Summary
Microbial secretory phospholipases A2 (sPLA2s) are among the last discovered and least known members of this functionally diverse family of enzymes. We analyzed here two sPLA2s, named sPlaA and sPlaB, of the filamentous ascomycete Aspergillus oryzae. sPlaA and sPlaB consist of 222 and 160 amino acids, respectively, and share the conserved Cys and catalytic His-Asp residues typical of microbial sPLA2s. Two sPLA2s differ in pH optimum, Ca2+ requirement and expression profile. The splaA mRNA was strongly upregulated in response to carbon starvation, oxidative stress and during conidiation, while splaB was constitutively expressed at low levels and was weakly upregulated by heat shock. Experiments with sPLA2-overexpressing strains demonstrated that two enzymes produce subtly different phospholipid composition variations and also differ in their subcellular localization: sPlaA is most abundant in hyphal tips and secreted to the medium, whereas sPlaB predominantly localizes to the ER-like intracellular compartment. Both sPLA2-overexpressing strains were defective in conidiation, which was more pronounced for sPlaB overexpressors. Although no major morphological abnormality was detected in either ΔsplaA or ΔsplaB mutants, hyphal growth of ΔsplaB, but not that of ΔsplaA, displayed increased sensitivity to H2O2 treatment. These data indicate that two A. oryzae sPLA2 enzymes display distinct, presumably non-redundant, physiological functions.
1. Introduction
Phospholipases A2 (PLA2) belong to an heterogeneous family of enzymes which catalyze the hydrolysis of the ester bond at the sn-2 position of glycerophospholipids, liberating free fatty acids and lysophospholipids. Based on their primary structure, enzymatic and subcellular localization features, these enzymes are classified into three major subfamilies, i.e. Ca2+-independent, cytosolic and secretory Ca2+-dependent PLA2s. The latter, designated sPLA2s, are Ca2+-requiring extracellular enzymes characterized by a relatively low molecular mass (13–19 kDa), containing a His-Asp catalytic dyad and at number of disulfide bonds ranging from zero to eight, (Murakami and Kudo, 2004; Schaloske and Dennis, 2006). sPLA2s are present in a wide range of organisms and tissues. In mammals, 10 catalytically-active sPLA2 enzymes displaying overlapping, but distinct tissue distributions, substrate specificities, interfacial properties and putative physiological function(s) have been identified (Murakami and Kudo, 2004). For example, mammalian group IB enzymes are abundant in pancreatic juice and are implicated in food lipid digestion (even though a ligand-like role is also recognized), whereas group IIA sPLA2s are expressed at high levels in various inflamed tissues and are induced by pro-inflammatory cytokines. In this role, group IIA sPLA2s are considered as key cell signaling components, but they have also been implicated in host-defense mechanisms through their anti-microbial activity, especially toward Gram-positive bacteria (Weinrauch et al., 1996; Weinrauch et al., 1998). In addition, group IB and IIA sPLA2s have been reported to exert neurotoxic effects (Yagami et al., 2001; Yagami et al., 2002), while we have recently documented an opposite, neuroprotective role for group X and possibly group V sPLA2s, that appears to be mediated by the release of lysophosphatidylcholine (Ikeno et al., 2005).
By comparison, very little is known about microbial, group XIV sPLA2s. They were first identified in the mycorrhizal ascomycete Tuber borchii (Soragni et al., 2001), where a secreted Ca2+-dependent PLA2 was found to be strongly upregulated by nutrient (both carbon and nitrogen, but not for example phosphate) starvation. Given the symbiotic capacity of Tuber, starvation-induced expression of TbSP1 as well as its inner cell wall localization and accumulation in mycorrhizal hyphae, which usually experience nutrient limitation, might be indicative of a membrane remodeling and/or signaling role of this enzyme during early stages of plant colonization. Indeed, a substantially enhanced expression of TbSP1 was observed during co-cultivation with a host plant and mycorrhiza development (Miozzi et al., 2005). Following the discovery of TbSP1 a few other microbial sPLA2s were identified and characterized biochemically. Most notably, an orthologous enzyme from the filamentous actinomycete Streptomyces violaceoruber, whose 3D structure has been determined (Matoba et al., 2002; Sugiyama et al., 2002), and an sPLA2 found in the culture medium of the saprotrophic ascomycete Helicosporium sp. (Hanada et al., 1996). Interestingly, no putative group XIV sPLA2 has been identified so far in non-filamentous bacteria, yeasts (both ascomycetes and basidiomycetes) or multicellular, wholly sequenced basidiomycetes.
Here, we report on the molecular characterization of two sPLA2s, named sPlaA and sPlaB, identified in the recently sequenced genome of the saprotrophic ascomycete Aspergillus oryzae (Kobayashi et al., 2007; Machida et al., 2005). We show that sPlaA and sPlaB display distinct properties, showing difference in a number of features, including optimal conditions for enzyme activity, expression profile, cellular localization and effect on conidia formation. As revealed by phospholipid profiling of sPLA2 overexpressing strains, the two enzymes also display distinct substrate preferences. Although neither enzyme was found to be essential under most of the examined growth conditions, they appear to be differentially involved in oxidative stress tolerance.
2. Materials and Methods
2.1. A. oryzae strains, growth conditions, and transformation
A. oryzae RIB40 was used as a wild-type strain source of the splaA and splaB genes and for expression analysis by RT-PCR. The niaD300 strain (niaD−) was used for overexpression of splaA and splaB under the control of improved glucoamylase promoter, PglaA142 (Minetoki et al., 1998), and for expressing EGFP-tagged sPlaA and sPlaB. NSR13 (niaD− sC− adeA−) (Jin et al., 2004a) and NS4 (niaD− sC−) (Yamada et al., 1997) strains were used for the disruption of splaA and splaB, respectively (see Table 1 for further details). DPY medium (2% dextrin, 1% polypeptone, 0.5% yeast extract, 0.5% KH2PO4, 0.05% MgSO4·7H2O, pH 5.5) was used as a standard medium unless otherwise stated. Czapek-Dox (CD) medium (0.3% NaNO3, 0.2% KCl, 0.1% KH2PO4, 0.05% MgSO4·7H2O, 0.002% FeSO4·7H2O, 2% carbon source, pH 5.5) was used to analyze niaD300 growth and transformation. Glucose was used as a carbon source unless otherwise indicated. M medium (0.2% NH4Cl, 0.1% (NH4)2SO4, 0.05% KCl, 0.05% NaCl, 0.1% KH2PO4, 0.05% MgSO4·7H2O, 0.002% FeSO4·7H2O, 2% glucose, pH 5.5) and MM medium (M medium plus 0.15% methionine) were used for gene disruption in the NS4 and NSR13 strains, respectively. Potato Dextrose Agar (PD) medium (Nissui) was used for phenotypic analysis of spa-gene disruptants. Transformation of A. oryzae was performed as described previously (Kitamoto, 2002).
Table 1.
A. oryzae strains used in this study
| Strain | Host strain | Genotype | Plasmid introduced | Source or reference |
|---|---|---|---|---|
| RIB40 | wild type | Murakami et al., 1967; Machida et al., 2005 | ||
| niaD300 | RIB40 | niaD− | Minetoki et al., 1996 | |
| NS4 | niaD300 | niaD−sC− | Yamada et al., 1997 | |
| NSR13 | NS4 | niaD−sC−adeA− | Jin et al., 2004 | |
| NGA142-1 | niaD300 | niaD− niaD+ | pNGA142 | This study |
| OESA-1 | niaD300 | niaD− PglaA142-splaA∷niaD+ | pNGA142/splaA | This study |
| OESB-1 | niaD300 | niaD− PglaA142-splaA∷niaD+ | pNGA142/splaB | This study |
| SAfG | niaD300 | niaD− PamyB-splaA-egfp∷niaD+ | pAmy-fAgfp | This study |
| SAcG | niaD300 | niaD− PamyB-splaA(ΔC)-egfp∷niaD+ | pAmy-cAgfp | This study |
| SBfG | niaD300 | niaD− PamyB-splaB-egfp∷niaD+ | pAmy-fBgfp | This study |
| SBcG | niaD300 | niaD− PamyB-splaB(ΔC)-egfp∷niaD+ | pAmy-cBgfp | This study |
| DSAvec-1 | NSR13 | niaD−sC−adeA− adeA+ | pAdeA | This study |
| DSA-1 | NSR13 | niaD−sC−adeA− ΔsplaA∷adeA+ | pSAadeA (linear) | This study |
| DSAB-1 | DSA-1 | niaD−sC−adeA− ΔsplaA∷adeA+ ΔsplaB∷sC+ | pSBsC (linear) | This study |
| DSBvec-1 | NS4 | niaD−sC−sC+ | pBSsC | This study |
| DSB-1, −2 | NS4 | niaD−sC− ΔsplaB sC+ | pSBsC (linear) | This study |
2.2. Cloning of splaA and splaB
splaA and splaB genes (DDBJ accession numbers AB126038 and AB126039, respectively) were amplified by PCR using Pfx DNA polymerase (Invitrogen) and genomic DNA from A. oryzae RIB40 as a template. The following oligonucleotides were used as amplification primers: 5’-cagcgaattcATGAAGAACATCTTCGTTGC-3’ and 5’-cagcgaattcCTACAGGTTTTCAATATCGT-3’ for splaA; 5’-cagcgaattcATGAAGGCTAACAGCTTTCT-3’ and 5’-gcgcgaattccaaggtctcatatatgtatc-3’ for splaB; capital letters correspond to the coding regions, double-underlined capital letters indicate start and stop codons, and lower-case underlined letters indicate the EcoR I sites utilized for cloning. In the case of splaB, the primers were designed to amplify a putative splaB ORF and corresponding downstream region (approximately 50 bp) because the location of the stop codon was not known precisely at the beginning of this work. Amplified DNA fragments were A-tailed and cloned into pT7Blue T (Novagen) to generate pT7-splaA and pT7-splaB, respectively.
2.3. Expression and purification of recombinant sPlaA and sPlaB
The putative mature regions of sPlaA and sPlaB were amplified using pT7-splaA and pT7-csplaB (carrying splaB cDNA) as templates and the following oligonucleotides as primers: 5’-tagtgaattcCCCTACACAACCCCTGTCAA-3’ and 5’-gcatgaattCTAGCCAAAGTGGCGGACAGC-3’ for splaA; 5’-tagtgaattcCCCCTCCCCACACCAAATGA-3’ and 5’-gtataagcttCTAACCATGCCGCCCCGAAG-3’ for splaB. The regions of sPlaA and sPlaB encoded by the amplified fragments were comprised between Pro20 and Gly157 for sPlaA and between Pro18 and Gly146 for sPlaB, respectively (Fig. 1). PCR fragments digested with EcoR I and Hind III were ligated to the pRSET B expression vector (Invitrogen) to generate pRSETB-msplaA and pRSETB-msplaB, which were used to express the recombinant proteins as an in-frame N-terminal fusions with a vector-encoded His6 tag sequence. These two plasmids were transformed into E. coli BL21(DE3) pLysS (Invitrogen). Protein expression was induced by adding 0.3 mM isopropyl-β-D-thiogalactopyranoside when OD600 reached 0.3–0.4, and was allowed to proceed for 3 h at 37 °C. After cell lysis, inclusion bodies containing recombinant sPlaA (rsPlaA) or sPlaB (rsPlaB) were dissolved in guanidine hydrochloride and bound to a metal affinity, Ni2+-NTA agarose resin (QIAGEN). Subsequent elution of rsPlaA and rsPlaB was carried out with urea-containing buffers as per manufacturer’s instructions. Renaturation was achieved by stepwise dialysis against Tris buffer (20 mM Tris-HCl, pH 8.0, 0.5 M NaCl) containing gradually decreasing concentrations of urea (4 M, 2 M, 1 M, none); each dialysis step was carried out for 3 h at 4 °C. The purity and concentration of recombinant proteins were analyzed by SDS-PAGE (15% acrylamide) followed by Coomassie-blue staining (Fig . S1).
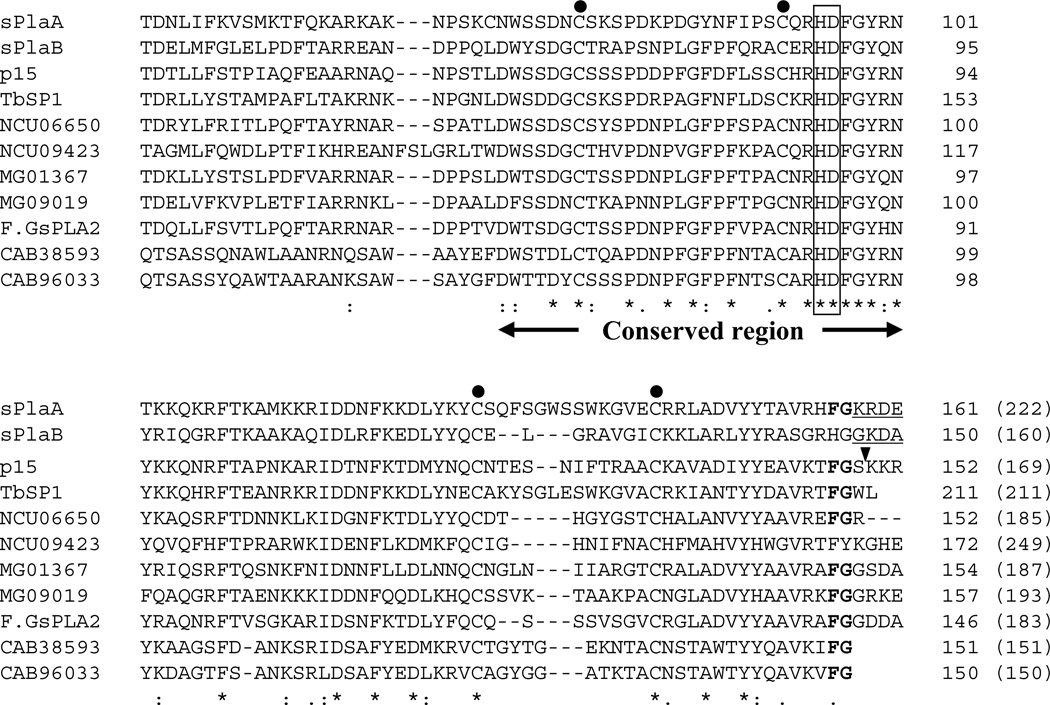
Fig. 1. Alignment of A. oryzae sPLA2s with related polypeptides from other microorganisms.
The phospholipase A2 domains of sPlaA and sPlaB predicted by SMART (Letunic et al., 2009) are aligned with those of other fungal and prokaryotic sPLA2s using ClustalW (Thompson et al., 1994). Species names and accession numbers of the aligned sPLA2 sequences are: TbSP1, T. borchii, AAF80454; p15, Helicosporium sp., BAB70714; NCU06650, N. crassa, XP_960883; NCU09423, N. crassa, XP_958163; MG01367, M. grisea XP_363441; MG09019, M. grisea, XP_364174; F.GsPLA2, Fusarium graminearum, XP_384087; CAB38593, S. coelicolor A3(2), NP_627436; CAB96033, S. coelicolor A3(2), NP_625343. The His-Asp catalytic dyad is boxed; conserved cysteine residues are marked by dots; the conserved FG sequence is in bold. Arrowhead indicates the C-terminal processing site for p15; the predicted C-terminal extensions which were not included in rsPlaA and rsPlaB are underlined. Positions and total numbers of amino acids (in parentheses) are shown in the right.
2.4. PLA2 activity assays
PLA2 activity was measured by quantifying the release of radioactivity in the assays carried out at 37 °C for 30 min in a 100 µl reaction mixture containing 455,000 dpm of [3H] oleic acid-labeled E. coli membranes and 10 µl of 1 mM rsPlaA or rsPlaB (Elsbach and Weiss, 1991); parallel assays in the absence of Ca2+ (plus 1 mM EGTA) or in the presence of increasing concentration of CaCl2 (0.01–100 mM) were conducted to determine the calcium-dependence of PLA2 activity. The pH optimum was determined by using 50 mM Na-acetate buffer for the pH range 4.5–5.5, 50 mM 2-N-morpholinoethanesulfonic acid (MES)-NaOH for pH 5.5–6.5, 50 mM 3-morpholinopropanesulfonic acid (MOPS) for pH 6.5–7.5, and 50 mM Tris-HCl for pH 7.5–9.5; all buffers were supplemented with 10 mM CaCl2. For the experiments in Fig. 4, the various A. oryzae strains were cultured in 20 ml DPY media at 30 °C for 3 days and the resulting culture media and mycelia were separated by filtration through Miracloth (Calbiochem). Mycelia were then washed with ddH2O, resuspended in 400 µl of phosphate-buffered saline (PBS: 13.7 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4) containing 1 mM PMSF, and lysed with a Multi-Beads Shocker (Yasui Kikai Corp., Osaka, Japan). A six cycles, two-step lysis procedure was utilized, in which the first step was carried out at 2,500 rpm for 20 sec using a metal cone, and the second step without the metal cone; the interval between the two steps was set at 60 sec. Protein concentration of cell lysates was adjusted to 0.5 mg/ml. PLA2 activity was measured using 10 µl of culture supernatant or cell lysate in a 100-µl reaction mixture as described above. A different, thin-layer chromatography-based PLA2 assay, utilizing double-labeled 1, 2-di[1-14C]palmitoyl L-α- phosphatidylcholine as substrate (Soragni et al., 2001) and carried out for 3 h at 30 °C under optimized reaction conditions (5 mM CaCl2 , 50 mM Na-acetate buffer pH 5.0 for rsPlaA; 1 mM CaCl2, 50 mM Tris-HCl buffer pH 8.0 for rsPlaB) was used to verify the lack of lysophospholipase activity in both recombinant sPLA2s from A. oryzae.
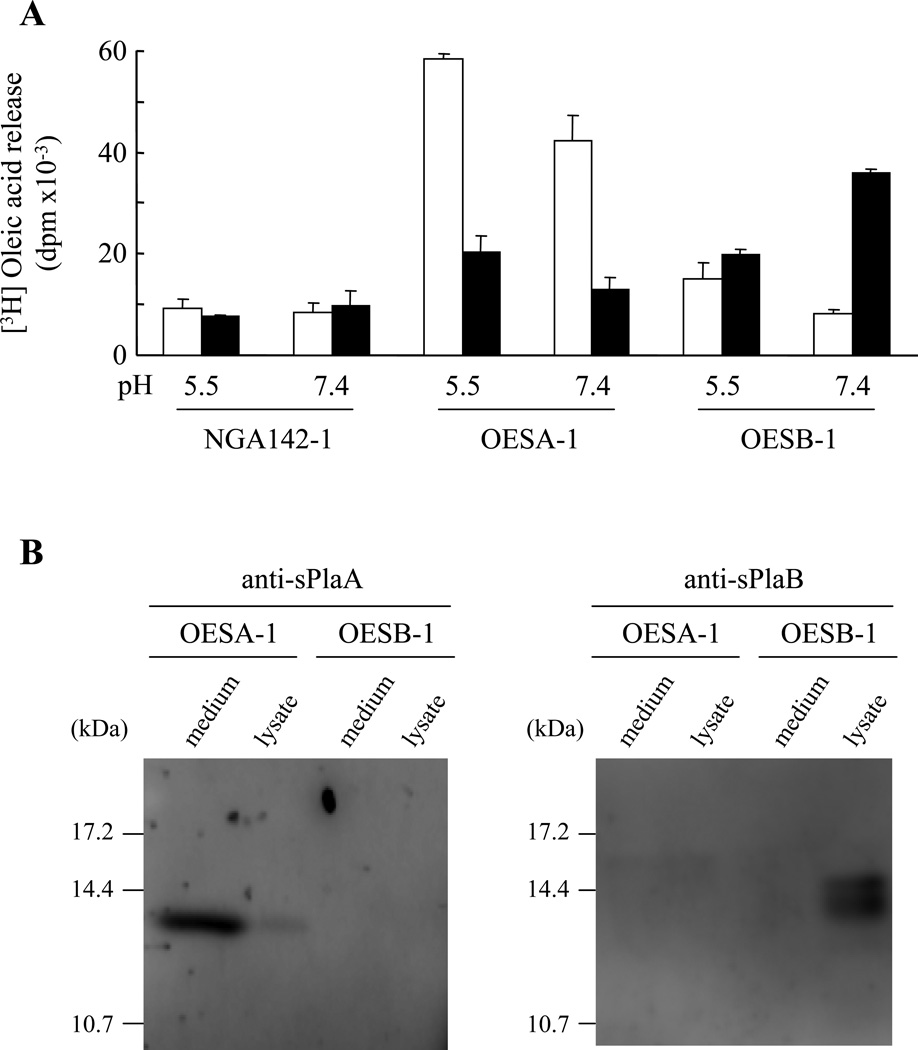
Fig. 4. Functional and immunological localization of sPlaA and sPlaB in overexpressing strains.
(A) Vector-transformed control strain NGA142-1, OESA-1, and OESB-1 were cultured in DPY medium for 3 days, followed by culture medium recovery and lysate preparation. PLA2 activity in each fraction (culture media: open bars; lysates: closed bars) was measured at both pH 5.5 and pH 7.4 in reaction mixtures supplemented with 10 mM Ca2+. (B) Immunoblot analysis of culture media and lysates prepared from OESA-1 and OESB-1 using the anti-sPlaA and the anti-sPlaB antibodies as indicated.
2.5. Reverse transcription-PCR analysis
splaA and splaB mRNA levels were determined by RT-PCR. Following growth under various conditions, mycelia (RIB40 strain) were frozen in liquid nitrogen, ground with a mortar and the resulting powder (ca. 500 µl) was transferred to a plastic tube, resuspended in 700 µl of the TRI reagent (Sigma) and subsequent treated according to the manufacturer’s instructions. Total RNA (2 µg in 6.5 µl of diethylpyrocarbonate-treated ddH2O) was then mixed with 1 µl of oligo dT primer (10 µM), 8 µl of dNTP mix (2.5 mM), 2 µl of 100 mM dithiothreitol, 0.5 µl reverse transcriptase and 4 µl of 5x buffer as specified in the PoweScript kit (Clontech), and incubated at 42°C for 1 h to synthesize first-strand cDNA. PCR reaction (30 cycles of 94°C, 30 sec, 50°C, 30 sec, and 72°C, 40 sec) was performed with the primers: 5’-cagcgaattcATGAAGAACATCTTCGTTGC-3’ and 5’-gcatgaattcTAGCCAAAGTGGCGGACAGC-3’ for splaA, and 5’-cagcgaattcATGAAGGCTAACAGCTTTCT-3’ and 5’-gtataagcttCTAACCATGCCGCCCCGAAG-3’ for splaB. The γ-actin cDNA, utilized as an internal control, was amplified with the primers 5’-GTTGCTGCTCTCGTCATTGAC-3’ and 5’-GTAATCGGTCAAATCACGGCC-3’. For expression profiling during development of aerial hyphae and conidia, A. oryzae was first cultured to early growth phase in liquid DPY medium for 24 h, the mycelial pellet was then collected, transferred to a Petri dish (10 cm diameter) containing 10 ml of DPY medium, and allowed to grow without shaking. Under these conditions, formation of aerial hyphae and conidia, utilized for RNA extraction, took place after about 24 h and 48 h, respectively.
2.6. Expression of splaA and splaB in A. oryzae
pT7-splaA and pT7-splaB were digested with EcoR I, and the resulting spa DNA fragments were ligated to the EcoR I site of pNGA142, which contains an expression cassette consisting of an improved glaA promoter (PglaA142) and an α-glucosidase terminator (TagdA) (Minetoki et al., 1998), to generate pNGA142/splaA and pNGA142/splaB. The niaD300 strain was transformed with the two plasmids and two sets of four phenotypically identical transformants were obtained; two transformants from each set, named OESA-1 and OESB-1, were chosen further analysis. Total RNA from the OESB-1 strain was used as template for RT-PCR cloning of the splaB cDNA, which was cloned into the pT7Blue T-vector to generate pT7-csplaB.
2.7. Subcellular localization of sPlaA and sPlaB
Subcellular localization of sPlaA and sPlaB was examined by indirect immunofluorescence analysis and microscopic observation of EGFP-fused proteins. In the former, OESA-1 and OESB-1 were immunostained with anti-sPlaA and anti-sPlaB antibodies as described by Takeshita et al. (Takeshita et al., 2005) with minor modifications. Conidia were incubated on glass cover slides at 30 °C for 12–15 h in CD medium supplemented with 2% dextrin instead of glucose. After washing with PBST (PBS plus 0.05% Tween 20), cells were fixed at room temperature (15–45 min) in PBS containing 3.7% formaldehyde, 5 mM MgSO4, and 2.5 mM EGTA. For cell wall digestion, cells were overlaid for 10 min at room temperature with a PBST-based digestion mix containing 3 mg/ml Yatalase (TaKaRa), 1 mg/ml Lysing enzyme (Sigma) and 10 mg/ml egg white (Sigma). Cover slides were then washed by PBST, immersed in methanol at −20 °C for 10 min and, after additional washings with PBST, they were incubated for 1 h at room temperature with anti-sPlaA or anti-sPlaB antibodies diluted 1:2,500 dilution in PBSTB (PBST containing 0.1 mg/ml bovine serum albumin). After several washings with PBST, cover slides were incubated for 1 h in the dark at room temperature with a FITC-conjugated anti-rabbit IgG secondary antibody (Sigma) diluted 1:500 in PBSTB; following one more round of PBST washing, they were visualized with an Olympus BX52 microscope. For in vivo fluorescence analysis, A. oryzae strains expressing sPlaA or sPlaB C-terminally fused with EGFP were generated and visualized by fluorescence microscopy. Since both sPLA2 proteins have putative C-terminal extension sequences, two types of EGFP fusion polypeptides were constructed: one bearing the full length proteins, the other bearing C-terminally-truncated versions of both proteins (ending with Gly157 and Ala151 for sPlaA and sPlaB, respectively) fused with EGFP. The corresponding sPla fragments were amplified by PCR and the resulting amplicons were inserted into pDONR221 (MultiSite Gateway system, Invitrogen) using the BP recombination reaction to generate center entry clones. The following primers were utilized: Ao1724s 5’-CAGCGAATTCATGAAGAACATCTTCGTTGC-3’ and splaA-full-egfp 5’-GAATTCCAGGTTTTCAATATCGTCGA-3’ for full-length splaA; Ao1724s 5’-CAGCGAATTCATGAAGAACATCTTCGTTGC-3’ and splaA-egfp 5’-GAATTCGCCAAAGTGGCGGACAGCAG-3’ for truncated splaA; Ao0940s 5’-CAGCGAATTCATGAAGGCTAACAGCTTTCT-3’ and splaB-full-egfp 5’-GAATTCAAGAAGTTCATCCAACTCCC-3’ for full-length splaB; Ao0940s 5’-CAGCGAATTCATGAAGGCTAACAGCTTTCT-3’ and splaB-egfp 5’-GAATTCCGCTGCATCTTTACCACCAT-3’ for truncated splaB (restriction sites are underlined). Four plasmids, named pAmy-fAgfp, pAmy-cAgfp, pAmy-fBgfp, and pAmy-cBgfp, were generated by the LR reaction using the following plasmids: 5’ entry clone carrying the amyB promoter for expression, center entry clone as described above, 3’ entry clone carrying the EGFP sequence and the amyB terminator, and a destination vector harboring the selectable niaD marker (Mabashi et al., 2006). The EGFP-fusion protein expressing strains SAfG, SAcG, SBfG, and SBcG (Table 1) were then generated by transforming niaD300 strain with the above plasmids. After culture in 2% dextrin supplemented-CD (16–20 h at 30 °C), individual strains were visualized by fluorescence microscopy. For endoplasmic reticulum (ER) staining, hyphae were treated with 1 µM ER-Tracker™ Blue-white DPX (Molecular Probes) for 30 min at 30 °C and, after washing and additional incubation for 15 min at 30 °C, they were examined microscopically.
2.8. Phospholipid profiling
OESA-1, OESAB-1 and the control NGA142-1 strains were cultured for three days in 20 ml of DPY medium under inducing conditions, starting from ∼104 conidia. Mycelia were collected by centrifugation, washed with ddH2O, immediately transferred to hot isopropanol (75 °C) supplemented with 0.01% butylated hydroxytoluene (3 ml/mycelial sample) and extracted as described (Welti et al., 2002). Following organic solvent extraction, residual mycelium was heated overnight at 105 °C and weighed; dry weights, subsequently utilized for data normalization, ranged from 100 to 400 mg. The combined extracts were washed once with 1 ml of 1 M KCl and once with 2 ml of ddH2O, prior to solvent evaporation and analysis. Individual samples (three biological replicates each), combined with the appropriate solvents and standards, were analyzed at the Kansas Lipidomics Research Center (Kansas State University) on a •triple• quadrupole tandem mass spectrometer (Applied Biosystems API 4000) equipped for electrospray ionization. Details on ESI-MS/MS analysis have been reported previously (Bartz et al., 2007) and can also be found at http://www.kstate.edu/lipid/lipidomics/profiling.htm. Data processing was performed with the Analyst software. Peak identification was primarily carried out against a yeast lipid template; peaks that did not match any yeast lipid were identified using an animal lipid template. The lipids in each class were quantified in comparison to internal standards belonging to the same class and were normalized with respect to the dry weight of each sample. Data were analyzed for statistical significance by t test analysis carried out with the Cyber-T program (Long et al., 2001). Statistically significant values were averaged and exported into an Excel file format, and ratios for the amount of individual lipid species in the overexpressing strains with respect to the control strain were calculated; only phospholipid species with an “overexpressor”/”control” ratio ≥2 or ≤0.5 were kept for further analysis.
2.9. Deletion of splaA and/or splaB
splaB-deleted strains were generated by gene replacement using the A. nidulans sC marker. To this end, the upstream and downstream regions of splaB (∼2 kb and 1.5 kb, respectively) were amplified and ligated to pUsC, thus generating pSBsC. This plasmid was linearized by Apa I/Not I digestion and transformed into NS4 (Table 1), to generate the splaB-deleted, DSB strains. Since the same method did not work for splaA, the A. oryzae marker adeA and the NSR13 strain ((Jin et al., 2004b); see also Table 1) were used for splaA disruption. The Pst I/EcoR I fragment of pAdeA, harboring the adeA gene of A. oryzae in the pT7Blue T-vector (Jin et al., 2004b), was ligated to Pst I/EcoR I digested pSAsC, carrying the upstream and downstream regions of splaA (∼1.5 kb each), thus generating pSAadeA. The latter plasmid was linearized by Apa I/Not I digestion and transformed into NSR13, to generate the splaA-disrupted DSA strain. The double disrupted ΔsplaA/ΔsplaB strain, DSAB, was obtained by transforming the disruption cassette for splaB into the DSA-1 strain. Initial disruptant validation was performed by PCR using primers annealing with the coding regions of the spla genes; primers annealing with the upstream served as controls. Subsequent Southern analysis was performed with the ECL Direct Nucleic Acid Labeling and Detection System (GE Healthcare). Genomic DNA (∼5 µg for each sample) was restriction-digested (see Fig. 8), the resulting fragments were separated by electrophoresis on a 0.8% agarose gel, transferred to Hybond N+ (GE Healthcare) and hybridized with the probes specified in Fig. 8. Hybridization, washing, and detection were performed as per manufacturer’s instruction; the LAS-1000 plus luminescent image analyzer (Fuji Photo Film, Japan) was used for detection. The deletion strains DSA-1, DSB-1, and DSAB-1, along with the control strains DSAvec-1 (DSA-1 transformed with pAdeA) and DSBvec-1 (DSB-1 transformed with pBSsC) were examined under the following growth conditions: carbon-deprived CD medium, nitrogen-deprived CD medium, CD medium containing either 2% glycerol, 0.5% oleic acid, or 100 ng/ml phosphatidylcholine as the sole carbon source, PD medium, CD medium adjusted to pH 8.0 or supplemented with 10 mM CaCl2, 1 M NaCl, or 1.2 M sorbitol, and wheat bran as solid state culture.
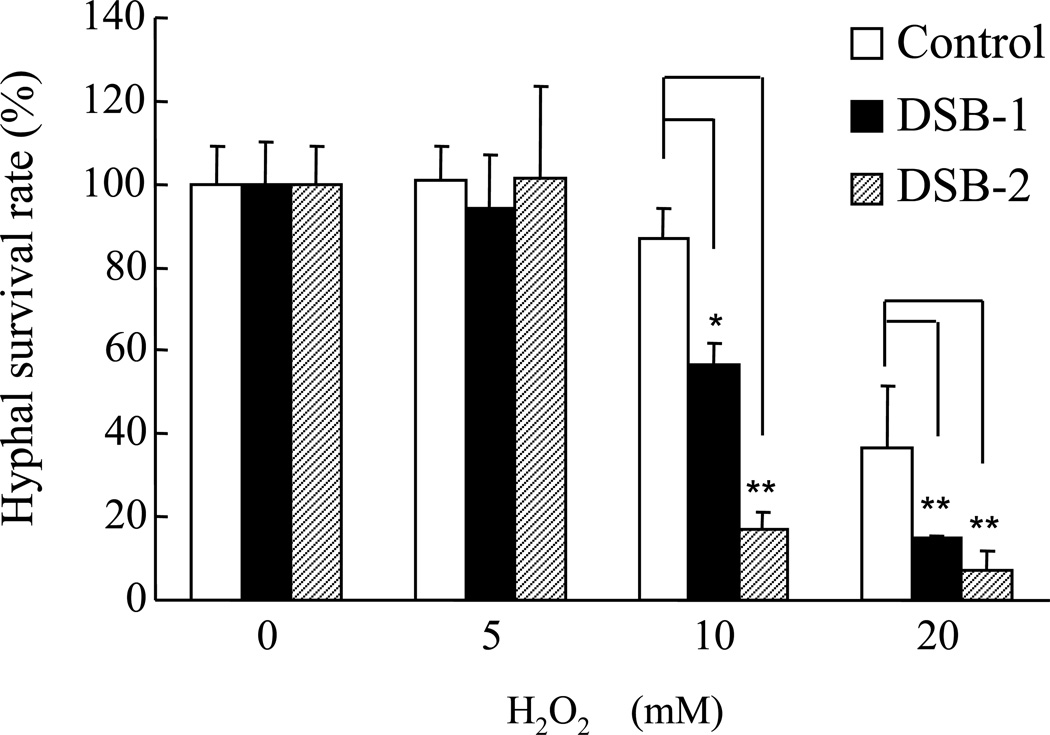
Fig. 8. Enhanced oxidative stress sensitivity of spa-disrupted strains.
Mycelia from the control DSBvec-1 strain (generated by transformation of NS4 with the pBSsC vector; open bars) and from two different ΔsplaB strains (DSB-1 and DSB-2; solid and hatched bars, respectively) were pre-cultured for 24 h at 30 °C on solid medium (PD) containing 0.25% Triton X-100, treated with 0, 5, 10, and 20 mM H2O2 for 10 min and allowed to grow for an additional 24 h at 30 °C prior to microscopic examination. The bar graph shows the relative survival rates of hyphae from the control, DSB-1, and DSB-2 strains. Data are expressed as in panel A.
2.10. Other procedures
The deduced amino acid sequences of fungal and bacterial sPLA2s were obtained by TBLASTN search of the databases using the amino acid sequence of sPlaA (for the sequences of M. grisea, N. crassa, and F. graminearum, http://www.broadinstitute.org/science/data#; for S. coelicolor A3(2), http://www.sanger.ac.uk/Projects/S_coelicolor/; other sequences were obtained from the DNA Data Bank of Japan (http://www.ddbj.nig.ac.jp/Welcome-j.html)). The positions of phospholipase A2 domain were predicted by SMART (http://smart.embl-heidelberg.de/), with the E-values for sPlaA and sPlaB being 1.3e−81 and 2.20e−45, respectively. N-terminal sequencing of sPlaA, electrophoretically purified from the culture supernatant of the OESA-1 strain and blotted onto a PVDF membrane, was done by Edman degradation using a 490 Procise (PE Applied Biosystems) amino acid sequencer. Endoglycosidase H treatment of sPlaA was carried out at 37 °C for 16 hr in a reaction mixture containing 20 µl of protein solution, 1 µl of 10 mU/µl endoglycosidase H (Seikagaku Kougyou, Japan) and 4 µl of 0.15 M citrate-phosphate buffer (pH 5.0). Polyclonal antibodies against A. oryzae sPLA2s were raised in rabbits (Tanpaku Seisei Kougyou, Japan) immunized with rsPlaB and with a modified version of rsPlaA, named rsPlaA-2, lacking an additional 17 amino acids from the N-terminus so to comply with the mature N-terminus of secreted sPlaA isolated from the culture medium of OESA-1. The primers utilized for constructing the rsPlaA-2 expression plasmid were 5’-gaattcGCGACAACATGCTCGGCCAA-3’ and 5’-gcatgaattCTAGCCAAAGTGGCGGACAGC-3’ (EcoR I sites underlined; coding sequences in capital letters). The reactivity and specificity of anti-Spa antibodies were verified by immunoblotting against rsPlaA-2 and rsPlaB. Culture media and cell lysates of OESA-1 and OESB-1 prepared as described in the “PLA2 activity assays” section (see above) were fractionated on SDS gels under reducing conditions, electro-blotted onto nitrocellulose membranes and analyzed with standard procedures (Ikeno et al., 2005; Soragni et al., 2001); anti-sPlaA and anti-sPlaB antibodies were used at a 1:1,000 dilution. H2O2 sensitivity of hyphae was evaluated as described by Ni et al. (Ni et al., 2005) with minor modifications. Briefly, conidia from the DSBvec-1, DSB-1, and DSB-2 strains were inoculated and incubated on solid PD medium containing 0.25% Triton X-100 (∼400 spores per plate) at 30 °C for 24 h. This incubation time was chosen as the one that allowed germination and formation of microscopically visible colonies from all strains without any appreciable conidiophore production. Hyphae were then overlaid with 10 ml of different solutions containing either zero, 5, 10, or 20 mM H2O2 and incubated at room temperature for 10 min. The H2O2 solution was then decanted, plates were washed three times with 10 ml of sterile ddH2O and incubated at 30 °C for an additional 24–48 h. The number of surviving colonies was finally counted and expressed as percentage of the zero H2O2 control. Data, expressed as the mean + standard deviation, were analyzed by Student’s t-test.
3. Results
3.1. Molecular cloning and sequence analysis of two sPLA2 genes from A. oryzae
A search in the A. oryzae genome database for nucleotide sequences displaying significant similarity to known microbial sPLA2s led to the identification of two novel, putative sPLA2 genes, named splaA (accession number AB126038) and splaB (AB126039). The splaA sequence, located on chromosome 2, was also found in the Expressed Sequence Tag (EST) database of A. oryzae (Akao et al., 2007) grown on the carbon source-deficient, Czapek-Dox (CD) medium, suggesting that splaA may be upregulated in response to carbon starvation. The deduced amino acid sequence of sPlaA is comprised of 222 amino acids with a 19 amino acid-long, putative N-terminal signal sequence and 6 cysteine residues (only 4 cyteines are shown in Fig. 1). In contrast, splaB, which is located on chromosome 6, was not found in any available EST database. As revealed by comparative analysis of genomic and cDNA sequences, and further corroborated by RT-PCR analysis of a splaB-overexpressing strain (see below), a 110 bp intron is present in splaB. Thus, the conceptual translation product of splaB is a 160 amino acid polypeptide, containing a 17 amino acid-long putative N-terminal signal sequence and 4 cysteines.
The deduced amino acid sequences of the phospholipase A2 domains of fungal and bacterial sPLA2s are aligned in Fig. 1. Interestingly, two sPLA2 genes are present in the genome database of Streptomyces coelicolor A3(2) and in most wholly sequenced filamentous ascomycetes, such as Neurospora crassa, Magnaporthe grisea, with the exception of the apparently single-copy sPLA2 genes found in Fusarium graminearum, Chaetomium globosum and Trichoderma reesei. In contrast, no sPLA2 homolog is present neither in any wholly sequenced yeast or filamentous basidiomycete, nor in Aspergillus nidulans, where at least two putative phospholipase B (PLB) encoding genes are present instead.
3.2. Biochemical characterization of recombinant sPlaA and sPlaB
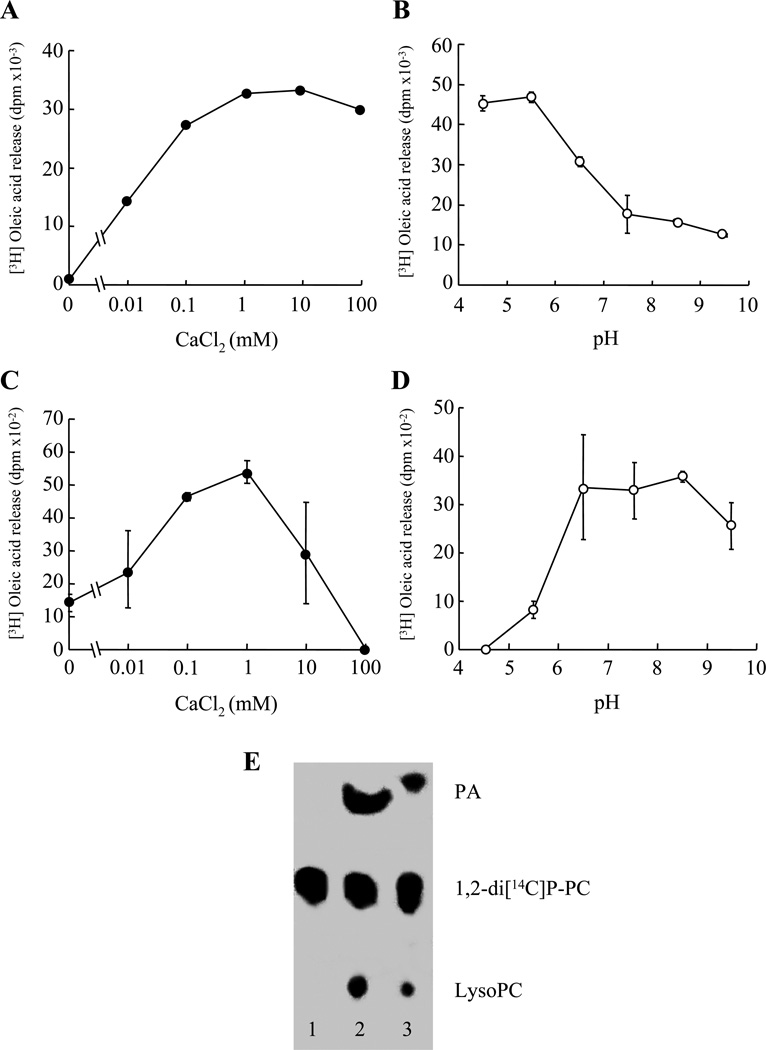
Polypeptides corresponding to the putative processed forms of sPlaA and sPlaB, i.e., lacking the predicted N-terminal signal sequence and C-terminal pro-sequence (see ‘Materials and Methods’ for details), were expressed in E. coli as N-terminal fusions with a metal-binding His6 tag. Both proteins turned out to be completely insoluble. They were thus extracted from inclusion bodies under denaturing conditions, followed by purification and refolding as described in ‘Materials and Methods’. As shown in Fig. 2, renatured recombinant proteins, rsPlaA and rsPlaB, displayed a calcium-dependent PLA2 activity against [3H] oleic acid-labelled bacterial membranes. The highest activity for rsPlaA was observed at 1–10 mM Ca2+ under acidic pH conditions (Fig. 2A and B), whereas rsPlaB was maximally active in the presence of 1 mM Ca2+ at a neutral to alkaline pH (Fig. 2C and D). Also, as revealed by a thin-layer chromatography-based assay utilizing double labeled 1, 2-di[1-14C]palmitoyl L-α-phosphatidylcholine as substrate, both enzymes led to the lysophospholipid accumulation typical of PLA2s (Fig. 2E).
Fig. 2. Enzymatic properties of rsPlaA and rsPlaB produced in E. coli.
(A and C) Calcium dependence of [3H] oleic acid release from radiolabeled bacterial membranes catalyzed by rsPlaA (A) and rsPlaB (C) at pH 5.5 and 8.5, respectively. (B and D) pH dependence of rsPlaA (B) and rsPlaB (D) PLA2 activity in buffers containing 10 mM Ca2+. (E) Thin-layer chromatography and phosphorimager visualization of radiolabeled hydrolysis products from a representative lipolytic assay, carried out under optimized reaction conditions, utilizing double-labeled 1, 2-di[1-14C]palmitoyl L-α-phosphatidylcholine as substrate and either rsPlaA (lane 2) or rsPlaB (lane 3) as enzyme sources (see ‘Materials and Methods’ for details). The substrate (1,2-di[14C]P-PC) and hydrolysis products (palmitic acid, PA; lysophosphatidylcholine, Lyso-PC) are indicated on the right; an enzyme unsupplemented, control reaction is shown in lane 1.
3.3. Expression analysis of splaA and splaB
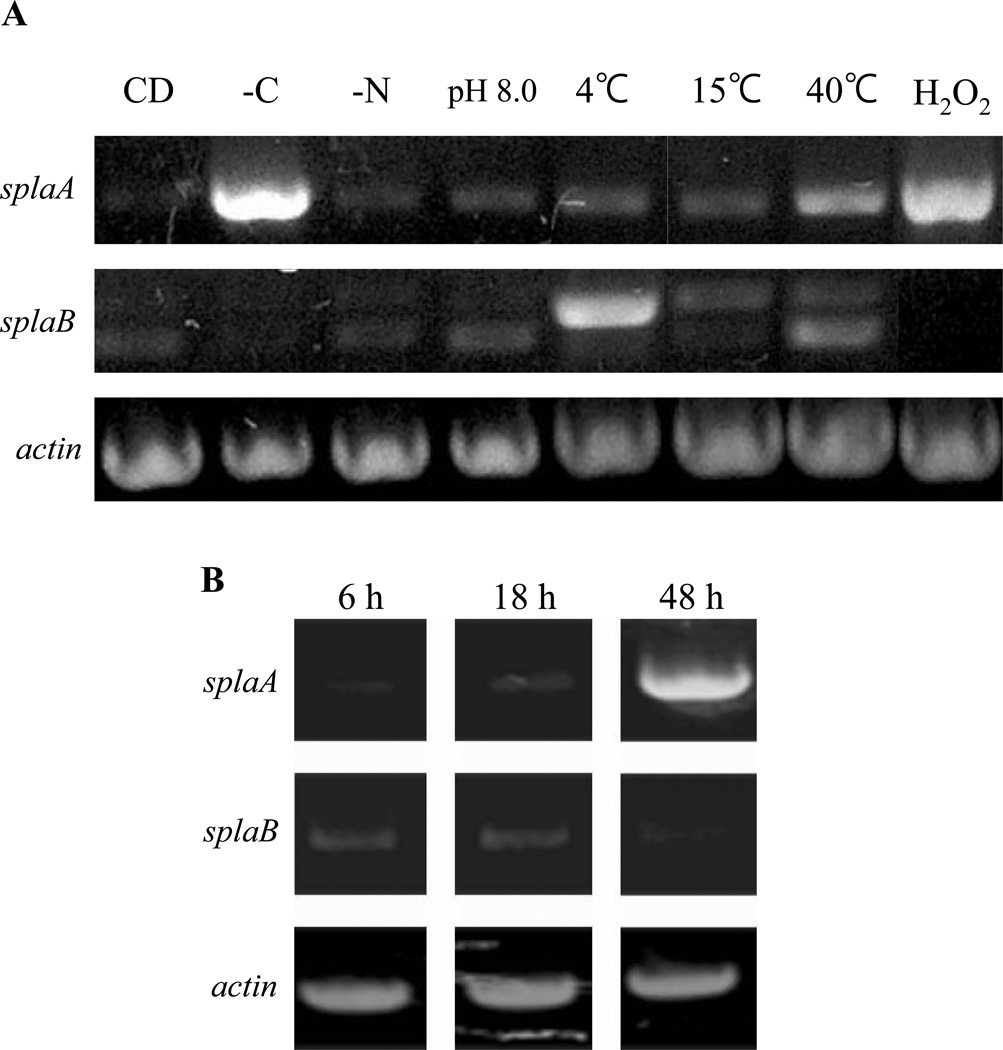
TbSP1, an sPLA2 from T. borchii, has been shown to be strongly upregulated following carbon or nitrogen source starvation (Soragni et al., 2001). As revealed by Northern analysis, splaA expression levels also increased in response to carbon, but not nitrogen deprivation, while the splaB mRNA could not be detected in either condition (data not shown). A more sensitive RT-PCR analysis and a larger set of perturbed or physiological growth conditions were utilized next (Fig. 3). Besides nutrient starvation, these included growth at alkaline pH (8.0), cold stress (4 °C and 15 °C), heat shock (40 °C), and oxidative stress (50 mM H2O2). As shown in Fig. 3A, splaA was strongly upregulated following carbon starvation, in line with the results of Northern analysis. splaA expression levels also increased upon oxidative stress and heat shock, while only a slight upregulation was observed in the other conditions. By comparison, the splaB transcript was barely detectable in most conditions and slightly upregulated in heat-shocked and cold-stressed mycelia. It should be noted, however, that the splaB transcript accumulating at low temperature might not be functional. In fact, as revealed by sequence analysis (not shown), only the lower, largely unchanged amplicon corresponds to the mature splaB transcript, while the seemingly upregulated upper amplicon corresponds to an intron-containing, incompletely spliced species. No amplification product was detected in control reactions in which the reverse transcription step was omitted (not shown), thus indicating that the observed amplicons specifically originate from the splaA and splaB transcripts.
Fig. 3. Expression analysis of splaA and splaB.
(A) splaA and splaB expression levels were determined by RT-PCR analysis of total RNA extracted from A. oryzae RIB40 pre-cultured in DPY liquid medium for 24 h, transferred to the following fresh media, and grown for 4 h at 30 °C unless stated otherwise: (left to right) CD medium, CD without any carbon source, CD without any nitrogen source, CD adjusted to pH 8.0, CD medium at 4 °C, 15 °C and 40 °C, and CD containing 50 mM H2O2. γ-actin, utilized as an internal control, is shown at the bottom. (B) RT-PCR analysis of splaA and splaB during aerial hyphae and conidia formation. A. oryzae RIB40 was pre-cultured in DPY liquid medium for 24 h and then transferred to a Petri dish containing 10 ml of DPY liquid medium, so to allow the formation of aerial hyphae and conidia. After 6 h (early aerial hyphae formation), 18 h (completion of aerial hyphae formation and early conidiation), and 48 h (full conidiation), as revealed by microscopic examination, samples were collected and processed for RT-PCR as above (see ‘Materials and Methods’ for details).
In an another set of assays, expression levels for both genes were comparatively analyzed during the development of aerial hyphae and conidia, upon culture on solid medium (Fig. 3B). Under these conditions (see ‘Materials and Methods’ for details), aerial hyphae and conidia, whose formation was verified by microscopic examination, typically accumulate after 24 h and 48 h from the onset of culture. As shown in Fig. 3B, splaA was strongly upregulated during or after conidiation, while the splaB transcript was steadily expressed until the very onset of conidiation and markedly decreased thereafter. The differential expression profiles of the two sPLA2 genes, along with the distinct properties displayed by rsPlaA and rsPlaB, provide strong circumstantial evidence for the non-redundant, physiologically distinct roles played by these enzymes in A. oryzae.
3.4. Expression and subcellular localization of splaA and splaB in A. oryzae
To examine the cellular functions of the two sPLA2s, A. oryzae strains expressing either splaA or splaB (named OESA-1 and OESB-1, respectively) under the control of starch-inducible PglaA142 promoter (Minetoki et al., 1998) were constructed and analyzed. PLA2 activity in the culture medium and in cell lysates from OESA-1, OESB-1 and a vector-transformed control strain (NGA142-1) grown under inducing conditions was determined (Fig. 4A). PLA2 activity in OESA-1 was highest in the culture medium, with lesser amounts in the cell lysate fraction. This distribution contrasts with that of OESB-1, in which PLA2 activity was more abundant in the lysate fraction than in the culture medium. Furthermore, the pH dependence of PLA2 activity in OESA-1- and OESB-1-derived fractions was similar to that previously determined for rsPlaA and rsPlaB, indicating that the activities measured in sPLA2-expressing strains genuinely reflect sPlaA and sPlaB, rather than unrelated phospholipase activities (e.g., PLBs) encoded by the A. oryzae genome.
Further analysis of the sPLA2-expressing strains took advantage of polyclonal antibodies raised against rsPlaA and rsPlaB (see ‘Materials and Methods’), selectively recognizing the two enzymes. As shown in Fig. 4B, a fairly abundant 14 kDa polypeptide was detected in the culture supernatant (and only in trace amounts in the cell lysate) of OESA-1, but neither in OESB-1, nor in the vector-transformed control strain (not shown). Conversely, a doublet of bands centered around 15 kDa was only detected by the anti-sPlaB antibody in the cell lysate fraction from the OESB-1 strain. As revealed by N-terminal sequencing (not shown), the sPlaA polypeptide found in the OESA-1 supernatant starts with the Ala residue at position 37 (ATTCS…). Also, the secreted sPlaA polypeptide appeared not to be glycosylated, since its molecular mass did not change upon endoglycosidase H treatment (not shown). Since the predicted molecular mass of a sPlaA polypeptide spanning the region comprised between Ala37 and the Gly157 residue of the conserved Phe-Gly sequence (14.1 kDa) best fits the experimentally determined mass of secreted sPlaA (14 kDa), it is most likely that the polypeptide detected by the anti-sPlaA antibody is sPlaA lacking both the N-terminally located secretion signal sequence as well as the C-terminal extension sequence. Based on similar evidence, the mature sPlaB polypeptides likely result from heterogeneous C-terminal cleavage at either Gly146 (14.8 kDa) or Arg153 (15.5 kDa), assuming that both polypeptides start at Pro18. Although the precise C-terminal ends remain to be determined experimentally, it is clear that the two mature sPLA2s have different subcellular localizations. sPlaA, whose N-terminal signal peptide sequence is more extended, is secreted extracellularly, with the lower amounts of seemingly intracellular activity (Fig. 4A) likely reflecting either sPlaA on the way of secretion or loosely associated to the cell wall. In contrast, sPlaB, with its shorter N-terminal signal sequence, is also likely internalized (and processed) into the endoplasmic reticulum, but maintains a predominantly (if not exclusively) intracellular localization.
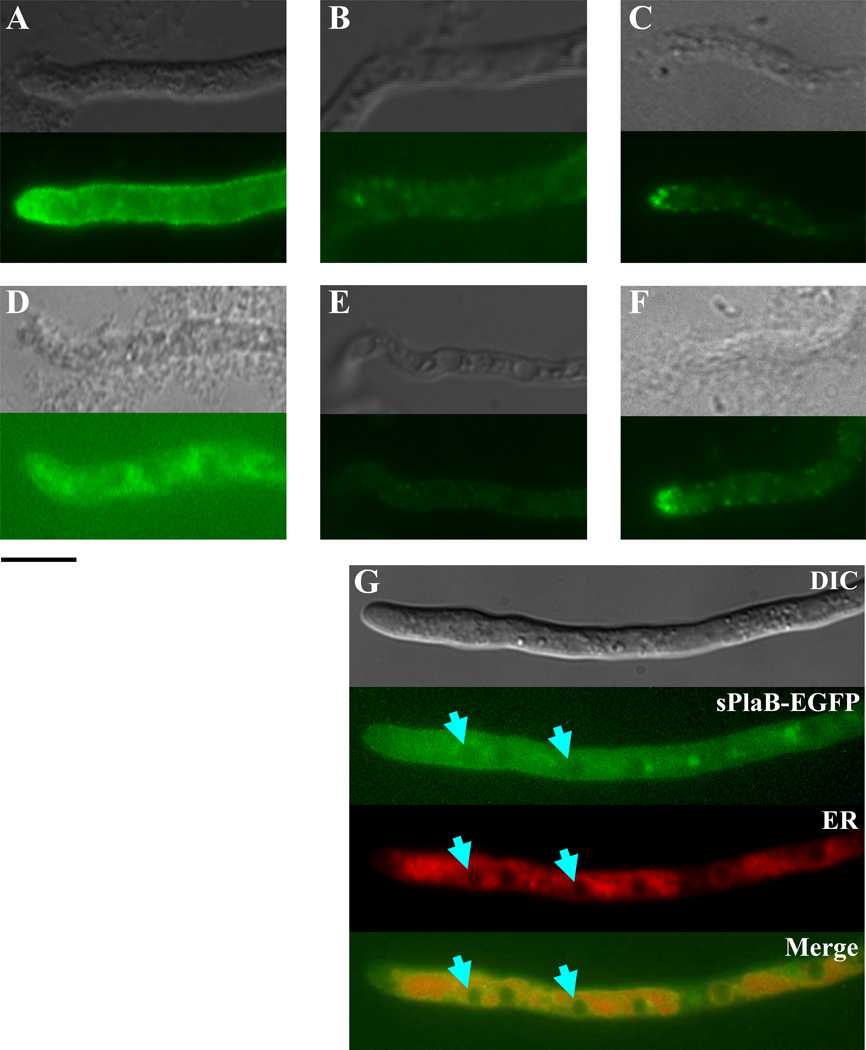
The latter conclusions regarding the different subcellular localization of sPlaA and sPlaB were confirmed and extended by the results of the indirect immunofluorescence analysis reported in Fig. 5. As revealed by this analysis, sPlaA accumulates in the cell surface layer with a marked preference for hyphal tips (Fig. 5A), a localization pattern closely resembling that of many secretory proteins in fungi (Masai et al., 2003). In contrast, sPlaB was mainly found within the mesh-like compartment around the nucleus with only a slight (if any) staining of the cell surface (Figs. 5D). None of the above distinctive staining patterns was observed in a double disrupted ΔsplaA/ΔsplaB strain (see below), which was used as a negative control for these experiments (Figs. 5B and E), whereas actin fine spots, typically accumulating in hyphal tips (Harris et al., 1994), were revealed by a positive control, anti-actin antibody (Figs. 5C and F).
Fig. 5. Immunocytochemical localization of sPlaA and sPlaB in A. oryzae.
Immunostaining of OESA-1 (A) and OESB-1 (D) with the anti-sPlaA and the anti-sPlaB antibodies, respectively. Immunostaining of a ΔsplaA/ΔsplaB double disruptant strain (DSAB-1, see Fig. 8), utilized as a negative control, with anti-sPlaA and anti-sPlaB is shown in panels B and E. Immunostaining of OESA-1 (C) and OESB-1 (F) with an anti-actin antibody, which served as a positive control, is also shown. Light transmission (top) and fluorescence (bottom) images are reported in each panel. (G) Immunofluorescence (sPlaB-EGFP), ER-tracker staining (ER) and merged images of the sPlaB-EGFP expressing strain, SBfG; arrowheads indicate sPlaB co-localization sites revealed by EGFP fluorescence and ER-tracker staining. A differential interference contrast image is shown at the top; scale bar, 10 µm.
The distinctive localization patterns revealed by immunofluorescence were essentially recapitulated by in vivo imaging of sPlaA-EGFP and sPlaB-EGFP transformants. The expression of both fusion proteins was driven by amyB (α-amylase) promoter, since the level of endogenous expression was quite low as evidenced from our microarray analysis (M.A., unpublished observation). As shown in Fig. 5G, we found that the mesh-like, ER-associated (perinuclear) distribution of sPlaB-EGFP closely overlapped that produced by the ER-Tracker dye, Blue-white DPX, thus further strengthening the notion that sPlaB localizes to the ER membrane (Maruyama et al., 2006).
3.5. Phospholipid profiling of sPLA2 overexpressing strains
To gain insight into the in vivo activity of sPlaA and sPlaB, a large scale phospholipid analysis was conducted in OESA-1 and OESB-1 mycelia grown under inducing conditions. Following verification of sPLA2 overexpression by immunoblot analysis (not shown), lipids were extracted, subjected to mass-spectrometric analysis, and individual phospholipid species, differing in either their polar head groups or fatty acyl moieties, were identified against pre-existing templates (see ‘Materials and Methods’ for details). As shown in Table 2, phospholipids bearing a choline (PC) or an ethanolamine (PE) polar head-group and a 36 or 34 carbon di-acyl moiety with two or four double bonds, are the most abundant phospholipid species in A. oryzae. No significant, strain-specific variations were observed neither in the type of fatty acyl chains, nor in their degree of unsaturation in the two overexpressing strains. Instead, the relative abundance of total phospholipids bearing a choline or an ethanolamine polar head group differentially decreased in OESA-1 and OESB-1, respectively. sn-2 ester bond hydrolysis by PLA2s can trigger further processing of the resulting lysophospholipid and phospholipid turnover (Brown et al., 2003). Thus, differential decrease of PC and PE may reflect the different polar head-group preference and/or the distinct subcellular localization of sPlaA and sPlaB. Also worth of note, is the overall increase in lysophosphatidic acid, and the preferential accumulation of lysophospholipids with saturated or mono-unsaturated, C14 or C16 sn-1 fatty acyl chains, observed in both strains.
Table 2.
Global phospholipid profiles of the sPLA2-expressing and control strains
| NGA 142-1a | OESA-1a | OESB-1a | |
|---|---|---|---|
| PHOSPHOLIPIDS | |||
| Polar head groupsb | |||
| Choline | 41.91 | 39.50 | 41.32 |
| Ethanolamine | 35.50 | 34.40 | 31.35 |
| Inositol | 12.71 | 14.20 | 14.60 |
| Serine | 7.74 | 9.72 | 10.33 |
| Phosphatidic Acid | 2.09 | 2.27 | 2.36 |
| Di-fatty acyl groups (N. of carbons)ce | |||
| 32 | 0.28 | 0.40 | 0.38 |
| 33 | 1.19 | 0.93 | 0.97 |
| 34 | 38.75 | 41.41 | 42.02 |
| 35 | 2.24 | 1.76 | 1.57 |
| 36 | 52.20 | 47.90 | 48.68 |
| 37 | 0.32 | 0.25 | 0.23 |
| 38 | 1.24 | 1.02 | 0.86 |
| 40 | 1.35 | 1.07 | 1.30 |
| 41 | 1.93 | 4.29 | 3.71 |
| 42 | 0.36 | 0.83 | 0.21 |
| Degree of unsaturation (N. of double bonds)de | |||
| 0 | 0.35 | 0.50 | 0.38 |
| 1 | 6.51 | 8.29 | 8.67 |
| 2 | 37.56 | 37.76 | 37.50 |
| 3 | 13.09 | 13.73 | 14.60 |
| 4 | 34.03 | 28.10 | 28.20 |
| 5 | 7.54 | 10.49 | 9.78 |
| 6 | 0.52 | 0.94 | 0.70 |
| 7 | 0.22 | 0.15 | 0.14 |
| LYSOPHOSPHOLIPIDS | |||
| Polar head groupsb | |||
| Choline | 64.61(0.52)f | 58.50 (0.46) | 61.09 (0.57) |
| Ethanolamine | 27.89 (0.22) | 26.50 (0.21) | 25.34 (0.24) |
| Lysophosphatidic acid | 7.50 (0.06) | 15.00 (0.12) | 13.57 (0.13) |
| Fatty acyl groups (N. of carbons)c, e | |||
| 14 | 0.05 | 0.34 | 0.31 |
| 15 | 0.56 | 0.57 | 0.57 |
| 16 | 28.24 | 34.29 | 30.75 |
| 17 | 1.40 | 1.32 | 0.85 |
| 18 | 69.17 | 62.71 | 67.14 |
| 20 | 0.58 | 0.78 | 0.39 |
| Degree of unsaturation (N. of double bonds)d, e | |||
| 0 | 31.06 | 37.10 | 34.17 |
| 1 | 9.82 | 11.47 | 11.80 |
| 2 | 54.95 | 47.09 | 50.38 |
| 3 | 4.12 | 4.29 | 3.62 |
The control (NGA 142-1), sPlaA- (OESA-1) and sPlaB- (OESB-1) expressing strains were subjected to PL, and LysoPL analysis.
Relative polar head group abundance (mol %) in the total PL or LysoPL pool.
Relative abundance (mol %) of the indicated di-fatty acyl or mono-fatty acyl groups in the total PL or LysoPL pool as indicated.
Relative abundance (mol %) of di-fatty acyl or mono-fatty acyl groups with different degrees of unsaturation in the total PL or LysoPL pool as indicated.
Only fatty acyl species with a relative abundance >0.1% are reported.
Indicated in brackets are LysoPL polar head group abundance values expressed as nmol/mg dry weight (see ‘Experimental procedures’ for details).
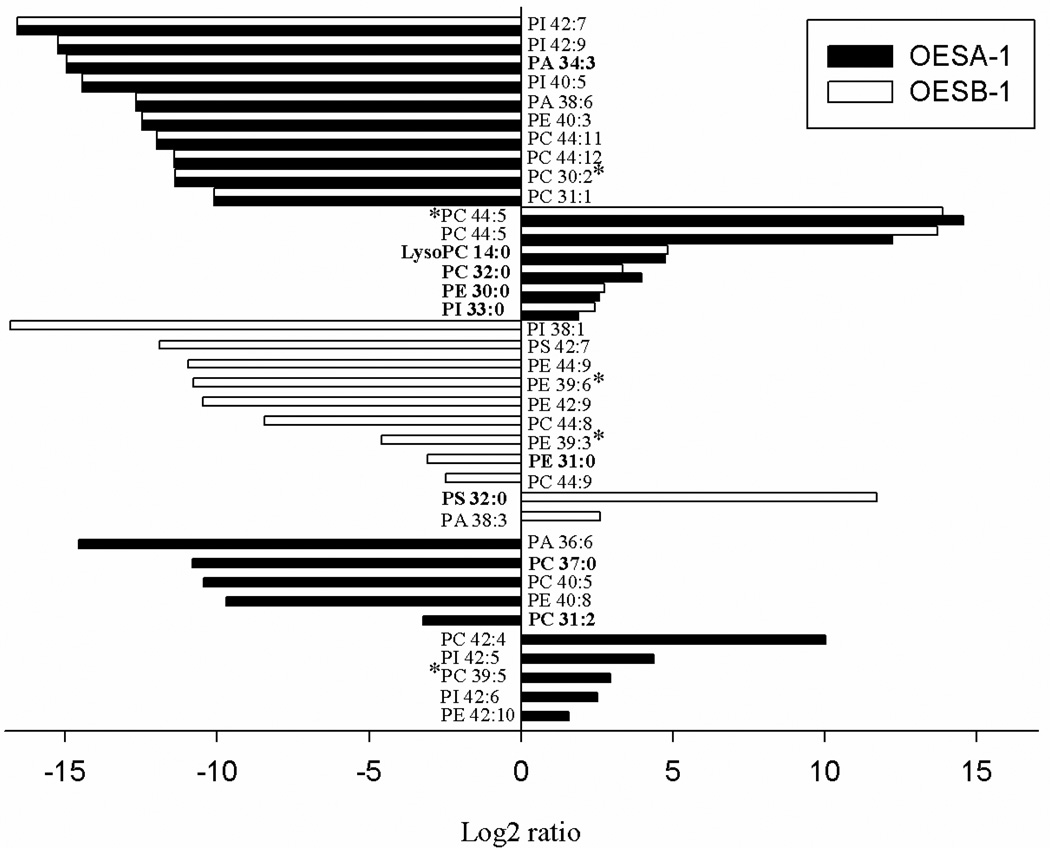
Individual phospholipids that, regardless of their relative abundance, are differentially represented in either OESA-1 or OESB-1, or in both overexpressing strains, compared to the control, were also identified (Fig. 6). Most notable among the latter species were lysoPC 14:0, which nearly equally increased in OESA-1 and OESB-1, and various PCs and phosphatidylinositols bearing unusually long polyunsaturated fatty acids, that were found to be significantly diminished in both overexpressing strains. On the other hand, and in keeping with the results of the previous global analysis (Table 2), other PC and PE species were found among the most frequently diminished phospholipids in OESA-1 and OESB-1, respectively. Also interesting to note is the occurrence, in both strains, of opposite sign variations involving different phospholipids sharing the same polar head group. One of them specifically involves two different phosphatidylserine species in OESB-1, while two reciprocal variations in the relative abundance of four distinct PC species were observed in OESA-1. Despite the inherent complexity of these phospholipid composition variations, which also reflect phospholipid remodeling events taking place downstream of sPLA2 action, a somewhat different substrate preference of sPlaA and sPlaB is apparent. This, coupled with the distinct subcellular localization of the two enzymes, might lead to the differential accumulation of distinct phospholipid and lysophospholipid (but also fatty acid) species.
Fig. 6. Changes in phospholipid abundance in sPLA2 overexpressing strains.
Summary of the phospholipids, whose levels were found to be consistently altered in OESA-1 (solid bars) and OESB-1 (open bars) compared to the control (NGA142-1) strain. Phospholipids that changed by at least two-fold in the overexpressing strains are reported; changes are annotated as the log2 ratio of the amount of each phospholipid in OESA-1 and OESB-1 relative to the control. Individual phospholipid and lysophospholipid species matching the available yeast lipid template are in bold, the remaining species were identified against a animal lipid template; lipid species that did not match neither the yeast nor the animal lipid template, were identified on the basis of their spectra and m/z values and are indicated with an asterisk. PI, phosphatidylinositol; PA, phosphatidic acid; PE, phosphatidylethanolamine; PC, phosphatidylcholine. Numbers are the length of the carbon chain: degree of unsaturation.
3.6. Phenotypic analysis of sPLA2 overexpressing strains
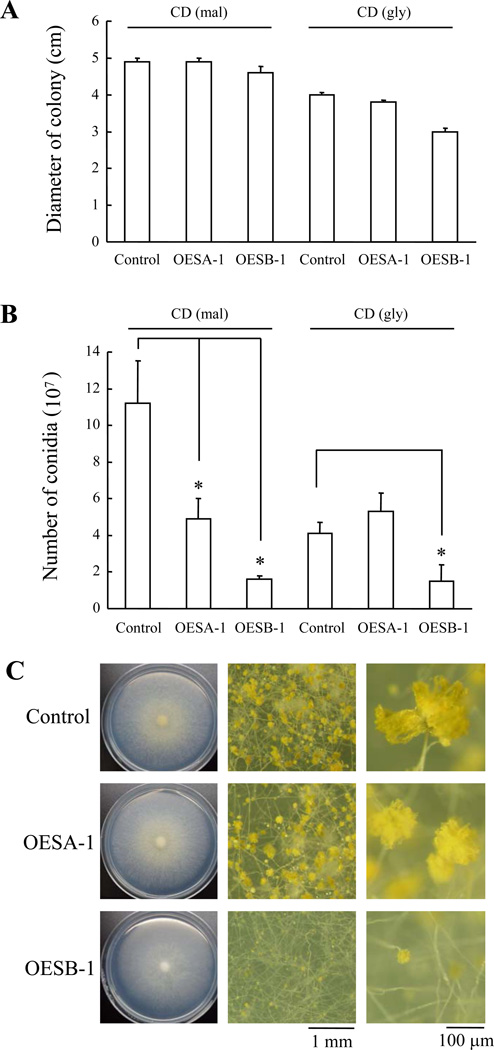
The phenotypes of sPLA2-overexpressing strains were examined next. In general, the PglaA142 promoter utilized for sPLA2 overexpression is turned on when starch, maltose, or glucose are used as the sole carbon source, while it is off (or minimally active) in glycerol-containing media. As shown in Fig. 7A, mycelial growth of OESA-1 was unaffected by sPLA2 overexpression and nearly the same as that of the vector-transformed control strain under both inducing and non-inducing conditions, while the growth of OESB-1 was slightly reduced. Interestingly, however, both sPLA2 overexpressing strains produced less conidia than the control strain under inducing conditions (Fig. 7B). The latter phenotype was especially pronounced in the case of OESB-1, which produced significantly less conidia even under non-inducing conditions -as if the low, basal levels of sPlaB produced on glycerol medium were sufficient to inhibit conidiation. As further documented by stereomicroscopic analysis (Fig. 7C), OESB-1 formed immature conidial heads at a much higher frequency than OESA-1 and the control strain. It thus appears that forced expression of sPLA2s, especially sPlaB, inhibits conidiation.
Fig. 7. Effect of splaA and splaB overexpression on conidiation.
Conidia from control, OESA-1 and OESB-1 strains (∼104 conidia in 5 µl of sterile ddH2O) were inoculated on inducing (CD-mal) or repressing medium (CD-gly) medium in agar plates and incubated for 6 days at 30 °C. (A) Diameter of the resulting mycelial colonies. (B) Conidial production. Data are expressed as the mean ± standard deviation of three independent replicates (*: P-value ≤ 0.05; ‘overexpressors’ vs. ‘control’). (C) Control and overexpressing strains cultured on CD-mal, visualized by stereomicroscopy; whole mycelia colonies (left panels) and higher magnification images of conidia (middle panels) and conidiophores (right panels) are shown.
3.7. sPLA2 gene disruptants
To further explore the functional roles of the two sPLA2s, mutants disrupted in either splaA or splaB were generated by using the disruption strategy outlined in Figs. S2A and S2C. Linear DNA fragments, in which the A. nidulans sC gene or the A. oryzae adeA gene are flanked by upstream and downstream sequences derived from splaA or splaB (1.5 kb each), were constructed and introduced into A. oryzae. One splaA-disruptant, named DSA-1, and eight splaB-disruptants, named DSB-1 to −8, were thus obtained. sPLA2 gene disruption was verified for all disruptants by PCR (data not shown) and by Southern analysis for DSA-1 and DSB-1 (Figs. S2B and S2D). None of the latter strains, however, showed any sign of phenotypic alteration under the previously examined mycelial growth conditions (listed in Fig. 3), and a similar result was obtained with a double disruptant, named DSAB-1, that was generated by knocking-out splaB within the splaA-disrupted DSA-1 strain (Figs. S2E and S2F; data not shown). What these negative results suggest is that sPlaA and sPlaB, although capable of causing defective conidiation when overexpressed, are dispensable for A. oryzae viability under most of the presently examined growth conditions. The only disruptant-specific defect was an increased sensitivity to oxidative stress. This was observed with proliferating hyphae from the ΔsplaB mutants (DSB-1 and DSB-2)-the growth of ΔsplaB mutants, but not ΔsplaA mutant, was more severely impaired when exposed to H2O2 (10 and 20 mM) compared to the control strain (Fig. 8).
4. Discussion
An increasingly growing number of biological functions is being documented for mammalian sPLA2s and the phospholipid hydrolysis products -unsaturated fatty acids and lysophospholipids- generated by their catalytic action (Murakami and Kudo, 2004; Schaloske and Dennis, 2006). As revealed by this work, a variety of roles, accompanied by distinct subcellular localizations, expression profiles and phospholipid substrate preferences, also appear to hold for the recently discovered group XIV fungal sPLA2s.
4.1. Conserved features of microbial sPLA2s
Microbial sPLA2s share a ∼30 amino acid-long conserved central region, containing the His-Asp dyad sequence responsible for the calcium-dependent catalytic activity of these enzymes. Another conserved feature of microbial sPLA2s, including sPlaA and sPlaB, is the relative position of Cys residues (dotted in Fig. 1). We previously showed that two disulfide-bonded cysteines are essential for enzymatic activity of the fungal sPLA2 p15 (Nakashima et al., 2003), and disulfide bonds at similar locations have been reported for the S. violaceoruber sPLA2 (Matoba et al., 2002). sPlaA and sPlaB, as well as other microbial sPLA2s, have cysteine residues at similar relative locations, suggesting that these disulfide bond features may also be conserved. In addition, there is a conserved Phe-Gly sequence at the C-terminal end of the two Streptomyces sPLA2s and upstream of the Kex2-recognition site (Lys-Arg) in various fungal sPLA2s including sPlaA. This observation, together with the experimentally documented C-terminal processing of p15 between the Ser and Lys residues following the Phe-Gly sequence (Wakatsuki et al., 1999), suggests that the presence of a C-terminal pro-sequence is a rather common feature in fungal sPLA2s.
4.2. Spa A and sPlaB are functionally divergent sPLA2s
sPlaA and sPlaB, the first pair of sPLA2s to be characterized in fungi, differ in a number of features, including pH optimum, Ca2+ requirement, expression profile and subcellular localization. sPlaA, which based on four distinct lines of evidence appears to accumulate on the surface of hyphal tips and to be secreted extracellularly, displays the highest enzymatic activity in vitro and is the most stress responsive. Similar to the TbSP1 sPLA2 from T. borchii (Soragni et al., 2001), sPlaA was strongly induced by carbon starvation. This suggests a potential role in carbon foraging, either through the attack of extracellular phospholipid substrates or through the autophagic breakdown of endogenous phospholipids and the release of fatty acids for gluconeogenesis. These shared surface membrane association and extracellular release properties, along with carbon starvation stress responsiveness is in line with the co-clustering of sPlaA and TbSP1 highlighted by phylogenetic analysis (unpublished observation). Without ruling out possible differences in signaling and/or abiotic stress responsiveness specifically related to the symbiotic (T. borchii) and the saprotrophic (A. oryzae) lifestyles, the above similarities support the notion of a true orthologous relationship between the two enzymes.
sPlaB, instead, is expressed at low levels in general and was poorly modulated in response to a variety of stimuli. It is less active in vitro and exhibited a strikingly different subcellular localization, with maximal accumulation within ER-like perinuclear structures. The distinct subcellular localizations of sPlaA and sPlaB are consistent with the different pH optima and Ca2+ requirements of the two recombinant enzymes. The acidic pH optimum of sPlaA parallels the pH of the culture medium, while the optimum pH and Ca2+ requirement of sPlaB are reminiscent of the neutral pH and sub-millimolar (0.25–0.60 mM) Ca2+ concentration previously reported for the ER lumen in mammals (Kim et al., 1998; Kneen et al., 1998; Demaurex and Frieden, 2003). Although sPlaB localization within ER-like structures is quite peculiar, the occurrence of sPLA2s within the intracellular compartment is not unprecedented. For example, a mammalian group IIA sPLA2 has been shown to localize to secretory granules (Enomoto et al., 2000) or to punctate and perinuclear structures that colocalize with caveolin, possibly through its association with glypican, a glycosylphosphatidylinositol-anchored heparan sulfate proteoglycan (Murakami et al., 1999). A similar localization pattern was displayed by a group IID sPLA2 (Murakami et al., 2001), whereas group V and X sPLA2s have been shown to be mainly localized within secretory granules and to shift to the plasma membrane (group V) or to the extracellular medium (group X) upon cell stimulation (Murakami et al., 2001). In another study (Balboa et al., 2003), evidence was presented as to a localization mechanism whereby secreted group IIA, IID, and V sPLA2s become localized intracellularly upon extracellular secretion and subsequent (re)internalization. Thus, intracellular localization (either direct or through secretion and re-internalization) and fatty acid/lysophospholipid release from intracellular membranes, in close proximity to diverse lipid-metabolizing enzymes, is likely a common mechanism underlying sPLA2-mediated signaling (Mounier et al., 2004). In this way, bioactive lipid precursors would be generated, in a stimulus-dependent manner, in close proximity to the downstream-acting enzymes (e.g., unsaturated fatty acid oxygenases, see below) that are ultimately responsible for bioactive lipid production.
4.3. sPLA2 overexpression and conidiation
sPLA2 overexpression, especially sPlaB, strongly interfered with conidiation, without any detectable effect on mycelial growth (Fig. 7). This, along with the lipid composition variations revealed by phospholipid profiling, suggests that increased production of phospholipid breakdown products as a consequence of sPLA2 overexpression may indeed trigger the downstream accumulation of conidiation-interfering lipid derivatives. Studies in A. nidulans have shown that hormone-like molecules derived from oleic, linoleic and linolenic acid, collectively designated as psi factors, influence the development of asexual conidiospores and asexual ascospores (Tsitsigiannis et al., 2004; Tsitsigiannis et al., 2005). Similar results as to the involvement of unsaturated fatty acid derivatives, also known as oxylipins, in sporulation has been reported in N. crassa, where two linoleic acid derivatives, psiBα and psiCα, stimulate sexual and inhibit asexual spore development, respectively, whereas a third one, psiAα, enhances asexual sporulation (Champe et al., 1987; Champe and el-Zayat, 1989). Although neither psi factor receptors, nor their transduction mechanisms have thus far been elucidated, it is clear that in two different filamentous ascomycetes the balance between distinct oxylipin species represents a key determinant of asexual vs. sexual sporulation. Also considering the well documented role of mammalian sPLA2s in the generation of arachidonate-derived lipid messengers, it is tempting to speculate that the conidiation impairment brought about by sPLA2 overexpression in A. oryzae is causally related to an unbalanced production of psi factor-like signaling molecules. In keeping with this view, at least six genes similar to the fatty acid oxygenases required for psi factor biosynthesis in A. nidulans (Tsitsigiannis et al., 2004; Tsitsigiannis et al., 2005) have been revealed by a survey of the A. oryzae genome database (M.A., unpublished observations). Therefore, even though no oxylipin-like molecule has thus far been identified in A. oryzae, it appears that fungal sPLA2s, similar to their animal counterparts, are also likely involved in intra- and intercellular communication through the release of unsaturated fatty acids. Along this view, the stronger conidiation interference effect exerted by sPlaB may reflect its intracellular localization and thus the in situ production of bioactive lipid precursors in closer proximity to their ultimate targets.
Further support to the existence of a functional link between sPLA2 and conidiation was provided by the observation that splaA and splaB differentially accumulate prior to or concomitant with conidia development, respectively. Similar results as to splaA expression during conidiation have been obtained by reporter assays monitoring the time course of EGFP expression driven by the splaA promoter (data not shown). In addition, as reported previously (Adams et al., 1998), we find that conidia-like structures also form under submerged culture conditions when cells are grown in carbon-deficient (CD) medium (T. N. and M. A., unpublished results). This further strengthens the link between splaA expression and conidiation and points to a causal relationship between carbon starvation and the initiation of conidia development.
4.4. sPLA2 disruption is phenotypically silent under unstressed conditions
Disruption of splaA and splaB did not cause any detectable defect in either mycelial growth or conidiation. This finding is not so surprising if one considers the natural lack of any recognizable sPLA2 homolog in the genome databases of the closely related species A. nidulans and A. fumigatus. Rather, it suggests that sPLA2 action is either largely dispensable or functionally replaceable by other phospholipases. In accordance with the latter hypothesis, a cPLA2 (Hong et al., 2005) as well as two putative PLBs are present in A. nidulans, and sequences homologous to both types of phospholipases are also present in the A. oryzae genome (M. A., unpublished observations).
An additional possibility is that one or both sPLA2s are specifically required during the sexual life cycle, a developmental phase not yet identified in A. oryzae, or under adverse growth conditions. Indeed, further search for phenotypic alterations that may become apparent under such conditions showed that splaB disruptant is more sensitive to oxidative stress compared to their isogenic wild-type strains; splaB deletion resulted in oxidative stress hypersensitivity in vegetative hyphae. The effect may be causally related to the well-known oxidation propensity of sn-2 position polyunsaturated fatty acids (Niki et al., 2005), which, as revealed by phospholipid profiling (Table 2), are highly represented also among A. oryzae phospholipids.
In mammals, two distinct pathways contribute to phospholipid hydroperoxide removal and prevention of free radical-induced membrane damage. One of them relies on direct reduction by hydroperoxide glutathione peroxidase (Imai et al., 1996), while the other proceeds through the release of fatty acid hydroperoxides by selected PLA2 isoforms (Cummings et al., 2000) and their subsequent detoxification by cytosolic glutathione peroxidase (Girotti, 1998). Since no hydroperoxide glutathione peroxidase homolog has been identified in fungal genome databases so far, it is conceivable that phospholipid hydroperoxide repair may represent a previously unknown function adding to the multifaceted and as yet largely unknown pathways supported by sPLA2s in filamentous ascomycetes.
Supplementary Material
Acknowledgements
We thank Angelo Bolchi (Department of Biochemistry and Molecular Biology, University of Parma) for critical reading of the manuscript, Riccardo Percudani for help with phylogenetic analysis, and Barbara Montanini for technical advice on phospholipid profiling data analysis. Phospholipid profiling was performed at the Kansas Lipidomics Research Center, which was supported by Kansas Technology Enterprise Corp., Kansas State University, NSF grants MCB 0455318, DBI 0521587, and EPS 0236913, and NIH grant P20 RR16475 from the National Center for Research Resources. This work was supported by a Grant-in-Aid for Scientific Research (No. 18580067 and No. 20580074) to M. A. from the Ministry of Education, Science, Sports and Culture of Japan, by a grant from Amano Enzyme Inc., and by grants from the Ministry of Education, University and Research of Italy, FIRB program “Genomica funzionale dell’interazione tra piante e microrganismi” and from the University of Parma, FIL 2005 program to S.O.
Abbreviations used in this paper
- His6
hexahistidine
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PLA2
phospholipase A2
- RT-PCR
reverse transcription-polymerase chain reaction
- sPLA2
secretory PLA2
References
- Adams TH, Wieser JK, Yu JH. Asexual sporulation in Aspergillus nidulans . Microbiol. Mol. Biol. Rev. 1998;62:35–54. doi: 10.1128/mmbr.62.1.35-54.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akao T, Sano M, Yamada O, Akeno T, Fujii K, Goto K, Ohashi Kunihiro S, Takase K, Yasukawa Watanabe M, Yamaguchi K, Kurihara Y, Maruyama J, Juvvadi PR, Tanaka A, Hata Y, Koyama Y, Yamaguchi S, Kitamoto N, Gomi K, Abe K, Takeuchi M, Kobayashi T, Horiuchi H, Kitamoto K, Kashiwagi Y, Machida M, Akita O. Analysis of expressed sequence tags from the fungus Aspergillus oryzae cultured under different conditions. DNA Res. 2007;14:47–57. doi: 10.1093/dnares/dsm008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboa MA, Shirai Y, Gaietta G, Ellisman MH, Balsinde J, Dennis EA. Localization of group V phospholipase A2 in caveolin-enriched granules in activated P388D1 macrophage-like cells. J. Biol. Chem. 2003;278:48059–48065. doi: 10.1074/jbc.M305904200. [DOI] [PubMed] [Google Scholar]
- Bartz R, Li WH, Venables B, Zehmer JK, Roth MR, Welti R, Anderson RG, Liu P, Chapman KD. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J. Lipid Res. 2007;48:837–847. doi: 10.1194/jlr.M600413-JLR200. [DOI] [PubMed] [Google Scholar]
- Brown WJ, Chambers K, Doody A. Phospholipase A2 (PLA2) enzymes in membrane trafficking: mediators of membrane shape and function. Traffic. 2003;4:214–221. doi: 10.1034/j.1600-0854.2003.00078.x. [DOI] [PubMed] [Google Scholar]
- Champe SP, el-Zayat AA. Isolation of a sexual sporulation hormone from Aspergillus nidulans . J. Bacteriol. 1989;171:3982–3988. doi: 10.1128/jb.171.7.3982-3988.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champe SP, Rao P, Chang A. An endogenous inducer of sexual development in Aspergillus nidulans . J. Gen. Microbiol. 1987;133:1383–1387. doi: 10.1099/00221287-133-5-1383. [DOI] [PubMed] [Google Scholar]
- Cummings BS, McHowat J, Schnellmann RG. Phospholipase A2s in cell injury and death. J. Pharmacol. Exp. Ther. 2000;294:793–799. [PubMed] [Google Scholar]
- Demaurex N, Frieden M. Measurements of the free luminal ER Ca2+ concentration with targeted “cameleon” fluorescent proteins. Cell Calcium. 2003;34:109–119. doi: 10.1016/s0143-4160(03)00081-2. [DOI] [PubMed] [Google Scholar]
- Elsbach P, Weiss J. Utilization of labeled Escherichia coli as phospholipase substrate. Methods Enzymol. 1991;197:24–31. doi: 10.1016/0076-6879(91)97130-q. [DOI] [PubMed] [Google Scholar]
- Enomoto A, Murakami M, Valentin E, Lambeau G, Gelb MH, Kudo I. Redundant and segregated functions of granule-associated heparin-binding group II subfamily of secretory phospholipases A2 in the regulation of degranulation and prostaglandin D2 synthesis in mast cells. J. Immunol. 2000;165:4007–4014. doi: 10.4049/jimmunol.165.7.4007. [DOI] [PubMed] [Google Scholar]
- Girotti AW. Lipid hydroperoxide generation, turnover, and effector action in biological systems. J. Lipid Res. 1998;39:1529–1542. [PubMed] [Google Scholar]
- Hanada T, Sato T, Arioka M, Uramoto M, Yamasaki M. Purification and characterization of a 15 kDa protein (p15) produced by Helicosporium that exhibits distinct effects on neurite outgrowth from cortical neurons and PC12 cells. Biochem. Biophys. Res. Commun. 1996;228:209–215. doi: 10.1006/bbrc.1996.1641. [DOI] [PubMed] [Google Scholar]
- Harris SD, Morrell JL, Hamer JE. Identification and characterization of Aspergillus nidulans mutants defective in cytokinesis. Genetics. 1994;136:517–532. doi: 10.1093/genetics/136.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Horiuchi H, Ohta A. Identification and molecular cloning of a gene encoding Phospholipase A2 (plaA) from Aspergillus nidulans . Biochim. Biophys. Acta. 2005;1735:222–229. doi: 10.1016/j.bbalip.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Ikeno Y, Konno N, Cheon SH, Bolchi A, Ottonello S, Kitamoto K, Arioka M. Secretory phospholipases A2 induce neurite outgrowth in PC12 cells through lysophosphatidylcholine generation and activation of G2A receptor. J. Biol. Chem. 2005;280:28044–28052. doi: 10.1074/jbc.M503343200. [DOI] [PubMed] [Google Scholar]
- Imai H, Sumi D, Sakamoto H, Hanamoto A, Arai M, Chiba N, Nakagawa Y. Overexpression of phospholipid hydroperoxide glutathione peroxidase suppressed cell death due to oxidative damage in rat basophile leukemia cells (RBL-2H3) Biochem. Biophys. Res. Commun. 1996;222:432–438. doi: 10.1006/bbrc.1996.0762. [DOI] [PubMed] [Google Scholar]
- Jin FJ, Maruyama J, Juvvadi PR, Arioka M, Kitamoto K. Development of a novel quadruple auxotrophic host transformation system by argB gene disruption using adeA gene and exploiting adenine auxotrophy in Aspergillus oryzae . FEMS Microbiol. Lett. 2004a;239:79–85. doi: 10.1016/j.femsle.2004.08.025. [DOI] [PubMed] [Google Scholar]
- Jin FJ, Maruyama J, Juvvadi PR, Arioka M, Kitamoto K. Adenine auxotrophic mutants of Aspergillus oryzae: development of a novel transformation system with triple auxotrophic hosts. Biosci. Biotechnol. Biochem. 2004b;68:656–662. doi: 10.1271/bbb.68.656. [DOI] [PubMed] [Google Scholar]
- Kim JH, Johannes L, Goud B, Antony C, Lingwood CA, Daneman R, Grinstein S. Noninvasive measurement of the pH of the endoplasmic reticulum at rest and during calcium release. Proc. Natl. Acad. Sci. USA. 1998;95:2997–3002. doi: 10.1073/pnas.95.6.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamoto K. Molecular biology of the Koji molds. Adv. Appl. Microbiol. 2002;51:129–153. doi: 10.1016/s0065-2164(02)51004-2. [DOI] [PubMed] [Google Scholar]
- Kneen M, Farinas J, Li Y, Verkman AS. Green fluorescent protein as a noninvasive intracellular pH indicator. Biophys. J. 1998;74:1591–1599. doi: 10.1016/S0006-3495(98)77870-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Abe K, Asai K, Gomi K, Juvvadi PR, Kato M, Kitamoto K, Takeuchi M, Machida M. Genomics of Aspergillus oryzae . Biosci. Biotechnol. Biochem. 2007;71:646–670. doi: 10.1271/bbb.60550. [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long AD, Mangalam HJ, Chan BY, Tolleri L, Hatfield GW, Baldi P. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. J. Biol. Chem. 2001;276:19937–19944. doi: 10.1074/jbc.M010192200. [DOI] [PubMed] [Google Scholar]
- Mabashi Y, Kikuma T, Maruyama J, Arioka M, Kitamoto K. Development of a versatile expression plasmid construction system for Aspergillus oryzae and its application to visualization of mitochondria. Biosci. Biotechnol. Biochem. 2006;70:1882–1889. doi: 10.1271/bbb.60052. [DOI] [PubMed] [Google Scholar]
- Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto K, Arima T, Akita O, Kashiwagi Y, Abe K, Gomi K, Horiuchi H, Kitamoto K, Kobayashi T, Takeuchi M, Denning DW, Galagan JE, Nierman WC, Yu J, Archer DB, Bennett JW, Bhatnagar D, Cleveland TE, Fedorova ND, Gotoh O, Horikawa H, Hosoyama A, Ichinomiya M, Igarashi R, Iwashita K, Juvvadi PR, Kato M, Kato Y, Kin T, Kokubun A, Maeda H, Maeyama N, Maruyama J, Nagasaki H, Nakajima T, Oda K, Okada K, Paulsen I, Sakamoto K, Sawano T, Takahashi M, Takase K, Terabayashi Y, Wortman JR, Yamada O, Yamagata Y, Anazawa H, Hata Y, Koide Y, Komori T, Koyama Y, Minetoki T, Suharnan S, Tanaka A, Isono K, Kuhara S, Ogasawara N, Kikuchi H. Genome sequencing and analysis of Aspergillus oryzae . Nature. 2005;438:1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- Maruyama J, Kikuchi S, Kitamoto K. Differential distribution of the endoplasmic reticulum network as visualized by the BipA-EGFP fusion protein in hyphal compartments across the septum of the filamentous fungus, Aspergillus oryzae . Fungal Genet. Biol. 2006;43:642–654. doi: 10.1016/j.fgb.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Masai K, Maruyama J, Nakajima H, Kitamoto K. In vivo visualization of the distribution of a secretory protein in Aspergillus oryzae hyphae using the RntA-EGFP fusion protein. Biosci. Biotechnol. Biochem. 2003;67:455–459. doi: 10.1271/bbb.67.455. [DOI] [PubMed] [Google Scholar]
- Matoba Y, Katsube Y, Sugiyama M. The crystal structure of prokaryotic phospholipase A2 . J. Biol. Chem. 2002;277:20059–20069. doi: 10.1074/jbc.M200263200. [DOI] [PubMed] [Google Scholar]
- Minetoki T, Kumagai C, Gomi K, Kitamoto K, Takahashi K. Improvement of promoter activity by the introduction of multiple copies of the conserved region III sequence, involved in the efficient expression of Aspergillus oryzae amylase-encoding genes. Appl. Microbiol. Biotechnol. 1998;50:459–467. doi: 10.1007/s002530051321. [DOI] [PubMed] [Google Scholar]
- Miozzi L, Balestrini R, Bolchi A, Novero M, Ottonello S, Bonfante P. Phospholipase A2 up-regulation during mycorrhiza formation in Tuber borchii . New Phytol. 2005;167:229–238. doi: 10.1111/j.1469-8137.2005.01400.x. [DOI] [PubMed] [Google Scholar]
- Mounier CM, Ghomashchi F, Lindsay MR, James S, Singer AG, Parton RG, Gelb MH. Arachidonic acid release from mammalian cells transfected with human groups IIA and X secreted phospholipase A2 occurs predominantly during the secretory process and with the involvement of cytosolic phospholipase A2-α. J. Biol. Chem. 2004;279:25024–25038. doi: 10.1074/jbc.M313019200. [DOI] [PubMed] [Google Scholar]
- Murakami M, Kambe T, Shimbara S, Yamamoto S, Kuwata H, Kudo I. Functional association of type IIA secretory phospholipase A2 with the glycosylphosphatidylinositol-anchored heparan sulfate proteoglycan in the cyclooxygenase-2-mediated delayed prostanoid-biosynthetic pathway. J. Biol. Chem. 1999;274:29927–29936. doi: 10.1074/jbc.274.42.29927. [DOI] [PubMed] [Google Scholar]
- Murakami M, Koduri RS, Enomoto A, Shimbara S, Seki M, Yoshihara K, Singer A, Valentin E, Ghomashchi F, Lambeau G, Gelb MH, Kudo I. Distinct arachidonate-releasing functions of mammalian secreted phospholipase A2s in human embryonic kidney 293 and rat mastocytoma RBL-2H3 cells through heparan sulfate shuttling and external plasma membrane mechanisms. J. Biol. Chem. 2001;276:10083–10096. doi: 10.1074/jbc.M007877200. [DOI] [PubMed] [Google Scholar]
- Murakami M, Kudo I. Secretory phospholipase A2 . Biol. Pharm. Bull. 2004;27:1158–1164. doi: 10.1248/bpb.27.1158. [DOI] [PubMed] [Google Scholar]
- Nakashima S, Ikeno Y, Yokoyama T, Kuwana M, Bolchi A, Ottonello S, Kitamoto K, Arioka M. Secretory phospholipases A2 induce neurite outgrowth in PC12 cells. Biochem. J. 2003;376:655–666. doi: 10.1042/BJ20030830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Rierson S, Seo JA, Yu JH. The pkaB gene encoding the secondary protein kinase A catalytic subunit has a synthetic lethal interaction with pkaA and plays overlapping and opposite roles in Aspergillus nidulans . Eukaryot. Cell. 2005;4:1465–1476. doi: 10.1128/EC.4.8.1465-1476.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki E, Yoshida Y, Saito Y, Noguchi N. Lipid peroxidation: mechanisms, inhibition, and biological effects. Biochem. Biophys. Res. Commun. 2005;338:668–676. doi: 10.1016/j.bbrc.2005.08.072. [DOI] [PubMed] [Google Scholar]
- Schaloske RH, Dennis EA. The phospholipase A2 superfamily and its group numbering system. Biochim. Biophys. Acta. 2006;1761:1246–1259. doi: 10.1016/j.bbalip.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Soragni E, Bolchi A, Balestrini R, Gambaretto C, Percudani R, Bonfante P, Ottonello S. A nutrient-regulated, dual localization phospholipase A2 in the symbiotic fungus Tuber borchii . EMBO J. 2001;20:5079–5090. doi: 10.1093/emboj/20.18.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama M, Ohtani K, Izuhara M, Koike T, Suzuki K, Imamura S, Misaki H. A novel prokaryotic phospholipase A2 . J. Biol. Chem. 2002;277:20051–20058. doi: 10.1074/jbc.M200264200. [DOI] [PubMed] [Google Scholar]
- Takeshita N, Ohta A, Horiuchi H. CsmA, a class V chitin synthase with a myosin motor-like domain, is localized through direct interaction with the actin cytoskeleton in Aspergillus nidulans . Mol. Biol. Cell. 2005;16:1961–1970. doi: 10.1091/mbc.E04-09-0761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsitsigiannis DI, Kowieski TM, Zarnowski R, Keller NP. Three putative oxylipin biosynthetic genes integrate sexual and asexual development in Aspergillus nidulans . Microbiology. 2005;151:1809–1821. doi: 10.1099/mic.0.27880-0. [DOI] [PubMed] [Google Scholar]
- Tsitsigiannis DI, Kowieski TM, Zarnowski R, Keller NP. Endogenous lipogenic regulators of spore balance in Aspergillus nidulans . Eukaryot. Cell. 2004;3:1398–1411. doi: 10.1128/EC.3.6.1398-1411.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsitsigiannis DI, Zarnowski R, Keller NP. The lipid body protein, PpoA, coordinates sexual and asexual sporulation in Aspergillus nidulans . J. Biol. Chem. 2004;279:11344–11353. doi: 10.1074/jbc.M310840200. [DOI] [PubMed] [Google Scholar]
- Wakatsuki S, Arioka M, Dohmae N, Takio K, Yamasaki M, Kitamoto K. Characterization of a novel fungal protein, p15, which induces neuronal differentiation of PC12 cells. J. Biochem. (Tokyo) 1999;126:1151–1160. doi: 10.1093/oxfordjournals.jbchem.a022561. [DOI] [PubMed] [Google Scholar]
- Weinrauch Y, Abad C, Liang NS, Lowry SF, Weiss J. Mobilization of potent plasma bactericidal activity during systemic bacterial challenge. J. Clin. Invest. 1998;102:633–638. doi: 10.1172/JCI3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinrauch Y, Elsbach P, Madsen LM, Foreman A, Weiss J. The potent anti-Staphylococcus aureus activity of a sterile rabbit inflammatory fluid is due to a 14-kD phospholipase A2 . J. Clin. Invest. 1996;97:250–257. doi: 10.1172/JCI118399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou HE, Rajashekar CB, Williams TD, Wang X. Profiling membrane lipids in plant stress responses. J. Biol. Chem. 2002;277:31994–32002. doi: 10.1074/jbc.M205375200. [DOI] [PubMed] [Google Scholar]
- Yagami T, Ueda K, Asakura K, Hayasaki-Kajiwara Y, Nakazato H, Sakaeda T, Hata S, Kuroda T, Takasu N, Hori Y. Group IB secretory phospholipase A2 induces neuronal cell death via apoptosis. J. Neurochem. 2002;81:449–461. doi: 10.1046/j.1471-4159.2002.00800.x. [DOI] [PubMed] [Google Scholar]
- Yagami T, Ueda K, Asakura K, Hori Y. Deterioration of axotomy-induced neurodegeneration by group IIA secretory phospholipase A2 . Brain Res. 2001;917:230–234. doi: 10.1016/s0006-8993(01)02994-8. [DOI] [PubMed] [Google Scholar]
- Yamada O, Lee BR, Gomi K. Transformation system for Aspergillus oryzae with double auxotrophic mutations, niaD and sC. Biosci. Biotech. Biochem. 1997;61:1367–1369. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.