Abstract
Epidemiological studies provide evidence that consumption of cruciferous vegetables, like broccoli, can reduce the risk of cancer development. Sulforaphane (SFN) is a phytochemical derived from cruciferous vegetables that induces anti-proliferative and pro-apoptotic responses in prostate cancer cells, but not in normal prostate cells. The mechanisms responsible for this cancer-specific cytotoxicity remain unclear. To examine this issue we utilized RNA sequencing and determined the transcriptomes of normal prostate epithelial cells, androgen-dependent prostate cancer cells, and androgen-independent prostate cancer cells treated with SFN. SFN treatment dynamically altered gene expression and resulted in distinct transcriptome profiles depending on prostate cell line. SFN also down-regulated the expression of genes that were up-regulated in prostate cancer cells. Network analysis of genes altered by SFN treatment revealed that the transcription factor Specificity protein 1 (Sp1) was present in an average of 90.5% of networks. Sp1 protein was significantly decreased by SFN treatment in prostate cancer cells and Sp1may be an important mediator of SFN-induced changes in expression. Overall, the data show that SFN alters gene expression differentially in normal and cancer cells with key targets in chemopreventive processes, making it a promising dietary anti-cancer agent.
Keywords: chemoprevention, prostate cancer, RNA sequencing, Specific protein 1, sulforaphane
Introduction
Globally, prostate cancer is the second most frequently diagnosed cancer in men [1]. In the United States, 1 in 6 men will develop prostate cancer during their lifetime, and prostate cancer is the second leading cause of cancer-related deaths in men [2]. This high prevalence translates to a significant societal and financial burden [2]. Previous studies have shown that increased consumption of cruciferous vegetables reduces the risk of developing prostate cancer [3, 4]. Sulforaphane (SFN) is an isothiocyanate produced during the consumption of cruciferous vegetables, like broccoli and broccoli sprouts, when the precursor glucoraphanin interacts with the enzyme myrosinase [4]. SFN has been shown to have chemopreventive and cancer suppressive properties in carcinogen-induced and genetic models of cancer including a model of prostate cancer [4, 5].
Classically SFN has been shown to inhibit the initiation of cancer by blocking damage caused by carcinogens, through the induction of phase 2 enzymes via Keap1-Nrf2 signaling and antioxidant response element driven gene expression [6-9]. SFN also inhibits the activity and/or expression of genes that regulate epigenetic mechanisms including histone deactylases (HDACs) and DNA methyltransferases (DNMTs) in cancer cells [10-12]. This is associated with the re-expression of epigenetically silenced tumor suppressor genes like p21 and Cyclin D2 in prostate cancer cells [10, 11]. Importantly, SFN exhibits cancer-specific cytotoxic and antiproliferative effects. More specifically, in androgen-dependent prostate cancer cells (LNCaP), 15 μM SFN treatment induces cell cycle arrest at the G1 phase and only modestly increases apoptosis [10]. In the more aggressive androgen-independent prostate cancer cells (PC-3), the same SFN treatment arrests cells at the G2/M checkpoint followed by a potent induction of apoptosis [10]. Normal prostate epithelial cells (PREC) do not undergo cell cycle arrest or apoptosis in response to this SFN treatment [10]. We currently do not have a full understanding of how SFN produces this notable differential response in prostate cells.
While multiple chemopreventive properties of SFN have been identified we do not have a clear understanding of how these mechanisms are inter-connected and if there are any key mediators that facilitate chemoprevention. While past microarray studies have addressed the effect of SFN on gene expression, the great majority of the microarray studies were analyzed with an Nrf2 bias and/or completed on only a subset of the genes found in the human genome [6, 8, 9, 13-15]. RNA-sequencing (RNA-seq) is a contemporary sequencing method that can be used to deeply interrogate transcriptomes to quantify changes in gene expression with many advantages over microarrays [16-19]. We utilized RNA-seq as an unbiased step towards identifying how SFN induces anti-proliferative and pro-apoptotic responses in prostate cancer cells, but not in normal prostate cells. We also characterized the transcriptomes of normal cells following SFN treatment to gain a more broad understanding of how normal prostate cells respond to SFN treatment.
Materials and Methods
Culturing and Treatment of Cells
PREC were obtained from Lonza (Basel, Switzerland) and cultured as recommended with 5% CO2 and 37°C in PREC basal media supplemented with PREC growth media singleQuots (Lonza). LNCaP and PC-3 cells were obtained from American Type Tissue Collection (Manassas, VA) and cultured as previously described [10]. All cells were confirmed to be mycoplasma free and have the expected allelic composition (Idexx Radil, Columbia, MO). Each cell line was treated at 50 – 70% confluent with SFN (LKT Laboratories, St. Paul, MN) dissolved in dimethylsulfoxide (DMSO) at a 15 μM concentration, or with an equal volume of DMSO (vehicle control) that was 0.03% of the total media volume. Adherent cells were harvested at 6 and 24 hours post-treatment. The concentration of SFN was chosen because μ15 μM of total SFN metabolites has been observed in the plasma of mice orally dosed with 20 μmol of SFN, and prostates have one of the highest accumulations of SFN among tissues examined [20]. The time points and concentration were also chosen based on previous reports showing HDAC inhibition and cell cycle arrest in prostate cancer cells but not normal prostate epithelial cells [10, 21, 22].
RNA sequencing (RNA-seq)
Cells were treated in triplicate and total RNA was isolated at each time point using a standard Trizol (Life Technologies) extraction with the exception that samples were allowed to dry for 1 hour while placed on ice, and then RNA secure reagent was added (Life Technologies). Contaminating DNA was removed by Turbo DNase treatment (Life Technologies), and RNA was purified using the RNeasy Mini Kit (Qiagen, Germantown, MD). RNA quality was measured on an Agilent Bioanalyzer 2100 (Santa Clara, CA) with a minimum RIN score of 9.6. RNA quantity was determined with a Qubit 2.0 Fluorometer (Life Technologies). To prepare cDNA libraries, 4 μg of high quality RNA for each sample was converted into an indexed library using the TruSeq RNA sample Preparation Kit v-2 according to the manufacturer’s protocol (Illumina, San Diego, CA). The quantity of indexed cDNA was finally quantified using the universal Kapa library quantification kit and the manufactures protocol (Kapa Biosystems, Wilmington, MA). The samples were multiplexed, 12 / lane, and sequenced using a single end cluster generation kit (Illumina) with a 50 cycle v3 sequencing kit (Illumina) on an Illumina HiSeq 2000 machine at the Center for Genome Research and Biocomputing core facility at Oregon State University. Reads were aligned to the human genome (GRCh37/hg19) and analyzed for significant differences in expression with the Tuxedo Suite and GENE-counter [17, 23]. Significant differences in gene expression (p <0.05) were determined using either the default parameters of Cuffdiff or the NBPseq software package, respectively. Differentially expressed genes were identified based on the criteria of being at the intersect of significant genes identified using both pipelines and exceeding a threshold of an average of 2 fragments per kilobase of exon per million fragments mapped (FPKM, Tuxedo suite pipeline), and 20 normalized reads (GENE-counter) in at least one treatment group. Fold changes that approached infinity were reassigned as 20. The raw RNA-seq reads and differential gene expression data have been deposited in NCBI Gene Expression Omnibus and are accessible through GEO Series accession number GSE48812 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE48812)
Quantitative Real-Time PCR (qPCR)
Cells were treated in triplicate and cDNA was synthesized using 1 μg of total RNA and SuperScript III First-Strand Synthesis SuperMix (Life Technologies). Real time PCR was done using primers that amplify all known transcript isoforms of each gene as a single product of expected size, between 150 and 300bp (Supporting Table S1). Reactions were performed using Fast SYBR Green Mastermix (Life Technologies) on 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA) as previously published [24]. Data were normalized to the expression of β-actin or GAPDH, as indicated in the results, and analyzed using the standard 2−ΔΔCT method (23).
Software
Graphs were generated using GraphPad Prism software (La Jolla, CA) unless otherwise indicated. Log2 fold distribution graphs were generated using the matplotlib Python package with a bin width of 0.1 [25]. Venn images were generated by BioVenn software [26]. Gene-annotation enrichment analysis was completed using the functional annotation clustering tool of the DAVID Bioinformatics Resources 6.7 with gene lists where the log2 fold change exceeded 0.5 or was less than −0.5 [27]. Pathway and network analysis was completed with MetaCore with all the default parameters (Thomson Reuters, New York, NY). The presence of Sp1 in each network was scored and Sp1 was considered a major regulator if it regulated five or more gene targets.
Sp1 bioinformatics analysis
A previously published model of putative Sp1 binding sites (UCSC Genome Browser, HMR conserved transcription factor binding sites), was used to identify genes that are likely regulated by the Sp1 transcription factor [28, 29]. This model was applied to the 2 kb regions upstream of transcriptional start sites for all protein coding genes in the human genome (GRCh37/hg19) [30]. A master list of 3,244 intergenic regions that contain at least one Sp1 binding site, and were on the same strand as the transcription start site, was created out of the 18,899 protein coding genes, representing 17.16% of the genes in the genome. This master list was compared to each of the gene lists from the RNA-seq data sets that were generated via Tuxedo suite using the same UCSC annotation of the genome. The percentage of genes that contained at least one Sp1 binding site was calculated. Between 22.7% and 25.9% of the genes that were significantly altered by SFN treatment, had at least one Sp1 putative binding sequence. To compare these percentages to what would be expected at random, we generated a thousand different lists composed of 3,000 random protein coding genes and calculated the percentage of genes related to Sp1. The percentages from the random gene lists were shown to have a normal distribution as determined by the Shapiro-Wilk normality test with a mean of 17.1%. The percentage of genes that were altered by SFN treatment, and had an Sp1 binding site, was then statistically tested against what is expected at random using one sample tests of proportion.
Immunoblot analysis
Nuclear protein was harvested using a nuclear extract kit (Active Motif, Carlsbad CA), as previously published [31]. Proteins were separated and detected as previously described [10] with anti-Sp1 (H-225), Sp3 (S-20) (Santa Cruz Biotechnology, Santa Cruz, CA) and USF2 (5E9, Novus Biologicals, Littleton, CO), followed by goat anti-rabbit, or goat anti-mouse secondary antibodies (Santa Cruz Biotechnology) using standard conditions.
Sp1 gene silencing
LNCaP cells were trypsinized and transfected with 30 nM Sp1-specific siRNA (Ambion s13319) or an equal concentration of negative control siRNA (Ambion) using siPORT NeoFX transfection agent (Life Technologies) and seeded in 6-well tissue culture plate. RNA was collected 48 h post transfection and live cell number was counted using trypan blue exclusion and a hemacytometer, 5 days post transfection.
Results
SFN Treatment Dynamically Alters Gene Expression and Results in Distinct Transcriptome Profiles Depending on Prostate Cell Line
We carried out an RNA-seq experiment to assess the effects of SFN on the transcriptome of normal and prostate cancer cells following 6 and 24 hours of 15 μM SFN treatment. A total of 573,504,958 reads, of 51 bases in length, were obtained totaling 29.2 giga bases of sequence. An average of μ15.9 million reads were obtained per sample and μ70% of the reads unambiguously mapped to a unique region in the human genome. To identify genes that were differentially expressed as a factor of cell-type, potentially resulting from the cancerous state, pairwise comparisons were made between normal and cancerous cell lines treated with the vehicle control. Between 7,362 and 9,698 genes were significantly differentially expressed in androgen-dependent LNCaP and androgen-independent PC-3 cells as compared to normal PREC cells (Cancer effect, Fig. 1A-B). To characterize the SFN-dependent effect on gene expression pairwise comparisons were also made between the SFN and vehicle control treatments within each cell line and time point. An average of 3,158 genes was significantly altered by SFN treatment in each prostate cell line at each time point (Fig. 1A and 1B). In order to validate the RNA-seq dataset we examined the expression of known SFN targets NAD(P)H:quinone oxidoreductase 1 (NQO1) and heme oxygenase 1 (HO1) [6, 8-10]. In all cell lines and time points, HO1 and NQO1 were identified as significantly upregulated by SFN both in the RNA-seq dataset and by qPCR (Supporting Information Figure S1 and online datasets). We also used qPCR to examine nine additional genes identified in the RNA-seq dataset. The genes were chosen because they showed either the greatest increase or decrease in response to SFN treatment, or are relevant to cancer development (Supporting Information Figure S2). Among all the genes and conditions examined by qPCR μ90% of the tests confirmed results identified by RNA-seq.
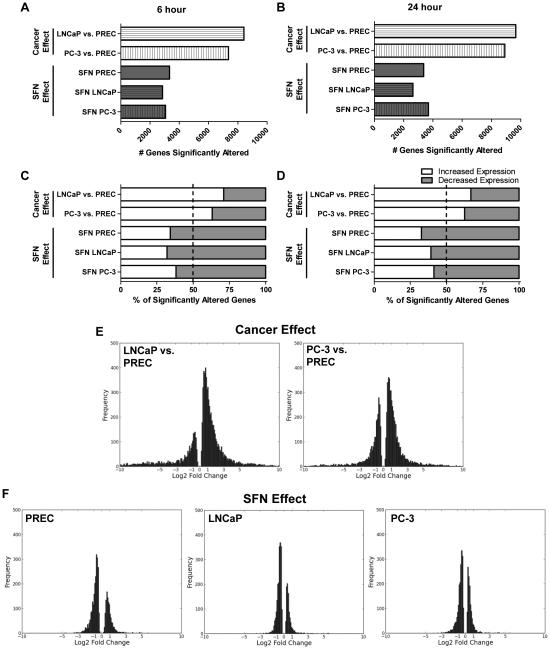
Figure 1. Genome-wide effects of cancer and SFN on the expression of mRNAs.
The cancer cancer effect was determined by comparing the mRNA levels found in LNCaP and PC-3 cells compared to normal PREC cells. The SFN effect was determined in each of the three cell lines by direct comparison of mRNA levels of samples treated with 15 μM SFN compared to their respective vehicle control at the same time point. A-B) Bars represent the number of genes that were significantly altered by prostate cancer development or by SFN treatment at the 6 or 24 hour time points. C-D) Data represent the percentage of genes that had a significant increase (white), or decrease in expression (grey) at the indicated time points at the C) 6 h and D) 24 h time points. E-F) Frequency plots of the amplitude of the changes in gene expression, expressed as log2 fold change, which was significantly altered under the indicated comparisons. Data are from the 6 hour time point although similar log2 fold distributions were also observed at the 24 hour time point (data not shown).
Comparisons of the transcriptomes in prostate cancer cells to normal cells showed that the majority of the genes (66%) were differentially upregulated in prostate cancer cell lines compared to normal cells (Fig. 1C-D). In stark contrast, SFN treatment resulted in a significant decrease in 64% of genes that were altered. This trend was observed in all prostate cell lines and at both time points showing that the majority of genes altered by SFN treatment undergo a decrease in gene expression (Fig 1C-D). A comparison of the cancer effect and SFN effect frequency plots shows a shift from increased gene expression in cancer cells to decreased gene expression with SFN treatment (Fig. 1E and 1F). The extent of changes in gene expression (ie. size of log2 fold values) was also more modest with SFN treatment as compared to the level of transcript differences found between normal and cancer cell lines (Fig. 1E-F).
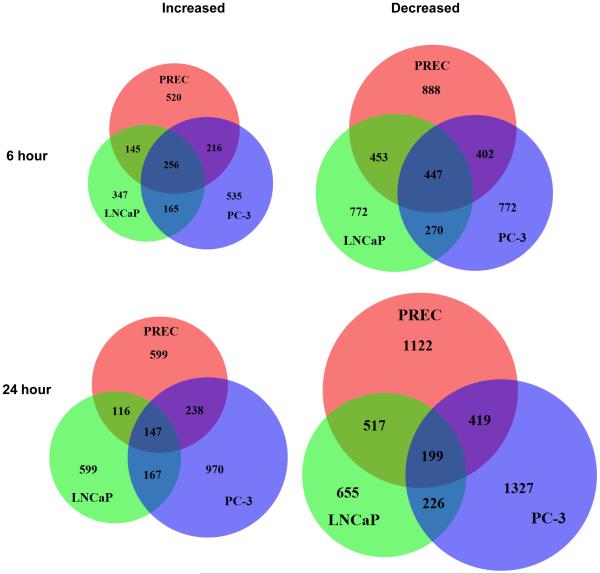
Venn diagrams show that the majority of genes altered with SFN treatment were specific to each cell line, which likely underlies the differential cellular response observed with SFN treatment (Fig. 2). There was a core set of genes affected by SFN in all three prostate cell lines but the number of core genes decreased in number over time, regardless of whether the genes were induced (from 256 to 147 genes), or repressed in expression (from 447 to 199 genes) (Fig. 2). We also determined the overlap of differentially expressed genes as a factor of time for each cell line (Supporting Information Figure S3). These data show that the effect of SFN in the cells is dynamic over time as the proportion of genes that are shared at both time points never exceeds 40%. In the most extreme example, only 197 genes (μ17%) were significantly upregulated by SFN treatment at both time points in PREC cells.
Figure 2. Comparison of SFN-induced changes expression in normal prostate epithelial cells and prostate cancer cells.
Venn diagrams showing the number and overlap of the genes whose mRNA expression levels were significantly altered in PREC, LNCaP, and PC-3 cells with 6 or 24 hours of SFN treatment.
SFN Treatment Produced Pleiotropic Chemopreventive Effects in Prostate Cells
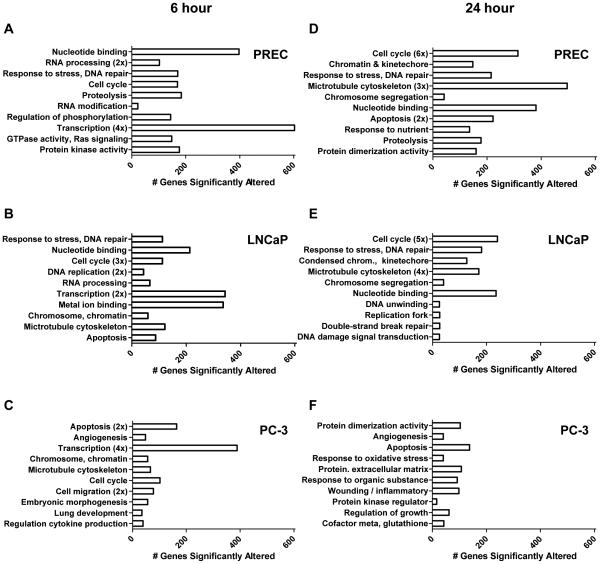
To begin characterizing how SFN induces anti-proliferative and pro-apoptotic responses in prostate cancer cells, but not in normal prostate cells, gene-annotation enrichment analysis was completed. At the 6 hour time point categories associated with gene regulation, such as the transcription category, were significantly enriched in all three cell lines treated with SFN suggesting a general effect of SFN treatment (Fig. 3A-C). The cell cycle category was also significantly enriched for in all cell lines but interestingly, different cell cycle-related genes were affected in the three cell lines. More specifically, in each cell line μ100 cell cycle-related genes were altered by SFN, but only 9 cell cycle genes were consistently increased, and 17 genes consistently decreased, in all three cell lines (Supporting Information Figure S4). The selective effects of SFN on distinct gene targets in each cell line likely contribute to the differential cellular response we observe between the various prostate cell lines. For example, the categories of apoptosis, chromosome/chromatin, and microtubule cytoskeleton were significantly enriched in both prostate cancer cell lines at the 6 hour time point but were absent from the normal prostate epithelial cells treated with SFN (Fig. 3A-C). Additionally, categories related to development, cell migration and angiogenesis were only enriched in PC-3 cells, at the 6 hour time point (Fig. 3C). The presence of the apoptosis category in LNCaP cells at the 6 h time point (Fig. 3B), but absence at the 24 h time point (Fig. 3E), corresponds with the cellular response to SFN treatment were a minor amount of apoptosis is detected with SFN and the predominant phenotype is cell cycle arrest.
Figure 3. Gene ontologies show global effects of SFN in normal and prostate cancer cells.
DAVID analysis of protein coding transcripts significantly altered by 6 (A-C) and 24 hour (D-F) SFN treatment in PREC (A,D), LNCaP (B,E), and PC-3 (C,F) cells. Bars indicate the number of genes that fall into the gene category, and numbers in parenthesis indicate if this category was represented multiple times when creating a top ten list. All gene categories shown are significantly enriched for, and the position of the category indicates the degree of significance. The category at the top of the y-axis is the most significant, and the category at the bottom of the y-axis is less significant.
To further identify key differences between SFN-mediated gene expression in normal versus prostate cancer cell lines gene-annotation enrichment analysis was also completed for the 24 hour time point. The 6 gene categories with the greatest significance, cell cycle, chromatin/kinetecore, response to stress/DNA repair, microtubule cytoskeleton, chromosome segregation, and nucleotide binding, were shared in PREC and LNCaP cells (Fig. 3D-E). These categories do not appear in the PC-3 cells at the 24 hour time point, where 9 out of the 10 most enriched categories are unique to PC-3 cells and likely contribute to the apoptosis associated with SFN treatment in these cells (Fig. 3F). Some of these unique categories include protein dimerization activity, angiogenesis, apoptosis, response to oxidative stress, regulation of growth, and cofactor metabolism/glutathione (Fig. 3F).
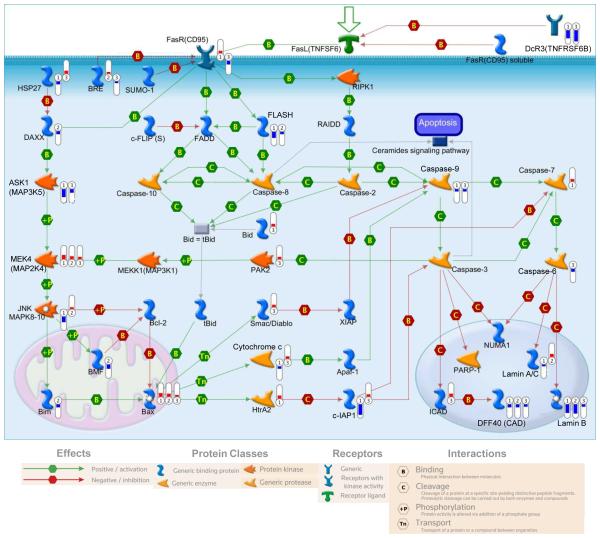
To further visualize how the SFN-induced changes in gene expression relate to each other both pathway and network analyses were completed with MetaCore software which contains a large body of known gene interactions, and well defined canonical pathways in human cells. Many pathways related to apoptosis and cell survival were significantly altered by SFN treatment as can be seen in a representative pathway (Fig. 4). Pathways involving cell cycle, cell signaling, DNA damage, cytoskeleton remodeling, development, immune response, and transcription, were also significantly enriched for with SFN treatment in prostate cells. Among the genes we examined in the validation of the RNA-seq dataset there are multiple genes that also contribute to these pathways including the oxidative stress induced growth inhibitor (OSGIN1) gene which encodes an oxidative stress response protein that regulates cell death (Supporting Information Figure S2). Additionally, BMX non-receptor tyrosine kinase (BMX), cyclin-dependent kinase 2 (CDK2), and polo-like kinase 1 (PLK1) had decreased expression with SFN treatment and regulate signal transduction pathways and cell cycle respectively (Supporting Information Figure S2).
Figure 4. SFN alters expression of genes related to apoptosis.
Representative image of an apoptosis pathway that was significantly enriched for in prostate cells treated with SFN for 24 hours. Significant and SFN-induced changes in gene expression are indicated by thermometers to the right of the gene name. Numbers in the thermometers indicate the cell line (1=PREC, 2=LNCaP, and 3=PC-3) in which SFN induced a significant change in expression. Colors of thermometers indicate a decrease (blue) and increase (red) in gene expression, and the scale of the thermometer indicates the log2 fold change.
SFN treatment also decreased the expression of a subset of genes that were upregulated in cancer cells (Fig 1E and 1F), and these genes encompassed a broad range of cellular functions which can be involved in cancer development and progression. More specifically, we examined the genes that were increased in prostate cancer cells, relative to normal PREC cells, and were also decreased by SFN in the prostate cancer cells. At the 6 hour time point, 744 genes meet these criteria in LNCaP cells, and 609 genes in PC-3 cells. When these gene lists were compared 117 common genes were increased in cancer cells and decreased by SFN treatment in both cell lines. A similar number of genes also meet these criteria at the 24 hour time point. The top 5 functional gene categories that were enriched with the genes increased in prostate cancer cells and decreased by SFN in both cancer cell lines, were nucleotide binding, tube development, chordate embryonic development, transcription, and chromatin binding at the 6 hour time point, and cellular response to stress, DNA replication, cell cycle, oxidation reduction, and organelle envelope at the 24 hour time point.
Sp1 is a Mediator of SFNs Effects in Prostate Cells
Beyond nrf2, little is known about what additional genes can act as key mediators that translate the initial signals from SFN treatment into chemopreventive effects. To address this issue MetaCore network analysis was used to generate an unbiased analysis of the data. This consisted of generating 30 gene networks that connected the greatest number of genes significantly altered by SFN treatment. Network analysis was undertaken for each cell line and time point of SFN treatment. The transcription factor Sp1 was present in an average of 90.6% of the networks and was considered a major regulator in 83.3% of networks (Table 1). Since Sp1 is a highly prevalent transcription factor we determined if Sp1 binding sites were over represented in the promoters of the genes that were significantly altered by SFN treatment. Between 22.7% and 25.9% of the genes that were significantly altered by SFN treatment, had at least one putative Sp1 binding sequence, and this was consistently and significantly higher (p < 0.0001) than what was found at random (Supporting Information Figure S5B). Networks involving c-Myc, NF-kB, c-Src, HNF4α, and G protein coupled receptors, were also noted as altered by SFN treatment but they were detected with a much lower frequency (data not shown). A representative network shows Sp1 at the center, and 10 Sp1 target genes with SFN-induced alterations in gene expression (Supporting Information Figure S5A). This network contains many genes relevant to cancer prevention including Cyclin E2, CDK4, CDC25A, E2F1 and p21. These genes are also a small subset of the genes that make up the cell cycle gene ontology found in figure 3B. Overall, the results from the bioinformatics approaches support the possibility that Sp1 related transcription is being altered under the condition of SFN treatment.
Table 1.
Sp1 Prevalence In SFN Networks
| PREC 6 hr |
LNCaP 6 hr |
PC-3 6 hr |
PREC 24 hr |
LNCaP 24 hr |
PC-3 24 hr |
|
|---|---|---|---|---|---|---|
| Contain Sp1 | 26/30 (86.7) |
26/30 (86.7) |
28/30 (93.3) |
29/30 (96.7) |
25/30 (83.3) |
29/30 (96.7) |
|
Sp1 as Major
Regulator |
25/30 (83.3) |
24/30 (80) |
26/30 (86.7) |
27/30 (90) |
24/30 (80) |
24/30 (80) |
(Numbers in parenthesis indicate a percentage of the networks for each category)
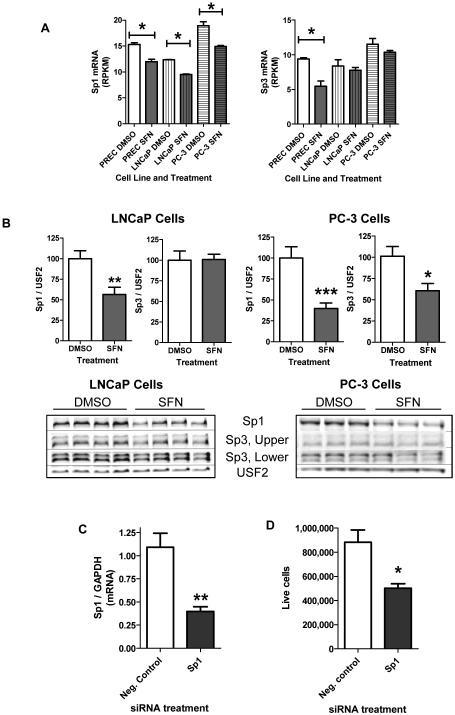
There is evidence that Sp1 can play an important role in cancer progression as Sp1 has been shown to be upregulated in several types of cancers and is associated with poor prognosis [32]. Since Sp1 could potentially regulate up to 25% of the genes altered by SFN treatment we examined Sp1 expression. Sp1 mRNA levels were significantly decreased in all three prostate cell lines with 24 hour SFN treatment (Fig. 5A). We confirmed these results in LNCaP cells by qPCR, where 24 hour SFN treatment was associated with μ50% decrease in Sp1 mRNA levels (data not shown). Sp3 mRNA levels were only significantly decreased in PREC cells with 24 hours of SFN exposure (Fig. 5A). At the protein level, 24 hours of SFN treatment significantly decreased (μ65%) the amount of nuclear Sp1 protein in prostate cancer cells (Fig. 5B). Nuclear Sp3 protein levels were not significantly altered by SFN treatment in LNCaP cells but were decreased in PC-3 cells following 24 hour SFN treatment (data not shown).
Figure 5. SFN-induced decrease in Sp1 mRNA and protein expression levels.
A-B) Prostate cells were treated with 15 μM SFN or DMSO for 24 hours. A) Sp1 and Sp3 mRNA expression levels as determined by the GENE-counter pipeline and expressed as reads per kilobase of transcript per million mapped reads (RPKM) where n =3. B) Densitometry results of Western blots for nuclear Sp1 and Sp3 protein levels normalized to USF2 and representative blots of Sp1, Sp3, and USF2 expression. Data represent an average of at least 10 replicates per treatment group, obtained from at least 3 independent experiments. C-D) LNCaP cells were treated with the indicated siRNA in quadruplicate and assayed for (C) Sp1 mRNA levels 48 h post transfection and (D) cell proliferation 5 days post transfection. B-D) Bars are indicative of the mean ± SEM and * , **, and *** indicates significant differences between the control and treatment group as calculated by either (A) GENE-counter or (B-D) an unpaired, two-tailed t test, where p < 0.05 , p < 0.01 and p <0.001 respectively.
Because SFN decreased the expression of Sp1, and to a lesser extent Sp3, we next asked if Sp1/Sp3 could act as a transcriptional mediator through which SFN may induced changes in expression relevant to chemoprevention. To address this question we used Metacore to examine what genes that were significantly altered by SFN treatment are upstream and downstream of the Sp1/Sp3 complex. Among all genes significantly altered by SFN in LNCaP cells, 38 genes were upstream of Sp1 including p53, NFκB, c-Myc, E2F1 and BRCA1 (Supporting Information Figure S6A). Downstream of Sp1 were 232 genes that were significantly altered by SFN treatment in LNCaP cells and this network also contained many genes highly relevant to cancer like EGFR, p21, and cyclin D1 (Supporting Information Figure S6B). When these 232 genes downstream of Sp1/Sp3 were examined by gene-annotation enrichment analysis the top 5 functional gene categories were regulation of apoptosis, cellular response to stress, cell cycle, transcription, and response to organic substance. This shows that some of the Sp1/Sp3 target genes that are affected by SFN have a documented link to cellular pathways that are important in cancer. Taken together the data suggests that Sp1 could be an important mediator through which SFN induces chemoprevention. To directly test this possibility we attempted to overexpress Sp1 in prostate cancer cells, with the intention of treating them with SFN and examining chemoprevention. Unfortunately overexpressing Sp1 in prostate cancer cells was cytotoxic. As an alternative approach, we significantly down regulated Sp1 expression in LNCaP cells with siRNA (Fig. 5C). Decreasing Sp1 mRNA expression was sufficient to significantly slow the growth of LNCaP cells (Fig. 5D). This data suggests that the SFN-induced decrease in Sp1 levels in prostate cancer cells may play a role in decreasing cancer cell proliferation and thus could contribute to chemoprevention.
Discussion
The genomic scale of RNA-seq facilitated a global analysis of the effect of SFN on gene expression at an unprecedented level. We show that SFN has broad chemopreventive actions in prostate cells and identified for the first time, both common and distinct effects of SFN depending on the cell line (normal, early or late-stage prostate cancer). We also show that the transcriptional response of prostate cells to SFN treatment is highly dynamic over time and this result is consistent with findings in rodents [9, 13]. Furthermore, our unbiased analysis of gene networks consistently identified that the transcription factors Sp1 and Sp3 may be important mediators of the SFN effect in prostate cells. We also showed that Sp1, and to a lesser extent Sp3, are downregulated by SFN treatment and suppression of Sp1 expression decreased prostate cancer cells proliferation.
One of our goals was to understand how SFN induces chemoprevention in normal prostate cells. The RNA-seq approach revealed that the expression of thousands of genes was significantly altered by SFN in normal prostate cells. Consistent with previous microarray studies that implicated the nrf2 pathway in SFN induced chemoprevention, we found that the nrf2 target genes HO1 and NQO1 were upregulated with SFN treatment [6-9, 13-15]. Furthermore, the response to oxidative stress category was found in our gene-annotation enrichment analyses of advance prostate cancer cells. Beyond the expected Nrf2 target genes, SFN also altered genes associated with the chromatin, kinetochore, and microtubule cytoskeleton structures. We also found significant enrichment of genes related to cell cycle, transcription, nucleotide binding, proteolysis, and signaling in normal cells. These results are supported by previous microarray studies on non-cancerous cells, where functional categories identified with SFN treatment included cell cycle/proliferation, metabolism, and transcription [14, 33]. Taken together, these data suggest that SFN has a broad pleitropic effect in normal cells and future work can delineate what role these categories of genes play in SFN-induced chemoprevention. Furthermore, a strength of the unbiased RNAseq approach is that we also identified that SFN induced changes in many additional processes and pathways, in prostate cancer cells, beyond those explored in detail here. This includes SFN-induced changes in genes associated with epigenetics and chromatin structure, long intergenic non coding RNAs, and changes in alternative splicing of RNA. Future work can determine if these processes also contribute to SFNs ability to prevent and suppress prostate cancer.
We expected SFN treatment to produce distinct transcriptomes in each of the cell types tested which represent various states of cancer progression. This is because SFN treatment results in different cellular endpoints that range from no visible change in normal cells, to apoptosis in advanced prostate cancer cells [10, 34, 35]. We do not have a complete understanding of why SFN has this specificity. Interestingly, we found that the cell cycle category was enriched for with SFN treatment in all three cell lines but a direct comparison of the gene lists revealed that the data was mostly cell line-dependent. Thus, overall SFN consistently alters the expression of cell cycle related genes but the specific effect was dependent on the cell line and presumably the state of cancer progression. We also paid particular attention to differences in the transcriptional response that may contribute to the robust induction of apoptosis in PC-3 cells [10]. Previous studies have shown that SFN targets genes in the Akt pathway that are dysregulated in prostate cancer, and this could contribute to the increased susceptibility of cancer cells to SFN [34, 35] , but this pathway alone cannot account for all of SFN effects in PC-3 cells [35]. Here we show that SFN treatment produced a significant increase in the expression of the apoptosis related genes Bid, Smac/Diablo, and ICAD only in PC-3 cells (Fig. 4). It also increased the expression of cytochrome c, c-IAP1, and HSP27 in PC-3 cells while it decreased expression in PREC cells. More upstream, SFN also induced a cell line specific transcriptional response for the genes FasR and BRE which also regulate apoptosis. More broadly, the gene categories related to development, cell migration, extracellular matrix, and angiogenesis were only enriched for in PC-3 cells. These genes and gene categories likely contribute to the selective induction of the pro-apoptotic response in aggressive prostate cancer cells.
Sp1 and Sp3 are transcription factors that enhance or repress the expression of genes involved in cell cycle progression and oncogenesis [36]. Sp1 levels are elevated in many different cancers and decreasing Sp1 levels is associated with decreased angiogenesis, and increased cancer cell death [36]. In prostate cancer patients, increased levels of Sp1 and Sp3 protein in the cancer, helped to predict the recurrence of the disease [37]. Previously, siRNA against Sp1 has been shown to decrease proliferation of DU145 prostate cancer cells [38]. We confirm those findings here in the LNCaP cell line and show that Sp1 protein, and to a lesser extent Sp3 protein, is decreased in SFN treated prostate cancer cells. We further show that the Sp1/Sp3 complex may integrate important upstream signals produced by SFN treatment (like alterations in the expression of NFκB, CDK2, c-Myc, and heat shock proteins) and translate this to downstream changes in gene expression in genes like GCLc, EGFR, p21 and the androgen receptor (Supporting Information Figure S6). As transcription factors Sp1 and Sp3 could directly regulate the expression of up to 25% of the genes that were altered following SFN treatment (Supporting Information Figure S5). This is a large portion of the genes that are influenced by SFN treatment and suggests that Sp1 may be an important mediator by which SFN induces chemoprevention. This is supported by a recent study in keratinocytes where Sp1 was shown to directly regulate the expression of p21 following SFN treatment [39].
While this is the first genomics scale study suggesting a role for Sp1 in SFN induced chemoprevention, there is a growing body of evidence that supports Sp1 as a target of natural chemopreventive phytochemicals in cancer cells [32]. Treatment of prostate cancer cells with the natural products epigallocatechin gallate, betulinic acid, artemisinin, and phenethyl isothiocyanate is also associated with inhibition of Sp1 activity and/or expression [40-43]. In these studies, inhibition of Sp1 is associated with inhibition of the cancer promoting genes survivin, CDK4, VEGF and the androgen receptor. Disruption of Sp1 and Sp3 binding to the promoters of p21, TERT and CDK6 has also been suggested as important chemopreventive mechanisms for the phytochemicals indole-3-carbinol, 3,3’-diindolylmethane, and butyrate in other cancer cell lines, often with an interconnection related to epigenetic modifications associated with the treatments [44-46]. Taken together, these data show that Sp1 is a common target of chemopreventive phytochemicals.
There are many mechanisms of action for SFN that contribute to chemoprevention including the well characterized Nrf2 pathway [6-9]. Our data show that SFN alters the expression of thousands of genes in a dynamic and cell type specific manner. This specificity likely contributes to the important differences in the cellular response to SFN treatment in normal and cancerous prostate cells. Furthermore, SFN reduces the expression of the transcription factor, Sp1, which may be an important mediator by which SFN induces a transcriptional response. This opens the door to a wide range of new scientific questions ranging from possible relationships between Sp1 and epigenetic targets of SFN to interconnections between Sp1 and Nrf-2 pathways. Overall the data presented here, and previous reports, show that SFN can target multiple steps in the carcinogenesis pathway and this makes it a promising cancer preventing agent.
Supplementary Material
Acknowledgments
We thank Jason S. Cumbie, Melanie L. Marine, Mark Dasenko, Christopher M. Sullivan, Matthew Peterson, and Drs. Richard Cronn, John Clarke, Anna Hsu and Adam Branscum for technical assistance and helpful conversations. This work was supported by NIH grants CA90890, CA65525, CA122906, CA122959, CA80176, R01GM104977, and by NIEHS Center grant P30 ES00210 and the Oregon State University general research fund.
Abbreviations
- DMSO
dimethylsulfoxide
- DNMTs
DNA methyltransferases
- FPKM
fragments per kilobase of exon per million fragments mapped
- HDACs
histone deacetylases
- HO1
heme oxygenase 1
- LNCaP
androgen-dependent prostate cancer cells
- NQO1
NAD(P)H:quinone oxidoreductase 1
- Nrf2
nuclear factor (erythroid-derived 2)-like 2
- PC-3
androgen-independent prostate cancer cells
- qPCR
quantitative Real-Time PCR
- RNA-seq
RNA sequencing
- SFN
sulforaphane
- Sp1
Specific protein 1
Footnotes
Conflict of Interest
The authors have no conflicts of interest to declare.
References
- [1].International Agency for Research on Cancer, GLOBOCAN Cancer Fact Sheet: Prostate Cancer. 2010 [Google Scholar]
- [2].American Cancer Society . Cancer Facts & Figures 2012. American Cancer Society Press; Atlanta: 2012. [Google Scholar]
- [3].Liu B, Mao Q, Cao M, Xie L. Cruciferous vegetables intake and risk of prostate cancer: a meta-analysis. Int J Urol. 2012;19:134–141. doi: 10.1111/j.1442-2042.2011.02906.x. [DOI] [PubMed] [Google Scholar]
- [4].Higdon JV, Delage B, Williams DE, Dashwood RH. Cruciferous vegetables and human cancer risk: epidemiologic evidence and mechanistic basis. Pharmacol Res. 2007;55:224–236. doi: 10.1016/j.phrs.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Singh SV, Warin R, Xiao D, Powolny AA, et al. Sulforaphane inhibits prostate carcinogenesis and pulmonary metastasis in TRAMP mice in association with increased cytotoxicity of natural killer cells. Cancer Res. 2009;69:2117–2125. doi: 10.1158/0008-5472.CAN-08-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Agyeman AS, Chaerkady R, Shaw PG, Davidson NE, et al. Transcriptomic and proteomic profiling of KEAP1 disrupted and sulforaphane-treated human breast epithelial cells reveals common expression profiles. Breast Cancer Res. Treat. 2012;132:175–187. doi: 10.1007/s10549-011-1536-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bhamre S, Sahoo D, Tibshirani R, Dill DL, Brooks JD. Temporal changes in gene expression induced by sulforaphane in human prostate cancer cells. Prostate. 2009;69:181–190. doi: 10.1002/pros.20869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, et al. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62:5196–5203. [PubMed] [Google Scholar]
- [9].Hu R, Xu C, Shen G, Jain MR, et al. Gene expression profiles induced by cancer chemopreventive isothiocyanate sulforaphane in the liver of C57BL/6J mice and C57BL/6J/Nrf2 (−/−) mice. Cancer Lett. 2006;243:170–192. doi: 10.1016/j.canlet.2005.11.050. [DOI] [PubMed] [Google Scholar]
- [10].Clarke JD, Hsu A, Yu Z, Dashwood RH, Ho E. Differential effects of sulforaphane on histone deacetylases, cell cycle arrest and apoptosis in normal prostate cells versus hyperplastic and cancerous prostate cells. Mol. Nutr. Food Res. 2011;55:999–1009. doi: 10.1002/mnfr.201000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hsu A, Wong CP, Yu Z, Williams DE, et al. Promoter de-methylation of cyclin D2 by sulforaphane in prostate cancer cells. Clin Epigenetics. 2011;3:3. doi: 10.1186/1868-7083-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ho E, Clarke JD, Dashwood RH. Dietary sulforaphane, a histone deacetylase inhibitor for cancer prevention. J Nutr. 2009;139:2393–2396. doi: 10.3945/jn.109.113332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hu R, Hebbar V, Kim BR, Chen C, et al. In vivo pharmacokinetics and regulation of gene expression profiles by isothiocyanate sulforaphane in the rat. J. Pharmacol. Exp. Ther. 2004;310:263–271. doi: 10.1124/jpet.103.064261. [DOI] [PubMed] [Google Scholar]
- [14].Khor TO, Hu R, Shen G, Jeong WS, et al. Pharmacogenomics of cancer chemopreventive isothiocyanate compound sulforaphane in the intestinal polyps of ApcMin/+ mice. Biopharm Drug Dispos. 2006;27:407–420. doi: 10.1002/bdd.522. [DOI] [PubMed] [Google Scholar]
- [15].Gross-Steinmeyer K, Stapleton PL, Tracy JH, Bammler TK, et al. Sulforaphane- and phenethyl isothiocyanate-induced inhibition of aflatoxin B1-mediated genotoxicity in human hepatocytes: role of GSTM1 genotype and CYP3A4 gene expression. Toxicol Sci. 2010;116:422–432. doi: 10.1093/toxsci/kfq135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Costa V, Angelini C, De Feis I, Ciccodicola A. Uncovering the complexity of transcriptomes with RNA-Seq. J Biomed Biotechnol. 2010;2010:853916. doi: 10.1155/2010/853916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cumbie JS, Kimbrel JA, Di Y, Schafer DW, et al. GENE-counter: a computational pipeline for the analysis of RNA-Seq data for gene expression differences. PLoS One. 2011;6:e25279. doi: 10.1371/journal.pone.0025279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Pflueger D, Terry S, Sboner A, Habegger L, et al. Discovery of non-ETS gene fusions in human prostate cancer using next-generation RNA sequencing. Genome Res. 2011;21:56–67. doi: 10.1101/gr.110684.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Clarke JD, Hsu A, Williams DE, Dashwood RH, et al. Metabolism and tissue distribution of sulforaphane in Nrf2 knockout and wild-type mice. Pharm Res. 2011;28:3171–3179. doi: 10.1007/s11095-011-0500-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Myzak MC, Hardin K, Wang R, Dashwood RH, Ho E. Sulforaphane inhibits histone deacetylase activity in BPH-1, LnCaP and PC-3 prostate epithelial cells. Carcinogenesis. 2006;27:811–819. doi: 10.1093/carcin/bgi265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Myzak MC, Karplus PA, Chung FL, Dashwood RH. A novel mechanism of chemoprotection by sulforaphane: inhibition of histone deacetylase. Cancer Res. 2004;64:5767–5774. doi: 10.1158/0008-5472.CAN-04-1326. [DOI] [PubMed] [Google Scholar]
- [23].Trapnell C, Roberts A, Goff L, Pertea G, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Beaver LM, Yu TW, Sokolowski EI, Williams DE, et al. 3,3'-Diindolylmethane, but not indole-3-carbinol, inhibits histone deacetylase activity in prostate cancer cells. Toxicol Appl Pharmacol. 2012;263:345–351. doi: 10.1016/j.taap.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hunter JD. Matplotlib: A 2D graphics environment. Comput Sci Eng. 2007;9:90–95. [Google Scholar]
- [26].Hulsen T, de Vlieg J, Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- [28].Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41:D64–69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Matys V, Kel-Margoulis OV, Fricke E, Liebich I, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lander ES, Linton LM, Birren B, Nusbaum C, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- [31].Wong CP, Bray TM, Ho E. Induction of proinflammatory response in prostate cancer epithelial cells by activated macrophages. Cancer Lett. 2009;276:38–46. doi: 10.1016/j.canlet.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sankpal UT, Goodison S, Abdelrahim M, Basha R. Targeting Sp1 transcription factors in prostate cancer therapy. Med Chem. 2011;7:518–525. doi: 10.2174/157340611796799203. [DOI] [PubMed] [Google Scholar]
- [33].Chambers KF, Bacon JR, Kemsley EK, Mills RD, et al. Gene expression profile of primary prostate epithelial and stromal cells in response to sulforaphane or iberin exposure. Prostate. 2009;69:1411–1421. doi: 10.1002/pros.20986. [DOI] [PubMed] [Google Scholar]
- [34].Traka MH, Spinks CA, Doleman JF, Melchini A, et al. The dietary isothiocyanate sulforaphane modulates gene expression and alternative gene splicing in a PTEN null preclinical murine model of prostate cancer. Mol Cancer. 2010;9:189. doi: 10.1186/1476-4598-9-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Melchini A, Needs PW, Mithen RF, Traka MH. Enhanced in vitro biological activity of synthetic 2-(2-pyridyl) ethyl isothiocyanate compared to natural 4-(methylsulfinyl) butyl isothiocyanate. J Med Chem. 2012;55:9682–9692. doi: 10.1021/jm300929v. [DOI] [PubMed] [Google Scholar]
- [36].Li L, Davie JR. The role of Sp1 and Sp3 in normal and cancer cell biology. Ann Anat. 2010;192:275–283. doi: 10.1016/j.aanat.2010.07.010. [DOI] [PubMed] [Google Scholar]
- [37].Bedolla RG, Gong J, Prihoda TJ, Yeh IT, et al. Predictive value of Sp1/Sp3/FLIP signature for prostate cancer recurrence. PLoS One. 2012;7:e44917. doi: 10.1371/journal.pone.0044917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lu S, Archer MC. Sp1 coordinately regulates de novo lipogenesis and proliferation in cancer cells. Int J. Cancer. 2010;126:416–425. doi: 10.1002/ijc.24761. [DOI] [PubMed] [Google Scholar]
- [39].Chew YC, Adhikary G, Wilson GM, Xu W, Eckert RL. Sulforaphane induction of p21(Cip1) cyclin-dependent kinase inhibitor expression requires p53 and Sp1 transcription factors and is p53-dependent. J Biol Chem. 2012;287:16168–16178. doi: 10.1074/jbc.M111.305292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ren F, Zhang S, Mitchell SH, Butler R, Young CY. Tea polyphenols down-regulate the expression of the androgen receptor in LNCaP prostate cancer cells. Oncogene. 2000;19:1924–1932. doi: 10.1038/sj.onc.1203511. [DOI] [PubMed] [Google Scholar]
- [41].Chintharlapalli S, Papineni S, Ramaiah SK, Safe S. Betulinic acid inhibits prostate cancer growth through inhibition of specificity protein transcription factors. Cancer Res. 2007;67:2816–2823. doi: 10.1158/0008-5472.CAN-06-3735. [DOI] [PubMed] [Google Scholar]
- [42].Willoughby JA, Sr., Sundar SN, Cheung M, Tin AS, et al. Artemisinin blocks prostate cancer growth and cell cycle progression by disrupting Sp1 interactions with the cyclin-dependent kinase-4 (CDK4) promoter and inhibiting CDK4 gene expression. J Biol Chem. 2009;284:2203–2213. doi: 10.1074/jbc.M804491200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wang LG, Liu XM, Chiao JW. Repression of androgen receptor in prostate cancer cells by phenethyl isothiocyanate. Carcinogenesis. 2006;27:2124–2132. doi: 10.1093/carcin/bgl075. [DOI] [PubMed] [Google Scholar]
- [44].Cram EJ, Liu BD, Bjeldanes LF, Firestone GL. Indole-3-carbinol inhibits CDK6 expression in human MCF-7 breast cancer cells by disrupting Sp1 transcription factor interactions with a composite element in the CDK6 gene promoter. J Biol. Chem. 2001;276:22332–22340. doi: 10.1074/jbc.M010539200. [DOI] [PubMed] [Google Scholar]
- [45].Hong C, Kim HA, Firestone GL, Bjeldanes LF. 3,3'-Diindolylmethane (DIM) induces a G(1) cell cycle arrest in human breast cancer cells that is accompanied by Sp1-mediated activation of p21(WAF1/CIP1) expression. Carcinogenesis. 2002;23:1297–1305. doi: 10.1093/carcin/23.8.1297. [DOI] [PubMed] [Google Scholar]
- [46].Davie JR. Inhibition of histone deacetylase activity by butyrate. J. Nutr. 2003;133:2485S–2493. doi: 10.1093/jn/133.7.2485S. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.