Abstract
SUMMARY
Understanding of the taxonomy and phylogeny of Cryptococcus gattii has been advanced by modern molecular techniques. C. gattii probably diverged from Cryptococcus neoformans between 16 million and 160 million years ago, depending on the dating methods applied, and maintains diversity by recombining in nature. South America is the likely source of the virulent C. gattii VGII molecular types that have emerged in North America. C. gattii shares major virulence determinants with C. neoformans, although genomic and transcriptomic studies revealed that despite similar genomes, the VGIIa and VGIIb subtypes employ very different transcriptional circuits and manifest differences in virulence phenotypes. Preliminary evidence suggests that C. gattii VGII causes severe lung disease and death without dissemination, whereas C. neoformans disseminates readily to the central nervous system (CNS) and causes death from meningoencephalitis. Overall, currently available data indicate that the C. gattii VGI, VGII, and VGIII molecular types more commonly affect nonimmunocompromised hosts, in contrast to VGIV. New, rapid, cheap diagnostic tests and imaging modalities are assisting early diagnosis and enabling better outcomes of cerebral cryptococcosis. Complications of CNS infection include increased intracranial pressure, severe neurological sequelae, and development of immune reconstitution syndrome, although the mortality rate is low. C. gattii VGII isolates may exhibit higher fluconazole MICs than other genotypes. Optimal therapeutic regimens are yet to be determined; in most cases, initial therapy with amphotericin B and 5-flucytosine is recommended.
INTRODUCTION
Cryptococcus gattii is a basidiomycetous yeast, which grows mainly as an asexual budding yeast in the environment and within human and animal hosts. Sexual reproduction can occur between cells of the opposite mating type or of the same mating type (1). During sexual development, yeast cells undergo a dimorphic transition to hyphal growth, generating a mycelium and forming basidiospores (2). It is not yet certain whether sexually produced blastospores, desiccated yeast cells, or sexually produced basidiospores are the infectious propagules.
C. gattii has long been recognized as an endemic pathogen in Australia. In the 1990s, it emerged and became established in British Columbia, Canada, and subsequently in the Pacific Northwest of the United States. In the last decade, major advances in molecular technology led to a revision of the taxonomy and phylogeny of this species and have enhanced our understanding of its ecology, epidemiology, and clinical associations. Furthermore, recognition of genotype-dependent differences within the species C. gattii has assisted studies of pathogenesis, which, while still limited, are revealing the complexity of transcriptional circuits, the phenotypic virulence composite, and host cellular responses. Significant advances in diagnosis, clinical epidemiology, and approaches to the management of human cryptococcosis have been made. This review places new information about C. gattii in a historical context.
TAXONOMY OF THE SPECIES C. GATTII
Historical Overview
The agents of cryptococcosis, Filobasidiella neoformans (teleomorph/sexual stage)-Cryptococcus neoformans (anamorph/asexual stage) and F. bacillispora-C. gattii, form a monophyletic group, which is placed taxonomically within the phylum Basidiomycota, subphylum Agaricomycotina, family Tremellales, and genus Filobasidiella, which contains only three additional, closely related sister taxa, Filobasidiella depauperata, F. lutea, and F. amylolenta. These species are distinct from the >80 species in the polyphyletic genus Cryptococcus, based on recent phylogenetic analysis of the D1/D2 region of the large-subunit (LSU) ribosomal DNA (rDNA) gene (3, 4).
Historically, several pathogens, Saccharomyces neoformans (5), Cryptococcus hominis (6), Torula neoformans (7), Torula histolytica (8), and Debaryomyces hominis (9), were identified by Benham as belonging to the same species, for which he proposed the name Cryptococcus neoformans (10, 11). Until 1949, C. neoformans was considered to be a homogeneous anamorph (asexual) species (12–14). In that year, three serotypes were described (serotypes A, B, and C) (15), and 20 years later, a fourth serotype (serotype D) was discovered (16). However, it was not until 1970 that Vanbreuseghem and Takashio described the production of distinct elliptical yeast cells by a cryptococcal strain isolated from an African patient with leukemia (RV20186, IHEM11796, CBS 6289, ATCC 32269, or MUCL 30449) and introduced the name Cryptococcus neoformans var. gattii (17). More recently, molecular analysis of a clinical isolate from France, reported by Curtis in 1896 as Saccharomyces subcutaneus tumefaciens (18), revealed that this was, in fact, the first reported case of human C. gattii infection (19).
Mating experiments conducted by Kwon-Chung in the early 1970s revealed two different teleomorph (sexual) species: Filobasidiella neoformans, which resulted from a cross between two serotype D strains of C. neoformans var. neoformans, B3501 (MATα) and B3502 (MATa) (20, 21), and the morphologically distinct Filobasidiella bacillispora, characterized by smooth-walled, bacilliform-shaped basidiospores, which resulted from a cross between two C. neoformans var. gattii strains, a serotype C MATa strain (NIH 191) and a serotype B MATα strain (NIH 444) (22). After the discovery of this new teleomorph species, C. neoformans var. gattii was raised to the species level and renamed C. bacillisporus (2). The teleomorph species F. neoformans corresponds to the two anamorphic (asexual) C. neoformans species (serotypes A and D), and F. bacillispora corresponds to C. bacillisporus (serotypes B and C). Further studies revealed that the C. neoformans var. gattii strain reported by Vanbreuseghem and Takashio and the C. bacillisporus strain (NIH 191, CBS 6955, or ATCC 32608) isolated from the cerebrospinal fluid (CSF) of a patient in California had the same biochemical, morphological, and serological characteristics (23).
Subsequently, it was revealed that strains of F. neoformans and F. bacillispora had intermediate DNA-DNA reassociation values compared to those of isolates of the same species (55 to 63% and 88 to 94%, respectively) (24). In addition, a MATα strain (CBS 6289), type culture of C. neoformans var. gattii (serotype B), when crossed with the MATa strain (CBS 6901) of C. neoformans (serotype D), produced viable basidiospores, and the F1 generation showed a mixture of both F. neoformans and F. bacillispora. Based on these observations and additional biochemical, morphological, serological, and genetic data, the species nomenclature C. bacillisporus was again reduced to a variety of C. neoformans. The older nomenclature C. neoformans var. gattii was given precedence over C. bacillisporus, and C. bacillisporus was made a synonym of C. neoformans var. gattii (23).
After the 1970s, there was increasing evidence of differences in morphological, biochemical, molecular, ecological, pathobiological, and clinical features between the two varieties of C. neoformans (2, 20, 22, 23, 25–29). Phylogenetic relationships between the two varieties were scrutinized by using DNA sequences of multiple genes, including the URA5, LAC1, CAP59, and CAP64 genes, as well as the intergenic spacer (IGS) and the internal transcribed spacer (ITS) regions of the rDNA gene cluster (4, 30–32). Two distinct monophyletic groups were identified, regardless of the genetic locus studied (Fig. 1). Population genetic analysis using either amplified fragment length polymorphism (AFLP) analysis or PCR fingerprinting approaches supported these two major groups (33–35). Since C. neoformans and C. gattii fulfilled the criteria for different species according to morphological, biological, and phylogenetic species concepts, they were designated as such in 2002. The name “gattii” was preserved, as it had been used preferentially over the previous 2 decades by the medical, veterinary, and scientific communities (19). Furthermore, genetic analysis of the F1 generation of a cross between strains of the two varieties showed no evidence of recombination. In addition, comparison of the entire genomes of B3501 (C. neoformans var. neoformans) and WM 276 (C. neoformans var. gattii) revealed only 87% identity between them (36).
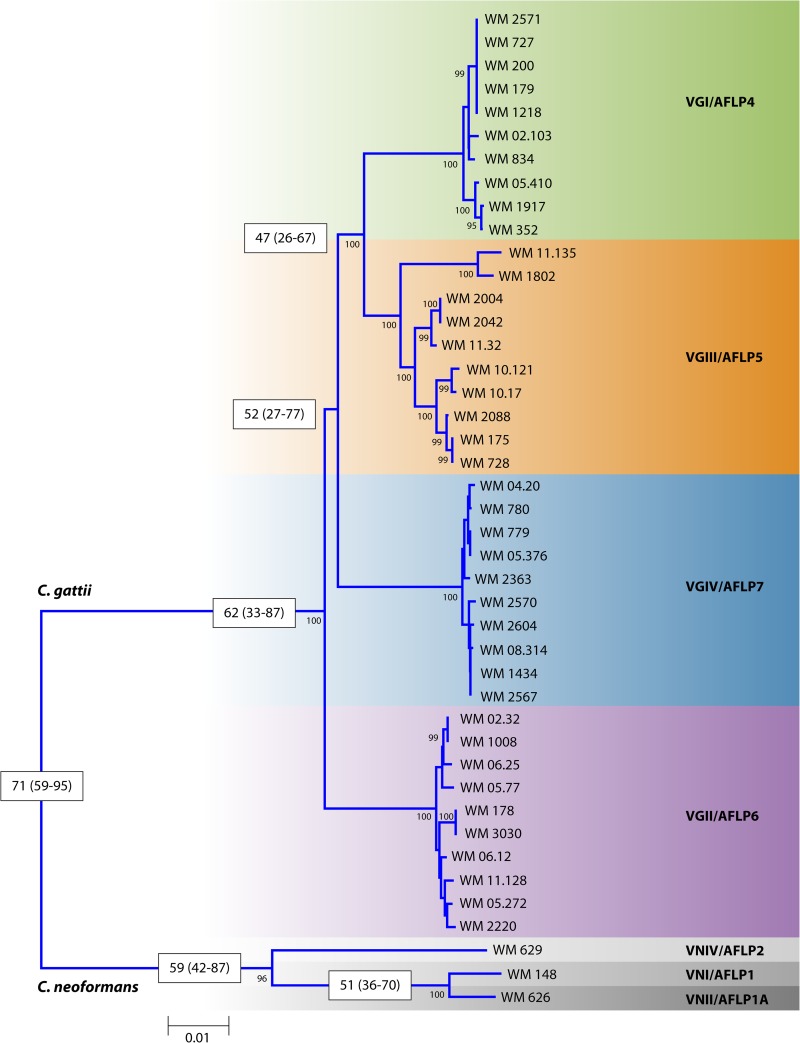
FIG 1.
Rooted neighbor-joining tree inferred from the concatenated MLST typing scheme sequences (CAP59, GPD1, LAC1, SOD1, URA5, and PLB1 genes and IGS) of 10 strains of each major C. gattii molecular type rooted with the standard strains of the three major C. neoformans molecular types. Numbers on branches are bootstrap support values obtained from 1,000 pseudoreplicates using the program MEGA version 5.05. Numbers in the white boxes indicate estimated diversion times based on the 10 genetic concatenated loci (ACT1, LAC1, IDE1, ITS1/2, IGS, PLB1, RPB1, RPB2, TEF1, and URA5), generated with the program BEAST.
The occurrence of outbreaks of cryptococcosis in new geographic locations over the last 2 decades (37–39) has stimulated detailed molecular studies of C. gattii.
Phylogenetic Studies and Molecular Subtyping
The application of PCR fingerprinting (34, 40), AFLP analysis (33), PLB1 restriction fragment length polymorphism (RFLP) analysis (41), URA5 RFLP analysis (40), and multilocus microsatellite type (MLMT) analysis (42, 43) revealed intraspecies genetic diversity within C. gattii and led to the discovery of interspecies hybrids of serotypes A and B and serotypes B and D (44, 45). C. neoformans-C. gattii is now considered a species complex, with the actual number of scientifically valid species being controversial (43).
Initial studies based on analyses of mitochondrial RNA (mtRNA), ITS1/2, and the OMP and DOX genes indicated that the two species separated 37 million years ago (32), with a subsequent analysis of the seven haploid molecular types, based on four genetic loci, ACT1, IDE, PLB1, and URA5, placing this separation at 49 million years ago (46). A Bayesian evolutionary analysis based on 10 genetic loci, ACT1, IDE, ITS1/2, IGS, LAC1, PLB1, RPB1, RPB2, TEF1, and URA5 (C. K. M. Tsui and W. Meyer, unpublished data) and a subsequent whole-genome sequence-based study (47) estimated that the two species diverged either 70 million years ago or 80 million years ago (range, 16 million to 160 million years ago).
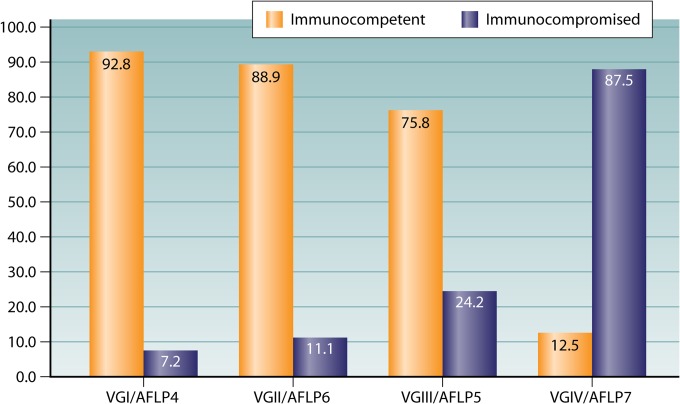
Four distinct genetic groups within C. gattii have been described, VGI/AFLP4, VGII/AFLP6, VGIII/AFLP5, and VGIV/AFLP7 (Fig. 1), each of which contains serotype C and B strains in different proportions (33, 48, 49). The phylogenetic relationships between these group were investigated in two major studies. In the first study, analysis of the major AFLP types was conducted by using six concatenated genetic loci (LAC1, ITS1/2, IGS, RPB1, RPB2, and TEF1). VGII/AFLP6 was identified as the lineage that is basal to the C. gattii clade, followed by VGIV, which itself is basal to the sister groups VGI/AFLP4 and VGIII/AFLP5 (49). However, a phylogenetic analysis of two mitochondrial loci (mtlrRNA and ATP6) revealed evidence of recombination (50), unlike the one with six nuclear loci. In the second study, analysis of the major molecular types obtained by PCR fingerprinting was performed by using either individual genetic loci or a combined analysis of four concatenated genetic loci (ACT1, IDE1, PLB1, and URA5). This analysis identified four major clades corresponding to the four major molecular types of C. gattii. This study confirmed that the VGII/AFLP6 clade is the basal lineage of C. gattii (32, 46). Given that the major molecular types within C. gattii contain both serotype B and C isolates, further studies using four concatenated genetic loci (ACT1, IDE1, PLB1, and URA5) were undertaken and revealed that the major molecular types of C. gattii are younger than those of C. neoformans. The VGI/AFLP4, VGIII/AFLP5, and VGIV/AFLP7 clades were estimated to have diverged from the basal VGII/AFLP6 clade approximately 12.5 million years ago, the VGIV/AFLP7 clade was estimated to have diverged from the VGIII/AFLP5 and VGI/AFLP4 sister clades 11.7 million years ago, and the VGIII/AFLP5 and VGI/AFLP4 clades were estimated to have diverged from each other 8.5 million years ago (46). A recent combined study of all 10 loci (ACT1, LAC1, IDE1, ITS1/2, IGS, PLB1, RPB1, RPB2, TEF1, and URA5) confirmed the ancestral position of VGII/AFLP6 (48). The initial divergence of the VGII/AFLP6 basal clade and the remaining major clades was estimated to have occurred 62 million years ago (range, 33 million to 87 million years ago), that of the VGIV/AFLP7 clade and the VGIII/AFLP5/VGI/AFLP4 clade was estimated to have occurred 52 million years ago (range, 27 million to 77 million years ago), and that of the VGIII/AFLP5 clade and the VGI/AFLP4 clade was estimated to have occurred 47 million years ago (range, 26 million to 67 million years ago) (C. K. M. Tsui and W. Meyer, unpublished) (Fig. 1).
Notably, whole-genome sequence typing (WGST) was recently employed to investigate the spread of highly virulent C. gattii VGII strains from the Pacific Northwest. The highest genetic diversity was present among VGIIa strains, followed by VGIIc and VGIIb strains. The WGST data showed clearly that the VGIIc genotype responsible for the most recent infections in the Pacific Northwest is genetically distinct from both VGIIa and VGIIb Vancouver Island outbreak subtypes and suggest that all three subtypes are equally distinct from each other (52). Similar findings based on multilocus sequence typing (MLST) and MLMT analyses, which have been confirmed based on WGST, have been reported recently (53–55). All studies suggest that the three subtypes have arisen as a result of recombination events that took place prior to their introduction into the Pacific Northwest.
The genetic variation between the four major molecular types of C. gattii is similar to that observed between C. neoformans var. grubii and C. neoformans var. neoformans and suggests that they warrant at least varietal if not species status. Several lines of evidence support this contention. The four molecular types have been distinguished clearly from each other by using multiple molecular techniques, including PCR fingerprinting (35, 40) (Fig. 2A), AFLP analysis (33) (Fig. 2B), PLB1 RFLP analysis (41) (Fig. 2C), URA5 RFLP analysis (40) (Fig. 2D), hyperbranched rolling-circle amplification (RCA) (56) (Fig. 2E), and matrix-assisted laser desorption ionization–time of flight mass spectroscopy (MALDI-TOF MS) (57, 58) (Fig. 2F) (see also Diagnostics, below). Furthermore, there is evidence for genotype-dependent differences in virulence in mouse and wax moth experimental models, for geographical localization of different molecular types, for differences in clinical epidemiology, and for reduced susceptibility to fluconazole among VGIII isolates. A detailed taxonomic description of the potential varieties and species within C. gattii is in preparation (T. Boekhout, K. J. Kwon-Chung, and W. Meyer, unpublished data).
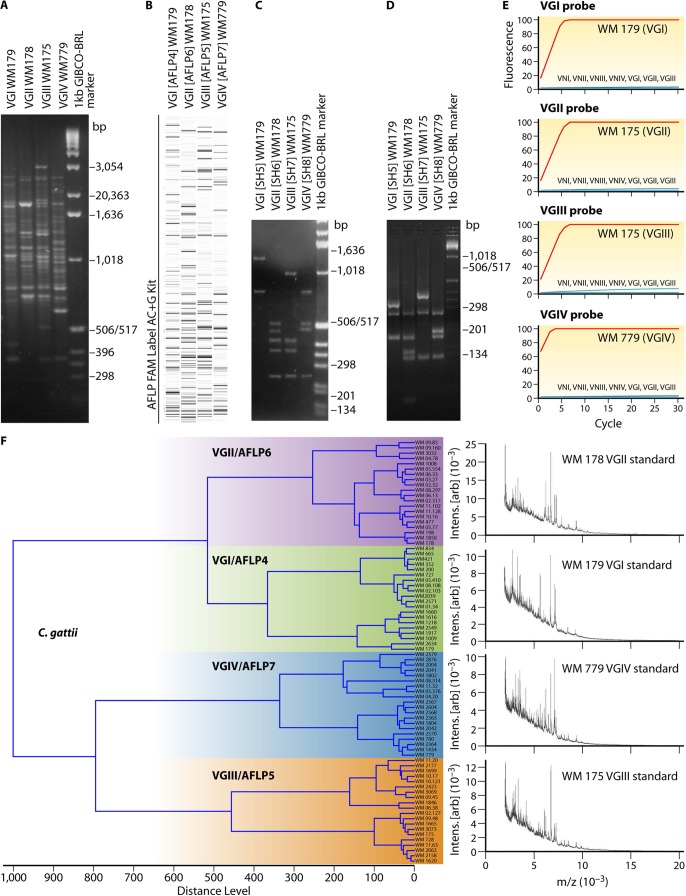
FIG 2.
Identification of the major molecular types within C. gattii. (A) PCR fingerprinting generated with the primer M13; (B) AFLP profiles generated with the 6-carboxyfluorescein (FAM) label AC+G kit; (C) PLB1 gene RFLP profiles identified via digestion with AvaI; (D) URA5 gene RFLP profiles identified via double digestion with Sau96I and HhaI; (E) PLB1 gene hyperbranched rolling-circle amplification; (F) MALDI-TOF MS profiles obtained from the reference strains of each major molecular type.
Standardization of Molecular Typing Systems
Many typing techniques have been applied to individual strains of C. gattii to gain insights into the molecular epidemiology and population structure of this species. Random amplified polymorphic DNA (RAPD) analysis (59), PCR fingerprinting (35) (Fig. 2A), AFLP analysis (33) (Fig. 2B), MLMT analysis (42), and MLST analysis (43) are in common use. More recently, WGST has been implemented (52).
RAPD analysis, based on the random amplification of DNA fragments using short arbitrary primers, was used initially for cryptococcal strain typing (59, 60). Advantages included high discriminatory power, simplicity, and speed (61). This method separated C. gattii into three major molecular types, designated VGI, VGII, and VGIII (60). Use of the primers ERIC1 and ERIC2 revealed variation in the geographic distribution of these molecular types (59). RAPD analysis has been superseded by other methods, since this technique was never standardized and its interlaboratory reproducibility is poor.
PCR fingerprinting, based on the amplification of DNA sequences flanked by hypervariable DNA repeats, uses single primers specific to either minisatellites (the core sequence of wild-type phage M13, 5′-GAGGTGGCGGTTCT-3′) or microsatellites [(GTG)5 and (GACA)4] (34). This technique distinguished four major molecular types of C. gattii, designated VGI, VGII, VGIII, and VGIV (35, 62, 63) (Fig. 2A). Although more reproducible than RAPD, banding patterns can vary with the source of laboratory reagents and different experimental conditions, making it unsuitable for multicenter typing studies.
AFLP analysis is based on the digestion of genomic DNA by using frequent (EcoRI)- and rare (MseI)-cutting restriction enzymes, followed by ligation with an adaptor and subsequent PCR amplification of specifically selected fragments. Reproducibility is excellent, especially with the inclusion of an internal standard in each lane and direct fragment detection by using an automated DNA sequencer. Three major AFLP groups were separated within a global collection of C. gattii, named AFLP4, AFLP5, and AFLP6 (33), with a subsequent study revealing a fourth group, AFLP7 (64) (Fig. 2B). Serotype B strains were distributed among all three AFLP types, but serotype C strains in this particular study were restricted to type AFLP5 (VGIII); strains of serotype VGIV, which also contains serotype C isolates, were not tested in this study (33). A drawback of AFLP is the potential for artificial variation in banding patterns due to incomplete digestion; thus, samples must be run in duplicate.
MLMT analysis is an even more discriminatory technique for strain typing. It is based on the identification of repeat variations within microsatellite sequences, short tandem repeats, or simple sequence repeats (SSRs) 1 to 6 bp in length following polyacrylamide gel electrophoresis (65). It is highly reproducible, and the primers used to amplify the microsatellites are species specific. This technique distinguishes all four major molecular types of C. gattii and has enabled the tracing of highly virulent strains (see below) (53, 66, 67).
MLST analysis is the most reproducible method and is readily standardized but expensive. Two main MLST schemes based on the amplification of highly variable partial sequences of 9 genetic loci (+22 additional loci) (1) or highly variable partial sequences of 7 genetic loci (43) within housekeeping genes have been developed. This method produces unambiguous, highly reproducible sequence data from which a sequence type is generated for each strain. Although both MLST schemes distinguish between the four major molecular types of C. gattii (1, 43, 68, 69), they do not yield comparable allele and sequence type numbers, thus precluding the possibility of combining them into a single global population genetic analysis.
WGST is the latest development in cryptococcal strain typing. It detects single nucleotide polymorphisms (SNPs) throughout the genome (52) and is the most accurate typing tool available. Currently, its use is limited by high costs and bioinformatic demands.
Despite the large number of typing tools available, understanding of the population structure of C. gattii is still fragmented. This is due mainly to the use of different nomenclatures and a lack of consensus between results obtained by using different systems. In 2007, a working group of the International Society for Human and Animal Mycology (ISHAM) was established to standardize genotyping of the C. neoformans-C. gattii species complex, thereby enabling the global tracking of highly virulent strains and, potentially, prediction of future outbreaks. Since the different typing methods yielded the same major cryptococcal molecular types (Table 1), it was agreed by this working group in 2007 that the VGI, VGII, VGIII, and VGIV nomenclature should be used to represent the global population structure of C. gattii (43). In addition, a set of standard strains was designated and deposited at major public culture collections, including the CBS Fungal Biodiversity Centre (CBS-KNAW), The Netherlands (http://www.cbs.knaw.nl/); the American Type Culture Collection (ATCC) (http://www.atcc.org/); the Fungal Genetic Stock Center (FGS) (http://www.fgsc.net/); the Westmead Hospital Collection (WM), Australia (http://www.mycologylab.org/); and Coleção de Micro-Organismos de Referência em Vigilância Sanitária (CMRVS), Brazil (http://cmrvs.fiocruz.br/). The respective culture collection strain numbers are listed in Table 2. Statistical analysis of all published MLST loci revealed that a minimum of seven loci is needed to type a given strain unambiguously. As a result, the ISHAM consensus MLST typing scheme for the C. neoformans-C. gattii species complex consists of the following seven variable unlinked genetic loci: CAP59, GPD1, LAC1, PLB1, SOD1, URA5, and the IGS1 region (43). Three of the seven loci encode major cryptococcal virulence factors: the polysaccharide capsule (CAP59), melanin synthesis (LAC1), and cell invasion (PLB1). The MLST data, including allele and sequence types, are stored in a centralized database at the Molecular Mycology Research Laboratory at the University of Sydney and can be accessed at http://mlst.mycologylab.org/. All research groups using this MLST scheme are encouraged to deposit their data in this database. Allele and sequence types can be retrieved online.
TABLE 1.
Concordance of different schemes used for molecular typing of Cryptococcus gattii
| C. gattii serotype | PCR fingerprinting molecular type (40, 77) | AFLP genotype (33) | URA5 RFLP type (40) | PLB1 RFLP type (41) | ITS genotype (342) | IGS genotype (31, 350) | MLST type (43) |
|---|---|---|---|---|---|---|---|
| B/C | VGI | AFLP4A/AFLP4B | VGI (SH5) | A5 | ITS3/ITS7 | 4 | VGI |
| B/C | VGII | AFLP6 | VGII (SH6) | A6 | ITS4 | 3 | VGII |
| B/C | VGIII | AFLP5A/AFLP5B/AFLP5C | VGIII (SH7) | A7 | ITS5 | 5 | VGIII |
| B/C | VGIV | AFLP7 | VGIV (SH8) | A8 | ITS6 | 6 | VGIV |
TABLE 2.
Standard/reference strains for Cryptococcus gattii strain typinga
| C. gattii PCR fingerprinting molecular type (reference) and AFLP genotype (reference) | CBS strain designation | ATCC strain designation | FGS strain designation | WM strain designation | CMRVS strain designation | Other strain designation(s) | MAT type and serotype | Description | Reference(s) |
|---|---|---|---|---|---|---|---|---|---|
| VGI (40) and AFLP4 (33) | CBS 10078 | ATCC MYA-4560 | 10419 | WM 179 | 70298 | Bryon, H33.1, MH56 | αB | 1993, Sydney, NSW, Australia; clinical, CSF, HIV negative; isolated by Sharon C.-A. Chen | 40, 154 |
| CBS 6289 | ATCC 32269 | WM 01.124 | MUCL 30449, RV 20186, CBS 8273 | aB | 1966, Kinshasa, Congo; clinical, CSF; isolated by E. Gatti and R. Eeckels; type strain of C. neoformans var. gattii | 199 | |||
| CBS 10510 | WM 276 | TCS, SC1 | αB | 1993, Mt. Annan National Park, NSW, Australia; environmental, Eucalyptus tereticornis woody debris; isolated by Tania C. Sorrell and Sharon C.-A. Chen; genome sequence strain | 40 | ||||
| VGII (40) and AFLP6 (22) | CBS 10082 | ATCC MYA-4561 | 10420 | WM 178 | 70302 | 49435, Colter, IFM 50894 | αB | 1991, Sydney, NSW Australia; clinical, CSF, HIV negative; isolated by Sharon C.-A. Chen | 40, 154 |
| CBS 10514 | WM 02.32 | CDC R265 | αB | 2001, Duncan, Vancouver Island, BC, Canada; clinical, bronchial wash specimen; isolated by British Columbia CDC; highly virulent Vancouver Island outbreak strain of VGIIa; genome sequence strain | 38 | ||||
| VGIII (40) and AFLP5 (22) | CBS 10081 | ATCC MYA-4562 | 10421 | WM 175 | 70299 | WM 161, E698, 689, TP 0689, D1.13H | αB | 1992, Blind Recreation Center/Park Boulevard UPAS Street, San Diego, CA, USA; environmental, Eucalyptus species woody debris; isolated by Tania Pfeiffer and David Ellis | 40, 100 |
| CBS 6955 | ATCC 32608 | 10424 | WM 01.125 | DBVPG 6225, WM 2220, MUCL 30454, NIH 191, CBS 6916 | aC | Before 1970, San Fernando, CA, USA; clinical, CSF | 2 | ||
| VGIV (40) and AFLP7 (56) | CBS 10101 | ATCC MYA-4563 | 10422 | WM 779 | 70300 | King Cheetah, IFM 50896 | αC | 1994, Johannesburg, South Africa; veterinary, cheetah; isolated by Valerie Davis | 40, 389 |
Adapted from reference 43.
Interspecies Hybrids
The application of PCR fingerprinting and AFLP typing revealed diploid or aneuploid hybrids in addition to the four major haploid molecular types in C. gattii. These hybrids may have originated from hybridization events between some of the major molecular types of C. gattii and its sister species C. neoformans. These hybrids include the VGI/VNIV (serotype B [VGI/AFLP4] and serotype D [VNIV/AFLP2]) hybrids (44), the VNI/VGI (serotype A [VNI/AFLP1] and serotype B [VGI/AFLP4]) hybrids (45, 70), and the VNI/VGII (serotype A [VNI/AFLP1] and serotype B [VGII/AFLP6]) hybrids (70). Virulence studies using the Galleria mellonella model revealed differences in the virulence of hybrids depending on the genome combinations present. The VNI/VGII (αABa) hybrids were found to be as virulent as haploid strains H99 and CDC R265. The VNI/VGI (αABa) and VNIII (αADa) hybrids were virulent but less so than strain H99 (M. Aminnejad and W. Meyer, unpublished data).
EPIDEMIOLOGY, ORIGIN, AND EVOLUTION
Global Distribution and Movement of C. gattii Genotypes
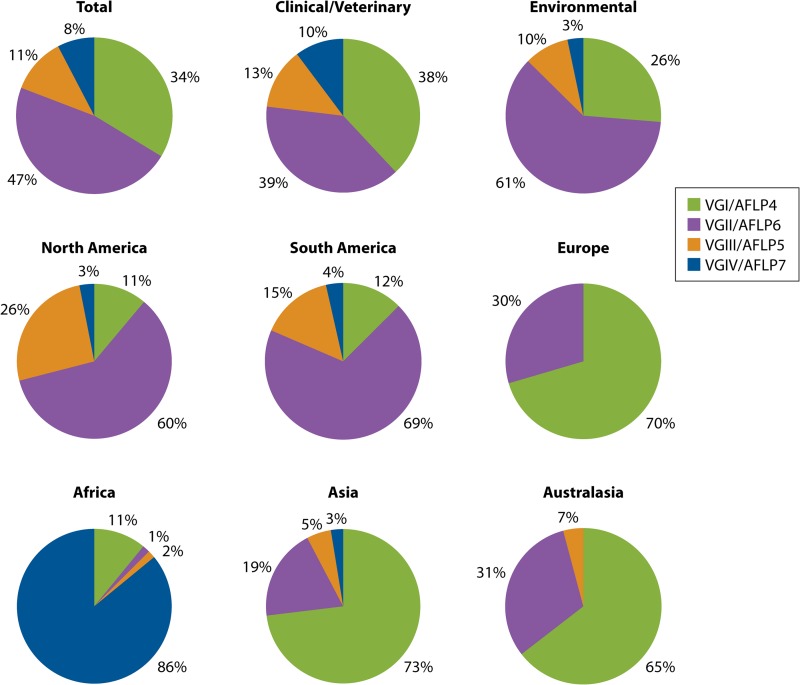
With the recognition of the emergence of VGII and VGIII infections in North America, there is clear evidence that C. gattii has spread to new temperate climatic zones outside the tropical and subtropical areas identified by Kwon-Chung and Bennett (71). It has always been present in the southern temperate zone of Australia. The global distribution of the four major molecular types of C. gattii has been determined by analyses of clinical, animal, and environmental isolates from several studies (33, 38, 40, 72–81; W. Meyer and L. Trilles, unpublished data). Acknowledging potential sample biases, in contrast to previous reports, in which the most frequent molecular type was VGI (48, 80), VGII is now more common (comprising 47% of all isolates), followed by VGI (34%), VGIII (11%), and VGIV (8%) (Fig. 3, top). However, among clinical isolates, VGI and VGII are represented in equal proportions, followed by VGIII and VGIV. Most environmental isolates are of molecular type VGII, although this may reflect sampling bias associated with attempts to determine the origin of the Vancouver Island outbreak, followed by VGI and VGIII. VGIV has rarely been isolated from the environment (Fig. 3, top). The geographic variation in the distributions of VGI, VGII, VGIII, and VGIV is striking.
FIG 3.
Distribution of the four major molecular types of C. gattii identified among a total of 980 clinical/veterinary and environmental global isolates. Shown is the distribution of the four major molecular types of C. gattii, combining clinical/veterinary (n = 615) and environmental (n = 365) isolates (top) from North America (including Mexico) (n = 162), South America (n = 367), and Europe (n = 44) (middle) and from Africa (n = 64), Asia (n = 78), and Australasia (n = 265) (bottom).
The Americas.
In the Americas, VGII and VGIII predominate among clinical, veterinary, and environmental isolates (Fig. 3, middle, and 4), with most VGII isolates being obtained from South America (40). In North America, this distribution of molecular types is commensurate with the establishment of VGII infections in British Columbia and the Pacific Northwest (38, 39, 82, 83) and the emergence of VGIII in southern California and Mexico (39, 84; W. Meyer and C. Firacative, unpublished data). The Canadian and Pacific Northwest outbreak strains are all of mating type α and comprise two subtypes of the major molecular type VGII/AFLP6, namely, VGIIa/AFLP6A and VGIIb/AFLP6B (38). As well as in British Columbia, Canada, molecular type VGII has now been reported in the United States (68, 85, 86) (Fig. 4). AFLP analysis has identified the same VGII molecular subtypes in other temperate regions (1, 38, 87). Closely related strains of a new highly virulent subtype, VGIIc, have been identified in the state of Oregon and, subsequently, in other regions of the Pacific Northwest (68, 69, 86).
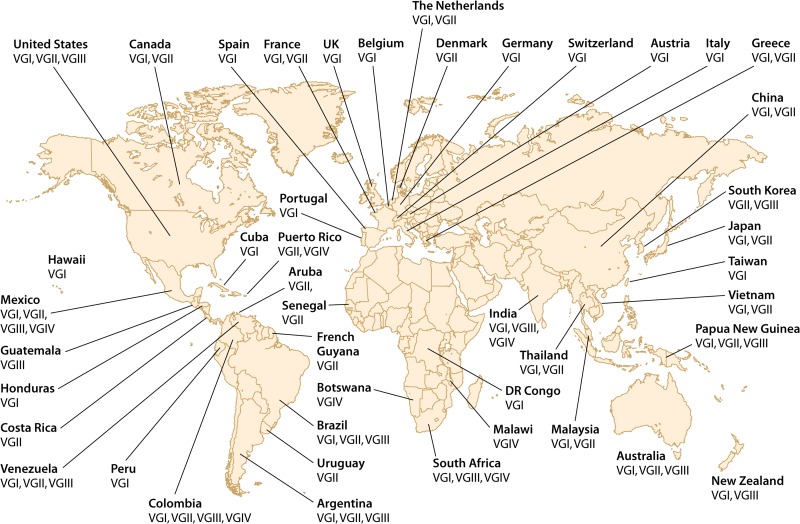
FIG 4.
Global distribution of the four major molecular types of C. gattii, based on 980 isolates.
Based on MLST of global isolates, Fraser et al. initially proposed that the Vancouver Island VGII outbreak strains originated in Australia or South America (1). More recent studies support the proposal that C. gattii molecular type VGII originated in South America (46, 72) and that Australia and Thailand are possible “stepping stones” in the global spread of VGIIb from South to North America (88, 89). The introduction of VGIIb from its ancestral origin in South America, via different parts of the world, has recently been confirmed by using WGST (54). These findings agree with a recent comprehensive, global study of VGII strains, which traced the origin of both outbreak subgenotypes VGIIa and VGIIb back to an ancestral C. gattii strain, LMM645, from the Amazon rainforest. This study revealed the presence of the VGIIa and VGIIb genotypes and MATa and MATα strains in Brazil and showed clear evidence for recombination among the Brazilian C. gattii population (53).
Studies in South America, including Argentina, Brazil, Chile, Colombia, Mexico, Peru, Venezuela, and Guatemala, found that VGII/AFLP6 is common in this part of the world, with 64% of all C. gattii isolates from Brazil being of molecular type VGII (40, 78, 79). The greatest genetic diversity among VGII/AFLP6 strains was found in Brazil (42, 78) and Colombia (74), where both MATa and MATα strains are present. Environmental studies conducted in South America show that VGII is particularly well adapted to biotopes associated with decaying wood (73, 78, 90). The Brazilian epidemiological data suggest that there is also a north-to-south geographical trend, with C. gattii being endemic in the north and northeast Brazilian states of Amazonas, Bahia, Pernambuco, Piauí, and Roraima. Here primary cryptococcosis is caused predominantly by VGII and affects immunocompetent patients, unlike in the southern states (Mato Grosso do Sul, Minas Gerais, Paraná, Rio de Janeiro, Rio Grande do Sul, and São Paulo) (78).
VGIII comprises 21% of Brazilian C. gattii isolates (78) and has been isolated from eucalypts in the Brazilian state of Rio Grande do Sul (72). A Mexican study using PCR fingerprinting with the primer M13 showed the presence of all four C. gattii molecular types, including VGIII (79). Molecular type VGIV is rarely found in the Americas (Fig. 3, middle, and 4).
Europe.
In Europe, C. gattii is isolated from patients and the environment only rarely, with the majority of human infections being imported and due to VGI (91). The occurrence of cases many years after travel to an area of endemicity is consistent with reactivation of dormant infection. In Spain, VGI caused an outbreak in goats (92). A small number of imported human cases of VGIIa/AFLP6A infection have occurred following travel to Vancouver Island (93–95).
Africa.
Although little is known about C. gattii in Africa, its epidemiology appears to differ significantly from that in the rest of the world. Most VGIV strains come from the southern part of Africa, including Botswana, Malawi, and South Africa (96–98). VGI and VGII are mainly from central Africa, for example, the Democratic Republic of Congo (formerly Zaire), Senegal, and South Africa (33, 97, 98) (Fig. 3, bottom, and 4).
Asia and Australasia.
In Asia and Australasia, the leading major molecular type is VGI, followed by VGII; VGIII has been isolated rarely (29, 60, 88) (Fig. 3, bottom, and 4). RAPD analysis of 62 clinical, 29 veterinary, and 45 environmental strains revealed a predominance of VGI among environmental and clinical isolates; two subgroups were identified within VGI and VGII (99, 100). RAPD analysis also identified environmental strains of molecular type VGII from the Northern Territory of Australia. Prior to that report, all C. gattii strains isolated from eucalypts in Australia were of molecular type VGI (29). A small number of VGI strains have been isolated from clinical and veterinary sources in China (75–77). VGIV was reported only from India; these isolates have been closely linked with VGIV isolates from South Africa, the major source of this molecular type. This association may represent an epidemiological link resulting from human migration between the two countries (81).
ECOLOGY
Environmental Studies
The first studies of the environmental niche of C. gattii identified the red gum species of eucalypt, Eucalyptus camaldulensis and E. tereticornis, as the major environmental sources of C. gattii VGI (25, 101), and seasonal flowering was linked with airborne dispersal (26, 102, 103). C. gattii concentrations in air samples obtained during the Vancouver Island outbreak were significantly higher in the warm, dry summer months, although potentially infectious propagules (<3.3 μm in diameter) were present throughout the year (104). Decomposing wood in tree hollows has been identified as an environmental source (22, 105). Eucalypts are in fact only one of >50 different species of tree that can provide an ecological niche for C. gattii (106–110). Environmental isolates have been obtained from Africa (33, 110–112), Asia (76, 77, 108), Australia (26, 110, 113–118), Europe (110, 119, 120), North America (38, 110, 121), and South America (122). In addition, it has been shown that plant material promotes fertility (108, 123–125) and may render C. gattii strains more pathogenic (125). Molecular types VGI and VGII are those most commonly associated with a wide variety of tree species (74, 110, 126). VGIII has been isolated from eucalypts and almond trees in Colombia and other trees in the greater Los Angeles area of California (74, 126–128), and both mating types of VGIV were found in the same two species in Colombia (127) and Tunisia (112).
Notably, both C. neoformans and C. gattii were isolated concurrently from a hollow in a pink shower tree in Piauí in northeast Brazil (73, 107, 129), and C. gattii molecular types VGI and VGII coexisted in the hollow of an Australian Eucalyptus tereticornis tree (115).
Reproduction in Nature
Initial studies of VGI isolates obtained from E. camaldulensis trees suggested that environmental populations of C. gattii are highly fragmented, have a limited ability to disperse, and are confined to individual tree hollows (114). AFLP analysis of clinical isolates revealed that they are geographically restricted, highly clonal, and of low fertility (53, 116, 117). Subsequent extensive testing of isolates from single tree hollows by AFLP analysis revealed that recombination is occurring in both MATa and MATα or MATα-only populations (118). Isolates of VGII from Australia are highly fertile and recombining (117), with a recent study also suggesting recombination within the MATα-MATa population, especially in Western Australia (88). Among a global collection of C. gattii clinical, veterinary, and environmental strains, representing molecular types VGI, VGII, VGIII, and VGIV, the topology of the four clades was noncongruent, suggesting the occurrence of recent recombination events within C. gattii (46). These observations suggest that C. gattii populations are likely to be sustained in nature by recombination between the same or opposite mating types.
PATHOGENESIS
This section is intended to provide a historical summary and to highlight the complexity revealed by the use of genomic and other “omics”-related technological advances. Rather than providing a comprehensive review of the literature, we provide insights from key studies and, in addition, those that focus on C. gattii.
To establish invasive disease, inhaled cryptococci must penetrate lung tissue, reproduce, enter the bloodstream, and thence disseminate to other organs, predominantly the central nervous system (CNS). Experimental data on the pathogenesis of C. gattii infection are limited. In animal models, as with the much more intensively studied C. neoformans, the outcome of exposure is determined by a complex set of interacting pathogen-derived and host factors. Broadly, these factors include the extent of exposure to cryptococci (inoculum size), the cryptococcal strain/genotype and its virulence composite, and the host immune system. In human cryptococcosis due to C. gattii, genotype-associated clinical and host differences have been observed. For example, VGI infection typically presents with CNS disease, including cryptococcomas, often with concurrent lung lesions (130), whereas VGII infection more commonly presents with pulmonary disease (131, 132). Both of these molecular types of C. gattii have a predilection for immunocompetent hosts. In contrast, molecular type VGIII was overrepresented in a series of HIV-infected patients in southern California (84, 133), and VGIV has been identified in Africa in association with HIV/AIDS (see Clinical Epidemiology of Human Infection, below).
Cryptococcal Virulence Determinants
C. gattii expresses the same suite of major virulence determinants as C. neoformans. These determinants include the polysaccharide capsule, the ability to grow at 37°C, and laccase activity, which is responsible for the production of melanin, phospholipase B (Plb1), urease, superoxide dismutase (Sod1), and trehalose. Some gene products, such as the capsule (134) and Plb1 (135, 136), have been well characterized in C. gattii, but in few instances have deletion mutants or overexpressing strains been created and tested for virulence in animal models. An exception is the laccase gene LAC1, created in R265, the virulent VGIIa strain from the Vancouver Island outbreak, the overexpression of which was associated with increased virulence in a mouse inhalation model (137). In that same study, disruption of CAS1, which is associated with the construction of the polysaccharide capsule backbone in C. neoformans (138), did not affect capsule size but, when overexpressed, was associated with hypervirulence in mice. Disruption of the signal transduction pathway molecule MPK1, which causes defective melanin synthesis and cell wall integrity, was associated with reduced virulence (137). Additional genes, transcription factors, and signaling molecules that have been linked to virulence in C. gattii are summarized in Table 3. A recent comprehensive review of stress signaling pathways and the pathogenicity of Cryptococcus was provided by Bahn and Jung (139). Other virulence-related functions in C. neoformans are likely to be conserved in C. gattii. These functions include lipid signaling pathways, adaptation to hypoxia, determinants of proliferation and metabolism in the host, intracellular trafficking and pH sensing, iron and copper regulation and uptake (140), and zinc metabolism (141).
TABLE 3.
Comparison of cellular functions and virulence of Cryptococcus gattii and Cryptococcus neoformans by using deletion mutantsa
| Gene(s) of interest, product | Function | Phenotype | Required for virulence, model | Reference(s) |
|---|---|---|---|---|
| SOD1, superoxide dismutase | Cytoplasmic antioxidant | Required for production of virulence factors urease, PLB, and laccase in C. gattii but not C. neoformans | Yes, in BALB/c mouse intravenous inoculation model and in A/JCr mouse inhalational model of C. neoformans | 390 |
| SOD2, superoxide dismutase | Mitochondrial antioxidant | Required for growth at 37°C in 20% but not in 1.3% oxygen | Yes, for C. gattii BALB/c mouse inhalation and i.v. inoculation model; C. neoformans was not tested | 391 |
| TPS1 and TPS2, trehalose-6-phosphate synthase | Trehalose biosynthesis; trehalose functions as an antioxidant and stress protectant | Required for thermotolerance, capsule and melanin production, mating, and cell wall integrity in C. gattii and thermotolerance in C. neoformans | Yes, for C. gattii Caenorhabditis elegans (worm) and A/JCr mouse inhalational models; TSP1 but not TSP2 is required for virulence in C. neoformans | 392, 393 |
| PKA1, cAMP-activated protein kinase A | Signal transduction pathway regulator | Required for capsule production in C. gattii and mating and capsule and melanin production in C. neoformans | C. gattii was not tested; yes, for virulence of C. neoformans in BALB/c mouse inhalational model and immunosuppressed rabbit CSF inoculation model | 394, 395 |
| PKA2, cAMP-activated protein kinase A | Signal transduction pathway regulator | Required for mating and capsule and melanin production in C. gattii but not C. neoformans | Not tested | 394, 395 |
| PLC1 | Signal transduction pathway regulator | Regulates growth at 37°C and melanin and PLB production in C. neoformans through the PKC/MAPK pathway (see below) | Yes, in C. neoformans BALB/c mouse inhalational model | 396 |
| MPK1 | Signal transduction pathway regulator | Regulates melanin, capsule production, and cell wall integrity in C. gattii and thermotolerance and cell wall integrity at 37°C in C. neoformans | Yes, in C. gattii (BALB/c inhalational model) and in C. neoformans i.v. DBA/2 complement-deficient mouse model | 397 |
| STE12α | Transcription factor | Regulates melanin, mating, and ecological fitness in C. gattii and mating and capsule size in C. neoformans | Yes, in C. gattii but not C. neoformans | 398, 399 |
| GAT1 | GATA transcription factor | Regulates nitrogen utilization | Yes, in C. gattii but not in C. neoformans BALB/c intrapharyngeal instillation model | 400 |
| CNA1, calcineurin catalytic subunit (A) | Subunit of the heterodimer calcineurin, a Ca2+ calmodulin-activated serine-threonine-specific protein phosphatase | In C. gattii, regulates thermotolerance (37°C) (strains differ) and is required for plasma membrane integrity, tolerance to fluconazole, and optimal growth in the presence of Ca2+ Li+, with no role in melanin and a minor role in capsule production; in C. neoformans, lesser effect of Ca2+ and not required for fluconazole tolerance in | Yes, in C. gattii (molecular type-dependent) G. mellonella (wax moth) larva and A/JCr mouse inhalational models and also in C. neoformans rabbit intracisternal inoculation and BALB/c mouse i.v. inoculation models | 156, 401, 402 |
Note the functional divergence between C. gattii and C. neoformans for all genes analyzed, with an evolutionary switch of functions of PKA1 and -2. cAMP, cyclic AMP; PKC, protein kinase C; MAPK, mitogen-activated protein kinase; PLB, phospholipase B.
Function of major virulence determinants.
The importance of tolerance to growth at physiological temperatures is self-evident. The polysaccharide capsule functions in evasion/suppression of the host immune response, laccase is responsible for the production of the oxidative stress protectant melanin, phospholipase B (Plb1) and urease promote invasion of host tissue, and superoxide dismutase (Sod1) and trehalose function as antioxidants (for a major review, see reference 142). In murine models of infection with C. neoformans strain H99 and deletion mutants, Plb1 and laccase are essential for the egress of cryptococci from the lung and dissemination via the blood to the CNS (143, 144). Plb1 and urease are required to establish cerebral cryptococcosis, urease promotes the migration of cryptococci across the blood-brain barrier (BBB) (143–146), and Plb1 is required for the extrusion of C. neoformans from macrophages (147). Urease has also been reported to promote an anti-inflammatory Th2 cellular response in the lung (148).
Virulence in experimental models.
Virulence per se is most often assessed in survival studies by using mouse inhalational models (to mimic the route of natural infection), invertebrates or insects such as Galleria mellonella (wax moth) larvae, or rates of proliferation in the mouse macrophage-like cell line J774, expressed as the intracellular proliferation rate (IPR) (142). Inoculation of the tail vein in mice has also been commonly used, and since intravenous (i.v.) inoculation bypasses the respiratory route, it is especially useful in elucidating determinants of pulmonary versus CNS infection. The invertebrate/insect and macrophage models are cheap and convenient, allow higher throughput, and yield results similar to those in mice. Although none of these animals are natural hosts, their relevance has been validated by observations of human cryptococcosis. Thus, in the recent C. gattii VGII outbreak in British Columbia, ∼90% of outbreak strains were of the VGIIa genotype, and ∼10% were of the VGIIb genotype (suggesting that in humans, it is more virulent than the minor genotype VGIIb). This clinical observation is consistent with the much decreased survival and, hence, greater virulence of C. gattii VGIIa in mice and a substantially increased ability to replicate within J774 macrophages (1, 38, 84, 149). A more extensive study of VGIIa and VGIIb isolates from around the world confirmed these findings (150). More recently, zebrafish larvae have been proposed as a tractable model in which to study cryptococcal interactions with the innate immune system (151).
Correlations between the molecular type of C. gattii and virulence have been made. In general, VGIIa is more virulent than VGIIb in mice, and VGII overall is more virulent than VGI (36, 38). However, such global assessments of virulence may not yield consistent results, suggesting that molecular type may be a marker, but not a determinant, of virulence. For example, in mice, the clinical type strain of C. neoformans, H99, was less virulent than the C. gattii VGIIa type strain, R265, from the Vancouver Island outbreak (1, 150) but was equally virulent in another study (152). Indeed, threshold inocula required for disease expression and, in some cases, organ burdens are strain dependent (153), and individual isolates of the same molecular (sero)type of C. gattii may be more or less virulent than isolates of C. neoformans (154). Subtype-dependent differences in virulence have also been described for VGIII strains isolated from patients with AIDS. In particular, strains of VGIIIa were more virulent than those of VGIIIb (84). In this context, a recent report is of interest (155). Assessment of hypervirulence markers (IPR and tubularization of mitochondria) during mating of VGII and VGIII strains and within VGII strains revealed that these traits were infrequently transferred in outgroup crosses but readily transferred during ingroup (within VGII) crosses.
Phenotypic and genotypic virulence correlates.
In early studies of C. gattii VGI, phospholipase production per se quantified in vitro was correlated with virulence in mice (154). Differences between the two closely related VGII Vancouver Island outbreak subtypes were noted; all VGIIa isolates were fertile, compared with only 17% of VGIIb isolates, and at 37°C, growth rates, melanin production, and capsule size were increased (150). In another study of Vancouver Island outbreak strains, capsule size; melanin, phospholipase, and proteinase production; and other enzyme activities were analyzed in 39 C. gattii strains of variable virulences and different molecular types. There was no correlation between virulence and individual virulence phenotypes. Despite this, expression of PLB1, CRG1, capsule-related genes, and genes on the mating-type locus correlated with the IPR in macrophages, suggesting that these genes contribute synergistically to virulence in the VGII (AFLP6) lineage (142). There is also evidence that the relative importance of particular virulence determinants varies between molecular types of C. gattii. For example, VGIIa was more tolerant to growth at 37°C in the presence of calcineurin inhibitors or in CNA1 deletion mutants than isolates of VGI, VGIIb, VGIIc, VGIII, and VGIV. Calcineurin was required for virulence in the WM 276 and R265 backgrounds but much less so in the R272 background, whereas melanin production levels were similar in all three strains (156).
These observations confirm that virulence, as measured by disease outcome, is not simply the sum of individually expressed determinants but rather the outcome of a complex set of interacting factors. The term fungal virulence composite was coined by Cutler in 1991 (157) to reflect this in Candida albicans. More recently, the deterministic nature of cryptococcal virulence was elucidated in G. mellonella and Acanthamoeba castellanii models, which confirmed that the cryptococcal phenotype does not necessarily correlate with disease-causing potential, nor does the expression of individual virulence factors correlate directly with pathogenicity (158). The complexity of the fungal virulence composite is further exemplified by the study of genomic and transcriptomic profiles in cryptococcal species.
Genomic and Transcriptomic Profiles
The advent of the omics revolution has revealed a complex array of genes and their transcripts that contribute to the virulence composite in Cryptococcus spp. Sequencing of a C. gattii VGI environmental strain (WM 276) and the VGIIa Vancouver outbreak strain (R265) revealed that the genomes are colinear for most of the 14 chromosomes, with minor rearrangements, but comparison of different strains showed considerable variation in gene content and overall sequence identity. There was extensive variation between strains of C. gattii VGII, precluding simple explanations of differences in virulence based on genome content (36). C. gattii strains that are very closely related by genome analysis can have widely different transcriptional circuits and phenotypic differences in virulence in experimental models of infection (159).
Transcriptomes of C. gattii have been analyzed in an attempt to fully characterize the C. gattii virulence composite. A recent whole-genome expression study identified mitochondrial regulation as an important factor in the enhanced pathogenicity of genotype VGIIa (149); notably, mitochondria were shown to be essential for cryptococcal viability and for protection against hypoxia and oxidative stress. As mentioned above, expression of genes encoding known virulence factors, for example, PLB1, CRG1, capsule-related genes, and genes on the mating-type locus, correlated with the IPR in J774 macrophages, suggesting a synergistic contribution to virulence in the VGII lineage. Previously uncharacterized genes shown subsequently to influence melanin and capsule production and, thus, cryptococcal infectivity (for example, the COP9 signalosome complex and ubiquitin carboxyl-terminal hydrolase) were also upregulated in VGIIa strains (149).
Comparison of the transcriptomes of R265 (VGIIa) and R272 (VGIIb) revealed increased expression levels of genes encoding melanin, capsule, and growth at 37°C (LAC1, LAC2, CAS3, and MPK1). Levels of proteins involved in cell wall assembly, membrane components, carbohydrate and lipid metabolism, transport, the stress response, and lignin degradation were all increased in the VGIIa strain, whereas genes involved in the regulation of mitosis, ergosterol biosynthesis, and drug resistance were downregulated (137). Differences in several other genes, including cell wall assembly and mitotic regulatory genes, were identified. Unlike the study by Ma et al. (149), the expression of the mitochondrial genes was not different in R265 and R272. The differences in the results from the two studies can be explained by the similarity in mitochondrial genomes of strains R265 and R272, since creation of congenic a and α strains by backcrossing of R265 with a nonoutbreak VGII strain indicated that neither the mating type nor the mitochondrial genotype was a major virulence determinant (407).
Observations In Vivo
In both rat and mouse models, there is evidence that C. gattii VGII strains cause significant lung disease but disseminate to the CNS infrequently (153, 160). In a rat model of C. gattii VGII infection, the five strains studied included MATα strains R265 (VGIIa) and R272 (VGIIb) from Vancouver Island, which caused progressive and ultimately fatal pneumonia, but consistent and substantial late dissemination to the CNS was noted only with a VGIIb strain of feline origin from Australia (153). In a rat intrapulmonary injection model, strain variation was again apparent, with an environmental isolate (VGI) and an Australian feline isolate (VGIIb) causing more severe pulmonary disease than 2 human clinical isolates of VGI. Increased virulence in rats was correlated directly with extensive infiltration of the lungs with budding cryptococci and inversely with the extent of the inflammatory response. Only the most virulent isolate disseminated to the brain; in these animals, microscopic examination revealed occasional single or budding yeasts in the meninges (153). Pulmonary responses in C57BL/6J mice infected with the H99 or R265 strain differed substantially, with proliferation of intra-alveolar yeast and reduced macrophage responses to infection with R265 and an intense nodular macrophage and multinucleated giant cell response to H99. Interestingly, mortality rates were similar in both groups (161, 162).
Another group found that mice infected by intrapharyngeal instillation of VGII strain R265 died from extensive pulmonary infection without significant brain involvement, whereas C. neoformans strain H99 disseminated readily to the brain and caused death from cerebral infection (160). Notably, strain R265 induced a relatively poor inflammatory response compared to that induced by strain H99, and both H99 and R265 crossed the blood-brain barrier to establish fatal brain infection upon intravenous inoculation. The predilection of R265 for the murine lung is consistent with the clinical observation that human infections with the C. gattii VGII genotype present more commonly with pulmonary than with neurological infection (131).
The influence of C. gattii on the host immune response is poorly understood, although studies with C. neoformans underscore the importance of the innate immune response in the pathogenesis of cryptococcosis. Cellular interactions between neutrophils, natural killer (NK) lymphocytes, monocytes, and macrophages have been investigated, predominantly in vitro, in order to understand the inflammatory/immune response in vivo. Cryptococcal genotype-, strain-, and species-dependent effects have been observed in different studies but have not necessarily correlated with the outcome of infection in vivo.
For example, a nuclear magnetic resonance (NMR) spectroscopic analysis of metabolites released by single strains of C. gattii and C. neoformans identified 23 strains with supernatant concentrations differing by at least 2-fold and two metabolites, acetoin and dihydroxyacetone, that were uniquely released by C. gattii. The effect of the supernatants from cultures of C. neoformans was generally more proinflammatory, causing increased neutrophil necrosis and phorbol myristate acetate (PMA)-induced neutrophil superoxide production and greater adhesion/migration of cryptococci through A549 lung epithelial cell monolayers. Despite these differences in vitro, these two strains produced similar neutrophil responses in the rat lung (162). In C57BL/6 mice infected with C. gattii VGII Vancouver Island outbreak strains R265 and R272, Australian environmental strain WM 276, and C. neoformans strain H99, early migration of neutrophils to sites of infection and production of protective cytokines were reduced in mice infected with the C. gattii strains, but strain R265 and C. neoformans strains were equally virulent in mice and more virulent than the other two C. gattii strains (152).
NK cells are the predominant lymphocytes involved in the innate immune response to cryptococcal infection. They exert a direct cytotoxic effect on cryptococcal cells via exocytosis of lytic granules containing the effector compound perforin (164). It was recently reported that perforin degranulation is increased in an acidic environment, at pH levels reported previously to be generated within cryptococcomas (163); this effect was observed with both C. gattii and C. neoformans (165).
Alveolar and pulmonary macrophages are a first line of defense against cryptococcal infection. Animal models reveal substantial numbers of cryptococci within pulmonary macrophages and an increase in the number of extracellular organisms during the progression of disease (for example, see reference 143). In C. neoformans models, cryptococci exit the lung and are transported in the blood in mononuclear phagocytes (143) and as free cells (146). The cryptococci cross the blood-brain barrier either via transendothelial cell transport of free cryptococci or by paracellular transport within the phagolysosome of mononuclear phagocytes (the so-called Trojan horse mechanism) (146, 166). Much is now known about phagocytosis, phagolysosomal inhibition, multiplication, and egress of C. neoformans from macrophages in the pathogenesis of disease (reviewed in reference 142), and the IPR of C. gattii VGIIa is enhanced in J774 murine macrophages and primary human monocyte-derived macrophages (149).
There are relatively few studies of host cell interactions with C. gattii that provide novel insights into pathogenesis, but a recent publication on the evasion of the adaptive immune response by C. gattii strains R265 and R272 is instructive, as similar evasion has not been demonstrated for C. neoformans (167). This group demonstrated that heat-killed C. gattii is efficiently bound to, and internalized by, human peripheral blood-derived dendritic cells (DCs), trafficked to late phagolysosomes, and killed. However, the DCs were not stimulated to mature into efficient antigen-presenting DCs, as measured by increased expression levels of major histocompatibility complex (MHC) class II and the costimulatory molecules CD86, CD80, CD83, and CCR7 (a chemokine associated with trafficking of DCs to lymph nodes) or decreased expression levels of CD11c and CD32. As a result, specific T cell responses were suboptimal. It was further determined that tumor necrosis factor alpha (TNF-α) levels were not increased, as would be expected following phagocytosis, but exogenous TNF-α or stimulation of its production restored DC maturation and normal T cell responses to cryptococci.
C. GATTII INFECTION IN ANIMALS
C. gattii is an emerging pathogen in a broad range of animals, including domestic cats, dogs, horses, sheep, cows, koalas, dolphins, gray squirrels, ferrets, birds, and marsupials. The epidemiology and clinical and pathological features of cryptococcosis, including that due to C. gattii, in various animals have been described in detail (168–171). This review focuses on more recent insights into C. gattii infection.
In North America, the prevalence of C. gattii infection relative to that of C. neoformans infection remains imprecise, since many veterinary isolates have not undergone identification to species level. Furthermore, the prevalence of each pathogen varies with the species of animal (168, 169). In two studies involving 82 cases of feline cryptococcosis in California (169) and western Canada (172), only 17 cryptococcal isolates were identified, 14 of which were C. gattii and 3 of which were C. neoformans. Much of our knowledge of species distribution comes from studies from Australia (99, 173). As companion dogs and cats are considered sentinel species, disease distribution may be expected to be similar to that in people (see Clinical Epidemiology of Human Infection, below).
In Australia, the majority of infections (and colonization) in native koalas and horses are due to C. gattii (174, 175), compared to about 33% in domestic cats and dogs (173). In Western Australia, C. gattii caused 5 of 9 feline and 11 of 22 canine infections (175). C. gattii causes disease in immunocompetent mammals and is considered a primary pathogen (168, 173). Different manifestations of cryptococcosis in different species may be due to a number of factors, including host anatomical differences, physiological differences (e.g., body temperature), ecological habitats, and species-specific differences in comorbidities, immunology, and behavior, which affect exposure and host reaction to the fungus. These aspects are not reviewed, but the salient features of C. gattii infection in various animals are presented.
Case reports and moderate-to-large case series of C. gattii in cats abound (37, 169, 171–173, 176–182). A review of the major studies where the etiologic cryptococcal species were reported indicates that there are no clinical features which reliably distinguish disease caused by C. neoformans from that due to C. gattii. As with C. neoformans infection, young to middle-aged cats (2 to 3 years of age) are typically affected (173), but the age range is wide. In Australia, Siamese, Birman, and Ragdoll breeds are overrepresented. Exposure to the outdoors is significantly associated with cryptococcosis, but indoor cats are also affected, presumably through exposure to aerosols, soil, or plant material (173, 178). Of note, one study found that cats in rural areas were significantly more likely to have C. gattii than C. neoformans infection (173). In the Vancouver Island outbreak, Duncan et al. reported that in a matched case-control study, cats that were active, had traveled on the island, or lived near a site of commercial environmental disturbance (including logging and soil disturbance) had a significantly increased risk of developing C. gattii infection (183). These findings pertained to the period between 2001 and 2003, and follow-up studies are worthwhile to determine if these activities still pose a risk 10 years later, since they have implications for pet owners. Interestingly, the majority of cats infected with the VGII genotype in two Australian studies (173) presented with more extensive disease than those infected with the VGI genotype, suggesting that VGII is more virulent than VGI. Studies of a larger numbers of cats with both types of infection, in other regions, are required to validate this hypothesis.
Cats present primarily with nasal and paranasal cavity disease. Other common sites of infection include the skin, lymph nodes, brain, meninges, and eyes. Typically, granulomatous lesions occur, especially in the head and neck regions, such as over the maxillary or frontal areas and the nares (173, 178, 179). Eye involvement, characterized by chorioretinitis and blindness, is not uncommon. Single or multiple cryptococcomas can occur in the brain and spinal cord (181). Unlike in koalas and horses (see below), lung manifestations, including pyothorax, are uncommon (180). However, sneezing, stertor, snuffling, and headshaking can occur (168, 169). In southwestern British Columbia, 27% of 78 feline cases had such respiratory symptoms (171). Nasal deformity with or without purulent, serous, or hemorrhagic nasal discharge, either unilateral or bilateral, is characteristic. Owners may report lethargy and poor appetite in their pets, but fever is uncommon (168, 169). Disseminated infection occurs in 8 to 16% of cases, and any organ may be affected, including the gastrointestinal and genitourinary tracts (169, 172, 173, 175, 178). In Canada, a multivariate survival analysis revealed that CNS infection but not respiratory disease or other medical history was a significant predictor of mortality (171). Retroviral infection in cats does not appear to predispose them to cryptococcosis (172). The prevalence of C. gattii infection in cats with feline immunodeficiency virus or feline leukemia virus infection is not known.
Collectively, young active dogs <6 years of age comprise the majority of cases, with no sex predilection. Large breeds (e.g., Border Collies, Boxers, Dalmatians, Doberman Pinschers, Great Danes, and German Shepherds) are overrepresented, presumably reflecting their active outdoor life-style (171, 172, 183). Outdoor walks through forested areas and accompanying humans on hiking activities were associated with C. gattii disease in the North American outbreaks (171, 183). In Australia, unlike with cats, a rural domicile was not associated with C. gattii infection (172, 173).
Dogs typically present with clinical signs involving more than one organ system (172, 184). In one study (173), although nasal cavity involvement was important, the canine cohort had a greater propensity to develop secondary CNS involvement and disseminated disease than feline cases. Neurological signs, including stumbling, paralysis, ataxia, hyperesthesia (often along the dorsal areas and neck), are most common, and seizures may occur. Blindness is not as common as in cats (168). Epistaxis, sneezing, and nasal discharge are present in ∼50% of cases of cryptococcosis (173, 175, 184, 185). The skin, kidney, and gastrointestinal tract may also be involved. As for cats, there are no clinical features that distinguish cryptococcosis caused by C. neoformans from that caused by C. gattii.
Cryptococcosis was first reported in koalas in 1960 (186) and is almost always due to C. gattii (174, 187). The koala habitat in the wild is the same as that of the species of eucalypt that has been identified as the environmental niche of C. gattii. Animals become heavily colonized with the fungus: colonization rates of 94 to 100% were found in koalas residing in the Coffs Harbor Wildlife Park, New South Wales, Australia, and the Sydney Taronga Zoo (174, 188). Koalas are susceptible to invasive infection and may act as reservoir hosts for humans by amplifying the environmental burden of C. gattii (187, 189). Of interest, in a recent Japanese study of 15 imported Australian koalas in a zoo in Yokohama, only 1 of 10 (66.7%) animals colonized with Cryptococcus was colonized with C. gattii (190). Koalas with invasive cryptococcosis most often present with pneumonia and meningoencephalitis, although nasal infection also occurs (187). Identification of C. gattii in asymptomatic koalas (188–190) has spawned studies of subclinical cryptococcosis, of the mechanisms that trigger clinical infection, and of the need for effective cryptococcal eradication measures to prevent the spread of infection.
Sheep often develop infections of the nasal cavity or CNS, while horses and goats present primarily with lung disease (92, 168, 175). In horses, the sinuses (animals present with rhinitis) are most consistently involved, but both pneumonia and CNS disease are also common. Horses with C. gattii infection in Western Australia (n = 9) had primarily pneumonia; cases of bone, lymph node, and bowel involvement were described, but the causative cryptococcal species was not specified (175). In birds, the presentation is variable, but upper and lower respiratory infections are relatively common. Granulomatous skin nodules on the neck or body and retrobulbar lesions also occur (172, 191). At least five confirmed cases of cryptococcosis due to C. gattii in ferrets, including cases from Spain, have been reported, two of which were autochthonous cases (192, 193); lesions were present in the nasopharynx, lung, lymph nodes, CNS, and bone.
In the United States and Canada, C. gattii infection in dolphins and porpoises has been reported (37). It is not clear how marine mammals become infected, but C. gattii can survive in freshwater and seawater for up to a year, suggesting that this may be a route of infection. Alternatively, these mammals may acquire infection through inhalation of aerosolized spores, as is the case for other mammals, since lung infections in dolphins and porpoises have been reported (37, 194). Norman et al. described a case of maternal-fetal transmission in a porpoise (195).
CLINICAL EPIDEMIOLOGY OF HUMAN INFECTION
In humans, C. gattii causes predominantly meningoencephalitis and other CNS and pulmonary diseases and is associated with substantial morbidity (27, 28, 130–132). It also poses a concern for veterinarians, and some animal infections, in particular those in feline and canine companion animals and Australian native koalas, represent sentinels for human exposure (37, 176, 188, 190). Shifts in the appreciation of the clinical epidemiology of C. gattii in the past decade include the recognition that it affects immunocompromised (including those with HIV/AIDS) as well as immunocompetent (130–133) hosts, the identification of multiple C. gattii molecular types (see Epidemiology, Origin, and Evolution, above) and their clinical implications, and the broadening of its geographic range beyond the tropical-subtropical locales of Northern Australia, Papua New Guinea (PNG), Southeast Asia, Africa, South America, and California (71) to the temperate regions of these and other countries (38, 69, 86, 91, 130, 132, 196–198). This review focuses on these changes and their possible associations with disease patterns in humans.
Epidemiology of Human Infection
The first clinical case of cryptococcosis in which mention of the name C. gattii was made was reported in 1970, involving a 7-year-old boy with leukemia from the Congo (formerly Zaire) who presented with meningitis (199), although in retrospect, the application of molecular methods revealed that an isolate recovered from a patient with infection due to Saccharomyces subcutaneous tumefaciens was, in fact, C. gattii (19). Since then, the literature has abounded with reports of this uncommon infection in single patients and small case series. The emergence of larger case clusters in 1999 in North America and reports of autochthonous cases outside known regions of endemicity has exponentially increased awareness of C. gattii as a pathogen. The major C. gattii reports (confirmed by culture- or molecular-based methods) in English are listed here or are referred to below (27–29, 38, 69, 86, 91, 94–96, 98, 106, 113, 130–133, 196–198, 200–253). Table 4 summarizes the pertinent epidemiological and clinical features in case series involving three or more patients. In addition, a number of reviews, editorials, and book chapters with substantive discussion on C. gattii clinical epidemiology and infection in humans have been published since 2000 (39, 85, 122, 254–265).
TABLE 4.
Epidemiology and clinical features of major case series of Cryptococcus gattii infectionc
| Reference | Location | No. of patients | Site(s) of infection | Host status(es) | Mortality rate (%) | Induction antifungal therapy | Complication(s) (no. of cases) | Sequela(e) (no. or % of survivors with sequela) |
|---|---|---|---|---|---|---|---|---|
| 218 | PNG | 3 (all children) | Meninges | IC | 33 | cAMB (6 wk) | Abnormal mentation (2), papilledema (2), retinal hemorrhage (1) | None |
| 213 | PNG | 7 (1 child) | Meninges | IC | 43 | cAMB + 5FC (mean, 7 wk) | Papilledema (3), blurred vision (3), deafness (1), seizures (1) | Not specified |
| 208 | PNG | 82 | Meninges | IC | Not stated | Not specified | Visual loss in 52.6% of survivors | Visual loss (52.6%) |
| 209 | PNG | 88 | Meninges | IC | 34.1 | Not specified | Not specified | Not specified |
| 211 | PNG | 49 | Meninges | IC | 22.4 | cAMB + 5FC; total dose of cAMB ranged from 890 mg to 2.4 g | Papilledema (26), blurred vision (25), abnormal mentation (12), cranial nerve palsy (10), seizures (7), raised ICP (3/6) | Visual loss (31%) |
| 204 | NT, Australia | 18 | CNS +/− lung | Primarily IC | Not specified for C. gattii but 9.1 overall | cAMB (range, 0.85 to 5 g) | Not specified | Not specified |
| 27 | Australia | 26 | CNS (26) | IC | 15 | cAMB + 5FC (median, 2.4 g cAMB) | Papilledema (13), abnormal mentation (8), focal signs (1), seizures (10), hydrocephalus (9) | Moderate-to-severe neurological sequelae (8 patients) |
| 28 | Australia | 20 | Meninges (17), brain (7), lungs (13) | IC | 0 | Not specified for C. gattii | Hydrocephalus (4), focal signs (5) | Neurological sequelae (7 patients) |
| 29 | Australia | 47 | Meninges (27), brain (10), lung (30), skin (3) | IC (41), IS (5) | Not specified | Hydrocephalus (5) | Not specified | |
| 215 | NT, Australia | 12 | CNS +/− lung (8), lung only (4) | Not specified | 33 | cAMB + 5FC (avg, 43 days) | Hydrocephalus (at least 3) | Not specified |
| 130 | Australia | 86 | CNS +/− lung (73), lung only (10) | IC (62), IS (24) | 13.6 | AMB + 5FC (57 patients) (6 wk), FLU (9) (4 wk), AMB + 5FC (7) (2 wk), FLU (2) (not specified) | Papilledema (9), abnormal mentation (18), cerebellar deficit (10), limb weakness (6), seizures (5), cranial nerve palsy (13), raised ICP (31; 42%), hydrocephalus (22; 30%) | Neurological sequelae (20; 27%)a |
| 98 | South Africa | 46 | Mostly meninges | HIV (29), non-HIV/unknown (18) | 35.6 | AMB (14), FLU (32), AMB + FLU (5) | ||
| 96 | Botswana | 29 | Meninges | HIV (29) | 17 | AMB (14 days) | ||
| 131 | Canada | 3 | Lung + CNS (1), lung only (2) | IC | 0 | AMB (1), FLU (2) | None | None |
| 198 | Canada | 218 | Lung only (167), CNS only (17), CNS + lung (22), other (2) | IC (148), IS (70) | 8.7 | Not specified | Not specified | |
| 230 | Canada | 38 in case-control studyb | Meninges (10), lung (28) | IC (not stated), IS (not stated) | ||||
| 132 | USA (Pacific Northwest states) | 96; 83 outbreak infections, 13 nonoutbreak infections | Meninges (29), Brain (6), lung (31) | IC (62), IS (34) | 33 | Not specified | Papilledema (3/49), blurred vision (9/49), seizures (4/49) | |
| 248 | USA (non-Pacific Northwest states) | 25 | CNS (19), lung (9), CNS only (12), lung only (12), blood (3), leg (1) | IC (20), IS (5) | 24 | AMB + 5FC (17 patients), AMB (1), FLU (1), unknown (2) | Papilloedema (2), blurred vision (8), seizures (1), cranial nerve palsy (5/13), hydrocephalus (4/18) | Not specified |
| 239 | Vietnam | 10 | Meninges | HIV negative | 10 | cAMB + 5FC (Vietnamese national guidelines) | Papilledema (5), abnormal mentation (1), blurred vision (4), focal signs (5) | Overall, blindness in 6 patients, deafness in 1 patient, and neurological deficits in 14 patients, but data were not stratified by infecting Cryptococcus species |
| 206 | Brazil | 8 | Unknown | IC (7), IS (HIV infection) (1) | Not stratified by cryptococcal species | Not stratified by cryptococcal species | ||
| 220 | Brazil | 11 | CNS alone (9), CNS and lung (2) | IC | 18.2 | AMB (7), AMB + 5FC (5) | Hydrocephalus (6) | Blindness (4 patients) |
| 222 | Brazil | 21 | Meninges | HIV (2), non-HIV (19) | ||||
| 221 | Brazil | 25 | Meninges | HIV (4), non-HIV (21) | 44 | |||
| 256 | Brazil | 7 (previously unreported cases only) | Meninges | HIV (1), IC (6) | 14.3 | AMB only (5), AMB + FLU (2) | ||
| 251 | Brazil | 4 | Meninges | HIV (2) | 50 |
Cranial nerve lesions, epilepsy, memory impairment, and visual field loss.
The case-control study identified oral steroid use, pneumonia, and other lung conditions to be associated with infection. In population comparisons, cases were more likely to be >50 years of age, to be current smokers, to have HIV infection, or to have a history of invasive cancer.
Abbreviations: AMB, amphotericin B formulation; cAMB, conventional amphotericin B deoxycholate; CNS, central nervous system; 5FC, 5-flucytosine; IC, immunocompetent; ICP, intracranial pressure; IS, immunosuppressed; PNG, Papua New Guinea.
Geographic distribution and incidence.
Knowledge regarding the global distribution of C. gattii cases assists in understanding its clinical epidemiology and, consequently, risk factors for infection (see below). However, this understanding is incomplete, in part because until recently, many laboratories did not distinguish between C. gattii and C. neoformans (247, 258). In Australia, where C. gattii infection is considered to be endemic, the two species have been distinguished routinely since the early 1990s by using canavanine-glycine-bromothymol blue (CGB) agar (266) (see Diagnostics, below). Earlier small (8 to 49 patients) case series of C. gattii infection were mainly from Australia (25, 27, 28, 99, 113, 204, 267), Papua New Guinea (208–213), and Brazil (206, 219). Incidence data from Brazil are imprecise, but compared to other parts of the world, children appear to be disproportionately affected by C. gattii. Prior to the emergence of AIDS in Papua New Guinea, C. gattii caused ∼95% of cases of cryptococcal meningitis, with an estimated annual population incidence in the Central Province of 42.8 cases per million population (214). In Australia, the annual incidence in the late 1990s was 0.94 cases per million population; this incidence varied with jurisdiction, being highest in the Northern Territory (8.5 cases per million) (29). The incidence has remained unchanged over the succeeding decade (0.61 per million) (130). Population-based rates are significantly higher in the indigenous Aboriginal population than in nonindigenous persons (10.4 versus 0.7 cases per million) (29). In the tropical Arnhemland region of the Northern Territory, the relative risk for cryptococcosis in Aboriginal people compared with non-Aboriginals is 20.6 (204).
C. gattii has traditionally been considered a “tropical or subtropical fungus” despite the fact that even prior to the North American outbreak, a large proportion of disease in Australia occurred in its southern temperate region (27–29, 99, 268). Before 1999, C. gattii infections were uncommon in North America, with small numbers of cases being reported in California and Hawaii (83, 269) and Europe (reviewed in reference 122). The spread of C. gattii infection into new geographic regions was heralded by an unprecedented outbreak of infection on Vancouver Island, British Columbia, in 1999, with subsequent clusters in British Columbia and the Pacific Northwest of the United States.
From 1999 to 2007, 218 patients acquired C. gattii infection as part of the Vancouver Island outbreak (198, 270). Between 2002 and 2006, the annual average incidence in British Columbia was 5.8 cases per million population, and that on Vancouver Island was 27.9 per million (270). It now appears that an endemic focus of C. gattii cryptococcosis was established in 2004 in the Pacific Northwest. As of 2012, >96 infections have been reported, mainly from the states of Washington and Oregon (132, 197, 258), although incidence data have not yet been reported (271). Only after these outbreaks and the consequent interest in species distinction has the complexity of the epidemiology been appreciated.
In parallel with the emergence of case clusters, an explosion of C. gattii cases were reported, some after travel to areas of western North America where the disease is “newly endemic,” including California, and others presenting as autochthonous cases (231, 238, 240, 247, 250). Single cases of infection have also been reported in the United States, including New England (85, 243, 247), New Mexico (272), and Florida (253); Europe (91, 95, 106, 245); Asia, including Japan and Singapore (227, 246, 249); and South America (223, 256). Most recently, Harris et al. reported 25 cases, inclusive of previously described patients (247, 253) diagnosed between 2009 and 2012, from Montana, Alabama, California, Hawaii, and Michigan; none of these patients had traveled to areas of C. gattii endemicity (251). In Australia, despite heightened awareness, no increase in the number cases has been observed (most large institutions care for 2 to 3 patients annually). Two reports (216, 217) highlighted the occurrence of immune reconstitution inflammatory syndrome (IRIS) in separate immunocompetent hosts with C. gattii meningitis (see Clinical Complications of Human Infection, below).
Central to the epidemiology of human infection is the appreciation of the distribution of C. gattii molecular types by geographic region (Table 1 and Fig. 4). Infection in Australia and PNG is caused largely by molecular type VGI (29, 99). VGII infections also occur but are, to date, geographically restricted to the Arnhemland region of the Northern Territory and the southwest of Western Australia (113, 117). In contrast, clonal VGII subtypes (referred to as outbreak strains) of C. gattii caused case clusters in British Columbia (subtypes VGIIa and VGIIb) and in the Pacific Northwest (subtypes VGIIa, VGIIb, and VGIIc) (38, 132, 198, 251); subtype VGIIc is a novel subtype not found outside the United States (68). Theories on how these VGII subtypes may have arisen in North America are discussed above (see Epidemiology, Origin, and Evolution, above). Infection outside the Pacific Northwest has been caused by VGI and VGIII (242, 243, 251, 272) as well as by subtype VGIIb and other, nonoutbreak subtypes of VGII (251).
The assignment of genotype is important not only for epidemiological purposes but also to track the origin of isolates. Pinto Junior et al. reported a child residing in Rio de Janeiro with C. gattii genotype VGII infection who had no history of travel outside the city (223). Given that VGII is the predominant molecular type (VGI is uncommon) in north/northeastern Brazil but not in the southern part of the country, those authors postulated that the ecological niche of this genotype may be expanding (223). Support for the notion that the niche of the VGII genotype is evolving is the observation of infection caused by molecular type VGIIa in a Japanese patient in Tokyo with no history of travel to an area of known endemicity; VGIIa genotype strains have not previously been documented in Japan (249). Further environmental surveillance is needed to determine new reservoirs of C. gattii. Obtaining a travel history from patients with suspected cryptococcosis in areas where C. gattii is not endemic should be routine. Hagen et al. described a Dutch patient who acquired infection due to molecular type VGIIb after visiting Vancouver Island (238). In a subsequent study, that same group of authors demonstrated that infections in Europe caused by molecular types VGIII and VGIV could be linked to exposure in regions of Africa (91).
Environmental exposure.
The ecology of C. gattii and its environmental molecular epidemiology are described above (see Ecology, above).
Historically, the first hint of an environmental source of C. gattii was reported in the early 1990s in Australia, when the distribution of human disease was noted to match the geographic location of two species of eucalypt: E. camaldulensis (river red gum) and E. tereticornis (forest red gum) (26, 273). The density of E. camaldulensis, in particular along water courses in rural/semirural Australia, and the association of rural-dwelling Aborigines with these trees are thought to partly explain the relatively high prevalence of infection in indigenous Australians. This link was confirmed by subsequent epidemiological investigations (29, 99), as was the association between C. gattii and residence, or employment, in a rural/semirural environment (29, 130, 215). Whether a similar association is evident in British Columbia and the U.S. Pacific Northwest is unknown; in the setting of an outbreak, the epidemiology may be different. Additional risk factors, such as host genetic determinants of susceptibility, have not been well studied.
As in Australia, C. gattii has been recovered from eucalypts in other countries with small numbers of human cases, including southern California, Italy, and Brazil (273, 274). However, infection also occurs in regions where eucalypts do not occur naturally, in particular Malaysia; South America; the Arnhemland region of the Northern Territory, Australia; and, most notably, Vancouver Island (38, 113, 274). In Arnhemland, the local reservoir of C. gattii remains uncertain; it is of interest that the predominant C. gattii molecular type in this location is VGII and not the VGI genotype that has been linked with eucalypts (99). While eucalypts are not indigenous to PNG, extensive sampling of imported trees in central PNG proved negative (212, 214). On Vancouver Island, in addition to imported eucalypts, trees native to the island have been implicated as being a niche for C. gattii and include Douglas fir, coastal western hemlock, alder, Garry oak, grand fir, and cedar (38, 258); C. gattii has also been found in soil.
Host factors.
A better understanding of the global distribution of C. gattii has been accompanied by an appreciation of evolving host risk factors. Unlike C. neoformans cryptococcosis, where HIV/AIDS, for example, is a major risk factor, risks for C. gattii infection are less well defined.
Cryptococcosis per se has occurred more frequently in males than in females, independent of the disproportionate representation of males with AIDS in data sets from Western countries. In one Australian study, male gender was a significant risk factor for cryptococcosis but only in patients with C. gattii infection (male-to-female ratio of 3.3:1) (29). Similar observations were reported in PNG and were attributed to an increased exposure of males to environmental sources of C. gattii (209). On Vancouver Island, however, only 55% of the outbreak cases affected males, and in the Pacific Northwest, 54% affected males (132, 198), suggesting that exposure is the predominant factor under these circumstances. Interestingly, 85% (21 of 25) of sporadic cases in the United States affected men (251). Population-based studies using uniform case definitions are needed to clarify whether there is a gender predisposition to C. gattii infection and, if so, whether there are additional genotype-dependent associations.
Data on host genetic factors as risks for C. gattii cryptococcosis are sparse. Indigenous Australians are consistently overrepresented in reports of C. gattii from rural Australia (29, 130, 204, 215), presumably due mainly to increased environmental exposure. However, the incidence of C. neoformans infection is also higher in Aborigines, suggesting that genetic and/or socioeconomic factors may be important. Likewise, exposure to C. gattii, even though the ecological niche has not been identified, has been linked to cryptococcosis in PNG, where the indigenous population is Melanesian. In another PNG study, neither leukocyte antigen class I nor class II phenotypes were associated with C. gattii infection, although there was a trend toward increased susceptibility in patients carrying the human leukocyte antigen B*5601 (275). Recently, high levels of certain anticytokine antibodies (e.g., anti-granulocyte-macrophage colony-stimulating factor [GM-CSF] antibodies) were associated with cryptococcosis in HIV-negative patients (276). These antibodies predispose individuals to opportunistic infections by impairing innate immunity and may be especially relevant among HIV-negative patients in Asia (277, 278). Saijo et al. most recently reported the presence of anti-GM-CSF autoantibodies in the plasma of seven apparently immunocompetent patients with C. gattii meningoencephalitis but, interestingly, not in the plasma of patients with C. neoformans disease (279). Their results, which suggest that anti-GM-CSF autoantibodies predispose otherwise immunocompetent individuals to meningoencephalitis caused by C. gattii, but not necessarily that caused by C. neoformans, are worthy of further study.
Distinct from C. neoformans, which typically causes disease in individuals with cell-mediated immune deficiencies (255, 260), historically, our understanding is that C. gattii causes disease predominantly in persons with apparently normal immune systems (72 to 100%) (27–29, 206, 211, 213, 215, 219). However, following the North American outbreaks and, more recently, outbreaks in Australia, new risk groups have been recognized. These groups include patients with underlying HIV infection, cancer, solid organ transplantation, and other immunodeficiencies (96, 98, 130, 132, 198, 230) (Table 4). In an Australian study of 86 patients, HIV was rare (1 of 79 patients tested for antibodies to HIV), but 7% of patients had underlying idiopathic CD4 lymphopenia, 3% had previously received kidney transplants, 8.1% had a malignancy, and 14% had received corticosteroids or immunosuppressive drugs (130). Notably, host immunocompromise was associated with increased mortality (29% versus 5%; P < 0.05). Overall, 72% of patients were immunocompetent, compared to 91.5% of patients in a previous study (29).
In British Columbia, only 62% of patients were considered previously healthy (198); a case-control analysis identified oral corticosteroid use, smoking, older age, HIV infection, and malignancy as risk factors (230). The proportion of patients who had predisposing conditions or who were taking immunosuppressive medications was even higher (76%) in U.S. cases (n = 76 patients) infected with “outbreak” strains. Here 4 of 59 patients were HIV positive, 50% of patients were immunocompromised, and 20% and 23% had had a solid organ transplant and cancer, respectively. These outbreak strains were defined as belonging to molecular types VGIIa, VGIIb, and VGIIc (132). There is increasing evidence of an association between the C. gattii molecular type and host status, although the pathogenic link between the two is not yet understood. Several studies have confirmed that molecular types VGI and VGII of C. gattii have a predilection for immunocompetent hosts, as discussed above (Fig. 5). In contrast, molecular type VGIII was overrepresented in a series of HIV-positive patients in southern California (84, 133), and VGIV has been identified in Africa in association with HIV/AIDS (Fig. 5) (29, 99). There are also data indicating an association between molecular type and clinical presentation (see Clinical Manifestations of Human Infection, below).
FIG 5.
Percentages of 256 clinical isolates obtained from immunocompetent and immunocompromised patients (HIV positive) and from patients with other risk factors per major molecular type, identified by URA5 RFLP analysis, PCR fingerprinting, or AFLP analysis, for which clinical data were available. Other risk factors are alcoholism, corticosteroid use, disorder T immunity, diabetes, leukemia, systemic lupus erythematosus, transplant, and tumor.
Patients with C. gattii infection may have subtle defects in immune function (276, 279). Marr et al. reported IgG2 antibody deficiency in one such patient (280). It is possible that selective antibody deficiencies are risk factors for C. gattii infection, rendering the host unable to mount a protective IgG2 antibody response to cryptococcal capsular polysaccharide. Idiopathic CD4 lymphopenia is a risk factor for C. neoformans infection (281), but its association with cryptococcosis due to C. gattii is new (130). Taken together with the identification of anticytokine antibodies in some patients with C. neoformans and C. gattii cryptococcosis (see above), these results suggest that immunological assessments, at least performing HIV antibody and T cell subset measurements, should be undertaken for patients with C. gattii infection.
Other comorbidities may also pose a risk for cryptococcosis caused by C. gattii. Almost 30% of patients in the Pacific Northwest outbreak had chronic lung disease (132). Concurrent corticosteroid use in such patients and/or changes in lung function associated with cigarette smoking may contribute to this risk. On Vancouver Island, case-control studies identified an association between underlying lung conditions, including chronic obstructive pulmonary disease (COPD), and C. gattii infection. However, in population analyses, only current smokers (P < 0.001), not those with a history of COPD or asthma, were more likely to develop infection (231). Smoking was also an independent risk factor, occurring in 36% of cases in a recent Australian study (130). Other cases of C. gattii cryptococcosis have been reported in smokers (231, 272). Pregnancy has been proposed to be a risk factor, and in two studies, 2 of 26 and 1 of 49 patients were pregnant (29, 130); however, the proportion of pregnant patients in a recent Australian study of C. gattii infections was not higher than that in the general female population of reproductive age. An association between diabetes mellitus and C. gattii has not been established. In one study, 6% of patients had diabetes, no higher than in the general population (29), but in another study, 20% were diabetic (132). Notably, C. gattii is overrepresented in Australian Aborigines (29), in whom diabetes is common. Diabetes was not a risk factor for C. gattii in British Columbia (230).
The possible clinical associations of C. gattii molecular types are further discussed below (see Clinical Manifestations of Human Infections).
Infection in children.
Cryptococcosis is uncommon in children, including those with HIV/AIDS (232, 282), and the same appears to be the case for C. gattii infection (213, 218, 232, 268), with the possible exception of children in Brazil. An earlier study from PNG showed that exposure to the fungus, as demonstrated by levels of serum IgG antibody directed against C. gattii, was significantly greater in adults, and especially adult males, than in children (209), suggesting that exposure increases with age and that it occurs away from the home environment. Antibodies to C. neoformans (including C. gattii) are uncommon in children <2 to 5 years old (268, 283).
In contrast, in the north and northeast regions of Brazil, particularly in the states of Para, Roriama, Piaui, Pernambuco, and Bahia, which include the Brazilian Amazon forest and savannah areas, a relatively high proportion of HIV-negative, otherwise healthy children present with cryptococcal meningitis (age range, 5 to 14 years; average age, 8.4 years) and are infected with C. gattii (219, 256, 284). Although sample sizes were small and only selected isolates were identified, Correa et al. reported that at least 47.3% of children with cryptococcal meningitis had C. gattii infection, while for 22 patients reviewed by Severo et al., C. gattii caused 33.3% of cases (219, 256). In that same study, 16 of 53 cases (30%) in adults and children were due to C. gattii (256); underlying conditions were not stratified by cryptococcal species, but at least 14 children were previously healthy. Small epidemiological studies suggest that C. gattii molecular genotype VGII predominates in infected children in north/northeast Brazil and that VGI is uncommon (221, 222). Infection in immunocompetent children in southern and southeastern Brazil has also been reported (223, 224, 256).
Among 16,192 episodes of cryptococcosis (due to both C. neoformans and C. gattii) in predominantly HIV-infected persons in South Africa from 2005 to 2007 (232), a bimodal distribution of infection was noted, with a peak in children <1 year old and a second peak in those aged 5 to 10 years. Although most pediatric infections were due to C. neoformans, children were significantly more likely than adults to be infected with C. gattii (9% versus 3%; P < 0.001).
CLINICAL MANIFESTATIONS OF HUMAN INFECTION
Incubation Period
The incubation period of C. gattii infection has been deduced from instances of exposure in persons visiting areas of endemicity or in an outbreak setting. Studies of travelers to Vancouver Island revealed a median time to clinical presentation of 6 to 7 months (range, 2 to 11 months) (229). However, in one instance, the incubation period was as short as 6 weeks (94), while other patients developed infection 13 to 35 months after visiting the island (95, 238, 250). Tsunemi et al. reported a Japanese national who developed infection 2 to 4 weeks after returning from Australia, where he had had close contact with koalas (246). Since the incubation period of C. gattii infection may be prolonged (>2 years), diagnosis may be delayed in an area where the disease is not endemic unless an appropriate travel history is obtained.
This estimated incubation period compares with 110 months for C. neoformans (serotype A) infection (285). The relatively long latent period of C. neoformans infection suggests that latency is established following primary infection and that reactivation of dormant infection can result from subsequent immunosuppression (286); the extent to which this happens in C. gattii infection is uncertain. Using MLST analysis, Hagen et al. described 24 C. gattii cases that were linked to exposure in regions where C. gattii is endemic in the United States, Brazil, Vancouver Island, and Africa; although the time to infection following exposure was not specified, the findings provide support for reactivation of dormant infection after many years (91).
Sites of Infection
CNS and lung infection.
C. gattii cryptococcosis most commonly affects the CNS and lung, although it can involve any body site. Importantly, there appear to be differences in clinical presentation and site(s) of infection between disease that occurs sporadically and that seen in the reported case clusters. Studies from Papua New Guinea indicate that CNS infection, particularly meningitis, is typical (208, 211, 213, 218). In Australia and New Zealand, meningoencephalitis and/or cerebral involvement occurred in 74% of patients with culture-confirmed C. gattii infection in the 1990s (29), while in a contemporary study, in which most patients underwent cerebral computerized tomography (CT) and magnetic resonance imaging (MRI), CNS infection was diagnosed in 85% of 86 patients; overall, 76% of patients had meningitis and 52% had brain infection, including cerebral cryptococcomas (130). Up to 97% of cases of C. gattii infection in patients with HIV/AIDS present as meningitis (96). Neurological disease was also common in non-outbreak-related infections in the United States, Brazil, and Europe (106, 223, 238, 242, 243, 248, 258).
In contrast, lung disease predominated in the North American outbreaks (131, 132, 252), with only 20 to 40% of patients having CNS infection. Among 218 patients in British Columbia, 189 (87%) presented with a respiratory illness, 22 of whom (10%) had concurrent CNS disease (198). In the U.S. outbreak, 31 of 57 (54%) patients developed pneumonia, and 20 of 61 (33%) patients had lung cryptococcomas (132). Patients considered to have acquired their infection in the northwest of North America also presented primarily with lung disease (231, 237, 240, 250). While lung disease is common in Australia, affecting 63 to 67% of cases in three population-based studies, it occurs mostly in association with CNS disease (81% of cases) (28, 29, 130). Notably, the pattern of disease in the Northern Territory differs from that in Australia as a whole, with isolated lung disease accounting for two-thirds of cases (215). Thus, there are regional differences in clinical presentation between, and within, countries.
Other sites of infection.
Other than pulmonary and neurological (including eye) disease, C. gattii infections may involve skin, soft tissues, bone, joint, larynx, bone marrow, and lymph nodes (29, 131, 206, 234, 287). Intra-abdominal infection is uncommon but is important, as it may be mistaken for underlying cancer (233). Primary cutaneous C. gattii infection has been reported for only a few patients, including patients from Australia and Brazil. It is typically preceded by traumatic inoculation and affects the scalp and limbs of immunocompetent patients (228, 235, 256, 287–290). Lesions may be solitary or present as extensive ulceration or cellulitis. Primary skin involvement is rare in Australia and British Columbia (130, 198). Skin lesions may herald disseminated disease, with papules, nodules, ulcers, or cellulitis occurring typically on the face and neck; these patients may be immunocompromised (236, 256, 290). Musculoskeletal infection has been reported. Byrnes et al. described a patient with a cryptococcoma in the thigh, which progressed to disseminated infection despite surgical resection and antifungal therapy (68). Another patient from Florida had disease in the femur together with lung and CNS infection (253). Finally, C. gattii fungemia was detected in 10 patients in one series (130), but its prevalence either alone or in the setting of CNS or other infection is unknown, since blood cultures are not taken routinely.
C. gattii molecular types, host risk factors, and disease patterns.
The distinctive features of the North American outbreaks have raised the possibility of an association between clinical presentation and C. gattii molecular type (see Clinical Epidemiology of Human Infection, above). Among patients in the Canadian outbreak, the VGIIa molecular type was responsible for 86.3% of cases, with VGIIb and (nonoutbreak strains of) VGI accounting for 7.3% and 6.5% of cases, respectively (198). In the U.S. Pacific Northwest, the distribution of molecular types was as follows: 50% VGIIa, 32% VGIIc, 10% VGIIb, 5% VGI, and 3% VGIII (86). It is possible that the VGII genotype has a propensity to cause respiratory disease (see above) and that it is associated with host immunocompromise or predisposing medical conditions (40 to 73% of patients) (86, 132, 230). Although features of infection differ between “outbreak” and “nonoutbreak” disease, the reason for the difference has not been determined. It is possible that there are genotype-dependent differences in the host response to the organism (see Pathogenesis, above) or an effect of concomitant illnesses. There is also debate as to whether these hypotheses are generalizable to the nonoutbreak setting. In Australia, the epidemiology has changed over the last 10 to 15 years, with an increase in the proportion of HIV-negative immunocompromised patients (130), although most infections were still caused by molecular type VGI (S.C.-A. Chen and T. C. Sorrell, unpublished data). As with VGI infections, the majority of infections due to the VGII molecular type occurred in nonimmunosuppressed hosts (Fig. 5) (29, 99). VGII genotype strains were isolated from otherwise healthy Brazilian patients and a single patient each from Japan, Holland, and Florida; all had CNS disease (222, 238, 249, 256).
Clinical Findings
Clinical findings depend on the site of infection. Systemic features such as chills, fever, and weight loss were reported in 17 to 47% of cases in the Pacific Northwest (132, 252). Fever was present in 54% of patients with HIV infection in South Africa (98) but was uncommon (10%) in an Australian survey (130); in the latter study, the median time from the onset of CNS symptoms to diagnosis was 45 days (interquartile range [IQR], 21 to 120 days), compared with 56.8 days (IQR, 1 to 180 days) in those with isolated lung infection. In a U.S. study outside the outbreak setting, the median time from symptom onset to diagnosis was 32 days (range, 2 to 263 days) (251).
Neurological disease.
Headache, sometimes with vomiting, and neck stiffness are common neurological presentations (130, 198, 211, 252). Abnormal neurological signs or deficits may be present or can evolve during illness. These include cranial nerve deficits, visual and hearing impairment or loss, cerebellar abnormalities, local limb weakness, seizures, and abnormal mentation (e.g., confusion, personality change, and coma) (27, 208, 211). Chen et al. reported that 73 patients had CNS infection, 43 had abnormal neurological manifestations at presentation, 18 had impaired consciousness (Glasgow coma scale range, 6 to 14) and 13 had cranial nerve abnormalities (1 patient with VII nerve palsy, 7 with VI nerve palsies, and 5 with optic atrophy) (130) (Table 4). In a recent U.S. series of cases outside the Pacific Northwest, headache occurred in 67% and blurred vision occurred in 62% of patients (251). Contemporary studies have reported seizures in 5.8 to 8% of cases (130, 132).
Importantly, papilledema was present in 15 to 53% of patients in earlier studies (29, 211, 213) but in only 6% of cases in the U.S. outbreak and in 12.4% of cases in a recent Australian study (130). Optic disc swelling is typically present, with loss of definition of the disc margins. Hemorrhages are found around the disc in severe cases and are accompanied by a loss of visual acuity.
Pulmonary disease.
Cough, dyspnea, chest pain, and hemoptysis are the most common symptoms of lung involvement, but asymptomatic pulmonary nodules or mass lesions (cryptococcomas) observed on chest X ray or CT scan (see Diagnostics, below) are not uncommon (130, 198, 240, 252). Very large lesions are typical of disease due to C. gattii (215, 231) and have been associated with Pancoast syndrome with respiratory stridor (205). Johannson et al. reported a patient with lung infection accompanied by systemic symptoms with an unproductive cough and dyspnea (250).
Clinical Manifestations in Children
Most children with C. gattii infection present with CNS involvement. In Brazil, symptoms of headache, fever nuchal rigidity, nausea, and vomiting were common (219, 221, 256). Cranial CT scans of 11 children with CNS disease revealed hypodense nodules in all patients, cerebral atrophy in 9, hydrocephalus in 6, and hydrocephalus with cerebral atrophy in 5 (220). Meningoencephalitis is also characteristic of most pediatric cases in PNG (218). Laboratory and treatment variables for outcome were cryptococcal species independent, but hospital stays were longer for children than for adults. Data from case reports and small case series in Brazil estimated the mortality rate to vary between 37.5 and 42.8% (256).
CLINICAL COMPLICATIONS OF HUMAN INFECTION
Neurological Complications and Sequelae
Compared with C. neoformans infection, neurological complications, including cranial nerve palsies, loss of visual acuity, blindness, seizures, ataxia, and focal long tract signs, have been frequently reported for disease due to C. gattii (27, 28, 211); significant disability can result. In one earlier report, 7 of 18 (39%) patients with C. gattii disease developed sequelae, including cranial nerve lesions, epilepsy, memory impairment, and visual field loss (28), while another report identified 8 of 26 (30.8%) patients with moderate-to-severe sequelae (27) (Table 4). In another study, 43 of 73 patients had a range of neurological complications at diagnosis, and of 63 survivors, 20 (31.7%) had persistent sequelae, including visual impairment (n = 8 patients; 3 became blind), deafness (n = 3), limb weakness (n = 2), and dysphasia (n = 2). The proportions of patients with concomitant CNS and lung infection or CNS infection with neurological sequelae were similar (24% versus 29%) (130). Cranial nerve palsies developed in 5 of 13 (38%) cases from the United States that were not associated with the outbreaks (251), but long-term sequelae (if any) from the C. gattii case clusters have not yet been reported. In a Vietnamese study, a large proportion of patients with cryptococcal meningitis had neurological sequelae (14 of 20; 70%); however, the frequency of sequelae was not specified for those with C. gattii infection (239). Whether there are species-dependent effects on neurological outcomes or whether these differences reflect differences between patients in seeking medical attention is uncertain.
Ocular complications of C. gattii infection are notable and include papilledema, especially in the presence of raised intracranial pressure (ICP) (see below). Although papilledema is the most common manifestation, blurred vision, visual field defects, extraocular muscle paresis, impaired pupillary/accommodative function, choroiditis, retinal hemorrhage, endophthalmitis, and optic atrophy have all been reported (27, 130, 208, 211) (Table 4). Visual loss is particularly disabling; in C. neoformans infection, it is thought to be due to direct invasion of the optic nerve by cryptococci or to the effects of raised ICP or from local arachnoiditis leading to choroidal infiltration with fungus (291, 292). The mechanism of eye involvement is likely to be similar in C. gattii infection. Visual loss has been reported in 1 to 9% of patients with C. neoformans infection (291, 293), much lower than the 18 to >50% of patients with blindness noted with C. gattii infection in PNG and Brazil, including children (208, 211, 219). These high rates have not been observed recently in Australia (28, 130). Chen et al. reported blindness in 3 of 73 patients, with significant visual impairment in a further 5 patients (130). Rates of visual sequelae in the North American case clusters have not been reported, but Harris et al. described papilledema in 9 of 49 patients with infection due to outbreak strains (132). In a later study of cases outside the Pacific Northwest, 8 of 25 patients reported blurred vision (251) (Table 4).
Elevated ICP (CSF pressure of ≥20 cm water) is common in C. gattii meningitis (50 to 60% of cases), as it is with C. neoformans (30 to 75%) (130, 210, 211, 282, 294). Early raised ICP was a poor prognostic indicator in PNG patients (208, 211). In a contemporary Australian study, although it was not a risk factor for mortality, elevated ICP predicted neurological sequelae and/or death at 12 months (P < 0.05) (130). Clinical manifestations, including severe headache, papilledema, loss of vision or hearing, other neurological deficits, and altered mental consciousness, are common, and death may result, presumably from cerebral ischemia (211, 291, 295). Cryptococcomas due to C. gattii have caused raised ICP or obstructive hydrocephalus via a mass effect (27, 296) but are not necessarily associated with measurably raised ICP (130). Patients with elevated pressures are more likely to present with abnormal neurological signs (P < 0.05) (130). Rapid diagnosis is the key to achieving good outcomes, and routine measurement of CSF pressures is essential whenever a lumbar puncture is performed (see Diagnostics, below).
Clinicians should also be vigilant regarding the presence or development of hydrocephalus, which is usually obstructive. Identification of dilated ventricles by cerebral imaging (CT or MRI) is required to establish diagnosis. Symptomatic patients with hydrocephalus present with ataxia and deteriorating mental status due to meningoencephalitis. Although the number of cases studied is small, the proportion of patients with hydrocephalus has ranged from 10.6 to 34.6% in Australia. In the largest series of 86 patients, hydrocephalus was present in 22 (30%) patients, typically at diagnosis, although in a small minority, it may not be detected for 2 to 12 months (130). Six of 11 children in Brazil with C. gattii CNS infection were diagnosed with hydrocephalus by cerebral imaging, while in one U.S. study, Harris et al. reported this complication in 4 of 18 patients (251). Notably, 25% of patients presenting with hydrocephalus had “silent” or unexpected findings of meningitis upon CSF examination (27). In C. neoformans infection, most patients with hydrocephalus have indolent meningitis (296). Factors that predispose patients with C. gattii meningitis to developing hydrocephalus remain unclear. As for C. neoformans, delays in diagnosis and treatment may lead to serious, permanent loss of cognitive function (282, 297). The management of raised ICP and hydrocephalus is discussed below (see Antifungal Therapy and Management, below).
Immune Reconstitution Inflammatory Syndrome
Immune reconstitution inflammatory syndrome (IRIS) is best studied in HIV-infected individuals with infections due to C. neoformans. In these patients, the development of IRIS is associated with immune reconstitution and typically presents with worsening of disease or recurrence in the same or new anatomical sites despite mycological evidence of effective antifungal therapy (reviewed in reference 298). IRIS has been described infrequently in association with C. gattii infection: occasional cases were reported in apparently healthy hosts in association with pregnancy and in immunocompromised individuals (216, 217, 241, 299). No cases were reported for 10 HIV-negative Vietnamese patients with C. gattii meningitis (239). However, in an Australian study, IRIS occurred in 9.4% of patients, two-thirds of whom had no underlying immunocompromise, although one patient was postpartum (299). In that study, IRIS was diagnosed when symptoms or radiological features consistent with inflammation worsened or appeared following a clinical and/or microbiological response to anticryptococcal therapy, and cultures were negative.
The pathogenesis of IRIS is incompletely understood. Clinical observations in patients with HIV/AIDS or organ transplantation indicate that it develops at the time of immune reconstitution or reduction in immunosuppression, respectively. Risk factors reported for HIV-infected patients include a poor baseline inflammatory response (indicated by low CSF white blood cell counts and protein levels and associated failure to sterilize the CSF rapidly) (300), rapid immune reconstitution from a low base, and a high organism or antigen burden at presentation (301) or at the time of initiation of antiretroviral therapy (ART) (302). The original hypothesis that IRIS results from dysregulation of the early Th2 (anti-inflammatory) and subsequent Th1 (proinflammatory) cytokine responses appears to be too simplistic, but dysregulation within the mononuclear phagocyte lineage is universal (303). Recent evidence from cases of HIV-associated IRIS suggests that IRIS results from the failure of a dysfunctional immune system to clear antigen during the early phase of immune recovery. This is followed by a supranormal proinflammatory response as immune reconstitution occurs during ART, which is not accompanied by appropriate effector responses (302, 303). Additional factors, such as cryptococcal genotype, may influence the development of IRIS (304). In healthy hosts, it is proposed that cryptococcal capsular polysaccharide induces an initial Th2 cytokine response, which is then replaced by a Th1 response during successful antifungal therapy, although the stimulus for an excessive response sufficient to cause IRIS in a small percentage of patients is unknown.
Risk factors for IRIS in C. gattii infection include female gender and initial presentation with brain involvement or brain, meningeal, and lung involvement. Notably, in that study, median CD4 counts were higher than those in patients who did not develop IRIS (299). Unlike in HIV-positive patients, neither initial CSF cryptococcal antigen (CRAG) titers nor CSF leukocyte counts and protein levels differed from those in patients who did not develop IRIS (299, 300). However, since patients with C. gattii who subsequently developed IRIS all presented with cryptococcomas in the brain, CSF parameters likely did not reflect the fungal load (or immune response) in this site.
In patients with C. gattii infection, IRIS develops following an initial clinical and microbiological response, 4 weeks to as long as 12 months after initiation of antifungal drugs (299). The clinical presentation ranges from headache or drowsiness to new or recurrent neurological manifestations, including blindness, ataxia, dysarthria, deafness, hemianesthesia, seizures, and falls. Neuroimaging studies reveal new or worsening inflammatory mass lesions, meningeal enhancement, hydrocephalus, or increased CSF inflammation. In the largest study to date, all patients presented with enlarging or new brain lesions, typically with surrounding edema; occasionally, patients also developed pulmonary lesions with or without subcarinal lymphadenopathy (299).
The recognition of IRIS is important, as it can be misdiagnosed as clinical failure and can lead to unnecessary reinduction antifungal therapy. In a series by Chen et al. (299), three of eight patients also received gamma interferon (IFN) for presumed failure of antifungal therapy. The use of gamma interferon in this setting carries a theoretical risk of exacerbating the severity of IRIS due its proinflammatory effect. Treatment of IRIS is discussed below (see Antifungal Therapy and Management, below).
Mortality from C. gattii CNS Cryptococcosis and Prognostic Determinants
Mortality data are summarized in Table 4. The mortality rate was high in early studies of C. gattii meningitis in PNG (22.4 to 43%) (210, 211, 213) and remains so, especially if diagnosis is delayed (218). In Brazil, the mortality rate has ranged from 18.2 to 50% between studies (219, 221, 248, 256), and in the Northern Territory of Australia, it has been as high as 33% (215). Lower death rates of 0 to 15% were reported in three other Australian studies conducted between the late 1980s and 2007 (27, 28, 299). In a recent Vietnamese study, 1 of 10 C. gattii-infected patients died (239). Mortality is likely to be influenced not only by patient factors but also by medical awareness and access to health resources, which vary between countries. The mortality rate in predominantly HIV/AIDS patients in Africa ranged from 17 to 35.6% (96, 98). In the outbreak setting, mortality has likewise varied substantially: in British Columbia, 8.7% of patients died, compared to 20 to 33% in the Pacific Northwest (132, 198, 252).
Determinants of mortality and patient outcomes, although well described in immunocompromised patients infected with C. neoformans, are less well defined in C. gattii infection. Seaton et al. studied 88 patients in PNG with C. gattii meningitis and found that the mortality rate was higher in men (P = 0.025), older patients (P = 0.39), those with altered consciousness (P < 0.001), and those with a history of convulsions prior to commencement of therapy (P = 0.002) (210). In a more recent study of 86 patients, immune compromise was associated with an increased risk of death by univariate analysis. Initial CSF CRAG titers of ≥256 independently predicted death and/or neurological sequelae (130); unlike C. neoformans infection in HIV-infected hosts (305), a high cryptococcal load (serum CRAG titer, ≥512) and abnormal neurological findings at presentation were not associated with increased mortality (130).
DIAGNOSTICS
Imaging
Imaging of the lungs, brain, and other clinically appropriate sites is essential for C. gattii infections because the type and duration of induction and total antifungal therapy are site dependent. Furthermore, surgical resection of selected mass lesions or CSF shunting may be required.
Lung infection.
Imaging of the chest may provide the first clue to the presence of cryptococcal disease, which most commonly manifests as one or more circumscribed cryptococcomas (Fig. 6). Pulmonary alveolar or interstitial infiltrates caused by C. gattii genotype VGI are uncommon in areas of endemicity such as Australia (130) and accounted for 14 to 17% of imaging abnormalities in the British Columbian VGII outbreak (198). In contrast, in patients immunosuppressed by HIV or other causes, alveolar and interstitial pulmonary infiltrates account for ≈70% of lung lesions.
FIG 6.
Chest X ray of a patient with a large mass lesion in the right lower lobe. Bronchoscopy and collection of bronchoalveolar lavage fluid showed multiple encapsulated yeasts upon cytological examination, and a culture grew Cryptococcus gattii. (Reprinted from reference 388 with permission of the publisher. Copyright 2013 UpToDate, Inc. [www.uptodate.com].)
Lung cryptococcomas are more often due to C. gattii than to C. neoformans, both in patients with isolated lung infection (odds ratio [OR], 12.2; 95% confidence interval [CI], 3.3 to 46; P < 0.001) and in those with combined lung and CNS disease (29, 130, 215). In British Columbia, 75% of patients had pulmonary nodules (198) of various sizes, compared with 33% in the Pacific Northwest (132). In a smaller U.S. study, cryptococcomas were present in 14/23 (61%) patients (251). Cryptococcomas can appear in any region of the lung and have ranged from 1 to 8 cm in diameter in a population-based survey (our unpublished data), although larger lesions (up to 10 cm in diameter) have been reported (231, 243). Uncommonly, pleural effusions, abscess, cavitation, and lymphadenopathy are observed, and one report described a patient with Pancoast syndrome associated with opacification of the entire upper lobe (205, 251). Both cryptococcomas and lung infiltrates can be present in the same patient (29, 132).
Neurological infection.
(i) Computerized tomography.
Cerebral imaging is required to exclude brain involvement, which may manifest as cryptococcomas, hydrocephalus, or abnormal foci of reduced attenuation, contrast enhancement, and edema. Mass lesions often resembling “abscesses” have been reported, most commonly in the basal ganglia, thalamus, and cerebellum, in cranial CT scans in 33 to 58% of cases of C. gattii disease (27–29, 130, 251) (Fig. 7). In one study, cerebral cryptococcomas were more common in patients with C. gattii infection than in those with C. neoformans infection but not when the analysis was confined to immunocompetent hosts (29). In contrast, concurrent pulmonary and cerebral cryptococcomas and hydrocephalus were more frequently associated with C. gattii independent of host status (27, 29). Correa et al. reported hypodense nodules on cerebral CT scans, often in the basal ganglia, in all 11 immunocompetent children studied (220). Obstructive hydrocephalus has been described in 17 to 50% of C. gattii infections (27–29, 130, 251).
FIG 7.
Computerized tomographic scan of brain showing a large mass lesion in the left cerebellar hemisphere (indicated by arrow). A provisional diagnosis of a cerebral neoplasm was made. Excision biopsy of the lesion showed encapsulated yeasts, and Cryptococcus gattii was isolated in culture.
(ii) Magnetic resonance imaging.
MRI technology is more sensitive than CT scans, especially for detection of small lesions. Cryptococcal infection spreads from the basilar cisterns, causing dilatation of the perivascular (Virchow-Robin) spaces as a result of accumulation of cryptococci and associated capsular material along the “walls” of these spaces. This abnormality is readily seen on MRI scans, most commonly near the basal ganglia. It is present in almost 50% of cases of HIV-associated cryptococcosis and is a diagnostic clue in such patients (306, 307). MRI scans are useful for delineating basilar meningeal enhancement after injection of gadolinium.
(iii) Newer techniques.
Nuclear magnetic resonance (NMR) spectroscopy is a technology that can be linked to MRI to identify the location of cryptococcomas 2 cm in diameter or more and their metabolic “fingerprint” in a single examination. This approach is already established for noninvasive diagnosis of brain tumors and pyogenic abscesses (308). By generating data through computerized pattern recognition methods, based on the chemical composition and metabolite profiles of various human and/or microbial cells, NMR spectra provide information on the content of a specific abnormality or lesion. Experimental studies showed that uniquely among pathogenic fungi, cryptococci cultured in vitro are characterized by an abundance of the disaccharide α,α-trehalose, which can therefore be used as a marker that contributes to their identification in vivo (309–311). In vivo, it has been demonstrated that identification of cerebral cryptococcomas and differentiation from cerebral tumor are feasible, although the acquisition of spectra from more cases is required to achieve an unambiguous diagnosis (308).
Microbiological Diagnosis
Conventional and antigen-antibody-based tests.
Almost all cases of cryptococcosis are readily diagnosed by visualization of the organism and its recovery in culture from appropriate clinical specimens and/or a positive serum or cerebrospinal fluid (CSF) cryptococcal antigen (CRAG) titer. Culture remains essential for definitive diagnosis since CRAG detection does not distinguish between C. neoformans and C. gattii.
C. gattii can be cultured from any body site. To diagnose lung infection, bronchoalveolar lavage (BAL) fluid, fine-needle aspirates, or lung biopsy sections are the preferred specimens, but sputum cultures can also be useful. Positive sputum cultures are indicative of invasive disease with few exceptions. As CNS disease is common, CSF should always be examined for cryptococci unless lumbar puncture cannot be performed; biopsy of other affected tissues may also be indicated. Positive blood cultures indicate disseminated disease, but as discussed above (see Clinical Manifestations of Human Infection, above), the frequency of recovery of C. gattii from blood is not known. Clinical specimens should be sent for both culture and histopathological examination.
(i) Histopathological stains.
Hematoxylin-and-eosin stains will detect cryptococci in tissue sections, although Gomori methenamine silver and calcofluor white are specific for the fungal cell wall. Mucicarmine stains the cryptococcal polysaccharide capsule in tissue samples, revealing organisms as bright carmine red structures with a spiny, scalloped appearance; the capsule can also be stained by using alcian blue and periodic acid-Schiff (PAS) stain. Cryptococci contain melanin in their cell wall (see Pathogenesis, above), which, upon staining with the Fontana-Masson reagent, appears brown to black. The use of more than one stain improves the sensitivity of detection of cryptococci in tissue samples. Either mucicarmine or alcian blue combined with Fontana-Masson stain is superior to any of these stains alone; the combination has the advantages of highlighting both the cell wall (Fontana-Masson stain) and mucin-positive capsule (mucicarmine/alcian blue) and of identifying rare capsule-deficient strains (summarized in reference 312).
(ii) Other stains.
India ink staining is a cheap and simple diagnostic test for CSF samples, becoming positive in <5 min. Cryptococci appear as encapsulated 5- to 7-μm-diameter structures with a halo, against a black background. The presence of budding cells distinguishes cryptococci from leukocytes. CSF India ink tests were positive in 70 to 95% of patients with C. gattii meningitis in three studies (29, 130, 211). It is a sensitive test in patients with C. gattii meningitis, in contrast to C. neoformans cryptococcosis, where the sensitivity is lower in HIV-negative patients (30 to 72%) than in those with AIDS (80%) (312, 313).
(iii) Culture.
Cryptococci grow readily on bacteriological agar (for example, blood agar) and standard mycological culture media such as Sabouraud's dextrose agar (SDA), Mycosel agar (BBL, Becton Dickinson, USA), and brain heart infusion agar. Enriched media, for example, brain heart infusion agar, may improve recovery. Cryptococci grow selectively on bird seed agar, which differentiates C. neoformans-C. gattii (brown colonies are formed due to melanin production) from other Cryptococcus species but which is time-consuming to prepare (266). Although two other cryptococcal species form brown colonies on bird seed agar (Cryptococcus podzolicus and Cryptotrichosporon anacardii), neither are human pathogens (314, 315). CGB agar reliably distinguishes between C. gattii (colonies produce a blue pigment) and C. neoformans (no change in colony color); the blue color results from the utilization of glycine, which releases ammonia, causing alkalinization and, hence, the blue color and susceptibility to canavanine (23, 266). True false-negative tests for C. gattii (the medium does not turn blue even after several days) are uncommon. Very recently, Candida famata was noted to give a “false-positive” blue color on CGB agar (316). Although it is unlikely that Candida strains would be mistaken for C. gattii or vice versa, laboratory scientists should be vigilant to carefully inspect and test all colonies that are suspected to be C. gattii.
(iv) Cryptococcal antigen.
CRAG has long been detected by incubating specific rabbit anti-C. neoformans antiserum with CSF and/or serum specimens. The test can be quantified (expressed as a titer) and is rapid (2 to 3 h). It is a sensitive, specific, and accurate diagnostic test for cryptococcosis in experienced hands.
There are two common commercial CRAG test formats: the latex agglutination test (LAT) and the enzyme-linked immunoassay (EIA). In general, agreement between the LAT and EIA results is excellent (92 to 98%) (summarized in reference 312). As the antibody used in these tests was developed from a strain of C. neoformans, in theory, occasional false-negative results may result from a lower affinity of the antibody for C. gattii. In clinical practice, the sensitivity of the test for C. gattii infection remains high, including for HIV-positive patients—90% for lung disease, with 87 to 100% sensitivity for CSF CRAG tests (29, 98, 130, 239). The level of detection of most LAT kits is at least 10 ng of polysaccharide/ml. On balance, the EIA is slightly more sensitive than the LAT (312), but it requires a plate reader and is more expensive.
Based primarily on studies of C. neoformans infection, false-negative results of the LAT are uncommon. They may result from prozone effects (where the amount of antigen in the sample is in excess of the amount of antibody in the assay mixture) or, occasionally, from a low organism burden or infection due to rare, capsule-deficient strains (131, 266, 317). In clinical practice, false-positive results are also uncommon but may be caused by Trichosporon beigelii, Stomatococcus mucilaginosus, Klebsiella spp., and Capnocytophaga canimorsus and by the presence of rheumatoid factor, collagen vascular disease, malignancy, and agar syneresis fluid (266, 312, 318). To minimize false-positive results, pretreatment of specimens with heat and proteolytic enzymes (pronase) is recommended, as is inclusion of a control reagent containing latex beads coated with normal rabbit globulin (319). Inclusion of IgM monoclonal antibody (MAb) effectively eliminates reactions with rheumatoid factor. CRAG testing by EIA does not require enzyme pretreatment of samples, nor is it associated with false-positive results in sera containing rheumatoid factor (320).
Low titers of CRAG may be found in the urine of patients with disseminated cryptococcosis and in bronchoalveolar lavage fluid and lung aspirates from those with pulmonary infection. These observations were based on small studies mainly in HIV positive patients (and, by inference, with C. neoformans infection) (321–323). The utility of CRAG testing of BAL fluid and urine in C. gattii infection is untested.
Recently, a lateral flow immunoassay (LFA) for CRAG (Immuno-Mycologics [IMMY], Norman, OK) was developed as a simple, rapid (10- to 15-min), inexpensive, point-of-care test that requires no sample preparation. Kits can be stored at room temperature. Each test contains an immunochromatographic dipstick-like strip impregnated with MAbs directed against epitopes on glucuronoxylomannan (GXM) of the four major serotypes of C. neoformans and C. gattii (324). Experience using the LFA to diagnose C. gattii infection is limited; in one study, CSF and serum specimens from four patients with C. gattii infection returned positive tests by the LFA, with no false-negative results (325). Testing of CSF/sera from subsequent C. gattii patients in the same laboratory has demonstrated no false-negative results (S.C.-A. Chen, unpublished data). The LFA is likely to perform similarly in C. gattii infection and C. neoformans infection (see below), but further evaluation of its utility in the diagnosis of C. gattii infections is needed, especially in patients with extra-CNS or lung infections.
The LFA has been extensively evaluated in C. neoformans infections, with excellent results. In two evaluations of its performance against culture and EIA, in HIV-positive patients with and without C. neoformans meningitis, LFA sensitivities when performed on serum were 90 to 99.8% (sensitivities of 70 to 92% for urine and 96 to 100% for CSF). Agreement between CRAG EIA values and LFA results was high (>93%), and dipstick titers correlated strongly with EIA results (326, 327). In another study of predominantly HIV-negative patients (n = 26) with CNS and lung cryptococcosis, the sensitivity of the LFA using both serum and CSF was 100%, with no false-negative results (325), but the correlation between LFA and LAT titers was poor (r = 0.85). Some LAT-negative samples had low LFA titers, and similar results were obtained in another study (324). Data from HIV-infected Ugandan patients showed that the LFA sensitivity for CSF specimens is higher than that of the LAT; likewise, Kabanda et al. and Rajasingham et al. reported both a sensitivity and specificity of 100% in their cohorts of HIV/AIDS patients (328, 329).
Until more data are available, titers obtained by the LFA and LAT should not be directly compared, and comparison of titers in the same patient should always be made by using the same test.
In contrast to their diagnostic value, serum or CSF CRAG titers are not helpful in monitoring the response to therapy, as the correlation between changes in titer and clinical progression or mycological responses is poor. Titers remain elevated for prolonged periods (months to >1 year) (130). Hence, CRAG results should always be interpreted in conjunction with clinical assessment. In one Australian study, CSF CRAG titers of ≥256 predicted death and/or neurological sequelae at 12 months after initiation of antifungal therapy (130). More data are required to support (or refute) this finding, which contrasts with observations of C. neoformans meningitis, where both high pretreatment LAT titers (≥512) and, more recently, LFA titers (≥1,280) were predictive of adverse outcomes (328, 330).
The place of the LFA in cryptococcosis diagnosis is yet to be clarified. Currently, it is recommended for use on serum, plasma, or CSF for diagnosis of cryptococcosis in symptomatic patients with established infection. Further evaluation of its utility in the diagnosis of C. gattii infections is needed, especially in patients with infection outside the CNS or lung. It is likely that the LFA will occupy an important role given its simplicity and accuracy thus far.
Globally, screening of asymptomatic patients for cryptococcosis in populations with a high prevalence of cryptococcal antigenemia has become a priority with endorsement by the World Health Organization. In countries with a high prevalence of HIV infection, the strategy of screening for antigenemia by the LFA and the use of targeted preemptive therapy is cost-effective; this may also be so in resource-rich countries (329, 331). Costs, including labor, shipping, and overheads, range from US$2.50 to US$5.50 per test in resource-poor settings to US$10 in higher-income countries (331).
Nucleic acid-based tests.
In clinical practice, molecular methods are seldom required to confirm the diagnosis of suspected cryptococcosis. Nevertheless, several assays have been evaluated for the rapid and sensitive detection of Cryptococcus in clinical specimens and to distinguish between C. neoformans and C. gattii. These assays may be useful when the organism does not grow, for example, where the fungus is visualized in tissue biopsy sections but is not cultured, or if a patient has had substantial anticryptococcal antifungal therapy. In the uncommon situation where phenotypic methods provide an ambiguous result, DNA sequencing of isolates for species assignment is helpful, but multiplex PCR and other molecular methods can also be used. Inclusion of primers specific for Cryptococcus in multiplex assays based on syndromic clusters (for example, “septicemia” and “CSF infection”) or for analysis of blood cultures or biopsy samples where the differential diagnosis includes fungal disease has been useful in some laboratories (C. Halliday and S.C.-A. Chen, unpublished data).
(i) Detection of C. gattii directly from clinical specimens.
PCR-based and non-PCR-based molecular techniques have demonstrated good sensitivity in identifying C. gattii in clinical specimens. Methods include single or multiplexed PCR assays, which may be nested or real time in format (332). Most approaches have exploited nucleotide polymorphisms in the ITS region or the large ribosomal subunit of the fungal rDNA complex, although other gene targets (e.g., URA5, CAP59, and M13) have also been used (see Taxonomy of the Species C. gattii, above). Positive broad-range PCR assays can also be followed by DNA sequencing of the amplicon.
(ii) Panfungal PCR.
Many panfungal PCR assays target the multicopy fungal rRNA gene, for example, the ITS region, to identify fungi. Such assays have detected DNA from members of the C. neoformans complex directly from clinical specimens, including CSF (333), tissue specimens (334), and blood (335). For laboratories choosing this approach, the entire ITS region should be amplified and sequenced to allow identification of C. gattii to the species level; in addition, querying the unknown sequence should be performed against a database which distinguishes C. gattii from C. neoformans (see below).
(iii) Nested PCR.
Nested PCR assays have successfully detected cryptococcal DNA in clinical specimens and have been validated for C. neoformans, but their utility in detecting C. gattii is largely untested. Rappelli et al. developed a nested ITS-targeted PCR protocol for detecting C. neoformans as well as C. gattii (serotype B) from CSF samples from patients with neurocryptococcosis (336). All 21 CSF samples from HIV/AIDS patients with confirmed cryptococcal meningitis yielded positive results. The analytical sensitivity of the assay was 10 cells/μl, and the specificity was 100%. Nested PCR has also been employed to detect C. neoformans in brain tissue from infected mice (337) and in the diagnosis of patients with primary lymphonodular abdominal cryptococcosis (338).
(iv) Real-time PCR.
Real-time PCR is attractive since it is rapid and provides much higher assay sensitivity without compromising specificity. Bialek et al. showed that real-time PCR was faster than nested PCR for the detection of C. neoformans in the brains of experimental mice (337). Amplification of the 18S and 28S rRNA genes using real-time PCR has been successfully employed to detect C. neoformans directly from pericardial fluid (339). Using a TaqMan real-time assay, other investigators showed that seven clinical samples that were culture positive for C. neoformans and C. gattii were confirmed to be positive by the real-time assay, with no false-negative, or false-positive, results (340).
The utility of panfungal, nested, and real-time assays to distinguish C. neoformans from C. gattii is uncertain, and where this is not possible, the organism should be identified as “C. neoformans complex.”
(v) DNA sequencing as an identification method.
As for other fungi, DNA sequencing has been considered to be the gold standard for accurate species identification. Apart from its high level of reproducibility and objectivity, data generated by sequencing methods are deposited in public central repositories and are readily accessible. It should, however, be noted that there are errors within unrefereed public gene databases, collectively comprising the International Nucleotide Sequence Database (INSD), containing GenBank (http://www.ncbi.nlm.nih.gov/); the European Molecular Biology Laboratory (EMBL) Nucleotide Sequence Database (http://www.ebi.ac.uk/embl/); and the DNA Data Bank of Japan (DDBJ) (http://www.ddbj.nig.ac.jp/). Error rates for fungal sequences within GenBank, for example, can be as high as 14 to 20%. The commercial MicroSeq D2 DNA sequencing kit system (Applied Biosystems, Rotkreuz, Switzerland) is unable to distinguish C. neoformans from C. gattii. However, the two species can be identified by using curated databases containing ITS sequences for pathogenic fungi, available through the Centraalbureau voor Schimmelcultures Fungal Biodiversity Centre (which also contains a proportion of GenBank data associated with CBS strain numbers) (www.cbs.knaw.nl/collections/) and at the University of Sydney, Australia (http://its.mycologylab.org/).
In addition to the ITS gene region, other cryptococcal genes may have utility for the identification of C. gattii (discussed in detail in Taxonomy of the Species C. gattii, above). While acknowledging the limitations of ITS sequence analysis, the ITS region remains one of the more informative regions for species identification. Sequencing of both the ITS1 and ITS2 (including the 5.8S rRNA gene) regions is required for species identification of C. gattii. Using this approach, Katsu et al. accurately differentiated members of the C. neoformans complex (341). ITS sequences delineated genetic polymorphisms that allowed the distinction of C. neoformans from C. gattii; furthermore, species-specific ITS “sequence types” correlated with PCR fingerprinting/random amplification of polymorphic DNA (RAPD) molecular types, and ITS sequencing was able to assign subtypes within C. gattii. In contrast, sequencing of the ITS1, ITS2, or D1/D2 amplicons alone identified C. neoformans complex isolates only as “C. neoformans” (334, 342). ITS sequence analysis is especially useful when combined with sequencing of other genes, for example, in MLST (see Taxonomy of the Species C. gattii, above). Even with automation of sequencing, proofreading of the sequences still constitutes a significant task in a busy, high-throughput laboratory.
(vi) Multiplex PCR.
Multiplex PCR has the advantage of simultaneously detecting a number of gene targets and can be performed quickly with a small amount of DNA compared to conventional PCR. It allows rapid identification to the species level and can distinguish between serotypes by using different combinations of primers. Assays can be coupled with real-time PCR (343). Multiplex PCR has also been used to determine mating-type profiles through the amplification of STE (sterile) gene sequences of C. neoformans, although there are no similar reports for C. gattii (344, 345). Leal et al. described a multiplex PCR protocol based on amplification of the ITS region with species-specific primers. The results allowed rapid differentiation between isolates of C. neoformans and C. gattii, and the PCR assay showed specificity and sensitivity comparable to those of the Crypto Check serotyping kit (Iatron Laboratories; no longer available commercially) and CGB agar (343). Recently, Feng et al. developed a uniplex assay for the rapid identification of C. neoformans and C. gattii. The assay targeted the putative sugar transporter (STR1) gene; amplified PCR fragments sizes for C. neoformans var. grubii and C. neoformans var. neoformans were 274 bp and 224 bp, respectively, while that for C. gattii was 170 bp (346). The identity of all amplicons was confirmed by DNA sequencing of representative strains. By PCR analysis, 253 of 255 (99.2%) isolates of the C. neoformans-C. gattii complex were correctly assigned to the respective species, including serotype AB, AD, and BD hybrid strains. Subsequently, those same authors developed a duplex PCR assay to differentiate isolates within the species C. gattii (346). Whether this technique detects C. gattii from clinical samples has not yet been tested.
(vii) Isothermal amplification method.
Isothermal amplification approaches, including hyperbranched rolling-circle amplification (RCA), targeting single nucleotide polymorphisms (SNPs) in the ITS region, have also successfully identified C. gattii (347). This technology uses circularizing padlock probes of approximately a 50- to 100-mer length that contain target complementary sequences by a linker. Specialized DNA polymerases are employed under isothermal conditions to replicate short single-stranded DNA circles. This technique is simple, robust, and highly specific, detecting differences in target nucleic sequences down to the single-nucleotide level (348). It can be performed within 2 h by using a water bath, heating block, or thermocycler. By using real-time PCR (RT-PCR) systems, an RCA signal is detected within 15 min of initiation of the RCA reaction. Studies examining fungal identification with this method have reported good results with cost-effectiveness in comparison with DNA sequencing (349). Based on this approach, Kaocharoen et al. (347) designed four padlock probes targeting cryptococcal species-specific SNPs. The assay was validated with 99 C. neoformans complex isolates; it clearly distinguished between C. neoformans and C. gattii. However, false-positive “C. neoformans var. grubii” signals were obtained when an RCA3-ITS cryptococcal probe was challenged with certain Candida species, including Candida albicans. Hence, careful design of padlock probes is essential for specificity. Further evaluation of RCA as a technique for identification of C. gattii is required.
(viii) Probe-based microarrays and the Luminex bead suspension array.
The position of probe-based microarrays in fungal identification is currently limited by the high cost of microarray technology, limited platform flexibility, slow hybridization kinetics, and the need for strict optimization of test parameters to achieve reproducibility (for methodological details, see reference 312). Nonetheless, a variation of this assay format, the high-throughput Luminex x-map technology system (Luminex Corp., Austin, TX), has been evaluated for its ability to identify both C. neoformans and C. gattii; the assay is based on detecting gene polymorphisms within the intergenic spacer (IGS) regions. This system employs flow cytometry principles to detect and identify DNA targets. It detects 5′-biotin-labeled PCR amplicons hybridized to specific probes of the target species and can utilize a direct DNA hybridization or solution-based enzymatic reaction capture format. As its name suggests, specificity is achieved by the use of uniquely color-coded beads or microspheres to capture “species-specific” combinations with red/infrared fluorescence, and it is capable of detecting up to 500 different analytes (representing up to 100 species) in a reaction tube/well. By using direct hybridization in the Luminex system, Diaz et al. described an 8-plex hybridization array for the simultaneous detection of species, varieties, and genotypes within the C. neoformans complex, which allowed discrimination of 1-bp mismatches with no apparent cross-reactivity (350). This assay was subsequently validated with 58 clinical and environmental isolates, and isolates were correctly identified to the species and genotype levels, including hybrids (serotypes AD and BD). Furthermore, the assay identified C. neoformans directly from stored CSF samples from eight patients with cryptococcal meningitis, although none of the samples evaluated contained C. gattii (351). Because of their potential for automation, bead-based technologies show promise for DNA-based diagnostics. Their current high acquisition costs restrict their wider use.
Proteomic approaches.
In parallel with advances in proteomic approaches in diagnostic microbiology, methods such as matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) have been evaluated for the identification of Cryptococcus spp. (57, 58, 352). In the largest evaluation to date, 81 clinical and environmental C. gattii isolates representing each of the four major molecular types (VGI, VGII, VGIII, and VGIV) were tested. Protein extracts were obtained by a standard formic acid extraction method (summarized in reference 353). MALDI-TOF MS using the Bruker Microflex system (Bruker Daltonik GmbH) and MALDI Biotyper software (version 3.0) correctly identified 100% of C. gattii isolates and correctly grouped each isolate according to its molecular type; furthermore, the technique was able to identify hybrids of C. neoformans and C. gattii. It is important to note that those authors first established reference spectra of all four molecular types by using well-characterized reference strains and subsequently added all spectra generated to an “in-house” supplementary MALDI Biotyper library (2011), thus extending a validated spectral library (57). Head-to-head comparisons of MALDI-TOF MS with traditional genotyping methods are essential in positioning MALDI-TOF MS as a typing tool.
In another study, McTaggart et al. compared MALDI-TOF MS analysis with DNA sequencing to identify Cryptococcus species. They studied 27 strains of C. gattii (23 clinical and 4 reference strains) and compared spectra against the MALDI Biotyper version 2.0.1 database (Bruker Daltonik). When the Bruker library was supplemented with in-house-generated spectral entries, all 27 strains were correctly identified to the species level (score, ≥2.0). However, when queried against the existing Bruker library, a substantially smaller proportion of isolates was identified. If C. neoformans plus C. gattii isolates were taken into consideration, only 58.4% of isolates were correctly assigned (352). One other evaluation, which also complemented the Bruker library (version 3.0) with in-house spectra, yielded correct species identification for all 10 strains studied (58). Very few studies have tested the ability of the current Bruker library alone to identify C. gattii. In a study in which only three isolates of C. gattii were encountered, two isolates yielded scores of ≥2.0, and one yielded scores of ≥1.7 to 1.99 (354). Ongoing construction of libraries containing reliable spectra from known isolates is essential. Until these extended libraries are more widely available, identification of C. gattii should not be reliant only on MALDI-TOF MS.
There is ongoing debate about the requirement for extraction of yeast protein for MALDI-TOF MS. A review of published studies (beyond the scope of this article) indicates that the use of the extraction procedure recommended by the manufacturer yields optimal and reproducible spectra (353). With respect to C. gattii, Firacative et al. reported that no spectra were obtained from intact yeast cells (57). The number of colonies used and the time of incubation did not affect results. Whether MALDI-TOF MS can be successfully applied to detect cryptococci in clinical specimens such as CSF is uncertain. In a small study, 11 CSF samples were inoculated in blood culture bottles, cultured, and then analyzed by MALDI-TOF MS for bacteria and yeasts, but none of the patients had cryptococcal meningitis (355).
ANTIFUNGAL SUSCEPTIBILITY
On casual reading, there appears to be little value in routinely determining the susceptibility of C. gattii to antifungal agents, given that firm therapeutic recommendations based on MICs cannot be made due to the lack of clinical breakpoints (CBPs). Only C. neoformans has been included in Clinical and Laboratory Standards Institute (CLSI) guidelines for testing of yeasts (356). There are as yet no data to indicate that adverse patient outcomes are correlated with raised MICs and, if so, at what level.
However, antifungal susceptibilities have been used to guide antifungal treatment, especially in patients failing to respond to antifungal therapy. MICs, mainly of amphotericin B, 5-flucytosine, and fluconazole, which have been the cornerstone of anticryptococcal treatment for decades and determined by CLSI broth microdilution methods, have been reported for both C. neoformans and C. gattii (see below; see also Antifungal Therapy and Management, below). There are also in vitro data for the newer azoles voriconazole, posaconazole, and isavuconazole.
Epidemiological Cutoff Values for C. gattii
Prior to the recent C. gattii outbreaks, there was evidence that MICs of antifungal drugs varied with the geographic origin of the isolates, although many of the isolates tested were derived from individual institutional culture collections. Subsequently, MICs of fluconazole were noted to be increased among isolates from C. gattii VGII case clusters (see discussion below). This prompted an ongoing large-scale study of MIC CBPs and of epidemiological MIC cutoff values (ECVs) based on MIC distributions of wild-type strains (357–359). ECVs for fluconazole were determined in an international consensus study of a large number of clinical isolates from Europe, the United States, Australia, Brazil, Canada, India, and South Africa, viz., 8 mg/liter for C. gattii molecular types VGI, VGIIa, and VGIII and nontyped strains; 16 mg/liter for molecular type VGIV; and 32 mg/liter for molecular type VGII (except for VGIIa). Although varying by genotype, the ECVs are within 1 to 2 dilutions of each other. Similar molecular type-specific ECVs have been reported for itraconazole, voriconazole, and posaconazole (358). Espinel-Ingroff et al. proposed that ECVs of 0.5 mg/liter for amphotericin B and 4 mg/liter for 5-flucytosine would encompass >99% and >95.7% of C. gattii strains, respectively (357). Lockhart et al. (359) also established azole ECVs for C. gattii based on MIC distribution, proposing an ECV of 32 mg/liter for fluconazole, but acknowledged that >50% of their isolates were from the Pacific Northwest (these isolates were predominantly of molecular type VGII [see below]). ECVs are useful adjuncts for identifying isolates with acquired or intrinsic resistance or those that need further study. Whether they can be universally accepted as surrogates for CBPs to define “susceptibility” or “resistance” is less clear (360, 361).
Antifungal Susceptibility
A number of studies have documented comparatively low MICs of standard antifungals against C. gattii (and in comparison with C. neoformans), which have not increased over time (29, 362–364); resistance to amphotericin B and 5-flucytosine was rare. Based on the fluconazole CBPs for Candida spp. reported in CLSI document M27-A3 (356), fluconazole “resistance” (MIC, ≥64 mg/liter) among Cryptococcus spp. was uncommon in Australia (29, 362). There are no data to suggest whether the new CBPs for Candida spp. (resistance defined by an MIC of ≥8 mg/liter [361]) are applicable to Cryptococcus. The apparently low frequency of fluconazole resistance appears to be the case in other regions, including Malaysia, India, Spain, and Taiwan (Table 5) (29, 365–368). In a Malaysian study, 2 of 48 (4.2%) isolates had fluconazole MICs of >64 mg/liter. In addition, there were no significant differences in MICs between Cryptococcus species (P > 0.05), indicating that C. gattii was as susceptible as C. neoformans var. grubii. Furthermore, no significant difference in the MICs for C. gattii isolates collected from 1980 to 1990 and 2002 to 2004 were observed (365). In a Brazilian study, eight C. gattii strains (fluconazole MIC range, 8 to 32 mg/liter) were susceptible to most drugs tested, and susceptibility profiles of C. gattii were similar to those of C. neoformans (369). Table 5 summarizes fluconazole susceptibility data from major clinical studies.
TABLE 5.
Major clinical studies reporting antifungal susceptibility testing of Cryptococcus gattii against fluconazolec
| Reference | Origin(s) of isolates | Total no. of isolates | Source (no.) of isolates | Modal MIC (mg/liter)a | MIC90 (mg/liter) | GM MIC (mg/liter) | MIC range (mg/liter) | % of isolates with MIC ≥ 32 mg/liter | Description |
|---|---|---|---|---|---|---|---|---|---|
| 29 | Australia | 18 | C | 4 | 16 | NS | 0.5–64 | 11 | Primarily VGI isolates |
| 75 | Taiwan | 21 | C | 8 | 16 | NS | 0.125–16 | 0 | C. gattii was less susceptible to AMB and 5FC than C. neoformans; all C. gattii isolates were FLU susceptible |
| 403 | South America | 11 | C | NS | NS | NS | 8–16 | 0 | High degree of susceptibility to AMB, 5FC, FLU, ITC |
| 370 | Spain | 57 | C (52) | NS | 32 | 9.54 | 1–64 | NS | C. gattii was less susceptible than C. neoformans to FLU but not AMB and 5FC |
| E (5) | |||||||||
| 368 | Spain | 30 | C (mainly) | NS | NS | 0.7–1 | 0.25–2 | 0 | Overall low-level resistance; MICs for FLU were higher for C. gattii than for C. neoformans (P = 0.007) |
| 404 | Brazil | 23 | C | NS | 25.6 | 15.52 | 4–>64 | NS | All strains had low MICs of AMB (0.03–0.25 mg/liter); among azoles, POS had greatest activity, followed by VOR, ITC, and FLU |
| 366 | USA | 35 | C (NS) | NS | 0.064 | 0.03 | 0.03–0.06 | 0 | Highly susceptible to isavuconazole |
| E (NS) | 0.06 | 0.027 | 0.015–0.25 | 0 | |||||
| 363 | Worldwide | 42 | C (NS) | NS | 4 | 2.36 | 0.25–64 | Few differences in susceptibility between C. gattii and C. neoformans | |
| E (NS) | |||||||||
| 371 | USA | 43 | C | 4 | 16 | 6.38 | 0.5–32 | 0.05 | VGII molecular type strains, especially those of VGIIc, had highest FLU MICs |
| 375 | Australia, USA, Canada | 45 | C (NS) and E (NS); study I | NS | 8 | 3.54 | 1–8 | 0 | C. gattii less susceptible than C. neoformans; strains of the VGII molecular type had significantly higher MICs for 5FC and all azoles, especially FLU |
| 103 | Extended study | NS | 8 | 4.96 | 1–32 | NS | |||
| 373 | Worldwide | 350 | C (215) | NS | 8 | 3.573 | 0.25–64 | NS | Veterinary isolates were least susceptible to ITC, POS, VRC, and ISA; MICs varied with molecular type for FLU and 5FC |
| 367 | India | 62 | C (2) | 1.9 | 1.55 | 1–16 | 0 | VGI; C. gattii less susceptible than C. neoformans to FLU, ITC, and VRC but not AMB and 5FC; E was less susceptible than C and more resistant than C | |
| E (60) | NS | 8 | 8 | 0 | |||||
| 405 | Brazil | 13 | C | NS | NS | NS | 16–>64 | 15 | VGII molecular type studied |
| 372 | Brazil | 49 | C (43) and E (6); VGII | 16 | 6.08 | 0.25–>64 | NS | VGII molecular type strains less susceptible to FLU, VRC, and 5FC than VGI strains | |
| VGI | 4 | 1.55 | 1–8 | 0 | |||||
| 359 | Global | 298 | C and E (NS) | 8 | 16 | 5.51 | 0.5–32 | 20 | Molecular type VGII strains had the highest FLU GM MICs, and molecular type VGI had the lowest (8.6 vs 1.7 mg/liter) |
| 406 | Brazil | 54 | C (50) | NS | 16 | NS | 1–64 | NS | Mainly molecular type VGII strains |
| E (4) | |||||||||
| 369 | Brazil | 8 | C | NS | 32 | 13.45 | 8–32 | 0 | Susceptible to most drugs; no differences in susceptibility between C. neoformans and C. gattii; ECV = 32 mg/liter |
| 374 | Canada, USA | 90 | C (75) | 4 | 8 | 4.5 | 2–64 | NS | Molecular type VGII studied, of which types IIb and IIc were less susceptible |
| E (15) | |||||||||
| 376b | Brazil | 24 | NS | ≤1 | 4 | NS | <0.12–32 | 3 | 3 isolates with MIC of 32 mg/liter; no molecular type stated |
In some instances, calculated by the authors.
Measured by flow cytometry.
Abbreviations: AMB, amphotericin B; C, clinical strains; E, environmental; 5FC, 5-flucytosine; FLU, fluconazole; GM, geometric mean; ITC, itraconazole; ISA, isavuconazole; POS, posaconazole; NS, not specified; VRC, voriconazole.
Nonetheless, there remains concern over whether C. gattii may be less susceptible to some antifungal agents. In a study from Taiwan, C. gattii was less susceptible to 5-flucytosine and amphotericin B (75), but the geometric means (GMs) of the MICs were still within the range considered “susceptible.” A group in Brazil reported significantly higher GMs of MICs for fluconazole, voriconazole, amphotericin B, and 5-flucytosine against C. gattii than against C. neoformans (370); likewise, Torres-Rodriguez et al. (368) reported fluconazole, voriconazole, and posaconazole MICs to be significantly higher for C. gattii, although the overall prevalence of strains with fluconazole resistance was still low (range, 0.25 to 2 mg/liter for C. gattii) (Table 5). Similarly, Chowdhary et al. noted that C. gattii strains from India were less susceptible than C. neoformans var. grubii to fluconazole, itraconazole, and voriconazole (P < 0.05) but not amphotericin B and 5-flucytosine (367). Elevated MICs of amphotericin B and 5-flucytosine were rare in the United States (371).
Of note, there are data to indicate that fluconazole susceptibility, and, to some extent, those of the other azoles, varies with the molecular type of C. gattii. Elevated MICs against fluconazole (and voriconazole) have been reported from the Pacific Northwest, with genotypes VGIIa and VGIIc having the highest MICs (359, 371–374). In one study of 43 clinical isolates from the Pacific Northwest, molecular type VGIIc had the highest fluconazole GM MIC (12.7 mg/liter), while types VGI and VGII had the lowest GM MICs (2.38 mg/liter). Among the azoles, fluconazole had the least activity (GM MIC 16- to 63-fold lower for all isolates) (371). Lockhart et al. also noted that molecular type VGII strains had the highest fluconazole GM MICs (8.6 mg/liter), with molecular type VGI isolates exhibiting the lowest GM MICs (1.7 μg/ml) (359). In a study of 350 clinical (human and veterinary) and environmental isolates, Datta and coworkers reported that clinical VGII isolates had significantly higher GM MICs for flucytosine and fluconazole than did clinical VGI isolates; isolates from veterinary sources were the least susceptible to the azoles (374). In another large study of clinical, veterinary, and environmental isolates (375) (Table 5), most of which originated from Australia and Canada, C. gattii genotype VGII isolates also had significantly higher MICs for all azole antifungals, particularly fluconazole, than did isolates of other genotypes. Notably, the inclusion of large numbers of environmental isolates may skew results, since there are data to suggest that these isolates may be less susceptible to antifungal agents (367). In addition, comparison of MIC results between studies is complicated by the fact that many studies were done by using different MIC testing methods. Although the implications of higher fluconazole MICs for therapeutic dosing and clinical outcomes have not been determined, there is major interest in this issue since C. gattii genotype VGII is the genotype associated with the ongoing C. gattii outbreak in North America.
Several studies have documented that voriconazole, posaconazole, and isavuconazole are more active in vitro against C. gattii than fluconazole (359, 366, 371–374), and hence, they may represent valuable treatment alternatives to fluconazole. In a study of 90 clinical and environmental isolates of C. gattii from the Pacific Northwest of the United States, fluconazole MICs against C. gattii VGII clinical and environmental isolates were higher than those of isavuconazole, voriconazole, and itraconazole (374). U.S. and Spanish data revealed GM MICs of voriconazole of 0.1 mg/liter and 0.03 mg/liter, respectively (371, 376). In another study that tested 298 strains, the modal MICs were 0.12 mg/liter for voriconazole, 0.5 mg/liter for itraconazole, and 0.5 mg/liter for posaconazole, compared with a GM fluconazole MIC of 8 mg/liter (359). Thompson et al. tested 35 C. gattii strains by using the Etest (AB Biodisk, Solna, Sweden) and CLSI broth microdilution (overall agreement, 97.8%). GM MICs of isavuconazole were 0.027 mg/liter by broth microdilution and 0.03 mg/liter by Etest; the corresponding MIC90 values (the concentration of an antimicrobial agent at which 90% of the organisms are inhibited) were 0.06 and 0.125 mg/liter, respectively, indicating a high level of susceptibility. That same group of investigators also showed that isavuconazole had the lowest MIC90 and GM MIC values, with the next most active azole being posaconazole, followed by voriconazole and then itraconazole (363).
In summary, despite the fact that there are no data on the relationship between MICs, drug dosing, and clinical outcome, susceptibility testing may be helpful for patients failing to respond to apparently appropriate therapy, when a high MIC result may favor substituting an alternative drug. Susceptibility testing is recommended to monitor epidemiological trends.
Mechanisms of Azole Resistance in C. gattii
The emergence of C. gattii strains with elevated MICs of fluconazole has sparked interest in the mechanisms of azole resistance in this organism. A few studies have examined the roles of the cryptococcal ERG11 gene and of the ATP-binding cassette transporters in resistance in C. neoformans (377–380); whether these observations are applicable to C. gattii are uncertain. Mutations in, or overexpression of, the ERG11 gene, encoding the azole target lanosterol 14-α demethylase, and overexpression of one or more plasma membrane proteins that pump azoles out of the cell are well recognized mechanisms by which fungi acquire resistance to azoles (379).
Rodero et al. studied five sequential C. neoformans isolates recovered from a patient with recurrent cryptococcal meningitis; four were fluconazole susceptible (MICs, 1.5 to 2 mg/liter), while the fifth isolate, associated with clinical relapse, was considered resistant (MIC, 32 mg/liter). The latter isolate contained a point mutation responsible for the amino acid substitution G484S (377). However, the possibility of another mechanism of resistance was not excluded. Sionov et al. identified a single missense mutation in a fluconazole-resistant strain of C. neoformans resulting in the replacement of tyrosine by phenylalanine at amino acid position 145 (Y145F); this conferred resistance to fluconazole and voriconazole but not itraconazole and posaconazole (378). Most recently, the role of ERG11 mutations in conferring azole resistance in C. gattii was studied in 25 U.S. strains with relatively high MICs of fluconazole (GM MIC of 20 mg/liter and MIC90 of 64 mg/liter) and 34 isolates with lower MICs (GM MIC of 9.9 mg/liter and MIC90 of 32 mg/liter). Amino acid substitutions in the deduced Erg11p sequences of strains with higher fluconazole MICs were not associated with differences in the susceptibilities of these proteins to inhibition by the azole (381). This conclusion was supported by experiments determining azole MICs for conditional Saccharomyces cerevisiae erg11 mutants expressing variant gene sequences. In comparison to the “wild-type” isolates, azole MICs were all within 1 to 2 dilutions of each other. Gast et al. also compared ERG11 mRNA levels in the C. gattii strains; ERG11/ACT1 mRNA ratios did not correlate with fluconazole MICs. They concluded that neither ERG11 overexpression nor variation in ERG11 coding sequences was responsible for the high fluconazole MICs observed in their study (381).
Enhanced activity of plasma membrane azole efflux pumps, which has been associated with azole resistance of C. neoformans, has not yet been studied in C. gattii (380, 382). Upregulation of the ATP-binding cassette transporter-encoding gene AFR1 in C. neoformans was associated with resistance to fluconazole in vitro and in mice with systemic cryptococcosis (380, 383).
An additional phenomenon of heteroresistance among C. neoformans strains in AIDS patients undergoing fluconazole maintenance therapy has been reported (384, 385). This is a form of intrinsic and adaptive resistance that has been associated with clinical failure. Researchers in the United States have established that C. neoformans strains are innately heteroresistant to fluconazole, with each strain producing a subpopulation that can survive concentrations of fluconazole well above the MICs (385). Sionov and colleagues have subsequently shown that, in vitro, strains adapt to high fluconazole concentrations by undergoing disomy (where the cryptococcal cell has 2 members of a pair of homologous chromosomes), first in chromosome 1 and then in multiple other chromosomes. The duplication of chromosome 1 parallels the duplication of the ERG11 and AFR1 genes and is reversed by removing the drug from the cultures. Whether a similar mechanism of heteroresistance involving chromosome duplication occurs in C. gattii exposed to azoles is uncertain but is an important area of future study.
ANTIFUNGAL THERAPY AND MANAGEMENT
Antifungal Therapy
Current recommendations for the management of infections caused by C. gattii are based largely on extrapolation from clinical trials in patients with infection caused by C. neoformans and on individual case reports, case series, and expert opinion (282). This review focuses only on the most recently reported data, as a comprehensive review and an article detailing practical management guidelines were published in 2013 (262, 386).
Infectious Diseases Society of America (IDSA) guidelines outline therapeutic approaches based on host status, site of infection, complications of cryptococcosis, and limitations placed on therapeutic options in resource-limited settings. In general, these guidelines recommend similar antifungal treatment strategies for C. gattii and C. neoformans (282). There is some evidence to support this approach from, for example, a subanalysis of a retrospective study of C. gattii and C. neoformans meningitis in immunocompetent hosts (27) and reports from Africa of outcomes of cryptococcal meningitis in patients with AIDS (96–98). Notably, the genotype of HIV-associated C gattii infections was not stated, and it is possible that disease caused by C. gattii VGIV, which has been isolated from HIV-associated infections in Africa (see Epidemiology, Origin, and Evolution, above), resembles that due to C. neoformans. Diagnostic distinction between C. gattii and C. neoformans by the simple laboratory test of growth on CGB agar has long been implemented in Australia due to the endemicity of C. gattii (now typed as VGI), its association with substantial pulmonary and cerebral cryptococcomas (29), and the attendant differences in the duration of induction and consolidation/maintenance therapy.
More recent and larger studies of C. gattii infection in Australia and of the outbreaks in Canada and the United States suggest that therapeutic recommendations may be refined by taking into account the genotype of the causative C. gattii strain and possibly its antimicrobial susceptibility (130, 131, 258, 299), although the latter is yet to be confirmed in vivo. Recently reported data indicate that for C. gattii VGI infection involving the lung and/or the CNS, induction therapy with amphotericin B plus 5-flucytosine is appropriate (∼6 weeks for cerebral disease and 2 weeks for isolated lung disease) (299). Shorter induction courses may be effective in patients with CNS disease and minimal brain involvement, but current data are insufficient to confirm this proposition. As recommended by IDSA guidelines, for lung infection not considered “serious” (282), single small cryptococcomas due to C. gattii can be treated initially with fluconazole (386). Consolidation, and then maintenance, therapy with fluconazole is recommended universally. In patients with C. gattii cryptococcosis, there is a compelling argument to use the term “eradication therapy” rather than “consolidation” and “maintenance” therapy, as infection is curable with adequate antifungal and ancillary therapies. The minimum duration required to achieve cure in an immunocompetent patient has not been defined, but durations of 6 to 12 months for isolated and circumscribed lung infection and 12 to 18 months for cerebral disease are commonly used (262, 282, 299, 386).
Surgery
There are few studies that address indications for surgery in cryptococcosis. Single large lesions, especially when encroaching on local structures in the lung or with mass effect and surrounding edema upon cerebral imaging, require resection if they are surgically accessible (262). Insertion of a lumbar drain or an early CSF shunt procedure may be required for raised ICP not controlled by repeated lumbar punctures, and shunting is necessary in the presence of symptomatic hydrocephalus (see “Management of Complications,” below).
Management of Complications
Raised intracranial pressure.
Control of raised ICP is a critical determinant of the outcome of CNS cryptococcosis. Repeated lumbar punctures or, if necessary, insertion of a lumbar drain or a CSF shunt is indicated to reduce the ICP to acceptable levels. Symptomatic hydrocephalus visible upon cerebral imaging likewise requires the insertion of a shunt to relieve the high ICP, regardless of the cryptococcal species responsible (262, 282, 299).
Immune reconstitution inflammatory syndrome.
In all cases, consolidation/maintenance antifungal therapy (in the case of C. gattii which can be cured, this is actually eradication therapy) should be continued. Patients with minor manifestations of IRIS can be managed with supportive care. Corticosteroids have been used successfully to control symptoms and reduce the size of inflammatory lesions (299). An early retrospective observational study in PNG suggested that corticosteroid use improved visual outcomes in patients with C. gattii meningitis (387). After initiation, a slow reduction in corticosteroid doses is needed to prevent a relapse of inflammation. Most cases of IRIS will resolve within days to weeks. In a small series of 8 patients with C. gattii neurological infection and IRIS, none died, but 4 had persistent neurological sequelae at 12 months (3 had received gamma IFN). Cerebral imaging abnormalities returned to pre-IRIS appearances within 4 to 6 months (299).
CONCLUSIONS
In conclusion, based on morphological, biological, and phylogenetic species concepts, C. gattii is recognized as a valid species in the C. neoformans-C. gattii species complex. This is congruent with its ecology, epidemiology, and clinical associations, which differ substantially from those of C. neoformans. Modern genetic techniques have determined the population genetic structure of C. gattii and include recombination studies, which have given us insight into how C. gattii is sustained in nature. There are four main genetic groups of C. gattii (VGI, VGII, VGIII, and VGIV, which are equivalent to AFLP4 through to AFLP7), with MLST typing superseding earlier methods of strain typing. The ISHAM consensus method is based on MLST using a minimum of seven predetermined genetic loci; this approach is recommended to enable strain pattern comparisons. Whole-genome sequencing, which is the most discriminatory strain typing method, is likely to become more common with time.
Clinically apparent cryptococcosis represents the outcome of a complex set of interactions that include the size of the inhaled inoculum, the individual C. gattii virulence composite (which is not simply the sum of individual virulence phenotypes), and the host status/response. Through the application of genomics and other omics technologies, it is increasingly apparent that the outcome of the host-cryptococcus interaction is regulated by complex sets of transcriptional circuits in both the fungus and host cells and that differences between human disease caused by C. gattii and that caused by C. neoformans can be elucidated in relevant animal models.
The ecology and epidemiology of infection vary with geographic region. Both humans and animals are susceptible to C. gattii, and in some circumstances, animals may be considered sentinels of human disease. Despite the emergence of C. gattii infections in new environments where 40 to 73% of patients have predisposing or immunosuppressive medical conditions, its propensity to cause disease in healthy hosts remains strong. Many infections, including those in new geographic locations, have occurred in apparently immunocompetent patients. However, the recent focus on identifying at least subgroups of patients with a genetic predisposition or immune dysfunction, for example, an association with anti-GM-CSF antibodies, may alter this thinking.
CNS and lung infections are the most common manifestations, although any body site may be affected. CNS infection is frequently encountered in the nonoutbreak setting; hence, routine lumbar puncture of patients with suspected cryptococcosis is recommended. CNS complications are common, particularly raised intracranial pressure, hydrocephalus, and severe neurological sequelae. New, rapid diagnostic tests and imaging modalities are assisting early diagnosis. Differentiation of C. gattii from C. neoformans is recommended as a routine and is achievable in most cases by simple phenotypic methods. Molecular techniques are definitive and provide subtype data. Despite the fact that there are no data on the relationship between MICs, drug dosing, and clinical outcome, antifungal susceptibility testing of azole drugs may be helpful for patients failing to respond to apparently appropriate therapy, and regular MIC surveillance in reference laboratories is recommended to monitor trends.
There are no antifungal drug trials to guide antifungal treatment of C. gattii infections. However, data from contemporary studies suggest that the site(s) of infection is the main influence on the choice of the antifungal drug regimen. For CNS infection and for severe pulmonary disease, intensive induction therapy with amphotericin B plus 5-flucytosine is necessary to optimize outcomes. The duration of induction therapy in patients with CNS disease is recommended to be at least 6 weeks, with a total duration of therapy of 18 to 24 months, although further evaluation of shorter induction courses is warranted. For isolated lung disease, induction therapy should be offered for 2 weeks, with a total duration of therapy of approximately 12 months. Fluconazole remains the preferred azole for use after induction therapy. Aggressive management of hydrocephalus and raised intracranial pressure is essential and may require early surgical drainage of CSF. Surgery may also be indicated for relief of anatomical complications caused by mass lesions. IRIS is an occasional complication that must be distinguished from failure of antifungal treatment.
ACKNOWLEDGMENTS
T.C.S. is a Sydney Medical School Foundation fellow.
The work of S.C.-A.C., W.M., and T.C.S. is supported by the National Health and Medical Research Council of Australia.
S.C.-A.C. and T.C.S. sit or have sat on advisory boards for Merck, Pfizer, and Gilead Sciences, and all three authors have received untied grants from one or more of these pharmaceutical companies in the past.
Biographies

Sharon C. A. Chen is the Director of the Centre for Infectious Diseases and Microbiology Laboratory Services, Westmead Hospital, Sydney, Australia, incorporating a reference laboratory in Mycology, and is an Associate Professor at the University of Sydney. She is also an Infectious Diseases physician with a research interest in medical mycology. She is also the Medical Mycologist for the Molecular Mycology Research Laboratory at Westmead. She is the immediate past scientific chair of the Australia and New Zealand Mycoses Interest Group (ANZMIG), a member of the ASID Clinical Trials Network Steering Committee, a member of National Antimicrobial Committee of Australia, and a member of the National Scientific Advisory Committee for the Australian Society for Microbiology. Her research interests include clinical and molecular epidemiology of fungal infections; infections in immunocompromised hosts, critically ill persons, and those with chronic lung disease; novel diagnostic methods for fungal and bacterial infections; and resistance to antifungal agents.

Wieland Meyer is the Head of the Molecular Mycology Research Laboratory at the Center for Infectious Diseases and Microbiology and Professor for Molecular Medical Mycology, Sydney Medical School-Westmead Hospital, Marie Bashir Institute for Infectious Diseases and Biosecurity, the University of Sydney, Westmead Millennium Institute, Sydney, Australia. He obtained his Ph.D. in 1992 from the Humboldt University of Berlin. His research focuses on the evolution, phylogeny, identification of species, population genetics, molecular epidemiology, and typing of human-pathogenic fungi; the development of fast, simple, and reliable molecular identification techniques for human/animal fungal pathogens; and the understanding of fungal pathogenesis on a molecular level. He has published more than 130 peer-reviewed research papers. He is Vice President of the International Society of Human and Animal Mycology and an associate editor of Personia—Molecular Phylogeny and Evolution of Fungi, Australasian Mycologist, Microbiology Australia, IMA Fungus, Japanese Medical Mycology Journal, and DNA Barcoding.

Tania C. Sorrell is the Director of the Marie Bashir Institute for Infectious Diseases and Biosecurity at the University of Sydney and a Senior Physician in Infectious Diseases at Westmead Hospital. She graduated in Medicine and obtained her research M.D. from the University of Adelaide, South Australia. She has longstanding interests in prevention, diagnosis, and treatment of infectious diseases, especially in immunocompromised hosts, and in the emergence of resistant microorganisms. Her research on the serious fungal infection cryptococcosis has provided new insights into host-microbe interactions and resulted in a new antifungal drug development program. She has developed new diagnostics for infectious diseases and led national studies on the epidemiology of invasive fungal infections. She is a previous President of the Australasian Society for Infectious Diseases and has served/serves on national advisory committees on Infectious Diseases and the Research and Human Ethics Committees of the National Health and Medical Research Council of Australia.
REFERENCES
- 1.Fraser JA, Giles SS, Wenink EC, Geunes-Boyer SG, Wright JR, Diezmann S, Allen A, Stajich JE, Dietrich FS, Perfect JR, Heitman J. 2005. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature 437:1360–1364. 10.1038/nature04220 [DOI] [PubMed] [Google Scholar]
- 2.Kwon-Chung KJ, Bennett JE, Theodore TS. 1978. Cryptococcus bacillisporus sp. nov.: serotype B-C of Cryptococcus neoformans. Int. J. Syst. Bacteriol. 28:616–620. 10.1099/00207713-28-4-616 [DOI] [Google Scholar]
- 3.Findley K, Sun S, Fraser JA, Hsueh YP, Averette AF, Li W, Dietrich FS, Heitman J. 2012. Discovery of a modified tetrapolar sexual cycle in Cryptococcus amylolentus and the evolution of MAT in the Cryptococcus species complex. PLoS Genet. 8:e1002528. 10.1371/journal.pgen.1002528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fell JW, Boekhout T, Fonseca A, Scorzetti G, Statzell-Tallman A. 2000. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. Evol. Microbiol. 50(Part 3):1351–1371. 10.1099/00207713-50-3-1351 [DOI] [PubMed] [Google Scholar]
- 5.Sanfelice F. 1895. Ueber einen neuen pathogenen Blastomyceten welcher innerhalb der Gewebe unter Bildung kalkartig aussehender Massen degeneriert. Zentralbl. Bakteriol. Parasit. Infect. Hyg. 18:521–526 [Google Scholar]
- 6.Vuilemin P. 1901. Les blastomycetes pathogens. Rev. Gen. Sci. Pures Appl. 12:732–751 [Google Scholar]
- 7.Weis JD. 1902. Four pathogenic torulae (Blastomycetes). J. Med. Res. 7:280–311 [PMC free article] [PubMed] [Google Scholar]
- 8.Stoddard JL, Cutler EC. 1916. Torula infection in man. Rockefeller Inst. Med. Res. Monogr. 6:1098 [Google Scholar]
- 9.Todd RL, Herrmann WW. 1936. The life cycle of the organism causing yeast meningitis. J. Bacteriol. 32:89–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benham RW. 1935. Cryptococci: their identification by morphology and serology. J. Infect. Dis. 57:255–274. 10.1093/infdis/57.3.255 [DOI] [Google Scholar]
- 11.Benham RW. 1950. Cryptococcosis and blastomycosis. Ann. N. Y. Acad. Sci. 50:1299–1314. 10.1111/j.1749-6632.1950.tb39828.x [DOI] [PubMed] [Google Scholar]
- 12.Buschke A. 1895. Ueber eine durch Coccidien hervorgerufene Krankheit des Menschen. Dtsch. Med. Wochenschr. 21:14 [Google Scholar]
- 13.Busse O. 1895. Ueber parasitaere Zelleinschluesse und ihre Zuechtung. Zentralbl. Bakteriol. 16:175–180 [Google Scholar]
- 14.Sanfelice F. 1894. Contributo alla morfologia e biologia dei blastomiceti che si sviluppano nei succhi di alcuni frutti. Ann. Igiene 4:463–495 [Google Scholar]
- 15.Evans EE. 1949. An immunologic comparison of twelve strains of Cryptococcus neoformans (Torula histolytica). Proc. Soc. Exp. Biol. Med. 71:644–646. 10.3181/00379727-71-17283 [DOI] [PubMed] [Google Scholar]
- 16.Vogel RA. 1966. The indirect fluorescent antibody test for the detection of antibody in human cryptococcal disease. J. Infect. Dis. 116:573–580. 10.1093/infdis/116.5.573 [DOI] [PubMed] [Google Scholar]
- 17.Vanbreuseghem R, Takashio M. 1970. An atypical strain of Cryptococcus neoformans (Sanfelice) Vuillemin, 1895, II. Cryptococcus neoformans var. gattii var. nov. Ann. Soc. Belg. Med. Trop. 50:692–702 [PubMed] [Google Scholar]
- 18.Curtis F. 1896. Contribution à l'étude de la saccharomycose humaine. Ann. Inst. Pasteur (Paris) 10:449–468 [Google Scholar]
- 19.Kwon-Chung KJ, Boekhout T, Fell JW, Diaz M. 2002. Proposal to conserve the name Cryptococcus gattii against C. hondurianus and C. bacillisporus (Basidiomycota, Hymenomycetes, Tremellomycetidae). Taxon 51:804–806. 10.2307/1555045 [DOI] [Google Scholar]
- 20.Kwon-Chung KJ. 1975. A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia 67:1197–1200. 10.2307/3758842 [DOI] [PubMed] [Google Scholar]
- 21.Kwon-Chung KJ. 1976. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia 68:821–833. 10.2307/3758800 [DOI] [PubMed] [Google Scholar]
- 22.Kwon-Chung KJ. 1976. A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia 68:943–946 [PubMed] [Google Scholar]
- 23.Kwon-Chung KJ, Polacheck I, Bennett JE. 1982. Improved diagnostic medium for separation of Cryptococcus neoformans var. neoformans (serotypes A and D) and Cryptococcus neoformans var. gattii (serotypes B and C). J. Clin. Microbiol. 15:535–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aulakh HS, Straus SE, Kwon-Chung KJ. 1981. Genetic relatedness of Filobasidiella neoformans (Cryptococcus neoformans) and Filobasidiella bacilispora (Cryptococcus bacillisporus). Int. J. Syst. Bacteriol. 31:97–103. 10.1099/00207713-31-1-97 [DOI] [Google Scholar]
- 25.Ellis DH. 1987. Cryptococcus neoformans var. gattii in Australia. J. Clin. Microbiol. 25:430–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ellis DH, Pfeiffer TJ. 1990. Natural habitat of Cryptococcus neoformans var. gattii. J. Clin. Microbiol. 28:1642–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell DH, Sorrell TC, Allworth AM, Heath CH, McGregor AR, Papanaoum K, Richards MJ, Gottlieb T. 1995. Cryptococcal disease of the CNS in immunocompetent hosts: influence of cryptococcal variety on clinical manifestations and outcome. Clin. Infect. Dis. 20:611–616. 10.1093/clinids/20.3.611 [DOI] [PubMed] [Google Scholar]
- 28.Speed B, Dunt D. 1995. Clinical and host differences between infections with the two varieties of Cryptococcus neoformans. Clin. Infect. Dis. 21:28–34; discussion 35–36. 10.1093/clinids/21.1.28 [DOI] [PubMed] [Google Scholar]
- 29.Chen S, Sorrell T, Nimmo G, Speed B, Currie B, Ellis D, Marriott D, Pfeiffer T, Parr D, Byth K, Australasian Cryptococcal Study Group 2000. Epidemiology and host- and variety-dependent characteristics of infection due to Cryptococcus neoformans in Australia and New Zealand. Clin. Infect. Dis. 31:499–508. 10.1086/313992 [DOI] [PubMed] [Google Scholar]
- 30.Fan M, Chen LC, Ragan MA, Gutell RR, Warner JR, Currie BP, Casadevall A. 1995. The 5S rRNA and the rRNA intergenic spacer of the two varieties of Cryptococcus neoformans. J. Med. Vet. Mycol. 33:215–221. 10.1080/02681219580000451 [DOI] [PubMed] [Google Scholar]
- 31.Diaz MR, Boekhout T, Theelen B, Fell JW. 2000. Molecular sequence analyses of the intergenic spacer (IGS) associated with rDNA of the two varieties of the pathogenic yeast, Cryptococcus neoformans. Syst. Appl. Microbiol. 23:535–545. 10.1016/S0723-2020(00)80028-4 [DOI] [PubMed] [Google Scholar]
- 32.Xu J, Vilgalys R, Mitchell TG. 2000. Multiple gene genealogies reveal recent dispersion and hybridization in the human pathogenic fungus Cryptococcus neoformans. Mol. Ecol. 9:1471–1481. 10.1046/j.1365-294x.2000.01021.x [DOI] [PubMed] [Google Scholar]
- 33.Boekhout T, Theelen B, Diaz M, Fell JW, Hop WC, Abeln EC, Dromer F, Meyer W. 2001. Hybrid genotypes in the pathogenic yeast Cryptococcus neoformans. Microbiology 147:891–907 [DOI] [PubMed] [Google Scholar]
- 34.Meyer W, Mitchell TG, Freedman EZ, Vilgalys R. 1993. Hybridization probes for conventional DNA fingerprinting used as single primers in the polymerase chain reaction to distinguish strains of Cryptococcus neoformans. J. Clin. Microbiol. 31:2274–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer W, Mitchell TG. 1995. Polymerase chain reaction fingerprinting in fungi using single primers specific to minisatellites and simple repetitive DNA sequences: strain variation in Cryptococcus neoformans. Electrophoresis 16:1648–1656. 10.1002/elps.11501601273 [DOI] [PubMed] [Google Scholar]
- 36.D'Souza CA, Kronstad JW, Taylor G, Warren R, Yuen M, Hu G, Jung WH, Sham A, Kidd SE, Tangen K, Lee N, Zeilmaker T, Sawkins J, McVicker G, Shah S, Gnerre S, Griggs A, Zeng Q, Bartlett K, Li W, Wang X, Heitman J, Stajich JE, Fraser JA, Meyer W, Carter D, Schein J, Krzywinski M, Kwon-Chung KJ, Varma A, Wang J, Brunham R, Fyfe M, Ouellette BF, Siddiqui A, Marra M, Jones S, Holt R, Birren BW, Galagan JE, Cuomo CA. 2011. Genome variation in Cryptococcus gattii, an emerging pathogen of immunocompetent hosts. mBio 2(1):e00342-10. 10.1128/mBio.00342-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephen C, Lester S, Black W, Fyfe M, Raverty S. 2002. Multispecies outbreak of cryptococcosis on southern Vancouver Island, British Columbia. Can. Vet. J. 43:792–794 [PMC free article] [PubMed] [Google Scholar]
- 38.Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, Fyfe M, Macdougall L, Boekhout T, Kwon-Chung KJ, Meyer W. 2004. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc. Natl. Acad. Sci. U. S. A. 101:17258–17263. 10.1073/pnas.0402981101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Byrnes EJ, Bartlett KH, Perfect JR, Heitman J. 2011. Cryptococcus gattii: an emerging fungal pathogen infecting humans and animals. Microbes Infect. 13:895–907. 10.1016/j.micinf.2011.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyer W, Castaneda A, Jackson S, Huynh M, Castaneda E, IberoAmerican Cryptococcal Study Group 2003. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg. Infect. Dis. 9:189–195. 10.3201/eid0902.020246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Latouche GN, Huynh M, Sorrell TC, Meyer W. 2003. PCR-restriction fragment length polymorphism analysis of the phospholipase B (PLB1) gene for subtyping of Cryptococcus neoformans isolates. Appl. Environ. Microbiol. 69:2080–2086. 10.1128/AEM.69.4.2080-2086.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanafy A, Kaocharoen S, Jover-Botella A, Katsu M, Iida S, Kogure T, Gonoi T, Mikami Y, Meyer W. 2008. Multilocus microsatellite typing for Cryptococcus neoformans var. grubii. Med. Mycol. 46:685–696. 10.1080/13693780802027062 [DOI] [PubMed] [Google Scholar]
- 43.Meyer W, Aanensen DM, Boekhout T, Cogliati M, Diaz MR, Esposto MC, Fisher M, Gilgado F, Hagen F, Kaocharoen S, Litvintseva AP, Mitchell TG, Simwami SP, Trilles L, Viviani MA, Kwon-Chung J. 2009. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med. Mycol. 47:561–570. 10.1080/13693780902953886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bovers M, Hagen F, Kuramae EE, Diaz MR, Spanjaard L, Dromer F, Hoogveld HL, Boekhout T. 2006. Unique hybrids between the fungal pathogens Cryptococcus neoformans and Cryptococcus gattii. FEMS Yeast Res. 6:599–607. 10.1111/j.1567-1364.2006.00082.x [DOI] [PubMed] [Google Scholar]
- 45.Bovers M, Hagen F, Kuramae EE, Hoogveld HL, Dromer F, St-Germain G, Boekhout T. 2008. AIDS patient death caused by novel Cryptococcus neoformans × C. gattii hybrid. Emerg. Infect. Dis. 14:1105–1108. 10.3201/eid1407.080122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ngamskulrungroj P, Gilgado F, Faganello J, Litvintseva AP, Leal AL, Tsui KM, Mitchell TG, Vainstein MH, Meyer W. 2009. Genetic diversity of the Cryptococcus species complex suggests that Cryptococcus gattii deserves to have varieties. PLoS One 4:e5862. 10.1371/journal.pone.0005862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharpton TJ, Neafsey DE, Galagan JE, Taylor JW. 2008. Mechanisms of intron gain and loss in Cryptococcus. Genome Biol. 9:R24. 10.1186/gb-2008-9-1-r24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meyer W, Gilgado F, Ngamskulrungroj P, Trilles L, Hagen F, Castaneda E, Boekhout T. 2011. Molecular typing of the Cryptococcus neoformans/Cryptococcus gattii species complex, p 327–357 In Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, Casadevall A. (ed), Cryptococcus: from human pathogen to model yeast. ASM Press, Washington, DC [Google Scholar]
- 49.Bovers M, Hagen F, Kuramae EE, Boekhout T. 2008. Six monophyletic lineages identified within Cryptococcus neoformans and Cryptococcus gattii by multi-locus sequence typing. Fungal Genet. Biol. 45:400–421. 10.1016/j.fgb.2007.12.004 [DOI] [PubMed] [Google Scholar]
- 50.Bovers M, Hagen F, Kuramae EE, Boekhout T. 2009. Promiscuous mitochondria in Cryptococcus gattii. FEMS Yeast Res. 9:489–503. 10.1111/j.1567-1364.2009.00494.x [DOI] [PubMed] [Google Scholar]
- 51. Reference deleted.
- 52.Gillece JD, Schupp JM, Balajee SA, Harris J, Pearson T, Yan Y, Keim P, DeBess E, Marsden-Haug N, Wohrle R, Engelthaler DM, Lockhart SR. 2011. Whole genome sequence analysis of Cryptococcus gattii from the Pacific Northwest reveals unexpected diversity. PLoS One 6:e28550. 10.1371/journal.pone.0028550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hagen F, Ceresini PC, Polacheck I, Ma H, van Nieuwerburgh F, Gabaldon T, Kagan S, Pursall ER, Hoogveld HL, van Iersel LJ, Klau GW, Kelk SM, Stougie L, Bartlett KH, Pryszcz KLP, Castaneda E, Lazera M, Meyer W, Deforce D, Meis JF, May RC, Klaassen CH, Boekhout T. 2013. Ancient dispersal of the human fungal pathogen Cryptococcus gattii from the Amazon rainforest. PLoS One 8:e71148. 10.1371/journal.pone.0071148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Engelthaler DM, Hicks ND, Gillece JD, Roe CC, Schupp JM, Driebe EM, Gilgado F, Carriconde F, Trilles L, Firacative C, Ngamskulrungroj P, Castañeda E, dos Santos Lazera M, Melhem MSC, Pérez-Bercoff A, Huttley G, Sorrell TC, Voelz K, May RC, Fisher MC, Thompson GR, Lockhart SR, Keim P, Meyer W. 2014. Cryptococcus gattii in North American Pacific Northwest: whole-population genome analysis provides insights into species evolution and dispersal. mBio 5(4):e01464-14. 10.1128/mBio.01464-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Billmyre RB, Croll D, Lin W, Mieczkowski P, Carter DA, Cuomo CA, Kronstad JW, Heitman J. 2014. Highly recombinant VGII Cryptococcus gattii population develops clonal outbreak clusters through both sexual macroevolution and asexual microevolution. mBio 5(4):e01494-14. 10.1128/mBio.01494-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trilles L, Wang B, Firacative C, dos Santos Lazéra M, Wanke B, Meyer W. 2014. Identification of the major molecular types of Cryptococcus neoformans and Cryptococcus gattii by hyperbranched rolling circle amplification. PLoS One 9:e0094648. 10.1371/journal.pone.0094648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Firacative C, Trilles L, Meyer W. 2012. MALDI-TOF MS enables the rapid identification of the major molecular types within the Cryptococcus neoformans/C. gattii species complex. PLoS One 7:e37566. 10.1371/journal.pone.0037566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Posteraro B, Vella A, Cogliati M, De Carolis E, Florio AR, Posteraro P, Sanguinetti M, Tortorano AM. 2012. Matrix-assisted laser desorption ionization-time of flight mass spectrometry-based method for discrimination between molecular types of Cryptococcus neoformans and Cryptococcus gattii. J. Clin. Microbiol. 50:2472–2476. 10.1128/JCM.00737-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boekhout T, van Belkum A. 1997. Variability of karyotypes and RAPD types in genetically related strains of Cryptococcus neoformans. Curr. Genet. 32:203–208. 10.1007/s002940050267 [DOI] [PubMed] [Google Scholar]
- 60.Ruma P, Chen SC, Sorrell TC, Brownlee AG. 1996. Characterization of Cryptococcus neoformans by random DNA amplification. Lett. Appl. Microbiol. 23:312–316. 10.1111/j.1472-765X.1996.tb00197.x [DOI] [PubMed] [Google Scholar]
- 61.Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 18:6531–6535. 10.1093/nar/18.22.6531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mitchell TG, Sandin RL, Bowman BH, Meyer W, Merz WG. 1994. Molecular mycology: probes and applications of PCR technology. J. Med. Vet. Mycol. 32(Suppl 1):S351–S366 [DOI] [PubMed] [Google Scholar]
- 63.Ellis D, Marriott D, Hajjeh RA, Warnock D, Meyer W, Barton R. 2000. Epidemiology: surveillance of fungal infections. Med. Mycol. 38:173–182. 10.1080/mmy.38.s1.173.182 [DOI] [PubMed] [Google Scholar]
- 64.Bovers M, Hagen F, Boekhout T. 2008. Diversity of the Cryptococcus neoformans-Cryptococcus gattii species complex. Rev. Iberoam. Micol. 25:S4–S12. 10.1016/S1130-1406(08)70019-6 [DOI] [PubMed] [Google Scholar]
- 65.Tautz D, Renz M. 1984. Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res. 12:4127–4138. 10.1093/nar/12.10.4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karaoglu H, Lee CMY, Meyer W. 2005. Survey of simple sequence repeats in completed fungal genomes. Mol. Biol. Evol. 22:639–649. 10.1093/molbev/msi057 [DOI] [PubMed] [Google Scholar]
- 67.Karaoglu H, Lee CMY, Carter D, Meyer W. 2008. Development of polymorphic microsatellite markers for Cryptococcus neoformans. Mol. Ecol. Resour. 8:1136–1138. 10.1111/j.1755-0998.2008.02196.x [DOI] [PubMed] [Google Scholar]
- 68.Byrnes EJ, Bildfell RJ, Frank SA, Mitchell TG, Marr KA, Heitman J. 2009. Molecular evidence that the range of the Vancouver Island outbreak of Cryptococcus gattii infection has expanded into the Pacific Northwest in the United States. J. Infect. Dis. 199:1081–1086. 10.1086/597306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Byrnes EJ, Li W, Lewit Y, Ma H, Voelz K, Ren P, Carter DA, Chaturvedi V, Bildfell RJ, May RC, Heitman J. 2010. Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog. 6:e1000850. 10.1371/journal.ppat.1000850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aminnejad M, Diaz M, Arabatzis M, Castaneda E, Lazera M, Velegraki A, Marriott D, Sorrell TC, Meyer W. 2012. Identification of novel hybrids between Cryptococcus neoformans var. grubii VNI and Cryptococcus gattii VGII. Mycopathologia 173:337–346. 10.1007/s11046-011-9491-x [DOI] [PubMed] [Google Scholar]
- 71.Kwon-Chung KJ, Bennett JE. 1984. Epidemiologic differences between the two varieties of Cryptococcus neoformans. Am. J. Epidemiol. 120:123–130 [DOI] [PubMed] [Google Scholar]
- 72.Casali AK, Goulart L, Rosa e Silva LK, Ribeiro AM, Amaral AA, Alves SH, Schrank A, Meyer W, Vainstein MH. 2003. Molecular typing of clinical and environmental Cryptococcus neoformans isolates in the Brazilian state Rio Grande do Sul. FEMS Yeast Res. 3:405–415. 10.1016/S1567-1356(03)00038-2 [DOI] [PubMed] [Google Scholar]
- 73.Trilles L, Lazera M, Wanke B, Theelen B, Boekhout T. 2003. Genetic characterization of environmental isolates of the Cryptococcus neoformans species complex from Brazil. Med. Mycol. 41:383–390. 10.1080/1369378031000137206 [DOI] [PubMed] [Google Scholar]
- 74.Escandon P, Sanchez A, Martinez M, Meyer W, Castaneda E. 2006. Molecular epidemiology of clinical and environmental isolates of the Cryptococcus neoformans species complex reveals a high genetic diversity and the presence of the molecular type VGII mating type a in Colombia. FEMS Yeast Res. 6:625–635. 10.1111/j.1567-1364.2006.00055.x [DOI] [PubMed] [Google Scholar]
- 75.Chen J, Varma A, Diaz MR, Litvintseva AP, Wollenberg KK, Kwon-Chung KJ. 2008. Cryptococcus neoformans strains and infection in apparently immunocompetent patients, China. Emerg. Infect. Dis. 14:755–762. 10.3201/eid1405.071312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Feng X, Yao Z, Ren D, Liao W. 2008. Simultaneous identification of molecular and mating types within the Cryptococcus species complex by PCR-RFLP analysis. J. Med. Microbiol. 57:1481–1490. 10.1099/jmm.0.2008/003665-0 [DOI] [PubMed] [Google Scholar]
- 77.Feng X, Yao Z, Ren D, Liao W, Wu J. 2008. Genotype and mating type analysis of Cryptococcus neoformans and Cryptococcus gattii isolates from China that mainly originated from non-HIV-infected patients. FEMS Yeast Res. 8:930–938. 10.1111/j.1567-1364.2008.00422.x [DOI] [PubMed] [Google Scholar]
- 78.Trilles L, dos Santos Lazera M, Wanke B, Oliveira RV, Barbosa GG, Nishikawa MM, Morales BP, Meyer W. 2008. Regional pattern of the molecular types of Cryptococcus neoformans and Cryptococcus gattii in Brazil. Mem. Inst. Oswaldo Cruz 103:455–462. 10.1590/S0074-02762008000500008 [DOI] [PubMed] [Google Scholar]
- 79.Olivares LR, Martinez KM, Cruz RM, Rivera MA, Meyer W, Espinosa RA, Martinez RL, Santos GM. 2009. Genotyping of Mexican Cryptococcus neoformans and C. gattii isolates by PCR-fingerprinting. Med. Mycol. 47:713–721. 10.3109/13693780802559031 [DOI] [PubMed] [Google Scholar]
- 80.Meyer W, Trilles L. 2010. Genotyping of the Cryptococcus neoformans/C. gattii species complex. Aust. Biochem. 41:11–15 [Google Scholar]
- 81.Cogliati M, Chandrashekar N, Esposto MC, Chandramuki A, Petrini B, Viviani MA. 2012. Cryptococcus gattii serotype-C strains isolated in Bangalore, Karnataka, India. Mycoses 55:262–268. 10.1111/j.1439-0507.2011.02082.x [DOI] [PubMed] [Google Scholar]
- 82.Byrnes EJ, Bildfell RJ, Dearing PL, Valentine BA, Heitman J. 2009. Cryptococcus gattii with bimorphic colony types in a dog in western Oregon: additional evidence for expansion of the Vancouver Island outbreak. J. Vet. Diagn. Invest. 21:133–136. 10.1177/104063870902100122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lockhart SR, Iqbal N, Harris JR, Grossman NT, DeBess E, Wohrle R, Marsden-Haug N, Vugia DJ. 2013. Cryptococcus gattii in the United States: genotypic diversity of human and veterinary isolates. PLoS One 8:e74737. 10.1371/journal.pone.0074737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Byrnes EJ, Li W, Ren P, Lewit Y, Voelz K, Fraser JA, Dietrich FS, May RC, Chaturvedi S, Chaturvedi V, Heitman J. 2011. A diverse population of Cryptococcus gattii molecular type VGIII in southern Californian HIV/AIDS patients. PLoS Pathog. 7:e1002205. 10.1371/journal.ppat.1002205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Byrnes EJ, Marr KA. 2011. The outbreak of Cryptococcus gattii in western North America: epidemiology and clinical issues. Curr. Infect. Dis. Rep. 13:256–261. 10.1007/s11908-011-0181-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.MacDougall L, Kidd SE, Galanis E, Mak S, Leslie MJ, Cieslak PR, Kronstad JW, Morshed MG, Bartlett KH. 2007. Spread of Cryptococcus gattii in British Columbia, Canada, and detection in the Pacific Northwest, USA. Emerg. Infect. Dis. 13:42–50. 10.3201/eid1301.060827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Meyer W. 2007. The emergence of Cryptococcus gattii VGII as a super killer? Microbiol. Aust. 5:70–71 [Google Scholar]
- 88.Carriconde F, Gilgado F, Arthur I, Ellis D, Malik R, van de Wiele N, Robert V, Currie BJ, Meyer W. 2011. Clonality and alpha-a recombination in the Australian Cryptococcus gattii VGII population—an emerging outbreak in Australia. PLoS One 6:e16936. 10.1371/journal.pone.0016936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kaocharoen S, Ngamskulrungroj P, Firacative C, Trilles L, Piyabongkarn D, Banlunara W, Poonwan N, Chaiprasert A, Meyer W, Chindamporn A. 2013. Molecular epidemiology reveals genetic diversity amongst isolates of the Cryptococcus neoformans/C. gattii species complex in Thailand. PLoS Negl. Trop. Dis. 7:e2297. 10.1371/journal.pntd.0002297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lazera MS, Salmito Cavalcanti MA, Londero AT, Trilles L, Nishikawa MM, Wanke B. 2000. Possible primary ecological niche of Cryptococcus neoformans. Med. Mycol. 38:379–383. 10.1080/mmy.38.5.379.383 [DOI] [PubMed] [Google Scholar]
- 91.Hagen F, Colom MF, Swinne D, Tintelnot K, Iatta R, Montagna MT, Torres-Rodriguez JM, Cogliati M, Velegraki A, Burggraaf A, Kamermans A, Sweere JM, Meis JF, Klaassen CH, Boekhout T. 2012. Autochthonous and dormant Cryptococcus gattii infections in Europe. Emerg. Infect. Dis. 18:1618–1624. 10.3201/eid1810.120068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Baro T, Torres-Rodriguez JM, De Mendoza MH, Morera Y, Alia C. 1998. First identification of autochthonous Cryptococcus neoformans var. gattii isloated from goats with predominantly severe pulmonary disease in Spain. J. Clin. Microbiol. 36:458–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Levy R, Pitout J, Long P, Gill MJ. 2007. Late presentation of Cryptococcus gattii meningitis in a traveller to Vancouver Island: a case report. Can. J. Infect. Dis. Med. Microbiol. 18:197–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lindberg J, Hagen F, Laursen A, Stenderup J, Boekhout T. 2007. Cryptococcus gattii risk for tourists visiting Vancouver Island, Canada. Emerg. Infect. Dis. 13:178–179. 10.3201/eid1301.060945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Georgi A, Schneemann M, Tintelnot K, Calligaris-Maibach RC, Meyer S, Weber R, Bosshard PP. 2009. Cryptococcus gattii meningoencephalitis in an immunocompetent person 13 months after exposure. Infection 37:370–373. 10.1007/s15010-008-8211-z [DOI] [PubMed] [Google Scholar]
- 96.Steele KT, Thakur R, Nthobatsang R, Steenhoff AP, Bisson GP. 2010. In-hospital mortality of HIV-infected cryptococcal meningitis patients with C. gattii and C. neoformans infection in Gaborone, Botswana. Med. Mycol. 48:1112–1115. 10.3109/13693781003774689 [DOI] [PubMed] [Google Scholar]
- 97.Litvintseva AP, Thakur R, Reller LB, Mitchell TG. 2005. Prevalence of clinical isolates of Cryptococcus gattii serotype C among patients with AIDS in sub-Saharan Africa. J. Infect. Dis. 192:888–892. 10.1086/432486 [DOI] [PubMed] [Google Scholar]
- 98.Morgan J, McCarthy KM, Gould S, Fan K, Arthington-Skaggs B, Iqbal N, Stamey K, Hajjeh RA, Brandt ME, Gauteng Cryptococcal Surveillance Initiative Group 2006. Cryptococcus gattii infection: characteristics and epidemiology of cases identified in a South African province with high HIV seroprevalence, 2002-2004. Clin. Infect. Dis. 43:1077–1080. 10.1086/507897 [DOI] [PubMed] [Google Scholar]
- 99.Sorrell TC, Chen SC, Ruma P, Meyer W, Pfeiffer TJ, Ellis DH, Brownlee AG. 1996. Concordance of clinical and environmental isolates of Cryptococcus neoformans var. gattii by random amplification of polymorphic DNA analysis and PCR fingerprinting. J. Clin. Microbiol. 34:1253–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sorrell TC, Brownlee AG, Ruma P, Malik R, Pfeiffer TJ, Ellis DH. 1996. Natural environmental sources of Cryptococcus neoformans var. gattii. J. Clin. Microbiol. 34:1261–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pfeiffer TJ, Ellis DH. 1992. Environmental isolation of Cryptococcus neoformans var. gattii from Eucalyptus tereticornis. J. Med. Vet. Mycol. 30:407–408. 10.1080/02681219280000541 [DOI] [PubMed] [Google Scholar]
- 102.Ellis DH, Pfeiffer TJ. 1990. Ecology, life cycle, and infectious propagule of Cryptococcus neoformans. Lancet 336:923–925. 10.1016/0140-6736(90)92283-N [DOI] [PubMed] [Google Scholar]
- 103.Pfeiffer T, Ellis D. 1991. Environmental isolation of Cryptococcus neoformans gattii from California. J. Infect. Dis. 163:929–930. 10.1093/infdis/163.4.929 [DOI] [PubMed] [Google Scholar]
- 104.Kidd SE, Bach PJ, Hingston AO, Mak S, Chow Y, MacDougall L, Kronstad JW, Bartlett KH. 2007. Cryptococcus gattii dispersal mechanisms, British Columbia, Canada. Emerg. Infect. Dis. 13:51–57. 10.3201/eid1301.060823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Madrenys N, De Vroey C, Raes-Wuytack C, Torres-Rodriguez JM. 1993. Identification of the perfect state of Cryptococcus neoformans from 195 clinical isolates including 84 from AIDS patients. Mycopathologia 123:65–68. 10.1007/BF01365081 [DOI] [PubMed] [Google Scholar]
- 106.Colom MF, Frases S, Ferrer C, Jover A, Andreu M, Reus S, Sanchez M, Torres-Rodriguez JM. 2005. First case of human cryptococcosis due to Cryptococcus neoformans var. gattii in Spain. J. Clin. Microbiol. 43:3548–3550. 10.1128/JCM.43.7.3548-3550.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fortes ST, Lazera MS, Nishikawa MM, Macedo RC, Wanke B. 2001. First isolation of Cryptococcus neoformans var. gattii from a native jungle tree in the Brazilian Amazon rainforest. Mycoses 44:137–140. 10.1046/j.1439-0507.2001.00651.x [DOI] [PubMed] [Google Scholar]
- 108.Grover N, Nawange SR, Naidu J, Singh SM, Sharma A. 2007. Ecological niche of Cryptococcus neoformans var. grubii and Cryptococcus gattii in decaying wood of trunk hollows of living trees in Jabalpur City of central India. Mycopathologia 164:159–170. 10.1007/s11046-007-9039-2 [DOI] [PubMed] [Google Scholar]
- 109.Hagen F, Boekhout T. 2010. The search for the natural habitat of Cryptococcus gattii. Mycopathologia 170:209–211. 10.1007/s11046-010-9313-6 [DOI] [PubMed] [Google Scholar]
- 110.Mitchell TG, Castaneda E, Nielson K, Wanke B, dos Santos Lazera M. 2011. Environmental niches for Cryptococcus neoformans and Cryptococcus gattii, p 237–260 In Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, Casadevall A. (ed), Cryptococcus: from pathogen to model yeast. ASM Press, Washington, DC [Google Scholar]
- 111.Mahmoud YA. 2000. First environmental isolation of Cryptococcus neoformans var. neoformans and var. gatti from the Gharbia Governorate, Egypt. Mycopathologia 148:83–86. 10.1023/A:1007166818993 [DOI] [PubMed] [Google Scholar]
- 112.Mseddi F, Sellami A, Jarboui MA, Sellami H, Makni F, Ayadi A. 2011. First environmental isolations of Cryptococcus neoformans and Cryptococcus gattii in Tunisia and review of published studies on environmental isolations in Africa. Mycopathologia 171:355–360. 10.1007/s11046-010-9381-7 [DOI] [PubMed] [Google Scholar]
- 113.Chen SC, Currie BJ, Campbell HM, Fisher DA, Pfeiffer TJ, Ellis DH, Sorrell TC. 1997. Cryptococcus neoformans var. gattii infection in northern Australia: existence of an environmental source other than known host eucalypts. Trans. R. Soc. Trop. Med. Hyg. 91:547–550. 10.1016/S0035-9203(97)90021-3 [DOI] [PubMed] [Google Scholar]
- 114.Halliday CL, Carter DA. 2003. Clonal reproduction and limited dispersal in an environmental population of Cryptococcus neoformans var. gattii isolates from Australia. J. Clin. Microbiol. 41:703–711. 10.1128/JCM.41.2.703-711.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kidd SE, Sorrell TC, Meyer W. 2003. Isolation of two molecular types of Cryptococcus neoformans var. gattii from insect frass. Med. Mycol. 41:171–176. 10.1080/mmy.41.2.171.176 [DOI] [PubMed] [Google Scholar]
- 116.Campbell LT, Currie BJ, Krockenberger M, Malik R, Meyer W, Heitman J, Carter D. 2005. Clonality and recombination in genetically differentiated subgroups of Cryptococcus gattii. Eukaryot. Cell 4:1403–1409. 10.1128/EC.4.8.1403-1409.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Campbell LT, Fraser JA, Nichols CB, Dietrich FS, Carter D, Heitman J. 2005. Clinical and environmental isolates of Cryptococcus gattii from Australia that retain sexual fecundity. Eukaryot. Cell 4:1410–1419. 10.1128/EC.4.8.1410-1419.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Saul N, Krockenberger M, Carter D. 2008. Evidence of recombination in mixed-mating-type and alpha-only populations of Cryptococcus gattii sourced from single eucalyptus tree hollows. Eukaryot. Cell 7:727–734. 10.1128/EC.00020-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Colom MF, Hagen F, Gonzalez A, Mellado A, Morera N, Linares C, Garcia DF, Penataro JS, Boekhout T, Sanchez M. 2012. Ceratonia siliqua (carob) trees as natural habitat and source of infection by Cryptococcus gattii in the Mediterranean environment. Med. Mycol. 50:67–73. 10.3109/13693786.2011.574239 [DOI] [PubMed] [Google Scholar]
- 120.Campisi E, Mancianti F, Pini G, Faggi E, Gargani G. 2003. Investigation in central Italy of the possible association between Cryptococcus neoformans var. gattii and Eucalyptus camaldulensis. Eur. J. Epidemiol. 18:357–362. 10.1023/A:1023652920595 [DOI] [PubMed] [Google Scholar]
- 121.Kidd SE, Chow Y, Mak S, Bach PJ, Chen H, Hingston AO, Kronstad JW, Bartlett KH. 2007. Characterization of environmental sources of the human and animal pathogen Cryptococcus gattii in British Columbia, Canada, and the Pacific Northwest of the United States. Appl. Environ. Microbiol. 73:1433–1443. 10.1128/AEM.01330-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Springer DJ, Chaturvedi V. 2010. Projecting global occurrence of Cryptococcus gattii. Emerg. Infect. Dis. 16:14–20. 10.3201/eid1601.090369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Botes A, Boekhout T, Hagen F, Vismer H, Swart J, Botha A. 2009. Growth and mating of Cryptococcus neoformans var. grubii on woody debris. Microb. Ecol. 57:757–765. 10.1007/s00248-008-9452-1 [DOI] [PubMed] [Google Scholar]
- 124.Xue C, Tada Y, Dong X, Heitman J. 2007. The human fungal pathogen Cryptococcus can complete its sexual cycle during a pathogenic association with plants. Cell Host Microbe 1:263–273. 10.1016/j.chom.2007.05.005 [DOI] [PubMed] [Google Scholar]
- 125.Springer DJ, Ren P, Raina R, Dong Y, Behr MJ, McEwen BF, Bowser SS, Samsonoff WA, Chaturvedi S, Chaturvedi V. 2010. Extracellular fibrils of pathogenic yeast Cryptococcus gattii are important for ecological niche, murine virulence and human neutrophil interactions. PLoS One 5:e10978. 10.1371/journal.pone.0010978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Escandon P, Quintero E, Granados D, Huerfano S, Ruiz A, Castaneda E. 2005. Isolation of Cryptococcus gattii serotype B from detritus of eucalyptus trees in Colombia. Biomedica (Bogota) 25:390–397 [PubMed] [Google Scholar]
- 127.Callejas A, Ordonez N, Rodriguez MC, Castaneda E. 1998. First isolation of Cryptococcus neoformans var. gattii, serotype C, from the environment in Colombia. Med. Mycol. 36:341–344. 10.1046/j.1365-280X.1998.00159.x, [DOI] [PubMed] [Google Scholar]
- 128.Springer DJ, Billmyre RB, Filler EE, Voelz K, Pursall R, Mieczkowski PA, Larsen RA, Dietrich FS, May RC, Filler SG, Heitman J. 2014. Cryptococcus gattii VGIII isolates causing infections in HIV/AIDS patients in Southern California: identification of the local environmental source as arboreal. PLoS Pathog. 10(8):e1004285. 10.1371/journal.ppat.1004285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Barreto de Oliveira MT, Boekhout T, Theelen B, Hagen F, Baroni FA, Lazera MS, Lengeler KB, Heitman J, Rivera IN, Paula CR. 2004. Cryptococcus neoformans shows a remarkable genotypic diversity in Brazil. J. Clin. Microbiol. 42:1356–1359. 10.1128/JCM.42.3.1356-1359.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chen SC, Slavin MA, Heath CH, Playford EG, Byth K, Marriott D, Kidd SE, Bak N, Currie B, Hajkowicz K, Korman TM, McBride WJ, Meyer W, Murray R, Sorrell TC, Australia New Zealand Mycoses Interest Group-Cryptococcus Study 2012. Clinical manifestations of Cryptococcus gattii infection: determinants of neurological sequelae and death. Clin. Infect. Dis. 55:789–798. 10.1093/cid/cis529 [DOI] [PubMed] [Google Scholar]
- 131.Galanis E, Hoang L, Kibsey P, Morshed M, Phillips P. 2009. Clinical presentation, diagnosis and management of Cryptococcus gattii cases: lessons learned from British Columbia. Can. J. Infect. Dis. Med. Microbiol. 20:23–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Harris JR, Lockhart SR, Debess E, Marsden-Haug N, Goldoft M, Wohrle R, Lee S, Smelser C, Park B, Chiller T. 2011. Cryptococcus gattii in the United States: clinical aspects of infection with an emerging pathogen. Clin. Infect. Dis. 53:1188–1195. 10.1093/cid/cir723 [DOI] [PubMed] [Google Scholar]
- 133.Chaturvedi S, Dyavaiah M, Larsen RA, Chaturvedi V. 2005. Cryptococcus gattii in AIDS patients, southern California. Emerg. Infect. Dis. 11:1686–1692. 10.3201/eid1111.040875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bose I, Reese AJ, Ory JJ, Janbon G, Doering TL. 2003. A yeast under cover: the capsule of Cryptococcus neoformans. Eukaryot. Cell 2:655–663. 10.1128/EC.2.4.655-663.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen SC, Wright LC, Golding JC, Sorrell TC. 2000. Purification and characterization of secretory phospholipase B, lysophospholipase and lysophospholipase/transacylase from a virulent strain of the pathogenic fungus Cryptococcus neoformans. Biochem. J. 347:431–439. 10.1042/0264-6021:3470431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Latouche GN, Sorrell TC, Meyer W. 2002. Isolation and characterisation of the phospholipase B gene of Cryptococcus neoformans var. gattii. FEMS Yeast Res. 2:551–561. 10.1111/j.1567-1364.2002.tb00122.x [DOI] [PubMed] [Google Scholar]
- 137.Ngamskulrungroj P, Price J, Sorrell T, Perfect JR, Meyer W. 2011. Cryptococcus gattii virulence composite: candidate genes revealed by microarray analysis of high and less virulent Vancouver Island outbreak strains. PLoS One 6:e16076. 10.1371/journal.pone.0016076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Moyrand F, Chang YC, Himmelreich U, Kwon-Chung KJ, Janbon G. 2004. Cas3p belongs to a seven-member family of capsule structure designer proteins. Eukaryot. Cell 3:1513–1524. 10.1128/EC.3.6.1513-1524.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bahn YS, Jung KW. 2013. Stress signaling pathways for the pathogenicity of Cryptococcus. Eukaryot. Cell 12:1564–1577. 10.1128/EC.00218-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kronstad JW, Attarian R, Cadieux B, Choi J, D'Souza CA, Griffiths EJ, Geddes JM, Hu G, Jung WH, Kretschmer M, Saikia S, Wang J. 2011. Expanding fungal pathogenesis: Cryptococcus breaks out of the opportunistic box. Nat. Rev. Microbiol. 9:193–203. 10.1038/nrmicro2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Schneider RO, Fogaca NS, Kmetzsch L, Schrank A, Vainstein MH, Staats CC. 2012. Zap1 regulates zinc homeostasis and modulates virulence in Cryptococcus gattii. PLoS One 7:e43773. 10.1371/journal.pone.0043773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ma H, May RC. 2009. Virulence in Cryptococcus species. Adv. Appl. Microbiol. 67:131–190. 10.1016/S0065-2164(08)01005-8 [DOI] [PubMed] [Google Scholar]
- 143.Santangelo R, Zoellner H, Sorrell T, Wilson C, Donald C, Djordjevic J, Shounan Y, Wright L. 2004. Role of extracellular phospholipases and mononuclear phagocytes in dissemination of cryptococcosis in a murine model. Infect. Immun. 72:2229–2239. 10.1128/IAI.72.4.2229-2239.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Noverr MC, Williamson PR, Fajardo RS, Huffnagle GB. 2004. LAC1 is required for extrapulmonary dissemination of Cryptococcus neoformans but not pulmonary persistence. Infect. Immun. 72:1693–1699. 10.1128/IAI.72.3.1693-1699.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Olszewski MA, Noverr MC, Chen GH, Toews GB, Cox GM, Perfect JR, Huffnagle GB. 2004. Urease expression by Cryptococcus neoformans promotes microvascular sequestration, thereby enhancing central nervous system invasion. Am. J. Pathol. 164:1761–1771. 10.1016/S0002-9440(10)63734-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shi M, Li SS, Zheng C, Jones GJ, Kim KS, Zhou H, Kubes P, Mody CH. 2010. Real-time imaging of trapping and urease-dependent transmigration of Cryptococcus neoformans in mouse brain. J. Clin. Invest. 120:1683–1693. 10.1172/JCI41963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chayakulkeeree M, Johnston SA, Oei JB, Lev S, Williamson PR, Wilson CF, Zuo X, Leal AL, Vainstein MH, Meyer W, Sorrell TC, May RC, Djordjevic JT. 2011. SEC14 is a specific requirement for secretion of phospholipase B1 and pathogenicity of Cryptococcus neoformans. Mol. Microbiol. 80:1088–1101. 10.1111/j.1365-2958.2011.07632.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Osterholzer JJ, Surana R, Milam JE, Montano GT, Chen GH, Sonstein J, Curtis JL, Huffnagle GB, Toews GB, Olszewski MA. 2009. Cryptococcal urease promotes the accumulation of immature dendritic cells and a non-protective T2 immune response within the lung. Am. J. Pathol. 174:932–943. 10.2353/ajpath.2009.080673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ma H, Hagen F, Stekel DJ, Johnston SA, Sionov E, Falk R, Polacheck I, Boekhout T, May RC. 2009. The fatal fungal outbreak on Vancouver Island is characterized by enhanced intracellular parasitism driven by mitochondrial regulation. Proc. Natl. Acad. Sci. U. S. A. 106:12980–12985. 10.1073/pnas.0902963106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ngamskulrungroj P, Serena C, Gilgado F, Malik R, Meyer W. 2011. Global VGIIa isolates are of comparable virulence to the major fatal Cryptococcus gattii Vancouver Island outbreak genotype. Clin. Microbiol. Infect. 17:251–258. 10.1111/j.1469-0691.2010.03222.x [DOI] [PubMed] [Google Scholar]
- 151.Tobin DM, May RC, Wheeler RT. 2012. Zebrafish: a see-through host and a fluorescent toolbox to probe host-pathogen interaction. PLoS Pathog. 8:e1002349. 10.1371/journal.ppat.1002349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cheng PY, Sham A, Kronstad JW. 2009. Cryptococcus gattii isolates from the British Columbia cryptococcosis outbreak induce less protective inflammation in a murine model of infection than Cryptococcus neoformans. Infect. Immun. 77:4284–4294. 10.1128/IAI.00628-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Krockenberger MB, Malik R, Ngamskulrungroj P, Trilles L, Escandon P, Dowd S, Allen C, Himmelreich U, Canfield PJ, Sorrell TC, Meyer W. 2010. Pathogenesis of pulmonary Cryptococcus gattii infection: a rat model. Mycopathologia 170:315–330. 10.1007/s11046-010-9328-z [DOI] [PubMed] [Google Scholar]
- 154.Chen SC, Muller M, Zhou JZ, Wright LC, Sorrell TC. 1997. Phospholipase activity in Cryptococcus neoformans: a new virulence factor? J. Infect. Dis. 175:414–420. 10.1093/infdis/175.2.414 [DOI] [PubMed] [Google Scholar]
- 155.Voelz K, Ma H, Phadke S, Byrnes EJ, Zhu P, Mueller O, Farrer RA, Henk DA, Lewit Y, Hsueh YP, Fisher MC, Idnurm A, Heitman J, May RC. 2013. Transmission of hypervirulence traits via sexual reproduction within and between lineages of the human fungal pathogen Cryptococcus gattii. PLoS Genet. 9:e1003771. 10.1371/journal.pgen.1003771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chen YL, Lehman VN, Lewit Y, Averette AF, Heitman J. 2013. Calcineurin governs thermotolerance and virulence of Cryptococcus gattii. G3 3:527–539. 10.1534/g3.112.004242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cutler JE. 1991. Putative virulence factors of Candida albicans. Annu. Rev. Microbiol. 45:187–218. 10.1146/annurev.mi.45.100191.001155 [DOI] [PubMed] [Google Scholar]
- 158.Garcia-Solache MA, Izquierdo-Garcia D, Smith C, Bergman A, Casadevall A. 2013. Fungal virulence in a lepidopteran model is an emergent property with deterministic features. mBio 4(3):e00100-13. 10.1128/mBio.00100-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Perfect JR. 2012. The triple threat of cryptococcosis: it's the body site, the strain, and/or the host. mBio 3(4):e00165-12. 10.1128/mBio.00165-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ngamskulrungroj P, Chang Y, Sionov E, Kwon-Chung KJ. 2012. The primary target organ of Cryptococcus gattii is different from that of Cryptococcus neoformans in a murine model. mBio 3(3):e00103-12. 10.1128/mBio.00103-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Okubo Y, Tochigi N, Wakayama M, Shinozaki M, Nakayama H, Ishiwatari T, Shimodaira K, Nemoto T, Ohno H, Kaneko Y, Makimura K, Uchida K, Miyazaki Y, Yamaguchi H, Shibuya K. 2013. How histopathology can contribute to an understanding of defense mechanisms against cryptococci. Mediators Inflamm. 2013:465319. 10.1155/2013/465319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Okubo Y, Wakayama M, Ohno H, Yamamoto S, Tochigi N, Tanabe K, Kaneko Y, Yamagoe S, Umeyama T, Shinozaki M, Nemoto T, Nakayama H, Sasai D, Ishiwatari T, Shimodaira K, Yamamoto Y, Kamei K, Miyazaki Y, Shibuya K. 2013. Histopathological study of murine pulmonary cryptococcosis induced by Cryptococcus gattii and Cryptococcus neoformans. Jpn. J. Infect. Dis. 66:216–221. 10.7883/yoken.66.216 [DOI] [PubMed] [Google Scholar]
- 163.Wright L, Bubb W, Davidson J, Santangelo R, Krockenberger M, Himmelreich U, Sorrell TC. 2002. Metabolites released by Cryptococcus neoformans var. neoformans and var. gattii differentially affect human neutrophil function. Microbes Infect. 4:1427–1438. 10.1016/S1286-4579(02)00024-2 [DOI] [PubMed] [Google Scholar]
- 164.Huston SM, Mody CH. 2011. Cryptococcus interactions with innate cytotoxic lymphocytes, p 417–427 In Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, Casadevall A. (ed), Cryptococcus: from human pathogen to model yeast. ASM Press, Washington, DC [Google Scholar]
- 165.Islam A, Li SS, Oykhman P, Timm-McCann M, Huston SM, Stack D, Xiang RF, Kelly MM, Mody CH. 2013. An acidic microenvironment increases NK cell killing of Cryptococcus neoformans and Cryptococcus gattii by enhancing perforin degranulation. PLoS Pathog. 9:e1003439. 10.1371/journal.ppat.1003439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Charlier C, Nielsen K, Daou S, Brigitte M, Chretien F, Dromer F. 2009. Evidence of a role for monocytes in dissemination and brain invasion by Cryptococcus neoformans. Infect. Immun. 77:120–127. 10.1128/IAI.01065-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Huston SM, Li SS, Stack D, Timm-McCann M, Jones GJ, Islam A, Berenger BM, Xiang RF, Colarusso P, Mody CH. 2013. Cryptococcus gattii is killed by dendritic cells, but evades adaptive immunity by failing to induce dendritic cell maturation. J. Immunol. 191:249–261. 10.4049/jimmunol.1202707 [DOI] [PubMed] [Google Scholar]
- 168.Lester SJ, Malik R, Bartlett KH, Duncan CG. 2011. Cryptococcosis: update and emergence of Cryptococcus gattii. Vet. Clin. Pathol. 40:4–17. 10.1111/j.1939-165X.2010.00281.x [DOI] [PubMed] [Google Scholar]
- 169.Trivedi SR, Malik R, Meyer W, Sykes JE. 2011. Feline cryptococcosis: impact of current research on clinical management. J. Feline Med. Surg. 13:163–172. 10.1016/j.jfms.2011.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sykes JE, Malik R. 2011. Cryptococcosis, p 621–634 In Greene CE. (ed), Infectious diseases of the dog and cat. Elsevier Saunders, Philadelphia, PA [Google Scholar]
- 171.Duncan C, Stephen C, Campbell J. 2006. Clinical characteristics and predictors of mortality for Cryptococcus gattii infection in dogs and cats of southwestern British Columbia. Can. Vet. J. 47:993–998 [PMC free article] [PubMed] [Google Scholar]
- 172.Lester SJ, Kowalewich NJ, Bartlett KH, Krockenberger MB, Fairfax TM, Malik R. 2004. Clinicopathologic features of an unusual outbreak of cryptococcosis in dogs, cats, ferrets, and a bird: 38 cases (January to July 2003). J. Am. Vet. Med. Assoc. 225:1716–1722. 10.2460/javma.2004.225.1716 [DOI] [PubMed] [Google Scholar]
- 173.O'Brien CR, Krockenberger MB, Wigney DI, Martin P, Malik R. 2004. Retrospective study of feline and canine cryptococcosis in Australia from 1981 to 2001: 195 cases. Med. Mycol. 42:449–460. 10.1080/13693780310001624547 [DOI] [PubMed] [Google Scholar]
- 174.Connolly JH, Krockenberger MB, Malik R, Canfield PJ, Wigney DI, Muir DB. 1999. Asymptomatic carriage of Cryptococcus neoformans in the nasal cavity of the koala (Phascolarctos cinereus). Med. Mycol. 37:331–338. 10.1046/j.1365-280X.1999.00236.x [DOI] [PubMed] [Google Scholar]
- 175.McGill S, Malik R, Saul N, Beetson S, Secombe C, Robertson I, Irwin P. 2009. Cryptococcosis in domestic animals in Western Australia: a retrospective study from 1995-2006. Med. Mycol. 47:625–639. 10.1080/13693780802512519 [DOI] [PubMed] [Google Scholar]
- 176.Duncan C, Stephen C, Lester S, Bartlett KH. 2005. Follow-up study of dogs and cats with asymptomatic Cryptococcus gattii infection or nasal colonization. Med. Mycol. 43:663–666. 10.1080/13693780500220076 [DOI] [PubMed] [Google Scholar]
- 177.Kluger EK, Karaoglu HK, Krockenberger MB, Della Torre PK, Meyer W, Malik R. 2006. Recrudescent cryptococcosis, caused by Cryptococcus gattii (molecular type VGII), over a 13-year period in a Birman cat. Med. Mycol. 44:561–566. 10.1080/13693780600582847 [DOI] [PubMed] [Google Scholar]
- 178.Malik R, Wigney DI, Muir DB, Gregory DJ, Love DN. 1992. Cryptococcosis in cats: clinical and mycological assessment of 29 cases and evaluation of treatment using orally administered fluconazole. J. Med. Vet. Mycol. 30:133–144. 10.1080/02681219280000181 [DOI] [PubMed] [Google Scholar]
- 179.Malik R, Martin P, Wigney DI, Church DB, Bradley W, Bellenger CR, Lamb WA, Barrs VR, Foster S, Hemsley S, Canfield PJ, Love DN. 1997. Nasopharyngeal cryptococcosis. Aust. Vet. J. 75:483–488. 10.1111/j.1751-0813.1997.tb14377.x [DOI] [PubMed] [Google Scholar]
- 180.Barrs VR, Allan GS, Martin P, Beatty JA, Malik R. 2005. Feline pyothorax: a retrospective study of 27 cases in Australia. J. Feline Med. Surg. 7:211–222. 10.1016/j.jfms.2004.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sykes JE, Sturges BK, Cannon MS, Gericota B, Higgins RJ, Trivedi SR, Dickinson PJ, Vernau KM, Meyer W, Wisner ER. 2010. Clinical signs, imaging features, neuropathology, and outcome in cats and dogs with central nervous system cryptococcosis from California. J. Vet. Intern. Med. 24:1427–1438. 10.1111/j.1939-1676.2010.0633.x [DOI] [PubMed] [Google Scholar]
- 182.Berthiaume DR, Kline KL. 2012. What is your neurologic diagnosis? C. gattii infection. J. Am. Vet. Med. Assoc. 241:1437–1439. 10.2460/javma.241.11.1437 [DOI] [PubMed] [Google Scholar]
- 183.Duncan CG, Stephen C, Campbell J. 2006. Evaluation of risk factors for Cryptococcus gattii infection in dogs and cats. J. Am. Vet. Med. Assoc. 228:377–382. 10.2460/javma.228.3.377 [DOI] [PubMed] [Google Scholar]
- 184.Malik R, Dill-Macky E, Martin P, Wigney DI, Muir DB, Love DN. 1995. Cryptococcosis in dogs: a retrospective study of 20 consecutive cases. J. Med. Vet. Mycol. 33:291–297. 10.1080/02681219580000601 [DOI] [PubMed] [Google Scholar]
- 185.Bowles DB, Fry DR. 2009. Nasal cryptococcosis in two dogs in New Zealand. N. Z. Vet. J. 57:53–57. 10.1080/00480169.2009.36869 [DOI] [PubMed] [Google Scholar]
- 186.Backhouse TC, Bolliger A. 1960. Cryptococcosis in the koala Phascolarctos cinereus. Aust. J. Sci. 23:86–87 [Google Scholar]
- 187.Krockenberger MB, Canfield PJ, Malik R. 2003. Cryptococcus neoformans var. gattii in the koala (Phascolarctos cinereus): a review of 43 cases of cryptococcosis. Med. Mycol. 41:225–234. 10.1080/369378031000137242 [DOI] [PubMed] [Google Scholar]
- 188.Krockenberger MB, Canfield PJ, Barnes J, Vogelnest L, Connolly J, Ley C, Malik R. 2002. Cryptococcus neoformans var. gattii in the koala (Phascolarctos cinereus): serological evidence for subclinical cryptococcosis. Med. Mycol. 40:273–282. 10.1080/mmy.40.3.273.282 [DOI] [PubMed] [Google Scholar]
- 189.Krockenberger MB, Canfield PJ, Malik R. 2002. Cryptococcus neoformans in the koala (Phascolarctos cinereus): colonization by C. n. var. gattii and investigation of environmental sources. Med. Mycol. 40:263–272. 10.1080/mmy.40.3.263.272 [DOI] [PubMed] [Google Scholar]
- 190.Kido N, Makimura K, Kamegaya C, Shindo I, Shibata E, Omiya T, Yamamoto Y. 2012. Long-term surveillance and treatment of subclinical cryptococcosis and nasal colonization by Cryptococcus neoformans and C. gattii species complex in captive koalas (Phascolarctes cinereus). Med. Mycol. 50:291–298. 10.3109/13693786.2011.594967 [DOI] [PubMed] [Google Scholar]
- 191.Malik R, Krockenberger MB, Cross G, Doneley R, Madill DN, Black D, McWhirter P, Rozenwax A, Rose K, Alley M, Forshaw D, Russell-Brown I, Johnstone AC, Martin P, O'Brien CR, Love DN. 2003. Avian cryptococcosis. Med. Mycol. 41:115–124. 10.1080/mmy.41.2.115.124 [DOI] [PubMed] [Google Scholar]
- 192.Morera N, Juan-Salles C, Torres JM, Andreu M, Sanchez M, Zamora MA, Colom MF. 2011. Cryptococcus gattii infection in a Spanish pet ferret (Mustela putorius furo) and asymptomatic carriage in ferrets and humans from its environment. Med. Mycol. 49:779–784. 10.3109/13693786.2011.564216 [DOI] [PubMed] [Google Scholar]
- 193.Malik R, Alderton B, Finlaison D, Krockenberger MB, Karaoglu H, Meyer W, Martin P, France MP, McGill J, Lester SJ, O'Brien CR, Love DN. 2002. Cryptococcosis in ferrets: a diverse spectrum of clinical disease. Aust. Vet. J. 80:749–755. 10.1111/j.1751-0813.2002.tb11343.x [DOI] [PubMed] [Google Scholar]
- 194.Miller WG, Padhye AA, van Bonn W, Jensen E, Brandt ME, Ridgway SH. 2002. Cryptococcosis in a bottlenose dolphin (Tursiops truncatus) caused by Cryptococcus neoformans var. gattii. J. Clin. Microbiol. 40:721–724. 10.1128/JCM.40.2.721-724.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Norman SA, Raverty S, Zabek E, Etheridge S, Ford JK, Hoang LM, Morshed M. 2011. Maternal-fetal transmission of Cryptococcus gattii in harbor porpoise. Emerg. Infect. Dis. 17:304–305. 10.3201/eid1702.101232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Tintelnot K, Lemmer K, Losert H, Schar G, Polak A. 2004. Follow-up of epidemiological data of cryptococcosis in Austria, Germany and Switzerland with special focus on the characterization of clinical isolates. Mycoses 47:455–464. 10.1111/j.1439-0507.2004.01072.x [DOI] [PubMed] [Google Scholar]
- 197.Datta K, Bartlett KH, Baer R, Byrnes E, Galanis E, Heitman J, Hoang L, Leslie MJ, MacDougall L, Magill SS, Morshed MG, Marr KA, Cryptococcus gattii Working Group of the Pacific Northwest 2009. Spread of Cryptococcus gattii into Pacific Northwest region of the United States. Emerg. Infect. Dis. 15:1185–1191. 10.3201/eid1508.081384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Galanis E, MacDougall L, Kidd S, Morshed M, British Columbia Cryptococcus gattii Working Group 2010. Epidemiology of Cryptococcus gattii, British Columbia, Canada, 1999-2007. Emerg. Infect. Dis. 16:251–257. 10.3201/eid1602.090900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Gatti F, Eeckels R. 1970. An atypical strain of Cryptococcus neoformans (San Felice) Vuillemin 1894. I. Description of the disease and of the strain. Ann. Soc. Belges Med. Trop. Parasitol. Mycol. 50:689–693 [PubMed] [Google Scholar]
- 200.Slobodniuk R, Naraqi S. 1980. Cryptococcal meningitis in the central province of Papua New Guinea. P. N. G. Med. J. 23:111–116 [PubMed] [Google Scholar]
- 201.Lehmann PF, Morgan RJ, Freimer EH. 1984. Infection with Cryptococcus neoformans var. gattii leading to a pulmonary cryptococcoma and meningitis. J. Infect. Dis. 9:301–306 [DOI] [PubMed] [Google Scholar]
- 202.Kohl KH, Hof H, Schrettenbrunner A, Seeliger HP, Kwon-Chung KJ. 1985. Cryptococcus neoformans var. gattii in Europe. Lancet i:1515. [DOI] [PubMed] [Google Scholar]
- 203.Dromer F, Ronin O, Dupont B. 1992. Isolation of Cryptococcus neoformans var. gattii from an Asian patient in France: evidence for dormant infection in healthy subjects. J. Med. Vet. Mycol. 30:395–397. 10.1080/02681219280000511 [DOI] [PubMed] [Google Scholar]
- 204.Fisher D, Burrow J, Lo D, Currie B. 1993. Cryptococcus neoformans in tropical northern Australia: predominantly variant gattii with good outcomes. Aust. N. Z. J. Med. 23:678–682. 10.1111/j.1445-5994.1993.tb04726.x [DOI] [PubMed] [Google Scholar]
- 205.Mitchell DH, Sorrell TC. 1992. Pancoast's syndrome due to pulmonary infection with Cryptococcus neoformans var. gattii. Clin. Infect. Dis. 14:1142–1144. 10.1093/clinids/14.5.1142 [DOI] [PubMed] [Google Scholar]
- 206.Rozenbaum R, Goncalves AJ. 1994. Clinical epidemiological study of 171 cases of cryptococcosis. Clin. Infect. Dis. 18:369–380. 10.1093/clinids/18.3.369 [DOI] [PubMed] [Google Scholar]
- 207.Lopez-Martinez R, Soto-Hernandez JL, Ostrosky-Zeichner L, Castanon-Olivares LR, Angeles-Morales V, Sotelo J. 1996. Cryptococcus neoformans var. gattii among patients with cryptococcal meningitis in Mexico. First observations. Mycopathologia 134:61–64 [DOI] [PubMed] [Google Scholar]
- 208.Seaton RA, Verma N, Naraqi S, Wembri JP, Warrell DA. 1997. Visual loss in immunocompetent patients with Cryptococcus neoformans var. gattii meningitis. Trans. R. Soc. Trop. Med. Hyg. 91:44–49. 10.1016/S0035-9203(97)90391-6 [DOI] [PubMed] [Google Scholar]
- 209.Seaton RA, Hamilton AJ, Hay RJ, Warrell DA. 1996. Exposure to Cryptococcus neoformans var. gattii—a seroepidemiological study. Trans. R. Soc. Trop. Med. Hyg. 90:508–512. 10.1016/S0035-9203(96)90297-7 [DOI] [PubMed] [Google Scholar]
- 210.Seaton RA, Naraqi S, Wembri JP, Warrell DA. 1996. Predictors of outcome in Cryptococcus neoformans var. gattii meningitis. QJM 89:423–428. 10.1093/qjmed/89.6.423 [DOI] [PubMed] [Google Scholar]
- 211.Lalloo D, Fisher D, Naraqi S, Laurenson I, Temu P, Sinha A, Saweri A, Mavo B. 1994. Cryptococcal meningitis (C. neoformans var. gattii) leading to blindness in previously healthy Melanesian adults in Papua New Guinea. QJM 87:343–349 [PubMed] [Google Scholar]
- 212.Laurenson I, Naraqi S, Howcroft N, Burrows I, Saulei S. 1993. Cryptococcal meningitis in Papua New Guinea: ecology and the role of eucalypts. Med. J. Aust. 158:213. [DOI] [PubMed] [Google Scholar]
- 213.Laurenson IF, Trevett AJ, Lalloo DG, Nwokolo N, Naraqi S, Black J, Tefurani N, Saweri A, Mavo B, Igo J, Warrell DA. 1996. Meningitis caused by Cryptococcus neoformans var. gattii and var. neoformans in Papua New Guinea. Trans. R. Soc. Trop. Med. Hyg. 90:57–60. 10.1016/S0035-9203(96)90479-4 [DOI] [PubMed] [Google Scholar]
- 214.Laurenson IF, Lalloo DG, Naraqi S, Seaton RA, Trevett AJ, Matuka A, Kevau IH. 1997. Cryptococcus neoformans in Papua New Guinea: a common pathogen but an elusive source. J. Med. Vet. Mycol. 35:437–440. 10.1080/02681219780001561 [DOI] [PubMed] [Google Scholar]
- 215.Jenney A, Pandithage K, Fisher DA, Currie BJ. 2004. Cryptococcus infection in tropical Australia. J. Clin. Microbiol. 42:3865–3868. 10.1128/JCM.42.8.3865-3868.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Lane M, McBride J, Archer J. 2004. Steroid responsive late deterioration in Cryptococcus neoformans var. gattii meningitis. Neurology 63:713–714. 10.1212/01.WNL.0000134677.29120.62 [DOI] [PubMed] [Google Scholar]
- 217.Einsiedel L, Gordon DL, Dyer JR. 2004. Paradoxical inflammatory reaction during treatment of Cryptococcus neoformans var. gattii meningitis in an HIV-seronegative woman. Clin. Infect. Dis. 39:e78–e82. 10.1086/424746 [DOI] [PubMed] [Google Scholar]
- 218.Laman M, Hwaihwanje I, Davis TM, Manning L. 2010. Cryptococcal meningitis in immunocompetent Papua New Guinean children. Trop. Doct. 40:61–63. 10.1258/td.2009.090333 [DOI] [PubMed] [Google Scholar]
- 219.Correa MP, Oliveira EC, Duarte RR, Pardal PP, Oliveira FM, Severo LC. 1999. Cryptococcosis in children in the State of Para, Brazil. Rev. Soc. Bras. Med. Trop. 32:505–508. 10.1590/S0037-86821999000500006 [DOI] [PubMed] [Google Scholar]
- 220.Correa MP, Severo LC, Oliveira FM, Irion K, Londero AT. 2002. The spectrum of computerized tomography (CT) findings in central nervous system (CNS) infection due to Cryptococcus neoformans var. gattii in immunocompetent children. Rev. Inst. Med. Trop. Sao Paulo 44:283–287. 10.1590/S0036-46652002000500010 [DOI] [PubMed] [Google Scholar]
- 221.Martins LM, Wanke B, Lazera MS, Trilles L, Barbosa GG, Macedo RC, Cavalcanti MA, Eulalio KD, Castro JA, Silva AS, Nascimento FF, Gouveia VA, Monte SJ. 2011. Genotypes of Cryptococcus neoformans and Cryptococcus gattii as agents of endemic cryptococcosis in Teresina, Piaui (northeastern Brazil). Mem. Inst. Oswaldo Cruz 106:725–730. 10.1590/S0074-02762011000600012 [DOI] [PubMed] [Google Scholar]
- 222.dos Santos WR, Meyer W, Wanke B, Costa SP, Trilles L, Nascimento JL, Medeiros R, Morales BP, Bezzera CC, Macêdo RC, Ferriera SO, Barbosa GG, Perez MA, Nishikawa MM, Lazéra MS. 2008. Primary endemic Cryptococcus gattii molecular type VGII in the state of Brasil. Mem. Inst. Oswaldo Cruz 103:813–818. 10.1590/S0074-02762008000800012 [DOI] [PubMed] [Google Scholar]
- 223.Pinto Junior VL, Pone MV, Pone SM, Campos JM, Garrido JR, de Barros AC, Trilles L, Barbosa GG, Morales BP, Bezerra CC, Lazera MS. 2010. Cryptococcus gattii molecular type VGII as agent of meningitis in a healthy child in Rio de Janeiro, Brazil: report of an autochthonous case. Rev. Soc. Bras. Med. Trop. 43:746–748. 10.1590/S0037-86822010000600032 [DOI] [PubMed] [Google Scholar]
- 224.Mora DJ, Pedrosa AL, Rodrigues V, Leite Maffei CM, Trilles L, Dos Santos Lazera M, Silva-Vergara ML. 2010. Genotype and mating type distribution within clinical Cryptococcus neoformans and Cryptococcus gattii isolates from patients with cryptococcal meningitis in Uberaba, Minas Gerais, Brazil. Med. Mycol. 48:561–569. 10.3109/13693780903358317 [DOI] [PubMed] [Google Scholar]
- 225.Grosse P, Tintelnot K, Sollner O, Schmitz B. 2001. Encephalomyelitis due to Cryptococcus neoformans var. gattii presenting as spinal tumour: case report and review of the literature. J. Neurol. Neurosurg. Psychiatr. 70:113–116. 10.1136/jnnp.70.1.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Taylor MB, Chadwick D, Barkham T. 2002. First reported isolation of Cryptococcus neoformans var. gattii from a patient in Singapore. J. Clin. Microbiol. 40:3098–3099. 10.1128/JCM.40.8.3098-3099.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Koh TH, Tan AL, Lo YL, Oh H. 2002. Cryptococcus neoformans var. gattii meningitis in Singapore. Med. Mycol. 40:221–223. 10.1080/mmy.40.2.221.223 [DOI] [PubMed] [Google Scholar]
- 228.Lingegowda BP, Koh TH, Ong HS, Tan TT. 2011. Primary cutaneous cryptococcosis due to Cryptococcus gattii in Singapore. Singapore Med. J. 52:e160–e162 [PubMed] [Google Scholar]
- 229.MacDougall L, Fyfe M. 2006. Emergence of Cryptococcus gattii in a novel environment provides clues to its incubation period. J. Clin. Microbiol. 44:1851–1852. 10.1128/JCM.44.5.1851-1852.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.MacDougall L, Fyfe M, Romney M, Starr M, Galanis E. 2011. Risk factors for Cryptococcus gattii infection, British Columbia, Canada. Emerg. Infect. Dis. 17:193–199. 10.3201/eid1702.101020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Garrett L, Marr K, West S, Allada G. 2011. 74-year-old man from the Pacific Northwest with fever and a lung mass. Chest 140:814–817. 10.1378/chest.10-2964 [DOI] [PubMed] [Google Scholar]
- 232.Meiring ST, Quan VC, Cohen C, Dawood H, Karstaedt AS, McCarthy KM, Whitelaw AC, Govender NP, Group for Enteric, Respiratory and Meningeal Disease Surveillance in South Africa 2012. A comparison of cases of paediatric-onset and adult-onset cryptococcosis detected through population-based surveillance, 2005-2007. AIDS 26:2307–2314. 10.1097/QAD.0b013e3283570567 [DOI] [PubMed] [Google Scholar]
- 233.Araujo BS, Bay M, Reichert R, Goldani LZ. 2012. Intra-abdominal cryptococcosis by Cryptococcus gattii: case report and review. Mycopathologia 174:81–85. 10.1007/s11046-011-9517-4 [DOI] [PubMed] [Google Scholar]
- 234.Mistry N, Tan K, Shokravi M, Hoang L. 2011. Cryptococcus gattii infections with cutaneous involvement. J. Cutan. Med. Surg. 15:236–237 [DOI] [PubMed] [Google Scholar]
- 235.Hamann ID, Gillespie RJ, Ferguson JK. 1997. Primary cryptococcal cellulitis caused by Cryptococcus neoformans var. gattii in an immunocompetent host. Australas. J. Dermatol. 38:29–32. 10.1111/j.1440-0960.1997.tb01095.x [DOI] [PubMed] [Google Scholar]
- 236.Bellissimo-Rodrigues F, Baciotti M, Zanatto MP, Silva JO, Martins MA, Martinez R. 2010. Cutaneous cryptococcosis due to Cryptococcus gattii in a patient on chronic corticotherapy. Rev. Soc. Bras. Med. Trop. 43:211–212. 10.1590/S0037-86822010000200022 [DOI] [PubMed] [Google Scholar]
- 237.Dewar GJ, Kelly JK. 2008. Cryptococcus gattii: an emerging cause of pulmonary nodules. Can. Respir. J. 15:153–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Hagen F, van Assen S, Luijckx GJ, Boekhout T, Kampinga GA. 2010. Activated dormant Cryptococcus gattii infection in a Dutch tourist who visited Vancouver Island (Canada): a molecular epidemiological approach. Med. Mycol. 48:528–531. 10.3109/13693780903300319 [DOI] [PubMed] [Google Scholar]
- 239.Chau TT, Mai NH, Phu NH, Nghia HD, Chuong LV, Sinh DX, Duong VA, Diep PT, Campbell JI, Baker S, Hien TT, Lalloo DG, Farrar JJ, Day JN. 2010. A prospective descriptive study of cryptococcal meningitis in HIV uninfected patients in Vietnam—high prevalence of Cryptococcus neoformans var. grubii in the absence of underlying disease. BMC Infect. Dis. 10:199. 10.1186/1471-2334-10-199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Upton A, Fraser JA, Kidd SE, Bretz C, Bartlett KH, Heitman J, Marr KA. 2007. First contemporary case of human infection with Cryptococcus gattii in Puget Sound: evidence for spread of the Vancouver Island outbreak. J. Clin. Microbiol. 45:3086–3088. 10.1128/JCM.00593-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Phillips P, Chapman K, Sharp M, Harrison P, Vortel J, Steiner T, Bowie W. 2009. Dexamethasone in Cryptococcus gattii central nervous system infection. Clin. Infect. Dis. 49:591–595. 10.1086/603554 [DOI] [PubMed] [Google Scholar]
- 242.Byrnes EJ, III, Li W, Lewit Y, Perfect JR, Carter DA, Cox GM, Heitman J. 2009. First reported case of Cryptococcus gattii in the Southeastern USA: implications for travel-associated acquisition of an emerging pathogen. PLoS One 4:e5851. 10.1371/journal.pone.0005851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.McCulloh RJ, Phillips R, Perfect JR, Byrnes EJ, III, Heitman J, Dufort E. 2011. Cryptococcus gattii genotype VGI infection in New England. Pediatr. Infect. Dis. J. 30:1111–1114. 10.1097/INF.0b013e31822d14fd [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Nagarathna S, Kumari HB, Arvind N, Divyalakshmi A, Chandramuki A, Satishchandra P, Ravi V. 2010. Prevalence of Cryptococcus gattii causing meningitis in a tertiary neurocare center from south India: a pilot study. Ind. J. Pathol. Microbiol. 53:855–856. 10.4103/0377-4929.72001 [DOI] [PubMed] [Google Scholar]
- 245.Iatta R, Hagen F, Fico C, Lopatriello N, Boekhout T, Montagna MT. 2012. Cryptococcus gattii infection in an immunocompetent patient from southern Italy. Mycopathologia 174:87–92. 10.1007/s11046-011-9493-8 [DOI] [PubMed] [Google Scholar]
- 246.Tsunemi T, Kamata T, Fumimura Y, Watanabe M, Yamawaki M, Saito Y, Kanda T, Ohashi K, Suegara N, Murayama S, Makimura K, Yamaguchi H, Mizusawa H. 2001. Immunohistochemical diagnosis of Cryptococcus neoformans var. gattii infection in chronic meningoencephalitis: the first case in Japan. Intern. Med. 40:1241–1244. 10.2169/internalmedicine.40.1241 [DOI] [PubMed] [Google Scholar]
- 247.Sellers B, Hall P, Cine-Gowdie S, Hays AL, Patel K, Lockhart SR, Franco-Paredes C. 2012. Cryptococcus gattii: an emerging fungal pathogen in the Southeastern United States. Am. J. Med. Sci. 343:510–511. 10.1097/MAJ.0b013e3182464bc7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Hasimoto e Souza LK, Costa CR, Fernandes OFL, Abrao FY, Silva TC, Tremea CM, Silva MR. 2013. Clinical and microbiological features of cryptococcal meningitis. Rev. Soc. Bras. Med. Trop. 46:343–347. 10.1590/0037-8682-0061-2012 [DOI] [PubMed] [Google Scholar]
- 249.Okamoto K, Hatakeyama S, Itoyama S, Nukui Y, Yoshino Y, Kitazawa T, Yotsuyanagi H, Ikeda R, Sugita T, Koike K. 2010. Cryptococcus gattii genotype VGIIa infection in man, Japan, 2007. Emerg. Infect. Dis. 16:1155–1157. 10.3201/eid1607.100106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Johannson KA, Huston SM, Mody CH, Davidson W. 2012. Cryptococcus gattii pneumonia. CMAJ 184:1387–1390. 10.1503/cmaj.111346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Harris JR, Lockhart SR, Sondermeyer G, Vugia DJ, Crist MB, D'Angelo MT, Sellers B, Franco-Paredes C, Makvandi M, Smelser C, Greene J, Stanek D, Signs K, Nett RJ, Chiller T, Park BJ. 2013. Cryptococcus gattii infections in multiple states outside the US Pacific Northwest. Emerg. Infect. Dis. 19:1620–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Centers for Disease Control and Prevention. 2010. Emergence of Cryptococcus gattii—Pacific Northwest, 2004-2010. MMWR Morb. Mortal. Wkly. Rep. 59:865–868 [PubMed] [Google Scholar]
- 253.Kunadharaju R, Choe U, Harris JR, Lockhart SR, Greene JN. 2013. Cryptococcus gattii, Florida, USA, 2011. Emerg. Infect. Dis. 19:519–521. 10.3201/eid1903.121399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Sorrell TC. 2001. Cryptococcus neoformans variety gattii. Med. Mycol. 39:155–168. 10.1080/mmy.39.2.155.168 [DOI] [PubMed] [Google Scholar]
- 255.Sorrell TC, Chen SCA, Phillips P, Marr K. 2011. Clinical perspectives on Cryptococcus neoformans and Cryptococcus gattii: implications for diagnosis and management, p 595–606 In Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, Casadevall A. (ed), Cryptococcus: from pathogen to model yeast. ASM Press, Washington, DC [Google Scholar]
- 256.Severo CB, Xavier MO, Gazzoni AF, Severo LC. 2009. Cryptococcosis in children. Paediatr. Respir. Rev. 10:166–171. 10.1016/j.prrv.2009.06.009 [DOI] [PubMed] [Google Scholar]
- 257.Harris J, Lockhart S, Chiller T. 2012. Cryptococcus gattii: where do we go from here? Med. Mycol. 50:113–129. 10.3109/13693786.2011.607854 [DOI] [PubMed] [Google Scholar]
- 258.Marr KA. 2012. Cryptococcus gattii as an important fungal pathogen of western North America. Expert Rev. Anti Infect. Ther. 10:637–643. 10.1586/eri.12.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Bartlett K, Byrnes E, Duncan C, Fyfe M, Galanis E, Heitman J, Hoand L, Kidd S, MacDougall L, Mak S, Marr K, Moshed M, West S, Kronstad J. 2011. The emergence of Cryptococcus gattii infections on Vancouver Island and expansion in the Pacific Northwest, p 313–325 In Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, Casadevall A. (ed), Cryptococcus: from pathogen to model yeast. ASM Press, Washington, DC [Google Scholar]
- 260.Pukkila-Worley R, Mylonakis E. 2008. Epidemiology and management of cryptococcal meningitis: developments and challenges. Expert Opin. Pharmacother. 9:551–560. 10.1517/14656566.9.4.551 [DOI] [PubMed] [Google Scholar]
- 261.Banerjee U, Datta K, Majumdar T, Gupta K. 2001. Cryptococcosis in India: the awakening of a giant? Med. Mycol. 39:51–67. 10.1080/mmy.39.1.51.67 [DOI] [PubMed] [Google Scholar]
- 262.McMullan BJ, Sorell TC, Chen SCA. 2013. Cryptococcus gattii infections: contemporary aspects of epidemiology, clinical manifestations and management of infection. Future Microbiol. 8:1613–1631. 10.2217/fmb.13.123 [DOI] [PubMed] [Google Scholar]
- 263.Pappas PG. 2013. Cryptococcal infections in non-HIV-infected patients. Trans. Am. Clin. Climatol. Assoc. 124:61–79 [PMC free article] [PubMed] [Google Scholar]
- 264.Bartlett KH, Cheng PY, Duncan C, Galanis E, Hoang L, Kidd S, Lee MK, Lester S, MacDougall L, Mak S, Morshed M, Taylor M, Kronstad J. 2012. A decade of experience: Cryptococcus gattii in British Columbia. Mycopathologia 173:311–319. 10.1007/s11046-011-9475-x [DOI] [PubMed] [Google Scholar]
- 265.Chaturvedi V, Chaturvedi S. 2011. Cryptococcus gattii: a resurgent fungal pathogen. Trends Microbiol. 19:564–571. 10.1016/j.tim.2011.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Howell SA. 2011. Candida, Cryptococcus, and other yeasts of medical importance, p 1793–1821 In Versalovic J, Carroll KC, Guido F, Jorgensen JH, Landry ML, Warnock DW. (ed), Manual of clinical microbiology, 19th ed. ASM Press, Washington, DC [Google Scholar]
- 267.Currie B, Vigus T, Leach G, Dwyer B. 1990. Cryptococcus neoformans var. gattii. Lancet 336:1442. 10.1016/0140-6736(90)93137-E [DOI] [PubMed] [Google Scholar]
- 268.Speed BR, Kaldor J. 1997. Rarity of cryptococcal infection in children. Pediatr. Infect. Dis. J. 16:536–537. 10.1097/00006454-199705000-00024 [DOI] [PubMed] [Google Scholar]
- 269.Brandt ME, Hutwagner LC, Klug LA, Baughman WS, Rimland D, Graviss EA, Hamill RJ, Thomas C, Pappas PG, Reingold AL, Pinner RW, Cryptococcal Disease Active Surveillance Group 1996. Molecular subtype distribution of Cryptococcus neoformans in four areas of the United States. J. Clin. Microbiol. 34:912–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.BC Center for Disease Control. 2011. British Columbia annual summary of reportable disease. BC Center for Disease Control, Vancouver, BC, Canada [Google Scholar]
- 271.Iverson SA, Chiller T, Beekmann S, Polgreen PM, Harris J. 2012. Recognition and diagnosis of Cryptococcus gattii infections in the United States. Emerg. Infect. Dis. 18:1012–1015. 10.3201/eid1806.111228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Walraven CJ, Gerstein W, Hardison SE, Wormley F, Lockhart SR, Harris JR, Fothergill A, Wickes B, Gober-Wilcox J, Massie L, Ku TS, Firacative C, Meyer W, Lee SA. 2011. Fatal disseminated Cryptococcus gattii infection in New Mexico. PLoS One 6:e28625. 10.1371/journal.pone.0028625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Ellis D, Pfeiffer T. 1994. Cryptococcosis and the ecology of Cryptococcus neoformans. Jpn. J. Med. Mycol. 35:111–122. 10.3314/jjmm.35.111 [DOI] [Google Scholar]
- 274.Montenegro H, Paula CR. 2000. Environmental isolation of Cryptococcus neoformans var. gattii and C. neoformans var. neoformans in the city of Sao Paulo, Brazil. Med. Mycol. 38:385–390. 10.1080/mmy.38.5.385.390 [DOI] [PubMed] [Google Scholar]
- 275.Van Dam MG, Seaton RA, Hamilton AJ. 1998. Analysis of HLA association in susceptibility to infection with Cryptococcus neoformans var. gattii in a Papua New Guinean population. Med. Mycol. 36:185–188. 10.1080/02681219880000281 [DOI] [PubMed] [Google Scholar]
- 276.Rosen LB, Freeman AF, Yang LM, Jutivorakool K, Olivier KN, Angkasekwinai N, Suputtamongkol Y, Bennett JE, Pyrgos V, Williamson PR, Ding L, Holland SM, Browne SK. 2013. Anti-GM-CSF autoantibodies in patients with cryptococcal meningitis. J. Immunol. 190:3959–3966. 10.4049/jimmunol.1202526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Kampitak T, Suwanpimolkul G, Browne S, Suankratay C. 2011. Anti-interferon-gamma autoantibody and opportunistic infections: case series and review of the literature. Infection 39:65–71. 10.1007/s15010-010-0067-3 [DOI] [PubMed] [Google Scholar]
- 278.Browne SK, Burbelo PD, Chetchotisakd P, Suputtamongkol Y, Kiertiburanakul S, Shaw PA, Kirk JL, Jutivorakool K, Zaman R, Ding L, Hsu AP, Patel SY, Olivier KN, Lulitanond V, Mootsikapun P, Anunnatsiri S, Angkasekwinai N, Sathapatayavongs B, Hsueh PR, Shieh CC, Brown MR, Thongnoppakhun W, Claypool R, Sampaio EP, Thepthai C, Waywa D, Dacombe C, Reizes Y, Zelazny AM, Saleeb P, Rosen LB, Mo A, Iadarola M, Holland SM. 2012. Adult-onset immunodeficiency in Thailand and Taiwan. N. Engl. J. Med. 367:725–734. 10.1056/NEJMoa1111160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Saijo T, Chen J, Chen SC, Rosen LB, Yi J, Sorrell TC, Bennett JE, Holland SM, Browne SK, Kwon-Chung KJ. 2014. Anti-granulocyte-macrophage colony-stimulating factor autoantibodies are a risk factor for central nervous system infection by Cryptococcus gattii in otherwise immunocompetent patients. mBio 5(2):e00912-14. 10.1128/mBio.00912-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Marr KA, Datta K, Pirofski LA, Barnes R. 2012. Cryptococcus gattii infection in healthy hosts: a sentinel for subclinical immunodeficiency? Clin. Infect. Dis. 54:153–154. 10.1093/cid/cir756 [DOI] [PubMed] [Google Scholar]
- 281.Zonios DI, Falloon J, Huang CY, Chaitt D, Bennett JE. 2007. Cryptococcosis and idiopathic CD4 lymphocytopenia. Medicine 86:78–92. 10.1097/md.0b013e31803b52f5 [DOI] [PubMed] [Google Scholar]
- 282.Perfect JR, Dismukes WE, Dromer F, Goldman DL, Graybill JR, Hamill RJ, Harrison TS, Larsen RA, Lortholary O, Nguyen MH, Pappas PG, Powderly WG, Singh N, Sobel JD, Sorrell TC. 2010. Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 50:291–322. 10.1086/649858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Goldman DL, Khine H, Abadi J, Lindenberg DJ, Pirofski L, Niang R, Casadevall A. 2001. Serologic evidence for Cryptococcus neoformans infection in early childhood. Pediatrics 107:E66. 10.1542/peds.107.5.e66 [DOI] [PubMed] [Google Scholar]
- 284.Pappalardo MC, Melhem MS. 2003. Cryptococcosis: a review of the Brazilian experience for the disease. Rev. Inst. Med. Trop. Sao Paulo 45:299–305. 10.1590/S0036-46652003000600001 [DOI] [PubMed] [Google Scholar]
- 285.Garcia-Hermoso D, Janbon G, Dromer F. 1999. Epidemiological evidence for dormant Cryptococcus neoformans infection. J. Clin. Microbiol. 37:3204–3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Dromer F, Casadevall A, Perfect JR, Sorrell TC. 2011. Cryptococcus neoformans: latency and disease, p 431–439 In Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, Casadevall A. (ed), Cryptococcus: from pathogen to model yeast. ASM Press, Washington, DC [Google Scholar]
- 287.Dora JM, Kelbert S, Deutschendorf C, Cunha VS, Aquino VR, Santos RP, Goldani LZ. 2006. Cutaneous cryptococccosis due to Cryptococcus gattii in immunocompetent hosts: case report and review. Mycopathologia 161:235–238. 10.1007/s11046-006-0277-5 [DOI] [PubMed] [Google Scholar]
- 288.Lacaz CS, Heins-Vaccari EM, Hernandez-Arriagada GL, Martins EL, Prearo CA, Corim SM, Martins MA. 2002. Primary cutaneous cryptococcosis due to Cryptococcus neoformans var. gattii serotype B, in an immunocompetent patient. Rev. Inst. Med. Trop. Sao Paulo 44:225–228. 10.1590/S0036-46652002000400008 [DOI] [PubMed] [Google Scholar]
- 289.Leao CA, Ferreira-Paim K, Andrade-Silva L, Mora DJ, da Silva PR, Machado AS, Neves PF, Pena GS, Teixeira LS, Silva-Vergara ML. 2011. Primary cutaneous cryptococcosis caused by Cryptococcus gattii in an immunocompetent host. Med. Mycol. 49:352–355. 10.3109/13693786.2010.530697 [DOI] [PubMed] [Google Scholar]
- 290.Tilak R, Prakash P, Nigam C, Tilak V, Gambhir IS, Gulati AK. 2009. Cryptococcal meningitis with an antecedent cutaneous cryptococcal lesion. Dermatol. Online J. 15:12. [PubMed] [Google Scholar]
- 291.Rex JH, Larsen RA, Dismukes WE, Cloud GA, Bennett JE. 1993. Catastrophic visual loss due to Cryptococcus neoformans meningitis. Medicine 72:207–224 [DOI] [PubMed] [Google Scholar]
- 292.Andreola C, Ribeiro MP, de Carli CR, Gouvea AL, Curi AL. 2006. Multifocal choroiditis in disseminated Cryptococcus neoformans infection. Am. J. Ophthalmol. 142:346–348. 10.1016/j.ajo.2006.03.024 [DOI] [PubMed] [Google Scholar]
- 293.Kestelyn P, Taelman H, Bogaerts J, Kagame A, Abdel Aziz M, Batungwanayo J, Stevens AM, Van de Perre P. 1993. Ophthalmic manifestations of infections with Cryptococcus neoformans in patients with the acquired immunodeficiency syndrome. Am. J. Ophthalmol. 116:721–727 [DOI] [PubMed] [Google Scholar]
- 294.Graybill JR, Sobel J, Saag M, van Der Horst C, Powderly W, Cloud G, Riser L, Hamill R, Dismukes W, NIAID Mycoses Study Group and AIDS Cooperative Treatment Groups 2000. Diagnosis and management of increased intracranial pressure in patients with AIDS and cryptococcal meningitis. Clin. Infect. Dis. 30:47–54. 10.1086/313603 [DOI] [PubMed] [Google Scholar]
- 295.Johnston SR, Corbett EL, Foster O, Ash S, Cohen J. 1992. Raised intracranial pressure and visual complications in AIDS patients with cryptococcal meningitis. J. Infect. 24:185–189. 10.1016/0163-4453(92)92954-H [DOI] [PubMed] [Google Scholar]
- 296.Park MK, Hospenthal DR, Bennett JE. 1999. Treatment of hydrocephalus secondary to cryptococcal meningitis by use of shunting. Clin. Infect. Dis. 28:629–633. 10.1086/515161 [DOI] [PubMed] [Google Scholar]
- 297.Liliang PC, Liang CL, Chang WN, Chen HJ, Su TM, Lu K, Lu CH. 2003. Shunt surgery for hydrocephalus complicating cryptococcal meningitis in human immunodeficiency virus-negative patients. Clin. Infect. Dis. 37:673–678. 10.1086/377208 [DOI] [PubMed] [Google Scholar]
- 298.Haddow LJ, Colebunders R, Meintjes G, Lawn SD, Elliott JH, Manabe YC, Bohjanen PR, Sungkanuparph S, Easterbrook PJ, French MA, Boulware DR, International Network for the Study of HIV-Associated IRIS 2010. Cryptococcal immune reconstitution inflammatory syndrome in HIV-1-infected individuals: proposed clinical case definitions. Lancet Infect. Dis. 10:791–802. 10.1016/S1473-3099(10)70170-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Chen SCA, Korman TM, Slavin MA, Marriott D, Byth K, Bak N, Currie BJ, Hajkowicz K, Heath CH, Kidd S, McBride WJ, Meyer W, Murray R, Playford EG, Sorrell TC, Australia New Zealand Mycoses Interest Group Cryptococcus Study 2013. Antifungal therapy and management of complications of cryptococcosis due to Cryptococcus gattii. Clin. Infect. Dis. 57:543–551. 10.1093/cid/cit341 [DOI] [PubMed] [Google Scholar]
- 300.Boulware DR, Bonham SC, Meya DB, Wiesner DL, Park GS, Kambugu A, Janoff EN, Bohjanen PR. 2010. Paucity of initial cerebrospinal fluid inflammation in cryptococcal meningitis is associated with subsequent immune reconstitution inflammatory syndrome. J. Infect. Dis. 202:962–970. 10.1086/655785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Longley N, Harrison TS, Jarvis JN. 2013. Cryptococcal immune reconstitution inflammatory syndrome. Curr. Opin. Infect. Dis. 26:26–34. 10.1097/QCO.0b013e32835c21d1 [DOI] [PubMed] [Google Scholar]
- 302.Boulware DR, Meya DB, Bergemann TL, Wiesner DL, Rhein J, Musubire A, Lee SJ, Kambugu A, Janoff EN, Bohjanen PR. 2010. Clinical features and serum biomarkers in HIV immune reconstitution inflammatory syndrome after cryptococcal meningitis: a prospective cohort study. PLoS Med. 7:e1000384. 10.1371/journal.pmed.1000384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Wiesner DL, Boulware DR. 2011. Cryptococcus-related immune reconstitution inflammatory syndrome (IRIS): pathogenesis and its clinical implications. Curr. Fungal Infect. Rep. 5:252–261. 10.1007/s12281-011-0064-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Wiesner DL, Moskalenko O, Corcoran JM, McDonald T, Rolfes MA, Meya DB, Kajumbula H, Kambugu A, Bohjanen PR, Knight JF, Boulware DR, Nielsen K. 2012. Cryptococcal genotype influences immunologic response and human clinical outcome after meningitis. mBio 3(5):e00196-12. 10.1128/mBio.00196-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Dromer F, Mathoulin-Pélissier S, Launay O, Lortholary O, French Cryptococcosis Study Group 2007. Determinants of disease presentation and outcome during cryptococcosis: the Crypto A/D study. PLoS Med. 4(2):e21. 10.1371/journal.pmed.0040021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Ostrow TD, Hudgins PA. 1994. Magnetic resonance imaging of intracranial fungal infections. Top. Magn. Reson. Imaging 6:22–31 [PubMed] [Google Scholar]
- 307.Smith AB, Smirniotopoulos JG, Rushing EJ. 2008. Central nervous system infections associated with human immunodeficiency virus infection: radiologic-pathologic correlation. Radiographics 28:2033–2058. 10.1148/rg.287085135 [DOI] [PubMed] [Google Scholar]
- 308.Sorrell TC, Himmelreich U. 2008. NMR in mycology. Curr. Fungal Infect. Rep. 2:149–156. 10.1007/s12281-008-0022-2 [DOI] [Google Scholar]
- 309.Himmelreich U, Dzendrowskyj TE, Allen C, Dowd S, Malik R, Shehan BP, Russell P, Mountford CE, Sorrell TC. 2001. Cryptococcomas distinguished from gliomas with MR spectroscopy: an experimental rat and cell culture study. Radiology 220:122–128. 10.1148/radiology.220.1.r01jl25122 [DOI] [PubMed] [Google Scholar]
- 310.Himmelreich U, Allen C, Dowd S, Malik R, Shehan BP, Mountford C, Sorrell TC. 2003. Identification of metabolites of importance in the pathogenesis of pulmonary cryptococcoma using nuclear magnetic resonance spectroscopy. Microbes Infect. 5:285–290. 10.1016/S1286-4579(03)00028-5 [DOI] [PubMed] [Google Scholar]
- 311.Dzendrowskyj TE, Dolenko B, Sorrell TC, Somorjai RL, Malik R, Mountford CE, Himmelreich U. 2005. Diagnosis of cerebral cryptococcoma using a computerized analysis of 1H NMR spectra in an animal model. Diagn. Microbiol. Infect. Dis. 52:101–105. 10.1016/j.diagmicrobio.2005.02.004 [DOI] [PubMed] [Google Scholar]
- 312.Diaz MR, Nguyen MH. 2011. Diagnostic approach based on capsular antigen, capsule detection, B-glucan and DNA analysis, p 547–564 In Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, Casadevall A. (ed), Cryptococcus: from human pathogen to model yeast. ASM Press, Washington, DC [Google Scholar]
- 313.Saha DC, Xess I, Jain N. 2008. Evaluation of conventional & serological methods for rapid diagnosis of cryptococcosis. Indian J. Med. Res. 127:483–488 [PubMed] [Google Scholar]
- 314.Petter R, Kang BS, Boekhout T, Davis BJ, Kwon-Chung KJ. 2001. A survey of heterobasidiomycetous yeasts for the presence of the genes homologous to virulence factors of Filobasidiella neoformans, LAC1 and CAP59. Microbiology 147:2029–2036 [DOI] [PubMed] [Google Scholar]
- 315.Okoli I, Oyeka CA, Kwon-Chung KJ, Theelen B, Robert V, Groenewald JZ, McFadden DC, Casadevall A, Boekhout T. 2007. Cryptotrichosporon anacardii gen. nov., sp. nov., a new trichosporonoid capsulate basidiomycetous yeast from Nigeria that is able to form melanin on niger seed agar. FEMS Yeast Res. 7:339–350. 10.1111/j.1567-1364.2006.00164.x [DOI] [PubMed] [Google Scholar]
- 316.Suwantarat N, Watkins T, Lee R, Carroll KC. 2014. False-positive reaction of L-canavanine glycine bromothymol blue medium with Candida famata. J. Clin. Microbiol. 52:1308–1309. 10.1128/JCM.00149-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Currie BP, Freundlich LF, Soto MA, Casadevall A. 1993. False-negative cerebrospinal fluid cryptococcal latex agglutination tests for patients with culture-positive cryptococcal meningitis. J. Clin. Microbiol. 31:2519–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Heelan JS, Corpus L, Kessimian N. 1991. False-positive reactions in the latex agglutination test for Cryptococcus neoformans antigen. J. Clin. Microbiol. 29:1260–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Hamilton JR, Noble A, Denning DW, Stevens DA. 1991. Performance of cryptococcus antigen latex agglutination kits on serum and cerebrospinal fluid specimens of AIDS patients before and after pronase treatment. J. Clin. Microbiol. 29:333–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Engler HD, Shea YR. 1994. Effect of potential interference factors on performance of enzyme immunoassay and latex agglutination assay for cryptococcal antigen. J. Clin. Microbiol. 32:2307–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Liaw YS, Yang PC, Yu CJ, Chang DB, Wang HJ, Lee LN, Kuo SH, Luh KT. 1995. Direct determination of cryptococcal antigen in transthoracic needle aspirate for diagnosis of pulmonary cryptococcosis. J. Clin. Microbiol. 33:1588–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Chapin-Robertson K, Bechtel C, Waycott S, Kontnick C, Edberg SC. 1993. Cryptococcal antigen detection from the urine of AIDS patients. Diagn. Microbiol. Infect. Dis. 17:197–201. 10.1016/0732-8893(93)90096-P [DOI] [PubMed] [Google Scholar]
- 323.Kralovic SM, Rhodes JC. 1998. Utility of routine testing of bronchoalveolar lavage fluid for cryptococcal antigen. J. Clin. Microbiol. 36:3088–3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Hansen J, Slechta ES, Gates-Hollingsworth MA, Neary B, Barker AP, Bauman S, Kozel TR, Hanson KE. 2013. Large-scale evaluation of the Immuno-Mycologics lateral flow and enzyme-linked immunoassays for detection of cryptococcal antigen in serum and cerebrospinal fluid. Clin. Vaccine Immunol. 20:52–55. 10.1128/CVI.00536-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.McMullan BJ, Halliday C, Sorrell TC, Judd D, Sleiman S, Marriott D, Olma T, Chen SCA. 2012. Clinical utility of the cryptococcal antigen lateral flow assay in a diagnostic mycology laboratory. PLoS One 7:e49541. 10.1371/journal.pone.0049541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Jarvis JN, Percival A, Bauman S, Pelfrey J, Meintjes G, Williams GN, Longley N, Harrison TS, Kozel TR. 2011. Evaluation of a novel point-of-care cryptococcal antigen test on serum, plasma, and urine from patients with HIV-associated cryptococcal meningitis. Clin. Infect. Dis. 53:1019–1023. 10.1093/cid/cir613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Lindsley MD, Mekha N, Baggett HC, Surinthong Y, Autthateinchai R, Sawatwong P, Harris JR, Park BJ, Chiller T, Balajee SA, Poonwan N. 2011. Evaluation of a newly developed lateral flow immunoassay for the diagnosis of cryptococcosis. Clin. Infect. Dis. 53:321–325. 10.1093/cid/cir379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Kabanda T, Siedner MJ, Klausner JD, Muzoora C, Boulware DR. 2014. Point-of-care diagnosis and prognostication of cryptococcal meningitis with the cryptococcal antigen lateral flow assay on cerebrospinal fluid. Clin. Infect. Dis. 58:113–116. 10.1093/cid/cit641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Rajasingham R, Meya DB, Boulware DR. 2012. Integrating cryptococcal antigen screening and pre-emptive treatment into routine HIV care. J. Acquir. Immune Defic. Syndr. 59:e85–e91. 10.1097/QAI.0b013e31824c837e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Brouwer AE, Teparrukkul P, Pinpraphaporn S, Larsen RA, Chierakul W, Peacock S, Day N, White NJ, Harrison TS. 2005. Baseline correlation and comparative kinetics of cerebrospinal fluid colony-forming unit counts and antigen titers in cryptococcal meningitis. J. Infect. Dis. 192:681–684. 10.1086/432073 [DOI] [PubMed] [Google Scholar]
- 331.Rajasingham R, Boulware DR. 2012. Reconsidering cryptococcal antigen screening in the U.S. among persons with CD4 <100 cells/mcL. Clin. Infect. Dis. 55:1742–1744. 10.1093/cid/cis725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Sidrim JJ, Costa AK, Cordeiro RA, Brilhante RS, Moura FE, Castelo-Branco DS, Neto MP, Rocha MF. 2010. Molecular methods for the diagnosis and characterization of Cryptococcus: a review. Can. J. Microbiol. 56:445–458. 10.1139/W10-030 [DOI] [PubMed] [Google Scholar]
- 333.Paschoal RC, Hirata MH, Hirata RC, Melhem MS, Dias AL, Paula CR. 2004. Neurocryptococcosis: diagnosis by PCR method. Rev. Inst. Med. Trop. Sao Paulo 46:203–207. 10.1590/S0036-46652004000400006 [DOI] [PubMed] [Google Scholar]
- 334.Lau A, Chen S, Sorrell T, Carter D, Malik R, Martin P, Halliday C. 2007. Development and clinical application of a panfungal PCR assay to detect and identify fungal DNA in tissue specimens. J. Clin. Microbiol. 45:380–385. 10.1128/JCM.01862-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Pryce TM, Palladino S, Kay ID, Coombs GW. 2003. Rapid identification of fungi by sequencing the ITS1 and ITS2 regions using an automated capillary electrophoresis system. Med. Mycol. 41:369–381. 10.1080/13693780310001600435 [DOI] [PubMed] [Google Scholar]
- 336.Rappelli P, Are R, Casu G, Fiori PL, Cappuccinelli P, Aceti A. 1998. Development of a nested PCR for detection of Cryptococcus neoformans in cerebrospinal fluid. J. Clin. Microbiol. 36:3438–3440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Bialek R, Weiss M, Bekure-Nemariam K, Najvar LK, Alberdi MB, Graybill JR, Reischl U. 2002. Detection of Cryptococcus neoformans DNA in tissue samples by nested and real-time PCR assays. Clin. Diagn. Lab. Immunol. 9:461–469. 10.1128/CDLI.9.2.461-469.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Zou CC, Yu ZS, Tang LF, Liang L, Zhao ZY. 2006. Primary abdominal lymphonodular cryptococcosis in children: 2 case reports and a literature review. J. Pediatr. Surg. 41:e11–e15. 10.1016/j.jpedsurg.2005.11.083 [DOI] [PubMed] [Google Scholar]
- 339.Levy PY, Habib G, Reynaud-Gaubert M, Raoult D, Rolain JM. 2008. Pericardial effusion due to Cryptococcus neoformans in a patient with cystic fibrosis following lung transplantation. Int. J. Infect. Dis. 12:452. 10.1016/j.ijid.2007.12.001 [DOI] [PubMed] [Google Scholar]
- 340.Veron V, Simon S, Blanchet D, Aznar C. 2009. Real-time polymerase chain reaction detection of Cryptococcus neoformans and Cryptococcus gattii in human samples. Diagn. Microbiol. Infect. Dis. 65:69–72. 10.1016/j.diagmicrobio.2009.05.005 [DOI] [PubMed] [Google Scholar]
- 341.Katsu M, Kidd S, Ando A, Moretti-Branchini ML, Mikami Y, Nishimura K, Meyer W. 2004. The internal transcribed spacers and 5.8S rRNA gene show extensive diversity among isolates of the Cryptococcus neoformans species complex. FEMS Yeast Res. 4:377–388. 10.1016/S1567-1356(03)00176-4 [DOI] [PubMed] [Google Scholar]
- 342.Chen YC, Eisner JD, Kattar MM, Rassoulian-Barrett SL, Lafe K, Bui U, Limaye AP, Cookson BT. 2001. Polymorphic internal transcribed spacer region 1 DNA sequences identify medically important yeasts. J. Clin. Microbiol. 39:4042–4051. 10.1128/JCM.39.11.4042-4051.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Leal AL, Faganello J, Bassanesi MC, Vainstein MH. 2008. Cryptococcus species identification by multiplex PCR. Med. Mycol. 46:377–383. 10.1080/13693780701824429 [DOI] [PubMed] [Google Scholar]
- 344.Esposto MC, Cogliati M, Tortorano AM, Viviani MA. 2004. Determination of Cryptococcus neoformans var. neoformans mating type by multiplex PCR. Clin. Microbiol. Infect. 10:1092–1094. 10.1111/j.1469-0691.2004.00972.x [DOI] [PubMed] [Google Scholar]
- 345.Carvalho VG, Terceti MS, Dias AL, Paula CR, Lyon JP, de Siqueira AM, Franco MC. 2007. Serotype and mating type characterization of Cryptococcus neoformans by multiplex PCR. Rev. Soc. Bras. Med. Trop. 49:207–210. 10.1590/S0036-46652007000400002 [DOI] [PubMed] [Google Scholar]
- 346.Feng X, Fu X, Ling B, Wang L, Liao W, Yao Z. 2013. Development of a singleplex PCR assay for rapid identification and differentiation of Cryptococcus neoformans var. grubii, Cryptococcus neoformans var. neoformans, Cryptococcus gattii, and hybrids. J. Clin. Microbiol. 51:1920–1923. 10.1128/JCM.00064-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Kaocharoen S, Wang B, Tsui KM, Trilles L, Kong F, Meyer W. 2008. Hyperbranched rolling circle amplification as a rapid and sensitive method for species identification within the Cryptococcus species complex. Electrophoresis 29:3183–3191. 10.1002/elps.200700903 [DOI] [PubMed] [Google Scholar]
- 348.Demidov VV. 2002. Rolling-circle amplification in DNA diagnostics: the power of simplicity. Expert Rev. Mol. Diagn. 2:542–548. 10.1586/14737159.2.6.542 [DOI] [PubMed] [Google Scholar]
- 349.Zhou X, Kong F, Sorrell TC, Wang H, Duan Y, Chen SCA. 2008. Practical method for detection and identification of Candida, Aspergillus, and Scedosporium spp. by use of rolling-circle amplification. J. Clin. Microbiol. 46:2423–2427. 10.1128/JCM.00420-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Diaz MR, Boekhout T, Kiesling T, Fell JW. 2005. Comparative analysis of the intergenic spacer regions and population structure of the species complex of the pathogenic yeast Cryptococcus neoformans. FEMS Yeast Res. 5:1129–1140. 10.1016/j.femsyr.2005.05.005 [DOI] [PubMed] [Google Scholar]
- 351.Bovers M, Diaz MR, Hagen F, Spanjaard L, Duim B, Visser CE, Hoogveld HL, Scharringa J, Hoepelman IM, Fell JW, Boekhout T. 2007. Identification of genotypically diverse Cryptococcus neoformans and Cryptococcus gattii isolates by Luminex xMAP technology. J. Clin. Microbiol. 45:1874–1883. 10.1128/JCM.00223-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.McTaggart LR, Lei E, Richardson SE, Hoang L, Fothergill A, Zhang SX. 2011. Rapid identification of Cryptococcus neoformans and Cryptococcus gattii by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 49:3050–3053. 10.1128/JCM.00651-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Clark AE, Kaleta EJ, Arora A, Wolk DM. 2013. Matrix-assisted laser desorption ionization–time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clin. Microbiol. Rev. 26:547–603. 10.1128/CMR.00072-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Pinto A, Halliday C, Zahra M, van Hal S, Olma T, Maszewska K, Iredell JR, Meyer W, Chen SCA. 2011. Matrix-assisted laser desorption ionization-time of flight mass spectrometry identification of yeasts is contingent on robust reference spectra. PLoS One 6:e25712. 10.1371/journal.pone.0025712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Calderaro A, Martinelli M, Motta F, Larini S, Arcangeletti MC, Medici MC, Chezzi C, De Conto F. 2014. Comparison of peptide nucleic acid fluorescence in situ hybridization assays with culture-based matrix-assisted laser desorption/ionization-time of flight mass spectrometry for the identification of bacteria and yeasts from blood cultures and cerebrospinal fluid cultures. Clin. Microbiol. Infect. 20:O468–O475. 10.1111/1469-0691.12490 [DOI] [PubMed] [Google Scholar]
- 356.CLSI. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts, CLSI document M27–A3. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 357.Espinel-Ingroff A, Chowdhary A, Cuenca-Estrella M, Fothergill A, Fuller J, Hagen F, Govender N, Guarro J, Johnson E, Lass-Florl C, Lockhart SR, Martins MA, Meis JF, Melhem MS, Ostrosky-Zeichner L, Pelaez T, Pfaller MA, Schell WA, Trilles L, Kidd S, Turnidge J. 2012. Cryptococcus neoformans-Cryptococcus gattii species complex: an international study of wild-type susceptibility endpoint distributions and epidemiological cutoff values for amphotericin B and flucytosine. Antimicrob. Agents Chemother. 56:3107–3113. 10.1128/AAC.06252-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Espinel-Ingroff A, Aller AI, Canton E, Castanon-Olivares LR, Chowdhary A, Cordoba S, Cuenca-Estrella M, Fothergill A, Fuller J, Govender N, Hagen F, Illnait-Zaragozi MT, Johnson E, Kidd S, Lass-Florl C, Lockhart SR, Martins MA, Meis JF, Melhem MS, Ostrosky-Zeichner L, Pelaez T, Pfaller MA, Schell WA, St-Germain G, Trilles L, Turnidge J. 2012. Cryptococcus neoformans-Cryptococcus gattii species complex: an international study of wild-type susceptibility endpoint distributions and epidemiological cutoff values for fluconazole, itraconazole, posaconazole, and voriconazole. Antimicrob. Agents Chemother. 56:5898–5906. 10.1128/AAC.01115-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Lockhart SR, Iqbal N, Bolden CB, DeBess EE, Marsden-Haug N, Worhle R, Thakur R, Harris JR, Cryptococcus gattii PNW Public Health Working Group 2012. Epidemiologic cutoff values for triazole drugs in Cryptococcus gattii: correlation of molecular type and in vitro susceptibility. Diagn. Microbiol. Infect. Dis. 73:144–148. 10.1016/j.diagmicrobio.2012.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Pfaller MA, Castanheira M, Diekema DJ, Messer SA, Jones RN. 2011. Wild-type MIC distributions and epidemiologic cutoff values for fluconazole, posaconazole, and voriconazole when testing Cryptococcus neoformans as determined by the CLSI broth microdilution method. Diagn. Microbiol. Infect. Dis. 71:252–259. 10.1016/j.diagmicrobio.2011.07.007 [DOI] [PubMed] [Google Scholar]
- 361.Pfaller MA, Castanheira M, Messer SA, Moet GJ, Jones RN. 2011. Echinocandin and triazole antifungal susceptibility profiles for Candida spp., Cryptococcus neoformans, and Aspergillus fumigatus: application of new CLSI clinical breakpoints and epidemiologic cutoff values to characterize resistance in the SENTRY Antimicrobial Surveillance Program (2009). Diagn. Microbiol. Infect. Dis. 69:45–50. 10.1016/j.diagmicrobio.2010.08.013 [DOI] [PubMed] [Google Scholar]
- 362.Ellis D, Sorrell T, Chen S. 2007. Impact of antifungal resistance in Australia. Microbiol. Aust. 28:171–173 [Google Scholar]
- 363.Thompson GR, Wiederhold NP, Fothergill AW, Vallor AC, Wickes BL, Patterson TF. 2009. Antifungal susceptibilities among different serotypes of Cryptococcus gattii and Cryptococcus neoformans. Antimicrob. Agents Chemother. 53:309–311. 10.1128/AAC.01216-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Pfaller MA, Messer SA, Boyken L, Rice C, Tendolkar S, Hollis RJ, Doern GV, Diekema DJ. 2005. Global trends in the antifungal susceptibility of Cryptococcus neoformans (1990 to 2004). J. Clin. Microbiol. 43:2163–2167. 10.1128/JCM.43.5.2163-2167.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Tay ST, Tanty Haryanty T, Ng KP, Rohani MY, Hamimah H. 2006. In vitro susceptibilities of Malaysian clinical isolates of Cryptococcus neoformans var. grubii and Cryptococcus gattii to five antifungal drugs. Mycoses 49:324–330. 10.1111/j.1439-0507.2006.01242.x [DOI] [PubMed] [Google Scholar]
- 366.Thompson GR, Fothergill AW, Wiederhold NP, Vallor AC, Wickes BL, Patterson TF. 2008. Evaluation of Etest method for determining isavuconazole MICs against Cryptococcus gattii and Cryptococcus neoformans. Antimicrob. Agents Chemother. 52:2959–2961. 10.1128/AAC.00646-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Chowdhary A, Randhawa HS, Sundar G, Kathuria S, Prakash A, Khan Z, Sun S, Xu J. 2011. In vitro antifungal susceptibility profiles and genotypes of 308 clinical and environmental isolates of Cryptococcus neoformans var. grubii and Cryptococcus gattii serotype B from north-western India. J. Med. Microbiol. 60:961–967. 10.1099/jmm.0.029025-0 [DOI] [PubMed] [Google Scholar]
- 368.Torres-Rodriguez JM, Alvarado-Ramirez E, Murciano F, Sellart M. 2008. MICs and minimum fungicidal concentrations of posaconazole, voriconazole and fluconazole for Cryptococcus neoformans and Cryptococcus gattii. Antimicrob. Agents Chemother. 62:205–206. 10.1093/jac/dkn132 [DOI] [PubMed] [Google Scholar]
- 369.Andrade-Silva L, Ferreira-Paim K, Mora DJ, Da Silva PR, Andrade AA, Araujo NE, Pedrosa AL, Silva-Vergara ML. 2013. Susceptibility profile of clinical and environmental isolates of Cryptococcus neoformans and Cryptococcus gattii in Uberaba, Minas Gerais, Brazil. Med. Mycol. 51:635–640. 10.3109/13693786.2012.761737 [DOI] [PubMed] [Google Scholar]
- 370.Trilles L, Fernandez-Torres B, Lazera MS, Wanke B, Guarro J. 2004. In vitro antifungal susceptibility of Cryptococcus gattii. J. Clin. Microbiol. 42:4815–4817. 10.1128/JCM.42.10.4815-4817.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Iqbal N, DeBess EE, Wohrle R, Sun B, Nett RJ, Ahlquist AM, Chiller T, Lockhart SR, Cryptococcus gattii Public Health Working Group 2010. Correlation of genotype and in vitro susceptibilities of Cryptococcus gattii strains from the Pacific Northwest of the United States. J. Clin. Microbiol. 48:539–544. 10.1128/JCM.01505-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Trilles L, Meyer W, Wanke B, Guarro J, Lazera M. 2012. Correlation of antifungal susceptibility and molecular type within the Cryptococcus neoformans/C. gattii species complex. Med. Mycol. 50:328–332. 10.3109/13693786.2011.602126 [DOI] [PubMed] [Google Scholar]
- 373.Hagen F, Illnait-Zaragozi MT, Bartlett KH, Swinne D, Geertsen E, Klaassen CH, Boekhout T, Meis JF. 2010. In vitro antifungal susceptibilities and amplified fragment length polymorphism genotyping of a worldwide collection of 350 clinical, veterinary, and environmental Cryptococcus gattii isolates. Antimicrob. Agents Chemother. 54:5139–5145. 10.1128/AAC.00746-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Datta K, Rhee P, Byrnes E, III, Garcia-Effron G, Perlin DS, Staab JF, Marr KA. 2013. Isavuconazole activity against Aspergillus lentulus, Neosartorya udagawae, and Cryptococcus gattii, emerging fungal pathogens with reduced azole susceptibility. J. Clin. Microbiol. 51:3090–3093. 10.1128/JCM.01190-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Chong HS, Dagg R, Malik R, Chen S, Carter D. 2010. In vitro susceptibility of the yeast pathogen Cryptococcus to fluconazole and other azoles varies with molecular genotype. J. Clin. Microbiol. 48:4115–4120. 10.1128/JCM.01271-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Morales BP, Neves Junior I, Trilles L, Bertho AL, Oliveira RD, Nishikawa MM, Elias MD, Wanke B, Lazera MD. 2014. Determination of the minimum inhibitory concentration of Cryptococcus neoformans and Cryptococcus gattii against fluconazole by flow cytometry. Med. Mycol. 52:90–98. 10.3109/13693786.2013.806827 [DOI] [PubMed] [Google Scholar]
- 377.Rodero L, Mellado E, Rodriguez AC, Salve A, Guelfand L, Cahn P, Cuenca-Estrella M, Davel G, Rodriguez-Tudela JL. 2003. G484S amino acid substitution in lanosterol 14-alpha demethylase (ERG11) is related to fluconazole resistance in a recurrent Cryptococcus neoformans clinical isolate. Antimicrob. Agents Chemother. 47:3653–3656. 10.1128/AAC.47.11.3653-3656.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Sionov E, Lee H, Chang YC, Kwon-Chung KJ. 2010. Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathog. 6:e1000848. 10.1371/journal.ppat.1000848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Morschhauser J. 2002. The genetic basis of fluconazole resistance development in Candida albicans. Biochim. Biophys. Acta 1587:240–248. 10.1016/S0925-4439(02)00087-X [DOI] [PubMed] [Google Scholar]
- 380.Sanguinetti M, Posteraro B, La Sorda M, Torelli R, Fiori B, Santangelo R, Delogu G, Fadda G. 2006. Role of AFR1, an ABC transporter-encoding gene, in the in vivo response to fluconazole and virulence of Cryptococcus neoformans. Infect. Immun. 74:1352–1359. 10.1128/IAI.74.2.1352-1359.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Gast CE, Basso LR, Jr, Bruzual I, Wong B. 2013. Azole resistance in Cryptococcus gattii from the Pacific Northwest: investigation of the role of ERG11. Antimicrob. Agents Chemother. 57:5478–5485. 10.1128/AAC.02287-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Cannon RD, Lamping E, Holmes AR, Niimi K, Baret PV, Keniya MV, Tanabe K, Niimi M, Goffeau A, Monk BC. 2009. Efflux-mediated antifungal drug resistance. Clin. Microbiol. Rev. 22:291–321. 10.1128/CMR.00051-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Posteraro B, Sanguinetti M, Sanglard D, La Sorda M, Boccia S, Romano L, Morace G, Fadda G. 2003. Identification and characterization of a Cryptococcus neoformans ATP binding cassette (ABC) transporter-encoding gene, CnAFR1, involved in the resistance to fluconazole. Mol. Microbiol. 47:357–371. 10.1046/j.1365-2958.2003.03281.x [DOI] [PubMed] [Google Scholar]
- 384.Mondon P, Petter R, Amalfitano G, Luzzati R, Concia E, Polacheck I, Kwon-Chung KJ. 1999. Heteroresistance to fluconazole and voriconazole in Cryptococcus neoformans. Antimicrob. Agents Chemother. 43:1856–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Sionov E, Chang YC, Garraffo HM, Kwon-Chung KJ. 2009. Heteroresistance to fluconazole in Cryptococcus neoformans is intrinsic and associated with virulence. Antimicrob. Agents Chemother. 53:2804–2815. 10.1128/AAC.00295-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Chen S, Marr KA, Sorrell TC. 2013. Cryptococcus gattii infection—treatment. In Kauffman CA. (ed), UpToDate. UpToDate, Inc, Waltham, MA: http://www.uptodate.com/contents/cryptococcus-gattii-infection-treatment [Google Scholar]
- 387.Seaton RA, Verma N, Naraqi S, Wembri JP, Warrell DA. 1997. The effect of corticosteroids on visual loss in Cryptococcus neoformans var. gattii meningitis. Trans. R. Soc. Trop. Med. Hyg. 91:50–52. 10.1016/S0035-9203(97)90393-X [DOI] [PubMed] [Google Scholar]
- 388.Chen S, Marr KA, Sorrell TC. 2013. Cryptococcus gattii infection: clinical features and diagnosis. In Basow DS. (ed), UpToDate. UpToDate, Inc, Waltham, MA [Google Scholar]
- 389.Bolton LA, Lobetti RG, Evezard DN, Picard JA, Nesbit JW, van Heerden J, Burroughs RE. 1999. Cryptococcosis in captive cheetah (Acinonyx jubatus): two cases. J. S. Afr. Vet. Assoc. 70:35–39 [DOI] [PubMed] [Google Scholar]
- 390.Narasipura SD, Ault JG, Behr MJ, Chaturvedi V, Chaturvedi S. 2003. Characterization of Cu,Zn superoxide dismutase (SOD1) gene knock-out mutant of Cryptococcus neoformans var. gattii: role in biology and virulence. Mol. Microbiol. 47:1681–1694. 10.1046/j.1365-2958.2003.03393.x [DOI] [PubMed] [Google Scholar]
- 391.Narasipura SD, Chaturvedi V, Chaturvedi S. 2005. Characterization of Cryptococcus neoformans var. gattii SOD2 reveals distinct roles of the two superoxide dismutases in fungal biology and virulence. Mol. Microbiol. 55:1782–1800. 10.1111/j.1365-2958.2005.04503.x [DOI] [PubMed] [Google Scholar]
- 392.Petzold EW, Himmelreich U, Mylonakis E, Rude T, Toffaletti D, Cox GM, Miller JL, Perfect JR. 2006. Characterization and regulation of the trehalose synthesis pathway and its importance in the pathogenicity of Cryptococcus neoformans. Infect. Immun. 74:5877–5887. 10.1128/IAI.00624-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Ngamskulrungroj P, Himmelreich U, Breger JA, Wilson C, Chayakulkeeree M, Krockenberger MB, Malik R, Daniel HM, Toffaletti D, Djordjevic JT, Mylonakis E, Meyer W, Perfect JR. 2009. The trehalose synthesis pathway is an integral part of the virulence composite for Cryptococcus gattii. Infect. Immun. 77:4584–4596. 10.1128/IAI.00565-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.D'Souza CA, Alspaugh JA, Yue C, Harashima T, Cox GM, Perfect JR, Heitman J. 2001. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol. Cell. Biol. 21:3179–3191. 10.1128/MCB.21.9.3179-3191.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Hicks JK, Heitman J. 2007. Divergence of protein kinase A catalytic subunits in Cryptococcus neoformans and Cryptococcus gattii illustrates evolutionary reconfiguration of a signaling cascade. Eukaryot. Cell 6:413–420. 10.1128/EC.00213-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Chayakulkeeree M, Sorrell TC, Siafakas AR, Wilson CF, Pantarat N, Gerik KJ, Boadle R, Djordjevic JT. 2008. Role and mechanism of phosphatidylinositol-specific phospholipase C in survival and virulence of Cryptococcus neoformans. Mol. Microbiol. 69:809–826. 10.1111/j.1365-2958.2008.06310.x [DOI] [PubMed] [Google Scholar]
- 397.Kraus PR, Fox DS, Cox GM, Heitman J. 2003. The Cryptococcus neoformans MAP kinase Mpk1 regulates cell integrity in response to antifungal drugs and loss of calcineurin function. Mol. Microbiol. 48:1377–1387. 10.1046/j.1365-2958.2003.03508.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Ren P, Springer DJ, Behr MJ, Samsonoff WA, Chaturvedi S, Chaturvedi V. 2006. Transcription factor STE12alpha has distinct roles in morphogenesis, virulence, and ecological fitness of the primary pathogenic yeast Cryptococcus gattii. Eukaryot. Cell 5:1065–1080. 10.1128/EC.00009-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Yue C, Cavallo LM, Alspaugh JA, Wang P, Cox GM, Perfect JR, Heitman J. 1999. The STE12alpha homolog is required for haploid filamentation but largely dispensable for mating and virulence in Cryptococcus neoformans. Genetics 153:1601–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Ngamskulrungroj P, Chang Y, Roh J, Kwon-Chung KJ. 2012. Differences in nitrogen metabolism between Cryptococcus neoformans and C. gattii, the two etiologic agents of cryptococcosis. PLoS One 7:e34258. 10.1371/journal.pone.0034258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Odom A, Muir S, Lim E, Toffaletti DL, Perfect J, Heitman J. 1997. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 16:2576–2589. 10.1093/emboj/16.10.2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Cruz MC, Sia RA, Olson M, Cox GM, Heitman J. 2000. Comparison of the roles of calcineurin in physiology and virulence in serotype D and serotype A strains of Cryptococcus neoformans. Infect. Immun. 68:982–985. 10.1128/IAI.68.2.982-985.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Calvo BM, Colombo AL, Fischman O, Santiago A, Thompson L, Lazera M, Telles F, Fukushima K, Nishimura K, Tanaka R, Myiajy M, Moretti-Branchini ML. 2001. Antifungal susceptibilities, varieties, and electrophoretic karyotypes of clinical isolates of Cryptococcus neoformans from Brazil, Chile, and Venezuela. J. Clin. Microbiol. 39:2348–2350. 10.1128/JCM.39.6.2348-2350.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Gomez-Lopez A, Zaragoza O, Dos Anjos Martins M, Melhem MC, Rodriguez-Tudela JL, Cuenca-Estrella M. 2008. In vitro susceptibility of Cryptococcus gattii clinical isolates. Clin. Microbiol. Infect. 14:727–730. 10.1111/j.1469-0691.2008.02021.x [DOI] [PubMed] [Google Scholar]
- 405.Matos CS, de Souza Andrade A, Oliveira NS, Barros TF. 2012. Microbiological characteristics of clinical isolates of Cryptococcus spp. in Bahia, Brazil: molecular types and antifungal susceptibilities. Eur. J. Clin. Microbiol. Infect. Dis. 31:1647–1652. 10.1007/s10096-011-1488-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Silva DC, Martins MA, Szeszs MW, Bonfietti LX, Matos D, Melhem MS. 2012. Susceptibility to antifungal agents and genotypes of Brazilian clinical and environmental Cryptococcus gattii strains. Diagn. Microbiol. Infect. Dis. 72:332–339. 10.1016/j.diagmicrobio.2011.11.016 [DOI] [PubMed] [Google Scholar]
- 407.Zhu P, Zhai B, Lin X, Idnurm A. 2013. Congenic strains for genetic analysis of virulence traits in Cryptococcus gattii. Infect. Immun. 81:2616–2625. 10.1128/IAI.00018-13 [DOI] [PMC free article] [PubMed] [Google Scholar]