Phagocytes deploy an arsenal of microbicidal defense mechanisms to combat M. tuberculosis (Mtb) infection. These mechanisms are part of coordinated intracellular programs that limit Mtb counterstrategies.
Abstract
Few pathogens run the gauntlet of sterilizing immunity like Mycobacterium tuberculosis (Mtb). This organism infects mononuclear phagocytes and is also ingested by neutrophils, both of which possess an arsenal of cell-intrinsic effector mechanisms capable of eliminating it. Here Mtb encounters acid, oxidants, nitrosylating agents, and redox congeners, often exuberantly delivered under low oxygen tension. Further pressure is applied by withholding divalent Fe2+, Mn2+, Cu2+, and Zn2+, as well as by metabolic privation in the form of carbon needed for anaplerosis and aromatic amino acids for growth. Finally, host E3 ligases ubiquinate, cationic peptides disrupt, and lysosomal enzymes digest Mtb as part of the autophagic response to this particular pathogen. It is a testament to the evolutionary fitness of Mtb that sterilization is rarely complete, although sufficient to ensure most people infected with this airborne bacterium remain disease-free.
Cells constitute the basic unit in biology. They safeguard and transmit genetic information, exchange gases, assimilate nutrients, and compartmentalize their enzymatic machinery to concentrate activities that share a common function. Beyond these servoregulatory duties, cells also deploy host defense factors to monitor their structural integrity and protect their inner sanctity from infection by the outside world. This universal system of self-defense is known as cell-autonomous immunity (Beutler et al. 2006; Kim et al. 2012a; MacMicking 2012; Randow et al. 2013).
Cell-autonomous immunity operates across all three domains of life where it defends against facultative and obligate intracellular pathogens (Randow et al. 2013). It is particularly effective against microorganisms engaged in accidental or fleeting interactions with their host; however, durable pathogens necessitate multiple intracellular defense programs to overcome microbial counterstrategies (Staskawicz et al. 2001). One such example is Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis (TB). It survives much of the microbicidal onslaught triggered inside human phagocytes via numerous adaptive and exaptative mechanisms (MacMicking 2008). These mechanisms reflect a close and long-standing association with its host. Indeed, paleogenetic evidence suggests Mtb has evolved alongside as well as within humans for at least 70,000 years (Comas et al. 2013).
Such tactics currently enable Mtb to persist within an estimated third of the world’s populace. Here it causes disease in ∼10% of people infected. For the remaining 90% of cases, however, host immunity likely contains and, in some cases, naturally eradicates Mtb (McKinney et al. 2001; Cobat et al. 2009). Autopsy studies conducted in the preantibiotic era, for example, found pulmonary lesions were often sterilized in latently infected individuals dying of nontuberculous causes (see McKinney et al. 2001 and references therein). Similar outcomes were recently reported for cynomolgus macaques (Lin et al. 2014), which together with Metchnikoff’s original observations in rat macrophages and Lurie’s classic studies of inbred rabbits encapsulate the idea that mycobactericidal activities may exist across several mammalian species (Metchnikoff 1905; Lurie 1964). How microbial killing is enacted and what molecular mechanisms are used for Mtb clearance or stasis remain central questions in mycobacterial pathogenesis. Potential antitubercular mechanisms are discussed below.
CELL-AUTONOMOUS DEFENSE AGAINST Mtb
Constitutive versus Inducible Programs
A basal repertoire of host defense factors provide some measure of protection against Mtb; however, resistance is often more effective in cells preactivated by cytokine or Toll-like receptor (TLR) signaling (Fig. 1). Here multiple defense genes are induced within 0.5–6 h of stimulation (MacMicking 2012). Notably, the order of activation and infection appears critical, because resting or naïve macrophages infected before immune stimulation allows Mtb time to interfere with host signaling events such as STAT1 binding its transcriptional coactivators, CBP and p300 (Ting et al. 1999), or blocking PI3K-dependent trafficking (Vergne et al. 2003). Both are needed to inhibit mycobacterial replication.
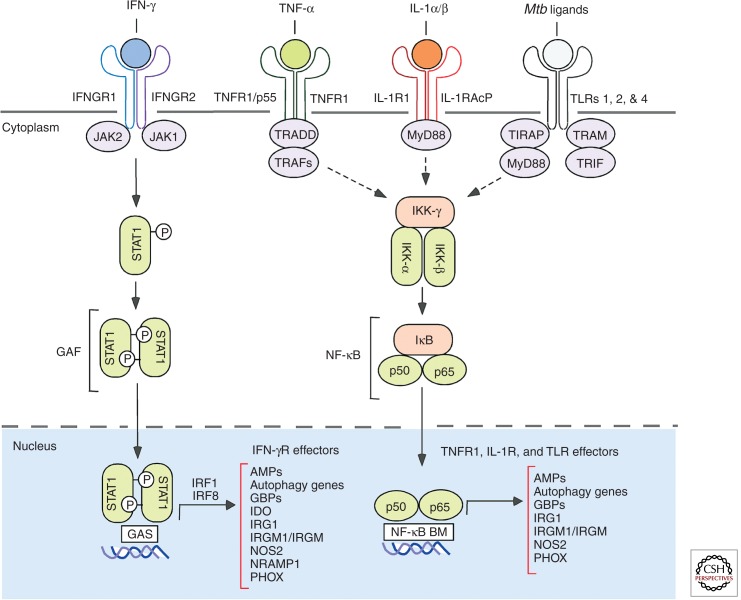
Figure 1.
Immunoreceptor signaling for mobilizing the major cell-autonomous effector programs against Mtb. In addition, new NLR, VDR-, and P2XR-induced activities are emerging (see text). Mtb ligands that elicit TLR signaling include mannosylated lipoarabinomann, phosphatidylinositol mannosides (PIMs), and the 19-kDa lipoprotein. These pathways synergize to induce distinct as well as overlapping signatures of effector proteins for bacterial restriction as shown. For schematic simplicity, receptors are depicted as single chains instead of their trimeric, tetrameric, and heteromeric forms (especially in the case of the TLRs). Adaptor and transcription factor abbreviations: IRF, interferon regulatory factor; JAK, Janus kinase; NF-κB BM, NF-κB binding motif; STAT1, signal transducer and activator of transcription 1; TIRAP, toll-interleukin 1 receptor (TIR) domain containing adaptor protein; TRADD, tumor necrosis factor receptor type 1–associated DEATH domain; TRAF, TNF receptor–associated factor; TRAM, TRIF-related adaptor molecule; TRIF, TIR-domain-containing adapter-inducing interferon-β.
Likewise, the magnitude and success of cell-autonomous immunity depends on which immunoreceptor is engaged. Stimulating C-type lectin receptors dectin-1 or Mincle with Mtb mannosylated lipoarabinomannan or trehelose dimycolate, respectively, induces a limited defense repertoire that fails to control the pathogen (Ishikawa et al. 2009; Betz et al. 2011; Marakalala et al. 2011; Das et al. 2013; Heitmann et al. 2013). In contrast, activation with interferon-γ (IFN-γ) or tumor necrosis factor-α (TNF-α), elicits hundreds of genes that can curtail mycobacterial growth (Ehrt et al. 2001; Nau et al. 2002; Tailleux et al. 2008; Vogt and Nathan 2011).
Mtb-dependent signaling further enriches the host defense repertoire, either through direct transcription of effector genes by pattern-recognition receptor (PRR) ligation or after autocrine TNF-α and interleukin-1 beta (IL-1β) signaling (Ehrt et al. 2001; Nau et al. 2002; Vogt and Nathan 2011). Human monocytes and mouse macrophages regulate approximately 300–700 genes within 12–24 h of Mtb exposure (Ehrt et al. 2001; Ragno et al. 2001; Nau et al. 2002). Similar transcriptional plasticity occurs in human alveolar macrophages (AMs) and dendritic cells (Tailleux et al. 2008). This expression profile may reach as many as 1300 genes when IFN-γ is also present (Ehrt et al. 2001). Thus Mtb plus classical Th1 cytokines like IFN-γ are potent synergistic stimuli for remodeling the transcriptome of phagocytic cells. In combination they enlarge the suite of host proteins available for cell-autonomous defense, a recurring theme for many inducible programs directed against major human pathogens (MacMicking 2012). Indeed, resistance is most effective when multiple activating stimuli are present, consistent with the idea that a single stimulus may be necessary although not always sufficient to eradicate Mtb (Shi et al. 2003; Fabri et al. 2011; Vogt and Nathan 2011).
IFN-γR-Induced Programs
Discovery of IFN-γ as the principal macrophage-activating factor (Nathan et al. 1983, 1984; Pace et al. 1983) soon led to reports of its effectiveness against Mtb in monotypic cell culture (Rook et al. 1986a; Flesch and Kaufmann 1987). These in vitro activities likely operate in vivo given loss-of-function mutations in the IFN-γ signaling cascade confer profound susceptibility to Mtb and, more commonly, disseminated nontuberculous mycobacteriosis in both humans and mice (Cooper et al. 1993; Flynn et al. 1993; Casanova and Abel 2002). A congenital syndrome termed Mendelian susceptibility to mycobacterial disease (MSMD) reveals impaired antimycobacterial immunity in patients with hypomorphic or nullizygous IFNGR, STAT1, and IRF8 alleles, as do mutations in loci that encode IFN-induced effectors such as ISG15 or CYBB (Zhang et al. 2008; Bustamante et al. 2011; Hambleton et al. 2011; Bogunovic et al. 2012).
Genetic lesions in the IFN-γ signaling cascade provide strong support for its role in antitubercular defense. Pharmacologic complementation further reinforces it. Here aerosolized recombinant IFN-γ can alleviate bacillary burdens in patients with antibiotic-resistant Mtb strains and mobilize bacteriostatic programs in human alveolar macrophages ex vivo (Condos et al. 1997; Bonecini-Almeida et al. 1998). Concordant results are seen in long-term differentiated human PBMCs activated with IFN-γ under low physiologic oxygen (5%–10%) that may begin to approach hypoxic conditions found in some pulmonary and extrapulmonary tissue granulomas (Vogt and Nathan 2011). Importantly, the tuberculostatic activities of IFN-γ can be augmented by other stimuli. These include TNF superfamily members (TNF-α, CD40L), IL-1R/TLR, and NLR (NOD-like receptor) agonists or bioactive metabolites such as vitamin D3 (Fabri et al. 2011; Vogt and Nathan 2011; Pilli et al. 2012; Jayaraman et al. 2013; Klug-Micu et al. 2013). Their synergistic effects are discussed below.
TNFR-Induced Programs
Besides their crucial role in granuloma formation (Flynn et al. 1995; Bean et al. 1999), members of the TNF superfamily directly stimulate cell-autonomous immunity to help contain Mtb (Chan et al. 1992; Klug-Micu et al. 2013). TNF-α enhances IFN-γ-induced macrophage programming for Mtb clearance, a synergy which provides several advantages: (1) diversifying the host gene repertoire through alternate transcription factor usage; (2) heightened gene expression from promoters harboring transcription factor-binding sites for both pathways; and (3) lowering the immune activation threshold, a change brought about by shared preexisting components (Fig. 1). This latter point is especially germane given the relative paucity of IFNGRs (4000–12,000/cell−1) on the human macrophage surface (Pace et al. 1983; Finbloom et al. 1985). TNFR1 is likewise expressed at low levels for capturing soluble TNF-α to restrict Mtb growth rather than the nonprotective TNFR2 isoform that instead engages membrane-bound ligand (Jayaraman et al. 2013).
The other two benefits of this synergy emerge when regulation of the antitubercular enzyme, inducible nitric oxide synthase (NOS2), is considered (Chan et al. 1992; MacMicking et al. 1997a; Bekker et al. 2001; Nathan 2006). NOS2 transcription requires IFN-γ-induced STAT1 binding to γ-activated sites (GASs) that is increased by NF-κB docking to its own sites within the NOS2 promoter after TNF-α or lipoarabinomannan stimulation (MacMicking et al. 1997a; Chan et al. 2001. In addition, TNF-α elicits GTP cyclohydrolase 1 that furnishes tetrahydrobiopterin (BH4) as an essential cofactor for NOS2 catalysis and induces argininosuccinate synthetase 1 and cationic amino acid transporter 2 (CAT-2) to regenerate and import the NOS2 substrate, l-arginine, respectively (Bogdan 2001; Qualls et al. 2012). Thus cooperation between TNF-α and IFN-γ regulates transcriptional and posttranslational events for antitubercular defense at the level of the individual cell.
Macrophage reprogramming by another TNF superfamily member, CD40L, also impacts Mtb survival. Human monocytes stimulated with CD40L, alone or in combination with IFN-γ, induce CYP27b1-hydroxylase, which converts the vitamin D3 precursor, 25-hydroxyvitamin D3 (25D3), into bioactive 1,25-dihydroxyvitamin D (1,25D3) (Klug-Micu et al. 2013). Subsequent 1,25D binding to the vitamin D receptor (VDR) elicits antimicrobial peptide expression and autophagy, both potent effector mechanisms targeting mycobacterial replication (Thoma-Uszynski et al. 2001; Gutierrez et al. 2004; Liu et al. 2006, 2007; Singh et al. 2006; Yuk et al. 2009; Ponpuak et al. 2010; Fabri et al. 2011; Pilli et al. 2012).
IL-1R-, TLR-, and NLR-Induced Programs
Members of the IL-1/TLR family stimulate distinct intracellular programs following cognate ligand interaction to combat TB. For IL-1 receptor (IL-1R) signaling, both IL-1β and IL-1α limit mycobacterial growth in murine macrophages via a MyD88-dependent pathway (Jayaraman et al. 2013). A similar IL-1β-dependent profile emerges within human monocyte-derived macrophages. Here IL-1β up-regulates TNFR1 and promotes autocrine signaling via TNF-α release, along with caspase 3–dependent apoptosis (Jayaraman et al. 2013). IL-1β can also elicit autophagic killing via a Tank-binding kinase-1 (TBK-1)-dependent pathway (Pilli et al. 2012), suggesting multiple effectors are deployed downstream of IL-1R to restrict Mtb growth. Such effects presumably operate within intact hosts because IL-1β- and IL-1R1-deficient mice display marked vulnerability to aerogenic Mtb challenge (Mayer-Barber et al. 2010).
Like the IL-1R cascade, TLRs 1, 2, and 4 mobilize multiple effectors as part of the antitubercular arsenal. Engagement of TLR1/TLR2 heterodimers with the 19-kDa Mtb-derived lipoprotein promotes a novel NOS2- and TNF-independent pathway in human macrophages, whereas the same lipopeptide induces a NOS2-dependent mechanism in mouse cells (Thoma-Uszynski et al. 2001). This novel human pathway involves CYP27b1-hydroxylase to produce 1,25D3 for binding the VDR, which in turn elicits cathelicidin (hCAP-18/LL-37) and defensin-4 (DEF4) antimicrobial peptides for tuberculostasis (Liu et al. 2006, 2007). Because the latter cascade also induces autophagy, it may overlap with signals provided by IFN-γ, CD40, and IL-1β at a common juncture downstream (Fabri et al. 2011; Klug-Micu et al. 2013).
Autophagic clearance of Mtb invoked by TLR4 stimulation in human and mouse macrophages requires TRIF (Toll-interleukin-1 receptor domain-containing adaptor-inducing interferon-β) adaptor signaling, unlike IL-1β that relies on MyD88 (Xu et al. 2007). Use of a different adaptor helps widen the TLR transcriptional response to encompass IRF3-dependent genes. In humans, genetic mapping has unearthed numerous TLR4 SNPs that segregate with TB in ethnically diverse subjects, suggesting this TLR4-TRIF pathway operates across a broad swathe of the global population (Velez et al. 2009; Shah et al. 2012). Genetic TB associations have also been noted for TLRs 1 and 2 along with their signaling adaptors (Hawn et al. 2006; Thuong et al. 2007).
Recent studies have shown members of the NLR family may mobilize intracellular immunity to control Mtb growth. NOD2 detects muramyl dipeptide (MDP) that is N-glycolylated in mycobacteria to elicit TNF-α secretion as part of the defense profile in mouse macrophages (Coulombe et al. 2009). Human AMs stimulated with MDP also elicit IL-1β and TNF-α plus the antimicrobial peptide cathelicidin and autophagy-related defense protein, immunity-related GTPase M (IRGM) (Juárez et al. 2012). Here MDP promotes IRGM, ATG16L, and LC3B recruitment to Mtb phagosomes, suggesting NOD2 signaling governs both transcriptional and autophagic defense to this pathogen. Gain-of-function NLRP3 and CARD8 mutations cooperatively limit Mtb in human macrophages (Eklund et al. 2014). Hence several NLRs and related adaptors contribute to bacterial restriction within infected cells.
VDR- and P2XR-Induced Programs
Besides classical cytokine and PRR signaling, two other inducible programs deserve mention within the context of cell-autonomous immunity to Mtb: VDR and purinergic receptor signaling. Early studies reported that 1,25D3 was crucial for IFN-γ-induced tuberculostasis in human macrophages; this effect was attributed to 1,25D3 promoting macrophage differentiation (Rook et al. 1986b; Crowle et al. 1987). Subsequent work, however, has delineated a circuitry whereby CYP27b1-hydroxylase expression and 1,25D3 production are induced by IL-15 following IFN-γ or TLR1/2 stimulation (Fabri et al. 2011). VDR signaling in turn elicits cathelicidin to activate Beclin-1 and Atg5 for autophagosomal capture of mycobacteria (Yuk et al. 2009). Such activities may impact TB outcomes in people because lower levels of serum 1,25D correlate with susceptibility, with VDR disease-related polymorphisms in linkage for some African and Asian populations (Bellamy et al. 1999; Wilkinson et al. 2000).
ATP-dependent stimulation of P2 purinergic receptor subtypes has likewise been strongly implicated in macrophage-mediated Mtb killing. In human macrophages, P2X7 activates cytosolic Ca2+ release and phospholipase D for maturation of Mtb phagosomes (Kusner and Adams 2000; Fairbairn et al. 2001; Kusner and Barton 2001). Subtypes other than P2X7, however, appear to stimulate NOS2-mediated killing in murine cells (Sikora et al. 1999). Because a number of P2X7R polymorphisms associate with TB susceptibility in the human population, this pathway likely influences disease development in its natural setting (Fernando et al. 2007; Sharma et al. 2010; Areeshi et al. 2013).
In sum, multiple receptors induce intracellular programs to protect the host cell from Mtb infection. These signals originate from IFN-γR, TNFR, IL-1R, TLR, NLR, VDR, and P2XR complexes after ligand binding primarily at the plasma membrane or in the cytosol. How their downstream effectors subsequently operate and the mechanisms they deploy are discussed below.
CELL-AUTONOMOUS ARSENAL FOR DEFENSE AGAINST Mtb
Cytotoxic Gases: Reactive Oxygen Species and Nitrogen-Centered Free-Radical Species
Production of cytotoxic gases is a major component of antimicrobial arsenals in all aerobic organisms (Nathan and Shiloh 2000). Reactive oxygen species (ROS) and nitrogen-centered free-radical species (RNS) show distinct chemical properties—oxidizing power, penetrative diffusion limits, lipophilicity—well-suited for killing a variety of facultative bacteria including Mtb (Klebanoff 1999; Nathan and Shiloh 2000). ROS and RNS typically possess low Stokes radii, are highly lipophilic, and electroneutral; thus they are small enough to bypass structural barriers that impede the access of larger host macromolecules and sufficiently permeable to diffuse across the bacterial membrane (Klebanoff 1999). Other chemical properties, notably extended diffusion limits for some RNS, allow them to protect neighboring cells (MacMicking 2012). Carbon monoxide (CO) has also been recently touted as an emerging bactericidal agent, although whether it is generated in sufficient amounts to act as a genuine mycobacterial poison in addition to its role as a sensory cue (Shiloh et al. 2008) has yet to be determined.
Oxidant Stress
Aerobic organisms use allotropic dioxygen as a substrate for generating superoxide (O2−) via oxidoreductases and as a by-product of electron (e−) transport in mitochondria to combat infection (Klebanoff 1999; MacMicking 2012). Single e− additions yield, respectively, hydrogen peroxide (H2O2) and the hydroxyl radical (OH·), both powerful oxidants capable of damaging mycobacterial DNA, lipids, and hemoproteins (Chan et al. 1992; Nathan and Shiloh 2000; Vilchèze et al. 2013). Moreover, within phagolysosomes, elevated levels of O2− (Podinovskaia et al. 2013) undergo spontaneous dismutation at low pH to generate H2O2 that enhances lysosomal killing. Hypochlorous acid (HOCl−) produced by myeloperoxidase-catalyzed oxidation of chloride by H2O2 in neutrophils could also target Mtb internalized from infected macrophages (Yang et al. 2012). Here Mtb expresses methionine sulfate reductases that may partly mitigate this damage (Lee et al. 2009). Indeed, Mtb detoxifying enzymes (e.g., catalases, SODs, peroxiredoxins) limit the overall effectiveness of ROS as a sole agent that could regain toxicity when other bactericidal agents help disable these antioxidant systems.
Large quantities of ROS are produced within phagocytic cells via the NADPH oxidase 2 (NOX2) isoform encoding the flavocytochromeb558 heavy chain subunit (gp91phox; CYBB gene) of the phagocyte oxidase, which generates the respiratory burst (Nathan and Shiloh 2000; MacMicking 2012) (Fig. 2). Congenital X-linked CYBB mutations lead to recurrent TB and “BCG-osis” in patients with defects preferentially manifest in macrophages rather than monocytes or granulocytes (Bustamante et al. 2011). Additional mutations in other NADPH oxidase subunits yield a collective syndrome—chronic granulomatous disease (CGD)—that also appears to predispose to TB (Lau et al. 1998). Mtb susceptibility is less pronounced in mice with targeted disruptions in either gp91phox or gp47phox subunits (Adams et al. 1997; Cooper et al. 2000); however, relative vulnerability increases when Mtb mutants lacking KatG are used, reinforcing the importance of pathogen detoxification systems (Ng et al. 2004).
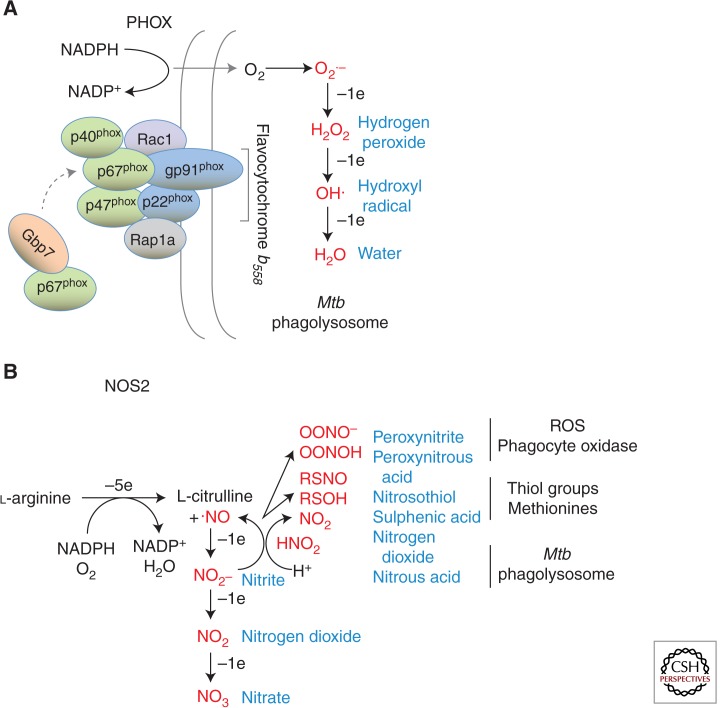
Figure 2.
Oxidative and nitrosative defense against Mtb. (A) Multisubunit complex of the phagocyte oxidase (NADPH oxidase) depicting the core membrane (gp91phox, p22phox; known as flavocytochrome b558) and cytosolic (p67phox, p47phox, p40phox) components. Also shown are immune (Gbp7) and nonimmune GTPases (Rac1, Rap1a) that regulate trafficking to the Mtb phagosome for membrane anchorage as well as complex assembly. The product superoxide (O2−) and oxidant derivatives are shown in red font. (B) Enzymatic production of nitric oxide (·NO) via 5-electron oxidation of the guanidino nitrogen of l-arginine by inducible nitric oxide synthase 2. Downstream intermediates or congeners are shown in red font.
NADPH oxidase is composed of two membrane-associated (gp91phox, p22phox) and three cytosolic subunits (p67phox, p47phox, p40phox). IFN-γ increases gp91phox and p22phox mRNA transcription and promotes NADPH oxidase assembly on mycobacterial phagosomes (Kim et al. 2011). An IFN-γ-induced GTPase, guanylate binding protein 7 (Gbp7), facilitates this assembly by recruiting cytosolic p67phox-p47phox heterodimers to gp91phox-p22phox membrane complexes surrounding the pathogen compartment (Kim et al. 2011, 2012) (Fig. 2). Whether natural mutations in this or other members of the human GBP family induced during TB (Berry et al. 2010; Maertzdorf et al. 2011) are linked to disease is a topic for future investigation.
TNF-induced ROS has also recently emerged in zebrafish models of TB (Roca and Ramakrishnan 2013). RIP1-RIP3 signaling elicits microbicidal activity that if left unabated can induce macrophage necroptosis for subsequent dissemination of bacilli. Notably, the primary source of ROS in this setting was mitochondria (Roca and Ramakrishnan 2013). It will therefore be interesting to see if mitochondrial ROS plays a defining role against Mtb like its reported anti-Salmonella activities (West et al. 2011).
Nitrosative Stress
RNS are a second class of oxidants capable of potent tuberculocidal activity. As little as 90 ppm of nitric oxide (·NO) gas—approaching the exhaled concentrations found for some TB patients (Wang et al. 1998)—kills >99% of plated organisms (Long et al. 1999). ·NO shows molar potencies approaching many of the current antibiotics used to treat TB, and the tuberculocidal activity of some new drugs like bicyclic nitroimidazoles are ascribed to release of ·NO (Singh et al. 2008). Because it is a water- and lipid-soluble diatomic gas, ·NO traverses biologic membranes to react with O2− or other ROS to yield stable nontoxic anions (NO2−, NO3−) as well as intermediates with marked bactericidal properties. The latter include unstable dinitrogen oxides (N2O3, N2O4), compound peroxides (ONOO−), and nitrosothiol adducts (RSNO) (Yu et al. 1999; Venketaraman et al. 2005) (Fig. 2).
Within acidified phagolysosomes, ·NO equivalents can be retrieved from stably oxidized forms (e.g., NO2−) by protonation to HNO2 and subsequent dismutation (Nathan and Shiloh 2000) (Fig. 2). Indeed, acidified NaNO2 was one of the earliest compounds used to show a tuberculostatic role for ·NO (Chan et al. 1992). Along with other RNS, ROS, and hypoxia, ·NO is sensed by Mtb to mobilize the dormancy regulon, underscoring the environmental pressure applied by both nitrosative and oxidative stress within tissue granulomas (Schnappinger et al. 2003; Voskuil et al. 2003). RNS-mediated growth arrest may also involve nitrogenous products from Mtb itself: Reduction of nitrate NO3− by the bacterial narG system yields nitrite (NO2−) that inhibits ATP consumption and induces a transcriptomic profile distinct from the ·NO dormancy regulon in human macrophages (Cunningham-Bussel et al. 2013).
The chief phagocytic source of RNS is the dimeric flavoenzyme, NOS2, which catalyzes the 5e− oxidation of l-arginine to l-citrulline plus ·NO (MacMicking et al. 1997a) (Fig. 2). Robust NOS2 expression is elicited via cytokine (notably IFN-γ, TNF-α, IL-1β) and microbial-based PRR signals (e.g., lipopeptides), although its induction in human monocytes may be more complex. Some human cells require at least three combined stimuli—IFN-γ, TNF-α, and IL-1β—to produce a relatively weak response caused by STAT1 and NF-κB acting at some distance (∼5–8 Kb) from the transcriptional start site (Ganster et al. 2001). Limited availability of the NOS2 cofactor, BH4, may also contribute to the generally lower RNS production of human versus rodent phagocytes in cell culture (Bertholet et al. 1999).
Priming signals or cofactors that may be missing in vitro nonetheless appear present in vivo because robust NOS2 expression within Mtb granulomas is observed and macrophages isolated from inflamed lungs or peripheral blood of TB patients, or AMs from healthy donors subsequently infected with Mtbex vivo, produce mycobactericidal amounts of ·NO (Nicholson et al. 1996; Fachetti et al. 1999; Dlugovitzky et al. 2000; Means et al. 2001; Wang et al. 2001; Choi et al. 2002; Mattila et al. 2013). Thus NOS2 is likely to be active under native TB settings. Other evidence supporting an in vivo role for RNS includes NOS2 loss-of-function mutations or use of NOS inhibitors that lead to acute susceptibility in experimental mouse and zebrafish models (Chan et al. 1995; MacMicking et al. 1997b, 2003; Flynn et al. 1998; Mogues et al. 2001; Scanga et al. 2001; Cambier et al. 2014). Finally, the acquisition of Mtb resistance proteins, including NoxR1, NoxR3, alkyl hydroperoxide reductase (AhpC), peptidyl methionine sulphoxide reductase (MsrA), dihydrolipoamide dehydrogenase (Lpd), and dihydrolipoamide succinyl-transferase (SucB), also suggest RNS have impacted the evolutionary course of TB (Nathan and Shiloh 2000; St. John et al. 2001; Bryk et al. 2002; Nathan 2012).
Acid Exposure and Lysosomal Killing
Acid as an antimicrobial defense was first proposed by Metchnikoff using litmus dyes on infected guinea pig macrophages (Metchnikoff 1905) and gained credence from two early observations: (1) that activated phagocytes acidify their phagolysosomes (Rous 1925), and (2) that achlorhydric hosts permit colonization of the stomach (Giannella et al. 1973).
For Mtb, additional evidence came from its marked sensitivity to pH < 6.5 in axenic media with growth arrest at pH 5.0, especially when examined under hypoxic or microaerophilic conditions (Chapman and Bernard 1962; Tan et al. 2010). Whether protonation itself or other components (e.g., low divalent cation concentrations) contribute to this axenic attenuation is unknown (Jackett et al. 1978; Chan et al. 1992; Piddington et al. 2000; Vandal et al. 2008). Mtb has evolved periplasmic mechanisms such as Rv3176c encoding a serine protease (Vandal et al. 2008) and putative transporters that could exclude protons (H+) acting directly on the bacterium (Pethe et al. 2004). Even so, because low pH embellishes both ROS and RNS (Fig. 2) coupled with the fact that lysosomal hydrolyses have pH optima in this range, acidification probably facilitates other tuberculocidal effectors to mediate killing inside the (auto)phagolysosomal compartment (Rohde et al. 2007).
IFN-γ- or TLR-stimulated macrophages rapidly acidify mycobacterial phagolysosomes to a pH of ∼4.5–5.5, where it remains for at least 24 h (Schaible et al.1998; MacMicking et al. 2003; Vandal et al. 2008). Sustained acidification requires proton-pumping V-ATPases and enables processing of lysosomal cathepsins; both correlate with diminished Mtb growth (Schaible et al. 1998; Gomes et al. 1999; Ullrich et al. 1999; MacMicking et al. 2003). Agents that inhibit the V-ATPase (e.g., omeprezole) increase Mtb replication in human AMs and murine bone marrow–derived macrophages (Suzuki et al. 2000; MacMicking et al. 2003). Notably, the low luminal pH within phagolysosomes approaches the pKa of nitrous acid (3.8), which may allow dismutation to ·NO and nitrogen dioxide (NO2) as a way of reusing protonated nitrite (Vandal et al. 2009) (Fig. 2). This intraluminal RNS also overcomes phagosomal arrest normally imposed by Mtb and envelope lipids like trehalose dimycolate (Axelrod et al. 2008). Thus acidification cooperates with RNS to target intravacuolar Mtb. Low pH also promotes the activity of lysosomal proteases, lipases, glycosidases, and antimicrobial peptides—estimated at more than 200 in all (Trost et al. 2009)—that may disrupt and digest Mtb once it is delivered to this compartment.
Immune and Nonimmune GTPases
Delivery of Mtb to (auto)lysosomes relies on vesicular trafficking by immune and nonimmune GTPases. Members of the immunity-related GTPase (IRG) and GBP families orchestrate assembly of fusogenic SNARE proteins and autophagy components on mycobacterial phagosomes in IFN-γ-activated macrophages (MacMicking et al. 2003; Gutierrez et al. 2004; Singh et al. 2006; Tiwari et al. 2009; Kim et al. 2011, 2012a) (Fig. 3). Irgm1 targets the nascent phagocytic cup surrounding mycobacteria through carboxy-terminal amphipathic helical interactions with PI3,4,5P3 and PI3,4P2 generated by host lipid kinases on the plasma membrane (Tiwari et al. 2009). Once recruited, Irgm1 binds the SNARE and dynein motor adaptor, Snapin, for ATG14L-mediated endolysosomal fusion (MacMicking et al. 2003; Tiwari et al. 2009; Kim et al. 2012b). Human IRGM similarly facilitates trafficking of the autophagic machinery to Mtb vacuoles for lysosomal fusion and acidification (Singh et al. 2006). Such cell-autonomous activities also manifest in vivo: Irgm1−/− mice are profoundly susceptible to Mtb infection (MacMicking et al. 2003; Feng et al. 2004) (Fig. 3), and IRGM polymorphic variants associate with human TB in Ghanese, African American, Korean, and Chinese populations (Intemann et al. 2009; Che et al. 2010; King et al. 2011; Song et al. 2014).
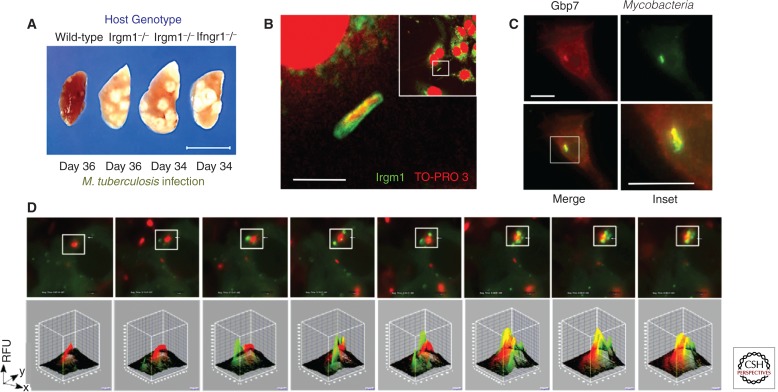
Figure 3.
IFN-γ- and TLR-induced immune GTPases involved in autophagic trafficking of Mtb and antimicrobial cargo to lysosomes. (A) Deletion of the Irgm1 gene leads to profound TB susceptibility after aerogenic challenge (MacMicking et al. 2003). Scale bar, 1.0 cm. (B) Targeting of endogenous Irgm1 to mycobacterial phagosomes in IFN-γ-activated macrophages. TO-PRO-3 detects nuclear and bacterial DNA. Original inset for higher magnification shown. Scale bar, 2 µm. (C) Recruitment of the 65-kDa guanylate binding protein, Gbp7, to mycobacterial phagosomes in IFN-γ-activated macrophages. Scale bar, 5 µm. Gbp7 helps recruit p67phox to assemble the phagocyte oxidase complex in this location (Kim et al. 2011). (D) Live imaging of Gbp7 vesicle recruitment (green) to internalized mycobacteria (red) over a 30.8-min data collection period. Image J software reveals fluorescence convergence within the chosen inset area (Kim et al. 2011).
For the GBPs, Gbp7 delivers antitubercular cargo in the form of NADPH oxidase to mycobacterial phagosomes for local ROS production (Kim et al. 2011) (Fig. 3). In addition, Gbp1 interacts with SQSTM1/p62 for transporting substrates that generate tuberculocidal peptides within lysosomes and Gbp1−/− mice are vulnerable to infection (Alonso et al. 2007; Kim et al. 2011). Last, RAB family proteins such as Rab20 recruit the Rab5 exchange factor Rabex-5 for transition to late endosomes in IFN- γ-activated macrophages infected with mycobacteria (Pei et al. 2014), and Rab8 directs TBK1-mediated autophagic clearance (Pilli et al. 2012). Together these different GTPase classes orchestrate lysosomal killing of Mtb following immune activation of host cells.
Ubiquitination and Autophagic Clearance
A second route to lysosomal degradation involves the detection of cytosolically exposed Mtb and subsequent sequestration by autophagy (Stanley and Cox 2013) (Fig. 4). It has been known for some time that Mtb communicates with the cytosol (Myrvik et al. 1984; McDonough et al. 1993). Access to or escape into this compartment departs from the historical view of bacilli being exclusively phagosome-bound inside human macrophages (Armstrong and Hart 1971). In fact both fates probably operate within a given cell where most Mtb destined for lysosomal killing remain “trapped” inside vacuoles, whereas a smaller number (12%–25%) may enter the cytosol, most notably after several days of infection (see Stanley and Cox 2013 and references therein).
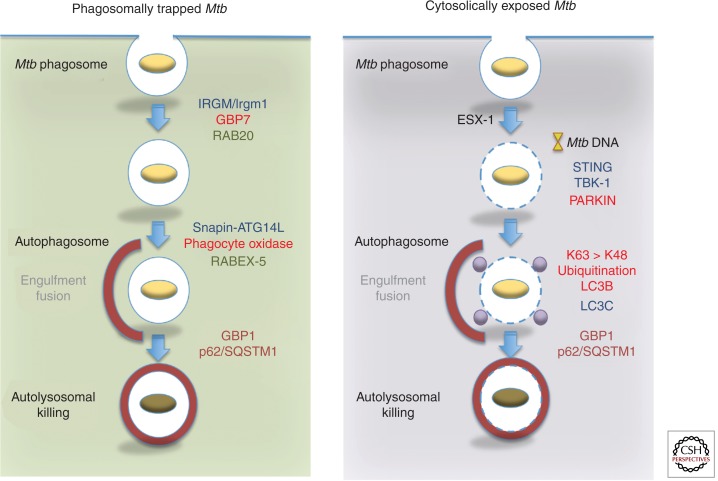
Figure 4.
Differential recruitment of effectors to “trapped” and “escaped” Mtb in immunologically activated macrophages. (Left) Delivery of phagosomal bacilli to autolysosomes after autophagic capture requires both immune and nonimmune GTPases. Their position in the pathway and corresponding interacting partners are color matched. (Right) Cytosolically exposed bacilli following permeabilization of the phagosomal membrane by ESX-1 releases DNA for detection by STING. This recruits TBK-1 and probably LC3C for Mtb capture. The E3 ligase PARKIN also ubiquitinates Mtb directly or its surrounding membrane remnants for detection by the autophagic pathway. A preferential requirement for K63 over K48 ubiquitin linkage is reported (Manzanillo et al. 2013).
Rupture or permeabilization of phagosomal membranes appears to rely on the type VII secretion system ESX-1 as first shown by examining Mtb RD1 mutants via electron microscopy (van der Wel et al. 2007; Houben et al. 2012). Permeabilization leads to mycobacterial DNA leakage detected by cytosolic surveillance pathways including the DNA and cyclic dinucleotide sensor, STING (Manzinillo et al. 2012; Watson et al. 2012). STING activation initiates a cascade involving TBK1 that engages ubiquitin-binding receptors downstream (Watson et al. 2012) (Fig. 4). Cytosolic mycobacteria are ubiquitinated primarily on K63 chains for detection by p62/SQSTM1 and NDP52, both of which contain LC3-interacting regions (LIRs) for binding to the autophagic membrane proteins, LC3B and LC3C, respectively (von Muhlinen et al. 2012; Watson et al. 2012).
The E3 ligase, PARKIN, mediates ubiquitination and recruitment of downstream Ub receptors (Fig. 4) and PARK2-deficient mice are moderately susceptible to Mtb challenge (Manzanillo et al. 2013). This differs from the highly susceptible phenotype of Atg5−/− animals, suggesting additional roles for some autophagy-related proteins (Watson et al. 2012). In this respect, SQSTM1/p62 also participates in delivering unbiquitinated substrates (e.g., ribosomal subunits) to autolysosomes for generating tuberculocidal peptides in IFN-γ-activated macrophages (Alonso et al. 2007; Ponpuak et al. 2010), a process that may require interaction with GBP1 (Kim et al. 2011). A human IRGMd isoform likewise induces mitochondrial Bax/Bak-dependent cell death during mycobacterial infection as an IFN-γ-induced mechanism distinct from its autophagic activities (Singh et al. 2010). Hence, unconventional roles may be subsumed by both autophagy-related and ubiquitin-binding proteins to confer antitubercular defense.
Antimicrobial Peptides
Antimicrobial peptides (AMPs) are evolutionarily ancient constituents of lysosomes, secretory granules, and inflammatory exudates (Ganz 2003; Zanetti 2004; Lai and Gallo 2009). At least 1700 AMPs exist in metazoans and unicellular organisms which show considerable sequence diversity (http://aps.unmc.edu/AP/). Nonetheless, AMPs share certain structural features, namely, an ∼15–30 amino acid amphipathic core composed largely of cationic and hydrophobic residues. Both features likely contribute to mycobacterial killing. Electrostatic interactions between positively charged amino acids and negatively charged Mtb cell wall phospholipids may provide initial contact, whereas hydrophobic residues integrate into the bacterial membrane for disruption (Ganz 2003). These features—size, amino acid composition, helical or extended loop conformations, and disulfide linkage—help classify the lytic intracellular AMPs into three major categories: α-defensins, β-defensins, and cathelicidins (Ganz 2003; Zanetti 2004; Lai and Gallo 2009).
More than 50 mammalian α-defensins, 90 β-defensins, and 30 cathelicidins are known. To date, evidence exists for β-defensins and cathelicidins in macrophage immunity to Mtb although neutrophil granules also kill bacilli via uncharacterized peptides (Tan et al. 2006). IFN-γ elicits β-defensin 4 and the single processed cathelicidin peptide, hCAP18/LL-37, in human macrophages to promote 1,25-dihydroxyvitamin D3-dependent autophagic killing of tubercle bacilli (Fabri et al. 2011). In mice, IFN-γ elicits ubiquicidin-like peptides, which, when isolated from macrophage lysosomal fractions or synthetically reconstituted, kill Mtb at low micrometer concentrations (Alonso et al. 2007). Like the AMPs, Ub-like peptides (e.g., Ub2) have been shown to insert into the bacterial membrane to disrupt integrity and equilibrate the transmembrane potential (Foss et al. 2012).
Nutritional Immunity: Competition for Cations
Macrophages and other phagocytic cells constitute rich nutritional sources of amino acids, lipids, sugars, and transition metals. Competition for cations is one of the main contests fought between Mtb and its host cell with the pathogen coming well-equipped in terms of siderophores (e.g., mycobactins, exochelins) that sequester transition metals like iron (Schaible and Kaufmann 2004). Divalent zinc (Zn2+), manganese (Mn2+), magnesium (Mg2+), and copper (Cu2+) are other cations nutritionally contested within phagocytes.
Iron
Iron is essential for Mtb growth, although the labile pool of free ferrous form (Fe2+) is low given that most Fe is complexed with transferrin, ferritin, lactoferrin, and other hemoproteins inside host cells (Schaible and Kaufmann 2004). Macrophage iron uptake proceeds via hemoglobin scavenger receptors (CD163) as well as capture via transferrin receptors (TFs) that internalize two Fe3+ moieties as part of the holoTF complex. Release of Fe3+ occurs in the endosome with which the Mtb phagosome intersects, thus allowing bacterial access to iron (Sturgill-Koszycki et al. 1996). IFN-γ-activated macrophages limit this availability by down-regulating TF expression and preventing accumulation of Fe2+ in its saturated storage form (as Fe3+-holoferritin complex) within the cytosol (Byrd and Horwitz 1993; Schaible et al. 1998).
Phagosomal Fe2+ (as well as Mn2+, Mg2+, and Zn2+) is concomittently lowered by approximately two- to sixfold in IFN-γ-activated macrophages infected with Mtb (Wagner et al. 2005), a reduction due in part to the proton-dependent Mn2+ and Fe2+ efflux pump, NRAMP1 (encoded by SLC11A1; Jabado et al. 2000). Nramp1 was originally identified via positional cloning in inbred mouse strains harboring G169A variants of the Bcg/Ity/Lsh allele (denoting Mycobacterium bovis BCG, Salmonella typhimurium, and Leishmania donovani susceptibility), and NRAMP1 polymorphisms are thought to predispose to human TB as well (see Fortin et al. 2007). NRAMP1 is a 12-transmembrane spanning protein found on lysosomes, tertiary granules (neutrophils), and phagosomes, where it aids mycobacterial killing (Jabado et al. 2000; Fortin et al. 2007). Genetic studies infer cationic competition between NRAMP1 and an Mtb homolog, Mramp, because Mn2+ and Fe2+ concentrations are elevated in phagosomes harboring Mramp-deficient Mtb (Wagner et al. 2005). Phagosomal recruitment of the Fe export protein ferriportin-1 also reduces intravacuolar iron after IFN-γ activation (Van Zandt et al. 2008), whereas lipocalin-2 directly sequesters iron to inhibit Mtb in macrophages, neutrophils, and airway epithelium (Martineau et al. 2007; Saiga et al. 2008; Johnson et al. 2010).
Other Cations
Magnesium, zinc, and copper are likewise sensed or used by Mtb; however, as with iron, high concentrations of these transitional metals are directly toxic or promote tuberculocidal ROS (Russell 2011; Samanovic et al. 2012). A requirement for Mg2+ in Mtb replication was identified under low cation and pH conditions akin to those found in the phagolysosome where bacterial growth was impaired (Piddington et al. 2000). This finding was reinforced by Mtb mutants lacking the mgtC (Mg2+ transporter) gene that perform poorly under the same conditions and are highly attenuated in human macrophages and mice (Buchmeier et al. 2000). For Zn2+, intracellular stores are pumped into Mtb phagosomes via a ROS-dependent pathway to promote killing in TLR-activated macrophages (Botella et al. 2011). Here Mtb attempts to counter Zn2+ toxicity by expressing a P-type ATPase efflux transporter encoded in the ctpC locus (Botella et al. 2011). For Cu2+, a host P-type ATPase pump ATP7A imports Cu2+ into the phagosome for generating intraluminal OH· radicals, which is countered to some extent by Mtb transport proteins CtpV and MctB (Ward et al. 2010; Wolschendorf et al. 2011; Samanovic et al. 2012). Divalent cations are thus selectively furnished or withheld to limit Mtb growth.
Nutritional Immunity: Carbon and Amino Acid Metabolism
ICLs and IRG1
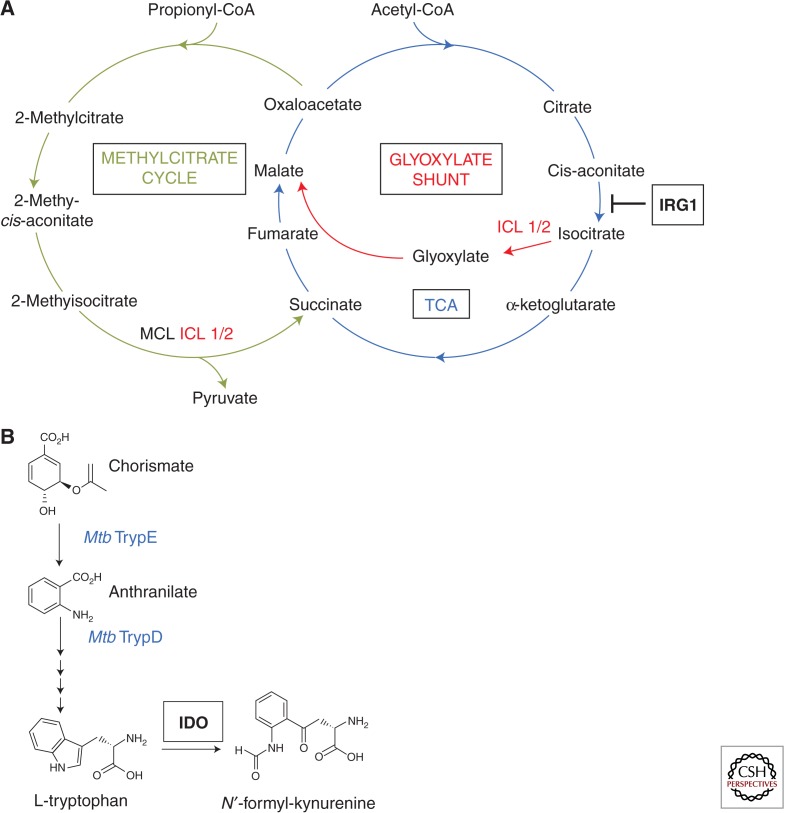
Carbon catabolism is critical for Mtb survival and persistence (Muñoz-Elías and McKinney 2006). Conserving C2 carbon during periods of nutrient restriction via anaplerosis helps maintain bacterial metabolism fueled by fatty acids. One metabolic target for cell-autonomous immunity is the Mtb glyoxylate shunt pathway that replenishes tricarboxylic acid (TCA) cycle intermediates normally diverted for biosynthetic purposes via successive carboxylation reactions (McKinney et al. 2000). Early work discovered a glycoxylate shunt enzyme, isocitrate lyase 1 (ICL1), was essential for Mtb persistence in IFN-γ-activated macrophages and mice (McKinney et al. 2000). Subsequent discovery of a second functional ICL isoform (ICL2) corroborates the strong metabolic pressure placed by cell-autonomous immunity on Mtb to use anaplerotic pathways (Muñoz-Elías and McKinney 2005).
The immune-responsive gene 1 (IRG1) has recently been identified as exerting part of that metabolic pressure in activated human and mouse macrophages (Michelucci et al. 2013) (Fig. 5). IRG1 is transcriptionally induced by autocrine IFN/STAT1 signaling in response to both virulent and environmental mycobacteria (Shi et al. 2005; Basler et al. 2006; Michelucci et al. 2013). Thereafter it localizes to mitochondria (DeGrandi et al. 2009), where it decarboxylates the TCA intermediate cis-aconitate needed for isocitrate synthesis, thus depriving ICL1 and ICL2 of their substrate to run the glyoxylate shunt (Michelucci et al. 2013) (Fig. 5). Decarboxylation of cis-aconitate also produces itaconic acid (alternatively known as methylenesuccinic acid) that may be directly toxic to bacilli (Michelucci et al. 2013). IRG1 could additionally promote β-oxidation-dependent mitochondrial ROS production for some of its bactericidal activities (Hall et al. 2013; Roca and Ramakrishnan 2013).
Figure 5.
Nutritional immunity to Mtb. (A) Depletion of tricarboxylic acid (TCA) cycle and potential glyoxylate shunt substrates via a cis-aconitate decarboxylase known as immunoresponsive gene-1 (IRG1) that resides in mitochondria. Position at which IRG1 blocks anaplerotic substrate production for Mtb is shown. Isocitrate lyases 1 and 2 (ICL1/2) that serve as important Mtb anaplerotic enzymes in the glycoxylate shunt and that are the functional Mtb paralogs of methylcitrate lyases (MCLs) in the methylcitrate pathway (Muñoz-Elias et al. 2006) are also depicted. (Adapted from data in Michelucci et al. 2013.) (B) Tryptophan depletion by the IFN-γ-induced enzyme, indolamine-2,3-dioxygenase (IDO), in activated macrophages. Mtb genes encoding the enzymes known to be involved in generating precursors of tryptophan biosynthesis are shown in blue font.
IDO
Other effector mechanisms target the production of amino acids catabolized or assimilated by Mtb for its growth (Zhang and Rubin 2013). Tryptophan degradation is an effective defense against obligate bacterial pathogens like Chlamydia and protozoan parasites including Toxoplasma and Leishmania spp. (MacMicking 2012). IFN-γ induces indoleamine-2,3-dioxygenase (IDO), a heme-containing oxidoreductase responsible for the initial rate-limiting step of the kynurenine pathway in which it degrades l-tryptophan to generate N-formyl-kynurenine (MacMicking 2012) (Fig. 5).
Although most laboratory Mtb strains are not tryptophan auxotrophs and IDO inhibition does not impact their growth within IFN-γ-activated macrophages (MacMicking et al. 2003), recent use of Mtb Trp− mutants has revealed tryptophan degradation by the host does exert metabolic pressure on Mtb in macrophages and infected mice (Zhang et al. 2013). Mtb synthesizes l-tryptophan via an operon containing TrpE, encoding an enzyme that converts chorismate to anthranilate, and TrpD, which encodes a ribosylating enzyme for the substrate (Zhang et al. 2013) (Fig. 5). Deletion of TrpE greatly attenuates Mtb growth in IFN-γ-activated macrophages that is restored in IDO-1-deficient cells; a similar outcome occurs in vivo. Moreover, fluorinated anthranilate (5-FABA and 6-FABA) inhibitors that block Mtb tryptophan synthesis render the bacillus susceptible to IDO-mediated killing. Thus drugs targeting bacterial counterimmune mechanisms allow the corresponding host effector to regain functionality inside phagocytic cells.
CONCLUDING REMARKS
This perspective has outlined the major cell-autonomous effectors deployed against Mtb. Because apoptosis and autophagy are covered extensively elsewhere in this collection, they have been given limited coverage or omitted altogether in this particular article. Furthermore, the list of effectors is by no means complete. Cell-autonomous immunity typically uses all the tools at its disposal—proteins, peptides, nucleotides, cations, metabolites, and physiological gases—to defend the host against infection. Hence, it can be anticipated that new pathways and effector molecules will emerge with antitubercular defense functions. IFN-γ- or PRR-independent pathways like GM-CSF-mediated killing (Rothchild et al. 2014) and unconventional activities such as mitochondrial immunity (via ROS, IRGM1, and IRG1) or autophagic engulfment of cytosolically exposed bacilli will be fertile areas of future investigation. Once understood, these novel defense factors may join established mechanisms as a platform from which to design host-directed therapies or adjunctive treatments that more fully expose Mtb to killing by the human immune system (Nathan 2012; Hawn et al. 2013; Wilkinson 2014). Both offer hope for shortening treatment as well as for tackling the problem of bacterial multidrug resistance (Singh et al. 2008; Zhang et al. 2013; Stanley et al. 2014).
ACKNOWLEDGMENTS
The author acknowledges colleagues inside and outside of the laboratory plus the following funding sources for work highlighted in this perspective: National Institutes of Health/National Institute of Allergy and Infectious Diseases (R01 AI068041–07), BWF Investigator in Pathogenesis of Infectious Disease Award (1007845), Cancer Research Institute Investigator Award Program (CRI06–10), Searle Foundation (05-F-114), and W.W. Winchester Award.
REFERENCES
- Adams LB, Dinauer MC, Morgenstern DE, Krahenbuhl JL. 1997. Comparison of the roles of reactive oxygen and nitrogen intermediates in the host response to Mycobacterium tuberculosis using transgenic mice. Tuber Lung Dis 78: 237–246 [DOI] [PubMed] [Google Scholar]
- Alonso S, Pethe K, Russell DG, Purdy GE. 2007. Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc Natl Acad Sci 104: 6031–6036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Areeshi MY, Mandal RK, Panda AK, Haque S. 2013. Association of P2X7 A1513C (rs3751143) gene polymorphism with risk of tuberculosis: Evidence from a meta-analysis. Genet Test Mol Biomarkers 17: 662–668 [DOI] [PubMed] [Google Scholar]
- Armstrong JA, Hart PD. 1971. Response of cultured macrophages to Mycobacterium tuberculosis, with observations on fusion of lysosomes with phagosomes. J Exp Med 134: 713–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod S, Oschkinat H, Enders J, Schlegel B, Brinkmann V, Kaufmann SH, Haas A, Schaible UE. 2008. Delay of phagosome maturation by a mycobacterial lipid is reversed by nitric oxide. Cell Microbiol 10: 1530–1545 [DOI] [PubMed] [Google Scholar]
- Basler T, Jeckstadt S, Valentin-Weigand P, Goethe R. 2006. Mycobacterium paratuberculosis, Mycobacterium smegmatis, and lipopolysaccharide induce different transcriptional and post-transcriptional regulation of the IRG1 gene in murine macrophages. J Leukoc Biol 79: 628–638 [DOI] [PubMed] [Google Scholar]
- Bean AG, Roach DR, Briscoe H, France MP, Korner H, Sedgewick JD, Britton WJ. 1999. Structural deficiencies in granuloma formation in TNF gene-targeted mice underlie the heightened susceptibility to aerosol Mycobacterium tuberculosis infection, which is not compensated for by lymphotoxin. J Immunol 162: 3504–3511 [PubMed] [Google Scholar]
- Bekker LG, Freeman S, Murray PJ, Ryffel B, Kaplan G. 2001. TNF-α controls intracellular mycobacterial growth by both inducible nitric oxide synthase-dependent and inducible nitric oxide synthase-independent pathways. J Immunol 166: 6728–6734 [DOI] [PubMed] [Google Scholar]
- Bellamy R, Ruwende C, Corrah R, McAdam KP, Whittle HC, Hill AV. 1999. Tuberculosis and chronic hepatitis B virus infection in Africans and variation in the vitamin D receptor gene. J Infect Dis 179: 721–724 [DOI] [PubMed] [Google Scholar]
- Bertholet S, Tzeng E, Felley-Bosco E, Mauel J. 1999. Expression of inducible nitric oxide synthase in human monocytic U937 cells allows high output nitric oxide synthase. J Leukoc Biol 65: 50–58 [DOI] [PubMed] [Google Scholar]
- Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T, Wilkinson KA, Banchereau R, Skinner J, Wilkinson RJ, et al. 2010. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 466: 973–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz BE, Azad AK, Morris JD, Rajaram MV, Schlesinger LS. 2011. β-Glucans inhibit intracellular growth of Mycobacterium bovis BCG but not virulent Mycobacterium tuberculosis in human macrophages. Microb Pathog 51: 233–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler B, Jiang Z, Georgel P, Crozat K, Croker B, Rutschmann S, Du X, Hoebe K. 2006. Genetic analysis of host resistance: Toll-like receptor signaling and immunity at large. Annu Rev Immunol 24: 353–389 [DOI] [PubMed] [Google Scholar]
- Blumenthal A, Nagalingam G, Huch JH, Walker L, Guillemin GJ, Smythe GA, Ehrt S, Britton WJ, Saunders BM. 2012. M. tuberculosis induces potent activation of IDO-1, but this is not essential for the immunological control of infection. PLoS ONE 7: e37314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdan C. 2001. Nitric oxide and the immune response. Nat Immunol 2: 907–916 [DOI] [PubMed] [Google Scholar]
- Bogunovic D, Byun M, Durfee LA, Abhyankar A, Sanal O, Mansouri D, Salem S, Radovanovic I, Grant AV, Adimi P, et al. 2012. Mycobacterial disease and impaired IFN-γ immunity in humans with inherited ISG15 deficiency. Science 337: 1684–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonecini-Almeida MG, Chitale S, Boutsikakis I, Geng J, Doo H, He S, Ho JL. 1998. Induction of in vitro human macrophage anti-Mycobacterium tuberculosis activity: Requirement for IFN-γ and primed lymphocytes. J Immunol 160: 4490–4499 [PubMed] [Google Scholar]
- Botella H, Peyron P, Levillain F, Poincloux R, Poquet Y, Brandli I, Wang C, Tailleux L, Tilleul S, Charrière GM, et al. 2011. Mycobacterial P1-type ATPases mediate resistance to zinc poisoning in human macrophages. Cell Host Microbe 10: 248–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk R, Lima CD, Erdjument-Bromage H, Tempst P, Nathan C. 2002. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science 295: 1073–1077 [DOI] [PubMed] [Google Scholar]
- Byrd TF, Horwitz MA. 1993. Regulation of transferrin receptor expression and ferritin content in human mononuclear phagocytes. Coordinate upregulation by iron transferrin and downregulation by interferon γ. J Clin Invest 91: 969–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmeier N, Blanc-Polard A, Erht S, Piddington D, Riley L, Groisman EA. 2000. A parallel intraphagosomal survival strategy shared by Mycobacterium tuberculosis and Salmonella enterica. Mol Microbiol 35: 1375–1382 [DOI] [PubMed] [Google Scholar]
- Bustamante J, Arias AA, Vogt G, Picard C, Galicia LB, Prando C, Grant AV, Marchal CC, Hubeau M, Chapgier A, et al. 2011. Germline CYBB mutations that selectively affect macrophages in kindreds with X-linked predisposition to tuberculous mycobacterial disease. Nat Immunol 12: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambier CJ, Takaki KK, Larson RP, Hernandez RE, Tobin DM, Urdahl KB, Cosma CL, Ramakrishnan L. 2014. Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature 505: 218–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova J-L, Abel L. 2002. Genetic dissection of immunity to mycobacteria: The human model. Ann Rev Immunol 20: 581–620 [DOI] [PubMed] [Google Scholar]
- Chan J, Xing Y, Magliozzo RS, Bloom BR. 1992. Killing of virulent Mycobacterium tuberculosis by reactive nitrogen intermediates produced by activated murine macrophages. J Exp Med 175: 1111–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J, Tanaka K, Carroll D, Flynn J, Bloom BR. 1995. Effects of nitric oxide synthase inhibitors on murine infection with Mycobacterium tuberculosis. Infect Immun 63: 736–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan ED, Morris KR, Belisle JT, Hill P, Remigio LK, Brennan PJ, Riches DW. 2001. Induction of inducible nitric oxide synthase-NO by lipoarabinomannan of Mycobacterium tuberculosis is mediated by MEK1-ERK, MKK7-JNK, and NFκB signaling pathways. Infect Immun 69: 2001–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J, Bernard J. 1962. The tolerances of unclassified mycobacteria. Am Rev Respir Dis 86: 582–583 [DOI] [PubMed] [Google Scholar]
- Che N, Li S, Gao T, Zhang Z, Han Y, Zhang X, Sun Y, Liu Y, Sun Z, Zhang J, et al. 2010. Identification of a novel IRGM promoter single nucleotide polymorphism associated with tuberculosis. Clin Chim Acta 411: 1645–1649 [DOI] [PubMed] [Google Scholar]
- Choi HS, Rai PR, Chu HW, Cool C, Chan ED. 2002. Analysis of nitric oxide synthase and nitrotyrosine expression in human pulmonary tuberculosis. Am J Respir Crit Care Med 166: 178–186 [DOI] [PubMed] [Google Scholar]
- Cobat A, Gallant CJ, Simkin L, Black GF, Stanley K, Hughes J, Doherty TM, Hanekom WA, Eley B, Jaïs JP, et al. 2009. Two loci control tuberculin skin test reactivity in an area hyperendemic for tuberculosis. J Exp Med 206: 2583–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas I, Coscolla M, Luo T, Borrell S, Holt KE, Kato-Maeda M, Parkhill J, Malla B, Berg S, Thwaites G, et al. 2013. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet 45: 1176–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condos R, Rom WN, Schluger NW. 1997. Treatment of multi-drug-resistant pulmonary tuberculosis with interferon-γ via aerosol. Lancet 349: 1513–1515 [DOI] [PubMed] [Google Scholar]
- Cooper AM, Dalton DK, Stewart TA, Griffin JP, Russell DG, Orme IM. 1993. Disseminated tuberculosis in interferon γ gene-disrupted mice. J Exp Med 178: 2243–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AM, Segal BH, Frank AA, Holland SM, Orme IM. 2000. Transient loss of resistance to pulmonary tuberculosis in p47phox−/− mice. Infect Immun 68: 1231–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe F, Divangahi M, Veyrier F, de Léséleuc L, Gleason JL, Yang Y, Kelliher MA, Pandey AK, Sassetti CM, Reed MB, et al. 2009. Increased NOD2-mediated recognition of N-glycolyl muramyl dipeptide. J Exp Med 206: 1709–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowle AJ, Ross EJ, May MH. 1987. Inhibition of 1,25(OH)2-vitamin D3 of the multiplication of virulent tubercle bacilli in cultured human macrophages. Infect Immun 55: 2945–2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham-Bussel A, Zhang T, Nathan CF. 2013. Nitrite produced by Mycobacterium tuberculosis in human macrophages in physiologic oxygen impacts bacterial ATP consumption and gene expression. Proc Natl Acad Sci 110: E4256–E4265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Koo MS, Kim BH, Jacob ST, Subbian S, Yao J, Leng L, Levy R, Murchison C, Burman WJ, et al. 2013. Macrophage migration inhibitory factor (MIF) is a critical mediator of the innate immune response to Mycobacterium tuberculosis. Proc Natl Acad Sci 110: E2997–E3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degrandi D, Hoffmann R, Beuter-Gunia C, Pfeffer K. 2009. The proinflammatory cytokine-induced IRG1 protein associates with mitochondria. J Interferon Cytokine Res 29: 55–67 [DOI] [PubMed] [Google Scholar]
- Dlugovitzky D, Bay ML, Rateni L, Fiorenza G, Vietti L, Farroni MA, Bottasso OA. 2000. Influence of disease severity on nitrite and cytokine production by peripheral blood monouclear cells (PBMC) from patients with pulmonary tuberculosis (TB). Clin Exp Immunol 122: 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund D, Welin A, Andersson H, Verma D, Söderkvist P, Stendahl O, Särndahl E, Lerm M. 2014. Human gene variants linked to enhanced NLRP3 activity limit intramacrophage growth of Mycobacterium tuberculosis. J Infect Dis 209: 749–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrt S, Schnappinger D, Bekiranov S, Drenkow J, Shi S, Gingeras TR, Gaasterland T, Schoolnik G, Nathan C. 2001. Reprogramming of the macrophage transcriptome in response to interferon-γ and Mycobacterium tuberculosis: Signaling roles of nitric oxide synthase-2 and phagocyte oxidase. J Exp Med 194: 1123–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabri M, Stenger S, Shin DM, Yuk JM, Liu PT, Realegeno S, Lee HM, Krutzik SR, Schenk M, Sieling PA, et al. 2011. Vitamin D is required for IFN-γ-mediated antimicrobial activity of human macrophages. Sci Transl Med 3: 104ra102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachetti F, Vermi W, Fiorentini S, Chilosi M, Caruso A, Duse M, Notarangelo LD, Badolato R. 1999. Expression of inducible nitric oxide synthase in human granulomas and histiocytic reactions. Am J Path 154: 145–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbairn IP, Stober CB, Kumararatne DS, Lammas DA. 2001. ATP-mediated killing of intracellular mycobacteria by macrophages is a P2X7-dependent process inducing bacterial death by phagosome-lysosome fusion. J Immunol 167: 3300–3307 [DOI] [PubMed] [Google Scholar]
- Feng CG, Collazo-Custodio CM, Eckhaus M, Hieny S, Belkaid Y, Elkins K, Jankovic D, Taylor GA, Sher A. 2004. Mice deficient in LRG-47 display increased susceptibility to mycobacterial infection associated with the induction of lymphopenia. J Immunol 172: 1163–1168 [DOI] [PubMed] [Google Scholar]
- Fernando SL, Saunders BM, Sluyter R, Skarratt KK, Goldberg H, Marks GB, Wiley JS, Britton WJ. 2007. A polymorphism in the P2X7 gene increases susceptibility to extrapulmonary tuberculosis. Am J Respir Crit Care Med 175: 360–366 [DOI] [PubMed] [Google Scholar]
- Finbloom DS, Hoover DL, Wahl LM. 1985. The characteristics of binding of human recombinant interferon-γ to its receptor on human monocytes and human monocyte-like lines. J Immunol 135: 300–305 [PubMed] [Google Scholar]
- Flesch I, Kaufmann SH. 1987. Mycobacterial growth inhibition by interferon-γ activated bone marrow macrophages and differential susceptibility among strains of Mycobacterium tuberculosis. J Immunol 138: 4408–4413 [PubMed] [Google Scholar]
- Flynn J, Chan J, Triebold K, Dalton D, Stewart T, Bloom B. 1993. An essential role for interferon γ in resistance to Mycobacterium tuberculosis infection. J Exp Med 178: 2249–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JL, Goldstein MM, Chan J, Triebold KJ, Pfeffer K, Lowenstein CJ, Schreiber R, Mak TW, Bloom BR. 1995. Tumor necrosis factor-α is required in the protective immune response against Mycobacterium tuberculosis in mice. Immunity 2: 561–572 [DOI] [PubMed] [Google Scholar]
- Flynn JL, Scanga CA, Tanaka KE, Chan J. 1998. Effects of aminoguanidine on latent murine tuberculosis. J Immunol 160: 1796–1803 [PubMed] [Google Scholar]
- Fortin A, Abel L, Casanova JL, Gros P. 2007. Host genetics of mycobacterial diseases in mice and men: Forward genetic studies of BCG-osis and tuberculosis. Annu Rev Genomics Hum Genet 8: 163–192 [DOI] [PubMed] [Google Scholar]
- Foss MH, Powers KM, Purdy GE. 2012. Structural and functional characterization of mycobactericidal ubiquitin-derived peptides in model and bacterial membranes. Biochemistry 51: 9922–9929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganster RW, Taylor BS, Shao L, Geller DA. 2001. Complex regulation of human inducible nitric oxide synthase gene transcription by Stat 1 and NF-κB. Proc Natl Acad Sci 98: 8638–8643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz T. 2003. Defensins: Antimicrobial peptides of innate immunity. Nat Rev Immunol 3: 710–720 [DOI] [PubMed] [Google Scholar]
- Giannella RA, Broitman SA, Zamcheck N. 1973. Influence of gastric acidity on bacterial and parasitic enteric infections. Ann Int Med 78: 271–276 [DOI] [PubMed] [Google Scholar]
- Gomes MS, Paul S, Moreira AL, Appelberg R, Rabinovitch M, Kaplan G. 1999. Survival of Mycobacterium avium and Mycobacterium tuberculosis in acidified vacuoles of murine macrophages. Infect Immun 67: 3199–3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. 2004. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119: 753–766 [DOI] [PubMed] [Google Scholar]
- Hall CJ, Boyle RH, Astin JW, Flores MV, Oehlers SH, Sanderson LE, Ellett F, Lieschke GJ, Crosier KE, Crosier PS. 2013. Immunoresponsive gene 1 augments bactericidal activity of macrophage-lineage cells by regulating β-oxidation-dependent mitochondrial ROS production. Cell Metab 18: 265–278 [DOI] [PubMed] [Google Scholar]
- Hambleton S, Salem S, Bustamante J, Bigley V, Boisson-Dupuis S, Azevedo J, Fortin A, Haniffa M, Ceron-Gutierrez L, Bacon CM, et al. 2011. IRF8 mutations and human dendritic-cell immunodeficiency. N Engl J Med 365: 127–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawn TR, Dunstan SJ, Thwaites GE, Simmons CP, Thuong NT, Lan NT, Quy HT, Chau TT, Hieu NT, Rodrigues S, et al. 2006. A polymorphism in Toll-interleukin 1 receptor domain containing adaptor protein is associated with susceptibility to meningeal tuberculosis. J Infect Dis 194: 1127–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawn TR, Matheson AI, Maley SN, Vandal O. 2013. Host-directed therapeutics for tuberculosis: Can we harness the host? Microbiol Mol Biol Rev 77: 608–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitmann L, Schoenen H, Ehlers S, Lang R, Hölscher C. 2013. Mincle is not essential for controlling Mycobacterium tuberculosis infection. Immunobiology 218: 506–516 [DOI] [PubMed] [Google Scholar]
- Houben D, Demangel C, van Ingen J, Perez J, Baldeón L, Abdallah AM, Caleechurn L, Bottai D, van Zon M, de Punder K, et al. 2012. ESX-1-mediated translocation to the cytosol controls virulence of mycobacteria. Cell Microbiol 14: 1287–1298 [DOI] [PubMed] [Google Scholar]
- Intemann CD, Thye T, Niemann S, Browne EN, Amanua Chinbuah M, Enimil A, Gyapong J, Osei I, Owusu-Dabo E, Helm S, et al. 2009. Autophagy gene variant IRGM -261T contributes to protection from tuberculosis caused by Mycobacterium tuberculosis but not by M. africanum strains. PLoS Pathog 5: e1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa E, Ishikawa T, Morita YS, Toyonaga K, Yamada H, Takeuchi O, Kinoshita T, Akira S, Yoshikai Y, Yamasaki S. 2009. Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin Mincle. J Exp Med 206: 2879–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabado N, Jankowski A, Dougaparsad S, Picard V, Grinstein S, Gros P. 2000. Natural resistance to intracellular infections: Natural resistance-associated macrophage protein 1 (Nramp1) functions as a pH-dependent manganese transporter at the phagosomal membrane. J Exp Med 192: 1237–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackett PS, Aber V, Lowrie DB. 1978. Virulence and resistance to superoxide, low pH and hydrogen peroxide among strains of Mycobacterium tuberculosis. J Gen Microbiol 107: 37–45 [DOI] [PubMed] [Google Scholar]
- Jayaraman P, Sada-Ovalle I, Nishimura T, Anderson AC, Kuchroo VK, Remold HG, Behar SM. 2013. IL-1β promotes antimicrobial immunity in macrophages by regulating TNFR signaling and caspase-3 activation. J Immunol 190: 4196–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EE, Srikanth CV, Sandgren A, Harrington L, Trebicka E, Wang L, Borregaard N, Murray M, Cherayil BJ. 2010. Siderocalin inhibits the intracellular replication of Mycobacterium tuberculosis in macrophages. FEMS Immunol Med Microbiol 58: 138–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juárez E, Carranza C, Hernández-Sánchez F, León-Contreras JC, Hernández-Pando R, Escobedo D, Torres M, Sada E. 2012. NOD2 enhances the innate response of alveolar macrophages to Mycobacterium tuberculosis in humans. Eur J Immunol 42: 880–889 [DOI] [PubMed] [Google Scholar]
- Kim BH, Shenoy AR, Kumar P, Das R, Tiwari S, MacMicking JD. 2011. A family of IFN-γ-inducible 65-kD GTPases protects against bacterial infection. Science 332: 717–721 [DOI] [PubMed] [Google Scholar]
- Kim BH, Shenoy AR, Kumar P, Bradfield CJ, MacMicking JD. 2012a. IFN-inducible GTPases in host cell defense. Cell Host Microbe 12: 432–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Zhong Q, Sheng ZH, Yoshimori T, Liang C, Jung JU. 2012b. Beclin-1-interacting autophagy protein Atg14L targets the SNARE-associated protein Snapin to coordinate endocytic trafficking. J Cell Sci 125: 4740–4750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King KY, Lew JD, Ha NP, Lin JS, Ma X, Graviss EA, Goodell MA. 2011. Polymorphic allele of human IRGM1 is associated with susceptibility to tuberculosis in African Americans. PLoS ONE 6: e16317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebanoff S. 1999. Oxygen metabolites from phagocytes. In Inflammation: Basic principles and clinical correlates, 3rd ed (ed. Gallin JI, Snyderman R, Fearon DT, et al. ), pp. 721–768. Lippincott Williams & Wilkins, Philadelphia [Google Scholar]
- Klug-Micu GM, Stenger S, Sommer A, Liu PT, Krutzik SR, Modlin RL, Fabri M. 2013. CD40 ligand and interferon-γ induce an antimicrobial response against Mycobacterium tuberculosis in human monocytes. Immunology 139: 121–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusner DJ, Adams J. 2000. ATP-induced killing of virulent Mycobacterium tuberculosis within human macrophages requires phospholipase D. J Immunol 164: 379–388 [DOI] [PubMed] [Google Scholar]
- Kusner DJ, Barton JA. 2001. ATP stimulates human macrophages to kill intracellular virulent Mycobacterium tuberculosis via calcium-dependent phagosome-lysosome fusion. J Immunol 167: 3308–3315 [DOI] [PubMed] [Google Scholar]
- Lai Y, Gallo RL. 2009. AMPed up immunity: How antimicrobial peptides have multiple roles in immune defense. Trends Immunol 30: 131–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau YL, Chan CF, Ha SY, Hui YF, Yuen KY. 1998. The role of phagocytic respiratory burst in host defense against Mycobacterium tuberculosis. Clin Infect Dis 26: 226–227 [DOI] [PubMed] [Google Scholar]
- Lee WL, Gold B, Darby C, Brot N, Jiang X, de Carvalho LP, Wellner D, St John G, Jacobs WR Jr, Nathan C. 2009. Mycobacterium tuberculosis expresses methionine sulphoxide reductases A and B that protect from killing by nitrite and hypochlorite. Mol Microbiol 71: 583–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PL, Ford CB, Coleman MT, Myers AJ, Gawande R, Ioerger T, Sacchettini J, Fortune SM, Flynn JL. 2014. Sterilization of granulomas is common in active and latent tuberculosis despite within-host variability in bacterial killing. Nat Med 20: 75–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PT, Stenger S, Li H, Wenzel L, Tan BH, Krutzik SR, Ochoa MT, Schauber J, Wu K, Meinken C, et al. 2006. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science 311: 1770–1773 [DOI] [PubMed] [Google Scholar]
- Liu PT, Stenger S, Tang DH, Modlin RL. 2007. Cutting edge: Vitamin D-mediated human antimicrobial activity against Mycobacterium tuberculosis is dependent on the induction of cathelicidin. J Immunol 179: 2060–2063 [DOI] [PubMed] [Google Scholar]
- Long R, Light B, Talbot JA. 1999. Mycobacteriocidal action of exogenous nitric oxide. Antimicrob Agents Chemother 43: 403–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurie MB. 1964. Resistance to tuberculosis: Experimental studies in native and acquired mechanisms. Harvard University Press, Cambridge, MA [Google Scholar]
- MacMicking JD. 2008. M. tuberculosis passes the litmus test. Nat Med 14: 809–810 [DOI] [PubMed] [Google Scholar]
- MacMicking JD. 2012. IFN-inducible effector mechanisms in cell-autonomous immunity. Nat Rev Immunol 12: 367–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMicking J, Xie Qw, Nathan C. 1997a. Nitric oxide and macrophage function. Ann Rev Immunol 15: 323–350 [DOI] [PubMed] [Google Scholar]
- MacMicking JD, North RJ, LaCourse R, Mudgett JS, Shah SK, Nathan CF. 1997b. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc Natl Acad Sci 94: 5243–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMicking JD, Taylor GA, McKinney JD. 2003. Immune control of tuberculosis by IFN-γ-inducible LRG-47. Science 302: 654–659 [DOI] [PubMed] [Google Scholar]
- Maertzdorf J, Repsilber D, Parida SK, Stanley K, Roberts T, Black G, Walzl G, Kaufmann SH. 2011. Human gene expression profiles of susceptibility and resistance in tuberculosis. Genes Immun 12: 15–22 [DOI] [PubMed] [Google Scholar]
- Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS. 2012. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe 11: 469–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanillo PS, Ayres JS, Watson RO, Collins AC, Souza G, Rae CS, Schneider DS, Nakamura K, Shiloh MU, Cox JS. 2013. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature 501: 512–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marakalala MJ, Guler R, Matika L, Murray G, Jacobs M, Brombacher F, Rothfuchs AG, Sher A, Brown GD. 2011. The Syk/CARD9-coupled receptor Dectin-1 is not required for host resistance to Mycobacterium tuberculosis in mice. Microbes Infect 13: 198–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martineau AR, Newton SM, Wilkinson KA, Kampmann B, Hall BM, Nawroly N, Packe GE, Davidson RN, Griffiths CJ, Wilkinson RJ. 2007. Neutrophil-mediated innate immune resistance to mycobacteria. J Clin Invest 117: 1894–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila JT, Ojo OO, Kepka-Lenhart D, Marino S, Kim JH, Eum SY, Via LE, Barry CE 3rd, Klein E, Kirschner DE, et al. 2013. Microenvironments in tuberculous granulomas are delineated by distinct populations of macrophage subsets and expression of nitric oxide synthase and arginase isoforms. J Immunol 191: 773–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer-Barber KD, Barber DL, Shenderov K, White SD, Wilson MS, Cheever A, Kugler D, Hieny S, Caspar P, Núñez G, et al. 2010. Caspase-1 independent IL-1β production is critical for host resistance to Mycobacterium tuberculosis and does not require TLR signaling in vivo. J Immunol 184: 3326–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough KA, Kress Y, Bloom BR. 1993. Pathogenesis of tuberculosis: Interaction of Mycobacterium tuberculosis with macrophages. Infect Immun 61: 2763–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney JD, Höner zu Bentrup K, Muñoz-Elías EJ, Miczak A, Chen B, Chan WT, Swenson D, Sacchettini JC, Jacobs WR Jr, Russell DG. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406: 735–738 [DOI] [PubMed] [Google Scholar]
- McKinney JD, Bloom BR, Modlin RL. 2001. Tuberculosis and leprosy. In Samter’s immunologic diseases, 6th ed (ed. Austen KF, Frank MM, Atkinson JP, et al. ), pp. 985–1002. Lippincott Williams & Wilkins, Philadelphia [Google Scholar]
- Means TK, Jones BW, Schromm AB, Shurtleff BA, Smith JA, Keane J, Golenbock DT, Vogel SN, Fenton MJ. 2001. Differential effects of a Toll-like receptor antagonist on Mycobacterium tuberculosis-induced macrophage responses. J Immunol 166: 4074–4082 [DOI] [PubMed] [Google Scholar]
- Metchnikoff E. 1905. Immunity in the infectious diseases. MacMillan, New York [Google Scholar]
- Michelucci A, Cordes T, Ghelfi J, Pailot A, Reiling N, Goldmann O, Binz T, Wegner A, Tallam A, Rausell A, et al. 2013. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc Natl Acad Sci 110: 7820–7825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogues T, Goodrich ME, Ryan L, LaCourse R, North RJ. 2001. The relative importance of T cell subsets in immunity and immunopathology of airborne Mycobacterium tuberculosis infection in mice. J Exp Med 193: 271–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Elías EJ, McKinney JD. 2005. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat Med 11: 638–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Elías EJ, McKinney JD. 2006. Carbon metabolism of intracellular bacteria. Cell Microbiol 8: 10–22 [DOI] [PubMed] [Google Scholar]
- Muñoz-Elias EJ, Upton AM, Cherian J, McKinney JD. 2006. Role of the methylcitrate cycle in Mycobacterium tuberculosis metabolism, intracellular growth, and virulance. Mol Microbiol 60: 1109–1122 [DOI] [PubMed] [Google Scholar]
- Myrvik QN, Leake ES, Wright MJ. 1984. Disruption of phagosomal membranes of normal alveolar macrophages by the H37Rv strain of Mycobacterium tuberculosis. A correlate of virulence. Am Rev Respir Dis 129: 322–328 [PubMed] [Google Scholar]
- Nathan C. 2006. Role of iNOS in human disease. Science 312: 1874–1875 [DOI] [PubMed] [Google Scholar]
- Nathan C. 2012. Fresh approaches to anti-infective therapies. Sci Transl Med 4: 140sr2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C, Shiloh MU. 2000. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc Natl Acad Sci 97: 8841–8848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan CF, Murray HW, Wiebe ME, Rubin BY. 1983. Identification of interferon-γ as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J Exp Med 158: 670–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan CF, Prendergast TJ, Wiebe ME, Stanley ER, Platzer E, Remold HG, Welte K, Rubin BY, Murray HW. 1984. Activation of human macrophages. Comparison of other cytokines with interferon-γ. J Exp Med 160: 600–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nau GJ, Richmond JF, Schlesinger A, Jennings EG, Lander ES, Young RA. 2002. Human macrophage activation programs induced by bacterial pathogens. Proc Natl Acad Sci 99: 1503–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng VH, Cox JS, Sousa AO, MacMicking JD, McKinney JD. 2004. Role of KatG catalase-peroxidase in mycobacterial pathogenesis: Countering the oxidative burst. Mol Microbiol 52: 1291–1302 [DOI] [PubMed] [Google Scholar]
- Nicholson S, Bonecini-Almeida Mda G, Lapa e Silva JR, Nathan C, Xie QW, Mumford R, Weidner JR, Calaycay J, Geng J, et al. 1996. Inducible nitric oxide synthase in pulmonary alveolar macrophages from patients with tuberculosis. J Exp Med 183: 2293–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace JL, Russell SW, Schreiber RD, Altman A, Katz DH. 1983. Macrophage activation: Priming activity from a T-cell hybridoma is attributable to interferon-γ. Proc Natl Acad Sci 80: 3782–3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei G, Repnik U, Griffiths G, Gutierrez MG. 2014. Identification of an immune regulated phagosomal Rab cascade in macrophages. J Cell Sci Feb 25. 10.1242/jcs.144923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pethe K, Swenson DL, Alonso S, Anderson J, Wang C, Russell DG. 2004. Isolation of Mycobacterium tuberculosis mutants defective in the arrest of phagosome maturation. Proc Natl Acad Sci 101: 13642–13647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddington DL, Kashkouli A, Buchmeier NA. 2000. Growth of Mycobacterium tuberculosis in a defined medium is very restricted by acid pH and Mg2+ levels. Infect Immun 68: 4518–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, Mandell MA, Dupont N, Ornatowski W, Jiang S, Bradfute SB, et al. 2012. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity 37: 223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podinovskaia M, Lee W, Caldwell S, Russell DG. 2013. Infection of macrophages with Mycobacterium tuberculosis induces global modifications to phagosomal function. Cell Microbiol 15: 589–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponpuak M, Davis AS, Roberts EA, Delgado MA, Dinkins C, Zhao Z, Virgin HW 4th, Kyei GB, Johansen T, Vergne I, et al. 2010. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity 32: 329–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualls JE, Subramanian C, Rafi W, Smith AM, Balouzian L, DeFreitas AA, Shirey KA, Reutterer B, Kernbauer E, Stockinger S, et al. 2012. Sustained generation of nitric oxide and control of mycobacterial infection requires argininosuccinate synthase 1. Cell Host Microbe 12: 313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragno S, Romano M, Howell S, Pappin DJ, Jenner PJ, Colston MJ. 2001. Changes in gene expression in macrophages infected with Mycobacterium tuberculosis: A combined transcriptomic and proteomic approach. Immunology 104: 99–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randow F, MacMicking JD, James LC. 2013. Cellular self-defense: How cell-autonomous immunity protects against pathogens. Science 340: 701–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca FJ, Ramakrishnan L. 2013. TNF dually mediates resistance and susceptibility to mycobacteria via mitochondrial reactive oxygen species. Cell 153: 521–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde K, Yates RM, Purdy GE, Russell DG. 2007. Mycobacterium tuberculosis and the environment within the phagosome. Immunol Rev 219: 37–54 [DOI] [PubMed] [Google Scholar]
- Rook GA, Steele J, Ainsworth M, Champion Br. 1986a. Activation of macrophages to inhibit proliferation of Mycobacterium tuberculosis: Comparison of the effects of recombinant γ interferon on human monocytes and murine peritoneal macrophages. Immunology 59: 333–338 [PMC free article] [PubMed] [Google Scholar]
- Rook GA, Steele J, Fraher L, Barker S, Karmali R, O’Riordan J, Stanford J. 1986b. Vitamin D3, γ interferon, and control of the proliferation of Mycobacterium tuberculosis by human macrophages. Immunology 57: 159–163 [PMC free article] [PubMed] [Google Scholar]
- Rothchild AC, Jayaraman P, Nunes-Alves C, Behar SM. 2014. iNKT cell production of GM-CSF controls Mycobacterium tuberculosis. PLoS Pathog 10: e1003805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rous P. 1925. The relative reaction within living mammalian tissues. II. On the mobilization of acid material within cells, and the reaction as influenced by the cell state. J Exp Med 41: 399–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DG. 2011. The galvanizing of Mycobacterium tuberculosis: An antimicrobial mechanism. Cell Host Microbe 10: 181–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiga H, Nishimura J, Kuwata H, Okuyama M, Matsumoto S, Sato S, Matsumoto M, Akira S, Yoshikai Y, Honda K, et al. 2008. Lipocalin 2-dependent inhibition of mycobacterial growth in alveolar epithelium. J Immunol 181: 8521–8527 [DOI] [PubMed] [Google Scholar]
- Samanovic MI, Ding C, Thiele DJ, Darwin KH. 2012. Copper in microbial pathogenesis: Meddling with the metal. Cell Host Microbe 11: 106–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanga CA, Mohan VP, Tanaka K, Alland D, Flynn JL, Chan J. 2001. The inducible nitric oxide synthase locus confers protection against aerogenic challenge of both clinical and laboratory strains of Mycobacterium tuberculosis in mice. Infect Immun 69: 7711–7717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaible UE, Kaufmann SH. 2004. Iron and microbial infection. Nat Rev Microbiol 2: 946–953 [DOI] [PubMed] [Google Scholar]
- Schaible UE, Sturgill-Koszycki S, Schlesinger PH, Russell DG. 1998. Cytokine activation leads to acidification and increases maturation of Mycobacterium avium–containing phagosomes in murine macrophages. J Immunol 160: 1290–1296 [PubMed] [Google Scholar]
- Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD, Nathan C, et al. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: Insights into the phagosomal environment. J Exp Med 198: 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah JA, Vary JC, Chau TT, Bang ND, Yen NT, Farrar JJ, Dunstan SJ, Hawn TR. 2012. Human TOLLIP regulates TLR2 and TLR4 signaling and its polymorphisms are associated with susceptibility to tuberculosis. J Immunol 189: 1737–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Kumar V, Khosla R, Kajal N, Sarin B, Sehajpal P. 2010. Association of P2X7 receptor +1513 (A→C) polymorphism with tuberculosis in a Punjabi population. Int J Tuberc Lung Dis 14: 1159–1163 [PubMed] [Google Scholar]
- Shi S, Nathan C, Schnappinger D, Drenkow J, Fuortes M, Block E, Ding A, Gingeras TR, Schoolnik G, Akira S, et al. 2003. MyD88 primes macrophages for full-scale activation by interferon-γ yet mediates few responses to Mycobacterium tuberculosis. J Exp Med 198: 987–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Blumenthal A, Hickey CM, Gandotra S, Levy D, Ehrt S. 2005. Expression of many immunologically important genes in Mycobacterium tuberculosis–infected macrophages is independent of both TLR2 and TLR4 but dependent on IFN-αβ receptor and STAT1. J Immunol 175: 3318–3328 [DOI] [PubMed] [Google Scholar]
- Shiloh MU, Manzanillo P, Cox JS. 2008. Mycobacterium tuberculosis senses host-derived carbon monoxide during macrophage infection. Cell Host Microbe 3: 323–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikora A, Lui J, Brosnan C, Buell G, Chessel I, Bloom BR. 1999. Purinergic signaling regulates radical-mediated bacterial killing mechanisms in macrophages through a P2X7-independent mechanism. J Immunol 163: 558–561 [PubMed] [Google Scholar]
- Singh SB, Davis AS, Taylor GA, Deretic V. 2006. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science 313: 1438–1441 [DOI] [PubMed] [Google Scholar]
- Singh R, Manjunatha U, Boshoff HI, Ha YH, Niyomrattanakit P, Ledwidge R, Dowd CS, Lee IY, Kim P, Zhang L, et al. 2008. PA-824 kills nonreplicating Mycobacterium tuberculosis by intracellular NO release. Science 322: 1392–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SB, Ornatowski W, Vergne I, Naylor J, Delgado M, Roberts E, Ponpuak M, Master S, Pilli M, White E, et al. 2010. Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nat Cell Biol 12: 1154–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JH, Kim SY, Chung KS, Moon CM, Kim SW, Kim EY, Jung JY, Park MS, Kim YS, Kim SK, et al. 2014. Association between genetic variants in the IRGM gene and tuberculosis in a Korean population. Infection 10.1007/s15010-014-0604-6 [DOI] [PubMed] [Google Scholar]
- St. John G, Brot N, Ruan J, Erdjument-Bromage H, Tempst P, Weissbach H, Nathan C. 2001. Peptide methionine sulfoxide reductase from Escherichia coli and Mycobacterium tuberculosis protects bacteria against oxidative damage from reactive nitrogen intermediates. Proc Natl Acad Sci 98: 9901–9906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SA, Cox JS. 2013. Host–pathogen interactions during Mycobacterium tuberculosis infections. Curr Top Microbiol Immunol 374: 211–241 [DOI] [PubMed] [Google Scholar]
- Stanley SA, Barczak AK, Silvis MR, Luo SS, Sogi K, Vokes M, Bray MA, Carpenter AE, Moore CB, Siddiqi N, et al. 2014. Identification of host-targeted small molecules that restrict intracellular Mycobacterium tuberculosis growth. PLoS Pathog 10: e1003946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskawicz BJ, Mudgett MB, Dangl JL, Galan JE. 2001. Common and contrasting themes of plant and animal diseases. Science 292: 2285–2289 [DOI] [PubMed] [Google Scholar]
- Sturgill-Koszycki S, Schlesinger PH, Chakraborty P, Haddix PL, Collins HL, Fok AK, Allen RD, Gluck SL, Heuser J, Russell DG. 1994. Lack of acidification in mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science 263: 678–681 [DOI] [PubMed] [Google Scholar]
- Sturgill-Koszycki S, Schaible UE, Russell DG. 1996. Mycobacterium-containing phagosomes are accessible to early endosomes and reflect a transitional state in normal phagosome biogenesis. EMBO J 15: 6960–6968 [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Tsuyuguchi K, Matsumoto H, Niimi A, Tanaka E, Amitani R. 2000. Effect of proton pump inhibitor alone or in combination with clathrinomycin on mycobacterial growth in human macrophages. FEMS Microbiol Lett 182: 69–72 [DOI] [PubMed] [Google Scholar]
- Tailleux L, Waddell SJ, Pelizzola M, Mortellaro A, Withers M, Tanne A, Castagnoli PR, Gicquel B, Stoker NG, Butcher PD, et al. 2008. Probing host pathogen cross-talk by transcriptional profiling of both Mycobacterium tuberculosis and infected human dendritic cells and macrophages. PLoS ONE 3: e1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BH, Meinken C, Bastian M, Bruns H, Legaspi A, Ochoa MT, Krutzik SR, Bloom BR, Ganz T, Modlin RL, et al. 2006. Macrophages acquire neutrophil granules for antimicrobial activity against intracellular pathogens. J Immunol 177: 1864–1871 [DOI] [PubMed] [Google Scholar]
- Tan MP, Sequeira P, Lin WW, Phong WY, Cliff P, Ng SH, Lee BH, Camacho L, Schnappinger D, Ehrt S, et al. 2010. Nitrate respiration protects hypoxic Mycobacterium tuberculosis against acid- and reactive nitrogen species stresses. PLoS ONE 5: e13356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuong NT, Hawn TR, Thwaites GE, Chau TT, Lan NT, Quy HT, Hieu NT, Aderem A, Hien TT, Farrar JJ, et al. 2007. A polymorphism in human TLR2 is associated with increased susceptibility to tuberculous meningitis. Genes Immun 8: 422–428 [DOI] [PubMed] [Google Scholar]
- Ting LM, Kim AC, Cattamanchi A, Ernst JD. 1999. Mycobacterium tuberculosis inhibits IFN-γ transcriptional responses without inhibiting activation of STAT1. J Immunol 163: 3898–3906 [PubMed] [Google Scholar]
- Tiwari S, Choi HP, Matsuzawa T, Pypaert M, MacMicking JD. 2009. Targeting of the GTPase Irgm1 to the phagosomal membrane via PtdIns(3,4)P2 and PtdIns(3,4,5)P3 promotes immunity to mycobacteria. Nat Immunol 10: 907–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma-Uszynski S, Stenger S, Takeuchi O, Ochoa M, Engele MT, Sieling PA, Barnes PF, Rollinghoff M, Bolcskei PL, Wagner M, et al. 2001. Induction of direct antimicrobial activity through mammalian Toll-like receptors. Science 291: 1544–1547 [DOI] [PubMed] [Google Scholar]
- Trost M, English L, Lemieux S, Courcelles M, Desjardins M, Thibault P. 2009. The phagosomal proteome in interferon-γ-activated macrophages. Immunity 30: 143–154 [DOI] [PubMed] [Google Scholar]
- Ullrich H-J, Beatty WL, Russell DG. 1999. Direct delivery of procathepsin D to phagosomes: Implications for phagosome biogenesis and parasitism by mycobacterium. Eur J Cell Biol 78: 739–748 [DOI] [PubMed] [Google Scholar]
- Vandal OH, Pierini LM, Schnappinger D, Nathan CF, Ehrt S. 2008. A membrane protein preserves intrabacterial pH in intraphagosomal Mycobacterium tuberculosis. Nat Med 14: 849–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandal OH, Nathan CF, Ehrt S. 2009. Acid resistance in Mycobacterium tuberculosis. J Bacteriol 191: 4714–4721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wel N, Hava D, Houben D, Fluitsma D, van Zon M, Pierson J, Brenner M, Peters PJ. 2007. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell 129: 1287–1298 [DOI] [PubMed] [Google Scholar]
- Van Zandt KE, Sow FB, Florence WC, Zwilling BS, Satoskar AR, Schlesinger LS, Lafuse WP. 2008. The iron export protein ferroportin 1 is differentially expressed in mouse macrophage populations and is present in the mycobacterial-containing phagosome. J Leukoc Biol 84: 689–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velez DR, Hulme WF, Myers JL, Weinberg JB, Levesque MC, Stryjewski ME, Abbate E, Estevan R, Patillo SG, Gilbert JR, et al. 2009. NOS2A, TLR4, and IFNGR1 interactions influence pulmonary tuberculosis susceptibility in African-Americans. Hum Genet 126: 643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venketaraman V, Dayaram YK, Talaue MT, Connell ND. 2005. Glutathione and nitrosoglutathione in macrophage defense against Mycobacterium tuberculosis. Infect Immun 73: 1886–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergne I, Chua J, Deretic V. 2003. Tuberculosis toxin blocking phagosome maturation inhibits a novel Ca2+/calmodulin-PI3K hVPS34 cascade. J Exp Med 198: 653–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilchèze C, Hartman T, Weinrick B, Jacobs WR Jr. 2013. Mycobacterium tuberculosis is extraordinarily sensitive to killing by a vitamin C–induced Fenton reaction. Nat Commun 4: 1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt G, Nathan C. 2011. In vitro differentiation of human macrophages with enhanced antimycobacterial activity. J Clin Invest 121: 3889–3901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Muhlinen N, Akutsu M, Ravenhill BJ, Foeglein Á, Bloor S, Rutherford TJ, Freund SM, Komander D, Randow F. 2012. LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol Cell 48: 329–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskuil MI, Schnappinger D, Visconti KC, Harrell MI, Dolganov GM, Sherman DR, Schoolnik GK. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med 198: 705–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D, Maser J, Lai B, Cai Z, Barry CE 3rd, Höner Zu Bentrup K, Russell DG, Bermudez LE. 2005. Elemental analysis of Mycobacterium avium–, Mycobacterium tuberculosis–, and Mycobacterium smegmatis–containing phagosomes indicates pathogen-induced microenvironments within the host cell’s endosomal system. J Immunol 174: 1491–500 [DOI] [PubMed] [Google Scholar]
- Wang C-h, Lui C-Y, Lin H-C, Yu C-T, Chung K, Kuo H. 1998. Increased exhaled nitric oxide in active pulmonary tuberculosis due to inducible nitric oxide synthase upregulation in alveolar macrophages. Eur J Respir 11: 809–815 [DOI] [PubMed] [Google Scholar]
- Wang C, Lin H, Lui C, Huang K, Huang T, Yu C, Kuo H. 2001. Upregulation of inducible nitric oxide synthase and cytokine secretion in peripheral blood monocytes from pulmonary tuberculosis patients. Int J Tuberc Lung Dis 5: 283–291 [PubMed] [Google Scholar]
- Ward SK, Abomoelak B, Hoye EA, Steinberg H, Talaat AM. 2010. CtpV: A putative copper exporter required for full virulence of Mycobacterium tuberculosis. Mol Microbiol 77: 1096–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson RO, Manzanillo PS, Cox JS. 2012. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell 150: 803–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, Brodsky IE, Rahner C, Woo DK, Erdjument-Bromage H, Tempst P, Walsh MC, Choi Y, Shadel GS, Ghosh S. 2011. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature 472: 476–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson RJ. 2014. Host-directed therapies against tuberculosis. Lancet Respir Med 85–87 [DOI] [PubMed] [Google Scholar]
- Wilkinson RJ, Llewelyn M, Toossi Z, Patel P, Pasvol G, Lalvani A, Wright D, Latif M, Davidson RN. 2000. Influence of vitamin D deficiency and vitamin D receptor polymorphisms on tuberculosis among Gujarati Asians in west London: A case-control study. Lancet 355: 618–621 [DOI] [PubMed] [Google Scholar]
- Wolschendorf F, Ackart D, Shrestha TB, Hascall-Dove L, Nolan S, Lamichhane G, Wang Y, Bossmann SH, Basaraba RJ, Niederweis M. 2011. Copper resistance is essential for virulence of Mycobacterium tuberculosis. Proc Natl Acad Sci 108: 1621–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. 2007. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity 27: 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CT, Cambier CJ, Davis JM, Hall CJ, Crosier PS, Ramakrishnan L. 2012. Neutrophils exert protection in the early tuberculous granuloma by oxidative killing of mycobacteria phagocytosed from infected macrophages. Cell Host Microbe 12: 301–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Mitchell C, Xing Y, Magliozzo RS, Bloom BR, Chan J. 1999. Toxicity of nitrogen oxides and related oxidants on mycobacteria: M tuberculosis is resistant to peroxynitrite anion. Tuberc Lung Dis 79: 191–198 [DOI] [PubMed] [Google Scholar]
- Yuk JM, Shin DM, Lee HM, Yang CS, Jin HS, Kim KK, Lee ZW, Lee SH, Kim JM, Jo EK. 2009. Vitamin D3 induces autophagy in human monocytes/macrophages via cathelicidin. Cell Host Microbe 6: 231–243 [DOI] [PubMed] [Google Scholar]
- Zanetti M. 2004. Cathelicidins, multifunctional peptides of the innate immunity. J Leukoc Biol 75: 39–48 [DOI] [PubMed] [Google Scholar]
- Zhang YJ, Rubin EJ. 2013. Feast or famine: The host–pathogen battle over amino acids. Cell Microbiol 15: 1079–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SY, Boisson-Dupuis S, Chapgier A, Yang K, Bustamante J, Puel A, Picard C, Abel L, Jouanguy E, Casanova JL. 2008. Inborn errors of interferon (IFN)-mediated immunity in humans: Insights into the respective roles of IFN-α/β, IFN-γ, and IFN-λ in host defense. Immunol Rev 226: 29–40 [DOI] [PubMed] [Google Scholar]
- Zhang YJ, Reddy MC, Ioerger TR, Rothchild AC, Dartois V, Schuster BM, Trauner A, Wallis D, Galaviz S, Huttenhower C, et al. 2013. Tryptophan biosynthesis protects mycobacteria from CD4 T-cell-mediated killing. Cell 155: 1296–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]