Abstract
Chronic alcohol consumption may induce gene expression alterations in brain reward regions such as the prefrontal cortex (PFC), modulating the risk of alcohol use disorders (AUDs). Transcriptome profiles of 23 AUD cases and 23 matched controls (16 pairs of males and 7 pairs of females) in postmortem PFC were generated using Illumina’s HumanHT-12 v4 Expression BeadChip. Probe-level differentially expressed genes and gene modules in AUD subjects were identified using multiple linear regression and weighted gene co-expression network analyses. The enrichment of differentially co-expressed genes in alcohol dependence-associated genes identified by genome-wide association studies (GWAS) was examined using gene set enrichment analysis. Biological pathways overrepresented by differentially co-expressed genes were uncovered using DAVID bioinformatics resources. Three AUD-associated gene modules in males [Module 1 (561 probes mapping to 505 genes): r=0.42, Pcorrelation=0.020; Module 2 (815 probes mapping to 713 genes): r=0.41, Pcorrelation=0.020; Module 3 (1,446 probes mapping to 1,305 genes): r=−0.38, Pcorrelation=0.030] and one AUD-associated gene module in females [Module 4 (683 probes mapping to 652 genes): r=0.64, Pcorrelation=0.010] were identified. Differentially expressed genes mapped by significant expression probes (Pnominal≤0.05) clustered in Modules 1 and 2 were enriched in GWAS-identified alcohol dependence-associated genes [Module 1 (134 genes): P=0.028; Module 2 (243 genes): P=0.004]. These differentially expressed genes, including ALDH2, ALDH7A1, and ALDH9A1, are involved in cellular functions such as aldehyde detoxification, mitochondrial function, and fatty acid metabolism. Our study revealed differentially co-expressed genes in postmortem PFC of AUD subjects and demonstrated that some of these differentially co-expressed genes participate in alcohol metabolism.
Keywords: Alcohol use disorders, postmortem prefrontal cortex, genome-wide gene expression, co-expression, gene set enrichment analysis, biological pathways
Introduction
Alcohol use disorders (AUDs), including alcohol abuse or dependence, cause substantial morbidity and mortality. Despite the high prevalence (around 8%) of AUDs (Grant et al. 2004), the molecular mechanism of AUDs is not well understood. Given its heterogeneous nature, AUDs are presumed to be caused by variation in multiple genes and by gene-gene and gene-environment interactions (Schuckit 2009). AUD-associated genetic variants, such as those situated in alcohol metabolizing genes ADH1B, ADH1C, and ALDH2 (Gelernter et al. 2014; Quillen et al. 2014), have been identified by candidate gene or genome-wide association studies (GWAS). On the other hand, environmental factors such as chronic alcohol consumption may also alter transcriptome patterns of subjects, leading to alcohol tolerance or dependence through neuroadaptations.
Chronic alcohol consumption drives broad changes in gene expression. Studies using animal or cell culture models have shown that alcohol exposure alters the expression of genes involved in numerous cellular functions, including catecholamine metabolism (Patterson-Buckendahl et al. 2004), signal transduction cascades (Fox et al. 1996), or mitochondrial function and oxidative stress response (Chu et al. 2007). Studies of postmortem brains from human AUD subjects demonstrated altered gene expression in specific brain regions, such as frontal and motor cortices (Lewohl et al. 2000; Liu et al. 2004; Liu et al. 2006; MacKay et al. 2011; Mayfield et al. 2002). Alcohol-responsive genes are involved in numerous cellular functions, such as myelination, ubiquitination, apoptosis, cell adhesion, and neurogenesis (Liu et al. 2006). These results suggest that genes with expression alterations in the brains of AUD subjects may participate in multiple biological pathways that are relevant to the development of AUDs. Additionally, AUD-associated gene expression changes may also help discover novel AUD-related pathways.
Although a number of genes appeared to have differential expression in the brains of AUD subjects, these changes in individual genes were usually small. For example, in the superior frontal cortex, gene expression differences between AUD cases and matched healthy controls ranged from 20% to 50% (Liu et al. 2006). This is congruent with genetic association study findings that the risk of AUDs is influenced by multiple genes, but each gene exerts only a small effect (Agrawal and Bierut 2012; Edenberg and Foroud 2006; Gelernter et al. 2014). Expression alterations in individual genes cannot reasonably account for the total cause of AUDs and are unable to predict the occurrence of AUDs. Thus, an integrative analysis of AUD-associated gene clusters (or modules) should be conducted. It is of particular interest to explore differential expression of co-expressed genes in the brains of AUD subjects because modules of co-expressed genes may reflect functional and structural organizations of brain regions. Recently, the weighted gene co-expression network analysis (WGCNA) method (Langfelder and Horvath 2008; Zhang and Horvath 2005) was applied to detect gene co-expression modules that were associated with psychiatric disorders such as bipolar disorders or schizophrenia (Chen et al. 2013; Torkamani et al. 2010). Ponomarev et al. (Ponomarev et al. 2012) performed WGCNA to obtain an integrative view of AUD-associated transcriptome alterations in three brain regions (the central and basolateral amygdala as well as the superior frontal cortex) by analyzing a small set of postmortem brain tissue samples from 17 AUD cases and 15 controls.
In the present study, we profiled the transcriptome patterns of 23 AUD cases and 23 matched controls in postmortem PFC using microarray technology, and identified AUD-associated co-expressed gene modules in male and female AUD subjects using WGCNA. We took the advantage of the probe-level information on expression arrays for considering the potential expression differences of transcript isoforms of genes. The PFC was selected to study because of its function in regulating cognitive function (Miller and Cohen 2001) and some AUD-related behaviors (Paszti-Gere and Jakus 2013; Walaas et al. 2011). Moreover, the PFC is closely related to the reward system, and alcohol has profound effects on the function of the PFC (Abernathy et al. 2010). To explore the function of differentially expressed genes in AUD subjects, we tested whether AUD-associated gene modules were enriched in genes that were identified to be associated with alcohol dependence by our recent GWAS (Gelernter et al. 2014). Additionally, AUD-relevant biological pathways enriched in a set of differentially co-expressed genes were analyzed.
Methods
Postmortem PFC Tissues
Autopsy brain tissue samples were obtained from the New South Wales Tissue Resource Centre (NSW TRC) at the University of Sydney. The NSW TRC is partially supported by the National Institute on Alcohol Abuse and Alcoholism (NIAAA). Its adherence to standards for ethical research is overseen by the Sydney Local Health Network and The University of Sydney. Fresh-frozen sections of Brodmann area 9 (BA9, mainly the dorsolateral PFC) were obtained from postmortem tissue from 23 European Australians with AUDs and 23 European Australian healthy control subjects. Cases and controls were matched by sex, age, brain weight, brain pH, and postmortem interval (PMI). Exclusion criteria included a history of consumption of illegal drugs of abuse or major psychotic disorders (such as schizophrenia and bipolar disorder) according to the criteria in the Diagnostic and Statistical Manual of Mental Disorders 4th Edition (DSM-IV) (American Psychiatric Association 1994). Comparison subjects also had no history of alcohol abuse or dependence. Characteristics of the two groups of samples (presence of AUDs, sex, age, alcohol daily use, PMI, brain weight, and brain pH) is summarized in Table 1.
Table 1.
Clinical information of postmortem prefrontal cortex (PFC) tissues
| AUD Cases (n = 23) |
Controls (n = 23) |
P-value (t-test) |
|
|---|---|---|---|
| Sex (Male), n (%) | 16 (69.6%) | 16 (69.6%) | |
| Age (years) (mean ± S.D.) | 56 ± 9 | 57 ± 9 | 0.706 |
| Alcohol daily use (gram) (mean ± S.D.) | 165 ± 81 | 11 ± 9 | 5.8×10-9 |
| Postmortem interval (hour) (mean ± S.D.) | 39.7 ± 15.0 | 32.7 ± 13.4 | 0.103 |
| Brain weight (gram) (mean ± S.D.) | 1,380 ± 139 | 1,412 ± 135 | 0.434 |
| Brain pH (mean ± S.D.) | 6.58 ± 0.20 | 6.64 ± 0.27 | 0.371 |
AUD: alcohol use disorder.
Genome-wide gene expression assay
Total RNA was extracted from postmortem PFC tissues of 23 AUD and 23 control subjects using the miRNeasy Mini Kit (QIAGEN, Valencia, CA, USA) and quantified with a NanoDrop 8000 spectrophotometer (Thermo Scientific). RNA Quality was determined with the Agilent 2100 Bioanalyser using the Agilent RNA 6000 Nano Kit (Agilent Technologies, Santa Clara, CA, USA). The RNA integrity number (RIN) ranged from 5.0 to 7.0 for the 46 RNA samples. Genome-wide gene expression was profiled using the Illumina HumanHT-12 v4 Expression BeadChip (Illumina, San Diego, CA, USA). Each array on the HumanHT-12 v4 Expression BeadChip contains >47,000 gene expression probes derived from the National Center for Biotechnology Information (NCBI) Reference Sequence RefSeq Release 38 (November 7, 2009) and other sources. Probe intensity and gene expression data were analyzed using the Illumina GenomeStudio software V2011.1 (Gene Expression Module V1.9.0). Low-level analysis of microarray data was performed in R 2.15.1 (http://www.r-project.org/) using the Bioconductor package lumi (Du et al. 2008). The variance stabilizing transformation (VST) method (Lin et al. 2008) and the robust spline normalization (RSN) (Bell et al. 2011) method were applied to all arrays. After normalization, expression probes with intensities indistinguishable from background noise (detection P value > 0.05) in more than half of the 46 RNA samples were removed. The ComBat function built-in R package sva (Johnson et al. 2007) was applied to remove batch effect (due to different chips being used). After the above data quality control process, a total of 21,521 expression probes remained for analysis. The microarray data were submitted to the NCBI GEO archive (accession #GSE49376).
To assess the reproducibility of the expression array-based assay, one RNA sample was measured in triplicate. The pair-wise correlation coefficient was ≥ 0.997 (Supplementary Fig. S1a-c). To evaluate the reliability of the expression array-based assays, expression levels of six genes (AGT, ALDH1L1, GABRA1, GRIN2C, PAX6, and SLC1A3) that showed differential expression in AUD subjects by paired t-tests, were validated by quantitative real-time PCR using the High Capacity RNA-to-cDNA Kit and the SYBR Green PCR Master Mix Kit (Applied Biosystems, Foster City, CA). The mean expression levels of the six genes measured by the two methods were highly correlated [correlation coefficient = -0.869; the negative correlation was due to that gene expression levels were denoted as probe intensities (by expression array-based assay) and numbers of threshold cycles (Ct) or crossing points (Cp) (by quantitative PCR)] (Supplementary Fig. S1d). Information on primers and conditions for quantitative PCRs is described in Supplementary Table S1.
Statistical analysis
Probe-level analysis of AUD-associated gene expression alterations in human postmortem PFC was implemented using the R package (version 3.0.2). Genome-wide gene expression patterns (at the probe level) were compared between 23 AUD cases and 23 controls using multiple linear regression models with adjustment for sex, age, PMI, and RIN. Because of the widespread sex differences in gene expression and splicing in the adult human brain (Trabzuni et al. 2013), we further analyzed the gene expression data in subgroups of men (16 male AUD cases vs. 16 matched male controls) and women (7 female AUD cases vs. 7 matched female controls), respectively. Differentially expressed genes (at the probe level) in male or female AUD subjects were identified. The q value was computed for each nominal P value by controlling the False Discovery Rate (FDR) at 0.05 using the q value package (Storey and Tibshirani 2003).
Bioinformatics analysis
To assess the inter-correlation of the intensities of the 21,521 expression probes (that remained after preprocessing), co-expression analysis was performed using weighted gene co-expression network analysis (WGCNA) R package (Langfelder and Horvath 2008). The overall intensity profile of the expression probes clustered in a module was represented by module eigengene (ME) (Langfelder and Horvath 2007), which was equivalent to the first principal component (PC1). AUD-associated expression probe clusters (or modules), in which the intensities of probes for interrogating gene expression were highly correlated, were identified. For each module, the correlation of two parameters [the gene significance (GS) and the module membership (MM)] was evaluated. GS stood for the magnitude of correlation between the intensities of individual expression probes in the module and AUDs, and MM meant the magnitude of correlation between the intensities of individual expression probes in the module and ME of the module. A significant correlation suggested that genes differentially expressed (at the probe level) in AUD subjects were also the most important (or central) elements of the module for AUDs. The hypergeometric-based test was then conducted to examine whether expression probes clustered in AUD-associated modules were enriched in those expression probes that showed differential intensities (Pnominal ≤ 0.05) in AUD subjects.
Gene set enrichment analysis (GSEA) (Mootha et al. 2003; Subramanian et al. 2005) was conducted to assess whether a set of differentially expressed genes mapped by expression probes included in AUD-associated modules were over-represented by alcohol dependence-associated genes that were identified by our recent GWAS (Gelernter et al. 2014). To generate the ranked gene list for performing GSEA, P values of 7,019,991 SNPs (imputed or genotyped) were retrieved from our recent alcohol dependence GWAS (Gelernter et al. 2014), which carried out both GWAS and meta-analysis by integrating genotype data from 9,915 European Americans (4,938 cases with alcohol dependence and 4,977 healthy controls). The genomic coordinates (or the starting and ending positions) of genes across the genome were retrieved from the UCSC Genome Browser (the GRCh37/hg19 assembly). Each gene was assigned the minimal P value of SNPs (included in GWAS) that were located from 20 Kb upstream to 20 Kb downstream of the gene. The P values of 17,571 genes (mapped by GWAS SNPs) were ranked by -log10 (P value) to form the ranked gene list.
To explore the function of genes interrogated by expression probes that were clustered in AUD-associated modules, a set of genes mapped by expression probes in the module was uploaded to the gene annotation web server DAVID (Huang da et al. 2009) to identify GO terms or KEGG pathways (PEASE < 0.1 indicated significant enrichment). The protein-protein interaction (PPI) network potentially involved in AUD-associated pathways was visualized using the program EnrichNet (Glaab et al. 2012), which is a web server for identifying and visualizing functional associations between a set of genes (mapped by expression probes clustered in AUD-associated modules) and genes in a known cellular pathway.
Results
Differentially expressed genes in AUD subjects
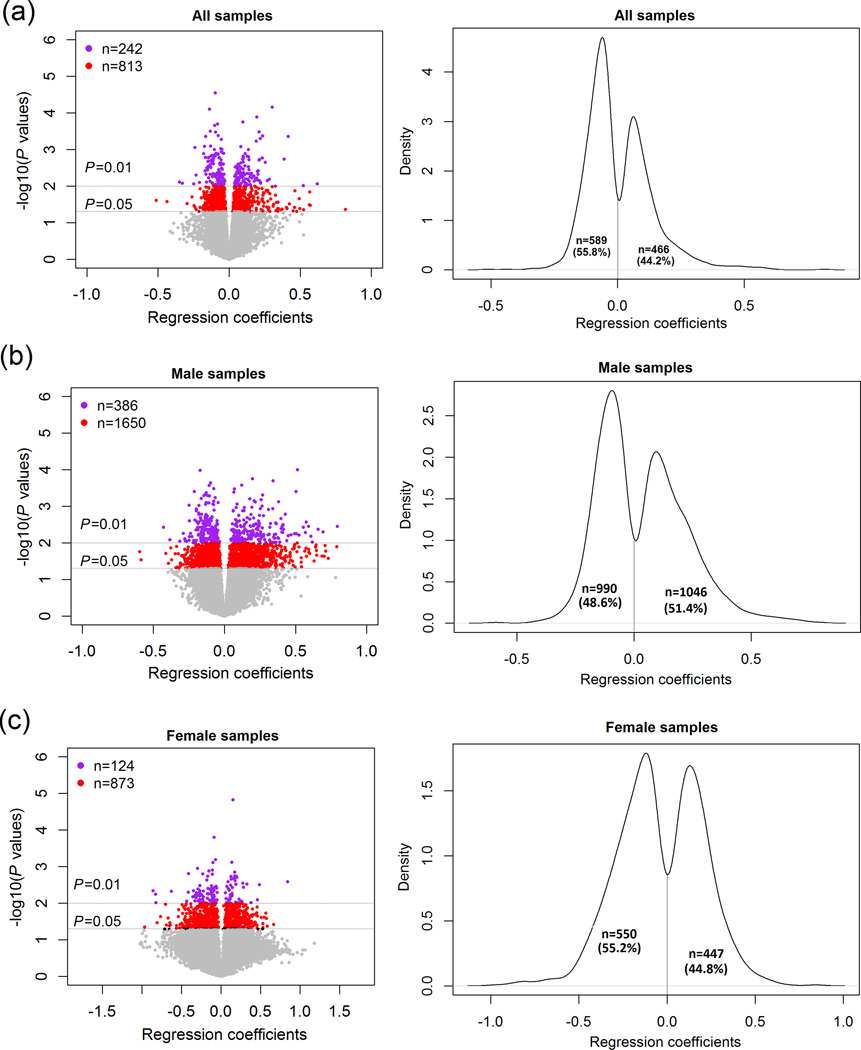
The overall intensity patterns of the 21,521 expression probes that passed quality control procedures were similar across the genomes of 23 AUD cases and 23 matched controls (i.e., no samples were found to be outliers that must be excluded in the analysis) (Supplementary Fig. S2). The intensities of 1,055 (1,055/21,521 = 4.9%) expression probes (mapped to 925 genes) were significantly different between AUD cases and healthy control (2.8×10−5 ≤ Pnominal (all) ≤ 0.05) (Fig. 1a, the volcano plot on the left), and 589 (55.8%) expression probes (mapped to 512 genes) showed a reduced intensity in AUD subjects (Fig. 1a, the Kernel density plot on the right). In a further analysis of the gene expression data from the 16 pairs of male subjects (i.e., 16 male AUD cases vs. 16 male controls), 2,036 (2,036/21,521 = 9.5%) expression probes (mapped to 1,822 genes) showed differences in intensities between male AUD cases and male controls (9.9×10−5 ≤ Pnominal (male) ≤ 0.05) (Fig. 1b, volcano plot on the left), and 990 (990/2,036 = 48.6%) expression probes (mapped to 884 genes) showed a reduced intensity in male AUD subjects (Fig. 1b, Kernel density plot on the right). Additionally, analysis of the expression data from the seven pairs of female subjects (7 female AUD cases vs. 7 match female controls) indicated that 997 (997/21,521 = 4.6%) expression probes (mapped to 914 genes) showed differences in intensity between female AUD cases and female controls (1.5×10−5 ≤ Pnominal (female) ≤ 0.05) (Fig. 1c, volcano plot on the left), and 550 (550/997 = 55.2%) expression probes (mapped to 494 genes) showed a reduced intensity in female AUD subjects (Fig. 1c, Kernel density plot on the right). Only 61 expression probes (mapped to 59 genes) displayed differential intensities in both male (3.4×10−4 ≤ Pnominal (male) ≤ 0.05) and female (1.6×10−4 ≤ Pnominal (female) ≤ 0.05) AUD subjects. The top significant expression probes (Pnominal ≤ 0.001) identified in either male or female AUD subjects are listed in Table 2. None of the expression probes showed genome-wide significant results, i.e., the findings did not survive multiple testing corrections at a false discovery rate (FDR) threshold of q = 0.05.
Figure 1.
Genome-wide gene expression differences (at the probe level) between alcohol use disorder (AUD) cases and matched healthy controls.
For each of the three panels, the volcano plot is on the left. It plots regression coefficients (or effect size) of 21,521 gene expression probes vs. significance [-log10(P values)]. Regression coefficients were obtained from multiple linear regression analysis. Purple dots represent expression probes with P ≤ 0.01, red dots represent expression probes with P ≤ 0.05, and grey dots represent expression probes with P > 0.05. The Kernel density plot is on the right. It plots the distribution of regression coefficients of expression probes with P ≤ 0.05. (a) 23 AUD cases vs. 23 healthy controls; (b) 16 male AUD cases vs. 16 male healthy controls; and (c) 7 female AUD cases vs. 7 female healthy controls.
Table 2.
Top expression probes (Pnominal < 0.001) showing differential intensities in subjects with alcohol use disorders (AUDs)
| Expression probes | Genes | Chr. | Probe intensity (in AUD cases) |
Probe intensity (in control) |
Beta | Pnominal | q-value |
|---|---|---|---|---|---|---|---|
| 34 expression probes (Pnominal < 0.001) identified in male AUD subjects | |||||||
| ILMN_1730229 | CGNL1 | 15 | 9.74 | 9.24 | 0.51 | 9.9E-05 | 0.38 |
| ILMN_2366710 | UCRC | 22 | 9.00 | 9.16 | −0.17 | 1.0E-04 | 0.38 |
| ILMN_1764266 | CKMT2 | 5 | 7.43 | 7.23 | 0.20 | 1.8E-04 | 0.38 |
| ILMN_1716461 | LGI4 | 19 | 8.57 | 8.23 | 0.34 | 2.0E-04 | 0.38 |
| ILMN_1695430 | DDB1 | 11 | 7.20 | 7.09 | 0.12 | 2.6E-04 | 0.38 |
| ILMN_2297373 | C14orf43 | 14 | 6.88 | 6.99 | −0.11 | 2.7E-04 | 0.38 |
| ILMN_2181867 | ANKRD20B | 2 | 6.79 | 6.86 | −0.08 | 3.3E-04 | 0.38 |
| ILMN_1669833 | SH2B2 | 7 | 6.75 | 6.69 | 0.07 | 3.4E-04 | 0.38 |
| ILMN_2198376 | PSMA4 | 15 | 9.90 | 10.12 | −0.21 | 3.9E-04 | 0.38 |
| ILMN_1696339 | ZIC2 | 13 | 8.28 | 7.82 | 0.50 | 4.0E-04 | 0.38 |
| ILMN_1654916 | KIF7 | 15 | 6.80 | 6.75 | 0.06 | 4.1E-04 | 0.38 |
| ILMN_1752967 | DHPS | 19 | 9.08 | 9.23 | −0.14 | 4.7E-04 | 0.38 |
| ILMN_2133675 | SGSH | 17 | 8.60 | 8.37 | 0.27 | 4.9E-04 | 0.38 |
| ILMN_1688730 | DNAJC18 | 5 | 7.86 | 8.00 | −0.14 | 5.1E-04 | 0.38 |
| ILMN_1681741 | C1orf31 | 1 | 6.95 | 7.05 | −0.12 | 5.1E-04 | 0.38 |
| ILMN_1784176 | ABCA5 | 17 | 7.10 | 7.20 | −0.11 | 5.2E-04 | 0.38 |
| ILMN_3263393 | LOC100128202 | 13 | 6.71 | 6.77 | −0.07 | 5.7E-04 | 0.38 |
| ILMN_1723021 | ICMT | 1 | 7.97 | 8.12 | −0.16 | 6.1E-04 | 0.38 |
| ILMN_2399769 | GPR177 | 1 | 7.29 | 7.06 | 0.26 | 6.5E-04 | 0.38 |
| ILMN_2107613 | RHOJ | 14 | 7.13 | 6.94 | 0.21 | 7.0E-04 | 0.38 |
| ILMN_1651902 | RASL11A | 13 | 6.92 | 6.99 | −0.08 | 7.3E-04 | 0.38 |
| ILMN_2172969 | STXBP6 | 14 | 10.05 | 9.66 | 0.38 | 7.5E-04 | 0.38 |
| ILMN_1684321 | CYB5B | 16 | 10.79 | 10.56 | 0.22 | 7.7E-04 | 0.38 |
| ILMN_1737563 | RS1 | NA | 6.68 | 6.72 | −0.05 | 7.9E-04 | 0.38 |
| ILMN_1659725 | EXOSC5 | 19 | 7.76 | 7.89 | −0.13 | 7.9E-04 | 0.38 |
| ILMN_1741300 | ZNF407 | 18 | 7.93 | 8.05 | −0.11 | 8.0E-04 | 0.38 |
| ILMN_1664243 | USE1 | 19 | 7.68 | 7.81 | −0.13 | 8.1E-04 | 0.38 |
| ILMN_1714848 | ZNF354A | 5 | 6.75 | 6.83 | −0.08 | 8.7E-04 | 0.38 |
| ILMN_1809537 | MASP1 | 3 | 6.77 | 6.68 | 0.09 | 8.7E-04 | 0.38 |
| ILMN_1791770 | SMARCC2 | 12 | 7.03 | 7.16 | −0.13 | 8.8E-04 | 0.38 |
| ILMN_1664242 | C20orf12 | 20 | 6.88 | 6.95 | −0.08 | 9.0E-04 | 0.38 |
| ILMN_2415926 | THOC3 | 5 | 7.60 | 7.90 | −0.25 | 9.2E-04 | 0.38 |
| ILMN_2152402 | BAT5 | 6 | 8.07 | 8.27 | −0.18 | 9.5E-04 | 0.38 |
| ILMN_1749081 | AUTS2 | 7 | 9.49 | 9.26 | 0.26 | 9.8E-04 | 0.38 |
| 3 expression probes (Pnominal < 0.001) identified in female AUD subjects | |||||||
| ILMN_2119421 | LOC143543 | 11 | 6.95 | 6.82 | 0.15 | 1.5E-05 | 0.19 |
| ILMN_1733441 | POGZ | 1 | 6.73 | 6.82 | −0.09 | 1.6E-04 | 0.42 |
| ILMN_1793643 | MRM1 | 17 | 6.69 | 6.73 | −0.07 | 6.4E-04 | 0.42 |
Differentially co-expressed gene modules in AUD subjects
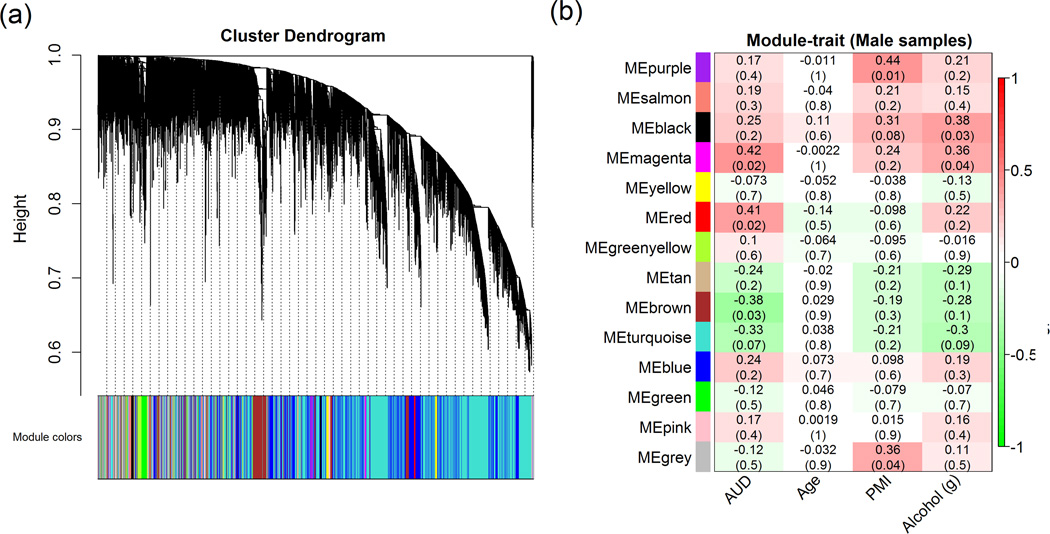
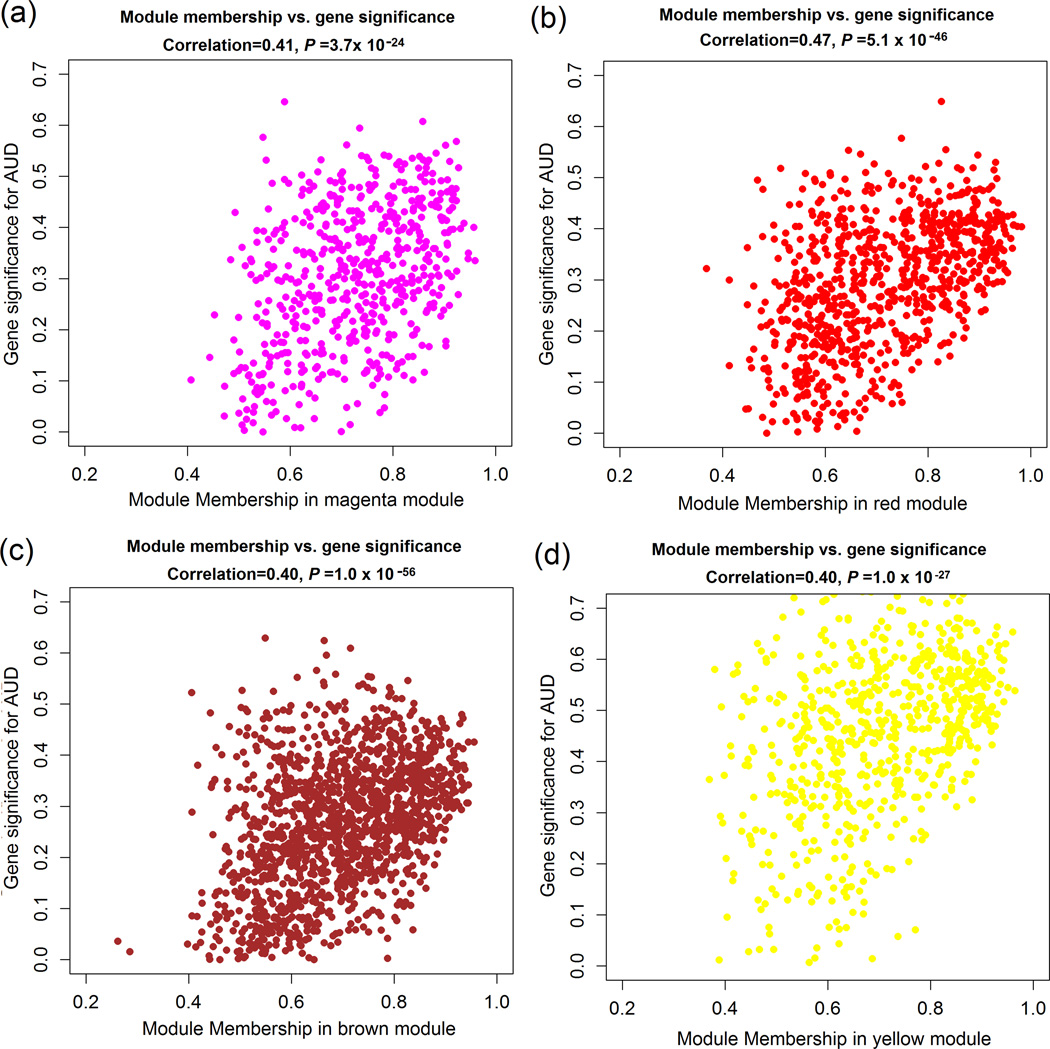
In all 23 pairs of subjects (i.e., 23 AUD cases vs. 23 healthy controls), 11 modules with co-expressed genes (at the probe level) were identified, but none of the 11 modules showed significant correlations with AUDs in overall expression patterns (represented by ME) (Supplementary Fig. S3). In the 16 pairs of male subjects (i.e., 16 male AUD cases vs. 16 male controls), 14 modules with co-expressed genes (at the probe level) were identified, and the overall expression patterns (or ME) of three modules were significantly correlated with AUDs [the magenta-colored module or Module 1 (consisting of 561 expression probes mapping to 505 genes): r = 0.42, Pcorrelation = 0.02; the red-colored module or Module 2 (consisting of 815 expression probes mapping to 713 genes): r = 0.41, Pcorrelation = 0.02; the brown-colored module or Module 3 (consisting of 1,446 expression probes mapping to 1,305 genes): r = −0.38, Pcorrelation = 0.03] (Fig. 2). In the seven pairs of female subjects (i.e., 7 female AUD cases vs. 7 female controls), 15 modules with co-expressed genes (at the probe level) were identified, and the overall expression pattern (or ME) of only one module was significantly correlated with AUDs [the yellow-colored module or Module 4 (consisting of 683 expression probes mapping to 652 genes): r = 0.64, Pcorrelation = 0.01) (Supplementary Fig. S4). As shown in Fig. 3a-3d, there was a significant positive correlation between GS and MM in the above four AUD-associated modules [the magenta module (or Module 1) in males: r = 0.41, Pcorrelation = 3.7×10−24; the red module (or Module 2) in males: r = 0.47, Pcorrelation = 5.1×10−46; the brown module (or Module 3) in males: r = 0.40, Pcorrelation = 1.1×10−56; and the yellow module (or Module 4) in females: r = 0.40, Pcorrelation = 1.3×10-27] . Information (including numbers of significant probes, correlation of expression probes with AUDs, and GSEA results) on the four specific modules is summarized in Supplementary Table S2.
Figure 2.
Genome-wide co-expression analysis in 16 pairs of male subjects.
Weighted gene co-expression network analysis (WGCNA) was performed to assess the inter-correlation of the intensities of 21,521 expression probes in the 32 male subjects (i.e., 16 male cases with alcohol use disorders or AUDs vs. 16 matched male healthy controls). (a) Dendrogram of 14 modules identified by WGCNA under default setting. (b) Association of 14 modules with clinical traits [AUDs, age, postmortem internal (PMI), or alcohol daily use in grams (AlcGram)].
Figure 3.
Scatter plotting of the correlation of the gene significance (GS) and the module membership (MM) for expression probes clustered in AUD-associated modules.
There was a highly significant correlation between GS and MM in three AUD-associated modules (in magenta, brown, or red colors) identified in the 16 pairs of males subjects (16 male AUD cases vs. 16 male controls) and one AUD-associated module (in yellow color) identified in the seven pairs of female subjects (7 female AUD cases vs. 7 female controls).
Of the three AUD-associated modules identified in the 16 pairs of male subjects, the magenta-colored module (or Module 1) had 177 expression probes with Pnominal (male) ≤ 0.05 [i.e., a 3.3-fold enrichment (177/561 = 31.6%) over background (2,036/2,1521 = 9.5%), Phypergenometic < 0.001], the red-colored module (or Module 2) had 326 expression probes with Pnominal (male) ≤ 0.05 [i.e., a 4.2-fold enrichment (326/815 = 40.0%) over background (2,036/21,521 = 9.5%), Phypergenometic < 0.001], and the brown-colored module (or Module 3) had 255 expression probes with Pnominal (male) ≤ 0.05 [i.e., a 1.8-fold enrichment (255/1,446 = 17.6%) over background (2,036/21,521 = 9.5%), Phypergenometic < 0.001]. The AUD-associated yellow-colored module (Module 4) identified in the seven pairs of female subjects harbored 62 expression probes with Pnominal (female) ≤ 0.05 [i.e., a 2.0-fold enrichment (62/683 = 9.1%) over background (997/21,521 = 4.6%), Phypergenometic < 0.001].
Differentially expressed genes enriched in alcohol dependence-associated genes
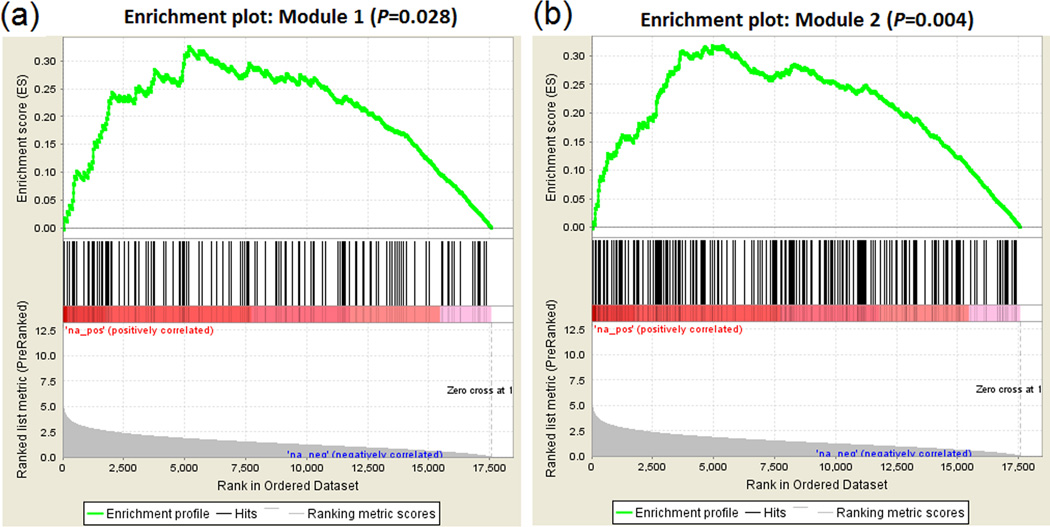
GSEA was performed using genes mapped by significant expression probes (Pnominal ≤ 0.05) clustered in the above four AUD-associated modules (Modules 1, 2 and 3 identified in males and Module 4 identified in females). Genes mapped by significant expression probes contained in two AUD-associated modules [colored in magenta (Module 1) or red (Module 2) in Fig. 2] identified in the 16 pairs of male subjects showed a significant enrichment. 134 annotated genes mapped by 177 significant expression probes included in Module 1 and 243 annotated genes mapped by 326 significant expression probes included in Module 2 were overrepresented by alcohol dependence-associated genes identified by our recent GWAS (Module 1: P = 0.028, q < 0.05; Module 2: P=0.004, q < 0.05) (Fig. 4). There were 54 (54/134 = 40.3%) leadings genes in Module 1 and 97 (97/243 = 39.9%) leading genes in Module 2 (Supplementary Tables S3 and S4).
Figure 4.
Significant enrichment of differentially expressed genes (from Modules 1 and 2) in alcohol dependence-associated genes.
The enrichment scores (shown in the upper curve) were calculated along the ranked genes (the bottom histogram, from left to right, i.e., from more to less significant). The vertical lines in the middle indicate the position of genes mapped by expression probes that showed differential intensities in male AUD subjects and were contained in the AUD-associated Modules 1 (colored in magenta) or 2 (colored in red). The ranked genes were obtained from our recent GWAS on alcohol dependence (Gelernter et al., 2014), and the P values for individual genes were assigned by minimal P values of SNPs nearby (±20 Kb of the gene).
Functional annotation of differentially expressed genes in AUD subjects
To explore the functional roles of genes mapped by significant expression probes (Pnominal (male) ≤ 0.05) contained in the above two AUD-associated modules (Modules 1 and 2 from male subjects), we uploaded 426 unique genes mapped by 503 expression probes (including 177 expression probes with Pnominal (male) ≤0.05 in Module 1 and 326 expression probes with Pnominal (male) ≤ 0.05 in Module 2) into the web-based functional annotation tool DAVID to identify biological processes or pathways. Biological Processes (such as Oxidation Reduction), Cellular Components (such as Mitochondrion), and KEGG Pathways (such as Fatty Acid Metabolism) were overrepresented in this set of genes (Table 3). Through protein-protein interaction (PPI) analysis, nine of the 426 genes, including ACAA2, ACOX1, GCDH, ALDH7A1, CPT2, ALDH2, HADH, ALDH9A1, and ACSL6 (3 of them are involved in aldehyde detoxification), were demonstrated to overlap with 32 genes involved in Fatty Acid Metabolism (Supplementary Fig. S5). Additionally, the expression of these 426 genes was found to be highly enriched in liver (PEASE = 1.2×10−7, PBH-adj=2.2×10−5), adipose tissue (PEASE = 3.0×10−5, PBH-adj = 3.0×10−3), and brain (PEASE = 4.0×10−4, PBH-adj = 0.026) by querying gene expression data using the DAVID tool.
Table 3.
Function annotation of 426 gene mapped by significant expression probes (Pnominal < 0.05) clustered in Modules 1 and 2 (from 16 pairs of male subjects)
| Number of gene |
Fold | PEASE | PBH-adj | ||
|---|---|---|---|---|---|
| Biological Processes | |||||
| GO:0055114 | oxidation reduction | 39 | 2.6 | 1.8E-07 | 4.0E-04 |
| GO:0006631 | fatty acid metabolic process | 16 | 3.4 | 7.9E-05 | 8.6E-02 |
| GO:0014031 | mesenchymal cell development | 8 | 6.6 | 1.9E-04 | 1.3E-01 |
| GO:0048762 | mesenchymal cell differentiation | 8 | 6.6 | 1.9E-04 | 1.3E-01 |
| GO:0060485 | mesenchyme development | 8 | 6.4 | 2.2E-04 | 1.2E-01 |
| GO:0016054 | organic acid catabolic process | 11 | 4.2 | 3.1E-04 | 1.3E-01 |
| GO:0046395 | carboxylic acid catabolic process | 11 | 4.2 | 3.1E-04 | 1.3E-01 |
| GO:0010035 | response to inorganic substance | 15 | 3.1 | 4.0E-04 | 1.4E-01 |
| GO:0045165 | cell fate commitment | 12 | 3.6 | 4.9E-04 | 1.5E-01 |
| GO:0003012 | muscle system process | 13 | 3.2 | 6.9E-04 | 1.8E-01 |
| Cellular Components | |||||
| GO:0005739 | mitochondrion | 56 | 2.1 | 1.9E-07 | 6.3E-05 |
| GO:0044429 | mitochondrial part | 35 | 2.4 | 4.2E-06 | 6.8E-04 |
| GO:0005740 | mitochondrial envelope | 27 | 2.6 | 1.5E-05 | 1.6E-03 |
| GO:0031966 | mitochondrial membrane | 24 | 2.5 | 1.2E-04 | 9.5E-03 |
| GO:0005743 | mitochondrial inner membrane | 20 | 2.6 | 2.1E-04 | 1.4E-02 |
| GO:0019866 | organelle inner membrane | 20 | 2.5 | 5.3E-04 | 2.8E-02 |
| GO:0044459 | plasma membrane part | 77 | 1.4 | 1.0E-03 | 4.7E-02 |
| GO:0031967 | organelle envelope | 29 | 1.9 | 1.5E-03 | 5.8E-02 |
| GO:0031975 | envelope | 29 | 1.9 | 1.6E-03 | 5.5E-02 |
| GO:0016323 | basolateral plasma membrane | 14 | 2.8 | 1.6E-03 | 5.0E-02 |
| KEGG Pathways | |||||
| hsa00071 | Fatty acid metabolism | 9 | 8.0 | 1.1E-05 | 1.5E-03 |
| hsa00310 | Lysine degradation | 8 | 6.5 | 1.8E-04 | 1.2E-02 |
| hsa00330 | Arginine and proline metabolism | 8 | 5.4 | 6.0E-04 | 2.7E-02 |
| hsa00620 | Pyruvate metabolism | 7 | 6.2 | 7.5E-04 | 2.5E-02 |
| hsa00010 | Glycolysis / Gluconeogenesis | 8 | 4.7 | 1.3E-03 | 3.4E-02 |
| hsa00561 | Glycerolipid metabolism | 7 | 5.5 | 1.4E-03 | 3.1E-02 |
| hsa00640 | Propanoate metabolism | 6 | 6.7 | 1.7E-03 | 3.3E-02 |
| hsa03320 | PPAR signaling pathway | 8 | 4.1 | 2.9E-03 | 4.8E-02 |
| hsa00380 | Tryptophan metabolism | 6 | 5.3 | 4.7E-03 | 6.9E-02 |
| hsa00280 | Valine, leucine and isoleucine degradation | 6 | 4.8 | 7.2E-03 | 9.2E-02 |
Discussion
To our knowledge, this is the largest study so far to have examined genome-wide gene expression alterations in postmortem PFC of AUD subjects. Besides analyzing AUD-associated expression changes in individual genes, we also examined AUD-associated clusters of genes that co-expressed in postmortem PFC. Since the abundance and stability of the transcripts of a gene may be different, we applied probe-level analysis to examine gene co-expression differences in postmortem PFC between AUD cases and matched controls. We found that the expression levels of a number of co-regulated genes in the PFC were significantly influenced by chronic alcohol abuse.
Our study demonstrated that AUD-associated gene modules in the PFC were sex-specific. When performing WGCNA in all 23 pairs of AUD cases and matched controls, no significant gene modules in association with AUDs were identified. Nevertheless, and despite the reduction in sample size from splitting the sample, three AUD-associated gene modules (Modules 1, 2, and 3) were obtained in the male subjects and one AUD-associated gene module (Module 4) was obtained in the female subjects (Fig. 3). Only a small number of expression probes in Module 4 overlapped with those in Module 1 (n = 16), 2 (n = 19), or 3 (n = 27). These findings suggest that chronic alcohol abuse may have a different impact on transcriptome patterns of males and females. A plausible explanation for these findings is that chronic alcohol abuse may influence the epigenomic status of males and females differently, as evidenced by our recent study that analyzed the methylome patterns of this same set of postmortem PFC tissue samples and demonstrated sex-specific DNA methylation changes in AUD subjects (unpublished data). Additionally, sex-biased transcriptomic and methylomic patterns in human PFC (Xu et al. 2014) may also contribute to the formation of sex-specific gene modules in association with AUDs.
The enrichment of the differentially co-expressed genes in alcohol dependence-associated signals suggests that these co-expressed genes may interact to influence susceptibility to AUDs. We observed that genes mapped by expression probes clustered in Module 1 or 2 were enriched in alcohol dependence-associated genes identified by our recent GWAS (Gelernter et al. 2014). About 40% of the differentially expressed genes (mapped by expression probes with Pnominal (male) ≤ 0.05) in Module 1 or 2 were the leading genes for achieving the maximum enrichment score (Supplementary Tables S3 and S4). Most published gene expression studies focused on genes with the highest statistical significance but ignored those genes with less significant results. Nonetheless, some of the neglected genes may play important roles in the etiology of diseases even though they only reveal small or moderate expression changes in patients. Since we presume that chronic alcohol abuse leads to widespread gene expression changes across the genome and some of these changes may contribute to the risk of AUDs, it is necessary to generate an integrative view of molecular changes underlying AUDs. Only one study is known to have used the co-expression analysis approach to identify AUD-associated gene modules in the amygdala and the superior frontal cortex of a small number of subjects (Ponomarev et al. 2012). The present study provides further evidence that chronic alcohol abuse may affect the expression of a group of correlated genes in the PFC and these co-expressed genes may have a combined effect on AUD risk.
Chronic alcohol abuse may affect the expression of multiple co-expressed genes participating in multiple functional systems. As show in Table 3, the most significant Biological Process potentially involved in AUDs is Oxidation Reduction, which was overrepresented by 39 differentially expressed genes mapped by significant expression probes in Modules 1 and 2. The primary enzymes involved in the oxidation/reduction reaction for metabolizing alcohol include the alcohol dehydrogenases (ADHs), aldehyde dehydrogenase (ALDHs), and cytochrome P450 (Zakhari 2006). Among the 39 differentially expressed genes identified in postmortem PFC of AUD subjects, one (ADHFE1) was from the ADH gene family, six (ALDH1L1, ALDH2, ALDH4A1, ALDH6A1, ALDH7A1, and ALDH9A1) were from the ALDH gen family, and four (CYP11A1, CYP26A1, CYP2U1, and CYP4F11) were from the cytochrome P450 gene family. These findings suggest that altered expression of alcohol metabolizing genes may either confer increased risk for AUDs, or reflect the physiological response to chronically high levels of alcohol intake. Moreover, the most significant Cellular Component potentially involved in AUDs is Mitochondrion. Mitochondria participate in alcohol metabolism with the conversion of acetaldehyde into acetate (Manzo-Avalos and Saavedra-Molina 2010). Chronic alcohol abuse may affect the function of mitochondria. Ethanol-induced deficits in mitochondrial function were reported (Hoek et al. 2002; Manzo-Avalos and Saavedra-Molina 2010). Similar to the functional annotation analysis results reported in a published study that analyzed gene expression changes in the superior frontal cortex of AUD subjects (Liu et al. 2006), we found that 56 genes participating in the function of mitochondria were differentially expressed in postmortem PFC of AUD subjects. In other words, expression alterations in this group of genes could either make subjects more vulnerable to AUDs, or reflect the results of that vulnerability. The most significant KEGG Pathway potentially involved in AUDs was Fatty Acid Metabolism, which was overrepresented by nine differentially expressed genes (ACAA2, ACOX1, ACSL6, ALDH2, ALDH7A1, ALDH9A1, CPT2, GCDH, and HADH) that were mapped by significant expression probes in Modules 1 or 2. There is evidence that fatty acid plays an important role in AUDs. Alcohol-induced tissue damages such as alcoholic fatty liver were associated with impaired fatty acid metabolism (Glen et al. 1987; You et al. 2002). Thus, some of the genes involved in the Fatty Acid Metabolism pathway, such as the above nine differentially expressed genes identified by the present study, are likely to be causal genes for AUD development or to reflect its development.
Additionally, the findings from the present study suggest that the gene module-based analysis approach is more powerful than the individual gene analysis approach. This is especially true for studying genome-wide gene expression changes associated with complex genetic disorders such as AUDs, on which each gene may only exert a minor or moderate effect. As presented in Table 2, the findings from even those top expression probes (or genes) identified in male or female AUD subjects could not withstand multiple testing corrections. Moreover, the implication of the most significant gene identified in the male AUD subjects (CGNL1 mapped by probe ILMN_1730229: Pnominal= 9.9×10−5) or the female AUD subjects (LOC143543 mapped by probe ILMN_2119421: Pnominal= 1.5×10−5) in AUDs is unknown. In other words, the individual probe (or gene) analysis approach may either generate a genome-wide negative result or identify genes that show significant results but play little role in diseases. The gene module analysis approach circumvents the need for multiple testing corrections and reveals a joint effect of co-expressed genes on diseases (which could be additive, multiplicative, synergistic, or via some other mechanism).
Although the present study was able to identify AUD-associated gene modules, it did not have the power to clarify whether the gene expression changes in AUD subjects reflected a pre-existing gene expression differences (possibly due to genetic variation) between AUD cases and healthy controls or an alteration that resulted from chronic alcohol abuse. Additionally, the present study is limited by studying AUD-associated transcriptome changes in only one brain region (i.e., the PFC) of subjects from only one population (with the European ancestry). Future studies should examine AUD-associated gene expression changes in other reward-related brain regions (such as the nucleus accumbens and the ventral tegmental area) and recruit subjects from other population groups to study.
In summary, the present study performed an integrative or co-expression analysis and provided evidence that a group of closely correlated genes could interact with each other to influence the function of specific biological pathways relevant to AUD development. These findings could help in the development of pharmacotherapies for AUDs by targeting the differentially expressed genes or biological pathways identified here.
Supplementary Material
Acknowledgements
This study was supported by the National Institutes of Health (NIH) Grants K99/R00 DA022891 (HZ) and P50 AA12870 (JG & HZ). The authors are grateful to the Australian Brain Donor Programs New South Wales Tissue Resource Centre for providing alcoholic and control brain tissues for this study. The centre is supported by the University of Sydney, the National Health and Medical Research Council of Australia, and the National Institute on Alcohol Abuse and Alcoholism. We also thank the deceased subjects’ next of kin for providing informed written consent for the studies.
Footnotes
Conflict of interest
The authors declare that there are no conflicts of interest.
References
- Abernathy K, Chandler LJ, Woodward JJ. Alcohol and the prefrontal cortex. Int Rev Neurobiol. 2010;91:289–320. doi: 10.1016/S0074-7742(10)91009-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Bierut LJ. Identifying genetic variation for alcohol dependence. Alcohol Res. 2012;34:274–281. doi: 10.35946/arcr.v34.3.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 4th ed. Washington, DC: 1994. [Google Scholar]
- Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, Gilad Y, Pritchard JK. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12:R10. doi: 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Cheng L, Grennan K, Pibiri F, Zhang C, Badner JA, Members of the Bipolar Disorder Genome Study C, Gershon ES, Liu C. Two gene co-expression modules differentiate psychotics and controls. Mol Psychiatry. 2013;18:1308–1314. doi: 10.1038/mp.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J, Tong M, de la Monte SM. Chronic ethanol exposure causes mitochondrial dysfunction and oxidative stress in immature central nervous system neurons. Acta Neuropathol. 2007;113:659–673. doi: 10.1007/s00401-007-0199-4. [DOI] [PubMed] [Google Scholar]
- Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T. The genetics of alcoholism: identifying specific genes through family studies. Addict Biol. 2006;11:386–396. doi: 10.1111/j.1369-1600.2006.00035.x. [DOI] [PubMed] [Google Scholar]
- Fox ES, Cantrell CH, Leingang KA. Inhibition of the Kupffer cell inflammatory response by acute ethanol: NF-kappa B activation and subsequent cytokine production. Biochem Biophys Res Commun. 1996;225:134–140. doi: 10.1006/bbrc.1996.1142. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, Anton R, Preuss UW, Ridinger M, Rujescu D, Wodarz N, Zill P, Zhao H, Farrer LA. Genome-wide association study of alcohol dependence:significant findings in African- and European-Americans including novel risk loci. Mol Psychiatry. 2014;19:41–49. doi: 10.1038/mp.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaab E, Baudot A, Krasnogor N, Schneider R, Valencia A. EnrichNet: network-based gene set enrichment analysis. Bioinformatics. 2012;28:i451–i457. doi: 10.1093/bioinformatics/bts389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glen I, Skinner F, Glen E, MacDonell L. The role of essential fatty acids in alcohol dependence and tissue damage. Alcohol Clin Exp Res. 1987;11:37–41. doi: 10.1111/j.1530-0277.1987.tb01257.x. [DOI] [PubMed] [Google Scholar]
- Grant BF, Dawson DA, Stinson FS, Chou SP, Dufour MC, Pickering RP. The 12-month prevalence and trends in DSM-IV alcohol abuse and dependence: United States, 1991-1992 and 2001-2002. Drug Alcohol Depend. 2004;74:223–234. doi: 10.1016/j.drugalcdep.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Hoek JB, Cahill A, Pastorino JG. Alcohol and mitochondria: a dysfunctional relationship. Gastroenterology. 2002;122:2049–2063. doi: 10.1053/gast.2002.33613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol. 2007;1:54. doi: 10.1186/1752-0509-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. C Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewohl JM, Wang L, Miles MF, Zhang L, Dodd PR, Harris RA. Gene expression in human alcoholism: microarray analysis of frontal cortex. Alcohol Clin Exp Res. 2000;24:1873–1882. [PubMed] [Google Scholar]
- Lin SM, Du P, Huber W, Kibbe WA. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 2008;36:e11. doi: 10.1093/nar/gkm1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Dodd PR, Randall PK, Harris RA, Mayfield RD. Gene expression profiling of individual cases reveals consistent transcriptional changes in alcoholic human brain. J Neurochem. 2004;90:1050–1058. doi: 10.1111/j.1471-4159.2004.02570.x. [DOI] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Harris RA, Iyer VR, Dodd PR, Randall PK, Mayfield RD. Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacology. 2006;31:1574–1582. doi: 10.1038/sj.npp.1300947. [DOI] [PubMed] [Google Scholar]
- MacKay RK, Colson NJ, Dodd PR, Lewohl JM. Differential expression of 14-3-3 isoforms in human alcoholic brain. Alcohol Clin Exp Res. 2011;35:1041–1049. doi: 10.1111/j.1530-0277.2011.01436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzo-Avalos S, Saavedra-Molina A. Cellular and mitochondrial effects of alcohol consumption. Int J Environ Res Public Health. 2010;7:4281–4304. doi: 10.3390/ijerph7124281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield RD, Lewohl JM, Dodd PR, Herlihy A, Liu J, Harris RA. Patterns of gene expression are altered in the frontal and motor cortices of human alcoholics. J Neurochem. 2002;81:802–813. doi: 10.1046/j.1471-4159.2002.00860.x. [DOI] [PubMed] [Google Scholar]
- Miller EK, Cohen JD. An integrative theory of prefrontal cortex function. Annu Rev Neurosci. 2001;24:167–202. doi: 10.1146/annurev.neuro.24.1.167. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Paszti-Gere E, Jakus J. Protein Phosphatases but not Reactive Oxygen Species Play Functional Role in Acute Amphetamine-Mediated Dopamine Release. Cell Biochem Biophys. 2013;66:831–841. doi: 10.1007/s12013-013-9608-6. [DOI] [PubMed] [Google Scholar]
- Patterson-Buckendahl P, Blakley G, Kubovcakova L, Krizanova O, Pohorecky LA, Kvetnansky R. Alcohol alters rat adrenomedullary function and stress response. Ann N Y Acad Sci. 2004;1018:173–182. doi: 10.1196/annals.1296.020. [DOI] [PubMed] [Google Scholar]
- Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci. 2012;32:1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillen EE, Chen XD, Almasy L, Yang F, He H, Li X, Wang XY, Liu TQ, Hao W, Deng HW, Kranzler HR, Gelernter J. ALDH2 is associated to alcohol dependence and is the major genetic determinant of"daily maximum drinks" in a GWAS study of an isolated rural chinese sample. Am J Med Genet B Neuropsychiatr Genet. 2014;165B:103–110. doi: 10.1002/ajmg.b.32213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA. Alcohol-use disorders. Lancet. 2009;373:492–501. doi: 10.1016/S0140-6736(09)60009-X. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9440. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torkamani A, Dean B, Schork NJ, Thomas EA. Coexpression network analysis of neural tissue reveals perturbations in developmental processes in schizophrenia. Genome Res. 2010;20:403–412. doi: 10.1101/gr.101956.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabzuni D, Ramasamy A, Imran S, Walker R, Smith C, Weale ME, Hardy J, Ryten M, North American Brain Expression C. Widespread sex differences in gene expression and splicing in the adult human brain. Nat Commun. 2013;4:2771. doi: 10.1038/ncomms3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walaas SI, Hemmings HC, Jr, Greengard P, Nairn AC. Beyond the dopamine receptor: regulation and roles of serine/threonine protein phosphatases. Front Neuroanat. 2011;5:50. doi: 10.3389/fnana.2011.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Wang F, Liu Y, Yu Y, Gelernter J, Zhang H. Sex-biased methylome and transcriptome in human prefrontal cortex. Hum Mol Genet. 2014;23:1260–1270. doi: 10.1093/hmg/ddt516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You M, Fischer M, Deeg MA, Crabb DW. Ethanol induces fatty acid synthesis pathways by activation of sterol regulatory element-binding protein (SREBP) J Biol Chem. 2002;277:29342–29347. doi: 10.1074/jbc.M202411200. [DOI] [PubMed] [Google Scholar]
- Zakhari S. Overview: how is alcohol metabolized by the body? Alcohol Res Health. 2006;29:245–254. [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.