Abstract
Accurate knowledge of a patient’s disease state and trajectory is critical in a clinical setting. Modern electronic healthcare records contain an increasingly large amount of data, and the ability to automatically identify the factors that influence patient outcomes stand to greatly improve the efficiency and quality of care.
We examined the use of latent variable models (viz. Latent Dirichlet Allocation) to decompose free-text hospital notes into meaningful features, and the predictive power of these features for patient mortality. We considered three prediction regimes: (1) baseline prediction, (2) dynamic (time-varying) outcome prediction, and (3) retrospective outcome prediction. In each, our prediction task differs from the familiar time-varying situation whereby data accumulates; since fewer patients have long ICU stays, as we move forward in time fewer patients are available and the prediction task becomes increasingly difficult.
We found that latent topic-derived features were effective in determining patient mortality under three timelines: inhospital, 30 day post-discharge, and 1 year post-discharge mortality. Our results demonstrated that the latent topic features important in predicting hospital mortality are very different from those that are important in post-discharge mortality. In general, latent topic features were more predictive than structured features, and a combination of the two performed best.
The time-varying models that combined latent topic features and baseline features had AUCs that reached 0.85, 0.80, and 0.77 for in-hospital, 30 day post-discharge and 1 year post-discharge mortality respectively. Our results agreed with other work suggesting that the first 24 hours of patient information are often the most predictive of hospital mortality. Retrospective models that used a combination of latent topic features and structured features achieved AUCs of 0.96, 0.82, and 0.81 for in-hospital, 30 day, and 1-year mortality prediction.
Our work focuses on the dynamic (time-varying) setting because models from this regime could facilitate an on-going severity stratification system that helps direct care-staff resources and inform treatment strategies.
1. INTRODUCTION
In a fragmented healthcare system of patients, doctors, caregivers, and specialists, an accurate knowledge of a patient’s disease state is critical. Electronic monitoring systems and health records facilitate the flow of information among these parties to effectively manage patient health. However, information is not knowledge, and often only some of the information will be relevant in the context of providing care. Expert physicians want to sift through these extensive records to discover the data most relevant to a patient’s current condition. As such, systems that can identify these patterns of relevant characteristics stand to improve the efficiency and quality of care.
This work focused on the task of on-going mortality prediction in the intensive care unit (ICU). The ICU is a particularly challenging environment because each patient’s severity of illness is constantly evolving. Further, modern ICUs are equipped with many independent measurement devices that often produce conflicting (and even false) alarms, adversely affecting the quality of care. Consequently, much recent work in ICU mortality models [8, 10, 17] has aimed to consolidate data from these devices (primarily structured data and physiological waveforms) and transform these information streams into knowledge. However, these works omit perhaps the most descriptive sources of medical information: free-text clinical notes and reports.
The narrative in the clinical notes, recorded by expert care staff, is designed to provide trained professionals a quick glance into the most important aspects of a patient’s physiology. Combining features extracted from these notations with standard physiological measurements results in a more complete representation of patients’ physiological states, thus affording improved outcome prediction. Unfortunately, free-text data are often more difficult to include in predictive models because they lack the structure required by most machine learning methods. To overcome the obstacles inherent in clinical text, latent variable models such as topic models [1, 2] may be used to infer intermediary representations that can in turn be used as structured features for a prediction task.
We demonstrate the value of incorporating information from clinical notes, via latent topic features, in the task of in-hospital mortality prediction as well as 30 day and 1 year post-discharge mortality prediction. Specifically, we evaluated mortality prediction under three prediction regimes: (1) baseline regime, which used structured data available on admission (2) time-varying regime, which used baseline features together with dynamically accumulated clinical text using increasigly large subsets of the patient’s narrative record, and (3) retrospective regime, which used all clinical text generated from a hospital stay to supplement the baseline features. In all targeted outcomes, we demonstrate that adding information from clinical notes improves predictions of mortality.
2. RELATED WORK
Mortality models for acute (i.e. ICU) settings constitute a broad area of research. Siontis et al. [16] reviewed 94 studies with 240 assessments of 118 mortality prediction tools from 2009 alone. Many of these studies evaluated established clinical decision rules for predicting mortality, such as APACHE [9], SAPS-II [10], and SOFA [17] (with median reported AUCs of 0.77, 0.77, and 0.84, respectively). Sionitis et al. also noted a large variability of these measures across various diseases and population subgroups. Other acuity scores have also been proposed, including the recent OASIS score [8] which uses machine-learning algorithms to identify the minimal set of variables capable of yielding an accurate severity of illness score (AUC 0.88).
Work by Hug et al. [7] used several hundred structured clinical variables to create a real-time ICU acuity score that reported an AUC of 0.88-0.89 for in-hospital mortality prediction. Notably, most of the predictive power of their models was from data gathered within the first 24 hours of the ICU stay. For example, their computed acuity score reported an AUC of 0.809 for in-hospital mortality prediction based on information during the first 24 hours of ICU stays in 1,954 patients.
Several recent works have used information from clinical notes in their model formulations. Saria et al. [15] combined structured physiological data with concepts from the discharge summaries to achieve a patient outcome classification F1 score of 88.3 with a corresponding reduction in error of 23.52%. Similarly, [5] described preliminary results indicating that topic models extracted from clinical text in a subgroup of ICU patients were valuable in the prediction of per-admission mortality. They found that common topics from the unlabeled clinical notes were predictive of mortality, and an RBF SVM achieved a retrospective AUC of 0.855 for in-hospital mortality prediction using only learned topics. Finally, Lehman et al. [11] applied Hierarchical Dirichlet Processes to nursing notes from the first 24 hours for ICU patient risk stratification. They demonstrated that unstructured nursing notes were enriched with clinically meaningful information, and this information could be used for clinical support. Using topic proportions, the average AUC for hospital mortality prediction was 0.78 (± 0.01). In combination with the SAPS-I variable, their average AUC for hospital mortality prediction was 0.82 (± 0.003).
3. METHODS
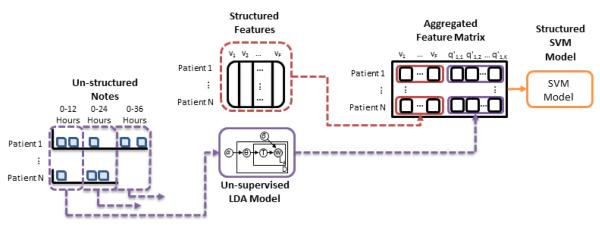
Figure 1 gives a general overview of our experimental process. First, we extract clinical baseline features, including age, sex, and SAPS-II score, from the database for every patient. We also extract each patient’s de-identified clinical notes. We use these notes as the observed data in an LDA topic model, and infer a total of 50 topics. We normalize the word counts associated with each note, so that each note is represented by a 50-dimension vector, summing to 1. These per-note topic distributions are then aggregated on a 12 hour semi-continuous timescale (e.g. notes within 0-12 hours, notes within 0-24 hours, etc.). A linear kernel SVM is trained to create classification boundaries with combinations of the structured clinical features and latent topic features to predict in-hospital mortality, 30 day post-discharge mortality, and 1 year post-discharge mortality.
Figure 1.
Overall flow of experiment. 1) Clinical baseline features are extracted from the database for every patient (e.g. age, sex, admitting SAPS-II score) and derived features are computed (e.g. maximum/minimum SAPS-II score) to form the Structured Features matrix v (vp,f is the value of feature f in the pth patient). 2) Each patient’s de-identified clinical notes are used as the observed data in an LDA topic model (i.e., Un-supervisted LDA Model), and a total of 50 topics are inferred to create the per-note topic proportion matrix q. 3) Per-note latent topic features are aggregated in extending 12 hour windows (e.g. notes within 0-12 hours, notes within 0-24 hours, etc.) and used to form matrix q’ where is the overall proportion of topic k in time-window m. 4) Depending on the model and time window being evaluated, subsets of the feature matrix v and matrix q’ are combined into an Aggregated Feature Matrix. 5) A linear kernel SVM is trained to create classification boundaries for three clinical outcomes: in-hospital mortality, 30 day post-discharge mortality, and 1 year post-discharge mortality (i.e. Structured SVM Model).
3.1 Data and Pre-Processing
We used ICU data from the MIMIC II 2.6 database [13], a publicly-available, de-identified medical corpus which includes electronic medical records (EMRs) for 26, 870 ICU patients at the Beth Israel Deaconess Medical Center (BIDMC) collected from 2001 to 2008. Patient age, sex, SAPS-II scores, International Classification of Diseases-Ninth Revision (ICD-9) diagnoses, and Disease-Related Group were extracted. Medical co-morbidities were represented by the Elixhauser scores (EH) for 30 co-morbidities as calculated from the ICD-9 codes. Patient mortality outcomes were also queried to determine which patients died in-hospital, or lived past the most recent query of Social Security records.
We extracted all clinical notes recorded prior to the patient’s first discharge, including notes from nursing, physicians, labs, and radiology. The discharge summaries themselves were excluded because they typically stated the patient’s outcome explicitly. Vocabularies for each note were generated by first tokenizing the free text and then removing stopwords using the Onix stopword list 1. A TF-IDF metric [14] was applied to determine the 500 most informative words in each patient’s notes, and we then limited our overall vocabulary to the union of the most informative words per-patient. This pre-processing step reduced the overall vocabulary down to 285,840 words from over 1 million terms while maintaining the most distinctive features of each patient.2
Patients were excluded if they had fewer than 100 non-stop words or were under the age of 18. Specific notes were excluded if they occurred after the the end of the day in which a patient died or was discharged (e.g. radiology or lab reports whose results were reported afterwards). The resulting cohort consisted of 19,308 patients with 473,764 notes. We held out a random 30% of the patients as a test set. The remaining 70% of patients were used to train our topic models and mortality predictors. Table 1 summarizes the number of notes and patients in the training and test sets.
Table 1.
Cohort Composition
| Train | Test | Total | |
|---|---|---|---|
| Patients | 13,524 | 5,784 | 19,308 |
| Notes | 331,635 | 142,129 | 473,764 |
3.2 Structured and Derived Features
In total, we extracted and derived 36 structured clinical variables for each patient: the age, gender, SAPS II score on admission, minimum SAPS II score, maximum SAPS II score, final SAPS II score, and the 30 EH comorbidities. Data were scaled to avoid the range of a feature impacting its classification importance. This formed a feature matrix v, where the element vp,f was the value of feature f in the pth patient.
3.3 Topic Inference
Instead of considering each note separately, we used the set of all of notes that occurred in a particular time period as features for that period. We examined the distribution of note times, and found three peaks in note entry for any given day in a patient’s stay (e.g. day 1, day 2, etc.): around 06:00, 18:00 and 24:00.3 Given this distribution, we used 12-hour windows for our time windows.
Topics were generated for each note using Latent Dirichlet Allocation [2,6]. Our initial experiments found no significant difference in held-out prediction accuracy across a range of 20 to 100 topics. We set hyperparameters on the Dirichlet priors for the topic distributions (α) and the topic-word distributions (β) as , . We used 50 topics in our final experiments, and topic distributions were sampled from an MCMC chain after 2,500 iterations. This topic-modeling step resulted in a 50-dimensional vector of topic proportions for each patient for each note.
We concatenated the topic vectors into a matrix q where the element qn,k was the proportion of topic k in the nth note. Of particular interest was whether certain topics were enriched for in-hospital mortality and long-term survival. We used an enrichment measure defined by Marlin et al [12], where the probability of mortality for each topic is calculated as , where yn is the noted mortality outcome (0 for a patient that lives, and 1 for a patient that dies). These enrichment measures are reported in section 4.1.
The time windows were used to construct feature vectors for each prediction task, where (at each step) we extended the period of consideration forward by 12 hours. From the previously constructed per-note matrix q that describes the distribution over topics in each note, we collapse into another matrix q’ where describes the overall proportion of topic k in time-window m. The element is given by the mean of that topic’s proportions of all the notes in time-window m: meann∈mqn,k.
3.4 Prediction
We considered three prediction regimes with the inferred topic distributions: baseline prediction, dynamic (time-varying) outcome prediction and retrospective outcome prediction for the outcomes of in-hospital, 30-day, and 1-year mortality.
A separate linear SVM [4] was trained for each of the three outcomes, and each set of model features evaluated. The loss and class weight parameters for the SVM were selected using five-fold cross-validation on the training data to determine the optimal values with AUC as an objective. The learned parameters were then used to construct a model for the entire training set, and make predictions on the test data.
All outcomes had large class-imbalance (mortality rates of 10.9% in-hospital, 3.7% 30 day post-discharge, and 13.7% 1 year post-discharge4). To address this issue, we randomly sub-sampled the negative class in the training set to produce a minimum 70%/30% ratio between the negative and positive classes. Test set distributions were not modified to reflect the reality of class imbalance during prediction, and reported performance reflects those distributions.
First, we established a static baseline model using only structured features present at admission (i.e. clinical baseline features and derived features thereof). We then ran dynamic outcome prediction in intervals of 12 hours at each step by including larger sets of patient notes in a step-wise manner. We finally performed retrospective outcome predictions, where we included structured features and all notes written during the stay as a static entity for prediction. Significantly, predictions of mortality with this type of feature set are a retrospective exercise only: it is not possible to first select all notes that occur before a patient’s death, and then predict in-hospital mortality, because the time of mortality is not known a-priori. The observer would have to “know” that the patient’s hospital record was about to finish (either by death or discharge). The following settings were evaluated:
Admission Baseline Model: A baseline model using the structured features of age, gender, and the SAPS II score at admission. These baseline features are extracted from the data present at patient admission only. (3 features total)
Time-varying Topic Model 1 - 20: Outcome prediction performed by including notes in a step-wise fashion, extending the period of consideration forward by 12 hours at each step. For example, Time-varying Topic Model 1 includes topic features derived from all notes written during the first 12 hours of a patient’s stay in the ICU, while Time-varying Topic Model 20 includes those derived from the first 240 hours. (50 features total)
Combined Time-varying Model 1 - 20: Outcome pre diction using the same setup as Time-varying Topic Model 1 - 20, but with the static structured features from Admission Baseline Model (gender, age, admitting SAPS score) included. (53 features total)
Retrospective Derived Features Model: A retrospective model using the structured features of age, gender, admitting SAPS II score, the minimum SAPS II score, the maximum SAPS II score, the final SAPS II score, and all EH comorbidities. (36 features total)
Retrospective Topic Model: A retrospective model using topics derived from all notes written during a patient’s stay in the ICU. (50 features total)
Retrospective Topic + Admission Model: A retrospective model combining structured features from Admission Baseline Model (gender, age, admitting SAPS scores) with latent topic features from Retrospective Topic Model. (53 features total)
Retrospective Topic + Derived Features Model: A retrospective model combining structured features from Retrospective Derived Features Model (gender, age, admitting/min/max/final SAPS scores, EH comorbidities) with latent topic features from Retrospective Topic Model. (86 features total)
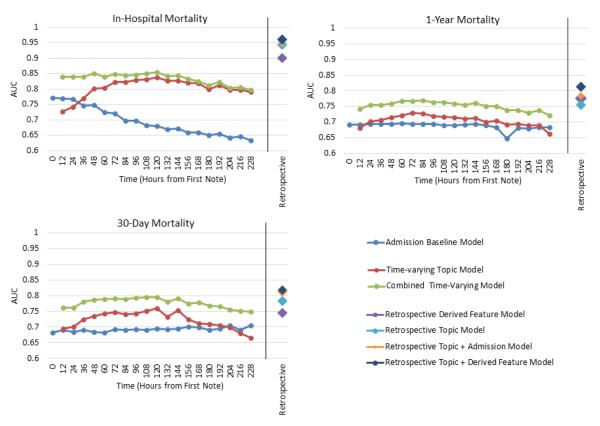
We compare the prediction results for all models on each of the outcomes in Figure 3 and Table A.2. We again emphasize that retrospective models are retrospective exercises only to establish the isolated and combined prediction ability of clinical notes and features. We also note that our Time-varying Topic Model is time-varying only in its application. We do not use other possible latent variable models such as “Dynamic topic models” [3], because we do not want to model the time evolution of topics, but rather the time evolution of membership to a given set of topics.
Figure 3.
Linear SVM model performance measured via AUC on three outcomes: in-hospital mortality, 30 day post-discharge mortality, and 1 year post-discharge mortality. In each case, the features used are described in detail in Section 3.4. Our prediction task is different from the usual situation where data is accumulated over time. Since fewer patients have long ICU stays, in this case, we actually lose data points as time goes on, making the prediction task harder. For example, at time 0 there are 5,784 patients (5,157 controls/627 positives for in-hospital mortality) in the test set. By 72 hours, this had dropped to 5,084 patients (4,591 controls/493 positives for in-hospital mortality) and at 144 hours to 3,496 patients (3,141 controls/355 positives for in-hospital mortality). (Table A.1)
4. RESULTS
4.1 Qualitative Enrichment
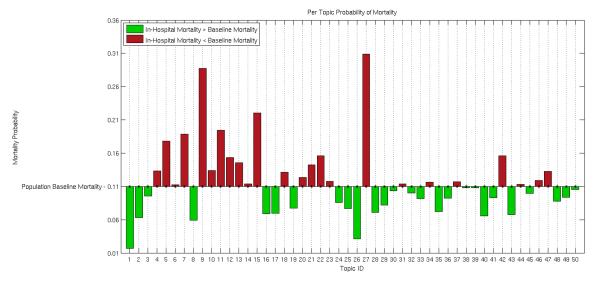
Table 2 lists the top words for the topics which had the largest enrichment for in-hospital mortality, the smallest enrichment for in-hospital mortality, and the highest enrichment for 1 year mortality. The relative distributions of the in-hospital mortality probabilities for each of the 50 topics are shown in Figure 2. There were a wide variation in the in-hospital mortality concentration for the different topics, ranging from 3% - 30%. (See Table A.3 for a listing of top ten words for all topics.)
Table 2.
Top ten words in topics enriched for in-hospital mortality, hospital survival (any number of days post-discharge), and 1 year post-discharge mortality.
| Topic | Top Ten Words | Possible Topic |
|
|---|---|---|---|
| In- hospital Mor- tality |
27 | name, family, neuro, care, noted, status, plan, stitle, dr, remains |
Discussion of end- of-life care |
|
|
|||
| 15 | intubated, vent, ett, secre- tions, propofol, abg, respi- ratory, resp, care, sedated |
Respiratory failure |
|
|
|
|||
| 7 | thick, secretions, vent, trach, resp, tf, tube, coarse, cont, suctioned |
Respiratory infection |
|
|
|
|||
| 5 | liver, renal, hepatic, ascites, dialysis, failure, ow, transplant, portal, ultrasound |
Renal Failure |
|
|
| |||
| Hospital Sur- vival |
1 | cabg, pain, ct, artery, coronary, valve, post, wires, chest, sp |
Cardio- vascular surgery |
|
|
|||
| 40 | left, fracture, ap, views, reason, clip, hip, distal, lat, report |
Fracture | |
|
|
|||
| 16 | gtt, insulin, bs, lasix, endo, monitor, mg, am, plan, iv |
Chronic diabetes |
|
|
| |||
| 1 Year Mor- tality |
3 | picc, line, name, proce- dure, catheter, vein, tip, placement, clip, access |
PICC5 line inser- tion |
|
|
|||
| 4 | biliary, mass, duct, metastatic, bile, cancer, left, ca, tumor, clip |
Cancer treatment |
|
|
|
|||
| 45 | catheter, name, pro- cedure, contrast, wire, french, placed, needle, advanced, clip |
Coronary catheteri- zation |
|
Figure 2.
The probability of in-hospital mortality for each topic, indicating that topics represent differences in outcome. Probabilities are calculated as (see section 3.3). Each bar shows the prevalence of a given topic k in the mortality category, as compared to the set of all patients. Bars are shown as above (in red) or below (in green) the baseline in-hospital mortality based on the value of θk for each topic k.
The topics enriched for in-hospital mortality presented a detailed view of the possible causes of death in the ICU. For example, patients in a modern ICU rarely die suddenly. Often patient life is sustained for some time in order for their family to express their wishes regarding terminal care and death. This could be one interpretation for Topic 27, which pertains to the discussion of end-of-life care options. Other topics with in-hospital mortality enrichment pertained to top causes of ICU mortality: respiratory infection (Topic 7), respiratory failure (Topic 15), and renal failure (Topic 5).
Hospital survival was also marked by topics which seem relevant to factors tied closely to the ability to recover from physiological insults: patients who are admitted for cardiovascular surgery (Topic 1) are often not allowed as surgical candidates until they are in very good health; patients who are able to respond to their care staff and the ICU environment (Topic 26, Table A.3) are adequately dealing with the known stress of ICU admission; patients with trauma-based injuries such as fracture and pneumothorax (Topics 8, 40); and patients with chronic conditions like diabetes (Topic 16).
The topics enriched for 1 year post-discharge mortality suggested that patients who are discharged but die within a year have conditions with a low chance of long-term survival. For example, cancer (Topic 4), the need for long-term IV access while in the ICU (Topic 3), and the use of coronary catheterization (Topic 45) to diagnose activity in coronary arteries or other valvular/cardiac issues.
4.2 Prediction
We evaluated the predictive power of each model and outcome pair. Figure 3 shows the AUCs achieved by each model for the three targeted outcomes. Table A.2 lists a more complete set of the SVM classification metrics.
As shown in Table A.2, the prevalent class imbalance resulted in a bias toward low specificities in the Admission Baseline Model. The balance between sensitivity and specificity generally leaned towards favoring higher specificities for in-hospital and 30 day mortality prediction as time moved forward in the Time-varying models, but this was not uniformaly true in all cases. In general, the Retrospective Derived Features Model had a high sensitivity and low specificity, the Retrospective Topic Model had good specificity, and the combined models tended to have a more even set of both measures.
For 30 day and 1 year post-discharge mortality prediction, the Admission Baseline Model was very steady, averaging an AUC of 0.68 over all time windows for both outcomes. The Combined Time-varying Model achieved an average/best performance of 0.77/0.8 for 30 day mortality and 0.75/0.77 for 1 year mortality. In both outcomes the Time-varying Topic Model performed strictly better than the Admission Baseline Model until the available patient subset became minimal (the 204*** -216 hour windows), and the Combined Time-varying Model was always better than either alone.
As expected, the four Retrospective models were generally more predictive than any of the Time-varying models. Retrospective models tended to increase performance as more features were added. For in-hospital and 30 day mortality prediction, the Retrospective Topic Model performed better than the Retrospective Derived Features Model (AUCs increased from 0.90 to 0.94 and 0.75 to 0.78 respectively). For 1 year mortality this was reversed (AUC decreased from 0.78 to 0.76).
In the in-hospital mortality setting, it seemed that admission features were not needed once latent topic features are known, but the derived features did provide extra information6. However, in the 30 day setting, latent topic features were similarly improved by either the admission features or the derived features7. This is likely because the derived features included EH comorbidities derived from the ICD-9 codes, and the ICD-9 codes themselves are often transcribed after a patient’s discharge with the most actionable (or billable) conditions a patient presented. It is possible that these features are most relevant to in-hospital mortality risks (e.g. EH scores for myocardial infarction, congestive heart failures, etc.).
5. DISCUSSION
Models that incorporated latent topic features were generally more predictive than those using only structured features, and a combination of the two feature types performed best. Notably, the combination provides a robustness that is able to perform well initially, leveraging primarily the structured information, and then continue to improve over the first 24 hours by incorporating the latent topic features. This resilience is particularly important since we observed that the first 24 hours of clinical notes appear to be the most meaningful toward predicting in-hospital mortality, while the baseline begins to steadily decrease.
Our observation of the importance of early data agrees with other reported results. Recall that, using topics derived from the first 24 hours of notes only, Lehman et al obtained an average AUC for in-hospital mortality prediction of 0.78 (± 0.01), and this was increased to 0.82 (± 0.003) with the SAPS-I variable. Further, Hug et al. obtained an AUC of 0.809 for in-hospital mortality prediction based on information during the first 24 hours of ICU. As such, we examined our results for in-hospital mortality when using topics derived from the first 24 hours of notes only (prediction time of 36 hours in Figure 3), and obtained corresponding AUCs of 0.77 for the Time-varying Topic Model, and 0.841 for the Combined Time-varying Model. Compared to Lehman et al’s result, this implies that (with enough data) neither the extra hierarchical machinery added with HDPs nor the knowledge-based cleansing of medical terms before modeling improve prediction results (i.e. an AUC of 0.78 vs. 0.77). Compared to Hug et al’s results, this implies that the addition of clinical text provides reasonable performance boosts to the power of gold-standard structured information like SAPS-II score (i.e. an AUC of 0.809 vs. 0.841).
Further, when predicting in-hospital mortality, we observed that the Admission Baseline Model’s predictive power (i.e. information acquired on admission) becomes much less valuable to predicting mortality as patients stay longer. This is likely because those who are not discharged within the first day of hospital admission are significantly sicker than those who are. Note that the average ICU stay time in the MIMIC II database is 3 days, and Figure 3 shows that after this time there was no additional predictive power gained by adding the structured admission information to the latent topic features (i.e., the Time-varying Topic Model and the Combined Time-varying Model converge).
This convergence draws attention to another interesting observation. Namely, both of the Time-varying models trended up in their ability to predict in-hospital mortality until 120 hours, and then trended down until the end of prediction. While initially counterintuitive, this is likely due to the loss of a significant number of patients (from both death and discharge) in the available patient cohort. For example, the test set population goes from 4,030 patients (3,626 control/404 positive for in-hospital mortality) to 3570 patients at this point (3,210 control/360 positive for in-hospital mortality).
Additionally, the predictive power of each topic changed depending on the target outcome. This appeals to intuition asIn a modern ICU, conditions that lead to in-hospital mortality are very different from those that would allow for a live discharge leading to a 30 day or 1 year mortality. As such, information about which topics tend to bias a patient towards any set of outcomes in useful for clinicians, when compared to the typical “black-box” approach to feature selection.
Finally, much work focuses on retrospective prediction of mortality outcomes. We also performed these predictions to compare the relative predictive power of different feature types and were able to achieve retrospective AUCs of 0.9, 0.94 and 0.96 for in-hospital mortality prediction using the Retrospective Derived Feature Model, Retrospective Topic Model, and combined Retrospective Topic + Dervied Features Model. However, we re-emphasize that predictions of mortality with retrospective feature sets are not helpful or relevant for clinical staff because statistical functions of signals or features (e.g. min/max) and other structured data (such as ICD-9 codes and EH comorbidities) are not known a-priori.
6. CONCLUSIONS
Modern electronic healthcare records contain an increasingly large amount of data including high-frequency signals from biomedical instrumentation, intermittent results from lab tests, and text from notes. Such voluminous records can make it difficult for care-sta to identify the information relevant to diagnose a patient’s condition and stratify patients with similar characteristics.
Standard approaches to hospital mortality prediction use features such as gender, age, SAPS and SOFA score. In this work, we examined the utility of augmenting these standard features with textual information—specifically in the form of topic-based features—for predicting mortality in the ICU. Features extracted by latent variable models are attractive in this clinical application because scientific understanding is as important as clinical utility.
Qualitatively, the discovered topics correlated with known causes of in-hospital and post-discharge death. Further, adding latent topic features to structured clincal features increased classification performance in a variety of prediction scenarios: in-hospital mortality, 30-day mortality, and 1-year mortality.
The models and results explored in this work could ultimately be useful for interpretable models of disease and mortality.
ACKNOWLEDGMENTS
This research was funded in part by the Intel Science and Technology Center for Big Data and the National Library of Medicine Biomedical Informatics Research Training grant (NIH/NLM 2T15 LM007092-22) which supported Marzyeh Ghassemi and Tristan Naumann.
The authors would like to thank Abbas Benjamin Munson-Ghassemi for many nights of heartburn and kick-fueled writing.
General Terms
- Primary
Data mining for social good.
- Secondary
Healthcare and medicine; Topic, graphical and latent variable models; Text; Support vector machines.
APPENDIX. A. PATIENT COHORT SIZES
Table A.1.
Patient cohort size at each time tested by time-varying models. Note that patients are removed from a prediction time if they are discharged or die prior to that time.
| Cohort Size (Control, Positive) |
||||
|---|---|---|---|---|
| Time (Hours) |
Total | In- Hospital |
30 Day | 1 Year |
| 0 | 5784 | 5157, 627 | 5597, 187 | 5058, 726 |
| 12 | 5784 | 5157, 627 | 5597, 187 | 5058, 726 |
| 24 | 5749 | 5128, 621 | 5563, 186 | 5026, 723 |
| 36 | 5563 | 4998, 565 | 5382, 181 | 4855, 708 |
| 48 | 5497 | 4937, 560 | 5318, 179 | 4795, 702 |
| 60 | 5161 | 4664, 497 | 4986, 175 | 4480, 681 |
| 72 | 5084 | 4591, 493 | 4911, 173 | 4407, 677 |
| 84 | 4691 | 4241, 450 | 4524, 167 | 4043, 648 |
| 96 | 4587 | 4140, 447 | 4421, 166 | 3945, 642 |
| 108 | 4116 | 3710, 406 | 3963, 153 | 3530, 586 |
| 120 | 4030 | 3626, 404 | 3877, 153 | 3448, 582 |
| 132 | 3570 | 3210, 360 | 3427, 143 | 3023, 547 |
| 144 | 3496 | 3141, 355 | 3354, 142 | 2956, 540 |
| 156 | 3026 | 2707, 319 | 2898, 128 | 2533, 493 |
| 168 | 2967 | 2652, 315 | 2840, 127 | 2479, 488 |
| 180 | 2580 | 2291, 289 | 2468, 112 | 2138, 442 |
| 192 | 2541 | 2254, 287 | 2431, 110 | 2109, 432 |
| 204 | 2215 | 1953, 262 | 2117, 98 | 1825, 390 |
| 216 | 2186 | 1925, 261 | 2090, 96 | 1802, 384 |
| 228 | 1925 | 1681, 244 | 1837, 88 | 1575, 350 |
B. MODEL PERFORMANCE RESULTS
Table A.2.
Detailed model prediction results for three outcomes: in-hospital mortality, 30 day post-discharge mortality, and 1 year post-discharge mortality. This also appears in Figure 3.
| Outcome Predicted | Model Used | AUC | Sens. | Spec. |
|---|---|---|---|---|
| In-Hospital Mortality | Admission Baseline Model | 0.771 | 0.999 | 0.010 |
| Time-varying Topic Model 1 | 0.728 | 0.858 | 0.471 | |
| … | … | |||
| Time-varying Topic Model 10 | 0.838 | 0.686 | 0.829 | |
| … | … | |||
| Time-varying Topic Model 20 | 0.791 | 0.525 | 0.853 | |
| Combined Time-varying Model 1 | 0.840 | 0.638 | 0.85 | |
| … | … | |||
| Combined Time-varying Model 10 | 0.854 | 0.666 | 0.844 | |
| … | … | |||
| Combined Time-varying Model 20 | 0.798 | 0.299 | 0.950 | |
| Retrospective Derived Features Model | 0.901 | 0.997 | 0.108 | |
| Retrospective Topic Model | 0.944 | 0.856 | 0.892 | |
| Retrospective Topic + Admission Model | 0.944 | 0.821 | 0.910 | |
| Retrospective Topic + Derived Features Model | 0.961 | 0.915 | 0.870 | |
| 30 Day Mortality | Admission Baseline Model | 0.683 | 0.995 | 0.075 |
| Time-varying Topic Model 1 | 0.695 | 0.150 | 0.944 | |
| … | … | |||
| Time-varying Topic Model 10 | 0.759 | 0.817 | 0.551 | |
| … | … | |||
| Time-varying Topic Model 20 | 0.665 | 0.602 | 0.579 | |
| Combined Time-varying Model 1 | 0.761 | 0.348 | 0.885 | |
| … | … | |||
| Combined Time-varying Model 10 | 0.796 | 0.641 | 0.770 | |
| … | … | |||
| Combined Time-varying Model 20 | 0.75 | 0.011 | 0.991 | |
| Retrospective Derived Features Model | 0.745 | 0.941 | 0.220 | |
| Retrospective Topic Model | 0.783 | 0.342 | 0.909 | |
| Retrospective Topic + Admission Model | 0.813 | 0.872 | 0.633 | |
| Retrospective Topic + Derived Features Model | 0.818 | 0.096 | 0.985 | |
| 1 Year Mortality | Admission Baseline Model | 0.692 | 0.997 | 0.021 |
| Time-varying Topic Model 1 | 0.681 | 0.218 | 0.907 | |
| … | … | |||
| Time-varying Topic Model 10 | 0.715 | 0.321 | 0.870 | |
| … | … | |||
| Time-varying Topic Model 20 | 0.662 | 0.834 | 0.379 | |
| Combined Time-varying Model 1 | 0.743 | 0.705 | 0.665 | |
| … | … | |||
| Combined Time-varying Model 10 | 0.760 | 0.512 | 0.812 | |
| … | … | |||
| Combined Time-varying Model 20 | 0.722 | 0.451 | 0.804 | |
| Retrospective Derived Features Model | 0.776 | 0.999 | 0.045 | |
| Retrospective Topic Model | 0.755 | 0.358 | 0.890 | |
| Retrospective Topic + Admission Model | 0.784 | 0.314 | 0.919 | |
| Retrospective Topic + Derived Features Model | 0.813 | 0.464 | 0.887 | |
C. LIST OF INFERRED TOPICS
Table A.3.
Top ten most probable words for all topics.
| Topic Number | Top Ten Words |
|---|---|
| 1 | cabg, pain, ct, artery, coronary, valve, post, wires, chest, sp |
| 2 | ccu, cath, mg, am, sp, groin, bp, cardiac, hr, cont |
| 3 | picc, line, name, procedure, catheter, vein, tip, placement, clip, access |
| 4 | biliary, mass, duct, metastatic, bile, cancer, left, ca, tumor, clip |
| 5 | liver, renal, hepatic, ascites, dialysis, failure, ow, transplant, portal, ultrasound |
| 6 | ct, contrast, pelvis, abdomen, uid, bowel, clip, free, wcontrast, iv |
| 7 | thick, secretions, vent, trach, resp, tf, tube, coarse, cont, suctioned |
| 8 | chest, pneumothorax, tube, reason, clip, sp, ap, left, portable, ptx |
| 9 | remains, family, gtt, line, map, cont, levophed, cvp, bp, levo |
| 10 | name, neo, gtt, stitle, dr, sbp, resp, cont, wean, aware |
| 11 | remains, increased, temp, hr, pt, cc, ativan, cont, mg, continues |
| 12 | micu, code, stool, hr, bp, social, note, id, received, cchr |
| 13 | chest, pulmonary, bilateral, edema, portable, clip, reason, ap, pleural, effusions |
| 14 | resp, cough, sats, mask, sob, wheezes, nc, status, mg, neb |
| 15 | intubated, vent, ett, secretions, propofol, abg, respiratory, resp, care, sedated |
| 16 | gtt, insulin, bs, lasix, endo, monitor, mg, am, plan, iv |
| 17 | drainage, pain, abd, uid, draining, drain, incision, sp, intact, pt |
| 18 | heparin, afib, ptt, am, gtt, mg, rate, hr, pvcs, iv |
| 19 | name, pacer, namepattern, placement, heart, pacemaker, ventricular, av, rate, chest |
| 20 | left, lung, effusion, lobe, pleural, lower, chest, upper, ct, opacity |
| 21 | skin, noted, care, left, applied, changed, draining, coccyx, wound, edema |
| 22 | tube, placement, tip, line, portable, ap, reason, position, chest, ng |
| 23 | noted, shift, name, pt, patent, patient, foley, agitated, soft, mg |
| 24 | hct, pt, gi, blood, bleeding, am, stable, unit, bleed, noted |
| 25 | name, am, mg, able, bp, time, night, times, doctor, confused |
| 26 | pain, co, denies, oriented, neuro, plan, diet, po, pt, floor |
| 27 | name, family, neuro, care, noted, status, plan, stitle, dr, remains |
| 28 | clip, reason, ro, medical, examination, evidence, impression, underlying, condition, normal |
| 29 | neuro, sbp, bp, commands, iv, cough, soft, status, lopressor, swallow |
| 30 | skin, stable, social, family, intact, tsicu, id, note, support, endo |
| 31 | woman, female, husband, name, pain, patient, pm, am, hospital, noted |
| 32 | diagnosis, admitting, name, reason, please, examination, yearold, eval, findings, underlying |
| 33 | name, neck, soft, patient, noted, anterior, epidural, level, posterior, namepattern |
| 34 | ct, contrast, chest, lymph, optiray, images, lesions, iv, nodes, lobe |
| 35 | left, stenosis, disease, clip, reason, carotid, severe, report, radiology, final |
| 36 | femoral, foot, left, leg, iliac, groin, lower, patent, graft, extremity |
| 37 | acute, reason, head, clip, evidence, eval, name, wo, status, ct |
| 38 | aortic, aorta, cta, wwo, dissection, recons, contrast, left, aneurysm, chest |
| 39 | left, ivc, filter, vein, pulmonary, veins, dvt, clip, inferior, upper |
| 40 | left, fracture, ap, views, reason, clip, hip, distal, lat, report |
| 41 | spine, cervical, spinal, clip, thoracic, fall, lumbar, vertebral, contrast, reason |
| 42 | hemorrhage, head, ct, left, frontal, contrast, subdural, hematoma, clip, bleed |
| 43 | ct, trauma, contrast, injury, fracture, fractures, pelvis, clip, wcontrast, sp |
| 44 | contrast, brain, head, left, mri, images, mra, stroke, clip, cerebral |
| 45 | catheter, name, procedure, contrast, wire, french, placed, needle, advanced, clip |
| 46 | artery, left, common, distal, catheter, internal, branches, ow, name, middle |
| 47 | vein, stent, catheter, name, mm, portal, tips, balloon, venous, sheath |
| 48 | service, distinct, procedural, artery, sel, carotid, left, cath, name, clip |
| 49 | catheter, name, performed, embolization, contrast, bleeding, procedure, mesenteric, extrava- sation, clip |
| 50 | artery, carotid, left, aneurysm, injection, vertebral, internal, evidence, clip, cerebral |
Footnotes
Onix Text Retrieval Toolkit, API Reference, http://www.lextek.com/manuals/onix
Some medical term canonicalization parsers were also examined, but we found their outputs to be fairly unreliable for this task.
The increases in note submission at 06:00 and 18:00 were likely due to the current 12 hour nursing shift cycle. The large number of notes submitted at end-of-day were likely due to a previously common 14:00 - midnight nursing shift.
This includes those who die within the first 30-days post-discharge, so two of the prediction targets have overlap.
Adding the admission features did not improve the Retrospective Topic Model, but adding the derived features boosted AUC slightly to 0.96.
Adding the admission features to the Retrospective Topic Model improved AUC to 0.81 but adding the derived features did not improve AUC further.
Permission to make digital or hard copies of all or part of this work for personal or classroom use is granted without fee provided that copies are not made or distributed for profit or commercial advantage and that copies bear this notice and the full citation on the first page. Copyrights for components of this work owned by others than the author(s) must be honored. Abstracting with credit is permitted. To copy otherwise, or republish, to post on servers or to redistribute to lists, requires prior specific permission and/or a fee. Request permissions from permissions@acm.org.
Contributor Information
Marzyeh Ghassemi, Massachusetts Institute of Technology 77 Massachusetts Ave. Cambridge, MA 02139 USA mghassem@mit.edu.
Tristan Naumann, Massachusetts Institute of Technology 77 Massachusetts Ave. Cambridge, MA 02139 USA tjn@mit.edu.
Finale Doshi-Velez, Harvard 10 Shattuck Street Boston, MA 02115 USA finale@alum.mit.edu.
Nicole Brimmer, Massachusetts Institute of Technology 77 Massachusetts Ave. Cambridge, MA 02139 USA nbrimmer@mit.edu.
Rohit Joshi, Massachusetts Institute of Technology 77 Massachusetts Ave. Cambridge, MA 02139 USArjoshi@mit.edu.
Anna Rumshisky, University of Massachusetts Lowell One University Ave. Lowell, MA 01854 USA arum@cs.uml.edu.
Peter Szolovits, Massachusetts Institute of Technology 77 Massachusetts Ave. Cambridge, MA 02139 USA psz@mit.edu.
8. REFERENCES
- [1].Arnold C, et al. AMIA Annual Symposium Proceedings. ume 2010. AMIA; 2010. Clinical case-based retrieval using latent topic analysis; p. 26. [PMC free article] [PubMed] [Google Scholar]
- [2].Blei D, Ng A, Jordan M. Latent dirichlet allocation. JMLR. 2003;3(5):993–1022. [Google Scholar]
- [3].Blei DM, Lafferty JD. Dynamic topic models. Proceedings of the 23rd international conference on Machine learning; ACM; 2006. pp. 113–120. [Google Scholar]
- [4].Chang C-C, Lin C-J. LIBSVM: A library for support vector machines. ACM TIST. 2011;2:27:1–27:27. [Google Scholar]
- [5].Ghassemi M, Naumann T, Joshi R, Rumshisky A. Topic models for mortality modeling in intensive care units. Proceedings of ICML 2012 (Machine Learning for Clinical Data Analysis Workshop), Poster Presentation; Edinburgh, UK. June 2012. [Google Scholar]
- [6].Griffiths T, Steyvers M. Finding scientific topics. PNAS. 2004;ume 101:5228–5235. doi: 10.1073/pnas.0307752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hug CW, Szolovits P. AMIA Annual Symposium Proceedings. ume 2009. American Medical Informatics Association; 2009. Icu acuity: real-time models versus daily models; p. 260. [PMC free article] [PubMed] [Google Scholar]
- [8].Johnson AE, Kramer AA, Clifford GD. A new severity of illness scale using a subset of acute physiology and chronic health evaluation data elements shows comparable predictive accuracy*. Critical care medicine. 2013;41(7):1711–1718. doi: 10.1097/CCM.0b013e31828a24fe. [DOI] [PubMed] [Google Scholar]
- [9].Knaus WA, Wagner D, Draper E. e. a., Zimmerman J, Bergner M, Bastos PG, Sirio C, Murphy D, Lotring T, Damiano A. The apache iii prognostic system. risk prediction of hospital mortality for critically ill hospitalized adults. CHEST Journal. 1991;100(6):1619–1636. doi: 10.1378/chest.100.6.1619. [DOI] [PubMed] [Google Scholar]
- [10].Le Gall J, Lemeshow S, Saulnier F. A new simplified acute physiology score (saps ii) based on a european/north american multicenter study. JAMA. 1993;270(24):2957–2963. doi: 10.1001/jama.270.24.2957. [DOI] [PubMed] [Google Scholar]
- [11].Lehman L.-w., Saeed M, Long W, Lee J, Mark R. AMIA Annual Symposium Proceedings. ume 2012. American Medical Informatics Association; 2012. Risk stratification of icu patients using topic models inferred from unstructured progress notes; p. 505. [PMC free article] [PubMed] [Google Scholar]
- [12].Marlin BM, Kale DC, Khemani RG, Wetzel RC. Unsupervised pattern discovery in electronic health care data using probabilistic clustering models. Proceedings of the 2nd ACM SIGHIT International Health Informatics Symposium; ACM; 2012. pp. 389–398. [Google Scholar]
- [13].Saeed M, et al. Multiparameter Intelligent Monitoring in Intensive Care II: A public-access intensive care unit database. Critical Care Medicine. 2011 May;39(5):952–960. doi: 10.1097/CCM.0b013e31820a92c6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Salton G, Yang CS. On the specification of term values in automatic indexing. Journal of Documentation. 1973;29(4):351–372. [Google Scholar]
- [15].Saria S, McElvain G, Rajani AK, Penn AA, Koller DL. AMIA Annual Symposium Proceedings. ume 2010. American Medical Informatics Association; 2010. Combining structured and free-text data for automatic coding of patient outcomes; p. 712. [PMC free article] [PubMed] [Google Scholar]
- [16].Siontis G, Tzoulaki I, Ioannidis J. Predicting death: an empirical evaluation of predictive tools for mortality. Archives of internal medicine. 2011 doi: 10.1001/archinternmed.2011.334. pages archinternmed–2011. [DOI] [PubMed] [Google Scholar]
- [17].Vincent J-L, Moreno R, Takala J, Willatts S, De Mendonça A, Bruining H, Reinhart C, Suter P, Thijs L. The sofa (sepsis-related organ failure assessment) score to describe organ dysfunction/failure. Intensive care medicine. 1996;22(7):707–710. doi: 10.1007/BF01709751. [DOI] [PubMed] [Google Scholar]