Abstract
Fifteen bacterial isolates from spotted fever group rickettsiosis in Brazil were genetically identified as Rickettsia rickettsii. In a phylogenetic analysis with other R. rickettsii isolates from GenBank, the Central/South American isolates showed low polymorphism and formed a clade distinct from two North American clades, with the North American clades having greater in-branch polymorphism.
TEXT
The tick-borne bacterium Rickettsia rickettsii is the etiologic agent of Rocky Mountain spotted fever (RMSF), the most severe rickettsiosis affecting humans in the Western Hemisphere (1). In Brazil, the disease has been referred to as Brazilian spotted fever (BSF). While RMSF fatality rates are usually 5% to 10% in the United States, general rates of 20% to 40% have been reported in Brazil (1).
In the state of São Paulo in southeastern Brazil, where BSF is a disease for which notification is compulsory, various spotted fever group (SFG) rickettsial isolates have been obtained from human clinical cases during the past few years (2, 3). Although all these isolates were confirmed to be SFG rickettsiae through indirect immunofluorescence assays using anti-R. rickettsii human sera (4), they were not genetically identified through molecular analysis. Herein, we performed genetic identification and molecular characterization of these rickettsial isolates and compared their genetic profiles with those of isolates from ticks in Brazil and with those of R. rickettsii isolates from other American countries.
Blood clot or skin lesion biopsy specimens from BSF-suspected cases were processed by the shell vial technique, as described previously (5). Once rickettsiae were visualized within Vero cells through an immunofluorescence assay using anti-R. rickettsii human polyclonal serum (4, 6), 1st-passage-infected cells were harvested, and one aliquot was used for DNA extraction through the PureLink genomic DNA kit (Invitrogen, Carlsbad, CA). Extracted DNA was assayed according to PCR protocols using primers targeting the rickettsial genes gltA, ompA, and ompB and the RR0155-rpmB, RR1240-tlc5b, and cspA-ksgA intergenic regions, as shown in Table 1. In addition, DNA extracted from 3rd-passage-infected cells of three R. rickettsii isolates, previously isolated from Amblyomma sculptum (reported as Amblyomma cajennense), Amblyomma aureolatum, and Rhipicephalus sanguineus ticks in Brazil (12–14), was tested according to intergenic region PCR protocols. PCR products were sequenced in an ABI automated sequencer (model ABI 3500 Genetic Analyzer; Applied Biosystems/Thermo Fisher Scientific, Foster City, CA) with the same primers used for PCR. The generated sequences were compared with each other and submitted to BLAST analyses (www.ncbi.nlm.nih.gov/blast) to infer the closest similarities available in GenBank.
TABLE 1.
Primer pairs used for amplification of rickettsial genes (gltA, ompA, and ompB) or intergenic regions (RR0155-rpmB, RR1240-tlc5b, and cspA-ksgA)
| Primer pair | Target | Primer | Primer sequence (5′ to 3′) | Amplicon size (bp) | Reference |
|---|---|---|---|---|---|
| 1 | gltA | CS-239 | GCTCTTCTCATCCTATGGCTATTAT | 834 | 7 |
| CS-1069 | CAGGGTCTTCGTGCATTTCTT | 7 | |||
| 2 | ompA | Rr190.70p | ATGGCGAATATTTCTCCAAAA | 530 | 8 |
| Rr190.602n | AGTGCAGCATTCGCTCCCCCT | 8 | |||
| 3 | ompB | 120-M59 | CCGCAGGGTTGGTAACTGC | 862 | 9 |
| 120–807 | CCTTTTAGATTACCGCCTAA | 9 | |||
| 4 | RR0155-rpmB | Forward | TTTCTAGCAGCGGTTGTTTTATCC | 290 | 10 |
| Reverse | TTAGCCCATGTTGACAGGTTTACT | 10 | |||
| 5 | RR1240-tlc5b | Forward | CGGGATAACGCCGAGTAATA | 357 | 11 |
| Reverse | ATGCCGCTCTGAATTTGTTT | 11 | |||
| 6 | cspA-ksgA | Forward | CATCACTGCTTCGCTTATTTT | 405 | 10 |
| Reverse | ATTTCTTTTCTTCCTCTTCATCAA | 10 |
Phylogenetic analyses were performed using PAUP version 4.0b10 (15) to maximum parsimony (MP); the confidence values for individual branches of the resulting tree were determined by bootstrap analysis with 1,000 replicates. Bayesian analysis (BA) was performed with MrBayes version 3.1.2 (16) software with 1,000,000 generations using the GTR+I+G substitution model. Partial DNA sequences obtained from the amplified PCR products (gltA, RR0155-rpmB, RR1240-tlc5b, cspA-ksgA, and ompB) were concatenated and aligned with corresponding sequences of different strains of R. rickettsii available in GenBank using CLUSTAL X (17) and adjusted manually using GeneDoc (18). Partial sequences of the ompA gene were not included because the region of the gene that was amplified in the present study showed no polymorphism among the R. rickettsii isolates. Corresponding sequences of R. rickettsii strain Hlp#2 (CP003311) and Rickettsia philipii strain 364D (CP003308) were used as outgroups.
Fifteen rickettsial isolates (designated IAL 1 to 15) from BSF patients (13 fatal cases) in the state of São Paulo, southeastern Brazil, were identified as R. rickettsii, since their gltA (737 nt), ompA (491 nt), and ompB (787 nt) DNA sequences were 100% identical to each other and to corresponding sequences on the genome of the R. rickettsii strain Brazil (GenBank accession no. CP003305). While the ompA partial sequences were also 100% identical to corresponding sequences of R. rickettsii strains from North America (e.g., CP000848 and CP000766), the ompB partial sequences were 100% identical to that of the North American Sheila Smith strain (CP000848) and, at the same time, differed from those of other North American strains (e.g., CP000766 and CP003307) by one single nucleotide polymorphism. The gltA sequences of the 15 Brazilian human isolates differed by an extra codon (CGG) compared to those of several North American strains, such as Sheila Smith (CP000848) and Bitterroot (U59729). This extra codon was also present in the R. rickettsii tick isolates (Taiaçu, Itu, and Rs1) from Brazil (12–14). The sequences of three intergenic regions (249 nucleotides [nt] for RR0155-rpmB, 315 nt for RR1240-tlc5b, and 362 nt for cspA-ksgA) were determined for the 15 human isolates and for the three tick isolates (Taiaçu, Itu, and Rs1) from Brazil. For each intergenic region, the sequences were 100% identical to each other (no polymorphisms were detected), and when subjected to BLAST analysis, they were 100% identical to the corresponding sequences of the R. rickettsii strain Brazil (CP003305).
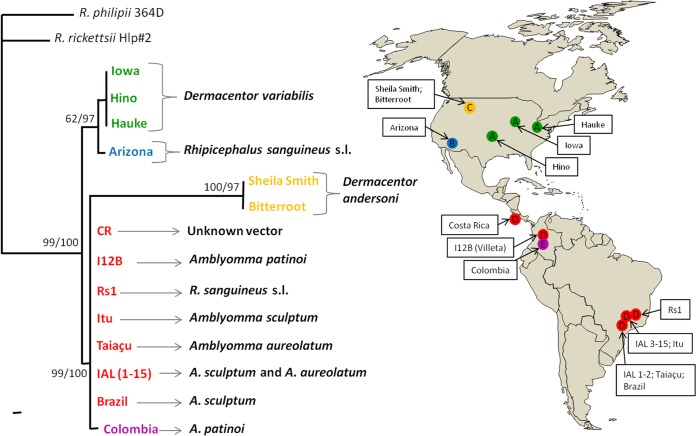
For the concatenated phylogenetic analysis, which included a total of 2,392 nt, the sequences of the 15 human isolates (IAL 1 to 15) and 3 tick isolates (Taiaçu, Itu, and Rs1) from Brazil were aligned with the corresponding sequences from 10 other R. rickettsii isolates available in GenBank (6 from the United States, 1 from Costa Rica, 2 from Colombia, and 1 from Brazil) (Table 2). The 15 human and 3 tick isolates of R. rickettsii from Brazil formed a clade under high bootstrap support (99% to 100%), with all three South American isolates available in GenBank (1 from Brazil and 2 from Colombia) and with the Central American isolate from Costa Rica (Fig. 1). This Central/South American clade, ecologically associated with 4 different tick species (namely, A. aureolatum, Amblyomma patinoi, A. sculptum, and R. sanguineus) (Table 2), had a sister group formed by the North American Sheila Smith and Bitterroot strains that were ecologically associated with the tick vector Dermacentor andersoni. A more divergent clade was composed of North American isolates that have been ecologically associated with the ticks Dermacentor variabilis and R. sanguineus.
TABLE 2.
Isolates of Rickettsia rickettsii used in the phylogenetic analysis of the present study
| Isolate | Isolation source | Clinical outcome | Source of DNA sequences for rickettsial genes (GenBank accession no. or reference no.) for: |
Haplotypea | Tickb | ||||
|---|---|---|---|---|---|---|---|---|---|
| gltA | RR0155-rpmB | RR1240-tlc5b | cspA-ksgA | ompB | |||||
| Iowa | Tick | CP000766 | CP000766 | CP000766 | CP000766 | CP000766 | A | Dermacentor variabilis | |
| Hino | Human | Fatal | CP003309 | CP003309 | CP003309 | CP003309 | CP003309 | A | D. variabilis |
| Hauke | Human | Fatal | CP003318 | CP003318 | CP003318 | CP003318 | CP003318 | A | D. variabilis |
| Arizona | Human | Fatal | CP003307 | CP003307 | CP003307 | CP003307 | CP003307 | B | Rhipicephalus sanguineus sensu lato |
| Sheila Smith | Human | Fatal | CP000848 | CP000848 | CP000848 | CP000848 | CP000848 | C | Dermacentor andersoni |
| Bitterroot | Tick | RRU59729 | EF216032 | EF215983 | EF215860 | X16353 | C | D. andersoni | |
| Costa Rica | Human | Fatal | 19, 27 | EF216038 | EF215987 | EF215872 | 27 | D | ? |
| I12B (Villeta) | Tick | KJ735644 | KJ735647 | KJ735648 | KJ735649 | KJ735646 | D | Amblyomma patinoi | |
| Rs1 | Tick | 13 | This study | This study | This study | 13 | D | Amblyomma sculptum | |
| Itu | Tick | KF742602 | This study | This study | This study | KF742604 | D | A. sculptum | |
| Taiaçu | Tick | DQ115890 | This study | This study | This study | 12 | D | Amblyomma aureolatum | |
| IAL 1–2c | Human | Fatal | This study | This study | This study | This study | This study | D | A. aureolatum |
| IAL 4, 9d | Human | Cure | This study | This study | This study | This study | This study | D | A. sculptum |
| IAL 3, 5–8, 10–15e | Human | Fatal | This study | This study | This study | This study | This study | D | A. sculptum |
| Brazil | Human | Unknown | CP003305 | CP003305 | CP003305 | CP003305 | CP003305 | D | A. sculptum |
| Colombia | Human | Fatal | CP003306 | CP003306 | CP003306 | CP003306 | CP003306 | E | A. patinoi |
Refers to the concatenated haplotypes shown in the phylogenetic tree (Fig. 1).
For tick isolates, refers to the tick species from which the isolate was obtained; for human isolates, refers to the incriminated vector of R. rickettsii to humans in the area of origin of the isolate, according to Ogrzewalska et al. (32) for Amblyomma aureolatum; A. A. Faccini-Martínez, F. B. Costa, T. E. Hayama-Ueno, A. Ramírez-Hernández, J. Cortés-Vecino, M. B. Labruna, and M. Hidalgo (submitted for publication), Nava et al. (33), and Pacheco et al. (13) for Amblyomma patinoi and Amblyomma sculptum; and Karpathy et al. (10) for Dermacentor variabilis, Dermacentor andersoni, and Rhipicephalus sanguineus sensu lato. The question mark represents an unknown vector, according to Hun et al. (27).
Geographic origins (municipalities in the state of São Paulo, Brazil) of these isolates: São Paulo, IAL 1; São Bernardo do Campo, IAL 2.
Geographic origins (municipalities in the state of São Paulo) of these isolates: Piracicaba, IAL 4; Valinhos, IAL 9.
Geographic origins (municipalities in the state of São Paulo) of these isolates: Valinhos, IAL 3, 5, 8, 11, 12, and 15; Campinas, IAL 6 and 13; Jaguariúna, IAL 7; Piracicaba, IAL 10; Limeira, IAL 14.
FIG 1.
Molecular phylogenetic analysis of Rickettsia rickettsii isolates from the United States, Costa Rica, Colombia, and Brazil. A total of 2,392 unambiguously aligned nucleotide sites of two rickettsial genes (gltA and ompB) and three intergenic regions (RR0155-rpmB, RR1240-tlc5b, and cspA-ksgA) were concatenated and subjected to analysis by maximum-parsimony and Bayesian methods. Corresponding sequences of Rickettsia philipii strain 364D and R. rickettsii strain Hlp#2 were used as an outgroup. Numbers at nodes are support values derived from bootstrap and posterior probability for MP and BA analyses (MP/BA). Sequence codes A to E, each with a different color, represent the five haplotypes generated from the 28 isolates of R. rickettsii described in Table 2. Gray braces or arrows at the clades indicate the tick species that has been ecologically associated with the R. rickettsii isolates. The geographical region of origin of the 28 isolates and their corresponding haplotypes (A to E) are indicated on the map of the American continents. (The map is reprinted from http://www.usgs.gov/.)
As reported in previous studies (10, 19–21), the North American isolates of R. rickettsii presented relatively high polymorphism compared to that of the Central/South American isolates. Our concatenated analysis showed the formation of 3 North American haplotypes (A, B, and C), each associated with a different tick species (Fig. 1, Table 2). Conversely, excluding the Colombia strain (haplotype E), there was a single haplotype (D) in Central/South America, although it was associated with 4 different tick species. Geographic distances cannot be inferred from this discrepancy because the distance between Costa Rica and southeastern Brazil is much higher than the distances between distinct North American isolates (Fig. 1). Interestingly, while low-, mild-, and high-virulent strains of R. rickettsii have been reported in the eastern and western parts of the United States (20, 22), only high-virulent strains, responsible for high fatality rates, have been reported in Central/South America, regardless of the tick vector (2, 23–29). While our results of no polymorphisms among Central/South American isolates was biased because most of these isolates were derived from fatal cases, a recent study reported the same clade distribution as shown in our Fig. 1 when analyzing the intergenic regions of R. rickettsii derived from fatal cases from the United States, Mexico, and Central/South America (21).
The relatively high level of polymorphism among North American isolates of R. rickettsii and the contrasting low level of polymorphism in Central/South America suggest that R. rickettsii radiated in North America and was introduced into South America during more recent periods. This scenario may also explain why there is a mixture of highly and less virulent R. rickettsii strains in North America (due to longer coevolving periods with vertebrates), while only highly virulent strains have been found in South America.
It has been suggested that the higher fatality rates of BSF, compared to those of RMSF in the United States, are related to delayed treatment, the use of less effective antibiotics, and more virulent R. rickettsii strains occurring in Brazil (2, 30, 31). The present study corroborates previous studies (10, 19–21) that provided genetic evidence for a very low level of polymorphism occurring among R. rickettsii isolates from South America. This fact should be a significant reason for the much higher fatality rates of BSF, although the others discussed above may also be contributing factors.
Nucleotide sequence accession numbers.
The GenBank nucleotide sequence accession numbers of the partial sequences of R. rickettsii generated in this study are KJ994337, KJ994338, and KJ994339 for the gltA, ompA, and ompB genes, respectively, and KJ994340, KJ994341, and KJ994342 for the RR0155-rpmB, RR1240-tlc5b, and cspA-ksgA intergenic regions, respectively.
ACKNOWLEDGMENTS
This work was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Coordenadoria de Apoio a Pesquisa e Desenvolvimento (CAPES), and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
Footnotes
Published ahead of print 30 July 2014
REFERENCES
- 1.Parola P, Paddock CD, Socolovschi C, Labruna MB, Mediannikov O, Kernif T, Abdad MY, Stenos J, Bitam I, Fournier PE, Raoult D. 2013. Update on tick-borne rickettsioses around the world: a geographic approach. Clin. Microbiol. Rev. 26:657–702. 10.1128/CMR.00032-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angerami RN, Nunes EM, Mendes Nascimento EM, Ribas Freitas A, Kemp B, Feltrin AF, Pacola MR, Perecin GE, Sinkoc V, Ribeiro Resende M, Katz G, Jacintho da Silva L. 2009. Clusters of Brazilian spotted fever in São Paulo State, southeastern Brazil: a review of official reports and the scientific literature. Clin. Microbiol. Infect. 15(Suppl 2):S202–S204. 10.1111/j.1469-0691.2008.02637.x [DOI] [PubMed] [Google Scholar]
- 3.Angerami RN, Câmara M, Pacola MR, Rezende RC, Duarte RM, Nascimento EM, Colombo S, Santos FC, Leite RM, Katz G, Silva LJ. 2012. Features of Brazilian spotted fever in two different endemic areas in Brazil. Ticks Tick Borne Dis. 3:346–348. 10.1016/j.ttbdis.2012.10.010 [DOI] [PubMed] [Google Scholar]
- 4.Nascimento E. 2006. Cultivo celular de riquétsias no Brasil, p 131–132 2nd Simposio Lationamericano de Ricketsioses. Colégio Brasileiro de Parasitologia Veterinária, Ribeirão Preto, Brazil [Google Scholar]
- 5.Marrero M, Raoult D. 1989. Centrifugation-shell vial technique for rapid detection of Mediterranean spotted fever rickettsia in blood culture. Am. J. Trop. Med. Hyg. 40:197–199 [DOI] [PubMed] [Google Scholar]
- 6.Melles HH, Colombo S, de Lemos ER. 1999. Isolamento de Rickettsia em cultura de células vero. Rev. Soc. Bras. Med. Trop. 32:469–473. 10.1590/S0037-86821999000500001 [DOI] [PubMed] [Google Scholar]
- 7.Labruna MB, Whitworth T, Horta MC, Bouyer DH, McBride JW, Pinter A, Popov V, Gennari SM, Walker DH. 2004. Rickettsia species infecting Amblyomma cooperi ticks from an area in the state of São Paulo, Brazil, where Brazilian spotted fever is endemic. J. Clin. Microbiol. 42:90–98. 10.1128/JCM.42.1.90-98.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Regnery RL, Spruill CL, Plikaytis BD. 1991. Genotypic identification of rickettsiae and estimation of intraspecies sequence divergence for portions of two rickettsial genes. J. Bacteriol. 173:1576–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roux V, Raoult D. 2000. Phylogenetic analysis of members of the genus Rickettsia using the gene encoding the outer membrane protein rOmpB (ompB). Int. J. Syst. Evol. Microbiol. 50:1449–1455. 10.1099/00207713-50-4-1449 [DOI] [PubMed] [Google Scholar]
- 10.Karpathy SE, Dasch GA, Eremeeva ME. 2007. Molecular typing of isolates of Rickettsia rickettsii by use of DNA sequencing of variable intergenic regions. J. Clin. Microbiol. 45:2545–2553. 10.1128/JCM.00367-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fournier PE, Zhu Y, Ogata H, Raoult D. 2004. Use of highly variable intergenic spacer sequences for multispacer typing of Rickettsia conorii strains. J. Clin. Microbiol. 42:5757–5766. 10.1128/JCM.42.12.5757-5766.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pinter A, Labruna MB. 2006. Isolation of Rickettsia rickettsii and Rickettsia bellii in cell culture from the tick Amblyomma aureolatum in Brazil. Ann. N. Y. Acad. Sci. 1078:523–529. 10.1196/annals.1374.103 [DOI] [PubMed] [Google Scholar]
- 13.Pacheco RC, Moraes-Filho J, Guedes E, Silveira I, Richtzenhain LJ, Leite RC, Labruna MB. 2011. Rickettsial infections of dogs, horses and ticks in Juiz de Fora, southeastern Brazil, and isolation of Rickettsia rickettsii from Rhipicephalus sanguineus ticks. Med. Vet. Entomol. 25:148–155. 10.1111/j.1365-2915.2010.00915.x [DOI] [PubMed] [Google Scholar]
- 14.Krawczak FS, Nieri-Bastos FA, Nunes FP, Soares JF, Moraes-Filho J, Labruna MB. 2014. Rickettsial infection in Amblyomma cajennense ticks and capybaras (Hydrochoerus hydrochaeris) in a Brazilian spotted fever-endemic area. Parasit. Vectors 7:7. 10.1186/1756-3305-7-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swofford DL. 2002. PAUP: phylogenetic analysis using parsimony. Beta Version 4.0b10 Sinauer and Associates, Sunderland, MA [Google Scholar]
- 16.Huelsenbeck JP, Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- 17.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882. 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicholas KB, Nicholas HB., Jr 1997. GeneDoc: analysis and visualization of genetic variation. EMBNEW News 4:14 [Google Scholar]
- 19.Eremeeva ME, Klemt RM, Santucci-Domotor LA, Silverman DJ, Dasch GA. 2003. Genetic analysis of isolates of Rickettsia rickettsii that differ in virulence. Ann. N. Y. Acad. Sci. 990:717–722. 10.1111/j.1749-6632.2003.tb07449.x [DOI] [PubMed] [Google Scholar]
- 20.Eremeeva ME, Dasch GA. 2009. Closing the gaps between genotype and phenotype in Rickettsia rickettsii. Ann. N. Y. Acad. Sci. 1166:12–26. 10.1111/j.1749-6632.2009.04526.x [DOI] [PubMed] [Google Scholar]
- 21.Paddock CD, Dension AM, Lash RR, Liu L, Batten BC, Dahlgren FS, Kanamura CT, Angerami RN, Pereira Dos Santos FC, Brasil Martines R, Karpathy SE. 23 June 2014. Phylogeography of Rickettsia rickettsii genotypes associated with fatal Rocky Mountain spotted fever. Am. J. Trop. Med. Hyg. 10.4269/ajtmh.14-0146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Topping NH. 1947. The epidemiology of Rocky Mountain spotted fever. N. Y. State J. Med. 47:1585–1587 [PubMed] [Google Scholar]
- 23.Patino-Camargo L. 1941. Nuevas observaciones sobre un tercer foco de fiebre petequial (maculosa) en el hemisferio americano. Bol. Oficina. Sanit. Panam. 20:1112–1124 [Google Scholar]
- 24.Dias E, Martins AV. 1939. Spotted fever in Brazil: a summary. Am. J. Trop. Med. 19:103–108 [Google Scholar]
- 25.de Rodaniche EC, Rodaniche A. 1950. Spotted fever in Panama; isolation of the etiologic agent from a fatal case. Am. J. Trop. Med. Hyg. 30:511–517 [DOI] [PubMed] [Google Scholar]
- 26.Hidalgo M, Orejuela L, Fuya P, Carrillo P, Hernandez J, Parra E, Keng C, Small M, Olano JP, Bouyer D, Castaneda E, Walker D, Valbuena G. 2007. Rocky Mountain spotted fever, Colombia. Emerg. Infect. Dis. 13:1058–1060. 10.3201/eid1307.060537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hun L, Cortés X, Taylor L. 2008. Molecular characterization of Rickettsia rickettsii isolated from human clinical samples and from the rabbit tick Haemaphysalis leporispalustris collected at different geographic zones in Costa Rica. Am. J. Trop. Med. Hyg. 79:899–902 [PubMed] [Google Scholar]
- 28.Hun-Opfer L. 2008. Las fiebres manchadas y su importancia en Costa Rica. Acta Méd. Costarric. 50:77–86 [Google Scholar]
- 29.Tribaldos M, Zaldivar Y, Bermudez S, Samudio F, Mendoza Y, Martinez AA, Villalobos R, Eremeeva ME, Paddock CD, Page K, Smith RE, Pascale JM. 2011. Rocky Mountain spotted fever in Panama: a cluster description. J. Infect. Dev. Ctries. 5:737–741. 10.3855/jidc.2189 [DOI] [PubMed] [Google Scholar]
- 30.Angerami RN, Resende MR, Feltrin AF, Katz G, Nascimento EM, Stucchi RS, Silva LJ. 2006. Brazilian spotted fever: a case series from an endemic area in southeastern Brazil: clinical aspects. Ann. N. Y. Acad. Sci. 1078:252–254. 10.1196/annals.1374.044 [DOI] [PubMed] [Google Scholar]
- 31.Walker D. 2013. The challenge of rickettsial diagnosis, research, and awareness in Latin America. Acta Méd. Costarric. 55(Suppl 1):S4–S6 [Google Scholar]
- 32.Ogrzewalska M, Saraiva DG, Moraes-Filho J, Martins TF, Costa FB, Pinter A, Labruna MB. 2012. Epidemiology of Brazilian spotted fever in the Atlantic Forest, state of São Paulo, Brazil. Parasitology 139:1283–1300. 10.1017/S0031182012000546 [DOI] [PubMed] [Google Scholar]
- 33.Nava S, Beati L, Labruna MB, Cáceres AG, Mangold AJ, Guglielmone AA. 2014. Reassessment of the taxonomic status of Amblyomma cajennense (Fabricius, 1787) with the description of three new species, Amblyomma tonelliae n. sp., Amblyomma interandinum n. sp. and Amblyomma patinoi n. sp., and reinstatement of Amblyomma mixtum, and Amblyomma sculptum (Ixodida: Ixodidae). Ticks Tick Borne Dis. 5:252–276. 10.1016/j.ttbdis.2013.11.004 [DOI] [PubMed] [Google Scholar]