Abstract
Conformational dynamics plays a critical role in the activation, deactivation, and open–close activities of ion channels in living cells. Such conformational dynamics is often inhomogeneous and extremely difficult to be directly characterized by ensemble-averaged spectroscopic imaging or only by single channel patch-clamp electric recording methods. We have developed a new and combined technical approach, single-molecule patch-clamp FRET microscopy, to probe ion channel conformational dynamics in living cell by simultaneous and correlated measurements of real-time single-molecule FRET spectroscopic imaging with single-channel electric current recording. Our approach is particularly capable of resolving ion channel conformational change rate process when the channel is at its electrically off states and before the ion channel is activated, the so-called “silent time” when the electric current signals are at zero or background. We have probed NMDA (N-methyl-d-aspartate) receptor ion channel in live HEK-293 cell, especially, the single ion channel open–close activity and its associated protein conformational changes simultaneously. Furthermore, we have revealed that the seemingly identical electrically off states are associated with multiple conformational states. On the basis of our experimental results, we have proposed a multistate clamshell model to interpret the NMDA receptor open–close dynamics.
Introduction
NMDA (N-methyl-d-aspartate) receptor ion channel is a heterotetrameric ligand-gated and voltage-gated ion channel composed of two NR1 and two NR2 subunits.1−33 The NMDA receptor ion channel is activated by the binding of two glycine (Gly) and two glutamate (Glu) molecules (agonist) at the ligand binding domains (LBD) of NR1 and NR2 subunits, respectively (Figure 1A).1−33 NMDA receptor ion channels are important for the development and function of neural systems, such as human brain and its signaling networks,1,8,11,12 having some unique biophysical properties, such as high unitary conductance, high Ca2+ permeability, and slow gating kinetics.1,16−18 The biological significance of an ion channel in neural system largely depends on the regulation, amplitude, and duration of the ion flux through that ion channel.1−7,11,12 At single channel level, NMDA receptor open–close activities can occur in bursts and clusters, capable of switching open–close temporal patterns associated with multistate reaction and gating mechanisms.5,6,16−18 On the basis of analysis of single channel current trajectories recorded from NMDA receptor, it has been proposed that after binding of Glu–Gly agonists at ligand binding domains, the conformation of NMDA receptor ion channel goes through intermediate states.1,2,5,6,16−18 For example, there are reported gating models of NMDA receptors according to the mean on–off duration and amplitude of single ion channel electric current.16−18 Overall, experimental evidence are limited to prove the presence of the proposed intermediate states, because the single channel patch-clamp electric recording measurements alone do not specifically and directly identify the ion channel open–close conformational intermediate states.
Figure 1.
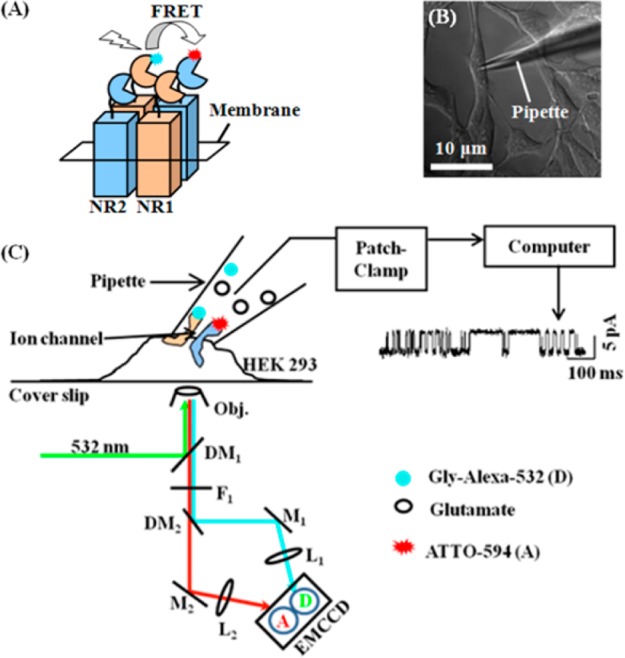
(A) Schematic representation of heterotetrameric NMDA receptor ion channel and single-molecule FRET experimental strategy. Glycine is covalently attached with Alexa-532 (cyan, donor). NR2b subunit is labeled with antibody which is previously attached with ATTO-594 (red, acceptor). NMDA receptor ion channel is activated by the binding of glycine and glutamate to the two NR1 and two NR2, respectively.5 (B) DIC image of HEK-293 cell at on-cell giga-ohm seal by patch-pipet. Note, the pipet tip makes the contact with the cell membrane at an incidence angle of ∼45°. (C) Schematic representation of single-molecule patch-clamp FRET microscope setup. Single NMDA receptor channel is patched for current measurement. The laser excitation is focused through a microscope objective onto the cell membrane. We note that the Alexa-532 labeled glycine ligands are confined in the pipet of 0.5–1 μm tip aperture size. Two-channel donor (D)–acceptor (A) fluorescence is recorded by a EMCCD camera simultaneously with patch-clamp electric current recording. DM1, dichroic mirror (Chroma Technology, z532rdc); DM2, dichroic mirror (Chroma Technology, 645dcxr); F1, 545 nm long-pass filter; M, mirror; and L, lens; Obj., 60× objective. Red and cyan lines are for acceptor and donor fluorescence signal, respectively. The excitation wavelength for the correlated experiment is 532 nm.
Structure and function relation of NMDA receptor ion channel has been reported extensively using different type of techniques, e.g., X-ray crystallography, electrophysiology and molecular dynamics simulation.6,7,23,26−33 Recently, Dai et al. have developed an atomistic model to study the gating of NMDA receptor and subunit specific contribution to the gating mechanism by targeted molecular dynamics simulation.32 They have shown that upon binding of agonist, lobe closure of ligand binding domain produce an outward pulling. The translational motion in ligand binding domain arising from the outward pulling of C-terminal results to the opening of ion channel. For NR2 subunit, this pulling is more than the NR1 subunit, and thus, NR2 subunit contributes more in NMDA receptor channel gating. This simulation model also predicts that a strong negative electrostatic potential attracts Ca2+ cation to the mouth of channel pore, and then the ion enters into the channel pore, and is thereafter solvated.32 The structure of isolated ligand binding domain monomer bound to agonist and antagonist has been reported previously.27,28 Those structures reveal that clamshell of ligand binding domain is open when bound with antagonists and closed when bound with agonists molecules. The structure of dimer of NR1 and NR2 bound to glycine and glutamate has also been reported previously.27−31 To understand the structure–function relation and dynamics of NMDA receptor, it is necessary to view the protein structure at the condition of some functions such as, membrane potential, ligand induced channel gating, Ca2+/Na+ ion permeability, interdomain interaction, etc. Very recently, a detailed view of intact heterotetrameric NMDA receptor ion channel composed of amino terminal domain, ligand binding domain and transmembrane domain has been reported by high resolution X-ray crystallography.23 It is reported that interdomain interaction between ligand binding domain and amino terminal domain plays a key role for function of NMDA receptor ion channel. The amino terminal domain and ligand binding domain are tightly packed to each other compared to AMPA receptor, and thus, amino terminal domain also plays a crucial role for NMDA receptor ion channel function. The transmembrane domain has a structural similarity with the voltage gated potassium channel, and thus, gating of NMDA receptor may involve M3 helices.23 Gouaux and co-workers have very recently reported the unprecedented 3-D view of extracellular domains and transmembrane domains of NMDA receptor by X-ray crystallography in the presence of ion channel blocker, partial agonist and inhibitor.33 They have also shown that close packed structure between amino terminal domain and ligand binding domain plays a crucial role in ion channel gating mechanism. The extracellular ends of the M3 helices of transmembrane domain form a pyramid-like shape and it may explain the Mg2+ blocking and Ca2+ permeability through the transmembrane domain.33
Single-molecule fluorescence resonance energy transfer (FRET) imaging is a powerful and sensitive approach for probing intermediate conformational states of protein and ion channels.34−66 Superresolution imaging of chemical synapses in brain, including imaging of NMDA receptors, has been demonstrated recently.56 There are a number of reports on correlated patch-clamp and fluorescence measurements in live cells as well as in artificial lipid membranes.34,67−84 For example, Lu and co-workers have demonstrated the existence of multiple intermediate states of gramicidin ion channel in artificial membrane by single-molecule patch-clamp fluorescence microscopy.34 Seemingly simple on–off two electric state signals are evidently associated with multiple intermediate conformational states identified by single-molecule fluorescence spectroscopic imaging at the electric close states of the ion channels.34,47,73,74
Here we demonstrate a new technical approach, single-molecule patch-clamp FRET imaging spectroscopy studies of NMDA receptor ion channel dynamics in HEK-293 cells (Figure 1). The single channel electric on–off trajectories have been correlated in real-time with simultaneous changes in single-molecule FRET trajectories to probe ion-channel open–close conformational dynamics. Our results suggest the occurrence of multiple close intermediate conformational states that are associated with the seemingly similar electric off states of the NMDA receptor ion channels. There are multiple conformations associated with a broad FRET efficiency distribution that are contributed to the ion channel electric off state, although the channel electrically on state is typically associated with a much narrowly distributed subset of open state conformations. Our experimental results reveal the complex relationship of the fluctuating structure and function, previously hidden from the conventional measurements, of the NMDA receptor ion channels.
Experimental Methods and Materials
1. Chemicals and Reagents
All reagents are purchased from Sigma and Invitrogen for single-molecule patch-clamp FRET experiments. They are used without further purification.
2. Heterogeneous Expression of NMDA Receptor Ion Channel on HEK-293 Cell Membrane
HEK-293 cells are cultured in EMEM (ATCC, 30–2003) supplemented with 10% fetal bovine serum (ATCC, 30-2020) and 1% penicillin–streptomycin (Gibco, 15070-063) at 37 °C in a 5% CO2 atmosphere. Cells are subcultured when they reach ∼75% confluence. Plasmid DNA encoding NR1a, NR1b, NR2a and NR2b are gift from Prof. David R. Lynch, University of Pennsylvania. Plasmids are used for heterogeneous expression without further purification.
For efficient gene transfer by transfection, we have followed standard protocol of lipid mediated transfection with Lipofectamine 2000 reagent (Invitrogen, 11668-019). Cells are incubated with 1 μg of cDNA (NR1a/NR2b/GFP = 1:1:1) for protein expression in 35 mm petri dish having a round shaped glass slide in it. Before transfection of plasmids, the cells are subcultured by EMEM without phenol red (Invitrogen, 21063-029) instead of EMEM with phenol red to minimize the autofluorescence from cells. Cells are also washed with PBS buffer solution before patch-clamp experiment. For only patch-clamp experiment, the GFP is used as marker of the NMDA receptor expressed in HEK-293 cells (Figure 2B), but for correlated experiments, we do not use GFP to avoid fluorescence signal contamination with donor.
Figure 2.
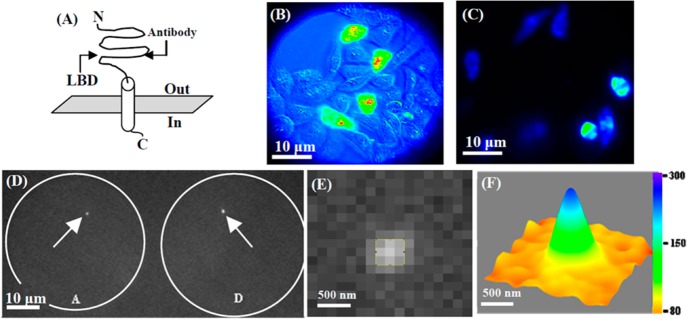
(A) Antibody binding site (Epitope: 323-337) at NR2b subunit of the NMDA receptor ion channel. (B) Simultaneously acquired differential interference contrast (DIC) and fluorescence image of HEK-293 cells coexpressed with GFP, NR1a, and NR2b plasmid DNA. Bright green colored cells have expressed NMDA receptor ion channel. (C) Fluorescence image of HEK-293 cells stained with antibody of NR2b subunits of NMDA receptor ion channel. The antibody is covalently attached with ATTO-594. The excitation wavelength for this cell imaging is 632 nm. (D) A fluorescence image frame recorded by EMCCD camera during correlated single-molecule FRET and patch-clamp experiment to capture donor (D) and acceptor (A) intensity fluctuation. The fluorescence signals from donor–acceptor are separated by a dichroic mirror (645dcxr) and then focused on a EMCCD camera as shown in Figure 1C. The artificial white circles are shown to indicate clearly the field of view focused on camera. The left and right circles are due to acceptor and donor, respectively. As donor molecule is inside the pipet, the corresponding acceptor molecule patched in the pipet only gives fluorescence signal due to FRET. The donor and acceptor fluorescence spots are indicated by white arrow. The excitation wavelength for the correlated experiment is 532 nm. Scale bar of panels B–D is 10 μm. (E) Single-molecule fluorescence image of donor molecule from panel D in a zoomed-in scale. (F) 3D surface plot of donor fluorescence spot in panel E fitted with 2D-Gaussian function. The green peak corresponds to fluorescence intensity of the single donor molecule.
3. Donor–Acceptor Labeling
NMDA receptor is activated by simultaneous binding of the glycine and glutamate molecule in the ligand binding domains of the NMDA receptor ion channel.5−33 Glycine is covalently labeled with Alexa-532 dye and then added in the extracellular solution for single-molecule patch-clamp FRET experiment. Multiple HPLC purification (C8 column with 80% methanol and 1% ACN) ensures the purity of the dye labeled glycine, which is further confirmed by mass spectroscopy. For our single-molecule FRET measurement, the Alexa-532 labeled glycine acts as donor.
After NMDA receptor expression on the HEK-293 cell membrane, the NR2b subunit of the NMDA receptor ion channel is labeled (according to protocol, Alomone Lab) with anti-NMDA receptor 2b which is covalently labeled with ATTO-594. The fluorescence imaging of the live HEK-293 cell (Figure 2C) by 632 nm laser evidently shows the staining of the antibody on NR2b subunit of NMDA receptor ion channel. Antibody is labeled on the epitope site 323–337 (Figure 2A).
4. Electrophysiology Experiment
Single channel currant is recorded with cell-attached patch clamp technique with applied voltage (100 mV) through the recording pipet. Borosilicate glass capillaries (Sutter Instruments Co., BF150-86-10) are pulled (Sutter Instruments Co., Model: P-2000) to make patch-pipettes, and then, the pipettes are fire polished to reach a high resistance of ∼15–20 MΩ. The currents are amplified by HEKA EPC-9 amplifier, and data is recorded by acquisition interface LIH-1600. The data is written into digital file by software “Pulse” and off-line data analysis is done by “PulseFit”. The patch-clamp pipet with an electrode is filled with extracellular solution containing 150 mM NaCl, 2.5 mM KCl, 1 mM EDTA, 10 mM HEPES, pH = 8.0, glucose, 1 mM glutamate and 0.1 mM glycine or Alexa-532 labeled glycine. Single channel current is recorded at very low concentration of base (pH = 8) and without presence of any divalent cations (1 mM EDTA). To maximize the chance of on–off activity from only single channel, we selected time-trajectories of currents which have no successive opening in current amplitude in the current–time trajectories. Control experiment is performed without any NMDA receptor (Figure 3) ion channel expression in HEK-293 cell, and no on–off current activity is observed. The single ion channel activity is tested with dye labeled glycine as ligand, and after antibody binding on the NR2b subunit of NMDA receptor, but in each case, single channel current activity is observed. The selection of a cell and the methods of attaching the pipet with motorized micromanipulator on cell membrane are followed as standard protocol.
Figure 3.
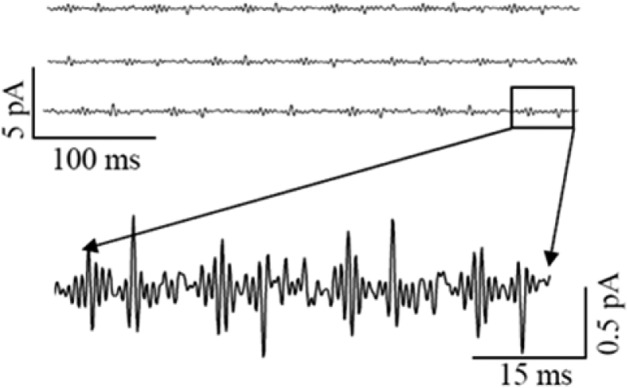
Control experiment of patch-clamp electric current measurement without expressing any NMDA receptor ion channel in HEK-293 cell. A part of current trace is shown in short time scale. No on–off electric current is observed except capacitance spikes.
5. Single-Molecule Fluorescence Microscopy
Fluorescence image and photon counting trajectories are acquired with an inverted microscope (Olympus IX71). The excitation laser (CrystaLaser) beam is reflected by a dichroic beam splitter (Chroma Technology, z532rdc) and focused onto cell membrane by a high numerical-aperture objective (Olympous, UPlanSApo 1.2 NA, 60×). In all correlated experiments, a 532 nm CW-laser is used, but for antibody labeled cell imaging, we have used 632 nm He–Ne laser (Figure 2C). To obtain the fluorescence images and intensity trajectories, the emission signals are passed through a 545 long pass filter and then the signals are separated by a dichroic beam splitter (645dcxr) into two colors to separate out the emission signal of donor, Alexa-532 and acceptor, ATTO- 594. The signals from donor and acceptor (Figure 1C) are then focused on electron multiplying charge coupled device (EMCCD) camera (Princeton Instruments, ProEM) for single-molecule fluorescence signal time-trajectory measurement.
Results and Discussion
Figure 1C shows the experimental technique to probe a single ion channel by simultaneous single-molecule FRET imaging and patch-clamp electric recording measurements. Detailed experimental methods are described elsewhere.34,47 Briefly, a single NMDA receptor ion channel on HEK-293 cell membrane is patched by on-cell method (Figure 1C). NR2b subunits of the NMDA receptor are labeled by antibody that is covalently labeled with ATTO-594, acting as the FRET acceptor in the single-molecule patch-clamp FRET imaging experiments.85,86 The agonist ligand, glycine, is covalently labeled with Alexa-532, serving as the FRET donor (Figure 1A). The patch-clamp pipet tip and the laser focus point of microscope are in an upper-under configuration with the HEK-293 cell at the same measurement point. Extracellular solution containing glutamate and glycine labeled with Alexa-532 are introduced to NMDA receptors through the patch-pipet in our experiments. A 532 nm CW-laser is focused by 60× objective onto the cell membrane that is patched with a pipet. The fluorescence signals from the donor and acceptor are separated out by a dual-color dichroic mirror and then focused on a donor–acceptor two-channel imaging EMCCD camera (Figures 1C and 2D). Electric current trajectories are recorded by on-cell patch-clamp electric recording method at the same time of the fluorescence imaging.
Figure 4A shows a typical NMDA receptor single ion channel electric current trajectory. The current amplitude histogram shows clearly two states, fully on and fully off with current amplitude ∼5 and 0 pA, respectively (Figure 4B). The decay of autocorrelation function, C(t) (Figure 4C), is deduced from current trace in Figure 4A using eq 2. Biexponential fitting gives a fast rate constant of 80 ± 5 s–1 and a slow rate constant of 1.2 ± 0.003 s–1. Figure 4D shows the distribution of the slow decay rate constants of the NMDA receptor electric current on–off dynamics. The histogram of off dwell time (Figure 4E) components is deduced from the trajectory in Figure 4A, showing two time components of 0.33 and 9.6 ms. Similarly, the on dwell time histogram gives the time components of 0.42 and 4.9 ms (Figure 4F). We have performed a control experiment to further confirm that the current trajectories measured are originated from individual NMDA ion channels.87 Control experiments performed on the HEK-293 cell without NMDA receptor ion-channel expression show no observed current signals beyond the background noises (Figure 3).
Figure 4.
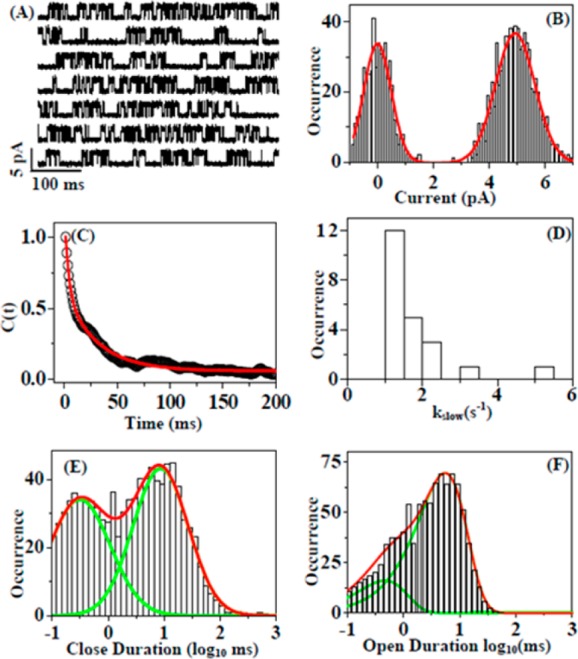
Single channel current analysis in single-molecule patch-clamp FRET microscopy. (A) A typical single channel current trajectory recorded from NMDA receptor ion channel in live HEK-293 cell. (B) Histogram of current amplitude calculated from current trajectory in panel A. Two peaks at 0 and 5 pA represent fully close state and fully open state of the ion channel, respectively. (C) Decay of autocorrelation function, C(t) = ⟨ΔI(0)ΔI(t)⟩, calculated from single channel current trajectory in panel A. Biexponential fitting gives two decay rate constant, kfast = 80 ± 5 s–1 and kslow = 1.2 ± 0.003 s–1. (D) Distribution of slow decay rate constants. (E) Histogram of close time components of NMDA receptor ion channel. The red line is for overall distribution, and the green line is for the distribution of individual time components, t1 = 0.33 ms, Amp1 = 42% and t2 = 9.6 ms, Amp2 = 58%. (F) Distribution of on time components derived from single channel current trace in panel A. The on dwell time histogram gives two time components of 0.42 and 4.9 ms.
For dissecting the conformational state changes associated with the seemingly two-state on–off electric current activities, we have also recorded and analyzed the NMDA receptor conformational fluctuation trajectories simultaneously measured with the electric on–off state change trajectories in our correlated single-molecule FRET imaging and patch-clamp electric recording experiments. Figure 5A shows a typical two-channel FRET donor {ID(t)} and acceptor {IA(t)} intensity trajectories. Typically, single-molecule donor–acceptor fluorescence intensity fluctuation trajectories recorded from a single protein molecule involve anticorrelated intensity fluctuation resulting from intramolecular FRET. We have performed multiple approaches of correlation function analysis, including second-order autocorrelation function and cross-correlation function calculated from two band fluctuation trajectories {ID(t)} and {IA(t)}.46−51,88 The correlation time between {ID(t)} and {IA(t)} is calculated by autocorrelation function Cauto(t) and cross-correlation function Ccross(t):
| 1 |
When IA(t) = ID(t), we have the autocorrelation function:
| 2 |
where IA(t) and ID(t) represent the signal variables measured in time trajectories {IA(t)} and {ID(t)}. In our experiment, {IA(t)} and {ID(t)} are the time trajectories of fluorescence photon counts or intensities.
Figure 5.
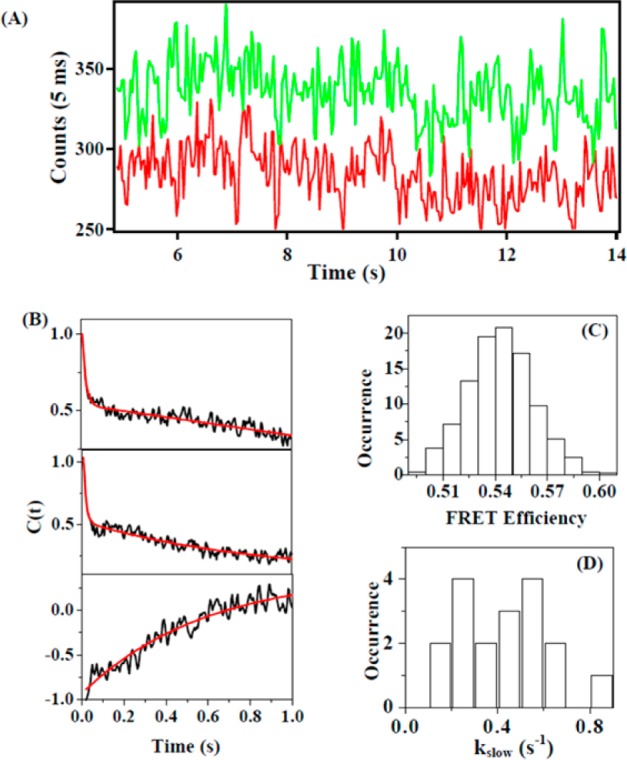
Single-molecule fluorescence recording from donor and acceptor due to conformational dynamics of NMDA receptor ion channel in live HEK-293 cell in real-time. (A) A part of single-molecule fluorescence intensity-time trajectories from donor Alexa-532 (red) and acceptor ATTO-594 (green). Anticorrelated fluctuation features are evident from the trajectories. (B) Decay of autocorrelation function of donor (top) and acceptor (middle). Decay of cross-correlation function of donor and acceptor (bottom). Biexponential fitting gives two decay rate constant, kfast = 64 ± 15 s–1 and kslow = 0.45 ± 0.05 s–1. (C) FRET efficiency histogram calculated from the intensity trajectories in panel A. Mean FRET efficiency = 0.55. (D) Distribution of the slow decay rate constants (kslow) of donor.
Figure 5B shows a typical autocorrelation function analysis of both donor and acceptor fluorescence trajectories, ⟨ΔID(0)ΔID(t)⟩ and ⟨ΔIA(0)ΔIA(t)⟩, and the analysis gives essentially the same correlation decay rate constants, a fast decay rate constant of 64 ± 15 s–1 and a slow decay rate constant of 0.45 ± 0.05 s–1. In contrast, the cross-correlation function between the donor and acceptor trajectories, ⟨ΔID(0)ΔIA(t)⟩, shows anticorrelated behavior with the same slow decay time (Figure 5B), which strongly indicates that the slow fluctuations of both donor and acceptor fluorescence intensity at a rate of 0.45 ± 0.05 s–1 are from the same origin, the FRET fluctuation associated with the conformational fluctuations of the NMDA receptor, especially, the conformational changes in the coordinate among the four subunits of the ion channel. The identified fast decay rate constant of 64 ± 15 s–1 is likely due to a different source of fluctuation that is orthogonal with the FRET fluctuation and with the associated protein conformational fluctuations based on the fluctuation dissipation theorems.46−51,88−90 Furthermore, the fast component is likely due to local environment fast fluctuation as the component only shows up in the autocorrelation function but averaged out in cross-correlation function. Nevertheless, it is the slow fluctuation rate of 0.45 ± 0.05 s–1 originated from the ion channel conformational fluctuation that is relevant to our correlated ion channel dynamics studies. On the basis of the FRET trajectories, we have also calculated the FRET efficiency histogram, using the equation,36−38,40,43,46EFRET = [IA/(ID + IA)], to evaluate the activity of the ion channels. The mean FRET efficiency is 0.55 (Figure 5C). On the basis of the histogram of the slow rate constants from the correlation function analysis of 18 single-molecule fluorescence time trajectories of donor, we obtained the mean of the rate constants to be 0.45 s–1 (Figure 5D). The average slow rate constant of electric on–off fluctuation is ∼3 times faster than the average slow rate constant from conformational open–close dynamics because the electric current measurements can only detect the on and off states of ion channel. In case of conformational change, the FRET measurement can identify the intermediate close states in addition to the open–close conformational states.
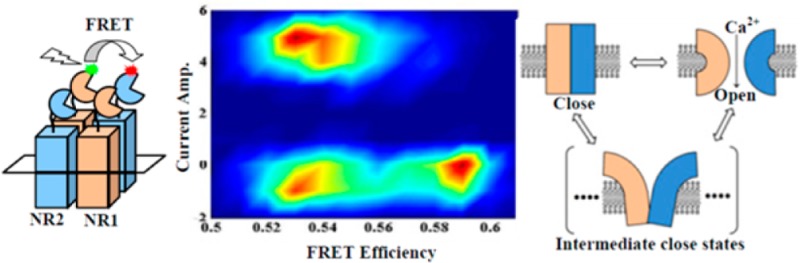
To further identify the correlation between the electric recording trajectories and the protein conformational fluctuation trajectories, especially, the open–close ion channel conformational states and the on–off electric current states, we have further explored the correlated relationship between the simultaneously recorded time trajectories from both electric recording and the FRET imaging, correlatively comparing the on–off current fluctuation states and the FRET efficiency fluctuation states time-bin to time-bin. Figure 6A,B shows a single ion channel current fluctuation time trajectory and the simultaneously recorded FRET efficiency time-trajectory for same recording time intervals. Figure 6C shows the fluorescence images of donor and acceptor channels that are acquired simultaneously with the electric current fluctuation trajectories. To resolve quantitatively that the ion channel conformational state changes are correlated with the ion channel electrical on–off states, we have plotted the single-molecule FRET efficiency time trajectory with the correlated electric on–off current trajectory, specifically to separate out the FRET efficiency distributions at the electrical on and off states (Figure 6D). This is possible because we are able to obtain temporally correlated single-molecule FRET time trajectory and electric recording time trajectory simultaneously. It is evident that the FRET efficiency is low in the range of 0.45–0.54 when the ion channel is in the electric on state, and the FRET efficiency is relatively high in the range of 0.54–0.6 when the ion channel is in the electric off state. This result supports the clamshell model of NMDA receptor ion channels activity.1,5,91−94 The four subunit domains of the NMDA receptor ion channel clam back at the channel open state and reform closer together at the channel close state, which consequentially give a low FRET efficiency at the channel open state and high FRET efficiency at the channel close state.
Figure 6.
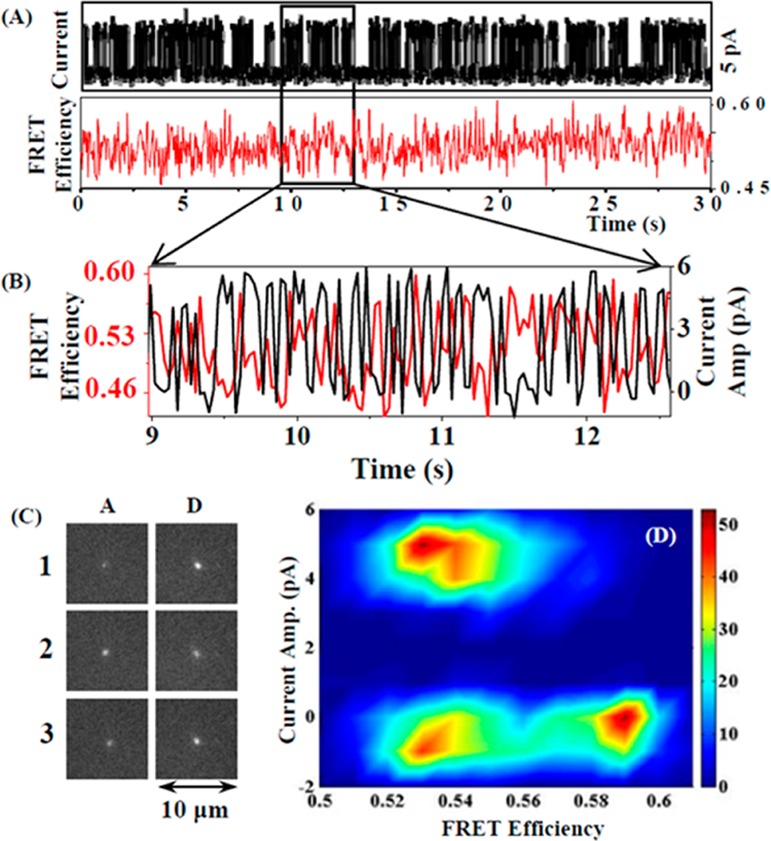
Correlated single channel current and fluorescence analysis in single-molecule patch-clamp FRET microscopy. (A) Single channel current and FRET efficiency time-trajectory at same time interval. (B) A part of the correlated current fluctuation and single-molecule FRET efficiency fluctuation time trajectory from the long time trajectories shown in panel A. (C) Fluorescence image frames of acceptor (A) and donor (D) at fully open state (1, 3) and fully close state (2). Each image size is 10 μm × 10 μm. (D) Distribution of FRET efficiency according to current amplitude. Low FRET efficiency is observed when ion channel is at fully open state, and high FRET efficiency is observed when ion channel is at fully close state. In addition to the fully open–close states, there are multiple intermediate close states that also show low FRET efficiency.
Furthermore, there is a significant broadening in the FRET distribution correlated with the electric off state comparing to the FRET distribution correlated with the electric on state (Figure 6D). It is also noticeable that there is a subset of FRET distribution with essentially the same FRET efficiency of the electric on state of the ion channel. Considering the total FRET efficiency change of 0.15, the total distance change is calculated to be 7 ± 2 Å, assuming the R0 (Förster distance36−40) value for the donor–acceptor pair is ∼50 Å.95 Sakmann and his co-workers have reported the single channel and ensemble currents from a recombinant NMDA receptor to estimate its channel pore size.24 Their result suggests that NMDA receptor is an extracellular to cytoplasomic cylindrical ion channel which has maximum and minimum mean diameter respectively 7.3 and 5.5 Å. This result is consistent with our measured and calculated distance change from single-molecule patch-clamp FRET microscopy. Jayaraman and co-workers have reported the specific configuration of four subunit arrangement of NMDA receptor, measuring the distance between the NR1 and NR2 subunits by luminescence resonance energy transfer (LRET).25 The reported distance change between the closed state and the open state among NR1 and NR2 subunits is from 6.3 to 8.2 Å, which is also consistent with our measured distance change by single-molecule FRET spectroscopy. In addition to the existence of the full open–close channel conformational states correlating to the on–off electric states, there is a subset of electric off states, about 10–15% of the total off states recorded, which have essentially the similar open conformations measured by the similar low FRET efficiency.
It is highly significant that the seemingly identical electronic off states in a clear two-state on–off dynamics of the ion channel actually associated with a broad conformational distribution across the entire range of ion channel open to close conformational states. The NMDA receptor kinetics has been extensively analyzed and reported over the last several decades,1,5−25 and intermediate conformational states other than the full open and close conformational states of the ion channel have been proposed and reported; however, to our knowledge, our experimental results from the correlated single-molecule FRET and channel electric recording results provide the first direct experimental evidence for the intermediate conformational states. On the basis of the information we get from the correlated single-molecule FRET and electric current measurements, we propose a multistate model of NMDA receptor channel dynamics (Figure 7). There are multiple intermediate conformational states that constitute the electric off states in addition to fully open and fully close state in the conformation dynamics, i.e., NMDA receptor goes through different close conformation states that all contribute to the seemingly identical electric off states, while the electric on states are essentially associated with the conformational open states, a much defined and narrow distribution of the conformational states.
Figure 7.
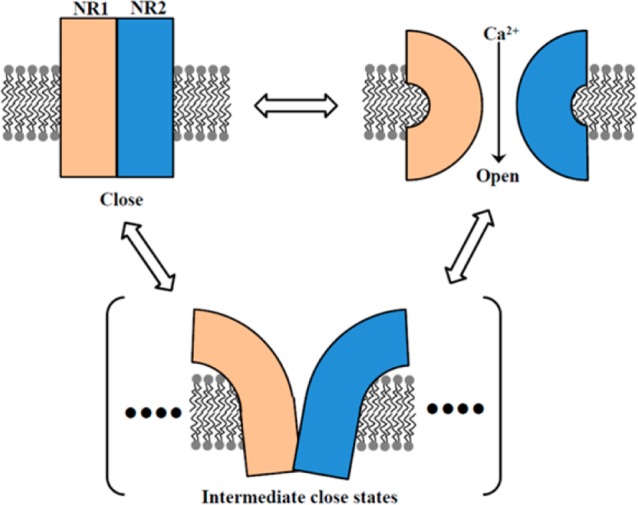
Proposed multistate clamshell model of NMDA receptor ion channel dynamics indicating presence of nonconducting or partially conducting ion channel conformation where donor and acceptor are largely separated. There are intermediate conformational states that constitute the electrically off states in addition to fully open and fully close states in the conformation dynamics.
The NMDA receptor, consisting of two NR1 and two NR2 domains, is presumably activated by binding two glutamate ligands to the two NR2 domains and two donor-labeled glycine ligands to the two NR1 domains. Furthermore, one or two antibodies with ATTO-594 (acceptor) labeling may bind to the two NR2b. Dye molecules self-quenching34,35 may complicate the FRET data analysis to identify the conformational closed state of NMDA should there be two acceptors binding to NMDA receptor. However, we were able to selectively focus our measurements on the NMDA receptors that only have one acceptor binding by identifying the anticorrelated donor–acceptor signal fluctuations through cross-correlation function analysis (Figure 5, for example). Furthermore, our single-molecule fluorescence imaging microscopic system is capable of imaging single-molecules that are immobilized on the cell membrane, such as the dye-labeled glycine ligands bound to the NMDA receptor in a HEK cell (Figures 1 and 2). Similar single-molecule fluorescence imaging measurements on cell membranes have been reported in recent years, utilizing the specific imaging background characteristic of a microsecond time scale for the free diffusion time of fluorescent molecules unbound to the cell membrane and in the background, whereas the protein motions probed are at millisecond and longer time scales.96−100 The low fluorescence background is essentially constant, exhibiting negligible fluctuation.96−100 The fluorescence background is also detected due to autofluorescence of living cell.96−100 In our imaging measurements, the intensity distribution of the donor fluorescence spot clearly shows the signal intensity above the background (Figure 2). We also note that the averaged distance of nonbounded donor molecules near the single NMDA channel patch is about a factor of 3 beyond the effective FRET distance. The rapid decay of illumination to the entrance of patch-pipet of size (0.5–1 μm) at an incidence angle of ∼45° with the imaging plane can provide a smaller than femtoliter observation volume, providing a helping condition for single-molecule imaging characterization.
We have proposed a mechanism based on the clamshell type of model1,5,91−94 for conformation change of NMDA receptor ion channel (Figure 7). The crystal structure of intact NMDA receptor has been reported recently.23,33 The amino-terminal domain and the ligand binding domain of NMDA receptor are more highly packed than those of the AMPA receptor. One of the major differences between NMDA and AMPA is that the function of NMDA receptor is regulated by the amino-terminal domain. It has also been reported that the NMDA receptor ion channel pore shows high structural similarity to that of K-ion channel (KcsA).23,33 According to crystal structure of the transmembrane (TMB) domain of NMDA receptor,23,33 it is postulated that the gating mechanism may involve the rearrangement of M3 helices, which is consistent and may shed light on the possible specific structures of the multiple closed states revealed from our correlated single-molecule patch-clamp FRET microscopic analysis.
Conclusions
In summary, we have developed a new technical approach of correlated single-molecule FRET imaging and path-clamp ion channel electric recording that can simultaneously record single-channel current fluctuation trajectories of NMDA receptor ion channels on live cell membrane and the correlated FRET efficiency time trajectories associated with the conformation changes. Correlating the optical imaging and electrophysiological recording of single ion channel NMDA receptor in live HEK-293 cells simultaneously, our experimental results suggest the existence of multiple conformation states during the open–close activity of the NMDA receptor ion channel, although the complex conformational dynamics gives apparently simple on–off two-state electric change dynamics. In particular, there are multiple conformational states that are associated with the electric off states, while the electric on states are associated with a much narrowly distributed conformational states. Technically, our new method has a great potential to provide new structure–function analysis for understanding the function, activity and mechanism of glutamate receptor ion-channels.
Acknowledgments
We acknowledge Prof. David R. Lynch, University of Pennsylvania, for providing us DNA plasmids of NMDA receptor. We also thank Dr. Gabriela K. Popescu, University of Buffalo, NY for giving us HEK-293 cells as gift and technical advice to perform electrophysiological experiments. We acknowledge Dr. David Giovannucci, University of Toledo, for technical advice and help in the early stage of our instrumental development. This work is supported by NIH/NIGMS.
The authors declare no competing financial interest.
The caption of Figure 2 and Equation 1 have been updated. The revised version was re-posted on September 17, 2014.
Funding Statement
National Institutes of Health, United States
References
- Dongen A. M. V.Biology of the NMDA Receptor; CRC Press: Boca Raton, FL; 2009. [Google Scholar]
- Sakmann B.; Neher E.. Single Channel Recordings; Plenum Press: New York, 1995. [Google Scholar]
- Hille B.Ion Channels of Excitable Membranes, 3rd eE=d.; Sinauer Associates, Inc.: Sunderland, MA, 2001. [Google Scholar]
- Lenaz G. Biosci. Rep. 1987, 7, 823. [DOI] [PubMed] [Google Scholar]
- Traynelis S. F.; Wollmuth L. P.; McBain C. J.; Menniti F. S.; Vance K. M.; Ogden K. K. Pharmacol. Rev. 2010, 62, 405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banke T. G.; Traynelis S. F. Nat. Neurosci. 2003, 6, 144. [DOI] [PubMed] [Google Scholar]
- Sobolevsky A. I.; Rosconi M. P.; Gouaux E. Nature 2009, 462, 745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. H.; Gouaux E. PLoS One 2011, 6, e19180. 10.1371/journal.pone.0019180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R.; Singh S. K.; Gu S.; Furukawa H.; Sobolevsky A. I.; Zhou J.; Jin Y.; Gouaux E. EMBO J. 2009, 28, 1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa H.; Singh S. K.; Mancusso R.; Gouaux E. Nature 2005, 438, 185. [DOI] [PubMed] [Google Scholar]
- Mayer M. L. Nature 2006, 440, 456. [DOI] [PubMed] [Google Scholar]
- Mayer M. L. Structure 2011, 19, 1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenmund C.; Stern-Bach Y.; Stevens C. F. Science 1998, 280, 1596. [DOI] [PubMed] [Google Scholar]
- Jespersen A.; Tajima N.; Fernandez-Cuervo G.; Garnier-Amblard E. C.; Furukawa H. Neuron 2014, 81, 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance K.; Simorowski N.; Traynelis S.; Furukawa H. Nat. Commun. 2011, 2, 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amico-Ruvio S. A.; Popescu G. A. Biophys. J. 2010, 98, 1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.; Howe J. R.; Popescue G. K. Nat. Neurosci. 2008, 11, 1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescue G. K. J. Physiol. 2012, 590.1, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledo A.; Barbosa R. M.; Gerhardt G. A.; Cadenas E.; Laranjinha J. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 17483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H.; Huganir R.; Liao D. Biochem. Biophys. Res. Commun. 2004, 316, 501. [DOI] [PubMed] [Google Scholar]
- Hansen K. B.; Burger P.; Vance K. M.; Snyder J. P.; Clausen R. P.; Traynelis S. F. Biophys. J. 2010, 98, 525A. [Google Scholar]
- Sobczyk A.; Scheuss V.; Svoboda K. J. Neurosci. 2005, 25, 6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkas E.; Furukawa H. Science 2014, 344, 992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarroel A.; Burnashev N.; Sakmann B. Biophys. J. 1995, 68, 866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambhadran A.; Gonzalez J.; Jayaraman V. J. Biol. Chem. 2010, 285, 15296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foucaud B.; Laube B.; Schemm R.; Kreimeyer A.; Goeldner M.; Betz H. J. Biol. Chem. 2003, 278, 24011. [DOI] [PubMed] [Google Scholar]
- Furukawa H.; Gouaux E. EMBO J. 2003, 22, 2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inanobe A.; Furukawa H.; Gouaux E. Neuron 2005, 47, 71. [DOI] [PubMed] [Google Scholar]
- MacDermott A. B.; Mayer M. L.; Westbrook G. L.; Smith S. J.; Barker J. L. Nature 1986, 321, 519. [DOI] [PubMed] [Google Scholar]
- McFeeters R. L.; Oswald R. E. FASEB J. 2004, 18, 428. [DOI] [PubMed] [Google Scholar]
- Yao Y.; Belcher J.; Berger A. J.; Mayer M. L.; Lau A. Y. Structure 2013, 21, 1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J.; Zhou H. X. Biophys. J. 2013, 104, 2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-H.; Lu W.; Michel J. C.; Goehring A.; Song X.; Gouaux E. Nature 2014, 511, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms G. S.; Orr G.; Montal M.; Thrall B. D.; Colson S. D.; Lu H. P. Biophy. J. 2003, 85, 1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X.; Ha T.; Kim H. D.; Centner T.; Labeit S.; Chu S. Proc. Natl. Acad. Sci. U.S.A. 2000, 97, 14241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowicz J. R.Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: New York, 2006. [Google Scholar]
- Roy R.; Hohng S.; Ha T. Nat. Methods 2008, 5, 507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvin P. R.; Ha T.. Single-Molecule Techniques: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, 2008. [Google Scholar]
- Ha T.; Ting A. Y.; Liang J.; Caldwell W. B.; Deniz A. A.; Chemla D. S.; Schultz P. G.; Weiss S. Proc. Natl. Acad. Sci. U.S.A. 1999, 96, 893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasselet S.; Peterman E. J. G.; Miyawaki A.; Moerner W. E. J. Phys. Chem. B 2000, 104, 3676. [Google Scholar]
- Moerner W. E.; Orrit M. Science 1999, 283, 1670. [DOI] [PubMed] [Google Scholar]
- Min W.; Xie X. S.; Bagchi B. J. Chem. Phys. 2009, 131, 065104. [DOI] [PubMed] [Google Scholar]
- Joo C.; Balci H.; Ishitsuka Y.; Buranachai C.; Ha T. Annu. Rev. Biochem. 2008, 77, 51. [DOI] [PubMed] [Google Scholar]
- Ishii Y.; Yanagida T. Single Mol. 2000, 1, 5. [Google Scholar]
- Sako Y.; Minoguchi S.; Yanagida T. Nat. Cell Biol. 2000, 2, 168. [DOI] [PubMed] [Google Scholar]
- Lu H. P. Chem. Soc. Rev. 2014, 43, 1118. [DOI] [PubMed] [Google Scholar]
- Lu H. P. Acc. Chem. Res. 2005, 38, 557. [DOI] [PubMed] [Google Scholar]
- Liu R. C.; Hu D. H.; Tan X.; Lu H. P. J. Am. Chem. Soc. 2006, 128, 10034. [DOI] [PubMed] [Google Scholar]
- Wang X. F.; Lu H. P. J. Phys. Chem. B 2008, 112, 14920. [DOI] [PubMed] [Google Scholar]
- Pan D.; Hu D. H.; Liu R. C.; Zeng X. H.; Kaplan S.; Lu H. P. J. Phys. Chem. C 2007, 111, 8948. [Google Scholar]
- He Y.; Li Y.; Mukherjee S.; Wu Y.; Yan H.; Lu H. P. J. Am. Chem. Soc. 2011, 133, 14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippitz M.; Kulzer F.; Orrit M. ChemPhysChem 2005, 6, 770. [DOI] [PubMed] [Google Scholar]
- Lu H. P.; Xun L. Y.; Xie X. S. Science 1998, 282, 1877. [DOI] [PubMed] [Google Scholar]
- Landes C. F.; Rambhadran A.; Taylor J. N.; Salatan F.; Jayaraman V. Nat. Chem. Biol. 2011, 7, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Liu Y.; DeBerg H. A.; Nomura T.; Hoffman M. T.; Rohde P. R.; Schulten K.; Martinac B.; Selvin P. R. eLife 2014, 3, e01834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani A.; Huang B.; Bergan J.; Dulac C.; Zhuang X. Neuron 2010, 68, 843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Chu X.; Longhi S.; Roche P.; Han W.; Wang E.; Wang J. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, E3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z. Z.; Lu Q.; Wang J. J. Phys. Chem. B 2011, 115, 4147. [DOI] [PubMed] [Google Scholar]
- Lu Q.; Wang J. J. Phys. Chem. B 2009, 113, 1517. [DOI] [PubMed] [Google Scholar]
- Lu Q.; Wang J. J. Am. Chem. Soc. 2008, 130, 4772. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Gan L. F.; Wang E. K.; Wang J. J. Chem. Theory Comput. 2013, 9, 84. [DOI] [PubMed] [Google Scholar]
- Zheng W.; Schafer N. P.; Davtyan A.; Papoian G. A.; Wolynes P. G. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 19244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyeon C.; Jennings P. A.; Adams J. A.; Onuchic J. N. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasmal D. K.; Mandal T.; Mojumdar S. S.; Choudhury A.; Banerjee R.; Bhattacharyya K. J. Phys. Chem. B 2011, 115, 13075. [DOI] [PubMed] [Google Scholar]
- Paul B. K.; Ghosh N.; Mukherjee S. Langmuir 2014, 30, 5921. [DOI] [PubMed] [Google Scholar]
- Hamil A. C.; Wang S. C.; Lee C. T. Jr. Biochemistry 2005, 44, 15139. [DOI] [PubMed] [Google Scholar]
- Ide T.; Yanagida T. Biochem. Biophys. Res. Commun. 1999, 265, 595. [DOI] [PubMed] [Google Scholar]
- Ide T.; Hirano M.; Takeuchi Y.. Ion Channels. In Single Molecule Dynamics in Life Science; Yanagida T.; Ishii Y., Eds.; Wiley-VCH: Weinheim, Germany, 2009, 87. [Google Scholar]
- Leptihn S.; Thompson J. R.; Ellory J. C.; Tucker S. J.; Wallace M. I. J. Am. Chem. Soc. 2011, 133, 9370. [DOI] [PubMed] [Google Scholar]
- Heron A. J.; Thompson J. R.; Cronin B.; Bayley H.; Wallace M. I. J. Am. Chem. Soc. 2009, 131, 1652. [DOI] [PubMed] [Google Scholar]
- Kong L.; Harrington L.; Li Q.; Cheley S.; Davis B. G.; Bayley H. Nat. Chem. 2013, 5, 651. [DOI] [PubMed] [Google Scholar]
- Rajapaksha S. P.; Wang X.; Lu H. P. Anal. Chem. 2013, 85, 8951. [DOI] [PubMed] [Google Scholar]
- Lu H. P. Methods Cell Biol. 2008, 90, 435. [DOI] [PubMed] [Google Scholar]
- Orr G.; Montal M.; Thrall B. D.; Colson S. D.; Lu H. P. Biophys. J. 2001, 80, 151a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene M. J.; Korlach J.; Turner S. W.; Foquet M.; Graighead H. G.; Webb W. W. Science 2003, 299, 682. [DOI] [PubMed] [Google Scholar]
- Borisenko S. V.; Kordyuk A. A.; Kim T. K.; Koitzsh A.; Knupfer M.; Fink J.; Golden M. S.; Eschrig M.; Berger H.; Follath Y. Phys. Rev. Lett. 2003, 90, 207001. [DOI] [PubMed] [Google Scholar]
- Sonnleitner A.; Mannuzzu L. M.; Terakawa S.; Isacoff E. Y. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 12759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms G. S.; Cognet L.; Lommerse P. H. M.; Blab G. A.; Kahr H.; Gamsjäger R.; Spaink H. P.; Soldatov N. M.; Romanin C.; Schmidt T. Biophys. J. 2001, 81, 2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J.; Zagotta W. N. Neuron 2000, 28, 369. [DOI] [PubMed] [Google Scholar]
- Demuro A.; Parker I. Biol. Res. 2004, 37, 675. [DOI] [PubMed] [Google Scholar]
- Zou H.; Lifshitz L. M.; Tuft R. A.; Fogarty T. K.; Singer J. J. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuro A.; Parker I. Cell Calcium 2006, 40, 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisenko V.; Lougheed T.; Hesse J.; Füreder-Kitzmüller E.; Fertig N.; Behrends J. C.; Woolley G. A.; Schütz G. J. Biophy. J. 2003, 84, 612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lougheed T.; Borisenko S.; Hand C. E.; Woolley G. A. Bioconjugate Chem. 2001, 12, 594. [DOI] [PubMed] [Google Scholar]
- Kusch J.; Biskup C.; Thon S.; Schulz E.; Nache V.; Zimmer T.; Schwede F.; Benndorf K. Neuron 2010, 67, 75. [DOI] [PubMed] [Google Scholar]
- Toro D. D.; Xifró X.; Pol A.; Humbert S.; Saudou F.; Canals J. M.; Alberch J. J. Neurochem. 2010, 115, 153. [DOI] [PubMed] [Google Scholar]
- Mosgaard L. A.; Heimburg T. Acc. Chem. Res. 2013, 46, 2966. [DOI] [PubMed] [Google Scholar]
- Lu H. P. Phys. Chem. Chem. Phys. 2011, 13, 6734. [DOI] [PubMed] [Google Scholar]
- Chandler D.Introduction to Modern Statistical Mechanics; Oxford University Press: New York, 1987. [Google Scholar]
- Zwanzig R.Nonequilibrial Statistical Mechanics; Oxford University Press: New York, 2001. [Google Scholar]
- Zhu S.; Stroebel D.; Yao C. A.; Taly A.; Paoletti P. Nat. Struct. Biol. 2013, 20, 477. [DOI] [PubMed] [Google Scholar]
- Gielen M.; Retchless B. S.; Mony L.; Johnson J. W.; Paoletti P. Nature 2009, 459, 703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryga A.; Chakrabarty S.; Vicatos S.; Warshel A. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. T.; Xu Y.; Tang P. J. Phys. Chem. B 2008, 114, 9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Hu D.; Vorpagel E. R.; Lu H. P. J. Phys. Chem. B 2003, 107, 7947. [Google Scholar]
- Cai L.; Friedman N.; Sie X. S. Nature 2006, 440, 358. [DOI] [PubMed] [Google Scholar]
- Elf J.; Li G. W.; Xie X. S. Science 2007, 316, 1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.; Xiao J.; Ren X.; Lao K.; Xie X. S. Science 2006, 311, 1600. [DOI] [PubMed] [Google Scholar]
- Choi P.; Long C.; Kirsten F.; Xie X. S. Science 2008, 332, 442.18927393 [Google Scholar]
- Deich J.; Judd E. M.; McAdams H. H.; Moerner W. E. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 15921. [DOI] [PMC free article] [PubMed] [Google Scholar]


