Abstract
Pluripotent stem cells are a potential source of various cell types for use in regenerative medicine. Despite accumulating knowledge, there is currently no efficient and reproducible protocol that does not require genetic manipulation for generation of myogenic cells from pluripotent stem cells. Here, we examined whether mouse embryonic stem (ES) cells are able to undergo myogenic differentiation and fusion in response to signals released by differentiating myoblasts. Using ES cells expressing the histone 2B-green fluorescent fusion protein, we were able to detect hybrid myotubes formed by ES cells and differentiating myoblasts. ES cells that fused with myoblasts downregulated the expression of pluripotency markers and induced the expression of myogenic markers, while unfused ES cells did not exhibit this expression pattern. Thus, the signals released by myoblasts were not sufficient to induce myogenic differentiation of ES cells. Although ES cells synthesize many proteins involved in myoblast adhesion and fusion, we did not observe any myotubes formed exclusively by ES cells. We found that ES cells lacked M-cadherin and vascular cell adhesion molecule-1, which may account for the low frequency of hybrid myotube formation in ES cell-myoblast co-cultures and the inability of ES cells alone to form myotubes.
Introduction
Pluripotent stem cells, such as embryonic stem (ES) cells and induced pluripotent stem cells (iPS cells), have the ability to self-renew and differentiate into all cell types within the mammalian body. For this reason, they are considered a valuable resource that could be used for transplantation into damaged or malfunctioning tissues or organs. However, the development of safe, efficient, and reproducible methods of stem cell differentiation into desired cell types should be preceded by detailed analysis of the molecular mechanisms involved. In particular, in vitro generation of ES- or iPS-derived myoblasts is crucial to the development of cell-based therapies of yet unresponsive skeletal muscle diseases, such as muscular dystrophies [1]. Progression of some diseases leads to the exhaustion of satellite cells (SC), muscle stem cells that play a key role in the growth and regeneration of skeletal muscle. Transplantation of cells that could replenish SC populations could lead to restoration of muscle structure and functionality, including its ability to regenerate. Unfortunately, despite accumulating knowledge, methods of generating myogenic cells from ES or iPS cells are still imperfect [2].
In vivo (eg, chimeric animals or teratomas), both ES and iPS cells can differentiate into skeletal muscle. In vitro, myogenic differentiation of pluripotent stem cells can be induced after overexpression of crucial myogenic factors that govern embryonic myogenesis, such as Pax3, Pax7, or MyoD [3–7]. Pax3 and Pax7 play pivotal roles in the formation of muscle precursor cells, while MyoD along with other muscle regulatory factors (MRFs; Myf-5, myogenin, Mrf4) are responsible for determining myogenic fate and differentiation of myoblasts into skeletal muscle myofibers [8]. In adult organisms, Pax7 is an SC marker and MyoD is a muscle master switch, which interacts with cell cycle machinery, epigenetic modulators, and muscle-specific genes and serves as the key regulator of myoblast proliferation and differentiation [9,10]. Thus, Pax7 and MyoD are involved not only in embryonic myogenesis, but also in the regulation of the identity and functionality of adult myogenic cells [10].
Almost 20 years ago, Rohwedel and co-workers were the first to describe cells expressing muscle-specific factors, such as Myf-5, MyoD, and Myog, which were produced by genetically unmodified ES cells differentiated into three-dimensional aggregates called embryoid bodies (EB) [11]. However, in this study, the myogenic potential of such cells was not verified in vivo. Moreover, while Rohwedel and co-workers detected myogenic factors in ES cells propagated in EB, others were only able to detect them in the presence of additional factors such as spermine or 5-azacytidyne (5-azaC) [12,13]. At present, the most efficient method of directing myogenic differentiation of pluripotent stem cells relies on the overexpression of Pax3 or Pax7. Darabi et al. [3–5] showed that transplantation of myogenic cells derived from ES or iPS cells overexpressing these factors significantly improved muscle function. However, since these cells were genetically manipulated, their therapeutic use in humans is very limited. Although different in vitro approaches have been proposed to promote myogenic differentiation of ES or iPS cells, methods that would avoid genetic manipulations and rely on extracellular factors to induce conversion of pluripotent stem cells into functional myogenic cells remain undeveloped [2].
Various lines of evidence indicate that myogenic differentiation of different cell types, such as cells derived from the stromal vascular fraction of adipose tissue, AC133+ cells, or vascular cells from the retina, can be induced by culturing in the presence of myoblasts or in routine myoblast culture media [14–16]. In the current study, we examined whether co-culture of ES cells with differentiating myoblasts and/or in media supporting myoblast differentiation would result in their myogenic differentiation. Co-culture enables us to assess the ability of ES cells to fuse and initiate myogenic differentiation. Since fusion is a prerequisite for proper development, growth, and repair of skeletal muscle, cells that do not fuse, despite their expression of myogenic markers (eg, Pax3, Pax7, or MRFs), cannot be considered functional myoblasts. Studies by Bedada and Braun [17] showed that such “partially committed” myogenic cells (ie, express myogenic markers but never form regular myotubes) are generated from bone marrow-derived cells cultured in the presence of Wnt protein. Since the ability for fusion of so-called “myogenic” cells derived from pluripotent stem cells in the previous studies has not been verified, the identity and quality of such cells remains questionable [2].
The ability to fuse is determined by the presence and activity of numerous factors, including integrins, cadherins, and tetraspanins [18]. Myoblasts lacking integrin β1 [19], neural cell adhesion molecule (NCAM) [20], or vascular cell adhesion molecule 1 (VCAM-1) [21] are unable to fuse and form myotubes. In contrast, overexpression of Ncam or Itga3 (integrin α3) in myoblasts enhances their fusibility [22,23]. Other studies have shown that ES cells lacking integrin β1 exhibit accelerated neuronal, but delayed cardiac and myogenic differentiation [24,25]. On the other hand, mesenchymal precursors expressing NCAM derived from human ES cells were shown to express myogenic markers and form contracting myotubes [26]. Global profiling studies have shown that both mouse and human ES cells express a large variety of cell surface proteins with a broad range of functions [27–30]. However, the significance and exact role of most of these factors in ES cells remains unknown. Moreover, only mRNAs have been identified for some of these factors, while the presence of cognate proteins in ES cells is unknown.
In the current study, we tested the ability of ES cells to fuse with differentiating myoblasts. We also focused on molecular factors that are crucial for the adhesion and fusion of myoblasts, including M-cadherin, NCAM, VCAM-1, integrin α3, integrin β1, A disintegrin and metalloproteinase 12 (ADAM12), CD9, and CD81. We analyzed whether the expression of adhesion molecules corresponds to the ability of mouse ES cells to fuse with each other or with differentiating myoblasts. Furthermore, using ES cells expressing the histone 2B-green fluorescent protein (H2B-GFP) fusion protein, we examined the frequency of hybrid myotube formation by the fusion of ES cells with myoblasts.
Materials and Methods
Animals
Animal care and all experimental procedures were approved by the First Warsaw Local Ethics Committee for Animal Experimentation (permit number 659/2006; Poland).
Cell culture
Mouse embryonic fibroblasts (MEFs) were isolated from 13- to 14 day embryos obtained after mating of C57Bl6N mice. Before use as a feeder layer for ES cells, MEFs were passaged and inactivated with mitomycin C (0.01 mg/mL; Sigma-Aldrich). ES cells constitutively expressing histone H2B-GFP (hereafter referred to as ES-GFP) were provided by Dr. Kat Hadjantonakis (Memorial Sloan Kettering Cancer Center, New York) [31].
Inactivated MEFs were plated in dishes coated with 1% gelatin (Sigma-Aldrich) in high-glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Life Technologies), 50 U/mL penicillin (Life Technologies), and 50 μg/mL streptomycin (Life Technologies). Next, ES-GFP cells were seeded onto MEFs and cultured under various experimental conditions. Standard culture was conducted in DMEM+LIF medium, consisting of Knockout DMEM (Life Technologies) supplemented with 15% ES-qualified FBS (Life Technologies), 0.1 mM nonessential amino acids (Sigma-Aldrich), 2 mM L-glutamine (Life Technologies), 0.1 mM β-mercaptoethanol (Sigma-Aldrich), 50 U/mL penicillin (Life Technologies), 50 μg/mL streptomycin (Life Technologies), and 500 U/mL leukaemia inhibitory factor (LIF; Chemicon). Next, ES-GFP cells were cultured in media commonly used for myoblast culture: DMEM+FBS medium (high-glucose DMEM supplemented with 10% FBS and antibiotics) or DMEM+HS (high-glucose DMEM supplemented with 2% horse serum [HS; Life Technologies] and antibiotics) or DMEM+FBS+HS (low-glucose DMEM [Life Technologies] supplemented with 20% FBS, 10% HS, 0.5% chicken embryo extract (Sera Laboratories), and antibiotics).
In additional experiments, ES cells were pretreated with 5-azaC (Sigma-Aldrich) before their culture in DMEM+LIF, DMEM+FBS, DMEM+HS, or DMEM+FBS+HS. For these experiments, ES cells were incubated with 0.5, 1, 3, 10, or 30 μM 5-azaC for 24 h and then cultured in the same medium but without 5-azaC. ES cells cultured on round cover slips (13 mm diameter; Menzel Gloezer) in 24-well plates (BD Biosciences) were used for cell counting or were fixed and processed for immunostaining, while ES cells cultured in 100 mm dishes (BD Biosciences) were collected for RNA isolation and reverse transcription (RT)-polymerase chain reaction (PCR) analyses. Before RNA isolation, ES cells were separated from MEFs by preplating, in which the cell suspension was plated on dishes coated with 1% gelatin and incubated for 20 min at 37°C. Next, the medium containing unattached cells was transferred to another gelatin-coated dish; this procedure was repeated thrice. Preplating enabled MEF attachment to the dish, while ES cells remained suspended in the medium. These “purified” ES cells were harvested and frozen at −80°C until they were used for RNA isolation. In other experiments, the ES cells were propagated on Matrigel-coated (BD Biosciences) 100 mm dishes (without MEFs) in DMEM+LIF, harvested, and frozen at −80°C for RNA isolation.
In another set of experiments, “MEF-purified” ES cells were used for EB formation. Eight hundred ES cells were placed in 30 μL of Knockout DMEM medium supplemented with 15% ES-qualified FBS, 0.1 mM nonessential amino acids, 2 mM L-glutamine, 0.1 mM β-mercaptoethanol, 50 U/mL penicillin, and 50 μg/mL streptomycin. The EB were cultured using the hanging-drop system [32,33] for 2 days and then transferred to a nonadhesive bacterial Petri dish (BD Biosciences). EB cultured for 2 (EB2), 5 (EB5), or 7 days (EB7) were collected and analyzed by fluorescence-activated cell sorting (FACS) or were frozen at −80°C and subsequently used for RNA isolation and qPCR. In additional experiments, EB7 were transferred to 24-well dishes and cultured for 3 days to obtain outgrowths. EB7 outgrowths were fixed with 4% paraformaldehyde for 10 min and immunostained or collected and used for RNA isolation and qPCR analysis.
C2C12 myoblasts (European Collection of Cell Cultures, ECACC no. 91031101, passage no. 13) were cultured in DMEM+FBS for 3 days under conditions supporting their proliferation. Differentiation and fusion of C2C12 myoblasts was promoted by culturing in DMEM+HS. Primary mouse myoblasts were obtained from SC isolated from flexor digitorum brevis (FDB) muscle, according to standard protocol [34]. Briefly, FDB muscles were dissected from the hind limb of 3 month-old male C57Bl6N mice. Single muscle fibers were obtained by gentle trituration of muscle in 0.2% type I collagenase (Sigma Aldrich) in low-glucose DMEM for 90 min at 37°C. Next, SC were liberated from muscle fibers with a 22G needle and plated onto dishes coated with Matrigel diluted 1:10 with low-glucose DMEM. SC-derived myoblasts were cultured on Matrigel-coated dishes in DMEM+FBS+HS under standard conditions (at 37°C with 5% CO2).
Co-culture experiments
Before beginning co-cultures, ES-GFP cells were separated from MEFs by preplating. Next, ES cells were harvested and seeded onto differentiating C2C12 (day 4 of culture) or SC-derived myoblasts (day 6 of culture); differentiating myoblasts co-cultured with either undifferentiated C2C12 myoblasts or MEFs served as a control. In these control experiments, the undifferentiated C2C12 myoblasts and MEFs were labeled with Cell Tracker™ Orange CMRA according to the manufacturer's instructions (Life Technologies) before seeding onto a layer of differentiating myoblasts. All co-cultures were conducted on 13 mm round cover slips in 24-well plates for 3, 6, and 9 days. Co-cultures were then fixed and stained with Giemsa or May–Grünwald staining or were immunostained.
In additional experiments, the ES cells were cultured in medium conditioned by differentiating myoblasts. Here, the ES cells were seeded on the 8.0 μm inserts (BD Biosciences) placed in the six-well dishes (BD Biosciences) with differentiating myoblasts, and cultured for 3, 6, and 9 days. Then, cells were fixed and processed either for Giemsa and May–Grünwald staining or for immunostaining.
Cytological staining
Control or experimental co-cultures were fixed in cold methanol for 10 min at 4°C. Next, cells were stained with Giemsa and May–Grünwald dyes according to the manufacturer's protocol (Merck) and examined using 10× objectives on an Eclipse TE200 microscope (Nikon) to determine the cell fusion index. The fusion index was calculated as the ratio between the number of nuclei localized within myotubes and the number of all nuclei in the field of view. Images were acquired with a DXM 1200 digital camera and analyzed using NIS Elements F 2.30 software (Nikon). Ten representative fields of view for each type of the co-culture were analyzed.
Immunostaining
Cells were fixed with 4% paraformaldehyde for 10 min, permeabilized for 3 min with 0.1% Triton X-100 (Sigma Aldrich) in phosphate-buffered saline (PBS), and incubated in 0.25% glycine (Sigma Aldrich) in PBS for 30 min. The nonspecific binding of antibodies was blocked by incubation in 3% bovine serum albumin (BSA; Fluka) in PBS for 1 h. Next, cells were incubated with primary antibodies at 4°C overnight. All but anti-Pax3 and anti-Pax7 antibodies were diluted 1:50 in 3% BSA in PBS. Anti-Pax3 and anti-Pax7 antibodies were diluted 1:10 in 3% BSA in PBS. The following primary antibodies were used: Oct-4 (sc-5279), Nanog (sc-30328), Sox2 (sc-20088), MyoD (sc-706), Myf-5 (sc-302), myogenin (sc-576), Mrf4 (sc-301), CD81 (sc-7102), integrin α3 (sc-7019), integrin β1 (sc-9936), NCAM (sc-1507), and VCAM-1 (sc-8304) from Santa Cruz Biotechnology; Pax3 and Pax7 from Developmental Studies Hybridoma Bank; ADAM12 (ab39155) and M-cadherin (ab78090) from Abcam; and CD9 (C9993) from Sigma-Aldrich. After washing, samples were incubated for 1 h at room temperature with appropriate AlexaFluor 594 secondary antibodies (donkey anti-mouse IgG A21203, goat anti-rabbit IgG A11037, and rabbit anti-goat IgG A11080; Molecular Probes, Life Technologies) diluted 1:200 in 3% BSA in PBS. Nuclei were visualized with DraQ5 (Biostatus Limited) diluted 1:1,000 in PBS for 5 min and then mounted with Fluorescent Mounting Medium (Dako). The specimens were analyzed using an Axiovert 100 M scanning confocal microscope (Zeiss) that was equipped with LSM 510 software. Primary antibody specificity was confirmed by incubation of cells with secondary antibodies only.
FACS analysis
Cells obtained after dissociation of EB2, EB5, and EB7 outgrowths with Cell Dissociation Buffer Enzyme-Free Hanks' Balanced Salt Solution (Life Technologies), as well as undifferentiated ES cells and differentiating C2C12 myoblasts cultured for 4 days, were incubated in staining medium (PBS with 1% FBS and 0.1% BSA) for 30 min and stained with a 1:50 dilution of anti-VCAM-1 (sc-8304; Santa Cruz Biotechnology) for 30 min. Next, cells were washed with staining medium, incubated with a 1:100 dilution of goat anti-rabbit IgG AlexaFluor 488 secondary antibody (A11034; Molecular Probes, Life Technologies) for 30 min, washed, fixed with 0.1% paraformaldehyde for 10 min, washed, and suspended in staining medium. In control experiments, incubation with primary antibody was omitted. Cells were analyzed with an FACSCalibur instrument (Becton Dickinson) and CellQuest Pro software (BD Biosciences).
RNA isolation and RT-PCR
Total RNA was isolated from ES-GFP, MEFs, proliferating or differentiating C2C12, and SC-derived myoblasts using a High Pure RNA Isolation Kit (Roche). RT-PCR was performed using 0.5 μg of total RNA, appropriate sets of primers, and the Titan One Tube RT-PCR Kit (Roche) according to the manufacturer's instructions. Three independent experiments were performed using two different sets of primers (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/scd). RT-PCR conditions were as follows: reverse transcription at 50°C for 30 min, 94°C initial denaturation for 2 min, and 35 cycles of 94°C denaturation for 30 s, annealing at primer-specific temperatures for 30 s, 68°C elongation for 45 s, and final elongation at 68°C for 7 min. RT-PCR products were separated in 1.5% agarose gel (Roche) and visualized with ethidium bromide (Sigma Aldrich). Agarose gels were analyzed with a GelDoc 2000 using Quantity One software (BioRad). All RT-PCR products were isolated with the Agarose Gel DNA Extraction Kit (Roche), sequenced by an outsourced company (Genomed), and verified using NCBI databases.
Quantitative polymerase chain reaction
Total RNA was isolated (High Pure RNA Isolation Kit, Roche) from undifferentiated ES cells, EB2, EB5, EB7, EB7 outgrowths, differentiating C2C12 myoblasts, and 13 day-old embryos obtained after mating of C57Bl6N mice. Reverse transcription was performed using 0.5 μg total RNA and a RevertAid First Strand cDNA kit (Thermo Scientific) according to the manufacturer's instructions. qPCR was performed using specific TaqMan® probes for VCAM-1 (Mm01320970), M-cadherin (Mm00483191), and β-actin (Mm01205647_g1; Life Technologies), the TaqMan Gene Expression Master Mix (Life Technologies), and a LightCycler 96 instrument (Roche). Data were collected and analyzed with LightCycler 96 SW1.1 software (Roche); three independent experiments were performed.
Statistical analysis
All experiments were performed at least thrice. Data are shown as mean±standard deviation (SD), and ANOVAs were used for statistical analysis; sP<0.05 was considered statistically significant.
Results
Media supporting myoblast differentiation have limited influence on in vitro cultured ES cells
In initial experiments, we characterized all cells used in this study: ES-GFP cells, inactivated feeder layer MEFs, and C2C12 and SC-derived myoblasts. Expression of pluripotency markers, such as Oct-4, Nanog, and Sox2, as well as myogenic markers Pax3, Pax7, MyoD, Myf-5, myogenin, and Mrf4, was determined at the level of both mRNA and protein. As the expression of myogenic markers changed during myoblast differentiation, we analyzed both proliferating (culture day 2 C2C12, culture day 4 SC-derived) and differentiating/fusing (culture day 4 C2C12, culture day 6 SC-derived) myoblasts.
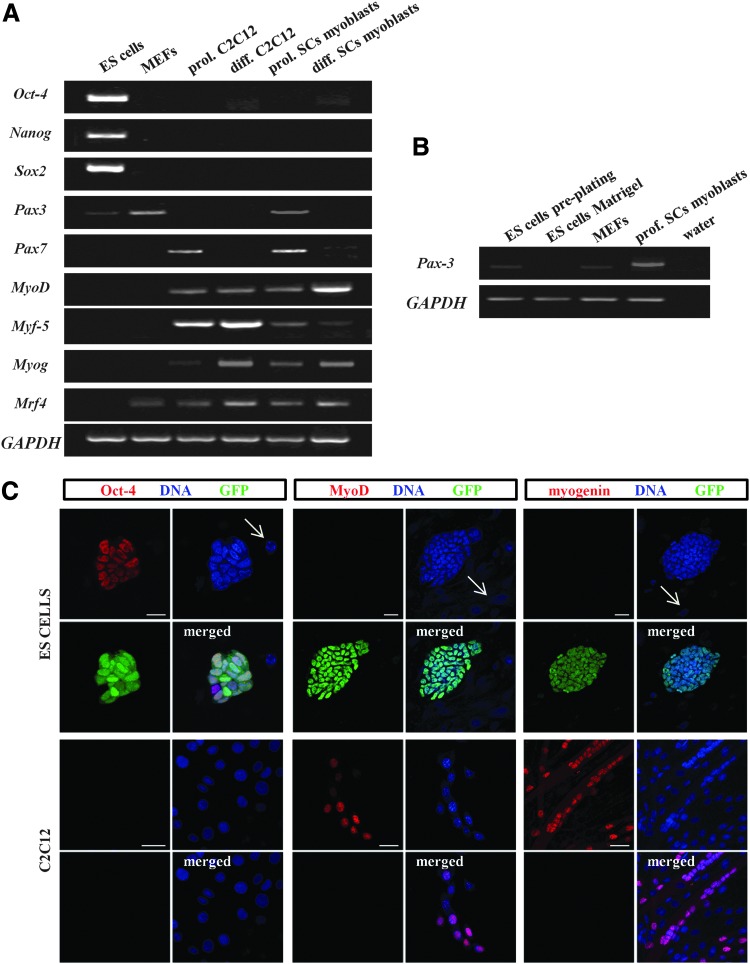
As expected, all pluripotency markers were easily detected in ES-GFP cells and were absent in MEFs, C2C12, and SC-derived myoblasts (Fig. 1A; Oct-4 protein shown in Fig. 1C). Pax7 and all analyzed MRFs (Myf-5, MyoD, myogenin, and Mrf4) were absent in ES-GFP cells but, as expected, detected in both types of myoblasts at mRNA and protein levels (Fig. 1A; MyoD and myogenin shown in Fig. 1C). Interestingly, weak expression of Pax3 was detectable in ES-GFP cells by RT-PCR, but Pax3 protein was not found in these cells (data not shown). Expression of Pax3 and Mrf4 was also observed in inactivated MEFs, but again only at the transcript level (Fig. 1A). We estimated that the average contribution of MEFs to all cells counted after preplating was 3.8%±0.6% (data acquired from 12 independent experiments). Additional RT-PCR experiments showed that ES cells propagated on Matrigel without MEFs did not express Pax3 (Fig. 1B). The identity of all RT-PCR products was confirmed.
FIG. 1.
Expression of pluripotency and myogenic markers in embryonic stem (ES)-green fluorescent protein (GFP) cells, mouse embryonic fibroblasts (MEFs), and myoblasts. (A) Reverse transcription (RT)-polymerase chain reaction (PCR) analysis of transcripts encoding pluripotency markers (Oct-4, Nanog, and Sox2) and myogenic markers (Pax3, Pax7, MyoD, Myf-5, Myog, and Mrf4) in ES-GFP cells, MEFs, C2C12, and satellite cell (SC)-derived myoblasts. GAPDH was used as a reference gene. Representative images of gels are shown. (B) RT-PCR analysis of the Pax3 transcript in ES-GFP cells collected after preplating, ES-GFP cells propagated on Matrigel without MEFs, MEFs alone, and SC-derived myoblasts. GAPDH was used as a reference gene. (C) Immunofluorescent detection of Oct-4, MyoD, and myogenin in ES-GFP cells and C2C12 myoblasts. Staining for Oct-4 and MyoD in proliferating C2C12 cells (day 2 of culture) and myogenin staining in differentiating/fusing C2C12 cells (day 4 of culture) are shown. Arrows indicate nuclei of MEFs, distinguishable from nuclei of ES-GFP cells by the absence of GFP. Scale bar, 20 μm. Color images available online at www.liebertpub.com/scd
Next, we investigated whether the expression of myogenic factors can be induced in ES-GFP cells by culturing them in media supporting myoblast proliferation and differentiation. In our study, we compared how ES-GFP cells maintained under standard conditions (ie, DMEM+LIF) differ from those cultured in three different types of media routinely used for myoblast culture (ie, DMEM+FBS, DMEM+HS, and DMEM+FBS+HS) [32]. In each experiment, ES-GFP cells were seeded onto inactivated MEFs and analyzed after 3, 6, 9, and 12 days of culture.
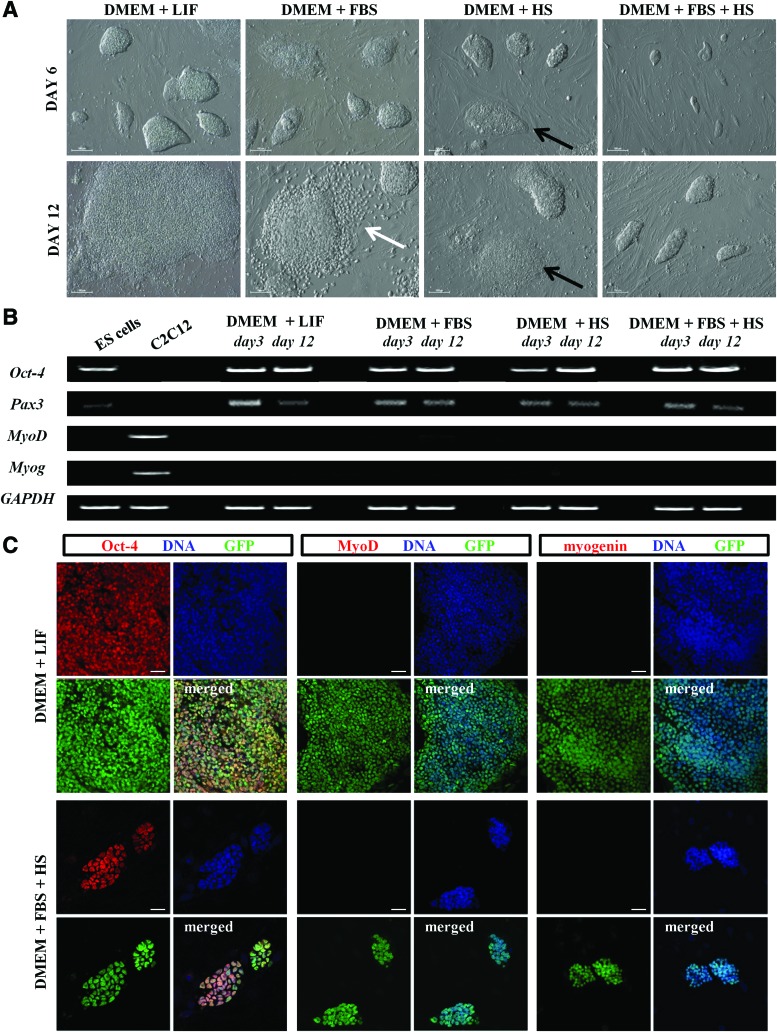
The effect of different media on the morphology of ES-GFP cell colonies was noticeable as early as culture day 3 and became more evident after 6, 9, and 12 days of culture (Fig. 2A). Control ES cells cultured in DMEM+LIF formed the biggest colonies of typical morphology, while those cultured in DMEM+FBS+HS formed the smallest (Fig. 2A). At first, flattened colonies and dispersed cells were observed after culture day 6 in all types of media routinely used for myoblasts; however, the most significant changes were noticeable in DMEM+FBS (Fig. 2A). Apart from morphology, we also compared the number of ES-GFP cells under the four indicated culture conditions (Supplementary Fig. S1A). After 12 days, the number of ES-GFP cells cultured with DMEM+LIF was twice to thrice higher than the number of cells in DMEM+FBS or DMEM+HS (Supplementary Fig. S1A; 389±12.1×103 versus 121±9.8×103 and 167±7.3×103, respectively), and as much as seven times higher compared with DMEM+FBS+HS cultures (Supplementary Fig. S1A; 389±12.1×103 versus 63±2.9×103). Thus, the different media significantly decreased the proliferation rate of mouse ES cells. This suggests that these media either inhibited cell proliferation or induced differentiation. This was assessed by analysis of the expression of pluripotency markers, such as Oct-4 and Nanog, as well as factors characteristic for myoblasts, such as Pax3, Pax7, and MRFs.
FIG. 2.
Comparison of ES-GFP cells under different culture conditions. (A) Morphology of ES-GFP cell colonies cultured in standard medium (DMEM+LIF) or in media supporting proliferation and differentiation of myoblasts (DMEM+FBS, DMEM+HS, and DMEM+FBS+HS) after 6 and 12 days of culture. White arrow indicates dispersed cells; black arrow indicates flattened colonies. Scale bar, 100 μm. (B) RT-PCR analysis of transcripts encoding pluripotency (Oct-4) and myogenic markers (Pax3, MyoD, and Myog) in ES-GFP cells cultured in different media for 3 or 12 days. Undifferentiated ES-GFP cells and C2C12 myoblasts were used as a control; GAPDH was used as a reference gene. Representative images of gels are shown. (C) Immunofluorescent detection of Oct-4, MyoD, and myogenin in ES-GFP cells cultured in DMEM+LIF or DMEM+FBS+HS for 12 days. Scale bar, 20 μm. Color images available online at www.liebertpub.com/scd
After culture day 12, Oct-4 was easily detectable in ES-GFP cells at both the mRNA and protein level, regardless of culture conditions (Fig. 2B, C). Pax3 was the only myogenic marker detected in ES-GFP cells (Pax3, MyoD, and myogenin shown in Fig. 2B). None of the myogenic markers were detected at the protein level in ES-GFP cells cultured in different conditions (Fig. 2C). Even though the media had a noticeable effect on ES cell proliferation, they did not induce myogenic differentiation. The disappearance of Oct-4 and the presence of MyoD was found only when ES-GFP cells were pretreated with 3 or 10 μM 5-azaC before their culture in DMEM+HS or DMEM+FBS+HS (Supplementary Fig. S1B). Although changes in the morphology of such cells were evident (cells became bipolar and elongated; Supplementary Fig. S1C), we did not observe myotubes. Colonies of cells treated with 5-azaC and cultured in DMEM+LIF or DMEM+FBS were smaller than control colonies, and some of these cells did not express Oct-4; however, any myogenic markers were detected (Supplementary Fig. S1B, C). The addition of ≤1 μM 5-azaC had negligible effects on ES cell morphology and gene expression, but resulted in decreased cell viability when used at concentrations >10 μM (Supplementary Fig. S1D).
Co-culture of ES cells with differentiating myoblasts results in formation of hybrid myotubes
The major goal of our study was to determine whether undifferentiated mouse ES cells co-cultured with differentiating myoblasts are able to undergo myogenic differentiation. We used two types of myoblasts that are commonly used in in vitro myogenic differentiation studies, C2C12 and primary SC-derived myoblasts. Since these two types of myoblasts differ in their timing of proliferation and differentiation [35], the signals they release in vitro may also differ. For this reason, we decided to co-culture ES-GFP cells with either C2C12 or SC-derived myoblasts in parallel.
After seeding and proliferation, C2C21 and SC-derived myoblasts start to differentiate and fuse, eventually forming multinucleated myotubes (apparent at days 6 and 8 of culture, respectively). Based on our preliminary results (data not shown), we chose to start ES-GFP cell-myoblast co-cultures just after the onset of myoblast differentiation and fusion (days 4 and 6 of myoblast culture, respectively). Co-cultures involving C2C12 cells were conducted using DMEM+FBS or DMEM+HS. ES-GFP and SC-derived myoblasts were cultured in DMEM+FBS+HS. Both types of co-culture were analyzed after 3, 6, and 9 days.
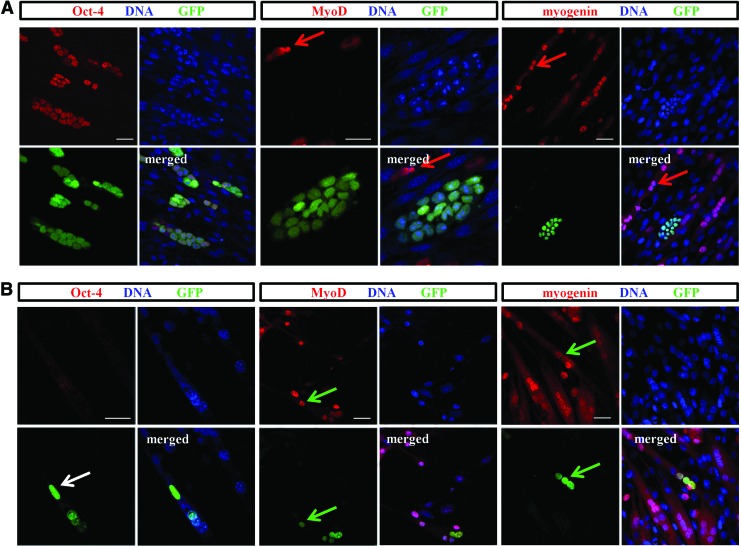
The presence of ES cells did not impact either type of myoblast differentiation. Myoblasts co-cultured with ES-GFP cells fused and formed multinucleated myotubes similar to control cultures without ES cells (Supplementary Fig. S2). In both co-cultures types, ES cell colonies gradually expanded but were easily distinguishable and visible among growing myotubes (Supplementary Fig. S2). Interestingly, ES-GFP cells expressed pluripotency markers even after 9 days of co-culture, as shown by immunolocalization of Oct-4 (Fig. 3A); the same was true for Nanog (data not shown). In contrast, myogenic markers Pax7, MyoD, and/or myogenin were present in myoblasts and myotubes (MyoD and myogenin shown in Fig. 3A) but absent in ES-GFP cell colonies. Importantly, we detected myotubes containing GFP-expressing nuclei, proving that ES-GFP participated in the formation of myotubes with both C2C12 and SC-derived myoblasts. We did not observe any significant differences between ES-GFP cell-C2C12 and ES-GFP cell-SC-derived myoblast co-cultures. Thus, only results obtained from ES cell-C2C12 myoblasts are shown (Fig. 3).
FIG. 3.
Co-culture of ES-GFP cells and C2C12 myoblasts. (A) Immunofluorescent detection of Oct-4, MyoD, and myogenin in ES-GFP cells and C2C12 myoblasts co-cultured for 9 days. Colonies of ES cells are visible between myoblasts and myotubes. Red arrows indicate the presence of myogenic factors in the nuclei of myoblasts and myotubes. Scale bar, 20 μm. (B) Immunolocalization of Oct-4, MyoD, and myogenin in hybrid myotubes formed by ES-GFP cells and C2C12 myoblasts co-cultured for 6 days. Arrows indicate nuclei derived from ES-GFP cells and present in the hybrid myotube; white arrow indicates nuclei devoid of Oct-4 (pluripotency marker); green arrows indicate nuclei containing myogenic markers. Scale bar, 20 μm. Color images available online at www.liebertpub.com/scd
None of the myotubes contained exclusively GFP-positive nuclei, suggesting that none of them originated solely from differentiating ES cells fusing with each other. Furthermore, under such conditions, ES cells were not able to differentiate into myotubes alone, but could only form hybrid myotubes by fusing with myoblasts. Hybrid myotubes were detected as early as after 3 days of co-culture regardless of the type of myoblast used. To exclude the possibility that ES cells remain adjacent to myotubes, each hybrid myotube was carefully examined to confirm that GFP nuclei were, indeed, incorporated into the myotube rather than being localized in close proximity. Importantly, the GFP-positive nuclei found in hybrid myotubes did not contain Oct-4, but were positive for myogenic markers MyoD and myogenin (Fig. 3B). However, Pax3 and Pax7 were not detected in such nuclei (data not shown). Our results support the idea that ES cells are not able to undergo myogenic differentiation and convert into myoblasts and myotubes in response to factors released by differentiating myoblasts, but are able to participate in the formation of hybrid myotubes.
To verify whether direct contact between ES cells and myoblasts is crucial for hybrid myotube formation, we cultured ES-GFP cells in medium conditioned by differentiating myoblasts, using inserts with pores to enable cells to migrate. Under these conditions, we did not observe any hybrid myotubes formed by ES-GFP cells and myoblasts (data not shown). This indicates that signals released by differentiating myoblasts and myotubes are not sufficient to trigger hybrid myotube formation.
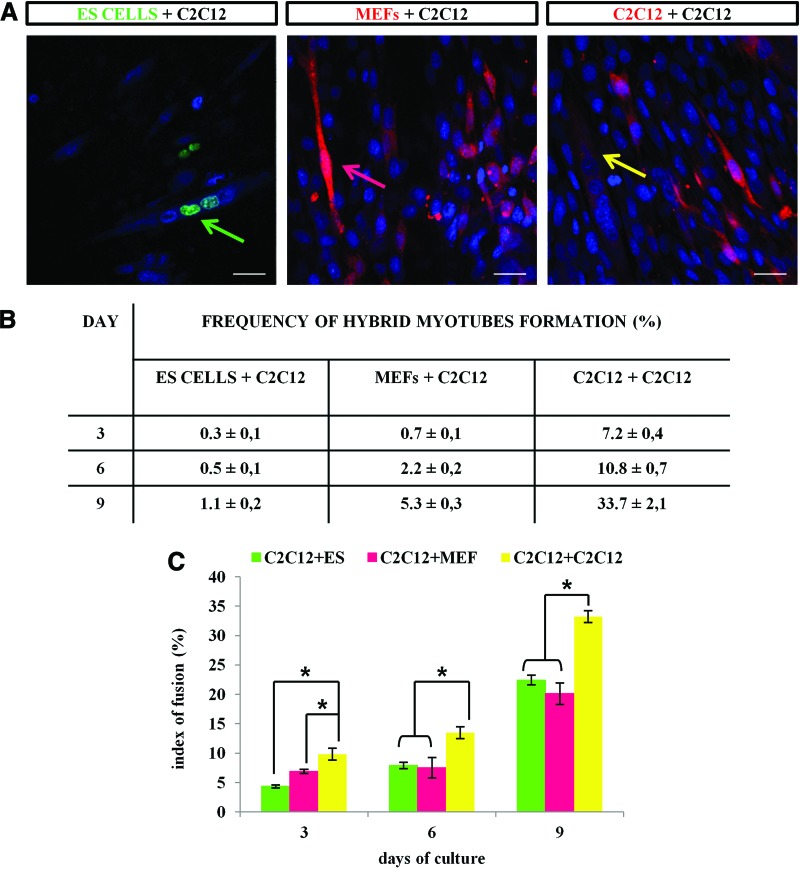
Next, we quantified the number of hybrid myotubes after 3, 6, and 9 days of co-culture. In control experiments, either MEFs or undifferentiated C2C12 myoblasts labeled with Cell Tracker Orange CMRA were seeded on the layer of unlabeled differentiating C2C12 or SC-derived myoblasts and cultured for 3, 6, or 9 days. At co-culture day 9, we detected 33.7% hybrid myotubes formed between labeled and unlabeled C2C12 myoblasts (Fig. 4A). Simultaneously, the co-culture of labeled MEFs and unlabeled C2C12 myoblasts contained 5.3% hybrid myotubes. Only 1.1% hybrid myotubes originated from ES-GFP cells and C2C12 myoblasts (Fig. 4A). Similar results were obtained for the co-culture of ES-GFP, MEFs, or C2C12 myoblasts with SC-derived myoblasts (data not shown). Next, we determined the index of fusion, which serves as an indicator of a cell's fusibility. We found that at all analyzed time points the fusion index was almost twice lower in the C2C12-MEF or C2C12–ES cell co-culture compared with the myoblast–myoblast co-culture (Fig. 4B). These results suggest that the fusion competence of ES cells is dramatically lower, in comparison to that of not only the myoblasts, but also the MEFs, which do not spontaneously fuse.
FIG. 4.
Hybrid myotubes in ES-GFP cell-C2C12 myoblast co-culture. (A) Formation of hybrid myotubes in three types of co-cultures. Arrows indicate hybrid myotubes formed in each type of the co-culture after 6 days of differentiation: green, ES-GFP cells and C2C12 myoblasts; pink, MEFs and C2C12 myoblasts; yellow, C2C12 and C2C12 myoblasts. Red corresponds to CMRA dye used for labeling MEFs and C212 cells in control co-cultures. Scale bar, 20 μm. (B) The table presents the frequency of hybrid myotube formation in all co-cultures types after 3, 6, and 9 days. Data from three independent experiments are shown as mean±SD. (C) Comparison of the cell fusion index in three types of co-culture: ES-GFP cells+C2C12; MEFs+C2C12; and C2C12+C2C12. The index of fusion was calculated after 3, 6, and 9 days of co-culture; *P<0.05. Color images available online at www.liebertpub.com/scd
ES cells express adhesion and fusion factors with the exception of M-cadherin and VCAM-1
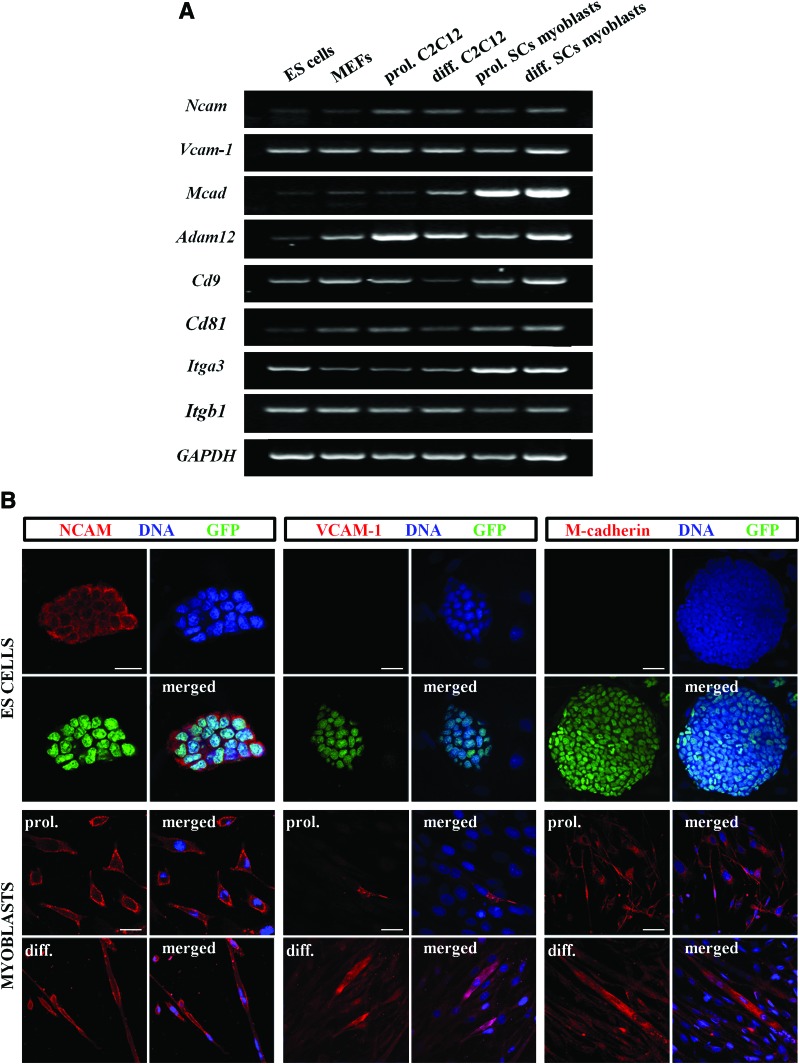
Since the ability of cells to fuse depends on the presence of numerous surface proteins, we speculated that limited competence of ES cells to fuse with myoblasts might result from insufficient expression or lack of these factors [19–23, 36–38]. Therefore, we investigated whether undifferentiated ES-GFP cells express factors crucial for proper adhesion and fusion of myoblasts, including integrin α3, integrin β1, CD9, CD81, VCAM-1, NCAM, M-cadherin, and ADAM12. We studied the presence of these factors in ES-GFP, MEF, C2C12, and SC-derived myoblasts at both the mRNA and protein level. Since the expression of adhesion and fusion factors may potentially change during myoblast differentiation, we analyzed both proliferating and fusing myoblasts.
We detected transcripts of Itga3 (integrin α3), Itgb1 (integrin β1), Cd9, Cd81, Vcam-1, Ncam, Mcad (M-cadherin), and Adam12 in ES-GFP cells, MEFs, C2C12, and SC-derived myoblasts (Fig. 5A). We also found proteins encoded by these transcripts in the membrane of myoblasts and myotubes (Fig. 5B and Supplementary Figs. S3 and S4). Most of the analyzed proteins were also detected in ES-GFP cells with the exception of M-cadherin and VCAM-1 (Fig. 5B and Supplementary Figs. S3 and S4). In ES cell colonies, the distribution of these proteins was uniform; we did not observe significant differences between cells localized at the periphery or in the middle of colonies (Fig. 5B and Supplementary Figs. S3 and S4). All these results suggest that the limited fusibility of ES cells may result from the lack of M-cadherin and VCAM-1.
FIG. 5.
Expression of adhesion and fusion factors in ES-GFP cells, MEFs, and myoblasts. (A) RT-PCR analysis of adhesion and fusion factor transcripts (Ncam, Vcam-1, Mcad, Adam12, Cd9, Cd81, Itga3, and Itgb1) in ES-GFP cells, MEFs, C2C12, and SC-derived myoblasts. GAPDH was used as a reference gene. Representative images of gels are shown. (B) Immunolocalization of NCAM, VCAM-1, and M-cadherin in ES-GFP cells and myoblasts. “prol.,” marks images showing localization of analyzed proteins in proliferating C2C12 or SC-derived myoblasts; “diff.,” indicates images showing presence of proteins in differentiating myoblasts. Scale bar, 20 μm. Color images available online at www.liebertpub.com/scd
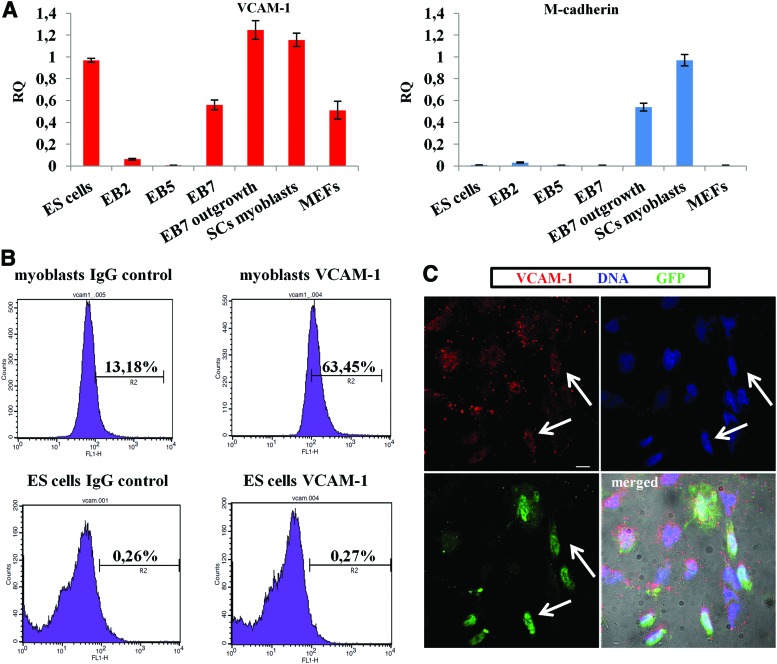
In additional experiments, we investigated whether expression of VCAM-1 and M-cadherin changes in ES cells differentiated in EB. qPCR showed that VCAM-1 expression decreases in EBs cultured for 2 (EB2) or 5 (EB5) days and then increases in EBs cultured for 7 days (EB7) as well as in the cell outgrowths obtained from EB7 (Fig. 6A). FACS analysis showed that cells obtained by dissociation of EB2, EB5, and EB7 do not synthesize VCAM-1 protein, similar to undifferentiated ES cells (Fig. 6B). However, single VCAM-1-positive cells were found in EB7 outgrowths (Fig. 6C). In addition, qPCR showed that the expression of M-cadherin increases in outgrowths derived from EB7 (Fig. 6A).
FIG. 6.
Expression of VCAM-1 and M-cadherin in undifferentiated and differentiating ES cells. (A) The level of VCAM-1 and M-cadherin transcripts in undifferentiated ES cells, ES cells differentiated in EB for 2 (EB2), 5 (EB5), or 7 (EB7) days, and outgrowths derived from EB7. RNA isolated from SC-derived myoblasts, MEFs, and 13 day embryos (used as a control sample) was also analyzed by qPCR. B-actin was used as a reference gene. Transcript levels in experimental samples were compared with the transcript level (100%) in the control (embryo) sample. (B) VCAM-1 analysis by FACS. ES cells, cells derived from EB2, EB5, EB7, and differentiating myoblasts were stained with anti-VCAM-1antibody and analyzed by flow cytometry. In control experiments, anti-VCAM-1 antibody was omitted. Data for cells derived from EB2, EB5, and EB7 were similar to results obtained for undifferentiated ES cells (lower row). (C) Immunolocalization of VCAM-1 in outgrowths derived from EB7. Arrows indicate cells that synthesize VCAM-1. Scale bar, 20 μm. Color images available online at www.liebertpub.com/scd
Discussion
Although ES cells possess the ability to undergo multidirectional differentiation, directed conversion of such cells into desired cell types is not always effective [39,40]. In the current study, we examined whether mouse ES cells were able to undergo myogenic differentiation and fusion in co-culture with differentiating myoblasts. In addition, we compared the behavior of ES cells cultured in standard conditions (ie, in the presence of LIF) and in media designed to support myoblast propagation. We were prompted to perform such experiments by the fact that different types of cells (eg, cells derived from the stromal vascular fraction of adipose tissue [14] or AC133+ cells [16]) were shown to undergo myogenic differentiation when cultured in media routinely used for myoblast propagation or/and when co-cultured with myoblasts. The results of our experiments clearly indicated that factors present in the media used for myoblast culture or released by differentiating myoblasts are not sufficient to induce myogenic conversion of ES cells. Despite noticeable changes in the morphology of ES cell colonies even after 2 weeks of culture, ES-GFP cells maintained expression of pluripotency markers and never expressed myogenic markers (with the exception of Pax3 detected at the mRNA level). Similarly, Mizuno et al. [41] showed the presence of Pax3 transcripts in undifferentiated mouse ES cells of the D3 cell line, while Tian et al. [42] detected a few Pax3-positive 129/Sv ES cells. It should be pointed out, however, that in many experiments, including the present, pluripotent stem cells were expanded using feeder layers of inactivated MEFs, cells known to express low levels of Pax3. Although we determined that preplating removes >95% of MEF cells, we believe that the Pax3 mRNA detected in ES cell samples resulted from MEF mRNA contamination. This is supported by the fact that the Pax3 transcript was never detected in ES cells propagated without MEFs.
Discrepancies in the ability to undergo myogenic differentiation observed between various stem cells, such as ES cells and cells derived from the stromal vascular fraction of adipose tissue, may be explained by the fact that myogenic and adipose cells have a common embryonic origin: both of them are derived from mesodermal progenitors. Competence of AC133+ cells to undergo myogenic differentiation may be explained by the fact that these cells already express some myogenic factors, including Pax7 and M-cadherin [16]. This suggests that only cells which share developmental origin and/or other myogenic cell features (eg, utilize the same signaling pathways or express similar factors) are competent to undergo myogenic differentiation in response to factors released by myoblasts or components present in the media. This explanation is also supported by results of studies on myogenic differentiation of mesodermal/myogenic precursors obtained from ES cells. For example, Darabi et al. showed that cells selected from differentiating ES cells on the basis of PDGFαR expression (paraxial mesoderm marker serving as a source of skeletal muscle during embryo development) are able to undergo myogenic differentiation and form myotubes in vitro [3,5]. Similarly, mesoderm/myogenic precursors selected from differentiating ES cells on the basis of SM/C-2.6 expression synthesize Pax7 and M-cadherin and form myotubes in vitro [43]. In both studies, ES cells were differentiated within EBs (ie, in three-dimensional aggregates, in which cells spontaneously form ecto-, endo-, and mesoderm) [44]. However, cellular differentiation within EBs is often asynchronous, and the formation efficiency of the mesoderm and its derivatives is rather low [2]. Thus, various supplements such as ITS (insulin, transferrin, and selenium), dexamethasone, bone morphogenetic protein 4, epidermal growth factor, or 5-azaC have been used to promote mesodermal/myogenic formation [2].
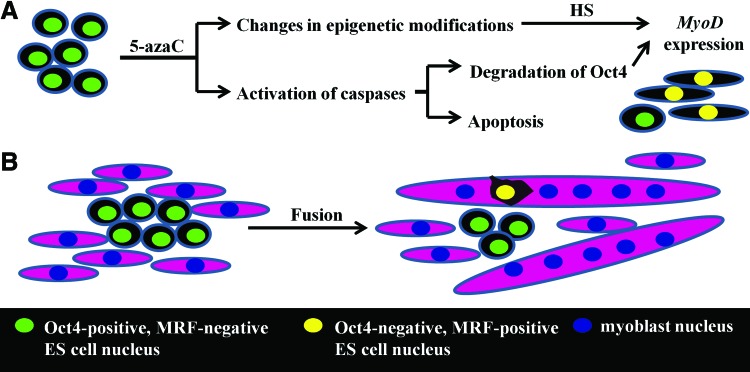
However, despite numerous attempts, direct in vitro differentiation of ES cells into mesodermal/myogenic precursors remains inefficient. Several studies showed that although cells started expressing myogenic genes in response to supplemental treatments, they were unable to form myotubes and convert into functional myoblasts [2]. Difficulties in efficient derivation of functional myogenic cells from ES cells emphasize the importance of the sequential steps involved in this process. Our current study showed that pretreatment of ES cells with 5-azaC before their culture in DMEM+HS or DMEM+FBS+HS resulted in the inhibition of Oct-4 and the induction of MyoD synthesis. 5-AzaC and 5-aza–2′-deoxycytidine are known epigenetic modifiers that have been used to induce myogenic conversion since the 1970s [45,46]. In our study, the ES cells treated with 5-azaC and cultured in the presence of HS resembled myoblasts, but they were not able to form myotubes. The disappearance of Oct-4 in ES cells treated with 5-azaC likely resulted from its degradation by caspases activated in response to 5-azaC [47]. Since Oct-4 prevents expression of MyoD by directly binding its gene, degradation of the Oct-4 protein may facilitate transcription of MyoD (Fig. 7) [48]. Expression of MyoD in 5-azaC-treated cells may be further promoted by epigenetic changes, such as inhibition of DNA methylation and histone deacetylation, caused by this reagent [49]. However, since in our study MyoD was detected only in 5-azaC-treated ES cells cultured in DMEM+HS or DMEM+FBS+HS, it is possible that MyoD expression was triggered by unidentified components present in HS (Fig. 7).
FIG. 7.
Changes occurring during the culture of ES cells in tested conditions. (A) Treatment of ES cells with 5-azaC leads to activation of caspases, which trigger Oct-4 degradation and dose-dependent apoptosis. Due to epigenetic modification, the cells become “sensitive” to environmental cues, such as factors present in horse serum. As a result, MyoD expression is induced in some ES cells. (B) Incorporation of ES cell nucleus into myotubes leads to disappearance of pluripotent markers and induction of myogenic markers. ES cells that are not recruited to myotubes retain pluripotent markers and do not express myogenic markers. Color images available online at www.liebertpub.com/scd
We also found myogenic markers in the nuclei of ES-GFP cells in hybrid myotubes. Although it is possible that myogenic marker mRNA or proteins could be delivered to myotubes by microvesicles, considering the fact that GFP and myogenic markers were present exclusively in the nuclei, this is very unlikely [50]. The presence of hybrid myotubes formed by ES-GFP cells and myoblasts contradicts previous studies showing that undifferentiated human ES cells co-cultured with C2C12 cells are unable to form myotubes. Barberi et al. [51] showed that only those ES cells which were cultured for 40 days under differentiation priming conditions and then selected on the basis of CD73 expression (paraxial mesoderm marker) were able to fuse with C2C12 cells and form hybrid myotubes [51]. This discrepancy may be the result of different ES cell origins (mouse versus human) and/or different culture conditions in these two studies. Since we detected the first hybrid myotubes after 3 days of co-culture, it seems unlikely that ES cells differentiated into fusion-competent cells, such as macrophages.
In the current study, we demonstrated that undifferentiated ES cells synthesize numerous surface proteins that are critical for proper adhesion and fusion of myoblasts, but lack VCAM-1 and M-cadherin. The latter observation agrees with the results of Rose and co-workers, who demonstrated that M-cadherin is detectable only in ES cells differentiating in EB [52]. However, since myoblasts derived from mice lacking M-cadherin are able to fuse [53], it seems that the limited fusibility of ES cells is not caused by the lack of M-cadherin but by the lack of VCAM-1. It has been shown that an inhibition of VCAM-1 protein in myoblasts completely hinders their fusion during in vitro culture [21]. Recently, VCAM-1 was shown to be robustly expressed in human ES and iPS cells differentiating into cardiomyocytes from culture day 7 [54]. Tian et al. [55] showed that VCAM-1 was present on the surface of undifferentiated mouse ES cells but disappeared rapidly after the removal of LIF from the medium [55]. In contrast, Heyward and co-workers showed that VCAM-1 expression was sustained during ES cell differentiation [56]. Our studies showed that undifferentiated ES-GFP cells and cells propagated in EB until day 7 of culture lacked the VCAM-1 protein. The first single cells synthesizing VCAM-1 were detected in outgrowths derived from EB7 and cultured for another 3 days; these cells also had elevated levels of Vcam-1 mRNA. However, it should be noted that Vcam-1 mRNA was also present in undifferentiated mouse ES-GFP cells. These discrepancies indicate that the significance of VCAM-1 in determining the fate of ES cells, including their myogenic differentiation, requires further study.
Data presented in the current study clearly demonstrate that ES cell nuclei start expression of myogenic factors, such as MyoD and myogenin, only after incorporation into myotubes (Fig. 7). Similar myogenic conversion preceded by fusion has been shown for mesenchymal stem cells [57] and vascular cells from retina [15] fusing with myotubes or myofibers. Pioneering experiments by Johnson and Rao demonstrated that fusion of two different cell types may lead to changes in their chromatin organization, epigenetic modifications, and, as a result, gene expression [58]. It has also been shown that the phenotype of the less-differentiated fusion partner may dominate over the phenotype of the more-differentiated partner [59]. However, since the hybrid myotubes analyzed in this study were formed predominantly by the myoblast component, the activity of factors present in myoblasts and myotubes probably prevailed over those introduced by ES cells. As a consequence, the nuclei of hybrid myotubes did not express pluripotency markers.
The current study provides new and important information about reciprocal interactions between pluripotent stem and myogenic cells. We report for the first time that undifferentiated mouse ES cells may participate in the formation of hybrid myotubes, although their fusibility is limited. Since myoblasts fuse very efficiently, a low frequency of hybrid myotube formation may result from competition between myoblasts and ES cells. Direct contact between both types of cells was crucial for participation of ES cells in myotube formation, as they were unable to form myotubes independently. We also found that undifferentiated ES cells express a variety of surface proteins, but they lack M-cadherin and VCAM-1, the factors involved in myoblast fusion. Since differentiation of pluripotent stem cells is affected by both cell–cell and cell–extracellular signal interactions, deciphering the role of those and other membrane proteins in the differentiation of ES cells into myogenic cells remains of great importance for the development of future therapeutic approaches.
Supplementary Material
Acknowledgments
The authors are very grateful to Dr. Kat Hadjantonakis (Memorial Sloan Kettering Cancer Center, New York, United States) for providing ES-GFP cells. This study was supported by a NN303308133 grant from the Ministry of Science and Higher Education of Poland and a grant funded by the Foundation for Polish Science (Parent Bridge Programme, grant number: POMOST/2012-5/1). K.A. was supported by a scholarship for young PhDs founded by the European Union within European Social Fund (POKL.04.03.00-00-060/12). The authors are grateful to Professor Małgorzata Kloc (the Houston Methodist Hospital Research Institute, Houston, United States) for her very careful reading of this article and helpful comments.
Author Disclosure Statement
The authors declare no conflict of interest. No competing financial interests exist.
References
- 1.Goyenvalle A, Seto JT, Davies KE. and Chamberlain J J. (2011). Therapeutic approaches to muscular dystrophy. Hum Mol Genet 20:69–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grabowska I, Archacka K, Czerwinska AM, Krupa M. and Ciemerych MA. (2012). Mouse and human pluripotent stem cells and the means of their myogenic differentiation. Results Probl Cell Differ 55:321–356 [DOI] [PubMed] [Google Scholar]
- 3.Darabi R, Gehlbach K, Bachoo RM, Kamath S, Osawa M, Kamm KE, Kyba M. and Perlingeiro RC. (2008). Functional skeletal muscle regeneration from differentiating embryonic stem cells. Nat Med 14:134–143 [DOI] [PubMed] [Google Scholar]
- 4.Darabi R, Pan W, Bosnakovski D, Baik J, Kyba M. and Perlingeiro RC. (2011). Functional myogenic engraftment from mouse iPS cells. Stem Cell Rev 7:948–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darabi R, Santos FN, Filareto A, Pan W, Koene R, Rudnicki MA, Kyba M. and Perlingeiro RC. (2011). Assessment of the myogenic stem cell compartment following transplantation of pax3/pax7-induced embryonic stem cell-derived progenitors. Stem Cells 29:777–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dekel I, Magal Y, Pearson-White S, Emerson CP. and Shani M. (1992). Conditional conversion of ES cells to skeletal muscle by an exogenous MyoD1 gene. New Biol 4:217–224 [PubMed] [Google Scholar]
- 7.Shani M, Faerman A, Emerson CP, Pearson-White S, Dekel I. and Magal Y. (1992).The consequences of a constitutive expression of MyoD1 in ES cells and mouse embryos. Symp Soc Exp Biol 46:19–36 [PubMed] [Google Scholar]
- 8.Tajbakhsh S. (2009). Skeletal muscle stem cells in developmental versus regenerative myogenesis. J Intern Med 266:372–389 [DOI] [PubMed] [Google Scholar]
- 9.Ciemerych MA, Archacka K, Grabowska I. and Przewoźniak M. (2011). Cell cycle regulation during proliferation and differentiation of Mammalian muscle precursor cells. Results Probl Cell Differ 53:473–527 [DOI] [PubMed] [Google Scholar]
- 10.Zammit PS. (2008). All muscle satellite cells are equal, but are some more equal than others? J Cell Sci 121:2975–2982 [DOI] [PubMed] [Google Scholar]
- 11.Rohwedel J, Maltsev V, Bober E, Arnold HH, Hescheler J. and Wobus AM. (1994). Muscle cell differentiation of embryonic stem cells reflects myogenesis in vivo: developmentally regulated expression of myogenic determination genes and functional expression of ionic currents. Dev Biol 164:87–101 [DOI] [PubMed] [Google Scholar]
- 12.Sasaki T, Matsuoka H. and Saito M. (2008). Generation of a multi-layer muscle fiber sheet from mouse ES cells by the spermine action at specific timing and concentration. Differentiation 76:1023–1030 [DOI] [PubMed] [Google Scholar]
- 13.Zheng JK, Wang Y, Karandikar A, Wang Q, Gai H, Liu AL, Peng C. and Sheng HZ. (2006). Skeletal myogenesis by human embryonic stem cells. Cell Res 16:713–722 [DOI] [PubMed] [Google Scholar]
- 14.Di Rocco G, Iachininoto MG, Tritarelli A, Straino S, Zacheo A, Germani A, Crea F. and Capogrossi MC.(2006). Myogenic potential of adipose-tissue-derived cells. J Cell Sci 119:2945–2952 [DOI] [PubMed] [Google Scholar]
- 15.Kirillova I, Gussoni E, Goldhamer DJ. and Yablonka-Reuveni Z.(2007). Myogenic reprogramming of retina-derived cells following their spontaneous fusion with myotubes. Dev Biol 311:449–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torrente Y, Belicchi M, Sampaolesi M, Pisati F, Meregalli M, D'Antona G, Tonlorenzi R, Porretti L, Gavina M, et al. (2004). Human circulating AC133(+) stem cells restore dystrophin expression and ameliorate function in dystrophic skeletal muscle. J Clin Invest 114:182–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bedada FB. and Braun T. (2008). Partial induction of the myogenic program in noncommitted adult stem cells. Cells Tissues Organs 188:189–201 [DOI] [PubMed] [Google Scholar]
- 18.Pavlath GK. (2010). Spatial and functional restriction of regulatory molecules during mammalian myoblast fusion. Exp Cell Res 316:3067–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwander M, Leu M, Stumm M, Dorchies OM, Ruegg UT, Schittny J. and Müller U. (2003). Beta1 integrins regulate myoblast fusion and sarcomere assembly. Dev Cell 4:673–685 [DOI] [PubMed] [Google Scholar]
- 20.Knudsen KA, McElwee SA. and Myers L. (1990). A role for the neural cell adhesion molecule, NCAM, in myoblast interaction during myogenesis. Dev Biol 138:159–168 [DOI] [PubMed] [Google Scholar]
- 21.Rosen GD, Sanes JR, LaChance R, Cunningham JM, Roman J. and Dean DC. (1992). Roles for the integrin VLA-4 and its counter receptor VCAM-1 in myogenesis. Cell 69:1107–1119 [DOI] [PubMed] [Google Scholar]
- 22.Brzóska E, Bello V, Darribère T. and Moraczewski J. (2006). Integrin alpha3 subunit participates in myoblast adhesion and fusion in vitro. Differentiation 74:5–18 [DOI] [PubMed] [Google Scholar]
- 23.Dickson G, Peck D, Moore SE, Barton CH. and Walsh FS. (1990). Enhanced myogenesis in NCAM-transfected mouse myoblasts. Nature 344:348–351 [DOI] [PubMed] [Google Scholar]
- 24.Fässler R, Rohwedel J, Maltsev VA, Bloch W, Lentini S, Guan K, Gullberg D, Hescheler J, Addicks K. and Wobus AM. (1996). Differentiation and integrity of cardiac muscle cells are impaired in the absence of b1 integrin. J Cell Sci 109:2989–2999 [DOI] [PubMed] [Google Scholar]
- 25.Rohwedel J, Guan K, Zuschratter W, Jin S, Ahnert-Hilger G, Furst D, Fassler R. and Wobus AM. (1998). Loss of beta1 integrin function results in a retardation of myogenic, but an acceleration of neuronal, differentiation of embryonic stem cells in vitro. Dev Biol 201:167–184 [DOI] [PubMed] [Google Scholar]
- 26.Barberi T, Bradbury M, Dincer Z, Panagiotakos G, Socci ND. and Studer L. (2007). Derivation of engraftable skeletal myoblasts from human embryonic stem cells. Nat Med 13:642–648 [DOI] [PubMed] [Google Scholar]
- 27.Gu B, Zhang J, Wang W, Mo L, Zhou Y, Chen L, Liu Y. and Zhang M. (2010). Global expression of cell surface proteins in embryonic stem cells. PLoS One 5:e15795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Intoh A, Kurisaki A, Yamanaka Y, Hirano H, Fukuda H, Sugino H. and Asashima M. (2009). Proteomic analysis of membrane proteins expressed specifically in pluripotent murine embryonic stem cells. Proteomics 9:126–137 [DOI] [PubMed] [Google Scholar]
- 29.Kolle G, Ho M, Zhou Q, Chy HS, Krishnan K, Cloonan N, Bertoncello I, Laslett AL. and Grimmond SM. (2009). Identification of human embryonic stem cell surface markers by combined membrane-polysome translation state array analysis and immunotranscriptional profiling. Stem Cells 27:2446–2456 [DOI] [PubMed] [Google Scholar]
- 30.Rugg-Gunn PJ, Cox BJ, Lanner F, Sharma P, Ignatchenko V, McDonald AC, Garner J, Gramolini AO, Rossant J. and Kislinger T. (2012). Cell-surface proteomics identifies lineage-specific markers of embryo-derived stem cells. Dev Cell 22:887–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hadjantonakis AK. and Papaioannou VE. (2004). Dynamic in vivo imaging and cell tracking using a histone fluorescent protein fusion in mice. BMC Biotechnol 24:4–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Archacka K, Pozzobon M, Repele A, Rossi CA, Campanella M. and De Coppi P. (2014). Culturing muscle fibres in hanging drop: a novel approach to solve an old problem. Biol Cell 106:72–82 [DOI] [PubMed] [Google Scholar]
- 33.Keller G. (1995). In vitro differentiation of embryonic stem cells. Curr Opin Cell Biol 7:862–869 [DOI] [PubMed] [Google Scholar]
- 34.Rosenblatt JD, Lunt AI, Parry DJ. and Partridge TA. (1995). Culturing satellite cells from living single muscle fiber explants. In Vitro Cell Dev Biol Anim 31:773–779 [DOI] [PubMed] [Google Scholar]
- 35.Grabowska I, Szeliga A, Moraczewski J, Czaplicka I, Brzóska E. (2011). Comparison of satellite cell-derived myoblasts and C2C12 differentiation in two- and three-dimensional cultures: changes in adhesion protein expression. Cell Biol Int 35:125–133 [DOI] [PubMed] [Google Scholar]
- 36.Donalies M, Cramer M, Ringwald M. and Starzinski-Powitz A. (1991). Expression of M-cadherin, a member of the cadherin multigene family, correlates with differentiation of skeletal muscle cells. Proc Natl Acad Sci U S A 88:8024–8028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tachibana I. and Hemler ME. (1999). Role of transmembrane 4 superfamily (TM4SF) proteins CD9 and CD81 in muscle cell fusion and myotube maintenance. J Cell Biol 146:893–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yagami-Hiromasa T, Sato T, Kurisaki T, Kamijo K, Nabeshima Y. and Fujisawa-Sehara A. (1995). A metalloprotease-disintegrin participating in myoblast fusion. Nature 377:652–656 [DOI] [PubMed] [Google Scholar]
- 39.Grivennikov IA. (2008). Embryonic stem cells and the problem of directed differentiation. Biochemistry 73:1438–1452 [DOI] [PubMed] [Google Scholar]
- 40.Murry CE. and Keller G. (2008). Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell 132:661–680 [DOI] [PubMed] [Google Scholar]
- 41.Mizuno Y, Chang H, Umeda K, Niwa A, Iwasa T, Awaya T, Fukada S, Yamamoto H, Yamanaka S, Nakahata T. and Heike T. (2010). Generation of skeletal muscle stem/progenitor cells from murine induced pluripotent stem cells. Faseb J 24:2245–2253 [DOI] [PubMed] [Google Scholar]
- 42.Tian C, Lu Y, Gilbert R. and Karpati G. (2008). Differentiation of murine embryonic stem cells in skeletal muscles of mice. Cell Transplant 17:325–335 [DOI] [PubMed] [Google Scholar]
- 43.Chang H, Yoshimoto M, Umeda K, Iwasa T, Mizuno Y, Fukada S, Yamamoto H, Motohashi N, Miyagoe-Suzuki Y, et al. (2009). Generation of transplantable, functional satellite-like cells from mouse embryonic stem cells. FASEB J 23:1907–1919 [DOI] [PubMed] [Google Scholar]
- 44.Czyz J. and Wobus A. (2001). Embryonic stem cell differentiation: the role of extracellular factors. Differentiation 68:167–174 [DOI] [PubMed] [Google Scholar]
- 45.Constantinides PG, Jones PA. and Gevers W. (1977). Functional striated muscle cells from non-myoblast precursors following 5-azacytidine treatment. Nature 267:364–366 [DOI] [PubMed] [Google Scholar]
- 46.Taylor SM. and Jones PA. (1979). Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell 17:771–779 [DOI] [PubMed] [Google Scholar]
- 47.Musch T, Oz Y, Lyko F. and Breiling A. (2010). Nucleoside drugs induce cellular differentiation by caspase-dependent degradation of stem cell factors. PLoS One 5:e10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watanabe S, Hirai H, Asakura Y, Tastad C, Verma M, Keller C, Dutton JR. and Asakura A. (2011). MyoD gene suppression by Oct4 is required for reprogramming in myoblasts to produce induced pluripotent stem cells. Stem Cells 29:505–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruau D, Ensenat-Waser R, Dinger TC, Vallabhapurapu DS, Rolletschek A, Hacker C, Hieronymus T, Wobus AM, Müller AM. and Zenke M. (2008). Pluripotency associated genes are reactivated by chromatin-modifying agents in neurosphere cells. Stem Cells 26:920–926 [DOI] [PubMed] [Google Scholar]
- 50.György B, Szabó TG, Pásztói M, Pál Z, Misják P, Aradi B, László V, Pállinger E, Pap E, et al. (2011). Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci 68:2667–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barberi T, Willis L, Socci ND. and Studer L. (2005). Derivation of multipotent mesenchymal precursors from human embryonic stem cells. PLoS Med 2:e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rose O, Rohwedel J, Reinhardt S, Bachmann M, Cramer M, Rotter M, Wobus A. and Starzinski-Powitz A. (1994). Expression of M-cadherin protein in myogenic cells during prenatal mouse development and differentiation of embryonic stem cells in culture. Dev Dyn 201:245–259 [DOI] [PubMed] [Google Scholar]
- 53.Hollnagel A, Grund C, Franke WW. and Arnold HH. (2002). The cell adhesion molecule M-cadherin is not essential for muscle development and regeneration. Mol Cell Biol 22:4760–4770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Uosaki H, Fukushima H, Takeuchi A, Matsuoka S, Nakatsuji N, Yamanaka S. and Yamashita JK. (2011). Efficient and scalable purification of cardiomyocytes from human embryonic and induced pluripotent stem cells by VCAM1 surface expression. PLoS One 6:e23657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tian L, Catt JW, O'Neill C. and King NJ. (1997). Expression of immunoglobulin superfamily cell adhesion molecules on murine embryonic stem cells. Biol Reprod 57:561–568 [DOI] [PubMed] [Google Scholar]
- 56.Heyward SA, Dubois-Stringfellow N, Rapoport R. and Bautch VL. (1995). Expression and inducibility of vascular adhesion receptors in development. FASEB J 9:956–962 [DOI] [PubMed] [Google Scholar]
- 57.Schulze M, Bedada FB, Technau A. and Braun T. (2005). Mesenchymal stem cells are recruited to striated muscle by NFAT/IL-4-mediated cell fusion. Genes Dev 19:1787–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson RT. and Rao RN. (1970). Mammalian cell fusion: induction of premature chromosome condensation in interphase nuclei. Nature 226:717–722 [DOI] [PubMed] [Google Scholar]
- 59.Hochedlinger K. and Jaenisch R. (2006). Nuclear reprogramming and pluripotency. Nature 441:1061–1067 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.