Abstract
SUMMARY
Surface-associated microbial communities, called biofilms, are present in all environments. Although biofilms play an important positive role in a variety of ecosystems, they also have many negative effects, including biofilm-related infections in medical settings. The ability of pathogenic biofilms to survive in the presence of high concentrations of antibiotics is called “recalcitrance” and is a characteristic property of the biofilm lifestyle, leading to treatment failure and infection recurrence. This review presents our current understanding of the molecular mechanisms of biofilm recalcitrance toward antibiotics and describes how recent progress has improved our capacity to design original and efficient strategies to prevent or eradicate biofilm-related infections.
INTRODUCTION
For centuries, humankind suffered from acute bacterial infections and life-threatening diseases caused by pathogens such as Streptococcus pneumoniae, Vibrio cholerae, and Yersinia pestis. The discovery and use of hygiene, antibiotics, and vaccines led to massive reductions in the burden of and mortality related to such infections, mainly caused by individualized pathogenic bacteria (1, 2). Following this “antibiotic golden age,” physicians confronted two major challenges: the occurrence and spread of antibiotic-resistant bacteria and the rise of chronic, difficult-to-eradicate infections (3). Indeed, D. Holsclaw, referring to cystic fibrosis (CF) patients in 1980, stated that “even with the use of large doses of parental antibiotics, Pseudomonas cannot be eradicated from the sputum” (4).
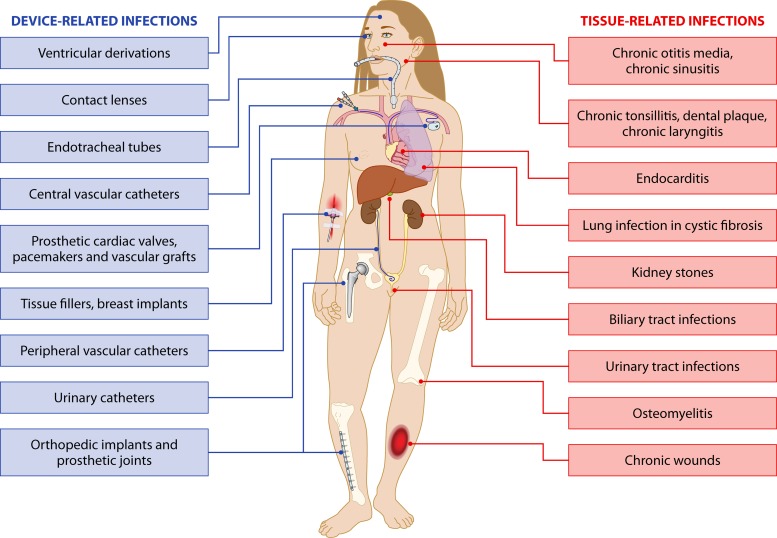
Meanwhile, environmental microbiologists have progressively established that surface-associated bacterial biofilm communities are widespread in all types of natural environments, where they often prevail, in contrast to individualized, planktonic bacteria (5–8). While biofilms display specific biological properties compared with planktonic bacteria, N. Høiby, J. W. Costerton, and their collaborators were the first to suspect a direct correlation between development of biofilms and persistent infections, notably in the case of Pseudomonas aeruginosa colonizing the lungs of CF patients (9, 10). Consistently, the decades that followed confirmed the role of biofilms in the pathophysiology of tissue-related infections (Fig. 1) (11). Furthermore, it was recognized that the widespread use of various types of indwelling medical devices implanted in humans could favor microorganism adhesion and cause colonization, leading to infection. In this regard, the first evidence of the involvement of biofilms in device-related infections was provided in 1982 by an electron microscopy study of a pacemaker lead in a patient with recurrent Staphylococcus aureus bloodstream infection (BSI) (12). Since then, almost all types of indwelling devices have been associated with the occurrence of bacterial or fungal biofilms (Fig. 1) (2).
FIG 1.
Biofilm-related infections. (Adapted from reference 365 with permission of the publisher and from reference 11.)
Due to their high tolerance toward antibiotics, these chronic tissue-related and device-related infections are difficult to treat and expose the patient to the risk of recurrence (13, 14). During a biofilm-related infection, planktonic bacteria originating from the biofilm can spread into the bloodstream or around the source of the infection (13, 14). Whereas planktonic bacteria can be eradicated via the combined action of antimicrobials and cellular and humoral host immune responses, a subset of highly tolerant biofilm bacteria frequently survive the treatment and can cause infection recurrence. In most cases, removal of the colonized device or surgical excision of infected tissue is the only efficient way to eradicate a biofilm-related infection (1, 13). Hence, the ability of biofilm bacteria to withstand antibiotics significantly influences the outcome and management of patients (1).
This review provides a description of the mechanisms involved in the capacity of bacterial biofilms to survive in the presence of antibiotics and presents recent therapeutic approaches developed to specifically target biofilm-related infections.
MECHANISMS OF BIOFILM RECALCITRANCE TOWARD ANTIBIOTICS
Once a biofilm is established, bacteria are able to survive after various types of physicochemical aggression, including UV light, heavy metals, acidity, changes in hydration or salinity, and phagocytosis (15–19). In addition, biofilm bacteria also display a characteristic ability to withstand antibiotic-mediated killing, which is directly responsible for a significant number of therapeutic difficulties encountered in clinical settings.
It is now clear that well-studied mechanisms involved in classical antibiotic resistance, such as efflux or antibiotic-modifying enzymes, play only a marginal role in the ability of biofilms to survive antibiotics (20, 21). Indeed, bacteria embedded in a biofilm are able to partly withstand high concentrations of bactericidal antibiotics even when these bacteria are fully susceptible to such antibiotics in vitro under planktonic conditions (22). This phenomenon, here named “recalcitrance of biofilm bacteria toward antibiotics,” is complex and is due to several tolerance and resistance mechanisms, as described below.
Tolerance and Resistance: Biofilm Recalcitrance Defined
The study of how in vitro planktonic bacteria escape antibiotic treatment led to the definition of two different concepts: resistance and tolerance.
Resistance: how to grow in the presence of an antibiotic.
Resistance can be defined as the ability of a microorganism to multiply in the presence of a toxic compound (antibiotic or antiseptic) and can be applied to both bacteriostatic and bactericidal antibiotics (14, 23–25). Resistance is usually tested by measuring the MIC of a compound, i.e., the lowest concentration inhibiting growth of a standardized inoculum of exponentially growing bacteria. A bacterium is more resistant toward an antibiotic if its MIC is higher than that of another bacterial strain. Numerous mechanisms explain this phenomenon, including antibiotic efflux, reduced permeability to antibiotics, activities of enzymes that modify or destroy antibiotics, and modification of the antibiotic target (through mutation, enzymatic action, or the presence of an alternate target) (21). Generally speaking, these resistance mechanisms avoid interactions between the antibiotic and its target, thereby allowing bacteria to multiply in the presence of the antibiotic (13). Resistance is often genetically inherited and therefore transmitted from mother to daughter bacteria, or it can be acquired through horizontal gene transfer.
Tolerance: how to avoid antibiotic-induced cell death.
In contrast to resistance, tolerance can only be associated with the use of bactericidal antibiotics, i.e., an antibiotic able to kill at least 99.9% of a bacterial population after overnight incubation (14). The lowest antibiotic concentration that enables reaching this threshold is called the minimal bactericidal concentration (MBC), according to the Clinical and Laboratory Standards Institute (CLSI) (14).
Thus, tolerance can be defined as the absence of growth but the existence of bacterial survival in the presence of a bactericidal antibiotic (13, 26, 27). Hence, a tolerant bacterial strain may be susceptible to a bactericidal antibiotic, as indicated by an unmodified MIC, while at the same time displaying increased survival, as defined by an MBC/MIC ratio of ≥32 or a kill rate of ≤99.9% after a 24-h challenge (28).
Two types of tolerance have been described: genotypic and phenotypic. In the first case, the presence of a genetic modification leads to a reduced ability of the antibiotic to kill the bacteria and can be transmitted to daughter cells. Examples have been described for Streptococcus pneumoniae and for small-colony variants (SCV) of Staphylococcus aureus (26, 29). In the case of phenotypic tolerance, the environment is not favorable to the action of antibiotics, thus leading to a decreased ability to kill. This is the case for nongrowing bacteria that are tolerant to β-lactams despite a normal MIC. Phenotypic tolerance is therefore rapidly reversible after the return to a growth-promoting medium (28).
Biofilm recalcitrance: a problematic mixture of resistance and tolerance.
In the study of biofilms, bacterial survival is often determined after an antibiotic challenge. This phenotype is therefore closer to the definition of tolerance than to that of resistance, as biofilm bacteria do not grow but a subset of them is able to survive in the presence of high concentrations of bactericidal antibiotics (up to 1,000× MIC) (14, 22, 27, 30). However, we will see that in addition to tolerance, resistance mechanisms sensu stricto also contribute to biofilm survival against antibiotics. Therefore, neither of these two concepts fully applies to biofilms. Recalcitrance, on the other hand, was previously proposed to characterize the capacity of biofilm bacteria to withstand treatment (31, 32). Since the word “recalcitrance” covers the notion of nonsusceptibility to (antimicrobial) control of refractory biofilms, we use it here to characterize the ability of a subset of biofilm bacteria to survive in the presence of antibiotics. Biofilm recalcitrance is reversible and mainly noninherited, and it disappears when the biofilm is disrupted and bacteria return to a planktonic state (20, 33).
Biofilm Recalcitrance Is Multifactorial
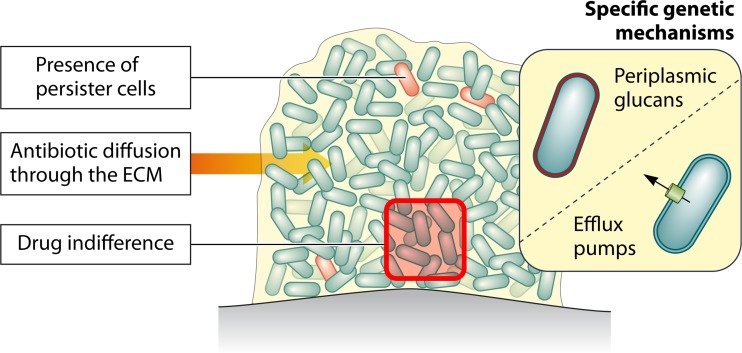
Recent studies on the ability of bacterial biofilms to survive high concentrations of antibiotics led to a complete shift in our understanding of mechanisms involved in biofilm recalcitrance. This phenomenon is multifactorial and, depending on the class of antibiotic used, involves different mechanisms, including impaired antibiotic diffusion, drug indifference, expression of biofilm-specific genetic mechanisms, and the presence of persister cells (Fig. 2).
FIG 2.
Summary of the main mechanisms involved in recalcitrance of biofilms toward antibiotics. (Adapted from reference 366 with permission of the publisher.)
Antibiotic penetration.
Historically, it was proposed that the extracellular matrix (ECM) surrounding bacteria was responsible for biofilm recalcitrance. Many reports suggested that mechanical and physicochemical properties of the biofilm matrix can reduce or delay penetration of numerous compounds, including antibiotics and antiseptics (34, 35). For instance, the effect of an antibiotic can be reduced after adsorption on the matrix due to electrical interactions with polymers surrounding biofilm bacteria (20, 36). Other studies reported slow penetration of positively charged aminoglycosides through negatively charged polymers of the biofilm matrix (37, 38). In this regard, the chemical structure of the biofilm matrix is important, and it has been shown that even for a single pathogen, different types of exopolysaccharides can be involved, depending on the environment surrounding the biofilm (39). Monitoring of antibiotic diffusion through cardiac vegetation in an in vivo model of endocarditis demonstrated that a diffusion gradient could be observed in the case of teicoplanin (a glycopeptide) and penicillin (40). In contrast, tobramycin was shown to be homogeneously distributed. Conversely, in the case of P. aeruginosa biofilm, tobramycin was shown to exhibit delayed and reduced diffusion in vitro (41, 42). Thus, experimental data regarding the diffusion of an antibiotic through the biofilm matrix cannot be extrapolated to another bacterial strain and should be interpreted carefully (Table 1).
TABLE 1.
Penetration of antibiotics through thebiofilm extracellular matrix
| Microorganism | Antibiotic | Penetration | Reference(s) |
|---|---|---|---|
| P. aeruginosa | Piperacillin | Reduced/yes | 367, 60 |
| Imipenem | Yes | 60 | |
| Ofloxacin | Yes | 60, 368 | |
| Ciprofloxacin | Yes | 45, 60, 369, 370, 42 | |
| Levofloxacin | Yes | 60, 369, 370 | |
| Sparfloxacin | Yes | 60 | |
| Gentamicin | Reduced | 60 | |
| Amikacin | Reduced | 60 | |
| Tobramycin | Reduced | 41, 42 | |
| Amoxicillin-clavulanic acid | Yes | 45 | |
| Fosfomycin | Yes | 45 | |
| Clarithromycin | Yes | 368 | |
| E. coli | Moxalactam | Yes | 371 |
| Fosfomycin | Yes | 45 | |
| Amoxicillin-clavulanic acid | Yes | 45 | |
| Ciprofloxacin | Yes | 45 | |
| K. pneumoniae | Ampicillin | No | 22 |
| Ciprofloxacin | Yes | 22 | |
| S. epidermidis | Rifampin | Yes | 372, 46 |
| Vancomycin | Yes | 373, 372 | |
| Ciprofloxacin | Yes | 374 | |
| Ofloxacin | Yes | 368 | |
| Clarithromycin | Yes | 368 | |
| Daptomycin | Yes | 228 | |
| Cefotaxime | Reduced | 374 | |
| Oxacillin | Reduced | 374 | |
| Cefotiam | Yes | 368 | |
| Amikacin | Yes | 374 | |
| S. aureus | Vancomycin | Yes/reduced | 48, 374 |
| Cefotaxime | Reduced | 374 | |
| Oxacillin | Reduced | 374 | |
| Ciprofloxacin | Yes | 374 | |
| Amikacin | Yes | 374 |
The study of chlorine antiseptic diffusion by use of microelectrodes showed that the chlorine concentration in the bulk of a mixed biofilm (P. aeruginosa and Klebsiella pneumoniae) represented only 20% of the concentration in the bulk liquid after 2 h (43). These results were confirmed using bacteria entrapped in alginate beads, with a time to reach 50% of the chlorine bulk concentration at the bead center of approximately 46 h (44).
However, many reports also suggested that reduction of antibiotic penetration cannot fully explain biofilm recalcitrance toward antibiotics. Indeed, antibiotics such as fluoroquinolones, rifampin, and ampicillin penetrate well through the matrix, even though they fail to eradicate 100% of biofilm bacteria (22, 42, 45, 46). Moreover, even in the case of compounds slowly diffusing within biofilms, most antibiotics ultimately reach all biofilm bacteria. For instance, P. aeruginosa and Escherichia coli 24-h in vitro biofilms were not eradicated by a 24-h treatment with fosfomycin or ciprofloxacin, whereas these drugs reached more than 50% of the bulk concentration within 6 h (45). The same observation was made concerning K. pneumoniae biofilms and ciprofloxacin (22). Studies using fluorescent tetracycline demonstrated that 2-day biofilms were less susceptible than planktonic bacteria, whereas tetracycline-mediated fluorescence was present throughout the biofilm within 10 min (47).
On the other hand, delayed antibiotic penetration may have important phenotypic consequences. For instance, bacterial cell physiology could adapt to the presence of antibiotics though metabolic or transcriptional adaptation induced by antibiotic stress (48). Furthermore, due to slow diffusion, biofilm bacteria could be transiently exposed to subinhibitory concentrations of antibiotics (see below). Limited diffusion can also protect biofilms from degradable antimicrobials. Indeed, P. aeruginosa produces AmpC β-lactamase, and it has been demonstrated that 2.5% of clinical isolates from CF patient sputa are totally derepressed, with a high basal level of enzyme production that can be increased further through β-lactam-mediated induction and β-lactamase accumulation in the biofilm matrix (49, 50). In clinical samples, insertion sequences inactivating the ampD gene have been described for CF patients with constitutively high expression of β-lactamase (51). The association of a diffusion barrier that slows down diffusion of β-lactams and the presence of a hydrolyzing enzyme may act synergistically, especially if the enzymes degrade antibiotics faster than they diffuse (52–54).
Drug indifference and an altered microenvironment.
Deep biofilm layers correspond to a particular physicochemical microenvironment due to various gradients of nutrients, waste, pH, oxygen, and metabolic by-products through the ECM (55). Since many antibiotics are more active against bacteria that are metabolically active and growing, the characteristic lack of nutrients or anoxia of these microenvironments can antagonize the effects of antibiotics (56–58). This is the case for β-lactam antibiotics, which target the bacterial membrane and are effective only against actively growing bacteria undergoing cell division (14, 59). Consistently, the effects of β-lactams against P. aeruginosa biofilms have been correlated with the metabolic activity of biofilm bacteria (60, 61). Similarly, the low oxygen concentrations found in deep layers of P. aeruginosa biofilms reduce tobramycin and ciprofloxacin bactericidal effects (42). Other physicochemical characteristics can impair the effects of antibiotics, such as low pH and the anaerobic environment, leading to decreased activities of aminoglycosides (20, 62–64).
However, a reduction in β-lactams or in aminoglycoside antibiotic efficacy in altered microenvironments does not fully account for the observed biofilm recalcitrance toward antibiotics that are active against nongrowing bacteria, such as fluoroquinolones (61, 65, 66).
Contributions of genetically determined mechanisms.
Many investigators have tried to identify genetic mechanisms of recalcitrance specifically activated during the biofilm lifestyle. To do so, Mah and collaborators used a random transposon insertion library screened for P. aeruginosa mutants, making biofilms more sensitive to tobramycin, and they identified 3 genes or operons: ndvB, PA1875 to PA1877 (PA1875–PA1877), and tssC1 (67). The first gene identified, ndvB, encodes a putative glucosyltransferase that was later shown to be required for synthesis of cyclic-β-(1,3)-glucans (68). Periplasmic glucans interact with and sequester aminoglycosides in the periplasm and keep them away from their intracellular target. Note that these glucans are also secreted into the biofilm matrix. An ndvB mutant was also more sensitive to ciprofloxacin, ofloxacin, gentamicin, and chloramphenicol, suggesting that induction of ndvB was also involved in recalcitrance toward other classes of antibiotics. In that case, periplasmic glucans inhibit the interaction between an antibiotic and its target. Interestingly, ndvB seemed also to contribute to P. aeruginosa antibiotic tolerance by an unknown mechanism involving increased ethanol oxidation (69), therefore suggesting that the ndvB action is the sum of pleiotropic effects. The second locus identified, PA1875–PA1877, corresponds to a 3-gene operon coding for an outer membrane protein, an ATP binding cassette transporter, and a membrane fusion protein (70). Deletion of these genes resulted in biofilms with increased sensitivity to tobramycin and ciprofloxacin. Moreover, a deletion mutant accumulated more tobramycin than the wild type, suggesting that the identified locus could code for an efflux pump. The last identified gene, tssC1, was shown to be an essential component of the type VI secretion (T6S) system potentially involved in cell-to-cell interactions (71). More recently, three other loci, i.e., PA0756-PA0757 (encoding a putative two-component regulatory system), PA2070, and PA5033 (both encoding hypothetical proteins of unknown function), were shown to contribute to the biofilm-specific antibiotic tolerance of P. aeruginosa (72).
Several studies performed in P. aeruginosa and E. coli suggested that efflux pumps, induced specifically under biofilm conditions and removing antibiotics from the bacterial intracytoplasmic space, could be involved in biofilm-specific recalcitrance. In P. aeruginosa biofilms, MexAB-OprM and MexCD-OprJ pumps are involved in the efflux of azithromycin, colistin, and ofloxacin, but in the latter case, only at low concentrations (73–75). Interestingly, MexEF-OprN and MexXY-OprM are upregulated in response to reactive oxygen species (ROS) and could help in removing cellular elements damaged by ROS (76). From the sputa of CF patients, highly tolerant P. aeruginosa strains with mutations in the mexZ repressor controlling expression of the MexXY-OprM pump have been isolated (77). Strikingly, deletions in the mexXY locus restored wild-type resistance but did not affect antibiotic tolerance, suggesting that MexXY-OprM plays only a marginal role in biofilm recalcitrance (77). Recently, the DNA-binding transcriptional regulator BrlR was shown to contribute to P. aeruginosa tolerance (78). When inactivated, biofilm bacteria are more susceptible to hydrogen peroxide and antibiotics of different classes, including tobramycin and norfloxacin. On the other hand, brlR overexpression increased P. aeruginosa tolerance toward antimicrobials. The same group identified BrlR as an activator of mexAB-oprM and mexEF-oprN (79) and the two-component hybrid protein SagS as a possible upstream regulator of BrlR (80). Recently, SagS was shown to contribute to BrlR activation and tolerance toward antibiotics through an increase of the level of the second messenger cyclic di-GMP (c-di-GMP) (81).
In E. coli, many efflux pumps, such as AcrAB-TolC, are upregulated in biofilms and may remove toxic compounds, including antibiotics (82). High-thoughput screening of E. coli mutants showed that rapA mutants displayed decreased resistance toward penicillin, norfloxacin, chloramphenicol, and gentamicin (83). It was demonstrated that rapA not only regulates yhcQ, a gene encoding a putative multidrug resistance (MDR) pump, but also yeeZ, a gene suspected to be involved in ECM production (83). Thus, a dual rapA-mediated action was proposed, with efflux through a pump and reduced penetration through an increase in polysaccharide production (83).
The hypothesis of biofilm-induced E. coli or P. aeruginosa efflux pumps preventing antibiotic action is attractive. However, these pumps play a role mostly at low antibiotic concentrations, and, to date, the involvement of biofilm-specific efflux pumps in biofilm recalcitrance has remained controversial (14, 70, 73, 74).
Bacterial persistence.
While the above-mentioned mechanisms play an important role in the inability of antibiotics to fully eradicate biofilm bacteria, they cannot fully explain biofilm recalcitrance. This is particularly clear in the case of fluoroquinolones: although these antibiotics are able to kill nondividing cells and diffuse easily throughout the biofilm matrix, experimental studies have demonstrated their inability to fully eradicate biofilms (27). The presence of an isogenic subpopulation of tolerant bacteria, called “persister cells” or “persisters,” is now considered to explain most of the biofilm recalcitrance toward antibiotics (31). The presence of persisters in the bacterial population has been known since the origin of the antibiotic era; indeed, Joseph Bigger identified them in a population of S. aureus planktonic bacteria 70 years ago. When Bigger analyzed what he called “variations in sterilizing power” by quantifying the precise number of surviving bacteria after 3 days of treatment, he recovered fewer than 100 surviving individuals from an initial population of 250,000,000 bacteria (0.00004% survival) (84). He then showed that this subpopulation of bacterial cells resumed growth after the end of antibiotic exposure and that they were not resistant mutants, since they exhibited the same survival pattern following another exposure to antibiotics. He concluded that “the only hypothesis which seems to explain the occurrence of a small number of survivors out of the millions of cocci originally present is that these differ from the majority of their fellows in that they are capable of surviving a concentration of penicillin which, in the time or action allowed, kills the others” (84). He called these survivors “persisters” and suggested that they were in a dormant and nondividing phase and that their production was not due to penicillin. Retrospectively, he described most of what constitutes our current knowledge of persisters (Fig. 3): (i) persisters make up less than 1% of a bacterial population and are equally present in late-stationary-phase cultures and biofilms; (ii) they do not multiply in the presence of antibiotics, and their phenotypic tolerance is not related to any genetic modifications but rather to a phenotypic state; (iii) they are isogenic toward nonpersisters; and (iv) once persisters resume growth, they display the original, nonpersister antibiotic tolerance profile.
FIG 3.
Main phenotypic characteristics of persister cells. (A) Persisters (red bacteria) are present under planktonic and biofilm conditions and account for only a small subset of the whole population (0.001% to 0.1%). (B) Persisters are not resistant mutants. After treatment of a bacterial population with a bactericidal antibiotic, all nonpersister cells die, giving a biphasic survival curve. After a rapid decrease, surviving cell fractions reach a plateau corresponding to persisters (red curve). After antibiotic removal and addition of rich medium, persisters resume growth. The population obtained displays a susceptible phenotype toward the antibiotic (blue curve). If a resistant mutant were present, it would be able to grow in the presence of the antibiotic (dotted line). Panel B was inspired by previous reports (13, 31, 94).
At the population level, the presence of persisters can be viewed as an insurance strategy (85). In case of intense stress for the community, persisters may survive and permit the survival of the community. It has also been proposed that persisters might be bacteria that escape antibiotic-induced programmed cell death (PCD). In that case, the antibiotic is able to interact with its target, thereby leading to growth inhibition; on the other hand, bacteria do not die because of inactivation of PCD (25).
Analysis of survival curves of antibiotic-treated biofilms suggested the presence of a subpopulation of tolerant bacteria surviving bactericidal antibiotics despite increased concentrations and times of exposure, leading to the hypothesis that persisters may play a part in biofilm recalcitrance toward antibiotics (73). Indeed, when exposed to increasing concentrations of fluoroquinolones (active against nondividing cells and without any diffusion impairment), biofilm bacteria are killed until reaching a survival plateau, thereby creating a biphasic curve such as those seen under planktonic conditions (Fig. 3). These tolerant bacteria are mostly persisters, and many in vitro, in vivo, and clinical studies support the idea that they are responsible for most of the antibiotic recalcitrance of biofilms.
Persisters Play a Central Role in Biofilm Recalcitrance toward Antibiotics
Persisters and clinical issues.
Several in vitro and in vivo studies demonstrated the presence of highly tolerant bacterial persisters in biofilms formed by Gram-positive and Gram-negative pathogens (86–89). Clinical demonstration of the presence of persisters can be inferred from the risk of infection recurrence during biofilm-related infections. For instance, in the case of catheter-related bloodstream infection (CRBSI), even after local treatment with high concentrations of antibiotics (up to 1,000-fold higher than the MIC) for 2 weeks, more than 20% of infections relapse because of the survival of persisters inside the biofilm (90). Thus, the currently proposed model to explain biofilm recalcitrance toward antibiotics relies mainly on the presence of persister cells (13, 14). For antibiotics such as fluoroquinolones, which freely diffuse through the matrix and kill nondividing bacteria, impaired antibiotic diffusion, drug indifference, and specific genetic mechanisms play minor roles in biofilm recalcitrance. Conversely, persisters are able to survive antibiotic-mediated bacterial cell death induced by any bactericidal antibiotics. Furthermore, persisters inside the biofilm matrix escape the effect of the host immune system. Once antibiotic treatment is withdrawn, persisters hiding in the matrix can resume growth, repopulate the biofilm, and cause infection recurrence.
The main mechanisms involved in persister generation.
Because persisters are isogenic and present prior to the introduction of antibiotics, they are now believed to appear through a phenotypic switch (91). Several factors and mechanisms have been described as playing important roles in the occurrence of this switch. Most studies on the molecular mechanisms involved in persister formation were conducted with planktonic rather than biofilm bacteria. It is now believed that the presence of persisters is related to both passive and active mechanisms, environmental factors, and stochastic gene expression.
(i) Contribution of dormancy to bacterial persistence.
Dormancy can be defined as a state of low metabolic activity during which bacteria do not proliferate without a resuscitation phase (92). Therefore, truly dormant cells do not display metabolic activity. Different lines of argument suggest a link between dormancy and persistence. By use of microfluid devices, E. coli persisters were shown to be nongrowing before the introduction of antibiotic (91). Furthermore, using an unstable fluorescent reporter gene associated with a ribosomal promoter (rrnBP1, which controls expression of rrnB genes expressed at high levels during growth), it was shown that a weakly fluorescent population (i.e., with low ribosomal activity) was enriched in persisters (93). Even when enrichment is significant, it is important that not all dormant bacteria are persisters; conversely, all persisters do not necessarily correspond to dormant cells (93).
Therefore, it is likely that passive dormancy per se is not entirely responsible for the persister phenotype (94). A recent study confirmed these findings by using flow cytometry sorting of E. coli cells based on their level of metabolic activity and/or cell division (95). The authors showed that bacteria that grow rapidly prior to antibiotic exposure can give rise to persisters, whereas low metabolic activity or a low growth rate only increases the odds of entry into persistence (95).
(ii) TA modules.
Expression of toxin-antitoxin (TA) modules often leads to a shutdown of bacterial cellular processes. Although the molecular nature of TA modules varies, from protein to RNA molecules, the toxin is usually a stable component that inhibits major cellular functions, such as translation and replication (96, 97). To keep a toxin in check, degradable antitoxin antagonizes the effect of the toxin through formation of an inactive complex. In the case of a TA module carried by a plasmid, after cell division, newly formed daughter cells die unless receiving the plasmid, as the antitoxin will be degraded through proteolysis, allowing the toxin to exert its deleterious effects in plasmid-free bacteria. Such a system allows maintenance of the plasmid and was previously referred to as an “addiction module” (98). In E. coli, at least 36 putative TA modules have been identified (96, 99). Since toxins halt growth and thus reduce the activity of the antibiotic target, they appear to be attractive effectors of the switch to the persister state (100, 101).
In E. coli, the first TA locus associated with an increased level of persister production was hip (for “high persister”), identified through random mutagenesis. In an hipA7 mutant, the persister level is increased 1,000-fold compared to that of the wild type, with increased tolerance toward β-lactams, fluoroquinolones, and aminoglycosides (102, 103). This hipA7 allele is associated with two point mutations resulting in a gain of function. As overexpression of HipA is toxic and leads to the arrest of cell division, it has been proposed that the locus carries a toxin-antitoxin module (104). The deletion of hipB is lethal because of HipA toxicity, suggesting that HipB is the repressor of the operon (105). Note that deletion of the complete hip locus has no effect on persister frequency in exponentially growing bacteria, possibly because of TA module redundancy (94, 105). Another explanation is that the HipBA module contributes to the persister switch only in cases of slow growth (stationary-phase cultures) (106). HipA was first thought to phosphorylate the translation factor EF-Tu, leading to persistence via cell stasis (107). However, it was recently shown that HipA more likely inhibits glutamyl-tRNA synthetase (GltX) through phosphorylation and thus triggers the synthesis of ppGpp (see below) (108).
Using an hipA7 E. coli mutant, the gene expression profile of persisters after lysis of nonpersisters by β-lactams demonstrated overexpression of genes coding for TA modules (dinJ/yafQ, yefM, relBE, and mazEF) and for proteins blocking critical cellular functions (Rmf, which inhibits translation; UmuDC, which inhibits replication; and SulA, which inhibits septation) (103). The study confirmed that overexpression of relE led to growth inhibition and increased the level of persisters. Note that, in E. coli, the MazEF chromosomal TA module and the RelE toxin are known to induce reversible stasis because of inhibition of translation and/or replication (109). Deletion of the hipBA locus leads to a decrease in the level of persisters in stationary-phase culture. Conversely, deletion of the other identified TA modules had no effect on the level of persisters in stationary-phase culture, suggesting a probable redundancy (103). Redundancy was later confirmed when Maisonneuve et al. showed that single mutations of 10 independent TA modules had no effect on persister formation but a combination of mutations increased susceptibility toward ampicillin and ciprofloxacin (110). The same group used a flow cytometer to sort E. coli cells with low ribosomal activity that had been demonstrated to be enriched in persisters (93). The study of gene expression identified overexpression of known TA modules (dinJ, yoeB, and yefM) and also of ygiU, part of the ygiUT operon, which resembles a TA module. Overexpression of ygiU led to growth inhibition and also increased the levels of tolerance toward ofloxacin and cefotaxime but not tobramycin. Recently, a new type of TA module, type V, was associated with persistence in E. coli. In this case, the antitoxin GhoS masks GhoT toxicity through specific cleavage of ghoT mRNA, thereby preventing its synthesis (111). Interestingly, the authors also identified a possible interaction between GhoST and MqsR, a toxin that, upon inactivation, decreases formation of persister cells (112). They showed that the ghoT-encoded toxin transcript is resistant to MqsR, the toxin RNase encoded by mqsRA. Thus, when MqsR is induced, ghoT is still expressed and can contribute to persistence. Indeed, deletion of ghoT decreases MqsR-mediated persistence, and mild production of the GhoT toxin leads to persistence upon ampicillin treatment. Lastly, expression of the F-plasmid-based CcdAB TA system increases the persister level and could constitute a transmissible persistence factor (see below) (113).
Therefore, it appears that various TA modules have different and cumulative effects under different conditions, suggesting a certain level of redundancy. Another way to link TA modules and persister genesis would be through degradation of the unstable antitoxin, which ultimately would lead to activation of the toxin. In this regard, recent studies on the effects of the stringent reponse and Lon protease led to establishment of new connections between starvation and persistence.
(iii) Nutrient limitation and the stringent response.
When a bacterial culture is kept in exponential phase with continuous dilution and constant medium renewal, persisters disappear (114). Conversely, at late stationary phase, the percentage of persisters increases and reaches a maximum, suggesting the importance of starvation in the genesis of persisters (114). This may be explained by indole production during stationary phase and nutrient limitation, leading to increased levels of E. coli persisters (115). Note that it was previously shown that indole production was increased in response to oxidative stress and antimicrobial exposure, through upregulation of the tnaA gene, which is responsible for indole synthesis (116).
Because the ppGpp-mediated stringent response is induced in cases of nutrient limitation, it was suspected of playing a role in the phenotypic switch of persisters 20 years ago. In 1995, ppGpp overexpression in E. coli was shown not only to increase antibiotic tolerance but also to inhibit peptidoglycan and phospholipid synthesis, thereby indicating a link between amino acid starvation, the stringent response, and antibiotic tolerance (117). Thus far, two major connections between the stringent response and persistence have been described: a defense against oxidative stress and an interaction with TA modules.
(a) The stringent response and oxidative stress defense.
In P. aeruginosa, it was shown that spoT and dksA mutants had higher levels of ppGpp and were more tolerant toward fluoroquinolones (118). Furthermore, in P. aeruginosa, the stringent response is required for optimal catalase activity and mediates H2O2 tolerance during both planktonic and biofilm growth. Upon amino acid starvation, induction of the stringent response upregulates catalase activity (119). The demonstration of a link between the stringent response and oxidative stress defense is interesting, as ROS have been proposed to explain antibiotic-induced bacterial cell death (120–122). This subject remains a matter of intense debate and controversy, as other scientists recently published conflicting results that contradict this theory (123–126). However, it might be envisaged that because of the stringent response, bacterial persisters will be less damaged by ROS and thus exhibit tolerance (Fig. 4A). For instance, it was shown that in P. aeruginosa biofilms, a starvation-induced stringent response induces antioxidant mechanisms, such as superoxide dismutase (SOD) and catalase production, thus reducing ROS-induced damage and preventing cell death, ultimately leading to tolerance toward bactericidal antibiotics (89).
FIG 4.
Main factors involved in generation of persisters. The stringent response (A) and the SOS response (B) are now considered pivotal in the generation of persisters. (C) Connection between stochasticity and persister genesis. In exponential-phase cultures, due to stochasticity, only a few bacteria reach the required threshold of a toxic molecule that is necessary to switch to the persister state (in red). Due to the factors described in panels A and B, there is an increased level of molecules inducing persistence; thus, more bacteria reach the threshold and become persisters. Note that most of these studies were conducted with planktonic bacteria. Panel C was inspired by a previous report (103).
(b) The stringent response and TA modules.
In the last 10 years, major studies have increased our understanding of the connection between the stringent reponse, TA modules, and persistence, and two main models have been proposed. For a comprehensive overview of this question, see reference 127.
The first model proposes that Lon protease plays a central role. The stringent response alarmone (p)ppGpp inhibits exopolyphosphatase, thus increasing the level of inorganic polyphosphate and ultimately inducing Lon protease activity (128, 129). It has been demonstrated that the Lon protease inactivates type II antitoxin molecules, including HipB (110, 130). Strikingly, all type II TA modules of E. coli encode mRNA endonucleases (mRNases) that degrade mRNA. The degradation of the related unstable antitoxin by Lon leads to an increased ratio of toxin to antitoxin, translation and replication arrest, and thus tolerance (Fig. 4A) (110). The same group demonstrated that (p)ppGpp stochastically triggers the activation of TA modules and thus controls the frequency of persisters (129).
Interestingly, a reverse model was proposed in 2003 for E. coli. It was suggested that free Hip toxin increases the level of ppGpp, thereby leading to altered gene expression and thus priming cells for the phenotypic switch (131). More recently, overexpression of HipA was shown to trigger growth arrest by inducing RelA-mediated synthesis of ppGpp (132). Suppression of ppGpp synthesis by use of chloramphenicol relieves Hip-mediated inhibition of DNA replication, thereby restoring vulnerability to β-lactam antibiotics (132).
These conflicting results were explained in a recent study in which the authors demonstrated that free HipA inactivates GltX (the glutamyl-tRNA synthetase) through phosphorylation. This event leads to the accumulation of uncharged tRNAGlu in the cell, which induces RelA-mediated activation of the stringent response (133). Ultimately, the level of ppGpp increases, leading to growth arrest and persister formation (Fig. 4A).
The second model to explain the connection between the stringent reponse, TA modules, and persistence suggests that the stringent response inhibits DNA supercoiling. Amato et al. studied the effects of carbon starvation on E. coli tolerance toward fluoroquinolones and demonstrated that, upon starvation, an increased level of cyclic AMP (cAMP) and/or a decrease in amino acid availability leads to an increase in ppGpp (100). Indeed, the interaction of cAMP and its receptor (cAMP receptor protein [CRP]) activates expression of relA and dksA. RelA and SpoT then synthesize ppGpp, which can repress the expression of stable RNA through an interaction with RNA polymerase (RNAP) and DksA, a small RNA polymerase binding protein (134, 135). This DksA-dependent repression of RNAP activity is associated with inhibition of DNA supercoiling, ultimately leading to inhibition of DNA gyrase and thus to tolerance toward fluoroquinolones (Fig. 4A) (100). Furthermore, the authors demonstrated that ppGpp-SpoT acted as a TA module on its own, with the following lines of argument: (i) the biochemical network of ppGpp suggests the possibility of bistability, and they confirmed this by using a kinetic model; (ii) ppGpp causes growth arrest through its interaction with RNAP; (iii) SpoT, the only known ppGpp hydrolase, cannot be knocked out in a relA+ background; and (iv) ppGpp in excess of its antitoxin increases the level of persistence. In this ppGpp-SpoT model, persisters are cells with a higher level of ppGpp. ppGpp inhibits transcription, DNA replication, and DNA gyrase negative-supercoiling activity, thereby leading to fluoroquinolone tolerance. The same group recently demonstrated that a ppGpp-dependent pathway is also involved under biofilm conditions (136). Strikingly, they identified specificities regarding the importance of each involved protein or enzyme (136).
Finally, these results could lead to the design of antibacterial agents targeting the stringent response, such as RelA inhibitors, in order to increase persister cell mortality (137).
(iv) The SOS response.
The SOS response, also called the DNA damage response, involves all the molecular mechanisms induced by chromosomal DNA damage caused by UV radiation or oxidative radicals. In 2004, a connection was established between the SOS system and tolerance. In that study, the authors demonstrated that inactivation of the ftsI gene product, penicillin-binding protein 3, by β-lactams induced SOS in E. coli, through the DpiBA two-component signal transduction system. This event, which requires the SOS-promoting recA and lexA genes as well as dpiA, transiently halts bacterial cell division, enabling survival upon otherwise lethal antibiotic exposure (138). A more recent study demonstrated that, in E. coli, ciprofloxacin at low concentrations triggered the SOS response system that leads to release of LexA-dependent repression of the tisB toxin gene (Fig. 4B). TisB can be inserted into the inner membrane and disrupt the proton motive force, which leads to a drop in the intracellular level of ATP. Subsequent shutdown of cellular processes is thought to be responsible for the observed higher level of persisters (139, 140).
Recently, it was shown that starvation and the SOS response can induce high biofilm-specific tolerance toward ofloxacin (141). In that study, a screen for E. coli mutants forming biofilms with increased tolerance toward antibiotics led to identification of amino acid auxotrophs displaying strong tolerance toward either ticarcillin or ofloxacin upon starvation. It was demonstrated that both functional RecA and cleavable LexA were essential for the starvation-induced biofilm-specific ofloxacin tolerance phenotype and that the SOS response was induced upon biofilm aging concomitantly with ofloxacin tolerance (Fig. 4B). Interestingly, a previous study from the same group showed that recA and other SOS response genes were significantly induced in mature biofilms compared to exponentially grown planktonic cells (142). Conversely, the ofloxacin tolerance of planktonic bacteria was likely due to a mechanism other than the SOS response, since a ΔrecA mutant did not significantly impair the overall tolerance of either nonstarving or starving populations. The latter results strengthen the notion that induction of ofloxacin tolerance in starving biofilms is likely to involve mechanisms different from those currently described for planktonic cells (143, 144).
It is noteworthy that the SOS system is also induced by conjugative DNA transfer, an event that is enhanced in biofilms (145).
(v) Oxidative stress defense.
Oxidative stress defense includes all bacterial mechanisms involved in protection against inadvertent by-products of aerobic metabolism, such as superoxide (O2−) and hydrogen peroxide (H2O2), which are partially reduced oxygen species (146). ROS ultimately lead to DNA, protein, and lipid damages. Different defense mechanisms can be activated depending on the type of ROS. For example, activated macrophages produce O2− and H2O2, which induce bacterial SoxRS and OxyR regulons, respectively. This leads to activation of genes involved in ROS elimination and DNA repair (147). As discussed above, antibiotic-induced oxidative stress might play an important role in bacterial cell death. Therefore, it was deemed plausible that a way for persisters to survive in the presence of bactericidal antibiotics was to protect themselves from oxidative stress. For instance, flow cytometer analysis demonstrated that in a population of antibiotic-treated E. coli cells, persisters did not overproduce hydroxyl radicals, whereas most bacteria killed had a high level of hydroxyl radicals (148). Alongside the previously described stringent response-mediated defense against oxidative stress damages, another group reported that antioxidant strategies could lead to tolerance of bactericidal antibiotics. Indeed, H2S has been demonstrated to increase the antioxidant capacity of Gram-positive and Gram-negative bacteria through suppression of the Fenton reaction and stimulation of SOD and catalase production (149).
Another group proposed a different scenario involving oxidative stress. They revealed that paraquat-induced oxidative stress led to an increase in the level of persisters surviving fluoroquinolone antibiotics, but not ampicillin or kanamycin (147). They showed that SoxRS induces overexpression of the AcrAB-TolC MDR pump, which can extrude fluoroquinolones (147). Thus, exposure to lower concentrations of fluoroquinolones may lead to persister formation. Furthermore, MDR pumps are also involved in protection against oxidative stress via the elimination of ROS (76).
Lastly, it was shown that oxidative stress was induced in biofilms independently of the presence of antibiotics (150). On the other hand, the SOS reponse is induced by ROS (151). Therefore, it can be envisaged that in biofilms, due to an increased level of oxidative stress, the SOS response is induced and increases the level of tolerance, as demonstrated in the case of ofloxacin (141).
(vi) Other cues leading to persistence.
Aside from the above-described genetic mechanisms, different genes or regulators are involved in the switch to the persister state or, less precisely, in an increase in bacterial tolerance. In most of the following cases, the precise links between these genes and tolerance are not known.
(a) E. coli.
In E. coli, the screening of a transposon mutagenesis library revealed that PhoU could play a major role in persistence (152). Inactivation of the phoU gene leads to decreased tolerance toward a wide range of antibiotics and various stresses, such as acidic pH, starvation, and heat. phoU expression is regulated by environmental changes, such as nutrient availability or the age of the culture, and its expression is decreased in rich media. phoU mutants exhibit upregulation of flagella, chemotaxis genes, and energy production enzymes, suggesting that the loss of the PhoU regulator renders the cells hyperactive. In case of starvation, phoU is expressed and affects genes involved in energy production and membrane transport. The precise effectors through which PhoU suppresses cellular metabolic activity are not known.
Another group used survival of ampicillin treatment as a screening method for an E. coli genomic mutant library and identified a hypertolerant clone with overexpression of the gene coding for the conserved aerobic sn-glycerol-3-phosphate dehydrogenase GlpD (153). Although deletion of glpD did not affect tolerance in exponential-phase cultures, it eliminated the majority of persisters in stationary phase. Two additional multidrug tolerance loci, glpABC and plsB, were identified through study of the pathway involving sn-glycerol-3-phosphate metabolism.
(b) P. aeruginosa.
The importance of quorum sensing (QS) signals in tolerance was demonstrated in P. aeruginosa biofilms; indeed, their inhibition through mutations (ΔlasR rhlR QS receptor mutants) or use of inhibitors (furanones C-30 and C-56) led to decreases in tolerance toward tobramycin, H2O2, and phagocytosis by polymorphonuclear cells (PMNs) (154). The difference in tolerance might be related partly to a different biofilm structure, as QS plays an important part in biofilm architecture. However, similar observations were made in P. aeruginosa exponential-phase culture, as the level of persisters was significantly increased through the adjunction of exogenous phenazine pyocyanin or 3-OC12-HSL (155). Pyocyanin, secreted by P. aeruginosa during stationary phase, reduced the growth of P. aeruginosa and exhibited an effect on persister formation, during both the exponential and stationary phases, in a dose-dependent manner. Another structurally related compound (paraquat) had a similar effect, whereas phenazine-1-carboxylic acid (PCA) did not, despite strong structural similarity. Since a mutant unable to produce phenazine (Δphz1 and Δphz2) had a similar level of persisters, it was suggested that redundant systems are present. The spectrum of action of each QS signal probably varies, as 3-OC12-HSL increased the level of persisters only in strain PAO1, not in PA14 (155). In addition, small volatile QS compounds, such as 2′-amino-acetophenone (2-AA), have also been shown to influence persister cell accumulation (156).
In P. aeruginosa, in addition to QS signals, another locus has been identified. AmgRS is a two-component regulator, and its mutation was identified through screening of tobramycin-susceptible mutants. Indeed, amgRS mutations reduce planktonic and biofilm tolerance toward aminoglycosides (157). Transcription profiles suggest that AmgRS controls an adaptive response to membrane stress, possibly caused by aminoglycoside-induced insertion of misfolded proteins (157). The possible effectors of AmgRS-induced tolerance may be membrane proteases (HtpX and NlpD) and a protease-associated factor (YccA), which would help to eliminate misfolded proteins.
(vii) Stochastic gene expression.
One mechanism that can be hypothesized for the persister switch is that of stochastic gene expression through fluctuations in transcription and translation rates despite stable environmental conditions (158). These variations result from two types of noise: (i) intrinsic noise, related to the nature of the process of gene expression and secondary to the rates of translation and mRNA and protein degradation; and (ii) extrinsic noise that varies from one cell to another and is caused by ribosome abundance or asymmetric distribution of proteins upon cell division. Indeed, even when all members of a planktonic culture are exposed to the same growth conditions, only a small fraction of them are persisters, suggesting the involvement of stochasticity (13). In this case, we speculate that at the population level, there exists a mean level of key persister regulatory protein expression associated with intracellular fluctuations due to the noises. For a small subset of bacteria, the level reaches a threshold, leading to the phenotypic switch (Fig. 4C). Then, when the population meets environmental triggers inducing stringent or SOS responses, the basal level of expression increases, leading to an increase in the percentage of cells reaching the threshold (94, 159). This hypothesis has been supported by experiments performed with TA modules that also demonstrate that the amount by which the threshold is exceeded determines the duration of dormancy (160).
(viii) Persister heterogeneity.
As demonstrated above, many pathways, molecular mechanisms, and environmental factors are involved in the phenotypic switch that leads a bacterium to become a persister. Furthermore, some of these pathways are interconnected. Therefore, it is very likely that depending on the conditions prevailing during the switch, different types of persisters may appear, possibly simultaneously, in the same culture (100, 161). The type of antibiotic used to eradicate nonpersisters is a striking example of this and can influence gene expression, SOS induction, and the oxidative stress defense.
Even with homogeneous stresses similarly affecting the whole population, it was demonstrated that both a growth-arrest-mediated pathway and ppGpp-dependent pathways can be activated, leading to different types of persisters (100).
Ten years ago, the study of persisters by use of a microfluid device led to the hypothesis that two main types of persisters were produced: type I persisters were generated during stationary phase, with a prolonged lag phase before resuming growth upon transfer to rich media, whereas type II persisters were continuously produced independently of the growth phase (91). It is now clear that this view caught only a glimpse of the complexity and diversity of persisters.
Biofilms as a relevant environment for persister generation.
Although most of the above-mentioned mechanisms were discovered under plankonic conditions, it is very likely that they are also involved in the generation of persisters in biofilms. For instance, due to nutrient limitations, the stringent response has already been shown to play a central role in P. aeruginosa biofilm recalcitrance (89). The SOS response is induced in biofilms and plays a role in biofilm recalcitrance toward antibiotics (141). On the other hand, due to the existence of biofilm-specific phenotypes and functions, caution should be taken in extrapolating persister data obtained under planktonic conditions to the biofilm lifestyle. Indeed, biofilm-specific mechanisms have been described and underline the complexity in the study of persisters (141).
The Biofilm Environment Favors Acquisition of Antibiotic Resistance
Patients suffering from biofilm-related infections are also exposed to nosocomial microorganisms present in their health care environment and selected by repeated antibiotic treatments. As a result, treatment of biofilm-related infections is difficult, not only due to biofilm recalcitrance toward antibiotics but also due to potential infection by multiresistent microorganisms carrying resistance genes, such as those encoding extended-spectrum β-lactamases (ESBLs) or methicillin resistance. In this case, biofilm formation and gene resistance issues can be additive as well as synergistic.
Biofilm formation favors horizontal gene transfer.
Biological processes involved in horizontal gene transfer, such as conjugation, transformation, and transduction, have been demonstrated to be increased in vitro in biofilms (for a comprehensive review of this issue, see reference 162). Furthermore, while the presence of conjugative plasmids promotes biofilm formation, the biofilm lifestyle also increases plasmid stability and the range of mobile genetic elements (163). Hence, the presence of a biofilm is expected to facilitate the transfer of resistance genes, as demonstrated in an in vitro study, with an increased rate of transfer of a plasmid encoding CTX-M-15 (an ESBL) in a K. pneumoniae biofilm compared to the case under planktonic conditions (164). Transfer of a conjugative transposon (Tn916) carrying antibiotic resistance might also be responsible for acquisition of resistance mechanisms in biofilm bacteria (165). Transferability of genetic mobile elements between bacteria belonging to a multispecies biofilm has been described for a medical device implanted in a patient (166).
Interestingly, many transmissible DNA elements encode biofilm-promoting factors, including adhesins, such as conjugative pili, fimbriae, and autotransporter adhesins, and persistence factors, such as toxin-antitoxin modules. For instance, the F-plasmid-based CcdAB TA system increases the persister level and thus constitutes a transmissible persistence factor (113).
Impaired antibiotic diffusion through the matrix leads to bacterial exposure to subinhibitory concentrations of antibiotics.
Due to biofilm architecture and drug diffusion issues, it is likely that some biofilm areas may be submitted at least transiently to subinhibitory concentrations of antibiotics. Exposure to subinhibitory concentrations of antibiotics is known to increase the likelihood of selecting resistant mutants (for a comprehensive review, see reference 167). Although it is generally assumed that selection of resistant bacteria occurs at antibiotic concentrations between the MIC of the susceptible wild-type population and that of the resistant bacteria, recent studies suggested that such selection could also occur at lower antibiotic concentrations (168). Furthermore, bacteria may produce hydroxyl radicals when exposed to sublethal concentrations of antibiotics (169). These hydroxyl radicals can induce the occurrence of mutations and help the organism to acquire resistance mechanisms. It has also been demonstrated that β-lactam antibiotics increase E. coli mutagenesis through RpoS-mediated reduction of replication fidelity (170). Similar findings have been made in P. aeruginosa during long-term experimental evolution, suggesting that CF patients who receive prolonged fluoroquinolone treatment might be exposed to this phenomenon (171).
Exposure to tobramycin at subinhibitory concentrations can increase the c-di-GMP level and biofilm formation, as demonstrated in E. coli and P. aeruginosa, through alterations in the level of c-di-GMP (172). Similar findings were made upon exposure of Corynebacterium diphtheriae to subinhibitory concentrations of erythromycin and, to a lesser extent, penicillin (173), but also for P. aeruginosa and imipenem or S. aureus and vancomycin or oxacillin (174, 175).
Recent studies also reported that antibiotics at subinhibitory concentrations can promote the transfer of mobile genetic elements, even though this has been demonstrated primarily under planktonic conditions. For instance, the fluoroquinolone-mediated SOS response may trigger expression, excision, and transfer of prophage genes (176). SOS induction may promote mobilization of various mobile elements, such as integrating conjugative elements (177). It has been shown that conjugation induces the SOS response and promotes antibiotic resistance through integron integration and activation in vitro (145, 178). More recently, an in vivo demonstration of this phenomenon was made through the identification of SOS-induced integrase expression ultimately leading to rearrangement of an integron gene cassette, full expression of a β-lactamase, and, thus, resistance toward ceftazidime (179).
Finally, as previously discussed, ciprofloxacin has been shown to increase the frequency of persisters through induction of SOS and, ultimately, production of the TisB toxin (139, 140). In general, preexposure to subinhibitory concentrations of antibiotics (0.2-fold MIC) increases the frequency of persisters with tolerance toward drugs belonging to different classes of antibiotics (ciprofloxacin, gentamicin, oxacillin, and vancomycin) (180). Although most of the data discussed here were generated with planktonic bacteria, it can be envisaged that this phenomenon is relevant in the case of reduced diffusion of antibiotics through the biofilm matrix.
Because biofilm persisters are more likely to survive antibiotic treatment, they are exposed to repeated rounds of different classes of antibiotics, inducing all the above-mentioned consequences and thereby amplifying the phenomenon (181). Although the interplay between biofilm recalcitrance, gene transfer, and spread of resistance could be of key importance in nosocomial settings, it remains to be demonstrated in clinical settings, or even in a relevant in vivo model of biofilm-related infections.
Genetic diversity within biofilms and its impact on biofilm recalcitrance.
Various examples of genetic diversity occurring in biofilms have been described as influencing biofilm tolerance toward antimicrobial agents.
(i) Hypermutability.
In P. aeruginosa, endogenous oxidative stress induces double-stranded DNA breaks in some cells within biofilms (150). Genetic variants arise when breaks are repaired by a mutagenic mechanism involving recombinatorial DNA repair genes. It was suggested that diversity and adaptability generated by this mechanism increase the ability of biofilm communities to adapt and survive in harsh environments; this mechanism is known as the “insurance effect” (150). Several genes, such as katA and sodB, also shown to be involved in protection against oxidative DNA damage, were downregulated under biofilm conditions (182). A similar mechanism has been described for the mucoid conversion of P. aeruginosa in CF patients. Indeed, free oxygen radicals, such as H2O2, released by PMNs can induce formation of mucoid variants through mutations in mucA, which encodes an anti-σ factor (183). This leads to deregulation of an alternative σ factor (σ22, AlgT, or AlgU) that is required for expression of the alginate biosynthetic operon (183). Hypermutators have been identified in clinical samples, and some of them are associated with specific mutations, such as mutS, belonging to the DNA mismatch repair (MMR) system (77). Aside from mutL and uvrD, which also belong to the MMR system, other genes were found to be mutated in hypermutators, such as mutT, mutY, and mutM, belonging to the DNA oxidative lesion repair system (184–186). Similar findings have been made in staphylococci, with mutability in biofilms that is 60-fold (S. aureus) and 4-fold (Staphylococcus epidermidis) higher than that under planktonic conditions (187). These mutations can lead to tolerance or resistance mechanisms.
(ii) Small-colony variants.
SCV constitute a subset of the bacterial population that has been identified in a wide range of bacteria, including S. aureus and P. aeruginosa (29). They are associated with many diseases, including biofilm-related infections, such as osteomyelitis, chronic pulmonary infections in CF patients, and device-related infections. It has been demonstrated that their slow growth originates mainly from mutations associated with two types of defect: a deficiency in electron transport and a deficiency in thymidine biosynthesis. These SCV are frequently auxotrophic and are less susceptible to various antibiotics, depending on the metabolic alterations they exhibit (for comprehensive reviews of these issues, see references 188 and 29). As SCV may be present in biofilms, they may be involved in the global recalcitrance of the bacterial community. For instance, P. aeruginosa SCV have increased piliation, biofilm formation ability, and better adhesion to respiratory cell lines (189). Aside from SCV associated with mutations, phenotypic SCV have been described for P. aeruginosa. For instance, rough SCV (RSCV) of P. aeruginosa can be found in vitro and in clinical samples from CF patients and are associated with increased biofilm formation and antibiotic resistance (190). When RSCV are grown on antibiotic-free agar, wild-type revertants with a large colony size and a smooth appearance arise on the edges of the variant colonies. The regulatory protein PvrR of the two-component system PvrSR has been found to control conversion between antibiotic-resistant and antibiotic-susceptible forms. Indeed, a PA14 ΔpvrR strain exhibited an increased frequency of resistant variants on kanamycin plates compared with the wild type. PvrR was later described as a phosphodiesterase modulating the c-di-GMP level in P. aeruginosa, suggesting the importance of c-di-GMP in controlling the onset of SCV (191).
ANTIBIOFILM STRATEGIES
Even prior to identification of the link between biofilms and human diseases, different therapeutic strategies were developed to prevent the occurrence of microbial colonization and to eradicate device-related infections, once established. However, most developments in the field of antimicrobial agents were based on planktonic studies, without taking into account the specificities of the bacterial biofilm lifestyle.
Currently Used Approaches Do Not Specifically Target Biofilm Bacteria
Hygiene, training, and reduction in the number of implanted devices.
(i) Hygiene and training.
Although hygiene is not a specific antibiofilm strategy, it prevents microbial contamination and thus adherence and subsequent biofilm formation. For almost all types of device-related infections, guidelines have been proposed to standardize procedures for device implantation and handling. For instance, the insertion of any central venous catheter (CVC) must be performed by trained personnel with maximum sterile barrier precautions, defined by the use of sterile gloves, cap, mask, sterile gown, and a sterile full-body drape (192, 193). The choice of skin disinfection solution and methods is also of key importance, and many reports suggest that alcohol-based antiseptics, such as alcohol-based chlorhexidine and alcohol-based povidone-iodine, are the most efficient solutions (192). Improvement of hygiene measures should always be attempted through definition and implementation of local clinical bundles for device insertion and handling, and in the case of CVC, dedicated infusion therapy teams have been developed for the education of patients and health care workers (192, 194, 195).
(ii) Early removal of an unnecessary device.
Once a device is removed, the risk of bacterial contamination drops to zero. Therefore, at any time, physicians must discuss the benefits of maintaining an indwelling foreign body. For instance, a meta-analysis reported that use of an automatic reminder system for the removal of useless urinary catheters significantly decreased the incidence of catheter-associated urinary tract infections (CAUTI) (196). Of course, this approach is more difficult in the case of mandatory devices, such as pacemakers.
(iii) Systemic antibiotic prophylaxis during device insertion.
Depending on the type of device, systemic antibiotic prophylaxis can be proposed in order to reduce the risk of microbial contamination. In that case, antibiotics are injected a few minutes before skin incision and are dedicated to eradicating any microorganisms that are not removed by skin disinfection. This approach is recommended in the case of surgically implanted devices, such as orthopedic and cardiac devices (197, 198).
Antibiotic coating of implanted devices.
The principle of antibiotic coating of implanted devices is to deliver a locally high concentration of antimicrobials at the site of potential colonization (199). Depending on the type of device and the length of implantation, these antibiotic-coated materials can efficiently reduce the rate of colonization. The example of CVC can be taken to illustrate the benefits and limits of the antibiotic coating strategy. Indeed, for short-term CVC (<10 to 14 days of expected dwelling time), use of a coated CVC significantly reduces the risk of catheter-related infections and can be proposed when the infection incidence is still high despite implementation of all other preventive measures (192). Two types of coating have been developed: minocycline-rifampin and chlorhexidine-silver sulfadiazine. Comparative studies concluded that the former is more efficient than the latter (200–204). However, the benefits of antibiotic coating for long-term intravenous catheters (LTIVC) have not yet been demonstrated. Indeed, as these devices dwell for longer periods, the surfaces of LTIVC will be covered by a conditioning film composed of host cells or proteins that might limit the effect of any active surface. Furthermore, as soon as the antibiotic contained in the device is exhausted, antibiotic delivery stops. Antibiotic-coated surfaces have also been studied in animal and clinical studies of urinary catheters, endotracheal tubes, orthopedic devices, and vascular grafts, with contrasting clinical benefits (205–216). Thus, development of a coated surface that prevents bacterial colonization for a long time remains a challenge.
Mechanical removal of the source of infection.
When clinicians are confronted with therapeutic difficulties or local and systemic complications, removal of the indwelling device may be required in the case of biofilm-related infection (194, 198, 212, 217). For short-term peripheral catheters, removal and replacement are easy, painless, and inexpensive. In contrast, removal of long-term catheters, pacemakers, or orthopedic prostheses is associated with complications for the patient, as well as with high costs. In the case of tissue-related infections, surgical removal of biofilm may be indicated for antibiotic failure. This is particularly the case for infective endocarditis (IE) and osteomyelitis, during which failure to cure the infection is an indication for surgery (218).
Optimization of the antibiotic regimen against biofilms.
As physicians and clinical microbiologists became more aware of the importance of biofilms in infectious diseases, they attempted to define the antibiotics that were most active against biofilms and how these antibiotics should be prescribed so as to increase the likelihood of infection eradication.
One famous example of this challenging process is that of the rifampin-containing regimen, demonstrated to significantly improve the outcome of foreign-body-related S. aureus infections, first in vivo and then in clinical studies. Furthermore, fosfomycin and daptomycin are currently being investigated and might be promising candidates in the fight against foreign-body-related infections (219–221). In the case of prosthetic joint-related infection (PJI), in vivo models led to the demonstration that fluoroquinolones exhibited more penetration into the site of infection (222). Furthermore, in vivo models of foreign-body-related infections demonstrated that fluoroquinolones were the most efficient molecules when associated with rifampin (223). Based on these findings, fluoroquinolones have now become one of the mainstay treatments of PJI (223). A more recent example of an antibiotic associated with a potent antibiofilm effect is that of daptomycin. This bactericidal cyclic lipopeptide has an in vitro spectrum against Gram-positive pathogens through calcium-dependent disruption of membrane function, leading to potassium ion leakage and inhibition of DNA, RNA, and protein synthesis (224, 225). In vitro studies suggested that daptomycin may quickly penetrate S. epidermidis biofilms, that it is effective against biofilms, and that it is bactericidal against stationary-phase and nondividing S. aureus (225–228). However, daptomycin alone was not able to cure the infection caused by methicillin-resistant S. aureus (MRSA) in tissue cage foreign body models, and its association with rifampin was not significantly better than a levofloxacin-rifampin association. Nevertheless, these 2 antibiotic combinations were more efficient than the previously recommended vancomycin-rifampin and linezolid-rifampin combinations (229). Using a similar methodology, another group demonstrated that daptomycin or rifampin as a single agent against MRSA was more effective than vancomycin or linezolid (221). Daptomycin has also been proposed for the treatment of catheter-related infections, and an in vivo study demonstrated that vancomycin and daptomycin were equally efficient at eradicating methicillin-resistant S. epidermidis (MRSE) catheter-related infections (230). Subsequently, a phase II clinical study was conducted using daptomycin antibiotic lock therapy (ALT) (see below), and it reported a cure rate of 85% (231). Comparative clinical studies are now expected to determine, for instance, whether daptomycin is more efficient and more rapid than vancomycin.
In the case of P. aeruginosa pneumonia in CF patients, optimized antibiotics may increase the likelihood of bacterial eradication, especially in early colonization. In that case, the early association of oral ciprofloxacin with inhaled colicin is associated with a reduced risk of chronic colonization (232, 233).
In addition to the choice of specific antibiotics, high dosages and prolonged treatment courses are required for biofilm-associated infections, as emphasized by cases of IE and osteomyelitis (218, 223).
Lock solutions to address the problem of biofilm recalcitrance.
ALT is a strategy that relies on the injection of a highly concentrated (100× to 1,000× MIC) antibiotic solution into the lumen of a CVC. This solution should dwell for an extended time (at least 12 h) in order to eradicate any incoming bacteria. The chosen volume must allow coverage of the entire internal surface and therefore depends on the type of device, but it is usually small (between 2 and 5 ml). ALT can be used for prevention and treatment of catheter-related infections, but in most cases, its use is restricted to LTIVC. Indeed, microbial contamination of LTIVC occurs on the inner side of the device, defining intraluminal colonization. Thus, the highly concentrated antibiotic solution will be in close contact with the biofilm. On the other hand, in case of short-term CVC, contamination occurs mainly on the external surface of the device, defining extraluminal contamination. In that case, ALT cannot access the biofilm and is therefore useless.
(i) ALT for prevention of catheter-related bloodstream infections.
As stated above, the ALT approach is restricted to prevention of LTIVC-related infections. A meta-analysis demonstrated that ALT composed of vancomycin reduced the risk of CRBSI (234). Other groups also assessed the combination of an antibiotic (minocycline) and a chelator, such as EDTA. Two studies in the pediatric oncology setting showed that minocyline-EDTA ALT was more effective than heparin for prevention of CRBSI (235, 236). Nevertheless, systematic use of ALT could lead to increased antibiotic resistance and should therefore be considered only for high-risk patients who have already experienced LTIVC-related infections (192, 237, 238). On the other hand, limited data are available concerning nonantibiotic lock solutions, such as ethanol and taurolidine, but they might also be used among high-risk patients (239, 240).
(ii) Conservative treatment of CRBSI with ALT.
In cases of uncomplicated LTIVC-related BSI, a conservative treatment can be used based on ALT (90, 194). Indeed, if the clinical situation allows, catheter salvage is indicated in cases of reduced venous access or the potential presence of coagulation disorders (194). Such conservative treatment could avoid risks and reduce costs associated with a new surgical procedure. However, LTIVC removal is mandatory in cases of local or distant complications or in cases of infection caused by S. aureus or Candida spp., based on the high failure rate of treatment when the colonized catheter is retained (241). In other cases, conservative treatment using a combination of systemic antimicrobials and ALT can be considered (90, 194). Despite several limitations, there is a growing body of evidence favoring the use of ALT. For instance, a randomized, placebo-controlled study showed that ALT plus systemic antimicrobial therapy is more effective than systemic antimicrobial therapy alone for treating LTIVC-related BSI, although the result did not reach statistical significance due to the small sample size (242). In addition, large uncontrolled studies demonstrated high cure rates in patients with uncomplicated LTIVC-related BSI due to coagulase-negative staphylococci (CoNS) (89%) or Gram-negative rods (GNR) (95%) (241, 243, 244). Thus, the current indication for ALT is uncomplicated LTIVC-related BSI caused by CoNS or GNR (90, 194). Aside from commonly used antimicrobials in ALT, ethanol and daptomycin have recently been used for conservative treatment (see the previous section for daptomycin data). However, clinical data are still needed in order to recommend ethanol as a first-line compound for ALT (245–247).
Targeting Biofilm Recalcitrance: Progress and Perspectives
Currently used strategies have clearly improved the management of patients with indwelling devices in terms of both prevention and treatment of biofilm-related infections. However, many challenges remain before we can decrease the risk of microbial contamination on a surface or increase the likelihood of biofilm eradication. It is very likely that specific targeting of mechanisms known to play a role in biofilm recalcitrance will be a relevant strategy.
Preventive strategies.
Within the limits of the different preventive approaches and the fact that most of them rely on the use of antibiotics, many efforts have been made to identify preventive strategies based on fundamental knowledge of mechanisms involved in bacterial adherence and biofilm formation (Fig. 5).
FIG 5.
Antibiofilm strategies arising from fundamental research. Approaches to preventing formation of biofilms are depicted in blue; approaches to eradicating an established biofilm are shown in red. Persister cells are shown in red. AG, aminoglycosides; c-diGMP, cyclic di-GMP; FQ, fluoroquinolones; NAC, N-acetylcysteine; QS, quorum sensing; ROS, reactive oxygen species.
(i) Inhibiting microbial adhesion.
Given the fact that without initial adhesion a biofilm cannot develop, the objective of inhibiting microbial adhesion is to impede the initial steps in biofilm formation.
(a) Material optimization, surface modifications, and biosurfactants.
Inhibition of microbial adhesion to surfaces has been discussed extensively in several reviews (248–251). Here we simply describe the main approaches and develop a relevant example of each.
Since initial adhesion implies bacterial and surface factors, the physicochemical characteristics of the surface are of key importance in prevention of device-related infections. The physical nature of the material is important, as illustrated by human models of dental implant-associated biofilm (252). This experimental approach was used to demonstrate that bacterial adhesion to implant surfaces is significantly lower with a zirconium oxide surface than with pure titanium (Ti) (253). The biomaterial manufacturing process can also modify roughness and physicochemical properties and thus affect bacterial adhesion. Indeed, electropolished stainless steel reduces bacterial adhesion compared to that with sandblasted steel (254). Furthermore, the choice of the polymeric material, even without any modification, is of key importance. Using a high-throughput microarray assay, bacterial adhesion was assessed on hundreds of polymeric materials and led to identification of materials comprising ester and cyclic hydrocarbon moieties (255). Coating of silicone with these materials significantly decreased S. aureus and E. coli adhesion in vitro and in vivo (255).
The physical architecture of the surface can also help to prevent microbial adhesion. For instance, the sharklet micropattern is a surface modification that mimics the microtopography of shark skin and has been shown to significantly reduce Gram-negative bacterial adhesion in vitro (256, 257).
Another major strategy for reducing bacterial adhesion is to modify the surface so it is protected by grafting antiadhesive molecules. One limitation to coated devices lies in the progressive coverage by a conditioning film made of proteins or cells from the patient. Thus, different attempts have been made to reduce not only microbial adhesion but also the deposition of host components or the occurrence of thrombosis. To do so, a peptide-based coating technology was proposed to modify the surface of Ti metal through noncovalent binding (258). In that study, a peptide (SHKHGGHKHGSSGK) possessing affinity for Ti was identified and coated with a pegylated analogue that efficiently blocked adsorption of fibronectin and S. aureus adhesion (258). Another group used lysozyme immobilized on polyethylene glycol monomethacrylate (PEGMA) to coat stainless steel surfaces and demonstrated that bacterial adhesion and albumin adsorption were reduced (259). Another surface modification using zwitterionic (a molecule with both positive and negative charges) nonleaching polymeric sulfobetaine (polySB) was associated with significant reductions in adherence and activation of platelets and white blood cells (260). This scaffold retains water on the surface of the catheter surface and reduces not only protein, host cell, and microbial adhesion but also thrombus formation in vitro and in vivo (260). Although these approaches have produced encouraging results, they still need to be evaluated in long-term settings.
Other surface modifications have been designed to kill bacteria once they stick to the surface, without using antibiotics. Two examples can be presented. First, poly(4-vinyl-N-alkylpyridinium bromide) covalently attached to glass slides and immobilizing polycationic chains (that have antibacterial properties) is able to kill airborne bacteria on contact (261). Second, single-walled carbon nanotube (SWNT) coatings were reported to have antimicrobial activities through cell membrane perturbation after an initial SWNT-bacterium interaction that ultimately leads to electronic structure-dependent bacterial oxidation and death (262).
Biosurfactants are surface-active molecules produced by many bacteria to inhibit adhesion of competitors. These compounds alter surface properties such as wettability and charge, thereby modifying bacterium-surface and/or bacterium-bacterium interactions (263, 264). Such molecules have therefore been studied as a possible surface modification in order to prevent bacterial adhesion. For instance, group 2 capsule and Ec300p, two hydrophilic high-molecular-weight polysaccharides produced by different E. coli strains, have been shown to prevent biofilm formation of Gram-positive and/or Gram-negative pathogens (265, 266). Other molecules have been tested, including surfactin, rhamnolipids, and other molecules produced by lactobacilli and Streptococcus thermophilus, although these have not been identified clearly (see the reviews in references 263 and 267).
(b) Other types of nonantibiotic coatings.
Because of limitations related to antibiotic coatings, such as their effect being restricted to nonresistant bacteria, different groups have tried to identify nonantibiotic coatings for preventing microbial adhesion. Use of antibody-releasing surfaces, such as a biomedical-grade polyurethane hydrogel coating containing solid dispersed bioactive antibodies, was proposed (268). The presence of antibodies reduced bacterial adhesion and enhanced bacterial killing during an in vitro opsonophagocytic assay using freshly isolated blood neutrophils (268). IgG opsonization was shown to inhibit bacterial adhesion by blocking cell surface attachment factors and altering the surface hydrophobicity of the bacterial cell (269). The main limitation of this approach was the short duration of antibody release (∼24 h) (199), and an in vivo assessment of the preventive efficacy of this approach is still lacking. Since nitric oxide (NO) has antibacterial properties, NO-releasing surfaces have been proposed. An in vivo model using a medical-grade silicone elastomer with an NO-storing film implanted in rats led to a reduction in bacterial colonization after S. aureus challenge (see below for the effects of NO on dispersal) (270). Other vascular catheter coatings have been studied, such as the association of triclosan (an antiseptic) and dispersin B (an antibiofilm enzyme) (see below) to prevent S. aureus colonization in vitro and in vivo (271). Triclosan-loaded urinary catheters have also been studied successfully in vivo for the prevention of Proteus mirabilis CAUTI (272). Another antiseptic-coated catheter containing gendine demonstrated significant reductions in E. coli adhesion both in vitro and in vivo in a CAUTI model (273).
(c) Inhibition of production of adhesins.
Different molecules have been designed to specifically inhibit the production of bacterial adhesins involved in biofilm formation. As an example, ring-fused 2-pyridones inhibit curli biogenesis in uropathogenic E. coli (UPEC) and prevent polymerization of the major curli subunit protein, CsgA. Some of them also have a pilicide effect, i.e., inhibition of the assembly of type 1 pili, which is required for pathogenesis during urinary tract infection via the FimH adhesin exposed at the tips of the pili. One molecule, FN075, has been demonstrated to block biogenesis of both curli and type 1 pilus, to inhibit biofilm formation, and to attenuate virulence in a mouse model of urinary tract infection (274).
(d) Blocking of interaction of adhesins with their receptors.
Another approach is to specifically target the FimH type 1 pilus lectin of UPEC, which mediates bacterial colonization, invasion, and formation of recalcitrant intracellular bacterial communities in the bladder epithelium (275). Low-molecular-weight mannose-derived compounds called mannosides were designed and adapted for oral administration. Indeed, the mannose binding pocket of FimH is composed of amino acids that are invariant in all strains of E. coli. When tested in a mouse model, the mannosides were able to prevent UTI when given prophylactically or to treat an established chronic urinary tract infection (275). These molecules have been demonstrated to prevent and treat UPEC CAUTI in a mouse model (276). Furthermore, synergistic action was noted between mannosides and trimethoprim-sulfamethoxazole, suggesting the utility of adjuvant approaches in this setting (276). Prophylactic administration of a mannoside molecule, compound ZFH-04269, was recently demonstrated to significantly reduce bacterial colonization of the bladder and to prevent acute UTI caused by an epidemic multidrug-resistant UPEC ST131 clone. Treatment of chronically infected mice with the same FimH inhibitor lowered their bladder bacterial burdens over 1,000-fold (277).
Aside from direct administration of an adhesin inhibitor, other authors proposed covering a surface with an adhesin inhibitor. For instance, coverage of a surface with multivalent galabiose derivatives significantly inhibits adhesion of E. coli through inhibition of P fimbriae in vitro (278). The main limitation of this approach is the multiplicity of structures involved in bacterial adhesion. However, one way to circumvent this issue is to use multivalent inhibitors linked to a scaffold of glycopolymers, glyconanoparticles that may permit inhibition of several adhesins at the same time (279–281).
(e) Use of lactoferrin.
Lactoferrin is a component of innate immunity found in numerous body fluids (tears, milk, and respiratory secretions) and is an iron chelator that has been demonstrated to inhibit irreversible adhesion of P. aeruginosa in vitro (282). Through iron chelation, lactoferrin stimulates twitching motility, during which bacteria wander across the surface instead of forming microcolonies and biofilms. Indeed, iron metabolism and transport are required for normal biofilm development (283). Interestingly, S. aureus biofilm production is induced under iron-restricted conditions and is repressed by iron via a Fur-independent mechanism, suggesting that the effect of lactoferrin may depend on the bacterial species (284). The effect of lactoferrin can be increased by the adjunction of xylitol, a rare sugar that inhibits the ability of the bacteria to produce siderophores under conditions of iron restriction (285). Such an association could be proposed in case of chronic wounds colonized by P. aeruginosa biofilm (286). Assessment of the antibiofilm efficacy of other known iron chelators and development of new iron chelators targeting biofilms might be future antibiofilm strategies to consider.
(f) Inhibition of c-di-GMP biosynthesis.
The inhibition of c-di-GMP biosynthesis by diguanylate cyclase (DGC) is also promising, in light of its importance in the shift from the planktonic to the biofilm lifestyle. Indeed, blocking c-di-GMP biosynthesis may keep bacteria in the planktonic state. Screening for DGC inhibitors identified sulfathiazole (287). Sulfathiazole inhibits formation of biofilms in vitro and indirectly inhibits DGC through inhibition of tetrahydrofolate biosynthesis, which affects the pool of thymidine, and DNA synthesis, rather than via enzymatic inhibition (287, 288). More direct inhibition of DGC was identified in V. cholerae, P. aeruginosa, and Acinetobacter baumanii through high-throughput screening (289, 290). Several molecules inhibiting DGC and biofilm formation of these three pathogens were identified; however, the tolerance and toxicity of most of these compounds remain to be assessed.
(g) Physical approaches.
Physical approaches have been developed to prevent biofilm formation on catheters, including low-energy surface acoustic waves and iontophoresis as preventive measures (291, 292). In the latter case, urethral catheters are modified in order to deliver a current to electrodes located on the catheter tip, leading to production of ions of soluble salts and allowing formation of a local biocide. After 3 weeks, this approach significantly reduced the bacterial burden in urine. Surface acoustic waves have also been proposed for the eradication of biofilms, in conjunction with antibiotics (293).
(ii) Jamming communication through inhibition of quorum sensing.
The objective of the jamming approach is to inhibit biofilm formation by altering the progression from initial attachment to microcolonies and development of a mature biofilm. As quorum sensing (QS) is a key component of biofilm communication, many authors have speculated that interfering with QS signals might alter biofilm maturation, thereby leading to easier eradication. However, the main limitation of QS inhibition is the spectrum of action, which depends on the type of QS system used by the microorganism responsible for the infection.
RNAIII-inhibiting peptide (RIP), a compound interfering with S. aureus QS, efficiently prevents CVC-related infection in vivo, alone or in association with antibiotics, in a rat model (294). Similar in vitro and in vivo data have been published for S. epidermidis (295). Aside from the CVC situation, RIP has been assessed in other types of biofilm-related infections, such as biofilms formed by S. epidermidis or S. aureus on chronic wounds, where it reduces the healing time in vivo (296).
In P. aeruginosa, different molecules have been developed to interfere with QS signals. Azithromycin has poor antimicrobial activity against P. aeruginosa, but it interferes with lasI-mediated QS signals (297–299). It was shown to inhibit P. aeruginosa biofilm formation and virulence factor expression both in vitro and in vivo (297, 300). Clinical studies in CF patients colonized by P. aeruginosa demonstrated an improvement in respiratory function and a reduced number of exacerbations compared with the placebo (301). Nevertheless, recent data suggest that the chronic use of azithromycin might be associated with side effects, such as ototoxicity and an increased level of bacterial resistance (302, 303). Possible cardiovascular toxicity has been described, with conflicting results (304). As acyl-homoserine lactones (AHLs) play a key role in the development of P. aeruginosa biofilms, inhibitors of these autoinducers have been developed (305). An N-acyl-homoserine lactone hydrolase (BpiB05) was identified through screening of a soil metagenome, and it inhibits P. aeruginosa biofilm formation in vitro (306). Along with AHLs, synthetic furanones derived from an algal metabolite now constitute potential prevention candidates, as they inhibit Gram-negative bacterial QS through their fixation to LasR and inhibition of the action of AHLs (288, 307). In vitro and in vivo studies reported reduced biofilm formation, virulence factor expression, and antibiotic tolerance of P. aeruginosa biofilms exposed to furanones (308, 309). However, furanones have a narrow spectrum of activity, as they are efficient only against bacteria that share this QS signaling pathway (309, 310). Furthermore, the use of halogenated furanones remains hampered by their carcinogenic effects as well as poor stability in aqueous solutions.
Through screening of chemical libraries, different QS inhibitors have been identified, such as garlic extract and 1-isothiocyanato-3-(methylsulfinyl)propane, also known as iberin, from horseradish (311, 312). The compound isolated from garlic, ajoene (4,5,9-trithiadodeca-1,6,11-triene-9-oxide), was shown to increase P. aeruginosa biofilm susceptibility to tobramycin and PMN activity in vitro (311, 313). In vivo, mice treated with garlic extract for 7 days, with the initial 2 days being given before P. aeruginosa instillation, had more severe initial inflammation but better bacterial clearance of the infection than placebo-treated mice (313). Another example of a QS inhibitor identified in vegetal matter is green tea epigallocatechin gallate, which was shown to reduce QS, biofilm development, and virulence factor production of P. aeruginosa through inhibition of the enoyl-acyl carrier protein reductase (ENR), ultimately leading to a reduction in 3OC12-HSL of the las QS system (314).
Lastly, different authors have proposed grafting enzymes able to digest QS signals, called quorum-quenching molecules, on the surface in order to inhibit bacterial adhesion (315, 316).
(iii) Vaccination.
The goal of vaccination is to induce the production of antibodies against bacterial biofilm antigens, such as structures involved in adhesion or biofilm maturation. This strategy requires predefining groups of patients about to be exposed to the risk of biofilm-associated infection and treating them before exposure. A relevant example is the scheduled implantation of devices such as heart prosthetic valves, pacemakers, and prosthetic joints. This strategy may also be relevant for patients exposed to chronic tissue-associated infections, such as CF patients or patients suffering from recurrent UTI. Ideally, biofilm-specific antigens should be used to increase the effect of vaccination. Choosing the right antigen remains an arduous task due to the obvious redundancy of bacterial appendages involved in adhesion and biofilm formation. Therefore, current strategies are aimed at using an antigenic cocktail (317, 318). For UTI, in vitro and in vivo studies demonstrated that immunization with FimH or components of the P pilus from UPEC reduced in vivo colonization of the bladder mucosa (319, 320). For CVC-related infections, a rat model enabled assessment of immunization of rats prior to catheter insertion, leading to a protective effect in bacterial colonization of the device by S. aureus or S. epidermidis (321). In that study, two different antigens were used: SERP0630 (MenD) (for S. epidermidis) and SACOL1138, or iron-regulated surface determinant B (IsdB) (for S. aureus). With S. aureus, a recent study reported that extracellular proteins found in the biofilm matrix could induce a protective immune response that prevented subsequent infections (322).
Aside from vaccination aimed at preventing bacterial adhesion, it has also been suggested that vaccination will increase the likelihood of biofilm eradication (323). Antigens were chosen (glucosaminidase, an ABC transporter lipoprotein, a conserved hypothetical protein, and a conserved lipoprotein) because they are upregulated in biofilms both in vitro and in vivo. In a rabbit osteomyelitis model, the association of antibiotics and vaccination significantly increased the rate of therapeutic success (323). In that model, vaccination was initiated 30 days prior to the onset of infection, thus reducing the impact of the findings.
(iv) Use of nonpathogenic bacteria to prevent colonization.
The use of nonpathogenic bacteria to prevent colonization relies on nonpathogenic bacteria that are able to efficiently colonize a surface and thus compete with other bacterial pathogens and prevent their adhesion (324). The best-documented case is the E. coli 83972 strain, which is responsible for asymptomatic bacteriuria (ABU). This strain lacks most virulence factors and UTI-associated adhesins and fails to induce bladder inflammation (325). It was observed that antibiotic treatment of patients with ABU led to a paradoxical increase in the risk of UTI by other bacteria, thus leading to the hypothesis that E. coli 83972 bladder colonization could prevent the occurrence of UTI. Since then, different clinical studies have demonstrated that bladder inoculation with E. coli 83972 has beneficial effects. For instance, in a clinical pilot study, patients with incomplete bladder emptying and recurrent UTI were randomized to receive blinded bladder inoculations with E. coli 83972 or saline (326). Inoculated patients experienced significantly fewer UTI during the 12 months following inoculation.
Hence, several promising strategies have been developed to prevent microbial adhesion and biofilm formation. Only a few of them have undergone in vivo efficacy tests, and for most of them, the precise mechanisms of action remain unknown (Table 2).
TABLE 2.
Antibiofilm strategies originating from fundamental research
| Mode of action | Commentsa |
Reference(s) | |
|---|---|---|---|
| In vitro | In vivo | ||
| Inhibition of microbial adhesion | |||
| Material optimization | Physical nature of the substrate (zirconium versus titanium) and its handling (electropolished versus sandblasted) reduce bacterial adhesion | Physical nature of the substrate (zirconium versus titanium) and its handling (electropolished versus sandblasted) reduce bacterial adhesion | 254, 252, 253, 375 |
| Specific polymeric material | Reduction of S. aureus and E. coli adhesion | Reduction of S. aureus and E. coli adhesion | 255 |
| Modify physical architecture of the surface (sharklet micropattern) | Reduction of E. coli, P. aeruginosa, A. baumannii, and K. pneumoniae adhesion | 256, 257 | |
| Grafting of antiadhesive molecules | Reduction of S. aureus adhesion and fibronectin adsorption on titanium grafted with specific peptides | Nonleaching polymeric sulfobetaine reduces bacterial adhesion and thrombus formation on the surface of a catheter | 258, 260, 259 |
| Killing of bacteria upon contact | Polycationic chains attached to glass slides or single-walled carbon nanotubes | 261, 262 | |
| Biosurfactants | Reduction of biofilm formation (E. coli, P. mirabilis, Candida spp., S. aureus) | 265, 267, 266 | |
| Other nonantibiotic coatings | |||
| Antibody-releasing surfaces | IgG opsonization inhibits bacterial adhesion and increases bacterial killing by neutrophils | 376, 268 | |
| Nitric oxide-releasing surfaces | NO-coated surfaces in rats show reductions in S. aureus colonization | 270 | |
| Association of antiseptic (triclosan) and dispersin B | Prevention of S. aureus colonization | Prevention of S. aureus colonization on vascular catheters | 271 |
| Gendine-coated surfaces | Reduction of E. coli adhesion | Reduction of E. coli adhesion on a urinary catheter model | 273 |
| Inhibition of production of adhesins (FN075, ring-fused 2-pyridones) | Inhibits curli biogenesis in uropathogenic E. coli and inhibits biofilm formation | Inhibits biofilm formation and attenuates virulence in a mouse model of urinary tract infection | 274 |
| Blocking the receptor of adhesins | |||
| Mannosides | Target the FimH type 1 pilus lectin of UPEC | Prevent UTI or treat an established chronic urinary tract infection | 275, 276, 277 |
| Coverage of a surface with multivalent galabiose derivatives | Inhibits adhesion of E. coli through inhibition of P fimbriae | 280, 278 | |
| Lactoferrin | Inhibits irreversible adhesion of P. aeruginosa (via iron chelation) | 282 | |
| EDTA (divalent cation chelator) | Prevents biofilm formation | When associated with minocycline, reduces the risk of catheter-related infections | 377 |
| Inhibition of c-di-GMP biosynthesis | |||
| DGC inhibitor (sulfathiazole) | Indirect effect through inhibition of tetrahydrofolate biosynthesis | 287, 288 | |
| Direct inhibitors of DGC | Inhibits DGC and biofilm formation of V. cholerae, P. aeruginosa, and A. baumannii | 289, 290 | |
| Physical approaches | |||
| Low-energy surface acoustic waves | Reduction of E. coli biofilm formation on urinary catheters | 292 | |
| Iontophoresis | Reduction of E. coli biofilm formation on urinary catheters | 291 | |
| Jamming communications | |||
| Quorum-sensing inhibitors | |||
| RNAIII-inhibiting peptide (RIP) | Reduces adhesion and virulence of S. aureus | With S. aureus and S. epidermidis, reduces colonization of CVC (rats) and improves healing of chronic wounds (mice) | 294, 296 |
| Azithromycin | Inhibits biofilm formation and virulence of P. aeruginosa | Inhibits biofilm formation and virulence of P. aeruginosa | 297, 301 |
| Acyl-homoserine lactone inhibitors or hydrolases | Reduce P. aeruginosa biofilm formation | 306, 305 | |
| Furanones | Inhibit biofilm formation and virulence of P. aeruginosa | Reduce P. aeruginosa virulence | 308, 288 |
| Garlic extract (ajoene) | Increases P. aeruginosa biofilm susceptibility to tobramycin and PMN activity | Increases bacterial clearance in a mouse P. aeruginosa infection model | 154, 311 |
| Green tea epigallocatechin gallate | Reduction of QS, biofilm development, and virulence factor production of P. aeruginosa | 314 | |
| Quorum-quenching molecules | Inhibit bacterial adhesion | 315, 316 | |
| Vaccination | |||
| Prevent initial adhesion | Prevents CVC colonization by S. epidermidis and S. aureus in rats | 321 | |
| Improve eradication of a biofilm-related infection | In a rabbit osteomyelitis model, the combination of antibiotic and vaccination significantly increased the rate of therapeutic success | 323 | |
| Other preventive measures | |||
| Using nonpathogenic bacteria (E. coli 83972) | Inoculated patients experienced fewer UTI during the 12 months following inoculation | 325, 326 | |
| Cerium nitrate, chitosan, and hamamelitannin | Prevent formation of biofilms of S. epidermidis, S. aureus, Acinetobacter baumannii, and Candida albicans | Prevention of formation of biofilms of S. epidermidis, S. aureus, Acinetobacter baumannii, and C. albicans in a subcutaneous CVC model | 378, 379 |
| Allicin from garlic | Inhibits PIA biosynthesis and biofilm development of S. epidermidis | 380 | |
| Sulfhydril compounds (dithiothreitol or cysteine) | Inhibit S. aureus biofilm formation through inhibition of ica | 381 | |
| Ginseng extract | Prevents and disperses P. aeruginosa biofilm; stimulates swimming and twitching motilities | Oral administration of ginseng enhances phagocytosis of P. aeruginosa | 382 |
| Blockade of DNA replication (5-fluorouracil [5-FU]) | Reduces the virulence and biofilm formation of P. aeruginosa and E. coli | 5-FU-coated catheters in clinics for prevention of catheter colonization | 383, 384, 385 |
| Favoring dispersal | |||
| Enzymes | |||
| DNase I | Favors dispersion of S. aureus, P. aeruginosa, or Gardnerella vaginalis | Inhibits Gardnerella vaginalis biofilm formation | 333, 330, 334 |
| Dispersin B (against PNAG) | Favors dispersal of S. epidermidis more than that of S. aureus | With triclosan (antiseptic), reduces S. aureus colonization in a rabbit CVC model | 329, 330 |
| Divalent cation chelator (EDTA) | Dispersal and lysis of P. aeruginosa biofilm bacteria | Gentamicin-EDTA combination eradicates Gram-positive and Gram-negative biofilms | 340, 87 |
| Modulation of quorum sensing | |||
| Autoinducing peptide | Induces S. aureus biofilm dispersal | 345 | |
| RIP | RIP plus teicoplanin was used against methicillin-resistant S. aureus biofilms in chronic wounds | 346 | |
| cis-2-Decenoic acid | Induces dispersal of S. aureus, E. coli, C. albicans, and Streptococcus pyogenes biofilms | 347 | |
| d-Amino acids | Induce B. subtilis biofilm dispersal and prevent biofilm formation by S. aureus and P. aeruginosa | 348 | |
| Norspermidine | Induces B. subtilis biofilm dispersal and prevents biofilm formation by S. aureus and E. coli; ongoing controversy | 351, 350 | |
| Nitric oxide | Induces dispersal of P. aeruginosa biofilm | 352, 353 | |
| Phages | Phage PT-6 produces an alginase that favors P. aeruginosa dispersal | 362 | |
| Antipersister compounds | |||
| Sugars (mannitol or fructose) plus aminoglycosides | Increase mortality among persisters (S. aureus, E. coli, and P. aeruginosa) | Increase aminoglycoside efficacy against E. coli biofilms (catheter-associated urinary tract infection) | 86, 355 |
| Silver plus antibiotics | Increases the effect of gentamicin, ofloxacin, or ampicillin against planktonic and biofilm persisters | Increases the effect of gentamicin, ofloxacin, or ampicillin against planktonic and biofilm persisters | 356 |
| ADEP4 plus rifampin | Eradicates S. aureus biofilms | Eradicates S. aureus biofilms | 88 |
| C10 plus norfloxacin | Increases mortality among E. coli persisters | 386 | |
| Farnesol | Reduces S. aureus tolerance toward gentamicin | 387 | |
| Bacteriophages | Phage T4 against E. coli or phage F116 against P. aeruginosa | Reduce mouse ileum colonization by E. coli | 358, 359, 388, 364, 361 |
| Other compounds for eradicating biofilms | |||
| N-Acetylcysteine | Eradication of biofilms formed by Gram-positive and Gram-negative pathogens | Associated with tigecycline in a clinical study | 389, 390, 391 |
| Honey | Antibiofilm (Enterobacter cloacae, P. mirabilis, P. aeruginosa, S. aureus, and S. pyogenes) and anti-inflammatory effects; inhibits QS signals and represses curli genes (csgBAC) or indole biosynthesis | Adjunct therapy for chronic wound care | 392, 393, 394, 395, 396, 397, 398, 399 |
| Cranberry or selenium, with or without ciprofloxacin | Treatment of chronic bacterial prostatitis | 400, 401 | |
| Catechin (extract from Chinese tea) plus ciprofloxacin | Treatment of chronic bacterial prostatitis | 402 | |
| Synthetic antimicrobial peptidomimetics (SAMP) | Active against staphylococcal biofilms | 403 | |
| Electrical current (in the presence of NaCl) | Active against S. epidermidis or P. aeruginosa biofilms, through generation of free chlorine and, ultimately, hypochlorous acid and hypochlorite | 404 | |
| Ultrasound-mediated microbubbles | Enhance vancomycin effect against S. epidermidis biofilms | 405 | |
CAUTI, catheter-associated urinary tract infection; CVC, central venous catheter; NO, nitric oxide; PNAG, poly-N-acetylglucosamine; QS, quorum sensing; RIP, RNAIII-inhibiting peptide; UPEC, uropathogenic E. coli; UTI, urinary tract infection.
New approaches to eradicating already formed biofilms.
Most currently used strategies for biofilm eradication were developed even before the identification of the importance of biofilms in human medicine. Many clinical studies have been conducted using robust endpoints, such as clinical and/or microbiological cures and infection recurrence. Therefore, major improvements have already been made in these fields. However, several therapeutic failures are still being observed, even when patients are managed at reference centers. First, cure rates never reach 100%, and the risk of treatment failure can even reach 50%, depending on host and pathogen factors. Furthermore, most currently used strategies rely on antibiotics, thereby increasing the selective pressure and the risk of antibiotic resistance. Finally, prolonged treatment is frequently required, leading to considerable medical cost and toxicity.
Nonantibiotic compounds, used alone or in combination with antibiotics to increase the likelihood of biofilm eradication or to reduce the length of treatment, are therefore viewed as modern “holy grails.” Recent breakthroughs in understanding biofilm recalcitrance have given rise to plausible therapeutic strategies.
(i) Induction of dispersal.
Inducing dispersal is a tempting strategy; indeed, biofilm bacteria lose some of their antibiotic tolerance when they return to a planktonic state and are exposed to the host immune system (14). However, the dispersal approach needs to be associated with the use of systemic antibiotics, as release of biofilm bacteria into the bloodstream can lead to severe sepsis (327, 328). Several strategies have been proposed to induce biofilm dispersal.
(a) Enzymes for dissociating components of the ECM.
Because ECM plays an important role in maintaining biofilm stability and structure, it has been speculated that use of an enzyme able to dissociate or digest ECM components would lead to dispersal of the biofilm. Two main targets have been identified: poly-N-acetylglucosamine (PNAG) and extracellular DNA. Dispersin B is a hexosaminidase produced by Aggregatibacter actinomycetemcomitans that hydrolyzes PNAG, an important component of S. epidermidis ECM. It is therefore effective against biofilms formed by this bacterial species (329, 330). It should be noted that PNAG is also produced by some S. aureus strains, as well as E. coli. On the other hand, given the important role played by extracellular DNA in the structure of the biofilm matrix (331, 332), DNase I, an enzyme that degrades DNA, was efficiently used to dissolve biofilms from a broad range of bacteria, including P. aeruginosa, S. aureus, and Gardnerella vaginalis (330, 333, 334).
However, the enzyme-based approach is associated with two limitations: (i) the restricted spectrum of action and (ii) the risk of immunization against these molecules.
(b) Divalent cation chelators.
Since divalent cations play a key role in maintaining biofilm ECM stability and cohesiveness, another approach is to use chelators such as EDTA and citrate (335, 336). For instance, calcium ions cross-link alginate, and the calcium concentration was shown to be critical for maintaining P. aeruginosa biofilm resistance toward compressive stresses (337). Iron has also been demonstrated to be an important cross-linker of the ECM (338). However, little is known about the mode of action and precise effects of chelators on biofilms. One study in 1983 reported that the addition of EGTA, a specific calcium chelator, led to immediate detachment of a mixed bacterial film from the walls of a recycle tube reactor (339). EDTA at 50 mM has been shown to induce dispersal and lysis of P. aeruginosa biofilm bacteria (340). Strikingly, addition of calcium, iron, or magnesium inhibited the phenotype. In that study, EGTA led to the same dispersal phenotype, but without inducing lysis. Furthermore, citrate and EDTA were also shown to exhibit direct bactericidal effects against planktonic bacteria (340, 341).
Many in vitro studies have reported a synergistic antibiofilm effect of EDTA combined with gentamicin or minocycline-25% ethanol (340, 342, 343). Using a rat model of biofilm-related infection with a totally implantable venous access port (TIVAP), it was recently shown that the gentamicin-EDTA combination was the most effective lock solution compared to gentamicin alone, EDTA alone, or ethanol (70%) (87, 344). Gentamicin-EDTA is therefore a potential lock solution able to cure highly tolerant biofilms and eradicate persistent bacteria, thereby preventing recurrence of Gram-positive as well as Gram-negative bacterial biofilms on TIVAP (344).
(c) QS signals.
While QS signaling can be targeted to interfere with biofilm formation, some QS signals can also be used to trigger dispersal of a biofilm. In S. aureus, the agr (accessory gene regulator) QS system is strongly expressed by the bacterial population at the moment of dispersion. Artificial stimulation of this system, through adjunction of autoinducing peptide (AIP), leads to S. aureus biofilm dispersal (345). In vivo murine models also helped to reveal the effect of RIP (a quorum sensing inhibitor) in combination with teicoplanin against methicillin-resistant S. aureus in the setting of chronic wound biofilm colonization (346). Lastly, with P. aeruginosa, analysis of spent medium led to the discovery of a short-chain fatty acid that is implicated in bacterium-bacterium communication (cis-2-decenoic acid) and is able to induce dispersal in a wide range of Gram-positive as well as Gram-negative bacteria (347).
Like preventative QS signal jamming, this strategy is limited by the spectrum of action of each QS molecule.
(d) Other strategies for inducing dispersal.
Bacillus subtilis produces a mixture of d-amino acids (d-leucine, d-methionine, d-tyrosine, and d-tryptophan) that disperse existing biofilms through release of TasA, an amyloid fiber that links biofilm bacteria together. d-Amino acids also prevent biofilm formation by S. aureus and P. aeruginosa (348). While the same group reported that, in fact, the effect of d-amino acids on B. subtilis biofilm dispersal is due to their misincorporation into proteins via a mutation in the dtd gene, encoding d-Tyr-tRNA deacylase (349), the mechanisms by which d-amino acids affect biofilm formation by S. aureus and P. aeruginosa remain to be elucidated. The same group reported that B. subtilis also produces an additional biofilm disassembly factor, norspermidine, that interacts directly and disrupts exopolysaccharides. Strikingly, norspermidine also prevents biofilm formation by B. subtilis, E. coli, and S. aureus (350). However, another group recently published conflicting results demonstrating that norspermidine is not involved in biofilm disassembly (351).
Nitric oxide (NO) can induce dispersal of P. aeruginosa biofilms through induction of phosphodiesterases and, ultimately, a reduction in c-di-GMP levels (352, 353). Exposure to the NO donor sodium nitroprusside (SNP) not only induces dispersal but also increases the activity of antimicrobial compounds, such as tobramycin, against established P. aeruginosa biofilms (352). Exposure to low levels of NO in P. aeruginosa biofilms induces genes involved in motility and energy metabolism and downregulation of adhesins and virulence factors (353). Notably, the chemotaxis transducer BdlA is involved in the NO-induced biofilm dispersal response.
(ii) Eradication of persisters.
Another straightforward approach to fighting biofilms is to increase the activity of antibiotics against biofilm bacteria. As persister cells play a key role in the recalcitrance of biofilms toward antibiotics, the identification of compounds or associations that are active against persisters is important.
An adjuvant approach was recently proposed, based on the association of aminoglycosides and a sugar, such as mannitol or fructose, in order to increase antibiotic uptake and action against persister cells. After the sugar is taken up, it stimulates glycolysis, leading to the production of NADH, which, in turn, stimulates enzymes such as NADH dehydrogenases (NDH) or quinol oxidase. The electron transport chain oxidizes NADH and extrudes H+ ions, thereby increasing the proton motive force (PMF). PMF, also called Δp, is composed of Δψ (the electrical potential across the membrane) and ΔpH (the transmembrane difference in H+ concentration) (63, 354). In the case of mannitol or fructose, the generation of NADH stimulates PMF through an increase in Δψ (86). This stimulation of PMF leads to increased aminoglycoside uptake, and thus increased mortality of E. coli and S. aureus persisters in vitro and of E. coli in vivo, in a model of CAUTI. Recently, mannitol was also demonstrated to increase the aminoglycoside tobramycin's efficacy against P. aeruginosa biofilm persister cells, therefore pointing to a promising adjuvant with a broad range of activity (355).
Silver, usually used as an antimicrobial agent to coat material, was also proposed as an adjuvant to antibiotics against Gram-negative bacteria (356). It was demonstrated that silver could increase ROS production and membrane permeability and thus increase the effect of gentamicin, ofloxacin, or ampicillin against planktonic and biofilm persisters both in vitro and in vivo (356). It was also recently shown that persisters can be killed through the association of an antibiotic (rifampin) and a compound (ADEP4, an acyldepsipeptide) able to activate ClpP (88). The activation of ClpP through ADEP4 results in the degradation of more than 400 proteins. This association has been shown to successfully eradicate in vitro and in vivo S. aureus biofilms.
Finally, irrespective of the fact that the issue is currently being debated, several groups have tried to decrease persister tolerance through an increase in ROS production (121, 125, 126). Different potential targets have been identified by use of genome-scale metabolic models to predict ROS production (357).
(iii) Bacteriophages.
The worldwide spread of multidrug-resistant bacteria and the shortage of new antibiotics are now leading to a revival of interest in phage therapy. Different authors have proposed the use of bacteriophages for eradication of biofilms.
One classical example is their use in lock therapy to treat catheter-related infections (237). Via this approach, microorganisms responsible for the infection should first be screened against a bank of phages so as to choose the phage strain associated with the greatest lytic capacity (237). Different phages have been used, including phage T4 against E. coli and phage F116 against P. aeruginosa (358–361). Note that some bacteriophages have also been reported to induce biofilm dispersal, such as PT-6, which induces P. aeruginosa alginase (362). The use of bacteriophages has also been proposed as a preventive measure, e.g., pretreated hydrogel-coated catheters with a cocktail of five P. aeruginosa bacteriophages were used in an in vitro model (363). Cocktails of phages are therefore a promising strategy for fighting biofilms. Recently, a cocktail of three phages was shown to reduce mouse ileum colonization by an O104:H4 enteroaggregative strain of E. coli (364). The onset of potential resistance and long-term innocuity must now be evaluated in order to validate these strategies.
In concluding this section, it is worth noting that many other compounds have been proposed or used to treat established biofilms, with as yet unknown mechanisms of action and untested clinical value. As a consequence, establishing a complete list of these antibiofilm, antibacterial, and sometimes immunomodulatory molecules is an arduous task (Table 2).
CONCLUDING REMARKS
Biofilm recalcitrance toward antibiotics is responsible for most of the difficulties encountered in the treatment of biofilm-related infections. Major advances have been made in the characterization of factors associated with this problematic biofilm property. Recognition of the role played by persister cells and the recent identification of several molecular mechanisms involved in the generation of persisters have already led to several potential antibiofilm treatments. Validation of these new approaches will likely require renewed interactions between fundamental research and clinical practice before these approaches can be included in future therapeutic arsenals for use against difficult-to-treat infections.
ACKNOWLEDGMENTS
This work was supported by an Institut Pasteur grant and by the French Government's Investissement d'Avenir Program, Integrative Biology of Emerging Infectious Diseases Laboratoire d'Excellence (grant ANR-10-LABX-62-IBEID). D.L. was supported by a grant from the AXA Research Fund.
Biographies

David Lebeaux earned his M.D. in 2010 from the University Paris VI, France, studying clinical features of catheter-related infections. He then spent 3 years in the Genetics of Biofilm Laboratory at the Pasteur Institute, where he investigated biofilm recalcitrance toward antibiotics. He earned his Ph.D. from the University of Paris VII in 2013. He is now a clinical fellow in the Infectious Diseases Unit at Necker Hospital, Paris, France, working on device-related infections and infections in immunocompromised hosts.

Jean-Marc Ghigo obtained his Ph.D. in 1994 at the Institut Pasteur, in the laboratory of Cécile Wandersman, on the subject of protein secretion and iron acquisition in Gram-negative bacteria. In 1996, as a postdoctoral fellow, he joined the laboratory of Jon Beckwith at Harvard Medical School to study bacterial cell division. In 1999, he returned to the Institut Pasteur to develop a project on bacterial biofilm formation. Since then, the studies undertaken in his laboratory have been aimed at revealing new and underexplored molecular aspects of the bacterial biofilm lifestyle. He is now Professor and Deputy Director of the Department of Microbiology.

Christophe Beloin received his Ph.D. in 1998 from the University of Paris XI, France, and his postdoctoral work was performed at the Moyne Institute of Preventive Medicine, Department of Microbiology, Trinity College, Dublin, Ireland. From 2001 to 2013, he worked as an Assistant Professor in the Department of Microbiology, Unit of Genetics of Biofilms, Institut Pasteur, Paris, France, and he is currently an Associate Professor in the same department. Dr. Beloin's research interests involve the identification and characterization of new bacterial adhesins and the understanding of molecular mechanisms beyond the extreme recalcitrance of bacterial biofilms toward antibiotics.
REFERENCES
- 1.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. 10.1126/science.284.5418.1318 [DOI] [PubMed] [Google Scholar]
- 2.Donlan RM, Costerton JW. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167–193. 10.1128/CMR.15.2.167-193.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies J. 2007. Microbes have the last word. A drastic re-evaluation of antimicrobial treatment is needed to overcome the threat of antibiotic-resistant bacteria. EMBO Rep. 8:616–621. 10.1038/sj.embor.7401022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holsclaw DS. 1980. Cystic fibrosis: overview and pulmonary aspects in young adults. Clin. Chest Med. 1:407–421 [PubMed] [Google Scholar]
- 5.Geesey GG, Mutch R, Costertin JW, Green RB. 1978. Sessile bacteria: an important component of the microbial population in small mountain streams. Limnol. Oceanogr. 23:1214–1223. 10.4319/lo.1978.23.6.1214 [DOI] [Google Scholar]
- 6.Geesey GG, Richardson WT, Yeomans HG, Irvin RT, Costerton JW. 1977. Microscopic examination of natural sessile bacterial populations from an alpine stream. Can. J. Microbiol. 23:1733–1736. 10.1139/m77-249 [DOI] [PubMed] [Google Scholar]
- 7.Henrici AT. 1933. Studies of freshwater bacteria. I. A direct microscopic technique. J. Bacteriol. 25:277–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zobell CE, Allen EC. 1935. The significance of marine bacteria in the fouling of submerged surfaces. J. Bacteriol. 29:239–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Høiby N. 1977. Pseudomonas aeruginosa infection in cystic fibrosis. Diagnostic and prognostic significance of Pseudomonas aeruginosa precipitins determined by means of crossed immunoelectrophoresis. A survey. Acta Pathol. Microbiol. Scand. 1977(Suppl):1–96 [PubMed] [Google Scholar]
- 10.Lam J, Chan R, Lam K, Costerton JW. 1980. Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect. Immun. 28:546–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lebeaux D, Chauhan A, Rendueles O, Beloin C. 2013. From in vitro to in vivo models of bacterial biofilm-related infections. Pathogens 2:288–356. 10.3390/pathogens2020288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marrie TJ, Nelligan J, Costerton JW. 1982. A scanning and transmission electron microscopic study of an infected endocardial pacemaker lead. Circulation 66:1339–1341. 10.1161/01.CIR.66.6.1339 [DOI] [PubMed] [Google Scholar]
- 13.Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5:48–56. 10.1038/nrmicro1557 [DOI] [PubMed] [Google Scholar]
- 14.Lewis K. 2001. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45:999–1007. 10.1128/AAC.45.4.999-1007.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Espeland EM, Wetzel RG. 2001. Complexation, stabilization, and UV photolysis of extracellular and surface-bound glucosidase and alkaline phosphatase: implications for biofilm microbiota. Microb. Ecol. 42:572–585. 10.1007/s00248-001-1023-7 [DOI] [PubMed] [Google Scholar]
- 16.Le Magrex-Debar E, Lemoine J, Gelle MP, Jacquelin LF, Choisy C. 2000. Evaluation of biohazards in dehydrated biofilms on foodstuff packaging. Int. J. Food Microbiol. 55:239–243. 10.1016/S0168-1605(00)00177-X [DOI] [PubMed] [Google Scholar]
- 17.Leid JG, Shirtliff ME, Costerton JW, Stoodley P. 2002. Human leukocytes adhere to, penetrate, and respond to Staphylococcus aureus biofilms. Infect. Immun. 70:6339–6345. 10.1128/IAI.70.11.6339-6345.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNeill K, Hamilton IR. 2003. Acid tolerance response of biofilm cells of Streptococcus mutans. FEMS Microbiol. Lett. 221:25–30. 10.1016/S0378-1097(03)00164-2 [DOI] [PubMed] [Google Scholar]
- 19.Teitzel GM, Parsek MR. 2003. Heavy metal resistance of biofilm and planktonic Pseudomonas aeruginosa. Appl. Environ. Microbiol. 69:2313–2320. 10.1128/AEM.69.4.2313-2320.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart PS, Costerton JW. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135–138. 10.1016/S0140-6736(01)05321-1 [DOI] [PubMed] [Google Scholar]
- 21.Walsh C. 2000. Molecular mechanisms that confer antibacterial drug resistance. Nature 406:775–781. 10.1038/35021219 [DOI] [PubMed] [Google Scholar]
- 22.Anderl JN, Franklin MJ, Stewart PS. 2000. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 44:1818–1824. 10.1128/AAC.44.7.1818-1824.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Craig WA. 1998. Pharmacokinetic/pharmacodynamic parameters: rationale for antibacterial dosing of mice and men. Clin. Infect. Dis. 26:1–10. 10.1086/516284 [DOI] [PubMed] [Google Scholar]
- 24.Craig WA, Redington J, Ebert SC. 1991. Pharmacodynamics of amikacin in vitro and in mouse thigh and lung infections. J. Antimicrob. Chemother. 27(Suppl C):29–40. 10.1093/jac/27.suppl_C.29 [DOI] [PubMed] [Google Scholar]
- 25.Lewis K. 2000. Programmed death in bacteria. Microbiol. Mol. Biol. Rev. 64:503–514. 10.1128/MMBR.64.3.503-514.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henriques Normark B, Normark S. 2002. Antibiotic tolerance in pneumococci. Clin. Microbiol. Infect. 8:613–622. 10.1046/j.1469-0691.2002.00477.x [DOI] [PubMed] [Google Scholar]
- 27.Spoering AL, Lewis K. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183:6746–6751. 10.1128/JB.183.23.6746-6751.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaudaux P. 1998. Phenotypic antibiotic tolerance of Staphylococcus aureus in implant-related infections: relationship with in vitro colonization of artificial surfaces. Drug Resist. Updat. 1:352–357. 10.1016/S1368-7646(98)80011-3 [DOI] [PubMed] [Google Scholar]
- 29.Proctor RA, von Eiff C, Kahl BC, Becker K, McNamara P, Herrmann M, Peters G. 2006. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 4:295–305. 10.1038/nrmicro1384 [DOI] [PubMed] [Google Scholar]
- 30.Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. 1999. The Calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 37:1771–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis K. 2008. Multidrug tolerance of biofilms and persister cells. Curr. Top. Microbiol. Immunol. 322:107–131 [DOI] [PubMed] [Google Scholar]
- 32.Mah TF, O'Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34–39. 10.1016/S0966-842X(00)01913-2 [DOI] [PubMed] [Google Scholar]
- 33.Williams I, Venables WA, Lloyd D, Paul F, Critchley I. 1997. The effects of adherence to silicone surfaces on antibiotic susceptibility in Staphylococcus aureus. Microbiology 143:2407–2413. 10.1099/00221287-143-7-2407 [DOI] [PubMed] [Google Scholar]
- 34.Davies D. 2003. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2:114–122. 10.1038/nrd1008 [DOI] [PubMed] [Google Scholar]
- 35.Stewart PS. 2003. Diffusion in biofilms. J. Bacteriol. 185:1485–1491. 10.1128/JB.185.5.1485-1491.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gordon CA, Hodges NA, Marriott C. 1988. Antibiotic interaction and diffusion through alginate and exopolysaccharide of cystic fibrosis-derived Pseudomonas aeruginosa. J. Antimicrob. Chemother. 22:667–674. 10.1093/jac/22.5.667 [DOI] [PubMed] [Google Scholar]
- 37.Kumon H, Tomochika K, Matunaga T, Ogawa M, Ohmori H. 1994. A sandwich cup method for the penetration assay of antimicrobial agents through Pseudomonas exopolysaccharides. Microbiol. Immunol. 38:615–619. 10.1111/j.1348-0421.1994.tb01831.x [DOI] [PubMed] [Google Scholar]
- 38.Nichols WW, Dorrington SM, Slack MP, Walmsley HL. 1988. Inhibition of tobramycin diffusion by binding to alginate. Antimicrob. Agents Chemother. 32:518–523. 10.1128/AAC.32.4.518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiens JR, Vasil AI, Schurr MJ, Vasil ML. 2014. Iron-regulated expression of alginate production, mucoid phenotype, and biofilm formation by Pseudomonas aeruginosa. mBio 5:e01010-13. 10.1128/mBio.01010-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cremieux AC, Maziere B, Vallois JM, Ottaviani M, Azancot A, Raffoul H, Bouvet A, Pocidalo JJ, Carbon C. 1989. Evaluation of antibiotic diffusion into cardiac vegetations by quantitative autoradiography. J. Infect. Dis. 159:938–944. 10.1093/infdis/159.5.938 [DOI] [PubMed] [Google Scholar]
- 41.Tseng BS, Zhang W, Harrison JJ, Quach TP, Song JL, Penterman J, Singh PK, Chopp DL, Packman AI, Parsek MR. 2013. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ. Microbiol. 15:2865–2878. 10.1111/1462-2920.12155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walters MC, 3rd, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. 2003. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 47:317–323. 10.1128/AAC.47.1.317-323.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Beer D, Srinivasan R, Stewart PS. 1994. Direct measurement of chlorine penetration into biofilms during disinfection. Appl. Environ. Microbiol. 60:4339–4344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu X, Stewart PS, Chen X. 1996. Transport limitation of chlorine disinfection of Pseudomonas aeruginosa entrapped in alginate beads. Biotechnol. Bioeng. 49:93–100. [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez-Martinez JM, Ballesta S, Pascual A. 2007. Activity and penetration of fosfomycin, ciprofloxacin, amoxicillin/clavulanic acid and co-trimoxazole in Escherichia coli and Pseudomonas aeruginosa biofilms. Int. J. Antimicrob. Agents 30:366–368. 10.1016/j.ijantimicag.2007.05.005 [DOI] [PubMed] [Google Scholar]
- 46.Zheng Z, Stewart PS. 2002. Penetration of rifampin through Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 46:900–903. 10.1128/AAC.46.3.900-903.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stone G, Wood P, Dixon L, Keyhan M, Matin A. 2002. Tetracycline rapidly reaches all the constituent cells of uropathogenic Escherichia coli biofilms. Antimicrob. Agents Chemother. 46:2458–2461. 10.1128/AAC.46.8.2458-2461.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jefferson KK, Goldmann DA, Pier GB. 2005. Use of confocal microscopy to analyze the rate of vancomycin penetration through Staphylococcus aureus biofilms. Antimicrob. Agents Chemother. 49:2467–2473. 10.1128/AAC.49.6.2467-2473.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ciofu O. 2003. Pseudomonas aeruginosa chromosomal beta-lactamase in patients with cystic fibrosis and chronic lung infection. Mechanism of antibiotic resistance and target of the humoral immune response. APMIS 2003(Suppl):1–47 [PubMed] [Google Scholar]
- 50.Giwercman B, Jensen ET, Høiby N, Kharazmi A, Costerton JW. 1991. Induction of beta-lactamase production in Pseudomonas aeruginosa biofilm. Antimicrob. Agents Chemother. 35:1008–1010. 10.1128/AAC.35.5.1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bagge N, Ciofu O, Hentzer M, Campbell JI, Givskov M, Høiby N. 2002. Constitutive high expression of chromosomal beta-lactamase in Pseudomonas aeruginosa caused by a new insertion sequence (IS1669) located in ampD. Antimicrob. Agents Chemother. 46:3406–3411. 10.1128/AAC.46.11.3406-3411.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dibdin GH, Assinder SJ, Nichols WW, Lambert PA. 1996. Mathematical model of beta-lactam penetration into a biofilm of Pseudomonas aeruginosa while undergoing simultaneous inactivation by released beta-lactamases. J. Antimicrob. Chemother. 38:757–769. 10.1093/jac/38.5.757 [DOI] [PubMed] [Google Scholar]
- 53.Nichols WW, Evans MJ, Slack MP, Walmsley HL. 1989. The penetration of antibiotics into aggregates of mucoid and non-mucoid Pseudomonas aeruginosa. J. Gen. Microbiol. 135:1291–1303 [DOI] [PubMed] [Google Scholar]
- 54.Stewart PS. 1996. Theoretical aspects of antibiotic diffusion into microbial biofilms. Antimicrob. Agents Chemother. 40:2517–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stewart PS, Franklin MJ. 2008. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6:199–210. 10.1038/nrmicro1838 [DOI] [PubMed] [Google Scholar]
- 56.Huang CT, Xu KD, McFeters GA, Stewart PS. 1998. Spatial patterns of alkaline phosphatase expression within bacterial colonies and biofilms in response to phosphate starvation. Appl. Environ. Microbiol. 64:1526–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sternberg C, Christensen BB, Johansen T, Toftgaard Nielsen A, Andersen JB, Givskov M, Molin S. 1999. Distribution of bacterial growth activity in flow-chamber biofilms. Appl. Environ. Microbiol. 65:4108–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wentland EJ, Stewart PS, Huang CT, McFeters GA. 1996. Spatial variations in growth rate within Klebsiella pneumoniae colonies and biofilm. Biotechnol. Prog. 12:316–321. 10.1021/bp9600243 [DOI] [PubMed] [Google Scholar]
- 59.Tuomanen E, Cozens R, Tosch W, Zak O, Tomasz A. 1986. The rate of killing of Escherichia coli by beta-lactam antibiotics is strictly proportional to the rate of bacterial growth. J. Gen. Microbiol. 132:1297–1304 [DOI] [PubMed] [Google Scholar]
- 60.Shigeta M, Komatsuzawa H, Sugai M, Suginaka H, Usui T. 1997. Effect of the growth rate of Pseudomonas aeruginosa biofilms on the susceptibility to antimicrobial agents. Chemotherapy 43:137–141. 10.1159/000239548 [DOI] [PubMed] [Google Scholar]
- 61.Tanaka G, Shigeta M, Komatsuzawa H, Sugai M, Suginaka H, Usui T. 1999. Effect of the growth rate of Pseudomonas aeruginosa biofilms on the susceptibility to antimicrobial agents: beta-lactams and fluoroquinolones. Chemotherapy 45:28–36. 10.1159/000007162 [DOI] [PubMed] [Google Scholar]
- 62.Baudoux P, Bles N, Lemaire S, Mingeot-Leclercq MP, Tulkens PM, Van Bambeke F. 2007. Combined effect of pH and concentration on the activities of gentamicin and oxacillin against Staphylococcus aureus in pharmacodynamic models of extracellular and intracellular infections. J. Antimicrob. Chemother. 59:246–253. 10.1093/jac/dkl489 [DOI] [PubMed] [Google Scholar]
- 63.Schlessinger D. 1988. Failure of aminoglycoside antibiotics to kill anaerobic, low-pH, and resistant cultures. Clin. Microbiol. Rev. 1:54–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tack KJ, Sabath LD. 1985. Increased minimum inhibitory concentrations with anaerobiasis for tobramycin, gentamicin, and amikacin, compared to latamoxef, piperacillin, chloramphenicol, and clindamycin. Chemotherapy 31:204–210. 10.1159/000238337 [DOI] [PubMed] [Google Scholar]
- 65.Ashby MJ, Neale JE, Knott SJ, Critchley IA. 1994. Effect of antibiotics on non-growing planktonic cells and biofilms of Escherichia coli. J. Antimicrob. Chemother. 33:443–452. 10.1093/jac/33.3.443 [DOI] [PubMed] [Google Scholar]
- 66.Zhao X, Malik M, Chan N, Drlica-Wagner A, Wang JY, Li X, Drlica K. 2006. Lethal action of quinolones against a temperature-sensitive dnaB replication mutant of Escherichia coli. Antimicrob. Agents Chemother. 50:362–364. 10.1128/AAC.50.1.362-364.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mah TF, Pitts B, Pellock B, Walker GC, Stewart PS, O'Toole GA. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306–310. 10.1038/nature02122 [DOI] [PubMed] [Google Scholar]
- 68.Sadovskaya I, Vinogradov E, Li J, Hachani A, Kowalska K, Filloux A. 2010. High-level antibiotic resistance in Pseudomonas aeruginosa biofilm: the ndvB gene is involved in the production of highly glycerol-phosphorylated beta-(1→3)-glucans, which bind aminoglycosides. Glycobiology 20:895–904. 10.1093/glycob/cwq047 [DOI] [PubMed] [Google Scholar]
- 69.Beaudoin T, Zhang L, Hinz AJ, Parr CJ, Mah TF. 2012. The biofilm-specific antibiotic resistance gene ndvB is important for expression of ethanol oxidation genes in Pseudomonas aeruginosa biofilms. J. Bacteriol. 194:3128-3136. 10.1128/JB.06178-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang L, Mah TF. 2008. Involvement of a novel efflux system in biofilm-specific resistance to antibiotics. J. Bacteriol. 190:4447–4452. 10.1128/JB.01655-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang L, Hinz AJ, Nadeau JP, Mah TF. 2011. Pseudomonas aeruginosa tssC1 links type VI secretion and biofilm-specific antibiotic resistance. J. Bacteriol. 193:5510–5513. 10.1128/JB.00268-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang L, Fritsch M, Hammond L, Landreville R, Slatculescu C, Colavita A, Mah TF. 2013. Identification of genes involved in Pseudomonas aeruginosa biofilm-specific resistance to antibiotics. PLoS One 8:e61625. 10.1371/journal.pone.0061625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brooun A, Liu S, Lewis K. 2000. A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 44:640–646. 10.1128/AAC.44.3.640-646.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gillis RJ, White KG, Choi KH, Wagner VE, Schweizer HP, Iglewski BH. 2005. Molecular basis of azithromycin-resistant Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 49:3858–3867. 10.1128/AAC.49.9.3858-3867.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pamp SJ, Gjermansen M, Johansen HK, Tolker-Nielsen T. 2008. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol. Microbiol. 68:223–240. 10.1111/j.1365-2958.2008.06152.x [DOI] [PubMed] [Google Scholar]
- 76.Poole K. 2008. Bacterial multidrug efflux pumps serve other functions. Microbes Environ. 3:179–185 [Google Scholar]
- 77.Mulcahy LR, Burns JL, Lory S, Lewis K. 2010. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J. Bacteriol. 192:6191–6199. 10.1128/JB.01651-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liao J, Sauer K. 2012. The MerR-like transcriptional regulator BrlR contributes to Pseudomonas aeruginosa biofilm tolerance. J. Bacteriol. 194:4823–4836. 10.1128/JB.00765-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liao J, Schurr MJ, Sauer K. 2013. The MerR-like regulator BrlR confers biofilm tolerance by activating multidrug efflux pumps in Pseudomonas aeruginosa biofilms. J. Bacteriol. 195:3352–3363. 10.1128/JB.00318-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gupta K, Marques CN, Petrova OE, Sauer K. 2013. Antimicrobial tolerance of Pseudomonas aeruginosa biofilms is activated during an early developmental stage and requires the two-component hybrid SagS. J. Bacteriol. 195:4975–4987. 10.1128/JB.00732-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gupta K, Liao J, Petrova OE, Cherny KE, Sauer K. 2014. Elevated levels of the second messenger c-di-GMP contribute to antimicrobial resistance of Pseudomonas aeruginosa. Mol. Microbiol. 92:488–506. 10.1111/mmi.12587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kvist M, Hancock V, Klemm P. 2008. Inactivation of efflux pumps abolishes bacterial biofilm formation. Appl. Environ. Microbiol. 74:7376–7382. 10.1128/AEM.01310-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lynch SV, Dixon L, Benoit MR, Brodie EL, Keyhan M, Hu P, Ackerley DF, Andersen GL, Matin A. 2007. Role of the rapA gene in controlling antibiotic resistance of Escherichia coli biofilms. Antimicrob. Agents Chemother. 51:3650–3658. 10.1128/AAC.00601-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bigger JW. 1944. Treatment of staphylococcal infections with penicillin. Lancet 244(6320):497–500. 10.1016/S0140-6736(00)74210-3 [DOI] [Google Scholar]
- 85.Kussell E, Kishony R, Balaban NQ, Leibler S. 2005. Bacterial persistence: a model of survival in changing environments. Genetics 169:1807–1814. 10.1534/genetics.104.035352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Allison KR, Brynildsen MP, Collins JJ. 2011. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473:216–220. 10.1038/nature10069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chauhan A, Lebeaux D, Ghigo JM, Beloin C. 2012. Full and broad-spectrum in vivo eradication of catheter-associated biofilms using gentamicin-EDTA antibiotic lock therapy. Antimicrob. Agents Chemother. 56:6310–6318. 10.1128/AAC.01606-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Conlon BP, Nakayasu ES, Fleck LE, LaFleur MD, Isabella VM, Coleman K, Leonard SN, Smith RD, Adkins JN, Lewis K. 2013. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 503:365–370. 10.1038/nature12790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986. 10.1126/science.1211037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lebeaux D, Fernandez-Hidalgo N, Chauhan A, Lee S, Ghigo JM, Almirante B, Beloin C. 2014. Management of infections related to totally implantable venous-access ports: challenges and perspectives. Lancet Infect. Dis. 14:146–159. 10.1016/S1473-3099(13)70266-4 [DOI] [PubMed] [Google Scholar]
- 91.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625. 10.1126/science.1099390 [DOI] [PubMed] [Google Scholar]
- 92.Kell DB, Young M. 2000. Bacterial dormancy and culturability: the role of autocrine growth factors. Curr. Opin. Microbiol. 3:238–243. 10.1016/S1369-5274(00)00082-5 [DOI] [PubMed] [Google Scholar]
- 93.Shah D, Zhang Z, Khodursky A, Kaldalu N, Kurg K, Lewis K. 2006. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 6:53. 10.1186/1471-2180-6-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kint CI, Verstraeten N, Fauvart M, Michiels J. 2012. New-found fundamentals of bacterial persistence. Trends Microbiol. 20:577–585. 10.1016/j.tim.2012.08.009 [DOI] [PubMed] [Google Scholar]
- 95.Orman MA, Brynildsen MP. 2013. Dormancy is not necessary or sufficient for bacterial persistence. Antimicrob. Agents Chemother. 57:3230–3239. 10.1128/AAC.00243-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hayes F, Van Melderen L. 2011. Toxins-antitoxins: diversity, evolution and function. Crit. Rev. Biochem. Mol. Biol. 46:386–408. 10.3109/10409238.2011.600437 [DOI] [PubMed] [Google Scholar]
- 97.Yamaguchi Y, Park JH, Inouye M. 2011. Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 45:61–79. 10.1146/annurev-genet-110410-132412 [DOI] [PubMed] [Google Scholar]
- 98.Hazan R, Sat B, Reches M, Engelberg-Kulka H. 2001. Postsegregational killing mediated by the P1 phage “addiction module” phd-doc requires the Escherichia coli programmed cell death system mazEF. J. Bacteriol. 183:2046–2050. 10.1128/JB.183.6.2046-2050.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yamaguchi Y, Inouye M. 2011. Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat. Rev. Microbiol. 9:779–790. 10.1038/nrmicro2651 [DOI] [PubMed] [Google Scholar]
- 100.Amato SM, Orman MA, Brynildsen MP. 2013. Metabolic control of persister formation in Escherichia coli. Mol. Cell 50:475–487. 10.1016/j.molcel.2013.04.002 [DOI] [PubMed] [Google Scholar]
- 101.Fasani RA, Savageau MA. 2013. Molecular mechanisms of multiple toxin-antitoxin systems are coordinated to govern the persister phenotype. Proc. Natl. Acad. Sci. U. S. A. 110:E2528–E2537. 10.1073/pnas.1301023110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Falla TJ, Chopra I. 1998. Joint tolerance to beta-lactam and fluoroquinolone antibiotics in Escherichia coli results from overexpression of hipA. Antimicrob. Agents Chemother. 42:3282–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186:8172–8180. 10.1128/JB.186.24.8172-8180.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Falla TJ, Chopra I. 1999. Stabilization of Rhizobium symbiosis plasmids. Microbiology 145:515–516. 10.1099/13500872-145-3-515 [DOI] [PubMed] [Google Scholar]
- 105.Black DS, Kelly AJ, Mardis MJ, Moyed HS. 1991. Structure and organization of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol. 173:5732–5739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Feng J, Kessler DA, Ben-Jacob E, Levine H. 2014. Growth feedback as a basis for persister bistability. Proc. Natl. Acad. Sci. U. S. A. 111:544–549. 10.1073/pnas.1320396110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schumacher MA, Piro KM, Xu W, Hansen S, Lewis K, Brennan RG. 2009. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science 323:396–401. 10.1126/science.1163806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Germain E, Castro-Roa D, Zenkin N, Gerdes K. 2013. Molecular mechanism of bacterial persistence by HipA. Mol. Cell 52:248–254. 10.1016/j.molcel.2013.08.045 [DOI] [PubMed] [Google Scholar]
- 109.Pedersen K, Christensen SK, Gerdes K. 2002. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol. Microbiol. 45:501–510. 10.1046/j.1365-2958.2002.03027.x [DOI] [PubMed] [Google Scholar]
- 110.Maisonneuve E, Shakespeare LJ, Jorgensen MG, Gerdes K. 2011. Bacterial persistence by RNA endonucleases. Proc. Natl. Acad. Sci. U. S. A. 108:13206–13211. 10.1073/pnas.1100186108 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 111.Wang X, Lord DM, Cheng HY, Osbourne DO, Hong SH, Sanchez-Torres V, Quiroga C, Zheng K, Herrmann T, Peti W, Benedik MJ, Page R, Wood TK. 2012. A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat. Chem. Biol. 8:855–861. 10.1038/nchembio.1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kim Y, Wood TK. 2010. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem. Biophys. Res. Commun. 391:209–213. 10.1016/j.bbrc.2009.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tripathi A, Dewan PC, Barua B, Varadarajan R. 2012. Additional role for the ccd operon of F-plasmid as a transmissible persistence factor. Proc. Natl. Acad. Sci. U. S. A. 109:12497–12502. 10.1073/pnas.1121217109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230:13–18. 10.1016/S0378-1097(03)00856-5 [DOI] [PubMed] [Google Scholar]
- 115.Vega NM, Allison KR, Khalil AS, Collins JJ. 2012. Signaling-mediated bacterial persister formation. Nat. Chem. Biol. 8:431–433. 10.1038/nchembio.915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lee HH, Molla MN, Cantor CR, Collins JJ. 2010. Bacterial charity work leads to population-wide resistance. Nature 467:82–85. 10.1038/nature09354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rodionov DG, Ishiguro EE. 1995. Direct correlation between overproduction of guanosine 3′,5′-bispyrophosphate (ppGpp) and penicillin tolerance in Escherichia coli. J. Bacteriol. 177:4224–4229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Viducic D, Ono T, Murakami K, Susilowati H, Kayama S, Hirota K, Miyake Y. 2006. Functional analysis of spoT, relA and dksA genes on quinolone tolerance in Pseudomonas aeruginosa under nongrowing condition. Microbiol. Immunol. 50:349–357. 10.1111/j.1348-0421.2006.tb03793.x [DOI] [PubMed] [Google Scholar]
- 119.Khakimova M, Ahlgren HG, Harrison JJ, English AM, Nguyen D. 2013. The stringent response controls catalases in Pseudomonas aeruginosa and is required for hydrogen peroxide and antibiotic tolerance. J. Bacteriol. 195:2011–2020. 10.1128/JB.02061-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kohanski MA, Dwyer DJ, Collins JJ. 2010. How antibiotics kill bacteria: from targets to networks. Nat. Rev. Microbiol. 8:423–435. 10.1038/nrmicro2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. 10.1016/j.cell.2007.06.049 [DOI] [PubMed] [Google Scholar]
- 122.Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. 2008. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell 135:679–690. 10.1016/j.cell.2008.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ezraty B, Vergnes A, Banzhaf M, Duverger Y, Huguenot A, Brochado AR, Su SY, Espinosa L, Loiseau L, Py B, Typas A, Barras F. 2013. Fe-S cluster biosynthesis controls uptake of aminoglycosides in a ROS-less death pathway. Science 340:1583–1587. 10.1126/science.1238328 [DOI] [PubMed] [Google Scholar]
- 124.Fang FC. 2013. Antibiotic and ROS linkage questioned. Nat. Biotechnol. 31:415–416. 10.1038/nbt.2574 [DOI] [PubMed] [Google Scholar]
- 125.Keren I, Wu Y, Inocencio J, Mulcahy LR, Lewis K. 2013. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 339:1213–1216. 10.1126/science.1232688 [DOI] [PubMed] [Google Scholar]
- 126.Liu Y, Imlay JA. 2013. Cell death from antibiotics without the involvement of reactive oxygen species. Science 339:1210–1213. 10.1126/science.1232751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Amato SM, Fazen CH, Henry TC, Mok WW, Orman MA, Sandvik EL, Volzing KG, Brynildsen MP. 2014. The role of metabolism in bacterial persistence. Front. Microbiol. 5:70. 10.3389/fmicb.2014.00070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gerdes K, Maisonneuve E. 2012. Bacterial persistence and toxin-antitoxin loci. Annu. Rev. Microbiol. 66:103–123. 10.1146/annurev-micro-092611-150159 [DOI] [PubMed] [Google Scholar]
- 129.Maisonneuve E, Castro-Camargo M, Gerdes K. 2013. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154:1140–1150. 10.1016/j.cell.2013.07.048 [DOI] [PubMed] [Google Scholar]
- 130.Hansen S, Vulic M, Min J, Yen TJ, Schumacher MA, Brennan RG, Lewis K. 2012. Regulation of the Escherichia coli HipBA toxin-antitoxin system by proteolysis. PLoS One 7:e39185. 10.1371/journal.pone.0039185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Korch SB, Henderson TA, Hill TM. 2003. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol. Microbiol. 50:1199–1213. 10.1046/j.1365-2958.2003.03779.x [DOI] [PubMed] [Google Scholar]
- 132.Bokinsky G, Baidoo EE, Akella S, Burd H, Weaver D, Alonso-Gutierrez J, Garcia-Martin H, Lee TS, Keasling JD. 2013. HipA-triggered growth arrest and β-lactam tolerance in Escherichia coli are mediated by RelA-dependent ppGpp synthesis. J. Bacteriol. 195:3173–3182. 10.1128/JB.02210-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kaspy I, Rotem E, Weiss N, Ronin I, Balaban NQ, Glaser G. 2013. HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat. Commun. 4:3001. 10.1038/ncomms4001 [DOI] [PubMed] [Google Scholar]
- 134.Ferullo DJ, Lovett ST. 2008. The stringent response and cell cycle arrest in Escherichia coli. PLoS Genet. 4:e1000300. 10.1371/journal.pgen.1000300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lemke JJ, Sanchez-Vazquez P, Burgos HL, Hedberg G, Ross W, Gourse RL. 2011. Direct regulation of Escherichia coli ribosomal protein promoters by the transcription factors ppGpp and DksA. Proc. Natl. Acad. Sci. U. S. A. 108:5712–5717. 10.1073/pnas.1019383108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Amato SM, Brynildsen MP. 2014. Nutrient transitions are a source of persisters in Escherichia coli biofilms. PLoS One 9:e93110. 10.1371/journal.pone.0093110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wexselblatt E, Oppenheimer-Shaanan Y, Kaspy I, London N, Schueler-Furman O, Yavin E, Glaser G, Katzhendler J, Ben-Yehuda S. 2012. Relacin, a novel antibacterial agent targeting the stringent response. PLoS Pathog. 8:e1002925. 10.1371/journal.ppat.1002925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Miller C, Thomsen LE, Gaggero C, Mosseri R, Ingmer H, Cohen SN. 2004. SOS response induction by beta-lactams and bacterial defense against antibiotic lethality. Science 305:1629–1631. 10.1126/science.1101630 [DOI] [PubMed] [Google Scholar]
- 139.Dorr T, Lewis K, Vulic M. 2009. SOS response induces persistence to fluoroquinolones in Escherichia coli. PLoS Genet. 5:e1000760. 10.1371/journal.pgen.1000760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dorr T, Vulic M, Lewis K. 2010. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 8:e1000317. 10.1371/journal.pbio.1000317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bernier SP, Lebeaux D, DeFrancesco AS, Valomon A, Soubigou G, Coppee JY, Ghigo JM, Beloin C. 2013. Starvation, together with the SOS response, mediates high biofilm-specific tolerance to the fluoroquinolone ofloxacin. PLoS Genet. 9:e1003144. 10.1371/journal.pgen.1003144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Beloin C, Valle J, Latour-Lambert P, Faure P, Kzreminski M, Balestrino D, Haagensen JA, Molin S, Prensier G, Arbeille B, Ghigo JM. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51:659–674. 10.1046/j.1365-2958.2003.03865.x [DOI] [PubMed] [Google Scholar]
- 143.Girgis HS, Harris K, Tavazoie S. 2012. Large mutational target size for rapid emergence of bacterial persistence. Proc. Natl. Acad. Sci. U. S. A. 109:12740–12745. 10.1073/pnas.1205124109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lewis K. 2010. Persister cells. Annu. Rev. Microbiol. 64:357–372. 10.1146/annurev.micro.112408.134306 [DOI] [PubMed] [Google Scholar]
- 145.Baharoglu Z, Bikard D, Mazel D. 2010. Conjugative DNA transfer induces the bacterial SOS response and promotes antibiotic resistance development through integron activation. PLoS Genet. 6:e1001165. 10.1371/journal.pgen.1001165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Imlay JA. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77:755–776. 10.1146/annurev.biochem.77.061606.161055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wu Y, Vulic M, Keren I, Lewis K. 2012. Role of oxidative stress in persister tolerance. Antimicrob. Agents Chemother. 56:4922–4926. 10.1128/AAC.00921-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kim JS, Heo P, Yang TJ, Lee KS, Jin YS, Kim SK, Shin D, Kweon DH. 2011. Bacterial persisters tolerate antibiotics by not producing hydroxyl radicals. Biochem. Biophys. Res. Commun. 413:105–110. 10.1016/j.bbrc.2011.08.063 [DOI] [PubMed] [Google Scholar]
- 149.Shatalin K, Shatalina E, Mironov A, Nudler E. 2011. H2S: a universal defense against antibiotics in bacteria. Science 334:986–990. 10.1126/science.1209855 [DOI] [PubMed] [Google Scholar]
- 150.Boles BR, Singh PK. 2008. Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc. Natl. Acad. Sci. U. S. A. 105:12503–12508. 10.1073/pnas.0801499105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Imlay JA, Linn S. 1987. Mutagenesis and stress responses induced in Escherichia coli by hydrogen peroxide. J. Bacteriol. 169:2967–2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Li Y, Zhang Y. 2007. PhoU is a persistence switch involved in persister formation and tolerance to multiple antibiotics and stresses in Escherichia coli. Antimicrob. Agents Chemother. 51:2092–2099. 10.1128/AAC.00052-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Spoering AL, Vulic M, Lewis K. 2006. GlpD and PlsB participate in persister cell formation in Escherichia coli. J. Bacteriol. 188:5136–5144. 10.1128/JB.00369-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bjarnsholt T, Jensen PO, Burmolle M, Hentzer M, Haagensen JA, Hougen HP, Calum H, Madsen KG, Moser C, Molin S, Høiby N, Givskov M. 2005. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology 151:373–383. 10.1099/mic.0.27463-0 [DOI] [PubMed] [Google Scholar]
- 155.Moker N, Dean CR, Tao J. 2010. Pseudomonas aeruginosa increases formation of multidrug-tolerant persister cells in response to quorum-sensing signaling molecules. J. Bacteriol. 192:1946–1955. 10.1128/JB.01231-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Que YA, Hazan R, Strobel B, Maura D, He J, Kesarwani M, Panopoulos P, Tsurumi A, Giddey M, Wilhelmy J, Mindrinos MN, Rahme LG. 2013. A quorum sensing small volatile molecule promotes antibiotic tolerance in bacteria. PLoS One 8:e80140. 10.1371/journal.pone.0080140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lee S, Hinz A, Bauerle E, Angermeyer A, Juhaszova K, Kaneko Y, Singh PK, Manoil C. 2009. Targeting a bacterial stress response to enhance antibiotic action. Proc. Natl. Acad. Sci. U. S. A. 106:14570–14575. 10.1073/pnas.0903619106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kaern M, Elston TC, Blake WJ, Collins JJ. 2005. Stochasticity in gene expression: from theories to phenotypes. Nat. Rev. Genet. 6:451–464. 10.1038/nrg1615 [DOI] [PubMed] [Google Scholar]
- 159.Veening JW, Smits WK, Kuipers OP. 2008. Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol. 62:193–210. 10.1146/annurev.micro.62.081307.163002 [DOI] [PubMed] [Google Scholar]
- 160.Rotem E, Loinger A, Ronin I, Levin-Reisman I, Gabay C, Shoresh N, Biham O, Balaban NQ. 2010. Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence. Proc. Natl. Acad. Sci. U. S. A. 107:12541–12546. 10.1073/pnas.1004333107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Allison KR, Brynildsen MP, Collins JJ. 2011. Heterogeneous bacterial persisters and engineering approaches to eliminate them. Curr. Opin. Microbiol. 14:593–598. 10.1016/j.mib.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Madsen JS, Burmolle M, Hansen LH, Sorensen SJ. 2012. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol. Med. Microbiol. 65:183–195. 10.1111/j.1574-695X.2012.00960.x [DOI] [PubMed] [Google Scholar]
- 163.Ghigo JM. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442–445. 10.1038/35086581 [DOI] [PubMed] [Google Scholar]
- 164.Hennequin C, Aumeran C, Robin F, Traore O, Forestier C. 2012. Antibiotic resistance and plasmid transfer capacity in biofilm formed with a CTX-M-15-producing Klebsiella pneumoniae isolate. J. Antimicrob. Chemother. 67:2123–2130. 10.1093/jac/dks169 [DOI] [PubMed] [Google Scholar]
- 165.Hannan S, Ready D, Jasni AS, Rogers M, Pratten J, Roberts AP. 2010. Transfer of antibiotic resistance by transformation with eDNA within oral biofilms. FEMS Immunol. Med. Microbiol. 59:345–349. 10.1111/j.1574-695X.2010.00661.x [DOI] [PubMed] [Google Scholar]
- 166.Weigel LM, Donlan RM, Shin DH, Jensen B, Clark NC, McDougal LK, Zhu W, Musser KA, Thompson J, Kohlerschmidt D, Dumas N, Limberger RJ, Patel JB. 2007. High-level vancomycin-resistant Staphylococcus aureus isolates associated with a polymicrobial biofilm. Antimicrob. Agents Chemother. 51:231–238. 10.1128/AAC.00576-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hughes D, Andersson DI. 2012. Selection of resistance at lethal and non-lethal antibiotic concentrations. Curr. Opin. Microbiol. 15:555–560. 10.1016/j.mib.2012.07.005 [DOI] [PubMed] [Google Scholar]
- 168.Gullberg E, Cao S, Berg OG, Ilback C, Sandegren L, Hughes D, Andersson DI. 2011. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 7:e1002158. 10.1371/journal.ppat.1002158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kohanski MA, DePristo MA, Collins JJ. 2010. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol. Cell 37:311–320. 10.1016/j.molcel.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Gutierrez A, Laureti L, Crussard S, Abida H, Rodriguez-Rojas A, Blazquez J, Baharoglu Z, Mazel D, Darfeuille F, Vogel J, Matic I. 2013. Beta-lactam antibiotics promote bacterial mutagenesis via an RpoS-mediated reduction in replication fidelity. Nat. Commun. 4:1610. 10.1038/ncomms2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Jorgensen KM, Wassermann T, Jensen PO, Wang H, Molin S, Høiby N, Ciofu O. 2013. Sublethal ciprofloxacin treatment leads to rapid development of high-level ciprofloxacin resistance during long-term experimental evolution of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 57:4215–4221. 10.1128/AAC.00493-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hoffman LR, D'Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. 2005. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436:1171–1175. 10.1038/nature03912 [DOI] [PubMed] [Google Scholar]
- 173.Gomes DL, Peixoto RS, Barbosa EA, Napoleao F, Sabbadini PS, dos Santos KR, Mattos-Guaraldi AL, Hirata R., Jr 2013. SubMICs of penicillin and erythromycin enhance biofilm formation and hydrophobicity of Corynebacterium diphtheriae strains. J. Med. Microbiol. 62:754–760. 10.1099/jmm.0.052373-0 [DOI] [PubMed] [Google Scholar]
- 174.Bagge N, Schuster M, Hentzer M, Ciofu O, Givskov M, Greenberg EP, Høiby N. 2004. Pseudomonas aeruginosa biofilms exposed to imipenem exhibit changes in global gene expression and beta-lactamase and alginate production. Antimicrob. Agents Chemother. 48:1175–1187. 10.1128/AAC.48.4.1175-1187.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Mirani ZA, Jamil N. 2011. Effect of sub-lethal doses of vancomycin and oxacillin on biofilm formation by vancomycin intermediate resistant Staphylococcus aureus. J. Basic Microbiol. 51:191–195. 10.1002/jobm.201000221 [DOI] [PubMed] [Google Scholar]
- 176.Zhang X, McDaniel AD, Wolf LE, Keusch GT, Waldor MK, Acheson DW. 2000. Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J. Infect. Dis. 181:664–670. 10.1086/315239 [DOI] [PubMed] [Google Scholar]
- 177.Beaber JW, Hochhut B, Waldor MK. 2004. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427:72–74. 10.1038/nature02241 [DOI] [PubMed] [Google Scholar]
- 178.Guerin E, Cambray G, Sanchez-Alberola N, Campoy S, Erill I, Da Re S, Gonzalez-Zorn B, Barbe J, Ploy MC, Mazel D. 2009. The SOS response controls integron recombination. Science 324:1034. 10.1126/science.1172914 [DOI] [PubMed] [Google Scholar]
- 179.Hocquet D, Llanes C, Thouverez M, Kulasekara HD, Bertrand X, Plesiat P, Mazel D, Miller SI. 2012. Evidence for induction of integron-based antibiotic resistance by the SOS response in a clinical setting. PLoS Pathog. 8:e1002778. 10.1371/journal.ppat.1002778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Johnson PJ, Levin BR. 2013. Pharmacodynamics, population dynamics, and the evolution of persistence in Staphylococcus aureus. PLoS Genet. 9:e1003123. 10.1371/journal.pgen.1003123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Cohen NR, Lobritz MA, Collins JJ. 2013. Microbial persistence and the road to drug resistance. Cell Host Microbe 13:632–642. 10.1016/j.chom.2013.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Driffield K, Miller K, Bostock JM, O'Neill AJ, Chopra I. 2008. Increased mutability of Pseudomonas aeruginosa in biofilms. J. Antimicrob. Chemother. 61:1053–1056. 10.1093/jac/dkn044 [DOI] [PubMed] [Google Scholar]
- 183.Mathee K, Ciofu O, Sternberg C, Lindum PW, Campbell JI, Jensen P, Johnsen AH, Givskov M, Ohman DE, Molin S, Høiby N, Kharazmi A. 1999. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology 145:1349–1357. 10.1099/13500872-145-6-1349 [DOI] [PubMed] [Google Scholar]
- 184.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. 2010. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 35:322–332. 10.1016/j.ijantimicag.2009.12.011 [DOI] [PubMed] [Google Scholar]
- 185.Mandsberg LF, Ciofu O, Kirkby N, Christiansen LE, Poulsen HE, Høiby N. 2009. Antibiotic resistance in Pseudomonas aeruginosa strains with increased mutation frequency due to inactivation of the DNA oxidative repair system. Antimicrob. Agents Chemother. 53:2483–2491. 10.1128/AAC.00428-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Oliver A, Sanchez JM, Blazquez J. 2002. Characterization of the GO system of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 217:31–35. 10.1111/j.1574-6968.2002.tb11452.x [DOI] [PubMed] [Google Scholar]
- 187.Ryder VJ, Chopra I, O'Neill AJ. 2012. Increased mutability of staphylococci in biofilms as a consequence of oxidative stress. PLoS One 7:e47695. 10.1371/journal.pone.0047695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Garcia LG, Lemaire S, Kahl BC, Becker K, Proctor RA, Denis O, Tulkens PM, Van Bambeke F. 2013. Antibiotic activity against small-colony variants of Staphylococcus aureus: review of in vitro, animal and clinical data. J. Antimicrob. Chemother. 68:1455–1464. 10.1093/jac/dkt072 [DOI] [PubMed] [Google Scholar]
- 189.Haussler S, Ziegler I, Lottel A, von Gotz F, Rohde M, Wehmhohner D, Saravanamuthu S, Tummler B, Steinmetz I. 2003. Highly adherent small-colony variants of Pseudomonas aeruginosa in cystic fibrosis lung infection. J. Med. Microbiol. 52:295–301. 10.1099/jmm.0.05069-0 [DOI] [PubMed] [Google Scholar]
- 190.Drenkard E, Ausubel FM. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416:740–743. 10.1038/416740a [DOI] [PubMed] [Google Scholar]
- 191.Meissner A, Wild V, Simm R, Rohde M, Erck C, Bredenbruch F, Morr M, Romling U, Haussler S. 2007. Pseudomonas aeruginosa cupA-encoded fimbriae expression is regulated by a GGDEF and EAL domain-dependent modulation of the intracellular level of cyclic diguanylate. Environ. Microbiol. 9:2475–2485. 10.1111/j.1462-2920.2007.01366.x [DOI] [PubMed] [Google Scholar]
- 192.O'Grady NP, Alexander M, Burns LA, Dellinger EP, Garland J, Heard SO, Lipsett PA, Masur H, Mermel LA, Pearson ML, Raad II, Randolph AG, Rupp ME, Saint S. 2011. Guidelines for the prevention of intravascular catheter-related infections. Clin. Infect. Dis. 52:e162–e193. 10.1093/cid/cir257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Pronovost P, Needham D, Berenholtz S, Sinopoli D, Chu H, Cosgrove S, Sexton B, Hyzy R, Welsh R, Roth G, Bander J, Kepros J, Goeschel C. 2006. An intervention to decrease catheter-related bloodstream infections in the ICU. N. Engl. J. Med. 355:2725–2732. 10.1056/NEJMoa061115 [DOI] [PubMed] [Google Scholar]
- 194.Mermel LA, Allon M, Bouza E, Craven DE, Flynn P, O'Grady NP, Raad II, Rijnders BJ, Sherertz RJ, Warren DK. 2009. Clinical practice guidelines for the diagnosis and management of intravascular catheter-related infection: 2009 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 49:1–45. 10.1086/599376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Raad I, Hanna H, Maki D. 2007. Intravascular catheter-related infections: advances in diagnosis, prevention, and management. Lancet Infect. Dis. 7:645–657. 10.1016/S1473-3099(07)70235-9 [DOI] [PubMed] [Google Scholar]
- 196.Meddings J, Rogers MA, Macy M, Saint S. 2010. Systematic review and meta-analysis: reminder systems to reduce catheter-associated urinary tract infections and urinary catheter use in hospitalized patients. Clin. Infect. Dis. 51:550–560. 10.1086/655133 [DOI] [PubMed] [Google Scholar]
- 197.Baddour LM, Epstein AE, Erickson CC, Knight BP, Levison ME, Lockhart PB, Masoudi FA, Okum EJ, Wilson WR, Beerman LB, Bolger AF, Estes NAM, Gewitz M, Newburger JW, Schron EB, Taubert KA, American Heart Association Rheumatic Fever, Endocarditis, and Kawasaki Disease Committee of the Council on Cardiovascular Disease in the Young, Council on Cardiovascular Surgery and Anesthesia, Council on Cardiovascular Nursing, Council on Clinical Cardiology, Interdisciplinary Council on Quality of Care and Outcomes Research 2010. Update on cardiovascular implantable electronic device infections and their management: a scientific statement from the American Heart Association. Circulation 121:458–477. 10.1161/CIRCULATIONAHA.109.192665 [DOI] [PubMed] [Google Scholar]
- 198.Zimmerli W. 2006. Infection and musculoskeletal conditions: prosthetic-joint-associated infections. Best Pract. Res. Clin. Rheumatol. 20:1045–1063. 10.1016/j.berh.2006.08.003 [DOI] [PubMed] [Google Scholar]
- 199.Hetrick EM, Schoenfisch MH. 2006. Reducing implant-related infections: active release strategies. Chem. Soc. Rev. 35:780–789. 10.1039/b515219b [DOI] [PubMed] [Google Scholar]
- 200.Casey AL, Mermel LA, Nightingale P, Elliott TSJ. 2008. Antimicrobial central venous catheters in adults: a systematic review and meta-analysis. Lancet Infect. Dis. 8:763–776. 10.1016/S1473-3099(08)70280-9 [DOI] [PubMed] [Google Scholar]
- 201.Darouiche RO, Raad II, Heard SO, Thornby JI, Wenker OC, Gabrielli A, Berg J, Khardori N, Hanna H, Hachem R, Harris RL, Mayhall G. 1999. A comparison of two antimicrobial-impregnated central venous catheters. Catheter Study Group. N. Engl. J. Med. 340:1–8 [DOI] [PubMed] [Google Scholar]
- 202.Falagas ME, Fragoulis K, Bliziotis IA, Chatzinikolaou I. 2007. Rifampicin-impregnated central venous catheters: a meta-analysis of randomized controlled trials. J. Antimicrob. Chemother. 59:359–369. 10.1093/jac/dkl522 [DOI] [PubMed] [Google Scholar]
- 203.Gilbert RE, Harden M. 2008. Effectiveness of impregnated central venous catheters for catheter related blood stream infection: a systematic review. Curr. Opin. Infect. Dis. 21:235–245. 10.1097/QCO.0b013e3282ffd6e0 [DOI] [PubMed] [Google Scholar]
- 204.Rupp ME, Lisco SJ, Lipsett PA, Perl TM, Keating K, Civetta JM, Mermel LA, Lee D, Dellinger EP, Donahoe M, Giles D, Pfaller MA, Maki DG, Sherertz R. 2005. Effect of a second-generation venous catheter impregnated with chlorhexidine and silver sulfadiazine on central catheter-related infections: a randomized, controlled trial. Ann. Intern. Med. 143:570–580. 10.7326/0003-4819-143-8-200510180-00007 [DOI] [PubMed] [Google Scholar]
- 205.Aboshady I, Raad II, Shah AS, Vela D, Dvorak T, Safi HJ, Buja LM, Khalil KG. 2012. A pilot study of a triple antimicrobial-bonded Dacron graft for the prevention of aortic graft infection. J. Vasc. Surg. 56:794–801. 10.1016/j.jvs.2012.02.008 [DOI] [PubMed] [Google Scholar]
- 206.Bernthal NM, Stavrakis AI, Billi F, Cho JS, Kremen TJ, Simon SI, Cheung AL, Finerman GA, Lieberman JR, Adams JS, Miller LS. 2010. A mouse model of post-arthroplasty Staphylococcus aureus joint infection to evaluate in vivo the efficacy of antimicrobial implant coatings. PLoS One 5:e12580. 10.1371/journal.pone.0012580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Berra L, De Marchi L, Yu ZX, Laquerriere P, Baccarelli A, Kolobow T. 2004. Endotracheal tubes coated with antiseptics decrease bacterial colonization of the ventilator circuits, lungs, and endotracheal tube. Anesthesiology 100:1446–1456. 10.1097/00000542-200406000-00017 [DOI] [PubMed] [Google Scholar]
- 208.Fuchs T, Stange R, Schmidmaier G, Raschke MJ. 2011. The use of gentamicin-coated nails in the tibia: preliminary results of a prospective study. Arch. Orthop. Trauma Surg. 131:1419–1425. 10.1007/s00402-011-1321-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Gahtan V, Esses GE, Bandyk DF, Nelson RT, Dupont E, Mills JL. 1995. Antistaphylococcal activity of rifampin-bonded gelatin-impregnated Dacron grafts. J. Surg. Res. 58:105–110. 10.1006/jsre.1995.1017 [DOI] [PubMed] [Google Scholar]
- 210.Gao H, Sandermann J, Prag J, Lund L, Lindholt JS. 2012. Rifampicin-soaked silver polyester versus expanded polytetrafluoro-ethylene grafts for in situ replacement of infected grafts in a porcine randomised controlled trial. Eur. J. Vasc. Endovasc. Surg. 43:582–587. 10.1016/j.ejvs.2012.02.011 [DOI] [PubMed] [Google Scholar]
- 211.Hickok NJ, Shapiro IM. 2012. Immobilized antibiotics to prevent orthopaedic implant infections. Adv. Drug Deliv. Rev. 64:1165–1176. 10.1016/j.addr.2012.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Hooton TM, Bradley SF, Cardenas DD, Colgan R, Geerlings SE, Rice JC, Saint S, Schaeffer AJ, Tambayh PA, Tenke P, Nicolle LE. 2010. Diagnosis, prevention, and treatment of catheter-associated urinary tract infection in adults: 2009 international clinical practice guidelines from the Infectious Diseases Society of America. Clin. Infect. Dis. 50:625–663. 10.1086/650482 [DOI] [PubMed] [Google Scholar]
- 213.Kollef MH, Afessa B, Anzueto A, Veremakis C, Kerr KM, Margolis BD, Craven DE, Roberts PR, Arroliga AC, Hubmayr RD, Restrepo MI, Auger WR, Schinner R. 2008. Silver-coated endotracheal tubes and incidence of ventilator-associated pneumonia: the NASCENT randomized trial. JAMA 300:805–813. 10.1001/jama.300.7.805 [DOI] [PubMed] [Google Scholar]
- 214.Lew W, Moore W. 2011. Antibiotic-impregnated grafts for aortic reconstruction. Semin. Vasc. Surg. 24:211–219. 10.1053/j.semvascsurg.2011.10.015 [DOI] [PubMed] [Google Scholar]
- 215.Olson ME, Harmon BG, Kollef MH. 2002. Silver-coated endotracheal tubes associated with reduced bacterial burden in the lungs of mechanically ventilated dogs. Chest 121:863–870. 10.1378/chest.121.3.863 [DOI] [PubMed] [Google Scholar]
- 216.Pickard R, Lam T, MacLennan G, Starr K, Kilonzo M, McPherson G, Gillies K, McDonald A, Walton K, Buckley B, Glazener C, Boachie C, Burr J, Norrie J, Vale L, Grant A, N′Dow J. 2012. Antimicrobial catheters for reduction of symptomatic urinary tract infection in adults requiring short-term catheterisation in hospital: a multicentre randomised controlled trial. Lancet 380:1927–1935. 10.1016/S0140-6736(12)61380-4 [DOI] [PubMed] [Google Scholar]
- 217.Baker PJ, Evans RT, Roopenian DC. 1994. Oral infection with Porphyromonas gingivalis and induced alveolar bone loss in immunocompetent and severe combined immunodeficient mice. Arch. Oral Biol. 39:1035–1040. 10.1016/0003-9969(94)90055-8 [DOI] [PubMed] [Google Scholar]
- 218.Habib G, Hoen B, Tornos P, Thuny F, Prendergast B, Vilacosta I, Moreillon P, de Jesus Antunes M, Thilen U, Lekakis J, Lengyel M, Muller L, Naber CK, Nihoyannopoulos P, Moritz A, Zamorano JL. 2009. Guidelines on the prevention, diagnosis, and treatment of infective endocarditis (new version 2009): the Task Force on the Prevention, Diagnosis, and Treatment of Infective Endocarditis of the European Society of Cardiology (ESC). Endorsed by the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) and the International Society of Chemotherapy (ISC) for Infection and Cancer. Eur. Heart J. 30:2369–2413. 10.1093/eurheartj/ehp285 [DOI] [PubMed] [Google Scholar]
- 219.Corvec S, Furustrand Tafin U, Betrisey B, Borens O, Trampuz A. 2013. Activities of fosfomycin, tigecycline, colistin, and gentamicin against extended-spectrum-beta-lactamase-producing Escherichia coli in a foreign-body infection model. Antimicrob. Agents Chemother. 57:1421–1427. 10.1128/AAC.01718-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Garrigos C, Murillo O, Lora-Tamayo J, Verdaguer R, Tubau F, Cabellos C, Cabo J, Ariza J. 2013. Fosfomycin-daptomycin and other fosfomycin combinations as alternative therapies in experimental foreign-body infection by methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 57:606–610. 10.1128/AAC.01570-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Murillo O, Garrigos C, Pachon ME, Euba G, Verdaguer R, Cabellos C, Cabo J, Gudiol F, Ariza J. 2009. Efficacy of high doses of daptomycin versus alternative therapies against experimental foreign-body infection by methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53:4252–4257. 10.1128/AAC.00208-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Cremieux AC, Mghir AS, Bleton R, Manteau M, Belmatoug N, Massias L, Garry L, Sales N, Maziere B, Carbon C. 1996. Efficacy of sparfloxacin and autoradiographic diffusion pattern of [14C]sparfloxacin in experimental Staphylococcus aureus joint prosthesis infection. Antimicrob. Agents Chemother. 40:2111–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Zimmerli W, Moser C. 2012. Pathogenesis and treatment concepts of orthopaedic biofilm infections. FEMS Immunol. Med. Microbiol. 65:158–168. 10.1111/j.1574-695X.2012.00938.x [DOI] [PubMed] [Google Scholar]
- 224.Bouza E. 2009. New therapeutic choices for infections caused by methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 15(Suppl 7):S44–S52. 10.1111/j.1469-0691.2009.03091.x [DOI] [PubMed] [Google Scholar]
- 225.Mascio CT, Alder JD, Silverman JA. 2007. Bactericidal action of daptomycin against stationary-phase and nondividing Staphylococcus aureus cells. Antimicrob. Agents Chemother. 51:4255–4260. 10.1128/AAC.00824-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Raad I, Hanna H, Jiang Y, Dvorak T, Reitzel R, Chaiban G, Sherertz R, Hachem R. 2007. Comparative activities of daptomycin, linezolid, and tigecycline against catheter-related methicillin-resistant Staphylococcus bacteremic isolates embedded in biofilm. Antimicrob. Agents Chemother. 51:1656–1660. 10.1128/AAC.00350-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Smith K, Perez A, Ramage G, Gemmell CG, Lang S. 2009. Comparison of biofilm-associated cell survival following in vitro exposure of meticillin-resistant Staphylococcus aureus biofilms to the antibiotics clindamycin, daptomycin, linezolid, tigecycline and vancomycin. Int. J. Antimicrob. Agents 33:374–378. 10.1016/j.ijantimicag.2008.08.029 [DOI] [PubMed] [Google Scholar]
- 228.Stewart PS, Davison WM, Steenbergen JN. 2009. Daptomycin rapidly penetrates a Staphylococcus epidermidis biofilm. Antimicrob. Agents Chemother. 53:3505–3507. 10.1128/AAC.01728-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.John AK, Baldoni D, Haschke M, Rentsch K, Schaerli P, Zimmerli W, Trampuz A. 2009. Efficacy of daptomycin in implant-associated infection due to methicillin-resistant Staphylococcus aureus: importance of combination with rifampin. Antimicrob. Agents Chemother. 53:2719–2724. 10.1128/AAC.00047-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Van Praagh AD, Li T, Zhang S, Arya A, Chen L, Zhang XX, Bertolami S, Mortin LI. 2011. Daptomycin antibiotic lock therapy in a rat model of staphylococcal central venous catheter biofilm infections. Antimicrob. Agents Chemother. 55:4081–4089. 10.1128/AAC.00147-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Del Pozo JL, Rodil R, Aguinaga A, Yuste JR, Bustos C, Montero A, Espinosa G, Garcia-Fernandez N. 2012. Daptomycin lock therapy for gram positive long-term catheter-related bloodstream infections. Int. J. Clin. Pract. 66:305–308. 10.1111/j.1742-1241.2011.02830.x [DOI] [PubMed] [Google Scholar]
- 232.Canton R, Cobos N, de Gracia J, Baquero F, Honorato J, Gartner S, Alvarez A, Salcedo A, Oliver A, Garcia-Quetglas E. 2005. Antimicrobial therapy for pulmonary pathogenic colonisation and infection by Pseudomonas aeruginosa in cystic fibrosis patients. Clin. Microbiol. Infect. 11:690–703. 10.1111/j.1469-0691.2005.01217.x [DOI] [PubMed] [Google Scholar]
- 233.Høiby N. 2011. Recent advances in the treatment of Pseudomonas aeruginosa infections in cystic fibrosis. BMC Med. 9:32. 10.1186/1741-7015-9-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Safdar N, Maki DG. 2006. Use of vancomycin-containing lock or flush solutions for prevention of bloodstream infection associated with central venous access devices: a meta-analysis of prospective, randomized trials. Clin. Infect. Dis. 43:474–484. 10.1086/505976 [DOI] [PubMed] [Google Scholar]
- 235.Chatzinikolaou I, Zipf TF, Hanna H, Umphrey J, Roberts WM, Sherertz R, Hachem R, Raad I. 2003. Minocycline-ethylenediaminetetraacetate lock solution for the prevention of implantable port infections in children with cancer. Clin. Infect. Dis. 36:116–119. 10.1086/344952 [DOI] [PubMed] [Google Scholar]
- 236.Ferreira Chacon JM, Hato de Almeida E, de Lourdes Simoes R, Lazzarin COV, Alves BC, Mello de Andrea ML, Santiago Biernat M, Biernat JC. 2011. Randomized study of minocycline and edetic acid as a locking solution for central line (port-a-cath) in children with cancer. Chemotherapy 57:285–291. 10.1159/000328976 [DOI] [PubMed] [Google Scholar]
- 237.Donlan RM. 2011. Biofilm elimination on intravascular catheters: important considerations for the infectious disease practitioner. Clin. Infect. Dis. 52:1038–1045. 10.1093/cid/cir077 [DOI] [PubMed] [Google Scholar]
- 238.Landry DL, Braden GL, Gobeille SL, Haessler SD, Vaidya CK, Sweet SJ. 2010. Emergence of gentamicin-resistant bacteremia in hemodialysis patients receiving gentamicin lock catheter prophylaxis. Clin. J. Am. Soc. Nephrol. 5:1799–1804. 10.2215/CJN.01270210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Bradshaw JH, Puntis JW. 2008. Taurolidine and catheter-related bloodstream infection: a systematic review of the literature. J. Pediatr. Gastroenterol. Nutr. 47:179–186. 10.1097/MPG.0b013e318162c428 [DOI] [PubMed] [Google Scholar]
- 240.Wolf J, Shenep JL, Clifford V, Curtis N, Flynn PM. 2013. Ethanol lock therapy in pediatric hematology and oncology. Pediatr. Blood Cancer 60:18–25. 10.1002/pbc.24249 [DOI] [PubMed] [Google Scholar]
- 241.Fernandez-Hidalgo N, Almirante B, Calleja R, Ruiz I, Planes AM, Rodriguez D, Pigrau C, Pahissa A. 2006. Antibiotic-lock therapy for long-term intravascular catheter-related bacteraemia: results of an open, non-comparative study. J. Antimicrob. Chemother. 57:1172–1180. 10.1093/jac/dkl103 [DOI] [PubMed] [Google Scholar]
- 242.Rijnders BJ, Van Wijngaerden E, Vandecasteele SJ, Stas M, Peetermans WE. 2005. Treatment of long-term intravascular catheter-related bacteraemia with antibiotic lock: randomized, placebo-controlled trial. J. Antimicrob. Chemother. 55:90–94. 10.1093/jac/dkh488 [DOI] [PubMed] [Google Scholar]
- 243.Del Pozo JL, Garcia Cenoz M, Hernaez S, Martinez A, Serrera A, Aguinaga A, Alonso M, Leiva J. 2009. Effectiveness of teicoplanin versus vancomycin lock therapy in the treatment of port-related coagulase-negative staphylococci bacteraemia: a prospective case-series analysis. Int. J. Antimicrob. Agents 34:482–485. 10.1016/j.ijantimicag.2009.06.020 [DOI] [PubMed] [Google Scholar]
- 244.Funalleras G, Fernandez-Hidalgo N, Borrego A, Almirante B, Planes AM, Rodriguez D, Ruiz I, Pahissa A. 2011. Effectiveness of antibiotic-lock therapy for long-term catheter-related bacteremia due to Gram-negative bacilli: a prospective observational study. Clin. Infect. Dis. 53:e129–e132. 10.1093/cid/cir551 [DOI] [PubMed] [Google Scholar]
- 245.Broom J, Woods M, Allworth A, McCarthy J, Faoagali J, Macdonald S, Pithie A. 2008. Ethanol lock therapy to treat tunnelled central venous catheter-associated blood stream infections: results from a prospective trial. Scand. J. Infect. Dis. 40:399–406. 10.1080/00365540701756953 [DOI] [PubMed] [Google Scholar]
- 246.McGrath EJ, Salloum R, Chen X, Jiang Y, Boldt-MacDonald K, Becker C, Chu R, Ang JY. 2011. Short-dwell ethanol lock therapy in children is associated with increased clearance of central line-associated bloodstream infections. Clin. Pediatr. (Phila.) 50:943–951. 10.1177/0009922811409568 [DOI] [PubMed] [Google Scholar]
- 247.Onland W, Shin CE, Fustar S, Rushing T, Wong WY. 2006. Ethanol-lock technique for persistent bacteremia of long-term intravascular devices in pediatric patients. Arch. Pediatr. Adolesc. Med. 160:1049–1053. 10.1001/archpedi.160.10.1049 [DOI] [PubMed] [Google Scholar]
- 248.Busscher HJ, van der Mei HC, Subbiahdoss G, Jutte PC, van den Dungen JJ, Zaat SA, Schultz MJ, Grainger DW. 2012. Biomaterial-associated infection: locating the finish line in the race for the surface. Sci. Transl. Med. 4:153rv110. 10.1126/scitranslmed.3004528 [DOI] [PubMed] [Google Scholar]
- 249.Campoccia D, Montanaro L, Arciola CR. 2013. A review of the clinical implications of anti-infective biomaterials and infection-resistant surfaces. Biomaterials 34:8018–8029. 10.1016/j.biomaterials.2013.07.048 [DOI] [PubMed] [Google Scholar]
- 250.El Habnouni S, Lavigne JP, Darcos V, Porsio B, Garric X, Coudane J, Nottelet B. 2013. Toward potent antibiofilm degradable medical devices: a generic method for the antibacterial surface modification of polylactide. Acta Biomater. 9:7709–7718. 10.1016/j.actbio.2013.04.018 [DOI] [PubMed] [Google Scholar]
- 251.Hasan J, Crawford RJ, Ivanova EP. 2013. Antibacterial surfaces: the quest for a new generation of biomaterials. Trends Biotechnol. 31:295–304. 10.1016/j.tibtech.2013.01.017 [DOI] [PubMed] [Google Scholar]
- 252.Grossner-Schreiber B, Teichmann J, Hannig M, Dorfer C, Wenderoth DF, Ott SJ. 2009. Modified implant surfaces show different biofilm compositions under in vivo conditions. Clin. Oral Implants Res. 20:817–826. 10.1111/j.1600-0501.2009.01729.x [DOI] [PubMed] [Google Scholar]
- 253.Scarano A, Piattelli M, Caputi S, Favero GA, Piattelli A. 2004. Bacterial adhesion on commercially pure titanium and zirconium oxide disks: an in vivo human study. J. Periodontol. 75:292–296. 10.1902/jop.2004.75.2.292 [DOI] [PubMed] [Google Scholar]
- 254.Arnold JW, Bailey GW. 2000. Surface finishes on stainless steel reduce bacterial attachment and early biofilm formation: scanning electron and atomic force microscopy study. Poult. Sci. 79:1839–1845. 10.1093/ps/79.12.1839 [DOI] [PubMed] [Google Scholar]
- 255.Hook AL, Chang CY, Yang J, Luckett J, Cockayne A, Atkinson S, Mei Y, Bayston R, Irvine DJ, Langer R, Anderson DG, Williams P, Davies MC, Alexander MR. 2012. Combinatorial discovery of polymers resistant to bacterial attachment. Nat. Biotechnol. 30:868–875. 10.1038/nbt.2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.May RM, Hoffman MG, Sogo MJ, Parker AE, O'Toole GA, Brennan AB, Reddy ST. 2014. Micro-patterned surfaces reduce bacterial colonization and biofilm formation in vitro: potential for enhancing endotracheal tube designs. Clin. Transl. Med. 3:8. 10.1186/2001-1326-3-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Reddy ST, Chung KK, McDaniel CJ, Darouiche RO, Landman J, Brennan AB. 2011. Micropatterned surfaces for reducing the risk of catheter-associated urinary tract infection: an in vitro study on the effect of sharklet micropatterned surfaces to inhibit bacterial colonization and migration of uropathogenic Escherichia coli. J. Endourol. 25:1547–1552. 10.1089/end.2010.0611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Khoo X, Hamilton P, O'Toole GA, Snyder BD, Kenan DJ, Grinstaff MW. 2009. Directed assembly of PEGylated-peptide coatings for infection-resistant titanium metal. J. Am. Chem. Soc. 131:10992–10997. 10.1021/ja9020827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Yuan S, Wan D, Liang B, Pehkonen SO, Ting YP, Neoh KG, Kang ET. 2011. Lysozyme-coupled poly(poly(ethylene glycol) methacrylate)-stainless steel hybrids and their antifouling and antibacterial surfaces. Langmuir 27:2761–2774. 10.1021/la104442f [DOI] [PubMed] [Google Scholar]
- 260.Smith RS, Zhang Z, Bouchard M, Li J, Lapp HS, Brotske GR, Lucchino DL, Weaver D, Roth LA, Coury A, Biggerstaff J, Sukavaneshvar S, Langer R, Loose C. 2012. Vascular catheters with a nonleaching poly-sulfobetaine surface modification reduce thrombus formation and microbial attachment. Sci. Transl. Med. 4:153ra132. 10.1126/scitranslmed.3004120 [DOI] [PubMed] [Google Scholar]
- 261.Tiller JC, Liao CJ, Lewis K, Klibanov AM. 2001. Designing surfaces that kill bacteria on contact. Proc. Natl. Acad. Sci. U. S. A. 98:5981–5985. 10.1073/pnas.111143098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Vecitis CD, Zodrow KR, Kang S, Elimelech M. 2010. Electronic-structure-dependent bacterial cytotoxicity of single-walled carbon nanotubes. ACS Nano 4:5471–5479. 10.1021/nn101558x [DOI] [PubMed] [Google Scholar]
- 263.Rendueles O, Ghigo JM. 24 January 2012. Multi-species biofilms: how to avoid unfriendly neighbors. FEMS Microbiol. Rev. 10.1111/j.1574-6976.2012.00328.x [DOI] [PubMed] [Google Scholar]
- 264.Zeraik AE, Nitschke M. 2010. Biosurfactants as agents to reduce adhesion of pathogenic bacteria to polystyrene surfaces: effect of temperature and hydrophobicity. Curr. Microbiol. 61:554–559. 10.1007/s00284-010-9652-z [DOI] [PubMed] [Google Scholar]
- 265.Rendueles O, Travier L, Latour-Lambert P, Fontaine T, Magnus J, Denamur E, Ghigo JM. 2011. Screening of Escherichia coli species biodiversity reveals new biofilm-associated antiadhesion polysaccharides. mBio 2:e00043-11. 10.1128/mBio.00043-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Valle J, Da Re S, Henry N, Fontaine T, Balestrino D, Latour-Lambert P, Ghigo JM. 2006. Broad-spectrum biofilm inhibition by a secreted bacterial polysaccharide. Proc. Natl. Acad. Sci. U. S. A. 103:12558–12563. 10.1073/pnas.0605399103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Rodrigues LR. 2011. Inhibition of bacterial adhesion on medical devices. Adv. Exp. Med. Biol. 715:351–367. 10.1007/978-94-007-0940-9_22 [DOI] [PubMed] [Google Scholar]
- 268.Rojas IA, Slunt JB, Grainger DW. 2000. Polyurethane coatings release bioactive antibodies to reduce bacterial adhesion. J. Control Release 63:175–189. 10.1016/S0168-3659(99)00195-9 [DOI] [PubMed] [Google Scholar]
- 269.Poelstra KA, van der Mei HC, Gottenbos B, Grainger DW, van Horn JR, Busscher HJ. 2000. Pooled human immunoglobulins reduce adhesion of Pseudomonas aeruginosa in a parallel plate flow chamber. J. Biomed. Mater. Res. 51:224–232. [DOI] [PubMed] [Google Scholar]
- 270.Nablo BJ, Prichard HL, Butler RD, Klitzman B, Schoenfisch MH. 2005. Inhibition of implant-associated infections via nitric oxide release. Biomaterials 26:6984–6990. 10.1016/j.biomaterials.2005.05.017 [DOI] [PubMed] [Google Scholar]
- 271.Darouiche RO, Mansouri MD, Gawande PV, Madhyastha S. 2009. Antimicrobial and antibiofilm efficacy of triclosan and DispersinB combination. J. Antimicrob. Chemother. 64:88–93. 10.1093/jac/dkp158 [DOI] [PubMed] [Google Scholar]
- 272.Cadieux PA, Chew BH, Knudsen BE, Dejong K, Rowe E, Reid G, Denstedt JD. 2006. Triclosan loaded ureteral stents decrease Proteus mirabilis 296 infection in a rabbit urinary tract infection model. J. Urol. 175:2331–2335. 10.1016/S0022-5347(06)00252-7 [DOI] [PubMed] [Google Scholar]
- 273.Hachem R, Reitzel R, Borne A, Jiang Y, Tinkey P, Uthamanthil R, Chandra J, Ghannoum M, Raad I. 2009. Novel antiseptic urinary catheters for prevention of urinary tract infections: correlation of in vivo and in vitro test results. Antimicrob. Agents Chemother. 53:5145–5149. 10.1128/AAC.00718-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Cegelski L, Pinkner JS, Hammer ND, Cusumano CK, Hung CS, Chorell E, Aberg V, Walker JN, Seed PC, Almqvist F, Chapman MR, Hultgren SJ. 2009. Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nat. Chem. Biol. 5:913–919. 10.1038/nchembio.242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Cusumano CK, Pinkner JS, Han Z, Greene SE, Ford BA, Crowley JR, Henderson JP, Janetka JW, Hultgren SJ. 2011. Treatment and prevention of urinary tract infection with orally active FimH inhibitors. Sci. Transl. Med. 3:109ra115. 10.1126/scitranslmed.3003021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Guiton PS, Cusumano CK, Kline KA, Dodson KW, Han Z, Janetka JW, Henderson JP, Caparon MG, Hultgren SJ. 2012. Combinatorial small-molecule therapy prevents uropathogenic Escherichia coli catheter-associated urinary tract infections in mice. Antimicrob. Agents Chemother. 56:4738–4745. 10.1128/AAC.00447-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Totsika M, Kostakioti M, Hannan TJ, Upton M, Beatson SA, Janetka JW, Hultgren SJ, Schembri MA. 2013. A FimH inhibitor prevents acute bladder infection and treats chronic cystitis caused by multidrug-resistant uropathogenic Escherichia coli ST131. J. Infect. Dis. 208:921–928. 10.1093/infdis/jit245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Salminen A, Loimaranta V, Joosten JA, Khan AS, Hacker J, Pieters RJ, Finne J. 2007. Inhibition of P-fimbriated Escherichia coli adhesion by multivalent galabiose derivatives studied by a live-bacteria application of surface plasmon resonance. J. Antimicrob. Chemother. 60:495–501. 10.1093/jac/dkm251 [DOI] [PubMed] [Google Scholar]
- 279.Barras A, Martin FA, Bande O, Baumann JS, Ghigo JM, Boukherroub R, Beloin C, Siriwardena A, Szunerits S. 2013. Glycan-functionalized diamond nanoparticles as potent E. coli anti-adhesives. Nanoscale 5:2307–2316. 10.1039/c3nr33826f [DOI] [PubMed] [Google Scholar]
- 280.Korea CG, Ghigo JM, Beloin C. 2011. The sweet connection: solving the riddle of multiple sugar-binding fimbrial adhesins in Escherichia coli: multiple E. coli fimbriae form a versatile arsenal of sugar-binding lectins potentially involved in surface-colonisation and tissue tropism. Bioessays 33:300–311. 10.1002/bies.201000121 [DOI] [PubMed] [Google Scholar]
- 281.Siriwardena A, Pulukuri KK, Kandiyal PS, Roy S, Bande O, Ghosh S, Garcia Fernandez JM, Martin FA, Ghigo JM, Beloin C, Ito K, Woods RJ, Ampapathi RS, Chakraborty TK. 2013. Sugar-modified foldamers as conformationally defined and biologically distinct glycopeptide mimics. Angew. Chem. Int. Ed. Engl. 52:10221–10226. 10.1002/anie.201304239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Singh PK, Parsek MR, Greenberg EP, Welsh MJ. 2002. A component of innate immunity prevents bacterial biofilm development. Nature 417:552–555. 10.1038/417552a [DOI] [PubMed] [Google Scholar]
- 283.Banin E, Vasil ML, Greenberg EP. 2005. Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. U. S. A. 102:11076–11081. 10.1073/pnas.0504266102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Johnson M, Cockayne A, Williams PH, Morrissey JA. 2005. Iron-responsive regulation of biofilm formation in Staphylococcus aureus involves fur-dependent and fur-independent mechanisms. J. Bacteriol. 187:8211–8215. 10.1128/JB.187.23.8211-8215.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Ammons MC, Ward LS, Dowd S, James GA. 2011. Combined treatment of Pseudomonas aeruginosa biofilm with lactoferrin and xylitol inhibits the ability of bacteria to respond to damage resulting from lactoferrin iron chelation. Int. J. Antimicrob. Agents 37:316–323. 10.1016/j.ijantimicag.2010.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Ammons MC, Ward LS, James GA. 2011. Anti-biofilm efficacy of a lactoferrin/xylitol wound hydrogel used in combination with silver wound dressings. Int. Wound J. 8:268–273. 10.1111/j.1742-481X.2011.00781.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Antoniani D, Bocci P, Maciag A, Raffaelli N, Landini P. 2010. Monitoring of diguanylate cyclase activity and of cyclic-di-GMP biosynthesis by whole-cell assays suitable for high-throughput screening of biofilm inhibitors. Appl. Microbiol. Biotechnol. 85:1095–1104. 10.1007/s00253-009-2199-x [DOI] [PubMed] [Google Scholar]
- 288.Landini P, Antoniani D, Burgess JG, Nijland R. 2010. Molecular mechanisms of compounds affecting bacterial biofilm formation and dispersal. Appl. Microbiol. Biotechnol. 86:813–823. 10.1007/s00253-010-2468-8 [DOI] [PubMed] [Google Scholar]
- 289.Sambanthamoorthy K, Luo C, Pattabiraman N, Feng X, Koestler B, Waters CM, Palys TJ. 2014. Identification of small molecules inhibiting diguanylate cyclases to control bacterial biofilm development. Biofouling 30:17–28. 10.1080/08927014.2013.832224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Sambanthamoorthy K, Sloup RE, Parashar V, Smith JM, Kim EE, Semmelhack MF, Neiditch MB, Waters CM. 2012. Identification of small molecules that antagonize diguanylate cyclase enzymes to inhibit biofilm formation. Antimicrob. Agents Chemother. 56:5202–5211. 10.1128/AAC.01396-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Davis CP, Shirtliff ME, Scimeca JM, Hoskins SL, Warren MM. 1995. In vivo reduction of bacterial populations in the urinary tract of catheterized sheep by iontophoresis. J. Urol. 154:1948–1953. 10.1016/S0022-5347(01)66832-0 [DOI] [PubMed] [Google Scholar]
- 292.Hazan Z, Zumeris J, Jacob H, Raskin H, Kratysh G, Vishnia M, Dror N, Barliya T, Mandel M, Lavie G. 2006. Effective prevention of microbial biofilm formation on medical devices by low-energy surface acoustic waves. Antimicrob. Agents Chemother. 50:4144–4152. 10.1128/AAC.00418-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Kopel M, Degtyar E, Banin E. 2011. Surface acoustic waves increase the susceptibility of Pseudomonas aeruginosa biofilms to antibiotic treatment. Biofouling 27:701–710. 10.1080/08927014.2011.597051 [DOI] [PubMed] [Google Scholar]
- 294.Cirioni O, Giacometti A, Ghiselli R, Dell'Acqua G, Orlando F, Mocchegiani F, Silvestri C, Licci A, Saba V, Scalise G, Balaban N. 2006. RNAIII-inhibiting peptide significantly reduces bacterial load and enhances the effect of antibiotics in the treatment of central venous catheter-associated Staphylococcus aureus infections. J. Infect. Dis. 193:180–186. 10.1086/498914 [DOI] [PubMed] [Google Scholar]
- 295.Balaban N, Giacometti A, Cirioni O, Gov Y, Ghiselli R, Mocchegiani F, Viticchi C, Del Prete MS, Saba V, Scalise G, Dell'Acqua G. 2003. Use of the quorum-sensing inhibitor RNAIII-inhibiting peptide to prevent biofilm formation in vivo by drug-resistant Staphylococcus epidermidis. J. Infect. Dis. 187:625–630. 10.1086/345879 [DOI] [PubMed] [Google Scholar]
- 296.Schierle CF, De la Garza M, Mustoe TA, Galiano RD. 2009. Staphylococcal biofilms impair wound healing by delaying reepithelialization in a murine cutaneous wound model. Wound Repair Regen. 17:354–359. 10.1111/j.1524-475X.2009.00489.x [DOI] [PubMed] [Google Scholar]
- 297.Hoffmann N, Lee B, Hentzer M, Rasmussen TB, Song Z, Johansen HK, Givskov M, Høiby N. 2007. Azithromycin blocks quorum sensing and alginate polymer formation and increases the sensitivity to serum and stationary-growth-phase killing of Pseudomonas aeruginosa and attenuates chronic P. aeruginosa lung infection in Cftr(−/−) mice. Antimicrob. Agents Chemother. 51:3677–3687. 10.1128/AAC.01011-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Tateda K, Ishii Y, Matsumoto T, Furuya N, Nagashima M, Matsunaga T, Ohno A, Miyazaki S, Yamaguchi K. 1996. Direct evidence for antipseudomonal activity of macrolides: exposure-dependent bactericidal activity and inhibition of protein synthesis by erythromycin, clarithromycin, and azithromycin. Antimicrob. Agents Chemother. 40:2271–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Wagner T, Soong G, Sokol S, Saiman L, Prince A. 2005. Effects of azithromycin on clinical isolates of Pseudomonas aeruginosa from cystic fibrosis patients. Chest 128:912–919. 10.1378/chest.128.2.912 [DOI] [PubMed] [Google Scholar]
- 300.Nalca Y, Jansch L, Bredenbruch F, Geffers R, Buer J, Haussler S. 2006. Quorum-sensing antagonistic activities of azithromycin in Pseudomonas aeruginosa PAO1: a global approach. Antimicrob. Agents Chemother. 50:1680–1688. 10.1128/AAC.50.5.1680-1688.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Saiman L, Marshall BC, Mayer-Hamblett N, Burns JL, Quittner AL, Cibene DA, Coquillette S, Fieberg AY, Accurso FJ, Campbell PW., 3rd 2003. Azithromycin in patients with cystic fibrosis chronically infected with Pseudomonas aeruginosa: a randomized controlled trial. JAMA 290:1749–1756. 10.1001/jama.290.13.1749 [DOI] [PubMed] [Google Scholar]
- 302.Albert RK, Connett J, Bailey WC, Casaburi R, Cooper JA, Jr, Criner GJ, Curtis JL, Dransfield MT, Han MK, Lazarus SC, Make B, Marchetti N, Martinez FJ, Madinger NE, McEvoy C, Niewoehner DE, Porsasz J, Price CS, Reilly J, Scanlon PD, Sciurba FC, Scharf SM, Washko GR, Woodruff PG, Anthonisen NR. 2011. Azithromycin for prevention of exacerbations of COPD. N. Engl. J. Med. 365:689–698. 10.1056/NEJMoa1104623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Coles CL, Mabula K, Seidman JC, Levens J, Mkocha H, Munoz B, Mfinanga SG, West S. 2013. Mass distribution of azithromycin for trachoma control is associated with increased risk of azithromycin-resistant Streptococcus pneumoniae carriage in young children 6 months after treatment. Clin. Infect. Dis. 56:1519–1526. 10.1093/cid/cit137 [DOI] [PubMed] [Google Scholar]
- 304.Svanstrom H, Pasternak B, Hviid A. 2013. Use of azithromycin and death from cardiovascular causes. N. Engl. J. Med. 368:1704–1712. 10.1056/NEJMoa1300799 [DOI] [PubMed] [Google Scholar]
- 305.Parsek MR, Greenberg EP. 2000. Acyl-homoserine lactone quorum sensing in gram-negative bacteria: a signaling mechanism involved in associations with higher organisms. Proc. Natl. Acad. Sci. U. S. A. 97:8789–8793. 10.1073/pnas.97.16.8789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Bijtenhoorn P, Schipper C, Hornung C, Quitschau M, Grond S, Weiland N, Streit WR. 2011. BpiB05, a novel metagenome-derived hydrolase acting on N-acylhomoserine lactones. J. Biotechnol. 155:86–94. 10.1016/j.jbiotec.2010.12.016 [DOI] [PubMed] [Google Scholar]
- 307.Hentzer M, Riedel K, Rasmussen TB, Heydorn A, Andersen JB, Parsek MR, Rice SA, Eberl L, Molin S, Høiby N, Kjelleberg S, Givskov M. 2002. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 148:87–102 [DOI] [PubMed] [Google Scholar]
- 308.Hentzer M, Givskov M. 2003. Pharmacological inhibition of quorum sensing for the treatment of chronic bacterial infections. J. Clin. Invest. 112:1300–1307. 10.1172/JCI200320074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Hentzer M, Wu H, Andersen JB, Riedel K, Rasmussen TB, Bagge N, Kumar N, Schembri MA, Song Z, Kristoffersen P, Manefield M, Costerton JW, Molin S, Eberl L, Steinberg P, Kjelleberg S, Høiby N, Givskov M. 2003. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 22:3803–3815. 10.1093/emboj/cdg366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Wu H, Song Z, Hentzer M, Andersen JB, Molin S, Givskov M, Høiby N. 2004. Synthetic furanones inhibit quorum-sensing and enhance bacterial clearance in Pseudomonas aeruginosa lung infection in mice. J. Antimicrob. Chemother. 53:1054–1061. 10.1093/jac/dkh223 [DOI] [PubMed] [Google Scholar]
- 311.Christensen LD, van Gennip M, Jakobsen TH, Alhede M, Hougen HP, Høiby N, Bjarnsholt T, Givskov M. 2012. Synergistic antibacterial efficacy of early combination treatment with tobramycin and quorum-sensing inhibitors against Pseudomonas aeruginosa in an intraperitoneal foreign-body infection mouse model. J. Antimicrob. Chemother. 67:1198–1206. 10.1093/jac/dks002 [DOI] [PubMed] [Google Scholar]
- 312.Rasmussen TB, Bjarnsholt T, Skindersoe ME, Hentzer M, Kristoffersen P, Kote M, Nielsen J, Eberl L, Givskov M. 2005. Screening for quorum-sensing inhibitors (QSI) by use of a novel genetic system, the QSI selector. J. Bacteriol. 187:1799–1814. 10.1128/JB.187.5.1799-1814.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Bjarnsholt T, Jensen PO, Rasmussen TB, Christophersen L, Calum H, Hentzer M, Hougen HP, Rygaard J, Moser C, Eberl L, Høiby N, Givskov M. 2005. Garlic blocks quorum sensing and promotes rapid clearing of pulmonary Pseudomonas aeruginosa infections. Microbiology 151:3873–3880. 10.1099/mic.0.27955-0 [DOI] [PubMed] [Google Scholar]
- 314.Yang L, Liu Y, Sternberg C, Molin S. 2010. Evaluation of enoyl-acyl carrier protein reductase inhibitors as Pseudomonas aeruginosa quorum-quenching reagents. Molecules 15:780–792. 10.3390/molecules15020780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Kim JH, Choi DC, Yeon KM, Kim SR, Lee CH. 2011. Enzyme-immobilized nanofiltration membrane to mitigate biofouling based on quorum quenching. Environ. Sci. Technol. 45:1601–1607. 10.1021/es103483j [DOI] [PubMed] [Google Scholar]
- 316.Ng FS, Wright DM, Seah SY. 2011. Characterization of a phosphotriesterase-like lactonase from Sulfolobus solfataricus and its immobilization for disruption of quorum sensing. Appl. Environ. Microbiol. 77:1181–1186. 10.1128/AEM.01642-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Nanra JS, Timofeyeva Y, Buitrago SM, Sellman BR, Dilts DA, Fink P, Nunez L, Hagen M, Matsuka YV, Mininni T, Zhu D, Pavliak V, Green BA, Jansen KU, Anderson AS. 2009. Heterogeneous in vivo expression of clumping factor A and capsular polysaccharide by Staphylococcus aureus: implications for vaccine design. Vaccine 27:3276–3280. 10.1016/j.vaccine.2009.01.062 [DOI] [PubMed] [Google Scholar]
- 318.Pflumm M. 2011. Caught on film. Nat. Med. 17:650–653. 10.1038/nm0611-650 [DOI] [PubMed] [Google Scholar]
- 319.Langermann S, Palaszynski S, Barnhart M, Auguste G, Pinkner JS, Burlein J, Barren P, Koenig S, Leath S, Jones CH, Hultgren SJ. 1997. Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science 276:607–611. 10.1126/science.276.5312.607 [DOI] [PubMed] [Google Scholar]
- 320.Pecha B, Low D, O'Hanley P. 1989. Gal-Gal pili vaccines prevent pyelonephritis by piliated Escherichia coli in a murine model. Single-component Gal-Gal pili vaccines prevent pyelonephritis by homologous and heterologous piliated E. coli strains. J. Clin. Invest. 83:2102–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Ebert T, Smith S, Pancari G, Wu X, Zorman J, Clark D, Cook J, Burns C, Antonello JM, Cope L, Nagy E, Meinke A, McNeely T. 2011. Development of a rat central venous catheter model for evaluation of vaccines to prevent Staphylococcus epidermidis and Staphylococcus aureus early biofilms. Hum. Vaccin. 7:630–638. 10.4161/hv.7.6.15407 [DOI] [PubMed] [Google Scholar]
- 322.Gil C, Solano C, Burgui S, Latasa C, Garcia B, Toledo-Arana A, Lasa I, Valle J. 2014. Biofilm matrix exoproteins induce a protective immune response against Staphylococcus aureus biofilm infection. Infect. Immun. 82:1017–1029. 10.1128/IAI.01419-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Brady RA, O'May GA, Leid JG, Prior ML, Costerton JW, Shirtliff ME. 2011. Resolution of Staphylococcus aureus biofilm infection using vaccination and antibiotic treatment. Infect. Immun. 79:1797–1803. 10.1128/IAI.00451-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Reid G, Howard J, Gan BS. 2001. Can bacterial interference prevent infection? Trends Microbiol. 9:424–428. 10.1016/S0966-842X(01)02132-1 [DOI] [PubMed] [Google Scholar]
- 325.Klemm P, Hancock V, Schembri MA. 2007. Mellowing out: adaptation to commensalism by Escherichia coli asymptomatic bacteriuria strain 83972. Infect. Immun. 75:3688–3695. 10.1128/IAI.01730-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Sunden F, Hakansson L, Ljunggren E, Wullt B. 2010. Escherichia coli 83972 bacteriuria protects against recurrent lower urinary tract infections in patients with incomplete bladder emptying. J. Urol. 184:179–185. 10.1016/j.juro.2010.03.024 [DOI] [PubMed] [Google Scholar]
- 327.Kong KF, Vuong C, Otto M. 2006. Staphylococcus quorum sensing in biofilm formation and infection. Int. J. Med. Microbiol. 296:133–139. 10.1016/j.ijmm.2006.01.042 [DOI] [PubMed] [Google Scholar]
- 328.O'Toole GA. 2004. Microbiology: Jekyll or hide? Nature 432:680–681. 10.1038/432680a [DOI] [PubMed] [Google Scholar]
- 329.Chaignon P, Sadovskaya I, Ragunah C, Ramasubbu N, Kaplan JB, Jabbouri S. 2007. Susceptibility of staphylococcal biofilms to enzymatic treatments depends on their chemical composition. Appl. Microbiol. Biotechnol. 75:125–132. 10.1007/s00253-006-0790-y [DOI] [PubMed] [Google Scholar]
- 330.Izano EA, Amarante MA, Kher WB, Kaplan JB. 2008. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl. Environ. Microbiol. 74:470–476. 10.1128/AEM.02073-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Gloag ES, Turnbull L, Huang A, Vallotton P, Wang H, Nolan LM, Mililli L, Hunt C, Lu J, Osvath SR, Monahan LG, Cavaliere R, Charles IG, Wand MP, Gee ML, Prabhakar R, Whitchurch CB. 2013. Self-organization of bacterial biofilms is facilitated by extracellular DNA. Proc. Natl. Acad. Sci. U. S. A. 110:11541–11546. 10.1073/pnas.1218898110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Okshevsky M, Meyer RL. 4 December 2013. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit. Rev. Microbiol. 10.3109/1040841X.2013.841639 [DOI] [PubMed] [Google Scholar]
- 333.Hymes SR, Randis TM, Sun TY, Ratner AJ. 2013. DNase inhibits Gardnerella vaginalis biofilms in vitro and in vivo. J. Infect. Dis. 207:1491–1497. 10.1093/infdis/jit047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. 10.1126/science.295.5559.1487 [DOI] [PubMed] [Google Scholar]
- 335.Huang J, Pinder KL. 1995. Effects of calcium on development of anaerobic acidogenic biofilms. Biotechnol. Bioeng. 45:212–218. 10.1002/bit.260450305 [DOI] [PubMed] [Google Scholar]
- 336.Turakhia MH, Characklis WG. 1989. Activity of Pseudomonas aeruginosa in biofilms: effect of calcium. Biotechnol. Bioeng. 33:406–414. 10.1002/bit.260330405 [DOI] [PubMed] [Google Scholar]
- 337.Korstgens V, Flemming HC, Wingender J, Borchard W. 2001. Influence of calcium ions on the mechanical properties of a model biofilm of mucoid Pseudomonas aeruginosa. Water Sci. Technol. 43:49–57 [PubMed] [Google Scholar]
- 338.Chen X, Stewart PS. 2002. Role of electrostatic interactions in cohesion of bacterial biofilms. Appl. Microbiol. Biotechnol. 59:718–720. 10.1007/s00253-002-1044-2 [DOI] [PubMed] [Google Scholar]
- 339.Turakhia MH, Cooksey KE, Characklis WG. 1983. Influence of a calcium-specific chelant on biofilm removal. Appl. Environ. Microbiol. 46:1236–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Banin E, Brady KM, Greenberg EP. 2006. Chelator-induced dispersal and killing of Pseudomonas aeruginosa cells in a biofilm. Appl. Environ. Microbiol. 72:2064–2069. 10.1128/AEM.72.3.2064-2069.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Weijmer MC, Debets-Ossenkopp YJ, Van De Vondervoort FJ, ter Wee PM. 2002. Superior antimicrobial activity of trisodium citrate over heparin for catheter locking. Nephrol. Dial. Transplant. 17:2189–2195. 10.1093/ndt/17.12.2189 [DOI] [PubMed] [Google Scholar]
- 342.Bookstaver PB, Williamson JC, Tucker BK, Raad II, Sherertz RJ. 2009. Activity of novel antibiotic lock solutions in a model against isolates of catheter-related bloodstream infections. Ann. Pharmacother. 43:210–219. 10.1345/aph.1L145 [DOI] [PubMed] [Google Scholar]
- 343.Raad I, Rosenblatt J, Reitzel R, Jiang Y, Dvorak T, Hachem R. 2013. Chelator-based catheter lock solutions in eradicating organisms in biofilm. Antimicrob. Agents Chemother. 57:586–588. 10.1128/AAC.01287-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Chauhan A, Lebeaux D, Decante B, Kriegel I, Escande MC, Ghigo JM, Beloin C. 2012. A rat model of central venous catheter to study establishment of long-term bacterial biofilm and related acute and chronic infections. PLoS One 7:e37281. 10.1371/journal.pone.0037281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Boles BR, Horswill AR. 2008. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4:e1000052. 10.1371/journal.ppat.1000052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Simonetti O, Cirioni O, Ghiselli R, Goteri G, Scalise A, Orlando F, Silvestri C, Riva A, Saba V, Madanahally KD, Offidani A, Balaban N, Scalise G, Giacometti A. 2008. RNAIII-inhibiting peptide enhances healing of wounds infected with methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 52:2205–2211. 10.1128/AAC.01340-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Davies DG, Marques CN. 2009. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 191:1393–1403. 10.1128/JB.01214-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Kolodkin-Gal I, Romero D, Cao S, Clardy J, Kolter R, Losick R. 2010. D-amino acids trigger biofilm disassembly. Science 328:627–629. 10.1126/science.1188628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Leiman SA, May JM, Lebar MD, Kahne D, Kolter R, Losick R. 2013. D-amino acids indirectly inhibit biofilm formation in Bacillus subtilis by interfering with protein synthesis. J. Bacteriol. 195:5391–5395. 10.1128/JB.00975-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Kolodkin-Gal I, Cao S, Chai L, Bottcher T, Kolter R, Clardy J, Losick R. 2012. A self-produced trigger for biofilm disassembly that targets exopolysaccharide. Cell 149:684–692. 10.1016/j.cell.2012.02.055 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 351.Hobley L, Kim SH, Maezato Y, Wyllie S, Fairlamb AH, Stanley-Wall NR, Michael AJ. 2014. Norspermidine is not a self-produced trigger for biofilm disassembly. Cell 156:844–854. 10.1016/j.cell.2014.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Barraud N, Hassett DJ, Hwang SH, Rice SA, Kjelleberg S, Webb JS. 2006. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J. Bacteriol. 188:7344–7353. 10.1128/JB.00779-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Barraud N, Schleheck D, Klebensberger J, Webb JS, Hassett DJ, Rice SA, Kjelleberg S. 2009. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J. Bacteriol. 191:7333–7342. 10.1128/JB.00975-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Taber HW, Mueller JP, Miller PF, Arrow AS. 1987. Bacterial uptake of aminoglycoside antibiotics. Microbiol. Rev. 51:439–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Barraud N, Buson A, Jarolimek W, Rice SA. 2013. Mannitol enhances antibiotic sensitivity of persister bacteria in Pseudomonas aeruginosa biofilms. PLoS One 8:e84220. 10.1371/journal.pone.0084220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Morones-Ramirez JR, Winkler JA, Spina CS, Collins JJ. 2013. Silver enhances antibiotic activity against gram-negative bacteria. Sci. Transl. Med. 5:190ra181. 10.1126/scitranslmed.3006276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Brynildsen MP, Winkler JA, Spina CS, MacDonald IC, Collins JJ. 2013. Potentiating antibacterial activity by predictably enhancing endogenous microbial ROS production. Nat. Biotechnol. 31:160–165. 10.1038/nbt.2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Doolittle MM, Cooney JJ, Caldwell DE. 1995. Lytic infection of Escherichia coli biofilms by bacteriophage T4. Can. J. Microbiol. 41:12–18. 10.1139/m95-002 [DOI] [PubMed] [Google Scholar]
- 359.Hanlon GW, Denyer SP, Olliff CJ, Ibrahim LJ. 2001. Reduction in exopolysaccharide viscosity as an aid to bacteriophage penetration through Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 67:2746–2753. 10.1128/AEM.67.6.2746-2753.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Hughes KA, Sutherland IW, Jones MV. 1998. Biofilm susceptibility to bacteriophage attack: the role of phage-borne polysaccharide depolymerase. Microbiology 144:3039–3047. 10.1099/00221287-144-11-3039 [DOI] [PubMed] [Google Scholar]
- 361.Sillankorva S, Oliveira R, Vieira MJ, Sutherland I, Azeredo J. 2004. Pseudomonas fluorescens infection by bacteriophage PhiS1: the influence of temperature, host growth phase and media. FEMS Microbiol. Lett. 241:13–20. 10.1016/j.femsle.2004.06.058 [DOI] [PubMed] [Google Scholar]
- 362.Glonti T, Chanishvili N, Taylor PW. 2010. Bacteriophage-derived enzyme that depolymerizes the alginic acid capsule associated with cystic fibrosis isolates of Pseudomonas aeruginosa. J. Appl. Microbiol. 108:695–702. 10.1111/j.1365-2672.2009.04469.x [DOI] [PubMed] [Google Scholar]
- 363.Fu W, Forster T, Mayer O, Curtin JJ, Lehman SM, Donlan RM. 2010. Bacteriophage cocktail for the prevention of biofilm formation by Pseudomonas aeruginosa on catheters in an in vitro model system. Antimicrob. Agents Chemother. 54:397–404. 10.1128/AAC.00669-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Maura D, Galtier M, Le Bouguenec C, Debarbieux L. 2012. Virulent bacteriophages can target O104:H4 enteroaggregative Escherichia coli in the mouse intestine. Antimicrob. Agents Chemother. 56:6235–6242. 10.1128/AAC.00602-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Lebeaux D, Ghigo JM. 2012. Management of biofilm-associated infections: what can we expect from recent research on biofilm lifestyles? Med. Sci. (Paris) 28:727–739. 10.1051/medsci/2012288015 [DOI] [PubMed] [Google Scholar]
- 366.Lebeaux D, Ghigo JM. 2013. Les biofilms: des adversaires intraitables? Biofutur 32:34–39 [Google Scholar]
- 367.Hoyle BD, Alcantara J, Costerton JW. 1992. Pseudomonas aeruginosa biofilm as a diffusion barrier to piperacillin. Antimicrob. Agents Chemother. 36:2054–2056. 10.1128/AAC.36.9.2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Yasuda H, Ajiki Y, Koga T, Kawada H, Yokota T. 1993. Interaction between biofilms formed by Pseudomonas aeruginosa and clarithromycin. Antimicrob. Agents Chemother. 37:1749–1755. 10.1128/AAC.37.9.1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Suci PA, Mittelman MW, Yu FP, Geesey GG. 1994. Investigation of ciprofloxacin penetration into Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 38:2125–2133. 10.1128/AAC.38.9.2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Vrany JD, Stewart PS, Suci PA. 1997. Comparison of recalcitrance to ciprofloxacin and levofloxacin exhibited by Pseudomonas aeruginosa bofilms displaying rapid-transport characteristics. Antimicrob. Agents Chemother. 41:1352–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Jouenne T, Tresse O, Junter GA. 1994. Agar-entrapped bacteria as an in vitro model of biofilms and their susceptibility to antibiotics. FEMS Microbiol. Lett. 119:237–242. 10.1111/j.1574-6968.1994.tb06894.x [DOI] [PubMed] [Google Scholar]
- 372.Dunne WM, Jr, Mason EO, Jr, Kaplan SL. 1993. Diffusion of rifampin and vancomycin through a Staphylococcus epidermidis biofilm. Antimicrob. Agents Chemother. 37:2522–2526. 10.1128/AAC.37.12.2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Darouiche RO, Dhir A, Miller AJ, Landon GC, Raad II, Musher DM. 1994. Vancomycin penetration into biofilm covering infected prostheses and effect on bacteria. J. Infect. Dis. 170:720–723. 10.1093/infdis/170.3.720 [DOI] [PubMed] [Google Scholar]
- 374.Singh R, Ray P, Das A, Sharma M. 2010. Penetration of antibiotics through Staphylococcus aureus and Staphylococcus epidermidis biofilms. J. Antimicrob. Chemother. 65:1955–1958. 10.1093/jac/dkq257 [DOI] [PubMed] [Google Scholar]
- 375.Scarano A, Piattelli M, Vrespa G, Caputi S, Piattelli A. 2003. Bacterial adhesion on titanium nitride-coated and uncoated implants: an in vivo human study. J. Oral Implantol. 29:80–85. [DOI] [PubMed] [Google Scholar]
- 376.Poelstra KA, Barekzi NA, Rediske AM, Felts AG, Slunt JB, Grainger DW. 2002. Prophylactic treatment of gram-positive and gram-negative abdominal implant infections using locally delivered polyclonal antibodies. J. Biomed. Mater. Res. 60:206–215. 10.1002/jbm.10069 [DOI] [PubMed] [Google Scholar]
- 377.Raad II, Fang X, Keutgen XM, Jiang Y, Sherertz R, Hachem R. 2008. The role of chelators in preventing biofilm formation and catheter-related bloodstream infections. Curr. Opin. Infect. Dis. 21:385–392. 10.1097/QCO.0b013e32830634d8 [DOI] [PubMed] [Google Scholar]
- 378.Cobrado L, Azevedo MM, Silva-Dias A, Ramos JP, Pina-Vaz C, Rodrigues AG. 2012. Cerium, chitosan and hamamelitannin as novel biofilm inhibitors? J. Antimicrob. Chemother. 67:1159–1162. 10.1093/jac/dks007 [DOI] [PubMed] [Google Scholar]
- 379.Cobrado L, Silva-Dias A, Azevedo MM, Pina-Vaz C, Rodrigues AG. 2013. In vivo antibiofilm effect of cerium, chitosan and hamamelitannin against usual agents of catheter-related bloodstream infections. J. Antimicrob. Chemother. 68:126–130. 10.1093/jac/dks376 [DOI] [PubMed] [Google Scholar]
- 380.Cruz-Villalon G, Perez-Giraldo C. 2011. Effect of allicin on the production of polysaccharide intercellular adhesin in Staphylococcus epidermidis. J. Appl. Microbiol. 110:723–728. 10.1111/j.1365-2672.2010.04929.x [DOI] [PubMed] [Google Scholar]
- 381.Wu X, Wang Y, Tao L. 2011. Sulfhydryl compounds reduce Staphylococcus aureus biofilm formation by inhibiting PIA biosynthesis. FEMS Microbiol. Lett. 316:44–50. 10.1111/j.1574-6968.2010.02190.x [DOI] [PubMed] [Google Scholar]
- 382.Wu H, Lee B, Yang L, Wang H, Givskov M, Molin S, Høiby N, Song Z. 2011. Effects of ginseng on Pseudomonas aeruginosa motility and biofilm formation. FEMS Immunol. Med. Microbiol. 62:49–56. 10.1111/j.1574-695X.2011.00787.x [DOI] [PubMed] [Google Scholar]
- 383.Attila C, Ueda A, Wood TK. 2009. 5-Fluorouracil reduces biofilm formation in Escherichia coli K-12 through global regulator AriR as an antivirulence compound. Appl. Microbiol. Biotechnol. 82:525–533. 10.1007/s00253-009-1860-8 [DOI] [PubMed] [Google Scholar]
- 384.Ueda A, Attila C, Whiteley M, Wood TK. 2009. Uracil influences quorum sensing and biofilm formation in Pseudomonas aeruginosa and fluorouracil is an antagonist. Microb. Biotechnol. 2:62–74. 10.1111/j.1751-7915.2008.00060.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Walz JM, Avelar RL, Longtine KJ, Carter KL, Mermel LA, Heard SO. 2010. Anti-infective external coating of central venous catheters: a randomized, noninferiority trial comparing 5-fluorouracil with chlorhexidine/silver sulfadiazine in preventing catheter colonization. Crit. Care Med. 38:2095–2102. 10.1097/CCM.0b013e3181f265ba [DOI] [PubMed] [Google Scholar]
- 386.Kim JS, Heo P, Yang TJ, Lee KS, Cho DH, Kim BT, Suh JH, Lim HJ, Shin D, Kim SK, Kweon DH. 2011. Selective killing of bacterial persisters by a single chemical compound without affecting normal antibiotic-sensitive cells. Antimicrob. Agents Chemother. 55:5380–5383. 10.1128/AAC.00708-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Jabra-Rizk MA, Meiller TF, James CE, Shirtliff ME. 2006. Effect of farnesol on Staphylococcus aureus biofilm formation and antimicrobial susceptibility. Antimicrob. Agents Chemother. 50:1463–1469. 10.1128/AAC.50.4.1463-1469.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Hughes KA, Sutherland IW, Clark J, Jones MV. 1998. Bacteriophage and associated polysaccharide depolymerases—novel tools for study of bacterial biofilms. J. Appl. Microbiol. 85:583–590. 10.1046/j.1365-2672.1998.853541.x [DOI] [PubMed] [Google Scholar]
- 389.Aslam S, Darouiche RO. 2011. Role of antibiofilm-antimicrobial agents in controlling device-related infections. Int. J. Artif. Organs 34:752–758. 10.5301/ijao.5000024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Aslam S, Trautner BW, Ramanathan V, Darouiche RO. 2007. Combination of tigecycline and N-acetylcysteine reduces biofilm-embedded bacteria on vascular catheters. Antimicrob. Agents Chemother. 51:1556–1558. 10.1128/AAC.00893-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Aslam S, Trautner BW, Ramanathan V, Darouiche RO. 2008. Pilot trial of N-acetylcysteine and tigecycline as a catheter-lock solution for treatment of hemodialysis catheter-associated bacteremia. Infect. Control Hosp. Epidemiol. 29:894–897. 10.1086/590192 [DOI] [PubMed] [Google Scholar]
- 392.Gethin G, Cowman S. 2009. Manuka honey vs. hydrogel—a prospective, open label, multicentre, randomised controlled trial to compare desloughing efficacy and healing outcomes in venous ulcers. J. Clin. Nurs. 18:466–474. 10.1111/j.1365-2702.2008.02558.x [DOI] [PubMed] [Google Scholar]
- 393.Kilty SJ, Duval M, Chan FT, Ferris W, Slinger R. 2011. Methylglyoxal: (active agent of manuka honey) in vitro activity against bacterial biofilms. Int. Forum Allergy Rhinol. 1:348–350. 10.1002/alr.20073 [DOI] [PubMed] [Google Scholar]
- 394.Leaper DJ, Schultz G, Carville K, Fletcher J, Swanson T, Drake R. 2012. Extending the TIME concept: what have we learned in the past 10 years?(*). Int. Wound J. 9(Suppl 2):S1–S19. 10.1111/j.1742-481X.2012.01097.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Lee JH, Park JH, Kim JA, Neupane GP, Cho MH, Lee CS, Lee J. 2011. Low concentrations of honey reduce biofilm formation, quorum sensing, and virulence in Escherichia coli O157:H7. Biofouling 27:1095–1104. 10.1080/08927014.2011.633704 [DOI] [PubMed] [Google Scholar]
- 396.Maddocks SE, Lopez MS, Rowlands RS, Cooper RA. 2012. Manuka honey inhibits the development of Streptococcus pyogenes biofilms and causes reduced expression of two fibronectin binding proteins. Microbiology 158:781–790. 10.1099/mic.0.053959-0 [DOI] [PubMed] [Google Scholar]
- 397.Majtan J, Bohova J, Horniackova M, Klaudiny J, Majtan V. 2014. Anti-biofilm effects of honey against wound pathogens Proteus mirabilis and Enterobacter cloacae. Phytother. Res. 28:69–75. 10.1002/ptr.4957 [DOI] [PubMed] [Google Scholar]
- 398.Molan PC. 1992. The antibacterial activity of honey. 1. The nature of the antibacterial activity. Bee World 73:5–28 [Google Scholar]
- 399.Truchado P, Gil-Izquierdo A, Tomas-Barberan F, Allende A. 2009. Inhibition by chestnut honey of N-acyl-l-homoserine lactones and biofilm formation in Erwinia carotovora, Yersinia enterocolitica, and Aeromonas hydrophila. J. Agric. Food Chem. 57:11186–11193. 10.1021/jf9029139 [DOI] [PubMed] [Google Scholar]
- 400.Kim HW, Ha US, Woo JC, Kim SJ, Yoon BI, Lee SJ, Cho YH. 2012. Preventive effect of selenium on chronic bacterial prostatitis. J. Infect. Chemother. 18:30–34. 10.1007/s10156-011-0276-4 [DOI] [PubMed] [Google Scholar]
- 401.Kim SH, Ha US, Lee HR, Sohn DW, Lee SJ, Kim HW, Han CH, Lee CB, Cho YH. 2011. Do Escherichia coli extract and cranberry exert preventive effects on chronic bacterial prostatitis? Pilot study using an animal model. J. Infect. Chemother. 17:322–326. 10.1007/s10156-010-0170-5 [DOI] [PubMed] [Google Scholar]
- 402.Lee YS, Han CH, Kang SH, Lee SJ, Kim SW, Shin OR, Sim YC, Cho YH. 2005. Synergistic effect between catechin and ciprofloxacin on chronic bacterial prostatitis rat model. Int. J. Urol. 12:383–389. 10.1111/j.1442-2042.2005.01052.x [DOI] [PubMed] [Google Scholar]
- 403.Flemming K, Klingenberg C, Cavanagh JP, Sletteng M, Stensen W, Svendsen JS, Flaegstad T. 2009. High in vitro antimicrobial activity of synthetic antimicrobial peptidomimetics against staphylococcal biofilms. J. Antimicrob. Chemother. 63:136–145. 10.1093/jac/dkn464 [DOI] [PubMed] [Google Scholar]
- 404.Sandvik EL, McLeod BR, Parker AE, Stewart PS. 2013. Direct electric current treatment under physiologic saline conditions kills Staphylococcus epidermidis biofilms via electrolytic generation of hypochlorous acid. PLoS One 8:e55118. 10.1371/journal.pone.0055118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Dong Y, Chen S, Wang Z, Peng N, Yu J. 2013. Synergy of ultrasound microbubbles and vancomycin against Staphylococcus epidermidis biofilm. J. Antimicrob. Chemother. 68:816–826. 10.1093/jac/dks490 [DOI] [PubMed] [Google Scholar]