Abstract
Metallo-β-lactamases (MBLs) in Enterobacteriaceae are an increasing problem worldwide. This report describes the isolation of Citrobacter freundii carrying IMP-8 MBL from three patients during the period from March 2012 until March 2013 in Germany. The blaIMP-8 enzyme is predominantly found in Asia, where IMP-8 has spread to various enterobacterial species causing serious infections. To our best knowledge, this is the first report of blaIMP-8 habouring Enterobacteriaceae in Europe.
Keywords: Antimicrobial resistance, IMP, infection control, laboratory surveillance, metallo-β-lactamases, multidrug resistance
Introduction
The emergence and spread of carbapenemase-producing Enterobacteriaceae is an increasing problem of global dimensions. Originally, metallo-β-lactamases (MBLs) were associated with resistance in Gram-negative non-fermenters, but they have become increasingly important regarding carbapenem resistance in Enterobacteriaceae. MBLs confer resistance to almost all β-lactam antibiotics and are not inactivated by β-lactamase inhibitors, hence limiting treatment options in the individual patient and presenting a major challenge for infection control within the hospital setting 1.
To date, the occurrence of carbapenem-resistant Enterobacteriaceae in Germany is still a rare event; however, outbreaks involving KPC and VIM-1 carrying Klebsiella pneumoniae isolates have been described 2,3. In the year 2012, VIM-1 was the most prevalent MBL detected in Enterobacteriaceae in Germany, followed by NDM-1 and GIM-1 2. IMP type MBLs were first identified in Pseudomonas aeruginosa in Japan 4 and have since been reported predominantly from Asia 5. With the exception of Italy, IMP-type enzymes have rarely been reported from other countries in Europe 1,5,6.
Here, we report the isolation of Citrobacter freundii harbouring MBL IMP-8 from three patients between March 2012 and March 2013. All isolates were obtained from rectal swabs. All three patients had underlying haematological conditions (acute myeloid leukaemia n = 2 and myelodysplastic syndrome n = 1) and underwent haematopoietic stem cell transplantation. They were screened from rectal swabs for colonization with multidrug resistant Gram-negative bacteria on a weekly routine schedule.
Laboratory Analysis
Identification of the isolates was performed using matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI TOF-MS) (AXIMA Assurance, bioMérieux SA, Marcy l’Etoile, France; Saramis Database Version 4.09) and the VITEK 2 identification system (bioMérieux SA). Antimicrobial susceptibility testing was initially performed with the VITEK 2 system (bioMérieux SA) and confirmed by Etest (bioMérieux SA). Results were interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines (http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/Breakpoint_table_v_3.1.pdf). The detection of extended-spectrum β-lactamase genes blaCTX-M, blaTEM, blaSHV, 7 and carbapenemase genes blaOXA-48 8, blaKPC 9, blaNDM 10, blaIMP and blaVIM 11 was performed as described previously. Sequencing of the IMP genes was performed using class 1 integron primer 3CS (5′-AAG CAG ACT TGA CCT GA-3′) in combination with primer IMP-A, and 5CS (5′-GGC ATC CAA GCA GCA AG-3′) in combination with primer IMP-B 11. Sequence identification was determined by comparison with the sequences available in GenBank (http://www.ncbi.nih.gov/BLAST) and with the reference sequences of the Lahey database (http://www.lahey.org/studies). Alignment was performed using BioEdit version 7.1.11 (Ibis Biosciences, Carlsbad, CA, USA). Genomic fingerprinting of the isolates was done by the enterobacterial repetitive intergenic consensus (ERIC) method using the ERIC2 primer as described previously 12. Plasmids were extracted using the Qiagen large construct kit (Qiagen, Hilden, Germany) and digested using EcoRI and BamHI to allow for size estimation of the plasmids. Southern blotting was performed following a standard protocol. Briefly, DNA was blotted onto positively charged nylon membranes (Roche Biochemicals, Basel, Switzerland) in denaturation solution (3 M NaCl, 0.4 M NaOH) by capillary transfer. High stringency hybridization was performed in accordance with the instructions given by the manufacturer of the digoxigenin labelling and detection kit (Roche Biochemicals). Digoxigenin-labelled DNA probes were generated with a digoxigenin-labelling PCR kit as described in the manufacturer’s instructions (Roche Biochemicals) using the oligonucleotides IMP-A and IMP-B.
Results
The characteristics of the isolates are summarized in Table1. All three isolates were resistant to piperacillin-tazobactam, cefuroxime and cefotaxime. The MIC of meropenem was >32 mg/L for the isolate of patients two and three, whereas meropenem MIC for patient one was intermediate with an MIC of 8 mg/L. All three strains were susceptible to tigecycline, colistin and amikacin. Molecular detection of ESBL genes blaCTX-M, blaTEM, blaSHV and carbapenemase genes blaOXA-48, blaKPC, blaNDM and blaVIM was negative in all three isolates. The IMP PCR gave a positive result and subsequent determination of the nucleotide sequence revealed an MBL of the IMP-8 type. The sequence was compared with the reference sequence (GenBank accession number AF322577) 13. The blaIMP-8 gene detected in the three isolates harboured a non-coding point mutation at position 18 (T→C) as shown in Fig.1. Additionally, an IMP-8 carrying plasmid of approximately 28 500 bp could be isolated from all C. freundii strains. Genomic fingerprinting by the ERIC method revealed indistinguishable PCR patterns in all isolates (data not shown).
Table 1.
Characteristics of IMP-8 carrying Citrobacter freundii isolates from rectal swabs of three hospitalized patients in Germanya
| Patient | Source | Isolation date | Etest results: MIC for the antimicrobial agents in mg/L and interpretation | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TZP | CXM | CTX | CAZ | FEP | ATM | ERT | MEM | TIG | CIP | AN | COL | |||
| 1 | Rectal swab | March 2012 | 128 R | >256 R | 16 R | >256 R | 16 R | 2 I | >32 R | 8 I | 0.75 S | 8 R | 2 S | 0.75 S |
| 2 | Rectal swab | June 2012 | 96 R | >256 R | 16 R | >256 R | 24 R | 3 I | >32 R | >32 R | 0.75 S | 16 R | 2 S | 1 S |
| 3 | Rectal swab | March 2013 | >256 R | >256 R | 24 R | >256 R | 24 R | 2 I | >32 R | >32 R | 0.75 S | 12 R | 2 S | 1 S |
I, intermediate; R, resistant; S, susceptible; AN, amikacin; ATM, aztreonam; CAZ, ceftazidime; CIP, ciprofloxacin; COL, colistin; CTX, cefotaxime; CXM, cefuroxime; ERT, ertapenem; MEM, meropenem; TIG, tigecycline; TZP, piperacillin-tazobactam.
Interpretation according to the EUCAST clinical breakpoints (http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/Breakpoint_table_v_3.1.pdf).
Figure 1.
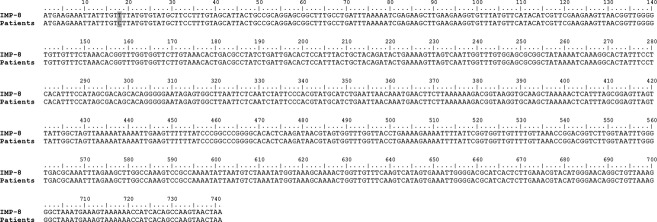
Sequence of the blaIMP-8 gene of three Citrobacter freundii strains isolated from hospitalized patients in Germany. Compared with the sequence of the IMP-8 reference strain (GenBank accession number AF322577 13), a non-coding point mutation at position 18 was identified in the IMP-8 from the three Citrobacter freundii strains (grey shaded).
Conclusion
This is the first report of IMP-8 MBL in Enterobacteriaceae in Germany. IMP-8 is very uncommon in Europe, only once reported from Portugal in a Pseudomonas mendocina strain 6,14. In contrast, IMP-8 is frequently encountered in Asia, especially in Taiwan, where IMP-8-producing Enterobacteriaceae are involved in serious infections. Yan et al. reported on a case series of 37 patients with bloodstream infections caused by a large variety of IMP-8-producing enterobacterial species including Escherichia coli, K. pneumoniae, Enterobacter cloacae and C. freundii 15. Furthermore, aggravating the issue, phenotypic screening for IMP-8-positive Enterobacteriaceae is extremely difficult because of the lack of distinctive phenotypes. Investigation of 95 IMP-8-positive Enterobacteriaceae revealed susceptibility to ertapenem in 21% and to meropenem in 45%, whereas phenotypic combined disk tests using EDTA and phenylboronic acid were positive in only 40% of the isolates 16. These observations from Taiwan suggest that IMP-8 is capable of spreading between enterobacterial species, causing serious problems in terms of infection control measures and limiting therapeutic options in critically ill patients.
The role of faecal carriage of MBL-producing Enterobacteriaceae remains unclear and has not been investigated in a large-scale epidemiological study. Faecal carriage of one C. freundii VIM-1 has been reported from Spain in an outpatient, who was also colonized with two different VIM-1-carrying K. pneumoniae isolates 17. In an Italian hospital, eight VIM-1-carrying C. freundii strains were isolated from rectal swab samples during active screening following the detection of a K. pneumoniae carbapenemase (KPC)-positive patient 18.
None of our three patients became infected by the IMP-8 C. freundii strains, but nevertheless it is alarming that IMP-8 MBL circulates in the gut flora of patients at high risk of nosocomial infections. Even more worrisome is the fact, that the IMP-8 gene is located on a plasmid, which might facilitate the transfer of the resistance gene within enterobacterial species. Our findings emphasize the importance of establishing screening schemes and laboratory diagnostic algorithms to ensure the implementation of efficient infection control measures and therapeutic strategies, not only in high-prevalence countries, but also in countries with a low incidence of MBL producing Enterobacteriaceae.
Acknowledgments
This work was supported by the German Centre for Infection Research (DZIF). We thank Nadine Hoffmann for expert technical assistance.
Conflict of Interest
None declared.
References
- Cornaglia G, Giamarellou H, Rossolini GM. Metallo-β-lactamases: a last frontier for β-lactams? Lancet Infect Dis. 2011;11:381–393. doi: 10.1016/S1473-3099(11)70056-1. [Review] [DOI] [PubMed] [Google Scholar]
- Nationales Referenzzentrum für gramnegative Krankenhauserreger (Kaase M. Zur aktuellen Situation bei Carbapenemase-bildenden gramnegativen Bakterien [current situation of carbapenemase producing Gram-negative]) Epidemiol Bull. 2013;19:167–171. [Google Scholar]
- Steinmann J, Kaase M, Gatermann S, et al. Outbreak due to a Klebsiella pneumoniae strain harbouring KPC-2 and VIM-1 in a German University Hospital, July 2010 to January 2011. Euro Surveill. 2011;16:19944. : pii: [PubMed] [Google Scholar]
- Watanabe M, Iyobe S, Inoue M, Mitsuhashi S. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1991;35:147–151. doi: 10.1128/aac.35.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao WH, Hu ZQ. IMP-type metallo-β-lactamases in Gram-negative bacilli: distribution, phylogeny, and association with integrons. Crit Rev Microbiol. 2011;37:214–226. doi: 10.3109/1040841X.2011.559944. [DOI] [PubMed] [Google Scholar]
- Grundmann H, Livermore DM, Giske CG, et al. Carbapenem-non-susceptible Enterobacteriaceae in Europe: conclusions from a meeting of national experts. Euro Surveill. 2010;15:19711. doi: 10.2807/ese.15.46.19711-en. : pii: [DOI] [PubMed] [Google Scholar]
- Cao V, Lambert T, Nhu DQ, et al. Distribution of extended-spectrum β-lactamases in clinical isolates of Enterobacteriaceae in Vietnam. Antimicrob Agents Chemother. 2002;46:3739–3743. doi: 10.1128/AAC.46.12.3739-3743.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L, Heritier C, Tolun V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48:15–22. doi: 10.1128/AAC.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit H, Queenan AM, Anderson GJ, et al. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaase M, Szabados F, Wassill L, Gatermann SG. Detection of carbapenemases in Enterobacteriaceae by a commercial multiplex PCR. J Clin Microbiol. 2012;50:3115–3118. doi: 10.1128/JCM.00991-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitout JD, Gregson DB, Poirel L, McClure JA, Le P, Church DL. Detection of Pseudomonas aeruginosa producing metallo-β-lactamases in a large centralized laboratory. J Clin Microbiol. 2005;43:3129–3135. doi: 10.1128/JCM.43.7.3129-3135.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JR, O’Bryan TT. Improved repetitive-element PCR fingerprinting for resolving pathogenic and nonpathogenic phylogenetic groups within Escherichia coli. Clin Diagn Lab Immunol. 2000;7:265–273. doi: 10.1128/cdli.7.2.265-273.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan JJ, Ko WC, Wu JJ. Identification of a plasmid encoding SHV-12, TEM-1, and a variant of IMP-2 metallo-β-lactamase, IMP-8, from a clinical isolate of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45:2368–2371. doi: 10.1128/AAC.45.8.2368-2371.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos C, Caetano T, Ferreira S, Mendo S. First description of bla IMP-8 in a Pseudomonas mendocina isolated at the Hospital Infante D. Pedro, Aveiro, Portugal. Res Microbiol. 2010;161:305–307. doi: 10.1016/j.resmic.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Yan JJ, Lee NY, Chen HM, et al. Bloodstream infections caused by IMP-8-producing Enterobacteriaceae isolates: the need for clinical laboratory detection of metallo-β-lactamases? Eur J Clin Microbiol Infect Dis. 2013;32:345–352. doi: 10.1007/s10096-012-1748-x. [DOI] [PubMed] [Google Scholar]
- Liao IC, Chen HM, Wu JJ, Tsai PF, Wang LR, Yan JJ. Metallo-β-lactamase-producing Enterobacteriaceae isolates at a Taiwanese hospital: lack of distinctive phenotypes for screening. APMIS. 2011;119:543–550. doi: 10.1111/j.1600-0463.2011.02772.x. [DOI] [PubMed] [Google Scholar]
- Gijon D, Curiao T, Baquero F, Coque TM, Canton R. Fecal carriage of carbapenemase-producing Enterobacteriaceae: a hidden reservoir in hospitalized and nonhospitalized patients. J Clin Microbiol. 2012;50:1558–1563. doi: 10.1128/JCM.00020-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaibani P, Ambretti S, Farruggia P, et al. Outbreak of Citrobacter freundii carrying VIM-1 in an Italian Hospital, identified during the carbapenemases screening actions, June 2012. Int J Infect Dis. 2013;17:e714–e717. doi: 10.1016/j.ijid.2013.02.007. [DOI] [PubMed] [Google Scholar]


