Abstract
Enterovirus 71 (EV71) is the major cause of hand-foot-and-mouth disease in children. In our study, using the complete genome sequences of 42 EV71 representing all three genotypes, we analyzed synonymous codon usage and the relative dinucleotide abundance in EV71 genome. The general correlation between base composition and codon usage bias suggests that mutational pressure rather than natural selection is the main factor that determines the codon usage bias in EV71 genome. Furthermore, we observed that the relative abundance of dinucleotides in EV71 is independent of the overall base composition but is still the result of differential mutational pressure, which also shapes codon usage. In addition, other factors, such as hydrophobicity and aromaticity, also influence the codon usage variation among the genomes of EV71. This study represents the most comprehensive analysis of EV71 codon usage patterns and provides a basic understanding of the mechanisms for codon usage bias.
Keywords: EV71, Synonymous codon usage, Mutational bias, Dinucleotide bias, Subgenotype
Introduction
Enterovirus 71 (EV71) is a member of the Enterovirus genus of the Picornaviridae family, which is the major cause of hand-foot-and-mouth disease (HFMD) in children. EV71 is a small, non-enveloped virus with a positive-stranded RNA genome size of about 7.4 kb. The virus has a single-stranded positive-sense RNA containing a single open reading frame (ORF) encoding a polyprotein that, following viral protease mediated co- and post-translational processing, gives rise to 4 capsid proteins (VP1, VP2, VP3, and VP4) and 7 nonstructural proteins (2A, 2B, 2C, 3A, 3B, 3C, and 3D) [26]. Studies on the phylogenetic relationship of EV71 have divided the viruses into genotypes A, B and C [2], which has been further divided into subtypes, designated A, B1-5, and C1-4 based on sequencing the region encoding the VP1 major capsid protein [16, 20]. Synonymous codons are not used randomly [14, 15]. Mutational pressure and translational selection were thought to be the main factors that account for codon usage variation among genes in some human RNA virus [12, 13, 29]. Studies the extent and causes of biases in codon usage is essential to the understanding of viral evolution, particularly the interplay between viruses and the immune response [4, 17]. Recently, recombination was found to play a more important role than positive selection in the formation of genetic diversity in EV71 virus. Positive selection was only detected at site 145 of VP1, but most amino acid sites of nonstructural proteins were under negative selection [5]. Previous studies of EV71 have mainly been limited to phylogenic analysis, and few synonymous codon usage analyses have been applied. In order to better understand the characteristics of the EV71 genome and to reveal more information about the viral genome, we have analyzed the codon usage and dinucleotide composition. In this report, we sought to address the issues concerning codon usage in EV71 virus.
A total of 42 publicly available complete human EV71 RNAs isolated from China, Taiwan, Malaysia, Singapore, Australia, Japan, South Korea, Viet Nam, America and Norway were download from GeneBank and sequence with >99 % sequence identities were excluded. The EV71 genome represented 3 genotypes (A, B and C). The serial number (SN), mononucleotide composition of each genome, GenBank accession numbers, genotype, and other detail information are listed in Table 1. Relative synonymous codon usage (RSCU) values are largely independent of amino acid composition and are particularly useful in comparing codon usage between genes, or sets of genes that differ in their size and amino acid composition. To examine synonymous codon usage without the confounding influence of amino acid composition of different EV71 virus, RSCU values of each codon in each ORF were used to measure the synonymous codon usage [18]. RSCU values of different codon in each ORF were calculated to investigate the extent of codon bias in EV71. The details of each ORF and the overall RSCU values of 59 codons in 42 EV71 genomes are, respectively, represented in Tables 1 and 2. The preferentially used codons were A-ended (3 ones), U-ended (6 ones) codons, C-ended (8 ones), codons G-ended (4 ones) codons. The average GC content of all EV71 viruses was 46.09 % (From 45.57 to 47.32 %, with a S.D. of 0.32 %), and the average GC3s content in codons was 48.05 % (From 46.62 to 51.98 %, with a S.D. of 0.94 %). This is consistent with our previous observations that EV71 viruses are GC-moderate genomes, and so it is expected that the third-ended codons are not preferentially used. In order to investigate whether these 42 coding sequences of EV71 display similar compositional features, ENC values were calculated (Table 1). The effective number of codons of a gene (ENC) is generally used to quantify the codon usage bias of a gene, which is essentially independent of gene length. The ENC values range from 20 to 61. The larger the extent of codon preference in a gene, the smaller the ENC value is. In an extremely biased gene where only one codon is used for each amino acid, this value would be 20; in an unbiased gene, it would be 61 [7]. The ENC values of different EV71 genes vary from 55.59 to 57.41, with a mean of 56.62 and S.D. of 0.4039. We found that all the ENC values for EV71 ORFs are much higher (ENC > 55). Based on this finding, together with published data on codon usage bias among some RNA viruses [10, 21, 29], we conclude that the codon usage bias in EV71 genome is slight.
Table 1.
List of EV71 strains used for analysis of synonymous codon usage in this study
| Genebank accession | Strain | Country | Date | Genotype | GC3s | ENC | Mononucleotide frequencies (%) | Reference | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| C | T | A | G | ||||||||
| AB204852 | BrCr-Tr | Japan | 2008 | A | 0.473 | 56.09 | 0.2172 | 0.2476 | 0.2795 | 0.2385 | [1] |
| AB204853 | BrCr-Ts | Japan | 2008 | A | 0.473 | 56.19 | 0.2174 | 0.2475 | 0.2795 | 0.2385 | [1] |
| AM396587 | UH1/PM/1997 | Malaysia | 1997 | B | 0.480 | 55.97 | 0.2201 | 0.2481 | 0.2744 | 0.2399 | [27] |
| U22522 | MS/7423/87 | America | 2001 | B | 0.478 | 55.94 | 0.2198 | 0.2488 | 0.2742 | 0.2394 | [3] |
| AM396586 | EV71/SAR/SHA66 | Malaysia | 2006 | B | 0.473 | 56.82 | 0.2212 | 0.2482 | 0.2754 | 0.2373 | [27] |
| DQ341362 | SB12736-SAR-03 | Malaysia | 2007 | B | 0.479 | 56.20 | 0.2218 | 0.2476 | 0.2739 | 0.2396 | Unpublished |
| DQ341365 | PP37-MAL-01 | Malaysia | 2007 | B | 0.484 | 56.55 | 0.2221 | 0.2465 | 0.2736 | 0.2409 | Unpublished |
| DQ341366 | SB2864-SAR-00 | Malaysia | 2007 | B | 0.477 | 56.19 | 0.2212 | 0.2470 | 0.2750 | 0.2397 | Unpublished |
| DQ341354 | 3799-SIN-98 | Singapore | 2007 | B | 0.470 | 56.63 | 0.2216 | 0.2478 | 0.2769 | 0.2356 | Unpublished |
| DQ341364 | 5511-SIN-00 | Singapore | 2007 | B | 0.479 | 55.59 | 0.2234 | 0.2449 | 0.2757 | 0.2388 | Unpublished |
| EU364841 | 26M/AUS/4/99 | Australia | 2008 | B | 0.479 | 56.80 | 0.2206 | 0.2485 | 0.2756 | 0.2374 | [6] |
| AB469183 | EV71-GFP | Malaysia | 2008 | B | 0.520 | 55.82 | 0.2319 | 0.2380 | 0.2715 | 0.2413 | Unpublished |
| EF373575 | E2002042-TW-CDC | Taiwan | 2008 | B | 0.473 | 56.19 | 0.2213 | 0.2468 | 0.2765 | 0.2380 | Unpublished |
| AF176044 | 1245a/98/tw | Taiwan | 1999 | C | 0.493 | 56.43 | 0.2207 | 0.2427 | 0.2762 | 0.2440 | Unpublished |
| AF119796 | TW/2086/98 | Taiwan | 2000 | C | 0.494 | 56.40 | 0.2209 | 0.2418 | 0.2748 | 0.2453 | [19] |
| AF304458 | Tainan/6092/98 | Taiwan | 2001 | C | 0.493 | 56.42 | 0.2210 | 0.2424 | 0.2763 | 0.2438 | [26] |
| AY465356 | SHZH03 | China | 2003 | C | 0.467 | 56.97 | 0.2210 | 0.2499 | 0.2751 | 0.2364 | Unpublished |
| DQ452074 | 804/NO/03 | Norway | 2003 | C | 0.479 | 57.07 | 0.2187 | 0.2455 | 0.2780 | 0.2414 | [22] |
| DQ133458 | 984 | Taiwan | 2004 | C | 0.478 | 56.96 | 0.2221 | 0.2482 | 0.2737 | 0.2377 | Unpublished |
| DQ133459 | 1235 | Taiwan | 2004 | C | 0.472 | 56.72 | 0.2206 | 0.2494 | 0.2756 | 0.2371 | Unpublished |
| DQ060149 | pinf7-54A | Taiwan | 2005 | C | 0.492 | 56.83 | 0.2199 | 0.2430 | 0.2763 | 0.2446 | Unpublished |
| JQ965759 | 540V/VNM/05 | Viet Nam | 2005 | C | 0.473 | 57.05 | 0.2222 | 0.2484 | 0.2756 | 0.2365 | [28] |
| AM396584 | ENT/PM/SHA52 | Malaysia | 2006 | C | 0.486 | 56.56 | 0.2204 | 0.2430 | 0.2785 | 0.2421 | [27] |
| DQ341357 | 7F-AUS-6-99 | Australia | 2007 | C | 0.497 | 57.22 | 0.2216 | 0.2421 | 0.2759 | 0.2447 | Unpublished |
| DQ341361 | 1M-AUS-12-00 | Australia | 2007 | C | 0.480 | 56.90 | 0.2192 | 0.2447 | 0.2783 | 0.2415 | Unpublished |
| DQ341358 | S40221-SAR-00 | Malaysia | 2007 | C | 0.482 | 56.76 | 0.2187 | 0.2449 | 0.2779 | 0.2426 | Unpublished |
| DQ341359 | S10862-SAR-98 | Malaysia | 2007 | C | 0.486 | 56.89 | 0.2209 | 0.2432 | 0.2783 | 0.2415 | Unpublished |
| DQ341360 | J115-MAL-01 | Malaysia | 2007 | C | 0.487 | 57.12 | 0.2198 | 0.2438 | 0.2777 | 0.2426 | Unpublished |
| DQ341355 | 06-KOR-00 | South Korea | 2007 | C | 0.484 | 56.41 | 0.2181 | 0.2465 | 0.2754 | 0.2434 | Unpublished |
| EF063152 | E2005125-TW | Taiwan | 2007 | C | 0.477 | 57.41 | 0.2158 | 0.2479 | 0.2762 | 0.2437 | unpublished |
| FJ194964 | EV71/GDFS/3/2008 | China | 2008 | C | 0.479 | 57.11 | 0.2207 | 0.2499 | 0.2733 | 0.2386 | Unpublished |
| FJ194965 | EV71/DGSC117/2008 | China | 2008 | C | 0.478 | 56.99 | 0.2236 | 0.2462 | 0.2756 | 0.2373 | Unpublished |
| FJ360544 | GZ08-01 | China | 2008 | C | 0.475 | 56.82 | 0.2224 | 0.2485 | 0.2739 | 0.2377 | Unpublished |
| FJ360545 | GZ08-02 | China | 2008 | C | 0.482 | 56.54 | 0.2239 | 0.2470 | 0.2745 | 0.2374 | Unpublished |
| FJ439769 | Fuyang-0805 | China | 2008 | C | 0.481 | 56.65 | 0.2260 | 0.2452 | 0.2751 | 0.2368 | [24] |
| EF373576 | E2004104-TW-CDC | Taiwan | 2008 | C | 0.478 | 56.96 | 0.2230 | 0.2478 | 0.2751 | 0.2368 | Unpublished |
| EU131776 | N3340-TW-02 | Taiwan | 2008 | C | 0.478 | 56.55 | 0.2236 | 0.2465 | 0.2771 | 0.2354 | [11] |
| FJ606447 | BJ08-Z004-3 | China | 2009 | C | 0.466 | 56.63 | 0.2199 | 0.2509 | 0.2757 | 0.2362 | [8] |
| FJ606449 | BJ08-Z020-1 | China | 2009 | C | 0.477 | 56.76 | 0.2250 | 0.2461 | 0.2759 | 0.2362 | [8] |
| FJ606450 | BJ08-Z025-5 | China | 2009 | C | 0.477 | 56.34 | 0.2244 | 0.2461 | 0.2756 | 0.2373 | [8] |
| FJ713137 | Shanghai036-2009 | China | 2009 | C | 0.477 | 56.91 | 0.2230 | 0.2478 | 0.2734 | 0.2385 | [25] |
| JQ806378 | 35/Jingdezhen/China/HFMD_Severe/2011 | China | 2011 | C | 0.479 | 56.79 | 0.2244 | 0.2473 | 0.2739 | 0.2373 | Unpublished |
Table 2.
Synonymous codon usage in EV71 viruses
| AA | Codon | N | RSCU | AA | Codon | N | RSCU |
|---|---|---|---|---|---|---|---|
| Phe | UUU | 1,918 | 0.99 | Ser | UCU | 1,168 | 1.01 |
| UUC | 1,938 | 1.01 | UCC | 1,294 | 1.12 | ||
| Leu | UUA | 1,004 | 0.77 | UCA | 1,502 | 1.30 | |
| UUG | 1,572 | 1.21 | UCG | 436 | 0.38 | ||
| Tyr | UAU | 1,654 | 0.91 | Cys | UGU | 868 | 0.99 |
| UAC | 1,970 | 1.09 | UGC | 880 | 1.01 | ||
| ter | UAA | 20 | 0.00 | ter | UGA | 1 | 0.00 |
| ter | UAG | 21 | 0.00 | Trp | UGG | 1,302 | 1.00 |
| Leu | CUU | 1,489 | 1.15 | Pro | CCU | 1,409 | 1.08 |
| CUC | 1,435 | 1.10 | CCC | 1,189 | 0.91 | ||
| CUA | 1,054 | 0.81 | CCA | 2,084 | 1.59 | ||
| CUG | 1,242 | 0.96 | CCG | 547 | 0.42 | ||
| His | CAU | 771 | 0.73 | Arg | CGU | 311 | 0.45 |
| CAC | 1,335 | 1.27 | CGC | 716 | 1.05 | ||
| Gln | CAA | 1,938 | 1.09 | CGA | 368 | 0.54 | |
| CAG | 1,608 | 0.91 | CGG | 276 | 0.40 | ||
| Ile | AUU | 2,220 | 1.25 | Thr | ACU | 1,874 | 1.18 |
| AUC | 1,887 | 1.06 | ACC | 1,953 | 1.23 | ||
| AUA | 1,233 | 0.69 | ACA | 1,894 | 1.20 | ||
| Met | AUG | 2,083 | 1.00 | ACG | 611 | 0.39 | |
| Asn | AAU | 1,924 | 0.92 | Ser | AGU | 1,373 | 1.19 |
| AAC | 2,277 | 1.08 | AGC | 1,136 | 0.99 | ||
| Lys | AAA | 2,145 | 0.91 | Arg | AGA | 1,261 | 1.84 |
| AAG | 2,576 | 1.09 | AGG | 1,170 | 1.71 | ||
| Val | GUU | 1,574 | 0.97 | Ala | GCU | 2,121 | 1.21 |
| GUC | 1,423 | 0.88 | GCC | 1,803 | 1.03 | ||
| GUA | 795 | 0.49 | GCA | 2,194 | 1.25 | ||
| GUG | 2,676 | 1.65 | GCG | 908 | 0.52 | ||
| Asp | GAU | 2,666 | 1.11 | Gly | GGU | 1,799 | 1.10 |
| GAC | 2,151 | 0.89 | GGC | 1,614 | 0.99 | ||
| Glu | GAA | 2,135 | 0.93 | GGA | 1,474 | 0.90 | |
| GAG | 2,479 | 1.07 | GGG | 1,637 | 1.00 |
The preferentially used codons for each amino acid are displayed in bold
AA Amino acids, N number of codons, RSCU cumulative relative synonymous codon usage
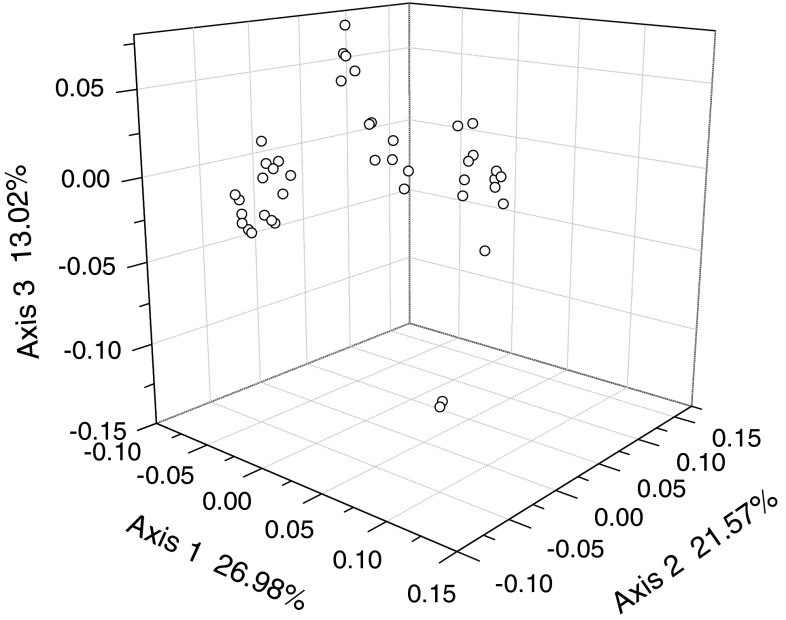
Spearman’s rank correlation analysis and multiple regression analysis were performed to determine the role of different factors in shaping the codon usage biases in the various EV71 viruses. All statistical analyses, as well as cluster analysis, were carried out using the statistical analysis software SPSS Version 15.0. To investigate synonymous codon usage variation among EV71 viruses, COA was implemented for all 42 EV71 ORFs selected for this study. Figure 1 depicts the position of each ORF on the plane defined by the first, second and third principal axes generated by COA on RSCU values of ORFs. The first principal axis accounts for 26.98 % of the total variation. The next three axes account for 21.57, 13.02, and 8.05 % of the variation, respectively. This observation indicates that although the first major axis explains a substantial amount of variation in trends in codon usage, the second major axis also has an appreciable impact on total variation in synonymous codon usage. If not specifically mentioned, the values of the first two axes of this COA were used for correlation and regression analysis hereafter.
Fig. 1.
A plot of value of the first, second and third axis of each ORF in COA. The first axis accounts for 26.98 % of all variation among ORFs, the second axis accounts for 21.57 % and third axis accounts for 13.02 % of total vibrations
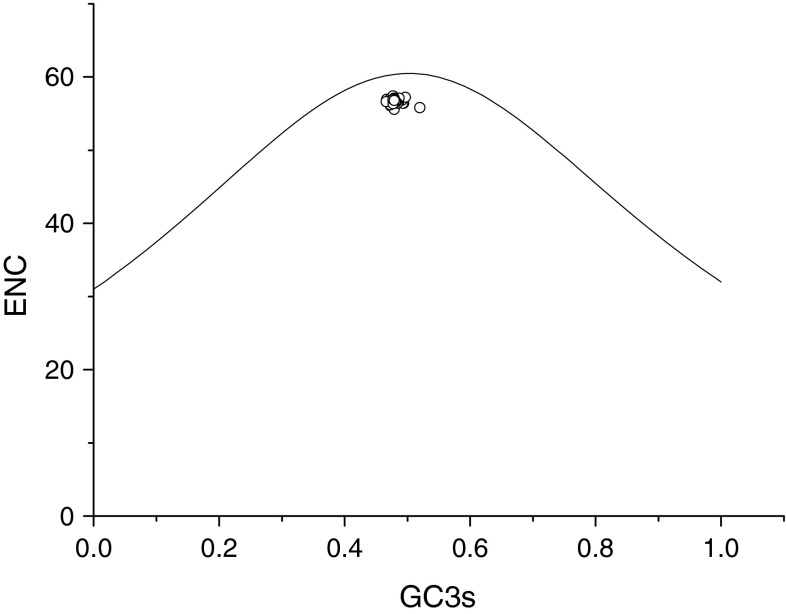
Mutational pressure and translational selection were thought to be the main factors that account for codon usage variation among genes in some human RNA virus [12, 13, 29]. Therefore, we compared the G+C content at the first and second codon positions (GC12s) with that at the synonymous third position (GC3s) to investigate which factor in EV71 can explain their codon usage. It was found that GC12s and GC3s are significantly correlated (r = 0.393, P < 0.01). This suggests that they are most likely the result of mutational pressure, as natural selection would be expected to act differently on different codon positions. The plot of ENC and GC3S is another effective way to explore codon usage variation among genes [23]. In order to further find whether codon usage variation among EV71 virus is determined by mutational bias, ENC values of each virus gene were plotted against its corresponding GC3s. Genes, whose codon choice is constrained only by a G+C mutational bias, will lie on or just below the curve of the predicted values. All of the spots lie below the expected curve as shown in Fig. 2. In addition, a significantly positive correlation between GC3s and ENC (r = −0.048, P < 0.001) values was observed. The results indicated that the codon usage bias in these 42 EV71 genomes is greatly influenced by the G+C mutation bias.
Fig. 2.
Effective number of codons used in each ORF plotted against the GC3s. The continuous curve plots the relationship between GC3s and NEC in the absence of selection. All of spots lie below the expected curve
Multivariate statistical analysis can be used to explore the relationships between variables and samples. The major trend in codon usage variation among ORFs can be investigated using correspondence analysis (COA). In order to minimize the effects of amino acid composition on codon usage, each ORF is represented as a 59-dimensional vector; each dimension corresponds to the RSCU value of one sense codon (excluding AUG, UGG, and stop codons). Major trends within this dataset can be determined using measures of relative inertia and genes ordered according to their positions along the axis of major inertia. We analyzed the correlation between the first or second axis values in COA and GC12s or GC3s values of each strain. The first axis value in COA of each selected genome, which contains most of the variation in synonymous codon usage bias between these genomes, is closely correlated with the GC composition at the first, second, and third codon position (Table 3). The second axis in the COA of each gene is also closely correlated with the GC12s. This analysis indicated that most of the codon usage bias among different ORFs is directly related to the nucleotide composition. Therefore, the compositional constraint is the main determinant of the variation in synonymous codon usage among different EV71 ORFs. These results were similar with previously study described by Liu et al. that the interaction between mutation pressure from virus and natural selection from host exists in the processes of evolution of EV71 [13]. However, other factors, such as hydrophobicity and aromaticity, whether influence the codon usage variation among the genomes of EV71 need to be elucidated.
Table 3.
Summary of correlation analysis between the first two axes in COA and GC12s, GC3s, GRAVY, or aromaticity in the selected 42 EV71 ORFs
| GRAVY | Aromaticity | GC3s | GC12s | |
|---|---|---|---|---|
| Axis 1 | ||||
| r | 0.450** | −0.104 | 0.342* | 0.628** |
| P | 0.003 | 0.513 | 0.027 | <0.001 |
| Axis 2 | ||||
| r | 0.446** | −0.653** | 0.208 | −0.373* |
| P | 0.003 | <0.001 | 0.186 | 0.018 |
| ENC | ||||
| r | −0.073 | 0.500** | −0.048** | |
| P | 0.644 | <0.001 | <0.001 | |
* P value ≤0.05; ** P value ≤0.01
The GC index was used to calculate the overall GC content in the ORF, while the index GC3s was used to calculate the fraction of GC nucleotides at the synonymous third codon position (excluding Met, Trp, and the termination codons). At the amino acid level, the general average hydrophobicity score (GRAVY) and the frequency of aromatic amino acids (Aromaticity) in the putative gene product were also analyzed. All the indices mentioned were calculated using the analysis program CodonW, version 1.4 [9]. As showed in Fig. 2, the majority of the actual ENC values are slightly lower than the expected ones, which indicated that other factors may also influence the codon usage in EV71 viruses. To test whether selection pressure contributes to the codon usage variation among EV71 viruses, we performed a correlation analysis to evaluate whether GRAVY and Aromaticity values were related to first two axes of COA and ENC values (Table 3). Our results showed that GRAVY was correlated with both axis 1 and axis 2, while Aromaticity was correlated with ENC and axis 2, indicating that the degree of hydrophobicity and the frequency of aromatic amino acids (Phe, Tyr, Trp) were also associated with the codon usage variation.
In our study, the synonymous codon usage biases in 42 EV71 genomes were analyzed. We found that both EV71 genes had low codon usage bias, and mutational pressure rather selection pressure is the main factor determining the codon usage biases. Moreover, aromaticity and hydrophobicity could be partially accounting for the codon usage variation.
Acknowledgments
This work is supported by National Natural Science Foundation of China (NSFC, Grant no 31300145, 31310103018 and 31200121), and the Technology Research Foundation of Education Department of HeiLongJiang Province, China (12541578).
References
- 1.Arita M, Shimizu H, Nagata N, Ami Y, Suzaki Y, Sata T, Iwasaki T, Miyamura T. Temperature-sensitive mutants of enterovirus 71 show attenuation in cynomolgus monkeys. J Gen Virol. 2005;86:1391–1401. doi: 10.1099/vir.0.80784-0. [DOI] [PubMed] [Google Scholar]
- 2.Brown BA, Oberste MS, Alexander JP, Kennett ML, Pallansch MA. Molecular epidemiology and evolution of enterovirus 71 strains isolated from 1970 to 1998. J Virol. 1999;73:9969–9975. doi: 10.1128/jvi.73.12.9969-9975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown BA, Pallansch MA. Complete nucleotide sequence of enterovirus 71 is distinct from poliovirus. Virus Res. 1995;39:195–205. doi: 10.1016/0168-1702(95)00087-9. [DOI] [PubMed] [Google Scholar]
- 4.Cao HW, Li DS, Zhang H. Analysis of synonymous codon usage in newcastle disease virus hemagglutinin-neuraminidase (HN) gene and fusion protein (F) gene. Virusdis. 2014;25:132–136. doi: 10.1007/s13337-013-0175-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen XM, Zhang Q, Li JH, Cao W, Zhang JX, Zhang L, Zhang WL, Shao ZJ, Yan YP. Analysis of recombination and natural selection in human enterovirus 71. Virology. 2010;398:251–261. doi: 10.1016/j.virol.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Chua BH, Phuektes P, Sanders SA, Nicholls PK, McMinn PC. The molecular basis of mouse adaptation by human enterovirus 71. J Gen Virol. 2008;89:1622–1632. doi: 10.1099/vir.0.83676-0. [DOI] [PubMed] [Google Scholar]
- 7.Comeron JM, Aguade M. An evaluation of measures of synonymous codon usage bias. J Mol Evol. 1998;47:268–274. doi: 10.1007/PL00006384. [DOI] [PubMed] [Google Scholar]
- 8.Ding NZ, Wang XM, Sun SW, Song Q, Li SN, He CQ. Appearance of mosaic enterovirus 71 in the 2008 outbreak of China. Virus Res. 2009;145:157–161. doi: 10.1016/j.virusres.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Grocock RJ, Sharp PM. Synonymous codon usage in cryptosporidium parvum: identification of two distinct trends among genes. Int J Parasitol. 2001;31:402–412. doi: 10.1016/S0020-7519(01)00129-1. [DOI] [PubMed] [Google Scholar]
- 10.Gu WJ, Zhou T, Ma JM, Sun X, Lu ZH. Analysis of synonymous codon usage in SARS coronavirus and other viruses in the nidovirales. Virus Res. 2004;101:155–161. doi: 10.1016/j.virusres.2004.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang SC, Hsu YW, Wang HC, Huang SW, Kiang D, Tsai HP, Wang SM, Liu CC, Lin KH, Su IJ, Wang JR. Appearance of intratypic recombination of enterovirus 71 in Taiwan from 2002 to 2005. Virus Res. 2008;131:250–259. doi: 10.1016/j.virusres.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Jenkins GM, Holmes EC. The extent of codon usage bias in human RNA viruses and its evolutionary origin. Virus Res. 2003;92:1–7. doi: 10.1016/S0168-1702(02)00309-X. [DOI] [PubMed] [Google Scholar]
- 13.Liu YS, Zhou JH, Chen HT, Ma LN, Pejsak Z, Ding YZ, Zhang J. The characteristics of the synonymous codon usage in enterovirus 71 virus and the effects of host on the virus in codon usage pattern. Infect Genet Evol. 2011;11:1168–1173. doi: 10.1016/j.meegid.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lloyd AT, Sharp PM. Evolution of codon usage patterns - the extent and nature of divergence between candida-albicans and saccharomyces-cerevisiae. Nucleic Acids Res. 1992;20:5289–5295. doi: 10.1093/nar/20.20.5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marin A, Bertranpetit J, Oliver JL, Medina JR. Variation in G+C-content and codon choice—differences among synonymous codon groups in vertebrate genes. Nucleic Acids Res. 1989;17:6181–6189. doi: 10.1093/nar/17.15.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McMinn P, Lindsay K, Perera D, Chan HM, Chan KP, Cardosa MJ. Phylogenetic analysis of enterovirus 71 strains isolated during linked epidemics in Malaysia, Singapore, and Western Australia. J Virol. 2001;75:7732–7738. doi: 10.1128/JVI.75.16.7732-7738.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shackelton LA, Parrish CR, Holmes EC. Evolutionary basis of codon usage and nucleotide composition bias in vertebrate DNA viruses. J Mol Evol. 2006;62:551–563. doi: 10.1007/s00239-005-0221-1. [DOI] [PubMed] [Google Scholar]
- 18.Sharp PM, Li WH. Codon usage in regulatory genes in Escherichia coli does not reflect selection for rare codons. Nucleic Acids Res. 1986;14:7737–7749. doi: 10.1093/nar/14.19.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shih SR, Ho MS, Lin KH, Wu SL, Chen YT, Wu CN, Lin TY, Chang LY, Tsao KC, Ning HC, Chang PY, Jung SM, Hsueh C, Chang KS. Genetic analysis of enterovirus 71 isolated from fatal and non-fatal cases of hand, foot and mouth disease during an epidemic in Taiwan, 1998. Virus Res. 2000;68:127–136. doi: 10.1016/S0168-1702(00)00162-3. [DOI] [PubMed] [Google Scholar]
- 20.Shimizu H, Utama A, Onnimala N, Chen L, Zhang LB, Ma YJ, Pongsuwanna Y, Miyamura T. Molecular epidemiology of enterovirus 71 infection in the Western Pacific Region. Pediatr Int. 2004;46:231–235. doi: 10.1046/j.1442-200x.2004.01868.x. [DOI] [PubMed] [Google Scholar]
- 21.Tao P, Dai L, Luo MC, Tang FQ, Tien P, Pan ZS. Analysis of synonymous codon usage in classical swine fever virus. Virus Genes. 2009;38:104–112. doi: 10.1007/s11262-008-0296-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Witso E, Palacios G, Ronningen KS, Cinek O, Janowitz D, Rewers M, Grinde B, Lipkin WI. Asymptomatic circulation of HEV71 in Norway. Virus Res. 2007;123:19–29. doi: 10.1016/j.virusres.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 23.Wright F. The Effective Number of Codons Used in a Gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- 24.Wu Z, Yang F, Zhao R, Zhao L, Guo D, Jin Q. Identification of small interfering RNAs which inhibit the replication of several Enterovirus 71 strains in China. J Virol Methods. 2009;159:233–238. doi: 10.1016/j.jviromet.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 25.Xia JF, Yan XF, Yu H, Qu D, Long JE. Simple and rapid detection of human enterovirus 71 by reverse-transcription and loop-mediated isothermal amplification: cryopreservation affected the detection ability. Diagn Microbiol Infect Dis. 2011;71:244–251. doi: 10.1016/j.diagmicrobio.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan JJ, Su IJ, Chen PF, Liu CC, Yu CK, Wang JR. Complete genome analysis of enterovirus 71 isolated from an outbreak in Taiwan and rapid identification of enterovirus 71 and coxsackievirus A16 by RT-PCR. J Med Virol. 2001;65:331–339. doi: 10.1002/jmv.2038. [DOI] [PubMed] [Google Scholar]
- 27.Yoke-Fun C, AbuBakar S. Phylogenetic evidence for inter-typic recombination in the emergence of human enterovirus 71 subgenotypes. BMC Microbiol. 2006;6:74–85. doi: 10.1186/1471-2180-6-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaini Z, McMinn P. A single mutation in capsid protein VP1 (Q145E) of a genogroup C4 strain of human enterovirus 71 generates a mouse virulent phenotype. J Gen Virol. 2012;93:1935–1940. doi: 10.1099/vir.0.043893-0. [DOI] [PubMed] [Google Scholar]
- 29.Zhou T, Gu WJ, Ma JM, Sun X, Lu ZH. Analysis of synonymous codon usage in H5N1 virus and other influenza A viruses. Biosystems. 2005;81:77–86. doi: 10.1016/j.biosystems.2005.03.002. [DOI] [PubMed] [Google Scholar]