Abstract
An essential step during the intracellular life cycle of many positive-strand RNA viruses is the rearrangement of host cell membranes to generate membrane-bound replication platforms. For example, Nidovirales and Flaviviridae subvert the membrane of the endoplasmic reticulum (ER) for their replication. However, the absence of conventional ER and secretory pathway markers in virus-induced ER-derived membranes has for a long time hampered a thorough understanding of their biogenesis. Recent reports highlight the analogies between mouse hepatitis virus-, equine arteritis virus-, and Japanese encephalitis virus-induced replication platforms and ER-associated degradation (ERAD) tuning vesicles (or EDEMosomes) that display nonlipidated LC3 at their cytosolic face and segregate the ERAD factors EDEM1, OS-9, and SEL1L from the ER lumen. In this Gem, we briefly summarize the current knowledge on ERAD tuning pathways and how they might be hijacked for viral genome replication. As ERAD tuning components, such as SEL1L and nonlipidated LC3, appear to contribute to viral infection, these cellular pathways represent novel candidate drug targets to combat positive-strand RNA viruses.
QUALITY CONTROL OPERATING IN THE ENDOPLASMIC RETICULUM
Secreted and membrane proteins are synthesized, folded, and assembled in the endoplasmic reticulum (ER). Acquisition of the native protein structure is assisted by a broad spectrum of resident molecular chaperones and folding enzymes that catalyze rate-limiting reactions, such as the formation of the correct configuration of disulfide and peptidyl-prolyl bonds. A dedicated quality control system ensures that only correctly folded and assembled proteins leave the ER and are transported along the secretory pathway to reach their final destination. Misfolded proteins are retrotranslocated (dislocated) into the cytosol, polyubiquitylated, and then degraded by 26S proteasomes, a process known as ER-associated degradation (ERAD) (1).
FOLDING AND ERAD IN THE BALANCE
The ERAD machinery can hardly distinguish nonnative intermediates of ongoing folding programs (which should be preserved) from nonnative side products of the folding process (which should be eliminated). As such, hyper-ERAD may result in loss-of-function phenotypes upon inappropriate degradation of folding intermediates, whereas hypo-ERAD may cause gain-of-toxic-function phenotypes upon accumulation of misfolded proteins. Therefore, a tight regulation of the ERAD capacity and its prompt adaptation to fluctuations in the ER cargo load is crucial to maintain cellular proteostasis. To large or prolonged variations of ER homeostasis (e.g., upon differentiation in highly secretory cells, exposure to drugs affecting sugar, calcium or redox homeostasis, widespread accumulation of misfolded polypeptides, challenges with pathogen) cells may respond by induction of the unfolded protein response (UPR) that consists in enhanced transcription/translation of ER-resident folding and degradation factors and expansion of the ER volume. Smaller or more transient variations may be dealt with by the immediate activation/inactivation of posttranslational pathways that rapidly adapt folding and ERAD activity to the cell's needs. The modulation of BiP activity by ADP-ribosylation (covered elsewhere [2]) and the client-dependent regulation of ERAD machinery assembly and function (Fig. 1, ERAD tuning) (3, 4) are examples thereof.
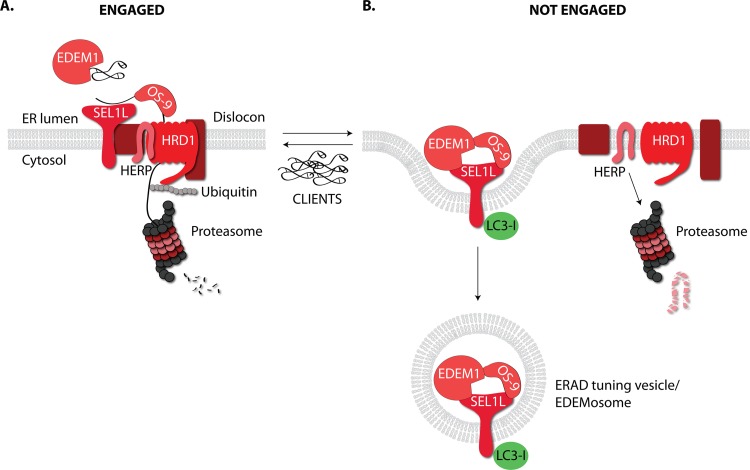
FIG 1.
ERAD tuning pathways. (A) EDEM1 and OS-9 contribute to the delivery of misfolded polypeptides to the adaptor protein SEL1L and the HRD1 dislocation machinery. The misfolded protein is dislocated across the ER membrane, polyubiquitylated, and degraded by cytosolic proteasomes. (B) HRD1 dislocons not engaged by clients are unstable. The triggering event of dislocon disassembly might be the polyubiquitylating activity of E3 ubiquitin ligases that, in the absence of client proteins, is directed to components of the dislocation machinery. The scaffold protein HERP is rapidly turned over by the proteasome. SEL1L is segregated into peripheral ER regions and/or into ERAD tuning vesicles/EDEMosomes with the associated luminal ERAD factors EDEM1 and OS-9 and the cytosolic LC3-I protein.
ERAD TUNING CONFERS ELASTICITY TO THE ERAD MACHINERY
Recent evidence has revealed that, at least in Escherichia coli and Saccharomyces cerevisiae, the synthesis of individual subunits of multimeric protein complexes is tightly controlled at the translational level (5). This so-called proportional synthesis tactic serves to restrict the energy-consuming process of protein synthesis to a minimum. In order to avoid the production of surplus complex components, the synthesis of the individual subunits is determined by the stoichiometry of the complex. This model is supported by the fact that in many cases, constituent subunits of multimeric complexes are characterized by half-lives that fall within a small range (6). In this scenario, posttranslational regulation of complex composition and function seems to play a minor role. At least in some cases, however, the level of clients of a given pathway (for example, the level of a misfolded protein that engages specific ERAD machinery) may determine the assembly, the composition, the stability, the modification, the subcellular localization and, eventually, the activity of supramolecular complexes. These posttranslational, client-regulated events might be a crucial strategy to allow more rapid, more readily reversible, and less energy-consuming responses to variations in the cellular environment than those involving changes of gene transcription/translation.
Our group recently described posttranslational mechanisms (collectively named ERAD tuning [7]) controlled by the misfolded protein load in the ER and determining the ERAD capacity (Fig. 1). An example thereof is the client-regulated assembly of the mammalian HRD1 dislocon complex, which comprises the HRD1 E3 ubiquitin ligase, SEL1L, DER1, and several other luminal, membrane, and cytosolic accessory proteins that operate to deliver ERAD substrates at the ER membrane and, eventually, to cytosolic proteasomes for degradation (Fig. 1A). This multiprotein complex contains one remarkably short-living protein, homocysteine-responsive endoplasmic reticulum-resident ubiquitin-like domain member 1 protein (HERP), which is conserved in yeast, where it regulates oligomerization and function of the HRD1 dislocon (8). Mammalian HERP has a half-life of about 1 h (9), 50 to 200 times shorter than that of most components of the HRD1 dislocation machinery and other conventional ER-resident proteins (6). Unengaged HRD1 dislocons are disassembled and HERP is rapidly turned over by the ubiquitin-proteasome system (Fig. 1B) (4). The expression of HRD1 clients maintains HRD1 dislocons in their fully assembled, functional state and inhibits the constitutively rapid HERP turnover. This autoadaptive, posttranslational mechanism preempts UPR activation upon misfolded protein accumulation by rapid enhancement of ERAD activity. The fate and the possible alternative functions of the components of unengaged and therefore disassembled HRD1 dislocons are ill-defined, with the exception of the luminal ERAD regulators EDEM1 and OS-9 that in the absence of clients and upon disassembly of the HRD1 dislocation complexes bind to SEL1L to be segregated from the ER into LC3-I-positive ER subregions or vesicles (ERAD tuning vesicles or EDEMosomes) (Fig. 1B) (3).
UNCONVENTIONAL ROLE OF NONLIPIDATED LC3 IN ERAD TUNING
LC3-I is the soluble precursor of the lipidated and membrane-associated protein LC3-II. The conversion of LC3-I to LC3-II occurs upon activation of macroautophagy. It consists of the covalent attachment of the membrane lipid phosphatidylethanolamine and promotes elongation of autophagosomal membranes (10). Much to our surprise, we found that a substantial pool of LC3-I is also associated with membranes. Unlike LC3-II, membrane association of LC3-I is noncovalent and occurs with the membranes of ERAD tuning vesicles/EDEMosomes that segregate the ERAD regulators EDEM1, OS-9, and SEL1L from the ER lumen (3, 11). ATG7, an essential autophagy gene implicated in conversion of LC3-I to LC3-II, is dispensable for ERAD tuning vesicle/EDEMosome formation, whereas small interfering RNA (siRNA)-mediated knockdown of LC3 substantially inhibits this process. Hence, the characterization of this ERAD tuning pathway revealed a novel, autophagy-independent role of nonlipidated LC3 (12).
ERAD TUNING VESICLES AS SCAFFOLD FOR POSITIVE-STRAND RNA VIRUS REPLICATION COMPLEXES
Positive-strand RNA viruses coopt intracellular membranes of the host cell in which the viral replication machineries are anchored and protected from degradative enzymes and from detection by the host's immune system. Members of the order Nidovirales induce formation of double-membrane vesicles (DMVs) and convoluted membranes (CMs) that are essential for viral replication and progeny production (13). Although ultrastructural analyses suggested that the ER is the source of the DMVs and CMs, no conventional ER markers could be detected, leaving the origin and composition of these structures mysterious.
In collaboration with the groups of Fulvio Reggiori and Cornelis de Haan, we found that mouse hepatitis virus (MHV), a coronavirus (CoV) belonging to the order Nidovirales, induces DMVs decorated with nonlipidated LC3 (14). LC3 is essential for DMV formation and viral replication, whereas components of the autophagic machinery required for LC3 lipidation are dispensable. The function of nonlipidated LC3-I during viral replication is yet to be determined and is one of the emerging nonautophagic roles of this and other autophagy factors (described elsewhere [12]).
The striking parallels between MHV DMV formation and the ERAD tuning segregation mechanism prompted us to test whether MHV replicates in ERAD tuning vesicles. Indeed, we found that EDEM1, OS-9, and SEL1L colocalize with viral double-stranded RNA and the viral proteins nsp2/3 (nonstructural proteins 2 and 3), components of the replication and transcription complexes. This suggests that MHV hijacks the ERAD tuning vesicles for replication (Fig. 2) (3, 14). Consistently, siRNA-mediated knockdown of SEL1L, an essential factor for the formation of ERAD tuning vesicles, substantially decreased MHV replication. Subsequently, equine arteritis virus (EAV) also has been reported to hijack the ERAD tuning vesicles in a manner similar to that of MHV, indicating that this mechanism may be conserved across different virus strains of the Nidovirales order (15). The strategy to subvert ERAD tuning vesicles for progeny production is not limited to Nidovirales, as it is also used by the flavivirus Japanese encephalitis virus (JEV) (16). How the viral subversion of ERAD tuning pathways affects host cell's ERAD activity and proteostasis remains to be established.
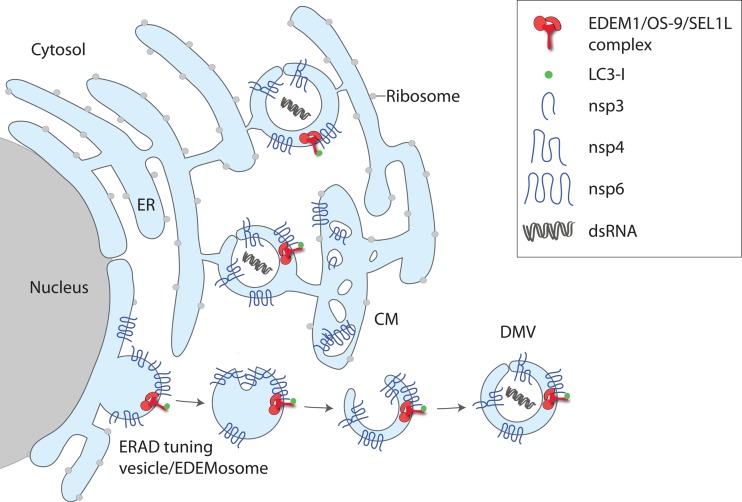
FIG 2.
Model for the subversion of the ERAD tuning pathway by MHV. The viral proteins nsp3, nsp4, and nsp6 are located in the ER membrane and induce the formation of DMVs that are interconnected with CMs in a reticular network and serve as replication platforms. As MHV-induced DMVs contain EDEM1, OS-9, and the SEL1L:LC3-I complex, they might arise from subversion of ERAD tuning vesicles/EDEMosomes by the actions of nsp3, nsp4, and nsp6. dsRNA, double-stranded RNA.
PUTATIVE ROLE OF VIRAL NONSTRUCTURAL PROTEINS IN THE SUBVERSION OF ERAD TUNING VESICLES
CoV and EAV express a set of nonstructural proteins (nsp) that are cleaved from a viral polyprotein precursor in a co- or posttranslational manner and are implicated in viral replication. As nsp3, -4, and -6 of CoVs and their EAV homologues nsp2, -3, and -5 are synthesized in the ER and are essential for DMV formation, it has been assumed that these proteins can cause ER membrane rearrangements. Individual expression of these nsp reveals DMV intermediates, such as proliferated membranes (nsp3), a vesiculated ER (nsp6), or paired ER membranes (nsp3 and nsp4) (13). Ultrastructural characterization of the paired ER membranes induced by a truncated version of nsp3 and nsp4 demonstrated that these proteins induce ER membrane curvature (17). Interestingly, the paired membrane structures form only when nsp3 and nsp4 are from the same virus strain. DMVs similar to those observed with infected cells require coexpression of CoV nsp3, -4, and -6. Thus, nsp6, which is a glycoprotein, is crucial for DMV formation. Since OS-9 and EDEM1 are sugar-binding and/or sugar-processing enzymes (18), it is possible that N-glycan-mediated interactions between MHV's nsp6 or EAV's nsp5 and the host cell ERAD factors promote DMV formation. Alternatively, nsp6 might be involved in the subversion of ERAD tuning vesicles at later stages. In this scenario, it might establish the contact between rearranged ER membranes (such as CMs) and ERAD tuning vesicles arising from the ER to form DMVs by inward budding events (19) (Fig. 2).
FUTURE PERSPECTIVES
Infections with positive-strand RNA viruses can lead to life-threatening diseases, such as severe acute respiratory syndrome (SARS), Middle East respiratory syndrome (MERS), Japanese encephalitis, and hepatitis C, to name but a few. In many cases, no FDA-approved vaccines or specific antiviral drugs for these viruses exist. The compound K22 has recently been reported to specifically inhibit MERS-CoV replication, possibly by targeting the viral protein nsp6 (20). The precise molecular mechanisms of K22 action remain to be established, but an appealing possibility is that K22 interferes with the viral subversion of ERAD tuning mechanisms. The viral subversion of ERAD tuning is yet another example of how pathogens may hijack host cell pathways to generate their progeny (or to escape immunosurveillance). For these reasons, it is of great interest to understand in molecular detail the cell invasion strategies adopted by these and other viruses.
ACKNOWLEDGMENTS
M.M. is supported by Signora Alessandra, the Foundation for Research on Neurodegenerative Diseases, the Swiss National Science Foundation, and the Comel, Gabriele, and Gelu Foundations.
Footnotes
Published ahead of print 2 July 2014
REFERENCES
- 1.Brodsky JL. 2012. Cleaning up: ER-associated degradation to the rescue. Cell 151:1163–1167. 10.1016/j.cell.2012.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chambers JE, Petrova K, Tomba G, Vendruscolo M, Ron D. 2012. ADP ribosylation adapts an ER chaperone response to short-term fluctuations in unfolded protein load. J. Cell Biol. 198:371–385. 10.1083/jcb.201202005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernasconi R, Galli C, Noack J, Bianchi S, de Haan CA, Reggiori F, Molinari M. 2012. Role of the SEL1L:LC3-I complex as an ERAD tuning receptor in the mammalian ER. Mol. Cell 46:809–819. 10.1016/j.molcel.2012.04.017 [DOI] [PubMed] [Google Scholar]
- 4.Bernasconi R, Galli C, Kokame K, Molinari M. 2013. Autoadaptive ER-associated degradation defines a preemptive unfolded protein response pathway. Mol. Cell 52:783–793. 10.1016/j.molcel.2013.10.016 [DOI] [PubMed] [Google Scholar]
- 5.Li GW, Burkhardt D, Gross C, Weissman JS. 2014. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157:624–635. 10.1016/j.cell.2014.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cambridge SB, Gnad F, Nguyen C, Bermejo JL, Kruger M, Mann M. 2011. Systems-wide proteomic analysis in mammalian cells reveals conserved, functional protein turnover. J. Proteome Res. 10:5275–5284. 10.1021/pr101183k [DOI] [PubMed] [Google Scholar]
- 7.Bernasconi R, Molinari M. 2011. ERAD and ERAD tuning: disposal of cargo and of ERAD regulators from the mammalian ER. Curr. Opin. Cell Biol. 23:176–183. 10.1016/j.ceb.2010.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horn SC, Hanna J, Hirsch C, Volkwein C, Schutz A, Heinemann U, Sommer T, Jarosch E. 2009. Usa1 functions as a scaffold of the HRD-ubiquitin ligase. Mol. Cell 36:782–793. 10.1016/j.molcel.2009.10.015 [DOI] [PubMed] [Google Scholar]
- 9.Hori O, Ichinoda F, Yamaguchi A, Tamatani T, Taniguchi M, Koyama Y, Katayama T, Tohyama M, Stern DM, Ozawa K, Kitao Y, Ogawa S. 2004. Role of Herp in the endoplasmic reticulum stress response. Genes Cells 9:457–469. 10.1111/j.1356-9597.2004.00735.x [DOI] [PubMed] [Google Scholar]
- 10.Mizushima N. 2007. Autophagy: process and function. Genes Dev. 21:2861–2873. 10.1101/gad.1599207 [DOI] [PubMed] [Google Scholar]
- 11.Cali T, Galli C, Olivari S, Molinari M. 2008. Segregation and rapid turnover of EDEM1 by an autophagy-like mechanism modulates standard ERAD and folding activities. Biochem. Biophys. Res. Commun. 371:405–410. 10.1016/j.bbrc.2008.04.098 [DOI] [PubMed] [Google Scholar]
- 12.Noack J, Bernasconi R, Molinari M. 2014. Non-lipidated LC3 is essential for mouse hepatitis virus infection, p 129–136 In Hayat MA. (ed), Autophagy: cancer, other pathologies, inflammation, immunity, infection and aging, vol 2 Academic Press, Amsterdam, Netherlands [Google Scholar]
- 13.Angelini MM, Neuman BW, Buchmeier MJ. 2014. Untangling membrane rearrangement in the nidovirales. DNA Cell Biol. 33:122–127. 10.1089/dna.2013.2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reggiori F, Monastyrska I, Verheije MH, Cali T, Ulasli M, Bianchi S, Bernasconi R, de Haan CA, Molinari M. 2010. Coronaviruses hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe 7:500–508. 10.1016/j.chom.2010.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monastyrska I, Ulasli M, Rottier PJ, Guan JL, Reggiori F, de Haan CA. 2013. An autophagy-independent role for LC3 in equine arteritis virus replication. Autophagy 9:164–174. 10.4161/auto.22743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma M, Bhattacharyya S, Nain M, Kaur M, Sood V, Gupta V, Khasa R, Abdin M, Vrati S, Kalia M. 2014. Japanese encephalitis virus replication is negatively regulated by autophagy and occurs on LC3-I- and EDEM1-containing membranes. Autophagy 10(9):53–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagemeijer MC, Monastyrska I, Griffith J, van der Sluijs P, Voortman J, van Bergen En Henegouwen PM, Vonk AM, Rottier PJM, Reggiori F, de Haan CA. 2014. Membrane rearrangements mediated by coronavirus nonstructural proteins 3 and 4. Virology 458–459:125–135. 10.1016/j.virol.2014.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aebi M, Bernasconi R, Clerc S, Molinari M. 2010. N-Glycan structures: recognition and processing in the ER. Trends Biochem. Sci. 35:74–82. 10.1016/j.tibs.2009.10.001 [DOI] [PubMed] [Google Scholar]
- 19.de Haan CA, Molinari M, Reggiori F. 2010. Autophagy-independent LC3 function in vesicular traffic. Autophagy 6:994–996. 10.4161/auto.6.7.13309 [DOI] [PubMed] [Google Scholar]
- 20.Lundin A, Dijkman R, Bergstrom T, Kann N, Adamiak B, Hannoun C, Kindler E, Jonsdottir HR, Muth D, Kint J, Forlenza M, Muller MA, Drosten C, Thiel V, Trybala E. 2014. Targeting membrane-bound viral RNA synthesis reveals potent inhibition of diverse coronaviruses including the middle East respiratory syndrome virus. PLoS Pathog. 10:e1004166. 10.1371/journal.ppat.1004166 [DOI] [PMC free article] [PubMed] [Google Scholar]