Abstract
HIV coinfection accelerates disease progression in chronic hepatitis C and reduces sustained antiviral responses (SVR) to interferon-based therapy. New direct-acting antivirals (DAAs) promise higher SVR rates, but the selection of preexisting resistance-associated variants (RAVs) may lead to virologic breakthrough or relapse. Thus, pretreatment frequencies of RAVs are likely determinants of treatment outcome but typically are below levels at which the viral sequence can be accurately resolved. Moreover, it is not known how HIV coinfection influences RAV frequency. We adopted an accurate high-throughput sequencing strategy to compare nucleotide diversity in HCV NS3 protease-coding sequences in 20 monoinfected and 20 coinfected subjects with well-controlled HIV infection. Differences in mean pairwise nucleotide diversity (π), Tajima's D statistic, and Shannon entropy index suggested that the genetic diversity of HCV is reduced in coinfection. Among coinfected subjects, diversity correlated positively with increases in CD4+ T cells on antiretroviral therapy, suggesting T cell responses are important determinants of diversity. At a median sequencing depth of 0.084%, preexisting RAVs were readily identified. Q80K, which negatively impacts clinical responses to simeprevir, was encoded by more than 99% of viral RNAs in 17 of the 40 subjects. RAVs other than Q80K were identified in 39 of 40 subjects, mostly at frequencies near 0.1%. RAV frequency did not differ significantly between monoinfected and coinfected subjects. We conclude that HCV genetic diversity is reduced in patients with well-controlled HIV infection, likely reflecting impaired T cell immunity. However, RAV frequency is not increased and should not adversely influence the outcome of DAA therapy.
INTRODUCTION
Coinfection with hepatitis C virus (HCV) is an important cause of morbidity and mortality among persons infected with HIV (1, 2). Disease progression is accelerated, and coinfected patients respond poorly to treatment with pegylated alpha interferon and ribavirin (Peg-IFN/RBV) compared to those infected only with HCV (monoinfection). The rate of sustained antiviral response (SVR) with Peg-IFN/RBV was only 19 to 22% in a large, international multicenter study of coinfected patients (3), less than half that observed in monoinfected patients. The addition of direct-acting antiviral (DAA) therapies to Peg-IFN/RBV significantly enhances SVR rates in monoinfected patients (4, 5). Four DAAs have been approved thus far for use in the United States, three of which (boceprevir, telaprevir, and simeprevir) target the viral NS3/4A protease (6 and http://www.hcvguidelines.org). Protease inhibitors show promise when used in combination with Peg-IFN for treatment of persons coinfected with HIV (7–9) and likely will be key components of all oral, interferon-sparing therapies for hepatitis C in the future. Nonetheless, experience with the use of DAAs in coinfected patients remains limited.
In monoinfected patients, HCV variants resistant to NS3/4A protease inhibitors emerge rapidly unless the drugs are used in combination with other inhibitors of HCV replication, such as Peg-IFN/RBV (10, 11). Even with concomitant Peg-IFN/RBV, however, virologic breakthrough or relapse occurred in as many as 11 to 14% of coinfected patients treated in phase IIa trials of HCV protease inhibitors (7, 12). Resistant HCV variants emerging during DAA therapy likely represent preexisting minor quasispecies that become predominant due to the selective pressure of the drugs (13). In part, this reflects the highly replicative nature of HCV infection, with large numbers of new virions produced each day (∼5 × 1012) in the typical infected individual (14), coupled with error-prone genome replication. Thus, the response to DAA therapy and the risk of emergence of resistant HCV variants may be influenced by the frequency and fitness of preexisting resistance-associated variants (RAVs) (15, 16). Preexisting RAVs likely will be particularly important in interferon-sparing treatment regimens consisting solely of combinations of DAAs.
An important question is whether the frequency of RAVs is increased in HCV treatment-naive, coinfected persons. This might be expected if the genetic diversity of HCV is enhanced in coinfection by impaired host responses with attendant reductions in the clearance of the virus. Indeed, some previous studies suggest increased diversity within the envelope-coding region of the HCV genome in coinfected versus monoinfected subjects (17–19). However, HIV diversity decreases with advancing immunosuppression (20), and other studies suggest that the genetic diversity of HCV is reduced in coinfected persons (21–23). Still other studies have simply failed to demonstrate a difference (24, 25). These conflicting results likely are related to variations in cohort size, sampling depth, the region of the HCV genome examined, and/or technical errors related to the sequencing strategy.
Efforts to define the population diversity of RNA viruses by high-throughput sequencing may be confounded by multiple sources of error (26, 27). Artifactual increases in sequence diversity may be imparted by the misincorporation of nucleotides during reverse transcription of RNA to cDNA or during cDNA amplification by PCR. Skewed views of viral population structure also can result from recombination during multiple rounds of PCR, nonuniform amplification of sequences, or oversampling of small numbers of genomes in the starting population. Each of these types of error may obscure the true frequency of minor variants within a viral population. However, by incorporating a random 8-nucleotide (nt) tag within the primer used for the initial reverse transcription of a population of viral RNAs while purposefully undersampling that population with the number of tagging combinations, it is possible to identify PCR amplicons derived from a single RNA template (28). This allows a consensus sequence to be determined for a cluster of sequencing reads originating from that one template and largely overcomes PCR-related errors in defining viral population diversity by high-throughput sequencing. We exploited this primer ID strategy to accurately sequence HCV quasispecies at an unprecedented depth.
Our goals in this study were 3-fold: (i) to adapt this high-throughput primer ID sequencing strategy to the study of HCV quasispecies; (ii) to compare the degree of genetic diversity of NS3 sequences in monoinfected, treatment-naive patients to that in coinfected patients who are HCV treatment naive but on stable antiretroviral therapy (ART); and (iii) to conduct an unbiased and accurate assessment of the frequency of protease inhibitor RAVs in HCV treatment-naive subjects and ascertain whether RAV frequencies are altered by HIV coinfection. Our results show that HCV diversity is decreased in coinfected subjects but that protease inhibitor RAVs are present at similar frequencies in these two groups.
MATERIALS AND METHODS
Patient samples.
Archived plasma or serum samples were collected between 2000 and 2011 from 20 monoinfected and 20 coinfected HCV treatment-naive subjects at University of North Carolina hospitals (Table 1). All were infected with genotype 1a HCV, with serum/plasma HCV RNA levels of ≥1 ×105 IU/ml. Coinfected subjects were on stable antiretroviral therapy (ART) with serum/plasma HIV RNA ratios of <50 copies/ml and a median CD4+ T cell count of 430 cells/mm3 (interquartile range [IQR], 284 to 665 cells/mm3). The study was approved by the Institutional Review Board of The University of North Carolina at Chapel Hill. All study subjects provided written informed consent prior to enrollment.
TABLE 1.
Demographic characteristics of study cohorts
| Characteristic | Value by infection type |
P valuea | |
|---|---|---|---|
| Monoinfected (n = 20) | Coinfected (n = 20) | ||
| Age, years (means ± SD) (median/IQR) | 52.0 ± 9.7 (54.0/49.5–56.0) | 50.0 ± 4.7 (51.0/48.0–52.5) | 0.08 |
| Gender (no. of males) | 14 | 17 | 0.45 |
| Race | |||
| African-American | 6 | 14 | 0.004 |
| Caucasian | 14 | 4 | |
| Other | 0 | 2 | |
| Serum/plasma HCV RNA, ×106 copies/ml (means ± SD) (median/IQR) | 17.5 ± 21.1 (5.79/2.34–33.7) | 22.4 ± 23.8 (11.2/3.06–40.1) | 0.44 |
| ALTb (U/liter; means ± SD) (median/IQR) | 98.3 ± 107 (69.0/57.5–91.5) | 58.2 ± 27.8 (54.5/36.5–71.0) | 0.08 |
| Bilirubin (mg/dl; means ± SD) (median/IQR) | 0.7 ± 0.4 (0.6/0.4–0.8) | 1.0 ± 0.9c (0.8/0.4–1.1) | 0.22 |
| Liver biopsy specimens (no. [%]) | 15 (75) | 13 (65) | |
| Grade activity (median/IQR)d | 2 (1/2) | 1 (1/1.5) | 0.45 |
| Stage fibrosis (median/IQR) | 1.5 (0/2) | 2 (1/3) | 0.26 |
| Serum/plasma HIV RNA (copies/ml) | NAe | <50 | |
| Serum HBsAgf positive (no.) | 0/20 | 0/20 | |
| ART daysg (no.; means ± SD) (median/IQR) | 3,082 ± 1,744 (3,360/1,482–4,442) | ||
| CD4+ cells/mm3 (means ± SD) (median/IQR) | NA | 515 ± 308 (430/284–665) | |
| ΔCD4h cells/mm3 (means ± SD) (median/IQR) | NA | 364 ± 216 (305/214–509) | |
Wilcoxon rank-sum (WRS) for continuous measures and Fisher's exact test for gender and African-American versus Caucasian race (excluding other races).
ALT, alanine aminotransferase.
Bilirubin data were available for 19 coinfected subjects.
Liver biopsy activity and fibrosis scored by the Metavir system: activity grade was reported for 12 coinfected biopsies only.
NA, not available.
HBsAg, hepatitis B virus surface antigen.
Duration of antiretroviral therapy (ART).
ΔCD4, the difference between the historic nadir in the CD4+ T cell count and that at the time of sampling.
Viral RNA extraction and cDNA synthesis.
Viral RNA was extracted using the QIAamp viral RNA minikit (Qiagen, Valencia, CA). Approximately 10,000 RNA copies were used as the template for reverse transcription with SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA) and a tagging primer with the consensus sequence 5′-ACCTTGCAAGCACGCTCTGGCCTTGAA-NNNNNNNN-CT-(barcode)-GAACACCGGGGACCTCATGGTTGTCTC-3′, in which an 8-nt degenerate primer ID tag (NNNNNNNN) was followed by a CT linker and a 3- to 4-nt barcode for sample identification. The 3′ end of the tagging primer (underlined sequence) was customized to provide an exact reverse complement of the HCV sequence downstream of codon 173 of NS3 in each subject (nt 3945 to 3971 in H77 virus; GenBank accession number AF011751) as determined by prior population sequencing. Primer sequences are shown in Table S1 in the supplemental material. Oligonucleotides were purchased from Integrated DNA Technologies (IDT, Coralville, IA) and purified by standard desalting.
Amplification of tagged cDNA.
Single-stranded cDNA was column purified using the PureLink PCR purification kit (Invitrogen, Carlsbad, CA), washing 3 times with binding buffer HC (high cutoff) to remove the cDNA primer. Primer removal was verified on a subset of samples by electropherogram analysis using an Experion HighSense RNA microfluidic chip (Bio-Rad Laboratories, Hercules, CA). Purified cDNA was amplified by nested PCR using the primer 5′-TAYTGCTYGGRCCRGCYGA-3′ (H77c nt 3370 to 3388) for first-round PCR and patient-matched primers with the consensus sequence 5′-AGTGGAGGGTGAGGTCCAGAT-3′ (H77c nt 3503 to 3523) for second-round PCR. Downstream primers for nested PCR targeted the 5′ end of the tagging primer: 5′-ACCTTGCAAGCACGCTCTGGC-3′ (first round) and 5′-CAAGCACGCTCTGGCCTTGAA-3′ (second round). Amplification was carried out in HS buffer premix (PrimerSTAR HS kit; TaKaRa, Japan). For first-round PCR, purified cDNA was divided between two 50-μl reaction mixes, each subjected to 20 amplification cycles at 98°C for 10 s followed by 68°C for 45 s. One μl of the combined first-round reaction products was subjected to the second round of PCR that involved amplification for 20 cycles at 98°C for 10 s and 68°C for 45 s. Reaction products were gel purified using a 1.5% agarose gel and the MinElute gel extraction kit (Qiagen) with incubation of the solubilization buffer at room temperature. PCR amplicons were quantified using the PicoGreen double-stranded DNA assay (Life Technologies, Grand Island, NY).
454 Junior pyrosequencing and bioinformatic processing of raw sequences.
Amplicons from groups of 4 patient samples, each with a unique barcode, were pooled in equal molar ratios. Library adaptors were added by blunt-end ligation using the Rapid Library preparation kit (Roche Diagnostics, Indianapolis, IN). Products were analyzed with a Bioanalyzer using a high-sensitivity DNA chip (Agilent Technologies, Santa Clara, CA). The resulting library (2 × 107 copies) was subjected to emulsion-based clonal amplification with the Lib L emPCR amplification kit (454 Life Sciences, Branford, CT) and sequenced on the 454 GS Junior platform using Titanium chemistry (454 Life Sciences). Raw sequencing reads were processed through the native amplicon pipeline using default settings. A custom bioinformatics pipeline was used to filter and parse raw 454 primer ID sequencing reads as previously described (28). In short, individual full-length reads were evaluated for the barcode and primer ID tag derived from the tagging primer used for cDNA synthesis. When 3 or more full-length reads from a single patient sample (identical barcode) contained identical primer IDs, a consensus sequence was constructed by majority rule.
Viral population diversity.
Diversity measures were computed for populations of unambiguous consensus sequences derived from 3 or more individual full-length reads as described above. The use of tagging primers containing numerically greater numbers of random tag sequences (in theory, 65,536) than viral RNA copies (∼10,000) in the reverse transcription reaction ensures that the large majority of consensus sequences are derived from a single RNA template molecule (28). Three different measures of genetic diversity were used to compare virus populations from different subjects: average pairwise nucleotide diversity (π) (29), Tajima's statistic (D) (30), and the Shannon index (31, 32). Nucleotide diversity was computed for the entire read length using a customized script according to the formula , in which xi and xj are the frequency of ith and jth sequences, respectively, and πij is the number of differences between them (29). Tajima's D and sliding-window analysis of π (100-nt window with 10-nt step size) were computed by DnaSP v.5.10.01 (33). The Shannon index was calculated using the VEGAN package of functions for R (34). Haplotypes (defined as unique combinations of sequence polymorphisms within a consensus sequence) and RAV occurrence at specific codons were enumerated with customized bioinformatics scripts in which consensus sequences in each population first were sorted by diversity, binned by unique variant, and then tallied to obtain variant frequencies. The RAVs that we searched for were V36A, V36L, or V36M; Q41R; F43S or F43I; T54A or T54S; V55A or V55I; Q80K or Q80R; V107I; R109K; S138T; R155G, R155K, R155M, or R155T; A156S, A156V, or A156D; D168E, D168N, or D168G; and V170T.
Statistical analysis.
Differences in diversity indices from monoinfected versus coinfected subjects were tested for statistical significance by a t test with Welch's adjustment for unequal variances; an exact Wilcoxon rank-sum (WRS) test was used as a sensitivity analysis. Multivariable associations between covariates and each diversity index were analyzed with linear regression. Correlations between diversity indices and clinical parameters were determined by Spearman's rank correlation. RAV frequency was modeled using negative binomial regression with coinfection status and codon nucleotide as predictor variables. This model was fit using generalized estimating equations (GEE) with an exchangeable working correlation structure to account for multiple within-subject codon nucleotide observations. The outcome variable was the number of occurrences of a given RAV, and the log-transformed number of consensus sequences was included as the offset term; associations were tested using a modified F-statistic. A binary sensitivity outcome, a RAV frequency of >0%, was evaluated using a logistic GEE model adjusted for sequencing depth. Analyses were conducted in two-sided format without adjustment for multiplicity, using Prism 5 for Mac OS X software (GraphPad Software, Inc.) or SAS version 9.3 (SAS Institute, Cary, NC).
RESULTS
Primer ID sequencing of HCV RNA.
We characterized sequence diversity within the segment of the HCV genome encoding the protease domain of NS3 (codons 36 to 173) isolated from serum or plasma from 40 HCV treatment-naive subjects, 20 of whom were infected solely with HCV and 20 of whom were coinfected with HIV (Table 1). All coinfected subjects were on stable ART with HIV RNA levels of <50 copies/ml. The use of tagged primers for cDNA synthesis eliminated several common sources of error in high-throughput sequencing populations of viral genomes (28). A random 8-nt sequence (termed primer ID) embedded in each primer labeled cDNA products derived from individual viral RNA molecules, allowing these molecules to be tracked through subsequent PCR amplification and high-throughput sequencing on the 454 Junior platform. Sequencing reads sharing a common primer ID were clustered, and a consensus sequence was developed for each cluster of 3 or more reads. Unambiguous consensus sequences are likely to represent the authentic sequence of a single RNA molecule given the ratio of possible primer ID sequences and starting RNA templates (see Materials and Methods).
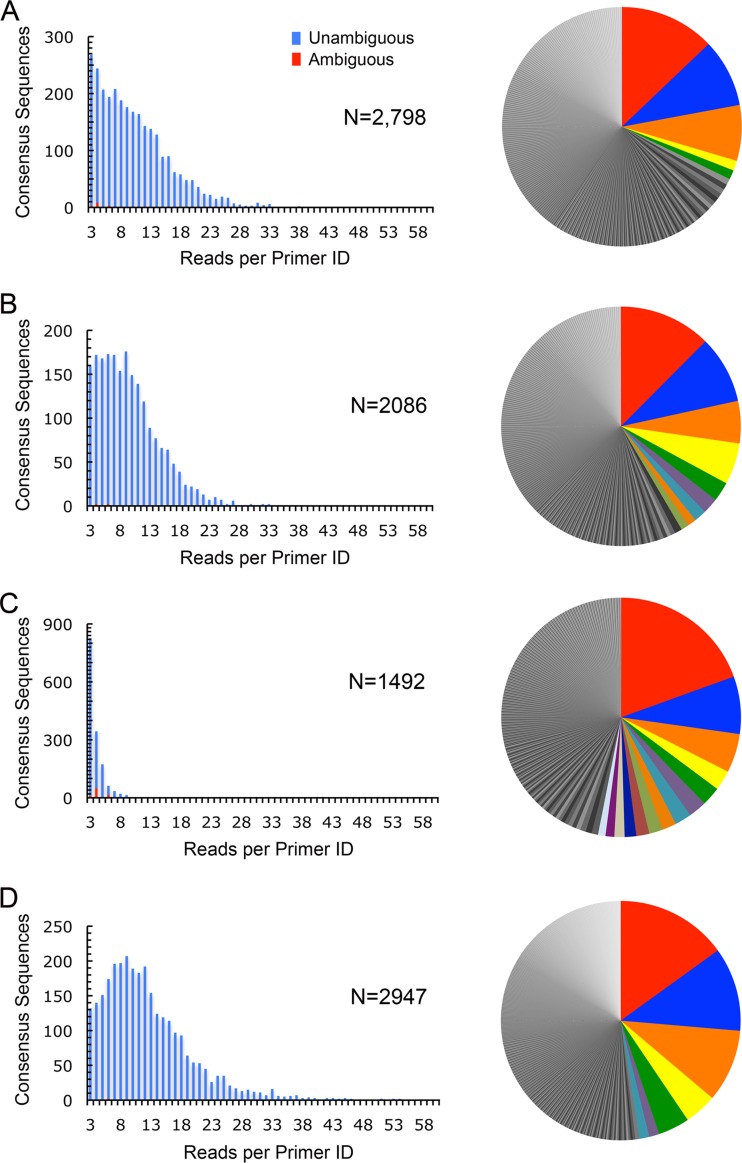
We used this primer ID strategy to sequence circulating HCV RNAs in all 40 study subjects. To illustrate the nature of the data obtained and how they were processed through the bioinformatics pipeline, we show in detail the representative results obtained with 2 monoinfected (M1 and M2) and 2 coinfected (C1 and C2) study subjects (Fig. 1). Between 14,461 and 37,864 sequence reads from each of these samples passed initial filtering, with the maximum number of reads sharing the same primer ID, varying from 19 to 77 (Table 2). This allowed the construction of between 1,492 and 2,798 consensus sequences per sample (Fig. 1A to D, left), most of which (94.3 to 99.7%) were unambiguous by majority call. Consistent with substantial diversity among virions circulating in individual subjects, numerous unique haplotypes (defined as described in Materials and Methods) were present within each population (Fig. 1A to D, right). The most frequent haplotype present comprised only 12.5 to 20.0% of all consensus sequences in any one subject. The number of unique haplotypes representing >1% of the consensus sequences ranged from 5 to 14. The most frequent 12 to 20 haplotypes comprised 50% of all sequences in the coinfected subjects. This compared with 36 to 80 haplotypes in the two monoinfected subjects, suggesting greater haplotype diversity in the absence of HIV coinfection.
FIG 1.
Results of primer ID sequencing of the NS3 protease region in HCV RNA from 4 representative study subjects. Approximately 10,000 copies of viral RNA from monoinfected (A and B) or coinfected (C and D) subjects were reverse transcribed into cDNA using patient virus strain-specific primers containing both a sample-specific barcode and an 8-nt random primer ID, amplified by PCR, pooled, and sequenced. From 1,492 to 2,798 consensus sequences were compiled from alignments of 3 or more 454 Junior reads sharing identical primer ID tags (left), most of which were nonambiguous. Pie charts on the right show the frequency distribution of unique haplotypes (with more frequent individual haplotypes represented by a unique color) within each sequenced virus population (range, 552 to 1,255 per sample).
TABLE 2.
Population sequencing throughput and depth in replicate independent primer ID sequencing runs
| Read parameter | Run |
|
|---|---|---|
| 1 | 2 | |
| Raw reads (no.) | 116,211 | 113,932 |
| Reads with >495 bases (%) | 98.81 | 99.05 |
| Preconsensus reads (no.) | ||
| M1 | 29,766 | 15,413 |
| M2 | 20,619 | 18,098 |
| C1 | 14,461 | 20,370 |
| C2 | 37,864 | 48,410 |
| Unique primer IDs (no.) | ||
| M1 | 3,736 | 4,536 |
| M2 | 2,492 | 3,582 |
| C1 | 8,111 | 11,071 |
| C2 | 3,663 | 4,872 |
| Consensus sequences (no. of clusters of ≥3 reads) | ||
| M1 | 2,798 | 2,445 |
| M2 | 2,086 | 2,571 |
| C1 | 1,492 | 2,150 |
| C2 | 2,947 | 3,589 |
| Maximum no. of reads per primer ID cluster | ||
| M1 | 58 | 22 |
| M2 | 37 | 26 |
| C1 | 19 | 27 |
| C2 | 77 | 71 |
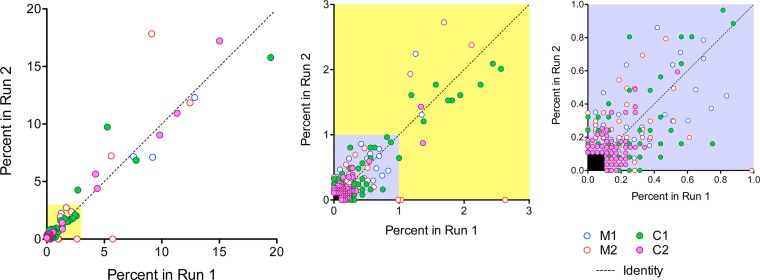
To ascertain reproducibility, we carried out replicate, independent cDNA syntheses and sequencing runs on these 4 subjects. The two pyrosequencing runs yielded similar numbers of consensus sequences from each subject (Table 2). Excluding several notable exceptions (all cytosine-rich primer ID tags; see Table S2 in the supplemental material), the most frequent primer IDs observed were unique to each run. The most prevalent primer IDs in reads from subject C1 were reproducibly enriched for cytosine (see Table S2), suggesting either poor randomization of the degenerate region in the tagging primer during synthesis or more efficient priming for cDNA synthesis. This was not observed with subsequent samples using different tagging primers for cDNA synthesis. Importantly, there was strong reproducibility in the frequency of individual haplotypes resolved in the two independent runs (Fig. 2). Overall, between 1,411 and 3,577 unambiguous consensus sequences were resolved per sample, resulting in population depths between 0.07% and 0.03%. Because the probability of sampling a low-abundance haplotype approximates the Poisson distribution, this depth of sampling would be expected to identify variants with frequencies as low as 0.2% to 0.08%. Collectively, these data show that primer ID sequencing is capable of accurately resolving genetic diversity in populations of RNA viruses.
FIG 2.
Reproducibility of primer ID quantitation of HCV haplotype frequency. Data shown represent the frequency of individual haplotypes in two independent sequencing runs carried out on samples from 2 monoinfected (M1 and M2) and 2 coinfected (C1 and C2) subjects. Haplotypes present at >0.1% in either run are included. The yellow inset (expanded in the middle) shows those haplotypes present between 0.1 and 3%, while the blue inset (expanded to the right) shows those between 0.1 and 1%. The dashed line represents the line of identity. Haplotype frequency was grouped into 4 categories: ≤0.5%, >0.5 to 1.0%, >1.0 to 3.0%, and >3.0%, and agreement was summarized with a weighted Cohen's kappa coefficient. There was substantial agreement between the two independent runs (kappa of 0.84). Most (87%, or 431/495) of the haplotypes included in the analysis were detected at a frequency of ≤0.5% in both runs. When haplotype frequencies of ≤1.0% were collapsed into a single group, only 6 of 495 observations were not in agreement (weighted kappa of 0.92).
Genetic diversity of HCV in monoinfection versus coinfection.
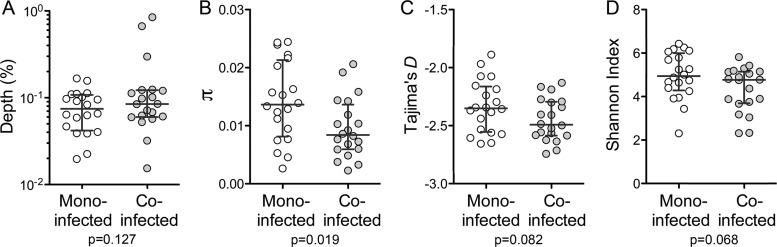
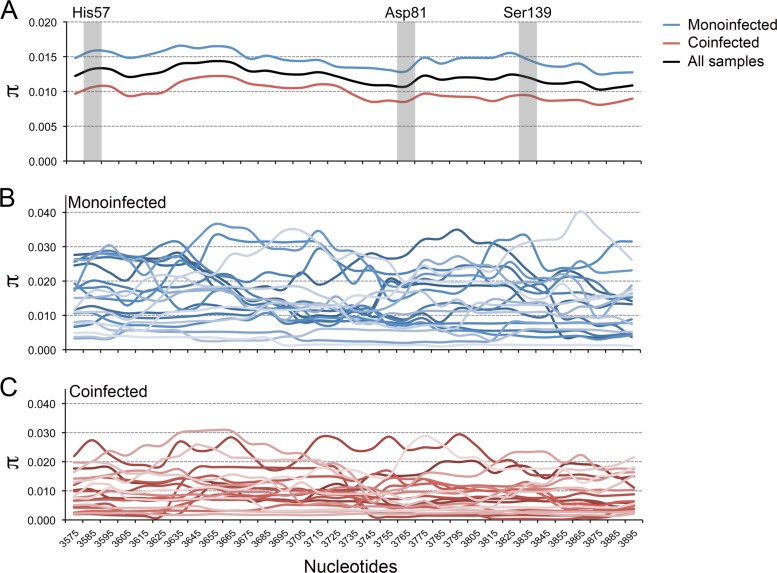
A similar primer ID analysis of NS3 coding sequence was carried out with RNA extracted from serum or plasma from each of the other study subjects, thereby providing data on a total of 40 subjects with genotype 1a HCV infection, 20 of whom were coinfected with HIV but on stable ART with nondetectable HIV load (<50 copies/ml). These cohorts were of comparable age (monoinfected median age, 54 years; coinfected median age, 51 years) and predominantly male, but coinfected subjects were more likely to be of African-American, rather than Caucasian, race (78% and 30%, respectively; P = 0.004) and had somewhat lower serum alanine aminotransferase (ALT) levels (Table 1). The results of the replicate sequencing runs described above were pooled and included in this analysis. Median sampling depth was 0.07% for monoinfected subjects and 0.08% for coinfected subjects (Fig. 3A). Each viral population was rich in allelic diversity, including numerous low-frequency haplotypes. Average pairwise nucleotide diversity (π) across the NS3 region sequenced varied by subject but was higher overall in monoinfected (π = 0.0143 ± 0.0069 [means ± standard deviations {SD}]) than coinfected (0.00954 ± 0.0051) subjects (P = 0.02; P = 0.03 by WRS) (Fig. 3B). A sliding-window analysis revealed localized regions of minimally increased or decreased mean nucleotide diversity within the linear sequence of the protease-coding region that were conserved in both patient cohorts (Fig. 4, top). This position dependence did not correlate with the location of active-site residues in the protease. In contrast, regions of maximum and minimum nucleotide diversity varied widely between individual subjects, suggesting host-specific differences in regional selective forces such as might be mediated by HLA-restricted T cell responses (Fig. 4, middle and lower). Nucleotide diversity was not significantly associated with sequencing depth, HCV RNA copy number, levels of ALT or bilirubin, age, or sex in univariate models (P > 0.05).
FIG 3.
NS3 sequence diversity in monoinfected and coinfected patient cohorts of 20 patients each. (A) Sequencing depth (1/n, where n is the number of nonambiguous consensus sequences constructed from 3 or more 454 Junior sequencing reads containing identical primer IDs). (B) Nucleotide diversity (π). (C) Tajima's D statistic. (D) Shannon entropy index. Bars indicate medians ± quartiles. Significance (P) was assessed by a two-sided Welch t test.
FIG 4.
Sliding-window analysis of pairwise nucleotide diversity (π) across the NS3 protease-coding region (NS3 residues 36 to 175). His57, Asp81, and Ser139 represent the catalytic triad within the active site of the protease. (A) Mean nucleotide diversity within 100-nt windows with a 10-nt step in monoinfected (blue; n = 20), coinfected (red; n = 20), and all (black; n = 40) subjects. The nucleotide numbering represents the midpoint in each window. Diversity was higher in monoinfected subjects in all regions of the sequence. (B and C) Nucleotide diversity calculated for virus populations in individual monoinfected and coinfected subjects (n = 20 each). A repeat analysis using 30-nt windows with a 10-nt step generated similar results, with no suggestion of reduced diversity surrounding the active-site residues (data not shown).
Tajima's D statistic reflects the deviation between the observed and expected distribution of allelic frequencies at mutation-drift equilibrium (30). Strongly negative values of D may reflect recent selective sweeps, population bottlenecks, and population expansion, whereas positive values can indicate balancing selection, migration, and population subdivision. Tajima's D was ≤−2.0 in all but two subjects, indicating an excess of low-frequency polymorphisms. While the mean value of Tajima's D for coinfected subjects (−2.45 ± 0.18) was lower than that for monoinfected subjects (−2.33 ± 0.23), this difference did not achieve statistical significance (P = 0.08; P = 0.15 by WRS) (Fig. 3C). We also computed the Shannon diversity index for each virus population. This measure captures both the number of variants and the evenness of their distribution (how similar their numbers are) within the population (31, 32). An increase in the number of unique haplotypes and/or an increase in the evenness of their distribution will result in an increased Shannon index. Consistent with other measures of diversity, the Shannon index showed a trend toward higher values in virus populations from monoinfected versus coinfected subjects (mean results of 4.96 ± 1.07 versus 4.34 ± 1.03; P = 0.07; P = 0.10 by WRS) (Fig. 3D).
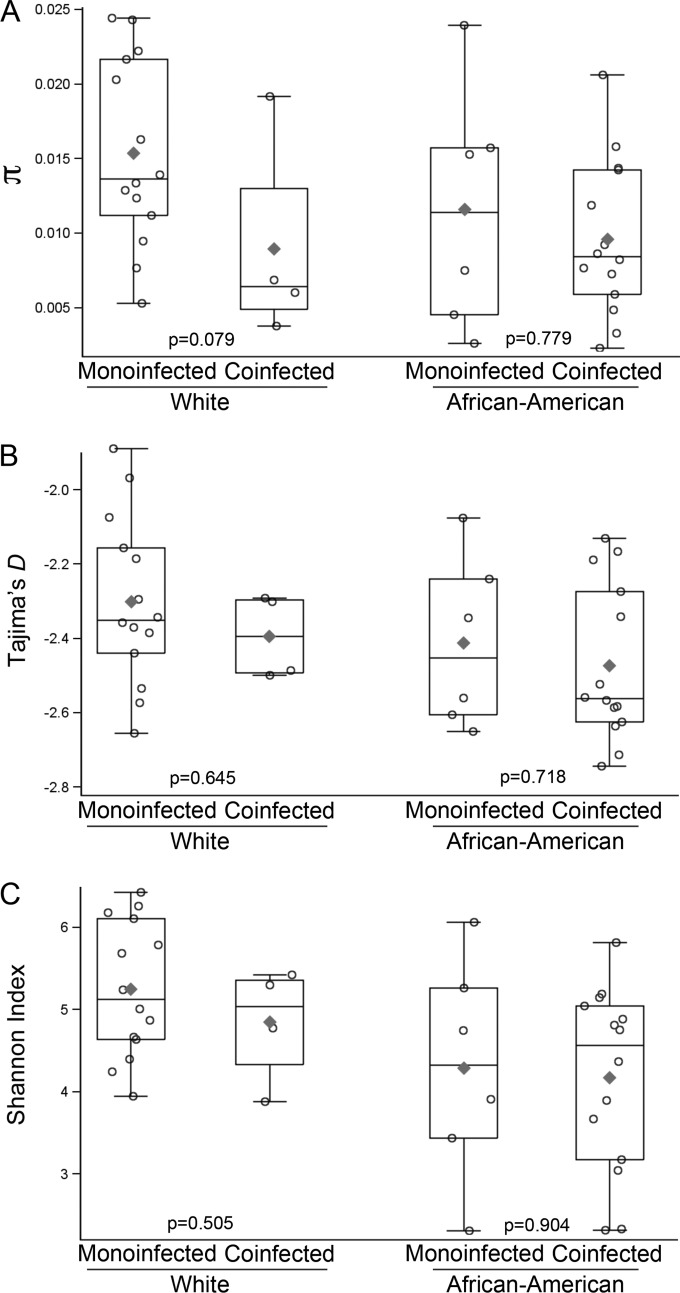
As significantly more coinfected subjects were African-American than monoinfected subjects, we explored the association between these measures of HCV diversity and race in a series of multivariable linear regression models. Nucleotide diversity was numerically greater in monoinfected subjects among both African-Americans and whites (Fig. 5). Moreover, in a model adjusted for sequencing depth and HCV RNA copy number but ignoring race, coinfection was associated with lower nucleotide diversity (P = 0.02), while in a comparable model ignoring coinfection status, no association with race was detected (P = 0.12). However, in a model that included coinfection, race, sequencing depth, and HCV RNA level, neither coinfection (P = 0.11) nor race (P = 0.52) was statistically significant, but interpretation of this model was limited by the colinearity of coinfection and race (see Table S3 in the supplemental material). None of the evaluated covariates were significantly associated with Tajima's D in multivariable models (see Table S4). In univariate analyses, lower Shannon indices were associated with African-American race (P = 0.006) and marginally associated with coinfection status (P = 0.07). The association between lower Shannon index and African-American race also was evident in a nonparametric sensitivity analysis (P = 0.01 by WRS), as well as a multivariable model including coinfection status (P = 0.53) and race (P = 0.03) (see Table S5). The biological significance of the association between race and Shannon index is uncertain (see Discussion).
FIG 5.
Exploratory analysis of HCV diversity measures by racial groups and coinfection status. Boxplots showing medians, interquartile ranges, and means (⧫) for average pairwise nucleotide diversity (π) (A), Tajima's D (B), and the Shannon index (C) of NS3 protease domain sequences from monoinfected and coinfected subjects of Caucasian (n = 14 and 4, respectively) and African-American (n = 6 and 14) race. An exact Wilcoxon rank-sum test was used to determine P values post hoc. This study was not designed to assess differences in HCV genetic diversity across racial groups.
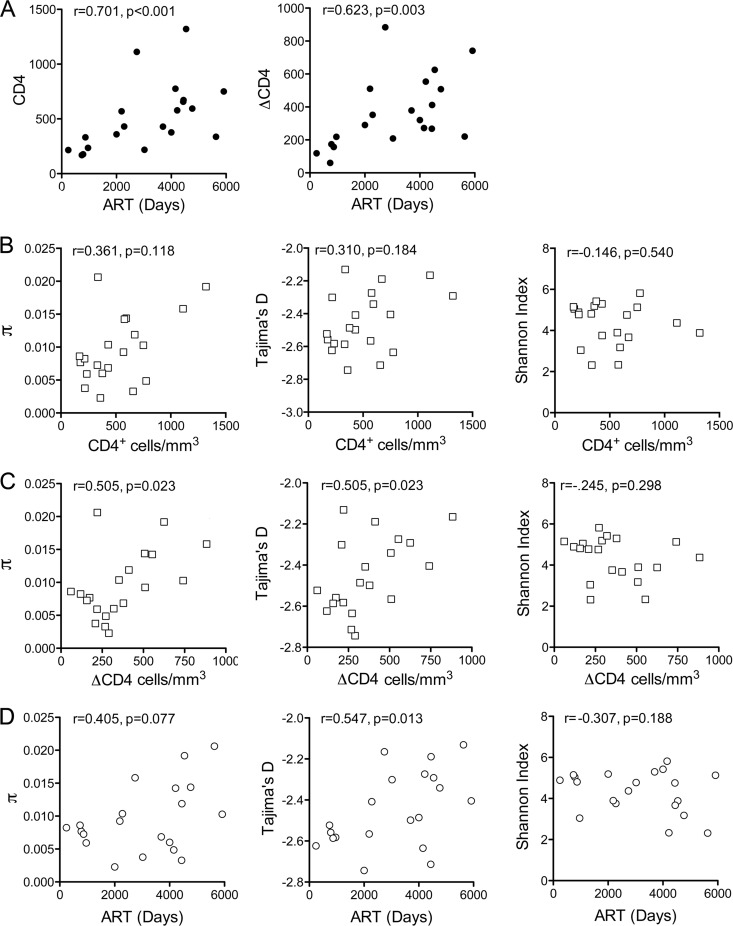
CD4+ T cell counts and HCV diversity in coinfected subjects.
While substantial overlap between coinfection status and race made it difficult to ascertain the individual impact of these variables, these analyses point collectively toward a reduction in the genetic diversity of the NS3 protease-coding sequence in coinfected subjects. Since the selective pressure of T cell responses acting only on certain variants within the replicating HCV quasispecies ensemble could generate greater diversity, we sought evidence for a correlation between the CD4+ T cell count at the time of sample collection and NS3 sequence diversity. The coinfected patients had been receiving ART for a median of 110 months (range, 8 to 194) when studied. As expected, both the CD4+ T cell count at the time of sampling and the difference between the historic nadir in the CD4+ T cell count (Spearman's r = 0.701; P < 0.001) and that at the time of sampling (ΔCD4) (r = 0.623 and P = 0.003) correlated strongly with the length of ART therapy (Fig. 6A). Nucleotide diversity and Tajima's D trended toward increases with increasing CD4+ count, but these correlations did not reach statistical significance (r = 0.361 and P = 0.12 for π and r = 0.310 and P = 0.18 for Tajima's D). However, both π (r = 0.505; P = 0.023) and Tajima's D (r = 0.505 and P = 0.023) were positively correlated with ΔCD4 (Fig. 6C). These diversity measures also tended to be higher in subjects with longer lengths of ART, but this relationship was statistically significant only for Tajima's D (r = 0.548; P = 0.01) (Fig. 6D). The Shannon index showed slight, statistically insignificant downward trends with both ΔCD4 and length of ART (Fig. 6C and D, right). Taken together, these analyses suggest that the reduced diversity of NS3 sequence in these subjects with well-controlled HIV coinfection likely is related to residual impaired cellular immunity (see Discussion).
FIG 6.
NS3 sequence diversity, CD4+ T cell counts, and length of ART in the coinfected patient cohort (n = 20). (A) CD4+ T cell count (left) and ΔCD4 (the difference between the historic nadir in the CD4+ T cell count and that at the time of sampling, right) increased with increased duration of ART. (B) CD4 cell count and nucleotide diversity (π), Tajima's D, and Shannon index of NS3 sequences from coinfected subjects (n = 20). (C) ΔCD4 and nucleotide diversity (π), Tajima's D, and Shannon index of NS3 sequences from coinfected subjects (n = 20). (D) Length of ART and nucleotide diversity (π), Tajima's D, and Shannon index of NS3 sequences in coinfected subjects (n = 20). Correlations were assessed by Spearman's rho.
Protease inhibitor RAVs.
Virologic failure in patients receiving potent DAA regimens typically is associated with the emergence of RAVs (35). Theoretical considerations suggest that these emergent variants represent preexisting minor quasispecies that become dominant under the selective pressure of the drugs (13). There is good experimental evidence to support this concept, but only limited information is available concerning the baseline frequencies of such variants in treatment-naive patients (36, 37). Since primer ID sequencing provides a uniquely accurate and unbiased approach to assessing the prevalence of low-frequency variants, we examined the consensus sequences recovered from monoinfected and coinfected subjects for the presence of known protease inhibitor RAVs (see Materials and Methods).
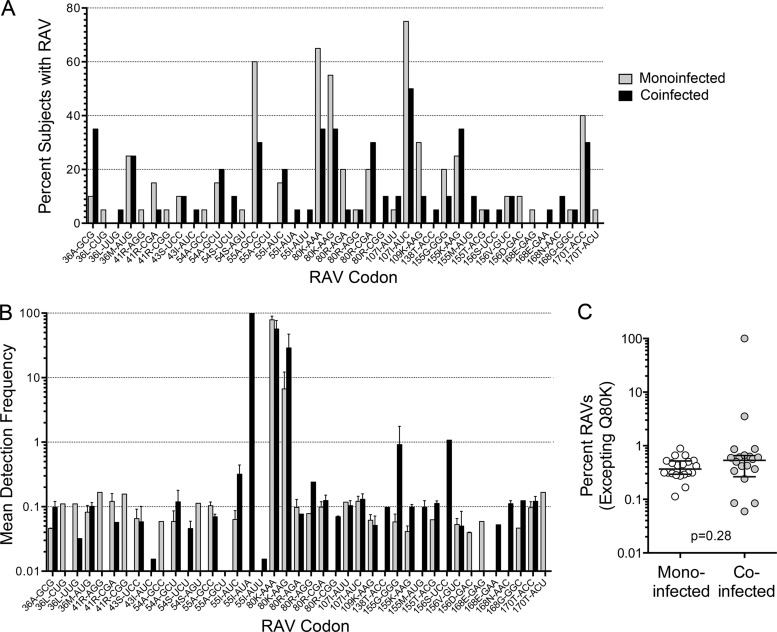
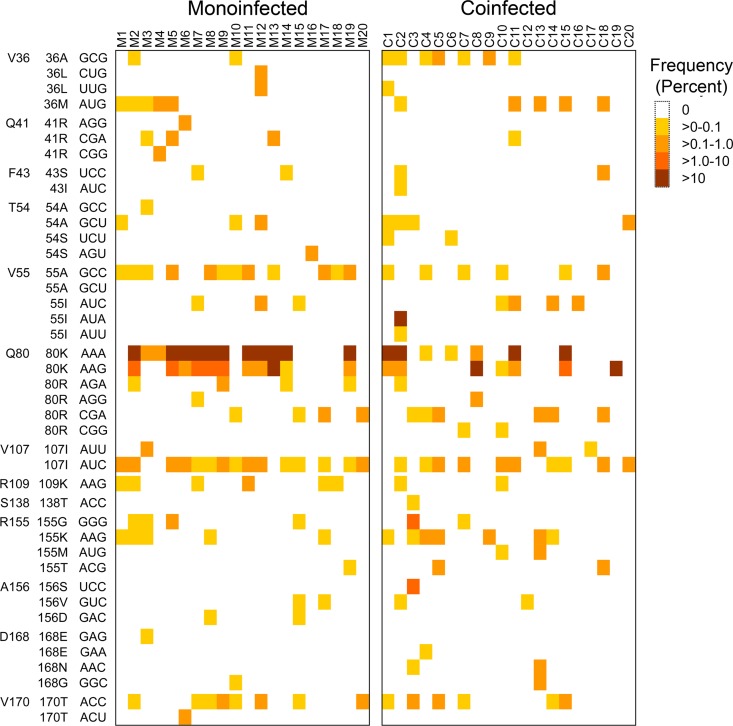
Q80K was the most prevalent RAV identified in both patient cohorts, both in terms of the proportion of subjects in which it was identified (Fig. 7A) and its frequency within the virus population when present (Fig. 7B). Q80K is associated with modest resistance to newer macrocyclic NS3/4A protease inhibitors, and it negatively impacts the therapeutic response to simeprevir (16, 38, 39). However, it is a common polymorphism in genotype 1a virus. Over 99% of the consensus sequences in 11/20 (55%) monoinfected and 6/20 (30%) coinfected subjects encoded Q80K. To focus on low-frequency RAVs, we excluded Q80K (both AAA and AAG codons) from further analyses, but not Q80R variants that are associated with similar resistance (38) but were much less prevalent (<0.25% in all subjects). Except for one coinfected individual (C19, who was sequenced to a depth of only 0.85%), one or more low-frequency RAVs were detected in all subjects. Importantly, the overall frequency of RAVs (excepting Q80K) was similar in monoinfected versus coinfected subjects, and the median cumulative frequency was 0.37% versus 0.53% (P = 0.31; P = 0.28 by WRS) (Fig. 7C) despite one coinfected subject (C2) with over 99% V55I. The distribution and frequency of RAVs involving different codons in individual subjects is shown as a heat map in Fig. 8. A negative binomial GEE model demonstrated that the frequency of RAVs varied significantly at different codons (P < 0.0001, excluding Q80K, V55I-AUA [high frequency], and one unobserved RAV, V55A-GCU), likely reflecting differences in the fitness of these variants (40). However, the frequency of RAVs across the codons was not significantly associated with coinfection status (P = 0.20). Results were similar when an RAV frequency of >0% was evaluated with a logistic GEE model (P = 0.004 for codon position; P = 0.64 for coinfection).
FIG 7.
NS3/4A inhibitor resistance-associated variants (RAVs; 41 codons) identified by primer ID sequencing in monoinfected and coinfected subjects. (A) Percentage of monoinfected and coinfected subjects in which individual RAVs were identified. (B) Mean detection frequency of RAVs when present, shown as the percentage of consensus reads in those subjects with the RAV. Bars indicate standard errors of the means. (C) Mean cumulative sum of all RAVs except those encoding Q80K (cumulative percentage of consensus reads) in individual study subjects. Bars represent medians and IQR. P = 0.28 by Wilcoxon rank-sum test.
FIG 8.
Heat map showing the presence and relative frequency of individual RAVs across virus populations from the two patient cohorts. Negative binomial generalized estimating equations demonstrated that RAV frequency varied significantly by codon (P < 0.0001, excluding Q80K, V55I-AUA, and one unobserved RAV, V55A-GCU), but no significant dependence on coinfection status was observed (P = 0.20).
Despite the remarkable depth and accuracy of the primer ID sequencing protocol (Fig. 2), most RAVs existed in these treatment-naïve subjects at frequencies sufficiently low to confound their unambiguous resolution. If the high-frequency Q80K and V55I (subject C2) RAVs are excluded, 122 of the 296 RAVs identified in the 64,338 consensus sequences we constructed from all 40 subjects were single occurrences within the viral population (termed singlets). While each of these consensus sequences was built from a minimum of three sequencing reads, such a singlet cannot be differentiated unambiguously from an error introduced during reverse transcription of the viral genome. Nonetheless, two lines of evidence suggest that many of these singlets represent actual variants in the viral population and not artifactual diversity. First, in the 4 sequencing runs that were independently replicated (Fig. 2), approximately half of the singlet RAVs identified in any one run were confirmed in one or more consensus sequences in the paired run. This accords with sampling probabilities predicted by the binomial distribution and a formal series of Monte Carlo simulations (not shown). Second, there was substantial association between occurrences of singlet and nonsinglet RAVs involving specific codons (see Fig. S1 in the supplemental material). This would not be expected were singlets to arise randomly from errors introduced during reverse transcription.
DISCUSSION
The emergence of resistant HCV variants is a significant concern for coinfected patients treated with interferon-sparing combinations of DAAs, as it is for those infected only with HCV. These variants likely exist as a minor fraction of the HCV quasispecies swarm prior to therapy and become dominant due to the selective pressure of DAAs on the viral population (13). The frequency of RAVs in treatment-naive individuals, coupled with their fitness, is likely to determine both the rapidity with which resistance emerges as well the probability of successfully eliminating the virus. Similarly, the extent to which such RAVs persist off treatment in patients who fail DAA therapy could determine the response to retreatment with drugs that have similar resistance profiles. Whether protease inhibitor RAVs are more or less frequent in patients with HIV coinfection is an important question that we have addressed here using a novel and extremely accurate high-throughput sequencing method in which sequence reads generated from a single viral RNA template are tagged (28). This strategy allows for the development of a consensus sequence for each RNA template and thereby obviates a variety of technical errors that may otherwise serve to artifactually enhance the diversity observed within a viral population. It also permits reliable detection of low-frequency haplotypes.
High-throughput pyrosequencing of RNA viruses can be confounded by multiple types of errors. Artifactual diversity can be imparted by base misincorporation during reverse transcription of viral RNA to cDNA or by DNA recombination during multiple rounds of PCR, biased PCR amplification of sequence, and oversampling of small numbers of genomes (26, 27). Each of these types of error may obscure the true frequency of minor variants within a viral population and provide an inaccurate view of the architecture of viral quasispecies. The primer ID strategy we adopted was designed to avoid many of these pitfalls and was adapted from a procedure developed to examine HIV diversity (28). In previous studies of HIV, 80% of the diversity present in individual sequencing reads from the Roche 454 platform was eliminated by clustering and developing a consensus sequence from 3 or more reads sharing a common primer ID tag (28). The single most important modification required to achieve reliable amplification of HCV sequence with random primer ID incorporation into cDNA was the use of tagging primers customized to provide an exact match with the patient's virus based on the results of prior population sequencing. This was mandated by the diversity of sequence 3′ to the NS3 coding region of interest in different genotype 1a viruses that precluded development of a universal primer.
Nonetheless, the primer ID approach cannot eliminate artifactual diversity resulting from errors in reverse transcription of RNA to cDNA, which may occur as often as 1 in 5,000 bases (0.02%) (28). We approached this limit in our analysis of some samples. The depth was 0.015% and 0.02% in subjects C2 and M1, respectively, achieved by pooling results from two independent runs. However, the median depth to which we sequenced was 0.08% overall, and the typical frequency of the RAVs we detected was substantially greater than 0.02% (Fig. 7B). While the identification of a RAV in a single consensus sequence from one sample does not unambiguously demonstrate the presence of that RAV in the original viral population, the single occurrences we observed likely represent true viral diversity for reasons discussed above. They do not appear randomly distributed across codons (see Fig. S1 in the supplemental material), as might be expected from base misincorporation during reverse transcription of viral RNAs. An alternative to the primer ID strategy that allows for highly accurate high-throughput sequencing independent of errors in reverse transcription is CirSeq, in which tandem repeats of viral sequence generated from circularized fragments of the genome are sequenced (41). This approach, while offering greater accurate depth of reads than the primer ID method, is limited to sequencing very small genome fragments; therefore, it provides less insight into linkage.
In this study, we focused exclusively on HCV sequence encoding the protease domain of NS3 protein. However, we anticipate that the primer ID approach will work equally well and reveal comparable degrees of genetic diversity and RAV frequencies in regions of the HCV genome that encode alternative DAA targets, such as the NS5A protein or the NS5B RNA-dependent RNA polymerase. Ongoing primer ID studies of the 5′ NS5A coding sequence provide preliminary evidence for this, and in four monoinfected subjects we have generated sequence reads with diversity comparable to what we observed in the NS3 region (comparable values for π and Tajima's D) (F. Hu, S. Zhou, and S. M. Lemon, unpublished data).
HIV coinfection profoundly influences the course of HCV infection in ways that remain incompletely understood (1, 2). Thus, one of the most interesting findings to emerge from this study was the lower diversity of HCV NS3 sequence in coinfected compared to monoinfected subjects (Fig. 3B to D). Relatively few previous studies have compared the population structure of HCV in coinfected and monoinfected subjects, and none have approached the depth and accuracy of the analysis or number of subjects in this study. Most previous studies have focused on hypervariable region 1 (HVR1) within the E2 envelope protein, which is known to be targeted by antibodies to the virus (42), and not HCV nonstructural proteins targeted by DAAs. Using a variety of methods, and not always achieving statistical significance, several previous studies suggested trends toward greater population diversity in coinfected versus monoinfected persons (17–19). On the other hand, other studies have suggested that HCV quasispecies diversity is reduced in coinfected subjects (21, 23, 43, 44), and that diversity within HVR1 is increased by successful ART (23, 45–48). Only one previous study examined the impact of HIV coinfection on NS3 sequence diversity. This involved a comparison of 3 monoinfected and 6 coinfected individuals by clonal sequencing and revealed no significant differences (21). Our study differs from these previous studies in several important respects, including both the greater accuracy provided by the primer ID method and much larger patient cohorts (20 subjects in each group).
The coinfected patients we studied had been receiving ART, most for many years, and all had an undetectable HIV load (<50 copies/ml). Nonetheless, average pairwise nucleotide diversity (π) was significantly lower in the coinfected than the monoinfected subjects (Fig. 3B). Trends toward reduced population diversity also were evident in Tajima's D statistic and in the Shannon entropy index (Fig. 3C and D). Nucleotide diversity and Tajima's D were significantly associated with the difference between CD4+ T cell count at the historic nadir prior to initiation of ART and that at the time of sample collection (Fig. 6C). These associations were not large but are consistent with the hypothesis that reduced immunological pressure on the virus limits the selection of escape mutants and reduces diversity (43, 44). ART enhances this pressure and results in an increase in diversity. It is notable that these effects on HCV population structure were observed with a median CD4+ T cell count of 430 cells/mm3 (IQR, 284 to 665 cells/mm3). Similar increases in sequence diversity within the hypervariable region of the E2 protein-coding region of HCV have been linked to immune reconstitution and increasing CD4+ T cell counts following institution of ART in coinfected patients (45). This interpretation is consistent with a large body of evidence indicating that CD4+ and CD8+ T cell responses shape HCV quasispecies evolution through selection of escape variants, particularly in the early weeks of HCV infection (49–52).
A significantly greater proportion of the coinfected subjects we studied were African-American, whereas the monoinfected subjects predominantly were Caucasian (Table 1). While the study was not designed to assess the influence of race on HCV genetic diversity, post hoc analyses suggested an association between lower Shannon entropy indices and African-American race that was evident in both univariate and multivariable analyses. In contrast, lower Shannon indices were only marginally associated with coinfection status in the univariate analysis (P = 0.07). This suggests that African-American subjects have fewer unique haplotypes and/or a less even distribution of these haplotypes among circulating HCV quasispecies. Why this should be so for African-Americans is not clear, but it is intriguing to consider that it reflects a greater prevalence of interleukin-28B (IL-28B) (interferon lambda 3 [IFN-λ3]) genetic polymorphisms unfavorable for immune control of HCV (53, 54). Unfortunately, data concerning IL-28B genotypes were not available for most of the subjects we studied. Other genetic polymorphisms also have been associated with reduced control of HCV infection, including polymorphisms in IFN-γ, human leukocyte antigen (HLA), or natural killer (NK) cell inhibitory receptor genes (55, 56).
Is the difference in the genetic diversity of HCV that we documented between monoinfected and coinfected subjects of clinical significance? Of the 41 specific protease inhibitor RAVs we searched for, one or more were present in all but 1 of the 40 patients we studied (Fig. 7). These data provide direct experimental evidence supporting the premise that such RAVs naturally exist in treatment-naive HCV quasispecies (13). Antiviral resistance likely results from the selection of these preexisting variants under drug pressure. Importantly, although one coinfected subject (C2) had a high proportion of V55I (>99.9%) and another patient (C3) had R155G (1.76%) and A156S (1.08%) variants, the overall frequency of RAVs was similar in coinfected and monoinfected subjects (Fig. 7C). Moreover, we observed a near-zero correlation between the frequency of RAVs and CD4+ cell count (not shown). We conclude that while pairwise nucleotide diversity is significantly reduced within the NS3 coding region of HCV in coinfected patients with well-controlled HIV infection, the frequency of preexisting RAVs is similar to that in monoinfected patients and will not by itself influence the outcome of NS3/4A protease inhibitor therapy. The extent to which the response to DAA therapy will be reduced by the subtle impairment of T cell responses to HCV that our data suggest exists in coinfected patients with well-controlled HIV infection remains to be determined.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge expert technical assistance provided by Lucinda Hensley, Sam Stinnette, and Sonia Napravnik.
This work was supported in part by a research grant from the Investigator-Initiated Studies Program of Merck Sharp & Dohme. Additional support was provided by grants from the National Institutes of Health, R01-CA164029 (S.M.L.), R37-AI44667 (R.S.), K24-DK066144 (M.W.F.), the University of North Carolina Center for AIDS Research (P30-AI50410), the UNC Center for Gastrointestinal Biology and Disease (P30-DK034987), the Lineberger Comprehensive Cancer Center (P30-CA16086), and the University Cancer Research Fund.
C.J. and R.S. are inventors of intellectual property related to the sequencing strategy used in this study; S.M.L. is a consultant to Merck, Sharp & Dohme, one of the sponsors of this study.
Footnotes
Published ahead of print 4 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.03466-14.
REFERENCES
- 1.Kim AY, Chung RT. 2009. Coinfection with HIV-1 and HCV–a one-two punch. Gastroenterology 137:795–814. 10.1053/j.gastro.2009.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Naggie S, Sulkowski MS. 2012. Management of patients coinfected with HCV and HIV: a close look at the role for direct-acting antivirals. Gastroenterology 142:1324–1334. 10.1053/j.gastro.2012.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodriguez-Torres M, Slim J, Bhatti L, Sterling R, Sulkowski M, Hassanein T, Serrao R, Sola R, Bertasso A, Passe S, Stancic S. 2012. Peginterferon alfa-2a plus ribavirin for HIV-HCV genotype 1 coinfected patients: a randomized international trial. HIV Clin. Trials 13:142–152. 10.1310/hct1303-142 [DOI] [PubMed] [Google Scholar]
- 4.Poordad F, McCone J, Jr, Bacon BR, Bruno S, Manns MP, Sulkowski MS, Jacobson IM, Reddy KR, Goodman ZD, Boparai N, DiNubile MJ, Sniukiene V, Brass CA, Albrecht JK, Bronowicki JP. 2011. Boceprevir for untreated chronic HCV genotype 1 infection. N. Engl. J. Med. 364:1195–1206. 10.1056/NEJMoa1010494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobson IM, McHutchison JG, Dusheiko G, Di Bisceglie AM, Reddy KR, Bzowej NH, Marcellin P, Muir AJ, Ferenci P, Flisiak R, George J, Rizzetto M, Shouval D, Sola R, Terg RA, Yoshida EM, Adda N, Bengtsson L, Sankoh AJ, Kieffer TL, George S, Kauffman RS, Zeuzem S. 2011. Telaprevir for previously untreated chronic hepatitis C virus infection. N. Engl. J. Med. 364:2405–2416. 10.1056/NEJMoa1012912 [DOI] [PubMed] [Google Scholar]
- 6.Scheel TK, Rice CM. 2013. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat. Med. 19:837–849. 10.1038/nm.3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sulkowski M, Pol S, Mallolas J, Fainboim H, Cooper C, Slim J, Rivero A, Mak C, Thompson S, Howe AY, Wenning L, Sklar P, Wahl J, Greaves W. 2013. Boceprevir versus placebo with pegylated interferon alfa-2b and ribavirin for treatment of hepatitis C virus genotype 1 in patients with HIV: a randomised, double-blind, controlled phase 2 trial. Lancet Infect. Dis. 13:597–605. 10.1016/S1473-3099(13)70149-X [DOI] [PubMed] [Google Scholar]
- 8.Martel-Laferriere V, Brinkley S, Bichoupan K, Posner S, Stivala A, Perumalswami P, Schiano T, Sulkowski M, Dieterich D, Branch A. 2014. Virological response rates for telaprevir-based hepatitis C triple therapy in patients with and without HIV coinfection. HIV Med. 15:108–115. 10.1111/hiv.12086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sulkowski MS, Sherman KE, Dieterich DT, Bsharat M, Mahnke L, Rockstroh JK, Gharakhanian S, McCallister S, Henshaw J, Girard PM, Adiwijaya B, Garg V, Rubin RA, Adda N, Soriano V. 2013. Combination therapy with telaprevir for chronic hepatitis C virus genotype 1 infection in patients with HIV: a randomized trial. Ann. Intern. Med. 159:86–96. 10.7326/0003-4819-159-2-201307160-00654 [DOI] [PubMed] [Google Scholar]
- 10.Kieffer TL, Sarrazin C, Miller JS, Welker MW, Forestier N, Reesink HW, Kwong AD, Zeuzem S. 2007. Telaprevir and pegylated interferon-alpha-2a inhibit wild-type and resistant genotype 1 hepatitis C virus replication in patients. Hepatology 46:631–639. 10.1002/hep.21781 [DOI] [PubMed] [Google Scholar]
- 11.Susser S, Welsch C, Wang Y, Zettler M, Domingues FS, Karey U, Hughes E, Ralston R, Tong X, Herrmann E, Zeuzem S, Sarrazin C. 2009. Characterization of resistance to the protease inhibitor boceprevir in hepatitis C virus-infected patients. Hepatology 50:1709–1718. 10.1002/hep.23192 [DOI] [PubMed] [Google Scholar]
- 12.Sulkowski MS. 2013. Current management of hepatitis C virus infection in patients with HIV co-infection. J. Infect. Dis. 207(Suppl 1):S26–S32. 10.1093/infdis/jis764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rong L, Dahari H, Ribeiro RM, Perelson AS. 2010. Rapid emergence of protease inhibitor resistance in hepatitis C virus. Sci. Transl. Med. 2:30ra32. 10.1126/scitranslmed.3000544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guedj J, Dahari H, Rong L, Sansone ND, Nettles RE, Cotler SJ, Layden TJ, Uprichard SL, Perelson AS. 2013. Modeling shows that the NS5A inhibitor daclatasvir has two modes of action and yields a shorter estimate of the hepatitis C virus half-life. Proc. Natl. Acad. Sci. U. S. A. 110:3991–3996. 10.1073/pnas.1203110110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarrazin C, Zeuzem S. 2010. Resistance to direct antiviral agents in patients with hepatitis C virus infection. Gastroenterology 138:447–462. 10.1053/j.gastro.2009.11.055 [DOI] [PubMed] [Google Scholar]
- 16.Fried MW, Buti M, Dore GJ, Flisiak R, Ferenci P, Jacobson I, Marcellin P, Manns M, Nikitin I, Poordad F, Sherman M, Zeuzem S, Scott J, Gilles L, Lenz O, Peeters M, Sekar V, De Smedt G, Beumont-Mauviel M. 2013. Once-daily simeprevir (TMC435) with pegylated interferon and ribavirin in treatment-naive genotype 1 hepatitis C: the randomized PILLAR study. Hepatology 58:1918–1929. 10.1002/hep.26641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanaka Y, Hanada K, Hanabusa H, Kurbanov F, Gojobori T, Mizokami M. 2007. Increasing genetic diversity of hepatitis C virus in haemophiliacs with human immunodeficiency virus coinfection. J. Gen. Virol. 88:2513–2519. 10.1099/vir.0.82974-0 [DOI] [PubMed] [Google Scholar]
- 18.Sherman KE, Andreatta C, O'Brien J, Gutierrez A, Harris R. 1996. Hepatitis C in human immunodeficiency virus-coinfected patients: increased variability in the hypervariable envelope coding domain. Hepatology 23:688–694. 10.1002/hep.510230405 [DOI] [PubMed] [Google Scholar]
- 19.Blackard JT, Hiasa Y, Smeaton L, Jamieson DJ, Rodriguez I, Mayer KH, Chung RT. 2007. Compartmentalization of hepatitis C virus (HCV) during HCV/HIV coinfection. J. Infect. Dis. 195:1765–1773. 10.1086/518251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Farzadegan H, Gupta P, Rinaldo CR, Learn GH, He X, Huang XL, Mullins JI. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489–10502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez-Labrador FX, Dove L, Hui CK, Phung Y, Kim M, Berenguer M, Wright TL. 2007. Trends for genetic variation of hepatitis C virus quasispecies in human immunodeficiency virus-1 coinfected patients. Virus Res. 130:285–291. 10.1016/j.virusres.2007.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartolini B, Giombini E, Zaccaro P, Selleri M, Rozera G, Abbate I, Comandini UV, Ippolito G, Solmone M, Capobianchi MR. 2013. Extent of HCV NS3 protease variability and resistance-associated mutations assessed by next generation sequencing in HCV monoinfected and HIV/HCV coinfected patients. Virus Res. 177:205–208. 10.1016/j.virusres.2013.08.001 [DOI] [PubMed] [Google Scholar]
- 23.Shuhart MC, Sullivan DG, Bekele K, Harrington RD, Kitahata MM, Mathisen TL, Thomassen LV, Emerson SS, Gretch DR. 2006. HIV infection and antiretroviral therapy: effect on hepatitis C virus quasispecies variability. J. Infect. Dis. 193:1211–1218. 10.1086/502974 [DOI] [PubMed] [Google Scholar]
- 24.Winters MA, Chary A, Eison R, Asmuth D, Holodniy M. 2010. Impact of highly active antiretroviral therapy on hepatitis C virus protease quasispecies diversity in HIV co-infected patients. J. Med. Virol. 82:791–798. 10.1002/jmv.21679 [DOI] [PubMed] [Google Scholar]
- 25.Netski DM, Mao Q, Ray SC, Klein RS. 2008. Genetic divergence of hepatitis C virus: the role of HIV-related immunosuppression. J. Acquir. Immune Defic. Syndr. 49:136–141. 10.1053/j.gastro.2009.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beerenwinkel N, Zagordi O. 2011. Ultra-deep sequencing for the analysis of viral populations. Curr. Opin. Virol. 1:413–418. 10.1016/j.coviro.2011.07.008 [DOI] [PubMed] [Google Scholar]
- 27.Di Giallonardo F, Zagordi O, Duport Y, Leemann C, Joos B, Kunzli-Gontarczyk M, Bruggmann R, Beerenwinkel N, Gunthard HF, Metzner KJ. 2013. Next-generation sequencing of HIV-1 RNA genomes: determination of error rates and minimizing artificial recombination. PLoS One 8:e74249. 10.1371/journal.pone.0074249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jabara CB, Jones CD, Roach J, Anderson JA, Swanstrom R. 2011. Accurate sampling and deep sequencing of the HIV-1 protease gene using a primer ID. Proc. Natl. Acad. Sci. U. S. A. 108:20166–20171. 10.1073/pnas.1110064108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nei M, Li WH. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. U. S. A. 76:5269–5273. 10.1073/pnas.76.10.5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shannon CE. 1948. A mathematical theory of communication. Bell Syst. Tech. J. 27:379–423. 10.1002/j.1538-7305.1948.tb01338.x [DOI] [Google Scholar]
- 32.Akhter S, Bailey BA, Salamon P, Aziz RK, Edwards RA. 2013. Applying Shannon's information theory to bacterial and phage genomes and metagenomes. Sci. Rep. 3:1033. 10.1038/srep01033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 34.Dixon P. 2003. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14:927–930. 10.1111/j.1654-1103.2003.tb02228.x [DOI] [Google Scholar]
- 35.Sullivan JC, De Meyer S, Bartels DJ, Dierynck I, Zhang EZ, Spanks J, Tigges AM, Ghys A, Dorrian J, Adda N, Martin EC, Beumont M, Jacobson IM, Sherman KE, Zeuzem S, Picchio G, Kieffer TL. 2013. Evolution of treatment-emergent resistant variants in telaprevir phase 3 clinical trials. Clin. Infect. Dis. 57:221–229. 10.1093/cid/cit226 [DOI] [PubMed] [Google Scholar]
- 36.Bartels DJ, Sullivan JC, Zhang EZ, Tigges AM, Dorrian JL, De Meyer S, Takemoto D, Dondero E, Kwong AD, Picchio G, Kieffer TL. 2013. Hepatitis C virus variants with decreased sensitivity to direct-acting antivirals (DAAs) were rarely observed in DAA-naive patients prior to treatment. J. Virol. 87:1544–1553. 10.1128/JVI.02294-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barnard RJ, McHale CM, Newhard W, Cheney CA, Graham DJ, Himmelberger AL, Strizki J, Hwang PM, Rivera AA, Reeves JD, Nickle D, Dinubile MJ, Hazuda DJ, Mobashery N. 2013. Emergence of resistance-associated variants after failed triple therapy with vaniprevir in treatment-experienced non-cirrhotic patients with hepatitis C-genotype 1 infection: a population and clonal analysis. Virology 443:278–284. 10.1016/j.virol.2013.05.013 [DOI] [PubMed] [Google Scholar]
- 38.Lenz O, Verbinnen T, Lin TI, Vijgen L, Cummings MD, Lindberg J, Berke JM, Dehertogh P, Fransen E, Scholliers A, Vermeiren K, Ivens T, Raboisson P, Edlund M, Storm S, Vrang L, de Kock H, Fanning GC, Simmen KA. 2010. In vitro resistance profile of the hepatitis C virus NS3/4A protease inhibitor TMC435. Antimicrob. Agents Chemother. 54:1878–1887. 10.1128/AAC.01452-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McPhee F, Friborg J, Levine S, Chen C, Falk P, Yu F, Hernandez D, Lee MS, Chaniewski S, Sheaffer AK, Pasquinelli C. 2012. Resistance analysis of the hepatitis C virus NS3 protease inhibitor asunaprevir. Antimicrob. Agents Chemother. 56:3670–3681. 10.1128/AAC.00308-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimakami T, Welsch C, Yamane D, McGivern D, Yi M, Zeuzem S, Lemon SM. 2011. Protease inhibitor-resistant hepatitis C virus mutants with reduced fitness from impaired production of infectious virus. Gastroenterology 140:667–675. 10.1053/j.gastro.2010.10.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Acevedo A, Brodsky L, Andino R. 2014. Mutational and fitness landscapes of an RNA virus revealed through population sequencing. Nature 505:686–690. 10.1038/nature12861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kato N, Sekiya H, Ootsuyama Y, Nakazawa T, Hijikata M, Ohkoshi S, Shimotohno K. 1993. Humoral immune response to hypervariable region 1 of the putative envelope glycoprotein (gp70) of hepatitis C virus. J. Virol. 67:3923–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mao Q, Ray SC, Laeyendecker O, Ticehurst JR, Strathdee SA, Vlahov D, Thomas DL. 2001. Human immunodeficiency virus seroconversion and evolution of the hepatitis C virus quasispecies. J. Virol. 75:3259–3267. 10.1128/JVI.75.7.3259-3267.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang GP, Sherrill-Mix SA, Chang KM, Quince C, Bushman FD. 2010. Hepatitis C virus transmission bottlenecks analyzed by deep sequencing. J. Virol. 84:6218–6228. 10.1128/JVI.02271-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang XP, Goodwin L, Kahn P, Gawel C, Cunha CB, Laser B, Sahn B, Kaplan MH. 2006. Influence of increased CD4 cell counts on the genetic variability of hepatitis C virus in patients co-infected with human immunodeficiency virus I. J. Biomol. Tech. 17:228–239 [PMC free article] [PubMed] [Google Scholar]
- 46.Bernini F, Ebranati E, De Maddalena C, Shkjezi R, Milazzo L, Lo Presti A, Ciccozzi M, Galli M, Zehender G. 2011. Within-host dynamics of the hepatitis C virus quasispecies population in HIV-1/HCV coinfected patients. PLoS One 6:e16551. 10.1371/journal.pone.0016551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Babik JM, Holodniy M. 2003. Impact of highly active antiretroviral therapy and immunologic status on hepatitis C virus quasispecies diversity in human immunodeficiency virus/hepatitis C virus-coinfected patients. J. Virol. 77:1940–1950. 10.1128/JVI.77.3.1940-1950.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Solmone M, Girardi E, Lalle E, Abbate I, D'Arminio Monforte A, Cozzi-Lepri A, Alessandrini A, Piscopo R, Ebo F, Cosco L, Antonucci G, Ippolito G, Capobianchi MR. 2006. Evolution of HVR-1 quasispecies after 1-year treatment in HIV/HCV-coinfected patients according to the pattern of response to highly active antiretroviral therapy. Antivir. Ther. 11:87–94 [PubMed] [Google Scholar]
- 49.Erickson AL, Kimura Y, Igarashi S, Eichelberger J, Houghton M, Sidney J, McKinney D, Sette A, Hughes AL, Walker CM. 2001. The outcome of hepatitis C virus infection is predicted by escape mutations in epitopes targeted by cytotoxic T lymphocytes. Immunity 15:883–895. 10.1016/S1074-7613(01)00245-X [DOI] [PubMed] [Google Scholar]
- 50.Kuntzen T, Timm J, Berical A, Lewis-Ximenez LL, Jones A, Nolan B, Schulze zur Wiesch J, Li B, Schneidewind A, Kim AY, Chung RT, Lauer GM, Allen TM. 2007. Viral sequence evolution in acute hepatitis C virus infection. J. Virol. 81:11658–11668. 10.1128/JVI.00995-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scotta C, Garbuglia AR, Ruggeri L, Spada E, Laurenti L, Perrone MP, Girelli G, Mele A, Capobianchi MR, Folgori A, Nicosia A, Del Porto P, Piccolella E. 2008. Influence of specific CD4+ T cells and antibodies on evolution of hypervariable region 1 during acute HCV infection. J. Hepatol. 48:216–228. 10.1016/j.jhep.2007.09.011 [DOI] [PubMed] [Google Scholar]
- 52.Bull RA, Luciani F, McElroy K, Gaudieri S, Pham ST, Chopra A, Cameron B, Maher L, Dore GJ, White PA, Lloyd AR. 2011. Sequential bottlenecks drive viral evolution in early acute hepatitis C virus infection. PLoS Pathog. 7:e1002243. 10.1371/journal.ppat.1002243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O'Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G, Goedert JJ, Kirk GD, Donfield SM, Rosen HR, Tobler LH, Busch MP, McHutchison JG, Goldstein DB, Carrington M. 2009. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 461:798–801. 10.1038/nature08463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ, Sulkowski M, McHutchison JG, Goldstein DB. 2009. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461:399–401. 10.1038/nature08309 [DOI] [PubMed] [Google Scholar]
- 55.Huang Y, Yang H, Borg BB, Su X, Rhodes SL, Yang K, Tong X, Tang G, Howell CD, Rosen HR, Thio CL, Thomas DL, Alter HJ, Sapp RK, Liang TJ. 2007. A functional SNP of interferon-gamma gene is important for interferon-alpha-induced and spontaneous recovery from hepatitis C virus infection. Proc. Natl. Acad. Sci. U. S. A. 104:985–990. 10.1073/pnas.0609954104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, Cheng J, Goedert JJ, Vlahov D, Hilgartner M, Cox S, Little AM, Alexander GJ, Cramp ME, O'Brien SJ, Rosenberg WM, Thomas DL, Carrington M. 2004. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science 305:872–874. 10.1126/science.1097670 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.