Abstract
Many insects are associated with heritable symbionts that mediate ecological interactions, including host protection against natural enemies. The cowpea aphid, Aphis craccivora, is a polyphagous pest that harbors Hamiltonella defensa, which defends against parasitic wasps. Despite this protective benefit, this symbiont occurs only at intermediate frequencies in field populations. To identify factors constraining H. defensa invasion in Ap. craccivora, we estimated symbiont transmission rates, performed fitness assays, and measured infection dynamics in population cages to evaluate effects of infection. Similar to results with the pea aphid, Acyrthosiphon pisum, we found no consistent costs to infection using component fitness assays, but we did identify clear costs to infection in population cages when no enemies were present. Maternal transmission rates of H. defensa in Ap. craccivora were high (ca. 99%) but not perfect. Transmission failures and infection costs likely limit the spread of protective H. defensa in Ap. craccivora. We also characterized several parameters of H. defensa infection potentially relevant to the protective phenotype. We confirmed the presence of H. defensa in aphid hemolymph, where it potentially interacts with endoparasites, and performed real-time quantitative PCR (qPCR) to estimate symbiont and phage abundance during aphid development. We also examined strain variation of H. defensa and its bacteriophage at multiple loci, and despite our lines being collected in different regions of North America, they were infected with a nearly identical strains of H. defensa and APSE4 phage. The limited strain diversity observed for these defensive elements may result in relatively static protection profile for this defensive symbiosis.
INTRODUCTION
Many, if not most, insect species are infected with maternally transmitted bacterial symbionts capable of exerting major effects on host biology (1–5). While some heritable symbiont infections are required for their insect host's survival and reproduction, e.g., the bacteriocyte-associated nutritional symbionts found in sap-feeding insects (6), the majority represent facultative infections (4). Many facultative symbionts mediate important ecological interactions, and there is an emerging awareness that infection can protect hosts from a range of environmental threats (3, 7–9). Both theory and experimental evidence indicate that protective benefits likely contribute to the spread of heritable symbionts within host populations (10–13). Heritable facultative symbiont infections, however, are often found at intermediate frequencies in natural populations, and infection costs and transmission failures are potentially important in limiting their spread (9).
The heritable symbionts of the pea aphid Acyrthosiphon pisum are among the best studied (14). All Ac. pisum aphids possess the obligate nutritional symbiont Buchnera aphidicola (1, 15), and many harbor one or more facultative symbionts (14). Aphids are ideal for studying the effects of heritable infections, because facultative symbionts can be manipulated among clonal lineages, resulting in experimental aphid lines that, for example, share the same genetic background but vary in terms of infection by particular symbionts (14). Such experimental studies with Ac. pisum have identified diverse benefits to infection with common facultative symbionts, including protection against heat stress (16–18), fungal pathogens (19–21), and parasitoid wasps (22–24). Despite these benefits, the heritable protective symbionts of Ac. pisum typically remain at intermediate frequencies among surveyed populations (for examples, see references 25 to 31). For example, a survey of Hamiltonella defensa, which defends against parasitoids, reported infection frequencies ranging from ca. 30 to 60% in three North American Medicago populations (27). In laboratory-reared Ac. pisum, vertical transmission rates of the common gammaproteobacterial facultative symbionts approach 100% (32–34). Inheritance failures under natural conditions and during the overwintering egg stage potentially influence symbiont frequencies, but rates of loss are unknown. Clear costs to infections with protective bacteria, including Hamiltonella defensa, have been difficult to identify for particular symbionts in Ac. pisum (9). Studies of standard fitness parameters (e.g., fecundity and development time) comparing aphids of the same genotype with and without H. defensa, which protects against parasitoids, find no consistent costs to infection and in fact often report benefits (for examples, see references 13 and 18). Population cage studies, however, identify benefits to infection in the presence of parasitoid wasps but costs in control cages lacking enemies (13), suggesting that fitness tradeoffs contribute to the maintenance of H. defensa at intermediate frequencies in natural populations.
Many of the symbionts infecting Ac. pisum are found in other aphid species and in other insects more generally, yet their roles in other hosts are less well known. H. defensa, for example, is estimated to occur in 14% of aphid species (14). Recent work has shown that this symbiont confers protection against parasitoids in two other aphid hosts: the black bean aphid, Aphis fabae (35), with which costs to infection have also been found (36), and the cowpea aphid, Aphis craccivora (37, 38). Interestingly, in Ap. craccivora, H. defensa protects only against some parasitoid species, completely eliminating parasitism by two Binodoxys species but having no effect on two other aphidiine braconids, Aphidius colemani and Lysiphlebus orientalis (37). These findings, together with results from Ac. pisum, indicate that host protection may be a common phenotype associated with H. defensa, although there have been strains identified in both the grain aphid, Sitobion avenae (39), and possibly Ac. pisum (40) that do not confer host protection.
Although the mechanisms underlying H. defensa-based protection are unknown, temperate bacteriophages, called APSEs (Ac. pisum secondary endosymbionts), are required to produce the protective phenotype in Ac. pisum (22, 41–43). Seven APSE variants have been described, including three from Ac. pisum (APSE1 to APSE3) and one from Ap. craccivora (APSE4) (41). While no in vitro assays confirm that phage toxins kill developing parasitoids, APSE2s, which encode a homolog of CdtB (cytolethal distending toxin), are associated with H. defensa strains conferring moderate levels of protection, and strains carrying YDp (putative toxin)-encoding APSE3s receive high levels of protection (22, 23, 41). A strain of H. defensa from Ap. craccivora was transferred into Ac. pisum, on which it conferred moderate protection against the wasp Aphidius ervi (23). The APSE4 haplotype from this H. defensa strain (5ATac) is the only Ap. craccivora APSE characterized to date and contains Shiga-like toxin homologs (as do APSE1 and APSE5, from Ac. pisum and Uroleucon rudbeckiae, respectively) including a possible functional correlate of StxA, the cytotoxic alpha unit (41).
The purpose of this study was to investigate infection costs and maternal transmission efficiency of H. defensa infecting Ap. craccivora as factors constraining the spread of this symbiont in natural populations, given that this symbiont confers protection against parasitoids (37) yet remains at intermediate frequencies (0 to 85%) in natural populations (44, 45). In addition, we sought to characterize certain attributes of H. defensa infecting Ap. craccivora that may be important for the protective phenotype. We determined phage variant and putative toxins associated with the strain already known to confer protection in the native host, verified that this strain also persists extracellularly in the hemolymph (which may be necessary to protect against endoparasitic wasps), and estimated symbiont and APSE abundances across aphid development.
MATERIALS AND METHODS
Experimental aphids.
Ap. craccivora is a cosmopolitan polyphagous crop pest of legumes (46–48). Though there have been reports of sexual morphs (49), Ap. craccivora aphids are thought to be primarily anholocyclic, i.e., only reproducing parthenogenetically, in northern latitudes, dying off during the winter and recolonizing temperate areas via migration each season (46, 50, 51).
We reared Ap. craccivora aphids field collected from alfalfa plants (Medicago sativa) on Vicia faba (fava bean), which is a suitable food source regardless of the host plant from which this aphid is collected (52). Clonal lines were initiated from single parthenogenetic Ap. craccivora females (Table 1), and each line was held in replicate cup cages (inverted Solo [Lake Forest, IL] clear plastic cups vented with a mesh top over a 3.5-in. plant pot) in biological incubators (Percival Scientific, Inc., Perry, IA) at both 20 and 25°C with a 16-h:8-h light-dark cycle. The long-day conditions imitate summer photoperiod to ensure maintenance of parthenogenetic reproduction.
TABLE 1.
Collection data for Ap. craccivora clones used in the present studya
| Clone | Location | Year |
|---|---|---|
| AC1 | Lexington, KY | 2010 |
| AC17 | Tucson, AZ | 2009 |
| AC21 | Tucson, AZ | 2009 |
| LL1 | Lexington, KY | 2011 |
| SV1 | Harrodsburg, KY | 2011 |
The host plant in all cases was Medicago sativa. DGGE confirmed the presence of H. defensa only in all clones.
Determination of infection status and creation and genotyping of experimental aphid lines.
Diverse and sometimes unexpected symbiont lineages have been discovered within field-collected aphids (for examples, see references 27 and 53 to 55). To ensure the presence of only H. defensa in our experimental lines, we used a universal 16S rRNA screening technique called denaturing gradient gel electrophoresis (DGGE) to detect all bacteria present. DNA extractions were conducted using the E.Z.N.A. insect DNA isolation kit (Omega Bio-Tek; Norcross, GA) by following the manufacturer's instructions, and DNA concentration was measured with a NanoDrop spectrophotometer (Thermo Scientific). Extractions with >15 ng/μl were stored at 4°C (if used for testing within 3 months) or −20°C.
All Ap. craccivora clones used in this study were screened with DGGE primers 356F, with a stabilizing GC-clamp (5′-GC CGC CCG CCG CGC CCC GCG CCC GTC CCG CCG CCC CCG CCC CCC TAC GGG AGG CAG CAG-3′), and 517R (5′-ATT ACC GCG GCT GCT GG-3′), which amplify the variable V3 region of 16S (56). PCRs with 20 μl were conducted with a cocktail of PCR-grade water, 10× PCR buffer (200 mM Tris-HCl [pH 8.4], 500 mM KCl), MgCl2 (1.5 mM), deoxynucleoside triphosphates (dNTPs; 200 μM), APEX HotStart Taq DNA polymerase (5 U/μl; Genesee Scientific, San Diego, CA), forward and reverse primers (5 μM each), and a DNA template (∼300 ng) under the following conditions: 95°C for 15 min to activate the enzyme; 20 cycles of 94°C for 1 min, a 65 to 55°C touchdown for 1 min (−0.5°C/cycle), and 72°C for 1 min; 25 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min; and, finally, 72°C for 8 min, followed by a 4°C hold (56). 16S PCR amplicons were run at 70 V for 16.5 h (60°C) on 6.5% acrylamide gels containing a 40 to 65% denaturing gradient of 7 M urea and 40% (vol/vol) formamide in a CBS Scientific DGGE rig (Del Mar, CA). Gel bands were excised, eluted in 50 μl of PCR-grade water, and incubated at 37°C for 30 min for template use for repeating the DGGE PCR. These PCR products were cleaned with the Omega Bio-Tek E.Z.N.A. Cycle Pure kit per the manufacturer's instructions and sequenced by Eurofins MWG Operon (Huntsville, AL).
We initially screened 10 Ap. craccivora lines and confirmed the presence of only H. defensa (i.e., no coinfections) with DGGE and diagnostic PCR. Five of these H. defensa lines were used for subsequent experimental assays (Table 1). Using a technique modified from reference 57, cohorts in three aphid lines (AC1, SV1, and LL1) were selectively cured of H. defensa. Briefly, 10 4th-instar aphids from each clonal line fed on an artificial diet (58) mixed with an antibiotic cocktail of 50 μg/ml each of gentamicin, cefotaxime, and ampicillin for 3 days. We pipetted the antibiotic-treated diet into a small polystyrene petri dish (35-mm diameter) covered with Parafilm M stretched thoroughly across the dish's opening. This dish was inverted and placed in a larger petri dish so that aphids could feed through the membrane via their stylet. Clonal lines were maintained on separate petri dishes, and after 3 days, aphids were individually placed on V. faba plants, offspring were collected, and infection absence was confirmed using PCR for H. defensa. All experimental lines were started from single parthenogenetic females, and those cured were designated with an “ab” following their clone names (e.g., AC1ab). Cured lines were not used in any experiment until at least 10 generations had passed to eliminate any residual effects from the antibiotic treatment (59), and lines were retested with diagnostic PCR prior to each experimental assay.
Given the anholocyclic reproduction of this aphid in North America, we conducted microsatellite genotyping using cross-species amplification of loci from related aphids (60, 61) to determine if discernible genetic variation was present among the five lines considered in this study. Eight microsatellite loci (see Table 2 for loci used and primer sequences) were PCR amplified with Dye Set-30 (DS-30) fluorescent primers using a touchdown reaction as follows: 94°C for 3 min; 45 cycles of 95°C for 30 s, 68 to 56°C touchdown for 13 cycles and then 55°C for 32 cycles, each cycle for 30 s, followed by 72°C for 30 s; and then 72°C for 6 min and a 4°C hold. The PCR samples were then sent to The Georgia Genomics Facility for fluorescent genotyping analysis on an Applied Biosystems 3730xl DNA analyzer, using the ROX500 size standard, and then analyzed using Geneious 6.1. We found at least one allelic difference per line, indicating that the lines comprise distinct clonal lineages (Table 3).
TABLE 2.
Oligonucleotide primers used in this study
| Locus | Primers (5′–3′)a | Reference(s) |
|---|---|---|
| Microsatellite loci | ||
| Ago53 | F: TGACGAACGTGGTTAGTCGT | 62 |
| R: TGACGAACGTGGTTAGTCGT | ||
| Ago89 | F: GAACAGTGCTCGCAGTCTAT | 62 |
| R: GACAGCGTAAACATCGCGGT | ||
| Ago59 | F: GCGAGTGGTATTCGCTTAGT | 62 |
| R: GTTACCCTCGACGATTGCGT | ||
| Ago66 | F: TCGGTTTGGCAACGTCGGGC | 62 |
| R: GACTAGGGAGATGCCGGCGA | ||
| Ago24 | F: TTTTCCCGGCACACCGAGT | 62 |
| R: GCCAAACTTTACACCCCGC | ||
| R5.10 | F: CGACTAAGCTTAATATTGTTTG | 63 |
| R: CGGTTCGGAGAACATAAGAG | ||
| S23 | F: GGTCCGAGAGCATTCATTAGG | 64 |
| R: CGTCGTTGTCATTGTCGTCG | ||
| s17b | F: TTCTGGCTTCATTCCGGTCG | 64 |
| R: CGTCGCGTTAGTGAACCGTG | ||
| APSE variable cassette region | ||
| APSE4 P5 to P8 | F: AGACATGGACCCCGAGGTGA | Modified from references 41 and 43 |
| R: TCGCCTACTACAATACCTACCTGGC | ||
| APSE4 P8 and P9 | F: GCGCTAGTGCTTGTTTTAGGCGG | Modified from references 41 and 43 |
| R: GCCGGGATCATCTGCTCTTTCGC | ||
| APSE4 P9 and P10 | F: ACCTGGACCCATCAAAGAGAGTTCA | Modified from references 41 and 43 |
| R: AACGTAGCAAGGCCAGGCGG | ||
F, forward; R, reverse.
TABLE 3.
Allele sizes for five aphid lines across eight microsatellite loci
| Line | Allele size(s) (bp) at each diploid locusa |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ago53 |
Ago89 |
Ago24 |
Ago59 |
R5.10 |
Ago66 |
S23 |
S17b |
|||||||||
| Allele 1 | Allele 2 | Allele 1 | Allele 2 | Allele 1 | Allele 2 | Allele 1 | Allele 2 | Allele 1 | Allele 2 | Allele 1 | Allele 2 | Allele 1 | Allele 2 | Allele 1 | Allele 2 | |
| AC1 | 114 | 171 | 148 | 144 | 246 | 134 | 164 | 137 | 141 | |||||||
| AC17 | 114 | 171 | 146 | 144 | 246 | 151 | 153 | 164 | 137 | 141 | ||||||
| AC21 | 114 | 171 | 148 | 144 | 246 | 151 | 153 | 164 | 137 | 141 | ||||||
| LL1 | 114 | 171 | 146 | 144 | 246 | 151 | 153 | 164 | 137 | 139 | ||||||
| SV1 | 114 | 171 | 144 | 246 | 151 | 153 | 164 | 137 | 141 | |||||||
Empty cells indicate homozygosity.
Estimating maternal transmission efficiency.
To estimate vertical transmission rates of H. defensa in Ap. craccivora, a total of 601 3rd-instar offspring from 57 mothers from four A. craccivora lines (AC1, AC21, AC17, and SV1) known to be infected were screened with diagnostic PCR for the presence of H. defensa. Most (n = 430) were reared at 25°C, but some (n = 171) were evaluated at 20°C to determine if temperature had large effects on transmission rates. A subset (n = 341) of offspring were also screened for APSE presence. All cohorts were started with 4th-instar aphids individually placed in petri dishes with freshly clipped V. faba leaves and allowed to reproduce for several days. Offspring were sampled every 2 to 3 days until the mother died. Third-instar offspring were collected in 0.2-μl test tubes, and DNA was prepared with the “squish extraction” method adapted from reference 65: individual whole aphids were ground, with a pipette tip, in a lysis buffer (10 mM Tris-Cl [pH 8.2], 1 mM EDTA, 25 mM NaCl) containing proteinase K (20 mg/ml) and incubated at 40°C for 35 min, then incubated at 95°C for 2 min 30 s, and held at 4°C until use. Diagnostic PCR was conducted within 1 week of squish extractions.
To assess H. defensa presence, we amplified a fragment of the H. defensa dnaK gene using primers T70F2 (5′-GGT TCA GAA AAA AGT GGC AG-3′) and T70R2 (5′-CGA GCG AAA GAG TGA-3′), and for APSE we amplified a portion of the P28 gene using primers APSEP28F (5′-TGA TAA AAG CGG ATA ATG CC-3′) and APSEP28R (5′-GCG TTT GTC ATA CTG AAA AGG-3′) (42). We confirmed extraction viability of any samples not found to be infected with H. defensa by amplifying fragments of the aphid gene ef1α (primers ACef1aF [5′-CCG TGG AGA TGC ACC ACG AAG C-3′] and ACef1aR [5′-AGC AGC TCC CTT GGG TGG GT-3′]) and Buchnera gene dnaK (primers 215F [5′-CCA ACA GCT GCG GCA CTT GC-3′] and 216R [5′-TCA CCT CCA AGA TGG GTG TCT CCA-3′]). All diagnostic PCRs were performed on a Roche LightCycler 480 II. Each 10-μl reaction mixture contained 5 μl of SYBR green I Master (Molecular Probes, Inc., Eugene, OR), 1 μl (5 μM) each of forward and reverse primers, 2 μl of PCR-grade water, and 1 μl of DNA template. PCR included 95°C for 5 min; 45 cycles at 95°C for 10 s, annealing temperature decreases from 68°C to 55°C at 18°C increments, and 72°C for 10 s; a melting curve at 95°C for 5 s, 65°C for 1 min, and then 97°C; and a final hold at 40°C. Each PCR run also included positive and negative controls.
Identifying potential fitness costs in H. defensa-infected Ap. craccivora.
Using genetically controlled experimental Ap. craccivora lines, we conducted fitness assays to determine whether H. defensa infection influenced development time, fecundity, size, and survival (as described in references 18 and 26). All fitness assays were performed at both 20 and 25°C, and within each incubator, cohort cup cages were kept on the same tray and routinely rotated to reduce potential positional effects. To further control for variation, all experiments were started with same-aged (±3 h) aphids on ∼2-week-old V. faba plants.
We first compared Ap. craccivora development time as measured from the time of birth (±3 h) to first reproduction (TFR) and adult fresh weight (FW) at first reproduction using three aphid lines with H. defensa relative to their uninfected counterparts sharing the same genetic background (AC1 versus AC1ab, SV1 versus SV1ab, and LL1 versus LL1ab). Starting 4 days after the onset of the assay, cup cages were monitored at 2-h intervals for reproduction, at which point the TFR was noted and individual adults were immediately weighed on a microscale.
We then assessed cumulative fecundity and survivorship for experimental lines (SV1 versus SV1ab and AC1 versus AC1ab). These assays were not conducted with the experimental pair LL1 and LL1ab due to loss of H. defensa infection in LL1, likely due to transmission failure (see “Maternal transmission loss in Ap. craccivora” below). Five nymphs (48 ± 3 h) from each aphid line were placed per V. faba cup cage (n = 8) at both 20 and 25°C. Once aphids reached adulthood, the numbers of offspring and adults (alive and dead) were monitored at 2- to 3-day intervals. All offspring were removed to prevent them from developing to adults and contributing to offspring total, and the best efforts were made to avoid disturbing adult aphids. Cage bottoms were lined with Fisher weighing paper (4 by 4 in.) to catch dropping aphids and prevent them from falling into the soil, reducing mortality for fallen aphids and resulting in more accurate counts and easier offspring monitoring. Due to plant senescence, adult aphids were carefully transferred to a fresh plant approximately 15 days into the assay.
Population cages.
To determine if costs to infection were more apparent in the presence of competition, we conducted population cage experiments. Due to the labor-intensive nature of population cage experiments, we used a single aphid background (AC1) with and without H. defensa since this is the line known to confer resistance to parasitoids. Three replicate cages (BugDorm; 25 cm by 25 cm by 25 cm) were maintained at both 20 and 25°C (n = 6 total), each containing four V. faba plants of similar ages and sizes. All cages were initiated with an equal number (80) of same-aged 3rd-instar aphid nymphs in one clonal background, but 50% were infected with H. defensa (AC1), and 50% were uninfected (AC1ab). By following protocols outlined in reference 13, we then estimated H. defensa infection frequency over time by sampling 60 3rd-instar nymphs at regular intervals. Care was taken to collect aphids from all plant parts within each cage to provide a representative sample. Due to the amount of time required to perform DNA extractions, sampling was staggered over 2 days where samples from 25°C cages were collected and extracted on day 1 and samples from 20°C cages were processed on day 2. DNA extractions were performed using the squish extraction protocol, and diagnostic PCR for H. defensa infection status was performed using the dnaK primers (T70F2 and T70R2), PCR cocktail, and reaction conditions outlined above. Also as described above, individuals testing negative for H. defensa were screened for Buchnera to ensure that extractions were viable. The 25°C cages were tested at eight time points, but the 20°C cages had lower population sizes and were tested at six time points so that sampling itself would not have large effects on aphid abundance.
Characterization of H. defensa strains and phage variants in experimental aphid lines.
To characterize the H. defensa strains and APSE variants used in this study, we adapted the multilocus approach developed by Degnan and Moran (66). We examined six H. defensa housekeeping genes (accD, dnaA, hrpA, recJ, ptsI, and rpoS) and two genes involved with the type three secretion system (invC and spaP) as well as APSE genes (P45, P3, P35, P41, and P51) in six aphid lines. Primer sequences and reaction parameters can be found in reference 66. We also PCR amplified and sequenced portions of the toxin-encoding APSE variable cassette regions (VCR) thought to contribute to wasp mortality. Primers were designed with Geneious Pro V.5.4.6 (Biomatters, Ltd.), which uses the Primer3 engine, to amplify portions flanking conserved core genes (e.g., P3) of the APSE genome (41). A portion of this VCR of APSE was sequenced from region P5 to P10 (see Table 2 for primer sequences).
We conducted PCRs with 30-μl mixtures containing 15 μl of 2× GoTaq Hot Start Colorless master mix (Promega, Madison, WI) (2× Colorless GoTaq reaction buffer [pH 8.5], 400 μM each dNTP, and 4 mM MgCl2), 5 μM (each) forward and reverse primers, 7.5 μl of PCR-grade water, and 1.5 μl of DNA template (∼9 to 15 ng/μl). For H. defensa and APSE core genes, PCR conditions included 1 cycle at 94°C for 2 min; 11 cycles at 94°C for 30 s; a touchdown from 58°C to 46°C for 50 s and 72°C for 1.5 min; then 25 cycles at 94°C for 30 s, 46°C for 50 s, and 72°C for 1.5 min; and 72°C for 5 min, followed by a hold at 4°C. APSE VCR primers spanning P5 to P8 and P9 and P10 were amplified for 1 cycle at 94°C for 4 min; 25 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 2 min; 72°C for 10 min; and a hold at 14°C. APSE4 P8 and P9 primers were denatured at 94°C for 2 min and then underwent 30 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 45 s, followed by 72°C for 2 min and a hold at 4°C. PCR products were cleaned with Fermentas (Glen Burnie, MD) FastAP (0.5 μl) and exonuclease I (0.3 μl) by incubation for 10 min at 37°C and 5 min at 75°C, and then they were Sanger sequenced (Eurofins MWG Operon, Huntsville, AL). The forward and reverse sequences were manually inspected for mismatched base pair assignments and ambiguities, and poor-quality ends were trimmed using Geneious Pro V.5.4.6 software (Biomatters Ltd.).
Estimating APSE phage and H. defensa abundance.
We estimated symbiont abundance in Ap. craccivora throughout aphid development using real-time quantitative PCR (qPCR). We estimated titers in two clonal lines (AC1 and SV1) at four time points over aphid development (48, 96, 144, and 216 h ± 2 h) corresponding to three nymphal samples and one in early adulthood (n = 8 aphids per line and age). Titers were taken at both 20 and 25°C, as temperature has been shown to influence within-host densities in other insect systems (for examples, see reference 67). Samples for each time point were processed using the whole-aphid squish extraction protocol outlined above. Single-copy bacterial (dnaK) and phage (P28) genes were amplified using a Roche LightCycler to estimate the number of bacterial cells or phage genomic copies present in each aphid using primers, PCR cocktail, and reaction conditions as described above. Amplifications were analyzed with an external “absolute” standard curve produced from serial dilutions of 1E02 to 1E09 (34). Each qPCR run contained three standards and two negative controls to calibrate the external curve for each targeted gene. H. defensa and phage values were corrected using the aphid ef1α gene (primers ACef1aF [5′-CCG TGG AGA TGC ACC ACG AAG C-3′] and ACef1aR [5′-AGC AGC TCC CTT GGG TGG GT-3′]) to account for differences in extraction efficiencies. A “relative” standard curve was created for the aphid ef1α gene with the experimental sample with the highest crossing point (Cp) value, which was serially diluted (5-fold) and run in triplicate to create an internal standard curve. We used the Pfaffl method (68) to correct for variation, as standard curve efficiency values differed (H. defensa dnaK, 2.02; APSE P28, 2.01; and ef1α, 1.91).
Is H. defensa present in Ap. craccivora hemolymph?
In Ac. pisum, H. defensa persists intracellularly in bacteriocytes or sheath cells but also extracellularly in the hemolymph (59, 69–72). H. defensa taken from Ap. craccivora hemolymph has been used to successfully transfect Ac. pisum (23), suggesting symbiont presence within the hemolymph. To confirm this for the strain infecting line AC1, which confers protection in its native Ap. craccivora host against some parasitoid species (37), hemolymph from surface-sterilized aphids was obtained by clipping legs and collecting clear hemolymph in 1× phosphate-buffered saline on a mounting slide. Samples were then fixed with 4% paraformaldehyde and incubated for ∼30 min at 4°C. Slides were then stained with ∼25 μl of Vectashield mounting medium (Vector Laboratories, Inc., Burlingame, CA) containing 4′-6-diamidino-2-phenylindole (DAPI; 1.5 μg/ml), and hemolymph was observed with an inverted microscope with a UV light cube. Previous diagnostics indicated that H. defensa was the only symbiont present in this line (see above) and that gut-associated bacteria or environmental contaminants are unlikely to be present in the hemolymph. However, we also performed qPCR on hemolymph samples to verify that titers for H. defensa roughly corresponded with microscopy observations.
Data analysis.
All statistical analyses were performed using the JMP v. 9.0.3 64-bit platform (SAS Institute Inc., Cary, NC). Development times, estimated by TFR, were nonnormally distributed, so a nonparametric Wilcoxon rank sum analysis was performed. Both fresh-weight (FW) and cumulative fecundity (per cup cage) data had log normal distributions, so all were log transformed prior to analyses of variance (ANOVA). Bonferroni corrections were applied to fecundity assays to control for familywise error rate. Our population cage experiment resulted in nonnormally distributed data, so all ratios were logit transformed, and a linear regression was performed to estimate selection for each infection type against time combined with a t test to evaluate whether the regression slopes significantly differed from zero. A likelihood ratio χ2 test was then used to evaluate differences between the start and the final time points in the population cages. We used logistic regression to analyze survivorship.
Nucleotide sequence accession number.
Newly determined sequence data for this study have been deposited in GenBank under accession number KM250079.
RESULTS
Maternal transmission loss in Ap. craccivora.
Overall, the maternal transmission rate of H. defensa in Ap. craccivora (Table 4) was high (99.33%, n = 601) but imperfect. Of the 601 offspring evaluated, only 4 were not infected with H. defensa. These negative values were confirmed with additional diagnostic PCR screens verifying that DNA extractions were viable. All four transmission failures occurred at 25°C, but given the small number of negatives it is not surprising that a Pearson χ2 test found no relationship between frequency of transmission and temperature (SV1 at 20 and 25°C, χ21,256 = 1.748 and P = 0.19). The vertical transmission rate for APSE was also very high, with 100% of the H. defensa-infected aphids testing positive for the bacteriophage (n = 341) (Table 4).
TABLE 4.
Vertical transmission rates of H. defensa and APSE in Ap. craccivora
| Organism | No. of offspring infected/no. screened |
||
|---|---|---|---|
| 20°C | 25°C | Total | |
| H. defensa | 167/167 | 430/434 | 597/601 |
| APSE | 166/166 | 175/175 | 341/341 |
Estimates of fitness parameters between H. defensa-infected and uninfected aphids.
Overall, we found little effect of H. defensa infection on aphid fresh weight at adulthood (Table 5). A single H. defensa line (LL1) exhibited significant reductions in fresh weight relative to the uninfected control (t76 = 5.104; P < 0.0001) at 20°C, suggesting an infection cost. We did not detect such a difference in this line at 25°C, indicating that if costs are present, they may vary with temperature. We also found that infection with H. defensa had little effect on development time. In our TFR assays (Table 5), H. defensa-infected line AC1 showed a significant increase (P = 0.0001) in medium development time relative to the uninfected control, but only at 25°C, consistent with infection costs.
TABLE 5.
Fitness assays of H. defensa-infected and uninfected aphid lines sharing the same genotype at 20 and 25°Ca
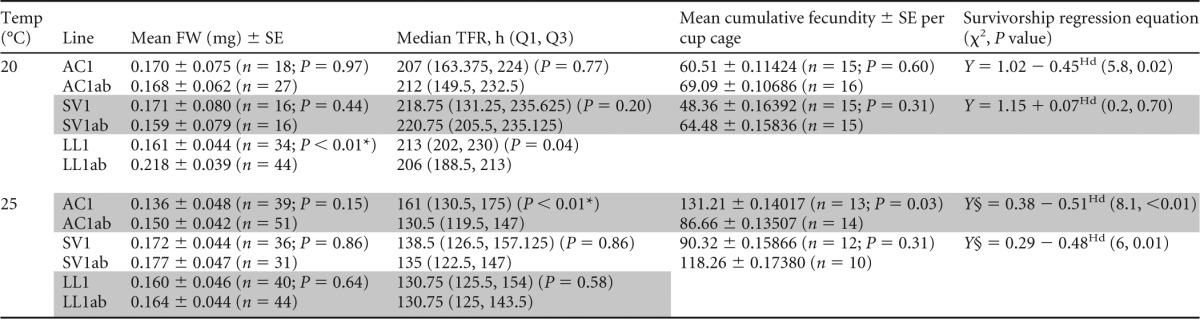
Means (back-transformed), SE, and t test outcomes are presented for fresh weights (FW). For development time (TFR, in hours), median, quartiles 1 and 3 (Q1 and Q3), and comparisons based on Wilcoxon rank sum test are presented. For both FW and TFR, n represents the number of aphids screened. Means for cumulative fecundity (back-transformed) per cage and ANOVA t test outcomes, a comparison of aphid survival (up to day 13; 5 aphids per cage) logistic regression equation, and results of the likelihood ratio test are presented. For cumulative fecundity and survivorship, n is the number of aphid cages per line and are the same for both assays, except survivorship differed for lines AC1ab (n = 15) and SV1ab (n = 11) at 25°C (§). *, significant result after Bonferroni correction.
A comparison of two H. defensa-infected and uninfected cohorts revealed no significant differences in the cumulative fecundity for either line at 20 or 25°C (Table 5). Tukey's honestly significant different (HSD) test found no differences among means at either 20°C (ANOVA, F3,61 = 0.89; P = 0.45) or 25°C (ANOVA, F3,48 = 1.85; P = 0.15). A logistic regression analysis of aphid survivorship revealed that both clonal lines at 25°C, and one at 20°C, were more likely to survive through day 13 when infected with H. defensa than were their uninfected clonal controls (Table 5). This suggests possible benefits to infection in the absence of wasps, or even thermal protection. The only exception was SV1 at 20°C, which showed no significant differences in life span associated with infection.
Population cages reveal clear costs to H. defensa infection in the absence of wasps.
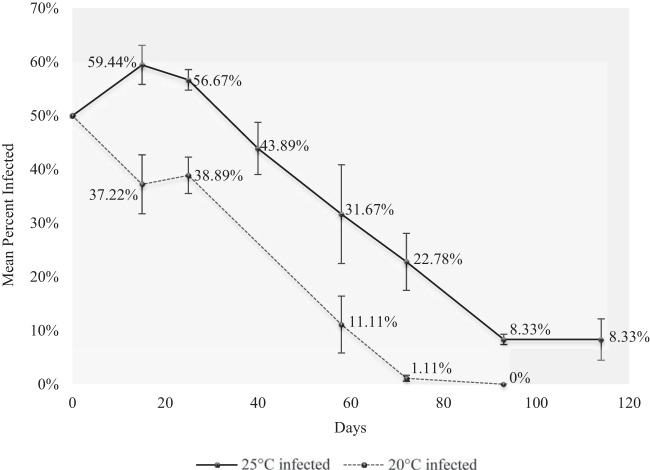
We found significant decreases in the proportion of Ap. craccivora (line AC1) infected with H. defensa in population cages over time at both 20 and 25°C (Fig. 1). Symbiont loss occurred earlier at 20°C and was complete for all 20°C cages. Final infection frequencies differed significantly from the starting frequencies (time zero) for both 20°C (likelihood ratio χ2 = 155.3; P value < 0.0001) and 25°C cages (likelihood ratio χ2 = 76.6; P value < 0.0001), and slopes differed significantly from null (i.e., no change in infection frequency) (see Table S1 in the supplemental material).
FIG 1.
Frequencies of H. defensa infection over time in population cages held at 20 (dashed line) and 25°C (solid line). At time zero, all cages started with 50% infected aphids. For each time point, mean infection frequencies (with standard error) are presented.
Characterization of H. defensa and APSE.
Our experimental lines (three from Kentucky and two from Arizona) were all infected with the same H. defensa strain, which were 100% identical at eight loci (totaling 5,465 bp) to one another and the previously published Ap. craccivora-associated H. defensa strain (5ATac), which was also collected in Arizona but >10 years earlier (66). All five phage strain-typing loci (totaling 3,704 bp) were also identical to the APSE4/5ATac previously characterized (66). Sequencing of the variable cassette region revealed that this APSE4 encodes the same putative toxin (Stx), with just a few changes (GenBank accession numbers KM250079 and EU794051): the P9 (putative stxA analog) coding regions from the strains used in our study were identical to one another, but all shared two single nucleotide polymorphisms (SNPs) relative to the APSE4/5ATac previously reported, including one that results in a change from glycine to aspartic acid (41).
H. defensa and phage abundances in Ap. craccivora.
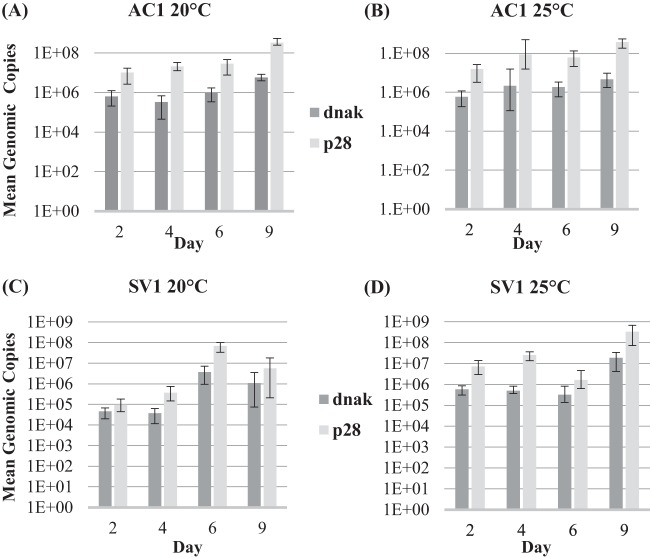
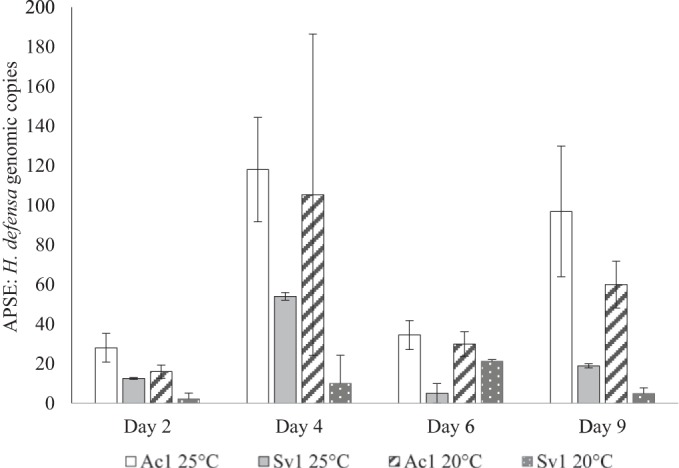
We estimated the abundances of APSE4 and H. defensa in aphids at several developmental stages using qPCR. Overall, titers of H. defensa and APSE increased from 48-h-old nymphs (∼2nd instar) to 9-day-old adults (Fig. 2). We detected more phage genomic copies (ratio range: 4.8 to 36.9 APSEs per H. defensa), with higher ratios often found at 25°C than at 20°C (back-transformed 20°C x̄ = 15.7039, 25°C x̄ = 29.6802; ANOVA, n = 128 and P = 0.0016). Also, ratios were lower in SV1 than in AC1 (back-transformed AC1 x̄ = 44.6284 and SV1 x̄ = 10.4439; ANOVA, n = 128 and P < 0.0001) (Fig. 3).
FIG 2.
H. defensa and APSE4 titers over time. Mean genomic copies (y axis) of H. defensa (dnaK) and APSE (P28) for AC1 (A and B) and SV1 (C and D) at 20 and 25°C are shown. Values represent whole aphid (n = 8). Bars represent ranges.
FIG 3.
Mean ratio of APSE copies to H. defensa copies. For each column, the mean ratio is shown (n = 8); bars represent 95% confidence intervals.
H. defensa present in Ap. craccivora hemolymph.
With microscopy of hemolymph collections combined with qPCR, we were able to confirm the extracellular presence of H. defensa in Ap. craccivora hemolymph (see Fig. S1 in the supplemental material). In line AC1, which harbors H. defensa and no other facultative symbiont, rod-shaped bacteria (∼2 μm long) were seen as individuals or in pairs throughout the hemolymph, similar to how H. defensa is described to occur in Ac. pisum (69, 71). qPCR with primers specific for H. defensa generated estimates in agreement (103 to 104 H. defensa per μl) with microscopy observations.
DISCUSSION
The cowpea aphid Ap. craccivora is infected with the heritable facultative symbiont H. defensa, which confers protection against some species of parasitic wasps (37, 38). Despite clear benefits to infection in the presence of wasps, however, this symbiont is found only at intermediate frequencies in natural populations (44, 45). In this study, we identified clear costs to infection and maternal transmission failures associated with this protective symbiont, which likely constrain H. defensa spread in natural populations.
Costs were most evident in population cage experiments, where the proportion of uninfected aphids increased over time relative to that of H. defensa-infected aphids sharing the same aphid genotype (Fig. 1). In cages held at 20°C, H. defensa-infected aphids were completely lost from the population in about 90 days from a starting frequency of 50%. However, when we investigated specific fitness parameters using cup cage assays in which aphids were not reared in direct competition, we failed to identify consistent costs among clones and treatments. In the aphid line SV1, for example, there were no significant differences between H. defensa-infected and uninfected aphids among most parameters other than an increase in survivorship for infected aphids at 25°C (Table 5). Other clones showed costs to infection in some fitness assays but only at one temperature; H. defensa-infected LL1, for example, exhibited delayed development time and reduced mass but only at 20°C (Table 5). Specific costs (e.g., fecundity and survival) have not been identified, but infection conceivably results in constitutive costs, as H. defensa likely relies on the aphid and the aphid's obligate symbiont Buchnera for nutrition (73).
Overall, fitness estimates for Ap. craccivora are similar to published findings for Ac. pisum, which also failed to identify specific and consistent costs to H. defensa infection in standard fitness assays but found costs in population cages (for examples, see references 13 and 18). The discrepancy between cup and population cage assays may be due to holding lines separately in cup cage assays, which can create statistical noise not present when different aphid lines are subjected to identical conditions and allowing small fitness differences to be detected (13). Costs (and benefits) may also vary across food plants of polyphagous species (52) and may differ on the natural host (Medicago sativa) relative to the experimental host (Vicia faba). Preliminary trials, however, indicate no consistent effect of host plant (M. sativa versus V. faba) on Ap. craccivora performance with and without H. defensa (J. A. White, unpublished data).
Cost to infection with H. defensa has also been reported for the black bean aphid, Ap. fabae, a third aphid host in which this symbiont has been shown to confer protection against parasitoids (35, 36, 74). In the latter case, costs to survival were identified (36), indicating that infection costs may vary across species. Across these aphids, however, it appears that benefits in the presence of parasitoids and costs in their absence likely contribute to the maintenance of intermediate frequencies of H. defensa in nature.
We also documented imperfect maternal transmission of H. defensa in Ap. craccivora (Table 4), which could influence H. defensa infection frequencies in natural populations. Occasional transmission failures in Ap. craccivora, when coupled with costs identified in population cages, are consistent with anecdotal observations of repeated losses of H. defensa that we encountered from subcultures started from single infected mothers. These subcultures go through severe bottlenecks (5 to 10 aphids) exacerbating effects. This is in contrast to laboratory-held lines of Ac. pisum carrying H. defensa in which vertical transmission rates approach 100%, and no instances of H. defensa loss have been recorded across numerous laboratory-held lines, many maintained for >10 years (33, 34). Of course, transmission rates for both species may be substantially lower in the field, due to high or low temperatures or other factors (for examples, see reference 75). It is unlikely, however, that transmission failure contributed substantially to the decline of H. defensa in our population cage studies given the very low rates of failure at 0.67%. A previous study (13) also considered inefficient transmission an unlikely factor influencing symbiont loss within their population. In field populations, however, transmission failure may be important to explain intermediate infection frequencies for species like Ap. craccivora, which do not undergo sexual reproduction or overwinter at higher latitudes. This is because new populations may be founded each season by one or a few H. defensa-infected aphids, requiring maternal transmission failure before relative fitness differences between infected and infected individuals can be acted on by selection. Once infection status is mixed among individuals, then fitness costs associated with infection probably constrain symbiont spread relatively more than transmission failure and potentially lead to the rapid spread of uninfected individuals when parasitism pressure is low (as in Fig. 1).
Transmission failure of APSE bacteriophages have been documented for Ac. pisum, in which loss of APSE3s, associated with high levels of protection against parasitoids, eliminates symbiont-based defense and culminates in the breakdown of the symbiosis (22, 34). In our transmission assay, however, we detected no losses of APSE4 in H. defensa-infected Ap. craccivora and have never detected an H. defensa-carrying cowpea aphid to be APSE free from any laboratory line.
All five Ap. craccivora lines used in this study were infected with H. defensa and APSE haplotypes that were genetically identical to one another at all examined loci (8 for H. defensa and 5 for APSE4) and nearly identical to the 5ATac strain shown to provide protective benefits to Ac. pisum after transfection (23, 66). Thus, despite our lines being collected >2,400 km apart and over a decade later than the previously characterized 5ATac strain (Table 1), all Ap. craccivora lines examined to date share the same strains of H. defensa and APSE. The limited diversity observed in these defensive elements potentially restricts the functional repertoire of this protective symbiosis. For example, this strain was found to confer protection against some, but not all, aphidiine parasitoid species attacking Ap. craccivora (37). Thus, limited strain diversity may leave this aphid vulnerable to attack depending on which wasp species are present, and the inability of H. defensa to protect against all parasitoid species may itself constrain symbiont spread. Despite the limited H. defensa strain diversity identified in Ap. craccivora, facultative symbionts may still comprise much of the ecologically relevant heritable genetic variation present in these populations given the likely anholocyclic nature of this aphid in North America (44). The overall lack of allelic diversity uncovered in our microsatellite analysis (Table 3) is consistent with limited sexual reproduction such that a few related clones, with and without particular symbiont infections, may recolonize the North American range seasonally from overwintering source populations.
We also performed microscopy and qPCR to confirm that H. defensa persists extracellularly in the hemolymph of Ap. craccivora as it does in Ac. pisum (71). The occurrence of free-living H. defensa throughout the hemolymph allows for direct contact with wasp tissue and may be necessary for the defensive phenotype. Using qPCR, we estimated phage and H. defensa abundances in two Ap. craccivora lines throughout aphid development at both 20 and 25°C. In all lines and treatments, we observed that H. defensa and APSE4 titers increase with aphid age with little variation between clones or temperatures (Fig. 2). We also found that APSE titers always exceeded those of H. defensa. In sum, patterns of in vivo infection were similar to those observed in Ac. pisum (for examples, see reference 40). In Ac. pisum, APSEs appear to play a role in regulating the protective symbiont's abundance (34), but more study is needed to determine if the APSE4s associated with Ap. craccivora influence H. defensa densities. While not directly comparable, H. defensa titers are generally lower at equivalent time points in Ap. craccivora relative to Ac. pisum. For example, 2-day-old Ac. pisum were found to have ∼1E07 H. defensa organisms per aphid (34), while Ap. craccivora values were all 2 orders of magnitude lower (6.41E05). The cowpea aphid, however, is much smaller, and titers may be limited by aphid size.
In conclusion, we characterized a protective strain of H. defensa and its associated APSE virus in the important aphid pest Aphis craccivora. We identified clear costs to infection and maternal transmission failure which likely limit symbiont spread in natural populations. Heritable symbionts infect a large number of insect species, including many that compete with humans for food or other resources. Understanding symbiont roles in mediating protection from natural enemies, as well as factors limiting their effectiveness and invasion potential, is of great interest given the effort in employing biological programs to control herbivore pest populations.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Nasreen Bano, Cristina Brady, Matthew Doremus, Maureen Egbosiuba, Kyungsun L. Kim, Jimmy Quijada, and Kassandra Sandoval for technical assistance.
This work was funded by USDA-AFRI grant 2009-65104-05983.
Footnotes
Published ahead of print 11 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01775-14.
REFERENCES
- 1.Buchner P. 1965. Animal symbiosis with plant microorganisms. Interscience, New York, NY [Google Scholar]
- 2.Duron O, Bouchon D, Boutin S, Bellamy L, Zhou L, Engelstaedter J, Hurst GD. 2008. The diversity of reproductive parasites among arthropods: Wolbachia do not walk alone. BMC Biol. 6:27. 10.1186/1741-7007-6-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feldhaar H. 2011. Bacterial symbionts as mediators of ecologically important traits of insect hosts. Ecol. Entomol. 36:533–543. 10.1111/j.1365-2311.2011.01318.x [DOI] [Google Scholar]
- 4.Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42:165–190. 10.1146/annurev.genet.41.110306.130119 [DOI] [PubMed] [Google Scholar]
- 5.Zug R, Hammerstein P. 2012. Still a host of hosts for Wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS One 7(6):e38544. 10.1371/journal.pone.0038544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumann P. 2005. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 59:155–189. 10.1146/annurev.micro.59.030804.121041 [DOI] [PubMed] [Google Scholar]
- 7.Brownlie JC, Johnson KN. 2009. Symbiont-mediated protection in insect hosts. Trends Microbiol. 17:348–354. 10.1016/j.tim.2009.05.005 [DOI] [PubMed] [Google Scholar]
- 8.Oliver KM, Moran NA. 2009. Defensive symbionts in aphids and other insects, p 129–147 In White JF, Torres MS. (ed), Mycology, vol 27 Defensive mutualism in microbial symbiosis. CRC Press, Boca Raton, FL [Google Scholar]
- 9.Oliver KM, Smith AH, Russell JA. 2014. Defensive symbiosis in the real world—advancing ecological studies of heritable, protective bacteria in aphids and beyond. Funct. Ecol. 28:341–355. 10.1111/1365-2435.12133 [DOI] [Google Scholar]
- 10.Bull JJ. 1983. Evolution of sex determining mechanisms. The Benjamin/Cummings Publishing Company, Reading, MA [Google Scholar]
- 11.Jaenike J. 2012. Population genetics of beneficial heritable symbionts. Trends Ecol. Evol. 27:226–232. 10.1016/j.tree.2011.10.005 [DOI] [PubMed] [Google Scholar]
- 12.Jaenike J, Unckless R, Cockburn SN, Boelio LM, Perlman SJ. 2010. Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science 329:212–215. 10.1126/science.1188235 [DOI] [PubMed] [Google Scholar]
- 13.Oliver KM, Campos J, Moran NA, Hunter MS. 2008. Population dynamics of defensive symbionts in aphids. Proc. Biol. Sci. 275:293–299. 10.1098/rspb.2007.1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oliver KM, Degnan PH, Burke GR, Moran NA. 2010. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu. Rev. Entomol. 55:247–266. 10.1146/annurev-ento-112408-085305 [DOI] [PubMed] [Google Scholar]
- 15.Douglas AE. 1998. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu. Rev. Entomol. 43:17–37. 10.1146/annurev.ento.43.1.17 [DOI] [PubMed] [Google Scholar]
- 16.Chen DQ, Montllor CB, Purcell AH. 2000. Fitness effects of two facultative endosymbiotic bacteria on the pea aphid, Acyrthosiphon pisum, and the blue alfalfa aphid, A. kondoi. Entomol. Exp. Appl. 95:315-323. 10.1046/j.1570-7458.2000.00670.x [DOI] [Google Scholar]
- 17.Montllor CB, Maxmen A, Purcell AH. 2002. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol. Entomol. 27:189–195. 10.1046/j.1365-2311.2002.00393.x [DOI] [Google Scholar]
- 18.Russell JA, Moran NA. 2006. Costs and benefits of symbiont infection in aphids: variation among symbionts and across temperatures. Proc. Biol. Sci. 273:603–610. 10.1098/rspb.2005.3348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Łukasik P, van Asch M, Guo HF, Ferrari J, Godfray HCJ. 2013. Unrelated facultative endosymbionts protect aphids against a fungal pathogen. Ecol. Lett. 16:214–218. 10.1111/ele.12031 [DOI] [PubMed] [Google Scholar]
- 20.Parker BJ, Spragg CJ, Altincicek B, Gerardo NM. 2013. Symbiont-mediated protection against fungal pathogens in pea aphids: a role for pathogen specificity? Appl. Environ. Microbiol. 79:2455–2458. 10.1128/AEM.03193-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scarborough CL, Ferrari J, Godfray HCJ. 2005. Aphid protected from pathogen by endosymbiont. Science 310:1781. 10.1126/science.1120180 [DOI] [PubMed] [Google Scholar]
- 22.Oliver KM, Degnan PH, Hunter MS, Moran NA. 2009. Bacteriophages encode factors required for protection in a symbiotic mutualism. Science 325:992–994. 10.1126/science.1174463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oliver KM, Moran NA, Hunter MS. 2005. Variation in resistance to parasitism in aphids is due to symbionts not host genotype. Proc. Natl. Acad. Sci. U. S. A. 102:12795–12800. 10.1073/pnas.0506131102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliver KM, Russell JA, Moran NA, Hunter MS. 2003. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl. Acad. Sci. U. S. A. 100:1803–1807. 10.1073/pnas.0335320100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferrari J, West JA, Via S, Godfray HCJ. 2012. Population genetic structure and secondary symbionts in host-associated populations of the pea aphid complex. Evolution 66:375–390. 10.1111/j.1558-5646.2011.01436.x [DOI] [PubMed] [Google Scholar]
- 26.Oliver KM, Moran NA, Hunter MS. 2006. Costs and benefits of a superinfection of facultative symbionts in aphids. Proc. Biol. Sci. 273:1273–1280. 10.1098/rspb.2005.3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russell JA, Weldon S, Smith AH, Kim KL, Hu Y, Lukasik P, Doll S, Anastopoulos I, Novin M, Oliver KM. 2013. Uncovering symbiont-driven genetic diversity across North American pea aphids. Mol. Ecol. 22:2045–2059. 10.1111/mec.12211 [DOI] [PubMed] [Google Scholar]
- 28.Sandström JP, Russell JA, White JP, Moran NA. 2001. Independent origins and horizontal transfer of bacterial symbionts of aphids. Mol. Ecol. 10:217–228. 10.1046/j.1365-294X.2001.01189.x [DOI] [PubMed] [Google Scholar]
- 29.Simon JC, Carre S, Boutin M, Prunier-Leterme N, Sabater-Munoz B, Latorre A, Bournoville R. 2003. Host-based divergence in populations of the pea aphid: insights from nuclear markers and the prevalence of facultative symbionts. Proc. Biol. Sci. 270:1703–1712. 10.1098/rspb.2003.2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsuchida T, Koga R, Sakurai M, Fukatsu T. 2006. Facultative bacterial endosymbionts of three aphid species, Aphis craccivora, Megoura crassicauda and Acyrthosiphon pisum, sympatrically found on the same host plants. Appl. Entomol. Zool. 41:129–137. 10.1303/aez.2006.129 [DOI] [Google Scholar]
- 31.Tsuchida T, Koga R, Shibao H, Matsumoto T, Fukatsu T. 2002. Diversity and geographic distribution of secondary endosymbiotic bacteria in natural populations of the pea aphid, Acyrthosiphon pisum. Mol. Ecol. 11:2123–2135. 10.1046/j.1365-294X.2002.01606.x [DOI] [PubMed] [Google Scholar]
- 32.Darby AC, Douglas AE. 2003. Elucidation of the transmission patterns of an insect-borne bacterium. Appl. Environ. Microbiol. 69:4403–4407. 10.1128/AEM.69.8.4403-4407.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moran NA, Dunbar HE. 2006. Sexual acquisition of beneficial symbionts in aphids. Proc. Natl. Acad. Sci. U. S. A. 103:12803–12806. 10.1073/pnas.0605772103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weldon SR, Strand MR, Oliver KM. 2013. Phage loss and the breakdown of a defensive symbiosis in aphids. Proc. Biol. Sci. 280:20122103. 10.1098/rspb.2012.2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmid M, Sieber R, Zimmermann YS, Vorburger C. 2012. Development, specificity and sublethal effects of symbiont-conferred resistance to parasitoids in aphids. Funct. Ecol. 26:207–215. 10.1111/j.1365-2435.2011.01904.x [DOI] [Google Scholar]
- 36.Vorburger C, Gouskov A. 2011. Only helpful when required: a longevity cost of harbouring defensive symbionts. J. Evol. Biol. 24:1611–1617. 10.1111/j.1420-9101.2011.02292.x [DOI] [PubMed] [Google Scholar]
- 37.Asplen MK, Bano N, Brady CM, Desneux N, Hopper KR, Malouines C, Oliver KM, White JA, Heimpel GE. Specialization of bacterial endosymbionts that protect aphids from parasitoids. Ecol. Entomol., in press [Google Scholar]
- 38.Desneux N, Barta RJ, Hoelmer KA, Hopper KR, Heimpel GE. 2009. Multifaceted determinants of host specificity in an aphid parasitoid. Oecologia 160:387–398. 10.1007/s00442-009-1289-x [DOI] [PubMed] [Google Scholar]
- 39.Łukasik P, Dawid MA, Ferrari J, Godfray HCJ. 2013. The diversity and fitness effects of infection with facultative endosymbionts in the grain aphid, Sitobion avenae. Oecologia 173:985–996. 10.1007/s00442-013-2660-5 [DOI] [PubMed] [Google Scholar]
- 40.Martinez AJ, Weldon SR, Oliver KM. 2014. Effects of parasitism on aphid nutritional and protective symbioses. Mol. Ecol. 23:1594–1607. 10.1111/mec.12550 [DOI] [PubMed] [Google Scholar]
- 41.Degnan PH, Moran NA. 2008. Diverse phage-encoded toxins in a protective insect endosymbiont. Appl. Environ. Microbiol. 74:6782–6791. 10.1128/AEM.01285-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moran NA, Degnan PH, Santos SR, Dunbar HE, Ochman H. 2005. The players in a mutualistic symbiosis: insects, bacteria, viruses, and virulence genes. Proc. Natl. Acad. Sci. U. S. A. 102:16919–16926. 10.1073/pnas.0507029102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Wilk F, Dullemans AM, Verbeek M, van den Heuvel J. 1999. Isolation and characterization of APSE-1, a bacteriophage infecting the secondary endosymbiont of Acyrthosiphon pisum. Virology 262:104–113. 10.1006/viro.1999.9902 [DOI] [PubMed] [Google Scholar]
- 44.Brady C, Asplen M, Desneux N, Heimpel G, Hopper K, Linnen C, Oliver K, Wulff J, White J. 2014. Worldwide populations of the aphid Aphis craccivora are infected with diverse facultative bacterial symbionts. Microb. Ecol. 67:195–204. 10.1007/s00248-013-0314-0 [DOI] [PubMed] [Google Scholar]
- 45.Brady CM, White JA. 2013. Cowpea aphid (Aphis craccivora) associated with different host plants has different facultative endosymbionts. Ecol. Entomol. 38:433–437. 10.1111/een.12020 [DOI] [Google Scholar]
- 46.Blackman RL, Eastop VF. 2000. Aphids on the world's crops: an identification and information guide, 2nd ed. John Wiley and Sons, Chichester, United Kingdom [Google Scholar]
- 47.Cammell M, Way M. 1983. Aphid pests, p 315–346 In Hebbethwaite P. (ed), The faba bean (Vicia faba L.). A basis for improvement. Butterworths, London, United Kingdom [Google Scholar]
- 48.Singh SR, Emden HFV. 1979. Insect pests of grain legumes. Annu. Rev. Entomol. 24:255–278. 10.1146/annurev.en.24.010179.001351 [DOI] [Google Scholar]
- 49.Van Emden HF, Harrington R. 2007. Taxonomic issues, p 1–22 In Van Emden HF, Harrington R. (ed), Aphids as crop pests. Cabi Publishing, Wallingford, United Kingdom [Google Scholar]
- 50.Blackman RL, Eastop VF. 2006. Aphids on the world's herbaceous plants and shrubs, vol 1 Host lists and keys. John Wiley and Sons, Chichester, United Kingdom [Google Scholar]
- 51.Gutierrez AP, Morgan DJ, Havenstein D. 1971. Ecology of Aphis craccivora Koch and subterranean clover stunt virus. 1. Phenology of aphid populations and epidemiology of virus in pastures in southeast Australia. J. Appl. Ecol. 8:699–721 [Google Scholar]
- 52.Berg GN. 1984. The effect of temperature and host species on the population-growth potential of the cowpea aphid, Aphis-craccivora Koch (Homoptera, Aphididae). Aust. J. Zool. 32:345–352. 10.1071/ZO9840345 [DOI] [Google Scholar]
- 53.Guay JF, Boudreault S, Michaud D, Cloutier C. 2009. Impact of environmental stress on aphid clonal resistance to parasitoids: role of Hamiltonella defensa bacterial symbiosis in association with a new facultative symbiont of the pea aphid. J. Insect Physiol. 55:919–926. 10.1016/j.jinsphys.2009.06.006 [DOI] [PubMed] [Google Scholar]
- 54.Haynes S, Darby AC, Daniell TJ, Webster G, van Veen FJF, Godfray HCJ, Prosser JI, Douglas AE. 2003. Diversity of bacteria associated with natural aphid populations. Appl. Environ. Microbiol. 69:7216–7223. 10.1128/AEM.69.12.7216-7223.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsuchida T, Koga R, Horikawa M, Tsunoda T, Maoka T, Matsumoto S, Simon J-C, Fukatsu T. 2010. Symbiotic bacterium modifies aphid body color. Science 330:1102–1104. 10.1126/science.1195463 [DOI] [PubMed] [Google Scholar]
- 56.Muyzer G, Dewaal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Douglas AE, Francois C, Minto LB. 2006. Facultative ‘secondary' bacterial symbionts and the nutrition of the pea aphid, Acyrthosiphon pisum. Physiol. Entomol. 31:262–269. 10.1111/j.1365-3032.2006.00516.x [DOI] [Google Scholar]
- 58.Koga R, Tsuchida T, Sakurai M, Fukatsu T. 2007. Selective elimination of aphid endosymbionts: effects of antibiotic dose and host genotype, and fitness consequences. FEMS Microbiol. Ecol. 60:229–239. 10.1111/j.1574-6941.2007.00284.x [DOI] [PubMed] [Google Scholar]
- 59.Koga R, Tsuchida T, Fukatsu T. 2003. Changing partners in an obligate symbiosis: a facultative endosymbiont can compensate for loss of the essential endosymbiont Buchnera in an aphid. Proc. Biol. Sci. 270:2543–2550. 10.1098/rspb.2003.2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harrison JS, Mondor EB. 2011. Evidence for an invasive aphid “superclone”: extremely low genetic diversity in oleander aphid (Aphis nerii) populations in the southern United States. PLoS One 6(3):e17524. 10.1371/journal.pone.0017524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilson ACC, Massonnet B, Simon JC, Prunier-Leterme N, Dolatti L, Llewellyn KS, Figueroa CC, Ramirez CC, Blackman RL, Estoup A, Sunnucks P. 2004. Cross-species amplification of microsatellite loci in aphids: assessment and application. Mol. Ecol. Notes 4:104–109. 10.1046/j.1471-8286.2004.00584.x [DOI] [Google Scholar]
- 62.Vanlerberghe-Masutti F, Chavigny P, Fuller SJ. 1999. Characterization of microsatellite loci in the aphid species Aphis gossypii Glover. Mol. Ecol. 8:693–695. 10.1046/j.1365-294x.1999.00876.x [DOI] [PubMed] [Google Scholar]
- 63.Simon JC, Leterme N, Delmotte F, Martin O, Estoup A. 2001. Isolation and characterization of microsatellite loci in the aphid species, Rhopalosiphum padi. Mol. Ecol. Notes 1:4–5. 10.1046/j.1471-8278.2000.00002.x [DOI] [Google Scholar]
- 64.Sunnucks P, DeBarro PJ, Lushai G, Maclean N, Hales D. 1997. Genetic structure of an aphid studied using microsatellites: cyclic parthenogenesis, differentiated lineages and host specialization. Mol. Ecol. 6:1059–1073. 10.1046/j.1365-294X.1997.00280.x [DOI] [PubMed] [Google Scholar]
- 65.Gloor GB, Preston CR, Johnson-Schlitz DM, Nassif NA, Phillis RW, Benz WK, Robertson HM, Engels WR. 1993. Type I repressors of P element mobility. Genetics 135:81–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Degnan PH, Moran NA. 2008. Evolutionary genetics of a defensive facultative symbiont of insects: exchange of toxin-encoding bacteriophage. Mol. Ecol. 17:916–929. 10.1111/j.1365-294X.2007.03616.x [DOI] [PubMed] [Google Scholar]
- 67.Hurst GDD, Johnson AP, von der Schulenburg JHG, Fuyama Y. 2000. Male-killing Wolbachia in Drosophila: a temperature-sensitive trait with a threshold bacterial density. Genetics 156:699–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29(9):e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Darby AC, Birkle LM, Turner SL, Douglas AE. 2001. An aphid-borne bacterium allied to the secondary symbionts of whitefly. FEMS Microbiol. Ecol. 36:43–50. 10.1111/j.1574-6941.2001.tb00824.x [DOI] [PubMed] [Google Scholar]
- 70.Fukatsu T, Nikoh N, Kawai R, Koga R. 2000. The secondary endosymbiotic bacterium of the pea aphid Acyrthosiphon pisum (Insecta: Homoptera). Appl. Environ. Microbiol. 66:2748–2758. 10.1128/AEM.66.7.2748-2758.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moran NA, Russell JA, Koga R, Fukatsu T. 2005. Evolutionary relationships of three new species of Enterobacteriaceae living as symbionts of aphids and other insects. Appl. Environ. Microbiol. 71:3302–3310. 10.1128/AEM.71.6.3302-3310.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tsuchida T, Koga R, Meng XY, Matsumoto T, Fukatsu T. 2005. Characterization of a facultative endosymbiotic bacterium of the pea aphid Acyrthosiphon pisum. Microb. Ecol. 49:126–133. 10.1007/s00248-004-0216-2 [DOI] [PubMed] [Google Scholar]
- 73.Degnan PH, Yu Y, Sisneros N, Wing RA, Moran NA. 2009. Hamiltonella defensa, genome evolution of protective bacterial endosymbiont from pathogenic ancestors. Proc. Natl. Acad. Sci. U. S. A. 106:9063–9068. 10.1073/pnas.0900194106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vorburger C, Ganesanandamoorthy P, Kwiatkowski M. 2013. Comparing constitutive and induced costs of symbiont-conferred resistance to parasitoids in aphids. Ecol. Evol. 3:706–713. 10.1002/ece3.491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hurst GDD, Jiggins FM, Robinson SJW. 2001. What causes inefficient transmission of male-killing Wolbachia in Drosophila? Heredity 87:220–226. 10.1046/j.1365-2540.2001.00917.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.