Abstract
Subtilomycin was detected from the plant endophytic strain Bacillus subtilis BSn5 and was first reported from B. subtilis strain MMA7. In this study, a gene cluster that has been proposed to be related to subtilomycin biosynthesis was isolated from the BSn5 genome and was experimentally validated by gene inactivation and heterologous expression. Comparison of the subtilomycin gene cluster with other verified related lantibiotic gene clusters revealed a particular organization of the genes apnI and apnT downstream of apnAPBC, which may be involved in subtilomycin immunity. Through analysis of expression of the apnI and/or apnT genes in the subtilomycin-sensitive strain CU1065 and inactivation of apnI and apnT in the producer strain BSn5, we showed that the single gene apnI, encoding a putative transmembrane protein, was responsible for subtilomycin immunity. To our knowledge, evidence for lantibiotic immunity that is solely dependent on a transmembrane protein is quite rare. Further bioinformatic analysis revealed the abundant presence of ApnI-like proteins that may be responsible for lantibiotic immunity in Bacillus and Paenibacillus. We cloned the paeI gene, encoding one such ApnI-like protein, into CU1065 and showed that it confers resistance to paenibacillin. However, no cross-resistance was detected between ApnI and PaeI, even though subtilomycin and paenibacillin share similar structures, suggesting that the protection provided by ApnI/ApnI-like proteins involves a specific-sequence recognition mechanism. Peptide release/binding assays indicated that the recombinant B. subtilis expressing apnI interacted with subtilomycin. Thus, ApnI represents a novel model for lantibiotic immunity that appears to be common.
INTRODUCTION
Bacillus subtilis is a Gram-positive, endospore-forming model bacterium. In general, B. subtilis grows in association with plants and promotes plant growth (1). However, a recent report revealed that B. subtilis is also a saprophytic organism in other environments, such as in an animal's gastrointestinal tract (2). Dozens of antibiotics and enzymes produced by various B. subtilis isolates are used in the food industry and agricultural fields. Because of its ability to take up DNA and integrate it into its genome, i.e., natural competence (3), its genetic sequences have been well characterized, including those related to the biosynthesis and regulation mode of most of the antibiotics discovered in this species to date.
B. subtilis is also known to produce lantibiotics (4), which are lanthipeptides with antimicrobial activity (5). Lanthipeptides are a class of ribosomally synthesized and posttranslationally modified peptides (6), which are also referred to as lanthionine (Lan) or 3-methyl-lanthionin (MeLan)-containing peptides. The ring-forming Lan and MeLan peptides are usually generated via a two-step modification process, involving dehydration of the threonine (Thr) and/or serine (Ser) in the precursor peptide to form the double bonds, followed by cyclization mediated by a Michael addition with the thiol bond of cysteine (Cys) in the proper site. This process involves the dehydratase and cyclase, which are, respectively, encoded by either two dependent genes, lanB and lanC, or a combined gene, lanM. Lanthipeptides are subdivided into four classes (I to IV) depending on the specific biosynthetic gene(s) involved. Structurally established lantibiotics can be further subdivided based on sequence similarity, such as the Pep5 group (7). Class I lantibiotics are synthesized as the precursor peptide LanA and are modified by two different enzymes, LanB enzyme (dehydratase) and LanC enzyme (cyclase), and are exported by LanT, and then the leading peptides are removed by the extracellular serine protease LanP.
A lantibiotic producer must possess one or two synergistic immunity mechanisms to protect itself against its own produced lantibiotics. The reported immunity mechanisms usually involve immunity proteins, including ABC transporters (LanFE[G]), lipoproteins or membrane-associated peptides (LanI), and transmembrane proteins (LanH) (8). For lantibiotic immunity, ABC transporters export lantibiotics to the extracellular space by ATP hydrolysis, and in some cases, the transmembrane protein LanH acts as an accessory protein for the ABC transporters involved in substrate recognition (9, 10). In addition, lipoproteins or membrane-associated proteins (LanI) can work either solely or synergistically with ABC transporters by interacting with lantibiotics (11, 12). Most of the LanI proteins thus far reported are located on the external surface of the membrane, such as NisI, SpaI, and PepI. Very few LanI proteins, especially those solely providing immunity, are located within the cell membrane.
Subtilomycin is a recently identified lantibiotic produced by B. subtilis strain MMA7 isolated from the marine sponge Haliclona simulans, and it shows a broad spectrum of activity against Gram-positive bacteria, including drug-resistant pathogens (13). The relationship between subtilomycin and its gene cluster was established based on the close match between the N-terminal amino acid sequence and the 5′-end sequence of the structural gene subA. Paenibacillin is a lantibiotic produced by the strain Paenibacillus polymyxa OSY-DF (14), and its structure has been elucidated (15); however, the paenibacillin biosynthetic gene cluster has not yet been reported. Since it shows high sequence similarity (54%) to subtilomycin, the structure of subtilomycin has been proposed using the structure of paenibacillin as a template (13). However, no molecular biological evidence has been provided to validate this proposed biosynthetic gene cluster.
In this report, we provide direct evidence to confirm the presence of a subtilomycin gene cluster, termed the apn cluster, in the complete genome of B. subtilis strain BSn5, which was previously isolated from the plant Amorphophallus konjac (16). We compared the gene organization of the apn cluster with those of other related gene clusters to determine its specificity and potential function. DNA fragments containing different regions of the gene cluster were expressed in the heterologous host B. subtilis strain CU1065 (W168 attSPβ trpC2) (17), and gene inactivation analysis was performed for the producer BSn5 strain to identify the gene(s) involved in conferring immunity. Our study thus provides valuable information of a rare molecular model underlying lantibiotic immunity.
MATERIALS AND METHODS
Bacterial strains, plasmids, and general methods.
The strains and plasmids used in this study are listed in Table 1. B. subtilis strains BSn5 and CU1065, along with their derivatives, were cultured in Luria-Bertani (LB) medium at 28°C, and Escherichia coli DH5α, which was used as a host for clones, was routinely cultured in LB medium (Invitrogen, Karlsruhe, Germany) at 37°C. For solid culture, LB medium was supplemented with 1.5% agar. For stock preparation, the culture was cultivated overnight at 28°C in LB medium mixed with sterile glycerol (final concentration of 17%) and stored at −80°C. For selective media, the following antibiotics were added: ampicillin, 100 μg/ml (E. coli); spectinomycin, 100 μg/ml (B. subtilis); and kanamycin, 20 μg/ml (B. subtilis). Isolation of plasmids and chromosomal DNA, restriction endonuclease digestion, agarose gel electrophoresis, PCR, and transformation of E. coli were performed by following established protocols for molecular biology techniques described by Sambrook and Russell (18).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli DH5α | Cloning host | 42 |
| Paenibacillus ATCC 842 | Wild type; paenibacillin producer strain | 28 |
| B. subtilis | ||
| BSn5 | Wild type; subtilomycin producer | 16 |
| B1201 (ΔapnB) | Strain BSn5 derivative; ΔapnB::spc | This study |
| B1202 (ΔapnT) | Strain BSn5 derivative; ΔapnT::kan | This study |
| B1203 (ΔybcL) | Strain BSn5 derivative; ΔybcL::kan | This study |
| B1204 (ΔybdG) | Strain BSn5 derivative; ΔybdG::kan | This study |
| B1205 (ΔapnI) | Strain BSn5 derivative; ΔapnI::kan | This study |
| CU1065 | Strain168 derivative; trpC2 attSPβ | 17 |
| B1301 | CU1065 amyE::(spc subtilomycin gene cluster) | This study |
| B1302 | CU1065 amyE::(spc fragment apnI) | This study |
| B1303 | CU1065 amyE::(spc fragment apnT) | This study |
| B1304 | CU1065 amyE::(spc fragment apnI-apnT) | This study |
| B1305 | CU1065 amyE::(spc fragment proapnA apnI) | This study |
| B1306 | CU1065 amyE::(spc fragment proapnA apnT) | This study |
| B1307 | CU1065 amyE::(spc fragment proapnA apnI-apnT) | This study |
| B1308 | CU1065 amyE::(spc fragment proapnA paeI) | This study |
| B1309 | CU1065 amyE::(spc fragment propaeA paeI) | This study |
| Plasmids | ||
| pDG780 | pBluescript KS+ harboring kan (amp kan) | 43 |
| pMD18T simple | Clone vector for E. coli (amp) | TaKaRa |
| pIC333 | Mini-Tn10 transposons delivery vector harboring spc and erm | 44 |
| pDG1730 | Integration vector harboring amyE::spc (amp erm) | 45 |
| pB1201 | pMD18T harboring mutation of ΔapnB::spc | This study |
| pB1202 | pDG780 harboring mutation of ΔapnT::kan | This study |
| pB1203 | pDG780 harboring mutation of ΔybcL::kan | This study |
| pB1204 | pDG780 harboring mutation of ΔybdG::kan | This study |
| pB1205 | pDG780 harboring mutation of ΔapnI::kan | This study |
| pB1301 | pDG1730 harboring amyE::(spc apn gene cluster) | This study |
| pB1302 | pDG1730 harboring amyE::(spc apnI) | This study |
| pB1303 | pDG1730 harboring amyE::(spc apnT) | This study |
| pB1304 | pDG1730 harboring amyE::(spc apnI-apnT) | This study |
| pB1305 | pDG1730 harboring amyE::(spc proapnA apnI) | This study |
| pB1306 | pDG1730 harboring amyE::(spc proapnA apnT) | This study |
| pB1307 | pDG1730 harboring amyE::(spc proapnA apnI-T) | This study |
| pB1308 | pDG1730 harboring amyE::(spc proapnA paeI) | This study |
| pB1309 | pDG1730 harboring amyE::(spc propaeA paeI) | This study |
Plasmid construction and manipulation of B. subtilis strains.
Gene disruption plasmids were constructed using two approaches. The first was establishment of the apnB mutant allele (apnB::spc) by subcloning of the spectinomycin resistance gene (spc) from pIC333 into the PCR-introduced ClaI and XbaI sites of the apnB gene in pMD18T-apnB, resulting in plasmid pB1201 (Table 1). The second approach was the establishment of apnT, ybcL, ybdG, and apnI mutant alleles (apnT::kan, ybcL::kan, ybdG::kan, and apnI::kan, respectively) by amplifying the upstream and downstream arms of each gene fragment into the upstream and downstream restriction sites of the integration vector pDG780, respectively, resulting in plasmids pB1202, pB1203, pB1204, and pB1205 (Table 1). The B. subtilis competent cell preparation was performed by following the classical nutritional downshift method described by Anagnostopoulos and Spizizen in 1960 (3), with modifications described by Yasbin et al. (19). A series of at least five 10-min interval grades were set to ensure that the t0 point (i.e., the point at which the culture leaves the logarithmic growth phase) could be captured at every cycle considering the lower transformation efficiency of wild-type strain BSn5 than of B. subtilis 168. Chromosomal gene replacement of wild-type apnB, apnT, ybcL, and ybdG with the corresponding constructed mutation alleles was carried out by transforming BSn5 with the constructed plasmids, resulting in the mutants B1201, B1202, B1203, B1204, and B1205. B1201 was selected for spectinomycin resistance evaluation, and the other mutants were selected for kanamycin resistance evaluation. Proper allelic replacement was confirmed by PCR and sequencing.
Different fragments containing the whole subtilomycin gene cluster, apnI, apnT, or apnI-apnT were amplified using the primers listed in Table 2. Then, these fragments were cut using proper restriction endonuclease and cloned into another integration vector, pDG1730, resulting in the heterologous expression alleles amyE::(spc subtilomycin gene cluster), amyE::(spc fragment apnI), amyE::(spc fragment apnT), and amyE::(spc fragment apnI-apnT) in plasmids pB1301, pB1302, pB1303, and pB1304, respectively. The fragment containing the predicted promoter region of apnA (proapnA) was amplified with primers containing BamHI restriction sites and cloned into all of the constructed vectors, except for that containing the whole apn cluster. PCR was used to identify the inserted direction of the proapnA fragment. The same procedure as described above was used to prepare the competent cells of B. subtilis CU1065. The heterologous expression recombinants were selected for spectinomycin resistance and were confirmed by PCR and sequencing.
TABLE 2.
Primer sequences used in this study
| Primer | Sequence (5′→3′)a | Use |
|---|---|---|
| ybcL UpF | CTAACTAGTCAGAGATCAGTAAGTCC | ybcL gene disruption |
| ybcL UpR | TGTCTGCAGAGGTTCCAACTAAAAAGC | ybcL gene disruption |
| ybcL DownF | GGACTCGAGAGTTTGGTTGGATCAAAG | ybcL gene disruption |
| ybcL DownR | TGAGGTACCCTCAGTAATCTTATTAGCC | ybcL gene disruption |
| ApnBUpF | TTTGCATGCTTAGTATTCTCGAGCAG | apnB gene disruption |
| ApnBDownR | TGCGGATCCATTAAATAGGGATAGTG | apnB gene disruption |
| ApnBUpR | CCTTCGTATCCTATTACTCGCATTT | apnB gene disruption |
| ApnBDownF | AGAATCTAGAGCAATCACCTATTCTAAGAGC | apnB gene disruption |
| SpcF | AGTATCGATGCGGTGCTACAGAGTTCTTG | Amplification of spectinomycin resistance gene |
| SpcR | GGGTCTAGAGTAAACGCTGAATATCGTGTT | Amplification of spectinomycin resistance gene |
| ApnTUpF | TTTGGATCCGGTTGTTGAAGACGCAG | apnT gene disruption |
| ApnTDownR | TCCTTTCCCATACTCTTC | apnT gene disruption |
| ApnTDownF | CAACTCGAGCGAGTTCTCTAAGTTA | apnT gene disruption |
| ApnTDownR | TTTGGTACCTGGTCAGAGACCGCATCT | apnT gene disruption |
| YbdGUpF | GGGACTAGTTCGAGATAG | ybdG gene disruption |
| YbdGUpR | CACCTGCAGATTATATACAGCGGAAG | ybdG gene disruption |
| YbdGDownF | TATCTCGAGGGTTCGCTAGGATGAAGG | ybdG gene disruption |
| YbdGDownR | TGAGGTACCGGTTTAAACATGCTGTC | ybdG gene disruption |
| ApnIUpF | GAAACTAGTTCTAGAGGCGGAACAAGAT | apnI gene disruption |
| ApnIDownR | GGGGAATTCATGAAAAGAACTGCGAACC | apnI gene disruption |
| ApnIDownF | TAACTCGAGAGGTTATACATATGGGC | apnI gene disruption |
| ApnIDownR | GCAACAAAGGTACCTGGTGAAATAG | apnI gene disruption |
| apnF | CCTGGATCCGCTCTGTTTCCGTTGTCG | Clone of apn cluster |
| apnR | TGAGGATCCTACCCAAGTGGTCTCATTATTT | Clone of apn cluster |
| ApnIF | TTCGGATCCGGTCAAGATTCGTGGGAC | Clone of fragment apnI-apnT (I) |
| ApnIR | GGCGAATTCGTTATTGCCCATATGTATAAC | Clone of fragment apnI |
| ApnTF | AGTGGATCCTAGCGAAAGTGTTCATTC | Clone of fragment apnT |
| ApnTR | GACGAATTCAAGCAAAACCAAAGCAGCC | Clone of fragment apnI-apnT (T) |
| ProapnAF | TTTGGATCCGCGAGCTGAAGTACAGTACG | Clone of the apnA promoter region |
| ProapnAR | CTTGGATCCTTCCATTTCCTCCTTTT | Clone of the apnA promoter region |
| ProapnAF2 | TTTGGATCCGCGAGCTGAAGTACAGTACG | Clone of the apnA promoter region |
| ProapnAR2 | CTTGAATTCCTTAGCTCGAGTTCCATTTCCTCCTTTT | Clone of the apnA promoter region |
| PropaeAF | AACGGATCCATCGGGACCAAAGTGATTCGACTCA | Clone of the paeA promoter region |
| PropaeAR | CTTGAATTCTGCCTCTCGAGTTTCCTCTTCCTCTCTGAATTAAAT | Clone of the paeA promoter region |
| PaeIF | TTTGGATCCCTTAGCTCGAGCAGATGCTGGTATTTAGCT | Clone of the paeI gene |
| PaeIR | CTCCGAATTCGAATGGGCCGTTTGTTTT | Clone of the paeI gene |
Restriction sites are underlined.
To express the gene paeI for putative paenibacillin immunity in CU1065, the fragments containing paeI and the predicted promoter regions of apnA and paeA (proapnA and propaeA, respectively) were amplified using the primers listed in Table 2 and were T/A cloned into the pMD18T Simple vector. Then, the paeI fragments were cut and inserted into the downstream regions of the proapnA and propaeA fragments, resulting in insertion of proapnA-paeI and propaeA-paeI, respectively, in the vector pMD18T Simple. Finally, the two fragments were cut using BamHI and EcoRI and cloned into the integration vector pDG1730, resulting in the heterologous expression alleles amyE::(spc fragment proapnA-paeI) and amyE::(spc fragment propaeA-paeI) in plasmids pB1308 and pB1309 (Table 1). The B. subtilis CU1065 transformation and the confirmation of recombinants were performed by following the procedures described above.
Isolation and purification of subtilomycin.
To prepare subtilomycin, a single colony of BSn5 was inoculated from the culture plate into 5 ml of LB medium with agitation, and then the overnight culture was transferred into a 500-ml flask containing 100 ml of LB medium at the ratio of 1/100 (vol/vol). The flask was incubated at 28°C for 12 h in a rotary shaker with agitation at 200 rpm. After centrifugation at 10,000 × g for 10 min at 4°C using the centrifuge Sorvall ST 16R (Thermo Fisher Scientific; USA), the cell-free supernatant was filtered through a 0.22-μm membrane filter. Ammonium sulfate was added intermittently to the cell-free supernatant to 30% saturation during stirring using a magnetic stirrer. The mixture was set at 4°C for 2 h. The resulting precipitate was collected by centrifugation at 12,000 × g for 30 min at 4°C, followed by dialyzation desalination. The extract was used as the active ammonium sulfate crude extract (ASCE) of the subtilomycin preparation for further bioassays, high-performance liquid chromatography (HPLC), and matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrum (MS) analysis. The subtilomycin extracts from BSn5 mutants were prepared by following the same protocol.
Crude subtilomycin extracts were condensed by filtering through a Millipore Amicon Ultra-15 3K device and adjusted to 25% acetonitrile (0.1% trifluoroacetic acid [TFA]), followed by centrifugation at 12,000 × g for 5 min to remove insoluble material. Subtilomycin was purified by preparative HPLC using a Waters 1528 binary HPLC pump, Waters 2489 UV/visible detector, and Waters 2707 auto-sample system equipped with an Agilent Eclipse XDB-C18 PrepHT column (5 μm, 21.2 by 150 mm). The mobile phase consisted of chromatography-grade water containing 0.1% trifluoroacetic acid (TFA) in pump A and 80% chromatography-grade acetonitrile containing 0.086% TFA in pump B. The elution step was performed under an acetonitrile gradient of 30 to 70% over 20 min at a flow rate of 9.0 ml/min. Subtilomycin was eluted within 14.3 min (see Fig. S4 in the supplemental material). The purified peptide was lyophilized and used for subtilomycin quantification by HPLC.
Bioassay for detection of subtilomycin.
Agar well diffusion assays were used to detect the antimicrobial activities of the isolates as previously described (20). Thirty milliliters of LB agar (1.0%, wt/vol) medium was poured into 100- by 100-mm2 plastic dishes and inoculated (1%, vol/vol) with the indicator suspension. After drying for 30 min, nine 6-mm wells were bored in each plate. Approximately 60 μl of crude extract samples from the supernatants of B. subtilis strains was pipetted into each well, and the plates were maintained at 4°C for 4 h for diffusion and then incubated overnight at 28°C. The diameter of the inhibition zones was measured to determine antimicrobial activity.
Detection of subtilomycin by HPLC and liquid chromatography (LC)-MS.
The same HPLC system as described above for purification was used for subtilomycin detection. The data were displayed and analyzed by using a Breeze system (Waters Corporation, USA). An Agilent HC-C18 reverse-phase column (250 by 4.6 mm; particle size of 5 μm) was employed (Agilent, USA). The mobile phase consisted of chromatography-grade water containing 0.1% TFA in pump A and chromatography-grade acetonitrile in pump B. The crude lantibiotic preparation was dissolved in 20% acetonitrile (TFA, 0.1%), and 5 to 20 μl was loaded. The elution step was performed under an acetonitrile gradient of 20 to 80% over 15 min at a flow rate of 1 ml/min. The peaks were detected by measuring the absorbance at 235 nm and 254 nm.
Liquid chromatography quadrupole-time of flight (LC-Q-TOF) MS was carried out using an Agilent 1260 LC device equipped with a C18 reverse-phase column (100 by 1.8 mm; particle size of 3.5 μm) under chromatography detection conditions as the flow rate was decreased by 0.3 ml/min; the injection volume was 1 μl, and the diode array detection was performed at 254 nm. MS was performed using the Q-TOF MS G6540A system (Agilent) equipped with a dual-source electrospray ionization (ESI) ion source and was operated in positive-ion mode. Calibration was carried out with standard references of mass 121.0509 and 922.0098. The source parameters were as follows: gas temperature of 350°C, gas flow rate of 9 liters/min, and nebulizer stress of 40 lb/in2 (gauge). The capillary, fragmenter, skimmer, and octopole radio frequency (RF) peak voltage were set at 4,000 V, 250 V, 65 V, and 750 V, respectively, for the scan source. The quadrupole was set to pass ions from m/z 200 to 2,000. The MS scan rate was set to 1.5 spectra/s.
After determination of the ions of subtilomycin at m/z 1,079.1666 by MS, target tandem MS (MS/MS) mode was chosen for acquisition of the fragmental ions of subtilomycin. The MS and MS/MS scan rates were set to 1.5 spectra/s and 0.8 spectrum/s, respectively. The MS and MS/MS scan ranges were set to m/z 200 to 2,000 and m/z 100 to 2,000, respectively. The target ions were isolated and fragmented in the collision cell by adding optimized collision energy at 34 eV. Data were analyzed using Agilent MassHunter qualitative analysis software.
Detection of subtilomycin by MALDI-TOF MS.
The MS analysis of the peptide preparations was performed by using the 4800 Plus MALDI TOF/TOF analyzer (Applied Biosystems, USA). Aliquots of 0.5-μl extracts were mixed with a 0.5-μl sample of matrix (α-cyano-4-hydroxycinnamic acid in acetonitrile and 0.1% TFA in water, 1:3). The samples were spotted onto the MALDI target and dried in air. Mass spectra were measured in positive ion-mode in the range of m/z 800 to 4,000 for searching and 2,000 to 5,000 Da for comparison and analyzed using ProteinPilot software.
Preparation of peptide antibiotics for sensitivity assay.
Vancomycin and nisin were purchased from Sigma-Aldrich, USA. By searching the National Center for Biotechnology Information (NCBI) database using the paenibacillin structural sequence, we found that a Bacillus Genetic Stock Center (BGSC) preserved isolate, Paenibacillus strain ATCC 842, with complete genome sequence information, may produce paenibacillin. Based on a previously reported isolation and detection strategy (14), we aimed to detect and prepare crude extracts of paenibacillin. Briefly, a single colony of ATCC 842 was activated in 5 ml of tryptic soy broth yeast extract and incubated at 28°C for 24 h. The resulting culture was inoculated into a 500-ml flask containing 100 ml of tryptic soy broth yeast extract. The flask was incubated at 28°C for 24 h in a rotary shaker with agitation at 200 rpm. After centrifugation at 9000 × g for 10 min, the cell-free supernatant was mixed with Amberlite XAD-7 resin (Sigma) at a 10% (wt/vol) ratio, and the mixture was set for static adsorption overnight at 4°C. The absorbed paenibacillin resin was washed sequentially with 1 liter of distilled water and 500 ml of 30% (vol/vol) ethanol. Finally, the resin was eluted with 250 ml of 75% (vol/vol) ethanol (pH 2.0). The fraction was condensed with a rotary evaporator at 35°C under vacuum, and the concentrate was adjusted to pH 7.0 with phosphate buffer and then was freeze-dried. The powder was resolved in distilled water, followed by centrifugation at 12,000 × g for 5 min. The resulting supernatant was considered to be the paenibacillin crude extract and was used to detect paenibacillin under LC-MS by following the same procedure as described above for subtilomycin detection.
Subtilomycin sensitivity assay.
The subtilomycin sensitivity assay of B. subtilis CU1065 and recombinants was performed using agar diffusion tests according to a modification of a previously described protocol (21). Stationary-phase-grown CU1065 and its recombinant cultures with an optical density at 600 nm (OD600) of 0.8 were each inoculated at 1/1,000 into diluted LB agar (1.0%, wt/vol) at 40°C and then poured into 90-mm petri dishes (20 ml) and dried for 30 min. Seven 6-mm wells were bored into each plate. Serially diluted (0.125 to 200 μM) subtilomycin extracts (60 μl) were loaded into the wells of test plates. The plates were maintained at 4°C for 4 h to allow sufficient diffusion and then incubated overnight at 28°C. The response was calculated as the mean area of the inhibition halos. Each test was independently replicated three times. The same procedure was employed for vancomycin, nisin, and paenibacillin sensitivity tests of B. subtilis CU1065 and the apnI-expressing recombinants (proapnAapnI).
Peptide-binding assay for subtilomycin.
The peptide-binding assay for subtilomycin was performed according to a previously described method, with some modifications (21). B. subtilis CU1065 and its recombinant expressing apnI were cultured overnight. The cells were harvested and washed twice with 50 mM sodium phosphate buffer (pH 7.0) containing 1% glucose. The cell concentration was adjusted to an OD600 of 10 with 1 ml of incubation buffer containing 50 mM sodium phosphate, 1% glucose, and 1 M NaCl. The cell suspension was incubated with subtilomycin at concentrations of 15 μg/ml and 25 μg/ml with shaking (150 rpm) for 30 min at 30°C. After incubation, the cells were centrifuged at 17,000 × g for 10 min. The supernatants were transferred to the HPLC system with the same parameters as described above. The harvested cell pellets were gently mixed with 1 ml of 20% acetonitrile in water containing 0.1% trifluoroacetic acid and incubated with shaking (150 rpm) for 5 min at 30°C. The cells were removed by centrifugation at 17,000 × g for 10 min, and the remaining supernatants were loaded into the HPLC system. Subtilomycin amounts in cell-free supernatants and in cell extracts were quantitatively determined.
RESULTS
Production of subtilomycin in B. subtilis strain BSn5.
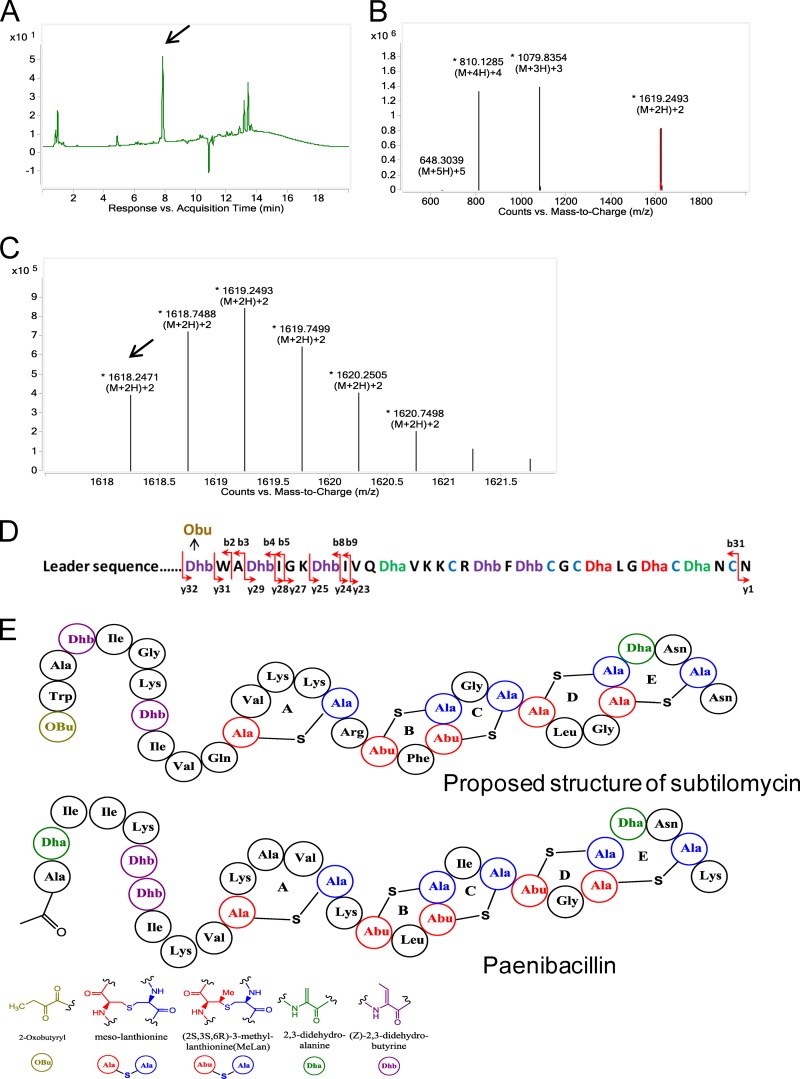
In accordance with a previously reported method for purifying subtilomycin (13), we prepared ASCEs to detect the production of subtilomycin from the 12-h supernatant of B. subtilis BSn5. LC-electrospray ionization MS of the crude extracts revealed that BSn5 can produce a compound with the same characteristics as subtilomycin. The corresponding main peak was detected at a retention time of 7.9 min under the UV monitor at 254 nm using similar mobile-phase conditions (Fig. 1A), and the MS extracted from this peak showed that the monoisotopic mass of the quadruply charged [M + 4H]4+, triply charged [M + 3H]3+, and doubly charged [M + 2H]2+ ions present in the sample were m/z 809.6273, m/z 1,079.1666, and m/z 1,618.2471, respectively, which are all equivalent to the mass of 3,234.48 Da, which corresponds extremely well to the theoretical mass of subtilomycin of 3,234.4806 Da (Fig. 1B and C).
FIG 1.
Detection of the production of subtilomycin by liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS). (A) Chromatography profile of active ammonium sulfate crude extracts of the Bacillus subtilis BSn5 culture supernatant. The target compound is indicated by an arrow. (B) ESI-MS of the compound indicated by the arrow in panel A. (C) Magnification of the doubly charged ions [M + 2H]2+ in panel B; the monoisotopic mass peak is indicated by an arrow. (D) Fragmental ions obtained from target tandem mass spectrometry of the peptide with a mass of 3,234.48 Da. Ions y and b indicate the fragmented peptides extended from the carboxyl terminus and amino terminus, respectively. (E) Predicted structure of subtilomycin and established structure of paenibacillin (15). The predicted structure motifs are shown in different colors. Obu refers to 2-oxobutyryl.
Based on the MS/MS results, we further verified the predicted N-terminal modification of 2-oxobutyrate. Similar to the reported N-terminal modification of Pep5 (22), the N-terminal Dhb (C4H6NO—) is spontaneously deaminated into 2-oxobutyrate (C4H5O2—) in subtilomycin, resulting in an accurate mass gain of 0.9843 Da. All the measured b ions (extending from the amino terminus) are consistent with the mass of peptides with an N-terminal 2-oxobutyrate modification (see Table S1 in the supplemental material). Previously, Li et al. used tandem mass spectrometric analysis to determine the ring topology of lanthipeptides that do not contain overlapping rings (23). As the proposed subtilomycin structure does contain overlapping rings, we could not directly determine the subtilomycin structure based on our MS/MS results. However, significantly increased fragmentation was observed at the N-terminal region, which contains no predicted thioether rings, thereby providing substantial support for the predicted subtilomycin structure (Fig. 1D and E).
Identification and cloning of the subtilomycin biosynthetic gene cluster.
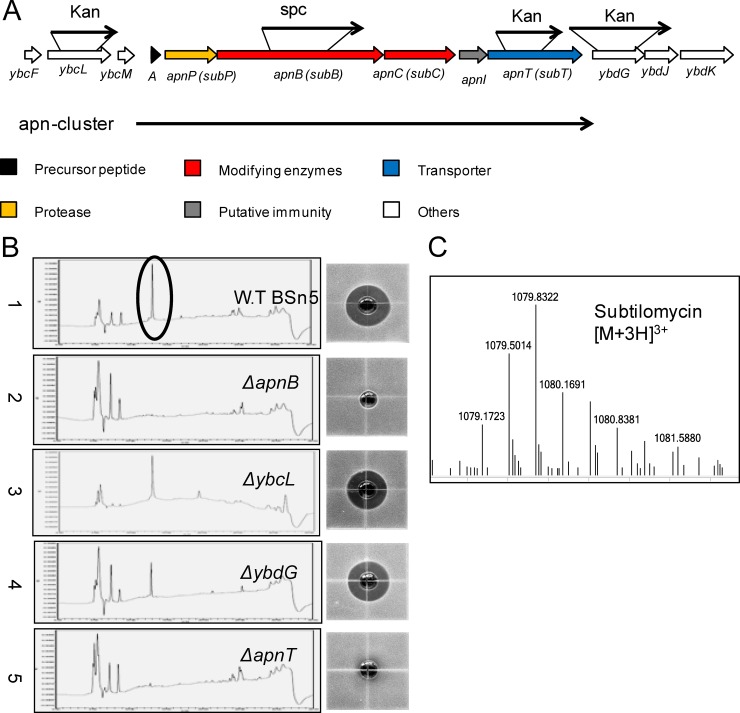
Through scanning the whole genome sequence of strain BSn5, a putative subtilomycin gene cluster (BSn5_12550 to BSn5_12575), termed the apn cluster, that spans positions 2428600 to 2436982 was identified (16). Although the relationship between subtilomycin and its biosynthetic gene cluster was recently established based on the single match between the primary amino acid sequence of the N-terminal region of subtilomycin and the 5′ end of the structural gene subA (13), no direct molecular biological evidence has yet been provided to validate the function of the proposed subtilomycin biosynthetic gene cluster. Therefore, we explored the effect of gene deletion of apnB, which encodes a LanB-like dehydratase, and apnT, which encodes a putative transporter, to identify the function of the gene cluster (Fig. 2A).
FIG 2.
The subtilomycin gene cluster and inactivation analysis of related genes. (A) The apn cluster genes are shown in different colors: the precursor peptide encoded by apnA is shown in black, the genes encoding modifying enzymes are shown in red, the gene encoding protease is shown in yellow, the gene involved in putative immunity is shown in gray, and the gene encoding the transporter is shown in blue. The flanking genes are shown in white and are numbered. The black arrow indicates related genes interrupted by spectinomycin (Spc) or kanamycin (Kan) resistance genes. (B) Bioassay and high-performance liquid chromatography results for detection of subtilomycin from active ammonium sulfate crude extracts of wild-type strain BSn5 and its mutants. 1, wild-type strain BSn5; 2, ΔapnB mutant; 3, ΔybcL mutant; 4, ΔybdG mutant; 5, ΔapnT mutant. The black ring refers to the respective target peaks of subtilomycin in high-performance liquid chromatography. (C) ESI-MS detection of subtilomycin production in the 12-h supernatant of the recombinant harboring the apn cluster. The triply charged ion [M + 3H]3+ is shown.
Bioassays and HPLC were used to detect the change in subtilomycin production in the mutants (Fig. 2B). In contrast to the wild-type strain, the culture supernatant from the ΔapnB mutants failed to inhibit the target indicator strain B. subtilis CU1065, and no production of subtilomycin was detected by HPLC from the ASCEs of the ΔapnB mutant (Fig. 2B). Therefore, apnB is necessary for subtilomycin production. The results from bioassays and HPLC showed that very little subtilomycin could be detected from the supernatant of the ΔapnT mutant. The apnT gene, which encodes a putative ABC transporter, was predicted to be a transporter for the export of subtilomycin. We prepared intracellular extracts from the 10-h cultured cells of the wild-type BSn5 and the ΔapnT mutant by liquid nitrogen grinding to detect the precursor peptide of subtilomycin by high-resolution LC-MS. However, no predicted mass (6,250.35 Da) of precursor peptide or significant accumulation of masses from 5,500 to 6,500 Da could be observed in BSn5 or the ΔapnT mutant (data not shown). In order to confirm the presence of the minimal subtilomycin gene cluster, two flanking genes of the cluster were deleted as described in Materials and Methods. The ybcL gene, which encodes a putative efflux transporter, and ybdG, which encodes a putative hydrolase, located upstream and downstream of the cluster, respectively, were disrupted (Fig. 2A). No significant change was observed with respect to the production of subtilomycin (Fig. 2B). The detection of subtilomycin production was also evaluated using MALDI-TOF MS, and the results were consistent with those from the bioassay and HPLC analyses (see Fig. S1 in the supplemental material).
In addition, the B. subtilis CU1065 recombinant harboring the whole apn cluster was constructed. Subtilomycin production was evaluated in the 12- to 16-h cultured supernatant of the recombinant by LC-MS (Fig. 2C; see also Fig. S2 in the supplemental material), which further verified that the minimal apn cluster was responsible for subtilomycin biosynthesis.
The putative transmembrane protein-encoding gene apnI confers subtilomycin immunity.
A structurally elucidated lantibiotic, paenibacillin, produced by Paenibacillus polymyxa OSY-DF was identified with the highest identity (54%) to subtilomycin; however, its biosynthetic gene cluster has not been published. In addition, other lantibiotics from the Pep5 group showed homology with subtilomycin, including epilancin 15X, epilancin K7, Pep5, and epicidin 280. We compared the subtilomycin biosynthetic cluster with four reported and related lantibiotic gene clusters from the Pep5 group (Fig. 3). Four genes homologous to lanA, lanP, lanB, and lanC were found to be relatively conserved in all of the gene clusters examined, including the subtilomycin cluster. However, the genes responsible for lantibiotic export, immunity, and N-terminal modification showed variations in terms of their arrangement. The genes responsible for lantibiotic export in the Pep5 group are located upstream of the genes lanAPBC in the direction opposite to that observed in the subtilomycin cluster. The transporter gene is missing in the gene cluster for epicidin 280, which was proposed to be replaced by other transporters in the host (24). The immunity genes pepI and eciI are located closely upstream of the structural genes pepA and eciA, respectively. The putative epilancin 15X immunity gene elxI is located downstream and next to elxC.
FIG 3.
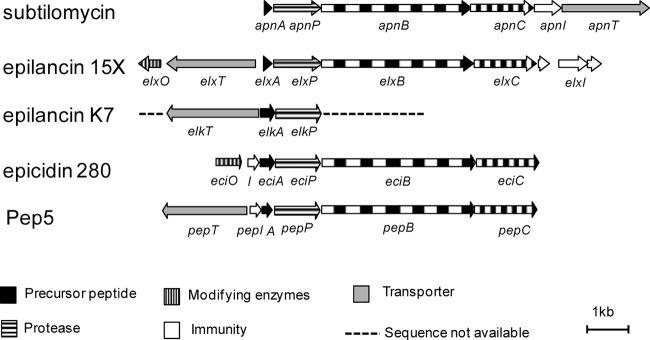
Comparison of the organization of subtilomycin gene clusters with biosynthetic gene clusters of Pep5 group lantibiotics, epilancin 15X (39), epilancin K7 (40), epicidin 280 (24), and Pep5 (41). Genes are labeled differently depending on the predicted function of their products: precursor peptides (black), proteases (horizontal stripes), modification enzymes (vertical stripes), immunity (white), and transporters (gray). The unavailable sequences in strain K7 are shown as a broken line.
In the apn cluster, the gene products of apnAPBC were predicted, respectively, as structural peptide ApnA, leader peptide-cleaving protease ApnP, and lanthionine synthetases ApnB and ApnC. The gene organization apnI-apnT, which is located downstream of apnAPBC, is different from that of the other reported gene clusters (Fig. 3). As deletion of apnT led to a considerable loss of subtilomycin production in the supernatant, apnT, which encodes an ABC transporter, likely plays a key role in subtilomycin export. Through protein BLAST using the sequence of the apnI-encoded protein, no functionally validated homolog could be found in the NCBI database. Membrane helix prediction using TMHMM (25), HMMTOP (26), and the DAS-TM filter (27) revealed the presence of five transmembrane domains in the putative protein ApnI. Immunity genes are usually present in lantibiotic biosynthetic clusters and are necessary for lantibiotic production. Therefore, the fact that no immunity-related gene was found in the subtilomycin gene clusters indicated that apnI and/or apnT is likely involved in subtilomycin immunity.
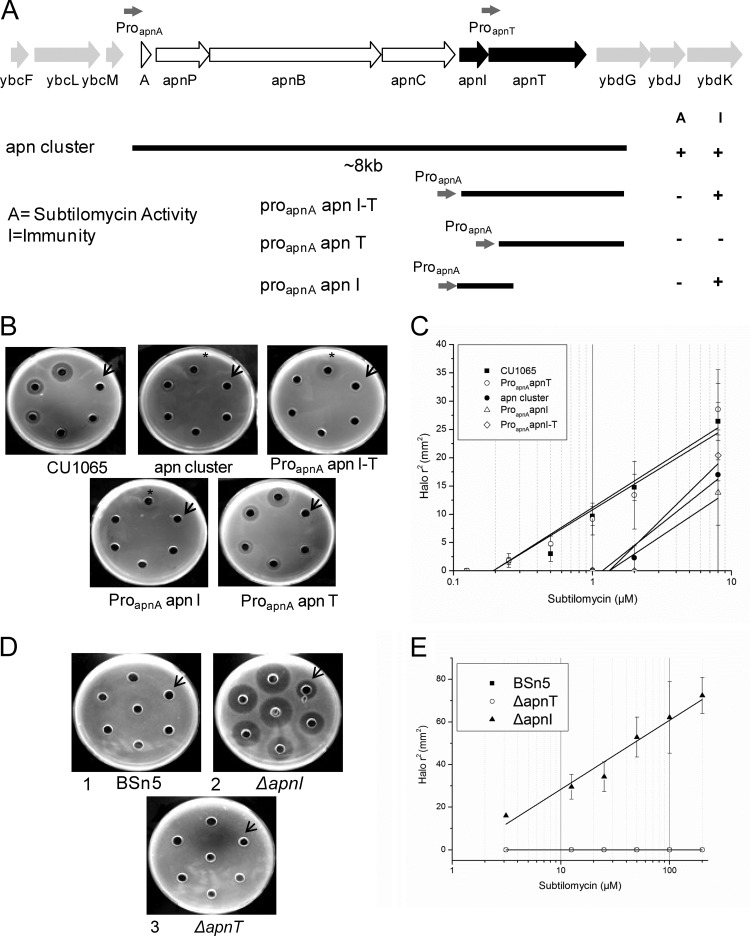
To investigate which genes, apnI and/or apnT, could confer resistance on the subtilomycin-sensitive host B. subtilis CU1065 (B. subtilis 168 background), we first predicted two putative promoters in the apn cluster, which are located on the upstream genes apnA and apnT. Different fragments that contained the whole cluster, apnI-apnT, apnT, or apnI were cloned into the integration vector pDG1730. The promoter region of apnA (proapnA) was cloned in front of each of the constructed fragments apnI-apnT, apnT, and apnI, resulting in the vectors proapnAapnI-T, proapnAapnT, and proapnAapnI, respectively, to ensure the expression of each gene (Fig. 4A). The constructed vectors were transformed and integrated into the amyE locus of the B. subtilis CU1065 chromosome, resulting in corresponding recombinants.
FIG 4.
Functional analysis of subtilomycin immunity by gene expression in B. subtilis CU1065 and gene interruption in B. subtilis BSn5. (A) Map displaying essential DNA fragments for subtilomycin immunity. Each construction was cloned into the vector pDG1730 and integrated into the amyE locus of the chromosome of the subtilomycin-sensitive strain B. subtilis CU1065. The predicted fragments containing the apnA and apnT promoter regions are indicated by gray arrows. (B) Subtilomycin sensitivities of B. subtilis CU1065 strains harboring different fragments of the subtilomycin gene cluster were investigated in agar diffusion tests. B. subtilis CU1065 and its recombinants were transformed with the vector containing proapnA-apnI-T, proapnA-apnI, proapnA-apnT, or the whole subcluster. Starting from the arrow and moving clockwise, the subtilomycin concentrations applied were as follows: 0.125 μM, 0.25 μM, 0.5 μM, 1 μM, 2 μM, and 8 μM. The asterisk indicates a blurry inhibition zone. (C) Linear dependencies between the square of the radius of the halos shown in panel B and the natural logarithm of the applied subtilomycin concentration with 60 μl of subtilomycin and diffusion for at least 4 h. Results for B. subtilis CU1065 transformed with vectors containing apnI, apnT, apnI-apnT, and subgene clusters are shown. (D) Subtilomycin immunity of strain BSn5 (plate 1) with interruption of apnI (plate 2) and apnT (plate 3). Starting from the arrow and moving clockwise, the applied concentrations of subtilomycin were 3.125 μM, 12.5 μM, 25 μM, 50 μM, 100 μM, and 200 μM. The well in the middle of the plate was not counted. (E) Subtilomycin immunity of BSn5 with interruption of apnT and apnI. The immunity response was calculated corresponding to the square of the radius of the halos shown in panel D.
Subtilomycin sensitivity tests were conducted to determine if the cloned fragments conferred resistance on different B. subtilis recombinants. The fragments containing apnI with proapnA resulted in the same CU1065 resistance level as that observed for fragments containing the whole cluster (Fig. 4B and C). The recombinants normally expressing apnT showed the same level of sensitivity to subtilomycin as wild-type CU1065 (Fig. 4B and C), which suggested that apnT does not confer subtilomycin resistance on the host. Our results indicated that the single gene apnI could confer protection against subtilomycin on the sensitive host strain B. subtilis CU1065.
To determine whether the apnI gene plays a role in immunity in a producer strain, subtilomycin sensitivity tests were conducted using wild-type BSn5 and its ΔapnI and ΔapnT mutants. The results showed that wild-type strain BSn5 and the ΔapnT mutant could tolerate high subtilomycin concentrations (up to approximately 0.3 mM), whereas the ΔapnI mutant was significantly sensitive to subtilomycin (Fig. 4D and E).
Discovery of the ApnI-like proteins for lantibiotic immunity.
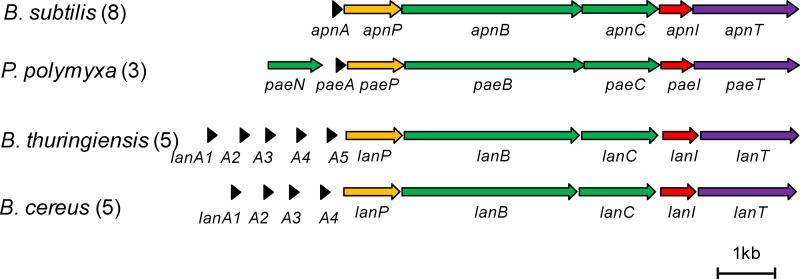
As subtilomycin shares structural similarity with paenibacillin, we thus wondered what gene does play a role in paenibacillin immunity. Using the reported draft genome sequence (28), we found that the annotated genes of the putative paenibacillin biosynthetic gene cluster are present in the order paeNAPBCIT. We then compared the proposed paenibacillin gene cluster to the apn cluster (Fig. 5). Interestingly, a putative transmembrane protein-encoding gene, paeI, is present downstream of the conserved lanAPBC gene organization. Despite the low sequence identity (27%) between ApnI and PaeI, their similar topological structures and the same gene arrangement of the cluster as for the apn cluster suggested that PaeI was responsible for paenibacillin immunity.
FIG 5.
The potential ApnI-like immunity protein-encoding gene from the uncharacterized lantibiotic gene clusters. Genes are color-coded by the predicted function of their products: genes for precursor peptides (black), genes for proteases (yellow), genes for modifying enzymes (green), genes for transporters (purple), and genes for immunity (red). The bar represents 1 kbp. The number of independent isolates that have the potential cluster is shown. Details for these sequences are shown in Table S2 in the supplemental material.
Furthermore, we obtained more uncharacterized putative lantibiotic gene clusters from the Bacillus cereus group (including 5 B. cereus strains and 5 Bacillus thuringiensis strains) based on running protein BLAST using the sequences of ApnA and ApnI on the NCBI website. Comparison of these gene clusters with the apn cluster showed that they shared the same gene arrangement, lanAPBC followed by lanI and lanT. The LanI proteins from the B. cereus group showed low similarity (27 to 29%) to ApnI. The LanIs from these gene clusters were predicted to have six transmembrane domains, which had similar topology with ApnI. Accordingly, we named these LanI proteins as ApnI-like proteins and predicted that they were responsible for the immunity of the corresponding lantibiotics.
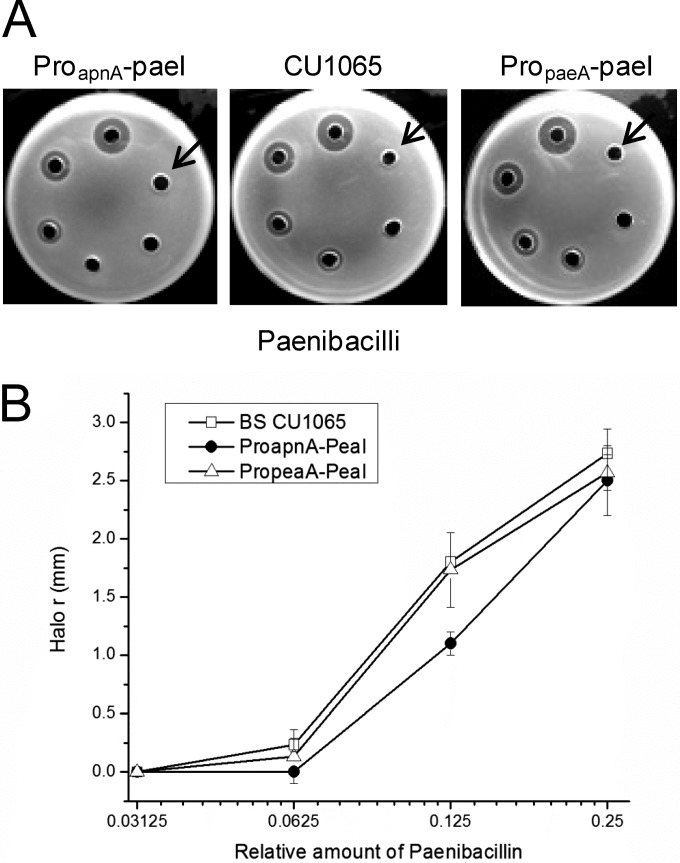
We predicted that the paenibacillin structural gene paeA would also exist in the genome sequence of Paenibacillus strain ATCC 842, which is the strain most closely related genetically to the paenibacillin producer strain P. polymyxa OSY-DF (28), and paenibacillin production was detected from the crude extracts of the 24-h-cultured supernatant of strain ATCC 842, according to a previously reported method (see Fig. S3 in the supplemental material) (14). We estimated that the purity of paenibacillin in the crude extracts was about 8% based on the relative abundance of the paenibacillin peak under the detection of UV 254 nm by LC-MS. To validate the function of paeI, which encodes an ApnI-like protein, we cloned the fragment containing the paeI and the downstream promoter regions of apnA (proapnA) and paeA (propaeA) into the integrated vector pDG1730, respectively, and transformed them into B. subtilis CU1065. A concentration gradient of crude paenibacillin was employed for the sensitivity test of the three strains. The results showed that the fragment containing paeI under the control of proapnA could confer certain resistance on CU1065 at the low concentration dilutions of paenibacillin (the 2nd and 3rd dilutions), compared with the other two strains (Fig. 6). At the increasing paenibacillin concentrations (the 5th and 6th dilutions), no significant difference could be observed among the three strains (Fig. 6A), which indicates that the paenibacillin resistance level conferred by PaeI was much lower than the ApnI-conferred subtilomycin resistance level in the background of B. subtilis CU1065.
FIG 6.
Functional analysis of the paeI gene in B. subtilis CU1065. (A) Paenibacillin sensitivity test results of B. subtilis CU1065 and the recombinants harboring fragments proapnA-paeI and propaeA-paeI. The concentrations of crude paenibacillin were applied with a series of 2-fold dilutions and were not quantified. The concentrations of paenibacillin increased starting from the arrow and moving clockwise. (B) The response as the length of the radii of the inhibition halos versus the corresponding relative amount of paenibacillin as shown in panel A. Two high concentrations of paenibacillin (the 5th and 6th dilutions) were removed. This experiment was independently performed at least 3 times, and the results were consistent.
Immunity provided by ApnI is specific toward subtilomycin.
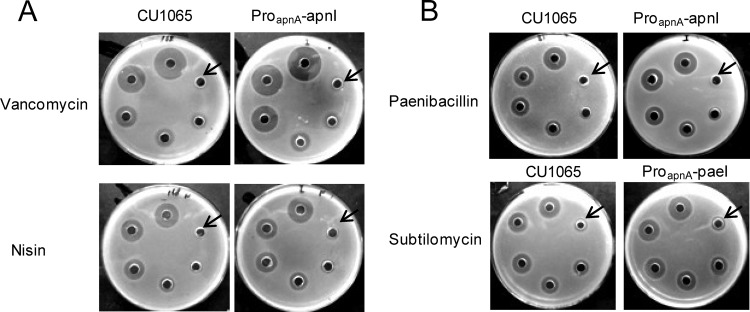
Since the sole putative transmembrane protein, ApnI, conferred significant resistance to the sensitive host CU1065, we next evaluated whether ApnI could confer cross-resistance against other bacteriocins. Toward this end, we selected the following peptide antibiotics based on their relationship with subtilomycin, listed from more distantly to closely related: vancomycin, nisin, and paenibacillin. We first carried out a sensitivity test with vancomycin and nisin. As shown in Fig. 7A, CU1065 with and without the integrated gene apnI (with the proapnA-apnI fragment) showed an inhibition zone of the same size with various vancomycin and nisin concentrations, suggesting that ApnI could not recognize vancomycin and nisin.
FIG 7.
(A) Specificity analysis of the subtilomycin immunity gene apnI by sensitive test in CU1065. Gradient amounts of antibiotics were added from the arrow and moving clockwise. Vancomycin concentrations were 0.3375 nM, 0.675 nM, 1.35 nM, 2.7 nM, 5.4 nM, and 10.8 nM; nisin concentrations were 0.41 μM, 0.82 μM, 1.64 μM, 3.28 μM, 6.54 μM, and 13.08 μM. (B) Detection of cross-resistance to paenibacillin and subtilomycin in CU1065 conferred by genes apnI and paeI, respectively. The amounts of crude paenibacillin were applied with diluted series 2 times and not quantified. Gradient amounts of subtilomycin were added from the arrow and moving clockwise: 0.5 μM, 1 μM, 2 μM, 4 μM, 8 μM, and 16 μM. Each assay was independently repeated at least three times.
Since subtilomycin and paenibacillin showed high similarity (54%) and have the same five-thioether ring backbone (Fig. 2B), we next evaluated whether ApnI and PaeI could confer cross-resistance against paenibacillin and subtilomycin, respectively. The sensitivity tests showed that despite the fact that these antibiotics are structurally similar, no cross-resistance was observed between the proapnA-apnI and the proapnA-paeI recombinants (Fig. 7B). Together, these results imply that ApnI provides immunity that may involve a specific sequence recognition mechanism.
The recombinant B. subtilis expressing ApnI could bind subtilomycin.
We characterized the function of ApnI by conducting a quantitative peptide-binding assay using 15 μg/ml and 25 μg/ml of subtilomycin (Fig. 8). As addition of NaCl to the assay solution could decrease the amount of cell-binding lantibiotics by reducing the lantibiotic affinity to the cytoplasmic membrane (21), to investigate the target specificity of ApnI against subtilomycin, the assay was performed in the presence of 1 M NaCl. For B. subtilis CU1065 expressing apnI, ∼7 μg of subtilomycin was found to be attached to the cells independently of the quantity of applied subtilomycin (15 or 25 μg/ml) (Fig. 8). Correspondingly, the amount of subtilomycin remaining in the supernatant of the apnI-expressing recombinant decreased significantly (Fig. 8). This result suggests that the apnI-expressing strain confers resistance by sequestering subtilomycin.
FIG 8.
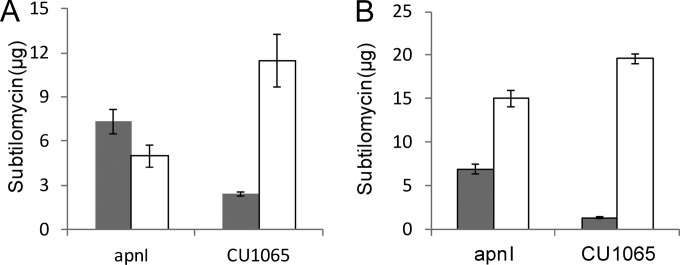
Quantitative peptide-binding assay for ApnI with subtilomycin. The cell suspensions (OD600 = 10) of stationary-phase-grown B. subtilis CU1065 and its recombinant expressing apnI were incubated with subtilomycin at concentrations of 15 μg/ml (A) and 25 μg/ml (B) in the presence of 1 M NaCl under the conditions described in Materials and Methods. The amounts of subtilomycin remaining in the supernatants (open bars) and the amounts remaining in the cell-associated samples (filled bars) were quantified by reverse-phase HPLC. Values represent means, with error bars representing standard deviations.
DISCUSSION
Although a link between subtilomycin and its gene cluster was proposed based on a match between the N-terminal region and a functional gene at the 5′ end, its biosynthetic gene cluster has not yet been functionally validated. In this study, we experimentally identified the apn cluster responsible for subtilomycin biosynthesis and provided evidence that apnI does indeed confer subtilomycin immunity on its host. When we BLAST searched the NCBI database using the ApnI amino acid sequence, no functionally validated homolog could be found. We thus first determined the mechanism of self-resistance of a lantibiotic producer toward Pep5, which is homologous with subtilomycin. The self-protection against Pep5 is solely dependent on functional expression of the 69-amino-acid PepI, which was demonstrated through a series of cloning experiments (29, 30). Further functional analysis indicated that PepI is a lipoprotein with an apolar N-terminal segment for its localization at the membrane-cell wall interface and a hydrophilic C terminus, which confers immunity (11). However, the predicted apnI gene product is a membrane protein with five transmembrane domains, which is vastly different from PepI.
Other known models for lantibiotic immunity usually involve the lanFE(G) genes, encoding an ABC transporter complex (Fig. 9). LanFE(G) can independently provide lantibiotic immunity, such as LctFEG for lacticin 481 immunity (31), or work synergistically with a membrane-associated protein, such as NisI (32), or a transmembrane protein, such as NukH (9, 10) and LtnI (33). Lantibiotic immunity that is solely provided by transmembrane proteins is very rare. We found only one report of such a protein, CylI, which is different from other transmembrane immunity proteins, as it contains a conserved zinc metalloprotease (MEROPS family M50A) domain, which suggests that it plays a role in immunity by cleaving one or both of the cytolysin subunits (34, 35).
FIG 9.
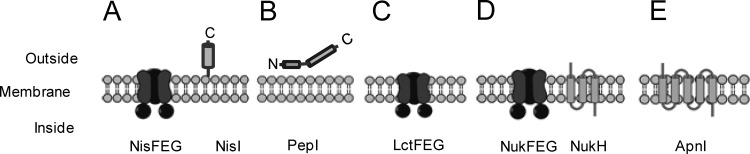
Models of lantibiotic immunity systems. A, NisFEG and NisI for nisin immunity; B, PepI for Pep5 immunity; C, LctFEG for lacticin 481 immunity; D, NukFEG and NukH for nukacin ISK-1 immunity; E, ApnI for subtilomycin immunity.
Through bioinformatics analysis, many ApnI-like proteins in the uncharacterized lantibiotic gene clusters that are related to the apn cluster were found in the NCBI database (see Table S2 in the supplemental material). Although low identity (27 to 29%) is present between the sequences of ApnI and the LanI proteins, they all are predicted to have a topological structure similar to that of ApnI, with five or six transmembrane domains. The LanA structural peptide genes in these gene clusters were also shown to be closely related, with a sequence identity of 47% to 58% to ApnA (Fig. 5; see also Table S2). In total, 21 strains of B. subtilis, Paenibacillus, B. cereus, and B. thuringiensis have the putative lantibiotic biosynthetic gene cluster with ApnI-like proteins. We successfully introduced one such ApnI-like protein, PaeI, in B. subtilis CU1065 from Paenibacillus strain ATCC 842 and found that PaeI indeed confers resistance to paenibacillin. These results indicated that the novel model represented by ApnI for lantibiotic immunity is widely present among the Bacillus and Paenibacillus taxa. In addition, in the B. subtilis background, paeI could be expressed under the control of the promoter of apnA derived from B. subtilis. However, it could not be expressed under the control of the promoter of paeA derived from Paenibacillus, indicating that expression of subtilomycin and paenibacillin might be involved in different regulation mechanisms. We found that B. subtilis became relatively resistant to paenibacillin when paeI was introduced into the sensitive strain CU1065. However, the paenibacillin resistance level afforded by PaeI was much lower than the ApnI-conferred subtilomycin resistance level (Fig. 4B and C and Fig. 6). In addition, we tested if paeI-expressing cells could bind paenibacillin in the peptide-binding assay. The result showed that no interaction between paenibacillin and the recombinant expressing paeI could be observed (data not shown). We inferred that the protein PaeI was not well expressed in CU1065 on the basis of its poor incorporation into the membrane of B. subtilis. On the other hand, other factors, like the protein PaeT, which was encoded by paeT immediately downstream of paeI, might be necessary for full immunity against paenibacillin.
Immunity genes often provide protection that is specific to corresponding lantibiotics and structurally related analogs. Conferred cross-protection against other bacteriocins is rare and has been found only among very closely related lantibiotics, such as Pep5 and epicidin 280 (24) as well as lacticin 3147 and C55 (36). The results of the sensitivity tests showed that PaeI and ApnI did not provide cross-protection against subtilomycin and paenibacillin, respectively, even though paenibacillin is structurally similar to subtilomycin (Fig. 2B). We inferred that the sequence similarity of 54% between paenibacillin and subtilomycin is not suitably high compared with that of lantibiotics that show cross-immunity, such as Pep5 and epicidin 280 (75% similarity) (24) and subunits LtnA1 and C55α (86% similarity) and LtnA2 and C55β (55% similarity), components of the lantibiotics lacticin 3147 and C55, respectively (36, 37). These results suggest that ApnI confers subtilomycin immunity through a specific sequence recognition mechanism. Using the peptide-binding assay (21), we detected the interaction of subtilomycin with the cells that expressed apnI, which suggested that ApnI proteins play a role in immunity by sequestering subtilomycin from its site of action to the cell membrane. Therefore, the fact that the transmembrane protein ApnI solely provided protection against subtilomycin indicates that this case represents a novel model for lantibiotic immunity (Fig. 9).
The lantibiotic subtilomycin is produced by B. subtilis strain MMA7, isolated from the marine sponge Haliclona simulans. The presence of the proposed subtilomycin gene cluster was detected from all sponge-derived B. subtilis strains by PCR screening (13). The proposed subtilomycin cluster was also determined to be present in B. subtilis strain BSn5, which was isolated from the plant Amorphophallus konjac, by mining the complete genome sequence (16). In the present study, we used high-resolution LC-MS analysis to confirm the production of subtilomycin in strain BSn5 and provided experimental evidence relating the production of subtilomycin to its biosynthetic gene cluster. The fact that the plant endophytic strain BSn5 can produce subtilomycin further suggests that the production of subtilomycin might be specific to B. subtilis strains originating from an animal or plant host. Therefore, it would be interesting to investigate subtilomycin production from more isolates from different hosts and nonhost environments. Subtilomycin may provide some benefits to the producer with respect to host colonization, which is in line with the concept of bacteriocin production as a probiotic trait (38). Therefore, further investigations of the relationship between host colonization ability and subtilomycin production are merited.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ying Zhou, Dong-Mei Han, and Hui-Fang Ban for their early fundamental work and Yi Luo, Er-Ning Yang, and Cheng-Chen Xu for suggestions. We thank the director of the BGSC, Daniel R. Zeigler, for providing tool vectors and Paenibacillus strain ATCC 842, as well as for his suggestions and discussion. We thank John D. Helmann for providing recipient B. subtilis strain CU1065.
This work was supported by grants from the National High Technology Research and Development Program (program 863) of China (grant 2011AA10A203), the China 948 Program of the Ministry of Agriculture (grant 2011-G25), the National Basic Research Program (program 973) of China (grants 2009CB118902 and 2013CB127504), the National Natural Science Foundation of China (grants 30970037 and 31170047), the International Scientific Cooperation of Hubei Province (grant 2011BFA019), and the Fundamental Research Fund for the Central University (grant 2011PY056).
Footnotes
Published ahead of print 1 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02280-14.
REFERENCES
- 1.Earl AM, Losick R, Kolter R. 2008. Ecology and genomics of Bacillus subtilis. Trends Microbiol. 16:269–275. 10.1016/j.tim.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tam NKM, Uyen NQ, Hong HA, Duc LH, Hoa TT, Serra CR, Henriques AO, Cutting SM. 2006. The intestinal life cycle of Bacillus subtilis and close relatives. J. Bacteriol. 188:2692–2700. 10.1128/JB.188.7.2692-2700.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anagnostopoulos C, Spizizen J. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stein T. 2005. Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol. Microbiol. 56:845–857. 10.1111/j.1365-2958.2005.04587.x [DOI] [PubMed] [Google Scholar]
- 5.Knerr PJ, van der Donk WA. 2012. Discovery, biosynthesis, and engineering of lantipeptides. Annu. Rev. Biochem. 81:479–505. 10.1146/annurev-biochem-060110-113521 [DOI] [PubMed] [Google Scholar]
- 6.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian KD, Fischbach MA, Garavelli JS, Goransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Muller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJ, Rebuffat S, Ross RP, Sahl HG, Schmidt EW, Selsted ME, et al. 2013. Ribosomally synthesized and translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30:108–160. 10.1039/c2np20085f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bierbaum G, Sahl HG. 2009. Lantibiotics: mode of action, biosynthesis and bioengineering. Curr. Pharm. Biotechnol. 10:2–18. 10.2174/138920109787048616 [DOI] [PubMed] [Google Scholar]
- 8.Okuda K, Sonomoto K. 2011. Structural and functional diversity of lantibiotic immunity proteins. Curr. Pharm. Biotechnol. 12:1231–1239. 10.2174/138920111796117274 [DOI] [PubMed] [Google Scholar]
- 9.Okuda K, Aso Y, Nakayama J, Sonomoto K. 2008. Cooperative transport between NukFEG and NukH in immunity against the lantibiotic nukacin ISK-1 produced by Staphylococcus warneri ISK-1. J. Bacteriol. 190:356–362. 10.1128/JB.01300-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aso Y, Okuda K, Nagao J, Kanemasa Y, Thi Bich Phuong N, Koga H, Shioya K, Sashihara T, Nakayama J, Sonomoto K. 2005. A novel type of immunity protein, NukH, for the lantibiotic nukacin ISK-1 produced by Staphylococcus warneri ISK-1. Biosci. Biotechnol. Biochem. 69:1403–1410. 10.1271/bbb.69.1403 [DOI] [PubMed] [Google Scholar]
- 11.Hoffmann A, Schneider T, Pag U, Sahl HG. 2004. Localization and functional analysis of PepI, the immunity peptide of Pep5-producing Staphylococcus epidermidis strain 5. Appl. Environ. Microbiol. 70:3263–3271. 10.1128/AEM.70.6.3263-3271.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ra R, Beerthuyzen MM, de Vos WM, Saris PE, Kuipers OP. 1999. Effects of gene disruptions in the nisin gene cluster of Lactococcus lactis on nisin production and producer immunity. Microbiology 145(Part 5):1227–1233 [DOI] [PubMed] [Google Scholar]
- 13.Phelan RW, Barret M, Cotter PD, O'Connor PM, Chen R, Morrissey JP, Dobson AD, O'Gara F, Barbosa TM. 2013. Subtilomycin: a new lantibiotic from Bacillus subtilis strain MMA7 isolated from the marine sponge Haliclona simulans. Mar. Drugs 11:1878–1898. 10.3390/md11061878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He Z, Kisla D, Zhang L, Yuan C, Green-Church KB, Yousef AE. 2007. Isolation and identification of a Paenibacillus polymyxa strain that coproduces a novel lantibiotic and polymyxin. Appl. Environ. Microbiol. 73:168–178. 10.1128/AEM.02023-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Z, Yuan C, Zhang L, Yousef AE. 2008. N-terminal acetylation in paenibacillin, a novel lantibiotic. FEBS Lett. 582:2787–2792. 10.1016/j.febslet.2008.07.008 [DOI] [PubMed] [Google Scholar]
- 16.Deng Y, Zhu Y, Wang P, Zhu L, Zheng J, Li R, Ruan L, Peng D, Sun M. 2011. Complete genome sequence of Bacillus subtilis BSn5, an endophytic bacterium of Amorphophallus konjac with antimicrobial activity for the plant pathogen Erwinia carotovora subsp. carotovora. J. Bacteriol. 193:2070–2071. 10.1128/JB.00129-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao M, Helmann JD. 2002. Regulation of the Bacillus subtilis bcrC bacitracin resistance gene by two extracytoplasmic function sigma factors. J. Bacteriol. 184:6123–6129. 10.1128/JB.184.22.6123-6129.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Russell DW. 2006. The condensed protocols from molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 19.Yasbin RE, Wilson GA, Young FE. 1975. Transformation and transfection in lysogenic strains of Bacillus subtilis: evidence for selective induction of prophage in competent cells. J. Bacteriol. 121:296–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo Y, Ruan LF, Zhao CM, Wang CX, Peng DH, Sun M. 2011. Validation of the intact zwittermicin A biosynthetic gene cluster and discovery of a complementary resistance mechanism in Bacillus thuringiensis. Antimicrob. Agents Chemother. 55:4161–4169. 10.1128/AAC.00111-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stein T, Heinzmann S, Solovieva I, Entian KD. 2003. Function of Lactococcus lactis nisin immunity genes nisI and nisFEG after coordinated expression in the surrogate host Bacillus subtilis. J. Biol. Chem. 278:89–94. 10.1074/jbc.M207237200 [DOI] [PubMed] [Google Scholar]
- 22.Sahl HG, Jack RW, Bierbaum G. 1995. Biosynthesis and biological activities of lantibiotics with unique post-translational modifications. Eur. J. Biochem. 230:827–853. 10.1111/j.1432-1033.1995.tb20627.x [DOI] [PubMed] [Google Scholar]
- 23.Li B, Sher D, Kelly L, Shi Y, Huang K, Knerr PJ, Joewono I, Rusch D, Chisholm SW, van der Donk WA. 2010. Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria. Proc. Natl. Acad. Sci. U. S. A. 107:10430–10435. 10.1073/pnas.0913677107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heidrich C, Pag U, Josten M, Metzger J, Jack RW, Bierbaum G, Jung G, Sahl HG. 1998. Isolation, characterization, and heterologous expression of the novel lantibiotic epicidin 280 and analysis of its biosynthetic gene cluster. Appl. Environ. Microbiol. 64:3140–3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 26.Tusnády GE, Simon I. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849–850. 10.1093/bioinformatics/17.9.849 [DOI] [PubMed] [Google Scholar]
- 27.Dosztányi Z, Magyar C, Tusnady GE, Cserzo M, Fiser A, Simon I. 2003. Servers for sequence-structure relationship analysis and prediction. Nucleic Acids Res. 31:3359–3363. 10.1093/nar/gkg589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeong H, Park SY, Chung WH, Kim SH, Kim N, Park SH, Kim JF. 2011. Draft genome sequence of the Paenibacillus polymyxa type strain (ATCC 842T), a plant growth-promoting bacterium. J. Bacteriol. 193:5026–5027. 10.1128/JB.05447-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reis M, Eschbach-Bludau M, Iglesias-Wind MI, Kupke T, Sahl HG. 1994. Producer immunity towards the lantibiotic Pep5: identification of the immunity gene pepI and localization and functional analysis of its gene product. Appl. Environ. Microbiol. 60:2876–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pag U, Heidrich C, Bierbaum G, Sahl HG. 1999. Molecular analysis of expression of the lantibiotic pep5 immunity phenotype. Appl. Environ. Microbiol. 65:591–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rincé A, Dufour A, Uguen P, Le Pennec JP, Haras D. 1997. Characterization of the lacticin 481 operon: the Lactococcus lactis genes lctF, lctE, and lctG encode a putative ABC transporter involved in bacteriocin immunity. Appl. Environ. Microbiol. 63:4252–4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuipers OP, Beerthuyzen MM, Siezen RJ, De Vos WM. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281–291 [DOI] [PubMed] [Google Scholar]
- 33.Draper LA, Deegan LH, Hill C, Cotter PD, Ross RP. 2012. Insights into lantibiotic immunity provided by bioengineering of LtnI. Antimicrob. Agents Chemother. 56:5122–5133. 10.1128/AAC.00979-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coburn PS, Hancock LE, Booth MC, Gilmore MS. 1999. A novel means of self-protection, unrelated to toxin activation, confers immunity to the bactericidal effects of the Enterococcus faecalis cytolysin. Infect. Immun. 67:3339–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Tyne D, Martin MJ, Gilmore MS. 2013. Structure, function, and biology of the Enterococcus faecalis cytolysin. Toxins (Basel) 5:895–911. 10.3390/toxins5050895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Draper LA, Grainger K, Deegan LH, Cotter PD, Hill C, Ross RP. 2009. Cross-immunity and immune mimicry as mechanisms of resistance to the lantibiotic lacticin 3147. Mol. Microbiol. 71:1043–1054. 10.1111/j.1365-2958.2008.06590.x [DOI] [PubMed] [Google Scholar]
- 37.O'Connor EB, Cotter PD, O'Connor P, O'Sullivan O, Tagg JR, Ross RP, Hill C. 2007. Relatedness between the two-component lantibiotics lacticin 3147 and staphylococcin C55 based on structure, genetics and biological activity. BMC Microbiol. 7:24. 10.1186/1471-2180-7-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dobson A, Cotter PD, Ross RP, Hill C. 2012. Bacteriocin production: a probiotic trait? Appl. Environ. Microbiol. 78:1–6. 10.1128/AEM.05576-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Velásquez JE, Zhang X, van der Donk WA. 2011. Biosynthesis of the antimicrobial peptide epilancin 15X and its N-terminal lactate. Chem. Biol. 18:857–867. 10.1016/j.chembiol.2011.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van de Kamp M, van den Hooven HW, Konings RN, Bierbaum G, Sahl HG, Kuipers OP, Siezen RJ, de Vos WM, Hilbers CW, van de Ven FJ. 1995. Elucidation of the primary structure of the lantibiotic epilancin K7 from Staphylococcus epidermidis K7. Cloning and characterisation of the epilancin-K7-encoding gene and NMR analysis of mature epilancin K7. Eur. J. Biochem. 230:587–600 [DOI] [PubMed] [Google Scholar]
- 41.Weil HP, Beck-Sickinger AG, Metzger J, Stevanovic S, Jung G, Josten M, Sahl HG. 1990. Biosynthesis of the lantibiotic Pep5. Isolation and characterization of a prepeptide containing dehydroamino acids. Eur. J. Biochem. 194:217–223 [DOI] [PubMed] [Google Scholar]
- 42.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580. 10.1016/S0022-2836(83)80284-8 [DOI] [PubMed] [Google Scholar]
- 43.Guérout-Fleury AM, Shazand K, Frandsen N, Stragier P. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335–336. 10.1016/0378-1119(95)00652-4 [DOI] [PubMed] [Google Scholar]
- 44.Steinmetz M, Richter R. 1994. Easy cloning of mini-Tn10 insertions from the Bacillus subtilis chromosome. J. Bacteriol. 176:1761–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guérout-Fleury AM, Frandsen N, Stragier P. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180:57–61. 10.1016/S0378-1119(96)00404-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.