Abstract
The cyclic AMP/protein kinase A (cAMP/PKA) and regulation of Ace2 and morphogenesis (RAM) pathways are important regulators of the yeast-to-hypha transition in Candida albicans that interact genetically during this process. To further understand this interaction, we have characterized the expression of ACE2 during morphogenesis. In normoxic, planktonic conditions, ACE2 expression is very low in stationary-phase cells at both the mRNA and protein levels. Upon shifting to Spider medium, ACE2/Ace2p levels increase. Although Ace2 is not absolutely required for hypha formation, ace2Δ/Δ mutants show delayed hypha formation in Spider medium (but not others) and morphological changes to the hyphal tip and lateral yeast. We also show that Efg1 directly binds the promoter of Ace2 in stationary phase, and ACE2 levels are increased in strains lacking Efg1 and the protein kinase A proteins Tpk1 and Tpk2, indicating that the PKA pathway directly regulates ACE2 expression. ACE2 expression is positively regulated by Tec1 and Brg1, which bind the promoters of ACE2 in hyphal cells but not in the yeast phase. Under embedded conditions, Efg1 is dispensable for filamentation and Ace2 is required. We have found that ACE2 expression is much higher in embedded cells than in planktonic cells, providing a potential rationale for this observation. Taken together, our observations indicate that the PKA pathway directly regulates the RAM pathway under specific conditions and are consistent with a model where the two pathways carry out similar functions that depend on the specific environmental context.
INTRODUCTION
Candida albicans is one of the most important human fungal pathogens and causes disease in both immunocompetent and immunocompromised people (1, 2). Mucocutaneous diseases, such as vulvovaginitis and diaper dermatitis, affect those with intact immune systems. Oropharyngeal candidiasis, a more serious mucocutaneous disease, is a common complication of HIV/AIDs. Invasive bloodstream and deep-organ infections occur almost exclusively in patients with defects in innate immunity related to cancer chemotherapy, transplantation, or prematurity. The pathogenesis of candidiasis has been the subject of intense interest for many years. One of the characteristics of C. albicans that is associated with virulence is its ability to transition between three distinct morphological states: yeast, pseudohyphae, and true hyphae (3). Yeast-phase cells display the familiar oval cell bodies typical of budding yeast, such as the model organism Saccharomyces cerevisiae. Pseudohyphae have elongated daughter cells with septa at the bud neck and nonparallel cell wall structures in the elongated cell. True hyphae have a septum within the filament and parallel cell wall structures (4). Based on histological examination of infected tissues, all three morphotypes exist in the host during infection (5).
The role of C. albicans morphotypes in infection and disease is complex, and a complete understanding of the correlation between morphology and pathogenesis has yet to be achieved (6, 7). The landmark study of Lo et al. showed that C. albicans strains that fail to form hyphae or pseudohyphae also are less virulent (7, 8). However, a variety of studies over the years have demonstrated that the role of morphogenesis in C. albicans pathogenesis is more complex than a simple “no hyphae/no virulence” model (5, 9, 10, 11). A recent example of such a study is the large-scale genetic study using a library of C. albicans deletion mutants which found that many strains with no defect in morphogenesis were infectious; conversely, strains with normal virulence showed reduced infectivity (12). In addition, C. albicans cells conditionally locked in yeast form are able to disseminate in the bloodstream of mice and establish an infection; however, the mice do not develop disease unless hypha repression is relieved, suggesting that yeast and filaments play distinct roles in the pathogenesis of C. albicans infections (13).
C. albicans morphogenesis has been studied extensively, and a large number of genes play direct and/or indirect roles in the ability of C. albicans to filament. These studies have shown that many signaling pathways and transcription factors modulate morphogenesis (3). Until recently, little was known about how the various individual regulatory pathways interacted to orchestrate this complex biological process. As previously reported, we carried out the first genetic interaction screen in C. albicans as an approach to investigating how specific signaling networks interact during morphogenesis (14). The screen was based on the concept of complex haploinsufficiency and was applied to the identification of genes that interact with CBK1, the key protein kinase in the regulation of Ace2 and morphogenesis (RAM) pathway, during hyphal morphogenesis in response to Spider medium. Among the set of CBK1 interacting genes identified in the screen, a core set of genes were either direct targets of the Cbk1-regulated transcription factor Ace2 or were related to the cyclic AMP/protein kinase A (cAMP/PKA) pathway, suggesting that the RAM and PKA pathways interact during morphogenesis (14). The goal of the work described here was to investigate the mechanism through which the RAM and PKA pathways interact as well as to further characterize the role of Ace2 during morphogenesis.
Ace2 is a daughter cell-specific zinc finger transcription factor in S. cerevisiae and regulates genes related to cell separation, cell wall integrity, and other processes (15, 16, 17). The localization of Ace2 to the nucleus is closely coordinated with the cell cycle and requires phosphorylation by Cbk1 (18). In C. albicans, Ace2 also is required for proper cell separation and, similar to S. cerevisiae, localizes to daughter nuclei in both yeast and filamentous cells (19). Under conditions that do not induce filamentation in wild-type cells, ace2Δ/Δ mutants form increased numbers of pseudohyphae relative to the wild type (19). In the presence of serum, ace2Δ/Δ cells form hyphae in a manner indistinguishable from that of the wild type (19), whereas the deletion of Cbk1, the kinase that regulates Ace2, results in a complete inability to form either hyphae or pseudohyphae regardless of the inducing medium (20, 21). In contrast, ace2Δ/Δ mutants are deficient for filamentation induced by hypoxia or by the respiratory chain inhibitor antimycin A (22). In addition, Ace2 appears to be required for normal filamentation when cells are embedded in solid agar medium. Transcriptional profiling of ace2Δ/Δ mutants during yeast and serum-induced hyphal phases indicated that genes involved in respiration were upregulated while glycolytic genes were downregulated, suggesting that Ace2 represses respiration and promotes glycolysis under certain conditions (22).
Interestingly, deletion of Efg1, the major transcriptional regulator for the cAMP-PKA pathway and a master regulator of filamentation (23), leads to phenotypes that are opposite those of ace2Δ/Δ under some conditions (22). Specifically, efg1Δ/Δ is hyperfilamentous under hypoxic or embedded conditions and is severely deficient for filamentation in any inducing condition in the presence of normoxia. Since Efg1 also has been implicated in the modulation of genes involved in metabolism (24), these observations suggest that Ace2 and Efg1 have overlapping functions and that one or the other transcription factor plays the dominant role based on the specific environmental context.
In addition to our genetic interaction study linking the RAM and PKA pathways during C. albicans morphogenesis (14), connections between the RAM and PKA pathways in yeast have been characterized previously. First, Schneper et al. showed that, in S. cerevisiae, overexpression of the PKA catalytic subunit TPK1 suppressed growth and budding defects in RAM mutants (25). Second, Wang et al. showed that Efg1 binds to the promoters of Ace2-regulated cell separation genes and suppresses their expression early in morphogenesis in a Cdc28-dependent fashion (26). Finally, Gutierrez-Escribano et al. have shown that Cdc28 not only regulates Efg1-mediated Ace2 target suppression but also modulates activity of the Mob2-Cbk1 complex through phosphorylation of Mob2 (27).
In order to probe the scope and basis of the interactions between the RAM and PKA pathways during morphogenesis, we have characterized the regulation of Ace2 expression during morphogenesis as well as its role in morphogenesis under a variety of conditions. Importantly, we have found that the PKA pathway directly represses ACE2 expression during stationary phase, which provides a mechanistic explanation for the interaction between the two pathways. In addition, Tec1 and Brg1, two important regulators of gene expression during morphogenesis, positively regulate ACE2. Since Tec1 also is positively regulated by Efg1, Efg1 both positively regulates ACE2 expression (indirectly) and negatively regulates ACE2 expression (directly) depending upon the stage of hyphal induction. We also provide evidence that Ace2 expression is elevated under embedded conditions relative to planktonic conditions, and that it functions to suppress respiratory activity and positively regulate hyphal morphogenesis.
MATERIALS AND METHODS
Strains, media, and materials.
The genotypes and sources of the strains used in this study are compiled in Table S1 in the supplemental material. New strains were generated using standard PCR-based methods, and correct integrations were confirmed by PCR. For epitope-tagged strains, candidate strains were evaluated by Western blotting to ensure the presence of a band at the expected molecular weight (MW). Yeast culture media were prepared according to published recipes (12, 28). Hyphal morphogenesis was induced by dilution of a starter culture (overnight growth in yeast-peptone-dextrose medium [YPD] starting from a single colony) into fresh inducing medium and incubation at 37°C unless otherwise stated. To assess the hypha-to-yeast transition, an overnight culture was harvested, washed with fresh media, and diluted 1/50 into 6-well plates containing various media. Cells were incubated at 37°C for ∼16 h, washed, and visualized by light microscopy. All chemical reagents were obtained from Sigma Chemicals (St. Louis, MO) and used without further purification. Oligonucleotides were synthesized by IDT Technologies (Coralville, IA) and used as received.
Characterization of colony respiratory activity.
Characterization of the respiratory activity of yeast colonies was done as described by Morales et al. (29). Strains were grown overnight in YPD at 30°C, washed, and spotted (5 μl) onto plates with various media. The plates were incubated at 30°C or 37°C for the time periods indicated in the text. Colorless triphenyl tetrazolium chloride (TTC) was added to a final concentration of 0.1% in overlay agar (1.5% Bacto agar, 0.067 M potassium phosphate, pH 7) and poured over the colonies after the incubation time indicated in the text. To analyze the respiratory activity of embedded colonies, strains were spotted on yeast extract-peptone plus 2% sucrose (YPS) media, overlaid with soft agar, and incubated at 37°C for ∼48 h. At that time, ∼250 μl 40% TTC solution was spread over the plate, and the plate was incubated for an additional 24 h. Plates were photographed with a digital camera and the images processed in PhotoShop; contrast and color levels were applied equally to all strains on a given plate.
Transcriptional analysis by quantitative reverse transcription-PCR (qRT-PCR).
RNA was isolated from C. albicans strains using the RiboPure yeast kit (AM1926; Ambion) and reverse transcribed using an iScript cDNA synthesis kit (170-8891; Bio-Rad). Changes in transcript levels were analyzed using iQ SYBR green supermix (170-8882; Bio-Rad) and normalized to the housekeeping gene GSP1 using the 2−ΔΔCT method (30). Data are reported as the means from 2 to 3 independent biological replicates performed in triplicate. Expression differences between reference and mutant strains were analyzed by Student's t test with significance set at P < 0.05.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation was preformed essentially as described in reference 31, with a few modifications. Briefly, stationary- or hyphal-phase cells were fixed with formaldehyde and quenched with glycine, followed by vortexing in lysis buffer (50 mM Tris-HCl, pH 7, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate) for 2 h at 4°C. Lysates were recovered and chromatin sonicated using a Branson sonifier 150 (3 times for 20 s each, sonicator power setting of 20%, with 1 min on ice between each pulse). Cell debris was pelleted for 5 min at 13,200 rpm at 4°C, and supernatant was transferred to a fresh tube. Antibody (2 μg monoclonal anti-Myc antibody; R95025; Invitrogen) was added and allowed to incubate at 4°C with rotating for ∼16 h. Washed protein-G Sepharose (10-1241; Invitrogen) in 50% slurry was added to each sample and allowed to rotate for an additional 2 h at 4°C. Beads were washed 2× with lysis buffer, 2× with lysis buffer (high-salt lysis buffer plus 500 mM NaCl), 2× with wash buffer (10 mM Tris-HCl, 250 mM LiCl, 0.5% NP-40, 0.5% Na-deoxycholate, 1 mM EDTA), and 1× with TE (10 mM Tris-HCl, 1 mM EDTA). Protein-DNA complexes were eluted from beads with the addition of elution buffer (50 mM Tris-HCl, pH 7, 10 mM EDTA, 1% SDS) and incubated for 15 min at 65°C (with vortexing every 2 min). The supernatant was transferred to a fresh tube. TE plus SDS (0.67%) solution was added to the beads and vortexed for an additional 30 s. After elution of DNA-protein complexes and digestion of protein with proteinase K, DNA was cleaned up using a Qiagen PCR purification kit (28104). Enrichment of promoter regions in tagged strains was compared to that from untagged strains by qRT-PCR using primers designed for the region of the consensus transcription factor binding sites and according to the methods described above.
One-dimensional Western blotting.
Cell pellets were resuspended in ∼200 μl lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) with added protease inhibitor cocktail (Roche), phosphatase inhibitor cocktails II and III (Sigma), and beads. Cells were lysed by 10 20-s bursts with a bead beater. Total protein was determined using a Bio-Rad protein assay dye concentrate according to the manufacturer's instructions, and 30 to 50 μg of total protein was fractionated by SDS-PAGE using 10% Mini-Protean TGX gels (Bio-Rad). Fractionated proteins were transferred to nitrocellulose membranes and blocked overnight at 4°C (5% nonfat milk in 50 mM Tris [pH 7.5], 150 mM NaCl, and 0.05% Tween 20 [TBST]). The membranes were probed using anti-Myc antibody (Invitrogen) followed by anti-mouse horseradish peroxidase (HRP)-conjugated secondary antibody (Bio-Rad). Anti-PSTAIRE antibody was used as a loading control (Upstate/Millipore). Blots were visualized with the ECL plus Western blotting detection system (GE Healthcare).
Two-dimensional Western blotting.
Stationary-phase or hyphal-phase C. albicans cells were lysed in buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7, 15 mM EDTA, 1% Triton X-100, 10% glycerol) plus phosphatase inhibitors (P5726 and P0044; Sigma) and protease inhibitors (complete miniprotease inhibitor cocktail tablets; 04693124001; Roche) with glass beads and placed in a bead beater for 10 cycles of 20 s each, with 1 min on ice between pulses. Cell extracts were precipitated with methanol-chloroform and resuspended in rehydration buffer (163-2105; Bio-Rad). Approximately 30 μg protein was loaded onto 7-cm immobilized pH gradient (IPG) strips (pH 3 to 10; 163-2000; Bio-Rad) and resolved using a Bio-Rad Protean i12 isoelectric focusing (IEF) system.
Microscopy.
Light and fluorescence microscopy was performed using a Nikon ES80 epifluorescence microscope equipped with a CoolSnap charge-coupled device (CCD) camera. Images were collected using NIS-Elements software and processed in PhotoShop. Indirect immunofluorescence for the localization of Efg1-Myc was performed as previously described (14) using anti-Myc (Invitrogen) primary and Texas red-conjugated (Molecular Probes) secondary antibodies. 4′,6-Diamidino-2-phenylindole (DAPI) staining was performed as described previously (28). The expression of ACE2 and EFG1 using the pEFG1-GFP and pACE2-mCherry reporter strains was determined using the methods described by Pierce and Kumamoto (32). Briefly, strains were grown to saturation in YPD. Light and fluorescence microscopy was performed, and images were processed as described above. ImageJ (NIH) software was used to quantify the fluorescence density of cells. For each condition, at least 100 cells were imaged, and experiments were performed in duplicate. For the embedded conditions, 100 μl of saturated culture was plated onto YPS, the culture was dried, ∼15 ml molten YPS was poured on top, and then the culture was incubated. For fluorescence microscopy, cells were extracted from the plate using a Pasteur pipette and vortexed with ∼500 μl PBS until agar appeared emulsified. Tubes were centrifuged briefly at 200 × g in a microcentrifuge tube to pellet agar. Cells were washed from the pellet and placed on slides for visualization.
RESULTS
Loss of Ace2 function delays filamentation in Spider medium and affects hyphal and lateral yeast morphology.
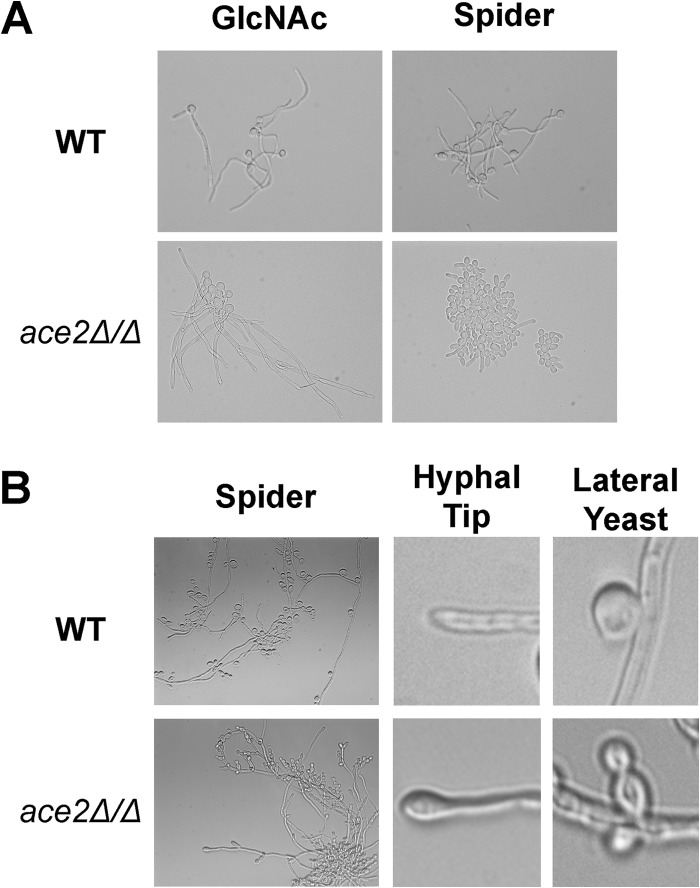
Previously, Ace2 was shown to be required for formation of true hyphae under hypoxic conditions, under embedded conditions, and in response to the respiratory chain inhibitor antimycin A (22). In contrast, ace2Δ/Δ cells form normal hyphae in response to serum and following phagocytosis by macrophages (19, 22). These observations suggested that the role of Ace2 in filamentation varies with the specific hypha-inducing conditions. To further characterize the role of Ace2 in the formation of true hyphae, we examined the ability of ace2Δ/Δ cells to filament in a variety of hypha-inducing conditions. We performed the experiments at 37°C using the following liquid media: YPD plus 10% serum, Spider medium, M199, RPMI-morpholinepropanesulfonic acid (MOPS), N-acetylglucosamine, and Casamino Acids medium (8). For each condition, an overnight culture of the strains (YPD, 30°C with shaking) was diluted (1:50) into the inducing medium and shifted to 37°C for 3 h, and the cells were examined by microscopy. For all but Spider medium, ace2Δ/Δ cells formed true hyphae to a similar extent and within a similar time frame as the reference strain (Fig. 1A; also see Fig. S1 in the supplemental material). In Spider medium at 3 h, ace2Δ/Δ cells were primarily pseudohyphal, while the vast majority of WT cells had formed hyphae (Fig. 1A). This is consistent with previous studies indicating that, on Spider medium agar, ace2Δ/Δ colonies have increased numbers of pseudohyphal cells relative to hyphal cells (14, 22). In liquid Spider medium, true hyphae were present in all samples, and after incubation overnight, the major morphotype was hyphae, indicating that Ace2 is not absolutely required for hyphal formation under these conditions. Taken together, these results indicate that Ace2 is dispensable for hyphal formation and mainly affects the rate of hyphal formation in Spider medium.
FIG 1.
Morphologies of hyphae and lateral yeast are altered in ace2Δ/Δ mutants. (A) Micrographs of WT (CAF2) and ace2Δ/Δ strains after 3 h of incubation in the indicated medium at 37°C. (B) Micrographs of WT and ace2Δ/Δ mutants after 18 h of incubation in Spider medium, showing general formation of hyphae with lateral yeast (left), the morphology of the hyphal tip (center), and the morphology of lateral yeast emerging from hyphae (right).
The filamentous cells formed by ace2Δ/Δ in Spider medium fit the definition of true hyphae in that they have parallel membranes and septa within the filament. However, the hyphae did display morphological differences from the wild type that were particularly apparent after overnight incubation. Under these conditions, the hyphal tip of ace2Δ/Δ cells was swollen or bulging relative to that of the wild type (Fig. 1B). This hyphal tip morphology has been reported previously in strains of C. albicans in which EFG1 is overexpressed from the inducible PCK1 promoter (33). We have previously reported EFG1 levels are elevated in ace2Δ/Δ (14); thus, this phenotype may be related to the increased expression of EFG1. The lateral yeast cells that emerge from ace2Δ/Δ hyphae also are altered from that observed with the wild-type strain (Fig. 1B). Specifically, WT lateral yeast forms generally are single cells that are nearly spherical. In contrast, lateral yeast cells in ace2Δ/Δ mutants are elongated, pseudohypha-like cells. In addition, multiple buds were present at most sites of lateral yeast emergence. The latter finding is consistent with the role of Ace2 in the regulation of genes involved in cell separation (22). The pseudohyphal morphology also is consistent with the reported increased pseudohyphal morphology of ace2Δ/Δ cells under non-filament-inducing conditions (14, 19). As with the bulging hyphal tip phenotype, pseudohypha-like lateral yeast cells also were observed in strains overexpressing EFG1. Thus, Ace2 does not appear to affect the ability of hyphae to transition to yeast cells; however, the hyphae and the lateral yeast cells that emerge from ace2Δ/Δ display morphological changes that are consistent with the effects of elevated EFG1 expression (14). As such, these data provide additional support for the interaction between the PKA-Efg1 pathway and the RAM pathway.
Characterization of Efg1 and Ace2 during morphogenesis.
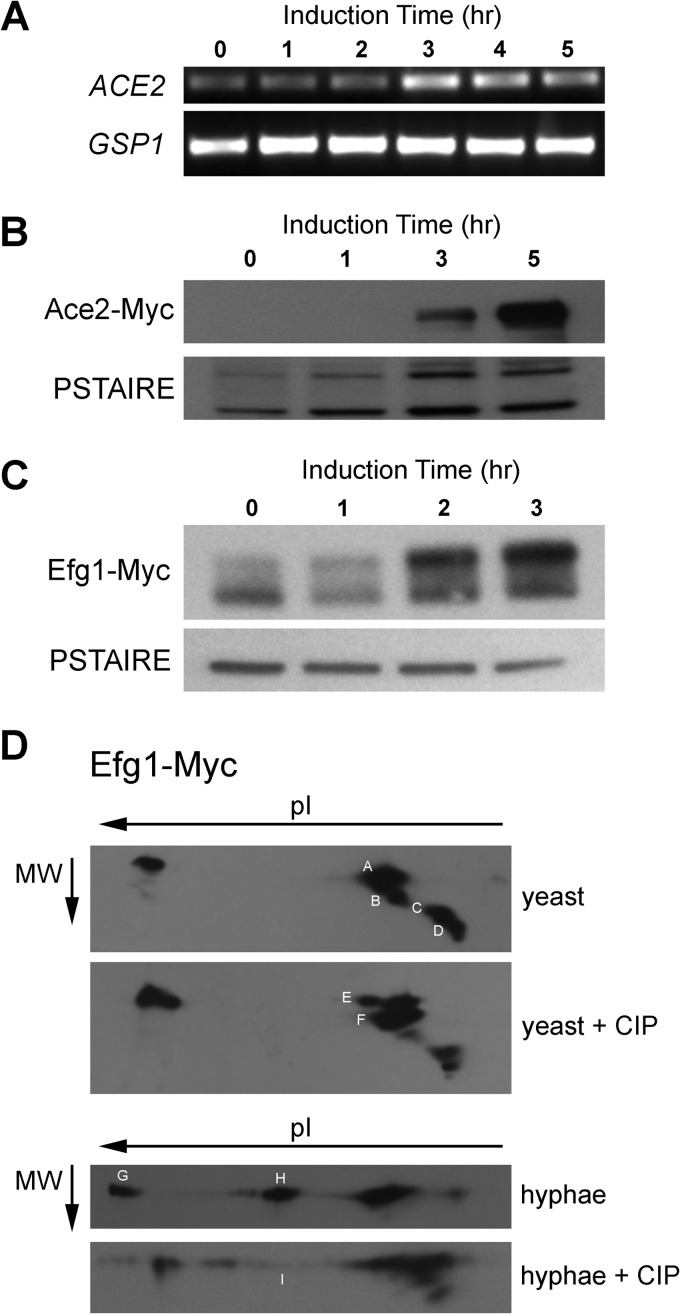
To further explore the relationship between the RAM and PKA pathways during morphogenesis, we characterized the dynamics of expression, localization, and modification of the RAM and PKA transcription factors, Ace2 and Efg1, respectively. To our knowledge, very little is known about the expression of Ace2 during hyphal morphogenesis (33). Therefore, we first examined the expression of ACE2 over a time course of hyphal induction in Spider medium. Stationary-phase cells were shifted to Spider medium at 37°C. Aliquots of cells were harvested every 1 h over a 5-h course of induction, and ACE2 levels were determined by semiquantitative RT-PCR. As shown in Fig. 2A, ACE2 expression is quite low in stationary-phase cells but increases as hyphal cells emerge. Previous studies have shown that EFG1 expression decreases slightly upon initiation of hyphal morphogenesis and that this decrease is attributable to an autoregulatory process; we observed expression changes consistent with these findings (data not shown). To determine if these changes in gene expression translated to changes in protein levels, we examined the levels of Ace2 and Efg1 proteins using strains with C-terminal fusions to 13×-MYC epitope tags. Consistent with the low levels of ACE2 in stationary-phase cells, Ace2-Myc was undetectable by Western blotting at the initiation of morphogenesis; however, Ace2 levels increased significantly over the time course of morphogenesis in Spider medium (Fig. 2B). Taken together, these data indicate that Ace2 is not expressed in stationary-phase cells but is induced as cells enter hyphal growth.
FIG 2.
Characterization of Ace2 and Efg1 expression and phosphorylation in yeast and hyphae. (A) Semiquantitative RT-PCR of ACE2 expression during hyphal induction in Spider medium. GSP1 expression is shown as a control. The time after the shift to inducing conditions is shown. (B and C) WT strains containing an allele of ACE2 (B) or EFG1 (C) with a C-terminal MYC epitope tag was shifted to Spider medium at 37°C. At the indicated time, samples were harvested, lysed, and processed for Western blotting with anti-Myc antibodies. PSTAIRE antibody was used as a loading control. (D) Lysates of yeast- and hypha-phase cells containing Efg1-Myc were treated with calf intestine phosphatase (CIP) or left untreated, fractionated by two-dimensional electrophoresis, transferred to nitrocellulose membranes, and subjected to immunoblotting with anti-Myc antibodies.
In Spider medium, Efg1 protein levels appear to decrease upon the shift to inducing conditions (Fig. 2C, 1-h time point), consistent with previous mRNA studies and with the notion that Efg1 undergoes negative autoregulation initially (33). By 2 h, Efg1 levels are restored and slightly exceed that in stationary phase (Fig. 2C). As observed by others (34), Efg1 has an electrophoretic mobility that is much slower than predicted by its molecular mass. In addition, multiple species are present in the immunoblots. In Spider medium, a higher-mobility species becomes predominant as the cells proceed through morphogenesis (Fig. 2C). Efg1 is a phosphoprotein that is modified by both Cdc28 (26) and PKA (35), suggesting that this shift in mobility is related to phosphorylation. Consistent with this notion, previous studies from the Ernst laboratory have shown that the mobility of Efg1 is altered by treatment with phosphatase (34).
To further characterize the phosphorylation state of Efg1 in stationary-phase yeast and hyphae, we performed two-dimensional electrophoresis followed by Western blotting using the Efg1-Myc-containing strain. As shown in Fig. 2D, Efg1-Myc is a complex mixture of species that have different apparent molecular weights and pIs; major bands are denoted A, B, C, and D in Fig. 2D. Treatment of stationary-phase samples with phosphatase decreases the apparent abundance of the higher-molecular-weight E band (phosphatase-treated sample) and increases the abundance of the lower-molecular-weight F band (phosphatase-treated sample). The pI of band D changes slightly, while that of band C decreases in apparent abundance relative to the sample that was not treated with phosphatase.
In hyphal-phase cells, the mixture is somewhat simpler with respect to the number of species; as such, it corresponds well with the one-dimensional Western blot shown in Fig. 2C. Relative to the yeast-phase blot, two new bands are seen at pIs that are suggestive of potential phosphorylated species (Fig. 2D, bands G and H). Consistent with this interpretation, treatment of hyphal-phase lysates with phosphatase leads to the disappearance of bands G and H as well as a shift in the major hyphal band to the less electronegative band I. These findings provide strong evidence that the phosphorylation state of Efg1 shifts significantly between stationary-phase and hyphal-phase cells. Attempts to correlate specific bands with the canonical Efg1-related kinases PKA (35) and Cdc28 (26) have not been successful to this point. These attempts were limited by the fact that Efg1 mutants or treatment with PKA inhibitors leads to changes in the mobility of nearly all of the bands. In addition, the extent and tempo of morphogenesis is drastically altered under those conditions and further prevents the interpretation of the experiments. Nevertheless, our results provide strong evidence indicating that Efg1 phosphorylation varies during morphogenesis.
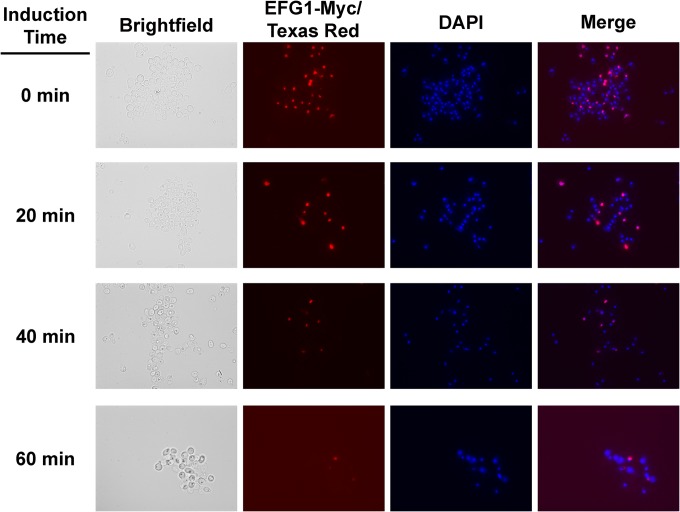
Previously, we reported that Efg1 is relatively abundant in the nuclei of stationary-phase cells and essentially undetectable in the nuclei of hyphal cells (14). To further characterize the nuclear localization patterns of Efg1, we performed a time course experiment in which samples of cells were collected every 20 min after the shift to hypha-inducing conditions, fixed, and processed for immunofluorescence to detect Efg1-Myc. As shown in Fig. 3, the number of cells with detectable amounts of Efg1 in the nuclei decreases rapidly upon the shift to inducing medium (Spider medium, 37°C). By 1 h of exposure to inducing conditions, only rare cells have detectable levels of Efg1 colocalized with nuclei; similar results are seen with cells shifted to serum-inducing conditions (data not shown). Importantly, yeast cells show a decrease in Efg1 nuclear localization, suggesting that the reduction is related to initiation of hyphae. This rapid decrease in nuclear abundance corresponds to the time frame when EFG1 expression and protein levels decrease (Fig. 2A). However, if the decrease in the amount of Efg1 detectable in the nucleus was directly related to total cellular Efg1, we would expect it to reaccumulate later during morphogenesis as Efg1 levels increase. Our previous results indicated that at 3 h, neither hyphae nor yeast cells have significant amounts of Efg1 detectable by immunofluorescence (14). Therefore, the decrease in nuclear Efg1 does not appear to be a simple function of protein levels.
FIG 3.
Amount of Efg1 in the nucleus of cells rapidly decreases upon shift to hypha-inducing conditions. WT cells containing an allele of EFG1 with a C-terminal MYC epitope tag was shifted to Spider medium at 37°C. Samples were harvested at the indicated time points and processed for immunofluorescence with anti-Myc primary and Texas red secondary antibodies. Nuclei were stained with DAPI as described in Materials and Methods.
We speculated that phosphorylation by PKA (35) or Cdc28 (26) regulates nuclear localization. However, treatment of stationary-phase cells with the substrate-based PKA inhibitor MyrPKI at concentrations that prevent hypha formation had no effect on the decrease in nuclear Efg1 upon shifting to Spider medium. In addition, the known PKA inhibitor farnesol had no effect (data not shown). Similarly, EFG1-MYC alleles containing mutations that either prevent (efg1T179A) or mimic (efg1T179D) phosphorylation by Cdc28 had no effect on the reduction in nuclear Efg1 (26). At this point, we have not identified the mechanism controlling Efg1 nuclear localization. However, it is important to emphasize that it is unlikely that Efg1 completely leaves the nucleus. Efg1 appears to be a relatively low-abundance protein. For example, we have been unable to detect Efg1-mCherry or Efg1-GFP fusion proteins by microscopy (D. Krysan, unpublished results); similarly, others have reported that Efg1 is a low-abundance protein in Spider medium (36). Other studies have clearly shown that Efg1 binds to the promoters of genes in hyphal-phase cells (34, 37). We feel it is more likely that the extent of nuclear Efg1 decreases during hyphal morphogenesis relative to levels of stationary-phase cells and that nuclear Efg1 is simply below levels of detection.
Efg1 represses ACE2 expression in a protein kinase A-dependent fashion.
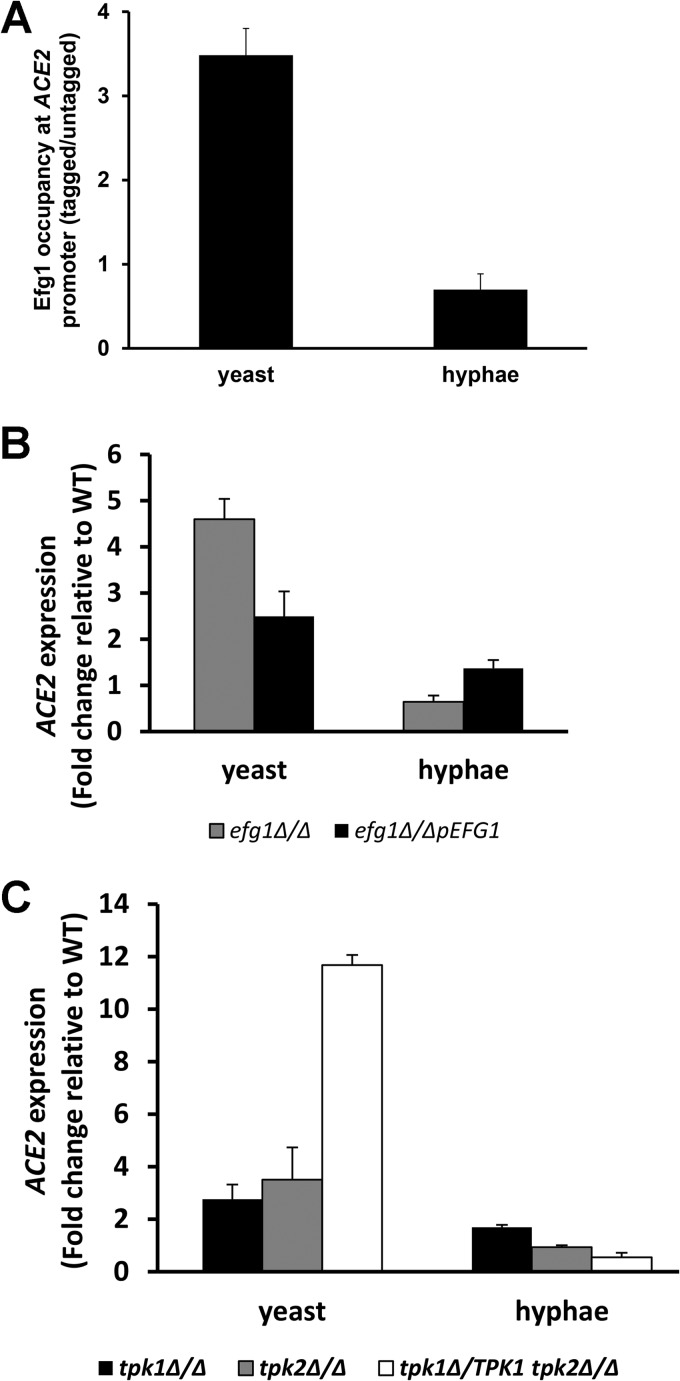
Our previously reported genetic interaction studies indicated that the RAM and PKA pathways interact during morphogenesis (14). We hypothesized that a direct interaction between the pathways occurs at the transcriptional level. Efg1 is a well-characterized transcriptional regulator that both activates and represses genes involved in morphogenesis, the white-opaque transition, biofilm formation, and essential cellular processes, such as glycolysis and respiration (24, 34, 37, 38). Supporting this hypothesis is the fact that the promoter region of ACE2 contains six putative Efg1 binding sites (E boxes; CANNTG) as well as a consensus binding site (TGACAT) recently formulated by Nobile et al. (38), suggesting that Efg1 directly regulates ACE2 expression. We performed chromatin immunoprecipitation experiments using both stationary-phase and hyphal cells derived from a strain containing an EFG1 allele with the C-terminal fusion of the 13×-MYC epitope tag (26). In stationary-phase cells, Efg1-Myc bound to the promoter of ACE2 (Fig. 4A). In contrast, there was no enrichment relative to an untagged control strain in hyphal cells. We then compared the expression of ACE2 in a homozygous EFG1 deletion mutant and its corresponding complement to the WT in stationary-phase cells and after 3 h of exposure to hypha-inducing conditions (Spider medium, 37°C) by quantitative RT-PCR. As shown in Fig. 4B, the expression of ACE2 is elevated in a homozygous deletion mutant in stationary-phase cells, while after shifting to hypha-inducing conditions, the levels are essentially identical to those of the wild type. The complemented strain has a reduced level of ACE2 expression relative to the homozygous deletion mutant. Increased ACE2 expression was observed for efg1Δ/Δ mutants in both the CAI and SN (12) genetic backgrounds (data not shown). These data suggest that Efg1 directly represses ACE2 expression in stationary-phase cells while having a less important role in its regulation after the formation of hyphae.
FIG 4.
Efg1 directly represses the expression of ACE2 in a manner dependent on the PKA kinases Tpk1 and Tpk2. (A) Chromatin immunoprecipitation with Efg1-Myc in yeast and hyphal cells. The presence of ACE2 in the precipitates was assayed by quantitative real-time PCR with primers spanning the consensus Efg1 binding sites. The bars represent the mean enrichment in tagged samples relative to that of untagged samples, and error bars indicate standard deviations from 3 biological replicates. (B and C) ACE2 expression was determined by RT-PCR for the indicated strains. Bars represent mean ACE2 expression in the mutant strain relative to that of the WT (fold change). Error bars represent standard deviations from 2 to 3 biological replicates assayed in triplicate. Yeast cells were from an overnight culture in YPD at 30°C (stationary phase), and hyphal cells were generated by shifting stationary-phase cells to Spider medium at 37°C for 3 h.
Efg1 is a well-characterized transcriptional regulator downstream of the cAMP/PKA pathway (35); therefore, we expected mutations of the PKA kinases TPK1 and TPK2 would affect ACE2 expression as well. To test this hypothesis, we utilized a strain with either TPK1 deleted or TPK2 deleted or a strain lacking both alleles of TPK1 and heterozygous at the TPK2 locus (tpk1Δ/Δ tpk2Δ/TPK2; generous gift of C. d'Enfert) (these strains were constructed independently in different laboratories and are not derivatives of one another). As shown in Fig. 4C, the expression of ACE2 is elevated relative to that of the WT in all mutants in stationary-phase cells. These data indicate that the PKA pathway represses ACE2 expression in stationary phase in a process dependent on both Efg1 and Tpk1/2, and that the repression is relieved as the cells transition to the hyphal stage. Although the promoter region of EFG1 contains sequences that correspond to the consensus binding sites for Ace2, we were unable to detect Ace2 at the promoter of EFG1 by chromatin immunoprecipitation (data not shown). Previously, we had shown that EFG1 expression is elevated in RAM pathway mutants such as ace2Δ/Δ, but this appears to be due to an indirect mechanism (14).
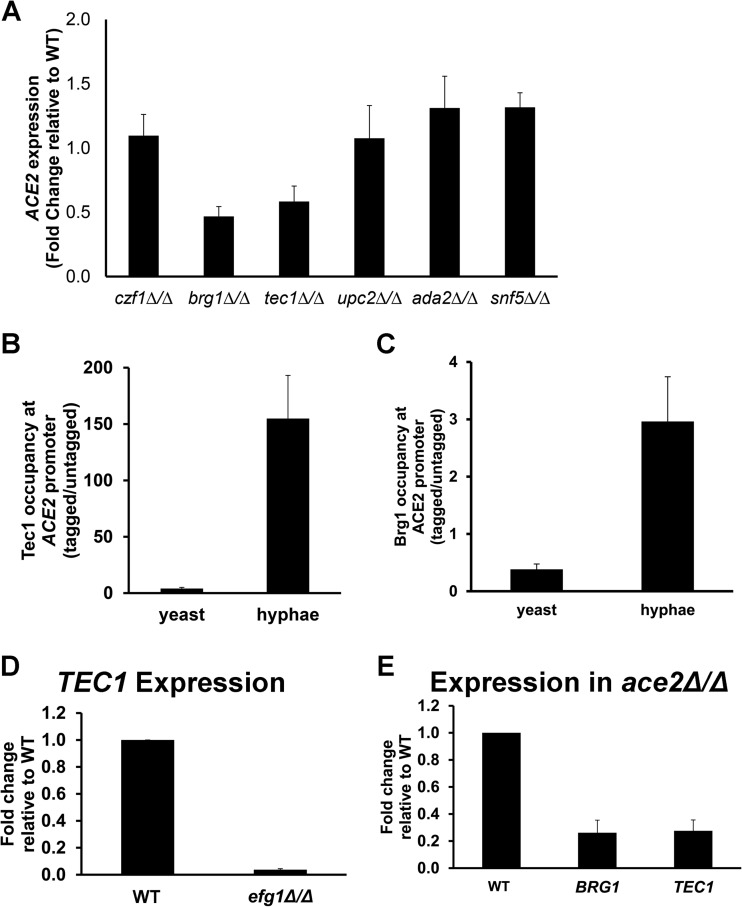
Tec1 and Brg1 positively regulate ACE2 expression during morphogenesis.
To further characterize the regulation of ACE2 transcription during morphogenesis, we sought to identify transcription factors that positively regulate its expression upon the shift to hypha-inducing conditions. An examination of the promoter region of ACE2 indicated the presence of consensus binding sites for a number of transcriptional regulators, including Ace2, Snf5, Czf1, Brg1, Tec1, and Ada2. The chromatin remodeling protein Snf5 recently has been shown to be required for Ace2 expression during biofilm formation (39). In addition, Czf1 is a transcriptional regulator that opposes the functions of Efg1 (40); accordingly, we speculated that it participates in ACE2 regulation. Therefore, we examined the expression of ACE2 in czf1Δ/Δ, snf5Δ/Δ, brg1Δ/Δ, tec1Δ/Δ, and ada2Δ/Δ mutants after 3 h of exposure to hypha-inducing conditions (Spider medium, 37°C). Loss of Czf1 function did not lead to a statistically significant decrease in ACE2 expression, as would be expected if it was required to relieve Efg1-mediated ACE2 suppression (Fig. 5A). ACE2 expression was significantly reduced (∼2-fold) in both tec1Δ/Δ and brg1Δ/Δ mutants relative to a genetically matched reference strain. Ada2, which has been shown to bind the ACE2 promoter by ChIP-Seq (41), did not have a statistically significant effect on ACE2 expression under these conditions. Somewhat surprisingly, Snf5 did not have a significant effect on ACE2 expression. This result suggests that the regulation of ACE2 expression during morphogenesis is distinct from its regulation during biofilm formation (39).
FIG 5.
ACE2 expression is positively regulated by Tec1 and Brg1 during hyphal morphogenesis. (A) ACE2 expression was determined by RT-PCR for the indicated strains after shifting to Spider medium for 3 h at 37°C. The bars indicate the mean expression of the mutant relative to that of the WT. Error bars indicate standard deviations from 2 to 3 biological replicates assayed in triplicate. Asterisks indicate strains with a statistically significant reduction from the WT level (P < 0.05, Student's t test).
To determine whether the effects of Tec1 and Brg1 on ACE2 expression were direct, we performed chromatin immunoprecipitation experiments with stationary-phase yeast cells and hyphal cells using strains harboring Myc-tagged Tec1 and Brg1. Tec1 does not bind to the ACE2 promoter in stationary-phase cells but is present after 3 h of induction in Spider medium (Fig. 5B). Similarly, Brg1 binds to the promoter of Ace2 in the hyphal phase but not during the yeast phase (Fig. 5C). Tec1 (42) and Brg1 (43) both are known regulators of gene transcription during morphogenesis, and expression of each has been shown to increase when cells are shifted to inducing media. Lane et al. had found that Efg1 directly regulates Tec1 expression during morphogenesis under serum and Lee's medium hyphal induction (44). We confirmed that Efg1 also regulates TEC1 expression during Spider medium induction, suggesting that Efg1 also positively regulates ACE2 through TEC1 during hyphal morphogenesis (Fig. 5D). Finally, both BRG1 and TEC1 expression is reduced in ace2Δ/Δ mutants, indicating that Ace2 may, in part, directly or indirectly regulate its own expression (Fig. 5E). Taken together, our results indicate that the expression of ACE2 during morphogenesis is regulated, at least in part, by a complex network of transcription factors. Prior to the induction of morphogenesis, Efg1 negatively regulates ACE2 expression directly. Following initiation of morphogenesis, Efg1 repression of ACE2 expression is relieved. Tec1 and Brg1 positively regulate ACE2 expression in a direct manner. In addition, it appears that Efg1 also contributes to the positive regulation of ACE2 through its direct regulation of Tec1 (44). Finally, Ace2 is required for full expression of TEC1 and BRG1, suggesting that a positive feedback loop contributes to ACE2 expression during the maintenance phase of morphogenesis.
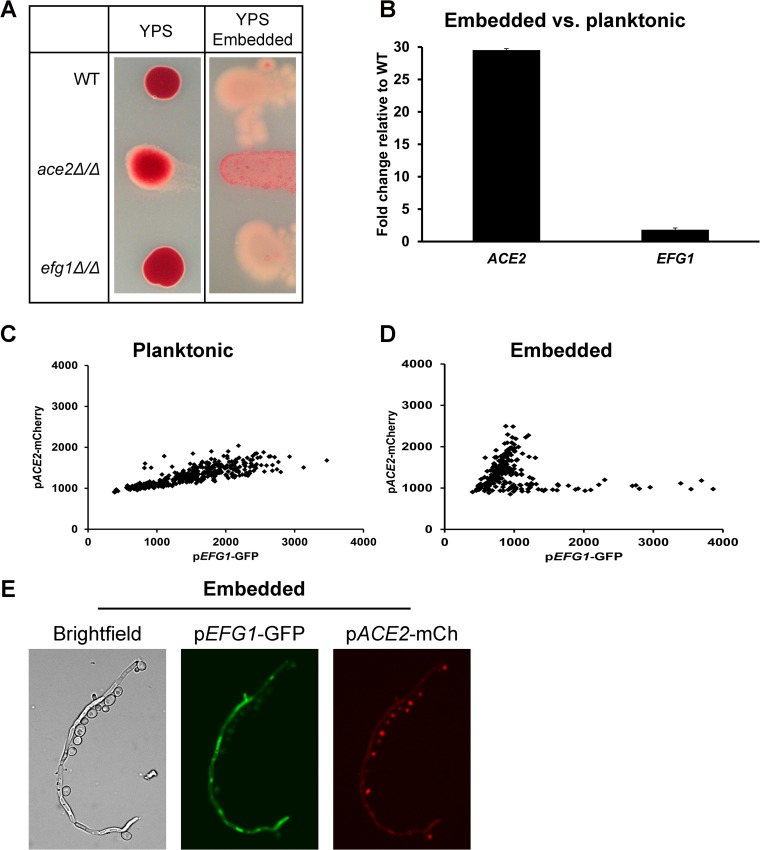
Ace2 expression is elevated in embedded yeast cells relative to planktonic cells and suppresses respiratory metabolism.
Transcriptional profiling studies of Ace2 (18) and Efg1 (24) indicate that these two transcription factors have similar direct and/or indirect effects on the expression of metabolic genes involved in glycolysis and respiration. Our finding that Efg1 negatively regulates Ace2 under some conditions suggested that Efg1 and Ace2 carry out similar functions under distinct cellular or environmental conditions. One setting where this proposed Efg1 and Ace2 dichotomy appeared to be operative is when C. albicans is embedded within a solid matrix, such as agar. Mulhern et al. have shown that under embedded conditions, Ace2 is required for filamentation while efg1Δ/Δ strains display increased filamentation (22). These phenotypes are the opposite of those shown by ace2Δ/Δ and efg1Δ/Δ strains in normoxic liquid medium or growth on top of solid agar: under these conditions, ace2Δ/Δ cells display increased pseudohyphal filamentation and efg1Δ/Δ cells fail to filament. Based on the increased expression of respiratory genes in ace2Δ/Δ mutants and their resistance to the respiratory chain inhibitor antimycin A, Mulhern et al. proposed that the inability of ace2Δ/Δ mutants to filament was due to their increased respiratory activity relative to that of wild-type cells under embedded conditions (22). To test this proposed mechanism further, we compared the respiratory activity of ace2Δ/Δ, efg1Δ/Δ, and WT cells under embedded conditions using a TTC overlay assay as described by Morales et al. (29). TTC is converted to a red color by respiratory metabolism. As shown in Fig. 6A, embedded colonies of WT and efg1Δ/Δ cells are much less red than ace2Δ/Δ cells, indicating that cells lacking Ace2 have increased respiration. In contrast, strains plated on the same medium under nonembedded conditions all had much higher levels of respiration (Fig. 6A), although ace2Δ/Δ colonies generated a slightly less intense color. These results are consistent with and support the model proposed by Mulhern et al. (22) and indicate that Ace2 suppresses the respiratory activity of embedded C. albicans but does not do so under nonembedded conditions.
FIG 6.
Ace2 represses respiration and is expressed at high levels under embedded conditions. (A) The indicated strains were incubated at 30°C in YP plus 2% sucrose either on top of the agar or embedded within the agar plate. The respiratory activity of the colonies was determined using the triphenyl tetrazolium chloride overlay assay as described in Materials and Methods. The increased respiratory activity of the colony is indicated by the deeper red color. (B) Expression of ACE2 and EFG1 by qRT-PCR was determined for WT strains in stationary-phase planktonic culture or incubated in embedded agar. The bars indicate the mean fold change between embedded cells and planktonic stationary-phase cells for three biological replicates assayed in triplicate. Standard deviations are indicated by the error bars. (C and D) A strain containing the EFG1 promoter fused to GFP and the ACE2 promoter fused to mCherry were examined under stationary-phase (C) and embedded (D) conditions by fluorescence microscopy, and the pixel density of the signal per cell for GFP and mCherry was determined as described in Materials and Methods (n ≥ 100). Each data point represents a single cell with the GFP and mCherry signal denoted in arbitrary units. (E) The pACE2-mCherry/pEFG1-GFP strain was incubated embedded in solid agar medium; the cells were scraped into a microcentrifuge tube and examined under bright-field, GFP, and mCherry filters.
These observations suggest that Ace2 plays an important role in the regulation of metabolism under embedded conditions compared to nonembedded and planktonic conditions. Accordingly, we hypothesized that ACE2 expression was higher in embedded cells than in planktonic cells. Therefore, we isolated RNA from planktonic and embedded WT cells and compared ACE2 levels by qRT-PCR. Consistent with this hypothesis, ACE2 expression is elevated ∼30-fold in embedded cells relative to planktonic cells in stationary phase (Fig. 6B). EFG1 expression is slightly elevated (1.8-fold) under embedded conditions compared to planktonic conditions.
To characterize the relationship between Efg1 and Ace2 under these conditions, we obtained a strain in which GFP expression is driven by the EFG1 promoter (generous gift of C. Kumamoto [32]) and fused the ACE2 promoter to mCherry to create a dual ACE2/EFG1 expression reporter strain. We then quantified promoter activity on a per-cell basis using quantitative microscopy as described by Pierce and Kumamoto (32). As expected, ACE2 promoter activity was low and consistent across the cell population under planktonic conditions (Fig. 6C). In contrast and in keeping with previously reported results (32), EFG1 promoter activity was higher and varied significantly on a cell-to-cell basis (Fig. 6C). ACE2 promoter activity was dramatically elevated in embedded cells, while EFG1 promoter activity was reduced overall (Fig. 6D); the wide distribution of EFG1 expression on a per-cell basis also was observed under embedded conditions. Strikingly, cells displaying the highest levels of EFG1 promoter activity were those with the lowest ACE2 promoter activity, supporting the notion that ACE2 and EFG1 are counterregulated. The promoter activity quantification was done only on yeast cells, because the topology and morphology of hyphae make quantitative image analysis problematic. Micrographs (Fig. 6E) of the embedded dual reporter strain show that EFG1 activity is much more apparent in hyphae than in the yeast forms, while ACE2 promoter activity is much higher in yeast than in hyphae. Taken together, these data suggest that Ace2 plays an important role in embedded yeast cells, and one of its functions is to repress respiratory activity. Our observations also suggest that Ace2 is more important in yeast-phase cells, while Efg1 functions mainly in hyphae under embedded conditions.
DISCUSSION
The RAM signaling pathway regulates a variety of cellular processes in both model and pathogenic fungi (4, 45). The best-characterized functions of the RAM pathway are those associated with the transcription factor Ace2. Studies of the function and regulation of Ace2 in S. cerevisiae have led to a detailed understanding of the mechanisms by which it executes its role in this model organism, particularly with respect to its role in the cell cycle and daughter cell-specific gene regulation (15, 16, 17, 18). Although a number of important and illuminating studies of Ace2 in pathogenic yeast, such as C. albicans, have been reported (4), our understanding of the role of Ace2 in C. albicans continues to evolve. As first characterized by Kelly et al. (19), deletion of ACE2 in C. albicans leads to constitutive pseudohyphal growth under noninducing conditions; additionally, and consistent with S. cerevisiae, C. albicans ace2Δ/Δ mutants also have a cell separation defect due to the key role that this transcription factor plays in the regulation of genes involved in the degradation of the septum. In liquid or solid serum-containing medium, ace2Δ/Δ cells were able to form hyphae (19). Subsequent studies found that although ace2Δ/Δ formed filaments in macrophages, it was deficient for filamentation in low-oxygen environments and under embedded conditions regardless of the medium employed (22). This suggested to us that the role of Ace2 in C. albicans morphogenesis was highly dependent on the inducing medium. Consistent with that hypothesis, we found that Spider medium was the only standard hypha-inducing medium in which ace2Δ/Δ was significantly deficient in morphogenesis; importantly, the requirement for Ace2 is not absolute, because true hyphae eventually were observed (Fig. 1D). Previously, we carried out a genetic interaction screen of the RAM pathway for heterozygous mutations that exacerbated the filamentation defects of the cbk1Δ/CBK1 mutant (14). We performed this screen on Spider medium, and the majority of the mutants isolated in the screen were transcriptional targets of Ace2 and filamented on serum-containing medium. Thus, our results are consistent with Ace2 function being required for normal morphogenesis on Spider medium and nicely explain why the set of interacting mutants that emerged from our complex haploinsufficiency screen were related to Ace2 (14).
Because Ace2 is well known to play an important role in daughter cell transcriptional regulation (17, 18, 45), we were interested in whether the loss of Ace2 function would have an effect on the emergence of yeast from hyphae. Interestingly, the number of emerging yeast from ace2Δ/Δ hyphae does not appear to differ from that of the wild type, indicating that Ace2 is not required for the budding process (Fig. 1B). However, the initial buds from WT cells generally were large, spherical cells, while those from ace2Δ/Δ mutants were elongated and pseudohypha-like (Fig. 1B). The pseudohyphal nature of the lateral yeast fits with the fact that planktonic ace2Δ/Δ mutants have increased numbers of pseudohyphal cells under noninducing conditions. Furthermore, the number of yeast-like cells attached to the hyphae generally was increased in the ace2Δ/Δ mutant, consistent with its role in the regulation of cell separation genes and the corresponding cell separation defect observed in planktonic cultures. We previously showed that ace2Δ/Δ mutants have elevated levels of PKA activity and Efg1 expression and that inhibition of PKA suppressed the pseudohyphal phenotype of ace2Δ/Δ mutants (14). Interestingly, the phenotype of ace2Δ/Δ hyphae at the stage of lateral yeast formation is identical to that of hyphae formed by strains overexpressing EFG1 from the strong PCK1 promoter (33). Specifically, pPCK1-EFG1 strains show both the elongated/pseudohyphal lateral yeast and the bulging hyphal tips displayed by ace2Δ/Δ. This suggests that disrupting the balance between Efg1 and Ace2 interferes with the hypha-to-yeast transition (46). Recently, Lindsay et al. has shown that decreased PKA pathway activity promotes the hypha-yeast transition (46). Combining our observations with this information suggests a model whereby PKA pathway activity decreases at the time of lateral yeast formation, leading to increased Ace2 expression and normal lateral yeast bud formation.
The genetic interaction screen previously performed in our laboratories indicated that the PKA and RAM pathways interact during morphogenesis (14). Accordingly, we were interested in exploring the mechanism of this interaction. The expression of ACE2 had previously been studied in S. cerevisiae in the context of the cell cycle (18). In C. albicans, the expression of ACE2 during biofilm formation has been shown to be dependent on the chromatin remodeling protein Snf5 (35), but its expression has not been characterized during morphogenesis. We found that ACE2 expression is very low in stationary-phase cells, the cell type used to initiate hyphal morphogenesis, and that this low expression is due, at least in part, to direct repression by the PKA-regulated transcriptional regulator Efg1. Supporting that conclusion is the fact that Efg1 directly binds the promoter of ACE2 and that ACE2 expression is elevated in efg1Δ/Δ mutants in stationary phase. As the cells transition through hyphal morphogenesis, Ace2 protein levels increase and Efg1 is no longer present at the promoter of ACE2. These data integrate well with the fact that Wang et al. have previously shown that Efg1 represses expression of Ace2-regulated cell separation genes such as CTS1 and SCW11 (26). Thus, it appears that Efg1 negatively regulates both Ace2 and its targets. Interestingly, Wang et al. showed that Efg1 repression of Ace2 targets was mediated by Cdc28 (26); in contrast, we found that Efg1-mediated repression of ACE2 was independent of Cdc28 (data not shown) but dependent on the PKA pathway. As such, our data are consistent with the model recently described by Lassak et al. in which Efg1 exerts a repressive function early in morphogenesis that also is dependent on the PKA pathway (34).
To further characterize the regulation of ACE2 expression, we tested the effect of transcriptional regulators for which consensus binding sequences were present in the 5′-upstream region of ACE2 and found that Tec1 and Brg1, two transcriptional regulators known to regulate hypha-associated genes, were required for full expression of ACE2 in hyphae. Both Tec1 and Brg1 bind to the promoter regions of ACE2 in hyphae but not in yeast, indicating that they directly regulate ACE2 expression. Tec1 is a well-characterized regulator of gene expression during morphogenesis, and its expression is dependent upon Efg1 (44). Thus, it appears that Efg1 negatively regulates ACE2 expression in stationary phase and early morphogenesis but then contributes to positive regulation during the maintenance phase of hyphal elongation. This model is also consistent with data from Lassak et al., who also showed that the binding patterns of Efg1 change soon after the induction of morphogenesis and that Efg1 is repressive early in morphogenesis (34).
At least one other transcriptional regulator, Brg1, is required for full expression of ACE2 during hypha formation (Fig. 5A). Like Tec1, Brg1 is an important regulator of gene expression during hypha formation (37, 43). Interestingly, we found that deletion of ACE2 reduced the expression of Tec1 and Brg1 (Fig. 5E). This suggests that Ace2 is regulated, in part, by a positive feedback mechanism. However, we have not determined if Ace2 directly binds to the promoters of TEC1 or BRG1; inspection of the 5′ untranslated regions of the two genes does not show the consensus binding site for Ace2. Consequently, the effect of Ace2 on TEC1 and BRG1 expression could be indirect.
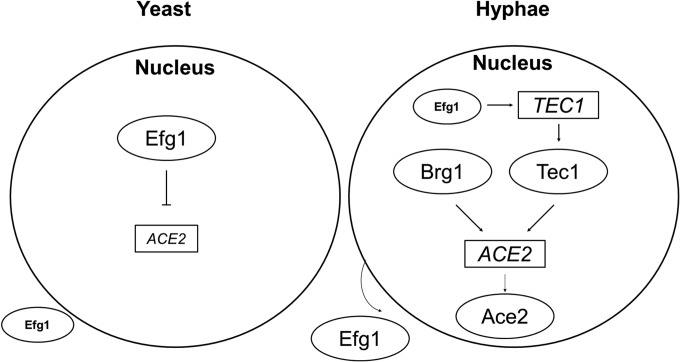
We have examined only the effect of transcription factors with characterized consensus binding sites in the promoter region of ACE2; therefore, we cannot exclude the possibility that other transcription factors also play a role in its regulation. As indicated above, the chromatin remodeling protein Snf5 was recently shown to be required for ACE2 expression during biofilm formation (39). Under the conditions we examined, deletion of SNF5 did not affect ACE2 expression during morphogenesis, suggesting that ACE2 is controlled by different transcriptional regulators depending on the environmental conditions. To summarize our data and synthesize it with that of others, Fig. 7 presents a schematic of the small network of transcriptional regulators of ACE2 as we currently understand it. The interconnected nature of these transcriptional regulators fits well with the transcriptional networks that have been identified in C. albicans during other growth states, such as biofilm formation (39).
FIG 7.
Model for the regulation of ACE2 expression in stationary-phase yeast cells at the initiation of hyphal morphogenesis and during the formation of hyphae. Boxes indicate the gene, and circles indicate the corresponding protein. Arrows indicate positive regulation, and lines with perpendicular crosses indicate negative regulation. The small-sized font for ACE2 and Efg1 indicates relative gene expression or protein levels.
In an attempt to gain insight into the mechanisms by which Efg1 is regulated during morphogenesis, we examined the levels of gene expression, protein expression, nuclear localization, and phosphorylation status of Efg1 during the time course of hyphal morphogenesis. Consistent with results reported previously by others, EFG1 gene expression rapidly decreases upon the shift to hypha-inducing conditions. Tebarth et al. have shown that downregulation likely is mediated by the PKA pathway (33). We observed a corresponding decrease in Efg1 protein levels 1 h after hyphal induction. However, Efg1 returns to preinduction levels by 2 h and is slightly elevated by 3 h. During the window during which Efg1 protein levels decrease, we found that the amount of nuclear Efg1 also decreases rapidly and that this occurs in yeast cells. However, it does not appear that this decrease in nuclear Efg1 is due to decreased protein levels, because we have previously shown that very few cells contain nuclear Efg1 at later time points when Efg1 levels have returned to preinduction levels. We must emphasize that Efg1 appears to be a relatively low-abundance protein. Therefore, while our immunofluorescence experiments clearly indicate there is a reduction in nuclear Efg1 early in morphogenesis, we cannot rule out the possibility that a proportion of the total Efg1 remains within the nucleus at levels below detection. Indeed, chromatin immunoprecipitation data from a number of laboratories indicate that Efg1 is present on the promoters of genes in hyphal-phase cells (34, 37), supporting the notion that our observations represent a change in the extent of Efg1 nuclear localization and not the absolute absence of the protein from the nucleus.
At least two protein kinases, Cdc28 (26) and PKA (35), are known to regulate the function of Efg1. Two-dimensional Western blotting indicates that at least two phosphorylated Efg1 species are present in hyphal-stage C. albicans that are not present in yeast-phase cells. In addition, these blots indicate that a complex mixture of Efg1 species with different isoelectric points is present in both the yeast and hypha phases. Because of the complexity of these mixtures, we cannot rule out the possibility that other phosphorylated species are present but were not clearly separable using this method. We also attempted to analyze samples from cultures that were taken at intermediate time points during morphogenesis; however, we were unable to obtain consistent results. We speculate that this situation is due to the fact that cultures at intermediate time points are morphologically heterogenous, while the cultures at the initiation and completion of morphogenesis are essentially all yeast or all hyphae. Since the development of these two phosphorylated species correlated with hyphae, we explored the possibility that Cdc28 or PKA regulated Efg1 localization but found no evidence that either kinase plays a role in the accumulation of Efg1 in the nucleus of stationary-phase cells or in the rapid decrease in nuclear Efg1 during morphogenesis. Since our genetic data indicate that PKA is required for Efg1-mediated repression of ACE2, it appears that relief of repression is not due to decreased nuclear Efg1 but is either directly or indirectly related to PKA activity. This is consistent with Lassak et al.'s finding that PKA regulates Efg1-mediated repression early in morphogenesis (34).
Taking our observations together with previous data regarding the function of Ace2 and Efg1, it appears that these two transcriptional regulators have a yin-and-yang relationship with respect to their roles during morphogenesis. Under standard normoxic planktonic conditions, Efg1 is (i) a positive regulator of filament formation, (ii) present in yeast forms at the initiation of filamentation, and (iii) absolutely required for filament formation. Under these conditions, Ace2 is (i) not absolutely required for filamentation, (ii) expressed at low levels in the yeast forms at the initiation of filamentation (partly due to direct repression by Efg1), and (iii) an indirect, negative regulator of filamentation (pseudohyphae). In contrast, the roles of these two transcription factors appear to be reversed under embedded conditions. Ace2 is now a positive regulator of filamentation, is expressed in yeast, and is required for filament formation. Efg1 is a negative regulator of filamentation in embedded conditions, expressed at low levels in yeast-phase cells, and expressed at higher levels in hyphal-phase cells. Thus, we propose that Ace2 and Efg1 play partially analogous roles during filamentation that vary according to the specific conditions inducing morphogenesis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joachim Ernst, Haoping Liu, Christophe d' Enfert, Aaron Mitchell, Alexander Johnson, Clarissa Nobile, Judy Berman, Carol Kumamoto, Deborah Hogan, Pete Sudbery, and Melanie Wellington for strains, protocols, and/or helpful discussions. We thank Thomas Murante for assistance with construction of C. albicans strains.
This work was supported by National Institute of Allergy and Infectious Diseases grant 1R01AI098450-02 (D.J.K.) and partially supported by NIH training grant T90-DE021985 (S.S.).
We also thank the Fungal Genetics Stock Center for providing strains and plasmids.
Footnotes
Published ahead of print 7 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00148-14.
REFERENCES
- 1.Moran GP, Coleman D, Sullivan D. 2012. An introduction to the medically important Candida species, p 11–25 In Calderone R, Clancy CJ. (ed), Candida and candidiasis, 2nd ed. ASM Press, Washington, DC [Google Scholar]
- 2.Vazquez JA, Sobel JD. 2003. Candidiasis, p 143–187 In Dismukes WE, Pappas PG, Sobel JD. (ed), Clinical mycology. Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 3.Sudbery PE. 2011. Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 9:737–748. 10.1038/nrmicro2636 [DOI] [PubMed] [Google Scholar]
- 4.Sudbery PE, Gow N, Sudbery PE. 2004. The distinct morphotypes of Candida albicans. Trends Microbiol. 12:317–324. 10.1016/j.tim.2004.05.008 [DOI] [PubMed] [Google Scholar]
- 5.Guarner J, Brandt ME. 2011. Histopathologic diagnosis of fungal infections in the 21st century. Clin. Microbiol. Rev. 24:247–280. 10.1128/CMR.00053-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Magee PT. 2010. Fungal pathogenesis and morphological switches. Nat. Genet. 42:560–561. 10.1038/ng0710-560 [DOI] [PubMed] [Google Scholar]
- 7.Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cucciapuoti A, Fink GR. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939–949. 10.1016/S0092-8674(00)80358-X [DOI] [PubMed] [Google Scholar]
- 8.Braun BR, Head WS, Wang MX, Johnson AD. 2000. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 156:31–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarthy AN, McGonigal T, Coen M, Frost DJ, Meulbroek JA, Goldman RC. 1997. Phenotype in Candida albicans of a disruption of the BGL2 gene encoding a 1,3-beta-glucosyltransferase. Microbiology 143:367–376. 10.1099/00221287-143-2-367 [DOI] [PubMed] [Google Scholar]
- 10.Lorenz MC, Fink GR. 2001. The glyoxylate cycle is required for fungal virulence. Nature 412:83–86. 10.1038/35083594 [DOI] [PubMed] [Google Scholar]
- 11.Gow NAR, van de Veerdonk FL, Brown AJP, Netea MG. 2012. Candida albicans morphogenesis and host defense: discriminating invasion from colonization. Nat. Rev. Microbiol. 10:112–122. 10.1038/nrmicro2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noble SM, French S, Kohn LA, Chen V, Johnson AD. 2010. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 42:590–598. 10.1038/ng.605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saville SP, Lazzell AL, Monteagudo C, Lopez-Ribot JL. 2003. Engineered control of cell morphology in vivo reveals distinct role for yeast and filamentous forms of Candida albicans during infection. Eukaryot. Cell 2:1053–1060. 10.1128/EC.2.5.1053-1060.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bharucha N, Chabrier-Roselló Y, Xu T, Johnson C, Sobcynski S, Song SQ, Dobry CJ, Anderson CP, Benjamin AJ, Kumar A, Krysan DJ. 2011. A large-scale complex haploinsufficiency-based genetic interaction screen in Candida albicans: analysis of the RAM Network during morphogenesis. PLoS Genet. 7:e1002058. 10.1371/journal.pgen.1002058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mazanka E, Weiss EL. 2010. Sequential counteracting kinases restrict an asymmetric gene expression program to early G1. Mol. Biol. Cell 21:2809–2820. 10.1091/mbc.E10-02-0174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurischko C, Weiss G, Ottey M, Luca FC. 2005. A role for the Saccharomyces cerevisiae regulation of Ace2 and polarized morphogenesis signaling network in cell integrity. Genetics 171:443–455. 10.1534/genetics.105.042101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiss EL. 2012. Mitotic exit and separation of mother and daughter cells. Genetics 192:1165–1202. 10.1534/genetics.112.145516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sbia M, Parnell EJ, Yu Y, Olsen AE, Kretschmann KL, Voth WP, Stillman DJ. 2008. Regulation of the yeast Ace2 transcription factor during the cell cycle. J. Biol. Chem. 283:11135–11145. 10.1074/jbc.M800196200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelly MT, MacCallum DM, Clancy SD, Odds FC, Brown AJP, Butler G. 2004. The Candida albicans CaACE2 gene affects morphogenesis, adherence, and virulence. Mol. Microbiol. 53:969–983. 10.1111/j.1365-2958.2004.04185.x [DOI] [PubMed] [Google Scholar]
- 20.McNemar MD, Fonzi WA. 2002. Conserved serine and threonine kinase encoded by CBK1 regulates expression of several hypha-associated transcripts and genes encoding cell wall proteins in Candida albicans. J. Bacteriol. 184:2058–2061. 10.1128/JB.184.7.2058-2061.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song Y, Cheon SA, Lee KE, Lee S-Y, Lee B-K, Oh D-B, Kang HA, Kim J-Y. 2008. Role of the RAM network in cell polarity and hyphal morphogenesis in Candida albicans. Mol. Biol. Cell 19:5456–5477. 10.1091/mbc.E08-03-0272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mulhern SM, Logue ME, Butler G. 2006. Candida albicans transcription factor Ace2 regulates metabolism and is required for filamentation in hypoxic conditions. Eukaryot. Cell 5:2001–2013. 10.1128/EC.00155-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stoldt VR, Sonneborn A, Leuker CE, Ernst JF. 1997. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH protein regulating morphogenetic processes in fungi. EMBO J. 16:1982–1991. 10.1093/emboj/16.8.1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doedt T, Krishnamurthy S, Bockmühl DP, Tebarth B, Stempel C, Russell CL, Brown AJ, Ernst JF. 2004. APES proteins regulate morphogenesis and metabolism in Candida albicans. Mol. Biol. Cell 15:3167–3180. 10.1091/mbc.E03-11-0782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneper L, Krauss A, Miyamoto R, Fang S, Broach JR. 2004. The ras/protein kinase A pathway acts in parallel with the Mob2/Cbk1 pathway to effect cell cycle progression and proper bud site selection. Eukaryot. Cell 3:108–120. 10.1128/EC.3.1.108-120.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang A, Raniga PP, Lane S, Lu Y, Liu H. 2009. Hyphal chain formation in Candida albicans: Cdc28-Hgc1 phosphorylation of Efg1 represses cell separation genes. Mol. Cell. Biol. 29:4406–4416. 10.1128/MCB.01502-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gutierrez-Escribano P, Gonzalez-Novo A, Belen Suarez M, Li C-R, Wang Y, Vazquez de Aldana CR, Correa-Bordes J. 2011. CDK-dependent phosphorylation of Mob2 is essential for hyphal development in Candida albicans. Mol. Biol. Cell 22:2458–2469. 10.1091/mbc.E11-03-0205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burke DJ, Dawson D, Stearns T. 2000. Methods in yeast genetics. CSHL Press, Woodbury, NY [Google Scholar]
- 29.Morales DK, Grahl N, Okegbe C, Dietrich LE, Jacobs NJ, Hogan DA. 2013. Control of Candida albicans metabolism and biofilm formation by Pseudomonas aeruginosa phenazines. mBio 4:e00526–12. 10.1128/mBio.00526-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3:1101–1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- 31.Hernday AD, Noble SM, Mitrovich QM, Johnson AD. 2010. Genetics and molecular biology of Candida albicans. Methods Enzymol. 470:737–758. 10.1016/S0076-6879(10)70031-8 [DOI] [PubMed] [Google Scholar]
- 32.Pierce JV, Kumamoto CA. 2012. Variation in Candida albicans EFG1 expression enables host-dependent changes in colonizing fungal populations. mBio 3:e00117–12. 10.1128/mBio.00117-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tebarth B, Doedt T, Krishnamurthy S, Weide M, Monterola F, Domingues A, Ernst JF. 2003. Adaptation of the Efg1p morphogenetic pathway in Candida albicans by negative autoregulation and PKA-dependent repression of the EFG1 gene. J. Mol. Biol. 329:949–962. 10.1016/S0022-2836(03)00505-9 [DOI] [PubMed] [Google Scholar]
- 34.Lassak T, Schneider E, Bussmann M, Kurtz D, Manak JR, Srkantha T, Soll DR, Ernst JF. 2011. Target specificity of the Candida albicans Efg1 regulator. Mol. Microbiol. 82:602–618. 10.1111/j.1365-2958.2011.07837.x [DOI] [PubMed] [Google Scholar]
- 35.Bockmuhl DP, Ernst JF. 2001. A potential phosphorylation site for an A-kinase in the Efg1 regulator protein contributes to hyphal morphogenesis of Candida albicans. Genetics 157:1523–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bastidas RJ, Heitman J, Cardenas ME. 2009. The protein kinase Tor1 regulates adhesin gene expression in Candida albicans. PLoS Pathog. 5:e1000294. 10.1371/journal.ppat.1000294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hnisz D, Bardet AF, Nobile CJ, Petryshyn A, Glaser W, Schöck U, Stark A, Kuchler K. 2012. A histone deacetylase adjusts transcription kinetics at coding sequences during Candida albicans morphogenesis. PLoS Genet. 8:e1003118. 10.1371/journal.pgen.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, Tuch BB, Andes DR, Johnson AD. 2012. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 148:126–138. 10.1016/j.cell.2011.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finkel JS, Xu W, Huang D, Hill EM, Desai JV, Woolford CA, Nett JE, Taff H, Norice CT, Andes DR, Lanni F, Mitchell AP. 2012. Portrait of Candida albicans adherence regulators. PLoS Pathog. 8:e1002525. 10.1371/journal.ppat.1002525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giusani AD, Vinces M, Kumamoto CA. 2002. Invasive filamentous growth of Candida albicans is promoted by Czf1p-dependent relief of Efg1p-mediated repression. Genetics 160:1749–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sellam A, Askew C, Epp E, Lavoie H, Whiteway M, Nantel A. 2009. Genome-wide mapping of the coactivator Ada2p yields insight into the functional roles of SAGA/ADA complex in Candida albicans. Mol. Biol. Cell 20:2389–2400. 10.1091/mbc.E08-11-1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schweizer A, Rupp S, Taylor BN, Röllinghoff M, Schröppel K. 2000. The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Mol. Microbiol. 38:435–445. 10.1046/j.1365-2958.2000.02132.x [DOI] [PubMed] [Google Scholar]
- 43.Lu Y, Su C, Liu H. 2012. A GATA transcription factor recruits Hda1 in response to reduced Tor1 signaling to establish a hyphal chromatin state in Candida albicans. PLoS Pathog. 8:e1002663. 10.1371/journal.ppat.1002663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lane S, Birse C, Zhou S, Matson R, Liu H. 2001. DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2, and Efg1 in Candida albicans. J. Biol. Chem. 276:48988–48996. 10.1074/jbc.M104484200 [DOI] [PubMed] [Google Scholar]
- 45.Saputo S, Chabrier-Rosello Y, Luca FC, Kumar A, Krysan DJ. 2012. The RAM network in pathogenic fungi. Eukaryot. Cell 11:708–717. 10.1128/EC.00044-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lindsay AK, Deveau A, Piispanen AE, Hogan DA. 2012. Farnesol and cyclic AMP signaling effects on the hypha-to-yeast transition in Candida albicans. Eukaryot. Cell 11:1219–1225. 10.1128/EC.00144-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.