Abstract
Mitochondrial synthesis of Cox1, the largest subunit of the cytochrome c oxidase complex, is controlled by Mss51 and Pet309, two mRNA-specific translational activators that act via the COX1 mRNA 5′-UTR through an unknown mechanism. Pet309 belongs to the pentatricopeptide repeat (PPR) protein family, which is involved in RNA metabolism in mitochondria and chloroplasts, and its sequence predicts at least 12 PPR motifs in the central portion of the protein. Deletion of these motifs selectively disrupted translation but not accumulation of the COX1 mRNA. We used RNA coimmunoprecipitation assays to show that Pet309 interacts with the COX1 mRNA in vivo and that this association is present before processing of the COX1 mRNA from the ATP8/6 polycistronic mRNA. This association was not affected by deletion of 8 of the PPR motifs but was undetectable after deletion of the entire 12-PPR region. However, interaction of the Pet309 protein lacking 12 PPR motifs with the COX1 mRNA was detected after overexpression of the mutated form of the protein, suggesting that deletion of this region decreased the binding affinity for the COX1 mRNA without abolishing it entirely. Moreover, binding of Pet309 to the COX1 mRNA was affected by deletion of Mss51. This work demonstrates an in vivo physical interaction between a yeast mitochondrial translational activator and its target mRNA and shows the cooperativity of the PPR domains of Pet309 in interaction with the COX1 mRNA.
Keywords: COX1 mRNA, cytochrome c oxidase, mitochondria, pentatricopeptide repeat, translation, yeast
Introduction
The biogenesis of mitochondria and chloroplasts is controlled by protein factors encoded in the nucleus, many of which interact with organellar RNAs. Among these protein factors are the pentatricopeptide repeat proteins (PPR), a major sequence-specific RNA binding family. PPR proteins contain degenerate 35-amino acid units that are usually present in tandem arrays of 2 to 26 repeats throughout the protein. The crystal structures of the mitochondrial RNA polymerase1 and ribonuclease P,2 and more recently the chloroplast THA8L 3, THA84 and PPR10,5 show that each PPR folds into a pair of antiparallel α-helices. The structure of PPR10 demonstrate that the stacking of 19 repeats form a right-handed superhelical structure. THA8 and PPR10 were crystalyzed in the presence of their cognate RNA.4,5 PPR proteins are mainly localized to mitochondria and chloroplasts and have been found in all eukaryotes,6 although the family is particularly expanded in land plants, with more than 450 members in Arabidopsis thaliana.7 The PPR proteins described to date have roles in transcription, RNA processing, intron splicing, RNA stability and editing, tRNA biogenesis, translation, and mRNA polyadenylation.8,9 Recent work has begun to shed light on the mechanisms of action of PPR proteins as well as the molecular basis of PPR-RNA interaction and recognition. Physical interactions of some PPR proteins with RNA have been demonstrated in vitro (for example, see10-16) and in vivo17-19. What is known is that some members of this family have no recognizable domains apart from the PPR motifs and are usually involved in mRNA stability.18,20,21 Others have motifs that are unrelated to PPRs, usually located at the C-terminal end, and in some cases have been implicated in the recruitment of catalytic factors for RNA processing.8,22
The genome of the yeast Saccharomyces cerevisiae predicts the presence of 15 PPR protein members.8,23 Some of these proteins are involved in general mitochondrial RNA maturation and translation (e.g., Cbp1, Rpm2, Rmd9, Dmr1, Aep3),24-28 whereas others (e.g., Msc6, Rmd9L, Yer077c, Sov1) have unknown functions. An important group of PPR proteins found in yeast are the mRNA-specific translational activators (Pet111, Pet309, Atp22, Aep1, Aep2) that function in translational regulation of mitochondrially-encoded mRNAs (reviewed in29,30). Pet309 is a translational activator specific for the mitochondrial COX1 mRNA, which encodes subunit 1 of the cytochrome c oxidase complex (CcO).31 Cox1 is the largest subunit of the complex and bears the metallic centers heme a, a3, and CuB for oxygen reduction. Synthesis of Cox1 is highly regulated and is coupled to CcO assembly.32,33 Together with Pet309, Mss51 activates translation of the COX1 mRNA.31,34 In addition, Mss51 is involved in coordination between Cox1 synthesis and CcO assembly through an interaction with newly synthesized Cox1 protein in high-molecular-weight complexes that are CcO assembly intermediates.33
Pet309 is a 965-residue, peripheral inner membrane protein that faces the matrix and acts on the COX1 mRNA 5′-UTR to activate its translation.31,35 This is the only known site of action of Pet309, based on the finding that replacement of the COX1 5′-UTR by the COX2, COX3, or COB 5′-UTRs abolished the requirement of Pet309 for Cox1 synthesis.31,34 In addition to its role in translation, Pet309 affects COX1 mRNA accumulation, and this function is particularly relevant in strains containing introns in COX1.31 The Pet309 ortholog in Schizosaccharomyces pombe, Ppr4, is also involved in COX1 mRNA translational activation and accumulation, although the target sequence in the mRNA remains elusive.36
We are interested in the mechanism of action of Pet309 and, in particular, in how the PPR motifs present in the protein participate in translation. Our previous data indicated that the 8 central PPR motifs are necessary for translation but not for accumulation of the COX1 mRNA.35 By site-directed mutagenesis it was observed that two basic amino acids that are predicted to be in the inner groove of the superhelical structure of Pet309 are necessary for translation, and it was proposed that this might be due to the presence of electrostatic interactions with the COX1 mRNA. In order to better understand the mechanism of action of Pet309, we analyzed if the protein could physically interact with the COX1 mRNA in vivo by immunoprecipitation of Pet309-RNA complexes from solubilized mitochondria and determined what conditions affect the interaction of Pet309 with the COX1 mRNA. Specifically, we investigated whether mutated forms of the protein lacking different numbers of PPR motifs or the presence of inactive ribosomes affected this interaction. We conclude that Pet309 physically interacts with the COX1 mRNA, that the PPR motifs act cooperatively to promote a high-affinity interaction with the COX1 mRNA, and that these PPR motifs are necessary for the activity of Pet309 in activating COX1 translation. This work contributes to our understanding of the mechanism of action of PPR proteins as well as of translational activation of yeast mitochondrial mRNAs.
Results
Pet309 interacts with the COX1 mRNA
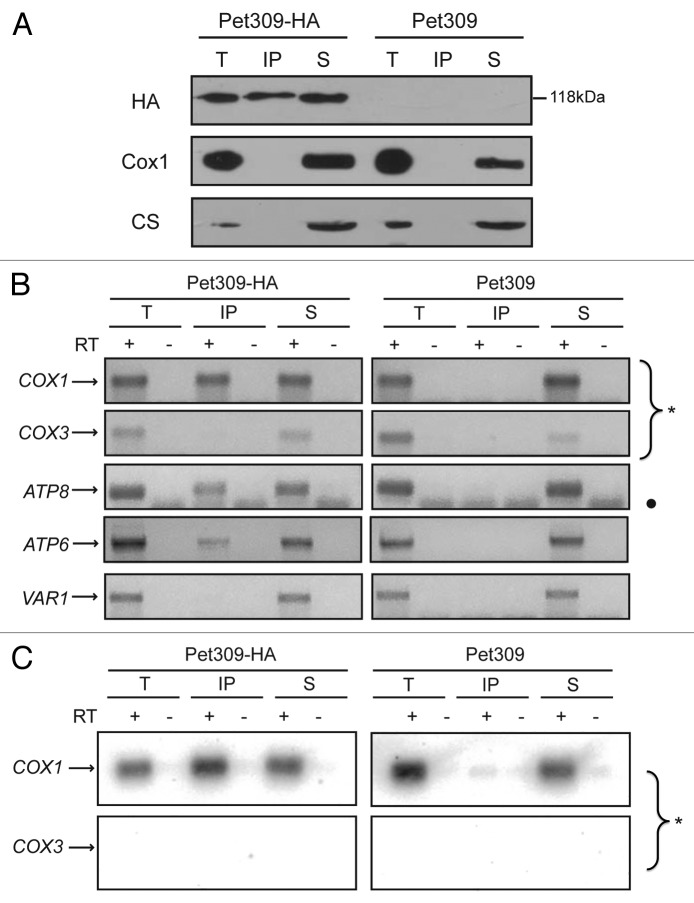
Genetic evidence suggests that mitochondrial translational activators specifically act on the 5′-UTRs of their target mRNAs. However, to date there is no experimental evidence that this group of proteins physically interacts with their target mRNAs in vivo. We therefore asked whether Pet309 could interact with the COX1 mRNA. To allow detection of Pet309, we added a sequence encoding three hemagglutinin epitopes at the C-terminal end of Pet309 (Pet309-HA). These epitopes did not affect respiratory growth of otherwise wild-type strains.35 In addition, in order to minimize RNA degradation, we deleted the NUC1 gene encoding a mitochondrial nuclease that is usually activated after solubilization; this is particularly important in the presence of divalent cations,37-39 (and our own unpublished observations). Deletion of NUC1 did not cause a respiratory phenotype in our strains. Mitochondria were purified and solubilized with dodecyl maltoside. Pet309-HA was immunoprecipitated with anti-HA antibodies, and RNA was isolated from the supernatant and immunoprecipitate fractions. Western blot analysis of the immunoprecipitate fractions indicated that Pet309-HA was precipitated with partial efficiency, whereas no HA signal was detectable in the precipitate in a control experiment using untagged Pet309 cells (Figure 1A). In contrast, the unrelated mitochondrial citrate synthase protein was exclusively present in the total and supernatant fractions, further demonstrating that the immunoprecipitation reaction was specific for Pet309-HA. Cox1 was also observed in the total and supernatant fractions but not in the anti-HA precipitate containing Pet309, as expected. With the aim to investigate if COX1 mRNA was associated with Pet309, RNA was purified from the total, supernatant, and precipitation fractions. The presence of the COX1, COX3, VAR1, ATP8, and ATP6 mRNA was analyzed by cDNA synthesis and PCR amplification. The primers used were designed against the 5′-UTR sequence of each gene. PCR amplification showed that COX1 mRNA was present in the Pet309-HA immunoprecipitation fraction, as well as in the total and supernatant fractions (Fig. 1B); however, it was absent from the immunoprecipitation fraction from untagged Pet309 mitochondria. As expected, COX3 and VAR1 mRNAs were absent from the immunoprecipitate, as translation of these mRNAs is independent of Pet309. Interestingly, we observed that the ATP8 and ATP6 genes, which are co-transcribed with COX1, are also enriched in the immunoprecipitate fraction (Fig. 1B). This indicates that Pet309 can interact with the COX1 mRNA before it is processed from the polycistronic mRNA.
Figure 1. Pet309 interacts with the COX1 mRNA. (A) Mitochondria were solubilized with dodecyl maltoside and Pet309-HA or untagged Pet309 were subjected to immunoprecipitation with anti-HA antibody. One fourth of the immunoprecipitate (IP) and the supernatant fraction representing non-bound proteins (S) were separated by SDS-PAGE and transferred to a PVDF membrane for western blot. The membrane was probed with anti-HA antibody (HA), with anti-Cox1 antibody, and with anti-citrate synthase antibody (CS) as a negative control for interaction. The total fraction (T) represents 5% of the mitochondrial extract used for immunoprecipitation. (B) RNA was extracted from the total (T), immunoprecipitate (IP) and supernatant (S) fractions. Each fraction was divided in two, and cDNA was prepared in the presence (+) or absence (-) of reverse transcriptase (RT) using primers for the COX1, COX3, ATP8, ATP6 or VAR1 5′-UTRs. The (-) RT lanes represent a negative control for DNA contamination. The PCR products were run on an agarose gel, and for clarity, the gel pictures were color inverted. The circle (•) indicates bands due to primer dimers. (C) The agarose gels from B) were transferred to a Nylon membrane and the portion of the membrane with the COX1 and COX3 amplification products (*) were hybridized with probes for COX1 and COX3. This experiment represents one of three independent repeats.
To confirm that the observed PCR amplification product was derived from COX1, the agarose gel from Figure 1B was transferred to a membrane and subjected to Southern blot analysis using a probe for COX1. As expected, the PCR amplification product of COX1 was detected with the radioactive probe, whereas the COX3 products did not give signal with this probe (Fig. 1C).
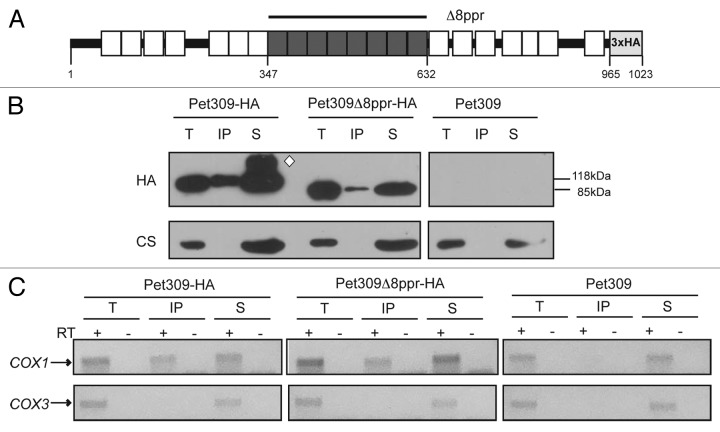
The sequence of Pet309 predicts up to 22 PPR motifs present throughout the protein23; however, the most strongly predicted PPR domains using the TPRpred software are located at the center of the protein.40 We investigated whether mutated forms of the protein lacking the central PPR motifs would retain the capacity to bind RNA. We previously demonstrated that the eight central motifs (residues 347 to 632, which include repeats 8 to 15 according to the model of Lipinski and collaborators; Figure 2A), are necessary for COX1 mRNA translation.35 In this case, cells transformed with plasmids expressing the pet309Δ8ppr-HA mutation at either low or high copy number did not grow on nonfermentable carbon sources (see below). To search for an interaction between Pet309Δppr8-HA and the COX1 mRNA, mitochondria were solubilized with dodecyl maltoside, and Pet309-HA was immunoprecipitated with an anti-HA antibody. Western blot analysis indicated that the immunoprecipitation was less efficient with pet309Δ8ppr-HA than with the wild-type control (Fig. 2B). The presence and absence, respectively, of COX1 and COX3 mRNAs was confirmed by cDNA synthesis and PCR amplification (Fig. 2C). Similar to what was observed for the wild-type strain, the mutated protein Pet309Δ8ppr was still able to interact with COX1 mRNA. Unfortunately, after several attempts we were unable to obtain quantitative data on the COX1 mRNA binding, so we cannot tell if the binding affinity of the mutated form of the protein is the same as in wild-type Pet309.
Figure 2. Deletion of the eight central PPR motifs do not interfere with the association of Pet309 with the COX1 mRNA. (A) Diagram representing Pet309 with the 22 predicted PPR motifs by the SCIPHER algorithm23 (boxes), as well as the 8 PPR central motifs that are most strongly predicted by the TPRpred software40 and that were deleted in this study (dark gray boxes). Residue numbers, as well as the triple hemagglutinin epitope used for detection and immunoprecipitation of Pet309 are indicated. (B) Mitochondrial extracts from strains bearing the wild-type Pet309-HA, the mutated Pet309∆8ppr-HA protein or the untagged Pet309 proteins were subjected to immunoprecipitation with anti-HA antibody as described in Figure 1. Immunoprecipitation fractions were analyzed by western blot. Wild-type Pet309-HA migrates with an apparent molecular weight of 118 KDa, and the mutated Pet309∆8ppr-HA protein with a molecular weight of 85 KDa. The white diamond indicates probable aggregation products of Pet309-HA. (C) The presence of the COX1 or COX3 mRNAs in the precipitate fractions was analyzed by cDNA synthesis and PCR amplification.
Together these data indicate that Pet309 physically interacts with the COX1 mRNA. This interaction occurs before COX1 is processed from the precursor transcript that includes the ATP8 and ATP6 genes. In addition, deletion of the 8 central PPR motifs does not abolish Pet309 interaction with the COX1 mRNA.
The 12 central PPR repeats of Pet309 are necessary for translation of the COX1 mRNA
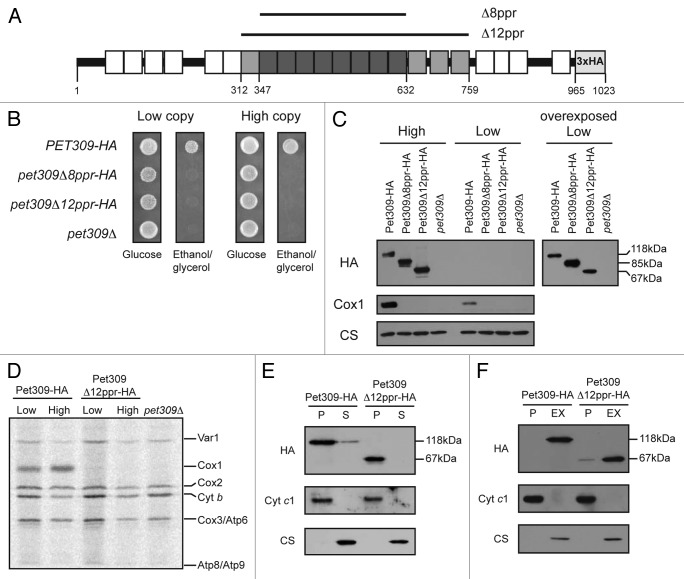
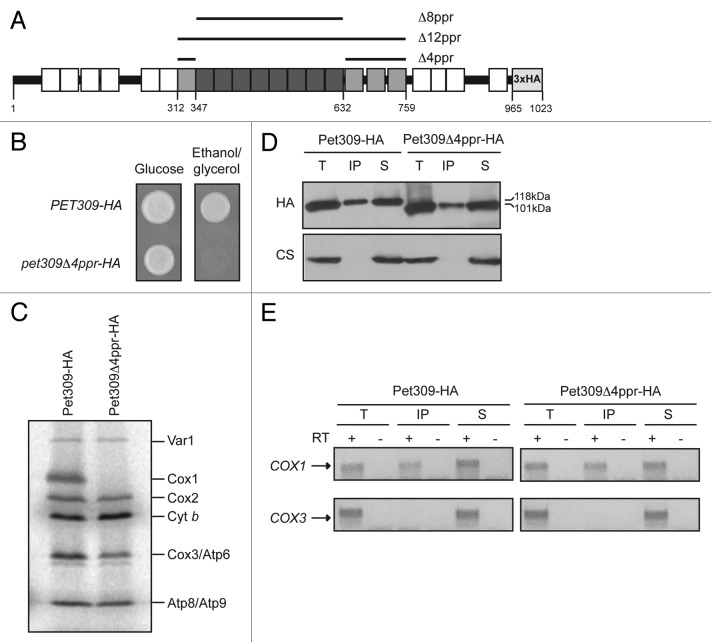
In addition to the eight central PPR motifs, the TPRpred server predicts another four repeats, one upstream and three downstream of this region, albeit with lower score (i.e., having higher P values). In order to understand the role of these additional PPR motifs, we created a deletion from residues 312 to 759 (Pet309Δ12ppr-HA), which corresponds to the 12 central PPR motifs. This construct included a triple HA epitope fused to the C-terminal end of the protein for immunopreciptations and western blot analysis (Fig. 3A).
Figure 3. Pet309∆12ppr-HA is a peripheral inner membrane protein and is impaired for COX1 mRNA translation. (A) Diagram representing Pet309 as in Figure 2. The 12 PPR central motifs that were deleted are indicated in gray boxes. The 8 PPR motifs that were deleted in Figure 2 are shown in dark gray boxes. Residue numbers are as indicated. (B) Δpet309 cells were transformed with low-copy or high-copy number plasmids containing the wild-type PET309-HA, the pet309Δ8ppr-HA, the pet309Δ12ppr-HA genes or empty vector (pet309Δ). Cells were spotted on synthetic complete medium lacking uracil with glucose or ethanol/glycerol, and incubated for 4 days at 30 °C. (C) 10 μg of mitochondrial protein from the wild-type (PET309-HA), pet309∆8ppr-HA, and pet309∆12ppr-HA constructs expressed from high-copy or low-copy number plasmids were analyzed by SDS-PAGE and western blot. The membrane was probed with anti-HA, anti-Cox1 or anti-citrate synthase (CS) antibodies. In order to visualize the HA signal from the low copy plasmids, the blot was overexposed (right panel). (D) Wild-type, pet309∆12ppr-HA and pet309∆ cells were labeled with 35S methionine in the presence of cycloheximide for 15 min at 30 °C, the proteins were analyzed by SDS-PAGE and autoradiography. Cytochrome c oxidase subunit 1, Cox1; subunit 2, Cox2; subunit 3, Cox3; cytochrome b, Cytb; ATPase subunit 6, Atp6; subunit 8, ATP8; subunit 9, Atp9; ribosomal protein, Var1. (E) 100 μg of mitochondrial protein from the wild-type and pet309∆12ppr-HA strains were sonicated and centrifuged to separate the membrane (P) and soluble (S) fractions, and were analyzed by western blot using antibodies to HA, citrate synthase as a soluble protein marker, and cytochrome c1 as a membrane protein control. (F) 100 μg of mitochondrial protein from the strains in (E) were incubated with alkaline, 100 mM Na2CO3. The integral membrane (P) and soluble, extracted (EX) proteins were analyzed by western blot as in (E).
Both, the wild-type PET309-HA and the pet309Δ12ppr-HA genes (as was previously done for the pet309Δ8ppr-HA gene35) were cloned in ARS/CEN and 2μ vectors to allow low-copy or multiple-copy dependent expression, respectively, in a yeast pet309Δ::LEU2 mutant strain. Neither the pet309Δ8ppr-HA-expressing strain nor the pet309Δ12ppr-HA-expressing strain could grow on a nonfermentable carbon source when it contained either single-copy or multiple-copy expression plasmids of these PPR deletion constructs, whereas the wild-type, HA-tagged construct was sufficient for growth (Fig. 3B). To investigate the basis of the non-respiratory phenotype of the pet309Δ12ppr-HA strain, we first investigated whether the mutated protein was stable. Western blot analysis of mitochondrial protein extracts showed that the Pet309-HA protein was recognized by the anti-HA antibody as a 118 KDa band for the wild-type gene or as a 67 KDa band corresponding to the pet309Δ12ppr-HA mutant (Fig. 3C). Pet309-HA signals for all constructs were more abundant under overexpression conditions. The same blot was probed with an antibody to Cox1, which showed no accumulation of Cox1 protein in the pet309Δ12ppr-HA mutant, even in the presence of high-copy expression, suggesting that Cox1 was not produced.
To investigate the effect of the pet309Δ12ppr-HA mutation on COX1 mRNA translation, we analyzed 35S methionine-labeled proteins from mitochondria carrying the pet309Δ12ppr-HA mutation in low-copy or high-copy-expression plasmids (Fig. 3D). The pet309Δ12ppr-HA mutation completely prevented Cox1 labeling even under overexpression conditions. As expected, labeling of Cox1 in strains expressing wild-type PET309-HA was normal, whereas a null mutation (pet309Δ) completely prevented Cox1 labeling. These results confirm that the 12 PPR domains of Pet309 are necessary for COX1 mRNA translation.
We next investigated whether Pet309Δ12ppr-HA was membrane bound or soluble. Mitochondria from strains bearing the PET309-HA or the pet309Δ12ppr-HA low-copy plasmids were sonicated and centrifuged. Both wild-type Pet309-HA and the mutated Pet309Δ12ppr-HA proteins were enriched in the membrane pellet (Fig. 3E). Alkaline Na2CO3 extraction of the mitochondrial membranes solubilized wild-type Pet309-HA and Pet309Δ12ppr-HA proteins (Fig. 3F), indicating that both behave as peripheral membrane proteins.
We conclude that the 12 PPR domains present in the central portion of Pet309 do not affect the association of the protein with the membrane but are necessary for translation of COX1 mRNA.
The 12 central PPR motifs of Pet309 are necessary for high-affinity interaction with the COX1 mRNA
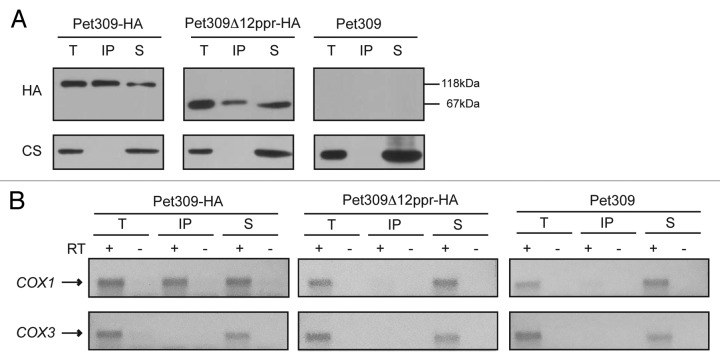
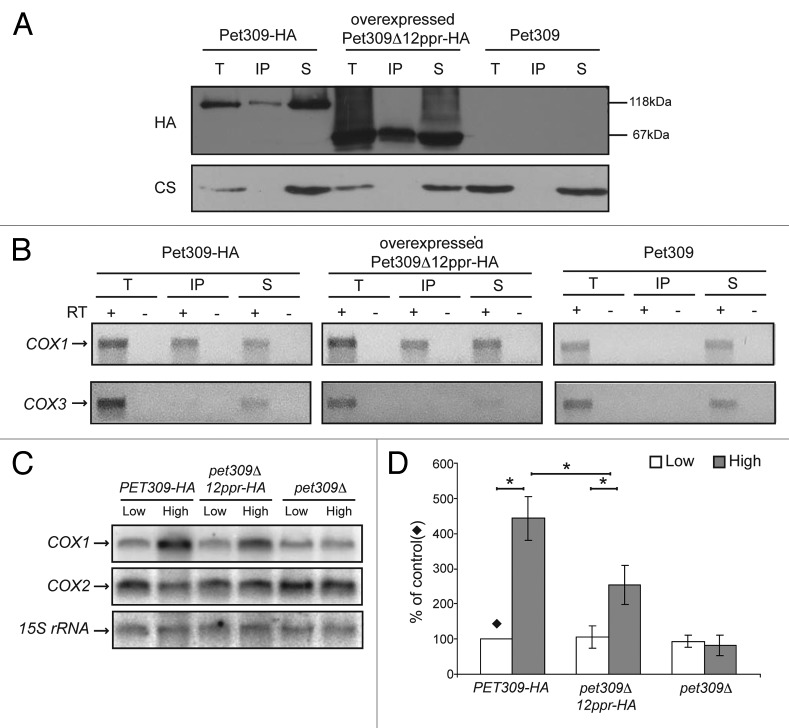
We sought to investigate whether the 12 PPR motifs on Pet309 were necessary to maintain interaction with the COX1 mRNA. Immunoprecipitation with anti-HA antibody was performed in mitochondrial extracts carrying either wild-type PET309-HA or pet309Δ12ppr-HA genes. Western blot analysis of the precipitate and supernatant fractions showed that Pet309Δ12ppr-HA immunoprecipitated with anti-HA antibody. The unrelated protein citrate synthase was present only in the total and supernatant fractions (Fig. 4A). We then looked for the presence of COX1 or COX3 mRNAs in the immunoprecipitate. None of these mRNAs were found to be associated with the Pet309Δ12ppr-HA protein, indicating that deletion of the 12 PPR motifs decreased the interaction between Pet309 and COX1 mRNA to undetectable levels (Fig. 4B).
Figure 4. No interaction was detected between the Pet309∆12ppr-HA protein and the COX1 mRNA. Mitochondrial extracts from strains bearing the wild-type Pet309-HA protein, the mutated Pet309∆12ppr-HA expressed from a low-copy number plasmid or the untagged Pet309 were subjected to immunoprecipitation with anti-HA antibody as described in Figure 1. (A) Western blot analysis of the immunoprecipitation fractions. (B) Agarose gel of the RT-PCR reactions from the immunoprecipitations with anti-HA antibody.
This result suggested that binding of Pet309 to COX1 mRNA might depend on the four additional PPR motifs that were deleted from Pet309Δ12ppr-HA relative to Pet309Δ8ppr-HA. Alternatively, binding of COX1 mRNA to Pet309 might be progressively decreased with deletion of more motifs. To try to understand if the interaction of Pet309 with COX1 mRNA is specifically determined by the four additional motifs surrounding the eight central PPR repeats, we created a mutant lacking only these four repeats (Pet309Δ4ppr-HA, Figure 5A). Even though respiratory growth and translation were abolished (Figs. 5B, C), interaction of the mutated Pet309Δ4ppr-HA protein with COX1 mRNA was detectable (Figs. 5D, E), indicating that these motifs are not essential for COX1 mRNA binding. This also suggests that the lack of interaction between Pet309Δ12ppr-HA and COX1 mRNA observed in Figure 4B might be because binding decreased below the threshold of detection in our experimental conditions.
Figure 5. Pet309∆4ppr-HA interacts with the COX1 mRNA. (A) Diagram showing Pet309 as in Figure 2. The 4 PPR motifs that were present in the Pet309∆8ppr-HA mutation but not in the Pet309∆12ppr-HA are indicated in light gray boxes. They were deleted to create the protein Pet309∆4ppr-HA. (B) Δpet309 cells were transformed with low-copy number plasmids containing the wild-type PET309-HA or the pet309Δ4ppr-HA constructs. Cells were spotted on synthetic complete medium lacking uracil with glucose or ethanol/glycerol, and incubated for 4 d at 30 °C. (C) Wild-type and Pet309∆4ppr-HA cells were labeled with 35S methionine in the presence of cycloheximide. The proteins were analyzed by SDS-PAGE and autoradiography. (D) Western blot analysis of the immunoprecipitation fractions. (E) Co-immunoprecipitation of the COX1 or COX3 mRNAs in the different fractions was analyzed by cDNA synthesis and PCR amplification.
We then tested if overexpressed Pet309Δ12ppr-HA protein could bind COX1 mRNA more efficiently. Mitochondrial extracts from a strain carrying a 2μ plasmid with pet309Δ12ppr-HA were immunoprecipitated with an antibody to HA (Fig. 6A). RT-PCR analysis of the isolated RNA showed that over-produced Pet309Δ12ppr-HA recovered detectable levels of COX1 mRNA binding (Fig. 6B).
Figure 6. The 12 central PPR motifs are necessary for efficient interaction between Pet309 and the COX1 mRNA. (A) Mitochondrial extracts from wild-type Pet309, Pet309Δ12ppr-HA expressed from a 2μ plasmid or untagged Pet309 were immunoprecipitated with an anti-HA antibody, and the fractions were analyzed by western blot. (B) The presence of the COX1 and COX3 mRNAs was analyzed by RT-PCR as previously indicated. (C) 10 μg of total RNA from the wild-type (PET309-HA) and pet309∆12ppr-HA constructs expressed from high-copy or low-copy number plasmids were analyzed by northern blot. The membrane was probed with 32P-labeled COX1, COX2 and 15S rRNA (as loading control). Total RNA from pet309∆ cells expressing low copy or high-copy, empty plasmids were used as negative control. (D) Quantification of the COX1 signals from 3 independent experiments like the one shown on C) were normalized to the 15S rRNA signals. The signal from the low copy, wild-type PET309-HA was taken as 100% (). The relevant significant differences between strains (*) were determined by ANOVA and Bonferroni post-test (P < 0.05) methods.
Pet309 has also been implicated in COX1 mRNA stability, as null mutants show reduced accumulation of the mature COX1 mRNA. This function is particularly relevant in the presence of introns.31 It was previously demonstrated that the pet309Δ8ppr-HA mutation did not affect COX1 mRNA accumulation.35 We analyzed whether deletion of the 12 PPR repeats present in Pet309 could affect COX1 mRNA accumulation. Levels of COX1 mRNA were analyzed by northern blot and normalized to the mitochondrial 15S rRNA (Figs. 6C, D). In wild-type cells expressing high-copy PET309-HA, COX1 mRNA signal was increased four times compared with low-copy PET309-HA cells. A similar pattern was obtained for pet309Δ12ppr-HA cells, except that COX1 accumulation was increased only 2-fold. This effect was specific for COX1, as COX2 mRNA levels were not affected in any sample. In contrast, the null pet309 mutant did not show accumulation of COX1 mRNA in the presence of the high-copy, empty plasmid (Fig. 6C,D). It has been suggested that high levels of translational activators could stabilize their target mRNAs.41 Our result indicated that Pet309 lacking the 12 PPR repeats still has the capacity to induce accumulation of COX1 mRNA when it is overexpressed. A similar behavior was previously observed for the protein Pet309Δ8ppr-HA, which under overexpression conditions accumulated COX1 transcript.35
In conclusion, our data indicate that Pet309Δ12ppr-HA still has the capacity to bind to COX1 mRNA but with decreased affinity. The fact that Pet309Δ8ppr-HA and Pet309Δ4ppr-HA proteins interact with the COX1 mRNA with apparent higher affinity (at least over the level of detection of the IP/RT-PCR assay) suggests that the number of PPR motifs present in the protein is important for RNA binding rather than specific repeats. However we cannot discard the possibility that the differences observed for RNA interaction of the Pet309Δ12ppr-HA protein could also be due to an effect on Pet309-folding.
Interaction of Pet309 with COX1 mRNA does not depend on active ribosomes
It has been observed that Pet309 interacts with the mitochondrial ribosome42 (Zamudio-Ochoa, Camacho-Villasana and Pérez-Martinez, unpublished data). Therefore, we investigated whether Pet309 was still able to bind to the COX1 mRNA in the presence of puromycin, which inhibits mitochondrial translation. This antibiotic causes premature chain termination and release.43 Purified mitochondria were incubated with puromycin prior to carrying out protein synthesis in the presence of 35S methionine (Fig. 7A). After Pet309-HA immunoprecipitation, we observed that COX1 mRNA co-precipitated with Pet309 regardless of the presence of puromycin (Figs. 7B, 7C). This indicated that the interaction of Pet309 with COX1 mRNA does not depend on whether ribosomes are translationally active.
Figure 7. The interaction of Pet309 with the COX1 mRNA is independent of active ribosomes. Purified mitochondria (1 mg) from a strain with tagged Pet309-HA were incubated with translation buffer in the absence (mock) or presence of 25 μg/μl of puromycin. Mitochondria from a strain with untagged Pet309 were also mock treated. (A) 50 μg of this sample were incubated with 35S methionine, and the translation products were separated by SDS-PAGE, transferred to a PVDF membrane and revealed by autoradiography. The membrane was probed with an antibody to citrate synthase (CS) as a loading control. (B) After incubation of mitochondria with translation buffer for 30 min, the dodecyl maltoside extracts were subjected to immunoprecipitation with anti-HA antibody as described in Figure 1. The total (T), supernatant (S) and immunoprecipitate (IP) fractions were analyzed by western blot with an antibody to HA and citrate synthase (CS). (C) Agarose gel of the RT-PCR reactions from the immunoprecipitations with anti-HA antibody.
Mss51 acts differently from Pet309 as a translational activator
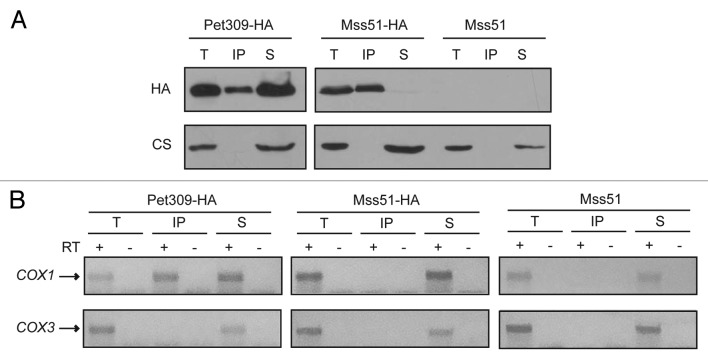
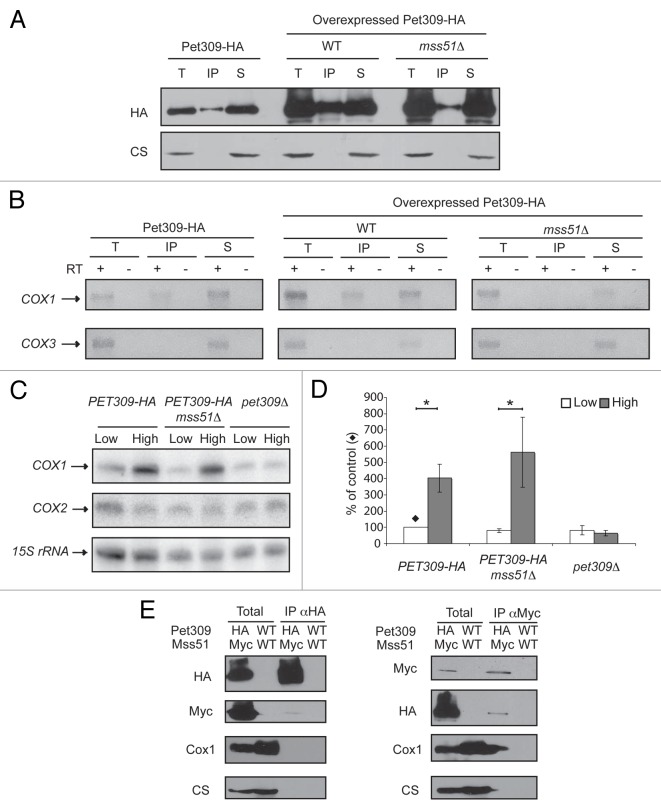
Pet309 and Mss51 are translational activators of the COX1 mRNA; however, very little is understood about their mechanisms of action as translational activators and about whether both proteins act in a similar way to cooperatively translate the COX1 mRNA. Both proteins have targets that map to the COX1 5′-UTR,31,34 independent of the COX1 coding region, as demonstrated by the fact that expression of the mitochondrial ARG8m reporter gene in place of the COX1 coding region (cox1Δ::ARG8m) depends on both proteins.34,44 To try to understand how Pet309 and Mss51 can cooperate to activate translation of the COX1 mRNA, we first analyzed whether Mss51 was able to interact with the COX1 mRNA as Pet309 does. We used a strain in which Mss51 is tagged at the C-terminal end with a triple epitope of hemagglutinin (Mss51-HA). This epitope does not interfere with the catalytic function of Mss51.44 Mss51-HA mitochondrial extracts were immunoprecipitated with an anti-HA antibody in a similar way as was done previously for Pet309-HA mitochondria. As a control for the quality of Mss51 immunoprecipitation we observed that in our experimental conditions Mss51 was coprecipitated with the Cox1 protein as previously reported44,45 (data not shown). COX1 and COX3 mRNAs were analyzed by RT-PCR. Even though immunoprecipitation of Mss51-HA was highly efficient (Fig. 8A), no evident interaction of Mss51-HA with the COX1 mRNA was detected (Fig. 8B). We next asked if the interaction of Pet309-HA with the COX1 mRNA depends on the presence of Mss51. Pet309-HA was immunoprecipitated from mitochondrial extracts of wild-type and mss51Δ cells. First, Pet309-HA was immunoprecipitated from mitochondria carrying a low-copy plasmid of this gene, however no COX1 mRNA presence was observed on the precipitated fraction with mss51Δ cells (data not shown). Next, Pet309-HA was expressed from a high-copy plasmid and was immunoprecipitated as previously described. Again, we were unable to detect an interaction of Pet309-HA with the COX1 mRNA on the mss51Δ strain (Fig. 9A, 9B). As observed in Figure 9A, Pet309 immunoprecipitation efficiency decreased by deletion of Mss51. However on mss51Δ cells, the amount of precipitated Pet309 when overexpressed was similar to the amount of Pet309 precipitated under low expression levels on MSS51 cells, yet no COX1 RNA was amplified. We performed a northern blot analysis to detect the COX1 mRNA levels on mss51Δ cells expressing PET309 from low-copy or high-copy plasmids. We observed that deletion of mss51 did not affect accumulation of the COX1 mRNA (Fig. 9C, 9D). This is consistent to what was previously reported.46 In addition we observed that, as in wild-type strains, expression of high-copy PET309 in mss51Δ cells increased the COX1 signal four times compared with low-copy PET309 cells. These results suggest that the interaction of Pet309-HA with the RNA was present but modified by the absence of Mss51 so we could not detect it under our experimental conditions.
Figure 8. The interaction of Mss51 with the COX1 mRNA is undetectable. (A) Mitochondrial extracts of Pet309-HA, Mss51-HA, and an untagged strain were immunoprecipitated as in Figure 1. Total (T), immunoprecipitate (IP) and supernatant (S) fractions were analyzed by western blot using the indicated antibodies. (B) RNA from the immunoprecipitation fraction was isolated and analyzed as in Figure 1.
Figure 9. The interaction of Pet309 with the COX1 mRNA is modified by the absence of Mss51. (A) pet309Δ cells were transformed with a low-copy expression plasmid expressing PET309-HA. In addition, pet309Δ (WT) or pet309Δ/mss51Δ cells (mss51Δ) were transformed with a high-copy plasmid expressing PET309-HA (overexpressed Pet309-HA). Mitochondria were solubilized with dodecyl maltoside, and Pet309-HA was immunoprecipitated with anti-HA antibody as described in Figure 1. The total (T), supernatant (S) and immunoprecipitate (IP) fractions were analyzed by western blot with an antibody to HA and citrate synthase (CS). (B) The presence of COX1 and COX3 mRNAs on the immunoprecipitated fractions was analyzed as in Figure 1B. (C) Total RNA from pet309Δ and pet309Δ/mss51Δ cells transformed with low-copy or high-copy plasmids expressing PET309-HA was isolated and analyzed by northern blot as in Figure 6C. (D) The COX1 signals from 3 independent experiments like the one shown in (C) were quantified as in Figure 6D. The relevant significant differences between strains (*) were determined by ANOVA and Bonferroni post-test (P < 0.05) methods. (E) Mitochondria (4mg/ml) of a strain bearing Pet309–3xHA (HA) and Mss51–3xMyc (Myc) or an untagged strain (WT) were solubilized with dodecyl maltoside. The extracts were immunoprecipitated with an antibody against the HA or the Myc epitopes as described in Figure 1. The Total and immunoprecipitate fractions were analyzed by western blot using antibodies as indicated. The total fraction represents 8% of the mitochondrial extract.
We sought to investigate if Pet309 and Mss51 physically interact by creating a strain where the PET309–3xHA and MSS51–3xMyc genes were integrated on their original chromosomal loci. The triple HA or Myc epitopes fused to the C-terminal end of Pet309 or Mss51, respectively, did not affect the respiratory growth of the cells (data not shown). Mitochondrial extracts from this strain and from the untagged strain were immunoprecipitated with antibodies against the HA or Myc epitopes under the same experimental conditions used before. After western blot, we detected a small fraction of Mss51 and Pet309 co-precipitating (Fig. 9E).
In conclusion, we observed that a small fraction of Pet309 and Mss51 physically interacted. Even though the site of action of Mss51 and Pet309 map to the COX1 5′-UTR,31,34 in our conditions no physical interaction of Mss51 with the COX1 mRNA was detected. Moreover, in the absence of Mss51 the interaction of Pet309-HA with the COX1 mRNA was not detected, indicating that the Pet309-RNA interaction was modified by Mss51.
Discussion
PPR-containing proteins are members of a eukaryote-specific RNA-binding family involved in genome expression in mitochondria and chloroplasts. More than ten years after their discovery,47,48 our understanding of the mechanisms underlying the functions of PPR proteins is starting to unravel. Mitochondrial translational activators like Pet309 are proposed to interact with their target mRNAs to localize the ribosome at the AUG start codon30,49; however, the details of the mechanism of action of these proteins have not been demonstrated experimentally. Our results demonstrate that Pet309 interacts with the COX1 mRNA and that this interaction is independent of the presence of active ribosomes. This is the first demonstration of a physical interaction in vivo between a yeast mitochondrial translational activator and its target mRNA and provides important in vivo validation of a physical interaction like that previously observed in vitro for the translational activator Pet54 with the target COX3 mRNA as well as with the aI5β intron of the COX1 transcript.50 The Pet309-COX1 mRNA interaction that we detected could be direct or mediated by a second protein. However a direct interaction is more plausible because there is increasing evidence by in vitro10-16 and crystallography4,5 experiments of the direct RNA binding capacity of proteins with PPR motifs. The immunoprecipitate of Pet309 was enriched not only with COX1 mRNA but also with ATP8 and ATP6 mRNAs, which are co-transcribed with COX1. This suggests that Pet309 interacts with the COX1 precursor transcript before processing. However, this interaction does not appear to regulate expression of ATP8 and ATP6, given that deletion of Pet309 does not affect translation of these mRNAs.31 It is possible that Pet309 interacts co-transcriptionally with the COX1 mRNA 5′-UTR. This would be consistent with the observation that Pet309 physically interacts with Nam1, which was previously found to couple transcription and translation,51 as well as with the mitochondrial RNA polymerase.52,53
We previously demonstrated that the absence of eight PPR motifs located in the middle portion of Pet309 abolished translational activity.35 Here, we showed that this mutated protein retains the capacity to bind to the COX1 mRNA. However, it was possible to detect interaction of a mutated form of the protein lacking the 12 most strongly predicted central PPR motifs with the mRNA only after overexpression of the protein, suggesting that 12 PPR motifs are necessary for high-affinity interaction of Pet309 with the COX1 mRNA. Cooperativity between PPR motifs has been observed in vitro for the human LRPPRC protein,15,54 although it is still possible that, in addition to binding cooperativity, specific PPR motifs are more important for RNA binding than others, as observed for the Arabidopsis PGR3 protein.55 In addition to the central motifs predicted by TPRpred, Lipinsky et. al. 23 predicted ten additional motifs; of these, we deleted the last four PPR motifs located at the C-terminal end of the protein, and found that even though translation of COX1 was abolished, the mutated Pet309 still retained capacity to bind the COX1 mRNA (our unpublished data). This result is similar to what we observed for the mutated forms of the protein reported here. Because our interaction experiments are not quantitative, it is possible that the mutated proteins lacking four or eight PPR motifs already have compromised interaction with the COX1 mRNA, explaining the lack of translational capacity. However, even if binding is compromised we were still able to detect this interaction. Interestingly, deletion of any one of the central PPR motifs abolished translation.35 This suggests that there could be another explanation for the observation that mutated forms of the protein lacking some of the central PPR motifs is translationally inactive even when the protein retains the capacity to bind RNA. In these mutants the binding of Pet309 to the mRNA could be sub-optimal for ribosome recognition of the AUG start codon. This is supported by the finding that the chloroplast PPR protein Atp4 enhances association of its target mRNA, ATPB/E, to the ribosome.56 It is possible that the external surface of Pet309 is a target for interaction of other proteins necessary for adequate ribosome localization and that deletion of a few PPR motifs affects this surface. This is similar to what has been described for the Pumilio family of proteins, whose activity is regulated by the proteins Nanos and Brat, which bind to the external surface of the Pumilio structure.57 However, only few examples exist of factors that interact with PPR proteins and seem to be important for their function,58-60 and their mechanisms of action remain to be described.
As for many other PPR proteins, in addition to its role on translation, Pet309 is proposed to be involved in stabilization of the COX1 transcript.31 It has been observed that overexpression of Pet309 or the mutated forms lacking some of the PPR motifs accumulate COX1 transcript (35; present study), although we cannot rule out the possibility that this accumulation is due to increased transcription. Growing evidence suggests that PPR proteins can bind to their target RNAs and block exonuclease cleavage from the 5′ and 3′ ends.12,16 Pet309 could bind to an RNase-sensitive site to prevent access by an endonuclease, making the mRNA stable, as observed for the yeast mitochondrial-targeted protein Cbp1, which is necessary to protect the COB mRNA from the nuclease Pet127.61 Another example is the chloroplast PPR5 protein, which stabilizes the trnG-UCC precursor by binding to an internal group II intron.17 The maize chloroplast PPR10 protein is involved in stabilization of the adjacent atpI and atpH RNAs, as well as in the stimulation of atpH mRNA translation.12,13 Through ribosome profiling experiments it was demonstrated that the primary function of PPR10 is to enhance atpH translation.62 It binds to the atpI-atpH intergenic region to block 5′ and 3′ exonuclease activities. It also recognizes a stem-loop structure which is located upstream of the AUG start codon and binds to single-stranded RNA to enhance translation.12,13 Pet309 could have a similar mechanism of action to accumulate and translate the COX1 mRNA.
Considerable progress has been made to understand and predict the RNA recognition code of plant PPR proteins.4,5,55,63-65 A polar amino acid (primarily N, T or S) at position 5 (according to amino acid positions in the PPR10 crystal structure) appears to be the determinant for base specific recognition.4,5,63,64 Positions 2 and 35 seem also to be important for RNA binding. Residues at position 2 of adjacent PPR motifs (primarily V, F, I, R) sandwich one nucleotide base mainly through van der Waals interactions.5 The residue at position 35 is close to the base, and could make hydrogen bonds with it through water molecules; in addition, some of them could stabilize the conformation of the residue at the fifth position of the same repeat.5 Some of the 22 PPR motifs predicted on Pet309 match this recognition code. For example, position 5 is an N, T or S in 8 of the 22 repeats, while position 35 seems to match one of the expected residues in 9 repeats. Mutagenesis studies are needed to understand if the recognition code for plants has the same relevance in yeast PPR proteins.
Binding of Pet309 to the RNA might not be the only requisite for correct binding and/or start site selection of the ribosome. Mss51, the second translational activator specific for the COX1 mRNA, could have an essential role in this step. In the present work we observed that in the absence of Mss51, the interaction of Pet309 with the COX1 mRNA was not detected after immunoprecipitation assays, yet the accumulation of the transcript was not affected. These results suggest that in the absence of Mss51, Pet309 retained the ability to bind the COX1 mRNA, but this interaction was modified so we could not detect it on our experimental conditions. It was previously demonstrated that, in the absence of Mss51, an aberrant peptide (mp15) derived from the COX1 gene is produced. Synthesis of mp15 depends on Pet309,66 suggesting that COX1 mRNA translation can proceed in the absence of Mss51; however, Pet309 could not have an appropriate interaction with the mRNA to activate translation, as the ribosome might not be able to find the correct AUG start codon on the mRNA. More studies are needed to unravel the mechanism by which Mss51 and Pet309 act together on translational activation of the COX1 mRNA. In addition, we searched for an interaction between Mss51 and the COX1 mRNA. However, even when immunoprecipitation of Mss51 was highly efficient, no association with the COX1 mRNA was observed, indicating that either Mss51 does not directly interact with the mRNA or that the interaction is transient. Zambrano and colleagues observed an interaction with the COX1 mRNA using a segment of Mss51 in a triple hybrid experiment.66 They proposed that the most probable region of interaction for Mss51 was around -97 to +23 nucleotides with respect to the COX1 AUG start codon. Here we detected a physical interaction between Mss51 and Pet309, which could be direct or mediated by the translational machinery to promote translation initiation. In this scenario, Mss51 would interact with the COX1 mRNA only transiently, whereas the interaction of Pet309 with the COX1 mRNA would be more stable, but regulated by Mss51 through a mechanism yet to be revealed.
Materials and Methods
Strains, media, and genetic methods
The S. cerevisiae strains used in this study are listed in Table 1. Standard genetic methods and media recipes were as described previously.67,68 Complete fermentable media were YPD or YPRaf (containing 2% glucose or 2% raffinose). Minimal media contained 0.67% yeast nitrogen base, 2% glucose or 3% glycerol/3% ethanol, and Complete Supplement Mixtures (CSMs) purchased from Bio 101 (Vista, CA) and ForMedium (UK). Gene deletion constructs with KanMX4, LEU2, or URA3 cassettes were generated by PCR.
Table 1. List of strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| FTC25 | Matα, ura3Δ, ade2 PET309–3xHA nuc1Δ::KanMX4 [ρ+]a | Faviola Tavares-Carreon (unpublished) |
| FTC26 | Matα, ura3–52, leu2–3, 112 lys2 arg8::hisG pet309Δ::LEU2 nuc1Δ::KanMX4 ρ+, ΔΣai]b | Faviola Tavares-Carreon (unpublished) |
| JPM13 | Matα, ura3Δ, ade2 PET309–3xHA, MSS51–3xMYC [ρ+] | Juan Pablo Mayorga-Juárez (unpublished) |
| XPM232 | Matα, ura3–52, leu2–3, 112 lys2 arg8::hisG pet309Δ::LEU2 [ρ+, ΔΣai] | 35 |
| YC128 | Matα, ura3–52, leu2–3, 112 lys2 arg8::hisG pet309Δ::LEU2, mss51Δ::KanMX4 [ρ+, ΔΣai] | This work |
| YC131 | Matα, ura3Δ, ade2, mss51Δ::URA3, PET309–3xHA nuc1Δ::KanMX4 [ρ+]a | This work |
| YC132 | Matα, ura3Δ, ade2 MSS51::3xHA nuc1Δ::KanMX4 [ρ+] | This work |
| YC133 | Matα, ura3Δ, ade2 nuc1Δ::KanMX4 [ρ+] | This work |
| AGG54 | Matα, ura3–52, ade2, arg8::hisG, lys2, leu2–3,112, mss51Δ::LEU2, PET309::3xHA nuc1Δ::KanMX4 [ρ+] | This work |
All strains are isogenic or congenic to D273–10B; a The mitochondrial phenotype is shown between brackets; b The mitochondrial COX1 gene has no introns.
Molecular biology methods
Standard molecular biology methods for cloning, northern and Southern blot analyses were as described previously.69 The plasmids used and generated during this work were derivatives of pXP96 (derivative of the pBluescript backbone), pXP97 (ARS/CEN plasmid derived from pRS416), and pXP104 (2μ plasmid derived from YEp352) and contain the PET309::HA sequence, including 310 and 205 nt of the PET309 5′ and 3′-UTRs, respectively.35 The pet309Δ8ppr was previously created,35 and lacks residues 347 to 632. During the cloning of this construct the TPRpred software40 did not predict the sequence F562 to D596 to be a PPR motif, so the construct was claimed to be a deletion of 7 PPR motifs. As the database was enriched with PPR motif sequences, the TPRpred software identified sequence F562 to D596 as a PPR motif. The pet309∆12ppr and pet309∆4ppr mutated forms of the protein were generated by fusion PCR70 using Accuzyme DNA polymerase (Bioline) and pXP97 as the DNA template. The construct pet309∆12ppr lacks residues N312 to N759, and the pet309∆4ppr construct lacks residues N312 to I346 and I638 to N759. PCR products were digested with PstI and EcoRI and cloned into similarly digested pXP96. The XbaI - XhoI DNA fragments from pet309∆12ppr and pet309∆4ppr were ligated into pXP97 to generate the ARS/CEN, low-copy-number plasmids. In addition, the XbaI – XhoI insert of pet309∆12ppr was ligated into a 2μ, high-copy-number plasmid. For northern blot analysis, total RNA was extracted using the RNeasy Mini kit (Qiagen) from yeast cultures grown to late log phase on raffinose-synthetic complete media lacking uracil. RNA was separated by agarose gel electrophoresis and blotted to Hybond XL membranes (GE Healthcare). Blots were probed with radioactively labeled probes recognizing COX1 exon 4, COX2, and 15S rRNA.71 15S rRNA signal was used to standardize loading. Blots were analyzed with a Typhoon 8600 PhosphorImager (GE Healthcare) and quantitated with ImageQuaNT. The probe for Southern blot hybridization was obtained from the HindIII fragment of pXPM19, which contains the full-length COX1 5′-UTR.44
RNA immunoprecipitation assay
This technique is based on chloroplast RNA IP assays that have been described previously.18,72 To reduce the risk of RNA degradation during solubilization of mitochondria, we deleted the NUC1 gene encoding a mitochondrial nuclease with a nuc1Δ::KanMX4 cassette. Mitochondria (1 mg) were lysed with 500 μl of 0.7% n-Dodecyl β-D-maltoside, 100 mM NaCl, 20 mM TRIS-HCl pH 7.4, 200 U of RNaseOUT (Invitrogen), and a cocktail of protease inhibitors (Roche). The solubilized fraction was subjected to a clarifying spin for 10 min at 12,000 g. The supernatant was incubated with an anti-HA high-affinity antibody (Roche or Pierce), which was coupled to protein A-sepharose (GE Healthcare) for two hours at 4 °C with constant rocking. After centrifugation, the supernatant was kept in a separate tube, and excess liquid was discarded with a microsyringe. The immunoprecipitate was washed twice with 500 μl of lysis buffer and twice with 1 ml of 20 mM HEPES-KOH pH 7.4 and then resuspended in 150 μl of the same buffer. One-fourth of the supernatant and precipitate fractions were saved for western blot analysis, and the remainder was used for RNA extraction. RNA from total, immunoprecipitated, and supernatant fractions was extracted by incubation with 1 ml of TRIzol® reagent (Invitrogen). Twenty ng of RNA were treated with 1 unit of DNase I (Invitrogen) for 15 min at 25 °C. After inactivation with 25 mM EDTA and incubation at 65 °C, the first strand of cDNA was prepared by addition of primers for COX1, COX3, VAR1 ATP8 or ATP6 in the presence of SuperScript III Reverse Transcriptase (Invitrogen). The reaction was incubated at 25 °C for 5 min, then at 50 °C for 60 min, and then at 70 °C for 15 min. The resulting cDNA was used as template for PCR reactions to amplify the COX1, COX3, VAR1, ATP8 or ATP6 5′-UTRs using 35 cycles. Note that under these conditions RT-PCR is not quantitative. For clarity, RT-PCR agarose gel pictures shown in the figures were color-inverted, resulting in dark bands on a white background.
Analysis of Mitochondrial Proteins
Yeast cells were grown in raffinose medium until late log phase. Mitochondria were isolated as previously described.73 Proteins were separated by SDS-PAGE.74 For western blotting, proteins were transferred to polyvinylidene difluoride (PVDF) membranes and probed with anti-Cox1 (Rodolfo García-Villegas), anti-hemagglutinin (HA) (Roche), anti c-MYC (Roche), anti-citrate synthase (Thomas D. Fox), or anti-cytochrome c1 (Diego González-Halphen and Miriam Vázquez- Acevedo) antibodies. Immune complexes were detected with either goat anti-rabbit immunoglobulin (IgG) or anti-mouse IgG conjugated to horseradish peroxidase (Invitrogen) and the enhanced chemiluminescence kit (Pierce), with the exception that the anti-HA signal was detected using the Immobilion substrate (Millipore) or WestPico Kit (Pierce). Mitochondrial translation products were radiolabeled with 35S methionine in whole cells in the presence of cycloheximide or in purified mitochondria as previously described.75 Mitochondrial separation into membrane and soluble fractions and alkaline carbonate extractions of membranes was as described.35,73
Acknowledgments
We thank Rodolfo García-Villegas, Diego González-Halphen, Miriam Vázquez-Acevedo and Thomas Fox for the gift of antisera, Gabriel del Río-Guerra and Teresa Lara-Ortiz for the gift of yeast deletion strains, and Faviola Tavares-Carreón for yeast strains and preliminary experiments. We are also indebted to Felix Recillas-Targa for critical review of the manuscript. This work was supported by research grants from Consejo Nacional de Ciencia y Tecnología (47514 to X. P-M, fellowship 298954 to A. Z-O, fellowship 255917 to AE. G-G), Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica, Universidad Nacional Autónoma de México (IN208711 and IN204414 to X. P-M), and Fundación Miguel Alemán, A.C. (to X. P-M). This manuscript is part of the Ph. D. thesis of A. Z-O from the Doctorado en Ciencias Biomédicas, Universidad Nacional Autónoma de México.
References
- 1.Ringel R, Sologub M, Morozov YI, Litonin D, Cramer P, Temiakov D. Structure of human mitochondrial RNA polymerase. Nature. 2011;478:269–73. doi: 10.1038/nature10435. [DOI] [PubMed] [Google Scholar]
- 2.Howard MJ, Lim WH, Fierke CA, Koutmos M. Mitochondrial ribonuclease P structure provides insight into the evolution of catalytic strategies for precursor-tRNA 5′ processing. Proc Natl Acad Sci U S A. 2012;109:16149–54. doi: 10.1073/pnas.1209062109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ban T, Ke J, Chen R, Gu X, Tan MH, Zhou XE, Kang Y, Melcher K, Zhu JK, Xu HE. Structure of a PLS-class pentatricopeptide repeat protein provides insights into mechanism of RNA recognition. J Biol Chem. 2013;288:31540–8. doi: 10.1074/jbc.M113.496828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ke J, Chen RZ, Ban T, Zhou XE, Gu X, Tan MH, Chen C, Kang Y, Brunzelle JS, Zhu JK, et al. Structural basis for RNA recognition by a dimeric PPR-protein complex. Nat Struct Mol Biol. 2013;20:1377–82. doi: 10.1038/nsmb.2710. [DOI] [PubMed] [Google Scholar]
- 5.Yin P, Li Q, Yan C, Liu Y, Liu J, Yu F, Wang Z, Long J, He J, Wang HW, et al. Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature. 2013;504:168–71. doi: 10.1038/nature12651. [DOI] [PubMed] [Google Scholar]
- 6.O’Toole N, Hattori M, Andres C, Iida K, Lurin C, Schmitz-Linneweber C, Sugita M, Small I. On the expansion of the pentatricopeptide repeat gene family in plants. Mol Biol Evol. 2008;25:1120–8. doi: 10.1093/molbev/msn057. [DOI] [PubMed] [Google Scholar]
- 7.Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16:2089–103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmitz-Linneweber C, Small I. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 2008;13:663–70. doi: 10.1016/j.tplants.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Aphasizheva I, Maslov D, Wang X, Huang L, Aphasizhev R. Pentatricopeptide repeat proteins stimulate mRNA adenylation/uridylation to activate mitochondrial translation in trypanosomes. Mol Cell. 2011;42:106–17. doi: 10.1016/j.molcel.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammani K, des Francs-Small CC, Takenaka M, Tanz SK, Okuda K, Shikanai T, Brennicke A, Small I. The pentatricopeptide repeat protein OTP87 is essential for RNA editing of nad7 and atp1 transcripts in Arabidopsis mitochondria. J Biol Chem. 2011;286:21361–71. doi: 10.1074/jbc.M111.230516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okuda K, Nakamura T, Sugita M, Shimizu T, Shikanai T. A pentatricopeptide repeat protein is a site recognition factor in chloroplast RNA editing. J Biol Chem. 2006;281:37661–7. doi: 10.1074/jbc.M608184200. [DOI] [PubMed] [Google Scholar]
- 12.Pfalz J, Bayraktar OA, Prikryl J, Barkan A. Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 2009;28:2042–52. doi: 10.1038/emboj.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prikryl J, Rojas M, Schuster G, Barkan A. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc Natl Acad Sci U S A. 2011;108:415–20. doi: 10.1073/pnas.1012076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams-Carrier R, Kroeger T, Barkan A. Sequence-specific binding of a chloroplast pentatricopeptide repeat protein to its native group II intron ligand. RNA. 2008;14:1930–41. doi: 10.1261/rna.1077708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu F, Addis JB, Cameron JM, Robinson BH. LRPPRC mutation suppresses cytochrome oxidase activity by altering mitochondrial RNA transcript stability in a mouse model. Biochem J. 2012;441:275–83. doi: 10.1042/BJ20110985. [DOI] [PubMed] [Google Scholar]
- 16.Zhelyazkova P, Hammani K, Rojas M, Voelker R, Vargas-Suárez M, Börner T, Barkan A. Protein-mediated protection as the predominant mechanism for defining processed mRNA termini in land plant chloroplasts. Nucleic Acids Res. 2012;40:3092–105. doi: 10.1093/nar/gkr1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beick S, Schmitz-Linneweber C, Williams-Carrier R, Jensen B, Barkan A. The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol Cell Biol. 2008;28:5337–47. doi: 10.1128/MCB.00563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmitz-Linneweber C, Williams-Carrier R, Barkan A. RNA immunoprecipitation and microarray analysis show a chloroplast Pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell. 2005;17:2791–804. doi: 10.1105/tpc.105.034454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell SF, Jain S, She M, Parker R. Global analysis of yeast mRNPs. Nat Struct Mol Biol. 2013;20:127–33. doi: 10.1038/nsmb.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamazaki H, Tasaka M, Shikanai T. PPR motifs of the nucleus-encoded factor, PGR3, function in the selective and distinct steps of chloroplast gene expression in Arabidopsis. Plant J. 2004;38:152–63. doi: 10.1111/j.1365-313X.2004.02035.x. [DOI] [PubMed] [Google Scholar]
- 21.Loiselay C, Gumpel NJ, Girard-Bascou J, Watson AT, Purton S, Wollman FA, Choquet Y. Molecular identification and function of cis- and trans-acting determinants for petA transcript stability in Chlamydomonas reinhardtii chloroplasts. Mol Cell Biol. 2008;28:5529–42. doi: 10.1128/MCB.02056-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rivals E, Bruyère C, Toffano-Nioche C, Lecharny A. Formation of the Arabidopsis pentatricopeptide repeat family. Plant Physiol. 2006;141:825–39. doi: 10.1104/pp.106.077826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lipinski KA, Puchta O, Surendranath V, Kudla M, Golik P. Revisiting the yeast PPR proteins--application of an Iterative Hidden Markov Model algorithm reveals new members of the rapidly evolving family. Mol Biol Evol. 2011;28:2935–48. doi: 10.1093/molbev/msr120. [DOI] [PubMed] [Google Scholar]
- 24.Stribinskis V, Gao GJ, Ellis SR, Martin NC. Rpm2, the protein subunit of mitochondrial RNase P in Saccharomyces cerevisiae, also has a role in the translation of mitochondrially encoded subunits of cytochrome c oxidase. Genetics. 2001;158:573–85. doi: 10.1093/genetics/158.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stribinskis V, Gao GJ, Sulo P, Ellis SR, Martin NC. Rpm2p: separate domains promote tRNA and Rpm1r maturation in Saccharomyces cerevisiae mitochondria. Nucleic Acids Res. 2001;29:3631–7. doi: 10.1093/nar/29.17.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nouet C, Bourens M, Hlavacek O, Marsy S, Lemaire C, Dujardin G. Rmd9p controls the processing/stability of mitochondrial mRNAs and its overexpression compensates for a partial deficiency of oxa1p in Saccharomyces cerevisiae. Genetics. 2007;175:1105–15. doi: 10.1534/genetics.106.063883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puchta O, Lubas M, Lipinski KA, Piatkowski J, Malecki M, Golik P. DMR1 (CCM1/YGR150C) of Saccharomyces cerevisiae encodes an RNA-binding protein from the pentatricopeptide repeat family required for the maintenance of the mitochondrial 15S ribosomal RNA. Genetics. 2010;184:959–73. doi: 10.1534/genetics.110.113969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee C, Tibbetts AS, Kramer G, Appling DR. Yeast AEP3p is an accessory factor in initiation of mitochondrial translation. J Biol Chem. 2009;284:34116–25. doi: 10.1074/jbc.M109.055350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pérez-Martínez X, Funes S, Camacho-Villasana Y, Marjavaara S, Tavares-Carreón F, Shingú-Vázquez M. Protein synthesis and assembly in mitochondrial disorders. Curr Top Med Chem. 2008;8:1335–50. doi: 10.2174/156802608786141124. [DOI] [PubMed] [Google Scholar]
- 30.Herrmann JM, Woellhaf MW, Bonnefoy N. Control of protein synthesis in yeast mitochondria: the concept of translational activators. Biochim Biophys Acta. 2013;1833:286–94. doi: 10.1016/j.bbamcr.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 31.Manthey GM, McEwen JE. The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 1995;14:4031–43. doi: 10.1002/j.1460-2075.1995.tb00074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fontanesi F, Soto IC, Barrientos A. Cytochrome c oxidase biogenesis: new levels of regulation. IUBMB Life. 2008;60:557–68. doi: 10.1002/iub.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perez-Martinez X, Butler CA, Shingu-Vazquez M, Fox TD. Dual functions of Mss51 couple synthesis of Cox1 to assembly of cytochrome c oxidase in Saccharomyces cerevisiae mitochondria. Mol Biol Cell. 2009;20:4371–80. doi: 10.1091/mbc.E09-06-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tavares-Carreón F, Camacho-Villasana Y, Zamudio-Ochoa A, Shingú-Vázquez M, Torres-Larios A, Pérez-Martínez X. The pentatricopeptide repeats present in Pet309 are necessary for translation but not for stability of the mitochondrial COX1 mRNA in yeast. J Biol Chem. 2008;283:1472–9. doi: 10.1074/jbc.M708437200. [DOI] [PubMed] [Google Scholar]
- 36.Kühl I, Dujeancourt L, Gaisne M, Herbert CJ, Bonnefoy N. A genome wide study in fission yeast reveals nine PPR proteins that regulate mitochondrial gene expression. Nucleic Acids Res. 2011;39:8029–41. doi: 10.1093/nar/gkr511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dake E, Hofmann TJ, McIntire S, Hudson A, Zassenhaus HP. Purification and properties of the major nuclease from mitochondria of Saccharomyces cerevisiae. J Biol Chem. 1988;263:7691–702. [PubMed] [Google Scholar]
- 38.Iqbal J, Hudson AP. An in vitro transcription assay for yeast mitochondria using organellar lysates. Anal Biochem. 1996;243:270–6. doi: 10.1006/abio.1996.0516. [DOI] [PubMed] [Google Scholar]
- 39.Miyakawa I, Fujimura R, Kadowaki Y. Use of the nuc1 null mutant for analysis of yeast mitochondrial nucleoids. J Gen Appl Microbiol. 2008;54:317–25. doi: 10.2323/jgam.54.317. [DOI] [PubMed] [Google Scholar]
- 40.Karpenahalli MR, Lupas AN, Söding J. TPRpred: a tool for prediction of TPR-, PPR- and SEL1-like repeats from protein sequences. BMC Bioinformatics. 2007;8:2. doi: 10.1186/1471-2105-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fiori A, Perez-Martinez X, Fox TD. Overexpression of the COX2 translational activator, Pet111p, prevents translation of COX1 mRNA and cytochrome c oxidase assembly in mitochondria of Saccharomyces cerevisiae. Mol Microbiol. 2005;56:1689–704. doi: 10.1111/j.1365-2958.2005.04658.x. [DOI] [PubMed] [Google Scholar]
- 42.Bauerschmitt H, Mick DU, Deckers M, Vollmer C, Funes S, Kehrein K, Ott M, Rehling P, Herrmann JM. Ribosome-binding proteins Mdm38 and Mba1 display overlapping functions for regulation of mitochondrial translation. Mol Biol Cell. 2010;21:1937–44. doi: 10.1091/mbc.E10-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sohmen D, Harms JM, Schlünzen F, Wilson DN. SnapShot: Antibiotic inhibition of protein synthesis I. Cell. 2009;138:e1. doi: 10.1016/j.cell.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 44.Perez-Martinez X, Broadley SA, Fox TD. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 2003;22:5951–61. doi: 10.1093/emboj/cdg566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barrientos A, Zambrano A, Tzagoloff A. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 2004;23:3472–82. doi: 10.1038/sj.emboj.7600358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Decoster E, Simon M, Hatat D, Faye G. The MSS51 gene product is required for the translation of the COX1 mRNA in yeast mitochondria. Mol Gen Genet. 1990;224:111–8. doi: 10.1007/BF00259457. [DOI] [PubMed] [Google Scholar]
- 47.Aubourg S, Boudet N, Kreis M, Lecharny A. In Arabidopsis thaliana, 1% of the genome codes for a novel protein family unique to plants. Plant Mol Biol. 2000;42:603–13. doi: 10.1023/A:1006352315928. [DOI] [PubMed] [Google Scholar]
- 48.Small ID, Peeters N. The PPR motif - a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci. 2000;25:46–7. doi: 10.1016/S0968-0004(99)01520-0. [DOI] [PubMed] [Google Scholar]
- 49.Fox TD. Genetics of mitochondrial translation. In: Hershey JWB, Matthews MB, Sonenberg N, eds. Translational Control. Cold Spring Harbor, NY: Cold Spring Harbor Press, 1996:733-58 [Google Scholar]
- 50.Kaspar BJ, Bifano AL, Caprara MG. A shared RNA-binding site in the Pet54 protein is required for translational activation and group I intron splicing in yeast mitochondria. Nucleic Acids Res. 2008;36:2958–68. doi: 10.1093/nar/gkn045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodeheffer MS, Boone BE, Bryan AC, Shadel GS. Nam1p, a protein involved in RNA processing and translation, is coupled to transcription through an interaction with yeast mitochondrial RNA polymerase. J Biol Chem. 2001;276:8616–22. doi: 10.1074/jbc.M009901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Markov DA, Savkina M, Anikin M, Del Campo M, Ecker K, Lambowitz AM, De Gnore JP, McAllister WT. Identification of proteins associated with the yeast mitochondrial RNA polymerase by tandem affinity purification. Yeast. 2009;26:423–40. doi: 10.1002/yea.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Naithani S, Saracco SA, Butler CA, Fox TD. Interactions among COX1, COX2, and COX3 mRNA-specific translational activator proteins on the inner surface of the mitochondrial inner membrane of Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:324–33. doi: 10.1091/mbc.E02-08-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mili S, Piñol-Roma S. LRP130, a pentatricopeptide motif protein with a noncanonical RNA-binding domain, is bound in vivo to mitochondrial and nuclear RNAs. Mol Cell Biol. 2003;23:4972–82. doi: 10.1128/MCB.23.14.4972-4982.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fujii S, Sato N, Shikanai T. Mutagenesis of individual pentatricopeptide repeat motifs affects RNA binding activity and reveals functional partitioning of Arabidopsis PROTON gradient regulation3. Plant Cell. 2013;25:3079–88. doi: 10.1105/tpc.113.112193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zoschke R, Kroeger T, Belcher S, Schöttler MA, Barkan A, Schmitz-Linneweber C. The pentatricopeptide repeat-SMR protein ATP4 promotes translation of the chloroplast atpB/E mRNA. Plant J. 2012;72:547–58. doi: 10.1111/j.1365-313X.2012.05081.x. [DOI] [PubMed] [Google Scholar]
- 57.Edwards TA, Pyle SE, Wharton RP, Aggarwal AK. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell. 2001;105:281–9. doi: 10.1016/S0092-8674(01)00318-X. [DOI] [PubMed] [Google Scholar]
- 58.Sasarman F, Brunel-Guitton C, Antonicka H, Wai T, Shoubridge EA, LSFC Consortium LRPPRC and SLIRP interact in a ribonucleoprotein complex that regulates posttranscriptional gene expression in mitochondria. Mol Biol Cell. 2010;21:1315–23. doi: 10.1091/mbc.E10-01-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bentolila S, Heller WP, Sun T, Babina AM, Friso G, van Wijk KJ, Hanson MR. RIP1, a member of an Arabidopsis protein family, interacts with the protein RARE1 and broadly affects RNA editing. Proc Natl Acad Sci U S A. 2012;109:E1453–61. doi: 10.1073/pnas.1121465109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boussardon C, Salone V, Avon A, Berthomé R, Hammani K, Okuda K, Shikanai T, Small I, Lurin C. Two interacting proteins are necessary for the editing of the NdhD-1 site in Arabidopsis plastids. Plant Cell. 2012;24:3684–94. doi: 10.1105/tpc.112.099507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fekete Z, Ellis TP, Schonauer MS, Dieckmann CL. Pet127 governs a 5′ -> 3′-exonuclease important in maturation of apocytochrome b mRNA in Saccharomyces cerevisiae. J Biol Chem. 2008;283:3767–72. doi: 10.1074/jbc.M709617200. [DOI] [PubMed] [Google Scholar]
- 62.Zoschke R, Watkins KP, Barkan A. A rapid ribosome profiling method elucidates chloroplast ribosome behavior in vivo. Plant Cell. 2013;25:2265–75. doi: 10.1105/tpc.113.111567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barkan A, Rojas M, Fujii S, Yap A, Chong YS, Bond CS, Small I. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 2012;8:e1002910. doi: 10.1371/journal.pgen.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yagi Y, Hayashi S, Kobayashi K, Hirayama T, Nakamura T. Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS One. 2013;8:e57286. doi: 10.1371/journal.pone.0057286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takenaka M, Zehrmann A, Brennicke A, Graichen K. Improved computational target site prediction for pentatricopeptide repeat RNA editing factors. PLoS One. 2013;8:e65343. doi: 10.1371/journal.pone.0065343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zambrano A, Fontanesi F, Solans A, de Oliveira RL, Fox TD, Tzagoloff A, Barrientos A. Aberrant translation of cytochrome c oxidase subunit 1 mRNA species in the absence of Mss51p in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:523–35. doi: 10.1091/mbc.E06-09-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guthrie C, Fink GR, eds. Guide to yeast genetics and molecular and cell biology. San Diego: Academic Press, 2002 [Google Scholar]
- 68.Burke D, Dawson D, Stearns T. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, 2000 [Google Scholar]
- 69.Sambrook J, Russell DW. Molecular cloning, a laboratory manual. Cold Spring Harbor Laboratory Press, 2001 [Google Scholar]
- 70.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–9. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 71.Shen ZH, Fox TD. Substitution of an invariant nucleotide at the base of the highly conserved ‘530-loop’ of 15S rRNA causes suppression of yeast mitochondrial ochre mutations. Nucleic Acids Res. 1989;17:4535–9. doi: 10.1093/nar/17.12.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ostheimer GJ, Williams-Carrier R, Belcher S, Osborne E, Gierke J, Barkan A. Group II intron splicing factors derived by diversification of an ancient RNA-binding domain. EMBO J. 2003;22:3919–29. doi: 10.1093/emboj/cdg372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Diekert K, de Kroon AI, Kispal G, Lill R. Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol. 2001;65:37–51. doi: 10.1016/S0091-679X(01)65003-9. [DOI] [PubMed] [Google Scholar]
- 74.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 75.Westermann B, Herrmann JM, Neupert W. Analysis of mitochondrial translation products in vivo and in organello in yeast. Methods Cell Biol. 2001;65:429–38. doi: 10.1016/S0091-679X(01)65025-8. [DOI] [PubMed] [Google Scholar]