Abstract
Amelogenesis Imperfecta (AI) is a clinical diagnosis that encompasses a group of genetic mutations, each affecting processes involved in tooth enamel formation and thus, result in various enamel defects. The hypomaturation enamel phenotype has been described for mutations involved in the later stage of enamel formation, including Klk4, Mmp20, C4orf26, and Wdr72. Using a candidate gene approach we discovered a novel Wdr72 human mutation in association with AI to be a 5-base pair deletion (c.806_810delGGCAG; p.G255VfsX294). To gain insight into the function of WDR72, we used computer modeling of the full-length human WDR72 protein structure and found that the predicted N-terminal sequence forms two beta-propeller folds with an alpha-solenoid tail at the C-terminus. This domain iteration is characteristic of vesicle coat proteins, such as beta′-COP, suggesting a role for WDR72 in the formation of membrane deformation complexes to regulate intracellular trafficking. Our Wdr72 knockout mouse model (Wdr72−/−), containing a LacZ reporter knock-in, exhibited hypomineralized enamel similar to the AI phenotype observed in humans with Wdr72 mutations. MicroCT scans of Wdr72−/− mandibles affirmed the hypomineralized enamel phenotype occurring at the onset of the maturation stage. H&E staining revealed a shortened height phenotype in the Wdr72−/− ameloblasts with retained proteins in the enamel matrix during maturation stage. H+/Cl− exchange transporter 5 (CLC5), an early endosome acidifier, was co-localized with WDR72 in maturation-stage ameloblasts and decreased in Wdr72−/− maturation-stage ameloblasts. There were no obvious differences in RAB4A and LAMP1 immunostaining of Wdr72−/− mice as compared to wildtype controls. Moreover, Wdr72−/− ameloblasts had reduced amelogenin immunoreactivity, suggesting defects in amelogenin fragment resorption from the matrix. These data demonstrate that WDR72 has a major role in enamel mineralization, most notably during the maturation stage, and suggest a function involving endocytic vesicle trafficking, possibly in the removal of amelogenin proteins.
Keywords: Amelogenesis imperfecta (AI), Wdr72 knockout mouse, Hypomaturation, Vesicle trafficking, Protein modeling, Maturation-stage ameloblasts
1. Introduction
Amelogenesis Imperfecta (AI) is a genetically linked disease affecting tooth enamel development, varying in phenotype and inheritance pattern. Several genes encoding enamel matrix proteins and associated proteases, namely Ambn (MIM 601259), AmelX (MIM 300391), Amtn (MIM 610912), Enam (MIM 606585), Klk4 (MIM 603767), and Mmp20 (MIM 604629), were among the first identified candidate genes for AI (Hu et al., 2007; Wright et al., 2011). Recently, this list has grown with the advent of genome-wide searches and whole-exome sequencing, identifying Fam20a (MIM 611062) (Kantaputra et al., 2014; Wang et al., 2014), Slc24a4 (MIM 609840) (Parry et al., 2013), and C4orf26 (MIM 614829) (Parry et al., 2012). Also included in this list are Fam83h (MIM 611927) (Ding et al., 2009) and Wdr72 (MIM 613214) (El-Sayed et al., 2009; Lee et al., 2010), which are thought to have vesicle-related functions in the enamel-producing ameloblasts, though their specific functions in vesicle trafficking remain unknown.
Six mutations in the Wdr72 gene have been previously identified in humans affected with AI, all displaying hypomineralized enamel phenotypes in which the unerupted tooth enamel forms a matrix of normal thickness, but is radiolucent and abrades easily from the underlying dentin upon tooth eruption (El-Sayed et al., 2009; Lee et al., 2010; El-Sayed et al., 2011; Wright et al., 2011; Kuechler et al., 2012). This describes a hypomaturation defect, suggesting WDR72 function occurs during the maturation stage of enamel formation. Indeed, a previous study showed WDR72 to be expressed in murine ameloblasts with an increased expression during maturation stage (El-Sayed et al., 2009); however, the specific temporal and spatial expression of WDR72 during enamel maturation has not been characterized.
No other syndromic effects have been reported in AI patients carrying a Wdr72 mutation, though one variant was reported to be associated with developmental problems in height, speech, respiration, and vision (Kuechler et al., 2012). Independent of AI, several Wdr72 single nucleotide polymorphisms (SNPs) have been associated with kidney, heart, pancreatic, and neural diseases (Vasan et al., 2007; Kottgen et al., 2010; Paterson et al., 2010; Hertel et al., 2011; LeBlanc et al., 2012; Franceschini et al., 2014). Therefore, elucidating WDR72 function is of great importance for understanding mechanisms for tooth enamel mineralization and potential risk factors for disease in patients carrying a Wdr72 mutation.
The function of WDR72 has been proposed to be vesicle-related, based on the known vesicle trafficking functions of WDR72's closest human homologue, WDR7 (El-Sayed et al., 2009; Lee et al., 2010). Both WDR72 and WDR7 are members of the WD40-repeat domain super family. Proteins in the WD40-repeat domain superfamily are defined by 4–8 repeating units of approximately 44–60 amino acids ending in tryptophan (W) and aspartic acid (D). WD40 proteins typically contain several repeat domains that encode a series of anti-parallel β-sheet blades that configure into a well-stabilized, non-catalytic propeller, called a “β-propeller”, for multi-unit complex docking (Stirnimann et al., 2010; Xu and Min, 2011). Proteins containing these β-propellers are observed in a broad range of cell processes, including signal transduction, cell cycle regulation, and vesicular trafficking; as such, they often contain other domain types that dictate specificities of function and pathway (Good et al., 2011). Our molecular modeling of WDR72 predicted a vesicle coat function, based on its conserved iteration of sub-structural domains and structural homology to β′-COP, a known protein structure that is a necessary component of the COPI vesicle coatomer.
To further investigate the function of WDR72 in enamel formation, we generated a Wdr72 knockout (Wdr72−/−) mouse model. The mice had a hypomineralized tooth defect, similar to what is observed in humans with Wdr72-associated AI. Our immunohistochemical studies of maturation-stage ameloblasts support a role for WDR72 in enamel maturation, possibly related to removal of amelogenin proteins from the matrix to complete mineralization of the tooth enamel.
2. Results
2.1. Identification of a novel 5-base pair deletion in exon 8 of the Wdr72 gene in a family with autosomal recessive Amelogenesis imperfecta (AI)
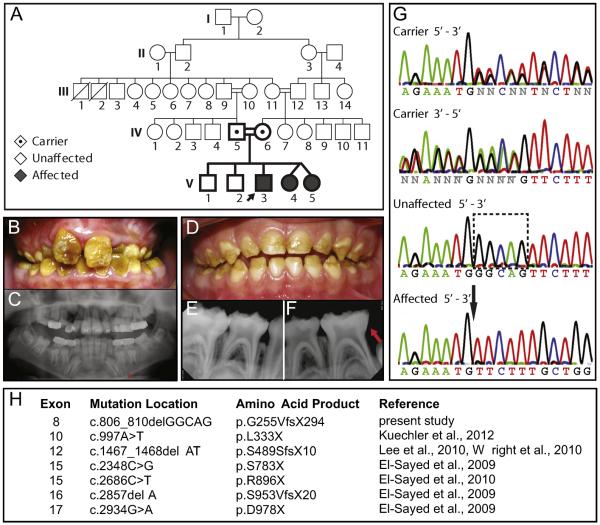
We identified the 7th reported mutation in Wdr72 to be associated with an autosomal recessive pattern of inheritance to result in AI (Fig. 1A). Among the affected individuals, the 10.5-year-old proband (V3) illustrated the most severe phenotype, exhibiting yellow-brown staining and hypomature enamel in the permanent dentition (Fig. 1B). His panoramic radiograph revealed unerupted tooth enamel with an apparently normal thickness and an indistinguishable radiopacity to dentin, which is typically less radiopaque than enamel and indicates hypomineralization (Fig. 1C). Primary teeth of the affected identical twin sisters (V4 & V5) at 4-years-old displayed less severe phenotypes, although enamel was largely absent on the occlusal third of all primary teeth (Fig. 1D). Their radiographic images showed erosion of erupted primary molar enamel with similar radiopacities to dentin (Fig. 1E & F). All affected children were highly sensitive to thermal and chemical stimuli. These enamel phenotypes are similar to those described in previous reports of patients with mutated copies of the Wdr72 gene (Lee et al., 2010; El-Sayed et al., 2011; Wright et al., 2011; Kuechler et al., 2012), all of which predict early stop codons (Fig. 1H).
Fig. 1. Identification of a novel 5-base pair deletion in exon 8 of the Wdr72 gene in patients affected with autosomal recessive AI of a pigmented and hypomaturation phenotype.
(A) Family pedigree with autosomal recessive inheritance of Amelogenesis imperfecta (AI). Double lines (=) symbolize consanguineous unions, black arrow denotes the proband, emboldened shapes identify family members analyzed for DNA sequencing. (B) Permanent dentition of the male proband (V3; 10.5 y.o.) was pigmented yellowish-brown with a loss of tooth enamel at the surface. (C) The proband's panoramic radiograph revealed hypomineralized enamel in unerupted teeth, missing first permanent molars (teeth #3, 19 and 30), and delayed eruption sequences in primary canines and molars (red asterisk). (D) Representative photograph of one affected twin sister (V4; 4 y.o.) showing primary dentition also exhibiting a hypomaturation enamel phenotype on the occlusal surfaces of erupted teeth. (E, F) Lower PAs also showed a loss of enamel in erupted teeth with similar radiopacity to dentin, indicating hypomineralization (red arrow). (G) Comparisons of representative chromatograms for carrier, unaffected, and affected individuals revealed a 5-base pair deletion in affected patients, which localized to exon 8 of the Wdr72 gene and matched with the AI phenotype. The dotted box encompasses the nucleotides absent in affected patients with a black arrow pointing to the deletion site. (H) List of reported Wdr72 mutations in association with AI.
The proband was also congenitally missing 3 out of 4 six-year-old permanent molars (teeth #3, 19 and 30) and exhibited delayed eruption of his mandibular primary canines and molars, indicating a 1-year delay in dental development (Fig. 1C). Similar tooth development phenotypes have been reported in association with only one other Wdr72 mutation, the second-most upstream mutation occurring at exon 10 (Fig. 1H) (Kuechler et al., 2012).
We used a candidate gene approach to compare DNA sequences of affected individuals and their immediate family members (Fig. 1A) (Supplementary Table 1). Chromatograms of affected, unaffected, and carrier individuals at exon 8 of the human Wdr72 gene (NM_182758. 2) revealed a 5-base pair deletion (c.806_810delGGCAG) that followed an autosomal recessive inheritance pattern of the AI phenotype (Fig. 1G). This exon 8 deletion mutation resulted in a frameshift and premature stop codon (c.806_810delGGCAG; p.G255VfsX294). In addition, single nucleotide polymorphisms (SNPs) were also found to segregate with the deletion mutation, occurring in exon 14 as a silent mutation (c.1865G > A, p.613 V > V; rs74018741) and in the intron between exons 12 and 13 (g.53994305A > G; rs74018741) (Supplementary Table 2). All other variations in sequenced candidate genes (Amtn, Ambn, Enam, Mmp20, and Klk4) of this family did not follow the disease inheritance (Supplementary Tables 1 & 2).
2.2. WDR72 structure modeling predicted a configuration unique to proteins found in membrane-deformation complexes with homology to the crystallized β′-COP structure
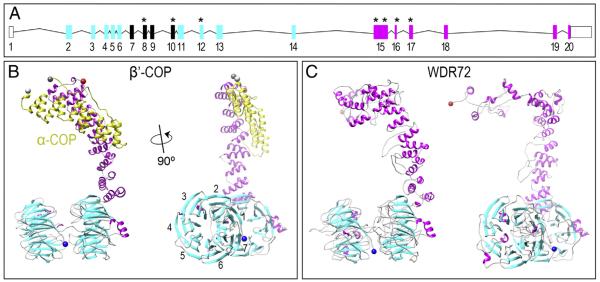
The human full-length WDR72 protein is 1102 amino acids in length and predicts two clusters of WD40 repeat domains at the N-terminus and a unique C-terminus with no identifiable homology domains (Fig. 2A; ENSG00000166415). To provide further insight into WDR72 function, we generated all-atom models of the human full-length WDR72 protein (Fig. 2A) and mutated variants (Supplementary Fig. 1). These models were generated using sequence similarity to annotated proteins using two comparative protein structure prediction pipelines: (1) I-TASSER (Zhang, 2007; Roy et al., 2010) and (2) HHsearch ported into MODELLER (Soding et al., 2005; Zhang, 2007). Among all known protein structures in the Protein Data Bank (PDB), both modeling pipelines showed full-length WDR72 with greatest structural homology to the known crystal structure of Saccharomyces cerevisiae β′-COP (Fig. 2B), an essential subunit in eukaryotic COPI vesicle coat assemblies (PDB identifier: 3mkq) and was thus used as the primary template for constructing the WDR72 models. All statistically reliable models of full-length WDR72 (zDOPE < – 1) showed a series of β-sheets folded into two β-propellers, followed by a succession of parallel α-helices twisting into a compact tail, referred to as an α-solenoid (Fig. 2C). This structural iteration of 1 or 2 β-propellers followed by an α-solenoid is unique to membrane-deformation complexes (Lee and Goldberg, 2010; Field et al., 2011), which include vesicle coatomers (i.e. Clathrin, COPII, COPI) and the nuclear pore complex (i.e. Nup).
Fig. 2. WDR72 adopts the β-propeller and α-solenoid fold configuration characteristic of membrane-deformation complexes.
(A) The human Wdr72 gene with predicted WD40 domains (cyan boxes) and sequence homology divergence to all other human genes (magenta boxes). Reported mutations associated with Amelogenesis Imperfecta are denoted with an asterisk (*), showing our identified exon 8 mutation in between two clusters of WD40 repeat domains (ENST00000360509). Mutation positions based on CCDC 10151.1; numbered box, Wdr72 exon; line, intron; empty box, untranslated region (UTR). (B) Monomeric structure of β′-COP protein interacting with the C-terminus of α-COP (yellow), another subunit of the crystallized Saccharomyces cerevisiae COPI triskelion vesicle coat complex (PDB identifier: 3mkq). The side view of β′-COP (right) depicts what is referred to as a βeta-propeller structure (cyan) and an α-solenoid tail (magenta) unique to proteins that interact with curved lipid bilayers. This β′-COP structure was used as a template for model building, as it showed the greatest similarity to WDR72 compared to any other solved protein structures. Numbers represent blades #1–7 that comprise a single β-propeller. (C) Constructed model of WDR72 showing two β-propeller domains and a curved α-solenoid tail. The side view of full-length WDR72 (right) depicts two β-propellers, each with 7 blades (cyan), and a series of α-helices (magenta) forming an α-solenoid tail. Blue sphere, N-terminus; red sphere, C-terminus. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To evaluate the number of blades present in each β-propeller, we applied the hidden Markov random field algorithm SMURF (Menke et al., 2010), built specifically to detect combinations of 6-, 7-, and 8-bladed double β-propellers. We found that full-length WDR72 was most likely composed of two 7-bladed β-propellers (P < 0.0001) rather than any other combinations of one or two 6-, 7- or 8-bladed β-propellers (P > 0.05), which is consistent with other membrane-deformation complex proteins. The 1st β-propeller blades were predicted as the following residues: 22–59, 67–104, 119–150, 168–198, 215–251, 260–292, and 301–353; and for the 2nd β-propeller: 359–400, 407–442, 466–494, 521–554, 562–627, 633–659, and 665–686; with α-helices thereafter (Fig. 2C).
We observed that both exon 8 and exon 10 mutations occurred between two predicted WD40-repeat domain clusters (Fig. 2A; ENSG00000166415). Multiple splice variants have been identified for the human Wdr72 gene, whose encoding is regulated by several different 5′UTRs and 3′UTRs (AceView) (Thierry-Mieg and Thierry-Mieg, 2006). This may suggest evasion of RNA decay in Wdr72 mutations observed in individuals with AI. Under this possibility, we modeled all putative WDR72 truncations resulting from AI-related mutations and found that these two most up-stream mutations formed a single β-propeller containing only 6 blades (P < 0.01), rather than 7, which suggests instability of the protein fold (Supplementary Fig. 1).
2.3. Successful knockout of Wdr72 in mice resulted in hypomaturation enamel phenotypes similar to AI patients with Wdr72 mutations
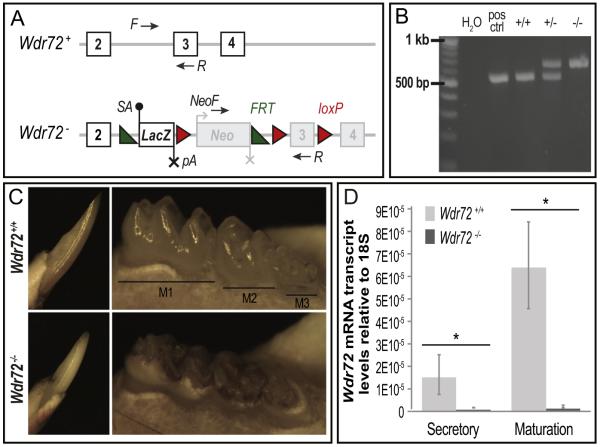
To further examine the role of Wdr72 in enamel formation, we established a functional knockout mouse model. The Wdr72 knockout mouse strain (Wdr72−/−) and wild-type littermate controls (Wdr72+/+) used in this study were created by breeding heterozygous mice (Wdr72+/−) obtained from the Knockout Mouse Project (KOMP) Repository and the Mouse Biology Program at the University of California, Davis. The Wdr72 mutant allele (Wdr72−) was generated using a `knockout first conditional ready' approach using previously published methods (Skarnes et al., 2011), which splices in a LacZ gene reporter cassette prior to the critical Wdr72 exon 3 to all identified Wdr72 transcripts and thus a functional knockout of Wdr72 (Fig. 3A).
Fig. 3. Successful Wdr72−/− mouse model exhibits hypomaturation enamel phenotypes.
(A) Wdr72 wildtype (+) and mutant (−) allelic variants. The Wdr72 mutant allele (Wdr72−) is a functional knockout through splicing in a LacZ gene reporter containing a high affinity splice acceptor (SA, black dot) and termination sequence (pA, black X) prior to the critical exon 3. The encoded mutant transcript is a truncated Wdr72 at exon 2 fused to LacZ. Neomycin (Neo) is independently regulated, containing its own promotor (gray bent arrow) and termination sequence (gray X). Numbered white boxes represent coded exons under the Wdr72 promotor, whereas numbered gray boxes represent exons not encoded under the Wdr72 promotor. Black arrows denote primer locations used for genotyping. Green triangles, frt sites; Red triangles, loxP sites; (B) PCR genotyping of isolated mouse DNA from Wdr72+/+, Wdr72+/−, and Wdr72−/− tail biopsies. Amplicon sizes of Wdr72+ allele, 520 bp; and Wdr72− allele, 633 bp. (C) Representative photographs of 6-week-old male mandibular incisors (left panels, buccal view) and molars (right panels, lingual view) after soft tissue removal. Wdr72−/− enamel was lost from the intact dentin surface when teeth erupted into the oral cavity. Wdr72−/− enamel also appeared opaque and stained as compared to the translucent Wdr72+/+ enamel. (D) Quantitative real-time PCR (qPCR) of micro-dissected secretory and maturation-stage ameloblasts. Wdr72 transcript is successfully knocked out in Wdr72−/− mice and is significantly up-regulated from secretory to maturation stages in Wdr72+/+ amelo-blasts (n=3, P < 0.05). Error bars represent ± SD about the mean.
Mandibular molars and incisors of 6-week-old male Wdr72+/+ and Wdr72−/− mice were evaluated for gross morphological differences (Fig. 3C) and exhibited an enamel phenotype similar to that observed in our affected AI patients (Fig. 1B & D). Wdr72−/− mice showed opaque and darkly stained enamel relative to the translucency observed in Wdr72+/+ mice (Fig. 3C). Wdr72−/− enamel was of normal thickness at the base of incisors and molar crowns but was lost at occlusal surfaces, while the exposed dentin remained relatively intact. Heterozygous mice (Wdr72+/−) appeared to have normal enamel akin to Wdr72+/+ mice (data not shown), which mimics the autosomal recessive inheritance pattern seen in AI.
No other obvious tooth phenotypes or differences in major organs known to express Wdr72 (ie. kidney and brain) were observed in Wdr72−/− mice (data not shown). Whole body weights of Wdr72−/− mice were significantly reduced (P < 0.05), beginning at postnatal day 21 (P21) in males and P24 in females compared to their Wdr72+/+ controls (Supplementary Fig. 2). Tooth eruption in mice occurs between ages P10 and P14, while weaning age is P21, indicating that these body weight differences were due to difficulties with chewing hard foods rather than systemic effects caused by a loss of Wdr72.
Quantitative real-time PCR (qPCR) of micro-dissected secretory and maturation-stage ameloblasts showed significantly reduced Wdr72 mRNA expression in Wdr72−/− secretory and maturation-stage ameloblasts to 4.4% and 1.8%, respectively, of Wdr72+/+ controls (P < 0.05). Such relatively low levels of Wdr72 transcript observed in our mouse model is consistent with leaky expression levels observed for gene trapping methodologies and indicate successful Wdr72 knockout (Fig. 3D) (Galy et al., 2004). In addition, relative amounts of Wdr72 transcript were significantly up-regulated from secretory to maturation stage in Wdr72+/+ ameloblasts (P < 0.05), suggesting a major function during enamel maturation (Fig. 3D).
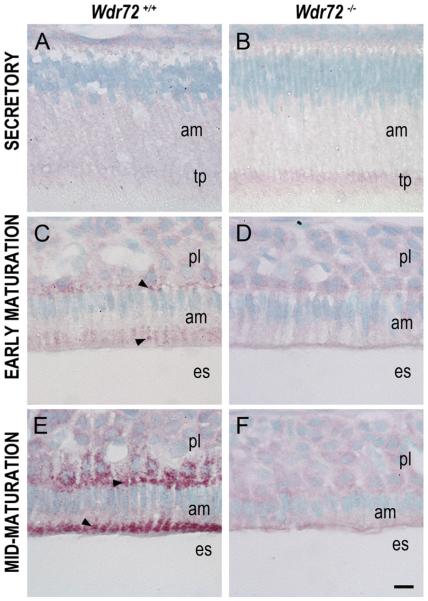
Similar to our qPCR findings, immunostaining with an antibody made to the CETGTLERHETGERA peptide sequence of the WDR72 protein (amino acids 587–600 plus an extra cysteine residue) illustrated little to no WDR72 present in Wdr72−/− ameloblasts (Fig. 4B, D, & F). Verification of our WDR72 antibody, which was made against the same peptide sequence as the one previously synthesized (El-Sayed et al., 2009), was performed on Wdr72+/+ and Wdr72−/− kidneys by western blot (Supplementary Fig. 3). In Wdr72+/+ mice, ameloblasts showed increased WDR72 immunoreactivity from secretory to maturation stage (Fig. 4A, C, & E). Using the continuously growing mouse incisor to visualize WDR72 expression on a time-scaled spectrum of ameloblast differentiation stages required for enamel formation, we further observed a specific increase at the onset of maturation stage and subcellular localization to distinct vesicle-like puncta at apical and basal regions of the cell (Fig. 4C & E), whereas, immunoreactivity in secretory ameloblasts appeared light and diffuse (Fig. 4A).
Fig. 4. WDR72 immunolocalizes to vesicle-like puncta in Wdr72+/+ maturation-stage ameloblasts and shows protein-level knockout in Wdr72−/− mice.
Representative sagittal sections of P10 male Wdr72+/+ and Wdr72−/− mandibles immunostained with a polyclonal rabbit antibody to WDR72 peptide. (A) In Wdr72+/+ mice, immunoreactivity (red) was diffusely positive in secretory ameloblasts and increased upon entry into the maturation stage (C, E), immunolocalizing to vesicle-like structures at the basal and apical ends (arrowheads). (B, D, F) Slight non-specific background immunoreactivity was present in Wdr72−/− ameloblasts. am, ameloblast; es, enamel space; pl, papillary layer; tp, Tomes' process. Scale bar, 10 μm.
2.4. Wdr72−/− mice had enamel phenotypes occurring specifically during the maturation stage, showing a hypomineralized enamel matrix and shortened heights in ameloblasts
Murine mandibles from 6-week-old male Wdr72+/+ (n = 3) and Wdr72−/− mice (n = 3) were scanned by micro-computed tomography (microCT) and analyzed for differences in relative intensities of the major mineralized tissue types (enamel, dentin, and alveolar bone). The process of normal enamel mineralization was observed in the continuously growing mouse incisor of Wdr72+/+ mice, which showed an increasing enamel radiopacity that coincided with the transition zone of differentiating ameloblasts from secretory to maturation-stage morphologies (Fig. 5A, arrow). In contrast, radiopacity of enamel, beginning at the maturation stage of Wdr72−/− molars and incisors, was significantly reduced compared to that of the Wdr72+/+mice (Fig. 5A & B). These observations were quantified in cross-sections of the mandibular incisor, taken at secretory and maturation stages, and averaged for gray scale values within a selected region of interest (Fig. 5C–F). We found enamel radiopacities of Wdr72+/+ and Wdr72−/− mice were not significantly different at secretory stage (P = 0.92) but were significantly reduced at maturation stage in Wdr72−/− mice (P < 0.05) (Fig. 5E). Interestingly, Wdr72−/− enamel from secretory to maturation stages showed significant differences (P < 0.05), suggesting that partial mineralization of the enamel matrix still occurred in the absence of Wdr72. Wdr72−/− dentin (P = 0.26) and alveolar bone (P = 0.22) did not significantly differ in gray scale values compared to Wdr72+/+ controls (Fig. 5E).
Fig. 5. Hypomineralized enamel phenotype in Wdr72−/− mice occurs at the onset of maturation stage.
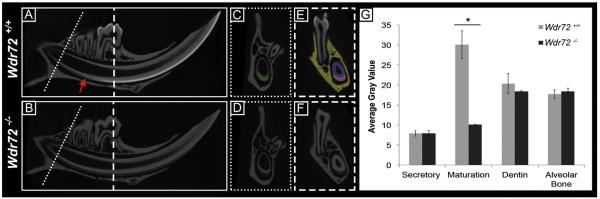
Representative sagittal (A, B) and cross sectional (C–F) microCT images of Wdr72+/+ and Wdr72−/− 6-week-old male mandibles. In the continuously growing incisor, Wdr72+/+ enamel mineralization increased at the onset of maturation stage to form an enamel layer of increasing relative intensity (A), which remained radiolucent in the Wdr72−/− mice (B). Red arrow marks transition stage. Similar relative intensity comparisons between the Wdr72+/+ and Wdr72−/− enamel were observed in fully formed molars (E, F). Dotted and dashed lines mark the sites along the continuously growing incisor where coronal planes were taken at secretory stage (C, D) and maturation (E, F) stages of enamel, respectively. Dentin and alveolar bone showed no differences (A–D). Green, red, blue, and yellow shading overlays on the Wdr72+/+ cross-section panels (C & E) represent the regions of interest that were used to quantify the average gray values seen in the bar graph; green, incisor enamel at secretory stage; red, incisor enamel at maturation stage; blue, dentin; yellow, alveolar bone. Quantification of these observations is depicted in the graph to the right (G). Error bars represent ± SD about the mean (n=3). Paired student t-tests between Wdr72+/+ and Wdr72−/− average gray scale values with a threshold for P-values at <0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
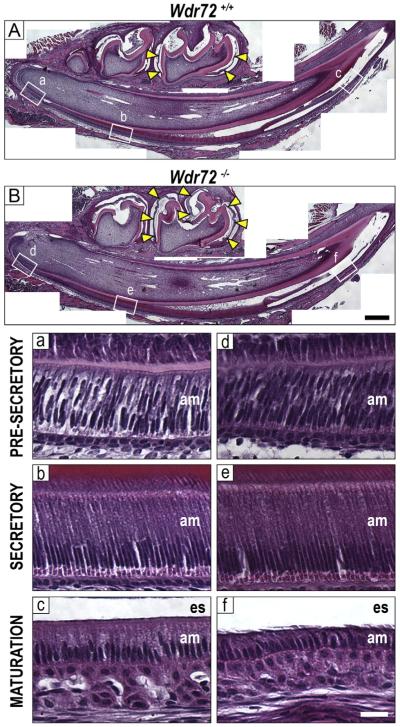
Hematoxylin and eosin (H&E) stains of Wdr72+/+ and Wdr72−/− male mandibles at P10 showed morphologically normal secretory ameloblasts in Wdr72−/− mice and shortened maturation-stage ameloblasts, (Fig. 6). In addition, Wdr72−/− mice exhibited retained non-mineralized, organic material in the enamel matrix during maturation stage (Fig. 6, yellow arrows), whereas the lack of this proteinaceous staining in Wdr72+/+ enamel matrices indicated a more mineralized enamel matrix (Fig. 6A & B).
Fig. 6. Enamel matrix and ameloblasts display maturation-stage specific phenotypes in Wdr72−/− mice.
Representative sagittal sections of Wdr72+/+ (A) and Wdr72−/− (B) P10 male mandibles stained with hematoxylin and eosin. Unerupted first molars are at maturation stage, showing a lack of organic material (yellow arrows) in the enamel space of the Wdr72+/+ mice but retention in the Wdr72−/−. Lettered white boxes correspond to enlarged images below. Black scale bar, 400 μm. Bottom panels show Wdr72+/+ (a–c) and Wdr72−/− (d–f) ameloblasts at different stages of differentiation, showing the spectrum of enamel development along the continuously growing incisor. During maturation stage, Wdr72−/− ameloblasts (f) are shorter in height compared to those of Wdr72+/+ mice (c), while earlier stages appear to have morphologically normal ameloblasts. am, ameloblasts; es, enamel space. White scale bar, 20 μm. (For interpretation of the references to color in this figure, the reader is referred to the web version of this article.)
2.5. Loss of Wdr72 resulted in decreased intracellular amelogenin proteins with no effect on transcript levels
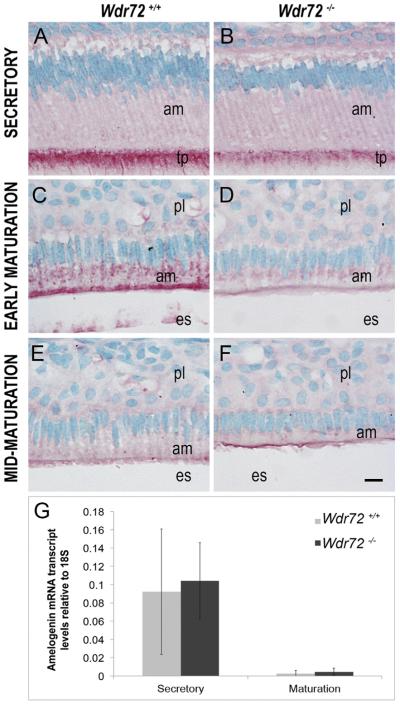
We next investigated the possibility that Wdr72 was needed for reuptake of amelogenin proteins from the matrix, since our Wdr72−/− mice showed retained proteinaceous material in the enamel matrix, and amelogenin is the predominant extracellular matrix protein secreted and hydrolyzed into fragments during enamel formation. Amelogenin immunoreactivity was slightly reduced in secretory stage ameloblasts at the Tomes' processes in Wdr72−/− mice (Fig. 7B) and was more obviously reduced at the apical border and the cytoplasm in Wdr72−/− ameloblasts at early maturation stage (Fig. 7D) as compared to ameloblasts from Wdr72+/+ mice (Fig. 7A and C, respectively). By mid-maturation, intracellular amelogenin staining was absent in both Wdr72+/+ and Wdr72−/− ameloblasts (Fig. 7E & F).
Fig. 7. Wdr72−/− ameloblast cells have decreased intracellular amelogenin proteins with no change in transcript levels.
Representative images of ameloblasts from P10 male Wdr72+/+ and Wdr72−/− mice immunostained with amelogenin antibody (red) and counterstained with methyl green. Amelogenin is immunoreactive in secretory ameloblasts at the Tomes' processes (A, B) and in early maturation-stage ameloblasts at the center and apical regions in small puncta (C, D), but was more reactive in Wdr72+/+ than Wdr72−/− mice. As enamel formation progressed to mid-maturation, intracellular amelogenin immunostaining was absent in both Wdr72+/+ (E) and Wdr72−/− (F) ameloblasts. am, ameloblast; es, enamel space; pl, papillary layer. Scale bar, 10 μm. (G) Relative amelogenin mRNA expression in microdissected enamel epithelia from 6-week-old male mice at secretory and maturation stages, showing no significant differences between Wdr72+/+ and Wdr72−/− mice at either secretory (P = 0.81) or maturation stages (P = 0.53). Gene copy numbers are relative to 18S. Error bars represent ± SD about the mean (n = 3). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To verify that the decreased intracellular amelogenins in Wdr72−/− mice was not due to a decrease in amelogenin mRNA production, we quantified amelogenin transcripts from microdissected enamel epithelia at secretory and maturation stages. Analyses by qPCR showed no significant differences between Wdr72+/+ and Wdr72−/− ameloblasts at either stage (n = 3; P = 0.81 secretory, P = 0.53 maturation) (Fig. 7G).
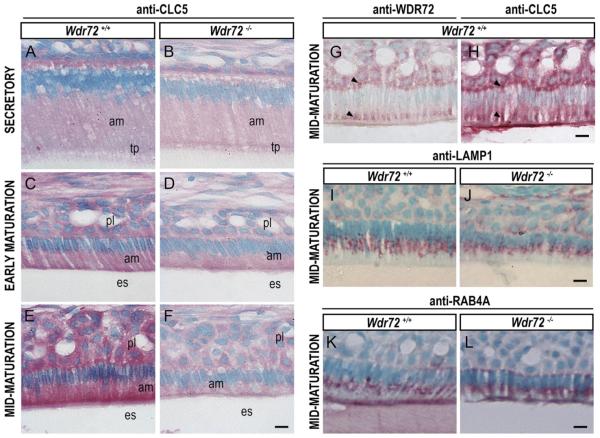
2.6. Wdr72−/− mice showed reduced immunostaining of the endosome protein CLC5 during the maturation stage compared to other vesicle markers
Consecutive sections of Wdr72+/+ maturation-stage ameloblasts showed WDR72 and CLC5 immunolocalization to the same supernuclear (basal) areas, as well as to apical regions slightly distal to the enamel matrix border (arrowheads, Fig. 8G & H). In addition, CLC5 was less immunoreactive in Wdr72−/− maturation-stage ameloblasts, as compared to Wdr72+/+ mice (Fig. 8B, D, & F). Unlike CLC5, immunostaining for RAB4A (a marker for endosome sorting and recycling) and LAMP1 (a marker for lysosomes) did not show obvious differences between Wdr72+/+ and Wdr72−/− maturation-stage ameloblasts (Fig. 8I–L).
Fig. 8. Immunolocalization of vesicle markers in Wdr72+/+ and Wdr72−/− ameloblasts.
Immunostaining of CLC5 in Wdr72+/+ and Wdr72−/− ameloblasts (A–F) showed that CLC5 is decreased in maturation-stage ameloblasts of Wdr72−/− mice as compared to Wdr72+/+. Serial sections of Wdr72+/+ enamel organs at maturation stage showed WDR72 and CLC5 both immunolocalizing in similar patterns at apical and basal regions of the ameloblasts, as well as the papillary layer (G & H, arrowheads). Further immunostaining of LAMP1 (I, J) and RAB4A (K, L) showed no differences between Wdr72+/+ and Wdr72−/− maturation-stage ameloblasts. am, ameloblast; es, enamel space; pl, papillary layer; tp, Tomes' process. Scale bars, 10 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3. Discussion
The human Wdr72 gene has a total of 20 exons that contribute to several different transcript variants (ENST00000360509) (Thierry-Mieg and Thierry-Mieg, 2006), the longest and most common of which encodes a protein 1,102 amino acids in length (CCDS 10151.1). Wdr72 was first found to be a candidate for autosomal recessive amelogenesis imperfecta (AI) in individuals affected with a pigmented and hypomaturation enamel phenotype (AI2A3; MIM 613211) through the use of a single nucleotide polymorphism (SNP) microarray (El-Sayed et al., 2009). Since this discovery, a total of seven Wdr72 mutations have been reported as causal links to AI, including the 5-base pair deletion that we identified in exon 8 (c.806_810delGGCAG). Previously reported mutations located farther downstream, spanning exons 10 through 17, are also predicted to disrupt the unique C-terminal domain (exons 15 to 20) by introducing premature stop codons that lead to truncated proteins (El-Sayed et al., 2009; Lee et al., 2010; El-Sayed et al., 2011; Wright et al., 2011; Kuechler et al., 2012). While it is unclear whether any of these mutated Wdr72 variants are indeed translated as defective proteins or degraded by nonsense-mediated decay, all mutations lead to similar hypomaturation enamel phenotypes.
In addition to a tooth enamel defect, we observed congenitally missing permanent molars and a one-year delay in tooth development in an individual with our identified exon 8 mutation (Fig. 1C). Similar phenotypes have been reported in only one other Wdr72 mutation (exon 10) (Kuechler et al., 2012), the second-most upstream to be found in AI (Fig. 1H), possibly linking early Wdr72 mutations to additional tooth-related defects.
To evaluate the potential functions of WDR72, we used bioinformatics to model a predicted protein structure of WDR72 by mapping the full-length human amino acid sequence onto all known structures within the Protein Data Bank (PDB). Our WDR72 structure model predicted a membrane-coating architecture composed of two 7-bladed β-propeller heads, followed by an α-solenoid fold, which are respectively encoded by two N-terminal WD40-repeat domain clusters and a unique C-terminus (residues #687–1102). This particular domain combination (1 or 2 β-propellers, then α-solenoid) is a highly conserved architecture among proteins that form membrane coat complexes, dating back to the early eukaryotic endomembrane system (Devos et al., 2004; Field et al., 2011). In addition, this group of membrane-coating complexed proteins has remained somewhat exclusive to (a) the scaffold-layer proteins of the nuclear pore complex (Nups) that stabilize the inner and outer nuclear envelope membranes at the nuclear pore edges and (b) the vesicle coatomers (COPI, COPII, and Clathrin) that assemble in cage-like lattices to initiate the budding step of vesicle formation (Devos et al., 2006). Structure domains of these membrane-coating proteins typically utilize their WD40 β-propeller folds to serve as docking sites for multiple protein–protein interactions (Stirnimann et al., 2010; Good et al., 2011; Xu and Min, 2011) and their α-solenoids, formed from anti-parallel stacked α-helices, to stabilize and induce membrane curvature (Devos et al., 2006; Field et al., 2011). Modeling WDR72 as a structure containing these domains, specifically in the β–β–α order, suggests it as a member of this membrane-coating group.
We found that when WDR72 was compared to all solved protein structures, the highest structural homology was to β′-COP (PDB identifier: 3mkq), an essential subunit in the COPI vesicle coatomer complex. This homology further suggests that WDR72 functions as a vesicle coat protein, and together these bioinformatic analyses are consistent with the previously proposed role for WDR72 in vesicle trafficking (El-Sayed et al., 2009; Lee et al., 2010). This previously proposed function for WDR72 was based on the high sequence homology to WDR7, a known regulator of Rab3A GTPase in Ca2+-dependent exocytosis at neural synapses (Nagano et al., 2002; Kawabe et al., 2003). Interestingly, WDR7 (under the alias “Rabconnectin-3β”) has another function in regulating vesicles, rather, in the endocytic pathway by directly monitoring vacuolar H+-ATPase (v-ATPase) activity in Drosophila (Yan et al., 2009; Sethi et al., 2010), indicating that WDR72 may too function in both exocytosis and endocytosis. Further evidence pointing to an endocytic vesicle coat function for WDR72 are the in vivo immunolocalization of AP2 (cargo-specifying adaptor component of Clathrin coatomers) found by Lacruz and co-workers (Lacruz et al., 2013). Together, these findings support a role for WDR72 in mediating vesicle formation, potentially as an endocytic and/or exocytic coat protein to help regulate transport between cell and matrix.
It is also worth noting that WDR72 and WDR7 diverge at residue #845 (exon 15) (Lee et al., 2010), and the remaining 60% of WDR72's C-terminus is unique. The C-terminus is predicted to form the α-solenoid region, and α-solenoids of canonical vesicle coatomers are structurally conserved but exhibit little similarity to one another at the sequence level (Field et al., 2011). These differences are thought to contribute to the diversity of lattice size and shape, which invariably dictate cargo specificities and vesicle type, suggesting that the unique C-terminal sequence of WDR72 specifies its function to enamel formation and its vesicle cargo. The importance of this α-solenoid is emphasized by our modeling of all putative WDR72 proteins containing mutations relevant to hypomaturation enamel AI phenotypes, all of which showed a shortened or absent α-solenoid tail (Supplementary Fig. 1).
To further define the function of WDR72, we generated a knockout mouse (Wdr72−/−), which exhibited a hypomaturation enamel phenotype similar to that observed in humans with Wdr72-associated AI (El-Sayed et al., 2009; Wright et al., 2011; Kuechler et al., 2012). Quantitative RT-PCR (qPCR) of Wdr72−/− secretory and maturation-stage ameloblasts showed low mRNA Wdr72 transcript levels in both tissue groups relative to the observed up-regulation from secretory to maturation stage in Wdr72+/+ control mice. Similarly, anti-WDR72 immunostaining of Wdr72−/− ameloblasts exhibited non-specific background staining at all stages of enamel formation, whereas Wdr72+/+ controls were intensely immunoreactive shortly after entering the maturation stage. Western blots of Wdr72+/+ and Wdr72−/− kidney samples paralleled these findings with low WDR72 protein levels in Wdr72−/− mice.
Whole body weights of both female and male Wdr72−/− mice were also decreased compared to Wdr72+/+ mice, however, this was observed only after weaning age (P21), suggesting that this difference was due to the tooth-related defect as opposed to other systemic effects. Overall, the Wdr72−/− mice paralleled the descriptions of individuals with Wdr72-related AI. Taken together, these initial characterization studies illustrate that Wdr72 is functionally knocked out in our Wdr72−/− mouse model, providing an excellent system for studying Wdr72-associated AI to elucidate its role in tooth development.
Similar to the radiolucent enamel phenotypes described for human Wdr72 mutations, our microCT imaging of Wdr72−/− mandibles showed stage-specific and tissue-specific hypomineralized defects in the tooth enamel. At secretory stage, relative intensities of Wdr72−/− enamel radiopacities were comparable to those of Wdr72+/+ controls but did not increase in radiopacity to the same extent as Wdr72+/+ enamel once the maturation stage began. Wdr72 loss also appeared to have tissue-specific effects, showing dentin and alveolar bone to be unchanged in Wdr72−/− mice.
Additional evidence in support of the stage-specificity of WDR72 during enamel maturation were our findings that Wdr72−/− ameloblasts appeared shorter in height only after differentiating into the maturation-stage cell type, and the Wdr72−/− enamel matrix differed only during the maturation stage, showing retained organic matter. These comparisons were made along the continuously growing mouse incisor, which presents the entire spectrum of enamel formation (Leblond and Warshawsky, 1979), thus providing time-scale insight into WDR72 function. The observed cell and matrix phenotypes further support a stage-specific role for WDR72 in regulating enamel mineralization and suggests a function in one of the major cell processes in maturation-stage ameloblasts.
A major function of maturation-stage ameloblasts that is often disrupted in AI hypomaturation phenotypes is the processing of amelogenin proteins, which constitute the majority of the organic extracellular matrix (Hart et al., 2004; Kim et al., 2005; Barron et al., 2010). Our observation that organic material was retained in Wdr72−/− enamel matrices during maturation suggests that WDR72 has a role in either the secretion of proteases into the mineralizing matrix or in removal of hydrolyzed protein fragments from the matrix to allow final matrix mineralization to occur. Amelogenins are secreted during early matrix formation and are subsequently hydrolyzed and removed from the enamel matrix during the maturation stage to allow thickening of the hydroxyapatite crystals that comprise the tooth enamel (Smith, 1979; Hu et al., 2007; Gibson, 2011). Removal of hydrolyzed matrix proteins, including amelogenins, may be due to either a passive diffusion between the open junctions of smooth-ended ameloblasts or an active transport mechanism through the cells (Nanci et al., 1998). As amelogenins are removed, calcium is transported through ameloblasts and deposited along with phosphate to form the mineralized enamel matrix (Hubbard, 2000).
We found that Wdr72−/− maturation-stage ameloblasts had less intracellular amelogenin protein with no change in its relative synthesis compared to Wdr72+/+ controls, suggesting that WDR72 influences the active uptake of amelogenin fragments into the cell, presumably by regulating endocytosis. Interestingly, we also observed decreased amelogenin immunoreactivity at the Tomes' processes in Wdr72−/− secretory ameloblasts with no effect on transcript levels. This was surprising, given that Wdr72 loss had so far manifested as a maturation stage phenotype; however, it is still consistent with the fact that Wdr72+/+ secretory ameloblasts express WDR72, albeit at low levels (El-Sayed et al., 2009). Tomes' processes, though largely functioning in the exocytosis of amelogenins, also have endosomes that contain hydrolyzed amelogenins (Smith, 1979; Sasaki, 1984; Nanci et al., 1996). It is also possible that WDR72 has more subtle roles at earlier stages of tooth development that involve intracellular amelogenin transport, which is supported by the tooth agenesis and delayed eruption observed in some AI individuals.
To further investigate the possible vesicle trafficking function of WDR72, we immunostained Wdr72+/+ and Wdr72−/− maturation-stage ameloblasts with anti-CLC5 as a marker of early endosomes, anti-RAB4A as a marker of early and recycling endosomes, and anti-LAMP1 as a marker of lysosomes. Our immunostains of WDR72 and CLC5 on consecutive sections of Wdr72+/+ maturation-stage ameloblasts suggest that WDR72 localizes to endosomes, which is supported by similar immunolocalizations observed for RAB4A and the endocytic coat protein, AP2 (Lacruz et al., 2013), but not the more basal immunolocalization of LAMP1 to lysosomes.
In Wdr72−/− maturation-stage ameloblasts, CLC5 showed decreased immunoreactivity relative to Wdr72+/+ controls, while RAB4A and LAMP1 appeared to be unaffected. CLC5 and RAB4A are both early endosome markers and are frequently associated with one another (Devuyst et al., 1999), although RAB4A also specifically functions in recycling endosomes. This suggests a unique function for WDR72 in mediating a specific subpopulation of endosomes, possibly related to other known functions of CLC5, such as exocytosis of ion channel antiporters (Lin et al., 2011), Ca2+ transport (Luyckx et al., 1998), or pH regulation (Hara-Chikuma et al., 2005; El-Sayed et al., 2011; Duan, 2014) that are integral to major processes carried out by maturation-stage ameloblasts. While it is also possible that CLC5 decreases observed in Wdr72−/− ameloblasts were attributed to secondary effects on the cell, these decreases coincided with the timing of normal WDR72 expression.
LAMP1 has been suggested to mediate amelogenin uptake into ameloblasts, and extracellular increases in amelogenin in vitro has been shown to upregulate LAMP1 (Le et al., 2007; Shapiro et al., 2007). Therefore, we would expect that if amelogenin uptake into the cell was decreased, LAMP1 immunostaining would also be relatively decreased in Wdr72−/− mice. The lack of any change in LAMP1 may suggest that WDR72 does not mediate amelogenin uptake into the cell, but rather is involved in degradation of amelogenins in the extra-cellular matrix, possibly through the secretion of enamel matrix proteinases. Reduced proteinase secretion at the maturation stage could inhibit the uptake of amelogenin protein fragments into the cell. Further analysis of matrix proteinases in Wdr72−/− as compared to Wdr72+/+ will further clarify the possible roles of WDR72 in amelogenin degradation and endocytosis.
In summary, these studies have reported the use of structural modeling to support a function of WDR72 in vesicle trafficking. The generation and initial analyses of the Wdr72−/− mouse confirmed the importance of WDR72 in enamel formation and illustrated a direct link between mutated WDR72 to hypomineralized enamel. We found reduced intracellular amelogenin in the Wdr72−/− ameloblasts with a similar reduction in the endosome marker protein ClC5, suggesting a role for WDR72 in the removal of amelogenins during enamel maturation. Further understanding of WDR72's function in amelogenesis will allow us to better understand this unique biomineralization process and how other tissues potentially regulate mineralization. These studies are also important in determining other yet unknown risk factors for AI patients affected with mutated Wdr72.
4. Methods
4.1. Mutation screening
A consanguineous family exhibiting hypomature phenotypes of autosomal recessive inheritance Amelogenesis Imperfecta (AI) was examined at the UCSF pediatric dental clinic and screened for mutations using a candidate gene approach. DNA samples of the probands and all immediate family members were obtained using Oragene®DNA sample collection kit (DNA Genotek, Inc., Ontario, Canada) with approval by the UCSF Committee on Human Research. Forward and reverse primers were designed within introns to flank coding regions and potential splice sites in ameloblastin (Ambn), amelogenin (AmelX), amelotin (Amtn), enamelin (Enam), kallekrein-related peptidase 4 (Klk4), matrix metalloproteinase-20 (Mmp20), and WD repeat-containing protein 72 (Wdr72) (Supplementary Table 1). Polymerase chain-reaction (PCR) amplifications of these candidate genes were performed using taq polymerase (Invitrogen, Carlsbad, CA), and sequencing was conducted at Elim Biopharmaceuticals (Hayward, CA). DNA sequence assemblies and SNP analyses (Supplementary Table 2) were performed and documented with CodonCode Aligner software (Centerville, MA).
4.2. Protein structure modeling
The WDR72 sequence was downloaded with accession identifier Q3MJ13 from the UniProt knowledgebase (Apweiler et al., 2004). To generate atomic models for WDR72 wild type and mutant structures, we applied the iterative threading assembler (I-TASSER) (Zhang, 2007) and the restraint-based comparative modeling program MODELLER-v9.10 (Sali and Blundell, 1993) with alignments generated by the profile alignment homology alignment algorithm HHpred (Soding et al., 2005). Models were generated with the automated modeling pipelines accessible through the I-TASSER webserver (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) (Zhang, 2007; Roy et al., 2010) and the bioinformatics toolkit (http://toolkit.tuebingen. mpg.de/hhpred) (Soding et al., 2005), respectively. These two pipelines model diverse crystallized proteins with an average structural alignment GDT-TS measure (range 0–100) (Zemla et al., 2001) of 70 in blinded assessments, outperforming all other automated protein structure prediction tools (Mariani et al., 2011). To verify double seven-bladed β propeller fold topology for WDR72, we applied the β-propeller blade structural motif algorithm SMURF (Menke et al., 2010).
WDR72 oligomer models were generated with the multi-scale modeling and structural comparison tools in Chimera (Yang et al., 2012) by comparison of monomer models to crystallographic structures of structurally homologous protein oligomers found by HHpred.
4.3. Knockout mouse generation
The Wdr72 knockout mouse strain used in this study was created from an ES cell clone (EPD0085_5_D06) generated by the Welcome Trust Sanger Institute and injected into a pregnant female mouse blastocyst by the KOMP Repository and the Mouse Biology Program at the University of California, Davis. The Wdr72 mutant allele (Wdr72−) was created using a `knockout first conditional ready' approach using previously published methods (Skarnes et al., 2011), generating a functional knockout through splicing in LacZ gene reporter and Neomycin (Neo) selection cassettes (Fig. 3A). The targeting vector incorporated an En2 splice acceptor (SA) and internal ribosome entry site (IRES) upstream of LacZ, followed by a polyadenylation (pA) signal. A loxP site separated the LacZ cassette from the subsequent Neo resistance cassette, which was driven by an autonomous promoter (hBactP) and pA signal. Flippase recognition target (FRT) sites flanked both LacZ and Neo cassettes, all of which was inserted between exons 2 and 3. Two additional loxP sites were introduced on either side of exon 3.
Heterozygous mice (Wdr72+/−) on a C57BL/6 genetic background were purchased and subsequently bred to generate the knockout (Wdr72−/−) and wild-type (Wdr72+/+) mice used in these experiments. Genotypes of mice were determined by standard and quantitative PCR (Transnetyx, Cordova, TN) using genomic DNA obtained from tail biopsies with forward primers: NeoF-GGGATCTCATGCTGGAGTTCTTCG, F-TCTTTCACCTAAGCAACACATGCGG, and reverse primer R-GAAACCCGGAGATGAAGGAATGTGC. Amplicon sizes of Wdr72+ and Wdr72− alleles were 520 and 633 bp, respectively.
4.4. RNA isolation and quantitative real-time PCR
Total RNA was isolated from dissected flash-frozen kidneys and from micro-dissected secretory and maturation-stage ameloblasts of Wdr72+/+ and Wdr72−/− mice at postnatal day 24 using RNeasy Mini kit (QIAGEN, Germantown, MD). Aliquot containing 20 μg of total RNA was reverse transcribed to cDNA using SuperScript® III Reverse Transcriptase and oligo dT primers (Invitrogen, Carlsbad, CA). For kidney samples, polymerase chain reaction amplification for standard PCR was performed with the Hot Start Taq kit (Qiagen) by first incubating the reaction mixture at 95° for 5 min, followed by 94 °C, 57 °C, and 72 °C for 1 min each for 35 cycles and then 72 °C for 10 min. The primers used flanked the exon–exon boundaries surrounding exon 14 of the Wdr72 transcript (ENST00000360509), targeting the epitope region to where our WDR72 antibody would recognize. Primers were designed using Frodo (http://frodo.wi.mit.edu/primer3/) (Koressaar and Remm, 2007; Untergasser et al., 2012). The products were visualized on a 1.7% agarose gel with SYBR GREEN staining (Invitrogen, Carlsbad, CA). Real-time PCR gene expression was characterized by quantitative PCR using the ABI 7500 system (Applied Biosystems, Carlsbad, CA). cDNA was amplified with the Fast Start SYBR Green master mix (Roche, Indianapolis, IN). Relative expression levels of target genes were analyzed by the delta-Ct method as published previously (Thomsen et al., 2010) using GAPDH or 18S as endogenous controls.
4.5. Micro-computed tomography (microCT)
Mineral density levels of 6-week-old undecalcified Wdr72+/+ (n = 3) and Wdr72−/− (n = 3) murine hemimandibles were scanned and compared by microCT (SkyScan1076; Bruker-microCT, Kontich, Belgium). X-ray source operating settings were set to 50 kV and 160 μA and image reconstitution was carried out with NRecon software (Bruker-microCT). Using Amira software (ver 1.4.1 SkyScan), coronal sections perpendicular to the curve of the incisor and in line with the midsagittal plane of the incisor were selected from 3D reconstructions for quantitative densitometry (g/cm2) analyses. We compared Wdr72+/+ and Wdr72−/− coronal sections landmarked at either the mesial root of the first molar (maturation stage) or the distal root of the third molar (secretory stage) with indexed and normalized gray scale levels (range 0–255). A region of interest (ie. enamel, dentin or alveolar bone) was selected from each coronal section and quantified for mineral density by measuring the average of relative gray values at each pixel within the selected region (ImageJ, ver 1.46r). These gray value outputs were then averaged across samples and compared by paired Students' t-tests with a statistical significance threshold of P < 0.05.
4.6. Histology
Dissected Wdr72+/+ and Wdr72−/− mandibles at P10 were immediately immerse-fixed in 4% paraformaldehyde (PFA)/0.06 M cacodylate buffer (pH 7.3) overnight and decalcified in 8% EDTA (pH 7.2) for two weeks at 4 °C. Samples were then dehydrated and paraffin-processed for routine embedding and sectioning. Sagittal incisor sections at 5 μm were utilized for standard hematoxylin and eosin (H&E) staining or for immunohistochemistry.
Immunostained tissue sections were deparaffinized and rehydrated, followed by incubation with 10% swine serum for blocking. Primary antibodies targeting WDR72, amelogenin, or CLC5 were incubated at 25 °C overnight, followed by a biotinylated secondary antibody (Dako, Carpinteria, CA) at 25 °C for 1 hr. Alkaline phosphatase conjugated to streptavidin (Vector Laboratories Inc., Burlingame, CA) was used to visualize the colorimetric reaction. Sections were then counterstained using methyl green (Dako, Carpinteria, CA).
The polyclonal rabbit anti-WDR72 antibody was synthesized by Genscript (Piscataway, NJ) from a synthetic peptide (CETGTLERHETGERA) as previously described (El-Sayed et al., 2009), and was used at a 2.7 μg/mL concentration. The rabbit anti-amelogenin antibody (1:500 dilution) was developed in our laboratory as previously described (Le et al., 2007; Li et al., 1998). Purchased antibodies used in this study included polyclonal rabbit anti-CLC5 (Novus Biologicals, Littleton, CO) (NBP1-70374) at a 1:800 dilution, polyclonal rabbit anti-RAB4A (Santa Cruz Biotechnology, Santa Cruz, CA) (sc-312) at 1 μg/mL concentration. The monoclonal rat anti-LAMP1 antibody (1D4B; 1 μg/mL concentration) developed by J. Thomas August was obtained from the Developmental Studies Hybridoma Bank under the auspices of the NICHD and maintained by the University of Iowa, Department of Biology (Iowa Cita, IA 52242). Negative controls were performed with normal rabbit IgG (Vector Laboratories Inc., Burlingame, CA) (I-1000) at matching concentrations to the experimental diluted primary antibodies. Histological images were taken with a Nikon Eclipse E3800 microscope (Melville, NY) using a digital camera (QImaging Inc., Surrey, Canada) and SimplePCI imaging software version 5.3.1.
Supplementary Material
Acknowledgments
Thank you to our AI patients and family members for contributing samples for this study. In addition, we would like to acknowledge Dr. Neil Katsura for providing the clinical evaluations of the AI patients' radiographs and Dr. Yuki Mochida for the comparative discussions of rodent Wdr72 knockout models. MicroCT imaging work was performed by Sabra Djomehri at the Division of Biomaterials and Bioengineering MicroCT Imaging Facility, UCSF, generously supported by the Department of Health and Human Services/NIH S10 Shared Instrumentation Grant (S10RR026645), and Departments of Preventive and Restorative Dental Sciences and Orofacial Sciences, School of Dentistry, UCSF. This research was funded by the UCSF Department of Oral and Craniofacial Sciences, NIDCR 1R03 DE019682 and UCSF Start-up Fund to T.L., and NIDCR T32 DE007306 to P.D.
Footnotes
Appendix A. Supplementary data Supplementary data to this article can be found online at http://dx. doi.org/10.1016/j.matbio.2014.06.005.
References
- Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O'Donovan C, Redaschi N, Yeh L-SL. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. http://dx.doi.org/10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron MJ, Brookes SJ, Kirkham J, Shore RC, Hunt C, Mironov A, Kingswell NJ, Maycock J, Shuttleworth CA, Dixon MJ. A mutation in the mouse Amelx tri-tyrosyl domain results in impaired secretion of amelogenin and phenocopies human X-linked amelogenesis imperfecta. Hum. Mol. Genet. 2010;19:1230–1247. doi: 10.1093/hmg/ddq001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2004;2:e380. doi: 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Williams R, Alber F, Eswar N, Chait BT, Rout MP, Sali A. Simple fold composition and modular architecture of the nuclear pore complex. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2172–2177. doi: 10.1073/pnas.0506345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devuyst O, Christie PT, Courtoy PJ, Beauwens R, Thakker RV. Intra-renal and subcellular distribution of the human chloride channel, CLC-5, reveals a pathophysiological basis for Dent's disease. Hum. Mol. Genet. 1999;8:247–257. doi: 10.1093/hmg/8.2.247. [DOI] [PubMed] [Google Scholar]
- Ding Y, Estrella MR, Hu YY, Chan HL, Zhang HD, Kim JW, Simmer JP, Hu JC. Fam83h is associated with intracellular vesicles and ADHCAI. J. Dent. Res. 2009;88:991–996. doi: 10.1177/0022034509349454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X. Ion channels, channelopathies, and tooth formation. J. Dent. Res. 2014;93:117–125. doi: 10.1177/0022034513507066. [DOI] [PubMed] [Google Scholar]
- El-Sayed W, Parry DA, Shore RC, Ahmed M, Jafri H, Rashid Y, Al-Bahlani S, Al Harasi S, Kirkham J, Inglehearn CF, Mighell AJ. Mutations in the beta propeller WDR72 cause autosomal-recessive hypomaturation amelogenesis imperfecta. Am. J. Hum. Genet. 2009;85:699–705. doi: 10.1016/j.ajhg.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sayed W, Shore RC, Parry DA, Inglehearn CF, Mighell AJ. Hypomaturation amelogenesis imperfecta due to WDR72 mutations: a novel mutation and ultrastructural analyses of deciduous teeth. Cells Tissues Organs. 2011;194:60–66. doi: 10.1159/000322036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field MC, Sali A, Rout MP. Evolution: On a bender—BARs, ESCRTs, COPs, and finally getting your coat. J. Cell Biol. 2011;193:963–972. doi: 10.1083/jcb.201102042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini N, Haack K, Almasy L, Laston S, Lee ET, Best LG, Fabsitz RR, MacCluer JW, Howard BV, Umans JG, Cole SA. Generalization of associations of kidney-related genetic loci to American Indians. Clin. J. Am. Soc. Nephrol. 2014;9:150–158. doi: 10.2215/CJN.02300213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy B, Ferring D, Benesova M, Benes V, Hentze MW. Targeted mutagenesis of the murine IRP1 and IRP2 genes reveals context-dependent RNA processing differences in vivo. RNA. 2004;10:1019–1025. doi: 10.1261/rna.7220704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson CW. The amelogenin proteins and enamel development in humans and mice. J. Oral Biosci. 2011;53:248–256. doi: 10.2330/joralbiosci.53.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good MC, Zalatan JG, Lim WA. Scaffold proteins: hubs for controlling the flow of cellular information. Science. 2011;332:680–686. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Chikuma M, Wang Y, Guggino SE, Guggino WB, Verkman AS. Impaired acidification in early endosomes of ClC-5 deficient proximal tubule. Biochem. Biophys. Res. Commun. 2005;329:941–946. doi: 10.1016/j.bbrc.2005.02.060. [DOI] [PubMed] [Google Scholar]
- Hart PS, Hart TC, Michalec MD, Ryu OH, Simmons D, Hong S, Wright JT. Mutation in kallikrein 4 causes autosomal recessive hypomaturation amelogenesis imperfecta. J. Med. Genet. 2004;41:545–549. doi: 10.1136/jmg.2003.017657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel JK, Johansson S, Raeder H, Platou CG, Midthjell K, Hveem K, Molven A, Njolstad PR. Evaluation of four novel genetic variants affecting hemoglobin A1c levels in a population-based type 2 diabetes cohort (the HUNT2 study) BMC Med. Genet. 2011;12:20. doi: 10.1186/1471-2350-12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JC, Chun YH, Al Hazzazzi T, Simmer JP. Enamel formation and amelogenesis imperfecta. Cells Tissues Organs. 2007;186:78–85. doi: 10.1159/000102683. [DOI] [PubMed] [Google Scholar]
- Hubbard MJ. Calcium transport across the dental enamel epithelium. Crit. Rev. Oral Biol. Med. 2000;11:437–466. doi: 10.1177/10454411000110040401. [DOI] [PubMed] [Google Scholar]
- Kantaputra PN, Kaewgahya M, Khemaleelakul U, Dejkhamron P, Sutthimethakorn S, Thongboonkerd V, Iamaroon A. Enamel–renal–gingival syndrome and FAM20A mutations. Am. J. Med. Genet. A. 2014;164A:1–9. doi: 10.1002/ajmg.a.36187. [DOI] [PubMed] [Google Scholar]
- Kawabe H, Sakisaka T, Yasumi M, Shingai T, Izumi G, Nagano F, Deguchi-Tawarada M, Takeuchi M, Nakanishi H, Takai Y. A novel rabconnectin-3-binding protein that directly binds a GDP/GTP exchange protein for Rab3A small G protein implicated in Ca(2+)-dependent exocytosis of neurotransmitter. Genes Cells. 2003;8:537–546. doi: 10.1046/j.1365-2443.2003.00655.x. [DOI] [PubMed] [Google Scholar]
- Kim JW, Simmer JP, Hart TC, Hart PS, Ramaswami MD, Bartlett JD, Hu JC. MMP-20 mutation in autosomal recessive pigmented hypomaturation amelogenesis imperfecta. J. Med. Genet. 2005;42:271–275. doi: 10.1136/jmg.2004.024505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289–1291. doi: 10.1093/bioinformatics/btm091. http://dx.doi.org/10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- Kottgen A, Pattaro C, Boger CA, Fuchsberger C, Olden M, Glazer NL, Parsa A, Gao X, Yang Q, Smith AV, O'Connell JR, Li M, Schmidt H, Tanaka T, Isaacs A, Ketkar S, Hwang SJ, Johnson AD, Dehghan A, Teumer A, Pare G, Atkinson EJ, Zeller T, Lohman K, Cornelis MC, Probst-Hensch NM, Kronenberg F, Tonjes A, Hayward C, Aspelund T, Eiriksdottir G, Launer LJ, Harris TB, Rampersaud E, Mitchell BD, Arking DE, Boerwinkle E, Struchalin M, Cavalieri M, Singleton A, Giallauria F, Metter J, de Boer IH, Haritunians T, Lumley T, Siscovick D, Psaty BM, Zillikens MC, Oostra BA, Feitosa M, Province M, de Andrade M, Turner ST, Schillert A, Ziegler A, Wild PS, Schnabel RB, Wilde S, Munzel TF, Leak TS, Illig T, Klopp N, Meisinger C, Wichmann HE, Koenig W, Zgaga L, Zemunik T, Kolcic I, Minelli C, Hu FB, Johansson A, Igl W, Zaboli G, Wild SH, Wright AF, Campbell H, Ellinghaus D, Schreiber S, Aulchenko YS, Felix JF, Rivadeneira F, Uitterlinden AG, Hofman A, Imboden M, Nitsch D, Brandstatter A, Kollerits B, Kedenko L, Magi R, Stumvoll M, Kovacs P, Boban M, Campbell S, Endlich K, Volzke H, Kroemer HK, Nauck M, Volker U, Polasek O, Vitart V, Badola S, Parker AN, Ridker PM, Kardia SL, Blankenberg S, Liu Y, Curhan GC, Franke A, Rochat T, Paulweber B, Prokopenko I, Wang W, Gudnason V, Shuldiner AR, Coresh J, Schmidt R, Ferrucci L, Shlipak MG, van Duijn CM, Borecki I, Kramer BK, Rudan I, Gyllensten U, Wilson JF, Witteman JC, Pramstaller PP, Rettig R, Hastie N, Chasman DI, Kao WH, Heid IM, Fox CS. New loci associated with kidney function and chronic kidney disease. Nat. Genet. 2010;42:376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuechler A, Hentschel J, Kurth I, Stephan B, Prott EC, Schweiger B, Schuster A, Wieczorek D, Ludecke HJ. A novel homozygous WDR72 mutation in two siblings with amelogenesis imperfecta and mild short stature. Mol. Syndromol. 2012;3:223–229. doi: 10.1159/000343746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacruz RS, Brookes SJ, Wen X, Jimenez JM, Vikman S, Hu P, White SN, Lyngstadaas SP, Okamoto CT, Smith CE, Paine ML. Adaptor protein complex 2-mediated, clathrin-dependent endocytosis, and related gene activities, are a prominent feature during maturation stage amelogenesis. J. Bone Miner. Res. 2013;28:672–687. doi: 10.1002/jbmr.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TQ, Zhang Y, Li W, Denbesten PK. The effect of LRAP on enamel organ epithelial cell differentiation. J. Dent. Res. 2007;86:1095–1099. doi: 10.1177/154405910708601114. [DOI] [PubMed] [Google Scholar]
- LeBlanc M, Kulle B, Sundet K, Agartz I, Melle I, Djurovic S, Frigessi A, Andreassen OA. Genome-wide study identifies PTPRO and WDR72 and FOXQ1-SUMO1P1 interaction associated with neurocognitive function. J. Psychiatr. Res. 2012;46:271–278. doi: 10.1016/j.jpsychires.2011.11.001. [DOI] [PubMed] [Google Scholar]
- Leblond CP, Warshawsky H. Dynamics of enamel formation in the rat incisor tooth. J. Dent. Res. 1979;58:950–975. doi: 10.1177/00220345790580024901. [DOI] [PubMed] [Google Scholar]
- Lee C, Goldberg J. Structure of coatomer cage proteins and the relationship among COPI, COPII, and clathrin vesicle coats. Cell. 2010;142:123–132. doi: 10.1016/j.cell.2010.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Seymen F, Lee KE, Kang HY, Yildirim M, Tuna EB, Gencay K, Hwang YH, Nam KH, De La Garza RJ, Hu JC, Simmer JP, Kim JW. Novel WDR72 mutation and cytoplasmic localization. J. Dent. Res. 2010;89:1378–1382. doi: 10.1177/0022034510382117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Mathews C, Gao C, DenBesten PK. Identification of two additional exons at the 3' end of the amelogenin gene. Arch. Oral Biol. 1998;43:497–504. doi: 10.1016/s0003-9969(98)00013-2. [DOI] [PubMed] [Google Scholar]
- Lin Z, Jin S, Duan X, Wang T, Martini S, Hulamm P, Cha B, Hubbard A, Donowitz M, Guggino SE. Chloride channel (Clc)-5 is necessary for exocytic trafficking of Na+/H + exchanger 3 (NHE3) J. Biol. Chem. 2011;286:22833–22845. doi: 10.1074/jbc.M111.224998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luyckx VA, Goda FO, Mount DB, Nishio T, Hall A, Hebert SC, Hammond TG, Yu AS. Intrarenal and subcellular localization of rat CLC5. Am. J. Physiol. 1998;275:F761–F769. doi: 10.1152/ajprenal.1998.275.5.F761. [DOI] [PubMed] [Google Scholar]
- Mariani V, Kiefer F, Schmidt T, Haas J, Schwede T. Assessment of template based protein structure predictions in CASP9. Proteins. 2011;79(Suppl 10):37–58. doi: 10.1002/prot.23177. http://dx.doi.org/10.1002/prot.23177. [DOI] [PubMed] [Google Scholar]
- Menke M, Berger B, Cowen L. Markov random fields reveal an N-terminal double beta-propeller motif as part of a bacterial hybrid two-component sensor system. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4069–4074. doi: 10.1073/pnas.0909950107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano F, Kawabe H, Nakanishi H, Shinohara M, Deguchi-Tawarada M, Takeuchi M, Sasaki T, Takai Y. Rabconnectin-3, a novel protein that binds both GDP/GTP exchange protein and GTPase-activating protein for Rab3 small G protein family. J. Biol. Chem. 2002;277:9629–9632. doi: 10.1074/jbc.C100730200. [DOI] [PubMed] [Google Scholar]
- Nanci A, Fortin M, Ghitescu L. Endocytotic functions of ameloblasts and odonto-blasts: immunocytochemical and tracer studies on the uptake of plasma proteins. Anat. Rec. 1996;245:219–234. doi: 10.1002/(SICI)1097-0185(199606)245:2<219::AID-AR9>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Nanci A, Zalzal S, Lavoie P, Kunikata M, Chen W, Krebsbach PH, Yamada Y, Hammarstrom L, Simmer JP, Fincham AG, Snead ML, Smith CE. Comparative immunochemical analyses of the developmental expression and distribution of ameloblastin and amelogenin in rat incisors. J. Histochem. Cytochem. 1998;46:911–934. doi: 10.1177/002215549804600806. [DOI] [PubMed] [Google Scholar]
- Parry DA, Brookes SJ, Logan CV, Poulter JA, El-Sayed W, Al-Bahlani S, Al Harasi S, Sayed J, Raif el M, Shore RC, Dashash M, Barron M, Morgan JE, Carr IM, Taylor GR, Johnson CA, Aldred MJ, Dixon MJ, Wright JT, Kirkham J, Inglehearn CF, Mighell AJ. Mutations in C4orf26, encoding a peptide with in vitro hydroxyapatite crystal nucleation and growth activity, cause amelogenesis imperfecta. Am. J. Hum. Genet. 2012;91:565–571. doi: 10.1016/j.ajhg.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry DA, Poulter JA, Logan CV, Brookes SJ, Jafri H, Ferguson CH, Anwari BM, Rashid Y, Zhao H, Johnson CA, Inglehearn CF, Mighell AJ. Identification of mutations in SLC24A4, encoding a potassium-dependent sodium/calcium exchanger, as a cause of amelogenesis imperfecta. Am. J. Hum. Genet. 2013;92:307–312. doi: 10.1016/j.ajhg.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AD, Waggott D, Boright AP, Hosseini SM, Shen E, Sylvestre MP, Wong I, Bharaj B, Cleary PA, Lachin JM, Below JE, Nicolae D, Cox NJ, Canty AJ, Sun L, Bull SB. A genome-wide association study identifies a novel major locus for glycemic control in type 1 diabetes, as measured by both A1C and glucose. Diabetes. 2010;59:539–549. doi: 10.2337/db09-0653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. http://dx.doi.org/10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Sasaki T. Endocytotic pathways at the ruffled borders of rat maturation amelo-blasts. Histochemistry. 1984;80:263–268. doi: 10.1007/BF00495775. [DOI] [PubMed] [Google Scholar]
- Sethi N, Yan Y, Quek D, Schupbach T, Kang Y. Rabconnectin-3 is a functional regulator of mammalian Notch signaling. J. Biol. Chem. 2010;285:34757–34764. doi: 10.1074/jbc.M110.158634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro JL, Wen X, Okamoto CT, Wang HJ, Lyngstadaas SP, Goldberg M, Snead ML, Paine ML. Cellular uptake of amelogenin, and its localization to CD63, and Lamp1-positive vesicles. Cell. Mol. Life Sci. 2007;64:244–256. doi: 10.1007/s00018-006-6429-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, Jackson D, Severin J, Biggs P, Fu J, Nefedov M, de Jong PJ, Stewart AF, Bradley A. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE. Ameloblasts: secretory and resorptive functions. J. Dent. Res. 1979;58:695–707. doi: 10.1177/002203457905800221011. [DOI] [PubMed] [Google Scholar]
- Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnimann CU, Petsalaki E, Russell RB, Muller CW. WD40 proteins propel cellular networks. Trends Biochem. Sci. 2010;35:565–574. doi: 10.1016/j.tibs.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7(Suppl. 1, S12):11–14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen RR, Sølvsten CAEC, Linnet TET, Blechingberg JJ, Nielsen ALA. Analysis of qPCR data by converting exponentially related Ct values into linearly related X0 values. J. Bioinform. Comput. Biol. 2010;8:885–900. doi: 10.1142/s0219720010004963. [DOI] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115–e115. doi: 10.1093/nar/gks596. http://dx.doi.org/10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasan RS, Larson MG, Aragam J, Wang TJ, Mitchell GF, Kathiresan S, Newton-Cheh C, Vita JA, Keyes MJ, O'Donnell CJ, Levy D, Benjamin EJ. Genome-wide association of echocardiographic dimensions, brachial artery endothelial function and treadmill exercise responses in the Framingham Heart Study. BMC Med. Genet. 2007;8(Suppl. 1):S2. doi: 10.1186/1471-2350-8-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SK, Reid BM, Dugan SL, Roggenbuck JA, Read L, Aref P, Taheri AP, Yeganeh MZ, Simmer JP, Hu JC. FAM20A mutations associated with enamel renal syndrome. J. Dent. Res. 2014;93:42–48. doi: 10.1177/0022034513512653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JT, Torain M, Long K, Seow K, Crawford P, Aldred MJ, Hart PS, Hart TC. Amelogenesis imperfecta: genotype-phenotype studies in 71 families. Cells Tissues Organs. 2011;194:279–283. doi: 10.1159/000324339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Min J. Structure and function of WD40 domain proteins. Protein Cell. 2011;2:202–214. doi: 10.1007/s13238-011-1018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Denef N, Schupbach T. The vacuolar proton pump, V-ATPase, is required for notch signaling and endosomal trafficking in Drosophila. Dev. Cell. 2009;17:387–402. doi: 10.1016/j.devcel.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Lasker K, Schneidman-Duhovny D, Webb B, Huang CC, Pettersen EF, Goddard TD, Meng EC, Sali A, Ferrin TE. UCSF Chimera, MODELLER, and IMP: an integrated modeling system. J. Struct. Biol. 2012;179:269–278. doi: 10.1016/j.jsb.2011.09.006. http://dx.doi.org/10.1016/j.jsb.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemla A, Venclovas, Moult J, Fidelis K. Processing and evaluation of predictions in CASP4. Proteins. 2001;(Suppl 5):13–21. doi: 10.1002/prot.10052. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins. 2007;69(Suppl. 8):108–117. doi: 10.1002/prot.21702. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.