SUMMARY
Clec16a has been identified as a disease susceptibility gene for type 1 diabetes, multiple sclerosis and adrenal dysfunction, but its function is unknown. Here we report that Clec16a is a membrane-associated endosomal protein that interacts with E3 ubiquitin ligase Nrdp1. Loss of Clec16a leads to an increase in the Nrdp1 target Parkin, a master regulator of mitophagy. Islets from mice with pancreas-specific deletion of Clec16a have abnormal mitochondria with reduced oxygen consumption and ATP concentration, both of which are required for normal β-cell function. Indeed, pancreatic Clec16a is required for normal glucose-stimulated insulin release. Moreover, patients harboring a diabetogenic SNP in the Clec16a gene have reduced islet Clec16a expression and reduced insulin secretion. Thus, Clec16a controls β-cell function and prevents diabetes by controlling mitophagy. This novel pathway could be targeted for prevention and control of diabetes and may extend to the pathogenesis of other Clec16a and Parkin associated diseases.
INTRODUCTION
Genome wide association studies (GWAS) are a powerful approach to the identification of genes involved in common human diseases, yet are limited by the identification of variants in the loci of genes with completely unknown functions. Further, many single nucleotide polymorphisms (SNPs) identified in GWAS are found in intergenic regions that affect the function of transcriptional enhancers located far from the disease-relevant gene. Thus, it is critical to directly examine the functional role of potential disease genes and to correlate gene variation in potential enhancers to expression of the putative associated gene. Molecular understanding of new disease loci may provide important insights into the pathogenesis of human diseases and reveal new therapeutic targets (Pociot et al., 2010).
C-type lectin domain family 16, member A (Clec16a; KIAA0350), a gene locus associated with type 1 diabetes mellitus (T1DM), multiple sclerosis, and adrenal dysfunction (Hakonarson et al., 2007; IMSGC, 2009; Skinningsrud et al., 2008; WTCCC, 2007) is a 24-exon gene that encodes a large protein (1053 amino acids) with evolutionary conservation of the N-terminus but no recognizable conserved motifs. Little is known of mammalian Clec16a function or of its role in disease pathogenesis.
Here we discover a key role for Clec16a in the regulation of mitophagy, a selective form of autophagy necessary for mitochondrial quality control (Ashrafi and Schwarz, 2013). Utilizing proteomics analyses, we determine that the E3 ubiquitin ligase Neuregulin receptor degradation protein 1 (Nrdp1 or RNF41) interacts with Clec16a and mediates Clec16a functions, through the Nrdp1 target Parkin, in multiple cell types. We find a key role for Clec16a in the maintenance of glucose homeostasis through its effect on the mitochondrial health of pancreatic β-cells and, consequently, glucose-stimulated insulin secretion. Lastly, we demonstrate that a diabetogenic SNP in the CLEC16A locus correlates with islet CLEC16A expression, β-cell function, and glycemic control in human subjects.
RESULTS
Identification of E3 ubiquitin ligase Nrdp1 as a specific partner of Clec16a
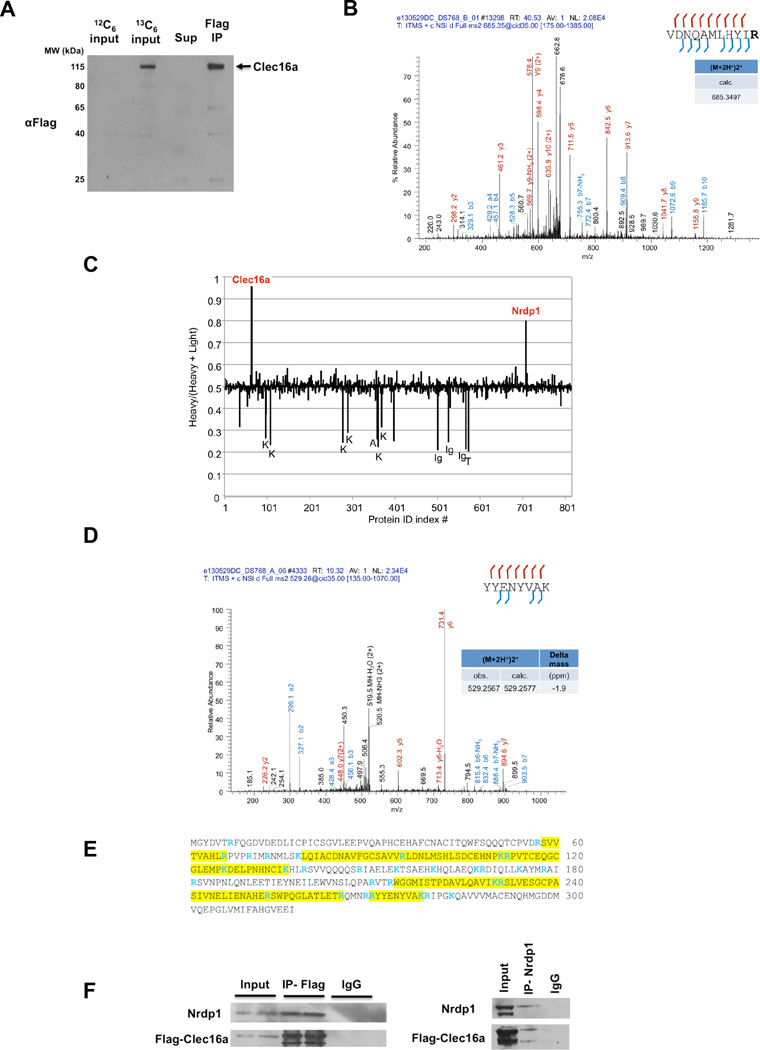
We hypothesized that Clec16a plays an important role in multiple tissues and that the identification of novel Clec16a interacting partners might shed light on its function. To this end, we utilized the I-DIRT (Isotopic Differentiation of Interactions as Random or Targeted) technique (Tackett et al., 2005) to identify specific Clec16a partners in NIH3T3 fibroblasts stably expressing an epitope tagged form of Clec16a (Flag-Clec16a). NIH3T3 fibroblasts containing Flag-tagged Clec16a were grown in heavy isotopic (13C6,15N2-lysine and 13C6,15N4-arginine), while wild-type cells were grown in light (12C, 14N), natural isotope abundance medium. Equal quantities of the two cell preparations were mixed, and the epitope-tagged protein complex was isolated (Figure 1A). With I-DIRT, specific protein interactions are identified by mass spectrometry (MS) as isotopically heavy, while nonspecific interactions appear as a mixture of isotopically light and heavy. Following immunoisolation, we verified enrichment of Clec16a peptides by tandem MS (Figure 1B). Of over 800 proteins that were isolated and sequenced by MS, the I-DIRT approach identified the E3 ubiquitin ligase Nrdp1 as the major specific partner of Clec16a (Figure 1C). Tandem MS data confirmed enrichment of Nrdp1 peptides (Figure 1D). Broad coverage of the Nrdp1 protein sequence was observed with MS identification of 9 unique Nrdp1 peptides from 6 independent I-DIRT experiments (Figure 1E). We then validated the interaction of Clec16a and Nrdp1 by co-immunoprecipitation with either Flag or Nrdp1 specific antisera, and confirmed pulldown of the Clec16a-Nrdp1 protein complex (Figure 1F).
Figure 1. I-DIRT identifies Clec16a interaction with E3 ubiquitin ligase Nrdp1.
(A) Representative Western blot (WB) of parental NIH3T3 fibroblast (12C6-labeled) or stably expressing Flag-Clec16a NIH3T3 fibroblast (13C6-labeled) input protein lysates, supernatant and Flag immunoprecipitation for Flag-Clec16a (n=6). (B) MS/MS map marked with b ions (blue) and y ions (red) for Clec16a specific peptide sequence with calculated m/z ratios. Heavy residue indicated in bold print in peptide sequence. (C) I-DIRT ratios for >800 proteins identified from 6 independent I-DIRT experiments with calculated non-specific value of 0.5. Clec16a and Nrdp1 peaks indicated. Contaminant peptides (human keratin (K), immunoglobulins (Ig), trypsin (T), and albumin (A)) indicated by peaks below non-specific interactors. (D) MS/MS map marked with b ions (blue) and y ions (red) for an Nrdp1 specific peptide sequence with observed and calculated m/z ratios. (E) Amino acid sequence of Nrdp1 with peptides identified by MS (highlighted residues). Trypsin cleavage sites demarcated in blue. (F) Representative WB (3 independent experiments) of lysates of Flag-Clec16a NIH3T3 fibroblasts following anti-Flag IP (left) or anti-Nrdp1 IP (right).
Clec16a regulation of the Nrdp1/Parkin pathway
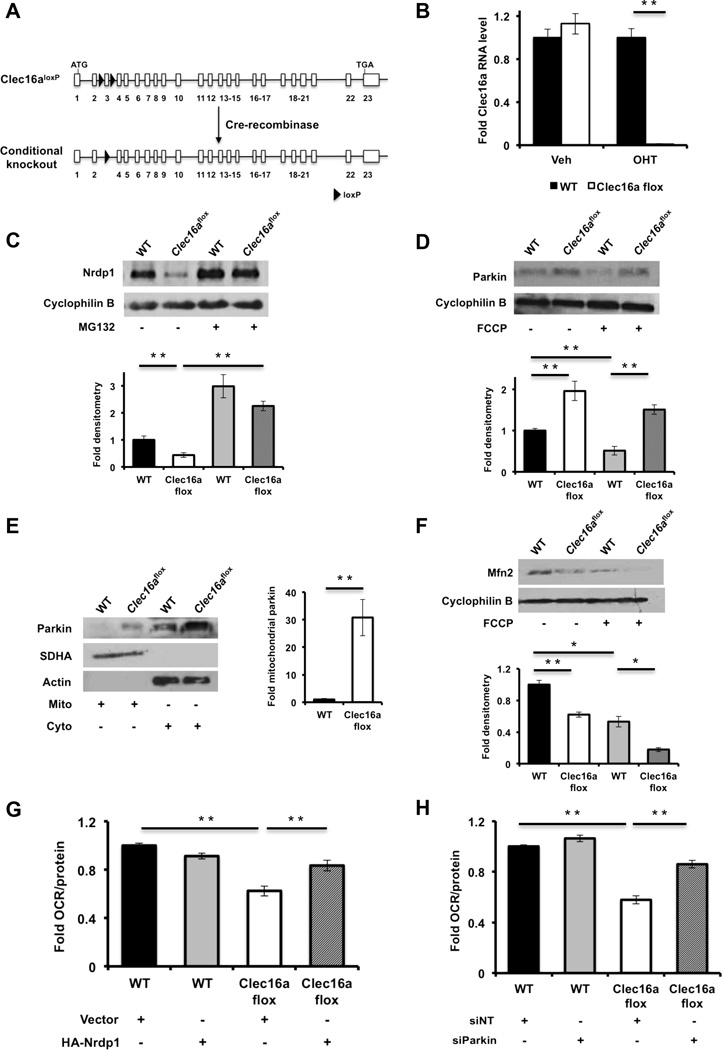
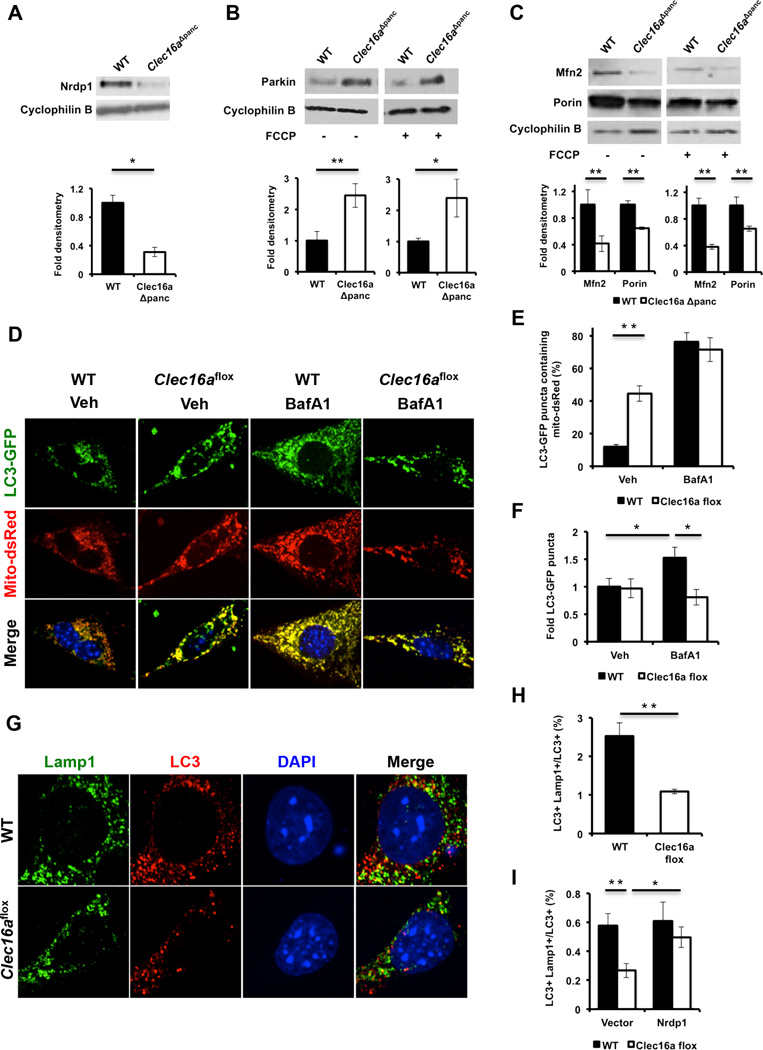
Our observation of Clec16a and Nrdp1 interaction led us to assess the role of Clec16a in the regulation of pathways downstream of Nrdp1 using a loss of function approach. We generated mice with loxP sites flanking exon 3 of the Clec16a gene (Clec16aloxP) and intercrossed them with UBC-Cre/ERT2 mice (Figure 2A). Primary dermal fibroblasts were isolated from Clec16aloxP/loxP;UBC-Cre/ERT2 mice and wild-type littermates. After treatment with hydroxytamoxifen (OHT), efficient recombination at the Clec16a locus in OHT-treated Clec16aloxP/loxP;UBC-Cre/ERT2 (Clec16aflox) fibroblasts was confirmed (Figure 2B). As an E3 ligase, Nrdp1 will self-ubiquitinate to regulate its own stability in part through interaction with deubiquitination enzymes (Wu et al., 2004). We observed a reduction in Nrdp1 in Clec16aflox fibroblasts compared to controls, and treatment with the proteasome inhibitor MG132 normalized expression between the groups (Figure 2C), suggesting that Clec16a protects Nrdp1 from proteosomal degradation.
Figure 2. Clec16a controls mitochondrial function through the Nrdp1/Parkin pathway.
(A) Model of Cre-mediated recombination of the Clec16a locus. (B) Clec16a expression by qRTPCR of RNA isolated from vehicle (EtOH) or OHT treated WT or Clec16aflox fibroblasts (n=3/group). (C) Nrdp1 protein expression by WB of WT or Clec16aflox fibroblasts following overnight treatment with vehicle (DMSO) or 10 µM MG132 (n=3/group). (D) Parkin expression by WB in WT or Clec16aflox fibroblasts following 3-hr treatment with vehicle (DMSO) or 10 µM FCCP (n=3/group). (E) Parkin subcellular localization by WB in WT or Clec16aflox fibroblasts following cell fractionation and enrichment of mitochondrial or cytosolic proteins. SDHA and actin serve as mitochondrial and cytosolic loading controls, respectively (n=3/group). Fold mitochondrial parkin as estimated by densitometry. (F) Mitofusin 2 expression by WB in WT or Clec16aflox fibroblasts following 3-hr treatment with vehicle (DMSO) or 10 µM FCCP (n=3/group). (G) Fold oxygen consumption rate (OCR) in WT or Clec16aflox fibroblasts, 96 hours after transient overexpression of empty vector (pcDNA3-3xHA) or 3xHA-Nrdp1 (n=3/group). (H) Fold OCR in WT or Clec16aflox fibroblasts, 96 hours after transient transfection of non-targeting (siNT) or Parkin (siParkin) siRNA (n=3/group). For all WBs, representative images chosen from among 3 independent experiments. Densitometry, shown adjacent to each WB, represents mean (±SEM) of all 3 independent experiments. qRT-PCR and OCR data expressed as mean (±SEM) of 3 experiments performed in triplicate. *p<0.05 **p<0.01. See also Figure S1.
Nrdp1 regulates proteosomal degradation of the E3 ligase Parkin (Zhong et al., 2005), a key regulator of mitochondrial autophagy or mitophagy (Jin and Youle, 2012). The initiation of mitophagy is mediated by Parkin translocation to the mitochondrial outer membrane, allowing recognition of its outer membrane substrates. At the mitochondrial surface, Parkin ubiquitinates Mitofusins 1 and 2 and Porin (Gegg et al., 2010; Geisler et al., 2010), thereby promoting their degradation. Finally, rapid turnover of Parkin-bound mitochondria occurs through the autophagosomal machinery (Jin and Youle, 2012). Following loss of Clec16a, we observed a significant increase in Parkin expression (Figure 2D). As expected, Parkin levels dropped following FCCP-stimulation of mitophagy in WT fibroblasts, while the increased Parkin levels in Clec16aflox fibroblasts were not reduced following FCCP treatment (Figure 2D). To determine if the increased basal expression of Parkin in Clec16aflox fibroblasts augments early events of mitophagy, we evaluated Parkin localization following cellular fractionation. As expected, Parkin did not significantly localize to mitochondria in WT fibroblasts; however, in Clec16aflox fibroblasts, Parkin was found in the mitochondrial fraction (Figure 2E). Importantly, mitofusin 2 (Mfn2) expression was reduced in Clec16aflox compared to WT fibroblasts (Figure 2F), consistent with increased Parkin activity in mitochondria.
Alterations in expression of Mfn2, which has been previously implicated in control of mitochondrial metabolism (Liesa and Shirihai, 2013; Zorzano et al., 2010), led us to assess mitochondrial respiration following Clec16a loss of function. We measured oxygen consumption in NIH3T3 fibroblasts stably expressing shRNA directed against Clec16a as well as Clec16aflox fibroblasts and their respective controls. Following confirmation of reduced Clec16a expression by two independent shRNA sequences (Figure S1A), we found that loss of Clec16a led to a reduced oxygen consumption rate (OCR) in NIH3T3 fibroblasts (Figure S1B). Similarly, we observed reduced OCR in Clec16aflox fibroblasts (Figure 2G-H). Defects in OCR were not due to effects on glycolysis, as glycolytic function reflected by the extracellular acidification rate (ECAR) was unchanged between the groups (Figure S1C). Transient overexpression of Nrdp1 partially rescued OCR in Clec16aflox fibroblasts as well as shClec16a-treated NIH3T3 cells (Figures S1D-E and 2G). A similar partial rescue of OCR was also observed following transient siRNA-mediated loss of function of Parkin in Clec16aflox fibroblasts (Figure 2H). In primary fibroblasts, both Nrdp1 overexpression and Parkin loss of function returned Mfn2 expression to similar levels between the groups (Figures S1E-F). Taken together, we find that Clec16a regulation of the Nrdp1/Parkin pathway restrains mitophagy and maintains normal mitochondrial oxygen consumption.
Pancreas-specific Clec16a deficiency impairs glucose tolerance by reducing β-cell function
While T1DM is an autoimmune disease, many T1DM-associated genes are expressed in pancreatic β-cells, suggesting functions directly in β-cells that may contribute to disease pathogenesis (Eizirik et al., 2009; Soleimanpour and Stoffers, 2013). We previously found that Clec16a is expressed in pancreatic islets and that its expression is reduced in Pdx1+/− islets (Sachdeva et al., 2009). Moreover, while previous reports suggest Clec16a is an NK cell enriched factor (Hakonarson et al., 2007), we observed that Clec16a is ubiquitously expressed and at similar levels in pancreatic islets and NK cells (Figure S2).
These observations led us to interrogate the role of Clec16a in pancreatic β-cell function in vivo. We generated pancreas-specific deletion of Clec16a (Clec16aloxP/loxP; Pdx1-Cre, hereafter referred to as Clec16aΔpanc). Clec16aloxP mice have normal glucose tolerance and body weight prior to Cre-mediated recombination (Figures S3A-B). Pdx1-Cre mice display efficient Cre-mediated recombination within pancreatic islets and mosaic recombination in the exocrine pancreas (Herrera, 2000) and little to no recombination within the hypothalamus when crossed to a Rosa-LacZ reporter (Rozo and Stoffers, unpublished data). Pdx1-Cre mice did not exhibit glucose intolerance compared to wild-type controls (data not shown).
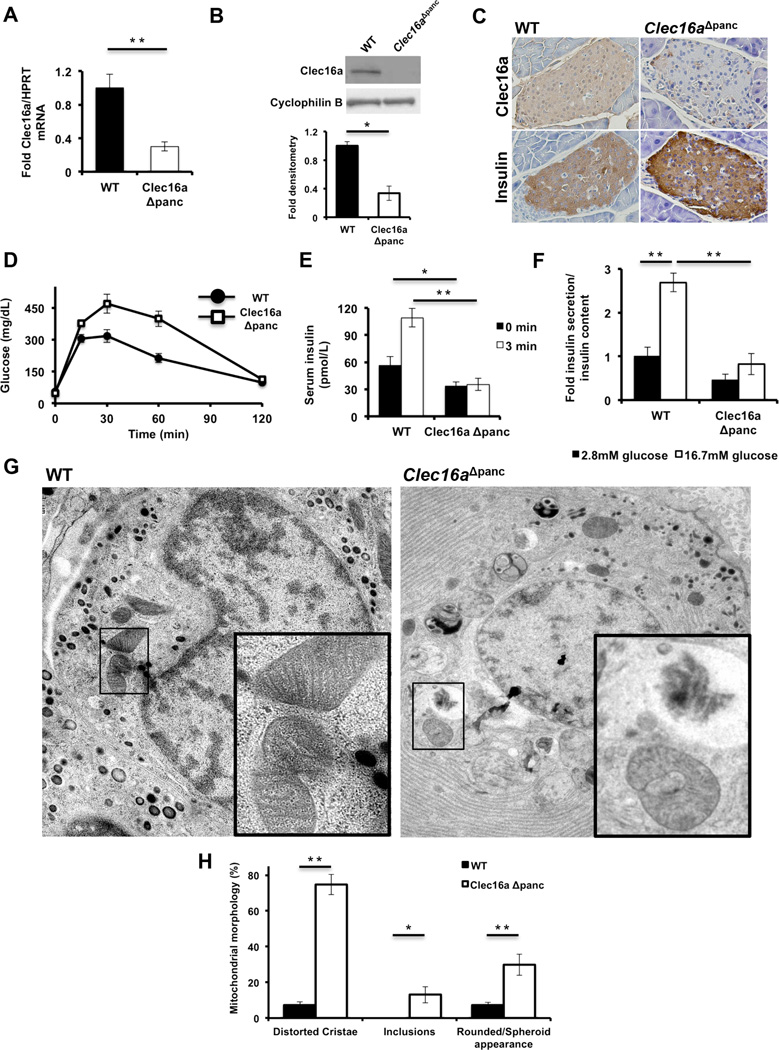
Clec16aΔpanc mice exhibited a 70% reduction in islet Clec16a transcript (Figure 3A), which correlated with reduced islet Clec16a protein expression (Figures 3B and S3C). Clec16a was expressed throughout the islet, including in β-cells, with efficient deletion in Clec16aΔpanc islets (Figure 3C). Intraperitoneal glucose tolerance testing (IPGTT) revealed that Clec16aΔpanc mice are significantly hyperglycemic compared to littermate controls (p<0.005 by ANOVA; Figure 3D). Further, Clec16aΔpanc mice have reduced basal and blunted insulin release at 0 and 3 minutes after glucose administration in vivo (Figure 3E) and in isolated islets (Figure 3F), indicating decreased pancreatic islet glucose-stimulated insulin release as the cause of the impaired glucose tolerance in vivo. There were no differences in peripheral insulin sensitivity between the groups (Figure S3D) and no evidence of exocrine pancreatic dysfunction, such as weight loss (Figure S3E), steatorrhea, or histologic abnormalities of acinar or ductal cells in Clec16aΔpanc mice.
Figure 3. Pancreas-specific Clec16a loss of function leads to hyperglycemia due to functional defects in β-cells.
(A) Clec16a mRNA expression by qRT-PCR from RNA isolated from WT or Clec16aΔpanc islets (n=10/group). (B) Clec16a expression by WB in WT or Clec16aΔpanc islets (n=4/group). (C) Immunohistochemistry of adjacent sections of WT and Clec16aΔpanc pancreata for Clec16a (top) and insulin (bottom). Image shown is representative of IHC performed in 3 mice/group. (D) Blood glucose concentrations measured during IPGTT of 5 week old WT and Clec16aΔpanc littermates. (p<0.005 by ANOVA; n=6–15/group) (E) Serum insulin (n=6–15/group) measured during in vivo glucose-stimulated insulin release testing in 8 week old WT and Clec16aΔpanc littermates. (F) Fold glucose-stimulated insulin secretion following static incubations in 2.8mM and 16.7 mM glucose performed in isolated WT or Clec16aΔpanc islets of 8 week old littermates (n=4/group). (G) Transmission EM images from 3-month-old WT and Clec16aΔpanc β-cells. Inset (lower right) – focused area of mitochondria on EM images. (H) Quantification of changes in mitochondrial morphology (% of total mitochondria) observed in WT and Clec16aΔpanc β-cells in transmission EM images (~250 independent mitochondria scored/animal). Images captured from 3–6 animals/group. Representative WBs chosen from among 4 independent experiments. Densitometry, shown adjacent to each WB, represents mean (±SEM) of 4 independent experiments. *p<0.05 **p<0.01. See also Figures S2-S4.
Given the association of Clec16a with T1DM, we assessed whether Clec16a deficiency was associated with immune infiltration and insulitis but found no evidence of infiltrating lymphocytes into the pancreatic islets of WT or Clec16aΔpanc mice (Figure S3E). Immunofluorescence for insulin and glucagon revealed normal islet architecture in Clec16aΔpanc mice (Figure S3F). β-cell mass was also normal following Clec16a loss of function (Figure S3G). The presence of normal islet architecture and β-cell mass in the context of an increase in pancreatic insulin content (Figure S3G) indicates a functional defect imposed by Clec16a deficiency accounts for the reduced insulin release observed in vivo. This is supported by observations in Min6 β-cells following transient reduction of Clec16a expression by siRNA, which resulted in reduced glucose-stimulated insulin release (Figures S4A-B). Normal insulin release following depolarization with KCl in Clec16a deficient Min6 β-cells (Figure S4B) suggests that Clec16a does not regulate exocytosis.
Examination of β-cells from Clec16aΔpanc and littermate control mice by transmission electron microscopy revealed a dramatic impact on β-cell ultrastructure. While β-cells from both groups had the expected appearance of electron dense insulin granules, Clec16aΔpanc β-cells displayed an accumulation of vacuolated structures, some containing partially degraded cytoplasmic material consistent with autophagic vacuoles, that were not seen in WT β-cells (Figures 3G). Further, Clec16aΔpanc β-cells displayed rounded mitochondria with dramatically disordered and amorphous structure (Figures 3G-H). These effects were also observed in Clec16aΔpanc α-cells (data not shown).
Clec16a deficiency promotes the accumulation of unhealthy mitochondria
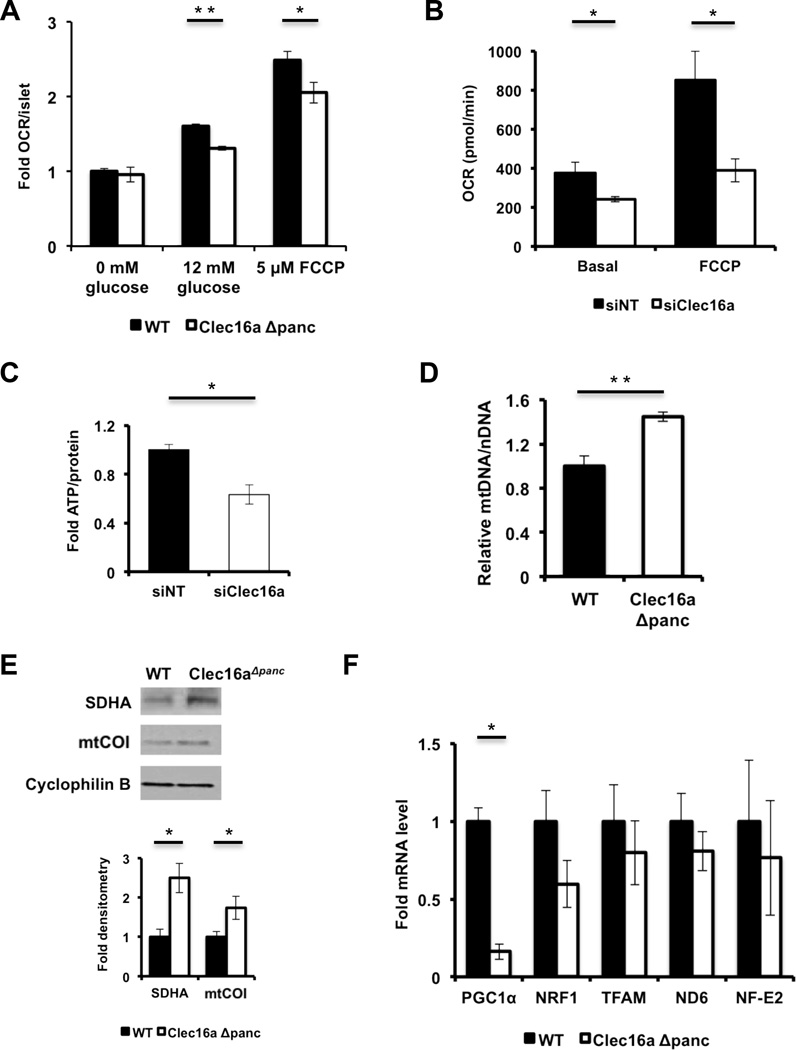
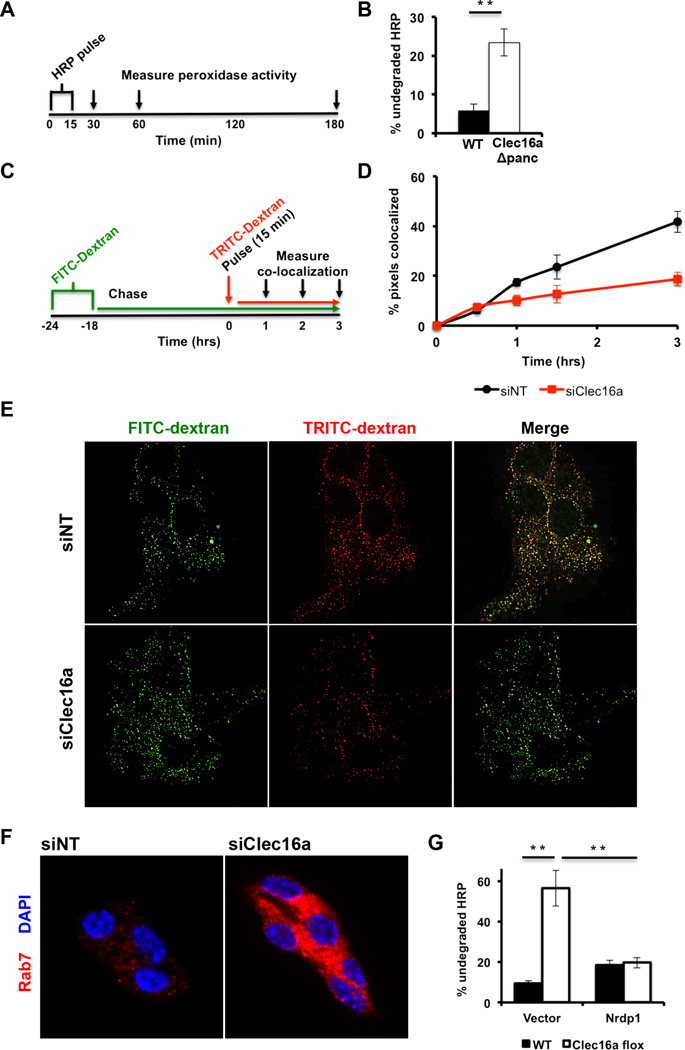
Mitochondrial function is a key regulator of glucose-stimulated insulin release (Supale et al., 2012). The appearance of defects in glucose tolerance, insulin secretion, and mitochondrial ultrastructure in Clec16aΔpanc β-cells suggests that, similar to the observations in primary fibroblasts, mitochondrial respiration is impacted in Clec16aΔpanc β-cells. Indeed, islets isolated from Clec16aΔpanc mice displayed reduced glucose-stimulated OCR as well as maximal OCR following FCCP treatment when compared to WT islets (Figure 4A), with similar results in siClec16a treated Min6 β-cells (Figure 4B). Accordingly, ATP level was reduced following glucose stimulation of siClec16a-treated Min6 β-cells (Figure 4C).
Figure 4. Clec16a loss of function results in the accumulation of unhealthy mitochondria.
(A) Relative OCR measured in isolated WT and Clec16aΔpanc islets (n=6/group). (B) OCR (n=4/group) measured at baseline or following 300 nM FCCP in Min6 β-cells following transfection with non-targeting (siNT) or Clec16a specific siRNA (siClec16a). (C) Fold ATP concentrations (normalized to protein content; n=3/group) in Min6 β-cells following transfection with siNT or siClec16a. (D) Relative mtDNA content measured by qPCR (normalized to nuclear DNA expression) in isolated WT and Clec16aΔpanc islets (n=3–4/group). (E) Expression of the mitochondrial proteins succinate dehydrogenase A (SDHA) and mitochondrial complex IV subunit I (mtCOI) in isolated WT and Clec16aΔpanc islets by WB. (F) Quantitative RT-PCR of markers of mitochondrial biogenesis (PGC1α, NRF1, TFAM) and ROS-induced genes (ND6 and NF-E2) from RNA isolated from WT and Clec16aΔpanc islets (n=5/group). For all WBs, representative images chosen from 3 independent experiments performed. WB densitometry represents mean (±SEM) of 3 independent experiments. *p<0.05 **p<0.01. See also Figures S5-S6.
Interestingly, mtDNA levels as well as levels of the mitochondrial proteins succinate dehydrogenase A (SDHA) and mitochondrial complex IV subunit I (mtCOI) were increased in Clec16aΔpanc islets (Figures 4D-E), indicating an increase in mitochondrial mass. Elevated mitochondrial content could arise through increases in mitochondrial biogenesis or diminished degradation. We measured mRNA expression of PGC1α, NRF1, and TFAM (Scarpulla, 2011) and found that they were not increased in Clec16aΔpanc islets (Figure 4F), suggesting that increased mitochondrial biogenesis does not contribute to increased mitochondrial mass. The dramatic reduction in PGC1α could reflect compensation for mitochondrial accumulation in Clec16aΔpanc islets. We also assessed expression of factors induced by reactive oxygen species, a potential regulator of mitochondrial biogenesis, and observed no differences between the groups (Figure 4F).
To examine islet mitochondrial quality control, we assessed the Nrdp1/Parkin pathway and found that, similar to Clec16aflox fibroblasts, Nrdp1 expression is reduced in Clec16aΔpanc islets (Figure 5A). We also observed increased Parkin levels (Figure 5B) associated with decreased expression of Parkin targets Mfn2 and Porin (Figure 5C). Nrdp1 and Parkin transcript levels were unaffected (data not shown). The accumulation of unhealthy mitochondria, coupled with dysregulation of the Nrdp1/Parkin pathway, implicates defective mitophagy/autophagy leading to impaired glucose-stimulated OCR and insulin secretion in Clec16aΔpanc mice.
Figure 5. Clec16a regulates mitophagy.
(A) Nrdp1 expression by WB of WT and Clec16aΔpanc islets. Representative WB chosen from 4 independent experiments performed. Each sample analyzed comprised islet lysates pooled from 8 mice (32 mice total/group). (B) Parkin expression by WB of WT and Clec16aΔpanc islets following 3-hr of vehicle (DMSO) or 10 µM FCCP (n=3/group). (C) Mfn2 and Porin expression by WB in WT and Clec16aΔpanc islets following 3-hr of vehicle (DMSO) or 10 µM FCCP (n=3/group). (D) Representative deconvolution image of WT and Clec16aflox fibroblasts, 72 hours after overexpression of LC3-GFP and mito-dsRed, treated with vehicle (DMSO) or 150 nM BafA1 for 6 hours. (E) Quantification of LC3-GFP/mito-dsRed co-localization in WT and Clec16aflox fibroblasts following vehicle (DMSO) or 150 nM BafA1 for 6 hours (n=5–8/group). (F) Fold LC3-GFP puncta quantified from WT and Clec16aflox fibroblasts, 72 hours after overexpression of LC3-GFP, treated with vehicle (DMSO) or 150 nM BafA1 for 6 hours. (G) Representative deconvolution image of WT and Clec16aflox fibroblasts treated with 10 µM FCCP for 60 minutes followed by indirect immunofluorescence for Lamp1 (green), LC3 (red), and DAPI (blue). (H) Quantification of LC3+ Lamp1+ co-localization in WT and Clec16aflox fibroblasts following 10 µM FCCP for 60 minutes (n=5/group). (I) Quantification of LC3+ Lamp1+ co-localization in non-FCCP treated WT and Clec16aflox fibroblasts, 72 hours after transient overexpression of empty vector (pcDNA3–3xHA) or 3xHANrdp1 (n=5–7/group). For Figure 5B and 5C, representative WBs chosen from 3 independent experiments performed. Densitometry, shown adjacent to each WB, represents mean (±SEM) of all experiments performed. *p<0.05 **p<0.01. See also Figures S5-S6 and Movies S1–S3.
The Clec16a-Nrdp1 pathway regulates autophagosome-lysosome fusion during late mitophagy
A role for Clec16a in autophagosomal trafficking is supported by the observation of autophagic vacuoles in in Clec16aΔpanc islets (Figure 3G) and of reduced Mfn2 in Clec16aflox fibroblasts and Clec16aΔpanc islets. Defective autophagosome-lysosome fusion has been reported following depletion of Mfn2 in cardiomyocytes (Zhao et al., 2012). To assess trafficking defects during mitophagy, we assessed co-localization of autophagosomes with mitochondria using fluorescent-labeled markers for autophagosomes (LC3-GFP) and mitochondria (mito-dsRed) in WT and Clec16aflox fibroblasts. We found increased LC3-GFP and mito-dsRed co localization in Clec16aflox fibroblasts (Figures 5D-E), indicating the autophagosomal machinery does not efficiently degrade autophagosomes containing mitochondria. As reduced autophagic flux could be a cause of mitochondrial accumulation in autophagosomes, we quantified LC3-GFP puncta with and without Bafilomycin A1 (BafA1), an inhibitor of the vacuolar H+ ATPase. As expected, LC3-GFP puncta accumulated in WT fibroblasts following BafA1 exposure, but this did not occur in Clec16aflox fibroblasts, suggesting a defect in autophagic flux (Figures 5D and 5F). Lastly, following FCCP-mediated depolarization to induce mitophagy, we found reduced co-localization of LC3 with the lysosomal marker Lamp1 (Figures 5G-H), indicating a defect in trafficking of autophagosomes to the lysosomal compartment during late mitophagy. Overexpression of Nrdp1 rescued the reduced co-localization of LC3/Lamp1 seen in Clec16aflox fibroblasts (Figure 5I), indicating that Clec16a regulation of autophagosomal trafficking is mediated at least in part by Nrdp1 and highlighting a new role for Nrdp1. Taken altogether, the Clec16a-Nrdp1 pathway modulates mitochondrial function through regulation of mitophagy/autophagy.
Reduced mitochondrial function due to Clec16a deficiency mediated by increased Parkin expression contrasts with previously reported beneficial effects of Parkin overexpression on lifespan and cell viability (Rana et al., 2013; Suen et al., 2010). We hypothesized that the simultaneous defect in clearance of unhealthy mitochondria caused by the defect in autophagosomal-lysosomal trafficking explains this unexpected result. To directly test this hypothesis, we overexpressed mCherry-Parkin in Min6 β-cells and then tested the importance of autophagosome-lysosome fusion to mitochondrial oxygen consumption utilizing two pharmacologic inhibitors of the vacuolar H+ ATPase (vATPase), the pleomacrolide Bafilomycin A1 and KM91104, which disrupts a subunit of the vATPase highly expressed in pancreatic β-cells (Kartner et al., 2010; Sun-Wada et al., 2006). We observed the expected inhibition of acidification of endolysosomal compartments by BafA1 and KM91104 (Figure S5A). Due to the reported off-target effect of BafA1 to disrupt mitochondrial function by acting as a K+ ionophore (Teplova et al., 2007), we assessed respiration of isolated mitochondria from Min6 β-cells treated with both inhibitors. While BafA1 did impair isolated mitochondrial function (Figure S5B; Veh RCR 5.78±0.60, BafA1 RCR 3.00±0.22, p<0.01), KM91104 did not, allowing us to use this inhibitor to selectively assess the role of endolysosomal function on whole cell respiration (Figure S5B; Veh RCR 4.40±0.36, KM91104 RCR 3.75±0.31, p=0.27). As expected, Parkin overexpression reduced porin expression, indicative of functional overexpression (Figure S5C). While overexpression of Parkin or KM91104 treatment alone did not cause marked reductions in oxygen consumption, the addition of vATPase inhibition to Parkin overexpression dramatically reduced oxygen consumption (Figure S5D). Echoing our findings with Clec16a loss of function, we conclude that increased early mitophagy by overexpressed Parkin in addition to impaired autophagosomal trafficking in late mitophagy together are necessary to cause deleterious effects on mitochondrial function in pancreatic β-cells.
Pancreas-specific Clec16a deficiency induces age-dependent β-cell ER stress
The effect of Clec16a on expression of Parkin and Mfn2 suggests that Clec16a could affect other Mfn2-regulated pathways. Beyond regulation of mitochondrial respiration and dynamics, Mfn2 regulates ER function and the unfolded protein response by mediating an interaction between ER and mitochondria (de Brito and Scorrano, 2008; Ngoh et al., 2012). Further, Mfn2 plays a vital role in ER homeostasis in POMC neurons (Schneeberger et al., 2013), a neuroendocrine secretory cell with similarity to β-cells. Notably, by EM we observed an accumulation of electron lucent vacuolated structures in Clec16aΔpanc β-cells that were not observed in WT β-cells (Figures S6A-B). By 3-dimensional EM reconstruction tomography, we attempted to determine the identity of these structures. While WT β-cells possess normal stacks of ER seen throughout the cytoplasm (Movie S1; Figure S6B), in Clec16aΔpanc β-cells, we observed two distinct populations of electron lucent vacuolated structures; large interconnected vacuoles, which likely represent dilated ER (Movie S2; Figure S6C) as well as smaller spherical vacuoles, which could represent endosomes (Movie S3; Figure S6D). We also found increased levels of IRE1α and spliced XBP1 in 6-month old Clec16aΔpanc islets (Figure S6E), consistent with ER stress. Interestingly, 6-week old Clec16aΔpanc islets did not express increased levels of sXBP1 (Figure S6E), suggesting the development of progressive ER stress with age. In contrast, Clec16aflox fibroblasts did not develop noticeably abnormal ER morphology, assessed by confocal microscopy of the ER marker protein disulfide isomerase (PDI) (Figure S6F). We speculate that high secretory demand promotes the development of ER stress in the context of defects caused by Clec16a deficiency. These studies implicate Clec16a as an upstream regulator of several pathways downstream of Nrdp1, Parkin, and Mfn2, including ER function, autophagy, and mitochondrial function.
Clec16a is a membrane-associated endolysosomal protein
Regulation of mitochondrial function by Clec16a led us to ask whether Clec16a is a mitochondrial protein. The subcellular localization of Clec16a has been in question, with recent studies suggesting Clec16a strictly localizes to endosomal, Golgi, or ER compartments (Kim et al., 2012; Kim et al., 2010; Zouk et al., 2013). We assessed Clec16a subcellular localization in Min6 β-cells stably expressing Flag-Clec16a following biochemical fractionation. Clec16a was not enriched in mitochondria but rather was equally expressed in cytosolic and membrane fractions (Figure S7A). Alkaline carbonate extraction displaced Clec16a from the membrane fraction, indicating that Clec16a is membrane-associated (Figure S7B). Overlay of membrane proteins on a non-continuous Nycodenz gradient demonstrated Clec16a enrichment in fractions similar to those of proteins in the endolysosomal pathway (including Rab5, Rab7, and Lamp1), and distinct from those of the endoplasmic reticulum, Golgi and secretory granules (Figure S7C). Similarly, we found Clec16a co-localized with the lysosomal marker Lamp1 by immunostaining (Figure S7D). Thus, Clec16a is a membrane-associated endolysosomal protein.
Localization of Clec16a to the endosomal compartment and the accumulation of small vacuoles on EM (Figure S6D) led us to examine endosomal trafficking Clec16a deficient pancreatic β-cells. We estimated endosomal trafficking utilizing fluid phase tracers in both dispersed pancreatic islets plated in monolayers and in Min6 β-cells. Following pulse-chase of horseradish peroxidase (HRP), 5.7% of HRP remained in WT islets. In contrast, Clec16aΔpanc islets could not degrade HRP as effectively, with 23.4% of HRP remaining after a 180-minute chase (Figure 6A-B). In a complementary approach, we also utilized fluorescent conjugated dextran tracers to estimate endolysosomal trafficking (Figure 6C and S7E) and found a reduction in FITC-and TRITC-dextran co-localization over time in siClec16a Min6 cells (p<0.05 by ANOVA; Figures 6D-E), indicating impaired endosomal trafficking. We noted no significant differences in total labeled-dextran uptake, ruling out a defect in endocytosis (Figure S7F). Rab7 immunoreactivity was also increased, suggesting an accumulation of late endosomes following Clec16a loss of function (Figure 6F). Interestingly, overexpression of Nrdp1 reduced the accumulation of HRP in Clec16aflox fibroblasts, indicating that Nrdp1 mediates the effect of Clec16a on endosomal trafficking (Figure 6G). Therefore, the endolysosomal protein Clec16a is a mediator of both endosomal and autophagosomal trafficking in part through Nrdp1.
Figure 6. Clec16a regulates β-cell endosomal trafficking.
(A) Study design for HRP (5 mg/mL) trafficking and degradation by a pulse-chase approach. (B) Quantification of undegraded HRP activity in dispersed WT and Clec16aΔpanc islets (n=6/group). (C) Study design for fluorescent-conjugated dextran trafficking and co-localization by pulse-chase in Min6 β-cells. (D) Quantification of FITC-dextran and TRITC-dextran co-localization over time in Min6 β-cells (p<0.05 by ANOVA; n=3/group) 72 hours after transient transfection with non-targeting or Clec16a-specific siRNA. (E) Representative deconvolution image of FITC-dextran and TRITC-dextran treated Min6 β-cells (3 hours post-TRITC-dextran chase) following transient transfection with non-targeting or Clec16a-specific siRNA. (F) Representative deconvolution image following immunofluorescence staining for Rab7 (red) and DAPI (DNA, blue) of Min6 β-cells following transient transfection with non-targeting or Clec16a-specific siRNA. (G) Quantification of undegraded HRP activity in WT and Clec16aflox fibroblasts, 72 hours after overexpression of empty vector (pcDNA3–3xHA) or 3xHA-Nrdp1, utilizing HRP pulse-chase approach (n=6/group). *p<0.05 **p<0.01. See also Figure S7 and Movies S1 and S3.
A diabetogenic CLEC16A SNP associated with reduced islet CLEC16A expression and β-cell function in humans
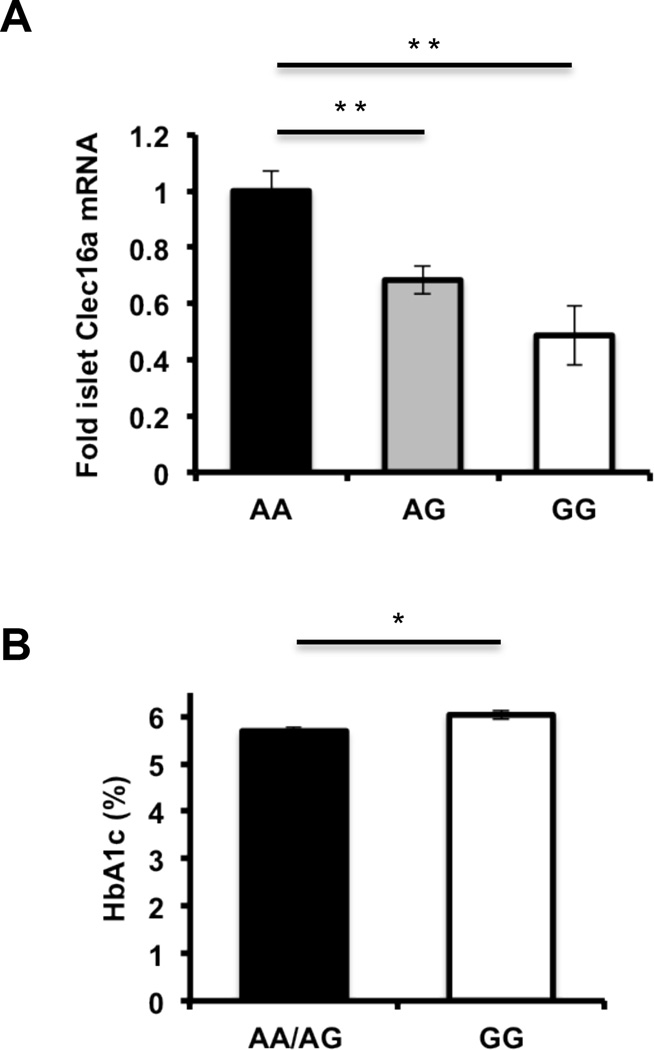
To determine whether CLEC16A regulates human pancreatic β-cell function and potentially contributes to disease pathogenesis in humans, we studied the impact of diabetogenic CLEC16A SNP, rs12708716 (Hakonarson et al., 2007; Todd et al., 2007; WTCCC, 2007), on islet CLEC16A expression, β-cell function, and glucose homeostasis. We assessed complementary high-throughput DNA sequencing and RNA sequencing data from >80 healthy human islet donors. In islet donors possessing one or two copies of the T1DM-associated SNP, we observed a dose-dependent decrease of CLEC16A mRNA expression (Figure 7A), indicating that rs12708716 acts as an expression quantitative trait locus for CLEC16A. Then, we accessed data collected by the Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) to correlate rs12708716 with β-cell function as measured by HOMA-β (Dupuis et al., 2010). Following adjustment for age, sex, study co-variates as well as natural log transformation of HOMA-β data, rs12708716 reduced HOMA-β measurements by 7.4×10−3 ± 3.4×10−3 (p<0.03; n=35,285) while not impairing peripheral insulin sensitivity as measured by HOMA-IR. Lastly, we found a small but significant elevation in hemoglobin A1c in non-diabetic human islet donors homozygous for the rs12708716 SNP (Figure 7B), thus linking a T1DM associated SNP to islet CLEC16A expression, β-cell function and glucose homeostasis in humans.
Figure 7. The diabetogenic CLEC16A SNP rs12708716 is associated with reduced islet CLEC16A expression and impaired glucose control.
(A) Human islet CLEC16A RNA expression measured by RNA-seq in islet donors stratified by DNA sequencing at position of rs12708716 A/G SNP. **p<0.001. (B) Hemoglobin A1c measurements in islet donors sequenced at position of rs12708716 A/G SNP. (AA n=31/group, AG n=42/group, GG n=8/group). *p<0.05.
DISCUSSION
Here we identify the diabetes susceptibility gene Clec16a as a novel regulator of mitophagy through upstream regulation of the Nrdp1/Parkin pathway. The E3 ubiquitin ligase Nrdp1 is a major interacting partner of Clec16a and Clec16a appears to protect Nrdp1 from proteosomal degradation. The mechanism by which Clec16a stabilizes Nrdp1 remains to be explored. Nrdp1 is efficiently degraded by the ubiquitin-proteosome system; mutations in the RING finger domain that disrupt its ubiquitin ligase activity enhance Nrdp1 stability (Wu et al., 2004). Nrdp1 stability is also maintained by interaction with the deubiquitinating enzyme USP8 (Avvakumov et al., 2006; Wu et al., 2004). Similar to Clec16a, USP8 controls endosomal trafficking as well as endosomal ubiquitin dynamics and cell surface receptor turnover (Mizuno et al., 2005; Row et al., 2006). Our observations regarding Clec16a localization, function, and interaction with Nrdp1, along with the role of Drosophila ortholog Ema in EGFR stability (Kim et al., 2010), suggest a tripartite regulation of membrane trafficking, receptor ubiquitination and degradation, and mitophagy, by Clec16a, Nrdp1, and USP8.
We describe an important and novel role for mitophagy in the regulation of pancreatic β-cell function via regulation of key proteins essential for both mitochondrial respiration and dynamics. It is well known that pancreatic β-cells rely heavily on mitochondrial respiration to maintain normal glucose stimulated insulin release. Recent reports also demonstrate the importance of mitochondrial dynamics to β-cell function (Stiles and Shirihai, 2012; Supale et al., 2012). Dysfunctional mitophagy is known lead to both mitochondrial respiratory dysfunction and defects in mitochondrial dynamics (under the control of mitofusins 1 and 2 which we show to be targeted by the Clec16a-Nrdp1-Parkin pathway) (Youle and Narendra, 2011).
Our observations of defective insulin release, impaired glucose homeostasis, and ER stress related to β-cell dysfunction in the context of Clec16a deficiency may provide insight into early non-immune related events in the course of T1DM. Amongst the earliest signs of T1DM in non-diabetic high-risk patients is the loss of first phase insulin release (FPIR), an early indicator of β-cell impairment (Chase et al., 2001; Defronzo, 2009), with only rare instances of insulitis (In't Veld et al., 2007), suggesting that β-cell dysfunction predates insulitis. Similarly, animal models of T1DM display loss of FPIR and β-cell ER stress prior to immune infiltration (Ize-Ludlow et al., 2011; Marhfour et al., 2012). ER stress in turn may serve as a trigger for autoimmunity (Tersey et al., 2012). The expression of ~60% of T1DM-associated genes in pancreatic islets further supports that the β-cell is more than an innocent bystander in its own demise, (Eizirik et al., 2012; Soleimanpour and Stoffers, 2013).
These studies demonstrate that Clec16a controls β-cell function by regulating mitophagy/autophagy. Regulation of insulin secretion by mitophagy has not been previously appreciated in diabetes. The Clec16a/Nrdp1/Parkin axis in β-cells could be targeted to prevent or control T1DM and serve as a basis for understanding the pathogenesis of other Clec16a and Parkin related diseases.
EXPERIMENTAL PROCEDURES
Please see Extended Experimental Procedures for additional details.
Proteomics studies
Stably expressing Flag-Clec16a or control NIH3T3 fibroblasts were maintained for at least 5 doublings in DMEM not containing lysine or arginine (Invitrogen) supplemented with 0.398 mM arginine and 0.798 mM lysine or 0.398 mM U-13C6,15N4-arginine and 0.798 mM U-13C6,15N2-lysine (Cambridge Isotope Laboratories). Cell lysis and immunoprecipitation were performed with Anti-Flag M2 affinity gel (Sigma). Samples were run in a 10% Bis Tris reducing gel (Invitrogen), stained with Coomassie Blue, and pixilated as described (Shevchenko et al., 1996). Tryptic digests were analyzed by LC-MS/MS on a hybrid LTQ Orbitrap Elite mass spectrometer (ThermoFisher) coupled with a nanoLC Ultra (Eksigent).
Animals
Animal studies were approved by the University of Pennsylvania Institutional Animal Care and Use Committee. To generate Clec16aloxP mice, a 15.3kb C57BL/6 genomic DNA fragment containing exons 2–4 and flanking intronic sequence was subcloned into plasmid FLSniper (Ozgene) containing an FRT-flanked PGK-driven Neomycin cassette. LoxP sites flanked exon 3. The linearized targeting vector was electroporated into B6 ES cells and clones that survived selection were screened for homologous recombination by Southern blot. Targeted clones were injected into C57BL/6-derived blastocysts and transferred to pseudopregnant females. Male offspring were mated to C57BL/6 females and ES cell-derived offspring were identified by PCR-based genotyping. Mice harboring the targeted insertion of the two loxP sites in the Clec16a gene locus were then crossed to the ACTB-FLPe line to achieve deletion of the FRT-flanked Neomycin cassette (Rodriguez et al., 2000). Clec16aloxP mice were mated to UBC-Cre/ERT2 mice (Ruzankina et al., 2007) or Pdx1-Cre (Herrera, 2000) to generate experimental groups. All animals were maintained on a 100% C57BL/6 background.
Oxygen consumption assays
Oxygen consumption was measured in isolated islets by a Clark-type O2 electrode (Strathkelvin Instruments) and in mitochondria or adherent cells using a XF24 Flux Analyzer (Seahorse Bioscience).
Human islet studies
Islets from non-diabetic cadaver donors (n=81) were provided by the Nordic Islet Transplantation Program (http://www.nordicislets.org). Purity of islets was assessed as described previously (Taneera et al., 2012). RNA-seq libraries were prepared using the standard Illumina mRNA-Seq protocol and sequencing was performed on an Illumina HiSeq 2000. Gene expression was considered as the normalized sum of expression of all exons and further normalization was applied by adjusting the expression to gene length as previously described (t Hart et al., 2013). GWAS on DNA from islet donors was performed using the Genome-Wide Human SNP array 6.0 containing 906,000 SNPs (Affymetrix) as described (Taneera et al., 2012). Hemoglobin A1c values were measured in banked donor serum samples obtained prior to death.
Statistical Analysis
Data are presented as mean ± SEM, unless otherwise noted. Statistical comparisons were performed using the student’s t-test or two-way ANOVA (Prism GraphPad). For 2×2 comparisons that reached statistical significance by ANOVA (p<0.05), a protected Fisher’s least significant difference test was utilized for post-hoc analysis.
Supplementary Material
Research Highlights.
Type 1 diabetes susceptibility gene Clec16a interacts with E3 ubiquitin ligase Nrdp1
Clec16a regulates Nrdp1, its target Parkin and effectors porin, Mfn1, and Mfn2
Clec16a via Nrdp1 regulates autophagosomal trafficking during late mitophagy
Clec16a regulates pancreatic β-cell function through control of mitophagy
ACKNOWLEDGMENTS
We acknowledge Dr. N. Doliba and Q. Wei of the Penn Islet Cell Biology Core of the University of Pennsylvania DRC (P30DK19525) for assistance with islet oxygen consumption studies, Dr. D.Williams of the Penn EM Resource Laboratory for imaging assistance, Dr. J. Florez for access to and assistance with MAGIC data, Drs. B. Keith and S. Reiner for immune cell samples, and Dr. P. Herrera for Pdx1-Cre mice. We thank Drs. M. Lazar, M. Birnbaum, M. Marks, and J. Baur for helpful advice. We acknowledge funding support from the Margaret Q. Landenberger Foundation, the Charles H. Humpton, Jr. Endowment, the JDRF and the NIH (K08-DK089117, 5-P01-DK049210-15, and 1DP3DK085708-01).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
S.A.S. conceived of, designed and performed experiments, interpreted results, drafted and reviewed the manuscript. A.G., A.M.F., D.N.G., L.A.S. designed and performed experiments and interpreted results. M.B. provided mice. J.F. performed and analyzed results of human islet studies. L.G. analyzed and interpreted human islet studies and reviewed the manuscript. J.A.K. designed the mouse model and reviewed the manuscript. S.H.S. analyzed and interpreted IDIRT studies and reviewed the manuscript. B.A.K. designed mitochondrial studies, interpreted results and reviewed the manuscript. H.H. provided mice, edited and reviewed the manuscript. D.A.S. conceived of and designed the studies, interpreted results, edited and reviewed the manuscript.
REFERENCES
- Ashrafi G, Schwarz TL. The pathways of mitophagy for quality control and clearance of mitochondria. Cell death and differentiation. 2013;20:31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avvakumov GV, Walker JR, Xue S, Finerty PJ, Jr, Mackenzie F, Newman EM, Dhe-Paganon S. Amino-terminal dimerization, NRDP1-rhodanese interaction, and inhibited catalytic domain conformation of the ubiquitin-specific protease 8 (USP8) J Biol Chem. 2006;281:38061–38070. doi: 10.1074/jbc.M606704200. [DOI] [PubMed] [Google Scholar]
- Chase HP, Cuthbertson DD, Dolan LM, Kaufman F, Krischer JP, Schatz DA, White NH, Wilson DM, Wolfsdorf J. First-phase insulin release during the intravenous glucose tolerance test as a risk factor for type 1 diabetes. J Pediatr. 2001;138:244–249. doi: 10.1067/mpd.2001.111274. [DOI] [PubMed] [Google Scholar]
- de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- Defronzo RA. Banting Lecture. From the triumvirate to the ominous octet: a new paradigm for the treatment of type 2 diabetes mellitus. Diabetes. 2009;58:773–795. doi: 10.2337/db09-9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eizirik DL, Colli ML, Ortis F. The role of inflammation in insulitis and beta-cell loss in type 1 diabetes. Nature reviews Endocrinology. 2009;5:219–226. doi: 10.1038/nrendo.2009.21. [DOI] [PubMed] [Google Scholar]
- Eizirik DL, Sammeth M, Bouckenooghe T, Bottu G, Sisino G, Igoillo-Esteve M, Ortis F, Santin I, Colli ML, Barthson J, et al. The human pancreatic islet transcriptome: expression of candidate genes for type 1 diabetes and the impact of pro-inflammatory cytokines. PLoS genetics. 2012;8:e1002552. doi: 10.1371/journal.pgen.1002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegg ME, Cooper JM, Chau KY, Rojo M, Schapira AH, Taanman JW. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum Mol Genet. 2010;19:4861–4870. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Hakonarson H, Grant SF, Bradfield JP, Marchand L, Kim CE, Glessner JT, Grabs R, Casalunovo T, Taback SP, Frackelton EC, et al. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature. 2007;448:591–594. doi: 10.1038/nature06010. [DOI] [PubMed] [Google Scholar]
- Herrera PL. Adult insulin-and glucagon-producing cells differentiate from two independent cell lineages. Development. 2000;127:2317–2322. doi: 10.1242/dev.127.11.2317. [DOI] [PubMed] [Google Scholar]
- IMSGC. The expanding genetic overlap between multiple sclerosis and type I diabetes. Genes Immun. 2009;10:11–14. doi: 10.1038/gene.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- In't Veld P, Lievens D, De Grijse J, Ling Z, Van der Auwera B, Pipeleers-Marichal M, Gorus F, Pipeleers D. Screening for insulitis in adult autoantibody-positive organ donors. Diabetes. 2007;56:2400–2404. doi: 10.2337/db07-0416. [DOI] [PubMed] [Google Scholar]
- Ize-Ludlow D, Lightfoot YL, Parker M, Xue S, Wasserfall C, Haller MJ, Schatz D, Becker DJ, Atkinson MA, Mathews CE. Progressive erosion of beta-cell function precedes the onset of hyperglycemia in the NOD mouse model of type 1 diabetes. Diabetes. 2011;60:2086–2091. doi: 10.2337/db11-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SM, Youle RJ. PINK1-and Parkin-mediated mitophagy at a glance. J Cell Sci. 2012;125:795–799. doi: 10.1242/jcs.093849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartner N, Yao Y, Li K, Crasto GJ, Datti A, Manolson MF. Inhibition of osteoclast bone resorption by disrupting vacuolar H+-ATPase a3-B2 subunit interaction. J Biol Chem. 2010;285:37476–37490. doi: 10.1074/jbc.M110.123281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Naylor SA, DiAntonio A. Drosophila Golgi membrane protein Ema promotes autophagosomal growth and function. Proc Natl Acad Sci U S A. 2012;109:E1072–E1081. doi: 10.1073/pnas.1120320109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Wairkar YP, Daniels RW, DiAntonio A. The novel endosomal membrane protein Ema interacts with the class C Vps-HOPS complex to promote endosomal maturation. J Cell Biol. 2010;188:717–734. doi: 10.1083/jcb.200911126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesa M, Shirihai OS. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab. 2013;17:491–506. doi: 10.1016/j.cmet.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhfour I, Lopez XM, Lefkaditis D, Salmon I, Allagnat F, Richardson SJ, Morgan NG, Eizirik DL. Expression of endoplasmic reticulum stress markers in the islets of patients with type 1 diabetes. Diabetologia. 2012;55:2417–2420. doi: 10.1007/s00125-012-2604-3. [DOI] [PubMed] [Google Scholar]
- Mizuno E, Iura T, Mukai A, Yoshimori T, Kitamura N, Komada M. Regulation of epidermal growth factor receptor down-regulation by UBPY-mediated deubiquitination at endosomes. Mol Biol Cell. 2005;16:5163–5174. doi: 10.1091/mbc.E05-06-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngoh GA, Papanicolaou KN, Walsh K. Loss of mitofusin 2 promotes endoplasmic reticulum stress. J Biol Chem. 2012;287:20321–20332. doi: 10.1074/jbc.M112.359174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pociot F, Akolkar B, Concannon P, Erlich HA, Julier C, Morahan G, Nierras CR, Todd JA, Rich SS, Nerup J. Genetics of type 1 diabetes: what's next? Diabetes. 2010;59:1561–1571. doi: 10.2337/db10-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana A, Rera M, Walker DW. Parkin overexpression during aging reduces proteotoxicity, alters mitochondrial dynamics, and extends lifespan. Proc Natl Acad Sci U S A. 2013;110:8638–8643. doi: 10.1073/pnas.1216197110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, Dymecki SM. High-efficiency deleter mice show that FLPe is an alternative to CreloxP. Nat Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- Row PE, Prior IA, McCullough J, Clague MJ, Urbe S. The ubiquitin isopeptidase UBPY regulates endosomal ubiquitin dynamics and is essential for receptor downregulation. J Biol Chem. 2006;281:12618–12624. doi: 10.1074/jbc.M512615200. [DOI] [PubMed] [Google Scholar]
- Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, Brown EJ. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell stem cell. 2007;1:113–126. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdeva MM, Claiborn KC, Khoo C, Yang J, Groff DN, Mirmira RG, Stoffers DA. Pdx1 (MODY4) regulates pancreatic beta cell susceptibility to ER stress. Proc Natl Acad Sci U S A. 2009;106:19090–19095. doi: 10.1073/pnas.0904849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla RC. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochimica et biophysica acta. 2011;1813:1269–1278. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger M, Dietrich MO, Sebastian D, Imbernon M, Castano C, Garcia A, Esteban Y, Gonzalez-Franquesa A, Rodriguez IC, Bortolozzi A, et al. Mitofusin 2 in POMC neurons connects ER stress with leptin resistance and energy imbalance. Cell. 2013;155:172–187. doi: 10.1016/j.cell.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass Spectrometric Sequencing of Proteins from Silver-Stained Polyacrylamide Gels. Analytical Chemistry. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Skinningsrud B, Husebye ES, Pearce SH, McDonald DO, Brandal K, Wolff AB, Lovas K, Egeland T, Undlien DE. Polymorphisms in CLEC16A and CIITA at 16p13 are associated with primary adrenal insufficiency. J Clin Endocrinol Metab. 2008;93:3310–3317. doi: 10.1210/jc.2008-0821. [DOI] [PubMed] [Google Scholar]
- Soleimanpour SA, Stoffers DA. The pancreatic beta cell and type 1 diabetes: innocent bystander or active participant? Trends in endocrinology and metabolism: TEM. 2013;24:324–331. doi: 10.1016/j.tem.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles L, Shirihai OS. Mitochondrial dynamics and morphology in beta-cells. Best practice & research Clinical endocrinology & metabolism. 2012;26:725–738. doi: 10.1016/j.beem.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen DF, Narendra DP, Tanaka A, Manfredi G, Youle RJ. Parkin overexpression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proc Natl Acad Sci U S A. 2010;107:11835–11840. doi: 10.1073/pnas.0914569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun-Wada GH, Toyomura T, Murata Y, Yamamoto A, Futai M, Wada Y. The a3 isoform of V-ATPase regulates insulin secretion from pancreatic beta-cells. J Cell Sci. 2006;119:4531–4540. doi: 10.1242/jcs.03234. [DOI] [PubMed] [Google Scholar]
- Supale S, Li N, Brun T, Maechler P. Mitochondrial dysfunction in pancreatic beta cells. Trends in endocrinology and metabolism: TEM. 2012;23:477–487. doi: 10.1016/j.tem.2012.06.002. [DOI] [PubMed] [Google Scholar]
- t Hart LM, Fritsche A, Nijpels G, van Leeuwen N, Donnelly LA, Dekker JM, Alssema M, Fadista J, Carlotti F, Gjesing AP, et al. The CTRB1/2 locus affects diabetes susceptibility and treatment via the incretin pathway. Diabetes. 2013 doi: 10.2337/db13-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tackett AJ, DeGrasse JA, Sekedat MD, Oeffinger M, Rout MP, Chait BT. I-DIRT, a general method for distinguishing between specific and nonspecific protein interactions. J Proteome Res. 2005;4:1752–1756. doi: 10.1021/pr050225e. [DOI] [PubMed] [Google Scholar]
- Taneera J, Lang S, Sharma A, Fadista J, Zhou Y, Ahlqvist E, Jonsson A, Lyssenko V, Vikman P, Hansson O, et al. A systems genetics approach identifies genes and pathways for type 2 diabetes in human islets. Cell Metab. 2012;16:122–134. doi: 10.1016/j.cmet.2012.06.006. [DOI] [PubMed] [Google Scholar]
- Teplova VV, Tonshin AA, Grigoriev PA, Saris NE, Salkinoja-Salonen MS. Bafilomycin A1 is a potassium ionophore that impairs mitochondrial functions. Journal of bioenergetics and biomembranes. 2007;39:321–329. doi: 10.1007/s10863-007-9095-9. [DOI] [PubMed] [Google Scholar]
- Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, Bailey R, Nejentsev S, Field SF, Payne F, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WTCCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yen L, Irwin L, Sweeney C, Carraway KL., 3rd Stabilization of the E3 ubiquitin ligase Nrdp1 by the deubiquitinating enzyme USP8. Mol Cell Biol. 2004;24:7748–7757. doi: 10.1128/MCB.24.17.7748-7757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, Narendra DP. Mechanisms of mitophagy. Nature reviews Molecular cell biology. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Huang X, Han L, Wang X, Cheng H, Zhao Y, Chen Q, Chen J, Cheng H, Xiao R, et al. Central role of mitofusin 2 in autophagosome-lysosome fusion in cardiomyocytes. J Biol Chem. 2012;287:23615–23625. doi: 10.1074/jbc.M112.379164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Tan Y, Zhou A, Yu Q, Zhou J. RING finger ubiquitin-protein isopeptide ligase Nrdp1/FLRF regulates parkin stability and activity. J Biol Chem. 2005;280:9425–9430. doi: 10.1074/jbc.M408955200. [DOI] [PubMed] [Google Scholar]
- Zorzano A, Liesa M, Sebastian D, Segales J, Palacin M. Mitochondrial fusion proteins: dual regulators of morphology and metabolism. Seminars in cell & developmental biology. 2010;21:566–574. doi: 10.1016/j.semcdb.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Zouk H, D'Hennezel E, Du X, Ounissi H, Piccirillo CA, Polychronakos C. Functional evaluation of the role of CLEC16A at the chromosome 16p13 locus. Clinical and experimental immunology. 2013 doi: 10.1111/cei.12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.