A combination of reagents and methodologies was developed that enables the rapid, definitive assessment of human bone formation in a mouse bone defect repair model, and a method was developed for inserting the Col2.3GFP reporter construct into a specific location in the human genome. Results indicate that this inserted Col2.3GFP construct provides a marker for differentiation of human embryonic stem cells, which will allow objective evaluation of preimplantation differentiation protocols for in vivo human bone formation.
Keywords: Zinc fingers, Gene targeting, Mesenchymal stem cell, Osteoblast, Bone
Abstract
The use of human embryonic stem cells (hESCs) and induced pluripotent stem cells (iPSCs) for study and treatment of bone diseases or traumatic bone injuries requires efficient protocols to differentiate hESCs/iPSCs into cells with osteogenic potential and the ability to isolate differentiated osteoblasts for analysis. We have used zinc finger nuclease technology to deliver a construct containing the Col2.3 promoter driving GFPemerald to the AAVS1 site (referred to as a “safe harbor” site), in human embryonic stem cells (H9Zn2.3GFP), with the goal of marking the cells that have become differentiated osteoblasts. In teratomas formed using these cells, we identified green fluorescent protein (GFP)-positive cells specifically associated with in vivo bone formation. We also differentiated the cells into a mesenchymal stem cell population with osteogenic potential and implanted them into a mouse calvarial defect model. We observed GFP-positive cells associated with alizarin complexone-labeled newly formed bone surfaces. The cells were alkaline phosphatase-positive, and immunohistochemistry with human specific bone sialoprotein (BSP) antibody indicates that the GFP-positive cells are also associated with the human BSP-containing matrix, demonstrating that the Col2.3GFP construct marks cells in the osteoblast lineage. Single-cell cloning generated a 100% Col2.3GFP-positive cell population, as demonstrated by fluorescence in situ hybridization using a GFP probe. The karyotype was normal, and pluripotency was demonstrated by Tra1-60 immunostaining, pluripotent low density reverse transcription-polymerase chain reaction array and embryoid body formation. These cells will be useful to develop optimal osteogenic differentiation protocols and to isolate osteoblasts from normal and diseased iPSCs for analysis.
Introduction
Developing reliable osteoprogenitor cells from human sources will be a critical step in the clinical application of cell-based skeletal repair. In some situations, cells derived from pluripotent stem cells such as human embryonic stem cells (hESCs) or induced pluripotent stem cells (iPSCs) could potentially have significant advantages over adult stem cell-derived bone marrow or other sources, because hESCs and iPSCs are essentially immortal and thus can be expanded indefinitely without loss of their differentiation potential [1]. This would allow repair of large skeletal defects and potentially correction of genetic defects in a patient’s iPSCs [2–4] followed by expansion from a single corrected cell to sufficient numbers to correct a systemic defect. Although there have been many reports of successful derivation of osteoblasts that produce bone in vivo from hESCs and iPSCs [5–9], providing unequivocal evidence of human osteoblasts producing bone matrix can be a challenge. Most studies have demonstrated the presence of human cells within a formed bone matrix by in situ hybridization with probes for primate-specific Alu repeat sequences, or staining with human specific antibodies. Using either of these approaches alone, it can be difficult to exclude the possibility that the human cells were bystanders and that the bone was produced by mouse osteoblasts that had infiltrated the implant.
Previously we found that osteoblast-restricted green fluorescent protein (GFP) reporters can provide essential histological evidence to discriminate an osteoblast from other cell types that can decorate the bone surface. Although ubiquitously expressed reporters do mark cells on the bone surface, nonosteoblastic cells can be present on the bone surface that are not associated with an underlying mineralization dye line and often have a histological marker of a mononuclear osteoclast (tartrate resistant acid phosphatase [TRAP]-positive). However, the osteoblast-associated reporter almost always overlies a mineralization dye line and is not positive for TRAP activity [10–12]. To replicate our success in the mouse system with human cells, a combination of reagents and methodologies was developed that enables the rapid, definitive assessment of the extent of human bone formation in a mouse bone defect repair model. Using the fluorescence-based cryohistology developed for murine histology, it is possible to identify human osteoblasts as cells that are adjacent to a newly formed bone surface, express alkaline phosphatase, stain for human specific mitochondrial antigen, and are associated with a bone matrix that contains human bone sialoprotein. In addition, a method was developed and validated for inserting the Col2.3GFP reporter construct, which we have previously shown to produce a strong osteoblast-specific signal in mice [12, 13], into a specific location in the human genome.
We chose to use a construct containing a fragment of the rat Col1a1 promoter rather than try to identify an equivalent fragment of the human COL1A1 promoter, because of our and others’ extensive experience using this promoter with transgenic mice. We have shown that the 2.3-kilobase (kb) fragment shows high expression in only a subset of the cell types that express the endogenous Col1a1 gene and shows strong expression in osteoblasts but not fibroblasts. Although the human COL1A1 promoter has significant homology to the rat gene, the regulatory sequences may not be organized the same way, so it may be difficult to identify a fragment with the same specificity for differentiated osteoblasts as the rat Col2.3 fragment. In addition, we have found that the rat Col2.3GFP reporter is expressed specifically in human osteoblasts when introduced into human bone marrow mesenchymal stem cells (MSCs) using a lentiviral vector [14].
Zinc finger nuclease technology was used to insert the construct into the AAVS1 locus, which has been previously shown to be a “safe harbor,” defined as a site where inserted sequences have no negative effects on human cell function, and which has an open chromatin structure in most cell types [3, 4, 15]. Our results indicate that the Col2.3GFP construct inserted at this locus provides a strong, specific marker for osteoblastic differentiation of hESCs. This will allow a rapid and objective evaluation of the effectiveness of preimplantation differentiation protocols for in vivo human bone formation.
Materials and Methods
hESC Culture and Preimplantation Differentiation Using EGM Medium
Human embryonic stem (hES) (H9) cells were obtained from the University of Connecticut/Wesleyan Stem Cell Core. Cells were maintained in irradiated mouse embryonic fibroblast conditioned medium supplemented with 4 ng/ml of basic fibroblast growth factor (conditioned medium [CM]) on Matrigel (BD Biosciences, San Diego, CA, http://www.bdbiosciences.com)-coated six-well tissue culture plates. Cells were passaged every 5–7 days using the cut/paste method to eliminate spontaneous differentiation.
We recently demonstrated a strategy for forming large volumes of human bone in a mouse calvarial model [16]. Following that approach, in vitro differentiation of the hES cells was carried out using a protocol described in [7]. Briefly, cells were harvested using Accutase (BD Biosciences) and replated on laminin-coated (catalog no. L2020, 1 μg/cm2; Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com) 100-mm tissue culture dishes in CM with 10 μM ROCK inhibitor (Y27632; Calbiochem, San Diego, CA, http://www.emdbiosciences.com). The cultures were switched to EGM medium (Lonza, Walkersville, MD, http://www.lonza.com; EBM2 basal medium, supplements EGM-2MV) when the cell density reached 90% confluence. The cells were maintained in EGM medium for 20–30 days and passaged into EGM medium when more than 90% of the cells demonstrated epithelial- or fibroblast-like morphology. Passage 2 or 3 cells were used for implantation into a mouse calvarial defect model.
Col2.3GFP pZDonor Vector Construction
The oligonucleotide 5′-GAT CAA GCT TTC CTT GAT GAT GTC ATA CTT ATC CTG TCC CTT TTT TTT CCA CAG CTC GCG GAG GGC AGA GGA AGT CTT CTA ACA TG-3′ containing a HindIII site plus a splice acceptor and T2A sequence was used in conjunction with oligonucleotide 5′-CTG AAA GCT TGA GCC CAC CGC ATC CCC AGC ATG-3′ (BGHPA Hind III) to amplify a construct containing the T2A, puromycin, and bovine growth hormone poly(A) sequences. Polymerase chain reaction (PCR) was performed using PFX polymerase (Life Technologies, Rockville, MD, http://www.lifetech.com). The resulting fragment was cloned into the HindIII site of the targeting construct pZDonor (Sigma). A fragment from pOBCol2.3GFPemd [13] containing the rat α1 collagen promoter linked to GFPemerald and SV 40 poly(A) (2.3 GFPemd PA) was released with Sal1 and cloned into pZDonor downstream of the bovine growth hormone poly(A) sequence. The resulting construct was approximately 9 kb in length.
Zinc Finger Nuclease Targeting and Colony Screening
One day prior to Amaxa Nucleofection, H9 cells were harvested and digested into a single-cell suspension using Accutase and replated on Matrigel-coated six-well plates. The cells were harvested, and 2 × 106 cells were transferred to a 1.5-ml microcentrifuge tube and pelleted by centrifugation. The cell pellet was resuspended in 100 μl of Nucleofection solution (82 μl of Solution V and 18 μl of supplement solution; catalog no. VCA-1003; Lonza). Five microliters per 14 μg of Col2.3GFP-pZDonor DNA and 5 μl of zinc finger nuclease (ZFN) mRNA (Sigma-Aldrich; catalog no. CTI1) were mixed with the cell suspension. The entire mixture was electroporated using program B-016 in Amaxa Nucleofector 2 (Lonza). The cells were replated and maintained in CM on Matrigel-coated six-well tissue culture plates. Puromycin (0.5 μg/ml) containing CM was applied to the cells 3 days after Nucleofection. Puromycin-resistant colonies were established by 5–7 days after selection. Colonies with high Col2.3GFP expression were selected by semi-quantitative PCR screening. AAVS1for (5′-GGCCCTGGCCATTGTCACTT-3′) and T2A.2 (5′-GTGGGCTTGTACTCGGTCAT-3′) were oligonucleotides used for PCR to test the correct 5′ insertion into embryonic stem (ES) cells from genomic DNA harvested from portions of colonies of cultured ES cells; the rest of the cells in the colonies were used to maintain the cultures. AAVS1rev (GGAACGGGGCTCAGTCTG) and GFP.1 3′ (GCGCGATCACATGGTCCTGCT) were likewise used to test the correct 3′ insertion into ES cells.
Karyotyping and Fluorescence In Situ Hybridization
Karyotyping and fluorescence in situ hybridization (FISH) (colonies C341 and C045) were performed to confirm the proper integration site and that the procedure did not change the karyotype (University of Connecticut Chromosome Core). FISH was performed with a GFP probe and demonstrated that only 30%–40% of cells were transgene-positive in these two colonies, indicating that puromycin selection was not sufficient to eliminate all Col2.3GFP-negative cells. After single-cell cloning described below, we obtained 100% transgene-positive colonies with a normal karyotype.
Single-Cell Cloning
Colony C341 cells were digested with Accutase to form a single-cell suspension and diluted to a density of 100 cells per milliliter of CM. Ten milliliters of cell suspension (1,000 cells) was seeded into one 100-mm dish precoated with Matrigel. After overnight attachment, single cells were identified microscopically and marked with an object marker (Nikon). After 7–10 days, colonies formed from the observed single cells were cut/pasted to Matrigel-coated fresh six-well plates and expanded for further experiments and storage.
Staining With Tra1-60
C341-6 cells (the cell line generated after single-cell cloning) were passaged on four–well glass chamber slides (Nalge Nunc) precoated with Matrigel and cultured for 5 days in CM. The cells were fixed with cold methanol for 15 minutes, rinsed three times with phosphate-buffered saline (PBS), and blocked with 5% bovine serum albumin (BSA)/PBS for 30 minutes. DyLight 448 mouse anti-human Tra1-60 antibody (1:100; Stemgent) was applied to the cells and incubated overnight in a humidified container. After three rinses with PBS, the chambers were removed, and the slides were mounted with ProLong Gold Antifade Reagent (Invitrogen, Carlsbad, CA, http://www.invitrogen.com). Images were taken with an Olympus IX50 fluorescence microscope and a ×10 objective.
Teratoma Test
C341-6 hESCs were cultured in 6-well plates to 85% density, and then colonies were detached using Dispase (Invitrogen; 1 mg/ml in DMEM/F12) for 30 minutes. Cell clumps were pelleted, rinsed twice with Dulbecco’s modified Eagle’s medium (DMEM)-F12 and broken into smaller pieces in 100 μl of DMEM-F12 basal medium by trituration with a P200 pipette. The cell suspension was injected into the left thigh of NOD/Scid/IL2rg null (NSG) mice (Jackson Laboratory, Bar Harbor, ME, http://www.jax.org). Tumors were harvested and analyzed by histology after 3–4 months. For teratoma bone analysis, a dye to label newly deposited mineral, alizarin complexone (AC) (Sigma-Aldrich; catalog no. 3882) was injected i.p. at 30 mg/kg, in 2% NaHCO3 (pH 7.4) 1 day before sample harvesting.
Calvarial Defect Implantation Model
NSG mice were anesthetized with ketamine/xylazine, and two 3.5-mm diameter calvarial bone discs on both sides of the suture were extracted without damaging the dura mater. Scaffold discs (3.5 mm × 0.5 mm) were cut from a hydroxyapatite/collagen matrix (HEALOS, DePuy Spine, Inc., Raynham, MA, http://www.depuy.com), loaded with cell samples (1 × 106), and placed onto the defect area. Implanted mice were maintained for 6 weeks. One day before sample harvesting, AC was injected as described above.
Bone Marrow Mesenchymal Stem Cell Culture
Bone marrow was obtained, with informed consent and approval from the UCHC IRB, from the humerus of a patient undergoing shoulder surgery. Bone marrow was centrifuged at 280g for 10 minutes to remove most of the red blood cells, and the supernatant was plated in α-minimum essential medium (phenol-free; Life Technologies) with 10% FBS at 2–4 × 106 cells per 100-mm plate and cultured in an incubator maintained at 5% O2 and 5% CO2. Bone marrow mesenchymal stem cell colonies became visible after 7 days, at which time the medium was changed, and the floating cells were removed. On days 11 and 14 of culture, the cells were transduced with a lentiviral vector, FUGW cherry, which expresses RFPcherry from the ubiquitin C promoter and is a derivative of FUGW [17, 18]. On day 19, the MSC colonies were large and had become confluent in the center, and the cultures were passaged using Accutase at a density of 0.5 × 106 cells per 100-mm dish.
Histology Sample Preparation and Immunohistochemistry
Teratoma or calvarial samples were harvested and fixed in 10% neutral buffered formalin (Sigma-Aldrich) at 4°C for 2–3 days and then imaged by digital x-ray (LX 60; Faxitron, Tucson, AZ, http://www.faxitron.com). Samples were soaked overnight in 30% sucrose/PBS solution then embedded in Cryomatrix (Thermo Fisher Scientific, Kalamazoo, MI, http://www.thermo.com/pathology). A nonautofluorescent adhesive film (Section Laboratory Co., Hiroshima, Japan, http://section-lab.jp) was used to capture the cut section (5 μm). The film was adhered, section side up, to a glass slide using a 0.2% chitosan (C3646; Sigma-Aldrich) solution in 0.25% acetic acid and allowed to dry for 48 hours at 4°C. The glass slide was soaked for 10 minutes in PBS, and a cover slide was put on with 50% glycerin in PBS prior to microscopy for the endogenous fluorescent signals (GFP and bone mineral). GFPemerald was detected using an enhanced GFP (eGFP) filter set (51019; Chroma), and the AC mineralization line was captured using a tetramethylrhodamine isothiocyanate (TRITC) filter (49005ET; Chroma). After a section was imaged for endogenous signals, the cover slide was removed by brief soaking in PBS and then processed for additional stains.
For immunohistochemistry, frozen sections were rinsed with PBS and blocked with 5% normal goat serum and 1% BSA in PBS for 1 hour at room temperature (RT). The sections were incubated overnight at 4°C with a 1:100 dilution of mouse anti-human mitochondria (catalog no. MAB1273; Millipore) or mouse anti-human bone sialoprotein (BSP) (catalog no. MAB1061; Millipore) in 1% normal goat serum and 1% BSA in PBS. After rinsing with PBS, the slides were incubated with 1:100 dilution fluorescein isothiocyanate (FITC)- or TRITC-conjugated goat anti-mouse IgG at RT for 1 hour. Then the slides were washed and mounted with 50% glycerin in PBS with 1:1,000 diluted Hoechst 33342 (catalog no. H-3570; Molecular Probes). The sections were imaged using a Zeiss Imager Z1 microscope (Carl Zeiss, Thornwood, NY, http://www.zeiss.com) using AxioVision Rel.4.7 (Carl Zeiss). The fluorescent signals were captured by a gray scale Zeiss Axiocam and pseudocolored to provide a visual contrast between the filters. FITC was captured with a YFP filter (49003; Chroma). The specificity of the BSP antibody for human and not mouse protein was confirmed in two ways. In one study, we stained a section of mouse calvaria that had been implanted with human bone marrow mesenchymal stem cells that had been transduced with a lentivirus vector FURW, which expresses RFPcherry ubiquitously (supplemental online Fig. 2). In a second study, we compared staining of human bone in a teratoma with embryonic mouse bone placed on the same slide and processed together (supplemental online Fig. 3). Both sections were stained with anti-BSP antibody as described above.
To stain for alkaline phosphatase (AP) activity, the cover slide was removed, and the section was incubated in the AP reaction buffer (100 mM Tris [pH 9.5], 50 mM MgCl2, 100 mM NaCl) for 10 minutes followed by reaction buffer containing 200 μg/ml Fast Red TR (catalog no. F8764-5G; Sigma-Aldrich) and 100 μg/ml naphthol AS-MX phosphate (catalog no. N-4875; Sigma-Aldrich) for 5 minutes. Hematoxylin only or hematoxylin and eosin (H&E) staining was performed on the same slides once the fluorescent staining and imaging was completed. To visualize cartilage in a teratoma section, a fresh frozen section was washed twice in water for 2 minutes, stained in Weigert’s iron hematoxylin (0.5% hematoxylin, 47.5% ethanol, 0.58% FeCl3) for 5 minutes, rinsed in water, stained in fast green (0.02% fast green FCF in H2O, rinsed in 1% acetic acid for 5 seconds, and stained in 0.1% safranin O for 5 minutes before mounting. Imaging was done using an RGB chromogenic filter set (Zeiss) and reconstructed to provide a visual color image.
Results
Targeting the Col2.3GFP Reporter Into the AAVS1 Locus of hES Cells
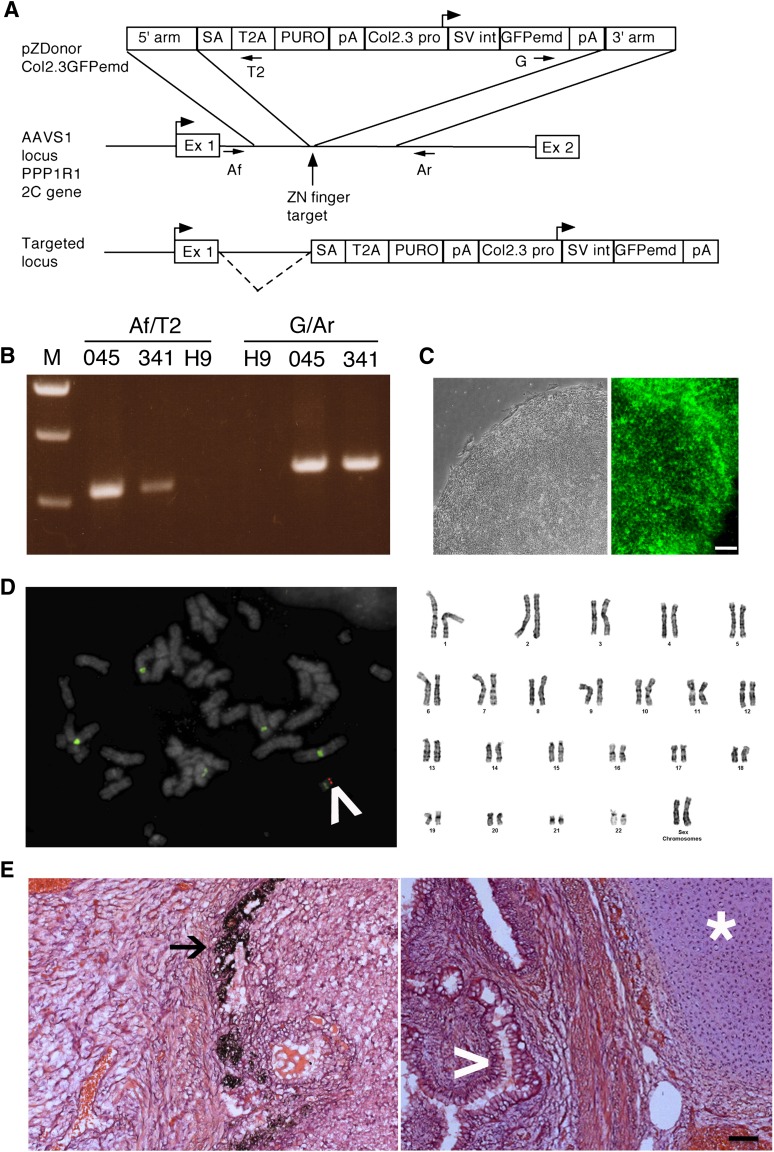
Our objective was to produce hESC lines that contain a transgene for the mature osteoblast marker Col2.3GFP to facilitate identification of differentiated osteoblasts in vivo. Zinc finger nuclease technology was used to promote insertion of the construct by homologous recombination into a specific location in the human genome [3, 4, 15]. There are several advantages of this method. Constructs can be inserted into a defined location with no known function and a generally open chromatin configuration (a “safe harbor”). The inserts carry no lentiviral or prokaryotic plasmid sequences, which can mediate transcriptional repression [19–21], and are single copy, avoiding potential repression of multiple tandem repeated inserts. The AAVS1 locus has been previously shown to be a safe harbor site, and a zinc finger nuclease that targets this location is commercially available. In the construct, puromycin resistance is driven by the endogenous PPP1R12C gene promoter, which made the construct smaller and avoided placing a constitutive promoter close to the Col2.3 promoter, minimizing promoter interference (Fig. 1A). The construct also contained an SV 40 intron and polyadenylation signal and GFPemerald, which has a similar excitation/emission spectrum to eGFP but is brighter [22]. The construct was electroporated into H9 hES cells, puromycin-resistant colonies were isolated, and DNA was extracted. PCR analysis to confirm proper targeting at the 3′ and 5′ ends was done using primer pairs with one primer in the AAVS1 locus outside of the homology arm and one in nonmammalian sequences within the targeting vector (Fig. 1A, 1B). We picked two PCR-positive colonies with good hESC morphology (clones 045 and 341) for further analysis.
Figure 1.
Pluripotency of human embryonic stem cells (hESCs) is maintained after zinc finger nuclease targeting and single-cell cloning. (A): Strategy for targeting Col2.3GFPemd to the PPP1R12C gene at the AAVS1 locus. The vertical arrow shows the cut site of the zinc finger nuclease, where the transgene is inserted. The top diagram shows the targeting vector. The 5′ arm is the 5′ homology arm; T2A is the ribosome skip sequence from Thosea asigna, which separates the peptide sequence of the first exon of the PPP1R12C gene from PURO, the puromycin resistance gene. The box on the left labeled pA indicates bovine growth hormone polyadenylation signal, whereas the box on the right labeled pA indicates the SV40 virus polyadenylation signal. The targeted locus diagram shows that the puromycin resistance gene is driven by the PPP1R12C gene promoter, and the dashed line indicates splicing from the PPP1R12C gene first exon to the splice acceptor in the inserted construct. (B): AAVS1 forward and T2A reverse primer pairs showing the correct integration of the 5′ end of the construct; green fluorescent protein (GFP) forward and AAVS1 reverse primer pairs showing correct integration of the 3′ end of the construct. (C): A typical hESC colony from the C341-6 clone with a well-defined edge was imaged with phase contrast optics (left), and the C341-6 clone was stained with Alexa 445-labeled Tra1-60 antibody (right). Scale bar = 100 μm. (D): The single correct targeting site was demonstrated by fluorescence in situ hybridization on C341-6 clone with Alexa Fluor 555-labeled GFPemd probe (left; red band indicated by white arrowhead). The green signal is fluorescein-labeled Aquarius Enumeration Probe that hybridizes to the centromeres of chromosomes 1, 5, and 19. Light 4′,6-diamidino-2-phenylindole stain (gray) allowed visualization of chromosome banding pattern to confirm integration of transgene at 19q13.3–13.4. The C341-6 clone with normal karyotyping is demonstrated by Giemsa chromosome banding (right). (E): Tissues from three germ layers were found in a teratoma derived from C341-6 cells: melanocytes (ectoderm, left, black arrow), cartilage (mesoderm, right, white star), and intestine-like (endoderm, right, white arrowhead). Scale bar = 100 μm. Abbreviations: 045, colony C045; 341, colony C341; Af, AAVS1 forward primer pair; Ar, AAVS1 reverse primer pair; Col2.3 pro, 2.3-kilobase rat Col1a1 promoter fragment; Ex 1, exon 1 of the PPP1R12C gene; Ex 2, exon 2 of the PPP1R12C gene; G, GFP forward primer pair; GFPemd, GFPemerald; H9, H9Zn2.3GFP cells; pA, polyadenylation signal; SA, artificial splice acceptor; SV int, SV40 virus late intron; T2, T2A reverse primer pair; ZN, zinc.
Transgene-Positive hESCs Maintain Pluripotency After Single-Cell Cloning
Both selected clones expressed pluripotency markers by low density array screening (Invitrogen; data not shown) and by immunostaining with Tra1-60 antibody (representative results for clone C341, that was selected for further study are shown in Fig. 1C). Karyotyping and FISH using a GFPemd probe showed a normal female chromosome complement and correct integration of the construct in 19q13.3–13.4 (Fig. 1D). FISH analysis of multiple nuclei showed that in the initial cell population that we isolated, approximately 30%–40% contain the transgene (data not shown); however, these cells were adequate to confirm the osteoblastic specificity of expression of GFP in the cells that contain the transgene. We isolated clones from line C341 that are 100% transgene-positive, and further studies were done on clone C341-6. Teratoma analysis showed that cells from all three germ layers were produced (Fig. 1E) by this clone. The experiments shown in Figures 4–6 were done using the original mixed cell population C341, whereas the experiments shown in Figures 3 and 7 were done with clone C341-6.
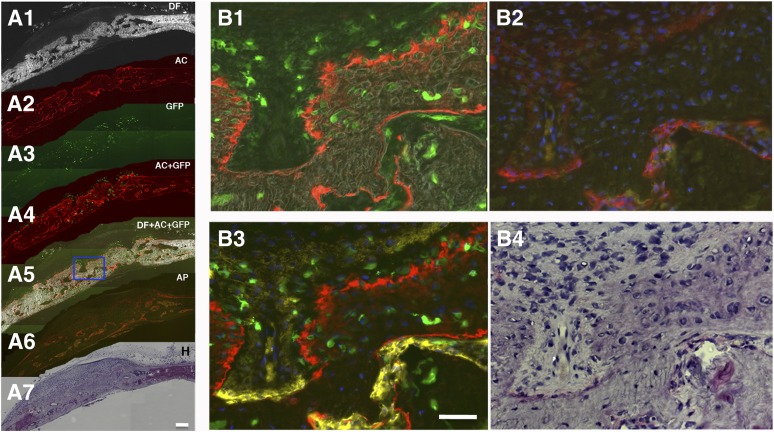
Figure 4.
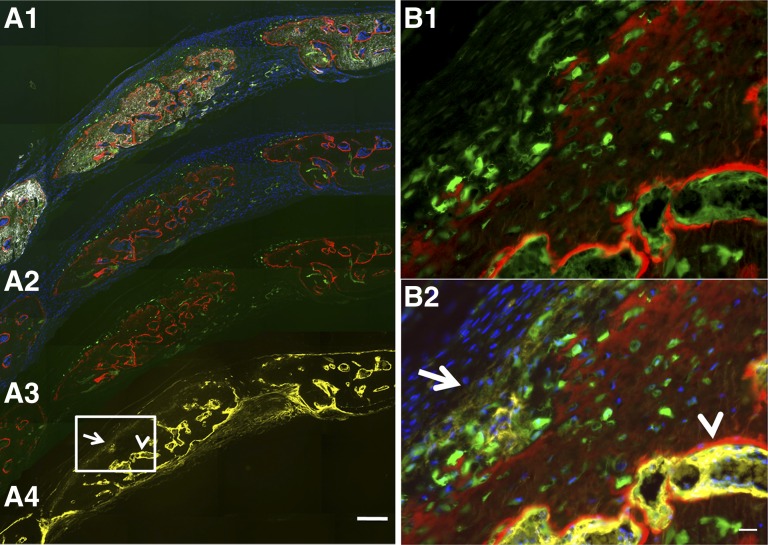
Col2.3 GFPemd in partially transgene-positive human embryonic stem cell function as an osteoblast reporter was confirmed by immunohistochemistry in mouse calvarial defect model. (A): Images collected from low magnification scan (×5, scale bar = 200 μm). Images (A1–A5) were scanned before decalcification and immunohistochemistry. DF optics reveal white or light gray areas in the image, indicating high-density bone matrix structures. Red staining by AC labels newly formed bone surfaces. Green fluorescence from GFP indicates Col2.3 promoter activity. Col2.3GFP (green) is expressed in the cells near and within the bone matrix. (A6) and (A7) were taken on the same section after decalcification, which removes AC labeling. Hematoxylin staining was performed after AP staining was imaged. Nuclei stained dark blue. Cytosol and extracellular matrices are stained pink/purple. (B): Images are from the boxed region in (A5). (B1): High magnification scanning with dark-field optics (gray in this image) demonstrated the bone matrix and the association of AC labeling (red) and Col2.3 promoter activity (green) near or within the bone matrix. (B2): After decalcification and AP staining, AC labeling and Col2.3GFP were diminished, and AP activity was detected by FAST-Red-labeled alkaline phosphatase substrate (red). Nuclei were stained by 4′,6-diamidino-2-phenylindole (blue). (B3): FAST-Red AP staining was converted to yellow before images in (B1) and (B2) were merged. (B4): High magnification image of hematoxylin-stained section. Scale bar = 50 μm. Abbreviations: AC, alizarin complexone; AC+GFP, Col2.3 GFP-expressing cells are associated with the AC labeling; AP, alkaline phosphatase (alkaline phosphatase activity detected by Fast Red TR/Naphthol AS-MX Phosphate [red]); DF, dark-field; DF+AC+GFP, dark-field red AC labeling outlining the bone surface (white or gray); GFP, green fluorescent protein; H, hematoxylin.
Figure 6.
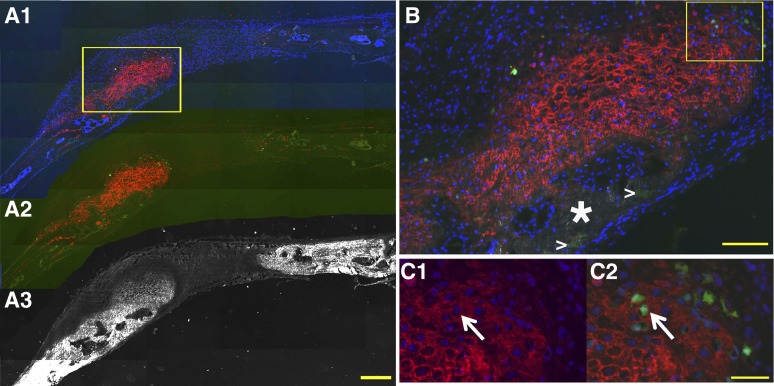
Col2.3 GFPemd cells are embedded in human bone sialoprotein (BSP) staining matrix. An adjacent section to those used in Figures 4 and 5 was scanned with dark-field optics to show the bone matrix ([A3], white). Human BSP immunostaining ([A1] and [A2], red) was applied to the section after decalcification. Col2.3GFPemd-positive cells were identified by green fluorescence ([A1] and [A2], green). Cell nuclei were shown by 4′,6-diamidino-2-phenylindole ([A1], blue). Scale bar = 200 μm. (B): Higher magnification of the boxed region in (A1) was imaged. The white star marks human BSP negative mouse bone. White arrowheads mark the hydroxyapatites within the remaining scaffold. Scale bar = 50 μm. (C): Higher magnification of boxed region in (B) was imaged with (C2) or without (C1) green fluorescence. The white arrows in (C1) and (C2) point to Col2.3GFPemd-positive cells (C2) embedded in human BSP staining (C1). Scale bar = 20 μm.
Figure 3.
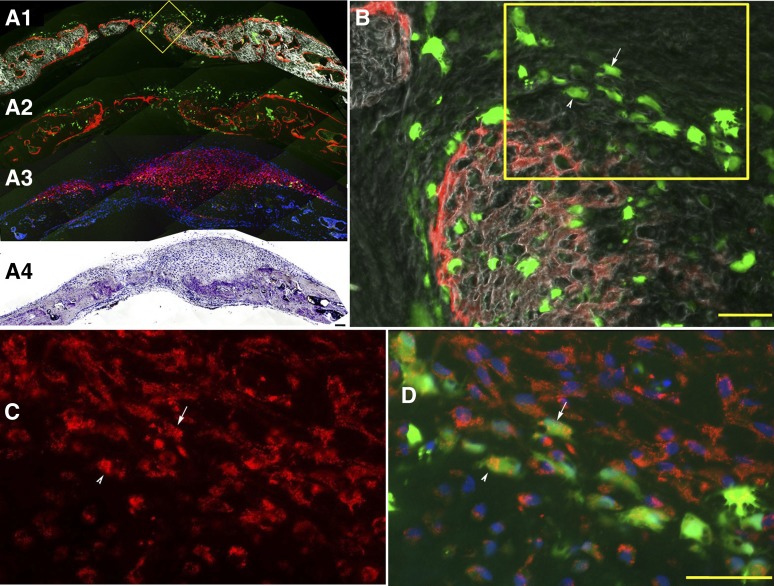
Col2.3GFPemd expression is restricted to alkaline phosphatase (AP)-positive cells near the bone surface or imbedded in matrix in teratoma bone formed by C341-6 cells. The presence of bone was shown by hematoxylin and eosin staining of the section after fluorescent imaging is completed (not shown). (A1): Bright green GFPemd-positive cells. No green fluorescent protein (GFP)-positive cells were detected in other parts of the teratoma. Scale bar = 200 μm. (A2): Alizarin complexone (AC) was injected 1 day prior to sample harvesting. Newly formed bone tissue is marked by AC labeling (red), and GFP-positive osteoblasts (green) were located adjacent to the AC labeling. Blue shows 4′,6-diamidino-2-phenylindole (DAPI) staining of cell nuclei. The boxed region is shown in B. (A3): The section shown in (A) and (B) was stained for AP activity, shown in yellow. DAPI staining again shows nuclei. (A4): Overlay of the images shown in (A2) and (A3). (B1): GFP and DAPI of the boxed portion of (A2). (B2): AC labeling was added to (B1). (B3): DAPI and AP (yellow)-positive cells were located adjacent to the AC labeling. (B4): Merged image of (B2) and (B3) shows that Col2.3GFP-positive cells are located in the band of AP-positive cells near the AC-labeled newly mineralized surface. AP is located primarily in the cell membrane and matrix vesicles that are released from the cells, whereas the GFP is cytoplasmic, so colocalization of the two signals is not precise. Because AP is expressed in preosteoblasts as well as mature osteoblasts, many of the AP-positive cells are not yet GFP-positive. The arrows indicate examples of GFP-positive cells that are also AP-positive. Scale bar = 20 μm. Abbreviation: B (in [A1] and [A3]), bone matrix.
Figure 7.
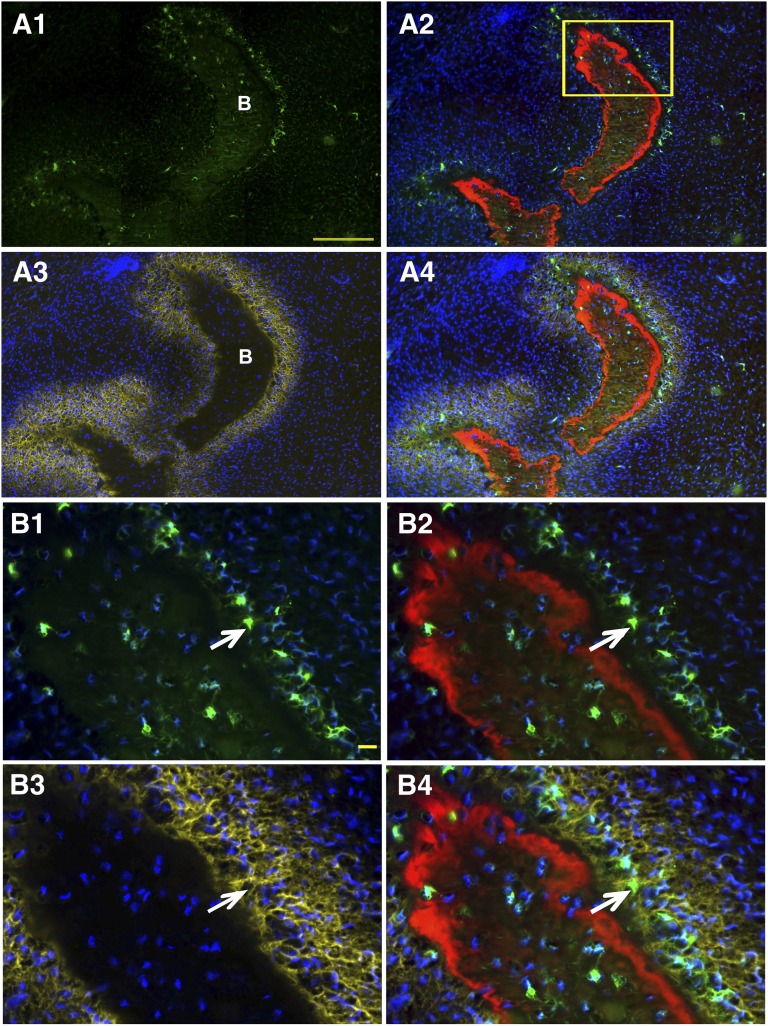
Col2.3GFPemd-positive cells were more enriched in defects implanted with C341-6-derived mesenchymal cells. (A): Col2.3GFPemd-positive human osteoblasts (green) were detected in calvarial defect implanted with C341-6 derived mesenchymal cells. Samples were processed as described in Figure 4. White: dark-field. Red: AC labeling. Blue: 4′,6-diamidino-2-phenylindole nuclei staining. Yellow: AP staining. Arrows in (A) and (B): AP staining of human osteoblasts. Arrowhead in (A) and (B): AP staining of mouse osteoblasts. Boxed region in (A) is shown in (B). Color assignments for (B1) and (B2) are the same as for Figure 4B1 and 4B3, respectively. Scale bars = 200 μm (A) and 50 μm (B).
Histological Evaluation of Human Cells in the Murine Bone Repair Environment
While the Col2.3GFP targeting studies were underway, histological methods were developed to conclusively distinguish human donor osteoblasts from recipient mouse osteoblasts in an implant. This has proved to be important because our unpublished observations have indicated that when human cells are implanted within a mouse bone repair defect, much of the newly formed bone can be derived from the surrounding mouse bone. Human cells may line the bone surface, but they are not functional osteoblasts because they are not associated with underlying mineralization dye label on the surface of newly formed bone, nor do they become embedded within the murine bone matrix.
Teratomas produced from hES cells by intramuscular injection were used to demonstrate whether antibodies can distinguish human bone lineage cells from mouse bone cells. Nondecalcified frozen sections and a tape transfer methodology was used that allows sectioning of mineralized tissue, visualization of all deposited mineral by dark-field optics, and detection by labeling of newly formed mineralized tissue with various injectable dyes that bind to newly deposited osteoid. This methodology also preserves enzyme activity and antigen accessibility. The use of fluorescent enzyme substrates and antibodies and of a microscope with a computer-controlled stage facilitates imaging with one or more methodologies, removal of the section and further processing, and visualizing the same region of the section. The sample shown was a fragment of mineralizing teratoma tissue that resembled an embryonic long bone with mineralizing hypertrophic chondrocytes and a bony collar, as shown in the H&E-stained image (Fig. 2A1). Note that the section used for H&E staining was adjacent to the section used in the rest of Figure 2, so there are some slight differences in the pattern of the mineralized tissue.
Figure 2.
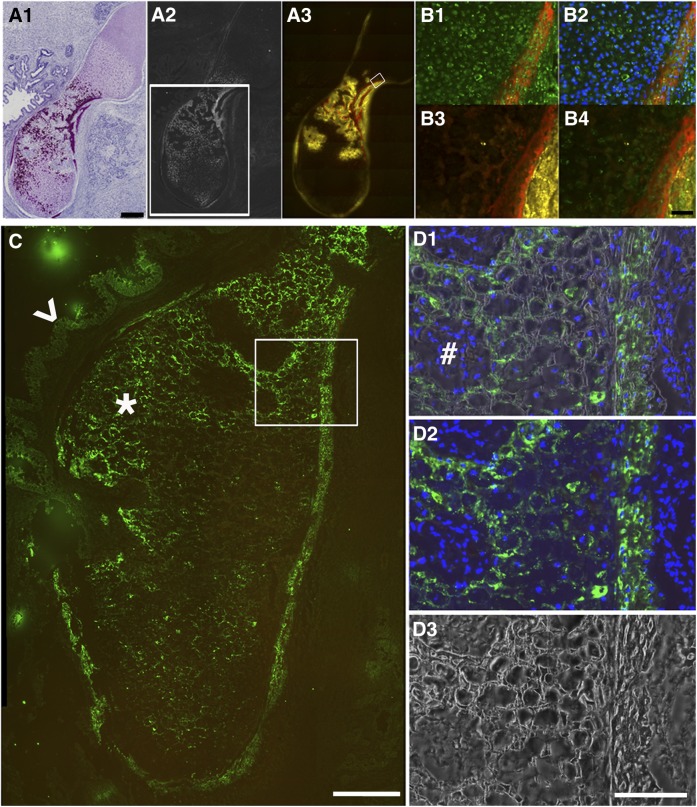
Establishing immunohistochemistry methods to demonstrate alkaline phosphatase (AP) activity and identify human cells in human embryonic stem cell teratoma bone. (A1): low magnification scanning was performed on the hematoxylin/eosin-stained section adjacent to the section for A2 and 3. Nuclei stain is dark blue, cytosol and extracellular matrix stain is pink, and high density mineralized bone matrix stain is purple. (A2): Dark-field scan imaged high density bone matrix (white). The white box shows an adjacent section in the region analyzed in C. (A3): AP activity demonstrated by ELF97-labeled AP substrate (yellow) adjacent to the red alizarin complexone (AC)-labeled newly formed bone surfaces. Scale bar = 500 μm. (B): High magnification image of the boxed region in (A3). (B1): Section was stained with mouse anti-human mitochondrial antibody (catalog no. MAB1273, 1:100; Millipore) and then fluorescein isothiocyanate (FITC) donkey anti-mouse secondary antibody (Jackson, 1:500, green). Red shows AC labeling. (B2): Merged images of B1 with 4′,6-diamidino-2-phenylindole (DAPI) nuclei. (B3): ELF97-labeled AP activity (yellow) was associated with AC labeling (red). (B4): Images (B1) and (B3) were merged. Scale bar = 100 μm. (C): Anti-human bone sialoprotein (BSP) antibody demonstrated human cell deposition of BSP into teratoma bone. A section adjacent to the one boxed in (A2) was used for BSP antibody staining and visualized with FITC donkey anti-mouse secondary antibody (green, star). The arrowhead points to a weaker signal in gut-like or glandular tissue structure. The boxed region is shown at high magnification in (D). Scale bar = 200 μm. (D1) shows the merged image of (D2) (immunostaining) and (D3) (dark field), demonstrating that BSP antibody staining (green) was specific to the cells (DAPI-stained cell nuclei, blue) surrounded by mineralized matrix (dark-field, white). Cells (DAPI-stained cell nuclei, blue) without mineralized matrix were BSP negative (#). Scale bar = 100 μm.
The dark-field imaging of the teratoma revealed a mineralized structure resembling embryonic bone. H&E staining showed cells with the appearance of resting, proliferating, and hypertrophic cartilage (Fig. 2A1). This was confirmed by safranin O staining of another section of the same teratoma (supplemental online Fig. 1). The H&E staining also showed regions of matrix with the appearance of cortical bone adjacent to some of the cells with the appearance of hypertrophic mineralized cartilage (Fig. 2A1) and also incipient trabecular bone forming around regions in which the mineralized cartilage matrix has been degraded (Fig. 2A2). Alkaline phosphatase staining showed activity in cells flanking the newly formed bone and also in some of the cells filling the spaces produced by degradation of the mineralized cartilage matrix (Fig. 2A3, yellow). Alizarin complexone, which was injected 24 hours before sacrifice, also diffusely stained the bone matrix and bone surface in a pattern that was distinct from the sharp mineralized lines characteristic for adult mouse bone. This pattern resembled immature woven bone, whereas the mouse bone was more mature and lamellar. The mineralized cartilage matrix was stained more faintly and diffusely, which is visible in a higher magnification image (Fig. 2B3, 2B4 marked with asterisks).
Two antibodies that were reported to be human-specific by the manufacturers were evaluated for their ability to demonstrate the presence of functional human osteoblasts within bone grown in a mouse host. An antibody to a human mitochondrial antigen, monoclonal antibody clone 113-1 (BD Biosciences), is commonly used to identify human cells in mouse implants, but it was not clear that it produces a detectable signal within all skeletal cell types. Staining the teratoma with this antibody produced a strong signal in chondrocytes, osteoblasts, and osteocytes (Figs. 2B1, 2B2, 4). AP staining of the same section showed cells that were positive for both human mitochondrial antigen and AP that were lining newly formed bone as shown by alizarin complexone staining.
To evaluate a second antibody for its specificity for human BSP, we implanted human adult bone marrow MSCs that had been transduced with a lentivirus vector containing an ubiquitin RFPcherry marker into a mouse calvarial defect and stained a coronal section through the defect and the suture (supplemental Fig. 2B, 2C). There is abundant green staining of bone matrix within the defect, which contained extensive human bone, as indicated by the RFPcherry fluorescent cells embedded in bone matrix and lining newly formed bone matrix, demonstrated by green calcein labeling (supplemental online Fig. 2A), but only faint background fluorescence in the adjacent mouse bone (supplemental online Fig. 2B). We also compared a high magnification image of staining of a mineralized portion of a human teratoma with a section of mouse embryonic bone (supplemental online Fig. 3). The human sample (supplemental online Fig. 3A1, 3A2) showed green matrix staining that colocalized with the mineralized matrix, shown as white by the dark-field optics in supplemental online Figure 3A1, whereas the mouse bone did not show staining above background.
We stained the human teratoma with this antibody and observed strong staining of bone matrix and weaker staining of mineralized chondrocyte matrix (green fluorescence in Fig. 2C and at higher magnification in Fig. 2D1–2D3). BSP is expressed in bone and hypertrophic chondrocytes. The colocalization of BSP (Fig. 2D2) and the corresponding mineral within the bone and cartilage matrix as revealed by dark-field optics (Fig. 2D3) is demonstrated in Figure 2D1. We believe that these histological findings provide convincing evidence for functional human osteoblasts and hypertrophic chondrocytes. Because the epitopes identified in the human cells were not present in the corresponding mouse tissues, these histological criteria will be useful in distinguishing human from mouse osteoblast and chondrocytes in human to mouse transplantation studies.
Evaluating Col2.3GFP as an Osteoblast Reporter in Teratomas
Line C341-6 of the H9 hESCs containing the Col2.3GFP construct in 100% of the cells was tested in the teratoma system. GFP signal was only detected in a small area of AP-positive cells associated with AC-labeled bone matrix that appeared to be forming an initial condensation of membranous bone (Fig. 3); the rest of the teratoma, that had not produced bone, was negative for GFP, indicating that the Col2.3 promoter was not expressed in nonosteoblastic cells.
Osteoblast Differentiation of Col2.3GFP hESCs In Vivo
Col2.3GFP hESCs were tested for in vivo osteoblast differentiation in a critical-sized mouse calvarial defect using a preimplantation differentiation protocol designed to enrich for a mesenchymal cell population with osteogenic potential. The methodology was originally described by Boyd et al. [7], and a detailed evaluation of the ability of cells produced by this method to form human bone in vivo has been described [16]. The differentiation procedure involves culturing hESCs on laminin-coated plates in an endothelial growth media (EGM-2), with 5% FBS and FGF, EGF, VEGF, and IGF-1. The cells are cultured under these conditions for 20–30 days, during which time they take on an epithelial appearance. They are then passaged in the same media, at which time they become mesenchymal in appearance. After passaging two or three times in the same medium, the cells are infiltrated into a disc of hydroxyapatite/collagen matrix (HEALOS) and implanted into 3.5-mm circular defects in the parietal bones of 3–4-month-old immunodeficient NSG mice. The results showed numerous strongly GFP-positive cells (Fig. 4A3–4A5) that stained positive for AP (Fig. 4A6; 4B2, red; 4B3, yellow) and were associated with AC stained newly formed bone (Fig. 5A2, 5A3, 5A6, 5B1, 5B3). In Figure 5B2, AP is false-colored red, whereas in Figure 5B3, it is false-colored yellow to allow display of GFP, AC, and AP in the same image. As mentioned above, the human osteoblast AP staining was less intense than the mouse AP. The AP-positive human osteoblasts, some of which were also GFP-positive, were found in regions that were a greater distance from the newly mineralized surface than is seen in mouse bone. The GFP-positive cells were found within the region of AP-positive cells, between the AP-positive cells and the mineralized bone surface, lining the mineralized bone surface, or embedded in the bone matrix. Only a fraction of the cells in the bone-forming area are GFP-positive, because most of the cells do not contain the transgene. In the regions with many GFP-positive human cells buried in bone matrix, the AC staining line is thicker, is more irregular, and fades gradually as it gets deeper into the matrix, whereas in regions of bone with no GFP-positive cells, which were produced by mouse osteoblasts, the AC stain is sharper, with little staining in the deeper areas of the matrix. The AP staining is also much brighter and closer to the AC-stained surface.
Figure 5.
Col2.3GFPemd-positive cells stain with human mitochondrial specific antibody. Cells from the same population of differentiated cells as was used in Figure 4 were implanted in the calvarial defect model. (A): Low magnification imaging. (A1–A3): Dark-field imaging ([A1], white) demonstrated the bone matrix, outlined by the AC labeling ([A1] and [A2], red). Col2.3GFPemd transgene-positive cells were identified by green fluorescence expression (A1–A3). (A3): The same section imaged in (A1) and (A2) was decalcified to remove mineral from the bone matrix and eliminated the AC labeling. Immunohistochemistry was performed with hMit antibody and visualized with Cy3-donkey anti-mouse IgG ([A3], red). Cell nuclei were detected by 4′,6-diamidino-2-phenylindole (DAPI) staining ([A3], blue). (B): Higher magnification image of yellow boxed region in A1. Arrow and arrowhead in (B) point to GFPemd cells that are shown to be hMit-positive in (C) and (D), which are higher magnification image of the yellow boxed region in (B). (C, D): (C) shows only the red signal of hMit antibody staining, whereas (D) is an overlay of green GFP, red hMit antibody staining, and blue DAPI. Scale bar in (B) and (D) = 50 μm.
Staining of a section from a different implant produced from the same differentiated cell population used in Figure 4 with human mitochondria specific antibody, using a red secondary antibody, showed many double-labeled red and green cells (Fig. 5C, 5D, arrows and arrowheads), confirming the human origin of the GFP-positive cells. In Figure 5A3, there are many GFP- and human mitochondrial antigen-negative mouse cells in the lower part of the implant. Staining with the human-specific BSP antibody showed that Col2.3GFP-positive cells are associated with human BSP-positive bone matrix (Fig. 6C1, 6C2, arrows).
After subcloning to obtain a 100% transgene-positive population, cells were again differentiated and tested in the calvarial defect model. As was seen previously, there are numerous GFP-positive cells associated with weak AP activity over an AC-positive newly formed bone surface, with many GFP-positive in the process of being imbedded, or completely imbedded, in the bone matrix (Fig. 7). As expected, the percentage of GFP-positive cells in the human bone forming area seems to be considerably higher. Also consistent with what was seen before, the implant has mostly mouse bone on the side nearest the dura mater, whereas the human bone is mostly on the outer surface. The connection between the human and mouse bone is seamless, and although there are distinct regions of mouse and human bone, there is also some mixture of human and mouse bone cells. In some areas that appear to be exclusively human bone, there are some 4′,6-diamidino-2-phenylindole (DAPI)-labeled nuclei that seem to be GFP-negative (Fig. 7B1, 7B2); however, higher magnification imaging without DAPI imaging shows weak GFP (data not shown), consistent with the general observation that GFP intensity is variable.
Discussion
The use of hESCs and iPSCs for study and treatment of genetic bone diseases and traumatic bone injuries requires efficient protocols to differentiate hESCs/iPSCs into cells with in vivo osteogenic potential and the ability to isolate differentiated osteoblasts for analysis. It is, however, not clear whether the bone formed in most in vivo osteoblast differentiation models is formed by the transplanted hES or induced pluripotent stem (iPS) cells. Numerous studies have reported protocols that are purported to produce cells that differentiate into osteoblasts in vivo (for example, [5, 6, 8, 9]). However, many studies either failed to distinguish between human and mouse cells in bone formed by an implant or, if they did identify human cells within a region where implant bone was formed, did not demonstrate that the human cells were functional osteoblasts. This is important because previous studies have shown that cells can be located on or near a bone surface and have an osteoblast-like morphology that are not in the osteoblast lineage and do not become osteocytes (for example, osteomacs [23]). Identification of human osteocytes based on the presence of human cells (as indicated by Alu repeat hybridization-positive or human-specific antibody-positive cells embedded in bone matrix) is not sufficient in the absence of evidence of human osteoblasts located on the bone surface and producing a bone matrix, which would be the anticipated source of the osteocytic cells. Decalcified paraffin section methodologies do not allow acquisition of the ancillary information that can demonstrate active osteogenesis from a bone surface cell, such as staining to reveal active bone matrix deposition. The chromogenic paraffin methods also limit the ability to colocalize multiple signals such as Alu in situ hybridization, immunohistochemistry, and enzymatic stains within the same section, which will be required to fully distinguish host and donor contributions to a model of bone repair.
In this paper, we used immunohistochemical methods for identification of human osteoblasts in mouse calvarial defects. These methods are applied to undecalcified frozen sections, eliminating the time required for decalcification, and they preserve fluorescent labeling of newly formed bone, enzymatic activity, and epitope exposure. They allow definitive identification of human osteoblasts in implant experiments using unmarked human cells. However, they require multiple time-consuming steps, and they also do not allow convenient isolation of pure osteoblasts. These methods also can be difficult to definitively interpret if small amounts of potential human bone are observed.
We wished to establish hESC lines containing a Col2.3-driven reporter construct, because we have extensive experience that this construct is robustly and specifically expressed in mature osteoblasts in transgenic mice [12, 13]. Initially retro- or lentiviral vectors were chosen to deliver the promoter-reporter construct, but we found that this and several other cell type-specific promoter constructs did not show appropriate marker gene induction during hESC differentiation (X. Xin, M. Stover, S. Zhan et al., unpublished results). In this study, ZFN technology was used to direct homologous recombination at the AAVS1 site, which is where the adeno-associated virus (AAV) integrates. This site has been referred to as a “safe harbor” because integration of AAV into this site has never been shown to be harmful. In addition, the integration site is in the first intron of a gene, PPP1R12C, which is widely expressed at moderate levels, so the locus retains an open chromatin structure, allowing consistent expression of integrated transgenes. A construct design was chosen in which expression of an antibiotic resistance gene, which is needed to select for integration of the construct in undifferentiated hES cells, is driven by the promoter of the PPP1R12C gene and is terminated by a polyadenylation site. The Col2.3-regulated promoter-reporter cassette is located further downstream of the polyadenylation site. This strategy provides a moderately but constitutively expressed host promoter driving the antibiotic resistance gene at some distance from the regulated promoter, avoiding placing a constitutive and a regulated promoter in close proximity. The polyadenylation signal helps to attenuate upstream transcription through the regulated promoter, thereby decreasing the possibility of promoter occlusion.
The construct containing Col2.3 promoter-driven GFPemerald was inserted into the AAVS1 site of human embryonic stem cells to mark the cells that differentiate into osteoblasts in vivo. In a teratoma formed using these cells, we identified GFP-positive cells specifically associated with bone. We also differentiated the cells into a mesenchymal stem cell population with osteogenic potential and implanted the cells into a mouse calvarial defect. We observed GFP-positive cells associated with AC-labeled newly formed bone surfaces. This bone showed a similar distinctive pattern of AC labeling and bone structure as was seen with human bone in teratomas, which was different from mouse bone. The GFP-positive cells were also AP-positive, and immunohistochemistry with human specific BSP antibody has indicated that the GFP-positive cells are also associated with human BSP-containing matrix. Therefore, we believed that our Col2.3GFP is marking osteogenic cells capable of producing a bone matrix. The drug selection method initially generated a mixed cell population with approximately 30% Col2.3GFP-targeted cells that upon cloning achieved a 100% Col2.3GFP-positive cell population, as confirmed by FISH using a GFP probe. The drug selection and recloning steps did not induce karyotypic alterations, and the pluripotency of these clonal lines was maintained. We expect that these cells will be useful to develop optimal in vivo and in vitro osteogenic differentiation protocols and provide a cross-laboratory standard for comparing the effectiveness of various preimplantation differentiation protocols for in vivo differentiation.
Conclusion
In more recent studies, we produced a version of the reporter construct that contains red fluorescent protein instead of GFP and found that the Col2.3 promoter fragment drives osteoblast-restricted expression of the reporter in iPS cells (data not shown). This indicates the reproducibility and wide applicability of the technology and suggests that it can be used for studying the impact of a human mutation on the osteogenic lineage in vivo. Using the GFP signal and single-cell laser capture technology, it should be feasible to examine the molecular profile of osteoblastic cells in vivo uncontaminated by mouse or nonosteogenic human cells. Furthermore, it provides a strategy for assessing in real time the extent of osteogenesis in vivo in living animals using either a luciferase or thymidine kinase reporter.
Supplementary Material
Acknowledgments
This study was supported by Department of Defense Grant W81XWH1110262, Connecticut Stem Cell Research Initiative 09SCBUCHC20, and NIH/NIDCR Grant R21 DE19892.
Author Contributions
X.X., X.J., and M.L.S.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; L.W.: conception and design, collection and/or assembly of data, data analysis and interpretation, final approval of manuscript; S.Z. and J.H.: collection and/or assembly of data; A.J.G. and Y.L.: conception and design, final approval of manuscript; L.K.: conception and design, data analysis and interpretation, final approval of manuscript; E.J.R.: financial support, final approval of manuscript; D.W.R.: conception and design, financial support, administrative support, data analysis and interpretation, manuscript writing, final approval of manuscript; A.C.L.: conception and design, financial support, data analysis and interpretation, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Yamanaka S. Induced pluripotent stem cells: Past, present, and future. Cell Stem Cell. 2012;10:678–684. doi: 10.1016/j.stem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Urnov FD, Miller JC, Lee YL, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 3.Hockemeyer D, Soldner F, Beard C, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soldner F, Laganière J, Cheng AW, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barberi T, Willis LM, Socci ND, et al. Derivation of multipotent mesenchymal precursors from human embryonic stem cells. PLoS Med. 2005;2:0554–0560. doi: 10.1371/journal.pmed.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bielby RCBA, Boccaccini AR, Polak JM, et al. In vitro differentiation and in vivo mineralization of osteogenic cells derived from human embryonic stem cells. Tissue Eng. 2004;10:1518–1525. doi: 10.1089/ten.2004.10.1518. [DOI] [PubMed] [Google Scholar]
- 7.Boyd NL, Robbins KR, Dhara SK, et al. Human embryonic stem cell-derived mesoderm-like epithelium transitions to mesenchymal progenitor cells. Tissue Eng Part A. 2009;15:1897–1907. doi: 10.1089/ten.tea.2008.0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arpornmaeklong P, Brown SE, Wang Z, et al. Phenotypic characterization, osteoblastic differentiation, and bone regeneration capacity of human embryonic stem cell-derived mesenchymal stem cells. Stem Cells Dev. 2009;18:955–968. doi: 10.1089/scd.2008.0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuznetsov SA, Cherman N, Robey PG. In vivo bone formation by progeny of human embryonic stem cells. Stem Cells Dev. 2011;20:269–287. doi: 10.1089/scd.2009.0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bilic-Curcic I, Kalajzic Z, Wang L, et al. Origins of endothelial and osteogenic cells in the subcutaneous collagen gel implant. Bone. 2005;37:678–687. doi: 10.1016/j.bone.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Wang L, Liu Y, Kalajzic Z, et al. Heterogeneity of engrafted bone-lining cells after systemic and local transplantation. Blood. 2005;106:3650–3657. doi: 10.1182/blood-2005-02-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang X, Kalajzic Z, Maye P, et al. Histological analysis of GFP expression in murine bone. J Histochem Cytochem. 2005;53:593–602. doi: 10.1369/jhc.4A6401.2005. [DOI] [PubMed] [Google Scholar]
- 13.Kalajzic I, Kalajzic Z, Kaliterna M, et al. Use of type I collagen green fluorescent protein transgenes to identify subpopulations of cells at different stages of the osteoblast lineage. J Bone Miner Res. 2002;17:15–25. doi: 10.1359/jbmr.2002.17.1.15. [DOI] [PubMed] [Google Scholar]
- 14.Yin D, Wang Z, Gao Q, et al. Determination of the fate and contribution of ex vivo expanded human bone marrow stem and progenitor cells for bone formation by 2.3ColGFP. Mol Ther. 2009;17:1967–1978. doi: 10.1038/mt.2009.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lombardo A, Genovese P, Beausejour CM, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 16.Kuhn LT, Liu Y, Boyd NL, et al. Developmental-like bone regeneration by human embryonic stem cell-derived mesenchymal cells. Tissue Eng Part A. 2014;20:365–377. doi: 10.1089/ten.tea.2013.0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lois C, Hong EJ, Pease S, et al. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 18.Shaner NC, Campbell RE, Steinbach PA, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 19.Ellis J, Yao S. Retrovirus silencing and vector design: Relevance to normal and cancer stem cells? Curr Gene Ther. 2005;5:367–373. doi: 10.2174/1566523054546233. [DOI] [PubMed] [Google Scholar]
- 20.Yao S, Sukonnik T, Kean T, et al. Retrovirus silencing, variegation, extinction, and memory are controlled by a dynamic interplay of multiple epigenetic modifications. Mol Ther. 2004;10:27–36. doi: 10.1016/j.ymthe.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki M, Kasai K, Saeki Y. Plasmid DNA sequences present in conventional herpes simplex virus amplicon vectors cause rapid transgene silencing by forming inactive chromatin. J Virol. 2006;80:3293–3300. doi: 10.1128/JVI.80.7.3293-3300.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 23.Chang MK, Raggatt LJ, Alexander KA, et al. Osteal tissue macrophages are intercalated throughout human and mouse bone lining tissues and regulate osteoblast function in vitro and in vivo. J Immunol. 2008;181:1232–1244. doi: 10.4049/jimmunol.181.2.1232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.