Abstract
Determining the extent and structure of intra-host genetic diversity and the magnitude and impact of population bottlenecks is central to understanding the mechanisms of viral evolution. To determine the nature of viral evolution following systemic movement through a plant, we performed deep sequencing of 23 leaves that grew sequentially along a single Cucurbita pepo vine that was infected with zucchini yellow mosaic virus (ZYMV), and on a leaf that grew in on a side branch. Strikingly, of 112 genetic (i.e. sub-consensus) variants observed in the data set as a whole, only 22 were found in multiple leaves. Similarly, only three of the 13 variants present in the inoculating population were found in the subsequent leaves on the vine. Hence, it appears that systemic movement is characterized by sequential population bottlenecks, although not sufficient to reduce the population to a single virion as multiple variants were consistently transmitted between leaves. In addition, the number of variants within a leaf increases as a function of distance from the inoculated (source) leaf, suggesting that the circulating sap may serve as a continual source of virus. Notably, multiple mutational variants were observed in the cylindrical Inclusion (CI) protein (known to be involved in both cell-to-cell and systemic movement of the virus) that were present in multiple (19/24) leaf samples. These mutations resulted in a conformational change, suggesting that they might confer a selective advantage in systemic movement within the vine. Overall, these data reveal that bottlenecks occur during systemic movement, that variants circulate in the phloem sap throughout the infection process, and that important conformational changes in CI protein may arise during individual infections.
Keywords: zucchini yellow mosaic virus, evolution, systemic movement, intra-host genetic diversity, cylindrical inclusion protein
1. Introduction
RNA viruses tend to harbor abundant genetic diversity through a combination of highly error-prone replication, short generation times and large population sizes (Duffy et al., 2008). This genetic diversity has been associated with the ability of RNA viruses to switch hosts (Jerzak et al., 2008; Holmes, 2009) overcome host resistance mechanisms (Feuer et al., 1999: Lech et al., 1996), and alter virulence (Acosta-Leal et al., 2011). However, viral populations also experience important bottlenecks within individual hosts and at inter-host transmission, which likely reduce standing genetic diversity and hence impact the patterns and dynamics of viral evolution. In the case of plant viruses, dramatic population bottlenecks (down to 1-20 virions) have been reported during both systemic (Fabre et al., 2012; French and Stenger, 2003; Li and Roossinck, 2004; Sacristan et al., 2003) and cell-to-cell movement (Gonzalez-Jara et al., 2009; Miyashita and Kishino, 2010; Tromas et al., 2014). Hence, although plant virus populations are expected to be large (for example, 1011 – 1012 virions per infected leaf have been reported in tobacco mosaic virus (TMV)) (Sacristan et al., 2003), population bottlenecks will mean that effective population sizes will be significantly smaller (Garcia-Arenal et al., 2001). This, in turn, will influence the respective strengths of natural selection and genetic drift, with the latter expected to dominate in populations with small effective sizes. Determining how genetic diversity changes as the virus moves systemically through individual plants therefore provides important insights into the strength of population bottlenecks and hence the nature of viral evolution.
Unlike animal viruses in which receptor mediated mechanisms allow use of the extracellular environment to move throughout the host, plant viruses are restricted to the intra-cellular environment, and cell-to-cell movement is restricted to the plasmodesmata (Maule and Wang, 1996). Systemic infection of the plant necessitates movement of the viral population both cell-to-cell through the plasmodesmata, as well as organ-to-organ through the phloem. For this process to occur successfully the virus must enter the vascular tissue. This requires movement from the mesophyll cells and through a series of cells: the perivascular parenchyma, the phloem parenchyma, the companion cells, and finally into the sieve tube elements (Niehl and Heinlein, 2011). Hence, movement of the virus through the plant follows the same path as the photoassimilates (Maule and Wang, 1996).
The Potyviridae is the largest family of plant viruses and comprises roughly 30% of known plant viruses (Ward and Shukla, 1991). This family, particularly the aphid-transmitted members, are among the most successful plant pathogens (Rybicki and Pietersen, 1999). Potyviruses possess single-stranded positive-sense RNA genomes (Berger, 2001), and harbour at least five proteins known to be involved in viral movement; the coat protein (CP), the helper component protein (HC-Pro), the cylindrical inclusion protein (CI), the P3N-PIPO and the viral genome-linked protein (VPg). The CP is necessary for long distance spread within the plant and may also facilitate cell-to-cell movement (Dolja et al., 1994; Dolja et al., 1995) via binding to the viral RNA and altering the exclusion size limit of the plasmodesmata. This is thought to follow the infection front in a transient fashion (Heinlein et al., 1995; Oparka et al 1997). Mutation analysis with wheat streak mosaic virus suggests that the C terminus of the CP may be required for cell-to-cell movement (Tatineni et al., 2014). The HC-Pro is thought to function in viral movement by increasing plasmodesmatal permeability (Rojas et al., 1997). The cylindrical inclusion protein (CI) protein is also involved in cell-to-cell movement (Shukla et al., 1991; Urcuqui-Inchima et al., 2001), and is believed to guide the CP-RNA complex to the plasmodesmata (Roberts et al., 1998; Rodriguez-Cerezo et al., 1997). Mutation analysis has revealed that mutations affecting the N terminus region of the CI were defective in cell-to-cell movement in both tobacco etch virus (TEV) (Carrington et al., 1998), and plum pox virus (PPV) (Gomez de Cedron et al., 2006). The P3N-PIPO modulates the plasmodesmatal localization of the CI, and the CI-P3N-PIPO complex is thought to be responsible for the plasmodesmatal associated structures that assist cell-to-cell movement (Wei et al., 2010). Although the role of the VPg is currently unknown, it has been demonstrated that mutated VPgs in turnip mosaic virus reduce both cell-to-cell and systemic movement (Dunoyer et al., 2004).
Zucchini yellow mosaic virus (ZYMV) is a member of the Potyviridae that infects Cucurbitacae (squash, melon and cucumber) globally. ZYMV is considered to be an emerging virus as it achieved a worldwide distribution within two decades of its discovery (Desbiez and Lecoq, 1997). The symptoms of ZYMV include severe stunting of the plant and fruits as well as a distinctive yellow mottling of the leave (Desbiez and Lecoq, 1997). Fruits harvested from ZYMV infected plants are often mottled and deformed and thus tend to be unmarketable, and ZYMV can reduce agricultural yields up to 94% (Blua and Perring, 1989). In the United States Cucurbitacae production is estimated to be 1.5 billion per annum and, given that these are among the 15 most important agricultural crops in the United States (Cantliffe et al., 2007), it is clear that ZYMV is a significant crop pathogen. ZYMV is primarily a vector-borne pathogen and is non-persistently transmitted by aphids. Experimentally, 26 aphid species have been shown to transmit ZYMV (Katis et al., 2006), and we previously determined a seed to seedling (vertical) transmission rate of 1.6% (Simmons et al., 2011).
To ascertain the extent and pattern of viral genetic diversity as it moves systemically through the plant, and how this might be impacted by population bottlenecks (as measured by changes in genetic diversity), we undertook deep sequencing of 23 sequential leaves on a Cucurbita pepo ssp. texana vine as well as an additional leaf that grew on a side branch. C. pepo is believed to be the progenitor of domestic squash (Decker and Wilson, 1987), and the optimal host for the maintenance of ZYMV (Gal-On, 2007).
2. Methods
2.1 Greenhouse experiment
The first true leaf of a C. pepo plant was mechanically inoculated in a greenhouse at The Pennsylvania State University in April 2011 with a ZYMV sample taken from the first diseased individual from an experimental field during the 2007 season. The inoculum used here was the same inoculum source as that used in Simmons et al. 2012, and was prepared from infected leaf tissue diluted in a phosphate buffer (0.1 M Na2H/KH2PO4 buffer) in a 1:3 v/v ratio. Carborundum powder (500gm) was then rubbed on the surface of the first true leaf, and the inoculum subsequently applied to the leaf surface with a pestle. Leaves along one vine were collected over a two-month period (Fig 1). It appears that removing the inoculated leaf, particularly early in infection, can effect systemic infection (unpublished data in (Zwart et al., 2012)). Accordingly, the first true leaf (i.e. the inoculated leaf) was not removed but was retained to serve as a “source” of viral material. Hence, all the leaves collected are in reference to the inoculated leaf, such that leaf one is the second true leaf.
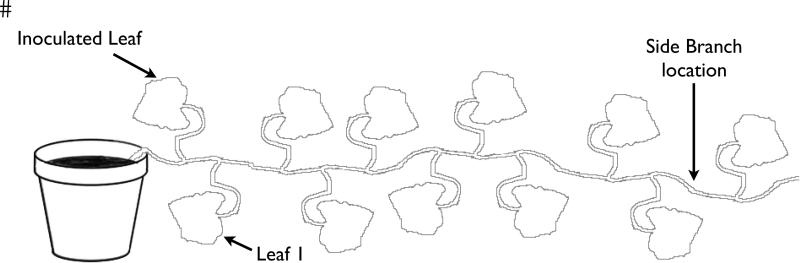
Figure 1.
Schematic of the plant depicting the leaves that were harvested from a growing vine over time. A total of 23 leaves were collected sequentially, while the 24th leaf (side branch) grew in between where leaves nine and ten had grown prior to harvesting. The side branch was harvested 21 days after leaf nine and 17 days after leaf 10 had been harvested.
A total of 24 leaves were collected. Of these, 1-23 were collected in sequential order as each leaf attained full size. The exception was the side branch leaf, which grew at the same time as leaf 16, but was physically located between where leaf nine and ten had grown with the effect that this leaf was harvested several weeks after its nearest neighbors were collected (21 days after leaf nine and 17 days after leaf 10).
2.2 RNA isolation, RT-PCR, qPCR and sequencing of seedlings
Fifty mg of leaf tissue was used for analysis. Frozen leaf samples were used with the E.Z.N.A.® RNA isolation kits (Omega bio-tek, GA) for the isolation of total RNA. First-strand synthesis of cDNA was generated from the total RNA using genome-specific primers. The RT-PCR was conducted using Superscript III First-Strand Synthesis kit (Life Technologies, CA) following the manufacturers protocol. The single stranded cDNA was then used as template for PCR amplification using Phusion High-Fidelity polymerase (New England Biolabs, MA). Manufacturers protocols were followed using HF PCR buffer and 15 μl of first-strand product was used in a 50 μl total reaction. The PCR conditions were: 98°C for 1 min, 98°C for 10 s, 58°C for 20 s, 72°C for 1 min 20 s, for a total of 20 cycles with a final 5 min 72°C extension and held at 4°C. The five primers pairs were synthesized (Integrated DNA Technologies, IA) with 787, 886, 759, and 842 bp overlap between amplicons across the genome. Primers: ZYMV_1/5 F1: (nt 27-50 of the reference strain NC_003224.1) AGAAATCAACGAACAAGCAGACGA, ZYMV_1/5 R1: (nt 2199-2219) GCAACATCCATCAAC GAAGGC, ZYMV_2/5 F1: (nt 1432-1453) CAGAACCATACAA GCAC TCACA, ZYMV_2.5 R1: (nt 4072-4096) GAAAAGCAAA TCCACTCGTCATC, ZYMV_3/5 F1: (nt 3210-3233) CGTGTGTTTG GTATTCTCCTTGGT, ZYMV_3/5 R1: (nt 5824-5847) TCCTTTTGT GCTGTTGTTTCCTTT, ZYMV_4/5 F1: (nt 5088-5108) TGAGAGCACACGCATA CCTTT, ZYMV_4/5 R1: (nt 7803-7823) CGACCCA CCAATCCTCCATA, ZYMV_5/5 F1: (nt 6981-7001) TGAAACACAGAGCAAGC GAGA, ZYMV_5/5 R5: (nt 9516-9534) CCGAC AGGACTACGG CATT. The lengths of each primer product were 2193, 2665, 2638, 2730, and 2553 bp respectively. The primer products were pooled per sample and gel extracted using Zymoclean Gel Recovery kit (Zymo Research, CA) for removal of non-specific amplification product. The library construction method used was as outlined in (Dunham and Friesen, 2013).
The primers used for qPCR used were those in Simmons et al. (2013), and the reference strain (GenBank accession no. NC_003224.1) was used to design them using Primer Express® Software version 3.0 (Applied Biosystems) and fell within the CI protein; forward primer: 5’-GGACAGTGCGACTATAGCTTCAA-3’ and reverse primer: 5’-TTTAACCGCGAATTGCGTATC-3’ (2013). The mix for the PCR reaction contained the following: 10.0μl SYBER green, 0.2μl of each primer, 7.6μl of PCR grade water and 2.0μl of template for a total reaction volume of 20μl. The PCR was carried out in triplicate in an Applied Biosystems StepOnePlus™ Real-Time PCR-System and the cycling conditions were as follows: holding temperature of 95oC for 5 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. The standard curve was produced by creating a dilution series of cDNA stock of ZYMV.
2.3 Alignment of raw reads and variant calling
Alignments were performed using the Burrows Wheeler aligner (BWA) version 0.6.2 allowing 10 mismatches (Li and Durbin, 2009). BAM to SAM file conversion and filtering was performed with Samtools version 0.1.18 (Li et al., 2009). The inoculum sequence from Simmons et al. (2012) was used as a reference sequence for read alignment and variant calls. Varscan (version 2.3.2) Koboldt et al., 2012) was used to call the minor mutational variants. To conservatively eliminate false positive intra-host mutations we only retained variants that occurred at greater than 100X coverage (although all the variant sites observed here had a coverage greater than 1000X; Table S1), had a frequency of 1% or greater, and had a quality score of 30 or greater. In addition, we simulated the false positive and false negative rates following the protocol of Goto et al. (2011), to determine the appropriate low frequency cut-off, which was 1%. The strand filter was also applied to eliminate any strand bias. Conformational protein changes were determined using Phyre 2 (Kelley and Sternberg, 2009) and confirmed using I-TASSER (Roy et al., 2010; Zhang, 2008).
All the variant consensus nucleotide sequences generated here (three in total) have been submitted to GenBank and assigned accession numbers KJ923767-69.
2.4 Statistical Analysis
Correlation analysis and a one-way ANOVA followed by post testing for a linear trend was performed on all 24 samples using GraphPad Prism version 6.0b for Mac OS X, GraphPad Software (La Jolla California USA). Additionally, figures depicting the number of mutations across the genome as well as the rain plot were generated using GraphPad Prism. All ZYMV sequences were manually aligned using Se-Al (2.0a11; kindly provided by Andrew Rambaut, University of Edinburgh) and a minimum spanning tree among them was estimated using the statistical parsimony approach available in the TCS 1.21 program (Clement et al., 2000).
3. Results
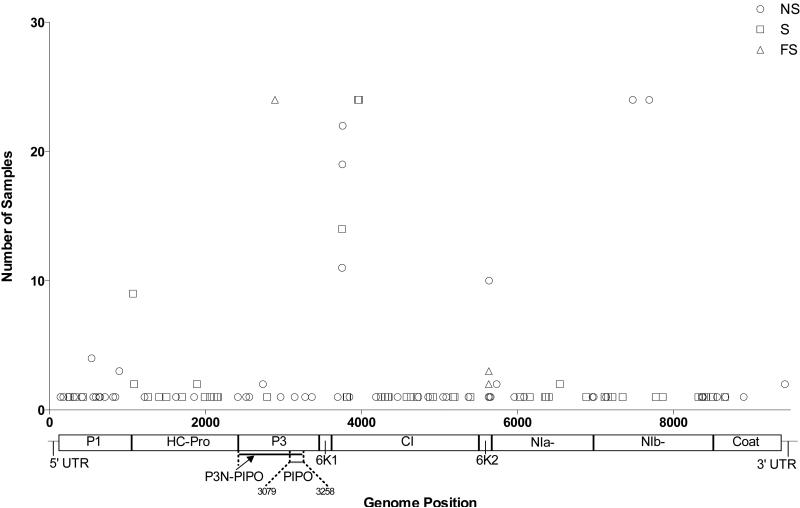
The average depth of sequencing coverage of ZYMV was 27,150X (range 14,671X – 42,764X) and the average amount of genome sequenced was 99.7%. In total, we observed 112 variants (excluding the 5’ untranslated region), 90 of which were found in a single sample only (Table S1) and 22 of which were shared between two or more samples (Table S2). 109 of the variants were SNPs and three were frameshifts (Fig 2). No stop codon mutations were observed. Of the 109 SNPs, 54 were synonymous and 56 non-synonymous. Four of the shared variants approached fixation (i.e. were at a frequency greater than 99%), at sites 3954, 3969, 7477 and 7688, and one other shared variant occurred at a frequency greater than 50% in some of the samples (position 1071 in leaves 17, 21 and 23). All other shared mutations were minor variants (i.e. occurred in less than 50% of the reads) (Fig 3). The variant that occurred at position 1071, in combination with a variant that occurred in one sample only (position 8899 in Leaf 17), resulted in three different consensus sequences. One of the variants that approached fixation was synonymous (3969), while the other three variants were non-synonymous 3954 (Q->K), 7477 (Y->H) and 7688 (R->K)). Interestingly, the last two mutations were found in the NIb gene that encodes the viral polymerase. Also of note was that all the frameshift mutations were shared among multiple samples, albeit at low frequency (~1-2%; Table S2). The exception was the frameshift at site 5633, which attained a frequency of 16.64% in sample 2, 9.3% in sample 10, and 14.34% in sample 12.
Figure 2.
Plot illustrating mutation position and type. The NS (circle) represents non-synonymous mutations, the S (square) synonymous mutations, and the FS (triangle) frame-shift mutations. The x-axis is the variant position and the y-axis is the number of samples that contained the each mutation, and below the x-axis is a schematic of a potyvirus genome.
Figure 3.
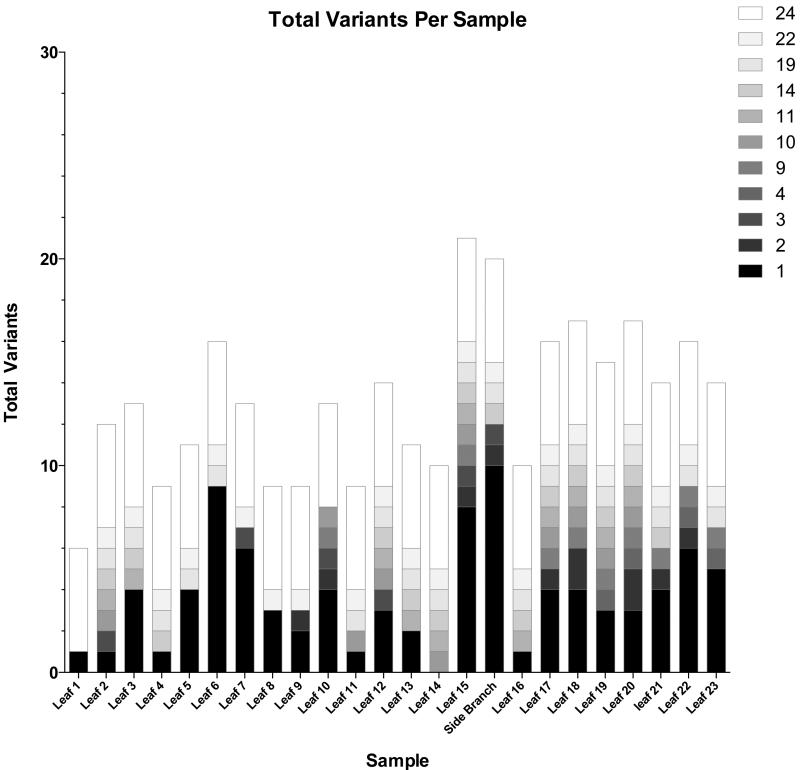
A plot of the number of variants found in each sample. Variants in each column are organized into blocks that correspond to specific group sizes (number of samples that share the variants within each group). The actual number of leaf samples that share a variant is denoted by the color of the block. The X-axis denotes the total number of variants found in each sample and the samples are denoted in the y-axis.
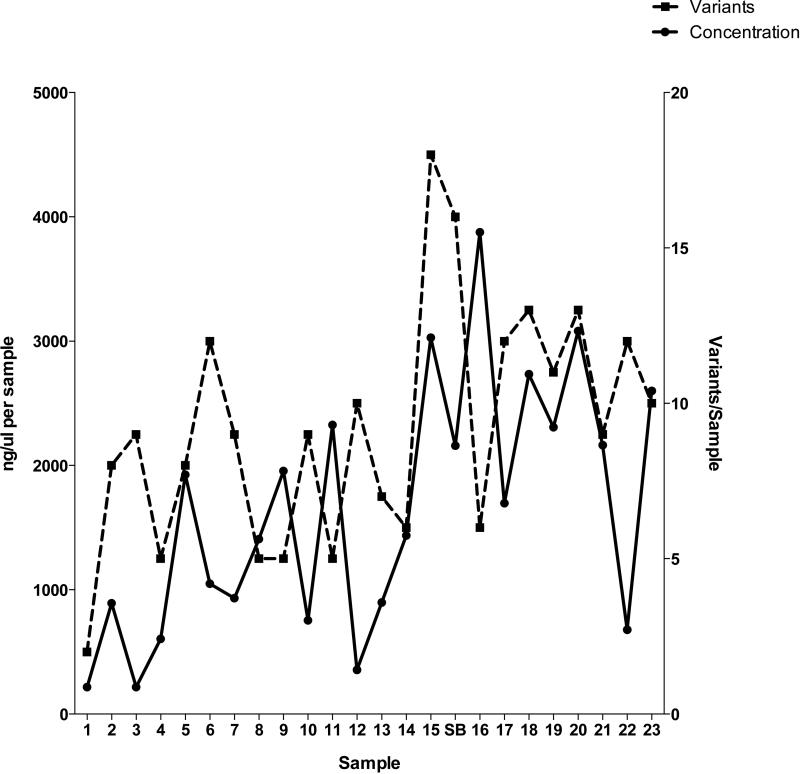
There was an average of 9.2 mutations per sample (range 2-18). Only one minor variant (2891) was found in all 24 samples. The starting inoculant population had a total of 13 variants, only three of which were present in the leaf-to-leaf samples: that at site 2891 and two that approached fixation (3954 and 7688). Interestingly, a significant positive correlation was found between the increase in the number of variants and time (r = 0.442; p = 0.031), and a significant linear trend was identified between the average variant frequency and time (p = 0.021). In addition as the number of variants increased over time we observed an overall increase in viral concentration (determined via qPCR) (Fig 4).
Figure 4.
The concentration of virus and number of variants found in each sample. The y-axes denote the concentration of virus per leaf sample as determined via qPCR (left hand side) and the number of variants per sample (right hand side), with the leaf samples given on the x-axis.
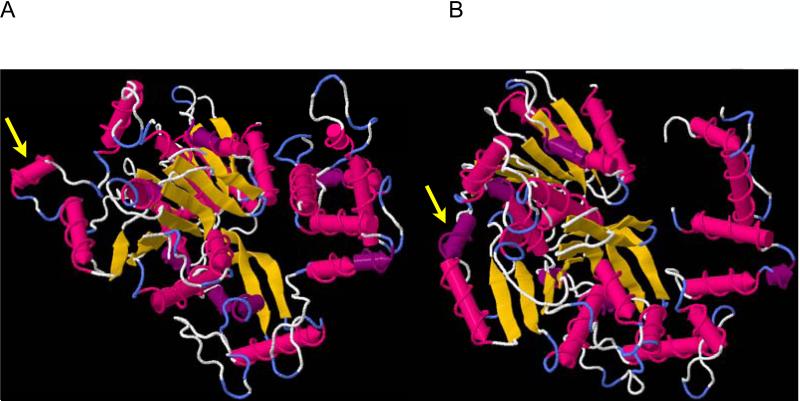
Four variants were in close proximity to each other in the CI protein: positions 3750, 3751, 3754 and 3758. Of these, position 3751, was a synonymous change (C->A), while the other three resulted in amino acid changes: position 3750 (N->K), 3754 (Q->K) and 3758 (L->Q). Eleven of the 24 leaf samples sequenced in this study exhibited all four changes, and an additional four exhibited three of the four variants. To determine if these variants might have resulted in a conformational change, we used two protein prediction platforms, Phyre 2 (Kelley and Sternberg, 2009) and I-TASSER (Roy et al., 2010; Zhang, 2008). Interestingly, two of the three changes (positions 3750 and 3754) resulted in the same conformational change (Fig 5A-B), in which an alpha helix contracted inwards towards the protein in the mutated form relative to the wild-type conformation. Nineteen of 24 samples had either one or both of these mutations that resulted in the conformational change.
Figure 5.
The Cylindrical Inclusion (CI) protein as predicted by Phyre2 (A) is the wild type conformation and (B) depicts the three amino acid substitutions that resulted in the conformational change. The yellow arrows highlight the alpha helix that altered as a result of the three mutations.
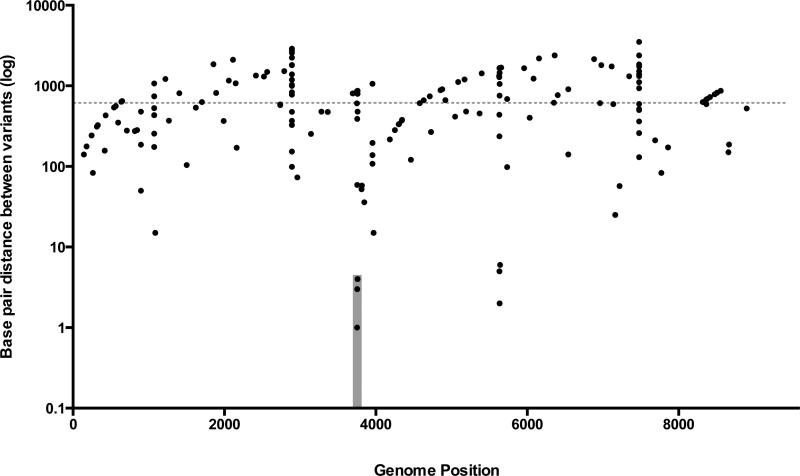
The spatial distribution of mutations did not differ significantly from that expected by chance alone in 10 of the 11 genes. The exception was 6K2 (a ZYMV protein that is believed to anchor the replication apparatus to ER-like membranes (Urcuqui-Inchima et al., 2001)) which harbored more mutations than expected by chance alone (p=0.05). To determine whether variants were spatially clustered within the genome, we determined that the average variant distance was 604.1 base pairs. We defined a cluster as a genomic region containing at least three variants, the variant distance was less then 50 bp, and the variants must be shared by at least five samples. A single region met these criteria – bases 3750-3758 of the CI gene (Fig 6). Two other regions, which did not meet all of our clustering criteria (i.e. that the region contain three or more variants and that at least five samples share these variants), were identified (regions 1071-1086 and 5631-5641) and these regions fell in the HC-Pro and 6K2, respectively.
Figure 6.
A rainfall plot was generated to illustrate mutation clustering. Each dot corresponds to an individual mutation, with the x-axis reflecting the genome position of each mutation, and the y-axis reflecting the distance between each mutation and its preceding mutation. The grey dotted line is the average distance between variants across all samples, with the cluster in the CI gene highlighted in the gray shaded box.
4. Discussion
Given that the bulk (80%) of the intra-plant mutational variants that we found were in one sample only, it is clear that the majority of mutants were not maintained in the viral population as it moved systemically through the plant. Whether this is due to selection (i.e. removing deleterious mutations) or genetic drift, including the action of population bottlenecks, or a combination of both is unclear. It is possible that the majority of the variants were deleterious and as such are expected to be purged from the population. Indeed, previous studies have shown that approximately 70% of the mutations found in RNA viruses are deleterious or slightly so (Carrasco et al., 2007; Domingo-Calap et al., 2009; Sanjuan et al., 2004). However, that many of the unique (one sample) variants were synonymous sites suggests that their loss is more likely associated with neutral evolutionary processes such as population bottlenecks. In this context it is notable that only 13 variants in the inoculant were found in the subsequent leaves, and 22 (of the total of 112) variants were shared amongst the samples. Hence, these data are compatible with the action of sequential population bottlenecks during systemic movement, although not so severe as to reduce the virus population to only a single virion as mutations were consistently shared among samples. Indeed, the presence of likely deleterious frameshift mutations in multiple samples, and at frequencies >10%, strongly suggests that complementation has occurred, which obviously requires the co-transmission of multiple viral genomes (Aaskov et al., 2006).
There have been variable estimates of the size of the population bottleneck following intra-host movement within plants, covering one to hundreds of virions, and it is likely that the extent and influence of population bottlenecks is not uniform across viral-host combinations. Indeed, previous work has shown that the magnitude of the genetic bottleneck is dependent on the size of the inoculant dose (Zwart et al., 2011). In addition, work with PPV in Prunus suggests that although the viral population within a host may harbour extensive genetic diversity, this diversity will be differentiated into sub-populations that reflect the physical structure of the tree (Jridi et al., 2006), and which will in turn influence the impact of any population bottleneck. The existence of relatively wide population bottlenecks, as noted here, has been reported in the case of some other plant viruses, (Fabre et al., 2014; Gutierrez et al., 2012) and may be of sufficient size to allow natural selection to proceed efficiently (Bergstrom et al., 1999).
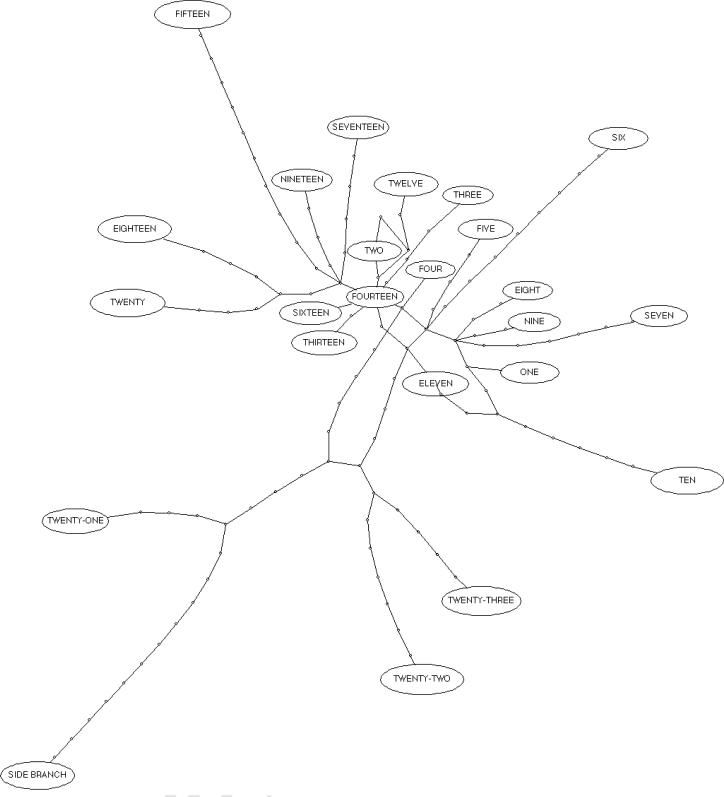
One of the most notable observations of our study was that the number of variants increased as a function of distance from the source inoculated leaf. This is in accord with previous studies with cauliflower mosaic virus (CaMV), at least in the early stages of infection (Gutierrez et al., 2012). However, contradictory data has also been reported. For instance, an examination of the movement of 12 experimental cucumber mosaic virus mutants in tobacco found that the number of mutants in successive leaves decreased as a function of distance from the source (an average of seven mutants were found in the eighth leaf and an average of five in the 15th leaf; (Li and Roossinck, 2004). Furthermore, the side branch leaf grew in several weeks after the nearest leaves (between leaves 9 and 10) had been harvested, and it appears to be more closely related to leaves further along the vine. This is supported by the minimum spanning tree in which the side branch clusters with leaves 21, 22, and 23 (Fig 7). Hence, these data suggest that the viral population in the phloem sap may serve as a constant source of genetic diversity as has been found in CaMV (Gutierrez et al., 2012).
Figure 7.
Minimum spanning trees depicting the population structure of ZYMV in each leaf. Each oval represents the viral population from each leaf. Each dot on the lines linking the ovals represents one mutation.
Of particular interest was the observation that four mutations in the CI protein found in 11 of the 24 samples resulted in a conformational change in the predicted protein structure. The CI is known to be involved in viral movement in planta, and two mutations in the N-terminus region of the TEV CI resulted in defects in cell-to-cell movement, although still able to replicate at wild type levels (Carrington et al., 1998). Similar results were found with PPV mutants located in the first 125 amino acids of the CI (Gomez de Cedron et al., 2006). Interestingly, two of the three changes resulted in the same predicted conformational change (positions 3750 and 3754) such that 19 out of the 24 leaf samples contain virions that contained these conformational changes (Fig 5B). Although tentative, these results suggest that there may be a selective advantage to these variants in systemic movement. Functional work is needed to determine the effects of these variants in systemic movement within the host and whether they are able to persist between hosts.
We had previously conducted a transmission experiment in which we inoculated a C. pepo plant and completed a series of eight mechanical inoculations between plants, after which we deep-sequenced these populations to determine the extent and pattern of viral genetic diversity at the inter-host scale (Simmons et al., 2012). As we used the same inoculant population for both experiments, the fifth leaf of the first plant of the serial inoculation corresponds to the leaf four population sequenced in this study (the first true leaf was not removed from the plant but rather was retained as a viral source). In the previous experiment there were 16 variants in the 5th true leaf and in the current experiment there were nine variants in the 5th true leaf and only one in common between the two (7688; one of the two variants that approached fixation in this study and the predominant variant in the inoculum). This suggests that either there is a substantial stochastic element to the viral population as it moves along the vine, or that there is structuring of the viral population within the same leaf, or a combination of both. Either way, it is possible that the portion of the leaf used to infect the plant in the previous experiment may have contained a vastly different population than the leaf portion that was used to inoculate in this experiment. However additional work is needed to ascertain the spatial structuring of the viral population within the leaf.
Evidently, the extent of the population bottleneck imposed on the viral population as it moves through the plant is not straightforward to determine, and is influenced by a variety of factors, including the size of the inoculum dose, the population structure of the virus, the timing of infection, as well as the host genotype/viral strain specific interactions. However, what is clear is that despite the marked population structure within leaves, the population bottlenecks imposed on the virus as it moves systemically through the plant are not particularly severe. This is shown by the multiple variants shared between leaf samples, including those likely producing conformational changes in the CI protein, as well as the frameshift mutations. Moreover, because the number of variants increased as a function of distance from the inoculum source, it is likely that the sap serves as a continual source of circulating virus.
Supplementary Material
Highlights.
ZYMV experiences population bottlenecks during systemic movement
Multiple variants are transmitted among leaves so the bottleneck may not be severe
Number of variants in a leaf increases as a function of distance from the source
Viral population may be highly structured within the leaf
A conformational change is observed in the Cylindrical Inclusion protein
Acknowledgments
This work was supported by the National Science Foundation Doctorial Dissertation Grant Program no. 1010881, the Biotechnology Risk Assessment Program Grant no. 2009-33120-20093 from the USDA National Institute of Food and Agriculture, and the USDA-FAS, Technical Assistance for Specialty Crops, grant #2011-23. JPD is supported by NIH RO1 EUREKA GM098741. ECH is supported by an NHMRC Australia Fellowship. We thank Tony Omeis for greenhouse assistance as well as the use of the Biology Greenhouse
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aaskov J, Buzacott K, Thu HM, Lowry K, Holmes EC. Long-term transmission of defective RNA viruses in humans and Aedes mosquitoes. Science. 2006;311(5758):236–238. doi: 10.1126/science.1115030. [DOI] [PubMed] [Google Scholar]
- Acosta-Leal R, Duffy S, Xiong Z, Hammond RW, Elena SF. Advances in plant virus evolution: translating evolutionary insights into better disease management. Phytopathol. 2011;101(10):1136–1148. doi: 10.1094/PHYTO-01-11-0017. [DOI] [PubMed] [Google Scholar]
- Berger PH. Potyviridae, eLS. John Wiley & Sons, Ltd.; 2001. [Google Scholar]
- Bergstrom CT, McElhany P, Real LA. Transmission bottlenecks as determinants of virulence in rapidly evolving pathogens. Proc Natl Acad Sci U S A. 1999;96(9):5095–5100. doi: 10.1073/pnas.96.9.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blua MJ, Perring TM. Effect of Zucchini Yellow Mosaic-Virus on Development and Yield of Cantaloupe (Cucumis-Melo). Plant Dis. 1989;73(4):317–320. [Google Scholar]
- Cantliffe DJ, Shaw NL, Stoffella PJ. Current trends in cucurbit production in the US. Proceedings of the IIIrd International Symposium on Cucurbits. 2007;(731):473–478. [Google Scholar]
- Carrasco P, de la Iglesia F, Elena SF. Distribution of fitness and virulence effects caused by single-nucleotide substitutions in tobacco etch virus. J Virol. 2007;81(23):12979–12984. doi: 10.1128/JVI.00524-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington JC, Jensen PE, Schaad MC. Genetic evidence for an essential role for potyvirus CI protein in cell-to-cell movement. The Plant J. 1998;14(4):393–400. doi: 10.1046/j.1365-313x.1998.00120.x. [DOI] [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9(10):1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Decker DS, Wilson HD. Allozyme variation in Cucurbita pepo complex: C. pep ovar. overifera vs. C. texana. Syst Bot. 1987;12:263–273. [Google Scholar]
- Desbiez C, Lecoq H. Zucchini yellow mosaic virus. Plant Pathol. 1997;46(6):809–829. [Google Scholar]
- Dolja VV, Haldeman R, Robertson NL, Dougherty WG, Carrington JC. Distinct Functions of Capsid Protein in Assembly and Movement of Tobacco Etch Potyvirus in Plants. Embo J. 1994;13(6):1482–1491. doi: 10.1002/j.1460-2075.1994.tb06403.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolja VV, Haldeman-Cahill R, Montgomery AE, Vandenbosch KA, Carrington JC. Capsid protein determinants involved in cell-to-cell and long distance movement of tobacco etch potyvirus. Virology. 1995;206(2):1007–1016. doi: 10.1006/viro.1995.1023. [DOI] [PubMed] [Google Scholar]
- Domingo-Calap P, Cuevas JM, Sanjuan R. The Fitness Effects of Random Mutations in Single-Stranded DNA and RNA Bacteriophages. Plos Genet. 2009;5(11) doi: 10.1371/journal.pgen.1000742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S, Shackelton LA, Holmes EC. Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genet. 2008;9(4):267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- Dunham JP, Friesen ML. A cost-effective method for high-throughput construction of illumina sequencing libraries. Cold Spring Harbor protocols. 2013;2013(9):820–834. doi: 10.1101/pdb.prot074187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P, Thomas C, Harrison S, Revers F, Maule A. A cysteine-rich plant protein potentiates Potyvirus movement through an interaction with the virus genome-linked protein VPg. J Virol. 2004;78(5):2301–2309. doi: 10.1128/JVI.78.5.2301-2309.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre F, Montarry J, Coville J, Senoussi R, Simon V, Moury B. Modelling the Evolutionary Dynamics of Viruses within Their Hosts: A Case Study Using High-Throughput Sequencing. PLoS Pathog. 2012;8(4) doi: 10.1371/journal.ppat.1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre F, Moury B, Johansen EI, Simon V, Jacquemond M, Senoussi R. Narrow Bottlenecks Affect Pea Seedborne Mosaic Virus Populations during Vertical Seed Transmission but not during Leaf Colonization. PLoS Pathog. 2014;10(1):e1003833. doi: 10.1371/journal.ppat.1003833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuer R, Boone JD, Netski D, Morzunov SP, St Jeor SC. Temporal and spatial analysis of Sin Nombre virus quasispecies in naturally infected rodents. J Virol. 1999;73(11):9544–9554. doi: 10.1128/jvi.73.11.9544-9554.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French R, Stenger DC. Evolution of wheat streak mosaic virus: Dynamics of population growth within plants may explain limited variation. Annual Review of Phytopathol. 2003;41:199–214. doi: 10.1146/annurev.phyto.41.052002.095559. [DOI] [PubMed] [Google Scholar]
- Gal-On A. Zucchini yellow mosaic virus: insect transmission and pathogenicity - the tails of two proteins. Mol Plant Pathol. 2007;8(2):139–150. doi: 10.1111/j.1364-3703.2007.00381.x. [DOI] [PubMed] [Google Scholar]
- Garcia-Arenal F, Fraile A, Malpica JM. Variability and genetic structure of plant virus populations. Annual Review of Phytopathol. 2001;39:157–186. doi: 10.1146/annurev.phyto.39.1.157. [DOI] [PubMed] [Google Scholar]
- Gómez de Cedrón M, Osaba L, López L, García JA. Genetic analysis of the function of the plum pox virus CI RNA helicase in virus movement. Virus Res. 2006;116(1–2):136–145. doi: 10.1016/j.virusres.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Jara P, Fraile A, Canto T, Garcia-Arenal F. The multiplicity of infection of a plant virus varies during colonization of its eukaryotic host. J Virol. 2009;83(15):7487–7494. doi: 10.1128/JVI.00636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez S, Yvon M, Pirolles E, Garzo E, Fereres A, Michalakis Y, Blanc S. Circulating Virus Load Determines the Size of Bottlenecks in Viral Populations Progressing within a Host. PLoS Pathog. 2012;8(11):e1003009. doi: 10.1371/journal.ppat.1003009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes EC. The evolutionary genetics of emerging viruses. Annu.Rev.Ecol.Evol.Syst. 2009;40:353–372. [Google Scholar]
- Heinlein M, Epel BL, Padgett HS, Beachy RN. Interaction of tobamovirus movement proteins with the plant cytoskeleton. Science. 1995;270(5244):1983–1985. doi: 10.1126/science.270.5244.1983. [DOI] [PubMed] [Google Scholar]
- Jerzak GV, Brown I, Shi PY, Kramer LD, Ebel GD. Genetic diversity and purifying selection in West Nile virus populations are maintained during host switching. Virology. 2008;374(2):256–260. doi: 10.1016/j.virol.2008.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jridi C, Martin JF, Marie-Jeanne V, Labonne G, Blanc S. Distinct viral Populations differentiate and evolve independently in a single perennial host plant. J Virol. 2006;80(5):2349–2357. doi: 10.1128/JVI.80.5.2349-2357.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katis NI, Tsitsipis JA, Lykouressis DP, Papapanayotou A, Margaritopoulos JT, Kokinis GM, Perdikis DC, Manoussopoulos IN. Transmission of Zucchini yellow mosaic virus by colonizing and non-colonizing aphids in Greece and new aphid species vectors of the virus. Journal of Phytopathol. 2006;154(5):293–302. [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat protoc. 2009;4(3):363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Koboldt D, Zhang Q, Larson D, Shen D, McLellan M, Lin L, Miller C, Mardis E, Ding L, Wilson R. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012 doi: 10.1101/gr.129684.111. DOI: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lech WJ, Wang G, Yang YL, Chee Y, Dorman K, McCrae D, Lazzeroni LC, Erickson JW, Sinsheimer JS, Kaplan AH. In vivo sequence diversity of the protease of human immunodeficiency virus type 1: presence of protease inhibitor-resistant variants in untreated subjects. J Virol. 1996;70(3):2038–2043. doi: 10.1128/jvi.70.3.2038-2043.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 1000 Genome Project Data Processing Subgroup. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Roossinck MJ. Genetic Bottlenecks Reduce Population Variation in an Experimental RNA Virus Population. J Virol. 2004;78(19):10582–10587. doi: 10.1128/JVI.78.19.10582-10587.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maule AJ, Wang D. Seed transmission of plant viruses: a lesson in biological complexity. Trends Microbiol. 1996;4(4):153–158. doi: 10.1016/0966-842x(96)10016-0. [DOI] [PubMed] [Google Scholar]
- Miyashita S, Kishino H. Estimation of the Size of Genetic Bottlenecks in Cell-to-Cell Movement of Soil-Borne Wheat Mosaic Virus and the Possible Role of the Bottlenecks in Speeding Up Selection of Variations in trans-Acting Genes or Elements. J Virol. 2010;84(4):1828–1837. doi: 10.1128/JVI.01890-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehl A, Heinlein M. Cellular pathways for viral transport through plasmodesmata. Protoplasma. 2011;248(1):75–99. doi: 10.1007/s00709-010-0246-1. [DOI] [PubMed] [Google Scholar]
- Oparka KJ, Prior DA, Santa Cruz S, Padgett HS, Beachy RN. Gating of epidermal plasmodesmata is restricted to the leading edge of expanding infection sites of tobacco mosaic virus (TMV). Plant J. 1997;12(4):781–789. doi: 10.1046/j.1365-313x.1997.12040781.x. [DOI] [PubMed] [Google Scholar]
- Roberts IM, Wang D, Findlay K, Maule AJ. Ultrastructural and Temporal Observations of the Potyvirus Cylindrical Inclusions (CIs) Show That the CI Protein Acts Transiently in Aiding Virus Movement. Virology. 1998;245(1):173–181. doi: 10.1006/viro.1998.9132. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Cerezo E, Findlay K, Shaw JG, Lomonossoff GP, Qiu SG, Linstead P, Shanks M, Risco C. The coat and cylindrical inclusion proteins of a potyvirus are associated with connections between plant cells. Virology. 1997;236(2):296–306. doi: 10.1006/viro.1997.8736. [DOI] [PubMed] [Google Scholar]
- Rojas MR, Zerbini FM, Allison RF, Gilbertson RL, Lucas WJ. Capsid protein and helper component proteinase function as potyvirus cell-to-cell movement proteins. Virology. 1997;237(2):283–295. doi: 10.1006/viro.1997.8777. [DOI] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protocols. 2010;5(4):725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybicki EP, Pietersen G. Plant virus disease problems in the developing world. Adv Virus Res. 1999;53:127–175. doi: 10.1016/s0065-3527(08)60346-2. [DOI] [PubMed] [Google Scholar]
- Sacristan S, Malpica JM, Fraile A, Garcia-Arenal F. Estimation of population bottlenecks during systemic movement of Tobacco mosaic virus in tobacco plants. J Virol. 2003;77(18):9906–9911. doi: 10.1128/JVI.77.18.9906-9911.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan R, Moya A, Elena SF. The distribution of fitness effects caused by single-nucleotide substitutions in an RNA virus. Proc Natl Acad Sci U S A. 2004;101(22):8396–8401. doi: 10.1073/pnas.0400146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla DD, Frenkel MJ, Ward CW. Structure and Function of the Potyvirus Genome with Special Reference to the Coat Protein Coding Region. Can. J. Plant Pathol.-Rev. Can. Phytopathol. 1991;13(2):178–191. [Google Scholar]
- Simmons HE, Dunham JP, Zinn KE, Munkvold GP, Holmes EC, Stephenson AG. Zucchini yellow mosaic virus (ZYMV, Potyvirus): Vertical transmission, seed infection and cryptic infections. Virus Research. 2013;176:259–264. doi: 10.1016/j.virusres.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons HE, Dunham JP, Stack JC, Dickins BJA, Pagan I, Holmes EC, Stephenson AG. Deep sequencing reveals persistence of intra- and inter-host genetic diversity in natural and greenhouse populations of zucchini yellow mosaic virus. J Gen Virol. 2012;93:1831–1840. doi: 10.1099/vir.0.042622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons HE, Holmes EC, Gildow FE, Bothe-Goralczyk MA, Stephenson AG. Experimental Verification of Seed Transmission of Zucchini yellow mosaic virus. Plant Dis. 2011;95(6):751–754. doi: 10.1094/PDIS-11-10-0843. [DOI] [PubMed] [Google Scholar]
- Tatineni S, Kovacs F, French R. Wheat streak mosaic virus infects systemically despite extensive coat protein deletions: Identification of virion assembly and cell-to-cell movement determinants. J Virol. 2014;88(2):1366–1380. doi: 10.1128/JVI.02737-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tromas N, Zwart MP, Lafforgue G, Elena SF. Within-host spatiotemporal dynamics of plant virus infection at the cellular level. Plos Genetics. 2014;10(2):e1004186. doi: 10.1371/journal.pgen.1004186. doi:10.1371/journal.pgen.1004186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urcuqui-Inchima S, Haenni AL, Bernardi F. Potyvirus proteins: a wealth of functions. Virus Res. 2001;74(1-2):157–175. doi: 10.1016/s0168-1702(01)00220-9. [DOI] [PubMed] [Google Scholar]
- Ward CW, Shukla DD. Taxonomy of potyviruses: current problems and some solutions. Intervirology. 1991;32(5):269–296. doi: 10.1159/000150211. [DOI] [PubMed] [Google Scholar]
- Wei T, Zhang C, Hong J, Xiong R, Kasschau KD, Zhou X, Carrington JC, Wang A. Formation of Complexes at Plasmodesmata for Potyvirus Intercellular Movement Is Mediated by the Viral Protein P3N-PIPO. PLoS Pathog. 2010;6(6):e1000962. doi: 10.1371/journal.ppat.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart MP, Daros JA, Elena SF. One Is Enough: In Vivo Effective Population Size Is Dose-Dependent for a Plant RNA Virus. PLoS Pathog. 2011;7(7) doi: 10.1371/journal.ppat.1002122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart MP, Daros JA, Elena SF. Effects of Potyvirus Effective Population Size in Inoculated Leaves on Viral Accumulation and the Onset of Symptoms. J Virol. 2012;86(18):9737–9747. doi: 10.1128/JVI.00909-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.