Abstract
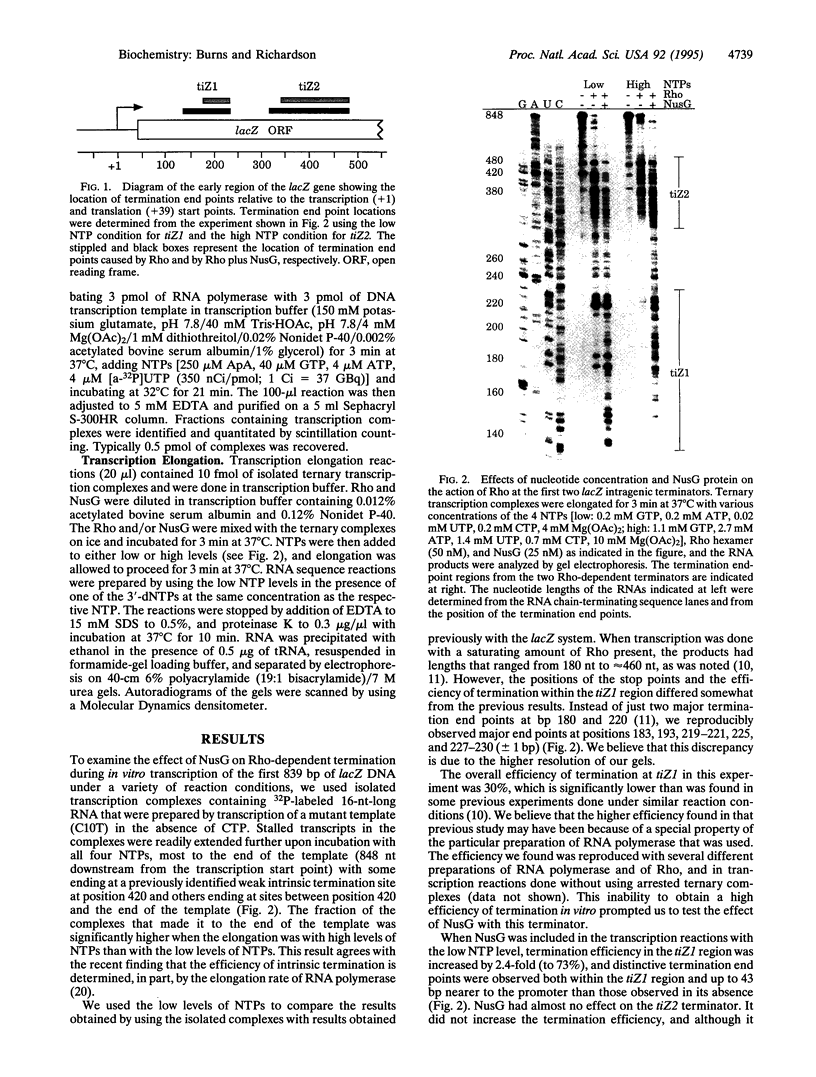
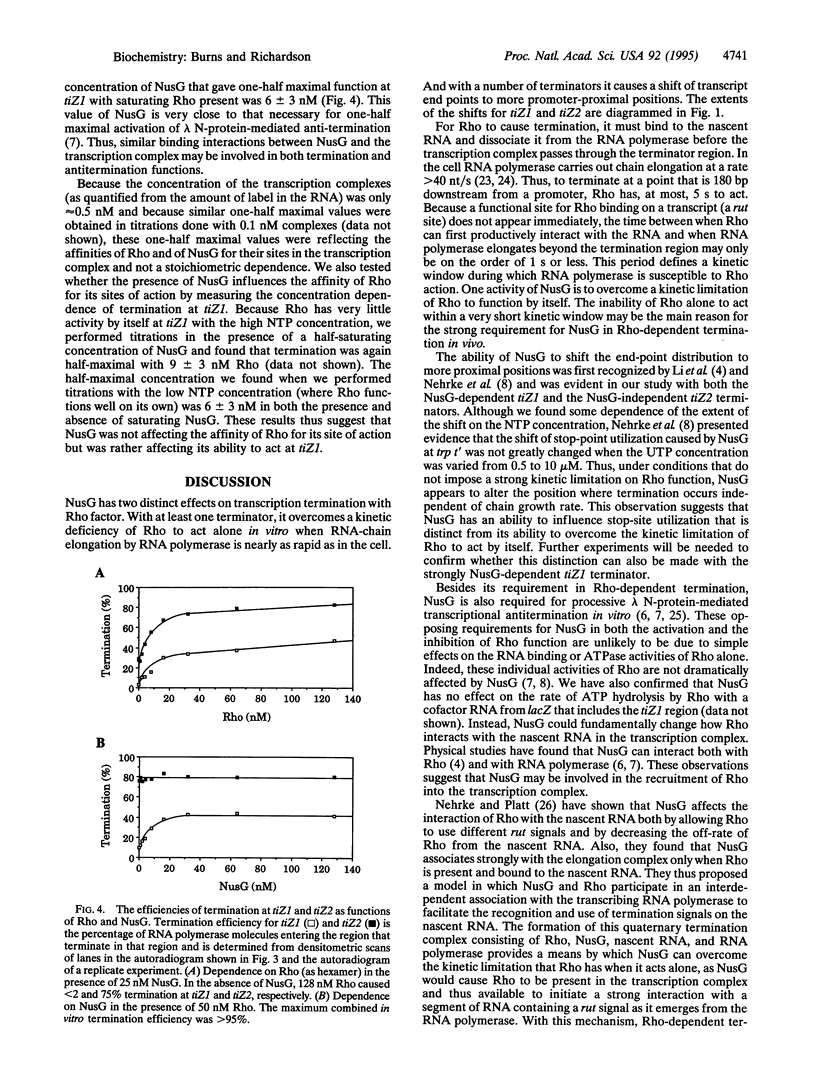
Rho-dependent transcription termination at certain terminators in Escherichia coli also depends on the presence of NusG [Sullivan, S. L. & Gottesman, M. E. (1992) Cell 68, 989-994]. We have found that termination at the first intragenic terminator in lacZ (tiZ1) is strongly dependent on NusG when transcription is done in vitro with the concentrations of NTPs found in vivo. With a lower level of NTPs, and consequently a slower rate of RNA-chain growth, Rho causes some termination by itself that is enhanced with NusG. These results suggest that NusG serves to overcome a kinetic limitation of Rho to function at certain terminators. At a second intragenic terminator within the lacZ reading frame (tiZ2) the efficiency of Rho-mediated termination was unaffected by either NusG or by RNA polymerase elongation kinetics. Thus, using purified components and intracellular levels of NTPs, we have confirmed the in vivo finding that certain Rho-dependent terminators also depend on NusG, whereas others do not.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Gottesman M. Control of transcription termination. Annu Rev Biochem. 1978;47:967–996. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- Burova E., Hung S. C., Sagitov V., Stitt B. L., Gottesman M. E. Escherichia coli NusG protein stimulates transcription elongation rates in vivo and in vitro. J Bacteriol. 1995 Mar;177(5):1388–1392. doi: 10.1128/jb.177.5.1388-1392.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calos M. P., Johnsrud L., Miller J. H. DNA sequence at the integration sites of the insertion element IS1. Cell. 1978 Mar;13(3):411–418. doi: 10.1016/0092-8674(78)90315-x. [DOI] [PubMed] [Google Scholar]
- DeVito J., Das A. Control of transcription processivity in phage lambda: Nus factors strengthen the termination-resistant state of RNA polymerase induced by N antiterminator. Proc Natl Acad Sci U S A. 1994 Aug 30;91(18):8660–8664. doi: 10.1073/pnas.91.18.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D. J., Burgess R. R., Richardson J. P., Gross C. A. Termination efficiency at rho-dependent terminators depends on kinetic coupling between RNA polymerase and rho. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1453–1457. doi: 10.1073/pnas.89.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin J. R., Krummel B., Chamberlin M. J. Isolation and properties of transcribing ternary complexes of Escherichia coli RNA polymerase positioned at a single template base. J Mol Biol. 1987 Jul 5;196(1):85–100. doi: 10.1016/0022-2836(87)90512-2. [DOI] [PubMed] [Google Scholar]
- Li J., Horwitz R., McCracken S., Greenblatt J. NusG, a new Escherichia coli elongation factor involved in transcriptional antitermination by the N protein of phage lambda. J Biol Chem. 1992 Mar 25;267(9):6012–6019. [PubMed] [Google Scholar]
- Li J., Mason S. W., Greenblatt J. Elongation factor NusG interacts with termination factor rho to regulate termination and antitermination of transcription. Genes Dev. 1993 Jan;7(1):161–172. doi: 10.1101/gad.7.1.161. [DOI] [PubMed] [Google Scholar]
- Linn T., St Pierre R. Improved vector system for constructing transcriptional fusions that ensures independent translation of lacZ. J Bacteriol. 1990 Feb;172(2):1077–1084. doi: 10.1128/jb.172.2.1077-1084.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason S. W., Greenblatt J. Assembly of transcription elongation complexes containing the N protein of phage lambda and the Escherichia coli elongation factors NusA, NusB, NusG, and S10. Genes Dev. 1991 Aug;5(8):1504–1512. doi: 10.1101/gad.5.8.1504. [DOI] [PubMed] [Google Scholar]
- McDowell J. C., Roberts J. W., Jin D. J., Gross C. Determination of intrinsic transcription termination efficiency by RNA polymerase elongation rate. Science. 1994 Nov 4;266(5186):822–825. doi: 10.1126/science.7526463. [DOI] [PubMed] [Google Scholar]
- Mosteller R. D., Yanofsky C. Transcription of the tryptophan operon in Escherichia coli: rifampicin as an inhibitor of initiation. J Mol Biol. 1970 Mar;48(3):525–531. doi: 10.1016/0022-2836(70)90064-1. [DOI] [PubMed] [Google Scholar]
- Nehrke K. W., Platt T. A quaternary transcription termination complex. Reciprocal stabilization by Rho factor and NusG protein. J Mol Biol. 1994 Nov 11;243(5):830–839. doi: 10.1006/jmbi.1994.1685. [DOI] [PubMed] [Google Scholar]
- Nehrke K. W., Zalatan F., Platt T. NusG alters rho-dependent termination of transcription in vitro independent of kinetic coupling. Gene Expr. 1993;3(2):119–133. [PMC free article] [PubMed] [Google Scholar]
- Richardson J. P., Macy M. R. Ribonucleic acid synthesis termination protein rho function: effects of conditions that destabilize ribonucleic acid secondary structure. Biochemistry. 1981 Mar 3;20(5):1133–1139. doi: 10.1021/bi00508a014. [DOI] [PubMed] [Google Scholar]
- Richardson J. P. Transcription termination. Crit Rev Biochem Mol Biol. 1993;28(1):1–30. doi: 10.3109/10409239309082571. [DOI] [PubMed] [Google Scholar]
- Richardson L. V., Richardson J. P. A vector for controlled, high-yield production of specifically mutated proteins in Escherichia coli: test of a putative cytidine-binding domain in Rho factor and its Thr16----Ala mutant. Gene. 1992 Sep 1;118(1):103–107. doi: 10.1016/0378-1119(92)90255-n. [DOI] [PubMed] [Google Scholar]
- Ruteshouser E. C., Richardson J. P. Identification and characterization of transcription termination sites in the Escherichia coli lacZ gene. J Mol Biol. 1989 Jul 5;208(1):23–43. doi: 10.1016/0022-2836(89)90085-5. [DOI] [PubMed] [Google Scholar]
- Stock J. B., Rauch B., Roseman S. Periplasmic space in Salmonella typhimurium and Escherichia coli. J Biol Chem. 1977 Nov 10;252(21):7850–7861. [PubMed] [Google Scholar]
- Sullivan S. L., Gottesman M. E. Requirement for E. coli NusG protein in factor-dependent transcription termination. Cell. 1992 Mar 6;68(5):989–994. doi: 10.1016/0092-8674(92)90041-a. [DOI] [PubMed] [Google Scholar]
- Sullivan S. L., Ward D. F., Gottesman M. E. Effect of Escherichia coli nusG function on lambda N-mediated transcription antitermination. J Bacteriol. 1992 Feb;174(4):1339–1344. doi: 10.1128/jb.174.4.1339-1344.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel U., Jensen K. F. The RNA chain elongation rate in Escherichia coli depends on the growth rate. J Bacteriol. 1994 May;176(10):2807–2813. doi: 10.1128/jb.176.10.2807-2813.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L. L., Richardson J. P. Enhancement of transcription termination factor rho activity with potassium glutamate. J Biol Chem. 1991 Jun 5;266(16):10201–10209. [PubMed] [Google Scholar]