Abstract
Virus-host interactions highlight key regulatory steps in the control of gene expression. MicroRNAs (miRNAs) are small noncoding RNAs that regulate protein production via base pairing with mRNAs. Both DNA and RNA viruses have evolved mechanisms to degrade, boost, or hijack cellular miRNAs to benefit the viral life cycle. This minireview focuses on recent discoveries in virus-host miRNA interactions.
INTRODUCTION
Viruses are masters of gene regulation. Their parasitic life style employs host machinery to carry out basic biological processes, from transcription to protein synthesis. Thus, viruses can dramatically downsize their genomes to the minimum number of genes essential for successful infection (1). Even though some viruses, such as pandoravirus (2) and mimivirus (3, 4), encode as many as several thousand proteins—more than some free-living bacteria—they are exceptions to the rule. Typical mammalian herpesviruses possess from 70 to 200 genes in 120,000 to 250,000 base pairs of viral genomic DNA (5). These viral genomes are remarkably compacted compared to those of the host. For example, of the 3.2 billion base pairs in the human genome, more than 98% do not encode proteins (6). In contrast, only ∼10 to 20% of a herpesviral genome is noncoding (Fig. 1). With miniature genomes that are several orders of magnitude smaller than those of their hosts, viruses produce proteins and noncoding RNAs (ncRNAs) to regulate key components in host gene networks to ensure successful infection. Therefore, a deeper understanding of virus-host interactions often reveals important mechanisms of host gene regulation.
FIG 1.
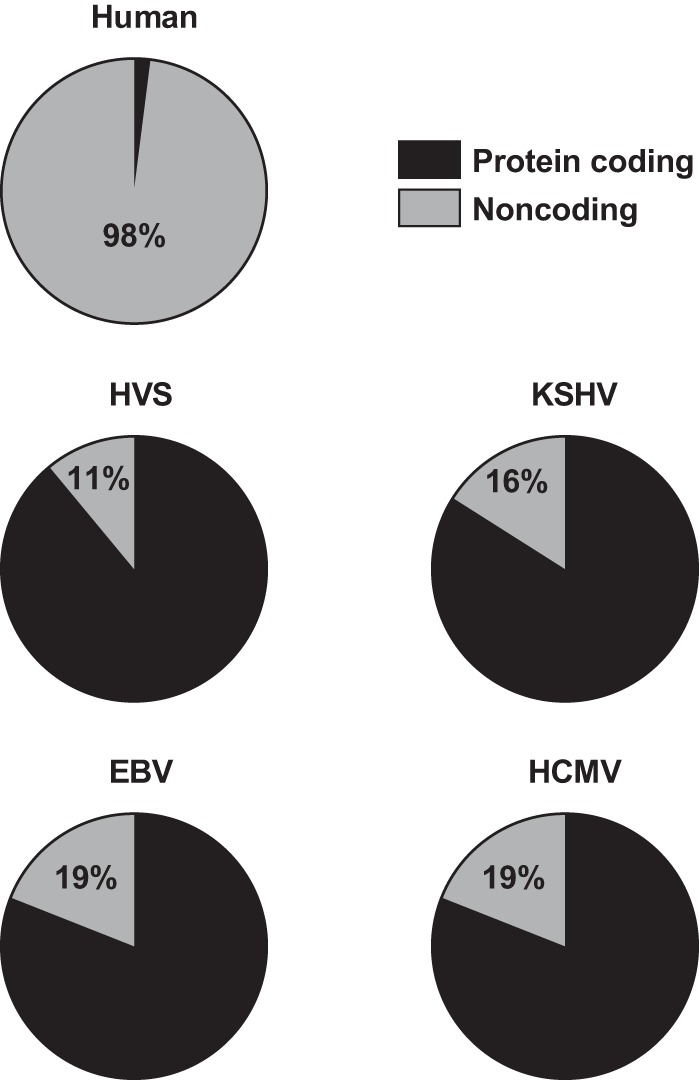
Protein coding potential of human and herpesviral genomes. HVS, herpesvirus saimiri; KSHV, Kaposi's sarcoma-associated herpesvirus; EBV, Epstein-Barr virus; HCMV, human cytomegalovirus.
MicroRNAs (miRNAs) are ∼22-nucleotide ncRNAs that play important roles in posttranscriptional regulation of gene expression. The specificity of target mRNA recognition is mostly determined by the seed region (nucleotides 2 to 8) at the 5′ end of a miRNA (7), although there are multiple experimentally validated mRNA targets that lack perfect base-pairing interactions with the miRNA seed region (8, 9). The relatively short seed sequence confers on a single miRNA the ability to regulate hundreds of mRNAs. One current hypothesis argues that miRNAs, like transcription factors, are master regulators of gene networks or pathways (10, 11). Given the importance and versatility of miRNAs, many viruses exploit this host pathway by destroying, boosting, or hijacking miRNAs to benefit the viral life cycle.
THE DESTROYER
miR-27 degradation by herpesvirus saimiri and murine cytomegalovirus.
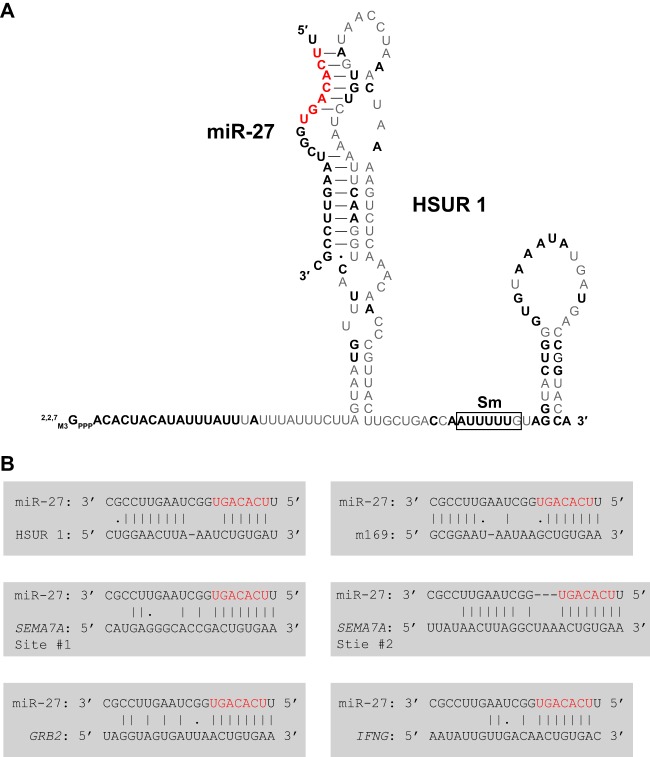
Herpesvirus saimiri (HVS) is an oncogenic gammaherpesvirus that transforms primate and human T cells (12). The most abundant viral transcripts in latently infected marmoset T cells are seven small U-rich Sm-class ncRNAs called HSURs (13–15). In these cells, HSUR 1 base pairs with the host miRNA 27 (miR-27), leading to miRNA degradation in a sequence-specific and binding-dependent manner (16) (Fig. 2), although the mechanism of miR-27 degradation is unknown. Recent high-throughput sequencing of RNA after cross-linking immunoprecipitation (HITS-CLIP) (17) analysis revealed that miR-27 interacts with mRNAs encoding components of the T-cell receptor (TCR) signaling pathway and downstream effectors in HVS-infected T cells (18). Specifically, miR-27 robustly decreases the levels of the cell surface signaling protein semaphorin 7A (SEMA7A), the adaptor protein growth factor receptor-bound protein 2 (GRB2), and the effector cytokine gamma interferon (IFN-γ) (18). This repressive role of miR-27 in T-cell activation explains the link between HSUR 1-induced miR-27 degradation (16) and activation of infected T cells conferred by expression of HSUR 1 and/or 2 (19). Unexpectedly, gammaherpesviruses distantly related to HVS, alcelaphine herpesvirus 1 (AlHV-1) and ovine herpesvirus 2 (OvHV-2), which cause similar T-lymphoproliferative disease, do not produce homologs of HSUR 1. Instead, AlHV-1 and OvHV-2 encode—in the syntenic regions of their genomes—viral homologs of miR-27 target proteins involved in T-cell activation (18) (Fig. 3).
FIG 2.
Base-pairing interactions between miR-27 and other RNAs. miR-27 seed nucleotides 2 to 8 are highlighted in red. (A) An HVS ncRNA, HSUR 1, base pairs with host miR-27, leading to its degradation. Nucleotides conserved between HVS strains and the related virus HVA (herpesvirus ateles) are in bold. The Sm protein binding site is boxed. (B) Complementarity of miR-27 with HSUR 1 is compared to that with various viral and host mRNAs, as discussed in the text.
FIG 3.
miR-27 is a pleiotropic repressor of T-cell activation that targets TCR signaling pathway components and downstream effector cytokines. In rhadinovirus (i.e., HVS)-infected cells, HSUR 1 upregulates expression of T-cell activation proteins by mediating degradation of miR-27. Distantly related macaviruses (i.e., AlHV-1 and OvHV-2) instead encode homologs of miR-27 target genes. Adapted from Molecular Cell (18) with permission of the publisher.
Murine cytomegalovirus (MCMV), a betaherpesvirus, also degrades host miR-27 using an antisense mechanism similar to that of HVS (20–22). Here the viral agent is not an ncRNA but a MCMV mRNA called m169, which contains a miR-27 target site in its 3′ untranslated region (3′ UTR). Interaction between m169 and miR-27 leads to 3′ tailing, trimming, and degradation of miR-27 (21, 22). In addition to rapid miRNA degradation, the level of the m169 transcript is reciprocally regulated by miR-27 (22). Like HSUR 1, m169 forms extensive base-pairing interactions not just with the seed sequence at the 5′ end but also with the 3′ end of miR-27. This interaction mode is different from the usual complementarity between cellular mRNAs and miR-27, which involves strong interaction only with the seed sequence (Fig. 2B). Presumably these more extensive base-pairing interactions lead to uridylation and degradation of miR-27 (23).
Why MCMV degrades miR-27 is still a mystery, even though it was reported that miR-27 degradation is important for efficient MCMV replication in mice (22). Because MCMV has a different cell tropism (such as macrophages, dendritic cells, fibroblasts, and hepatocytes [24]) than the T-lymphotropic gammaherpesvirus HVS, it is unlikely that MCMV degrades miR-27 to promote T-cell activation. One miR-27 target that might be important for both beta- and gammaherpesviruses is interleukin 10 (IL-10) (18). Many herpesviruses encode viral homologs of IL-10 that function similarly to cellular IL-10; these viral homologs modulate the host immune response and are thus important for the viral life cycle (25).
In contrast to MCMV, human cytomegalovirus (HCMV) does not regulate miR-27 levels. It instead encodes a IL-10 homolog in its genome. This approach may be functionally equivalent to upregulation of cellular IL-10 via degradation of miR-27 by MCMV. Together, these observations suggest that herpesviruses employ two alternative mechanisms—antisense-RNA-mediated miRNA degradation and acquisition of host protein-coding gene targets of miRNAs—to modify the host cell expression program to the benefit of the virus.
Since miRNAs usually target hundreds of mRNAs, one question regarding the biological function of miRNAs is whether the repression of the entire target network (with only moderate effects on individual targets) is important or whether only a few targets are dominant (7). The degradation of miR-27 by HVS versus the acquisition of the miR-27 target gene homologs by AlHV-1 and OvHV-2 sheds light on this critical question. Even though cellular miR-27 targets many mRNAs in the TCR signaling network, one target, SEMA7A, seems to be most important for T-lymphotropic gammaherpesviruses. Indeed, cellular SEMA7A mRNA contains two highly conserved target sites among the most robust miR-27–Argonaute binding sites revealed by HITS-CLIP, and the magnitude of SEMA7A protein repression by miR-27 is the largest for all validated targets (18).
miR-17∼92 degradation by human cytomegalovirus.
HCMV clinical strains contain a 15-kilobase region in the viral genome that is required for host cell tropism, latency, host cytokine regulation, and immune response (26). This region, which is absent in attenuated HCMV laboratory strains (26), produces a bicistronic mRNA UL144-145 that mediates degradation of cellular miR-17/miR-20a family miRNAs in a base-pairing-dependent and sequence-specific manner, similar to miR-27 degradation by HVS and MCMV (26). The host miR-17 ∼92 cluster generates six mature miRNAs: miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92a-1. This cluster and its paralogs (mir-106a∼363 and mir-106b∼25 clusters) act as oncogenes by promoting cell proliferation and tumor angiogenesis, while suppressing apoptosis (27). However, because HCMV is not a cancer-causing virus, degradation of these host miRNAs is likely beneficial to the virus for reasons other than oncogenesis. HCMV degrades only two of the miRNAs in the cluster, miR-17 and miR-20a, which differ by only two nucleotides and share the same seed sequence (26). Among the few direct mRNA targets that are currently known for miR-17 and miR-20a are the E2F-family transcription factors E2F1, E2F2, and E2F3 (28). HCMV strains carrying mutations in the single miR-17/miR-20a binding site show reduced synthesis of viral DNA and delayed viral production during lytic infection (26). It is currently unclear how miR-17 and miR-20a degradation affects HCMV DNA synthesis.
Degradation of host microRNAs by poxviruses.
Poxviruses are complex DNA viruses that replicate in the cytosol of infected cells, causing human diseases such as smallpox (5). Insect and mammalian poxviruses, Amsacta moorei entomopoxvirus (AMEV) and vaccinia virus (VACV), respectively, use viral poly(A) polymerase (VP55) to add A tails to all host miRNAs, which induces miRNA degradation (29). There are two possible explanations for why the virus-induced global degradation of host miRNAs might be beneficial to the viral life cycle. First, host miRNAs may target the long 3′ UTRs of viral mRNAs. Therefore, degradation of host miRNAs could reduce possible host miRNA-mediated downregulation of viral proteins—a mechanism that poxviruses may have evolved to evade the host miRNA defense system (29). Second, global downregulation of miRNAs is a common feature of many tumor and cancer cells, providing a growth advantage to these cells (30). Perhaps poxviruses reduce host miRNA levels globally (∼30-fold) to enhance proliferation of infected cells.
THE BOOSTER
miR-155 upregulation by Epstein-Barr virus.
Epstein-Barr virus (EBV) is a gammaherpesvirus associated with several human cancers, such as Hodgkin's disease, Burkitt's lymphoma, and nasopharyngeal carcinoma (5). In latently infected human B cells, EBV induces the expression of many host miRNAs; miR-155, which is induced ∼1,000-fold, is the most highly expressed miRNA (31–33). The miR-155 host gene is located in the B-cell integration cluster (BIC), which is transcriptionally activated by insertion of retroviruses in virally induced B-cell lymphomas (34). miR-155 plays important immunomodulatory roles in B cells, T cells, macrophages, and dendritic cells (35–37). Importantly, miR-155 is an oncogenic miRNA. Transgenic mice constitutively expressing high levels of mouse miR-155 (about 10-fold compared to wild-type animals) develop B-cell leukemia and lymphoma (38); transient miR-155 induction in the bone marrow leads to pathology similar to acute myeloid leukemia (39). miR-155 target mRNAs are implicated in hematopoietic development and disease (39), transcription regulation (31), the NK-κB signaling pathway (32), B-cell proliferation, and lymphocyte homeostasis (40). Recent genome-wide high-throughput studies, such as mRNA sequencing (mRNA-seq) analyses, stable isotope labeling by amino acids in cell culture (SILAC) proteomics, and HITS-CLIP, have confirmed some of these targets, as well as identified additional ones (40–42). In EBV-positive lymphoblastoid cell lines (LCLs) and diffuse large B-cell lymphomas (DLBCLs), inhibition or deletion of miR-155 reduces growth and promotes apoptosis (33). Therefore, EBV-induced upregulation of host miR-155 may be key to virus-induced oncogenesis.
miR-155 mimicry by Kaposi's sarcoma-associated herpesvirus, Marek's disease virus type 1, and simian foamy virus.
Kaposi's sarcoma-associated herpesvirus (KSHV) is a gammaherpesvirus that is the causative agent of several human B-cell cancers, including Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease (5). KSHV encodes 12 precursor miRNAs (pre-miRNAs), which produce 25 mature miRNAs (one miRNA is edited) (43, 44). Among them, mature miR-K12-11 (also known as miR-K11) shares its seed sequence (nucleotides 1 to 8) with host miR-155 (45). Two groups independently showed that miR-K12-11 and miR-155 regulate a common set of host mRNA targets, including transcription factors (e.g., BACH-1, FOS, and HIVEP2), a B-cell regulator (i.e., SLA), innate immunity modulators (e.g., PIK2CA and IKBKE), and proapoptotic proteins (e.g., LDOC-1, BIRC4BP, and XAF1) (46, 47). More recently, genome-wide Argonaute photoactivatable-ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP) and HITS-CLIP studies confirmed that ∼20 to 40% of miR-155 targets (121 and 151, respectively) are also targeted by miR-K12-11 (48, 49). This interesting observation suggests that KSHV produces a miRNA that mimics a miRNA master regulator to hijack an existing host gene network. Furthermore, host miR-155 is not expressed at detectible levels in KSHV-infected BC1 cells and primary effusion lymphoma (PEL) cells; both cell types instead express high levels of miR-K12-11 (46, 47, 49). Moreover, KSHV appears to use a similar miRNA mimicry strategy on other host miRNAs. For instance, KSHV miRNA miR-K12-10a shares seed nucleotides 2 to 8 with the abundant hematopoietic-cell-specific host miR-142-3p (48); similarly, miR-K12-3 and its isoform miR-K12-3+1 share a seed sequence identical to that of cellular miR-23 (50).
One of the limitations in studies of KSHV mimicry of host miR-155 is that no direct experimental evidence shows that miR-K12-11 is functionally equivalent to miR-155 in the context of the virus. However, several studies of another oncogenic herpesvirus, Marek's disease virus type 1 (MDV-1), shed light on this important question. MDV-1 is an alphaherpesvirus that causes aggressive T-cell lymphomas in chickens, known as Marek's disease (51). Interestingly, MDV-1 encodes a viral homolog of miR-155, called miR-M4, that shares seed nucleotides 1 to 8 with the host miRNA (52). Deletion of miR-M4 from MDV-1 or a 2-nucleotide mutation in the miR-M4 seed sequence abolishes MDV-1-induced oncogenicity and inhibits induction of lymphomas. This oncogenic phenotype can be rescued by revertant viruses expressing either miR-M4 or host miR-155 (53). Such studies provide direct evidence that the viral homolog and host miR-155 function similarly and argue that different viruses express miR-155 homologs to regulate the oncogenic gene network targeted by miR-155.
In addition to herpesviruses, retroviruses, such as simian foamy virus (SFV) and bovine leukemia virus (BLV), use similar mimicry strategies to boost levels of certain host miRNAs. SFV of African green monkeys (SFVagm) produces at least 6 mature miRNAs from its long terminal repeat (LTR) regions by RNA polymerase III transcription (54). Two of these viral miRNAs—SFVagm-miR-S4-3p and SFVagm-miR-S6-3p—exhibit seed sequence homology with host lymphoproliferative miR-155 and immunosuppressive miR-132, respectively (54). Similarly, one viral miRNA produced by B-lymphotropic BLV, BLV-miR-B4, shares its seed sequence with host B-cell tumorigenic miR-29a (55). All of these viral miRNA mimics target the same set of genes as their host counterparts (54, 55).
THE HIJACKER
miR-122 positively regulates hepatitis C virus replication.
Hepatitis C virus (HCV) is a positive-strand RNA virus associated with many liver diseases (5). miR-122 is a very abundant liver-specific miRNA (56) which regulates fatty acid and cholesterol biosynthesis (57, 58). miR-122 facilitates the replication, but not the translation or stability, of the HCV genomic RNA via base-pairing interactions with two conserved binding sites in the internal ribosome entry site (IRES) near the 5′ terminus of the genomic RNA (59, 60). Insertion of the HCV miR-122 binding site sequences into the 3′ UTR of a reporter gene instead causes downregulation, suggesting that the context of the miR-122 sites, as well as other virus and host factors, is important for miR-122-facilitated replication of viral RNA (60). The molecular mechanism by which miR-122 facilitates replication is still unknown, but it is critical for the HCV life cycle. miR-122 expression is sufficient to convert nonhepatic cells to become HCV permissive (61, 62). On the other hand, silencing of miR-122 with an antisense locked nucleic acid oligonucleotide leads to long-lasting suppression of HCV in infected primate models (63).
miR-138 and miR-200 promote latency of herpes simplex virus type 1 and human cytomegalovirus, respectively.
Herpes simplex virus type 1 (HSV-1) is an alphaherpesvirus that replicates in epithelial cells but establishes latency in sensory neurons (5). HSV-1 expresses several immediate early proteins, including ICP0, which plays important roles in both latent and lytic infection (64). Host miR-138 is abundant in the brain (65), where it regulates dendritic spine morphogenesis by downregulation of APT1 (66). The ICP0 mRNA contains two miR-138 target sites (confirmed by PAR-CLIP analysis [67]); consequently, the production of ICP0 proteins is repressed by miR-138. The biological significance of this regulation was tested in vivo: mice infected with HSV-1 containing three point mutations in each of the two miR-138 seed binding sites had higher levels of ICP0 and lytic gene expression during establishment of latency (67). However, no difference in virus replication was detected between wild-type and miR-138 target site-mutated HSV-1 either in culture or in mice (67). Similarly, host miR-200 family miRNAs repress HCMV immediate early protein UL122 (IE2) via a miR-200 seed binding site in the IE2 3′ UTR, and recombinant virus carrying mutations in the miR-200 binding site produces lytic rather than latent infection in primary cells (68). Because UL122 is a key transcription factor regulating HCMV lytic genes (5), miR-200 plays a critical role in viral latency. Furthermore, the host miR-17 family miRNAs regulate EBV latent transcripts LMP1 and BHRF1 (69). Therefore, critical roles for host miRNAs in controlling the lytic-to-latent switch are not uncommon in herpesvirus-infected cells.
miR-142 restricts cell type specificity of North American eastern equine encephalitis virus.
North American eastern equine encephalitis virus (EEEV) has a single-stranded sense RNA genome and can cause fatal infections in humans (5). miR-142-3p is a hematopoietic-cell-specific miRNA (65) which is involved in specification, formation, and differentiation of hematopoietic stem cells (70, 71), macrophage differentiation (72), proliferation and differentiation of mesenchymal cells in lungs (73), proliferation of CD25+ CD4 T cells (74), and the migration of CD4 T cells (75). miR-142-3p binds three highly conserved target sites in the 3′ UTR of the EEEV genomic RNA, thereby potently restricting EEEV in myeloid-lineage cells by blocking viral translation and subsequent replication (76). This miRNA-mediated restriction is important for efficient infection because it suppresses host cell type I interferon-induced antiviral immunity (76). miR-142-3p-mediation of innate immune suppression was reported in an earlier study where miR-142-3p binding sites were introduced into the influenza virus genome, leading to a reduced type I interferon response (77). Perhaps other viruses also hijack miR-142-3p to suppress innate immunity.
OTHER VIRUS-HOST miRNA INTERACTIONS
In addition to the examples discussed above, some host miRNAs carry out antiviral functions by suppressing viral infection in host cells. For example, miR-32 base pairs with mRNA of primate foamy virus type 1 (PFV-1, a retrovirus) and thereby restricts the accumulation of viral RNAs in human cells (78). Similarly, miR-24 and miR-93 mediate antiviral defense against vesicular stomatitis virus (VSV, a negative-sense RNA virus) infection in mice by targeting the viral large protein (L protein) and phosphoprotein (P protein) genes, respectively (79). In human T lymphocytes, miR-29a sequence-specifically targets the 3′ UTR of human immunodeficiency virus type 1 (HIV-1, a retrovirus) mRNA, reducing HIV-1 production and infectivity (80). miR-145 directly targets human papillomavirus (HPV, a DNA virus) E1 and E2 open reading frames to reduce HPV genome amplification and late gene expression (81). However, it is still debatable whether host miRNAs play antiviral roles in the physiological context of infection (43). For example, by global depletion or downregulation of host miRNAs, two recent studies show that the miRNA pathway does not counteract infection by a wide range of viruses in mammalian cells (82, 83).
FINAL REMARKS
In the past few years, there has been an explosion in discovery of novel virus-host miRNA interactions (Table 1). In most cases, the biological significance and the underlying mechanisms remain to be elucidated. More such interactions are likely to be discovered. Therefore, future work in this area will provide molecular insights not only into viral infection but also into host gene regulation mechanisms and viral evolution.
TABLE 1.
Summary of virus-host miRNA interactions
| Virus | Host miRNA | Mechanism | Function | Reference(s) |
|---|---|---|---|---|
| Herpesvirus saimiri | miR-27 | Antisense-RNA-mediated degradation | Promotes T-cell activation | 16, 18 |
| Mouse cytomegalovirus | miR-27 | Antisense-RNA-mediated degradation | Important for efficient viral replication | 20–22 |
| Human cytomegalovirus | miR-17/20 | Antisense-RNA-mediated degradation | Accelerates viral production | 26 |
| Vaccinia virus | All miRNAs | Tailing and degradation initiated by viral poly(A) polymerase | Counteracts host antiviral defense | 29 |
| Epstein-Barr virus | miR-155 | Expression increased by virus | Oncogenesis | 31–33 |
| Kaposi's sarcoma-associated herpesvirus | miR-155, miR-142-3p, miR-23 | Mimicry by viral miR-K12-11, miR-K12-10a, miR-K12-3 | Oncogenesis | 46–48, 50 |
| Marek's disease virus type 1 virus | miR-155 | Mimicry by miR-M4 | Oncogenesis | 52, 53 |
| Simian foamy virus | miR-155, miR-132 | Mimicry by SFVagm-miR-S4–3p, SFVagm-miR-S6–3p | Lymphoproliferation and innate immune suppression | 54 |
| Bovine leukemia virus | miR-29a | Mimicry by BLV miR-B4 | Oncogenesis | 55 |
| Hepatitis C virus | miR-122 | Binds 5′ UTR of viral RNA genome | Facilitates viral genome replication | 59, 60 |
| Herpes simplex virus 1 | miR-138 | Represses ICP0 expression | Promotes latency | 67 |
| Human cytomegalovirus | miR-200 | Represses UL122 expression | Promotes latency | 68 |
| Eastern equine encephalitis virus | miR-142-3p | Binds 3′ UTR of viral RNA genome | Restricts host tropism and suppresses innate immunity | 76 |
ACKNOWLEDGMENTS
We thank members of the Steitz laboratory for helpful discussions, K. Tycowski for critical commentary on the manuscript, and A. Miccinello for editorial assistance.
This work was supported by grant CA16038 from the NIH. J.A.S is an investigator of the Howard Hughes Medical Institute.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Biographies

Yang Eric Guo received a B.S. in chemical biology from the Department of Chemistry at the University of California, Berkeley, where he carried out undergraduate research on mechanisms of eukaryotic transcription in Drosophila cells. He is currently a graduate student in the Department of Cell Biology at Yale University. His Ph.D. thesis focuses on the regulatory functions of cellular and viral noncoding RNAs in mammalian cells.

Joan A. Steitz received a B.S. in chemistry from Antioch College and a Ph.D. in biochemistry and molecular biology from Harvard University. She is Sterling Professor of Molecular Biophysics and Biochemistry and an investigator of the Howard Hughes Medical Institute at Yale School of Medicine. She is interested in the multiple roles played by noncoding RNA-protein complexes in gene expression in vertebrate cells.
Footnotes
Published ahead of print 21 July 2014
REFERENCES
- 1.DiMaio D. 2012. Viruses, masters at downsizing. Cell Host Microbe 11:560–561. 10.1016/j.chom.2012.05.004 [DOI] [PubMed] [Google Scholar]
- 2.Philippe N, Legendre M, Doutre G, Coute Y, Poirot O, Lescot M, Arslan D, Seltzer V, Bertaux L, Bruley C, Garin J, Claverie JM, Abergel C. 2013. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 341:281–286. 10.1126/science.1239181 [DOI] [PubMed] [Google Scholar]
- 3.La Scola B, Audic S, Robert C, Jungang L, de Lamballerie X, Drancourt M, Birtles R, Claverie JM, Raoult D. 2003. A giant virus in amoebae. Science 299:2033. 10.1126/science.1081867 [DOI] [PubMed] [Google Scholar]
- 4.Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, La Scola B, Suzan M, Claverie JM. 2004. The 1.2-megabase genome sequence of mimivirus. Science 306:1344–1350. 10.1126/science.1101485 [DOI] [PubMed] [Google Scholar]
- 5.Fields BN, Knipe DM, Howley PM. 2007. Fields virology, 5th ed. Wolters Kluwer Health/Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 6.Lander ES. 2011. Initial impact of the sequencing of the human genome. Nature 470:187–197. 10.1038/nature09792 [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136:215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brodersen P, Voinnet O. 2009. Revisiting the principles of microRNA target recognition and mode of action. Nat. Rev. Mol. Cell Biol. 10:141–148. 10.1038/nrm2619 [DOI] [PubMed] [Google Scholar]
- 9.Chi SW, Hannon GJ, Darnell RB. 2012. An alternative mode of microRNA target recognition. Nat. Struct. Mol. Biol. 19:321–327. 10.1038/nsmb.2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hobert O. 2008. Gene regulation by transcription factors and microRNAs. Science 319:1785–1786. 10.1126/science.1151651 [DOI] [PubMed] [Google Scholar]
- 11.Ebert MS, Sharp PA. 2012. Roles for microRNAs in conferring robustness to biological processes. Cell 149:515–524. 10.1016/j.cell.2012.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ensser A, Fleckenstein B. 2005. T-cell transformation and oncogenesis by gamma2-herpesviruses. Adv. Cancer Res. 93:91–128. 10.1016/S0065-230X(05)93003-0 [DOI] [PubMed] [Google Scholar]
- 13.Lee SI, Murthy SC, Trimble JJ, Desrosiers RC, Steitz JA. 1988. Four novel U RNAs are encoded by a herpesvirus. Cell 54:599–607. 10.1016/S0092-8674(88)80004-7 [DOI] [PubMed] [Google Scholar]
- 14.Wassarman DA, Lee SI, Steitz JA. 1989. Nucleotide sequence of HSUR 5 RNA from herpesvirus saimiri. Nucleic Acids Res. 17:1258. 10.1093/nar/17.3.1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albrecht JC, Fleckenstein B. 1992. Nucleotide sequence of HSUR 6 and HSUR 7, two small RNAs of herpesvirus saimiri. Nucleic Acids Res. 20:1810. 10.1093/nar/20.7.1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cazalla D, Yario T, Steitz JA. 2010. Down-regulation of a host microRNA by a herpesvirus saimiri noncoding RNA. Science 328:1563–1566. 10.1126/science.1187197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chi SW, Zang JB, Mele A, Darnell RB. 2009. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460:479–486. 10.1038/nature08170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo YE, Riley KJ, Iwasaki A, Steitz JA. 2014. Alternative capture of noncoding RNAs or protein-coding genes by herpesviruses to alter host T cell function. Mol. Cell 54:67–79. 10.1016/j.molcel.2014.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cook HL, Lytle JR, Mischo HE, Li MJ, Rossi JJ, Silva DP, Desrosiers RC, Steitz JA. 2005. Small nuclear RNAs encoded by Herpesvirus saimiri upregulate the expression of genes linked to T cell activation in virally transformed T cells. Curr. Biol. 15:974–979. 10.1016/j.cub.2005.04.034 [DOI] [PubMed] [Google Scholar]
- 20.Buck AH, Perot J, Chisholm MA, Kumar DS, Tuddenham L, Cognat V, Marcinowski L, Dolken L, Pfeffer S. 2010. transcriptional regulation of miR-27 in murine cytomegalovirus infection. RNA 16:307–315. 10.1261/rna.1819210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Libri V, Helwak A, Miesen P, Santhakumar D, Borger JG, Kudla G, Grey F, Tollervey D, Buck AH. 2012. Murine cytomegalovirus encodes a miR-27 inhibitor disguised as a target. Proc. Natl. Acad. Sci. U. S. A. 109:279–284. 10.1073/pnas.1114204109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcinowski L, Tanguy M, Krmpotic A, Radle B, Lisnic VJ, Tuddenham L, Chane-Woon-Ming B, Ruzsics Z, Erhard F, Benkartek C, Babic M, Zimmer R, Trgovcich J, Koszinowski UH, Jonjic S, Pfeffer S, Dolken L. 2012. Degradation of cellular mir-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo. PLoS Pathog. 8:e1002510. 10.1371/journal.ppat.1002510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. 2010. Target RNA-directed trimming and tailing of small silencing RNAs. Science 328:1534–1539. 10.1126/science.1187058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu KM, Pratt JR, Akers WJ, Achilefu SI, Yokoyama WM. 2009. Murine cytomegalovirus displays selective infection of cells within hours after systemic administration. J. Gen. Virol. 90:33–43. 10.1099/vir.0.006668-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slobedman B, Barry PA, Spencer JV, Avdic S, Abendroth A. 2009. Virus-encoded homologs of cellular interleukin-10 and their control of host immune function. J. Virol. 83:9618–9629. 10.1128/JVI.01098-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee S, Song J, Kim S, Kim J, Hong Y, Kim Y, Kim D, Baek D, Ahn K. 2013. Selective degradation of host MicroRNAs by an intergenic HCMV noncoding RNA accelerates virus production. Cell Host Microbe 13:678–690. 10.1016/j.chom.2013.05.007 [DOI] [PubMed] [Google Scholar]
- 27.Mendell JT. 2008. miRiad roles for the miR-17-92 cluster in development and disease. Cell 133:217–222. 10.1016/j.cell.2008.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olive V, Jiang I, He L. 2010. mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int. J. Biochem. Cell Biol. 42:1348–1354. 10.1016/j.biocel.2010.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Backes S, Shapiro JS, Sabin LR, Pham AM, Reyes I, Moss B, Cherry S, ten Oever BR. 2012. Degradation of host microRNAs by poxvirus poly(A) polymerase reveals terminal RNA methylation as a protective antiviral mechanism. Cell Host Microbe 12:200–210. 10.1016/j.chom.2012.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. 2005. MicroRNA expression profiles classify human cancers. Nature 435:834–838. 10.1038/nature03702 [DOI] [PubMed] [Google Scholar]
- 31.Yin Q, McBride J, Fewell C, Lacey M, Wang X, Lin Z, Cameron J, Flemington EK. 2008. MicroRNA-155 is an Epstein-Barr virus-induced gene that modulates Epstein-Barr virus-regulated gene expression pathways. J. Virol. 82:5295–5306. 10.1128/JVI.02380-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu F, Weidmer A, Liu CG, Volinia S, Croce CM, Lieberman PM. 2008. Epstein-Barr virus-induced miR-155 attenuates NF-kappaB signaling and stabilizes latent virus persistence. J. Virol. 82:10436–10443. 10.1128/JVI.00752-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linnstaedt SD, Gottwein E, Skalsky RL, Luftig MA, Cullen BR. 2010. Virally induced cellular microRNA miR-155 plays a key role in B-cell immortalization by Epstein-Barr virus. J. Virol. 84:11670–11678. 10.1128/JVI.01248-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tam W, Ben-Yehuda D, Hayward WS. 1997. bic, a novel gene activated by proviral insertions in avian leukosis virus-induced lymphomas, is likely to function through its noncoding RNA. Mol. Cell. Biol. 17:1490–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haasch D, Chen YW, Reilly RM, Chiou XG, Koterski S, Smith ML, Kroeger P, McWeeny K, Halbert DN, Mollison KW, Djuric SW, Trevillyan JM. 2002. T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene, BIC. Cell. Immunol. 217:78–86. 10.1016/S0008-8749(02)00506-3 [DOI] [PubMed] [Google Scholar]
- 36.O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. 2007. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. U. S. A. 104:1604–1609. 10.1073/pnas.0610731104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. 2007. Regulation of the germinal center response by microRNA-155. Science 316:604–608. 10.1126/science.1141229 [DOI] [PubMed] [Google Scholar]
- 38.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. 2006. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 103:7024–7029. 10.1073/pnas.0602266103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D. 2008. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J. Exp. Med. 205:585–594. 10.1084/jem.20072108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loeb GB, Khan AA, Canner D, Hiatt JB, Shendure J, Darnell RB, Leslie CS, Rudensky AY. 2012. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Molecular Cell 48:760–770. 10.1016/j.molcel.2012.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455:58–63. 10.1038/nature07228 [DOI] [PubMed] [Google Scholar]
- 42.Xu G, Fewell C, Taylor C, Deng N, Hedges D, Wang X, Zhang K, Lacey M, Zhang H, Yin Q, Cameron J, Lin Z, Zhu D, Flemington EK. 2010. Transcriptome and targetome analysis in MIR155 expressing cells using RNA-seq. RNA 16:1610–1622. 10.1261/rna.2194910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skalsky RL, Cullen BR. 2010. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 64:123–141. 10.1146/annurev.micro.112408.134243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu Y, Haecker I, Yang Y, Gao SJ, Renne R. 2013. gamma-Herpesvirus-encoded miRNAs and their roles in viral biology and pathogenesis. Curr. Opin. Virol. 3:266–275. 10.1016/j.coviro.2013.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nair V, Zavolan M. 2006. Virus-encoded microRNAs: novel regulators of gene expression. Trends Microbiol. 14:169–175. 10.1016/j.tim.2006.02.007 [DOI] [PubMed] [Google Scholar]
- 46.Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JT, Braich R, Manoharan M, Soutschek J, Ohler U, Cullen BR. 2007. A viral microRNA functions as an orthologue of cellular miR-155. Nature 450:1096–1099. 10.1038/nature05992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC, Baker HV, Renne R. 2007. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J. Virol. 81:12836–12845. 10.1128/JVI.01804-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gottwein E, Corcoran DL, Mukherjee N, Skalsky RL, Hafner M, Nusbaum JD, Shamulailatpam P, Love CL, Dave SS, Tuschl T, Ohler U, Cullen BR. 2011. Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines. Cell Host Microbe 10:515–526. 10.1016/j.chom.2011.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haecker I, Gay LA, Yang Y, Hu J, Morse AM, McIntyre LM, Renne R. 2012. Ago HITS-CLIP expands understanding of Kaposi's sarcoma-associated herpesvirus miRNA function in primary effusion lymphomas. PLoS Pathog. 8:e1002884. 10.1371/journal.ppat.1002884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manzano M, Shamulailatpam P, Raja AN, Gottwein E. 2013. Kaposi's sarcoma-associated herpesvirus encodes a mimic of cellular miR-23. J. Virol. 87:11821–11830. 10.1128/JVI.01692-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Osterrieder N, Kamil JP, Schumacher D, Tischer BK, Trapp S. 2006. Marek's disease virus: from miasma to model. Nat. Rev. Microbiol. 4:283–294. 10.1038/nrmicro1382 [DOI] [PubMed] [Google Scholar]
- 52.Morgan R, Anderson A, Bernberg E, Kamboj S, Huang E, Lagasse G, Isaacs G, Parcells M, Meyers BC, Green PJ, Burnside J. 2008. Sequence conservation and differential expression of Marek's disease virus microRNAs. J. Virol. 82:12213–12220. 10.1128/JVI.01722-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao Y, Xu H, Yao Y, Smith LP, Kgosana L, Green J, Petherbridge L, Baigent SJ, Nair V. 2011. Critical role of the virus-encoded microRNA-155 ortholog in the induction of Marek's disease lymphomas. PLoS Pathog. 7:e1001305. 10.1371/journal.ppat.1001305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kincaid RP, Chen Y, Cox JE, Rethwilm A, Sullivan CS. 2014. Noncanonical microRNA (miRNA) biogenesis gives rise to retroviral mimics of lymphoproliferative and immunosuppressive host miRNAs. mBio 5:e00074–14. 10.1128/mBio.00074-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kincaid RP, Burke JM, Sullivan CS. 2012. RNA virus microRNA that mimics a B-cell oncomiR. Proc. Natl. Acad. Sci. U. S. A. 109:3077–3082. 10.1073/pnas.1116107109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R, Zaret KS, Taylor JM. 2004. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 1:106–113. 10.4161/rna.1.2.1066 [DOI] [PubMed] [Google Scholar]
- 57.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. 2005. Silencing of microRNAs in vivo with ‘antagomirs'. Nature 438:685–689. 10.1038/nature04303 [DOI] [PubMed] [Google Scholar]
- 58.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. 2006. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metabol. 3:87–98. 10.1016/j.cmet.2006.01.005 [DOI] [PubMed] [Google Scholar]
- 59.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 309:1577–1581. 10.1126/science.1113329 [DOI] [PubMed] [Google Scholar]
- 60.Jopling CL, Schutz S, Sarnow P. 2008. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe 4:77–85. 10.1016/j.chom.2008.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fukuhara T, Kambara H, Shiokawa M, Ono C, Katoh H, Morita E, Okuzaki D, Maehara Y, Koike K, Matsuura Y. 2012. Expression of microRNA miR-122 facilitates an efficient replication in nonhepatic cells upon infection with hepatitis C virus. J. Virol. 86:7918–7933. 10.1128/JVI.00567-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kambara H, Fukuhara T, Shiokawa M, Ono C, Ohara Y, Kamitani W, Matsuura Y. 2012. Establishment of a novel permissive cell line for the propagation of hepatitis C virus by expression of microRNA miR122. J. Virol. 86:1382–1393. 10.1128/JVI.06242-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. 2010. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327:198–201. 10.1126/science.1178178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boutell C, Everett RD. 2013. Regulation of alphaherpesvirus infections by the ICP0 family of proteins. J. Gen. Virol. 94:465–481. 10.1099/vir.0.048900-0 [DOI] [PubMed] [Google Scholar]
- 65.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129:1401–1414. 10.1016/j.cell.2007.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJ, Kane C, Hubel K, Dekker F, Hedberg C, Rengarajan B, Drepper C, Waldmann H, Kauppinen S, Greenberg ME, Draguhn A, Rehmsmeier M, Martinez J, Schratt GM. 2009. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat. Cell Biol. 11:705–716. 10.1038/ncb1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pan D, Flores O, Umbach JL, Pesola JM, Bentley P, Rosato PC, Leib DA, Cullen BR, Coen DM. 2014. A neuron-specific host microRNA targets herpes simplex virus-1 ICP0 expression and promotes latency. Cell Host Microbe 15:446–456. 10.1016/j.chom.2014.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O'Connor CM, Vanicek J, Murphy EA. 2014. Host microRNA regulation of human cytomegalovirus immediate early protein translation promotes viral latency. J. Virol. 88:5524–5532. 10.1128/JVI.00481-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Riley KJ, Rabinowitz GS, Yario TA, Luna JM, Darnell RB, Steitz JA. 2012. EBV and human microRNAs co-target oncogenic and apoptotic viral and human genes during latency. EMBO J. 31:2207–2221. 10.1038/emboj.2012.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lu X, Li X, He Q, Gao J, Gao Y, Liu B, Liu F. 2013. miR-142-3p regulates the formation and differentiation of hematopoietic stem cells in vertebrates. Cell Res. 23:1356–1368. 10.1038/cr.2013.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nimmo R, Ciau-Uitz A, Ruiz-Herguido C, Soneji S, Bigas A, Patient R, Enver T. 2013. miR-142-3p controls the specification of definitive hemangioblasts during ontogeny. Dev. Cell 26:237–249. 10.1016/j.devcel.2013.06.023 [DOI] [PubMed] [Google Scholar]
- 72.Sonda N, Simonato F, Peranzoni E, Cali B, Bortoluzzi S, Bisognin A, Wang E, Marincola FM, Naldini L, Gentner B, Trautwein C, Sackett SD, Zanovello P, Molon B, Bronte V. 2013. miR-142-3p prevents macrophage differentiation during cancer-induced myelopoiesis. Immunity 38:1236–1249. 10.1016/j.immuni.2013.06.004 [DOI] [PubMed] [Google Scholar]
- 73.Carraro G, Shrestha A, Rostkovius J, Contreras A, Chao CM, El Agha E, Mackenzie B, Dilai S, Guidolin D, Taketo MM, Gunther A, Kumar ME, Seeger W, De Langhe S, Barreto G, Bellusci S. 2014. miR-142-3p balances proliferation and differentiation of mesenchymal cells during lung development. Development 141:1272–1281. 10.1242/dev.105908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou Q, Haupt S, Prots I, Thummler K, Kremmer E, Lipsky PE, Schulze-Koops H, Skapenko A. 2013. miR-142-3p is involved in CD25+ CD4 T cell proliferation by targeting the expression of glycoprotein A repetitions predominant. J. Immunol. 190:6579–6588. 10.4049/jimmunol.1202993 [DOI] [PubMed] [Google Scholar]
- 75.Liu J, Li W, Wang S, Wu Y, Li Z, Wang W, Liu R, Ou J, Zhang C, Wang S. 2014. miR-142-3p attenuates the migration of CD4(+) T cells through regulating actin cytoskeleton via RAC1 and ROCK2 in arteriosclerosis obliterans. PLoS One 9:e95514. 10.1371/journal.pone.0095514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Trobaugh DW, Gardner CL, Sun C, Haddow AD, Wang E, Chapnik E, Mildner A, Weaver SC, Ryman KD, Klimstra WB. 2014. RNA viruses can hijack vertebrate microRNAs to suppress innate immunity. Nature 506:245–248. 10.1038/nature12869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Langlois RA, Varble A, Chua MA, Garcia-Sastre A, ten Oever BR. 2012. Hematopoietic-specific targeting of influenza A virus reveals replication requirements for induction of antiviral immune responses. Proc. Natl. Acad. Sci. U. S. A. 109:12117–12122. 10.1073/pnas.1206039109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O. 2005. A cellular microRNA mediates antiviral defense in human cells. Science 308:557–560. 10.1126/science.1108784 [DOI] [PubMed] [Google Scholar]
- 79.Otsuka M, Jing Q, Georgel P, New L, Chen J, Mols J, Kang YJ, Jiang Z, Du X, Cook R, Das SC, Pattnaik AK, Beutler B, Han J. 2007. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity 27:123–134. 10.1016/j.immuni.2007.05.014 [DOI] [PubMed] [Google Scholar]
- 80.Nathans R, Chu CY, Serquina AK, Lu CC, Cao H, Rana TM. 2009. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol. Cell 34:696–709. 10.1016/j.molcel.2009.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gunasekharan V, Laimins LA. 2013. Human papillomaviruses modulate microRNA 145 expression to directly control genome amplification. J. Virol. 87:6037–6043. 10.1128/JVI.00153-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Backes S, Langlois RA, Schmid S, Varble A, Shim JV, Sachs D, ten Oever BR. 2014. The Mammalian Response to Virus Infection Is Independent of Small RNA Silencing. Cell Rep. 8:114–125. 10.1016/j.celrep.2014.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bogerd HP, Skalsky RL, Kennedy EM, Furuse Y, Whisnant AW, Flores O, Schultz KL, Putnam N, Barrows NJ, Sherry B, Scholle F, Garcia-Blanco MA, Griffin DE, Cullen BR. 2014. Replication of many human viruses is refractory to inhibition by endogenous cellular microRNAs. J. Virol. 88:8065–8076. 10.1128/JVI.00985-14 [DOI] [PMC free article] [PubMed] [Google Scholar]