Abstract
Pantoea stewartii subsp. stewartii is a proteobacterium that causes Stewart's wilt disease in corn plants. The bacteria form a biofilm in the xylem of infected plants and produce capsule that blocks water transport, eventually causing wilt. At low cell densities, the quorum-sensing (QS) regulatory protein EsaR is known to directly repress expression of esaR itself as well as the genes for the capsular synthesis operon transcription regulator, rcsA, and a 2,5-diketogluconate reductase, dkgA. It simultaneously directly activates expression of genes for a putative small RNA, esaS, the glycerol utilization operon, glpFKX, and another transcriptional regulator, lrhA. At high bacterial cell densities, all of this regulation is relieved when EsaR binds an acylated homoserine lactone signal, which is synthesized constitutively over growth. QS-dependent gene expression is critical for the establishment of disease in the plant. However, the identity of the full set of genes controlled by EsaR/QS is unknown. A proteomic approach previously identified around 30 proteins in the QS regulon. In this study, a whole-transcriptome, next-generation sequencing analysis of rRNA-depleted RNA from QS-proficient and -deficient P. stewartii strains was performed to identify additional targets of EsaR. EsaR-dependent transcriptional regulation of a subset of differentially expressed genes was confirmed by quantitative reverse transcription-PCR (qRT-PCR). Electrophoretic mobility shift assays demonstrated that EsaR directly bound 10 newly identified target promoters. Overall, the QS regulon of P. stewartii orchestrates three major physiological responses: capsule and cell envelope biosynthesis, surface motility and adhesion, and stress response.
INTRODUCTION
Stewart's wilt is a disease affecting maize cultivars caused by the proteobacterium Pantoea stewartii subsp. stewartii. Transmission to plants is possible through an insect vector, the corn flea beetle Chaetocnema pulicaria, which carries the bacterium in its alimentary canal and transmits it during feeding (1). P. stewartii possesses a hypersensitivity response and pathogenicity (hrp-hrc) type III secretion system (T3SS) which deploys Wts (water-soaking) effectors that are responsible for water-soaked lesions in leaves during the early stages of infection (2). In the later stages of infection, the bacteria colonize the xylem and form a biofilm by secreting the exopolysaccharide stewartan. The formation of biofilm blocks water flow and leads to wilting, necrosis, and even death, causing severe reductions in crop yield (3). Stewart's wilt is a serious concern for susceptible sweet corn hybrids (1). Bacterial wilt diseases that cause disease by blockage of the vascular systems are seen in many other economically important plant species, such as the bacterial wilt of cucurbit by Erwinia tracheiphila (4), black rot of cruciferous plants by Xanthomonas campestris (5), Pierce disease of grapes caused by Xylella fastidiosa (6), and the broad-host-range plant pathogen Ralstonia solanacearum (7).
In P. stewartii, the production of stewartan is temporally controlled by a cell density-dependent quorum-sensing (QS) system regulated by the transcription factor EsaR, a homologue of LuxR from Vibrio fischeri (8). (EsaR is the only LuxR homologue in the P. stewartii genome besides SdiA [9].) EsaR controls stewartan production at low cell densities by repressing expression of rcsA, which encodes a transcriptional activator of the capsular synthesis operon (10). EsaR senses changes in cell density by recognizing an acylated homoserine lactone (AHL) signal, N-3-oxo-hexanoyl-l-homoserine lactone, which is synthesized by the cognate AHL synthase, EsaI. Production of AHL is constitutive during growth in P. stewartii; thus the concentration of AHL in the surrounding medium serves as an indicator of population density. In the absence of AHL, EsaR binds to specific 20-bp regulatory sequences in promoter regions called esa boxes and represses or activates transcription of downstream genes. At high cell density, AHL binds to EsaR and hinders its ability to bind to DNA, leading to derepression or deactivation of gene expression (8).
The quorum-sensing system of P. stewartii DC283 (SS104) (11), isolated in Illinois during 1967, has been intensively studied. Both esaR and esaI were rendered inactive via a 521-bp deletion that overlaps the ends of the two convergently transcribed genes (12). This DC283 ESΔIR strain is considered to be QS deficient. However, when complemented with esaR expressed from its native promoter (pSVB60) (8), the strain becomes constitutively QS proficient since there is no AHL production to inactivate EsaR. Using DC283 ESΔIR(pSVB60) as a QS-proficient strain and DC283 ESΔIR(pBBR1MCS-3) with the vector control as a QS-deficient strain, a proteomic analysis by Ramachandran and Stevens has shown that the QS regulon in P. stewartii consists of at least 30 proteins (13). In addition to RcsA, the production of three other proteins was shown to be directly regulated by EsaR, DkgA (2,5-diketogluconate reductase), which is repressed at low cell densities, and LrhA (a repressor of chemotaxis, adhesion, and motility) and GlpFKX (glycerol utilization proteins), which are activated at low cell densities (13). However, proteomic approaches suffer from a limitation in the detection of the protein product. Thus, in this study high-throughput sequencing of RNA transcripts (RNA-Seq) analysis was used as a screen for the identification of additional genes in the QS-regulon of P. stewartii. Bioinformatic tools, quantitative reverse transcription-PCR (qRT-PCR), and electrophoretic mobility shift assays (EMSAs) were employed during the analysis of select targets in the regulon. Ten newly identified promoters were found to be directly bound and regulated by EsaR. Importantly, regulation of three physiological functions, capsule production, surface motility/adhesion, and stress response, appears to be coordinately controlled by EsaR.
MATERIALS AND METHODS
RNA purification and rRNA depletion for RNA-Seq.
Cultures of P. stewartii, DC283 ESΔIR(pSVB60), a QS-proficient esaR+ ΔesaI strain, and DC283 ESΔIR(pBBR1MCS-3), a QS-deficient ΔesaR ΔesaI strain (13), grown overnight at 30°C were used to inoculate 5 ml of RM medium (1× M9 salts, 2% Casamino Acids, 1 mM MgCl2 [8]) supplemented with tetracycline (10 μg/ml) and grown to an optical density at 600 nm (OD600) of 0.5. Ten milliliters of RNA Protect reagent (Qiagen, Valencia, CA) was added and incubated with cells for 5 min at room temperature to stabilize mRNA before samples were centrifuged for 10 min at 5,000 × g. Cell pellets were resuspended in 200 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.0) supplemented with 15 mg/ml lysozyme (Qiagen) and 30 mAU/ml (where AU is arbitrary units) proteinase K (Qiagen). QIAzol lysis reagent was then added, and the total RNA was harvested using an miRNeasy kit (Qiagen) per the manufacturer's protocol. The quantity and quality of total RNA were assessed on an Agilent Bioanalyzer 2100 at the Virginia Bioinformatics Institute (VBI, Blacksburg, VA). Total RNA of each sample was depleted for rRNA using a Ribo-Zero rRNA removal kit for Gram-negative bacteria (Epicentre, Madison, WI) per the manufacturer's protocol. The rRNA-depleted sample was assessed for quality, including rRNA removal, prior to sample processing for Illumina sequencing (VBI) with single paired-end 50-bp reads.
Processing of Illumina sequencing data.
The P. stewartii subsp. stewartii transcriptome sequencing read files in FASTQ format were transformed into FASTA format using the SeqIO module of BioPython (14), version 1.63 (www.biopython.org), through Python, version 2.7.6. Then the stand-alone BLAST+ suite (15, 16), version 2.2.29 (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/2.2.29/), was used to align the reads to the partially assembled version 5b (v5b) P. stewartii DC283 genome sequence provided on the ASAP website (9). First, a BLAST database and its index were created using the FASTA files for each of the samples with the makeblastdb and makembindex BLAST+ routines. Second, for each sample, stand-alone blastn (task megablast) queries were performed using the nucleotide sequences for the P. stewartii DC283 protein-coding genes against the BLAST databases for each sample with a maximum E value of 1.e−10 for alignment. The table format output (outfmt 6) of each blastn query was subsequently processed using an awk-based shell script to count and list the total number of BLAST hits for each of the protein-coding genes in the P. stewartii subspecies stewartii data set. Although the BLAST alignment is slower than other alignment algorithms (such as Burrows-Wheeler Aligner [17]), every step uses analysis tools that can repeatedly be used for each protein-coding gene and for any other transcribed sequence (18). Since read hits are counted only if the alignments to the reference sequences are sufficiently close (E value, <1.e−10), no quality filtering is required, and no biases are introduced by preprocessing the reads using quality scores. The one caveat with this pipeline is that reads can be counted as hits to multiple genes so that differential expression levels for paralogues with sufficient sequence similarity will be difficult to determine without further analysis. Subsequent data processing was done using Microsoft Excel and Lasergene (DNASTAR, Inc., Madison, WI). Analysis of +1 transcription start sites was performed by viewing the alignment on reads for the presence of a sharp cliff of at least 10 reads when they were viewed in the Seqman Pro (Lasergene) browser. The analysis described above was repeated for the updated genome, version 8 (v8), made available on the ASAP website. Normalized gene expression levels in the two samples were determined by first removing all of the potentially repetitive transposases, insertion element sequences, and unnamed hypothetical proteins and normalizing the read counts to the total number of reads aligned to the remaining genes in each sample. The error bars for the ratios of normalized gene expression levels were conservatively estimated using the standard deviation of the log (base 2) ratios across all genes. This error estimate assumes that the overall difference in expression levels between the two samples is a good surrogate for biological replicates and that a multiplicative biological/sample preparation error dominates over the purely technical sampling errors (18).
A CKS number given for each gene in the text refers to the locus tag by which the genes are identified in the P. stewartii DC283 genome at the ASAP website (asap.ahabs.wisc.edu/asap/home.php) (9).
EMSAs.
FAM (6-carboxyfluorescein)-labeled forward and unlabeled reverse primers (IDT-DNA, Inc., Coralville, IA) (Table 1) were used to generate FAM-labeled DNA fragments. A His-maltose binding protein (MBP)-glycine linker-tagged EsaR protein (HMGE) was purified as previously described (19) with the following modifications. To ensure only DNA-binding-competent protein was used in the EMSAs, the purified HMGE was passed through a HiTrap Heparin HP column (GE Healthcare Life Sciences, Piscataway, NJ) and eluted using 10 mM NaH2PO4 buffer with a gradient of 0 to 800 mM NaCl. HMGE was then transferred into the EMSA binding buffer (20 mM HEPES, 1 mM EDTA, 30 mM KCl and 10% glycerol, pH 7.4) using a 5-ml desalting column (GE Healthcare Life Sciences). HMGE (0 to 100 nM) was incubated with 5 nM FAM-labeled DNA for 30 min in EMSA reaction buffer [20 mM HEPES, 1 mM EDTA, 30 mM KCl, 0.2% Tween 20, 10 mM (NH4)2SO4, 50 ng/μl poly(dI-dC), 150 μg/ml acetylated bovine serum albumin (BSA), 10% glycerol, pH 7.4] in a total reaction volume of 20 μl. The postincubation reaction products were separated on prechilled 4.5% Tris-glycine-EDTA native PAGE minigels (Bio-Rad, Hercules, CA) at 80 V for 2 h. Visualization was achieved through a Typhoon Trio Variable Mode Imager (GE Healthcare, Life Sciences) using the blue laser.
TABLE 1.
Primers utilized for amplification of DNA probes for EMSAs
| Primer name | Sequence (5′ to 3′) | Promoter amplified (reference) | Size of product (bp) |
|---|---|---|---|
| PDKGA-F-FAM | 56-FAM-GCTGACACAGTAAGTCGTGG | PdkgA (13) | 181 |
| PDKGA-F | GCTGACACAGTAAGTCGTGG | ||
| PDKGA-R | CGCTGTGCTTAAGTCTAGC | ||
| PUC-TMP-F-FAM | 56-FAM-TACGGACGTCCAGCTGAGATCTCCTAGGGGCC | pUC18-MCS | 32 |
| PUC-TMP-F | TACGGACGTCCAGCTGAGATCTCCTAGGGGCC | ||
| PUC-CDN-R | GGCCCCTAGGAGATCTCAGCTGGACGTCCGTA | ||
| PESAR28F-FAM | 56-FAM-TCTTGCCTGTACTATAGTGCAGGTTAAG | PesaR28 (8) | 28 |
| PESAR28F | TCTTGCCTGTACTATAGTGCAGGTTAAG | ||
| PESAR28R | CTTAACCTGCACTATAGTACAGGCAAGA | ||
| PUSPB-TMP-F-FAM | 56-FAM-TTCTGCACTGTTCTGCGGTGCCGGTAAGGAG | PuspB | 134 |
| PUSPBR | GAACCACCAGCAGCGCACGCAACG | ||
| PUSPA-F | GCAGAATTTGCCAACAACC | PuspA | 368 |
| PUSPA-R-FAM | 56-FAM-CCATGATATAACTCCTTCCAC | ||
| PWCE-F-FAM | 56-FAM-TCAGAAACGCTTCAGGCTCAA | PwceG1 | 108 |
| PWCE-R | ACGTAAAATGGTTATCGCGCAT | ||
| PYRR-TMP-F-FAM | 56-FAM-AAAATGCCCAGTATTGCCGGGCAGGGCGCCAT | PCKS-0678 | 176 |
| PPYRR-R | GGAGGCTGCCATACCGTCAGGATC | ||
| ENDO-F | ATTCGTGAGCCATACGTTAATG | PCKS-1289 | 121 |
| ENDO-FAM-R | 56-FAM-TTGTCTGTTATACCGGAGTTA | ||
| PHRPN-F-FAM | 56-FAM-ATTTCAGACAGGAACCAGCACC | PhrpN | 139 |
| PHRPN-R | CCAGCGGACTCGTATTCATACTC | ||
| P9766-F-FAM | 56-FAM-AGGCGTTACCTCTCTGAACG | Pspy | 318 |
| P9766-R | AGTTTACGCATAACAGTGTCC | ||
| P0526-F-FAM | 56-FAM-ACAGACAGGCAAAACAGTTCTG | PCKS-0458 | 313 |
| P0526-R | GCTTGAATTTCATAACCATTCC | ||
| YJBE-F | GGTATTTTGTTGCCCTATACTGG | PyjbE | 176 |
| YJBE-R-FAM | 56-FAM-AAGAGACGGCGACGAGTAAT | ||
| P8505-F-FAM | 56-FAM-AGACCGTAACTGGCAAATTTC | PCKS-0881 | 173 |
| P8505-R | CCTTCATTTGTCTACCCTCATTC | ||
| CONI-F-FAM | 56-FAM-AATATCCCGCTCCGCTTGC58. | PCKS-1103 | 254 |
| CONI-R | TTTGAAATTACCGCCGCCGC | ||
| P8895-F-FAM | 56-FAM-TGGACGGTAACCAGACATAAC | PCKS-4689 | 212 |
| P8895-R | GCAGGGAAAACCAGAACGGAA | ||
| YCIF-F-FAM | 56-FAM-AGGTGAGGCGGAGCATTTA | PyciF | 219 |
| YCIF-R | TTGACAGTCATAAAAGAGCCTC | ||
| PWCEL-F-FAM | 56-FAM-ACGAAAAGCTAAGCGCTAAG | PwceL | 268 |
| PWCEL-R | ATGGAGCCATGGTGTGATTC | ||
| PLYSP-F-FAM | 56-FAM-CACGCTTTATCGCATCC | PlysP | 300 |
| PLYSP-R | CGCAGGTTGTTGTGTTG | ||
| POSMY-F | CGCGTTTTCAGGGCCATTCTC | PosmY | 197 |
| POSMY-R-FAM | 56-FAM-CAACGCTACTGCGGCACAGG | ||
| PWCEG2-F-FAM | 56-FAM-CGTAAGCCCGGAGGATTAC | PwceG2 | 396 |
| PWCEG2-R | GTACTTGCCATAAAACCTGCTCTC | ||
| ELAB-F-FAM | 56-FAM-TCAGGTGAAACAGCTGGTAAT | PelaB | 306 |
| ELAB-R | GACTTCACCCCATTGTTGC |
qRT-PCR.
P. stewartii DC283 ESΔIR(pSVB60) QS-proficient and DC283 ESΔIR(pBBR1MCS-3) QS-deficient cells were grown, treated with RNA Protect (Qiagen), and harvested as described above for RNA-Seq. RNA was quantified using a NanoPhotometer (Implen, Westlake Village, CA) and checked for quality using an Agilent Bioanalyzer 2100 (VBI). All RNA integrity number (RIN) values were above 9.5. The extracted RNA was converted to cDNA using a High Capacity cDNA reverse transcription kit (Life Technologies, Grand Island, NY) per the manufacturer's instructions. The cDNA was quantified using a NanoPhotometer (Implen), checked for purity by measuring absorbance ratios at 260/280 nm and 260/230 nm, and used as the template in a 7300 Real-Time PCR System (Applied Biosystems/Life Technologies). The primer pairs (Table 2) for qRT-PCR analysis were designed using Primer Express, version 3.0 (Life Technologies), and optimized to 100% ± 10% efficiency using cloned coding regions of each gene as the template. Primers used for cloning control template regions into pGEM-T (Promega, Fitchburg, WI) are listed in Table 2. Parameters for qRT-PCR primer design were set as the following: 18 to 24 bp in length, a melting temperature (Tm) of 64°C, and amplicon length of 80 to 120 bp. Template DNA (either plasmid or cDNA) was used at concentrations ranging from 0.001 ng to 80 ng per 25-μl reaction mixture containing 300 nM (each) specific forward and reverse primer and 2× SYBR green PCR master mix (Life Technologies) diluted to a 1× concentration with distilled H2O (dH2O). Reactions were carried out in triplicate in a MicroAmp Optical 96-well reaction plate (Life Technologies) on samples from two separate experiments. The thermal cycler settings were programmed for 95°C for 10 min and 45 cycles of 95°C for 15 s and either 60°C, 62°C, or 64°C for 1 min, depending on the primer pair (Table 2). This last step was also set as the data collection point. A dissociation stage was added at the end of the PCR run to confirm specific product amplification. The data obtained were analyzed through 7300 System SDS RQ software, version 1.4 (Life Technologies), using an automated cycle threshold, and relative expression was calculated using the Pfaffl method (20).
TABLE 2.
Primers utilized for template cloning and qRT-PCR
| Primer name | Sequence (5′ to 3′) | Function (annealing temp [°C]) |
|---|---|---|
| 9766CLONINGF | CAAGTAAAAAAGGACACTGTTATGCG | Cloning spy coding region |
| 9766CLONINGR | CCGCTTTGTCATGCATCG | |
| 9766RTF2 | ATTATCGCCTCTGACAGCTTCGA | qRT-PCR (62) |
| 9766RTR2 | GGCGTTGGCGGTCATTTT | |
| 0526CLONINGF | TTTAATGTTATTTTTTCGAGGTTTTTGC | Cloning CKS-0458 coding region |
| 0526CLONINGR | AAACAGGTGACCGTCTTAC | |
| 0526RTFWD3 | CGGCACCAATTACGCGATCTAT | qRT-PCR (62) |
| 0526RTREV3 | GCCGGGATCACCGGATT | |
| YJBECLONING F | CCAATGTAGGTGTTGTTATGAAAAAACG | Cloning yjbE coding region |
| YJBECLONINGR | GTTGTGGTCGTGGTAGTTG | |
| YJBERTF | GCATTACTCGTCGCCGTCTCT | qRT-PCR (60) |
| YJBERTR | TGCTGCGGCACCAGCTT | |
| 8505CLONINGF | GAATGAGGGTAGACAAATGAAGG | Cloning CKS-0881 coding region |
| 8505CLONINGR | AGGATGTGATCAGAGGTGAG | |
| 8505RTF2 | GTGCAGAAGATGAATCTGACCAAAA | qRT-PCR (60) |
| 8505RTR2 | CATCCATTGGCGAAGTGGTTT | |
| CONIRTFWD | GAGCATCGTGGTGATTCAGGTAAT | Cloning CKS-1103 coding region |
| CONIRTREV | GCTGCTCTTTCCGCCTTTTTT | |
| 7180FWD2 | GCAGAGCATCGTGGTGATTCAG | qRT-PCR (60) |
| 7180REV2 | TGCTCTTTCCGCCTTTTTTACC | |
| 8895 CLONING F | CAAATGGCAAAATTAACCGTCCC | Cloning CKS-4689 coding region |
| 8895 CLONING R | TCAGAAGTTAAATCCGAGTTGC | |
| 8895RTF2 | CCCGGCGTTAATGACCAAA | qRT-PCR (64) |
| 8895RTR2 | CGCACTGGGAAAAATGATGCT | |
| FWDYCIFCLONING | ATGAGGCTCTTTTATGACTGTC | Cloning yciF coding region |
| REVYCIFCLONING | TCATTTCGCAACTCCTTCAG | |
| YCIFRTFWD | CCCTGGAAGGCCTGGTAGAA | qRT-PCR (60) |
| YCIFRTREV | GCGTACTTCACCCTTTTCAACAGA | |
| WCELCLONINGF | GGAATCACACCATGGCTCC | Cloning wceL coding region |
| WCELCLONINGR | CAACCAAAACTCCTGCGC | |
| WCELRTF | GGGTCGCTGGGTGAAAGG | qRT-PCR (62) |
| WCELRTR | CTGCCATCCATTGCGTAGGA | |
| LYSPCLONINGF | TCGCGCCTTATTTTTACCCAG | Cloning lysP coding region |
| LYSPCLONINGR | GCAGGCCAATATAGGTCGC | |
| LYSPRTF | GCCGTTTGCCGGAGGTT | qRT-PCR (64) |
| LYSPRTR | CCTGGAAAGAAAAGCCGACAA | |
| OSMYCLONINGF | GATGATATCGATGCACACAACTAAGC | Cloning osmY coding region |
| OSMYCLONINGR | TTTAACACTTTTTACGCCTTCGATAGC | |
| OSMYRTFWD | CGAAAATCGACAGCTCAATGAAGA | qRT-PCR (60) |
| OSMYRTREV | CCACCAGCGCCGCTTT | |
| WCEG2CLONINGF | CATTAAAGAGAGCAGGTTTTATGGC | Cloning wceG2 coding region |
| WCEG2CLONINGR | GGCAATATCCGTCCACAGC | |
| WCEG2RTF | GCGCGTCGCGAATGG | qRT-PCR (60) |
| WCEG2RTR | CCCCCGATGCGGGTAAT | |
| ELABCLONINGF | ATGTCAGGAAAAATTGAAGATGCCG | Cloning elaB coding region |
| ELABCLONINGR | TTACTTACGACCGAGCAGGAAG | |
| ELABRTF | CCCGCCGCTACACCAATC | qRT-PCR (62) |
| ELABRTR | CAAACGGGTTAGACTGCATCTGAT | |
| 27F | AGAGTTTGATCATGGCTCAG | Cloning 16S rRNA coding region |
| 1429RLONG | ACCTTGTTACGACTTCACC | |
| 16S-RTF | GCCAGCAGCCGCGGTAAT | qRT-PCR (60) |
| 16S-RTR | CGCTTTACGCCCAGTAATTCC |
Accession numbers.
The read data for the P. stewartii DC283 ESΔIR(pSVB60) QS-proficient esaR+ ΔesaI strain and DC283 ESΔIR(pBBR1MCS-3) QS-deficient ΔesaR ΔesaI strain have been deposited in the NCBI Sequence Read Archive (SRA) under accession numbers SRX529441 and SRX530931, respectively. An Excel file summarizing differential gene expression in total counts and normalized read counts per million aligned reads (RPM), using both versions of the P. stewartii DC283 genome (v5b and v8) annotation, has been deposited in the NCBI Gene Expression Omnibus (GEO) database under accession number GSE57635.
RESULTS
RNA-Seq analysis of the QS regulon in P. stewartii.
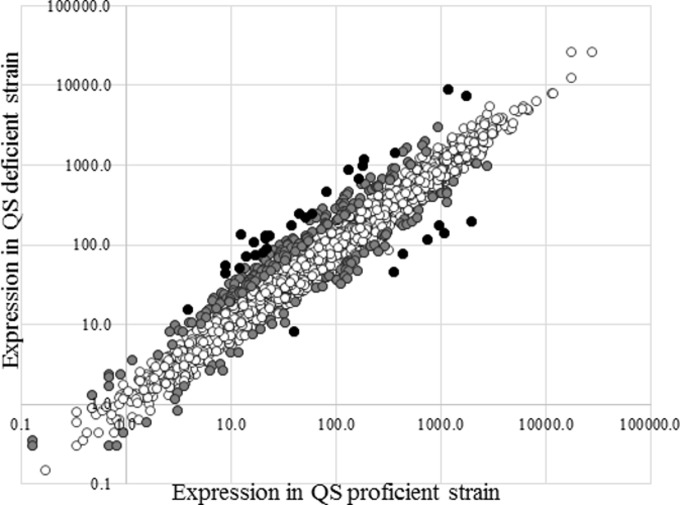
An RNA-Seq analysis was performed as a screen to identify genes in the EsaR regulon using rRNA depleted RNA from the QS-proficient esaR+ ΔesaI strain and the QS-deficient ΔesaR ΔesaI strain described above (13). Thirty million reads of 50 bp in length were obtained for each sample that aligned with nearly 100× coverage for most genes when aligned to the 5,353 protein-coding sequences in the v5b annotation of the P. stewartii DC283 genome available at the ASAP database (9). Since the draft genome of P. stewartii contains many identical (or nearly identical) transposases and insertion sequences (e.g., IS66), these were excluded, along with all unnamed hypothetical proteins, from the analysis during normalization of reads. From the refined data set of 3,683 protein-coding genes, 260 genes were differentially regulated 2-fold or more in the two strains (see Table S1 in the supplemental material). Of these, 184 genes were repressed by EsaR, and 76 genes were activated (Fig. 1; see also Table S1).
FIG 1.
Differential mRNA expression during QS. Whole-transcriptome data of the P. stewartii DC283 ESΔIR(pSVB60) QS-proficient esaR+ ΔesaI strain and DC283 ESΔIR(pBBR1MCS-3) QS-deficient ΔesaR ΔesaI strain were obtained from RNA sequencing. The RPM change of normalized expression for most genes (unfilled circles) falls around a line with a slope of 1, indicating that they are equally expressed in both strains. Genes with 2-fold and 4-fold or more change in expression levels are indicated by gray and black circles, respectively.
The mRNA transcripts for the EsaR-repressed rcsA gene (CKS-2570) were 5.8-fold higher in the QS-deficient strain. In addition, two previously identified EsaR direct targets, dkgA (CKS-0482) and lrhA (CKS-2075), were repressed 6.8-fold and activated 3.2-fold, respectively, consistent with published results (13). However, the data did not show any significant change in expression of the glpFKX operon (CKS-1089, -1090, and -1091), which had been previously identified (13) as a directly activated target of EsaR using the same growth medium and conditions. Of the previously identified QS regulon proteins found using a proteomic analysis, the genes for four others also showed a 2-fold or more change in mRNA expression in the presence of EsaR: CKS-2200 (ahpF) activated 2.5-fold, CKS-5250 (degP) activated 2.7-fold, CKS-0369 (osmC) repressed 2.0-fold, and CKS-5070 (osmY) repressed 4.3-fold. The gene expression levels for the remaining proteins identified in the proteomic analysis of the EsaR regulon showed changes less than 2-fold. Thus, while there was some overlap in the QS-controlled genes identified, the proteomic and transcriptomic approaches collectively identified a larger regulon than either method individually.
To complete the survey of differentially expressed transcripts, the changes in expression levels of 1,219 unnamed hypothetical protein-encoding genes in the v5b P. stewartii genome were also analyzed. The expression levels of an additional 190 hypothetical proteins were found to be changed 2-fold or greater (see Table S2 in the supplemental material). However, the analysis of these uncharacterized open reading frames was beyond the scope of this study.
When all of the reads were mapped to the 58 scaffolds for the partially assembled v5b P. stewartii DC283 genome, some mRNA reads were seen to align to noncoding regions of the genome, indicating the presence of possible noncoding small RNAs (sRNAs) and additional regulatory systems. For example, the gene for EsaS, an sRNA activated by EsaR (19), was found to be activated 15.2-fold in the QS-proficient strain (data not shown). The mapped transcriptome also allows visualization of 5′ untranslated regions, aiding in the prediction of transcriptional start sites, upstream regulatory elements, and genome assembly.
Bioinformatics approach to identify direct targets of EsaR.
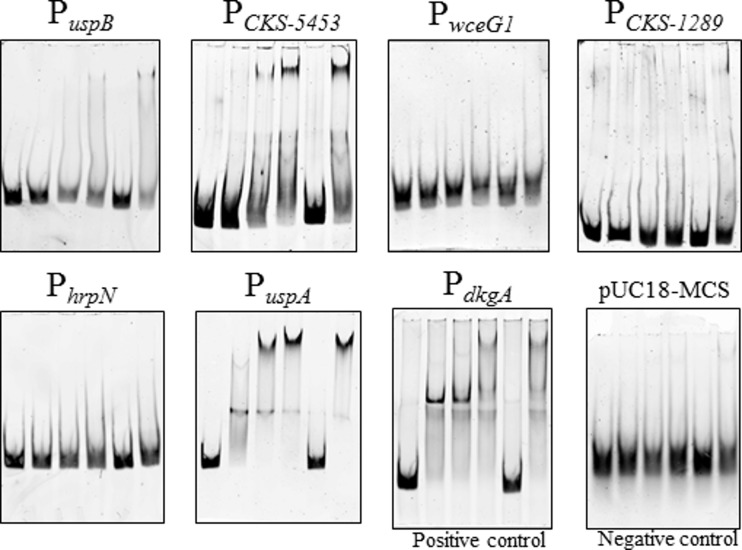
To identify additional genes that are directly regulated by the master QS regulator EsaR, a bioinformatics approach was first attempted. Such an approach was attractive as it helps in reducing the number of possible targets in the large amounts of data available from high-throughput analyses like RNA-Seq. A position-specific weight matrix (PSWM) was constructed using the alignment of eight published 20-bp EsaR binding sites (esa boxes) (13). Using the PATSER program (21), the entire P. stewartii database was scanned to identify promoters that possess an esa box with similarity to the PSWM. This list was next cross-referenced with the list of differentially expressed mRNAs determined through RNA-Seq to identify genes that were regulated at least 2-fold. Eight genes were identified, including three previously known genes, esaR, dkgA, and lrhA, which possessed high-scoring esa boxes in the PATSER program (Table 3). EMSAs were carried out on the promoters of the five remaining genes. Only one promoter, PCKS-0678, was found to be bound by the His-MBP-glycine linker-EsaR fusion protein (HMGE) (19), a biologically active fusion protein of EsaR, and the shift was successfully competed by the PesaR28-specific competitor, a 28-bp DNA fragment that includes the esaR box (8) (Fig. 2). As positive and negative controls, the 181-bp PdkgA promoter probe (13) and a 32-bp multiple cloning site (MCS) from pUC18 generated by annealing complementary primers were used, respectively (Fig. 2). Of the remaining five promoters, four were not bound by EsaR, despite possessing very high-scoring predicted esa boxes. However, one predicted promoter for CKS-0970 (uspB), PuspB, was bound very weakly by EsaR at the highest concentration of the protein used. Analysis of the region upstream of uspB revealed a divergently transcribed gene, CKS-0971 (uspA), repressed 2.3 times by EsaR in the RNA-Seq data (Table 3) and possessing a lower-scoring esa box in its promoter, possibly shared with uspB. An EMSA revealed that this promoter, PuspA, is strongly bound by EsaR, possibly regulating both uspA and uspB (Fig. 2). Taken together, these findings suggest that the scoring and sorting of potential targets primarily based on predicted PSWM scores were not highly effective, possibly due to the limitations of the bioinformatics tools applied or the consensus sequence utilized.
TABLE 3.
Genes in the RNA-Seq data with high-scoring esa boxes
| ASAP no. | Locus tag (gene name) | Protein productd | Fold regulationa | Predicted EsaR binding sequence | esa box scoreb | Promoter bound by EsaR (reference) |
|---|---|---|---|---|---|---|
| ACV-0290094c | CKS-2903 (esaR) | Quorum-sensing transcriptional activator EsaR | 5.4 (A) | GCCTGTACTATAGTGCAGGT | 14.9 | Yes (8) |
| ACV-0290502c | CKS-0482 (dkgA) | 2,5-Diketo-d-gluconate reductase A | 6.7 (R) | AGCTAGACTTAAGCACAGCG | 11.6 | Yes (13) |
| ACV-0288416 | CKS-0970 (uspB) | Universal stress protein B | 2.4 (R) | ACCGGCACCGCAGAACAGTG | 10.6 | Weakly |
| ACV-0285895c | CKS-2075 (lrhA) | LysR family transcriptional regulator LrhA | 3.1 (A) | AGAGCATCTTTAGTACAGGT | 8.9 | Yes (13) |
| ACV-0289544 | CKS-2241 (wceG1) | Undecaprenol-phosphate galactose-phosphotransferase/O-antigen transferase | 3.3 (R) | GGCTCAACATCAGTACTGTT | 8.8 | No |
| ACV-0290308 | CKS-0678 | C-terminal fragment of a PLP-dependent catabolic threonine dehydratase | 2.6 (A) | CCCTGCCCGGCAATACTGGG | 8.5 | Yes |
| ACV-0291078 | CKS-1289 | Endoribonuclease l-PSP | 2.1 (A) | ACCTATCCGGTATTGCAGAT | 8.4 | No |
| ACV-0286411 | CKS-3275 (hrpN) | Elicitor of the hypersensitivity reaction HrpN | 2.1 (A) | AGCGGCACTTAATAAAAGCT | 8.1 | No |
| ACV-0288415 | CKS-0971 (uspA) | Universal stress global response regulator | 2.3 (R) | GCATTGCCTGTATTGCAGGT | 7.4 | Yes |
Twofold or more regulation by EsaR according to RNA-Seq data. A, activation; R, repression.
Score generated using PATSER program (http://rsat.ulb.ac.be/rsat/patser_form.cgi) and published consensus (21).
Previously identified direct targets of EsaR, not analyzed by EMSAs in this study.
PLP, pyridoxal 5-phosphate.
FIG 2.
EMSAs of promoters identified using bioinformatics tools. Promoters of potential direct targets of EsaR identified via a primarily bioinformatics approach were analyzed for binding by the EsaR fusion protein, HMGE. The promoter of dkgA (13) was used as a positive control, and a 32-bp pUC18 MCS DNA fragment was used as a negative control. The concentration of FAM-labeled DNA probe in all lanes is 5 nM. The lanes within each panel consist of the following (left to right): DNA probe, DNA probe with 100 nM HMGE, DNA probe with 200 nM HMGE, DNA probe with 400 nM HMGE, DNA probe with 400 nM HMGE and 1 μM specific competitor PesaR28 DNA fragment, and DNA probe with 400 nM HMGE and 1 μM nonspecific pUC18 MCS DNA fragment.
Validating transcriptional control and identifying highly regulated putative EsaR direct targets.
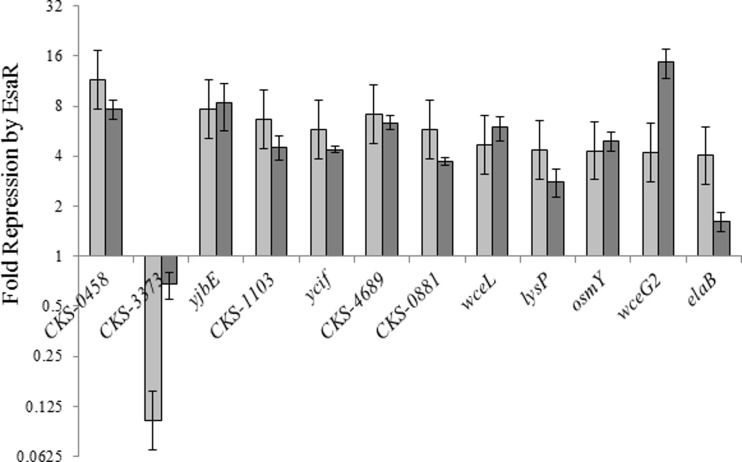
A different set of criteria was established to improve the potential list of predicted direct targets of EsaR starting with the differentially expressed genes seen in the RNA-Seq analysis. Promoters of the most differentially regulated genes that possessed a clear transcriptional start site (+1) and a possible EsaR binding site in or around the +1 site were analyzed. This approach placed less emphasis on the scoring of the possible esa boxes. Sequences annotated as transposases, insertion elements, or hypothetical or putative proteins were excluded from the analysis. Twenty-six fully annotated genes that were differentially expressed 4-fold or more in the presence of EsaR were studied for predicted +1 sites. A potential +1 site was characterized by a sharp rise in the number of aligned reads at a particular nucleotide residue upstream of the ATG start codon in the mapping of reads to the assembled v5b scaffold containing the gene. The +1 site of previously known direct targets of EsaR showed similar sharp rises. The putative EsaR binding sites were then identified upstream of the +1 sites using the PATSER program as described above (21). Of the 26 genes regulated 4-fold or higher, eight genes did not possess a clear +1 site and were not analyzed further. Four other genes were targets seen previously, esaI, esaR, rcsA, and dkgA. The remaining 14 genes, putatively controlled by 12 promoters, were analyzed further (Table 4). Quantitative RT-PCR was first performed to validate the changes in gene expression observed in the Illumina sequencing. Two separate RNA samples extracted independently of the sample generated for RNA-Seq analysis were used. The 16S rRNA coding region was used as a reference to normalize expression of other genes. All genes showed levels of regulation fairly similar to those seen in RNA-Seq analysis, except CKS-3373 and CKS-2413 (elaB), which appeared to be minimally regulated in qRT-PCR, and CKS-2708 (wceG2), which was downregulated 14.7-fold in the presence of EsaR in the qRT-PCR assays but only 4.2-fold in RNA-Seq analysis. Although the ratios of change in expression obtained via RNA-Seq and qRT-PCR differed slightly, all genes displayed similar trends in regulation (activation versus repression) (Fig. 3).
TABLE 4.
Highly regulated genes in the RNA-Seq data with a possible esa boxa
| ASAP no. | Locus tag (gene name) | Protein product | Fold regulationb | Promoter bound by EsaR |
|---|---|---|---|---|
| ACV-0290526 | CKS-0458 | Sigma-fimbria uncharacterized paralogous subunit | 11.5 (R) | Yes |
| ACV-0289766 | CKS-3373 (spy) | Envelope stress-induced periplasmic protein | 9.5 (A) | No |
| ACV-0288878 | CKS-4672 (yjbE) | Secreted protein | 7.7 (R) | Weakly |
| ACV-0287180 | CKS-1103 | Conidiation-specific protein, stress induced | 6.7 (R) | Yes |
| ACV-0290380 | CKS-0606 (yciF) | Uncharacterized DUF892 family protein | 5.7 (R) | Yes |
| ACV-0288895 | CKS-4689 | Conjugative transfer protein | 5.7 (R) | No |
| ACV-0288505 | CKS-0881 | Uncharacterized DUF1471 family protein | 5.7 (R) | Yes |
| ACV-0289538 | CKS-2247 (wceL) | Exopolysaccharide biosynthesis protein | 4.7 (R) | Yes |
| ACV-0289613 | CKS-2172 (lysP) | Lysine-specific permease | 4.4 (R) | No |
| ACV-0287610 | CKS-5070 (osmY) | Osmotically inducible protein | 4.3 (R) | Yes |
| ACV-0286879 | CKS-2708 (wceG2) | Undecaprenyl-phosphate galactose phosphotransferase | 4.2 (R) | Yes |
| ACV-0287347 | CKS-2413 (elaB) | Protein of unknown function DUF883 | 4.0 (R) | Yes |
Well-defined +1 site with a possible esa box nearby.
Fourfold or more regulation as determined by RNA-Seq data: A, activation; R, repression.
FIG 3.
Validating transcriptional control of putative EsaR direct targets. Changes in gene expression were compared between RNA-Seq analysis (light gray) and qRT-PCR assays (dark gray) of the 12 potential targets of EsaR assayed for direct regulation. The y axis represents the fold repression (>1) or activation (<1) in the presence of EsaR. CKS-3373 is the only gene seen to be activated in the subset. Data represent two experimental samples analyzed in triplicate. Error bars for qRT-PCR values denote standard error. Error bars for RNA-Seq ratios represent an estimate of the standard deviation across all genes in shown in Fig. 1.
EMSAs of putative EsaR direct targets.
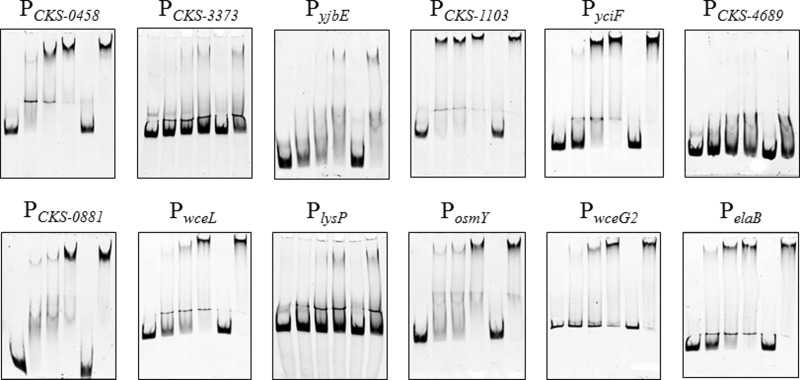
The 12 putative EsaR-controlled promoters (Table 4) were subsequently analyzed for direct binding using EMSAs with the same positive and negative controls described above. Eight promoters were shown to bind the His-MBP-glycine linker–EsaR fusion protein (HMGE) (19), and the shift was successfully outcompeted in the presence of a specific competitor, the 28-bp PesaR28 DNA fragment (8) (Fig. 4). The gene promoters that were directly bound are upstream of CKS-0458, CKS-1103, CKS-0606 (yciF), CKS-0881, CKS-2247 (wceL), CKS-5070 (osmY), CKS-2708 (wceG2), and CKS-2413 (elaB). Three promoters for the genes CKS-3373, CKS-4689, and CKS-2172 (lysP) did not bind HMGE. However, a fourth promoter for CKS-4672 (yjbE), PyjbE, did exhibit weak binding at the highest concentration of HMGE used. The promoter of CKS-3373, which failed to show any regulation in the qRT-PCR assays, also did not seem to be bound by EsaR. However, the promoter of elaB showed strong binding to the EsaR fusion protein, despite the lack of transcriptional regulation seen in qRT-PCR assays. This difference in assay output between EMSA and qRT-PCR had been observed previously for glpF (13).
FIG 4.
EMSAs of promoters most highly regulated by EsaR. EMSAs were performed with the EsaR fusion protein, HMGE. The promoter of dkgA (13) was used as a positive control, and a 32-bp pUC18 MCS DNA fragment was used as a negative control (Fig. 2). The concentration of FAM-labeled DNA probe in all lanes is 5 nM. The lanes within each panel consist of the following (left to right): DNA probe, DNA probe with 100 nM HMGE, DNA probe with 200 nM HMGE, DNA probe with 400 nM HMGE, DNA probe with 400 nM HMGE and 1 μM specific competitor PesaR28 DNA fragment, DNA probe with 400 nM HMGE and 1 μM nonspecific pUC18 MCS DNA fragment.
DISCUSSION
RNA-Seq analysis was performed on strains of P. stewartii DC283 proficient and deficient in QS to broaden the defined EsaR regulon. Transcriptome data consisting of 30 million reads for each strain were generated in a single RNA-Seq experiment and aligned to 58 scaffolds with 5,353 protein coding sequences in the v5b partial assembly of the P. stewartii genome. The complete DC283 genome is believed to consist of one chromosome and an estimated 11 plasmids (11). The alignment of reads to the annotated coding sequences was used to survey the quantitative levels of gene expression in the two strains, and the mapping of the reads to the genome scaffolds provided a visualization of the coverage of overlapping reads for each mRNA fragment. In particular, the alignment of reads to the genome allowed for estimation of putative +1 transcriptional start sites and the presence of any 3′ or 5′ untranslated sequence. The 260 annotated genes (see Table S1 in the supplemental material) and the 190 hypothetical genes (see Table S2) of the 5,353 protein coding genes that were differentially regulated 2-fold via QS represent approximately 8% of the annotated genome. This number may be low in comparison to some other plant pathogens (22), but it is perhaps not unsurprising, given that P. stewartii appears to use a limited arsenal of virulence factors (3).
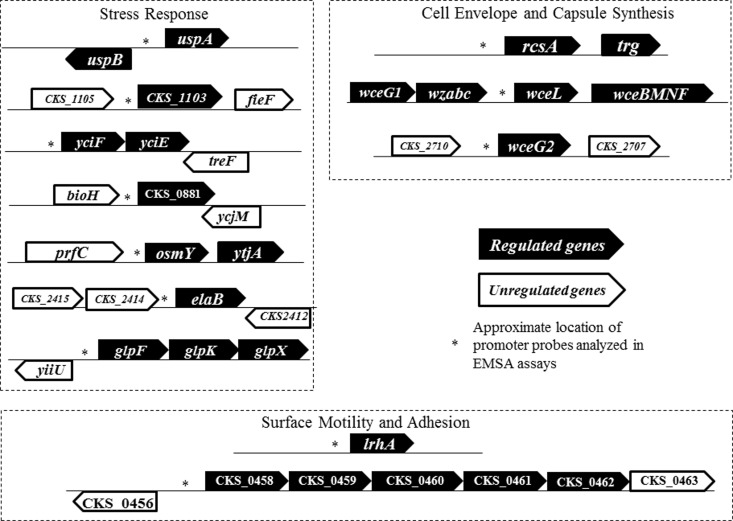
Subsequent validation and confirmation of genes directly regulated by EsaR, the master QS regulator in P. stewartii, were performed using qRT-PCR and EMSAs, respectively. In total, 10 promoters were newly identified as direct targets of EsaR. All but one of the promoters are repressed by EsaR, which could be the result of a bias during the selection process for putative targets. The genes putatively controlled by nine of these promoters can be broadly classified into the functional categories of cell envelope and capsule biosynthesis (WceL and WceG2), surface motility and adhesion (CKS-0458, CKS-0459, and CKS-0461), and stress response (CKS-1103, YciF, CKS-0881, OsmY, ElaB, UspA, and UspB) (Fig. 5). The remaining promoter, PCKS-0678, appears to regulate expression of the C-terminal domain of a threonine dehydratase, and the function and QS-dependent regulation of this partial enzyme are unclear.
FIG 5.
Genetic organization of the gene loci of P. stewartii directly regulated by EsaR. Gene architectures near nine of the promoters demonstrated to be directly bound by the EsaR fusion protein in this study and three promoters identified in previous studies (glpF, lrhA, and rcsA) are shown. Three EsaR direct targets with unknown physiological function, CKS-0678 encoding the C-terminal fragment of a threonine dehydratase, esaS encoding a small RNA, and dkgA encoding a 2,5-diketo gluconate reductase, are not included in the diagram. Also not included in the figure is the yjbEFGH operon, whose promoter is only weakly bound by EsaR. Differentially regulated genes from the RNA-Seq data are shown in black, and unregulated genes are shown in white.
With the addition of UspA as a directly regulated target, EsaR controls three regulators, RcsA, LrhA, and UspA, for each of the three broad functional classifications under which the majority of the directly regulated genes lie. Two putative 20-bp EsaR binding sites, those in the promoter of uspA and CKS-0678, were predicted using a position-specific weight matrix (PSWM) and the previously published consensus (Table 3). Multiple sequence alignments of the promoter regions for other direct targets did not yield a conclusive consensus sequence for EsaR binding, due to the complication of more than one possible binding site being identified in each promoter. Further experimental analysis of the binding site would aid in elucidating the exact bases necessary for binding of EsaR; however, such experiments are beyond the scope of this study.
EsaR indirectly represses the expression of stewartan via the RcsA-dependent pathway (23). Here, it is shown that EsaR directly regulates two additional genes involved in synthesis of capsule, wceL and wceG2, and possibly yjbE, whose promoter was only weakly bound by EsaR. WceL, a UDP-glucose-4-epimerase, and WceG2, undecaprenyl-phosphate UDP-galactose phosphotransferase, are two proteins expressed by genes in the stewartan I cluster (24) by promoters independent of the RcsA-controlled promoter. P. stewartii mutants of wceL are deficient in EPS production and proficient in adhesion (25). Synthesis of stewartan begins with WceG2, which has redundant function with WceG1 (26). Interestingly, wceG2 possesses a nearly 100-bp untranslated region in the RNA-Seq read alignment, with considerably higher expression than the coding region (data not shown), suggesting the presence of a small regulatory RNA that would add to the complexity of regulation by EsaR. Regulation of capsule production in P. stewartii appears to be an example of a coherent feed-forward loop, in which a master transcription factor, EsaR, regulates a downstream regulator, RcsA, and genes further down the cascade concurrently. Such kinetics are critical for filtering out noisy inputs in an environment while yet achieving rapid response (27). In P. stewartii, this multilayered control could help in varying the amounts of capsule produced by fine-tuning the expression of each target when appropriate environmental cues are received. The promoter of yjbE, although only weakly bound by EsaR, is still interesting to study (Fig. 4). YjbE is a predicted protein thought to play a role in exopolysaccharide production and overexpression of the yjbEFGH operon. These genes influence colony morphology and production of a non-colanic acid polysaccharide (28). P. stewartii possesses an intact yjbEFGH operon which is seen to be repressed by EsaR in our RNA-Seq experiments. Coincidentally, in Escherichia coli, the heterodimer RcsAB also binds to and activates expression from PyjbE, further suggesting a possible coherent feed-forward regulatory mechanism adopted by EsaR (28).
With regard to surface motility and adhesion, three of the most highly regulated genes seen in the RNA-Seq data, CKS-0458, CKS-0459, and CKS-0461, belong to a sigma-fimbriae operon whose expression is repressed in the presence of EsaR (Fig. 5). It is interesting that EsaR activates lrhA, encoding a predicted repressor of chemotaxis, motility, and fimbria expression (13). The timed expression of fimbriae may be critical to adhesion or biofilm formation in the xylem since both LrhA and EsaR seem to work together to ensure repression at low cell density. P. stewartii utilizes surface motility at high cell densities to form a biofilm and spread systematically in the xylem. Its motility is dependent on expression of flagella and stewartan (29). Perhaps the expression of the sigma-fimbria operon at high cell density within the xylem aids in adhesion of the biofilm to the lumen. Of the many flagellum-associated proteins that P. stewartii possesses, levels of fliC (flagellin subunit), fliD (flagellar hook-associated protein), and fliR (flagellar biosynthesis protein) are repressed more than 2-fold at low cell densities in the presence of EsaR in the RNA-Seq data, further supporting this hypothesis.
The majority of the genes directly regulated by EsaR are likely involved in stress response. UspA, the universal stress global response regulator, was repressed by EsaR, along with five other proteins. Based on studies in E. coli, CKS-1103, yciF, osmY, and elaB may be part of a regulon controlled by the alternative sigma factor for stress, σS (30). How EsaR affects the expression of σS in P. stewartii is not known. However, rpoS was seen to be repressed 1.9-fold in the RNA-Seq data set. CKS-1103 is a 55-amino-acid-long peptide that is induced in response to stress and shares 71% identity with YciG of E. coli (30). YciF is an uncharacterized protein belonging to the DUF892 family that is putatively involved in the stress response and is part of the σS regulon (30). Expression of yciF was repressed in a mutant of the master QS regulator MqsR in E. coli, although via an autoinducer 2-dependent pathway which P. stewartii is not known to possess (31). OsmY is a periplasmic protein that is induced under hyperosmotic stress and is under complex regulation by many global regulators in E. coli (32). EsaR was shown to repress production of OsmY in previous proteomic studies (13). ElaB, another protein thought to be in the σS regulon, is an inner membrane protein associated with the ribosomes whose function is unknown (33). Interestingly, elaB expression is under regulation by QS in another plant-associated microbe. A moderate change in expression of elaB has been observed in response to N-octanoylhomoserine lactone (C8-HSL)-induced QS regulation in Burkholderia ambifaria, a bacterium found normally in the pea rhizosphere (34).
Independent of the RpoS regulon, the remaining two proteins that could be involved in stress response in P. stewartii are CKS-0881 and UspA. CKS-0881 is a stress-induced protein that shares 35% identity with YhcN, a hydrogen peroxide stress-induced protein of E. coli that is involved in biofilm formation (35). The gene uspA codes for the universal stress global response regulator and is divergently transcribed from uspB, encoding the predicted universal stress response protein B. Universal stress proteins (USPs) are overproduced in response to a variety of stresses and afford the organism a mechanism to survive under unsuitable conditions, though by unknown pathways (36). EsaR represses production of UspA and other stress response genes at low cell density, tamping down the apparent stress response of P. stewartii when it is present in the leaf tissue during early stages of infection.
The studies described in the manuscript were performed using the then-available version 5b (v5b) of the P. stewartii DC283 genome on the ASAP website (9). During the course of the analysis, a newer, more complete version of the P. stewartii genome was released. Version 8 (v8) consists of 4,901 protein-coding DNA sequences (CDS), which includes 19 newly annotated CDS and removes 471 previously annotated CDS (mostly hypothetical proteins or pseudogenes). In addition, the annotation of a number of hypothetical proteins has been updated. Upon repeating the analysis with the newly available v8 of the genome, a few additional putative EsaR direct targets were identified that this study may have missed. For example, CKS-1810 (yebE), an uncharacterized DUF533 family protein that was previously annotated as a hypothetical protein, was upregulated 7.8-fold in the QS-proficient strain. In addition, three putative proteins were recognized to be regulated 4-fold or higher and may be of future interest. CKS-4376, coding for a putative transcriptional regulator, and CKS-2108, coding for a putative type I secretion protein, are repressed by EsaR, and CKS-2806, coding for a putative formate dehydrogenase oxidoreductase protein, was activated by EsaR.
In summary, genes relevant to three physiological responses, capsule and cell envelope biosynthesis, surface motility and adhesion, and stress response, were seen to be regulated by QS in P. stewartii in culture medium. Ten promoters were identified to be direct targets of EsaR, considerably broadening the defined QS-regulon of P. stewartii. Three second-tier regulators may also be involved in the three major physiological responses that are directly regulated by EsaR, suggesting a possible coherent feed-forward mechanism of regulation control that may permit a fine-tuned response to different environmental inputs. Analysis of the QS-controlled transcriptome in planta and of the function of the EsaR direct and indirect targets will further elucidate the role of these regulatory networks in plant pathogenesis. Such information could be useful in the development of disease prevention strategies in maize and perhaps be adaptable to other important crops affected by bacterial wilt diseases.
Supplementary Material
ACKNOWLEDGMENTS
We thank Susanne von Bodman for providing strains and plasmids, members of the Pantoea stewartii subsp. stewartii DC283 whole-genome shotgun (WGS) project at the University of Wisconsin for making the genome sequence available prior to publication, and Blakely Sproles for technical assistance in plasmid construction.
This work was supported by National Science Foundation grant MCB-0919984 (A.M.S.), Virginia Tech Graduate Student Association Graduate Research Development Program award (R.R.), and a Biological Sciences Graduate Student Development Award fellowship (R.R.).
Footnotes
Published ahead of print 11 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01489-14.
REFERENCES
- 1.Pataky J. 2003. Stewart's wilt of corn. APSnet Feature 2003:0703. 10.1094/APSnetFeature-2003-0703 [DOI] [Google Scholar]
- 2.Ham JH, Majerczak D, Ewert S, Sreerekha M-V, Mackey D, Coplin D. 2008. WtsE, an AvrE-family type III effector protein of Pantoea stewartii subsp. stewartii, causes cell death in non-host plants. Mol. Plant Pathol. 9:633–643. 10.1111/j.1364-3703.2008.00489.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roper MC. 2011. Pantoea stewartii subsp. stewartii: lessons learned from a xylem-dwelling pathogen of sweet corn. Mol. Plant Pathol. 12:628–637. 10.1111/j.1364-3703.2010.00698.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sasu MA, Seidl-Adams I, Wall K, Winsor JA, Stephenson AG. 2010. Floral transmission of Erwinia tracheiphila by cucumber beetles in a wild Cucurbita pepo. Environ. Entomol. 39:140–148. 10.1603/EN09190 [DOI] [PubMed] [Google Scholar]
- 5.Crossman L, Dow JM. 2004. Biofilm formation and dispersal in Xanthomonas campestris. Microbes Infect. 6:623–629. 10.1016/j.micinf.2004.01.013 [DOI] [PubMed] [Google Scholar]
- 6.Nunney L, Yuan X, Bromley R, Hartung J, Montero-Astúa M, Moreira L, Ortiz B, Stouthamer R. 2010. Population genomic analysis of a bacterial plant pathogen: novel insight into the origin of Pierce's disease of grapevine in the U.S. PLoS One 5:e15488. 10.1371/journal.pone.0015488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peeters N, Guidot A, Vailleau F, Valls M. 2013. Ralstonia solanacearum, a widespread bacterial plant pathogen in the post-genomic era. Mol. Plant Pathol. 14:651–662. 10.1111/mpp.12038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minogue TD, Wehland-von Trebra M, Bernhard F, von Bodman SB. 2002. The autoregulatory role of EsaR, a quorum-sensing regulator in Pantoea stewartii ssp. stewartii: evidence for a repressor function. Mol. Microbiol. 44:1625–1635. 10.1046/j.1365-2958.2002.02987.x [DOI] [PubMed] [Google Scholar]
- 9.Glasner JD. 2003. ASAP, a systematic annotation package for community analysis of genomes. Nucleic Acids Res. 31:147–151. 10.1093/nar/gkg125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beck von Bodman S, Farrand SK. 1995. Capsular polysaccharide biosynthesis and pathogenicity in Erwinia stewartii require induction by an N-acylhomoserine lactone autoinducer. J. Bacteriol. 177:5000–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coplin DL, Rowan RG, Chisholm DA, Whitmoyer RE. 1981. Characterization of plasmids in Erwinia stewartii. Appl. Environ. Microbiol. 42:599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.von Bodman SB, Majerczak DR, Coplin DL. 1998. A negative regulator mediates quorum-sensing control of exopolysaccharide production in Pantoea stewartii subsp. stewartii. Proc. Natl. Acad. Sci. U. S. A. 95:7687–7692. 10.1073/pnas.95.13.7687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramachandran R, Stevens AM. 2013. Proteomic analysis of the quorum-sensing regulon in Pantoea stewartii and identification of direct targets of EsaR. Appl. Environ. Microbiol. 79:6244–6252. 10.1128/AEM.01744-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJL. 2009. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25:1422–1423. 10.1093/bioinformatics/btp163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 16.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mane SP, Evans C, Cooper KL, Crasta OR, Folkerts O, Hutchison SK, Harkins TT, Thierry-Mieg D, Thierry-Mieg J, Jensen RV. 2009. Transcriptome sequencing of the microarray quality control (MAQC) RNA reference samples using next generation sequencing. BMC Genomics 10:264. 10.1186/1471-2164-10-264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schu DJ, Carlier AL, Jamison KP, von Bodman SB, Stevens AM. 2009. Structure/function analysis of the Pantoea stewartii quorum-sensing regulator EsaR as an activator of transcription. J. Bacteriol. 191:7402–7409. 10.1128/JB.00994-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hertz GZ, Stormo GD. 1999. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 15:563–577. 10.1093/bioinformatics/15.7.563 [DOI] [PubMed] [Google Scholar]
- 22.Liu H, Coulthurst SJ, Pritchard L, Hedley PE, Ravensdale M, Humphris S, Burr T, Takle G, Brurberg MB, Birch PR, Salmond GP, Toth IK. 2008. Quorum sensing coordinates brute force and stealth modes of infection in the plant pathogen Pectobacterium atrosepticum. PLoS Pathog. 4:e1000093. 10.1371/journal.ppat.1000093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carlier AL, von Bodman SB. 2006. The rcsA promoter of Pantoea stewartii subsp. stewartii features a low-level constitutive promoter and an EsaR quorum-sensing-regulated promoter. J. Bacteriol. 188:4581–4584. 10.1128/JB.00211-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Yang F, von Bodman SB. 2012. The genetic and structural basis of two distinct terminal side branch residues in stewartan and amylovoran exopolysaccharides and their potential role in host adaptation. Mol. Microbiol. 83:195–207. 10.1111/j.1365-2958.2011.07926.x [DOI] [PubMed] [Google Scholar]
- 25.Koutsoudis MD, Tsaltas D, Minogue TD, von Bodman SB. 2006. Quorum-sensing regulation governs bacterial adhesion, biofilm development, and host colonization in Pantoea stewartii subspecies stewartii. Proc. Natl. Acad. Sci. U. S. A. 103:5983–5988. 10.1073/pnas.0509860103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carlier A, Burbank L, von Bodman SB. 2009. Identification and characterization of three novel EsaI/EsaR quorum-sensing controlled stewartan exopolysaccharide biosynthetic genes in Pantoea stewartii ssp. stewartii. Mol. Microbiol. 74:903–913. 10.1111/j.1365-2958.2009.06906.x [DOI] [PubMed] [Google Scholar]
- 27.Mangan S, Zaslaver A, Alon U. 2003. The coherent feedforward loop serves as a sign-sensitive delay element in transcription networks. J. Mol. Biol. 334:197–204. 10.1016/j.jmb.2003.09.049 [DOI] [PubMed] [Google Scholar]
- 28.Ferrieres L, Aslam SN, Cooper RM, Clarke DJ. 2007. The yjbEFGH locus in Escherichia coli K-12 is an operon encoding proteins involved in exopolysaccharide production. Microbiology 153:1070–1080. 10.1099/mic.0.2006/002907-0 [DOI] [PubMed] [Google Scholar]
- 29.Herrera CM, Koutsoudis MD, Wang X, Bodman SB von. 2008. Pantoea stewartii subsp. stewartii exhibits surface motility, which is a critical aspect of Stewart's wilt disease development on maize. Mol. Plant Microbe Interact. 21:1359–1370. 10.1094/MPMI-21-10-1359 [DOI] [PubMed] [Google Scholar]
- 30.Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187:1591–1603. 10.1128/JB.187.5.1591-1603.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.González Barrios AF, Zuo R, Hashimoto Y, Yang L, Bentley WE, Wood TK. 2006. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022). J. Bacteriol. 188:305–316. 10.1128/JB.188.1.305-316.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yim HH, Brems RL, Villarejo M. 1994. Molecular characterization of the promoter of osmY, an rpoS-dependent gene. J. Bacteriol. 176:100–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshida H, Maki Y, Furuike S, Sakai A, Ueta M, Wada A. 2012. YqjD is an inner membrane protein associated with stationary-phase ribosomes in Escherichia coli. J. Bacteriol. 194:4178–4183. 10.1128/JB.00396-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chapalain A, Vial L, Laprade N, Dekimpe V, Perreault J, Déziel E. 2013. Identification of quorum sensing-controlled genes in Burkholderia ambifaria. Microbiologyopen 2:226–242. 10.1002/mbo3.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee J, Hiibel SR, Reardon KF, Wood TK. 2010. Identification of stress-related proteins in Escherichia coli using the pollutant cis-dichloroethylene. J. Appl. Microbiol. 108:2088–2102. 10.1111/j.1365-2672.2009.04611.x [DOI] [PubMed] [Google Scholar]
- 36.Tkaczuk KL, Shumilin IA, Chruszcz M, Evdokimova E, Savchenko A, Minor W. 2013. Structural and functional insight into the universal stress protein family. Evol. Appl. 6:434–449. 10.1111/eva.12057 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.