Abstract
The genetic organization of the aioBA operon, encoding the arsenite oxidase of the moderately acidophilic and facultative chemoautotrophic bacterium Thiomonas arsenitoxydans, is different from that of the aioBA operon in the other arsenite oxidizers, in that it encodes AioF, a metalloprotein belonging to the ArsR/SmtB family. AioF is stabilized by arsenite, arsenate, or antimonite but not molybdate. Arsenic is tightly attached to AioF, likely by cysteine residues. When loaded with arsenite or arsenate, AioF is able to bind specifically to the regulatory region of the aio operon at two distinct positions. In Thiomonas arsenitoxydans, the promoters of aioX and aioB are convergent, suggesting that transcriptional interference occurs. These results indicate that the regulation of the aioBA operon is more complex in Thiomonas arsenitoxydans than in the other aioBA containing arsenite oxidizers and that the arsenic binding protein AioF is involved in this regulation. On the basis of these data, a model to explain the tight control of aioBA expression by arsenic in Thiomonas arsenitoxydans is proposed.
INTRODUCTION
All organisms have to get rid of the metals or metalloids considered poisonous, such as cadmium, chromate, mercury, or arsenic. On the other hand, they require some metal(loid)s, including iron, copper, and zinc, as structural components of proteins or as cofactors for enzymatic catalysis for several vital functions. In addition, some prokaryotes use them as energy sources, as electron donors or electron acceptors, as in the case of iron (reviewed in reference 1) or arsenic (reviewed in reference 2). They must take up the essential ones into the cell and insert them where needed or utilize them as an energy source while maintaining an optimal bioavailable intracellular concentration to avoid detrimental effects. Prokaryotes face various environmental conditions and must therefore sense the metal(loid)s present and tightly regulate the transcription of genes encoding the proteins responsible for metal(loid) homeostasis (3). Different families of metal-sensing transcriptional regulators have been described in bacteria, with ArsR/SmtB being the largest and the most extensively studied (reviewed in references 4 to 8).
Prokaryotes have evolved a variety of mechanisms of resistance to arsenic (reviewed in reference 2). Furthermore, even if this metalloid is considered a notorious poison, some of them have the ability to oxidize arsenite [As(III)] or to reduce arsenate [As(V)] to gain energy for growth. They must therefore delicately balance arsenic intracellular trafficking by controlling the expression of the genes involved in these mechanisms. Until now, only three regulators have been proposed to be involved in arsenic sensing: the ArsR and ArsD repressors and the three-component As(III) binding protein/sensor/regulator signal transduction system AioXSR (2, 9, 10). ArsR and ArsD act together to repress the basal and maximal levels of expression of the arsenical resistance ars operon of the Escherichia coli R773 plasmid. ArsR is one of the founding members of the metal-responsive transcriptional regulator ArsR/SmtB family (11) and has been extensively reviewed (see references 2 and 4 to 8 for among the most recent reviews). ArsD functions primarily as a chaperone by binding and transferring As(III) or Sb(III) to the ATPase ArsA (12), but it also exhibits a weak repressor activity (13). In the absence of metalloids, ArsR blocks transcriptional initiation by binding as a homodimer to an imperfect inverted repeat proximal to or overlapping the −35 element of the ars promoter. Coordination of As(III) or Sb(III) at low concentrations to ArsR induces a conformational change, leading to DNA release and to the initiation of transcription of the ars operon. To prevent the toxic overproduction of the integral membrane protein ArsB, ArsD represses ars expression as soon as the ArsD concentration reaches a threshold value (14). In spite of the absence of sequence similarities, ArsR and ArsD bind the same operator site in the ars regulatory region but with different affinities for the inducer and for DNA, leading to arsenic concentration-dependent regulation, with ArsR functioning at low inducer concentrations and ArsD functioning at high concentrations (15). Both repressors coordinate As(III) or Sb(III) to three clustered cysteine thiolate ligands per monomer (11, 13, 16–20). In ArsR, the spatial location of this trigonal S3 cysteine complex is generally within or near the first helix of the DNA binding domain (α3 helix) but can be at the dimer interface of each monomer (α5 helix), as in Acidithiobacillus ferrooxidans (21), or at an intermolecular location between the subunits of the homodimer just before the α4 of the helix-turn-helix (HTH) DNA binding domain, as in Corynebacterium glutamicum (22). The third arsenic-sensing regulator described is the AioXRS system, which was proposed to induce the arsenite oxidase-encoding aioBA operon in the presence of As(III) in Agrobacterium tumefaciens (23), Herminiimonas arsenitoxydans (24), and Rhizobium sp. strain NT-26 (10). The aioXSR genes have been detected upstream from the aioBA operon in several bacteria, including Thiomonas arsenitoxydans (see Fig. S1 in the supplemental material) (25, 26). The regulator AioR belongs to the NtrC σ54 RNA transcriptional activator family. It is likely to be a specific coactivator with σ54 in the initiation of aioBA operon transcription (24). In Rhizobium NT-26, AioR has been shown to be phosphorylated by AioS (10). However, no or only one cysteine residue is present within the putative sensory domain of AioS. It was recently shown in A. tumefaciens that the protein that binds As(III) is, in fact, AioX, and it was proposed that this binding results in a conformational change of AioX, allowing its interaction with AioS (9).
In the acidophilic chemoautotrophic betaproteobacterium Thiomonas arsenitoxydans, the aioBA-cyc1-aioF-cyc2 operon (here named the aioBA operon) is expressed in the presence of As(III) or As(V) but not Sb(III) (26–28). The aioXSR genes are located upstream from this operon, but unlike in A. tumefaciens and Rhizobium NT-26, it is divergently transcribed (see Fig. S1 in the supplemental material). A noteworthy finding is that, until now, a gene predicted to encode a member of the metal-responsive transcriptional regulator ArsR/SmtB family has been detected only in T. arsenitoxydans and has been shown to be cotranscribed with aioBA (see Fig. S1 in the supplemental material) (26). The purpose of this paper is to characterize this regulator, here termed AioF, and to determine whether it is involved in aioBA operon regulation.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
T. arsenitoxydans strain 3As was used throughout this study (27, 29). Escherichia coli TG1 supE hsdΔ5 thi Δ(lac–proAB) [F′ traD36 proAB lacIq lacZΔM15] was used for plasmid propagation and for the production of recombinant AioF with a hexahistidine tag fused to its C terminus (AioF-His tag). For multifactorial expression screening conditions, we used several E. coli strains [BL21(DE3), C43(DE3)/pLysS, and Rosetta (DE3)/pLysS from Novagen], plasmids (pET21 [Novagen] and pJF119EH [30]), and growth media (Luria-Bertani [LB], ON Express Instant TB medium, Turbo broth, LB broth supplemented or not with Augmedium, and M9 minimal medium); different isopropyl β-d-1-thiogalactopyranoside (IPTG) concentrations (0.2, 0.5, 1, and 2 mM); various induction temperatures (20, 30, and 37°C or a heat shock at 42°C for 30 min followed by induction at 20°C); and various induction times (2, 4, 6, 18, or 24 h).
T. arsenitoxydans was routinely grown at 30°C with vigorous shaking on CSM6-TYE medium containing basal salts, 10 mM thiosulfate, and 0.05% (wt/vol) yeast extract, as described in reference 27. When necessary, 2 mM As(III) was added. E. coli was grown at 37°C with vigorous shaking in LB medium (31) with 50 μg ml−1 ampicillin when necessary.
Cellular growth was followed by determination of the optical density at 600 nm (OD600).
General DNA manipulations.
Genomic DNA was prepared as previously described (27). Plasmid DNA was obtained using a Wizard Plus SV DNA purification system from Promega.
For aioF cloning, PCR amplifications were carried out with Platinum Taq DNA polymerase (Invitrogen) on genomic DNA. Otherwise, GoTaq polymerase (Promega) was used following the manufacturer's instructions. Primers are provided in Table S1 in the supplemental material.
DNA digestion with restriction enzymes and ligation with T4 DNA ligase were performed according to New England BioLabs' recommendations.
DNA products were analyzed on a 1% agarose gel as described previously (31). Amplicons were purified with a QIAquick PCR purification kit (Qiagen) or with Montage PCR centrifugal filter devices (Millipore). The linear vector was cut from the agarose gel and purified with the QIAquick gel extraction kit (Qiagen). Recombinant plasmids were introduced into E. coli competent cells as described previously (32).
The nucleotide sequences were determined by Cogenics Biotechnologies (Grenoble, France) or Eurofins MWG Operon (Germany).
RNA manipulations.
T. arsenitoxydans total RNA was extracted as described in reference 33 and treated once with the reagents from a Turbo DNA-free kit (Applied Biosystems). The lack of DNA contamination was checked by PCR of each RNA sample. The RNA integrity was controlled on an agarose gel.
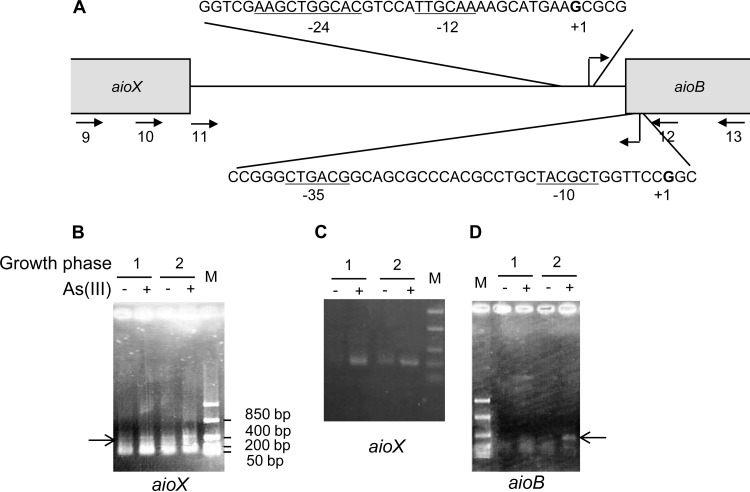
The transcriptional start site of aioX and aioB was determined with a 5′ rapid amplification of cDNA ends (RACE) system (Invitrogen) according to the instructions provided. The cDNA corresponding to the 5′ end of the mRNA for the gene of interest was synthesized using a gene-specific antisense oligonucleotide (see Table S1 in the supplemental material) and elongated with a dC tail at its 3′ end. Then, this tailed cDNA was amplified with an abridged anchor primer (AAP; see Table S1 in the supplemental material) hybridizing to the dC tail and a gene-specific antisense oligonucleotide located upstream from the previous one (see Fig. 5A and Table S1 in the supplemental material).
FIG 5.
Transcriptional start sites of aioB and aioX operons determined by 5′ RACE. (A) Schematic representation of the aioX-aioB intergenic region. The oligonucleotides used are indicated as arrows. Transcriptional start sites are in bold, and the corresponding σ-dependent promoter motifs are underlined. (B to D) Amplification of cDNA corresponding to the 5′ end of aioX determined in T. arsenitoxydans grown in the presence (lanes +) or the absence (lanes −) of As(III) during the first (lanes labeled 1) and the second (lanes labeled 2) growth phases with oligonucleotide aoxX5′-R2 (arrow labeled 10 in panel A) (B) or oligonucleotide aoxA11 (arrow labeled 11 in panel A) (C). (D) Amplification of cDNA corresponding to the 5′ end of aioB mRNA determined under the same conditions with oligonucleotide aoxA div9 (arrow labeled 12 in panel A). Lanes M, FastRuler low-range DNA ladder (Fermentas).
Cloning of aioF and overexpression and purification of the recombinant AioF-His tag.
Several attempts were performed to get the recombinant AioF-His tag peptide soluble (see the multifactorial conditions described above). The only construction that gave a soluble and stable recombinant AioF-His tag peptide was that with the pJF119EH plasmid in the TG1 strain with 0.1 mM As(III) at the time of IPTG induction. In this construction, the DNA fragment corresponding to the AioF peptide was amplified by PCR with the ArsR1-EcoRI and the ArsR1C-BamHI oligonucleotides (see Table S1 in the supplemental material). The ArsR1C-BamHI oligonucleotide contained the sequence encoding a hexahistidine tag followed by a translational termination site. The amplified product was cloned into EcoRI- and BamHI-digested pJF119EH to give pJF119EH-AioF-His tag. The construction was checked by nucleotide sequencing with the pJF119EHup and pJF119EHrev oligonucleotides (see Table S1 in the supplemental material).
A soluble recombinant AioF-6His protein was obtained from pJF119EH-AioF-His tag under the following conditions. Cells were initially grown at 37°C in LB medium supplemented with ampicillin to an OD600 of between 0.6 and 0.8. At this stage, 1 mM IPTG in the presence of 0.1 mM As(III), As(V), or Sb(III) was added and the temperature was reduced to 30°C. The cells were further incubated for 2 h. The cells were harvested by centrifugation and washed once with 40 mM Tris-HCl, pH 7.6. The pellets were resuspended in 20 mM sodium phosphate, 500 mM NaCl, pH 7.4 (5 ml g−1), in the presence of 1 mM phenylmethanesulfonyl fluoride (Thermo Scientific) and then passed three times through a French press (Thermo Electron Corporation) at 20,000 lb/in2. The lysate was centrifuged at 13,000 × g for 30 min at 4°C to remove unbroken cells and cellular debris. The supernatant was ultracentrifuged at 77,000 × g for 1 h at 4°C to remove the membrane fractions. The soluble protein fraction was loaded onto a nickel column (HisTrap; GE Healthcare) according to the manufacturer's instructions. The fractions containing the recombinant AioF-His tag were eluted with 500 mM imidazole, 500 mM NaCl, 20 mM sodium phosphate, pH 7.4 according to the manufacturer's guidelines. Solvent exchange of the eluate was performed with Amicon Ultra4 centrifugal filter devices (Millipore) following the manufacturer's instructions. The purified protein was kept in 300 mM NaCl, 20 mM sodium phosphate, pH 7.4, at 4°C.
General analytical procedures.
The protein concentration was determined by the modified Bradford method (Bio-Rad protein assay) according to the manufacturer's instructions. The purity of the preparation was checked by 16% SDS-PAGE with staining of the gels with Coomassie blue and by immunodetection with antibodies directed against the hexahistidine tag using a SuperSignal West Hisprobe kit (Thermo Scientific), as described in the manufacturer's guidelines.
Chemical treatment of AioF-His tag-As(III) or AioF-His tag-As(V) with EDTA, dithionite, or dithiothreitol was performed as follows. The recombinant protein (29 μM) was treated with 100 mM EDTA, a few crystals of dithionite, or 1 mM dithiothreitol (DTT) to release As and then dialyzed (Spectra/Por membrane; molecular weight cutoff, 6,000 to 8,000) against 100 mM KCl–20 mM sodium phosphate (pH 7.4) buffer at 4°C for 12 h to remove the chemical agent and arsenic.
Mass spectrometry.
Molecular mass was determined by matrix-assisted laser desorption ionization (MALDI)–time of flight (TOF) mass spectrometry for the recombinant protein AioF-His tag when produced in the presence of As(III), As(V), or Sb(III). The proteins were directly spotted onto a MALDI stainless steel target plate (17 pmol for each spot), and an equal volume of a saturated solution of the matrix sinapinic acid (40% CH3CN in water, 0.1% trifluoroacetic acid [vol/vol]) was added. The mixtures were allowed to dry at room temperature. Data were acquired on a MALDI-TOF mass spectrometer (Microflex II) from Bruker Daltonics using Flex control software in the linear positive mode. External mass calibration was carried out with a protein calibration standard I solution (Bruker Daltonics), and mass spectra were treated by Gaussian smoothing.
For identification of AioF-His tag-As(III), the purified recombinant protein (about 0.42 μg in 2 μl) was reduced with 2 μl 10 mM dithiothreitol (DTT) in 50 mM NH4HCO3 for 45 min at 56°C in the dark and alkylated with 4 μl 55 mM iodoacetamide (IAA) in 50 mM NH4HCO3 for 30 min at room temperature and was then submitted to trypsin digestion (16.5 μg of sequencing-grade trypsin [Sigma, St. Louis, MO]) for 3 h at 37°C. The sample was vacuum dried, and the tryptic peptides were diluted in 5% formic acid; 1 μl of this peptide solution was directly spotted onto the MALDI plate with 1 μl of a saturated solution of the matrix alpha-cyano-4-hydroxycinnamic acid (Sigma) in 70% CH3CN in water, 0.1% (vol/vol) trifluoroacetic acid. Mass spectra were obtained on a MicroflexII MALDI-TOF mass spectrometer in a positive reflectron mode using internal and external calibrations (monoisotopic [M + H]+ from the peptide calibration standard). A peak list was generated by a peptide mass fingerprint (PMF) method from FlexAnalysis software and manually checked, and the experimentally measured peptide masses were compared with the masses of the theoretical tryptic peptides calculated from the sequence of AioF-His tag, including sequences with variable modifications of the cysteine residues (carbamidomethylated) and the methionine residues (oxidation), by use of the BioTools program (Bruker Daltonics). Searches were performed with a maximum peptide mass tolerance of 150 ppm. Sequence recovery of 73% to 93% was obtained with various sample preparations of AioF-His tag. For identification of the cysteine residues not involved in disulfide bridges, AioF-His tag-As(III) was directly treated with IAA [200 pmol AioF-His tag-As(III), 275 nmol IAA in 50 mM NH4HCO3, 30 min at room temperature] and submitted to trypsin digestion as described above to detect carbamidomethyl cysteine-containing peptides.
Electrophoretic mobility shift assays (EMSAs).
DNA substrates for band shift assays were produced by PCR amplification using a 5′ Cy5-labeled reverse oligonucleotide (Eurogentec) (see Table S1 in the supplemental material). The reaction mix (15 μl) contained 6 to 20 ng Cy5-labeled DNA, depending on the PCR fragment, in 40 mM Tris-HCl, pH 8.8, 100 mM KCl, 1 mM EDTA, and 0.2 mM sucrose. Herring sperm DNA (10 ng μl−1) was used as nonspecific carrier DNA. Recombinant AioF-His tag was added at the concentrations indicated in the appropriate figures, and the binding reaction mix was incubated at room temperature for 30 min. Samples were loaded on a native 6% PAGE that had been prerun for 20 min and run for an additional 1 to 2 h in 0.5× TBE (70 mM Tris-HCl, pH 8.3, 90 mM boric acid, 2 mM EDTA; Euromedex) at 30 mA and 4°C. The gel was then scanned using a 635-nm laser and a long-pass red (LPR) filter (FLA5100; Fujifilm).
Bioinformatics analysis.
The complete genomes of 2,722 prokaryotic (2,560 bacterial and 162 archaeal) organisms available in November 2013 were downloaded from the NCBI FTP site (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/) and constituted the primary data source. Multiple-sequence alignments of the structural AioF homologs were constructed with the ClustalW (version 2.1) program (34) and BLOSUM as protein weight matrices. The localization of the helix-turn-helix (HTH) region within a protein was performed by using the helixturnhelix program (from the EMBOSS package; http://emboss.sourceforge.net/) with the minimum standard deviation set to 1. The fuzzpro program (from the EMBOSS package; http://emboss.sourceforge.net/) was used to search for the CXXXC motif within the protein sequences. The BLAST package (35) and self-written Perl scripts were then used to search for the first 50 closely related AioF homologs (with AioF from T. arsenitoxydans 3As serving as the query) among AioF protein homologs.
RESULTS
AioF is stabilized by metalloids.
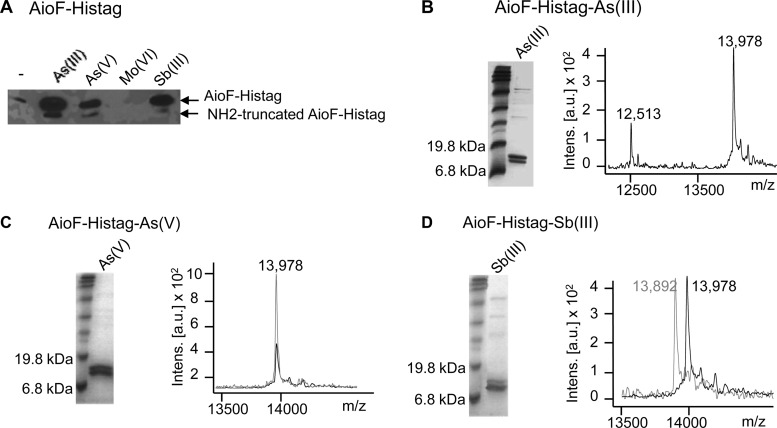
The ArsR/SmtB family member AioF is encoded by the aioBA operon (see Fig. S1 in the supplemental material) (26) and therefore is likely involved in its regulation. To check this hypothesis, AioF with a hexahistidine tag fused to its C terminus (AioF-His tag) was produced in Escherichia coli. Surprisingly, whatever the conditions used (see the Materials and Methods), AioF-His tag was found in the inclusion bodies but at a level so low that it was barely detected by immunodetection with antibodies directed against the hexahistidine tag. Furthermore, when solubilized with 6 M urea, AioF-His tag formed aggregates and precipitated as soon as the urea concentration was lowered by successive dialyses. These results suggest that AioF-His tag was unstable likely because it did not fold correctly under the conditions tested. Because AioF belongs to the ArsR/SmtB metal-sensing transcriptional regulator family, it likely binds metal(loid)s. Therefore, arsenic as As(III) or As(V), antimony as Sb(III), or molybdenum as Mo(VI) at a 0.1 mM concentration was added together with IPTG at the time of induction. As shown in Fig. 1A, for the same amount of cells, more AioF-His tag could be detected in the soluble fraction by immunodetection when As(III), As(V), or Sb(III) was added but not when Mo(VI) was added. These results suggest that As(III), As(V), or Sb(III), but not Mo(VI), binds to AioF, allowing its correct folding.
FIG 1.
Analysis of recombinant AioF-His tag produced in Escherichia coli. (A) Total cell extracts obtained in the presence or in the absence of As(III), As(V), Mo(VI), and Sb(III) added at the time of induction with IPTG. AioF-His tag was revealed by immunodetection with antibodies raised against the hexahistidine tag. (B to D) Purified AioF-His tag obtained after addition of As(III) (B), As(V) (C), and Sb(III) (D) at the time of induction with IPTG was analyzed by SDS-PAGE (left) with Coomassie blue staining and MALDI-TOF mass spectrometry (right). Black spectra, reference AioF-His tag-As(III) spectrum (m/z 13,978); gray spectra, reference AioF-His tag-As(V) spectrum (m/z 13,978) in panel C and AioF-His tag-Sb(III) spectrum (m/z 13,892) in panel D. Intens., intensity; a.u., arbitrary units.
The recombinant AioF proteins produced in the presence of As(III) [AioF-His tag-As(III)], As(V) [AioF-His tag-As(V)], and Sb(III) [AioF-His tag-Sb(III)] were purified on an affinity nickel column. The analysis by MALDI-TOF mass spectrometry of AioF-His tag-As(III) digested with trypsin after reduction by DTT and alkylation by iodoacetamide confirmed that the purified protein was AioF-His tag-As(III) (90.3% sequence coverage; Table 1). The same two peptides observed in Fig. 1A and recognized by anti-hexahistidine tag antibodies could be visualized on Coomassie blue-stained SDS-polyacrylamide gels (Fig. 1B to D, left). The global mass of the smaller peptide determined by MALDI-TOF mass spectrometry was 12,513 Da (Fig. 1B, right) and corresponded to a truncated form with a deletion of the first 13 amino acids (aa), as determined by N-terminal sequencing. The global mass of the larger peptide was 13,978 Da when it was produced in the presence of As(III) or As(V) (Fig. 1B and C, right) and 13,892 Da when it was produced in the presence of Sb(III) (Fig. 1D, right). Given that the theoretical molecular mass of AioF is 13,905 Da and the theoretical molecular masses of As and Sb are 75 and 122 Da, respectively, it appears that AioF-His tag-As(III) and AioF-His tag-As(V) monomers contained one atom of As, while the AioF-His tag-Sb(III) monomer was devoid of Sb. Inductively coupled plasma mass spectrometry (ICP-MS) experiments not only confirmed that one atom of As was present per AioF-His tag-As(III) monomer (see Fig. S2A, left, in the supplemental material) but also detected one Sb atom per AioF-His tag-Sb(III) monomer (see Fig. S2A, right, in the supplemental material; the two peaks correspond to the two stablest isotopes, 121Sb and 123Sb). These results indicate that the link between As and AioF is tight, while that between Sb and AioF is loose and broken by the laser shots during MALDI ionization experiments. To confirm that AioF-His tag bound As(V), AioF-His tag-As(III) was oxidized with 2 mM ferricyanide to get AioF-His tag-As(V). Its molecular mass determined by MALDI-TOF mass spectrometry was 13,987 Da, indicating that As was not released and therefore that AioF-His tag is indeed able to bind As(V) (see Fig. S2B in the supplemental material).
TABLE 1.
Peptides obtained after trypsin digestion of AioF-His tag-As(III) after reduction by DTT and alkylation by iodoacetamide, without treatment, and after direct alkylation by iodoacetamide
| Theoretical mass (Da) | Sequencea | AioF-His tag-As(III) trypsin digestionb |
||
|---|---|---|---|---|
| After reduction by DTT (90.3%) | Without treatment (73.4%) | After direct alkylation by iodoacetamide (89.5%) | ||
| 1,482,667 | MHHELPFSSGADR | + | + | + |
| 1,498,662 | MHHELPFSSGADR (oxidation of methionine) | + | − | + |
| 1,638,769 | MHHELPFSSGADRR | + | + | − |
| 1,654,763 | MHHELPFSSGADRR (oxidation of methionine) | + | + | − |
| 2,712,259 | STDEDDVAAVFEAAAELFAALSSPMR | + | + | + |
| 2,728,254 | STDEDDVAAVFEAAAELFAALSSPMR (oxidation of methionine) | − | − | + |
| 939,532 | LSIVCHLR (SH cysteine) | + | + | + |
| 996,554 | LSIVCHLR (carbamidomethyl cysteine) | + | − | + |
| 1,444,673 | EQDMNVQQIANR | + | + | + |
| 1,460,668 | EQDMNVQQIANR (oxidation of methionine) | + | − | + |
| 1,408,742 | IGSSQPNTSLHLR | + | + | + |
| 1,177,657 | QLHQIGIVDR | + | + | + |
| 896,435 | SGQSVTYR | + | + | + |
| 1,009,49 | NTFVADLCK (SH cysteine) | + | − | + |
| 1,066,512 | NTFVADLCK (carbamidomethyl cysteine) | − | − | + |
| 1,446,659 | IVCPGHHHHHHH (SH cysteine) | − | − | − |
| 1,503,68 | IVCPGHHHHHHH (carbamidomethyl cysteine) | + | − | + |
| Disulfide bridges | ||||
| 1,947,022 | NTFVADLCK + LSIVCHLR (S-S cysteine) | − | − | + |
| 2,384,191 | LSIVCHLR + IVCPGHHHHHHH (S-S cysteine) | − | − | + |
| 2,454,149 | NTFVADLCK + IVCPGHHHHHHH (S-S cysteine) | − | − | + |
Cysteine residues are in bold.
The percent sequence coverage is given in parentheses. +, peptides detected; −, peptides not detected.
Given that no oligomers could be detected by MALDI-TOF mass spectrometry, other approaches were attempted to determine the native molecular mass of AioF-His tag-As(III). SDS-PAGE, run under denaturing but nonreducing conditions, indicated a monomer of AioF-His tag-As(III) migrating at the predicted molecular mass of 13,900 Da (see Fig. S3A in the supplemental material), indicating the absence of intermolecular sulfide bridges between AioF monomers. Size exclusion chromatography confirmed the monomeric structure of AioF-His tag-As(III) since it was eluted at a 17.87-ml retention volume (see Fig. S3B in the supplemental material), corresponding to an estimated molecular mass of approximately 13,800 Da. These data confirmed the findings of the mass spectrometry experiments, i.e., that there are no covalent bonds between AioF-His tag-As(III) monomers. By dynamic light scattering, AioF-His tag-As(III) was shown to be polydisperse with two populations. The first population has an average hydrodynamic radius of 2.794 nm (σ, 0.7385 nm) (see Fig. S3C, left, in the supplemental material), and the second population has an average hydrodynamic radius of 106.6 nm (σ, 76.56 nm). Nevertheless, the molecule with an average hydrodynamic radius of 2.794 nm is the protein that represents 98.8% of the population. AioF-His tag-As(III) oscillated between a monomer, a dimer, and a tetramer throughout the experiment (see Fig. S3C, left, in the supplemental material). This result demonstrates that AioF-His tag-As(III) is soluble and folds with minor aggregates present in the solution. To determine if AioF-His tag-As(III) unfolds in the absence of arsenic, we added 20 μM DTT to remove As(III). We observed an irreversible shift to large aggregated oligomers in the presence of DTT (see Fig. S3C, right, in the supplemental material). This indicated that when AioF-His tag-As(III) was reduced and was no longer bound to As(III), it unfolded, multimerized, and aggregated.
To determine whether recombinant AioF-His tag-As(III) was correctly folded, circular dichroism and one-dimensional nuclear magnetic resonance (1D-NMR) spectroscopies were performed. The far-UV spectra of AioF-His tag-As(III) showed a typical circular dichrogram of a helical protein with negative maxima at 208 and 222 nm (see Fig. S3D in the supplemental material). In addition, a positive maximum at 195 nm may indicate that when the protein is bound to As(III) it belongs to the class of proteins with a mix of α helices and β turns but with a predominant α-helical content. The composition of the secondary structure determined using the K2D (Kohonen neural network with a two-dimensional output layer) deconvolution method showed that the protein has a 27% helical content, a 25% beta sheet content, and a 48% random coil content. The ratio of the ellipticity at 222 and 208 nm (ϴ222/208) of 1.0 may indicate the presence of interacting helices. These results were corroborated by the 1D-NMR results (see Fig. S3E in the supplemental material) and support the suggestion that AioF-His tag-As(III) is soluble, correctly folded, and mainly constituted of α helices, as predicted (26).
Cysteine residues are likely involved in As(III) coordination in AioF.
The tight binding of As to AioF-His tag suggests that cysteine residues are involved in the coordination, as already shown in other As binding proteins, such as ArsR or ArsD (for a review, see reference 2 and references therein). The three cysteine residues detected in AioF (Cys53, Cys111, and Cyc115) (26) are present in the 12,513-Da truncated form that still binds As. To find out whether these cysteine residues are involved in intramolecular disulfide bridges or not, AioF-His tag-As(III) was directly digested with trypsin without DTT reduction and iodoacetamide treatment. As can be seen in Table 1, in which the peptides obtained by MALDI-TOF mass spectrometry (73.4% sequence coverage) are listed, no disulfide bridges were observed. The three cysteine residues could therefore coordinate arsenic. Two experiments were set up to check this hypothesis. In the first one, AioF-His tag-As(III) was directly treated with iodoacetamide, which has a high affinity for cysteine residues not involved in disulfide bridges. Analysis of the peptides obtained after trypsin digestion (89.5% sequence coverage) indicated that the three cysteines of AioF could be detected in their carbamidomethylated form (Table 1). These data show that (i) the iodoacetamide treatment partially releases As as a result of iodoacetamide binding to the cysteine residues and (ii) the three cysteine residues are accessible to iodoacetamide and therefore not involved in intra- or intermolecular disulfide bridge formation (in agreement with the results for the peptides obtained after trypsin digestion and in the absence of multimers, which were not detected by mass spectrometry analysis, nonreducing SDS-PAGE, and size exclusion chromatography). In a second experiment, AioF-His tag-As(III) was incubated at 90°C for 10 min, and the temperature was then progressively decreased to room temperature with or without As(III) to allow the protein to reach its initial conformation. By MALDI-TOF mass spectrometry, the main mass observed after incubation in the absence of As(III) at [M + H]+ 13,913 corresponded to the mass of AioF-His tag devoid of As, while after As(III) addition, a new mass appeared at [M + H]+ 13,976, corresponding to the mass of the protein with one As atom bound per monomer (see Fig. S4 in the supplemental material). These results are typically observed with cytochromes c, in which the heme C is coordinated to the protein through cysteine residues. It can be concluded that (i) the cysteines are not involved in a disulfide bridge and, therefore, the environmental signal detected by AioF is not modulated by the redox status of the cysteines, unlike BigR (36), a member of the ArsR/SmtB family, and (ii) the cysteine residues are likely involved in As binding.
AioF binds specifically upstream from aioB in the presence of As(III) and As(V).
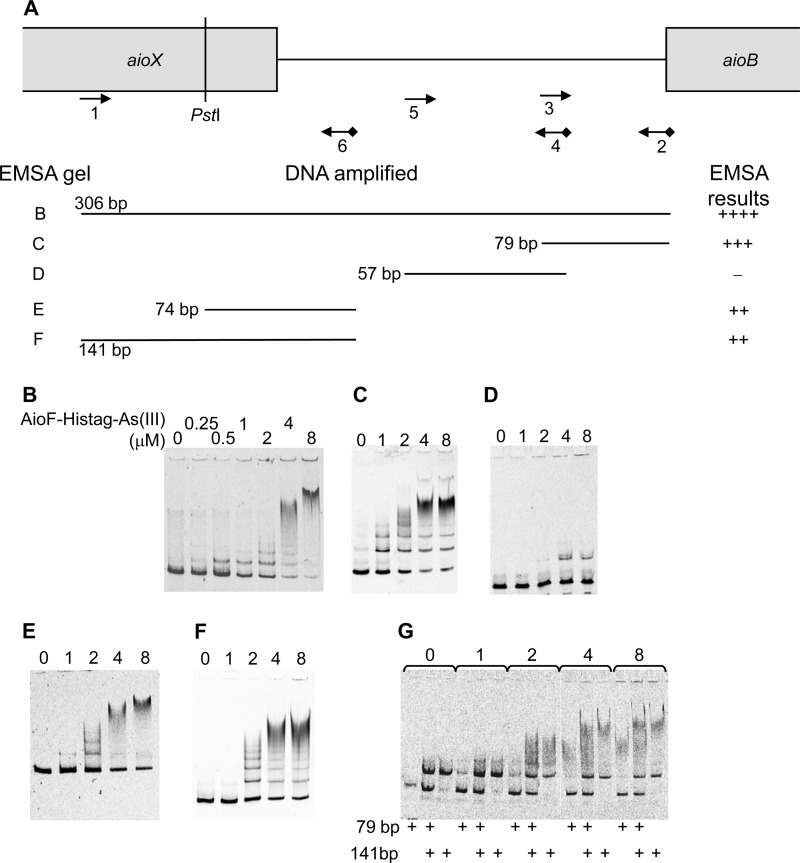
Because AioF is predicted to belong to the ArsR/SmtB metal-sensing family and tightly binds As(III) and As(V) but not Sb(III) and because the expression of the aioBA operon is higher in the presence than in the absence of As(III) or As(V) but not of Sb(III) (26; this paper), we hypothesized that AioF could be involved in the As regulation of the aioBA operon. Binding reactions between AioF and the aioBA regulatory region were determined by electrophoretic mobility shift assay (EMSA). For this purpose, a cyanine 5-labeled 306-bp fragment encompassing the intergenic region between aioX and aioB (see Fig. 4A) was amplified by PCR and incubated with increasing concentrations of AioF-His tag-As(III) (0 to 8 μM). In spite of an excess of nonspecific DNA competitor in the reaction mix, a retarded band could clearly be detected with 0.25 μM AioF-His tag-As(III) (Fig. 2A). At higher concentrations (from 1 μM), several retarded bands were detected, indicating a high-order complex formed between AioF-His tag-As(III) and DNA due to either (i) the binding of AioF-His tag-As(III) to multiple sites or (ii) AioF-His tag-As(III) oligomerization at a single site. To test the specificity of the binding of AioF-His tag-As(III) to the aio regulatory region, two experiments were set up. In the first one, increasing concentrations of the same unlabeled DNA were added as a competitor. As shown in Fig. S5A in the supplemental material, the shift was lost when the competitor was present. In the second experiment, AioF-His tag-As(III) was incubated with a DNA fragment unrelated to arsenic (the internal region of the 16S rRNA gene from T. arsenitoxydans). A shift was faintly observed with high concentrations of the protein (see Fig. S5B in the supplemental material). These two experiments showed that AioF-His tag-As(III) binding to the aio regulatory region is specific.
FIG 4.
Analysis of AioF-His tag-As(III) binding to different regions of the aioX-aioB intergenic region by gel mobility shift assays. (A) Schematic representation of the DNA fragments analyzed. DNA substrates for band shift assays were produced by PCR amplification using a 5′ Cy5-labeled reverse oligonucleotide (Eurogentec) (see Table S1 in the supplemental material), except that the 74-bp fragment was generated by PstI digestion of the 141-bp amplicon. The locations of the oligonucleotides used to amplify the different DNA fragments are indicated as arrows. The cyanide 5 is indicated as a black diamond. The size of the amplicon, as well as the EMSA gel reference and the EMSA results, is given. The sizes of the different DNA fragments incubated with AioF-His tag-As(III) (0, 0.25, 0.5, 1, 2, 4, or 8 μM) and tested by EMSAs were as follows: 306 bp (B), 79 bp (C), 57 bp (D), 74 bp (E), 141 bp (F), and both 79 and 141 bp (G).
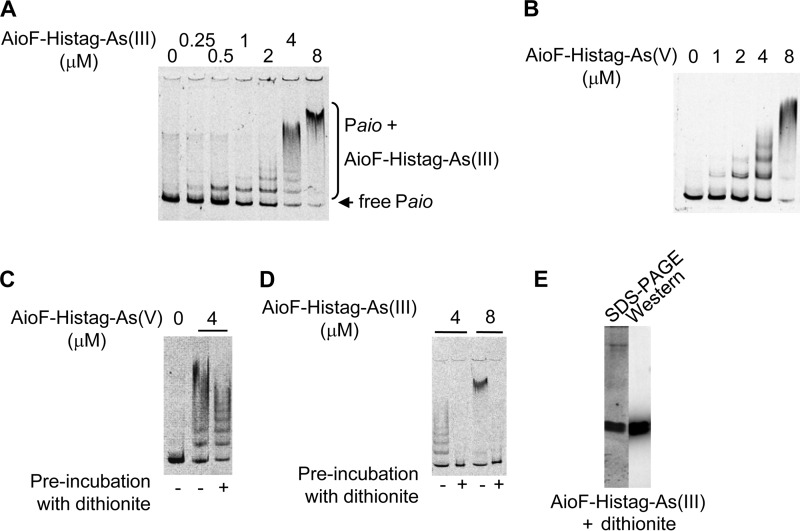
FIG 2.
Binding of AioF-His tag loaded with As to the aioX-aioB intergenic region analyzed by gel mobility shift assays. A 306-bp DNA fragment encompassing the aioX-aioB intergenic region (A to D) was incubated in the presence of recombinant AioF-His tag-As(III) (A and D) or AioF-His tag-As(V) (B and C). Recombinant AioF-His tag-As(V) (C) and AioF-His tag-As(III) (D) proteins were preincubated with dithionite. (E) SDS-PAGE and Western immunodetection with antibodies against the hexahistidine tag of AioF-His tag-As(III) after dithionite treatment.
Until now, with the exception of the BigR subfamily, the members of which do not sense metal, all the ArsR/SmtB family members bind to their cognate DNA in the absence of the metal(loid) and release it in their presence. However, contrary to these metalloregulators, AioF-His tag-As(III), which contains one As atom per monomer (see Fig. 1B and S2A in the supplemental material), bound specifically to the aioBA regulatory region (Fig. 2A). To confirm that AioF was able to bind its target DNA in the presence of As(III) and that the effect observed was not due to a low concentration of apo-AioF-His tag in the preparation, AioF-His tag-As(III) was preincubated with 0.1 mM As(III) for 15 and 30 min. The gel shift was the same with or without preincubation with As(III) (see Fig. S5C in the supplemental material), indicating that As(III) does not inhibit the binding of AioF to DNA.
AioF-His tag was able to bind not only As(III) but also As(V) (Fig. 1C; see also Fig. S2B in the supplemental material). To determine whether AioF-His tag binds the aioBA regulatory region in the presence of As(V), AioF-His tag-As(V) and AioF-His tag-As(III) oxidized with 2 mM ferricyanide, to form AioF-His tag-As(V), were incubated with the labeled 306-bp fragment encompassing the intergenic region between aioX and aioB. In both cases, retarded bands could be observed (Fig. 2B; see also Fig. S5D in the supplemental material). Therefore, AioF specifically bound its cognate DNA in the presence of As(V).
When AioF-His tag-As(V), produced by IPTG induction in the presence of As(V), was reduced to AioF-His tag-As(III) with sodium dithionite, it was able to bind the aioBA regulatory region (Fig. 2C), corroborating our data (Fig. 2A; see also Fig. S5C in the supplemental material) and indicating that dithionite treatment has no effect on the formation of the AioF-DNA complex. However, when AioF-His tag-As(III) was treated in the same way by dithionite to get AioF-His tag-As(0), no retarded bands were observed (Fig. 2D). After dithionite treatment, the proteins remained stable, as shown by SDS-PAGE and immunodetection (Fig. 2E). These data suggest that when AioF-His tag is coordinated with As(III) or As(V), it specifically binds its cognate DNA but not As(0).
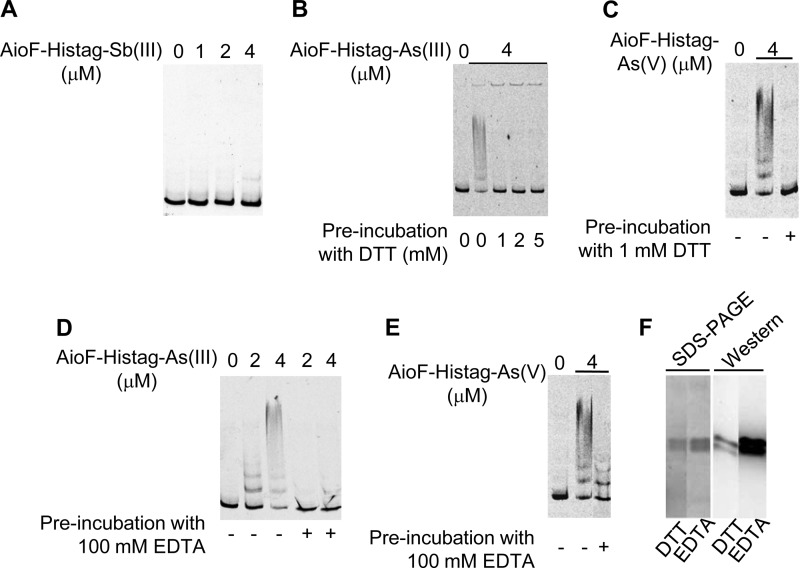
AioF does not bind upstream from aioB in the absence of As.
The subsequent task was to determine whether AioF is able to bind its target DNA in the absence of As. We have been unable to get AioF-His tag in the absence of metalloid (see above), likely because the protein did not fold correctly in the absence of the metalloid and was unstable. To circumvent this problem, we tested in EMSAs (i) AioF-His tag-Sb(III) and (ii) AioF-His tag-As(III) or AioF-His tag-As(V) treated with a reducing agent or with a di- and trivalent cation chelating agent. Dithiothreitol (DTT), by reducing the cysteine residues of AioF, was expected to remove As(III) (Table 1) and As(V) from AioF-His tag-As(III) and AioF-His tag-As(V), respectively, to produce AioF-His tag free of As. EDTA was expected to chelate As(III) but not As(V) and, therefore, to generate AioF-His tag from AioF-His tag-As(III) but not from AioF-His tag-As(V). AioF-His tag-Sb(III) (Fig. 3A), AioF-His tag-As(III) and AioF-His tag-As(V) preincubated with DTT (Fig. 3B and C), and AioF-His tag-As(III) preincubated with 100 mM EDTA (Fig. 3D) were unable to bind DNA. As expected, AioF-His tag-As(V) preincubated with EDTA still bound its target DNA (Fig. 3E). This result also indicates that the EDTA treatment has no effect on the protein AioF-His tag or on the EMSAs under the conditions described in Materials and Methods. The loss of interaction between AioF-His tag and the DNA fragment was also not due to protein instability following DTT and EDTA treatment, as shown by SDS-PAGE and immunodetection with antibodies raised against the hexahistidine tag (Fig. 3F). These data suggest that the decrease of AioF-His tag DNA binding was indeed due to the removal of As(III) or As(V) by EDTA or DTT and that AioF does not bind the aioBA regulatory region in the absence of As (Fig. 3B to D) or in the presence of Sb(III) (Fig. 3A).
FIG 3.
Binding of AioF-His tag free of arsenic to the aioX-aioB intergenic region analyzed by gel mobility shift assays. A 306-bp DNA fragment encompassing the aioX-aioB intergenic region was incubated in the presence of the recombinant AioF-His tag-Sb(III) (A), AioF-His tag-As(III) treated with DTT (B) or EDTA (D), or AioF-His tag-As(V) treated with DTT (C) or EDTA (E). (F) SDS-PAGE and Western immunodetection with antibodies against the hexahistidine tag of AioF-His tag-As(III) after DTT or EDTA treatment.
AioF binds to two distinct regions upstream from aioB.
To determine more precisely where AioF-His tag binds in the aio intergenic region, EMSAs were repeated using shorter probes obtained by PCR amplification or restriction digestion of labeled PCR products (Fig. 4A; see also Fig. S6A in the supplemental material). Retarded complexes were detected with all the fragments analyzed (Fig. 4B, C, E, and F; see also Fig. S6B to F in the supplemental material), with the exception of the central 125- and 57-bp fragments, for which many fewer shifts (125 bp; see also Fig. S6G in the supplemental material) or nearly no shifts (57 bp; Fig. 4D) were observed. These results suggest that AioF-His tag binds two regions adjacent to the 57-bp central fragment. To test whether AioF acts as an intermolecular bridge to link these two regions to form a DNA loop (37), EMSAs were performed using two DNA fragments flanking the 57-bp central region, i.e., the 141- and 79-bp fragments (fragments F and C, respectively, in Fig. 4A). No intermediate retarded bands were observed between the bands obtained from the individual fragments alone (Fig. 4G), indicating that no sandwich structures were formed. These data suggest that AioF-His tag alone does not interact with both fragments to assume a looped (intramolecular bridge) configuration.
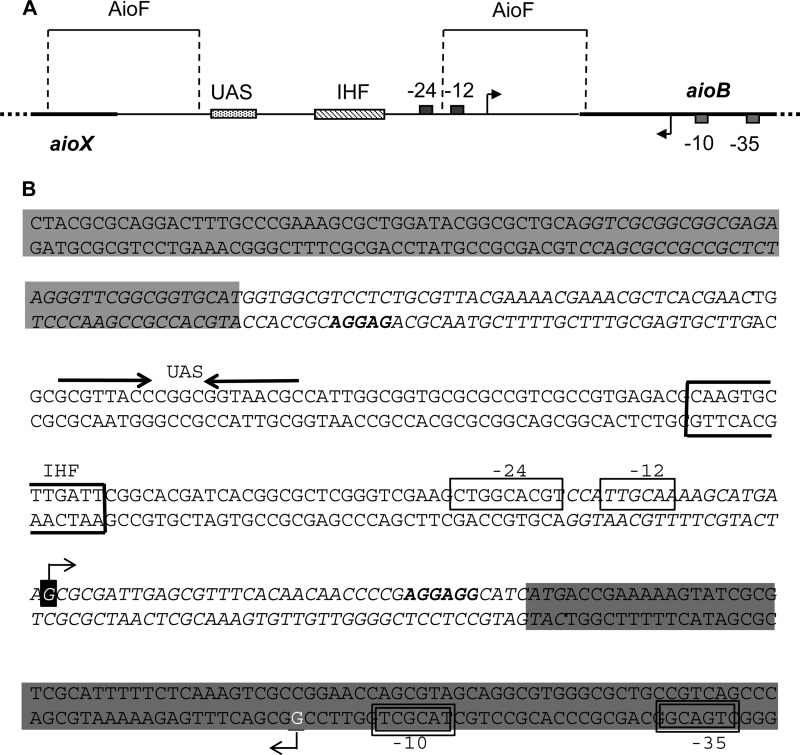
The aioB and aioX promoters are convergent.
The transcriptional start sites of the aioX and aioBA operons were determined by 5′ RACE with the oligonucleotides depicted in Fig. 5A and listed in Table S1 in the supplemental material. Total RNA exempt of genomic DNA was extracted from T. arsenitoxydans cells grown with thiosulfate in the presence or absence of As(III) during the first and the second phases of growth, corresponding to a hierarchy in the use of the electron donors (26). Since the same concentration of total RNA was used in each 5′ RACE experiment, the intensity of the amplified cDNA reflected the amount of mRNA corresponding to the gene of interest under the conditions analyzed. The data obtained confirmed the regulation of the aioX gene and of the aioBA operon (26), as shown by a slight induction of aioX by As(III) whatever the growth phase (Fig. 5B and C) and high As(III) induction of aioB during the second growth phase (Fig. 5D). The tailed cDNA was sequenced, leading to the identification of a single transcriptional start site for each operon. The aioB transcriptional start site corresponds to the predicted σ54-dependent promoter located at position −39 bp from the translational start site of aioB (Fig. 5A) (26). That of aioX is located at position −247 bp from the translational start site of aioX (Fig. 5A), inside aioB. Upstream from the aioX transcriptional start site, a cTGACg-(17 bp)-TAcgcT putative σ70-dependent promoter was identified (nucleotides different from the consensus sequence are in lowercase) (Fig. 5A).
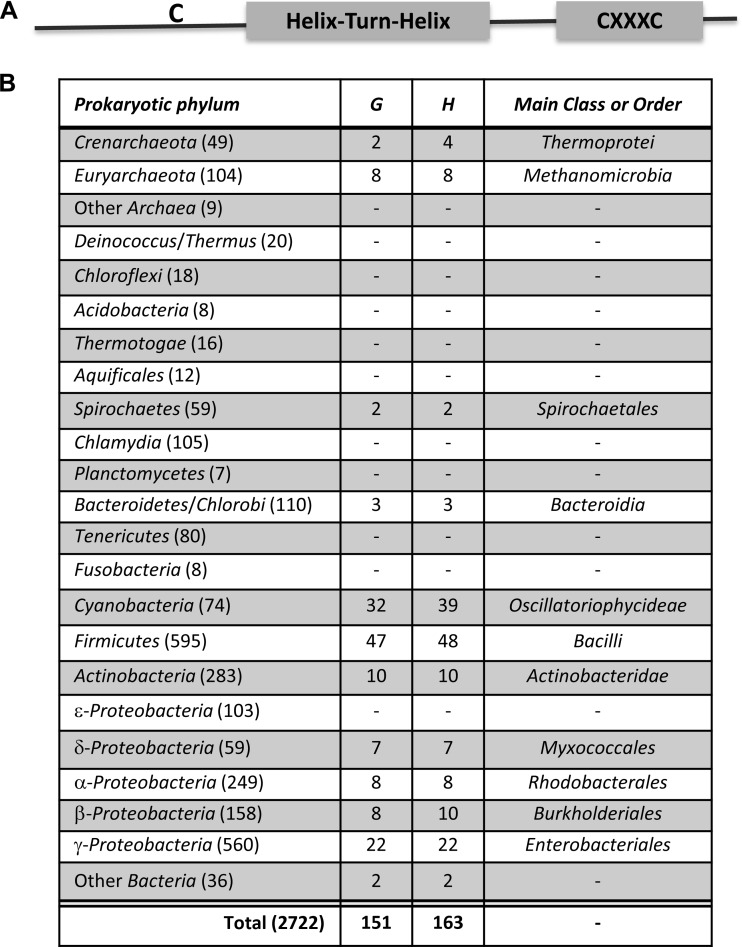
Occurrence of predicted structural AioF proteins in prokaryotic genomes.
To address the general relevance of the AioF specific transcriptional regulatory proteins, a large-scale in silico search for AioF homologs in the complete prokaryotic genomes available in November 2013 was performed. An outline of the in silico search is described in Materials and Methods. The AioF protein sequence (118 aa) of T. arsenitoxydans is known to contain one helix-turn-helix (HTH) region (positions 60 to 85) as well as three cysteine residues: one (position 53) located before the HTH region and two in a CXXXC motif (positions 111 to 115) located in the C-terminal region of the protein (26). Cysteine residues are known to be involved in As(III) coordination in ArsR and ArsD (11, 13, 16–20), and the same likely holds true in AioF, as shown above (Table 1; see also Fig. S4 in the supplemental material), even if their spatial location is different from that of the known ArsR (26). Therefore, we defined a structural model for the AioF homologs to be a 75- to 150-aa protein containing one HTH region followed by the CXXXC motif (Fig. 6A). In addition, as in the AioF reference protein, the HTH region has to be preceded by at least one Cys residue and no Cys residue may be found downstream of the CXXXC motif. Based on these structural and functional constraints, we detected 163 AioF protein homologs within 2,722 complete prokaryotic genomes, which constitute the primary data set used in this study (Fig. 6B; see also Table S2 in the supplemental material). These AioF homologs are present in 151 genomes (5.55% of the overall genomes), with most of them harboring only one copy per genome. However, there can be up to 3 copies in cyanobacterial strains (e.g., Cyanothece sp. strain ATCC 51142), which indicated the possibility of gene expansion and duplication. The number of organisms harboring AioF was fairly low in all prokaryotic phyla, but the abundance was high in the Firmicutes, Cyanobacteria, and Gammaproteobacteria (Fig. 6B), indicating extensive variations in the distribution of AioF. In most prokaryotic phyla, a proportion bias in the distribution of AioF was also observed within each class or within each order (Fig. 6B). These observations indicate that these genes probably originated from horizontal gene transfer (HGT) events from an unknown ancestor during evolution and subsequently disseminated by the same process in other organisms. On the basis of BLAST comparisons between AioF (from T. arsenitoxydans) and each of the AioF homologs, the 50 most closely related homologs (i.e., E value, <e−10) were retrieved. Interestingly, most of them were from the Cyanobacteria and Betaproteobacteria phyla (37/50 = 74% and 8/50 = 16%, respectively), suggesting that these homologs may have similar evolutionary histories. Multiple-sequence alignment of the closely related AioF homologs (see Fig. S7 in the supplemental material) clearly showed the HTH region associated with each protein and the cysteine residues (upstream of the HTH region and within the CXXXC motif). In addition, well-conserved amino acids were identified: Arg and Leu residues (at positions 52 and 53 of the alignment, respectively) and two Leu residues (at positions 84 and 87 within the HTH region). These residues are therefore likely very important for the structure and/or the function of the AioF-like proteins. Except for T. intermedia, gene synteny (seven genes downstream and seven genes upstream from aioF homologs) was not conserved. More surprisingly, these neighboring genes of aioF homologs are not related to metal(loid) resistance (data not shown). In addition, since AioF-like genes were detected in prokaryote genomic sequences in which no aioBA genes were found, we cannot exclude the possibility that these AioF-like genes can regulate metalloenzymes other than arsenite oxidase. These results are in agreement with the HGT dissemination of aioF in prokaryotic organisms.
FIG 6.
Predicted structural AioF protein homologs in prokaryotic genomes. (A) Structural and functional model of the AioF protein with Cys residues and the CXXXC motif located upstream and downstream of the HTH region, respectively. (B) Distribution of aioF gene homologs within prokaryotic taxonomic groups. For each main archaeal and bacterial phylum, the numbers in parentheses indicate the number of organisms for which complete genomes were available for this study. G and H, the number of genomes harboring at least one aioF and the total number aioF genes in the phylum, respectively.
DISCUSSION
The aioF gene, which belongs to the arsenite oxidase aioBA operon and which is absent from the other arsenite oxidizers, was predicted to encode an ArsR/SmtB metal(loid)-responsive regulator (26). Indeed, AioF has all the characteristics of the members of this family: (i) it contains the typical winged helix-turn-helix motif DNA binding motif (26), (ii) it tightly coordinates As(III) (Fig. 1B; see also Fig. S2A and S4 in the supplemental material), and (iii) it binds DNA and, more particularly, the aioX-aioB intergenic region (Fig. 2 and 4; see also Fig. S5 and S6 in the supplemental material). However, AioF presents a number of features distinctive from those of ArsR, the repressor of the arsenical resistance ars operon. AioF tightly binds not only As(III) but also As(V) and loosely binds Sb(III) (Fig. 1C and D; see also Fig. S2A and B in the supplemental material), while ArsR binds As(III) and Sb(III) but not As(V) (19, 38). While As(III) and As(V) binding is likely coordinated by cysteine residues, as suggested by iodoacetamide and heating treatments (Table 1; see also Fig. S4 in the supplemental material), the spatial location of the three cysteine residues in the predicted structure of AioF is different from that of the characterized ArsR: in AioF, one cysteine is located upstream from the α3 helix and two cysteines are located at the C terminus of the α5 helix rather than within or near the α3 helix, as in most ArsR proteins (26). Interestingly, As binding sites in Acidithiobacillus ferrooxidans ArsR are located at the end of the α5 helix (21), and the induction of the ars genes in this bacterium takes place in the presence of As(III) and As(V), even in the absence of the arsenate reductase-encoding gene, arsC (39). It is therefore possible that the close localization of the two cysteine residues at the C terminus allows the binding not only of As(III) but also of As(V). As opposed to ArsR, binding of As(III) and As(V) to apo-AioF stabilizes it (Fig. 1A; see also Fig. S3C in the supplemental material). The stabilization of AioF by arsenic is substantiated by the sequential arrangement of two of the cysteine residues (Cys111 and Cys115, interval + 4), which was shown to enhance the α-helical structure (40). Therefore, arsenic binding to AioF likely stabilizes the α5 helix forming the dimerization domain between the two subunits. In that case, AioF binding to DNA is expected to be facilitated by arsenic, in contrast with ArsR, which releases it under these conditions (38). Indeed, our results do not support AioF binding to its target DNA in the absence of As(III) or As(V): (i) apo-AioF, obtained by removing As by EDTA or DTT treatment without destabilizing the protein, is unable to bind DNA, since no more DNA-AioF complex was detected (Fig. 3B to D), and (ii) AioF-His tag-Sb(III) (Fig. 3A) and AioF-His tag-As(0) (Fig. 2D) are not able to form a stable complex with the aioX-aioB intergenic region DNA. In addition, the transcription of the aioF gene in T. arsenitoxydans is completely turned off in the absence of arsenic (26), and therefore, AioF cannot repress the aioBA operon since it is not synthesized under this condition. Furthermore, AioF is unstable when produced in the absence of arsenic (Fig. 1A), and when As(III) was released by DTT treatment in vitro, AioF-His tag multimerized and aggregated (see Fig. S3C in the supplemental material), suggesting that removal of arsenic induces a conformational change of AioF leading to its aggregation and degradation in vivo. Therefore, as opposed to the other ArsR/SmtB family members that bind DNA in the absence of their cognate metal(loid) (5, 7, 41), AioF does not seem to repress the aio cluster in the absence of arsenic. On the contrary, evidence that AioF is implicated in aio cluster regulation by arsenic seems to be obvious since it is stabilized by As(III) and As(V) but not Sb(III) (Fig. 1; see also Fig. S2 in the supplemental material) and since it specifically binds the intergenic region between aioX and aioB only when loaded with As(III) or As(V) (Fig. 2; see also Fig. S5 in the supplemental material). This fits perfectly well with the expression of the aioBA operon, which is higher in the presence than in the absence of As(III) or As(V) but not of Sb(III) (26; this paper). On the basis of the arguments presented above and in agreement with our previous proposition (26), while AioF clearly belongs to the ArsR/SmtB family, it presents some peculiarities and may constitute the first member of a new subgroup. Through a large-scale in silico analysis, we have identified several members of this new subgroup in a very few number of organisms, suggesting their acquisition by HGT during evolution. Closely related members of the AioF subgroup also show additional conserved amino acid residues that may be useful for the regulatory activity of these proteins. To our knowledge, none of the members of this new subgroup have been characterized experimentally.
As in the other arsenite oxidizers (10, 24, 42–44), the T. arsenitoxydans aioBA operon is transcribed from a σ54-dependent promoter (26) (Fig. 5). The alternate σ54-RNA polymerase holoenzyme is known to remain bound to the −24 and −12 consensus sequences in a stable inactive conformation (45), and transcription is turned off. It has been proposed that AioR is phosphorylated in the presence of arsenic (9, 10) and uses the energy released by ATP hydrolysis to remodel the initial closed σ54-RNA polymerase-promoter DNA complex to a transcriptionally proficient open complex (46–50). A perfect palindrome (GCGTTACC—GGTAACGC) positioned 99 bp upstream from the aioB transcriptional start site (26) (Fig. 7) is likely the binding site of this transcriptional activator. Surprisingly, the aioX transcriptional start site is located inside the aioB gene (Fig. 5). A correctly positioned σ70-dependent promoter was identified upstream from this site (Fig. 5). Therefore, in T. arsenitoxydans the aioB and aioX promoters are convergent (Fig. 5 and 7). This unusual topology with overlapping gene organization has been proposed to provide different levels of regulation (51, 52): (i) steric hindrance by RNA polymerase bound to DNA (but this is unlikely because the two promoters are 79 bp apart), (ii) formation of antisense RNA-RNA complexes between complementary transcripts that are subjected to degradation or that sequester the mRNA and prevent their translation (53, 54), and (iii) transcriptional interference due to the prolonged occlusion by paused RNA polymerase or to the dislodgement of the preinitiation complex from a promoter (the so-called weak promoter) by an elongating RNA polymerase from another promoter (the so-called strong promoter) (52).
FIG 7.
Schematic representation (A) and genetic organization (B) of the aioX-aioB intergenic region. aioX and aioB are shaded in light and dark gray, respectively. The ribosome binding sites are in bold letters. The −24 and −12 boxes of the σ54-dependent promoter of aioB are boxed, and the upstream activating sequence (UAS) likely corresponding to the AioR binding site is indicated above the sequence with converging arrows. The −35 and −10 boxes of the σ70-dependent promoter of aioX are boxed with double lines. Transcriptional start sites are indicated in reverse white lettering. The IHF binding site is boxed in bold. The regions where AioF binds are indicated in italics.
What could be the role of AioF in the regulation of the aio cluster? By comparing the EMSA results obtained and shown in Fig. 4 (see also Fig. S6 in the supplemental material), AioF binds to two different regions: (i) between positions −197 and −124 and (ii) between positions −18 and +42 relative to the aioBA transcriptional start site, which is upstream from the putative AioR binding site and downstream from the −24 box (Fig. 7). No obvious similarities between these two regions are detected, and no motif for AioF binding can be deduced. The interaction of the transcriptional activator with the σ54-RNA polymerase complex requires DNA looping, which is often facilitated by DNA bending proteins, such as the integration host factor (IHF) (46, 47). In T. arsenitoxydans, the region between the putative binding sites of AioR and σ54 is relatively short and GC rich (47/69) and therefore is not flexible. However, a putative IHF binding site (55) was detected in this region (Fig. 7). A possibility is that the AioF bound upstream from AioR and the AioF bound downstream from σ54 (Fig. 7) interact, acting as pliers and helping IHF to bend the DNA and thus improving the interaction between phosphorylated AioR and σ54. Although no intramolecular bridge mediated by AioF could be detected between these two regions (Fig. 4F), the possibility of AioF involvement in a looped DNA configuration assisting IHF cannot be excluded. Another hypothesis is that AioF plays a role in transcriptional interference between aioXSR and aioBA. Transcriptional interference is often asymmetric, leading to the increased expression of the gene with a strong promoter and reduced expression of the weak promoter. In the absence of arsenic, aioBA operon transcription, which is σ54 dependent, is turned off, whereas aioXSR (σ70 dependent) is expressed (Fig. 5B) (26). In that case, the aioBA promoter (Paio) is the weak one. In the presence of arsenic, the situation is reversed and PaioBA becomes the strong promoter. Arsenic is detected in the periplasm by the AioXS system, which activates AioR, allowing the expression of the aioBA operon, including aioF. Some arsenic is transported into the cytoplasm by aquaglyceroporins (see the references in references 2 and 56) or other transporters (57, 58), where it stabilizes AioF. Given the localization of the AioF binding sites (Fig. 7), AioF could likely come into contact with AioR on one site and σ54 on the other. An argument for this cooperative binding of AioF with AioR and σ54 bound to nearby sites is that AioF-His tag-As(III) [or AioF-His tag-As(V)] alone interacts with its target DNA with a relatively low affinity (low-μM range; Fig. 2 and 4; see also Fig. S6 in the supplemental material) (59). We propose that AioF binding to its target DNA sequence leads to the correct positioning of AioR and σ54 and to their productive specific interaction to form a stable nucleoprotein complex. Once AioR is engaged with the σ54-RNA polymerase complex, its ATPase activity allows the remodeling and relocalization of the closed (inactive) complex to an open (active) complex (45, 47).
In conclusion, our data provide evidence that the control of arsenite oxidation in T. arsenitoxydans is complex and involves a transcriptional factor, AioF, absent in the other arsenite oxidizers studied so far (26; this paper). Interestingly, AioX, AioR, and AioS from T. arsenitoxydans clearly did not cluster with their orthologs from the other arsenite oxidizers (60; T. R. McDermott, personal communication), in agreement with our proposition that the arsenic regulation by the AioXSR system of the aioBA operon in this bacterium may present some dissimilarities. This complex regulation may allow a fine-tuning of arsenite oxidase production in T. arsenitoxydans. In the absence of arsenic, the transcription of the aioBA operon is completely silent. Nevertheless, everything is in place to respond immediately to the presence of arsenic in the environment: σ54 and AioR are bound to the DNA but are in an inactive conformation. As soon as the metalloid is detected, AioR is phosphorylated and the aioBA is transcribed at a low level, allowing the production of the metallofactor AioF. Then, in the presence of both AioR and AioF, aioBA operon expression is optimal. This leads to aioBA transcription in two steps, a basal level with AioR functioning as soon as arsenic is detected and an optimal level with both AioR and AioF functioning together. In conclusion, tight control over aioBA expression in T. arsenitoxydans is exerted by arsenic through (i) the phosphorylation cascade of AioS and AioR, (ii) the stability of AioF, and (iii) the binding of AioF to its target DNA.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge V. Mejean (LCB, Marseille, France) for fruitful scientific discussions and critical reading of the manuscript. We thank F. Chaspoul from P. Gallice's lab (Laboratoire de Chimie Générale, Faculté de Pharmacie, Marseille, France) for total arsenic concentration determination by ICP-MS and Olivier Bornet and Françoise Guerlesquin (Laboratoire d'Ingénierie des Systèmes Macromoléculaires, IMM, Marseille, France) for the 1D-NMR spectroscopy experiments. We thank T. R. McDermott for sharing unpublished data and stimulating discussion.
D.S. acknowledges MENRT for Ph.D. support. This work was partly performed in the framework of the Groupement de Recherche Métabolisme de l'Arsenic chez les Prokaryotes: de la Résistance à la Détoxication (GDR2909-CNRS).
Footnotes
Published ahead of print 8 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01771-14.
REFERENCES
- 1.Bird LJ, Bonnefoy V, Newman DK. 2011. Bioenergetic challenges of microbial iron metabolisms. Trends Microbiol. 19:330–340. 10.1016/j.tim.2011.05.001 [DOI] [PubMed] [Google Scholar]
- 2.Slyemi D, Bonnefoy V. 2012. How prokaryotes deal with arsenic. Environ. Microbiol. Rep. 4:571–586. 10.1111/j.1758-2229.2011.00300.x [DOI] [PubMed] [Google Scholar]
- 3.Finney LA, O'Halloran TV. 2003. Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science 300:931–936. 10.1126/science.1085049 [DOI] [PubMed] [Google Scholar]
- 4.Pennella MA, Giedroc DP. 2005. Structural determinants of metal selectivity in prokaryotic metal-responsive transcriptional regulators. Biometals 18:413–428. 10.1007/s10534-005-3716-8 [DOI] [PubMed] [Google Scholar]
- 5.Osman D, Cavet JS. 2010. Bacterial metal-sensing proteins exemplified by ArsR-SmtB family repressors. Nat. Prod. Rep. 27:668–680. 10.1039/b906682a [DOI] [PubMed] [Google Scholar]
- 6.Ma Z, Jacobsen FE, Giedroc DP. 2009. Coordination chemistry of bacterial metal transport and sensing. Chem. Rev. 109:4644–4681. 10.1021/cr900077w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Busenlehner LS, Pennella MA, Giedroc DP. 2003. The SmtB/ArsR family of metalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 27:131–143. 10.1016/S0168-6445(03)00054-8 [DOI] [PubMed] [Google Scholar]
- 8.Rensing C. 2005. Form and function in metal-dependent transcriptional regulation: dawn of the enlightenment. J. Bacteriol. 187:3909–3912. 10.1128/JB.187.12.3909-3912.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu G, Liu M, Kim EH, Maaty WS, Bothner B, Lei B, Rensing C, Wang G, McDermott TR. 2012. A periplasmic arsenite-binding protein involved in regulating arsenite oxidation. Environ. Microbiol. 14:1624–1634. 10.1111/j.1462-2920.2011.02672.x [DOI] [PubMed] [Google Scholar]
- 10.Sardiwal S, Santini JM, Osborne TH, Djordjevic S. 2010. Characterization of a two-component signal transduction system that controls arsenite oxidation in the chemolithoautotroph NT-26. FEMS Microbiol. Lett. 313:20–28. 10.1111/j.1574-6968.2010.02121.x [DOI] [PubMed] [Google Scholar]
- 11.Shi W, Wu J, Rosen BP. 1994. Identification of a putative metal binding site in a new family of metalloregulatory proteins. J. Biol. Chem. 269:19826–19829 [PubMed] [Google Scholar]
- 12.Ajees AA, Yang J, Rosen BP. 2011. The ArsD As(III) metallochaperone. Biometals 24:391–399. 10.1007/s10534-010-9398-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li S, Chen Y, Rosen BP. 2001. Role of vicinal cysteine pairs in metalloid sensing by the ArsD As(III)-responsive repressor. Mol. Microbiol. 41:687–696. 10.1046/j.1365-2958.2001.02546.x [DOI] [PubMed] [Google Scholar]
- 14.Wu J, Rosen BP. 1993. The arsD gene encodes a second trans-acting regulatory protein of the plasmid-encoded arsenical resistance operon. Mol. Microbiol. 8:615–623. 10.1111/j.1365-2958.1993.tb01605.x [DOI] [PubMed] [Google Scholar]
- 15.Chen Y, Rosen BP. 1997. Metalloregulatory properties of the ArsD repressor. J. Biol. Chem. 272:14257–14262. 10.1074/jbc.272.22.14257 [DOI] [PubMed] [Google Scholar]
- 16.Li S, Rosen BP, Borges-Walmsley MI, Walmsley AR. 2002. Evidence for cooperativity between the four binding sites of dimeric ArsD, an As(III)-responsive transcriptional regulator. J. Biol. Chem. 277:25992–26002. 10.1074/jbc.M201619200 [DOI] [PubMed] [Google Scholar]
- 17.Lin YF, Walmsley AR, Rosen BP. 2006. An arsenic metallochaperone for an arsenic detoxification pump. Proc. Natl. Acad. Sci. U. S. A. 103:15617–15622. 10.1073/pnas.0603974103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J, Rawat S, Stemmler TL, Rosen BP. 2010. Arsenic binding and transfer by the ArsD As(III) metallochaperone. Biochemistry 49:3658–3666. 10.1021/bi100026a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi W, Dong J, Scott RA, Ksenzenko MY, Rosen BP. 1996. The role of arsenic-thiol interactions in metalloregulation of the ars operon. J. Biol. Chem. 271:9291–9297. 10.1074/jbc.271.16.9291 [DOI] [PubMed] [Google Scholar]
- 20.Ye J, Ajees AA, Yang J, Rosen BP. 2010. The 1.4 Å crystal structure of the ArsD arsenic metallochaperone provides insights into its interaction with the ArsA ATPase. Biochemistry 49:5206–5212. 10.1021/bi100571r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin J, Fu HL, Ye J, Bencze KZ, Stemmler TL, Rawlings DE, Rosen BP. 2007. Convergent evolution of a new arsenic binding site in the ArsR/SmtB family of metalloregulators. J. Biol. Chem. 282:34346–34355. 10.1074/jbc.M706565200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ordonez E, Thiyagarajan S, Cook JD, Stemmler TL, Gil JA, Mateos LM, Rosen BP. 2008. Evolution of metal(loid) binding sites in transcriptional regulators. J. Biol. Chem. 283:25706–25714. 10.1074/jbc.M803209200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kashyap DR, Botero LM, Franck WL, Hassett DJ, McDermott TR. 2006. Complex regulation of arsenite oxidation in Agrobacterium tumefaciens. J. Bacteriol. 188:1081–1088. 10.1128/JB.188.3.1081-1088.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koechler S, Cleiss-Arnold J, Proux C, Sismeiro O, Dillies MA, Goulhen-Chollet F, Hommais F, Lievremont D, Arsene-Ploetze F, Coppee JY, Bertin PN. 2010. Multiple controls affect arsenite oxidase gene expression in Herminiimonas arsenicoxydans. BMC Microbiol. 10:53. 10.1186/1471-2180-10-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arsene-Ploetze F, Koechler S, Marchal M, Coppee JY, Chandler M, Bonnefoy V, Brochier-Armanet C, Barakat M, Barbe V, Battaglia-Brunet F, Bruneel O, Bryan CG, Cleiss-Arnold J, Cruveiller S, Erhardt M, Heinrich-Salmeron A, Hommais F, Joulian C, Krin E, Lieutaud A, Lievremont D, Michel C, Muller D, Ortet P, Proux C, Siguier P, Roche D, Rouy Z, Salvignol G, Slyemi D, Talla E, Weiss S, Weissenbach J, Medigue C, Bertin PN. 2010. Structure, function, and evolution of the Thiomonas spp. genome. PLoS Genet. 6:e1000859. 10.1371/journal.pgen.1000859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slyemi D, Moinier D, Talla E, Bonnefoy V. 2013. Organization and regulation of the arsenite oxidase operon of the moderately acidophilic and facultative chemoautotrophic Thiomonas arsenitoxydans. Extremophiles 17:911–920. 10.1007/s00792-013-0573-1 [DOI] [PubMed] [Google Scholar]
- 27.Duquesne K, Lieutaud A, Ratouchniak J, Muller D, Lett MC, Bonnefoy V. 2008. Arsenite oxidation by a chemoautotrophic moderately acidophilic Thiomonas sp.: from the strain isolation to the gene study. Environ. Microbiol. 10:228–237. 10.1111/j.1462-2920.2007.01447.x [DOI] [PubMed] [Google Scholar]
- 28.Slyemi D, Ratouchniak J, Bonnefoy V. 2007. Regulation of the arsenic oxidation encoding genes of a moderately acidophilic, facultative chemolithoautotrophic Thiomonas sp. Adv. Mat. Res. 20–21:427–430. 10.4028/www.scientific.net/AMR.20-21.427 [DOI] [Google Scholar]
- 29.Slyemi D, Moinier D, Brochier-Armanet C, Bonnefoy V, Johnson DB. 2011. Characteristics of a phylogenetically ambiguous, arsenic-oxidizing Thiomonas sp., Thiomonas arsenitoxydans strain 3AsT sp. nov. Arch. Microbiol. 193:439–449. 10.1007/s00203-011-0684-y [DOI] [PubMed] [Google Scholar]
- 30.Furste JP, Pansegrau W, Frank R, Blocker H, Scholz P, Bagdasarian M, Lanka E. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48:119–131. 10.1016/0378-1119(86)90358-6 [DOI] [PubMed] [Google Scholar]
- 31.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1998. Current protocols in molecular biology. Greene Publishing, New York, NY [Google Scholar]
- 32.Chung CT, Miller RH. 1988. A rapid and convenient method for the preparation and storage of competent bacterial cells. Nucleic Acids Res. 16:3580. 10.1093/nar/16.8.3580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guiliani N, Bengrine A, Borne F, Chippaux M, Bonnefoy V. 1997. Alanyl-tRNA synthetase gene of the extreme acidophilic chemolithoautotrophic Thiobacillus ferrooxidans is highly homologous to alaS genes from all living kingdoms but cannot be transcribed from its promoter in Escherichia coli. Microbiology 143(Pt 7):2179–2187 [DOI] [PubMed] [Google Scholar]
- 34.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 35.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 36.Guimaraes BG, Barbosa RL, Soprano AS, Campos BM, de Souza TA, Tonoli CC, Leme AF, Murakami MT, Benedetti CE. 2011. Plant pathogenic bacteria utilize biofilm growth-associated repressor (BigR), a novel winged-helix redox switch, to control hydrogen sulfide detoxification under hypoxia. J. Biol. Chem. 286:26148–26157. 10.1074/jbc.M111.234039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lane D, Prentki P, Chandler M. 1992. Use of gel retardation to analyze protein-nucleic acid interactions. Microbiol. Rev. 56:509–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu J, Rosen BP. 1993. Metalloregulated expression of the ars operon. J. Biol. Chem. 268:52–58 [PubMed] [Google Scholar]
- 39.Butcher BG, Rawlings DE. 2002. The divergent chromosomal ars operon of Acidithiobacillus ferrooxidans is regulated by an atypical ArsR protein. Microbiology 148:3983–3992 [DOI] [PubMed] [Google Scholar]
- 40.Cline DJ, Thorpe C, Schneider JP. 2003. Effects of As(III) binding on alpha-helical structure. J. Am. Chem. Soc. 125:2923–2929. 10.1021/ja0282644 [DOI] [PubMed] [Google Scholar]
- 41.Xu C, Shi W, Rosen BP. 1996. The chromosomal arsR gene of Escherichia coli encodes a trans-acting metalloregulatory protein. J. Biol. Chem. 271:2427–2432. 10.1074/jbc.271.5.2427 [DOI] [PubMed] [Google Scholar]
- 42.Santini JM, Kappler U, Ward SA, Honeychurch MJ, vanden Hoven RN, Bernhardt PV. 2007. The NT-26 cytochrome c552 and its role in arsenite oxidation. Biochim. Biophys. Acta 1767:189–196. 10.1016/j.bbabio.2007.01.009 [DOI] [PubMed] [Google Scholar]
- 43.Branco R, Francisco R, Chung AP, Morais PV. 2009. Identification of an aox system that requires cytochrome c in the highly arsenic-resistant bacterium Ochrobactrum tritici SCII24. Appl. Environ. Microbiol. 75:5141–5147. 10.1128/AEM.02798-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang YS, Bothner B, Rensing C, McDermott TR. 2012. Involvement of RpoN in regulating bacterial arsenite oxidation. Appl. Environ. Microbiol. 78:5638–5645. 10.1128/AEM.00238-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bush M, Dixon R. 2012. The role of bacterial enhancer binding proteins as specialized activators of σ54-dependent transcription. Microbiol. Mol. Biol. Rev. 76:497–529. 10.1128/MMBR.00006-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wigneshweraraj S, Bose D, Burrows PC, Joly N, Schumacher J, Rappas M, Pape T, Zhang X, Stockley P, Severinov K, Buck M. 2008. Modus operandi of the bacterial RNA polymerase containing the σ54 promoter-specificity factor. Mol. Microbiol. 68:538–546. 10.1111/j.1365-2958.2008.06181.x [DOI] [PubMed] [Google Scholar]
- 47.Ghosh T, Bose D, Zhang X. 2010. Mechanisms for activating bacterial RNA polymerase. FEMS Microbiol. Rev. 34:611–627. 10.1111/j.1574-6976.2010.00239.x [DOI] [PubMed] [Google Scholar]
- 48.Shingler V. 2011. Signal sensory systems that impact σ54-dependent transcription. FEMS Microbiol. Rev. 35:425–440. 10.1111/j.1574-6976.2010.00255.x [DOI] [PubMed] [Google Scholar]
- 49.Joly N, Zhang N, Buck M, Zhang X. 2012. Coupling AAA protein function to regulated gene expression. Biochim. Biophys. Acta 1823:108–116. 10.1016/j.bbamcr.2011.08.012 [DOI] [PubMed] [Google Scholar]
- 50.Francke C, Groot Kormelink T, Hagemeijer Y, Overmars L, Sluijter V, Moezelaar R, Siezen RJ. 2011. Comparative analyses imply that the enigmatic sigma factor 54 is a central controller of the bacterial exterior. BMC Genomics 12:385. 10.1186/1471-2164-12-385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Callen BP, Shearwin KE, Egan JB. 2004. Transcriptional interference between convergent promoters caused by elongation over the promoter. Mol. Cell 14:647–656. 10.1016/j.molcel.2004.05.010 [DOI] [PubMed] [Google Scholar]
- 52.Palmer AC, Egan JB, Shearwin KE. 2011. Transcriptional interference by RNA polymerase pausing and dislodgement of transcription factors. Transcription 2:9–14. 10.4161/trns.2.1.13511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chatterjee A, Drews L, Mehra S, Takano E, Kaznessis YN, Hu WS. 2011. Convergent transcription in the butyrolactone regulon in Streptomyces coelicolor confers a bistable genetic switch for antibiotic biosynthesis. PLoS One 6:e21974. 10.1371/journal.pone.0021974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chatterjee A, Johnson CM, Shu CC, Kaznessis YN, Ramkrishna D, Dunny GM, Hu WS. 2011. Convergent transcription confers a bistable switch in Enterococcus faecalis conjugation. Proc. Natl. Acad. Sci. U. S. A. 108:9721–9726. 10.1073/pnas.1101569108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goodrich JA, Schwartz ML, McClure WR. 1990. Searching for and predicting the activity of sites for DNA binding proteins: compilation and analysis of the binding sites for Escherichia coli integration host factor (IHF). Nucleic Acids Res. 18:4993–5000. 10.1093/nar/18.17.4993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosen BP, Tamas MJ. 2010. Arsenic transport in prokaryotes and eukaryotic microbes. Adv. Exp. Med. Biol. 679:47–55. 10.1007/978-1-4419-6315-4_4 [DOI] [PubMed] [Google Scholar]
- 57.Kashyap DR, Botero LM, Lehr C, Hassett DJ, McDermott TR. 2006. A Na+:H+ antiporter and a molybdate transporter are essential for arsenite oxidation in Agrobacterium tumefaciens. J. Bacteriol. 188:1577–1584. 10.1128/JB.188.4.1577-1584.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kang YS, Shi Z, Bothner B, Wang G, McDermott TR. 27 March 2014. Involvement of the Acr3 and DctA antiporters in arsenite oxidation in Agrobacterium tumefaciens 5A. Environ. Microbiol. 10.1111/1462-2920.12468 [DOI] [PubMed] [Google Scholar]
- 59.Carey MF, Peterson CL, Smale ST. 2012. Experimental strategies for the identification of DNA-binding proteins. Cold Spring Harbor Protoc. 2012:18–33. 10.1101/pdb.top067470 [DOI] [PubMed] [Google Scholar]
- 60.Andres J, Arsene-Ploetze F, Barbe V, Brochier-Armanet C, Cleiss-Arnold J, Coppee JY, Dillies MA, Geist L, Joublin A, Koechler S, Lassalle F, Marchal M, Medigue C, Muller D, Nesme X, Plewniak F, Proux C, Ramirez-Bahena MH, Schenowitz C, Sismeiro O, Vallenet D, Santini JM, Bertin PN. 2013. Life in an arsenic-containing gold mine: genome and physiology of the autotrophic arsenite-oxidizing bacterium Rhizobium sp. NT-26. Genome Biol. Evol. 5:934–953. 10.1093/gbe/evt061 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.