentiation. PLoS ONE. 2012;7:e40728. doi: 10.1371/journal.pone.0040728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Merrill R.A., Ahrens J.M., Kaiser M.E., Federhart K.S., Poon V.Y., Clagett-Dame M. All-trans retinoic acid-responsive genes identified in the human SH-SY5Y neuroblastoma cell line and their regulated expression in the nervous system of early embryos. Biol. Chem. 2004;385:605–614. doi: 10.1515/BC.2004.075. [DOI] [PubMed] [Google Scholar]
- 108.Ishikawa T., Kramer R.H. Sdc1 negatively modulates carcinoma cell motility and invasion. Exp. Cell Res. 2010;316:951–965. doi: 10.1016/j.yexcr.2009.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yang Y., Shi J., Tolomelli G., Xu H., Xia J., Wang H., Zhou W., Zhou Y., Das S., Gu Z., et al. RARalpha2 expression confers myeloma stem cell features. Blood. 2013;122:1437–1447. doi: 10.1182/blood-2013-02-482919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Barber N., Belov L., Christopherson R.I. All-trans retinoic acid induces different immunophenotypic changes on human HL60 and NB4 myeloid leukaemias. Leuk. Res. 2008;32:315–322. doi: 10.1016/j.leukres.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 111.Chen Q., Mosovsky K.L., Ross A.C. Retinoic acid and alpha-galactosylceramide regulate the expression of costimulatory receptors and transcription factors responsible for B cell activation and differentiation. Immunobiology. 2013;218:1477–1487. doi: 10.1016/j.imbio.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gooch J.L., Christy B., Yee D. STAT6 mediates interleukin-4 growth inhibition in human breast cancer cells. Neoplasia. 2002;4:324–331. doi: 10.1038/sj.neo.7900248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dawson H.D., Collins G., Pyle R., Key M., Weeraratna A., Deep-Dixit V., Nadal C.N., Taub D.D. Direct and indirect effects of retinoic acid on human Th2 cytokine and chemokine expression by human T lymphocytes. BMC immunol. 2006;7:27. doi: 10.1186/1471-2172-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Litterst C.M., Pfitzner E. Transcriptional activation by STAT6 requires the direct interaction with NCoA-1. J. Biol. Chem. 2001;276:45713–45721. doi: 10.1074/jbc.M108132200. [DOI] [PubMed] [Google Scholar]
- 115.Reijntjes S., Francis-West P., Mankoo B.S. Retinoic acid is both necessary for and inhibits myogenic commitment and differentiation in the chick limb. Int. J. Dev. Biol. 2010;54:125–134. doi: 10.1387/ijdb.082783sr. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


