Abstract
The badnavirus, piper yellow mottle virus (PYMoV) is known to infect black pepper (Piper nigrum), betelvine (P. betle) and Indian long pepper (P. longum) in India and other parts of the world. Occurrence of PYMoV or other badnaviruses in other species of Piper and its variability is not reported so far. We have analysed sequence variability in the conserved putative reverse transcriptase (RT)/ribonuclease H (RNase H) coding region of the virus using specific badnavirus primers from 13 virus isolates of black pepper collected from different cultivars and regions and one isolate each from 23 other species of Piper. Of these, four species failed to produce expected amplicon while amplicon from four other species showed more similarities to plant sequences than to badnaviruses. Of the remaining, isolates from black pepper, P. argyrophyllum, P. attenuatum, P. barberi, P. betle, P. colubrinum, P. galeatum, P. longum, P. ornatum, P. sarmentosum and P. trichostachyon showed an identity of >85 % at the nucleotide and >90 % at the amino acid level with PYMoV indicating that they are isolates of PYMoV. On the other hand high sequence variability of 21–43 % at nucleotide and 17–46 % at amino acid level compared to PYMoV was found among isolates infecting P. bababudani, P. chaba, P. peepuloides, P. mullesua and P. thomsonii suggesting the presence of new badnaviruses. Phylogenetic analyses showed close clustering of all PYMoV isolates that were well separated from other known badnaviruses. This is the first report of occurrence of PYMoV in eight Piper spp and likely occurrence of four new species in five Piper spp.
Keywords: Piper yellow mottle virus, Sequence variability, Piper spp. RT/RNase region
The genus Badnavirus, which belongs to the family Caulimoviridae, have bacilliform shaped virions and a circular non-covalently closed double stranded DNA genome of 7–8 kb. The genomes of all described badnaviruses have a similar organization and contain 3–6 open reading frames (ORFs) [7, 10, 16]. Badnaviruses are mainly transmitted by vegetative propagation, mealy bug and through seeds [10, 12, 16]. There are more than 30 badnaviruses reported all around the world, infecting a wide range of economically important tropical crops including banana, black pepper, cacao, citrus and sugarcane. Many of the badnaviruses were shown to be highly variable at the genomic level [6, 11, 13, 16] while a few such as citrus yellow mosaic virus (CYMV) were shown to be less variable [1, 2]. The guidelines for demarcation of species within the genus Badnavirus have been reported. The criteria for defining species are <80 % identity (in the nucleotide sequence) or <89 % identity (in the amino acid sequence) in the conserved reverse transcriptase (RT)/ribonuclease H (RNase H) coding region [3].
Black pepper (Piper nigrum L.), which originated in the tropical evergreen forests of the Western Ghats in India is used for a variety of purposes including medicinal applications [14]. The first badnavirus reported on black pepper was Piper yellow mottle virus (PYMoV) causing yellow mottle disease, from Malaysia, the Philippines, Sri Lanka and Thailand based on electron microscopy, mealybug transmission studies, genomic characterization and partial genomic sequencing [12]. Subsequently PYMoV was also reported from India [9]. Yellow mottle disease of black pepper is characterized by mottled and small leaves, short internodes, and stunted vines. Recently complete genome sequence of an isolate of PYMoV showed that it possesses four ORFs [8]. Like other badnaviruses, the ORF 3 of PYMoV encodes a polyprotein consisting of viral movement protein, trimeric dUTPase, Zinc finger, retropepsin, RT-LTR and RNase H [8]. In addition to black pepper, occurrence of PYMoV in other two economically important Piper species such as betelvine (Piper betle) and Indian long pepper (P. longum) are also known [12, 15].
Studies on variability in PYMoV or occurrence of other badnaviruses infecting black pepper, betelvine and Indian long pepper is not available so far. Similarly there is no report of occurrence of badnaviruses in other Piper species. In the present study we have analysed sequence variability in the conserved putative RT/RNase H coding region of the virus using specific badnavirus primers from 13 isolates of black pepper collected from different cultivars and regions and one isolate each from 23 other species.
Diseased leaves from black pepper showing mild to severe virus-like symptoms were collected from different cultivars/varieties and regions of India (Table 1). Isolates from 23 related species originally collected from different regions of India (Table 1) and maintained at the Experimental Farm of Indian Institute of Spices Research, Kozhikode, Kerala, India were also used. Total nucleic acid was extracted from leaf tissue using the method described previously [9]. Two degenerate primers, Badna FP (5′ ATGCCITTYGGIITIAARAAYGCICC 3′) and Badna RP (5′ CCAYTTRCAIACISCICCCCAICC 3′) [16] were synthesized to amplify 580 bp region comprising of reverse transcriptase (RT)/ribonuclease H (RNase H) coding region of badnaviruses. The PCR reaction contained 1 × PCR buffer, 2.5 mM MgCl2, 200 µM dNTPs, 50 µM of each primer, 2.5 U Taq polymerase, 1 µl of template DNA and sterile water to a final volume of 50 µl. The thermal cycler was programmed for initial denaturation at 94 °C for 3 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 50 °C for 30 s, synthesis at 72 °C for 30 s and a final extension for 10 min at 72 °C. Total DNA isolated from a known healthy plant was used as negative control in PCR. Total DNA isolation and PCR tests were repeated for all plants that gave negative results. The reaction products were analysed on 0.8 % agarose gel along with 1 kb DNA ladder. The DNA bands were visualized and photographed using a UV transilluminator and a gel documentation apparatus (Cell Biosciences, Santa Clara, USA).
Table 1.
Geographical origin and accession numbers of the badnavirus isolates used in the study
| Sl.no | Piper species | Geographical origin | Cultivar/Variety | Accession number | Abbreviation used |
|---|---|---|---|---|---|
| 1 | P. nigrum L. | Kozhikode, Kerala | Subhakara | DQ836227 | nigrum-1 |
| 2 | P. nigrum L. | Idukki, Kerala | Jeerakamundi | DQ836229 | nigrum-2 |
| 3 | P. nigrum L. | Wynad, Kerala | Wynadan | DQ836231 | nigrum-3 |
| 4 | P. nigrum L. | Kodagu, Karnataka | Panniyur 1 | DQ836232 | nigrum-4 |
| 5 | P. nigrum L. | Kozhikode, Kerala | Karimunda | KC808712 | nigrum-5 |
| 6 | P. nigrum L. | Hassan, Karnataka | Panniyur 1 | KJ195468a | nigrum-6 |
| 7 | P. nigrum L. | Visakhapatnam, Andhra Pradesh | Sreekara | KJ195469a | nigrum-7 |
| 8 | P. nigrum L. | Kassargod, Kerala | Karimunda | KJ195470a | nigrum-8 |
| 9 | P. nigrum L. | Kannur, Kerala | Karimunda | KJ195471a | nigrum- 9 |
| 10 | P. nigrum L. | Udupi, Karnataka | Panniyur 1 | KJ195472a | nigrum-10 |
| 11 | P. nigrum L. | Chikkmagalur, Karnataka | Panniyur 1 | KJ195473a | nigrum-11 |
| 12 | P. nigrum L. | Palakkad, Kerala | Arakulamunda | KJ195474a | nigrum-12 |
| 13 | P. nigrum L. | Kozhikode, Kerala | Subhakara | KJ195475a | nigrum-13 |
| 14 | P. nigrum L. | Kozhikode, Kerala | IISR Girimunda | KJ195476a | nigrum-14 |
| 15 | P. nigrum L. | Kozhikode, Kerala | IISR-Thevam | KJ195477a | nigrum-15 |
| 16 | P. nigrum L. | Kozhikode, Kerala | Pournami | KJ195478a | nigrum-16 |
| 17 | P. nigrum L. | Kozhikode, Kerala | IISR-Shakthi | KJ195479a | nigrum-17 |
| 18 | P. betle L. | Kasaragod, Kerala | – | DQ836235 | betle -1 |
| 19 | P. betle L. | Kozhikode, Kerala | – | KJ195480a | betle-2 |
| 20 | P. longum L. | Kozhikode, Kerala | – | DQ836237 | longum-1 |
| 21 | P. longum L. | Kozhikode, Kerala | – | KJ195481a | longum-2 |
| 22 | P. argyrophyllum Miq | Palakkad, Kerala | – | KJ195482a | argyrophyllum |
| 23 | P.attenuatum Herb.ex Link | Tirunelveli, Tamil Nadu | – | KJ195483a | attenuatum |
| 24 | P. barberi Gamble | Nagarcoil, Tamil Nadu | – | KJ195484a | barberi |
| 25 | P. colubrinum Link | Kannur, Kerala | – | KJ195485a | colubrinum |
| 26 | P. galeatum (Miq.) C.DC | Palakkad, Kerala | – | KJ195486a | galeatum |
| 27 | P. ornatum N.E.Br | Wynad, Kerala | – | KJ195487a | ornatum |
| 28 | P. sarmentosum Roxb. | Andamans | – | KJ195488a | sarmentosum |
| 29 | P. trichostachyon (Miq) C.DC | Idukki, Kerala | – | KJ195489a | trichostachyon |
| 30 | P. bababudani | Chikkmagalur, Karnataka | – | KJ195490a | bababudani |
| 31 | P. chaba Hunter | Kozhikode, Kerala | – | KJ195491a | chaba |
| 32 | P. peepuloides Roxb. | Jalpaiguri, West Bengal | – | KJ195492a | peepuloides |
| 33 | P. mullesua Buch. Ham ex D.Don | Ooty, Tamil Nadu | KJ195493a | mullesua | |
| 34 | P. thomsonii (C.DC) Hook.f | Jalpaiguri, West Bengal | – | KJ195494a | thomsonii |
aSequences obtained in this study
The PCR products obtained was eluted from the gel using GenElute Gel Elution kit (Sigma-Aldrich, Bangalore, India), cloned into pTZ57R/T cloning vector (Fermentas, Glen Burnie, USA) and transformed into competent E. coli strain DH5α using InsTAclone PCR cloning Kit (Fermentas, Glen Burnie, USA) following manufacturer’s instructions. Recombinant clones were identified by PCR as well as restriction endonuclease digestion and selected clones were sequenced in both orientations at the automated DNA sequencing facility available at Chromous Biotech, Bangalore, India. Percent nucleotide and amino acid identities were determined using Bioedit program version 5.0.9. The corresponding nucleotide and amino acid sequences of other badnavirus isolates used for comparison were obtained from GenBank. The BLAST programme was used to identify related sequences available from GenBank database. Multiple sequence alignments were made using ClustalW2 programme. Phylogenetic analysis was performed using different methods such as Bayesian analysis (MrBayes version 3.1), maximum parsimony (MEGA5), maximum likelihood (GARLI version 2.0) and neighbor- joining method (MEGA5) each for a bootstrap analysis of 10000 replicates and the consensus tree was identified using consense package of Phylip 3.69. Rice tungro bacilliform virus (RTBV) with an accession no. X57924 was designated as outgroup for the selected sequences.
Using primers Badna FP and Badna RP, products of an approximately size of 580 bp were amplified from all virus isolates collected from black pepper, and there was no amplification from negative (badna-free) control. Of the 23 related species used, all except four (P. arboreum, P. magnificum, P. silentvalleyense and P. sugandhi) gave expected amplicon of 580 bp. All the amplicons were cloned and their nucleotide sequence determined. BLAST analysis of the sequence from P. hamiltoni, P. hapnium, P. hymenophyllum and P. sylvaticum showed highest identity with different plant genomes including retrotransposon (Ty3-gypsy) while their identity with known badnaviruses was quite low (43–55 %). This suggests that they may represent either host genome or retrotransposon and hence were not used in the present analysis. When the nucleotide sequence of all remaining isolates was compared including the published sequence of six PYMoV isolates (four from black pepper, one each from betelvine to Indian long pepper), a high identity of 85–100 % was observed among all 17 virus isolates from black pepper and many Piper spp indicating that they are isolates of PYMoV (Table 2). On the other hand, sequences from P. bababudani, P. chaba, P. mullesua, P. peepuloides and P. thomsonii showed low nucleotide sequence identity with PYMoV isolates (57–79 %) and other badnavirus species (57–69 %) suggesting the occurrence of new badnaviruses in these Piper species. Of these, virus isolates from P. bababudani and P. chaba showed an identity of 99 % among each other suggesting that they belong to one badnavirus species (Table 2). BLAST analysis of nucleotide sequence from P. bababudani and P. chaba showed high identity with PYMoV (77–79 %) suggesting that they are closely related to PYMoV. The virus isolates from P. mullesua,P. peepuloides and P. thomsonii indicated their distinctiveness as they shared only <71 % identity in the nucleotide sequence between them. The BLAST analysis of nucleotide sequences from P. mullesua, P. peepuloides and P. thomsonii showed highest identity (68 %) with different badnaviruses such as Musa acuminata endogenous badnavirus, Taro bacilliform virus and Canna streak virus respectively with sequence coverage up to 99–100 % suggesting occurrence of new badnaviruses in each of these species.
Table 2.
Percent nucleotide and deduced amino acid sequence (values shown in bracket) identities in the 579 bp coding region of the RT/RNase H of badnaviral isolates from different species of Piper from India
| Isolatesa | 1–29 | 30–31 | 34 | 32 | 33 | Other badnavirusesb |
|---|---|---|---|---|---|---|
| 1–29 | 85–100 (90–100) | 77–79 | 61–65 | 60–64 | 57–60 | 59–68 |
| 30–31 | (79–83) | 99 (99) | 64 | 62 | 59 | 59–69 |
| 34 | (60–65) | (65–66) | 100 (100) | 71 | 63 | 60–65 |
| 32 | (59–65) | (64–65) | (77) | 100 (100) | 59 | 58–66 |
| 33 | (54–59) | (57–58) | (60) | (58) | 100 (100) | 57–63 |
| Other badnavirusesb | (54–73) | (58–72) | (59–69) | (57–67) | (52–62) | 57–69 (52–73) |
Designation used for each of the isolates is given in Table 1
aFor details of isolates, refer to the Sl. No in Table 1. Isolates 1–17: nigrum-1 to nigrum-17; 18–19: betle-1 to betle-2; 20–21: longum-1 to longum-2; 22: attenuatum; 23: barberi; 24: colubrinum; 25: argyrophyllum; 26: galeatum; 27: ornatum; 28: sarmentosum; 29: trichostachyon; 30: bababudani; 31: chaba; 32: peepuloides; 33: mullesua and 34: thomsoni
bRetrieved from GenBank, their acronyms, accession numbers and full names are provided in Fig. 1
To examine whether each of the sequences contained functional protein coding sequence, they were translated. Cloned region of all isolates except the P. peepuloides was 579 bp long (predicted to code for 193 amino acids), the expected size for a badnavirus. The virus sequence from P. peepuloides was 573 bp long coding for 191amino acids. Analysis of the deduced amino acid sequences showed conserved region (YILDDILV) present in badnaviruses. As observed with nucleotide sequence, high levels of amino acid sequence identity (90–100 %) with PYMoV were detected between the 17 virus isolates of PYMoV from black pepper and many other Piper spp confirming that they are isolates of PYMoV (Table 2). On the other hand, isolates from P. bababudani, P. chaba, P. mullesua, P. peepuloides, and P. thomsonii showed low amino acid sequence identity with PYMoV isolates (54–83 %) and other badnavirus species (52–72 %) suggesting the presence of new badnaviruses. Amino acid sequence identities confirmed high identity between virus isolates from P. bababudani and P. chaba suggesting the occurrence of a new badnavirus in these species which is most closely related to PYMoV isolates. Further, amino acid sequence identities also suggested likely occurrence of three new badnaviruses in P. mullesua, P. peepuloides, and P. thomsonii as they shared only 57–77 % identity between them (Table 2). The alignment of deduced amino acid sequence of RT/RNase H region of all isolates produced 159 variable and 34 conserved sites.
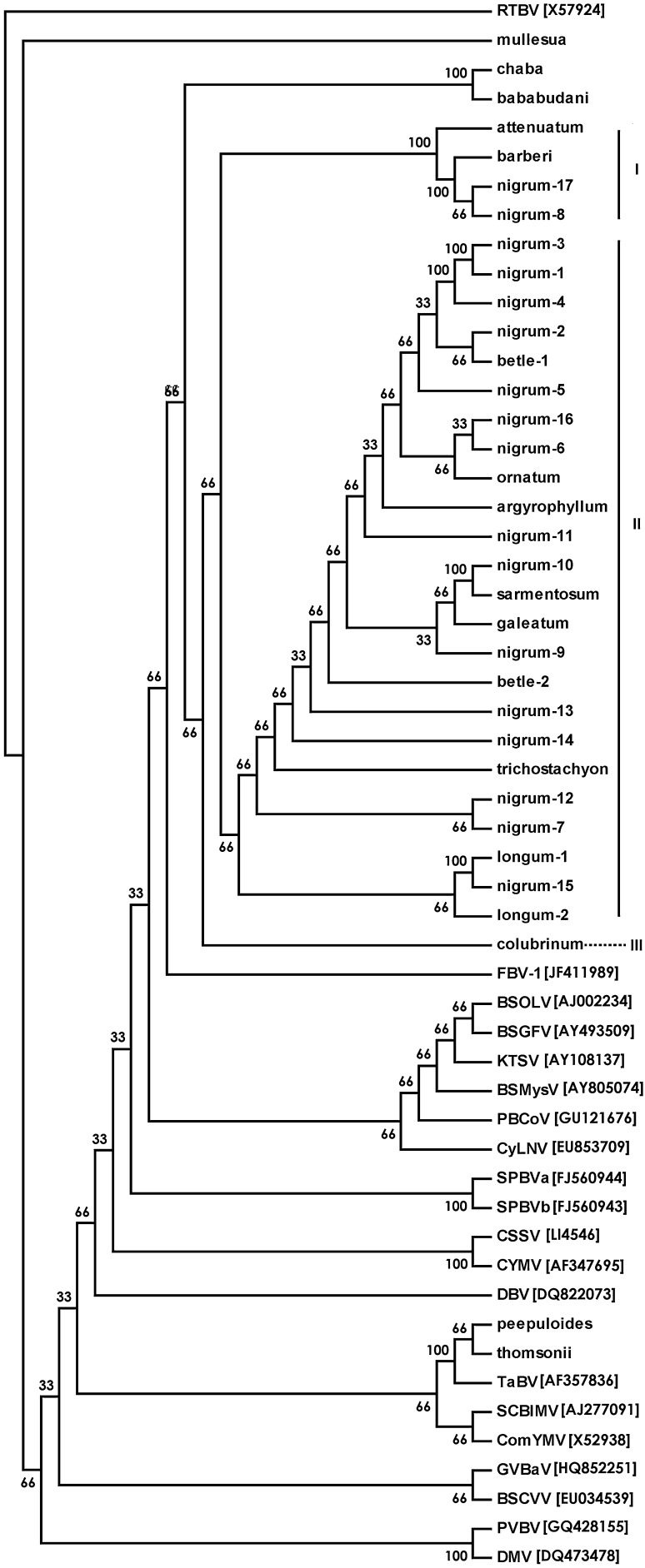
The consensus phylogenetic tree of the fifty-four sequences including one outgroup based on four methods confirmed results of sequence identity and showed distinctiveness of all PYMoV isolates from black pepper and related species that formed a group well separated from other known badnavirus species (Fig. 1). The relationships between the sequences were supported by high bootstrap values. Phylogenetic analysis of PYMoV isolates revealed that the 29 isolates formed three subgroups; subgroup 1 consisting of virus isolates from P. attenuatum, P. barberi and two isolates from P. nigrum (nigrum-17 and nigrum-8); subgroup 2 consisting 24 isolates and subgroup 3 consisting of only one isolate from P. colubrinum. The subgroup 2 was further branched into several subsubgroups (Fig. 1). The virus isolates within subgroup 1 showed 97–99 % amino acid identity while it was 92–100 % among isolates of subgroup 2. The only isolate in subgroup 3 showed an amino acid sequence identity of 94–96 % and 93–95 % respectively with subgroup 1 and subgroup 2 isolates. The badnavirus isolates from P. bababudani and P. chaba showed very close relationship with PYMoV isolates while badnavirus isolates from P. mullesua, P. peepuloides and P. thomsonii showed very distant relationship with PYMoV (Fig. 1). Fig badnavirus 1 (FBV-1) was found to be the closest badnavirus species to PYMoV isolates. Badnavirus isolate from P. peepuloides and P. thomsonii showed close phylogenetic relationship with Taro bacilliform virus (TaBV) while virus isolate from P. mullesua showed close relationship with Pelargonium vein banding virus (PVBV) and Dracaena mottle virus (DMV).
Fig. 1.
Consensus tree generated using consense in the Phylip package with Neighbor joining, Bayesian, Maximum Likelihood and Maximum Parsimony methods based on the multiple alignment of the deduced amino acid sequences of RT/RNaseH region of badnavirus isolates from black pepper and related species with known badnaviruses. Details of the badnavirus isolates from Piper species and their GenBank accession number is provided in Table 1. Corresponding sequences from other badnavirus species used were retrieved from GenBank, their acronym and accession number is indicated in the figure. BSGFV Banana streak GF virus, BSMysV Banana streak Mysore virus, BSOLV Banana streak OL virus, BSCVV Bougainvillea spectabilis chlorotic vein-banding virus, CSSV Cacao swollen shoot virus, CYMV Citrus yellow mosaic virus, ComYMV Commelina yellow mottle virus, CyLNV Cycad leaf necrosis virus, DBV Dioscorea bacilliform virus, DMV Dracaena mottle virus, FBV-1 Fig badnavirus 1, GVBaV Gooseberry vein banding virus, KTSV Kalanchoe top-spotting virus, PVBV Pelargonium vein banding virus, SCBIMV Sugarcane bacilliform IM virus, SPBVa Sweetpotato badnavirus A, SPBVb Sweetpotato badnavirus B, TaBV Taro bacilliform virus. Rice tungro bacilliform virus (RTBV) was taken as outgroup. Bootstrap values are shown at the nodes
The presence of conserved region observed in the amino acid sequences of the RT/RNaseH region of the badnavirus isolates infecting black pepper and related species in this study confirms that all virus isolates in this study belong to badnavirus genome [5]. The 34 virus isolates from black pepper and other Piper species from India analysed in this study can clearly be distinguished into five virus species. Of these, 29 virus isolates collected from P. nigrum (black pepper), P. longum (long pepper) and P. betle (betelvine), P. argyrophyllum, P. attenuatum, P. barberi, P. colubrinum, P. galeatum, P. ornatum, P. sarmentosum and P. trichostachyon belong to PYMoV, a known badnavirus species as they showed >90 % identity in the amino acid sequence in the coding region of RT/RNaseH [4, 8, 9, 15]. The remaining five isolates collected from P. bababudani, P. chaba, P. mullesua, P. peepuloides and P. thomsonii did not show any significant sequence identity with any of the known badnaviruses described so far suggesting the likely occurrence of new badnaviruses in these species. Occurrence of PYMoV in black pepper, betelvine and Indian long pepper is known [4, 8, 9, 12, 15] while its occurrence in other Piper spp and its variability was not known so far. The present study for the first time clearly showed the occurrence of PYMoV in eight species of Piper. Of the different species, P. nigrum, P. betle and P. longum are commercially cultivated on a large scale in India, Brazil and many south-east Asian countries while P. colubrinum is used as root stock for grafting black pepper as it is known for its resistance to Phytophthora capsici, an important fungus causing foot rot disease of black pepper [14]. A sequence variability of 0–15 % and 0–10 % in the nucleotide and amino acid respectively was observed among PYMoV isolates infecting black pepper and other Piper species. The sequence variability observed among the isolates may be attributed to the inaccurate replication by reverse transcription reported for members of the badnavirus. Similar kind of sequence variability was reported among isolates of Citrus yellow mosaic virus (CYMV) infecting different citrus species such as Citrus aurantifolia, C. grandis, C. jambhiri, and C. sinensis [1, 2] and different species of Dioscorea [5]. As per the criteria for classification of badnaviruses [3], the present study also suggested likely occurrence of four new badnaviruses in P. bababudani, P. chaba, P. mullesua, P. peepuloides and P. thomsonii. Occurrence of more than one badnaviruses infecting banana, cacao, dioscorea, sugarcane and taro were reported [5, 6, 11, 13, 16]. This forms the first report of likely occurrence of four new badnaviruses in different Piper spp. Further studies on complete genome sequencing of these virus isolates are needed to confirm whether or not they represent new badnavirus species.
Acknowledgments
Authors are thankful to Mr K. P. Premachandran for technical assistance; Distributed Information Sub Centre (DISC) and Director, Indian Institute of Spices Research, Kozhikode for providing necessary facilities.
Footnotes
Sequences reported in this paper have been submitted to the GenBank database and have been assigned the accession numbers KJ195468 to KJ195494.
References
- 1.Anthony-Johnson AM, Borah BK. Sai Gopal DV, Dasgupta I. Analysis of full-length sequences of two Citrus yellow mosaic badnavirus isolates infecting Citrus jambhiri (Rough Lemon) and Citrus sinensis L. Osbeck (Sweet Orange) from a nursery in India. Virus Genes. 2012;45:600–605. doi: 10.1007/s11262-012-0808-8. [DOI] [PubMed] [Google Scholar]
- 2.Borah BK, Johnson AM. Sai Gopal DV, Dasgupta I. Sequencing and computational analysis of complete genome sequences of Citrus yellow mosaic badnavirus from acid lime and pummelo. Virus Genes. 2009;39:137–140. doi: 10.1007/s11262-009-0367-9. [DOI] [PubMed] [Google Scholar]
- 3.Bousalem M, Durand O, Scarcelli N, Lebas BS, Kenyon L, Marchand JL, Lefort F, Seal SE. Dilemmas caused by endogenous pararetroviruses regarding the taxonomy and diagnosis of yam (Dioscorea spp.) badnaviruses: analyses to support safe germplasm movement. 2009. Arch Virol 154:297-314. [DOI] [PubMed]
- 4.de Silva DPP, Jones P, Shaw MW. Identification and transmission of Piper yellow mottle virus and Cucumber mosaic virus infecting black pepper (Piper nigrum L.) in Sri Lanka. Plant Pathol. 2002;51:537–545. doi: 10.1046/j.0032-0862.2002.00757.x. [DOI] [Google Scholar]
- 5.Eni AO, Hughes Jd A, Asiedu R, Rey MEC. Sequence diversity among badnavirus isolates infecting yam (Dioscorea spp.) in Ghana, Togo, Benin and Nigeria. Arch Virol. 2008;153:2263–2272. doi: 10.1007/s00705-008-0258-8. [DOI] [PubMed] [Google Scholar]
- 6.Geering AD, McMichael LA, Dietzgen RG, Thomas JE. Genetic diversity among Banana streak virus isolates from Australia. Phytopathol. 2000;90:921–927. doi: 10.1094/PHYTO.2000.90.8.921. [DOI] [PubMed] [Google Scholar]
- 7.Hagen LS, Jacquemond M, Lepingle A, Lot H, Tepfer M. Nucleotide sequence and genomic organization of cacao swollen shoot virus. Virology. 1993;196:619–628. doi: 10.1006/viro.1993.1518. [DOI] [PubMed] [Google Scholar]
- 8.Hany U, Adams IP, Glover R, Bhat AI, Boonham N. The complete genome sequence of Piper yellow mottle virus (PYMoV) Arch Virol. 2014;159:385–388. doi: 10.1007/s00705-013-1824-2. [DOI] [PubMed] [Google Scholar]
- 9.Hareesh PS, Bhat AI. Detection and partial nucleotide sequence analysis of Piper yellow mottle virus infecting black pepper (Piper nigrum L.) in India. Indian. J Virol. 2008;19:160–167. [Google Scholar]
- 10.Huang Q, Hartung JS. Cloning and sequence analysis of an infectious clone of Citrus yellow mosaic virus that can infect sweet orange via Agrobacterium mediated inoculation. J Gen Virol. 2001;82:2549–2558. doi: 10.1099/0022-1317-82-10-2549. [DOI] [PubMed] [Google Scholar]
- 11.Kenyon L, Lebas BS, Seal SE. Yams (Dioscorea spp.) from the South Pacific Islands contains many novel badnaviruses: implications for international movement of yam germplasm. Arch Virol. 2008;153:877–889. doi: 10.1007/s00705-008-0062-5. [DOI] [PubMed] [Google Scholar]
- 12.Lockhart BEL, Kirtisak KA, Jones P, Padmini DS, Olsziewski NE, Lockhart N, Nuarchan D, Sangalang J. Identification of Piper yellow mottle virus, a mealybug–transmitted badnavirus infecting Piper spp in southeast- Asia. Eur J Plant Pathol. 1997;103:303–311. [Google Scholar]
- 13.Muller E, Dupuy V, Blondin L, Bauffe F, Daugrois JH, Nathalie L, Iskra-Caruana ML. High molecular variability of sugarcane bacilliform viruses in Guadeloupe implying the existence of at least three new species. Virus Res. 2011;160:414–419. doi: 10.1016/j.virusres.2011.06.016. [DOI] [PubMed] [Google Scholar]
- 14.Ravindran PN. Black pepper (Piper nigrum L.) The Netherlands: Harwood Academic Publishers; 2000. [Google Scholar]
- 15.Siju S, Bhat AI, Hareesh PS. Identification and characterization of a Badnavirus infecting betel vine and Indian long pepper. J Plant Biochem Biotech. 2008;17:73–76. [Google Scholar]
- 16.Yang IC, Hafner GJ, Revill PA, Dale JL, Harding RM. Sequence diversity of South Pacific isolates of Taro bacilliform virus and the development of a PCR-based diagnostic test. Arch Virol. 2003;148:1957–1966. doi: 10.1007/s00705-003-0163-0. [DOI] [PubMed] [Google Scholar]