Background: RNase 1 is an RNA-degrading enzyme conserved in mammals and with unknown biological function.
Results: RNase 1 homologs in human and cow display different biochemical and biological properties, implying divergent physiology.
Conclusion: Bovine brain ribonuclease, not RNase A, is the functional homolog of human RNase 1.
Significance: Fundamental insight into the biology and evolution of human RNase 1 is attained from analyses of homologous proteins.
Keywords: Double-stranded RNA (dsRNA), Enzyme Catalysis, Heparin-binding Protein, Ribonuclease, RNA
Abstract
Mounting evidence suggests that human pancreatic ribonuclease (RNase 1) plays important roles in vivo, ranging from regulating blood clotting and inflammation to directly counteracting tumorigenic cells. Understanding these putative roles has been pursued with continual comparisons of human RNase 1 to bovine RNase A, an enzyme that appears to function primarily in the ruminant gut. Our results imply a different physiology for human RNase 1. We demonstrate distinct functional differences between human RNase 1 and bovine RNase A. Moreover, we characterize another RNase 1 homolog, bovine brain ribonuclease, and find pronounced similarities between that enzyme and human RNase 1. We report that human RNase 1 and bovine brain ribonuclease share high catalytic activity against double-stranded RNA substrates, a rare quality among ribonucleases. Both human RNase 1 and bovine brain RNase are readily endocytosed by mammalian cells, aided by tight interactions with cell surface glycans. Finally, we show that both human RNase 1 and bovine brain RNase are secreted from endothelial cells in a regulated manner, implying a potential role in vascular homeostasis. Our results suggest that brain ribonuclease, not RNase A, is the true bovine homolog of human RNase 1, and provide fundamental insight into the ancestral roles and functional adaptations of RNase 1 in mammals.
Introduction
Pancreatic ribonuclease (RNase) 1 is a small, secreted, RNA-degrading enzyme conserved in mammals (EC 3.1.27.5). Its biological purpose is unknown. Until recently, all assumptions about its physiology were based on studies of a well known bovine homolog, RNase A. This protein is secreted primarily from the bovine exocrine pancreas, and is believed to degrade mRNA from symbiotic bacteria in the rumen, a harsh environment with a normal pH range of 5.8–6.4 and temperatures from 38 to 42 °C (1, 2). Indeed, RNase A seems well suited for this role, possessing extremely high catalytic activity against single-stranded (ss)4 RNA at acidic pH, as well as remarkable thermostability and acid tolerance (3).
Studies of RNase 1 in other species suggest a biological function apart from digestion. Observations in rats demonstrated that RNase 1 levels do not change following periods of fasting or consumption, as with other digestive enzymes (4). In humans, pancreatectomy does not affect circulating RNase 1 levels (5), and we now recognize the primary source of RNase 1 in human blood to be the vascular endothelium (6). Recent work suggests that RNase 1 degrades extracellular RNA, potentially regulating hemostasis, inflammation, and innate immunity (7–11). Data in vitro (12, 13) and in vivo (14–16) have implicated human RNase 1 as having an endogenous anti-cancer function, and clinical trials for a variant of this enzyme are underway (17, 18). Taken together, these data imply a much broader physiological role for mammalian RNase 1 than digestion.
The discrepancies between bovine RNase A and mammalian homologs might reside in the peculiar evolution of RNase 1 in ruminants. Whereas most mammals possess a single RNASE1 gene, evolutionary analyses predict that bovine RNASE1 underwent two major gene duplication events around 30 million years ago, resulting in paralogous genes encoding three distinct proteins: RNase A, seminal ribonuclease (BSR), and brain ribonuclease (BRB). Intriguingly, BSR naturally dimerizes upon folding, whereas all other homologs exist as monomers. Orthologs of these three ribonucleases have been identified in many ruminant species. Although many BSR genes show pseudogene features (including stop-codon insertion, loss of catalytic residues, or loss of dimerization), BRB genes do not, implying a necessary function for the BRB protein (19–21).
Of the three bovine ribonucleases, only BRB is not well characterized. Apart from classic work on RNase A (3), D'Alessio (22), Matoušek (23), and others have established that BSR possesses interesting biological functions not associated with digestion. Indeed, BSR has cytotoxic, aspermagenic, and immunosuppressive activity, likely related to the need to protect sperm cells from the female immune system. BSR is, however, only expressed in the seminal vesicles and testes of Bos taurus, limiting the potential to extrapolate its functions and properties to other mammalian RNase 1 homologs. In contrast, BRB (which was named for its initial discovery and purification from bovine brain (24, 25)) is expressed not only in brain, but in all tissues examined, including endometrium, lymph node, small intestine, liver, and kidney (26). The widespread expression pattern of BRB closely resembles that for human and mouse RNASE1 genes (27). Furthermore, phylogenetic analyses imply that BRB is evolutionarily older than both RNase A and BSR, suggesting greater similarity to the ancestral form of RNase 1 in ruminants (28). RNase A shares greater overall sequence identity with human RNase 1 than does BRB (Table 1). Nevertheless, conclusions based on sequence similarity are not nearly as powerful or as precise as those based on protein function.
TABLE 1.
Biochemical properties of human and bovine ribonucleases
| Ribonuclease | Molecular mass | Za | Identity to RNase 1 | Similarity to RNase 1b | Tmc | Kdd |
|---|---|---|---|---|---|---|
| kDa | % | °C | fm | |||
| RNase 1 (Homo sapiens) | 14.7 | +6 | 100 | 100 | 55.5 ± 0.5 | 0.12 ± 0.1 |
| BRB (B. taurus) | 15.8 | +11 | 61 | 72 | 52.2 ± 0.4 | 0.35 ± 0.21 |
| mBSR (B. taurus) | 13.7 | +9 | 70 | 80 | 60.1 ± 0.5 | 1.94 ± 0.72 |
| dBSR (B. taurus) | 27.5 | +18 | 70 | 80 | 62e | >2 × 109f |
| RNase A (B. taurus) | 13.7 | +4 | 68 | 82 | 63.9 ± 0.4 | 0.16 ± 0.12 |
a Value is for the net molecular charge: Arg + Lys − Asp − Glu.
b For % similarity calculations: K = R; D = E; C = G = H = N = Q = S = T = Y; A = F = I = L = M = P = V = W.
c Value is the temperature at the midpoint of thermal denaturation, determined by incorporation of a hydrophobic dye and quantitation by differential scanning fluorimetry (33).
d Value is for the complex with bovine ribonuclease inhibitor, determined as described (31, 35, 36).
e Value was determined with circular dichroism spectroscopy (75).
f Value is for the complex with human ribonuclease inhibitor (46).
We have performed the first detailed biochemical characterization of BRB. Our data upend the relationship between human RNase 1 and bovine RNase A; instead, the true functional homolog of human RNase 1 in the cow appears to be BRB. Moreover, our findings support the hypothesis that mammalian RNase 1 is not merely a digestive enzyme, but rather an evolutionarily honed vascular regulator.
EXPERIMENTAL PROCEDURES
Equipment
All fluorescence and absorbance measurements were made with a Tecan M1000 fluorescence plate reader, unless stated otherwise. All data were fitted and analyzed with the program Prism 5 (GraphPad), unless stated otherwise.
Cloning, Expression, and Purification of Proteins
DNA fragments encoding human RNase 1, P19C human RNase 1, and H12A human RNase 1; BRB and S19C BRB, RNase A and A19C RNase A; BSR, C31A/C32A mBSR, and P19C/C31A/C32A mBSR; and bovine RI were inserted into the pET22b (Novagen) expression vector for tagless expression in Escherichia coli strain BL21(DE3). All mutations were generated using site-directed mutagenesis. Ribonucleases were purified as inclusion bodies, and variants containing a free cysteine residue were labeled with either 2′,7′-diethylfluorescein (29) or BODIPY FL (Molecular Probes) as described (30). Bovine RI was purified via RNase A affinity chromatography as described (31). Dimeric BSR was isolated as a monomer and allowed to dimerize upon refolding as described (32). Following purification, protein solutions were dialyzed against PBS and filtered prior to use. The molecular mass of each ribonuclease conjugate was confirmed by MALDI-TOF mass spectrometry. Protein concentration was determined by using a bicinchoninic acid assay kit (Pierce) with wild-type RNase A as a standard.
Tm Determination
Thermal unfolding of ribonucleases was monitored in the presence of a fluorescent dye using differential scanning fluorimetry. Differential scanning fluorimetry was performed using a ViiA 7 Real-time PCR machine (Applied Biosystems) as described (33, 34). Briefly, a solution of protein (30 μg) was placed in the wells of a MicroAmp optical 96-well plate, and SYPRO Orange dye (Sigma) was added to a final dye dilution of 1:166 in relationship to the stock solution of the manufacturer. The temperature was increased from 20 to 96 °C at 1 °C/min in steps of 1 °C. Fluorescence intensity was measured at 578 nm, and the resulting data were analyzed with Protein Thermal Shift software (Applied Biosystems). A solution with no protein was used for background correction. Values of Tm were calculated from ∂fluorescence/∂T and are the mean of three independent experiments.
Inhibitor Dissociation Rate
The equilibrium dissociation rates of the RI·ribonuclease complexes were determined as described (31, 35, 36). Briefly, RI and 2′,7′-diethylfluorescein-labeled ribonucleases were mixed in equimolar ratios, and the resulting solution was incubated at 25 °C for 5 min. A 50-fold molar excess of human RNase 1 was added to scavenge dissociated RI. Complex dissociation was measured by monitoring the increasing fluorescence of dissociated ribonuclease over time. Values of Kd are the mean of at least three independent experiments.
pH Dependence of Enzyme Activity
The pH dependence of ribonucleolytic activity with a ssRNA substrate was determined by measuring the initial velocity of cleavage of 6-FAM-dArU(dA)2–6-TAMRA (IDT) (37) (0.2 μm) at pH 4.0–9.0. Assays were carried out in 96-well plates (Corning) at 25 °C in various ribonuclease-free buffers: 0.10 m NaOAc, 0.10 m NaCl (pH 4.0–5.5); 0.10 m BisTris, 0.10 m NaCl (pH 6.0–6.5); 0.10 m Tris, 0.10 m NaCl (pH 7.0–9.0). All assays were performed in triplicate with three different enzyme preparations. Values of optimal pH were calculated by fitting of normalized initial velocity data from solutions of various pH to a bell-shaped distribution. Values of kcat/Km at the optimal pH were determined from initial velocity data, as described (37).
Double-stranded RNA Degradation
Steady-state kinetic parameters for a double-stranded (ds) RNA substrate were determined by following changes in absorbance upon enzymatic degradation, as described (38). Poly(A:U) (Sigma) was dissolved in reaction buffer (0.10 m Tris-HCl, 0.10 m NaCl, pH 7.4) and serially diluted by 2-fold in a 96-well plate (Corning). After equilibration at 25 °C, a baseline at A260 was established, and the initial substrate concentration was determined using ϵ260 = 6.5 mm−1 cm−1 for poly(A:U). Ribonucleases were added to solutions of varying substrate concentrations, and after mixing the change in absorbance at 260 nm was monitored over time. Initial reaction velocities were determined using Δϵ260 = 3.4 mm−1 cm−1 for poly(A:U). All assays were performed in triplicate with three different enzyme preparations. Values of Vmax and Km were calculated by fitting data to the Michaelis-Menten equation.
dsRNA degradation was also assessed with a stable fluorescent hairpin substrate with the sequence: 5,6-FAM-CGATC(rU)ACTGCAACGGCAGTAGATCG (IDT). This substrate had a single RNA nucleotide near the fluorophore-labeled 5′ end. The substrate was dissolved in water and annealed by first heating to 95 °C and then cooling slowly to room temperature. A solution of substrate (50 nm) was added to a solution of ribonuclease (1 μm), and the resulting mixture was incubated for 5 min. The reaction was quenched by the addition of 40 units of rRNasin (Promega), and the products were subjected to electrophoresis on a 20% (w/v) native acrylamide gel at 10 mAmp. Formation of cleavage product was monitored by excitation at 495 nm and emission at 515 nm with a Typhoon FLA 9000 scanner (GE Healthcare), and band density was quantified with ImageQuant software (GE Healthcare). The gel was then incubated in SYBR Gold (Invitrogen) and imaged for total nucleic acid. All assays were performed in triplicate with three different enzyme preparations.
Binding of Ribonucleases to Glycans
Soluble glycans, including heparin, chondroitin sulfate A, chondroitin sulfate B, and chondroitin sulfate C (Sigma), were diluted across a 96-well plate in 5-fold dilutions in 1× PBS (pH 7.4). Ribonuclease-BODIPY conjugates were added to a final concentration of 50 nm, and the resulting solutions were incubated for 30 min at room temperature. Polarization was monitored by excitation at 470 nm and emission at 535 nm, and data were normalized to a solution lacking carbohydrate and fitted to a binding isotherm by nonlinear regression.
Liposomal Disruption Assay
Liposomes were constructed as described (39) using 1,2-dioleoyl-sn-glycero-3-ethylphosphocholine (Avanti Polar Lipids). Lyophilized lipids were resuspended in 25 mm Tris-HCl buffer (pH 7.0), containing NaCl (80 mm), 8-aminonaphthalene-1,3,6-trisulfonic acid (12.5 mm), and p-xylene bis(pyridinium bromide) (45 mm) (40). The lipid suspension was subjected to five freeze-thaw cycles and extruded through polycarbonate filters to form unilamilar vesicles of ∼100–150 nm diameter as determined by dynamic light scattering. Liposomes were diluted to 700 μm and incubated with 5 μm ribonuclease in a 96-well plate. Ribonuclease-induced leakage of the entrapped vesicle content was monitored by measuring the de-quenching of the fluorescence of 8-aminonapthalene-1,3,6-trisulfonic acid over time (40). Percent leakage was calculated by normalizing to liposome disruption by Triton X-100.
Cellular Internalization of Ribonucleases
The uptake of BODIPY-labeled ribonucleases into nonadherent mammalian cells was monitored by flow cytometry, as described (35). Human K-562 cells were grown in RPMI media (Invitrogen) containing FBS (10% v/v) and penicillin/streptomycin (Invitrogen). Cells were maintained at 37 °C in 5% CO2. Cells were plated at 2 × 106 cells/ml in a 96-well plate. Ribonucleases in PBS were added to 5 μm, and the resulting solution was incubated for 4 h. Cells were collected by centrifugation at 1000 × g for 5 min, washed twice with PBS, exchanged into fresh medium, and collected on ice. The total fluorescence of live cells was measured using a FacsCalibur flow cytometer (BD Bioscience). Fluorescence data between experiments were normalized by calibrating each run with fluorescent beads. Data were analyzed with FlowJo software (Tree Star).
The uptake of BODIPY-labeled recombinant ribonucleases into adherent human umbilical vein endothelial cells (Lonza) and transformed bovine brain endothelial cells (TBBEC) (C. Czuprynski, University of Wisconsin-Madison) was monitored with confocal microscopy (Nikon). Human umbilical vein endothelial cells were grown in EGM-2 medium (Lonza); TBBEC were grown in RPMI medium containing FBS (10% v/v) and penicillin/streptomycin (Invitrogen). Cells were maintained at 37 °C in 5% CO2. Cells were plated at 2 × 106 cells/ml in a 96-well plate. Ribonucleases in PBS were added to 1.25 μm, and the resulting medium was incubated for 4 h. The outer membrane was stained with WGA-594, and the nucleus was stained with Hoechst 33342 (Invitrogen). Cells were then washed 3 times with PBS. Cells were imaged with an Eclipse TE2000-U laser scanning confocal microscope (Nikon) equipped with an AxioCam digital camera (Carl Zeiss).
Ribonucleolytic Assay of Conditioned Media
After serum starvation for 4 h, cells were treated with PBS or poly(C) (Sigma), poly(I:C) (InvivoGen), or poly(A:U) (Sigma) to 25 μg/ml. After 20 min, conditioned medium was collected, and a protease inhibitor mixture (1×) (Sigma), EDTA (0.10 m), and Triton X-100 (1% v/v) were added immediately. Medium (10 μl) was assayed for ribonucleolytic activity using 6-FAM-dArU(dA)2–6-TAMRA in 0.10 m Tris-HCl buffer (pH 7.4), containing NaCl (0.10 m) as described above.
Zymogram of Conditioned Media
Conditioned medium obtained as described above was concentrated using 5K MWCO spin concentrators (Corning). Samples were treated with peptide:N-glycosidase F (50 units) (New England Biolabs) in reaction buffer overnight at 37 °C. Samples were diluted with 2× Laemmli buffer (Bio-Rad) and loaded into a polyacrylamide gel (15% w/v) containing poly(C) (Sigma). Loaded samples were subjected to electrophoresis for 1.5 h at 100 V. Subsequent washing, refolding, and staining with toluidine blue were performed as described (9, 41).
Sequence Alignment and Phylogram Construction
Protein sequence alignments were made using MUSCLE (42) with manual adjustments. A maximum-likelihood phylogram was generated in MEGA5.2 using the Jones-Taylor-Thornton substitution model with uniform site substitution rates (43) and 1000 bootstrap replicates.
Statistical Analyses
Numerical data from experiments in vitro and in cellulo were analyzed by using unpaired and paired t tests, respectively, to determine p values.
RESULTS
Recombinant BRB had never been characterized prior to our work. Moreover, only preliminary studies had been performed on isolated enzyme (44, 45). To enable relevant comparisons, we also analyzed well known homologs: human RNase 1, bovine RNase A, and both monomeric (mBSR) and dimeric (dBSR) forms of BSR. Until now, these enzymes had never been compared in a single, controlled study using the same methods and substrates. We chose to include a monomeric form of BSR (C31A/C32A) (32) to establish any differences in biochemical properties conferred by dimerization.
Initial Characterizations of BRB
We began our study by analyzing biochemical properties of BRB that had been investigated for its homologs (Table 1). Analysis of aligned sequences demonstrated that BRB has less overall sequence identity and similarity to human RNase 1 than to either RNase A or BSR. Yet, when the divergent, 17-residue C-terminal tail of BRB was excluded from analysis, the ensuing BRBΔ125–141 displayed identity (70%) and similarity (82%) to human RNase 1 as high or higher than those of RNase A and BSR. Like human RNase 1, BRB was found to be less thermostable than either RNase A or BSR. BRB was found to bind tightly to its endogenous inhibitor, bovine RI, similar to other homologous ribonucleases (21, 31, 46, 47).
Human RNase 1 and BRB Show a Pronounced Shift in Catalytic pH Optimum
Previous studies have shown that orthologous ribonucleases can exhibit different pH optima for catalysis (48). Our results reveal similar contrasts. Although RNase A had its highest activity at pH 6.1, both human RNase 1 and BRB had their highest activity at pH 7.2 (Fig. 1A). Additionally, RNase A was ∼5-fold more active against ssRNA at optimal pH than either human RNase 1 or BRB (Fig. 1B). Interestingly, we also found a distinct shift in pH optimum between monomeric and dimeric forms of BSR: mBSR had its highest activity at pH 6.5, whereas dBSR shows its highest activity at pH 7.1 (Fig. 1A). Furthermore, we noted a drastic drop in catalytic efficiency for dBSR over its monomeric form (Fig. 1B).
FIGURE 1.
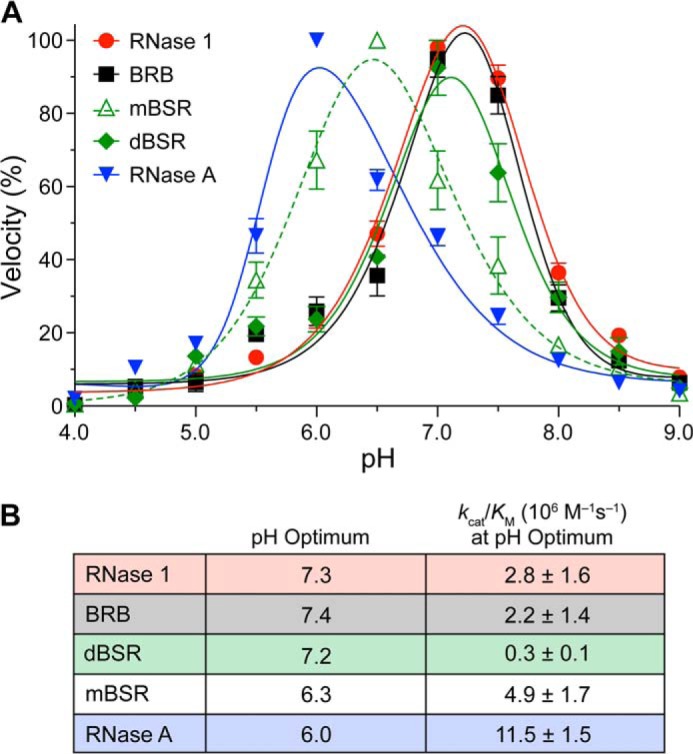
Effect of pH on catalysis of single-stranded RNA cleavage by human and bovine ribonucleases. A, pH rate profiles using the normalized initial velocity for cleavage of single-stranded RNA. Values (mean ± S.E.) are from at least three independent experiments. B, pH optima for catalysis as calculated from the data in panel A, and values (mean ± S.E.) of kcat/Km at that pH.
Human RNase 1 and BRB Can Degrade Double-stranded RNA with High Efficiency
Although all pancreatic-type ribonucleases can degrade ssRNA substrates, a small subset display high activity toward dsRNA (38). We examined the ability of ribonucleases to degrade dsRNA using poly(A:U) as substrate. We found that human RNase 1 degraded this dsRNA substrate with >2000-fold higher efficiency than that of RNase A. Tellingly, we found that BRB degraded poly(A:U) with efficiency ∼200-, 7-, and 2-fold higher than those of RNase A, mBSR, and dBSR, respectively (Fig. 2A). The trend for human RNase 1 and dBSR agrees with previous reports (38, 49, 50). An active site variant, H12A RNase 1, demonstrated little measurable activity against the substrate. We also assessed the ability of RNase B (Sigma), which is a naturally occurring glycoform of RNase A, to degrade poly(A:U), and found no significant change in activity from RNase A (data not shown).
FIGURE 2.
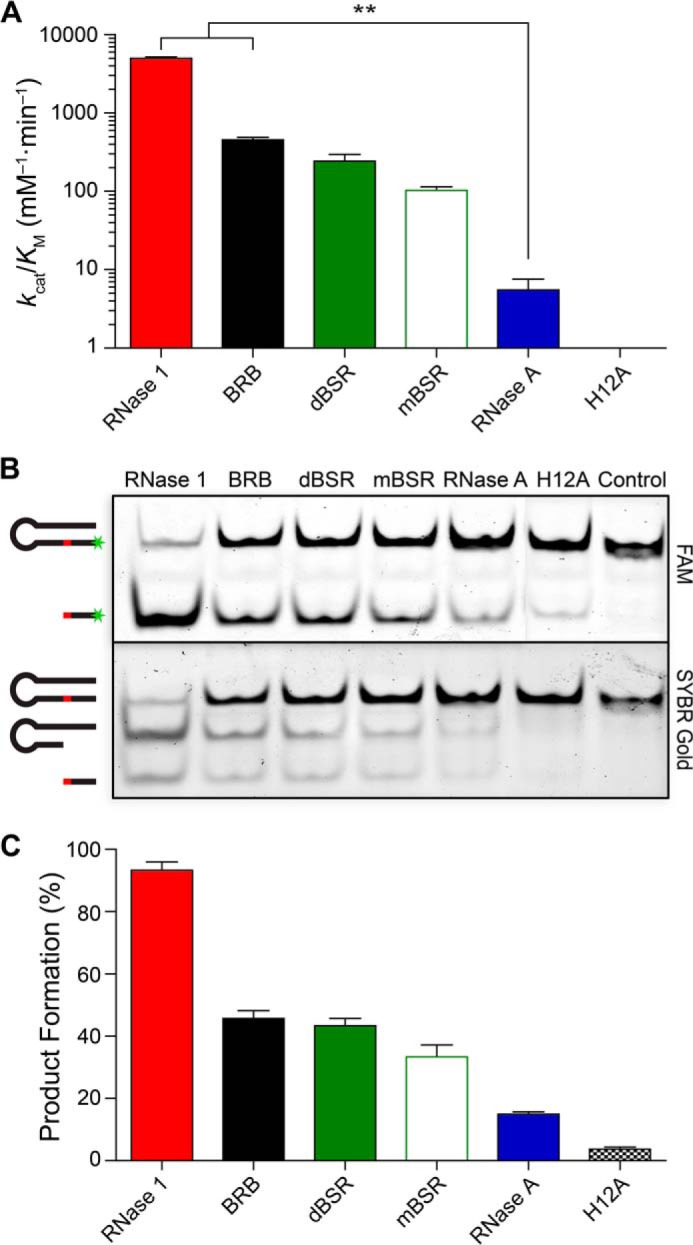
Catalysis of double-stranded RNA cleavage by human and bovine ribonucleases. A, values kcat/Km for the cleavage of poly(A:U). Values (mean ± S.E.) are from at least three independent experiments. **, p < 0.01. B, native polyacrylamide gel showing cleavage of a DNA hairpin containing a single RNA residue (red) and labeled on the 5′ end with FAM (green). SYBR Gold enables imaging of all nucleic acids. C, extent of FAM-labeled product formation for the data in panel B. Values (mean ± S.E.) are from four native gels.
As the heterogeneous nature of poly(A:U) does not allow for controlled secondary structure, we sought to create a novel dsRNA substrate to confirm our findings with poly(A:U). We designed a simple hairpin containing a single ribonucleotide embedded within a DNA oligonucleotide and labeled on the 5′ end with a fluorophore. Successful cleavage of this ribonucleolytic substrate necessitates unwinding of the DNA duplex formed by the hairpin. We monitored the formation of the fluorescent 6-mer cleavage product of ribonuclease catalysis by electrophoresis using a native polyacrylamide gel (Fig. 2B). Densitometric analysis of substrate and cleavage products mirrored the same trend observed with the poly(A:U) substrate. Specifically, human RNase 1 demonstrated the most product formation, followed by BRB, then dBSR, mBSR, and RNase A (Fig. 2C). Again, H12A RNase 1 demonstrated little activity.
Human RNase 1 and BRB Bind Cell Surface Molecules and Disrupt Liposomes
We used fluorescence polarization to compare the affinity of human and bovine ribonucleases toward common cell surface glycans. Representative data are shown in Fig. 3A. We found that both human RNase 1 and BRB had significantly higher affinity for all glycans tested than did either mBSR or RNase A. Average Kd values determined from at least three independent fluorescence polarization experiments are displayed as a heat map (Fig. 3B), and show that human RNase 1 and BRB exhibited nanomolar affinity for various carbohydrates.
FIGURE 3.
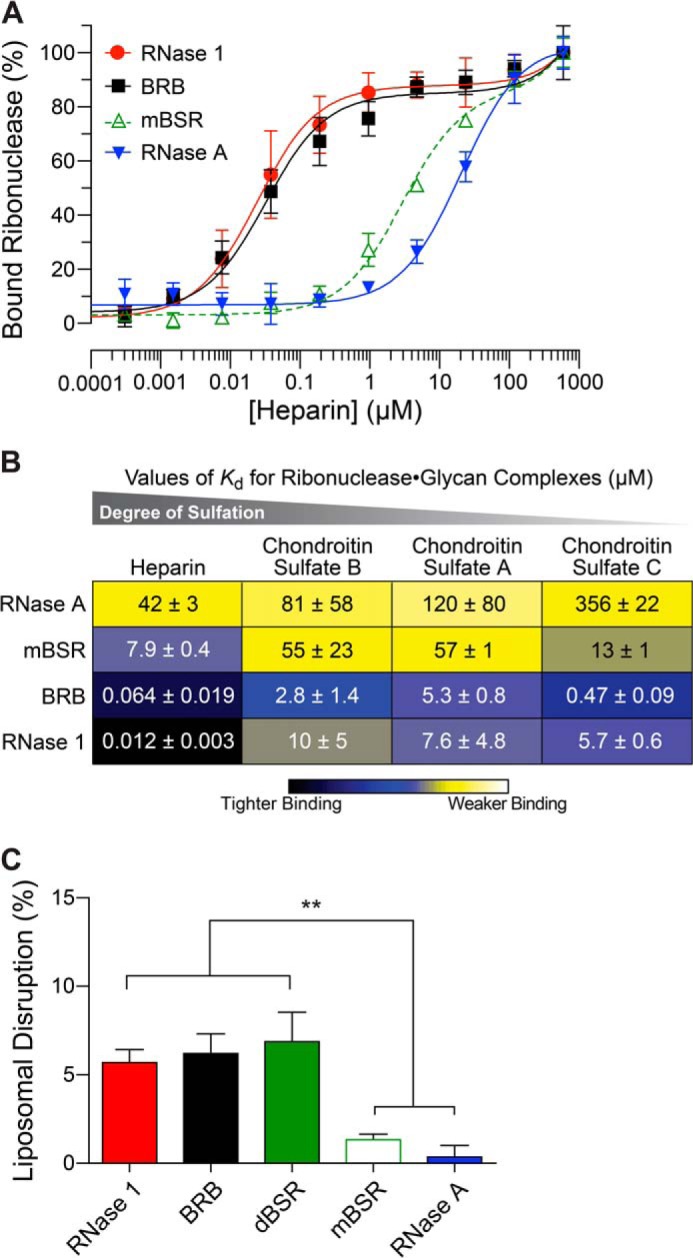
Interaction of human and bovine ribonucleases with membranes. A, representative isotherms for binding of BODIPY-labeled ribonucleases to heparin as determined by fluorescence polarization. B, heat map indicating the relative affinity for various cell surface glycans, determined as in panel A. Values (mean ± S.E.) are from at least three independent experiments. Blue tones represent nm affinity; yellow tones represent μm affinity. C, disruption of phosphatidylcholine liposomes by ribonucleases, as measured by the release of an encapsulated dye. Values (mean ± S.E.) are from at least three independent experiments. **, p < 0.01.
We sought to determine whether the increased affinity of ribonucleases for glycans correlated with their ability to disrupt lipid membranes. We observed significant differences between human RNase 1, BRB, and dBSR as compared with RNase A and mBSR (Fig. 3C). Still, the rates of liposomal disruption were relatively low compared with enzymes such as lysozyme, which exhibits ∼12-fold higher disruption efficiency than human RNase 1 (data not shown). The hydrophobic C-terminal tail did not endow BRB with a marked ability to disrupt liposomes.
Human RNase 1 and BRB Readily Enter Mammalian Cells
Next, we determined if greater cell surface glycan association enhanced the uptake of human RNase 1 and BRB into nonadherent human cells. A representative sample of raw fluorescence data acquired by flow cytometry is shown in Fig. 4A. Averaged, normalized data from three independent experiments indicated that both human RNase 1 and BRB were internalized into K-562 cells a significantly greater extent than either mBSR or RNase A (Fig. 4B).
FIGURE 4.
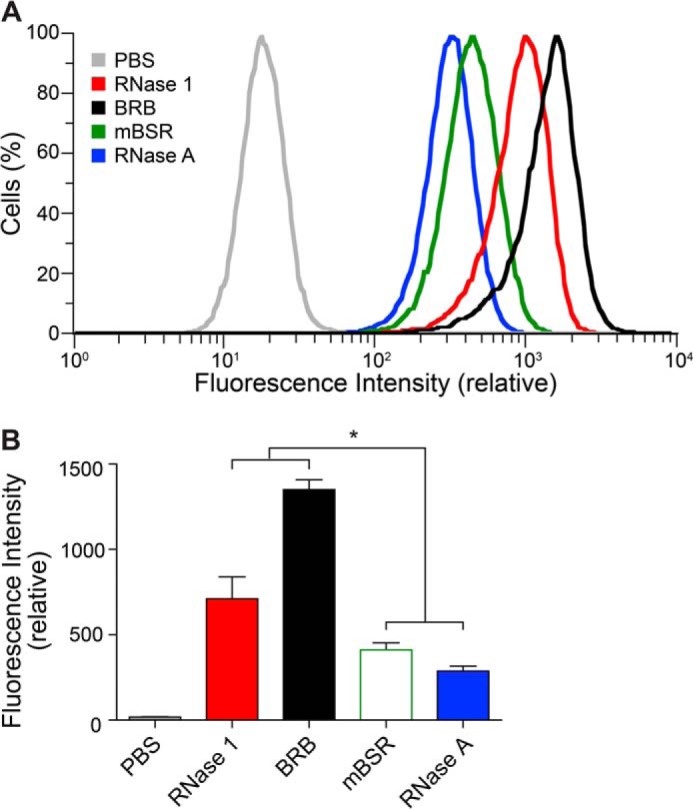
Mammalian cell internalization of human and bovine ribonucleases. A, representative flow cytometry data for the uptake of BODIPY-labeled ribonucleases into K-562 cells after 4 h. B. Values (mean ± S.E.) are from four independent experiments. *, p < 0.05.
Human RNase 1 is known to circulate freely in all bodily fluids, including blood (6, 9, 51). Hence, we were curious to know if ribonucleases could interact with the vascular endothelium, cells known to play dynamic regulatory roles in host defense and vascular homeostasis (52). We probed the internalization of ribonucleases into both human and bovine endothelial cells. Interestingly, we found that RNase 1 and BRB were taken up to a far greater extent by both human and bovine endothelial cells than either mBSR or RNase A (Fig. 5, A and D).
FIGURE 5.
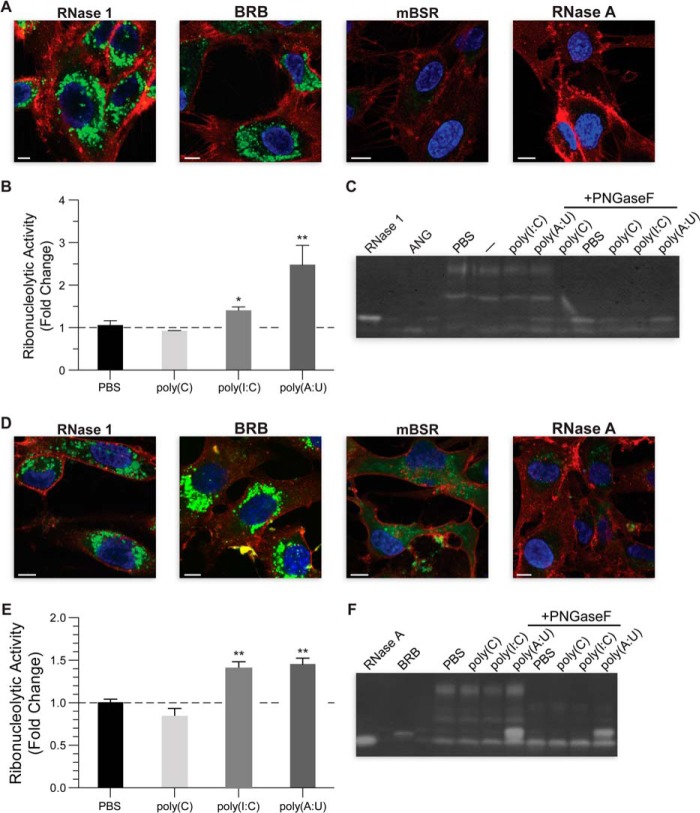
Endothelial cell internalization and release of human and bovine ribonucleases. A–C, human umbilical vein endothelial cells; D–F, TBBEC cells. A and D, cellular internalization of exogenous ribonucleases (1.25 μm) after 4 h. Ribonuclease label, BODIPY (green); outer membrane stain: WGA-594 (red); nuclear stain, Hoechst 33342 (blue). Scale bar, 5 μm. B and E, ribonucleolytic activity of cell-conditioned media after treatment with ssRNA or dsRNA. Values (mean ± S.E.; n = 6) are normalized to basal activity. *, p < 0.05; **, p < 0.01. C and F, zymogram of concentrated conditioned medium before or after treatment with peptide:N-glycosidase F. ANG, human angiogenin (76).
Human RNase 1 and BRB Are Released from Cells in Response to dsRNAs
Previous studies have demonstrated that human RNase 1 is secreted profusely from endothelial cells (6, 9, 51). Based on our findings that human RNase 1 and BRB can readily degrade dsRNA, we hypothesized that extracellular dsRNA in the blood might act as an agonist that promotes RNase 1 secretion. Within 20 min of exposure to the double-stranded RNA substrates poly(I:C) or poly(A:U), conditioned medium from both human and bovine endothelial cells displayed significantly higher levels of ribonucleolytic activity (Fig. 5, B and E). Surprisingly, this phenomenon did not occur upon exposure to single-stranded RNA (poly(C)), or DNA (data not shown). Zymogram analysis of cell-conditioned media provide a qualitative indication that the increased RNase activity in samples was due to human RNase 1 and BRB produced by human and bovine endothelial cells, respectively (Fig. 5, C and F). The ribonucleases secreted by these cells have N-linked glycans, which are removed by treatment with peptide:N-glycosidase F.
DISCUSSION
Mounting evidence suggests that mammalian RNase 1 plays important roles in vivo, ranging from regulating blood clotting and inflammation to directly counteracting tumorigenic cells. We believe that progress toward understanding these putative roles has been hindered by continual comparisons of human RNase 1 to bovine RNase A. Although RNase A is perhaps the most important model protein in biological chemistry (3, 53, 54), RNase A is the product of but one of three RNASE1 duplicates in the bovine genome. Its expression is limited in vivo, and its evolution is recent. Despite these shortcomings, RNase A has been considered the archetypal RNase 1 enzyme, with its properties ascribed globally to all homologous proteins. Thus, the prevailing view of RNase 1 has been of a digestive enzyme possessing little importance beyond the ruminant gut.
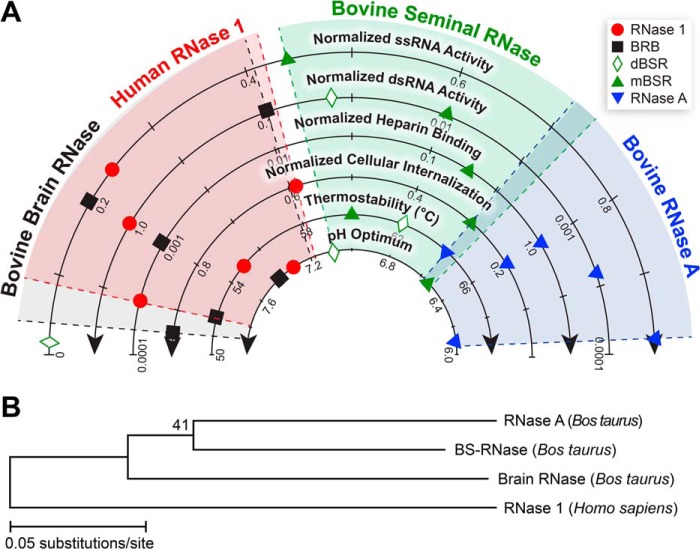
Our data stand in stark contrast to this hypothesis. We find distinct differences between human RNase 1 and RNase A. Moreover, we have characterized an additional bovine variant, BRB, and find pronounced similarities between this unappreciated protein and human RNase 1. We have demonstrated that human RNase 1 and BRB share similar biochemical properties, including strong catalytic activity against double-stranded RNA, as well as the intriguing ability to enter mammalian cells readily. These clustered attributes set human RNase 1 and BRB apart from either BSR or RNase A (Fig. 6A). Coupled with previous reports of the widespread tissue expression of BRB in cows (26), our data suggest that BRB, not RNase A, is the true functional homolog of human RNase 1. Moreover, our data support the existence of an important biological role for both human RNase 1 and BRB unrelated to digestion.
FIGURE 6.
Schematic summaries of data for human and bovine ribonucleases. A, schematic representation of normalized biochemical parameters from Table 1 and Figs. 1–4. Symbols indicate data obtained from assays; shaded regions indicate the range of all values for a particular ribonuclease. B, phylogram indicating bootstrap values >40.
Our treatise is consistent with a phylogenetic analysis (Fig. 6B), which suggests BRB resulted from an earlier genetic duplication than either BSR or RNase A and thus resembles more closely the ancestral form of RNase 1 in ruminants (28). Laboratory reconstructions of proposed “ancient” bovine ribonucleases support this claim, showing that “ancestral” forms of bovine RNase 1 display properties more similar to BRB than RNase A, including decreased thermostability and increased activity toward dsRNA (55, 56). Compellingly, the timeline of the divergence of RNase A corresponds to the Oligocene cooling epoch, which resulted in the rise of grasslands and the emergence of ruminant digestion. Hence, RNase A most likely represents a specialized digestive form of RNase 1 that arose simultaneously with foregut fermentation (21, 55). A similar phenomenon is known to have occurred in colobine monkeys, where a secondary form of RNase 1 with distinct properties evolved to participate in ruminant-like digestion (48). Taken together, extant evidence indicates that RNase A is not the prototype for mammalian RNase 1 in terms of function.
The question remains: if not digestion, what is the biological purpose of RNase 1? A conclusive answer to this question hinges upon future analysis of in vivo models. Still, our work does provide a basis for speculation. The ability of human RNase 1 and BRB to degrade dsRNA is of special interest because of its immunological implications. Most viruses produce dsRNA at some point during their replication. In mammalian cells, dsRNA is a potent antigen recognized by sensors such as Toll-like receptor (TLR) 3, through which dsRNA can trigger the transcription-based antiviral interferon response (57, 58). We found that stimulating endothelial cells with the dsRNA substrate poly(A:U), as well as with the synthetic viral dsRNA analog poly(I:C), increased the secretion of both RNase 1 and BRB significantly, whereas treatment with ssRNA or DNA did not. RNase 1 has been shown to be released spontaneously from endothelial cells upon treatment with various vascular agonists (9). Potentially, the presence of extracellular dsRNA provides a signal to direct the spontaneous release of stored, latent RNase 1. By degrading antigenic stimulants like extracellular RNAs, RNase 1 could play a crucial role in regulating antiviral immunity and inflammation.
Whereas the potential importance of dsRNA degradation by RNase 1 is clear, the mechanism of catalysis is not. The RNase 1 active site cannot accommodate two nucleic acid strands simultaneously; thus, the putative mechanism invokes the unwinding of the double helix by cationic residues near the enzymic active site. Arg-32 (20, 38) and Lys-102 (49) have been implicated, in particular. Both of these cationic residues are present in human RNase 1 and BRB, but absent in BSR and RNase A. Yet, other residues that could be important for dsRNA degradation by human RNase 1, including Arg-4, Lys-6, Lys-62, and Lys-74 (49, 50), are not found in BRB, leaving unknown the precise basis for dsRNA degradation.
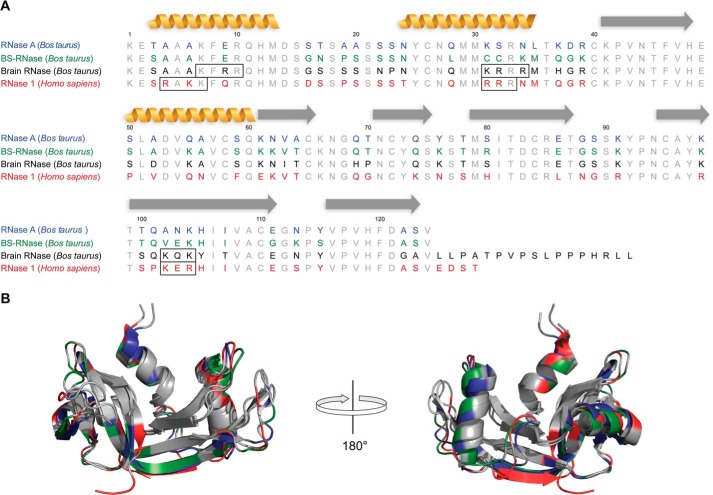
Intriguingly, the Toll-like receptors that respond to ssRNA and dsRNA (TLR7/TLR8 and TLR3, respectively) are all localized within endosomes (59–61). Our current work demonstrates that both human RNase 1 and BRB internalize into mammalian cells significantly better either RNase A or mBSR (Figs. 4 and 5, A and D). We have shown previously that RNase 1 internalization involves endocytosis (62–64); thus, human RNase 1 and BRB might be especially well adapted to enter endosomes, where they could degrade antigenic RNA and regulate signaling cascades. Their increased cellular uptake could hinge upon increased interactions with anionic cell surface glycans. Indeed, our data show that human RNase 1 and BRB bind much more tightly to an assortment of sulfated glycans, especially heparin, than does RNase A. This interaction is not merely based on Coulomb's law, as mBSR (Z = +9) binds much more weakly than human RNase 1 (Z = +6). Accordingly, we posit that human RNase 1 and BRB contain putative heparan sulfate-binding motifs. For example, the BBXB motif, where B represents a basic residue, has been shown to be a common heparan sulfate-binding motif in proteins (65–67). Human RNase 1 and BRB both contain three cationic regions that are similar to a BBXB motif and are absent from both mBSR and RNase A (Fig. 7). These unique areas of positive charge might account for many of the distinct properties shared by these enzymes, including dsRNA degradation, increased lipid disruption, and enhanced cellular internalization.
FIGURE 7.
Sequence and structural alignment of human and bovine ribonucleases. A, gray residues indicate residues conserved in all proteins; colored residues indicate divergence. Black boxes indicate putative heparan sulfate-binding domains. Yellow coils denote α-helices; gray arrows denote β-sheets. B, backbone overlay of human RNase 1 (red; PDB code 1z7x), mBSR (green; PDB code 1bsr), and RNase A (blue; PDB code 1fs3).
An unexpected result from our study is the pronounced divergence in pH optimum for catalysis among ribonucleases (Fig. 1). We found RNase A to have a pH optimum of 6.0, a value that closely reflects classic studies (68) and makes RNase A well suited for the acidic environment of the bovine rumen. Conversely, human RNase 1 and BRB had pH optimums of 7.3 and 7.4, respectively, which are close to the pH of many bodily fluids, including blood (pH 7.365). These data correlate with observations that human RNase 1 circulates freely throughout the body in all fluids tested (27). We speculate that differences in pH optimum between homologs could be due to slight perturbations in the pKa values of the two active site histidine residues. We were surprised to observe a large difference in pH optimum (∼1.3 pH units) for catalysis by the native dimeric form of BSR and the artificial monomer. The dimeric structure of BSR is also necessary for its other putative biological functions, including its immunosuppressive and antitumor activity (22, 46). The dimer is known to swap its N-terminal helices (69, 70), thereby forming a chimeric active site that could have higher histidine pKa values. Thus, its unique quaternary structure appears to equip BSR for catalysis in the cytosol as well as in bovine seminal fluid, where the typical pH is 6.8–7.2 (71). This functional imperative for a dimeric form could explain the existence of the BSR gene as a pseudogene in species where the cysteine residues required for dimerization have been lost.
Many questions remain regarding the biology of mammalian RNase 1, and BRB in particular. An ongoing mystery is how glycosylation of RNase 1 influences its endogenous functions. Analyses of human tissues and fluids indicate that various tissue sources produce differentially glycosylated forms of RNase 1 (6, 72); BRB has also been shown to have N-linked glycans that are highly heterogeneous and distinct from those attached to RNase A (73). We too found secreted ribonucleases that were N-glycosylated (Fig. 5, C and F). A second perplexity surrounds BRB: what is the purpose of its extended, hydrophobic C-terminal tail? Although all ruminant brain ribonucleases possess a similar tail, the amino acid sequences of these regions are not conserved, and seem to have arisen through multiple substitutions and deletions (19, 44). The tail is known to be O-glycosylated at two sites (25), but the significance of these oligosaccharide chains is not known. We speculated that the hydrophobic tail allows BRB to preferentially disrupt lipid membranes, but our data showed that BRB did not have significantly different activity toward liposomes than either human RNase 1 or dBSR (Fig. 3C). Previous studies have shown that proline-rich motifs can be associated with facilitating protein-protein interactions, specifically transient interactions such as recruitment of multiple factors (74). Thus, the proline-rich C-terminal tail of BRB could act as a protein scaffold to recruit other proteins.
In conclusion, we have presented data that establish functional relationships between human and bovine homologs of mammalian RNase 1. Our data provide fundamental insight into the biological role of RNase 1 in mammals, suggesting a physiology not associated with digestion. Further studies, including analyses of mammalian animal models, are necessary for a complete description of the most significant biological functions of RNase 1 in humans and other mammals. Currently, mouse studies probing the therapeutic potential of RNase 1 against various pathologies have utilized RNase A as their treatment of choice (7, 8, 10, 11, 15, 77). We speculate that the use of recombinant human RNase 1, BRB, or mouse RNase 1 would provide a more robust and relevant phenotype. Finally, we note that our understanding of BRB has been hindered by its appellation, which incorrectly implies an association only with the brain, just as our understanding of human RNase 1 has been obfuscated by its undue association with the pancreas.
Acknowledgments
We are grateful to Prof. C. J. Czuprynski (University of Wisconsin-Madison) for providing TBBEC cells, and T. T. Hoang for human angiogenin.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 CA073808.
- ssRNA
- single-stranded RNA
- BRB
- bovine brain ribonuclease
- BSR
- transformed ribonuclease
- mBSR
- monomeric BSR
- dBSR
- dimeric BSR
- TBBEC
- transformed bovine brain endothelial cell
- TLR
- Toll-like receptor
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- 6-TAMRA
- N,N,N′,N′-tetramethyl-6-carboxyrhodamine
- 6-FAM
- 6-carboxyfluorescein
- RI
- ribonuclease inhibitor.
REFERENCES
- 1. Barnard E. A. (1969) Biological function of pancreatic ribonuclease. Nature 221, 340–344 [DOI] [PubMed] [Google Scholar]
- 2. AlZahal O., Kebreab E., France J., Froetschel M., McBride B. (2008) Ruminal temperature may aid in the detection of subacute ruminal acidosis. J. Dairy Sci. 91, 202–207 [DOI] [PubMed] [Google Scholar]
- 3. Raines R. T. (1998) Ribonuclease A. Chem. Rev. 98, 1045–1066 [DOI] [PubMed] [Google Scholar]
- 4. Snook J. T. (1965) Dietary regulation of pancreatic enzyme synthesis, secretion and inactivation in the rat. J. Nutr. 87, 297–305 [DOI] [PubMed] [Google Scholar]
- 5. Peterson L. M. (1979) Serum RNase in the diagnosis of pancreatic carcinoma. Proc. Natl. Acad. Sci. U.S.A. 76, 2630–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barrabés S., Pagès-Pons L., Radcliffe C. M., Tabarés G., Fort E., Royle L., Harvey D. J., Moenner M., Dwek R. A., Rudd P. M., De Llorens R., Peracaula R. (2007) Glycosylation of serum ribonuclease 1 indicates a major endothelial origin and reveals an increase in core fucosylation in pancreatic cancer. Glycobiology 17, 388–400 [DOI] [PubMed] [Google Scholar]
- 7. Kannemeier C., Shibamiya A., Nakazawa F., Trusheim H., Ruppert C., Markart P., Song Y., Tzima E., Kennerknecht E., Niepmann M., von Bruehl M. L., Sedding D., Massberg S., Günther A., Engelmann B., Preissner K. T. (2007) Extracellular RNA constitutes a natural procoagulant cofactor in blood coagulation. Proc. Natl. Acad. Sci. U.S.A. 104, 6388–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Walberer M., Tschernatsch M., Fischer S., Ritschel N., Volk K., Friedrich C., Bachmann G., Mueller C., Kaps M., Nedelmann M., Blaes F., Preissner K. T., Gerriets T. (2009) RNase therapy assessed by magnetic resonance imaging reduces cerebral edema and infarction size in acute stroke. Curr. Neurovasc. Res. 6, 12–19 [DOI] [PubMed] [Google Scholar]
- 9. Fischer S., Nishio M., Dadkhahi S., Gansler J., Saffarzadeh M., Shibamiyama A., Kral N., Baal N., Koyama T., Deindl E., Preissner K. T. (2011) Expression and localisation of vascular ribonucleases in endothelial cells. Thromb. Haemost. 105, 345–355 [DOI] [PubMed] [Google Scholar]
- 10. Sun X., Wiedeman A., Agrawal N., Teal T. H., Tanaka L., Hudkins K. L., Alpers C. E., Bolland S., Buechler M. B., Hamerman J. A., Ledbetter J. A., Liggitt D., Elkon K. B. (2013) Increased ribonuclease expression reduces inflammation and prolongs survival in TLR7 transgenic mice. J. Immunol. 190, 2536–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Simsekyilmaz S., Cabrera-Fuentes H. A., Meiler S., Kostin S., Baumer Y., Liehn E. A., Weber C., Boisvert W. A., Preissner K. T., Zernecke A. (2014) Role of extracellular RNA in atherosclerotic plaque formation in mice. Circulation 129, 598–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Johnson R. J., McCoy J. G., Bingman C. A., Phillips G. N., Jr., Raines R. T. (2007) Inhibition of human pancreatic ribonuclease by the human ribonuclease inhibitor protein. J. Mol. Biol. 368, 434–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rutkoski T. J., Raines R. T. (2008) Evasion of ribonuclease inhibitor as a determinant of ribonuclease cytotoxicity. Curr. Pharm. Biotechnol. 9, 185–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rutkoski T. J., Kink J. A., Strong L. E., Raines R. T. (2013) Human ribonuclease with a pendant poly(ethylene glycol) inhibits tumor growth in mice. Transl. Oncol. 6, 392–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fischer S., Gesierich S., Griemert B., Schänzer A., Acker T., Augustin H. G., Olsson A. K., Preissner K. T. (2013) Extracellular RNA liberates tumor necrosis factor-α to promote tumor cell trafficking and progression. Cancer Res. 73, 5080–5089 [DOI] [PubMed] [Google Scholar]
- 16. D'Avino C., Paciello R., Riccio G., Coppola M., Laccetti P., Maurea N., Raines R. T., De Lorenzo C. (2014) Effects of a second-generation human anti-ErbB2 immunoRNase on trastuzumab-resistant tumors and cardiac cells. Protein Eng. Des. Sel. 27, 83–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Strong L. E., Kink J. A., Pensinger D., Mei B., Shahan M., Raines R. T. (2012) Efficacy of ribonuclease QBI-139 in combination with standard of care therapies. Cancer Res. 72, 1838 [Google Scholar]
- 18. Strong L. E., Kink J. A., Mei B., Shahan M. N., Raines R. T. (2012) First in human phase I clinical trial of QBI-139, a human ribonuclease variant, in solid tumors. J. Clin. Oncol. 30, TPS3113 [Google Scholar]
- 19. Confalone E., Beintema J. J., Sasso M. P., Carsana A., Palmieri M., Vento M. T., Furia A. (1995) Molecular evolution of genes encoding ribonucleases in ruminant species. J. Mol. Evol. 41, 850–858 [DOI] [PubMed] [Google Scholar]
- 20. Kleineidam R. G., Pesole G., Breukelman H. J., Beintema J. J., Kastelein R. A. (1999) Inclusion of cetaceans within the order Artiodactyla based on phylogenetic analysis of pancreatic ribonuclease genes. J. Mol. Evol. 48, 360–368 [DOI] [PubMed] [Google Scholar]
- 21. Beintema J. J. (2002) Ribonucleases in ruminants. Science 297, 1121–1122 [DOI] [PubMed] [Google Scholar]
- 22. D'Alessio G., Di Donato A., Parente A., Piccoli R. (1991) Seminal RNase: a unique member of the ribonuclease superfamily. Trends Biochem. Sci. 16, 104–106 [DOI] [PubMed] [Google Scholar]
- 23. Matoušek J. (2001) Ribonucleases and their antitumor activity. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 129, 175–191 [DOI] [PubMed] [Google Scholar]
- 24. Elson M., Glitz D. G. (1975) Characterization of a ribonuclease from bovine brain. Biochemistry 14, 1471–1476 [DOI] [PubMed] [Google Scholar]
- 25. Watanabe H., Katoh H., Ishii M., Komoda Y., Sanda A., Takizawa Y., Ohgi K., Irie M. (1988) Primary structure of a ribonuclease from bovine brain. J. Biochem. 104, 939–945 [DOI] [PubMed] [Google Scholar]
- 26. Wheeler T. T., Maqbool N. J., Gupta S. K. (2012) Mapping, phylogenetic and expression analysis of the RNase (RNaseA) locus in cattle. J. Mol. Evol. 74, 237–248 [DOI] [PubMed] [Google Scholar]
- 27. Su A. I., Wiltshire T., Batalov S., Lapp H., Ching K. A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Cooke M. P., Walker J. R., Hogenesch J. B. (2004) A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. U.S.A. 101, 6062–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang J. (2003) Parallel functional changes in the digestive RNases of ruminants and colobines by divergent amino acid substitutions. Mol. Biol. Evol. 20, 1310–1317 [DOI] [PubMed] [Google Scholar]
- 29. Lavis L. D., Rutkoski T. J., Raines R. T. (2007) Tuning the pKa of fluorescein to optimize binding assays. Anal. Chem. 79, 6775–6782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sundlass N. K., Eller C. H., Cui Q., Raines R. T. (2013) Contribution of electrostatics to the binding of pancreatic-type ribonucleases to membranes. Biochemistry 52, 6304–6312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Johnson R. J., Lavis L. D., Raines R. T. (2007) Intraspecies regulation of ribonucleolytic activity. Biochemistry 46, 13131–13140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee J. E., Raines R. T. (2005) Cytotoxicity of bovine seminal ribonuclease: monomer versus dimer. Biochemistry 44, 15760–15767 [DOI] [PubMed] [Google Scholar]
- 33. Niesen F. H., Berglund H., Vedadi M. (2007) The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2, 2212–2221 [DOI] [PubMed] [Google Scholar]
- 34. Menzen T., Friess W. (2013) High-throughput melting-temperature analysis of a monoclonal antibody by differential scanning fluorimetry in the presence of surfactants. J. Pharm. Sci. 102, 415–428 [DOI] [PubMed] [Google Scholar]
- 35. Lomax J. E., Eller C. H., Raines R. T. (2012) Rational design and evaluation of mammalian ribonuclease cytotoxins. Methods Enzymol. 502, 273–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lomax J. E., Bianchetti C. M., Chang A., Phillips G. N., Jr., Fox B. G., Raines R. T. (2014) Functional evolution of ribonuclease inhibitor: Insights from birds and reptiles. J. Mol. Biol., 426, 3041–3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kelemen B. R., Klink T. A., Behlke M. A., Eubanks S. R., Leland P. A., Raines R. T. (1999) Hypersensitive substrate for ribonucleases. Nucleic Acids Res. 27, 3696–3701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Libonati M., Sorrentino S. (2001) Degradation of double-stranded RNA by mammalian pancreatic-type ribonucleases. Methods Enzymol. 341, 234–248 [DOI] [PubMed] [Google Scholar]
- 39. Esbjörner E. K., Oglecka K., Lincoln P., Gräslund A., Nordén B. (2007) Membrane binding of pH-sensitive influenza fusion peptides: positioning, configuration, and induced leakage in a lipid vesicle model. Biochemistry 46, 13490–13504 [DOI] [PubMed] [Google Scholar]
- 40. Ladokhin A. S., Wimley W. C., Hristova K., White S. H. (1997) Mechanism of leakage of contents of membrane vesicles determined by fluorescence requenching. Methods Enzymol. 278, 474–486 [DOI] [PubMed] [Google Scholar]
- 41. Bravo J., Fernández E., Ribó M., de Llorens R., Cuchillo C. M. (1994) A versatile negative-staining ribonuclease zymogram. Anal. Biochem. 219, 82–86 [DOI] [PubMed] [Google Scholar]
- 42. Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jones D. T., Taylor W. R., Thornton J. M. (1992) The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282 [DOI] [PubMed] [Google Scholar]
- 44. Sasso M. P., Carsana A., Confalone E., Cosi C., Sorrentino S., Viola M., Palmieri M., Russo E., Furia A. (1991) Molecular cloning of the gene encoding the bovine brain ribonuclease and its expression in different regions of the brain. Nucleic Acids Res. 19, 6469–6474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhao W., Confalone E., Breukelman H. J., Sasso M. P., Jekel P. A., Hodge E., Furia A., Beintema J. J. (2001) Ruminant brain ribonucleases: expression and evolution. Biochim. Biophys. Acta 1547, 95–103 [DOI] [PubMed] [Google Scholar]
- 46. Antignani A., Naddeo M., Cubellis M. V., Russo A., D'Alessio G. (2001) Antitumor action of seminal ribonuclease, its dimeric structure, and its resistance to the cytosolic ribonuclease inhibitor. Biochemistry 40, 3492–3496 [DOI] [PubMed] [Google Scholar]
- 47. Lomax J. E. (2014) Ph.D. Thesis, University of Wisconsin, Madison [Google Scholar]
- 48. Zhang J., Zhang Y. P., Rosenberg H. F. (2002) Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nat. Genet. 30, 411–415 [DOI] [PubMed] [Google Scholar]
- 49. Sorrentino S., Naddeo M., Russo A., D'Alessio G. (2003) Degradation of double-stranded RNA by human pancreatic ribonuclease: crucial role of noncatalytic basic amino acid residue. Biochemistry 42, 10182–10190 [DOI] [PubMed] [Google Scholar]
- 50. Dey P., Islam A., Ahmad F., Batra J. K. (2007) Role of unique basic residues of human pancreatic ribonuclease in its catalysis and structural stability. Biochem. Biophys. Res. Commun. 360, 809–814 [DOI] [PubMed] [Google Scholar]
- 51. Landré J. B., Hewett P. W., Olivot J. M., Friedl P., Ko Y., Sachinidis A., Moenner M. (2002) Human endothelial cells selectively express large amounts of pancreatic-type ribonuclease (RNase 1). J. Cell Biochem. 86, 540–552 [DOI] [PubMed] [Google Scholar]
- 52. Danese S., Dejana E., Fiocchi C. (2007) Immune regulation by microvascular endothelial cells: directing innate and adaptive immunity, coagulation, and inflammation. J. Immunol. 178, 6017–6022 [DOI] [PubMed] [Google Scholar]
- 53. Marshall G. R., Feng J. A., Kuster D. J. (2008) Back to the future: ribonuclease A. Biopolymers 90, 259–277 [DOI] [PubMed] [Google Scholar]
- 54. Cuchillo C. M., Nogués M. V., Raines R. T. (2011) Bovine pancreatic ribonuclease: fifty years of the first enzymatic reaction mechanism. Biochemistry 50, 7835–7841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jermann T. M., Opitz J. G., Stackhouse J., Benner S. A. (1995) Reconstructing the evolutionary history of the artiodactyl ribonuclease superfamily. Nature 374, 57–59 [DOI] [PubMed] [Google Scholar]
- 56. Benner S. A., Caraco M. D., Thomson J. M., Gaucher E. A. (2002) Planetary biology: paleontological, geological, and molecular histories of life. Science 296, 864–868 [DOI] [PubMed] [Google Scholar]
- 57. Haller O., Kochs G., Weber F. (2006) The interferon response circuit: induction and suppression by pathogenic viruses. Virology 344, 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Satkunanathan S., Kumar N., Bajorek M., Purbhoo M. A., Culley F. J. (2014) Respiratory syncytial virus infection, TLR3 ligands, and proinflammatory cytokines induce CD161 ligand LLT1 expression on the respiratory epithelium. J. Virol. 88, 2366–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Alexopoulou L., Holt A. C., Medzhitov R., Flavell R. A. (2001) Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413, 732–738 [DOI] [PubMed] [Google Scholar]
- 60. Lund J. M., Alexopoulou L., Sato A., Karow M., Adams N. C., Gale N. W., Iwasaki A., Flavell R. A. (2004) Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl. Acad. Sci. U.S.A. 101, 5598–5603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Akira S., Uematsu S., Takeuchi O. (2006) Pathogen recognition and innate immunity. Cell 124, 783–801 [DOI] [PubMed] [Google Scholar]
- 62. Haigis M. C., Raines R. T. (2003) Secretory ribonucleases are internalized by a dynamin-independent endocytic pathway. J. Cell Sci. 116, 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chao T.-Y., Lavis L. D., Raines R. T. (2010) Cellular uptake of ribonuclease A relies on anionic glycans. Biochemistry 49, 10666–10673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chao T.-Y., Raines R. T. (2011) Mechanism of ribonuclease A endocytosis: Analogies to cell-penetrating peptides. Biochemistry 50, 8374–8382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cardin A. D., Weintraub H. J. (1989) Molecular modeling of protein: glycosaminoglycan interactions. Arteriosclerosis 9, 21–32 [DOI] [PubMed] [Google Scholar]
- 66. Hileman R. E., Fromm J. R., Weiler J. M., Linhardt R. J. (1998) Glycosaminoglycan-protein interactions: definition of consensus sites in glycosaminoglycan binding proteins. Bioessays 20, 156–167 [DOI] [PubMed] [Google Scholar]
- 67. Proudfoot A. E., Fritchley S., Borlat F., Shaw J. P., Vilbois F., Zwahlen C., Trkola A., Marchant D., Clapham P. R., Wells T. N. (2001) The BBXB motif of RANTES is the principal site for heparin binding and controls receptor selectivity. J. Biol. Chem. 276, 10620–10626 [DOI] [PubMed] [Google Scholar]
- 68. del Rosario E. J., Hammes G. G. (1969) Kinetic and equilibrium studies of the ribonuclease-catalyzed hydrolysis of uridine 2′,3′-cyclic phosphate. Biochemistry 8, 1884–1889 [DOI] [PubMed] [Google Scholar]
- 69. Mazzarella L., Mattia C. A., Capasso S., Di Lorenzo G. (1987) Composite active sites in bovine seminal ribonuclease. Gaz. Chim. Ital. 117, 91–97 [Google Scholar]
- 70. Piccoli R., Tamburrini M., Piccialli G., Di Donato A., Parente A., D'Alessio G. (1992) The dual-mode quaternary structure of seminal RNase. Proc. Natl. Acad. Sci. U.S.A. 89, 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Raps G., Cannon C. Y. (1947) The influence of management, breed and season upon the pH of bull semen. J. Dairy Sci. 30, 933–938 [Google Scholar]
- 72. Ribó M., Beintema J. J., Osset M., Fernández E., Bravo J., De Llorens R., Cuchillo C. M. (1994) Heterogeneity in the glycosylation pattern of human pancreatic ribonuclease. Biol. Chem. Hoppe-Seyler 375, 357–363 [PubMed] [Google Scholar]
- 73. Katoh H., Ohgi K., Irie M., Endo T., Kobata A. (1990) The structure of the asparagine-linked sugar chains of bovine brain ribonuclease. Carbohydr. Res. 195, 273–293 [DOI] [PubMed] [Google Scholar]
- 74. Kay B. K., Williamson M. P., Sudol M. (2000) The importance of being proline: The interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 14, 231–241 [PubMed] [Google Scholar]
- 75. D'Errico G., Ercole C., Lista M., Pizzo E., Falanga A., Galdiero S., Spadaccini R., Picone D. (2011) Enforcing the positive charge of N-termini enhances membrane interaction and antitumor activity of bovine seminal ribonuclease. Biochim. Biophys. Acta 1808, 3007–3015 [DOI] [PubMed] [Google Scholar]
- 76. Vallee B. L., Riordan J. F. (1997) Organogenesis and angiogenin. Cell Mol. Life Sci. 53, 803–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chen C., Feng Y., Zou L., Wang L., Chen H. H., Cai J. Y., Xu J. M., Sosnovik D. E., Chao W. (2014) Role of extracellular RNA and TLR3-Trif signaling in myocardial ischemia-reperfusion injury. J. Am. Heart Assoc. 3, e000683. [DOI] [PMC free article] [PubMed] [Google Scholar]