Abstract
Nucleic acid-based analytical methods, ranging from species-targeted PCRs to metagenomics, have greatly expanded our understanding of microbiological diversity in natural samples. However, these methods provide only limited information on the activities and physiological states of microorganisms in samples. Even the most fundamental physiological state, viability, cannot be assessed cross-sectionally by standard DNA-targeted methods such as PCR. New PCR-based strategies, collectively called molecular viability analyses, have been developed that differentiate nucleic acids associated with viable cells from those associated with inactivated cells. In order to maximize the utility of these methods and to correctly interpret results, it is necessary to consider the physiological diversity of life and death in the microbial world. This article reviews molecular viability analysis in that context and discusses future opportunities for these strategies in genetic, metagenomic, and single-cell microbiology.
INTRODUCTION
Yet it hath happened that the veritable body without the spirit hath walked.
—Ambrose Bierce, The Death of Halpin Frayser
Microbiologists, like characters in zombie fiction, quickly learn the critical importance of distinguishing the living from the dead. In addition to characterizing the numbers and species of microorganisms in samples, it is important to collect data on their physiological states. The most fundamental physiological state of microbial cells is their viability, defined here as the capacity to form progeny. For ecologists, pathobiologists, metagenomicists, food or water safety analysts, infectious-disease clinicians, and virtually every other stripe of microbiologist, the observation of a viable microorganism in a sample means something entirely different from the observation of a dead one. Despite its importance, this distinction remains extremely challenging by current microbiological methods.
Microbiological culture meets this requirement, as it selectively detects viable organisms. However, because only a small percentage of species can be cultured, this strategy underestimates microbial diversity (1–6). In contrast, nucleic acid-based methods, ranging from species-specific PCR to metagenomic methods, have greatly advanced our ability to detect diverse microorganisms independently of microbiological culture (7, 8). However, these methods provide only limited information on the physiology of microorganisms in samples. They can assess microbial viability retrospectively, by measuring quantitative changes over time, but they cannot discern viability cross-sectionally (in single measurements). Traditional PCR is notoriously poor at differentiating DNA associated with a viable bacterial cell from DNA associated with an inactivated one or from a free DNA fragment. All of these analytes register as “hits” in PCR, despite their very distinct meanings.
In order to address this limitation, alternative PCR-based strategies have been developed. This article reviews two complementary strategies. One strategy, termed viability PCR, or vPCR, correlates viability with cell envelope impermeability (9, 10). In viability PCR, microbes in samples are incubated with a membrane-impermeative reagent such as propidium monoazide (PMA). Upon photoactivation, PMA binds tightly to exposed DNA and interferes with PCR amplification. Nonviable cells with damaged membranes, and free nucleic acids, are not protected from the reagent, and their amplification is inhibited after the reagent-DNA complex is photoactivated. In contrast, viable cells with intact cell membranes exclude PMA, enabling strong quantitative PCR (qPCR) signals in the presence of the reagent. The operating principle is similar to microscopy-based live/dead staining, in which a membrane-impermeative DNA stain (typically, propidium iodide [PI]) is excluded from intact cells but penetrates and stains the DNA of membrane-compromised cells. In live/dead staining, inactivated cells are quantified relative to total cell counts by fluorescence microscopy or flow cytometry (11) rather than by PCR.
The second strategy, termed “molecular viability testing” (MVT), correlates viability with the ability to rapidly synthesize a macromolecule (a species-specific rRNA precursor, or pre-rRNA) in response to a brief nutritional stimulus (12–14). Pre-rRNA synthesis upon nutritional stimulation is detected by reverse transcriptase-qPCR (RT-qPCR) measurement of species-specific pre-rRNA molecules. Pre-rRNAs in inactivated cells, and free nucleic acids, do not increase in numbers upon nutritional stimulation and therefore are excluded.
The two methods have complementary applications which are best understood in the context of the physiology of microbial viability and inactivation.
Diversity in death.
It is tempting to define microbial viability on the level of a single cell. We can state that a cell becomes nonviable when it loses the capacity to form progeny. A broader definition of viability might extend to cells that retain homeostasis and metabolic activity, even if they can no longer divide over the near term under specific conditions. Such cells are sometimes termed “viable but nonculturable” (15–18). Either way, a single-cell view can be limiting, because most microbiological activities of practical interest are exerted by populations of cells. Within a population of cells of a given species, there can be tremendous physiological diversity due to microenvironments and to stochastic variations (17, 19). Therefore, a population exhibiting microbial viability might be defined as one that includes a detectable number of proliferation-competent cells. The “detectable” threshold, of course, depends on the analytical method used.
The definition of death is also important, and far from straightforward, because microbial cells are inactivated by diverse pathways (11). Table 1 illustrates four (of numerous) possible scenarios for inactivation of bacterial cells. In the first scenario, cationic surfactants such as quaternary ammonium compounds (QACs) disrupt the cell membrane, resulting in immediate and catastrophic loss of nearly all metabolic and catabolic activities (20). In this “one-step” situation, all measures of bacterial viability correlate well with culturability. In the second, more complex scenario, disinfectants such as ethanol, phenol, and chlorine act broadly on membranes, internal proteins, and/or nucleic acids, in hierarchies that differ between agents, doses, microbial species, and conditions. Under nearly all conditions, however, these effects follow each other in rapid succession, such that a few short minutes of exposure can result in broad damage. As a result, there can be good correlation between different measures of viability in this scenario, as in the first scenario (20–24).
TABLE 1.
Examples of bacterial inactivation
| Scenario | Description | Examples of causes |
|---|---|---|
| 1 | Rapid loss of cytoplasmic membrane integrity, resulting in immediate and catastrophic loss of homeostasis, cellular functions, and culturability | Surfactants (e.g., quaternary ammonium compounds) |
| 2 | Rapid and nearly simultaneous oxidation or denaturation of multiple targets, including the cytoplasmic membrane, proteins, ribosomes, and/or DNA | Oxidative disinfectants (e.g., chlorine, peroxide), organic solvents (e.g., ethanol, phenol), heat |
| 3 | Rapid physical inactivation of a narrow range of targets, resulting in rapid loss of viability followed by slower decay (over hours or days) of cellular components, including the cytoplasmic membrane | UV, solar disinfection, low-temp pasteurization |
| 4 | Inactivation of a specific and essential target (e.g., DNA, RNA, and protein biosynthetic enzymes), resulting in rapid loss of viability followed by slower decay (over hours or days) of cellular components, including the cytoplasmic membrane | Antibiotics such as rifamycins, macrolides, aminoglycosides, and quinolones |
The third and fourth scenarios in Table 1 differ from the first two in that the immediate effects are relatively narrow. Viability is lost rapidly due to damage to one or more critical cellular components, but other cellular components are left intact. Examples include shortwave UV damage to DNA (25), inactivation of the respiratory chain by solar disinfection (26, 27), denaturation of critical catabolic or metabolic machinery by low-temperature pasteurization (28), and inhibition of DNA, RNA, or protein synthesis by antibiotics that bind tightly to specific enzymatic targets within one of these pathways. In these situations, bacterial cells can remain visibly intact and impermeable to PMA and PI for hours or days after viability is lost.
In some situations, “cadaver” cells can persist for very long times before environmental damage and innate senescence processes (17, 29) finally take their tolls on cell envelopes. In one study (26), Salmonella enterica serovar Typhimurium cells were exposed to a transient dose of artificial UV A light (1,500 kJ m−2, equivalent to a half day of solar disinfection), followed by a 48-h “chase” in darkness. Over the course of the chase period, multiple physiological parameters, including culturability, glucose uptake, ATP content, ATP synthesis (proton pumping), membrane polarization, and membrane permeability (PI staining), were measured. ATP depletion and loss of ATP synthetic capacity were immediate, followed rapidly by loss of glucose uptake capacity. Loss of membrane polarity and culturability was complete at 24 h in darkness. However, most cells were still structurally intact and impermeable to PI after 48 h postexposure (26). In a related study, continuous UV A exposure impacted Escherichia coli cells in a stepwise fashion, such that membrane permeabilization required nearly twice the dosage required for loss of viability (27). Under scenarios such as these, there can be marked divergence between different measures of viability.
While it is convenient to define viability as the ability to form progeny either in the laboratory or in nature, this view is complicated by the fact that most microbial species are not easily cultured experimentally. Therefore, molecular correlates of viability such as viability PCR, LIVE/DEAD staining, and MVT are useful and even necessary. However, such correlates must always be used and interpreted with an eye to the diverse ways that microorganisms can die. A physiological definition of death such as the irreversible loss of all brain functions in humans seems not in sight for microorganisms.
Viability PCR.
Of the two methods discussed in this review, viability PCR is by far the more extensively evaluated and vetted. In one of several reviews (9, 30–33), Elizaquível et al. (31) cataloged over 30 published studies that applied the method to food safety models alone. Viability PCR has also been extensively optimized. The most significant optimization was the replacement of a first-generation membrane-impermeative reagent, ethidium monoazide (EMA), with the next-generation PMA reagent (10). EMA was found to penetrate viable cells of many species, resulting in signal reduction in the presence of viable cells. PMA was found to be more membrane impermeative and more specific in its ability to differentiate intact from permeabilized cells, possibly due to its higher charge relative to EMA (10).
In outline, viability PCR involves splitting a sample into two aliquots and incubating (“treating”) one of the aliquots with PMA at concentrations that are usually optimized in preliminary experiments. A “control” aliquot is left untreated. After an appropriate incubation period, the treated aliquot is subjected to photoactivation, which catalyzes stable cross-linking between PMA and any DNA molecules to which it has access. Both aliquots are subsequently subjected to DNA purification and qPCR amplification. If the two aliquots exhibit similar qPCR signals, then target microorganisms in the sample are interpreted to be mostly viable (membranes intact). If the PMA-treated aliquot exhibits a measurably weaker signal than the control, then the target microorganisms are interpreted to be mostly inactivated. The difference in qPCR signal between the treated and control aliquot is often expressed as “ΔCT,” which refers to the difference in qPCR threshold cycles (CT). The extent of signal reduction or ΔCT correlates with the portion of the target DNA in the sample that is associated with inactivated cells (34).
The utility of viability PCR has been expanded by modifications to this basic strategy (31). Dithiothreitol cotreatment was reported to facilitate PMA penetration of inactivated Bacillus subtilis spores, thereby improving the ability to discern the viability of spores (35). Deoxycholate (DOC) cotreatment helped PMA penetrate E. coli cells that were inactivated but not disrupted by pasteurization at low temperature (between 52°C and 70°C) (36). However, this approach may be restricted to Gram-negative bacteria due to the effects of bile salts such as DOC on Gram-positive cell walls (37). Additional strategies to increase PMA treatment efficiency include incubation at elevated temperature (10°C above the optimal growth temperature) to maximize dye penetration into damaged cells (37) and the amplification of longer DNA sequences (38–40). The latter strategy is thought to increase the probability that at least one dye molecule would bind to the targeted DNA stretch in damaged cells (38–40).
In theory, viability PCR can function with any cellular or viral organism whose nucleic acid is enclosed by a lipid membrane or other PMA-impermeable structure. In addition to vegetative cells of commonly studied bacteria, it has been applied to fastidious bacteria (41, 42), bacterial spores (35, 43), protozoa (44, 45), fungi (46, 47), and some viruses (48–51). Moreover, the method can be applied to any genetic target that can be amplified by PCR or by other DNA amplification procedures (52). Therefore, in addition to analyzing species-defining DNA sequences, viability PCR can test the association of specific genetic functions (e.g., virulence, antibiotic resistance) with viable microbial cells. In theory, it can be integrated into virtually any nucleic acid amplification-based molecular microbiological operation, ranging from pathogen-targeted microbiological safety testing to metagenomic analyses.
In addition to the extensive published information about viability PCR, collective experience with PI as a viability stain (reviewed in reference 11) translates well to PMA-based methods. PMA is PI with an amino group on the phenanthridine ring replaced by an azide group that enables photoinduced cross-linking to DNA. Thus, the two compounds are likely to have similar membrane-permeability characteristics (10). Microbiologists who consider the use of these methods can draw from this wealth of published information.
Viability PCR has limitations for certain applications. Membrane integrity is a conservative correlate of microbial viability. The vast majority of cells with PMA-permeable membranes are likely to be nonviable; however, not all nonviable cells have compromised membranes (9, 34). Inactivation under conditions such as scenarios 3 and 4 in Table 1 can lead to an overestimation of the viable cell population. Reported examples include photoinactivation of Enterococcus faecalis (53), UV inactivation of E. coli O157:H7, Salmonella, and Campylobacter (34, 38), low-temperature pasteurization of Listeria innocua (54), and aminoglycoside treatment of Staphylococcus species (55). There can be challenges even under scenario 2 (Table 1). For example, in one study (34), hypochlorite exposures required for maximal permeability effects were significantly higher than those required for inactivation as measured by culture. Experience with PI and live/dead staining predicts additional challenges, for example, cryoinjury of Mycobacterium tuberculosis cells (56).
ΔCT values derive from the fraction of amplifiable DNA fragments that undergo at least one cross-linking event upon incubation with PMA and photoactivation. Often, at least a few fragments associated with inactivated cells escape PMA binding within the amplified sequences, enabling qPCR signal activity in PMA-treated samples. Conversely, at least a few fragments associated with viable cells undergo cross-linking due to “leakage” of PMA into intact cells (although this problem was far more pronounced with EMA [10]) and/or breakage of intact cells during the experimental procedure. As a result, ΔCT values are rarely zero even when bacteria in a sample are mostly viable. These factors can differ between target organisms, inactivation mechanisms, and sample types (34, 55). Thus, it is often necessary to conduct preliminary laboratory-based experiments to establish a threshold ΔCT value for specific organisms and treatments of interest. This process can be complicated by a third variable, namely, the composition of natural samples. Liang and Keeley (57) successfully applied viability PCR to cryptosporidium oocysts under defined medium conditions but had less success applying the method to natural-water samples, due to suspended solids that interfered with PMA uptake and/or cross-linking.
Despite these caveats, viability PCR meets important needs in molecular microbiology. With ongoing development and refinement, the method's applications continue to expand. The only insurmountable limitation is the innate inability to detect inactivation under conditions that do not impact membrane permeability. For such conditions, the alternative MVT approach should be considered.
Pre-rRNA analysis (MVT).
Because MVT is newer than viability PCR, published experience and information are much more limited. Thus, there remains greater uncertainty as to the breadth of its applicability. However, there are enough data to demonstrate its value as a complementary approach for certain applications.
MVT detects an innate biosynthetic activity of viable bacterial cells, namely, the synthesis of rRNA precursors (pre-rRNA) that occurs in bacteria immediately upon exposure to fresh nutrients (12–14). Pre-rRNAs are intermediates in rRNA synthesis whose leader and tail fragments are enzymatically removed to yield mature rRNA (Fig. 1). In growing or nutritionally stimulated bacteria, pre-rRNAs can account for ≥25% of total cellular rRNA (58). This translates to hundreds or thousands of copies per cell, making them orders of magnitude easier to detect by RT-qPCR than even the most strongly expressed mRNA. When growth slows, pre-rRNA synthesis stops but maturation continues, resulting in active and substantial drainage of pre-rRNA pools (58–63). Pre-rRNA is rapidly replenished when growth-limited cells are given fresh nutrients (12–14). As a result of these active functions of drainage and replenishment, growth-related fluctuations in pre-rRNA copy numbers far exceed those of DNA and mature rRNA (58, 59, 63, 64) and are readily resolved by RT-qPCR.
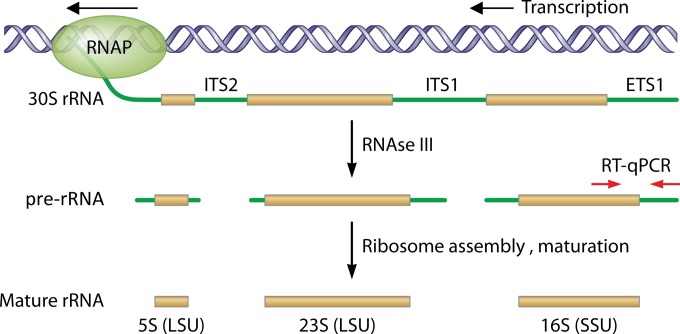
FIG 1.
rRNA synthesis and maturation in bacteria. RNA polymerase (RNAP), reading from right to left in this diagram, produces a long transcript (30S rRNA) containing 3 rRNA subunits interspersed with external transcribed spacers (ETS) and internal transcribed spacers (ITS). This transcript is rapidly converted by RNase activity and other endonucleolytic activities to pre-RNA subunits with leader and tail sequences. The leaders and tails are trimmed in exonucleolytic processes closely tied to ribosome assembly and the initiation of protein synthesis. RT-qPCRs can be designed to target the pre-rRNA exclusively, or to straddle a pre-rRNA-mature rRNA junction as shown here, such that intact pre-rRNA is needed for successful amplification. The 5′ leader region (ETS1) is especially useful for MVT because of its species specificity and relative abundance when transcription is active; however, other pre-rRNA sequences (e.g., ITS1) can also be targeted by RT-qPCR primers.
Because rRNA biosynthetic and regulatory pathways are evolutionarily conserved in bacteria, biosynthesis of pre-rRNA is consistently seen in nutritionally stimulated cells of all or nearly all bacterial species. Table 2 lists phylogenetically and physiologically diverse bacteria in which this response has been observed in our research (unpublished data) (12–14). Despite this functional conservation, the nucleotide sequences of pre-rRNA leaders and tails are hypervariable and highly species specific. Therefore, RT-qPCR tests can be designed to detect pre-rRNA synthesis in virtually any bacterial species and to differentiate this activity from that seen with other organisms' nucleic acids in complex samples (12, 58, 63, 66).
TABLE 2.
Diversity of bacteria shown to exhibit MVT response
| Species | Bacterial phyluma | Physiological characteristic(s) |
|---|---|---|
| Acinetobacter baumannii | Proteobacteria | Gram-negative aerobe |
| Aeromonas hydrophila | Proteobacteria | Gram-negative facultative anaerobe |
| Burkholderia cepacia | Proteobacteria | Gram-negative anaerobe |
| Chlamydia pneumoniae | Chlamydiae | Obligate intracellular organismb |
| Escherichia coli | Proteobacteria | Gram-negative facultative anaerobe |
| Filifactor alocis | Firmicutes | Fastidious Gram-positive anaerobe |
| Haemophilus influenzae | Proteobacteria | Gram-negative facultative anaerobe |
| Listeria monocytogenes | Firmicutes | Gram-positive facultative anaerobe |
| Mycobacterium avium | Actinobacteria | Slow-growing mycobacterium |
| Mycobacterium tuberculosis | Actinobacteria | Slow-growing mycobacterium |
| Porphyromonas gingivalis | Bacteriodetes | Gram-negative anaerobe |
| Pseudomonas aeruginosa | Proteobacteria | Gram-negative facultative anaerobe |
| Salmonella enterica | Proteobacteria | Gram-negative facultative anaerobe |
| Serratia marcescens | Proteobacteria | Gram-negative facultative anaerobe |
| Staphylococcus aureus | Firmicutes | Gram-positive facultative anaerobe |
| Stenotrophomonas maltophilia | Proteobacteria | Gram-negative aerobe |
In MVT, a sample is split into two aliquots, one of which is nutritionally stimulated by addition of bacteriological culture medium. If viable cells of a targeted species are present in the sample, then pre-rRNA (measured by RT-qPCR; Fig. 1) levels are seen to increase in the stimulated aliquot relative to the control (nonstimulated) aliquot. Because nonviable cells cannot catalyze this increase, the method selectively detects viable bacteria. Pre-rRNA stimulation is very rapid. One to 2 h of nutrient exposure is sufficient for consistent pre-rRNA upshift in most organisms. Slow-growing mycobacteria such as M. tuberculosis (G = ∼24 h) need 4 to 6 h of stimulation (12–14). In most cases, these time periods are 1 to 2 generation times or less. Thus, although MVT assesses an early step in bacterial cell division, it is not bacteriological culture. Steps in bacterial proliferation that occur after initial rRNA synthesis (including but not limited to DNA replication and cell growth, septation, and division, repeated through many cycles) are not required for MVT positivity. Therefore, it may be possible to use MVT to assess the viability of at least some “unculturable” species.
In addition to membrane integrity, a cell needs to be able to sense and respond to its environment and to catalyze the energy-expensive process of rRNA synthesis in order to be positive by MVT. Therefore, MVT does not depend entirely on membrane integrity and is capable of assessing microbial inactivation under some, if not all, of the scenario 3 and 4 conditions listed in Table 1. The method successfully detected inactivation of several Gram-negative and Gram-positive bacterial species by low-temperature pasteurization (13). It also detected inactivation of Staphylococcus cells by tobramycin, an aminoglycoside antibiotic that specifically targets protein synthesis (unpublished results). Even hypochlorite treatment, a scenario 2 condition in Table 1, can present challenges to viability PCR under some conditions. Whereas the hypochlorite exposures required for maximal permeability effects were higher than those required for inactivation as measured by culture (34); this was not seen when MVT was used in a separate study to assess hypochlorite inactivation of Aeromonas hydrophila and Mycobacterium avium cells (12). A caveat is that no studies to our knowledge have applied the two methods side by side to identical bacteria under identical conditions.
MVT is very sensitive. The method has been shown to increase, by factors of 5-fold to 10-fold relative to standard (static) DNA detection by qPCR, the analytical sensitivity of detection of diverse bacterial species spiked into water, serum, and dairy milk (13). Sensitivity comes in part from the elevated copy number of pre-rRNAs in stimulated bacteria. Upon lysis, a single cell can release hundreds or thousands of copies for detection. Sensitivity is also bolstered by the dynamic nature of the method. In contrast to static qPCR, MVT measures a bacterial “movement” in the form of a physiological change in response to a stimulus. In this sense, it is analogous to grouse hunting, in which the quarry is “flushed” to render it more visible. This quality could help to resolve borderline samples in diagnostic and other microbiological tasks (13).
An additional strength of MVT is the possibility of using a uniform (nonstimulated minus stimulated) threshold ΔCT value for diverse species and sample types. For example, a ΔCT threshold of 1 (corresponding to approximately 2- to 3-fold more pre-rRNA of the targeted species in the stimulated aliquot) was used to confirm serum and aminoglycoside inactivation of Pseudomonas aeruginosa and Acinetobacter baumannii cells and to confirm successful pasteurization (63°C for 45 min) of milk spiked with Listeria monocytogenes, M. avium, and S. enterica (13, 14). This may mitigate the need for preliminary experiments to define threshold ΔCT values prior to application in the field.
Many environments are rich in nutrients that could potentially confound MVT by elevating steady-state pre-rRNA pools and thereby muting pre-rRNA upshift upon nutritional stimulation. However, most natural environments are limited in at least some nutrients. Provision of limiting nutrients in vitro appears to consistently stimulate pre-rRNA synthesis in bacteria derived from diverse environments, as seen with lake and tap water (12), human serum (13, 14), and dairy milk (13). Given the physiological heterogeneity of bacteria in nature, there is a risk that large numbers of inactivated target cells could mute pre-rRNA upshift in viable target cells. However, viable cells of A. hydrophila (12) and Staphylococcus aureus (14) were detectable even when outnumbered ≥1,000-fold by chlorine- or serum-inactivated cells.
Although characteristics such as these are promising, MVT also has important limitations, the most significant of which is the relatively small amount of experience with and published information on the method. Inevitably, as use of MVT expands, its “warts” will emerge and conditions will be identified that confound it. Examples might include conditions that require varying ΔCT thresholds or natural conditions that permit full-throttle growth of a bacterial species, such that pre-rRNA synthesis cannot be further stimulated. Although such conditions have yet to be identified, it is unlikely that no such conditions exist and we cannot yet predict where they might arise.
Even if such problems are never observed, MVT has conceptual limitations. It cannot be applied to viruses, which have no ribosomes, and its applicability to eukaryotic cells has yet to be demonstrated. In contrast to bacteria, at least some eukaryotic protists have been reported to retain stable pools of pre-rRNA irrespective of growth physiology (66, 67). Such species may not respond to nutritional stimulation as needed for MVT. An additional and significant limitation is that MVT is confined to a single type of genetic target, namely, pre-rRNA. Although pre-rRNA is very useful for detecting individual species, it does not confer information on specific genetic traits of interest such as virulence or drug resistance. In contrast, viability PCR can be applied to virtually any genetic target. Another limitation is the requirement for RNA amplification by use of RT-qPCR, which adds complexity and vulnerability to sample effects. Finally, we do not yet know whether dormant and viable-but-nonculturable cells make pre-rRNA in response to nutritional stimulation. These responses may depend on conditions and cell type.
Viability analysis in metagenomic and single-cell microbiology.
Just as viability is an important consideration when assessing individual microbial species in samples, it is also important when studying microbial communities. When an environment changes or is subjected to stress, some species might be inactivated in large numbers while the activity of others might be unaffected or even bolstered. These distinctions can be difficult to discern in real time by standard DNA-based methods. In some situations, they can be detected retrospectively, but only after sufficient time has elapsed for the degradation and removal of DNA associated with inactivated cells. This makes it difficult to distinguish immediate environmental impacts on organisms, such as stimulation of the growth of specific species, from alternative effects that merely impact the persistence of residual DNA. Viability PCR and MVT have the potential to address these challenges.
Nocker et al. (68) combined viability PCR with 454 pyrosequencing of amplified small-subunit (SSU) rRNA genes. Evidence was presented that the rRNA gene profiles of PMA-resistant cells changed dramatically when environmental water samples were heated to 50°C or 60°C. Similar results were seen when denaturing gradient gel electrophoresis was used as an analytical method in place of 454 pyrosequencing (69). Lee and Levin (70) combined EMA viability PCR with amplification of rRNA gene sequences using primers complementary to conserved prokaryotic small-subunit rRNA gene sequences. They applied this approach not to raw samples but to suspensions of mixed bacterial cells cultured from fish fillets. However, the experiments demonstrated some aspects of a strategy that could be applied to microbial populations. They observed good correlations between EMA viability PCR and culture results when the cell suspensions were inactivated under some but not all conditions (70). The discrepancies could be related to the use of EMA rather than PMA or, alternatively, to limitations of the “gold standard” culture method, which might not have provided an accurate picture of the true viable cell population due to the presence of species that did not grow under the culture conditions used. In view of this caveat, the feasibility of viable microbiome analysis might best be assessed independently of culture results.
Although MVT has not yet been evaluated as a tool for assessing viability on population or microbiomic scales, such strategies may be possible. Pre-rRNAs have regions such as the 5′ external transcribed spacer upstream of the SSU rRNA (ETS1) and the internal transcribed spacer region (ITS1) between the SSU and large-subunit (LSU) rRNAs, which are bordered by highly conserved sequences within the mature rRNA (71–73). It is conceivable that RT-qPCR protocols can be designed to generate cDNA primed from “universal” prokaryotic sequences within the mature rRNA, reading into species-specific pre-rRNA spacers. These cDNAs might then be characterized and measured quantitatively in stimulated versus nonstimulated samples.
In addition to population and microbiomic studies, microbiologists increasingly seek to characterize their subjects on the single-cell level (74, 75). Viability staining using PI, combined with fluorescence microscopy or flow cytometry, was designed precisely for such applications (11). This is less true of viability PCR and MVT, and we are not aware of studies that have applied these approaches to isolated single microbial cells. However, given the physiological heterogeneity of microbial cells in nature, such applications can and should be designed. In an intriguing study, Oerther et al. (58) conducted fluorescence in situ hybridization (FISH) experiments with oligonucleotide probes targeting both total 16S rRNA and precursor 16S rRNA of Acinetobacter calcoaceticus. By measuring ratios of the latter to the former, they showed that individual cells of this species expressed highly variable levels of precursor 16S rRNA when adapting to various conditions in culture media and filtered sewage. Such approaches could in theory be used to detect pre-rRNA production after nutritional stimulation, thereby enabling the assessment of viability of individual cells.
To conclude, molecular viability analyses can distinguish the living from the dead in natural samples, without relying on microbiological culture with its many limitations. Although each strategy has limitations, MVT and viability PCR have complementary advantages that make them useful additions to the molecular microbiological toolbox. They fill critical needs, and they can help to advance our understanding of life and death in the microbial world.
ACKNOWLEDGMENTS
We are grateful to Kris Weigel of the University of Washington and Julie Do of AttoDx, Inc., for helpful input. We also thank Andreas Nocker of Cranfield University for his critical review of the manuscript prior to submission.
We are both inventors listed on patent applications related to molecular viability testing, one of the technologies discussed here. The technology has been licensed by the University of Washington to a commercial entity, AttoDx, Inc.
Biographies

Gerard A. Cangelosi earned his Ph.D. in Microbiology at the University of California, Davis, in 1984. He is currently a Professor of Environmental and Occupational Health Sciences and an Adjunct Professor of Global Health and Epidemiology, School of Public Health, University of Washington. His research focuses on molecular diagnostics, epidemiology, and the biology of infectious diseases. His work in the public and private sectors has generated 8 patents and over 70 publications in relevant areas, including tuberculosis, environmental mycobacteria, waterborne disease, and health care-associated infections.

John S. Meschke earned his Ph.D. in Environmental Sciences and Engineering at the University of North Carolina in 2001. He is currently an Associate Professor of Environmental and Occupational Health Sciences, School of Public Health, University of Washington. He is also the Director of the Environmental Health graduate program and maintains an active research program in environmental virology and microbiology. His research focuses on the fate, transport, detection, and control of pathogenic viruses and bacteria in environmental media (air, water, food, and surfaces).
Footnotes
Published ahead of print 18 July 2014
REFERENCES
- 1.Kesmen Z, Kacmaz N. 2011. Determination of lactic microflora of kefir grains and kefir beverage by using culture-dependent and culture-independent methods. J. Food Sci. 76:M276–M283. 10.1111/j.1750-3841.2011.02191.x [DOI] [PubMed] [Google Scholar]
- 2.Montalvo NF, Davis J, Vicente J, Pittiglio R, Ravel J, Hill RT. 2014. Integration of culture-based and molecular analysis of a complex sponge-associated bacterial community. PLoS One 9:e90517. 10.1371/journal.pone.0090517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pham VHT, Kim J. 2012. Cultivation of unculturable soil bacteria. Trends Biotechnol. 30:475–484. 10.1016/j.tibtech.2012.05.007 [DOI] [PubMed] [Google Scholar]
- 4.Rappé MS, Giovannoni SJ. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369–394. 10.1146/annurev.micro.57.030502.090759 [DOI] [PubMed] [Google Scholar]
- 5.Siqueira JF, Jr, Rôças IN. 2013. As-yet-uncultivated oral bacteria: breadth and association with oral and extra-oral diseases. J. Oral Microbiol. 2013:5. 10.3402/jom.v5i0.21077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomic-Canic M, Perez-Perez GI, Blumenberg M. 2014. Cutaneous microbiome studies in the times of affordable sequencing. J. Dermatol. Sci. 75:82–87. 10.1016/j.jdermsci.2014.05.001 [DOI] [PubMed] [Google Scholar]
- 7.Harwood C, Buckley M. 2008. The uncharted microbial world: microbes and their activities in the environment, 2008. American Academy of Microbiology, Washington, DC [Google Scholar]
- 8.Medini D, Serruto D, Parkhill J, Relman DA, Donati C, Moxon R, Falkow S, Rappuoli R. 2008. Microbiology in the post-genomic era. Nat. Rev. Microbiol. 6:419–430. 10.1038/nrmicro1901 [DOI] [PubMed] [Google Scholar]
- 9.Nocker A, Camper AK. 2009. Novel approaches toward preferential detection of viable cells using nucleic acid amplification techniques. FEMS Microbiol. Lett. 291:137–142. 10.1111/j.1574-6968.2008.01429.x [DOI] [PubMed] [Google Scholar]
- 10.Nocker A, Cheung CY, Camper AK. 2006. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods 67:310–320. 10.1016/j.mimet.2006.04.015 [DOI] [PubMed] [Google Scholar]
- 11.Davey HM. 2011. Life, death, and in-between: meanings and methods in microbiology. Appl. Environ. Microbiol. 77:5571–5576. 10.1128/AEM.00744-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cangelosi GA, Weigel KM, Lefthand-Begay C, Meschke JS. 2010. Molecular detection of viable bacterial pathogens in water by ratiometric pre-rRNA analysis. Appl. Environ. Microbiol. 76:960–962. 10.1128/AEM.01810-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Do JS, Weigel KM, Meschke JS, Cangelosi GA. 17 January 2014. Biosynthetic enhancement of the detection of bacteria by the polymerase chain reaction. PLoS One. 10.1371/journal.pone.0086433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weigel KM, Jones KL, Do JS, Melton Witt J, Chung JH, Valcke C, Cangelosi GA. 2013. Molecular viability testing of bacterial pathogens from a complex human sample matrix. PLoS One 8:e54886. 10.1371/journal.pone.0054886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allegra S, Berger F, Berthelot P, Grattard F, Pozzetto B, Riffard S. 2008. Use of flow cytometry to monitor Legionella viability. Appl. Environ. Microbiol. 74:7813–7816. 10.1128/AEM.01364-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Byrd JJ, Xu HS, Colwell RR. 1991. Viable but nonculturable bacteria in drinking water. Appl. Environ. Microbiol. 57:875–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nyström T. 2001. Not quite dead enough: on bacterial life, culturability, senescence, and death. Arch. Microbiol. 176:159–164. 10.1007/s002030100314 [DOI] [PubMed] [Google Scholar]
- 18.Rollins DM, Colwell RR. 1986. Viable but nonculturable stage of Campylobacter jejuni and its role in survival in the natural aquatic environment. Appl. Environ. Microbiol. 52:531–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kell DB, Kaprelyants AS, Weichart DH, Harwood CR, Barer MR. 1998. Viability and activity in readily culturable bacteria: a review and discussion of the practical issues. Antonie Van Leeuwenhoek 73:169–187. 10.1023/A:1000664013047 [DOI] [PubMed] [Google Scholar]
- 20.McDonnell G, Russell AD. 1999. Antiseptics and disinfectants: activity, action, and resistance. Clin. Microbiol. Rev. 12:147–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lisle JT, Pyle BH, McFeters GA. 1999. The use of multiple indices of physiological activity to access viability in chlorine disinfected Escherichia coli O157:H7. Lett. Appl. Microbiol. 29:42–47. 10.1046/j.1365-2672.1999.00572.x [DOI] [PubMed] [Google Scholar]
- 22.McKenna SM, Davies KJ. 1988. The inhibition of bacterial growth by hypochlorous acid. Possible role in the bactericidal activity of phagocytes. Biochem. J. 254:685–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nocker A, Caspers M, Esveld-Amanatidou A, van der Vossen J, Schuren F, Montijn R, Kort R. 2011. Multiparameter viability assay for stress profiling applied to the food pathogen Listeria monocytogenes F2365. Appl. Environ. Microbiol. 77:6433–6440. 10.1128/AEM.00142-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu FP, McFeters GA. 1994. Physiological responses of bacteria in biofilms to disinfection. Appl. Environ. Microbiol. 60:2462–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santos AL, Oliveira V, Baptista I, Henriques I, Gomes NC, Almeida A, Correia A, Cunha Â. 2013. Wavelength dependence of biological damage induced by UV radiation on bacteria. Arch. Microbiol. 195:63–74. 10.1007/s00203-012-0847-5 [DOI] [PubMed] [Google Scholar]
- 26.Bosshard F, Berney M, Scheifele M, Weilenmann HU, Egli T. 2009. Solar disinfection (SODIS) and subsequent dark storage of Salmonella typhimurium and Shigella flexneri monitored by flow cytometry. Microbiology 155:1310–1317. 10.1099/mic.0.024794-0 [DOI] [PubMed] [Google Scholar]
- 27.Bosshard F, Bucheli M, Meur Y, Egli T. 2010. The respiratory chain is the cell's Achilles' heel during UVA inactivation in Escherichia coli. Microbiology 156:2006–2015. 10.1099/mic.0.038471-0 [DOI] [PubMed] [Google Scholar]
- 28.Lee J, Kaletunç G. 2002. Evaluation of the heat inactivation of Escherichia coli and Lactobacillus plantarum by differential scanning calorimetry. Appl. Environ. Microbiol. 68:5379–5386. 10.1128/AEM.68.11.5379-5386.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nyström T. 2003. Conditional senescence in bacteria: death of the immortals. Mol. Microbiol. 48:17–23. 10.1046/j.1365-2958.2003.03385.x [DOI] [PubMed] [Google Scholar]
- 30.Cenciarini-Borde C, Courtois S, La Scola B. 2009. Nucleic acids as viability markers for bacteria detection using molecular tools. Future Microbiol. 4:45–64. 10.2217/17460913.4.1.45 [DOI] [PubMed] [Google Scholar]
- 31.Elizaquível P, Aznar R, Sánchez G. 2014. Recent developments in the use of viability dyes and quantitative PCR in the food microbiology field. J. Appl. Microbiol. 116:1–13. 10.1111/jam.12365 [DOI] [PubMed] [Google Scholar]
- 32.Fittipaldi M, Nocker A, Codony F. 2012. Progress in understanding preferential detection of live cells using viability dyes in combination with DNA amplification. J. Microbiol. Methods 91:276–289. 10.1016/j.mimet.2012.08.007 [DOI] [PubMed] [Google Scholar]
- 33.van Frankenhuyzen JK, Trevors JT, Lee H, Flemming CA, Habash MB. 2011. Molecular pathogen detection in biosolids with a focus on quantitative PCR using propidium monoazide for viable cell enumeration. J. Microbiol. Methods 87:263–272. 10.1016/j.mimet.2011.09.007 [DOI] [PubMed] [Google Scholar]
- 34.Nocker A, Sossa KE, Camper AK. 2007. Molecular monitoring of disinfection efficacy using propidium monoazide in combination with quantitative PCR. J. Microbiol. Methods 70:252–260. 10.1016/j.mimet.2007.04.014 [DOI] [PubMed] [Google Scholar]
- 35.Rawsthorne H, Dock CN, Jaykus LA. 2009. PCR-based method using propidium monoazide to distinguish viable from nonviable Bacillus subtilis spores. Appl. Environ. Microbiol. 75:2936–2939. 10.1128/AEM.02524-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang X, Badoni M, Gill CO. 2011. Use of propidium monoazide and quantitative PCR for differentiation of viable Escherichia coli from E. coli killed by mild or pasteurizing heat treatments. Food Microbiol. 28:1478–1482. 10.1016/j.fm.2011.08.013 [DOI] [PubMed] [Google Scholar]
- 37.Nkuipou-Kenfack E, Engel H, Fakih S, Nocker A. 2013. Improving efficiency of viability-PCR for selective detection of live cells. J. Microbiol. Methods 93:20–24. 10.1016/j.mimet.2013.01.018 [DOI] [PubMed] [Google Scholar]
- 38.Banihashemi A, Van Dyke MI, Huck PM. 2012. Long-amplicon propidium monoazide-PCR enumeration assay to detect viable Campylobacter and Salmonella. J. Appl. Microbiol. 113:863–873. 10.1111/j.1365-2672.2012.05382.x [DOI] [PubMed] [Google Scholar]
- 39.Contreras PJ, Urrutia H, Sossa K, Nocker A. 2011. Effect of PCR amplicon length on suppressing signals from membrane-compromised cells by propidium monoazide treatment. J. Microbiol. Methods 87:89–95. 10.1016/j.mimet.2011.07.016 [DOI] [PubMed] [Google Scholar]
- 40.Soejima T, Schlitt-Dittrich F, Yoshida Si. 2011. Polymerase chain reaction amplification length-dependent ethidium monoazide suppression power for heat-killed cells of Enterobacteriaceae. Anal. Biochem. 418:37–43. 10.1016/j.ab.2011.06.027 [DOI] [PubMed] [Google Scholar]
- 41.Agustí G, Codony F, Fittipaldi M, Adrados B, Morató J. 2010. Viability determination of Helicobacter pylori using propidium monoazide quantitative PCR. Helicobacter 15:473–476. 10.1111/j.1523-5378.2010.00794.x [DOI] [PubMed] [Google Scholar]
- 42.Kralik P, Nocker A, Pavlik I. 2010. Mycobacterium avium subsp. paratuberculosis viability determination using F57 quantitative PCR in combination with propidium monoazide treatment. Int. J. Food Microbiol. 141(Suppl 1):S80–S86. 10.1016/j.ijfoodmicro.2010.03.018 [DOI] [PubMed] [Google Scholar]
- 43.Cattani F, Ferreira CAS, Oliveira SD. 2013. The detection of viable vegetative cells of Bacillus sporothermodurans using propidium monoazide with semi-nested PCR. Food Microbiol. 34:196–201. 10.1016/j.fm.2012.12.007 [DOI] [PubMed] [Google Scholar]
- 44.Brescia CC, Griffin SM, Ware MW, Varughese EA, Egorov AI, Villegas EN. 2009. Cryptosporidium propidium monoazide-PCR, a molecular biology-based technique for genotyping of viable Cryptosporidium oocysts. Appl. Environ. Microbiol. 75:6856–6863. 10.1128/AEM.00540-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fittipaldi M, Pino Rodriguez NJ, Adrados B, Agustí G, Peñuela G, Morató J, Codony F. 2011. Discrimination of viable Acanthamoeba castellani trophozoites and cysts by propidium monoazide real-time polymerase chain reaction. J. Eukaryot. Microbiol. 58:359–364. 10.1111/j.1550-7408.2011.00557.x [DOI] [PubMed] [Google Scholar]
- 46.Agustí G, Fittipaldi M, Morató J, Codony F. 2013. Viable quantitative PCR for assessing the response of Candida albicans to antifungal treatment. Appl. Microbiol. Biotechnol. 97:341–349. 10.1007/s00253-012-4524-z [DOI] [PubMed] [Google Scholar]
- 47.Andorrà I, Esteve-Zarzoso B, Guillamón JM, Mas A. 2010. Determination of viable wine yeast using DNA binding dyes and quantitative PCR. Int. J. Food Microbiol. 144:257–262. 10.1016/j.ijfoodmicro.2010.10.003 [DOI] [PubMed] [Google Scholar]
- 48.Fittipaldi M, Pino Rodriguez NJ, Codony F, Adrados B, Peñuela GAG, Morató J. 2010. Discrimination of infectious bacteriophage T4 virus by propidium monoazide real-time PCR. J. Virol. Methods 168:228–232. 10.1016/j.jviromet.2010.06.011 [DOI] [PubMed] [Google Scholar]
- 49.Kim SY, Ko G. 2012. Using propidium monoazide to distinguish between viable and nonviable bacteria, MS2 and murine norovirus. Lett. Appl. Microbiol. 55:182–188. 10.1111/j.1472-765X.2012.03276.x [DOI] [PubMed] [Google Scholar]
- 50.Parshionikar S, Laseke I, Fout GS. 2010. Use of propidium monoazide in reverse transcriptase PCR to distinguish between infectious and noninfectious enteric viruses in water samples. Appl. Environ. Microbiol. 76:4318–4326. 10.1128/AEM.02800-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sánchez G, Elizaquível P, Aznar R. 2012. Discrimination of infectious hepatitis A viruses by propidium monoazide real-time RT-PCR. Food Environ. Virol. 4:21–25. 10.1007/s12560-011-9074-5 [DOI] [PubMed] [Google Scholar]
- 52.Chen S, Wang F, Beaulieu JC, Stein RE, Ge B. 2011. Rapid detection of viable Salmonellae in produce by coupling propidium monoazide with loop-mediated isothermal amplification. Appl. Environ. Microbiol. 77:4008–4016. 10.1128/AEM.00354-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sassoubre LM, Nelson KL, Boehm AB. 2012. Mechanisms for photoinactivation of Enterococcus faecalis in seawater. Appl. Environ. Microbiol. 78:7776–7785. 10.1128/AEM.02375-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Løvdal T, Hovda MB, Björkblom B, Møller SG. 2011. Propidium monoazide combined with real-time quantitative PCR underestimates heat-killed Listeria innocua. J. Microbiol. Methods 85:164–169. 10.1016/j.mimet.2011.01.027 [DOI] [PubMed] [Google Scholar]
- 55.Kobayashi H, Oethinger M, Tuohy MJ, Hall GS, Bauer TW. 2010. Distinction between intact and antibiotic-inactivated bacteria by real-time PCR after treatment with propidium monoazide. J. Orthop. Res. 28:1245–1251. 10.1002/jor.21108 [DOI] [PubMed] [Google Scholar]
- 56.Shu Z, Weigel KM, Soelberg SD, Lakey A, Cangelosi GA, Lee KH, Chung JH, Gao D. 2012. Cryopreservation of Mycobacterium tuberculosis complex cells. J. Clin. Microbiol. 50:3575–3580. 10.1128/JCM.00896-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liang Z, Keeley A. 2012. Comparison of propidium monoazide-quantitative PCR and reverse transcription quantitative PCR for viability detection of fresh Cryptosporidium oocysts following disinfection and after long-term storage in water samples. Water Res. 46:5941–5953. 10.1016/j.watres.2012.08.014 [DOI] [PubMed] [Google Scholar]
- 58.Oerther DB, Pernthaler J, Schramm A, Amann R, Raskin L. 2000. Monitoring precursor 16S rRNAs of Acinetobacter spp. in activated sludge wastewater treatment systems. Appl. Environ. Microbiol. 66:2154–2165. 10.1128/AEM.66.5.2154-2165.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cangelosi GA, Brabant WH. 1997. Depletion of pre-16S rRNA in starved Escherichia coli cells. J. Bacteriol. 179:4457–4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cutter MR, Stroot PG. 2008. Determination of specific growth rate by measurement of specific rate of ribosome synthesis in growing and nongrowing cultures of Acinetobacter calcoaceticus. Appl. Environ. Microbiol. 74:901–903. 10.1128/AEM.01899-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Licht TR, Tolker-Nielsen T, Holmstrom K, Krogfelt KA, Molin S. 1999. Inhibition of Escherichia coli precursor-16S rRNA processing by mouse intestinal contents. Environ. Microbiol. 1:23–32. 10.1046/j.1462-2920.1999.00001.x [DOI] [PubMed] [Google Scholar]
- 62.Lu T, Stroot PG, Oerther DB. 2009. Reverse transcription of 16S rRNA to monitor ribosome-synthesizing bacterial populations in the environment. Appl. Environ. Microbiol. 75:4589–4598. 10.1128/AEM.02970-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stroot PG, Oerther DB. 2003. Elevated precursor 16S rRNA levels suggest the presence of growth inhibitors in wastewater. Water Sci. Technol. 47:241–250 [PubMed] [Google Scholar]
- 64.Oerther DB, van Loosdrecht MC, Raskin L. 2002. Quantifying the impact of wastewater micronutrient composition on in situ growth activity of Acinetobacter spp. Water Sci. Technol. 46:443–437 [PubMed] [Google Scholar]
- 65.Woese CR. 1987. Bacterial evolution. Microbiol. Rev. 51:221–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cangelosi GA, Hamlin AM, Marin R, Scholin CA. 1997. Detection of stable pre-rRNA in toxigenic Pseudo-nitzschia species. Appl. Environ. Microbiol. 63:4859–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eckert WA, Kaffenberger W. 1980. Regulation of rRNA metabolism in Tetrahymena pyriformis. I. Nutritional shift-down. Eur. J. Cell Biol. 21:53–62 [PubMed] [Google Scholar]
- 68.Nocker A, Richter-Heitmann T, Montjin R, Schuren F, Kort R. 2010. Discrimination between live and dead cells in bacterial communities from environmental water samples analyzed by 454 pyrosequencing. Int. Microbiol. 13:59–65 [DOI] [PubMed] [Google Scholar]
- 69.Nocker A, Sossa-Fernandez P, Burr MD, Camper AK. 2007. Use of propidium monoazide for live/dead distinction in microbial ecology. Appl. Environ. Microbiol. 73:5111–5117. 10.1128/AEM.02987-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee JL, Levin RE. 2006. Use of ethidium bromide monoazide for quantification of viable and dead mixed bacterial flora from fish fillets by polymerase chain reaction. J. Microbiol. Methods 67:456–462. 10.1016/j.mimet.2006.04.019 [DOI] [PubMed] [Google Scholar]
- 71.Nocker A, Burr M, Camper A. 2007. Genotypic microbial community profiling: a critical technical review. Microb. Ecol. 54:276–289. 10.1007/s00248-006-9199-5 [DOI] [PubMed] [Google Scholar]
- 72.Schmid M, Schmitz-Esser S, Jetten M, Wagner M. 2001. 16S-23S rDNA intergenic spacer and 23S rDNA of anaerobic ammonium-oxidizing bacteria: implications for phylogeny and in situ detection. Environ. Microbiol. 3:450–459. 10.1046/j.1462-2920.2001.00211.x [DOI] [PubMed] [Google Scholar]
- 73.van Dorst J, Bissett A, Palmer AS, Brown M, Snape I, Stark JS, Raymond B, McKinlay J, Ji M, Winsley T, Ferrari BC. 1 March 2014. Community fingerprinting in a sequencing world. FEMS Microbiol. Ecol. 10.1111/1574-6941.12308 [DOI] [PubMed] [Google Scholar]
- 74.Blainey PC. 2013. The future is now: single-cell genomics of bacteria and archaea. FEMS Microbiol. Rev. 37:407–427. 10.1111/1574-6976.12015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lasken RS. 2012. Genomic sequencing of uncultured microorganisms from single cells. Nat. Rev. Microbiol. 10:631–640. 10.1038/nrmicro2857 [DOI] [PubMed] [Google Scholar]