Abstract
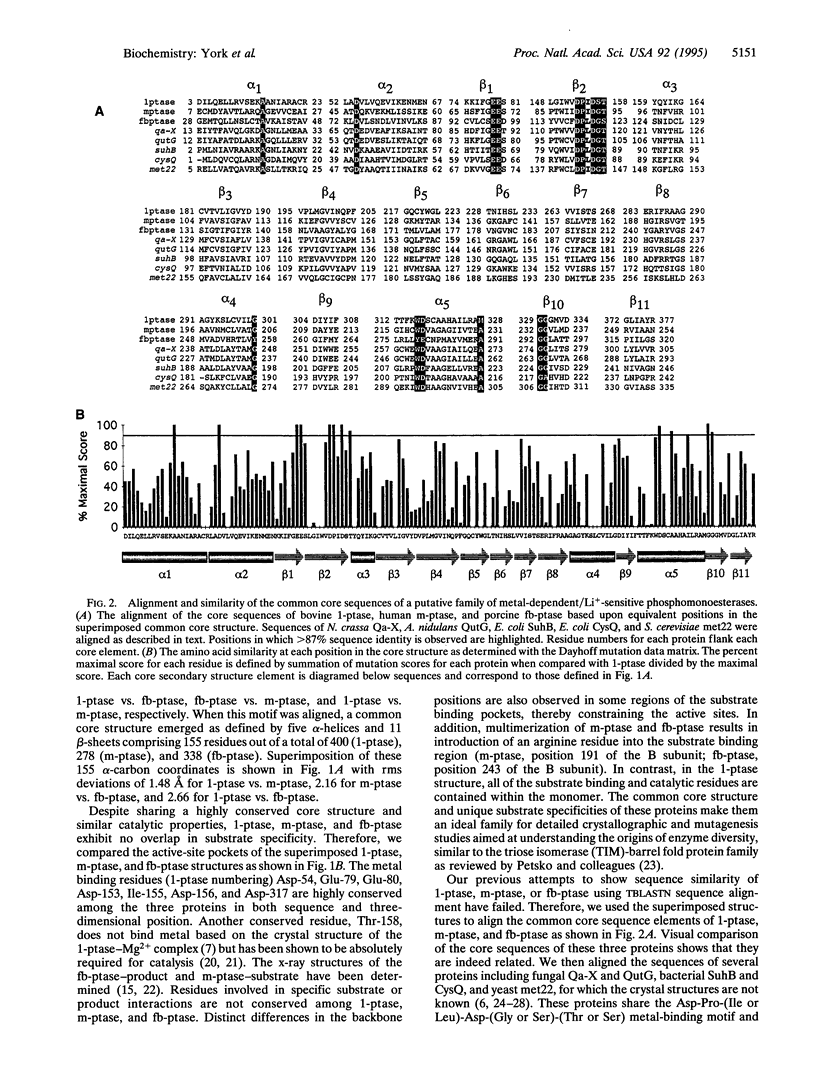
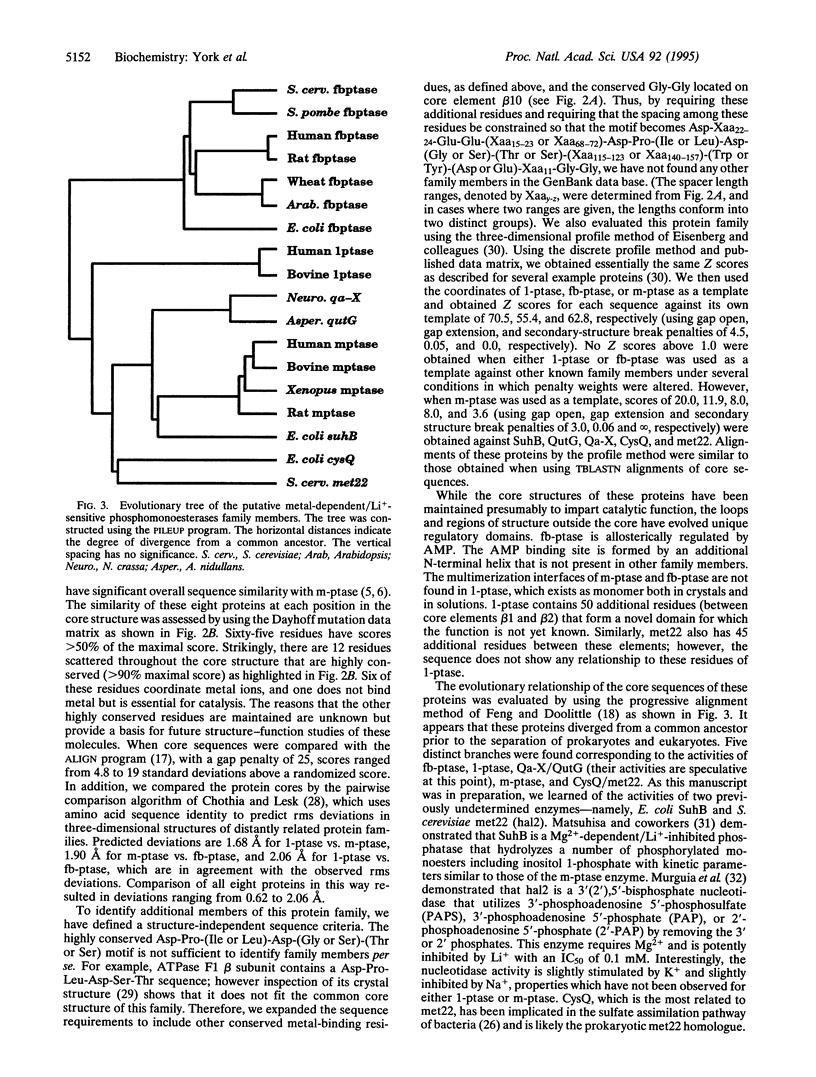
Inositol polyphosphate 1-phosphatase, inositol monophosphate phosphatase, and fructose 1,6-bisphosphatase share a sequence motif, Asp-Pro-(Ile or Leu)-Asp-(Gly or Ser)-(Thr or Ser), that has been shown by crystallographic and mutagenesis studies to bind metal ions and participate in catalysis. We compared the six alpha-carbon coordinates of this motif from the crystal structures of these three phosphatases and found that they are superimposable with rms deviations ranging from 0.27 to 0.60 A. Remarkably, when these proteins were aligned by this motif a common core structure emerged, defined by five alpha-helices and 11 beta-strands comprising 155 residues having rms deviations ranging from 1.48 to 2.66 A. We used the superimposed structures to align the sequences within the common core, and a distant relationship was observed suggesting a common ancestor. The common core was used to align the sequences of several other proteins that share significant similarity to inositol monophosphate phosphatase, including proteins encoded by fungal qa-X and qutG, bacterial suhB and cysQ (identical to amtA), and yeast met22 (identical to hal2). Evolutionary comparison of the core sequences indicate that five distinct branches exist within this family. These proteins share metal-dependent/Li(+)-sensitive phosphomonoesterase activity, and each predicted tree branch exhibits unique substrate specificity. Thus, these proteins define an ancient structurally conserved family involved in diverse metabolic pathways including inositol signaling, gluconeogenesis, sulfate assimilation, and possibly quinone metabolism. Furthermore, we suggest that this protein family identifies candidate enzymes to account for both the therapeutic and toxic actions of Li+ as it is used in patients treated for manic depressive disease.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abrahams J. P., Leslie A. G., Lutter R., Walker J. E. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994 Aug 25;370(6491):621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- Allison J. H., Stewart M. A. Reduced brain inositol in lithium-treated rats. Nat New Biol. 1971 Oct 27;233(43):267–268. doi: 10.1038/newbio233267a0. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Asch D. K., Orejas M., Geever R. F., Case M. E. Comparative studies of the quinic acid (qa) cluster in several Neurospora species with special emphasis on the qa-x-qa-2 intergenic region. Mol Gen Genet. 1991 Dec;230(3):337–344. doi: 10.1007/BF00280289. [DOI] [PubMed] [Google Scholar]
- Baraban J. M. Toward a crystal-clear view of lithium's site of action. Proc Natl Acad Sci U S A. 1994 Jun 21;91(13):5738–5739. doi: 10.1073/pnas.91.13.5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkovic S. J., deMaine M. M. Mechanism of action of fructose 1,6-bisphosphatase. Adv Enzymol Relat Areas Mol Biol. 1982;53:45–82. doi: 10.1002/9780470122983.ch2. [DOI] [PubMed] [Google Scholar]
- Berridge M. J., Downes C. P., Hanley M. R. Neural and developmental actions of lithium: a unifying hypothesis. Cell. 1989 Nov 3;59(3):411–419. doi: 10.1016/0092-8674(89)90026-3. [DOI] [PubMed] [Google Scholar]
- Bone R., Frank L., Springer J. P., Pollack S. J., Osborne S. A., Atack J. R., Knowles M. R., McAllister G., Ragan C. I., Broughton H. B. Structural analysis of inositol monophosphatase complexes with substrates. Biochemistry. 1994 Aug 16;33(32):9460–9467. doi: 10.1021/bi00198a011. [DOI] [PubMed] [Google Scholar]
- Bone R., Springer J. P., Atack J. R. Structure of inositol monophosphatase, the putative target of lithium therapy. Proc Natl Acad Sci U S A. 1992 Nov 1;89(21):10031–10035. doi: 10.1073/pnas.89.21.10031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie J. U., Lüthy R., Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991 Jul 12;253(5016):164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- CADE J. F. J. Lithium salts in the treatment of psychotic excitement. Med J Aust. 1949 Sep 3;2(10):349–352. doi: 10.1080/j.1440-1614.1999.06241.x. [DOI] [PubMed] [Google Scholar]
- Chothia C., Lesk A. M. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986 Apr;5(4):823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayhoff M. O., Barker W. C., Hunt L. T. Establishing homologies in protein sequences. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D. F., Doolittle R. F. Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J Mol Evol. 1987;25(4):351–360. doi: 10.1007/BF02603120. [DOI] [PubMed] [Google Scholar]
- Gläser H. U., Thomas D., Gaxiola R., Montrichard F., Surdin-Kerjan Y., Serrano R. Salt tolerance and methionine biosynthesis in Saccharomyces cerevisiae involve a putative phosphatase gene. EMBO J. 1993 Aug;12(8):3105–3110. doi: 10.1002/j.1460-2075.1993.tb05979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallcher L. M., Sherman W. R. The effects of lithium ion and other agents on the activity of myo-inositol-1-phosphatase from bovine brain. J Biol Chem. 1980 Nov 25;255(22):10896–10901. [PubMed] [Google Scholar]
- Hawkins A. R., Lamb H. K., Smith M., Keyte J. W., Roberts C. F. Molecular organisation of the quinic acid utilization (QUT) gene cluster in Aspergillus nidulans. Mol Gen Genet. 1988 Oct;214(2):224–231. doi: 10.1007/BF00337715. [DOI] [PubMed] [Google Scholar]
- Hodgkinson S., Sherrington R., Gurling H., Marchbanks R., Reeders S., Mallet J., McInnis M., Petursson H., Brynjolfsson J. Molecular genetic evidence for heterogeneity in manic depression. 1987 Feb 26-Mar 4Nature. 325(6107):805–806. doi: 10.1038/325805a0. [DOI] [PubMed] [Google Scholar]
- Inhorn R. C., Majerus P. W. Properties of inositol polyphosphate 1-phosphatase. J Biol Chem. 1988 Oct 5;263(28):14559–14565. [PubMed] [Google Scholar]
- MCGILVERY R. W., MOKRASCH L. C. Purification and properties of fructose-1, 6-diphosphatase. J Biol Chem. 1956 Aug;221(2):909–917. [PubMed] [Google Scholar]
- Majerus P. W. Inositol phosphate biochemistry. Annu Rev Biochem. 1992;61:225–250. doi: 10.1146/annurev.bi.61.070192.001301. [DOI] [PubMed] [Google Scholar]
- Marcus F., Hosey M. M. Purification and properties of liver fructose 1,6-bisphosphatase from C57BL/KsJ normal and diabetic mice. J Biol Chem. 1980 Mar 25;255(6):2481–2486. [PubMed] [Google Scholar]
- Matsuhisa A., Suzuki N., Noda T., Shiba K. Inositol monophosphatase activity from the Escherichia coli suhB gene product. J Bacteriol. 1995 Jan;177(1):200–205. doi: 10.1128/jb.177.1.200-205.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murguía J. R., Bellés J. M., Serrano R. A salt-sensitive 3'(2'),5'-bisphosphate nucleotidase involved in sulfate activation. Science. 1995 Jan 13;267(5195):232–234. doi: 10.1126/science.7809627. [DOI] [PubMed] [Google Scholar]
- Nakashima K., Tuboi S. Size-dependent allosteric effects of monovalent cations on rabbit liver fructose-1,6-bisphosphatase. J Biol Chem. 1976 Jul 25;251(14):4315–4321. [PubMed] [Google Scholar]
- Neuwald A. F., Krishnan B. R., Brikun I., Kulakauskas S., Suziedelis K., Tomcsanyi T., Leyh T. S., Berg D. E. cysQ, a gene needed for cysteine synthesis in Escherichia coli K-12 only during aerobic growth. J Bacteriol. 1992 Jan;174(2):415–425. doi: 10.1128/jb.174.2.415-425.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald A. F., York J. D., Majerus P. W. Diverse proteins homologous to inositol monophosphatase. FEBS Lett. 1991 Dec 2;294(1-2):16–18. doi: 10.1016/0014-5793(91)81332-3. [DOI] [PubMed] [Google Scholar]
- Petsko G. A., Kenyon G. L., Gerlt J. A., Ringe D., Kozarich J. W. On the origin of enzymatic species. Trends Biochem Sci. 1993 Oct;18(10):372–376. doi: 10.1016/0968-0004(93)90091-z. [DOI] [PubMed] [Google Scholar]
- Pollack S. J., Knowles M. R., Atack J. R., Broughton H. B., Ragan C. I., Osborne S., McAllister G. Probing the role of metal ions in the mechanism of inositol monophosphatase by site-directed mutagenesis. Eur J Biochem. 1993 Oct 1;217(1):281–287. doi: 10.1111/j.1432-1033.1993.tb18244.x. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S. G., Jakoby W. B. (2')3',5'-Bisphosphate nucleotidase. J Biol Chem. 1987 Jul 25;262(21):10044–10047. [PubMed] [Google Scholar]
- Thomas D., Barbey R., Surdin-Kerjan Y. Gene-enzyme relationship in the sulfate assimilation pathway of Saccharomyces cerevisiae. Study of the 3'-phosphoadenylylsulfate reductase structural gene. J Biol Chem. 1990 Sep 15;265(26):15518–15524. [PubMed] [Google Scholar]
- York J. D., Majerus P. W. Isolation and heterologous expression of a cDNA encoding bovine inositol polyphosphate 1-phosphatase. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9548–9552. doi: 10.1073/pnas.87.24.9548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York J. D., Ponder J. W., Chen Z. W., Mathews F. S., Majerus P. W. Crystal structure of inositol polyphosphate 1-phosphatase at 2.3-A resolution. Biochemistry. 1994 Nov 15;33(45):13164–13171. doi: 10.1021/bi00249a002. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liang J. Y., Lipscomb W. N. Structural similarities between fructose-1,6-bisphosphatase and inositol monophosphatase. Biochem Biophys Res Commun. 1993 Feb 15;190(3):1080–1083. doi: 10.1006/bbrc.1993.1159. [DOI] [PubMed] [Google Scholar]



