Abstract
Picobirnavirus (PBV) is a small, non-enveloped, bisegmented double-stranded RNA (dsRNA) virus of vertebrate hosts. The name ‘Picobirnavirus’ derives from the prefix ‘pico’ (latin for ‘small’) in reference to the small virion size, plus the prefix ‘bi’ (latin for ‘two’) and the word ‘RNA’ to indicate the nature of the viral genome. The serendipitous discovery of PBV dates back to 1988 from Brazil, when human fecal samples collected during the acute gastroenteritis outbreaks were subjected for routine rotavirus surveillance by polyacrylamide gel electrophoresis (PAGE) and silver straining (S/S). The PAGE gels after silver staining showed a typical ‘two RNA band’ pattern, and it was identified as Picobirnavirus. Likewise, the feces of wild black-footed pigmy rice rats (Oryzomys nigripes) subjected for PAGE assay by the same research group in Brazil reported the presence of PBV (Pereira et al., J Gen Virol 69:2749–2754, 1988). PBVs have been detected in faeces of humans and wide range of animal species with or without diarrhoea, worldwide. The probable role of PBV as either a ‘primary diarrhoeal agent’ in ‘immunocompetent children’; or a ‘potential pathogen’ in ‘immunocompromised individuals’ or an ‘innocuous virus’ in the intestine remains elusive and needs to be investigated despite the numerous reports of the presence of PBV in fecal samples of various species of domestic mammals, wild animals, birds and snakes; our current knowledge of their biology, etiology, pathogenicity or their transmission characteristics remains subtle. This review aims to analyse the veterinary and zoonotic aspects of animal Picobirnavirus infections since its discovery.
Keywords: Picobirnavirus, Genogroup I PBV, Genogroup II PBV, Human PBV, Porcine PBV, Zoonotic potential
Introduction
Picobirnaviruses (PBVs) are novel, small, bisegmented double-stranded RNA (dsRNA) virus of vertebrate hosts. PBVs have been detected in faeces of humans and wide range of animal species with or without diarrhoea, worldwide. The virion is non-enveloped, small, spherical, 33–41 nm in diameter, with bisegmented dsRNA and consists of a simple core capsid with a distinctive icosahedral arrangement [21, 63, 68, 82]. Based on the migration pattern of the bisegmented dsRNA, PBV appears either with large genome profile (2.3–2.6 kbp for the larger and 1.5–1.9 kbp for the smaller segment, respectively) [34, 68] or small genome profile (1.75 and 1.55 kbp for segments 1 and 2, respectively) [6, 26, 27, 34] in PAGE experiments.
PBVs have also been associated with acute watery diarrhoea and gastroenteritis in humans both as ‘sole pathogen’ [7, 14] as well as with other known enteric pathogens [5, 6, 27–29, 40]. Similarly, PBVs have also been detected as sole pathogens in diarrhoeic animals [9, 60, 67] and with other known and established enteric pathogens [2, 4, 83]. They are still classified as ‘opportunistic diarrhoeagenic pathogen’, since they have been detected from immunocompromised patients and patients with prolonged immunosuppressive treatment who suffered from diarrhoea than non-diarrhoeic controls [18, 38, 39, 41, 42, 44, 75].
In recent times, with the advent of advanced molecular technologies such as high-throughput sequencing, massive parallel pyrosequening, sequence-independent amplification, etc., under the umbrella of ‘Metagenomics’ a relatively new branch of genetic research has revealed the existence of vast majority of microbial diversity in humans, animals, avian feces as well as environmental samples. It includes, detecting previously characterized viruses, as well as genetically diverse, novel subtypes or genotypes of viruses including picobirnaviruses from different geographical locations around the world [19, 22, 78, 87].
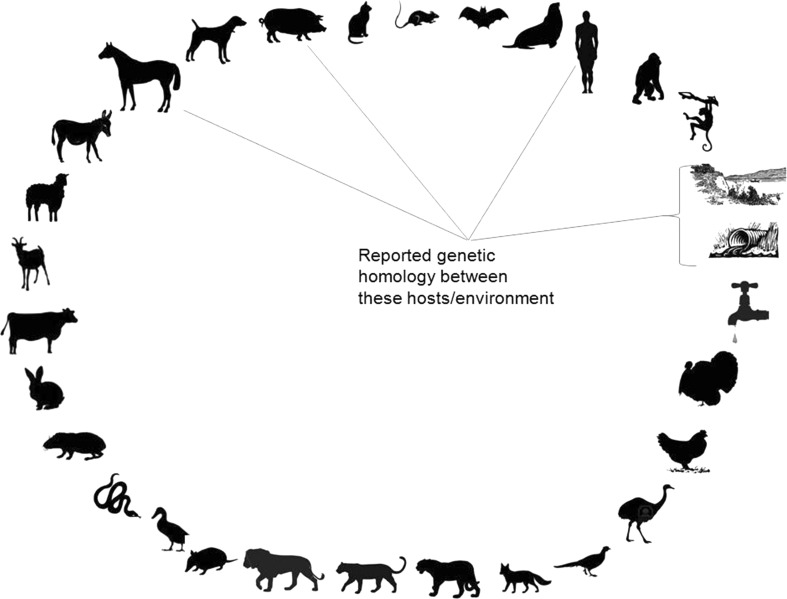
The schematic representation of PBV detection in various hosts and environmental samples is depicted in Fig. 1. The information published on the PBV research studies from across the globe, from various host species, and environmental samples were listed and recorded in Tables 1, 2, 3 and 4.
Fig. 1.
The schematic representation of PBV detection in various hosts and environmental samples
Table 1.
The epidemiological study reports of PBV detection in humans
| Country | Year of publication | Authors | PAGE | Sequence analyses |
|---|---|---|---|---|
| Humans | ||||
| Brazil | 1988a | Pereira et al. | ✓ | NA |
| UK | 1995a, b | Gallimore et al. | ✓ | NA |
| China | 2000 | Rosen et al. | ✓ | ✓ |
| Argentina | 2003 | Martinez et al. | ✓ | NA |
| Hungary | 2003 | Banyai et al. | ✓ | ✓ |
| Children | ||||
| Brazil | 1988a | Pereira et al. | ✓ | NA |
| Venezuela | 1993 | Ludert et al. | ✓ | NA |
| Italy | 1996 | Cascio et al. | ✓ | NA |
| Russia | 2003 | Novikova et al. | ✓ | NA |
| Thailand | 2005 | Wakuda et al. | ✓ | ✓ |
| Brazil | 2006 | Barreto et al. | ✓ | NA |
| India | 2006a,b; 2007 | Bhattacharya et al. | ✓ | ✓ |
| USA/Australia | 2008 | Finkbeiner et al. | ✓ | ✓ |
| Argentina | 2008 | Giordano et al. | ✓ | NA |
| South Asia/Australia | 2009 | Victoria et al. | NA | ✓ |
| India | 2010; 2011 | Ganesh et al. | ✓ | ✓ |
| Netherlands | 2010 | van Leeuwen et al. | NA | ✓ |
| Immunocompromised human hosts | ||||
| USA | 1993 | Grohmann et al. | ✓ | NA |
| USA | 1996 | Cunliffe and Glass | ✓ | NA |
| Venezuela | 1998 | Gonzalez et al. | ✓ | NA |
| Argentina | 1998; 1999 | Giordano et al. | ✓ | NA |
| USA | 2000 | Rosen et al. | ✓ | ✓ |
| Argentina | 2003 | Martinez et al. | ✓ | ✓ |
| Kidney transplant patients under immunosuppressive treatment | ||||
| Argentina | 2001 | Valle et al. | ✓ | NA |
| Argentina | 2010 | Giordano et al. | ✓ | ✓ |
| Asymptomatic human hosts | ||||
| USA | 2006 | Zhang et al. | ✓ | NA |
Table 2.
The epidemiological study reports of PBV detection in domestic animals
| Country | Year of publication | Authors | PAGE | Sequence analyses |
|---|---|---|---|---|
| Pig (Porcine) | ||||
| Brazil | 1989 | Gatti et al. | ✓ | NA |
| UK | 1990 | Chasey et al. | ✓ | NA |
| Venezuela | 1991 | Ludert et al. | ✓ | NA |
| Brazil | 1993 | Alfieri et al. | ✓ | NA |
| Thailand | 1996 | Pongsuwanna et al. | ✓ | NA |
| Hungary | 2008 | Banyai et al. | ✓ | ✓ |
| Venezuela | 2009 | Carruyo et al. | ✓ | ✓ |
| Argentina | 2010 | Martinez et al. | ✓ | ✓ |
| Argentina | 2010 | Giordano et al. | ✓ | ✓ |
| India | 2012 | Ganesh et al. | ✓ | ✓ |
| Calves (Bovine) | ||||
| Russia | 1989 | Vanopdenbosch and Wellemans | ✓ | NA |
| Belgium | 1991 | Villacorta et al. | ✓ | NA |
| UK | 1997 | Chandra | ✓ | NA |
| Brazil | 2003 | Buzinaro et al. | ✓ | NA |
| Russia | 2003 | Novikova et al. | ✓ | NA |
| India | 2009 | Ghosh et al. | ✓ | ✓ |
| India | 2011 | Malik et al. | ✓ | NA |
| India | 2013 | Malik et al. | ✓ | ✓ |
| Foals (Equine) | ||||
| Britain and Ireland | 1991 | Browning et al. | ✓ | NA |
| India | 2011 | Ganesh et al. | ✓ | ✓ |
| Lambs (Ovine) | ||||
| Spain | 1996 | Munoz et al. | ✓ | ✓ |
| Rabbits (Lapine) | ||||
| UK | 1993 | Gallimore et al. | ✓ | NA |
| Venezuela | 1995 | Ludert et al. | ✓ | NA |
| UK | 1999 | Green et al. | ✓ | ✓ |
| Guniea pigs (Caviinae) | ||||
| Brazil | 1989 | Periera et al. | ✓ | NA |
| Birds chicken (Avian) | ||||
| Brazil | 1989 | Alfieri et al. | ✓ | NA |
| Brazil | 1990 | Leite et al. | ✓ | NA |
| Brazil | 1991 | Monteiro et al. | ✓ | NA |
| Brazil | 2003 | Tamehiro et al. | ✓ | NA |
| USA (Turkeys) | 2010 | Day et al. | NA | ✓ |
| Brazil | 2012 | Bezerra et al. | ✓ | ✓ |
| Brazil | 2014 | Silva et al. | ✓ | ✓ |
| Dogs (Canine) | ||||
| Brazil | 2001 | Volotao et al. | ✓ | NA |
| Brazil | 2004 | Costa et al. | ✓ | NA |
| Brazil | 2009a | Fregolente et al. | ✓ | ✓ |
| Rats (Murine) | ||||
| Brazil | 1988b | Pereira et al. | ✓ | NA |
| Brazil | 2009a | Fregolente et al. | ✓ | ✓ |
Table 3.
The epidemiological study reports of PBV detection in wild animals
| Country | Year of publication | Authors | PAGE | Sequence analyses |
|---|---|---|---|---|
| Monkey (Simian) China |
2012 | Wang et al. | ✓ | NA |
| Giant anteater Brazil |
1999 | Haga et al. | ✓ | NA |
| Snake (Serpentine) Brazil |
2009a | Fregolente et al. | ✓ | ✓ |
| California Sea Lion [Otarine PBV] Hong Kong |
2012 | Woo et al. | ✓ | ✓ |
| (Giant Cats) Panthera leo Panthera onca Puma concolor Oncifelis geoffroyi Uruguay |
2013 | Gillman et al. | ✓ | NA |
| Red Fox Netherlands |
2013 | Bodewes et al. | ✓ | ✓ |
| Animal species kept in collective corrals | ||||
| Argentina | 2007 | Masachessi et al. | ✓ | NA |
| Donkey (Equus asinus) (Equine) | 2007 | Masachessi et al. | ✓ | NA |
| Choiques adult (Pterocnemia pennata) (Avian) | 2007 | Masachessi et al. | ✓ | NA |
| Chinese goose (Melegeris sp.) (Avian) | 2007 | Masachessi et al. | ✓ | NA |
| American ostrich (Rhea americana) (Avian) | 2007, 2012 | Masachessi et al. | ✓ | NA |
| Pelican (Pelecanus sp.) (Avian) | 2007 | Masachessi et al. | ✓ | NA |
| Animals kept in individual cages | ||||
| Orangutan (Pongo pygmaeus) | 2007 | Masachessi et al. | ✓ | NA |
| Armadillo (Chaetophractus vellerosus) | 2007 | Masachessi et al. | ✓ | NA |
| Gloomy pheasant (Phasianus sp.) | 2007 | Masachessi et al. | ✓ | NA |
Table 4.
The environmental study reports of PBV detection in raw sewage/river, surface and waste water
| Country | Year of publication | Authors | PAGE | Sequence analyses |
|---|---|---|---|---|
| Raw sewage and effluents | ||||
| USA | 2009 | Symonds et al. | ✓ | ✓ |
| River/surface and waste water | ||||
| Germany | 2010 | Hamza et al. | NA | NA |
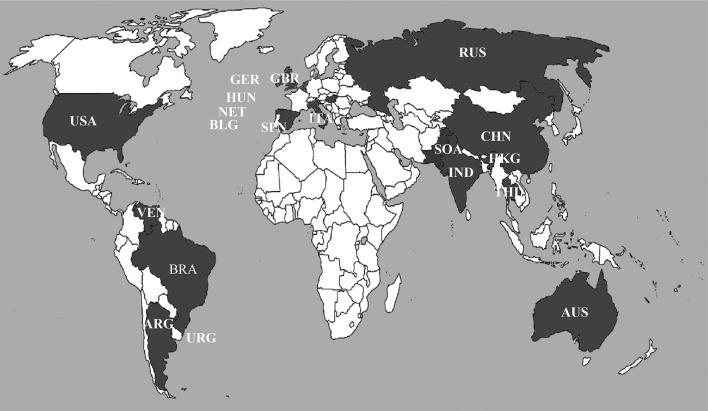
The countries that have reported PBVs is depicted in the World map based on scientific research publications to date (December’ 2013) since its discovery in 1988 (Fig. 2).
Fig. 2.
The countries that have reported PBV is depicted in the World map (darkly shaded) based on scientific research publications to date (January 2014) since its discovery in Brazil in the year 1988. USA United States of America, ARG Argentina, BLG Belgium, BRA Brazil, VEN Venezuela, ITA Italy, GER Germany, HUN Hungary, RUS Russia, CHN China, IND India, THL Thailand, AUS Australia, NET Netherlands, URG Uruguay, HKG Hong Kong, SPN Spain, GBR Great Britain and Ireland, SOA South Asia
This shows that the PBVs are genetically distinct, rapidly evolving and spreading worldwide. This warrants a global surveillance for PBVs among humans, domestic farm mammals, companion animals and other wildlife to corroborate, elaborate, and to fully understand the origin, adaptation, evolution, the natural and reservoir hosts of picobirnaviruses. In addition, further research is needed to establish the precise role of humans and domestic animals living close to one another in developing countries in the PBV transmission pattern among various host species.
The detection of PBVs in faeces of diarrhoeic and healthy free-living mammals, farm, companion animals and a wide range of wild birds and zoo animals has increased concern over the public health aspects regarding the transmission of these viruses. Similarly, the detection of genetically related PBVs found in humans and porcine hosts has raised the concern over these viruses about their potential zoonotic transmission. The genetic relatedness is also hypothesized as these viruses might have undergone cross over events during their evolution. This was substantiated by the results of the French research group’s report describing that the Picobirnavirus particles are capable of disrupting biological membranes in vitro, indicating that its simple 120-subunits capsid has evolved animal cell invasion properties [21]. Likewise, as per the published reports on porcine PBVs, in a few cases complete and almost-complete sequence identities were identified among clones derived from distinct animals, suggesting that PBV strains can be easily transmitted from one host to another [4].
Discovery of Picobirnavirus
Picobirnaviruses were first detected in the human fecal specimens collected during acute gastroenteritis outbreaks and from feces of black-footed pigmy rice rats (Oryzomysnigripes) in 1988 from Brazil [63, 64, 66]. Later, PBVs have been identified in faecal specimens of humans practically worldwide [3, 6, 7, 14, 26–29, 51, 55, 56, 61, 68, 76].
PBVs were also detected in faeces of a wide variety of farm mammals, birds, wild animals and birds kept in captivity, etc., viz., pigs [2, 4, 16, 31, 34, 50, 56, 67], calves [10, 15, 35, 53, 54, 61, 77, 79], rabbits and guinea pigs [25, 43, 52, 65], bats [86], red fox [8], avian such as chickens & poults [1, 49, 59, 69, 73], and other wild animals kept in captivity like Giant Anteaters [45]; giant cats like Lion, Puma, Jaguar and Geoffroy’s cat [37], sea lion [84], human primates such as Orangutan, wild birds such as American Ostrich, gloomy pheasant, Chinese goose [57], goat kids and lambs [60], donkeys [57, 58], foals [9, 30], laboratory non-human primates such as rhesus, pigtailed macaques and cynomolgus monkeys [83] and dogs [17, 23, 81], rats [23, 64], snakes [23] and turkeys [19] in fecal samples with or without diarrhea.
Recent environmental monitoring studies conducted in USA and Germany [46, 72] detected PBVs at high frequencies in the communal sewage, final effluent after wastewater treatment and natural surface water samples. Genogroup I PBVs were detected in 12/12 (100 %) of raw sewage and 4/12 (33 %) of final effluents of the wastewater treatment facilities at coastal Metropolitan areas of the United States of America viz. Alabama, California, Connecticut, Louisiana, Maine, Maryland, New Jersey, North Carolina, Oregon, and Washington. The partial RNA-dependent RNA polymerase (RdRp) gene (~200 bp) fragments were sequenced and sequence analyses revealed 83–100 % sequence identities to known human and porcine picobirnaviruses [72]. Another study in Germany was taken up to analyze the river waters, revealed that the concentrations of the PBVs ranged between 4.0 × 102 to 2.9 × 104 gen.equ./l [46]. The primary objective of these environmental studies was to evaluate PBV as an indicator of fecal contamination, the potential health risk of the presence of PBVs in sewages and natural water resources should not be neglected and this finding deserves further investigations.
Studies in immuncompromised and immunosuppressed hosts indicated that these viruses might be ‘opportunistic pathogens’ [38, 39, 41, 42, 44, 75, 80].
Thus, the probable role of PBV as either a primary diarrhoeal agent in immunocompetent children [6, 7], a potential pathogen in immunocompromised individuals [38, 39, 44] or an innocuous virus in the intestine remains elusive and needs to be investigated despite the numerous reports of the presence of PBV in fecal samples of various hosts species; our current knowledge of their biology, etiology, pathogenicity or their transmission characteristics remains subtle [15, 32, 33, 57], mainly because of their non-cultivable status (in vivo and in vitro) that is, the absence of a cell culture system and suitable animal model for propagating the virus. Further studies in gnotobiotic animals may shed light on PBV pathogenic potential [15].
Laboratory diagnosis mainly relies upon the detection of dsRNA bisegmented genome of PBVs by PAGE and silver staining [48]. Based on RT-PCR experiments, which specifically amplify region RdRp gene encoded by segment 2, PBV strains have been classified into two genogroups. The RT-PCR assay with two pairs of primers targeted to genomic segment 2 of 1-CHN-97 (prototype of genogroup I) and 4-GA-91 (prototype of genogroup II) PBV strains, isolated in China and USA, respectively, greatly improved the detection and molecular characterization of PBV worldwide [68].
Molecular epidemiological data presented in different reports [6, 28–30, 55, 68] showed a limited efficacy for the sets of primers available to detect PBVs circulating in USA, Argentina, and India. This suggests that human PBVs may be present with wide genetic diversity. On the other hand, although the PAGE technique has limited sensitivity, it allows unveiling the circulating PBV genotypes independent of PBV genomic sequences. This allows to study the incidence and prevalence of PBV infections and to compare the PBV detection rate by RT-PCR assays. As a consequence of the limited diagnostic methodology available, PBV is only detected in research laboratories with specific interest in this agent or during rotavirus surveillance by PAGE assay. Therefore, what is known about the epidemiology of PBV reflects more the ease of laboratory detection than the true epidemiology of infection with the agent itself [32, 33, 40].
In many laboratories both in developed and developing nations, for the purpose of routine surveillance, ELISA technique was the method of choice for detection of Group A rotaviruses from bulk or whole-stool specimen obtained from children with acute gastroenteritis as per the recommendations in the WHO generic protocol for hospital and community-based surveillance [85] as it has high sensitivity, and has a built-in control for non-specific reaction, and serves as a reliable diagnostic tool for virus detection for a large number of samples in a cost-effective way. However, if the PAGE assay is implemented for global rotavirus surveillance, the incidence of Picobirnavirus might increase drastically.
It has been suggested that segmented nature of Picobirnavirus genome renders them as potential candidates for segment reassortment, which could explain their highly heterogenous nature [3, 4]. In another study, it was stated that the segmented viruses have the tendency to reassort, and often such reassortments may lead to emergence of virulent progeny [62]. In general, mutation, recombination and genome segment reassortment and combination of these molecular events act as mechanisms of viral emergence. These mechanisms results in complex and phenotypically diverse populations of viruses. Similarly, the majority of emerging viral diseases of humans have a zoonotic origin [20].
A hypothesis is that PBV may also exhibit similar pattern of evolutionary events. Genogroup I PBVs those were detected and sequenced from pigs in Hungary [4], Venezuela and Argentina [11, 41], showed close relationship to human genogroup I PBVs. The PBV strains detected among children in Kolkata, India were reported to be closely related to porcine PBV strains [28, 29]. Similarly, the equine PBV strains detected and sequenced from feces of domestic foals in Kolkata, India [30] showed genetic relationship with earlier reported human PBV strains of the same region [28] as well as PBV strains of environmental samples (raw sewage, treated effluent and river water/surface water) reported from the USA [72].
The results of other studies also suggest that porcine PBV strains that are genetically diverse, are related to human strains, and cause frequent and persistent infections among young pigs with or without diarrhea or any sign of illness [11, 56]. So, stringent surveillance is required to monitor the genotype diversity and to determine the possible selective pressures under which PBV variants adapt to different hosts and environmental conditions.
A research team while working on the protection afforded by colostrum feeding to calves against rotavirus infection at the Compton Laboratory, UK, observed that 16 out of 108 faecal extracts from 5 calves were positive for PBVs [15]. Some of the faecal samples screened by them showed the presence of mixed infections, as two larger and two smaller, well separated segments in PAGE experiments.
It has been reported [4] that in swine, genetic diversity was also observed among PBV strains identified in mixed infections. Single point mutations and deleterious mutations within highly related strains suggested that PBVs exist as quasispecies in the swine alimentary tract. Clones with complete sequence identities originating from different animals suggested effective animal-to-animal transmission of the virus.
Similarly, in another study in pigs [11] PBV positive samples exhibiting single electrophoretic patterns in polyacrylamide gels, but containing multiple strains have been reported. The possible explanations they quoted were the degenerated primers were able to recognize several strains with the identical electrophoretic pattern present in the sample, and the presence of strains in the sample in viral loads below the detection limit of the PAGE technique.
Likewise, multiple strains of PBV infection were recently reported in a diarrhoeic child in Kolkata, India [29]. The PAGE assay showed well separated three pairs of bands with slight variation in migration pattern of PBV large genome profile. The RT-PCR experiments resulted in amplicons for both genogroup I and II PBVs. The direct sequencing of the genogroup II amplicon resulted in clear sequence, whereas genogroup I sequence was mixed and undistinguishable. To get clear sequence, the genogroup I amplicon was cloned and sequenced. This resulted in the presence of four different strains of genogroup I PBVs in the sample. The genogroup II PBV showed genetic resemblance to the genogroup II PBV prototype strain 4-GA-91 (USA). Our observation together with earlier findings on genetic relatedness between human and animal strains warrant further studies on zoonotic potential of PBVs and also the presence of multiple strains and both genogroup strains, suggests that the virus may have crossed host–species barrier and it was also observed that any specific genogroup need not be restricted to specific host.
Morphology
Virion
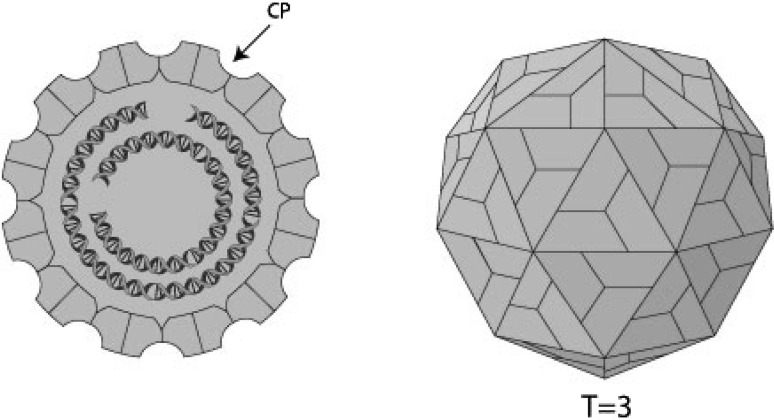
Picobirnaviruses are a group of small, spherical viruses. The virion is non-enveloped, with a diameter of 33–41 nm without any apparent surface morphology showing characteristic bisegmented double-stranded RNA (dsRNA) genome of two types viz. large profile and small profile, respectively based on the electrophoretic migration of bands in PAGE experiments (Fig. 3).
Fig. 3.
The schematic representation of PBV virion. Referred from the webpage: http://viralzone.expasy.org/all_by_species/602.html
In 2009, Duquerroy et al., the French research group using 3.4 Å X-ray diffraction crystallography first time reported the structure of Picobirnavirus in detail. PBV has a simple core capsid with a distinctive icosahedral arrangement. The coat protein has a new 3D-fold, having 60 two-fold symmetric dimers around it. Their data also show that the PBV particles are capable of disrupting biological membranes in vitro.
This study also showed that the structure of the PBV virion-like particles (VLPs) is not related to birnaviruses. They display a different capsid architecture made of 60 symmetric dimers, with the closest relatives appearing to be viruses infecting unicellular eukaryotes and plants, like the partitiviruses.
The autoproteolytic processing of the PBV capsid protein, together with the liposome-perforating capacity of the particles provide strong weightage to the interpretation that PBVs are indeed animal viruses, and not viruses of some eukaryotic parasite present in the intestinal flora of vertebrates. In contrast to viruses infecting plants or unicellular eukaryotes, animal viruses need a means of translocating across the cell membrane for entry.
The variability of the projecting domain is another feature presented by PBVs in common with other animal viruses. More information can be inferred from the comparison between the rabbit and human PBV sequences. These domains are the most exposed, and would be targeted by the adaptive immune system of vertebrates, as well as being responsible for interaction with an entry receptor that may vary from species to species. Serological studies of patients in which PBV has been detected are needed to understand the antigenicity of the virus [21].
The structural relation of PBVs with partitiviruses suggests that PBVs may have crossed the species barrier from putative unicellular eukaryotic organism to infect vertebrates [74]. Given the large number of microorganisms present in the intestinal flora of vertebrates, such species-crossing events are likely to be more common than anticipated earlier. Similarly, as long as the resulting viruses remain apathogenic/asymptomatic for their new host, they are essentially unnoticed [21].
With more and more outbreaks of new and emerging viral infections, with indirect signals indicating that PBVs may be involved in pathogenic symptoms, at least in immunocompromised patients, it becomes important to gather as much knowledge as possible on all the different viruses dwelling in the human environment, and to understand the adaptation mechanisms at play [21].
Taxonomy and phylogeny
The Picobirnavirus is the only genus [24], belong to the family Picobirnaviridae [http://www.ictvonline.org/virusTaxonomy.asp?version=2008] under the proposed order ‘Diplornavirales’. In 2008, the International Committee on Taxonomy of Viruses (ICTV) ratified ‘Human picobirnavirus’ as the ‘type species’ and ‘Rabbit picobirnavirus’ as a ‘designated species’ of Picobirnavirus [12, 13].
Molecular biology
Genome
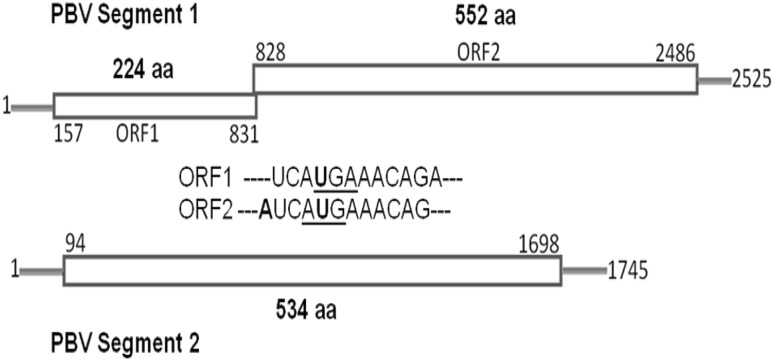
In 2005, the first complete nucleotide sequence of the two genome segments of Picobirnavirus was determined for a human strain (Hy005102) isolated from an 6 year-old male child with acute non-bacterial gastroenteritis in Thailand during the year 2002 [82]. The genomic segment 1 was found to encode two open reading frames (ORF1 and ORF2); ORF1 encodes a hydrophilic protein with 224 amino acids of unknown function. ORF2 encodes the capsid protein with 552 amino acids. The smaller segment 2 has a single ORF of 534 amino acids and encodes the viral RNA-dependent RNA polymerase (RdRp) (Fig. 4).
Fig. 4.
Segmented dsRNA linear genome: 2 segments encode for a total of 3–4 proteins. Segments range in size from 1.7 to 2.5 kb. Total genome size is about 4.2 kb. Genomic organization of segments 1 and 2 of human picobirnavirus. The termination codon (UGA) for ORF1 and the initiation codon for (AUG) ORF2 in segment 1 are overlapped.
Recently, additional sequence information of full or nearly full genome segments has become available in the public DNA databases [http://www.ncbi.nlm.nih.gov/sites/entrez?db=nuccore&cmd=search&term=picobirnavirus] for other human strains [68, 76] as well as a lapine [43], a bovine PBV strain [35] and recently a California sea lion PBV (Otarine PBV) [84]. The increasing genomic information of PBVs along with the available data will be useful to know more about PBVs at the molecular level.
Genetic relatedness among heterologous PBV strains
Based on the existing published reports, it is inferred that interspecies transmission of PBVs might have occurred. The PBVs detected from porcine hosts from Europe [4] and Latin America [11, 41] showed to be closely related to human PBVs. Similarly, the PBVs detected from diarrhoeic children in an urban slum in Kolkata, India [28] showed to be genetically closely related to porcine PBV strains reported from Hungary, Venezuela and Argentina. These reports raised the issue of zoonotic potential of PBV infections. For more than a decade, the primer sets designed by Rosen et al. [68] was used for molecular characterization of PBV strains collected from humans but presently remains a milestone to be reached successfully for characterizing PBV strains worldwide as these primer pairs have some limitations as reported from India [6, 7], Argentina, Venezuela [11], and Hungary [4].
Recently, the Venezuelan research group has designed and reported that the primer sets worked well in detecting the porcine PBV strains detected from porcine faecal samples [11]. The human PBV primer sets were used for characterization of porcine strains that were genetically related to human PBV strains. This report proved the interspecies transmission of PBV strains between humans and porcine hosts [4]. This report also confirms that the PBV strains undergo zoonotic transmission. The Hungarian research group also suggested that PBV strains might have undergone crossing over events during the process of evolution between human and porcine hosts.
PBV diversity and tropism
Sequence-independent amplification and specific reverse transcription PCRs identified genogroup I and II picobirnaviruses in respiratory tracts of pigs as well as humans [70, 71]. These data expand knowledge of Picobirnavirus diversity and tropism. Genetic relationships between porcine respiratory and human enteric picobirnaviruses suggest cross-species transmission of picobirnaviruses between pigs and humans.
The pathogenic potential
In farm animals, the first report of the association of Picobirnavirus with disease came from a study involving pigs in an organized farm in Brazil [34]. These authors reported a higher proportion of PBV detection in animals with diarrhea than in those without diarrhea (15.3 vs 9.6 %). This suggested that it would be a new causative agent of diarrhea in these animals. In contrast, Ludert et al. [50] detected the virus in equal frequency among pigs with and without diarrhea in farms in Venezuela (10 vs 12.3 %). In agreement, Carruyo et al. [11] in Venezuela and Martinez et al., (2010) in Argentina detected PBV in similar frequency among diarrheic and non-diarrheic pigs.
Similar results were reported in recent studies conducted by the same research group in Argentina in two different sites were 29.6 % [41] and 21.13 % [56]. In studies carried out in poultry, PBV was detected in 17 (14.16 %) fecal samples of 25–30 days old chickens at Parana and Minas Gerais state, Brazil [1]. Similarly, intestines of 257 chickens slaughtered for marketing at the age of 38–43 days when subjected for PAGE assay resulted in detection of 44 (17.1 %) PBVs in the State of Rio de Janeiro, Brazil [49]. In another study, intestines from a total of 227 chickens slaughtered in Rio de Janeiro, Brazil by PAGE assay showed 30 (13.21 %) of PBVs [59]. A total of 378 fecal samples from 1 to 7 weeks old broiler chickens analyzed by PAGE assay resulted in detection of 13 (3.4 %) PBVs at Parana state, Brazil [73] and statistically significant correlation was seen between PBV shedding and altered consistency of faeces (diarrheic and pasty) with clinical signs of the enteritis [73].
The correlation of PBV diagnosis in feces with altered consistency (diarrheic and pasty) collected from these animals with clinical signs of the enteritis was significant (P < 0.05) when compared with normal (control) feces [73]. Costa et al. [17] also detected the virus in low-frequency in samples from dogs with gastroenteritis (3/163 vs 1.84 %). On the other hand, studies carried out in chickens [1, 49], foals [9], dogs [23] and calves [77] did not find any association between PBV detection and disease.
The first report of PBV in feces of animals in captivity was conducted by Haga et al. [45] in three giant anteaters. The animals had no clinical signs of diarrhea or other evidence of enteric disease. Likewise, in a primate colony from China, PBV was detected in the course of research on epidemiology, prevalence and causal role of viral agents in diarrheal disease among monkeys. The prevalence of PBV was 2 %. These authors did not reach any conclusion about the virus and diarrhea [83]. On the other hand, Fregolente et al. [23] reported the detection of PBV in snakes (8.5 %) and rats (25 %) kept in captivity. The stool samples did not show any signs of diarrhea. In 2007, Masachessi et al., described a systematic sampling of fecal specimens from 150 different host species in captivity from a Zoo of Córdoba, Argentina. In this study, picobirnaviruses were identified in 19 (3.70 %) of the 513 fecal samples examined by PAGE assay among birds (Gloomy pheasant, Pelican and Chinese goose) and mammals (armadillo, donkey and Orangutan). None of these animals showed signs of diarrhea, and the authors suggested a lack of etiological relation of PBV with the disease.
Despite the numerous reports of the presence of PBV in fecal samples from a large list of animal species, the results are controversial and do not allow us to conclude on the etiological relationship of the PBV and the diarrhoeal syndrome. In research studies conducted in humans, the results are also contradictory. These authors suggest that PBV might not have etiological relation with diarrhea, but many factors including induced or physiologic immunosuppression might establish the necessary conditions for viral excretion instead.
In animals tested in captivity, Masachessi et al. [57] did not find clinical animal data which could suggest state of immunosupression, but these authors concluded that another factor such as stress due to captivity and/or isolation might favor viral replicative cycles. In agreement, in farm animals, Martinez et al., (2010) reported PBV excretion at dissimilar frequencies in fecal samples of pigs of different ages and also with different physiological characteristics, viz. highest frequency of virus excretion in pigs was observed during the lactation period and during the final stage of pregnancy. These observations suggest that a particular physiological status, such as farrowing and lactation might establish the necessary conditions for PBV excretion. The authors conclude that stress conditions generated by pig farming practices during these stages may be the basis for this observation. Further research studies are needed to establish and to gain deeper understanding of the significance of PBV detection in infected hosts.
The infection pattern
Three research studies carried out in animals attempted to describe the natural history of PBV infection, resulted in similar observations. Haga et al. [45] described weekly PBV detection for at least 4 months in a facility with three giant anteaters (Myrmecophaga tridactyla) in captivity in a Brazilian zoo. Although the authors could not obtain individual sampling, they related the prolonged period of virus shedding to chronic PBV infection and provided evidence that adult animals infected with PBVs could be asymptomatic carriers persistently infected and could serve as reservoirs of infection. In addition, Masachessi et al. [57] analyzed individual sampling from animals kept in single corrals (Orangutan and armadillo) in a Zoo at Argentina, where the animals excreted PBV during prolonged periods, for at least 6 months (armadillo) and 7 months (Orangutan).
PBV negative results were revealed in samples obtained from both animal species between periods of PBV positive detection. This could be due to drastic change in quantity of viral load, in the fecal samples and PAGE sensitivity. Based on the result, the authors concluded that this standard resembles a model of persistent infection. In contrast with those reported by Haga et al. [45 where the three giant anteaters were in the same farmyard, the animals in the study of Masachessi et al. [57], had no contact with each other and they were individually supplied with water and food. Due to this reason, it is not possible to hypothesize about PBV re-infections in these animals.
It is important to note that the above mentioned studies were carried out through the PAGE technique. Although this technique is very useful, it has its limitations because it is cumbersome and relatively insensitive.
In the year 2000, Rosen et al., developed a reverse transcription-polymerase chain reaction (RT-PCR) assay for the detection of human PBV and later, Carruyo et al. [11] designed new sets of primer pairs for molecular characterization of porcine PBV strains. Through the amplification of a conserved region of genome segment 2 of PBV, containing motifs 1 and 2 of the viral RNA-dependent RNA polymerase, these techniques allowed, the partial molecular and phylogenetic characterization of PBV strains circulating in humans and pigs.
Based on the PAGE screening and the RT-PCR assay developed for pigs by Carruyo et al. [11], and Martinez et al., (2010) obtained preliminary results about the ecological pattern of PBV circulation in a pig population in a breeding farm in Argentina. A follow-up study was carried out for a 29-month period in a female pig. The study period spanned from weaning (26 days after birth) up to its fourth reproductive cycle for a period of 2 years and 5 months (898 days old). This study allowed them to determine the viral excretion pattern in pigs. Negative samples seen by PAGE and silver staining (PAGE S/S) were positive by RT-PCR during the early first week after weaning, and 2 months later, the virus could also be detected by PAGE S/S. Thereafter, PBV excretion was detected frequently by RT-PCR and sporadically by PAGE S/S. This period was followed by almost 6 months without virus detection by PAGE S/S and only occasional detection by RT-PCR. The next period of active virus excretion, when the virus could be detected not only by RT-PCR but also both by PAGE; silver staining and RT-PCR techniques, was determined by the first gestation and farrowing cycle. During this period, a continuous PBV excretion pattern was identified.
A similar continuous PBV excretion pattern was again identified during the third and fourth reproductive cycles, but positive samples could only be detected by RT-PCR, suggesting shedding of lower viral loads than in the first reproductive cycle. Eleven of 33 RT-PCR products obtained in this study were sequenced. The amplicons showed 100 % homology in their nucleotide sequence (~280 bp). In summary, PBV infection was characterized by periods of high and low active viral excretion (detected by PAGE and RT-PCR, respectively) interspaced by silent periods. The sequence analysis suggested that the pigs possibly excreted Picobirnavirus of the same strain during its lifetime.
On the other hand, it is important to note that Martinez et al., (2010) found PBV excretion in highest frequency in pigs during the lactogenic period. This observation was similar in previous studies carried out in pigs [50], where the authors found a higher prevalence in samples from population of piglets 15–35 days old. In rabbits, Gallimore et al. [25] detected PBV more commonly in newly weaned rabbits than in young adults. Tamehiro et al. [73] detected excretion of PBV in broiler chickens 2–7 weeks old and Masachessi et al. [57] detected PBV in fecal specimens obtained from asymptomatic rheas (American ostrich: Rhea americana) approximately 3 weeks old. According to the data obtained from all these studies mentioned above, the primary PBV infection could take place between the first few weeks of life in animals and birds.
In summary, the results herein suggest that PBV infection can be acquired early in life and then establishes a persistent infection, with periods of high viral activity intermingled with periods of silence. That is, adult infected animals could be asymptomatic carriers of PBV, or may be persistently infected and serve as reservoirs of viruses and not infection.
PBV infection in animals, birds and reptiles
Recently, PBV were detected in faeces of cow calves and buffalo calves with or without signs of diarrhea from subtropical and temperate Himalayan foothills of India [53]. PAGE screening resulted in 3.67 % PBV positivity. These findings indicate the sporadic incidences of PBV in bovine calves and also emphasize the need for the development of better diagnostics to monitor the emerging etiologies of farm animals of economic significance.
Along with identification of the virus in stool, research studies were carried out to determine the significance of the virus to understand the possible association between PBV and pathology, which resulted in contradictory observations. Further investigation on molecular characterization of emerging PBV strains from various animals and birds across the globe will yield very valuable data regarding the origin, transmission, spread and genetic diversity of these rapidly evolving novel dsRNA viruses with emerging zoonotic or anthroponotic potential.
Tamehiro et al. [73] detected excretion of PBV in broiler chickens 2–7 weeks old and Masachessi et al. [57] detected PBV in fecal specimens obtained from asymptomatic rheas viz. American ostrich (Rhea americana) of approximately 3 weeks old. According to the data obtained from all these studies mentioned above, the primary PBV infection could take place between the first weeks of life in birds. Recently, through metagenomic approach, many viruses that were previously characterized as well as new, genetically divergent and novel viruses have been identified including picobirnaviruses [19]. This reveals that the putative turkey-origin for PBVs is unique among the available PBV sequences so far from different hosts; PBVs are genetically diverse and rapidly evolving worldwide in different hosts. PBVs cross the host species barrier, multiple PBV strains have also been reported both in humans [76] and farm mammals [4, 11].
Fregolente et al. [23] reported detection of PBV in snakes (8.5 %) and rats (25 %) kept in captivity. The stool samples did not show any signs of diarrhea. The presence of PBVs in the serpentine hosts may be due to the ingested food having the virus or innocuous agent of the gut flora remains elusive. More detailed study is required to further understand the evolution and transmission pattern of the virus in reptilian hosts from different geographical locations that will shed light on the nature and spread of the virus.
Diagnostic issues
The detection of bisegmented dsRNA genome of PBV by PAGE and silver staining [48] is a standard method for reliable laboratory diagnoses. Broadly reactive primer pairs utilized in RT-PCR assay have served as an alternative to PAGE, in molecular detection and characterization of PBVs [68]. The two sets of primer pairs, that specifically amplify small fragments within the RdRp gene have been developed; these primer pairs are also capable of differentiating two major PBV genogroups, genogroup I represented by the prototype strain 1-CHN-97 and genogroup II, represented the prototype strain 4-GA-91.
Since attempts to culture PBV in vitro have not been made to date and no animal model of infection and disease exists, laboratory diagnosis relies upon the detection of dsRNA bisegmented genome by PAGE and silver staining. The RT-PCR detection assay with two pairs of primers targeted to genomic segment 2 of 1-CHN-97 (GGI) and 4-GA-91(GGII) PBV strains, isolated in China and USA, respectively, greatly improved the detection and molecular characterization of PBV worldwide [68]. These authors reported that PBV related to the Chinese strain was the predominant virus detected in stool samples and the primers from the 1-CHN-97 strain had the broadest reactivity. However, these primer sets could not successfully amplify many of the PAGE positive PBV samples detected from humans as well as domestic farm animals [3–7, 28, 40].
Therefore, the information known about the incidence and infections of PBVs reflects more the ease of laboratory detection than the true epidemiology of infections with the agent itself [4]. Despite the numerous reports of the presence of PBV in fecal samples from vertebrates, the pathogenicity of these viruses remains elusive and requires detailed/elaborate studies.
Future research direction
Picobirnaviruses are generally considered to be viruses of vertebrates, but their pathogenic host has not yet been confirmed. In 1999, it was reported that, the sequences of three ORFs of genomic segment 1 of a rabbit PBV were unrelated to the sequence of atypical Picobirnavirus (APBV), indicating that there are two distinct groups of bisegmented dsRNAs found in faecal samples [43]. In the year 2000, it was reported that few similarities were noted during comparison of RdRp-specific motifs of the genomic segment 2 of human picobirnaviruses with a unique group of closely related non-infectious dsRNA viruses of lower eukaryotes [68] (Fig. 5).
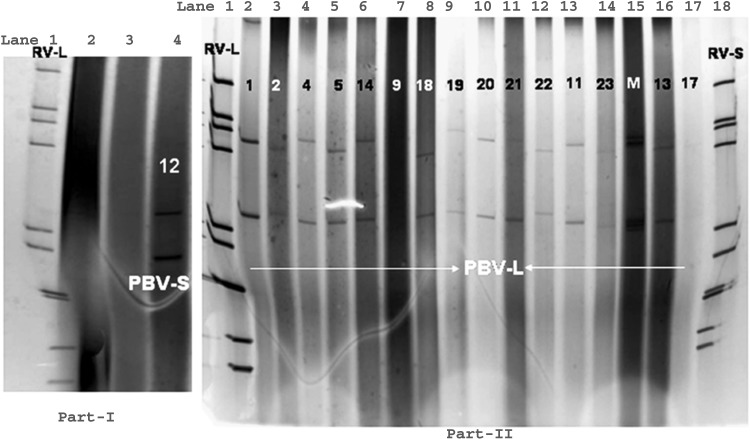
Fig. 5.
One small genome profile PBV (Lane 4) alongside a long electropherotype pattern of eleven segmented, genomic dsRNA profile of human group A rotaviruses (Lane 1) in Part-I PAGE gel. Polyacrylamide gel electrophoresis of Human picobirnavirus (large genome profile) (Lanes 2–6, 8–16) alongside a long electropherotype pattern of eleven segmented, genomic dsRNA profile of human group A rotavirus (Lane 1) and short electropherotype pattern of group A rotavirus (Lane 18) in part-II PAGE gel. Samples negative for rotavirus or PBV in (Lanes 2, 3) in part-I and (Lanes 7, 17) in Part-II PAGE gel
PBVs have been detected in the stools of guinea pigs [65] and suckling piglets [50] suggesting that passive ingestion with food and excretion of the virus does not occur and virus replication occurs in animal hosts. Astroviruses, picobirnaviruses and adenoviruses were each significantly correlated with the presence of diarrhea, and small round structured viruses (SRSVs) were more likely to be found in patients with diarrhea than in controls. Finally, picobirnaviruses were also identified in several patients with chronic diarrhea; hence prolonged shedding of virus could be documented [44].
In the year 2000, the development of the RT-PCR assay [68] was a new milestone that showed 100-fold increased sensitivity in the detection of Picobirnavirus in clinical samples compared to detection of PBV dsRNA in PAGE gels. PBVs related to the genogroup I (1-CHN-97 strain) were the predominant viruses detected in RT-PCR assays of stool samples collected from people in four countries on three continents [68]. Primers derived from the 4-GA-91 strain produced RT-PCR products only from stool samples collected from a single patient who shed the virus from which the clone was derived. The strain-specific reactivity of the primers, the nucleotide and the amino acid sequence differences observed between the genomic segment 2 of the 4-GA-91 and 1-CHN-97 PBV strains respectively, suggested that these PBV strains may be the representative of two distinct genogroups [68].
As reported from Hungary [4], Venezuela and Argentina [11] the clones they had isolated showed complete sequence identities originating from different animals that suggested effective, easy animal to animal transmission of the virus. It has been suggested that segmented nature of human PBV genome renders it as a potential candidate for segment reassortment, which could explain the highly heterogenous nature [3].
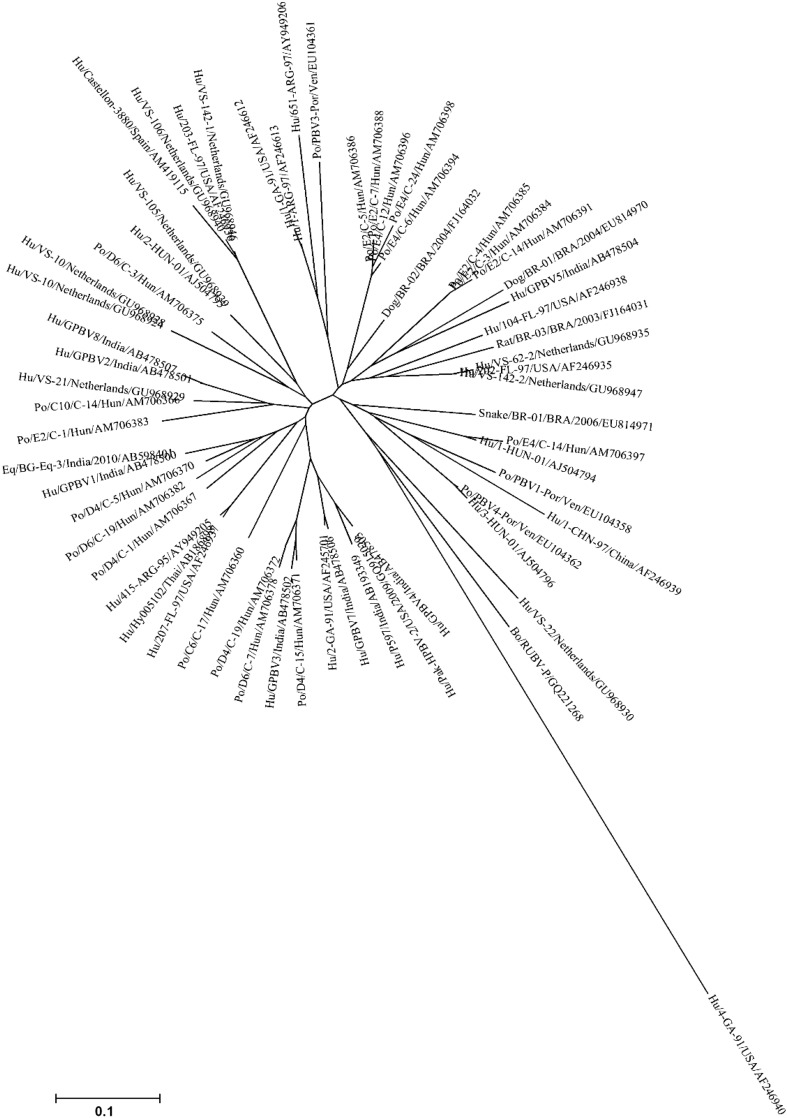
These results strongly suggest that PBV strains may circulate between humans and pigs. The other animal hosts, which are also sharing close genetic relationship with human PBVs requires further investigations (Fig. 6).
Fig. 6.
The phylogenetic tree showing the overall genetic diversity and evolutionary differences of genogroup I PBV strains detected from different hosts and from different geographical locales. The genogroup I PBV picobirnavirus strains reported from human, porcine, bovine, canine, murine and serpentine hosts, based on partial amino acid sequence [56 amino acids (aa)] of genomic segment 2 were taken from the public DNA Databank NCBI/DDBJ/EMBL. The tree was rooted with cognate stretch of gene segment 2 of genogroup II prototype strain Hu/4-GA-91 (USA) defined as the outgroup strain. The phylogenetic tree was inferred using the MEGA software (Version 4.1) program in combination with the neighbor-joining method, with evolutionary distances computed using the p-distance method. A bootstrap value >90% was indicated for the corresponding nodes based on a resampling analysis of 1000 replicates. The scale bar represents 0.1 substitution per 100 nucleotide residues
The zoonotic potential of PBV
The zoonotic potential of PBV is an interesting question that deserves further attention. These findings have initiated our quest to investigate the zoonotic potential of PBVs and to gain a better understanding of emerging, novel, enteric picobirnaviruses.
A recent report of molecular characterization of calf PBV detected in Kolkata and a bubaline PBV detected from Maharashtra, Central India have shown that the bovine genogroup I strains are distinct from PBVs of human and other hosts [35, 54].
By comparing the research findings from this study with other reported studies [4, 11, 28, 31] the question arises whether certain human and porcine PBVs shared crossing points in their genetic evolution. Repeated exposures of humans to heterologous, but genetically related and rapidly evolving viruses, shed in large amounts from domestic animals, might be an occupational health risk that needs attention and thorough investigations in future. In conclusion, the detection of human isolates of PBVs that are closely related to Genogroup I porcine Picobirnavirus strains reported earlier from Hungary, Venezuela and Argentina indicates that these viruses have zoonotic potential which may assist their rapid spread to new geographical locales across the globe.
Further studies in gnotobiotic animals may shed light on PBV pathogenic potential [15]. Similarly, the full genomic analyses of the PBVs detected from different host species from different geographical locations may explain the complex interspecies transmission dynamics and circulation pattern of PBVs.
A research study on animal rotaviruses (dsRNA virus), one of the major etiology of gastroenteritis in young ones of various host species including humans has shown that the virus can cross the host–species barrier. As an example, it is evident that one of the equine rotavirus strain H-1, showed antigenic and genetic relatedness in many of its genome segments with SG I porcine rotavirus strain [36] isolated from different geographical locations around the globe viz., the United States, Venezuela, Argentina and Australia. This proves that the dsRNA virus has the capability to move and spread rapidly between animal to animal as well as interspecies transmission from pigs to horses [31, 32].
The method of applying metagenomic approach of studying the human and avian faeces has really shed light on the existence of vast majority of microbial diversity in the gut system. It also revealed that the viruses replicating in mucosal cells were undetectable in stools from healthy hosts. But in inflammatory conditions, when diarrhoeal symptoms arise, these viruses are shed abundantly in faeces [20, 87]. It was also found that the RNA viral community in human faeces was dynamic in nature. Similarly, there was substantial variation in the representation of RNA viruses found in a single individual over a time period or even in different individuals.
The detection of PBVs in faeces of diarrhoeic and healthy free-living mammals, farm animals, companion animals and a wide range of wild birds and zoo animals increased concern over the public health aspects of transmission and circulation of these viruses posing a zoonotic and/or anthroponotic threat, since the natural host as well as the reservoir of this virus remains elusive. Similarly, the detection of genetically closely related PBVs in humans and pigs has shown that these viruses are one of the potential emerging zoonotic agents. Such valuable information about the PBV (dsRNA bisegmented virus) being reported from humans, animals and various environmental samples worldwide, largely focus on the emerging interdisciplinary field of conservation medicine as an emerging zoonosis [47]. It is also important to emphasize that regularly monitoring and increased vigilance during surveillance is essential to detect the emergence of new zoonotic pathogens in our settings.
Although, PBV were first detected in feces of children and animals with signs of diarrhea, later numerous studies have failed to establish a clear association between PBV excretion and disease. Thus, the concept that PBVs were diarrheal etiology is benevolent approach to the perception that PBV establishes persistent infection in the host without any sign of disease. However, there are numerous factors that favor viral activity in establishing persistent infections in hosts which includes immune status, stress syndrome, clinical illness, age and environmental conditions. Thus the role of PBV as an opportunistic pathogen associated with diarrhea cannot be ruled out. In addition, recent report on detection of PBV in respiratory tract of pigs could extend previous directions of PBV research [70, 71].
Most of the evidence of genetic diversity of PBVs was based molecular characterization of conserved region of the partial RdRp gene fragments. Additional full-length genome sequences are required for comprehensive sequence and phylogenetic analyses to understand more about the driving forces of the marked genetic diversity and the zoonotic potential of PBVs. The observation that PBV establishes persistent infections, with periods of silence intermingled with periods of viral excretion, in healthy animals without causing any signs of disease, suggests that PBV is a virus well-adapted to vertebrates, with a peculiar host–virus interactions. Thus, the establishment of animal models or adaptation of the virus to grow in tissue culture will be essential to better describe the pathobiology, the host–virus relationships and the unique infection pattern of PBVs. An equally important part of research is still awaited on the characterization of the immune response elicited by PBV infections in hosts.
Acknowledgments
We sincerely acknowledge and thank all the members of different research groups across the globe involved in various aspects of research on picobirnaviruses, for sharing their observations in their publications that helped us to concisely compile this review manuscript. We sincerely acknowledge all our colleagues for their constant encouragement and whole-hearted support. The study was financially supported by the Indian Council of Medical Research (ICMR, Govt. of India).
References
- 1.Alfieri AF, Resende JS, Alfieri A, Resende M. A new bi-segmented double-stranded RNA virus in avian faeces. Arq Bras Med Vet Zootec. 1989;40:437–440. [Google Scholar]
- 2.Alfieri AA, Alfieri AF, Freitas JC, Silva CA, Freire RL, Barros AR, Barreiros MAB, Muller EE. Occurrence of Escherichiacoli, rotavirus, picobirnavirus and Cryptosporidiumparvum in a post weaning diarrhoea focus in swine. Semina Ci Agr. 1994;15:5–7. [Google Scholar]
- 3.Bányai K, Jakab F, Reuter G, Bene J, Uj M, Melegh B, Szücs G. Sequence heterogeneity among human picobirnaviruses detected in a gastroenteritis outbreak. Arch Virol. 2003;148:2281–2291. doi: 10.1007/s00705-003-0200-z. [DOI] [PubMed] [Google Scholar]
- 4.Bányai K, Martella V, Bogdán A, Forgách P, Jakab F, Meleg E, Bíró H, Melegh B, Szucs G. Genogroup I picobirnaviruses in pigs: evidence for genetic diversity and relatedness to human strains. J Gen Virol. 2008;89:534–539. doi: 10.1099/vir.0.83134-0. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharya R, Sahoo GC, Nayak MK, Ghosh S, Dutta P, Bhattacharya MK, Mitra U, Gangopadhyay D, Dutta S, Niyogi SK, Saha DR, Naik TN, Bhattacharya SK, Krishnan T. Molecular epidemiology of human astrovirus infections in Kolkata, India. Infect Genet Evol. 2006;6:425–435. doi: 10.1016/j.meegid.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Bhattacharya R, Sahoo GC, Nayak MK, Saha DR, Sur D, Naik TN, Bhattacharya SK, Krishnan T. Molecular epidemiology of human picobirnaviruses among children of a slum community in Kolkata, India. Infect Genet Evol. 2006;6:453–458. doi: 10.1016/j.meegid.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharya R, Sahoo GC, Nayak MK, Rajendran K, Dutta P, Mitra U, Bhattacharya MK, Naik TN, Bhattacharya SK, Krishnan T. Detection of genogroup I and II human picobirnaviruses showing small genomic RNA profile causing acute watery diarrhoea among children in Kolkata, India. Infect Genet Evol. 2007;7:229–238. doi: 10.1016/j.meegid.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 8.Bodewes R, van der Giessen J, Haagmans BL, Osterhaus AD, Smits SL. Identification of multiple novel viruses, including a parvovirus and a hepevirus, in feces of red foxes. J Virol. 2013;87:7758–7764. doi: 10.1128/JVI.00568-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Browning GF, Chalmers RM, Snodgrass DR, Batt RM, Hart CA, Ormarod SE, Leadon D, Stoneham SJ, Rossdale PD. The prevalence of enteric pathogens in diarrhoeic thoroughbred foals in Britain and Ireland. Equine Vet J. 1991;23:405–409. doi: 10.1111/j.2042-3306.1991.tb03751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buzinaro MG, Freitas PP, Kisiellius JJ, Ueda M, Jerez JA. Identification of a bisegmented double-stranded RNA virus (picobirnavirus) in calf faeces. Vet J. 2003;166:185–187. doi: 10.1016/S1090-0233(03)00031-5. [DOI] [PubMed] [Google Scholar]
- 11.Carruyo GM, Mateu G, Martínez LC, Pujol FH, Nates SV, Liprandi F, Ludert JE. Molecular characterization of porcine picobirnaviruses and development of a specific reverse transcription-PCR assay. J Clin Microbiol. 2008;46:2402–2405. doi: 10.1128/JCM.00655-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carstens EB. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2009) Arch Virol. 2010;155:133–146. doi: 10.1007/s00705-009-0547-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carstens EB, Ball LA. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2008) Arch Virol. 2009;154:1181–1188. doi: 10.1007/s00705-009-0400-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cascio A, Bosco M, Vizzi E, Giammanco A, Ferraro D, Arista S. Identification of Picobirnavirus from faeces of Italian children suffering from acute diarrhea. Eur J Epidemiol. 1996;12:545–547. doi: 10.1007/BF00144011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandra R. Picobirnavirus, a novel group of undescribed viruses of mammals and birds: a minireview. Acta Virol. 1997;41:59–62. [PubMed] [Google Scholar]
- 16.Chasey D. Porcine Picobirnavirus in UK? Vet Rec. 1990;126:465. [PubMed] [Google Scholar]
- 17.Costa AP, Cubel Garcia RCN, Labarthe NV, Leite JPG. Detection of double-stranded RNA viruses in fecal samples of dogs with gastroenteritis in Rio de Janeiro, Brazil. Arq Bras Med Vet Zootec. 2004;56:554–557. doi: 10.1590/S0102-09352004000400020. [DOI] [Google Scholar]
- 18.Cunliffe NA, Glass RI. Gastrointestinal manifestations of HIV infection. Lancet. 1996;348:1037. doi: 10.1016/S0140-6736(05)64970-7. [DOI] [PubMed] [Google Scholar]
- 19.Day JM, Ballard LL, Duke MV, Scheffler BE, Zsak L. Metagenomic analysis of the turkey gut RNA virus community. Virol J. 2010;7:313. doi: 10.1186/1743-422X-7-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Domingo E. Mechanisms of viral emergence. Vet Res. 2010;41:38. doi: 10.1051/vetres/2010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duquerroy S, Da Costa B, Henry C, Vigouroux A, Libersou S, Lepault J, Navaza J, Delmas B, Rey FA. The Picobirnavirus crystal structure provides functional insights into virion assembly and cell entry. EMBO J. 2009;28:1655–1665. doi: 10.1038/emboj.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, Wang D. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog. 2008;4:e1000011. doi: 10.1371/journal.ppat.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fregolente MC, de Castro-Dias E, Martins SS, Spilki FR, Allegretti SM, Gatti MS. Molecular characterization of picobirnaviruses from new hosts. Virus Res. 2009;143:134–136. doi: 10.1016/j.virusres.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 24.Fregolente MC, Gatti MS. Nomenclature proposal for Picobirnavirus. Arch Virol. 2009;154:1953–1954. doi: 10.1007/s00705-009-0537-z. [DOI] [PubMed] [Google Scholar]
- 25.Gallimore C, Lewis D, Brown D. Detection and characterization of a novel bisegmented double-stranded RNA virus (Picobirnavirus) from rabbit faeces. Arch Virol. 1993;133:63–73. doi: 10.1007/BF01309744. [DOI] [PubMed] [Google Scholar]
- 26.Gallimore CI, Green J, Casemore DP, Brown DW. Detection of a Picobirnavirus associated with Cryptosporidium positive stools from humans. Arch Virol. 1995;140:1275–1278. doi: 10.1007/BF01322752. [DOI] [PubMed] [Google Scholar]
- 27.Gallimore CI, Appleton H, Lewis D, Green J, Brown DW. Detection and characterisation of bisegmented double-stranded RNA viruses (picobirnaviruses) in human faecal specimens. J Med Virol. 1995;45:135–140. doi: 10.1002/jmv.1890450204. [DOI] [PubMed] [Google Scholar]
- 28.Ganesh B, Nataraju SM, Rajendran K, Ramamurthy T, Kanungo S, Manna B, Nagashima S, Sur D, Kobayashi N, Krishnan T. Detection of closely related picobirnaviruses among diarrhoeic children in Kolkata: evidence of zoonoses? Infect Genet Evol. 2010;10:511–516. doi: 10.1016/j.meegid.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 29.Ganesh B, Nagashima S, Ghosh S, Nataraju SM, Rajendran K, Manna B, Ramamurthy T, Niyogi SK, Kanungo S, Sur D, Kobayashi N, Krishnan T. Detection and molecular characterization of multiple strains of Picobirnavirus causing mixed infection in a diarrhoeic child: emergence of prototype genogroup II-like strain in Kolkata, India. Int J Mol Epidemiol Genet. 2011;2:61–72. [PMC free article] [PubMed] [Google Scholar]
- 30.Ganesh B, Banyai K, Masachessi G, Mladenova Z, Nagashima S, Ghosh S, Nataraju SM, Pativada M, Kumar R, Kobayashi N. Genogroup I Picobirnavirus in diarrhoeic foals: can the horse serve as a natural reservoir for human infection? Vet Res. 2011;42:52. doi: 10.1186/1297-9716-42-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ganesh B, Banyai K, Kanungo S, Sur D, Malik YS, Kobayashi N. Detection and molecular characterization of porcine Picobirnavirus in feces of domestic pigs from Kolkata, India. Ind J Virol. 2012;23:387–391. doi: 10.1007/s13337-012-0106-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ganesh B, Banyai K, Martella V, Jakab F, Masachessi G, Kobayashi N. Picobirnavirus infections: viral persistence and zoonotic potential. Rev Med Virol. 2012;22:245–256. doi: 10.1002/rmv.1707. [DOI] [PubMed] [Google Scholar]
- 33.Ganesh B, Banyai K, Kobayashi N. Picobirnaviruses. In: Malik YPS, Pandey AB, Kataria JM, editors. Trends in diagnosis of animal viral diseases. Prasant Book Agency, New Delhi; 2013. p. 37–50; ISBN 978-8192229010.
- 34.Gatti MS, de Castro AF, Ferraz MM, Fialho AM, Pereira HG. Viruses with bisegmented double-stranded RNA in pig faeces. Res Vet Sci. 1989;47:397–398. [PubMed] [Google Scholar]
- 35.Ghosh S, Kobayashi N, Nagashima S, Naik TN. Molecular characterization of full-length genomic segment 2 of a bovine Picobirnavirus strain: evidence for high genetic diversity with genogroup I picobirnaviruses. J Gen Virol. 2009;90:2519–2524. doi: 10.1099/vir.0.013987-0. [DOI] [PubMed] [Google Scholar]
- 36.Ghosh S, Shintani T, Kobayashi N. Evidence for the porcine origin of equine rotavirus strain H-1. Vet Microbiol. 2012;158:410–414. doi: 10.1016/j.vetmic.2012.02.037. [DOI] [PubMed] [Google Scholar]
- 37.Gillman L, Sánchez AM, Arbiza J. Picobirnavirus in captive animals from Uruguay: identification of new hosts. Intervirology. 2013;56:46–49. doi: 10.1159/000338275. [DOI] [PubMed] [Google Scholar]
- 38.Giordano MO, Martinez LC, Rinaldi D, Gúinard S, Naretto E, Casero R, Yacci MR, Depetris AR, Medeot SI, Nates SV. Detection of Picobirnavirus in HIV-infected patients with diarrhea in Argentina. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;18:380–383. doi: 10.1097/00042560-199808010-00010. [DOI] [PubMed] [Google Scholar]
- 39.Giordano MO, Martinez LC, Rinaldi D, Espul C, Martinez N, Isa MB, Depetris AR, Medeot SI, Nates SV. Diarrhea and enteric emerging viruses in HIV-infected patients. AIDS Res Hum Retroviruses. 1999;15:1427–1432. doi: 10.1089/088922299309937. [DOI] [PubMed] [Google Scholar]
- 40.Giordano MO, Masachessi G, Martinez LC, Barril PA, Ferreyra LJ, Isa MB, Nates SV. Two instances of large genome profile Picobirnavirus occurrence in Argentinian infants with diarrhea over a 26-year period (1977–2002) J Infect. 2008;56:371–375. doi: 10.1016/j.jinf.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 41.Giordano MO, Martinez LC, Masachessi G, Barril PA, Ferreyra LJ, Isa MB, Valle MC, Massari PU, Nates SV. Evidence of closely related Picobirnavirus strains circulating in humans and pigs in Argentina. J Infect. 2011;62:45–51. doi: 10.1016/j.jinf.2010.09.031. [DOI] [PubMed] [Google Scholar]
- 42.González GG, Pujol FH, Liprandi F, Deibis L, Ludert JE. Prevalence of enteric viruses in human immunodeficiency virus seropositive patients in Venezuela. J Med Virol. 1998;55:288–292. doi: 10.1002/(SICI)1096-9071(199808)55:4<288::AID-JMV6>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 43.Green J, Gallimore CI, Clewley JP, Brown DW. Genomic characterisation of the large segment of a rabbit Picobirnavirus and comparison with the atypical Picobirnavirus of Cryptosporidium parvum. Arch Virol. 1999;144:2457–2465. doi: 10.1007/s007050050658. [DOI] [PubMed] [Google Scholar]
- 44.Grohmann GS, Glass RI, Pereira HG, Monroe SS, Hightower AW, Weber R, Bryan RT. Enteric viruses and diarrhea in HIV-infected patients. Enteric Opportunistic Infections Working Group. N Engl J Med. 1993;329:14–20. doi: 10.1056/NEJM199307013290103. [DOI] [PubMed] [Google Scholar]
- 45.Haga IR, Martins SS, Hosomi ST, Vicentini F, Tanaka H, Gatti MS. Identification of a bisegmented double-stranded RNA virus (Picobirnavirus) in faeces of giant anteaters (Myrmecophaga tridactyla) Vet J. 1999;158:234–236. doi: 10.1053/tvjl.1999.0369. [DOI] [PubMed] [Google Scholar]
- 46.Hamza IA, Jurzik L, Uberla K, Wilhelm M. Evaluation of pepper mild mottle virus, human Picobirnavirus and Torque teno virus as indicators of fecal contamination in river water. Water Res. 2011;45:1358–1368. doi: 10.1016/j.watres.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 47.Hart CA, Bennett M, Begon ME. Zoonoses. J Epidemiol Community Health. 1999;53:514–515. doi: 10.1136/jech.53.9.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herring AJ, Inolis NF, Ojeh CK, Snodgrass DR, Menzies JD. Rapid diagnosis of rotavirus infection by direct detection of viral nucleic acid in silver-stained polyacrylamide gels. J Clin Microbiol. 1982;16:473–477. doi: 10.1128/jcm.16.3.473-477.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leite JP, Monteiro SP, Fialho AM, Pereira HG. A novel avian virus with trisegmented double-stranded RNA and further observations on previously described similar viruses with bisegmented genome. Virus Res. 1990;16:119–126. doi: 10.1016/0168-1702(90)90016-5. [DOI] [PubMed] [Google Scholar]
- 50.Ludert JE, Hidalgo M, Gil F, Liprandi F. Identification in porcine faeces of a novel virus with a bisegmented double stranded RNA genome. Arch Virol. 1991;117:97–107. doi: 10.1007/BF01310495. [DOI] [PubMed] [Google Scholar]
- 51.Ludert JE, Liprandi F. Identification of viruses with bi- and trisegmented double-stranded RNA genome in faeces of children with gastroenteritis. Res Virol. 1993;144:219–224. doi: 10.1016/S0923-2516(06)80032-4. [DOI] [PubMed] [Google Scholar]
- 52.Ludert JE, Abdul-Latiff L, Liprandi A, Liprandi F. Identification of Picobirnavirus, viruses with bisegmented double stranded RNA, in rabbit faeces. Res Vet Sci. 1995;59:222–225. doi: 10.1016/0034-5288(95)90006-3. [DOI] [PubMed] [Google Scholar]
- 53.Malik YS, Chandrashekar KM, Sharma K, Haq AA, Vaid N, Chakravarti S, Batra M, Singh R, Pandey AB. Picobirnavirus detection in bovine and buffalo calves from foothills of Himalaya and Central India. Trop Anim Health Prod. 2011;43:1475–1478. doi: 10.1007/s11250-011-9834-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malik YS, Kumar N, Sharma K, Sharma AK, Sircar S, Jeena LM, Singh NK, Mondal A, Joardar SN, Balasubramanian G. Molecular characterization of a genetically diverse bubaline Picobirnavirus strain, India. Thai J Vet Med. 2013;43:609–613. [Google Scholar]
- 55.Martínez LC, Giordano MO, Isa MB, Alvarado LF, Paván JV, Rinaldi D, Nates SV. Molecular diversity of partial-length genomic segment 2 of human Picobirnavirus. Intervirology. 2003;46:207–213. doi: 10.1159/000072429. [DOI] [PubMed] [Google Scholar]
- 56.Martínez LC, Masachessi G, Carruyo G, Ferreyra LJ, Barril PA, Isa MB, Giordano MO, Ludert JE, Nates SV. Picobirnavirus causes persistent infection in pigs. Infect Genet Evol. 2010;10:984–988. doi: 10.1016/j.meegid.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 57.Masachessi G, Martínez LC, Giordano MO, Barril PA, Isa BM, Ferreyra L, Villareal D, Carello M, Asis C, Nates SV. Picobirnavirus (PBV) natural hosts in captivity and virus excretion pattern in infected animals. Arch Virol. 2007;152:989–998. doi: 10.1007/s00705-006-0900-2. [DOI] [PubMed] [Google Scholar]
- 58.Masachessi G, Martinez LC, Ganesh B, Giordano MO, Barril PA, Isa MB, Ibars A, Pavan JV, Nates SV. Establishment and maintenance of persistent infection by Picobirnavirus in greater rhea (Rhea americana) Arch Virol. 2012;157:2075–2082. doi: 10.1007/s00705-012-1400-1. [DOI] [PubMed] [Google Scholar]
- 59.Monteiro SP, Fialho AM, Pereira HG, Leite JPG. Viruses with segmented double-stranded RNA in chickens. Arq Bras Med Vet Zootec. 1991;42:141–146. [Google Scholar]
- 60.Munoz M, Alvarez M, Lanza I, Cármenes P. Role of enteric pathogens in the aetiology of neonatal diarrhoea in lambs and goat kids in Spain. Epidemiol Infect. 1996;117:203–211. doi: 10.1017/S0950268800001321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Novikova NA, Epifanova NV, Fedorova OF, Golitsyna LN, Kupriianova NV. Detection of picobirnaviruses by electrophoresis of RNA in polyacrylamide gel. Vopr Virusol. 2003;48:41–43. [PubMed] [Google Scholar]
- 62.Parrish CR, Holmes EC, Morens DM, Park EC, Burke DS, Calisher CH, Laughlin CA, Saif LJ, Daszak P. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol Mol Biol Rev. 2008;72:457–470. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pereira HG, Fialho AM, Flewett TH, Teixeira JM, Andrade ZP. Novel viruses in human faeces. Lancet. 1988;2:103–104. doi: 10.1016/S0140-6736(88)90032-3. [DOI] [PubMed] [Google Scholar]
- 64.Pereira HG, Flewett TH, Candeias JA, Barth OM. A virus with a bisegmented double-stranded RNA genome in rat (Oryzomys nigripes) intestines. J Gen Virol. 1988;69:2749–2754. doi: 10.1099/0022-1317-69-11-2749. [DOI] [PubMed] [Google Scholar]
- 65.Pereira HG, de Araujo HP, Fialho AM, de Castro L, Monteiro SP. A virus with bi-segmented double-stranded RNA genome in guinea pig intestines. Mem Inst Oswaldo Cruz. 1989;84:137–140. doi: 10.1590/S0074-02761989000100025. [DOI] [PubMed] [Google Scholar]
- 66.Pereira HG, Linhares AC, Candeias JA, Glass RI. National laboratory surveillance of viral agents of gastroenteritis in Brazil. Bull Pan Am Health Organ. 1993;27:224–233. [PubMed] [Google Scholar]
- 67.Pongsuwanna Y, Taniguchi K, Chiwakul M, Urasawa T, Wakasugi F, Jayavasu C, Urasawa S. Serological and genomic characterization of porcine rotaviruses in Thailand: detection of a G10 porcine rotavirus. J Clin Microbiol. 1996;34:1050–1057. doi: 10.1128/jcm.34.5.1050-1057.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rosen BI, Fang ZY, Glass RI, Monroe SS. Cloning of human Picobirnavirus genomic segments and development of an RT-PCR detection assay. Virology. 2000;277:316–329. doi: 10.1006/viro.2000.0594. [DOI] [PubMed] [Google Scholar]
- 69.Silva R, Bezerra D, Kaiano J, Oliveira D, Silvestre R, Gabbay Y, Ganesh B, Mascarenhas J. Genogroup I avian Picobirnavirus detected in Brazilian broiler chickens: a molecular epidemiology study. J Gen Virol. 2014;95:117–122. doi: 10.1099/vir.0.054783-0. [DOI] [PubMed] [Google Scholar]
- 70.Smits SL, Poon LL, van Leeuwen M, Lau PN, Perera HK, Peiris JS, Simon JH, Osterhaus AD. Genogroup I and II picobirnaviruses in respiratory tracts of pigs. Emerg Infect Dis. 2011;17:2328–2330. doi: 10.3201/eid1712.110934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smits SL, van Leeuwen M, Schapendonk CM, Schürch AC, Bodewes R, Haagmans BL, Osterhaus AD. Picobirnaviruses in the human respiratory tract. Emerg Infect Dis. 2012;18:1539–1540. doi: 10.3201/eid1809.120507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Symonds EM, Griffin DW, Breitbart M. Eukaryotic viruses in wastewater samples from the United States. Appl Environ Microbiol. 2009;75:1402–1409. doi: 10.1128/AEM.01899-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tamehiro CY, Alfieri AF, Medici KC, Alfieri AA. Segmented double-stranded genomic RNA viruses in fecal samples from broiler chicken. Bras J Microbiol. 2003;34:349–353. [Google Scholar]
- 74.Tang J, Ochoa WF, Li H, Havens WM, Nibert ML, Ghabrial SA, Baker TS. Structure of Fusarium poae virus 1 shows conserved and variable elements of partitivirus capsids and evolutionary relationships to Picobirnavirus. J Struct Biol. 2010;172:363–371. doi: 10.1016/j.jsb.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Valle MC, Martínez LC, Ferreyra LJ, Giordano MO, Isa MB, Paván JV, De Boccardo G, Massari PU, Nates SV. Viral agents related to diarrheic syndrome in kidney transplanted patients. Medicina. 2001;61:179–182. [PubMed] [Google Scholar]
- 76.van Leeuwen M, Williams MM, Koraka P, Simon JH, Smits SL, Osterhaus AD. Human picobirnaviruses identified by molecular screening of diarrhea samples. J Clin Microbiol. 2010;48:1787–1794. doi: 10.1128/JCM.02452-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vanopdenbosch E, Wellemans G. Birna-type virus in diarrhoeic calf faeces. Vet Rec. 1989;125:610. [PubMed] [Google Scholar]
- 78.Victoria JG, Kapoor A, Li L, Blinkova O, Slikas B, Wang C, Naeem A, Zaidi S, Delwart E. Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J Virol. 2009;83:4642–4651. doi: 10.1128/JVI.02301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Villacorta I, Peeters JE, Vanopdenbosch E, Ares-Mazás E, Theys H. Efficacy of halofuginone lactate against Cryptosporidiumparvum in calves. Antimicrob Agents Chemother. 1991;35:283–287. doi: 10.1128/AAC.35.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vizzi E, Angulo Medina LA. Enteropathogens responsible for gastrointestinal disorders in HIV patients. Invest Clin. 2013;54:90–108. [PubMed] [Google Scholar]
- 81.Volotäo EM, Soares CC, Albuquerque MC, Da Silva FM, Carvalho TR, Marins LP, D’Oliveira IC, Santos N. First evidence of a trisegmented double-stranded RNA virus in canine faeces. Vet J. 2001;161:205–207. doi: 10.1053/tvjl.2000.0532. [DOI] [PubMed] [Google Scholar]
- 82.Wakuda M, Pongsuwanna Y, Taniguchi K. Complete nucleotide sequences of two RNA segments of human Picobirnavirus. J Virol Methods. 2005;126:165–169. doi: 10.1016/j.jviromet.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 83.Wang Y, Tu X, Humphrey C, McClure H, Jiang X, Qin C, Glass RI, Jiang B. Detection of viral agents in fecal specimens of monkeys with diarrhea. J Med Primatol. 2007;36:101–107. doi: 10.1111/j.1600-0684.2006.00167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Woo PC, Lau SK, Bai R, Teng JL, Lee P, Martelli P, Hui SW, Yuen KY. Complete genome sequence of a novel Picobirnavirus, otarine Picobirnavirus, discovered in California sea lions. J Virol. 2012;86:6377–6378. doi: 10.1128/JVI.00686-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.World Health Organization (WHO). Generic protocols for (i) hospital-based surveillance to estimate the burden of rotavirus gastroenteritis in children and (ii) a community-based survey on utilization of health care services for gastroenteritis in children. Field test version. Department of Vaccines and Biologicals, World Health Organization, Geneva, Switzerland. 2002. http://www.who.int/vaccines-documents/DocsPDF02/www698.pdf. Accessed 25 Oct 2010.
- 86.Yang F, Wang Y, Zheng W, He B, Jiang T, Li Y, Xia L, Feng Y, Fan Q, Tu C. Metagenomic analysis of bat virome in several Chinese regions. Sheng Wu Gong Cheng Xue Bao. 2013;29:586–600. [PubMed] [Google Scholar]
- 87.Zhang T, Breitbart M, Lee WH, Run JQ, Wei CL, Soh SW, Hibberd ML, Liu ET, Rohwer F, Ruan Y. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2006;4:e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]