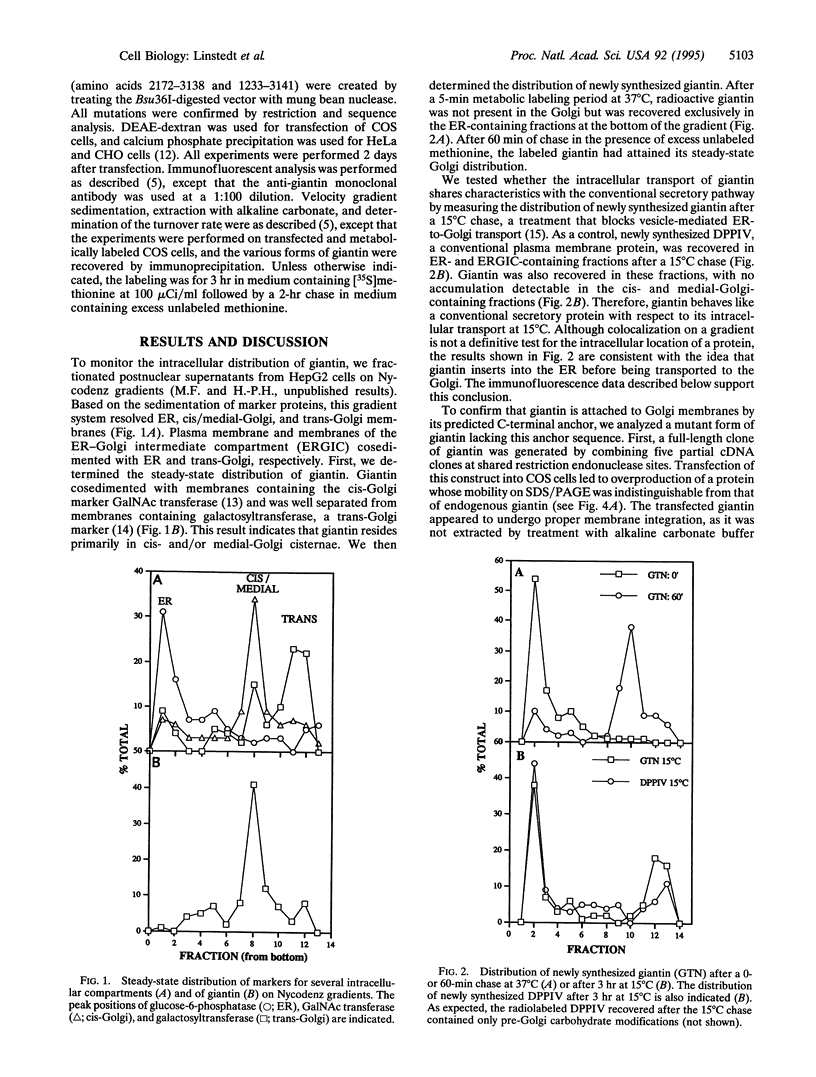
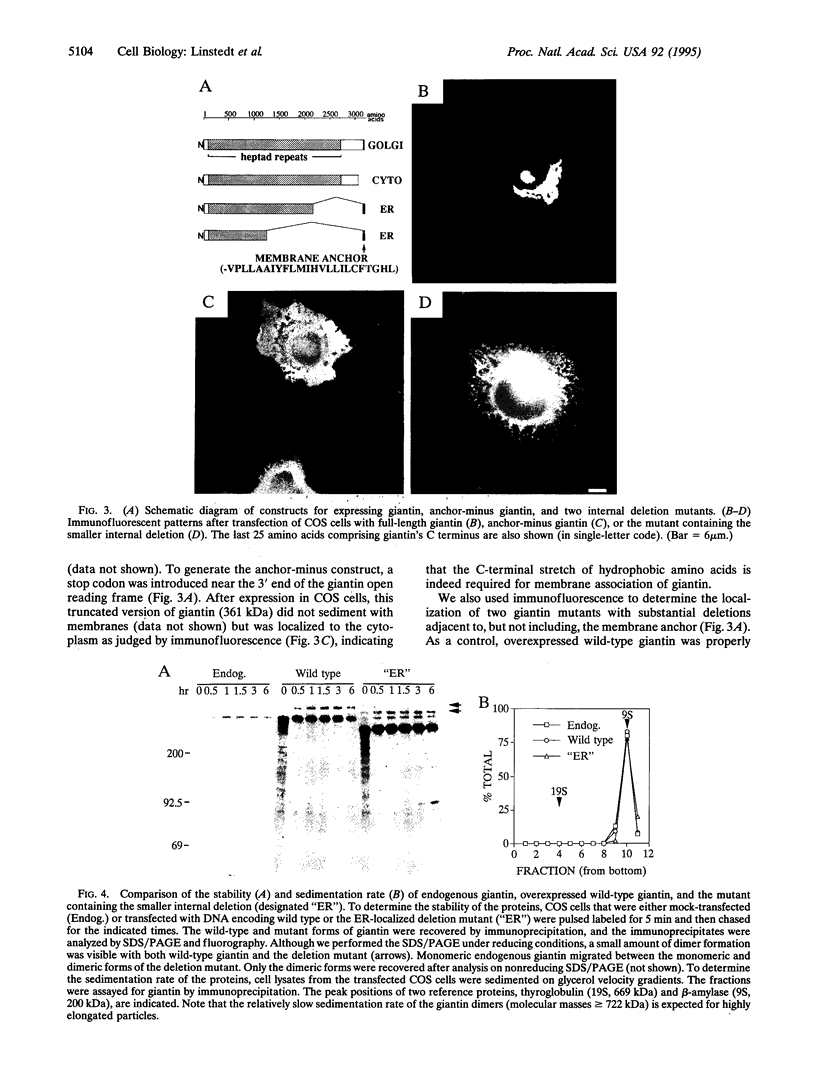
Abstract
Unlike conventional membrane proteins of the secretory pathway, proteins anchored to the cytoplasmic surface of membranes by hydrophobic sequences near their C termini follow a posttranslational, signal recognition particle-independent insertion pathway. Many such C-terminally-anchored proteins have restricted intracellular locations, but it is not known whether these proteins are targeted directly to the membranes in which they will ultimately reside. Here we have analyzed the intracellular sorting of the Golgi protein giantin, which consists of a rod-shaped 376-kDa cytoplasmic domain followed by a hydrophobic C-terminal anchor sequence. Unexpectedly, we find that giantin behaves like a conventional secretory protein in that it inserts into the endoplasmic reticulum (ER) and then is transported to the Golgi. A deletion mutant lacking a portion of the cytoplasmic domain adjacent to the membrane anchor still inserts into the ER but fails to reach the Golgi, even though this mutant has a stable folded structure. These findings suggest that the localization of a C-terminally-anchored Golgi protein involves at least three steps: insertion into the ER membrane, controlled incorporation into transport vesicles, and retention within the Golgi.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. J., Mostov K. E., Blobel G. Mechanisms of integration of de novo-synthesized polypeptides into membranes: signal-recognition particle is required for integration into microsomal membranes of calcium ATPase and of lens MP26 but not of cytochrome b5. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7249–7253. doi: 10.1073/pnas.80.23.7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson N. N., Jr, Touster O. Isolation of rat liver plasma membrane fragments in isotonic sucrose. Methods Enzymol. 1974;31:90–102. doi: 10.1016/0076-6879(74)31009-9. [DOI] [PubMed] [Google Scholar]
- Balch W. E., Rothman J. E. Characterization of protein transport between successive compartments of the Golgi apparatus: asymmetric properties of donor and acceptor activities in a cell-free system. Arch Biochem Biophys. 1985 Jul;240(1):413–425. doi: 10.1016/0003-9861(85)90046-3. [DOI] [PubMed] [Google Scholar]
- Banfield D. K., Lewis M. J., Rabouille C., Warren G., Pelham H. R. Localization of Sed5, a putative vesicle targeting molecule, to the cis-Golgi network involves both its transmembrane and cytoplasmic domains. J Cell Biol. 1994 Oct;127(2):357–371. doi: 10.1083/jcb.127.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colley K. J., Lee E. U., Paulson J. C. The signal anchor and stem regions of the beta-galactoside alpha 2,6-sialyltransferase may each act to localize the enzyme to the Golgi apparatus. J Biol Chem. 1992 Apr 15;267(11):7784–7793. [PubMed] [Google Scholar]
- Gilmore R. The protein translocation apparatus of the rough endoplasmic reticulum, its associated proteins, and the mechanism of translocation. Curr Opin Cell Biol. 1991 Aug;3(4):580–584. doi: 10.1016/0955-0674(91)90026-u. [DOI] [PubMed] [Google Scholar]
- Kutay U., Hartmann E., Rapoport T. A. A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 1993 Mar;3(3):72–75. doi: 10.1016/0962-8924(93)90066-a. [DOI] [PubMed] [Google Scholar]
- Linstedt A. D., Hauri H. P. Giantin, a novel conserved Golgi membrane protein containing a cytoplasmic domain of at least 350 kDa. Mol Biol Cell. 1993 Jul;4(7):679–693. doi: 10.1091/mbc.4.7.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machamer C. E. Targeting and retention of Golgi membrane proteins. Curr Opin Cell Biol. 1993 Aug;5(4):606–612. doi: 10.1016/0955-0674(93)90129-E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S. Sequences within and adjacent to the transmembrane segment of alpha-2,6-sialyltransferase specify Golgi retention. EMBO J. 1991 Dec;10(12):3577–3588. doi: 10.1002/j.1460-2075.1991.tb04924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T., Hoe M. H., Slusarewicz P., Rabouille C., Watson R., Hunte F., Watzele G., Berger E. G., Warren G. Kin recognition between medial Golgi enzymes in HeLa cells. EMBO J. 1994 Feb 1;13(3):562–574. doi: 10.1002/j.1460-2075.1994.tb06294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T., Lucocq J. M., Mackay D., Warren G. The membrane spanning domain of beta-1,4-galactosyltransferase specifies trans Golgi localization. EMBO J. 1991 Dec;10(12):3567–3575. doi: 10.1002/j.1460-2075.1991.tb04923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T., Slusarewicz P., Hoe M. H., Warren G. Kin recognition. A model for the retention of Golgi enzymes. FEBS Lett. 1993 Sep 6;330(1):1–4. doi: 10.1016/0014-5793(93)80906-b. [DOI] [PubMed] [Google Scholar]
- Oprins A., Duden R., Kreis T. E., Geuze H. J., Slot J. W. Beta-COP localizes mainly to the cis-Golgi side in exocrine pancreas. J Cell Biol. 1993 Apr;121(1):49–59. doi: 10.1083/jcb.121.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickwood D., Ford T., Graham J. Nycodenz: a new nonionic iodinated gradient medium. Anal Biochem. 1982 Jun;123(1):23–31. doi: 10.1016/0003-2697(82)90618-2. [DOI] [PubMed] [Google Scholar]
- Roth J., Berger E. G. Immunocytochemical localization of galactosyltransferase in HeLa cells: codistribution with thiamine pyrophosphatase in trans-Golgi cisternae. J Cell Biol. 1982 Apr;93(1):223–229. doi: 10.1083/jcb.93.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J., Wang Y., Eckhardt A. E., Hill R. L. Subcellular localization of the UDP-N-acetyl-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase-mediated O-glycosylation reaction in the submaxillary gland. Proc Natl Acad Sci U S A. 1994 Sep 13;91(19):8935–8939. doi: 10.1073/pnas.91.19.8935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo R. N., Shaper N. L., Taatjes D. J., Shaper J. H. Beta 1,4-galactosyltransferase: a short NH2-terminal fragment that includes the cytoplasmic and transmembrane domain is sufficient for Golgi retention. J Biol Chem. 1992 May 5;267(13):9241–9247. [PubMed] [Google Scholar]
- Schindler R., Itin C., Zerial M., Lottspeich F., Hauri H. P. ERGIC-53, a membrane protein of the ER-Golgi intermediate compartment, carries an ER retention motif. Eur J Cell Biol. 1993 Jun;61(1):1–9. [PubMed] [Google Scholar]
- Schweizer A., Clausen H., van Meer G., Hauri H. P. Localization of O-glycan initiation, sphingomyelin synthesis, and glucosylceramide synthesis in Vero cells with respect to the endoplasmic reticulum-Golgi intermediate compartment. J Biol Chem. 1994 Feb 11;269(6):4035–4041. [PubMed] [Google Scholar]
- Seelig H. P., Schranz P., Schröter H., Wiemann C., Griffiths G., Renz M. Molecular genetic analyses of a 376-kilodalton Golgi complex membrane protein (giantin) Mol Cell Biol. 1994 Apr;14(4):2564–2576. doi: 10.1128/mcb.14.4.2564. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tartakoff A. M. Temperature and energy dependence of secretory protein transport in the exocrine pancreas. EMBO J. 1986 Jul;5(7):1477–1482. doi: 10.1002/j.1460-2075.1986.tb04385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teasdale R. D., D'Agostaro G., Gleeson P. A. The signal for Golgi retention of bovine beta 1,4-galactosyltransferase is in the transmembrane domain. J Biol Chem. 1992 Feb 25;267(6):4084–4096. [PubMed] [Google Scholar]