Abstract
Diffusion-weighted imaging coupled with tractography is the only method for in vivo mapping of human white-matter fascicles. Tractography takes diffusion measurements as input and produces a large collection of white-matter fascicles as output; the connectome. We introduce a method to evaluate the evidence supporting connectomes. Linear Fascicle Evaluation (LiFE) takes any connectome as input and predicts diffusion measurements as output, using the difference between the measured and predicted diffusion signals to measure prediction error. Finally, we introduce two metrics that use the prediction error to evaluate the evidence supporting properties of the connectome. One metric compares the mean prediction error between alternative hypotheses, and the second metric compares full distributions of prediction error. We use these metrics to (1) compare tractography algorithms, and (2) test hypotheses about tracts and connections.
Keywords: Myelin, Brain connectivity, Glia, Connection, Cortex, Cortical, Computational neuroanatomy, Neuroanatomy, Anatomy, Brain map, Forward modeling, Connectomics, Brain Networks, Mapping the brain, Connectivity matrix, Optimization, cross-validation, statistics, nonparametric
Introduction
In a remarkable advance over the last decade, MR diffusion imaging methods and tractography algorithms now estimate the trajectories of white-matter fascicles (tracts) in the living human brain1–3. Because these measurements are in the living human brain, they can be used to clarify the relationship between the tract tissue properties and behavior, cognition, and development as well as to identify disease biomarkers. Experimental measurements combining behavior and diffusion show that tissue properties of specific tracts are correlated with a range of cognitive abilities. These tracts are living wires whose properties change during development and in response to experience. For example, changes in tissue property are correlated with developmental progress, language, cognition, decision making, disease and trauma4–9.
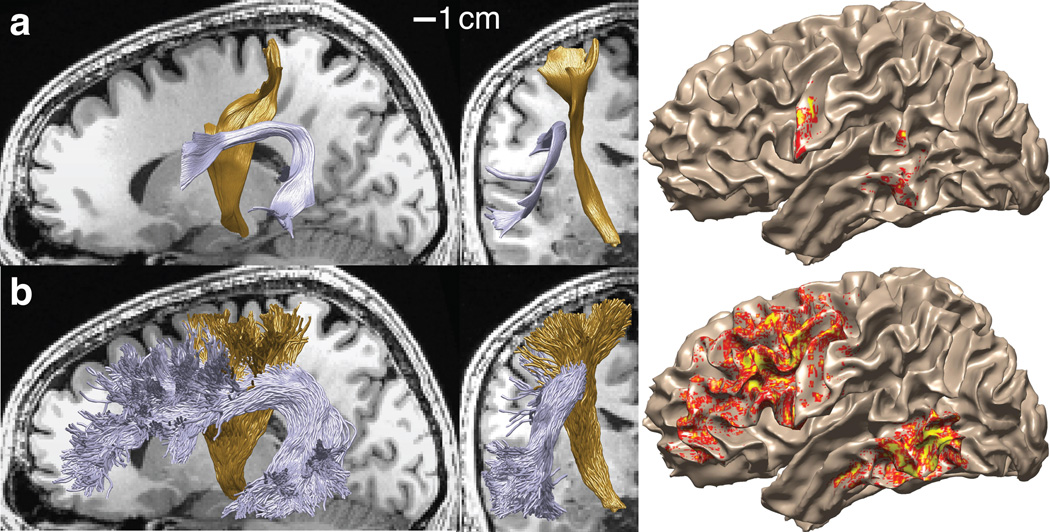
The complete collection of white matter tracts and connections in a large volume is called a connectome10,11. A key limitation of current practice concerns how to establish confidence in the connectome and specific fascicles within the connectome. Establishing confidence is important because in current practice selecting different parameters or different algorithms produces substantially different candidate connectomes (Fig. 1). The estimates differ in ways that are very meaningful to neuroscientists12–14. Investigators are interested in measuring the strength of the evidence supporting each of the models for this particular individual with this particular instrument.
Figure 1.
Tract trajectory estimates and cortical projection zones differ markedly between tractography algorithms. (a) Deterministic tractography. Sagittal and coronal views of the arcuate fasciculus (purple) and cortico-spinal tract (gold) in a candidate connectome generated using deterministic tractography. The density with which the fiber projects onto the cortical surface are indicated by the color overlay. Yellow indicates highest fiber density. (b) Probabilistic tractography. As in part (a) except for a candidate connectome generated using probabilistic tractography. Supplementary Fig. 1 shows another example of the same problem.
We introduce a method that reduces a candidate connectome containing many fascicles to an optimized connectome. Fascicles are retained for the optimized connectome only if they are needed to predict the diffusion data. We then present two sets of results that rely upon this method. The first set shows that the optimized connectome predicts the diffusion data accurately and analyzes the properties of the optimized connectome. The second set introduces methods that evaluate the strength of the evidence concerning specific tracts and connections. The methods that create the optimized connectome and evaluate the evidence in support of connectome properties are the main contributions of this paper. Software implementation of the method provided at https://francopestilli.github.io/life. Data provided at http://purl.stanford.edu/cs392kv3054.
Results
LiFE: Linear Fascicle Evaluation
Deriving the optimized connectome
Tractography algorithms use diffusion-weighted images to derive many fascicles that comprise the candidate connectome. Conventional tractography generates fascicles one at the time; these algorithms do not assess how well the full connectome predicts the diffusion data.
The Linear Fascicle Evaluation (LiFE) algorithm evaluates how well the entire connectome fits the white-matter diffusion data. The method solves a set of simultaneous linear equations (see Supplementary Fig. 2 and Methods) to estimate a weight for each fascicle; this weight describes the fascicle’s contribution towards predicting the data. These equations are solved by non-negative linear least squares algorithms15. Only fascicles with positive weight are retained (see Methods and Supplementary Fig. 2). This connectome evaluation is global: The fascicles in the optimized connectome all contribute to predict the diffusion data measured in the full white matter volume.
The LiFE method can be applied to a candidate connectome created with any number of fascicles, parameter settings and tractography algorithm; the method identifies the subset of fascicles that are supported by the diffusion data.
Prediction accuracy of the optimized connectome
We evaluated the prediction accuracy using cross-validation (Fig. 2). In the examples here and below, we began with candidate connectomes generated using constrained spherical deconvolution based probabilistic tractography16 (Lmax= 10, 5×105 seeds). We analyzed data from two different data sets. Dataset STN150 was obtained using 150 angles and 2 mm isotropic spatial resolution, b-value of 2000s/mm2. Dataset STN96 includes 96 diffusion directions at 1.5mm3, b-value 2000s/mm2. Both of these are whole-brain connectomes. In each case, the candidate connectome contains 500,000 fascicles.
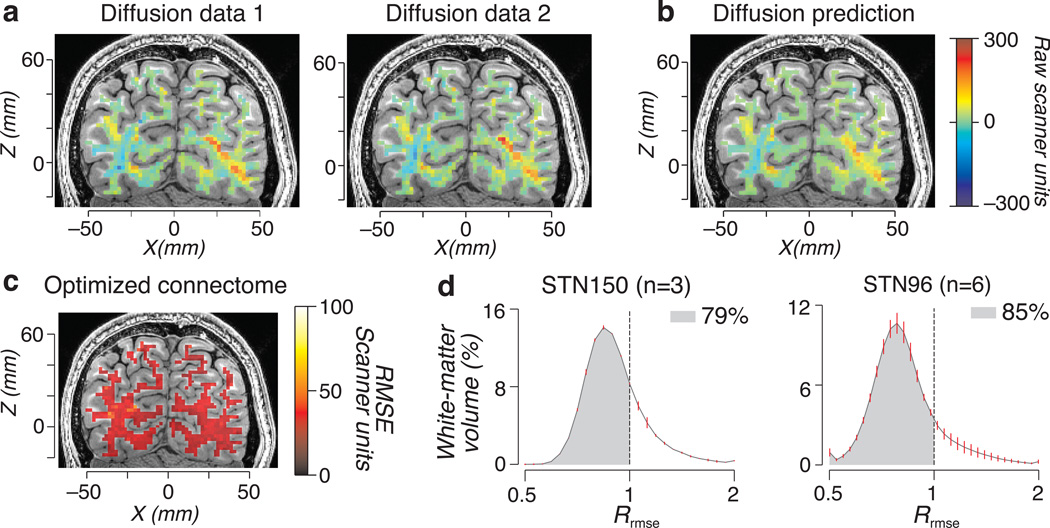
Figure 2.
Measured and predicted diffusion and LiFE model prediction accuracy. (a) Maps of measured diffusion modulation (Diffusion data 1 and 2, D1 and D2 respectively in the main text) in a typical coronal brain slice and for a single diffusion direction. (b) Map of predicted diffusion modulation by LiFE (synthetic MRI signal; Eq. 7) in for the same brain slice and diffusion direction. Connectome generated with CSD-based probabilistic tractography with Lmax=10 STN150 data set (n=1). (c) Distribution of LiFE model RMSE. Representative coronal brain slice; the color overlay shows the cross-validated model error (Mrmse) for the CSD-based probabilistic tractography with Lmax=10 using the STN150 data set. The Mrmse is uniformly distributed across the white-matter tissue. (d) Histograms of the percent of white-matter volume with a certain Rrmse for the STN150 (left, n=3) and STN96 (right, n=6) directions data sets. The voxel-wise Rrmse model predicts better than data (< 1) in more than 70% of the total white-matter volume. Error bars indicate ±1 s.e.m. across brains. Supplementary Fig. 2 shows a visual representation of LiFE model and extends the results to other data sets and connectomes.
Fig. 2 (and Supplementary Fig. 2) shows the measured and predicted diffusion modulation, i.e., the diffusion relative to the mean isotropic signal in each voxel for one diffusion direction. Data were divided into two parts, D1 and D2 (Fig. 2a). We tracked and estimated the fascicle weights that best fit D1. We used the connectome and the estimated weights from D1 to predict the diffusion signal in D2 (Fig. 2b). We use the root mean squared error (RMSE; Fig. 2c) between the prediction and D2 to measure prediction error. The RMSE is uniformly distributed across the white matter (Fig. 2c).
The optimized connectome performs better than test-retest reliability
We compared the model prediction to the test-retest reliability of the data, which is the RMSE between the two data sets, Drmse. The model prediction error (Mrmse) is the RMSE between the connectome prediction and D2. We compared the model and test-retest reliability at each voxel using the ratio: Rrmse = Mrmse / Drmse. When Rrmse is below one, the model predicts D2 more accurately than D1 predicts D2. For most voxels Rrmse is less than 1 (Mrmse is lower than Drmse). More than 70% of the voxels have an Rrmse less than 1 (Fig. 2d and Supplementary Fig. 2). In summary, the connectome model predicts the second data set slightly more accurately than assuming that the second data set equals the first.
Fascicle lengths and weights
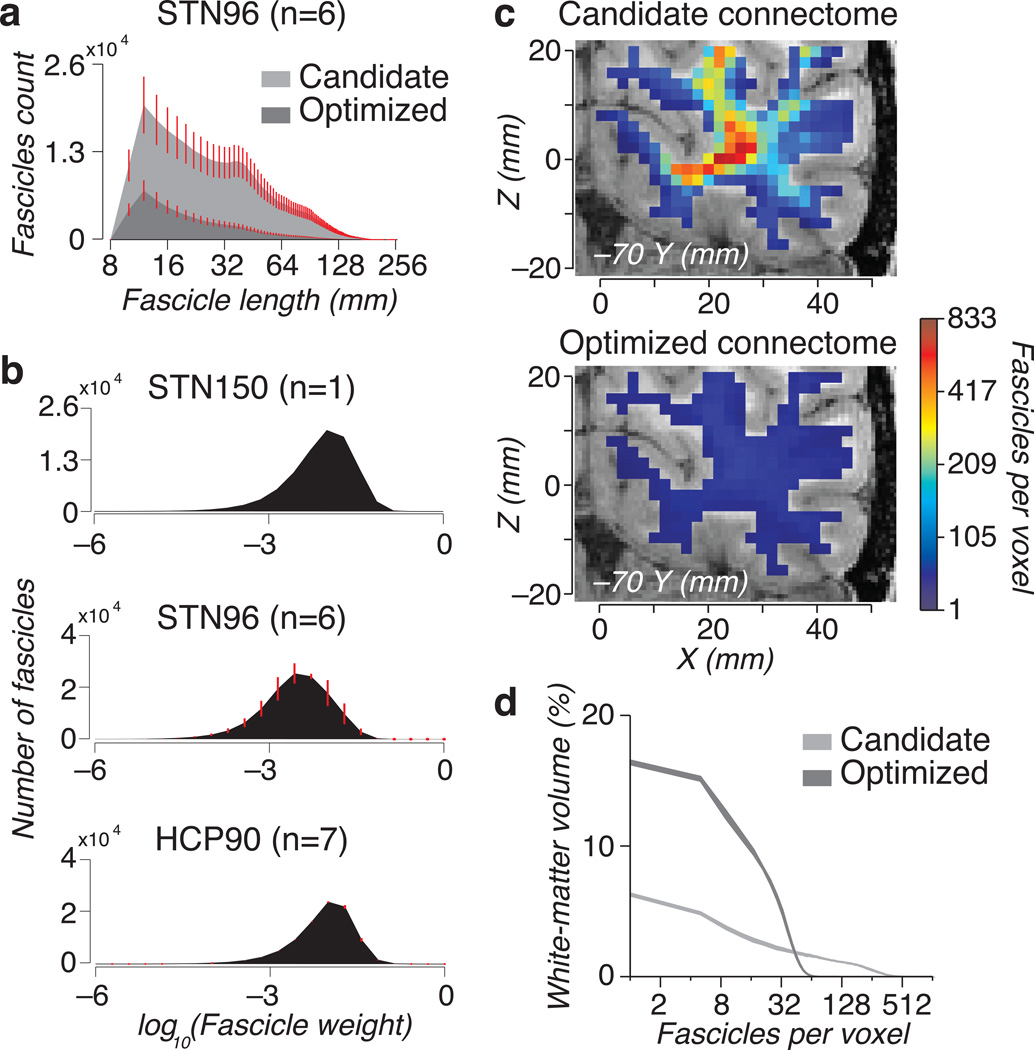
The candidate and optimized connectomes include many more short (1–5 cm) than long (10 cm) fibers (Fig. 3a and Supplementary Fig. 3). Histology shows17,18 that there are vastly more short fibers (< 1 cm), but these are not picked up by the tractography. Optimizing the connectome reduced the count in long and short fascicles approximately equally.
Figure 3.
Properties of the LiFE solution. (a) Fascicle length distribution. Histogram of fascicles length for candidate and optimized connectomes (STN96 data set, Probabilistic CSD Lmax=10) averaged across six brains. (b) Distribution of fascicles weights. Top. Weights for a single connectome for the STN150 data set. Middle, weights averaged across six connectomes for the STN96 dataset. Bottom. Weights averaged across seven connectomes for the HCP90 dataset, these connectomes contain two large portions of the left a right occipital and temporal lobes (see Methods). Error bars indicate ±1 s.e.m. across brains. (c) Fiber density maps. Maps for the candidate (left) and optimized (right) connectome are overlaid on a coronal brain slice (STN150). Fiber density near the gray matter is similar. The optimization reduces the density in the core of the white matter by a factor of 10. Optimizing the connectome equalizes fascicle density across the white matter. The optimized connectome fascicle density is below 100 across the whole-slice and the color appears as a uniform blue. (d) Candidate and optimized connectome fascicle density histogram. Fiber density histogram for the candidate (light gray) and optimized (dark gray) connectomes (STN96). Fascicle density and dynamic range are reduced by the LiFE model. Line width indicates ± 1 standard deviations across six brains. Supplementary Fig. 3 extends the results to other data sets and reports additional analyses of the number of fascicles as a function of number of measured diffusion directions.
The number and values of the positive weights in the optimized connectome will depend on the spatial resolution, angular resolution, and signal-to-noise in the acquired data. The number of positive weights for STN96 and STN150 were both about 95,000. For all three data sets (STN150, STN96 and HCP90), the fascicle weight distribution is approximately symmetric on a log-weight axis (Fig. 3b). There is a large range of assigned weights. Some fascicles contribute more to the predictions than others, and their weights can be as much as two orders of magnitude greater.
Fascicle density
Axon density varies across the white matter. For example, in the corpus callosum the axon density range is about 1.3:119. Examining only the axons with a diameter greater than three microns, the range can be as large as 30:119. Yet, typical candidate connectomes can have ranges on the order of 300:1 or more20 (Fig. 3c,d and Supplementary Fig. 3). This range exceeds the range observed in biological material. Recently, investigators considered principled methods to bring the dynamic range of the fascicles into biologically plausible ranges20–22.
The optimized connectome fascicle density is close to a biologically plausible level (Fig. 3d). The optimized connectome has a much smaller dynamic range because many fibers in the core of the white matter are eliminated. The number of fascicles per voxel in the white matter adjacent to cortex remains stable.
Naturally, any change in the power of the data - such as reducing the number of directions, making the sample size coarser, or decreasing the SNR - will impact how many of the fascicles are supported by the measurements and thus retained in the optimized connectome. This is an important characteristic of the procedure as it allows researchers to evaluate the acquisition parameters that are needed to measure specific tracts (Supplementary Fig. 3c).
Statistical inference on models, tracts and connections
Comparing connectome models
Using LiFE, we can compare the prediction error of optimized connectomes derived from different tractography methods. There will no single tractography method optimal for all data acquisitions (e.g., 30-direction on a 1.5T scanner vs. 256-direction on a 3T scanner). Hence it is crucial to have a method to select the optimal algorithm for particular data sets and research question.
We illustrate how to use LiFE to compare connectome models using two tractography methods16. In one case we used a tensor-based deterministic algorithm16,23,24. In the second case we used probabilistic tractography based on constrained spherical deconvolution (Lmax=1016,25,26). The resulting connectomes are very different (Fig. 1 and Supplementary Fig. 1). One important difference is that the probabilistic candidate connectome spans the entire white-matter volume; the deterministic connectome spans about 80% of the voxels in the white matter volume.
Next, we compared the voxel-wise prediction error (RMSE) from the optimal probabilistic and deterministic connectomes (Fig. 4a). In the voxels without any fascicles from the deterministic connectome, we set the diffusion signal prediction to an isotropic diffusion signal. For most voxels the RMSE is higher for the tensor-based deterministic algorithm than the CSD-based probabilistic algorithm (Fig. 4b). This quantifies the widely held belief that tensor-based deterministic algorithms fail to capitalize on all the information present in the data.
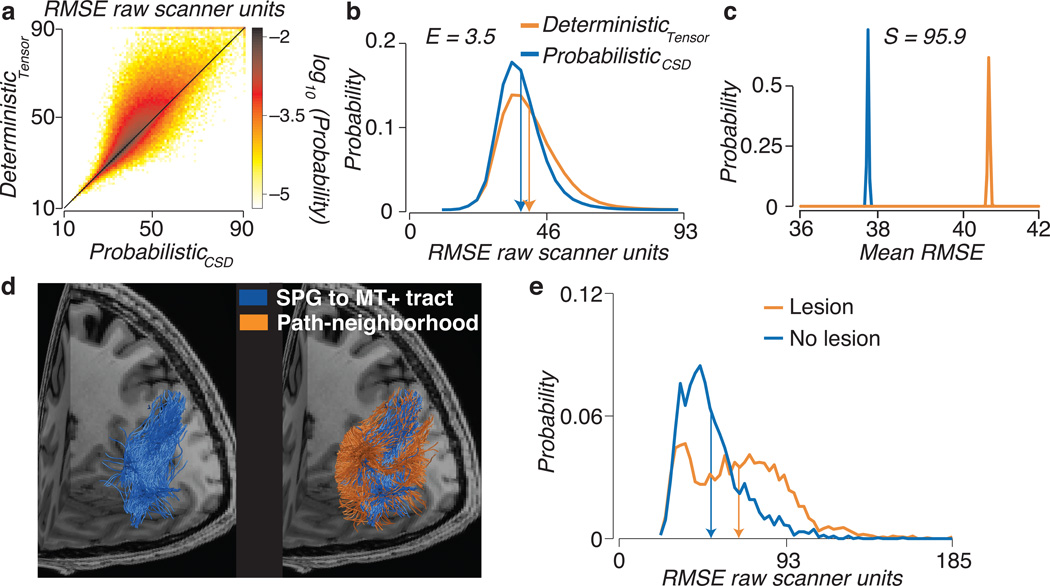
Figure 4.
Comparison of connectome models and virtual lesion method. (a) Voxel-wise prediction error. Error is show for whole-brain probabilistic (CSD, Lmax=10) and tensor-based deterministic optimized connectomes. Each white matter voxel has a cross-validated RMSE for both the optimal probabilistic and deterministic connectomes. The scatter-density plot compares these two values and shows that in a large majority of the white matter voxels the deterministic errors exceed the probabilistic errors. (b) Distribution of RMSE for probabilistic (CSD, Lmax=10) and tensor-based deterministic optimized connectomes. Histogram of RMSE for the two tractography models. The mean of each distribution is shown as vertical arrow. The Earth Movers Distance (E) quantifies the lower error for the Probabilistic model. (c) Distribution of mean RMSE. Representative bootstrap distribution of the mean RMSE. The Mean RMSE is higher for the deterministic than probabilistic connectome. There is very strong evidence favoring the probabilistic connectome over the deterministic connectome, S=95.9. All plots are for one brain data set STN96. See Supplementary Fig. 4 for additional information about S and E. (d) A novel pathway connecting hMT+ and the superior parietal gyrus (SPG) identified using a virtual lesion. Left. White matter fascicles intersecting hMT+ and SPG in the left hemisphere of one individual brain. Right. Path-neighborhood, fascicles sharing voxels with the hMT+ / SPG tract. (e) Distribution of RMSE for lesioned and unlesioned models. The RMSE increases (orange) as the hMT+ to SPG tract is removed. See Supplementary Fig. 4 for additional subjects and data sets.
We used a bootstrap method27 to quantify the strength of the evidence showing that the mean RMSE of the probabilistic connectome is smaller than the mean RMSE of the deterministic connectome (Fig. 4c). We resampled (with replacement) the distribution of RMSE values and computed the mean RMSE of each resample. The distribution of mean RMSE values for the deterministic algorithm is higher than the mean RMSE from the probabilistic algorithm. We summarize the strength of the evidence for one model versus the other by using a measure of distance between these two distributions, S.. We calculate the distance as the difference in the two means (μ) divided by their pooled standard deviations (σ).
The values and are the bootstrapped means and variances of the RMSE for deterministic and probabilistic connectomes, respectively. The denominator is a conservative upper-bound estimate of the standard deviation of the distribution of mean RMSE pooled between deterministic and probabilistic connectomes28. The distance between the means, S, is on the order of 90 standard deviation units, a measure that is equivalent to d-prime. This indicates that the evidence is very strong that the mean RMSE of the probabilistic connectome is smaller than that of the deterministic connectome for this data set and instrument. This result is consistent across data sets (Supplementary Fig. 4) suggesting that in many measurement conditions probabilistic tracking is superior.
In addition to the mean RMSE, we can compare the complete RMSE distributions of the two models (Fig. 4b). We evaluated three additional metrics to compare RMSE distributions: the Earth Mover’s Distance29, Kullback-Leibler divergence, and Jeffrey’s divergence (Supplementary Fig. 4). The strength of evidence (S) and Earth Movers Distance (E) are both informative and reliable across a range of experimentally plausible conditions. Hence, we use these two measures for statistical evidence.
The virtual lesion method: evidence for a brain connection
In this section we explain how to use the connectome model to measure the strength of the evidence supporting the existence of specific tracts connecting different brain regions.
The LiFE algorithm requires that fascicles in the optimized connectome contribute to the data prediction; removing any fascicle from the optimized connectome increases model prediction error. The impact on error depends on the number and weights of the removed fascicles. Removing very few fascicles, or only removing fascicles with small weights, increases error slightly. But removing a large number of fascicles or fascicles with large weights increases error greatly. We use the magnitude of the error increase to measure the strength of the evidence supporting the existence of any specific set of fascicles. We describe the analysis of fascicle removal as a virtual lesion.
Here illustrate a virtual lesion using an example tract connecting the Superior Parietal Gyrus (SPG) and human MT+30,31. First, we created a candidate connectome using MRtrix (Lmax=10, 1,500,000 fibers). Second, we identified all fascicles terminating in the SPG and hMT+. Third, we identified the path-neighborhood of the fascicle, a useful concept introduced by Wedeen and colleagues13. Suppose that f is the set of fascicles under test in the optimized connectome, for example the SPG-hMT+ tract (Fig. 4d). The fascicle passes through a set of white matter voxels, v(f). The collection of all the other fascicles that pass through at least one of the voxels in v(f) is the path-neighborhood of f. We refer to the path-neighborhood with the symbol F (Fig. 4d). Only the fascicles in F contribute to the model prediction of the diffusion signal in v(f).
We measured the strength of the evidence in favor of the existence of the fascicles, f, using a bootstrap method27. We calculated the RMSE of the predicted diffusion signal in each voxel, v(f), using the path-neighborhood, F. We then calculated the RMSE using a model in which we remove the fascicles in f, F’ = F − f. The histograms in Fig. 4e compare the RMSE distribution for the path-neighborhood v(f) for the lesioned and unlesioned model. Supplementary Fig. 4 shows similar results for additional subjects and reports the mean values for S and E across subjects and datasets STN96, HCP90.
The analysis supports the existence of a white-matter tract between the SPG and hMT+, which has not been reported in humans. The tract is contained within the larger vertical occipital fasciculus32,33. This anatomical evidence in the living brain is consistent with functional data and post-mortem dissections in human34–36 and macaque30,31,37.
LiFE confirms twenty major white-matter tracts
We calculated the strength of the evidence in favor of the existence of each of twenty long tracts that are known to exist in the human brain3 (n=5). The S and E values strongly support the existence of all twenty major tracts (Fig. 5 and Supplementary Fig. 5).
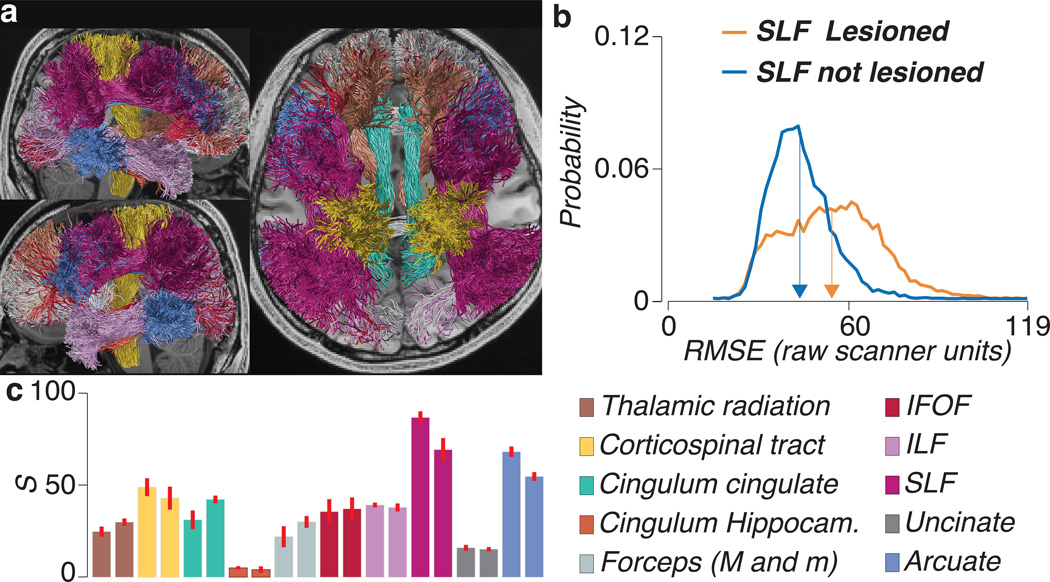
Figure 5.
Major white-matter fascicles are supported by LiFE. (a) Twenty major human white-matter tracts the optimized connectome (STN96, CSD probabilistic Lmax=10). (b) Distribution of cross-validated RMSE of the optimized connectome (F), and the lesioned optimized connectome without the SLF (F’ = F − f). RMSE is larger for F’. (c) Mean strength of the evidence (S) across five brains supporting the existence of each of the twenty major tracts shown in a. Bar location matches hemisphere (left and right respectively). Error bars indicate ±1 s.e.m. across brains. See Supplementary Fig. 5a for the results using the Earth Movers distance. Supplementary Fig. 5b reports additional analyses showing that LiFE alters the matrix of brain connections.
Further, these twenty tracts comprise twelve left-right pairs. In each case, the strength of the evidence supporting the corresponding left and right tract is very similar, and approximately consistent with their size. The SLF and arcuate are both large tracts and there is more data supporting their existence. The Uncinate and Cingulum projecting to the hippocampus are somewhat smaller and the S values for these tracts are smaller. Whereas the strength of evidence computed using S depends on the size of the tract, the characteristics of the data acquisition (e.g., signal-to-noise), and the effect of the lesion, E depends only on the data and the lesion effect size (see Supplementary Fig. 4 and 5). This analysis validates LiFE by showing that it confirms the existence of known white matter tracts.
Supplementary Fig. 5 also shows that LiFE can alter the matrix of connectivity between brain regions.
Discussion
Connectome generation
Investigators can choose from a large set of diffusion measurements and tractography algorithms to generate white-matter fascicles (Supplementary Table 1). Each algorithm generates connectomes using its own set of theoretical principles and heuristics, and the choice of parameters and algorithms has a significant impact on the tract estimates (Fig. 1). LiFE is a method that is applied to the white matter tracts from any tractography algorithm. The method helps the investigator measure the strength of the evidence supporting the existence of specific candidate tracts.
Tractography estimates can fail if they miss a real fascicle or generate a fascicle that does not exist (false alarm). The LiFE method reduces false alarms (Fig. 3). But it is not a tractography algorithm, and thus LiFE does not supply missed tracts. Consequently, when using LiFE investigators should begin with large candidate connectomes that are created to minimize misses but tolerate false alarms.
Global tractography
The first algorithms for tractography were deterministic, local and greedy38. Following this early work, probabilistic formulations were introduced to account for uncertainty16,25,26. Also, non-local algorithms that operate on more than a single voxel at a time were proposed39–43. These and most tractography algorithms combine connectome generation with some form of indirect evaluation39–41.
Sherbondy et al.43,44 suggested separating connectome generation and evaluation algorithms. This separation enables investigators to measure the accuracy of predictions derived from different global tractography algorithms and to measure the accuracy of connectomes in predicting diffusion data. The principle of separating connectome generation and evaluation is adopted in several recent publications45.
Global tractography algorithms can be further subdivided into two types, fascicle-global and connectome-global. Most global tractography algorithms are global in the sense that they consider constrains to path generation computed over entire fascicles; but they do not constrain path generation over the complete connectome39–41,44. The principal goal of fascicle-global algorithms is to impose some degree of smoothness on the fascicle path. Connectome-global algorithms generate paths by constraining entire connectomes21,43. The goal of these algorithms is path generation; they do not provide mechanisms for evaluating tractography solution. LiFE provides a computational algorithm and a statistical inference framework that can evaluate virtually any connectome against the measured diffusion data.
Connectome model validation
One approach to validation asks whether the tract estimates conform to measurements made using another method12,14,46,47. LiFE makes a different and equally important assessment: How much support exists for the tract estimates in the present data? We consider the utility of these two different types of validation.
The most common approach to connectome validation is to compare tractography with another method. First, validation has been performed qualitatively by comparing tract estimates in individual ex-vivo brains using both diffusion tractography and histology48,49. This method has various limitations. It is extremely time consuming; the neural tissue is distorted and shrunk during the histological preparation; and the method can only be applied to a few cubic millimeters of white-matter volume of a single pathway50. Second, tractography estimates from probabilistic tractography in-vivo also can be compared with blunt dissection in ex-vivo specimens. For example, the overall shape and length of the optic radiation obtained with probabilistic tractography in living brains agree well with estimates obtained using ex-vivo data44. Third, tractography algorithms are evaluated using artificially constructed physical or simulated phantoms of the fibers51–53.
Using a separate method to confirm the existence of tracts is helpful for establishing general confidence in tractography. But this approach fails to inform us about the strength of the evidence in a specific data set. There is value in knowing that a tract estimated from a child at 3T is also found in an ex-vivo brain measurement at 7T. But it is fundamental to measure the degree of support for the presence or absence of a specific tract with specific anatomy in the 3T data from that child. What if this child has developed a tumor that changed the shape of the tract? Or the same child undergone radiation therapy for a cancer treatment that affected the white-matter in the tract54? We would like to know the strength of the evidence given the data in this particular subject using this particular instrumentation. LiFE establishes confidence about tracts and data obtained with specific instruments and subjects, rather than relying on related measurements made with other instruments, methods and specimens.
Using LiFE investigators can compare the accuracy of different connectomes and quantify the support in the data for specific tracts. New computational methods in quantitative MRI will clarify much more about the tissue properties of these tracts55. Taken together, the measurements and algorithms are helping us to build a complete model of the locations, organization, and tissue properties of the human connectome. Because this work is being done in human, we have an excellent opportunity to see the relationship between the connectome and the human mind.
Online Methods
STN96 and STN150 data sets: Diffusion-weighted MRI acquisition
Magnetic Resonance Imaging diffusion-weighted data (DWI) were collected at Stanford’s Center for Cognitive and Neurobiological Imaging (www.cni.stanford.edu). We collected data in six males, age 37 – 39 using a 3T General Electric Discovery 750 (General Electric Healthcare, Milwaukee, WI) equipped with a 32-channel head coil (Nova Medical, Wilmington, MA). Data collection procedures were approved by the Stanford University Institutional Review Board. Written consent was collected from each participant. Stanford 96 diffusion directions data set (STN96): For six subjects we acquired two diffusion weighted scans within a single scan session. Water diffusion was measured at 96 different directions across the surface of a sphere as determined by the electro-static repulsion algorithm of Jones, Horsfield, & Simmons56. In all subjects, data were acquired at 1.5 mm3 spatial resolution and diffusion gradient strength was set to 2000 s/mm2 (TE 96.8 msec). We used a dual-spin echo diffusion-weighted sequences with full head coverage. Individual data sets were acquired with using two excitations (nex = 2) that were averaged in k-space. We obtained 10 non-diffusion weighted (b=0) images at the beginning of each data set. The signal-to-noise-ratio calculated over repeats of the non-diffusion images was greater than 20 in all data sets. Stanford 150 diffusion directions data set (STN150): For one subject we acquired multiple data sets with 150 directions at 2 mm3 spatial resolution and b values of 1000, 2000 and 4000 s/mm2 (TE 83.1/93.6/106.9 msec).
MRI images for STN96 and STN150 were corrected for spatial distortions due to B0 field inhomogeneity. To do so we measured the B0 magnetic field maps. Field maps were collected in the same slices as the functional data using a 16-shot, gradient-echo spiral-trajectory pulse sequence. Two volumes were successively acquired one with TE set to 9.091 ms and the other with TE increased by 2.272 ms, and the phase difference between the volumes was used as an estimate of the magnetic field. To track slow drifts in the magnetic field (e.g. due to gradient heating), field maps were collected before, after and between the two diffusion scans.
Subjects’ motion was corrected using a rigid body alignment algorithm57. Diffusion-gradients were adjusted to account for the rotation applied to the measurements during motion correction. The dual-spin echo sequence we used does not require eddy current correction because it has a relatively long delay between the RF excitation pulse and image acquisition. This allows for sufficient time for the eddy currents to dephase. Pre-processing are publicly available as part of the vistasoft software distribution (http://github.com/vistalab/vistasoft/mrDiffusion).
HCP90 data set
We used 7 brains with DWI data downloaded from https://www.humanconnectome.org/data/58. Measurements from the 2000 s/mm2 shell were extracted from the original data and were used for further analyses. Processing methods described in the following articles are applied to all HCP open access pre-processed diffusion data59.
Anatomical MRI acquisition and tissue segmentation
The white- and gray-matter border was defined using a 0.7 mm3 T1-weighted FSPGR image. White/gray matter tissue contrast was increased by averaging two T1 measurements acquired in the same scan session. An initial segmentation was performed using an automated procedure (Freesurfer60) and refined manually (http://www.itksnap.org/pmwiki/pmwiki.php).
Generating whole-brain connectomes and tracts
Fiber tracking was performed using MRtrix61. Diffusion-weighted images were motion compensated and aligned to the high-resolution T1-weighted anatomical. The total white-matter volume was identified from the cortical segmentation (see above) and resampled at the resolution of the diffusion data. The white-matter volume was used as seed region for fiber tracking. We tested three tracking methods implemented within MRtrix: (1) Tensor-based deterministic tractography61–63, this methods requires fitting a tensor at each voxel and tracking using the principal diffusion direction identified from the tensor. (2) Constrained-spherical deconvolution (CSD) based deterministic tractography, this method requires modelling the fiber orientation distribution function in each voxel using a constrained-spherical deconvolution method61 and using the fiber orientation direction within voxels as direction of tracking. (3) Finally we used CSD-based probabilistic tracking61,64,65. We tested a range of maximum harmonic order (Lmax=6–12), which determines the maximum number of deconvolution kernels utilized to estimated the fODF at each voxel by the CSD model (step size: 0.2 mm; minimum radius of curvature: 1 mm; maximum length: 200 mm; minimum length: 10 mm; fODF amplitude cutoff: 0.1). Results were qualitatively similar across the Lmax values. In the text we compare results between two tracking methods. The highly used tensor-based deterministic tracking and the more recent probabilistic tractography based on CSD (Lmax=10).
For each tractography method, data set (STN150: b-value 1000, 2000 or 4000 s/mm2 with 150 directions, STN96 b-value of 2000 s/mm2 with 96 directions and HCP90 b-value 2000 s/mm2 with 90 directions) and subject we created one whole-brain connectomes with 500,000 fascicles each. We repeated the analysis independently for each brain in each data set. Connectomes for the HCP90 data set were restricted to the posterior portion of each brain (occipital, parietal and temporal lobe as well as the cerebellum). For some analysis on the STN150 data set (Supplementary Fig. 4 and 7) we used three connectomes restricted to the left occipital lobe to build independent LiFE models and repeated our analyses in each one of these three connectomes. This allowed us to test the robustness of the results within a single brain given by (1) The stochasticity of the placement of the seed within the total white-matter volume to initiate fiber tracking. (2) The stochasticity introduced by the probabilistic tracking algorithm in generating a connectome. As reported in the main text Results and in the Supplementary Figs. 3, 4 and 7 (see error bars), results were quantitatively indistinguishable across repeated tracking, and fitting of LiFE models.
Segmenting the connections terminating in specific regions of interest
To identify connections between different brain regions (hMT+, superior parietal gyrus, and primary visual cortex, Fig. 8 and 9), we: (1) performed a whole-brain automatic parcellation using FreeSurfer66. (2) We used the cortical regions for primary visual cortex (V1) and hMT+ using the cortical regions provided by FreeSurfer67 by transforming them to the space of the diffusion data. (2) We expanded these regions to cover portions of the white matter adjacent to each cortical area by applying a 3 dimensional Gaussian smoothing with a spatial kernel of 3 mm3. (3) We identified the fascicles in the whole-brain connectome with termination (endpoints) inside these expanded ROIs. (4) The anterior commissure was identified manually in each subject from the high-resolution anatomical image. We selected fascicles that intersected a 5 mm-diameter sphere centered at the location of the anterior commissure.
Segmenting the major white-matter fascicles
We segmented the major white-matter fascicles using AFQ68.
Visualization of white-matter fascicles and brain anatomy
Tracts and brain images were generated using Matlab Brain Anatomy (MBA) https://github.com/francopestilli/mba.
Predicting the diffusion signal within a voxel
A complete diffusion MRI experiment measures brain volumes with and without diffusion sensitization. Diffusion sensitization (a combination of diffusion gradient strength, duration of the diffusion gradient and the interval between the pulses of the diffusion gradient) is denoted as b. In a typical experiment one might measure in 100 diffusion directions, θ, and 10 non-diffusion measurements (b = 0)69. We represent direction, as a three-dimensional, unit length, column vector.
Suppose the non-diffusion signal at a voxel is S0 and the diffusion signal in the direction θ and the presence of a gradient, b, is S(θ, b). The diffusion signal in a particular direction using a specific gradient strength (b) is specified using the following equation70
| (1) |
where A(θ) is the apparent diffusion coefficient in the direction θ. For a simple shape, such as an idealized cylinder that represents a short segment of a fascicle, f, the apparent diffusion coefficients in different directions can be summarized by the quadratic formula71
| (2) |
Equation (2) states that for a single small segment of a fascicle the apparent diffusion in any direction can be computed using a matrix Q, whose entries depend on the local fascicle orientation. The matrix is a 3×3 positive-definite quadratic form, which means that there is an invertible matrix M such that Q = Mt M. It follows that Q is symmetric and θt Qθ > 0.
The expected diffusion in a specific voxel, ν, completely filled by one fascicle, f, is
| (3) |
An important special case is the purely isotropic compartment, A0, (i.e., equal diffusion in all directions, θ). This corresponds, say, to the portion of the voxel containing cerebrospinal fluids, astrocytes and other tissue. A typical voxel is likely to contain a combination of fascicles and these isotropic tissues. We express the predicted diffusion signal from a single voxel as the weighted sum of the contributions from the fascicles in the compartment and the isotropic term We can rewrite Equation (4), the diffusion in a voxel, as the sum of an isotropic term and the sum of orientation dependent functions from each fascicle
| (5) |
The isotropic term, Iν, is simply the mean diffusion signal in the voxel
| (6) |
The fascicle-specific function, Of(θ), is anisotropic with zero mean. It describes the modulation of the diffusion signal around its mean.
| (7) |
The fascicle weights in a voxel are estimated by first subtracting Iν from the diffusion signal and then solving the linear equation for the values, wf, which minimize
| (8) |
The difference between the measured diffusion signal and the mean diffusion signal is
| (9) |
The predicted signal modulation based on the fascicles in v is
| (10) |
Estimating fascicle weights from the connectome
Finally, we solve for the fascicle weights by minimizing the error across all the voxels in the connectome, C. Specifically, we find wf that minimize the expression
or equivalently
| (11) |
Supplementary Fig. 2b shows Equation 11 in matrix tableau. We solve for the non-negative weights using the algorithm defined in72.
There are many possible variants of this formulation. For example, it is possible to impose additional minimization constraints (e.g., sparsity or uniformity on the weights), or even to allow the weights to vary along the fascicle path. We are experimenting with several of these alternatives, but the formulation introduced here has proved to be a useful starting point for further development.
In summary, the connectome model is expressed as a minimization with respect to a large set of linear equations (Eq. 11). The matrix representing the connectome model is sparse, and for our high-resolution data sets the matrix row size is (Nν × Nθ), about 7,000,000 for the STN150 data set and 40,000,000 for the STN96 data set). The matrix begins with a column size (Nf) of about 500,000 fascicles in the candidate connectome. The column dimension is reduced when the optimized connectome is reached. See Supplementary Fig. 2 for details on the matrix representation of the linear model for the connectome.
Supplementary Material
Acknowledgements
We thank M.L. Perry for assistance with collecting and preprocessing the data, M. Ben Schachar, R. Dougherty, J. Gardner, S. Ling, A. Mezer, A. Sherbondy, J. Winawer, H. Takemura, J. Solomon, L. Guibas, A. Butscher, C.Y. Zheng, R. Tibshirani and T. Hastie for useful comments and discussions. NEI F32 EY022294 US National Institute of Health to A.R. & US Natioanl Science Foundation BCS1228397 to B.W. Data were provided in part by the Human Connectome Project, WU-Minn Consortium (Van Essen, D. and Ugurbil, K., 1U54MH091657 US National Institute of Health).
Footnotes
Author contributions: FP, JY and BW designed the general approach. FP designed the statistical approach. FP implemented the software, with help from AR and BW. AR, FP, and JY collected the data. KK advised on fitting algorithms and statistics. FP and BW wrote the manuscript.
Competing financial interests
Authors declare no competing financial interests.
References
- 1.Zhang Y, et al. Atlas-guided tract reconstruction for automated and comprehensive examination of the white matter anatomy. NeuroImage. 2010;52:1289–1301. doi: 10.1016/j.neuroimage.2010.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yendiki A, et al. Automated probabilistic reconstruction of white-matter pathways in health and disease using an atlas of the underlying anatomy. Frontiers in neuroinformatics. 2011;5:23. doi: 10.3389/fninf.2011.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yeatman JD, Dougherty RF, Myall NJ, Wandell BA, Feldman HM. Tract profiles of white matter properties: automating fiber-tract quantification. PloS one. 2012;7:e49790. doi: 10.1371/journal.pone.0049790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeatman JD, Dougherty RF, Ben-Shachar M, Wandell BA. Development of white matter and reading skills. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E3045–E3053. doi: 10.1073/pnas.1206792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samanez-Larkin GR, Levens SM, Perry LM, Dougherty RF, Knutson B. Frontostriatal white matter integrity mediates adult age differences in probabilistic reward learning. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:5333–5337. doi: 10.1523/JNEUROSCI.5756-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomason ME, Thompson PM. Diffusion imaging, white matter, and psychopathology. Annual review of clinical psychology. 2011;7:63–85. doi: 10.1146/annurev-clinpsy-032210-104507. [DOI] [PubMed] [Google Scholar]
- 7.Lebel C, Walker L, Leemans A, Phillips L, Beaulieu C. Microstructural maturation of the human brain from childhood to adulthood. NeuroImage. 2008;40:1044–1055. doi: 10.1016/j.neuroimage.2007.12.053. [DOI] [PubMed] [Google Scholar]
- 8.Scholz J, Klein MC, Behrens TEJ, Johansen-Berg H. Training induces changes in white-matter architecture. Nature neuroscience. 2009;12:1370–1371. doi: 10.1038/nn.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoeft F, et al. Neural systems predicting long-term outcome in dyslexia. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:361–366. doi: 10.1073/pnas.1008950108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sporns O, Tononi G, Kotter R. The human connectome: A structural description of the human brain. PLoS computational biology. 2005;1:e42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hagmann P, et al. MR connectomics: Principles and challenges. Journal of neuroscience methods. 2010;194:34–45. doi: 10.1016/j.jneumeth.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 12.Craddock RC, et al. Imaging human connectomes at the macroscale. Nature Methods. 2013;10:524–539. doi: 10.1038/nmeth.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wedeen VJ, et al. The geometric structure of the brain fiber pathways. Science (New York, N.Y.) 2012;335:1628–1634. doi: 10.1126/science.1215280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Catani M, Bodi I, Dell'acqua F. Comment on "The geometric structure of the brain fiber pathways". Science (New York, N.Y.) 2012;337:1605. doi: 10.1126/science.1223425. [DOI] [PubMed] [Google Scholar]
- 15.Dongmin K, Suvrit S, Inderjit SD. A non-monotonic method for large-scale nonnegative least squares. Optimization Methods and Software. 2012;28:1–17. [Google Scholar]
- 16.Tournier J-D, Calamante F, Connelly A. MRtrix: Diffusion tractography in crossing fiber regions. International Journal of Imaging Systems and Technology. 2012;22:53–66. [Google Scholar]
- 17.Markov NT, et al. Cortical High-Density Counterstream Architectures. Science. 2013;342:1238406–1238406. doi: 10.1126/science.1238406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braitenberg V, Sch uz. A. Cortex: Statistics and Geometry of Neuronal Connectivity. Springer Berlin Heidelberg: 1998. [Google Scholar]
- 19.Aboitiz F, Scheibel AB, Fisher RS, Zaidel E. Fiber composition of the human corpus callosum. Brain research. 1992;598:143–153. doi: 10.1016/0006-8993(92)90178-c. [DOI] [PubMed] [Google Scholar]
- 20.Sherbondy AJ, Rowe MC, Alexander DC. MicroTrack: an algorithm for concurrent projectome and microstructure estimation. Medical image computing and computer-assisted intervention : MICCAI … International Conference on Medical Image Computing and Computer-Assisted Intervention. 2010;13:183–190. doi: 10.1007/978-3-642-15705-9_23. [DOI] [PubMed] [Google Scholar]
- 21.Smith RE, Tournier J-D, Calamante F, Connelly A. SIFT: Spherical-deconvolution informed filtering of tractograms. NeuroImage. 2013;67:298–312. doi: 10.1016/j.neuroimage.2012.11.049. [DOI] [PubMed] [Google Scholar]
- 22.Bastiani M, Shah NJ, Goebel R, Roebroeck A. Human cortical connectome reconstruction from diffusion weighted MRI: the effect of tractography algorithm. NeuroImage. 2012;62:1732–1749. doi: 10.1016/j.neuroimage.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 23.Basser PJ, Pajevic S, Pierpaoli C, Duda J, Aldroubi A. In vivo fiber tractography using DT-MRI data. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2000;44:625–632. doi: 10.1002/1522-2594(200010)44:4<625::aid-mrm17>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 24.Lazar M, et al. White matter tractography using diffusion tensor deflection. Human brain mapping. 2003;18:306–321. doi: 10.1002/hbm.10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Behrens TEJ, et al. Non-invasive mapping of connections between human thalamus and cortex using diffusion imaging. Nature neuroscience. 2003;6:750–757. doi: 10.1038/nn1075. [DOI] [PubMed] [Google Scholar]
- 26.Parker GJM, Haroon HA, Wheeler-Kingshott CAM. A framework for a streamline-based probabilistic index of connectivity (PICo) using a structural interpretation of MRI diffusion measurements. Journal of magnetic resonance imaging : JMRI. 2003;18:242–254. doi: 10.1002/jmri.10350. [DOI] [PubMed] [Google Scholar]
- 27.Efron B, Tibshirani R. An Introduction to the Bootstrap. Chapman \& Hall: 1993. [Google Scholar]
- 28.Green DM, Swets JA. Signal Detection Theory and Psychophysics. 1966 @doi.apa.org. [Google Scholar]
- 29.Rubner Y, Tomasi C, Guibas LJ. The earth mover's distance as a metric for image retrieval. International Journal of Computer Vision. 2000;40:99–121. [Google Scholar]
- 30.Maunsell JH, van Essen DC. The connections of the middle temporal visual area (MT) and their relationship to a cortical hierarchy in the macaque monkey. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1983;3:2563–2586. doi: 10.1523/JNEUROSCI.03-12-02563.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boussaoud D, Ungerleider LG, Desimone R. Pathways for motion analysis: cortical connections of the medial superior temporal and fundus of the superior temporal visual areas in the macaque. The Journal of comparative neurology. 1990;296:462–495. doi: 10.1002/cne.902960311. [DOI] [PubMed] [Google Scholar]
- 32.Greenblatt SH. Alexia without agraphia or hemianopsia. Anatomical analysis of an autopsied case. Brain : a journal of neurology. 1973;96:307–316. doi: 10.1093/brain/96.2.307. [DOI] [PubMed] [Google Scholar]
- 33.Yeatman JD, Rauschecker AM, Wandell BA. Brain & Language. Brain and Language. 2013;125:146–155. doi: 10.1016/j.bandl.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kriegeskorte N, et al. Human Cortical Object Recognition from a Visual Motion Flowfield. J. Neurosci. 2003;23:1451–1463. doi: 10.1523/JNEUROSCI.23-04-01451.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orban GA. A Motion Area in Human Visual Cortex. Proceedings of the National Academy of Sciences. 1995;92:993–997. doi: 10.1073/pnas.92.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braddick OJ, O'Brien JM, Wattam-Bell J, Atkinson J, Turner R. Form and motion coherence activate independent, but not dorsal/ventral segregated, networks in the human brain. Current biology : CB. 2000;10:731–734. doi: 10.1016/s0960-9822(00)00540-6. [DOI] [PubMed] [Google Scholar]
- 37.Colby CL, Duhamel JR, Goldberg ME. Ventral intraparietal area of the macaque: anatomic location and visual response properties. Journal of neurophysiology. 1993;69:902–914. doi: 10.1152/jn.1993.69.3.902. [DOI] [PubMed] [Google Scholar]
- 38.Conturo TE, et al. Tracking neuronal fiber pathways in the living human brain. Proceedings of the National Academy of Sciences. 1999;96:10422–10427. doi: 10.1073/pnas.96.18.10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mangin JF, et al. Toward global tractography. NeuroImage. 2013;80:290–296. doi: 10.1016/j.neuroimage.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 40.Aganj I, et al. Medical Image Analysis. Medical image analysis. 2011;15:414–425. doi: 10.1016/j.media.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neher PF, et al. MITK global tractography. In: Haynor DR, Ourselin S, editors. SPIE Medical Imaging. Vol. 8314. SPIE; 2012. p. 83144D. [Google Scholar]
- 42.Jbabdi S, Woolrich MW, Andersson JLR, Behrens TEJ. A Bayesian framework for global tractography. NeuroImage. 2007;37:116–129. doi: 10.1016/j.neuroimage.2007.04.039. [DOI] [PubMed] [Google Scholar]
- 43.Sherbondy AJ, Dougherty RF, Ananthanarayanan R, Modha DS, Wandell BA. Think global, act local; projectome estimation with BlueMatter. Medical image computing and computer-assisted intervention : MICCAI International Conference on Medical Image Computing and Computer-Assisted Intervention. 2009;12:861–868. doi: 10.1007/978-3-642-04268-3_106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sherbondy AJ, Dougherty RF, Ben-Shachar M, Napel S, Wandell BA. ConTrack: finding the most likely pathways between brain regions using diffusion tractography. Journal of vision. 2008;8:15.1–16. doi: 10.1167/8.9.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schreiber J, Riffert T, Anwander A, Knösche TR. Plausibility Tracking: A method to evaluate anatomical connectivity and microstructural properties along fiber pathways. NeuroImage. 2014;90:163–178. doi: 10.1016/j.neuroimage.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 46.Sporns O. Making sense of brain network data. Nature Methods. 2013;10:491–493. doi: 10.1038/nmeth.2485. [DOI] [PubMed] [Google Scholar]
- 47.Uğurbil K, et al. Pushing spatial and temporal resolution for functional and diffusion MRI in the Human Connectome Project. NeuroImage. 2013;80:80–104. doi: 10.1016/j.neuroimage.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parker GJM, et al. Initial demonstration of in vivo tracing of axonal projections in the macaque brain and comparison with the human brain using diffusion tensor imaging and fast marching tractography. NeuroImage. 2002;15:797–809. doi: 10.1006/nimg.2001.0994. [DOI] [PubMed] [Google Scholar]
- 49.Seehaus AK, et al. Histological validation of DW-MRI tractography in human postmortem tissue. Cerebral cortex (New York, NY.: 1991) 2013;23:442–450. doi: 10.1093/cercor/bhs036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones DK, Knosche TR, Turner R. White matter integrity, fiber count, and other fallacies: the do‘s and don’ts of diffusion MRI. NeuroImage. 2013;73:239–254. doi: 10.1016/j.neuroimage.2012.06.081. [DOI] [PubMed] [Google Scholar]
- 51.Fillard P, et al. NeuroImage Quantitative evaluation of 10 tractography algorithms on a realistic diffusion MR phantom. NeuroImage. 2011;56:220–234. doi: 10.1016/j.neuroimage.2011.01.032. [DOI] [PubMed] [Google Scholar]
- 52.Assaf Y, et al. The CONNECT project: Combining macro- and micro-structure. NeuroImage. 2013;80:273–282. doi: 10.1016/j.neuroimage.2013.05.055. [DOI] [PubMed] [Google Scholar]
- 53.Close TG, et al. A software tool to generate simulated white matter structures for the assessment of fibre-tracking algorithms. NeuroImage. 2009;47:1288–1300. doi: 10.1016/j.neuroimage.2009.03.077. [DOI] [PubMed] [Google Scholar]
- 54.Rauschecker AM, et al. Reading impairment in a patient with missing arcuate fasciculus. Neuropsychologia. 2009;47:180–194. doi: 10.1016/j.neuropsychologia.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mezer A, et al. Quantifying the local tissue volume and composition in individual brains with magnetic resonance imaging. Nature Medicine. 2013;19:1667–1672. doi: 10.1038/nm.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 56.Jones DK, Horsfield MA, Simmons A. Optimal Strategies for Measuring Diffusion in Anisotropic Systems by Magnetic Resonance Imaging. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 1999;42:515–525. [PubMed] [Google Scholar]
- 57.Friston KJ, et al. Spatial Registration and Normalization of Images. Human brain mapping. 1995;2:165–189. [Google Scholar]
- 58.van Essen DC, et al. The Human Connectome Project: A data acquisition perspective. NeuroImage. 2012;62:2222–2231. doi: 10.1016/j.neuroimage.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barch DM, Behrens T, Yacoub E, Ugurbil K. The WU-Minn human connectome project: an overview. NeuroImage. 2013 doi: 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fischl B. FreeSurfer. NeuroImage. 2012;62:774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tournier J-D, Calamante F, Connelly A. MRtrix: Diffusion tractography in crossing fiber regions. International Journal of Imaging Systems and Technology. 2012;22:53–66. [Google Scholar]
- 62.Lazar M, et al. White matter tractography using diffusion tensor deflection. Human brain mapping. 2003;18:306–321. doi: 10.1002/hbm.10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Basser PJ, Pajevic S, Pierpaoli C, Duda J, Aldroubi A. In vivo fiber tractography using DT-MRI data. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2000;44:625–632. doi: 10.1002/1522-2594(200010)44:4<625::aid-mrm17>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 64.Behrens TEJ, et al. Non-invasive mapping of connections between human thalamus and cortex using diffusion imaging. Nature neuroscience. 2003;6:750–757. doi: 10.1038/nn1075. [DOI] [PubMed] [Google Scholar]
- 65.Parker GJM, Haroon HA, Wheeler-Kingshott CAM. A framework for a streamline-based probabilistic index of connectivity (PICo) using a structural interpretation of MRI diffusion measurements. Journal of magnetic resonance imaging : JMRI. 2003;18:242–254. doi: 10.1002/jmri.10350. [DOI] [PubMed] [Google Scholar]
- 66.Destrieux C, Fischl B, Dale A, Halgren E. Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature. NeuroImage. 2010;53:1–15. doi: 10.1016/j.neuroimage.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fischl B, et al. Cortical Folding Patterns and Predicting Cytoarchitecture. Cerebral cortex (New York, N.Y.: 1991) 2008;18:1973–1980. doi: 10.1093/cercor/bhm225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yeatman JD, Dougherty RF, Myall NJ, Wandell BA, Feldman HM. Tract profiles of white matter properties: automating fiber-tract quantification. PloS one. 2012;7:e49790. doi: 10.1371/journal.pone.0049790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tuch DS, Reese TG, Wiegell MR, Wedeen VJ. Diffusion MRI of complex neural architecture. Neuron. 2003;40:885–895. doi: 10.1016/s0896-6273(03)00758-x. [DOI] [PubMed] [Google Scholar]
- 70.Stejskal EO, Tanner JE. Spin Diffusion Measurements: Spin Echoes in the Presence of a Time-Dependent Field Gradient. The Journal of Chemical Physics. 1965;42:288. [Google Scholar]
- 71.Basser PJ, Pierpaoli C. Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor MRI. Journal of magnetic resonance. Series B. 1996;111:209–219. doi: 10.1006/jmrb.1996.0086. [DOI] [PubMed] [Google Scholar]
- 72.Dongmin K, Suvrit S, Inderjit SD. A non-monotonic method for large-scale nonnegative least squares. Optimization Methods and Software. 2012;28:1–17. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.