Abstract
Chromatin dynamics crucially contributes to gene regulation. Studies of the yeast PHO5 promoter were key to establish this nowadays accepted view and continuously provide mechanistic insight in chromatin remodeling and promoter regulation, both on single locus as well as on systems level. The PHO5 promoter is a context independent chromatin switch module where in the repressed state positioned nucleosomes occlude transcription factor sites such that nucleosome remodeling is a prerequisite for and not consequence of induced gene transcription. This massive chromatin transition from positioned nucleosomes to an extensive hypersensitive site, together with respective transitions at the co-regulated PHO8 and PHO84 promoters, became a prime model for dissecting how remodelers, histone modifiers and chaperones co-operate in nucleosome remodeling upon gene induction. This revealed a surprisingly complex cofactor network at the PHO5 promoter, including five remodeler ATPases (SWI/SNF, RSC, INO80, Isw1, Chd1), and demonstrated for the first time histone eviction in trans as remodeling mode in vivo. Recently, the PHO5 promoter and the whole PHO regulon were harnessed for quantitative analyses and computational modeling of remodeling, transcription factor binding and promoter input-output relations such that this rewarding single-locus model becomes a paradigm also for theoretical and systems approaches to gene regulatory networks.
Chromatin structure and dynamics contribute importantly to the regulation of gene expression in eukaryotes (1). This view is nowadays broadly accepted and inspires an ever growing number of studies, in part also as chromatin is seen as a major mechanism of epigenetic information (2–4). However, such consensus was not always the case, and many molecular mechanisms, even most basic ones like the determinants for nucleosome positioning (5–7), are still controversial. Nonetheless, our understanding of the relationship between chromatin and gene expression matured to an amazing extent during the last four decades. We review here how this development was pioneered and continues to be inspired by studies of the budding yeast PHO5 promoter (8).
CLASSICAL STUDIES: PAST AND PRESENT
The PHO regulon and PHO induction
PHO5 is one of ∼20 PHO regulon genes in Saccharomyces cerevisiae that are regulated by the availability of phosphate and were first identified genetically by the Oshima group (9). PHO regulated genes are repressed if inorganic phosphate (Pi) is abundant and induced upon phosphate starvation. Importantly, this refers to intracellular Pi levels. Just removal of extracellular phosphate is not sufficient for PHO induction (10) as yeast cells have ample phosphate stores, especially in the form of polyphosphate in the vacuole (11–14). Only if this storage is used up, e.g. by DNA and phospholipid synthesis during proliferation or cell growth, without repletion from extracellular sources, physiological PHO induction will occur. So the classical induction protocol is incubation of replicating yeast cells in phosphate-depleted medium. Induction kinetics depends on the amount of residual extracellular phosphate—this is why phosphate-free medium is preferred to obtain faster and better defined induction conditions—and the extent of intracellular storage, which can be impaired by mutating enzymes of the polyphosphate storage pathway (14). The actual sensor of intracellular phosphate is still unclear, but signal transduction eventually leads to activation of the principal PHO regulator, the transactivator Pho4 (9,15). Pho4 is a basic helix-turn-helix protein (16) that specifically binds to E-box (CACGTG) based sequence elements upstream of Pho4-regulated genes (UASp = Upstream Activating Sequence phosphate) (17–19). Many UASp elements are close to binding sites of the pleiotropic homeobox-type transcription cofactor Pho2, which binds co-operatively with Pho4 at the PHO5 promoter and can increase both the binding affinity as well as the transactivation potential of Pho4 (19–21). Pho4 itself is constitutively expressed (22), but regulated by phosphorylation through the cyclin/cyclin-dependent-kinase pair Pho80/Pho85, which is under negative control of its inhibitor Pho81 (Figure 1; (11,23–28)). Pho81 binds constitutively to Pho80/Pho85, but is turned into an inhibitor only during phosphate starvation 27. This is mediated by binding of a certain inositol polyphosphate (4/6-IP7), which is increasingly produced upon phosphate starvation (29,30). Inositol polyphosphates also have a role in PHO5 repression (31). Pho4 Phosphorylation expedites nuclear export, blocks nuclear import and inhibits the Pho4–Pho2 interaction and consequently decreases Pho4 binding to the promoter. Therefore, interference with this phosphorylation allows circumvention of the physiological signal transduction pathway and PHO induction despite the presence of phosphate. For example, the deletion (32), chemically (33) or temperature-induced (10,32) inactivation of Pho80 or Pho85, or simply the out-titration of the Pho80/Pho85 activity by overexpression of PHO4 (34) will induce PHO genes, although not always to full induction levels. As intracellular phosphate and polyphosphate levels are transiently depleted during the cell cycle, also PHO5 expression oscillates with the cell cycle (35). Accordingly, medium with intermediate phosphate levels, e.g. standard yeast extract-peptone-dextrose (YPD) medium, may allow weak induction of PHO genes, particularly during mitosis. The Kladde group found that this mitotic induction is weaker than that upon phosphate starvation and is regulated not only through Pho4 and Pho2, but also by Mcm1 and Fkh2 (14,36).
Figure 1.
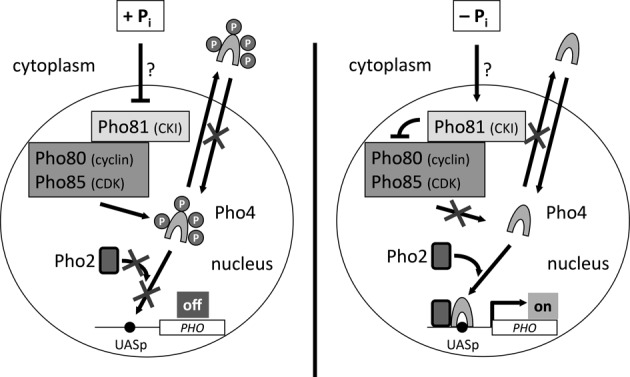
Schematics of PHO regulon signal transduction. For details see the main text (The PHO regulon and PHO induction).
Choosing the PHO5 promoter as model due to its massive chromatin transition upon activation
PHO5 encodes a secreted and glycosylated acidic phosphatase that scavenges phosphate from diverse extracellular substrates. As the Pho5 enzyme is located in the periplasm between the plasma membrane and cell wall in soluble form, its activity can be easily monitored with whole cells (37–39). It is non-essential and mutations do not matter under standard laboratory growth conditions making it amenable to extensive mutational analysis. The PHO5 promoter stood out due to extremely high induction levels (∼20–40 fold on mRNA and Pho5 enzyme activity level (40)) and was therefore even considered as a prospective biotechnology tool for recombinant protein production. However, the main interest in PHO5 was kindled after analysis of the PHO5 promoter chromatin structure in the repressed versus induced state (Figure 2a) (41–44). In the repressed state, there are five nucleosomes upstream of the transcription start site (TSS), numbered -1 to -5, with a short hypersensitive site of about 80 bp length between nucleosomes -2 and -3. This long linker was missed at first (44) but is of key importance as it harbors the low affinity UASp1 element 41and one Pho2 binding site (20,21). These nucleosomes are classic examples of well-positioned nucleosomes in yeast and were used as benchmark for genome-wide nucleosome mapping (45–47). As generally true, even these are not perfectly positioned but represent Gaussian distributions in ∼10 bp increments around an average midpoint (48,49) which probably explains why restriction enzyme accessibility in their linker regions reaches only ∼50% (41). Nonetheless, nucleosome positioning is precise enough such that the TATA box/TSS is covered by nucleosome -1 and a high affinity UASp2 element and two Pho2 sites by nucleosome -2. This ordered organization, especially nucleosomes -1 to -4, is remodeled into an extensive hypersensitive site (∼600 bp) upon promoter activation. Such a massive chromatin transition is rather exceptional. For most yeast genes promoter chromatin perturbations upon expression changes are less extensive, maybe involving just one nucleosome (46,48,50–51). Of note, hypersensitive sites have been recently renamed in the course of genome-wide nucleosome mapping as ‘nucleosome-free regions’ (NFR) or ‘nucleosome-depleted regions’ (NDR) (e.g. (45,52)).
Figure 2.
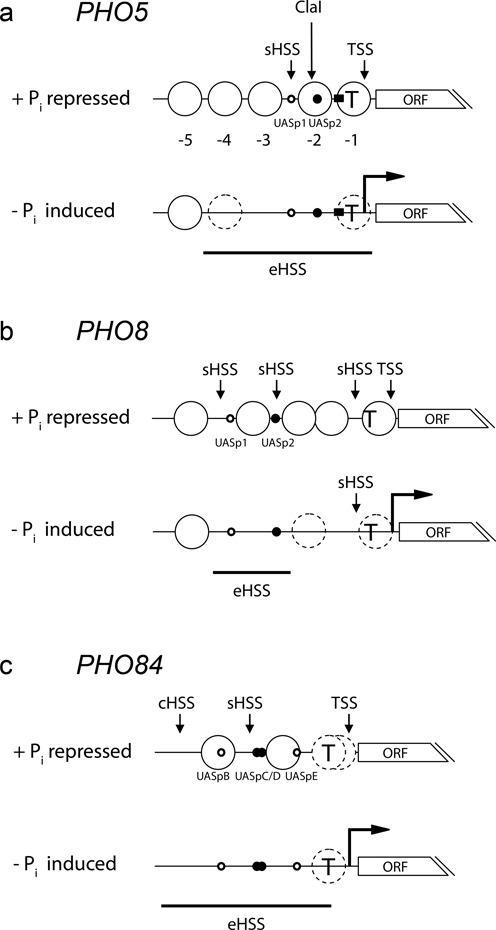
Nucleosome organization schematics for the repressed (+Pi) and induced (−Pi) states of PHO promoters. (a) PHO5 promoter organization. Canonical and well-positioned nucleosomes that largely inhibit nuclease access are symbolized by large solid circles and regions of intermediate nuclease accessibility or unclear nucleosome positioning with large stippled circles. Pho4 binding sites (UASp elements) of low and high affinity are represented by open or closed small circles, respectively. Closed squares denote Mcm1 and Fkh2 binding sites and broken arrows induced transcription. sHSS, eHSS and cHSS stand for short, extended and constitutive hypersensitive site, respectively. TSS: transcription start site; ORF: open reading frame; T: TATA box; ClaI: ClaI restriction site. All relative positions approximately to scale. (b), (c) as (a) but for the PHO8 and PHO84 promoter, respectively. See main text for references.
Overarching mechanistic questions
Since the discovery of the nucleosome (53–56) and the development of nucleosome mapping, especially via DNaseI indirect endlabeling (57,58), it became clear that nuclease hypersensitive sites were linked to functional DNA elements, like enhancers, promoters and replication origins, and that their appearance changed with biological conditions, e.g. during gene induction or differentiation (59). So there was a keen interest in the following mechanistic questions: is chromatin regulative, i.e. are chromatin changes cause or consequence of changes in DNA templated processes like transcription or replication? What is a hypersensitive site in molecular terms (‘anatomy of a hypersensitive site’ (60,61))? How are hypersensitive sites generated? For all these questions, the PHO5 promoter turned out to be one of the most instructive models.
Early basics on PHO5 promoter chromatin remodeling: chromatin regulates
Mainly Hörz and colleagues established the first basics of PHO5 promoter chromatin and its transition. The principal transcription regulator Pho4 turned out to be also the principal inducer of PHO5 promoter chromatin opening (34). Lack of Pho2 was compensated by overexpression of PHO4 and even the whole physiological context of PHO induction was irrelevant as the full chromatin transition was observed also at usually repressive high phosphate conditions if PHO4 was overexpressed (34). Further, the PHO5 promoter could be moved to a plasmid, i.e. to a different nuclear location, and even truncated until only nucleosomes -3 to -1 were left, flanked by prokaryotic sequences (vector backbone upstream, lacZ reporter downstream), without losing the positioned nucleosomes of the repressed state and the Pho4-triggered chromatin transition (43,62–63). So the PHO5 promoter is a compact chromatin switch module, independent of the physiological, nuclear and DNA sequence context that is essentially triggered by a single factor, Pho4. This allows mechanistic studies without confounding peripherals.
In the beginning, the massive chromatin transition seemed likely to be a consequence of the strongly activated PHO5 transcription. However, a series of studies from the Grunstein group showed that reducing histone levels in vivo led to up-regulation of many genes, including prominently PHO5, under otherwise repressive conditions (64–66). This suggested that nucleosomes are repressive and need to be removed prior to transcription initiation. Indeed, both at the SUC2 (67) and at the PHO5 promoter (62) a full chromatin transition was still possible upon induction even if gene transcription was abolished by deletion of the TATA box. Conversely, there is never substantial Pho5 activity without PHO5 promoter chromatin opening. This clearly established that chromatin remodeling is a prerequisite and not consequence of gene transcription. Just recently this basic finding was confirmed again for the PHO5 promoter by elaborate single molecule in vivo analysis and computational modeling (68).
Consistent with the pioneering Grunstein studies, impairment of histone reassembly by ablation of the histone chaperone Spt6 (69), which leads to reduced histone occupancy at the PHO5 locus, sustained elevated PHO5 transcription even after return to repressive high phosphate conditions (70). Similarly, a whole range of mutations that interfere with proper nucleosome assembly over coding regions, either affecting histone occupancy, modification, turnover or nucleosome positioning, alleviate the repressive effect of chromatin and allow transcription initiation from otherwise silent, so called ‘cryptic’ promoters (71,72).
Excluding other early concepts of nucleosome remodeling: not replication, not factor binding competition
In the late 1980s and early 1990s, it was unclear how nucleosomes could be remodeled in vivo. Until chromatin remodeling enzymes were recognized, polymerases were prominent candidates for nucleosome disruption and remodeling. Besides the process of transcription, it was proposed that DNA polymerase passage established a window of opportunity for DNA binding factors, like transcription factors, to occupy their binding sites, prevent nucleosome re-assembly and thereby generate a hypersensitive site (73). As physiological PHO induction required at least one round of replication—to deplete the intracellular phosphate stores—PHO5 promoter opening may have seemed as an example for this mechanism. However, by use of a temperature-sensitive pho80ts allele the Hörz group showed PHO5 promoter chromatin switching between the closed and open state in the absence of DNA replication (10).
Another prominent hypothesis was that factors with high affinity to DNA sites could compete away nucleosomes by sheer mass action. There was evidence from in vitro (74) and in vivo (75) studies where positioned nucleosomes containing one or multiple binding sites for the transactivator Gal4 were disassembled upon binding of Gal4, even only by binding the Gal4-DNA binding domain. However, the analogous experiment did not work for the PHO5 promoter. At this physiological example, in contrast to the engineered Gal4 site containing nucleosomes, it was rigorously shown that nucleosome -2 prevents Pho4 binding (76), and that this nucleosome was not displaced by mere binding competition even if the Pho4 DNA binding domain was overexpressed. The Pho4 activation domain was absolutely required to trigger chromatin opening (77). Nonetheless, this was not specific solely to the Pho4 activation domain. PHO5 promoter opening also worked with the VP16 (77) or the glucocorticoid receptor (78) activation domain fused to the Pho4 DNA binding domain, or if the Pho4 DNA binding domain was fused to the mediator subunit Gal11/Med15 (79), even though mediator has only a minor role otherwise (80) or with a PHO5 promoter variant activated by Gal4 instead of Pho4 (81,82). Attempts to dissect the Pho4 activation domain into parts responsible for either chromatin remodeling or for transactivation of transcription failed (83). Even though activation domains are known to interact with a surprising variety of cofactors it is still the common view that they cannot be subdivided into different parts that would target different interaction partners, probably because activation domains and their targets form so called ‘fuzzy complexes’ (84).
It should be noted that this series of clear negative evidence regarding the PHO5 promoter chromatin remodeling mechanism—not signaling/nuclear/chromosomal context, not transcription, not replication, not factor binding competition, not specific activator—paved the way to the recognition of the actual chromatin remodeling enzymes (reviewed for yeast in (8)). “It was Wolfram [Hörz] who kept pointing out to us that all our mechanisms did not work for his PHO5 promoter.” (Jerry Workman, 2008, personal communication to PK) The ATP-dependent nucleosome remodeling activity of the yeast SWI/SNF complex (85,86) and its activation domain dependent targeting (87), explicitly shown also for Pho4, could explain in principle the transcription-, replication- and context-independent PHO5 promoter chromatin remodeling. Soon many homologs to the SWI/SNF ATPase subunit Snf2 were recognized in eukaryotic genomes, already 17 in yeast (88), and all of these were tested with regard to PHO5 promoter opening (14,79,89–96).
What remodels PHO5 promoter chromatin? The hunt for involved chromatin cofactors
After identification of numerous Snf2-type remodeler ATPases and especially the characterization of the bona fide transcription factor Gcn5 as histone acetyl transferase (97–99) the field of transcription regulation finally acknowledged nucleosome remodeling and modification as an important level of gene regulation. Now the stage was set to call on the cofactors responsible for a given chromatin transition. Again the PHO5 promoter was a most useful model and whichever chromatin cofactor mutant available was tested for effects on PHO5 promoter opening, also in the context of genetic screens (93,100). At first this seemingly straight forward approach was somewhat frustrating as no cofactor's absence seemed to prevent PHO5 promoter chromatin remodeling. Especially the first candidate mutations, snf2 and gcn5, still showed full remodeling upon full induction (79,101). This was even more surprising as at the same time opening of the PHO8 promoter, which is coregulated by Pho4 and also displays a substantial chromatin transition upon induction (18), was completely abolished in snf2 and confined to a minor local alteration in gcn5 cells (102).
Nonetheless, lack of Gcn5 did impair PHO5 promoter chromatin remodeling but only under sub-optimal induction conditions at high phosphate in pho80 cells (101) or at early time points of induction kinetics (82). Especially the latter so called ‘kinetic effect’ made the important point that chromatin effects are often negligible under steady state conditions but very pronounced during the transition from one biological state to the other, e.g. from repression to induction. Just recently this concept was highlighted in a comprehensive study of many chromatin cofactor mutants and their effects during oxidative stress dynamics in yeast (103).
Consequently, cofactors involved in PHO5 promoter opening were rather identified at suboptimal induction, e.g. at high phosphate conditions upon inactivation of Pho80/Pho85, or overexpression of PHO4, or early during induction kinetics upon phosphate starvation (14,82,91,93–94,100–101,104–105). As in all these cases mutation effects become stronger with decreasing induction strength (94) there were controversies about how important or even essential a cofactor is for PHO5 promoter opening. For example, under suboptimal induction conditions an asf1 or isw1 chd1 mutant is unable to remodel PHO5 promoter chromatin making these cofactors seem essential or others, like RSC, unimportant (89,106). However, upon full induction these mutants show full remodeling (95,104) demonstrating that other cofactors can compensate and are involved, too. Moreover, even if a remodeler is essential, e.g. SWI/SNF for opening the PHO8 promoter (102), this does not preclude that yet others contribute, too, like INO80 is also involved at the PHO8 promoter (91). We advocate the view that all cofactors with significant effects at weak or strong induction are part of the physiological remodeling mechanism, but only if lack of a cofactor prevents remodeling under fully inducing conditions, e.g. laboratory conditions of no phosphate, this factor is truly essential for chromatin opening. This is a purely mechanistic view and does not deny that it is the rapid adaptation to changed conditions which is ultimately important from a physiological point of view and in this sense ‘essential’ for yeast survival in the wild.
It is crucial to show that effects due to mutations in vivo are direct and not indirect. For example, effects on promoter chromatin remodeling may be exerted via effects on signal transduction and therefore induction strength. For the PHO5 promoter, especially the use of the Gal4-driven promoter variant was very instructive as this variant recapitulated the same chromatin cofactor dependencies even though the entire physiological context of promoter induction, including the transactivator, were altered leaving the promoter nucleosomes as common denominator and therefore likely direct target of the identified cofactors (82,91,95,104). In addition, for SWI/SNF, RSC, INO80, SAGA and Rpd3 binding to the PHO5 promoter was shown by chromatin immunoprecipitation (ChIP) (92,94,107–110) or mass spectrometry of isolated promoter minicircles (111) supporting their direct roles, but not necessarily excluding indirect effects. We caution that the assay monitoring the nuclear localization of Pho4-GFP (26) saturates soon and may not sufficiently discriminate weak from strong induction conditions.
Starting point for promoter opening: the dynamics of the repressed state
The collective efforts of several groups identified a surprisingly complex network of chromatin cofactors involved in the mechanism of PHO5 promoter chromatin opening (Table 1 and Figure 3). At this point we consider first the repressed state and switch from a largely chronological review to a summary of present day knowledge.
Table 1. Chromatin cofactors involved in chromatin remodeling at the PHO5 promoter.
| Cofactor type | Role in PHO5 promoter opening | References |
|---|---|---|
| Remodeling enzyme (name of ATPase subunit) | SWI/SNF (Snf2), RSC (Sth1), INO80 (Ino80), Isw1, Chd1 | (14,79,89–96) |
| Histone acetylase (name of acetylase subunit) | SAGA (Gcn5), Rtt109, NuA4 (Esa1) | (82,93–94,101,105,114–115) |
| Histone deacetylase | Rpd3 | (110) |
| Histone chaperone | Asf1, HIR, Spt6 | (70,104,106) |
| Histone methylase | Set1 | (110) |
Figure 3.
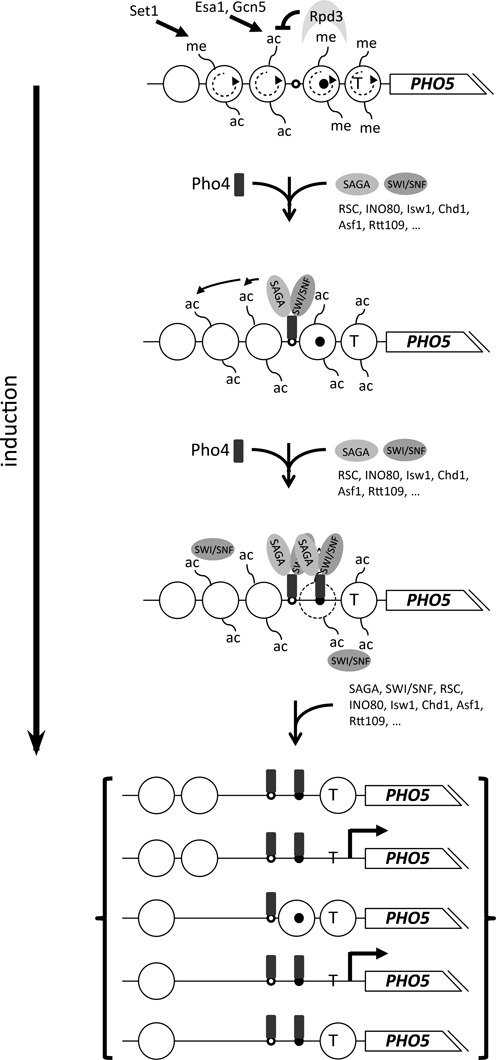
Mechanistic model for PHO5 promoter chromatin opening upon induction. Top panel: the PHO5 promoter nucleosomes of the repressed state (see also Figure 2a and corresponding symbols) are methylated (me) by Set1 at H3K4 (only two histone tails per nucleosome as proxy for all possible tails are shown), which recruits the histone deacetylase Rpd3 (gray arc). This curbs acetylation (ac) set by Esa1 and Gcn5 and keeps histone turnover (circular stippled arrows) low. Second panel: upon induction, Pho4 (rounded rectangle) becomes nuclear, binds first the constitutively accessible UASp1 element (small open circle) and recruits the SAGA and SWI/SNF complex. Only the direct recruitment for SAGA and SWI/SNF, for which there are clear Pho4 interaction data, is shown, but the other listed cofactors, and presumably even more, are involved too (see Table 1). The RSC complex, e.g. is present already under +Pi conditions (108). SAGA and possibly other acetylases hyperacetylate promoter nucleosome histones. Third panel: SWI/SNF and/or other remodelers remodel nucleosome -2 such that UASp2 (small closed circle) becomes accessible for Pho4. At first this need not require complete nucleosome disassembly (stippled outline of nucleosome circle), although complete removal of nucleosome -2 is a hallmark of the activated PHO5 promoter. Bracketed panels: continued recruitment of cofactors through bound Pho4 and/or acetylated histones (e.g. SWI/SNF through bromodomains 112) leads to increased nucleosome disassembly and an ensemble of nucleosome configurations with different combinations of nucleosomes absent or present. Bold brackets include just five examples out of up to 16 possible configurations (68,113). Recruited cofactors and histone tail modifications are not shown for simplicity. Transitions between states are possible by nucleosome assembly, disassembly and sliding. Only configurations without nucleosome -2 are conducive for promoter activation and only configurations with accessible TATA box (T) allow bursts of PHO5 transcription (bold broken arrow). See main text for further references.
The determinants for nucleosome positioning at the repressed PHO5 promoter (Figure 2a) are still unknown, despite considerable progress in the identification of factors responsible for genome-wide nucleosome patterns (5–7). The genes encoding the principal factors binding at the PHO5 promoter, i.e. Pho2 and Pho4 or Cbf1, which can bind to Pho4 sites (116), and even the long linker between nucleosomes -2 and -3, which would be a prime candidate region for binding sites of some organizing factors, can be deleted without losing proper nucleosome positioning ((34,62) and our own unpublished data regarding the cbf1 mutant).
In general, positioned nucleosomes at yeast promoters are not set in stone but their histones undergo a rather rapid turnover (117–119). Such histone dynamics can be influenced by histone acetylation levels and determines basal promoter activity. In the case of PHO5, histone turnover is reduced by histone deacetylation through Rpd3, which is recruited via histone H3 K4 methylation by Set1 (110). This explains increased basal PHO5 transcription in set1, rpd3 and also rpd3 pho4 cells (110,120–122). The latter Pho4-independence is again consistent with the early Grunstein studies showing elevated transcription just because of reduced nucleosome occupancy. The NuA4 complex bearing the histone acetyltransferase Esa1 is constitutively recruited via Pho2 bound close to the constitutively accessible UASp1 and seems important for efficient Pho4 binding during activation (114). In addition, the histone variant H2A.Z (Htz1 in budding yeast), which is preferentially found at promoters (49,123–124) and may facilitate promoter activation by increasing histone turnover (125), is acetylated by Gcn5 and Esa1 and was implicated in nucleosome reassembly at the PHO5 promoter (126). Nonetheless, promoter opening is hardly affected by the absence of Htz1 (105), and lack of Pho2 or NuA4 can be compensated by overexpression of Pho4 (34,114). It was proposed that a certain level of basal promoter nucleosome dynamics is important for timely PHO5 promoter opening and ensured by antisense transcription through the PHO5 promoter (127). While the role of antisense transcription may need further validation, it is often true that mutations affecting cofactors involved in PHO5 promoter opening, e.g. gcn5 or snf2 (Table 1 and references therein), also show reduced levels of basal transcription and promoter accessibility. This means that at least some of these cofactors constitutively keep PHO5 promoter nucleosomes dynamic, it supports the Svejstrup notion (127) that such dynamics are important for efficient and timely remodeling, and shows that the molecular machinery for basal nucleosome dynamics is linked to that for promoter opening upon induction.
Molecular mechanism for generating an extended hypersensitive site: a surprisingly complex chromatin cofactor network remodels PHO5 promoter chromatin
Upon induction, Pho4 translocates into the nucleus, binds co-operatively with Pho2 at UASp1 (20–21,128) and recruits via its activation domain the SAGA complex, which leads to hyperacetylation of promoter histones (90), and recruitment of TATA box binding protein (TBP) via its Spt3 subunit (109). Hyperacetylated histones stabilize recruitment of remodelers with bromodomains, like SWI/SNF (112). SWI/SNF is also directly targeted by the Pho4 activation domain (87). The accessible UASp1 may serve as ‘pioneer’ site from where Pho4-recruited cofactors can induce remodeling of the -2 nucleosome and generate access to UASp2 (62). A ΔUASp1 PHO5 promoter variant is usually not opened at all (62,128), but PHO4 overexpression restores full remodeling (76). In contrast, a ΔUASp2 variant is not fully remodeled (128), not even with PHO4 overexpression (129). This makes Pho4 binding to UASp2 not only necessary, as recently confirmed (68), but also sufficient for promoter opening and induction. Consequently, there is the circular condition that nucleosome remodeling is required for Pho4 binding (76) and Pho4 binding for nucleosome remodeling. However, as mentioned above, the rapid turnover of promoter nucleosomes (117–119) and an almost equally rapid accessibility of both nucleosomal and non-nucleosomal regions, explicitly at the PHO5 promoter, to photolyase DNA repair (130) show that nucleosomes do not pose a rigid barrier in vivo but that there are always windows of opportunity for factors to access the DNA. The question is if nucleosome disassembly as triggered by a given factor can eventually overcome nucleosome re-assembly. In the case of Pho4 this is not a matter of binding competition but only possible if an activation domain is involved 77, presumably as it recruits chromatin remodeling and modifying factors that tip the balance toward nucleosome disassembly and stable Pho4 binding (68). Even though binding competition to an intranucleosomal UASp element alone is not sufficient for remodeling, it does help the remodeling process (129).
Recently, the interplay of nucleosomes, Pho4, Pho2 and TBP at the PHO5 promoter was systematically recapitulated (128) by the chromatin endogenous cleavage (ChEC) method (131) where MNase was fused to Pho4 and chromatin accessibility monitored after MNase activation by calcium. These and earlier (76) studies as well as X-ray crystallographical data on DNA binding by Pho4 (132) leave no doubt that Pho4 cannot bind to UASp2 if this site is within a canonical nucleosome. Nonetheless, there are probably non-canonical nucleosome intermediates during histone turnover and on the pathway to nucleosome disassembly that allow Pho4 binding. Such intermediates could explain why Pho4 binding kinetics need not become slower in the same way as histone eviction kinetics in an asf1 and other mutants (106–107,133).
While Pho4 binding is the essential trigger, it is the co-operation between chromatin cofactors that actually removes the nucleosomes. The main lesson from PHO5 here is the surprising complexity of this cofactor network (Table 1). This network is redundant (91) in the sense that chromatin can be opened even in the absence of many members of this network under full induction conditions. Nonetheless, the complete set is required to achieve full opening under suboptimal conditions and wt kinetics upon strong induction. It was a recent surprise that five remodelers from four major remodeler families are involved in PHO5 promoter opening, including the Remodels the Structure of Chromatin (RSC) complex (95). RSC was a prime candidate for the remodeler at the PHO5 promoter since all other Snf2-type ATPases could be deleted without preventing PHO5 promoter opening (91,134). RSC is the only remodeler essential for viability in budding yeast and was therefore difficult to test in vivo due to indirect effects in growth arrested or dying cells. A role for the RSC complex was initially suggested, at least indirectly (135,136), then dismissed based on in vitro studies (89,137), but now clearly demonstrated in vivo (95). Of note, the combined absence of RSC and Snf2 or RSC and Isw1/Chd1 represent the first in vivo cases where PHO5 promoter opening was not possible even under strong induction conditions. Whether RSC is not only a major remodeler but maybe even truly essential in our sense (see 'What remodels PHO5 promoter chromatin? The hunt for involved chromatin cofactors' section) remains to be established. To our knowledge, the PHO5 promoter chromatin transition is the only one so far where the full complement of remodeler ATPases encoded in a genome was tested for involvement.
We expect that the co-operation of many remodelers for a given chromatin remodeling process will be the rule rather than the exception. Indeed, a recent study in mouse showed that several remodelers are present and involved at many loci undergoing chromatin remodeling (138). Potentially, even more remodeler ATPases could be involved at the PHO5 promoter given the example of Isw1 and Chd1. Both remodelers at first seemed not involved as isw1 and chd1 single mutations did not have effects on PHO5 induction, neither on the final level nor on the kinetics (91–93,134,139). However, the combined isw1 chd1 double mutation showed a clear effect on PHO5 induction (89) and promoter opening (95). Therefore these remodelers have a truly redundant role that is only detected in their combined absence. As not all combinations of remodeler ATPase gene deletions were tested so far, or can even be tested with view of yeast viability, there remains some uncertainty as to how many remodeler ATPases can be involved in PHO5 promoter opening.
‘Anatomy of a hypersensitive site’: PHO5 promoter remodeling via histone eviction in trans
From the very beginning, it was clear that the molecular make up of hypersensitive sites does not correspond to canonical nucleosomes. Since the discovery of the nucleosome (53,56) the protection of ∼140–150 bp of DNA from limited MNase digestion and relative inaccessibility to other endonucleases, like DNaseI and restriction enzymes, is still a valid operational definition of a canonical nucleosome. Accordingly, the first detailed study on PHO5 promoter opening (41) demonstrated by several nuclease assays involving DNaseI, DNaseII, MNase and restriction enzymes that the extended hypersensitive site at the induced PHO5 promoter cannot harbor the same full complement of canonical nucleosomes as in the repressed state. Nonetheless, there was the possibility that few nucleosomes remained, possibly delocalized, and/or that nucleosomes were remodeled into a non-canonical state (140–142) that allows nuclease cleavage and Pho4 binding but keeps histones bound to the DNA. The latter possibility necessitates the often subsumed or simply overlooked, but mechanistically important distinction if remodeling results merely in ‘nucleosome eviction’ or also in ‘histone eviction’.
Chromatin remodeling at the PHO5 promoter was the first demonstrated case of histone eviction in vivo (90,143). Up to now, the only assay for monitoring histone-DNA contactsin vivo is anti-histone ChIP. All other assays (nuclease accessibility, topology, methyltransferase accessibility, hydroxyl radical cleavage, psoralen crosslinking) can only monitor the presence of a canonical nucleosome but cannot or not strictly distinguish non-canonical nucleosomes from histone-free DNA. Similar to Pho4 binding and histone eviction, nucleosome remodeling as monitored by increased nuclease accessibility can be uncoupled from histone eviction. Steady-state high phosphate conditions in gcn5 pho80 cells lead to altered nuclease accessibility due to randomized nucleosome positions at the PHO5 promoter (101) but not to nucleosome loss (134). Further, histone loss kinetics during induction at the PHO84 promoter is much delayed in gcn5 cells while chromatin remodeling kinetics monitored by HhaI restriction enzyme accessibility is not (105). The RNR3, CHA1 and other promoters are further examples where loss of cofactors inhibits histone eviction more than nuclease sensitivity (80,144).
Histone eviction is now firmly established as a global phenomenon across the genome and across species (1,145–146), and the test for histone eviction, i.e. monitoring histone occupancy as such (usually an anti-H3-C-terminus ChIP, which was first used at the PHO5 promoter (90)), has become an essential normalization procedure for assays monitoring histone modifications. Indeed, histone acetylation decreases over the PHO5 promoter during induction, but this observation is deceiving due to overall histone loss. If remodeling was slowed down in an snf2 mutant, a transient peak of histone hyperacetylation could be observed (90). As opening of the PHO8 promoter essentially depends on Snf2, this promoter becomes locked in the hyperacetylated state upon shift of snf2 cells to inducing conditions (147,148). Given that such promoter chromatin is still fully closed, this was an important demonstration that hyperacetylation per se does not lead to increased DNA accessibility. This is in contrast to the still somewhat common notion that chromatin opening in the sense of chromatin fiber unfolding would be the same as chromatin opening in the sense of increased accessibility to DNA that was part of a nucleosome core. While charge neutralization by lysine acetylation, especially at histone H4 lysine 16, does lead to fiber unfolding, it is mostly not sufficient to destabilize nucleosome cores (149).
Histone eviction raised the question if histones were lost from the PHO5 promoter region in cis due to lateral nucleosome sliding (150,151) or in trans due to nucleosome disassembly. Topology and nuclease assays on excised chromatin mini-circles (134) or on tiny plasmids bearing the PHO5 promoter (152) demonstrated histone eviction in trans. This does not exclude nucleosome sliding in cis at early remodeling stages as shown, e.g. for histone eviction by SWI/SNF in vitro (153). We note that this is to our knowledge the only physiological case where the cis-trans distinction is clearly demonstrated so far. It is therefore only by extrapolation from the PHO5 promoter when an observation of histone eviction is interpreted as eviction in trans, i.e. ‘nucleosome disassembly’. The PHO5 promoter was also the first model where histone exchange using differentially tagged histone isoforms (‘histone double tag strategy’) was monitored (154). By this approach, it was clearly established that histones return from a source in trans upon PHO5 promoter repression, and it prompted a fruitful series of histone exchange studies that shaped our view of global histone dynamics (117–119,155).
As histones outside the nucleosome structure are notoriously aggregation prone both in vivo as well as in vitro, histone eviction in trans calls for a histone acceptor. Histone chaperones are the prime candidates. Indeed, there is a role for the histone chaperones Asf1 and HIR observed at the PHO5 promoter under suboptimal induction conditions (104,106). However, Asf1 is also essential for H3K36 acetylation by the Rtt109 histone acetyl transferase (156–158) that also affects PHO5 promoter opening (105,115). It is therefore unresolved if Asf1 contributes to PHO5 promoter opening truly as histone acceptor or mainly as cofactor for Rtt109.
The question if few canonical, maybe delocalized, nucleosomes may still be present after PHO5 promoter chromatin remodeling received different answers over the years but appears to be resolved just recently. Nuclease digestion and topology analyses suggested that not all nucleosomes, especially the -1 and -4 nucleosomes, are completely remodeled, but that the open state corresponds to an heterogeneous ensemble of nucleosome configurations and that about one out of three nucleosomes remains at the promoter (41,134–135,143,152,159). The Kladde group developed nucleosome mapping in vivo based on accessibility to DNA methylases (160) that allows single molecule analysis due to a cloning step. First applied to PHO5 promoter opening kinetics (161), this technique revealed that opening leads to a heterogeneous population with variable numbers of nucleosomes per PHO5 promoter molecule. There was the general trend that nucleosomes -2 and -3 are remodeled first and more often, while nucleosomes -1 and -4 were often still present, supporting the idea that remodeling spreads bidirectionally from UASp1 (161). Some templates showed remodeling even of the -5 nucleosome, but only very few templates lacked all four PHO5 promoter nucleosomes, at least up to the last kinetic time point measured. All these initial observations suggested that hardly ever all PHO5 promoter nucleosomes were removed (‘nucleosome retention hypothesis’ (135)). This made the PHO5 promoter a favorite in vivo example for the highly intriguing concept that remodelers of the SWI/SNF family, SWI/SNF and RSC in yeast, remodel nucleosomes by ‘sliding mediated disassembly’ (135,153,162–163). The remodeler is thought to ‘ride on top’ of a nucleosome and slide it into neighboring nucleosomes leading to their disassembly. Accordingly, always one nucleosome should remain at the promoter. The remodeling concept as such was recently substantiated by domain swaps between remodelers of the SWI/SNF and the CHD family (164) and incorporated earlier into elaborate theoretical modeling by the Boeger group (135). However, the Boeger group very recently revisited the issue for the PHO5 promoter using single molecule analysis via psoralen crosslinking. They refuted their stable nucleosome retention hypothesis as they found the zero nucleosome state as the most abundant (∼25%) of eight possible states of nucleosome removal combinations and adapted their model accordingly (68).
Collectively, the molecular anatomy of induced PHO5 promoter chromatin does not correspond to a single well-defined state, but to a heterogeneous ensemble of states where a variable and relative to the repressed state increased number of nucleosomes is disassembled, even all of them, such that histones are evicted in trans. This allows Pho4 and the transcription machinery to bind more frequently to the critical and otherwise occluded UASp2 and TATA box elements, respectively, leading to more frequent transcription initiation than in the repressed state and therefore higher expression levels.
The coregulated PHO8 and PHO84 promoters: differential cofactor requirements due to different intrinsic nucleosome stabilities
The PHO8 and PHO84 promoters are two members of the PHO regulon, i.e. coregulated by Pho4, that were characterized in detail regarding their promoter chromatin alterations upon induction (Figure 2b and c; (18,105,165)). Both show pronounced chromatin transitions, including histone eviction (105,106), and critically depend on Pho4. Nonetheless, they demonstrated a different stringency of cofactor requirements for chromatin opening compared to the PHO5 promoter. Neither of these two promoters requires the RSC complex for chromatin opening (95), even though RSC is necessary for generating proper nucleosome positioning at the repressed PHO8 promoter (166). Complete remodeling at the PHO8 promoter strictly requires SWI/SNF and Gcn5 (102), while the PHO84 promoter nucleosomes appear like a hybrid between those of the PHO5 and PHO8 promoters. The nucleosome downstream of the hypersensitive site bearing UASpE (Figure 2c) has relaxed cofactor requirements reminiscent of the PHO5 promoter but the upstream nucleosome containing UASpB strictly depends on SWI/SNF like those at the PHO8 promoter (96,105). Especially the close proximity of these two neighboring nucleosomes at the PHO84 promoter and their similar distance to the Pho4 binding sites make it unlikely that their differential cofactor requirements are due to differences in cofactor recruitment strength or mode. Instead, intrinsic differences in nucleosome stability correlate with the differential requirement as shown in vitro for both the PHO8 (167) and PHO84 promoters (105) by using a reconstitution system at limiting histone concentrations. The causality was also demonstrated in vivo by introducing destabilizing poly(dA:dT) stretches into the Snf2-dependent nucleosome at the PHO84 promoter such that remodeling of this nucleosome became possible even in an snf2 mutant (105). Notably, also at the PHO8 and PHO84 promoters not just one but at least two remodelers, SWI/SNF and INO80 (91,105), participate in chromatin opening confirming again our notion regarding the pervasiveness of complex remodeler networks.
NEW TRICKS FOR OLD DOGS: PHO PROMOTERS GO IN VITRO, IN SILICO AND TO SYSTEMS LEVEL
In vitro reconstitution of nucleosome positioning and remodeling at PHO promoters
The exceedingly rewarding in vivo approach to chromatin remodeling mechanisms at the PHO promoters called for a corresponding in vitro system in order to further address mechanistic questions biochemically. So far PHO5 promoter chromatin was either isolated ex vivo (89,168) or reconstituted de novo (129,169–170), and then Pho4-induced generation of the hypersensitive site was aspired. In some cases Pho4-, ATP- and extract-dependent nucleosome remodeling was observed, but the extent of remodeling was considerably less than in vivo, maybe due to irreversibly aggregated templates during in vitro handling or due to inherent difficulties in reconstituting the proper time course of histone modifications and cofactor interactions. Curiously, reconstituted PHO8 promoter chromatin could not be remodeled at all under the same conditions (129). For whichever reason, there is still no in vitro system established that adequately recapitulates the in vivo chromatin transition of any PHO promoter.
Nonetheless, it is quite amazing to what degree of similarity the nucleosome positioning pattern of the repressed states of the PHO5 (167,170), PHO8 (167) and PHO84 (105) promoters can be reconstituted de novo using a combination of salt gradient dialysis and incubation with yeast extract and ATP (171). This enabled comparisons of intrinsic stability of promoter nucleosomes at their proper position ((105,167)) and identified a necessary, direct and specific role for the RSC complex in nucleosome positioning at the repressed PHO8 promoter (166). Such in vitro reconstitution of in vivo-like nucleosome positioning was achieved also on the genome level (172,173) and should allow the biochemical dissection of global nucleosome positioning mechanisms.
Theoretical modeling for the PHO promoters
Especially the O'Shea and Boeger groups combined quantitative measurements, including single cell and single molecule techniques, with theoretical modeling of PHO5 promoter properties. PHO5 (also PHO84) is a prime example for stochasticity in gene expression, also called ‘intrinsic noise’ of expression (174). Two identical copies of PHO5 promoters but each driving a different reporter gene, YFP and CFP, showed considerably different expression levels in the same cell. This intrinsic noise scaled inversely with expression level and was exacerbated if chromatin remodeling was compromised (snf6, gcn5, arp8 or UASp mutations). Modeling of such data suggested that a slow and stochastic PHO5 promoter activation step, presumably chromatin opening, precedes transcription activation. A series of studies from the Boeger group elaborated on this topic. Modeling of quantitative data on nucleosome occupancy at the induced PHO5 promoter confirmed that nucleosome disassembly is rate limiting for transcription (135) although this dataset erroneously assumed stable retention of one nucleosome at all times. Modeling the correlation between an extensive range of Pho4 activation domain mutations and the corresponding degrees of promoter opening and PHO5 expression revealed a dual role for Pho4 in both increasing nucleosome disassembly and transcription activation (159) and confirmed the earlier finding (83) that both roles are structurally inseparable. Nonetheless, equivalent studies at the PHO8 promoter showed a role for Pho4 in mainly accelerating nucleosome disassembly (96). These conclusions are consistent, regarding PHO8, but not, regarding PHO5, with the maintenance of full transcription levels even in the absence of Pho4 and Pho2 upon return to repressive conditions if chromatin reassembly was inhibited by Spt6 ablation (70). Maybe such Spt6 ablation conditions represent unusually open chromatin and do not represent the physiological state of the activated PHO5 promoter. The general view is that, whereas lack of nucleosomes is conducive for elevated transcription, a transactivator is still required for full activation (175).
The recent modeling of single molecule psoralen crosslinking nucleosome mapping at induced PHO5 promoter molecules combined with intrinsic noise data (68) suggests the following mechanistic picture and explains bursty transcription. Nucleosome configurations at the PHO5 promoter are intrinsically stochastic as nucleosomes continuously assemble, disassemble and slide. The mechanistic involvement of both (dis)assembly and sliding is consistent with a role for remodeling enzymes in PHO5 promoter opening (Table 1) of both the SWI/SNF family, i.e. SWI/SNF and RSC, as well as the ISWI and CHD families, i.e. Isw1 and Chd1, that rather mediate the former and latter mode of remodeling, respectively (176) (even though also nucleosome disassembly by SWI/SNF involves initial sliding (153)). Pho4 accelerates nucleosome removal via chromatin cofactor recruitment but does not inhibit nucleosome reassembly by mere steric exclusion. Removal of the -1 nucleosome requires disassembly of nucleosome -2, consistent with earlier population (41) and single molecule (161) measurements. As nucleosomes are occlusive to transcription factor binding (76,128), and as the stochastic movement of nucleosomes always yields configurations both conducive and non-conducive for transcription activation, transcription occurs in bursts. Importantly, the model poses that increased transcription burst frequency rather than longer burst duration underlies the increased expression level.
While this Survey and Summary article was under review, another single molecule PHO5 promoter study was published (113). The authors again monitored promoter nucleosome occupancy by protection from DNA methylation, but single molecule resolution was achieved by truly working with single cells rather than through a cloning step (161). This demonstrated in unprecedented resolution the variability of nucleosome positions, especially for nucleosome -3, that underlies the canonical patterns of the repressed and induced state (Figure 2a). Using a GFP reporter, cells with activated versus inactive PHO5 promoter were sorted and their respective promoter nucleosome configurations were directly measured for the first time. This directly confirms all earlier conclusions regarding that promoter activation requires remodeling of the -1 and -2 nucleosomes and directly visualizes the chromatin basis for cell-to-cell expression differences (bursty expression). In contrast to the Boeger study (68), the authors did not detect completely nucleosome-free promoters under equivalent inducing conditions. Such nucleosome-free promoters were only detected upon introduction of poly(dA:dT) elements in the PHO5 promoter. These shifted the distribution of nucleosome configurations toward open promoters and caused derepression under high phosphate conditions. Conversely, point mutations predicted to increase the intrinsic stability of the -3 nucleosome could even prevent PHO5 promoter opening upon induction consistent with our conclusion that intrinsic nucleosome stability determines remodeling cofactor requirements (105).
Systems biology with the PHO regulon
The PHO5 promoter and the PHO regulon were harnessed to derive quantitative and predictive models that are required for a systems approach to gene regulation. Importantly, such models explicitly accommodate the regulatory input of chromatin. Lam et al. (165) correlated the position of high and low affinity UASp elements relative to nucleosomes, i.e. intra- versus internucleosomal, of seven Pho4 regulated genes and eight PHO5 promoter variants with the corresponding transcription induction kinetics during phosphate starvation. Consistent with the classical knowledge that nucleosomes are occlusive and necessitate remodeling prior to Pho4 binding (76–77,128), this study comprehensively showed that the affinity of the constitutively accessible UASp elements determines early or late onset of gene induction (threshold), but that all UASp elements that become accessible after promoter chromatin remodeling contribute in the end to the full expression level (dynamic range). So nucleosome positioning fine tunes the input-output correlation (gene regulatory function) of a promoter and allows independent regulation of threshold and dynamic range. This was followed up and modeled accordingly using an elegant system where Pho4 levels were controlled by a doxycycline inducible promoter and Pho4 levels as well as PHO5 promoter output monitored via YFP tag and CFP reporter, respectively (177).
Nucleosome positioning also importantly contributes to the selection of functional transcription factor binding sites. The number of potential transcription factor binding sites encoded in eukaryotic genomes is often larger than that of biological function. Yeast Pho4 or Leu3 transactivator binding sites are good examples. A comparison of genome-wide Leu3 binding in vivo and to free DNA in vitro with in vivo nucleosome positioning (178) suggested that non-functional sites are mainly covered by nucleosomes. However, just the intra- versus internucleosomal position is clearly not sufficient to distinguish functional and non-functional Pho4 binding sites. UASp2 at the PHO5 promoter is intranucleosomal and highly functional (41,76) while the constitutively accessible UASp1 at the PHO8 and UASpA at the PHO84 promoter are not (17,105,179). Genome-wide analysis of this issue (180) suggested that the competition between Pho4 and the alternative UASp binding factor Cbf1 (116) co-determines binding to internucleosomal, while the co-operation between Pho4 and its binding helper Pho2 (20,21) contributes to the decision about functionality of intranucleosomal UASp elements.
Just recently, an extensive set of 209 PHO5 promoter variants bearing mutations of the UASp elements was tested both for Pho4 binding affinity in vitro (181) as well as PHO5 expression output in vivo (182). This latter study underscored the importance of just the binding affinity in predicting promoter output but also found several examples, including the wt promoter, where nucleosome remodeling had to be included in the model to match the data. In contrast to the conclusions by Lam et al. (165), also intranucleosomal binding sites contributed to setting the threshold of induction, but the overall notion was confirmed that the wt PHO5 promoter is optimized with regard to limiting activation at low while providing maximal output at full induction conditions. Such a PHO5 promoter library or the set of Pho4 activation domain mutations (159) or the doxycycline inducible Pho4 system (177) together with a sizeable number of Pho4 regulated genes and detailed knowledge of nucleosome positioning and remodeling mechanisms provide a rich tool box for further studies of the PHO regulon as model for eukaryotic transcriptional networks.
RETROSPECTIVE AND OUTLOOK
Confirmation of classical findings with new methodology
It is highly affirmative how robust a model system the PHO promoters are. Despite the long history of research on these models, none of the classical conclusions was refuted, but all of them independently confirmed in several groups and with new techniques as noted above throughout our review. This includes, e.g. the nucleosome organization in the repressed states, that Pho4 is the trigger, that binding to UASp2 is critical but impeded by nucleosome -2 making nucleosome remodeling rate-limiting, that remodeling is mediated by activation domain dependent recruitment of chromatin cofactors and not through steric exclusion, and that histones are evicted in trans.
Open questions
Besides confirming classical tenets, it is quite amazing how the PHO promoters keep turning up new insights and there are still numerous, even very basic questions unanswered regarding chromatin remodeling, for which PHO promoters continue to be promising models. Just to name a few: what determines nucleosome positioning in the repressed state? What determines the extent of remodeling in the induced state, i.e. why nucleosomes -1 to -4 at the PHO5 and the respective ones at the PHO8, PHO84 or other promoters? Why is remodeling of the TATA-box occluding nucleosome comparatively incomplete, even though this should be the most important one to remove? What is so repressive (179) and intrinsically stable (129,167) about PHO8 promoter nucleosomes? What causes the strict dependencies on chromatin cofactors, e.g. the Snf2-dependency of PHO8 and PHO84 promoter nucleosomes? What is the role of H2A.Z at promoters in yeast? What is necessary for the maintenance of the open promoter state? Indeed, the whole process of promoter closing is relatively understudied, besides the demonstration of histone re-assembly from a source in trans (154) and a role for Spt6 (70) hardly anything is known about its mechanism.
A special case turned into a general paradigm
In retrospective, it is maybe surprising how the PHO5 promoter became such a general paradigm even though it is a rather special case in many ways. For yeast standards, its extent of chromatin remodeling and induction is extremely high and its promoter chromatin organization is very much the exception rather than the rule. Genome-wide studies in yeast revealed that the vast majority of promoters harbor a broad hypersensitive site (NFR) just upstream of the TSS, drive constitutive expression of little intrinsic noise, are not much dependent on chromatin cofactors and do not rely on a canonical TATA box but on TATA-like elements (183–185). The PHO5 promoter does not fit this stereotype but has its own individualistic architecture. Nonetheless, this is due to the unusual biology of yeast in having a mostly active genome most of the time. Larger eukaryotes require switching genes on and off to a much larger extent, e.g. during differentiation. In this regard, the PHO5 promoter is a paradigm for a highly regulated promoter. With its large dynamic changes in chromatin structure and gene induction it therefore was and continues to be an experimentally very well accessible model for regulated chromatin transitions.
Acknowledgments
We thank Peter Becker for his support of this work and Corinna Lieleg for critical reading of the manuscript.
This article is dedicated to the memory of Wolfram Hörz (1944–2005), who would have turned 70 this year and who was a major promoter of chromatin research in yeast and beyond by his focus on PHO promoters (186).
FUNDING
The authors are funded through their positions at their respective institutions. Personal exchange between the authors was greatly facilitated through visiting researcher funds from the German Research Community (DFG, CRC1064). Funding for open access charge: German Research Community [DFG, CRC1064].
Conflict of interest statement. None declared.
REFERENCES
- 1.Bell O., Tiwari V.K., Thoma N.H., Schubeler D. Determinants and dynamics of genome accessibility. Nat. Rev. Genet. 2011;12:554–564. doi: 10.1038/nrg3017. [DOI] [PubMed] [Google Scholar]
- 2.Korber P., Becker P.B. Nucleosome dynamics and epigenetic stability. Essays Biochem. 2010;48:63–74. doi: 10.1042/bse0480063. [DOI] [PubMed] [Google Scholar]
- 3.Allis C.D., Jenuwein T., Reinberg D. In: Epigenetics. 1st edn. Allis CD, Jenuwein T, Reinberg D, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 23–61. [Google Scholar]
- 4.Becker P.B., Workman J.L. Nucleosome remodeling and epigenetics. Cold Spring Harb. Perspect. Biol. 2013;5 doi: 10.1101/cshperspect.a017905. doi:10.10.1/cshperspect.a017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Struhl K., Segal E. Determinants of nucleosome positioning. Nat. Struct. Mol. Biol. 2013;20:267–273. doi: 10.1038/nsmb.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hughes A.L., Rando O.J. Mechanisms underlying nucleosome positioning in vivo. Ann. Rev. Biophys. 2014;43:41–63. doi: 10.1146/annurev-biophys-051013-023114. [DOI] [PubMed] [Google Scholar]
- 7.Korber P. Active nucleosome positioning beyond intrinsic biophysics is revealed by in vitro reconstitution. Biochem. Soc. Transact. 2012;40:377–382. doi: 10.1042/BST20110730. [DOI] [PubMed] [Google Scholar]
- 8.Rando O.J., Winston F. Chromatin and transcription in yeast. Genetics. 2012;190:351–387. doi: 10.1534/genetics.111.132266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oshima Y. Regulatory Circuits for Gene Expression: The Metabolism of Galactose and Phosphate. In: Strathern JN, Jones EW, Broach JR, editors. The molecular biology of the yeast Saccharomyces cerevisiae: Metabolism and gene expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1982. pp. 159–180. [Google Scholar]
- 10.Schmid A., Fascher K.D., Hörz W. Nucleosome disruption at the yeast PHO5 promoter upon PHO5 induction occurs in the absence of DNA replication. Cell. 1992;71:853–864. doi: 10.1016/0092-8674(92)90560-y. [DOI] [PubMed] [Google Scholar]
- 11.Wykoff D.D., O'Shea E.K. Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics. 2001;159:1491–1499. doi: 10.1093/genetics/159.4.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Auesukaree C., Homma T., Tochio H., Shirakawa M., Kaneko Y., Harashima S. Intracellular phosphate serves as a signal for the regulation of the PHO pathway in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:17289–17294. doi: 10.1074/jbc.M312202200. [DOI] [PubMed] [Google Scholar]
- 13.Thomas M.R., O'Shea E.K. An intracellular phosphate buffer filters transient fluctuations in extracellular phosphate levels. Proc. Natl. Acad. Sci. U.S.A. 2005;102:9565–9570. doi: 10.1073/pnas.0501122102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neef D.W., Kladde M.P. Polyphosphate loss promotes SNF/SWI- and Gcn5-dependent mitotic induction of PHO5. Mol. Cell. Biol. 2003;23:3788–3797. doi: 10.1128/MCB.23.11.3788-3797.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oshima Y. The phosphatase system in Saccharomyces cerevisiae. Genes Genet. Syst. 1997;72:323–334. doi: 10.1266/ggs.72.323. [DOI] [PubMed] [Google Scholar]
- 16.Ogawa N., Oshima Y. Functional domains of a positive regulatory protein, PHO4, for transcriptional control of the phosphatase regulon in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990;10:2224–2236. doi: 10.1128/mcb.10.5.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogawa N., Saitoh H., Miura K., Magbanua J.P.V., Bunya M., Harashima S., Oshima Y. Structure and distribution of specific cis-elements for transcriptional regulation of PHO84 in Saccharomyces cerevisiae. Mol. Gen. Genet. 1995;249:406–416. doi: 10.1007/BF00287102. [DOI] [PubMed] [Google Scholar]
- 18.Barbaric S., Fascher K.D., Hörz W. Activation of the weakly regulated PHO8 promoter in S. cerevisiae: chromatin transition and binding sites for the positive regulator protein Pho4. Nucleic Acids Res. 1992;20:1031–1038. doi: 10.1093/nar/20.5.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogel K., Hörz W., Hinnen A. The two positively acting regulatory proteins PHO2 and PHO4 physically interact with PHO5 upstream activation regions. Mol. Cell. Biol. 1989;9:2050–2057. doi: 10.1128/mcb.9.5.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barbaric S., Münsterkötter M., Svaren J., Hörz W. The homeodomain protein Pho2 and the basic-helix-loop-helix protein Pho4 bind DNA cooperatively at the yeast PHO5 promoter. Nucleic Acids Res. 1996;24:4479–4486. doi: 10.1093/nar/24.22.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barbaric S., Münsterkötter M., Goding C., Hörz W. Cooperative Pho2-Pho4 interactions at the PHO5 promoter are critical for binding of Pho4 to UASp1 and for efficient transactivation by Pho4 at UASp2. Mol. Cell. Biol. 1998;18:2629–2639. doi: 10.1128/mcb.18.5.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshida K., Kuromitsu Z., Ogawa N., Oshima Y. Mode of expression of the positive regulatory genes PHO2 and PHO4 of the phosphatase regulon in Saccharomyces cerevisiae. Mol. Gen. Genet. 1989;217:31–39. doi: 10.1007/BF00330939. [DOI] [PubMed] [Google Scholar]
- 23.Jeffery D.A., Springer M., King D.S., O'Shea E.K. Multi-site phosphorylation of Pho4 by the cyclin-CDK Pho80-Pho85 is semi-processive with site preference. J. Mol. Biol. 2001;306:997–1010. doi: 10.1006/jmbi.2000.4417. [DOI] [PubMed] [Google Scholar]
- 24.Komeili A., O'Shea E.K. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science. 1999;284:977–980. doi: 10.1126/science.284.5416.977. [DOI] [PubMed] [Google Scholar]
- 25.Kaffman A., Rank N.M., O'Neill E.M., Huang L.S., O'Shea E.K. The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature. 1998;396:482–486. doi: 10.1038/24898. [DOI] [PubMed] [Google Scholar]
- 26.O'Neill E.M., Kaffman A., Jolly E.R., O'Shea E.K. Regulation of Pho4 nuclear localization by the Pho80-Pho85 cyclin-CDK complex. Science. 1996;271:209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- 27.Schneider K.R., Smith R.L., O'Shea E.K. Phosphate-regulated inactivation of the kinase PH080-PH085 by the CDK inhibitor PH081. Science. 1994;266:122–126. doi: 10.1126/science.7939631. [DOI] [PubMed] [Google Scholar]
- 28.Kaffman A., Herskowitz I., Tjian R., O'Shea E.K. Phosphorylation of the transcription factor Pho4 by a cyclin-CDK complex, Pho80-Pho85. Science. 1994;263:1153–1156. doi: 10.1126/science.8108735. [DOI] [PubMed] [Google Scholar]
- 29.Lee Y.S., Huang K., Quiocho F.A., O'Shea E.K. Molecular basis of cyclin-CDK-CKI regulation by reversible binding of an inositol pyrophosphate. Nat. Chem. Biol. 2008;4:25–32. doi: 10.1038/nchembio.2007.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee Y.S., Mulugu S., York J.D., O'Shea E.K. Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science. 2007;316:109–112. doi: 10.1126/science.1139080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El Alami M., Messenguy F., Scherens B., Dubois E. Arg82p is a bifunctional protein whose inositol polyphosphate kinase activity is essential for nitrogen and PHO gene expression but not for Mcm1p chaperoning in yeast. Mol. Microbiol. 2003;49:457–468. doi: 10.1046/j.1365-2958.2003.03562.x. [DOI] [PubMed] [Google Scholar]
- 32.Madden S.L., Creasy C.L., Srinivas V., Fawcett W., Bergman L.W. Structure and expression of the PHO80 gene of Saccharomyces cerevisiae. Nucleic Acids Res. 1988;16:2625–2637. doi: 10.1093/nar/16.6.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carroll A.S., Bishop A.C., DeRisi J.L., Shokat K.M., O'Shea E.K. Chemical inhibition of the Pho85 cyclin-dependent kinase reveals a role in the environmental stress response. Proc. Natl. Acad. Sci. U.S.A. 2001;98:12578–12583. doi: 10.1073/pnas.211195798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fascher K.D., Schmitz J., Hörz W. Role of trans-activating proteins in the generation of active chromatin at the PHO5 promoter in S. cerevisiae. EMBO J. 1990;9:2523–2528. doi: 10.1002/j.1460-2075.1990.tb07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spellman P.T., Sherlock G., Zhang M.Q., Iyer V.R., Anders K., Eisen M.B., Brown P.O., Botstein D., Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pondugula S., Neef D.W., Voth W.P., Darst R.P., Dhasarathy A., Reynolds M.M., Takahata S., Stillman D.J., Kladde M.P. Coupling phosphate homeostasis to cell cycle-specific transcription: mitotic activation of Saccharomyces cerevisiae PHO5 by Mcm1 and Forkhead proteins. Mol. Cell. Biol. 2009;29:4891–4905. doi: 10.1128/MCB.00222-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arnold W.N. Location of acid phosphatase and -fructofuranosidase within yeast cell envelopes. J. Bacteriol. 1972;112:1346–1352. doi: 10.1128/jb.112.3.1346-1352.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Rijn H.J., Boer P., Steyn-Parve E.P. Biosynthesis of acid phosphatase of baker's yeast. Factors influencing its production by protoplasts and characterization of the secreted enzyme. Biochim. Biophys. Acta. 1972;268:431–441. doi: 10.1016/0005-2744(72)90339-7. [DOI] [PubMed] [Google Scholar]
- 39.Mildner P., Barbaric S., Golubic Z., Ries B. Purification of protoplast-secreted acid phosphatase from baker's yeast. Effect on adenosine triphosphatase activity. Biochim. Biophys. Acta. 1976;429:274–282. doi: 10.1016/0005-2744(76)90050-4. [DOI] [PubMed] [Google Scholar]
- 40.Lemire J.M., Willcocks T., Halvorson H.O., Bostian K.A. Regulation of repressible acid phosphatase gene transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 1985;5:2131–2141. doi: 10.1128/mcb.5.8.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Almer A., Rudolph H., Hinnen A., Hörz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986;5:2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Almer A., Hörz W. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J. 1986;5:2681–2687. doi: 10.1002/j.1460-2075.1986.tb04551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bergman L.W., Stranathan M.C., Preis L.H. Structure of the transcriptionally repressed phosphate- repressible acid phosphatase gene (PHO5) of Saccharomyces cerevisiae. Mol. Cell. Biol. 1986;6:38–46. doi: 10.1128/mcb.6.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bergman L.W., Kramer R.A. Modulation of chromatin structure associated with derepression of the acid phosphatase gene of Saccharomyces cerevisiae. J. Biol. Chem. 1983;258:7223–7227. [PubMed] [Google Scholar]
- 45.Yuan G.C., Liu Y.J., Dion M.F., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 46.Shivaswamy S., Bhinge A., Zhao Y., Jones S., Hirst M., Iyer V.R. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 2008;6:e65. doi: 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rizzo J.M., Mieczkowski P.A., Buck M.J. Tup1 stabilizes promoter nucleosome positioning and occupancy at transcriptionally plastic genes. Nucleic Acids Res. 2011;39:8803–8819. doi: 10.1093/nar/gkr557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cole H.A., Howard B.H., Clark D.J. Activation-induced disruption of nucleosome position clusters on the coding regions of Gcn4-dependent genes extends into neighbouring genes. Nucleic Acids Res. 2011;39:9521–9535. doi: 10.1093/nar/gkr643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Albert I., Mavrich T.N., Tomsho L.P., Qi J., Zanton S.J., Schuster S.C., Pugh B.F. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 50.Huebert D.J., Kuan P.F., Keles S., Gasch A.P. Dynamic changes in nucleosome occupancy are not predictive of gene expression dynamics but are linked to transcription and chromatin regulators. Mol. Cell. Biol. 2012;32:1645–1653. doi: 10.1128/MCB.06170-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zawadzki K.A., Morozov A.V., Broach J.R. Chromatin-dependent transcription factor accessibility rather than nucleosome remodeling predominates during global transcriptional restructuring in Saccharomyces cerevisiae. Mol. Biol. Cell. 2009;20:3503–3513. doi: 10.1091/mbc.E09-02-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee W., Tillo D., Bray N., Morse R.H., Davis R.W., Hughes T.R., Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 53.Kornberg R.D., Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 54.Olins D.E., Olins A.L. Chromatin history: our view from the bridge. Nat. Rev. Mol. Cell. Biol. 2003;4:809–814. doi: 10.1038/nrm1225. [DOI] [PubMed] [Google Scholar]
- 55.Olins A.L., Olins D.E. Spheroid chromatin units (v bodies) Science. 1974;183:330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- 56.Kornberg R.D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 57.Wu C. The 5’ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980;286:854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- 58.Nedospasov S.A., Georgiev G.P. Non-random cleavage of SV40 DNA in the compact minichromosome and free in solution by micrococcal nuclease. Biochem. Biophys. Res. Commun. 1980;92:532–539. doi: 10.1016/0006-291x(80)90366-6. [DOI] [PubMed] [Google Scholar]
- 59.Elgin S.C. DNAase I-hypersensitive sites of chromatin. Cell. 1981;27:413–415. doi: 10.1016/0092-8674(81)90381-0. [DOI] [PubMed] [Google Scholar]
- 60.Reinke H., Hörz W. Anatomy of a hypersensitive site. Biochim. Biophys. Acta. 2004;1677:24–29. doi: 10.1016/j.bbaexp.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 61.Elgin S.C. Anatomy of hypersensitive sites. Nature. 1984;309:213–214. doi: 10.1038/309213a0. [DOI] [PubMed] [Google Scholar]
- 62.Fascher K.D., Schmitz J., Hörz W. Structural and functional requirements for the chromatin transition at the PHO5 promoter in Saccharomyces cerevisiae upon PHO5 activation. J. Mol. Biol. 1993;231:658–667. doi: 10.1006/jmbi.1993.1317. [DOI] [PubMed] [Google Scholar]
- 63.Bergman L.W. A DNA fragment containing the upstream activator sequence determines nucleosome positioning of the transcriptionally repressed PHO5 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 1986;6:2298–2304. doi: 10.1128/mcb.6.7.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Han M., Grunstein M. Nucleosome loss activates yeast downstream promoters in vivo. Cell. 1988;55:1137–1145. doi: 10.1016/0092-8674(88)90258-9. [DOI] [PubMed] [Google Scholar]
- 65.Han M., Kim U.J., Kayne P., Grunstein M. Depletion of histone H4 and nucleosomes activates the PHO5 gene in Saccharomyces cerevisiae. EMBO J. 1988;7:2221–2228. doi: 10.1002/j.1460-2075.1988.tb03061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim U.J., Han M., Kayne P., Grunstein M. Effects of histone H4 depletion on the cell cycle and transcription of Saccharomyces cerevisiae. EMBO J. 1988;7:2211–2219. doi: 10.1002/j.1460-2075.1988.tb03060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hirschhorn J.N., Brown S.A., Clark C.D., Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 68.Brown C.R., Mao C., Falkovskaia E., Jurica M.S., Boeger H. Linking stochastic fluctuations in chromatin structure and gene expression. PLoS Biol. 2013;11:e1001621. doi: 10.1371/journal.pbio.1001621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kaplan C.D., Laprade L., Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- 70.Adkins M.W., Tyler J.K. Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Mol. Cell. 2006;21:405–416. doi: 10.1016/j.molcel.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 71.Smolle M., Workman J.L. Transcription-associated histone modifications and cryptic transcription. Biochim. Biophys. Acta. 2012;1829:84–97. doi: 10.1016/j.bbagrm.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Owen-Hughes T., Gkikopoulos T. Making sense of transcribing chromatin. Curr. Opin. Cell. Biol. 2012;24:296–304. doi: 10.1016/j.ceb.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Svaren J., Chalkley R. The structure and assembly of active chromatin. Trends Genet. 1990;6:52–56. doi: 10.1016/0168-9525(90)90074-g. [DOI] [PubMed] [Google Scholar]
- 74.Workman J.L., Kingston R.E. Nucleosome core displacement in vitro via a metastable transcription factor nucleosome complex. Science. 1992;258:1780–1784. doi: 10.1126/science.1465613. [DOI] [PubMed] [Google Scholar]
- 75.Morse R.H. Nucleosome disruption by transcription factor binding in yeast. Science. 1993;262:1563–1566. doi: 10.1126/science.8248805. [DOI] [PubMed] [Google Scholar]
- 76.Venter U., Svaren J., Schmitz J., Schmid A., Hörz W. A nucleosome precludes binding of the transcription factor Pho4 in vivo to a critical target site in the PHO5 promoter. EMBO J. 1994;13:4848–4855. doi: 10.1002/j.1460-2075.1994.tb06811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Svaren J., Schmitz J., Hörz W. The transactivation domain of Pho4 is required for nucleosome disruption at the PHO5 promoter. EMBO J. 1994;13:4856–4862. doi: 10.1002/j.1460-2075.1994.tb06812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Then Bergh F., Flinn E.M., Svaren J., Wright A.P., Hörz W. Comparison of nucleosome remodeling by the yeast transcription factor Pho4 and the glucocorticoid receptor. J. Biol. Chem. 2000;275:9035–9042. doi: 10.1074/jbc.275.12.9035. [DOI] [PubMed] [Google Scholar]
- 79.Gaudreau L., Schmid A., Blaschke D., Ptashne M., Hörz W. RNA polymerase II holoenzyme recruitment is sufficient to remodel chromatin at the yeast PHO5 promoter. Cell. 1997;89:55–62. doi: 10.1016/s0092-8674(00)80182-8. [DOI] [PubMed] [Google Scholar]
- 80.Ansari S.A., Paul E., Sommer S., Lieleg C., He Q., Daly A.Z., Rode K.A., Barber W.T., Ellis L.C., Laporta E., et al. Mediator, TATA-Binding Protein, and RNA Polymerase II contribute to low histone occupancy at active gene promoters in yeast. J. Biol. Chem. 2014;289:14981–14995. doi: 10.1074/jbc.M113.529354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ertinger G. Ph.D Thesis. Universität München; 1998. [Google Scholar]
- 82.Barbaric S., Walker J., Schmid A., Svejstrup J.Q., Hörz W. Increasing the rate of chromatin remodeling and gene activation - a novel role for the histone acetyltransferase Gcn5. EMBO J. 2001;20:4944–4951. doi: 10.1093/emboj/20.17.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McAndrew P.C., Svaren J., Martin S.R., Hörz W., Goding C.R. Requirements for chromatin modulation and transcription activation by the Pho4 acidic activation domain. Mol. Cell. Biol. 1998;18:5818–5827. doi: 10.1128/mcb.18.10.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hahn S., Young E.T. Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics. 2011;189:705–736. doi: 10.1534/genetics.111.127019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Côté J., Quinn J., Workman J.L., Peterson C.L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 86.Laurent B.C., Treich I., Carlson M. The yeast SNF2/SWI2-protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 1993;7:583–591. doi: 10.1101/gad.7.4.583. [DOI] [PubMed] [Google Scholar]
- 87.Neely K.E., Hassan A.H., Brown C.E., Howe L., Workman J.L. Transcription activator interactions with multiple SWI/SNF subunits. Mol. Cell. Biol. 2002;22:1615–1625. doi: 10.1128/MCB.22.6.1615-1625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Flaus A., Martin D.M., Barton G.J., Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ehrensberger A.H., Kornberg R.D. Isolation of an activator-dependent, promoter-specific chromatin remodeling factor. Proc. Natl. Acad. Sci. U.S.A. 2011;108:10115–10120. doi: 10.1073/pnas.1101449108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reinke H., Hörz W. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell. 2003;11:1599–1607. doi: 10.1016/s1097-2765(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 91.Barbaric S., Luckenbach T., Schmid A., Blaschke D., Hörz W., Korber P. Redundancy of chromatin remodeling pathways for the induction of the yeast PHO5 promoter in vivo. J. Biol. Chem. 2007;282:27610–27621. doi: 10.1074/jbc.M700623200. [DOI] [PubMed] [Google Scholar]
- 92.Steger D.J., Haswell E.S., Miller A.L., Wente S.R., O'Shea E.K. Regulation of chromatin remodeling by inositol polyphosphates. Science. 2003;299:114–116. doi: 10.1126/science.1078062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huang S., O'Shea E.K. A systematic high-throughput screen of a yeast deletion collection for mutants defective in PHO5 regulation. Genetics. 2005;169:1859–1871. doi: 10.1534/genetics.104.038695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dhasarathy A., Kladde M.P. Promoter occupancy is a major determinant of chromatin remodeling enzyme requirements. Mol. Cell. Biol. 2005;25:2698–2707. doi: 10.1128/MCB.25.7.2698-2707.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Musladin S., Krietenstein N., Korber P., Barbaric S. The RSC chromatin remodeling complex has a crucial role in the complete remodeler set for yeast PHO5 promoter opening. Nucleic Acids Res. 2014;42:4270–4282. doi: 10.1093/nar/gkt1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brown C.R., Mao C., Falkovskaia E., Law J.K., Boeger H. In vivo role for the chromatin-remodeling enzyme SWI/SNF in the removal of promoter nucleosomes by disassembly rather than sliding. J. Biol. Chem. 2011;286:40556–40565. doi: 10.1074/jbc.M111.289918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Grant P.A., Duggan L., Côté J., Roberts S.M., Brownell J.E., Candau R., Ohba R., Owen-Hughes T., Allis C.D., Winston F., et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt-Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 98.Kuo M.H., Brownell J.E., Sobel R.E., Ranalli T.A., Cook R.G., Edmondson D.G., Roth S.Y., Allis C.D. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 99.Brownell J.E., Zhou J.X., Ranalli T., Kobayashi R., Edmondson D.G., Roth S.Y., Allis C.D. Tetrahymena histone acetyltransferase a: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 100.Steger D.J., O'Shea E.K. Genetic analysis of chromatin remodeling using budding yeast as a model. Methods Enzymol. 2004;377:55–60. doi: 10.1016/S0076-6879(03)77002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gregory P.D., Schmid A., Zavari M., Lui L., Berger S.L., Hörz W. Absence of Gcn5 HAT activity defines a novel state in the opening of chromatin at the PHO5 promoter in yeast. Mol. Cell. 1998;1:495–505. doi: 10.1016/s1097-2765(00)80050-7. [DOI] [PubMed] [Google Scholar]
- 102.Gregory P.D., Schmid A., Zavari M., Münsterkötter M., Hörz W. Chromatin remodelling at the PHO8 promoter requires SWI-SNF and SAGA at a step subsequent to activator binding. EMBO J. 1999;18:6407–6414. doi: 10.1093/emboj/18.22.6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Weiner A., Chen H.V., Liu C.L., Rahat A., Klien A., Soares L., Gudipati M., Pfeffner J., Regev A., Buratowski S., et al. Systematic dissection of roles for chromatin regulators in a yeast stress response. PLoS. Biol. 2012;10:e1001369. doi: 10.1371/journal.pbio.1001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Korber P., Barbaric S., Luckenbach T., Schmid A., Schermer U.J., Blaschke D., Hörz W. The histone chaperone Asf1 increases the rate of histone eviction at the yeast PHO5 and PHO8 promoters. J. Biol. Chem. 2006;281:5539–5545. doi: 10.1074/jbc.M513340200. [DOI] [PubMed] [Google Scholar]
- 105.Wippo C.J., Krstulovic B.S., Ertel F., Musladin S., Blaschke D., Sturzl S., Yuan G.C., Hörz W., Korber P., Barbaric S. Differential cofactor requirements for histone eviction from two nucleosomes at the yeast PHO84 promoter are determined by intrinsic nucleosome stability. Mol. Cell. Biol. 2009;29:2960–2981. doi: 10.1128/MCB.01054-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Adkins M.W., Howar S.R., Tyler J.K. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 Genes. Mol. Cell. 2004;14:657–666. doi: 10.1016/j.molcel.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 107.Adkins M.W., Williams S.K., Linger J., Tyler J.K. Chromatin disassembly from the PHO5 promoter is essential for the recruitment of the general transcription machinery and coactivators. Mol. Cell. Biol. 2007;27:6372–6382. doi: 10.1128/MCB.00981-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Venters B.J., Pugh B.F. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 2009;19:360–371. doi: 10.1101/gr.084970.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Barbaric S., Reinke H., Hörz W. Multiple mechanistically distinct functions of SAGA at the PHO5 promoter. Mol. Cell. Biol. 2003;23:3468–3476. doi: 10.1128/MCB.23.10.3468-3476.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang S.S., Zhou B.O., Zhou J.Q. Histone H3 lysine 4 hypermethylation prevents aberrant nucleosome remodeling at the PHO5 promoter. Mol. Cell. Biol. 2011;31:3171–3181. doi: 10.1128/MCB.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hamperl S., Brown C.R., Garea A.V., Perez-Fernandez J., Bruckmann A., Huber K., Wittner M., Babl V., Stoeckl U., Deutzmann R., et al. Compositional and structural analysis of selected chromosomal domains from Saccharomyces cerevisiae. Nucleic Acids Res. 2014;42 doi: 10.1093/nar/gkt891. doi:10.1093/nar/gkt891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hassan A.H., Prochasson P., Neely K.E., Galasinski S.C., Chandy M., Carrozza M.J., Workman J.L. Function and selectivity of bromodomains in anchoring chromatin- modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- 113.Small E.C., Xi L., Wang J.P., Widom J., Licht J.D. Single-cell nucleosome mapping reveals the molecular basis of gene expression heterogeneity. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E2462–E2471. doi: 10.1073/pnas.1400517111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nourani A., Utley R.T., Allard S., Cote J. Recruitment of the NuA4 complex poises the PHO5 promoter for chromatin remodeling and activation. EMBO J. 2004;23:2597–2607. doi: 10.1038/sj.emboj.7600230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Williams S.K., Truong D., Tyler J.K. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9000–9005. doi: 10.1073/pnas.0800057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fisher F., Goding C.R. Single amino acid substitutions alter helix-loop-helix protein specificity for bases flanking the core CANNTG motif. EMBO J. 1992;11:4103–4109. doi: 10.1002/j.1460-2075.1992.tb05503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dion M.F., Kaplan T., Kim M., Buratowski S., Friedman N., Rando O.J. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 118.Jamai A., Imoberdorf R.M., Strubin M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol. Cell. 2007;25:345–355. doi: 10.1016/j.molcel.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 119.Rufiange A., Jacques P.E., Bhat W., Robert F., Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol. Cell. 2007;27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 120.Wongwisansri S., Laybourn P.J. Disruption of histone deacetylase gene RPD3 accelerates PHO5 activation kinetics through inappropriate Pho84p recycling. Eukaryotic Cell. 2005;4:1387–1395. doi: 10.1128/EC.4.8.1387-1395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Colina A.R., Young D. Raf60, a novel component of the Rpd3 histone deacetylase complex required for Rpd3 activity in Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:42552–42556. doi: 10.1074/jbc.M511561200. [DOI] [PubMed] [Google Scholar]
- 122.Carvin C.D., Kladde M.P. Effectors of lysine 4 methylation of histone H3 in Saccharomyces cerevisiae are negative regulators of PHO5 and GAL1-10. J. Biol. Chem. 2004;279:33057–33062. doi: 10.1074/jbc.M405033200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Millar C.B., Grunstein M. Genome-wide patterns of histone modifications in yeast. Nat. Rev. Mol. Cell. Biol. 2006;7:657–666. doi: 10.1038/nrm1986. [DOI] [PubMed] [Google Scholar]
- 124.Marques M., Laflamme L., Gervais A.L., Gaudreau L. Reconciling the positive and negative roles of histone H2A.Z in gene transcription. Epigenetics. 2010;5:267–272. doi: 10.4161/epi.5.4.11520. [DOI] [PubMed] [Google Scholar]
- 125.Watanabe S., Radman-Livaja M., Rando O.J., Peterson C.L. A histone acetylation switch regulates H2A.Z deposition by the SWR-C remodeling enzyme. Science. 2013;340:195–199. doi: 10.1126/science.1229758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Millar C.B., Xu F., Zhang K., Grunstein M. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 2006;20:711–722. doi: 10.1101/gad.1395506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Uhler J.P., Hertel C., Svejstrup J.Q. A role for noncoding transcription in activation of the yeast PHO5 gene. Proc. Natl. Acad. Sci. U.S.A. 2007;104:8011–8016. doi: 10.1073/pnas.0702431104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mao C., Brown C.R., Griesenbeck J., Boeger H. Occlusion of regulatory sequences by promoter nucleosomes in vivo. PLoS One. 2011;6:e17521. doi: 10.1371/journal.pone.0017521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ertel F., Dirac-Svejstrup A.B., Hertel C.B., Blaschke D., Svejstrup J.Q., Korber P. In vitro reconstitution of PHO5 promoter chromatin remodeling points to a role for activator-nucleosome competition in vivo. Mol. Cell. Biol. 2010;30:4060–4076. doi: 10.1128/MCB.01399-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bucceri A., Kapitza K., Thoma F. Rapid accessibility of nucleosomal DNA in yeast on a second time scale. EMBO J. 2006;25:3123–3132. doi: 10.1038/sj.emboj.7601196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schmid M., Durussel T., Laemmli U.K. ChIC and ChEC; genomic mapping of chromatin proteins. Mol. Cell. 2004;16:147–157. doi: 10.1016/j.molcel.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 132.Shimizu T., Toumoto A., Ihara K., Shimizu M., Kyogoku Y., Ogawa N., Oshima Y., Hakoshima T. Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition. EMBO J. 1997;16:4689–4697. doi: 10.1093/emboj/16.15.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ransom M., Williams S.K., Dechassa M.L., Das C., Linger J., Adkins M., Liu C., Bartholomew B., Tyler J.K. FACT and the proteasome promote promoter chromatin disassembly and transcriptional initiation. J. Biol. Chem. 2009;284:23461–23471. doi: 10.1074/jbc.M109.019562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Boeger H., Griesenbeck J., Strattan J.S., Kornberg R.D. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol. Cell. 2004;14:667–673. doi: 10.1016/j.molcel.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 135.Boeger H., Griesenbeck J., Kornberg R.D. Nucleosome retention and the stochastic nature of promoter chromatin remodeling for transcription. Cell. 2008;133:716–726. doi: 10.1016/j.cell.2008.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lorch Y., Maier-Davis B., Kornberg R.D. Chromatin remodeling by nucleosome disassembly in vitro. Proc. Natl. Acad. Sci. U.S.A. 2006;103:3090–3093. doi: 10.1073/pnas.0511050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lorch Y., Griesenbeck J., Boeger H., Maier-Davis B., Kornberg R.D. Selective removal of promoter nucleosomes by the RSC chromatin-remodeling complex. Nat. Struct. Mol. Biol. 2011;18:881–885. doi: 10.1038/nsmb.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Morris S.A., Baek S., Sung M.H., John S., Wiench M., Johnson T.A., Schiltz R.L., Hager G.L. Overlapping chromatin-remodeling systems collaborate genome wide at dynamic chromatin transitions. Nat. Struct. Mol. Biol. 2014;21:73–81. doi: 10.1038/nsmb.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kent N.A., Karabetsou N., Politis P.K., Mellor J. In vivo chromatin remodeling by yeast ISWI homologs Isw1p and Isw2p. Genes Dev. 2001;15:619–626. doi: 10.1101/gad.190301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Schnitzler G., Sif S., Kingston R.E. Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell. 1998;94:17–27. doi: 10.1016/s0092-8674(00)81217-9. [DOI] [PubMed] [Google Scholar]
- 141.Shukla M.S., Syed S.H., Montel F., Faivre-Moskalenko C., Bednar J., Travers A., Angelov D., Dimitrov S. Remosomes: RSC generated non-mobilized particles with approximately 180 bp DNA loosely associated with the histone octamer. Proc. Natl. Acad. Sci. U.S.A. 2010;107:1936–1941. doi: 10.1073/pnas.0904497107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Côté J., Peterson C.L., Workman J.L. Perturbation of nucleosome core structure by the SWI/SNF complex persists after its detachment, enhancing subsequent transcription factor binding. Proc. Natl. Acad. Sci. U.S.A. 1998;95:4947–4952. doi: 10.1073/pnas.95.9.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Boeger H., Griesenbeck J., Strattan J.S., Kornberg R.D. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell. 2003;11:1587–1598. doi: 10.1016/s1097-2765(03)00231-4. [DOI] [PubMed] [Google Scholar]
- 144.Zhang H., Kruk J.A., Reese J.C. Dissection of coactivator requirement at RNR3 reveals unexpected contributions from TFIID and SAGA. J. Biol. Chem. 2008;283:27360–27368. doi: 10.1074/jbc.M803831200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bernstein B.E., Liu C.L., Humphrey E.L., Perlstein E.O., Schreiber S.L. Global nucleosome occupancy in yeast. Genome Biol. 2004;5:R62. doi: 10.1186/gb-2004-5-9-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lee C.K., Shibata Y., Rao B., Strahl B.D., Lieb J.D. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- 147.Reinke H., Gregory P.D., Hörz W. A transient histone hyperacetylation signal marks nucleosomes for remodeling at the PHO8 promoter. Mol. Cell. 2001;7:529–538. doi: 10.1016/s1097-2765(01)00200-3. [DOI] [PubMed] [Google Scholar]
- 148.Krebs J.E., Fry C.J., Samuels M.L., Peterson C.L. Global role for chromatin remodeling enzymes in mitotic gene expression. Cell. 2000;102:587–598. doi: 10.1016/s0092-8674(00)00081-7. [DOI] [PubMed] [Google Scholar]
- 149.Pepenella S., Murphy K.J., Hayes J.J. Intra- and inter-nucleosome interactions of the core histone tail domains in higher-order chromatin structure. Chromosoma. 2014;123:3–13. doi: 10.1007/s00412-013-0435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Längst G., Bonte E.J., Corona D.F., Becker P.B. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell. 1999;97:843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- 151.Mueller-Planitz F., Klinker H., Becker P.B. Nucleosome sliding mechanisms: new twists in a looped history. Nat. Struct. Mol. Biol. 2013;20:1026–1032. doi: 10.1038/nsmb.2648. [DOI] [PubMed] [Google Scholar]
- 152.Korber P., Luckenbach T., Blaschke D., Hörz W. Evidence for histone eviction in trans upon induction of the yeast PHO5 promoter. Mol. Cell. Biol. 2004;24:10965–10974. doi: 10.1128/MCB.24.24.10965-10974.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dechassa M.L., Sabri A., Pondugula S., Kassabov S.R., Chatterjee N., Kladde M.P., Bartholomew B. SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes. Mol. Cell. 2010;38:590–602. doi: 10.1016/j.molcel.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Schermer U.J., Korber P., Hörz W. Histones are incorporated in trans during reassembly of the yeast PHO5 promoter. Mol.Cell. 2005;19:279–285. doi: 10.1016/j.molcel.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 155.Radman-Livaja M., Verzijlbergen K.F., Weiner A., van Welsem T., Friedman N., Rando O.J., van Leeuwen F. Patterns and mechanisms of ancestral histone protein inheritance in budding yeast. PLoS Biol. 2011;9:e1001075. doi: 10.1371/journal.pbio.1001075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tsubota T., Berndsen C.E., Erkmann J.A., Smith C.L., Yang L., Freitas M.A., Denu J.M., Kaufman P.D. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol. Cell. 2007;25:703–712. doi: 10.1016/j.molcel.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Han J., Zhou H., Horazdovsky B., Zhang K., Xu R.M., Zhang Z. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 2007;315:653–655. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- 158.Driscoll R., Hudson A., Jackson S.P. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315:649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mao C., Brown C.R., Falkovskaia E., Dong S., Hrabeta-Robinson E., Wenger L., Boeger H. Quantitative analysis of the transcription control mechanism. Mol. Syst. Biol. 2010;6:431. doi: 10.1038/msb.2010.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kilgore J.A., Hoose S.A., Gustafson T.L., Porter W., Kladde M.P. Single-molecule and population probing of chromatin structure using DNA methyltransferases. Methods. 2007;41:320–332. doi: 10.1016/j.ymeth.2006.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jessen W.J., Hoose S.A., Kilgore J.A., Kladde M.P. Active PHO5 chromatin encompasses variable numbers of nucleosomes at individual promoters. Nat. Struct. Mol. Biol. 2006;13:256–263. doi: 10.1038/nsmb1062. [DOI] [PubMed] [Google Scholar]
- 162.Cairns B.R. Chromatin remodeling: insights and intrigue from single-molecule studies. Nat. Struct. Mol. Biol. 2007;14:989–996. doi: 10.1038/nsmb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Engeholm M., de Jager M., Flaus A., Brenk R., van Noort J., Owen-Hughes T. Nucleosomes can invade DNA territories occupied by their neighbors. Nat. Struct. Mol. Biol. 2009;16:151–158. doi: 10.1038/nsmb.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Patel A., Chakravarthy S., Morrone S., Nodelman I.M., McKnight J.N., Bowman G.D. Decoupling nucleosome recognition from DNA binding dramatically alters the properties of the Chd1 chromatin remodeler. Nucleic Acids Res. 2013;41:1637–1648. doi: 10.1093/nar/gks1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lam F.H., Steger D.J., O'Shea E.K. Chromatin decouples promoter threshold from dynamic range. Nature. 2008;453:246–250. doi: 10.1038/nature06867.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wippo C.J., Israel L., Watanabe S., Hochheimer A., Peterson C.L., Korber P. The RSC chromatin remodelling enzyme has a unique role in directing the accurate positioning of nucleosomes. EMBO J. 2011;30:1277–1288. doi: 10.1038/emboj.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hertel C.B., Längst G., Hörz W., Korber P. Nucleosome stability at the yeast PHO5 and PHO8 promoters correlates with differential cofactor requirements for chromatin opening. Mol. Cell. Biol. 2005;25:10755–10767. doi: 10.1128/MCB.25.24.10755-10767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Haswell E.S., O'Shea E.K. An in vitro system recapitulates chromatin remodeling at the PHO5 promoter. Mol. Cell. Biol. 1999;19:2817–2827. doi: 10.1128/mcb.19.4.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Terrell A.R., Wongwisansri S., Pilon J.L., Laybourn P.J. Reconstitution of nucleosome positioning, remodeling, histone acetylation, and transcriptional activation on the PHO5 promoter. J. Biol. Chem. 2002;277:31038–31047. doi: 10.1074/jbc.M204662200. [DOI] [PubMed] [Google Scholar]
- 170.Korber P., Hörz W. In vitro assembly of the characteristic chromatin organization at the yeast PHO5 promoter by a replication-independent extract system. J. Biol. Chem. 2004;279:35113–35120. doi: 10.1074/jbc.M405446200. [DOI] [PubMed] [Google Scholar]
- 171.Wippo C.J., Korber P. In vitro reconstitution of in vivo-like nucleosome positioning on yeast DNA. Methods Mol. Biol. 2012;833:271–287. doi: 10.1007/978-1-61779-477-3_17. [DOI] [PubMed] [Google Scholar]
- 172.Krietenstein N., Wippo C.J., Lieleg C., Korber P. Genome-wide in vitro reconstitution of yeast chromatin with in vivo-like nucleosome positioning. Methods Enzymol. 2012;513:205–232. doi: 10.1016/B978-0-12-391938-0.00009-4. [DOI] [PubMed] [Google Scholar]
- 173.Zhang Z., Wippo C.J., Wal M., Ward E., Korber P., Pugh B.F. A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science. 2011;332:977–980. doi: 10.1126/science.1200508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Raser J.M., O'Shea E.K. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Weake V.M., Workman J.L. Inducible gene expression: diverse regulatory mechanisms. Nat. Rev. Genet. 2010;11:426–437. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- 176.Clapier C.R., Cairns B.R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 177.Kim H.D., O'Shea E.K. A quantitative model of transcription factor-activated gene expression. Nat. Struct. Mol. Biol. 2008;15:1192–1198. doi: 10.1038/nsmb.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Liu X., Lee C.K., Granek J.A., Clarke N.D., Lieb J.D. Whole-genome comparison of Leu3 binding in vitro and in vivo reveals the importance of nucleosome occupancy in target site selection. Genome Res. 2006;16:1517–1528. doi: 10.1101/gr.5655606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Münsterkötter M., Barbaric S., Hörz W. Transcriptional regulation of the yeast PHO8 promoter in comparison to the coregulated PHO5 promoter. J. Biol. Chem. 2000;275:22678–22685. doi: 10.1074/jbc.M001409200. [DOI] [PubMed] [Google Scholar]
- 180.Zhou X., O'Shea E.K. Integrated approaches reveal determinants of genome-wide binding and function of the transcription factor Pho4. Mol. Cell. 2011;42:826–836. doi: 10.1016/j.molcel.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Maerkl S.J., Quake S.R. A systems approach to measuring the binding energy landscapes of transcription factors. Science. 2007;315:233–237. doi: 10.1126/science.1131007. [DOI] [PubMed] [Google Scholar]
- 182.Rajkumar A.S., Denervaud N., Maerkl S.J. Mapping the fine structure of a eukaryotic promoter input-output function. Nat. Genet. 2013;45:1207–1215. doi: 10.1038/ng.2729. [DOI] [PubMed] [Google Scholar]
- 183.Tirosh I., Barkai N. Two strategies for gene regulation by promoter nucleosomes. Genome Res. 2008;18:1084–1091. doi: 10.1101/gr.076059.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Cairns B.R. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 185.Rhee H.S., Pugh B.F. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012;483:295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Becker P.B. Wolfram Horz 1944–2005. Cell. 2006;124:13–14. doi: 10.1016/j.cell.2005.12.021. [DOI] [PubMed] [Google Scholar]


