Abstract
Epidermal growth factor (EGF) receptor (EGFR) has been implicated in tumor development and invasion. Dimerization and autophosphorylation of EGFR are the critical events for EGFR activation. However, the regulation of EGF-dependent and EGF-independent dimerization and phosphorylation of EGFR has not been fully understood. Here, we report that cytoplasmic protein plakophilin-2 (PKP2) is a novel positive regulator of EGFR signaling. PKP2 specifically interacts with EGFR via its N-terminal head domain. Increased PKP2 expression enhances EGF-dependent and EGF-independent EGFR dimerization and phosphorylation. Moreover, PKP2 knockdown reduces EGFR phosphorylation and attenuates EGFR-mediated signal activation, resulting in a significant decrease in proliferation and migration of cancer cells and tumor development. Our results indicate that PKP2 is a novel activator of the EGFR signaling pathway and a potential new drug target for inhibiting tumor growth.
INTRODUCTION
Epidermal growth factor receptor (EGFR) (also known as ErbB-1) is a member of the ErbB family of transmembrane cell surface receptor tyrosine kinases (RTKs), which also includes ErbB-2 (HER2), ErbB-3, and ErbB-4 (1–6). EGFR is activated upon binding to its ligands, such as epidermal growth factor (EGF), and aberrant EGFR function is a hallmark of many human cancers (1–6). The tyrosine autophosphorylation sites on the carboxyl terminus of EGFR serve as binding sites for molecules containing Src homology 2 (SH2) domains (SHC), growth factor receptor-bound protein 2 (Grb2), SH2-domain-containing protein tyrosine phosphatase 1 (SHP1), and a guanine nucleotide-exchanging factor (GEP100) for ARF6. These molecules in turn initiate EGFR downstream signaling pathways, such as the PI3K/AKT, Ras/Raf/MEK/extracellular signal-regulated kinase (ERK), and STAT1/3 pathways. These pathways activate NF-κB and AP-1 transcriptional activity, triggering cell proliferation, resistance to apoptosis, angiogenesis, and invasion (7–10). Recent studies discovered that GEP100 is recruited by phosphorylated tyrosine 1068 (pTyr1068, pY1068) of EGFR and functions to promote migration and invasion, as opposed to proliferation (9). Despite our current understanding of EGFR activation, the detailed mechanisms of the regulation of EGFR-mediated cytoplasmic signal transduction are not fully understood, since receptor dimerization can occur continuously and reversibly even in the absence of ligand (11).
Plakophilin 1 (PKP1), PKP2, and PKP3 form a small family of proteins. They are composed of a basic N-terminal head domain, followed by a series of 42-amino-acid armadillo repeats (arm-repeat) and a short C-terminal tail (12, 13). Although the N-terminal head domains of PKPs are quite diversified, a consensus sequence termed the HR2 motif is shared by all the PKP head domains (12, 13). PKP2 was originally isolated as a desmosomal protein, but additional studies have shown that it is also found in the cytoplasm and nucleus (14). Mutations of PKP2 are related to the cardiac disorder arrhythmogenic right ventricular cardiomyopathy/dysplasia (ARVC/D), and PKP2-deficient mice showed early-stage embryonic lethality due to abnormal heart morphogenesis, demonstrating that PKP2 is important for normal heart development and function (15–17). Although the mechanisms have not yet been identified, PKP2 overexpression is also associated with cancer malignancy as determined on the basis of several clinical and immunohistochemical analyses (14, 18–21).
Here, we unravel a novel function of PKP2 in EGFR function. Our findings demonstrate that PKP2 enhances EGFR dimerization and autophosphorylation, leading to activation of EGFR-mediated signaling pathways. More importantly, our results from in vivo xenotransplantation studies demonstrate a similar stimulatory effect of PKP2 on breast cancer development, suggesting PKP2 as a novel target of cancer therapy.
MATERIALS AND METHODS
Mice and tumor monitoring.
For spontaneous-metastasis assays, 2 × 106 MDA-MB-231 cells (single clones from a short hairpin control [sh-Control], sh-PKP2-2, and sh-PKP2-4) were injected into the fourth mammary fat pad of 6-to-8-week-old NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) female mice in 1:1 Matrigel plus phosphate-buffered saline (PBS). Primary tumor growth upon orthotopic injection was examined by weekly measurements. Approximate tumor volumes were calculated by the formula 4/3 × 3.14 × [(long diameter/2) (short diameter/2)2]. All dissected lungs were fixed in 4% paraformaldehyde, paraffin embedded, sectioned (10th serial, 10-μm-long space), and stained with hematoxylin and eosin (H&E), and the number of metastatic nodules per lung was counted by microscope.
All animals were maintained in the Laboratory Animal Center at the Moores Cancer Center of the University of California, San Diego (UCSD). All animal experiments were performed in accordance with NIH policies on the use of laboratory animals and were approved by the Animal Research Committee of the Office for the Protection of Research Subjects at the University of California, San Diego.
Mass spectrometry database.
As a tool for searching for candidate molecules that interact with EGFR, we used the EMBL-EBI IntAct database (EBI-702413).
Cell culture, transfection, retroviral infection, and luciferase reporter assays.
293T, A431, and MDA-MB-468 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with glutamine, penicillin-streptomycin, and 10% fetal bovine serum (FBS). MDA-MB-231, HT29, and HCT116 cells were further supplemented with 5% MEM (where MEM is minimum essential medium) nonessential amino acid solution (Gibco). 32D cells were grown in RPMI 1640 medium supplemented with 15% FBS and 5% conditional media from WEHI-3B cells. Plasmids were transfected using polyethyleneimine (PEI). For the transient expression of mouse PKP2 to 32D cells, Nucleofector kits for 32D cells (Lonza) were used. Small interfering RNA (siRNA) was transfected using Lipofectamine 2000 (Invitrogen) at an interval of 24 h. At 72 h after the first siRNA transfection, cell lysates were prepared for examination. For the retrovirus infection, spin infection (3,000 rpm, 3 h) was performed. Analysis of AP-1 and NF-κB dual-luciferase (Luc) assays were carried out using Promega's luciferase assay kit. As an internal control, pRL-SV was transfected.
Antibodies and reagents.
Antibodies were purchased from commercial sources as follows: anti-EGFR (Abcam), immunoprecipitation (IP)-specific anti-EGFR (Cell Signaling), anti-phospho-EGFR (Tyr845, -992, -1045, -1068, and -1173 [Cell Signaling] and Tyr 1086 [Invitrogen]), anti-PKP2 (ARP; American Research Products, Inc.), anti-ERK (BD Bioscience), anti-phospho-ERK (Cell Signaling), anti-phospho-MEK1 (Cell Signaling), anti-Grb2 (Cell Signaling), anti-SHP1 (Cell Signaling), anti-GEP100 (Sigma), anti-green fluorescent protein (anti-GFP), and anti-α-tubulin (Sigma). Antibodies against FLAG (anti-FLAG; M2), Myc (9E10), and hemagglutinin (HA) (12CA5 or 3F10) were purchased from Sigma, Santa Cruz, and Roche, respectively. Recombinant human EGF was purchased from PeproTech. Lapatinib (S1028) was purchased from Selleck.
cDNA construction.
Human PKP2 cDNA was cloned into C-terminally tagged pCAG vector, pcDNA3.1 vector, and glutathione S-transferase (GST) (6p-1) vector. Murine PKP2 cDNA was cloned into C-terminal FLAG pCAG vector and murine stem cell virus (MSCV)-internal ribosome entry site (IRES)-Puro (MIP) retroviral vector. Human pLNCX2-EGFR-GFP was kindly provided by Frank Furnari. Human EGFR and HER2 were cloned into pCMV5-FLAG and pcDNA3-Myc-His vector. Human PKP1 and PKP3 cDNAs were cloned into C-terminally tagged pCAG vector. Human Grb2 and Shc cDNAs were cloned into pcDNA3.1 vector. Deletion mutants of PKP2 and EGFR and Sh-PKP2-4-resistant PKP2 (T249A/T252A/G255A) were made by standard PCR procedures.
Knockdown, RNA isolation, and quantitative reverse transcription-PCR (qRT-PCR) analysis.
For knockdown of PKP2, the following siRNAs were used: si-PKP2-1 (5′-GAGACUACCCAAAAGCAAAUU-3′ [Ambion]); si-PKP2-2 (5′-GCUCCUAAAAGUUCAGAAUUU-3′ [Ambion]); and si-PKP2-3 (5′-CAAAAGUAUUGGAUGUUUUUU-3′ [Ambion]).
Control siRNA was purchased from Ambion.
Short hairpin RNA (shRNA) plasmids for PKP2 and control experiments were purchased from the SuperArray Bioscience Corporation. Insertion sequences for shRNAs were as follows: for sh-PKP2-1, GCACACACGGAACTGCATCAT; for sh-PKP2-2, TGACTCACTGGTCCATTATGT; for sh-PKP2-3, CACGAGTGCTTCCAGAAATCT; for sh-PKP2-4, CAGCAGTGTTCCTGAGTATGT; and for the short hairpin control (sh-Control), GGAATCTCATTCGATGCATAC.
RNA was extracted with an RNeasy microkit (Qiagen). For qRT-PCR analyses, equal amounts of RNA were reverse transcribed by the use of qScript (Quanta Biosciences) and the resulting cDNA templates were subjected to qRT-PCR using a SYBR green detection system on a CFX96 thermal cycler (Bio-Rad).
Primer sequences for PKP2 were as follows: for RT-PKP2-Fw, GAT GTT TTG GCA GTC GAA GCA G; and for RT-PKP2-Rev, AAT GGA ATG CCA CAG CCA CTC.
Western blotting and immunoprecipitation.
All samples were denatured in 1× sample buffer (50 mM Tris-HCl [pH 6.8], 2% SDS, 2-mercaptethanol, 10% glycerol, and 1% bromophenol blue) for 5 min at 100°C. Cells were lysed in a radioimmunoprecipitation assay (RIPA) buffer composed of 25 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol (DTT), 0.1% SDS, 1% Nonidet P-40, and 0.5% sodium deoxycholate. To analyze immune complexes, cells were lysed in binding buffer containing 25 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, and 0.5% Nonidet P-40 for coimmunoprecipitation assays. The cell lysates were centrifuged (10,000 × g) at 4°C for 5 min. All lysis buffers in this study contained proteinase and phosphatase inhibitors (Roche). Soluble fractions were precleared by the use of protein G-Sepharose at 4°C for 15 min. Precleared cell lysates were immunoprecipitated for ∼1 to ∼4 h with the indicated antibodies. Immunocomplexes were adsorbed to the protein G-Sepharose and, after three washes, were eluted by boiling for 5 min. FLAG-tagged proteins were immunoprecipitated with anti-FLAG M2-agarose (Sigma). For quantification, Fujifilm MultiGauge V3.0 software was used.
GST pulldown assay.
To purify full-length EGFR or the cytoplasmic region of EGFR (EGFR amino acids [aa] 644 to 1210), 293T cells expressing EGFR-FLAG or EGFR aa 644 to 1210 (cytoplasmic region)-FLAG were lysed in RIPA buffer with 1× proteinase inhibitor mixture, and cell lysates were immunoprecipitated with anti-FLAG M2 agarose in RIPA buffer overnight at 4°C. After stringent washing with RIPA buffer, the bound protein was eluted using 50 μg/ml FLAG peptide (Sigma). For GST pulldown, bacterial cell lysates expressing GST or GST-PKP2 were incubated with GST-Sepharose 4B beads (Roche) in binding buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1% Nonidet P-40, and 1× proteinase inhibitor mixture) for 3 h at 4°C and washed 5 times with binding buffer. Purified EGFR-FLAG or EGFR (aa 644 to 1210)-FLAG was then incubated in GST or GST-PKP2 columns. After washing with binding buffer, bound proteins were eluted and analyzed by Western blotting with the indicated antibodies.
Chemical cross-linking.
Twenty-four hours after transfection under starvation conditions, monolayer cells were washed twice with ice-cold phosphate-buffered saline containing 0.33 mM MgCl2 and 0.9 mM CaCl2 [PBS(+)] and chemically cross-linked for 20 min at room temperature with freshly prepared 1.5 mM bis(sulfosuccinimidyl) suberate (BS3; Pierce, Rockford, IL). To terminate the reaction, a final concentration of 20 mM glycine was added for an additional 5 min. For immunoblot analysis, the cells were washed twice with ice-cold PBS(+) and lysed with 10 mM iodoacetamide containing RIPA buffer. Samples were separated using 6% SDS-PAGE gels and transferred at 20 V overnight with standard transfer buffer (48 mM Tris, 39 mM glycine, 0.0375% SDS, and 20% methanol).
Proliferation assay.
Cell growth was monitored by trypan blue counting or an MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] assay. For the trypan blue counting, cells were seeded in 12-well plates at a density of 5 × 104 cells/well in triplicate. The MTS assay was conducted using CellTiter 96 AQueous One Solution reagent (MTS assay reagent) according to the instructions of the manufacturer (Promega). A total of 5 × 103 cells were seeded in each well of a 96-well plate on day 0.
Invasion assay.
A Matrigel invasion assay was performed with BD Biocoat Matrigel chambers (BD Bioscience). A layer of dried Matrigel insertions was rehydrated with DMEM for 2 h at 37°C. Aliquots (0.5 ml) of DMEM containing 10% FBS were added to each of the lower chambers of the 24-well Matrigel invasion chambers, and 1 × 105 cells (MDA-MB-468 and MDA-MB-231 cells, each cell line processed in duplicate) diluted in 0.5-ml aliquots of DMEM containing 0.5% FBS were then added to each of the upper chambers. After incubation for 22 h, cells were fixed by the use of ice-cold methanol, stained using Giemsa, and counted using light microscopy.
Statistical methods.
Statistical significance was determined by Student's t test using statcel2 software.
RESULTS
PKP2 interacts with EGFR.
In efforts to gain mechanistic insight into EGFR signaling, we utilized a mass spectrometry database (EMBL EBI-702413) to identify proteins that coimmunoprecipitated with EGFR. We found that a cytoplasmic protein, PKP2, interacted with EGFR. Several clinical studies have indicated that PKP2 expression is abnormally high in various cancers and that PKP2 is a potential biomarker for cancer diagnosis (14, 18–21). However, the molecular basis for PKP2 expression in cancer is unclear. We therefore examined the potential role of PKP2 in mediating EGFR signaling and tumor growth.
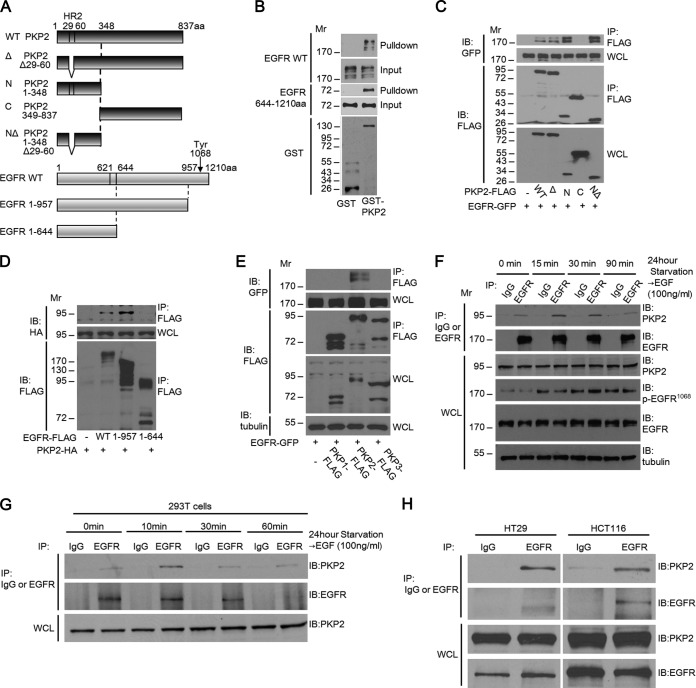
We first verified the interaction between PKP2 and EGFR. A schematic diagram of PKP2, EGFR, and their mutants used in this report is shown in Fig. 1A. Pulldown analysis revealed that GST-PKP2 purified from Escherichia coli interacted with both full-length EGFR-FLAG and the EGFR cytoplasmic domain (aa 644 to 1210)-FLAG purified from 293T cells (Fig. 1B). In order to determine the regions involved in this interaction, truncation and deletion mutants of PKP2 and EGFR were used for coimmunoprecipitation studies. Deletion of the (N-terminal) head domain of PKP2 abolished binding, indicating that it is essential for this interaction, and the homology region of PKP family members (HR2 motif; aa 29 to 60) was not required for this interaction (Fig. 1A and C). In addition, the cytoplasmic (C-terminal; aa 644 to 957) domain of EGFR is sufficient for its interaction with PKP2 (Fig. 1A and D). Interestingly, among the plakophilin family members, only PKP2 specifically associated with EGFR (Fig. 1E). To test the association between PKP2 and EGFR endogenously, coimmunoprecipitation studies were done in A431, 293T, HT29, and HCT116 cells (Fig. 1F, G, and H). A ligand-independent interaction between endogenous PKP2 and EGFR was also detected in these cells.
FIG 1.
PKP2 interacts with EGFR. (A) Schematic drawings of PKP2, EGFR, and the derivatives used in this work. A capital delta indicates an HR2 motif (aa 29 to 60) deletion. The location of EGFR Tyr1068 is indicated. Since the first 24-aa sequence of EGFR is a signal peptide sequence, the actual amino acid sequence of Tyr1068 is 1,092 aa. C, C terminus; N, N terminus; WT, wild type. (B) PKP2 directly interacted with the cytoplasmic region of EGFR. FLAG-tagged full-length EGFR or the cytoplasmic domain (aa 644 to 1210) of EGFR expressed in 293T cells was purified using immunoprecipitation and tested for binding to bacterially synthesized GST or GST-PKP2 on GST binding columns. Bound proteins were analyzed using Western blotting. (C) The N terminus of PKP2 is required for EGFR-PKP2 interaction. 293T cells were cotransfected with plasmids encoding EGFR-GFP and PKP2-FLAG or its mutants. Cell lysates were immunoprecipitated (IP) with anti-FLAG antibody and immunoblotted (IB) with GFP or FLAG. Whole-cell lysates (WCL) were also immunoblotted with the indicated antibodies. (D) The EGFR cytoplasmic domain interacted with PKP2 in 293T cells. 293T cells were cotransfected with plasmids encoding PKP2-HA and EGFR-FLAG or its mutants. Cell lysates were immunoprecipitated with anti-FLAG antibody and immunoblotted with HA or FLAG. Whole-cell lysates were also immunoblotted with HA. (E) PKP2 specifically interacts with EGFR among the members of the PKP family. 293T cells were cotransfected with plasmids encoding EGFR-GFP and PKP1, PKP2, or PKP3-FLAG. Cell lysates were immunoprecipitated with anti-FLAG antibody and immunoblotted with GFP or FLAG. Whole-cell lysates were also immunoblotted using the indicated antibodies. (F) Interaction of endogenous PKP2 and endogenous EGFR in A431 cells. A431 cells that had been serum starved (0.5% FBS) for 24 h were treated with EGF (100 ng/ml) at the indicated times, followed by immunoprecipitation with IgG or EGFR and immunoblotting with PKP2 (top rows) or EGFR (bottom rows) antibodies. Whole-cell lysates were also immunoblotted with the indicated antibodies. (G) 293T cells that had been serum starved (0.5% FBS) for 24 h were treated with EGF (100 ng/ml) at the indicated times, followed by immunoprecipitation with IgG or EGFR and immunoblotting with PKP2 (top rows) or EGFR (bottom rows) antibodies. Whole-cell lysates were also immunoblotted with the indicated antibodies. (H) Interaction of endogenous PKP2 and endogenous EGFR in HT29 (left column) and HCT116 (right column) cells. HT29 or HCT116 cells that had been serum starved (0.5% FBS) for 24 h were immunoprecipitated with IgG or EGFR and immunoblotted with PKP2 (top rows) or EGFR (bottom rows) antibodies. Whole-cell lysates were also immunoblotted with the indicated antibodies. Molecular weights are in thousands.
Our data demonstrate that PKP2 associates with EGFR endogenously and is the only member of the plakophilin family to do so.
PKP2 expression enhances EGFR phosphorylation.
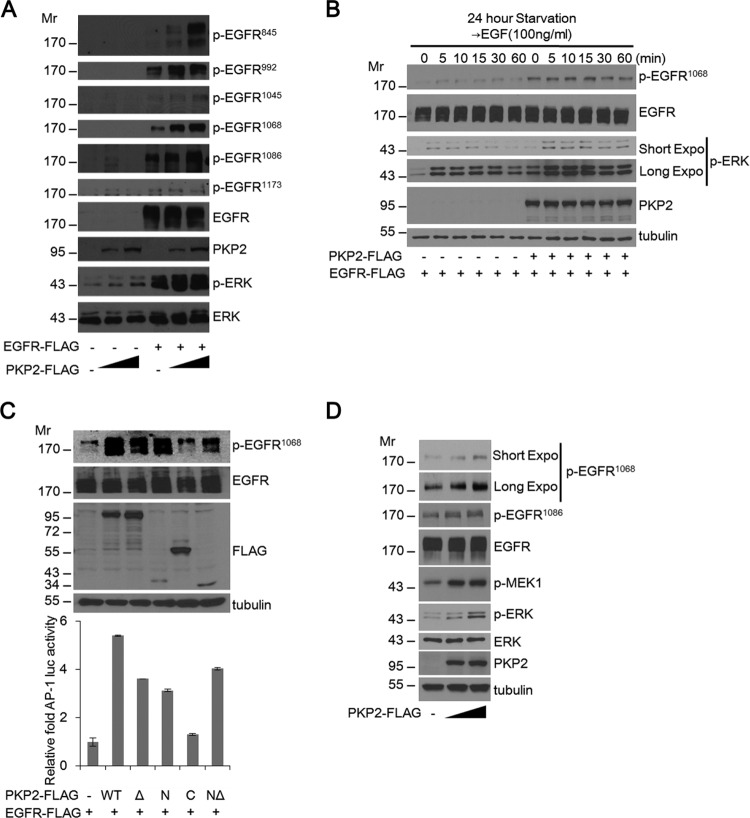
To test whether PKP2 can regulate EGFR signaling, PKP2 was coexpressed with EGFR. Interestingly, levels of phosphorylated EGFR were substantially enhanced by the increasing amounts of PKP2 under serum starvation conditions (Fig. 2A). Furthermore, the level of phosphorylated EGFR (Tyr1068) after EGF treatment in PKP2-expressing 293T cells was enhanced compared to the level in mock-transfected cells (Fig. 2B). Through the use of PKP2 deletion mutants, it was observed that the HR2 motif deleted the N terminus (NΔ) and not the C terminus of PKP2, which corresponds to the fact that the EGFR-interacting region (Fig. 1A and C) could enhance both the phosphorylation of EGFR and the activation of the downstream target AP-1 promoter-luciferase activity (Fig. 2C). Furthermore, ectopic expression of PKP2 increased phosphorylation of endogenous EGFR, MEK1, and ERK dose dependently (Fig. 2D).
FIG 2.
PKP2 expression enhances EGFR phosphorylation. (A) Serum-starved (1% FBS for 24 h) 293T cells were cotransfected with plasmids encoding empty vector or EGFR-FLAG and plasmid (0, 1, and 2 μg) expressing PKP2-FLAG as indicated. At 24 h after transfection, the cell lysates were then subjected to Western blotting using the indicated antibodies. (B) Expression of PKP2 increased EGFR phosphorylation and was enhanced after EGF stimulation in 293T cells. Mock-transfected or PKP2-expressing 293T cells were starved (1% FBS for 24 h), and EGF (100 ng/ml) was added for the indicated times. The cell lysates were then immunoblotted with the indicated antibodies. Expo, exposure. (C) N-terminal PKP2 expression enhanced EGFR phosphorylation and AP-1 luciferase activity. Serum-starved (1% FBS for 24 h) 293T cells were cotransfected with plasmids encoding AP-1-luc, EGFR-FLAG, and PKP2-FLAG and its mutants as indicated. At 24 h after transfection, AP-1 luciferase activities were analyzed. The protein levels of tubulin, EGFR, p-EGFR1068, and PKP2 and its mutants in the lysates are also shown. (D) PKP2 enhanced endogenous EGFR phosphorylation and increased downstream signal. Serum-starved (0.5% FBS for 24 h) A431 cells were transfected with plasmids (0 to 5 μg) encoding PKP2-FLAG. At 24 h after transfection, the cell lysates were examined using the indicated antibodies. Molecular weights are in thousands.
PKP2 is the only member of the plakophilin family capable of specifically activating EGFR signaling.
To examine if other plakophilin family proteins can regulate EGFR function, we coexpressed EGFR and PKP family proteins. PKP2 expression readily enhanced EGFR phosphorylation; however, the other two PKP family proteins, PKP1 and PKP3, had no effect on EGFR phosphorylation (Fig. 3A), which agrees with their lack of interaction with EGFR (Fig. 1E). Furthermore, PKP2 expression increased the level of ERK phosphorylation dose dependently, while neither PKP1 nor PKP3 had an effect on the phosphorylation of ERK (Fig. 3B). Consistent with the PKP2 levels, AP-1 and NF-κB reporter-luciferase activity was increased upon PKP2 expression (Fig. 3C, top and bottom, respectively). However, PKP3 increased NF-κB activity (Fig. 3C, bottom), suggesting that PKP3 has a novel effect on NF-κB that is independent of EGFR phosphorylation and activation.
FIG 3.
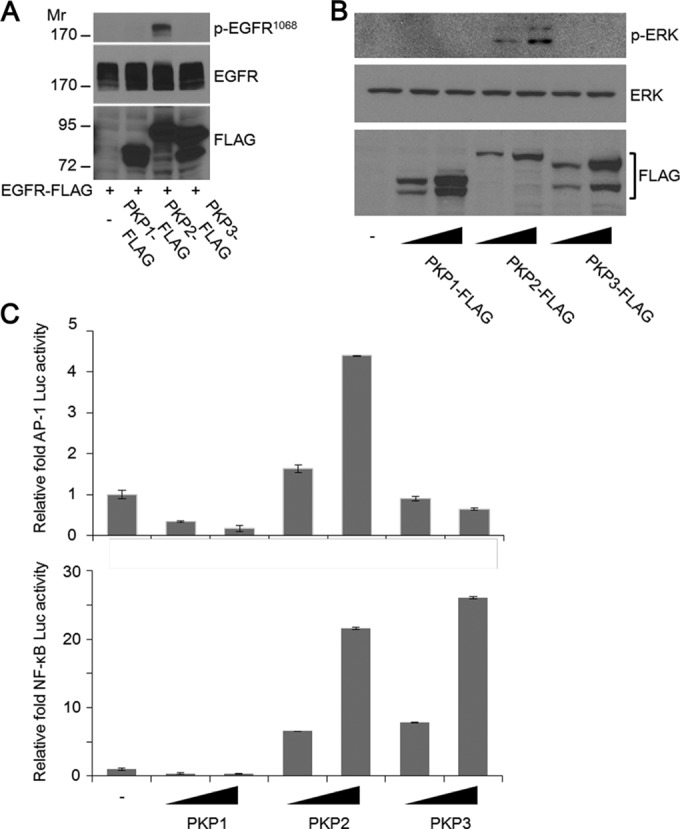
PKP2 is the only member of the plakophilin family capable of specifically activating EGFR signaling. (A) PKP2 specifically enhanced EGFR phosphorylation. Serum-starved (1% FBS for 24 h) 293T cells were cotransfected with plasmids encoding EGFR-FLAG and PKP1, PKP2, or PKP3-FLAG. At 24 h after transfection, the cell lysates were immunoblotted with the indicated antibodies. Molecular weights are in thousands. (B) At 24 h, serum-starved (1% FBS-containing DMEM) 293T cells were transfected with plasmids encoding increasing amounts of PKP1, PKP2, or PKP3-FLAG as indicated. At 24 h after transfection, the cell lysates were immunoblotted with the indicated antibodies. (C) 293T cells were transfected with plasmids encoding AP-1-Luc (upper panel) or NF-κB-Luc (lower panel) (50 ng), together with plasmids expressing PKP1, PKP2, or PKP3 (100 and 200 ng), as indicated. At 24 h after transfection, the luciferase activities were examined. Error bars represent the standard deviations (SD) of the means of the results of triplicate experiments.
Thus, these results indicate that PKP2 specifically interacts with EGFR and activates EGFR signaling.
PKP2 activates the EGFR signaling pathway via enhancement of EGFR dimerization followed by autophosphorylation.
Because EGFR autophosphorylation requires EGFR dimerization, we analyzed whether PKP2 expression affects the dimerization of EGFR. First, we found that expression of PKP2 clearly enhanced the EGFR-EGFR interaction (Fig. 4A and B) and markedly increased the dimer/monomer ratio of EGFR under starvation conditions (Fig. 4C). Interestingly, PKP2 also interacted with HER2 (ErbB2) and strengthened the level of interaction between EGFR and HER2, suggesting that PKP2 is able to facilitate both EGFR-EGFR and EGFR-HER2 dimerization (Fig. 4D and E).
FIG 4.
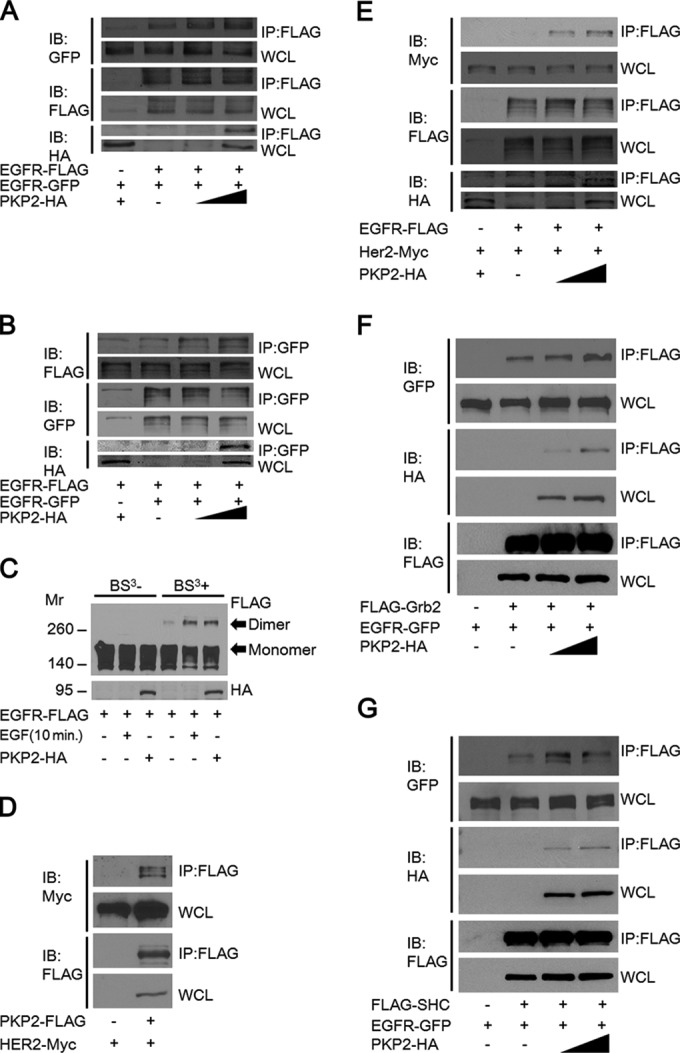
EGFR dimerization is increased by PKP2 overexpression. (A and B) 293T cells were cotransfected with EGFR-GFP, EGFR-FLAG, and increasing amounts of PKP2-HA as indicated. Cell lysates were immunoprecipitated with anti-FLAG antibody (A) or anti-GFP antibody (B) and immunoblotted with antibodies as indicated. Whole-cell lysates were also immunoblotted with the indicated antibodies. (C) The level of EGFR dimerization was increased by PKP2 expression. Serum-starved (1% FBS for 24 h) 293T cells were cotransfected with EGFR-FLAG and PKP2-HA as indicated. As a control, EGF (10 ng/ml) was added for 10 min. Cell lysates with or without BS3 cross-linking were analyzed by Western blotting. Molecular weights are in thousands. (D) PKP2 interacted with HER2. 293T cells were cotransfected with HER2-Myc and PKP2-FLAG. Cell lysates were immunoprecipitated with anti-FLAG antibody and immunoblotted with FLAG or Myc antibodies. Whole-cell lysates were also immunoblotted with FLAG or Myc antibodies. (E) The level of EGFR-HER2 interaction was increased by PKP2 overexpression. 293T cells were cotransfected with EGFR-FLAG, HER2-Myc, and increasing amounts of PKP2-HA as indicated. Cell lysates were immunoprecipitated with anti-FLAG antibody and immunoblotted with antibodies as indicated. Whole-cell lysates were also immunoblotted with the indicated antibodies. (F) The level of EGFR-Grb2 interaction was increased by PKP2 overexpression. 293T cells were cotransfected with EGFR-GFP, FLAG-Grb2, and increasing amounts of PKP2-HA as indicated. Cell lysates were immunoprecipitated with anti-FLAG antibody and immunoblotted with antibodies as indicated. Whole-cell lysates were also immunoblotted with the indicated antibodies. (G) The level of EGFR-SHC interaction was increased by PKP2 overexpression. 293T cells were cotransfected with EGFR-GFP, FLAG-SHC, and increasing amounts of PKP2-HA as indicated. Cell lysates were immunoprecipitated with anti-FLAG antibody and immunoblotted with antibodies as indicated. Whole-cell lysates were also immunoblotted with antibodies as indicated.
Phosphotyrosines on EGFR recruit cytosolic signaling molecules such as SHC and Grb2 and trigger a series of intracellular pathways, culminating in cellular proliferation, migration, invasion, and tumorigenicity (7–10). To examine whether PKP2 expression is involved in the recruitment of these molecules, we analyzed the interaction between EGFR and SHC or Grb2 in the presence of increasing amounts of PKP2 expression. Their levels of interaction increased dose dependently with PKP2 expression (Fig. 4F and G).
From this data, we found that PKP2 enhances EGFR dimerization and its subsequent autophosphorylation, as well as downstream target activation.
PKP2 knockdown results in a substantial reduction in EGF-induced EGFR activation.
To examine the physiological roles of PKP2 in EGFR-mediated signaling, we assessed phosphorylation of EGFR and its downstream signaling molecules in PKP2 knockdown cells. Upon siRNA and shRNA transfection, PKP2 knockdown efficiency was confirmed at both the mRNA and protein levels (Fig. 5A). PKP2 reduction by siRNA and two different targeting shRNAs substantially impaired EGF-induced phosphorylation of EGFR in A431 cells (Fig. 5B and C, top rows). Quantification of the ratio of phosphorylated EGFR (Y1068) to total EGFR clearly correlated with the expression level of PKP2 (Fig. 5D). Similar results were also observed in HCT116 cells (Fig. 5E). Moreover, in accordance with the levels of phosphorylated EGFR in control and sh-PKP2-4-expressing cells, the levels of interactions between EGFR and Grb2, GEP100, and SHP-1 were substantially reduced in PKP2 knockdown MDA-MB-468 cells (Fig. 5F). Similar results were also observed in A431 cells (Fig. 5G). In addition, increased expression of PKP2 in sh-PKP2-4-expressing A431 cells restored the interactions between EGFR and Grb2, GEP100, and SHP-1 (Fig. 5H). Furthermore, the levels of phosphorylated EGFR in sh-PKP2-4-expressing A431 cells were restored by expression of sh-PKP2-4-resistant PKP2 (PKP2shR) (Fig. 5I). Importantly, EGFR-EGFR interaction was substantially reduced in sh-PKP2-4-expressing A431 cells compared with short hairpin control-expressing A431 cells, further suggesting that PKP2 has the ability to facilitate EGFR dimerization (Fig. 5J).
FIG 5.
PKP2 knockdown impairs EGF-induced responses. (A) Efficiency of PKP2 knockdown using siRNA and shRNA in 293T cells. The mRNA levels of PKP2 were visualized using RT-PCR. To assess the protein levels of PKP2 to test the efficiency of siRNAs and shRNAs, plasmids encoding PKP1 and PKP2 with siRNAs or shRNAs were transfected to 293T cells. At 48 h after transfection, the cell lysates were subjected to Western blot analysis. si-Control, control siRNA; sh-Control, control shRNA. (B) A431 cells were treated with control siRNA or si-PKP2-2 (twice, at 0 and 48 h). At 72 h after the first siRNA transfection, the media were changed to 0.5% FBS-containing DMEM. EGF was added at 98 h after the first siRNA transfection and was harvested at the indicated times. (C) A431 cells were transfected with plasmids encoding control shRNA, shPKP2-2, or shPKP2-4, followed by selection by puromycin. Selected cells were serum starved with 0.5% FBS-containing DMEM for 24 h, and EGF was added and harvested at the indicated times. (D) The ratios of tyrosine 1068-phosphorylated EGFR to total EGFR presented in panel C were quantified. (E) HCT116 cells were transfected with plasmids encoding control shRNA or shPKP2-4, followed by selection with puromycin. Selected cells were serum starved in DMEM with 1% FBS for 24 h, and then EGF was added and harvested at the indicated times. (F) MDA-MB-468 cells were transfected with plasmids encoding control shRNA or shPKP2-4, followed by selection with puromycin. Selected cells were serum starved with 0.5% FBS DMEM for 24 h, and EGF was added and harvested at the indicated times. Cell lysates were immunoprecipitated with anti-EGFR antibody and immunoblotted with antibodies as indicated. (G) A431 cells were transfected with plasmids encoding control shRNA or sh-PKP2-4, followed by selection with puromycin. Selected cells were serum starved with 0.5% FBS DMEM for 24 h, and EGF was added and harvested at the indicated times. Cell lysates were immunoprecipitated with anti-EGFR antibody and immunoblotted with antibodies as indicated. Whole-cell lysates were also immunoblotted as indicated. (H) Expression of PKP2 increased the interactions between EGFR and adopter molecules. Sh-PKP2-4-expressing A431 cells were transfected with plasmids encoding empty PKP2 or increasing amounts of Sh-PKP2-4-resistant PKP2 as indicated. Cell lysates were immunoprecipitated with IgG or anti-EGFR antibody and immunoblotted with antibodies as indicated. Whole-cell lysates were also immunoblotted as indicated. (I) The sh-PKP2-4-expressing A431 cells were transfected with plasmids encoding empty vector (EV) or sh-PKP2-4-resistant PKP2 (PKP2shR). At 24 h after starvation (0.5% FBS-containing DMEM), EGF was added and harvested at the indicated times. (J) Suppression of PKP2 expression reduces the EGFR dimerization rate. The short hairpin control or sh-PKP2-4-expressing A431 cells were transiently transfected with EGFR-GFP and empty vector or EGFR-FLAG as indicated. At 24 h after transfection, cell lysates were immunoprecipitated with FLAG and immunoblotted with antibodies as indicated. Whole-cell lysates were also immunoblotted as indicated. Molecular weights are in thousands.
PKP2 knockdown suppresses cell proliferation and migration.
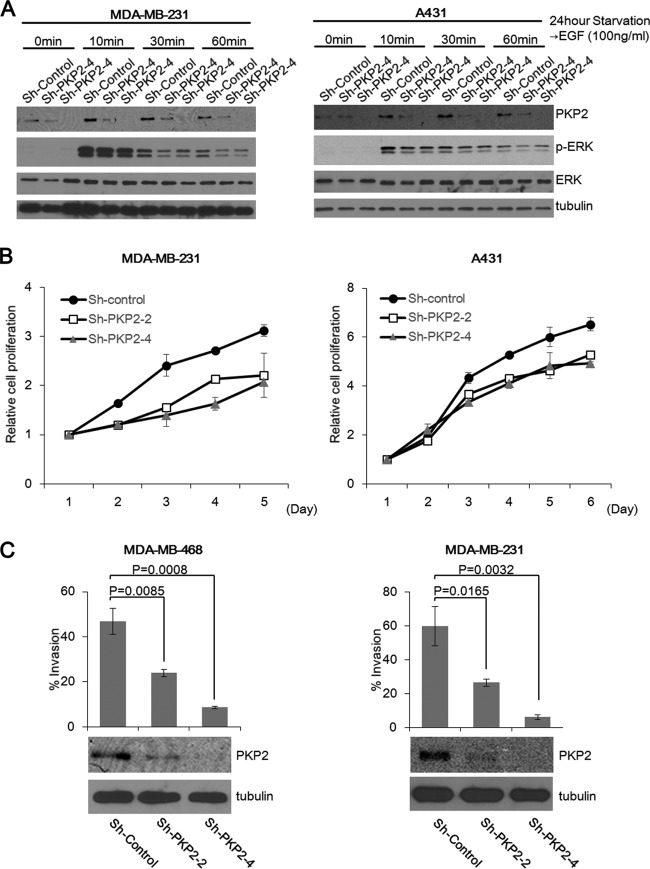
Because EGFR autophosphorylation-mediated signal activation is involved in cell proliferation and migration, we conducted experiments to examine the role of PKP2 in these functions. PKP2 knockdown MDA-MB-231 and A431 cells showed reduced phosphorylation of ERK and decreased proliferation activity (Fig. 6A and B, respectively). In addition, a clear decrease in cell invasion was observed in PKP2 knockdown MDA-MB-468 and MDA-MB-231 cells (Fig. 6C, left and right, respectively).
FIG 6.
Suppression of PKP2 expression reduces cell proliferation and invasion. (A) The short hairpin control (sh-Control)-, sh-PKP2-2-, or sh-PKP2-4-expressing MDA-MB-231 cells or A431 cells (left panel or right panel, respectively) were serum starved with 0.5% FBS-containing DMEM for 24 h. At the indicated times after EGF (100 ng/ml) treatment, cells were lysed and analyzed using the indicated antibodies. (B) The cell growth of short hairpin control (sh-Control)-, sh-PKP2-2-, or sh-PKP2-4-expressing MDA-MB-231 or MDA-MB-468 was monitored by an MTS assay (left panel or right panel, respectively). (C) Knockdown of PKP2 substantially impaired cell invasion. MDA-MB-468 cells (left) or MDA-MB-231 cells (right) were transfected with the short hairpin control (sh-Control), sh-PKP2-2, or sh-PKP2-4 and selected using puromycin. The selected cells were analyzed using the Matrigel invasion assay. The protein levels of PKP2 are also shown (bottom).
Thus, we have established that reduction of PKP2 clearly decreases EGFR signaling, resulting in suppression of cell proliferation and migration.
PKP2 functions directly through EGFR, and its effects are abolished in EGFR null cells.
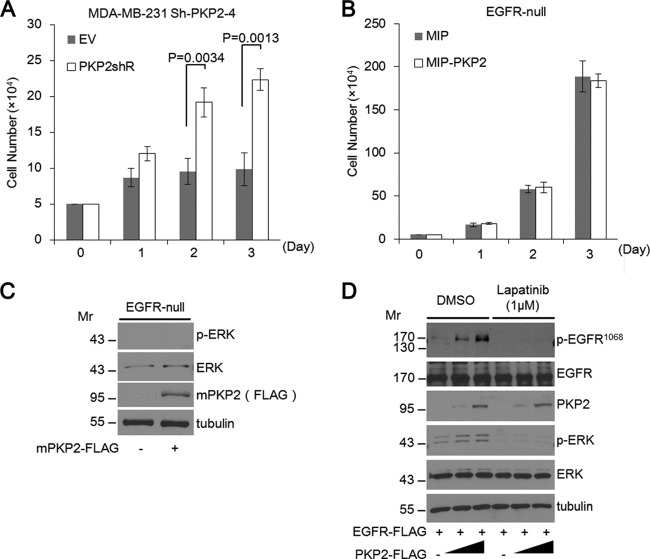
Consistent with our results reported above, restoring PKP2 expression in PKP2 Sh-PKP2-4 knockdown MDA-MB-231 cells resulted in enhanced cell proliferation (Fig. 7A). To investigate whether PKP2 can affect cell growth or EGFR downstream signal activation in EGFR null cells, we stably and transiently expressed PKP2 in 32D murine hematopoietic cells, which do not express EGFR and are unresponsive to EGFR ligands (22). As a result, 32D cells expressing PKP2 displayed cell growth profiles similar to those of control cells (Fig. 7B). Moreover, PKP2 expression in 32D cells did not affect ERK phosphorylation (Fig. 7C).
FIG 7.
PKP2 functions directly through EGFR, and its effects are abolished in EGFR null cells. (A) The sh-PKP2-4-expressing MDA-MB-231 cells were transfected with plasmids encoding empty vector (EV) or sh-PKP2-4-resistant PKP2 (PKP2shR). After selection, these cells were monitored by trypan blue counting. (B) 32D cells stably expressing MIP or MIP-mouse PKP2 were monitored by trypan blue counting. (C) Mouse PKP2 was transiently expressed in 32D cells as indicated. Twenty-four hours after transfection, cell lysates were immunoblotted with the indicated antibodies. (D) Lapatinib treatment completely impaired the enhancement of PKP2-mediated EGFR phosphorylation. Serum-starved (1% FBS for 24 h) 293T cells were cotransfected with plasmids encoding EGFR-FLAG and PKP2-FLAG as indicated. The cells were treated with DMSO or lapatinib (final 1 μM) for 24 h. At 24 h after transfection, the cell lysates were subjected to Western blotting using the indicated antibodies. Molecular weights are in thousands.
Finally, to test whether PKP2 is capable of enhancing EGFR signal activation through alternative EGFR-independent mechanisms, we examined the level of EGFR phosphorylation with increasing amounts of PKP2 in cells treated with the EGFR/HER2 kinase inhibitor lapatinib under starvation conditions. The levels of phosphorylated EGFR, as well as ERK, were increased in PKP2-expressing cells treated with dimethyl sulfoxide (DMSO). Conversely, EGFR and ERK phosphorylation was substantially reduced in PKP2-expressing cells treated with lapatinib. These results clearly indicate that PKP2-mediated activation of EGFR signaling functions directly through EGFR and requires EGFR kinase activity (Fig. 7D).
PKP2 regulates breast cancer development in vivo.
It was previously established that the reduction of EGFR expression or EGFR phosphorylation caused by either EGFR knockdown or EGFR inhibitors results in reduced cell proliferation and migration in MDA-MB-231 cells (23, 24). Furthermore, a recent report showed that EGFR knockdown in MDA-MB-231 cells reduced tumor growth in the mammary fat pad (25).
To corroborate that PKP2 is involved in EGFR signaling-mediated cancer development, we orthotopically injected single clones of MDA-MB-231 cells expressing control shRNA or one of two different PKP2 shRNAs into the fourth (abdominal) mammary fat pads of immune-deficient NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) mice. Mice were monitored for tumor growth weekly and were sacrificed at 10 weeks after injection. MDA-MB-231 tumor tissues were subjected to Western blotting to confirm protein levels of PKP2. As expected, short hairpin control (sh-Control) tumors still expressed PKP2. Tumor cells expressing sh-PKP2-2 expressed half the amount of PKP2 expressed by the short hairpin control, and shPKP2-4-expressing tumors had nearly no expression of PKP2 (Fig. 8A). The results indicate that sh-PKP2-2-expressing cells reduced the growth rate of tumors by around 40% compared to the control and that Sh-PKP2-4-expressing cells were severely impaired in their tumor growth capacity in NSG mice (Fig. 8B and C). In direct correlation with their effect on primary tumor growth, sh-PKP2-2- and sh-PKP2-4-expressing cells formed 65% and 98% fewer metastatic nodules than short hairpin control cells, respectively (Fig. 8D and E).
FIG 8.
PKP2 regulates breast cancer development in a mouse model. (A) Pieces of tumors were homogenized and subjected to Western blotting and analyzed using PKP2 and tubulin antibodies. Molecular weights are in thousands. (B) All tumors analyzed in this assay are shown. (n = 5; short hairpin control [sh-Control], sh-PKP2-2, and sh-PKP2-4). (C) Primary tumor growth was measured upon orthotopic injection of MDA-MB-231 cells with expression of the short hairpin control (sh-Control), sh-PKP2-2, or sh-PKP2-4. The experiment was terminated 10 weeks after injection (n = 5 per group per time point). Each time point shows the means ± standard errors of the means (SEM) of the results. (D) At 10 weeks after orthotopic implantation of MDA-MB-231 cells with or without PKP2 knockdown, lungs were stained by H&E. Representative H&E stains of lungs are shown. Arrows indicate metastatic foci. (E) Quantitative results of the lung metastasis analysis described for panel D.
Taking the findings together, we conclude that PKP2 plays a novel and important role in facilitating breast cancer cell growth and metastasis.
DISCUSSION
In this report, we are the first to identify PKP2 as a novel regulator of both EGF-independent and -dependent activation of EGFR. Notably, we found that PKP2 is the only member of the plakophilin family that can stimulate EGFR phosphorylation. This stimulation by PKP2 is also seen in the absence of EGF (0.5% to 1% serum), indicating that misregulation of PKP2 expression, as is often observed in cancer, can have functional consequences even in the absence of ligand. We showed that PKP2-induced enhancement of EGFR and ERK phosphorylation was completely abolished in the presence of lapatinib, suggesting that PKP2 can directly activate EGFR signaling in EGFR-expressing cancer cells. We also confirmed that expression of PKP2 did not activate downstream EGFR signaling in EGFR null cells. We demonstrated that knockdown of PKP2 significantly impaired EGF-induced EGFR autophosphorylation and recruitment of adaptor molecules to EGFR. Furthermore, diminished expression of PKP2 in MDA-MB-231 breast cancer cells reduced their proliferation and metastatic potential and ultimately decreased tumor development in vivo.
Previous work by others has identified other cytoplasmic activators of EGF receptor tyrosine kinases, such as YES (Yamaguchi sarcoma virus oncogene) and cytohesins (26–28). However, their roles in EGFR activation are limited. YES-mediated EGFR phosphorylation occurs only on the endosome, mildly increases phosphorylation on limited sites, and requires EGF stimulation and pIgA-pIgR transcytosis (26, 27). Furthermore, cytohesins do not influence receptor dimerization but function as conformational activators of dimerized receptors after EGF stimulation (26, 27). In contrast, PKP2 has a broader effect on EGFR activation and can enhance receptor dimerization and autophosphorylation even in the absence of ligand stimulation. The function of PKP2 in EGFR signaling does not overlap that of YES and cytohesins, suggesting that PKP2 functions as a novel “cytoplasmic EGF.” The issue of whether PKP2 can dimerize mutant EGFRs which cannot dimerize should be addressed in the future.
Our findings show that the N-terminal region of PKP2 directly interacts with the cytoplasmic domain of EGFR, resulting in an increase in EGFR autophosphorylation and EGFR-mediated signal transduction. Previous studies indicate that the N-terminal region of PKP2 is sufficient for its localization to the membrane near areas of cell-to-cell attachment (12, 13). This may suggest that PKP2 is involved in EGFR signal activation at regions of cell attachment. Desmosome proteins, including PKP2, are intercellular key junctions that confer strong cell-cell adhesion (29, 30). They are located at the cell membrane, where they act as anchors for intermediate filaments (29). Lack of PKP2 dissociates desmoplakin from the cardiac adhering desmosomal junctions and results in disruption of heart morphogenesis (15–17), suggesting that PKP2 plays a major role in cell-to-cell junctions in cardiomyocytes. Desmosome proteins have been reported to both promote and inhibit cancer development (31). Interestingly, PKP2 was shown to interact with β-catenin and modestly enhance LEF/TCF-mediated transactivation (13); however, the mechanism is not fully understood. Additionally, it has been reported that dimerized EGFR phosphorylates β-catenin and activates LEF/TCF promoters (32, 33). Our current findings may identify a novel mechanism for PKP2 in the activation of β-catenin which is mediated by PKP2-induced EGFR dimerization and activation and results in activation of the β-catenin pathway to promote cell proliferation and metastasis.
Although we demonstrate that PKP2 promotes the dimerization of EGFR and consequent activation of EGFR signaling, it remains unclear how PKP2 affects the conformational change of EGFR during activation and termination of PKP2-mediated EGFR signaling. Interestingly, upon EGF treatment, the interaction between endogenous PKP2 and EGFR increased within 30 min but decreased after 90 min, which may be part of the PKP2-mediated regulation of EGFR signaling. Recently, PKP2 was identified as one of the phosphorylated targets after EGF treatment (34). Therefore, it is possible that phosphorylation of PKP2 contributes to the regulation of PKP2-EGFR interaction. These issues should be addressed in future studies.
EGFR activation is well known to be involved in cancer development and progression. Although numerous EGFR inhibitors have been developed and are currently being used in the clinic, many patients eventually develop resistance (35, 36). Therefore, it is critical to explore alternative mechanisms of inhibiting receptor activation and downstream signaling. PKP2 expression is increased in many cancers, such as breast, prostate, colorectal, pancreatic, oropharyngeal, and bladder cancers (14, 18–21). Increased PKP2 expression also correlates with the elevated malignancy of many cancers. In addition, as shown by the Oncomine (Compendia Bioscience, Ann Arbor, MI) analysis, the PKP2 expression levels in cancers of breast (37), lung (38), bladder (39), and thyroid gland (40) were significantly higher than the PKP2 expression levels in the normal tissues of these organs. Our studies demonstrated that PKP2 enhances EGFR-mediated signaling by facilitating the dimerization of EGFR and activating downstream signaling pathways, as well as promoting cancer cell proliferation and tumor metastasis. The newly identified role of PKP2 in EGFR activation described in this report identifies a clear and novel molecular mechanism to explain the clinical correlations with PKP2. Development of a small molecule that disrupts protein-protein interactions by binding with high affinity to “hot spots,” which contribute most to the protein-protein interaction, could be applied to preventing the association between the N terminus of PKP2 and the cytoplasmic domain of EGFR (41). Altogether, our findings demonstrate a novel role of PKP2 in EGFR signaling and suggest PKP2 as a potential target for therapeutic intervention.
ACKNOWLEDGMENTS
We thank Paul S. Mischel and Frank Furnari (Ludwig Institute for Cancer Research, UCSD) and Gen-Sheng Feng (Department of Pathology, UCSD) for critical reading of the manuscript. We thank Kunitada Shimotohno (National Center for Global Health and Medicine, Japan) for plasmid vectors (C-terminally tagged pCAG vector, GST [6p-1], and C-terminally Myc-tagged pcDNA3.1 vector), Jo Minji (Steven Gonias laboratory, UCSD) for MDA-MB-468 cells, and David Cheresh (Moores Cancer Center, UCSD) for MDA-MB-231 cells. We thank all members of the D.-E.Z. laboratory and Keun Il Kim (Sookmyung Women's University, South Korea) for helpful discussion. We also thank Chuyi Cheng and Yue Zhang (Division of Biological Sciences, UCSD) for their technical assistance in this study.
This work was supported by funding from the U.S. National Institutes of Health (R01CA177305 and R01HL091549). Kei-ichiro Arimoto is a Japan Society for the Promotion of Science (JSPS) Postdoctoral Fellow for Research Abroad.
Footnotes
Published ahead of print 11 August 2014
REFERENCES
- 1.Blume-Jensen P, Hunter T. 2001. Oncogenic kinase signalling. Nature 411:355–365. 10.1038/35077225 [DOI] [PubMed] [Google Scholar]
- 2.Bogdan S, Klambt C. 2001. Epidermal growth factor receptor signaling. Curr. Biol. 11:R292–R295. 10.1016/S0960-9822(01)00167-1 [DOI] [PubMed] [Google Scholar]
- 3.Hynes NE, Lane HA. 2005. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat. Rev. Cancer 5:341–354. 10.1038/nrc1609 [DOI] [PubMed] [Google Scholar]
- 4.Hynes NE, MacDonald G. 2009. ErbB receptors and signaling pathways in cancer. Curr. Opin. Cell Biol. 21:177–184. 10.1016/j.ceb.2008.12.010 [DOI] [PubMed] [Google Scholar]
- 5.Yarden Y, Pines G. 2012. The ERBB network: at last, cancer therapy meets systems biology. Nat. Rev. Cancer 12:553–563. 10.1038/nrc3309 [DOI] [PubMed] [Google Scholar]
- 6.Lemmon MA, Schlessinger J. 2010. Cell signaling by receptor tyrosine kinases. Cell 141:1117–1134. 10.1016/j.cell.2010.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Batzer AG, Rotin D, Urena JM, Skolnik EY, Schlessinger J. 1994. Hierarchy of binding sites for Grb2 and Shc on the epidermal growth factor receptor. Mol. Cell. Biol. 14:5192–5201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu JM, Chen CT, Chou CK, Kuo HP, Li LY, Lin CY, Lee HJ, Wang YN, Liu M, Liao HW, Shi B, Lai CC, Bedford MT, Tsai CH, Hung MC. 2011. Crosstalk between Arg 1175 methylation and Tyr 1173 phosphorylation negatively modulates EGFR-mediated ERK activation. Nat. Cell Biol. 13:174–181. 10.1038/ncb2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morishige M, Hashimoto S, Ogawa E, Toda Y, Kotani H, Hirose M, Wei S, Hashimoto A, Yamada A, Yano H, Mazaki Y, Kodama H, Nio Y, Manabe T, Wada H, Kobayashi H, Sabe H. 2008. GEP100 links epidermal growth factor receptor signalling to Arf6 activation to induce breast cancer invasion. Nat. Cell Biol. 10:85–92. 10.1038/ncb1672 [DOI] [PubMed] [Google Scholar]
- 10.Rozakis-Adcock M, McGlade J, Mbamalu G, Pelicci G, Daly R, Li W, Batzer A, Thomas S, Brugge J, Pelicci PG, Schlessinger J, Pawson T. 1992. Association of the Shc and Grb2/Sem5 SH2-containing proteins is implicated in activation of the Ras pathway by tyrosine kinases. Nature 360:689–692. 10.1038/360689a0 [DOI] [PubMed] [Google Scholar]
- 11.Chung I, Akita R, Vandlen R, Toomre D, Schlessinger J, Mellman I. 2010. Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature 464:783–787. 10.1038/nature08827 [DOI] [PubMed] [Google Scholar]
- 12.Bass-Zubek AE, Godsel LM, Delmar M, Green KJ. 2009. Plakophilins: multifunctional scaffolds for adhesion and signaling. Curr. Opin. Cell Biol. 21:708–716. 10.1016/j.ceb.2009.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X, Bonne S, Hatzfeld M, van Roy F, Green KJ. 2002. Protein binding and functional characterization of plakophilin 2. Evidence for its diverse roles in desmosomes and beta-catenin signaling. J. Biol. Chem. 277:10512–10522. 10.1074/jbc.M108765200 [DOI] [PubMed] [Google Scholar]
- 14.Mertens C, Kuhn C, Moll R, Schwetlick I, Franke WW. 1999. Desmosomal plakophilin 2 as a differentiation marker in normal and malignant tissues. Differentiation 64:277–290. 10.1046/j.1432-0436.1999.6450277.x [DOI] [PubMed] [Google Scholar]
- 15.Gerull B, Heuser A, Wichter T, Paul M, Basson CT, McDermott DA, Lerman BB, Markowitz SM, Ellinor PT, MacRae CA, Peters S, Grossmann KS, Drenckhahn J, Michely B, Sasse-Klaassen S, Birchmeier W, Dietz R, Breithardt G, Schulze-Bahr E, Thierfelder L. 2004. Mutations in the desmosomal protein plakophilin-2 are common in arrhythmogenic right ventricular cardiomyopathy. Nat. Genet. 36:1162–1164. 10.1038/ng1461 [DOI] [PubMed] [Google Scholar]
- 16.Grossmann KS, Grund C, Huelsken J, Behrend M, Erdmann B, Franke WW, Birchmeier W. 2004. Requirement of plakophilin 2 for heart morphogenesis and cardiac junction formation. J. Cell Biol. 167:149–160. 10.1083/jcb.200402096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacRae CA, Birchmeier W, Thierfelder L. 2006. Arrhythmogenic right ventricular cardiomyopathy: moving toward mechanism. J. Clin. Invest. 116:1825–1828. 10.1172/JCI29174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng AS, Culhane AC, Chan MW, Venkataramu CR, Ehrich M, Nasir A, Rodriguez BA, Liu J, Yan PS, Quackenbush J, Nephew KP, Yeatman TJ, Huang TH. 2008. Epithelial progeny of estrogen-exposed breast progenitor cells display a cancer-like methylome. Cancer Res. 68:1786–1796. 10.1158/0008-5472.CAN-07-5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papagerakis S, Shabana AH, Depondt J, Gehanno P, Forest N. 2003. Immunohistochemical localization of plakophilins (PKP1, PKP2, PKP3, and p0071) in primary oropharyngeal tumors: correlation with clinical parameters. Hum. Pathol. 34:565–572. 10.1016/S0046-8177(03)00174-6 [DOI] [PubMed] [Google Scholar]
- 20.Schwarz J, Ayim A, Schmidt A, Jager S, Koch S, Baumann R, Dunne AA, Moll R. 2006. Differential expression of desmosomal plakophilins in various types of carcinomas: correlation with cell type and differentiation. Hum. Pathol. 37:613–622. 10.1016/j.humpath.2006.01.013 [DOI] [PubMed] [Google Scholar]
- 21.Takahashi H, Nakatsuji H, Takahashi M, Avirmed S, Fukawa T, Takemura M, Fukumori T, Kanayama H. 2012. Up-regulation of plakophilin-2 and down-regulation of plakophilin-3 are correlated with invasiveness in bladder cancer. Urology 79:240.e1–240.e8. 10.1016/j.urology.2011.08.049 [DOI] [PubMed] [Google Scholar]
- 22.Wang LM, Kuo A, Alimandi M, Veri MC, Lee CC, Kapoor V, Ellmore N, Chen XH, Pierce JH. 1998. ErbB2 expression increases the spectrum and potency of ligand-mediated signal transduction through ErbB4. Proc. Natl. Acad. Sci. U. S. A. 95:6809–6814. 10.1073/pnas.95.12.6809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson NG, Ahmad T, Chan K, Dobson R, Bundred NJ. 2001. ZD1839 (Iressa), a novel epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor, potently inhibits the growth of EGFR-positive cancer cell lines with or without erbB2 overexpression. Int. J. Cancer 94:774–782. 10.1002/ijc.1557 [DOI] [PubMed] [Google Scholar]
- 24.Hirsch DS, Shen Y, Wu WJ. 2006. Growth and motility inhibition of breast cancer cells by epidermal growth factor receptor degradation is correlated with inactivation of Cdc42. Cancer Res. 66:3523–3530. 10.1158/0008-5472.CAN-05-1547 [DOI] [PubMed] [Google Scholar]
- 25.Nickerson NK, Mohammad KS, Gilmore JL, Crismore E, Bruzzaniti A, Guise TA, Foley J. 2012. Decreased autocrine EGFR signaling in metastatic breast cancer cells inhibits tumor growth in bone and mammary fat pad. PLoS One 7:e30255. 10.1371/journal.pone.0030255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arteaga CL. 2011. ERBB receptors in cancer: signaling from the inside. Breast Cancer Res. 13:304. 10.1186/bcr2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bill A, Schmitz A, Albertoni B, Song JN, Heukamp LC, Walrafen D, Thorwirth F, Verveer PJ, Zimmer S, Meffert L, Schreiber A, Chatterjee S, Thomas RK, Ullrich RT, Lang T, Famulok M. 2010. Cytohesins are cytoplasmic ErbB receptor activators. Cell 143:201–211. 10.1016/j.cell.2010.09.011 [DOI] [PubMed] [Google Scholar]
- 28.Su T, Bryant DM, Luton F, Verges M, Ulrich SM, Hansen KC, Datta A, Eastburn DJ, Burlingame AL, Shokat KM, Mostov KE. 2010. A kinase cascade leading to Rab11-FIP5 controls transcytosis of the polymeric immunoglobulin receptor. Nat. Cell Biol. 12:1143–1153. 10.1038/ncb2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chidgey M, Dawson C. 2007. Desmosomes: a role in cancer? Br. J. Cancer 96:1783–1787. 10.1038/sj.bjc.6603808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Desai BV, Harmon RM, Green KJ. 2009. Desmosomes at a glance. J. Cell Sci. 122:4401–4407. 10.1242/jcs.037457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dusek RL, Attardi LD. 2011. Desmosomes: new perpetrators in tumour suppression. Nat. Rev. Cancer 11:317–323. 10.1038/nrc3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krejci P, Aklian A, Kaucka M, Sevcikova E, Prochazkova J, Masek JK, Mikolka P, Pospisilova T, Spoustova T, Weis M, Paznekas WA, Wolf JH, Gutkind JS, Wilcox WR, Kozubik A, Jabs EW, Bryja V, Salazar L, Vesela I, Balek L. 2012. Receptor tyrosine kinases activate canonical WNT/beta-catenin signaling via MAP kinase/LRP6 pathway and direct beta-catenin phosphorylation. PLoS One 7:e35826. 10.1371/journal.pone.0035826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee CH, Hung HW, Hung PH, Shieh YS. 2010. Epidermal growth factor receptor regulates beta-catenin location, stability, and transcriptional activity in oral cancer. Mol. Cancer 9:64. 10.1186/1476-4598-9-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heibeck TH, Ding SJ, Opresko LK, Zhao R, Schepmoes AA, Yang F, Tolmachev AV, Monroe ME, Camp DG, II, Smith RD, Wiley HS, Qian WJ. 2009. An extensive survey of tyrosine phosphorylation revealing new sites in human mammary epithelial cells. J. Proteome Res. 8:3852–3861. 10.1021/pr900044c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seshacharyulu P, Ponnusamy MP, Haridas D, Jain M, Ganti AK, Batra SK. 2012. Targeting the EGFR signaling pathway in cancer therapy. Expert Opin. Ther. Targets 16:15–31. 10.1517/14728222.2011.648617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamaguchi H, Chang SS, Hsu JL, Hung MC. 1 April 2013. Signaling cross-talk in the resistance to HER family receptor targeted therapy. Oncogene 10.1038/onc.2013.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, Hallett M, Park M. 2008. Stromal gene expression predicts clinical outcome in breast cancer. Nat. Med. 14:518–527. 10.1038/nm1764 [DOI] [PubMed] [Google Scholar]
- 38.Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens JA, Hoogsteden HC, Grosveld F, Philipsen S. 2010. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One 5:e10312. 10.1371/journal.pone.0010312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, Cordon-Cardo C. 2006. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J. Clin. Oncol. 24:778–789. 10.1200/JCO.2005.03.2375 [DOI] [PubMed] [Google Scholar]
- 40.He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, Kloos RT, Croce CM, de la Chapelle A. 2005. The role of microRNA genes in papillary thyroid carcinoma. Proc. Natl. Acad. Sci. U. S. A. 102:19075–19080. 10.1073/pnas.0509603102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wells JA, McClendon CL. 2007. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature 450:1001–1009. 10.1038/nature06526 [DOI] [PubMed] [Google Scholar]