The blood group Vel was discovered 60 years ago1, but the underlying gene is unknown. Individuals negative for the Vel antigen are rare and are required for the safe transfusion of patients with immune Vel antibodies2. To identify the gene we sequenced the exome of 5 individuals negative for the Vel-antigen and found that four were homozygous and one was heterozygous for a low-frequency 17-nucleotide frameshift deletion in the gene encoding the 78 amino-acid transmembrane protein SMIM1. A follow-up study showing that 59 of 64 Vel-negative individuals were homozygous for the same deletion and expression of the Vel antigen on SMIM1-transfected cells confirm SMIM1 as the gene underlying the Vel blood group. An expression-quantitative trait locus (eQTL), the common SNP rs1175550, contributes to variable expression of the Vel antigen (P=0.003) and influences the mean hemoglobin concentration of red blood cells (P=8.6×10−15, ref 3). In vivo zebrafish smim1 knock down studies showed a mild reduction in the number of red blood cells, identifying SMIM1 as a novel regulator of red blood cell formation. Our findings are of immediate relevance as homozygous presence of the deletion allows the unequivocal identification of Vel-negative blood donors.
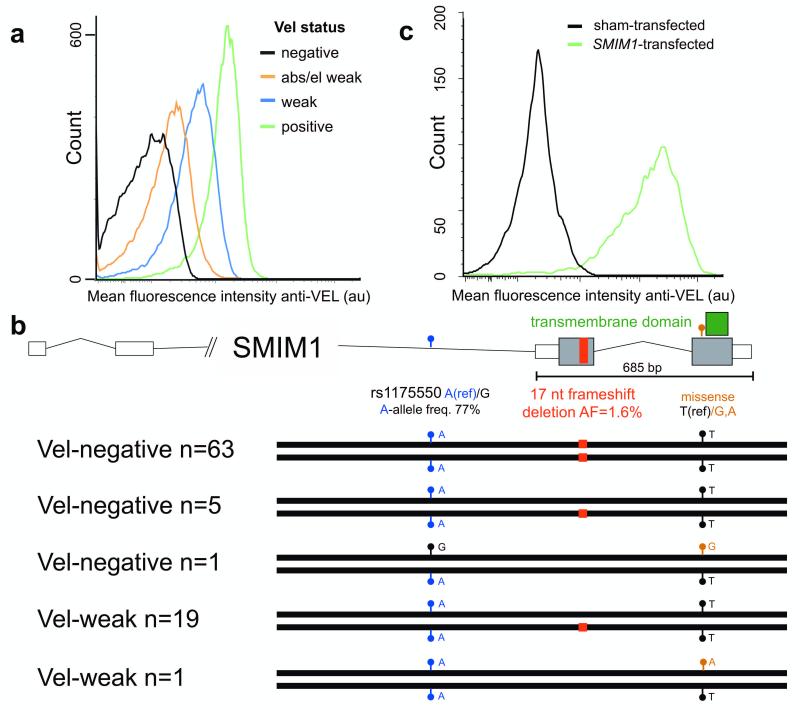
We screened nearly 350,000 blood donors for absence of expression of the Vel-antigen, showing less than 1 in 4000 to be negative (see Supplementary Fig. 1). Establishing the absence of the Vel antigen is challenging due to its low and variable abundance (Fig. 1a). To identify the gene underlying the Vel blood group, we sequenced 5 individuals negative for the Vel antigen (see Supplementary Fig. 1) on the Illumina HiSeq2000 platform following targeted enrichment with the Roche Nimblegen SeqCap EZ Human Exome v3.0 protocol (Online methods). All but one were homozygous for a 17 nucleotide frameshift deletion (hg19, chr1:Δ3691998-3692014:GTCAGCCTAGGGGCTGT/-) in SMIM1, and the remaining one was heterozygous for this deletion (Fig 1b, Supplementary Fig. 2). The deletion has a low frequency of 1.6% (119/7562) in the UK10K cohort, which has been whole-genome sequenced at mean coverage of 6X. We next followed up by Sanger sequencing an additional 84 unrelated individuals (see Supplementary Table 1) who were either Vel-negative (n=64) or showed weak expression for the antigen (Vel-weak, n=20). First, a total of 63 out of 69 Vel-negative individuals were found to be homozygous for the deletion (Fig. 1b, Supplementary Table 1 and Supplementary Fig. 1). Importantly, all 16 Vel-negative clinical cases (Supplementary Fig. 3) who carried immune antibodies against Vel, confirming their Vel-negative status with extremely high confidence, were homozygous for the deletion. Given the population frequency of the deletion, this replication is highly significant (P=1×10−58). Second, 19 out of 20 Vel-weak individuals were heterozygous for the deletion (Fig. 1b); the remaining one was heterozygous for a novel M51K missense mutation. This shows that heterozygous deletion of SMIM1 underlies weak expression of Vel. Last, 6 individuals classified as Vel-negative were heterozygous for the deletion or a second, but different missense mutation, this time replacing residue 51 by an arginine. The heterozygosity for the null allele in these individuals is most likely explained by misclassification of extremely weak Vel expression as Vel-negative (see Online methods). For one case (Unique Study Number (USN) 48) we were able to retrieve red blood cells and this case was indeed weakly positive for the Vel antigen (see Supplementary Fig. 1). Last, from the extremely weak Vel expression for the individuals with missense mutations we infer that these mutations may lead to failure of membrane incorporation4,5, or, alternatively, it is possible that these mutations only modify the epitope leading to a strongly reduced binding of the polyclonal anti-Vel. Further experiments are needed to determine the functional consequences of these missense mutations.
Figure 1. The gene SMIM1 encodes the Vel blood group.
a) Red blood cell membrane expression of the Vel antigen measured by flow-cytometry for respectively a Vel-negative, Vel-weak by adsorption/elution, Vel-weak, and a Vel-positive individual.
b) Homozygous inheritance of a 17 nucleotide frameshift deletion (hg19, chr1:Δ3691998-3692014:GTCAGCCTAGGGGCTGT/-) in SMIM1 underlies the Vel-negative phenotype. The major allele of the common SNP rs1175550, indicated by the blue circles, was associated with reduced expression of SMIM1 in whole blood (see Fig. 2a) and decreased mean red blood cell hemoglobin concentration (P=8×10−15) in a large meta-analysis of genome-wide association studies (GWAS) of red blood cell parameters3 (see main text).
c) Overexpression of human SMIM1 cDNA in HEK293T cells. Nearly all of the transfected cells showed expression of the Vel antigen as determined with human polyclonal antibodies against Vel and flow-cytometry, confirming SMIM1 as the gene encoding the Vel blood group.
We next validated this finding in vitro by overexpressing human SMIM1 cDNA in HEK293T cells (Fig. 1c, Online methods). The Vel antigen was revealed by flow cytometry using human immune-purified anti-Vel. Nearly all of the SMIM1-transfected cells expressed the Vel antigen. Taken together with our genetic findings, this identifies SMIM1 as the gene underlying the Vel blood group.
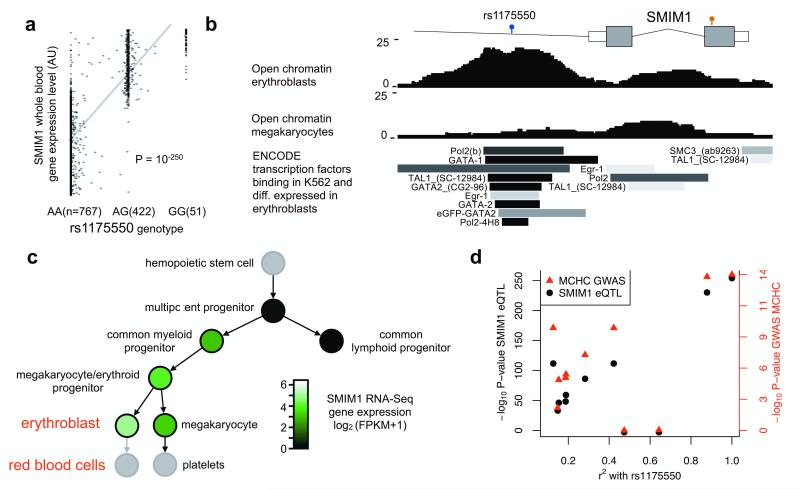
The individuals classified as Vel-weak generally have extremely low expression levels of the Vel-antigen. We therefore hypothesized that these individuals carry a modifier allele that lowers the transcription of SMIM1 on the copy not disrupted by the deletion. The major allele (the reference allele A, allele frequency 77% in the UK10K cohort) of the common variant rs1175550, located in the second intron of SMIM1, was strongly associated (P=10−250) with decreased SMIM1 transcript level in a gene expression-quantitative trait locus (expression-QTL) study in whole blood in 1240 individuals6 (Fig. 2a), and explains more than 60% of the variation in SMIM1 transcript level in the population. This SNP is located in a regulatory region in erythroblasts, the precursor cells of red blood cells, with clear transcription factor binding at this position in a myeloiderythroid cell line (Fig. 2b) but not in a lymphoid cell line, which is compatible with its pattern of expression as determined by the sequencing of RNA from primary human blood cell progenitors and precursors (Fig. 2c, Supplementary Fig. 4). Increased binding of nuclear proteins to the major (A) allele compared to the minor (G) allele of this variant (Supplementary Fig. 5) suggests that repressive factors reduce the level of SMIM1 transcription. All 24 Vel-weak or Vel-negative individuals heterozygous for the deletion were homozygous for the major allele of rs1175550; the Vel-negative individual heterozygous for rs1175550 and the missense mutation carried the A allele of rs1175550 on the wildtype haplotype without the missense mutation (Fig. 1b). Based on its frequency in the UK10K cohort, the probability of observing the major allele of rs1175550 on the non-deletion chromosome in 24 individuals by chance is P=0.003. This therefore suggests that the SMIM1-expression-QTL rs1175550 contributes to variable expression of the Vel-antigen SMIM1; we hypothesize that individuals heterozygous for the deletion but carrying the minor allele of rs1175550 on the non-deletion haplotype are normal Vel-positive.
Figure 2. Common SNP rs1175550 is an expression-QTL for SMIM1 and is associated with red blood cell traits.
a) rs1175550 is a gene expression-quantitative trait locus (eQTL) for SMIM1 transcript in whole blood in 1420 individuals6.
b) Open chromatin determined by FAIRE-Seq in erythroblasts, the precursor cell of red blood cells, indicates that the rs1175550 SNP is located in a regulatory element. This is further supported by the binding in the myeloid-erythroid cell line K562 of the subset of transcription factors assayed by the ENCODE Project9 that are differentially expressed in erythroblasts10.
c) Expression of the SMIM1 transcript based on RNA-Sequencing in the hematological lineage. SMIM1 is not transcribed in the lymphoid progenitor, but is highly expressed in red blood cell precursor cells, named erythroblasts. FPKM, fragments per kilobase per million
d) rs1175550 was also associated with mean corpuscular hemoglobin concentration (MCHC) in red blood cells in the large meta-analysis with P=8.6×10−15 (see ref3). Each SNP has both a SMIM1 gene expression-QTL association P-value (black, left axis) and a MCHC association P-value from the meta-analysis of GWAS of red blood cell parameters (red, right axis). The gene expression association signal and the meta-analysis association signal co-localize (i.e., their P-values are correlated), strongly suggesting that changes in SMIM1 expression mediate the GWAS association. A second SNP, rs1184341, which is in strong LD with rs1175550 (r2=0.92) but which was not directly tested in the gene expression study and the GWAS, also showed strong evidence for co-localization (Supplementary Fig. 6), further supporting SMIM1 as the gene underlying the GWAS association peak.
SMIM1 was only recently annotated as a bona-fide protein-coding gene and has no known function. The SMIM1 protein contains a single stretch of 22 hydrophobic residues (from Val 53 to Val 74), which is of sufficient length to act as a transmembrane domain. To assign a possible biological role to SMIM1, we considered a recent meta-analysis of genome-wide association studies (GWAS) in nearly 72,000 individuals of six red blood cell parameters3. Indeed, the major allele of the common variant rs1175550 is associated with decreased mean hemoglobin concentration of red blood cells (P=8.6×10−15), and is associated at nominal significance levels with red blood cell count (P=0.005), hemoglobin (P=0.001) and mean red blood cell volume (4.5×10−6) which all are correlated parameters3. Interestingly, the major allele was also strongly associated (P=10−250) with decreased SMIM1 transcript levels as noted above (Fig. 2a). We found that the association signal for SMIM1 in the gene expression-QTL study and the association signal for red blood cell hemoglobin levels co-localized (Fig. 2d, Supplementary Fig. 6). Additionally, we did not identify other target expression-QTL genes for rs1175550 (false discovery rate 0.05). These results strongly suggest that SMIM1 mediates the effect of the GWAS signal. Based on this finding we conclude that SMIM1 affects the mean hemoglobin concentration of red blood cells, although the effect size is likely to be small based on the GWAS result and the precise causative variant(s) remains to be identified. The role for SMIM1 in the formation and hemoglobinisation of red cells was further supported by a large-scale gene co-expression analysis (Supplementary Table 2).
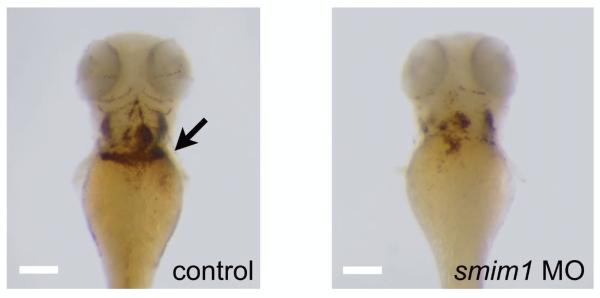
To establish the biological relevance of SMIM1 in red blood cell formation in vivo we performed a morpholino (MO) mediated knock down in zebrafish. Over the years zebrafish has proven its suitability as a model system for furthering our understanding of hematopoiesis in humans7,8. SMIM1 has evolved under strong purifying selection (Supplementary Fig. 7). The second protein-coding exon of human SMIM1, which encodes the transmembrane domain, is well conserved with the zebrafish ortholog smim1 we identified (Supplementary Fig. 7). Knock down of smim1 resulted in mild but consistent reduction of the number of red blood cells when compared with control embryos (Fig. 3 and Supplementary Fig. 8). This observation is consistent with the effect of reduced SMIM1 transcript levels on the average hemoglobin content of human red blood cells, since hemoglobin availability is a critical determinant of the number of red blood cells.
Figure 3. Zebrafish knock down of smim1.
Whole mount o-Dianisidine staining for hemoglobin at 3 days post-fertilization showed mild reduction in the total number of mature primitive erythrocytes (black arrow) in the zebrafish smim1 depleted embryos when compared to control. Scalebar, ~100μm
We investigated red blood cell and iron homeostasis parameters in 12 Vel-negative blood donors and observed a weak but non-significant trend suggestive of depletion in iron reserves (data not shown). Low iron is the most common cause of low hemoglobin levels and it should be taken into account that females and males with hemoglobin levels below 12.5 and 13.5 g/dL are removed from the donor pool. This selection may have reduced the power to observe the true effect of the SMIM1 deletion on iron reserves and hemoglobin concentrations; an independent follow-up study in a large unselected population is required to obtain an unbiased estimate of the effect of the homozygous frameshift deletion on the formation of red blood cells. However, the apparently limited phenotypic consequences of homozygous deletion, the small effect size of the common SNP rs1175550 on mean red blood cell hemoglobin concentration (MCHC), and the mild phenotype in zebrafish, suggest that the role for the SMIM1 protein in the regulation of red blood cell parameters is limited. Further studies are necessary to establish whether the frameshift deletion indeed results in complete loss of function (the frameshift occurs upstream of the transmembrane domain) and to more fully characterize the biological and biochemical function of SMIM1.
In summary, we have identified SMIM1 as the gene encoding the Vel blood group. Our finding that homozygosity for a low-frequency 17-nucleotide deletion polymorphism underlies Vel-negative status is of direct clinical relevance, as it allows the unequivocal identification of Vel-negative blood donors, thereby preventing erroneous Vel typing and reducing the risk of severe, and sometimes life-threatening destruction of incompatible donor red blood cells by immune Vel antibodies. Integrative analysis of a gene expression study and a meta-analysis of genetic association studies indicates that the SMIM1 expression-QTL rs1175550 (or a variant in strong linkage disequilibrium with rs1175550) contributes to variable expression of the Vel antigen in heterozygous carriers of the deletion and that SMIM1 affects the mean hemoglobin concentration of red blood cells. The mildly reduced red cell formation observed in zebrafish further supports a role for SMIM1 in the regulation of red blood cell hemoglobin parameters.
Online methods
Phenotyping of red cells for Vel blood group
The red blood cells (RBCs) from nearly 350,000 non-remunerated donors in the Netherlands and England were tested for the Vel blood group by a haemagglutination (HA) screening test (Test 1) using a single polyclonal human antiserum containing immune anti-Vel antibodies (anti-Vel hereafter) (Supplementary Fig. 1). The Test 1 is prone to high levels of false positive and false-negative results. Negative samples were therefore tested by a more sensitive HA confirmatory test (Test 2) with at least three different potent anti-Vel’s, however, this does not always detect extremely low levels of the Vel antigen (Supplementary Fig. 1). It is therefore reasonable to assume that a fraction of samples with negative Test 2 results are actually Vel-weak. An adsorption-elution of anti-Vel followed by the titration of the eluted anti-Vel in the HA test on Vel-positive cells (Test 3) has a forensic level of sensitivity. A number of samples with SMIM1 genotype-Vel phenotype discordance (Supplementary Fig. 1), for which cryopreserved RBCs were available, were investigated by Test 3. In addition, the RBCs from tens of thousands of blood donors in Denmark (no precise records on the number tested were maintained) were screened by Test 1 using different lots of anti-Vel then the antisera used in the Netherlands and UK, respectively. The six Test 1-negative samples (Unique Study Number 45-50, Supplementary Table 1) were confirmed by Test 2, but generally with not more than two anti-Vel sera. To have access to samples from individuals definitively Vel-negative we enrolled 16 patients (Supplementary Table 1 and Supplementary Fig. 3) who formed immune anti-Vel. In addition another 5 clinical cases with anti-Vel were enrolled but no DNA was available. The RBCs from 7 sibs from 7 different Vel-negative cases with anti-Vel were investigated by Test 2. Two cases were found to be Vel-weak and the remaining 5 to be Vel-negative, respectively (see Supplementary Table 1 and Supplementary Fig. 3).
The results obtained with the HA tests with anti-Vel are compatible with the notion that the abundance of the Vel antigen on RBCs is low and varies substantially in the normal population. This assumption was supported by the measurement by flow cytometry of Vel abundance on the RBC membrane with the most potent immune-purified IgG anti-Vel revealed by Alexa-488 labelled goat anti-human-IgG (Molecular Probes, Leiden, The Netherlands) (see Fig. 1a).
Genetic analysis
DNA was extracted from blood or saliva samples using standard laboratory procedures. The DNA samples from the 5 individuals (USN1-5) were analysed by exome sequencing (as described below) and all 96 samples (see Supplementary Table 1) were used for genetic analysis by Sanger sequencing. The SNP rs1175550 was typed either by Sanger sequencing or with a variant-specific TaqMan probe using standard conditions. The coding fraction and intronic flanking regions of the SMIM1 gene were Sanger sequenced using primers listed in Supplementary Table 3.
Exome sequencing and data analysis
Libraries were prepared and indexed using Illumina TruSeq library prep kit. Sequence capture was performed using Roche Nimblegen SeqCap EZ Human Exome v3.0. The 5 samples were sequenced on the Illumina HiSeq2000 platform. Reads were aligned with BWA v 0.6.111, duplicates were marked with Picard (see URLs), realignment around indels, base-quality recalibration, and variant calling were performed with the GATK12. On average 5.9 Gb of sequence was generated per sample and 92% (range 90-95%) of the 64 Mb capture target sequence was covered by at least 10-fold read depth. Variants were annotated using the Ensembl Variant Effect Predictor13, and allele frequencies were obtained from the European population in the Phase1 release of the 1000 Genomes Project14. We considered all variants predicted to disrupt protein-coding sequence with an allele frequency below 5% as potential causative variants underlying the Vel-negative status.
Transcript profiling of blood precursor cells
A compendium of transcripts was generated as part of the BluePrint project15 (see URLs) by sequencing RNA samples obtained from highly pure preparations (>95%) of 5 different primary progenitor and precursor blood cells (see Supplementary Table 4). The precursor cells for RBCs and platelets, the erythroblast (EB) and megakaryocyte (MK) respectively were generated by a 10-12 day culture of CD34+ haematopoietic stem cells (HSCs) as described previously16. Both the HSCs and other progenitor/precursor cells were isolated from the mononuclear cell fraction of donations of human cord blood by fluorescence-activated cell sorting using monoclonal antibodies against Cluster of Differentiation (CD) markers specific for certain stages of differentiation and lineage-commitment (Supplementary Table 4). The number of sorted cells, which ranged from 20,000 to 100,000, were directly collected into Trizol (Invitrogen) and stored at −80°C. For paired-end sequencing total RNA was isolated following the manufacturer’s protocol, followed by an additional purification and concentration step using the RNeasy mini kit (Qiagen). The transcriptome analysis libraries were prepared and amplified with the SMARTer Ultra Low Input RNA for Illumina sequencing and Advantage 2 PCR kit (Clontech) using 100 pg total RNA as input. For multiplexing purposes Biooscientific’s NEXTflex ChIP Adapters were ligated to the library fragments. Libraries were paired-end sequenced on a HiSeq 2000 system with TruSeq reagents (Illumina). All reagents were used according to manufacturer’s specifications.
Sequence reads were aligned to the February 2009 high coverage assembly using GSNAP17 version 2012-07-20 with trimming disabled. A maximum of 5 mismatches and identification of novel splice sites at genomic distances up to 100 kb were allowed. Quantification of gene expression was performed using Cufflinks18 v.1.3.0 allowing for multi-map correction. Sequencing coverage over genomic loci was visualised in IGV19.
SMIM1 Transfections
HEK293T cells (7.5×105) were plated in 6cm2 plates and lentivirally transduced with mLwpRRLsSMIM1itNGFR. Forty-eight hours after transfection cells were harvested and screened by flow cytometry using a Fortessa instrument (Beckman Coulter, Breda, the Netherlands). Vel expression in the transfectants was assessed by using human immune-purified IgG anti-Vel (immune-purified was by adsorption/elutionfrom Vel-positive RBCs) and Alexa488-labeled antihuman IgG (Molecular Probes, Leiden, The Netherlands) and Nerve Growth Factor Receptor expression was assessed by anti-CD 271-APC (Milteny Biotec Leiden, The Netherlands).
Electrophoretic mobility shift assay (EMSA)
Nuclear extracts were prepared from K562 cells using NE-PER Nuclear and Cytoplasmic extraction reagents Pierce (Rockford, IL, USA). Oligonucleotides (Biolegio, The Netherlands) were designed based on the genomic sequence flanking variant rs1175550. The forward probes were fluoresencent labeled with IRDye 700 tags on the 5′-end (see Supplementary Table 3). The labeled probes were annealed to excess unlabeled PCR products using standard protocol. The DNA-protein binding reaction was performed by mixing 2 to 10μg nuclear extract with 0.2 pM annealed oligonucleotides in binding buffer (10 mM Tris (pH7.5), 50 mM KCl, 1 mM DTT), 2.5% glycerol and 75 ng/uL poly(dIdC) in a final volume of 15 μl. The mixture was incubated for 30 min at room temperature, followed by polyacrylamide gel electrophoresis at 25°C on a 4.5% polyacrylamide gel. Fluorescence was visualized with the Odyssey Infrared Imaging System (Li-Cor Biosciences).
Zebrafish studies
General maintenance, collection, and staging of the wild type and transgenic Tg(cd41:EGFP) zebrafish were carried out according to the Zebrafish Book20. Embryos were maintained in egg water (60 mg/l Red Sea salts) at 28°C until the appropriate stage.
O-Dianisidine staining for hemoglobin was performed as previously described21. Photomicrographs were taken with a Zeiss camera AxioCam HRC attached to a LeicaMZ16 FA dissecting microscope.
Morpholinos (MO) targeting zebrafish smim1 and standard control oligo (see Supplementary Table 3) were obtained from GeneTools LLC. The oligos were resuspended in sterile water and approximately 1 nl was injected in zebrafish embryos, at the one cell stage. For both smim1 splice blocking MO and the standard control oligo a concentration of 6.4 μg/μl was used.
The efficiency of the splice-site MOs mediated gene knockdown was determined by reverse transcription (RT) of RNA followed by polymerase chain reaction (PCR) amplification of template using gene specific forward and reverse primers (see Supplementary Table 3). The following program of cycling was used for the KOD Hot Start PCR: 95°C 2 minutes, 95°C 20 s, 61°C 10 s, 70°C 12 s (40 cycles).
Evolutionary analysis
We retrieved genomic regions with flanking sequences of length 300 for orthologs of human SMIM1 (ENSG00000235169). human SMIM2 (ENSG00000139656) and human SMIM3 (ENSG00000256235) genes from Ensembl22. In addition to the 16 orthologs listed in Ensembl for SMIM1, we added one identified manually by syntheny in zebrafish (ENSDARG00000075500). For this particular ortholog, we found 7 SMIM3 zebrafish paralogs.
webPRANK23 was used (“genomic model”) to create 3 sequence alignments. (1) SMIM1 sequences were aligned using the species tree available from Ensembl as a guide. (2) SMIM1 and SMIM2 sequences were grouped for the second alignment, and (3) SMIM1, SMIM2 and SMIM3 sequences were grouped for the third alignment.
We fetched the protein translations of the canonical transcripts of the SMIM genes mentioned above (if existing) from Ensembl. We used StatAlign 1.1 (ref. 24) to perform statistical alignments of the protein sequences (“WAG substitution model, burn-in cycles: 50000, cycles after burn-in: 500000, sampling rate: 1000, 3 replications”).
We projected the canonical transcript of exons in human SMIM1 across all sequences in the genomic alignments produced above to generate protein-coding sequence alignments. We analysed the SMIM1 alignment using the M0 (one ratio) and M3 (discrete) site models available in the codeml program of PAML25.
Gene co-expression network analysis
We used a “guilt-by-association” approach to predict likely functions for genes based on gene co-expression. However, important to realize is that some phenomena exert very strong transcriptomic effects and therefore will overshadow more subtle effects. In order to be able to identify such subtle relationships as well, we conducted a principal component analysis (PCA) on an unprecedented scale (manuscript in preparation): We collected gene expression data for three different species (homo sapiens, mus musculus and rattus norvegicus) from the Gene Expression (GO) Omnibus. We confined analyses to 4 different Affymetrix expression platforms (Affymetrix Human Genome U133A Array, Affymetrix Human Genome U133 Plus 2.0 Array, Affymetrix Mouse Genome 430 2.0 Array and Affymetrix Rat Genome 230 2.0 Array). For each of these platforms we downloaded the raw CEL files (20,108, 43,278, 18,639 and 6,124 arrays, respectively), and used RMA for normalization. We could run RMA on all samples at once for the 20,108 Human Genome U133A Array, 18,639 Mouse Genome 430 2.0 Array and 6,123 Rat Genome 230 2.0 Array. For the 43,278 Human Genome U133 Plus 2.0 Array samples we ran RMA in 8 batches due to its size, by randomly assigning the samples to one of these batches. We subsequently conducted quality control (QC) on the data. We first removed duplicate samples, and subsequently conducted a PCA on the sample correlation matrix. The first principal component (PCqc) on such a matrix describes nearly always a constant pattern (dominating the data) which explains around 80-90% of the total variance26,27. This pattern can be regarded as probe-specific variance, independent of the biological sample hybridized to the array. The correlation of each individual microarray with this PCqc can be used to detect outliers, as arrays of lesser quality will have a lower correlation with the PCqc. We removed samples that had a correlation R < 0.75. After QC in total 77,840 different samples remained for downstream analysis (54,736 human samples, 17,081 mouse samples, 6,023 rat samples). Although this QCed dataset can be well used for the aforementioned guilt-by-association co-expression analysis, we reasoned that the presence of profound effects on many genes will make it difficult to identify the more subtle relationships that exist between genes. Therefore we conducted a PCA on the probe correlation matrix, resulting in the identification of in total 2,206 robustly estimated principal components (377 for Human Genome U133A, 777 for Human Genome U133 Plus 2.0, 677 for Mouse Genome 430 2.0 and 375 for Rat Genome 230 2.0) by requiring a Cronbach’s alpha > 0.70 for each individual PC. Jointly these components explain between 79% and 90% of the variance in the data per Affymetrix expression platform, and many of these are well conserved across the 3 species.
Subsequent Gene Set Enrichment Analysis (GSEA) revealed that each of these 2,206 components are significantly enriched (False discovery rate < 0.05) for at least one GO term, KEGG, BioCarta or Reactome pathway, indicating that these components are describing biologically relevant but often diverse phenomena. While per species the very first components describe profound effects on expression (i.e. many enriched pathways and GO terms), the other components are potentially equally biologically relevant, as each of the components describe certain biological phenomena. We therefore used the individual components and integrated the different platforms and species by collapsing the probe identifiers to human Ensembl genes and used orthology information from Ensembl for the mouse and rat platform, resulting in a harmonized matrix of 19,997 unique Ensembl genes × 2,206 principal components.
We subsequently predicted the most likely GO biological process using the following strategy: We first ascertained each individual GO term and assessed per PC whether the genes that were explicitly annotated with this GO term showed a significant difference from the genes that were not annotated with this GO term using a T-Test. We converted the resulting P-Value into an ‘enrichment’ Z-Score (to ensure normality). We subsequently investigated SMIM1 and correlated the 2,206 PC eigenvector coefficients of SMIM1 with each GO term by taking the 2,206 ‘enrichment’ Z-Scores as the expression profile for that GO term. A significant positive correlation means SMIM1 has an expression profile that is comparable to the GO term. We have visualized this method at Gene Network website (see URLs, click on “Method”). In order to correct for multiple testing, we permuted Ensembl gene identifiers: Using permuted data we repeated the ‘enrichment’ Z-score calculation and investigated how strong SMIM1 correlated with permuted pathway. We repeated this analysis 100 times, and observed that a P-value cut-off of 1.18×10−5 corresponded to a false discovery rate of 0.05. This resulted in significant prediction of 14 GO Biological Process functions.
Supplementary Material
Acknowledgments
We thank the individuals who participated in this study. We thank Dr Anthony Rogers and Dr Ilenia Simeoni from the Eastern Sequencing and Informatics Hub for performing the enrichment for the exome sequencing. We thank Mr Steven Garner from NHS Blood and Transplant Cambridge and Dr Kate Downes from the University of Cambridge BluePrint team and Prof Wendy Erber from the University of Western Australia for support with blood cell flow cytometry and morphology, Henk Moes, Geert Mesander and Roelof Jan van der Lei for help with cell sorting; Dr J J Erich and Dr A van Loon and colleagues (Departments of Obstetrics, University Medical Center Groningen and Martini Hospital Groningen) for collecting cord blood and Peter Ligthart (Department of Erytrocyte Serology, Sanquin Blood Supply, Amsterdam) for help with immune haemagglutination. This study makes use of data generated by the UK10K Consortium, derived from samples from the TwinsUK cohort. A full list of the investigators who contributed to the generation of the data is available from www.UK10K.org. Vel-negative and Vel-weak donors in England were enrolled via the Cambridge BioResource and the National Institute for Health Research (NIHR) BioResource for Rare Diseases. Jointly these resources have an in excess of 10,000 research volunteers and the resources are funded by the NIHR Cambridge Biomedical Research Centre. The study was supported by grants from the NIHR (RP-PG-0310-1002 to PAS, GK & WHO), the British Heart Foundation (RG/09/12/28096 to CAA and AR), the Wellcome Trust (084183/Z/07/Z and 082597/Z/07/Z to JS and AC), Cambridge BioResource (HLJ and JGS), the European Commission (BluePrint grant, 201110-201603, no 282510 to SF, MF, HHDK, HS, WHO), Bloddonorernes Forskningsfond Denmark (to KR), Cancer Research UK (C45041/A14953 to AC), EMBL (to PB), The Netherlands Organisation for Scientific Research (NWO VENI grant 916.761.70 to PvdH, NWO VENI grant 916.111.05 to HS, NWO VENI grant 916.10.135 to LF), the Netherlands Genomics Initiative (Horizon Breakthrough grant 92519031 to LF), European Community’s Health Seventh Framework Programme (FP7, 259867 to LF), the Dutch Interuniversity Cardiology Institute Netherlands (ICIN), and the Landsteiner Foundation for Blood Transfusion Research (LSBR, grant 1133).
Footnotes
URLs Picard Tools, http://picard.sourceforge.net
Gene co-expression network analysis, www.genenetwork.nl/genenetwork
BluePrint Project, www.blueprint-epigenome.eu
References
- 1.Sussman L, Miller E. Un nouveau facteur sanguine “Vel”. Rev Hémat. 1952;7:368–71. [PubMed] [Google Scholar]
- 2.Daniels G. Human Blood Groups. Wiley-Blackwell; 2002. [Google Scholar]
- 3.Van der Harst P, et al. Seventy-five genetic loci influencing the human red blood cell. Nature. 2012;492:369–75. doi: 10.1038/nature11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Körmöczi GF, et al. Genetic diversity of KELnull and KELel: a nationwide Austrian survey. Transfusion. 2007;47:703–714. doi: 10.1111/j.1537-2995.2007.01174.x. [DOI] [PubMed] [Google Scholar]
- 5.Wester ES, et al. KEL*02 alleles with alterations in and around exon 8 in individuals with apparent KEL:1,-2 phenotypes. Vox Sanguinis. 2010;99:150–157. doi: 10.1111/j.1423-0410.2010.01334.x. [DOI] [PubMed] [Google Scholar]
- 6.Fehrmann RSN, et al. Trans-eQTLs Reveal That Independent Genetic Variants Associated with a Complex Phenotype Converge on Intermediate Genes, with a Major Role for the HLA. PLoS Genet. 2011;7:e1002197. doi: 10.1371/journal.pgen.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsia N, Zon LI. Transcriptional regulation of hematopoietic stem cell development in zebrafish. Experimental hematology. 2005;33:1007–1014. doi: 10.1016/j.exphem.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 8.De Jong JLO, Zon LI. Use of the Zebrafish System to Study Primitive and Definitive Hematopoiesis. Annual Review of Genetics. 2005;39:481–501. doi: 10.1146/annurev.genet.39.073003.095931. [DOI] [PubMed] [Google Scholar]
- 9.The ENCODE Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watkins NA, et al. A HaemAtlas: characterizing gene expression in differentiated human blood cells. Blood. 2009;113:e1–9. doi: 10.1182/blood-2008-06-162958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLaren W, et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams D, et al. BLUEPRINT to decode the epigenetic signature written in blood. Nat Biotech. 2012;30:224–226. doi: 10.1038/nbt.2153. [DOI] [PubMed] [Google Scholar]
- 16.Gieger C, et al. High biological connectivity between genetic determinants of platelet biology. Nature [Google Scholar]
- 17.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts A, Pimentel H, Trapnell C, Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011 doi: 10.1093/bioinformatics/btr355. doi:10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- 19.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Westerfield M. The Zebrafish Book. University of Oregon Press; Eugene, OR: 1994. [Google Scholar]
- 21.Detrich HW, et al. Intraembryonic hematopoietic cell migration during vertebrate development. Proceedings of the National Academy of Sciences. 1995;92:10713–10717. doi: 10.1073/pnas.92.23.10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flicek P, et al. Ensembl 2012. Nucleic Acids Research. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loytynoja A, Goldman N. webPRANK: a phylogeny-aware multiple sequence aligner with interactive alignment browser. BMC Bioinformatics. 2010;11:579. doi: 10.1186/1471-2105-11-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novák Á, Miklós I, Lyngsø R, Hein J. StatAlign: an extendable software package for joint Bayesian estimation of alignments and evolutionary trees. Bioinformatics. 2008;24:2403–2404. doi: 10.1093/bioinformatics/btn457. [DOI] [PubMed] [Google Scholar]
- 25.Yang Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Molecular Biology and Evolution. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 26.Sherlock G. Analysis of large-scale gene expression data. Current Opinion in Immunology. 2000;12:201–205. doi: 10.1016/s0952-7915(99)00074-6. [DOI] [PubMed] [Google Scholar]
- 27.Alter O, Brown PO, Botstein D. Singular value decomposition for genome-wide expression data processing and modeling. Proceedings of the National Academy of Sciences. 2000;97:10101–10106. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.