Abstract
Cytophaga hutchinsonii is a widely distributed cellulolytic bacterium in the phylum Bacteroidetes. It can digest crystalline cellulose rapidly without free cellulases or cellulosomes. The mechanism of its cellulose utilization remains a mystery. We developed an efficient method based on a linear DNA double-crossover and FLP-FRT recombination system to obtain unmarked deletions of both single genes and large genomic fragments in C. hutchinsonii. Unmarked deletion of CHU_3237 (porU), an ortholog of the C-terminal signal peptidase of a type IX secretion system (T9SS), resulted in defects in colony spreading, cellulose degradation, and protein secretion, indicating that it is a component of the T9SS and that T9SS plays an important role in cellulose degradation by C. hutchinsonii. Furthermore, deletions of four large genomic fragments were obtained using our method, and the sizes of the excised fragments varied from 9 to 19 kb, spanning from 6 to 22 genes. The customized FLP-FRT method provides an efficient tool for more rapid progress in the cellulose degradation mechanism and other physiological aspects of C. hutchinsonii.
INTRODUCTION
Cytophaga hutchinsonii is a widely distributed Gram-negative cellulolytic bacterium which belongs to the phylum Bacteroidetes (1–3). C. hutchinsonii can digest crystalline cellulose rapidly in a contact-dependent way and exhibits gliding motility over surfaces. The strategies for microbial cellulose degradation are usually divided into two types, the free-cellulase mechanism used by many aerobic microorganisms and the multienzyme cellulosomes used by many anaerobic microorganisms (4–7). Cellulose degradation by C. hutchinsonii needs direct contact with cellulose, and most of the cellulase activity seems to be cell associated (2, 8). The analysis of the C. hutchinsonii genome shows that it does not encode any proteins containing dockerin or cohesion domains, which are the characteristics of cellulosomes (3). Thus, the mechanism of cellulose degradation by C. hutchinsonii is novel and still unknown (6, 9).
Due to the lack of genetic manipulation tools, the study of the cellulose degradation mechanism of C. hutchinsonii remained stagnant until transposon-mediated mutagenesis by conjugation was developed (10). Recently, many other genetic manipulation tools have been developed for C. hutchinsonii to obtain targeted gene disruptions, complementation with replicative plasmids, and unmarked deletions (11–15).
The FLP-FRT recombination system from Saccharomyces cerevisiae is one of the most useful tools for efficient genetic engineering and has proven functional in diverse bacterial species (16–24). The site-specific FLP recombinase recognizes the FRT (FLP recognition target) sites and excises the DNA fragment between them when the FRT sites are in the same orientation. Combined with minitransposons, the FLP-FRT recombination system can be used to excise large DNA segments from the genome randomly and cyclically (25). To our knowledge, this recombination system has not yet been used in the phylum Bacteroidetes. More than 40 percent of C. hutchinsonii genes remain unannotated, and characterization of them with the current genetic tools would require substantial effort and time. An efficient method is required for rapid characterization of unknown genes.
Recent studies have shown that some outer membrane proteins may play an important role in cellulose degradation by C. hutchinsonii (12, 26), a finding supported by the isolation of the C. hutchinsonii endoglucanase Cel5A (ChCel5A) from the outer membrane (14). Thus, a protein secretion system across the outer membrane may play a role in cellulose degradation. Analysis of the C. hutchinsonii genome showed only a few homologs of a type II secretion system and a full set of orthologs of a novel protein secretion system recently discovered in Porphyromonas gingivalis and Flavobacterium johnsoniae, named the Por secretion system (PorSS) or type IX secretion system (T9SS) (27, 28). The T9SS in P. gingivalis is used for secretion of many proteins including gingipains (27, 29). Protein substrates of the T9SS have conserved C-terminal domains (CTDs) which are important for secretion, posttranslational modification, and cell surface attachment (30–34). A novel C-terminal signal peptidase has been identified to be responsible for the cleavage of CTDs of the T9SS substrates in P. gingivalis (30). The deletion of one of the T9SS genes, CHU_0170 (sprP), causes defects in gliding motility and cellulose utilization (15). However, functions and physiological roles of other components of the T9SS in C. hutchinsonii remain unknown.
In this study, a method based on the FLP-FRT recombination system and linear DNA double-crossover recombination was developed for C. hutchinsonii to generate unmarked deletions of both single genes and large genomic fragments. Unmarked deletion of CHU_3237 (porU), an ortholog of the C-terminal signal peptidase from P. gingivalis, caused defects in colony spreading, cellulose degradation, and protein secretion, indicating that it is a component of the T9SS and that the T9SS has an important role in cellulose degradation by C. hutchinsonii. Furthermore, several deletions of different large genomic fragments were obtained using our method, and the sizes of the excised fragments varied from 9 to 19 kb, spanning from 6 to 22 genes. The customized FLP-FRT method thus appears to be an efficient tool for targeting large genomic fragments in C. hutchinsonii.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1, and primers are listed in Table S1 in the supplemental material. Cytophaga hutchinsonii ATCC 33406 was kindly provided by Mark J. McBride. Escherichia coli strains were grown at 37°C in Luria-Bertani medium. C. hutchinsonii was grown at 30°C in PY6 medium (6 g of peptone, 0.5 g of yeast extract, 4 g of glucose per liter, pH 7.3), modified from PY2 medium (35). PY6 agar contained 10 g/liter agar unless indicated otherwise.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α | Strain used for gene cloning | Clontech |
| BL21(DE3)pLysS | F− ompT hsdSB (rB- mB-) gal dcm (DE3) pLysS; Cmr | Seebio Biotech |
| C. hutchinsonii strains | ||
| ATCC 33406 | Wild type | ATCC |
| Δ3237 strain | Unmarked deletion mutant of CHU_3237 | This study |
| C3237 strain | Complementation of Δ3237 mutant with pCH3237 | This study |
| Δ3202–3190 strain | Deletion mutant of large genomic fragment from CHU_3202 to CHU_3190 | This study |
| Δ0804–0819 strain | Deletion mutant of large genomic fragment from CHU_0804 to CHU_0819 | This study |
| Δ0834–0841 strain | Deletion mutant of large genomic fragment from CHU_0834 to CHU_0841 | This study |
| Δ0428–0449 strain | Deletion mutant of large genomic fragment from CHU_0428 to CHU_0449 | This study |
| 0344::cat strain | Disruption mutant of CHU_0344 made with pSJHC as template | This study |
| Plasmids | ||
| pEP4351 | Cmr Tcr (Emr) Mob+ | 49 |
| pLYL03 | Apr (Emr) Mob+ | 50 |
| pKD3 | Plasmid carrying two FRT sites; Cmr Apr | 19 |
| pCP20 | Plasmid carrying FLP recombinase; Cmr Apr | 38 |
| pBJ113 | Plasmid carrying galK | 37 |
| pSKSO8 | oriC; Apr (Emr) | 13 |
| pSK1284gfp | Plasmid carrying gfp and promoter of CHU_1284; Apr (Emr) | 13 |
| pEPLO293cfxA | Plasmid carrying cfxA | 12 |
| pET-His | Used for recombinant CHU_0344 expression; Apr | Seebio Biotech |
| pSJHS | Gene-targeting template plasmid carrying ermF; Apr (Emr) | This study |
| pSJHC | Gene-targeting template plasmid carrying cat under the control of the ompA promoter from F. johnsoniae; Apr (Cmr) | This study |
| pTKS | Unmarked deletion template plasmid carrying ermF flanked by two FRT sites cloned from pKD3 with primers KSfrtF and KSfrtR; Apr (Emr) | This study |
| pTSK | Similar to pTKS except for cloning the FRT sites with primers frtSKF and frtSKR; Apr (Emr) | This study |
| pCFXKS | Similar to pTKS except for carrying cfxA instead of ermF; Apr (Cfr) | This study |
| pCFXSK | Similar to pTSK except for carrying cfxA instead of ermF; Apr (Cfr) | This study |
| pSKSO8TG | Promoterless gfp and a Rho-independent terminator inserted into pSKSO83; oriC; Apr (Emr) | This study |
| pCHF | Plasmid carrying flp cotranscribed with cat under the control of the ompA promoter from F. johnsoniae; promoter of CHU_1284 and galK inserted and gfp evicted from pSKSO8TG; oriC; Apr (Cmr) | This study |
| pCH | Similar to pCHF except for the flp gene; oriC; Apr (Cmr) | This study |
| pCH3237 | A 4.4-kbp fragment spanning CHU_3237 amplified with primers C3237F and C3237R and ligated into SacI and SalI sites of pCH; oriC; Apr (Cmr) | This study |
Antibiotic resistance phenotypes: Apr, ampicillin; Cmr, chloramphenicol; Emr, erythromycin; Cfr, cefoxitin; Kmr, kanamycin. Phenotypes in parentheses are expressed in C. hutchinsonii, and phenotypes not in parentheses are expressed in E. coli.
To observe colony spreading, C. hutchinsonii was grown on PY2 medium with 5 g/liter agar supplemented with 2 g/liter glucose at 30°C. To analyze digestion of filter paper, C. hutchinsonii was grown on Stanier medium (2) with 10 g/liter agar. A cellulose plate was prepared as described by Ji et al. (11). Briefly, Avicel PH-101 (Sigma-Aldrich, MO, USA) was ball milled for 7 days and added to the PY2 medium with 5 g/liter agar at a final concentration of 1% (wt/vol) as the top layer of the plate. To analyze cellulase activity and protein secretion, cells were grown in Stanier medium supplemented with 2 g/liter glucose at 30°C. Antibiotics were used at the following concentrations when needed: ampicillin (Ap), 100 μg/ml; chloramphenicol (Cm), 15 μg/ml; erythromycin (Em), 30 μg/ml; cefoxitin (Cf), 15 μg/ml; kanamycin (Km), 30 μg/ml.
Construction of the double-crossover template plasmids pSJHS and pSJHC.
pLYL03 was digested with ScaI and EcoRI, and the 2.5-kbp fragment containing oriT and a partial bla gene was selected as a backbone for plasmid construction. The remaining portion of bla was amplified by PCR from the plasmid pLYL03 with primers blaF and blaR, digested with ScaI and EcoRI, and ligated with the backbone, yielding plasmid pOT. The DNA fragment containing ermF was PCR amplified with primers isermF and isermR using pLYL03 as the template, digested with BamHI and SphI, and inserted into the corresponding sites of pOT to create pSJH. To reduce the size of template plasmid pSJH, ermF including 259 bp upstream of the start codon was PCR amplified with primers sermF and sermR, digested with SacI and KpnI, and ligated into the corresponding sites of pSJH to replace the IS4351-ermF fragment, yielding pSJHS. Plasmid pSJHS was used as the template plasmid for linear fragment double-crossover recombination, and the homologous arms were inserted into the restriction sites on the flanks of ermF.
To generate a chloramphenicol-resistant plasmid, the fusion of the promoterless chloramphenicol acetyltransferase gene (cat) from pEP4351 and the ompA promoter (36) from F. johnsoniae was generated by overlap extension PCR with primers catF, catR, ompAF, and ompAR. The cat expression cassette was digested with SacI and KpnI and inserted into the corresponding sites of pSJH to generate the chloramphenicol-resistant plasmid pSJHC.
Construction of the unmarked deletion template plasmids.
DNA fragments containing cat and FRT sites were PCR amplified from pKD3 (19) with two sets of primers (frtSKF/frtSKR and KSfrtF/KSfrtR), digested with SacI and KpnI, and ligated into the corresponding sites of pSJHS. The resulting plasmids were digested with BssHII, treated by alkaline phosphatase, and ligated with PCR-generated and BssHII-digested ermF and cfxA (12), respectively, yielding four template plasmids named pTSK, pTKS, pCFXSK, and pCFXKS. pTSK and pTKS had the erythromycin resistance gene ermF flanked by two FRT sites, while pCFXSK and pCFXKS were cefoxitin resistant. SK/KS represent the different orientations of the FRT sites against multiple cloning sites (MCS), which is important for deletion of large genomic fragments and served as markers for convenience.
Construction of the FLP recombinase helper plasmid pCHF.
The ompAp-cat cassette cloned from pSJHC with primers cat-PstIF and cat-SDR (introducing an SphI site and a putative ribosome binding site from C. hutchinsonii) was digested with PstI and KpnI and inserted into the corresponding sites of pSKSO8TG (described in supplemental methods). The resulting plasmid was digested with SacI and XbaI and ligated with a galK expression cassette (the galactokinase gene from pBJ113 [37] under the control of CHU_1284 promoter [13]), which was amplified with primers P1284galK-F and P1284galK-R and digested with the same restriction enzymes, generating pCG. The pCG plasmid was digested with XbaI and SalI to excise gfp, blunted with Fast Pfu polymerase (TransGen, Beijing, China), and self-ligated to generate pCHSD. The FLP recombinase was amplified with primers flpF and flpR from pCP20 (38), digested with SphI and KpnI, and inserted into the corresponding sites of pCHSD, yielding the helper plasmid pCHF.
Construction of the CHU_3237 deletion mutant.
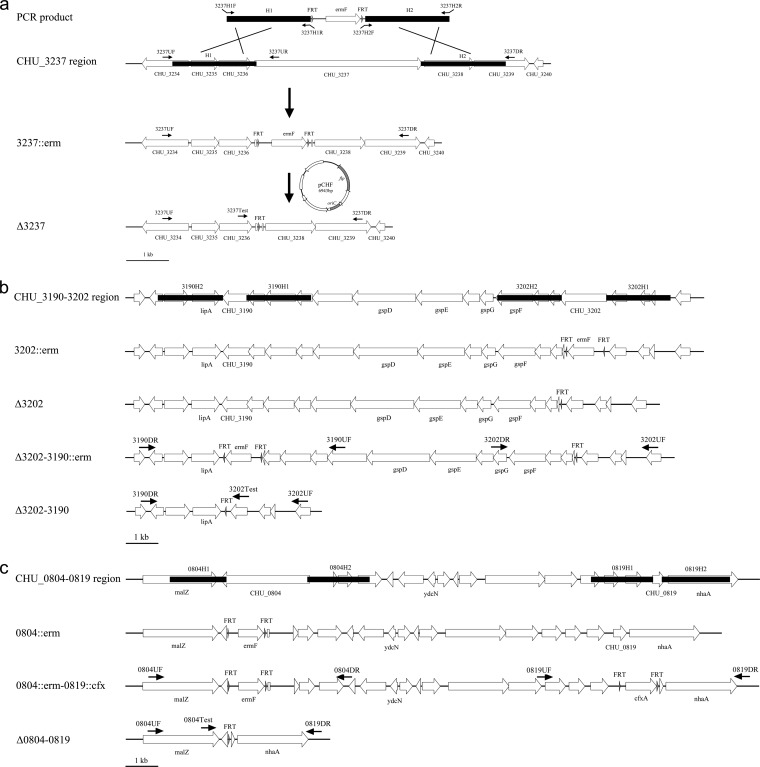
The unmarked deletion of CHU_3237 (yielding the Δ3237 mutant) was performed as follows (Fig. 1a). A 2.0-kbp fragment spanning the three flanking genes (CHU_3234, CHU_3235, and CHU_3236) and the first 43 bp of CHU_3237 were amplified with primers 3237H1F and 3237H1R. The fragment referred to as H1 was digested and ligated into the corresponding sites of pTKS. A 2.0-kbp fragment spanning the two flanking genes (CHU_3238 and CHU_3239) and the last 30 bp of CHU_3237 were amplified with primers 3237H2F and 3237H2R. The fragment referred to as H2 was digested and ligated into the corresponding sites of pTKS which flanked the ermF-FRT cassette opposite H1. The gene-targeting cassette was amplified by PCR with primers 3237H1F and 3237H2R and purified with a Cycle Pure kit (Omega, GA, USA). A total of 1.5 μg of PCR product was transformed into 100 μl of competent cells of C. hutchinsonii by electroporation as described previously (13) and grown on PY6 agar containing erythromycin at 30°C for 4 to 5 days. The transformants were grown in PY6 liquid containing erythromycin and streaked on selective medium to eliminate nonselected cells, followed by PCR verification with two sets of primers, 3237UF/3237DR and 3237UF/3237UR. The cells with an erythromycin resistance cassette replacing CHU_3237 were selected as parent cells for transforming the helper plasmid pCHF by electroporation. After incubation on PY6 agar containing chloramphenicol at 30°C for 15 to 20 days, colonies were inoculated into PY6 liquid without any antibiotics to allow the loss of pCHF. The cells were screened by PCR with primers 3237UF and 3237DR, and the products were sequenced with the primer 3237Test to verify the scar sequences after excision of the ermF gene. To confirm the loss of the resistance gene and pCHF, the cells were streaked on PY6 agar containing no antibiotics or containing erythromycin or chloramphenicol.
FIG 1.
Schematic representation of the unmarked deletion of CHU_3237 and large genomic fragments by FLP-FRT recombination. (a) The unmarked deletion of CHU_3237. Two homologous arms of CHU_3237 were amplified from the genome of C. hutchinsonii with two sets of primers 3237H1F/3237H1R and 3237H2F/3237H2R, digested, and ligated into the template plasmid pTKS. The gene-targeting cassette was amplified with primers 3237H1F and 3237H2R. The PCR product was transformed into C. hutchinsonii by electroporation, and transformants were selected by erythromycin resistance. The 3237::erm disruption mutant was subsequently selected for the helper plasmid pCHF transformation. The FLP recombinase excised the fragment between two FRT sites containing ermF. (b and c) Deletions of the fragments between CHU_3190 and CHU_3202 and between CHU_0804 and CHU_0819. Black arrows show approximate locations and orientations of primers; black filled boxes indicate homologous arms; open arrowheads show arrangements and orientations of genes; open boxes indicate residual genes; gray arrowheads indicate FRT sites and their relative orientations. The gray arrowhead in the plasmid pCHF indicates the flp gene.
Complementation of the CHU_3237 deletion mutant.
pCH was constructed from pSKSO8TG following a procedure similar to that for pCHF, except that the ompAp-cat cassette was amplified with primers cat-PstIF and cat-KpnIR, and no FLP recombinase gene was inserted. When used for complementation, pCH was digested with SacI and SalI to excise the galK cassette. A 4.4-kbp fragment spanning CHU_3237, 344 bp upstream of the start codon and 118 bp downstream of the stop codon, was amplified with primers C3237F and C3237R. The fragment was digested with SacI and SalI and ligated into the linearized pCH plasmid to generate pCH3237. Plasmid pCH3237 was electroporated into the Δ3237 mutant and selected by chloramphenicol resistance. C3237 refers to complemented strain of the Δ3237 mutant with pCH3237.
Deletion of large genomic fragments.
The deletions of specific genes located in the terminals of large genomic fragments were carried out similarly as done with CHU_3237. The primers used were named accordingly and are listed in Table S1 in the supplemental material. Disruption mutants were streaked at least once on selective medium to eliminate nonselected cells before they were transformed with pCHF.
Measurement of growth rates in glucose culture.
C. hutchinsonii strains were grown in Stanier medium supplemented with 2 g/liter glucose to late exponential phase and inoculated into 150 ml of Stanier medium supplemented with 2 g/liter glucose in 500-ml flasks with an inoculum concentration of 3% (vol/vol). The flasks were incubated with shaking (160 rpm) at 30°C. Growth was monitored by the optical density at 600 nm of the culture using a Unico UV-2000 spectrophotometer.
Cellulose degradation assay.
Cellulose degradation assays were carried out as described by Ji et al. (11, 12). Equivalent amounts of concentrated cells from PY6 medium were spotted on cellulose plates or Whatman number 1 filter paper overlaid on Stanier agar. The plates were incubated at 30°C to observe cellulose degradation.
Observation of colony spreading.
Colony spreading on soft agar was observed as described previously (11, 12). Briefly, cells were grown to mid-exponential phase in PY6 medium. Equivalent amounts of concentrated cells were spotted on the PY2 medium with 5 g/liter agar and 2 g/liter glucose, followed by incubation at 30°C.
Cellulase activity assay.
Cells were grown in Stanier medium supplemented with 2 g/liter glucose to mid-exponential phase. Cultures were centrifuged at 5,100 × g for 5 min to obtain cell pellets. For intact-cell samples, cell pellets were washed with Na2HPO4-KH2PO4 buffer (100 mM, pH 6.8) and resuspended in the same buffer. The protein concentration was quantified as described by Bradford (39). To measure carboxymethyl cellulase (CMCase) activity, a mixture of 500 μl of resuspended intact cells and 500 μl of 1% (wt/vol) sodium carboxymethyl cellulose (CMC-Na) in distilled water was incubated for 30 min at 30°C. The reducing sugars were measured using 3,5-dinitrosalicylic acid as previously described (12, 40). The measurements were carried out in triplicate.
SDS-PAGE of extracellular proteins and Western blot analysis.
C. hutchinsonii strains were grown in Stanier medium supplemented with 2 g/liter glucose at 30°C to mid-exponential phase. The cultures were centrifuged at 5,100 × g for 10 min, and the supernatants were filtered through a 0.22-μm-pore-size polyvinylidene difluoride (PDVF) filter (Sangon, Shanghai, China). The cell-free supernatants were concentrated using Amicon 10-kDa Ultra-15 centrifugal filter units (Millipore, MA, USA). Equal amounts of concentrated extracellular proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie brilliant blue R-250, and bands were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) in PTM Biolabs (Hangzhou, China) (as described in the supplemental methods).
For Western blot analysis, proteins in an SDS-PAGE gel were transferred onto 0.45-μm-pore-size PVDF membranes (Immobilon-P; Millipore, MA, USA) using a semidry electrophoretic transfer cell (Bio-Rad, CA, USA), according to the manufacturer's instructions. Membranes were blocked with skim milk and probed with anti-r0344 rabbit antiserum. Anti-r0344 was raised to the 301-amino-acid region of CHU_0344 (from Gly476 to Tyr776). After incubation with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (Cowin Biotech, Beijing, China) as a secondary antibody, proteins were detected by chemiluminescent HRP substrate (Immobilon Western, Millipore, MA, USA) according to the manufacturer's instructions, and the film was processed by an automatic X-ray film processor (SMPIC 2600C-1; Shanghai, China).
RESULTS
A new unmarked deletion method based on a linear DNA transformation and FLP-FRT recombination system for C. hutchinsonii.
In order to target genes independent of plasmid transformation, we tried to introduce linear DNA into C. hutchinsonii as done in other species (19, 24, 41). We constructed template plasmids with resistance genes flanked by multiple cloning sites for inserting homologous arms. The template plasmids were named pSJHS and pSJHC with ermF and cat as the resistance genes, respectively. Homologous arms were cloned from C. hutchinsonii and ligated into the restriction sites of the template plasmid. The double-crossover cassette was amplified by PCR, purified, and transformed into C. hutchinsonii by electroporation. After trying different fragment sizes and linear DNA concentrations, we found that ∼1 μg of linear PCR product with at least 1.5 kbp of homologous sequence for each arm transformed to approximately 109 electrocompetent cells was adequate for obtaining transformants. Using this approach, we have disrupted dozens of genes (data not shown).
To apply the FLP-FRT recombination system to unmarked mutagenesis, several plasmids were constructed to customize the system for C. hutchinsonii. Two FRT sites from pKD3 were introduced into pSHJS to generate unmarked template plasmids pTKS and pTSK. As no inducible promoter has been reported to be functional in C. hutchinsonii, the helper plasmid carrying flp should be cured under specific conditions for successive gene targeting. The helper plasmid was constructed from a replicative oriC plasmid, pSKSO8TG, with insertion of the functional counterselectable marker galK under the control of the CHU_1284 promoter (13) to generate pCHSD. The pCHSD plasmid was chloramphenicol resistant, and the flp gene was ligated directly downstream of a putative ribosome binding site (RBS) (42) behind the stop codon of cat, resulting in pCHF. The flp gene was cotranscribed with cat under the control of the F. johnsoniae ompA promoter (36) and was expressed with a putative RBS sequence 9 nucleotides (nt) upstream of the initiator AUG. At first, the helper plasmid pCHF was designed to be eliminated by growth in culture medium supplemented with 2-deoxy-galactose (DOG) (43). However, we found that the transformants grown on chloramphenicol-selective plates after pCHF electroporation retained weak resistance to chloramphenicol. Cells grown in fresh liquid medium without chloramphenicol were screened by PCR and streaked on plates containing chloramphenicol, and the results showed that pCHF could be easily cured from nonselective culture (data not shown).
CHU_3237, an ortholog of porU, was disrupted by linear DNA transformation (Fig. 1a). The erythromycin resistance gene was evicted with the transformation of the FLP recombinase helper plasmid pCHF. PCR tests using primers 3237UF/3237DR and 3237UF/3237UR showed that all transformants picked had the expected band sizes (see Fig. S1a in the supplemental material). The scar sequences were verified by sequencing with primer 3237Test, and all had the exact predicted sequences (data not shown). The cells of the Δ3237 mutant were streaked on PY6 agar without antibiotics and PY6 agar with erythromycin or chloramphenicol, and the results showed that the ermF gene was completely excised and that the helper plasmid was eliminated (see Fig. S1b in the supplemental material). We obtained several other unmarked deletions of genes using this method (data not shown), indicating that it is an efficient method for unmarked deletion in C. hutchinsonii.
Implementation of the new method for deletion of large genomic fragments.
Once the FLP-FRT recombination system was proved to be efficient for unmarked deletion in C. hutchinsonii, we decided to apply this method to the deletions of large genomic fragments. Two cefoxitin-resistant template plasmids, pCFXKS and pCFXSK, were constructed according to method described in the Materials and Methods section. We used pTKS, pTSK, pCFXKS, and pCFXSK as templates to carry out the deletions of large genomic fragments. The deletion of the fragment region from CHU_3190 to CHU_3202 (yielding the Δ3202-3190 mutant) was begun with disruption of CHU_3202 using pTKS as the template, followed by transformation of pCHF to obtain the unmarked CHU_3202 mutant (Δ3202 mutant) (Fig. 1b). Then, CHU_3190 was disrupted using pTSK as the template in the Δ3202 mutant, after which Emr colonies were isolated for another round of pCHF transformation. The resultant colonies were tested by PCR, and the new junction fragments were sequenced following PCR amplification. The verified colonies were streaked on PY6 agar with or without erythromycin to test their antibiotic sensitivity (see Fig. S2 in the supplemental material). All of the Δ3202-3190 mutants obtained had the expected junction fragments and were sensitive to erythromycin, indicating that the large fragment between CHU_3190 and CHU_3202 had been excised.
Other deletions of large genomic fragments were carried out as shown in Fig. 1c and in Fig. S3 in the supplemental material. For example, to delete the region from CHU_0804 to CHU_0819 (yielding the Δ0804-0819 mutant) we used Emr and Cfr templates, and the deletion was achieved by only three transformation steps (Fig. 1c). Six of the resultant colonies were randomly picked, tested, and verified similarly as described above. The Δ0804-0819 mutants obtained had the expected junction fragments and were sensitive to both erythromycin and cefoxitin (see Fig. S4), indicating that the large fragment between CHU_0804 and CHU_0819 and the resistance genes were eliminated successfully. Similarly, deletions of regions from CHU_0428 to CHU_0449 and from CHU_0834 to CHU_0841 were obtained and verified by PCR and sequencing (see Fig. S3).
However, excision between CHU_1075 and CHU_1107 had been tried several times and resulted in no colonies on the selection plates, while the electroporation with the control plasmid pCHSD obtained hundreds of colonies following the same procedure (see Fig. S5 in the supplemental material), suggesting that the deletion of this region might be lethal. Searching for potential essential genes covered in this part of genome in the Database of Essential Genes (DEG 10.02 [http://www.essentialgene.org/]) (51, 52) showed that CHU_1088 (murI, glutamate racemase) is an essential gene in 15 strains (out of 31 strains in all) and that CHU_1085 (fabG, 3-oxoacyl-ACP reductase) is present in 20 strains in rich medium. This might explain why no colony was obtained on the Cmr selection plate with electroporation of pCHF. Thus, the method developed for large-genomic-fragment deletion may be a useful tool for identifying conditionally essential genes of C. hutchinsonii.
During the process of testing deletion mutants of large genomic fragments, all colonies picked had the expected structures, and no colony was ever found such that the resistance genes had been evicted from the genome while the large fragment between the two distant FRT sites still remained in the genome. The results indicated that three or four FRT sites in the same orientation had recombined into one and that the helper plasmid could be cured by nonselective culture.
Preliminary phenotypic study showed that the Δ3202–3190, Δ0804–0819, Δ0834–0841, and Δ0428–0449 mutants with the large-fragment deletions had no obvious defects in cell growth and filter paper degradation, suggesting that these genes were not essential for cellulose degradation. Further studies on other phenotypes are now in progress.
Phenotypic properties of the unmarked Δ3237 deletion mutant.
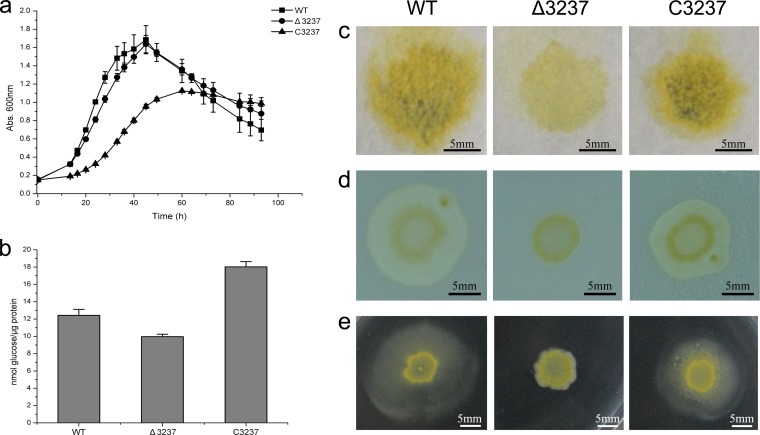
Deletion of CHU_3237 (porU) resulted in defects in many properties. The Δ3237 mutant had a little lower growth rate than the wild-type strain in both Stanier and PY6 media (Fig. 2a; see also Fig. S6 in the supplemental material) and was partially defective for cellulose utilization (Fig. 2c and d). The deletion mutant failed to digest filter paper completely even after incubation for 20 days. On cellulose plates, the mutant formed only a very small semitransparent circle around the inoculant while the wild type grew and spread quickly to form a transparent hydrolysis circle. Intact cells of the mutant had lower levels of specific CMCase activity than cells of the wild type (Fig. 2b).
FIG 2.
Phenotypic characteristics of the wild-type strain, the Δ3237 mutant, and the C3237 complemented strain. (a) Growth of the wild-type strain, the Δ3237 mutant, and the C3237 complemented strain. Cells were grown in Stanier medium with 2 g/liter glucose at 30°C (15 μg/ml chloramphenicol added for the C3237 strain), and growth was monitored by the absorbance at 600 nm. Abs, absorbance. (b) Specific endoglucanase activity of intact cells of the wild-type strain, the Δ3237 mutant, and the C3237 complemented strain. Endoglucanase activity was determined using CMC-Na as the substrate, and the reducing sugars were measured using the 3,5-dinitrosalicylic acid procedure. Error bars indicate standard errors. (c) Filter paper degradation of the cells on Stanier agar at 30°C for 12 days. (d) Cellulose utilization of the cells on a cellulose plate at 30°C for 10 days. (e) Colony spreading of the cells on soft agar (PY2 medium with 2 g/liter glucose and 5 g/liter agar) at 30°C for 10 days. WT, wild-type strain; Δ3237, Δ3237 unmarked deletion mutant; C3237, Δ3237 mutant complemented with pCH3237.
For colony spreading on soft agar, cells of the Δ3237 mutant dispersed only a little from the edge of the inoculant while the wild-type strain formed a large, spreading circle around the inoculant (Fig. 2e). This is in accordance with the small circle formed on the cellulose soft layer. The arrangement of cells on filter paper fibers showed no obvious differences between the mutant and the wild-type strain (see Fig. S7 in the supplemental material). This suggested that the deletion of CHU_3237 had no impact on the motility of cells on cellulose fiber.
Complementation of the Δ3237 mutant with pCH3237 resulted in recoveries of both cellulose utilization and colony spreading on soft agar (Fig. 2c to e). However, the growth rate of the complemented strain was lower than that of both the wild-type strain and the Δ3237 mutant (Fig. 2a), possibly because of the replicative oriC plasmid used and the supplementation of chloramphenicol to the culture.
The T9SS is a novel protein secretion system reported to secrete a number of cell surface and extracellular proteins linked to the gliding motility and pathogenesis of Bacteroidetes (27, 29). The CTDs of the protein substrates are crucial for their secretion and localization. CHU_3237 is predicted to be an ortholog of a CTD cleavage peptidase of the T9SS (27, 30). The extracellular proteins were extracted from the mid-exponential-phase cultures of both the wild type and the mutant and separated by SDS-PAGE (Fig. 3a). The result showed that the band patterns of the extracellular proteins of the mutant were obviously different from the band pattern of the wild type. CHU_0344 is one of the dominant extracellular proteins, and two bands of CHU_0344 with different molecular masses were identified from PYG (peptone-yeast-glucose) culture by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (12, 13). CHU_0344 (Fig. 3a, arrow) which was predicted to possess a CTD sequence, was absent from the Δ3237 mutant (the 0344::cat mutant was used as a negative control). This finding was confirmed by Western blot analysis (Fig. 3b). In addition, a blurry band (Fig. 3b, asterisk) was observed from the Δ3237 mutant at the position above 90 kDa, while the main band of the mature CHU_0344 was a little above the 75-kDa band marker. The blurry band with a molecular mass similar to that of the full-length CHU_0344 was probably the propeptide of the CHU_0344 with its CTD. Due to the loss of CHU_3237, the CTD of the CHU_0344 was unable to be cleaved, and most of the propeptide might be subsequently degraded.
FIG 3.
SDS-PAGE and Western blot of the extracellular proteins of the wild-type strain and the Δ3237 mutant. (a) SDS-PAGE of the extracellular proteins. (b) Western blot of the extracellular proteins with anti-r0344. WT, wild-type strain; Δ3237, Δ3237 unmarked deletion mutant; 0344::cat, disruption mutant of CHU_0344; M, molecular mass marker. Arrows indicate the bands of CHU_0344; the asterisk indicates the immature propeptide of CHU_0344.
LC-MS/MS analysis of extracellular proteins of the Δ3237 mutant and the wild-type strain showed that many proteins with putative CTDs were absent from the Δ3237 mutant (see Table S2 in the supplemental material). Proteins identified from the cell-free samples of both the Δ3237 mutant and the wild-type strain contained dozens of cytoplasmic and periplasmic proteins, suggesting that the autolysis of C. hutchinsonii cells happened along with their growth. The deletion of CHU_3237 resulted in defects in colony spreading, cellulose degradation, and protein secretion, which indicated the important role of CHU_3237 in C. hutchinsonii physiology.
DISCUSSION
C. hutchinsonii is a widely distributed cellulolytic bacterium, and the analysis of its genomic sequence suggests that it may use a novel mechanism for cellulose degradation (3, 6, 9). Recently, much progress in its cellulose degradation and cell motility has been made due to improved tools for genetic manipulation (11–15). All of these methods are based on plasmid transformation. Here, a method based on a linear DNA double-crossover recombination and site-specific FLP-FRT recombination system was developed for C. hutchinsonii to generate unmarked gene deletions. In our study, linear DNA fragments containing a resistance gene flanked by two homologous arms were feasible for gene replacement by electroporation into C. hutchinsonii. Combined with the FLP-FRT recombination system, unmarked deletions were obtained without counterselectable markers. Compared with the rpsL-mediated mutagenesis described by Zhu and McBride, which requires the use of a streptomycin-resistant mutant (15), our method could obtain unmarked gene deletions in the wild-type strain. The eviction of resistance genes is mediated by the FLP-FRT recombination system which is site specific and effective, while the eviction step of the counterselection method depends on intrinsic homologous recombination and usually results in half the colonies being wild type and half the colonies being deletion mutants. Furthermore, our method could also be used in deletion of large fragments from the genome. Use of a customized FLP-FRT recombination system for gene targeting in the phylum of Bacteroidetes has not been reported yet. Our study surely will provide one more choice for researchers to obtain unmarked deletions of other species in the phylum of Bacteroidetes, whose study has been hampered by the lack of genetic tools.
Unmarked deletions of several genes of C. hutchinsonii were obtained using our method. One of them is CHU_3237, which is predicted to be an ortholog of the C-terminal signal peptidase of the P. gingivalis T9SS (27, 30). The Δ3237 mutant showed partial defects in cellulose degradation and colony spreading. The results of SDS-PAGE and LC-MS/MS analysis of extracellular proteins showed that dozens of extracellular proteins were absent from the culture fluid of the Δ3237 mutant, and many of them were putative substrates of the C. hutchinsonii T9SS. CHU_0344 is found to be one of the main extracellular proteins (13) and can bind to crystalline cellulose in vitro (12). It contains a C-terminal sorting domain of the T9SS. The result of SDS-PAGE showed that CHU_0344 was absent from the supernatant of the Δ3237 mutant, and Western blot analysis of the extracellular proteins probed with anti-r0344 confirmed this. However, a trace of CHU_0344 with a molecular mass approximate to that of CHU_0344 with its CTD intact was detected in the supernatant of the Δ3237 mutant, suggesting that the CTD of CHU_0344 was unable to be cleaved and that the immature propeptide might be digested by proteases. These results showed that CHU_3237 played an important role in protein secretion and appeared to function as a CTD peptidase in C. hutchinsonii.
CHU_3238, which shows 39% identity in protein sequence to PG27/LptO of Porphyromonas gingivalis W83, was located downstream of CHU_3237. PG27/LptO is an outer membrane protein, which is essential for the O-deacylation of lipopolysaccharide (LPS) and secretion of CTD-containing proteins (30, 44, 45). Recently, the CTD peptidase PG0026 has been found to interact with PG27/LptO, and the PG27-PG0026 complex is stabilized by hemagglutinin A (HagA) (46). Complementation results showed that the defects of the Δ3237 mutant were not due to disturbance of the adjacent CHU_3238 gene. The function of CHU_3238 and its relationship to CHU_3237 need to be elucidated in further study. Previous work reported that the deletion of one of the T9SS genes, CHU_0170 (sprP), causes defects in gliding motility and cellulose utilization (15). Our work supported the idea that the T9SS existed in C. hutchinsonii and played important roles in cellulose utilization and protein secretion.
The fragment from CHU_3190 to CHU_3202 contains 13 genes, among which CHU_3195, CHU_3196, CHU_3198, and CHU_3199 are annotated as gspD, gspE, gspG, and gspF, respectively. These four genes belong to the type II secretion pathway, which is usually composed of 12 to 15 proteins, depending on species, and is related to the type IV pilus biogenesis system (47, 48). These are the only annotated genes related to those of the defined secretion systems across the outer membrane in C. hutchinsonii besides the T9SS. In addition to these four genes, other genes in the fragment are all described to encode hypothetical proteins. Among them, CHU_3197 showed similarity to the type IV leader peptidase of Solitalea canadensis DSM 3403 (34% identities over 146 amino acids [aa]), suggesting that it may also be involved in the type II general secretion pathway of C. hutchinsonii. Deletion of this putative operon showed no defects in cell growth and filter paper degradation, suggesting that these genes were not essential for cellulose degradation. Further study of the mutant would help us to explore the function of these genes in C. hutchinsonii.
More than 40 percent of C. hutchinsonii genes are unannotated, and many genes have their paralogs. Once one gene is targeted, C. hutchinsonii may activate an alternative genetic route to recover its phenotype. The identification of functions of these genes would consume a lot of time and effort. Using our method, genes flanked by their paralogs or gene clusters can be deleted together more efficiently. In this study, we obtained four mutants of large-fragment deletions by only three or four transformation steps, and the sizes of the excised fragments varied from 9 to 19 kb, spanning from 6 to 22 genes. Theoretically, this procedure could be cyclically repeated to gain larger fragment deletions. Although the size of the deletion fragment between the FRT sites would be minimized by the presence of essential genes, the FLP-FRT system could be applied to obtain fairly large deletions in C. hutchinsonii.
The results of large-fragment deletions indicated that three or four FRT sites in the same orientation had recombined into one, and the fragments between the furthest FRT sites were excised. Therefore, it appeared that the customized FLP-FRT deletion system might not be used to prepare consecutive unmarked deletions in C. hutchinsonii as done in Mycobacterium smegmatis (23). One possible explanation for the high efficiency of the large-fragment deletions was that the replicative plasmid pCHF functioned in the C. hutchinsonii cells much longer than inducible expression of flp. The efficiency of the FLP-mediated recombination between multiple FRT sites may change if the FLP recombinase is integrated into the chromosome under the control of an inducible promoter.
The efficiency of homologous recombination with linear DNA transformation was low in C. hutchinsonii as each homologous arm needed to be at least 1.5 kbp to obtain recombinant transformants. This resulted in the insertion of FRT sites into the genome as the rate-limiting step of our method. An alternative strategy is combining the FLP-FRT system with transposons to select for random deletion, as described in Pseudomonas putida (25). This would be helpful for genome streamlining but not for targeting specific fragments of the genome. The novel mechanism of cellulose degradation by C. hutchinsonii remains a mystery. The method developed for unmarked deletions of genomic fragments in C. hutchinsonii provides an efficient tool for quickly screening the unannotated genes, which may promote the study of its cellulose degradation mechanism and other physiological aspects.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Basic Research Program of China (2011CB707402) and the National Natural Science Foundation of China (31170051 and 31371262).
We sincerely thank Mark J. McBride (University of Wisconsin—Milwaukee, Milwaukee, WI, USA) for providing C. hutchinsonii ATCC 33406 and Qingsheng Qi (Shandong University, Jinan, China) for providing plasmids pKD3 and pCP20.
Footnotes
Published ahead of print 25 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01785-14.
REFERENCES
- 1.Walker E, Warren FL. 1938. Decomposition of cellulose by Cytophaga. I. Biochem. J. 32:31–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stanier RY. 1942. The Cytophaga group: a contribution to the biology of myxobacteria. Bacteriol. Rev. 6:143–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie G, Bruce DC, Challacombe JF, Chertkov O, Detter JC, Gilna P, Han CS, Lucas S, Misra M, Myers GL, Richardson P, Tapia R, Thayer N, Thompson LS, Brettin TS, Henrissat B, Wilson DB, McBride MJ. 2007. Genome sequence of the cellulolytic gliding bacterium Cytophaga hutchinsonii. Appl. Environ. Microbiol. 73:3536–3546. 10.1128/AEM.00225-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayer EA, Belaich J-P, Shoham Y, Lamed R. 2004. The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58:521–554. 10.1146/annurev.micro.57.030502.091022 [DOI] [PubMed] [Google Scholar]
- 5.Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506–577. 10.1128/MMBR.66.3.506-577.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson DB. 2008. Three microbial strategies for plant cell wall degradation. Ann. N. Y. Acad. Sci. 1125:289–297. 10.1196/annals.1419.026 [DOI] [PubMed] [Google Scholar]
- 7.Wilson DB. 2011. Microbial diversity of cellulose hydrolysis. Curr. Opin. Microbiol. 14:259–263. 10.1016/j.mib.2011.04.004 [DOI] [PubMed] [Google Scholar]
- 8.Chang WT, Thayer DW. 1977. The cellulase system of a Cytophaga species. Can. J. Microbiol. 23:1285–1292. 10.1139/m77-192 [DOI] [PubMed] [Google Scholar]
- 9.Wilson DB. 2009. Evidence for a novel mechanism of microbial cellulose degradation. Cellulose 16:723–727. 10.1007/s10570-009-9326-9 [DOI] [Google Scholar]
- 10.McBride MJ, Baker SA. 1996. Development of techniques to genetically manipulate members of the genera Cytophaga, Flavobacterium, Flexibacter, and Sporocytophaga. Appl. Environ. Microbiol. 62:3017–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji X, Bai X, Li Z, Wang S, Guan Z, Lu X. 2013. A novel locus essential for spreading of Cytophaga hutchinsonii colonies on agar. Appl. Microbiol. Biotechnol. 97:7317–7324. 10.1007/s00253-013-4820-2 [DOI] [PubMed] [Google Scholar]
- 12.Ji X, Xu Y, Zhang C, Chen N, Lu X. 2012. A new locus affects cell motility, cellulose binding, and degradation by Cytophaga hutchinsonii. Appl. Microbiol. Biotechnol. 96:161–170. 10.1007/s00253-012-4051-y [DOI] [PubMed] [Google Scholar]
- 13.Xu Y, Ji X, Chen N, Li P, Liu W, Lu X. 2012. Development of replicative oriC plasmids and their versatile use in genetic manipulation of Cytophaga hutchinsonii. Appl. Microbiol. Biotechnol. 93:697–705. 10.1007/s00253-011-3572-0 [DOI] [PubMed] [Google Scholar]
- 14.Zhu Y, Zhou H, Bi Y, Zhang W, Chen G, Liu W. 2013. Characterization of a family 5 glycoside hydrolase isolated from the outer membrane of cellulolytic Cytophaga hutchinsonii. Appl. Microbiol. Biotechnol. 97:3925–3937. 10.1007/s00253-012-4259-x [DOI] [PubMed] [Google Scholar]
- 15.Zhu Y, McBride MJ. 2014. Deletion of the Cytophaga hutchinsonii type IX secretion system gene sprP results in defects in gliding motility and cellulose utilization. Appl. Microbiol. Biotechnol. 98:763–775. 10.1007/s00253-013-5355-2 [DOI] [PubMed] [Google Scholar]
- 16.Barrett AR, Kang Y, Inamasu KS, Son MS, Vukovich JM, Hoang TT. 2008. Genetic tools for allelic replacement in Burkholderia species. Appl. Environ. Microbiol. 74:4498–4508. 10.1128/AEM.00531-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiang SL, Mekalanos JJ. 2000. Construction of a Vibrio cholerae vaccine candidate using transposon delivery and FLP recombinase-mediated excision. Infect. Immun. 68:6391–6397. 10.1128/IAI.68.11.6391-6397.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cox MM. 1983. The FLP protein of the yeast 2-μm plasmid: expression of a eukaryotic genetic recombination system in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 80:4223–4227. 10.1073/pnas.80.14.4223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. 10.1016/S0378-1119(98)00130-9 [DOI] [PubMed] [Google Scholar]
- 21.Ishikawa M, Hori K. 2013. A new simple method for introducing an unmarked mutation into a large gene of non-competent Gram-negative bacteria by FLP/FRT recombination. BMC Microbiol. 13:86. 10.1186/1471-2180-13-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schweizer HP. 2003. Applications of the Saccharomyces cerevisiae Flp-FRT system in bacterial genetics. J. Mol. Microbiol. Biotechnol. 5:67–77. 10.1159/000069976 [DOI] [PubMed] [Google Scholar]
- 23.Stephan J, Stemmer V, Niederweis M. 2004. Consecutive gene deletions in Mycobacterium smegmatis using the yeast FLP recombinase. Gene 343:181–190. 10.1016/j.gene.2004.08.028 [DOI] [PubMed] [Google Scholar]
- 24.Tracy E, Ye F, Baker BD, Munson RS. 2008. Construction of non-polar mutants in Haemophilus influenzae using FLP recombinase technology. BMC Mol. Biol. 9:101. 10.1186/1471-2199-9-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leprince A, de Lorenzo V, Völler P, van Passel MW, Martins dos Santos VA. 2012. Random and cyclical deletion of large DNA segments in the genome of Pseudomonas putida. Environ. Microbiol. 14:1444–1453. 10.1111/j.1462-2920.2012.02730.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji X, Wang Y, Zhang C, Bai X, Zhang W, Lu X. 16 May 2014. Novel outer membrane protein involved in cellulose and cellooligosaccharide degradation by Cytophaga hutchinsonii. Appl. Environ. Microbiol. 10.1128/AEM.00687-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sato K, Naito M, Yukitake H, Hirakawa H, Shoji M, McBride MJ, Rhodes RG, Nakayama K. 2010. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 107:276–281. 10.1073/pnas.0912010107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McBride MJ, Zhu Y. 2013. Gliding motility and Por secretion system genes are widespread among members of the phylum bacteroidetes. J. Bacteriol. 195:270–278. 10.1128/JB.01962-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato K, Yukitake H, Narita Y, Shoji M, Naito M, Nakayama K. 2013. Identification of Porphyromonas gingivalis proteins secreted by the Por secretion system. FEMS Microbiol. Lett. 338:68–76. 10.1111/1574-6968.12028 [DOI] [PubMed] [Google Scholar]
- 30.Glew MD, Veith PD, Peng B, Chen Y-Y, Gorasia DG, Yang Q, Slakeski N, Chen D, Moore C, Crawford S, Reynolds EC. 2012. PG0026 is the C-terminal signal peptidase of a novel secretion system of Porphyromonas gingivalis. J. Biol. Chem. 287:24605–24617. 10.1074/jbc.M112.369223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seers CA, Slakeski N, Veith PD, Nikolof T, Chen Y-Y, Dashper SG, Reynolds EC. 2006. The RgpB C-terminal domain has a role in attachment of RgpB to the outer membrane and belongs to a novel C-terminal-domain family found in Porphyromonas gingivalis. J. Bacteriol. 188:6376–6386. 10.1128/JB.00731-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shoji M, Sato K, Yukitake H, Kondo Y, Narita Y, Kadowaki T, Naito M, Nakayama K. 2011. Por secretion system-dependent secretion and glycosylation of Porphyromonas gingivalis hemin-binding protein 35. PLoS One 6:e21372. 10.1371/journal.pone.0021372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slakeski N, Seers CA, Ng K, Moore C, Cleal SM, Veith PD, Lo AW, Reynolds EC. 2011. C-terminal domain residues important for secretion and attachment of RgpB in Porphyromonas gingivalis. J. Bacteriol. 193:132–142. 10.1128/JB.00773-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veith PD, Nor Muhammad NA, Dashper SG, Likić VA, Gorasia DG, Chen D, Byrne SJ, Catmull DV, Reynolds EC. 2013. Protein substrates of a novel secretion system are numerous in the Bacteroidetes phylum and have in common a cleavable C-terminal secretion signal, extensive post-translational modification, and cell-surface attachment. J. Proteome Res. 12:4449–4461. 10.1021/pr400487b [DOI] [PubMed] [Google Scholar]
- 35.Agarwal S, Hunnicutt DW, McBride MJ. 1997. Cloning and characterization of the Flavobacterium johnsoniae (Cytophaga johnsonae) gliding motility gene, gldA. Proc. Natl. Acad. Sci. U. S. A. 94:12139–12144. 10.1073/pnas.94.22.12139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen S, Bagdasarian M, Kaufman MG, Bates AK, Walker ED. 2007. Mutational analysis of the ompA promoter from Flavobacterium johnsoniae. J. Bacteriol. 189:5108–5118. 10.1128/JB.00401-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ueki T, Inouye S, Inouye M. 1996. Positive-negative KG cassettes for construction of multi-gene deletions using a single drug marker. Gene 183:153–157. 10.1016/S0378-1119(96)00546-X [DOI] [PubMed] [Google Scholar]
- 38.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. 10.1016/0378-1119(95)00193-A [DOI] [PubMed] [Google Scholar]
- 39.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 40.Miller GL. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31:426–428. 10.1021/ac60147a030 [DOI] [Google Scholar]
- 41.Metzgar D, Bacher JM, Pezo V, Reader J, Döring V, Schimmel P, Marlière P, de Crécy-Lagard V. 2004. Acinetobacter sp. ADP1: an ideal model organism for genetic analysis and genome engineering. Nucleic Acids Res. 32:5780–5790. 10.1093/nar/gkh881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen S, Bagdasarian M, Kaufman MG, Walker ED. 2007. Characterization of strong promoters from an environmental Flavobacterium hibernum strain by using a green fluorescent protein-based reporter system. Appl. Environ. Microbiol. 73:1089–1100. 10.1128/AEM.01577-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33:e36. 10.1093/nar/gni035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Y-Y, Peng B, Yang Q, Glew MD, Veith PD, Cross KJ, Goldie KN, Chen D, O'Brien-Simpson N, Dashper SG, Reynolds EC. 2011. The outer membrane protein LptO is essential for the O-deacylation of LPS and the co-ordinated secretion and attachment of A-LPS and CTD proteins in Porphyromonas gingivalis. Mol. Microbiol. 79:1380–1401. 10.1111/j.1365-2958.2010.07530.x [DOI] [PubMed] [Google Scholar]
- 45.Ishiguro I, Saiki K, Konishi K. 2009. PG27 is a novel membrane protein essential for a Porphyromonas gingivalis protease secretion system. FEMS Microbiol. Lett. 292:261–267. 10.1111/j.1574-6968.2009.01489.x [DOI] [PubMed] [Google Scholar]
- 46.Saiki K, Konishi K. 2014. Porphyromonas gingivalis C-terminal signal peptidase PG0026 and HagA interact with outer membrane protein PG27/LptO. Mol. Oral Microbiol. 29:32–44. 10.1111/omi.12043 [DOI] [PubMed] [Google Scholar]
- 47.Peabody CR. 2003. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 149:3051–3072. 10.1099/mic.0.26364-0 [DOI] [PubMed] [Google Scholar]
- 48.Sandkvist M. 2001. Biology of type II secretion. Mol. Microbiol. 40:271–283. 10.1046/j.1365-2958.2001.02403.x [DOI] [PubMed] [Google Scholar]
- 49.Cooper AJ, Kalinowski AP, Shoemaker NB, Salyers AA. 1997. Construction and characterization of a Bacteroides thetaiotaomicron recA mutant: transfer of Bacteroides integrated conjugative elements is RecA independent. J. Bacteriol. 179:6221–6227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li LY, Shoemaker NB, Salyers AA. 1995. Location and characteristics of the transfer region of a Bacteroides conjugative transposon and regulation of transfer genes. J. Bacteriol. 177:4992–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang R, Ou H-Y, Zhang C-T. 2004. DEG: a database of essential genes. Nucleic Acids Res. 32:D271–D272. 10.1093/nar/gkh024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luo H, Lin Y, Gao F, Zhang C-T, Zhang R. 2014. DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucleic Acids Res. 42:D574–D580. 10.1093/nar/gkt1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.