Abstract
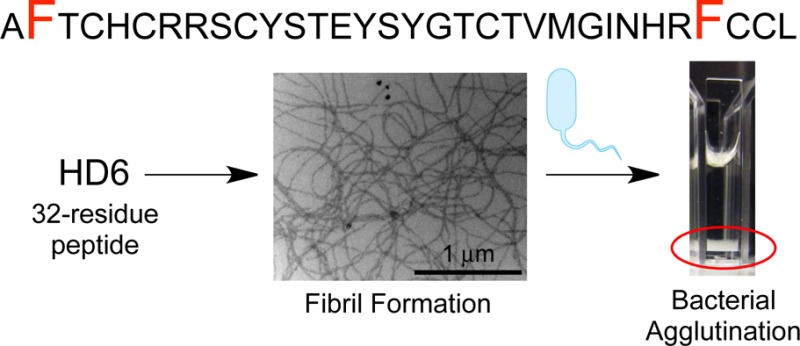
Human α-defensin 6 (HD6) is a 32-aa cysteine-rich peptide of the innate immune system. Although HD6 is a member of an antimicrobial peptide family, it exhibits negligible antibacterial activity in vitro. Rather, HD6 possesses a unique innate immune mechanism whereby it self-assembles into oligomers that capture pathogens to prevent microbial invasion of the intestinal epithelium and subsequent dissemination. Molecular-level understanding for why HD6 functions differently from other human defensins remains unclear. To further elucidate the HD6 self-assembly process and its biological activity, we developed robust protocols for obtaining native and mutant HD6 in high purity from overexpression in Escherichia coli. We combined biophysical characterization with biological assays to probe HD6 structure and function. We report that native HD6 readily self-assembles into elongated fibrils observable by transmission electron microscopy, agglutinates both Gram-negative and -positive bacteria, and prevents the human gastrointestinal pathogen Listeria monocytogenes from invading cultured mammalian cells. Mutation of hydrophobic residues (F2A, I22T, V25T, F29A) perturbs self-assembly and results in attenuated biological activity. In particular, the F2A and F29A mutants do not form fibrils under our experimental conditions and neither agglutinate bacteria nor prevent L. monocytogenes invasion. In total, our results demonstrate that the hydrophobic effect is essential for promoting HD6 self-assembly and innate immune function, and indicate that HD6 may provide host defense against Listeria in the gut. This investigation provides a timely description of how variations in amino acid sequence confer diverse physiological functions to members of the defensin family.
Introduction
The innate immune system provides the first line of defense for the detection of and response to microbial invasion. One of the key contributors to innate immunity is a family of small (2–5 kDa), cysteine-rich host-defense peptides called defensins.1−5 These biomolecules are typically described as antimicrobial peptides with broad-spectrum microbicidal activity. In the oxidized forms, mammalian defensins contain three conserved and regiospecific intramolecular disulfide bonds that stabilize a three-stranded β-sheet fold.5 The regiospecific disulfide-bond patterns divide these defensins into three subclasses called α-, β-, and θ-defensins. Humans produce and utilize α- and β-defensins in the battles against invading microbial pathogens.
The six α-defensins identified in humans are human neutrophil peptides 1–4 (HNP1–4) and human enteric defensins 5 and 6 (HD5 and HD6).6 These peptides exhibit CysI–CysVI, CysII–CysIV, CysIII–CysV linkages following oxidative folding. HNP1–4 are stored in the azurophilic granules of neutrophils,7,8 human monocytes,9 and natural killer cells.10 HD5 and HD6 are abundant in the granules of Paneth cells,11−14 a type of secretory cell located at the bases of the crypts of Lieberkühn in the small intestine.15,16 Paneth cells protect the intestinal epithelium against infection and colonization of opportunistic and pathogenic microbes by secreting a mixture of antimicrobial peptides and proteins that includes HD5 and HD6.15,16
Although the six human α-defensins display a common tertiary structure that results from conserved cysteine positioning and the regiospecific disulfide array,2,5 the primary sequences are highly variable and several recent studies demonstrated that α-defensins possess different structural and functional attributes.17−24 A comparison of the 32-residue Paneth cell defensins HD5 and HD6 exemplifies this notion.19−24 HD5 forms dimers and tetramers in aqueous solutions20,25 and provides broad-spectrum antibacterial activity in vitro.22,23 The HD5 transgenic mouse exhibits resistance to oral Salmonella challenge.24 In contrast, HD6 displays negligible antibacterial activity in vitro;21,22 however, studies of the HD6 transgenic mouse revealed that this peptide provides defense against Salmonella challenge by an unprecedented mechanism.21 In the current working model, HD6 forms higher-order structures described as “nanonets” that entrap bacteria in the intestinal lumen and thereby prevent bacterial invasion of the host epithelium and subsequent dissemination to other organ systems.21 This remarkable observation gives rise to a number of questions about HD6 at the molecular level. Why does HD6 function differently from other human α-defensins, including its Paneth cell congener HD5? What is the molecular basis for HD6 self-assembly that affords “nanonets” from 32-residue monomeric units?
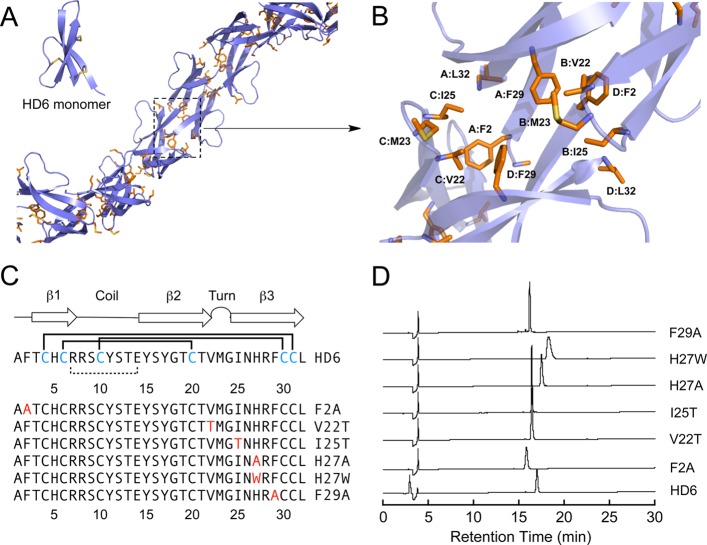
Several recent studies support the importance of the hydrophobic effect for the in vitro biological activities of α-defensins, including HNP1, HD5, and murine cryptdin-4.25−28 The amino acid sequence alignment of the six human α-defensins reveals that the distribution of hydrophobic residues in the HD6 primary sequence is distinct from those in HD5 and the HNPs (Supporting Information Figure S1). Prior X-ray crystallographic structural studies indicate that several hydrophobic residues of HD5 reside on the β-sheets and facilitate side-to-side dimerization (PDB: 1ZMP,19Supporting Information Figure S2), whereas the hydrophobic residues of HD6 (PDB: 1ZMQ,19Supporting Information Figure S2) are located in the loop (V22, M23, I25) and in the N- (F2) and C-terminal regions (F29, L32). Moreover, the hydrophobic residues of HD6 define a hydrophobic pocket that forms at the interface between four monomers.19,25 In each hydrophobic pocket, two monomers each contribute F2, F29, and L32, and the other two monomers each contribute V22, M23, and I25. Although each HD6 monomer exhibits the canonical α-defensin fold in the solid state, the monomers assemble to form an elongated fibril-like chain that is unique among structurally characterized defensins (Figure 1A).19,21 We reasoned that formation of this hydrophobic pocket allows HD6 monomers to form the elongated fibril-like structures observed crystallographically (Figure 1A,B), and hypothesized that the hydrophobic effect also contributes to formation of the nanonet structures observed in biological systems.
Figure 1.
(A) Previously reported extended crystal structure of HD6 (PDB: 1ZMQ).19 Hydrophobic residues are shown in orange. Inset: a HD6 monomer unit illustrates the three-stranded β-sheet fold. The disulfide bonds are shown in yellow. (B) Close-up view of the hydrophobic pocket located among HD6 monomers. Individual HD6 monomers are labeled A–D. (C) Amino acid sequences of HD6 and mutants evaluated in this work. The numbers represent amino acid position. The Cys residues in blue comprise the Cys4–Cys31, Cys6–Cys20, and Cys10–Cys30 disulfide linkages (solid lines). The salt-bridge between Arg7 and Glu14 is indicated as a dashed line. The mutated residues are shown in red. The secondary structure depiction is based on the crystal structure. (D) Analytical HPLC traces of purified HD6 and mutants (oxidized forms) dissolved in Milli-Q water (30 μM × 80 μL). Absorbance at 220 nm was monitored with a reference wavelength of 500 nm. Method: 10–60% B over 30 min at 1 mL/min.
In the present work, we describe biochemical and biophysical studies designed to investigate the self-assembly and biological function of HD6. We report that native HD6 forms micron-sized fibril structures in aqueous solution, agglutinates bacteria, and prevents Listeria monocytogenes invasion into cultured human cells. We demonstrate that hydrophobic residues, especially F2 and F29, are essential for self-assembly under the experimental conditions utilized in this work and provide HD6 with the ability to entrap bacteria and prevent invasive microbes from entering human cells. In total, our investigations provide important new molecular-level insight into a host-defense peptide with an unprecedented physiological activity and support a model whereby the disposition of hydrophobic residues along the canonical α-defensin fold tunes innate immune function. Moreover, the gut is a primary site for L. monocytogenes infection and our results suggest that HD6 may confer host defense against this pathogen in the gut.
Results and Discussion
Design and Preparation of a HD6 Mutant Family
We designed and prepared a six-membered HD6 mutant family (Figure 1C, Supporting Information Table S1) to evaluate the role of select residues in HD6 self-assembly and host-defense function. The mutants F2A, F29A, V22T, and I25T were prepared to probe the consequences of decreased hydrophobicity for each of these residues. We also evaluated H27W and H27A. This residue abuts the hydrophobic pocket and, in prior work, a role for H27 in self-assembly was proposed from evaluation of the H27W and H27A mutants.21 The X-ray crystal structure of H27W (PDB: 3QTE)21 differs from that of HD6 and supports a dimer of dimers arrangement and the absence of elongated structures. An electrostatic interaction between H27 of one monomer (A or D, Supporting Information Figure S3) and the C-terminal carboxylic acid of L32 of another monomer (B or C, Supporting Information Figure S3) was hypothesized to enable formation of the extended oligomers observed for native HD6.21 The X-ray crystal structure of HD6 also suggests that a π–π stacking interaction between the side chains of F2 of one monomer (A or D) and H27 of another monomer (B or C) may occur and contribute to self-assembly (Supporting Information Figure S3).
In prior studies of HD5, we overexpressed His6-Met-HD5 and employed cyanogen bromide to cleave the affinity tag and afford native HD5.29 HD6 contains a Met residue at position 23 and is not compatible with this approach. The HNPs and HD5 are transcribed and translated as prepropeptides that undergo maturation,30−32 and mRNA analysis indicates that HD6 is translated as a prepropeptide (100-aa), as well.12 We therefore expressed proHD6 (81-aa) harboring a N-terminal His6 tag and released native HD6 (oxidized form) by protease digestion (Supporting Information). For peptide overexpression, an Escherichia coli-optimized synthetic gene for proHD6 was ligated into the NdeI and XhoI sites of pET28b, and His6-proHD6 was overexpressed in E. coli BL21(DE3) cells and isolated by using Ni-NTA column chromatography. The oxidized form of His6-proHD6 was obtained by following an oxidative folding protocol,29 and native HD6 was isolated following treatment of His6-proHD6 with trypsin and preparative HPLC purification (Supporting Information Figure S4). We employed the same procedure to prepare the six HD6 mutants. Analytical HPLC indicated that HD6 and mutants were obtained in high purity (Figure 1D, Supporting Information Figure S5). The peptide yields ranged from 1.8 (F2A) to 4.5 (H27W) mg/L of culture (Supporting Information Table S2). Thiol quantification indicated that each peptide contained no free thiol moieties (Supporting Information Table S2), and the peptide identities were confirmed by LC–MS (Supporting Information Table S2). For each mutant, the canonical α-defensin disulfide bond pattern exhibited by native HD6 (Cys4–Cys31, Cys6–Cys20, Cys10–Cys30) was confirmed by manual Edman degradation (Supporting Information Table S3 and Scheme S1). All experiments were performed with the oxidized species and in the absence of reducing agents.
Biophysical Characterization Reveals That F2 and F29 Are Essential for HD6 Self-Assembly
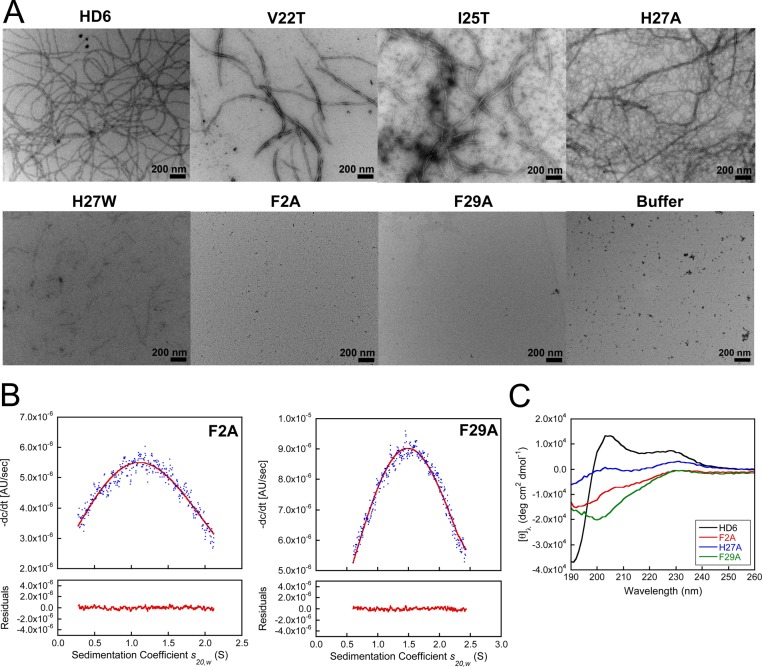
Transmission electron microscopy (TEM) revealed that HD6 spontaneously self-assembles into higher-order oligomers in aqueous solution. When we incubated HD6 (2 or 20 μM) at pH 7.4 in 10 mM sodium phosphate buffer and stained the samples with 2% uranyl acetate, we observed elongated fibril-like structures with lengths on the micron scale (Figure 2A, Supporting Information Figure S6). HD6 also formed micron-sized fibrils at pH 6.4 in 10 mM Tris-maleate buffer (Supporting Information Figure S7). The Tris-maleate buffer was employed in a prior investigation where Salmonella enterica serovar Typhimurium entrapped by HD6 nanonets in vitro was visualized by scanning electron microscopy (SEM).21 A comparison of the TEM images obtained in both buffers revealed a buffer effect on fibril morphology. In particular, the Tris-maleate buffer afforded HD6 fibrils that are relatively short and in relatively low abundance when compared to same experiment performed in sodium phosphate buffer.
Figure 2.
Biophysical characterization of native and mutant HD6 (10 mM sodium phosphate buffer, pH 7.4). (A) Transmission electron micrographs of 20 μM HD6 and mutants. Scale bar = 200 nm. (B) Analytical ultracentrifugation of 100 μM F2A and 100 μM F29A. The blue dots are the −dc/dt data obtained from sedimentation velocity experiments (absorbance at 280 nm). The red lines are the single Gaussian fits obtained using DCDT+. The summaries of the fits are provided in Supporting Information Tables S4 and S5. (C) CD spectra of 20 μM HD6 (black), F2A (red), H27A (blue), and F29A (green).
TEM of the HD6 mutants provided evidence for varying degrees of fibril formation under the same conditions (Figure 2A, Supporting Information Figure S7). Notably, mutation of hydrophobic residues that comprise the hydrophobic pocket had pronounced effects on fibril formation and morphology. No fibrils were observed for the F2A or F29A mutants in either buffer, and the V22T and I25T mutants afforded shorter and thicker fibrils. These observations indicate that these hydrophobic residues are important for self-assembly, and that mutation of either F2 or F29 to Ala prevents fibril formation. H27A afforded an extensive fibril network characterized by both thick and thin fibrils whereas fewer and significantly shorter fibril-like structures were observed for H27W. The latter result is in general agreement with prior studies of H27W, which reported defective oligomerization properties for this mutant as ascertained by surface plasmon resonance (SPR) and X-ray crystallography (PDB: 3QTE).21 However, fibril formation for H27A contrasts with conclusions drawn from a SPR investigation of H27A where this mutant was reported to exhibit the same behavior as H27W.21 From comparison of H27A and H27W by TEM under our experimental conditions, as well as in other experiments described below, we conclude that (i) the nature of the self-assembly varies from mutation to mutation and (ii) the presence of the bulky Trp residue at position 27 results in greater perturbation to self-assembly than loss of H27 and hence loss of either the electrostatic interaction between this residue and L32 or the putative π–π stacking interaction with F2.
To further evaluate the quaternary structures of F2A and F29A, we employed analytical ultracentrifugation (AUC) and determined the sedimentation coefficients (s20,w) for each peptide (Supporting Information Tables S4 and S5). At pH 7.4 in 10 mM sodium phosphate buffer, single peaks at ca. 0.8 and 1.2 S are observed for F2A (≤160 μM) and F29A (≤160 μM), respectively, over the range s20,w = 0–2.5 S in the Gaussian fits of the −dc/dt distributions obtained using DCDT+ (Figure 2B, Supporting Information Figures S8 and S9, Tables S4 and S5). The Gaussian fits support the predominance of a single species for both F2A and F29A over a concentration range of 40–160 μM at pH 7.4. Substitution of phosphate buffer with Tris-maleate buffer at pH 6.4 had negligible effect on the s values for both F2A and F29A (Supporting Information Figures S8 and S9, Tables S4 and S5). HYDROPRO33 estimated the sedimentation coefficients of HD6 to be 0.75 S (monomer), 1.17 S (dimer), and 1.83 S (tetramer) using the X-ray crystal structure of HD619 as a model (Supporting Information Table S6). A comparison between the experimental and estimated s values suggests that F2A and F29A predominantly exist as a monomer and a dimer, respectively, under the conditions employed for these experiments.
Prior to TEM imaging of the HD6 fibrils, we attempted to perform a sedimentation velocity experiment with HD6. This experiment failed because the peptide sedimented within 15 min, even at the lowest possible rotor speed (3000 rpm), and formed a white coat on the AUC cell attributed to rapid peptide aggregation. In total, the AUC results support the findings from TEM and confirm that F2 and F29 are essential for HD6 self-assembly in aqueous buffer.
Structural differences between native and mutant HD6 are also apparent in the CD spectroscopic signatures (Figure 2C, Supporting Information Figure S10). Native HD6 exhibits relatively intense features defined by a negative peak at ca. 190 nm and positive peaks centered at 205 and 230 nm. This CD spectrum differs from those reported for other defensins, including HD5.29 The HD6 mutants exhibit less intense CD features than native HD6, and the CD spectra of some mutants (e.g., F29A) resemble that of HD5. The relatively intense features in the CD spectrum of native HD6 may provide a fingerprint of high-order oligomerization and therefore be used to identify other defensins that display similar self-assembly.
Mutation of HD6 Does Not Confer Antibacterial Activity
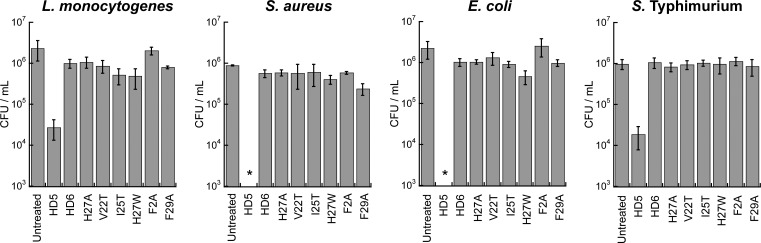
We questioned whether any of the HD6 mutants employed in this work, and F2A and F29A in particular, exhibit antibacterial activity as a result of disrupted oligomerization that allows smaller, potentially bioactive, species to exist. We compared the antibacterial activity of HD6 and the six mutants against four bacterial species. L. monocytogenes ATCC 19115 and Staphylococcus aureus ATCC 25923 were chosen as representative Gram-positive organisms, whereas E. coli ATCC 25922 and S. Typhimurium ATCC 14028 were selected as representative Gram-negative microbes. In these assays, HD5, which has broad-spectrum antibacterial activity, was employed as a positive control. The results presented in Figure 3 clearly delineate that neither HD6 nor the mutants exhibit significant antibacterial activity against any of the evaluated strains at a concentration of 20 μM. Thus, disruption of quaternary structure that affords lower-order oligomers (e.g., F2A, F29A) of the HD6 scaffold does not “turn on” a potent bactericidal killing activity for HD6 against these microbes.
Figure 3.
Antibacterial activity assays against L. monocytogenes ATCC 19115, S. aureus ATCC 25923, E. coli ATCC 25922, and S. Typhimurium ATCC 14028 (mean ± SDM, n = 3). The peptide concentrations were 20 μM. The asterisks indicate no colony formation.
Mutation of Hydrophobic Residues Alters the Bacterial Agglutination Behavior of HD6
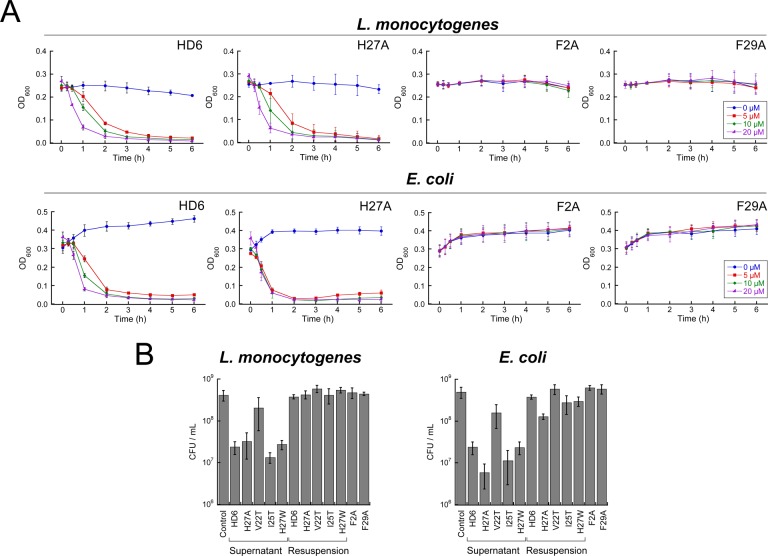
In seminal studies of HD6 host-defense function, SEM revealed that HD6 entangles S. Typhimurium.21 We reasoned that point mutations that disrupt HD6 self-assembly likely exhibit a reduced propensity to entrap and agglutinate bacteria. Thus, we established a simple bacterial agglutination assay based on solution turbidity and we employed this assay to probe the ability of HD6 and its mutants to entrap bacteria and cause sedimentation (Figure 4, Supporting Information Figures S11–S13). When we introduced 20 μM HD6 into a bacterial culture (108 CFU/mL) housed in a sterile plastic cuvette, we observed bacterial clumping by the eye within 5 min of mixing. These clumps sedimented to the bottom of the cuvette within about 2 h and clarified the culture medium. This agglutination phenomenon was observed for both Gram-negative E. coli and Gram-positive L. monocytogenes. In contrast, agglutination and sedimentation were not observed for the untreated control; these cultures remained homogeneous over the course of the assay. We defined the two phases of the heterogeneous mixture obtained from HD6 treatment as “supernatant” for the clarified medium solution in the top portion of the cuvette and “agglutinate” for the sedimented material at the bottom of the cuvette (Supporting Information Figure S13). We defined “resuspension” as the mixture that results from thoroughly agitating the heterogeneous mixture containing the sedimented material at the bottom of the cuvette. After incubating the bacteria and HD6 for 6 h, we determined the CFU/mL of the supernatant and observed a ca. 1.3-fold log reduction in CFU/mL relative to the bacteria-only control. Following agitation, the CFU/mL of the resuspension was comparable to that of the untreated control (Figure 4B). As expected, the reduction of CFU/mL in the supernatant is attributed to sedimentation of bacteria in the cuvette, and not to a bactericidal activity of HD6.
Figure 4.
Bacterial agglutination assay for native and mutant HD6. (A) Agglutination of L. monocytogenes ATCC 19115 and E. coli ATCC 25922 treated with HD6 and mutants at different concentrations. (B) Plots of colony forming units (CFU/mL) of L. monocytogenes ATCC 19115 and E. coli ATCC 25922 after treatment with 20 μM peptide for 6 h (mean ± SDM, n = 3).
We evaluated the ability of the HD6 mutants (0–20 μM) to agglutinate and sediment L. monocytogenes and E. coli over a 6 h period (Figure 4, Supporting Information Figures S11 and S12). On the basis of the resulting sedimentation profiles, we defined three different types of behavior for native and mutant HD6 and grouped the peptides accordingly. One group is composed of native HD6 and H27A, both of which afforded similar and relatively rapid bacterial agglutination with the OD600 values approaching zero in the presence of peptide. The second group includes mutants that caused bacterial agglutination, but to a lesser degree than native HD6 and H27A (Supporting Information Figures S11 and S12). These peptides are V22T, I25T and H27W, which agglutinated the bacteria only at relatively high (≥10 μM) peptide concentrations. The oligomerization-deficient F2A and F29A mutants define the third group. The bacterial cultures containing these peptides remained homogeneous after 6 h even at the highest peptide concentration evaluated, indicating that these mutants cannot promote bacterial agglutination and sedimentation under these conditions. These profiles correlate with the results from TEM where HD6 and H27A formed elongated fibrils; V22T, I25T and H27W formed fibrils that are smaller and with variable morphologies; and F2A and F29A did not form any observable oligomer.
To verify that the decreases in OD600 values observed for V22T, I25T, H27A and H27W correlated with bacterial agglutination and sedimentation at the bottom of the cuvette and not to bacterial cell death, we ascertained the CFU count for the supernatant and resuspension of each culture treated with 20 μM peptide for 6 h (Figure 4B). As expected based on the results of the antibacterial activity assays (Figure 3), this experiment confirmed that the bacteria remained alive over the course of the experiment. Moreover, the CFU/mL reductions observed for the supernatants correlate with the differences in the OD600 values before and after sedimentation took place. A smaller change in OD600 as observed for V22T correlates with a smaller decrease in CFU/mL for the supernatant. For I25T, H27A and H27W, each of which caused OD600 to approach zero at 20 μM of peptide, a CFU/mL reduction similar in magnitude to that of HD6 was observed.
Hydrophobic Residues Are Needed for HD6 To Prevent Listeria Invasion of Human T84 Cells
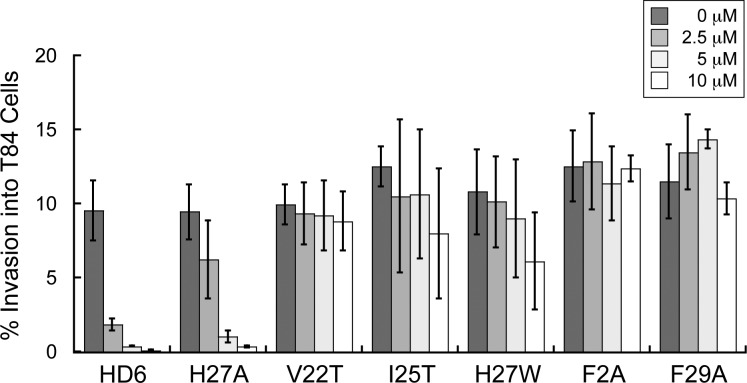
L. monocytogenes is a pathogenic Gram-positive bacterium that can colonize the human gastrointestinal tract and cause foodborne illness in adults as well as meningitis which is a serious threat to fetuses and newborns.34,35 This species binds to and invades host cells. We performed a series of L. monocytogenes invasion assays and determined that HD6 in the culture medium blocks Listeria invasion into human T84 intestinal epithelial cells. The percentage of Listeria invasion dropped from ca. 10% to <2% when ≥2.5 μM HD6 was included in the culture medium (Figure 5). HD6 was previously shown to block invasion of mammalian cells by two different Gram-negative species, S. Typhimurium and Yersinia enterocolitica.21 Taken together with the current results obtained for a Gram-positive organism, we conclude that the ability of HD6 to prevent bacterial invasion, at least in vitro, is broad-spectrum with no apparent selectivity for Gram-negative or Gram-positive organisms. Moreover, our results indicate that HD6 has the capacity to provide host defense against the gastrointestinal pathogen L. monocytogenes.
Figure 5.
Invasion of human T84 colon epithelial cells by L. monocytogenes ATCC 19115 in the presence of native and mutant HD6. The bacteria (2 × 106 CFU/mL) were incubated with the indicated peptides for 30 min prior to infection of the T84 cells (mean ± SDM, n = 3).
Like native HD6, H27A provided ≥4-fold reduction in Listeria invasion over the concentration range evaluated. In contrast, oligomerization-deficient HD6 mutants F2A and F29A did not inhibit Listeria invasion over this concentration range. Moreover, the V22T, I25T and H27W mutants exhibited attenuated abilities to prevent Listeria invasion compared to HD6. On the basis of the trends depicted in Figure 5, these mutants may prevent Listeria invasion at higher peptide concentrations. Our invasion results for H27W are in good agreement with previous work where this mutant exhibited attenuated ability to prevent Salmonella and Yersinia invasion into mammalian cells.21 We conclude that the hydrophobic residues and hence self-assembly are important for HD6 to prevent invasive microbes from entering mammalian cells.
Summary and Conclusions
In this study, we demonstrate that hydrophobic residues in HD6 drive its self-assembly and afford innate immune function. Our results highlight the importance of primary sequence for defensin function and, in particular, how variable amino acid sequences between α-defensin family members afford different biophysical properties and biological activity. Of the four hydrophobic residues evaluated in this work, we discovered that F2 and F29 are particularly important for both HD6 self-assembly and biological function.
The results from our solution and TEM experiments, as well as prior X-ray crystallographic characterization,19 establish that HD6 forms higher-order structures in the absence of bacteria or other biomolecules. How the structures observed by TEM relate to the structure and composition of the HD6 nanonets entangling bacteria observed21 both in vitro and in vivo requires further exploration. Along these lines, SEM studies of wild-type and mutant S. Typhimurium treated with HD6 indicated that certain cell surface proteins contribute to formation of HD6 nanonets in vitro.21 This observation suggests that bacterial surface proteins provide a nucleation site. It will be informative to ascertain whether different bacterial species and different bacterial proteins affect the morphology and network of HD6 nanonets, as well as precisely how HD6 interacts with such proteins. Nanonet maturation as well as the physiological fate of the nanonet and the entrapped microbes are additional questions that must be addressed. From a functional standpoint, we reason that entrapment of bacterial pathogens by HD6 in the lumen not only prevents bacterial species that potentially reside and proliferate inside host cells from reaching this destination and causing infection, but also allows for other host-defense factors that operate in the intestinal lumen, such as other Paneth cell antimicrobials (e.g., HD5) and recruited neutrophils, to kill them in the extracellular space.
In closing, the hydrophobic effect plays a crucial role in biological processes that include cell membrane formation,36 protein folding and stabilization,37 and blood coagulation.38 It also contributes to pathologies associated with protein misfolding42 as exemplified by the Aβ-peptide (Alzheimer’s disease)39,40 and α-synuclein (Parkinson’s disease).41,42 HD6 provides a novel example of how Nature employs hydrophobicity for a beneficial outcome. Indeed, creating a biomolecular self-assembly from a 32-residue cysteine-rich defensin peptide to capture pathogens is a remarkable strategy for combating infection.
Experimental Section
General Materials and Methods
All solvents, reagents, and chemicals were purchased from commercial suppliers and used as received unless noted otherwise. HD5 was overexpressed and purified as previously described.29 All buffers, aqueous solutions, and peptide/oligonucleotide stock solutions were prepared in Milli-Q water (18.2 MΩ cm–1) after it was passed through a 0.22-μm filter. Oligonucleotide primers were synthesized by Integrated DNA technologies and used as received (standard desalting protocol). A BioRad MyCycler thermocycler was employed for all polymerase chain reactions (PCR). Chemically competent E. coli TOP10 and BL21(DE3) cells were prepared in-house via standard protocols. PfuTurbo DNA polymerase was purchased from Agilent Technologies. T4 DNA ligase and all restriction enzymes were purchased from New England Biolabs. DNA sequencing was performed by dedicated staff in the MIT Biopolymers Laboratory.
Subcloning, Overexpression, and Purification of His6-proHD6
An E. coli-optimized synthetic gene encoding preproHD6 was obtained from DNA 2.0 in the pJ201 vector (Supporting Information) The proHD6 nucleotide (333 bp) was PCR amplified using pJ201-preproHD6 as a template and the forward and reverse primers 5′-GAATTCCATATGGAGCCGC TGCAAGCAG-3′ and 5′-GATCCTCGAGTTACAGACAACAAAAGCGATG-3′, respectively (restriction site, underlined; stop codon, bold). The PCR reactions were analyzed by 1% (w/v) agarose gel and a GE Healthcare illustra GFX PC DNA and Gel Band Purification Kit was employed to purify the PCR products. The products were subsequently digested with NdeI and XhoI. The resulting fragments were purified by 1% (w/v) agarose gel and ligated into the NdeI and XhoI sites of pET28b using T4 DNA ligase (2 h, rt). The resulting plasmids were transformed into chemically competent E. coli TOP10 cells and the pET28b-proHD6 plasmid was isolated by using a QIAprep spin miniprep kit (Qiagen). The plasmid identity was confirmed by DNA sequencing.
The overexpression and purification of His6-proHD6 were modified from the literature.29 The pET28b-proHD6 plasmid was transformed into chemically competent E. coli BL21(DE3) cells. Overnight cultures were prepared by inoculating LB medium containing 50 μg/mL of kanamycin with single colonies. These cultures were grown to saturation (37 °C, 150 rpm, 16–18 h) and used to prepare glycerol freezer stocks. The freezer stocks containing a 1:1 ratio of the overnight culture and sterile-filtered 50% glycerol in Milli-Q water were stored at −80 °C. For a given His6-proHD6 overexpression, 50 mL of LB medium containing 50 μg/mL of kanamycin in a 250 mL baffled flask was inoculated with the freezer stock and grown to saturation (37 °C, 150 rpm, 16–18 h). The resulting culture was diluted 1:100 into 2 L of fresh LB medum containing 50 μg/mL of kanamycin in a 4-L baffled flask, and the culture was incubated at 37 °C, 150 rpm until OD600 reached ∼0.6. A 400-μL aliquot of 0.5-mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) was then added to the 2-L culture, and the culture was incubated for an additional 3–4 h (37 °C, 150 rpm) until an OD600 of 1.2–1.5 was achieved. The culture was centrifuged (3000 rpm for 15 min, 4 °C) and the cell pellets were collected. Typically, overexpression of His6-proHD6 was performed on a 12-L scale and the cell pellets from 6 L of culture were combined in preweighed 50-mL polypropylene tubes (∼2 g/L wet cell weight), flash frozen in liquid N2, and stored at −80 °C for a period of 1–2 months.
For purification of His6-proHD6, each 6-L cell pellet was thawed on ice and resuspended in 30 mL of cold lysis buffer (100 mM Tris-HCl, 6 M GuHCl, pH 8.0). Subsequently, a 1-mL aliquot of phenylmethyl sulfonyl fluoride (PMSF, 100 mM in EtOH) was added and the resuspension was transferred to a prechilled stainless steel beaker and lysed by two rounds of sonication (10% amplitude with pulse on for 1 s and pulse off for 4 s for a total of 1 min, on ice, Branson sonicator). A second 1-mL aliquot of PMSF (100 mM in EtOH) was added, and the cell lysate was clarified by centrifugation (13 000 rpm for 30 min, 4 °C). The resulting supernatant was incubated with prewashed Ni-NTA resin (Qiagen, from 9 mL of Ni-NTA slurry that was prewashed with Milli-Q water (3 × 30 mL)) with gentle shaking for 1.5 h at 4 °C. The resulting mixture was loaded onto a fritted column, and the resin was washed with 40 mL of cold wash buffer (20 mM Tris-HCl, 300 mM NaCl, 6 M GuHCl, pH 8.0). His6-proHD6 was eluted with 30 mL of cold elution buffer (10 mM Tris-HCl, 300 mM NaH2PO4, 200 mM NaCl, 1 M imidazole, 6 M GuHCl, pH 6.5). The eluent was transferred into a dialysis bag (3500 MWCO, Spectrum Laboratories) and dialyzed against 5% acetic acid (4 L for 12 h, 4 °C) and then 0.1% acetic acid (4 L for 12 h, 4 °C). The dialyzed solution was subsequently lyophilized to dryness to afford His6-proHD6 as a white fluffy powder, which was stored at −20 °C. The yield was 26 mg/L culture. The purity of His6-proHD6 was routinely determined by analytical HPLC (10–60% B over 30 min, 1 mL/min). The HPLC retention time and results from LC–MS are shown in Supporting Information Table S1.
Oxidative Folding
His6-proHD6 was folded by a modified literature procedure.29,43 A 120-mg portion of His6-proHD6 was dissolved in 15 mL of 8 M GuHCl containing 3 mM of glutathione and 0.3 mM of glutathione disulfide. Then, 45 mL of 250 mM NaHCO3 was added to the solution to adjust the pH to ∼8.3 and afford a final peptide concentration of ∼2 mg/mL. The solution was incubated at room temperature overnight. The resulting solution was analyzed by HPLC and mass spectrometry to ensure that His6-proHD6 was completely folded. The solution was transferred into a dialysis bag (3500 MWCO) and dialyzed against 10 mM Tris-HCl pH 8.2 (4 L for 12 h, 4 °C) and then 100 mM Tris-HCl pH 8.2 containing 20 mM CaCl2 (4 L for 12 h, 4 °C). The dialyzed solution was adjusted to a concentration of 1 mg/mL with 100 mM Tris-HCl pH 8.2 containing 20 mM CaCl2 and was then transferred to 50 mL polypropylene centrifuge tubes and subjected to trypsin-catalyzed cleavage without further purification.
Preparation and Purification of Native HD6
An aliquot of a 1-mg/mL stock solution of TPCK-treated trypsin (Worthington) in Milli-Q water was added to the solution of His6-proHD6 (1 mg/mL), which was obtained from oxidative folding as described above, to afford a final trypsin concentration of 0.01 mg/mL. The reaction was incubated at room temperature for 15 min and subsequently quenched by addition of 6% TFA/H2O (10% v/v). The resulting solution was immediately vortexed, flash frozen in liquid N2, and lyophilized to dryness. The resulting powder was resuspended in 25 mL of 6 M GuHCl for 15 min and passed through a 0.22-μm filter. HD6 was purified by preparative HPLC using a solvent gradient of 25–33% B over 18 min. HD6 eluted at 13.4 min and the corresponding fractions were lyophilized to dryness to provide a white fluffy powder. The yield was 1.9 mg/L culture. The HPLC retention time, and results from LC–MS are listed in Supporting Information Table S2.
Negative-Staining Transmission Electron Microscopy
For each sample, a 5-μL aliquot of peptide solution (20 μM in 10 mM sodium phosphate pH 7.4 or 10 mM Tris-maleate pH 6.4) was placed onto the carbon-coated surface of a copper grid (400 square mesh, Electron Microscopy Sciences). After 1 min, the grid was stained with a 5-μL aliquot of 2% uranyl acetate (UA, Electron Microscopy Sciences) in Milli-Q water three times and air-dried for at least 15 min before imaging. A FEI Technai Spirit Transmission Electron Microscope was employed to collect all transmission electron micrographs (W.M. Keck Microscopy Facility, Whitehead Institute, Cambridge, MA). TEM images were obtained with at least two independent batches of each peptide and representative images are presented.
Sedimentation Velocity Experiments
Sedimentation velocity (SV) experiments were performed to determine the sedimentation coefficients of the F2A and F29A mutants. A Beckman XL-I Analytical Ultracentrifuge equipped with an An-50 Ti rotor was used for all SV experiments. The rotor housed double-sector charcoal-filled Epon centerpieces within the sample cells and contained quartz windows. All SV sample cells contained either 410 μL of buffer reference or 400 μL of peptide solution. In one set of experiments, peptide stock solutions in Milli-Q water were lyophilized to dryness, dissolved in 400 μL of 10 mM sodium phosphate pH 7.4 or 10 mM Tris-maleate pH 6.4 to achieve the desired concentrations (40, 50, 100, and 160 μM), and transferred to SV sample cells. The pH of each solution was measured to confirm that it remained unchanged. The samples were centrifuged at 42 000 rpm and 20 °C until sedimentation was complete. The absorption wavelength used for optical detection was 280 nm. All SV experiments were conducted with at least two independently prepared and purified samples of each peptide and in two independent trials.
The details of data analysis are reported elsewhere.20 The buffer viscosity (η), buffer density (ρ), and the partial specific volume (v̅) values of F2A and F29A at 20 °C were calculated by employing SEDNTERP.44 The reported HD6 crystal structure (PDB: 1ZMQ)19 was used in HYDROPRO33 hydrodynamic modeling to calculate sedimentation coefficients of the HD6 monomer, dimer, and tetramer (Supporting Information Table S6). It was assumed that the F2A and F29A mutations would have negligible impact on the calculated sedimentation coefficients. The buffer viscosity (η) and buffer density (ρ) values for water at 20 °C, and a partial specific volume (v̅) value of 0.6994 mL/g for HD6 were used in all HYDROPRO calculations. The experimental sedimentation coefficients of the HD6 mutants were calculated by fitting the time derivative of the sedimentation velocity (−dc/dt) data by using DCDT+.45,46 The −dc/dt distribution was generated from 26 to 34 scans with a peak-broadening limit of 50 kDa by using DCDT+. The results are reported in Supporting Information Tables S4 and S5.
Antimicrobial Activity Assays
Bacteria from freezer stocks were grown to saturation with shaking (37 °C, 16 h) in 5 mL of the indicated medium (L. monocytogenes ATCC 19115 in Brain Heart Infusion medium (BHI); S. aureus ATCC 25923, E. coli ATCC 25922, or S. Typhimurium ATCC 14028 in Tryptic Soy Broth medium (TSB) without dextrose). The overnight culture was diluted 1:100 into 5 mL of fresh BHI or TSB and incubated at 37 °C until OD600 of ∼0.6 was achieved. The resulting culture was centrifuged (3500 rpm for 10 min, 4 °C) and the supernatant was removed. The bacterial pellet was resuspended in 5 mL of AMA buffer (10 mM sodium phosphate, 1% TSB, pH 7.4). The cell suspension was centrifuged (3500 rpm for 10 min, 4 °C) and the supernatant was discarded. The resulting cell pellet was resuspended in 5 mL of AMA buffer and diluted with AMA buffer to obtain an OD600 value of 0.5 for L. monocytogenes, E. coli, and S. Typhimurium or 0.6 for S. aureus. For L. monocytogenes ATCC 19115, the bacterial suspension was further diluted 1:500 with AMA buffer in three steps (1:10 × 1:10 × 1:5). For S. aureus ATCC 25923, the bacterial suspension was further diluted 1:100 with AMA buffer in two steps (1:10 × 1:10). For E. coli ATCC 25922, the bacterial suspension was further diluted 1:250 with AMA buffer in three steps (1:10 × 1:10 × 1:2.5). For S. Typhimurium ATCC 14028, the bacterial suspension was further diluted 1:250 with AMA buffer in three steps (1:10 × 1:10 × 1:2.5). The diluted cultures were used immediately.
The assays were conducted in 96-well plates. To each well was added 10 μL of a 10× concentrated aqueous peptide solution (200 μM) or sterile Milli-Q water as a no-peptide control. A 90-μL aliquot of the diluted bacterial culture was added to each well and the plate was incubated for 1 h (37 °C, 150 rpm). A 50-μL aliquot from each well was subsequently added to 450 μL of AMA buffer (10–1 dilution). The resulting solution was vortexed gently and further diluted serially from 10–2 to 10–4 in 10-fold increments by adding a 100-μL aliquot from each dilution to 900 μL of AMA buffer. A 100-μL aliquot from each dilution was manually plated on BHI agar plates for L. monocytogenes or TSB agar plates for S. aureus, E. coli, and S. Typhimurium. The plates were then incubated at 37 °C for 15 h for E. coli and S. Typhimurium or 20 h for L. monocytogenes and S. aureus. The number of colony forming units obtained for each peptide sample was determined by colony counting. Only plates with 30–200 colonies were considered in these assays. These assays were performed with at least two independently prepared and purified samples of each peptide and in three independent trials. The resulting averages and standard deviations are reported.
Bacterial Agglutination Assays
A 5 mL portion of BHI medium for L. monocytogenes ATCC 19115 or Luria Broth (LB) for E. coli ATCC 25922 was inoculated with the bacteria from freezer stocks and grown to saturation with shaking (37 °C, 16 h). The overnight culture was diluted 1:100 into 10 mL of fresh BHI or LB medium and incubated at 37 °C until OD600 reached ∼0.6. The resulting culture was centrifuged (3500 rpm for 10 min, 4 °C) and the supernatant was removed. The bacterial pellet was resuspended in 10 mL of 50% Mueller-Hinton Broth pH 7.4 (MHB, Fluka). The cell suspension was centrifuged (3500 rpm for 10 min, 4 °C) and the supernatant was discarded. The resulting bacterial pellet was resuspended in 10 mL of 50% MHB and further diluted with 50% MHB to obtain an OD600 value of 0.25. The diluted bacterial cultures were immediately employed for the agglutination assays.
For all agglutination assays, a 450-μL aliquot of the bacterial culture (OD600 ∼ 0.25) was added to 50 μL of a 10× concentrated aqueous peptide solution (50, 100, or 200 μM) or sterile Milli-Q water as a no-peptide control and immediately transferred to an EtOH-sterilized two-sided disposable polystyrene cuvette (VWR International). The OD600 values were measured at the indicated time points and plotted versus time to afford a sedimentation curve. After 6 h, a 100-μL aliquot from each cuvette containing 20 μM of each peptide was diluted with 900 μL of 11 mM sodium phosphate pH 7.4, vortexed gently, and serially diluted from 10–2 to 10–7 in 10-fold increments by adding a 100-μL aliquot from each dilution to 900 μL of the buffer. If there was bacterial sedimentation in the cuvette, a 100-μL aliquot was taken from the supernatant for serial dilutions. Then, 100 μL of fresh 50% MHB was added to the cuvette and the bacterial aggregate was resuspended. Another 100-μL aliquot was subsequently taken from the resulting suspension for serial dilutions. To determine the number of colony forming units for each peptide treatment, a 100-μL aliquot from each dilution was manually plated on BHI agar plates for L. monocytogenes or LB agar plates for E. coli and incubated at 37 °C for 20 or 15 h, respectively. Only plates with 30–200 colonies were considered in these assays. All assays were performed with at least two independently prepared and purified samples of each peptide and in three independent trials. The resulting averages with standard deviations are reported.
General Cell Culture Methods
Human colon epithelial T84 cells (ATCC CCL-248) were obtained from the American Type Culture Collection (ATCC) and grown in 1:1 Dulbecco’s modified Eagle medium (DMEM) and Ham’s F12 medium containing 2.5 mM l-glutamine, 15 mM HEPES, 0.5 mM sodium pyruvate, and 1.2 g/L sodium bicarbonate, and supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Cells were cultured at 37 °C and 5% CO2 according to the instructions from ATCC. The growth medium and all supplements were purchased from ATCC.
Listeria Invasion Assays
These assays were conducted by modifying reported literature protocols.21,47,48L. monocytogenes ATCC 19115 from freezer stocks were grown overnight with shaking (37 °C, 16 h) in 5 mL of BHI medium. The overnight culture was diluted 1:100 into 5 mL of fresh BHI medium and incubated at 37 °C until OD600 of ∼0.6 was achieved. The resulting culture was centrifuged (3500 rpm for 10 min, 4 °C) and the supernatant was removed. The bacterial pellet was resuspended in 5 mL of serum- and antibiotic-free 1:1 DMEM/F12. The resulting suspension was centrifuged (3500 rpm for 10 min, 4 °C) and the supernatant was removed. The bacterial pellet was resuspended in 5 mL of serum- and antibiotic-free 1:1 DMEM/F12 and diluted with the medium to afford an OD600 value of 0.5. The resulting suspension was further diluted 1:150 into 3 mL of the fresh medium. A 190-μL aliquot of the diluted bacterial culture was immediately added to 10 μL of a 20× concentrated aqueous peptide solution (50, 100, or 200 μM) or sterile Milli-Q water as a no-peptide control and incubated at room temperature for 30 min.
T84 cells between passages 59 and 73 were cultured in 75 cm2 rectangular canted neck cell culture flasks (Corning) to approximately 95% confluency and treated with 3 mL of trypsin–EDTA (Corning). A 12-mL portion of fresh media was added to the detached cells, and the T84 cell suspension was centrifuged (600 rpm for 5 min, 37 °C). The supernatant was discarded and the cell pellet was resuspended in 6 mL of the fresh culture medium. The concentration of cells was quantified by using a manual hemocytometer (VWR International) and adjusted to 2 × 105 cells/mL. A 500-μL aliquot of T84 cells was then added to 24-well Costar tissue culture plates (Corning) and incubated at 37 °C and 5% CO2 for 24 h. With this cell density, the monolayers were ∼80% confluent at the time of the experiment. With cells at this confluency, the medium was discarded and the cells were washed twice with 500 μL of phosphate buffered saline (PBS) without calcium or magnesium (ATCC) and bathed in 500 μL of serum- and antibiotic-free 1:1 DMEM/F12. After a 2-h equilibration in serum- and antibiotic-free medium, the medium was discarded. The T84 cells were washed twice with 500 μL of PBS and infected with 200 μL of peptide-treated L. monocytogenes (multiplicity of infection, MOI = 10) at 37 °C and 5% CO2. MOI is a ratio of the number of bacterial cells to mammalian cells when infection is initiated. A 100-μL aliquot of the diluted bacterial culture was used to determine the number of inoculum by colony counting as described below. After 1.5 h of infection, the medium was removed and the T84 cells were washed twice with 500 μL of PBS. To kill any remaining extracellular bacteria, the T84 cells were subsequently incubated in 200 μL of serum-free 1:1 DMEM/F12 containing 100 μg/mL of gentamycin for 1.5 h at 37 °C and 5% CO2. After 1.5 h, the medium was removed and the T84 cells were washed twice with 500 μL of PBS. The T84 cells were then incubated in 200 μL of sterile-filtered 1% Triton X-100 (EMD) in PBS for 10 min at rt.
A 50-μL aliquot from each well was diluted with 450 μL of 11 mM sodium phosphate pH 7.4, vortexed gently, and serially diluted from 10–2 to 10–4 in 10-fold increments by adding a 100-μL aliquot from each dilution to 900 μL of the fresh buffer. To determine the number of invading bacteria for each peptide treatment, a 100-μL aliquot from each dilution was manually plated on BHI agar plates and incubated at 37 °C for 20 h. Only plates with 30–200 colonies were considered in these assays. The percentage of invasion represents the ratio of the number of invading bacteria to the number of inoculated bacteria. All assays were performed with at least two independently prepared and purified samples of each peptide and in three independent trials. The resulting averages and standard deviations are reported.
Acknowledgments
We gratefully acknowledge the NIH (Grant DP2OD007045 from the Office of the Director) and the Royal Thai Government (PC) for financial support. The Biophysical Instrumentation Facility for the Study of Complex Macromolecular Systems at MIT is supported by Grant NSF-007031. We thank Ms. D. Pheasant and Dr. A. J. Wommack for assistance with the AUC experiments and data analysis, and Dr. E. J. Brignole for helpful advice on TEM sample preparation and data collection.
Supporting Information Available
Additional experimental section that includes instrumentation, site-directed mutagenesis, purification of mutant peptides, CD spectroscopy, thiol quantification assays, disulfide bond connectivity determination by manual Edman degradation, design of the synthetic gene for preproHD6. Tables of amino acid sequences, peptide characterization, MS results from manual Edman degradation, calculated sedimentation coefficients for F2A and F29A, HYDROPRO analysis, primers and primer pairings employed in site-directed mutagenesis. Scheme for disulfide bond connectivity determination for F2A. Figures of α-defensin amino acid sequence alignment, HD5 and HD6 structural comparison, HD6 crystal structure, analytical HPLC traces of His6-proHD6 and HD6, analytical HPLC traces of peptides, additional TEM, CD spectra, agglutination assays and image of agglutinated bacteria. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Zhao L.; Lu W. Curr. Opin. Hematol. 2014, 21, 37–42. [DOI] [PubMed] [Google Scholar]
- Hazlett L.; Wu M. Cell Tissue Res. 2011, 343, 175–188. [DOI] [PubMed] [Google Scholar]
- Lehrer R. I.; Lu W. Immunol. Rev. 2012, 245, 84–112. [DOI] [PubMed] [Google Scholar]
- Doss M.; White M. R.; Tecle T.; Hartshorn K. L. J. Leukocyte Biol. 2010, 87, 79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette A. J. Cell. Mol. Life Sci. 2011, 68, 2215–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selsted M. E.; Ouellette A. J. Nat. Immunol. 2005, 6, 551–557. [DOI] [PubMed] [Google Scholar]
- Gabay J. E.; Scott R. W.; Campanelli D.; Griffith J.; Wilde C.; Marra M. N.; Seeger M.; Nathan C. F. Proc. Natl. Acad. Sci. U.S.A. 1989, 86, 5610–5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde C.; Gabay J. E.; Marra M. N.; Snable J. L.; Scott R. W. J. Biol. Chem. 1989, 264, 11200–11203. [PubMed] [Google Scholar]
- Mackewicz C. E.; Yuan J.; Tran P.; Diaz L.; Mack E.; Selsted M. E.; Levy J. A. AIDS 2003, 17, F23–F32. [DOI] [PubMed] [Google Scholar]
- Chalifour A.; Jeannin P.; Gauchat J.-F.; Blaecke A.; Malissard M.; N’Guyen T.; Thieblemont N.; Delneste Y. Blood 2004, 104, 1778–1783. [DOI] [PubMed] [Google Scholar]
- Jones D. E.; Bevins C. L. J. Biol. Chem. 1992, 267, 23216–23225. [PubMed] [Google Scholar]
- Jones D. E.; Bevins C. L. FEBS Lett. 1993, 315, 187–192. [DOI] [PubMed] [Google Scholar]
- Mallow E. B.; Harris A.; Salzman N.; Russell J. P.; DeBerardinis R. J.; Ruchelli E.; Bevins C. L. J. Biol. Chem. 1996, 271, 4038–4045. [DOI] [PubMed] [Google Scholar]
- Porter E. M.; Liu L.; Oren A.; Anton P. A.; Ganz T. Infect. Immun. 1997, 65, 2389–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter E. M.; Bevins C. L.; Ghosh D.; Ganz T. Cell. Mol. Life Sci. 2002, 59, 156–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. C.; Bevins C. L. Annu. Rev. Physiol. 2013, 75, 289–311. [DOI] [PubMed] [Google Scholar]
- Wei G.; de Leeuw E.; Pazgier M.; Yuan W.; Zou G.; Wang J.; Ericksen B.; Lu W.-Y.; Lehrer R. I.; Lu W. J. Biol. Chem. 2009, 284, 29180–29192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C. P.; Yee J.; Selsted M. E.; Eisenberg D. Science 1991, 251, 1481–1485. [DOI] [PubMed] [Google Scholar]
- Szyk A.; Wu Z.; Tucker K.; Yang D.; Lu W.; Lubkowski J. Crystal Structures of Human α-Defensins HNP4, HD5, and HD6. Protein Sci. 2006, 15, 2749–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wommack A. J.; Robson S. A.; Wanniarachchi Y. A.; Wan A.; Turner C. J.; Wagner G.; Nolan E. M. Biochemistry 2012, 51, 9624–9637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H.; Pazgier M.; Jung G.; Nuccio S.-P.; Castillo P. A.; de Jong M. F.; Winter M. G.; Winter S. E.; Wehkamp J.; Shen B.; Salzman N. H.; Underwood M. A.; Tsolis R. M.; Young G. M.; Lu W.; Lehrer R. I.; Bäumler A. J.; Bevins C. L. Science 2012, 337, 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericksen B.; Wu Z.; Lu W.; Lehrer R. I. Antimicrob. Agents Chemother. 2005, 49, 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter E. M.; van Dam E.; Valore E. V.; Ganz T. Infect. Immun. 1997, 65, 2396–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman N. H.; Ghosh D.; Huttner K. M.; Paterson Y.; Bevins C. L. Nature 2003, 442, 522–526. [DOI] [PubMed] [Google Scholar]
- Rajabi M.; Ericksen B.; Wu X.; de Leeuw E.; Zhao L.; Pazgier M.; Lu W. J. Biol. Chem. 2012, 287, 21615–21627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazgier M.; Wei G.; Ericksen B.; Jung G.; Wu Z.; de Leeuw E.; Yuan W.; Szmacinski H.; Lu W.-Y.; Lubkowski J.; Lehrer R. I.; Lu W. J. Biol. Chem. 2012, 287, 8944–8953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gounder A. P.; Weins M. E.; Wilson S. S.; Lu W.; Smith J. G. J. Biol. Chem. 2012, 287, 24554–24562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai K. P.; Le V. V.; Selsted M. E.; Ouellette A. J. Infect. Immun. 2014, 82, 2195–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanniarachchi Y. A.; Kaczmarek P.; Wan A.; Nolan E. M. Biochemistry 2011, 50, 8005–8017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.; Ganz T. Blood 1995, 85, 1095–1103. [PubMed] [Google Scholar]
- Valore E. V.; Ganz T. Blood 1992, 79, 1538–1544. [PubMed] [Google Scholar]
- Ghosh D.; Porter E.; Shen B.; Lee S. K.; Wilk D.; Drazba J.; Yadav S. P.; Crabb J. W.; Ganz T.; Bevins C. L. Nature 2002, 3, 583–590. [DOI] [PubMed] [Google Scholar]
- Ortega A.; Amorós D.; García de la Torre J. G. Biophys. J. 2011, 101, 892–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart P.; Pizarro-Cerdá J.; Lecuit M. Trends Cell Biol. 2003, 13, 23–31. [DOI] [PubMed] [Google Scholar]
- Vivant A.-L.; Garmyn D.; Piveteau P. Front. Cell. Infect. Microbiol. 2013, 3, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanford C. Science 1978, 200, 1012–1018. [DOI] [PubMed] [Google Scholar]
- Doig A. J.; Williams D. H. J. Mol. Biol. 1991, 217, 389–398. [DOI] [PubMed] [Google Scholar]
- Krishnan A.; Cha P.; Liu Y.-H.; Allara D.; Vogler E. A. Biomaterials 2006, 27, 3187–3194. [DOI] [PubMed] [Google Scholar]
- Wurth C.; Guimard N. K.; Hecht M. H. J. Mol. Biol. 2002, 319, 1279–1290. [DOI] [PubMed] [Google Scholar]
- Kim W.; Hecht M. H. J. Mol. Biol. 2008, 377, 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giasson B. I.; Murray I. V. J.; Trojanowski J. Q.; Lee V. M.-Y. J. Biol. Chem. 2001, 276, 2380–2386. [DOI] [PubMed] [Google Scholar]
- Chiti F.; Dobson C. M. Annu. Rev. Biochem. 2006, 75, 333–366. [DOI] [PubMed] [Google Scholar]
- Wu Z.; Ericksen B.; Tucker K.; Lubkowski J.; Lu W. J. Peptides Res. 2004, 64, 118–125. [DOI] [PubMed] [Google Scholar]
- Laue M.; Shah B. D.; Ridgeway T. M.; Pelletier S. L.. Analytical Ultracentrifugation in Biochemistry and Polymer Science; Harding S., Rowe A., Eds.; Royal Society of Chemistry: Cambridge, U.K., 1992; pp 90–125. [Google Scholar]
- Schuck P. Anal. Biochem. 2003, 320, 104–124. [DOI] [PubMed] [Google Scholar]
- Philo J. S. Anal. Biochem. 2000, 279, 151–163. [DOI] [PubMed] [Google Scholar]
- Jaradat Z. W.; Bhunia A. K. Appl. Environ. Microbiol. 2003, 69, 3640–3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang P.; Foubister V.; Pucciarelli M. G.; Finlay B. B. J. Microbiol. Methods 1993, 18, 227–240. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.