Abstract
Bile acids are the end products of cholesterol catabolism. Hepatic bile acid synthesis accounts for a major fraction of daily cholesterol turnover in humans. Biliary secretion of bile acids generates bile flow and facilitates hepatobiliary secretion of lipids, lipophilic metabolites, and xenobiotics. In the intestine, bile acids are essential for the absorption, transport, and metabolism of dietary fats and lipid-soluble vitamins. Extensive research in the last 2 decades has unveiled new functions of bile acids as signaling molecules and metabolic integrators. The bile acid–activated nuclear receptors farnesoid X receptor, pregnane X receptor, constitutive androstane receptor, vitamin D receptor, and G protein–coupled bile acid receptor play critical roles in the regulation of lipid, glucose, and energy metabolism, inflammation, and drug metabolism and detoxification. Bile acid synthesis exhibits a strong diurnal rhythm, which is entrained by fasting and refeeding as well as nutrient status and plays an important role for maintaining metabolic homeostasis. Recent research revealed an interaction of liver bile acids and gut microbiota in the regulation of liver metabolism. Circadian disturbance and altered gut microbiota contribute to the pathogenesis of liver diseases, inflammatory bowel diseases, nonalcoholic fatty liver disease, diabetes, and obesity. Bile acids and their derivatives are potential therapeutic agents for treating metabolic diseases of the liver.
I. Introduction
Bile acids are amphipathic molecules synthesized from cholesterol exclusively in the liver. Hepatic bile acid synthesis accounts for a major fraction of daily cholesterol turnover in humans. Biliary secretion of bile acids generates bile flow, and facilitates hepatobiliary secretion of lipids, endogenous metabolites, and xenobiotics. In bile, bile acids form mixed micelles with phospholipids and cholesterol, and bile acids are released into the small intestine after meal ingestion. Bile acids facilitate the intestinal digestion and absorption of dietary fat, steroids, drugs, and lipophilic vitamins. Most bile acids are efficiently reabsorbed in the ileum and transported back to the liver via portal circulation for resecretion into the gallbladder. Recent findings show that bile acids are also important signaling molecules that are involved in the regulation of lipid, glucose, and energy metabolism, drug metabolism, and the modulation of immune response. Basic research in the past 2 decades showed that such regulatory function of bile acids is mainly a result of bile acid activation of various intracellular ligand-activated nuclear receptors, such as the farnesoid X receptor (FXR), pregnane X receptor (PXR), and vitamin D receptor (VDR), and cell surface G protein–coupled receptors (GPCRs), such as the G protein–coupled bile acid receptor (TGR5 and Gpbar-1). Direct modulation of bile acid receptor activity by synthetic and natural receptor agonists or antagonists has shown promise in treating human diseases related to metabolic perturbations and inflammation. This review summarizes recent advances in the understanding of bile acid signaling and its regulation of metabolic homeostasis. In addition, this review covers emerging roles of circadian rhythm in bile acid regulation of metabolic diseases and drug metabolism, bile acid and the gut microbiome in metabolic diseases, microRNA (miRNA) in bile acid and drug metabolism, and the development of new bile acid–based therapeutics for the treatment of metabolic diseases.
A. Bile Acid Synthesis
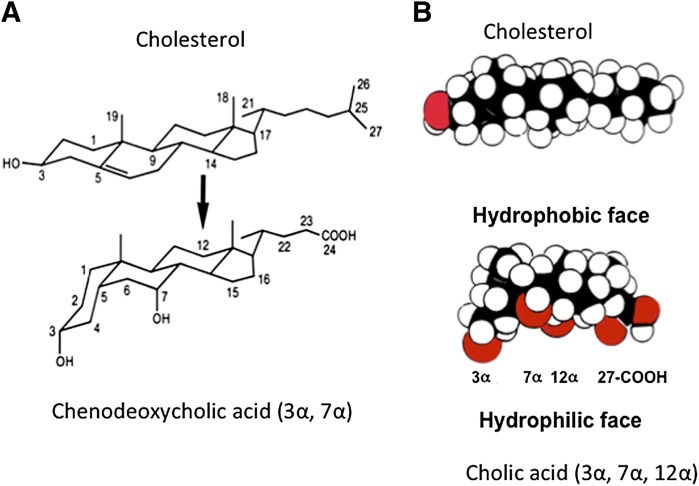
Bile acids are cholesterol derivatives. The steroid nucleus of cholesterol has four fused carbon rings consisting of three 6-carbon rings and one 5-carbon ring. The conversion of cholesterol to bile acids involves hydroxylation, saturation of the double bond at C5-C6, epimerization of the 3-hydroxyl group, and oxidative cleavage of a 3-carbon unit. Most bile acids have a 5β-hydrogen group and a cis-configuration along the plane of the fused A and B ring (Fig. 1A). In mammals, most bile acids are C24-5β-bile acids (5β-cholanoic acid). Most bile acids are conjugated to the amino acids glycine or taurine to form sodium salts (bile salts), which become negatively charged molecules with increased solubility under most physiologic pH ranges. Many bile acids have hydroxyl groups and the carboxyl group facing one side of the carbon skeleton to form a hydrophilic face opposing the other highly hydrophobic face. Thus, bile acids are amphipathic molecules with powerful detergent properties (Fig. 1B).
Fig. 1.
Bile acid structure. (A) Conversion of cholesterol to bile acid alters the stereo-configuration of the steroid ring structure. Saturation of the C5-C6 double bond changed the hydrogen group in C5 from α to β and a cis-configuration along the plane of the fused A and B ring, and caused a kink along the steroid plane in CDCA. (B) Space-filling models of cholesterol and CA. All three hydroxyl groups and the carboxyl group are faced to one side of the carbon skeleton to form a hydrophilic face, which is opposite to the hydrophobic face of the carbon skeleton.
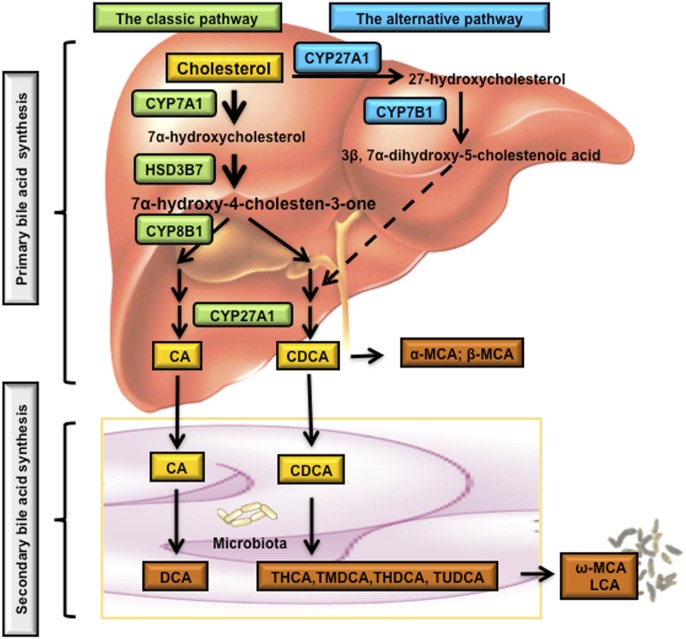
The human bile acid pool consists of the primary bile acids cholic acid (CA) and chenodeoxycholic acid (CDCA), and the secondary bile acid deoxycholic acid (DCA) and trace amount of lithocholic acid (LCA). Primary bile acids are synthesized from cholesterol in the liver. Some primary bile acids are converted to secondary bile acids by enzyme activities in gut bacteria. In the liver, primary bile acids are synthesized through two major bile acid synthetic pathways: the classic pathway (also called the neutral pathway) and the alternative pathway (also called the acidic pathway due to the production of acidic intermediates) (Fig. 2). Bile acid synthesis is a multistep reaction that involves enzymes localized in the endoplasmic reticulum (ER), mitochondria, cytosol, and peroxisomes (Russell, 2003).
Fig. 2.
Bile acid biosynthetic pathways. Two major bile acid biosynthetic pathways are shown. In the classic pathway, cholesterol is converted to 7α-hydroxycholesterol by the rate-limiting enzyme CYP7A1, which is located in the ER. The 3β-hydroxysteroid dehydrogenase (3βHSD, HSD3B7) converts 7α-hydroxycholesterol to 7α-hydroxy-4-cholesten-3-one (C4), which is converted to 7α,12α-dihydroxy-4-cholesten-3-one by a sterol 12α-hydroxylase (CYP8B1) leading to synthesis of CA. Without 12α-hydroxylation by CYP8B1, C4 is eventually converted to CDCA. The mitochondrial sterol 27-hydroxylase (CYP27A1) catalyzes the steroid side chain oxidation in both CA and CDCA synthesis. In the alternative pathway, cholesterol is first converted to 27-hydroxycholesterol by CYP27A1. Oxysterol 7α- hydroxylase (CYP7B1) catalyzes hydroxylation of 27-hydroxycholesterol to 3β,7α-dihydroxy-5-cholestenoic acid, which eventually is converted to CDCA. Oxysterol 7α-hydroxylase (CYP7B1) is nonspecific and can also catalyze hydroxylation of 25-hydroxycholesterol to 5-cholesten-3β,7α,25-triol. In the large intestine, bacterial 7α-dehydroxylase removes a hydroxyl group from C-7 and converts CA to DCA and CDCA to LCA. In mouse liver, most of CDCA is converted to α- and β-MCA. In the intestine, bacterial 7α-dehydroxylase activity convers CA and CDCA to DCA and LCA, respectively. CYP3A1 and epimerase also convert CDCA to the secondary bile acids, including THCA, TMDCA, ω-MCA, THDCA, and TUDCA. LCA and ω-MCA are excreted into feces. THCA, taurohyocholic acid; THDCA, taurohyodeoxycholic acid; TMDCA, tauromurideoxycholic acid.
1. The Classic Pathway.
In humans, the classic pathway accounts for more than 90% of total bile acid production and thus is considered the major bile acid biosynthetic pathway. The classic pathway is initiated by a microsomal cholesterol 7α-hydroxylase (CYP7A1), which converts cholesterol to 7α-hydroxycholesterol and is the rate-limiting step in the classic pathway (Myant and Mitropoulos, 1977). Then 3β-hydroxy-Δ5-C27-steroid dehydroxylase (3β-HSD) converts 7α-hydroxycholesterol to 7α-hydroxy-4-cholestene-3-one (C4), which is the common precursor for CA and CDCA, and has been used as a serum marker for the rate of bile acid synthesis (Axelson et al., 1988; Honda et al., 2007). The microsomal sterol 12α-hydroxylase (CYP8B1) mediates the hydroxylation of C4 at the C-12 position, followed by several reactions including the steroid side chain cleavage by the mitochondrial sterol 27-hydroxylase (CYP27A1), leading to the synthesis of CA. CYP7A1 regulates the overall rate of bile acid production, whereas CYP8B1 regulates the CA/CDCA ratio in the bile acid pool. In mice, the majority of CDCA is converted to α-muricholic acid (MCA) and β-MCA; therefore, CA and α- and β-MCAs are the major primary bile acids in the mouse bile acid pool.
2. The Alternative Pathway.
In humans, the alternative pathway is thought to produce less than 10% of the total bile acids under normal physiologic conditions. In this pathway, the mitochondrial CYP27A1 catalyzes the first hydroxylation reaction of cholesterol to 27-hydroxycholesterol and 3β-hydroxy-5-cholestenoic acid, which is subsequently hydroxylated at the C-7 position by oxysterol 7α-hydroxylase (CYP7B1) to form 3β,7α-dihydroxy-5-cholestenoic acid. CYP7B1 catalyzes many hydroxylation reactions in steroid synthesis in steroidogenic tissues and oxysterol synthesis in peripheral tissues. The oxysterol intermediates formed in the peripheral tissues could be transported to the liver and mainly converted to CDCA. The alternative pathway may be responsible for the synthesis of about 50% of bile acids in rodents.
B. The Enterohepatic Circulation of Bile Acids
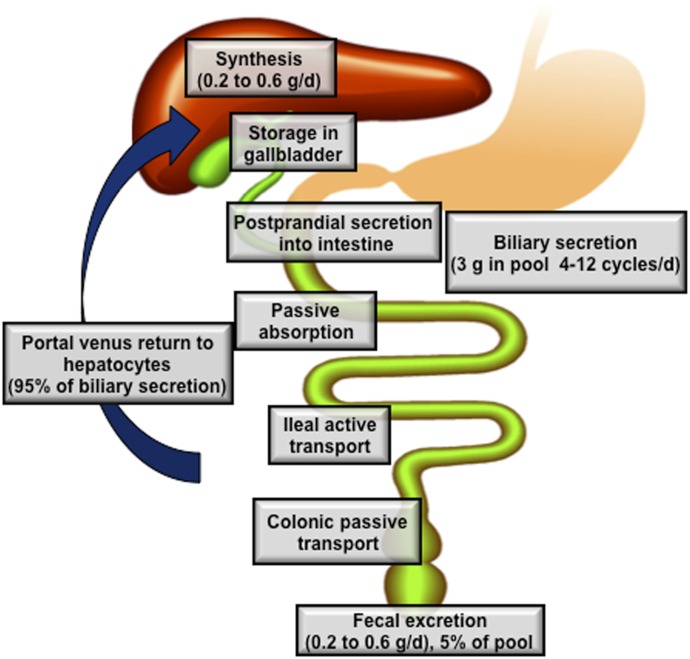
Conjugated bile acids synthesized in the hepatocytes are secreted across the canalicular (apical) membrane into the bile and stored in the gallbladder. After a meal, the duodenum secretes cholecystokinin, which stimulates gallbladder contraction and the release of bile acids into the intestinal tract. Along the small intestinal tract, micellar bile acids act as effective detergents to facilitate the solubilization of fatty acids and monoacylglycerols, digestion, and absorption of dietary lipids and fat-soluble vitamins. Bile acids are efficiently reabsorbed in the ileum, and transported back to the liver via portal blood for resecretion into the bile. This process is referred to as enterohepatic circulation of bile acids (Fig. 3). Only 5% of the total bile acid (approximately 0.5 g/day) is excreted into the feces, and this is replenished by the de novo synthesis in the liver. Bile acid transport across the plasma membrane is an active transport process that requires high-affinity bile acid transporters in the liver and the intestine (Trauner and Boyer, 2003). The bile acid pool size is defined as the total amount of bile acids in the enterohepatic circulation. It should be emphasized that bile acid compositions in humans and mice are very different. In humans, the highly hydrophobic bile acid pool consists of about 40% each of CA and CDCA, and 20% DCA. In mice, the highly hydrophilic bile acid pool consists of about 50% CA and 50% α- and β-MCAs.
Fig. 3.
Enterohepatic circulation of bile acids. The human bile acid pool consists of approximately 3 g of bile acids. Food intake stimulates the gallbladder to release bile acids into the small intestine. An average man produces approximately 0.5 g bile acid per day by synthesis in the liver, and secretes approximately 0.5 g/day. Conjugated bile acids are efficiently reabsorbed in the ileum by active transport, whereas a small amount of unconjugated bile acids is reabsorbed by passive diffusion in the small and large intestines. The first-pass extraction of bile acids from the portal blood by the liver is very efficient. Small amounts of bile acids that spilled over into the systemic circulation are recovered in kidney. The bile acids in the pool are recycled 4–12 times a day.
1. Hepatic Bile Acid Transport.
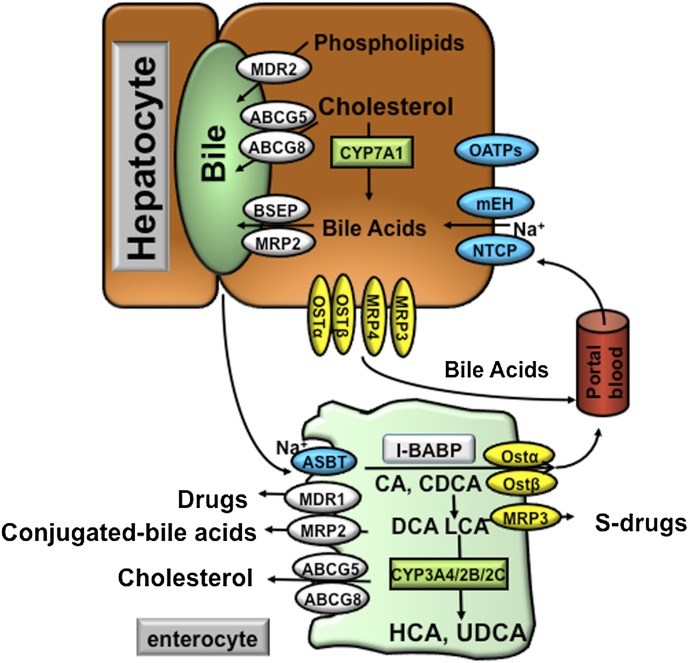
Hepatocytes are polarized cells with basolateral (sinusoidal) and apical (canalicular) membrane domains (Fig. 4). Hepatocytes take up circulating bile acids through the basolateral membrane that is in direct contact with the portal plasma. This process is highly efficient with an approximately 80–90% first-pass extraction rate for conjugated bile acids. Bile acids cannot across the hepatocyte membrane, but require active transport systems (Meier and Stieger, 2002). The majority of circulating bile acids are taken up by hepatocytes via Na+-dependent cotransport systems. The Na+-dependent taurocholate transporter (NTCP; SLC10A1) has been identified as the major bile acid uptake transporter in the basolateral (sinusoidal) membrane of hepatocytes (Ananthanarayanan et al., 1988) (Fig. 4). Several earlier studies suggested a role of the microsomal epoxide hydrolase (mEH) in mediating the basolateral Na+-dependent bile acid uptake (von Dippe et al., 1993, 1996; Fretland and Omiecinski, 2000). Mice lacking mEH showed no apparent alteration in bile acid homeostasis (Miyata et al., 1999). By contrast, a point mutation that resulted in significantly decreased mEH expression in a human individual led to hypercholanemia, a condition of increased plasma bile acid levels in the absence of hepatocyte injury, suggesting impaired basolateral bile acid uptake rather than intrahepatic bile acid accumulation (Zhu et al., 2003). It is estimated that about 25% of bile acid uptake by hepatocytes is mediated by Na+-independent mechanisms (Trauner and Boyer, 2003). This pathway is mainly responsible for the uptake of unconjugated bile acids. Several organic anion transporters (OATPs), including OATP1A2, OATP1B1, and OATP1B3, have been identified as Na+-independent bile acid transporters.
Fig. 4.
Bile acid transporters in the hepatocytes and enterocytes. At the basolateral membrane of the hepatocytes, the NTCP and mEH may be responsible for Na+-dependent uptake of conjugated bile acids, whereas OATPs show substrate specificity for unconjugated bile acids. At the canalicular membrane of the hepatocytes, the BSEP plays a major role in biliary secretion of bile acids, whereas the MRP2 mediates secretion of organic substrates including bile acids, bilirubin, and glutathione. ABCG5 and ABCG8 heterodimers transport cholesterol into the bile, whereas MDR2 is responsible for biliary secretion of phospholipids. At the basolateral membrane of the hepatocytes, organic solute transporters OSTα and OSTβ heterodimers, MRP3, and MRP4 mediate secretion of bile acids into the circulation. With cholestasis, both basolateral bile acid efflux and renal bile acid excretion are increased. After bile acids are released from the gallbladder into the intestine, ileal bile acid uptake is mediated by the ASBT. Intracellular bile acids are bound to the intestinal bile acid binding protein (IBABP). At the basolateral membrane, bile acid efflux is mediated by the OSTα and OSTβ heterodimers. At the apical membrane of the enterocytes, ABCG5 and ABCG8 heterodimers transport cholesterol back into the intestinal lumen, a process that limits intestine cholesterol absorption. CYP3A4, CYP2B, and CYP2C are involved in the metabolism and detoxification of LCA in the intestine. In the apical membrane of intestine, MDR1 effluxes drugs and MRP2 effluxes conjugated bile acids. In the sinusoidal membrane, MRP3 effluxes sulfur-conjugated drugs for renal excretion.
The concentration of bile acids in the bile can be approximately 100- to 1000-fold higher than that in the hepatocytes. Canalicular bile acid transport against the concentration gradient thus represents the rate-limiting step in bile formation. The ATP-binding cassette (ABC) transporter bile salt export pump (BSEP; ABCB11), also known as the sister of P-glycoprotein, effluxes bile acids across the canalicular membrane (Childs et al., 1995). Patients with progressive familial intrahepatic cholestasis (PFIC) subtype 2 due to mutations in the BSEP gene show markedly elevated plasma bile acid levels with only less than 1% of normal biliary bile acid concentration, suggesting that BSEP is the major canalicular bile acid transport system in the hepatocytes (Strautnieks et al., 1997). The multidrug resistance–associated protein MRP2 (ABCC2), which mediates the transport of a wide range of organic substrates including bilirubin conjugates, glutathione, drugs, and so forth, also shows substrate specificity for divalent bile acids, such as sulfate-conjugated or glucuronidated N-acetylamidated bile acids. Bile acids, phospholipids, and cholesterol are three major organic solutes of bile and once secreted, form mixed micelles to increase cholesterol solubility and lower the monomeric concentration of bile acids, thereby reducing their cytotoxicity to the bile duct. Cholesterol secretion into the bile is mediated by the ABCG5 and ABCG8 heterodimer at the canalicular membrane. The major phospholipid in the bile is phosphatidylcholine, which is excreted via the phospholipid flippase, multidrug-resistant protein MDR2 (ABCB4) (Smit et al., 1993).
2. Intestinal Bile Acid Transport.
It is estimated that about 90–95% of bile acids are reabsorbed in the intestine with minimal daily loss in the feces. Therefore, the amount of bile acid synthesized by the liver to compensate the daily fecal loss and thus to maintain a constant bile acid pool size is also low under physiologic conditions. Intestinal bile acid reabsorption mainly occurs at the terminal ileum by the apical sodium-dependent bile salt transporter (ASBT; SLC10A2) (Wong et al., 1994; Shneider et al., 1995) (Fig. 4). Once absorbed into the enterocytes, bile acids bind to the intestinal bile acid binding protein and are transported to the basolateral membrane for secretion (Gong et al., 1994). The heterodimer organic solute transporters OSTα and OSTβ appeared to be the major basolateral bile acid transport system in the intestine (Ballatori et al., 2005; Dawson et al., 2005). Overexpression of OSTα and OSTβ in mice enhanced basolateral efflux of taurocholate, whereas mice lacking ostα showed significantly decreased intestinal bile acid absorption, plasma bile acids, and total bile acid pool (Rao et al., 2008). OSTα and OSTβ are also expressed in the human liver, and at relatively low levels in mouse liver (Ballatori et al., 2005). Both OSTα and OSTβ are localized at the basolateral membrane of the hepatocytes. OSTα and OSTβ are induced during cholestasis and thus mediate bile acid efflux into the circulation for renal excretion (Boyer et al., 2006; Cui et al., 2009). Interestingly, recent evidence suggests that OSTα/OSTβ also plays a role in mediating bile acid reuptake in the kidney (Soroka et al., 2010).
In the intestine, bile acids undergo multistep biotransformation usually catalyzed by enzyme activities in gut bacteria (Chiang, 1998; Ridlon et al., 2006). In the small and large intestine, some conjugated bile acids are deconjugated by bacterial bile salt hydrolases (BSHs) to become free bile acids. Unconjugated bile acids can cross the plasma membrane via passive diffusion, a process that accounts for a small fraction of bile acids recycled along the small and large intestines. In the large intestine, bacterial 7α-dehydroxylase removes a hydroxyl group from C-7 and converts CA to DCA and CDCA to LCA, respectively. These secondary bile acids are cytotoxic. DCA and, to a much less extent, LCA are reabsorbed in the large intestine via passive absorption. In humans, LCA is sulfated and N-acylamidated in the liver and secreted into bile. In mice, LCA is detoxified by hydroxylation at C-6 and/or C-7 in the intestine for fecal excretion. A small amount of LCA (approximately 1%) circulated to the liver is sulfated and efficiently secreted into the circulation for renal excretion. In the intestine, CYP3A4, CYP2B, CYP2C, and epimerases are involved in detoxification of LCA to more soluble hyocholic acid and ursodeoxycholic acid (UDCA) in humans. In mouse intestine LCA can be 7α-hydroxylated to CDCA, which is converted to α-MCA, β-MCA, ω-MCA, UDCA, hyodeoxycholic acid, and hyocholic acid (Fig. 2).
C. Regulation of Bile Acid Synthesis
It has been shown that feeding rats with bile acids resulted in a strong reduction of hepatic CYP7A1 enzyme activity and bile acid synthesis, whereas interruption of bile acid reabsorption in the intestine by bile acid binding resin, and subsequent reduction of bile acids returning to the liver, stimulated hepatic CYP7A1 enzyme activity and bile acid synthesis. This early evidence suggests that hepatic CYP7A1 and bile acid synthesis are under negative feedback regulation by bile acids. It is now clear that bile acid synthesis is mainly controlled by bile acids via the transcriptional repression of the CYP7A1 gene (Chiang, 2009). This bile acid feedback repression mechanism allows the liver to efficiently stimulate or inhibit bile acid synthesis in response to changes in bile acid levels and thus to maintain bile acid homeostasis. During cholestasis, repression of bile acid synthesis is a protective mechanism against hepatic bile acid accumulation and bile acid cytotoxicity.
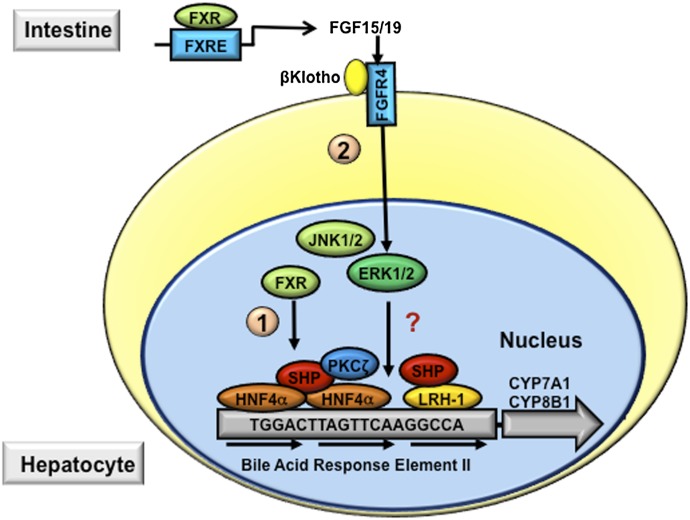
The mechanisms by which bile acids inhibit hepatic CYP7A1 have been extensively investigated in the past decades (Chiang, 2009). These studies have shown that the nuclear receptor FXR plays a key role in mediating the bile acid feedback inhibition of CYP7A1 in the hepatocytes. It was first shown that upon activation by bile acids, hepatic FXR transcriptionally induced a nuclear receptor small heterodimer partner (SHP), which acted as a corepressor to inhibit the transcriptional activity of a nuclear receptor liver-related homolog-1 (LRH-1) or hepatocyte nuclear factor (HNF) 4α bound to the CYP7A1 gene promoter, and resulted in inhibition of CYP7A1 gene transcription (pathway 1, Fig. 5) (Chiang et al., 2000; Goodwin et al., 2000; Lu et al., 2000). However, the repression of CYP7A1 by bile acids and an FXR agonist was still observed in shp null mice, which implied that the FXR/SHP/LRH-1 cascade may not likely be the only pathway mediating bile acid feedback inhibition of CYP7A1 (Kerr et al., 2002; Wang et al., 2002). It was reported earlier that intraduodenal, but not intravenous, infusion of taurocholic acid (TCA) repressed CYP7A1 mRNA expression in rats (Pandak et al., 1995), suggesting that bile acids might induce an intestinal factor that mediates bile acid feedback inhibition of bile acid synthesis. In addition to the liver, FXR is also highly expressed in the intestine. Inagaki et al. (2005) reported that activation of FXR by bile acids or synthetic agonists in mice induced an intestinal fibroblast growth factor FGF15, which activates a liver fibroblast growth factor receptor FGFR4 to inhibit CYP7A1 and bile acid synthesis (pathway 2, Fig. 5). The FGF15/FGFR4 signaling requires a transmembrane protein called β-Klotho (Lin et al., 2007). Mice lacking either FGFR4 or β-Klotho had elevated hepatic CYP7A1 mRNA, enlarged bile acid pool size, and impaired bile acid feedback inhibition of CYP7A1 (Yu et al., 2000, 2005a). Thus, this intestine-to-liver bile acid sensing mechanism may play a critical role in maintaining overall bile acid homeostasis.
Fig. 5.
Mechanisms of bile acid feedback inhibition of bile acid synthesis. Bile acid–activated signaling inhibits CYP7A1 and CYP8B1 and therefore reduces hepatic bile acid synthesis. The bile acid response element (BARE) located in the CYP7A1 gene promoter contains AGGTCA-like direct repeats. HNF4α and LRH1 bind to the BARE and stimulate CYP7A1 gene transcription. In hepatocytes, bile acids activate FXR, which induces the repressor SHP. SHP interacts with and represses the transactivating action of HNF4α and LRH, a process that involves the recruitment of corepressor complex and chromatin remodeling enzymes (indicated as pathway 1). In the intestine, bile acid–activated FXR induces FGF15 (FGF19 in humans), which binds and activates FGFR4 on the hepatocytes. FGFR4 activates intracellular signaling pathways, such as ERK, protein kinase Cζ (PKCζ), and c-Jun N-terminal kinase (JNK), which leads to the repression of CYP7A1 gene transcription (indicated in pathway 2).
Human FGF19 shares approximately 51% amino acid sequence identity with mouse FGF15, and is considered to be the mouse FGF15 ortholog. FGF19 has been shown to repress CYP7A1 in human hepatocytes (Holt et al., 2003; Song et al., 2009). In contrast with FGF15 in mice, FGF19 mRNA is detectable in human livers and human hepatocytes, as well as gallbladder (Schaap et al., 2009; Song et al., 2009), and FGF19 protein is detectable in human blood circulation (Lundåsen et al., 2006). Cholestyramine treatment reduced plasma FGF19 levels in humans (Lundåsen et al., 2006). Circulating FGF19 levels were elevated in human patients with obstructive cholestasis, indicating that human hepatocytes produce FGF19 (Schaap et al., 2009). In human hepatocytes, FGF19 is highly inducible by bile acids or FXR agonists (Holt et al., 2003; Song et al., 2009). Such evidence suggests that in humans, both liver and intestine secrete FGF19 into the blood circulation, and that the FXR/FGF19 pathway is involved in bile acid sensing and regulation in both human liver and intestine, either via an autocrine or an endocrine manner.
The intracellular events after FGFR4 activation to repress CYP7A1 are incompletely understood (Fig. 5). In human hepatocytes, FGF19 repression of CYP7A1 was shown to be largely dependent on the activation of extracellular signal-regulated kinase (ERK) signaling, but the downstream target is still not clear (Song et al., 2009). In mice, the possible involvement of ERK, c-Jun N-terminal kinase, and SHP in mediating FGF15 repression of CYP7A1 has been suggested (Inagaki et al., 2005). It was shown that the repressive effect of CYP7A1 by adenovirus-mediated FGF15 overexpression in the liver was attenuated in shp knockout mice (Inagaki et al., 2005). On the other hand, the FGF15 repression of CYP7A1 was still very strong in shp knockout mice (Kong et al., 2012), suggesting that SHP is unlikely to be the major mediator of FGF15 repression of CYP7A1. Further studies are needed to delineate the intracellular mechanisms mediating the FGF15/FGF19 repression of CYP7A1 in hepatocytes.
FGF15 was also implicated in the regulation of gallbladder refilling in mice (Choi et al., 2006). Mice lacking FGF15 had almost empty gallbladders, and the gallbladder volume was restored by the administration of recombinant FGF15. By contrast, human gallbladders secrete high levels of FGF19 into the bile, and gallbladder FGF19 concentrations are about 100-fold higher than that in the blood circulation (Zweers et al., 2012). Both FGFR4 and β-Klotho were expressed in the epithelial cells of the gallbladder, suggesting that the gallbladder should also respond to FGF19-activated intracellular signaling. The role of FGF19 signaling in the human gallbladder remains to be defined.
II. Bile Acid Receptor Signaling in Liver Metabolism
A. Bile Acid–Activated Nuclear Receptors
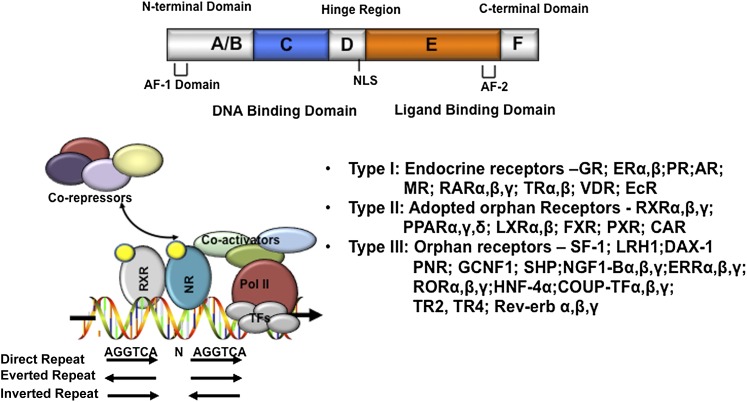
Nuclear receptors are a group of ligand-activated transcription factors that play important roles in embryogenesis, development, and metabolism (Mangelsdorf and Evans, 1995; Mangelsdorf et al., 1995; Chiang, 2002). Nuclear receptors play multiple roles in liver physiology and pathophysiology (Karpen and Trauner, 2010). The general structure of a nuclear receptor consists of an N-terminal DNA binding domain and a C-terminal ligand-binding domain (LBD) (Fig. 6). The N-terminal DNA binding domain is the most conserved region, containing two Zinc finger motifs that allow the nuclear receptor to bind to a consensus AGGTCA-like DNA sequence (termed the hormone response element) as a homodimer or heterodimer with the retinoid X receptor. A few nuclear receptors (e.g., HNF4 and LRH) bind DNA as monomers. The LBD is involved in ligand binding, nuclear receptor dimerization, and coregulator (coactivators or corepressors) interaction. Ligand binding causes a conformational change in the LBD, which leads to a shift of the helix 12 to allow the LBD to interact with the coactivators via the LXXLL motif–containing nuclear receptor box on the coactivators. Upon recruitment to the nuclear receptor, coactivators displace corepressors, and further recruit chromatin remodeling proteins to facilitate the assembly of general transcriptional complex that induces gene transcription. There are 48 nuclear receptor genes in the human genome and 49 in the mouse genome (Mangelsdorf et al., 1995).
Fig. 6.
Nuclear receptors. The domain structure of nuclear receptors is shown on the top. The putative nuclear receptor response element binding sequences, arranged in direct repeat, everted repeat, and inverted repeat, are shown. Ligand-activated receptors recruit coactivators to replace corepressors, which results in transactivation of target gene expression. Nuclear receptors are classified into three types: type I endocrine receptors, type II adapted orphan receptors, and type III orphan receptors. Refer to Chawla et al. (2001) for details on the nuclear receptor superfamily and nomenclature. AF-1, activation function-1. AF-2, activation function-2; NLS, nuclear localization sequence.
Bile acids directly activate three nuclear receptors: FXR (Forman et al., 1995; Makishima et al., 1999; Parks et al., 1999), PXR (Xie et al., 2001), and VDR (Makishima et al., 2002). FXR and PXR are highly expressed in tissues that are exposed to bile acids, including the liver and the intestine (Forman et al., 1995; Kliewer et al., 1998), whereas VDR is widely expressed in most tissues. FXR can be activated by both free and conjugated bile acids; the hydrophobic bile acid CDCA is the most efficacious ligand of FXR (EC50 = approximately 10 μM), followed by LCA, DCA, and CA, whereas hydrophilic bile acids UDCA and MCA do not activate FXR. The secondary bile acid LCA and its metabolite 3-keto-LCA are the most potent activators of PXR and VDR, with an EC50 of approximately 100 nM. These nuclear receptors act as sensors of bile acid levels in the hepatocytes and the enterocytes, and mediate the pleiotropic effects of bile acids in the regulation of metabolic homeostasis.
1. Farnesoid X Receptor Regulation of Bile Acid Homeostasis.
FXR regulates a network of genes in hepatic bile acid synthesis, biliary bile acid secretion, intestinal bile acid absorption, and hepatic bile acid uptake, and thus plays a key role in the regulation of bile acid homeostasis (Table 1). In addition to mediating the bile acid inhibition of bile acid synthesis, FXR activation by bile acids prevents hepatic bile acid accumulation via transcriptional induction of the apical bile acid efflux transporters BSEP (Ananthanarayanan et al., 2001) and MRP2 (Kast et al., 2002), the phosphatidylcholine transporter MDR2 (Liu et al., 2003), and the cholesterol transporters ABCG5 and ABCG8 (Li et al., 2011b). It seems that FXR activation coordinates the biliary secretion of bile acids, cholesterol, and phospholipids, which form mixed micelles in the bile to prevent gallstone formation and bile acid damage to the bile duct epithelium (Liu et al., 2003; Wittenburg et al., 2003; Moschetta et al., 2004). At the basolateral membrane of hepatocytes, the expression of bile acid uptake transporter NTCP is repressed by FXR, presumably via induction of SHP that represses the transactivation of NTCP by retinoic acid receptor and HNF4α (Denson et al., 2001). In conditions associated with hepatic bile acid accumulation, there is a compensatory induction of transporters at the basolateral membrane of the hepatocytes to efflux bile acids into the blood circulation, resulting in elevated plasma bile acid concentration and renal excretion of bile acids. FXR activation induces the OSTα and OSTβ expression at the sinusoidal membrane to efflux bile acids to the systemic circulation (Lee et al., 2006). Several basolateral drug transporters (MRP1, MRP3, and MRP4) are able to transport conjugated bile acids (Trauner and Boyer, 2003). These MRPs are induced during cholestasis in both mice and humans, but other studies suggest that induction of these basolateral MRPs may be due to activation of PXR rather than FXR (Zollner et al., 2007).
TABLE 1.
FXR-regulated genes in metabolism
| Metabolism | Target Gene | Physiologic Function |
|---|---|---|
| Bile acid | CYP7A1 | Bile acid synthesis |
| CYP8B1 | Bile acid synthesis | |
| BSEP | Biliary bile acid secretion | |
| NTCP | Basolateral bile acid uptake | |
| SHP | Bile acid synthesis | |
| OSTα/β | Basolateral bile acid secretion | |
| IBABP | Intracellular bile acid transport | |
| FGF15/19 | Bile acid synthesis | |
| Glucose | PEPCK | Gluconeogenesis |
| G6Pase | Gluconeogenesis | |
| CREB | Gluconeogenesis | |
| FoxO1 | Gluconeogenesis | |
| HNF4α | Gluconeogenesis | |
| FGF15/FGF19 | gluconeogenesis | |
| Insulin | Glucose metabolism | |
| Triglyceride | SREBP-1 | Lipogenesis |
| ChREBP | Lipogenesis | |
| PPARα | Fatty acid oxidation | |
| CES1 | Fatty acid oxidation | |
| FGF21 | Fatty acid oxidation | |
| ApoCII | Triglyceride clearance | |
| ApoCIII | Triglyceride clearance | |
| ApoA5 | Triglyceride clearance | |
| Cholesterol | PCSK9 | LDL uptake |
| SR-BI | HDL uptake | |
| ApoAI | HDL biogenesis | |
| ABCG5/G8 | Cholesterol secretion |
CES1, carboxylesterase 1; CREB, cAMP response element binding protein; FoxO1, forkhead box protein O1; IBABP, intestinal bile acid binding protein.
Similar to the effect of FXR activation in the hepatocytes, activation of intestine FXR by bile acids limits bile acid uptake and promotes basolateral bile acid secretion to decrease intracellular bile acid concentration. Bile acids inhibit mouse and human ASBT, but not rat ASBT (Arrese et al., 1998; Chen et al., 2003; Neimark et al., 2004). FXR induces the major intestinal bile acid efflux transporters OSTα and OSTβ located in the sinusoidal membrane (Lee et al., 2006).
Numerous studies have been carried out in fxr knockout mice to evaluate the role of FXR in regulating bile acid homeostasis and detoxification under bile duct ligation (BDL), bile acid feeding or drug-induced intrahepatic cholestasis (Wagner et al., 2003, 2011; Cui et al., 2009). Mice lacking FXR were phenotypically normal but showed elevated CYP7A1 mRNA expression and enlarged bile acid pool size (Sinal et al., 2000; Kok et al., 2003). When fed a CA-containing diet, fxr null mice showed more severe hepatotoxicity, which was accompanied by the lack of CYP7A1 repression and induction of FXR regulated transporters in the liver and the intestine. The fxr null mice were also more susceptible to bile acid–induced liver injury after BDL despite induction of the PXR-regulated bile acid detoxification network.
2. Farnesoid X Receptor Regulation of Triglyceride Metabolism.
Bile acids are known to have lipid-lowering effects and control liver triglycerides by mechanisms that are not fully understood. It has long been known that treating human gallstone patients with CDCA decreases hepatic very low-density lipoprotein (VLDL) production and plasma triglyceride levels (Schoenfield and Lachin, 1981), whereas treating hypercholesterolemia patients with bile acid–binding resins raised plasma triglyceride levels (Angelin et al., 1978). Studies conducted in fxr knockout mice further revealed that FXR deletion resulted in hepatic lipid accumulation and elevation of circulating total cholesterol and total triglycerides, as well as a proatherogenic lipoprotein profile. Conversely, activation of FXR by bile acids or FXR agonists decreased liver and plasma cholesterol and triglycerides in mice (Zhang et al., 2006a).
One mechanism by which FXR activation reduces hepatic fat accumulation and plasma triglyceride levels is the inhibition of hepatic de novo lipogenesis and VLDL overproduction. Obesity and diabetes are associated with hepatic fat accumulation, VLDL overproduction, and hypertriglyceridemia. It is generally thought that central obesity and insulin resistance lead to increased free fatty acid release from the adipose tissues and free fatty acid uptake by the liver. It is estimated that adipose-derived free fatty acid could account for up to 75% of hepatic triglycerides in nonalcoholic fatty liver associated with obesity and diabetes (Cohen et al., 2011). In addition, de novo lipogenesis from glycolysis is often induced in obesity and diabetes, which is accompanied by increased hepatic expression of steroid response element binding protein (SREBP)-1c. SREBPs are basic helix-loop-helix leucine-zipper transcription factors that are regulated by cholesterol/oxysterol and play a key role in the regulation of fatty acid and cholesterol synthesis (Brown and Goldstein, 1997; Horton et al., 2002; Shao and Espenshade, 2012). Two SREBP genes encode SREBP-1c and SREBP-2. SREBP-1c mainly regulates fatty acid synthesis, whereas SREBP-2 regulates cholesterol synthesis. SREBPs are synthesized as inactive precursors (125 kDa) in the ER. When intracellular oxysterol levels are high, SREBP/SREBP cleavage activation protein complex interacts with the insulin-induced gene (INSIG) and is retained in the ER. When intracellular oxysterol levels decrease, SREBP/SREBP cleavage activation protein escorted SREBP to the Golgi apparatus, where two sterol-sensitive proteases cleave an N terminus fragment (68 kDa). This processed or matured SREBP is translocated to the nucleus to bind to gene promoters containing the sterol response element and to activate target gene transcription (Wang et al., 1994). SREBP-1c induces the expression of a number of genes involved in de novo lipogenesis including acetyl-CoA carboxylase (ACC), fatty acid synthase (FAS), acetyl-CoA synthetase, ATP-citrate lyase, malic enzyme, glucose-6-phosphate dehydrogenase, 6-phosphogluconate dehydrogenase, and stearoyl-CoA desaturase-1 (Brown and Goldstein, 1997; Horton et al., 2002). Insulin is known to induce both SREBP-1c mRNA expression by the liver orphan receptor (LXR) and proteolytic cleavage (Repa et al., 2000; Shao and Espenshade, 2012). It was suggested that the FXR/SHP cascade might repress LXRα induction of SREBP-1c and lipogenesis (Watanabe et al., 2004). More recently, it was reported that FXR activation inhibits another lipogenic transcriptional factor called the carbohydrate response element binding protein (ChREBP) (Caron et al., 2013). High levels of glucose initiate the nuclear translocation of ChREBP, which induces a set of genes that promote the conversion of glucose into fatty acids (Uyeda et al., 2002; Postic et al., 2007). It was shown that FXR directly interacted with ChREBP and inhibited ChREBP transactivation of its target gene L–pyruvate kinase. Numerous studies have shown that inhibition of ChREBP alleviated hepatic steatosis in mice (Dentin et al., 2004; Iizuka et al., 2004). ChREBP is also a transcriptional target of LXR, and thus may mediate the lipogenic effects of LXR activation (Cha and Repa, 2007). Taken together, these studies suggest that bile acids activate FXR to reduce hepatic lipogenesis, and thus reduce hepatic steatosis and VLDL overproduction.
Bile acid sequestrants decrease bile acid activation of FXR signaling to stimulate CYP7A1 gene expression, which reduces intracellular cholesterol to activate SREBP and induce the low-density lipoprotein receptor (LDLR) to reduce serum cholesterol. Transgenic overexpression of CYP7A1 in mice increased hepatic expression of SREBP and lipogenic genes (Miyake et al., 2001). Importantly, Li et al. (2010) reported that CYP7A1 transgenic mice fed a chow diet had increased hepatic VLDL secretion and elevated plasma VLDL triglyceride levels even in the presence of a significantly enlarged bile acid pool. Although the endogenous CYP7A1 mRNA was repressed more than 90% by an enlarged bile acid pool, none of the FXR target genes such as FAS, ACC, and stearoyl-CoA desaturase-1 in fatty acid synthesis were repressed. Microarray analysis showed that hepatic CYP7A1 overexpression was associated with higher induction of SREBP-2 target genes in cholesterol synthesis than SREBP-1 target genes in lipogenesis (Li et al., 2013b). These studies show that increasing bile acid synthesis stimulates de novo cholesterol synthesis to provide cholesterol substrate for CYP7A1. This may shift acetyl-CoA from fatty acid synthesis to cholesterol synthesis and result in reducing lipogenesis.
FXR has been shown to induce peroxisome proliferator–activated receptor (PPAR)α, suggesting that FXR activation may promote hepatic fatty acid oxidation (Pineda Torra et al., 2003). FXR also induces hepatic expression and secretion of FGF21, a fasting-induced hormone involved in hepatic lipid oxidation and ketogenesis (Badman et al., 2007; Inagaki et al., 2007; Cyphert et al., 2012). This new evidence suggests that the triglyceride-lowering effect of FXR activation can also be attributed to increased hepatic fatty acid oxidation.
Plasma triglyceride is cleared after VLDL triglyceride is hydrolyzed by lipoprotein lipase (LPL) lining the endothelial cells of the peripheral tissues. Obesity and diabetes are reported to associate with impaired peripheral triglyceride clearance, contributing to diabetic hypertriglyceridemia (Pruneta-Deloche et al., 2004). Activation of FXR induces apolipoprotein (Apo) CII and ApoA5, which are LPL activators carried by VLDL, and represses ApoCIII, which is an LPL inhibitor (Kast et al., 2001; Claudel et al., 2003; Prieur et al., 2003). Another study showed that the VLDL receptor is induced upon FXR activation (Sirvent et al., 2004). The VLDL receptor is expressed at low levels in the liver but is also expressed in peripheral tissues including heart, skeletal muscle, adipose, and blood vessels and mediates peripheral triglyceride clearance. In summary, these studies support that activation of FXR may lower plasma triglyceride by both decreasing hepatic triglyceride production and promoting plasma triglyceride clearance.
3. Farnesoid X Receptor Regulation of Glucose Metabolism.
The first evidence linking FXR to glucose metabolism came from a study showing that hepatic FXR expression was decreased in streptozotocin-induced diabetic rats, which was corrected with insulin supplements (Duran-Sandoval et al., 2004). In fxr knockout mice, hepatic expressions of glucose metabolism genes showed altered response to refeeding (Duran-Sandoval et al., 2005). Bile acid synthesis was markedly increased during the postprandial period likely due to increased glucose influx and activation of insulin signaling (Li et al., 2012). A more recent study showed that FXR was O-Glc-N-acylated under high glucose concentration to stimulate ligand-dependent FXR transactivating activity and increase cellular glucose flux and hexoseamine biosynthetic pathway (Berrabah et al., 2014). These studies indicate that FXR may be activated during the postprandial period to play a role in the regulation of glucose homeostasis (Kir et al., 2011; Potthoff et al., 2011; Li et al., 2012). Thus, activation of FXR by bile acid feeding or administration of the FXR agonist GW4064 (3-[2-[2-chloro-4-[[3-(2,6-dichlorophenyl)-5-(1-methylethyl)-4-isoxazolyl]methoxy]phenyl]ethenyl]benzoic acid) lowered fasting plasma glucose and improved insulin sensitivity in obese and diabetic db/db mice (Ma et al., 2006; Zhang et al., 2006a), whereas FXR-deficient mice had insulin resistance and hyperglycemia (Ma et al., 2006).
Current antidiabetic drugs used to control fasting hyperglycemia act to either directly inhibit hepatic gluconeogenesis or to promote insulin secretion (Radziuk et al., 2003). The possibility that FXR activation inhibits hepatic gluconeogenesis to lower fasting plasma glucose has been extensively investigated in in vitro and in vivo models, but to date the role of FXR in the regulation of hepatic glucose production remains controversial. Bile acids and FXR repressed gluconeogenic genes phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase (G6Pase) in primary hepatocytes and in liver cell lines by inducing SHP, which inhibits the transactivating activity of many transcription factors known to regulate these genes, including fasting-induced cAMP responsive element binding protein, forkhead box protein O1, CCAAT/element binding protein, glucocorticoid receptor, HNF3, and HNF4α (Yamagata et al., 2004). By contrast, one study showed that FXR bound to the promoter of PEPCK and FXR agonism induced PEPCK mRNA expression and glucose output in human and rat hepatocytes and in mouse liver (Stayrook et al., 2005). It should be noted that, despite such discrepancies, a majority of studies concluded that FXR activation and bile acid administration decreased fasting plasma glucose levels in diabetic mice. The effect of FXR in improving plasma glucose homeostasis may not be solely dependent on the direct inhibition of hepatic gluconeogenesis.
Several studies suggest that FXR regulates peripheral insulin sensitivity. Hyperinsulinemic euglycemic clamp studies demonstrated that FXR deficiency was associated with decreased whole body glucose disposal in mice (Cariou et al., 2006; Ma et al., 2006). Fxr knockout mice had elevated circulating and muscle free fatty acids, which explains the peripheral insulin-resistant phenotype observed in these mice (Ma et al., 2006). Consistently, FXR activation decreased free fatty acid levels and increased insulin sensitivity in mice (Zhang et al., 2006a). It is noticed that fxr knockout mice had smaller adipocytes, and FXR agonist GW4064 treatment increased adipose differentiation and insulin-dependent glucose uptake in 3T3-L1 cells in vitro (Cariou et al., 2006). Another study suggests that the FXR agonist INT-747 (6α-ethyl-3α,7α-dihydroxy-5β-cholan-24-oic acid) induced adipose differentiation via induction of the expression of adipocyte-related genes including CCAAT/element binding protein-α and PPARγ (Rizzo et al., 2006). FXR is not expressed in skeletal muscle and is expressed in white adipose at a very low level, which raised the possibility that some insulin-sensitizing and glucose-reducing effects observed in FXR-deficient mice and in mice treated with FXR agonists or bile acids may be indirect or independent of FXR. Two recent studies showed that FXR deficiency in mice protected against genetic and diet-induced obesity and insulin resistance (Prawitt et al., 2011), whereas chronic administration of the FXR agonist GW4064 caused more weight gain when mice were fed a high-fat diet (Watanabe et al., 2011). Some of these effects may likely be due to an altered bile acid pool and signaling because fxr knockout may result in increased hepatic CYP7A1 and enlarged bile acid pool and tissue bile acid signaling, whereas the use of a nonbile acid–like FXR agonist such as GW4064 will likely diminish the bile acid pool.
FXR induces FGF15 in the intestine, which is secreted into the blood circulation and acts as a postprandial factor that promotes glycogen synthesis as a mechanism controlling postprandial glucose metabolism (Kir et al., 2011). It has been shown that plasma FGF19 increases during the postprandial period in humans, presumably due to increased bile acid signaling (Lundåsen et al., 2006). Interestingly, a few studies showed that FGF19 transgenic mice were resistant to diet-induced obesity and insulin resistance (Tomlinson et al., 2002; Fu et al., 2004). In addition, FGF19 has been shown to repress hepatic glucose production (Potthoff et al., 2011), promote glycogen synthesis (Kir et al., 2011), repress lipogenesis (Bhatnagar et al., 2009; Miyata et al., 2011a), and increase metabolic rate (Tomlinson et al., 2002; Fu et al., 2004). Fgf15 knockout mice showed glucose intolerance, increased hepatic gluconeogenesis, and reduced hepatic glycogen storage but unaltered overall insulin sensitivity. Decreased fasting FGF19 levels or impaired hepatic response to FGF19 have been reported in humans with fatty liver and insulin resistance (Schreuder et al., 2010; Wojcik et al., 2012). These studies suggest that bile acid regulation of hepatic glucose metabolism involves complex cross-talk between FXR-dependent pathways and FXR-independent signaling pathways. Some metabolic alteration observed in FXR loss-of-function or gain-of-function models may be due to indirect modulation of bile acid metabolism.
More recently, a few studies showed that FXR is also expressed in human and murine pancreatic β cells and may positively regulate glucose-dependent insulin secretion (Popescu et al., 2010; Renga et al., 2010; Düfer et al., 2012). The underlying mechanisms of this beneficial effect of FXR activation in the β cells are not clear and both genomic and nongenomic actions have been suggested. For example, FXR activation was shown to stimulate insulin gene transcription (Renga et al., 2010). It was also shown that bile acids, such as CDCA, rapidly increased cytosolic Ca2+ concentrations and membrane depolarization in β cells (Düfer et al., 2012). Another study showed that FXR activation was associated with increased AKT phosphorylation and translocation of glucose transporter 2 to the cell membrane, and thus enhanced glucose uptake into pancreatic β cells (Renga et al., 2010). Whether these effects are mediated via nongenomic FXR action or via FXR-independent signaling pathways is still not clear.
4. Farnesoid X Receptor Regulation of Cholesterol Metabolism.
The role of FXR in the development and progression of atherosclerosis has been extensively investigated (Hageman et al., 2010). It was shown that the synthetic FXR agonist INT-747 reduced aortic plaque formation in apoe knockout mice (Mencarelli et al., 2009). Similar antiatherogenic effects were also observed in ldlr knockout mice and in apoe knockout mice fed the synthetic FXR agonist WAY-362450 [3-(3,4-difluorobenzoyl)-1,2,3,6-tetrahydro-1,1-dimethylazepino[4,5-b]indole-5-carboxylic acid 1-methylethyl ester] (Hartman et al., 2009). Thus, current studies consistently support that FXR activation is antiatherogenic. By contrast, studies evaluating the role of FXR in the development of atherosclerosis in FXR loss-of-function models yielded inconsistent results. Hanniman et al. (2005) reported that FXR deletion in the apoe knockout mice fed a high-fat/high-cholesterol diet resulted in a more atherogenic lipoprotein profile with increased VLDL cholesterol, low-density lipoprotein-cholesterol (LDL-C), and decreased high-density lipoprotein (HDL)-cholesterol (HDL-C) and increased atherosclerotic lesion size. Opposing the findings from this study, two other studies found reduced atherosclerosis development when FXR was deleted in ldlr knockout mice and/or in apoe knockout mice (Guo et al., 2006; Zhang et al., 2006b). Reduced macrophage CD36 expression and low-density lipoprotein (LDL) uptake into macrophages have been reported in these mice, suggesting possible underlying mechanisms.
Mice lacking FXR had elevated LDL in the circulation when challenged with a high-cholesterol diet (Sinal et al., 2000). Activation of FXR by bile acids and synthetic FXR agonists mainly caused a reduction of HDL-C in wild-type mice, and decreased both LDL-C and HDL-C in hypercholesterolemia mice (Zhang et al., 2006a). How FXR activation reduced plasma LDL-C is still not fully understood. Plasma LDL-C is cleared via LDLR-mediated uptake. FXR activation was shown to repress proprotein convertase subtilisin/kexin type 9 (PCSK9), an LDLR inhibitor, in vitro (Langhi et al., 2008). Recent studies demonstrated that secreted PCSK9 promoted LDLR internalization and intracellular degradation and thus decreased LDLR recycling back to the plasma membrane (Maxwell et al., 2005). Gain-of-function mutation of PCSK9 was associated with premature atherosclerosis, whereas loss-of-function mutation of PCSK9 resulted in lower plasma LDL-C and reduced risk of coronary heart disease (Abifadel et al., 2003; Timms et al., 2004). However, activation of FXR significantly decreased plasma non-HDL cholesterol in ldlr−/− mice, suggesting that the cholesterol-lowering effects of FXR activation are not solely mediated by LDLR-mediated clearance (Hartman et al., 2009). FXR activation reduced hepatic lipogenesis and VLDL production, increased plasma triglyceride clearance, improved insulin sensitivity, and decreased circulating fatty acid levels, all of which could play a role in reducing plasma LDL-C upon FXR activation.
Plasma HDL transports cholesterol from peripheral tissues to the liver for catabolism to bile acids and biliary secretion. This process is called reverse cholesterol transport and plays a critical role in preventing the development of atherosclerosis (Brufau et al., 2011). The role of FXR in regulating HDL metabolism is still under debate. Mice lacking FXR showed either unaltered HDL-C (Sinal et al., 2000) or increased HDL-C (Claudel et al., 2002), whereas activation of FXR decreased HDL-C (Zhang et al., 2010). FXR inhibits the hepatic production of ApoAI, a key structural component of HDL (Claudel et al., 2002), suggesting that FXR may inhibit HDL biogenesis. It has been demonstrated that pharmacological activation of FXR reduced plasma HDL-C in a number of hyperlipidemia mouse models (Evans et al., 2009b; Hartman et al., 2009). It should be noted that although HDL is generally considered to be the major mediator of reverse cholesterol transport, the macrophage-derived cholesterol only accounts for a small portion of the total plasma HDL-C. Importantly, it was shown that activation of FXR promoted macrophage-to-feces reverse cholesterol transport in mice by inducing hepatic expression of the HDL receptor, scavenger receptor B1 (SR-BI) (Zhang et al., 2010). Fxr knockout mice showed a reduced rate of plasma HDL-C clearance (Lambert et al., 2003). SR-BI deficiency caused higher plasma HDL and increased atherosclerosis in mice (Rigotti et al., 1997; Trigatti et al., 2003). Human subjects with a heterozygous SR-BI mutation showed an approximately 50% increase in plasma HDL (Vergeer et al., 2006, 2011). Studies suggest that the antiatherogenic function of SR-BI may be mainly attributed to its role in mediating the cholesterol efflux from macrophages and hepatic HDL uptake (Ji et al., 1997; Covey et al., 2003; Zhang et al., 2005). Therefore, it is also possible that decreased plasma HDL upon FXR activation may partially result from increased hepatic HDL uptake (Mencarelli et al., 2009). The last step of reverse cholesterol transport is biliary secretion of cholesterol or bile acids for fecal excretion. FXR activation represses CYP7A1 and thus does not stimulate hepatic cholesterol catabolism. Mice lacking functional ABCG5/ABCG8 showed reduced macrophage-to-feces reverse cholesterol transport when stimulated by an LXR agonist (Calpe-Berdiel et al., 2008). It is shown that cholate feeding induced hepatic ABCG5 and ABCG8 expression, which was lower in fxr knockout mice (Repa et al., 2002; Yu et al., 2005b). Bile acid activation of FXR induced hepatic expression of ABCG5 and ABCG8 through a common FXR-responsive element located in the intergenic promoter shared by ABCG5 and ABCG8 (Li et al., 2011b). Thus, activation of FXR may simultaneously promote hepatic uptake of HDL-C and biliary secretion of free cholesterol via ABCG5/ABCG8 (Lambert et al., 2003). In addition to the classic peripheral-to-liver reverse cholesterol transport route, recent studies in mice revealed the existence of a transintestinal cholesterol excretion (TICE) pathway that is quantitatively significant in fecal cholesterol secretion (van der Velde et al., 2010; Brufau et al., 2011). TICE may also be present in humans, but the quantitative contribution to overall fecal neural sterol excretion is not clear (van der Velde et al., 2010). The molecular mechanisms for TICE have not been fully defined. FXR is highly expressed in the intestine but its potential role in the regulation of TICE remains unclear.
5. Anti-Inflammatory Function of Farnesoid X Receptor.
FXR is expressed in vascular smooth muscle cells (VSMCs) (Bishop-Bailey et al., 2004; Zhang et al., 2008). Recent evidence suggests that FXR activation may play a role in decreasing inflammation of the vasculature, which is critically involved in the progression of atherosclerosis (Bishop-Bailey et al., 2004; Li et al., 2007). In addition, FXR plays an anti-inflammatory role during liver injury (Wang et al., 2008; Zhang et al., 2009b) and in experimental models of inflammatory bowel disease (IBD) (Vavassori et al., 2009; Gadaleta et al., 2011b). The underlying molecular mechanism by which FXR reduces inflammation is not clear. It was reported that FXR induced SHP to inhibit the expression of cyclooxygenase 2 and inducible nitric oxide (NO) synthase, which are involved in vascular inflammation and VSMC migration. FXR activation may also antagonize nuclear factor-κB (NF-κB) signaling to decrease proinflammatory cytokine production in the liver (Wang et al., 2008). Although some studies did not detect FXR expression in macrophages, others reported that FXR was expressed in macrophages and activation of FXR repressed lipopolysaccharide (LPS)-induced proinflammatory cytokine expression, an effect that was abolished in fxr−/− macrophages (Mencarelli et al., 2009). The potential anti-inflammatory role of FXR in macrophages remains to be further defined.
6. Bile Acid/Xenobiotic Receptors in Bile Acid and Drug Metabolism and Detoxification.
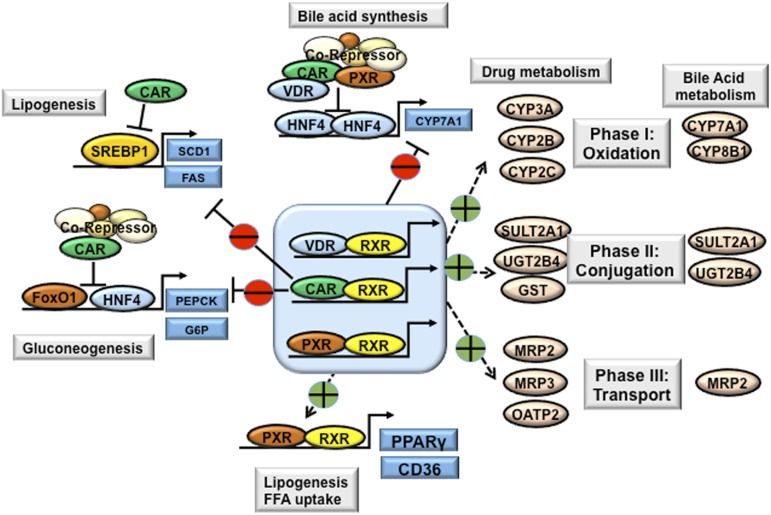
PXR, constitutive androstane receptor (CAR), and VDR play important roles in the regulation of all three phases of drug metabolism and detoxification, as well as in bile acid metabolism and detoxification, including phase I drug/bile acid hydroxylation/oxidation, phase II drug/bile acid conjugation, and phase III drug/bile acid transport (Fig. 7) (Li and Chiang, 2013). In cholestasis, bile acids activate these nuclear receptors to induce expression of the genes involved in bile acid and drug metabolism, conjugation, and transport in the liver and intestine as an adaptive response to protect against injury and inflammation.
Fig. 7.
Xenobiotic nuclear receptors in bile acid, drug, lipid, and glucose metabolism. The xenobiotic nuclear receptors PXR and CAR are highly expressed in the liver and intestine. VDR is highly expressed in the intestine, but is expressed at low levels in the liver. Activation of xenobiotic nuclear receptors by drugs, bile acids, and xenobiotics induces a network of genes involved in phase I, phase II, and phase III drug and bile acid metabolism and detoxification. CAR, PXR, and VDR inhibit CYP7A1 and thus bile acid synthesis via interaction with HNF4α and inhibition of HNF4α transactivation of CYP7A1. Similarly, CAR inhibits PEPCK and G6Pase involved in gluconeogenesis and inhibits SREBP-1c in lipogenesis. Activation of CAR decreases plasma glucose levels and improves hepatic steatosis in obesity and diabetes. By contrast, activation of PXR induces hepatic expression of PPARγ and CD36, leading to hepatic steatosis. FFA, free fatty acid; FoxO1, forkhead box protein O1; SCD1, stearoyl-CoA desaturase-1.
a. Pregnane X receptor.
Treating rodents with prognenolone-16α-carbonitrile, a mouse PXR agonist, repressed hepatic CYP7A1 activity (Ståhlberg, 1995), suggesting that PXR may regulate bile acid metabolism. This was subsequently confirmed by studies showing that prognenolone-16α-carbonitrile repressed CYP7A1 mRNA expression and biliary bile acid secretion, but failed to do so in pxr knockout mice (Staudinger et al., 2001). Two groups showed that rifampicin-activated PXR repressed human CYP7A1 gene transcription via inhibition of HNF4α and peroxisome proliferator–activated receptor γ coactivator-1α (PGC-1α) transactivation of CYP7A1 gene transcription (Bhalla et al., 2004; Li and Chiang, 2005). Activation of PXR in the intestine also induced FGF15 or FGF19 expression, and a PXR response element was identified in the promoter of the fgf19 gene (Wistuba et al., 2007; Wang et al., 2011a). PXR induces CYP3A enzymes, which play a major role in the hydroxylation of bile acids (Staudinger et al., 2001). PXR also induces the bile acid conjugation enzymes, sulfotransferase SULT2A1 and UDP-glucuronosyl N-transferase UGT1A1, the canalicular transporter MRP2, and the basolateral transporter OATP2 (Kliewer and Willson, 2002). Mice treated with LCA or subjected to BDL showed induction of PXR target genes involved in bile acid detoxification, whereas pxr knockout mice were more susceptible to hepatotoxicity caused by LCA treatment or BDL, supporting the significance of the adaptive activation of PXR in the prevention of bile acid toxicity during cholestasis (Staudinger et al., 2001; Stedman et al., 2005). In addition, numerous studies have demonstrated that pharmacological activation of PXR protected against bile acid–induced liver injury in experimental cholestasis models (Stedman et al., 2005). Rifampicin has been used to reduce pruritus (itch) associated with cholestasis in humans (Hofmann, 2002). The pathophysiological cause of pruritus is not completely known (Kremer et al., 2012), but may be associated with high tissue and plasma bile acid accumulation (Bunchorntavakul and Reddy, 2012). A recent study linked lysophospholipase D (autotaxin) and its product, lysophosphatidic acid, as mediators of cholestatic pruritus (Kremer et al., 2012). Rifampicin significantly reduced itch intensity and autotaxin activity in patients not responding to bile acid sequestrants. Rifampicin inhibited autotaxin expression in HepG2 cells overexpressed with PXR. This explains the antipruritic action of rifampicin by PXR-dependent transcriptional inhibition of autotaxin expression.
b. Constitutive androstane receptor.
CAR and PXR can bind to the same xenobiotic response elements in the target gene promoters and thus it is not unexpected that PXR and CAR regulate an overlapping set of target genes in xenobiotic and bile acid metabolism. There is no evidence that bile acids bind or activate CAR, but CAR agonists have been shown to repress the CYP7A1 gene in hepatocytes (Miao et al., 2006). A number of studies showed that activation of CAR was beneficial for protecting against bile acid toxicity during cholestasis in mice (Guo et al., 2003; Saini et al., 2004; Stedman et al., 2005; Beilke et al., 2009). Mice lacking CAR had higher degree of liver injury than wild-type mice upon LCA treatment or in BDL-induced cholestasis, and such liver damage was aggravated in car/pxr double knockout mice, suggesting that PXR and CAR may play complementary roles in bile acid detoxification (Stedman et al., 2005). CAR may play an important role in inducing sulfation of LCA as demonstrated by the resistance of CAR transgenic mice to LCA toxicity and increased sulfated LCA without the induction of CYP3A (Saini et al., 2004). In a study by Guo et al. (2003), fxr/pxr double knockout mice treated with phenobarbital and synthetic analog TCPOBOP (1,4-bis[2-(3,5-dichloropyridyloxy)]benzene) were protected from CA feeding-induced bile acid toxicity, which was accompanied by induction of CAR target genes carnitine palmitoyltransferase 2b, Cyp3a, Mrp2, Ugt1a1, and glutathione-S-transferase a.
c. Vitamin D receptor.
The VDR acts as a bile acid sensor in the intestine and protects the gut from bile acid toxicity (Nagpal et al., 2005). Activation of VDR by 1α,25-dihydroxyvitamin D3 induces CYP3A4, CYP2B, and CYP2C in intestinal cells, suggesting a role for VDR in drug and bile acid metabolism (Schmiedlin-Ren et al., 2001). Activation of VDR also induced SULT2A1 and thus simultaneously stimulated bile acid sulfation (Chatterjee et al., 2005). Furthermore, two bile acid transporters (MRP3 and ASBT) were shown to be VDR targets in the intestine (McCarthy et al., 2005; Chen et al., 2006). Accumulation of LCA in the gut may activate the VDR to convert LCA to less toxic intermediates for excretion. VDR mRNA and protein were detected at very low levels in primary human hepatocytes (Han et al., 2010). Treating primary human hepatocytes with 1α,25-dihydroxyvitamin D3 induced CYP3A, CYP2B, and CYP2C (Drocourt et al., 2002). During cholestasis, LCA levels may increase significantly in the liver. It was first shown that treating primary human hepatocytes with 1α,25-dihydroxyvitamin D3 repressed CYP7A1 mRNA expression (Han and Chiang, 2009). Consistently, a recent study showed that vdr−/− mice had higher hepatic cyp7a1 gene expression and enlarged bile acid pool size, whereas 1α,25-dihydroxyvitamin D3 treatment repressed hepatic cyp7a1 gene expression in mice (Schmidt et al., 2010). It was shown that vdr knockout mice had lower intestinal FGF15 expression, whereas 1α,25-dihydroxyvitamin D3 treatment increased intestinal FGF15 in mice. A VDR binding site was identified in the fgf15 gene promoter (Schmidt et al., 2010). In addition, 1α,25-dihydroxyvitamin D3 did not repress CYP7A1 in fgf15−/− mice, supporting that activation of intestinal VDR represses hepatic CYP7A1 via FGF15 signaling in mice. One study showed that 1α,25-dihydroxyvitamin D3 treatments had no effect on hepatic or plasma bile acid levels in mice after BDL, suggesting a minimal role of VDR in modulating bile acid metabolism in cholestasis (Ogura et al., 2009). However, this study showed that VDR activation in BDL mice was associated with reduced proinflammatory cytokine expression, consistent with the known role of VDR in immunity. These results suggest that the anti-inflammatory properties of VDR may provide certain benefits during cholestasis. The role of VDR in regulating innate and adaptive immunity is reviewed in detail elsewhere (Nagpal et al., 2005). It was also shown that 1α,25-dihydroxyvitamin D3 treatment was accompanied by increased renal MRP2, MRP3, and MRP4 mRNA expression and increased renal bile acid secretion (Nishida et al., 2009). However, this study also showed that 1α,25-dihydroxyvitamin D3 treatment markedly increased hepatic CYP7A1 mRNA without altering total bile acid pool size or serum bile acid concentration in chow-fed mice. Further studies are needed to evaluate the beneficial effects of pharmacological activation of VDR in cholestasis.
7. Bile Acid/Xenobiotic Receptors in Glucose and Lipid Metabolism.
Recent evidence suggests that the bile acid/xenobiotic receptors PXR and CAR also act as metabolic regulators (referred to as endobiotic receptors) under physiologic and pathologic conditions, and thus may be implicated in the development and treatment of metabolic diseases. It has long been known that phenobarbital reduces plasma fasting glucose and improves insulin sensitivity in diabetic patients (Lahtela et al., 1984). A few recent studies showed that activation of CAR by TCPOBOP improved hyperglycemia and insulin sensitivity in obese and diabetic ob/ob mice and high-fat diet–induced obese mice (Dong et al., 2009; Gao et al., 2009; Masuyama and Hiramatsu, 2012). It was reported that phenobarbital decreased the expression of PEPCK and G6Pase in both primary hepatocytes and mouse liver, suggesting that phenobarbital may inhibit hepatic gluconeogenesis (Kodama et al., 2004; Miao et al., 2006). On the other hand, the effect of CAR activation on plasma glucose levels may also be attributed to increased peripheral glucose clearance, as suggested by glucose tolerance testing and euglycemic clamp studies in both mice and humans (Lahtela et al., 1984).
A few recent studies have reported antiobesity and lipid-lowering effects of CAR agonists in mice (Gao et al., 2009; Sberna et al., 2011; Masuyama and Hiramatsu, 2012). The mechanisms by which CAR activation decreases weight gain is not clear, but reduced body weight gain in mice likely contributes to reduced hepatic steatosis and hepatic VLDL production and promotes the antiatherogenic lipid profile in mice treated with CAR agonists. In the liver, CAR activation decreased both SREBP-1c mRNA expression and the mature form of SREBP-1c, presumably via inducing Insig-1 involved in SREBP retention in the ER (Shao and Espenshade, 2012). CAR activation also increased reverse cholesterol transport, which may be due to increased hepatic bile acid excretion (Sberna et al., 2011).
Despite the fact that CAR and PXR can recognize common DNA response elements and regulate an overlapping set of target genes, activation of these two xenobiotic receptors surprisingly resulted in completely opposite outcomes in lipid metabolism. Liver-specific transgenic expression of a constitutively active PXR resulted in marked hepatic lipid accumulation, and treating “humanized” PXR transgenic mice with rifampicin also resulted in hepatic lipid accumulation (Zhou et al., 2006). Mechanistic studies suggest that PXR activation induces hepatic steatosis via a combined effect of enhanced lipogenesis and repressed fatty acid oxidation. It was shown that PXR increased the expression of the hepatic fatty acid uptake transporter fatty acid translocase FAT/CD36 (Zhou et al., 2008). This study also showed that PXR induced hepatic expression of PPARγ. Activation of PPARγ has been shown to cause hepatic fat accumulation. In addition, PXR activation was shown to promote an atherogenic lipoprotein profile in wild-type mice and increased the development of atherosclerosis in apoe−/− mice (Zhou et al., 2009). Consistently, PXR deficiency attenuated the development of atherosclerosis in apoe knockout mice (Sui et al., 2011). Studies so far suggest that the proatherogenic effect of PXR may be due to PXR induction of CD36 in the macrophages, which leads to increased uptake of oxidized LDL in atherogenic prone mouse models.
B. Bile Acid–Activated G Protein–Coupled Receptors
1. The G Protein–Coupled Bile Acid Receptor Is a Bile Acid–Activated Membrane Receptor.
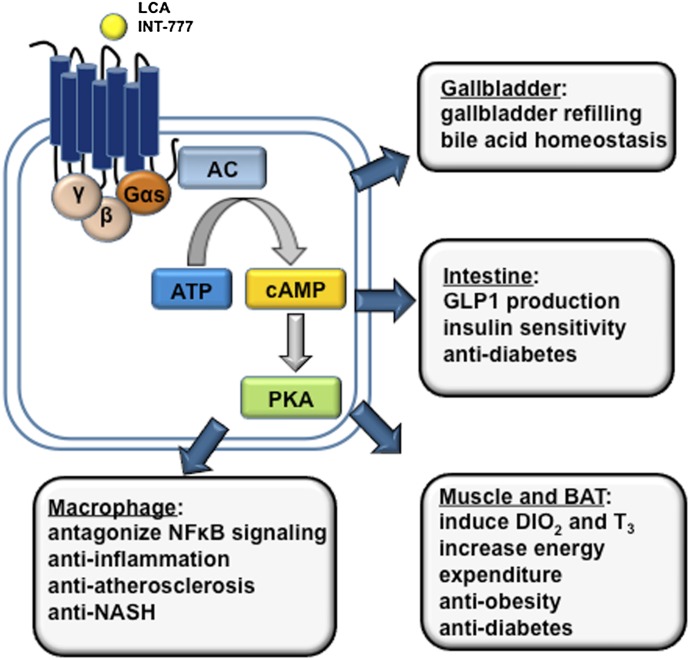
Bile acids are known to activate intracellular signaling pathways via nongenomic actions. Bile acid was shown to stimulate cAMP production in colonic cells, suggesting the existence of a bile acid–activated GPCR. Gαs protein–coupled receptor TGR5 was recently identified as a bile acid–activated membrane receptor (Maruyama et al., 2002; Kawamata et al., 2003). Activation of TGR5 by bile acids stimulated adenylate cyclase, rapid intracellular cAMP production, and protein kinase A activation (Fig. 8). Both conjugated bile acids and free bile acids are known to bind and activate TGR5. Among all bile acids, TLCA is the most potent TGR5 agonist with an EC50 value of 0.53 μM, followed by taurodeoxycholic acid (TDCA), taurochenodeoxycholic acid, and TCA with EC50 values of approximately 1.0, 4.4, and 7.7 μM, respectively. TGR5 is highly expressed along the intestinal tract, with the highest expression found in the ileum and colon (Kawamata et al., 2003). Despite the liver being a major bile acid target organ, TGR5 expression in the liver is very low. TGR5 is expressed in liver sinusoidal endothelial cells, gallbladder epithelial cells, and Kupffer cells (Keitel et al., 2007, 2008). TGR5 is also expressed in nontraditional bile acid target organs including white and brown adipose, spleen, kidney, pancreas, lung, macrophages, and the central nervous system (Kawamata et al., 2003; Keitel et al., 2010). It is generally recognized that TGR5 signaling plays important roles in energy and glucose metabolism as well as anti-inflammation in the digestive system.
Fig. 8.
Bile acid–activated TGR5 signaling in metabolism and inflammation. TGR5 is activated by secondary LCA and synthetic agonists (e.g., INT-777). TGR5 is a Gαs GPCR that induces cAMP/PKA signaling. TGR5 is expressed in brown adipocytes, macrophages/monocytes and hepatic Kupffer cells, gallbladder epithelium, and intestine, with high levels found in the colon. TGR5 is not expressed in hepatocytes. In brown adipose tissue, TGR5 activation stimulates energy expenditure; in the intestine, TGR5 activation stimulates GLP-1 production from L cells. These metabolic effects underlie the antiobesity and antidiabetic properties of TGR5 agonists. TGR5 activation shows anti-inflammatory effects, and TGR5 activation may protect against colitis, Crohn’s disease, and atherosclerosis. TGR5 in the gallbladder epithelium regulates gallbladder refilling. AC, adenylate cyclase; DIO2, type 2 deiodinase; PKA, protein kinase A; T3, 3,5,3′-triiodothyronine.
a. The G protein–coupled bile acid receptor in bile acid metabolism.
Maruyama et al. (2006) previously reported that the total bile acid pool size is decreased in tgr5−/− mice, suggesting that TGR5 might regulate bile acid metabolism. Tgr5−/− mice had normal fecal bile acid secretion, suggesting that decreased bile acid pool may not be due to a role of TGR5 in regulating intestinal bile acid reabsorption. In addition, hepatic expression of CYP7A1 was unaltered in tgr5−/− mice. It has been reported that tgr5−/− mice are protected from lithogenic diet–induced cholesterol gallstone and bile acid feedback regulation may be altered (Vassileva et al., 2006). TGR5 is highly expressed in the epithelium of mouse and human gallbladder and plays a role in stimulating gallbladder filling (Keitel et al., 2009; Li et al., 2011a). A recent study showed that tgr5−/− mice had a more hydrophobic bile acid composition with increased CYP8B1 and a decreased MCA/CA ratio, as well as more severe liver injury upon bile acid feeding or BDL (Péan et al., 2013). It was demonstrated that tgr5−/− mice had reduced liver regeneration capacity and TGR5 might protect the liver from bile acid overload during liver regeneration in mice. The mechanism by which tgr5 gene ablation alters bile acid metabolism is still largely unknown.
b. The G protein–coupled bile acid receptor in energy expenditure and glucose metabolism.
Feeding mice a lithogenic diet (containing 0.5% CA and 0.2% cholesterol) prevented diet-induced weight gain (Watanabe et al., 2006). It seems that the antiobesity effect of bile acids is mediated through the activation of TGR5 in the brown adipose tissue in mice. Bile acid activation of TGR5, via cAMP signaling, induced type 2 deiodinase and the conversion of thyroxine to the active 3,5,3′-triiodothyronine, which is known to stimulate mitochondrial oxidative phosphorylation and energy expenditure. A similar effect was also seen when mice were given a potent synthetic TGR5 agonist INT-777 [6α-ethyl-23(S)-methylcholic acid]. The effect of bile acids on body weight loss was independent of FXR activation because the FXR agonist GW4064 did not increase energy expenditure or reduce body weight in mice. Furthermore, it was later shown that chronic feeding of GW4064 to mice resulted in increased susceptibility to diet-induced weight gain, which was associated with a decreased bile acid pool and reduced energy expenditure (Watanabe et al., 2011). The role of TGR5 in energy metabolism in humans has not been studied.
TGR5 is highly expressed in the small and large intestines, which are exposed to high levels of bile acids. The colon is exposed to LCA, the most potent TGR5 agonist among all bile acids. Katsuma et al. (2005) first showed that bile acid–activated TGR5 stimulated glucagon-like peptide-1 (GLP-1) production in an enteroendocrine cell line. GLP-1 is known to promote insulin secretion and thus regulate glucose homeostasis. Thomas et al. (2009) further showed that administration of a potent TGR5 agonist INT-777 raised the intracellular ATP/ADP ratio and calcium influx, which leads to enhanced GLP-1 secretion. Both INT-777–treated mice and tgr5 transgenic mice showed improved glucose tolerance. Because GLP-1 mimetic and receptor agonists have shown promise in improving glucose homeostasis in diabetes, bile acid–based TGR5 agonists may be used to stimulate GLP-1 secretion in diabetic patients (Nauck, 2011). In contrast with these results obtained from TGR5 gain-of-function models, studies in tgr5−/− mice yielded inconsistent results. One study reported higher weight gain in female, but not male, tgr5−/− mice (Maruyama et al., 2006). Another study showed that neither under chow condition nor under a high-fat diet feeding condition did tgr5−/− mice show higher body weight than wild-type controls (Vassileva et al., 2010). The high-fat diet feeding effect on insulin sensitivity showed sex-specific differences, with male tgr5−/− mice showing impaired, but female tgr5−/− mice showing improved, insulin sensitivity (Vassileva et al., 2010). Thus, it is possible that TGR5 is not highly activated by the physiologic concentration of circulating bile acids outside of the enterohepatic system, which may explain why loss of TGR5 did not have a major impact on weight gain and glucose metabolism. By contrast, pharmacological activation of TGR5 showed a clear beneficial effect on body weight reduction and glucose homeostasis. The physiologic role of TGR5 in the regulation of metabolic homeostasis needs further study.
c. G protein–coupled bile acid receptor modulation of immune response.
TGR5 is highly expressed in monocytes and macrophages and in human spleen, and may play an anti-inflammatory role in the immune system. Thus far, TGR5 activation has shown protective effects in various inflammation-related diseases in experimental models (Kawamata et al., 2003; Keitel et al., 2008). In Kupffer cells, activation of TGR5 reduced LPS-stimulated proinflammatory cytokine production (Keitel et al., 2008). The specific TGR5 agonist INT-777 attenuated liver damage associated with high-fat diet–induced steatosis. Tgr5−/− mice challenged with LPS showed higher plasma liver enzymes and elevated cytokine expression, whereas 23(S)-mCDCA, a highly selective TGR5 agonist, antagonized LPS-induced cytokine expression in mouse liver (Wang et al., 2011b). Since TGR5 is not expressed in hepatocytes, such effects of TGR5 activation may be attributed to its activation in Kupffer cells. The anti-inflammatory role of TGR5 in the liver is supported by recent studies demonstrating a protective role of TGR5 activation in cholestasis and nonalcoholic steatohepatitis (NASH) (McMahan et al., 2013; Péan et al., 2013). However, the underlying mechanism of TGR5 signaling in anti-inflammation in liver is completely unknown. In the vasculature, TGR5 activation by INT-777 attenuated atherosclerosis in ldlr−/− mice, but not in ldlr/tgr5 double knockout mice. Importantly, it was shown that INT-777 failed to attenuate atherosclerosis in ldlr−/− mice transplanted with bone marrow of tgr5−/− mice, demonstrating that the antiatherogenic effect of the TGR5 agonist was due to TGR5 activation in macrophages. The anti-inflammatory function of TGR5 in protection against IBD is mediated by inhibition of NF-κB–dependent proinflammatory cytokine production. It was demonstrated that a TGR5 selective agonist protected the integrity of intestinal barrier function, immune response, and proinflammatory cytokine production in experimental colitis models (Cipriani et al., 2011; Yoneno et al., 2013).
d. Novel functions of the G protein–coupled bile acid receptor.
Two studies unveiled novel functions of TGR5 signaling in the brain. TGR5 is expressed in astrocytes and neurons, and is activated by neurosteroids, and functions as a neurosteroid receptor (Keitel et al., 2010). Another study showed that TGR5 may mediate bile acid–induced itch and analgesia associated with cholestasis (Alemi et al., 2013a). In this study, the investigators identified TGR5 in peptidergic neurons that transmit itch and pain in mouse dorsal root ganglia and in the spinal cord as well as in dermal macrophage–containing opioids. Alemi et al. (2013a) observed that scratching was reduced in tgr5−/− mice but increased in TGR5 transgenic mice exhibiting spontaneous pruritus. Bile acids activate TGR5 on sensory nerves and stimulate the release of neuropeptides in the spinal cord that transmits itch and analgesia. TGR5 is highly expressed in the colon and has been linked to gut motor function. Tgr5 knockout mice have higher transit time and lower defecation, suggesting that TGR5 mediates the prokinetic action of bile acids in the intestine and is required for normal defecation (Alemi et al., 2013b). A single nucleotide polymorphism in the TGR5 gene has been associated with irritable bowel syndrome (Camilleri et al., 2011). TLCA increased endothelial NO synthase phosphorylation and NO production, and TGR5-mediated NO production significantly reduced tumor necrosis factor α–induced monocyte adhesion in vascular endothelial cells (Kida et al., 2013).
2. Sphingosine-1-Phosphate Receptor 2.
A recent study identified the sphingosine-1-phosphate receptor 2 (S1PR2) as a bile acid–activated GPCR (Studer et al., 2012). S1PR2 is a Gαi class of GPCRs and is expressed in hepatocytes and many other tissues. It was previously reported that conjugated bile acids activated the ERK1/2 and AKT signaling pathways, which were sensitive to pertussis toxin inhibition in rat primary hepatocytes (Dent et al., 2005). Screening GPCRs in the lipid-activated family identified that TCA, TDCA, tauroursodeoxycholic acid (TUDCA), glycocholic acid, and glycodeoxycholic acid specifically activated S1PR2, but not other GPCRs in the family. Sphingosine-1-phosphate (S1P) is a potent pleiotropic lipid mediator that has been shown to activate at least five different S1PRs. S1P is the phosphorylated product of the membrane lipid sphingosine by sphingosine kinases SphK1 and SphK2. Extracellular signals activate SphK1, which is translocated from the cytosol to the plasma membrane to convert membrane sphingosine to S1P. SphK2 is located in the nucleus and is also activated by ERK1/2-mediated phosphorylation. S1P activates S1P receptors by an autocrine/paracrine manner. Molecular modeling shows that TCA is able to dock into the S1P binding site on S1PR2 (Studer et al., 2012). Sphingosine is also derived from ceramide or synthesized from serine and palmitoyl-CoA. It has been reported that DCA activates acidic sphingomyelinase to generate ceramide from sphingomyelin (Gupta et al., 2004). Both ceramide and S1P levels are increased in obese humans (Chavez and Summers, 2012). Ceramide production and signaling have been linked to the pathogenesis of insulin resistance and other obesity- and diabetes-associated metabolic disorders; in addition, promoting the conversion of ceramide to S1P may attenuate the deleterious effects of ceramide and high-fat diet–induced insulin resistance (Bruce et al., 2012). On the other hand, S1P has been implicated in the regulation of inflammation, cell death, and insulin sensitivity via activation of different S1P receptors in liver, muscle, and pancreas and immune cells (Adada et al., 2013). Interestingly, it was reported that SphK2 is associated with histone deacetylase histone deacetylase (HDAC)1/2, and S1P inhibits HDAC1/2 activity and stimulates histone acetylation and transcription of cyclin-dependent kinase inhibitor p21 and cFos (Hait et al., 2009).
III. Bile Acids and Gut Microbiota
Bile acids and gut microbiota are closely linked. Gut microbiota are involved in the biotransformation of bile acids through deconjugation, dehydroxylation, and reconjugation of bile acids (Ridlon et al., 2006). Bile acids have antimicrobial activity by damaging the bacterial cell membrane and thus inhibiting bacteria outgrowth (Kurdi et al., 2006). Obstruction of bile flow by BDL caused bacterial proliferation and mucosal injury in the small intestine and bile acid treatment reduced bacteria outgrowth. It has been reported recently that bile acid activation of FXR induced genes involved in enter protection and inhibited bacterial overgrowth and mucosal injury (Inagaki et al., 2006).
Gut microbiota play important roles in pathogen defense, immunity, and nutrient harvest. Recent evidence suggests that there is a regulatory relationship between the development of obesity and altered gut microbiota (Aron-Wisnewsky et al., 2013). Obesity, caloric intake, and macronutrients of the diet affect gut microbial communities (Ley et al., 2005, 2006; Parks et al., 2013). It is suggested that intestinal microbes generate short chain fatty acids from dietary carbohydrates that otherwise cannot be used as energy (Topping and Clifton, 2001; Gill et al., 2006; Turnbaugh et al., 2006). Therefore, gut microbiota composition directly affects energy metabolism, leading to remarkable alterations of lipid, glucose and energy metabolism in the liver, muscle, and adipose. Most of such evidence has been obtained by studying the metabolism in germ-free mice. A number of studies showed that germ-free mice were protected against diet-induced obesity compared with the conventionally raised counterparts (Bäckhed et al., 2004; Turnbaugh et al., 2006). Interestingly, germ-free mice receiving caecal microbiota from ob/ob mice had higher energy absorption from food and more weight gain than germ-free mice harboring caecal microbiota from lean mice (Turnbaugh et al., 2006). Importantly, introducing gut microbiota from the cecum of conventionally raised mice to germ-free mice caused rapid weight gain and insulin resistance without increasing food intake (Bäckhed et al., 2004). The gut microbiota affected the release of gut hormones, such as peptide YY and GLP1 (Reimer and McBurney, 1996; Kok et al., 1998; Cani et al., 2004), and low-grade inflammation (Noverr and Huffnagle, 2004; Cani et al., 2007), which linked gut microbiota to the development of obesity and diabetes (Qin et al., 2012; Joyce and Gahan, 2014).
Figure 9 illustrates the interaction between bile acid metabolism and gut microbiota and the effects of high-fat diets, drugs, and circadian rhythm on liver and intestine inflammation and related metabolic diseases. Bile acids, through activation of FXR and TGR5 signaling, control microbiota overgrowth and composition and protect liver and intestine against inflammation. On the other hand, gut microbiota regulate bile acid biotransformation in the intestine, which alter bile acid composition and modulate FXR and TGR5 signaling. Bile acids appear to play a major role in regulation of the gut microbiome (Ridlon et al., 2014). Recent evidence suggests that increased biliary secretion of bile acids upon high-fat diet feeding may reshape the gut microbiota in obesity. Both Western diet–fed mice and genetically obese ob/ob mice showed a reduction in abundance of Bacteroidetes and an increase in abundance of Firmicutes (Clarke et al., 2012). Feeding rats with a CA-containing diet resulted in increased gut Firmicutes-to-Bacteroidetes ratio, which was also seen in obese mice (Islam et al., 2011). DCA, escaped from reabsorption in the small intestine, enters the colon and exhibits the most potent antimicrobial activity and selective inhibition of gut bacteria growth, leading to altered composition of gut microbiota (Islam et al., 2011). Expansion of Firmicutes results in significant expension of DCA-producing bacteria (Ridlon et al., 2014). Another recent study showed that dietary saturated fats induced TCA, which promotes expansion of low-abundance sulfur-reducing pathobiont, Bilophila wasdworthia, increases proinflammatory cytokines and incidence of colitis in il-10−/− mice (Devkota et al., 2012). In a recent human study, the animal-based diet rapidly altered the gut microbiome, increasing the abundance of bile-tolerant microorganisms (B. wasdworthia and Bacteroides) and decreasing Firmicutes (David et al., 2014). This study supports a link between dietary fat, bile acids, and the outgrowth of micro-organisms in IBDs.
Fig. 9.
Bile acid and gut microbiota. In the intestine, bacteria overgrowth damages intestine barrier function and causes IBD, diarrhea, and impaired drug metabolism, detoxification, and absorption. Bile acids control gut bacteria overgrowth and protect against inflammation. Gut microbiota also play a role in biotransformation of bile acids and affected bile acid composition and metabolism via FXR and TGR5 signaling in the liver. In the liver, high levels of bile acids cause liver injury. Bile acids also have anti-inflammatory functions by activating FXR and TGR5 signaling in hepatocytes to protect against metabolic diseases such as NAFLD, diabetes, and obesity. High-fat diets and drugs alter bile acid biotransformation and gut microbiota, and contribute to pathogenesis of intestinal inflammatory disease and liver-related metabolic diseases.
Several early studies showed that the gut microbiota impacted bile acid metabolism and signaling. It was shown that germ-free rats had increased biliary bile acid concentration, increased intestinal fractional cholesterol absorption, and hyercholesterolemia (Wostmann, 1973). All bile acids are conjugated in germ-free rats, whereas β-MCA is converted to hyodeoxycholic acid and ω-MCA in conventionally raised rats (Madsen et al., 1976). In the intestine, bacterial BSHs deconjugate conjugated bile acids and are enriched in the human gut microbiome (Jones et al., 2008). More recent studies show that in germ-free mice and antibiotic-treated mice, tauro-conjugated bile acids are predominant in liver, kidney, plasma, and heart, especially tauro-β-muricholic acid (T-β-MCA) (Swann et al., 2011). Ampicillin increased hepatic primary bile acid synthesis and suppressed ileal FGF15 expression (Miyata et al., 2009). The marked decrease in intestine FGF15 expression reflects reduced intestine FXR activation by bile acids. In ampicillin-treated mice, fecal bile acid excretion rates were markedly reduced, portal bile acid concentrations increased, and ileal ASBT expression levels were increased (Miyata et al., 2011b). Taurodeoxycholic acid or CA administration to ampicillin-treated mice decreased ASBT expression. Analysis of the bile acid composition in the bile of germ-free mice showed an increased MCA/CA ratio. Because MCAs are hydrophilic bile acids and poor FXR agonists, the reduced FGF15 expression in the intestine resulted in increased CYP7A1. This is confirmed by a recent study of germ-free mice that T-α- and β-MCA were strong FXR antagonists (Sayin et al., 2013). Another study showed that an antioxidant, tempol, inhibited intestinal Lactobacilli and their BSH activity and resulted in increasing T-β-MCA, which inhibits FXR signaling and decreases obesity in mice (Li et al., 2013a). Tempol feeding reversed the Firmicutes/Bacteroides ratio and resulted in increasing T-β-MCA levels in the intestine. Interestingly, high-fat diet–fed intestine-specific fxr−/− mice had lower diet-induced obesity than wild-type mice. Another study shows that cyp8b1−/− mice, bile duct–ligated mice, antibiotic-treated mice, and germ-free mice all share similar bile acid phenotypes: increased bile acid synthesis, reduced bile acid reabsorption, and enlarged bile acid pool size (Hu et al., 2014). These data are consistent with the hypothesis that increasing T-α-MCA and T-β-MCA as well as TUDCA antagonize FXR activity. Increasing T-α-MCA and T-β-MCA may be a positive feedback mechanism to stimulate bile acid synthesis and increase bile acid pool size.
Bacterial dysbiosis has been associated with IBDs, obesity, type 2 diabetes mellitus (T2DM), nonalcoholic fatty liver disease (NAFLD), liver cirrhosis, and cancer (Kakiyama et al., 2013; Ridlon et al., 2013, 2014; Duseja and Chawla, 2014; Joyce and Gahan, 2014). Although strong evidence supports that gut microbiota can impact whole body metabolic homeostasis via the modulation of bile acid signaling, it is not clear how this could explain the observed metabolic changes and the development of metabolic disorders seen in germ-free mice. However, there seems to be a coordination between energy metabolism and bile acid signaling. Gut microbiota increased energy harvest from the diet, which may contribute to the development of obesity. At the same time, gut microbes may regulate the bile acid pool and composition, leading to increased hydrophobic bile acid content and activation of FXR and TGR5 signaling, which regulates lipid and glucose homeostasis in response to increased energy intake from the intestine. Similarly, it was shown that the total bile acid pool and the CA/MCA ratio were increased in obese mice (Li et al., 2012). Intestinal cholesterol absorption was also increased in obese mice. It was suggested that the increased bile acid pool may be a response to higher nutrient availability in obese mice (Li et al., 2012). Because altered gut microbiota in obese mice favor higher energy intake in the intestine, the altered bile acid metabolism and signaling in obese mice may be deemed as an adaptive response to handle such changes by increasing the bile acid–regulated network. Although a clear role of microbes in the regulation of bile acid metabolism has been demonstrated by comparing germ-free mice and conventionally raised mice, the contribution of altered microbiota to altered bile acid metabolism in obesity remains to be investigated. Dysbiosis is the major environmental factor that affects bile acid metabolism and contributes to diseases not only in the gastrointestinal system but also in chronic diseases, such as NAFLD, diabetes, and obesity (Jones et al., 2014). Treating dysbiosis by modulating bile acid metabolism by gut microbiota may be a therapeutic approach for improving health and preventing NAFLD and T2DM. Indeed, a recent study reported that probiotics modified microbiota to increase bile acid synthesis by deconjugation of bile acids for fecal excretion and antagonizing the FXR-FGF15 axis by increased T-MCAs in mice (Degirolamo et al., 2014).
IV. Circadian Rhythm in Bile Acid and Drug Metabolism
The diurnal rhythm of bile acid synthesis in humans was recognized more than 30 years ago (Duane et al., 1979). Circadian rhythms play critical roles in many physiologic processes, including metabolism in most tissues and organ systems (Rutter et al., 2002; Laposky et al., 2008). Bile acid metabolism is among the first identified direct molecular output of the central clock (Bass and Takahashi, 2010). Circadian rhythms are disturbed by diets, eating patterns, shift work, chronic jet leg, and sleep deprivation and affect metabolism by contributing to the pathogenesis of metabolic diseases, diabetes, and obesity (Damiola et al., 2000; Laposky et al., 2008; Bass and Takahashi, 2010; Froy, 2010; Shi et al., 2013). Diurnal regulation of bile acid metabolism also affects drug metabolism as well as efficacy and pharmacokinetics of drug therapy.
A. Clock Genes and Nuclear Receptors
Circadian rhythms are regulated by the central clock located in the suprachiasmatic nucleus of the anterior hypothalamus in the brain (Fig. 10). External signals, most importantly the light and dark cycle, reset the brain central clock, which synchronizes with the peripheral clocks in nearly every tissue type that is influenced by environmental factors including fasting/feeding cycles and nutrient availability (Bass and Takahashi, 2010). The output of the brain clock, such as sleep/wake phases and eating behavior, affects peripheral clock outputs, such as energy metabolism (Froy, 2010; Asher and Schibler, 2011). The molecular circadian clock is driven by a transcriptional-translational feedback loop. In the brain and peripheral tissues, the primary clock genes circadian locomotor output cycles kaput (CLOCK) and brain and muscle ARNT-like protein-1 (BMAL1) regulate transcription of period 1 (Per1) and period 2 (Per2) and cryptochrome 1 (Cry1) and cryptochrome 2 (Cry2). Per/Cry complexes then inhibit BMAL1 and CLOCK expression in a negative feedback loop; this loop occurs with a period of approximately 24 hours (Fig. 10). In the clock output loop, the Bmal1/Clock or neuronal PAS domain protein 2 (Npas2) complex regulates clock-controlled genes (CCGs) by binding to the E-boxes (CANNTG) in the gene promoter. The connection of circadian rhythm to lipid metabolism was unveiled by the finding that reverse-erythroblastosis α (Rev-erbα) regulates the oscillation of SREBP (Le Martelot et al., 2009) and miRNA122 (Gatfield et al., 2009) in lipid metabolism. Rev-erbα and retinoic acid–related orphan receptor α (RORα) regulate Bmal1 and Clock by binding to a ROR response element in the gene promoters. In liver cells, the BMAL1/CLOCK complex binds to the E-box sequences in the Rev-erbα and RORα gene promoters, and regulates the circadian rhythm of Rev-erbα and RORα expression. Rev-erbα is activated by heme and plays a key role in connecting energy metabolism to lipid metabolism (Preitner et al., 2002; Le Martelot et al., 2009; Feng et al., 2011). Rev-erbα and RORα regulate clock output genes containing ROREs, such as PEPCK, G6Pase in gluconeogenesis, CYP7A1 and CYP8B1 in bile acid synthesis, SREBP-1c, ACC, FAS, and microsomal triglyceride transfer protein in lipogenesis. Primary CCGs are D-site binding protein (DBP), E4 promoter binding protein 4 (E4BP4), transcription elongation factor (Tef), and hairly and enhancer of split 6 (Hes6). Disruption of primary clock genes, CCGs, and clock output genes exacerbates the development of metabolic diseases including fatty liver disease, diabetes, and obesity (Bass and Takahashi, 2010; Feng et al., 2011).
Fig. 10.
Circadian rhythms in liver metabolism. The central clock in the SCN synchronizes with the peripheral clock to regulate liver metabolisms. Eating behavior, sleep/wake cycle, and obesity affect central clock and liver clock functions and their synchronization. Hormones such as insulin, glucagon, and glucocorticoids, and nutrients including glucose, fatty acids, and bile acids affect circadian rhythms and liver metabolism. Bmal1 and Clock are primary clock products that bind to the E-box sequences in the Per and Cry gene promoters. Per and Cry complexes inhibit the Bmal/Clock complex in a negative loop to inhibit Per and Cry transcription. Bmal1 and Clock (also Npas2) are regulated by a negative regulator Rev-erbα, and positive regulator RORα, which bind to the same ROR response element (RORE) in the promoters. Rev-erbα recruits HDAC3 and NcoR to inhibit gene transcription and ultimately the circadian rhythms of many CCGs, such as PEPCK and G6Pase in gluconeogenesis, CYP7A1 and CYP8B1 in bile acid synthesis, and SREBP-1c and MTTP in lipogenesis in the liver. Alteration in synchronization of the central clock and liver clock contributes to the pathogenesis of fatty liver diseases, diabetes, and obesity, as well as fibrosis and hepatocellular carcinoma. HCC, hepatocellular carcinoma; MTTP, microsomal triglyceride transfer protein; NCOR, nuclear receptor corepressor; SCN, suprachiasmatic nucleus.
B. Diurnal Rhythm of Bile Acid Synthesis
Earlier studies in the 1990s observed that CYP7A1 activity, mRNA, and protein expression exhibited a strong diurnal rhythm, which peaked at the middle of the dark phase and decreased at the middle of the light phase in rats (Chiang et al., 1990; Noshiro et al., 1990; Falvey et al., 1995). More recent studies show that serum total bile acid levels were the lowest in the middle of the light phase and rapidly increased at the onset of the dark phase and also at the end of the dark cycle, indicating that mice have two episodes of eating during the dark phase (Zhang et al., 2011). Circadian profiles of bile acids show enhanced bacterial dehydroxylation during the fasting phase and reconjugation of bile acids in the fed phase. Cholestyramine feeding abolished the diurnal rhythm of serum bile acids, decreased α-MCA, β-MCA, ω-MCA, and UDCA, but increased DCA and CDCA. In free-fed mice, liver CYP7A1 mRNA expression rapidly increased to the highest level at the onset of the dark phase, and gradually decreased to the lowest level at 10:00 AM. SHP mRNA expression was the highest at 10:00 AM and the lowest at 10:00 PM, which is the inverse of the CYP7A1 expression pattern and is consistent with the negative regulation of CYP7A1 by the FXR/SHP pathway. Opposite to CYP7A1 diurnal rhythm, intestinal FGF15 mRNA showed a strong peak at 10:00 PM and a minor peak at 10:00 AM. Cholestyramine feeding markedly reduced intestinal FGF15 mRNA expression, but did not affect circadian rhythm of SHP and CYP7A1 mRNA. In intestine-specific FXR−/− mice, the circadian rhythm of CYP7A1 expression is similar to wild-type mice but FGF15 mRNA levels are markedly reduced and show no diurnal rhythm (Stroeve et al., 2010). Thus, the diurnal rhythm of SHP, not FGF15, regulates the diurnal rhythm of bile acid synthesis.
Bile acid synthesis as indicated by serum C4 levels in humans shows two peaks during the day, the first at 1:00 PM and the second at 9:00 PM (Gälman et al., 2005). During the night, C4 levels decreased and returned to basal levels the next morning, completely opposite to that in rodents. In humans, the majority of bile acid synthesis occurs during the postprandial period (Duane et al., 1983). Circulating FGF19 levels also exhibit a pronounced diurnal rhythm with peaks 90–120 minutes after the postprandial rise in serum bile acids (Lundåsen et al., 2006). The FGF19 peaks preceded the decline of bile acid synthesis. Fasting abolished the diurnal rhythm of FGF19. Therefore, in humans, the diurnal rhythm of circulating FGF19 is controlled by the transintestinal bile acid flux. An inverse relationship between the fasting serum FGF19 and C4 levels is consistent with the hypothesis that FGF19 exerts feedback inhibition of CYP7A1 in humans (Gälman et al., 2011).
Circadian rhythm of CYP7A1 is regulated by a positive clock-controlled gene DBP (Lavery and Schibler, 1993) and negative clock genes DEC2 and Rev-erbα (Noshiro et al., 2004, 2007; Le Martelot et al., 2009). DBP expression follows a stringent circadian rhythm and is a key liver clock-controlled gene (Wuarin and Schibler, 1990). DBP stimulates CYP7A1 expression and regulates its diurnal rhythm (Lavery and Schibler, 1993; Preitner et al., 2002). Interestingly, Rev-erbα–deficient mice had a lower bile acid synthesis rate and reduced CYP7A1 expression, whereas SHP and E4BP4 expression were increased. It was suggested that Rev-erbα induced CYP7A1 by inhibiting SHP and E4BP4 (Duez et al., 2008; Le Martelot et al., 2009). It is likely that Rev-erbα expression levels determine the extent of SHP and E4BP4 in inhibition of CYP7A1. Rev-erbα is an unstable protein with a very short half-life of less than 1 hour, and is activated by intracellular heme to recruit nuclear receptor corepressor 1 and HDAC3 to inhibit target gene transcription (Yin and Lazar, 2005; Yin et al., 2006).
Fasting and restricted feeding strongly affected the circadian rhythm of CYP7A1 and bile acid synthesis. Fasting strongly reduced CYP7A1 expression and blunted the circadian rhythm of CYP7A1, whereas restricted feeding rapidly stimulated CYP7A1 expression in mice (Li et al., 2012). Restricted feeding altered the circadian rhythm of bile acid synthesis and increased the bile acid pool by approximately 20%. The circadian rhythm of CYP8B1 is opposite to that of CYP7A1. Fasting-induced RORα induced CYP8B1 expression in mice (Pathak et al., 2013). RORα and Rev-erbα bind to the same response element on the CYP8B1 gene promoter and regulate the diurnal rhythm of CYP8B1 expression. Disruption of per1/per2 increases serum bile acids and results in hepatic cholestasis (Ma et al., 2009). In per1/per2 knockout mice, circadian expression of clock genes, such as Rev-erbα, Clock, E4BP4, and DBP, were altered, which altered CYP7A1 circadian rhythm and bile acid synthesis.
C. Circadian Rhythm in Metabolic Diseases
The linking of clock genes to metabolic syndrome was first observed in clock mutant mice (Turek et al., 2005). Fasting and refeeding as well as the time of feeding have profound effects on circadian rhythms of hepatic lipid and glucose metabolism (Vollmers et al., 2009; Asher and Schibler, 2011; Hatori et al., 2012; Tong and Yin, 2013). During fasting, glucagon stimulates fatty acid mobilization from adipose tissues to the liver for generation of energy. In the fed state, insulin signaling stimulates lipogenesis and secretes VLDL for delivery of triglycerides to adipose tissue for storage. Serum triglyceride levels show a circadian rhythm (Rudic et al., 2004; Pan and Hussain, 2007). Clock mutant mice have hypertriglyceridemia and altered diurnal rhythm of serum triglycerides (Pan et al., 2010). Expression of PPARα shows a circadian rhythm in liver and is abolished in clock mutant mice. PPARγ also exhibits a diurnal rhythm, which is regulated by E4BP4. Clock binds to the SHP promoter and induces SHP, which inhibits the microsomal triglyceride transfer protein involved in VLDL assembly. In Rev-erbα−/− mice, lipogenesis is stimulated (Feng et al., 2011). HDAC3 and Rev-erbα are colocalized in the promoter of genes in lipid metabolism, and deletion of either HDAC3 or Rev-erbα in mouse liver causes hepatic steatosis. Rev-erbα is also involved in the circadian rhythm of SREBP and modulates circadian rhythm of lipid, cholesterol, and bile acid synthesis (Le Martelot et al., 2009). Bmal1 and Clock are involved in glucose metabolism (Rudic et al., 2004). Gluconeogenesis is abolished in bmal1−/− mice and depressed in clock mutant mice. It was suggested that Bmal1 and Clock synchronize with peripheral clocks to control lipid and glucose metabolism. Clock mutant and per2−/− mice have increased food intake during the light cycle. A high-fat diet modulates glucose metabolism and amplifies circadian rhythm in glucose tolerance and insulin sensitivity. Interestingly, mice fed a high-fat diet only during the light phase (sleep period) gained significantly more weight than mice fed the same diet only during the dark phase (active period). Caloric intake and locomotor activity did not vary significantly between treatments, although day-fed mice trended toward being less active, which could contribute to decreased energy expenditure (Arble et al., 2009). Conversely, night-time–restricted feeding without reducing caloric intake has been shown to prevent obesity, insulin resistance, hepatic steatosis, and inflammation in mice fed a high-fat diet (Hatori et al., 2012). In this study, CYP7A1 expression shows a robust diurnal rhythm in time-restricted feeding but is blunted in free-fed mice. Serum cholesterol is lower and bile acids are higher in mice fed a time-restricted high-fat diet than in free-fed mice. Thus, a time-restricted feeding enhances diurnal rhythm of bile acid synthesis and may prevent high-fat diet–induced metabolic syndrome in mice.
In obese and diabetic mice, rhythmicity of the clock genes was attenuated (Ando et al., 2005a). Common genetic variations in the clock gene have been linked to NAFLD (Sookoian et al., 2007). Interestingly, a recent study shows that epigenetic regulation of insulin resistance was linked to NAFLD (Sookoian et al., 2010). The methylated/nonmethylated DNA ratios in CpG islands in the PGC-1α gene promoter in diabetic patients is correlated with serum insulin and a homeostasis model of assessment of insulin resistance. Chronic alcohol consumption disrupts clock genes and diurnal rhythms of liver metabolic genes (Filiano et al., 2013). However, the mechanism of ethanol effect on circadian clock expression is not clear. The ethanol diet used in chronic ethanol feeding is equivalent to a high-fat content of approximately 30% and ethanol feeding may alter eating behaviors and patterns as well as locomotor activity, thus possibly contributing to the observed disruption of circadian rhythm. Chronic alcohol consumption is known to alter lipid metabolism by stimulating lipogenesis and inhibiting fatty acid oxidation to contribute to alcoholic fatty liver disease (Gao and Bataller, 2011). Chronic alcohol feeding altered circadian rhythm in wild-type mice by delaying phase onset of activity and shortening the free-running period (Seggio et al., 2009). Alcohol treatment increased hepatic triglycerides in clock mutant mice more than in wild-type mice and altered lipogenic gene expression in clock mutant mouse liver (Kudo et al., 2009). The diurnal rhythm of many primary clock genes (Bmal1, Clock, Cry1/2, and Per1/2) and CCGs (Dbp, Npas2, and Rev-erbα) were altered in alcohol-fed mouse livers (Filiano et al., 2013) and caused phase shifting of diurnal rhythm of hepatic triglycerides, cholesterol, and bile acid levels (Zhou et al., 2014). Alcohol also alters the NAD/NADH ratio, which regulates sirtuin SIRT1 activity and downstream target genes Bmal1/Npas2/Clock (Nakahata et al., 2009). It was suggested that alcohol affects the diurnal rhythm through alteration of the redox state (NAD/NADH), reducing heme levels, and cellular energetics such as AMP/ATP and the AMP-activated kinase (AMPK).
D. Circadian Rhythm in Drug Metabolism
All three phases of drug metabolism exhibit circadian rhythm regulated by RORα and Rev-erbα, as well as by several CCGs (Gachon and Firsov, 2011; Zmrzljak and Rozman, 2012). Circadian regulation of drug metabolism and detoxification is complicated by drug induction of drug-metabolizing enzymes. In general, mRNA expression of phase I drug oxidation cytochrome P450 enzymes is increased during the dark phase, whereas that of phase II drug conjugation enzymes and phase III drug transporters reached maximal levels during the light cycle. Circadian rhythms of the CYP genes are regulated by PAS (Per-Arnt-Sim) domain basic-leucine-zipper transcription factors, DBP, TEF, and hepatic leukemia factor (Hirao et al., 2006; Froy, 2009; Zhang et al., 2009c; Gachon and Firsov, 2011; Košir et al., 2013). Rev-erbα, RORs, PPARα, CAR, and PXR are involved in circadian expression of drug metabolism enzymes (Gachon and Firsov, 2011; Košir et al., 2013). In RORα and RORγ double knockout mice, expression of several phase I and II drug metabolism enzymes are downregulated, suggesting that RORα and RORγ play a role in transcriptional regulation of circadian rhythm of drug-metabolizing enzyme expression (Kang et al., 2007). It appears that circadian rhythm of heme synthesis may link nuclear receptors to regulation of the circadian rhythms of drug metabolism. Heme is a ligand of Rev-erbα and Rev-erbβ that stimulates recruitment of corepressors (nuclear receptor corepressor 1/HDAC3) to inhibit transcription of Bmal1 and other target genes by decreasing histone acetylation (Yin et al., 2006, 2007; Burris, 2008). Heme binds to Npas2, a PAS domain protein, by inhibiting DNA binding activity of Bmal1/Npas2 complex (Crumbley et al., 2010). Heme also regulates Per2 by stimulating the Bmal1/Npas2 complex and regulates the circadian rhythm of δ-aminolevulinic acid synthase 1 (Alas1), the rate-limiting enzyme in heme synthesis (Kaasik and Lee, 2004). PXR and CAR induce alas1 gene expression (Podvinec et al., 2004), which affects PXR and CAR regulation of drug metabolism and circadian rhythm. Alas1 is also regulated by PGC-1α, which is induced by fasting (Handschin et al., 2005), thus linking nutritional status to heme synthesis and circadian rhythm of drug metabolism.
Diurnal variations in drug absorption, distribution, metabolism, and excretion are important for designing therapeutic drugs for optimal efficacy and dosing, as well as for minimizing side effects and injury to the tissues and organ systems. Absorption of many drugs is sensitive to the time of day of drug administration (Musiek and Fitzgerald, 2013). Most drugs are absorbed more efficiently in the morning and after a meal when bile acid synthesis reaches its peaks. Bile acids facilitate intestinal drug absorption and transport. Circadian rhythms and fasting and feeding cycles affect drug transporter MDR1 (ABCB1) in the intestinal brush border membrane, and efflux transporters (MRP3, ABCC3) in the sinusoidal membrane (Ando et al., 2005b). MDR1 exhibits a circadian rhythm regulated by hepatic leukemia factor and E4BP4 (Ando et al., 2005b; Murakami et al., 2008). Other transporters such as MRP2 and breast cancer–related protein also show circadian rhythms (Stearns et al., 2008). The pharmacodynamics of drugs is also regulated by the circadian rhythm of drug transporters, receptors, and enzymes and affects the efficacy of drugs, especially for chemotherapies (Musiek and Fitzgerald, 2013), as well as the detoxification of drugs (Zmrzljak and Rozman, 2012).
V. MicroRNA in Lipid and Glucose Metabolism
miRNAs are small noncoding RNAs that, after base paring with complementary sequences in the target mRNAs, promote mRNA degradation or inhibit protein synthesis. Recent studies identified several miRNAs involved in hepatic lipid metabolism (Krützfeldt and Stoffel, 2006; Moore et al., 2010, 2011; Najafi-Shoushtari et al., 2010; Fernández-Hernando et al., 2011; Rottiers and Näär, 2012; Sacco and Adeli, 2012; Ramírez et al., 2013b), metabolic diseases (Fernández-Hernando et al., 2013), and atherosclerosis (Aryal et al., 2014). The most abundant miRNA in liver, miR-122a, plays a key role in regulating cholesterol biosynthesis and lipid metabolism in mice (Krützfeldt et al., 2005; Esau et al., 2006; Elmén et al., 2008). Inhibition of miR-122a expression in mice using antagomirs or antisense oligonucleotides have uncovered the phenotypes of reduced serum cholesterol and triglyceride levels, increased fatty acid oxidation and decreased hepatic fatty acid and cholesterol synthesis rate (Krützfeldt et al., 2005; Esau et al., 2006; Elmén et al., 2008). Inhibition of miR-122a resulted in reducing plasma cholesterol and improving steatosis in a diet-induced obese mouse model (Esau et al., 2006). In NASH patients, miR-122a expression was reduced 63%, implicating a role for miR-122a in inhibiting fatty acid synthesis and preventing inflammation, proliferation, and steatosis (Cheung et al., 2008). The mechanism of miR-122a in regulation of cholesterol metabolism is unknown. miR-122a antagomirs do not affect miR-122 expression, and the miR-122a binding site is not present in the 3′-untranslated region (UTR) of mRNAs encoding 3-hydroxy-3-methylglutaryl-CoA reductase and the LDLR in mouse livers. It was speculated that miR-122a might indirectly induce genes by inhibiting the expression of a repressor. A putative miR-122a binding sequence has been identified in the 3′-UTR of CYP7A1 mRNA (Song et al., 2010). Results from this study provide a plausible mechanism that miR-122a antagomirs may induce CYP7A1 and result in reducing serum cholesterol and triglyceride levels.
MiR-33a, which is encoded by intron 16 of the SREBP-2 gene, was recently shown to regulate cellular cholesterol homeostasis (Najafi-Shoushtari et al., 2010), biliary bile acid secretion (Allen et al., 2012), fatty acid oxidation, and cell proliferation (Cirera-Salinas et al., 2012). It was shown that when cellular cholesterol levels decrease, miR-33a expression is coinduced with SREBP-2 mRNA. miR-33a inhibits ABCA1 and ABCG1 to reduce cellular cholesterol efflux, thus retaining cellular cholesterol. Studies in mice treated with an anti–miR-33a or in genetic miR-33a–deficient mice showed that antagonism of miR-33 induced ABCA1 in macrophages and in the liver, increased serum HDL levels, and promoted macrophage-to-feces reverse cholesterol transport. In addition, antagonism of miR-33a was shown to promote regression of atherosclerosis in ldlr−/− mice and nonhuman primates (Rayner et al., 2011). These studies suggest that miR-33a acts in a synergistic manner with SREBP-2 to regulate cellular cholesterol homeostasis. Study of transgenic mice overexpressing CYP7A1 in the liver (CYP7A1-tg mice) showed a parallel increase of SREBP-2 and miR-33a expression levels in liver. Overexpression of miR-33a in the liver decreased bile acid pool, increased hepatic cholesterol, and decreased serum cholesterol in these mice. This study suggests that coinduction of miR-33a with SREBP-2 may provide a negative feedback loop to maintain cholesterol and bile acid homeostasis (Li et al., 2013b). The cardioprotective effects of miR-33a antagonism can be attributed to not only HDL biogenesis in the liver, but also to cholesterol efflux from the macrophages, which is the first step in reverse cholesterol transport (Rayner et al., 2011). In addition, miR-33a and miR-33b inhibit the genes in fatty acid β-oxidation, such as carnitine O-octanoyltransferase, hydroxyacyl-coenzyme A dehydrogenase, and carnitine palmitoyl transferase 1A, and the energy sensors AMPKα1 and NAD+-dependent SIRT6. Antagonism of miR-33 may be a therapeutic strategy to induce CYP7A1 activity and bile acid synthesis to treat fatty liver diseases, diabetes, and obesity.
Interestingly, miR-33b, encoded by intron 17 of the SREBP-1a gene, regulates glucose metabolism (Ramírez et al., 2013a). It is interesting to note that miR-33b is expressed in human liver, but not in mouse liver. Overexpression of miR-33b inhibits PEPCK and G6PaseC3 expression and reduces glucose production in hepatocytes. miR-33b also inhibits glucose induction of cAMP responsive element binding protein 1, SRC1, RORα, and PGC-1α expression in human hepatocytes. SIRT6 and AMPKα are also miR-33b target genes. In glycolysis, miR-33b induces glucokinase but not other glycolytic genes. In glycogenolysis, miR-33b reduces glycogen phosphorylase and phosphoglucomutase mRNA expression in HepG2 cells. However, miR-33b binding sites are not present in mouse SIRT6, PEPCK1, or G6PaseC3. In rhesus monkeys, glucose infusion, but not insulin, increases SREBP-1c and miR-33b levels and decreases PEPCK and G6PC3 mRNA and protein levels in the liver. SREBP-1c and miR-33b may coordinately regulate hepatic glucose production.
miR-34a and miR-24 have emerged as key regulators of hepatic lipid metabolism (Rottiers and Näär, 2012). MiR-34a binding sites have been identified in the 3′-UTR and miR-24 binding sites in the coding region of HNF4α mRNA (Takagi et al., 2010). Overexpression of these two miRNAs downregulated HNF4α mRNA levels and decreased CYP7A1 and CYP8B1 expression in HepG2 cells. miR-24a expression is increased in high-fat diet–fed mice and in human hepatocytes incubated with fatty acids (Ng et al., 2014). Insig1 was identified as a target of miR-24. Inhibition of miR-24 increased Insig1 and subsequently decreased hepatic lipid accumulation. Antagonizing miR-24 and miR-34a may be used as a therapy to stimulate conversion of cholesterol to bile acids and to reduce high-fat diet–induced hyperlipidemia for NAFLD and atherosclerosis.
A high-throughput small mRNA sequencing detected 150 miRNAs in mouse liver. High-fat diet treatment revealed that 50 miRNAs were increased more than 2-fold. Unbiased in silico analysis identified miR-27b as a regulatory hub in lipid metabolism in humans and mice (Vickers et al., 2013). miR-27b downregulated mRNA expression of PPARγ, angiopoietin-like protein 3 (ANGPTL3), and glycerol-3-phosphate acyltransferase 1 (GPAM), all involved in lipid metabolism. In apoe−/− mice on a high-fat diet, miR-27a is upregulated in the liver. ANGPTL3 is highly expressed in the liver and is an inhibitor of LPL, which hydrolyzes triglycerides in VLDL and chylomicrons. Plasma levels of ANGPTL3 correlate with high serum lipids. GPAM is highly expressed in the liver and catalyzes the first and rate-limiting step in de novo triglyceride synthesis. ANGPTL3 and GPAM genetic polymorphisms are significantly associated with plasma lipid levels (Musunuru et al., 2010; Teslovich et al., 2010). miR-27b oligomers may be used to inhibit ANGPTL3 to stimulate LPL as a therapy to promote VLDL metabolism and prevent NAFLD. In CYP7A1-tg mouse livers, ANGPTL3 mRNA levels were significantly reduced, suggesting that bile acids may be used to inhibit ANGPTL3 and stimulate LPL.
VI. Bile Acid Signaling in Metabolic Diseases and Therapeutics
Bile acids are known to increase cell proliferation and apoptosis in the liver and intestine (Solá et al., 2006; Amaral et al., 2009). Accumulation of toxic endobiotics and xenobiotics causes damage to cells and organs in the digestive tract. Inborn errors in bile acid metabolism in pediatric human patients have been identified in almost every gene in bile acid synthesis and conjugation and have been extensively reviewed (Heubi et al., 2007; Ng et al., 2014). This section reviews bile acid signaling in inflammatory diseases of the digestive system including cholestatic liver diseases, IBDs, and fatty liver diseases as well as bile acid–based therapies.
A. Cholestatic Liver Diseases
1. Cholestasis.
Cholestasis is caused by a disruption of bile flow, which results in a lack of bile in the intestine, accumulation of toxic bile acids and other metabolites in the liver, and increased bile acids in the systemic circulation (Trauner et al., 1998). Congenital or acquired defects in bile flow can cause obstructive cholestasis. Obstruction of the bile ducts by tumors or stones, genetic mutations of bile acid transporter genes, and acquired dysregulation of bile transport system by drugs, pregnancy, and pathophysiological conditions causes intrahepatic and extrahepatic cholestasis (Wagner et al., 2009). Congenital or acquired defects in canalicular membrane transporters are the major cause of intrahepatic cholestasis. Congenital cholestasis occurs early in life and patients have jaundice, pruritus, low absorption of fat-soluble vitamins leading to slow growth, and progressive liver damage by increased hepatic and serum levels of bile acids. PFIC and benign recurrent intrahepatic cholestasis are autosomal recessive diseases. PFIC1 (also known as Byler disease) is linked to mutations in the ATP8B1 gene, which codes a P-type ATPase, functioning as an aminophospholipid flippase that maintains membrane asymmetry by inward flipping of phosphatidylserine from the outer leaflet of the plasma membrane. PFIC2 is linked to BSEP mutations and polymorphisms that have been linked to intrahepatic cholestasis of pregnancy (ICP) (Pauli-Magnus et al., 2004; Noe et al., 2005; Lang et al., 2006) and drug-induced liver injury (Lang et al., 2007). ICP is a reversible form of intrahepatic cholestasis associated with adverse pregnancy outcomes (Eloranta and Kullak-Ublick, 2008). PFIC3 is linked to MDR3 mutations and patients have high levels of γ-glutamyl transpeptidase activity, progressive cholestasis, and bile duct damage, and may require liver transplant. Phospholipids in bile are required for mixed micelle formation with bile acids and cholesterol (Jacquemin, 2001; Pauli-Magnus et al., 2004). Without forming mixed micelles, bile acids damage the canalicular membrane and cholangiocytes. MDR3 mutations may cause cholesterol gallstone diseases and have been linked to ICP (Jacquemin, 2001; Wasmuth et al., 2007). Genetic polymorphisms and heterozygote mutations of the PFIC1, PFIC2, and PFIC3 genes may increase susceptibility to acquired cholestasis in adults including ICP, drug-induced liver injury, primary biliary cirrhosis, and primary sclerosing cholangitis. FXR variants have been identified in ICP patients (Van Mil et al., 2007) and affect FXR target genes ABCB11 and ABCB4 as well as ATP8B1 expression in ICP (Müllenbach et al., 2005; Painter et al., 2005).
Mutations in the MRP2 gene have been linked to Dubin-Johnson syndrome, a disease characterized by chronic hyperbilirubinemia (Keitel et al., 2003). MRP2 excretes conjugated bile acids, bilirubin, and other organic anions. Patients have elevated bile acids and cholestasis. MRP2 mutations have also been linked to ICP. The FXR gene has been identified as a candidate gene for the cholesterol gallstone susceptibility locus Lith7 in mice (Wittenburg et al., 2003). A study of obstructive cholestasis in human patients shows that bile acid synthesis is suppressed but CYP7A1 expression is not altered (Bertolotti et al., 2001). By contrast, it was reported that CYP7A1 mRNA expression is repressed in human patients with obstructive extrahepatic cholestasis, likely due to increased FGF19 expression in hepatocytes (Schaap et al., 2009).
2. Bile Acids as Therapeutic Agents.
CDCA and UDCA have been used for gallstone dissolution and inborn errors of bile acid synthesis for many decades (Chiang, 2009). UDCA (ursodiol) is an approved drug for treating primary biliary cirrhosis. Recently, therapies targeted to FXR, TGR5, and bile acid metabolism have been developed for treatment of metabolic diseases and cholestatic liver diseases (reviewed in Claudel et al., 2004; Trauner and Halilbasic, 2011; Fiorucci et al., 2012; Porez et al., 2012; Chiang, 2013; Li and Chiang, 2013; Swanson et al., 2013; Schaap et al., 2014).
Activation of FXR inhibits CYP7A1 to reduce bile acid synthesis, and inhibits NTCP and OATPs to reduce sinusoidal uptake of bile acids. FXR also may upregulate MRP3/MRP4 and OSTα/β in the sinusoidal membrane as an adaptive response to efflux bile acids into systemic circulation in obstructive cholestasis. Bile acids also play a protective role in controlling bacterial overgrowth in the intestine (Inagaki et al., 2006). Thus, obstruction of bile flow or knockout of the fxr gene in mice increases bacterial growth and mucosal injury in the intestine, and bile acid administration reduces bacterial growth in obstructive cholestasis. CDCA, CA, and UDCA have been used for effective gallstone dissolution for many years. CDCA and CA are not toxic to humans. CDCA has been used to treat bile acid–deficient patients as a replacement of bile acids in the pool. CA is converted to DCA, which is highly toxic and is a colon cancer promoter. CA is more efficient in intestinal absorption of cholesterol than other bile acids and may cause gallstones in human patients (Wang et al., 2006). UDCA has been used in traditional Chinese medicine for treating digestive disease for several centuries. UDCA is a highly soluble and nontoxic bile acid. UDCA reduces the cytotoxicity of the circulating bile acid pool, and thus protects cholangiocytes, stimulates hepatobiliary secretion, and inhibits liver cell apoptosis (Solá et al., 2006). UDCA may also activate PXR and induce PXR target genes, including CYP3A4, SULTs, UDP-glucuronosyl N-transferases, BSEP, MDR3, and MRP4, for detoxification of bile acids. norUDCA is a side chain-shortened C23 homolog of UDCA that cannot be conjugated and is secreted into bile, reabsorbed by cholangiocytes, and returned to liver. Cholehepatic shunt of norUDCA leads to an increase in bicarbonate in bile and hypercholeresis. This bile acid derivative reverses sclerosing cholangitis in the Mdr2−/− (Abcb4) model of cholangiopathy by reducing bile acid hydrophobicity, increasing bile flow, and inducing bile acid detoxification, and reducing inflammation and fibrosis progression (Moustafa et al., 2012).
B. Inflammatory Gastrointestinal Diseases
IBDs are chronic condition of inflammation in intestinal mucosa. IBDs include Crohn’s disease of the gastrointestinal tract and ulcerative colitis in the colon. Intestinal microbiota, epithelial dysfunction, and aberrant mucosal immune response all contribute to IBD (Vavassori et al., 2009; Cipriani et al., 2011; Gadaleta et al., 2011b). In an early study of dextrin sodium sulfate– or trinitrobenzene sulphonic acid–induced colitis, CYP7A1 expression, bile acid secretion, bile acid content in gut and liver, and bile acid pool size were increased; by contrast, in indomethacin-induced small bowel inflammation, CYP7A1 expression, bile acid secretion, and bile acid pool were reduced but the hepatic acute phase and cytokine response were increased (Dikopoulos et al., 2007). A recent study of TGR5 knockout mice showed abnormal morphology of colonic mucous and altered epithelial tight junctions as well as increased intestinal permeability and susceptibility to develop dextrin sodium sulfate–induced colitis. As described in section III, a diet high in saturated fat induces TCA and organic sulfur to promote sulfite-reducing pathobiont B. wadsworthia and results in increased susceptibility to colitis in il-10−/− mice (Devkota et al., 2012). Interestingly, feeding a low-fat diet supplemented with TCA, but not glycocholic acid, increased B. wadsworthia abundance and colitis in il-10−/− mice. This study demonstrated a high-fat diet/bile acid/microbiota axis in pathogenesis of IBD. Several studies show that activation of FXR and TGR5 signaling protects against IBD by antagonizing Toll-like receptor 4/tumor necrosis factor α and NF-κB signaling and intestinal barrier structures and functions (Cipriani et al., 2011; Gadaleta et al., 2011a). A recent study show that bile acid activation of TGR5 signaling can induce macrophage differentiation toward interleukin-12–producing dendritic cells (Ichikawa et al., 2012) and inhibit the production of proinflammatory cytokines by intestinal macrophages in Crohn’s disease (Yoneno et al., 2013). An IBD patient study showed a link of genetic variation in FXR to IBD and ileal SHP mRNA levels were reduced in Crohn’s colitis, but not in ulcerative colitis (Nijmeijer et al., 2011). It was concluded that FXR activation in the ileum is reduced in Crohn’s patients, secondary to altered enterohepatic circulation of bile acids. TGR5 and FXR agonists may have therapeutic benefit to Crohn’s disease patients (Lian et al., 2011; Pols et al., 2011b; Stojancevic et al., 2012; Stepanov et al., 2013).
C. Fatty Liver Disease, Diabetes, and Obesity
Diabetes and fatty liver diseases are inflammatory diseases associated with dyslipidemia and hepatic insulin resistance. NAFLD is the most common chronic liver disease affecting approximately 20–40% of the adult population in the United States, and has a high prevalence of T2DM and obesity (Farrell and Larter, 2006; Brunt, 2010; Cohen et al., 2011). Accumulation of cytotoxic bile acids causes inflammation and injury to the liver. In diabetic patients, serum 12α-hydroxylated bile acids and their conjugates are increased and correlated with insulin resistance (Thomas et al., 2008; Haeusler et al., 2013). Increasing CA facilitates intestinal absorption of dietary cholesterol and fats and contributes to cholesterol gallstone diseases, NAFLD, and diabetes.
Metabolic syndrome is a collection of five clinical symptoms including hypertension, hyperglycemia, hypertriglyceridemia, insulin resistance, and central obesity (Reaven, 1988). Metabolic syndrome contributes to chronic heart disease, atherosclerosis, T2DM, and NAFLD. Dyslipidemia causes insulin resistance and inflammation, and pathogenesis of NAFLD (Ginsberg et al., 2006; Cohen et al., 2011). NAFLD is a spectrum of chronic liver abnormalities from simple steatosis to NASH to liver cirrhosis (Yeh and Brunt, 2007). In hepatic steatosis, more than 5% of hepatocytes accumulate lipid droplets. Hepatic steatosis is caused by alcohol, drugs, hepatitis, insulin resistance, high-fat diet, and other factors, and is reversible. About 30% of steatotic patients develop NASH, which involves hepatic ballooning, inflammatory infiltrates, and cell death. About 10–29% of NASH patients will progress to cirrhosis and hepatocellular carcinoma, the end stage of liver disease. NAFLD is closely associated with obesity, insulin resistance, and hepatic steatosis (Farrell and Larter, 2006; Tiniakos et al., 2010). The progression of hepatic steatosis to NASH requires at least “two hits”: hepatic steatosis is the first hit, followed by inflammation as the second hit (Day and James, 1998). Other factors, such as insulin resistance and oxidative stress, accelerate the progression from hepatic steatosis to NASH. Excessive accumulation of free fatty acids and triglycerides in hepatocytes may cause lipotoxicity and hepatocyte apoptosis (Trauner et al., 2010). Free fatty acids cause liver injury by increasing inflammation, the unfolded protein response or ER stress, and damaging mitochondria and increasing reactive oxidizing species. Autophagy is a critical lysosomal mechanism for degradation of cell organelles and maintenance of normal cell function and prevention of diseases in NASH (Czaja et al., 2013). Lipophagy (autophagy of lipids) degrades cellular fats to maintain lipid homeostasis (Singh et al., 2009). Excessive fat accumulation in the liver may alter autophagy and cause liver injury.
Based on the phenotypes of CYP7A1-tg mouse model (Li et al., 2010, 2011b), increasing bile acid synthesis with reduced CA in the pool may be a strategy for preventing high-fat diet–induced NAFLD and improving insulin resistance. A recent lipidomic analysis identified seven lipid markers, including lysophosphatidylcholine, phosphatidylcholine, sphingomyelin, and ceramide, that were significantly decreased in serum of CYP7A1-tg mice fed a high-fat diet. Metabolomics analysis identified 13 metabolites in bile acid synthesis, including taurochenodeoxycholic acid, TDCA, TUDCA, TCA, and T-β-MCA, that differed between CYP7A1-tg and wild-type mice. Interestingly, T-β-MCA was significantly increased only in intestine of CYP7A1-tg mice. This study suggest that reducing 12α-hydroxylated bile acids and increasing T-β-MCA, an FXR antagonist, may reduce the high-fat diet–induced increase of phospholipid, sphingomyelin, and ceramide and may ameliorate diabetes and obesity (Qi et al., 2014). CA facilitates dietary cholesterol absorption and contributes to cholesterol gallstone disease, and may also cause insulin resistance and NAFLD.
The therapeutic potential of bile acids and derivatives for treating hepatic and biliary diseases, NAFLD, and metabolic syndrome has been extensively reviewed (Thomas et al., 2008; Fiorucci and Baldelli, 2009; Zollner and Trauner, 2009; Lian et al., 2011; Pols et al., 2011a,b; Adorini et al., 2012; Hollman et al., 2012; Stojancevic et al., 2012; McMahan et al., 2013; Mudaliar et al., 2013; Stepanov et al., 2013; Duboc et al., 2014) and will be briefly summarized. Bile acid sequestrants have long been used for treating gallstone disease. The second-generation bile acid sequestrants are currently in clinical trials for metabolic diseases (Davidson et al., 1999; Aldridge and Ito, 2001; Bays et al., 2008; Brufau et al., 2010; Shang et al., 2010; Nwose and Jones, 2013). Gastric bypass surgeries are effective in reducing weight and improving insulin resistance and are increasingly popular for treatment of T2DM (Patti et al., 2009; Simonen et al., 2012; Gerhard et al., 2013).
1. Farnesoid X Receptor Agonists.
Many studies have shown that activation of FXR inhibits inflammation in liver and intestine and FXR agonists are potential therapeutic drugs for metabolic and inflammatory diseases. A synthetic bile acid derivative, obeticholic acid (OCA, 6-ethyl-CDCA or INT-747) is a potent and selective FXR agonist that has anticholestatic effects (Pellicciari et al., 2004). In animal studies, OCA increases insulin sensitivity, inhibits gluconeogenesis, inhibits lipogenesis, and has anti-inflammatory and antifibrotic properties (Adorini et al., 2012). OCA ameliorates high-fat diet–induced obesity and insulin resistance in mice, as well as insulin resistance and fatty liver in Zucker rats (fa/fa) (Cipriani et al., 2010). OCA antagonizes NF-κB–stimulated inflammation in liver (Wang et al., 2008), modulates innate immunity in animal models of colitis (Vavassori et al., 2009), inhibits and preserves the intestinal barrier in IBD (Nijmeijer et al., 2011), and inhibits VSMC inflammation and migration. A phase II clinical trial of OCA for NAFLD and T2DM patients showed improved insulin sensitivity, reduced γ-glutamyl-transpeptidase levels (a marker of NASH), and weight loss (Mudaliar et al., 2013).
2. G Protein–Coupled Bile Acid Receptor Agonists.
TGR5 signaling inhibits the production of proinflammatory cytokines in Crohn’s disease (Yoneno et al., 2013). Drugs targeting to TGR5 have the potential to treat metabolic and inflammatory diseases. Oleanolic acid extracted from olive leaves has a structure similar to bile acids and has been shown to be a specific TGR5 agonist (Sato et al., 2008). A bile acid derivative INT-777 is a selective and potent TGR5 agonist (Pellicciari et al., 2007, 2009). In animal studies, INT-777 improves glucose tolerance, stimulates GLP-1 secretion from enteroendocrine L cells and improves insulin sensitivity, and increases intracellular ATP/ADP ratio and calcium mobilization in obese mice (Thomas et al., 2009). Activation of TGR5 also reduces atherosclerotic lesions by reducing macrophage inflammation and lipid loading (Pols et al., 2011a). TGR5 agonists also reduce inflammation in liver and intestine to prevent liver inflammation and IBD (Pols et al., 2011b). The FXR and TGR5 dual agonist INT-767 (6α-ethyl-3α,7α,23-trihydroxy-24-nor-5β-cholan-23-sulphate) has been shown to improve NAFLD by modulating hepatocyte monocyte activity (McMahan et al., 2013). Other non-bile acid compounds have been reported as specific TGR5 agonists (Evans et al., 2009a; Herbert et al., 2010).
3. Bile Acid Sequestrants.
Many studies in animals and human diabetic patients showed that bile acids improved glycemic control (Claudel et al., 2005; Prawitt et al., 2014). In diabetes, bile acid pool size is enlarged, likely due to the altered circadian rhythm of CYP7A1 and the fasting to refeeding response (Li et al., 2012). Disruption of enterohepatic circulation of bile acids by bile fistula or feeding of bile acid sequestrants reduces bile acid pool size and stimulates hepatic bile acid synthesis. Increasing hepatic bile acid synthesis may inhibit gluconeogenesis and lipogenesis but stimulate glycolysis, fatty acid oxidation, and glycogenesis. Bile acid sequestrants are known to reduce serum cholesterol by stimulating bile acid synthesis, increasing LDLR-mediated uptake of serum LDL cholesterol for treating hypercholesterolemia. Cholestyramine and colestipol are classic bile acid sequestrants that have been used for gallstone dissolution and lipid lowering in humans. Colesevelam, a second-generation bile acid sequestrant, has been used in combination with statins or antidiabetic drugs for increasing glycemic control and improving insulin resistance (Davidson et al., 1999; Aldridge and Ito, 2001; Bays et al., 2008; Brufau et al., 2010; Shang et al., 2010). The underlying mechanism of the glucose-lowering effect of bile acid sequestrants is not completely known. Recent reports show that colesevelam may mediate its action through induction of GLP-1 secretion in rats (Shang et al., 2010). Colesevelam may reduce hepatic glucose production and improve hyperglycemia and hyperinsulinemia in diet-induced obese mice (Potthoff et al., 2013). This effect may be mediated through activation of TGR5 and GLP-1 release (Nwose and Jones, 2013). A recent study shows that colesevelam increases GLP-1 levels and improves β-cell function, but has no effect on insulin sensitivity (Beysen et al., 2012). Colesevelam improves glycemic control in T2DM patients, but is not correlated with changes in bile acid metabolism (Brufau et al., 2010). In these patients, CA synthesis is increased and colesevelam treatment preferentially increases CA but decreases CDCA and DCA, thus decreasing the hydrophobicity of the bile acid pool without changing the total bile acid pool size. There is no correlation between bile acid kinetics and glucose metabolism in these patients.
4. Gastric Bypass Surgery.
Gastric bypass is currently being used as a treatment for reducing weight and T2DM. Laparoscopic adjustable gastric banding, laparoscopic sleeve gastrectomy, and the Roux-en-Y gastric bypass (RYGB) are the most common bariatric surgeries for reducing weight of obese and diabetic patients (Bradley et al., 2012). These procedures decrease energy intake and improve energy balance. RYGB surgery creates a small pouch in the stomach and connects it to the proximal jejunum to form the Roux limb, which is anastomosed to the duodenal limb to form a Y configuration (Bradley et al., 2012; Mells and Anania, 2013). After RYGB surgery, bile acids are secreted to the empty duodenum and directly into the ileum, bypassing a large portion of the stomach and duodenum and 150 cm of the jejunum. Several recent studies report that serum bile acids are increased after RYGB surgery in obese and diabetic patients and are correlated with improved glycemic control, lipid oxidation, and weight loss (Patti et al., 2009; Simonen et al., 2012; Gerhard et al., 2013). RYGB surgery significantly increases bile flow, fasting serum bile acids, and FGF19 levels (Pournaras et al., 2012). Before gastric bypass, diabetic RYGB patients have lower levels of FGF19 and higher bile acids than nondiabetic patients (Patti et al., 2009; Gerhard et al., 2013). After RYGB surgery, serum FGF19 levels are correlated with increased CYP7A1 and bile acid synthesis in diabetic patients, and serum FGF19 and bile acid levels are increased more in diabetic patients with remission than nondiabetic patients and diabetic patients with no remission (Gerhard et al., 2013). It was concluded that the FGF19/CYP7A1/bile acid synthesis pathway might be involved in remission of T2DM after RYGB surgery. The mechanism for improving insulin resistance and glycemic control is not clear. A recent study of vertical sleeve gastrectomy (VSG) in mice showed that increased circulating bile acids after VSG might change gut microbial communities (Ryan et al., 2014). Gastric bypass changes gut microbiota and is associated with weight loss and inflammation (Zhang et al., 2009a; Furet et al., 2010; Graessler et al., 2013). In FXR knockout mice, the ability of VSG to reduce body weight and improve glucose tolerance was significantly reduced, suggesting that FXR signaling may be involved in weight loss and improving glycemic control in bariatric surgery (Zhang et al., 2009a; Furet et al., 2010; Graessler et al., 2013). After gastric bypass surgery, partially digested food and bile acids are mixed at the distal small intestine and bile acids may spill over to the large intestine, where they activate FXR to induce FGF19 and TGR5 in the large intestine to induce GLP-1 release (Kir et al., 2011). GLP-1 stimulates insulin secretion from the pancreas and improves insulin sensitivity, whereas FGF19 stimulates glycogen synthesis in hepatocytes and reduces serum glucose (Kir et al., 2011; Schaap et al., 2014). Further study using TGR5 knockout and FXR and TGR5 double knockout mice may provide more information about the underlying molecular mechanisms.
VII. Future Perspectives
Basic research in bile acid metabolism and signaling in the last 20 years has unveiled an important role for bile acids in integration of hepatic lipid, glucose, and energy metabolism. Maintaining bile acid homeostasis is essential for protection against bile acid toxicity and inflammation in the gastrointestinal tract. Circadian rhythms of bile acids are critical in maintaining metabolic homeostasis and preventing metabolic disorders. Much is to be learned about the liver-gut microbiota axis and host metabolism. Understanding bile acid–microbiome interaction is critical for treatment and prevention of inflammatory gastrointestinal diseases, NAFLD, diabetes, and obesity. Progress in translation of basic research in bile acid metabolism to clinical applications for patient treatment and drug therapies for liver diseases and diabetes has been made in recent years. Bile acids and their derivatives have the unwanted side effect of pruritus, which may cause low patient compliance. Nonbile acid–based agonists specific for FXR and TGR5 may be developed for treating NAFLD and IBD. miRNA therapy has great potential for treating liver-related diseases. Current research in bile acid metabolism relies on genetically modified mouse models. Future research using proteomics, metabolomics, lipidomics, and megagenomic sequencing will identify biomarkers for diagnosis of these diseases. It is anticipated that new treatment strategies for inflammatory metabolic diseases of the digestive system will be developed.
Abbreviations
- 3β-HSD
3β-hydroxy-Δ5-C27-steroid dehydroxylase
- ABC
ATP-binding cassette
- ACC
acetyl-CoA carboxylase
- AMPK
AMP-activated kinase
- Apo
apolipoprotein
- ASBT
apical sodium-dependent bile salt transporter
- BDL
bile duct ligation
- BMAL-1
brain and muscle ARNT-like protein-1
- BSH
bile salt hydrolase
- CA
cholic acid
- CAR
constitutive androstane receptor
- CCG
clock-controlled gene
- CDCA
chenodeoxycholic acid
- ChREBP
carbohydrate response element binding protein
- CLOCK
circadian locomotor output cycles kaput
- Cry
cryptochrome
- CYP7A1-tg mice
transgenic mice overexpressing CYP7A1 in the liver
- C4
7α-hydroxy-4-cholesten-3-one
- DBP
D-site binding protein
- DCA
deoxycholic acid
- E4BP4
E4 promoter binding protein 4
- ER
endoplasmic reticulum
- ERK
extracellular signal-regulated kinase
- FAS
fatty acid synthase
- FGF
fibroblast growth factor
- FGFR
fibroblast growth factor receptor
- FXR
farnesoid X receptor
- G6Pase
glucose-6-phosphatase
- GLP-1
glucagon-like peptide-1
- GPAM
glycerol-3-phosphate acyltransferase 1
- GPCR
G protein–coupled receptor
- GW4064
3-[2-[2-chloro-4-[[3-(2,6-dichlorophenyl)-5-(1-methylethyl)-4-isoxazolyl]methoxy]phenyl]ethenyl]benzoic acid
- HDAC
histone deacetylase
- HDL
high-density lipoprotein
- HDL-C
high-density lipoprotein cholesterol
- HNF
hepatic nuclear factor
- IBD
inflammatory bowel disease
- ICP
intrahepatic cholestasis of pregnancy
- INT-747
6α-ethyl-3α,7α-dihydroxy-5β-cholan-24-oic acid
- INT-767
6α-ethyl-3α,7α,23-trihydroxy-24-nor-5β-cholan-23-sulphate
- INT-777
6α-ethyl-23(S)-methylcholic acid
- LBD
ligand-binding domain
- LCA
lithocholic acid
- LDL
low-density lipoprotein
- LDL-C
low-density lipoprotein cholesterol
- LDLR
low-density lipoprotein receptor
- LPL
lipoprotein lipase
- LPS
lipopolysaccharide
- LRH-1
liver-related homolog-1
- LXR
liver orphan receptor
- MCA
muricholic acid
- MDR
multidrug resistance protein
- mEH
microsomal epoxide hydrolase
- miRNA
microRNA
- MRP
multidrug resistance-associated protein
- NAFLD
nonalcoholic fatty liver disease
- NASH
nonalcoholic steatohepatitis
- NF-κB
nuclear factor-κB
- NO
nitric oxide
- Npas2
neuronal PAS domain protein 2
- NTCP
Na+-taurocholate cotransport peptide
- OATP
organic anion transporter
- OCA
obeticholic acid
- OST
organic solute transporter
- PCSK9
proprotein convertase subtilisin/kexin type 9
- PEPCK
phosphoenolpyruvate carboxykinase
- Per
period
- PFIC
progressive familial intrahepatic cholestasis
- PGC-1α
peroxisome proliferator–activated receptor γ coactivator-1α
- PPAR
peroxisome proliferator–activated receptor
- PXR
pregnane X receptor
- Rev-erbα
reverse-erythroblastosis α
- RORα
retinoic acid–related orphan receptor α
- RYGB
Roux-en-Y gastric bypass
- S1P
sphingosine-1-phosphate
- S1PR2
sphingosine-1-phosphate receptor 2
- SHP
small heterodimer partner
- SIRT
sirtuin
- SphK
sphingosine kinase
- SR-BI
scavenger receptor B1
- SREBP
steroid response element binding protein
- SULT
sulfotransferase
- T2DM
type 2 diabetes mellitus
- T-β-MCA
tauro-β-muricholic acid
- TCA
taurocholic acid
- TCPOBOP
1,4-bis[2-(3,5-dichloropyridyloxy)]benzene
- TDCA
taurodeoxycholic acid
- TGR5
G protein–coupled bile acid receptor (Gpbar)
- TICE
transintestinal cholesterol excretion
- TUDCA
tauroursodeoxycholic acid
- UDCA
ursodeoxycholic acid
- UTR
untranslated region
- VDR
vitamin D receptor
- VLDL
very low-density lipoprotein
- VSG
vertical sleeve gastrectomy
- VSMC
vascular smooth muscle cell
- WAY-362450
3-(3,4-difluorobenzoyl)-1,2,3,6-tetrahydro-1,1-dimethylazepino[4,5-b]indole-5-carboxylic acid 1-methylethyl ester
Authorship Contributions
Wrote or contributed to the writing of the manuscript: Li, Chiang.
Footnotes
This research is supported by the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases [Grants R37-DK58379 and R01-DK44442].
References
- Abifadel M, Varret M, Rabès JP, Allard D, Ouguerram K, Devillers M, Cruaud C, Benjannet S, Wickham L, Erlich D, et al. (2003) Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet 34:154–156 [DOI] [PubMed] [Google Scholar]
- Adada M, Canals D, Hannun YA, Obeid LM. (2013) Sphingosine-1-phosphate receptor 2. FEBS J 280:6354–6366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adorini L, Pruzanski M, Shapiro D. (2012) Farnesoid X receptor targeting to treat nonalcoholic steatohepatitis. Drug Discov Today 17:988–997 [DOI] [PubMed] [Google Scholar]
- Aldridge MA, Ito MK. (2001) Colesevelam hydrochloride: a novel bile acid-binding resin. Ann Pharmacother 35:898–907 [DOI] [PubMed] [Google Scholar]
- Alemi F, Kwon E, Poole DP, Lieu T, Lyo V, Cattaruzza F, Cevikbas F, Steinhoff M, Nassini R, Materazzi S, et al. (2013a) The TGR5 receptor mediates bile acid-induced itch and analgesia. J Clin Invest 123:1513–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alemi F, Poole DP, Chiu J, Schoonjans K, Cattaruzza F, Grider JR, Bunnett NW, Corvera CU. (2013b) The receptor TGR5 mediates the prokinetic actions of intestinal bile acids and is required for normal defecation in mice. Gastroenterology 144:145–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen RM, Marquart TJ, Albert CJ, Suchy FJ, Wang DQ, Ananthanarayanan M, Ford DA, Baldán A. (2012) miR-33 controls the expression of biliary transporters, and mediates statin- and diet-induced hepatotoxicity. EMBO Mol Med 4:882–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral JD, Viana RJ, Ramalho RM, Steer CJ, Rodrigues CM. (2009) Bile acids: regulation of apoptosis by ursodeoxycholic acid. J Lipid Res 50:1721–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananthanarayanan M, Balasubramanian N, Makishima M, Mangelsdorf DJ, Suchy FJ. (2001) Human bile salt export pump promoter is transactivated by the farnesoid X receptor/bile acid receptor. J Biol Chem 276:28857–28865 [DOI] [PubMed] [Google Scholar]
- Ananthanarayanan M, von Dippe P, Levy D. (1988) Identification of the hepatocyte Na+-dependent bile acid transport protein using monoclonal antibodies. J Biol Chem 263:8338–8343 [PubMed] [Google Scholar]
- Ando H, Yanagihara H, Hayashi Y, Obi Y, Tsuruoka S, Takamura T, Kaneko S, Fujimura A. (2005a) Rhythmic messenger ribonucleic acid expression of clock genes and adipocytokines in mouse visceral adipose tissue. Endocrinology 146:5631–5636 [DOI] [PubMed] [Google Scholar]
- Ando H, Yanagihara H, Sugimoto K, Hayashi Y, Tsuruoka S, Takamura T, Kaneko S, Fujimura A. (2005b) Daily rhythms of P-glycoprotein expression in mice. Chronobiol Int 22:655–665 [DOI] [PubMed] [Google Scholar]
- Angelin B, Einarsson K, Hellström K, Leijd B. (1978) Effects of cholestyramine and chenodeoxycholic acid on the metabolism of endogenous triglyceride in hyperlipoproteinemia. J Lipid Res 19:1017–1024 [PubMed] [Google Scholar]
- Arble DM, Bass J, Laposky AD, Vitaterna MH, Turek FW. (2009) Circadian timing of food intake contributes to weight gain. Obesity (Silver Spring) 17:2100–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron-Wisnewsky J, Gaborit B, Dutour A, Clement K. (2013) Gut microbiota and non-alcoholic fatty liver disease: new insights. Clin Microbiol Infect 19:338–348 [DOI] [PubMed] [Google Scholar]
- Arrese M, Trauner M, Sacchiero RJ, Crossman MW, Shneider BL. (1998) Neither intestinal sequestration of bile acids nor common bile duct ligation modulate the expression and function of the rat ileal bile acid transporter. Hepatology 28:1081–1087 [DOI] [PubMed] [Google Scholar]
- Aryal B, Rotllan N, Fernández-Hernando C. (2014) Noncoding RNAs and atherosclerosis. Curr Atheroscler Rep 16:407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Schibler U. (2011) Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab 13:125–137 [DOI] [PubMed] [Google Scholar]
- Axelson M, Aly A, Sjövall J. (1988) Levels of 7 alpha-hydroxy-4-cholesten-3-one in plasma reflect rates of bile acid synthesis in man. FEBS Lett 239:324–328 [DOI] [PubMed] [Google Scholar]
- Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. (2004) The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA 101:15718–15723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badman MK, Pissios P, Kennedy AR, Koukos G, Flier JS, Maratos-Flier E. (2007) Hepatic fibroblast growth factor 21 is regulated by PPARalpha and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metab 5:426–437 [DOI] [PubMed] [Google Scholar]
- Ballatori N, Christian WV, Lee JY, Dawson PA, Soroka CJ, Boyer JL, Madejczyk MS, Li N. (2005) OSTalpha-OSTbeta: a major basolateral bile acid and steroid transporter in human intestinal, renal, and biliary epithelia. Hepatology 42:1270–1279 [DOI] [PubMed] [Google Scholar]
- Bass J, Takahashi JS. (2010) Circadian integration of metabolism and energetics. Science 330:1349–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bays HE, Goldberg RB, Truitt KE, Jones MR. (2008) Colesevelam hydrochloride therapy in patients with type 2 diabetes mellitus treated with metformin: glucose and lipid effects. Arch Intern Med 168:1975–1983 [DOI] [PubMed] [Google Scholar]
- Beilke LD, Aleksunes LM, Holland RD, Besselsen DG, Beger RD, Klaassen CD, Cherrington NJ. (2009) Constitutive androstane receptor-mediated changes in bile acid composition contributes to hepatoprotection from lithocholic acid-induced liver injury in mice. Drug Metab Dispos 37:1035–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrabah W, Aumercier P, Gheeraert C, Dehondt H, Bouchaert E, Alexandre J, Ploton M, Mazuy C, Caron S, Tailleux A, et al. (2014) Glucose sensing O-GlcNAcylation pathway regulates the nuclear bile acid receptor farnesoid X receptor (FXR). Hepatology 59:2022–2033 [DOI] [PubMed] [Google Scholar]
- Bertolotti M, Carulli L, Concari M, Martella P, Loria P, Tagliafico E, Ferrari S, Del Puppo M, Amati B, De Fabiani E, et al. (2001) Suppression of bile acid synthesis, but not of hepatic cholesterol 7alpha-hydroxylase expression, by obstructive cholestasis in humans. Hepatology 34:234–242 [DOI] [PubMed] [Google Scholar]
- Beysen C, Murphy EJ, Deines K, Chan M, Tsang E, Glass A, Turner SM, Protasio J, Riiff T, Hellerstein MK. (2012) Effect of bile acid sequestrants on glucose metabolism, hepatic de novo lipogenesis, and cholesterol and bile acid kinetics in type 2 diabetes: a randomised controlled study. Diabetologia 55:432–442 [DOI] [PubMed] [Google Scholar]
- Bhalla S, Ozalp C, Fang S, Xiang L, Kemper JK. (2004) Ligand-activated pregnane X receptor interferes with HNF-4 signaling by targeting a common coactivator PGC-1alpha. Functional implications in hepatic cholesterol and glucose metabolism. J Biol Chem 279:45139–45147 [DOI] [PubMed] [Google Scholar]
- Bhatnagar S, Damron HA, Hillgartner FB. (2009) Fibroblast growth factor-19, a novel factor that inhibits hepatic fatty acid synthesis. J Biol Chem 284:10023–10033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop-Bailey D, Walsh DT, Warner TD. (2004) Expression and activation of the farnesoid X receptor in the vasculature. Proc Natl Acad Sci USA 101:3668–3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer JL, Trauner M, Mennone A, Soroka CJ, Cai SY, Moustafa T, Zollner G, Lee JY, Ballatori N. (2006) Upregulation of a basolateral FXR-dependent bile acid efflux transporter OSTalpha-OSTbeta in cholestasis in humans and rodents. Am J Physiol Gastrointest Liver Physiol 290:G1124–G1130 [DOI] [PubMed] [Google Scholar]
- Bradley D, Magkos F, Klein S. (2012) Effects of bariatric surgery on glucose homeostasis and type 2 diabetes. Gastroenterology 143:897–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. (1997) The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89:331–340 [DOI] [PubMed] [Google Scholar]
- Bruce CR, Risis S, Babb JR, Yang C, Kowalski GM, Selathurai A, Lee-Young RS, Weir JM, Yoshioka K, Takuwa Y, et al. (2012) Overexpression of sphingosine kinase 1 prevents ceramide accumulation and ameliorates muscle insulin resistance in high-fat diet-fed mice. Diabetes 61:3148–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brufau G, Groen AK, Kuipers F. (2011) Reverse cholesterol transport revisited: contribution of biliary versus intestinal cholesterol excretion. Arterioscler Thromb Vasc Biol 31:1726–1733 [DOI] [PubMed] [Google Scholar]
- Brufau G, Stellaard F, Prado K, Bloks VW, Jonkers E, Boverhof R, Kuipers F, Murphy EJ. (2010) Improved glycemic control with colesevelam treatment in patients with type 2 diabetes is not directly associated with changes in bile acid metabolism. Hepatology 52:1455–1464 [DOI] [PubMed] [Google Scholar]
- Brunt EM. (2010) Pathology of nonalcoholic fatty liver disease. Nat Rev Gastroenterol Hepatol 7:195–203 [DOI] [PubMed] [Google Scholar]
- Bunchorntavakul C, Reddy KR. (2012) Pruritus in chronic cholestatic liver disease. Clin Liver Dis 16:331–346 [DOI] [PubMed] [Google Scholar]
- Burris TP. (2008) Nuclear hormone receptors for heme: REV-ERBalpha and REV-ERBbeta are ligand-regulated components of the mammalian clock. Mol Endocrinol 22:1509–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calpe-Berdiel L, Rotllan N, Fiévet C, Roig R, Blanco-Vaca F, Escolà-Gil JC. (2008) Liver X receptor-mediated activation of reverse cholesterol transport from macrophages to feces in vivo requires ABCG5/G8. J Lipid Res 49:1904–1911 [DOI] [PubMed] [Google Scholar]
- Camilleri M, Vazquez-Roque MI, Carlson P, Burton D, Wong BS, Zinsmeister AR. (2011) Association of bile acid receptor TGR5 variation and transit in health and lower functional gastrointestinal disorders. Neurogastroenterol Motil 23:995–999, e458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, et al. (2007) Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56:1761–1772 [DOI] [PubMed] [Google Scholar]
- Cani PD, Dewever C, Delzenne NM. (2004) Inulin-type fructans modulate gastrointestinal peptides involved in appetite regulation (glucagon-like peptide-1 and ghrelin) in rats. Br J Nutr 92:521–526 [DOI] [PubMed] [Google Scholar]
- Cariou B, van Harmelen K, Duran-Sandoval D, van Dijk TH, Grefhorst A, Abdelkarim M, Caron S, Torpier G, Fruchart JC, Gonzalez FJ, et al. (2006) The farnesoid X receptor modulates adiposity and peripheral insulin sensitivity in mice. J Biol Chem 281:11039–11049 [DOI] [PubMed] [Google Scholar]
- Caron S, Huaman Samanez C, Dehondt H, Ploton M, Briand O, Lien F, Dorchies E, Dumont J, Postic C, Cariou B, et al. (2013) Farnesoid X receptor inhibits the transcriptional activity of carbohydrate response element binding protein in human hepatocytes. Mol Cell Biol 33:2202–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha JY, Repa JJ. (2007) The liver X receptor (LXR) and hepatic lipogenesis. The carbohydrate-response element-binding protein is a target gene of LXR. J Biol Chem 282:743–751 [DOI] [PubMed] [Google Scholar]
- Chatterjee B, Echchgadda I, Song CS. (2005) Vitamin D receptor regulation of the steroid/bile acid sulfotransferase SULT2A1. Methods Enzymol 400:165–191 [DOI] [PubMed] [Google Scholar]
- Chavez JA, Summers SA. (2012) A ceramide-centric view of insulin resistance. Cell Metab 15:585–594 [DOI] [PubMed] [Google Scholar]
- Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. (2001) Nuclear receptors and lipid physiology: opening the X-files. Science 294:1866–1870 [DOI] [PubMed] [Google Scholar]
- Chen F, Ma L, Dawson PA, Sinal CJ, Sehayek E, Gonzalez FJ, Breslow J, Ananthanarayanan M, Shneider BL. (2003) Liver receptor homologue-1 mediates species- and cell line-specific bile acid-dependent negative feedback regulation of the apical sodium-dependent bile acid transporter. J Biol Chem 278:19909–19916 [DOI] [PubMed] [Google Scholar]
- Chen X, Chen F, Liu S, Glaeser H, Dawson PA, Hofmann AF, Kim RB, Shneider BL, Pang KS. (2006) Transactivation of rat apical sodium-dependent bile acid transporter and increased bile acid transport by 1alpha,25-dihydroxyvitamin D3 via the vitamin D receptor. Mol Pharmacol 69:1913–1923 [DOI] [PubMed] [Google Scholar]
- Cheung O, Puri P, Eicken C, Contos MJ, Mirshahi F, Maher JW, Kellum JM, Min H, Luketic VA, Sanyal AJ. (2008) Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology 48:1810–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang JY. (2002) Bile acid regulation of gene expression: roles of nuclear hormone receptors. Endocr Rev 23:443–463 [DOI] [PubMed] [Google Scholar]
- Chiang JY. (2009) Bile acids: regulation of synthesis. J Lipid Res 50:1955–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang JY. (2013) Bile acid metabolism and signaling. Compr Physiol 3:1191–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang JY, Kimmel R, Weinberger C, Stroup D. (2000) Farnesoid X receptor responds to bile acids and represses cholesterol 7alpha-hydroxylase gene (CYP7A1) transcription. J Biol Chem 275:10918–10924 [DOI] [PubMed] [Google Scholar]
- Chiang JY, Miller WF, Lin GM. (1990) Regulation of cholesterol 7 alpha-hydroxylase in the liver. Purification of cholesterol 7 alpha-hydroxylase and the immunochemical evidence for the induction of cholesterol 7 alpha-hydroxylase by cholestyramine and circadian rhythm. J Biol Chem 265:3889–3897 [PubMed] [Google Scholar]
- Chiang JYL. (1998) Regulation of bile acid synthesis. Front Biosci 3:d176–d193 [DOI] [PubMed] [Google Scholar]
- Childs S, Yeh RL, Georges E, Ling V. (1995) Identification of a sister gene to P-glycoprotein. Cancer Res 55:2029–2034 [PubMed] [Google Scholar]
- Choi M, Moschetta A, Bookout AL, Peng L, Umetani M, Holmstrom SR, Suino-Powell K, Xu HE, Richardson JA, Gerard RD, et al. (2006) Identification of a hormonal basis for gallbladder filling. Nat Med 12:1253–1255 [DOI] [PubMed] [Google Scholar]
- Cipriani S, Mencarelli A, Chini MG, Distrutti E, Renga B, Bifulco G, Baldelli F, Donini A, Fiorucci S. (2011) The bile acid receptor GPBAR-1 (TGR5) modulates integrity of intestinal barrier and immune response to experimental colitis. PLoS ONE 6:e25637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipriani S, Mencarelli A, Palladino G, Fiorucci S. (2010) FXR activation reverses insulin resistance and lipid abnormalities and protects against liver steatosis in Zucker (fa/fa) obese rats. J Lipid Res 51:771–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirera-Salinas D, Pauta M, Allen RM, Salerno AG, Ramírez CM, Chamorro-Jorganes A, Wanschel AC, Lasuncion MA, Morales-Ruiz M, Suarez Y, et al. (2012) Mir-33 regulates cell proliferation and cell cycle progression. Cell Cycle 11:922–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke SF, Murphy EF, Nilaweera K, Ross PR, Shanahan F, O’Toole PW, Cotter PD. (2012) The gut microbiota and its relationship to diet and obesity: new insights. Gut Microbes 3:186–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claudel T, Inoue Y, Barbier O, Duran-Sandoval D, Kosykh V, Fruchart J, Fruchart JC, Gonzalez FJ, Staels B. (2003) Farnesoid X receptor agonists suppress hepatic apolipoprotein CIII expression. Gastroenterology 125:544–555 [DOI] [PubMed] [Google Scholar]
- Claudel T, Staels B, Kuipers F. (2005) The Farnesoid X receptor: a molecular link between bile acid and lipid and glucose metabolism. Arterioscler Thromb Vasc Biol 25:2020–2030 [DOI] [PubMed] [Google Scholar]
- Claudel T, Sturm E, Duez H, Torra IP, Sirvent A, Kosykh V, Fruchart JC, Dallongeville J, Hum DW, Kuipers F, et al. (2002) Bile acid-activated nuclear receptor FXR suppresses apolipoprotein A-I transcription via a negative FXR response element. J Clin Invest 109:961–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claudel T, Sturm E, Kuipers F, Staels B. (2004) The farnesoid X receptor: a novel drug target? Expert Opin Investig Drugs 13:1135–1148 [DOI] [PubMed] [Google Scholar]
- Cohen JC, Horton JD, Hobbs HH. (2011) Human fatty liver disease: old questions and new insights. Science 332:1519–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covey SD, Krieger M, Wang W, Penman M, Trigatti BL. (2003) Scavenger receptor class B type I-mediated protection against atherosclerosis in LDL receptor-negative mice involves its expression in bone marrow-derived cells. Arterioscler Thromb Vasc Biol 23:1589–1594 [DOI] [PubMed] [Google Scholar]
- Crumbley C, Wang Y, Kojetin DJ, Burris TP. (2010) Characterization of the core mammalian clock component, NPAS2, as a REV-ERBalpha/RORalpha target gene. J Biol Chem 285:35386–35392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui YJ, Aleksunes LM, Tanaka Y, Goedken MJ, Klaassen CD. (2009) Compensatory induction of liver efflux transporters in response to ANIT-induced liver injury is impaired in FXR-null mice. Toxicol Sci 110:47–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyphert HA, Ge X, Kohan AB, Salati LM, Zhang Y, Hillgartner FB. (2012) Activation of the farnesoid X receptor induces hepatic expression and secretion of fibroblast growth factor 21. J Biol Chem 287:25123–25138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaja MJ, Ding WX, Donohue TM, Jr, Friedman SL, Kim JS, Komatsu M, Lemasters JJ, Lemoine A, Lin JD, Ou JH, et al. (2013) Functions of autophagy in normal and diseased liver. Autophagy 9:1131–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. (2000) Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev 14:2950–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. (2014) Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson MH, Dillon MA, Gordon B, Jones P, Samuels J, Weiss S, Isaacsohn J, Toth P, Burke SK. (1999) Colesevelam hydrochloride (cholestagel): a new, potent bile acid sequestrant associated with a low incidence of gastrointestinal side effects. Arch Intern Med 159:1893–1900 [DOI] [PubMed] [Google Scholar]
- Dawson PA, Hubbert M, Haywood J, Craddock AL, Zerangue N, Christian WV, Ballatori N. (2005) The heteromeric organic solute transporter alpha-beta, Ostalpha-Ostbeta, is an ileal basolateral bile acid transporter. J Biol Chem 280:6960–6968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day CP, James OF. (1998) Steatohepatitis: a tale of two “hits”? Gastroenterology 114:842–845 [DOI] [PubMed] [Google Scholar]
- Degirolamo C, Rainaldi S, Bovenga F, Murzilli S, Moschetta A. (2014) Microbiota modification with probiotics induces hepatic bile acid synthesis via downregulation of the Fxr-Fgf15 axis in mice. Cell Reports 7:12–18 [DOI] [PubMed] [Google Scholar]
- Denson LA, Sturm E, Echevarria W, Zimmerman TL, Makishima M, Mangelsdorf DJ, Karpen SJ. (2001) The orphan nuclear receptor, shp, mediates bile acid-induced inhibition of the rat bile acid transporter, ntcp. Gastroenterology 121:140–147 [DOI] [PubMed] [Google Scholar]
- Dent P, Fang Y, Gupta S, Studer E, Mitchell C, Spiegel S, Hylemon PB. (2005) Conjugated bile acids promote ERK1/2 and AKT activation via a pertussis toxin-sensitive mechanism in murine and human hepatocytes. Hepatology 42:1291–1299 [DOI] [PubMed] [Google Scholar]
- Dentin R, Pégorier JP, Benhamed F, Foufelle F, Ferré P, Fauveau V, Magnuson MA, Girard J, Postic C. (2004) Hepatic glucokinase is required for the synergistic action of ChREBP and SREBP-1c on glycolytic and lipogenic gene expression. J Biol Chem 279:20314–20326 [DOI] [PubMed] [Google Scholar]
- Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. (2012) Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature 487:104–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikopoulos N, Schmid RM, Bachem M, Buttenschoen K, Adler G, Chiang JY, Weidenbach H. (2007) Bile synthesis in rat models of inflammatory bowel diseases. Eur J Clin Invest 37:222–230 [DOI] [PubMed] [Google Scholar]
- Dong B, Qatanani M, Moore DD. (2009) Constitutive androstane receptor mediates the induction of drug metabolism in mouse models of type 1 diabetes. Hepatology 50:622–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drocourt L, Ourlin JC, Pascussi JM, Maurel P, Vilarem MJ. (2002) Expression of CYP3A4, CYP2B6, and CYP2C9 is regulated by the vitamin D receptor pathway in primary human hepatocytes. J Biol Chem 277:25125–25132 [DOI] [PubMed] [Google Scholar]
- Duane WC, Gilberstadt ML, Wiegand DM. (1979) Diurnal rhythms of bile acid production in the rat. Am J Physiol 236:R175–R179 [DOI] [PubMed] [Google Scholar]
- Duane WC, Levitt DG, Mueller SM, Behrens JC. (1983) Regulation of bile acid synthesis in man. Presence of a diurnal rhythm. J Clin Invest 72:1930–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboc H, Tache Y, Hofmann AF. (2014) The bile acid TGR5 membrane receptor: From basic research to clinical application. Dig Liver Dis 46:302–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duez H, van der Veen JN, Duhem C, Pourcet B, Touvier T, Fontaine C, Derudas B, Baugé E, Havinga R, Bloks VW, et al. (2008) Regulation of bile acid synthesis by the nuclear receptor Rev-erbalpha. Gastroenterology 135:689–698 [DOI] [PubMed] [Google Scholar]
- Düfer M, Hörth K, Wagner R, Schittenhelm B, Prowald S, Wagner TF, Oberwinkler J, Lukowski R, Gonzalez FJ, Krippeit-Drews P, et al. (2012) Bile acids acutely stimulate insulin secretion of mouse β-cells via farnesoid X receptor activation and K(ATP) channel inhibition. Diabetes 61:1479–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran-Sandoval D, Cariou B, Percevault F, Hennuyer N, Grefhorst A, van Dijk TH, Gonzalez FJ, Fruchart JC, Kuipers F, Staels B. (2005) The farnesoid X receptor modulates hepatic carbohydrate metabolism during the fasting-refeeding transition. J Biol Chem 280:29971–29979 [DOI] [PubMed] [Google Scholar]
- Duran-Sandoval D, Mautino G, Martin G, Percevault F, Barbier O, Fruchart JC, Kuipers F, Staels B. (2004) Glucose regulates the expression of the farnesoid X receptor in liver. Diabetes 53:890–898 [DOI] [PubMed] [Google Scholar]
- Duseja A, Chawla YK. (2014) Obesity and NAFLD: The role of bacteria and microbiota. Clin Liver Dis 18:59–71 [DOI] [PubMed] [Google Scholar]
- Elmén J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjärn M, Hansen JB, Hansen HF, Straarup EM, et al. (2008) Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res 36:1153–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eloranta JJ, Kullak-Ublick GA. (2008) The role of FXR in disorders of bile acid homeostasis. Physiology (Bethesda) 23:286–295 [DOI] [PubMed] [Google Scholar]
- Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, et al. (2006) miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 3:87–98 [DOI] [PubMed] [Google Scholar]
- Evans KA, Budzik BW, Ross SA, Wisnoski DD, Jin J, Rivero RA, Vimal M, Szewczyk GR, Jayawickreme C, Moncol DL, et al. (2009a) Discovery of 3-aryl-4-isoxazolecarboxamides as TGR5 receptor agonists. J Med Chem 52:7962–7965 [DOI] [PubMed] [Google Scholar]
- Evans MJ, Mahaney PE, Borges-Marcucci L, Lai K, Wang S, Krueger JA, Gardell SJ, Huard C, Martinez R, Vlasuk GP, et al. (2009b) A synthetic farnesoid X receptor (FXR) agonist promotes cholesterol lowering in models of dyslipidemia. Am J Physiol Gastrointest Liver Physiol 296:G543–G552 [DOI] [PubMed] [Google Scholar]
- Falvey E, Fleury-Olela F, Schibler U. (1995) The rat hepatic leukemia factor (HLF) gene encodes two transcriptional activators with distinct circadian rhythms, tissue distributions and target preferences. EMBO J 14:4307–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell GC, Larter CZ. (2006) Nonalcoholic fatty liver disease: from steatosis to cirrhosis. Hepatology 43 (Suppl 1):S99–S112 [DOI] [PubMed] [Google Scholar]
- Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA. (2011) A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331:1315–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Hernando C, Ramírez CM, Goedeke L, Suárez Y. (2013) MicroRNAs in metabolic disease. Arterioscler Thromb Vasc Biol 33:178–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Hernando C, Suárez Y, Rayner KJ, Moore KJ. (2011) MicroRNAs in lipid metabolism. Curr Opin Lipidol 22:86–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filiano AN, Millender-Swain T, Johnson R, Jr, Young ME, Gamble KL, Bailey SM. (2013) Chronic ethanol consumption disrupts the core molecular clock and diurnal rhythms of metabolic genes in the liver without affecting the suprachiasmatic nucleus. PLoS ONE 8:e71684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorucci S, Baldelli F. (2009) Farnesoid X receptor agonists in biliary tract disease. Curr Opin Gastroenterol 25:252–259 [DOI] [PubMed] [Google Scholar]
- Fiorucci S, Mencarelli A, Distrutti E, Zampella A. (2012) Farnesoid X receptor: from medicinal chemistry to clinical applications. Future Med Chem 4:877–891 [DOI] [PubMed] [Google Scholar]
- Forman BM, Goode E, Chen J, Oro AE, Bradley DJ, Perlmann T, Noonan DJ, Burka LT, McMorris T, Lamph WW, et al. (1995) Identification of a nuclear receptor that is activated by farnesol metabolites. Cell 81:687–693 [DOI] [PubMed] [Google Scholar]
- Fretland AJ, Omiecinski CJ. (2000) Epoxide hydrolases: biochemistry and molecular biology. Chem Biol Interact 129:41–59 [DOI] [PubMed] [Google Scholar]
- Froy O. (2009) Cytochrome P450 and the biological clock in mammals. Curr Drug Metab 10:104–115 [DOI] [PubMed] [Google Scholar]
- Froy O. (2010) Metabolism and circadian rhythms—implications for obesity. Endocr Rev 31:1–24 [DOI] [PubMed] [Google Scholar]
- Fu L, John LM, Adams SH, Yu XX, Tomlinson E, Renz M, Williams PM, Soriano R, Corpuz R, Moffat B, et al. (2004) Fibroblast growth factor 19 increases metabolic rate and reverses dietary and leptin-deficient diabetes. Endocrinology 145:2594–2603 [DOI] [PubMed] [Google Scholar]
- Furet JP, Kong LC, Tap J, Poitou C, Basdevant A, Bouillot JL, Mariat D, Corthier G, Doré J, Henegar C, et al. (2010) Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes 59:3049–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon F, Firsov D. (2011) The role of circadian timing system on drug metabolism and detoxification. Expert Opin Drug Metab Toxicol 7:147–158 [DOI] [PubMed] [Google Scholar]
- Gadaleta RM, Oldenburg B, Willemsen EC, Spit M, Murzilli S, Salvatore L, Klomp LW, Siersema PD, van Erpecum KJ, van Mil SW. (2011a) Activation of bile salt nuclear receptor FXR is repressed by pro-inflammatory cytokines activating NF-κB signaling in the intestine. Biochim Biophys Acta 1812:851–858 [DOI] [PubMed] [Google Scholar]
- Gadaleta RM, van Erpecum KJ, Oldenburg B, Willemsen EC, Renooij W, Murzilli S, Klomp LW, Siersema PD, Schipper ME, Danese S, et al. (2011b) Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut 60:463–472 [DOI] [PubMed] [Google Scholar]
- Gälman C, Angelin B, Rudling M. (2005) Bile acid synthesis in humans has a rapid diurnal variation that is asynchronous with cholesterol synthesis. Gastroenterology 129:1445–1453 [DOI] [PubMed] [Google Scholar]
- Gälman C, Angelin B, Rudling M. (2011) Pronounced variation in bile acid synthesis in humans is related to gender, hypertriglyceridaemia and circulating levels of fibroblast growth factor 19. J Intern Med 270:580–588 [DOI] [PubMed] [Google Scholar]
- Gao B, Bataller R. (2011) Alcoholic liver disease: pathogenesis and new therapeutic targets. Gastroenterology 141:1572–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, He J, Zhai Y, Wada T, Xie W. (2009) The constitutive androstane receptor is an anti-obesity nuclear receptor that improves insulin sensitivity. J Biol Chem 284:25984–25992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield D, Le Martelot G, Vejnar CE, Gerlach D, Schaad O, Fleury-Olela F, Ruskeepää AL, Oresic M, Esau CC, Zdobnov EM, et al. (2009) Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev 23:1313–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhard GS, Styer AM, Wood GC, Roesch SL, Petrick AT, Gabrielsen J, Strodel WE, Still CD, Argyropoulos G. (2013) A role for fibroblast growth factor 19 and bile acids in diabetes remission after Roux-en-Y gastric bypass. Diabetes Care 36:1859–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. (2006) Metagenomic analysis of the human distal gut microbiome. Science 312:1355–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg HN, Zhang YL, Hernandez-Ono A. (2006) Metabolic syndrome: focus on dyslipidemia. Obesity (Silver Spring) 14 (Suppl 1):41S–49S [DOI] [PubMed] [Google Scholar]
- Gong YZ, Everett ET, Schwartz DA, Norris JS, Wilson FA. (1994) Molecular cloning, tissue distribution, and expression of a 14-kDa bile acid-binding protein from rat ileal cytosol. Proc Natl Acad Sci USA 91:4741–4745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME, et al. (2000) A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell 6:517–526 [DOI] [PubMed] [Google Scholar]
- Graessler J, Qin Y, Zhong H, Zhang J, Licinio J, Wong ML, Xu A, Chavakis T, Bornstein AB, Ehrhart-Bornstein M, et al. (2013) Metagenomic sequencing of the human gut microbiome before and after bariatric surgery in obese patients with type 2 diabetes: correlation with inflammatory and metabolic parameters. Pharmacogenomics J 13:514–522 [DOI] [PubMed] [Google Scholar]
- Guo GL, Lambert G, Negishi M, Ward JM, Brewer HB, Jr, Kliewer SA, Gonzalez FJ, Sinal CJ. (2003) Complementary roles of farnesoid X receptor, pregnane X receptor, and constitutive androstane receptor in protection against bile acid toxicity. J Biol Chem 278:45062–45071 [DOI] [PubMed] [Google Scholar]
- Guo GL, Santamarina-Fojo S, Akiyama TE, Amar MJ, Paigen BJ, Brewer B, Jr, Gonzalez FJ. (2006) Effects of FXR in foam-cell formation and atherosclerosis development. Biochim Biophys Acta 1761:1401–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Natarajan R, Payne SG, Studer EJ, Spiegel S, Dent P, Hylemon PB. (2004) Deoxycholic acid activates the c-Jun N-terminal kinase pathway via FAS receptor activation in primary hepatocytes. Role of acidic sphingomyelinase-mediated ceramide generation in FAS receptor activation. J Biol Chem 279:5821–5828 [DOI] [PubMed] [Google Scholar]
- Haeusler RA, Astiarraga B, Camastra S, Accili D, Ferrannini E. (2013) Human insulin resistance is associated with increased plasma levels of 12α-hydroxylated bile acids. Diabetes 62:4184–4191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hageman J, Herrema H, Groen AK, Kuipers F. (2010) A role of the bile salt receptor FXR in atherosclerosis. Arterioscler Thromb Vasc Biol 30:1519–1528 [DOI] [PubMed] [Google Scholar]
- Hait NC, Allegood J, Maceyka M, Strub GM, Harikumar KB, Singh SK, Luo C, Marmorstein R, Kordula T, Milstien S, et al. (2009) Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science 325:1254–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Chiang JY. (2009) Mechanism of vitamin D receptor inhibition of cholesterol 7alpha-hydroxylase gene transcription in human hepatocytes. Drug Metab Dispos 37:469–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Li T, Ellis E, Strom S, Chiang JY. (2010) A novel bile acid-activated vitamin D receptor signaling in human hepatocytes. Mol Endocrinol 24:1151–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschin C, Lin J, Rhee J, Peyer AK, Chin S, Wu PH, Meyer UA, Spiegelman BM. (2005) Nutritional regulation of hepatic heme biosynthesis and porphyria through PGC-1alpha. Cell 122:505–515 [DOI] [PubMed] [Google Scholar]
- Hanniman EA, Lambert G, McCarthy TC, Sinal CJ. (2005) Loss of functional farnesoid X receptor increases atherosclerotic lesions in apolipoprotein E-deficient mice. J Lipid Res 46:2595–2604 [DOI] [PubMed] [Google Scholar]
- Hartman HB, Gardell SJ, Petucci CJ, Wang S, Krueger JA, Evans MJ. (2009) Activation of farnesoid X receptor prevents atherosclerotic lesion formation in LDLR-/- and apoE-/- mice. J Lipid Res 50:1090–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JA, et al. (2012) Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab 15:848–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert MR, Siegel DL, Staszewski L, Cayanan C, Banerjee U, Dhamija S, Anderson J, Fan A, Wang L, Rix P, et al. (2010) Synthesis and SAR of 2-aryl-3-aminomethylquinolines as agonists of the bile acid receptor TGR5. Bioorg Med Chem Lett 20:5718–5721 [DOI] [PubMed] [Google Scholar]
- Heubi JE, Setchell KD, Bove KE. (2007) Inborn errors of bile acid metabolism. Semin Liver Dis 27:282–294 [DOI] [PubMed] [Google Scholar]
- Hirao J, Arakawa S, Watanabe K, Ito K, Furukawa T. (2006) Effects of restricted feeding on daily fluctuations of hepatic functions including p450 monooxygenase activities in rats. J Biol Chem 281:3165–3171 [DOI] [PubMed] [Google Scholar]
- Hofmann AF. (2002) Rifampicin and treatment of cholestatic pruritus. Gut 51:756–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollman DA, Milona A, van Erpecum KJ, van Mil SW. (2012) Anti-inflammatory and metabolic actions of FXR: insights into molecular mechanisms. Biochim Biophys Acta 1821:1443–1452 [DOI] [PubMed] [Google Scholar]
- Holt JA, Luo G, Billin AN, Bisi J, McNeill YY, Kozarsky KF, Donahee M, Wang DY, Mansfield TA, Kliewer SA, et al. (2003) Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev 17:1581–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda A, Yamashita K, Numazawa M, Ikegami T, Doy M, Matsuzaki Y, Miyazaki H. (2007) Highly sensitive quantification of 7alpha-hydroxy-4-cholesten-3-one in human serum by LC-ESI-MS/MS. J Lipid Res 48:458–464 [DOI] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. (2002) SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest 109:1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Bonde Y, Eggertsen G, Rudling M. (2014) Muricholic bile acids are potent regulators of bile acid synthesis via a positive feedback mechanism. J Intern Med 275:27–38 [DOI] [PubMed] [Google Scholar]
- Ichikawa R, Takayama T, Yoneno K, Kamada N, Kitazume MT, Higuchi H, Matsuoka K, Watanabe M, Itoh H, Kanai T, et al. (2012) Bile acids induce monocyte differentiation toward interleukin-12 hypo-producing dendritic cells via a TGR5-dependent pathway. Immunology 136:153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka K, Bruick RK, Liang G, Horton JD, Uyeda K. (2004) Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc Natl Acad Sci USA 101:7281–7286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, Luo G, Jones SA, Goodwin B, Richardson JA, et al. (2005) Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab 2:217–225 [DOI] [PubMed] [Google Scholar]
- Inagaki T, Dutchak P, Zhao G, Ding X, Gautron L, Parameswara V, Li Y, Goetz R, Mohammadi M, Esser V, et al. (2007) Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metab 5:415–425 [DOI] [PubMed] [Google Scholar]
- Inagaki T, Moschetta A, Lee YK, Peng L, Zhao G, Downes M, Yu RT, Shelton JM, Richardson JA, Repa JJ, et al. (2006) Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci USA 103:3920–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam KB, Fukiya S, Hagio M, Fujii N, Ishizuka S, Ooka T, Ogura Y, Hayashi T, Yokota A. (2011) Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 141:1773–1781 [DOI] [PubMed] [Google Scholar]
- Jacquemin E. (2001) Role of multidrug resistance 3 deficiency in pediatric and adult liver disease: one gene for three diseases. Semin Liver Dis 21:551–562 [DOI] [PubMed] [Google Scholar]
- Ji Y, Jian B, Wang N, Sun Y, Moya ML, Phillips MC, Rothblat GH, Swaney JB, Tall AR. (1997) Scavenger receptor BI promotes high density lipoprotein-mediated cellular cholesterol efflux. J Biol Chem 272:20982–20985 [DOI] [PubMed] [Google Scholar]
- Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. (2008) Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci USA 105:13580–13585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones ML, Martoni CJ, Ganopolsky JG, Labbé A, Prakash S. (2014) The human microbiome and bile acid metabolism: dysbiosis, dysmetabolism, disease and intervention. Expert Opin Biol Ther 14:467–482 [DOI] [PubMed] [Google Scholar]
- Joyce SA, Gahan CG. (2014) The gut microbiota and the metabolic health of the host. Curr Opin Gastroenterol 30:120–127 [DOI] [PubMed] [Google Scholar]
- Kaasik K, Lee CC. (2004) Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature 430:467–471 [DOI] [PubMed] [Google Scholar]
- Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, Takei H, Muto A, Nittono H, Ridlon JM, et al. (2013) Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol 58:949–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HS, Angers M, Beak JY, Wu X, Gimble JM, Wada T, Xie W, Collins JB, Grissom SF, Jetten AM. (2007) Gene expression profiling reveals a regulatory role for ROR alpha and ROR gamma in phase I and phase II metabolism. Physiol Genomics 31:281–294 [DOI] [PubMed] [Google Scholar]
- Karpen SJ, Trauner M. (2010) The new therapeutic frontier—nuclear receptors and the liver. J Hepatol 52:455–462 [DOI] [PubMed] [Google Scholar]
- Kast HR, Goodwin B, Tarr PT, Jones SA, Anisfeld AM, Stoltz CM, Tontonoz P, Kliewer S, Willson TM, Edwards PA. (2002) Regulation of multidrug resistance-associated protein 2 (ABCC2) by the nuclear receptors pregnane X receptor, farnesoid X-activated receptor, and constitutive androstane receptor. J Biol Chem 277:2908–2915 [DOI] [PubMed] [Google Scholar]
- Kast HR, Nguyen CM, Sinal CJ, Jones SA, Laffitte BA, Reue K, Gonzalez FJ, Willson TM, Edwards PA. (2001) Farnesoid X-activated receptor induces apolipoprotein C-II transcription: a molecular mechanism linking plasma triglyceride levels to bile acids. Mol Endocrinol 15:1720–1728 [DOI] [PubMed] [Google Scholar]
- Katsuma S, Hirasawa A, Tsujimoto G. (2005) Bile acids promote glucagon-like peptide-1 secretion through TGR5 in a murine enteroendocrine cell line STC-1. Biochem Biophys Res Commun 329:386–390 [DOI] [PubMed] [Google Scholar]
- Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, Miwa M, Fukusumi S, Habata Y, Itoh T, Shintani Y, et al. (2003) A G protein-coupled receptor responsive to bile acids. J Biol Chem 278:9435–9440 [DOI] [PubMed] [Google Scholar]
- Keitel V, Cupisti K, Ullmer C, Knoefel WT, Kubitz R, Häussinger D. (2009) The membrane-bound bile acid receptor TGR5 is localized in the epithelium of human gallbladders. Hepatology 50:861–870 [DOI] [PubMed] [Google Scholar]
- Keitel V, Donner M, Winandy S, Kubitz R, Häussinger D. (2008) Expression and function of the bile acid receptor TGR5 in Kupffer cells. Biochem Biophys Res Commun 372:78–84 [DOI] [PubMed] [Google Scholar]
- Keitel V, Görg B, Bidmon HJ, Zemtsova I, Spomer L, Zilles K, Häussinger D. (2010) The bile acid receptor TGR5 (Gpbar-1) acts as a neurosteroid receptor in brain. Glia 58:1794–1805 [DOI] [PubMed] [Google Scholar]
- Keitel V, Nies AT, Brom M, Hummel-Eisenbeiss J, Spring H, Keppler D. (2003) A common Dubin-Johnson syndrome mutation impairs protein maturation and transport activity of MRP2 (ABCC2). Am J Physiol Gastrointest Liver Physiol 284:G165–G174 [DOI] [PubMed] [Google Scholar]
- Keitel V, Reinehr R, Gatsios P, Rupprecht C, Görg B, Selbach O, Häussinger D, Kubitz R. (2007) The G-protein coupled bile salt receptor TGR5 is expressed in liver sinusoidal endothelial cells. Hepatology 45:695–704 [DOI] [PubMed] [Google Scholar]
- Kerr TA, Saeki S, Schneider M, Schaefer K, Berdy S, Redder T, Shan B, Russell DW, Schwarz M. (2002) Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev Cell 2:713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida T, Tsubosaka Y, Hori M, Ozaki H, Murata T. (2013) Bile acid receptor TGR5 agonism induces NO production and reduces monocyte adhesion in vascular endothelial cells. Arterioscler Thromb Vasc Biol 33:1663–1669 [DOI] [PubMed] [Google Scholar]
- Kir S, Beddow SA, Samuel VT, Miller P, Previs SF, Suino-Powell K, Xu HE, Shulman GI, Kliewer SA, Mangelsdorf DJ. (2011) FGF19 as a postprandial, insulin-independent activator of hepatic protein and glycogen synthesis. Science 331:1621–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterström RH, et al. (1998) An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 92:73–82 [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Willson TM. (2002) Regulation of xenobiotic and bile acid metabolism by the nuclear pregnane X receptor. J Lipid Res 43:359–364 [PubMed] [Google Scholar]
- Kodama S, Koike C, Negishi M, Yamamoto Y. (2004) Nuclear receptors CAR and PXR cross talk with FOXO1 to regulate genes that encode drug-metabolizing and gluconeogenic enzymes. Mol Cell Biol 24:7931–7940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok NN, Morgan LM, Williams CM, Roberfroid MB, Thissen JP, Delzenne NM. (1998) Insulin, glucagon-like peptide 1, glucose-dependent insulinotropic polypeptide and insulin-like growth factor I as putative mediators of the hypolipidemic effect of oligofructose in rats. J Nutr 128:1099–1103 [DOI] [PubMed] [Google Scholar]
- Kok T, Hulzebos CV, Wolters H, Havinga R, Agellon LB, Stellaard F, Shan B, Schwarz M, Kuipers F. (2003) Enterohepatic circulation of bile salts in farnesoid X receptor-deficient mice: efficient intestinal bile salt absorption in the absence of ileal bile acid-binding protein. J Biol Chem 278:41930–41937 [DOI] [PubMed] [Google Scholar]
- Kong B, Wang L, Chiang JY, Zhang Y, Klaassen CD, Guo GL. (2012) Mechanism of tissue-specific farnesoid X receptor in suppressing the expression of genes in bile-acid synthesis in mice. Hepatology 56:1034–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Košir R, Španinger K, Rozman D. (2013) Circadian events in human diseases and in cytochrome P450-related drug metabolism and therapy. IUBMB Life 65:487–496 [DOI] [PubMed] [Google Scholar]
- Kremer AE, van Dijk R, Leckie P, Schaap FG, Kuiper EM, Mettang T, Reiners KS, Raap U, van Buuren HR, van Erpecum KJ, et al. (2012) Serum autotaxin is increased in pruritus of cholestasis, but not of other origin, and responds to therapeutic interventions. Hepatology 56:1391–1400 [DOI] [PubMed] [Google Scholar]
- Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. (2005) Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438:685–689 [DOI] [PubMed] [Google Scholar]
- Krützfeldt J, Stoffel M. (2006) MicroRNAs: a new class of regulatory genes affecting metabolism. Cell Metab 4:9–12 [DOI] [PubMed] [Google Scholar]
- Kudo T, Tamagawa T, Shibata S. (2009) Effect of chronic ethanol exposure on the liver of Clock-mutant mice. J Circadian Rhythms 7:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdi P, Kawanishi K, Mizutani K, Yokota A. (2006) Mechanism of growth inhibition by free bile acids in lactobacilli and bifidobacteria. J Bacteriol 188:1979–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahtela JT, Särkkä P, Sotaniemi EA. (1984) Phenobarbital treatment enhances insulin mediated glucose metabolism in man. Res Commun Chem Pathol Pharmacol 44:215–226 [PubMed] [Google Scholar]
- Lambert G, Amar MJ, Guo G, Brewer HB, Jr, Gonzalez FJ, Sinal CJ. (2003) The farnesoid X-receptor is an essential regulator of cholesterol homeostasis. J Biol Chem 278:2563–2570 [DOI] [PubMed] [Google Scholar]
- Lang C, Meier Y, Stieger B, Beuers U, Lang T, Kerb R, Kullak-Ublick GA, Meier PJ, Pauli-Magnus C. (2007) Mutations and polymorphisms in the bile salt export pump and the multidrug resistance protein 3 associated with drug-induced liver injury. Pharmacogenet Genomics 17:47–60 [DOI] [PubMed] [Google Scholar]
- Lang T, Haberl M, Jung D, Drescher A, Schlagenhaufer R, Keil A, Mornhinweg E, Stieger B, Kullak-Ublick GA, Kerb R. (2006) Genetic variability, haplotype structures, and ethnic diversity of hepatic transporters MDR3 (ABCB4) and bile salt export pump (ABCB11). Drug Metab Dispos 34:1582–1599 [DOI] [PubMed] [Google Scholar]
- Langhi C, Le May C, Kourimate S, Caron S, Staels B, Krempf M, Costet P, Cariou B. (2008) Activation of the farnesoid X receptor represses PCSK9 expression in human hepatocytes. FEBS Lett 582:949–955 [DOI] [PubMed] [Google Scholar]
- Laposky AD, Bass J, Kohsaka A, Turek FW. (2008) Sleep and circadian rhythms: key components in the regulation of energy metabolism. FEBS Lett 582:142–151 [DOI] [PubMed] [Google Scholar]
- Lavery DJ, Schibler U. (1993) Circadian transcription of the cholesterol 7 alpha hydroxylase gene may involve the liver-enriched bZIP protein DBP. Genes Dev 7:1871–1884 [DOI] [PubMed] [Google Scholar]
- Le Martelot G, Claudel T, Gatfield D, Schaad O, Kornmann B, Lo Sasso G, Moschetta A, Schibler U. (2009) REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol 7:e1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Zhang Y, Lee FY, Nelson SF, Gonzalez FJ, Edwards PA. (2006) FXR regulates organic solute transporters alpha and beta in the adrenal gland, kidney, and intestine. J Lipid Res 47:201–214 [DOI] [PubMed] [Google Scholar]
- Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. (2005) Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 102:11070–11075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. (2006) Microbial ecology: human gut microbes associated with obesity. Nature 444:1022–1023 [DOI] [PubMed] [Google Scholar]
- Li F, Jiang C, Krausz KW, Li Y, Albert I, Hao H, Fabre KM, Mitchell JB, Patterson AD, Gonzalez FJ. (2013a) Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat Commun 4:2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Chiang JY. (2005) Mechanism of rifampicin and pregnane X receptor inhibition of human cholesterol 7 alpha-hydroxylase gene transcription. Am J Physiol Gastrointest Liver Physiol 288:G74–G84 [DOI] [PubMed] [Google Scholar]
- Li T, Chiang JY. (2013) Nuclear receptors in bile acid metabolism. Drug Metab Rev 45:145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Francl JM, Boehme S, Chiang JY. (2013b) Regulation of cholesterol and bile acid homeostasis by the cholesterol 7α-hydroxylase/steroid response element-binding protein 2/microRNA-33a axis in mice. Hepatology 58:1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Francl JM, Boehme S, Ochoa A, Zhang Y, Klaassen CD, Erickson SK, Chiang JY. (2012) Glucose and insulin induction of bile acid synthesis: mechanisms and implication in diabetes and obesity. J Biol Chem 287:1861–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Holmstrom SR, Kir S, Umetani M, Schmidt DR, Kliewer SA, Mangelsdorf DJ. (2011a) The G protein-coupled bile acid receptor, TGR5, stimulates gallbladder filling. Mol Endocrinol 25:1066–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Matozel M, Boehme S, Kong B, Nilsson LM, Guo G, Ellis E, Chiang JY. (2011b) Overexpression of cholesterol 7α-hydroxylase promotes hepatic bile acid synthesis and secretion and maintains cholesterol homeostasis. Hepatology 53:996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Owsley E, Matozel M, Hsu P, Novak CM, Chiang JY. (2010) Transgenic expression of cholesterol 7alpha-hydroxylase in the liver prevents high-fat diet-induced obesity and insulin resistance in mice. Hepatology 52:678–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YT, Swales KE, Thomas GJ, Warner TD, Bishop-Bailey D. (2007) Farnesoid x receptor ligands inhibit vascular smooth muscle cell inflammation and migration. Arterioscler Thromb Vasc Biol 27:2606–2611 [DOI] [PubMed] [Google Scholar]
- Lian F, Xing X, Yuan G, Schäfer C, Rauser S, Walch A, Röcken C, Ebeling M, Wright MB, Schmid RM, et al. (2011) Farnesoid X receptor protects human and murine gastric epithelial cells against inflammation-induced damage. Biochem J 438:315–323 [DOI] [PubMed] [Google Scholar]
- Lin BC, Wang M, Blackmore C, Desnoyers LR. (2007) Liver-specific activities of FGF19 require Klotho beta. J Biol Chem 282:27277–27284 [DOI] [PubMed] [Google Scholar]
- Liu Y, Binz J, Numerick MJ, Dennis S, Luo G, Desai B, MacKenzie KI, Mansfield TA, Kliewer SA, Goodwin B, et al. (2003) Hepatoprotection by the farnesoid X receptor agonist GW4064 in rat models of intra- and extrahepatic cholestasis. J Clin Invest 112:1678–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ. (2000) Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell 6:507–515 [DOI] [PubMed] [Google Scholar]
- Lundåsen T, Gälman C, Angelin B, Rudling M. (2006) Circulating intestinal fibroblast growth factor 19 has a pronounced diurnal variation and modulates hepatic bile acid synthesis in man. J Intern Med 260:530–536 [DOI] [PubMed] [Google Scholar]
- Ma K, Saha PK, Chan L, Moore DD. (2006) Farnesoid X receptor is essential for normal glucose homeostasis. J Clin Invest 116:1102–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K, Xiao R, Tseng HT, Shan L, Fu L, Moore DD. (2009) Circadian dysregulation disrupts bile acid homeostasis. PLoS ONE 4:e6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen D, Beaver M, Chang L, Bruckner-Kardoss E, Wostmann B. (1976) Analysis of bile acids in conventional and germfree rats. J Lipid Res 17:107–111 [PubMed] [Google Scholar]
- Makishima M, Lu TT, Xie W, Whitfield GK, Domoto H, Evans RM, Haussler MR, Mangelsdorf DJ. (2002) Vitamin D receptor as an intestinal bile acid sensor. Science 296:1313–1316 [DOI] [PubMed] [Google Scholar]
- Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, Hull MV, Lustig KD, Mangelsdorf DJ, Shan B. (1999) Identification of a nuclear receptor for bile acids. Science 284:1362–1365 [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Evans RM. (1995) The RXR heterodimers and orphan receptors. Cell 83:841–850 [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. (1995) The nuclear receptor superfamily: the second decade. Cell 83:835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Miyamoto Y, Nakamura T, Tamai Y, Okada H, Sugiyama E, Nakamura T, Itadani H, Tanaka K. (2002) Identification of membrane-type receptor for bile acids (M-BAR). Biochem Biophys Res Commun 298:714–719 [DOI] [PubMed] [Google Scholar]
- Maruyama T, Tanaka K, Suzuki J, Miyoshi H, Harada N, Nakamura T, Miyamoto Y, Kanatani A, Tamai Y. (2006) Targeted disruption of G protein-coupled bile acid receptor 1 (Gpbar1/M-Bar) in mice. J Endocrinol 191:197–205 [DOI] [PubMed] [Google Scholar]
- Masuyama H, Hiramatsu Y. (2012) Treatment with constitutive androstane receptor ligand during pregnancy prevents insulin resistance in offspring from high-fat diet-induced obese pregnant mice. Am J Physiol Endocrinol Metab 303:E293–E300 [DOI] [PubMed] [Google Scholar]
- Maxwell KN, Fisher EA, Breslow JL. (2005) Overexpression of PCSK9 accelerates the degradation of the LDLR in a post-endoplasmic reticulum compartment. Proc Natl Acad Sci USA 102:2069–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy TC, Li X, Sinal CJ. (2005) Vitamin D receptor-dependent regulation of colon multidrug resistance-associated protein 3 gene expression by bile acids. J Biol Chem 280:23232–23242 [DOI] [PubMed] [Google Scholar]
- McMahan RH, Wang XX, Cheng LL, Krisko T, Smith M, El Kasmi K, Pruzanski M, Adorini L, Golden-Mason L, Levi M, et al. (2013) Bile acid receptor activation modulates hepatic monocyte activity and improves nonalcoholic fatty liver disease. J Biol Chem 288:11761–11770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier PJ, Stieger B. (2002) Bile salt transporters. Annu Rev Physiol 64:635–661 [DOI] [PubMed] [Google Scholar]
- Mells JE, Anania FA. (2013) The role of gastrointestinal hormones in hepatic lipid metabolism. Semin Liver Dis 33:343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencarelli A, Renga B, Distrutti E, Fiorucci S. (2009) Antiatherosclerotic effect of farnesoid X receptor. Am J Physiol Heart Circ Physiol 296:H272–H281 [DOI] [PubMed] [Google Scholar]
- Miao J, Fang S, Bae Y, Kemper JK. (2006) Functional inhibitory cross-talk between constitutive androstane receptor and hepatic nuclear factor-4 in hepatic lipid/glucose metabolism is mediated by competition for binding to the DR1 motif and to the common coactivators, GRIP-1 and PGC-1alpha. J Biol Chem 281:14537–14546 [DOI] [PubMed] [Google Scholar]
- Miyake JH, Doung XD, Strauss W, Moore GL, Castellani LW, Curtiss LK, Taylor JM, Davis RA. (2001) Increased production of apolipoprotein B-containing lipoproteins in the absence of hyperlipidemia in transgenic mice expressing cholesterol 7alpha-hydroxylase. J Biol Chem 276:23304–23311 [DOI] [PubMed] [Google Scholar]
- Miyata M, Kudo G, Lee YH, Yang TJ, Gelboin HV, Fernandez-Salguero P, Kimura S, Gonzalez FJ. (1999) Targeted disruption of the microsomal epoxide hydrolase gene. Microsomal epoxide hydrolase is required for the carcinogenic activity of 7,12-dimethylbenz[a]anthracene. J Biol Chem 274:23963–23968 [DOI] [PubMed] [Google Scholar]
- Miyata M, Sakaida Y, Matsuzawa H, Yoshinari K, Yamazoe Y. (2011a) Fibroblast growth factor 19 treatment ameliorates disruption of hepatic lipid metabolism in farnesoid X receptor (Fxr)-null mice. Biol Pharm Bull 34:1885–1889 [DOI] [PubMed] [Google Scholar]
- Miyata M, Takamatsu Y, Kuribayashi H, Yamazoe Y. (2009) Administration of ampicillin elevates hepatic primary bile acid synthesis through suppression of ileal fibroblast growth factor 15 expression. J Pharmacol Exp Ther 331:1079–1085 [DOI] [PubMed] [Google Scholar]
- Miyata M, Yamakawa H, Hamatsu M, Kuribayashi H, Takamatsu Y, Yamazoe Y. (2011b) Enterobacteria modulate intestinal bile acid transport and homeostasis through apical sodium-dependent bile acid transporter (SLC10A2) expression. J Pharmacol Exp Ther 336:188–196 [DOI] [PubMed] [Google Scholar]
- Moore KJ, Rayner KJ, Suárez Y, Fernández-Hernando C. (2010) microRNAs and cholesterol metabolism. Trends Endocrinol Metab 21:699–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KJ, Rayner KJ, Suárez Y, Fernández-Hernando C. (2011) The role of microRNAs in cholesterol efflux and hepatic lipid metabolism. Annu Rev Nutr 31:49–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschetta A, Bookout AL, Mangelsdorf DJ. (2004) Prevention of cholesterol gallstone disease by FXR agonists in a mouse model. Nat Med 10:1352–1358 [DOI] [PubMed] [Google Scholar]
- Moustafa T, Fickert P, Magnes C, Guelly C, Thueringer A, Frank S, Kratky D, Sattler W, Reicher H, Sinner F, Gumhold J, Silbert D, Fauler G, Hofler G, Lass A, Zechner R, Trauner M. (2012) Alterations in lipid metabolism mediate inflammation, fibrosis, and proliferation in a mouse model of chronic cholestatic liver injury. Gastroenterology 142:140–151.e12 [DOI] [PubMed] [Google Scholar]
- Mudaliar S, Henry RR, Sanyal AJ, Morrow L, Marschall HU, Kipnes M, Adorini L, Sciacca CI, Clopton P, Castelloe E, Dillon P, Pruzanski M, Shapiro D. (2013) Efficacy and safety of the farnesoid X receptor agonist obeticholic acid in patients with type 2 diabetes and nonalcoholic fatty liver disease. Gastroenterology 145:574–582.e571 [DOI] [PubMed] [Google Scholar]
- Müllenbach R, Bennett A, Tetlow N, Patel N, Hamilton G, Cheng F, Chambers J, Howard R, Taylor-Robinson SD, Williamson C. (2005) ATP8B1 mutations in British cases with intrahepatic cholestasis of pregnancy. Gut 54:829–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Higashi Y, Matsunaga N, Koyanagi S, Ohdo S. (2008) Circadian clock-controlled intestinal expression of the multidrug-resistance gene mdr1a in mice. Gastroenterology 135:1636–1644.e1633 [DOI] [PubMed] [Google Scholar]
- Musiek ES, Fitzgerald GA. (2013) Molecular clocks in pharmacology. Handbook Exp Pharmacol 217:243–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musunuru K, Pirruccello JP, Do R, Peloso GM, Guiducci C, Sougnez C, Garimella KV, Fisher S, Abreu J, Barry AJ, et al. (2010) Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N Engl J Med 363:2220–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myant NB, Mitropoulos KA. (1977) Cholesterol 7 alpha-hydroxylase. J Lipid Res 18:135–153 [PubMed] [Google Scholar]
- Nagpal S, Na S, Rathnachalam R. (2005) Noncalcemic actions of vitamin D receptor ligands. Endocr Rev 26:662–687 [DOI] [PubMed] [Google Scholar]
- Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Näär AM. (2010) MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 328:1566–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. (2009) Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324:654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nauck MA. (2011) Incretin-based therapies for type 2 diabetes mellitus: properties, functions, and clinical implications. Am J Med 124 (Suppl):S3–S18 [DOI] [PubMed] [Google Scholar]
- Neimark E, Chen F, Li X, Shneider BL. (2004) Bile acid-induced negative feedback regulation of the human ileal bile acid transporter. Hepatology 40:149–156 [DOI] [PubMed] [Google Scholar]
- Ng R, Wu H, Xiao H, Chen X, Willenbring H, Steer CJ, Song G. (2014) Inhibition of microRNA-24 expression in liver prevents hepatic lipid accumulation and hyperlipidemia. Hepatology DOI: 10.1002/hep.27153 [published ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijmeijer RM, Gadaleta RM, van Mil SW, van Bodegraven AA, Crusius JB, Dijkstra G, Hommes DW, de Jong DJ, Stokkers PC, Verspaget HW, et al. Dutch Initiative on Crohn, Colitis (ICC) (2011) Farnesoid X receptor (FXR) activation and FXR genetic variation in inflammatory bowel disease. PLoS ONE 6:e23745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida S, Ozeki J, Makishima M. (2009) Modulation of bile acid metabolism by 1alpha-hydroxyvitamin D3 administration in mice. Drug Metab Dispos 37:2037–2044 [DOI] [PubMed] [Google Scholar]
- Noe J, Kullak-Ublick GA, Jochum W, Stieger B, Kerb R, Haberl M, Müllhaupt B, Meier PJ, Pauli-Magnus C. (2005) Impaired expression and function of the bile salt export pump due to three novel ABCB11 mutations in intrahepatic cholestasis. J Hepatol 43:536–543 [DOI] [PubMed] [Google Scholar]
- Noshiro M, Kawamoto T, Furukawa M, Fujimoto K, Yoshida Y, Sasabe E, Tsutsumi S, Hamada T, Honma S, Honma K, et al. (2004) Rhythmic expression of DEC1 and DEC2 in peripheral tissues: DEC2 is a potent suppressor for hepatic cytochrome P450s opposing DBP. Genes Cells 9:317–329 [DOI] [PubMed] [Google Scholar]
- Noshiro M, Nishimoto M, Okuda K. (1990) Rat liver cholesterol 7 alpha-hydroxylase. Pretranslational regulation for circadian rhythm. J Biol Chem 265:10036–10041 [PubMed] [Google Scholar]
- Noshiro M, Usui E, Kawamoto T, Kubo H, Fujimoto K, Furukawa M, Honma S, Makishima M, Honma K, Kato Y. (2007) Multiple mechanisms regulate circadian expression of the gene for cholesterol 7alpha-hydroxylase (Cyp7a), a key enzyme in hepatic bile acid biosynthesis. J Biol Rhythms 22:299–311 [DOI] [PubMed] [Google Scholar]
- Noverr MC, Huffnagle GB. (2004) Does the microbiota regulate immune responses outside the gut? Trends Microbiol 12:562–568 [DOI] [PubMed] [Google Scholar]
- Nwose OM, Jones MR. (2013) Atypical mechanism of glucose modulation by colesevelam in patients with type 2 diabetes. Clin Med Insights Endocrinol Diabetes 6:75–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura M, Nishida S, Ishizawa M, Sakurai K, Shimizu M, Matsuo S, Amano S, Uno S, Makishima M. (2009) Vitamin D3 modulates the expression of bile acid regulatory genes and represses inflammation in bile duct-ligated mice. J Pharmacol Exp Ther 328:564–570 [DOI] [PubMed] [Google Scholar]
- Painter JN, Savander M, Ropponen A, Nupponen N, Riikonen S, Ylikorkala O, Lehesjoki AE, Aittomäki K. (2005) Sequence variation in the ATP8B1 gene and intrahepatic cholestasis of pregnancy. Eur J Hum Genet 13:435–439 [DOI] [PubMed] [Google Scholar]
- Pan X, Hussain MM. (2007) Diurnal regulation of microsomal triglyceride transfer protein and plasma lipid levels. J Biol Chem 282:24707–24719 [DOI] [PubMed] [Google Scholar]
- Pan X, Zhang Y, Wang L, Hussain MM. (2010) Diurnal regulation of MTP and plasma triglyceride by CLOCK is mediated by SHP. Cell Metab 12:174–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandak WM, Heuman DM, Hylemon PB, Chiang JY, Vlahcevic ZR. (1995) Failure of intravenous infusion of taurocholate to down-regulate cholesterol 7 α-hydroxylase in rats with biliary fistulas. Gastroenterology 108:533–544 [DOI] [PubMed] [Google Scholar]
- Parks BW, Nam E, Org E, Kostem E, Norheim F, Hui ST, Pan C, Civelek M, Rau CD, Bennett BJ, et al. (2013) Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab 17:141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, Willson TM, Zavacki AM, Moore DD, et al. (1999) Bile acids: natural ligands for an orphan nuclear receptor. Science 284:1365–1368 [DOI] [PubMed] [Google Scholar]
- Pathak P, Li T, Chiang JY. (2013) Retinoic acid-related orphan receptor α regulates diurnal rhythm and fasting induction of sterol 12α-hydroxylase in bile acid synthesis. J Biol Chem 288:37154–37165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti ME, Houten SM, Bianco AC, Bernier R, Larsen PR, Holst JJ, Badman MK, Maratos-Flier E, Mun EC, Pihlajamaki J, et al. (2009) Serum bile acids are higher in humans with prior gastric bypass: potential contribution to improved glucose and lipid metabolism. Obesity (Silver Spring) 17:1671–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli-Magnus C, Lang T, Meier Y, Zodan-Marin T, Jung D, Breymann C, Zimmermann R, Kenngott S, Beuers U, Reichel C, et al. (2004) Sequence analysis of bile salt export pump (ABCB11) and multidrug resistance p-glycoprotein 3 (ABCB4, MDR3) in patients with intrahepatic cholestasis of pregnancy. Pharmacogenetics 14:91–102 [DOI] [PubMed] [Google Scholar]
- Péan N, Doignon I, Garcin I, Besnard A, Julien B, Liu B, Branchereau S, Spraul A, Guettier C, Humbert L, et al. (2013) The receptor TGR5 protects the liver from bile acid overload during liver regeneration in mice. Hepatology 58:1451–1460 [DOI] [PubMed] [Google Scholar]
- Pellicciari R, Costantino G, Camaioni E, Sadeghpour BM, Entrena A, Willson TM, Fiorucci S, Clerici C, Gioiello A. (2004) Bile acid derivatives as ligands of the farnesoid X receptor. Synthesis, evaluation, and structure-activity relationship of a series of body and side chain modified analogues of chenodeoxycholic acid. J Med Chem 47:4559–4569 [DOI] [PubMed] [Google Scholar]
- Pellicciari R, Gioiello A, Macchiarulo A, Thomas C, Rosatelli E, Natalini B, Sardella R, Pruzanski M, Roda A, Pastorini E, et al. (2009) Discovery of 6alpha-ethyl-23(S)-methylcholic acid (S-EMCA, INT-777) as a potent and selective agonist for the TGR5 receptor, a novel target for diabesity. J Med Chem 52:7958–7961 [DOI] [PubMed] [Google Scholar]
- Pellicciari R, Sato H, Gioiello A, Costantino G, Macchiarulo A, Sadeghpour BM, Giorgi G, Schoonjans K, Auwerx J. (2007) Nongenomic actions of bile acids. Synthesis and preliminary characterization of 23- and 6,23-alkyl-substituted bile acid derivatives as selective modulators for the G-protein coupled receptor TGR5. J Med Chem 50:4265–4268 [DOI] [PubMed] [Google Scholar]
- Pineda Torra I, Claudel T, Duval C, Kosykh V, Fruchart JC, Staels B. (2003) Bile acids induce the expression of the human peroxisome proliferator-activated receptor alpha gene via activation of the farnesoid X receptor. Mol Endocrinol 17:259–272 [DOI] [PubMed] [Google Scholar]
- Podvinec M, Handschin C, Looser R, Meyer UA. (2004) Identification of the xenosensors regulating human 5-aminolevulinate synthase. Proc Natl Acad Sci USA 101:9127–9132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pols TW, Nomura M, Harach T, Lo Sasso G, Oosterveer MH, Thomas C, Rizzo G, Gioiello A, Adorini L, Pellicciari R, et al. (2011a) TGR5 activation inhibits atherosclerosis by reducing macrophage inflammation and lipid loading. Cell Metab 14:747–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pols TW, Noriega LG, Nomura M, Auwerx J, Schoonjans K. (2011b) The bile acid membrane receptor TGR5 as an emerging target in metabolism and inflammation. J Hepatol 54:1263–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu IR, Helleboid-Chapman A, Lucas A, Vandewalle B, Dumont J, Bouchaert E, Derudas B, Kerr-Conte J, Caron S, Pattou F, et al. (2010) The nuclear receptor FXR is expressed in pancreatic beta-cells and protects human islets from lipotoxicity. FEBS Lett 584:2845–2851 [DOI] [PubMed] [Google Scholar]
- Porez G, Prawitt J, Gross B, Staels B. (2012) Bile acid receptors as targets for the treatment of dyslipidemia and cardiovascular disease. J Lipid Res 53:1723–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postic C, Dentin R, Denechaud PD, Girard J. (2007) ChREBP, a transcriptional regulator of glucose and lipid metabolism. Annu Rev Nutr 27:179–192 [DOI] [PubMed] [Google Scholar]
- Potthoff MJ, Boney-Montoya J, Choi M, He T, Sunny NE, Satapati S, Suino-Powell K, Xu HE, Gerard RD, Finck BN, et al. (2011) FGF15/19 regulates hepatic glucose metabolism by inhibiting the CREB-PGC-1α pathway. Cell Metab 13:729–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potthoff MJ, Potts A, He T, Duarte JA, Taussig R, Mangelsdorf DJ, Kliewer SA, Burgess SC. (2013) Colesevelam suppresses hepatic glycogenolysis by TGR5-mediated induction of GLP-1 action in DIO mice. Am J Physiol Gastrointest Liver Physiol 304:G371–G380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pournaras DJ, Glicksman C, Vincent RP, Kuganolipava S, Alaghband-Zadeh J, Mahon D, Bekker JH, Ghatei MA, Bloom SR, Walters JR, et al. (2012) The role of bile after Roux-en-Y gastric bypass in promoting weight loss and improving glycaemic control. Endocrinology 153:3613–3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prawitt J, Abdelkarim M, Stroeve JH, Popescu I, Duez H, Velagapudi VR, Dumont J, Bouchaert E, van Dijk TH, Lucas A, et al. (2011) Farnesoid X receptor deficiency improves glucose homeostasis in mouse models of obesity. Diabetes 60:1861–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prawitt J, Caron S, Staels B. (2014) Glucose-lowering effects of intestinal bile acid sequestration through enhancement of splanchnic glucose utilization. Trends Endocrinol Metab 25:235–244 [DOI] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. (2002) The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110:251–260 [DOI] [PubMed] [Google Scholar]
- Prieur X, Coste H, Rodriguez JC. (2003) The human apolipoprotein AV gene is regulated by peroxisome proliferator-activated receptor-alpha and contains a novel farnesoid X-activated receptor response element. J Biol Chem 278:25468–25480 [DOI] [PubMed] [Google Scholar]
- Pruneta-Deloche V, Sassolas A, Dallinga-Thie GM, Berthezène F, Ponsin G, Moulin P. (2004) Alteration in lipoprotein lipase activity bound to triglyceride-rich lipoproteins in the postprandial state in type 2 diabetes. J Lipid Res 45:859–865 [DOI] [PubMed] [Google Scholar]
- Qi Y, Jiang C, Cheng J, Krausz KW, Li T, Ferrell JM, Gonzalez FJ, Chiang JY. (2014) Bile acid signaling in lipid metabolism: Metabolomic and lipidomic analysis of lipid and bile acid markers linked to anti-obesity and anti-diabetes in mice. Biochim Biophys Acta DOI: 10.1016/j.bbalip.2014.04.008 [published ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, et al. (2012) A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490:55–60 [DOI] [PubMed] [Google Scholar]
- Radziuk J, Bailey CJ, Wiernsperger NF, Yudkin JS. (2003) Metformin and its liver targets in the treatment of type 2 diabetes. Curr Drug Targets Immune Endocr Metabol Disord 3:151–169 [DOI] [PubMed] [Google Scholar]
- Ramírez CM, Goedeke L, Rotllan N, Yoon JH, Cirera-Salinas D, Mattison JA, Suárez Y, de Cabo R, Gorospe M, Fernández-Hernando C. (2013a) MicroRNA 33 regulates glucose metabolism. Mol Cell Biol 33:2891–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez CM, Rotllan N, Vlassov AV, Dávalos A, Li M, Goedeke L, Aranda JF, Cirera-Salinas D, Araldi E, Salerno A, et al. (2013b) Control of cholesterol metabolism and plasma high-density lipoprotein levels by microRNA-144. Circ Res 112:1592–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A, Haywood J, Craddock AL, Belinsky MG, Kruh GD, Dawson PA. (2008) The organic solute transporter alpha-beta, Ostalpha-Ostbeta, is essential for intestinal bile acid transport and homeostasis. Proc Natl Acad Sci USA 105:3891–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner KJ, Sheedy FJ, Esau CC, Hussain FN, Temel RE, Parathath S, van Gils JM, Rayner AJ, Chang AN, Suarez Y, et al. (2011) Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest 121:2921–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaven GM. (1988) Banting lecture 1988. Role of insulin resistance in human disease. Diabetes 37:1595–1607 [DOI] [PubMed] [Google Scholar]
- Reimer RA, McBurney MI. (1996) Dietary fiber modulates intestinal proglucagon messenger ribonucleic acid and postprandial secretion of glucagon-like peptide-1 and insulin in rats. Endocrinology 137:3948–3956 [DOI] [PubMed] [Google Scholar]
- Renga B, Mencarelli A, Vavassori P, Brancaleone V, Fiorucci S. (2010) The bile acid sensor FXR regulates insulin transcription and secretion. Biochim Biophys Acta 1802:363–372 [DOI] [PubMed] [Google Scholar]
- Repa JJ, Berge KE, Pomajzl C, Richardson JA, Hobbs H, Mangelsdorf DJ. (2002) Regulation of ATP-binding cassette sterol transporters ABCG5 and ABCG8 by the liver X receptors alpha and beta. J Biol Chem 277:18793–18800 [DOI] [PubMed] [Google Scholar]
- Repa JJ, Liang G, Ou J, Bashmakov Y, Lobaccaro JM, Shimomura I, Shan B, Brown MS, Goldstein JL, Mangelsdorf DJ. (2000) Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev 14:2819–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon JM, Alves JM, Hylemon PB, Bajaj JS. (2013) Cirrhosis, bile acids and gut microbiota: unraveling a complex relationship. Gut Microbes 4:382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon JM, Kang DJ, Hylemon PB. (2006) Bile salt biotransformations by human intestinal bacteria. J Lipid Res 47:241–259 [DOI] [PubMed] [Google Scholar]
- Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. (2014) Bile acids and the gut microbiome. Curr Opin Gastroenterol 30:332–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigotti A, Trigatti BL, Penman M, Rayburn H, Herz J, Krieger M. (1997) A targeted mutation in the murine gene encoding the high density lipoprotein (HDL) receptor scavenger receptor class B type I reveals its key role in HDL metabolism. Proc Natl Acad Sci USA 94:12610–12615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo G, Disante M, Mencarelli A, Renga B, Gioiello A, Pellicciari R, Fiorucci S. (2006) The farnesoid X receptor promotes adipocyte differentiation and regulates adipose cell function in vivo. Mol Pharmacol 70:1164–1173 [DOI] [PubMed] [Google Scholar]
- Rottiers V, Näär AM. (2012) MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol 13:239–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. (2004) BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol 2:e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DW. (2003) The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem 72:137–174 [DOI] [PubMed] [Google Scholar]
- Rutter J, Reick M, McKnight SL. (2002) Metabolism and the control of circadian rhythms. Annu Rev Biochem 71:307–331 [DOI] [PubMed] [Google Scholar]
- Ryan KK, Tremaroli V, Clemmensen C, Kovatcheva-Datchary P, Myronovych A, Karns R, Wilson-Pérez HE, Sandoval DA, Kohli R, Bäckhed F, et al. (2014) FXR is a molecular target for the effects of vertical sleeve gastrectomy. Nature 509:183–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco J, Adeli K. (2012) MicroRNAs: emerging roles in lipid and lipoprotein metabolism. Curr Opin Lipidol 23:220–225 [DOI] [PubMed] [Google Scholar]
- Saini SP, Sonoda J, Xu L, Toma D, Uppal H, Mu Y, Ren S, Moore DD, Evans RM, Xie W. (2004) A novel constitutive androstane receptor-mediated and CYP3A-independent pathway of bile acid detoxification. Mol Pharmacol 65:292–300 [DOI] [PubMed] [Google Scholar]
- Sato H, Macchiarulo A, Thomas C, Gioiello A, Une M, Hofmann AF, Saladin R, Schoonjans K, Pellicciari R, Auwerx J. (2008) Novel potent and selective bile acid derivatives as TGR5 agonists: biological screening, structure-activity relationships, and molecular modeling studies. J Med Chem 51:1831–1841 [DOI] [PubMed] [Google Scholar]
- Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall HU, Bamberg K, Angelin B, Hyötyläinen T, Orešič M, Bäckhed F. (2013) Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab 17:225–235 [DOI] [PubMed] [Google Scholar]
- Sberna AL, Assem M, Gautier T, Grober J, Guiu B, Jeannin A, Pais de Barros JP, Athias A, Lagrost L, Masson D. (2011) Constitutive androstane receptor activation stimulates faecal bile acid excretion and reverse cholesterol transport in mice. J Hepatol 55:154–161 [DOI] [PubMed] [Google Scholar]
- Schaap FG, Trauner M, Jansen PL. (2014) Bile acid receptors as targets for drug development. Nat Rev Gastroenterol Hepatol 11:55–67 [DOI] [PubMed] [Google Scholar]
- Schaap FG, van der Gaag NA, Gouma DJ, Jansen PL. (2009) High expression of the bile salt-homeostatic hormone fibroblast growth factor 19 in the liver of patients with extrahepatic cholestasis. Hepatology 49:1228–1235 [DOI] [PubMed] [Google Scholar]
- Schmidt DR, Holmstrom SR, Fon Tacer K, Bookout AL, Kliewer SA, Mangelsdorf DJ. (2010) Regulation of bile acid synthesis by fat-soluble vitamins A and D. J Biol Chem 285:14486–14494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiedlin-Ren P, Thummel KE, Fisher JM, Paine MF, Watkins PB. (2001) Induction of CYP3A4 by 1 alpha,25-dihydroxyvitamin D3 is human cell line-specific and is unlikely to involve pregnane X receptor. Drug Metab Dispos 29:1446–1453 [PubMed] [Google Scholar]
- Schoenfield LJ, Lachin JM. (1981) Chenodiol (chenodeoxycholic acid) for dissolution of gallstones: the National Cooperative Gallstone Study. A controlled trial of efficacy and safety. Ann Intern Med 95:257–282 [DOI] [PubMed] [Google Scholar]
- Schreuder TC, Marsman HA, Lenicek M, van Werven JR, Nederveen AJ, Jansen PL, Schaap FG. (2010) The hepatic response to FGF19 is impaired in patients with nonalcoholic fatty liver disease and insulin resistance. Am J Physiol Gastrointest Liver Physiol 298:G440–G445 [DOI] [PubMed] [Google Scholar]
- Seggio JA, Fixaris MC, Reed JD, Logan RW, Rosenwasser AM. (2009) Chronic ethanol intake alters circadian phase shifting and free-running period in mice. J Biol Rhythms 24:304–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Q, Saumoy M, Holst JJ, Salen G, Xu G. (2010) Colesevelam improves insulin resistance in a diet-induced obesity (F-DIO) rat model by increasing the release of GLP-1. Am J Physiol Gastrointest Liver Physiol 298:G419–G424 [DOI] [PubMed] [Google Scholar]
- Shao W, Espenshade PJ. (2012) Expanding roles for SREBP in metabolism. Cell Metab 16:414–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi SQ, Ansari TS, McGuinness OP, Wasserman DH, Johnson CH. (2013) Circadian disruption leads to insulin resistance and obesity. Curr Biol 23:372-381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shneider BL, Dawson PA, Christie DM, Hardikar W, Wong MH, Suchy FJ. (1995) Cloning and molecular characterization of the ontogeny of a rat ileal sodium-dependent bile acid transporter. J Clin Invest 95:745–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonen M, Dali-Youcef N, Kaminska D, Venesmaa S, Käkelä P, Pääkkönen M, Hallikainen M, Kolehmainen M, Uusitupa M, Moilanen L, et al. (2012) Conjugated bile acids associate with altered rates of glucose and lipid oxidation after Roux-en-Y gastric bypass. Obes Surg 22:1473–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. (2000) Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 102:731–744 [DOI] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. (2009) Autophagy regulates lipid metabolism. Nature 458:1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirvent A, Claudel T, Martin G, Brozek J, Kosykh V, Darteil R, Hum DW, Fruchart JC, Staels B. (2004) The farnesoid X receptor induces very low density lipoprotein receptor gene expression. FEBS Lett 566:173–177 [DOI] [PubMed] [Google Scholar]
- Smit JJ, Schinkel AH, Oude Elferink RP, Groen AK, Wagenaar E, van Deemter L, Mol CA, Ottenhoff R, van der Lugt NM, van Roon MA, et al. (1993) Homozygous disruption of the murine mdr2 P-glycoprotein gene leads to a complete absence of phospholipid from bile and to liver disease. Cell 75:451–462 [DOI] [PubMed] [Google Scholar]
- Solá S, Amaral JD, Aranha MM, Steer CJ, Rodrigues CM. (2006) Modulation of hepatocyte apoptosis: cross-talk between bile acids and nuclear steroid receptors. Curr Med Chem 13:3039–3051 [DOI] [PubMed] [Google Scholar]
- Song KH, Li T, Owsley E, Chiang JY. (2010) A putative role of micro RNA in regulation of cholesterol 7α-hydroxylase expression in human hepatocytes. J Lipid Res 51:2223–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song KH, Li T, Owsley E, Strom S, Chiang JY. (2009) Bile acids activate fibroblast growth factor 19 signaling in human hepatocytes to inhibit cholesterol 7alpha-hydroxylase gene expression. Hepatology 49:297–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sookoian S, Castaño G, Gemma C, Gianotti TF, Pirola CJ. (2007) Common genetic variations in CLOCK transcription factor are associated with nonalcoholic fatty liver disease. World J Gastroenterol 13:4242–4248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sookoian S, Rosselli MS, Gemma C, Burgueño AL, Fernández Gianotti T, Castaño GO, Pirola CJ. (2010) Epigenetic regulation of insulin resistance in nonalcoholic fatty liver disease: impact of liver methylation of the peroxisome proliferator-activated receptor γ coactivator 1α promoter. Hepatology 52:1992–2000 [DOI] [PubMed] [Google Scholar]
- Soroka CJ, Mennone A, Hagey LR, Ballatori N, Boyer JL. (2010) Mouse organic solute transporter alpha deficiency enhances renal excretion of bile acids and attenuates cholestasis. Hepatology 51:181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ståhlberg D. (1995) Effects of pregnenolone-16 alpha-carbonitrile on the metabolism of cholesterol in rat liver microsomes. Lipids 30:361–364 [DOI] [PubMed] [Google Scholar]
- Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, Liu Y, Klaassen CD, Brown KK, Reinhard J, et al. (2001) The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci USA 98:3369–3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stayrook KR, Bramlett KS, Savkur RS, Ficorilli J, Cook T, Christe ME, Michael LF, Burris TP. (2005) Regulation of carbohydrate metabolism by the farnesoid X receptor. Endocrinology 146:984–991 [DOI] [PubMed] [Google Scholar]
- Stearns AT, Balakrishnan A, Rhoads DB, Ashley SW, Tavakkolizadeh A. (2008) Diurnal rhythmicity in the transcription of jejunal drug transporters. J Pharmacol Sci 108:144–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman CA, Liddle C, Coulter SA, Sonoda J, Alvarez JG, Moore DD, Evans RM, Downes M. (2005) Nuclear receptors constitutive androstane receptor and pregnane X receptor ameliorate cholestatic liver injury. Proc Natl Acad Sci USA 102:2063–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanov V, Stankov K, Mikov M. (2013) The bile acid membrane receptor TGR5: a novel pharmacological target in metabolic, inflammatory and neoplastic disorders. J Recept Signal Transduct Res 33:213–223 [DOI] [PubMed] [Google Scholar]
- Stojancevic M, Stankov K, Mikov M. (2012) The impact of farnesoid X receptor activation on intestinal permeability in inflammatory bowel disease. Can J Gastroenterol 26:631–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strautnieks SS, Kagalwalla AF, Tanner MS, Knisely AS, Bull L, Freimer N, Kocoshis SA, Gardiner RM, Thompson RJ. (1997) Identification of a locus for progressive familial intrahepatic cholestasis PFIC2 on chromosome 2q24. Am J Hum Genet 61:630–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroeve JH, Brufau G, Stellaard F, Gonzalez FJ, Staels B, Kuipers F. (2010) Intestinal FXR-mediated FGF15 production contributes to diurnal control of hepatic bile acid synthesis in mice. Lab Invest 90:1457–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer E, Zhou X, Zhao R, Wang Y, Takabe K, Nagahashi M, Pandak WM, Dent P, Spiegel S, Shi R, et al. (2012) Conjugated bile acids activate the sphingosine-1-phosphate receptor 2 in primary rodent hepatocytes. Hepatology 55:267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui Y, Xu J, Rios-Pilier J, Zhou C. (2011) Deficiency of PXR decreases atherosclerosis in apoE-deficient mice. J Lipid Res 52:1652–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swann JR, Want EJ, Geier FM, Spagou K, Wilson ID, Sidaway JE, Nicholson JK, Holmes E. (2011) Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc Natl Acad Sci USA 108 (Suppl 1):4523–4530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson HI, Wada T, Xie W, Renga B, Zampella A, Distrutti E, Fiorucci S, Kong B, Thomas AM, Guo GL, et al. (2013) Role of nuclear receptors in lipid dysfunction and obesity-related diseases. Drug Metab Dispos 41:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi S, Nakajima M, Kida K, Yamaura Y, Fukami T, Yokoi T. (2010) MicroRNAs regulate human hepatocyte nuclear factor 4alpha, modulating the expression of metabolic enzymes and cell cycle. J Biol Chem 285:4415–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, et al. (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466:707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, Macchiarulo A, Yamamoto H, Mataki C, Pruzanski M, et al. (2009) TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab 10:167–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. (2008) Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov 7:678–693 [DOI] [PubMed] [Google Scholar]
- Timms KM, Wagner S, Samuels ME, Forbey K, Goldfine H, Jammulapati S, Skolnick MH, Hopkins PN, Hunt SC, Shattuck DM. (2004) A mutation in PCSK9 causing autosomal-dominant hypercholesterolemia in a Utah pedigree. Hum Genet 114:349–353 [DOI] [PubMed] [Google Scholar]
- Tiniakos DG, Vos MB, Brunt EM. (2010) Nonalcoholic fatty liver disease: pathology and pathogenesis. Annu Rev Pathol 5:145–171 [DOI] [PubMed] [Google Scholar]
- Tomlinson E, Fu L, John L, Hultgren B, Huang X, Renz M, Stephan JP, Tsai SP, Powell-Braxton L, French D, et al. (2002) Transgenic mice expressing human fibroblast growth factor-19 display increased metabolic rate and decreased adiposity. Endocrinology 143:1741–1747 [DOI] [PubMed] [Google Scholar]
- Tong X, Yin L. (2013) Circadian rhythms in liver physiology and liver diseases. Compr Physiol 3:917–940 [DOI] [PubMed] [Google Scholar]
- Topping DL, Clifton PM. (2001) Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev 81:1031–1064 [DOI] [PubMed] [Google Scholar]
- Trauner M, Arrese M, Wagner M. (2010) Fatty liver and lipotoxicity. Biochim Biophys Acta 1801:299–310 [DOI] [PubMed] [Google Scholar]
- Trauner M, Boyer JL. (2003) Bile salt transporters: molecular characterization, function, and regulation. Physiol Rev 83:633–671 [DOI] [PubMed] [Google Scholar]
- Trauner M, Halilbasic E. (2011) Nuclear receptors as new perspective for the management of liver diseases. Gastroenterology 140:1120–1125e1-12 [DOI] [PubMed] [Google Scholar]
- Trauner M, Meier PJ, Boyer JL. (1998) Molecular pathogenesis of cholestasis. N Engl J Med 339:1217–1227 [DOI] [PubMed] [Google Scholar]
- Trigatti BL, Krieger M, Rigotti A. (2003) Influence of the HDL receptor SR-BI on lipoprotein metabolism and atherosclerosis. Arterioscler Thromb Vasc Biol 23:1732–1738 [DOI] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. (2005) Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308:1043–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027–1031 [DOI] [PubMed] [Google Scholar]
- Uyeda K, Yamashita H, Kawaguchi T. (2002) Carbohydrate responsive element-binding protein (ChREBP): a key regulator of glucose metabolism and fat storage. Biochem Pharmacol 63:2075–2080 [DOI] [PubMed] [Google Scholar]
- van der Velde AE, Brufau G, Groen AK. (2010) Transintestinal cholesterol efflux. Curr Opin Lipidol 21:167–171 [DOI] [PubMed] [Google Scholar]
- Van Mil SW, Milona A, Dixon PH, Mullenbach R, Geenes VL, Chambers J, Shevchuk V, Moore GE, Lammert F, Glantz AG, et al. (2007) Functional variants of the central bile acid sensor FXR identified in intrahepatic cholestasis of pregnancy. Gastroenterology 133:507–516 [DOI] [PubMed] [Google Scholar]
- Vassileva G, Golovko A, Markowitz L, Abbondanzo SJ, Zeng M, Yang S, Hoos L, Tetzloff G, Levitan D, Murgolo NJ, et al. (2006) Targeted deletion of Gpbar1 protects mice from cholesterol gallstone formation. Biochem J 398:423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassileva G, Hu W, Hoos L, Tetzloff G, Yang S, Liu L, Kang L, Davis HR, Hedrick JA, Lan H, et al. (2010) Gender-dependent effect of Gpbar1 genetic deletion on the metabolic profiles of diet-induced obese mice. J Endocrinol 205:225–232 [DOI] [PubMed] [Google Scholar]
- Vavassori P, Mencarelli A, Renga B, Distrutti E, Fiorucci S. (2009) The bile acid receptor FXR is a modulator of intestinal innate immunity. J Immunol 183:6251–6261 [DOI] [PubMed] [Google Scholar]
- Vergeer M, Hovingh K, Vissers MN, Kastelein JJ, Kuivenhoven JA. (2006) Heterozygosity for a mutation in the extracellular domain of SR-BI is associated with high HDL cholesterol levels in a family of Caucasian descent. Circulation 114:II_254 [Google Scholar]
- Vergeer M, Korporaal SJ, Franssen R, Meurs I, Out R, Hovingh GK, Hoekstra M, Sierts JA, Dallinga-Thie GM, Motazacker MM, et al. (2011) Genetic variant of the scavenger receptor BI in humans. N Engl J Med 364:136–145 [DOI] [PubMed] [Google Scholar]
- Vickers KC, Shoucri BM, Levin MG, Wu H, Pearson DS, Osei-Hwedieh D, Collins FS, Remaley AT, Sethupathy P. (2013) MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology 57:533–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S. (2009) Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci USA 106:21453–21458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dippe P, Amoui M, Alves C, Levy D. (1993) Na(+)-dependent bile acid transport by hepatocytes is mediated by a protein similar to microsomal epoxide hydrolase. Am J Physiol 264:G528–G534 [DOI] [PubMed] [Google Scholar]
- von Dippe P, Amoui M, Stellwagen RH, Levy D. (1996) The functional expression of sodium-dependent bile acid transport in Madin-Darby canine kidney cells transfected with the cDNA for microsomal epoxide hydrolase. J Biol Chem 271:18176–18180 [DOI] [PubMed] [Google Scholar]
- Wagner M, Fickert P, Zollner G, Fuchsbichler A, Silbert D, Tsybrovskyy O, Zatloukal K, Guo GL, Schuetz JD, Gonzalez FJ, et al. (2003) Role of farnesoid X receptor in determining hepatic ABC transporter expression and liver injury in bile duct-ligated mice. Gastroenterology 125:825–838 [DOI] [PubMed] [Google Scholar]
- Wagner M, Zollner G, Trauner M. (2009) New molecular insights into the mechanisms of cholestasis. J Hepatol 51:565–580 [DOI] [PubMed] [Google Scholar]
- Wagner M, Zollner G, Trauner M. (2011) Nuclear receptors in liver disease. Hepatology 53:1023–1034 [DOI] [PubMed] [Google Scholar]
- Wang H, Venkatesh M, Li H, Goetz R, Mukherjee S, Biswas A, Zhu L, Kaubisch A, Wang L, Pullman J, et al. (2011a) Pregnane X receptor activation induces FGF19-dependent tumor aggressiveness in humans and mice. J Clin Invest 121:3220–3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gåfvels M, Rudling M, Murphy C, Björkhem I, Einarsson C, Eggertsen G. (2006) Critical role of cholic acid for development of hypercholesterolemia and gallstones in diabetic mice. Biochem Biophys Res Commun 342:1382–1388 [DOI] [PubMed] [Google Scholar]
- Wang L, Lee YK, Bundman D, Han Y, Thevananther S, Kim CS, Chua SS, Wei P, Heyman RA, Karin M, et al. (2002) Redundant pathways for negative feedback regulation of bile acid production. Dev Cell 2:721–731 [DOI] [PubMed] [Google Scholar]
- Wang X, Sato R, Brown MS, Hua X, Goldstein JL. (1994) SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell 77:53–62 [DOI] [PubMed] [Google Scholar]
- Wang YD, Chen WD, Wang M, Yu D, Forman BM, Huang W. (2008) Farnesoid X receptor antagonizes nuclear factor kappaB in hepatic inflammatory response. Hepatology 48:1632–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YD, Chen WD, Yu D, Forman BM, Huang W. (2011b) The G-protein-coupled bile acid receptor, Gpbar1 (TGR5), negatively regulates hepatic inflammatory response through antagonizing nuclear factor κ light-chain enhancer of activated B cells (NF-κB) in mice. Hepatology 54:1421–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth HE, Glantz A, Keppeler H, Simon E, Bartz C, Rath W, Mattsson LA, Marschall HU, Lammert F. (2007) Intrahepatic cholestasis of pregnancy: the severe form is associated with common variants of the hepatobiliary phospholipid transporter ABCB4 gene. Gut 56:265–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Horai Y, Houten SM, Morimoto K, Sugizaki T, Arita E, Mataki C, Sato H, Tanigawara Y, Schoonjans K, et al. (2011) Lowering bile acid pool size with a synthetic farnesoid X receptor (FXR) agonist induces obesity and diabetes through reduced energy expenditure. J Biol Chem 286:26913–26920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, Messaddeq N, Harney JW, Ezaki O, Kodama T, et al. (2006) Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature 439:484–489 [DOI] [PubMed] [Google Scholar]
- Watanabe M, Houten SM, Wang L, Moschetta A, Mangelsdorf DJ, Heyman RA, Moore DD, Auwerx J. (2004) Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J Clin Invest 113:1408–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wistuba W, Gnewuch C, Liebisch G, Schmitz G, Langmann T. (2007) Lithocholic acid induction of the FGF19 promoter in intestinal cells is mediated by PXR. World J Gastroenterol 13:4230–4235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenburg H, Lyons MA, Li R, Churchill GA, Carey MC, Paigen B. (2003) FXR and ABCG5/ABCG8 as determinants of cholesterol gallstone formation from quantitative trait locus mapping in mice. Gastroenterology 125:868–881 [DOI] [PubMed] [Google Scholar]
- Wojcik M, Janus D, Dolezal-Oltarzewska K, Kalicka-Kasperczyk A, Poplawska K, Drozdz D, Sztefko K, Starzyk JB. (2012) A decrease in fasting FGF19 levels is associated with the development of non-alcoholic fatty liver disease in obese adolescents. J Pediatr Endocrinol Metab 25:1089–1093 [DOI] [PubMed] [Google Scholar]
- Wong MH, Oelkers P, Craddock AL, Dawson PA. (1994) Expression cloning and characterization of the hamster ileal sodium-dependent bile acid transporter. J Biol Chem 269:1340–1347 [PubMed] [Google Scholar]
- Wostmann BS. (1973) Intestinal bile acids and cholesterol absorption in the germfree rat. J Nutr 103:982–990 [DOI] [PubMed] [Google Scholar]
- Wuarin J, Schibler U. (1990) Expression of the liver-enriched transcriptional activator protein DBP follows a stringent circadian rhythm. Cell 63:1257–1266 [DOI] [PubMed] [Google Scholar]
- Xie W, Radominska-Pandya A, Shi Y, Simon CM, Nelson MC, Ong ES, Waxman DJ, Evans RM. (2001) An essential role for nuclear receptors SXR/PXR in detoxification of cholestatic bile acids. Proc Natl Acad Sci USA 98:3375–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata K, Daitoku H, Shimamoto Y, Matsuzaki H, Hirota K, Ishida J, Fukamizu A. (2004) Bile acids regulate gluconeogenic gene expression via small heterodimer partner-mediated repression of hepatocyte nuclear factor 4 and Foxo1. J Biol Chem 279:23158–23165 [DOI] [PubMed] [Google Scholar]
- Yeh MM, Brunt EM. (2007) Pathology of nonalcoholic fatty liver disease. Am J Clin Pathol 128:837–847 [DOI] [PubMed] [Google Scholar]
- Yin L, Lazar MA. (2005) The orphan nuclear receptor Rev-erbalpha recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol Endocrinol 19:1452–1459 [DOI] [PubMed] [Google Scholar]
- Yin L, Wang J, Klein PS, Lazar MA. (2006) Nuclear receptor Rev-erbalpha is a critical lithium-sensitive component of the circadian clock. Science 311:1002–1005 [DOI] [PubMed] [Google Scholar]
- Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, et al. (2007) Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science 318:1786–1789 [DOI] [PubMed] [Google Scholar]
- Yoneno K, Hisamatsu T, Shimamura K, Kamada N, Ichikawa R, Kitazume MT, Mori M, Uo M, Namikawa Y, Matsuoka K, et al. (2013) TGR5 signalling inhibits the production of pro-inflammatory cytokines by in vitro differentiated inflammatory and intestinal macrophages in Crohn’s disease. Immunology 139:19–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Wang F, Jin C, Huang X, McKeehan WL. (2005a) Independent repression of bile acid synthesis and activation of c-Jun N-terminal kinase (JNK) by activated hepatocyte fibroblast growth factor receptor 4 (FGFR4) and bile acids. J Biol Chem 280:17707–17714 [DOI] [PubMed] [Google Scholar]
- Yu C, Wang F, Kan M, Jin C, Jones RB, Weinstein M, Deng CX, McKeehan WL. (2000) Elevated cholesterol metabolism and bile acid synthesis in mice lacking membrane tyrosine kinase receptor FGFR4. J Biol Chem 275:15482–15489 [DOI] [PubMed] [Google Scholar]
- Yu L, Gupta S, Xu F, Liverman AD, Moschetta A, Mangelsdorf DJ, Repa JJ, Hobbs HH, Cohen JC. (2005b) Expression of ABCG5 and ABCG8 is required for regulation of biliary cholesterol secretion. J Biol Chem 280:8742–8747 [DOI] [PubMed] [Google Scholar]
- Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, Parameswaran P, Crowell MD, Wing R, Rittmann BE, et al. (2009a) Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci USA 106:2365–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, He F, Kuruba R, Gao X, Wilson A, Li J, Billiar TR, Pitt BR, Xie W, Li S. (2008) FXR-mediated regulation of angiotensin type 2 receptor expression in vascular smooth muscle cells. Cardiovasc Res 77:560–569 [DOI] [PubMed] [Google Scholar]
- Zhang S, Wang J, Liu Q, Harnish DC. (2009b) Farnesoid X receptor agonist WAY-362450 attenuates liver inflammation and fibrosis in murine model of non-alcoholic steatohepatitis. J Hepatol 51:380–388 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Da Silva JR, Reilly M, Billheimer JT, Rothblat GH, Rader DJ. (2005) Hepatic expression of scavenger receptor class B type I (SR-BI) is a positive regulator of macrophage reverse cholesterol transport in vivo. J Clin Invest 115:2870–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lee FY, Barrera G, Lee H, Vales C, Gonzalez FJ, Willson TM, Edwards PA. (2006a) Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc Natl Acad Sci USA 103:1006–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang X, Vales C, Lee FY, Lee H, Lusis AJ, Edwards PA. (2006b) FXR deficiency causes reduced atherosclerosis in Ldlr-/- mice. Arterioscler Thromb Vasc Biol 26:2316–2321 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yin L, Anderson J, Ma H, Gonzalez FJ, Willson TM, Edwards PA. (2010) Identification of novel pathways that control farnesoid X receptor-mediated hypocholesterolemia. J Biol Chem 285:3035–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YK, Guo GL, Klaassen CD. (2011) Diurnal variations of mouse plasma and hepatic bile acid concentrations as well as expression of biosynthetic enzymes and transporters. PLoS ONE 6:e16683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YK, Yeager RL, Klaassen CD. (2009c) Circadian expression profiles of drug-processing genes and transcription factors in mouse liver. Drug Metab Dispos 37:106–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, King N, Chen KY, Breslow JL. (2009) Activation of PXR induces hypercholesterolemia in wild-type and accelerates atherosclerosis in apoE deficient mice. J Lipid Res 50:2004–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Febbraio M, Wada T, Zhai Y, Kuruba R, He J, Lee JH, Khadem S, Ren S, Li S, et al. (2008) Hepatic fatty acid transporter Cd36 is a common target of LXR, PXR, and PPARgamma in promoting steatosis. Gastroenterology 134:556–567 [DOI] [PubMed] [Google Scholar]
- Zhou J, Zhai Y, Mu Y, Gong H, Uppal H, Toma D, Ren S, Evans RM, Xie W. (2006) A novel pregnane X receptor-mediated and sterol regulatory element-binding protein-independent lipogenic pathway. J Biol Chem 281:15013–15020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Ross RA, Pywell CM, Liangpunsakul S, Duffield GE. (2014) Disturbances in the murine hepatic circadian clock in alcohol-induced hepatic steatosis. Sci Rep 4:3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QS, Xing W, Qian B, von Dippe P, Shneider BL, Fox VL, Levy D. (2003) Inhibition of human m-epoxide hydrolase gene expression in a case of hypercholanemia. Biochim Biophys Acta 1638:208–216 [DOI] [PubMed] [Google Scholar]
- Zmrzljak UP, Rozman D. (2012) Circadian regulation of the hepatic endobiotic and xenobitoic detoxification pathways: the time matters. Chem Res Toxicol 25:811–824 [DOI] [PubMed] [Google Scholar]
- Zollner G, Trauner M. (2009) Nuclear receptors as therapeutic targets in cholestatic liver diseases. Br J Pharmacol 156:7–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zollner G, Wagner M, Fickert P, Silbert D, Gumhold J, Zatloukal K, Denk H, Trauner M. (2007) Expression of bile acid synthesis and detoxification enzymes and the alternative bile acid efflux pump MRP4 in patients with primary biliary cirrhosis. Liver Int 27:920–929 [DOI] [PubMed] [Google Scholar]
- Zweers SJ, Booij KA, Komuta M, Roskams T, Gouma DJ, Jansen PL, Schaap FG. (2012) The human gallbladder secretes fibroblast growth factor 19 into bile: towards defining the role of fibroblast growth factor 19 in the enterobiliary tract. Hepatology 55:575–583 [DOI] [PubMed] [Google Scholar]