Abstract
From January 2011 to December 2013, a total of 262 samples, from 188 patients suspected of having syphilis were tested for the presence of treponemal DNA by PCR amplification of five chromosomal loci, including the polA (TP0105), tmpC (TP0319), TP0136, TP0548, and 23S rRNA genes. Altogether, 146 samples from 103 patients were PCR positive for treponemal DNA. A set of 81 samples from 62 PCR-positive patients were typeable, and among them, nine different genotypes were identified. Compared to a previous study in the Czech Republic during 2004 to 2010, the number of genotypes detected among syphilis patients in a particular year increased to six in both 2012 and 2013, although they were not the same six. The proportion of macrolide-resistant clinical isolates in this 3-year study was 66.7%.
INTRODUCTION
Syphilis is a sexually transmitted multistage disease caused by Treponema pallidum subsp. pallidum. Syphilis is usually diagnosed on the basis of clinical symptoms, serological findings, and other methods such as the observation of treponemes by dark-field microscopy. In the 1990s, molecular detection of syphilis treponemes based on PCR amplification was introduced (1). In addition to diagnostics, PCR methods have allowed molecular typing and therefore provided epidemiological information on T. pallidum subsp. pallidum isolates and allowed the determination of the prevalence of macrolide-resistant T. pallidum subsp. pallidum strains (2–6).
Most of the molecular typing studies of treponemal DNA were performed with the CDC typing system, which is based on the determination of the number of 60-bp repeats in the arp gene and on restriction fragment length polymorphism analysis of the tprE, tprG, and tprJ genes (2). Recently, this typing system has been improved by the additional determination of the number of repeats in the rpsA gene (3) and by additional sequence analysis of the TP0548 locus (4), which has been used for sequencing-based molecular typing of syphilis treponemes since 2006 (5). The reproducibility of the CDC typing system was recently questioned by the identification of multiple T. pallidum subsp. pallidum subtypes in clinical samples isolated from a single patient (7). In parallel with the CDC typing scheme, sequencing-based molecular typing based on sequencing of the TP0136, TP0548, and 23S rRNA genes has been introduced and tested (5, 8).
During the last decades, macrolide antibiotics have been used in the Czech Republic for the treatment of syphilis in patients allergic to penicillin, as well as for the treatment of other diseases (National Institute for Drug Control, unpublished data). At the same time, syphilis treponemes have developed resistance to macrolide antibiotics and their resistance has been found to be linked to an A2058G or A2059G mutation in the 23S rRNA gene (9, 10). Therefore, clinical isolates harboring this mutation are considered to be macrolide resistant.
In this study, we genotyped syphilis-causing clinical isolates collected in the Czech Republic from 2011 to 2013 by sequencing-based molecular typing. Increases in the variability of T. pallidum subsp. pallidum genotypes and also in the prevalence of macrolide resistance-encoding alleles at the 23S rRNA gene locus (A2058G and A2059G mutations) were noted compared to those in a previous study in our laboratory of clinical isolates from the same geographical region (5).
MATERIALS AND METHODS
Collection of clinical samples.
Clinical samples were collected in the Czech Republic between January 2011 and December 2013. Four clinical departments were involved in sample collection and serology testing, including (i) the Department of Dermatolovenereology, 1st Faculty of Medicine, Charles University in Prague; (ii) the National Reference Laboratory for Diagnostics of Syphilis, National Institute for Public Health; (iii) the Department of Dermatovenereology, St. Anne's Faculty Hospital Brno; and (iv) the Department of Medical Microbiology, Faculty of Medicine, St. Anne's Hospital and Masaryk University, Brno. The National Reference Laboratory for Diagnostics of Syphilis, National Institute for Public Health also provided samples from the Outpatient STI Clinic Medicentrum, Prague, and the Department of Dermatolovenereology, St. Anne's University Hospital, provided samples from regional hospitals and outpatient clinics in and around Brno. All other procedures (including DNA isolation, amplification, and sequencing of treponemal loci) were performed in the laboratory of the Department of Biology, Faculty of Medicine, Masaryk University.
Syphilis was diagnosed on the basis of a combination of observed clinical symptoms and the results of several serological tests. Each of the above laboratories performed the following syphilis serology tests: (i) rapid plasma reagin (RPR), T. pallidum hemagglutination (TPHA), enzyme-linked immunosorbent assay (ELISA) for IgM and IgG; (ii) RPR, T. pallidum particle agglutination (TPPA), fluorescent treponemal antibody absorption (FTA-ABS) for IgG and solid-phase hemadsorption (SPHA) for IgM; (iii) RPR, TPHA, and ELISA for IgM and IgG; and (iv) RPR, TPHA, and ELISA for IgM and IgG and Western blot assay for IgM and IgG. Serological tests were provided by OMEGA Diagnostics (Reinbek, Germany), TEST-LINE (Brno, Czech Republic), and MARDX (Carlsbad, CA, USA). No statistically significant differences between the laboratories with respect to the number of seronegative patients or the tests used in the different laboratories were found.
All clinical samples were obtained after patients signed an informed-consent form. The design of the study was approved by the ethics committee of the Faculty of Medicine, Masaryk University. Patients were considered syphilis positive when they showed positive clinical symptoms combined with a positive syphilis serology result or the detection of treponemal DNA by PCR.
Isolation of DNA.
Swab extracts (prepared by submersion of swabs in 1.5 ml of sterile water and agitation for 5 min at room temperature), whole blood (0.2 ml), cerebrospinal fluid (CSF; 0.2 ml), and tissue samples (up to 25 mg) were used for isolation of DNA with a QIAamp DNA minikit (tissue samples) and a QIAamp DNA Blood minikit (Qiagen, Hilden, Germany) (swab, whole-blood, and CSF samples) according to the manufacturer's recommendations. A QIAcube (Qiagen) robotic workstation was used for DNA isolation.
Molecular detection of treponemal DNA.
Five loci, including the polA (TP0105), tmpC (TP0319), TP0136, TP0548, and 23S rRNA genes were amplified by nested PCR as described previously (10). Briefly, each PCR mixture contained 0.5 μl of a 10 mM deoxynucleoside triphosphate (dNTP) mixture, 2.5 μl of ThermoPol Reaction buffer, 0.25 μl of each primer (100 pmol/μl), 0.05 μl of Taq polymerase (5,000 U/ml; New England BioLabs, Ipswich, MA), and 1 or 10 μl of DNA. The reaction mixture was supplemented with PCR-grade water to a final volume of 25 μl. All sets of specimens were first PCR tested with 1 μl of isolated DNA. PCR-negative reaction mixtures were repeatedly tested with 10 μl of isolated DNA from a total volume of 100 μl. PCR amplification of polA and tmpC was performed under the following cycling conditions: 94°C (1 min); 94°C (30 s), 58°C (30 s), and 72°C (1 min) for 30 cycles; and 72°C (10 min). The second step of nested PCR was performed under the same conditions but with an increase to 40 cycles. PCR amplification of TP0136, TP0548, and 23S rRNA gene loci was performed under the following cycling conditions: 94°C (1 min); 94°C (30 s), 48°C (30 s), and 72°C (1 min, 45 s) for 40 cycles; and 72°C (7 min). A DNA sample of the reference T. pallidum subsp. pallidum strain Nichols served as a positive control for amplification of treponemal loci. To test whether the samples contained PCR inhibitors, the human gene that encodes the enzyme methylenetetrahydrofolate reductase (MTHFR) was amplified in PCR syphilis-negative samples previously supplemented with 30 ng of human DNA. MTHFR is an enzyme involved in the methyl cycle. In humans, the enzyme is encoded by the MTHFR gene located on chromosome 1 (location p36.3) (11). A 30-μl PCR mixture contained 1 μl of a 10 mM dNTP mixture, 3 μl of ThermoPol Reaction buffer, 1.5 μl of each primer (100 pmol/μl), 19 μl of PCR-grade water, 0.1 μl of Taq polymerase (5,000 U/ml; New England BioLabs, Ipswich, MA), 3 μl of DNA, and 1 μl of amplifiable human DNA. Primers Mthfr1 (5′TGAAGGAGAAGGTGTCTGCGGGA-3′) and Mthfr2 (5′-AGGACGGTGCGGTGAGAGTG-3′) were used. PCR was performed under the following cycling conditions: 94°C (3 min); 94°C (20 s), 62°C (20 s), and 72°C (20 s) for 30 cycles; and 72°C (2 min).
DNA sequencing.
Sequencing of PCR products was performed by Microsynth AG, Wolfurt, Austria, and GATC Biotech AG, Constance, Germany, and sequence analyses were performed with Lasergene software (DNASTAR v. 7.1.0.; DNASTAR, Madison, WI). Sequences of 23S rRNA gene amplicons were evaluated only at positions corresponding to positions 2058 and 2059 in the 23S rRNA gene of Escherichia coli (accession no. V00331), where A→G mutations lead to macrolide resistance.
Statistical methods.
Analysis of statistical significance was performed with Fisher's exact two-tailed test with STATISTICA software v. 12 (StatSoft, Tulsa, OK). P values of <0.05 were considered to be statistically significant.
RESULTS
Characteristics of patients and clinical isolates.
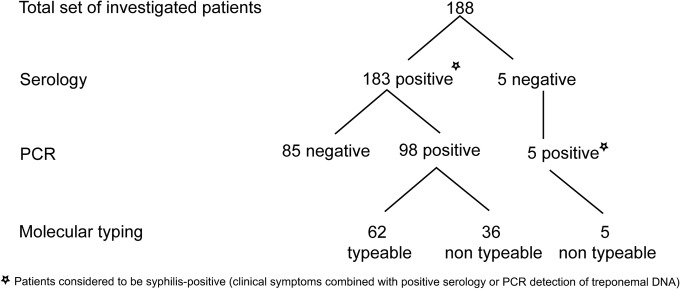
The clinical presentations of the patients enrolled in this study are shown in Table 1. During a 3-year period (from January 2011 to December 2013), a total of 262 samples from 188 patients suspected of having syphilis were tested for the presence of treponemal DNA by PCR amplification of five chromosomal loci (polA, tmpC, TP0136, TP0548, and 23S rRNA genes). Samples were considered PCR positive when PCR amplification was twice positive for at least one treponemal locus. A total of 146 samples from 103 patients were PCR positive for treponemal DNA (Fig. 1). In addition, all samples that were negative for amplification of treponemal DNA were positive for amplification of the human MTHFR gene, which had been externally added to the sample to indicate the absence of PCR inhibitors. Since most (84%) of the samples that were negative for amplification of treponemal DNA were also positive for internal human DNA, it follows that most of the samples contained undegraded human DNA and undetectable amounts of treponemal DNA (data not shown).
TABLE 1.
Clinical characteristics of patients with syphilis
| Variable | All patients (n = 188) | PCR-positive patients (n = 103) | PCR-negative patients (n = 85) |
|---|---|---|---|
| Mean age (yr)a | 35.4 | 35.3 | 35.5 |
| No. (%) of males/females | 166 (88.3)/22 (11.7) | 91(88.4)/12 (11.6) | 75 (88)/10 (22) |
| No. (%) of MSMb | 96 (51) | 50 (48.5) | 46 (54) |
| No. (%) HIV infected | 37 (19.6) | 21 (20.4) | 16 (18.8) |
| No. (%) with: | |||
| Primary syphilis | 92 (49) | 63 (61) | 29 (34) |
| Secondary syphilis | 59 (31.4) | 28 (27) | 31 (36.5) |
| Latent syphilis | 30 (16) | 8 (7.8) | 22 (25.9) |
| Otherc | 7 (3.7) | 4 (4.9) | 3 (3.5) |
A case of congenital syphilis (0 years) was excluded from analysis.
No data on the sexual orientation of 37 male patients were available.
Congenital syphilis or undetermined stage.
FIG 1.
Flow chart of the patients investigated in this study. Seronegative samples that were PCR negative were excluded from this study. We analyzed 262 samples from 188 patients. Altogether, we analyzed 153 whole-blood samples, 103 swabs, 3 CSF samples, and 3 tissue samples. Among this set of samples, we detected 146 PCR-positive samples from 103 patients.
All five of the genes tested were detected in samples from 36 patients. Of the remaining samples, 14, 13, 24, and 16 patients were PCR positive at four, three, two, and one of the treponemal loci, respectively. PCR-positive patients with negative serological test results (n = 5) were positive at a single locus only (three for polA, one for tmpC, and one for the 23S rRNA gene locus). The highest number of PCR-positive samples was found for the polA locus (122 out of 146; 83.6%), followed by the tmpC (120 out of 146; 82.2%), 23S rRNA gene (90 out of 146; 61.6%), TP0548 (66 out of 146; 45.2%), and TP0136 (60 out of 146; 41.1%) loci. Similar detection efficiency was also found when clinical isolates were separately evaluated with swabs and whole-blood samples (data not shown).
The highest number of PCR-positive samples was found among swab specimens (79 out of 103 positive; 76.7%), followed by whole-blood samples (64 out of 153 positive; 42%). Other samples included CSF samples (two out of three positive) and tissue samples (three out of three positive).
From patients with primary syphilis, CSF (n = 2), whole-blood (n = 57), and swab (n = 70) samples were collected. Patients with secondary syphilis provided whole-blood (n = 65) and swab (n = 24) samples. In cases of latent syphilis, whole-blood (n = 30) samples were collected. In one case of congenital syphilis, CSF (n = 1) and tissue (n = 3) samples were taken. Information regarding the syphilis stage was not available for six patients (10 samples). No statistically significant difference between PCR detection and the stage of syphilis was found.
PCR detection and molecular typing of syphilis treponemes.
Amplified treponemal DNA from 81 samples from 62 PCR-positive patients was typeable. Samples were considered typeable when at least one typing locus (either TP0136 or TP0548) was sequenced. When sequences of all three loci, including the TP0136, TP0548, and 23S rRNA genes, were determined, treponemal DNA was considered to be completely typed. Samples from 40 patients (out of 103 PCR-positive patients) were completely typed while samples from an additional 22 patients were partially typed. In cases where we analyzed parallel samples taken from the same patient at the same time (altogether, 30 samples from 12 patients), the treponemal genotypes identified were always identical.
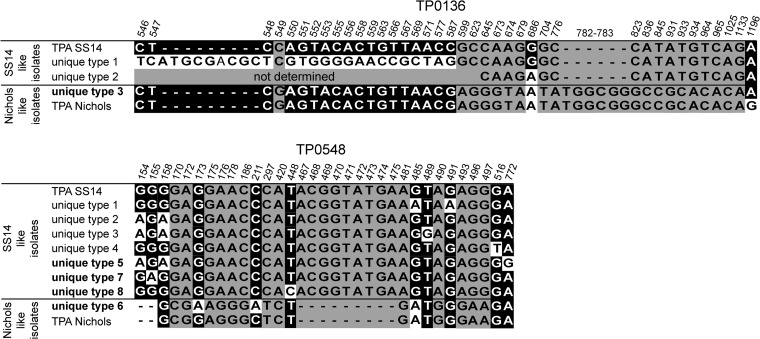
Altogether, nine different genotypes were found; eight were related to the sequence of T. pallidum subsp. pallidum strain SS14, and one was related to the sequence of T. pallidum subsp. pallidum strain Nichols (the nucleotide sequence similarity to either strain SS14 or Nichols was >99.7%). All identified T. pallidum subsp. pallidum genotypes found in the Czech Republic from 2011 to 2013 are shown in Table 2. Five new genotypes (SU2S, SU5R8, SU7S, XU8R9, and U3U6R8) not found in the previous study by our laboratory (5) were detected among the syphilis patients in this study in the Czech Republic (Fig. 2). The prevailing genotype (SU2R8) was closely related to strain SS14. While the sequence of the portion of the TP0136 locus tested was identical to the strain SS14 sequence, the part of the TP0548 locus tested was unique although very similar to the strain SS14 sequence (differing at only two nucleotide positions); the 23S rRNA genes sequence encoded resistance to macrolide antibiotics because of the A2058G mutation. This genotype was found in 15 (37.5%) out of 40 completely typed strains. The second most common genotype (SSS) was found in 9 (22.5%) out of 40 strains and showed sequences at the TP0136 and TP0548 loci identical to those of strain SS14; however, unlike strain SS14, these strains were sensitive to macrolides. In addition to train SS14-related genotypes, a strain Nichols-like genotype, U3U6R8, was found in six patients infected with syphilis. One unique sequence of the TP0136 locus (U3) and several unique sequences of TP0548 including U5, U6, U7, and U8 were seen for the first time in this study; the U2 sequence has been detected in the Czech Republic since 2008 (5). However, the U2 sequence was previously reported exclusively in a resistant strain (SU2R8) (5), while in this study it was also found in a macrolide-susceptible isolate (SU2S).
TABLE 2.
Definition of individual T. pallidum subsp. pallidum genotypes and their prevalence in typeable clinical isolates
| Genotypea | TP0136 | TP0548 | 23S rRNA genee | Strain similarity | No. of patients genotypedf |
|---|---|---|---|---|---|
| Completely typed patients (n = 40) | |||||
| SSS | Identical to SS14 | Identical to SS14 | Sensitive | SS14-likeb | 9 |
| SSR8 | Identical to SS14 | Identical to SS14 | A2058G | SS14-like | 2 |
| SSR9 | Identical to SS14 | Identical to SS14 | A2059G | SS14-like | 2 |
| SU2R8 | Identical to SS14 | Unique 2 | A2058G | SS14-like | 15 |
| SU2Sd | Identical to SS14 | Unique 2 | Sensitive | SS14-like | 1 |
| SU5R8d | Identical to SS14 | Unique 5 | A2058G | SS14-like | 6 |
| SU7Sd | Identical to SS14 | Unique 7 | Sensitive | SS14-like | 3 |
| U3U6R8d | Unique 3 | Unique 6 | A2058G | Nichols-likec | 2 |
| Partially typed patients (n = 22) | |||||
| SSX | Identical to SS14 | Identical to SS14 | Not determined | SS14-likeb | 1 |
| SXS | Identical to SS14 | Not determined | Sensitive | SS14-like | 3 |
| SXX | Identical to SS14 | Not determined | Not determined | SS14-like | 2 |
| XSS | Not determined | Identical to SS14 | Sensitive | SS14-like | 3 |
| XSX | Not determined | Identical to SS14 | Not determined | SS14-like | 1 |
| SXR8 | Identical to SS14 | Not determined | A2058G | SS14-like | 1 |
| XSR9 | Not determined | Identical to SS14 | A2059G | SS14-like | 2 |
| SU2X | Identical to SS14 | Unique 2 | Not determined | SS14-like | 1 |
| XU2X | Not determined | Unique 2 | Not determined | SS14-like | 1 |
| XU2R8 | Not determined | Unique 2 | A2058G | SS14-like | 2 |
| XU8R9d | Not determined | Unique 8 | A2059G | SS14-like | 1 |
| U3XR8 | Unique 3 | Not determined | A2058G | Nichols-likec | 1 |
| U3XX | Unique 3 | Not determined | Not determined | Nichols-like | 1 |
| XU6R8 | Not determined | Unique 6 | A2058G | SS14-like | 2 |
Genotypes were assigned according to a previous study in the Czech Republic (5) by a three-letter code, where the first letter stands for the TP0136 sequence (S, identical to SS14; U3, unique sequence 3; X, not determined), the second letter stands for the TP0548 sequence (S, identical to SS14; U2 and U5 to U8, unique sequences; X, not determined), and the third letter stands for sensitivity to macrolide antibiotics (S, sensitive; R8, mutation A2058G; R9, mutation A2059G).
Sequences of the TP0136 and TP0548 loci are identical or related to those of T. pallidum subsp. pallidum strain SS14.
Sequences of the TP0136 and TP0548 loci are related to those of T. pallidum subsp. pallidum strain Nichols.
New genotypes that were not detected in the Czech Republic in previous years.
We analyzed macrolide resistance data separately as described in the last paragraph of Results.
Total n = 62.
FIG 2.
Alignment of unique sequences of the TP0136 and TP0548 loci of clinical isolates collected in the Czech Republic between 2004 and 2013 (5; this study). Only positions showing nucleotide differences (indels, substitutions) are shown. Coordinates correspond to TP0136 and TP0548 in the T. pallidum subsp. pallidum (TPA) SS14 genome (accession no. CP004011) (37). Nucleotide differences shown on a gray background indicate differences between the SS14-like and Nichols-like groups of strains. Nucleotide changes shown on black and white backgrounds represent unique differences found in clinical isolates.
Association of T. pallidum subsp. pallidum genotypes with patient characteristics.
The relationship between genotypes and patient characteristics (i.e., the association between genotypes and patient age and sex, geographical origin, MSM [males who have sex with males] status, HIV status, or stage of syphilis) was tested (Table 3). The SU2R8 genotype was more common among older patients and samples from Prague. The SU5R8 genotype appeared only in samples from the Brno area. MSM patients were found to be infected more often with the SU2R8 genotype and less frequently with the SSS genotype. Additionally, MSM patients were infected more often than non-MSM patients with treponemes containing a macrolide resistance-associated mutation (A2058G or A2059G) (Table 3).
TABLE 3.
Associations between genotypes/alleles and patient age, origin of samples, and MSM statusa
| Parameter | No. of patients with following genotype/total no. analyzed (P value): |
|||
|---|---|---|---|---|
| SU2R8 | SU5R8 | SSS | A2058G or A2059G | |
| Age (yr) | ||||
| <35 | 7/49 | |||
| ≥35 | 14/40 (0.0263) | |||
| Origin | ||||
| Brno | 5/39 | 6/39 | ||
| Prague | 16/49 (0.0433) | 0/49 (0.006) | ||
| MSM statusb | ||||
| MSM | 10/15 | 1/15 | 26/29 | |
| Non-MSMc | 2/13 (0.0093) | 6/13 (0.0286) | 7/21 (<0.0001) | |
The data presented here are data from this study combined with previously published data (5). Only statistically significant results are shown.
Association of MSM status and other characteristics; only samples collected from 2011 to 2013 were used.
The non-MSM group comprised heterosexual men and women.
Prevalence of macrolide resistance-causing mutations in syphilis-causing treponemes and association of macrolide resistance with TP0136 and TP0548 alleles.
The 23S rRNA gene locus was amplified in samples isolated from 69 patients, and in 46 of them (66.7%), either the A2058G (39 patients, 56.5%) or the A2059G (7 patients, 10.1%) mutation was found. No strains harbored both the A2058G and A2059G mutations. Only four of the isolates tested were unique at the TP0136 locus (U3), and no associations were found. The U2, U5, and U6 alleles of TP0548 were found to be associated with macrolide resistance mutations (P > 0.0001, P = 0.0022, and P = 0.0286, respectively). In contrast, the S allele of TP0548 was associated with macrolide susceptibility. The A2059G mutation was detected in only two genotypes (SSR9 and XU8R9) and only in MSM patients.
DISCUSSION
As in our previous study (5), the relatively high number of PCR-negative specimens (45%) from syphilis-seropositive patients likely reflects undetectable amounts of treponemal DNA in biological material, especially in blood samples. However, the use of larger volumes of whole blood for isolation of DNA could potentially help increase the rate of PCR-positive blood samples. In addition, the possibility of self-medication by patients with syphilis cannot be excluded as an explanation for the failure to find T. pallidum subsp. pallidum DNA (5). No statistically significant differences between the RPR titers of the PCR-positive and PCR-negative samples were observed among syphilis-seropositive patients. However, a trend toward higher RPR titers in PCR-negative patients was found, suggesting a stronger humoral response in PCR-negative patients. The highest rate of treponemal DNA detection in this study was found in genital skin swabs (76.7%). This finding is in agreement with our previous study (5) and with the observations of others (12–14). The highest number of PCR-positive samples was found at the polA locus, followed by the tmpC, 23S rRNA, TP0548, and TP0136 loci; PCR amplification efficacy likely reflects the length of the PCR product in the first step of the nested PCR (637, 773, 1,666/1,658, 1,567, and 1,786 bp, respectively), indicating that smaller fragments are amplified more easily. The only exception was the 23S rRNA gene; however, this gene is present twice in the T. pallidum subsp. pallidum genome. Therefore, improved versions of detection systems should try to keep the target DNA length to a minimum. The most variable region among clinical isolates was found in the TP0548 gene between coordinates 170 and 208 and coordinates 154 and 772 of the Nichols-like and SS14-like isolates, respectively (15). The enhanced CDC typing system covers the entire highly variable region of TP0548 of Nichols-like isolates but only a portion of the highly variable regions of SS14-like isolates. These regions should therefore be included in improved versions of molecular typing schemes.
Until now, the enhanced CDC typing system identified 10 different versions of TP0548 (4, 16) among 809 treponemal samples tested (4, 16–21). Combined with the results of the previously published study, nine different sequence versions of the TP0548 have been identified in the Czech Republic since 2004 among 119 treponemal samples tested (5; this study). In contrast to the enhanced CDC typing system, which determines an 83-bp sequence of the TP0548 locus (positions 130 to 212 in TP0548 of T. pallidum subsp. pallidum Nichols [accession no. CP004010]), the sequencing-based typing system relies on the sequencing of 1,065 bp of the TP0548 locus. With our sequencing-based typing system, the most common enhanced CDC strain, type f, can be further divided into four separate types (S, U1, U4, U8). Similarly, strain type g can be divided into three groups (U2, U3, U5). The U6 type is identical to strain type d. In addition, unique sequence 7 (U7) corresponded to a new, previously undescribed type. On the basis of the enhanced CDC typing system nomenclature, this type can be classified under the next available letter in the alphabet, which in this case makes it strain type k. The relationships between enhanced CDC TP0548 types and sequencing-based types are shown in Table 4.
TABLE 4.
TP0548 types according to enhanced CDC typing and sequencing-based molecular typing
| TP0548 typea | TP0548 type(s)b | Relationship to either T. pallidum subsp. pallidum Nichols or SS14 |
|---|---|---|
| a | —c | Nichols-like |
| b | — | |
| c | — | |
| d | U6 | |
| e | — | SS14-like |
| f | S, U1, U4, U8 | |
| g | U2, U3, U5 | |
| h | — | Nichols-like |
| i | — | SS14-like |
| j | — | TPE-liked |
| ke | U7 | SS14-like |
According to sequencing-based molecular typing. The data are from reference 5 and this study.
—, not found in the Czech Republic.
Similar to T. pallidum subsp. pertenue strains.
The U7 type can be classified as a new strain of type k according to the enhanced CDC typing system.
Analyses of TP0136 in this study revealed one sequence identical to that of T. pallidum subsp. pallidum SS14 and one unique sequence (U3) similar to the sequence of T. pallidum subsp. pallidum Nichols. When combined with previous results, four different alleles (S, U1 to U3) of TP0136 have been found in the Czech Republic since 2004. Compared to the TP0548 locus, which revealed nine different alleles (S, U1 to U8) since 2004, TP0136 appears to have lower discrimination power than previously suggested (5, 8, 22, 23). A lower discrimination power of this locus was also found by Marra et al. (4). However, the overall discrimination power of the TP0136 locus likely depends on the particular geographical area under study. Therefore, to comprehensively asses its suitability for typing, samples isolated from different geographical regions should also be tested.
Several genotypes were found to be associated with patient age, origin of samples, and MSM status. While the genotypes associated with patient age and MSM status likely reflect the existence of several epidemiologically distinct subpopulations in the Czech Republic, genotypes associated with the geographic origin of samples likely represent local epidemics caused by the rapid spread of a particular strain. The U2, U5, and U6 alleles of TP0548 were found to be associated with macrolide resistance, and the S allele was associated with macrolide susceptibility. The association between the TP0548 gene variant and resistance to macrolides was not exclusive, since we also found the U2 variant, which has been reported in both resistant (SU2R8) (5; this study) and sensitive (SU2S) (this study) isolates. Similarly, the S variant of the TP0548 gene was found with 23S rRNA gene sequences encoding both macrolide susceptibility (SSS) and resistance (SSR8, SSR9).
Recently, syphilis-causing (T. pallidum subsp. pallidum) strains were described as forming two genetically distinct groups, including Nichols-like and SS14-like strains (15). An analysis of a number of independent studies showed that more than 94% of clinical isolates belong to the SS14 group while about half of reference T. pallidum subsp. pallidum strains are Nichols-like strains. Similarly, most (>90%) of the sequences of the clinical isolates investigated in this study were related to those of the SS14-like strain. The Nichols-like strain was first detected in the Czech Republic in 2012, and since then, six Nichols-like isolates have been identified, which is in contrast to a previous study (5) in which no Nichols-like isolates were noticed. Interestingly, one of the patients infected with a Nichols-like strain reported sexual contacts during his stay in Mexico, which suggests that this treponemal strain may have been imported from abroad.
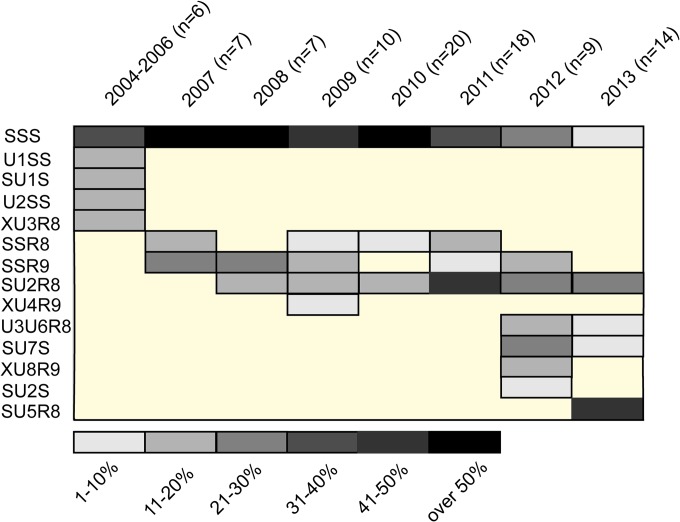
Several studies have shown that predominant treponemal strains in a particular population can change over time (4, 5, 18, 19). The same pattern was also observed in this study (Fig. 3). While the SSS genotype was predominant from 2004 to 2010, it was replaced by the SU2R8 genotype in 2011 (Fig. 3). Another change in the distribution of strains was observed in 2013, when the most prevalent genotype became SU5R8. Moreover, the number of genotypes detected increased in 2012 and 2013 to six, the largest number of genotypes since 2004. It is not clear if this finding correlates with increased sexual tourism or with other factors. During the entire 2004-to-2013 period, 14 different T. pallidum subsp. pallidum genotypes were identified. Several genotypes were detected only in a particular year and have never been detected again, while other genotypes (i.e., SSR8 and SSR9) declined and disappeared over a period of years. In general, the spectrum of treponemal strains present in a particular population seems to be surprisingly dynamic (Fig. 3).
FIG 3.
Distribution of T. pallidum subsp. pallidum genotypes in the Czech Republic from 2004 to 2013. Data for 2004 to 2010 are from a previous study performed in the Czech Republic (5). Eleven completely typed genotypes and three partially typed genotypes (XU3R8, XU4R9, XU8R9) with unique TP0548 sequences are shown. Allele numbering reflects not the year of isolation but the date of sequence analysis; therefore, genotype XU3R8 appeared earlier than genotype SU2R8. Similarly, genotype SU5R8 appeared later than genotypes U3U6R8, SU7S, and XU8R9. Note that 14 different genotypes of T. pallidum subsp. pallidum isolates were identified in the Czech Republic from 2004 to 2013. The number of genotypes detected in a particular year increased over time, while some genotypes disappeared over time. The relative abundance of genotypes in a particular year is color coded. Three of the 91 genotyped T. pallidum subsp. pallidum isolates represented were from patients in the early latent stage of syphilis. All of these genotypes are shown 1 year before the diagnosis of the disease to reflect the prevalence of the genotypes in a particular year.
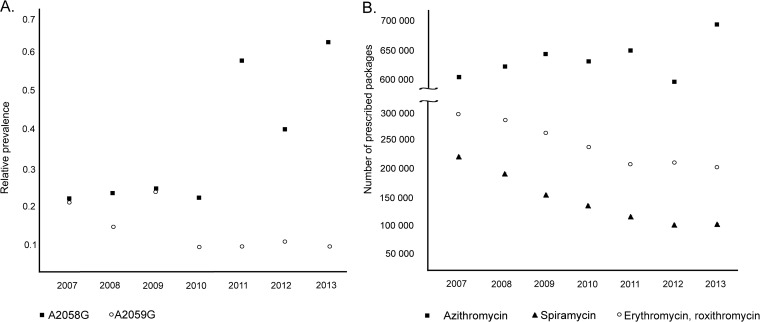
A rapid increase in the prevalence of T. pallidum strains resistant to macrolide antibiotics was found in the Czech Republic from 2011 to 2013 (66.7%) compared to previous years (2004 to 2010; 37%) (5). While the A2058G mutation was observed more frequently, the prevalence of the A2059G mutation appears to be relatively low and to be starting to decrease (Fig. 4A). A high prevalence of resistance to macrolides has been reported by a number of reports from China; the United States (especially in San Francisco and Seattle); Dublin, Ireland; and London, the United Kingdom (3, 6, 17, 19, 24–26). Moreover, an increasing trend in macrolide resistance has also been described in studies by Grimes et al. (19), Marra et al. (27), and Mitchell et al. (28). In contrast to these data, no macrolide resistance-causing mutations were found during analyses of the 23S rRNA gene locus in Madagascar (29) and Taiwan (21). As first predicted by Marra et al. (27), macrolide-resistant isolates appear in infected populations because of antibiotic pressure. Since that time, additional studies have reported that the prevalence of macrolide resistance mutations in T. pallidum is higher among patients who have taken macrolide antibiotics before and during syphilis treatment than among those who have not (19, 24).
FIG 4.
(A) Prevalence of the A2058G and A2059G mutations in treponemal 23S rRNA genes isolated from clinical samples from syphilis patients in the Czech Republic from 2007 to 2013. Data from 2007 to 2010 are from a previous study in the Czech Republic (5). While the prevalence of A2058G appears to be increasing, the prevalence of A2059G is relatively low and appears to be decreasing. The relative prevalence of mutations in the 23S rRNA gene locus since 2007 is shown because no isolates harboring these mutations were found in 2004 or 2005. In 2006, only one isolate harboring A2058G was identified. (B) Macrolide antibiotic use in the Czech Republic from 2007 to 2013. Data were provided by the State Institute for Drug Control in the Czech Republic. Antibiotic use is shown as the number of medical product packages representing the dose most frequently used for the treatment of a single patient.
The A2058G mutation encodes resistance to azithromycin, clarithromycin, erythromycin, and roxithromycin but not resistance to spiramycin (30–32). In contrast, A2059G is likely associated with resistance to all commonly used macrolides, including spiramycin (10). According to data provided by the Czech Republic State Institute for Drug Control (J. Židlická, personal communication), spiramycin use is consistently low and decreasing (Fig. 4B). In contrast, azithromycin use increased from 2007 to 2013 in the Czech Republic (Fig. 4B). Since it has been shown that mutations causing resistance to macrolides occur in multiple genotypes (19, 27, 33; this study), the occurrence of the A2058G and A2059G mutations could reflect the opposite trends in spiramycin and azithromycin use in the Czech Republic. Several macrolide-resistant bacterial strains have been shown to lose their macrolide resistance during subcultivation without antibiotics (34); nevertheless, this finding has not yet been reported in T. pallidum. Moreover, the decreasing occurrence of the A2059G mutation could reflect its higher fitness cost than that of the A2058G mutation in treponemes. Different fitness costs imposed by the A2058C and A2059C mutations in the 23S rRNA gene have been observed in Chlamydia caviae (35).
While the macrolide resistance phenotypes appear to be stable (36), the stability of the wild-type 23S rRNA gene is not known in treponemes. In our previous typing study, we documented transfers of macrolide-sensitive strains in five individual transmissions of T. pallidum strains from one patient to another during a time period of up to 31 days (5). In all of the cases analyzed, no changes were found in the 23S rRNA gene locus. Further analysis of the genotype associated with either the A2058G or the A2059G mutation could allow an estimate of the genetic stability of the wild-type 23S rRNA gene.
The study described in this communication has several limitations. Although the percentage of PCR-positive swab and whole-blood specimens from syphilis-seropositive patients (55%) was higher than in a previous study (44.7%) (5), there is still a reasonably high number of PCR-negative samples from those who were syphilis seropositive. In addition, the sexual contacts of patients remained unidentified since most of the patients claimed to have had anonymous sex. Moreover, there was a low but reasonable chance of a repeated collection of a clinical isolate from patients with relapsed syphilis.
In summary, nine different genotypes were identified among 81 typeable samples from 62 PCR-positive patients within the time scope of this study (2011 to 2013). Compared to a previous study in the Czech Republic, the number of genotypes detected among syphilis patients climbed to six in both 2012 and 2013, although they were not the same six. The distribution of genotypes has changed over time. In 2012, the first treponemal strain related to T. pallidum subsp. pallidum Nichols was identified. In addition, the number of macrolide-resistant clinical isolates also increased and was 66.7% at the time of this study.
ACKNOWLEDGMENTS
We thank Jitka Židlická for providing data regarding antibiotic consumption in the Czech Republic.
This work was supported by the Ministry of Health of the Czech Republic (NT11159-5/2010 to D.Š.) and the Grant Agency of the Czech Republic (310/07/0321 to D.Š.).
Footnotes
Published ahead of print 6 August 2014
REFERENCES
- 1.Hay PE, Clarke JR, Strugnell RA, Taylor-Robinson D, Goldmeier D. 1990. Use of the polymerase chain reaction to detect DNA sequences specific to pathogenic treponemes in according to the manufacturer fluid. FEMS Microbiol. Lett. 56:233–238 [DOI] [PubMed] [Google Scholar]
- 2.Pillay A, Liu H, Ebrahim S, Chen CY, Lai W, Fehler G, Ballard RC, Steiner B, Sturm AW, Morse SA. 2002. Molecular typing of Treponema pallidum in South Africa: cross-sectional studies. J. Clin. Microbiol. 40:256–258. 10.1128/JCM.40.1.256-258.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katz KA, Pillay A, Ahrens K, Kohn RP, Hermanstyne K, Bernstein KT, Ballard RC, Klausner JD. 2010. Molecular epidemiology of syphilis—San Francisco, 2004-2007. Sex. Transm. Dis. 37:660–663. 10.1097/OLQ.0b013e3181e1a77a [DOI] [PubMed] [Google Scholar]
- 4.Marra CM, Sahi SK, Tantalo LC, Godornes C, Reid T, Behets F, Rompalo A, Klausner JD, Yin Y, Mulcahy F, Golden MR, Centurion-Lara A, Lukehart SA. 2010. Enhanced molecular typing of Treponema pallidum: geographical distribution of strain types and association with neurosyphilis. J. Infect. Dis. 202:1380–1388. 10.1086/656533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flasarová M, Pospíšilová P, Mikalová L, Vališová Z, Dastychová E, Strnadel R, Kuklová I, Woznicová V, Zákoucká H, Šmajs D. 2012. Sequencing-based molecular typing of Treponema pallidum strains in the Czech Republic: all identified genotypes are related to the sequence of the SS14 strain. Acta Derm. Venereol. 92:669–674. 10.2340/00015555-1335 [DOI] [PubMed] [Google Scholar]
- 6.Lukehart SA, Godornes C, Molini BJ, Sonnett P, Hopkins S, Mulcahy F, Engelman J, Mitchell SJ, Rompalo AM, Marra CM, Klausner JD. 2004. Macrolide resistance in Treponema pallidum in the United States and Ireland. N. Engl. J. Med. 351:154–158. 10.1056/NEJMoa040216 [DOI] [PubMed] [Google Scholar]
- 7.Mikalová L, Pospíšilová P, Woznicová V, Kuklová I, Zákoucká H, Šmajs D. 2013. Comparison of CDC and sequence-based molecular typing of syphilis treponemes: tpr and arp loci are variable in multiple samples from the same patient. BMC Microbiol. 13:178. 10.1186/1471-2180-13-178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flasarová M, Šmajs D, Matĕjková P, Woznicová V, Heroldová-Dvořáková M, Votava M. 2006. Molecular detection and subtyping of Treponema pallidum subsp. pallidum in clinical specimens. Epidemiol. Mikrobiol. Immunol. 55:105–111 (In Czech.) [PubMed] [Google Scholar]
- 9.Stamm LV, Bergen HL. 2000. A point mutation associated with bacterial macrolide resistance is present in both 23S rRNA genes of an erythromycin-resistant Treponema pallidum clinical isolate. Antimicrob. Agents Chemother. 44:806–807. 10.1128/AAC.44.3.806-807.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matĕjková P, Flasarová M, Zákoucká H, Borek M, Kremenová S, Arenberger P, Woznicová V, Weinstock GM, Šmajs D. 2009. Macrolide treatment failure in a case of secondary syphilis: a novel A2059G mutation in the 23S rRNA gene of Treponema pallidum subsp. pallidum. J. Med. Microbiol. 58:832–836. 10.1099/jmm.0.007542-0 [DOI] [PubMed] [Google Scholar]
- 11.Goyette P, Sumner JS, Milos R, Duncan AM, Rosenblatt DS, Matthews RG, Rozen R. 1994. Human methylenetetrahydrofolate reductase: isolation of cDNA, mapping and mutation identification. Nat. Genet. 7:195–200. 10.1038/ng0694-195 [DOI] [PubMed] [Google Scholar]
- 12.Bruisten SM. 2012. Protocols for detection and typing of Treponema pallidum using PCR methods. Methods Mol. Biol. 903:141–167. 10.1007/978-1-61779-937-2_9 [DOI] [PubMed] [Google Scholar]
- 13.Gayet-Ageron A, Ninet B, Toutous-Trellu L, Lautenschlager S, Furrer H, Piguet V, Schrenzel J, Hirschel B. 2009. Assessment of a real-time PCR test to diagnose syphilis from diverse biological samples. Sex. Transm. Infect. 85:264–269. 10.1136/sti.2008.034314 [DOI] [PubMed] [Google Scholar]
- 14.Gayet-Ageron A, Lautenschlager S, Ninet B, Perneger TV, Combescure C. 2013. Sensitivity, specificity and likelihood ratios of PCR in the diagnosis of syphilis: a systematic review and meta-analysis. Sex. Transm. Infect. 89:251–256. 10.1136/sextrans-2012-050622 [DOI] [PubMed] [Google Scholar]
- 15.Nechvátal L, Pĕtrošová H, Grillová L, Pospíšilová P, Mikalová L, Strnadel R, Kuklová I, Kojanová M, Kneidlová M, Vaňousová D, Procházka P, Zákoucká H, Krchňáková A, Šmajs D. 2014. Syphilis-causing strains belong to separate SS14-like or Nichols-like groups as defined by multilocus analysis of 19 Treponema pallidum strains. Int. J. Med. Microbiol. 14:43–45. 10.1016/j.ijmm.2014.04.007 [DOI] [PubMed] [Google Scholar]
- 16.Grange PA, Allix-Beguec C, Chanal J, Benhaddou N, Gerhardt P, Morini JP, Deleuze J, Lassau F, Janier M, Dupin N. 2013. Molecular subtyping of Treponema pallidum in Paris, France. Sex. Transm. Dis. 40:641–644. 10.1097/OLQ.0000000000000006 [DOI] [PubMed] [Google Scholar]
- 17.Tipple C, McClure MO, Taylor GP. 2011. High prevalence of macrolide resistant Treponema pallidum strains in a London centre. Sex. Transm. Infect. 87:486–488. 10.1136/sextrans-2011-050082 [DOI] [PubMed] [Google Scholar]
- 18.Dai T, Li K, Lu H, Gu X, Wang Q, Zhou P. 2012. Molecular typing of Treponema pallidum: a 5-year surveillance in Shanghai, China. J. Clin. Microbiol. 50:3674–3677. 10.1128/JCM.01195-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grimes M, Sahi SK, Godornes BC, Tantalo LC, Roberts N, Bostick D, Marra CM, Lukehart SA. 2012. Two mutations associated with macrolide resistance in Treponema pallidum: increasing prevalence and correlation with molecular strain type in Seattle, Washington. Sex. Transm. Dis. 39:954–958. 10.1097/OLQ.0b013e31826ae7a8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng RR, Yin YP, Wei WH, Wang HC, Zhu BY, Liu QZ, Zheng HP, Zhang JP, Huang SJ, Chen XS. 2012. Molecular typing of Treponema pallidum causing early syphilis in China: a cross-sectional study. Sex. Transm. Dis. 39:42–45. 10.1097/OLQ.0b013e318232697d [DOI] [PubMed] [Google Scholar]
- 21.Wu H, Chang SY, Lee NY, Huang WC, Wu BR, Yang CJ, Liang SH, Lee CH, Ko WC, Lin HH, Chen YH, Liu WC, Su YC, Hsieh CY, Wu PY, Hung CC. 2012. Evaluation of macrolide resistance and enhanced molecular typing of Treponema pallidum in patients with syphilis in Taiwan: a prospective multicenter study. J. Clin. Microbiol. 50:2299–2304. 10.1128/JCM.00341-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brinkman MB, McGill MA, Pettersson J, Rogers A, Matĕjková P, Šmajs D, Weinstock GM, Noriss SJ, Palzkill T. 2008. A novel Treponema pallidum antigen, TP0136, is an outer membrane protein that binds human fibronectin. Infect. Immun. 76:1848–1857. 10.1128/IAI.01424-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woznicová V, Šmajs D, Wechsler D, Matĕjková P, Flasarová M. 2007. Detection of Treponema pallidum subsp. pallidum from skin lesions, serum, and according to the manufacturer fluid in an infant with congenital syphilis after clindamycin treatment of the mother during pregnancy. J. Clin. Microbiol. 45:659–661. 10.1128/JCM.02209-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X-S, Yin Y-P, Wei W-H, Wang H-C, Peng R-R, Zheng HP, Zhang JP, Zhu BY, Liu QZ, Huang SJ. 2013. High prevalence of azithromycin resistance to Treponema pallidum in geographically different areas in China. Clin. Microbiol. Infect. 19:975–979. 10.1111/1469-0691.12098 [DOI] [PubMed] [Google Scholar]
- 25.Muldoon EG, Walsh A, Crowley B, Mulcahy F. 2012. Treponema pallidum azithromycin resistance in Dublin, Ireland. Sex. Transm. Dis. 39:784–786. 10.1097/OLQ.0b013e318269995f [DOI] [PubMed] [Google Scholar]
- 26.A2058G Prevalence Workgroup. 2012. Prevalence of the 23S rRNA A2058G point mutation and molecular subtypes in Treponema pallidum in the United States, 2007 to 2009. Sex. Transm. Dis. 39:794–798 [DOI] [PubMed] [Google Scholar]
- 27.Marra CM, Colina AP, Godornes C, Tantalo LC, Puray M, Centurion-Lara A, Lukehart SA. 2006. Antibiotic selection may contribute to increases in macrolide-resistant Treponema pallidum. J. Infect. Dis. 194:1771–1773. 10.1086/509512 [DOI] [PubMed] [Google Scholar]
- 28.Mitchell SJ, Engelman J, Kent CK, Lukehart SA, Godornes C, Klausner JD. 2006. Azithromycin-resistant syphilis infection: San Francisco, California, 2000-2004. Clin. Infect. Dis. 42:337–345. 10.1086/498899 [DOI] [PubMed] [Google Scholar]
- 29.Van Damme K, Behets F, Ravelomanana N, Godornes C, Khan M, Randrianasolo B, Rabenja NL, Lukehart SA, Cohen M, Hook E. 2009. Evaluation of azithromycin resistance in Treponema pallidum specimens from Madagascar. Sex. Transm. Dis. 36:775–776. 10.1097/OLQ.0b013e3181bd11dd [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stamm LV, Stapleton JT, Bassford PJ. 1988. In vitro assay to demonstrate high-level erythromycin resistance of a clinical isolate of Treponema pallidum. Antimicrob. Agents Chemother. 32:164–169. 10.1128/AAC.32.2.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stamm LV, Parrish EA. 1990. In-vitro activity of azithromycin and CP-63,956 against Treponema pallidum. J. Antimicrob. Chemother. 25:11–14. 10.1093/jac/25.suppl_A.11 [DOI] [PubMed] [Google Scholar]
- 32.Woznicová V, Matĕjková P, Flasarová M, Zákoucká H, Valisová Z, Šmajs D, Dastychová E. 2010. Clarithromycin treatment failure due to macrolide resistance in Treponema pallidum in a patient with primary syphilis. Acta Derm. Venereol. 90:206–207. 10.2340/00015555-0774 [DOI] [PubMed] [Google Scholar]
- 33.Martin IE, Gu W, Yang Y, Tsang RS. 2009. Macrolide resistance and molecular types of Treponema pallidum causing primary syphilis in Shanghai, China. Clin. Infect. Dis. 49:515–521. 10.1086/600878 [DOI] [PubMed] [Google Scholar]
- 34.Alarcón T, Domingo D, Prieto N, Lópéz-Brea B. 2000. Clarithromycin resistance stability in Helicobacter pylori: influence of the MIC and type of mutation in the 23S rRNA. J. Antimicrob. Chemother. 46:613–616. 10.1093/jac/46.4.613 [DOI] [PubMed] [Google Scholar]
- 35.Binet R, Bowlin AK, Maurelli AT, Rank RG. 2010. Impact of azithromycin resistance mutations on the virulence and fitness of Chlamydia caviae in guinea pigs. Antimicrob. Agents Chemother. 54:1094–1101. 10.1128/AAC.01321-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stamm LV. 2010. Global challenge of antibiotic-resistant Treponema pallidum. Antimicrob. Agents Chemother. 54:583–589. 10.1128/AAC.01095-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pĕtrošová H, Pospíšilová P, Strouhal M, Čejková D, Zobaníková M, Mikalová L, Sodergren E, Weinstock GM, Šmajs D. 2013. Resequencing of Treponema pallidum ssp. pallidum strains Nichols and SS14: correction of sequencing errors resulted in increased separation of syphilis treponeme subclusters. PLoS One 8(9):74319. 10.1371/journal.pone.0074319 [DOI] [PMC free article] [PubMed] [Google Scholar]