ABSTRACT
The genus Potyvirus comprises a large group of positive-strand RNA plant viruses whose genome encodes a large polyprotein processed by three viral proteinases. P1 protein, the most amino-terminal product of the polyprotein, is an accessory factor stimulating viral genome amplification whose role during infection is not well understood. We infected plants with Tobacco etch virus (TEV; genus Potyvirus) clones in which P1 was tagged with a fluorescent protein to track its expression and subcellular localization or with an affinity tag to identify host proteins involved in complexes in which P1 also takes part during infection. Our results showed that TEV P1 exclusively accumulates in infected cells at an early stage of infection and that the protein displays a dynamic subcellular localization, trafficking in and out of the nucleus and nucleolus during infection. Inside the nucleolus, P1 particularly targets the dense granular component. Consistently, we found functional nucleolar localization and nuclear export signals in TEV P1 sequence. Our results also indicated that TEV P1 physically interacts with the host 80S cytoplasmic ribosomes and specifically binds to the 60S ribosomal subunits during infection. In vitro translation assays of reporter proteins suggested that TEV P1 stimulates protein translation, particularly when driven from the TEV internal ribosome entry site. These in vitro assays also suggested that TEV helper-component proteinase (HC-Pro) inhibits protein translation. Based on these findings, we propose that TEV P1 stimulates translation of viral proteins in infected cells.
IMPORTANCE In this work, we researched the role during infection of tobacco etch virus P1 protease. P1 is the most mysterious protein of potyviruses, a relevant group of RNA viruses infecting plants. Our experiments showed that the viral P1 protein exclusively accumulates in infected cells at an early stage of infection and moves in and out of the nucleus of infected cells, particularly targeting the nucleolus. Our experiments also showed that P1 protein binds host ribosomes during infection. Based on these findings and other in vitro experiments we propose that P1 protein stimulates translation of viral proteins during infection.
INTRODUCTION
Plant viruses have evolved as a combination of genes encoding proteins displaying a limited series of catalytic activities but with the ability to interact with specific cellular factors to successfully achieve virus genome replication and gene expression, virion assembly, and virus movement and to counter host defense systems. Tobacco etch virus (TEV) is a member of the genus Potyvirus (one of the nine currently accepted genera within the family Potyviridae), which by itself includes approximately 30% of known species of plant viruses. The genome of potyviruses consists of a single-stranded, positive-sense RNA of approximately 10,000 nucleotides (nt) that is linked at the 5′ end to a viral protein, genome-linked (VPg), and contains a poly(A) tail at the 3′ end. This genomic RNA includes a long open reading frame encoding a polyprotein that, when translated, is cleaved by three viral proteases into at least 10 mature proteins: P1, helper-component proteinase (HC-Pro), P3, 6K1, cylindrical inclusion (CI), 6K2, nuclear inclusion a (NIa; a polyprotein which is further processed to produce VPg and NIaPro), nuclear inclusion b (NIb), and the coat protein (CP) (1). Additionally, the protein P3N-PIPO is produced through a translational frameshift in the P3 cistron (2).
Most potyviral proteins have been assigned important roles during the infectious cycle (3). HC-Pro, a cysteine proteinase that self-cleaves from the viral polyprotein, suppresses the host defensive RNA silencing pathways (4). It is also involved in aphid transmission (5). P3N-PIPO and CI play roles in virus cell-to-cell movement (6). 6K2 anchors the viral replication complex to intracellular membranes (7, 8). VPg acts as a primer during viral RNA synthesis (9), establishes crucial interaction with host eukaryotic initiation factor 4E (eIF4E) or eIF(iso)4E (10), and is involved in translation (11). NIaPro is a serine protease that in cis and in trans cleaves most of the potyviral polyprotein (12). NIa is also involved in virus replication (13). NIb is the viral RNA-dependent RNA polymerase (14), and CP, in addition to virion assembly, is involved in aphid transmission (15) and viral movement (16). In contrast, P1, the first product of potyviral polyprotein, is a rather mysterious protein whose functions during the infectious cycle are mostly unknown (17). The P1 cistron is the most variable in size and sequence in the various species of the genus (18). The P1 protein contains a carboxy-terminal serine protease domain that catalyzes its own cleavage from the viral polyprotein (19) and exhibits a strong RNA binding activity (20, 21). Although P1 contributes to the virus infectious cycle, it is not essential because a virus deletion mutant completely lacking the P1 cistron, although debilitated, is still viable (22, 23). Although P1 protein enhances viral suppression of RNA silencing mediated by HC-Pro (24, 25), it has been recently shown that more HC-Pro accumulates if HC-Pro is translated as a P1–HC-Pro fusion than alone (26). However, it was recently shown that P1 protein is also important in defining virus host range (18, 27).
In this work we investigated the roles of P1 protein in TEV infection. Toward this end, we analyzed P1 expression and subcellular localization during infection. We also identified host proteins that form complexes with P1 during infection. We found that P1 accumulates in infected cells at an early stage of the infectious process and then mostly disappears. We also found that P1 initially localizes in the nucleolus and then traffics back to the cytoplasm. We demonstrated that TEV P1 contains a functional nucleolar localization signal (NoLS) and a nuclear export signal (NES). In infected plant tissues, we encountered P1 mostly associated with ribosomal proteins. We demonstrated that P1 binds in vivo to host 80S cytoplasmic ribosomes and, more specifically, to the 60S ribosome subunits. Based on this interaction, we investigated a possible role of P1 in translation and found that, in a wheat germ system, P1 stimulates translation of reporter proteins in vitro, particularly when translation is driven from the TEV internal ribosome entry site (IRES). Based on these findings, we propose a model in which potyviral P1 protein binds 60S ribosomal subunits to subvert the host translation machinery during infection.
MATERIALS AND METHODS
Recombinant TEV clones and plant inoculation.
Various recombinant TEV clones were constructed using common molecular biology techniques and starting from pGTEVa (28), a binary plasmid with a cassette to agroinoculate wild-type TEV (TEV-wt) (GenBank accession number DQ986288 including silent mutations G273A and A1119G) under the control of the Cauliflower mosaic virus (CaMV) 35S promoter and terminator. TEV-VenusP1 contained the fluorescent protein Venus, a green-yellow derivative of Aequorea victoria green fluorescent protein (GFP) (29), fused to the amino terminus of P1 (VenusP1) (Fig. 1A). TEV-GFP contained the enhanced GFP between the P1 and HC-Pro cistrons (Fig. 1A). In this clone, the GFP is released from the viral polyprotein through the proteolytic activities of P1 and NIaPro proteinases (28). In TEV-VenusP1-mCherryNIb, in addition to the Venus fusion to P1, the red fluorescent protein mCherry (30) was fused to the amino terminus of NIb (Fig. 1A). Finally, in TEV-TSTP1, a twin-Strep-tag (TST), commonly used for protein purification by affinity chromatography (31), was fused to the amino terminus of P1 (Fig. 1A). Sequencing of the corresponding plasmids confirmed the correct construction of all of these recombinant TEV clones. Their exact sequences are specified in the supplemental material (see Fig. S1 in the supplemental material). Nicotiana benthamiana Domin plants were kept in a growth chamber with a photoperiod of 12 h of light at 25°C and 12 h of dark at 23°C. Plant agroinoculation was performed using Agrobacterium tumefaciens C58C1 (harboring the helper plasmid pCLEAN-S48) at an optical density of 0.5 at 600 nm as described previously (32). TEV virions were partially purified from infected tissues and used for mechanical inoculation of N. benthamiana leaves (32).
FIG 1.
Expression of P1 protein during TEV infectious cycle. (A) Schematic representation of recombinant TEV infectious clones: TEV-wt, TEV-VenusP1, TEV-GFP, and TEV-VenusP1-mCherryNIb and TEV-TSTP1. Lines represent viral 5′ and 3′ UTRs, and white boxes represent viral cistrons P1, HC-Pro, P3, 6K1, CI, 6K2, VPg, NIaPro, NIb, and CP, as indicated. P3N-PIPO is represented by white (P3N) and dashed (PIPO) boxes. Light green, dark green, and red boxes represent Venus, GFP, and mCherry cDNAs, as indicated. The blue box represents twin-Strep-tag (TST) cDNA. (B) Fluorescence images taken under a stereomicroscope of N. benthamiana leaves showing TEV-GFP and TEV-VenusP1 infection foci at 3 dpi. Scale bars correspond to 500 and 50 μm at low and high magnification, respectively. (C) Fluorescence images taken under a stereomicroscope of a TEV-VenusP1-mCherryNIb infection focus on an N. benthamiana leaf at 3 dpi. Scale bar, 50 μm. (D) Time course analysis of TEV TSTP1 and CP accumulation in the third leaf of N. benthamiana plants above the leaf agroinoculated with TEV-TSTP1. Samples taken at different days postinoculation (dpi), as indicated, were separated by SDS-PAGE and analyzed by Western blotting. (E) Plots of TEV TSTP1 and CP accumulation (measured by Western blot analysis in arbitrary units [au]) versus days postinoculation in the third leaf of N. benthamiana plants above the leaf agroinoculated with TEV-TSTP1. Bars indicate the standard deviations of the measures taken in triplicate plants.
Protein transient expression in N. benthamiana leaves.
For transient expression, leaves of N. benthamiana plants were infiltrated with cultures of A. tumefaciens C58C1 (harboring the helper plasmid pCLEAN-S48) as described previously (33). To express Arabidopsis thaliana fibrillarin 2 (AtFib2) (34, 35) fused to a monomeric red fluorescent protein (AtFib2mRFP), A. tumefaciens was transformed with a previously described binary plasmid (35). To express A. thaliana ribosomal protein L24B (AtRPL24B) (36) fused to mCherry (AtRPL24BmCherry), the AtRPL24B cDNA was amplified from A. thaliana Col-0 by reverse transcription-PCR (RT-PCR) and cloned in a modified version of pEarlyGate101 (Invitrogen). To express Venus, VenusP1, and derivatives of VenusP1, A. tumefaciens was transformed with a series of binary plasmids based on a modified version (32) of pCLEAN-G181 (GenBank accession number EU186083) containing, between the left and right borders of the T-DNA, expression cassettes consisting of the CaMV 35S promoter, a modified version of the Cowpea mosaic virus (CPMV) RNA-2 5′ untranslated region (UTR) (37), the cDNA of the corresponding protein, CPMV RNA-2 3′ UTR, and the CaMV 35S terminator. The exact sequence of each construct is specified in the supplemental material (see Fig. S2).
Analysis of fluorescent proteins.
Expression of fluorescent proteins was analyzed by confocal laser scanning microscopy using a Leica TCS SL with an HCX PL APO 40× oil lens (numerical aperture, 1.25 to 0.75). Venus and mCherry were detected with excitation lasers of 488 and 543 nm and detection windows of 520 to 550 and of 610 to 670 nm, respectively. Optical section was 1 μm. Also, a Leica MZ16 F fluorescence stereomicroscope equipped with filters DSR and GFP2 (Leica) was used. Fluorescent infection foci were analyzed using ImageJ software.
Infectivity assays.
TEV recombinant clones containing mutations in the P1 cistron (see Fig. S3 in the supplemental material) and the transcription factor Rosea1 (Ros1) as a reporter marker (28) were agroinoculated in two different leaves of batches of 10 N. benthamiana plants. Infection symptoms in these plants were recorded over time by visual inspection. Viral load was estimated from the anthocyanin accumulation induced by the Ros1 reporter marker activity. Anthocyanins in infected tissues from three different plants for each viral construct were extracted in acidified methanol and quantified spectrophotometrically at 530 nm (28).
Protein analysis.
Proteins were separated by denaturing 0.5% sodium dodecyl sulfate–12.5% polyacrylamide gel electrophoresis (SDS-PAGE), electroblotted to polyvinylidene difluoride (PVDF) membranes (GE Healthcare), and analyzed by Western blotting as described previously (38). A monoclonal antibody against the TST tag (StrepMAB-Classic horseradish peroxidase; IBA) was used at a 1:5,000 dilution, and a polyclonal antibody against the TEV CP (conjugated to alkaline phosphatase; Agdia) was used at a 1:10,000 dilution. Immunoblots were quantified with a luminescent image analyzer (LAS-3000; Fujifilm) as described previously (28). Polysomes were isolated from 2 g of TEV-TSTP1-infected N. benthamiana plants, harvested at 5 days postinoculation (dpi), as described previously (39). Tissue was ground in a mortar in the presence of liquid N2 and homogenized with 10 volumes of extraction buffer (200 mM Tris-HCl, pH 9.0, 400 mM KCl, 200 mM sucrose, 35 mM MgCl2, 5 mM dithiothreitol [DTT], 30 mM EDTA, and a cocktail of protease inhibitors [Complete; Roche Applied Sciences]). Extract was clarified by centrifugation at 25,000 × g for 10 min. Five-milliliter aliquots of the supernatants were centrifuged through 4-ml layers of 1.75 M sucrose in 40 mM Tris-HCl, pH 9.0, 200 mM KCl, 30 mM MgCl2, 5 mM DTT, and either 30 mM or 5 mM EDTA to separate or not separate ribosomal particles and centrifuged at 340,000 × g for 1 h. Sediments were resuspended in a total of 1 ml of 40 mM Tris-HCl, pH 8.5, 200 mM KCl, 30 mM MgCl2, 5 mM DTT, and either 30 mM or 5 mM EDTA and layered on continuous 15 to 60% sucrose gradients in 40 mM Tris-HCl, pH 8.5, 20 mM KCl, 10 mM MgCl2, 5 mM DTT, and 10 mM EDTA. Samples were centrifuged for 3 h at 4°C in a SW 40 Ti rotor (Beckman Coulter) at 40,000 rpm (285,000 × g), and 16 fractions were collected.
Purification of protein complexes containing TEV TSTP1 and protein identification by mass spectrometry analysis.
Symptomatic N. benthamiana leaf tissues (15 g) infected by TEV-wt or TEV-TSTP1 (Fig. 1A) were harvested at 5 dpi, ground in a mortar with liquid N2, and homogenized with 45 ml of extraction buffer (100 mM Tris-HCl, pH 8.0, 150 mM KCl, 10 mM MgCl2, 10 mM DTT, 1 mM EDTA, 1% Nonidet P-40) containing a cocktail of protease inhibitors (Complete). The crude extracts were clarified twice by centrifugation at 4°C, first at 12,000 × g for 15 min and then at 95,000 × g for 30 min. The TST-tagged P1 protein was purified by chromatography using a 1-ml Strep-Tactin Superflow column (IBA) with an ÄKTA Prime Plus liquid chromatography system (GE Healthcare) operated at 4°C at a flow rate of 1 ml/min. After equilibration with 10 ml of extraction buffer, the column was loaded with the clarified extract and washed with 20 ml of extraction buffer. Bound protein complexes were eluted with 20 ml of extraction buffer containing 10 mM d-desthiobiotin, and 0.5-ml fractions were collected. Fractions from the TEV-TSTP1-infected tissues were analyzed by Western blotting with the anti-TST antibody; those containing substantial amounts of TSTP1 were pooled, and the protein was precipitated by adding 4 volumes of 12.5% trichloroacetic acid and 10 mM DTT in acetone. The same process was followed with the corresponding fractions eluted in the control purification process from tissues infected by TEV-wt. Protein preparations were separated by SDS-PAGE (12.5% polyacrylamide, 0.05% SDS), and the gel was stained with Coomassie blue. Whole lanes corresponding to each sample were excised from the gel and cut in pieces, and the proteins were subjected to in-gel digestion with sequencing-grade trypsin (Promega) as described previously (40). Peptides were eluted from the gel pieces and analyzed by liquid chromatography and tandem mass spectrometry as previously described (33).
In vitro translation assays.
A TNT Coupled Wheat Germ Extract System (Promega) was used to translate proteins in vitro in the presence of l-[35S]methionine (1,000 Ci/mmol). Reactions were started for 15 min at 30°C with 0.2 pmol of a series of plasmids with cassettes to translate the firefly luciferase (Fluc), TEV truncated P1, P1, HC-Pro, and P1–HC-Pro. Then, 0.2 pmol of plasmids with monocistronic or bicistronic reporter cassettes was added to all reaction mixtures, and incubation continued for 40 min (monocistronic reporter) or 2 h 15 min (bicistronic reporter). The exact sequences of the cassettes subjected to in vitro translation are in the supplemental material (see Fig. S4). Translation products were separated by SDS-PAGE (12.5% polyacrylamide, 0.05% SDS), and the gel was fixed in 20% methanol and 10% acetic acid for 30 min, dried under vacuum, and analyzed by phosphorimetry (Fujifilm FLA-5100).
RESULTS
P1 expression during TEV infectious cycle.
To learn about the roles of P1 protein in the TEV infectious cycle, we infected plants with a recombinant TEV clone in which the P1 protein was tagged with a fluorescent protein to track its expression and subcellular localization. We inserted a cDNA coding for Venus as an amino-terminal fusion to P1 in an infectious TEV clone, obtaining TEV-VenusP1 (Fig. 1A). We agroinoculated N. benthamiana plants with TEV-VenusP1 and, as controls, with TEV-wt and TEV-GFP (Fig. 1A). Plants agroinoculated with TEV-VenusP1 became infected showing mild disease symptoms with a 1-day delay with respect to TEV-wt and TEV-GFP (Fig. 2A). However, since P1 is a nonessential protein for the virus, it is difficult to evaluate the effect of the tag on P1 function.
FIG 2.
N. benthamiana plants inoculated with various recombinant TEV clones. (A) Pictures taken at 8 dpi of representative N. benthamiana plants mock inoculated and inoculated with TEV-wt, TEV-VenusP1, TEV-GFP, TEV-VenusP1-mCherryNIb, and TEV-TSTP1, as indicated. (B) Analysis by RT-PCR of the viral progeny in plants infected by various recombinant TEV clones. Viral cDNAs were amplified by RT-PCR from RNA preparations from infected tissues at 10 dpi. Amplification products were analyzed by electrophoresis in 1% agarose gels, followed by ethidium bromide staining. Products of the upper and lower gels were amplified with primer pairs flanking the P1 (primer I, 5′-TTATTCGCATGCCTAAGGATTTCCC-3′; and primer II, 5′-AGGAACGCCTCTCTATTAAGTCGAC-3′) and the NIb (primer III, 5′-CTATTGCAGCAATTTAAATCATTTC-3′; and primer IV, 5′-CTCTTGCCATGGGTGAGCGCGCGAC-3′) cistrons, respectively. Lane 1, RT-PCR negative control; lanes 2 to 11, RT-PCR products from RNA preparations from tissues from individual plants infected by TEV-wt (lane 2), TEV-TSTP1 (lanes 3 to 5), TEV-VenusP1 (lanes 6 to 8), and TEV-VenusP1-mCherryNIb (lanes 9 to 11); lane 12, DNA marker ladder with the size (in kbp) of the components indicated on the right.
Analysis with a fluorescence stereomicroscope of symptomatic noninoculated (systemic) leaves of plants infected by TEV-VenusP1 showed faint green fluorescence exclusively at the infection front. In contrast, systemic leaves of plants infected by TEV-GFP displayed a strong green fluorescence in all symptomatic tissues. We checked the stability of the TEV-VenusP1 clone by reverse transcription-PCR (RT-PCR) amplification of viral cDNAs from RNA preparations obtained from infected tissues (Fig. 2B). Sequence analysis confirmed the presence and correct insertion of the Venus cDNA in the genome of the viral progeny. Furthermore, new plants infected with virions purified from plants initially agroinoculated with TEV-VenusP1 exhibited the same pattern of green fluorescence in the periphery of symptomatic areas. In contrast to agroinoculation, mechanical inoculation of purified virions allows analysis of infection foci in inoculated leaves. Whereas infection foci of TEV-GFP appeared uniformly fluorescent when observed under a fluorescence stereomicroscope, infection foci of TEV-VenusP1 exhibited only a green fluorescent ring at the periphery (Fig. 1B).
We constructed a new recombinant TEV clone in which, in addition to the Venus fusion to P1, we also tagged NIb with the red fluorescent protein mCherry. In this new clone (TEV-VenusP1-mCherryNIb) (Fig. 1A), mCherry was inserted as an amino-terminal fusion to NIb. Agroinoculation of N. benthamiana plants with TEV-VenusP1-mCherryNIb demonstrated that this recombinant clone was also infectious although mild disease symptoms appeared with a 3-day delay with respect to TEV-wt (Fig. 2). This clone allowed observation of the green fluorescence of VenusP1 and the red fluorescence of mCherryNIb in the same infection foci. Whereas red fluorescence was detected throughout the foci, green fluorescence was observed only in the peripheral rings (Fig. 1C). Taken together, these results suggest a transient accumulation of the fusion protein VenusP1 at an early stage of viral infection. Since potyviruses express their proteins (except for P3N-PIPO) from a single large polyprotein, potyvirus proteins are synthesized in equimolar amounts, and relative changes in accumulation must result from different degradation rates. Therefore, these results suggest that TEV P1, in contrast to other viral proteins, reaches the highest level in the infected cells at an early stage of infection, and then at late stages, P1 is efficiently degraded.
To obtain further support for the above hypothesis, we constructed a new recombinant TEV clone in which P1 was tagged at the amino terminus with TST. The new recombinant clone (TEV-TSTP1) (Fig. 1A) was agroinoculated into N. benthamiana plants alongside plants inoculated with TEV-wt. TEV-TSTP1 was infectious, and agroinoculated plants displayed, again, mild symptoms with a 1-day delay with respect to TEV-wt-inoculated plants (Fig. 2). In plants infected by TEV-TSTP1, the third leaf above the agroinoculated one was harvested in triplicate on various days postinoculation. Proteins were extracted, and the accumulation of viral TSTP1 and CP was determined by SDS-PAGE, followed by Western blot analysis with anti-TST and anti-CP antibodies. A representative Western blot corresponding to a time course of one of the three sample replicates is shown in Fig. 1D. Figure 1E shows the dynamics of TSTP1 and CP accumulation in systemic leaves of infected plants. Whereas TEV CP accumulates continuously in the infected tissue from 4 to 14 dpi, TSTP1 increases accumulation from 4 to 6 dpi and then quickly falls. This result supports a transient accumulation of P1 during the early stage of infection and subsequent effective degradation at later stages.
P1 subcellular localization in infected cells.
To investigate P1 subcellular localization during infection, we used the infectious TEV-VenusP1-mCherryNIb clone (Fig. 1A). NIb subcellular localization is well known (41, 42) and served as an internal control in these experiments. N. benthamiana plants were mechanically inoculated with TEV-VenusP1-mCherryNIb virions, and infection foci were analyzed by confocal laser scanning microscopy. Green and red fluorescence reported VenusP1 and mCherryNIb subcellular localizations, respectively. We analyzed the accumulation of both fluorescent proteins from the periphery to the center of infection foci (Fig. 3A). Localization of VenusP1 differed across the infection foci (Fig. 3B). As already mentioned, green fluorescence was brightest at the periphery of the infection foci, just at the viral replication front. In these cells, VenusP1 was mostly localized in a large subnuclear body, presumably the nucleolus, and also more diffusely in the nucleus and cytoplasm (Fig. 3B, row I). mCherryNIb was still not detectable in these cells (Fig. 3B, row I). In adjacent cells toward the center of the infection foci, VenusP1 was again detected in the nucleolus, nucleus, and cytoplasm, whereas mCherryNIb was detected in the nucleus and cytoplasm (Fig. 3B, row II). The intensity of green fluorescence progressively decreased in cells toward the epicenter of the infection foci. Nonetheless, it was still possible to observe VenusP1 in the nucleus and cytoplasm but no longer in the nucleolus (Fig. 3B, rows III and IV). Inside the nucleus, VenusP1 displayed a more perinuclear localization than mCherryNIb (Fig. 3B, rows II to IV). At the center of the infection foci, VenusP1 was no longer detected (Fig. 3B, row V). mCherryNIb was always detected in the nucleus and cytoplasm, as expected (Fig. 3B, rows II to V) (41, 42). These results indicate that TEV P1 displays a dynamic intracellular localization during the cellular infectious cycle. At the beginning of the infection, the protein localizes in the cytoplasm and nucleus, particularly targeting the nucleolus. Then, the protein seems to exit the nucleolus.
FIG 3.
P1 and NIb subcellular localization during the TEV infectious cycle. (A) Schematic representation of an infection focus with cells (I to V) at different stages of infection. (B) Green and red fluorescence images taken under a confocal microscope of N. benthamiana leaf cells infected by TEV-VenusP1-mCherryNIb at 3 dpi. Rows I to V correspond to the positions of the cells in the infection focus, from the periphery to the epicenter. Each series includes the green and red fluorescence images at low and high magnifications and a merged image at high magnification. Scale bars correspond to 16 and 8 μm at low and high magnifications, respectively.
To confirm and further investigate P1 nucleolar localization, we performed colocalization experiments between VenusP1 and well-known nucleolar proteins: AtFib2 (34, 35) and AtRPL24B (36). AtFib2 and AtRPL24B were transiently expressed in N. benthamiana leaves by means of infiltration with A. tumefaciens cultures. Infiltrated leaves were mechanically inoculated with TEV-VenusP1 virions 6 h before infiltration. AtFib2 and AtRPL24B were expressed as amino-terminal fusions to monomeric red fluorescent protein (mRFP; AtFib2mRFP) and mCherry (AtRPL24BmCherry), respectively. Whereas AtFib2 localizes to the nucleolus dense fibrillar component and Cajal bodies, AtRPL24B localizes to nucleolus dense fibrillar and granular components, as well as the nucleus and cytoplasm. Three days after the inoculation and subsequent infiltration, we could observe under the confocal microscope both the red fluorescence of the transiently expressed proteins in the whole agroinfiltrated area and the green fluorescence of the viral VenusP1 at the periphery of the infection foci. Nucleolar localizations of marker AtFib2mRFP and AtRPL24BmCherry proteins in infected cells were the same as in uninfected cells. The intense green fluorescence of VenusP1 colocalized with the red fluorescence of AtFib2mRFP in the large subnuclear body, confirming that this actually was the nucleolus (Fig. 4A). However, the merged image with the two fluorescence signals indicated very little intranucleolar colocalization (Fig. 4A). Comparison with AtFib2mRFP showed that VenusP1 was also absent from the Cajal bodies (Fig. 4A). In contrast, the merged image indicated a better colocalization between VenusP1 and AtRPL24BmCherry at the periphery of the nucleolus (Fig. 4B), corresponding to the granular component (Fig. 4C). These results support the nucleolar localization of TEV P1 during the early stage of viral infection; more specifically, our results suggest that the protein traffics to the nucleolus granular component.
FIG 4.
Colocalization of A. thaliana AtFib2 and AtRPL24B nucleolar marker proteins and P1 during TEV infection. N. benthamiana leaves were inoculated with TEV-VenusP1 virions and infiltrated with A. tumefaciens cultures to transitorily express AtFib2mRFP and AtRPL24BmCherry. Green and red fluorescence images were taken under a confocal microscope 3 days later. A merged image is also shown. (A) Transient expression of AtFib2mRFP in a cell infected by TEV-VenusP1. Arrows point to the nucleolus (No) and a Cajal body (CB). (B) Transient expression of AtRPL24B in a cell infected by TEV-VenusP1. (C) Schematic representation of a plant nucleolus for a better interpretation of results: nucleolar cavity (NC), fibrillar component (FC), dense fibrillar component (DFC), and granular component (GC). Scale bar, 8 μm.
Nucleolar localization and nuclear export signals in P1 sequence.
The results shown above about P1 subcellular localization during the TEV infectious cycle suggest that this protein may traffic from the cytoplasm to the nucleus, particularly targeting the nucleolus, and then back to cytoplasm, like the ribosomal protein AtRPL24B. We searched for putative NoLS and NES in TEV P1 sequence using the NoD (43) and NetNES (44) algorithms, respectively. NoD predicted a single NoLS between TEV P1 amino acids 89 and 109 (LTHGKRRKVSVNNKRNRRRKV) (Fig. 5). NetNES predicted a single NES between amino acids 251 and 260 (LTFGSSGLVL) (Fig. 5). We tested the functionality of these motifs by A. tumefaciens-mediated transient expression in N. benthamiana leaves of a series of VenusP1 constructs containing deletions and amino acid substitutions in the P1 coding sequence (Fig. 6A; see also Fig. S2 in the supplemental material).
FIG 5.
Identification of a nucleolar localization signal (NoLS) and a nuclear export signal (NES) in TEV P1 sequence (GenBank accession number ABJ16044). (A) NoLS prediction per residue displayed by the NoD algorithm. (B) NES prediction in the 65 carboxy-terminal amino acids of TEV P1 displayed by the NetNES algorithm. HMM, hidden Markov model. (C) Amino acid sequence of TEV P1 with the predicted NoLS and NES underlined and highlighted over light and dark gray backgrounds, respectively.
FIG 6.
Transient expression of Venus, VenusP1 and a series of VenusP1 mutants in N. benthamiana. Leaves were infiltrated with A. tumefaciens cultures to express the different constructs. (A) Schematic representation of the expressed constructs (I to X). The green and white boxes represent Venus and TEV P1 cDNAs, respectively. The gray and black boxes represent TEV P1 NoLS and NES, as indicated. The black arrow and hexagon represent the CaMV 35S promoter (P35S) and terminator (t35S), respectively. Black lines represent the Cowpea mosaic virus (CPMV) RNA-2 5′ and 3′ UTRs, as indicated. Mutations in NoLS and NES sequences are represented in red. (B) Green fluorescence images of selected cells taken under a confocal microscope at 3 days after infiltration with constructs I to X. Some cells were imaged at two different magnifications. Scale bars correspond to 20 and 8 μm at low and high magnifications, respectively.
When wild-type VenusP1 (Fig. 6A, construct I) was transiently expressed in N. benthamiana leaves, analysis by confocal laser scanning microscopy of the infiltrated tissues 3 days postinfiltration showed an intense green fluorescence in the nucleolus. This fluorescence was less intense in the nucleus and even less so in the cytoplasm (Fig. 6B, construct I). In contrast, transient expression of a control construct consisting of Venus alone (Fig. 6A, construct II) showed similar green fluorescence intensity levels in both nucleus and cytoplasm but largely excluding the nucleolus (Fig. 6B, construct II). We split P1 in two halves: the amino-terminal half from amino acids 1 to 115 containing the presumed NoLS and the carboxy-terminal half from amino acids 116 to 304 (Fig. 6A, constructs III and IV). Transient expression of each half fused to the carboxy terminus of Venus showed green fluorescence in the nucleolus exclusively in the case of the construct containing amino acids 1 to 115 (Fig. 6B, compare constructs III and IV). Therefore, the first 115 amino acids of P1 are necessary and sufficient for its nucleolar localization. Next, we fused the 21 amino acids of the putative NoLS (from P1 amino acids 89 to 109) to the amino terminus of Venus (Fig. 6A, construct V). This polypeptide was sufficient to transport Venus inside the nucleolus (Fig. 6B, construct V), indicating that it contains a functional NoLS. This sequence contains two stretches of basic amino acids: 93-KRRK-96 and 102-KRNRRRK-108, the second of which is interrupted by Asn-104. We mutated wild-type VenusP1 to obtain three different constructs. In the first, each amino acid of the motif 93-KRRK-96 was mutated to Ala (Fig. 6A, construct VII). In the second, each basic amino acid of the motif 102-KRNRRRK-108 was mutated to Ala (Fig. 6A, construct VIII). In the third, both mutated motifs were combined (Fig. 6A, construct IX). No mutant reached the nucleolus (Fig. 6B, constructs VII to IX). Taken together, these results demonstrated the existence of a NoLS in TEV P1 constituted by two motifs of basic amino acids, both of them required for entering the nucleolus.
Next, we deleted the putative NES (251-LTFGSSGLVL-260) from the P1 sequence in the VenusP1 construct (Fig. 6A, construct VI). Inspection of the infiltrated leaves showed green fluorescence in the nucleus and nucleolus, but in contrast to wild-type VenusP1, no fluorescence was detected in the cytoplasm (Fig. 6B, compare constructs I and VI). We next mutated the three carboxy-terminal amino acids of the presumed NES (258-LVL-260) to three consecutive Ala residues (Fig. 6A, construct X) in the VenusP1 construct. Again, green fluorescence was detected in the nucleus and nucleolus but not in the cytoplasm. These results support the notion that TEV P1 contains a functional NES involved in protein export from the nucleus to the cytoplasm.
Effect of P1 mutations on viral infection.
We tested the effect of some of the P1 mutations described above on TEV infectivity and viral load. To this aim, we introduced a series of mutations in the P1 cistron of a TEV recombinant clone that contains the transcription factor Ros1 as a reporter marker (28). The resulting mutant viral clones (see Fig. S3 in the supplemental material), along with the control including the wild-type P1, were agroinoculated in batches of N. benthamiana plants. The number of infected plants was recorded over time for the different mutants. Viral load was estimated by measuring the anthocyanin accumulation induced by the Ros1 reporter at 3 days after symptoms first emerged in each infected plant. In TEV clones tagged with Ros1, anthocyanin accumulation correlates with viral load (28). As already reported (22, 23), the whole deletion of the P1 cistron in the TEV genome yields a viable virus although it reduces infectivity and viral load with respect to wild-type virus (Fig. 7A and B, mutant Mt-ΔP1). This is in contrast to what occurs with mutation of the His 214 to Ala in the P1 catalytic triad of the serine protease domain that abolishes infectivity (Fig. 7A, Mt-H214A). Also as reported previously (22, 23), this defect was partially rescued by inserting a subrogate NIaPro cleavage site (Fig. 7A and B, mutant Mt-H214+NIaPro). Mutation of the LVL motif in the P1 NES (Fig. 6A, construct X) yielded a noninfectious virus (Fig. 7A, Mt-X). As this motif is present in the serine protease domain, we also assayed this mutant but inserted a subrogate NIaPro cleavage site (Mt-X+NIaPro) to distinguish the nuclear export and the proteolytic functions. In this case, infectivity and viral load were recovered but only in part (Fig. 7A and B, Mt-X+NIaPro). Mutation of the basic amino acids in the P1 NoLS (Fig. 6A, constructs VII, VIII, and IX) had little effect on TEV infectivity but reduced viral load in infected plants (Fig. 7A and B, Mt-VII, Mt-VIII, and Mt-IX).
FIG 7.
Effect of mutations in the P1 cistron on TEV infectivity and accumulation. (A) Number of symptomatic N. benthamiana plants versus day postagroinoculation of wild-type (WT) and P1 mutant TEV clones including the Ros1 marker. (B) Viral load at 3 days after symptoms emerged, measured as anthocyanin accumulation (absorbance at 530 nm), in plants infected by wild-type and P1 mutant TEV clones including the Ros1 marker. Error bars indicate the standard deviations of the three sampled plants. Wild-type and mutant P1 (Mt-VII, Mt-VIII, Mt-IX, Mt-X, Mt-X+NIaPro, Mt-H214A, and Mt-H214A+NIaPro) sequences are shown in Fig. S3 in the supplemental material.
Host proteins associated to P1 during TEV infection.
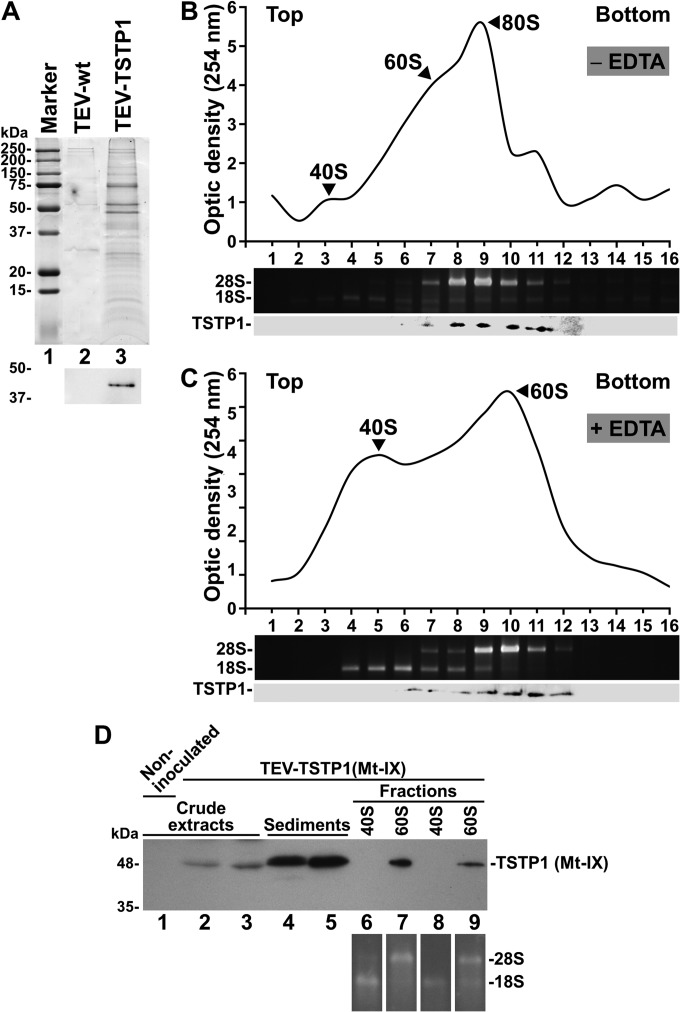
To further investigate the roles of P1 during TEV infection, we wished to identify host proteins involved in complexes in which P1 is also a component during infection. For this purpose, we inoculated N. benthamiana plants with the infectious TEV-TSTP1 clone (Fig. 1A). Control plants were also inoculated with TEV-wt. Symptomatic tissues were harvested at 5 dpi (an early stage of infection when P1 concentration in infected tissue is high) (Fig. 1E) and homogenized. The tagged P1 (TSTP1) was purified from the plant extract by liquid chromatography under native conditions using a Strep-Tactin column. This tag has previously been used to successfully purify complexes involving potyviral proteins from infected tissues (45). The same purification process was applied to an extract from tissues infected by TEV-wt as a negative control. Electrophoretic analysis of the chromatographic fractions showed enrichment of a number of proteins from the tissues infected by TEV-TSTP1 relative to those infected by TEV-wt (Fig. 8A, compare lanes 2 and 3). Moreover, the successful purification of TSTP1 was validated by Western blot analysis using an anti-TST antibody (Fig. 8A, lower panel). Purified proteins were digested in gel with trypsin, and the resulting peptides were extracted and characterized by liquid chromatography separation followed by tandem mass spectrometry analysis. To obtain a comprehensive list of host proteins actually associated to TEV P1 during infection and to eliminate possible false interactors, this analysis was performed independently on two different samples purified from two different batches of tissues infected by TEV-TSTP1 and on one sample purified from tissues infected by TEV-wt (see Table S1 in the supplemental material). Table 1 shows the refined list of 32 protein hits common to both TEV-TSTP1-infected samples and absent from the TEV-wt-infected control. Surprisingly, most of the hits corresponded to ribosomal proteins, particularly components of the cytosolic 60S ribosomal subunit (15 proteins). The list also contained two protein components of the 40S ribosomal subunit, one protein involved in translation and four chaperones. In addition, TEV P1, HC-Pro, and CP were also identified. These results suggest that TEV P1 interacts with the plant ribosome during infection.
FIG 8.
Identification of host proteins physically associated to P1 during TEV infection. TSTP1 was purified from N. benthamiana tissues infected with TEV-TSTP1 by liquid chromatography using a Strep-Tactin column under native conditions. (A) Proteins eluting from the column were precipitated, separated by SDS-PAGE, and the gel stained with Coomassie blue. Lane 1, protein standards with their molecular masses in kDa on the left; lane 2, negative control of proteins purified from tissues infected by TEV-wt; lane 3, proteins purified from tissues infected by TEV-TSTP1. The lower panel corresponds to Western blot analysis using an anti-TST antibody. (B, C, and D) Fractionation of polysomal preparations from N. benthamiana tissues infected by TEV-TSTP1 (B and C) or TEV-TSTP1 (mutant Mt-IX) (D). A polysomal preparation from tissue infected with TEV-TSTP1 was nontreated (B) or treated with EDTA (C) and subjected to centrifugation in sucrose gradients. The fractionated gradient profiles were obtained by measuring the optical density of the different fractions at 254 nm. Ribosomal 18S and 28S RNAs were detected in the fractions of the gradients by agarose electrophoresis and staining with ethidium bromide. The presence of TSTP1 was revealed by SDS-PAGE separation and Western blot analysis. The positions of the 80S ribosomes and the 60S and 40S ribosomal subunits are indicated in the profile. The positions of the viral TSTP1 and the ribosomal 28S and 18S RNAs are indicated. (D) Two independent polysomal preparations from tissues infected with TEV-TSTP1 (Mt-IX) were treated with EDTA and fractionated by centrifugation in sucrose gradients. Ribosomal RNAs and TSTP1 (Mt-IX) were detected as indicated for panels B and C. Lanes 1 to 3, crude extracts from a noninoculated plant and two independent plants infected with TEV-TSTP1 (Mt-IX), respectively; lanes 4 and 5, high-speed sediments previous to fractionation from both infected tissues; lanes 6 to 9, peak fractions with the ribosomal 40S (lanes 6 and 8) and 60S (lanes 7 and 9) subunits for both infected samples. The positions of the viral TSTP1 (Mt-IX) and the ribosomal 28S and 18S RNAs are indicated.
TABLE 1.
Refined list of identified host and viral proteins associated with TEV P1 protein in N. benthamiana infected tissue
| Functional category | Protein hit(s)a |
|---|---|
| Structural constituent of ribosome (60S) | Ribosomal proteins L3, L4, L6, L7, L10, L11, L13, L18, L21, L23, L24, L27, L32, L35, and P0 |
| Structural constituent of ribosome (40S) | Ribosomal proteins S6 and S23 |
| Translation | G-binding protein |
| Protein folding | HSP70-2, HSP70-3, CPHSC70-2, and BiP5 |
| Structural constituent of cytoskeleton | Actin, alpha and beta tubulin |
| Photosystem | ATP synthase, alpha and beta subunits |
| Metabolic process | GAPDH |
| Response to stress | Tudor-SNb |
| Viral | TEV P1, HC-Pro, and CP |
Scores, GenBank accession numbers, and full names of the proteins are in Table S1 in the supplemental material.
Tudor-SN, Tudor and staphylococcal nuclease domains.
To test this hypothesis, we purified polysomes from N. benthamiana tissues infected by TEV-TSTP1. The polysome preparation was either treated or not with EDTA to separate the ribosomal subunits and subsequently centrifuged on sucrose gradients (39, 46). After centrifugation, the gradients were fractionated, and the optical densities at 254 nm of the different fractions were measured. The presence of the two large ribosomal RNAs (18S and 28S) in the gradient fractions was verified by an electrophoretic analysis in an agarose gel, followed by ethidium bromide staining. Finally, the presence of TSTP1 in the fractions was examined by SDS-PAGE and Western blot analysis. When the polysome preparation was not treated with EDTA, TSTP1 was mainly detected in the fractions containing the complete 80S ribosomes (Fig. 8B). When the preparation was treated with EDTA, TSTP1 was exclusively detected in the fractions containing the 60S ribosomal subunit (Fig. 8C). These results support the physical association of TEV P1 with the large subunit of cytosolic ribosomes in actively translating ribosomes during the infectious process.
We investigated whether the nucleolar localization of P1 is required for binding the host 60S ribosome subunits. To this end, we inserted the mutations that abolish nucleolar localization of TEV P1 (Fig. 6A and B, construct IX) into the TEV-TSTP1 infectious clone and infected plants. Ribosomal 60S subunits were purified from infected tissue at 5 dpi by sucrose gradient centrifugation. SDS-PAGE and Western blot analysis of the fractions showed that P1 mutant IX is still able to specifically bind the ribosomal 60S subunits (Fig. 8D, Mt-IX), indicating that lack of nucleolar localization does not preclude P1 binding of the host 60S particles.
Effect of P1 on protein translation in vitro.
The results shown above may suggest an unexpected role of TEV P1 in protein translation during the infectious cycle. To get some insight into this hypothesis, we performed in vitro translation assays. We tried to produce recombinant TEV P1 in Escherichia coli to study its effect on in vitro translation, but the expressed protein accumulated in inclusion bodies, and we were unable to purify it as a soluble form under native conditions. For this reason, we chose a coupled in vitro transcription-translation system (47, 48) to coexpress TEV P1 or control proteins with reporter constructs to search for a possible effect of P1 on translation. In vitro translations were carried out using wheat germ extract containing bacteriophage SP6 RNA polymerase to couple in vitro transcription and translation of the desired cDNAs and with [35S]methionine to track protein synthesis. The in vitro translation extract was programmed with reporter plasmids containing the bacteriophage SP6 promoter and a poly(A) tail, designed to express the fluorescent protein Venus tagged at the amino terminus with the human influenza virus hemagglutinin tag and flanked with a modified version of CPMV RNA-2 5′ and 3′ UTRs (construct I, Fig. 9A; see also Fig. S4 in the supplemental material). We used CPMV RNA-2 5′ and 3′ UTRs because these elements very efficiently promote translation of heterologous proteins in plants, particularly the modified version of the 5′ UTR used here that contains a specific mutation eliminating an internal ATG codon (37). Fifteen minutes before the addition of the reporter plasmid, aliquots of the wheat germ extract were programmed with normalized amounts of plasmids containing expression cassettes to produce the following: (i) Fluc, taken as a negative control, (ii) a truncated version of TEV P1 (155 initial amino acids), (iii) wild-type TEV P1, (iv) wild-type TEV HC-Pro (the second protein in TEV polyprotein, though not involved in translation) initially selected as a negative control, and (v) a polyprotein consisting of TEV P1–HC-Pro. These expression cassettes consisted of the SP6 phage promoter, modified CPMV RNA-2 5′ UTR, the cDNAs coding for the corresponding protein, and CPMV RNA-2 3′ UTR followed by a poly(A) tail (Fig. 9A, constructs III to VII; see also Fig. S4 in the supplemental material). Reactions were allowed to continue for 40 more minutes. A prospective time course experiment showed that in vitro translation reactions were not saturated at this point (data not shown).
FIG 9.
Effect of TEV P1 on in vitro translation of reporter systems. (A) Schematic representation of the monocistronic and bicistronic reporter cassettes and the cassettes to express firefly luciferase (Fluc), a truncated form of TEV P1 (ΔP1; 155 initial amino acids), P1, HC-Pro and the P1–HC-Pro polyprotein. The black and gray rectangles represent the SP6 bacteriophage promoter and the poly(A) tail, respectively. Light and dark gray boxes represent Fluc and hemagglutinin-tagged Venus cDNAs, respectively. White boxes represent cDNAs corresponding to ΔP1 (the nontranslated part due to an in-frame insertion of three stop codons inserted in frame is indicated in a dashed box), P1, and HC-Pro, as indicated. Black lines represent the CPMV RNA-2 5′ and 3′ UTRs, as indicated. The TEV 5′ UTR is represented by a black rectangle, as indicated. (B) Production of Venus in in vitro translation reactions in which Fluc, TEV ΔP1, P1, HC-Pro, and P1–HC-Pro were cotranslationally produced. In vitro translation products in the presence of [35S]Met were separated by SDS-PAGE and quantified by phosphorimager analysis. Venus amounts were normalized by those obtained in the reaction cotranslating Fluc. (C) Production of upstream Fluc (light gray bars) and downstream Venus (dark gray bars) in in vitro translation reactions in which TEV ΔP1, P1, HC-Pro, and P1–HC-Pro were cotranslationally produced. The Fluc and Venus amounts were normalized by those obtained in the reaction cotranslating TEV ΔP1. In panels B and C, error bars indicate the standard deviations in three independent experiments, and the different letters over the columns indicate a significant statistical difference (least significant difference [LSD] test, P < 0.05). (D and E) Analysis of proteins and reporter mRNA produced during the coupled in vitro transcription-translation experiment. Two aliquots of the coupled in vitro transcription-translation reaction products were taken at 40 min for protein and reporter mRNA analysis. (D) Proteins were separated by PAGE (12.5% polyacrylamide, 0.05% SDS), the gel was dried, and the proteins were visualized by autoradiography. (E) RNAs were separated by electrophoresis in an agarose gel under denaturing conditions and electroblotted to a positively charged nylon membrane, and the Venus mRNA was detected with a cRNA probe labeled with 32P. Reactions were programmed to produce Venus reporter and Fluc (lane 1), TEV P1 (lane 2), a truncated form of TEV P1 (ΔP1, lane 3), TEV HC-Pro (lane 4), or the P1–HC-Pro polyprotein (lane 5). The positions of the different proteins are indicated on the left and right of panel D. Note that the P1–HC-Pro polyprotein partially self-cleaves during the reaction. The position of the Venus reporter mRNA is indicated on the right of panel E.
Three independent replicates of the experiments were performed. Translation products were separated by SDS-PAGE and quantified by phosphorimager analysis. Figure 9B and C show the effect of the expressed proteins on translation. Figure 9D shows the autoradiogram corresponding to one of the three experimental replicates. We took Fluc as a negative control to normalize the results in the three independent experiments. P1 significantly enhanced translation of the reporter protein (Fig. 9B). In contrast, the truncated version of P1 had no effect on translation (Fig. 9B). HC-Pro had an unexpected, significantly negative effect on translation that was even more intense when it was expressed as a P1–HC-Pro polyprotein (Fig. 9B). This result, remarkable by itself, showed that TEV HC-Pro possesses translation inhibition activity. In retrospect, HC-Pro was not a good choice as a negative control in these experiments. Northern blot analysis of in vitro-transcribed RNAs demonstrated that differences in reporter protein accumulation were due to differences in translation efficiency because mRNA concentration was not significantly different in the reaction mixtures (Fig. 9E). These results suggest that TEV P1 and HC-Pro have some stimulatory and inhibitory effects on translation, respectively.
TEV translation depends on the IRES activity present in the viral 5′ UTR (49). To further investigate the effect of P1 and HC-Pro proteins on viral translation, we used a bicistronic reporter construct as such constructs are usually used to analyze IRES activity (50–52). The bicistronic cassette consisted of the modified CPMV RNA-2 5′ UTR, the cDNA coding for Fluc, the TEV 5′ UTR, the cDNA coding for Venus, and the CPMV RNA-2 3′ UTR followed by a poly(A) tail (Fig. 9A, construct II; see Fig. S4 in the supplemental material). Translation of the downstream cistron (Venus) depended on the IRES activity of the TEV 5′ UTR. TEV P1 enhanced translation of upstream and downstream reporter proteins (Fig. 9C), indicating that the protein is able to stimulate translation driven by both UTRs, CPMV and TEV. In contrast, HC-Pro inhibited translation of both reporters, particularly the upstream but also the downstream promoter. Finally, when both P1 and HC-Pro were present in the reaction mixture, translation of the upstream reporter was inhibited, but interestingly the presence of P1 compensated the inhibitory effect of HC-Pro on translation of the downstream reporter. Taken together, these results suggest that TEV P1 enhances protein translation, particularly when it is driven from the viral 5′ UTR IRES, and that TEV HC-Pro inhibits translation.
DISCUSSION
In our work, we infected plants with a series of TEV recombinant clones in which the P1 protein was tagged with either a fluorescent protein to track P1 expression and localization during infection or with an affinity tag for specific purification under native conditions of protein complexes in which P1 is also a component. The proteomic analysis of these complexes fundamentally identified proteins from the ribosome 60S subunit (Table 1) and also some proteins from the 40S subunit, such as ribosomal protein S6 previously shown to be strictly required for turnip mosaic potyvirus infection (53). Analysis of polysomes purified from infected tissues corroborated the physical association between TEV P1 and the plant cytoplasmic 80S ribosomes during infection (Fig. 8B) and, more specifically, with the 60S ribosomal subunits (Fig. 8C). In addition to proteins involved in translation, some other host proteins were also identified in association with TEV P1 in infected tissues (Table 1). It is worth noting heat shock protein 70 chaperones, previously shown associated to potyviral replication complexes (41, 45), and the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) involved in replication of other positive-stand RNA viruses, like tomato bushy stunt tombusvirus (54).
Our experiments also demonstrate that TEV P1 is a nucleocytoplasmic protein that displays dynamic intracellular localization during the infectious cycle (Fig. 3). At an early stage, the protein enters the nuclei and targets the nucleoli of infected cells. Later, P1 protein is exported from the nucleus to cytoplasm. These findings are supported by our identification of functional nucleolar localization and nuclear export signals in TEV P1 (Fig. 5 and 6). Moreover, our analysis of TEV mutants in the P1 cistron showed the in vivo relevance of these signals (Fig. 7). Inside the nucleolus, TEV P1 specifically localizes in the granular component, where final processing of preribosomal particles takes place (55). In fact, TEV P1 nucleolar localization is similar to that of a genuine ribosomal protein (Fig. 4): A. thaliana RPL24B, a structural component of 60S ribosomal subunits. RNA viruses, including plant viruses, interact with the nucleolus to usurp host cell functions, diverting nucleolar proteins to perform novel roles in the virus infectious cycle (56, 57). Potyviral NIa protein, more specifically the VPg domain, also contains nuclear and nucleolar localization signals. And nuclear and nucleolar localization of potato potyvirus A NIa was demonstrated to be essential for completion of the infectious cycle (58).
These findings suggest that potyviral P1 protein may play an unsuspected role in translation during the infectious cycle. TEV genomic RNA differs from a conventional host mRNA because it does not contain a 5′ cap structure. In addition, the TEV 5′ UTR has been shown to possess IRES activity (49, 59). The rate-limiting step in the initiation of translation of eukaryotic mRNA is the recognition of cap structure by eIF4E, the small subunit of eIF4F. In cap-independent translation of TEV, recruitment of the translation preinitiation complex is not required because the viral IRES is able to directly bind the eIF4G and facilitates binding of the 40S subunit through interaction with other initiation factors (59–61). Sequences in the 3′ UTR of the pea enation mosaic tombusvirus have been shown to bind host ribosomal subunits assisting cap-independent translation of the viral mRNA (62). A model that may explain our experimental data is that, in the early stage of infection, potyviral P1 associates to the plant 60S ribosomal subunits to make them more competent in viral translation. This association may occur in the nucleolus during biogenesis of preribosomal particles, but not necessarily because a TEV P1 mutation (Mt-IX) that abolishes nucleolar localization of the protein (Fig. 6B, construct IX) does not preclude binding to ribosomal 60S subunits during infection (Fig. 8D). P1 may mediate the recruitment of the 60S subunits to the viral translation initiation complex. CaMV P6 protein has also been shown to localize in the nucleolus and interact with structural components of the 60S ribosome subunit (46, 63, 64). CaMV P6 is thought to transactivate translation of the viral polycistronic pregenomic RNA and its spliced versions. Recently, potyviral VPg has also been implicated in viral translation. Experiments based on transient expression of potato potyvirus A VPg in N. benthamiana plants have suggested that this viral protein stimulates translation of the viral mRNA and, at the same time, represses translation of the host mRNAs (11).
In support of the above model, in vitro expressed TEV P1 stimulated translation of reporter proteins (Fig. 9). These experiments led to the serendipitous observation that TEV HC-Pro inhibited translation (Fig. 9). This inhibition may result from the recently reported eIF4E and eIF(iso)4E binding activity of potyviral HC-Pro that may prevent the formation of functional cap-dependent translation complexes required for host protein production, resulting in selective inhibition of host cell protein synthesis (65). By incorporating the inhibitory effect of potyviral HC-Pro to our model, the coordinated action of P1 and HC-Pro during infection may stimulate IRES-dependent translation of the viral mRNA and suppress the cap-dependent translation of the host mRNAs. However, it should be taken into consideration that support for the model comes from in vitro translation experiments using RNAs with naked 5′ ends, which may not accurately reflect the situation occurring in vivo during virus infection.
When a potyvirus first enters a plant cell, after de-encapsidation, the viral genomic RNA needs to be efficiently translated to produce the proteins that will take part in the different steps of the infectious cycle. The virus may gain translation efficiency in a critical early stage of infection by the stimulating effect of viral P1. This is consistent with our observation that TEV P1 accumulates transitorily at the early stage of infection (Fig. 1). Since gene expression in potyviruses produces equimolar amounts of most proteins, this observation suggests a protein degradation mechanism to regulate P1 expression. The role we propose for potyviral P1 in our model is compatible with previous observations indicating that this cistron is dispensable for viral infection and that null P1 mutants can be rescued by expressing P1 protein in trans. Nonetheless, many viral proteins are multifunctional, and P1 most probably plays multiple roles during infection. A recent work shows that the hypervariable amino terminus of plum pox potyvirus P1 protein modulates replication and the host defense response (66). Also, a transient suppression of host gene expression was reported exclusively at the infection front of pea seed-borne mosaic potyvirus (67). As P1 shows the same accumulation dynamics at the infection front, this protein may also be related to this process.
Supplementary Material
ACKNOWLEDGMENTS
We thank Verónica Aragonés and Teresa Cordero for excellent technical assistance. We thank C. Douglas Grubb (Leibniz-Institut für Pflanzenbiochemie, Germany) for critical review of the manuscript. Proteomic analysis was performed in the proteomics laboratory of Centro de Investigación Príncipe Felipe de Valencia, a member of Spanish ProteoRed.
This work was supported by grant BIO2011-26741 from the Spanish Ministerio de Economía y Competitividad. F.M. was the recipient of a predoctoral fellowship from Universidad Politécnica de Valencia.
Footnotes
Published ahead of print 2 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00928-14.
REFERENCES
- 1.Riechmann JL, Laín S, García JA. 1992. Highlights and prospects of potyvirus molecular biology. J. Gen. Virol. 73:1–16. 10.1099/0022-1317-73-1-1 [DOI] [PubMed] [Google Scholar]
- 2.Chung BYW, Miller WA, Atkins JF, Firth AE. 2008. An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. U. S. A. 105:5897–5902. 10.1073/pnas.0800468105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Urcuqui-Inchima S, Haenni AL, Bernardi F. 2001. Potyvirus proteins: a wealth of functions. Virus Res. 74:157–175. 10.1016/S0168-1702(01)00220-9 [DOI] [PubMed] [Google Scholar]
- 4.Kasschau KD, Carrington JC. 1998. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95:461–470. 10.1016/S0092-8674(00)81614-1 [DOI] [PubMed] [Google Scholar]
- 5.Guo B, Lin J, Ye K. 2011. Structure of the autocatalytic cysteine protease domain of potyvirus helper-component proteinase. J. Biol. Chem. 286:21937–21943. 10.1074/jbc.M111.230706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei T, Zhang C, Hong J, Xiong R, Kasschau KD, Zhou X, Carrington JC, Wang A. 2010. Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N-PIPO. PLoS Pathog. 6:e1000962. 10.1371/journal.ppat.1000962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaad MC, Jensen PE, Carrington JC. 1997. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 16:4049–4059. 10.1093/emboj/16.13.4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei T, Zhang C, Hou X, Sanfacon H, Wang A. 2013. The SNARE protein Syp71 is essential for turnip mosaic virus infection by mediating fusion of virus-induced vesicles with chloroplasts. PLoS Pathog. 9:e1003378. 10.1371/journal.ppat.1003378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puustinen P, Mäkinen K. 2004. Uridylylation of the potyvirus VPg by viral replicase NIb correlates with the nucleotide binding capacity of VPg. J. Biol. Chem. 279:38103–38110. 10.1074/jbc.M402910200 [DOI] [PubMed] [Google Scholar]
- 10.Charron C, Nicolaï M, Gallois JL, Robaglia C, Moury B, Palloix A, Caranta C. 2008. Natural variation and functional analyses provide evidence for co-evolution between plant eIF4E and potyviral VPg. Plant J. 54:56–68. 10.1111/j.1365-313X.2008.03407.x [DOI] [PubMed] [Google Scholar]
- 11.Eskelin K, Hafrén A, Rantalainen KI, Mäkinen K. 2011. Potyviral VPg enhances viral RNA Translation and inhibits reporter mRNA translation in planta. J. Virol. 85:9210–9221. 10.1128/JVI.00052-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carrington JC, Dougherty WG. 1987. Small nuclear inclusion protein encoded by a plant potyvirus genome is a protease. J. Virol. 61:2540–2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daròs JA, Schaad MC, Carrington JC. 1999. Functional analysis of the interaction between VPg-proteinase (NIa) and RNA polymerase (NIb) of tobacco etch potyvirus, using conditional and suppressor mutants. J. Virol. 73:8732–8740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong Y, Hunt AG. 1996. RNA polymerase activity catalyzed by a potyvirus-encoded RNA-dependent RNA polymerase. Virology 226:146–151. 10.1006/viro.1996.0639 [DOI] [PubMed] [Google Scholar]
- 15.López-Moya JJ, Wang RY, Pirone TP. 1999. Context of the coat protein DAG motif affects potyvirus transmissibility by aphids. J. Gen. Virol. 80:3281–3288 [DOI] [PubMed] [Google Scholar]
- 16.Dolja VV, Haldeman-Cahill R, Montgomery AE, Vandenbosch KA, Carrington JC. 1995. Capsid protein determinants involved in cell-to-cell and long distance movement of tobacco etch potyvirus. Virology 206:1007–1016. 10.1006/viro.1995.1023 [DOI] [PubMed] [Google Scholar]
- 17.Rohozková J, Navrátil M. 2011. P1 peptidase—a mysterious protein of family Potyviridae. J. Biosci. 36:189–200. 10.1007/s12038-011-9020-6 [DOI] [PubMed] [Google Scholar]
- 18.Valli A, López-Moya JJ, García JA. 2007. Recombination and gene duplication in the evolutionary diversification of P1 proteins in the family Potyviridae. J. Gen. Virol. 88:1016–1028. 10.1099/vir.0.82402-0 [DOI] [PubMed] [Google Scholar]
- 19.Verchot J, Koonin EV, Carrington JC. 1991. The 35-kDa protein from the N terminus of the potyviral polyprotein functions as a third virus-encoded proteinase. Virology 185:527–535. 10.1016/0042-6822(91)90522-D [DOI] [PubMed] [Google Scholar]
- 20.Soumounou Y, Laliberté JF. 1994. Nucleic acid-binding properties of the P1 protein of turnip mosaic potyvirus produced in Escherichia coli. J. Gen. Virol. 75:2567–2573. 10.1099/0022-1317-75-10-2567 [DOI] [PubMed] [Google Scholar]
- 21.Brantley JD, Hunt AG. 1993. The N-terminal protein of the polyprotein encoded by the potyvirus tobacco vein mottling virus is an RNA-binding protein. J. Gen. Virol. 74:1157–1162. 10.1099/0022-1317-74-6-1157 [DOI] [PubMed] [Google Scholar]
- 22.Verchot J, Carrington JC. 1995. Evidence that the potyvirus P1 proteinase functions in trans as an accessory factor for genome amplification. J. Virol. 69:3668–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verchot J, Carrington JC. 1995. Debilitation of plant potyvirus infectivity by P1 proteinase-inactivating mutations and restoration by second-site modifications. J. Virol. 69:1582–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajamäki ML, Kelloniemi J, Alminaite A, Kekarainen T, Rabenstein F, Valkonen JP. 2005. A novel insertion site inside the potyvirus P1 cistron allows expression of heterologous proteins and suggests some P1 functions. Virology 342:88–101. 10.1016/j.virol.2005.07.019 [DOI] [PubMed] [Google Scholar]
- 25.Valli A, Martín-Hernández AM, López-Moya JJ, García JA. 2006. RNA silencing suppression by a second copy of the P1 serine protease of Cucumber vein yellowing ipomovirus, a member of the family Potyviridae that lacks the cysteine protease HCPro. J. Virol. 80:10055–10063. 10.1128/JVI.00985-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tena Fernández F, González I, Doblas P, Rodríguez C, Sahana N, Kaur H, Tenllado F, Praveen S, Canto T. 2013. The influence of cis-acting P1 protein and translational elements on the expression of Potato virus Y helper-component proteinase (HCPro) in heterologous systems and its suppression of silencing activity. Mol. Plant Pathol. 14:530–541. 10.1111/mpp.12025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salvador B, Saénz P, Yangüez E, Quiot JB, Quiot L, Delgadillo MO, García JA, Simón-Mateo C. 2008. Host-specific effect of P1 exchange between two potyviruses. Mol. Plant Pathol. 9:147–155. 10.1111/j.1364-3703.2007.00450.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bedoya LC, Martínez F, Orzáez D, Daròs JA. 2012. Visual tracking of plant virus infection and movement using a reporter MYB transcription factor that activates anthocyanin biosynthesis. Plant Physiol. 158:1130–1138. 10.1104/pp.111.192922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20:87–90. 10.1038/nbt0102-87 [DOI] [PubMed] [Google Scholar]
- 30.Subach OM, Gundorov IS, Yoshimura M, Subach FV, Zhang J, Grüenwald D, Souslova EA, Chudakov DM, Verkhusha VV. 2008. Conversion of red fluorescent protein into a bright blue probe. Chem. Biol. 15:1116–1124. 10.1016/j.chembiol.2008.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt TG, Skerra A. 2007. The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat. Protoc. 2:1528–1535. 10.1038/nprot.2007.209 [DOI] [PubMed] [Google Scholar]
- 32.Bedoya LC, Daròs JA. 2010. Stability of Tobacco etch virus infectious clones in plasmid vectors. Virus Res. 149:234–240. 10.1016/j.virusres.2010.02.004 [DOI] [PubMed] [Google Scholar]
- 33.Nohales MA, Flores R, Daròs JA. 2012. Viroid RNA redirects host DNA ligase 1 to act as an RNA ligase. Proc. Natl. Acad. Sci. U. S. A. 109:13805–13810. 10.1073/pnas.1206187109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barneche F, Steinmetz F, Echeverría M. 2000. Fibrillarin genes encode both a conserved nucleolar protein and a novel small nucleolar RNA involved in ribosomal RNA methylation in Arabidopsis thaliana. J. Biol. Chem. 275:27212–27220. 10.1074/jbc.M002996200 [DOI] [PubMed] [Google Scholar]
- 35.Kim SH, Ryabov EV, Kalinina NO, Rakitina DV, Gillespie T, MacFarlane S, Haupt S, Brown JW, Taliansky M. 2007. Cajal bodies and the nucleolus are required for a plant virus systemic infection. EMBO J. 26:2169–2179. 10.1038/sj.emboj.7601674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pendle AF, Clark GP, Boon R, Lewandowska D, Lam YW, Andersen J, Mann M, Lamond AI, Brown JW, Shaw PJ. 2005. Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol. Biol. Cell 16:260–269. 10.1091/mbc.E04-09-0791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sainsbury F, Lomonossoff GP. 2008. Extremely high-level and rapid transient protein production in plants without the use of viral replication. Plant Physiol. 148:1212–1218. 10.1104/pp.108.126284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bedoya L, Martínez F, Rubio L, Daròs JA. 2010. Simultaneous equimolar expression of multiple proteins in plants from a disarmed potyvirus vector. J. Biotechnol. 150:268–275. 10.1016/j.jbiotec.2010.08.006 [DOI] [PubMed] [Google Scholar]
- 39.Jackson AO, Larkins BA. 1976. Influence of ionic strength, pH, and chelation of divalent metals on isolation of polyribosomes from tobacco leaves. Plant Physiol. 57:5–10. 10.1104/pp.57.1.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shevchenko A, Jensen ON, Podtelejnikov AV, Sagliocco F, Wilm M, Vorm O, Mortensen P, Boucherie H, Mann M. 1996. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc. Natl. Acad. Sci. U. S. A. 93:14440–14445. 10.1073/pnas.93.25.14440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dufresne PJ, Thivierge K, Cotton S, Beauchemin C, Ide C, Ubalijoro E, Laliberté JF, Fortin MG. 2008. Heat shock 70 protein interaction with Turnip mosaic virus RNA-dependent RNA polymerase within virus-induced membrane vesicles. Virology 374:217–227. 10.1016/j.virol.2007.12.014 [DOI] [PubMed] [Google Scholar]
- 42.Wei T, Huang TS, McNeil J, Laliberté JF, Hong J, Nelson RS, Wang A. 2010. Sequential recruitment of the endoplasmic reticulum and chloroplasts for plant potyvirus replication. J. Virol. 84:799–809. 10.1128/JVI.01824-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scott MS, Troshin PV, Barton GJ. 2011. NoD: a nucleolar localization sequence detector for eukaryotic and viral proteins. BMC Bioinformatics 12:317. 10.1186/1471-2105-12-317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.la Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K, Brunak S. 2004. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 17:527–536. 10.1093/protein/gzh062 [DOI] [PubMed] [Google Scholar]
- 45.Hafrén A, Hofius D, Rönnholm G, Sonnewald U, Mäkinen K. 2010. HSP70 and its cochaperone CPIP promote potyvirus infection in Nicotiana benthamiana by regulating viral coat protein functions. Plant Cell 22:523–535. 10.1105/tpc.109.072413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park HS, Himmelbach A, Browning KS, Hohn T, Ryabova LA. 2001. A plant viral “reinitiation” factor interacts with the host translational machinery. Cell 106:723–733. 10.1016/S0092-8674(01)00487-1 [DOI] [PubMed] [Google Scholar]
- 47.Ren Q, Wang QS, Firth AE, Chan MM, Gouw JW, Guarna MM, Foster LJ, Atkins JF, Jan E. 2012. Alternative reading frame selection mediated by a tRNA-like domain of an internal ribosome entry site. Proc. Natl. Acad. Sci. U. S. A. 109:E630–E639. 10.1073/pnas.1111303109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koh DC, Wong SM, Liu DX. 2003. Synergism of the 3′-untranslated region and an internal ribosome entry site differentially enhances the translation of a plant virus coat protein. J. Biol. Chem. 278:20565–20573. 10.1074/jbc.M210212200 [DOI] [PubMed] [Google Scholar]
- 49.Carrington JC, Freed DD. 1990. Cap-independent enhancement of translation by a plant potyvirus 5′ nontranslated region. J. Virol. 64:1590–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niepel M, Gallie DR. 1999. Identification and characterization of the functional elements within the tobacco etch virus 5′ leader required for cap-independent translation. J. Virol. 73:9080–9088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martínez-Salas E. 1999. Internal ribosome entry site biology and its use in expression vectors. Curr. Opin. Biotechnol. 10:458–464. 10.1016/S0958-1669(99)00010-5 [DOI] [PubMed] [Google Scholar]
- 52.Hellen CU, Sarnow P. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15:1593–1612. 10.1101/gad.891101 [DOI] [PubMed] [Google Scholar]
- 53.Yang C, Zhang C, Dittman JD, Whitham SA. 2009. Differential requirement of ribosomal protein S6 by plant RNA viruses with different translation initiation strategies. Virology 390:163–173. 10.1016/j.virol.2009.05.018 [DOI] [PubMed] [Google Scholar]
- 54.Wang RY, Nagy PD. 2008. Tomato bushy stunt virus co-opts the RNA-binding function of a host metabolic enzyme for viral genomic RNA synthesis. Cell Host Microbe 3:178–187. 10.1016/j.chom.2008.02.005 [DOI] [PubMed] [Google Scholar]
- 55.Shaw P, Brown J. 2012. Nucleoli: composition, function, and dynamics. Plant Physiol. 158:44–51. 10.1104/pp.111.188052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hiscox JA. 2007. RNA viruses: hijacking the dynamic nucleolus. Nat. Rev. Microbiol. 5:119–127. 10.1038/nrmicro1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taliansky ME, Brown JW, Rajamäki ML, Valkonen JP, Kalinina NO. 2010. Involvement of the plant nucleolus in virus and viroid infections: parallels with animal pathosystems. Adv. Virus Res. 77:119–158. 10.1016/B978-0-12-385034-8.00005-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rajamäki ML, Valkonen JP. 2009. Control of nuclear and nucleolar localization of nuclear inclusion protein a of picorna-like Potato virus A in Nicotiana species. Plant Cell 21:2485–2502. 10.1105/tpc.108.064147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khan MA, Yumak H, Goss DJ. 2009. Kinetic mechanism for the binding of eIF4F and tobacco Etch virus internal ribosome entry site RNA: effects of eIF4B and poly(A)-binding protein. J. Biol. Chem. 284:35461–35470. 10.1074/jbc.M109.038463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ray S, Yumak H, Domashevskiy A, Khan MA, Gallie DR, Goss DJ. 2006. Tobacco etch virus mRNA preferentially binds wheat germ eukaryotic initiation factor (eIF) 4G rather than eIFiso4G. J. Biol. Chem. 281:35826–35834. 10.1074/jbc.M605762200 [DOI] [PubMed] [Google Scholar]
- 61.Gallie DR. 2001. Cap-independent translation conferred by the 5′ leader of tobacco etch virus is eukaryotic initiation factor 4G dependent. J. Virol. 75:12141–12152. 10.1128/JVI.75.24.12141-12152.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gao F, Gulay SP, Kasprzak W, Dinman JD, Shapiro BA, Simon AE. 2013. The kissing-loop T-shaped structure translational enhancer of Pea Enation Mosaic Virus can bind simultaneously to ribosomes and a 5′ proximal hairpin. J. Virol. 87:11987–12002. 10.1128/JVI.02005-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bureau M, Leh V, Haas M, Geldreich A, Ryabova L, Yot P, Keller M. 2004. P6 protein of Cauliflower mosaic virus, a translation reinitiator, interacts with ribosomal protein L13 from Arabidopsis thaliana. J. Gen. Virol. 85:3765–3775. 10.1099/vir.0.80242-0 [DOI] [PubMed] [Google Scholar]
- 64.Haas M, Geldreich A, Bureau M, Dupuis L, Leh V, Vetter G, Kobayashi K, Hohn T, Ryabova L, Yot P, Keller M. 2005. The open reading frame VI product of Cauliflower mosaic virus is a nucleocytoplasmic protein: its N terminus mediates its nuclear export and formation of electron-dense viroplasms. Plant Cell 17:927–943. 10.1105/tpc.104.029017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ala-Poikela M, Goytia E, Haikonen T, Rajamäki ML, Valkonen JP. 2011. Helper component proteinase of the genus Potyvirus is an interaction partner of translation initiation factors eIF(iso)4E and eIF4E and contains a 4E binding motif. J. Virol. 85:6784–6794. 10.1128/JVI.00485-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pasín F, Simón-Mateo C, García JA. 2014. The hypervariable amino terminus of P1 protease modulates potyviral replication and host defense responses. PLoS Pathog. 10:e1003985. 10.1371/journal.ppat.1003985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang D, Maule AJ. 1995. Inhibition of host gene expression associated with plant virus replication. Science 267:229–231. 10.1126/science.267.5195.229 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.