Abstract
The CRISPR-associated protein Cas9 is an RNA-guided endonuclease that cleaves double-stranded DNAs bearing sequences complementary to a 20-nucleotide segment in the guide RNA1,2. Cas9 has emerged as a versatile molecular tool for genome editing and gene expression control3. RNA-guided DNA recognition and cleavage strictly require the presence of a protospacer adjacent motif (PAM) in the target DNA1,4-6. Here, we report a crystal structure of Streptococcus pyogenes Cas9 complexed with a single-molecule guide RNA (sgRNA) and a target DNA containing a canonical 5′-NGG-3′ PAM. The structure reveals that the PAM motif resides in a base-paired DNA duplex. The non-complementary strand GG dinucleotide is read out via major groove interactions with conserved arginine residues from the C-terminal domain of Cas9. Interactions with the minor groove of the PAM duplex and the phosphodiester group at the +1 position in the target DNA strand contribute to local strand separation of the target DNA duplex immediately upstream of the PAM. These observations suggest a mechanism for PAM-dependent target DNA melting and RNA-DNA hybrid formation. Furthermore, this study establishes a framework for the rational engineering of Cas9 enzymes with novel PAM specificities.
In type II CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)-Cas (CRISPR-associated) systems, the endonuclease Cas9 associates with a dual-RNA guide structure consisting of a CRISPR RNA (crRNA) and a trans-activating crRNA (tracrRNA) to cleave double-stranded DNAs using its HNH and RuvC nuclease domains1,2,7. Cas9 has been exploited in numerous gene targeting applications, in which its sequence specificity is programmed by either dual-RNA guides or chimeric single-molecule guide RNAs (sgRNAs)8-19. PAM recognition is a critical aspect of Cas9-mediated DNA targeting, being a prerequisite for ATP-independent strand separation and guide RNA-target DNA heteroduplex formation6. Recent crystal structures and electron microscopic reconstructions of Cas9 and its RNA- and DNA-bound complexes revealed that Cas9 undergoes a dramatic RNA-induced conformational rearrangement that facilitates target DNA binding20,21. Although two tryptophan residues have been implicated in PAM binding20, how PAM recognition occurs at the molecular level remains unclear.
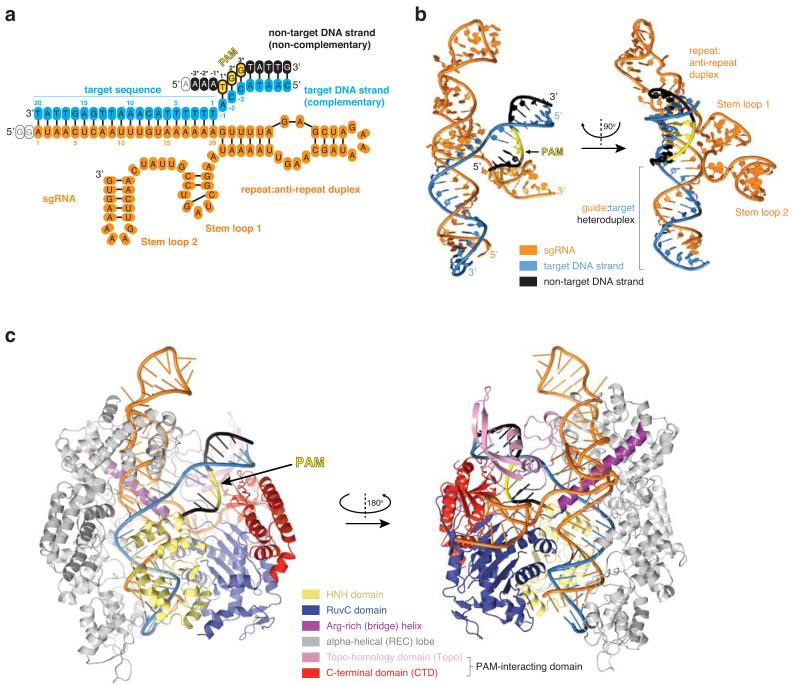
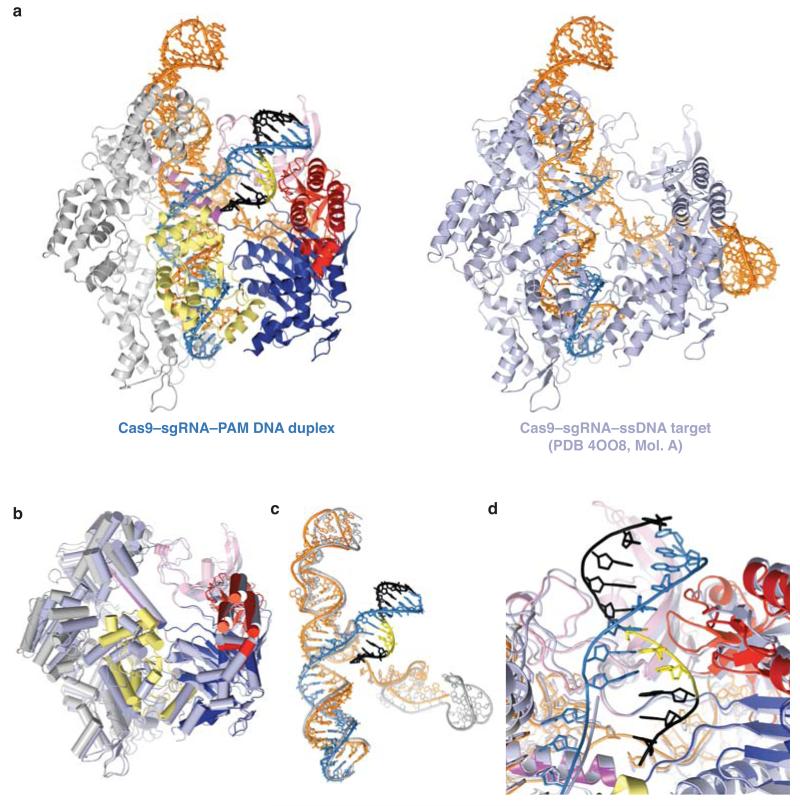
To illuminate the molecular mechanism of PAM recognition in Cas9, we determined the crystal structure of Streptococcus pyogenes Cas9 in complex with an 83-nucleotide sgRNA and a partially duplexed target DNA containing a 5′-TGG-3′ PAM sequence (Fig. 1a, Extended Data Table 1). Due to an inactivating mutation (H840A) in the Cas9 HNH nuclease domain, the structure reveals an intact target (complementary) DNA strand, while the non-target (non-complementary) DNA strand is captured as cleaved product that has dissociated from the RuvC domain active site (Fig. 1 a-c). In the complex, the bound nucleic acids are enclosed by the nuclease and helical recognition lobes of Cas9 and form a four-way junction that straddles the arginine-rich bridge helix (Fig. 1b, c). The entire PAM-containing region of the target DNA (target strand nucleotides −1 to −8 and non-target strand nucleotides +1* to +8*) is base-paired. Strand separation occurs only at the first base pair of the target sequence (the +1 position). Here, the target strand exhibits a pronounced kink as it hybridizes with the sgRNA. The PAM duplex is nestled in a positively charged groove between the Topo-homology and C-terminal domains (collectively referred to as the PAM-interacting domain21) (Fig. 2a, Extended Data Fig. 1). Comparison with the crystal structure of the Cas9-sgRNA complex bound to a single-stranded DNA target21 reveals a slight tightening of an otherwise pre-structured PAM binding cleft upon PAM duplex binding (Extended Data Fig. 2).
Figure 1. Crystal structure of Cas9 in complex with a sgRNA and a PAM-containing target DNA.
a, Schematic diagram of guide and target nucleic acids. Guide RNA is coloured orange, target DNA strand in light blue, and non-target DNA strand in black. The 5′-NGG-3′ PAM trinucleotide in the non-target strand is highlighted in yellow. The colour code is used throughout. Empty ovals denote nucleotides not observed in electron density. b, Orthogonal views of the sgRNA–target DNA four way junction. c, Front and rear views of the Cas9–sgRNA–DNA complex.
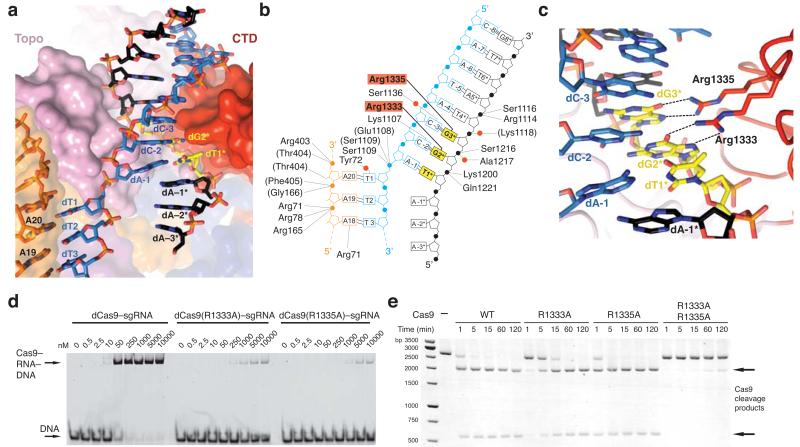
Figure 2. The GG dinucleotide of the PAM is read out by major groove interactions.
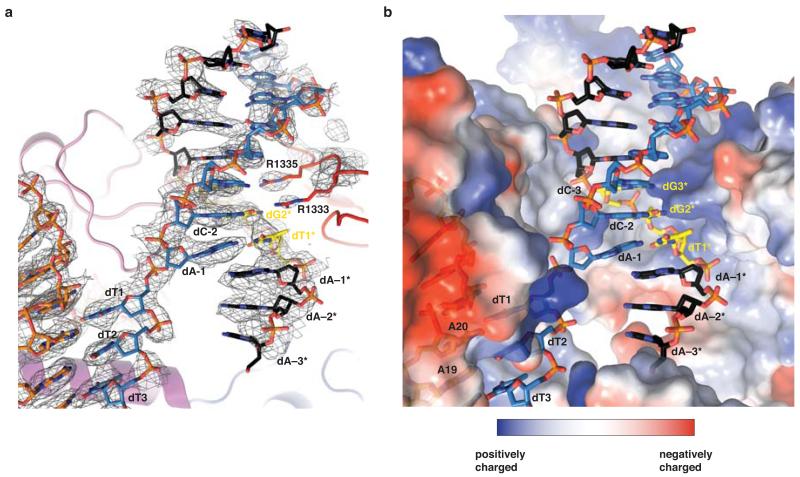
a, Zoomed-in view of the PAM binding region in Cas9. b, Schematic of Cas9 interactions with the PAM duplex. Red circles denote bridging water molecules. c, Detailed view of the major groove. Sequence-specific hydrogen-bonding interactions with the GG PAM dinucleotide are indicated with dashed lines. d, Electrophoretic mobility shift assay using catalytically inactive dCas9–sgRNA complexes and fluorophore-labelled target DNA duplex. e, Endonuclease activity assay of wild type (WT) and mutant Cas9 proteins using a linearized plasmid DNA containing a target sequence fully complementary to the sgRNA in Fig. 1a. 2104 and 598 bp bands correspond to Cas9 cleavage products.
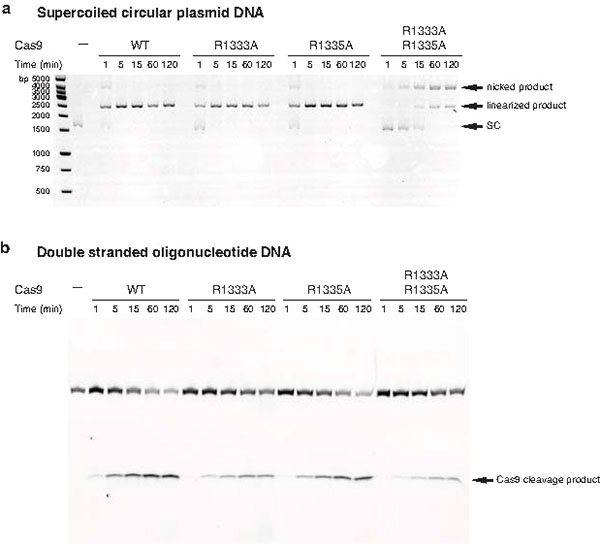
The deoxyribose-phosphate backbone of the non-target DNA strand is engaged in numerous ionic and hydrogen-bonding interactions (Fig. 2b). Conserved tryptophan residues Trp476 and Trp1126, previously implicated in PAM recognition by crosslinking experiments20, are not in direct contact with the PAM, suggesting that the crosslinks may have originated from a transient intermediate in the PAM recognition mechanism, or from non-specifically bound DNA. Instead, the guanine nucleobases of dG2* and dG3* in the non-target strand are read out in the major groove by base-specific hydrogen-bonding interactions with Arg1333 and Arg1335, respectively, provided by a beta-hairpin from the C-terminal domain of Cas9 (Fig. 2c). The target strand nucleotides complementary to the PAM are not recognized by major groove interactions (Fig. 2b,c), rationalizing previous observations that Cas9-mediated DNA cleavage requires the 5′-NGG-3′ trinucleotide in the non-target strand, but not its target strand complement1,6. The lack of interactions with the target strand backbone also explains why mismatches in the PAM are tolerated provided that a GG dinucleotide is present in the non-target strand1. In agreement with the observed role of the arginine residues in PAM recognition, substitutions of Arg1333 or Arg1335 with alanine residues resulted in substantially reduced target dsDNA binding in vitro (Fig. 2d). Furthermore, alanine substitution of both Arg1333 and Arg1335 nearly abolished cleavage of linearized plasmid DNA, and substantially reduced cleavage of supercoiled circular plasmid DNA and short dsDNA oligonucleotides in vitro (Fig. 2e and Extended Data Figure 3). Individual arginine substitutions yielded modest reductions of cleavage activity (Fig. 2e and Extended Data Figure 3).
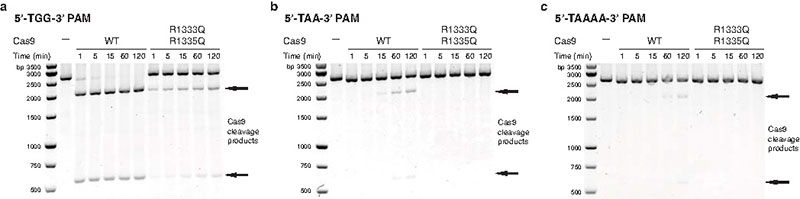
The Cas9 sequence motif containing the PAM-interacting arginine residues (D1332RKRY1336) is conserved in other type II-A Cas9 proteins known to recognize 5′-NGG-3′ PAMs (Extended Data Fig. 4 and Supplementary Information). Similar arginine-containing motifs are found in Cas9 from Francisella novicida (S1608RYPD1612) and from Streptococcus thermophilus CRISPR3 locus (P1350RYRDY1356), which recognize 5′-NG-3′ and 5′-NGGNG-3′ PAMs, respectively, but are notably absent from type II-C Cas9 proteins that are known to recognize distinct PAM sequences4,22,23 (Extended Data Fig. 4). Whereas arginine residues are commonly utilized by DNA binding proteins to recognize guanines, major groove read-out of adenines typically involves glutamine residues24. Interestingly, a Cas9 ortholog from Lactobacillus buchneri, predicted to recognize a 5′-NAAAA-3′ PAM25, contains glutamine residues (Q1338LQ1340) at the positions equivalent to Arg1333 and Arg1335 in S. pyogenes Cas9. Together, these observations suggest that at least in a subset of Cas9 proteins, PAM binding may be governed by a major groove base recognition code. Substitutions of Arg1333 and Arg1335 in S. pyogenes Cas9 with glutamine residues did not produce a specificity switch towards A-rich PAMs (Extended Data Figure 5). Reprogramming PAM specificity might thus require more extensive remodelling of the PAM-interacting motif by directed evolution and/or computational design, as has been done previously for homing endonucleases26,27.
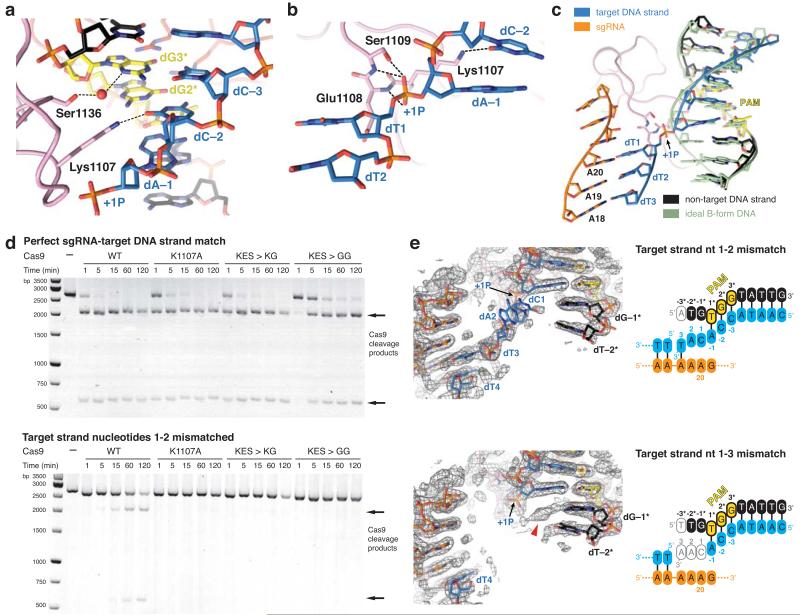
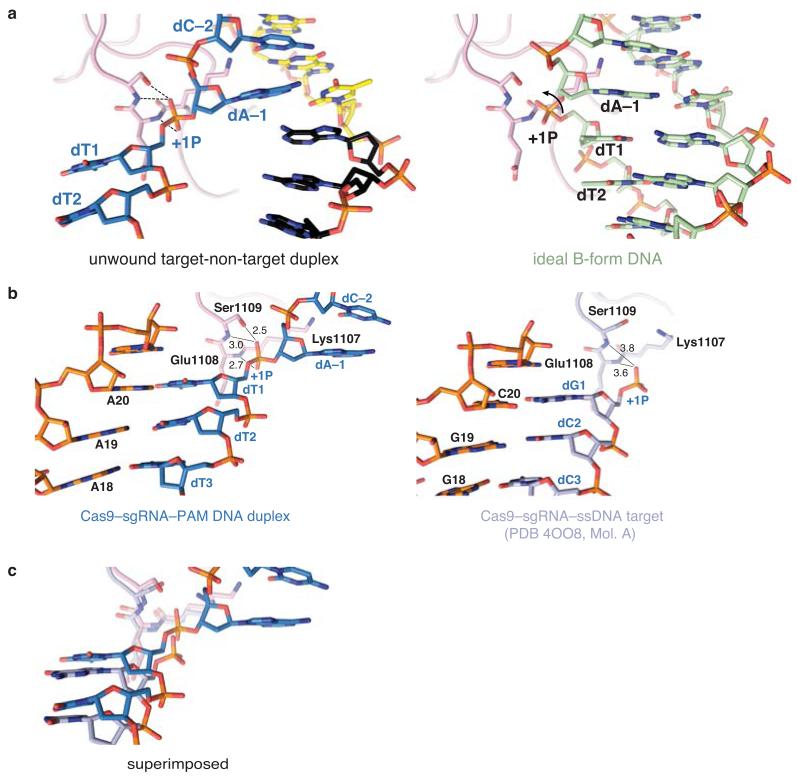
The PAM-interacting domain of Cas9 makes further contacts with the minor groove of the PAM duplex (Fig. 3a). Ser1136 interacts with the non-target strand dG3* through a water-mediated hydrogen bond, while Lys1107 contacts dC−2 of the target strand (Fig. 3a). This interaction enforces a pyrimidine at this position, explaining why 5′-NAG-3′ PAMs are weakly permissive for S. pyogenes Cas919,28,29. The minor groove interactions with the PAM duplex position the target DNA strand for base pairing with the guide RNA. Downstream of Lys1107, residues Glu1108–Ser1109 interact with the phosphodiester group linking dA−1 and dT1 in the target DNA strand (the +1 phosphate). The non-bridging phosphate oxygen atoms are hydrogen bonded to the backbone amide groups of Glu1108 and Ser1109, and to the side chain of Ser1109 (Fig. 3b). Due to its interaction with the Lys1107-Ser1109 loop (the “phosphate lock” loop), the +1 phosphate group is rotated (Fig. 3c, Extended Data Figure 6), which coincides with a distortion in the target DNA strand that allows the nucleobase of dT1 to base pair with A20 of the guide RNA. Furthermore, comparison with the structure of Cas9-sgRNA bound to a single-stranded DNA21 suggests that interaction between the +1 phosphate and the loop is PAM-dependent (Extended Data Fig. 6).
Figure 3. Interactions with the +1 phosphodiester group orient the target strand for guide RNA binding.
a, Detailed view of the minor groove of the PAM region. b, Hydrogen-bonding interactions (dashed lines) of the +1 phosphate (+1P) with the Lys1107-Ser1109 (phosphate lock) loop. c, Superposition of the unwound target DNA strand with an ideal B-form DNA duplex (green) d, Endonuclease activity assays using linearized plasmid DNA containing a fully complementary target sequence (top) or a target sequence mismatched to the sgRNA at positions 1–2 (bottom). e, Crystal structures of dCas9-sgRNA bound to DNA substrates containing mismatches to the sgRNA at positions 1–2 (top) and 1–3 (bottom), overlaid with refined 2mFo-DFc electron density maps (grey mesh, contoured at 1σ). The sgRNA is identical to that in Fig. 1a. In both structures, the target DNA strand is provided in two fragments, as indicated in the schematics. Residual electron density corresponding to the +1 base pair is indicated with a red arrowhead.
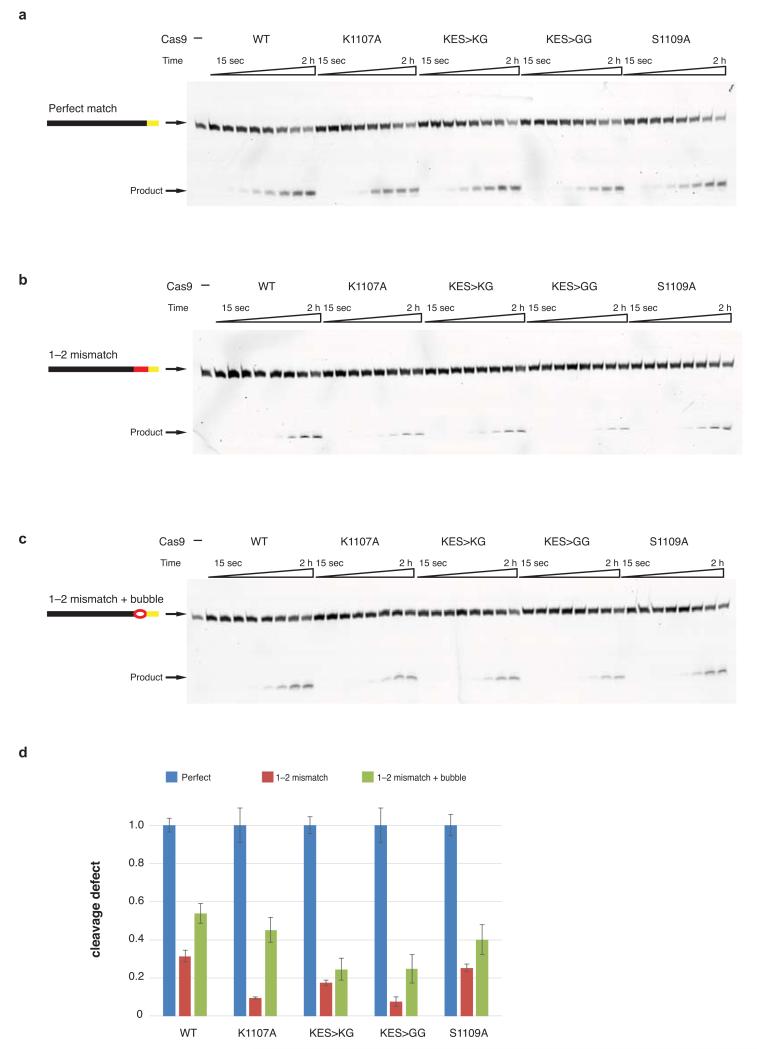
Previous biochemical studies indicated that PAM recognition is concomitant with local destabilization of the adjacent sequence and directional target DNA unwinding from the PAM-proximal end6. Our structural observations suggest that the interaction between the +1 phosphate and the phosphate lock loop might stabilize target DNA immediately upstream of the PAM in an unwound conformation, thereby linking PAM recognition with local strand separation. In agreements with this hypothesis, alanine substitution of Lys1107 or replacement of the Lys1107-Ser1109 loop with a Lys-Gly or Gly-Gly dipeptide yielded Cas9 proteins with modestly reduced cleavage activities towards linearized plasmid DNA containing a perfectly complementary sequence, but almost no activity towards DNA containing mismatches to the guide RNA at positions 1-2 (Fig. 3d). Moreover, the phosphate lock loop mutations also disproportionately impaired cleavage of an oligonucleotide duplex containing the same mismatch, but the defect was partially relieved with a duplex in which the mismatched nucleotides were themselves unpaired (Extended Data Fig. 7).
To provide additional support for the hypothesis, we determined two crystal structures of Cas9(D10A/H840A)-sgRNA bound to DNAs containing mismatches to the guide RNA (Fig. 3e, Extended Data Table 1). To discount the possibility that duplex melting in these target DNAs is driven from the unpaired PAM-distal end, the target strand was supplied in two fragments and interrupted by a gap at the scissile phosphate (+4) position. The structure of the complex containing mismatches at positions 1–2 reveals a fully melted duplex, with nucleotides +1 and +2 unpaired, and nucleotide dT3 base paired to A18 of the sgRNA. In the complex containing mismatches at positions 1–3, the target strand upstream of the PAM is disordered and we observe only residual electron density for the +1 base pair that cannot be modelled with full occupancy (Fig. 3e), suggesting that the DNA is unpaired in a substantial fraction of molecules in the crystal. Together, these structures reveal that even in the absence of compensatory base pairing to the guide RNA, target DNA binding by Cas9–RNA results in local strand separation immediately upstream of the PAM. Importantly, the interaction of the +1 phosphate with the phosphate lock loop is maintained in both structures, supporting the hypothesis that the loop contributes to stabilizing the target DNA strand in the unwound state.
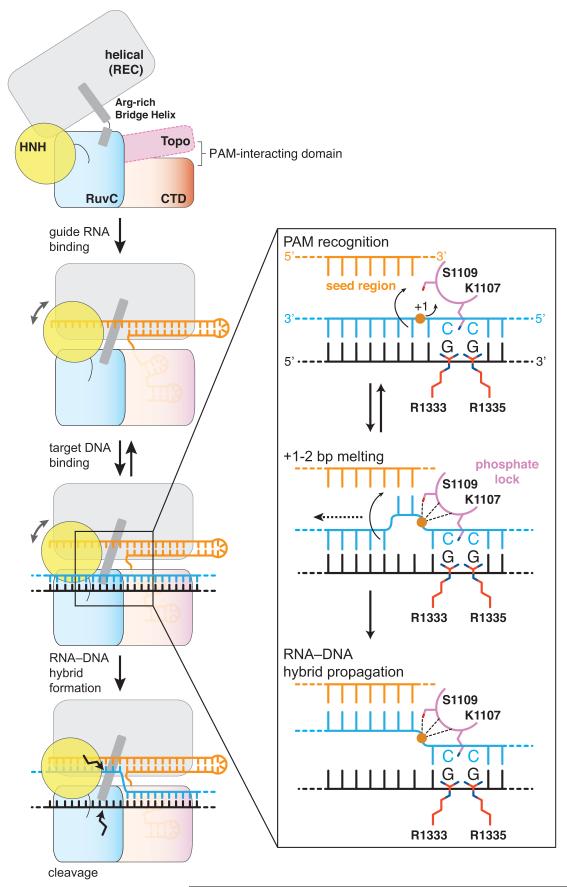
In this study, we highlight the central importance of PAM recognition in Cas9 function, both as a critical determinant of initial target DNA binding and as a licensing element in subsequent strand separation and guide RNA–target DNA hybridization. Based on our structural and biochemical observations, we propose a model for PAM-dependent target dsDNA recognition and unwinding (Fig. 4). Sequence-specific PAM readout by Arg1333 and Arg1335 in Cas9 positions the DNA duplex such that the +1 phosphate group of the target strand interacts with the phosphate lock loop. This promotes local duplex melting, allowing the Cas9–RNA complex to probe the identity of the nucleotides immediately upstream of the PAM. Base pairing between the seed region of the guide RNA and the target DNA strand subsequently drives further stepwise destabilization of the target DNA duplex and directional formation of the guide RNA-target DNA heteroduplex.
Figure 4. Model for PAM-dependent target DNA unwinding and recognition by Cas9.
Guide RNA binding to Cas9 results in the formation of the PAM binding site. Cas9-RNA engages the PAM GG dinucleotide using Arg1333 and Arg1335, and positions the target DNA duplex such that the +1 phosphate (orange circle) interacts with the phosphate lock loop, resulting in local strand separation immediately upstream of the PAM. Base-pairing between displaced target DNA strand and the seed region of the guide RNA promotes further stepwise strand displacement and propagation of the guide-target heteroduplex.
Methods
In vitro transcription and purification of sgRNA
The sequences of RNA oligonucleotides used in the study are provided in Extended Data Table 2. sgRNAs were prepared by in vitro transcription using recombinant T7 RNA polymerase as described6, except that for sgRNA 1, a fully double-stranded DNA template was used instead. Transcription template for sgRNA 1 was generated by PCR using the following oligonucleotides: PCR template 5′-CACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAACTTTTTTACAAATTGAGTTATCCTATAGTGAGTCGTATTA-3′; forward primer 5′-TAATACGACTCACTATA-3′; reverse primer 5′-CACTTTTTCAAGTTGA-3′. sgRNA 2 was transcribed directly from single-stranded oligonucleotide template (5′-CACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAG CTCTAAAACGCGTCTCATCTTTATGCGTCCCTATAGTGAGTCGTATTA-3′), that was hybridized to the forward primer listed above. Transcribed RNAs were purified by gel electrophoresis on an 8% denaturing (7 M urea) polyacrylamide gel and subsequently ethanol precipitated.
DNA oligonucleotides
The sequences of DNA oligonucleotides used in the study are provided in Extended Data Table 2. All DNA oligonucleotides were synthesized by Microsynth AG (Balgach, Switzerland). For crystallization, target strand (5′ CAATACCATTTTTTACAAATTGAGTTAT-3′) and non-target strand (5′-AAAATGGTATTG-3′) were used without further purification. Prior to complex formation, the DNA oligonucleotides were mixed in a 1:1 molar ratio (final concentration 100 μM) and hybridized by heating to 75 °C for 5 min, followed by slow cooling to room temperature. The target strand oligonucleotides used for cleavage assays contained a 5′-linked ATTO532 fluorophore (ATTO532-5′-GCGCAATACCATTTTTTACAAATTGAGTTAT-3′ and ATTO532-5′-GCGCAATACCAAATTTTACAAATTGAGTTAT-3′) and were PAGE-purified before use. The non-target strand oligonucleotides (5′-ATAACTCAATTTGTAAAAAATGGTATTG-3′ and 5′-ATAACTCAATTTGTAAAATTTGGTATTG-3′) were used without further purification. For electrophoretic mobility shift assays, the non-target strand oligonucleotide contained a 3′-linked ATTO532 fluorophore (5′-ATAACTCAATTTGTAAAAAATGGTATTGCGC-3′-ATTO532) and was PAGE-purified before use.
Cas9 expression, purification, and Cas9-sgRNA-DNA complex reconstitution
Streptococcus pyogenes Cas9 was expressed and purified essentially as described1,20. Point mutations were introduced by inverse PCR and verified by DNA sequencing and mutant proteins were purified as for wild-type Cas9. Briefly, Cas9 was expressed in Escherichia coli BL21 (DE3) Rosetta 2 (Novagen) fused to an N-terminal fusion protein containing a hexahistidine affinity tag, the maltose binding protein (MBP) polypeptide sequence, and the tobacco etch virus (TEV) protease cleavage site. Cells were lysed in 20 mM Tris pH 8.0, 250 mM NaCl, 5 mM Imidazole pH 8.0. Clarified lysate was applied to a 10 ml Ni-NTA (Sigma Aldrich) affinity column. The column was washed with 20 mM Tris pH 8.0, 250 mM NaCl, 10 mM Imidazole pH 8.0, and bound protein was eluted by increasing imidazole concentration to 250 mM. Eluted protein was dialysed against 20 mM HEPES pH 7.5, 150 mM KCl, 10 % glycerol, 1 mM dithiothreitol (DTT), 1 mM ethylenediaminetetraacetic acid (EDTA) overnight at 4 °C in the presence of TEV protease to remove the His6-MBP affinity tag. Cleaved protein was further purified by cation exchange chromatography (HiTrap SP FF, GE Healthcare), eluting with a linear gradient of 0.1–1.0 M KCl. For purification of Cas9-sgRNA complexes, purified Cas9 and in vitro transcribed sgRNA were mixed in a 1:1 molar ratio, concentrated to 6-10 mg ml−1 in a 50,000 MWCO centrifugal filter (Merck Millipore), applied to a Superdex 200 16/600 column (GE Healthcare) and eluted with 20 mM HEPES pH 7.5, 500 mM KCl. Reconstitution of Cas9-sgRNA-DNA complex was carried out by mixing purified Cas9 and in vitro transcribed sgRNA in a 1:1 molar ratio. The sample was slowly exchanged to a buffer containing 20 mM HEPES pH 7.5, 500 mM KCl, 5 mM MgCl2 during concentration in a 50,000 MWCO centrifugal filter. At a final concentration of 4-6 mg ml−1, pre-hybridized target DNA was added in 1.5-fold molar excess. The complex was applied to a Superdex 200 16/600 column and eluted with 20 mM HEPES pH 7.5, 500 mM KCl, 5 mM MgCl2. Purified complex was concentrated to 4-8 mg ml−1 , flash frozen in liquid nitrogen, and stored at −80 °C. For crystallization of complexes bound to mismatch-containing DNAs, purified Cas9-sgRNA complex was diluted with 20 mM HEPES pH 7.5, 500 mM KCl, 5 mM MgCl2 to a final concentration of 5 mg ml−1 and was supplemented with DNA oligonucleotides in a 1:2 molar ratio. Selenomethionine (SeMet)-substituted Cas9 was expressed and purified as for native protein, with the following modifications. The expression was carried out in M9 minimal media supplemented with 1 μg ml−1 biotin and 1 μg ml−1 thiamine. At OD600 of 0.8, the following amino acids were added to allow SeMet incorporation: 50 mg l−1 Leu, Ile, Val, 100 mg l−1 Phe, Lys, Thr, 75 mg l−1 SeMet. After 30 min, the temperature was reduced to 18 °C and 200 μM isopropyl-β-D-thiogalactopyranoside (IPTG) was added for induction. For purification, all buffers except for the final size exclusion buffer were supplemented with 1 mM DTT.
Complex crystallization and structure determination
Cas9-sgRNA-DNA crystals were grown at 20 °C using the hanging drop vapor diffusion method. The drops were composed of equal volumes (1 μl + 1 μl) of purified complex (diluted to 1.25–2 mg ml−1 in 20 mM HEPES pH 7.5, 250 mM KCl, 5 mM MgCl2) and reservoir solution (0.1 M Tris-acetate, pH 8.5, 200-400 mM KSCN, 14-18 % PEG 3350). Iterative microseeding was used to improve crystal nucleation, growth and morphology. For cryoprotection, crystals were transferred into 0.1 M Tris-acetate, pH 8.5, 200 mM KSCN, 30 % PEG 3350, 10 % ethylene glycol, and flash-cooled in liquid nitrogen. Diffraction data were measured at beamlines PXI and PXIII of the Swiss Light Source (Paul Scherer Institute, Villigen, Switzerland) and processed using XDS30. The crystals belonged to space group C2 and contained one complex in the asymmetric unit. Native data extended to a resolution of 2.59 Å. Data collection statistics are summarized in Extended Data Table 1. Phases were obtained from a single-wavelength anomalous diffraction (SAD) experiment using complex crystals containing SeMet-substituted Cas9 and measured at Se K-edge wavelength (0.97965 Å). Three data sets were measured by exposing different parts of the same crystal, rotating the crystal through 360° in each dataset. Selenium sites were located using the Hybrid Substructure Search (HySS) module of the Phenix package31. Additional phasing information came from a SAD experiment carried out with complex crystals soaked with 5 mM iridium hexamine chloride for 2 h and measured at the Ir L-III edge (1.10501 Å). The native and SAD data sets were combined for substructure refinement and phase calculations using the MIRAS procedure in AutoSHARP32, yielding readily interpretable electron density maps. Fragments of apo-Cas9 structure20 were docked using MOLREP33. Model building was completed in COOT34. The atomic model was refined using Phenix.refine35. The final model includes Cas9 residues 4–710, 719–764, 776–1012, 1030–1050, 1059–1241, 1253–1364; nucleotides 1–81 of the sgRNA; nucleotides (−8)–20 of the target DNA strand and nucleotides (−3)–8 of the non-target DNA strand. The structures of dCas9–sgRNA–mismatch DNA complexes were solved by molecular replacement using the Phaser module of the Phenix package31 using the atomic structures of Cas9 and sgRNA as separate search models and omitting DNA from the initial search.
Endonuclease activity assays
Plasmid cleavage assays were carried out using plasmids pMJ879, pMJ891, pMJ992, pMJ993, and pMJ994 (Extended Data Table 2), as described previously1,6. Equimolar quantities of Cas9 and sgRNA (final concentration of 1.5 μM) were pre-incubated in 20 mM HEPES pH 7.5, 100 mM KCl, 5% glycerol, 1 mM DTT, 0.5 mM EDTA, 2 mM MgCl2 at room temperature for 5 min. 400 ng of circular or SspI-linearized plasmid were added and the cleavage reactions (40 μl total volume) were incubated at 37 °C for 2 h. 7 μl aliquots were taken at the indicated time points, quenched by addition of EDTA (50 mM final concentration) and treated with 14 μg Proteinase K for 30 min at room temperature. Cleavage products were resolved by gel electrophoresis on 1% agarose gel stained with GelRed (Biotium) and visualized using a Typhoon FLA 9500 scanner (GE Healthcare). Cleavage assays on double-stranded oligonucleotides were performed as described above with minor changes. Cas9 and sgRNA were used at a final concentration of 5 μM. Target strand oligonucleotides (400 nM) and non-target strand oligonucleotides (435 nM) were mixed to a final molar ratio of 2:3, annealed, and added to a final concentration of 20 nM and 30 nM to start the cleavage reaction (105 μl total volume). 12.5 μl aliquots were taken at the indicated time points and treated as described above. Cleavage products were resolved by electrophoresis on 16 % denaturing polyacrylamide gel at room temperature in 0.5x TBE running buffer and were detected using a Typhoon FLA 9500 scanner and quantified using ImageQuant software (GE Healthcare).
Electrophoretic mobility shift assays
Electrophoretic mobility shift assays were carried out using catalytically inactive Cas9 (D10A/H840A) protein (dCas9) and its point mutants. dCas9, sgRNA and DNA concentrations were determined with a NanoDrop spectrophotometer (Thermo Scientific) using the calculated extinction coefficients at 280 nm for Cas9 (120450 M−1 cm−1) and at 260 nm for sgRNA (829800 M−1 cm−1), target DNA (276100 M−1 cm−1) and non-target DNA (332100 M−1 cm−1). Target and non-target DNA strands were hybridized in a 1.5:1 molar ratio. The resulting target DNA duplex (final concentration of 10 nM) was titrated with increasing concentrations of dCas9 reconstituted with a two-fold molar excess of sgRNA in 20 mM HEPES pH 7.5, 100 mM KCl, 5 % glycerol, 1 mM DTT and 5 mM MgCl2 in a total volume of 40 μl. All binding reactions were incubated at 37 °C for 10 min and 20 μl were subsequently resolved on a native 8 % polyacrylamide gel at room temperature using 0.5x TBE running buffer supplemented with 1 mM MgCl2. Bound and unbound fractions were detected using a Typhoon FLA 9500 scanner.
Extended Data
Extended Data Figure 1. The PAM duplex binds in a positively charged cleft on the C-terminal PAM-interacting domain.
a, Zoomed-in view of the PAM binding site in Cas9. Nucleic acids are shown in stick format, coloured according to the scheme in Fig. 1a and overlaid with refined experimentally phased, solvent flattened electron density map (grey mesh, contoured at 1σ). b, PAM binding site in Cas9, shown in the same orientation as in panel a. The molecular surface of Cas9 is coloured according to electrostatic potential.
Extended Data Figure 2. The PAM binding site is preordered in the Cas9–RNA complex.
a, Comparison of the structures of Cas9–sgRNA bound to a PAM-containing target DNA duplex (left) and single-stranded DNA target21 (right). The target DNA strands of the complexes were superimposed using a least-squares algorithm in Coot34 and the complexes are shown in identical orientations. Bound nucleic acids are shown in stick format and coloured according to the scheme in Fig. 1a. b, Superimposed Cas9 molecules from the PAM-containing and ssDNA bound complexes. The colour scheme is the same as in panel a. In both complexes, the HNH domain is in an inactive conformation, with the active site located approximately 40 Å away from the scissile phosphate in the target DNA strand, suggesting that the domain undergoes a conformational rearrangement upon target strand cleavage. c, Superimposed nucleic acid ligands. sgRNA and target DNA from the single-stranded target complex are coloured grey. d, Detailed view of the PAM binding site in the superimposed complexes, indicating a slight tightening of the PAM binding cleft.
Extended Data Figure 3. Endonuclease activities of Cas9 proteins containing mutations in the PAM binding motif.
a, Endonuclease activity assay of WT and mutant Cas9 proteins using supercoiled circular (SC) plasmid DNA containing a target sequence fully complementary to the sgRNA in Fig. 1a. Nucleotide sequences of target sites are provided in Extended Data Table 2. b, Endonuclease activity assay of WT and mutant Cas9 proteins using an oligonucleotide duplex containing a target sequence fully complementary to the sgRNA in Fig. 1a.
Extended Data Figure 4. PAM binding motifs in Cas9 orthologs.
a, Cas9 orthologs with known PAM sequences4,19,22,23. The PAM of Cas9 from Lactobacillus buchneri has been inferred from known protospacer sequences, but has not been experimentally validated25. b, Alignment of the amino acid sequences of the major groove interacting regions of Cas9 orthologs. Primary sequences of type II-A Cas9 proteins from Streptococcus pyogenes (GI 15675041), Listeria innocua Clip 11262 (GI 16801805), Streptococcus mutans UA159 (GI 24379809), Streptococcus thermophilus LMD-9 (S.thermophilus A, GI 11662823; S.thermophilus B, GI 116627542), Lactobacillus buchneri NRRL B-30929 (GI 331702228), Treponema denticola ATCC 35405 (GI 42525843), type II-B Cas9 from Francisella novicida U112 (GI 118497352), and type II-C Cas9 proteins from Campylobacter jejuni subsp. Jejuni NCTC 11168 (GI 218563121), Pasteurella multocida subsp. multocida str. Pm70 (GI 218767588) and Neisseria meningitidis Zs491 (GI 15602992) were aligned using MAFFT36. Amino acids are coloured in shades of blue according to their degree of conservation. The red boxes denote amino acid residues inferred to be involved in PAM recognition in type II-A and type II-B Cas9 proteins based on the sequence alignment and the crystal structure of the Cas9-sgRNA-DNA complex elucidated in this study.
Extended Data Figure 5. Glutamine substitution of Arg1333 and Arg1335 in S. pyogenes Cas9.
a, Endonuclease activity assay of WT and mutant Cas9 proteins using a linearized plasmid containing a target sequence fully complementary to sgRNA 2 and a 5′-TGG-3′ PAM (Extended Data Table 2). 2014 and 598 bp bands correspond to Cas9 cleavage products. b, Endonuclease activity assay as in panel a using linearized plasmid containing a 5′-TAA-3′ PAM. c, Endonuclease activity assay as in panel a using linearized plasmid containing an extended 5′-TAAAA-3′ PAM.
Extended Data Figure 6. PAM-dependent interaction of the +1 phosphate with the phosphate lock loop.
a, Comparison of the bound target DNA (left) and the modelled B-form DNA (right). Docking of the ideal B-form duplex yields a steric clash with the phosphate lock loop. The arrow indicates the rotation of the +1 phosphate group (+1P) needed for interaction with the phosphate lock loop. b, Comparison of the phosphate lock loop and the +1 phosphate positions in the Cas9-sgRNA-DNA complex containing a PAM (left) and the Cas9-sgRNA-ssDNA target complex21 (right). Molecule A from the crystallographic asymmetric unit of the Cas9-sgRNA–ssDNA complex is shown. In molecule B, the nucleotides upstream of the +1 phosphate are structurally ordered due to crystal packing interactions, and the +1 phosphate is positioned within hydrogen-bonding distance as a result. Numbers indicate interatomic distances in Å. c, Superposition of the two structures shown in panel b.
Extended Data Figure 7. Endonuclease activity of phosphate lock loop Cas9 mutants against mismatch- and bubble-containing DNA substrates.
a, Endonuclease activity assay of WT and mutant Cas9 proteins using double-stranded oligonucleotide DNA containing a target sequence fully complementary to sgRNA used in Fig. 1a. Samples were taken after 15 sec, 30 sec, 1 min, 2 min, 5 min, 15 min, 1 h, and 2 h. b, Endonuclease activity assay as in panel a but using an oligonucleotide duplex containing mismatches to the sgRNA at positions 1–2. c, Endonuclease activity assay as in panel a but using an oligonucleotide duplex consisting of a target sequence with mismatches to the sgRNA at positions 1–2 and a non-target sequence mismatched to the target strand at positions 1–2. d, Quantification of cleavage defects observed with mismatch- and bubble-containing substrates from a–c. For each protein, the amount of cleaved product obtained after 2 h was normalized to the amount of product obtained from perfectly complementary DNA substrate.
Extended Data Table 1. X-ray crystallographic data collection and refinement statistics.
Values in parentheses denote the highest resolution shell.
| Dataset | Native | SeMet | Iridium hexammine | 1–2 mismatch | 1–3 mismatch |
|---|---|---|---|---|---|
| Cas9 | H840A | H840A | H840A | D10A/H840A | D10A/H840A |
|
| |||||
| DNA | target strand 1 + non-target strand 1 (product) | target strand 1 + non-target strand 1 (product) | target strand 1 + non-target strand 1 (product) | target strand 2 MM + target strand 1 (distal) + non-target strand 2 MM | target strand 3 MM + target strand 1 (distal) + non-target strand 3 MM |
| X-ray source | SLS PXIII | SLS PXI | SLS PXIII | SLS PXI | SLS PXI |
| Space group | C2 | C2 | C2 | C2 | C2 |
| Cell dimensions | |||||
| a, b, c(Å) | 177.7, 68.1, 188.2 | 178.0, 68.1, 188.2 | 178.3, 66.5, 186.5 | 177.78, 66.82, 186.83 | 180.2, 68.07, 190.26 |
| α, β, γ (°) | 90.0, 111.2, 90.0 | 90.0, 11.3, 90.0 | 90.0, 110.7, 90.0 | 90.0, 111.39, 90.0 | 90.0, 111.37, 90.0 |
| Wavelength (Å) | 1.00000 | 0.97965 | 1.10501 | 1.00000 | 1.00000 |
| Resolution (Å)* | 48.15–2.59 (2.69–2.59) | 48.12–2.81 (2.91–2.81) | 47.5–3.30 (3.39–3.30) | 47.49–2.37 (2.46–2.37) | 48.35–2.40 (2.49–2.40) |
| Rsym (%)* | 10.7 (74.6) | 16.1 (78.4) | 13.5 (62.2) | 9.73 (80.59) | 8.97 (36.87) |
| Rpim (%)* | 4.4 (31.4) | 5.3 (31.2) | 5.5 (25.4) | 4.0 (33.7) | 4.1 (25.6) |
| I/σI* | 16.3 (2.4) | 11.1 (2.2) | 14.6 (4.2) | 16.5 (2.2) | 15.3 (4.0) |
| Completeness (%)* | 99.3 (93.5) | 99.5 (96.1) | 99.5 (99.0) | 99.1 (91.5) | 99.9 (100.0) |
| Redundancy* | 6.8 (6.3) | 9.8 (6.1) | 7.0 (6.9) | 6.8 (6.3) | 6.6 (6.7) |
| Refinement | |||||
| Resolution (Å) | 48.15–2.59 | 47.49–2.37 | 48.35–2.40 | ||
| No. reflections | 126432 | 160249 | 164018 | ||
| Rwork/Rfree | 0.221 / 0.249 | 0.209 / 0.245 | 0.216 / 0.247 | ||
| No. atoms | |||||
| Protein | 10686 | 10712 | 10712 | ||
| Nucleic acid | 2526 | 2503 | 2447 | ||
| Ion | 8 | 8 | 8 | ||
| Water | 396 | 695 | 758 | ||
| B-factors | |||||
| mean | 45.4 | 42.1 | 41.3 | ||
| Protein | 45.6 | 41.7 | 41.2 | ||
| Nucleic acid | 46.4 | 46.3 | 43.5 | ||
| Ion | 29.8 | 22 | 27.3 | ||
| Water | 33.5 | 33.2 | 35.8 | ||
| R.m.s. deviations | |||||
| Bond lengths (Å) | 0.003 | 0.004 | 0.003 | ||
| Bond angles (°) | 0.78 | 0.82 | 0.69 | ||
| Ramachandran plot | |||||
| % favoured | 96.0 | 97.0 | 96.0 | ||
| %allowed | 4.0 | 3.0 | 4.0 | ||
| % outliers | 0.0 | 0.0 | 0.0 | ||
| Molprobity | |||||
| Clashscore | 8.6 | 7.1 | 5.8 | ||
Values in parentheses denote highest resolution shell
Extended Data Table 2. DNA and RNA sequences used in the study.
Please refer to web version for coloured text.
| Description | Sequence* | Used in |
|---|---|---|
| Forward primer for PCR amplification of ssDNA templates, T7 promoter † | 5′–TAATACGACTCACTATA–3′ | in vitro transcription of sgRNA |
| ssDNA template sgRNA 1 † | 5′–CACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC TTTTTTACAAATTGAGTTATCCTATAGTGAGTCGTATTA–3′ |
in vitro transcription of sgRNA 1 |
| sgRNA 1 | 5′–GGAUAACUCAAUUUGUAAAAAAGUUUUAGAGCUAGAAAUAGCAAGUUAAAAUAAGG CUAGUCCGUUAUCAACUUGAAAAAGUG–3′ |
Fig. 1, 2a-c, , 3c,e, ED Fig. 1, 2a,c,d, 6b (crystallography) Fig. 2d,e, ED Fig. 3, 7 |
| Reverse primer for PCR amplification of sgRNA1 ssDNA template | 5′–CACTTTTTCAAGTTGA–3′ | in vitro transcription of sgRNA 1 |
| target strand 1 | 5′–CAATACCATTTTTTACAAATTGAGTTAT–3′ | Fig. 1, 2a-c, 3a-c, ED Fig. 1, 2a,c,d, 6 (crystallography) Fig. 2d |
| target strand 1 ATTO532-5′ | ATTO532–5′-GCGCAATACCATTTTTTACAAATTGAGTTAT–3′ | ED Fig. 3b, 7a,d |
| target strand 2 MM FL ATTO532-5′‡ | ATTO532–5′–GCGCAATACCAAATTTTACAAATTGAGTTAT–3′ | ED Fig. 7b,c,d |
| target strand 2 MM | 5′–CAATACCACAT–3′ | Fig. 3e (crystallography) |
| target strand 3 MM | 5′–CAATACCACAA–3′ | Fig. 3e (crystallography) |
| target strand 1 distal | 5′–TTTACAAATTGAGTTAT–3′ | Fig. 3e (crystallography) |
| non-target strand 1 | 5′–ATAACTCAATTTGTAAAAAATGGTATTG–3′ | ED Fig. 3b, 7a,c,d |
| non-target strand 1 3‘-ATTO532 | 5′–ATAACTCAATTTGTAAAAAATGGTATTGCGC-3′–ATTO532 | Fig. 2d |
| non-target strand 1 2 MM ‡ | 5′–ATAACTCAATTTGTAAAATTTGGTATTG–3′ | ED Fig. 7b,d |
| non-target strand 1 (product) | 5′–AAAATGGTATTG–3′ | Fig. 1, 2a-c, 3a,c, ED Fig. 1, 2a,c,d, 6a (crystallography) |
| non-target strand 2 MM | 5′–ATGTGGTATTG–3′ | Fig. 3e (crystallography) |
| non-target strand 3 MM | 5′–TTGTGGTATTG–3′ | Fig 3e (crystallography) |
| pMJ879 § (target plasmid 1) | 5′–….GAATTCCTAATGCGCCATTTTTTACAAATTGAGTTATGGATCC….-3′ 3′–….CTTAAGGATTACGCGGTAAAAAATGTTTAACTCAATACCTAGG….-5′ |
Fig. 2e, 3d, ED Fig. 3a |
| pMJ891 § (target plasmid 1, 2 MM) | 5′–….GAATTCCTAATGCGCCAAATTTTACAAATTGAGTTATGGATCC….-3′ 3′–….CTTAAGGATTACGCGGTTTAAAATGTTTAACTCAATACCTAGG–5′ |
Fig. 3d |
| ssDNA template sgRNA 2 † | 5′–CACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC GCGTCTCATCTTTATGCGTCCCTATAGTGAGTCGTATTA–3′ |
in vitro transcription of sgRNA 2 |
| sgRNA 2 | 5′–GGGACGCAUAAAGAUGAGACGCGUUUUAGAGCUAGAAAUAGCAAGUUAAAAUAAGGC UAGUCCGUUAUCAACUUGAAAAAGUG–3′ |
ED Fig 5 |
| pMJ992 § (target plasmid 2, NGG PAM) | 5′–…. GAATTCGTGAGACCAGCGTCTCATCTTTATGCGTCGGATCC–3′ 3′–….CTTAAGCACTCTGGTCGCAGAGTAGAAATACGCAGCCTAGG….-5′ |
ED Fig. 5a |
| pMJ993 § (target plasmid 2, NAA PAM) | 5′–…. GAATTCGTGAGATTAGCGTCTCATCTTTATGCGTCGGATCC–3′ 3′–….CTTAAGCACTCTAATCGCAGAGTAGAAATACGCAGCCTAGG….–5′ |
ED Fig. 5b |
| pMJ994 § (target plasmid 2, NAAAA PAM) | 5′–…. GAATTCGTGATTTTAGCGTCTCATCTTTATGCGTCGGATCC–3′ 3′–….CTTAAGCACTAAAATCGCAGAGTAGAAATACGCAGCCTAGG….–5′ |
ED Fig. 5c |
sgRNA guide sequences and complementary DNA target strand sequences are shown in red. Non-target strand PAM sites (5′-NGG-3′, 5′-NAA-3′, 5′-NAAA-3′) are highlighted in bold italics. Target-strand complement of the PAM (5′-CCN-3′, 5′-TTN-3′, 5′-TTTTN-3′) is denoted in italic.
The sequence corresponding to the T7 promoter is coloured blue.
Nucleotide positions with a mismatch (MM) to the sgRNA guide sequence are underlined.
All inserts shown are ligated into pUC19 vector between EcoRI and BamHI restriction (shown in grey).
Supplementary Material
Acknowledgements
We are grateful to J. Doudna for agreement on research directions, fruitful discussion and encouragement throughout the project. We thank B. Blattmann and C. Stutz-Ducommun for crystallization screening, N. Ban and M. Leibundgut for the gift of iridium hexamine, and R. Dutzler for sharing synchrotron beamtime and crystallographic advice. We thank E. Charpentier, I. Fonfara, S. Sternberg, P. Sledz, A. May and S. Kassube for critical reading of the manuscript. Part of this work was performed at the Swiss Light Source at the Paul Scherrer Institute, Villigen, Switzerland. We thank T. Tomizaki, V. Olieric and M. Wang for assistance with X-ray data collection. This work was supported by the European Research Council Starting Grant no. 337284 ANTIVIRNA and by start-up funds from the University of Zurich.
References
- 1.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109:E2579–86. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 5.Sapranauskas R, et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. doi:10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jinek M, et al. RNA-programmed genome editing in human cells. elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bassett AR, Tibbit C, Ponting CP, Liu J-L. Highly Efficient Targeted Mutagenesis of Drosophila with the CRISPR/Cas9 System. Cell Rep. 2013;4:220–228. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gratz SJ, et al. Genome Engineering of Drosophila with the CRISPR RNA-Guided Cas9 Nuclease. Genetics. 2013;194:1029–1035. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedland AE, et al. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods. 2013;10:741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qi LS, et al. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bikard D, et al. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41:7429–7437. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilbert LA, et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013 doi: 10.1038/nbt.2675. doi:10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jinek M, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishimasu H, et al. Crystal Structure of Cas9 in Complex with Guide RNA and Target DNA. Cell. 2014 doi: 10.1016/j.cell.2014.02.001. doi:10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fonfara I, et al. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt1074. doi:10.1093/nar/gkt1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esvelt KM, et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. doi:10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luscombe NM, Laskowski RA, Thornton JM. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001;29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Briner AE, Barrangou R. Lactobacillus buchneri genotyping on the basis of clustered regularly interspaced short palindromic repeat (CRISPR) locus diversity. Appl. Environ. Microbiol. 2014;80:994–1001. doi: 10.1128/AEM.03015-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Redondo P, et al. Molecular basis of xeroderma pigmentosum group C DNA recognition by engineered meganucleases. Nature. 2008;456:107–111. doi: 10.1038/nature07343. [DOI] [PubMed] [Google Scholar]
- 27.Ashworth J, et al. Computational reprogramming of homing endonuclease specificity at multiple adjacent base pairs. Nucleic Acids Res. 2010;38:5601–5608. doi: 10.1093/nar/gkq283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013 doi: 10.1038/nbt.2508. doi:10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013 doi: 10.1038/nbt.2647. doi:10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 33.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 34.Emsley P, Emsley P, Cowtan K, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katoh K, Standley DM. MAFFT: Iterative Refinement and Additional Methods. Methods Mol Biol. 2014;1079:131–146. doi: 10.1007/978-1-62703-646-7_8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.