Abstract
Candidate small RNAs (sRNAs) have recently been identified in Enterococcus faecalis, a Gram-positive opportunistic pathogen, and six of these candidate sRNAs with unknown functions were selected for a functional study. Deletion mutants and complemented strains were constructed, and their virulence was tested. We were unable to obtain the ef0869-0870 mutant, likely due to an essential role, and the ef0820-0821 sRNA seemed not to be involved in virulence. In contrast, the mutant lacking ef0408-0409 sRNA, homologous to the RNAII component of the toxin-antitoxin system, appeared more virulent and more able to colonize mouse organs. The three other mutants showed reduced virulence. In addition, we checked the responses of these mutant strains to several stresses encountered in the gastrointestinal tract or during the infection process. In parallel, the activities of the sRNA promoters were measured using transcriptional fusion constructions. To attempt to identify the regulons of these candidate sRNAs, proteomics profiles of the mutant strains were compared with that of the wild type. This showed that the selected sRNAs controlled the expression of proteins involved in diverse cellular processes and the stress response. The combined data highlight the roles of certain candidate sRNAs in the adaptation of E. faecalis to environmental changes and in the complex transition process from a commensal to a pathogen.
INTRODUCTION
It has now become evident that RNAs are not just involved in transmitting genetic information or have only structural and catalytic roles, like those of mRNA, rRNA, or tRNA, but also act as central regulatory molecules in numerous biological processes. Members of the bacterial small RNA (sRNA) family, including functional cis- and trans-acting sRNAs, have begun to be well characterized (1–7). Some of these molecules play important roles in bacterial physiology, affecting iron homeostasis; sugar metabolism; responses to stress, such as oxidative stress and conditions induced by DNA damage; stationary phase; cell surface composition; and pathogenesis (8–15). While mechanisms of action, messenger targets, and functions are now partially understood for species like Escherichia coli, similar investigations are needed for numerous other bacteria, including Enterococcus faecalis (16). Enterococci are normal inhabitants of the gastrointestinal (GI) tracts of humans and animals. While they are typically harmless to healthy individuals, one species, E. faecalis, is one of the leading microorganisms involved in hospital-acquired infections, such as catheter-associated urinary tract infections, endocarditis, and surgical and burn wound infections (17–19). Notably, the risk of death for patients infected with multidrug-resistant strains, (i.e., vancomycin-resistant enterococci [VRE]), is considerably higher than for those infected with antibiotic-susceptible strains (20, 21). Unlike Gram-positive pathogens, like Streptococcus pyogenes and Staphylococcus aureus, E. faecalis lacks classical virulence factors, such as proinflammatory toxins or immune modulators, but instead has “opportunism factors.” These factors are directly associated with the organism's ability to survive hostile conditions, including intrinsic resistance to antibiotics, host immune responses, antagonistic molecules produced by competing microbial flora, and the stresses imposed by its environment (17–19). Several transcriptional regulators in E. faecalis that are involved in the stress response, as well as in the virulence of the bacterium, have been characterized (22–27). However, no investigation of the role of sRNAs has been undertaken. Some studies have been conducted on the roles of sRNAs in the expression of the plasmid-encoded toxin-antitoxin (TA) system and of the CRISPR systems and on the riboswitch regulation of the expression of ethanolamine operon regulation (28–32). In 2008, Livny and coworkers identified 17 putative sRNAs in E. faecalis using a bioinformatics approach, followed in 2011 by a study that combined selected in silico sRNAs and the 5′ tag rapid amplification of cDNA ends (RACE) method (33, 34). In the same year, the first experimental identification of sRNA in E. faecalis, using tiling microarray experiments with probes covering intergenic regions, 5′ and 3′ RACE-PCR, and Northern blot analysis, was published (35). In the last study, around 100 potential sRNA candidates were identified, 11 of which were analyzed further.
In the present study, we focused on 6 of these 11 candidate sRNAs (5 members of the core genome and 1 located in the pathogenicity island [PAI]) by constructing mutants and complemented strains in order to determine whether they were implicated in pathogenesis and the stress response in E. faecalis. It appeared that the ef0869-0870 sRNA might be essential, since construction of the corresponding mutant strain always failed. The five other candidate sRNA mutant strains (Δef0408-0409, Δef0605-0606, Δef0820-0821, Δef1368-1369, and Δef3314-3315) were tested in the Galleria mellonella, urinary tract infection, and intracellular-macrophage survival models. For those that were involved in the virulence of E. faecalis, we looked at their phenotypes in response to several stress conditions potentially encountered by the bacteria in their niches or during the infection process. Lastly, to identify the regulons, we performed a 2-dimensional (2D) polyacrylamide gel electrophoresis (PAGE) technique in combination with mass spectrometry (MS) to compare the cytoplasmic proteomes of sRNA mutants with those of their corresponding wild-type strains. This study highlights for the first time the implication of certain candidate sRNAs in the virulence and the stress response of the opportunistic pathogen E. faecalis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains were cultured with shaking at 37°C in LB medium with chloramphenicol (10 μg/ml), erythromycin (100 μg/ml), or tetracycline (10 μg/ml) when required. E. coli DH5α (36) was used as the recipient for cloning. Mutants were constructed from the parental E. faecalis strain EryS, an erythromycin-sensitive strain derived from the vancomycin-resistant clinical isolate V583 (37). E. faecalis EryS and its derivates were grown without shaking at 37°C in M17 medium supplemented with 0.5% glucose (M17glu). Growth of the E. faecalis strains was also examined in horse serum under oxidative-stress conditions (with shaking in M17glu supplemented with 1.5 mM H2O2 or in CCM17 MOPS [morpholinepropanesulfonic acid] medium without carbon source supplemented with 0.5% glycerol), under osmotic-stress conditions (growth in M17glu supplemented with 8% sodium chloride), or under detergent conditions (growth in M17glu supplemented with 0.08% bile salts). For the acid challenge assays, overnight cultures of the wild type, the sRNA mutants, and the complemented strains were diluted to a final optical density at 600 nm (OD600) of 0.01 in fresh medium and allowed to grow to an OD600 of 0.5. Then, cells were harvested by centrifugation and resuspended in the same volume of M17/7, pH 2.8. Determination of the number of CFU was performed by plating on M17glu after serial dilution and counting colonies after incubation at 37°C for 24 h. The survival rates (the number of CFU after exposure to the different conditions divided by the number of CFU under control conditions) were determined. Each survival experiment was performed at least three times.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant properties | Reference or source |
|---|---|---|
| Strains | ||
| E. faecalis | ||
| EryS | Strain V583 (clinical isolate); erythromycin sensitive | 37 |
| EryS derivatives harboring | ||
| Δ0408-0409 | EryS isogenic derivative sRNA ef0408-ef0409 mutant | This study |
| Δ0605-0606 | EryS isogenic derivative sRNA ef0605-ef0606 mutant | This study |
| Δ0820-0821 | EryS isogenic derivative sRNA ef0820-ef0821 mutant | This study |
| Δ1368-1369 | EryS isogenic derivative sRNA ef1368-ef1369 mutant | This study |
| Δ3314-3315 | EryS isogenic derivative sRNA ef3314-ef3315 mutant | This study |
| Δ0408-0409C | Δ0408-0409 mutant strain derivative complemented | This study |
| Δ0605-0606C | Δ0605-0606 mutant strain derivative complemented | This study |
| Δ0820-0821C | Δ0820-0821 mutant strain derivative complemented | This study |
| Δ1368-1369Cgene | Δ1368-1369 mutant strain with pMSP3535 carrying ef1368-ef1369gene; Emr | This study |
| Δ1368-1369CsRNA | Δ1368-1369 mutant strain with pMSP3535 carrying ef1368-ef1369sRNA; Emr | This study |
| Δ3314-3315C | Δ3314-3315 mutant strain derivative complemented | This study |
| EryS pVEPhoZ | EryS strain with pVEPhoZ integrated in the phoZ locus; Tetr | 43 |
| EryS pMSP3535 | EryS strain with pMSP3535; Emr | 42 |
| pVEPhoZ-Pef0408-0409 | pVEPhoZ carrying sRNA ef0408-0409 promoter; Tetr | This study |
| pVEPhoZ-Pef0605-0606 | pVEPhoZ carrying sRNA ef0605-0606 promoter; Tetr | This study |
| pVEPhoZ-Pef1368-1369 | pVEPhoZ carrying sRNA ef1368-1369 promoter; Tetr | This study |
| pVEPhoZ-Pef3314-3315 | pVEPhoZ carrying sRNA ef3314-3315 promoter; Tetr | This study |
| E. coli | ||
| DH5α | recA endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 lacZΔM15 | Invitrogen |
| Plasmids | ||
| pLT06 | lacZ P-pheS from pCJK47 cat from pGB354, orfB orfC repA(Ts) orfD from pCASPER | |
| pLT06-Δ0408-0409 | pLT06 derivative carrying a 2-kb DNA fragment from E. faecalis EryS without sRNA ef0408-0409 | This study |
| pLT06wtef0408-0409 | pLT06 derivative carrying a 2.1-kb DNA fragment from E. faecalis EryS with sRNA ef0408-0409 | This study |
| pLT06-Δ0605-0606 | pLT06 derivative carrying a 2-kb DNA fragment from E. faecalis EryS without sRNA ef0605-0606 | This study |
| pLT06wtef0605-0606 | pLT06 derivative carrying a 2.3-kb DNA fragment from E. faecalis EryS with sRNA ef0605-0606 | This study |
| pLT06-Δ0820-0821 | pLT06 derivative carrying a 2-kb DNA fragment from E. faecalis EryS without sRNA ef0820-0821 | This study |
| pLT06wtef0820-0821 | pLT06 derivative carrying a 2.1-kb DNA fragment from E. faecalis EryS with sRNA ef0820-0821 | This study |
| pLT06-Δ1368-1369 | pLT06 derivative carrying a 1.8-kb DNA fragment from E. faecalis EryS without sRNA ef1368-1369 | This study |
| pLT06-Δ3314-3315 | pLT06 derivative carrying a 1.7-kb DNA fragment from E. faecalis EryS without sRNA ef3314-3315 | This study |
| pLT06wtef3314-3315 | pLT06 derivative carrying a 2.1-kb DNA fragment from E. faecalis EryS with sRNA ef3314-3315 | This study |
| pVEPhoZ | pVEPhoZ carrying the 5′ part of the phoZ gene; Tetr | 43 |
| pVEPhoZ-Pef0408-0409 | pVEPhoZ carrying the sRNA ef0408-0409 promoter; Tetr | This study |
| pVEPhoZ-Pef0605-0606 | pVEPhoZ carrying the sRNA ef0605-0606 promoter; Tetr | This study |
| pVEPhoZ-Pef1368-1369 | pVEPhoZ carrying the sRNA ef1368-1369 promoter; Tetr | This study |
| pVEPhoZ-Pef3314-3315 | pVEPhoZ carrying the sRNA ef3314-3315 promoter; Tetr | This study |
| pMSP3535 | 8.35 kb; nisR nisK PnisA (nisin-inducible promoter); Emr | 42 |
| pMSP3535-ef1368-ef1369 gene | pMSP3535 carrying ef1368-ef1369gene; Emr | This study |
Em, erythromycin; Tet, tetracycline; Cm, chloramphenicol.
General molecular methods.
PCR was performed with Phusion High-fidelity DNA polymerase (Finenzymes, Vantaa, Finland). The primers used for this work are listed in Table S1 in the supplemental material. PCR products and plasmids were purified using a Nucleospin plasmid kit (Macherey-Nagel, Dünen, Germany). Restriction endonucleases and T4 DNA ligase were purchased from Promega (Madison, WI, USA) and used according to the manufacturers' instructions. Genomic-DNA extraction and other standard methods were carried out as described by Sambrook (38).
Construction of sRNA mutants and complemented strains and transcriptional fusion.
For the construction of the sRNA mutant strain, allelic replacements were carried out as previously described (39, 40). Briefly, DNA fragments (obtained using PCR with chromosomal DNA of E. faecalis EryS as the template) containing ligated upstream and downstream sequences of the desired deletion were cloned into the plasmid pLT06 (41) (Table 1), and 1 μg of recombinant plasmid was used to transform E. faecalis competent cells. For the construction of the Δef0408-0409, Δef0605-0606, Δef0820-0821, Δef0869-0870, Δef1368-1369, and Δef3314-3315 mutant strains, fusion PCR products of the respective upstream (955, 989, 1,056, 1,020, 765, and 836 bp, respectively) and downstream (1,066, 1,030, 967, 958, 996, and 869 bp, respectively) fragments were constructed. Single-crossover transformants (dark-blue and chloramphenicol-resistant colonies) were used for temperature shifts in order to release the plasmid. Candidate clones resulting from a double-crossover event were isolated on M17glu agar with or without chloramphenicol. In antibiotic-susceptible clones, the loss of the plasmid and the deletion of the respective sRNA were verified by PCR and sequencing.
For construction of complemented mutant strains, the pLT06 plasmid containing the intact sRNA was introduced into the corresponding sRNA mutant. Double-recombination events allowed the restoration of intact sRNA. For the complementation of the antisense sRNA ef1368-1369, a DNA fragment containing the entire ef1369 gene or the sRNA antisense to the ef1369 gene of E. faecalis EryS was obtained by PCR using primers listed in Table S1 in the supplemental material. The PCR product was cloned into the pMSP3535 plasmid downstream of the nisin-inducible promoter (Pnis) (42). The recombinant plasmid was first transformed into E. coli DH5α cells, amplified, and finally used to transform competent cells of the E. faecalis Δef1368-1369 mutant. After electroporation, the colonies were analyzed by sequencing for the presence of the plasmid containing an intact ef1369 gene or corresponding antisense sRNA.
For the transcriptional fusion constructions, the 201-bp, 135-bp, 98-bp, 194-bp, and 163-bp putative promoter regions of sRNAs corresponding to ef0408-0409, ef0605-0606, ef0820-0821, ef1368-1369, and ef3314-3315, respectively, were cloned into the pVEPhoZ plasmid (43). This integrative plasmid was then introduced into the E. faecalis EryS chromosome by single crossover in the phoZ locus, as described by Le Jeune et al. (43).
Infection experiments and survival in macrophages.
Infection of G. mellonella larvae with E. faecalis was accomplished as previously described by Lebreton et al. (21). Briefly, using a syringe pump (KD Scientific, Holliston, MA, USA), larvae (about 0.3 g and 3 cm in length) were infected subcutaneously with washed E. faecalis wild-type sRNA mutants and the complemented strains from an overnight culture in M17glu, with 1.2 × 105 ± 0.5 × 105 CFU per larva administered in 10 μl of sterile saline buffer. In each test, 15 insects were infected, and the experiments were repeated at least five times. Larval killing was then monitored at 20 h postinfection. The results were analyzed by an unpaired t test, and all comparisons with a P value of less than 0.05 were considered significant.
The virulences of the wild-type strain and the sRNA deletion mutants were also assessed in a urinary tract infection (UTI), essentially as previously described by Lebreton et al. (21). Aliquots of 100 μl from each strain (∼108 cells) were used to inject each of 10 female BALB/c mice (10 weeks old; Harlan Italy S.r.l., San Pietro al Natisone, Udine, Italy) transurethrally. The mouse experiments were performed under a protocol approved by the Institutional Animal Use and Care Committee at the Catholic University of the Sacred Heart, Rome, Italy, and authorized by the Italian Ministry of Health (permit number Z21, 13 March 2012). Infection experiments were repeated three times. Mice were monitored twice daily and were euthanized by CO2 asphyxiation 7 days after infection. For determination of the numbers of CFU present in organs, serial homogenate dilutions were plated onto Enterococcus selective agar (Fluka Analytical, Switzerland). CFU counts were analyzed by an unpaired t test.
Survival of E. faecalis in mouse peritoneal macrophages was tested using an in vivo-in vitro infection model, as described previously (44). Bacterial cells were pelleted and resuspended in an adequate volume of phosphate-buffered saline (PBS) for injection. Male BALB/c mice (10 weeks old; Harlan Italy S.r.l., San Pietro al Natisone, Udine, Italy) were infected with 4.8 × 107 ± 0.3 × 107 cells of each strain (estimated by CFU determination) by intraperitoneal injection of 200 μl of the PBS-enterococcus suspension. After a 6-h infection period, the peritoneal macrophages were collected by peritoneal lavage, centrifuged, and suspended in Dulbecco's modified Eagle's medium (DMEM) containing 10 mM HEPES, 2 mM glutamine, 10% fetal bovine serum, and 1× nonessential amino acids, supplemented with vancomycin (10 μg/ml) and gentamicin (150 μg/ml). The cell suspension was dispensed into 24-well tissue culture plates and incubated at 37°C under 5% CO2 for 2 h. After exposure to antibiotics (i.e., 8 h postinfection) to kill extracellular bacteria, the infected macrophages were washed, and triplicate wells of macrophages were lysed with detergent. After dilution with brain heart infusion (BHI) broth, the lysates were plated on BHI agar to quantitate the viable intracellular bacteria. The remaining wells of infected macrophages were maintained in DMEM with the antibiotics for the duration of the experiment. The same procedure was performed at 24, 48, and 72 h postinfection. In the meantime, at 8, 24, 48, and 72 h postinfection, supernatant fluids from each well were removed, and extracellular bacteria were quantitated by counting on BHI agar plates. To assess their viability, macrophages were detached from tissue culture wells with cell scrapers and stained with trypan blue, and viable macrophages were counted with a hemacytometer. The procedure was repeated three times, and the results were analyzed using one-way analysis of variance with a Bonferroni correction posttest with SPSS statistical software (SPSS, Chicago, IL, USA). All statistical analyses were performed using Prism software (version 5.00) for Windows (GraphPad Software, San Diego, CA, USA). For all comparisons, a P value of less than 0.05 was considered significant.
Alkaline phosphatase assays.
For alkaline phosphatase (AP) assays, overnight cultures grown in M17glu were diluted in fresh medium to an OD600 of 0.01. At an OD600 of 0.5, the stressing agents mentioned above for growth conditions were added to the cultures, and after 30, 60, and 90 min of incubation at 37°C, 10 ml of culture was harvested. AP activity measurements were performed as described by Le Jeune et al. and were expressed in Miller units (MU) by the following formula: MU = (OD405 × 1,000)/(OD600 × volume [ml] × time [min]) (43).
RNA isolation and reverse transcriptase-quantitative PCR (RT-qPCR).
In order to assess comparative transcriptional gene expression, we used the EryS wild-type strain and its sRNA derivative mutants cultured on M17glu at 37°C in exponential phase. Total RNA from three biological replicates was extracted by a method based on the protocol of Toledo-Arana et al. and modified as described by Michaux et al. (40, 44).
For RT-PCR experiments, 2 micrograms of RNA was reverse transcribed with random-hexamer primers and QuantiTect enzyme (Qiagen, Valencia, CA) according to the manufacturer's recommendations. cDNAs were then used as templates for PCRs using primers listed in Table S1 in the supplemental material. Quantification of 23S rRNA and gyrA (encoding the A subunit of the DNA gyrase enzyme) mRNA provided internal controls. Amplification (using 5 μl of a 1/100 cDNA dilution), detection (with automatic calculation of the threshold value), and real-time analysis were performed three times for each cDNA sample, using the iCycler iQ detection system (Bio-Rad Laboratories). Relative mRNA levels of each gene in each sample were calculated using comparative cycle time, as described previously by Meijerink et al. (45).
Protein extraction, 2D PAGE technique, protein identification, and Western blotting.
Cytoplasmic proteins were prepared as described by Becher et al. (46). In brief, 50 ml of bacterial cell culture was centrifuged (7,000 × g) for 10 min at 4°C. The cell pellets were washed twice with 1 ml of ice-cold Tris-EDTA (TE) buffer and transferred into screw-top tubes containing 500 μl of glass beads. The cells were disrupted using a Fast Prep instrument (MP Biomedical LLC, Santa Ana, CA, USA) for 30 s at 6.5 m/s. The lysate was centrifuged for 25 min at 21,000 × g at 4°C to remove the cell debris, and the supernatant was transferred into a new tube and centrifuged for 45 min at 21,000 × g at 4°C in order to remove insoluble and aggregated proteins. Proteins were prepared from three biological replicates.
2D PAGE was performed using an immobilized pH gradient (IPG) technique as described previously (47). In the first dimension, proteins were separated on IPG strips (GE Healthcare, Uppsala, Sweden) in a linear pH range of 4 to 7. One hundred micrograms of protein extracts was loaded onto the IPG strips. The second dimension was performed as described by Engelmann and Hecker (48). The resulting gels were stained with the fluorescent dye Krypton according to the manufacturer's instructions (Thermo Scientific, Waltham, MA, USA) and scanned using a Typhoon 9400 Variable Mode Imager. For quantitation of proteins, the 2D gel image analysis was performed with the software DELTA 2D version 4.3 (Decodon GmbH, Greifswald, Germany) as described previously (49). Identified protein spots were labeled with the respective protein names or locus numbers on the gel. Multiple spots of the same protein were numbered consecutively. Gel-to-gel variations of spot positions on 2D gel images were first compensated for by warping (exact mode) of the images. Subsequently, they were fused intraexperimentally to a virtual fusion gel (union mode). This recomposed gel was used for protein spot detection. Automatically calculated spot borders were corrected and optimized manually if necessary. The final spot mask was transferred from the fusion gel to each gel image of the corresponding experiment. Spot intensities were normalized using total normalization. For each protein spot, the amounts of protein in the mutant sample and in the corresponding protein spot of the wild-type sample were calculated and presented as the protein ratio. For statistical analyses, standardized data were loaded into the J. Craig Venter Institute Multiexperiment Viewer 4.4.1. Each experiment was analyzed separately. To test whether the amount of protein in a given spot changed significantly in the mutant, we used t test analysis (Welch approximation; P values based on permutation and based on all possible permutations; adjusted Bonferroni correction). Only protein spots whose amount changed at least 2-fold were considered differentially expressed.
For protein identification, protein spots of interest were excised from the gel with an Ettan spot picker (GE Healthcare) with a picker head of 2 mm. In-gel digestion was performed with sequencing grade trypsin (Promega, Madison, WI), and extraction of peptides was carried out with the Ettan Spot Handling Workstation (GE Healthcare) using the protocol described by Eymann et al. (50). Identification of E. faecalis proteins by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) and MALDI-TOF MS was carried out as described previously (51). MALDI-TOF MS analyses of spotted peptide solutions were carried out on a Proteome Analyzer 4800 (Applied Biosystems, Foster City, CA, USA) (for details, see reference 52). For database searches, the Mascot search engine version 2.1.04 (Matrix Science Ltd., London, United Kingdom) was used, with an E. faecalis sequence database extracted from the National Center for Biotechnology Information (NCBI) bacterial genomes. Peptide mixtures that yielded a Mowse (molecular weight search) score of 59 at least twice were regarded as positive identifications. Details of the mass spectrometry results for the identified proteins are presented in Table S2 in the supplemental material.
For Western blot analysis, the proteins were transferred after electrophoresis onto a polyvinyl difluoride membrane, which was then stained with Coomassie blue in order to verify that equal amounts of protein were present in all lanes. Blocking, incubation with antibodies against E. faecalis Ers and E. coli GroEL, and enhanced-chemiluminescence detection (GE Healthcare, Little Chalfont, United Kingdom) were carried out as previously described by Riboulet-Bisson et al. (53).
In order to predict target genes for the identified sRNAs, the RNA Predator (http://rna.tbi.univie.ac.at/RNApredator2/target_search.cgi) (54) and CopraRNA (http://rna.informatik.uni-freiburg.de/CopraRNA/Input.jsp) (55) servers were used.
RESULTS AND DISCUSSION
Selection of sRNAs and construction of corresponding mutants.
In previous work performed in our laboratory, we characterized 11 candidate sRNAs highly expressed in E. faecalis V583 using tiling microarray, 5′ and 3′ RACE-PCR, and Northern blot analysis (35). The absence of a start codon and a putative ribosome binding site (RBS) in a consensus distance (8 nucleotides [nt]) from the start codon within the studied transcripts strongly suggests the lack of a putative open reading frame. ef1097-1098, which corresponded to E. faecalis transfer-messenger RNA (tmRNA) (ssrA), a unique bifunctional RNA acting as both a tRNA and an mRNA (56, 57); ef2205-2206 sRNA, which corresponded to the RNA component of the signal recognition particle (SRP) (58); and efA0080-A0081 and efB0062-B0063, the two plasmidic RNAII components of the TA system already identified in E. faecalis V583 (29, 30), were not analyzed further. Note that tmRNA is essential for some species, such as Neisseria gonorrhoeae, Haemophilus influenzae, and Shigella flexneri (56, 59). Similar observations were made here, where all attempts to construct a knockout mutant in E. faecalis failed (data not shown). To obtain functional insights into the other previously identified candidates, members of the core genome or located in the PAI with unknown roles, deletions were introduced into the genes corresponding to ef0408-0409, ef0605-0606, ef0820-0821, ef0869-0870, ef1368-1369, and ef3314-3315 sRNAs. The deletion of the sRNA region, depending on the candidate, was between 30% and the entire sRNA (see Fig. S1 in the supplemental material). Note that, except for ef0605-0606, located on the PAI (not systematically observed E. faecalis genomes), the presence of selected candidate sRNAs has been shown in the 54 sequenced strains of E. faecalis available (35).
In the case of ef0869-0870, we tried to construct a mutant strain using the double-crossover strategy with a thermosensitive replication plasmid. Unfortunately, all antibiotic-sensitive clones obtained after the second recombination (several hundred were analyzed) harbored the wild-type gene. This strongly suggests that the sRNA ef0869-0870 may be essential for E. faecalis.
Because ef1368-1369 likely acts as an antisense RNA of the ef1369 gene, two different complemented strains were constructed using the replicative vector pMSP3535: the first contained the gene ef1369, and the second expressed the sRNA. With these two strains, we would be able to verify whether any phenotypes observed for the mutant (see below) were due to lack of the gene or the sRNA. Before virulence assays or stress challenges, growth assays under standard conditions (M17glu at 37°C) were performed for the wild type and strains with sRNAs deleted without any obvious difference between them.
Implication of sRNAs in virulence, colonization, and survival within macrophages.
In order to determine the impacts of the sRNA deletions on the virulence of E. faecalis and their roles in the process of infection, different infection models and assays were employed. First, we compared the survival of larvae of G. mellonella infected with the sRNA mutant and the corresponding complemented strains with those infected with the wild type. Infection of G. mellonella worms has emerged as a reliable model system to study the pathogenesis of numerous human pathogens (60–66). Indeed, the growth of the larvae at 37°C and the high degree of structural and functional homology between the innate immune systems of Galleria and mammals make the model pertinent (67, 68). The nonpathogenic Gram-positive bacterium Lactococcus lactis IL1403 was used as a control, and all larvae infected with the organism survived, using the same infection protocol as for E. faecalis (data not shown). This model of virulence was used as the first screen for our functional analysis. The mutant strain showing a phenotype was then used for further studies (urinary tract infection, survival in macrophages, stress response, and proteomic study), whereas in the case of the nonvirulent phenotype, the corresponding strain was not further studied. The figures (and supplemental data) present the results when a difference between the mutant and the wild-type strains was observed.
ef0408-0409 sRNA is homologous to the RNAII component of the TA system (also named the par system) described previously (29, 30). These systems, also called postsegregation killing systems, first identified on bacterial plasmids, encode a stable toxin and an unstable antitoxin. After cellular division, the loss of the plasmid leads to degradation of the antitoxin and corresponding activation of the toxin. In most of the systems, both components are proteins, but in some cases, as in the well-studied par system in E. faecalis, the antitoxin is a small regulatory RNA that represses the translation of the toxin mRNA (69). In E. faecalis V583, three of these systems were identified in previous studies, two on plasmids and one on the chromosome (35). It has been shown that the 0408-0409 par addiction module is located on the bacterial chromosome, which raises the question of its cellular role (69, 70). Of note, two direct-repeat sequences allowing the base pairing of the two components of the toxin-antitoxin have been found in the sRNA and ef0409 sequences. In addition, the two loci showed the same bidirectional terminator (35). As shown in Fig. 1, the rate of killing of the larvae infected with the Δef0408-0409 mutant strain was significantly higher than the rate for those infected with the wild type. After 20 h of infection, 15% of the caterpillars infected with the wild-type strain were still alive, whereas less than 3% of the animals infected with the Δef0408-0409 mutant strain had survived. The “hypervirulence” phenotype was attenuated back to wild-type level in the complemented Δef0408-0409 mutant strain, demonstrating that the observed phenotype was due to the deletion of the sRNA. It was surprising that we were able to create a mutant lacking the ef0408-0409 sRNA, which is supposed to act as an antitoxin through interaction with RNAI (ef0409), encoding the Fts toxin (30, 69). Nevertheless, it is worth noting that our transcriptional analysis showed that RNAI (ef0409) was transcribed at a very low level (near background) in both the wild-type and Δef0408-0409 mutant strains.
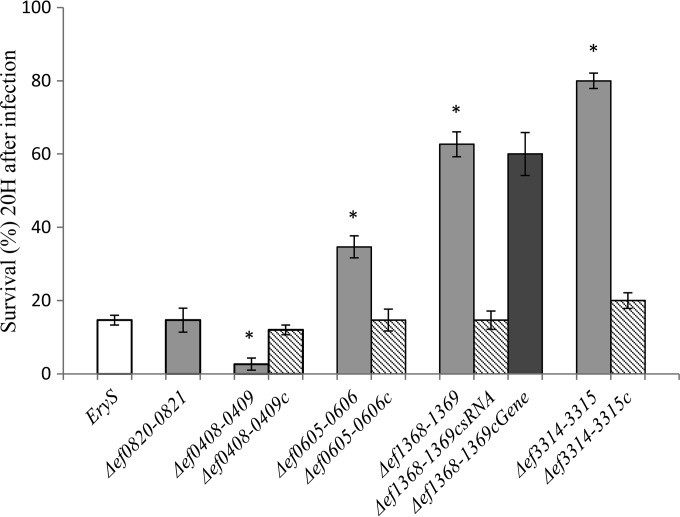
FIG 1.
Effects of inactivation of selected sRNAs on virulence. Shown is the percent survival of G. mellonella at 20 h after infection with around 1.2 × 105 CFU per larva of E. faecalis EryS V583 (wild type); the Δef0408-0409, Δef0605-0606, Δef0820-0821, Δef1368-1369, and Δef3314-3315 sRNA mutants; and the corresponding complemented strains when required. Note the presence of two complemented strains for the Δef1368-1369 mutant, Δef1368-1369csRNA, which represents the small-RNA strand complementation, and Δef1368-1369cGene, the gene strand complementation. Experiments were repeated at least five times, and the results represent the means ± standard deviations. P values of less than 0.001 (*) for the survival of the sRNA mutant strains compared to the wild type were considered to be significant.
The Δef0605-0606, Δef1368-1369, and Δef3314-3315 mutant strains were all less virulent than the wild type. At 20 h postinfection, 35, 63, and 80% of the larvae, respectively, were still alive versus 15% of the larvae infected with the wild type (Fig. 1). Virulence was restored to the wild-type level with the corresponding sRNA-complemented strains. Due to the antisense nature of the Δef1368-1369 sRNA, complementation was also tested with the construction containing the ef1369 gene, encoding a putative transcriptional regulator. In this case, no complementation was observed (Fig. 1). In addition, no difference in caterpillar survival was observed for the wild type with or without the empty pMSP3535 vector (data not shown).
The sRNA ef0820-0821 seems not to be involved in the virulence of E. faecalis, since the survival of larvae infected with the mutant strain was comparable to that of larvae infected with the wild type (Fig. 1). Consequently, we did not further investigate this sRNA.
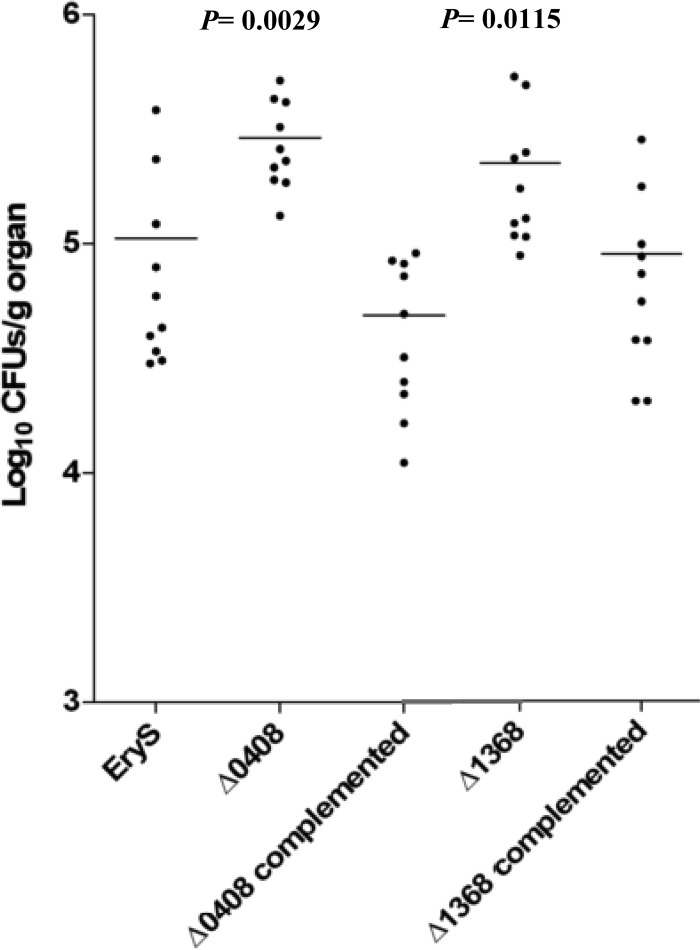
E. faecalis is one of the leading etiological agents of UTI (71). Therefore, we tested the fate of the Δef0408-0409, Δef0605-0606, Δef1368-1369, and Δef3314-3315 mutants, as well as the corresponding complemented and wild-type strains, in kidneys after urinary tract infection of mice. As shown in Fig. 2, the Δef0408-0409 and Δef1368-1369 mutants were around 0.5 log units more abundant in the kidneys than the wild-type and the complemented strains (with P values of 0.0029 and 0.0115, respectively). The UTI experiments performed with the Δef0605-0606 sRNA and the Δef3314-3315 sRNA mutant strains did not show any difference between the wild type and the mutants (data not shown).
FIG 2.
Bacterial persistence within the kidney in urinary tract infection. Enterococcal tissue burdens in kidneys from BALB/c mice infected intravenously with 1 × 109 bacteria/ml of the EryS wild type, the Δef0408-0409 and Δef1368-1369 mutants, and the corresponding complemented strains are shown. Kidney pair homogenates were obtained from groups of 10 mice sacrificed and necropsied at day 7 postinfection. The results represent values recorded separately for each of the 10 mice. The horizontal bars represent the geometric means. P values of less than 0.05 were considered to be significant.
Because of the higher virulence phenotype of the Δef0408-0409 mutant observed in G. mellonella and its better colonization than the wild type in the urinary tract, these strains were also tested in a systemic infection model. We observed that the tissue burden in the liver and kidneys provoked by Δef0408-0409 mutant cells was significantly increased compared to the wild type (see Fig. S2 in the supplemental material).
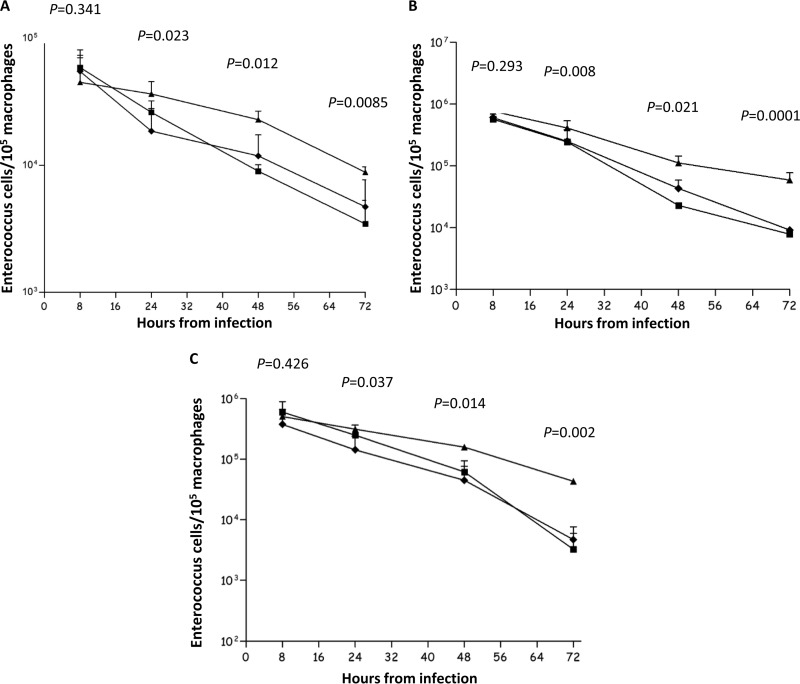
Phagocytes constitute an important part of the innate immunity against pathogens (72). Thus, we were interested in studying the survival of the sRNA mutant strains in an in vivo-in vitro macrophage infection model. As shown in Fig. 3, 3 of the 4 sRNA mutants tested (Δef0408-0409, Δef0605-0606, and Δef1368-1369) survived better within murine peritoneal macrophages than the wild-type or complemented strains over a 72-h period. Of note, no difference was observed between the cell counts in the first hours postinfection, suggesting that phagocytosis levels of the wild-type, mutant, and corresponding complemented strains were similar. No significant difference in survival was observed in the case of the Δef3314-3315 mutant (data not shown).
FIG 3.
Time course of intracellular macrophage survival of the Δef0408-0409 (A), Δef0605-0606 (B), and Δef1368-1369 (C) mutant strains. Diamonds, E. faecalis EryS; triangles, sRNA mutants; squares, complemented strains. The results represent the mean numbers and standard deviations of viable intracellular bacteria per 105 macrophages from three independent experiments.
A link between sRNA and virulence has also been observed for other Gram-positive pathogens, like S. aureus, Streptococcus, and Listeria. For example, the RNAIII of S. aureus activates the translation of hla, encoding hemolysin, and represses the synthesis of major cell surface virulence factors like coagulase and protein A (73–75). In Listeria monocytogenes, the lhrA sRNA regulates the expression of chitinases ChiA and ChiB, which contribute to pathogenesis (76, 77). This is also the case for fasX, pel, and rivX of streptococci, which affect the expression of several virulence factors (78–80). Taken together, our results show for the first time that candidate sRNAs play important roles in the virulence of E. faecalis.
The role of sRNAs in stress response.
To study the functional role of the candidate sRNAs implicated in the virulence process, we selected conditions that might be relevant in the gastrointestinal tract or during the infection process. For that purpose, the wild-type, mutant, and complemented strains were cultured under oxidative-, osmotic-, or detergent stress conditions, as well as in serum (Table 2; see Fig. S3 to S6 in the supplemental material). In the case of acid stress, survival was monitored at a killing pH of 2.8. Under standard growth conditions (in M17glu at 37°C), the growth rates of the sRNA mutants and the parental strain were similar (Table 2). Under all conditions, the phenotypes of the complemented strains were very similar to those of the wild type. This proved that the behavior of the mutants was due only to the deletion of the respective sRNA and not to an additional mutation.
TABLE 2.
Growth phenotypes and promoter activities under different stress conditions
| Growth conditions | Δef0408-0409 strain |
Δef0605-0606 strain |
Δef1368-1369 strain |
Δef3314-3315 strain |
||||
|---|---|---|---|---|---|---|---|---|
| Growth effecta | Promoter activityb | Growth effect | Promoter activity | Growth effect | Promoter activity | Growth effect | Promoter activity | |
| M17glu | NDc | ND | ND | ND | ND | ND | ND | ND |
| 1.5 mM H2O2 | + | −13.0 | + | −3.0 | ND | ND | ND | ND |
| cc17 MOPS glycerol | + | −2.0 | + | −8.0 | ND | ND | ND | ND |
| 8% NaCl | + | −1.3 | ND | ND | − | ND | ND | ND |
| 0.08% bile salts | + | −3.0 | + | −3.0 | ND | ND | + | −2.0 |
| 100% horse serum | ND | ND | ND | ND | + | ND | ND | ND |
+, higher, and −, lower growth rates of the sRNA mutant than of the wild-type strain. All the values are available in Fig. S2 to S5 in the supplemental material.
Ratio of the promoter activities (in Miller units) of the pVEPhoZPsRNAs strains (where phoZ transcription is under the control of sRNA promoters) at T90 (90 min in the presence of the stress) to the measure at T0 (just before the stress). All the values are available in Fig. S7 in the supplemental material.
ND, no difference.
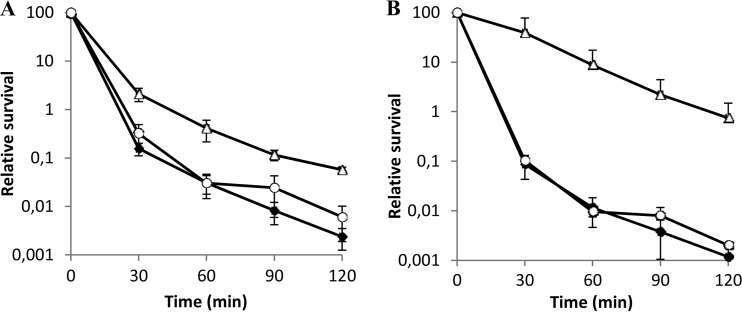
The growth of the Δef0408-0409 strain in serum was comparable to that of the parental strain (Table 2.). However, the mutant grew significantly better under different stress conditions (Table 2; see Fig. S3 in the supplemental material). In the case of oxidative stress, the growth rate of the mutant (μΔ = 0.51) was significantly higher in the presence of 1.5 mM H2O2 than that of the wild type (μwt = 0.17) or the complemented strain (μc = 0.21). It has recently been shown for E. faecalis that aerobic growth with glycerol as the energy source leads to the intracellular accumulation of H2O2 (81). Under these conditions, the wild-type and mutant strains showed similar growth rates during the first 6 h of growth, but the mutant reached a higher final OD600 after 24 h of incubation (0.344 and 0.867, respectively) (see Fig. S3 in the supplemental material). The mutant also showed an increased growth rate in the presence of 0.08% bile salts (μΔ = 0.41, μwt = 0.24, and μc = 0.21) or 8% NaCl (μΔ = 0.38, μwt = 0.2, and μc = 0.22) (Table 2; see Fig. S3 in the supplemental material). Finally, as shown in Fig. 4A, the ef0408-0409 sRNA mutant was significantly more resistant to acidity (M17, pH 2.8) than the wild-type strain (from 13-fold at 30 min to 23-fold at 120 min after the challenge). The ef0408-0409 deletion mutant showed a strong effect in the insect and mouse models, where it appeared more virulent and more able to colonize organs and survived better inside macrophages. These observations might be correlated with the ability of the mutant to better cope with stresses, especially oxidative stress. This showed that the chromosomal TA system was not cryptic and was involved in the virulence of E. faecalis, likely because of its ability to affect the cellular stress response, as well as its impact on the expression of metabolic enzymes (see below). It is tempting, then, to speculate that this system, likely from a plasmid, has been selected to promote colonization rather than virulence.
FIG 4.
Survival under acid stress conditions (M17/7 medium adjusted to pH 2.8) of the sRNA mutant strains. The E. faecalis wild type, the Δef0408-409 mutant (A) or the Δef1368-1369 mutant (B), and the complemented strains are represented by diamonds, triangles, and circles, respectively. At the indicated times, the viable-cell count was determined by plating dilutions onto M17glu; 100% survival corresponds to the viable-cell count determined just prior to exposure to the indicated stress. For all assays, each data point represents the mean ± standard error (SE) of 3 independent experiments.
It has recently been demonstrated that several sRNA deletion mutants of the haloarchaeon Haloferax volcanii showed a gain-of-function phenotype (82). Taken together, it may be suggested that the biological role of some sRNAs can be to stabilize the homeostasis of cells, especially when they are confronted with the multiple environmental changes that occur in real life.
The Δef0605-0606 mutant was exposed to the same stresses, and the results are summarized in Table 2. These cells grew better in the presence of 1.5 mM H2O2 (μΔ = 0.37, μwt = 0.18, and μc = 0.19) or during growth with glycerol than the wild-type strain (see Fig. S4 in the supplemental material). Under the latter condition, the mutant strain reached a much higher final OD600 (4.2) than the parental strain (0.344) (see Fig. S4 in the supplemental material). The Δef0605-0606 mutant also grew somewhat better in the presence of 0.08% bile salts (see Fig. S4 in the supplemental material), which is mostly due to a reduced lag phase compared to the wild-type and complemented strains. Lastly, no difference compared to the wild-type strain has been observed under acid and osmotic stress or in serum (see Fig. S4 in the supplemental material). The ef0605-0606 sRNA is present in only 9 strains of the 54 E. faecalis genomes analyzed, due to its localization in the PAI (35). It has been recently found, using 5′ RACE, that it is also induced under aerobic growth conditions (33, 35). The corresponding mutant was less virulent in the Galleria model but survived better inside macrophages, which may be correlated with its greater fitness in the presence of H2O2. The decreased virulence model of the mutant in the worm could be due to greater sensitivity to antimicrobial molecules present in the hemolymph of the larvae.
The Δef1368-1369 mutant also showed interesting phenotypes under stress conditions (Table 2). It had a somewhat reduced growth rate in the presence of 8% NaCl (μΔ = 0.16 and μwt = 0.28) (see Fig. S5 in the supplemental material) but had remarkably better survival (more than 2 orders of magnitude) in an acid environment (pH 2.8) than the wild-type and the complemented strains (Fig. 4B). Its better ability to cope with this stress may be due to its better survival inside macrophages and ability to colonize mouse organs. Furthermore, the strain lacking ef1368-1369 sRNA grew to a higher final OD600 than the wild type (OD600 = 0.5 and 0.24, respectively) when cultured in horse serum (see Fig. S5 in the supplemental material). Finally, the sRNA mutant lacking ef3314-3315 showed a reduced lag phase when grown in the presence of 0.08% bile salts compared to the wild type and the complemented strain (Table 2; see Fig. S6 in the supplemental material). However, no difference in growth between the wild type and the mutant was observed with the other stresses used in this study (data not shown).
Of note, the sensitivities of the wild-type strain and all the sRNA mutants to cold (8°C) and heat (62°C) shock, autolysin treatment, and exposure to antibiotics were tested, but no difference was observed (data not shown). In addition, the profiles of carbohydrate fermentation (using bioMérieux API test kits [Craponne, France] for bacterial identification of 50 vials of dehydrated substrates) were the same for all strains.
sRNA promoter activities under different stress conditions.
Transcription of sRNA is usually affected by specific environmental conditions (83). Therefore, we were interested in analyzing the promoter activities of the candidate sRNAs. The corresponding promoter regions were fused to the AP gene of E. faecalis, and AP activities, expressed in MU, were then determined under those conditions in which the sRNA mutants showed the most pronounced phenotypes.
Without a promoter fused to AP, only low activity of the enzyme (from 4 to 10 MU) was detected regardless of the growth condition analyzed (data not shown). The promoter activities of Pef0408-0409 and Pef0605-0606 were comparable under nonstress growth conditions (between 193 and 225 MU, respectively). However, both promoters showed 3-fold-decreased activity in the presence of bile salts, 3-fold (Pef0605-0606) and 13-fold (Pef0408-0409) decreases in the presence of 1.5 mM H2O2, and 2-fold (Pef0408-0409) and 8-fold (Pef0605-0606) reductions in expression during growth on glycerol in comparison to nonstress growth conditions (Table 2; see Fig. S7 in the supplemental material). The activity of the ef0408-0409 promoter also showed a slight decrease after 90 min under NaCl conditions (1.3-fold). Furthermore, 2-fold reduction in comparison to the expression under nonstress growth conditions (166 versus 122 MU) was measured for Pef3314-3315 in the presence of bile salts (Table 2; see Fig. S7 in the supplemental material). As previously mentioned, analysis of the different mutant strains revealed that the lack of the transcripts led to better growth under stress conditions. Thus, in the wild-type strain, it seems appropriate to observe that the stressing agents provoked reduced activities of the corresponding sRNA promoters, allowing cells to cope better with these challenges.
Finally, although the Δef1368-1369 mutant strain showed a slightly reduced growth rate under osmotic stress, no change in the AP activity of the Pef1368-1369 construct was found under these conditions (Table 2; see Fig. S7 in the supplemental material). In serum, the growth rates for the mutant and the wild type were similar, and no modification in the activity of Pef1368-1369 has been detected (Table 2; see Fig. S7 in the supplemental material). However, the difference observed in the final OD600 under serum growth conditions suggests that ef1368-1369 sRNA could mainly act during stationary phase.
Identification of the sRNA regulon.
Using quantitative real-time PCR, we checked the effects on the transcription of the flanking genes of the deleted candidate sRNAs. For the two mutant strains Δef0408-0409 and Δef3314-3315, no relevant impact on the transcription of neighboring genes was observed (Table 3). On the other hand, the ef0605 (encoding a hypothetical protein) and ef0606 (encoding a Dps protein) genes were, respectively, 4.4- and 2.7-fold overexpressed in the Δef0605-0606 mutant strain (Table 3). The increased transcription of the ef0606 gene, encoding an enzyme involved in the protection of DNA against oxidative stress, in the sRNA mutant appears consistent with its phenotype toward H2O2 (84). Dps is a DNA-binding protein that protects the genome against oxidative damage and also acts as an iron chelator, reducing its free cellular concentration and consequently the formation of reactive oxygen species (ROS). Indeed, Fe2+ ions are involved in the Fenton reaction that converts H2O2 into OH·, which is a highly deleterious ROS. These results showed that this sRNA is implicated in the regulation of the expression of the adjacent genes that correspond to the first member of the ef0605-0606 sRNA regulon and that ef0605-0606 may act as a repressor.
TABLE 3.
Transcriptional modifications of genes neighboring the sRNA deletions
| Neighboring sRNA gene | Function | Fold change in expressiona (P < 0.01) |
|---|---|---|
| sRNA ef0408-0409 | ||
| ef0407 | Transcriptional regulator | ND |
| ef0408 | Phosphotransferase (PTS) system, IIA component | ND |
| ef0409 | Hypothetical protein | ND |
| ef0411 | PTS system, mannitol-specfic IIBC component | ND |
| sRNA ef0605-0606 | ||
| ef0604 | Gls24 protein | ND |
| ef0605 | Conserved hypothetical protein | +4.4 |
| ef0606 | Dps protein family | +2.7 |
| sRNA ef1368-1369 | ||
| ef1368 | Conserved hypothetical protein | ND |
| ef1369 | Transcriptional regulator, Cro/CI family | −113.8 |
| ef1370 | Drug resistance transporter, EmrB/QacA family protein | −3.3 |
| sRNA ef3314-3315 | ||
| ef3311 | Glucose-inhibited division protein A | ND |
| ef3312 | tRNA modification GTPase TrmE | ND |
| ef3313 | Hypothetical protein | ND |
| ef3314 | Cell wall surface anchor family protein | ND |
| ef3315 | CitG family protein | ND |
The expression values are the averages of at least 3 different RNA extractions from logarithmic-growth-phase cells. All the gene expression values were normalized to the level of gyrA and 23S RNA, and those that differed by more than 2-fold with P values of <0.01 were considered significant. ND, no difference.
Since Δef1368-1369 also corresponds to the deletion of ef1368, as expected, no expression of the latter gene was observed. However, ef1370 (encoding an EmrB drug resistance transporter) appeared to be 3.3-fold repressed in the mutant strain (Table 3). Also, the Δef1368-1369 mutant is not more sensitive to all the antibiotics tested than the wild-type strain (data not shown).
In general, sRNAs act at the posttranscriptional level of regulation by interacting with mRNA (3, 85). Therefore, in order to identify members of sRNA regulons in E. faecalis, i.e., targets whose expression is directly or indirectly controlled by a given sRNA, a global proteomics approach was used to compare the profiles of the sRNA mutant and the parental strains. Using two antibodies (anti-GroEL and anti-Ers), we performed Western blot experiments and confirmed that some proteins were deregulated according to the mutant strain. Thus, GroEL was induced in the Δef0408-409 mutant and repressed in the mutant Δef3314-3315, and Ers was induced in the Δef0408-409 and Δef0605-0606 mutant strains (see Fig. S8 in the supplemental material).
The proteomics experiments revealed that, in general, induced proteins were twice as abundant as repressed proteins in the sRNA mutant strains (112 induced targets versus 61 repressed [at least 2-fold-change]). In the Δef0408-0409, Δef0605-0606, Δef1368-1369, and Δef3314-3315 strains, we observed, respectively, 1.7-, 2-, 3-, and 1.5-fold more induced proteins than repressed polypeptides (see Table S2 in the supplemental material). The putative direct or indirect targets of the candidate sRNAs studied seemed to belong to a large number of cellular-role categories, including cellular process, energy metabolism, regulatory functions, and transcription (Table 4). Of note, the regulation of metabolic pathways (i.e., entry into stationary phase, energy production, and biofilm formation) is necessary for bacterial adaptation to adverse conditions and for pathogenicity. Thus, the fact that several genes encoding enzymes linked to different metabolic pathways were members of the candidate sRNA regulons illustrates their roles in the fitness of bacterial cells, which can be correlated with a role in colonization and/or virulence.
TABLE 4.
Cellular role categories of putative targets up- and downregulated on the sRNA mutant strains
| Cellular role category | No. of targetsa |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
ef0408-0409 |
ef0605-0606 |
ef1368-1369 |
ef3314-3315 |
|||||||||
| T | R | I | T | R | I | T | R | I | T | R | I | |
| Amino acid biosynthesis | 3 | 0 | 3 | 2 | 2 | |||||||
| Biosynthesis of cofactors, prosthetic groups, and carriers | 1 | 0 | 1 | 2 | 2 | |||||||
| Cell envelope | 6 | 3 | 3 | 2 | 1 | 1 | 1 | 0 | 1 | |||
| Cellular processes | 5 | 2 | 3 | 4 | 2 | 2 | 1 | 1 | 0 | 3 | 2 | 1 |
| Central intermediary metabolism | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | |||
| DNA metabolism | 3 | 3 | 0 | 2 | 0 | 2 | ||||||
| Energy metabolism | 10 | 3 | 7 | 4 | 0 | 4 | 5 | 1 | 4 | 5 | 2 | 3 |
| Fatty acid and phospholipid metabolism | 1 | 0 | 1 | 2 | 0 | 2 | 2 | 0 | 2 | 1 | 0 | 1 |
| Hypothetical proteins | 4 | 0 | 4 | 3 | 0 | 3 | ||||||
| Mobile- and extrachromosomal-element functions | 1 | 0 | 1 | 1 | 0 | 1 | 2 | 0 | 2 | 1 | 0 | 1 |
| Protein fate | 5 | 2 | 3 | 3 | 1 | 2 | 1 | 0 | 1 | 2 | 1 | 1 |
| Protein synthesis | 7 | 4 | 3 | 6 | 4 | 2 | 2 | 1 | 1 | 4 | 2 | 2 |
| Purines, pyrimidines, nucleosides, and nucleotides | 8 | 4 | 4 | 10 | 6 | 4 | 2 | 1 | 1 | 1 | 1 | 0 |
| Regulatory functions | 5 | 2 | 3 | 3 | 0 | 3 | 1 | 1 | 0 | |||
| Transcription | 3 | 1 | 2 | 3 | 2 | 1 | 1 | 0 | 1 | 2 | 0 | 2 |
| Transport and binding proteins | 1 | 1 | 0 | |||||||||
| Unknown function | 11 | 3 | 8 | 8 | 2 | 6 | 2 | 0 | 2 | 2 | 0 | 2 |
| Total | 75 | 28 | 47 | 53 | 18 | 35 | 20 | 5 | 15 | 25 | 10 | 15 |
T, total; R, repressed; I, induced.
Some of the identified polypeptides, such as enzymes known to be involved in the oxidative-stress response and/or virulence that may, at least in part, explain the phenotypes of the mutant strains, are of particular interest. This may be true for proteins encoded by the arc operon, overproduced around 2-fold in the Δef0408-0409 mutant and more than 4-fold in the Δef0605-0606 mutant strain. Proteins encoded by the arc operon are enzymes of the arginine deiminase pathway but may also be involved in the oxidative-stress response. In Lactococcus lactis, the disruption of the arcA or arcB gene abolished H2O2 resistance (86). Other proteins linked to oxidative stress were observed. Indeed, the transcriptional regulator Ers, known to play roles in virulence, H2O2 resistance, and glycerol metabolism, has been found to be induced more than 3-fold and 4-fold in the Δef0408-0409 and Δef0605-0606 mutant strains, respectively (24, 53). Interestingly, in E. faecalis, Ers has been shown to participate in the regulation of the arc operon (53). It should be noted that ers (ef0074) and ef0605-0606 sRNA are both located near an operon structure that includes a gls24 paralogue encoding a general stress protein and virulence factor in E. faecalis (87–89).
In addition, the NADH peroxidase enzyme (Npr) was overexpressed 2-fold in the Δef0408-0409 mutant strain. Npr is implicated in the oxidative-stress response in E. faecalis and has been shown to be the most important peroxidase for protecting cells against external or internal H2O2 stress (39, 90, 91).
Expression of some polypeptides belonging to the category “cellular processes,” likely related to pathogenicity, seemed to be controlled by sRNAs. For example, hemolysin A (EF0982) was overproduced around 3-fold in the Δef0408-0409 and Δef0605-0606 mutant strains; the EF2150 protein, a member of the FemAB family, is induced around 5-fold in the two strains; and the superoxide dismutase SodA is induced 2-fold in the Δef3314-3315 mutant (see Table S2 in the supplemental material).
ef1368-1369 sRNA likely acts as an antisense sRNA. However, the expression of 20 proteins was modified in the corresponding mutant strain, showing that the sRNA may have other targets than ef1369 mRNA. We did not find EF1369 in our proteomics data, likely because of its small size (around 13 kDa). Indeed, the 2D gel electrophoresis conditions we used allowed a clear separation of proteins from 15 to 140 kDa.
Computational predictions for mRNA targets of candidate sRNAs wer performed using two software programs (Predator and Copra) (54, 55). For each candidate sRNA, around 100 putative targets were found. Among them, only a few were members of the regulons identified by proteomics (Table 5). Of note, as expected, ef0409, homologous to RNAI of the TA system, could be a target of ef0408-0409 sRNA. Bioinformatics approaches to predict the cellular targets of sRNAs are still plagued by false results, because all the parameters involved in the interaction are not known (2). This may explain the low coverage with genes encoding proteins deregulated in the sRNA mutants, as well as the different data obtained with the two informatics tools tested. Moreover, the members of the regulons identified by proteomics may result from a cascade of regulation events. Nevertheless, further study combining all the approaches and experimental data will provide insight into how these sRNAs function.
TABLE 5.
Computational identification of putative cellular targets of sRNAs that were member of the candidate sRNA regulons obtained by a proteomics approach
| Candidate sRNA | Putative target |
|||
|---|---|---|---|---|
| Predator |
Copra |
|||
| Gene | Function | Gene | function | |
| ef0408-0409 | ef0409a | Hypothetical protein | ef0409a | Hypothetical protein |
| ef3303b | Myosin cross-reactive antigen | |||
| ef2353b | Acetyltransferase | |||
| ef0605-0606 | ef0982b | Hemolysin A | ||
| ef0684a | cmp binding protein; putative | |||
| ef1368-1369 | ef0185b | Phosphopentomutase | ||
| ef2353b | Acetyltransferase | |||
| ef3314-3315 | ef1613a | Formate acetyltransferase | ||
CopraRNA and RNA Predator results with P values lower than 0.01.
CopraRNA and RNA Predator results with P values lower than 0.05.
Conclusions.
These results constitute the first functional analysis of candidate sRNA molecules in enterococci. Some sRNA mutant strains were less virulent, whereas another was more virulent. Commensal bacterial cells, such as E. faecalis, need to find equilibrium between behavior favorable to colonization (repressing virulence) and pathogenicity according to the host environment. Therefore, it is not surprising to observe that the transcription of certain sRNAs was modulated by stresses likely encountered in the different bacterial niches or during infection and that mutants were affected in their growth or survival under stress conditions compared to the wild-type strain. The proteomics approach allowed the identification of some members of the candidate sRNA regulons without distinction between direct and indirect interaction. Nevertheless, these results showed that for E. faecalis, an opportunistic pathogen, the implication of sRNAs in virulence appears to be mainly correlated with global metabolism and the stress response. The fact that the expression of some enzymes was altered in different mutant backgrounds underscores the cross talk that may exist between regulatory networks orchestrated by sRNAs. Because sRNAs are “cost-effective” molecules for cell adaptation to environmental changes, they appear here as key actors for the transition from a commensal relationship with the host to a virulence trait.
The selected candidate sRNAs for this study are among the most highly expressed during the exponential and/or stationary growth phase but correspond to a small fraction of sRNAs present in bacteria (more than 70) (35). We are thus at the first step of the identification of the roles of the sRNAs, which constitutes one of the main future challenges for a better understanding of genetic regulation.
Supplementary Material
ACKNOWLEDGMENTS
The expert technical assistance of Isabelle Rincé, Marie-Jeanne Pigny, Evelyne Marchand, and Thomas Meier is greatly appreciated. We thank A. Benachour, E. Riboulet-Bisson, C. Muller, A. Rincé, and N. Sauvageot for helpful discussions.
C.M. is funded by a thesis grant from the Ministère de l'Enseignement Supérieur et de la Recherche and cosupervised by N.V. and J.-C.G. This study was partly supported by grants from the Agence Nationale de la Recherche in the frame of a transnational ERA-NET PathoGenoMics program (ANR-08-PATH-008-01) and the BMBF (ERANET Pathogenomics Network sncRNAomics project).
Footnotes
Published ahead of print 9 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01900-14.
REFERENCES
- 1.Romby P, Charpentier E. 2010. An overview of RNAs with regulatory functions in gram-positive bacteria. Cell Mol. Life Sci. 67:217–237. 10.1007/s00018-009-0162-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Storz G, Vogel J, Wassarman KM. 2011. Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell 43:880–891. 10.1016/j.molcel.2011.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waters LS, Storz G. 2009. Regulatory RNAs in bacteria. Cell 136:615–628. 10.1016/j.cell.2009.01.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winkler WC, Breaker RR. 2005. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 59:487–517. 10.1146/annurev.micro.59.030804.121336 [DOI] [PubMed] [Google Scholar]
- 5.Zhang J, Lau MW, Ferré-D'Amaré AR. 2010. Ribozymes and riboswitches: modulation of RNA function by small molecules. Biochemistry 49:9123–9131. 10.1021/bi1012645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Irnov Kertsburg A, Winkler WC. 2006. Genetic control by cis-acting regulatory RNAs in Bacillus subtilis: general principles and prospects for discovery. Cold Spring Harbor Symp. Quant. Biol. 71:239–249. 10.1101/sqb.2006.71.021 [DOI] [PubMed] [Google Scholar]
- 7.Breaker RR. 2011. Prospects for riboswitch discovery and analysis. Mol. Cell 43:867–879. 10.1016/j.molcel.2011.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerdes K, Wagner EGH. 2007. RNA antitoxins. Curr. Opin. Microbiol. 10:117–124. 10.1016/j.mib.2007.03.003 [DOI] [PubMed] [Google Scholar]
- 9.Massé E, Salvail H, Desnoyers G, Arguin M. 2007. Small RNAs controlling iron metabolism. Curr. Opin. Microbiol. 10:140–145. 10.1016/j.mib.2007.03.013 [DOI] [PubMed] [Google Scholar]
- 10.Valentin-Hansen P, Johansen J, Rasmussen AA. 2007. Small RNAs controlling outer membrane porins. Curr. Opin. Microbiol. 10:152–155. 10.1016/j.mib.2007.03.001 [DOI] [PubMed] [Google Scholar]
- 11.Vanderpool CK. 2007. Physiological consequences of small RNA-mediated regulation of glucose-phosphate stress. Curr. Opin. Microbiol. 10:146–151. 10.1016/j.mib.2007.03.011 [DOI] [PubMed] [Google Scholar]
- 12.Vogel J, Papenfort K. 2006. Small non-coding RNAs and the bacterial outer membrane. Curr. Opin. Microbiol. 9:605–611. 10.1016/j.mib.2006.10.006 [DOI] [PubMed] [Google Scholar]
- 13.Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. 1997. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell 90:43–53. 10.1016/S0092-8674(00)80312-8 [DOI] [PubMed] [Google Scholar]
- 14.Toledo-Arana A, Repoila F, Cossart P. 2007. Small noncoding RNAs controlling pathogenesis. Curr. Opin. Microbiol. 10:182–188. 10.1016/j.mib.2007.03.004 [DOI] [PubMed] [Google Scholar]
- 15.Romby P, Vandenesch F, Wagner EGH. 2006. The role of RNAs in the regulation of virulence-gene expression. Curr. Opin. Microbiol. 9:229–236. 10.1016/j.mib.2006.02.005 [DOI] [PubMed] [Google Scholar]
- 16.Gottesman S. 2004. The small RNA regulators of Escherichia coli: roles and mechanisms*. Annu. Rev. Microbiol. 58:303–328. 10.1146/annurev.micro.58.030603.123841 [DOI] [PubMed] [Google Scholar]
- 17.Arias CA, Murray BE. 2012. The rise of the Enterococcus: beyond vancomycin resistance. Nat. Rev. Microbiol. 10:266–278. 10.1038/nrmicro2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray BE. 1990. The life and times of the Enterococcus. Clin. Microbiol. Rev. 3:46–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hollenbeck BL, Rice LB. 2012. Intrinsic and acquired resistance mechanisms in enterococcus. Virulence 3:421–433. 10.4161/viru.21282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bearman GML, Wenzel RP. 2005. Bacteremias: a leading cause of death. Arch. Med. Res. 36:646–659. 10.1016/j.arcmed.2005.02.005 [DOI] [PubMed] [Google Scholar]
- 21.Lebreton F, Riboulet-Bisson E, Serror P, Sanguinetti M, Posteraro B, Torelli R, Hartke A, Auffray Y, Giard J-C. 2009. ace, which encodes an adhesin in Enterococcus faecalis, is regulated by Ers and is involved in virulence. Infect. Immun. 77:2832–2839. 10.1128/IAI.01218-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michaux C, Martini C, Hanin A, Auffray Y, Hartke A, Giard J-C 2011. SlyA regulator is involved in bile salts stress response of Enterococcus faecalis. FEMS Microbiol. Lett. 324:142–146. 10.1111/j.1574-6968.2011.02390.x [DOI] [PubMed] [Google Scholar]
- 23.Michaux C, Sanguinetti M, Reffuveille F, Auffray Y, Posteraro B, Gilmore MS, Hartke A, Giard J-C 2011. SlyA is a transcriptional regulator involved in the virulence of Enterococcus faecalis. Infect. Immun. 79:2638–2645. 10.1128/IAI.01132-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riboulet-Bisson E, Le Jeune A, Benachour A, Auffray Y, Hartke A, Giard J-C. 2009. Ers a Crp/Fnr-like transcriptional regulator of Enterococcus faecalis. Int. J. Food Microbiol. 131:71–74. 10.1016/j.ijfoodmicro.2008.06.022 [DOI] [PubMed] [Google Scholar]
- 25.Verneuil N, Rincé A, Sanguinetti M, Auffray Y, Hartke A, Giard J-C. 2005. Implication of hypR in the virulence and oxidative stress response of Enterococcus faecalis. FEMS Microbiol. Lett. 252:137–141. 10.1016/j.femsle.2005.08.043 [DOI] [PubMed] [Google Scholar]
- 26.Verneuil N, Rincé A, Sanguinetti M, Posteraro B, Fadda G, Auffray Y, Hartke A, Giard J-C. 2005. Contribution of a PerR-like regulator to the oxidative-stress response and virulence of Enterococcus faecalis. Microbiology 151:3997–4004. 10.1099/mic.0.28325-0 [DOI] [PubMed] [Google Scholar]
- 27.Verneuil N, Sanguinetti M, Le Breton Y, Posteraro B, Fadda G, Auffray Y, Hartke A, Giard J-C. 2004. Effects of the Enterococcus faecalis hypR gene encoding a new transcriptional regulator on oxidative stress response and intracellular survival within macrophages. Infect. Immun. 72:4424–4431. 10.1128/IAI.72.8.4424-4431.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palmer KL, Gilmore MS. 2010. Multidrug-resistant enterococci lack CRISPR-cas. mBio 1:e00227–10. 10.1128/mBio.00227-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weaver KE, Jensen KD, Colwell A, Sriram SI. 1996. Functional analysis of the Enterococcus faecalis plasmid pAD1-encoded stability determinant par. Mol. Microbiol. 20:53–63. 10.1111/j.1365-2958.1996.tb02488.x [DOI] [PubMed] [Google Scholar]
- 30.Weaver KE. 2007. Emerging plasmid-encoded antisense RNA regulated systems. Curr. Opin. Microbiol. 10:110–116. 10.1016/j.mib.2007.03.002 [DOI] [PubMed] [Google Scholar]
- 31.Garsin DA. 2010. Ethanolamine utilization in bacterial pathogens: roles and regulation. Nat. Rev. Microbiol. 8:290–295. 10.1038/nrmicro2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramesh A, DebRoy S, Goodson JR, Fox KA, Faz H, Garsin DA, Winkler WC. 2012. The mechanism for RNA recognition by ANTAR regulators of gene expression. PLoS Genet. 8:e1002666. 10.1371/journal.pgen.1002666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fouquier d'Hérouel A, Wessner F, Halpern D, Ly-Vu J, Kennedy SP, Serror P, Aurell E, Repoila F. 2011. A simple and efficient method to search for selected primary transcripts: non-coding and antisense RNAs in the human pathogen Enterococcus faecalis. Nucleic Acids Res. 39:e46. 10.1093/nar/gkr012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livny J, Teonadi H, Livny M, Waldor MK. 2008. High-throughput, kingdom-wide prediction and annotation of bacterial non-coding RNAs. PLoS One 3:e3197. 10.1371/journal.pone.0003197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shioya K, Michaux C, Kuenne C, Hain T, Verneuil N, Budin-Verneuil A, Hartsch T, Hartke A, Giard J-C 2011. Genome-wide identification of small RNAs in the opportunistic pathogen Enterococcus faecalis V583. PLoS One 6:e23948. 10.1371/journal.pone.0023948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hancock LE, Gilmore MS. 2002. The capsular polysaccharide of Enterococcus faecalis and its relationship to other polysaccharides in the cell wall. Proc. Natl. Acad. Sci. U. S. A. 99:1574–1579. 10.1073/pnas.032448299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rigottier-Gois L, Alberti A, Houel A, Taly J-F, Palcy P, Manson J, Pinto D, Matos RC, Carrilero L, Montero N, Tariq M, Karsens H, Repp C, Kropec A, Budin-Verneuil A, Benachour A, Sauvageot N, Bizzini A, Gilmore MS, Bessières P, Kok J, Huebner J, Lopes F, Gonzalez-Zorn B, Hartke A, Serror P. 2011. Large-scale screening of a targeted Enterococcus faecalis mutant library identifies envelope fitness factors. PLoS One 6:e29023. 10.1371/journal.pone.0029023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 39.La Carbona S, Sauvageot N, Giard J-C, Benachour A, Posteraro B, Auffray Y, Sanguinetti M, Hartke A. 2007. Comparative study of the physiological roles of three peroxidases (NADH peroxidase, Alkyl hydroperoxide reductase and Thiol peroxidase) in oxidative stress response, survival inside macrophages and virulence of Enterococcus faecalis. Mol. Microbiol. 66:1148–1163. 10.1111/j.1365-2958.2007.05987.x [DOI] [PubMed] [Google Scholar]
- 40.Michaux C, Martini C, Shioya K, Ahmed Lecheheb S, Budin-Verneuil A, Cosette P, Sanguinetti M, Hartke A, Verneuil N, Giard J-C 2012. CspR, a cold shock RNA-binding protein involved in the long-term survival and the virulence of Enterococcus faecalis. J. Bacteriol. 194:6900–6908. 10.1128/JB.01673-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thurlow LR, Thomas VC, Hancock LE. 2009. Capsular polysaccharide production in Enterococcus faecalis and contribution of CpsF to capsule serospecificity. J. Bacteriol. 191:6203–6210. 10.1128/JB.00592-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bryan EM, Bae T, Kleerebezem M, Dunny GM. 2000. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44:183–190. 10.1006/plas.2000.1484 [DOI] [PubMed] [Google Scholar]
- 43.Le Jeune A, Touchet F, Zhao C, Hartke A, Auffray Y, Benachour A. 2010. Construction of a new sensitive molecular tool for the study of gene expression in Enterococcus faecalis. J. Mol. Microbiol. Biotechnol. 19:159–168. 10.1159/000321663 [DOI] [PubMed] [Google Scholar]
- 44.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, Barthelemy M, Vergassola M, Nahori M-A, Soubigou G, Régnault B, Coppée J-Y, Lecuit M, Johansson J, Cossart P. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950–956. 10.1038/nature08080 [DOI] [PubMed] [Google Scholar]
- 45.Meijerink J, Mandigers C, van de Locht L, Tönnissen E, Goodsaid F, Raemaekers J. 2001. A novel method to compensate for different amplification efficiencies between patient DNA samples in quantitative real-time PCR. J. Mol. Diagn. 3:55–61. 10.1016/S1525-1578(10)60652-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Becher D, Hempel K, Sievers S, Zühlke D, Pané-Farré J, Otto A, Fuchs S, Albrecht D, Bernhardt J, Engelmann S, Völker U, van Dijl JM, Hecker M. 2009. A proteomic view of an important human pathogen—towards the quantification of the entire Staphylococcus aureus proteome. PLoS One 4:e8176. 10.1371/journal.pone.0008176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bernhardt J, Büttner K, Scharf C, Hecker M. 1999. Dual channel imaging of two-dimensional electropherograms in Bacillus subtilis. Electrophoresis 20:2225–2240 [DOI] [PubMed] [Google Scholar]
- 48.Engelmann S, Hecker M. 2008. Proteomic analysis to investigate regulatory networks in Staphylococcus aureus. Methods Mol. Biol. 431:25–45. 10.1007/978-1-60327-032-8_3 [DOI] [PubMed] [Google Scholar]
- 49.Reiss CS. 2013. The complexities of producing proteins. DNA Cell Biol. 32:285. 10.1089/dna.2013.2529 [DOI] [PubMed] [Google Scholar]
- 50.Eymann C, Dreisbach A, Albrecht D, Bernhardt J, Becher D, Gentner S, Tam LT, Büttner K, Buurman G, Scharf C, Venz S, Völker U, Hecker M. 2004. A comprehensive proteome map of growing Bacillus subtilis cells. Proteomics 4:2849–2876. 10.1002/pmic.200400907 [DOI] [PubMed] [Google Scholar]
- 51.Wolf C, Hochgräfe F, Kusch H, Albrecht D, Hecker M, Engelmann S. 2008. Proteomic analysis of antioxidant strategies of Staphylococcus aureus: diverse responses to different oxidants. Proteomics 8:3139–3153. 10.1002/pmic.200701062 [DOI] [PubMed] [Google Scholar]
- 52.Fuchs S, Zühlke D, Pané-Farré J, Kusch H, Wolf C, Reiß S, Binh LTN, Albrecht D, Riedel K, Hecker M, Engelmann S. 2013. Aureolib, a proteome signature library: towards an understanding of Staphylococcus aureus pathophysiology. PLoS One 8:e70669. 10.1371/journal.pone.0070669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Riboulet-Bisson E, Sanguinetti M, Budin-Verneuil A, Auffray Y, Hartke A, Giard J-C. 2008. Characterization of the Ers regulon of Enterococcus faecalis. Infect. Immun. 76:3064–3074. 10.1128/IAI.00161-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eggenhofer F, Tafer H, Stadler PF, Hofacker IL. 2011. RNApredator: fast accessibility-based prediction of sRNA targets. Nucleic Acids Res. 39:W149–W154. 10.1093/nar/gkr467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wright PR, Richter AS, Papenfort K, Mann M, Vogel J, Hess WR, Backofen R, Georg J. 2013. Comparative genomics boosts target prediction for bacterial small RNAs. Proc. Natl. Acad. Sci. U. S. A. 110:E3487–E3496. 10.1073/pnas.1303248110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hayes CS, Keiler KC. 2010. Beyond ribosome rescue: tmRNA and co-translational processes. FEBS Lett. 584:413–419. 10.1016/j.febslet.2009.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dulebohn D, Choy J, Sundermeier T, Okan N, Karzai AW. 2007. Trans-translation: the tmRNA-mediated surveillance mechanism for ribosome rescue, directed protein degradation, and nonstop mRNA decay. Biochemistry 46:4681–4693. 10.1021/bi6026055 [DOI] [PubMed] [Google Scholar]
- 58.Keenan RJ, Freymann DM, Stroud RM, Walter P. 2001. The signal recognition particle. Annu. Rev. Biochem. 70:755–775. 10.1146/annurev.biochem.70.1.755 [DOI] [PubMed] [Google Scholar]
- 59.Keiler KC. 2007. Physiology of tmRNA: what gets tagged and why? Curr. Opin. Microbiol. 10:169–175. 10.1016/j.mib.2007.03.014 [DOI] [PubMed] [Google Scholar]
- 60.Champion OL, Cooper IAM, James SL, Ford D, Karlyshev A, Wren BW, Duffield M, Oyston PCF, Titball RW. 2009. Galleria mellonella as an alternative infection model for Yersinia pseudotuberculosis. Microbiology 155:1516–1522. 10.1099/mic.0.026823-0 [DOI] [PubMed] [Google Scholar]
- 61.Fuchs BB, O'Brien E, Khoury JBE, Mylonakis E. 2010. Methods for using Galleria mellonella as a model host to study fungal pathogenesis. Virulence 1:475–482. 10.4161/viru.1.6.12985 [DOI] [PubMed] [Google Scholar]
- 62.Jackson JC, Higgins LA, Lin X. 2009. Conidiation color mutants of Aspergillus fumigatus are highly pathogenic to the heterologous insect host Galleria mellonella. PLoS One 4:e4224. 10.1371/journal.pone.0004224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mukherjee K, Altincicek B, Hain T, Domann E, Vilcinskas A, Chakraborty T. 2010. Galleria mellonella as a model system for studying Listeria pathogenesis. Appl. Environ. Microbiol. 76:310–317. 10.1128/AEM.01301-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mylonakis E. 2008. Galleria mellonella and the study of fungal pathogenesis: making the case for another genetically tractable model host. Mycopathologia 165:1–3. 10.1007/s11046-007-9082-z [DOI] [PubMed] [Google Scholar]
- 65.Peleg AY, Jara S, Monga D, Eliopoulos GM, Moellering RC, Jr, Mylonakis E. 2009. Galleria mellonella as a model system to study Acinetobacter baumannii pathogenesis and therapeutics. Antimicrob. Agents Chemother. 53:2605–2609. 10.1128/AAC.01533-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Olsen RJ, Watkins ME, Cantu CC, Beres SB, Musser JM. 2011. Virulence of serotype M3 Group A Streptococcus strains in wax worms (Galleria mellonella larvae). Virulence 2:111–119. 10.4161/viru.2.2.14338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brennan M, Thomas DY, Whiteway M, Kavanagh K. 2002. Correlation between virulence of Candida albicans mutants in mice and Galleria mellonella larvae. FEMS Immunol. Med. Microbiol. 34:153–157. 10.1111/j.1574-695X.2002.tb00617.x [DOI] [PubMed] [Google Scholar]
- 68.Jander G, Rahme LG, Ausubel FM. 2000. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J. Bacteriol. 182:3843–3845. 10.1128/JB.182.13.3843-3845.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weaver KE, Reddy SG, Brinkman CL, Patel S, Bayles KW, Endres JL. 2009. Identification and characterization of a family of toxin-antitoxin systems related to the Enterococcus faecalis plasmid pAD1 par addiction module. Microbiology 155:2930–2940. 10.1099/mic.0.030932-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weaver KE. 2012. The par toxin-antitoxin system from Enterococcus faecalis plasmid pAD1 and its chromosomal homologs. RNA Biol. 9:1498–1503. 10.4161/rna.22311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Svanborg C, Godaly G. 1997. Bacterial virulence in urinary tract infection. Infect. Dis. Clin. North Am. 11:513–529. 10.1016/S0891-5520(05)70371-8 [DOI] [PubMed] [Google Scholar]
- 72.Sarantis H, Grinstein S. 2012. Subversion of phagocytosis for pathogen survival. Cell Host Microbe 12:419–431. 10.1016/j.chom.2012.09.001 [DOI] [PubMed] [Google Scholar]
- 73.Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, Gaspin C, Vandenesch F, Romby P. 2007. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 21:1353–1366. 10.1101/gad.423507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huntzinger E, Boisset S, Saveanu C, Benito Y, Geissmann T, Namane A, Lina G, Etienne J, Ehresmann B, Ehresmann C, Jacquier A, Vandenesch F, Romby P. 2005. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 24:824–835. 10.1038/sj.emboj.7600572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morfeldt E, Taylor D, von Gabain A, Arvidson S. 1995. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 14:4569–4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mraheil MA, Billion A, Kuenne C, Pischimarov J, Kreikemeyer B, Engelmann S, Hartke A, Giard J-C, Rupnik M, Vorwerk S, Beier M, Retey J, Hartsch T, Jacob A, Cemič F, Hemberger J, Chakraborty T, Hain T. 2010. Comparative genome-wide analysis of small RNAs of major Gram-positive pathogens: from identification to application. Microb. Biotechnol. 3:658–676. 10.1111/j.1751-7915.2010.00171.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mraheil MA, Billion A, Mohamed W, Mukherjee K, Kuenne C, Pischimarov J, Krawitz C, Retey J, Hartsch T, Chakraborty T, Hain T. 2011. The intracellular sRNA transcriptome of Listeria monocytogenes during growth in macrophages. Nucleic Acids Res. 39:4235–4248. 10.1093/nar/gkr033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Klenk M, Koczan D, Guthke R, Nakata M, Thiesen H-J, Podbielski A, Kreikemeyer B. 2005. Global epithelial cell transcriptional responses reveal Streptococcus pyogenes Fas regulator activity association with bacterial aggressiveness. Cell. Microbiol. 7:1237–1250. 10.1111/j.1462-5822.2005.00548.x [DOI] [PubMed] [Google Scholar]
- 79.Mangold M, Siller M, Roppenser B, Vlaminckx BJM, Penfound TA, Klein R, Novak R, Novick RP, Charpentier E. 2004. Synthesis of group A streptococcal virulence factors is controlled by a regulatory RNA molecule. Mol. Microbiol. 53:1515–1527. 10.1111/j.1365-2958.2004.04222.x [DOI] [PubMed] [Google Scholar]
- 80.Roberts SA, Scott JR. 2007. RivR and the small RNA RivX: the missing links between the CovR regulatory cascade and the Mga regulon. Mol. Microbiol. 66:1506–1522. 10.1111/j.1365-2958.2007.06015.x [DOI] [PubMed] [Google Scholar]
- 81.Bizzini A, Zhao C, Budin-Verneuil A, Sauvageot N, Giard J-C, Auffray Y, Hartke A. 2010. Glycerol is metabolized in a complex and strain-dependent manner in Enterococcus faecalis. J. Bacteriol. 192:779–785. 10.1128/JB.00959-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jaschinski K, Babski J, Lehr M, Burmester A, Benz J, Heyer R, Dörr M, Marchfelder A, Soppa J. 2014. Generation and phenotyping of a collection of sRNA gene deletion mutants of the haloarchaeon Haloferax volcanii. PLoS One 9:e90763. 10.1371/journal.pone.0090763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Repoila F, Darfeuille F. 2009. Small regulatory non-coding RNAs in bacteria: physiology and mechanistic aspects. Biol. Cell 101:117–131. 10.1042/BC20070137 [DOI] [PubMed] [Google Scholar]
- 84.Shankar J, Walker RG, Wilkinson MC, Ward D, Horsburgh MJ. 2012. SalB inactivation modulates culture supernatant exoproteins and affects autolysis and viability in Enterococcus faecalis OG1RF. J. Bacteriol. 194:3569–3578. 10.1128/JB.00376-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Storz G, Opdyke JA, Zhang A. 2004. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 7:140–144. 10.1016/j.mib.2004.02.015 [DOI] [PubMed] [Google Scholar]
- 86.Rochat T, Boudebbouze S, Gratadoux J-J, Blugeon S, Gaudu P, Langella P, Maguin E. 2012. Proteomic analysis of spontaneous mutants of Lactococcus lactis: involvement of GAPDH and arginine deiminase pathway in H2O2 resistance. Proteomics 12:1792–1805. 10.1002/pmic.201100465 [DOI] [PubMed] [Google Scholar]
- 87.Giard JC, Rince A, Capiaux H, Auffray Y, Hartke A. 2000. Inactivation of the stress- and starvation-inducible gls24 operon has a pleiotrophic effect on cell morphology, stress sensitivity, and gene expression in Enterococcus faecalis. J. Bacteriol. 182:4512–4520. 10.1128/JB.182.16.4512-4520.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Giard J-C, Verneuil N, Auffray Y, Hartke A. 2002. Characterization of genes homologous to the general stress-inducible gene gls24 in Enterococcus faecalis and Lactococcus lactis. FEMS Microbiol. Lett. 206:235–239. 10.1111/j.1574-6968.2002.tb11015.x [DOI] [PubMed] [Google Scholar]
- 89.Teng F, Nannini EC, Murray BE. 2005. Importance of gls24 in virulence and stress response of Enterococcus faecalis and use of the Gls24 protein as a possible immunotherapy target. J. Infect. Dis. 191:472–480. 10.1086/427191 [DOI] [PubMed] [Google Scholar]
- 90.Claiborne A, Ross RP, Parsonage D. 1992. Flavin-linked peroxide reductases: protein-sulfenic acids and the oxidative stress response. Trends Biochem. Sci. 17:183–186. 10.1016/0968-0004(92)90263-9 [DOI] [PubMed] [Google Scholar]
- 91.Claiborne A, Yeh JI, Mallett TC, Luba J, Crane EJ, III, Charrier V, Parsonage D. 1999. Protein-sulfenic acids: diverse roles for an unlikely player in enzyme catalysis and redox regulation. Biochemistry 38:15407–15416. 10.1021/bi992025k [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.