Abstract
Xanthomonas axonopodis pv. citri (Xac) is the causal agent of citrus bacterial canker (CBC) and is a serious problem worldwide. Like CBC, several important diseases in other fruits, such as mango, pomegranate, and grape, are also caused by Xanthomonas pathovars that display remarkable specificity toward their hosts. While citrus and mango diseases were documented more than 100 years ago, the pomegranate and grape diseases have been known only since the 1950s and 1970s, respectively. Interestingly, diseases caused by all these pathovars were noted first in India. Our genome-based phylogenetic studies suggest that these diverse pathogens belong to a single species and these pathovars may be just a group of rapidly evolving strains. Furthermore, the recently reported pathovars, such as those infecting grape and pomegranate, form independent clonal lineages, while the citrus and mango pathovars that have been known for a long time form one clonal lineage. Such an understanding of their phylogenomic relationship has further allowed us to understand major and unique variations in the lineages that give rise to these pathovars. Whole-genome sequencing studies including ecological relatives from their putative country of origin has allowed us to understand the evolutionary history of Xac and other pathovars that infect fruits.
INTRODUCTION
Xanthomonas is a complex group of plant-pathogenic bacteria that infect diverse fruit crops in a host-specific manner (1). Interestingly, several of the diseases caused by them on diverse fruits were first noticed in India (2–4). Xanthomonas axonopodis pv. citri (Xac), the causal agent of canker in citrus, is a well-known and serious pathogen worldwide (5), with billions of dollars spent in mass eradication programs in developed countries, such as the United States and Australia (6). X. citri pv. mangiferaindicae (Xmi), the causal agent of black spot in mango, is an endemic problem in mango-growing areas of the world, such as in Asia, Africa, and South America (7, 8). Both Xac and Xmi have been known of for the last 2 centuries.
X. axonopodis pv. punicae (Xap), the causal agent of oily spot disease in pomegranate, was first reported in the 1950s in India (3) and in recent decades has emerged as a devastating pathogen of this fruit (9). In the future, this pathogen might emerge as a huge threat to other pomegranate-growing countries, as it is also being reported outside India (10, 11). In addition, during the 1970s, a new bacterial disease of grape was reported for the first time from India (2), and the causal agent was later identified as X. campestris pv. viticola (Xvt) (12). In the late 1990s, this disease was also reported from Brazil, and in 2005, it was reported from Africa. It also could develop as a big problem in other grape-growing areas, such as in America, Australia, and Europe.
India is one of the largest cultivators, producers, and exporters of these fruits. Moreover, the Indian subcontinent also happens to be a center of diversity of their host plants, thereby providing excellent climatic conditions for the emergence and establishment of these hosts and pathogens. Hence, there is an urgent need to establish the accurate relationship of the xanthomonads that infect fruits to understand their origin, evolution, and management. However, taxonomically, the species status of these pathogens is not clear (13). Historically, all the Xanthomonas pathogens were assigned to unique species based on their host specificities (12) but later underwent reclassifications. For example, in the beginning, Xac, the Xanthomonas pathogen that infects citrus, was classified as Xanthomonas citri but later was reclassified as X. campestris pv. citri (12), X. axonopodis pv. citri (14), X. smithii pv. citri (15), and back again to X. citri subsp. citri (16–18). This highlights the great disconnect between taxonomy, pathology, ecology, and phylogeny of a complex group of bacteria like Xanthomonas.
Genomics is revolutionizing the way we do microbial research. Whole-genome information will potentially enable one to understand the variation and evolution of these pathogens at an unprecedented level (13). Further, whole-genome sequences are a rich resource for developing markers for quarantine purposes and epidemiological surveys. The Xac genome was one of the first plant-pathogenic bacterial genomes to be sequenced (19). We have previously deposited and announced the whole-genome sequences of reference strains of pathovars Xmi and Xap (20, 21). In this study, we sequenced the whole genome of the reference strain of Xvt. Here we report our detailed findings on comparative genomics of these pathogens, which have provided fine insights into their relationships and differences.
MATERIALS AND METHODS
Sequencing, assembly, and annotation.
The complete genome sequence of Xanthomonas axonopodis pv. citri strain 306 (Xac) is available under GenBank accession no. AE008923.1. Reference strains of X. axonopodis pv. punicae LMG 859 (Xap), X. citri pv. mangiferaindicae LMG 941 (Xmi), and X. campestris pv. viticola LMG 965 (Xvt) were obtained from Belgian Coordinated Collection of Microorganisms (BCCM), Belgium. DNA isolation was done using the Zymo Research fungal/bacterial DNA isolation kit. Sequencing of the genomic DNA of Xvt was obtained using the Roche 454 GS (FLX+ Titanium) pyrosequencing platform (Macrogen, Republic of Korea). Assembly of the reads was done using GS De Novo Assembler (version 2.3). Rapid Annotation using Subsystem Technology (RAST) and RNAmmer (version 1.2) were used to annotate the Xvt genome (22, 23). After automated assigning of the genes by the pipeline, further manual inspection and curation of the genome were done using NCBI ORF Finder (http://www.ncbi.nlm.nih.gov/projects/gorf/).
Phylogenomic analysis.
Phylogenetic analysis of the 31 broadly conserved housekeeping genes involved in informational processing (24) of strains belonging to genus Xanthomonas and Stenotrophomonas was done using MEGA version 5.1 (25). A maximum likelihood tree was constructed on the basis of the general time reversible model using gamma distributed with invariant sites (G+I) with 500 bootstrap replications. Average nucleotide identity (ANI) was calculated using JSpecies version 1.2.1 software (26).
Clonal analysis.
Clonal analysis of 31 conserved housekeeping genes was done using Clonal Frame version 1.1 (27). The run parameters were adjusted as described by Oh et al. (28). To obtain a tree without taking recombination into consideration, the value of r was set at zero. A Gelman and Rubin test was performed to check for satisfactory convergence of the five replicates.
Pangenome analysis.
A circular genome map was generated using BRIG-0.95 (29). Orthologous genes were identified using Pan-Genome Analysis Pipeline (30). This multiparanoid-based algorithm searches for homologs and orthologs in multiple genomes considering local matched regions to be not less than 25% of the longer gene protein sequence and global matched regions not less than 50% of the longer gene protein sequence. A minimum score value of 50 and an E value of less than 1 × 10−8 were used as cutoffs. We further characterized unique genes on the basis of their function using RAST (22).
LPS cluster analysis.
The sequences of highly conserved metB and etfA genes, which flank the lipopolysaccharide (LPS) locus in Xanthomonas, were used to retrieve the complete gene cluster from the genomic data of the strains under study. RAST was used to annotate sequences which were not already annotated (22). We could not find complete cluster sequences for Xap, Xanthomonas fuscans subsp. aurantifolii strain ICPB 11122 (Xf1), strain ICPB 10535 (Xf2), or X. axonopodis pv. malvacearum strain GSPB1386 (Xml1386). For Xap, we obtained the complete sequence of the LPS locus using junction PCR and primer walking on contigs 11 and 38 (see Table S2 in the supplemental material).
Integron cassette analysis.
The ilvD gene from Xac was used to retrieve homologs of ilvD from the other strains. For Xap, Xvt, and Xmi, the ilvD gene was found at the end of contigs, so after reordering of contigs with Mauve (31) using Xac as the reference genome, we amplified the missing part of the integron cassette by designing primers (see Table S2) and sequenced the amplified fragment by primer walking. An attI site was recognized by multiple-sequence alignment of the sequences downstream of the ilvD gene and the integron cassette of X. axonopodis pv. citri strain DAR73889 (GenBank accession no. AY928783). Repeat elements of 59 and 60 bp were identified by searching for the conserved 7-bp motifs RYYYAAC and GTTRRRY at left and right sides of the element, respectively (32). To find 59- and 60-bp repeat elements which may not share these conserved sequences, virtual PCR was run to find alternate binding sites of already identified elements.
CRISPR.
CRISPRfinder (http://crispr.u-psud.fr/Server/) was used to identify clustered regularly interspaced short palindromic repeats (CRISPR). Genes encoding CAS proteins were identified using Genome Property Search of JCVI CMR (http://cmr.jcvi.org/tigr-Scripts/CMR/shared/MakeFrontPages.cgi?page=genome_property).
NRPS/PKS.
Prediction of the nonribosomal peptide/polyketide synthetase cluster (NRPS/PKS) pathway was done using antiSMASH software (33). To further identify the acyl carrier protein (ACP) domain of the PKS pathway, structure-based sequence analysis of polyketide synthases (SBSPKS) was used (34). Phylogenetic analysis of the ketosynthase (KS) and condensation (C) domains was done using Natural Product Domain Seeker (NaPDoS) (35).
Xanthomonadin cluster analysis.
A well-characterized xanthomonadin cluster of X. oryzae pv. oryzae (Xoo) (GenBank accession no.AY010120) was used to retrieve homologs in all four strains. For Xmi, we were not able to find the complete length cluster, so primers were designed at the ends of both contig 31 and contig 32, in which cluster homologs were found (see Table S2 in the supplemental material), and the missing part of the cluster was completed by primer walking and was annotated using NCBI ORF Finder (http://www.ncbi.nlm.nih.gov/projects/gorf/). The xanthomonadin coding cluster from Xac was compared with that of Xmi using webACT (36).
Nucleotide sequence accession numbers.
Sequences of the whole-genome shotgun project of Xvt are available under accession numbers CBZT010000001 to CBZT010000050 in the EMBL and GenBank databases. Similarly, we also sequenced and deposited the whole-genome sequences of Xap and Xmi (GenBank accession numbers CAGJ01000001 to CAGJ01000217 and CAHO01000001 to CAHO01000195, respectively) (20, 21). All this information is consolidated for easy reference in Table S1 in the supplemental material. The LPS cluster of Xap has been submitted to GenBank under accession number KF991096. Sequences of integron cassettes of Xmi, Xap, and Xvt have been submitted to GenBank under accession numbers KF991093, KF991094, and KF991095, respectively. The xanthomonadin cluster of Xmi has been submitted to GenBank under accession number KF991092.
RESULTS
Sequencing and assembly.
The shotgun sequencing of Xvt yielded 726,914 reads amounting to 295,838,728 bases with ∼56-fold coverage. Assembly of the reads resulted in 50 contigs (over 500 bp) having an average contig size of 102.1 kb; the largest contig was 672.1 kb. The proportion of bases having a quality score of 40 or above was found to be 99.99%. The similar primary details of genome sequencing and coverage data for Xap and Xmi are available in the announcements (20, 21) and the NCBI database. Overall, all three genomes were sequenced using the Roche platform, with similar coverages and qualities (see Table S1 in the supplemental material).
Comparison of genomic features of Xanthomonas pathovars infecting mango, pomegranate, and grape fruits.
The genomes of Xvt, Xap, and Xmi are similar in size and have similar average GC contents, as shown in Table 1. The three genomes also have comparable numbers of coding sequences and tRNA-encoding genes. These salient genome features also match those of Xac, whose complete sequence is available in the public domain.
TABLE 1.
General features of the Xanthomonas pathovars infecting different fruit plants
| Parameter | Value for indicated strain |
|||
|---|---|---|---|---|
| Xac | Xap | Xvt | Xmi | |
| Size (bp) | 5,175,554 | 4,946,642 | 5,105,036 | 5,111,537 |
| GC content (%) | 64.7 | 64.9 | 64.5 | 64.8 |
| No. of protein-coding sequences | 4,312 | 4,385 | 4,431 | 4,521 |
| No. of rRNA-coding genes/operon | 6/2 | 3/1 | 3/1 | 3/1 |
| No. of tRNA-coding genes | 56 | 50 | 52 | 51 |
Phylogenomic analysis of Xanthomonas pathovars infecting diverse fruit plants. (i) Multilocus sequence analysis.
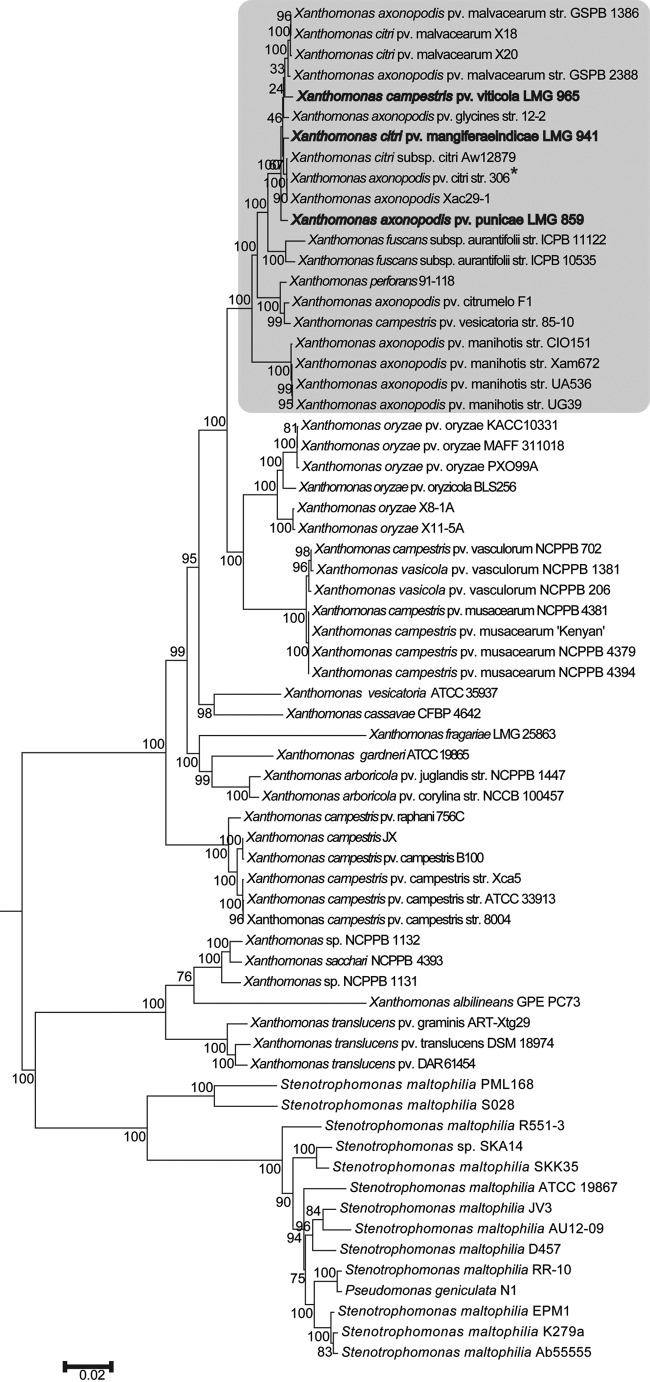
A phylogenetic tree constructed using a broad set of conserved housekeeping genes (Fig. 1) clusters all the fruit pathovars under study in one distinct phylogenetic group. Further, Xac, Xap, Xvt, and Xmi seem to have emerged from a common ancestor and possibly belong to same species despite being classified as different species, i.e., X. axonopodis, X. axonopodis, X. campestris, and X. citri, respectively.
FIG 1.
Phylogenomic tree of Xanthomonas pathovars that infect diverse fruit crops. The scale bar shows the number of nucleotide substitutions per site. The bootstrap value with 500 replications is shown on nodes. The Xanthomonas axonopodis group is highlighted by a gray background. Strains under study are shown in bold, and the reference strain is marked with an asterisk.
(ii) ANI.
To support the finding of their close phylogenetic relationship, we carried out ANI analysis of these pathovars. The ANI values for each pair in the study are listed in Table 2. For all pairs the ANI was found to be >98.8%, suggesting that these pathovars may just be specialized strains.
TABLE 2.
ANI among Xanthomonas pathovars infecting different fruit plants
| Strain | ANI (%) with: |
|||
|---|---|---|---|---|
| Xac | Xmi | Xap | Xvt | |
| Xac | NAa | 99.0 | 98.89 | 98.84 |
| Xmi | NA | 98.92 | 98.92 | |
| Xap | NA | 98.87 | ||
| Xvt | NA | |||
NA, not applicable.
(iii) Clonal frame analysis of X. axonopodis strains infecting different fruit crops.
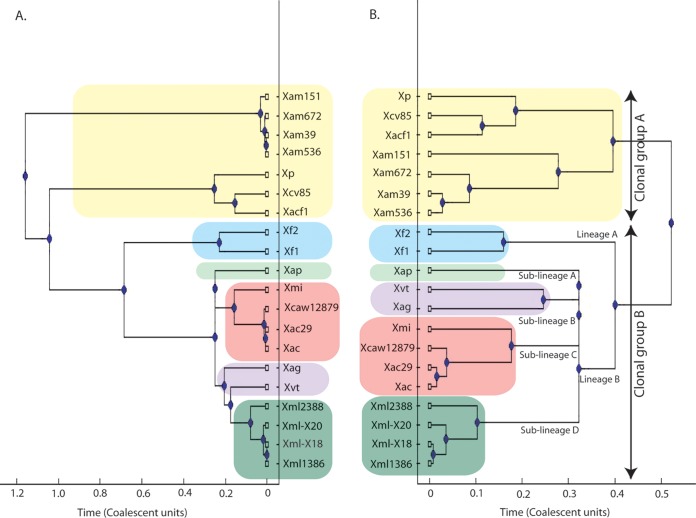
As phylogenomics revealed that these pathovars may be specialized strains, we carried out clonal analysis of fruit pathovars under study and of the members of the X. axonopodisgroup. Clonal frame is a coalescent model-based Bayesian approach which assumes that recombination events introduce a constant rate of substitutions to a contiguous region of sequence (27). We selected the 20 strains belonging to the X. axonopodis clade for this analysis, for which whole-genome sequences are available in the NCBI database. Figure 2 shows the clonal frame tree obtained with and without taking recombination into consideration. The clonal frame tree for the X. axonopodis group divided the 20 strains into two clonal groups as shown in Fig. 2. Clonal group A consists of X. perforans, X. axonopodis pv. citrumelo F1 (Xacf1), X. campestris pv. vesicatoria 85-10 (Xcv85), and X. axonopodis pv. manihotis strain CIO151 (Xam151), strain Xam672 (Xam672), strain UG39 (Xam39), and strain UA536 (Xam536). In clonal group B there are further two lineages: one comprises Xf1 and Xf2, and the second consists of four different sublineages. Sublineage A consists of Xap, sublineage B of X. axonopodis pv. glycines strain 12-2 (Xag) and Xvt, sublineage C of Xmi and Xanthomonas citrus pathovars (i.e., Xac, X. citri subsp. citri Aw12879 [Xcaw12879], and X. axonopodis Xac29-1 [Xac29]), and sublineage D of Xml1386, X. axonopodis pv. malvacearum strain GSPB2388 (Xml2388), and X. citri pv. malvacearum strains X20 (Xml-X20) and X18 (Xml-X18). Interestingly, Xac, Xmi, Xap, and Xvt form one clonal lineage in which Xac and Xmi form the same sublineage. There is no major change in the topology of the tree considering the recombining regions, except for separation of sublineages B and D. Table S3 in the supplemental material shows the values of recombination to mutation (r/m) and recombination to mutation (ρ/θ) for each run. The scale on the x axis shows the evolutionary time in coalescent units, showing evolutionary distance between strains. For X. axonopodis strains, the relative frequency of ρ/θ was 0.181, which reveals that recombination is taking place at a much slower pace than mutation, or ∼5.5 mutational events occur for each recombination event. The relative impact of r/m was found to be 1.611, suggesting a substantial impact of recombination on the evolution of these strains.
FIG 2.
X. axonopodis strain genealogy inferred by clonal frame tree. The clonal frame tree was obtained on the broad set of housekeeping genes using Clonal Frame version 1.1. A 50% majority rule consensus tree was computed without taking recombination into consideration (A) and with taking recombination into consideration (B). The scale on the x axis shows the evolutionary time in coalescent units. Different colors show the conserved groups, lineages, and sublineages considering recombination. Xp, X. perforans.
Genome variations in X. axonopodis pathovars infecting diverse fruit crops.
As there is no drastic change in size or reductive evolution in the genome, different sequence conservations and sequence variations could be having an impact on the diversification of different pathovars. As the phylogenomic and clonal relationship is based on the conserved regions of the genome, we wanted to study the variable parts of the genome. For that, we generated a circular map to identify sequence variations in the genome.
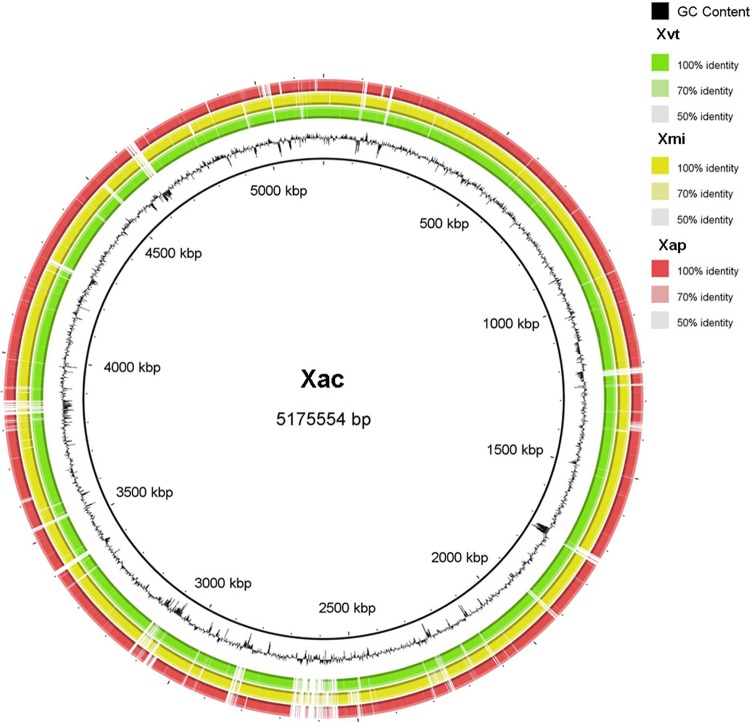
A circular map was generated taking Xac as the reference genome, and Xvt, Xap, and Xmi were aligned upon it (Fig. 3). The circular map shows that most of the genomic regions are conserved among the four strains under study, but there are unique regions in each pathogen. These variable regions also have differences in GC content suggesting the occurrence of horizontal gene transfer (HGT). To examine the impact of HGT, we computed the pangenome and systematically studied the loci known to be hypervariable in Xanthomonas and variations unique to each of the pathovars.
FIG 3.
Circular representation of whole-genome sequences of Xanthomonas campestris pv. viticola LMG 965 (Xvt), Xanthomonas citri pv. mangiferaeindicae LMG 941 (Xmi), and Xanthomonas axonopodis pv. punicae LMG 859 (Xap). The circular chromosome of Xanthomonas axonopodis pv. citri strain 306 (Xac) was taken as a reference. Moving outward, the black circle shows the percent G+C, followed by whole-genome sequences of Xvt (in green), Xmi (in yellow), and Xap (in red). Variation in intensity of each color correlates with approximate identity to the Xac genome.
(i) Pangenome analysis of X. axonopodis strains infecting diverse fruit crops.
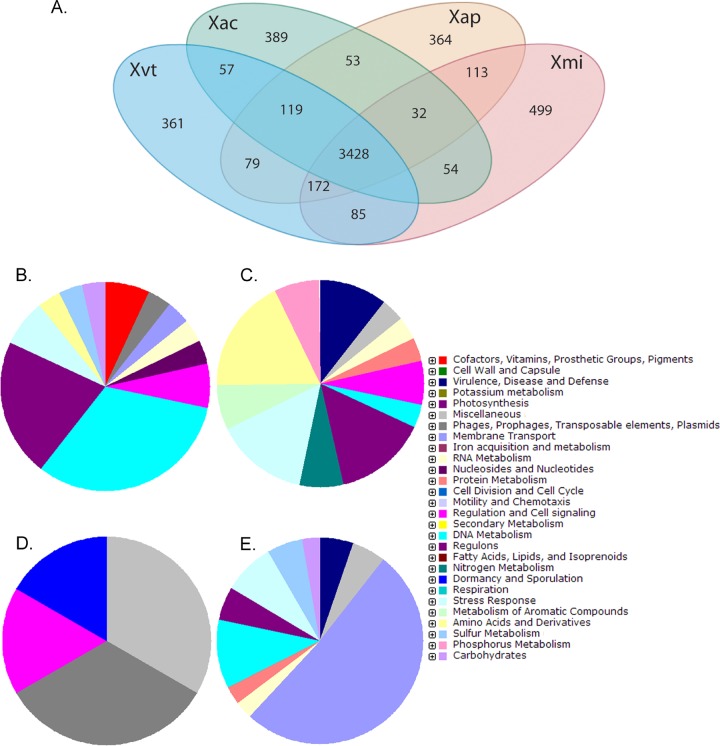
Figure 4A shows a Venn diagram illustrating the sharing of orthologous genes by the four genomes. Xac, Xap, Xvt, and Xmi have 389, 364, 361, and 499 unique gene clusters, respectively, the number being comparatively higher in Xmi. The unique genes identified for each genome are listed along with their GC contents in the supplemental material. The percentages of unique genes having GC content within 64.5% ± 2.5% are quite low (29.8, 42.6, 30.7, and 42.7 for Xac, Xap, Xvt, and Xmi, respectively). A functional characterization of the unique gene clusters is depicted in Fig. 4B to E. Figure 4 shows that a major portion of the unique genes in Xac are genes coding for DNA metabolism and regulation-associated genes. While in Xmi, a major portion of the unique genes are membrane transport genes, Xap has unique genes that are predicted to encode the stress response and genes for amino acids and their derivatives and regulons; however, in Xvt, the major portion of the unique genes code for phage-related proteins.
FIG 4.
(A) Venn diagram showing the sharing of orthologous genes among Xanthomonas axonopodis pv. citri (Xac), Xanthomonas campestris pv. viticola (Xvt), Xanthomonas citri pv. mangiferaeindicae (Xmi), and Xanthomonas axonopodis pv. punicae (Xap). (B to E) Pie charts showing functional characterization of unique genes of Xac (B), Xap (C), Xvt (D), and Xmi (E).
(ii) Hypervariations at a lipopolysaccharide locus.
The LPS locus in Xanthomonas is a hypervariable region which has a role in virulence (37). In Xanthomonas, the LPS cluster is confined between two highly conserved housekeeping genes, metB and etfA, encoding the cystathionine gamma lyase and electron transport flavoprotein, respectively (38). We were particularly interested in the pathovars that lie in clonal group B (Fig. 2). The schematic arrangement of LPS loci in different pathovars is shown in Fig. 5. Apart from the gene content, the length of the LPS clusters is variable from 19.5 to 24 kb, while the G+C content varies from 56% to 60%. There are four types of LPS loci. Type 1 is present in Xac and Xap, and type 2 is present in Xag and Xml. Types 3 and 4 are hybrid; while type 3 is present in Xvt and Xmi, type 4 is present in XcAw12879. Comparison of the LPS cluster and clonal frame tree (see Fig. S1 in the supplemental material) reveals incongruence with their genealogical relationship. For example, Xmi and Xac are from the same sublineage, but only half of their LPS cassettes is related. On the other hand, the Xac LPS cassette matches with that of Xap and the Xmi LPS cassette matches with that of Xvt.
FIG 5.
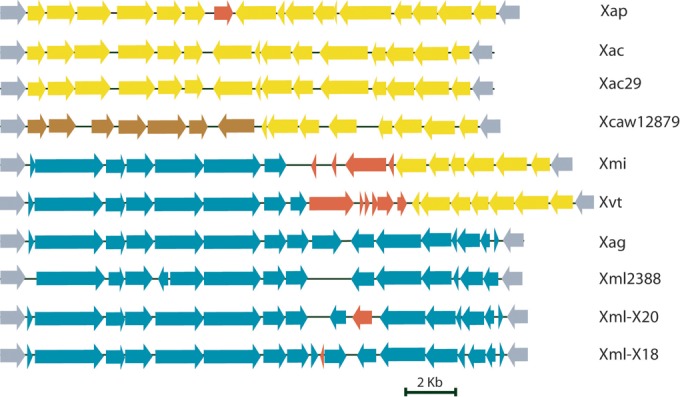
Comparison of nucleotide sequences encoding LPS in X. axonopodis pv. punicae LMG 859 (Xap), X. axonopodis pv. citri strain 306 (Xac), X. axonopodis Xac29-1 (Xac29), X. citri subsp. citri Aw12879 (Xcaw12879), X. citri pv. mangiferaeindicae LMG 941 (Xmi), X. campestris pv. viticola LMG 965 (Xvt), X. axonopodis pv. glycines strain 12-2 (Xag), X. axonopodis pv. malvacearum strain GSPB2388 (Xml2388), and X. citri pv. malvacearum strains X20 (Xml-X20) and X18 (Xml-X18). Different LPS cassettes are colored differently. All maps are approximately to scale. Red ORFs represent transposable elements. The color code of ORFs indicates homology among different LPS cassettes.
(iii) Variation in integron cassettes.
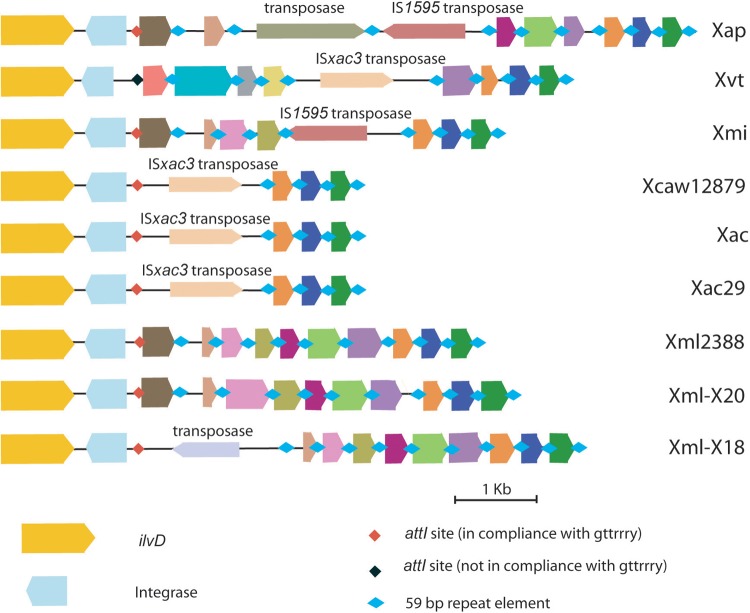
Integrons are genetic capture elements that allow site-specific incorporation of foreign DNA without affecting the functionality of the core genes. The integron cassette array which is known to be present downstream of ilvD, coding for acid dehydratase, in Xanthomonas (39) is not identical among the different pathovars under study, except for the distal ends comprising three open reading frames (ORFs) (Fig. 6). All the pathovars have variable numbers of 59- or 60-bp repeat elements (4 to 10). The sizes of the integron cassettes are variable in different pathovars; the cassette is smallest for citrus (4.5 kb) and largest in the pomegranate pathovar (8.6 kb). The GC content of the cassette array (52 to 59%) is lower than those of the ilvD and int genes (62 to 64%) and also lower than the average GC content of the Xanthomonas genome, which is 64.5 to 65%. While the three citrus pathovars harbor same type of integron cassettes, their close relative Xmi has a different one. Xvt has an integron cassette of which the proximal half does not share sequence homology with any of the cassettes of the other pathovars under study. Also, Xvt has a point mutation in the intI gene, leading to a frameshift in the coding sequence and thus a shorter ORF than the other four. The length of the integrase gene in all the strains (369 to 423 bp) is much shorter than that of Xanthomonas campestris pv. campestris ATCC 33913 (Xcc33913), i.e., 984 bp. The attI site in all the pathovars under study has the sequence GTTATCC, while in Xvt there is a slight variation yielding the sequence GTTAGGC, which is in compliance with the conserved motif GTTRRRY. Interestingly, the cassettes are also variable due to the type of transposable elements they encode. For example, Xap possess two transposable elements and the other pathovars harbor only a single insertion sequence (IS) element. IS1595 is same in the Xap and Xmi integron cassettes. Similarly, both Xac (including other two citrus pathovars) and Xvt have the same type of IS element, ISXac3, within their integron cassettes.
FIG 6.
Schematic diagram of integron cassette in X. axonopodis pv. punicae LMG 859 (Xap), X. campestris pv. viticola LMG 965 (Xvt), X. citri pv. mangiferaeindicae LMG 941 (Xmi), X. citri subsp. citri Aw12879 (Xcaw12879), X. axonopodis pv. citri strain 306 (Xac), X. axonopodis Xac29-1 (Xac29), X. axonopodis pv. malvacearum strain GSPB2388 (Xml2388), and X. citri pv. malvacearum strains X20 (Xml-X20) and X18 (Xml-X18). ilvD, integrase, attI (recombination site), 59-bp elements (secondary recombination site), and gene cassette arrays are shown. Different gene cassettes are colored differently, while genes with similar colors are homologs. All maps are approximately to scale.
(iv) CRISPRs.
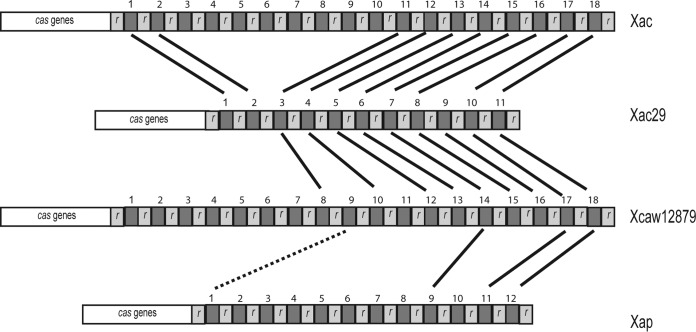
In these pathovars we were able to identify three different CRISPRs, CRISPR I (present in all strains under study), CRISPR II (present in Xac, Xac29, Xcaw12879, and Xap), and CRISPR III (present only in Xcaw12879) (see Table S4 in the supplemental material). In the case of CRISPR I, the number of direct repeats is 5 or 6 and the direct repeat length is 23 for all except X. fuscans, which has longer direct repeats (24 to 30 bp), but the starting 23 bp matches the consensus direct repeat sequence of Xmi. CRISPR I regions in all the pathovars were also found to have similar spacer regions.
In the case of CRISPR II, the direct repeat is 31 bp in length and identical in all four strains, while spacer regions are variable in length, ranging around 34 to 37 bp. The numbers of spacer regions were variable in the four strains: 18 in Xac, 11 in Xac29, 18 in Xcaw12879, and 12 in Xap. We were not able to recognize cas genes associated with CRISPR I, while for CRISPR II, Xac and Xap have seven cas genes, similar to Xanthomonas oryzae pv. oryzae PXO99A. Xac29 has six cas genes, with a frameshift in place of gene 3. Xcaw12879 also has six cas genes, with gene 7 replaced by a transposase gene and a dnaC gene before the repeats.
Figure 7 shows the CRISPR II repeats of the four X. axonopodis strains. Spacers are numbered by increasing age. Xac29 has least number of spacer regions. Spacers 10 and 11 are common in all four strains under study, which also include pathovar punicae along with citrus pathovars. Spacers 3 to 11 are present in all the three citrus pathovars, except for spacer 9 in the case of Xac.
FIG 7.
Comparison of CRSIPR II repeats (r) in X. axonopodis pv. punicae (Xap), X. citri subsp. citri Aw12879 (Xcaw12879), X. axonopodis pv. citri strain 306 (Xac), and X. axonopodis Xac29-1 (Xac29). Solid black bars connect the spacer regions that are identical among the strains, and the dotted black line shows spacer regions that show some similarity but are not identical. Numbers 1 to 18 show the arrangement of younger to older spacers.
(v) NRPS/PKS.
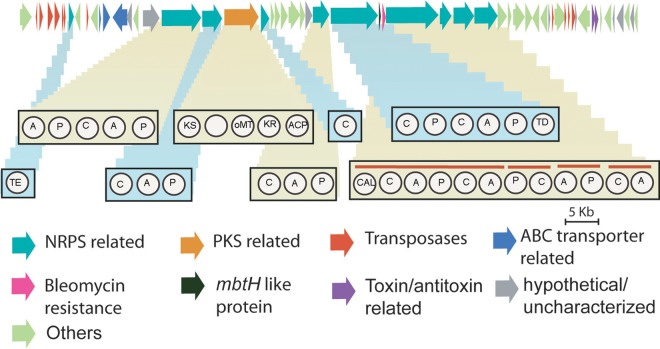
In silico analysis of the Xac genome predicted two NRPS clusters, which were similarly present in Xap, Xvt, and Xmi, but in Xmi we found the presence of a third NRPS cluster encompassing a genomic region of 89,742 bp in contig 13. This cluster did not show homology to any of other Xanthomonas organisms, while its close relative for a homologous gene cluster was predicted to be present in chromosome II of Burkholderia pseudomallei 668 and B. pseudomallei 1710b, which is, interestingly, a member of the class Betaproteobacteria. This contig has transposable elements associated at the start of the cluster, in the end of predicted NRPS domains, and then at the end of contig (see the supplemental material). Artemis comparison of contig 13 with the Xac genome shows the cluster being unique in Xmi, as only end regions of the contig share similarity with Xac (see Fig. S2 in the supplemental material). There is a large hybrid NRPS-PKS cluster containing 58 distinct genes (Fig. 8). Ten of these genes were found to be associated with NRPS, and one was associated with the PKS pathway. In the NRPS pathway we were able to predict an adenylation (A) domain, peptidyl carrier protein (PCP) domain, condensation (C) domain, and thioesterase (TE) domain. In the PKS pathway four different domains were predicted, ketosynthase (KS), methyltransferase (MT), ketoreductase (KR), and acyl carrier protein (ACP). However, we were not able to recognize an acyltransferase (AT) domain. Phylogenomic analysis of conserved KS domain suggested it to belong to a hybrid NRPS-PKS pathway type, while phylogenomic analysis of the conserved domain of 10 predicted C domains suggested that 2 of them to belong to a hybrid pathway type and other 8 C domains to an NRPS type. Table S5 in the supplemental material shows the closest matches of C and KS domains to other NRPS/PKS and hybrid NRPS-PKS pathways. KS and two C domains showed similarity to a bleomycin-encoding pathway, which is also a hybrid NRPS-PKS cluster, while other C domains showed closest matches to bacitracin, lychenocin, cyclosporine, and other antibiotic synthesis pathways. But the percent identity for all homologous domains was very low except for that for bleomycin, for which it was around 50%. There was also the presence of mbtH-like genes, genes related to an ABC transporter, and a bleomycin resistance-related gene. Analysis of six adenylation domains predicted the coded amino acid monomers to be Asn-His, hydroxyphenylglycine (Hpg), Cys, and Ser-Asn, while for three adenylation domains amino acid monomers could not be predicted. At the end of the cluster following the NRPS domain and transposable elements, there are also two genes encoding putative toxin and antitoxin.
FIG 8.
Schematic representation of hybrid NRPS-PKS system in X. citri pv. mangiferaeindicae LMG 941. Different genes are colored differently as per the predicted function. ORF maps are approximately to scale. The architecture of NRPS and PKS domains is shown in the rectangular boxes. A, adenylation domain; C, condensation domain; P, peptidyl carrier protein; TE, thioesterase; KS, ketosynthase; oMT, o-methyltransferase; KR, ketoreductase; ACP, acyl carrier protein; TD, terminal reductase domain; CAL, coenzyme A ligase domain.
Evolution of xanthomonadin coding cluster in X. citri pv. mangiferaindicae and X. campestris pv. viticola.
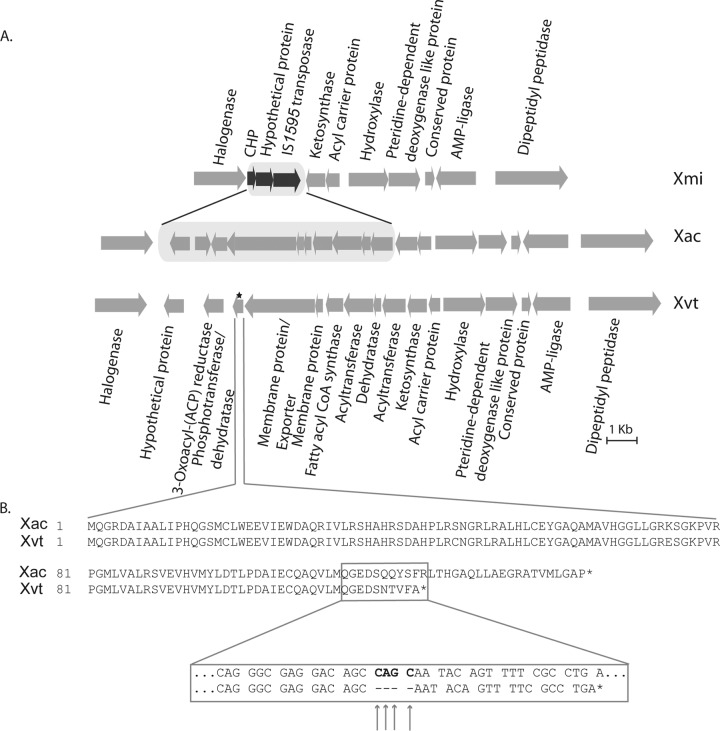
Despite belonging to the genus Xanthomonas, Xmi and Xvt produce white colonies, unlike other pathovars, which produce colonies with the yellow color characteristic of the genus Xanthomonas. To learn the genomic reason behind this unlikely phenotype of these pathovars, we studied the cluster of genes responsible for the production of pigment xanthomonadin. The gene cluster responsible for the xanthomonadin pigment production has already been identified in X. campestris pv. campestris (40) and Xoo (41). A similar gene cluster was found in the genomes of pathovars Xac, Xap, and Xvt, while in Xmi, we were not able to find the complete cluster. After amplification and primer walking of the missing sequence (see Materials and Methods), we compared the nucleotide sequences of the clusters from Xac and Xmi. A comparison between Xac and Xmi indicated that a 7,978-bp nucleotide sequence in Xac does not show identity with 2,128 bp of Xmi nucleotide sequence. We found this new 2,128-bp fragment to contain an IS element. Figure 9 shows the arrangement of genes in Xac, Xmi, and Xvt. In Xmi, the 10 ORFs of the xanthomonadin cluster are replaced by 3 ORFs, corresponding to two hypothetical proteins and IS1595 transposase.
FIG 9.
(A) Schematic diagram of xanthomonadin coding cluster in Xanthomonas citri pv. mangiferaeindicae (Xmi), Xanthomonas axonopodis pv. citri (Xac), and Xanthomonas campestris pv. viticola (Xvt). The location of frameshift mutation in Xvt is marked by a star. (B) Sequence alignment of protein phosphotransferase/dehydratase in Xac and Xvt. The enlarged box shows the nucleotide sequence alignment at the position of the frameshift mutation. The arrows mark the locations of deletions of four nucleotides in Xvt, and stop codons are marked by asterisks.
The xanthomonadin cluster arrangement in Xvt is similar to those in Xap and Xac. But Xvt was found to have a frameshift mutation because of a deletion of four nucleotides in the gene coding for phosphotransferase/dehydratase (already reported as necessary for pigment production [41]) at position 352 which has resulted in the formation of a truncated protein. The length of the protein in Xac is 143 amino acids, but in the case of Xvt the protein is terminated earlier, at 122 amino acids, because of the deletion of four nucleotides (bp 352 to 355).
DISCUSSION
Xac and its eco-relatives: are they just specialized strains?
Traditional studies on phylogeny of Xanthomonas were difficult because of limited DNA markers (42–44) and their characteristic host specialization (1). The advent of the next-generation sequencing era is enabling researchers to resolve the relationship of bacteria up to the strain and even the single-cell level (45, 46). In the present study, whole-genome sequencing and analysis have enabled us to determine evolutionary relationships and also the species status of Xanthomonas pathovars that infect citrus and its ecological relatives at a fine level. However, these pathogens have previously been classified several times into different species. Even though these pathovars infect diverse fruits, genome-based analysis revealed that they belong to same species and that they form a monophyletic group. The ANI among them is around 99%, much above the cutoff of 94% for designating different species (47).
Diverse pathovars from same clonal (sub)lineage.
Phylogenomic grouping of four fruit pathovars into a distinct cluster suggests their common ancestry, and their high genome identities as revealed by ANI indicate their recent evolutionary origins. Further clonal analysis suggests that all these fruit pathovars belong to one clonal lineage, as phylogenetic grouping of these pathogens is not affected by recombination events. Corroborating these findings is the fact that all these diseases were first noticed in India, which may be center of diversity of these pathovars. While Xac and Xmi were first identified more than 100 years ago (4), Xap was first identified in the 1950s (3) and Xvt was first reported in the1970s (2). In addition, Xap and Xvt form their own sublineages, while Xac and Xmi form one sublineage.
Genome plasticity and strain diversification: unique genes to large-scale variations.
A clear understanding of the phylogenetic and evolutionary relationship of a complex group of bacteria like Xanthomonas is necessary to make sense of the variations occurring during the diversification of its members into different lineages and sublineages. Our genome-based approach further enabled us to study the variable regions in the genome that are unique to or under selection in each of the pathovars.
(i) Localized and large-scale variations.
An LPS locus that is located between conserved housekeeping genes metB and etfA is known to be under diversifying selection at the species, pathovar, and strain levels (48). Even though the pathovars are from the same clonal lineage, they display hypervariation in size and organization of the LPS gene cluster. The altered GC content suggests that the large-scale variations are due to ancestral and multiple recent horizontal gene transfer events. Xac and Xap seem to have retained the LPS cassette that is the ancestral type in X. axonopodis pathovars, while Xmi and Xvt, which belong to different sublineages, have independently acquired a similar set of genes at the metB side. The relevance of these to strain virulence is worthy of further investigation.
Interestingly, Xcaw12879 is a citrus pathogen, but its LPS locus matches completely that of Xanthomonas oryzae pv. oryzicola BLS256 (Xoc), which is a rice pathogen, except for the IS elements in Xoc. However, only half the LPS cassette of Xcaw12879 has homologs in Xac and Xac29, which are phylogenetically close relatives. This hybrid LPS locus in Xcaw12879, similar to Xoc, could be a result of horizontal gene transfer. Similar observations have also been reported by Jalan and coworkers (44).
Xanthomonas strains are also known to harbor an integron whose location in the genome is conserved but for which the gene cassette array (particularly the proximal portion) is highly variable at the species, pathovar, and strain levels (39). Phylogenetic evidence suggested that it was acquired by the ancestor that gave rise to Xanthomonas, and its activity since then has contributed not only to the diversity of its members but also to host-specific pathogenesis. Different X. axonopodis pathovars possess different integron cassettes with various sizes, cassette arrays, and numbers of repeat elements, supporting the observation of Gillings and coworkers (39). The GC content of the cassette is also quite lower than that of the rest of the genome, and also the ilvD and int genes in particular, suggesting that the cassette arrays have been acquired by horizontal gene transfer. Gillings and coworkers proposed that the strains that have an identical deletion or frameshift mutation in the int gene also possess identical integron cassettes (39). However, in the present study, all strains except Xvt have similar int genes and attI sites, but they still show the presence of different integron cassettes.
Much of the variation seen in these cassettes is due to numbers and types of IS elements. High IS element activity suggests that the integron locus is highly dynamic even after the acquisition of novel genes. It is possible that the cassette present in Xml2388 may represent an ancestral one which has not undergone any change or has undergone a change similar to all others and that all the other strains except Xvt have cassettes similar to that in Xml2388 that may have undergone the replacement of some gene cassettes by transposases. It would be reasonable to infer that variation in these cassettes, having similar int genes and attI sites, would have been a result of transpositional events that occurred after integrase mutation and integron cassette fixation. However, the variations seen in the Xvt integron cassette suggest that it may have diversified from the ancestor before integron cassette fixation and its integrase gene may have evolved independently into a pseudogene. Three citrus pathovars have similar cassettes, suggesting that the transposition event may have occurred before their diversification from the common citrus ancestor.
Most hypervariations, like the ones in the LPS and integron cassettes, apart from their possible role in host-specific pathogenesis can also be due to selective pressure or genomic activity of specific phages. In this context it is important to know the variation in DNA loci, like CRISPR, that confer resistance to phages and act as the genetic memory of cell to form the adaptive immune system in bacteria (49). There is no CRIPR system that is present and variable in all these pathovars, which suggests minimal selection at these loci. CRISPR I repeats, although present in all strains under study, showed the existence of similar spacers. CRISPR II, which has also been previously reported for X. albilineans (50), X. oryzae pv. oryzae PXO99A, X. oryzae pv. oryzae MAFF 311018, X. oryzae pv. oryzae KACC1033 (51), and X. oryzae strains 21 and 604 (52), was also found in Pseudoxanthomonas spadix BD-a59, Rhodospirillum rubrum ATCC 11170, and Rhodospirillum rubrum F11. The presence of similar CRISPR II regions in phylogenetically distant bacteria and their absence in closely related bacteria like Xmi, Xvt, and other members of the X. axonopodis group support the notion that horizontal gene transfer may be playing a role in dispersal of these repeats, as suggested by previous studies (53).
(ii) Novel NRPS/PKS gene cluster.
Apart from the hypervariations mentioned above, we also found an 89-kb genomic region encoding an NRPS/PKS that is exclusive to the Xmi genome, probably the best example in our study showing the nature and size of unique variations. Nonribosomal peptidyl synthase/polyketide synthases are often involved in the production of bioactive peptides or small molecules. Xanthomonas has been considered a reservoir for the production of bioactive compounds by NRPS pathways (54). Two NRPS pathways identified in Xac have already been explored computationally for their putative functions. Xac3922 has been reported to function as a putative siderophore, as it carries an enzyme having similarity to Escherichia coli enterobactin synthase component F (EntF). The second NRPS pathway, in Xac2097, is a pathogenicity island (PAI) proposed to encode a tripeptide related to the phytotoxin syringomycin produced by Pseudomonas syringae (55). Recent reports refer to this NRPS as a pseudogene (54). Third, an NRPS identified in this study that is specific to Xmi is also possibly a PAI, as transposases are found on both sides of the cluster, and there is also a relatively high GC content of the NRPS and PKS genes (see the supplemental material). Toxin and antitoxin genes were also identified in this cluster apart from the NRPS/PKS modules. So these genes could have also been acquired in the genome by HGT along with this cluster, as they have also transposable elements on both sides and the GC content is greater than 65%.
The acyltransferase domain of the PKS pathway may have a diverse structure, as reported for Alteromonas macleodii (56), or it could be a trans-acting domain, as reported for X. albilineans and Clostridium spp. (57, 58). The presence of a module for the PKS pathway suggests that the pathway is a hybrid type, and it is also supported by the tree obtained for the conserved region of the KS domain, where it is grouped with hybrid-type KS (see Fig. S3 in the supplemental material). Proteins like that encoded by mbtH are considered to activate the adenylation domain in the NRPS system (59). ABC transporters were also identified, suggesting the product to be a secretory molecule. KS and C domains showed homologs to domains in the bleomycin-encoding NRPS-PKS pathway, and this cluster also has the gene for resistance to bleomycin. This suggests that this cluster could probably have a role in encoding a bleomycin-like antibiotic, as a cell has to be resistant to the antibiotic it produces. But low identity values of the domain suggest the molecule produced to be a variant of the known ones. This PAI was probably acquired in the sublineage that gave rise to Xmi. So this pathway could be a source of a novel antibiotic and also a putative drug target.
The presence of this unique NRPS/PKS cluster in Xmi and its absence from the closely related citrus pathovars suggest that it may have a role in the emergence and virulence of this pathogen. In a recent study, the emergence of the plant pathogen Dickeya solani was connected to a unique set of NRPS/PKS clusters (60).
(iii) How to be white Xanthomonas: from deletions to frameshifts.
The nomenclature of the genus Xanthomonas is based on the characteristic yellow colonies produced by its members. This color is due to a pigment known as xanthomonadin. Interestingly, some pathovars, like Xmi, Xvt, and Xanthomonas axonopodis pv. manihotis (Xam), are known to produce atypical white colonies. The biosynthesis of this pigment is known to occur by a type II PKS (41). All of the constituent enzymes required for this pathway have been identified in the xanthomonadin cluster of Xac, i.e., ketosynthase, ketoreductase (3-oxoacyl-ACP-reductase), dehydratase, acyl carrier protein, and acyltransferase (61). Among all these proteins, phosphotransferase/dehydratase, acyltransferase, dehydratase, and acyl carrier protein have been found to be required for pigment production in Xoo (41). In the case of Xam, it has already been reported that the absence of pigment production could be due to a frameshift in the acyl carrier protein-coding gene (62). We also found that Xmi and Xvt also carry mutations in this cluster, and this may result in decreased xanthomonadin production and white colonies. Interestingly, Xvt carries a frameshift mutation in the gene coding for phosphotransferase/dehydratase, but in the case of Xmi, three out of four necessary pigment-producing genes have been replaced with an IS element belonging to the IS1595 family (63). IS1595 has already been reported to occur in Xmi, and its sequence is available under GenBank accession number AF249895. It is also present in genomes of Xap and Xvt but not in Xac. The action of IS1595 has probably led to the deletion of the genes of the PKS pathway in Xmi, and thus, it could be the possible reason for the organism's white phenotype. Hence, Xmi, Xvt, and Xam have evolved independently to be nonpigmented. The presence of a distinct type of mutation indicates that Xmi and Xvt indeed belong to different sublineages. At the same time, deletion specific to Xmi might have occurred after the emergence of Xac pathovars. However, the ecological basis and adaptive role of these mutations need to be investigated in further genetic studies.
Xac and Xmi share a close relationship, as revealed by the maximum likelihood tree, and its robustness is also clear in a tree considering recombination. This evolutionary closeness is also shared by their host plants, Citrus and Mangifera, as they belong to same order, Sapindales (64). But still, the existence of a large NRPS/PKS cluster in Xmi, the absence of CRISPR II, the large deletion in the xanthomonadin cluster, the variation in integron cassettes, and the difference in its LPS cassette from that of Xac reveal the dynamicity and genomic flux these host-specific pathogens are undergoing. On the other hand, Xac and Xap, which belong to different sublineages and were reported at different periods, have similar LPS cassettes and CRISPR elements. Both of these facts reveal the plasticity in the genome of these pathovars. The Xanthomonas strains may be in genomic flux by continuously being exposed to a large gene pool, leading to increased virulence, invasiveness, host switches, etc. This can be further seen in the GC content variation and functional diversity of several hundreds of unique genes possessed by each pathovar. However, what is more puzzling and a probable clue for their host range and emergence is that the functions encoded by unique genes are distinct these pathovars.
In this context, the recent reports of new Xanthomonas pathovars of other important crops from India (e.g., Xanthomonas axonopodis causes gumming of Commiphora wightii) (65, 66) demonstrate the enormous potential for diversity of these pathogens and their genes in their probable center of origin. Functional characterization of unique genes and gene clusters and also hypervariable regions in these pathogens would further enable us to understand their ecology and host-specific pathogenesis.
Supplementary Material
ACKNOWLEDGMENTS
Samriti Midha is supported by fellowship from the Council of Scientific and Industrial Research (CSIR). We acknowledge funding from IMTECH and CSIR projects (OLP-62 and BSC-117/PMSI).
We acknowledge the help of the Microbial Type Culture Collection, IMTECH, during this study. We thank Girish Sahni, the director of IMTECH, for encouragement and support.
Footnotes
Published ahead of print 1 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01654-14.
REFERENCES
- 1.Hayward A. 1993. The hosts of Xanthomonas, p 1–119 In Swings J, Civerolo E. (ed), Xanthomonas. Chapman & Hall, London, United Kingdom [Google Scholar]
- 2.Nayudu M. 1972. Pseudomonas viticola sp. nov., incitant of a new bacterial disease of grape vine. J. Phytopathol. 73:183–186. 10.1111/j.1439-0434.1972.tb02539.x [DOI] [Google Scholar]
- 3.Hingorani M, Singh N. 1960. Xanthomonas punicae sp. nov. on Púnica granatum L. Indian J. Agric. Res. 29:45–48 [Google Scholar]
- 4.Raychaudhuri S, Verma J, Nariani T, Sen B. 1972. The history of plant pathology in India. Annu. Rev. Phytopathol. 10:21–36. 10.1146/annurev.py.10.090172.000321 [DOI] [Google Scholar]
- 5.Brunings AM, Gabriel DW. 2003. Xanthomonas citri: breaking the surface. Mol. Plant Pathol. 4:141–157. 10.1046/j.1364-3703.2003.00163.x [DOI] [PubMed] [Google Scholar]
- 6.Graham JH, Gottwald TR, Cubero J, Achor DS. 2004. Xanthomonas axonopodis pv. citri: factors affecting successful eradication of citrus canker. Mol. Plant Pathol. 5:1–15. 10.1046/j.1364-3703.2004.00197.x [DOI] [PubMed] [Google Scholar]
- 7.Patel MK, Moniz L, Kulkarni YS. 1948. A new bacterial disease of Mangifera indica L. Curr. Sci. 17:189. [PubMed] [Google Scholar]
- 8.Doidge E. 1915. A bacterial disease of the mango. Bacillus mangiferae n. sp. Ann. Appl. Biol. 2:1–45 [Google Scholar]
- 9.Chand R, Kishun R. 1991. Studies on bacterial blight (Xanthomonas campestris pv. punicae) of pomegranate. Indian Phytopathol. 44:370–372 [Google Scholar]
- 10.Akhtar MA, Bhatti MHR. 1992. Occurrence of bacterial leaf spot of pomegranate in Pakistan. Pak. J. Agric. Res. 13:95–97 [Google Scholar]
- 11.Petersen Y, Mansvelt E, Venter E, Langenhoven W. 2010. Detection of Xanthomonas axonopodis pv. punicae causing bacterial blight on pomegranate in South Africa. Australas. Plant Pathol. 39:544–546. 10.1071/AP10034 [DOI] [Google Scholar]
- 12.Young JM, Dye DW, Bradbury JF, Panagopoulos CG, Robbs CF. 1978. A proposed nomenclature and classification for plant pathogenic bacteria. N. Z. J. Agric. Res. 21:153–177. 10.1080/00288233.1978.10427397 [DOI] [Google Scholar]
- 13.Midha S, Patil PB. 2014. Genomic flux in Xanthomonas group of plant pathogenic bacteria, p 131–153 In Katsy EI. (ed), Plasticity in plant-growth-promoting and phytopathogenic bacteria. Springer, New York, NY [Google Scholar]
- 14.Vauterin L, Hoste B, Kersters K, Swings J. 1995. Reclassification of Xanthomonas. Int. J. Syst. Bacteriol. 45:472–489. 10.1099/00207713-45-3-472 [DOI] [Google Scholar]
- 15.Schaad NW, Postnikova E, Lacy GH, Sechler A, Agarkova I, Stromberg PE, Stromberg VK, Vidaver AK. 2005. Reclassification of Xanthomonas campestris pv. citri (ex Hasse 1915) Dye 1978 forms A, B/C/D, and E as X. smithii subsp. citri (ex Hasse) sp. nov. nom. rev. comb. nov., X. fuscans subsp. aurantifolii (ex Gabriel 1989) sp. nov. nom. rev. comb. nov., and X. alfalfae subsp. citrumelo (ex Riker and Jones) Gabriel et al., 1989 sp. nov. nom. rev. comb. nov.; X. campestris pv. malvacearum (ex smith 1901) Dye 1978 as X. smithii subsp. smithii nov. comb. nov. nom. nov.; X. campestris pv. alfalfae (ex Riker and Jones, 1935) Dye 1978 as X. alfalfae subsp. alfalfae (ex Riker et al., 1935) sp. nov. nom. rev.; and “var. fuscans” of X. campestris pv. phaseoli (ex Smith, 1987) Dye 1978 as X. fuscans subsp. fuscans sp. nov. Syst. Appl. Microbiol. 28:494–518. 10.1016/j.syapm.2005.03.017 [DOI] [PubMed] [Google Scholar]
- 16.Schaad NW, Postnikova E, Lacy G, Sechler A, Agarkova IV, Stromberg PE, Stromberg VK, Vidaver AM. 2006. Emended classification of xanthomonad pathogens on citrus. Syst. Appl. Microbiol. 29:690–695. 10.1016/j.syapm.2006.08.001 [DOI] [PubMed] [Google Scholar]
- 17.Gabriel D, Kingsley M, Hunter J, Gottwald T. 1989. Reinstatement of Xanthomonas citri (ex Hasse) and X. phaseoli (ex Smith) to species and reclassification of all X. campestris pv. citri strains. Int. J. Syst. Bacteriol. 39:14–22. 10.1099/00207713-39-1-14 [DOI] [Google Scholar]
- 18.Ah-You N, Gagnevin L, Grimont PAD, Brisse S, Nesme X, Chiroleu F, Ngoc LBT, Jouen E, Lefeuvre P, Vernière C, Pruvost O. 2009. Polyphasic characterization of xanthomonads pathogenic to members of the Anacardiaceae and their relatedness to species of Xanthomonas. Int. J. Syst. Evol. Microbiol. 59:306–318. 10.1099/ijs.0.65453-0 [DOI] [PubMed] [Google Scholar]
- 19.da Silva AC, Ferro JA, Reinach FC, Farah CS, Furlan LR, Quaggio RB, Monteiro-Vitorello CB, Van Sluys MA, Almeida NF, Alves LM, do Amaral AM, Bertolini MC, Camargo LE, Camarotte G, Cannavan F, Cardozo J, Chambergo F, Ciapina LP, Cicarelli RM, Coutinho LL, Cursino-Santos JR, El-Dorry H, Faria JB, Ferreira AJ, Ferreira RC, Ferro MI, Formighieri EF, Franco MC, Greggio CC, Gruber A, Katsuyama AM, Kishi LT, Leite RP, Lemos EG, Lemos MV, Locali EC, Machado MA, Madeira AM, Martinez-Rossi NM, Martins EC, Meidanis J, Menck CF, Miyaki CY, Moon DH, Moreira LM, Novo MT, Okura VK, Oliveira MC, Oliveira VR, Pereira HA, Rossi A, Sena JA, Silva C, de Souza RF, Spinola LA, Takita MA, Tamura RE, Teixeira EC, Tezza RI, Trindade dos Santos M, Truffi D, Tsai SM, White FF, Setubal JC, Kitajima JP. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459–463. 10.1038/417459a [DOI] [PubMed] [Google Scholar]
- 20.Midha S, Ranjan M, Sharma V, Pinnaka AK, Patil PB. 2012. Genome sequence of Xanthomonas citri pv. mangiferaeindicae strain LMG 941. J. Bacteriol. 194:3031. 10.1128/JB.00433-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma V, Midha S, Ranjan M, Pinnaka AK, Patil PB. 2012. Genome sequence of Xanthomonas axonopodis pv. punicae strain LMG 859. J. Bacteriol. 194:2395. 10.1128/JB.00181-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lagesen K, Hallin P, Rødland EA, Stærfeldt H-H, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35:3100–3108. 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu M, Eisen JA. 2008. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 9:R151. 10.1186/gb-2008-9-10-r151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar S, Nei M, Dudley J, Tamura K. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9:299–306. 10.1093/bib/bbn017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richter M, Rosselló-Móra R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U. S. A. 106:19126–19131. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Didelot X, Falush D. 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175:1251–1266. 10.1534/genetics.106.063305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oh PL, Benson AK, Peterson DA, Patil PB, Moriyama EN, Roos S, Walter J. 2010. Diversification of the gut symbiont Lactobacillus reuteri as a result of host-driven evolution. ISME J. 4:377–387. 10.1038/ismej.2009.123 [DOI] [PubMed] [Google Scholar]
- 29.Alikhan N-F, Petty NK, Zakour NLB, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Y, Wu J, Yang J, Sun S, Xiao J, Yu J. 2012. PGAP: pan-genomes analysis pipeline. Bioinformatics 28:416–418. 10.1093/bioinformatics/btr655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14:1394–1403. 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stokes H, O'gorman D, Recchia GD, Parsekhian M, Hall RM. 1997. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol. Microbiol. 26:731–745. 10.1046/j.1365-2958.1997.6091980.x [DOI] [PubMed] [Google Scholar]
- 33.Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R. 2011. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 39:W339–W346. 10.1093/nar/gkr466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anand S, Prasad M, Yadav G, Kumar N, Shehara J, Ansari MZ, Mohanty D. 2010. SBSPKS: structure based sequence analysis of polyketide synthases. Nucleic Acids Res. 38:W487–W496. 10.1093/nar/gkq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ziemert N, Podell S, Penn K, Badger JH, Allen E, Jensen PR. 2012. The natural product domain seeker NaPDoS: a phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS One 7:e34064. 10.1371/journal.pone.0034064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abbott J, Aanensen DM, Rutherford K, Butcher S, Spratt BG. 2005. WebACT—an online companion for the Artemis Comparison Tool. Bioinformatics 21:3665–3666. 10.1093/bioinformatics/bti601 [DOI] [PubMed] [Google Scholar]
- 37.Dharmapuri S, Yashitola J, Vishnupriya MR, Sonti RV. 2001. Novel genomic locus with atypical G+C content that is required for extracellular polysaccharide production and virulence in Xanthomonas oryzae pv. oryzae. Mol. Plant Microbe Interact. 14:1335–1339. 10.1094/MPMI.2001.14.11.1335 [DOI] [PubMed] [Google Scholar]
- 38.Patil PB, Sonti RV. 2004. Variation suggestive of horizontal gene transfer at a lipopolysaccharide (lps) biosynthetic locus in Xanthomonas oryzae pv. oryzae, the bacterial leaf blight pathogen of rice. BMC Microbiol. 4:40. 10.1186/1471-2180-4-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gillings MR, Holley MP, Stokes HW, Holmes AJ. 2005. Integrons in Xanthomonas: a source of species genome diversity. Proc. Natl. Acad. Sci. U. S. A. 102:4419–4424. 10.1073/pnas.0406620102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poplowsky A, Kawalek M, Schaad N. 1993. A xanthomonadin-encoding gene cluster for the identification of pathovars of Xanthomonas campestris. Mol. Plant Microbe Interact. 6:545–552 [Google Scholar]
- 41.Goel AK, Rajagopal L, Nagesh N, Sonti RV. 2002. Genetic locus encoding functions involved in biosynthesis and outer membrane localization of xanthomonadin in Xanthomonas oryzae pv. oryzae. J. Bacteriol. 184:3539–3548. 10.1128/JB.184.13.3539-3548.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parkinson N, Cowie C, Heeney J, Stead D. 2009. Phylogenetic structure of Xanthomonas determined by comparison of gyrB sequences. Int. J. Syst. Evol. Microbiol. 59:264–274. 10.1099/ijs.0.65825-0 [DOI] [PubMed] [Google Scholar]
- 43.Mhedbi-Hajri N, Hajri A, Boureau T, Darrasse A, Durand K, Brin C, Fischer-Le Saux M, Manceau C, Poussier S, Pruvost O. 2013. Evolutionary history of the plant pathogenic bacterium Xanthomonas axonopodis. PLoS One 8:e58474. 10.1371/journal.pone.0058474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jalan N, Kumar D, Andrade MO, Yu F, Jones JB, Graham JH, White FF, Setubal JC, Wang N. 2013. Comparative genomic and transcriptome analyses of pathotypes of Xanthomonas citri subsp. citri provide insights into mechanisms of bacterial virulence and host range. BMC Genomics 14:551. 10.1186/1471-2164-14-551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nasser W, Beres SB, Olsen RJ, Dean MA, Rice KA, Long SW, Kristinsson KG, Gottfredsson M, Vuopio J, Raisanen K. 2014. Evolutionary pathway to increased virulence and epidemic group A Streptococcus disease derived from 3,615 genome sequences. Proc. Natl. Acad. Sci. U. S. A. 111:E1768–E1776. 10.1073/pnas.1403138111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maiden MC, van Rensburg MJJ, Bray JE, Earle SG, Ford SA, Jolley KA, McCarthy ND. 2013. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat. Rev. Microbiol. 11:728–736. 10.1038/nrmicro3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Konstantinidis KT, Tiedje JM. 2005. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. U. S. A. 102:2567–2572. 10.1073/pnas.0409727102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patil PB, Bogdanove AJ, Sonti RV. 2007. The role of horizontal transfer in the evolution of a highly variable lipopolysaccharide biosynthesis locus in xanthomonads that infect rice, citrus and crucifers. BMC Evol. Biol. 7:243. 10.1186/1471-2148-7-243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marraffini LA, Sontheimer EJ. 2010. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 11:181–190. 10.1038/nrg2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pieretti I, Royer M, Barbe V, Carrere S, Koebnik R, Couloux A, Darrasse A, Gouzy J, Jacques MA, Lauber E, Manceau C, Mangenot S, Poussier S, Segurens B, Szurek B, Verdier V, Arlat M, Gabriel DW, Rott P, Cociancich S. 2012. Genomic insights into strategies used by Xanthomonas albilineans with its reduced artillery to spread within sugarcane xylem vessels. BMC Genomics 13:658. 10.1186/1471-2164-13-658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salzberg SL, Sommer DD, Schatz MC, Phillippy AM, Rabinowicz PD, Tsuge S, Furutani A, Ochiai H, Delcher AL, Kelley D, Madupu R, Puiu D, Radune D, Shumway M, Trapnell C, Aparna G, Jha G, Pandey A, Patil PB, Ishihara H, Meyer DF, Szurek B, Verdier V, Koebnik R, Dow JM, Ryan RP, Hirata H, Tsuyumu S, Won Lee S, Seo YS, Sriariyanum M, Ronald PC, Sonti RV, Van Sluys MA, Leach JE, White FF, Bogdanove AJ. 2008. Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics 9:204. 10.1186/1471-2164-9-204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Semenova E, Nagornykh M, Pyatnitskiy M, Artamonova II, Severinov K. 2009. Analysis of CRISPR system function in plant pathogen Xanthomonas oryzae. FEMS Microbiol. Lett. 296:110–116. 10.1111/j.1574-6968.2009.01626.x [DOI] [PubMed] [Google Scholar]
- 53.Godde JS, Bickerton A. 2006. The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J. Mol. Evol. 62:718–729. 10.1007/s00239-005-0223-z [DOI] [PubMed] [Google Scholar]
- 54.Royer M, Koebnik R, Marguerettaz M, Barbe V, Robin GP, Brin C, Carrere S, Gomez C, Hügelland M, Völler GH. 2013. Genome mining reveals the genus Xanthomonas to be a promising reservoir for new bioactive non-ribosomally synthesized peptides. BMC Genomics 14:658. 10.1186/1471-2164-14-658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Etchegaray A, Silva-Stenico ME, Moon DH, Tsai SM. 2004. In silico analysis of nonribosomal peptide synthetases of Xanthomonas axonopodis pv. citri: identification of putative siderophore and lipopeptide biosynthetic genes. Microbiol. Res. 159:425–437. 10.1016/j.micres.2004.09.009 [DOI] [PubMed] [Google Scholar]
- 56.Mizuno CM, Kimes NE, López-Pérez M, Ausó E, Rodriguez-Valera F, Ghai R. 2013. A hybrid NRPS-PKS gene cluster related to the bleomycin family of antitumor antibiotics in Alteromonas macleodii strains. PLoS One 8:e76021. 10.1371/journal.pone.0076021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Royer M, Costet L, Vivien E, Bes M, Cousin A, Damais A, Pieretti I, Savin A, Megessier S, Viard M. 2004. Albicidin pathotoxin produced by Xanthomonas albilineans is encoded by three large PKS and NRPS genes present in a gene cluster also containing several putative modifying, regulatory, and resistance genes. Mol. Plant Microbe Interact. 17:414–427. 10.1094/MPMI.2004.17.4.414 [DOI] [PubMed] [Google Scholar]
- 58.Behnken S, Hertweck C. 2012. Cryptic polyketide synthase genes in non-pathogenic Clostridium spp. PLoS One 7:e29609. 10.1371/journal.pone.0029609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Herbst DA, Boll Zocher BG, Stehle T, Heide L. 2013. Structural basis of the interaction of MbtH-like proteins, putative regulators of nonribosomal peptide biosynthesis, with adenylating enzymes. J. Biol. Chem. 288:1991–2003. 10.1074/jbc.M112.420182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garlant L, Koskinen P, Rouhiainen L, Laine P, Paulin L, Auvinen P, Holm L, Pirhonen M. 2013. Genome sequence of Dickeya solani, a new soft rot pathogen of potato, suggests its emergence may be related to a novel combination of non-ribosomal peptide/polyketide synthetase clusters. Diversity 5:824–842. 10.3390/d5040824 [DOI] [Google Scholar]
- 61.Hopwood DA. 1997. Genetic contributions to understanding polyketide synthases. Chem. Rev. 97:2465–2498. 10.1021/cr960034i [DOI] [PubMed] [Google Scholar]
- 62.Arrieta-Ortiz ML, Rodríguez-R LM, Pérez-Quintero AL, Poulin L, Díaz AC, Rojas NA, Trujillo C, Benavides MR, Bart R, Boch J, Boureau T, Darrasse A, David P, Bernonville TDD, Fontanilla P, Gagnevin L, Guerin F, Jacques M, Lauber E, Lefeuvre P, Medina C, Medina E, Montenegro N, Bodnar AM, Noel LD, Quinones JFO, Osorio D, Pardo C, Patil PB, Poussier S, Pruvost O, Robene-Soustrade I, Ryan RP, Tabima J, Morales OGU, Verniere C, Carrere S, Verdier V, Szurek B, Restrepo S, Lopez C, Koebnik R, Bernal A. 2013. Genomic survey of pathogenicity determinants and VNTR markers in the cassava bacterial pathogen Xanthomonas axonopodis pv. manihotis strain CIO151. PLoS One 8:e79704. 10.1371/journal.pone.0079704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Siguier P, Gagnevin L, Chandler M. 2009. The new IS1595 family, its relation to IS1 and the frontier between insertion sequences and transposons. Res. Microbiol. 160:232–241. 10.1016/j.resmic.2009.02.003 [DOI] [PubMed] [Google Scholar]
- 64.Jansen RK, Cai Z, Raubeson LA, Daniell H, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, Chumley TW. 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. U. S. A. 104:19369–19374. 10.1073/pnas.0709121104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Samanta JN, Mandal K, Maiti S. 2013. A novel pathovar of Xanthomonas axonopodis causes gumming of Guggal (Commiphora wightii). Eur. J. Plant Pathol. 135:115–125. 10.1007/s10658-012-0070-x [DOI] [Google Scholar]
- 66.Papdiwal P, Deshpande K. 1978. Bacterial leaf necrosis of custard apple. Geobios 5:285–286 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.