Abstract
Membrane recruitment of the COPI vesicle coat is fundamental to its function and contributes to compartment identity in the early secretory pathway. COPI recruitment is triggered by guanine nucleotide exchange activating the Arf1 GTPase, but the key exchange factor, GBF1, is a peripheral membrane component whose membrane association is dependent on another GTPase, Rab1. Inactive Rab GTPases are in a soluble complex with guanine nucleotide dissociation inhibitor (GDI) and activation of Rab GTPases by exchange factors can be enhanced by GDI dissociation factors (GDFs). In the present study, we investigated the vesicle docking protein p115 and it’s binding to the Rab1 isoform Rab1b. Inhibition of p115 expression induced dissociation of Rab1b from Golgi membranes. Rab1b bound the cc2 domain of p115 and p115 lacking this domain failed to recruit Rab1b. Further, p115 inhibition blocked association of the COPI coat with Golgi membranes and this was suppressed by constitutive activation of Rab1b. These findings show p115 enhancement of Rab1b activation leading to COPI recruitment suggesting a connection between the vesicle docking machinery and the vesicle coat complex during the establishment of post-ER compartment identity.
Keywords: vesicle tether, vesicle coat complex, Rab GTPase, p115, COPI
Introduction
In mammalian cells, the early secretory pathway consists of membrane-enclosed compartments including the endoplasmic reticulum (ER), the intermediate compartment (IC) and the Golgi. These compartments maintain distinct compositions and functional activities despite continuous material exchange along interconnecting transport routes. Remarkably, the IC and Golgi can arise de novo from the ER1raising the question of how the unique identity of these compartments is established. Post-ER compartment identity may be initiated by the coat protein II (COPII) complex as it sorts cargo molecules and recruits cytosolic factors to the domain of the ER membrane that will exit and move to the IC and Golgi. Concomitant with its recruitment activity, the COPII complex generates vesicles that undergo homotypic fusion forming the IC.2 Dissociation of the COPII complex is then accompanied by assembly of the coat protein I (COPI) complex marking the change in identity of the membrane from ER exit site to IC.3 COPI forms vesicles from the IC and Golgi that carry recycling proteins in a retrograde direction to maintain proper targeting of proteins to their respective compartments.4 Thus, COPI function strongly contributes to establishing post-ER compartment identity. Indeed, the packaging into COPI vesicles of the cycling SNARE proteins that will mediate the specific fusion of vesicles with their target membranes is a fundamental reaction in the establishment and maintenance of intracellular compartments.5
Membrane recruitment of COPI is surprisingly complicated involving a network of peripheral membrane proteins. The final step is via direct COPI interactions including those with the activated form of the small GTPase, Arf1.6,7 Arf1 is recruited and activated principally by the guanine nucleotide exchange factor, GBF1.8 But GBF1 is also a peripheral component and its membrane recruitment depends on another GTPase, Rab1b.9,10 Reversible association of Rab GTPases with their target membranes is coupled to their GDP- and GTP-bound states.11 GDP-bound Rab proteins are sequestered in soluble form in the cytoplasm via binding to guanine disassociation inhibitor (GDI), which masks their isoprenyl lipid anchor.12 Membrane association of Rabs can be promoted by GDI dissociation factors (GDFs), which catalyze the dissociation of the Rab-GDI complex thereby allowing the lipid anchor to be inserted into membranes.12 To date only a few GDFs have been described.11 Better known is the role of guanine nucleotide exchange factors (GEFs), which specifically interact with particular Rab proteins causing dissociation of the bound GDP. GTP binding then activates the Rab promoting Rab interactions with various effectors. Activation is terminated by GTP hydrolysis by the Rab accelerated by specific GTPase activating proteins (GAPs). The inactive GDP-bound Rabs are extracted from membranes by GDI.12 Interestingly, some Rab effectors play upstream roles in Rab activation by recruiting or modulating GEF activity, thereby forming feedback loops and cascades.12-15 Thus, the membrane association of Rab1 can be regulated by its GDF and GEF, and possibly also by Rab1 effectors.
The vesicle tethering protein p115 is one of several known Rab1 effectors16 and mediates vesicular trafficking between the ER and the Golgi.16-20 p115 has been shown in vitro to promote SNARE complex formation and stimulate docking of COPI-coated vesicles to Golgi membranes.21 The yeast homolog of p115, Uso1p, is required for docking of COPII vesicles to Golgi membranes22,23 and for sorting of GPI-anchored proteins into vesicles upon ER exit.24 p115 is a homodimer composed of an N-terminal globular head domain followed by an elongated tail containing four predicted coiled-coils (cc1-cc4) and a C-terminus enriched in acidic residues. Several regions on p115 directly interact with transport factors and are essential for p115 function.19,25-28 The binding site in p115 for Rab1 has been mapped to the cc1-cc2 region29but also to a conserved sequence in the head domain.30 Interestingly, Rab1 binding to p115 at the cc1-cc2 site is influenced by binding of the golgins GM130 or giantin to the p115 acidic C-terminus.29 Rab1 mediates membrane association of p115 in vitro16,23 but may not be required in vivo because p115 remains membrane associated after Rab1 depletion by siRNA.10 The head region of p115 also interacts with GBF1, but this interaction is not required for membrane association of GBF1 or p115.31
Herein, we report that inhibiting p115 expression induced dissociation of the Rab1 isoform, Rab1b, from Golgi membranes. Rab1b bound the cc2 domain of p115 and p115 lacking this domain failed to recruit Rab1b and failed to function in biogenesis of the Golgi apparatus. These results suggest that the interaction between p115 and Rab1b has an upstream role in conferring Rab1b membrane localization. Indeed, the cc2 domain promoted disassociation of Rab1b from GDI. Further, p115 acted upstream of Rab1b in that p115 inhibition blocked association of the COPI coat with Golgi membranes and this was suppressed by constitutive activation of Rab1b. Thus, p115 regulates Rab1b activation in a pathway controlling assembly of COPI vesicle coat on membranes, suggesting that vesicle docking machinery may contribute to establishing compartmental identity during Golgi biogenesis by regulating COPI membrane recruitment.
Materials and Methods
Constructs and reagents
Small interfering RNA (siRNA) targeting AAGACCGGCAATTGTAGTACT in p11519 was purchased from Xeragon (Valencia, CA). Hexahistidine-tagged Rab1b (His-Rab1b) and HA-tagged Rab1b were a kind give of Dr. T.H. Lee (Carnegie Mellon University). Variants of p115 and Rab1b were generated by restriction digests and by using the QuikChange site-directed mutagenesis kit (Stratagene,CA). Constructs were confirmed by restriction analysis and sequencing.
Immunofluorescence and image analysis
HeLa cells stably expressing N-acetylgalactosaminyl transferase-2tagged with the green fluorescent protein (GalNAcT2-GFP) were maintained and analyzed by immunofluorescence as described previously.1 Blocking buffer (PBS containing 0.05% saponin, 2.5% fetal bovine serum, 0.2M glycine) was used for staining endogenous Rab1b. The antibodies and their dilutions were rabbit anti-Rab1b (1:100) (Santa Cruz Biotechnology, CA), rabbit anti-p115 (1:500),32 mouse anti-GBF1 (1:1000) (BD Biosciences, CA), mouse anti-βCOP (1:1000) (Sigma-Aldrich, MO), and mouse anti-bovine p115 (1:500).17Microscopy was performed as described previously.33 Single optical sections were acquired using ImagingSuite software (PerkinElmer). Individual experiments were performed with identical laser output levels, exposure times, and scaling. To quantify the average signal intensity of βCOP, GBF1 or Rab1, the GalNAcT2-GFP positive pixels were selected using the Photoshop magic wand tool and the average pixel value of the corresponding region in the βCOP, GBF1 or Rab1 channel was calculated using the Photoshop histogram function. The average signal intensity of the cytoplasmic background of GBF1 or Rab1 staining was quantified by calculating the average pixel value of a random area in the cytoplasmic region through the Photoshop histogram function and subsequently, the average pixel value of the cytoplasmic background staining was subtracted from the average pixel value of βCOP, GBF1 or Rab1 staining.
RNA interference and replacement
RNA Interference was performed as described previously.19 For gene replacement, 48 h after siRNA transfection, the cells were transfected using Transfectol (Siegen, CA) or Turbo Fec (Invitrogen) with plasmids encoding Myc-tagged bovine p115 wildtype, myc-tagged bovine p115 Δcc2-C (Δ742–767), bovine p115 Δcc1(Δ652–715),19 HA-tagged Rab1b wildtype or HA-tagged Rab1b (Q67L). Cells were analyzed 24 h after transient transfection with the plasmid. Staining level was used to insure that cells expressed corresponding constructs at approximately equal levels and the results obtained were consistent across these expression levels.
Rab1b release assay
HeLa cells stably expressing GalNAcT2-GFP were plated in a 35mm dish and were either untreated or treated with siRNA against p115. At day 3 after knockdown, cells were washed 3 x 1 ml with cold MEM, and washed2 x 1 ml with cold KOAc buffer (115 mM KOAc, 2.5 mM MgOAc,25 mM Hepes, pH 7.2, and 1 mM dithiothreitol). The washed cells were permeabilized 6 min at RT in 1 ml 0.03 mg/ml digitonin in KOAc buffer containing 1 mM phenylmethylsulfonyl fluoride, 10 mg/ml leupeptin, 10 mg/ml pepstatin A. The proteins released into the KOAc buffer after permeablization were precipitated using trichloroacetic acid and analyzed by immunoblotting34 using enhanced chemiluminescence (Pierce Chemical Co.) with acquisition by the LAS-3000 imaging system (Fujifilm). Antibodies and their dilutions used for immunoblotting were rabbit anti-Rab1b at 1:1000, mouse anti-tubulin at 1:5000 (Sigma), peroxidase-conjugated anti-rabbit or anti-mouse at 1:5000 (Bio-Rad, Hercules, CA).
Protein purification and binding assays
GST fusion protein purification was performed as described previously26for the various p115 constructs: cc2–4 (710–959), cc2 with flanking sequence (710–772), N-terminal half of cc2 (710–741), C-terminal half of cc2 (742–772), cc2 without flanking sequence (726–767)and also full-length Rab1b. His-Rab1b purification was performed according to the protocol provided by QIAGEN (Valencia, CA). Binding assays were performed as described previously.26
Co-immunoprecipitation
Co-immunoprecipitation was performed as described.26 Briefly, HeLa cells transiently transfected with HA-Rab1b were lysed in HKMD buffer (20mM Hepes, pH 7.4, 100mM KCl, 5mM MgCl2, 2% digitonin, 1 mM phenylmethylsulfonyl fluoride, 10 mg/ml leupeptin, and 10 mg/ml pepstatin A, 1 mM dithiothreitol). After 1 h on ice the homogenate was passed through 25 g needle followed by centrifugation in a microfuge for 10 min at 4 °C. The supernatants were incubated at 4 °C for 90 min with 10 µl protein A beads (CL-4B, GE Healthcare) bearing mouse anti-HA antibody. The beads were then washed with HKM buffer (20mM Hepes, pH 7.4, 100mM KCl, 5mM MgCl2, 1 mM dithiothreitol) and analyzed by immunoblotting.
GDI dissociation assay
Prenylated His tagged Rab1b (His-ScRab1b) was isolated from Saccharomyces cerevisiae FY834 (MATα his3Δ200 ura3–52 leu2Δ1 lys2Δ202trp1Δ63) coexpressing His-ScRab1b and FLAG-GDI2.35 Cells were grown at 30 °C in minimal medium (0.67% yeast nitrogen base without amino acids, 2% dextrose, amino acid supplement without uracil and tryptophan) and recombinant protein production was induced by shifting cells to minimal medium containing 2% galactose.35 The cells in 2 L cell culture were harvested and resuspended in 10ml PBS containing 1mM β-mercaptoethanol and 1mM MgCl2, transferred to 15 ml tubes containing 1/2 volume glass beads, and vortexed 8 x 15 s. The cell lysate was sonicated three times for 30 s each, and centrifuged at 50,000 rpm for 30 min in a TLA 100.3 rotor (BeckmanCoulter, Fullerton, CA). The cleared lysate was collected and incubated with 0.5 ml nickel-nitrilotriaceticacid agarose beads (Invitrogen). After incubation, the beads were washed with 100x volume of PBS containing 200mM immidazole, 1mM β-mercaptoethanol, 1mM MgCl2 and were stored on ice for future analysis. Glutathione-agarose beads (4 µl) coated with approximately 3 µg of His-ScRab1b were incubated with approximately 2μg of cc2–4 (710–959) or cc2 (710–772) in 40 μl reaction buffer (PBS, 2 mM MgCl2, 1 mM β-mercaptoethanol). After a 2-h incubation at 4 °C, the supernatant containing unbound proteins was removed, beads were washed 5x in reaction buffer and resuspended in SDS sample buffer. Proteins in the supernatant or bound to beads were detected by immunoblotting using polyclonal anti- FLAG antibody (M2, Sigma, 1:1000).
GEF assay
The GEF assay was performed as described.36 GDP-loaded His-Rab1b wildtype (0.4nmol) was added to the GEF assay buffer (20 mM TRIS-HCl, pH 7.5, 100 mM NaCl, 5 mM MgCl2, 1 mM DTT, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, 10 mg/ml leupeptin,10 mg/ml pepstatin A, and 400 nM N-methylanthraniloyl-GTP [mant-GTP, Invitrogen]) in 200μl reaction volume, and equilibrated for 5 min at RT. The reaction was initiated by the addition of 62 pmol GST-p115 CC2 construct (710–772), and fluorescence was monitored at RT by using a Perkin Elmer LS-55 spectrophotometer (λex = 360 nm; λem = 440 nm; slits = 20 nm/20 nm).
Results
P115 knockdown displaces Rab1b from Golgi membranes
To investigate the role of Rab1b binding by p115 we analyzed Rab1b localization in cells after inhibition of p115 expression using RNA interference. The experiments were performed in HeLa cells stably expressing N-acetylgalactosaminyl transferase-2 tagged with the green fluorescent protein (GalNAcT2-GFP). As previously reported,19 knockdown of p115 yielded a clear fragmented Golgi phenotype and the cells lacked detectable p115 indicating successful knockdown (Fig. 1A-D). Control cells exhibited a characteristic Golgi ribbon pattern and, as expected, Rab1b was localized to these membranes as well as throughout the cytoplasm (Fig. 1E-G). Surprisingly, p115 knockdown caused loss of Rab1b Golgi membrane association (Fig. 1H-J). Analysis of the mean signal intensity of Rab1b that colocalized with GalNAcT2-GFP confirmed a significant reduction in Golgi-associated Rab1b after p115 knockdown (Fig. 1K). Neither cytosolic Rab1b nor total cellular Rab1b was reduced as determined by immunofluorescence and immunoblotting, respectively (not shown). Because p115 knockdown reduces p115 levels by about 90%,19 the residual Golgi-associated Rab1b likely reflects the participation of other mechanisms. To assess Rab1b membrane association using a distinct assay, we permeabilized control and p115 knockdown cells and used immunoblotting to measure the amount of Rab1b released during the permeabilization. The amount of tubulin released was used to normalize the data. There was a 1.6-fold increase in released Rab1b after p115 knockdown (Fig. 1L). Taken together, these results indicate that p115 levels influence Golgi membrane association of Rab1b.
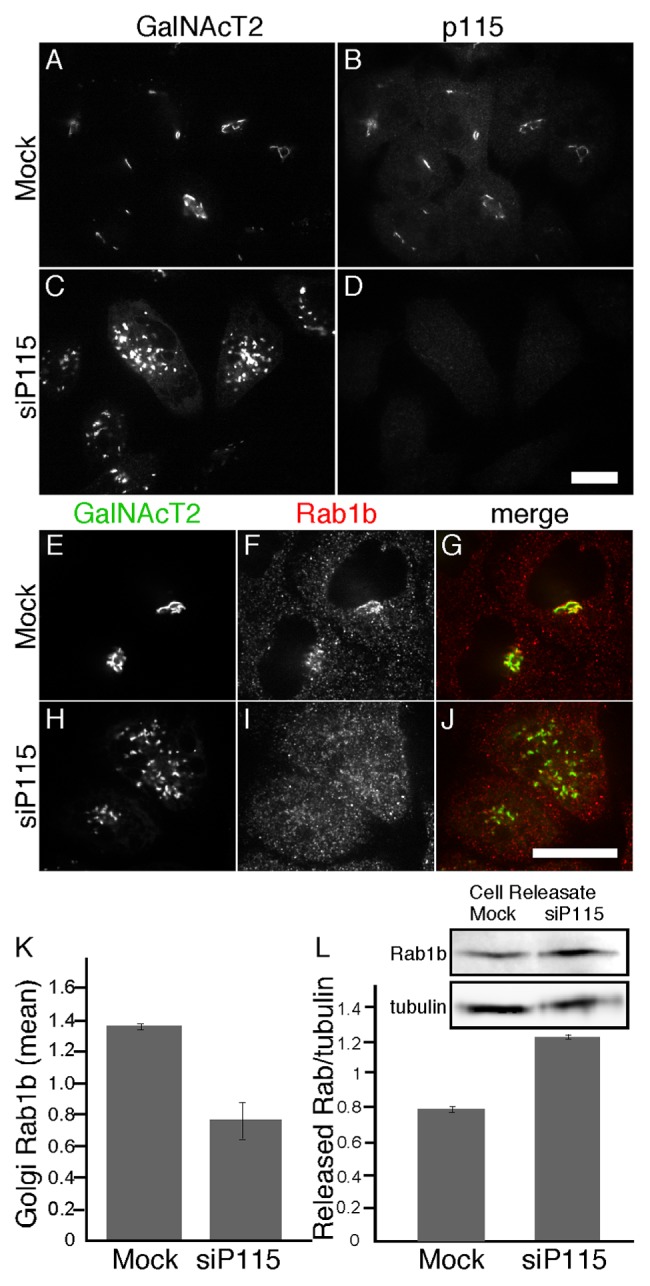
Figure 1. p115 knockdown displaces Rab1b from the Golgi. HeLa cells expressing GalNAcT2-GFP were either mock transfected or transfected with siRNA against p115. GalNacT2-GFP and either p115 (A-D) or Rab1b (E-J) were visualized 72 h later. Bar = 10µm. (K) Quantified average signal intensity of Golgi membrane associated Rab1b is shown for mock and p115 knockdown cells (mean ± SD; n = 2; > 15 cells each). To allow direct comparison of distinct experiments, values were normalized by dividing by the mean values of the entire data set for a given experiment. (L) HeLa cells expressing GalNAcT2-GFP, plated on 35-mm dishes, were either mock transfected or transfected with siRNA against p115. After 72 h, the cells were digitonin permeabilized and the amount of released Rab1b and tubulin was determined by immunoblotting with anti-Rab1b and anti-tubulin antibodies (mean ± SD; n = 2).
P115-mediated Rab1b membrane association impacts COPI recruitment
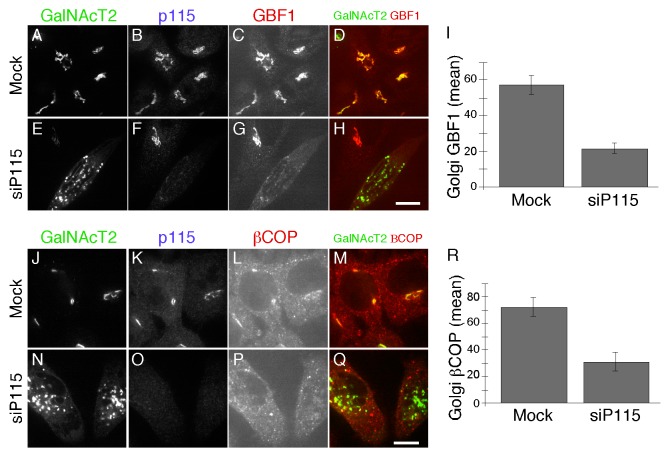
Because previously published work indicates that Rab1b is required for COPI membrane association9 our result showing Rab1b’s dependence on p115 suggested that p115 knockdown might interfere with membrane recruitment of the COPI coat complex. Rab1b promotes Arf1-dependent COPI recruitment through its interaction with GBF1, the GEF for Arf.37 Thus, we assayed Golgi membrane association of GBF1 and the β subunit of the COPI coat complex after depleting p115. Indeed, we detected a clear decrease in membrane associated GBF1 (Fig. 2A-I) and βCOP (Fig. 2J-R) in cells depleted of p115. The average signal intensity of Golgi membrane associated GBF1 and βCOP was reduced by more than half compared with control cells (Fig. 2I, P = 0.003 and Figure 2R, P = 0.007). Although we cannot exclude the possibility that these proteins have reduced expression under these conditions, it is likely that they redistributed to the cytosol.
Figure 2. p115 knockdown displaces GBF1 and COPI from the Golgi. HeLa cells expressing GalNAcT2-GFP were either mock transfected or transfected with siRNA against p115. GalNacT2-GFP, p115 and either GBF1 (A-H) or βCOP (J-Q) were visualized 72 h later. Bar = 10µm. The quantified average signal intensity of Golgi membrane associated GBF1 (I) and βCOP (R) is shown (mean ± SEM; > 20 cells).
Direct interaction between p115 and Rab1b regulates Rab1b membrane association
P115 directly interacts with Rab1 in vitro16 and we observed co-immunoprecipitation of p115 with HA-tagged Rab1b out of HeLa cell lysates, confirming the interaction in vivo (Fig. S1). To determine if Rab1b Golgi membrane association depends on Rab1b binding to p115 we sought to map and then delete the Rab1b interaction site in p115. Based on the information available at the time, we focused on the region containing the first and second predicted coiled-coil domains (cc1 and cc2, Fig. 3A), which had been shown to bind Rab1a.29 A series of GST-tagged p115 constructs were purified after expression in bacteria and tested for interaction with His-tagged Rab1b, also purified after expression in bacteria. It was observed that a construct containing cc2 and its flanking sequence specifically bound Rab1b (Fig. 3B, lane 2). The cc2 construct also bound HA-tagged Rab1b, but not GFP-tagged Rab27b, in HeLa cell lysates confirming the interaction and its specificity (Fig. S2). To further narrow the Rab1b interaction domain on p115, the cc2 construct was bisected. A construct containing the N-terminal half did not bind (Fig. 3B, lane 4) whereas a construct containing the C-terminal half did bind (Fig. 3B, lane 5). The short flanking sequences were not required because binding occurred in their absence (Fig. 3B, lane 6). Thus, a binding site for Rab1b mapped to the C-terminal part of cc2.
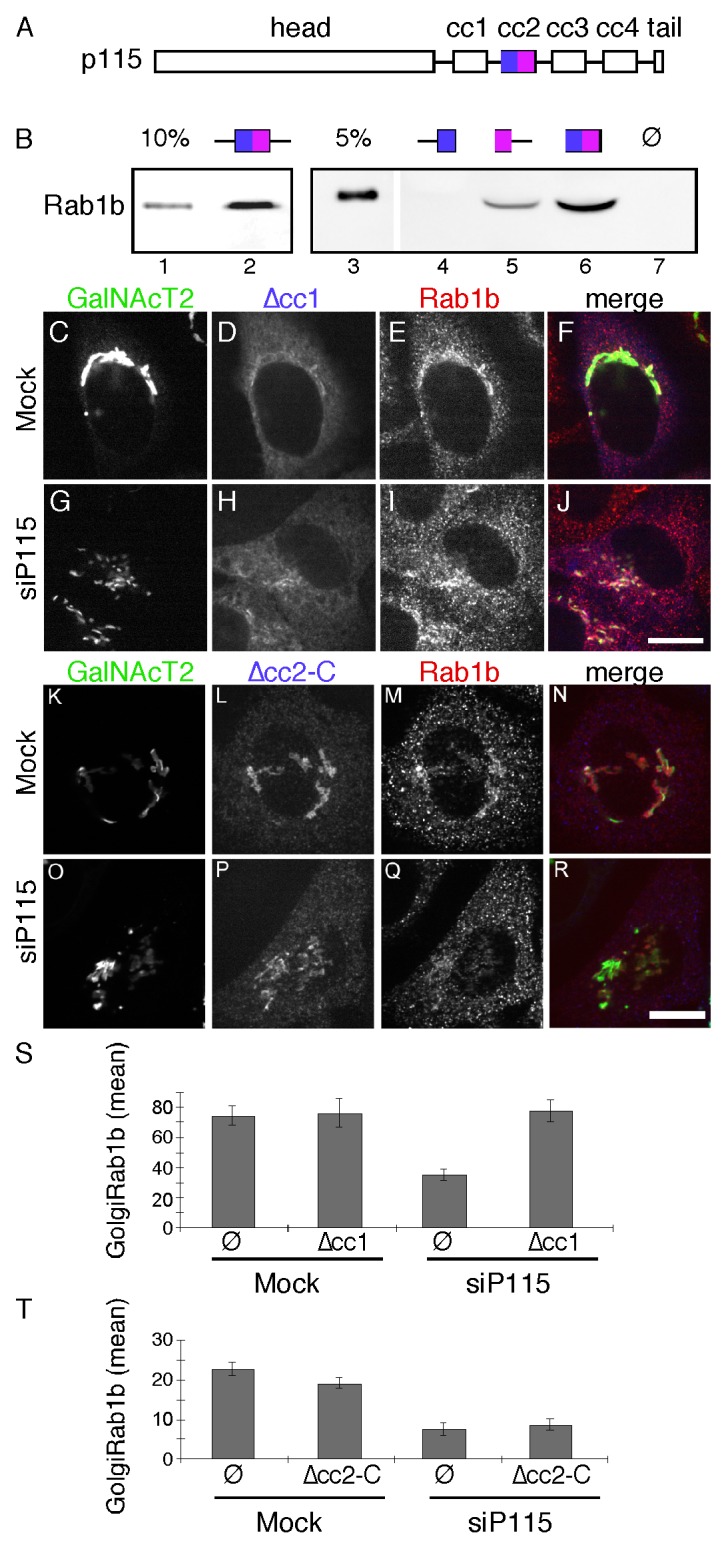
Figure 3. The cc2 domain of p115 binds Rab1b and is required to rescue Rab1b membrane association. (A) Schematic diagram of p115 domains. (B) Equivalent amounts (approximately 5 µg) of bead-bound GST (lane 7) and the indicated p115 constructs (see Materials and Methods) were incubated with 0.6 µg of purified hexahistidine-tagged Rab1b in the absence of GDP or GTPγS. Recovery of the Rab1b construct was determined by immunoblotting with an anti-Rab1b antibody and compared with the loading controls as indicated. (C-R) Cells were mock transfected or transfected with p115 siRNA and re-transfected after 48 h with plasmid encoding bovine p115Δcc1 (C-J) or Myc-tagged bovine p115Δcc2-C (K-R). After an additional 24 h, the cells were stained using rabbit anti-Rab1b antibody and either anti-bovine p115 antibody or anti-Myc antibody. Bar = 10µm. (S-T) The average signal intensity of Golgi membrane associated Rab1b for mock or p115 knockdown cells in the absence or presence of expressed p115Δcc1 (S) (mean ± SEM; > 20 cells) or p115Δcc2-C (T) is shown (mean ± SD; n = 2; > 20 cells for each experiment).
Having mapped an interaction site, we next tested its functional role by carrying out rescue after p115 knockdown using Myc-tagged bovine p115 rescue constructs corresponding to wild-type p115 and a version of p115 in which the C-terminal half of cc2 was deleted (p115Δcc2-C). As another control, we also performed rescue with p115 carrying a deletion of the cc1 domain, which interacts with SNARE proteins.21 This construct is Golgi-localized yet fails to support Golgi assembly.19 As expected expression of the wildtype construct rescued Golgi assembly (Fig. S3), whereas p115∆cc1 did not (Fig. 3G). Further, Golgi membrane association of Rab1b was rescued by both wildtype (Fig. S3) and p115∆cc1 (Fig. 3I), presumably because each construct contains the Rab1b-binding cc2 domain. Strikingly, p115 Δcc2-C failed to rescue the membrane association of Rab1b and it also failed to rescue Golgi assembly (Fig. 3O-R). Analysis of the mean signal intensity of Rab1b that was colocalized with GalNAcT2-GFP confirmed that p115∆cc2-C failed to recruit Rab1b whereas p115∆cc1 restored normal levels of Rab1b on the membrane (Fig. 3S and T). These results map a Rab1b binding site in p115 and show that this site is required for Rab1b membrane localization and maintenance of an intact Golgi ribbon.
P115 promotes dissociation of the Rab1-GDI complex
The membrane association of Rab1b is coupled to its GDP- and GTP-bound status. If p115 mediates Rab1b membrane association, p115 might facilitate nucleotide exchange of Rab1b or promote GDI release. As tests, we assayed nucleotide exchange and GDI dissociation activity of the purified cc2 domain of p115. Nucleotide exchange of GDP-loaded Rab1b in the presence of cc2 was similar to the intrinsic rate supporting an absence of GEF activity (Fig. 4A-B). To assay whether cc2 could facilitate GDI release, His-tagged human Rab1b was purified from Saccharomyces cerevisiae under conditions where it is prenylated and a fraction is bound to co-expressed FLAG-tagged human GDI2.35 The purified preparation, immobilized on beads, was incubated with cc2 and the release of GDI2 was determined. Although there was basal release, presumably reflecting the dissociation rate of GDI, the extent of GDI release was enhanced by the presence of the p115 fragment (Fig. 4C). This result shows that the p115/Rab1b interaction within the cc2 domain might facilitate membrane association of Rab1b by promoting dissociation of the cytosolic Rab1b/GDI complex.
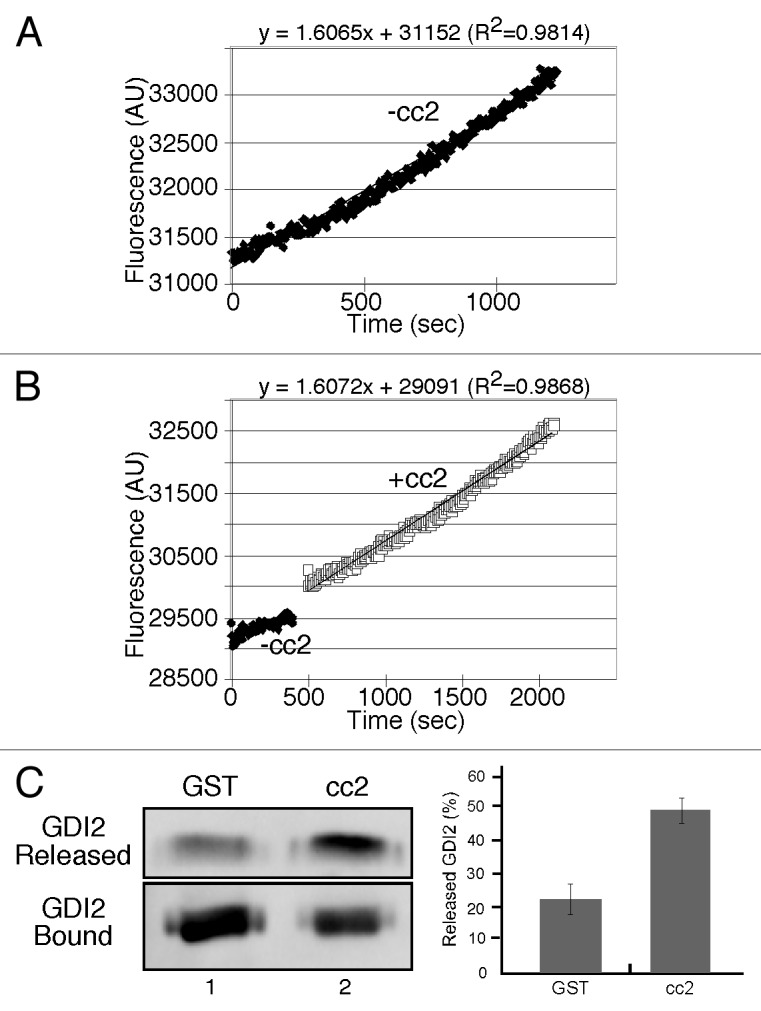
Figure 4. The p115 cc2 domain promotes dissociation of the Rab1b-GDI2 complex but fails to enhance nucleotide exchange. (A-B) Rates of MANT-GTP incorporation into His-Rab1b in the presence of GST or GST-p115-cc2. (C) Equivalent amounts (approximately 3 µg) of bead-bound His-ScRab1b, a fraction of which was associated with FLAG-GDI2, was incubated with approximately 2μg of cc2. FLAG-GDI2 proteins in the unbound fraction or bound to beads were detected by immunoblotting using a polyclonal anti-FLAG antibody and the percentage of released FLAG-GDI2 was quantified (mean ± SD; n = 3).
Rab1b activation bypasses p115 requirement
These observations indicate that p115 is an upstream component in a pathway involving successive membrane recruitment of Rab1b, GBF1, Arf1 and finally the COPI coat complex. Although previously unsuspected in mammalian cells, a role for p115 upstream of rab1b is consistent with the finding that Ypt1, yeast Rab1, suppresses the growth defect of a yeast strain deficient for Uso1p.38 If p115 acts upstream of Rab1b in HeLa cells then activation of Rab1b by other means should rescue membrane recruitment of GBF1 and the COPI coat in cells lacking p115. As a test, we analyzed the membrane recruitment of GBF1 in p115 knockdown cells transfected with Rab1b wildtype or the GTP-restricted form Rab1b (Q67L).9 In p115 knockdown cells expressing Rab1b wildtype, Golgi associated GBF1 was relatively low (Fig. 5A-H) whereas expression of Rab1b (Q67L) restored GBF1 membrane association (Fig. 5I-P). While residual GBF1 on the Golgi may reflect other mechanisms, these results are consistent with p115 regulating Rab1b-mediated GBF1 recruitment. Quantification showed that the average signal intensity of GBF1 in p115 knockdown cells expressing Rab1b (Q67L) was similar to mock-treated control cells (Fig. 5Q).
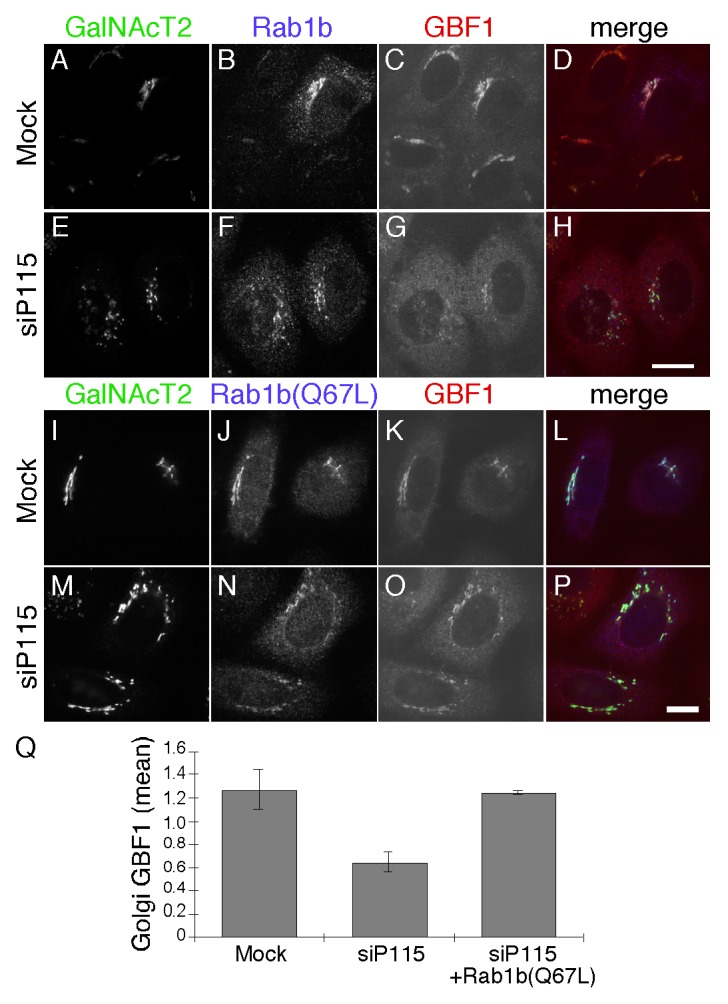
Figure 5. Activated Rab1b (Q67L) rescues GBF1 membrane association in p115 knockdown cells. Mock or p115 siRNA-transfected cells were retransfected after 48 h with plasmid encoding Rab1b (A-H) or Rab1b (Q67L) (I-P). GalNacT2-GFP, Rab1b and GBF1 were visualized 24 h later. Bar = 10µm. The quantified average signal intensity of Golgi membrane associated GBF1 (Q) is shown (mean ± SD; n = 2; > 15 cells each). To allow direct comparison of distinct experiments, values were normalized by dividing by the mean values of the entire data set for a given experiment.
Discussion
In addition to several other interactions, the vesicle tethering factor p115 binds Rab1 and has been considered a Rab1 effector.16,29 By mapping and mutating the Rab1b binding site, the present study demonstrates that a version of p115 defective for Rab1b binding failed to support Golgi biogenesis. Further, a surprising mechanism is described in which p115 promoted Rab1b release from GDI. Absence of p115 binding to Rab1b led to a loss of Rab1b recruitment and, as a consequence, inhibition of Rab1-dependent recruitment of GBF1, which in turn blocked COPI recruitment. Thus, the experiments reveal a GTPase cascade from vesicle tether to vesicle coat.
Establishing the identity of the intermediate compartment and Golgi apparatus depends, in part, on recruitment of the COPI coat complex so that it can carry out sorting reactions specific to these compartments. COPI coat recruitment is triggered by activation of the Arf1 GTPase and the key activity for this is nucleotide exchange catalyzed by the Arf GEF, GBF1.8 Given its central role in specifying the membranes to which recruitment of the COPI coat takes place, it makes sense that GBF1 is a peripheral membrane component. If it were a transmembrane containing protein its presence in the ER (e.g., during its synthesis and during de novo assembly of the Golgi out of the ER) could lead to improper COPI recruitment and a breakdown of compartment identity. Nevertheless, as a peripheral component, it must be recruited and this leaves the identity specifying reaction to an upstream component. Interestingly, it is the Rab1 GTPase that recruits GBF110 implying that the exchange factor for Rab1 plays a critical role. In yeast the TRAPP1 and TRAPP2 vesicle-tethering complexes are the GEFs for Ypt1, the Rab1 homolog39 and the mammalian homolog of TRAPPII subunit, Trs130p, is a GEF for Rab1 in mammalian cells.40 Thus, a tether controls activation of this key GTPase. There is a striking parallel in the present study. The vesicle tether p115 aids in Rab1b activation. This is indicated by its GDF activity and by the observation that p115 knockdown reduces the membrane-associated, that is activated, pool of Rab1b. Therefore, via its activity toward Rab1b, p115 may play a role in specifying IC and Golgi membrane identity.
Of course, this raises questions concerning p115’s recruitment to the membrane in terms of its mechanism and cellular location. Binding of p115 to Golgi membranes is still poorly understood. P115 binds the golgin GM13041 and GM130 is required for Golgi localization of p115.41,42 However, GM130 is a peripheral component and its targeting mechanism is also unclear. Further, the GM130 binding site on p115 is not required for p115 Golgi localization42 implying that, in addition to binding GM130, p115 binds another GM130-dependent factor for its Golgi localization. One possibility is that Rab1 plays a role establishing a positive feedback loop in which Rab1 first promotes p115 recruitment and then p115 promotes further Rab1 activation. The head domain site30 may be used for p115 recruitment causing a conformational change of p115, which activates GDF activity of the cc2 domain of p115.
The mechanistic details of GDF-catalyzed dissociation of Rab-GDI complexes remain unknown. The only well characterized endogenous Rab GDF is the integral membrane protein Yip3/PRA1, which functions to dissociate the Rab9-GDI complex leading to Rab9 recruitment to endosomal membranes.43,44 Interestingly, during infection the SidM/DrrA protein from the intracellular pathogen Legionella pneumophila acts as both GEF and GDF for Rab1 to mediate membrane recruitment of Rab1 to Legionella-containing vacuoles.35,45 Moreover, the GEF activity of SidM/DrrA is sufficient to displace Rab1 from the Rab1:GDI complex.46 In contrast, we observed that thep115 cc2 domain promoted dissociation of the Rab1b-GDI2 complex but failed to enhance nucleotide exchange. One possible mechanism underlying this process is that p115 binds directly to the Rab1b-GDI complex causing a conformational change that promotes the release of GDI. Another possibility is that the p115 cc2 domain competes with GDI for the same binding site on Rab1b. An important area of future work will be to determine the exact mechanism of GDI release.
In addition to its role in membrane recruitment of COPI, Rab1b regulates COPII dynamics and cargo sorting at the ER exit sites.47,48 P115 and Rab1 interactions take place during exit from the ER on the membranes of nascent COPII vesicles where p115 also interacts with SNARE proteins.16 SNARE binding by p115 is direct21 and required for p115 function.19 Several SNARE proteins also bind directly to the COPII coat. These observations suggest that both p115/SNARE interactions and p115/Rab1 interactions may facilitate incorporation of p115 into COPII vesicles. Thus, p115 and Rab1 recruitment takes place at precisely the time and place of establishment of post-ER identity and appears to also involve interactions with the SNARE proteins that will specify fusion of the released COPII vesicles.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Yang Yang and Cassandra Priddy for critical technical assistance and also thank Collin Bachert, Tina Lee, and Debrup Sengupta for comments on the manuscript. We thank Dr. RR Isberg (Tufts Univ. School of Medicine) and Dr. MP Machner (NIH) for kindly providing yeast strain FY834. Funding was provided by the NIH grant GM-56779 to ADL.
References
- 1.Puri S, Linstedt AD. Capacity of the golgi apparatus for biogenesis from the endoplasmic reticulum. Mol Biol Cell. 2003;14:5011–8. doi: 10.1091/mbc.E03-06-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu D, Hay JC. Reconstitution of COPII vesicle fusion to generate a pre-Golgi intermediate compartment. J Cell Biol. 2004;167:997–1003. doi: 10.1083/jcb.200408135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aridor M, Bannykh SI, Rowe T, Balch WE. Sequential coupling between COPII and COPI vesicle coats in endoplasmic reticulum to Golgi transport. J Cell Biol. 1995;131:875–93. doi: 10.1083/jcb.131.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–66. doi: 10.1016/S0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 5.Puthenveedu MA, Linstedt AD. Subcompartmentalizing the Golgi apparatus. Curr Opin Cell Biol. 2005;17:369–75. doi: 10.1016/j.ceb.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 6.Palmer DJ, Helms JB, Beckers CJ, Orci L, Rothman JE. Binding of coatomer to Golgi membranes requires ADP-ribosylation factor. J Biol Chem. 1993;268:12083–9. [PubMed] [Google Scholar]
- 7.Beck R, Rawet M, Wieland FT, Cassel D. The COPI system: molecular mechanisms and function. FEBS Lett. 2009;583:2701–9. doi: 10.1016/j.febslet.2009.07.032. [DOI] [PubMed] [Google Scholar]
- 8.Lee MC, Miller EA, Goldberg J, Orci L, Schekman R. Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- 9.Alvarez C, Garcia-Mata R, Brandon E, Sztul E. COPI recruitment is modulated by a Rab1b-dependent mechanism. Mol Biol Cell. 2003;14:2116–27. doi: 10.1091/mbc.E02-09-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monetta P, Slavin I, Romero N, Alvarez C. Rab1b interacts with GBF1 and modulates both ARF1 dynamics and COPI association. Mol Biol Cell. 2007;18:2400–10. doi: 10.1091/mbc.E06-11-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seabra MC, Wasmeier C. Controlling the location and activation of Rab GTPases. Curr Opin Cell Biol. 2004;16:451–7. doi: 10.1016/j.ceb.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 12.Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc Natl Acad Sci U S A. 2006;103:11821–7. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wurmser AE, Sato TK, Emr SD. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J Cell Biol. 2000;151:551–62. doi: 10.1083/jcb.151.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lippé R, Miaczynska M, Rybin V, Runge A, Zerial M. Functional synergy between Rab5 effector Rabaptin-5 and exchange factor Rabex-5 when physically associated in a complex. Mol Biol Cell. 2001;12:2219–28. doi: 10.1091/mbc.12.7.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Medkova M, France YE, Coleman J, Novick P. The rab exchange factor Sec2p reversibly associates with the exocyst. Mol Biol Cell. 2006;17:2757–69. doi: 10.1091/mbc.E05-10-0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allan BB, Moyer BD, Balch WE. Rab1 recruitment of p115 into a cis-SNARE complex: programming budding COPII vesicles for fusion. Science. 2000;289:444–8. doi: 10.1126/science.289.5478.444. [DOI] [PubMed] [Google Scholar]
- 17.Waters MG, Clary DO, Rothman JE. A novel 115-kD peripheral membrane protein is required for intercisternal transport in the Golgi stack. J Cell Biol. 1992;118:1015–26. doi: 10.1083/jcb.118.5.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alvarez C, Fujita H, Hubbard A, Sztul E. ER to Golgi transport: Requirement for p115 at a pre-Golgi VTC stage. J Cell Biol. 1999;147:1205–22. doi: 10.1083/jcb.147.6.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puthenveedu MA, Linstedt AD. Gene replacement reveals that p115/SNARE interactions are essential for Golgi biogenesis. Proc Natl Acad Sci U S A. 2004;101:1253–6. doi: 10.1073/pnas.0306373101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sztul E, Lupashin V. Role of vesicle tethering factors in the ER-Golgi membrane traffic. FEBS Lett. 2009;583:3770–83. doi: 10.1016/j.febslet.2009.10.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shorter J, Beard MB, Seemann J, Dirac-Svejstrup AB, Warren G. Sequential tethering of Golgins and catalysis of SNAREpin assembly by the vesicle-tethering protein p115. J Cell Biol. 2002;157:45–62. doi: 10.1083/jcb.200112127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barlowe C. Coupled ER to Golgi transport reconstituted with purified cytosolic proteins. J Cell Biol. 1997;139:1097–108. doi: 10.1083/jcb.139.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao X, Ballew N, Barlowe C. Initial docking of ER-derived vesicles requires Uso1p and Ypt1p but is independent of SNARE proteins. EMBO J. 1998;17:2156–65. doi: 10.1093/emboj/17.8.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morsomme P, Riezman H. The Rab GTPase Ypt1p and tethering factors couple protein sorting at the ER to vesicle targeting to the Golgi apparatus. Dev Cell. 2002;2:307–17. doi: 10.1016/S1534-5807(02)00133-8. [DOI] [PubMed] [Google Scholar]
- 25.Sohda M, Misumi Y, Yoshimura S, Nakamura N, Fusano T, Ogata S, Sakisaka S, Ikehara Y. The interaction of two tethering factors, p115 and COG complex, is required for Golgi integrity. Traffic. 2007;8:270–84. doi: 10.1111/j.1600-0854.2006.00530.x. [DOI] [PubMed] [Google Scholar]
- 26.Guo Y, Punj V, Sengupta D, Linstedt AD. Coat-tether interaction in Golgi organization. Mol Biol Cell. 2008;19:2830–43. doi: 10.1091/mbc.E07-12-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grabski R, Hay J, Sztul E. Tethering factor P115: A new model for tether-SNARE interactions. Bioarchitecture. 2012;2:2. doi: 10.4161/bioa.21702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grabski R, Balklava Z, Wyrozumska P, Szul T, Brandon E, Alvarez C, Holloway ZG, Sztul E. Identification of a functional domain within the p115 tethering factor that is required for Golgi ribbon assembly and membrane trafficking. J Cell Sci. 2012;125:1896–909. doi: 10.1242/jcs.090571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beard M, Satoh A, Shorter J, Warren G. A cryptic Rab1-binding site in the p115 tethering protein. J Biol Chem. 2005;280:25840–8. doi: 10.1074/jbc.M503925200. [DOI] [PubMed] [Google Scholar]
- 30.An Y, Chen CY, Moyer B, Rotkiewicz P, Elsliger MA, Godzik A, Wilson IA, Balch WE. Structural and functional analysis of the globular head domain of p115 provides insight into membrane tethering. J Mol Biol. 2009;391:26–41. doi: 10.1016/j.jmb.2009.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.García-Mata R, Sztul E. The membrane-tethering protein p115 interacts with GBF1, an ARF guanine-nucleotide-exchange factor. EMBO Rep. 2003;4:320–5. doi: 10.1038/sj.embor.embor762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puthenveedu MA, Linstedt AD. Evidence that Golgi structure depends on a p115 activity that is independent of the vesicle tether components giantin and GM130. J Cell Biol. 2001;155:227–38. doi: 10.1083/jcb.200105005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo Y, Linstedt AD. COPII-Golgi protein interactions regulate COPII coat assembly and Golgi size. J Cell Biol. 2006;174:53–63. doi: 10.1083/jcb.200604058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Linstedt AD, Hauri HP. Giantin, a novel conserved Golgi membrane protein containing a cytoplasmic domain of at least 350 kDa. Mol Biol Cell. 1993;4:679–93. doi: 10.1091/mbc.4.7.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Machner MP, Isberg RR. A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science. 2007;318:974–7. doi: 10.1126/science.1149121. [DOI] [PubMed] [Google Scholar]
- 36.Otani T, Ichii T, Aono S, Takeichi M. Cdc42 GEF Tuba regulates the junctional configuration of simple epithelial cells. J Cell Biol. 2006;175:135–46. doi: 10.1083/jcb.200605012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Claude A, Zhao BP, Kuziemsky CE, Dahan S, Berger SJ, Yan JP, Armold AD, Sullivan EM, Melançon P. GBF1: A novel Golgi-associated BFA-resistant guanine nucleotide exchange factor that displays specificity for ADP-ribosylation factor 5. J Cell Biol. 1999;146:71–84. [PMC free article] [PubMed] [Google Scholar]
- 38.Sapperstein SK, Lupashin VV, Schmitt HD, Waters MG. Assembly of the ER to Golgi SNARE complex requires Uso1p. J Cell Biol. 1996;132:755–67. doi: 10.1083/jcb.132.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang W, Sacher M, Ferro-Novick S. TRAPP stimulates guanine nucleotide exchange on Ypt1p. J Cell Biol. 2000;151:289–96. doi: 10.1083/jcb.151.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamasaki A, Menon S, Yu S, Barrowman J, Meerloo T, Oorschot V, Klumperman J, Satoh A, Ferro-Novick S. mTrs130 is a component of a mammalian TRAPPII complex, a Rab1 GEF that binds to COPI-coated vesicles. Mol Biol Cell. 2009;20:4205–15. doi: 10.1091/mbc.E09-05-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakamura N, Lowe M, Levine TP, Rabouille C, Warren G. The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell. 1997;89:445–55. doi: 10.1016/S0092-8674(00)80225-1. [DOI] [PubMed] [Google Scholar]
- 42.Puthenveedu MA, Bachert C, Puri S, Lanni F, Linstedt AD. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat Cell Biol. 2006;8:238–48. doi: 10.1038/ncb1366. [DOI] [PubMed] [Google Scholar]
- 43.Sivars U, Aivazian D, Pfeffer SR. Yip3 catalyses the dissociation of endosomal Rab-GDI complexes. Nature. 2003;425:856–9. doi: 10.1038/nature02057. [DOI] [PubMed] [Google Scholar]
- 44.Pfeffer S, Aivazian D. Targeting Rab GTPases to distinct membrane compartments. Nat Rev Mol Cell Biol. 2004;5:886–96. doi: 10.1038/nrm1500. [DOI] [PubMed] [Google Scholar]
- 45.Ingmundson A, Delprato A, Lambright DG, Roy CR. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature. 2007;450:365–9. doi: 10.1038/nature06336. [DOI] [PubMed] [Google Scholar]
- 46.Schoebel S, Oesterlin LK, Blankenfeldt W, Goody RS, Itzen A. RabGDI displacement by DrrA from Legionella is a consequence of its guanine nucleotide exchange activity. Mol Cell. 2009;36:1060–72. doi: 10.1016/j.molcel.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 47.Slavin I, García IA, Monetta P, Martinez H, Romero N, Alvarez C. Role of Rab1b in COPII dynamics and function. Eur J Cell Biol. 2011;90:301–11. doi: 10.1016/j.ejcb.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 48.García IA, Martinez HE, Alvarez C. Rab1b regulates COPI and COPII dynamics in mammalian cells. Cell Logist. 2011;1:159–63. doi: 10.4161/cl.1.4.18221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.