Abstract
Background
Schlafen proteins have previously been linked to leukocyte and intestinal epithelial differentiation. We hypothesized that Schlafen 12 (SLFN12) overexpression in prostate epithelial cells would modulate expression of prostate-specific antigen (PSA) and dipeptidyl peptidase-4 (DPP4), markers of prostatic epithelial differentiation.
Materials and Methods
Differentiation of the prostate cancer cell line LNCaP and PC-3 was compared after infection with an adenoviral vector coding for SLFN12-GFP (Ad-SLFN12) or GFP only expressing virus (control). Transcript levels of SLFN12, PSA and DPP4 were evaluated by RT-PCR and protein levels by Western blotting. Because Mixed Lineage Kinase (MLK) and one of its downstream effectors (ERK) have previously been implicated in some aspects of prostate epithelial differentiation, we conducted further studies in which LNCaP cells were co-treated with DMSO (control), PD98059 (ERK inhibitor) or MLK inhibitor during transfection with Ad-GFP-SLFN12 for 72 hours.
Results
Treatment of LNCaP or PC-3 cells with Ad-SLFN12 reduced PSA expression by 56.6±4.6% (p<0.05) but increased DPP4 transcript level by 4.8±1.0 fold (p<0.05) vs. Ad-GFP-treated controls. Further studies in LNCaP cells showed that Ad-SLFN12 overexpression increased the ratio of the mature E-cadherin protein to its precursor protein. Furthermore, SLFN12 overexpression promoted DPP4 expression either when MLK or ERK were blocked. ERK inhibition did not reverse SLFN12-induced changes in PSA, E-cadherin or DPP4.
Conclusions
SLFN12 may regulate differentiation in prostate epithelial cells, at least in part independently of ERK or MLK. Understanding how SLFN12 influences prostatic epithelial differentiation may ultimately identify targets to influence the phenotype of prostatic malignancy.
Keywords: LNCaP, PSA, PC-3, E-cadherin, DPP4, metastasis, proliferation
Introduction
Prostatic epithelium undergoes constant differentiation and proliferation during normal prostatic functions (1). Inhibiting prostate epithelial differentiation supports initiation and maintenance of oncogenic transformations (2). Indeed, increasing in vivo and in vitro evidence suggests that induction of differentiation in prostate cancer cells can slow or reverse carcinogenic malignant cells, inducing cell cycle arrest, and triggering apoptosis (3–5). In vitro, such stimuli not only induce differentiation directly but stimulate lipogenesis leading to reduced carcinogenesis by Peroxisome proliferatoractivated receptor gamma 2 (PPARγ2) (6) and increased Insulin-like growth factor (IGF) axis activity (7). Reduced levels of PSA and induction of differentiation have been observed in prostate cancer cells after treatment with flavonoids (8), genistein, seleno- L-methionine, DL-alpha-tocopherol (9), dietary components proposed for chemoprevention of prostate cancer. Restoration of DPP4 expression in prostate cancer cells reduces the malignant phenotype by blocking the bFGF pathway (10).
The Schlafen superfamily can be divided in a short (rodent Slfn1, Slfn2), intermediate (rodent Slfn3, Slfn4 and human SLFN12) as well as long (rodent Slfn5, Slfn8, Slfn9, Slfn10, Slfn14 and human SLFN5, SLFN11, SLFN13, SLFN 14) groups of loosely structurally related proteins(11). While their functions are poorly understood, the rodent short Schlafen protein Schlafen 1 (Slfn1) have previously been implicated in T cell development (12) while Schlafen 2 (Slfn2) and Schlafen 4 (Slfn4) modulate the differentiation of monocytes and macrophages to osteoclasts (13, 14). Schlafen 3 (Slfn3) may mediate rodent enterocyte differentiation (15, 16). Since the differentiated phenotype of prostate cancer can be critical for its eventual prognosis (17), it therefore became of interest to determine whether Schlafen proteins contribute to prostate epithelial differentiation. With regard to human epithelial cancers, Schlafen 12 seemed of particular interest since it is the only intermediate Schlafen protein expressed in human tissues without known function. We therefore sought to test the hypothesis that Schlafen 12 might modulate the differentiation of human prostate cancer cells. We used proliferation, prostate specific antigen (PSA), dipeptidyl-peptidase 4 (DPP4) and E-Cadherin as markers of prostate epithelial phenotype. Both PSA and DPP4 are serine proteases capable regulating peptide factors (18, 19). PSA is an androgen-dependent protease that is exclusively released into the lumen where it alters components of the seminal plasma. PSA is often used as a marker of prostate cancer progression (20). DPP4 is a brush-border enzyme expressed during differentiation of epithelial cells (21). E-cadherin is an adhesion molecule regulating cell growth by Wnt signaling (22) that is induced by differentiating agents in prostate cells (23). In some studies, we used sucrase-isomaltase (SI) and glucose transporter 2 (GLUT2) as negative controls because they are markers of intestinal epithelial differentiation (as opposed to prostatic epithelial differentiation) that can be induced in some epithelial cells by a different Schlafen protein (Slfn3) (16). Since Mixed Lineage Kinase (MLK) and Extracellularsignal- Regulated Kinases (ERK) can mediate growth factor and cytokine induced differentiation (24, 25) and since ERK signaling has been shown to regulate the differentiation of prostate cancer cells independently of cell cycle (26), we then evaluated the possible roles of MLK and ERK signaling in Schlafen 12 induced effects.
Methods
Cell culture
LNCaP (CRL-1740, American type Culture Collection, Manassas, VA) and PC-3 (CRL-1435, ATCC) human prostate cancer cells were maintained at 37° C 5% CO2 in RPMI 1640 media supplemented with 10% fetal bovine serum as previously described (23). LNCaP or PC-3 cells were seeded at 250,000 cells/well on 6-well culture plates.
Adenovirus vector construction
SLFN12 (Ad-GFP-SLFN12) and control (Ad-GFP) adenoviruses containing green fluorescent protein (GFP) were purchased from Applied Biological Material (Richmond, BC, Canada). Adenovirus vector purification, propagation and characterization were performed as described previously (27).
Proliferation and apoptosis
Subconfluent (50–60%) LNCaP, PC-3 cells were treated with 400 virus particles of Ad-GFP (Control) per cell or Ad-GFP- SLFN12 for 72 hours. In some experiments LNCaP cells were incubated in normal growth medium containing 0.1% DMSO or CEP-1347 inhibitor (800 nM) or PD98059 (20 nM) dissolved in DMSO for 72 h before trypsinization and cell counting. Cell number was determined in each of the 6 wells independently with a MTT proliferation assay (Promega, Madison, WI) using the manufacturer’s protocol. Apoptotic cells were quantified in LNCaP cells using Vybrant apoptosis assay kit (Life Technologies, Carlsbad, CA) following the protocol provided by the manufacturer. For the time course experiment, LNCaP cells were treated with Ad-GFP or Ad-GFP-SLFN12 virus for 24, 48, 72 and 96 hours. Each experiment was carried out at least twice in triplicate with similar results.
RNA isolation and qRT-PCR
Total RNA was isolated from the cells and tissues using Tri-Reagent (Molecular Research Center, Inc., Cincinnati, OH) in accordance with the manufacturer's instructions. cDNA was prepared from RNA samples as described previously (23). cDNA samples were analyzed by RT-PCR analysis using the BioRad MyiQ Real-Time PCR system and the BioRad SYBR Green supermix (BioRad Laboratories, Hercules, CA). Expression levels were determined from the threshold cycle (Ct) values using the method of 2−ΔΔCt (23) using 18S expression as the reference control gene. Primer information is as follows: The ribosomal protein S18 (18S) 5’- CGCCGGTCCAAGAATTTCACCTCT-3’ (upstream) and 5’- CCCCTCGATGCTCTTAGCTGAGTCT-3’ (downstream). GLUT2 forward 5’- TTAGCAACTGGGTCTGCAAT-3’, reverse 5’-GGTGTAGTCCTACACTCATG-3’; SI, forward – 5’-AAACCTACATGTCCGTGGTGGTCA-3’, reverse – 5’-AACAGAGAACCCTGTGCCATCTGA-3’, DPP4, forward – 5’-TTTGGGGCTGGTCATATGGAGGG-3’, reverse – 5’-ACTCCCACCGGGATACAGGCG-3’. PSA, forward – 5’- GGGCCCACTTGTCTGTAA-3’, reverse – 5’-GATGGTGTCCTTGATCCACT-3’. Androgen Receptor (AR), forward – 5’-GCCCTATCCCAGTCCCACTT-3’, reverse – 5’- TGGTCCCTGGCAGTCTCC-3’. SLFN12, forward – 5’- ATCTGGGCTTGCAAGAGAAG-3’, reverse – 5’-TTTTTGCCAGCTTCTGCTTT-3’. The cycle conditions for the PCR were 1 cycle of 3 minutes at 95°C and 40 cycles of 30 seconds at 95°C, 30 seconds at the annealing temperature (57°C), and 30 seconds at 72°C.
Western Blot Analysis
Cultured LNCaP cells were lysed in lysis buffer, centrifuged at 15,000g for 10 minutes at 4°C, resolved by SDS-PAGE and tra nsferred to Hybond ECL nitrocellulose membrane (Amersham Pharmacia Biotech, Piscataway, NJ) as previously described (28). Nonspecific binding sites were blocked for 1 h at room temperature using Odyssey Blocking Buffer (Licor, Lincoln, NE). Membranes were probed with antibodies to PSA, DPP4, E-cadherin (CDH1) (Santa Cruz Biotechnology, Santa Cruz, CA), phosphorylated pERK (pERK, Thr202/Tyr204), phosphorylated c-Jun N-terminal kinase (JNK) (pJNK), full-length Caspase 3, Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Cell Signaling, Danvers, MA) as well as appropriate secondary antibodies. Bands were visualized using the Odyssey imaging system (Licor, Lincoln, NE) and analyzed with the Kodak Image Station 440CF. All exposures used for densitometric analysis were within the linear range.
Data analysis
Values are reported as group means ± standard error of the mean (SEM) of the non-transformed data. Prior to analysis, all data were checked to ensure they fit a normal distribution using the plot of predicted values vs. residuals, as well as by the Shapiro-Wilk, Kolmogorov-Smirnov tests for normality. Two-tailed Student’s t-test or ANOVA were used when appropriate. Skewed or non-normally distributed data was log transformed prior to analysis and the correction to a normal distribution was confirmed using the tests described above. Differences between means were considered significant at p<0.05.
Results
Overexpression of rat SLFN12 suppressed PSA but increased DPP4 mRNA level in LNCaP human prostate cancer cells
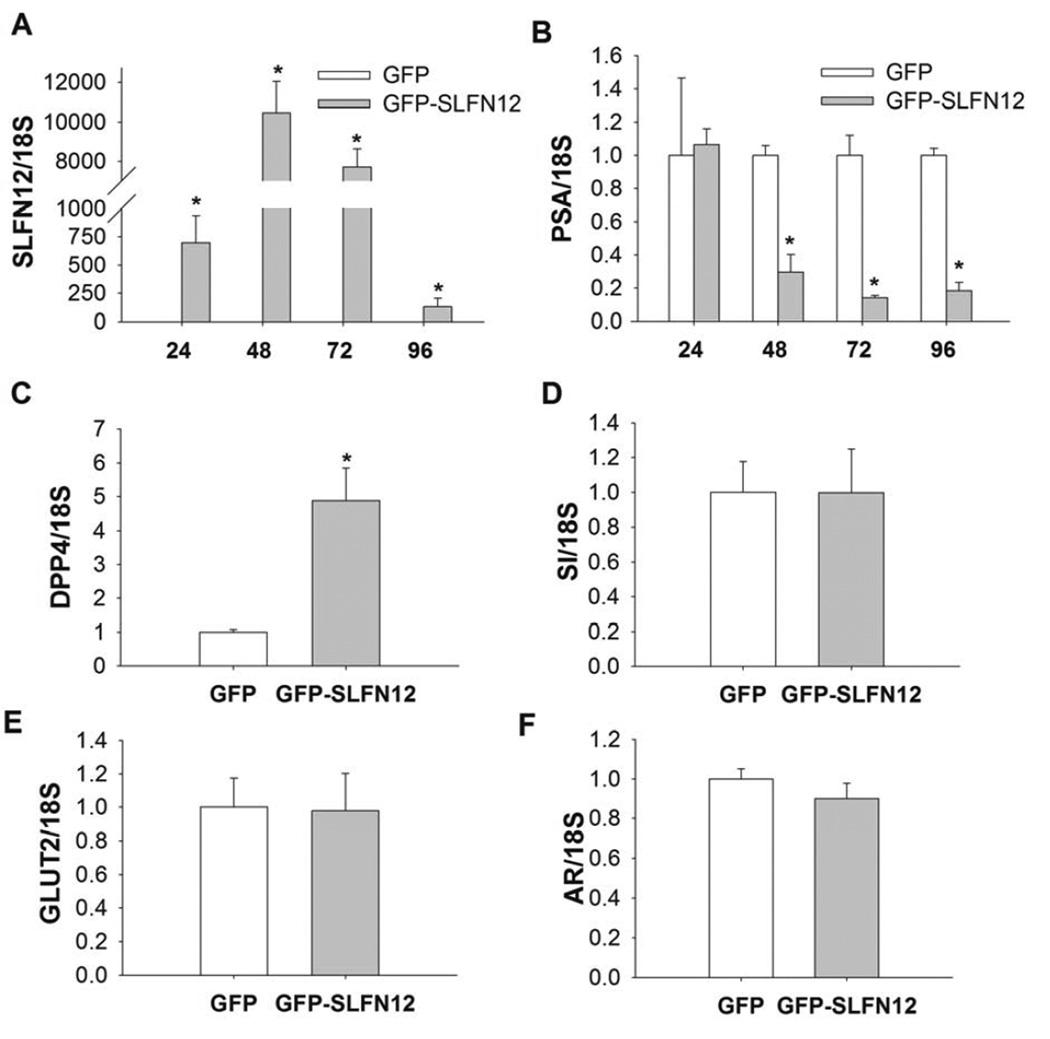
To first validate the function of the Ad-GFP-SLFN12 construct, we investigated whether infection of Ad-GFP-Slfn12 increases SLFN12 mRNA in LNCaP human prostate cancer cells. We subjected 50–60% confluent LNCaP cells to 400 vp/cell Ad GFP-SLFN12 for 24, 48, 72 and 96 hours. Ad-GFP-SLFN12 infection of LNCaP cells resulted in substantial measured SLFN12 transcript expression compared to Ad-GFPtreated cells at 48 and 72 hours (Fig. 1A, n=6, p<0.05). Ad-GFP-SLFN12 infection of LNCaP cells also reduced the level of PSA expression compared to that in Ad-GFP treated controls (Fig. 1B, n=6, p<0.01). Since other studies in vivo have indicated that expression of adenoviral target genes peaks at approximately 72 – 96 hours (29) we selected the 72 hour time point to analyze the effects of SLFN12 overexpression on other markers of differentiation in LNCaP and PC3 prostate cancer cells. Indeed, Ad-GFP- SLFN12 infection of LNCaP cells stimulated expression of DPP4 but did not change the expression of SI, GLUT2 or that of Androgen Receptor (AR) compared to control (Fig. 1C–F, n=6). Thus, we demonstrated that exogenous overexpression of SLFN12 by direct infection of an Ad vector coding for SLFN12 cDNA would promote not only SLFN12 expression but also an expression of specific differentiation markers in LNCaP cells.
Figure 1. SLFN12 induction modulates DPP4 and PSA but not SI, GLUT2 or AR transcript levels in LNCaP cells.
60% confluent LNCaP cells were treated with Ad-GFP (GFP) or Ad-GFP-SLFN12 (GFPSLFN12) for 24, 48, 72 and 96 hours (n=12). Transcript levels of SLFN12 (A), PSA (B), were determined by quantitative reverse transcriptase-polymerase chain reaction at all time points and transcript levels of DPP4 (C), SI (D), GLUT2 (E), AR (F) at the 72 hour time point. Error bars indicate standard errors. * - p<0.05
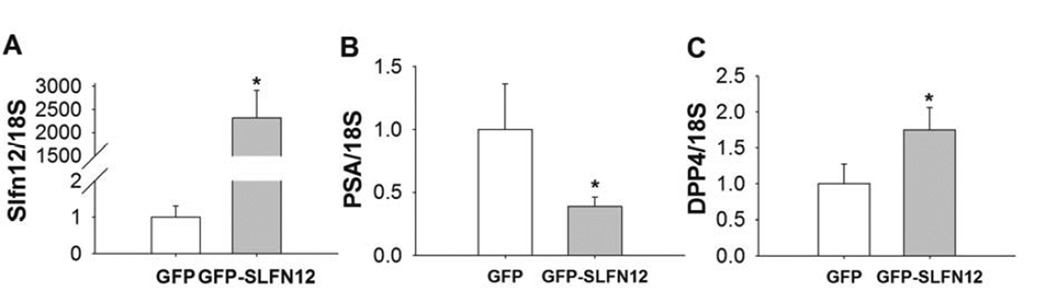
To confirm that the effects of SLFN12 were not restricted to a single cell line, we treated human PC-3 prostate cancer cells with Ad-GFP or Ad-GFP-SLFN12 adenovirus for 72 hours. Ad-GFP-SLFN12 infection of PC-3 cells resulted in significantly increased transcript levels of SLFN12 compared to Ad-GFP-treated cells (Fig. 2A n=6, p<0.05). Ad-GFP- SLFN12 infection of PC-3 cells also significantly reduced the level of PSA expression (Fig. 2B, n=6, p<0.05) and also significantly increased expression of the differentiation marker DPP4 (Fig. 2C, n=6, p<0.05).
Figure 2. SLFN12 induction modulates DPP4 and PSA transcript levels in PC-3 cells.
60% confluent PC-3 cells were treated with Ad-GFP (GFP) or Ad-GFP-SLFN12 (GFPSLFN12) for 72 hours (n=12). Transcript levels of SLFN12 (A), PSA (B), DPP4 (C) of PC-3 cells were determined by quantitative reverse transcriptase-polymerase chain reaction. Error bars indicate standard errors. * - p<0.05
SLFN12 overexpression reduces proliferation of LNCaP cells
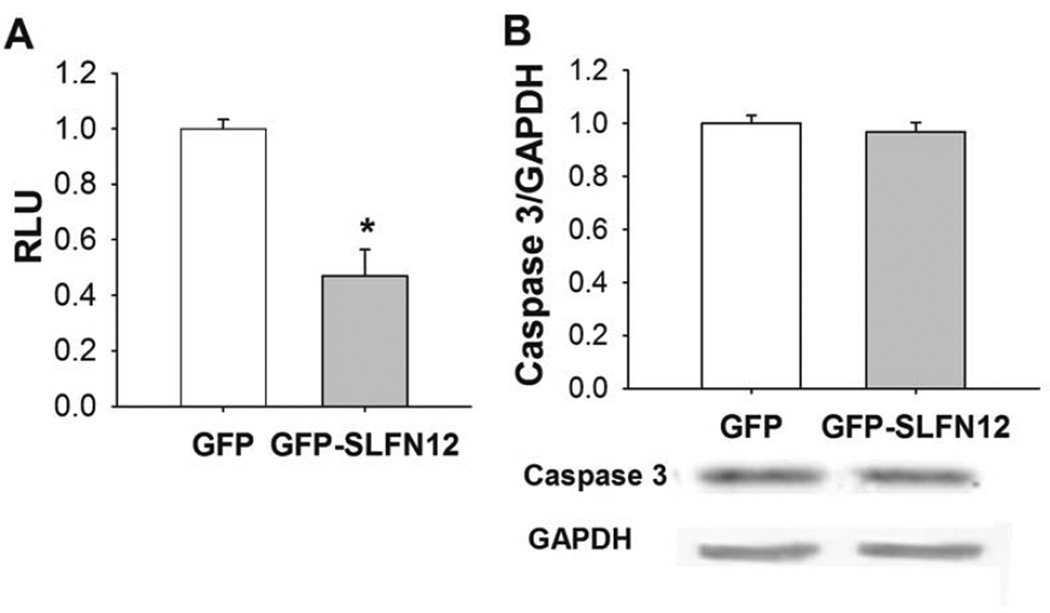
To determine whether Slfn12 overexpression influences cell proliferation, we measured LNCaP proliferation by MTT assay after 72 hours of Ad-GFP (control) or Ad-GFP-SLFN12 treatment. Cell number was significantly reduced after treatment of Ad-GFP-SLFN12 compared to controls (Fig 3A, n=6, p<0.05). There was no significant change in the level of full length Caspase 3 in LNCaP cells between treatments (Fig 3B, n=4). We performed parallel studies using the Vybrant apoptosis assay kit to assess aptoptosis in order to determine whether Ad-GFP-SLFN12 infection modulates apoptosis in LNCaP cells. The number of apoptotic cells was not significantly different between control and Ad-GFP-SLFN12 treated cells (data not shown). Taken together, these results suggest that SLFN12 overexpression may inhibit proliferation in LNCaP cells.
Figure 3. SLFN12 overexpression changes proliferation of LNCaP cells.
A. Treatment of 50–60% confluent LNCaP with Ad-GFP-SLFN12 for 72 h reduced proliferation compared to Ad-GFP treated control cells. B. Full length Caspase 3 in Ad-GFP and Ad-GFP-SLFN12 treated LNCaP cells. Error bars indicate standard errors. * - p<0.05.
SLFN12 mediated induction of DPP4 does not require MLK or ERK signaling
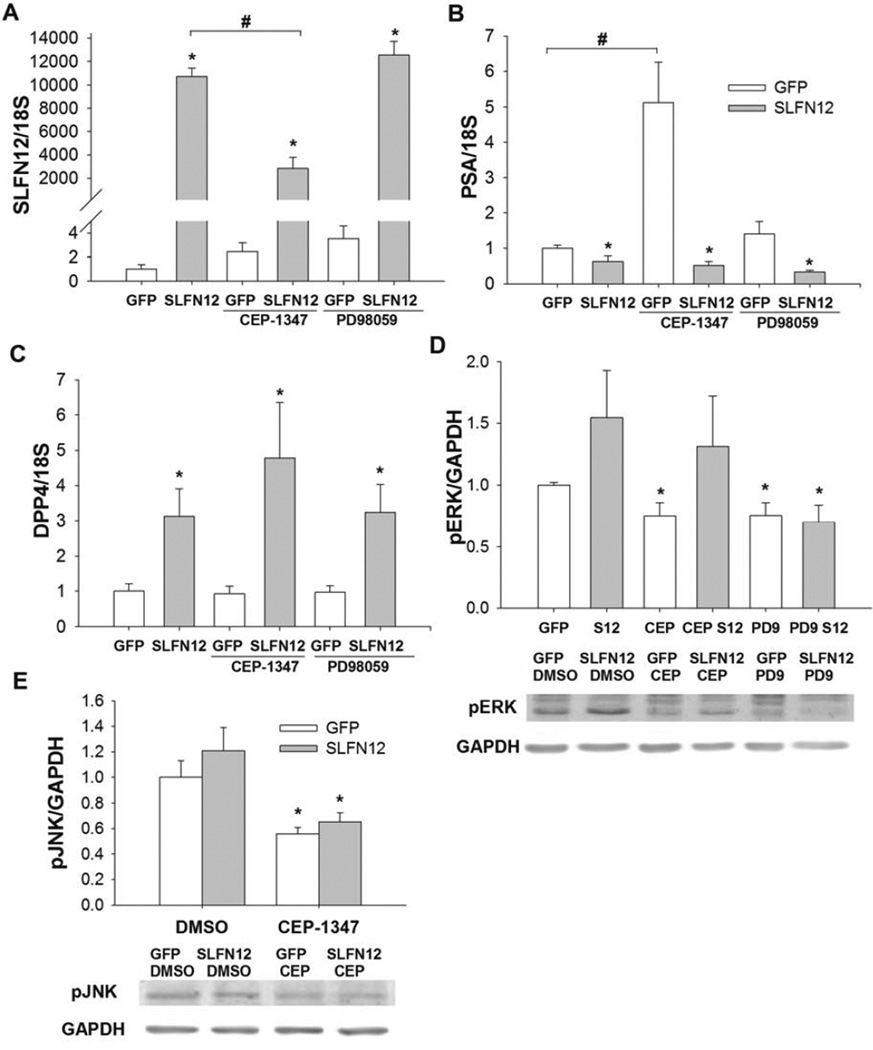
We tested whether ERK or MLK signaling is involved in SLFN12-induced differentiation of LNCaP cells by first pretreating these cells with ERK and MLK inhibitors. Suppression of ERK by PD98059 or MLK by CEP-1347 did not reverse the Ad-GFP-SLFN12 induction of SLFN12 or DPP4 in LNCaP cells (Fig 4A,C, n=9, p<0.05) or Ad-GFP-SLFN12 suppression of PSA (Fig 4C, n=9, p<0.05). Although SLFN12 overexpression was slightly reduced in MLK-inhibited cells, Ad-GFP-SLFN12 infection still resulted in marked and likely supramaximal overexpression of SLFN12 even in the MLK-inhibited cells (Fig 4A). We confirmed that both inhibitors were active in parallel studies that assessed level of ERK and jnk phosphorylation since ERK and jnk are downstream of MLK (30) The ratio of phosphorylated ERK (pERK) to GAPDH in the absence of SLFN12 overexpression was substantially reduced by treatment with either inhibitor (Fig 4D, p<0.05, n=8). Treatment with Ad-GFP-SLFN12 increased ERK phosphorylation in control (DMSO) and CEP-1347 treated groups but not in PD98059-treated cells compared to GFP treated controls (Fig 4D), suggesting that SLFN12 overexpression induces ERK activation in a fashion that requires MEK but not MLK. In order to confirm the efficacy of the MLK inhibitor we tested the level of phosphorylated JNK (pJNK) in DMSO and CEP-1347 treated LNCaP cells. Although CEP-1347 failed to prevent SLFN12 induction of ERK phosphorylation (Figure 4D, n=8, p<0.05), we found that the ratio of pJNK to GAPDH was significantly reduced after treatment with CEP-1347 compared to control (Figure 4E, n=8, p<0.05), suggesting that MLK inhibition did occur in the CEP-treated cells, but that the effect of SLFN12 overexpression on ERK phosphorylation occurs independently of or perhaps downstream of MLK.
Figure 4. Modulation of PSA and DPP4 transcript levels by SLFN12 does not depend on ERK or MLK signaling.
ERK (PD9) and MLK (CEP) inhibitors do not reverse SLFN12 induced expression of SLFN12 (A). MLK inhibitor increases PSA but similarly to ERK inhibitor does not reverse SLFN12-mediated suppression of PSA (B) (*-significant compared to GFP-treated group, #-significant to indicated control (DMSO) group). ERK (PD9) and MLK (CEP) inhibitors do not reverse SLFN12 induced expression of DPP4 (C). MEK inhibitor suppression of ERK was confirmed by measurement of protein levels of phosphorylated ERK (D), in Ad-GFP (GFP) and Ad-GFP-SLFN12 (SLFN12, S12) after treatment with DMSO (control), MLK (CEP) or ERK (PD9) inhibitors for 72 hours. Similarly, the efficacy of the MLK inhibitor was confirmed by the demonstration that CEP significantly reduced pJNK levels in Ad-GFP and Ad-GFP-SLFN12 treated cells (E). *-significant compared to Ad-GFP treated control (DMSO) group. Error bars indicate standard errors. * - p<0.05.
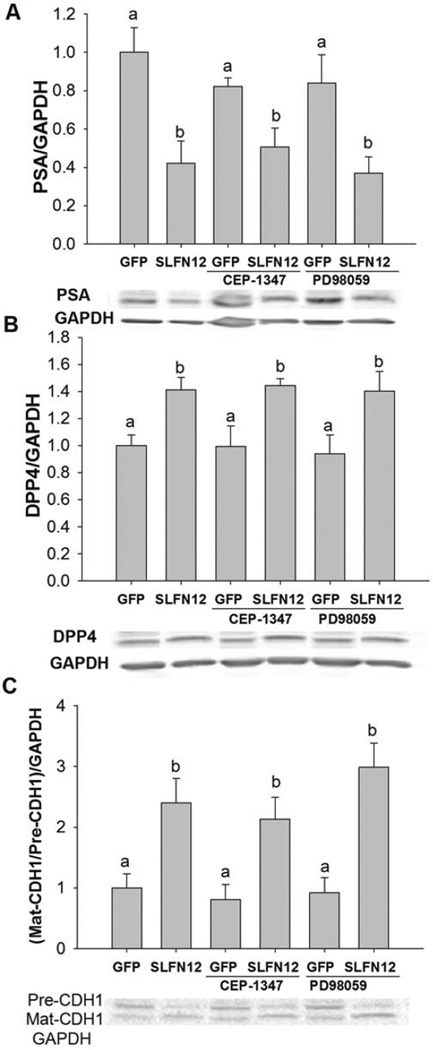
We further measured protein levels of DPP4 and PSA in cells treated with Ad-GFP or Ad-GFP-SLFN12. As in our qRT-PCR studies, we found that protein levels of PSA were significantly suppressed and DPP4 were increased after SLFN12 overexpression (Fig 5A,B, n=4–7, p<0.05). The ratio of the mature E-cadherin (CDH1) (80 kDa) to its precursor form (130 kDa) was significantly increased after overexpression of SLFN12 compared to the ratio in control cells (Fig 5B, n=6, p<0.05.). Notably, MLK inhibition also did not significantly increase PSA protein levels compared to DMSO-treated controls, as we had observed for PSA transcript levels (Fig 4C and Fig 5B). These results suggest that SLFN12 modulates PSA, DPP4 and E-cadherin expression independently of ERK.
Figure 5. Modulation of PSA, DPP4 and E-cadherin protein levels by SLFN12 does not depend on ERK or MLK signaling.
Protein levels of PSA (A), DPP4 (B) and Mature (Mat-CDH1) and precursor (Pre-CDH1) forms of E-Cadherin (C) were measured in Ad-GFP and Ad-GFP-SLFN12 after treatment with DMSO (control), MLK (CEP) or ERK (PD9) inhibitors for 72 hours. Error bars indicate standard errors. * - p<0.05. Bars with different letters are significantly different (p<0.05).
Discussion
Disruption of prostate epithelial differentiation promotes carcinogenesis. Our results suggest that SLFN12 increases DPP4 and mature E-cadherin in prostate cells while suppressing PSA expression independently of MLK or ERK. SLFN12 also inhibits LNCaP proliferation. Other Schlafen proteins have been studied in rodents (31, 32), where some tissue-specific effects on differentiation and proliferation have previously been ascribed to Slfn1 (proliferation in T-lymphocytes (33)), Slfn2 (osteoclast differentiation (13)), and Slfn3 (enterocyte differentiation (15)). Slfn4 is increased during macrophage activation by LPS, but suppressed during differentiation induced by colony-stimulating factor (CSF-1) (14). Slfn5 deletion enhances human melanoma invasion in vitro (34). Although a role for SLFN12 has not been elucidated, our results suggest SLFN12 may influence human prostatic epithelial differentiation. How Schlafen proteins act is poorly understood, but our present results may offer a few hints.
PSA is an androgen-regulated serine protease expressed only in differentiated prostate epithelial cells (20). PSA secretion in normal prostate correlates with androgen levels (20) but PSA is also induced in prostate cancer cells by growth factors or cytokines (35, 36). PSA may facilitate prostate cancer invasion by cleaving extracellular matrix proteins (37). Although proliferation, PSA, and DPP4 are all reduced in androgen-deprived LNCaP cells (38), SLFN12 affected them differently. SLFN12 reduced proliferation and PSA but induced DPP4. The reduction in PSA we observed is consistent with the reduction in PSA caused by differentiating agents like butyrate (39).
Our data suggest that, similarly to Slfn3 in rodent enterocytes (16), SLFN12 stimulates DPP4 expression in human prostatic epithelial cells. DPP4 is a cell surface serine protease that regulates peptide factors by cleaving NH2-terminal peptides from polypeptides, is expressed in multiple cells and tissues. Potential substrates for DPP4 include matrix interaction proteins, growth factors, cytokines, and hormones (18). SLFN12 induced DPP4 but not GLUT2 or SI transcripts in prostate cancer cells. This differs from the effects of SLFN3 in intestinal epithelial cells, suggesting that the effects of Schlafens on these genes vary with cell type and Schlafen protein. Taken together, these results suggest that SLFN12 and androgen regulate PSA and DPP4 in prostate cancer cells by different mechanisms.
E-cadherin is a calcium-dependent cell-cell adhesion glycoprotein that regulates Wnt signaling. Loss of E-cadherin in prostate cancer correlates with increased Gleason score and reduced survival (40). In contrast, treatment of prostate cancer cells with vitamin D analogs (41) or a disulfiram-sunitinib combination (42) increase E-cadherin and other differentiation markers and reduce proliferation. Slfn3 overexpression in human HCT-116 colon cancer cells similarly drives differentiation and increases E-cadherin levels (43). SLFN12 increased mature E-cadherin in LNCaP cells. These results therefore suggest the further hypothesis that Schlafen protein effects on E-cadherin may be involved in the growth inhibitory effects of Schlafen proteins.
Differentiation induced by butyrate, flavonoids or vitamin D reduces proliferation of prostate cancer and normal cells (5, 8, 39). Schlafen proteins involved in differentiation may also modulate proliferation in some cell types. Slfn1, implicated in myeloid differentiation and proliferation (31) in murine epithelial cells, reduces proliferation by regulating cyclin D1 expression (33). Slfn3-mediated proliferation and differentiation correlated with TGFβ expression in colon cancer cells (44) but Slfn3 does not change proliferation of normal enterocytes in vitro (15) or in vivo (16). Our results support an antiproliferative effect of Schlafen proteins in cancer cells, although the mechanisms responsible for these effects may vary among cell types.
ERK activation correlates with progression of androgen-independent prostate cancer (45), while DPP4 overexpression inhibits ERK in prostate cancer cells (10). However, neither ERK inhibition by MEK blockade nor MLK inhibition abolished SLFN12 effects on DPP4 or PSA, suggesting that SLFN12 acts in prostate cancer cells independently of ERK and MLK. Although SLFN12 may not require MLK to exert its effects, our data suggest that adenoviral over-expression of SLFN12 may itself be influenced by MLK signaling. The reduction of SLFN12 overexpression by the MLK inhibitor likely did not diminish the downstream effects of SLFN12 overexpression because the levels of SLFN12 achieved by the viral infection were already supramaximal, so that even the somewhat reduced SLFN12 levels achieved in the setting of MLK inhibition were sufficient to cause the SLFN12 downstream effects.
Other Schlafen proteins have been implicated in rodent immune regulation (12), and CD3, CD28, and CD25 may be important for the expression of Schlafen transcripts in regulatory T cells (46). Another short Schlafen protein, Slfn2, controls hematopoietic colony formation, but its deletion increases murine fibroblast proliferation and reduces IFNα-dependent growth inhibition (47). Potential mechanisms of Schlafen 12 in the regulation of immune functions and how they would interact with the effects that we study here are beyond the scope of the current investigation and await further study.
SLFN12 overexpression increases DPP4 expression at both the transcript and protein level, increases mature E-cadherin, and reduces both PSA expression and proliferation, consistent with a more differentiated, less aggressive phenotype for prostate cancer. While its mechanism awaits elucidation, the differentiating effects of SLFN12 seem independent of MLK or ERK. SLFN12 or the pathways by which it acts may be targets to regulate the proliferation or phenotype of prostate cancer cells.
Acknowledgments
Supported in part by NIH 1 R56 DK096137-01 (MDB)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors contributions:
P.L.K and M.D.B. are responsible for conception and design of the research; M.D.B obtained funding for the research; P.L.K performed the experiments, collected and analyzed the data; P.L.K and M.D.B. interpreted the results of the experiments; P.L.K. prepared the figures and drafted the manuscript; P.L.K and M.D.B. edited revised the manuscript and approved the final version of the manuscript.
References
- 1.Zenzmaier C, Untergasser G, Berger P. Aging of the prostate epithelial stem/progenitor cell. Exp Gerontol. 2008;43:981–985. doi: 10.1016/j.exger.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Frank SB, Miranti CK. Disruption of Prostate Epithelial Differentiation Pathways and Prostate Cancer Development. Front Oncol. 2013;3:273. doi: 10.3389/fonc.2013.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walczak J, Wood H, Wilding G, Williams T, Jr, Bishop CW, Carducci M. Prostate cancer prevention strategies using antiproliferative or differentiating agents. Urology. 2001;57:81–85. doi: 10.1016/s0090-4295(00)00947-x. [DOI] [PubMed] [Google Scholar]
- 4.Laurenzana A, Balliu M, Cellai C, Romanelli MN, Paoletti F. Effectiveness of the histone deacetylase inhibitor (S)-2 against LNCaP and PC3 human prostate cancer cells. PLoS One. 2013;8:e58267. doi: 10.1371/journal.pone.0058267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kovalenko PL, Zhang Z, Yu JG, Li Y, Clinton SK, Fleet JC. Dietary vitamin d and vitamin d receptor level modulate epithelial cell proliferation and apoptosis in the prostate. Cancer Prev Res (Phila) 2011;4:1617–1625. doi: 10.1158/1940-6207.CAPR-11-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strand DW, Jiang M, Murphy TA, Yi Y, Konvinse KC, Franco OE, Wang Y, Young JD, Hayward SW. PPARgamma isoforms differentially regulate metabolic networks to mediate mouse prostatic epithelial differentiation. Cell Death Dis. 2013;3:e361. doi: 10.1038/cddis.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Massoner P, Ladurner Rennau M, Heidegger I, Kloss-Brandstatter A, Summerer M, Reichhart E, Schafer G, Klocker H. Expression of the IGF axis is decreased in local prostate cancer but enhanced after benign prostate epithelial differentiation and TGF-beta treatment. Am J Pathol. 2011;179:2905–2919. doi: 10.1016/j.ajpath.2011.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zi X, Agarwal R. Silibinin decreases prostate-specific antigen with cell growth inhibition via G1 arrest, leading to differentiation of prostate carcinoma cells: implications for prostate cancer intervention. Proc Natl Acad Sci U S A. 1999;96:7490–7495. doi: 10.1073/pnas.96.13.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peternac D, Klima I, Cecchini MG, Schwaninger R, Studer UE, Thalmann GN. Agents used for chemoprevention of prostate cancer may influence PSA secretion independently of cell growth in the LNCaP model of human prostate cancer progression. Prostate. 2008;68:1307–1318. doi: 10.1002/pros.20795. [DOI] [PubMed] [Google Scholar]
- 10.Wesley UV, McGroarty M, Homoyouni A. Dipeptidyl peptidase inhibits malignant phenotype of prostate cancer cells by blocking basic fibroblast growth factor signaling pathway. Cancer Res. 2005;65:1325–1334. doi: 10.1158/0008-5472.CAN-04-1852. [DOI] [PubMed] [Google Scholar]
- 11.Neumann B, Zhao L, Murphy K, Gonda TJ. Subcellular localization of the Schlafen protein family. Biochem Biophys Res Commun. 2008;370:62–66. doi: 10.1016/j.bbrc.2008.03.032. [DOI] [PubMed] [Google Scholar]
- 12.Schwarz DA, Katayama CD, Hedrick SM. Schlafen, a new family of growth regulatory genes that affect thymocyte development. Immunity. 1998;9:657–668. doi: 10.1016/s1074-7613(00)80663-9. [DOI] [PubMed] [Google Scholar]
- 13.Lee NK, Choi HK, Yoo HJ, Shin J, Lee SY. RANKL-induced schlafen2 is a positive regulator of osteoclastogenesis. Cell Signal. 2008;20:2302–2308. doi: 10.1016/j.cellsig.2008.08.019. [DOI] [PubMed] [Google Scholar]
- 14.van Zuylen WJ, Garceau V, Idris A, Schroder K, Irvine KM, Lattin JE, Ovchinnikov DA, Perkins AC, Cook AD, Hamilton JA, Hertzog PJ, Stacey KJ, Kellie S, Hume DA, Sweet MJ. Macrophage activation and differentiation signals regulate schlafen-4 gene expression: evidence for Schlafen-4 as a modulator of myelopoiesis. PLoS One. 2011;6:e15723. doi: 10.1371/journal.pone.0015723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan L, Yu Y, Sanders MA, Majumdar AP, Basson MD. Schlafen 3 induction by cyclic strain regulates intestinal epithelial differentiation. Am J Physiol Gastrointest Liver Physiol. 2010;298:G994–G1003. doi: 10.1152/ajpgi.00517.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kovalenko PL, Yuan L, Sun K, Kunovska L, Seregin S, Amalfitano A, Basson MD. Regulation of epithelial differentiation in rat intestine by intraluminal delivery of an adenoviral vector or silencing RNA coding for schlafen 3. PLoS One. 2013;8:e79745. doi: 10.1371/journal.pone.0079745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sagnak L, Topaloglu H, Ozok U, Ersoy H. Prognostic significance of neuroendocrine differentiation in prostate adenocarcinoma. Clin Genitourin Cancer. 2011;9:73–80. doi: 10.1016/j.clgc.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 18.Vanhoof G, Goossens F, De Meester I, Hendriks D, Scharpe S. Proline motifs in peptides and their biological processing. FASEB J. 1995;9:736–744. [PubMed] [Google Scholar]
- 19.Ban Y, Wang MC, Watt KW, Loor R, Chu TM. The proteolytic activity of human prostate-specific antigen. Biochem Biophys Res Commun. 1984;123:482–488. doi: 10.1016/0006-291x(84)90256-0. [DOI] [PubMed] [Google Scholar]
- 20.Balk SP, Ko YJ, Bubley GJ. Biology of prostate-specific antigen. J Clin Oncol. 2003;21:383–391. doi: 10.1200/JCO.2003.02.083. [DOI] [PubMed] [Google Scholar]
- 21.Pageot LP, Perreault N, Basora N, Francoeur C, Magny P, Beaulieu JF. Human cell models to study small intestinal functions: recapitulation of the crypt-villus axis. Microsc Res Tech. 2000;49:394–406. doi: 10.1002/(SICI)1097-0029(20000515)49:4<394::AID-JEMT8>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 22.De Wever O, Derycke L, Hendrix A, De Meerleer G, Godeau F, Depypere H, Bracke M. Soluble cadherins as cancer biomarkers. Clin Exp Metastasis. 2007;24:685–697. doi: 10.1007/s10585-007-9104-8. [DOI] [PubMed] [Google Scholar]
- 23.Kovalenko PL, Zhang Z, Cui M, Clinton SK, Fleet JC. 1,25 dihydroxyvitamin D-mediated orchestration of anticancer, transcript-level effects in the immortalized, non-transformed prostate epithelial cell line, RWPE1. BMC Genomics. 2010;11:26. doi: 10.1186/1471-2164-11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katz M, Amit I, Yarden Y. Regulation of MAPKs by growth factors and receptor tyrosine kinases. Biochim Biophys Acta. 2007;1773:1161–1176. doi: 10.1016/j.bbamcr.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furler RL, Uittenbogaart CH. Signaling through the P38 and ERK pathways: a common link between HIV replication and the immune response. Immunol Res. 2010;48:99–109. doi: 10.1007/s12026-010-8170-1. [DOI] [PubMed] [Google Scholar]
- 26.Kim J, Adam RM, Freeman MR. Activation of the Erk mitogen-activated protein kinase pathway stimulates neuroendocrine differentiation in LNCaP cells independently of cell cycle withdrawal and STAT3 phosphorylation. Cancer Res. 2002;62:1549–1554. [PubMed] [Google Scholar]
- 27.Seregin SS, Aldhamen YA, Appledorn DM, Hartman ZC, Schuldt NJ, Scott J, Godbehere S, Jiang H, Frank MM, Amalfitano A. Adenovirus capsid-display of the retrooriented human complement inhibitor DAF reduces Ad vector-triggered immune responses in vitro and in vivo. Blood. 2010;116:1669–1677. doi: 10.1182/blood-2010-03-276949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kovalenko PL, Basson MD. Changes in morphology and function in small intestinal mucosa after Roux-en-Y surgery in a rat model. J Surg Res. 2012;177:63–69. doi: 10.1016/j.jss.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nemunaitis J, Cunningham C, Buchanan A, Blackburn A, Edelman G, Maples P, Netto G, Tong A, Randlev B, Olson S, Kirn D. Intravenous infusion of a replication-selective adenovirus (ONYX-015) in cancer patients: safety, feasibility and biological activity. Gene Ther. 2001;8:746–759. doi: 10.1038/sj.gt.3301424. [DOI] [PubMed] [Google Scholar]
- 30.Shen YH, Godlewski J, Zhu J, Sathyanarayana P, Leaner V, Birrer MJ, Rana A, Tzivion G. Cross-talk between JNK/SAPK and ERK/MAPK pathways: sustained activation of JNK blocks ERK activation by mitogenic factors. J Biol Chem. 2003;278:26715–26721. doi: 10.1074/jbc.M303264200. [DOI] [PubMed] [Google Scholar]
- 31.Geserick P, Kaiser F, Klemm U, Kaufmann SH, Zerrahn J. Modulation of T cell development and activation by novel members of the Schlafen (slfn) gene family harbouring an RNA helicase-like motif. Int Immunol. 2004;16:1535–1548. doi: 10.1093/intimm/dxh155. [DOI] [PubMed] [Google Scholar]
- 32.Kovalenko PL, Basson MD. The correlation between the expression of differentiation markers in rat small intestinal mucosa and the transcript levels of schlafen 3. JAMA Surg. 2013;148:1013–1019. doi: 10.1001/jamasurg.2013.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brady G, Boggan L, Bowie A, O'Neill LA. Schlafen-1 causes a cell cycle arrest by inhibiting induction of cyclin D1. J Biol Chem. 2005;280:30723–30734. doi: 10.1074/jbc.M500435200. [DOI] [PubMed] [Google Scholar]
- 34.Katsoulidis E, Mavrommatis E, Woodard J, Shields MA, Sassano A, Carayol N, Sawicki KT, Munshi HG, Platanias LC. Role of interferon {alpha} (IFN{alpha})-inducible Schlafen-5 in regulation of anchorage-independent growth and invasion of malignant melanoma cells. J Biol Chem. 2010;285:40333–40341. doi: 10.1074/jbc.M110.151076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Culig Z, Hobisch A, Cronauer MV, Hittmair A, Radmayr C, Bartsch G, Klocker H. Activation of the androgen receptor by polypeptide growth factors and cellular regulators. World J Urol. 1995;13:285–289. doi: 10.1007/BF00185971. [DOI] [PubMed] [Google Scholar]
- 36.Hobisch A, Eder IE, Putz T, Horninger W, Bartsch G, Klocker H, Culig Z. Interleukin-6 regulates prostate-specific protein expression in prostate carcinoma cells by activation of the androgen receptor. Cancer Res. 1998;58:4640–4645. [PubMed] [Google Scholar]
- 37.Webber MM, Waghray A, Bello D. Prostate-specific antigen, a serine protease, facilitates human prostate cancer cell invasion. Clin Cancer Res. 1995;1:1089–1094. [PubMed] [Google Scholar]
- 38.D'Antonio JM, Ma C, Monzon FA, Pflug BR. Longitudinal analysis of androgen deprivation of prostate cancer cells identifies pathways to androgen independence. Prostate. 2008;68:698–714. doi: 10.1002/pros.20677. [DOI] [PubMed] [Google Scholar]
- 39.Ellerhorst J, Nguyen T, Cooper DN, Estrov Y, Lotan D, Lotan R. Induction of differentiation and apoptosis in the prostate cancer cell line LNCaP by sodium butyrate and galectin-1. Int J Oncol. 1999;14:225–232. doi: 10.3892/ijo.14.2.225. [DOI] [PubMed] [Google Scholar]
- 40.Whiteland H, Spencer-Harty S, Thomas DH, Davies C, Morgan C, Kynaston H, Bose P, Fenn N, Lewis PD, Bodger O, Jenkins S, Doak SH. Putative prognostic epithelial-to-mesenchymal transition biomarkers for aggressive prostate cancer. Exp Mol Pathol. 2013;95:220–226. doi: 10.1016/j.yexmp.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 41.Campbell MJ, Elstner E, Holden S, Uskokovic M, Koeffler HP. Inhibition of proliferation of prostate cancer cells by a 19-nor-hexafluoride vitamin D3 analogue involves the induction of p21waf1, p27kip1 and E-cadherin. J Mol Endocrinol. 1997;19:15–27. doi: 10.1677/jme.0.0190015. [DOI] [PubMed] [Google Scholar]
- 42.Ketola K, Kallioniemi O, Iljin K. Chemical biology drug sensitivity screen identifies sunitinib as synergistic agent with disulfiram in prostate cancer cells. PLoS One. 2012;7:e51470. doi: 10.1371/journal.pone.0051470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patel VB, Yu Y, Das JK, Patel BB, Majumdar AP. Schlafen-3: a novel regulator of intestinal differentiation. Biochem Biophys Res Commun. 2009;388:752–756. doi: 10.1016/j.bbrc.2009.08.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patel BB, Yu Y, Du J, Rishi AK, Sarkar FH, Tarca AL, Wali A, Majumdar AP. Schlafen 3, a novel gene, regulates colonic mucosal growth during aging. Am J Physiol Gastrointest Liver Physiol. 2009;296:G955–G962. doi: 10.1152/ajpgi.90726.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gioeli D, Mandell JW, Petroni GR, Frierson HF, Jr, Weber MJ. Activation of mitogen-activated protein kinase associated with prostate cancer progression. Cancer Res. 1999;59:279–284. [PubMed] [Google Scholar]
- 46.Condamine T, Le Luduec JB, Chiffoleau E, Beriou G, Louvet C, Heslan M, Tilly G, Cuturi MC. Characterization of Schlafen-3 expression in effector and regulatory T cells. J Leukoc Biol. 2010;87:451–456. doi: 10.1189/jlb.0609410. [DOI] [PubMed] [Google Scholar]
- 47.Katsoulidis E, Carayol N, Woodard J, Konieczna I, Majchrzak-Kita B, Jordan A, Sassano A, Eklund EA, Fish EN, Platanias LC. Role of Schlafen 2 (SLFN2) in the generation of interferon alpha-induced growth inhibitory responses. J Biol Chem. 2009;284:25051–25064. doi: 10.1074/jbc.M109.030445. [DOI] [PMC free article] [PubMed] [Google Scholar]