Abstract
The hepatic circadian clock plays a pivotal role in regulating major aspects of energy homeostasis and lipid metabolism. In this study, we show that RORγ robustly regulates the rhythmic expression of several lipid metabolic genes, including the insulin-induced gene 2a, Insig2a, elongation of very long chain fatty acids-like 3, Elovl3 and sterol 12α-hydroxylase, Cyp8b1, by enhancing their expression at ZT20-4. The time-dependent increase in their expression correlates with the rhythmic expression pattern of RORγ. The enhanced recruitment of RORγ to ROREs in their promoter region, increased histone acetylation, and reporter and mutation analysis support the concept that RORγ regulates the transcription of several lipid metabolic genes directly by binding ROREs in their promoter regulatory region. Consistent with the disrupted expression of a number of lipid metabolic genes, loss of RORγ reduced the level of several lipids in liver and blood in a ZT-preferred manner. Particularly the whole-body bile acid pool size was considerably reduced in RORγ−/− mice in part through its regulation of several Cyp genes. Similar observations were made in liver-specific RORγ-deficient mice. Altogether, our study indicates that RORγ functions as an important link between the circadian clock and the transcriptional regulation of several metabolic genes.
INTRODUCTION
The retinoic acid-related orphan receptor γ (RORγ, NR1F3), a member of the ROR subfamily of nuclear receptors, has been implicated in the control of a variety of physiological processes (1–3). By alternative promoter usage, the RORγ gene generates two isoforms, RORγ1 and RORγ2 (RORγt). RORγt is restricted to several distinct immune cell types and has a critical role in a number of immune processes (1–3). However, the physiological functions of RORγ1, which is expressed in various peripheral tissues, including liver, adipose tissue, and kidney, but not brain (4,5), are still poorly understood.
The diurnal oscillations in behavioral activity and physiology are strictly controlled by the circadian clock (6–10). Disruption of the circadian rhythm has been linked to an increased risk for metabolic diseases, including obesity, diabetes, liver steatosis and atherosclerosis (11,12). In several peripheral tissues, including the liver, RORγ1 exhibits a robust oscillatory pattern of expression with a peak at ZT16–20 that is controlled by Clock/Bmal1 heterodimers and Rev-Erb nuclear receptors (13–18). Loss of RORγ reduced peak expression of Npas2, Cry1 and Rev-Erbα, but had little effect on the hepatic expression of Bmal1 and Clock (7,14,16,19). Although several studies indicated a connection between RORγ and the regulation of certain clock and metabolic genes, the precise role of RORγ is not yet clearly understood. The robust oscillatory regulation of RORγ1 expression by the clock machinery raised the possibility that RORγ might regulate the transcription of certain target genes in a ZT-dependent manner and as such mediates the diurnal regulation of metabolic genes by the clock machinery (16,19–22). However, little is known about physiological functions controlled by hepatic RORγ particularly in lipid metabolism.
To study this hypothesis further, we examined the effect of the loss of RORγ on the rhythmic expression of a number of genes involved in several lipid metabolic pathways. Our study provides evidence indicating that RORγ regulates the expression of a number of lipid metabolic genes by multiple mechanisms that involve ZT-(in)dependent as well as (in)direct regulation by RORγ. We demonstrate that the loss of RORγ reduced peak expression of several lipid metabolic genes linked to fatty acid and cholesterol metabolism. In addition, the loss of RORγ induced changes in cholesterol, bile acid, triglyceride (TG) and fatty acid metabolism. RORγ cistrome and promoter analysis demonstrated that the transcription of several of these metabolic genes was regulated directly by RORγ. These data indicated that RORγ regulates the diurnal expression of several lipid metabolic genes in liver by a mechanism that involves ZT-dependent recruitment of RORγ to ROREs in the regulatory regions of these genes. Together, these data support our hypothesis that RORγ plays an integral role in mediating the transcriptional regulation of certain hepatic metabolic genes downstream of the circadian clock and thereby functions as a link between the circadian clock and its regulation of hepatic metabolism.
MATERIALS AND METHODS
Experimental Animals
Heterozygous C57BL/6 staggerer (RORα+/sg) were obtained from the Jackson Laboratory (Bar Harbor, ME). RORγ−/− and RORαsg/sgRORγ−/− double knockout (DKO) mice were described previously (16,23). Liver-specific RORγ knockout mice, referred to as RORγfx/fxAlb-Cre+, were described previously (22). Mice were supplied ad libitum with NIH-A31 formula (normal diet, ND) and water, and maintained at 23°C on a constant 12 h light:12 h dark cycle. Two month-old male mice were fed with a high fat diet (40% kcal fat) (HFD: D12079B Research Diets Inc., New Brunswick, NJ) for 6 weeks. Littermate wild-type (WT) mice were used as controls. All animal protocols followed the guidelines outlined by the NIH Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee at the NIEHS.
RNA isolation and QRT-PCR
To investigate the circadian patterns of gene expression, liver tissues were collected from WT, RORγ−/−, RORαsg/sg and RORαsg/sgRORγ−/− DKO mice on a ND every 4 or 6 h over a period of 24 h, and processed overnight in RNAlater® solution (Ambion, Austin, TX) at 4°C, and stored at −80°C until use. Total RNA was then extracted using RNeasy Mini kit (Qiagen) according to the manufacturer's instructions. Liver tissues were also collected from RORγ−/− mice at ZT8 after retro-orbital injection with empty adenovirus (control) or RORγ-expressing adenovirus (n = 6), from RORγfx/fxAlb-Cre− and RORγfx/fxAlb-Cre+ mice at ZT8 and ZT20, and from WT and RORγ−/− mice (n = 5) fed with a HFD for 6 weeks at the ZT indicated. QRT-PCR analysis was performed using SYBR Green I (Applied Biosystems, Foster City, CA, USA) as described previously (16). All the results were normalized by the amount of Gapdh mRNA. All primer sequences for QPCR are listed in Supplementary Table S1.
Other molecular, biochemical and histological methods are described in Supplementary Data.
RESULTS
RORγ regulates the hepatic expression of several lipid metabolic genes
In this study, we demonstrate that the loss of RORγ significantly impacts the diurnal expression pattern of a number of genes linked to several different lipid metabolic pathways. Loss of RORγ significantly affected the circadian expression of insulin induced gene 2a (Insig2a), ELOVL fatty acid elongase 3 (Elovl3) and the sterol 12α-hydroxylase (Cyp8b1). In WT liver, these genes exhibited a robust rhythmic pattern of expression with peak expression at ZT20–4, a few hours after the optimum expression of RORγ at ZT20 (Figure 1A) (16), while peak expression was dramatically repressed in RORγ−/− mice, blunting their circadian oscillation. In contrast, little difference in the diurnal expression of these genes was observed between WT and RORαsg/sg mice. Although a recent study provided evidence for direct transcriptional regulation of Cyp8b1 by RORα in HepG2 cells (24), our study shows that loss of RORα had no effect on the diurnal expression of Cyp8b1. Insig2a and Elovl3 were repressed to a somewhat greater extent in RORαsg/sgRORγ−/− DKO mice than RORγ single KO mice (Figure 1A and C). These data suggest that these genes are regulated in a highly RORγ-selective manner. In contrast to the loss of RORγ, exogenous expression of RORγ in RORγ−/− liver tissue by adenovirus significantly increased the expression of Insig2a, Elovl3 and Cyp8b1 (Figure 1D and Supplementary Figure S1A and B). Reduced hepatic expression of Insig2a and Elovl3 was also observed in RORγ−/− mice after 16-h fasting (Supplementary Figure S1C). Together, these results indicate that RORγ plays a critical role in the diurnal regulation of Insig2a, Elovl3 and Cyp8b1 expression in liver.
Figure 1.
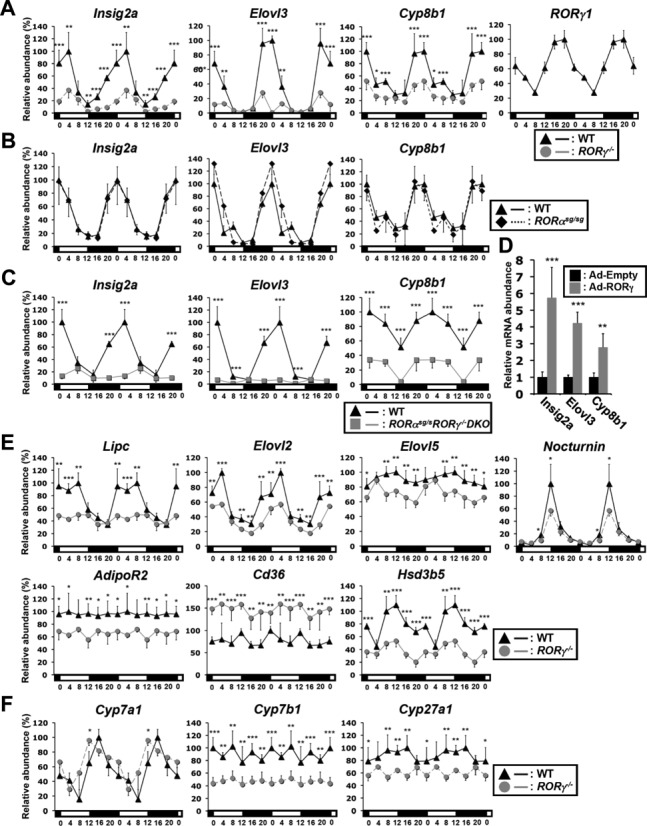
RORγ regulates the circadian expression of Insig2a, Elovl3, Cyp8b1 and other lipid metabolic genes. Rhythmic expression patterns of Insig2a, Elovl3 and Cyp8b1 mRNA in the liver of RORγ−/− (A), RORαsg/sg (B) and DKO mice (C). The diurnal pattern of RORγ expression in WT liver is shown for comparison (16). Livers (n = 4) were collected every 4 or 6 h over a period of 24 h. (D) Adenovirus mediated expression of RORγ in the liver of RORγ−/− mice (n = 6) enhanced the Insig2a, Elovl3 and Cyp8b1 mRNA levels. Circadian expression patterns of several other genes involved in fatty acid/triglycerides metabolism (E) and bile acid synthesis (F) in RORγ−/− mice. The 24 h expression patterns were double-plotted. Data represent mean ±SD, *P < 0.05, **P < 0.01, ***P < 0.001 by ANOVA.
The loss of RORγ also significantly reduced the circadian expression of hepatic TG lipase (Lipc) and Elovl2, which oscillatory patterns are very much in phase with those of Insig2a, Elovl3, Cyp8b1 and RORγ (Figure 1E and Supplementary Figure S1D) (16,21,25). The expression of Elovl5, which exhibits a weak oscillation, and peak expression of the clock output gene, Nocturnin (Ccrn4l) at ZT12 were also repressed in RORγ−/− liver, while the expression of the arrhythmic genes, adiponectin receptor 2 (AdipoR2), the fatty acid transporter, Cd36, and hydroxy-delta-5-steroid dehydrogenase, Hsd3b5 was down- or up-regulated at all ZTs (Figure 1E).
In addition to Cyp8b1, several other genes involved in bile acid synthesis, including Cyp7b1 and Cyp27a1, were down-regulated in RORγ−/− liver (Figure 1F). Little or no circadian oscillation was observed for Cyp7b1 and Cyp27a1 expression, both of which were repressed in RORγ−/− liver at all or most ZTs indicating that these genes are regulated in a ZT-independent manner. The latter suggests that their transcription may be regulated indirectly by RORγ. Although RORγ is recruited to the Cyp7a1 promoter, loss of RORγ had little effect on its level of expression; however, a small shift in circadian phase could be observed (Figure 1F). Exogenous expression of RORγ by adenovirus enhanced the hepatic expression of Cyp7b1, but not that of Cyp7a1 (Supplementary Figure S1E). Loss of RORα strongly reduced the hepatic expression of Cyp7b1 at all ZTs and affected both the level and phase of Cyp7a1 expression (Supplementary Figure S1F). The repression of Cyp7b1 in RORαsg/sg liver is consistent with a previous report showing that Cyp7b1 is a direct target gene of RORα (26). Together, these results suggest that although RORα and RORγ exhibit a certain degree of redundancy, they largely regulate different sets of Cyp genes.
Insig2a is directly regulated by RORγ through RORE
The regulation of the rhythmic expression of metabolic genes by RORγ might involve both direct and indirect mechanisms. ChIP-Seq analysis showed that RORγ was recruited to three sites, referred to as Insig2a (A, B, C), that are respectively at (−491/+177), (−3829/−2985) and (−17428/−15521) in the promoter of Insig2a (Figure 2A), a gene playing an important regulatory role in lipid metabolism (27,28). Reporter gene analysis revealed that RORγ increased the transactivation of the reporter via Insig2a(A) and Insig2a(B) sites, respectively, 14× and 5× (Figure 2A and B), while no activation was observed through the distal site C (not shown). Deletion analysis of Insig2a(A) and Insig2a(B) suggested that the activation was mediated through, respectively, the −160/−53 and −3592/−3452 region, which include a RORE-like motif. Mutation of the putative RORE sequence within Insig2a(A) and Insig2a(B) resulted in a significant reduction in RORγ-mediated transactivation, suggesting that the transcriptional activation is mediated through direct binding of RORγ to these ROREs. Addition of an RORγ-selective antagonist inhibited Insig2a(A) and Insig2a(B) activation by RORγ in a dose-responsive manner (Figure 2C). A previous report showed that Rev-Erbα, a transcriptional repressor that binds RORE-like motifs, regulates the expression of Insig2a (29). Figure 2D shows that Rev-Erbα repressed the RORγ-mediated as well as the endogenous activation of Insig2a(B) by about 55%, but had no effect on Insig2a(A)-mediated activation. These results suggest that the circadian regulation of Insig2a by Rev-Erbα may involve competition with RORγ for the binding to Insig2a(B) RORE.
Figure 2.
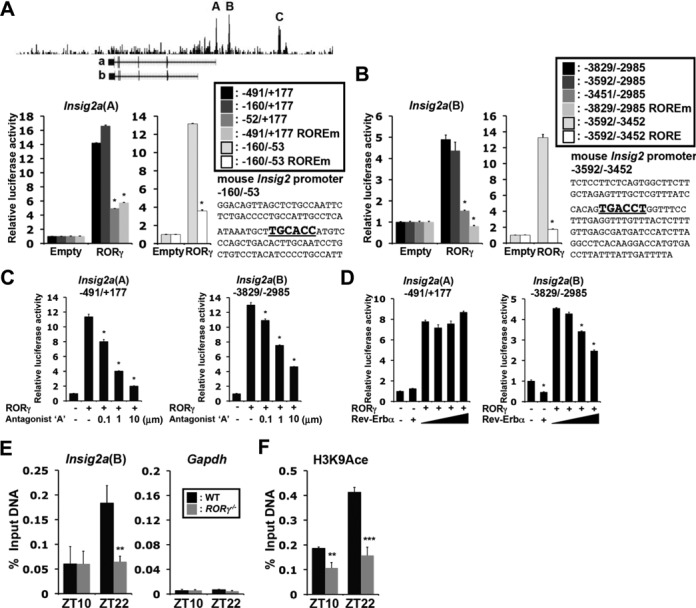
RORγ directly regulates the transcription of Insig2a. (A and B) ChIP-Seq analysis using anti-RORγ antibody and chromatin prepared from mouse liver tissue identified several RORγ-binding sites, A–C, in the Insig2a gene. Huh-7 cells were co-transfected with p3xFlag-CMV10-RORγ and a pGL4 reporter plasmid containing the WT or mutated Insig2a(A) (−491/+177) or Insig2a(B) (−3829/−2985). The ROREs are shown in bold. (C) The RORγ-selective antagonist ‘A’ inhibited the Insig2a(A)- and Insig2a(B)-mediated transactivation. (D) Rev-Erbα repressed the Insig2a(B)-mediated activation by RORγ, but not the Insig2a(A)-mediated activation. (E) ChIP-QPCR was performed using an anti-RORγ antibody and chromatin prepared from WT and RORγ−/− liver tissues (n = 4) collected at ZT10 (low expression of RORγ) and ZT22 (high expression of RORγ). Amplification of Gapdh gene was used as a negative control. (F) ChIP-QPCR was performed using anti-H3K9Ace.
ChIP-QPCR analysis indicated that the association of RORγ with the Insig2a(B) RORE was greater at ZT22 than at ZT10. The increased recruitment at ZT22 correlates with the time at which RORγ most robustly regulated Insig2a (Figure 2E). The high expression of Insig2a at ZT22 was also associated with increased H3K9 acetylation (Figure 2F). Both the recruitment of RORγ and H3K9 acetylation at Insig2a(B) were significantly reduced in the liver of RORγ−/− mice. Thus, the increased association of RORγ and H3K9 acetylation, a marker for transcriptionally active chromatin, at ZT22 correlated with enhanced Insig2a transcription.
Because Insig2a regulates the proteolytic activation of the sterol regulatory element binding transcription factor 1c (Srebp-1c) and its subsequent translocation to the nucleus (27,28,30), the effect of the down-regulation of Insig2a in RORγ−/− liver on Srebp-1c processing was examined. Both the precursor (135 kDa) and proteolytic (55–60 kDa) forms of Srebp-1c were detected in whole liver lysates prepared at ZT4; however, the level of the processed form was consistently increased in RORγ−/− liver (Figure 3A) as well as nuclear Srebp-1 protein (Figure 3B). The increase in nuclear Srebp-1 protein at ZT4 in RORγ−/− mice was associated with a temporal increase in the expression of the Srebp-1 target genes, Fasn and Elovl6, at ZT4 (Figure 3C). Loss of RORγ reduced Sreb-1c expression at ZT12–16. This was associated with a down-regulation of Fasn and Elovl6 expression. Thus, loss of RORγ appears to affect Srebp-1c at several levels.
Figure 3.
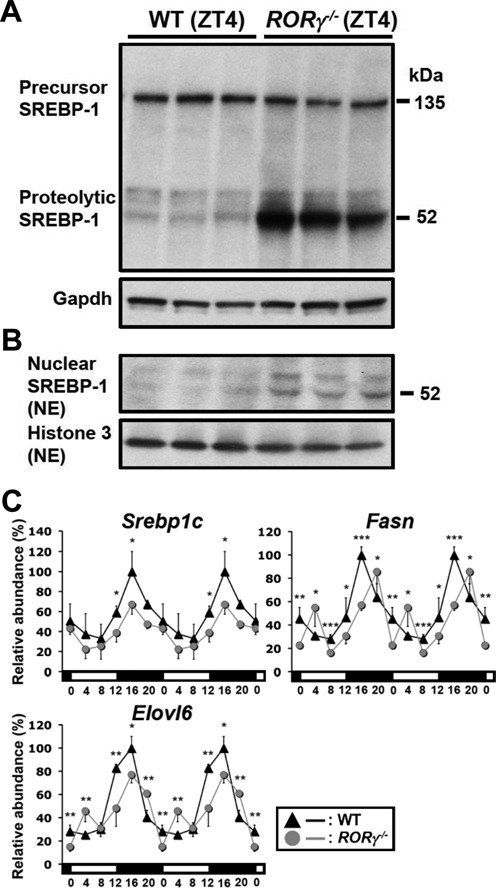
Effects of the loss of RORγ on Srebp-1. (A and B) Comparison of precursor Srebp-1 (135 kDa) and processed Srebp-1 (52–60 kDa) levels in whole liver lysates and nuclear extracts from WT and RORγ−/− mice collected at ZT4. (C) Circadian expression profiles of Srebp-1c, and the target genes, Fasn and Elovl6, in the liver of WT and RORγ−/− mice. Data represent mean ±SD, *P < 0.05, **P < 0.01, ***P < 0.001 by ANOVA.
Elovl3 is directly regulated by RORγ through RORE
Elovl3, which circadian expression was also strongly regulated by RORγ (Figure 1A), contains a putative RORE located at −836 bp in its proximal promoter (Figure 4A). Reporter analysis showed that RORγ caused a 5-fold increase in the activation of the Elovl3(−919/−1) promoter. Mutation (Elovl3(ROREm)) and deletion (Elovl3(−504/−1) and Elovl3(−42/−1)) of the putative RORE significantly reduced the activation by RORγ. Rev-Erbα was able to inhibit the activation of the Elovl3(−919/−1) promoter by competing with RORγ for RORE binding (Figure 4B). The RORγ-mediated transactivation was also repressed by the RORγ-selective antagonist (Figure 4C). ChIP analysis showed that the association of RORγ with the Elovl3 proximal promoter was increased particularly at ZT22 along with enhanced H3K9 acetylation (Figure 4D and E). This association was greatly reduced in RORγ−/− liver. Together, these observations suggest that RORγ regulates the diurnal expression of Elovl3 directly through RORE interaction.
Figure 4.
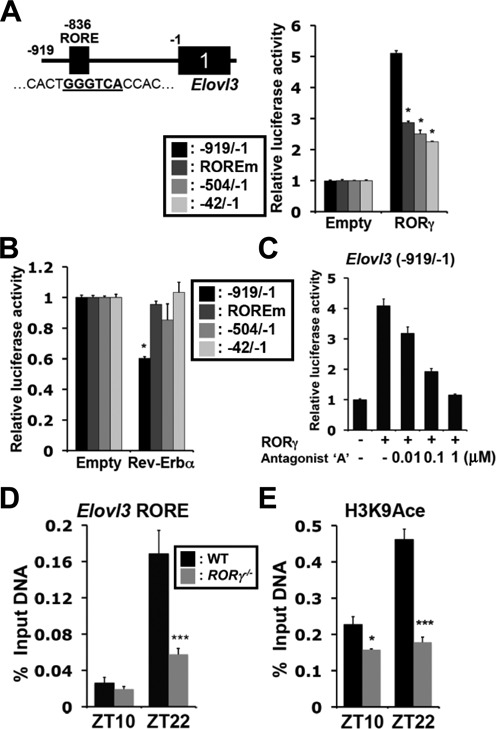
RORγ directly regulates the transcription of Elovl3. (A) Huh-7 cells were co-transfected with p3xFlag-CMV10-RORγ and pGL4 reporter plasmid containing the WT or the RORE-mutated Elovl3(−1 to −919) promoter region. (B) Rev-Erbα repressed the Elovl3 promoter through the RORE. (C) Inhibition of the RORγ-induced activation of the Elovl3 promoter by the RORγ-selective antagonist. (D and E) ChIP analysis. The increased association of RORγ and elevated H3K9 acetylation at the Elovl3 promoter is ZT-dependent. Data represent mean ±SEM. *P < 0.05, **P < 0.01, ***P < 0.001 by ANOVA.
Analysis of hepatic fatty acids by GC-MS showed that the composition of palmitoleic acid was increased at ZT4 and decreased at ZT20 in RORγ−/− mice (Supplementary Figure S2). The percentage of palmitic acid, oleic acid and linoleic acid at ZT4 was either increased or decreased, while that of docosahexaenoic acid (DHA) was significantly lower in RORγ−/− liver at both ZT4 and ZT20. Fatty acids were also analyzed in RORγ−/− serum collected at ZT4 (Supplementary Table S2). Consistent with the changes in liver, palmitic acid, palmitoleic acid, oleic acid were increased in RORγ−/− mice, while DHA was reduced. These results indicate that loss of RORγ significantly affects the composition of fatty acids, which may at least in part be related to changes in the regulation of several Elovl family genes.
Loss of RORγ causes changes in triglyceride and bile acid levels
We next examined whether the changes in the hepatic expression of lipid metabolic genes in RORγ−/− mice were associated with alterations in lipid homeostasis. Serum TG levels tended to be lower in RORγ−/−(ND) mice particularly after fasting (Supplementary Figure S3A and B). RORγ−/− mice fed a HFD for 6 weeks exhibited considerable changes in TGs without a significant change in their body weights (Supplementary Figure S3C). Serum TGs levels were significantly reduced at all ZTs in RORγ−/−(HFD) mice (Figure 5A), whereas hepatic TGs were reduced particularly at ZT4 and ZT8 (Figure 5B). The level of circulating free fatty acids, glycerol and total ketone bodies were lower particularly at ZT0 and ZT4 (Figure 5C), whereas that of hepatic free fatty acids was increased in RORγ−/−(HFD) mice at ZT4 (Figure 5D), both of which indicates lower fatty acid consumption in RORγ−/−(HFD) mice (31). Serum and hepatic TGs were similarly reduced in RORγ−/−(HFD) mice at ZT4 after 16-h fasting and so were levels of free fatty acids and glycerol (Supplementary Figure S3D and E). Adenovirus expression of RORγ in RORγ−/−(HFD) mice did not rescue serum TG levels, suggesting that longer term expression of RORγ as well as proper circadian pattern of RORγ expression might be required to impact lipid homeostasis (Supplementary Figure S3F). Similarly to RORγ−/−(ND) mice, the hepatic expression of several genes, including Insig2a, Elovl3 and Lipc was reduced in RORγ−/−(HFD) mice (Supplementary Figure S4A–C).
Figure 5.
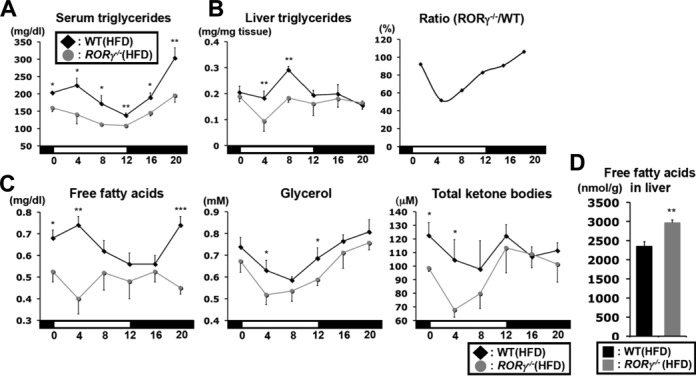
Changes in triglyceride and fatty acid levels in RORγ−/−(HFD) mice. Serum (A) and hepatic (B) triglyceride levels were examined WT(HFD) and RORγ−/−(HFD) mice (n = 5) every 4 h over a period of 24 h. Liver triglycerides at each ZT were plotted as a ratio between RORγ−/−(HFD) and WT(HFD) mice. (C) Daily patterns of free fatty acids, glycerol and total ketone bodies were measured in serum from WT(HFD) and RORγ−/−(HFD) mice (n = 5). (D) Free (unesterified) fatty acids extracted from liver tissue at ZT4 (n = 8) were quantified by LC-MS/MS. Data represent mean ±SEM, *P < 0.05, **P < 0.01, **P < 0.001 by ANOVA.
Figure 6A shows that serum cholesterol and circulating high and low density lipoprotein (HDL and LDL, respectively) levels were significantly reduced in RORγ−/−(HFD) mice at all ZTs analyzed, but the reduction was more pronounced at ZT0–4. Hepatic cholesterol levels were also reduced at all ZTs (Figure 6B). However, the loss of RORγ caused an increase rather than a decrease in peak (around ZT16) expression of HMG-CoA reductase, which encodes a rate-limiting enzyme in cholesterol synthesis in the liver (Supplementary Figure S4D). Hepatic bile acids were reduced 40–50% in RORγ−/−(HFD) mice at all ZTs, whereas levels of serum bile acids, which reached a peak at ZT0, were greatly reduced at ZT0 and ZT4 in RORγ−/−(HFD) mice (Figure 6C). Inversely, expression of exogenous RORγ in RORγ−/− liver by adenovirus injection increased hepatic bile acids (Figure 6D). Levels of total bile acids were also reduced in feces and gallbladder of RORγ−/−(HFD) mice as well as the whole-body pool size (the sum of bile acids in liver, gallbladder and small intestines) (Figure 6E). These observations suggest that not increased lipid excretion (Figure 6F), but lower cholesterol levels in the liver combined with a reduction in the expression of genes regulating bile acid synthesis, including Cyp8b1, Cyp27a1 and Cyp7b1, might be responsible for reduced bile acid generation in RORγ-deficient mice. Together these results indicate that RORγ plays an important role in the diurnal regulation of fatty acid, cholesterol and TG homeostasis. Several other nuclear receptors, particularly FXR and LXR, have been implicated in the regulation of cholesterol and bile acid metabolism (32–34). To examine whether the changes in cholesterol and bile acid metabolism in RORγ−/− mice were related to alterations in the expression of other nuclear receptors, we analyzed the expression of Fxr, Lxr, Car and Pxr in RORγ−/− liver. As shown in Supplemental Figure S4E, loss of RORγ had little effect on the diurnal pattern of expression of these receptors.
Figure 6.
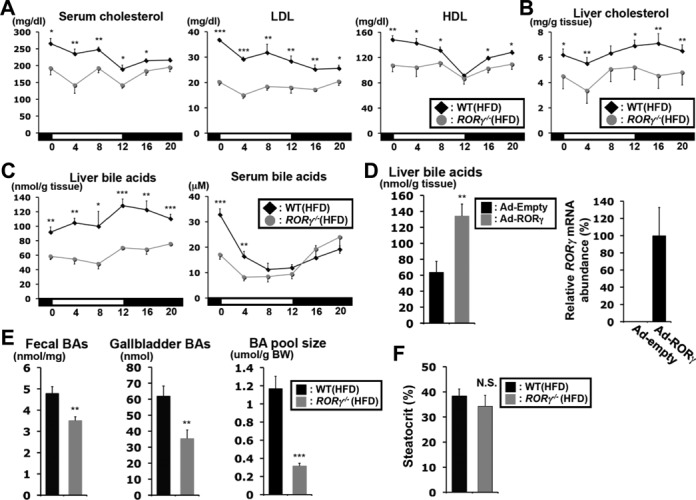
Loss of RORγ affects the hepatic and circulating levels of cholesterol and bile acids. (A) Comparison of the cholesterol, LDL and HDL levels in serum collected from WT(HFD) and RORγ−/−(HFD) mice (n = 5) every 4 h over a period of 24 h. (B) Comparison of the hepatic cholesterol levels (n = 5). (C) Total bile acids in the liver and serum from WT(HFD) and RORγ−/−(HFD) mice (n = 5). (D) Total bile acids were increased in the liver of RORγ−/−(HFD) mice (n = 6) injected with adenovirus expressing RORγ. Exogenous expression of RORγ was detected by QRT-PCR. Data represent mean ±SD. (E) Total bile acids in feces, gallbladder and the whole-body (whole liver, gallbladder and small intestines) were measured in WT(HFD) and RORγ−/−(HFD) mice (n = 7–9). (F) Comparison of steatocrit in feces (n = 8). Data represent mean ±SEM, *P < 0.05, **P < 0.01, ***P < 0.001 by ANOVA.
In addition to the reduced hepatic TGs and increased free fatty acid levels (Figure 5) in RORγ−/−(HFD) mice at daytime, histological examination of liver sections showed that WT(HFD) mice exhibited more of a microvesicular steatosis phenotype, while RORγ−/−(HFD) mice exhibited a more macrovesicular form of steatosis (Supplementary Figure S5A and Table S3). RORγ−/− mice also exhibited perivascular inflammation. This was supported by increased expression of various inflammatory genes (Supplementary Figure S5B and Table S4). Blood urea nitrogen was not changed, suggesting that kidney function was not impaired (Supplementary Figure S4F).
Changes in lipid metabolism in liver-specific RORγ knockout mice
To determine whether the effects on gene expression were based on the hepatocyte-specific loss of RORγ rather than the loss of RORγ in other tissues, we analyzed the expression of several lipid metabolic genes in liver-specific RORγ-deficient mice (RORγfx/fxAlb-Cre+). Consistent with our observations in ubiquitous RORγ knockout mice, the expression of Insig2a, Elovl3, Lipc and Hsd3b5 was reduced particularly at ZT20, while the expression of Cyp8b1, Cyp7b1 and Cyp27a1 was decreased both at ZT8 and ZT20 (Figure 7A). TG, cholesterol and bile acid levels in serum and liver were significantly reduced at ZT0 or ZT4 in RORγfx/fxAlb-Cre+ mice fed a HFD (Figure 7B), although the reductions were smaller than in ubiquitous RORγ−/− mice. These observations indicate that the changes in gene expression and lipid metabolism were directly related to the loss of hepatic RORγ expression.
Figure 7.
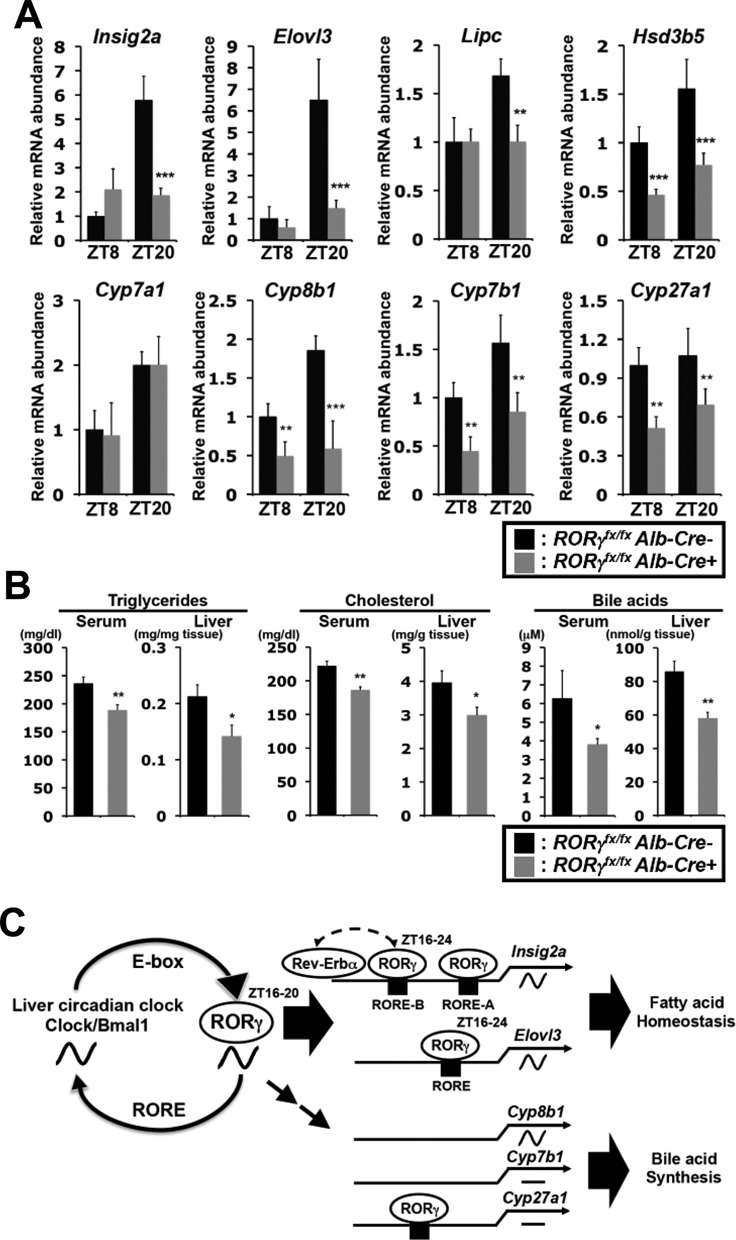
Analysis of liver-specific RORγ-deficient mice. (A) Comparison of the expression of several lipid metabolic genes in liver from RORγfx/fxAlb-Cre negative and positive mice (n = 4–5) at ZT8 and ZT20. Data represent mean ±SD. (B) Triglycerides, Cholesterol and bile acids levels in serum and liver collected from RORγfx/fxAlb-Cre negative and positive mice on HFD (n = 8) at ZT0 or ZT4. Data represent mean ±SEM, *P < 0.05, **P < 0.01, ***P < 0.001 by ANOVA. (C) Regulation of lipid metabolic genes by RORγ in liver involves multiple mechanisms. RORγ, which circadian expression is under direct control of the clock machinery, strongly regulates the peak expression of Insig2a, Elovl3 and Cyp8b1 in phase with the optimal expression of RORγ. ChIP-Seq and promoter analyses indicated that the diurnal expression of some metabolic genes, such as Insig2a and Elovl3, are regulated directly by RORγ. This is consistent with the concept that RORγ functions as a link between the circadian clock and its regulation of several metabolic genes and downstream metabolic outputs. However, certain metabolic genes, including Cyp7b1 and Cyp27a1, are regulated by RORγ in a ZT-independent manner likely involving indirect mechanisms. Association of RORγ with Cyp27a1 may suggest regulation by both direct and indirect mechanisms. Although, RORγ regulates Cyp8b1 in a ZT-dependent manner, no evidence was obtained to support regulation through a direct mechanism.
DISCUSSION
The regulation of many metabolic functions in the liver are linked to the daily timing of feeding and under the control of the circadian clock machinery (8,9,35). This includes transcriptional regulation of many metabolic genes by clock proteins, including Bmal1 and Clock (6,10,36,37), as well as by other transcription factors and, as we show in this study, RORγ1 (7,14,16,17). Our data indicate that the regulation of lipid metabolic genes by RORγ is complex and mediated by multiple mechanisms that involve ZT-(in)dependent as well as (in)direct regulation by RORγ (Figure 7C). The loss of RORγ results in significantly reduced peak expression of several hepatic genes, including Insig2a, Elovl3, Elovl2, Cyp8b1 and Lipc, without affecting their circadian phase (Figure 1) suggesting that their regulation is mediated by a ZT-dependent mechanism. ChIP-Seq analysis showing an association of RORγ with the regulatory region of several of these genes, including Insig2a and Elovl3, suggested that these genes are regulated directly by RORγ. Studies showing that the diurnal expression of RORγ is directly regulated by the circadian clock machinery (15,16,18), together with our findings that the phase of the diurnal pattern of expression of several of these genes closely follows that of RORγ, suggest a connection between the diurnal regulation of these metabolic genes, the rhythmic expression of RORγ, and the circadian clock. These observations are further consistent with our hypothesis that RORγ functions as a transcriptional mediator that regulates the rhythmic expression of certain metabolic genes downstream of the clock machinery (Figure 7C). Although the expression of certain genes, including Cyp7b1, was reduced in both RORγ−/− and RORαsg/sg mice suggesting a certain degree of redundancy, loss of RORα had little effect on the expression of Insig2a and Elovl3, suggesting that these genes are regulated in a highly RORγ-selective manner. This, together with our recent ChIP-Seq analysis (22), indicates that RORα and RORγ largely regulate different sets of metabolic genes and therefore control different metabolic pathways.
Analysis of the RORγ cistrome suggested that RORγ directly regulates the transcription of a number of lipid metabolic genes (Figure 2 and Supplementary Figure S1D). This was supported by our promoter analyses showing that RORγ was able to activate the Insig2a and Elovl3 promoters through ROREs in their respective upstream promoter region. The enhanced recruitment of RORγ to ROREs within Insig2a and Elovl3 promoters at ZT22 and the higher level of histone H3K9 acetylation are consistent with the increased activation of these promoters at ZT20–4 (Figures 2 and 4). Together, these data are consistent with the concept that RORγ positively regulates the transcription of these genes by binding ROREs in their promoter region in a ZT-dependent manner and around the time when RORγ is most highly expressed. These results support our hypothesis that RORγ functions as an intermediary regulator between the circadian clock machinery and the transcriptional regulation of the rhythmic expression of certain lipid metabolic genes (Figure 7C). However, some of these genes might be directly regulated Bmal1/Clock in conjunction with direct regulation by RORγ.
The hepatic expression of several arrhythmic genes, including Cyp7b1, Elovl5, AdipoR2, Cd36 and Hsd3b5, was reduced in RORγ−/− liver at all ZTs suggesting that they are regulated by RORγ in a ZT-independent manner. Moreover, our ChIP-Seq analysis showed no evidence for the recruitment of RORγ to some of these genes such as Cyp7b1, Cd36 and Hsd3b5, suggesting that they may be regulated by RORγ through an indirect mechanism (Figure 7C). Although peak expression of Cyp8b1 is reduced in RORγ−/− mice, ChIP-Seq (Supplementary Figure S1D), ChIP and reporter analyses (Takeda, Y., unpublished data) did not support direct regulation of Cyp8b1 by RORγ suggesting that it may also be regulated by an indirect mechanism.
The regulation of lipid metabolic genes is complex and other nuclear receptors, including LXRs and FXR, PXR and CAR (32,33), have been implicated in the regulation of lipid metabolism. Several studies have provided evidence for crosstalk between different nuclear receptor signaling pathways at multiple levels that include competition for the same DNA binding site, synergistic activation through adjacent binding sites, as well as regulation of the expression of one receptor by another (22). Thus, genes indirectly regulated by RORγ might be regulated through changes in the expression of other nuclear receptors or transcription factors. However, the loss of RORγ expression did not significantly change the diurnal pattern of expression of Lxr, Pxr, Fxr and Car (Supplementary Figure S4E) suggesting that changes in lipid metabolism did not relate to alterations in the level of expression of these receptors. Since the activity of these nuclear receptors is regulated by their interaction with (ant)agonists, including various oxysterols, bile acids and xenobiotics, an interesting alternative possibility is that metabolic changes in RORγ−/− liver might alter the level of certain nuclear receptor (ant)agonists thereby changing the activation of these receptors and subsequently the transactivation of their target genes. Crosstalk between receptor pathways is further complicated by the observations showing that certain oxysterol function as ligands for both LXR and ROR receptors (2,32,38).
Crosstalk between RORs and Rev-Erb receptors has been well established (1,7). Rev-Erbs, which function as transcriptional repressors, can bind similar ROREs as RORs and compete with RORs for binding thereby antagonizing their action. A recent study reported that the loss of Rev-Erbα eliminated the repression of Insig2a expression at ZT8–12, the time of optimal Rev-Erbα expression, thereby almost totally abolishing the rhythmic expression of Insig2a (29). Our data showing that Rev-Erbα was able to repress Insig2a(B)- and Elovl3(RORE)-dependent activation by RORγ is consistent with this. Together these observations suggest that the rhythmic expression of Insig2a and Elovl3 involves repression by Rev-Erbα at ZT8–12, whereas the optimal expression of Insig2a and Elovl3 at ZT20-4 is mediated in part by RORγ. These observations support the conclusion that RORγ plays a critical role in the positive transcriptional regulation of the Insig2a and Elovl3 expression at ZT16–24 (Figure 7C).
Insig2a regulates hepatic fatty acid synthesis by inhibiting proteolytic activation of Srebp-1c (27,28,39). The down-regulation of Insig2a in RORγ−/− liver resulted in increased processing and a higher level of nuclear Srebp-1c and a small temporal increase in Srepb-1c target gene expression at ZT4 (Figure 3A-C). However, the relative increase in processed Srebp-1c had only a limited effect on the expression of its target genes suggesting that additional mechanisms of regulation might be involved, such as regulation by Lipin-1 (40). Srebp-1c expression and activation is controlled at multiple levels and further study is needed to determine the precise role of Srebp-1c in the regulation of lipid homeostasis by RORγ.
The altered expression of lipid metabolic genes in RORγ−/− mice was associated with changes in serum and hepatic lipid composition and hepatic histology. These changes were more pronounced and distinct in RORγ−/−(HFD) mice than in mice fed an ND. The latter might be related to different mechanisms of regulation of lipid metabolism between ND and HFD mice and the higher lipid levels in serum and liver in mice fed an HFD. In addition, differences in the type and amount of lipids between WT(HFD) and RORγ−/−(HFD) livers may be linked to the increase in macrovesicular steatosis observed in RORγ−/− mice.
The most distinctive change seen in RORγ−/− mice was a reduction in bile acid levels in liver and serum as well as the whole body pool size. This reduction might at least in part be due to the repression of the hepatic expression of several genes involved in bile acid synthesis pathways, including Cyp8b1, Cyp7b1 and Cyp27a1. These findings suggest that RORγ positively regulates hepatic bile acid synthesis. This is supported by data showing that exogenous expression of RORγ in liver enhanced the level of hepatic bile acids and that liver-specific RORγ-deficient mice also showed a reduction in bile acid levels (Figures 6D and 7B). Recent studies showed that RORα and RORγ regulate the expression of several sulfotransferases, including Sult2A1, which has been implicated in bile acid synthesis (41), through their binding to RORE in their promoter (42,43). Thus, the combined regulation of several sterol metabolic genes by RORγ is likely responsible for the changes in cholesterol and bile acid homeostasis observed in RORγ−/− mice.
We further show that RORγ deficiency causes a change in fatty acid composition in liver and serum (Supplementary Figure S2 and Table S2), which might be in part related to the observed alterations in the expression of several Elovl genes (Figure 1A and E) and likely involves changes in the expression of other lipid metabolic genes. Although TGs in serum and liver were reduced in RORγ−/− mice (Figure 5A and B), it is not likely due to increased fatty acid consumption, but rather due to reduced rate of fatty acid/TG synthesis because the reduction of serum free fatty acids, glycerol and total ketone bodies indicates lower fatty acid consumption (Figure 5C). In addition, the reduced blood levels of TGs and free fatty acids in RORγ−/−(HFD) mice might be a contributory factor in the improved insulin sensitivity observed in these mice (22,44,45).
In summary, our study indicates that the transcriptional regulation of lipid metabolic genes by RORγ is complex and involves multiple mechanisms. The regulation by RORγ can be ZT-dependent and -independent and occur through a direct or indirect mechanism as shown for Insig2a and Cyp7b1, respectively. The circadian regulation of RORγ expression by the clock machinery together with our observations that RORγ directly regulates the transcription of a number of lipid metabolic genes, support our hypothesis that RORγ acts downstream of clock proteins and functions as an important link between the circadian clock machinery and its regulation of certain metabolic genes. Our study indicates that RORγ is an important participant in the diurnal regulation of lipid metabolism in the liver.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors would like to thank Dr Xiaoling Li for her comments on the manuscript and Laura Miller (NIEHS) for her assistance with the mice.
FUNDING
Intramural Research Program of the National Institute of Environmental Health Sciences (NIEHS); National Institutes of Health (NIH) [Z01-ES-101586, Z01-ES-050167]; Japanese Society for the Promotion of Science. Funding for open access charge: NIEHS; NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Jetten A.M. Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl. Recept. Signal. 2009;7:e003. doi: 10.1621/nrs.07003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Solt L.A., Burris T.P. Action of RORs and their ligands in (patho)physiology. Trends Endocrinol. Metab. 2012;23:619–627. doi: 10.1016/j.tem.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huh J.R., Littman D.R. Small molecule inhibitors of RORgammat: targeting Th17 cells and other applications. Eur. J. Immunol. 2012;42:2232–2237. doi: 10.1002/eji.201242740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bookout A.L., Jeong Y., Downes M., Yu R.T., Evans R.M., Mangelsdorf D.J. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126:789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Medvedev A., Yan Z.H., Hirose T., Giguere V., Jetten A.M. Cloning of a cDNA encoding the murine orphan receptor RZR/ROR gamma and characterization of its response element. Gene. 1996;181:199–206. doi: 10.1016/s0378-1119(96)00504-5. [DOI] [PubMed] [Google Scholar]
- 6.Bass J., Takahashi J.S. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duez H., Staels B. Nuclear receptors linking circadian rhythms and cardiometabolic control. Arterioscler. Thromb. Vasc. Biol. 2010;30:1529–1534. doi: 10.1161/ATVBAHA.110.209098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckel-Mahan K., Sassone-Corsi P. Metabolism and the circadian clock converge. Physiol. Rev. 2013;93:107–135. doi: 10.1152/physrev.00016.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asher G., Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 10.Rey G., Reddy A.B. Connecting cellular metabolism to circadian clocks. Trends Cell Biol. 2013;23:234–241. doi: 10.1016/j.tcb.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Maury E., Hong H.K., Bass J. Diabetes Metab. 2014. Circadian disruption in the pathogenesis of metabolic syndrome. doi:10.1016/j.diabet.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 12.Hatori M., Vollmers C., Zarrinpar A., DiTacchio L., Bushong E.A., Gill S., Leblanc M., Chaix A., Joens M., Fitzpatrick J.A., et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15:848–860. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang X., Downes M., Yu R.T., Bookout A.L., He W., Straume M., Mangelsdorf D.J., Evans R.M. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 14.Liu A.C., Tran H.G., Zhang E.E., Priest A.A., Welsh D.K., Kay S.A. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008;4:e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ueda H.R., Hayashi S., Chen W., Sano M., Machida M., Shigeyoshi Y., Iino M., Hashimoto S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 16.Takeda Y., Jothi R., Birault V., Jetten A.M. RORgamma directly regulates the circadian expression of clock genes and downstream targets in vivo. Nucleic Acids Res. 2012;40:8519–8535. doi: 10.1093/nar/gks630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho H., Zhao X., Hatori M., Yu R.T., Barish G.D., Lam M.T., Chong L.W., DiTacchio L., Atkins A.R., Glass C.K., et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mongrain V., Ruan X., Dardente H., Fortier E.E., Cermakian N. Clock-dependent and independent transcriptional control of the two isoforms from the mouse Rorgamma gene. Genes Cells. 2008;13:1197–1210. doi: 10.1111/j.1365-2443.2008.01237.x. [DOI] [PubMed] [Google Scholar]
- 19.Guillaumond F., Dardente H., Giguere V., Cermakian N. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J. Biol. Rhythms. 2005;20:391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- 20.Jetten A.M., Kang H.S., Takeda Y. Retinoic acid-related orphan receptors alpha and gamma: key regulators of lipid/glucose metabolism, inflammation, and insulin sensitivity. Front. Endocrinol. 2013;4:1–8. doi: 10.3389/fendo.2013.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takeda Y., Kang H.S., Angers M., Jetten A.M. Retinoic acid-related orphan receptor gamma directly regulates neuronal PAS domain protein 2 transcription in vivo. Nucleic Acids Res. 2011;39:4769–4782. doi: 10.1093/nar/gkq1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takeda Y., Kang H.S., Freudenberg J., DeGraff L.M., Jothi R., Jetten A.M. Retinoic acid-rrelated orphan receptor gamma (RORgamma): a novel participant in the diurnal regulation of hepatic gluconeogenesis and insulin sensitivity. PLoS Genet. 2014;10:e1004331. doi: 10.1371/journal.pgen.1004331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurebayashi S., Ueda E., Sakaue M., Patel D.D., Medvedev A., Zhang F., Jetten A.M. Retinoid-related orphan receptor gamma (RORgamma) is essential for lymphoid organogenesis and controls apoptosis during thymopoiesis. Proc. Natl. Acad. Sci. U.S.A. 2000;97:10132–10137. doi: 10.1073/pnas.97.18.10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pathak P., Li T., Chiang J.Y. Retinoic acid-related orphan receptor alpha regulates diurnal rhythm and fasting induction of sterol 12alpha-hydroxylase in bile acid synthesis. J. Biol. Chem. 2013;288:37154–37165. doi: 10.1074/jbc.M113.485987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeda Y., Jetten A.M. Prospero-related homeobox 1 (Prox1) functions as a novel modulator of retinoic acid-related orphan receptors alpha- and gamma-mediated transactivation. Nucleic Acids Res. 2013;41:6992–7008. doi: 10.1093/nar/gkt447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wada T., Kang H.S., Angers M., Gong H., Bhatia S., Khadem S., Ren S., Ellis E., Strom S.C., Jetten A.M., et al. Identification of oxysterol 7alpha-hydroxylase (Cyp7b1) as a novel retinoid-related orphan receptor alpha (RORalpha) (NR1F1) target gene and a functional cross-talk between RORalpha and liver X receptor (NR1H3) Mol. Pharmacol. 2008;73:891–899. doi: 10.1124/mol.107.040741. [DOI] [PubMed] [Google Scholar]
- 27.Dong X.Y., Tang S.Q. Insulin-induced gene: a new regulator in lipid metabolism. Peptides. 2010;31:2145–2150. doi: 10.1016/j.peptides.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 28.Shao W., Espenshade P.J. Expanding roles for SREBP in metabolism. Cell Metab. 2012;16:414–419. doi: 10.1016/j.cmet.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Martelot G., Claudel T., Gatfield D., Schaad O., Kornmann B., Sasso G.L., Moschetta A., Schibler U. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7:e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engelking L.J., Liang G., Hammer R.E., Takaishi K., Kuriyama H., Evers B.M., Li W.P., Horton J.D., Goldstein J.L., Brown M.S. Schoenheimer effect explained–feedback regulation of cholesterol synthesis in mice mediated by Insig proteins. J. Clin. Invest. 2005;115:2489–2498. doi: 10.1172/JCI25614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clugston R.D., Jiang H., Lee M.X., Piantedosi R., Yuen J.J., Ramakrishnan R., Lewis M.J., Gottesman M.E., Huang L.S., Goldberg I.J., et al. Altered hepatic lipid metabolism in C57BL/6 mice fed alcohol: a targeted lipidomic and gene expression study. J. Lipid Res. 2011;52:2021–2031. doi: 10.1194/jlr.M017368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calkin A.C., Tontonoz P. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nature Rev. Mol. Cell Biol. 2012;13:213–224. doi: 10.1038/nrm3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Handschin C., Meyer U.A. Regulatory network of lipid-sensing nuclear receptors: roles for CAR, PXR, LXR, and FXR. Arch. Biochem. Biophys. 2005;433:387–396. doi: 10.1016/j.abb.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 34.Kidani Y., Bensinger S.J. Liver X receptor and peroxisome proliferator-activated receptor as integrators of lipid homeostasis and immunity. Immunol. Rev. 2012;249:72–83. doi: 10.1111/j.1600-065X.2012.01153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bass J. Circadian topology of metabolism. Nature. 2012;491:348–356. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- 36.Kitazawa M. Circadian rhythms, metabolism, and insulin sensitivity: transcriptional networks in animal models. Curr. Diab. Rep. 2013;13:223–228. doi: 10.1007/s11892-012-0354-8. [DOI] [PubMed] [Google Scholar]
- 37.Koike N., Yoo S.H., Huang H.C., Kumar V., Lee C., Kim T.K., Takahashi J.S. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin L., Martynowski D., Zheng S., Wada T., Xie W., Li Y. Structural basis for hydroxycholesterols as natural ligands of orphan nuclear receptor RORgamma. Mol. Endocrinol. 2010;24:923–929. doi: 10.1210/me.2009-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yellaturu C.R., Deng X., Park E.A., Raghow R., Elam M.B. Insulin enhances the biogenesis of nuclear sterol regulatory element-binding protein (SREBP)-1c by posttranscriptional down-regulation of Insig-2A and its dissociation from SREBP cleavage-activating protein (SCAP).SREBP-1c complex. J. Biol. Chem. 2009;284:31726–31734. doi: 10.1074/jbc.M109.050914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peterson T.R., Sengupta S.S., Harris T.E., Carmack A.E., Kang S.A., Balderas E., Guertin D.A., Madden K.L., Carpenter A.E., Finck B.N., et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Radominska A., Comer K.A., Zimniak P., Falany J., Iscan M., Falany C.N. Human liver steroid sulphotransferase sulphates bile acids. Biochem. J. 1990;272:597–604. doi: 10.1042/bj2720597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang H.S., Angers M., Beak J.Y., Wu X., Gimble J.M., Wada T., Xie W., Collins J.B., Grissom S.F., Jetten A.M. Gene expression profiling reveals a regulatory role for ROR alpha and ROR gamma in phase I and phase II metabolism. Physiol. Genomics. 2007;31:281–294. doi: 10.1152/physiolgenomics.00098.2007. [DOI] [PubMed] [Google Scholar]
- 43.Ou Z., Shi X., Gilroy R.K., Kirisci L., Romkes M., Lynch C., Wang H., Xu M., Jiang M., Ren S., et al. Regulation of the human hydroxysteroid sulfotransferase (SULT2A1) by RORalpha and RORgamma and its potential relevance to human liver diseases. Mol. Endocrinol. 2013;27:106–115. doi: 10.1210/me.2012-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meissburger B., Ukropec J., Roeder E., Beaton N., Geiger M., Teupser D., Civan B., Langhans W., Nawroth P.P., Gasperikova D., et al. Adipogenesis and insulin sensitivity in obesity are regulated by retinoid-related orphan receptor gamma. EMBO Mol. Med. 2011;3:637–651. doi: 10.1002/emmm.201100172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tinahones F.J., Moreno-Santos I., Vendrell J., Chacon M.R., Garrido-Sanchez L., Garcia-Fuentes E., Macias-Gonzalez M. The retinoic acid receptor-related orphan nuclear receptor gamma1 (RORgamma1): a novel player determinant of insulin sensitivity in morbid obesity. Obesity. 2012;20:488–497. doi: 10.1038/oby.2011.267. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


